Abstract
PURPOSE
Large-scale analysis of real-world evidence is often limited to structured data fields that do not contain reliable information on recurrence status and disease sites. In this report, we describe a natural language processing (NLP) framework that uses data from free-text, unstructured reports to classify recurrence status and sites of recurrence for patients with breast and hepatocellular carcinomas (HCC).
METHODS
Using two cohorts of breast cancer and HCC cases, we validated the ability of a previously developed NLP model to distinguish between no recurrence, local recurrence, and distant recurrence, based on clinician notes, radiology reports, and pathology reports compared with manual curation. A second NLP model was trained and validated to identify sites of recurrence. We compared the ability of each NLP model to identify the presence, timing, and site of recurrence, when compared against manual chart review and International Classification of Diseases coding.
RESULTS
A total of 1,273 patients were included in the development and validation of the two models. The NLP model for recurrence detects distant recurrence with an area under the curve of 0.98 (95% CI, 0.96 to 0.99) and 0.95 (95% CI, 0.88 to 0.98) in breast and HCC cohorts, respectively. The mean accuracy of the NLP model for detecting any site of distant recurrence was 0.9 for breast cancer and 0.83 for HCC. The NLP model for recurrence identified a larger proportion of patients with distant recurrence in a breast cancer database (11.1%) compared with International Classification of Diseases coding (2.31%).
CONCLUSION
We developed two NLP models to identify distant cancer recurrence, timing of recurrence, and sites of recurrence based on unstructured electronic health record data. These models can be used to perform large-scale retrospective studies in oncology.
INTRODUCTION
With the increasingly widespread use of electronic health records (EHRs), data on real-world patient outcomes are more readily available. Use of these real-world data allows for analysis of the majority of adult patients with cancer who are treated outside of clinical trials. The current method of obtaining information on cancer disease status involves manual chart abstraction, which is labor-intensive, time-consuming, and is not feasible for large-scale data analysis. Population-based cancer registries such as SEER collect information on disease status at initial diagnosis only and are not funded to capture information on a patient's longitudinal disease course.1
CONTEXT
Key Objective
Can we identify patients with cancer who have developed distant recurrence, the timing of recurrence, and sites of recurrence using clinical information available in the electronic health record?
Knowledge Generated
Natural language processing was used to parse free text from clinician notes, pathology reports, and radiology reports, and then fed into machine learning models to identify cases of recurrence. Our data show that it is possible to distinguish between local versus distant recurrence cases and sites of recurrence and that machine learning models can capture more recurrence cases compared with use of International Classification of Diseases coding alone.
Relevance
Our natural language processing model can be applied to large data sets to create patient cohorts with recurrent disease and allow for retrospective, real-world analysis of outcomes based on recurrence status and sites of recurrence. The code for this model will be publically available for all investigators to use for research purposes.
Many studies have tried to automate chart abstraction using information available in the EHR. Use of International Classification of Diseases (ICD) codes and claims data have low sensitivity for identifying cancer disease status with sensitivity between 50% and 60%.2-4 Similar limitations in the identification of recurrence status also apply to assessment of sites of recurrence. ICD codes exist for various secondary sites of metastatic involvement but are inconsistently used in practice.4,5 Use of ICD coding and claims data fails to capture up to 17% of patients with bone metastasis5 and limits the analysis of outcomes based on sites of metastatic disease. Improvements in the aggregation and analysis of EHR data are necessary to enhance the use of real-world data and allow for replication and validation of randomized controlled trials.6
Natural language processing (NLP) techniques allow for the extraction of valuable information on disease progression contained within free text notes to determine recurrence status. Rule-based approaches can have high sensitivity of 92%-94% for detecting recurrence; however, these algorithms have limited generalizability because of differences in style and formatting between institutions and providers.7,8 Additionally, many earlier studies used selected pathology reports or imaging reports that limit sensitivity.9,10 More recently, using a combination of pathology reports, imaging reports, and clinician notes, Carrell et al11 were able to identify 92% of recurrent breast cancer cases. Use of NLP and deep machine learning has greatly improved sensitivity for detection of recurrence, but no studies thus far have algorithms that can distinguish between local recurrence and distant recurrence as well as timeline of recurrence or have been applied across multiple tumor types.
In our previous work, we developed a neural network-based NLP approach to extract breast cancer recurrence timeline information from progress notes and radiology and pathology reports, and were able to achieve a sensitivity of 83%, specificity of 73%, and AUROC (Area Under Receiver operating characteristic) of 0.9 for recurrence detection.12 However, this model lacks the ability to distinguish between local recurrence and distant recurrence, and its ability to detect cancer recurrence in other solid tumor types is unknown.
In this current study, we set out to answer three questions: (1) Can an NLP model be developed to identify distant sites of recurrence, rather than local recurrence? (2) Can an NLP model that was trained to detect recurrence in patients with breast cancer generalize well to a different solid tumor type? (3) Can an NLP model be developed to identify sites of recurrence using unstructured data?
METHODS
Cohort Development
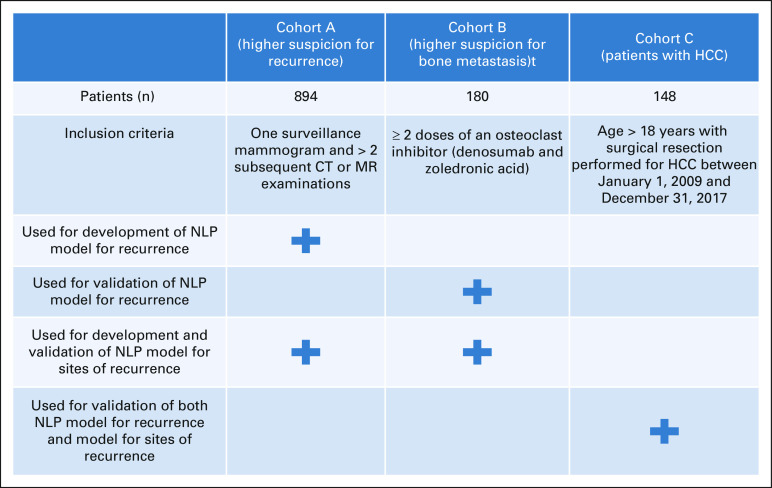
We used two data sets from breast and hepatocellular carcinoma (HCC) cohorts to develop and validate the NLP algorithms. Breast cancer was selected as the primary disease for initial development and testing since patients can often have long disease courses with late recurrence, and the specific site of distant recurrence can affect prognosis. HCC cases were selected for validation because of dissimilar distant recurrence sites with more local intrahepatic recurrence or peritoneal metastasis seen with HCC compared with bone and CNS metastatic sites seen in breast cancer. This would allow assessment of generalizability of these NLP models to various solid tumors.
