Abstract
CDK4/6 inhibitors (CDK4/6i) are effective in metastatic breast cancer, but they have been only modestly effective in most other tumor types. Here we show that tumors expressing low CDK6 rely on CDK4 function, and are exquisitely sensitive to CDK4/6i. In contrast, tumor cells expressing both CDK4 and CDK6 have increased reliance on CDK6 to ensure cell cycle progression. We discovered that CDK4/6i and CDK4/6 degraders potently bind and inhibit CDK6 selectively in tumors in which CDK6 is highly thermo-unstable and strongly associated with the HSP90/CDC37 complex. In contrast, CDK4/6i and CDK4/6 degraders are ineffective in antagonizing tumor cells expressing thermostable CDK6, due to their weaker binding to CDK6 in these cells. Thus, we uncover a general mechanism of intrinsic resistance to CDK4/6i and CDK4/6i-derived degraders and the need for novel inhibitors targeting the CDK4/6i-resistant, thermostable form of CDK6 for application as cancer therapeutics.
Cyclin-Dependent Kinases 4 and 6 (CDK4/6) regulate cell cycle progression by phosphorylating and inactivating the tumor suppressor Retinoblastoma protein (Rb) and thus have been targeted by small molecule inhibitors for cancer therapy1,2. Dissociation of hyper-phosphorylated Rb alleviates transcriptional repression of E2F promoters and allows initiation of DNA synthesis- and mitosis-related gene transcription2,3. Recently, CDK4/6 inhibitors (CDK4/6i) in combination with hormonal therapy showed significant clinical activity in Rb-proficient metastatic ER positive breast cancers4,5, and three CDK4/6i, palbociclib (PB), abemaciclib and ribociclib, are now FDA-approved for this indication4,6-8. Since the activity of CDK4/6 requires a functional RB protein, tumors that do not express functional Rb are resistant to these drugs9. However, in many tumor types predominantly expressing wild-type RB1 (lung adenocarcinomas, melanomas, colon cancers, and others) preclinical and clinical studies have shown only modest effectiveness of CDK4/6i 10-12, suggesting that other mechanisms limit their efficacy in these tumor types. In ER+ breast cancer, CDK6 amplification has been reported to confer acquired resistance to CDK4/6 inhibitors13. However, CDK4/6i are potent inhibitors of CDK6 in vitro, and they show effectiveness in some, mostly liquid, tumor types predominantly expressing CDK614. In solid tumors, a comprehensive analysis of CDK6 expression at baseline and its potential role in tumor response to CDK4/6i has been lacking. Thus, there is a need for better understanding of the role of CDK6 as a potential contributor to tumor resistance to CDK4/6i.
Similarly to most kinases, CDK4 and, to a lesser extent, CDK6 have been previously shown to require the HSP90/CDC37 chaperoning complex for their maturation15-17. Closely related kinases may show largely different dependencies on HSP90/CDC37, for example, CDK4 is highly dependent on HSP90, but the closely related CDK2 is almost completely independent18. Further, activating mutations have been found to increase the dependency of kinases19,20, but overall, dependence of a given kinase on HSP90/CDC37 is thought to be uniform across cells and to depend exclusively on its amino-acid sequence. Finally, small molecule inhibitors have been shown to compete with HSP90/CDC37 for kinase binding18 and there is evidence from primarily in vitro studies for a connection between the strength of the interaction of CDKs with the HSP90/CDC37 complex and their affinity for inhibitors21. However whether the kinase interaction with the HSP90/CDC37 complex affects tumor response to small-molecule inhibitors remains unknown.