With the approval of the Stanford University institutional review board, we trained and validated the NLP models using two cohorts of patients from the Oncoshare breast cancer research database. The Oncoshare database contains retrospective EHR data from Stanford Healthcare (SHC) that is linked on an individual patient level to data from the California Cancer Registry, a SEER registry.13 The Oncoshare database contains structured fields, including diagnostic codes, procedure codes, laboratory data, medications administered, and prescription data.14,15 Additionally, unstructured fields are available, including free-text clinician notes, radiology reports, and pathology reports. To complement this EHR data, registry data from California Cancer Registry contains demographic information, tumor characteristics at initial breast cancer diagnosis, and survival data.
Among 7,116 SHC patients within the Oncoshare database, we selected two cohorts of patients (Appendix Fig A1). The first cohort (cohort A) was previously identified for training and validation of an NLP model to detect recurrence.12 This cohort was selected based on patients who had a surveillance mammogram followed by > 2 computed tomography or magnetic resonance examinations to identify a population of patients with high suspicion for recurrence. The second cohort (cohort B) was selected based on patients having received two or more doses of osteoclast inhibitors to identify a population of patients with higher suspicion for distant metastasis. Patients included in cohort A who met inclusion criteria for cohort B were excluded to avoid patient overlap between the two cohorts. For the evaluation of model generalizability to patients with HCC, we used a cohort of 248 patients who underwent surgical resection for HCC at SHC between January 1, 2009, and December 31, 2017 (cohort C).
Manual Chart Review
Cases in cohort A were manually curated for recurrence status, earliest date of recurrence, and sites of initial recurrence using progress notes, pathology reports, and radiology reports, and the methods are described in our previous work. Cases in cohort B were manually curated by an expert oncologist for recurrence status, earliest date of recurrence, and sites of recurrence using NLP-assisted curation of progress notes, pathology reports, and radiology reports. Recurrence was defined as locoregional or distant metastatic disease that was newly documented during the follow-up period after the initial diagnosis and completion of initial definitive local therapy. Locoregional recurrence was defined as ipsilateral breast or ipsilateral regional lymph node disease (axillary, supraclavicular, infraclavicular, and internal mammary nodes). All other sites of recurrence or distant organ involvement were characterized as distant recurrence. Cancers in the contralateral breast were considered second primary cancers rather than recurrences. The recurrence date was recorded as the quarter during which there was either (1) a new radiographic finding of disease, (2) pathology report with tissue diagnosis of recurrence, or (3) clinician note documenting progression. If there was uncertainty in documentation, such as may represent or concern for, these were not considered to be definitive documentation of recurrence during that timeframe.
Cases in the HCC cohort were manually curated for recurrence status, date of recurrence, and sites of recurrence, using NLP-assisted curation of progress notes, pathology reports, and radiology reports. Local recurrence was defined as intrahepatic recurrence, whereas distant recurrence was defined as any extrahepatic nodal or distant organ involvement.
NLP Model Development and Evaluation
Figure 1 depicts the overall workflow of the proposed NLP pipeline, which contains two core processing modules—(1) recurrence timeline detection and (2) recurrence site detection. The recurrence timeline detection model is a neural network-based NLP algorithm that analyzes physician notes and pathology and radiology reports in Stanford's breast cancer database, Oncoshare (cohort A), to identify no recurrence, local recurrence, or distant recurrence by parsing the quarterly clinical notes documented in neutral language. We used vectorized clinical notes of each 3-month period of patient visits as model input, with the model computing the probabilistic output of recurrence via multiple nonlinear transformations. In the current study, we validated this algorithm on a distinct Oncoshare cohort (cohort B) where the cases were manually curated for longitudinal development of local recurrence or distant recurrence and recurrence sites by an expert oncologist.
FIG 1.