Lately, Proteolysis-Targeted Chimeras (PROTACs) i.e. hetero-bifunctional small molecules that are aimed on achieving selective degradation of the target proteins have been developed against a number of target kinases, including CDK4/622-24. We developed a potent and selective CDK4/6-directed PROTAC (CDK4/6 degrader) and we used it as tool to elucidate mechanisms of CDK4/6 regulation and response to CDK4/6i, by monitoring target degradation by CDK4/6 degrader as a surrogate for compound binding to target in cells. Use of this approach provided us with the first evidence of a critical role of the expression state of CDK6 in affecting tumor response to CDK4/6i.
Results
Intrinsic resistance to CDK4/6i is associated with incomplete inhibition of Rb/E2F and expression of CDK6
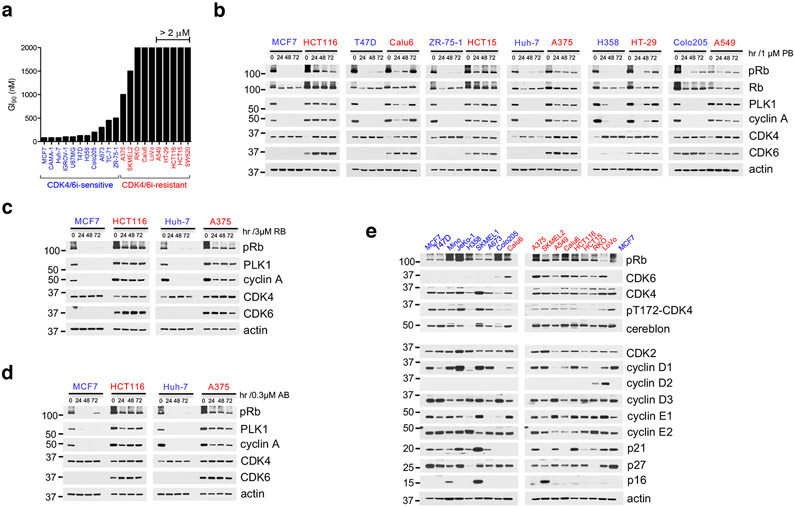
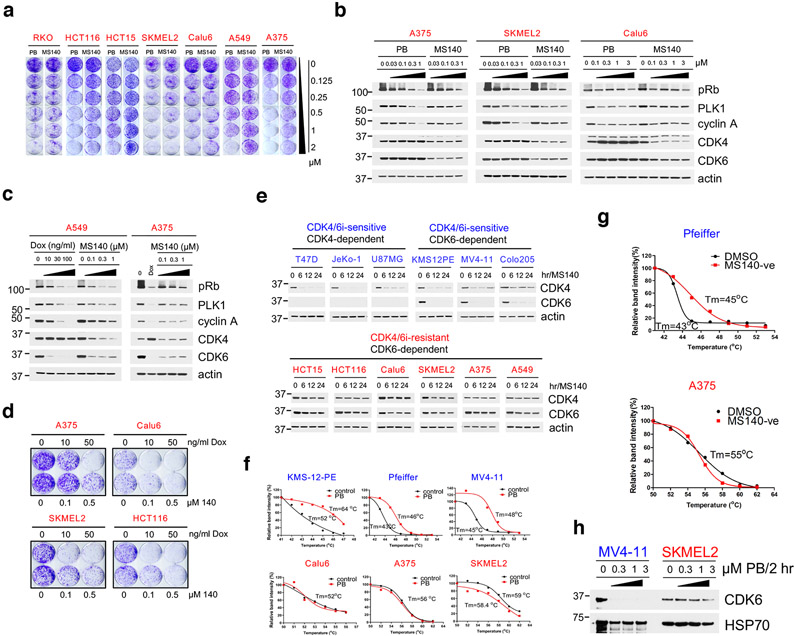
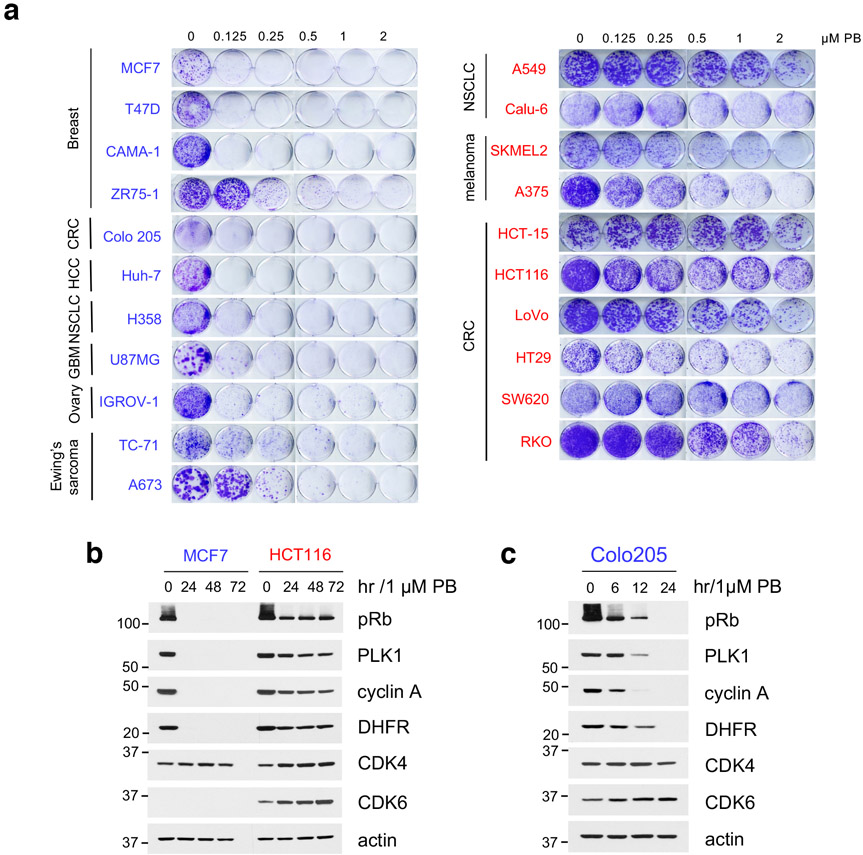
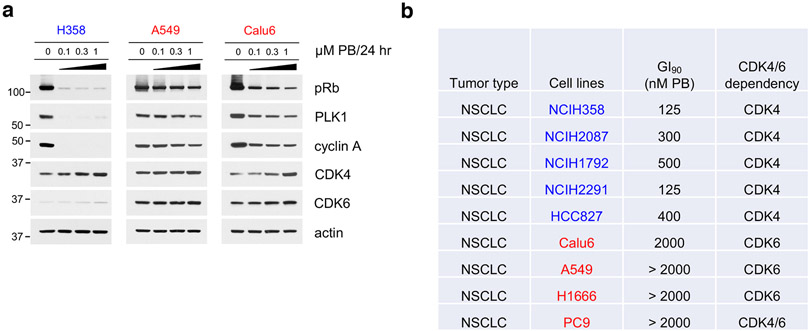
To gain insight into mechanisms of intrinsic resistance or sensitivity to CDK4/6i, we assessed the concentration-dependent effects of PB on the growth of cancer cell lines derived from a variety of Rb-proficient tumor types. We observed large variations in cell line response to CDK4/6i, consistent with previous reports11,12,25. In the "CDK4/6i-sensitive" (CDK4/6i-S) group, PB concentrations under 500 nM and in many cases as low as 80 nM were sufficient for substantial (over 90%) growth inhibition. In contrast, in the "CDK4/6i-resistant" (CDK4/6i-R) group, PB concentrations even as high as 2 μM had only modest effects on cell growth (Fig. 1a and Extended Data Fig. 1a). The CDK4/6i-S group included ER+ positive breast cancer cell lines, as expected26, as well as cancer cell lines from several other tumor types. However, most Rb-proficient cell lines derived from common solid tumor types including non-small cell lung carcinoma (NSCLC), melanoma and colorectal carcinoma (CRC) showed a CDK4/6i-R phenotype (Fig. 1a and Extended Data Fig. 1a).
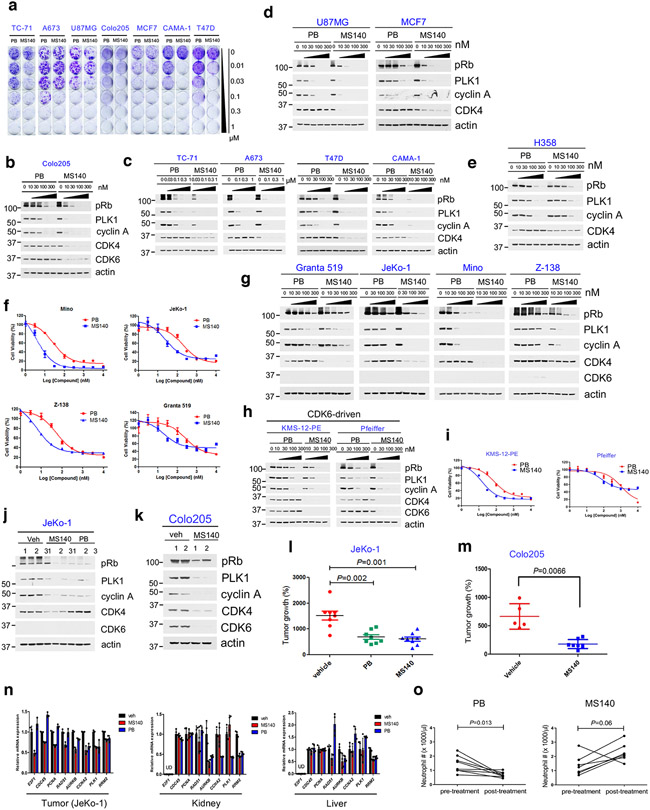
Fig. 1. Intrinsic resistance to CDK4/6i is associated with incomplete inhibition of Rb/E2F and expression of CDK6.
a, Bar graph of GI90 values based on cell growth crystal violet assays for the indicated cell lines treated with increasing concentrations of PB for 10-15 days. CDK4/6i-sensitive cell lines were highlighted in blue, CDK4/6i-resistant cell lines were in red.
b, The indicated cell lines were treated with 1 μM PB for 24, 48 and 72 hr and lysates were immunoblotted with the indicated antibodies.
c, The indicated cell lines were treated with 3 μM Ribociclib (RB) for 24, 48 and 72 hr and lysates were immunoblotted with the indicated antibodies.
d, The indicated cell lines were treated with 0.3 μM Abemaciclib (AB) for 24, 48 and 72 hr and lysates were immunoblotted with the indicated antibodies.
e, Expression of known cell cycle regulators were detected by immunoblotting with the indicated antibodies.
All data represent single experiments.
We next assessed whether the observed differences of tumor cell sensitivity to PB were associated with the extent of suppression of Rb/E2F output by the drug. Indeed, suppression of both Rb phosphorylation and of downstream E2F target proteins, PLK1, cyclin A and DHFR upon PB treatment (Rb/E2F output) were consistently of greater magnitude in CDK4/6i-S compared to CDK4/6i-R tumor cells (Fig. 1b and Extended Data Fig. 1b, c). Other FDA-approved CDK4/6i, including Ribociclib8 and Abemaciclib7, showed a similar profile (Fig. 1c, d). When we examined expression of several known CDK4/6 signaling-related proteins, including members of the cyclin D and cyclin E, INK4 and CIP/KIP families, we did not detect any consistent difference in expression between the two groups (Fig. 1e). However, we noticed that most CDK4/6i-S cell lines expressed CDK6 at very low levels, whereas the CDK4/6i-R cell lines in our panel, expressed both CDK4 and CDK6 (Fig. 1e).