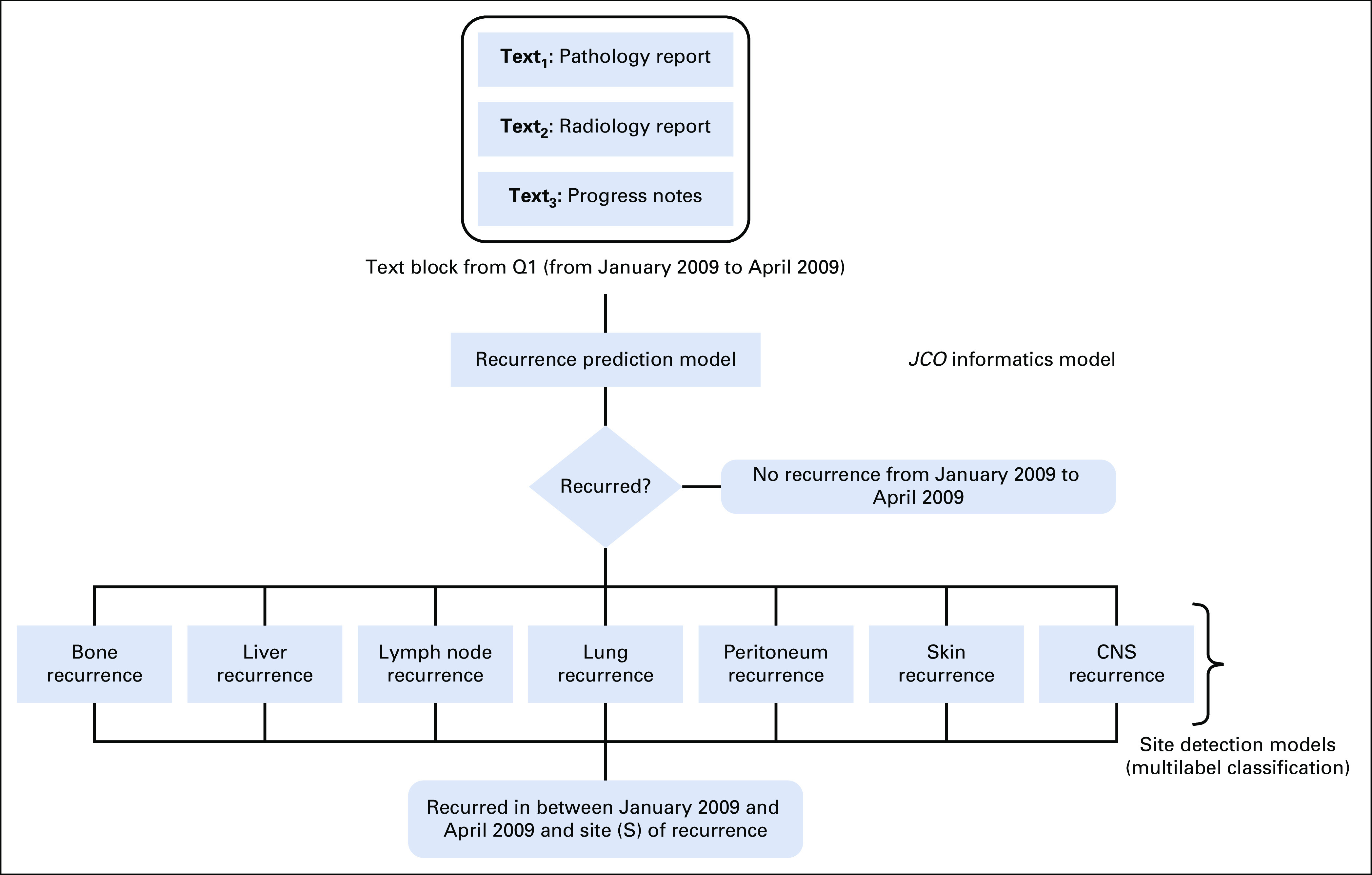
Model development: Step 1: recurrence prediction model no recurrence within quarter OR predicted probability of recurrence > .2. Step 2: sites of recurrence prediction model site of recurrence within quarter.
For the development of the NLP model for recurrence site detection, we used a hybrid approach where we combined the semantic sentence selection and machine learning to detect primarily seven anatomical regions for distant recurrence: bone, lymph node, liver, CNS, lung, peritoneum, and skin. We designed a one versus all classifier approach where for the seven anatomical sites, we generated the seven-binary XGBoost classifier models and trained them separately for each site. The model first takes input from the text block16 that is classified as distant recurrence by the recurrence detection model and then extracts relevant sentences based on a curated dictionary for anatomical terms or synonyms for the targeted sites. The sentence selection step not only allows the model to focus on important words, but also helps to reduce noise in the input data by removing uncertain or vague recurrence information about other sites. The dictionary of anatomical terms was generated based on consultation with domain experts and biomedical ontologies. Our site detection model also allows us to detect recurrence to multiple sites within the same period (more technical details of the NLP methods is in the Data Supplement).
The first validation of the recurrence timeline detection model in cohort B was performed using standard statistical measures (AUROC, sensitivity, and specificity) used in our previous publication.12 Using the mean probability of recurrence for distant recurrence in the breast and HCC cohorts, we calculated the sensitivity, specificity, accuracy, and f1-score for the NLP model's ability to detect distant recurrence, when compared with manual curation. A two-sided t-test was used to compare mean probabilities between local and distant recurrence cases. Next, we combined cases in cohorts A and B to train and validate the NLP classifiers that detect distant recurrence site. These cohorts were combined because of fewer recurrence cases in cohort A, with less variation in sites of recurrence, as these cases were originally curated only for distant recurrence sites at first recurrence. The combined cohort was randomly divided into 80% training and 20% validation sets. Sensitivity, specificity, and accuracy were calculated for the NLP model's ability to detect distant recurrence sites compared with manual curation. Last, we applied the models for recurrence and identification of recurrence site to patients with HCC (cohort C) and calculated sensitivity, specificity, and f1 scores for the model's ability to detect recurrence and sites of recurrence.
Comparison of NLP Classifications With ICD Coding
To compare clinical utility of this model for detection of any distant recurrence and bone recurrence against information available using only structured data fields, we compared NLP model predictions against ICD codes for all patients available in the Oncoshare Database. For coding of distant recurrence, we included ICD 9 or 10 codes of distant metastasis, including C77, C78, C79, and C80.0 (ICD 10 codes) and 196, 197, and 198 (ICD 9 codes). We assessed the first date of ICD coding for metastatic disease in patients with two or more metastatic codes on separate dates within 30 days. For coding of bone metastasis, we included the ICD 9 or 10 codes for secondary malignant neoplasm of bone C79.51 (ICD 10 code) and 198.5 (ICD 9 code) and similarly assessed for the first date of ICD code in patients with two or more codes on separate dates within 30 days.