Low expression of CDK6 predicts for sensitivity to CDK4/6i in NSCLC
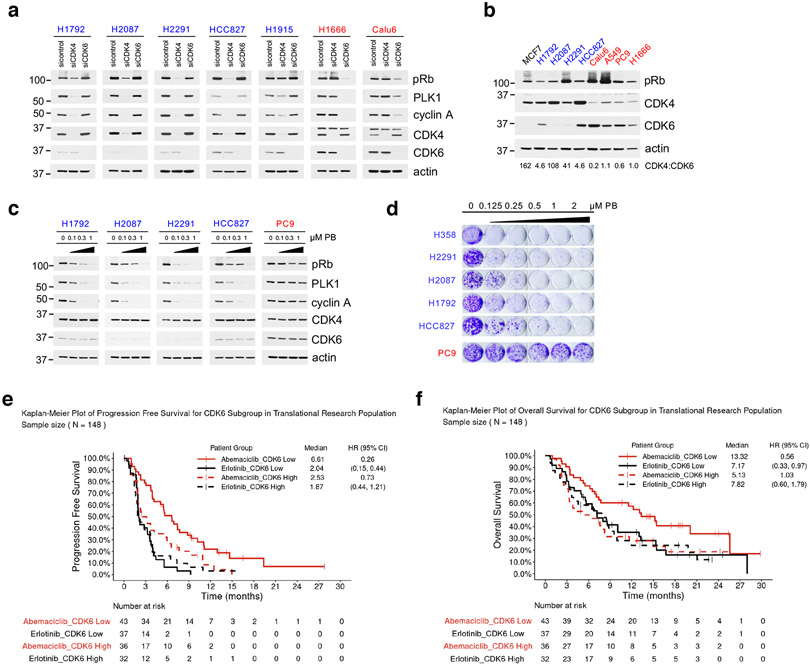
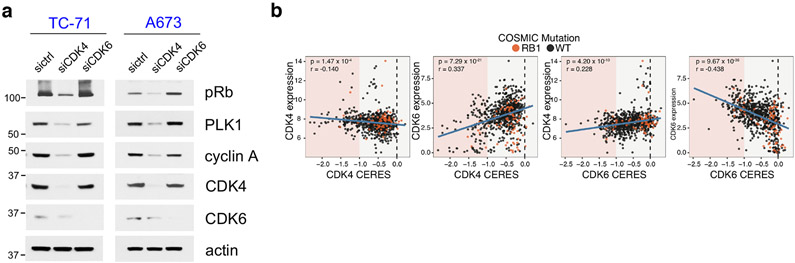
The observation that low CDK6 expression correlated with sensitivity to CDK4/6i, prompted us to assess whether we could identify subsets of CDK4/6i-sensitive cell lines within larger, predominantly Rb-proficient, tumor types. We focused on NSCLC, in which previous studies have shown clinical activity of these drugs, but failed to show a significant increase in either Progression Free Survival (PFS) or Overall Survival (OS), over current standard of care27,28. In addition to the NSCLC lines we examined initially (Extended Data Fig. 1a), we assessed five additional NSCLC lines with a high score for CDK4 dependence (i.e. H1792, H2087, H2291, HCC827 and H1915, in addition to H358) based on Achilles RNAi DEMETER scores29. For comparison, we examined two additional NSCLC CDK6-dependent lines (PC9 and H1666, in addition to A549, Calu6). We first confirmed dependence of this NSCLC panel on either CDK4 or CDK6. In the CDK4-dependent cells, siRNA-mediated knockdown of CDK4, but not CDK6 suppressed Rb/E2F output, whereas downregulation of CDK6, but not of CDK4 suppressed pRb/E2F output in CDK6-dependent cells (Fig. 2a). Further, although expression of CDK4 was variable across our panel of cell lines, CDK4-dependent cells expressed low levels of CDK6 compared to CDK4 (Fig. 2b).
Fig. 2. Low expression of CDK6 predicts for sensitivity to CDK4/6i in NSCLC.
a, The indicated NSCLC cell lines were transfected with control siRNA, siCDK4 and siCDK6 for 72 hr and lysates were immunoblotted with the indicated antibodies.
b, Total cell lysates of the indicated NSCLC cell lines were subjected to immunoblotting with the indicated antibodies. The ratio of CDK4/CDK6 was calculated by band intensity using Image J. The samples are derived from the same experiment and the images were processed in parallel.
c, The indicated NSCLC cell lines were treated with increasing concentrations of PB for 24 hr and lysates were immunoblotted with the indicated antibodies.
d, Cell growth crystal violet assay for the indicated NSCLC cell lines treated with increasing concentrations of PB for 10-15 days and stained with crystal violet. CDK4/6i-sensitive cell lines were in blue, CDK4/6i-resistant cell lines were in red.
e, Progression-free survival analysis of NSCLC/RAS-mutant patients that received abemaciclib in the JUNIPER trial based on CDK6-low versus CDK6-high tumors. (Abemaciclib: n=79 patients. n=43 patients in CDK6-low arm, n=36 patients in CDK6-high arm; Erlotinib: n=69 patients. n=37 patients in CDK6-low arm, n=32 patients in CDK6-high arm).
f, Overall survival analysis of NSCLC/RAS-mutant patients that received abemaciclib in the JUNIPER trial based on CDK6-low versus CDK6-high tumors. (Abemaciclib: n=79 patients. n=43 patients in CDK6-low arm, n=36 patients in CDK6-high arm; Erlotinib: n=69 patients. n=37 patients in CDK6-low arm, n=32 patients in CDK6-high arm).
All data represent single experiments.
We next assessed whether response to CDK4/6i correlates with low expression of CDK6 in NSCLC tumor cells. In all cases, we observed significantly higher sensitivity of CDK6-low cells to CDK4/6i, both in terms of Rb/E2F output (Fig. 2c and Extended Data Fig. 2a) and of cell growth (Fig. 2d) compared to CDK4/6i-R cells with high levels of CDK6 (H358 and A549, Calu6 data are included in Fig. 1a, b and Extended Data Fig. 1a, and a list of the NSCLC cell lines used in our study is in Extended Data Fig. 2b).
To assess the clinical relevance of our observations, we analyzed RNA expression data from tumors from the phase III JUNIPER trial (NCT02152631), in which abemaciclib was evaluated in KRAS-mutated, advanced NSCLC patients27. In a retrospective assessment, tumors with low CDK6 expression correlated with significantly longer PFS (Fig. 2e) and OS (Fig. 2f), compared to tumors expressing high levels of CDK6. These data confirm in a large clinical data set a critical role of CDK6 expression in the tumor in determining outcome of cancer patients treated with CDK4/6i and suggest that CDK6 expression in the tumor might be used a biomarker to stratify NSCLC patients for CDK4/6i treatment.