RESULTS
Baseline Characteristics of Cohorts
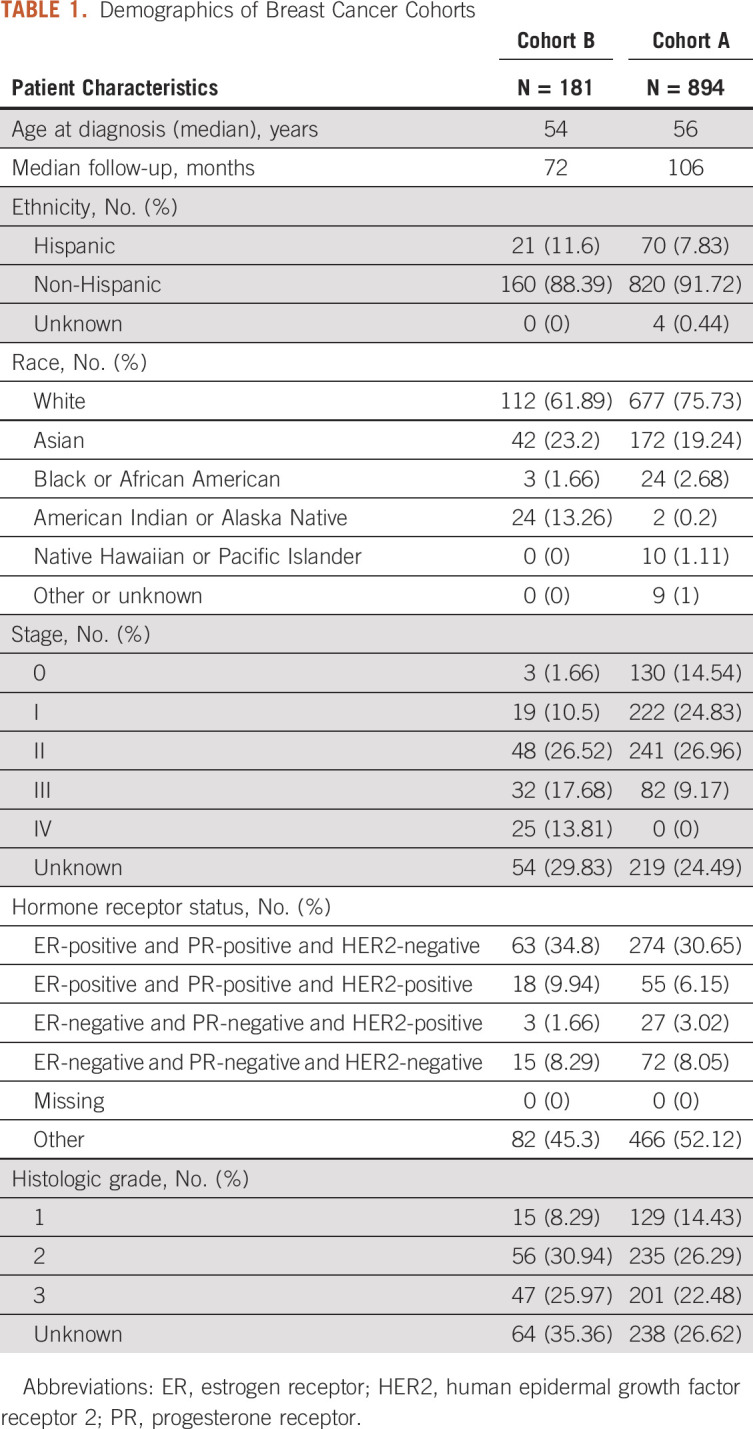
The validation breast cancer cohort (cohort B) curated for local versus distant recurrence consisted of 180 patients and 350 three-month periods (quarters) of patient visits. In cohort B, the median age at breast cancer diagnosis was 54 years, with a median follow-up time of 72 months (Table 1). Cases included in this validation cohort were mutually exclusive of the 894 cases initially used to design and validate the previously published convolutional neural network model for recurrence (cohort A).12
TABLE 1.
Demographics of Breast Cancer Cohorts
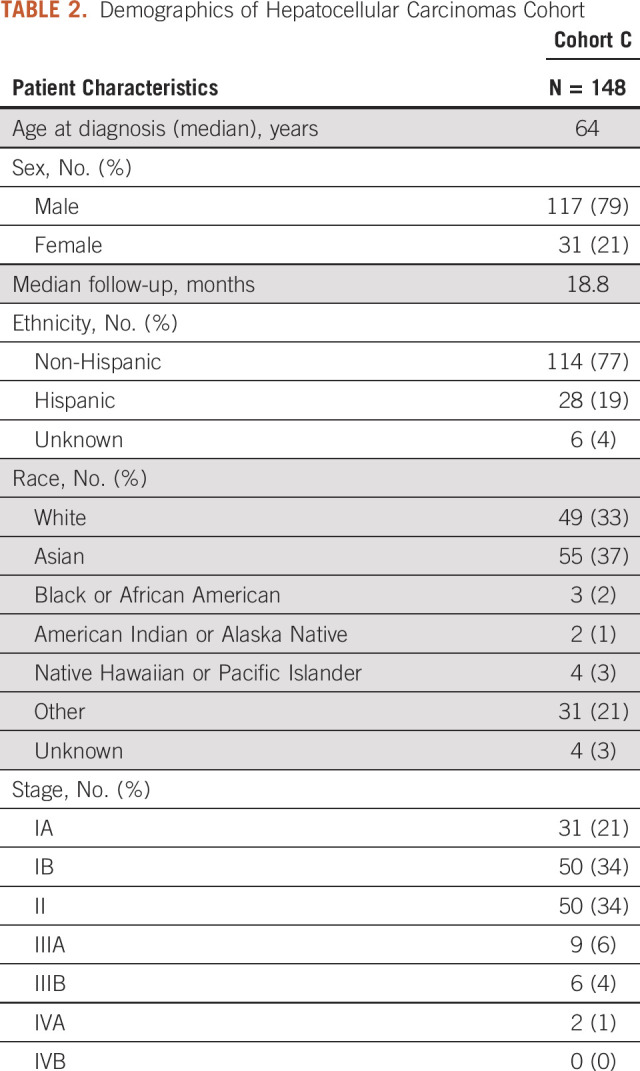
There were 248 patients in the HCC cohort (cohort C). Forty-seven patients were excluded because their surgical resection was performed for a recurrent HCC lesion. Two were excluded because of a lack of follow-up data after surgical resection. The analytic cohort was therefore composed of N = 199 patients. Median age at diagnosis was 64 years, predominantly male (79%) with a median follow-up time of 18.8 months (Table 2).