Tumors expressing both CDK4 and CDK6 depend selectively on CDK6
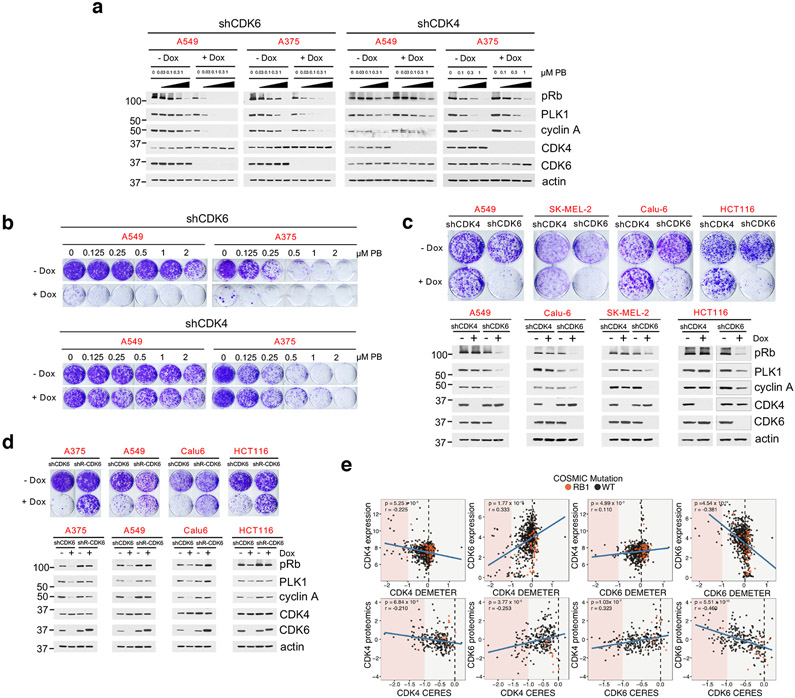
We next focused on understanding the basis of resistance of tumor cells expressing both CDK4 and CDK6 to CDK4/6i. Overexpression of a target is a mechanism that can drive resistance to small-molecule inhibitors. We thus assessed whether shRNA-mediated knockdown of CDK4 or CDK6 would sensitize PB-unresponsive cells to CDK4/6i. As expected, in cell lines expressing predominantly CDK4 and low levels of CDK6, knockdown of CDK4, but not of CDK6, reduced Rb/E2F output (Extended Data Fig. 3a). In CDK4/6i-R tumor cells, inducible CDK6 knockdown suppressed Rb/E2F output (Fig. 3a), as well as cell growth (Fig. 3b), which were further suppressed by treatment with PB. Surprisingly, knock-down of CDK4 in these same cells had no effect on either pRb levels or sensitivity to PB (Fig. 3a, b). CDK6 (but not CDK4) knockdown also suppressed Rb/E2F output as well as growth in additional CDK4/6i-R cell lines (Fig. 3c). Moreover, ectopic expression of an shRNA-resistant mutant of CDK6 rescued cell growth and Rb/E2F output inhibition promoted by CDK6 shRNA-mediated knock-down (Fig. 3d), thus establishing the specific role of CDK6 in driving Rb/E2F signaling and cell growth in these cells. Finally, phosphorylation of CDK4-T172, an established marker of CDK4 activation30, was readily detected in the CDK4/6i-S cells, but was substantially lower in the CDK4/6i-R cells (Fig. 1e). All of these findings suggested that despite expression of both CDK4 and CDK6, Rb/E2F signaling was driven selectively by CDK6 in CDK4/6i-R tumor cells.
Fig. 3. Tumors expressing both CDK4 and CDK6 depend selectively on CDK6.
a, A549 and A375 expressing Dox-inducible shCDK4 or shCDK6 were treated with 0.1 μg/ml doxycycline for 36 hr, followed by PB for 24 hr. Cell lysates were immunoblotted with the indicated antibodies.
b, Cell growth crystal violet assay for A549 and A375 expressing Dox-inducible shCDK4 or shCDK6 in the presence or absence of 0.1 μg/ml doxycycline and increasing concentrations of PB for 10 days.
c, Top panel: The indicated cell lines expressing Dox-inducible shCDK4 or shCDK6 were treated with or without 0.1 μg/ml doxycycline for 10 days followed by crystal violet staining. Bottom panel: cell lines were treated with or without 0.1 μg/ml doxycycline for 72 hr and cell lysates were subjected to immunoblotting with the indicated antibodies.
d, Top panel: The indicated cell lines co-expressing Dox-inducible shCDK6 and shRNA-resistant form of V5-CDK6 were treated with or without 0.05 μg/ml doxycycline for 10 days followed by crystal violet staining. Bottom panel: cell lines were treated with or without 0.05 μg/ml doxycycline for 72 hr and cell lysates were subjected to immunoblotting with the indicated antibodies.
e, Top panel: Scatterplots of DEMETER score (DepMap RNAi; DEMETER2 Data v5) and expression for CDK4 and CDK6. Bottom panel: Scatterplots of CERES (DepMap CRISPR; Public 20Q1) score and the mass spectrometry-based proteomics levels of CDK4 and CDK6 (PMID:31978347). All expression values are in log2(TPM +1). Proteomic levels are shown as normalized log2-transformed ratios to the bridge sample in each Tandem Mass Tags (TMT) 10-plex as previously described (BIORXIV: doi:10.1101/2020.02.03.932384). Cell lines harboring COSMIC hotspot mutations to RB1 are annotated in orange. P-values were calculated based on linear regression analysis.
All data represent single experiments.
To assess the generality of our observation, we analyzed the AVANA Dependency Map (DepMap) dataset to look at the correlation between the expression and protein levels to the dependency profiles of CDK4 and CDK631-36. First, similar to the cell lines used in our study, CDK4 expression was relatively uniform across cancer cell lines, whereas CDK6 expression was highly variable. Further, CDK6 expression and protein levels were correlated with dependence on CDK6, however CDK4 protein or mRNA expression levels were not predictive of dependence on CDK4 (Fig. 3e and Extended Data Fig. 3b). The data were similar whether dependency was interrogated using shRNA or sgRNA mediated loss-of-function31. Thus, the analysis revealed that our observation applies across tumor lines of various cancer types.
Development of MS140, a potent and selective CDK4/6-degrader (PROTAC)
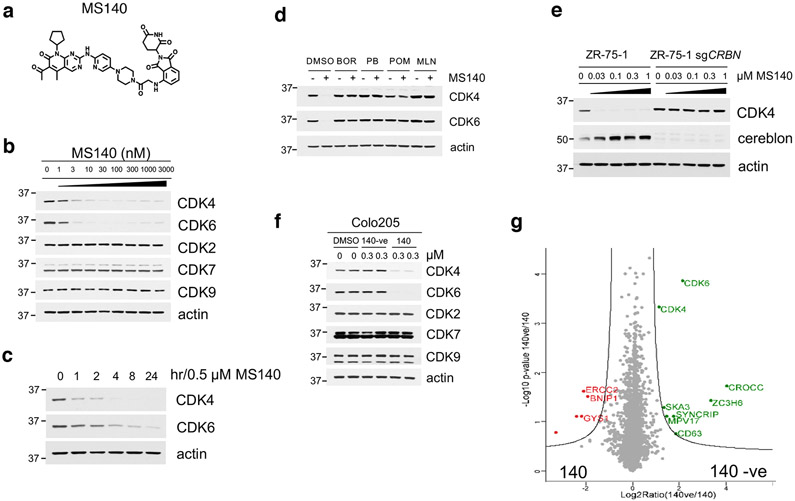
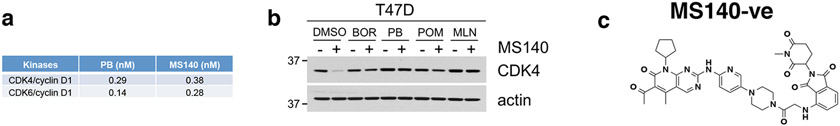
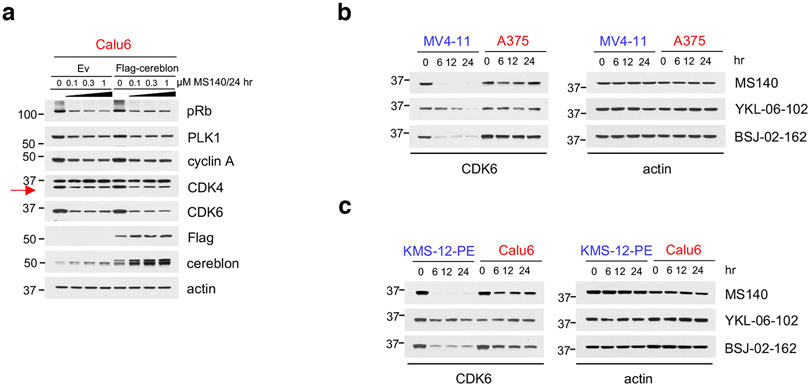
The inactivity of CDK4/6i in cells expressing both CDK4 and CDK6, led us to postulate that targeted degradation22 of CDK4/6 might result in more potent inhibition of Rb/E2F output and growth suppression of CDK4/6i-R tumor cells. As an approach, we developed hetero-bifunctional small-molecules37 that would both inhibit CDK4/6 kinase activity as well as target CDK4/6 proteins for degradation. Based on the crystal structure of CDK6 in complex with PB (PDB: 5L2I)38, we used the solvent-exposed piperazine as the linker attachment point. We synthesized heterobifunctional potential CDK4/6-directed PROTACs by linking PB to pomalidomide, a moiety with high affinity for the E3 ligase cereblon (CRBN), a component of a cullin-RING ubiquitin ligase complex39-43, using linkers of various lengths and types. By screening and optimizing synthesized compounds for their ability to degrade CDK4/6 and inhibit Rb/E2F output in cells, we identified MS140 (Fig. 4a), as a highly potent CDK4/6 kinase inhibitor in vitro (Extended Data Fig. 4a) that markedly suppressed Rb/E2F signaling and reduced CDK4 and CDK6 protein levels in a concentration and time-dependent manner (Fig. 4b, 4c). Degradation of CDK4/6 proteins by MS140 was specific, as shown by its abrogation upon pre-treatment with excess PB, or pomalidomide (Fig. 4d and Extended Data Fig. 4b). We also confirmed that CDK4/6 protein degradation by MS140 was specifically mediated by the proteasome by its abrogation upon pre-treatment with the proteasome inhibitor bortezomib, or with the Nedd8-activating enzyme inhibitor MLN4924 (Fig. 4d and Extended Data Fig. 4b). Further, MS140 failed to degrade CDK4 in cells in which CRBN had been knocked-out using CRISPR/Cas9 technology (Fig. 4e), confirming that target degradation by MS140 was specifically mediated by CRBN. As a negative control, we designed and synthesized MS140-ve (Extended Data Fig. 4c), a methyl analog of MS140 which is predicted not to bind CRBN42,44. Treatment with MS140-ve did not reduce CDK4/6 protein levels (Fig. 4f), further confirming that MS140 degrades its targets by linking them to the ubiquitin-proteasome machinery. Although PB has been reported to bind a number of kinases in addition to CDK4/6, including CDK945, we observed that MS140 did not degrade CDK7 or CDK9 (Fig. 4b, 4f). Finally, global analysis of protein degradation using mass spectrometry revealed an impressively selective protein downregulation profile of MS140, with very few hits other than CDK4 and CDK6 (Fig. 4g). Thus, MS140 potently and selectively both inhibited and degraded CDK4/6 kinases in CDK4/6i-S tumor cells by targeting them to the CRL4-CRBN-E3 ubiquitin complex.
Fig. 4. Development of MS140, a potent and selective CDK4/6-degrader (PROTAC).
a, Chemical structure of the bifunctional CDK4/6 inhibitor-degrader MS140.
b, Colo205 cells were treated with increasing concentrations of MS140 for 5 hr. Lysates were immunoblotted with the indicated antibodies.
c, Colo205 cells were treated with MS140 (0.5 μM) for the indicated time points. Lysates were immunoblotted with the indicated antibodies.
d, Colo205 cells were pretreated with either the proteasome inhibitor 100 nM bortezomib (BOR), 10 μM PB, 10 μM pomalidomide (POM) or 1 μM MLN4924 (MLN) for 4 hr, followed by treatment with MS140 (100 nM/3 hr). Lysates were subjected to immunoblotting with the indicated antibodies.
e, ZR-75-1 wild-type and CRBN-deficient cells were treated with the indicated concentrations of MS140 for 5 hr. Lysates were immunoblotted with the indicated antibodies.
f, Colo205 cells were treated with DMSO, MS140-ve and MS140 for 5 hr. Lysates were immunoblotted with the indicated antibodies.
g, Volcano plot of the protein log2 ratios represent the quantitative dynamics of 4,822 proteins in 140 and 140-ve (negative control of 140) treated Colo205 samples (0.3 μM, 5 hr) in duplicate.
All data represent single experiments.
CDK4/6-directed degradation is more effective than CDK4/6i in CDK4/6i-S tumor cells
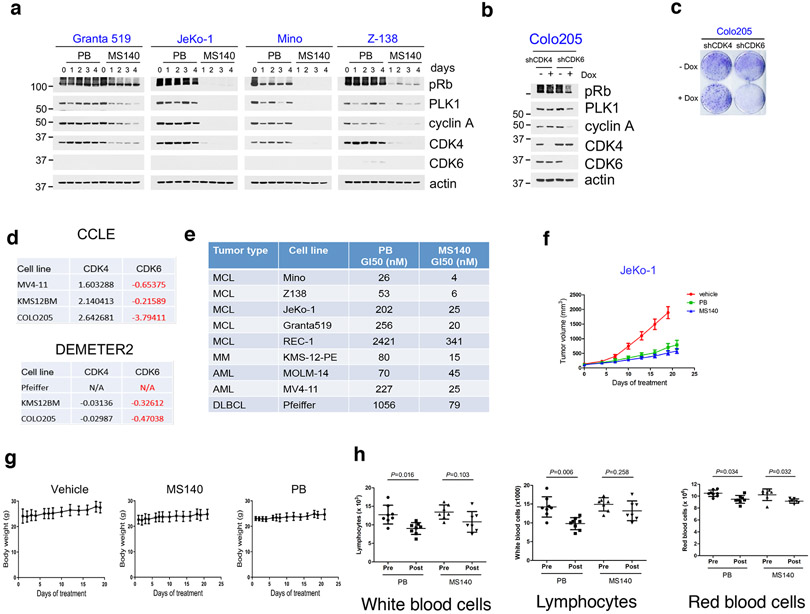
Treatment of various Rb-proficient tumor cell lines with MS140 resulted in 3 to 30-fold greater suppression of both Rb/E2F signaling and cell growth, compared to PB, in the CDK4/6i-S cells identified in our first screen (Fig. 5a-d). A notable exception was H358, in which MS140 only modestly decreased CDK4 levels or inhibited Rb signaling as compared to PB (Fig. 5e). However, this cell line exhibited very low endogenous levels of CRBN (Fig. 1e), consistent with the increased effectiveness of MS140 being CRBN-dependent. We next assayed additional cell lines of hematopoetic origin. All Mantle Cell Lymphoma (MCL) lines tested showed sensitivity to PB and higher sensitivity to MS140 (Fig. 5f), associated with more potent and durable inhibition of Rb/E2F signaling by MS140 (Fig. 5g, Extended Data Fig. 5a), consistent with these cells being predominantly CDK4-driven and expressing low levels of CDK6.