TABLE 2.
Demographics of Hepatocellular Carcinomas Cohort
Validation of NLP Algorithm for Local Versus Distant Recurrence
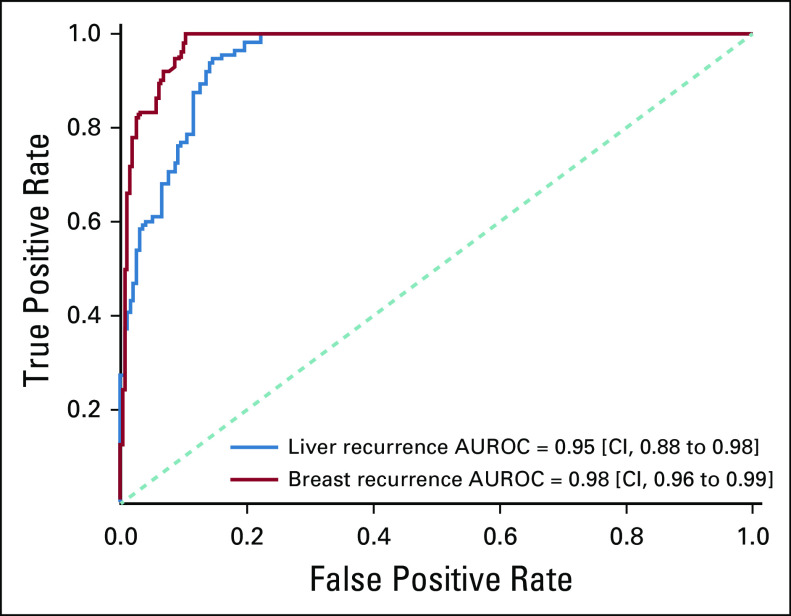
In Appendix Figure A2, we present the receiver operating characteristic curve for both breast and HCC, where the NLP-generated probability is compared against the clinical expert's annotations. The AUROCs were 0.98 (95% CI, 0.96 to 0.99) and 0.95 (95% CI, 0.88 to 0.98) for the breast (cohort B) and HCC cohorts, respectively. In cohort B, the model's mean predicted probability of recurrence was .42 versus .79 for patients with local versus distant recurrence (P < .001) and the median probability of recurrence was .43 and .96 for patients with local versus distant recurrence (P < .001) (Fig 2A). In cohort C (patients with HCC), mean and median probabilities of recurrence were .46 versus .70 (P < .001) and .44 versus .76 (P < .05) for local versus distant recurrence, respectively (Fig 2B). To reduce the number of missed cases for distant recurrence, we selected quartile 1 in the box plots of Figure 2, instead of the median, for optimizing the specificity. At a probability cutoff of .64 for breast cancer, the sensitivity, specificity, and accuracy for distant recurrence were 0.98, 0.75, and 0.87, respectively. At a predicted probability cutoff of .44 for HCC, sensitivity, specificity, and accuracy for distant recurrence were 0.91, 0.74, and 0.85, respectively.
FIG 2.
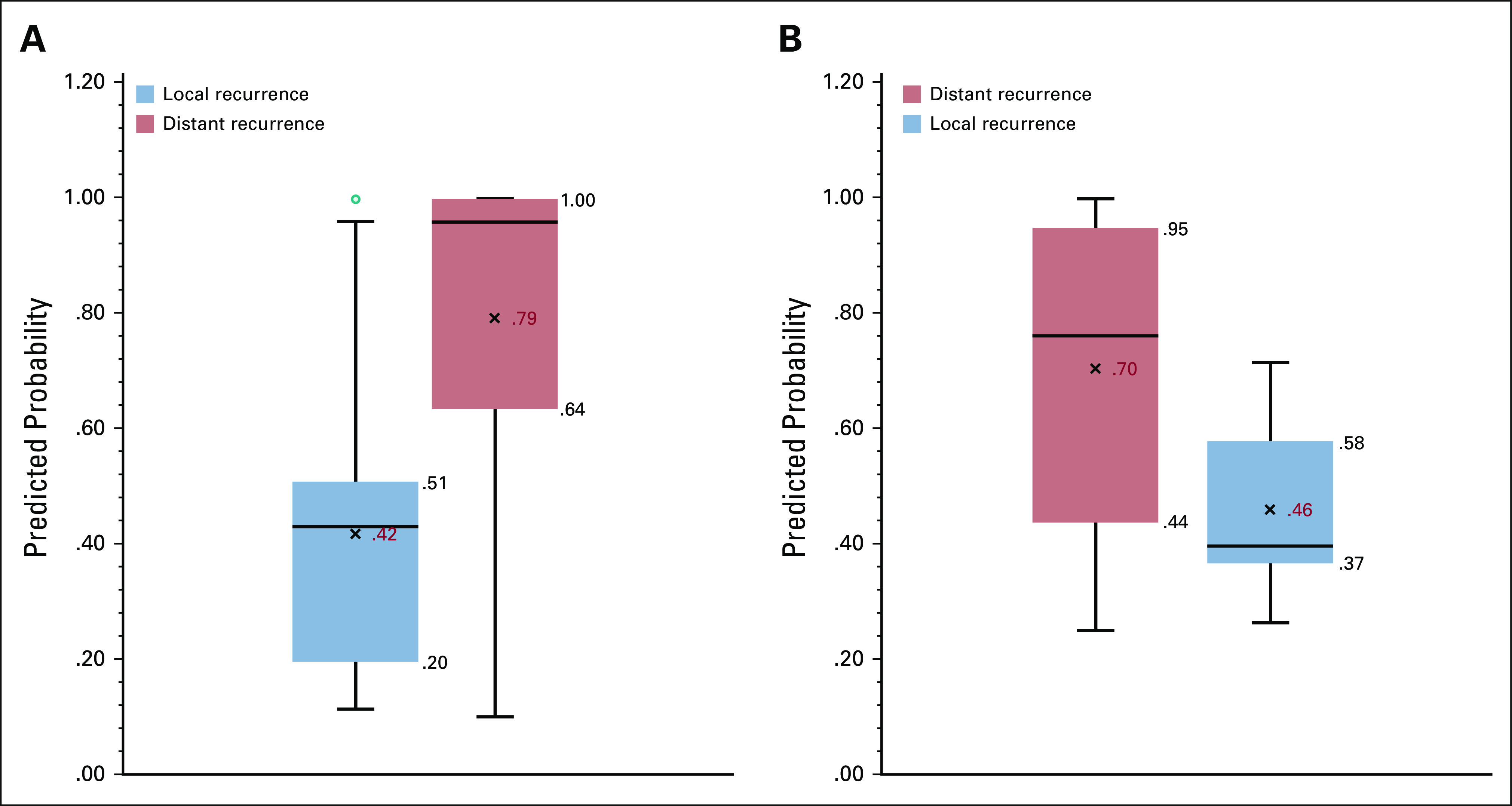
(A) Breast cohort local versus distant recurrence probability predicted by the natural language processing (NLP) models (cohort B). (B) Hepatocellular carcinomas cohort local versus distant recurrence probability predicted by the NLP models (cohort C).