Fig. 5. CDK4/6-directed degradation is more effective than CDK4/6i in CDK4/6i-S tumor cells.
a, The indicated cell lines were subjected to cell growth crystal violet assay in the presence of PB or MS140 for 10-15 days.
b, Colo205 cells were treated with increasing concentrations of PB or MS140 for 24 hr. Lysates were immunoblotted with the indicated antibodies.
c, The indicated cell lines were treated with increasing concentrations of PB or MS140 for 24 hr. Lysates were immunoblotted with the indicated antibodies.
d, U87MG or MCF7 cells were treated with increasing concentrations of PB or MS140 for 24 hr. Lysates were immunoblotted with the indicated antibodies.
e, H358 cells were treated with increasing concentrations of MS140 for 24 hr. Lysates were immunoblotted with the indicated antibodies.
f, Mantle Cell Lymphoma (MCL) cells were treated with PB and MS140 for 72-96 hr. Cell viability was assayed using 0.1 mg/ml resazurin solution. IC50 values were determined by nonlinear regression curve fit in Graphpad Prism (n=6 independent experiments). Data are presented as mean ± S.D.
g, MCL cell lines were treated with increasing concentrations of PB or MS140 for 24 hr. Lysates were immunoblotted with the indicated antibodies.
h, KMS-12-PE or Pfeiffer cells were treated with increasing concentrations of PB or MS140 for 24 hr. Lysates were immunoblotted with the indicated antibodies.
i. The indicated cells were treated with PB or MS140 for 72-96 hr. Cell viability was assayed using 0.1 mg/ml resazurin solution. IC50 values were determined by nonlinear regression curve fit in Graphpad Prism (n=6 independent experiments). Data are presented as mean ± S.D.
j, Mice carrying JeKo-1 xenografts were treated with vehicle or MS140 (25 mg/kg, b.i.d) or PB (50 mg/kg, q.d.) for 3 days. Tumor samples were lysed and immunoblotted with the indicated antibodies.
k, Mice carrying Colo205 xenografts were treated with vehicle or MS140 (30 mg/kg, b.i.d) for 3 days. Tumor samples were lysed and immunoblotted with the indicated antibodies.
l, Scatter plot of fold change for an efficacy assay in JeKo-1 tumor xenografts in mice treated with vehicle or MS140 (25 mg/kg, b.i.d) or PB (50 mg/kg, q.d.) for 21 days. Each treatment contained 8 animals (n=8). Data are presented as mean ± S.E.M. Statistical significance was determined by paired two-tailed Student’s t-test.
m, Scatter plot of fold change for an efficacy assay in Colo205 tumor xenografts in nude mice treated with vehicle or MS140 (30 mg/kg, b.i.d) for 21 days. Vehicle treatment contained 5 animals (n=5). MS140 treatment contained 8 animals (n=8). Data are presented as mean ± S.D. Statistical significance was determined by unpaired two-tailed Student’s t-test.
n, qPCR analysis of expression of known Rb/E2F target genes in tumors, kidney and liver from mice bearing JeKo-1 tumors treated with vehicle or MS140 (25 mg/kg, b.i.d) or PB (50 mg/kg, q.d.) for 3 days. Data are presented as mean ± S.D. (Tumor: n=1 mouse with three independent experiments; kidney and liver: n=2 mice with two independent experiments for each mouse).
o, Neutrophil counts in C57BL/6 mice before treatment and post treatment with PB (50 mg/kg, q.d., n=8 mice) or MS140 (25 mg/kg, b.i.d., n=7 mice) for 21 days. Data are presented as mean ± S.D. Statistical significance was determined by paired two-tailed Student’s t-test.
All data represent single experiments.
Of note, a smaller group of cancer cell lines expressing high levels of CDK6 compared to CDK4 are sensitive to CDK4/6i and highly sensitive to MS140 (Fig. 5h, i). These are mostly cells from hematopoetic origin, but also include the colorectal cancer cell line Colo205. These tumor cells dependent on CDK612,29 (Extended Data Fig. 5b-d), indicating that in this subgroup of tumor cells, CDK6 is sensitive to CDK4/6i. Our data on growth response of hematopoetic tumor lines to PB and MS140 are summarized in Extended Data Fig. 5e. Thus, MS140 was more effective than PB in inhibiting Rb/E2F signaling and cell growth of many CDK4/6i-S tumor cell lines. CDK4/6i-S cells are predominantly those expressing CDK4 and low CDK6, but we also identified a smaller subset of CDK6-dependent tumor cells, that are also sensitive to CDK4/6i and more sensitive to the CDK4/6 degrader.
The increased effectiveness of MS140, as compared to PB in CDK4/6i-S cells, prompted us to assess its biochemical and antitumor effects in vivo. Treatment of mice with MS140 promoted degradation of CDK4/6 kinases and suppression of Rb/E2F signaling in JeKo-1 (MCL) and Colo25 (CRC) xenografts in vivo (Fig. 5j, 5k), resulting in equivalent or greater suppression of tumor growth compared to PB administered at the same total daily concentration (Fig. 5l, 5m and Extended Data Fig. 5f) without significant body weight loss (Extended Data Fig. 5g), or other apparent toxicities.
The increased potency of MS140 in tumor cells compared to PB, prompted us to assess the effect of MS140 on normal tissue to obtain some evidence on its potential therapeutic index. To this end, we determined the effect of treatment of the same daily dose of either MS140 or PB on expression of known E2F targets in tumor and normal tissue (kidney and liver). In fact, the analysis revealed that treatment with MS140 resulted in more potent inhibition in the tumor than in normal tissue of several E2F-target transcripts, compared to PB (Fig. 5n), suggesting that the strategy of combined inhibition and degradation of CDK4/6 may show higher therapeutic index than current clinical CDK4/6i. Finally, as a known toxicity induced by CDK4/6i is a reduction on white blood cells and neutropenia4, we performed complete blood counts in mice after three weeks of treatment with either PB or MS140 at the same total daily concentration. Consistent with the clinical experience4 and previous studies in mice46, treatment with PB resulted in a severe (over 50%) reduction in neutrophil count (Fig. 5o). Total white blood cell count and lymphocytes were also reduced in response to PB treatment, whereas red blood cells were slightly reduced (Extended Data Fig. 5h). However, neither the white blood cell nor lymphocytes count was decreased after treatment with MS140 (Fig. 5o), suggesting that the compound can be better tolerated at doses that are comparably effective with PB. Thus, our results indicate that CDK4/6-directed degraders may be both a highly effective and well-tolerated therapeutic option for patients with CDK4-driven tumors, as well as for patients with tumors dependent on the CDK4/6i-sensitive CDK6.
In CDK4/6-R cells, CDK4/6 degraders fail to degrade CDK6 due to weak binding of compound
Since MS140 both inhibits and degrades CDK4/6, we would expect it to show increased potency compared to the parent CDK4/6i in all cell lines. Surprisingly however, in all CDK4/6i-R tumor lines tested, treatment with MS140 suppressed Rb/E2F signaling and cell growth less effectively than PB (Fig. 6a, b). This result was unexpected, since shRNA-mediated knockdown of CDK6 sensitized these same tumor cells to PB (Fig. 3a, b) suggesting that MS140 downregulated CDK6 expression and activity with different potency in the two groups of cells lines. As MS140 is a bifunctional compound, the difference could be the result of either differential binding to CDK6, or difference in CRBN expression between the two groups of cell lines. As shown in Fig. 1e, with the exception of H358, CRBN is expressed at comparable levels in CDK4/6i-S and CDK4/6-R cell lines. Further, ectopic expression of CRBN in a CDK4/6i-R cell line did not increase CDK6 protein degradation or inhibition of downstream Rb/E2F output by compound (Extended Data Fig. 6a). We thus concluded that differences in CRBN expression cannot be the underlying basis for the difference in sensitivity to MS140, and focused on the effect of MS140 directly on CDK6.
Fig. 6. In CDK4/6-R cells, CDK4/6 degraders fail to degrade CDK6 due to weak binding of compound.
a, Cell growth crystal violet assay for the indicated cell lines treated with increasing concentrations of PB or MS140 for 10-12 days.
b, A375, SKMEL2 and Calu6 cells were treated with increasing concentrations of PB or MS140 for 24 hr. Lysates were immunoblotted with the indicated antibodies.
c, A549 or A375 expressing Dox-inducible shCDK6 were treated with either the indicated concentrations of doxycycline (A375: 0.1 μg/ml) or MS140 for 72 hr. Lysates were subjected to immunoblotting with the indicated antibodies.
d, Cell growth crystal violet assay for the indicated cell lines treated with either doxycycline or MS140 for 10 days. Colonies were stained with crystal violet.
e, Rb-proficient cell lines were treated with MS140 (3 nM) for the indicated time points. Cell lysates were immunoblotted with the indicated antibodies.
f, The indicated cell lines were treated with PB (1 μM/2 hr) followed by CETSA assay. The graphs are derived by immunoblot analysis of CDK6 and actin expression using Image J. The samples derive from the same experiment and the blots were processed in parallel.
g, The indicated cell lines were treated with MS140-ve (15 μM/2 hr) followed by CETSA assay. The graphs are derived by immunoblot analysis of CDK6 and actin expression using Image J. The samples derive from the same experiment and the blots were processed in parallel.
h, Desthiobiotin-ADP enrichment assay for CDK6 in MV4-11 and SKMEL2 cells pretreated with increasing concentrations of PB for 2 hr. Elute were subjected to immunoblotting with the indicated antibodies.
All data represent single experiments.
We first compared the extent of reduction of CDK6 protein expression in response to shRNA-mediated CDK6 knock-down or MS140 treatment. As shown in Fig. 6c, d, MS140 was less potent in promoting CDK6 protein degradation and in suppressing cell growth, compared to shRNA-mediated CDK6 knockdown. These results prompted us to directly compare the extent of CDK4/6 degradation by MS140 in CDK4/6i-S and CDK4/6i-R cell lines. Treatment with MS140 resulted in potent degradation of CDK4/6 in both CDK4-driven and CDK6-driven CDK4/6i-S cells, but promoted only minimal CDK4/6 degradation in CDK6-driven CDK4/6i-R cells (Fig. 6e). To assess whether this difference applied more generally to CDK4/6 degraders, we tested two recently reported CDK4/6-directed PROTACs, YKL-06-10223,24 and BSJ-02-16224. We found that, similarly to MS140, treatment with either compound resulted in more potent degradation of CDK6 in CDK4/6i-S as compared to CDK4/6i-R cells (Extended Data Fig. 6b, c). Of note, one of these compounds has been reported to be a CDK6-selective inhibitor, compared to CDK4. We confirmed that, as reported, that compound was relatively more potent degrader of CDK6 than CDK4 in CDK4/6-S cells, compared to other CDK4/6 degraders (not shown). However, as shown in Extended Data Fig. 6b, c, YKL-06-102 was similarly ineffective to the rest of CDK4/6-degraders in degrading CDK6 in the CDK4/6-R cells. Thus, failure to degrade CDK4/6 in CDK4/6i-R cells is a general property of CDK4/6 degraders.
To directly assess binding of CDK4/6i to CDK6 in cells, we compared the shift of the CDK6 Tm promoted by CDK4/6i in CDK4/6i-S versus CDK4/6i-R tumor cells (all driven by CDK6) by Cellular Thermal Shift Assay (CETSA)47,48. Treatment of CDK4/6i-S, but not CDK4/6i-R, cells with PB resulted in a shift in Tm, indicating strong inhibitor binding to CDK6 in these cells (Fig. 6f). Similar results were obtained with MS140-ve, used instead of MS140 to make sure there is no CRBN-mediated protein degradation that could confound the experiment (Fig. 6g). Finally, to assess inhibitor binding in cells using a second method, we employed a probe-based chemoproteomic method using biotinylated phosphates of ATP or ADP that irreversibly react with protein kinases on conserved lysine residues in the ATP binding pocket49, followed by western blot analysis. We found that PB prevented binding of the probe in CDK4/6i-S cells but not in CDK4/6i-R tumor cells (Fig. 6h). Together these data show that CDK6-dependent tumor cells can be divided in two groups. In most tumor cells that express CDK6, CDK4/6i bind weakly to CDK6 and these cells are resistant to CDK4/6i (and consequently to CDK4/6 degraders). In a smaller group of CDK6-dependent cells, CDK6 binds strongly to CDK4/6i, and they are thus sensitive to CDK4/6i and degraders.
CDK4/6i-resistant cells express CDK6 as a thermostable, weak HSP90 client protein
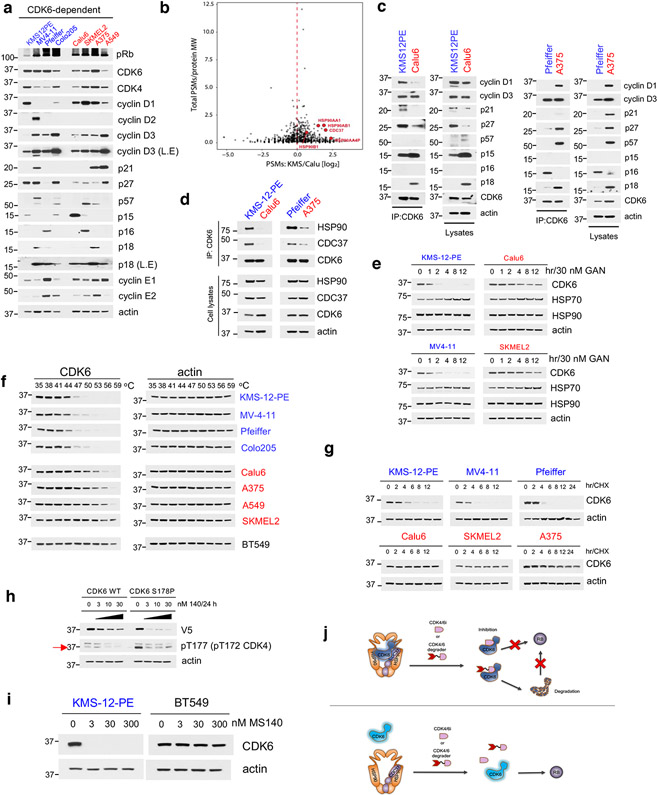
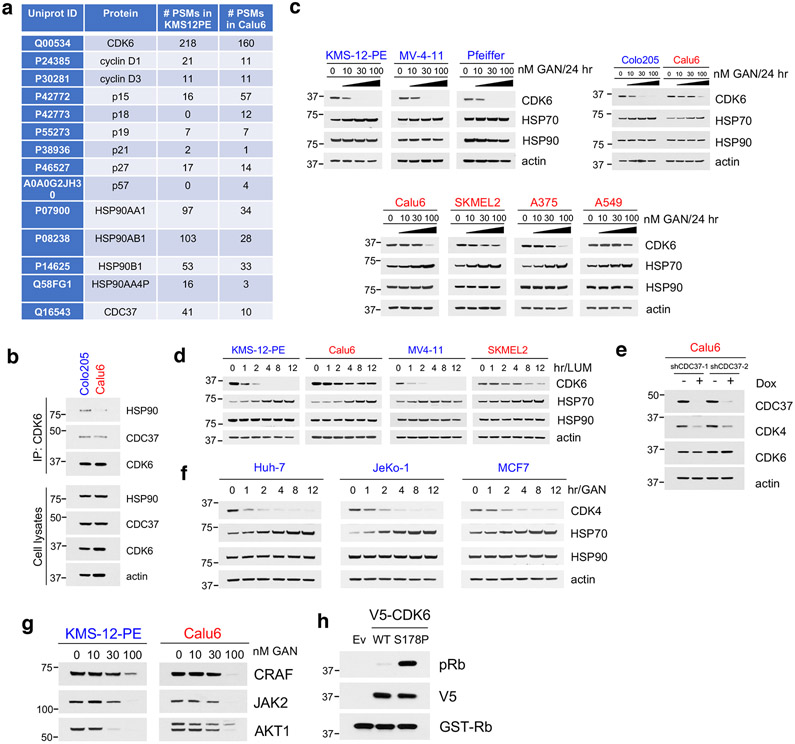
To gain mechanistic insight on the basis of the difference in binding of CDK6 to CDK4/6i, we carried out protein expression analysis of the main known CDK4/6-related regulators (Fig. 7a), as well as immunoprecipitation of CDK6 followed by mass spectrometry analysis of interacting partners in a CDK4/6-S (KMS-12-PE) and in a CDK4/6i-R (Calu6) cell line (both expressing high levels of and dependent on CDK6) (Fig. 7b). Association of CDK6 with known CDK6 interactors, including members of the cyclin, INK4 or CIP/KIP families correlated well with their relative basal expression in the two cell lines (Fig. 7c and Extended Data Fig. 7a), suggesting that additional factors affect tumor cell sensitivity to CDK4/6i. For example, p27 was found to be expressed at high levels and associate strongly with CDK6 in a CDK4/6i-S cell line (KMS12PE) as well as in a CDK4/6i-R line (A375) (Fig. 7c). We however noticed a 3-5 fold higher interaction of CDK6 with multiple components of the HSP90/CDC37 chaperoning complex in CDK4/6i-S compared to CDK4/6i-R cells (Fig. 7b and Extended Data Fig. 7a). The observation was further confirmed by direct co-immunoprecipitation experiments showing a much stronger interaction of CDK6 with HSP90 and CDC37 in CDK4/6i-S as compared to CDK4/6i-R tumor cells (Fig. 7d and Extended Data Fig. 7b).