Development of NLP Algorithm for Detection of Sites of Distant Recurrence
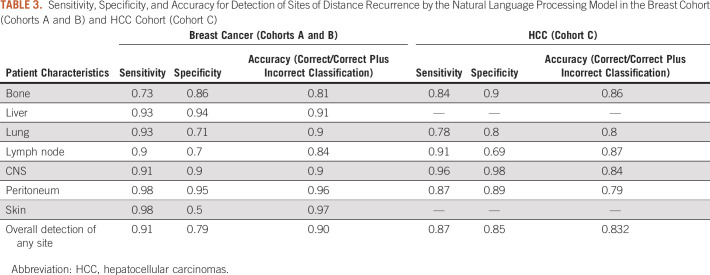
Six hundred thirty-two mutually exclusive breast cancer cases were combined from cohorts A and B to train and validate an NLP algorithm for identification of sites of distant recurrence in unstructured data fields. 80% of cases (n = 506) were used to train the model, and 20% of cases (n = 126) were used to calculate the sensitivity, specificity, and accuracy of the algorithm. Accuracy was highest for identification of liver and peritoneal disease, and lowest for bone and lymph node disease (Table 3). This model was then applied to patients with HCC (cohort C) to identify sites of distant recurrence, where accuracy was highest for identification of lymph node and bone sites of disease (Table 3).
TABLE 3.
Sensitivity, Specificity, and Accuracy for Detection of Sites of Distance Recurrence by the Natural Language Processing Model in the Breast Cohort (Cohorts A and B) and HCC Cohort (Cohort C)
Comparison of NLP Predicted Recurrence to ICD Coding
We compared the proportion of patients with an NLP predicted probability of recurrence > .2 with the proportion of patients identified as having recurrence based on ICD codes available in the Oncoshare database (Fig 3). Using the NLP model for recurrence, of those patients with > 2 encounters after 2008 in the Oncoshare database (N = 7,116), we identified 790 (11.1%) patients with a predicted probability of recurrence > .2. Using ICD codes for metastatic disease, 165 of 7,116 (2.31%) patients in the Oncoshare database were identified to have two or more codes for distant metastatic disease at any site.
FIG 3.
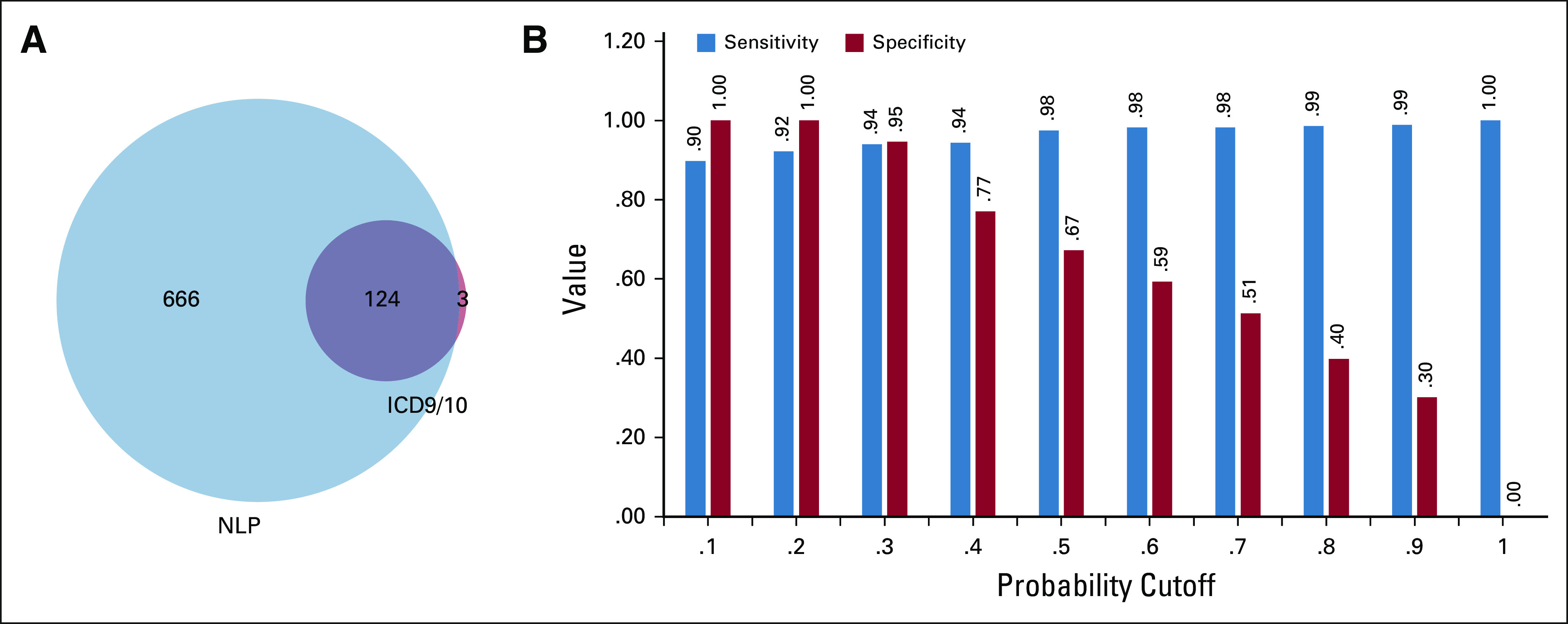
(A) Venn diagram showing overlap between patients who were found to have distant recurrence as predicted by NLP versus metastatic disease as predicted by ICD codes. (B) Sensitivity or specificity tradeoffs for distant recurrence detection in breast cancer cohort B. ICD, International Classification of Diseases; NLP, natural language processing.
Similarly, we compared NLP predicted bone recurrence to ICD coding for bone metastasis in the Oncoshare database. Among the 790 patients with > 20% probability of distant recurrence, there were 533 (67%) patients with predicted bone involvement using our NLP model. This corresponds to 7.49% patients identified as having recurrence in bone of the total Oncoshare population. Using two or more ICD codes of bone metastasis, 139 (1.95%) of 7,116 patients in the Oncoshare database were identified to have bone metastasis at any point during their disease course.
DISCUSSION
In this study, we have validated our recently developed NLP model for detection of cancer recurrence on an independent breast cancer cohort and extended the model to identify anatomic sites of cancer recurrence. In addition, we have demonstrated that this model can identify distant cancer recurrence and is generalizable to a solid tumor type on which it was not originally trained. To our knowledge, there have been no other published models that can identify all three features including (1) presence of recurrence, (2) site of recurrence, and (3) the date of recurrence. Our model generated an average accuracy of 0.9 for detecting any site of distant recurrence in breast cancer cases and average of 0.83 when this model was applied to HCC cases. This accuracy, as well as high discrimination for recurrence as shown with an AUROC of 0.98 and 0.95 for breast and liver cancer distant recurrence, respectively, compares favorably to others reported in the literature. Previous work for detection of recurrence and site of recurrence have shown accuracy of 0.85,10 and models identifying presence of recurrence and timing of recurrence have published accuracy of 0.59.11
These two NLP models for the detection of recurrence status and site show that NLP can be used to identify clinically relevant information from unstructured data fields and help generate large-scale cancer recurrence cohorts for further analysis with minimal human effort. Although we have reported model performance data using the mean probability of recurrence for the breast and HCC cohorts, any recurrence probability cutoff can be used for cohort identification, with varying sensitivities and specificities (Fig 3). This flexibility will allow investigators to determine what threshold probability of recurrence they prefer to use to generate cohorts that are appropriate for their study.
The strengths of these current models are that (1) we combine unstructured data from radiology, pathology, and clinician notes, rather than relying on a single source for evidence of recurrence; (2) these models capture the timing of recurrence and thus allow for correlation of outcomes based on approximate recurrence dates; and (3) the NLP model that was originally trained on patients with breast cancer was validated in a distinctly different tumor type (HCC).
The limitations are that this was a single-institution study and may require adaptation for differences in terminology used at other institutions. In addition, the NLP prediction is not perfect. The most common source of error stemmed from limited documentation of recurrence in clinical notes. However, we showed that NLP models are able to capture more patients in comparison to ICD code data. In the Oncoshare breast cancer patient database, the NLP model identifies 790 (11.1%) patients in the data set who are predicted to have distant recurrence. In comparison, while using ICD codes alone, only 165 patients (2.31%) are identified as having distant recurrence.
These NLP models have broad applicability. They can aid in cohort selection for metastatic patients to enable retrospective and real-world research studies. Additionally, these NLP models can allow for improved delivery of guideline-concordant care by rapidly analyzing unstructured patient-level data. Some EHR platforms now allow for integration and continuous deployment of NLP models. We are currently evaluating the utility of these NLP models for identifying patients with bone recurrence requiring therapy with osteoclast inhibitors, with a goal to ensure appropriate guideline-based therapy for these patients. Integration of NLP models with EHR platforms have the potential to improve the quality of care and allow for real-time, patient-centered approaches to oncologic care.
In conclusion, we have presented an NLP system that can simultaneously extract probability of recurrence, recurrence timeline, and sites of distant recurrence for two very different solid tumors, while having been trained on only one tumor type (breast cancer). The model is also able to distinguish between local versus distant recurrence based on predicted probability. Such NLP systems can unlock the potential of EHR-based data in generating valuable insights regarding distant cancer recurrence. Important next steps will include performance validation in diverse healthcare settings, both within the United States and internationally. To support reproducibility, we are publishing the models developed in this study with open-source licenses.
Appendix
FIG A1.
Inclusion criteria and use of each cohort in the development and validation of the NLP models. CT or MR, computed tomography or magnetic resonance; HCC, hepatocellular carcinomas; NLP, natural language processing.
FIG A2.
Area under the curves for timing and presence of distant recurrence.
Douglas W. Blayney
Leadership: Artelo Biosciences
Stock and Other Ownership Interests: Artelo Biosciences, Madorra
Consulting or Advisory Role: Creare, Daiichi Sankyo, Embold Health, Lilly, Google, Ipsen
Research Funding: Amgen, BeyondSpring Pharmaceuticals
Open Payments Link: https://openpaymentsdata.cms.gov/physician/728442 https://openpaymentsdata.cms.gov/physician/728442
Allison W. Kurian
Research Funding: Myriad Genetics
Other Relationship: Ambry Genetics, Color Genomics, GeneDx/BioReference, InVitae, Genentech
Daniel Rubin
Consulting or Advisory Role: Roche/Genentech
Research Funding: GE Healthcare, Philips Healthcare
Patents, Royalties, Other Intellectual Property: Several pending patents on AI algorithms
No other potential conflicts of interest were reported.
DISCLAIMER
The ideas and opinions expressed herein are those of the authors and do not necessarily reflect the opinions of the State of California, Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their contractors and subcontractors.
SUPPORT
Supported by the Koo Foundation Sun Yat Sen Cancer Center, the Breast Cancer Research Foundation, the Susan and Richard Levy Gift Fund, the Suzanne Pride Bryan Fund for Breast Cancer Research, the Jan Weimer Junior Faculty Chair in Breast Oncology, the Regents of the University of California's California Breast Cancer Research Program (16OB-0149 and 19IB-0124), the ASCO Conquer Cancer Foundation, and the BRCA Foundation. The collection of cancer incidence data used in this study was supported by the California Department of Public Health pursuant to California Health and Safety Code Section 103885; the Centers for Disease Control and Prevention’s National Program of Cancer Registries, under Cooperative Agreement No. 5NU58DP006344; and the National Cancer Institute's SEER Program under Contract No. HHSN261201800032I awarded to the University of California, San Francisco, Contract No. HHSN261201800015I awarded to the University of Southern California, and Contract No. HHSN261201800009I awarded to the Public Health Institute, Cancer Registry of Greater California.
DATA SHARING STATEMENT
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/CCI.20.00165.
AUTHOR CONTRIBUTIONS
Conception and design: Yasmin H. Karimi, Douglas W. Blayney, Allison W. Kurian, Imon Banerjee
Financial support: Douglas W. Blayney, Allison W. Kurian
Provision of study materials or patients: Allison W. Kurian, Jeanne Shen, Imon Banerjee
Collection and assembly of data: Yasmin H. Karimi, Douglas W. Blayney, Allison W. Kurian, Jeanne Shen, Rikiya Yamashita, Imon Banerjee
Data analysis and interpretation: Yasmin H. Karimi, Douglas W. Blayney, Allison W. Kurian, Daniel Rubin, Imon Banerjee
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Douglas W. Blayney
Leadership: Artelo Biosciences
Stock and Other Ownership Interests: Artelo Biosciences, Madorra
Consulting or Advisory Role: Creare, Daiichi Sankyo, Embold Health, Lilly, Google, Ipsen
Research Funding: Amgen, BeyondSpring Pharmaceuticals
Open Payments Link: https://openpaymentsdata.cms.gov/physician/728442 https://openpaymentsdata.cms.gov/physician/728442
Allison W. Kurian
Research Funding: Myriad Genetics
Other Relationship: Ambry Genetics, Color Genomics, GeneDx/BioReference, InVitae, Genentech
Daniel Rubin
Consulting or Advisory Role: Roche/Genentech
Research Funding: GE Healthcare, Philips Healthcare
Patents, Royalties, Other Intellectual Property: Several pending patents on AI algorithms
No other potential conflicts of interest were reported.
REFERENCES
- 1.Adamo MB, Johnson CH, Ruhl JL, Dickie LA, editors. SEER Program Coding and Staging Manual 2013. Bethesda, MD: National Cancer Institute; Surveillance Systems Branch Surveillance Research Program Division of Cancer Control and Population Sciences. [Google Scholar]
- 2.Lamont EB, Herndon JE, Weeks JC, et al. Measuring disease-free survival and cancer relapse using Medicare claims from CALGB breast cancer trial participants (companion to 9344) J Natl Cancer Inst 981335–13382006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chubak J, Yu O, Pocobelli G, et al. Administrative data algorithms to identify second breast cancer events following early-stage invasive breast cancer J Natl Cancer Inst 104931–9402012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whyte JL, Engel-Nitz NM, Teitelbaum A, et al. An evaluation of algorithms for identifying metastatic breast, lung, or colorectal cancer in administrative claims data Med Care 53e49–e572015 [DOI] [PubMed] [Google Scholar]
- 5.Liede A, Hernandez RK, Roth M, et al. Validation of International Classification of Diseases coding for bone metastases in electronic health records using technology-enabled abstraction Clin Epidemiol 7441–4482015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartlett VL, Dhruva SS, Shah ND, et al. Feasibility of using real-world data to replicate clinical trial evidence. JAMA Netw Open. 2019;2:e1912869. doi: 10.1001/jamanetworkopen.2019.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strauss JA, Chao CR, Kwan ML, et al. Identifying primary and recurrent cancers using a SAS-based natural language processing algorithm J Am Med Inform Assoc 20349–3552013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haque R, Shi J, Schottinger JE, et al. A hybrid approach to identify subsequent breast cancer using pathology and automated health information data Med Care 53380–3852015 [DOI] [PubMed] [Google Scholar]
- 9.Cheng LTE, Zheng J, Savova GK, et al. Discerning tumor status from unstructured MRI reports-completeness of information in existing reports and utility of automated natural language processing J Digit Imaging 23119–1322010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soysal E, Warner JL, Denny JC, et al. Identifying metastases-related information from pathology reports of lung cancer patients AMIA Jt Summits Transl Sci Proc 2017268–2772017 [PMC free article] [PubMed] [Google Scholar]
- 11.Carrell DS, Halgrim S, Tran DT, et al. Using natural language processing to improve efficiency of manual chart abstraction in research: The case of breast cancer recurrence Am J Epidemiol 179749–7582014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banerjee I, Bozkurt S, Caswell-Jin J, et al. Natural language processing approaches to detect the timeline of metastatic recurrence of breast cancer JCO Clin Cancer Inform 31–122019 [DOI] [PubMed] [Google Scholar]
- 13.California—SEER registries. https://seer.cancer.gov/registries/california.html
- 14.Kurian AW, Mitani A, Desai M, et al. Breast cancer treatment across health care systems: Linking electronic medical records and state registry data to enable outcomes research Cancer 120103–1112014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber SC, Seto T, Olson C, et al. Oncoshare: Lessons learned from building an integrated multi-institutional database for comparative effectiveness research AMIA Annu Symp Proc 2012970–9782012 [PMC free article] [PubMed] [Google Scholar]
- 16.Jones KS.A statistical interpretation of term specificity and its application in retrieval J Doc 6011–212004 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/CCI.20.00165.