Fig. 7. CDK4/6i-resistant cells express CDK6 as a thermostable, weak HSP90 client protein.
a, Immunoblots for cell cycle regulators in CDK6-dependent cell lines. L.E = long exposure.
b, Volcano plot of the CDK6-interacting proteins in KMS-12-PE and Calu6. Proteins in red were annotated as HSP90/CDC37-related.
c, Cell lysates from the indicated cell lines were either subjected to Co-IP with a CDK6 antibody followed by immunoblotting with known cell cycle regulators, or immunoblotted with the indicated antibodies.
d, Cell lysates from the indicated cell lines were either subjected to Co-IP with a CDK6 antibody followed by immunoblotting with HSP90, CDC37 and CDK6, or immunoblotted with the indicated antibodies.
e, Cell lines were treated with the HSP90 inhibitor Ganetespib (GAN, 30 nM) at the indicated time points. Cell lysates were immunoblotted with the indicated antibodies.
f, Immunoblots for CDK6 thermal stability assay (CETSA) in CDK4/6i-sensitive and CDK4/6i-resistant cell lines heat-treated at increasing temperature end points.
g, The indicated cell lines were treated with 100 μg/ml CHX at the indicated time points. Cell lysates were immunoblotted with the indicated antibodies.
h, A375 cells were transfected with either WT or CDK6(S178P) followed by treatment with increasing concentrations of MS140 for 24 h. Cell lysates were immunoblotted with the indicated antibodies.
i, KMS-12-PE and BT549 cells were treated with increasing concentrations of MS140 for 24 hr. Lysates were subjected to immunoblotting with the indicated antibodies.
j, Model of CDK6 association with the HSP90 complex affecting tumor cell sensitivity to CDK4/6 inhibitors and degraders. Top: In CDK4/6 inhibitor and degrader-sensitive cells CDK6 is associated with the HSP90 complex. CDK4/6 inhibitors or CDK6 degraders binds strongly CDK6, and promote CDK6 inhibition or both CDK6 inhibition and degradation respectively. Bottom: In CDK4/6 inhibitor and degrader-resistant cells, CDK6 is weakly associated with the HSP90 complex. In these cells, CDK4/6 inhibitors and degraders bind CDK6 weakly, and thus fail to promote CDK6 inhibition or both inhibition and degradation, respectively.
All data represent single experiments.
Association of a kinase with the HSP90/CDC37 complex has been shown to correlate with kinase dependence on HSP90 for its folding and stability15. In fact, treatment with the HSP90 inhibitor Ganetespib (GAN) resulted in much less potent CDK6 degradation in CDK4/6i-R cells, than in CDK4/6i-S cells (Fig. 7e and Extended Data Fig. 7c). A structurally distinct HSP90 inhibitor, Luminespib showed the same results (Extended Data Fig. 7d). Moreover, CDK6 protein levels remained unchanged after siRNA-mediated knockdown of CDC37 in a CDK4/6-R line (Extended Data Fig. 7e), further confirming that the HSP90/CDC37 chaperoning activity is not required for CDK6 protein stability in these cells. As expected, treatment of CDK4 only-expressing cells with GAN resulted in potent degradation of CDK4 (Extended Data Fig. 7f), a known strong HSP90 client18,50. Other known HSP90 clients were degraded to a similar degree upon GAN treatment (Extended Data Fig. 7g), indicating that the HSP90/CDC37 chaperone system operates similarly in the two settings, but CDK6 in CDK4/6i-S cells is expressed as a strong HSP90-client, whereas in CDK4/6i-R cells it is predominantly expressed as a weak HSP90-client.
Strong HSP90 client kinases have been shown to be highly thermo-unstable15. We compared the thermostability of CDK6 in CDK6-driven CDK4/6i-S versus CDK4/6i-R tumor cells using CETSA. Consistent with the relative association of the two CDK6 forms with HSP90, we found much lower thermostability (lower Tm) of CDK6 protein in CDK4/6i-S cells as compared to that of CDK6 expressed in CDK4/6i-R cells (Fig. 7f). CDK6 in the two cellular settings also differed remarkably in its overall stability. Cycloheximide treatment resulted in significantly faster degradation of CDK6 in CDK4/6i-S compared to CDK4/6i-R cells (Fig. 7g).
Stronger association of a kinase with HSP90 has been reported to shift the conformational ensemble of different activation states towards a more active distribution18,51. Crystallographic data show that CDK4/6i are Type I in that they bind the active conformation of CDK4/6 (αC-IN/DFG-IN))52. Our findings support a model by which, in CDK6-driven and CDK4/6i-S tumor cells, the CDK6 protein population is enriched in highly active conformations that have strong binding affinity for current CDK4/6i (and consequently for CDK4/6 degraders). In contrast, in CDK4/6i-R cells, CDK6 is expressed as thermostable, weak HSP90-client, with lower affinity for the CDK4/6i. To further test this idea, we inserted a previously reported activating mutation in CDK6 (S178P)53 and we confirmed that it activated CDK6 (Extended Data Fig. 7h). Treatment of cells expressing either wild-type (WT) or mutationally activated CDK6 with MS140 resulted in significantly more potent degradation of CDK6(S178P) compared to CDK6(WT) (Fig. 7h), consistent with a highly active conformation of CDK6 being more sensitive to degradation by MS140. Finally, a previous report using biotinylated phosphates of ATP or ADP followed by mass spectrometry has shown that in Rb-null cells, PB does not bind CDK4/6 kinases54. Consistent with those results, we found that in BT549, Rb-null cells, CDK6 is also highly thermostable and resistant to degradation by MS140, similar to the CDK6 state in PB-resistant cells (Fig. 7i and not shown).
Discussion
We show here that the expression state of CDK6 is a critical regulator of tumor response to CDK4/6i. Cells universally express CDK4, but CDK6 expression varies significantly across tumor types as well as within the same tumor type. We found that tumors with low CDK6 expression relative to CDK4 are sensitive to CDK4/6i. Such tumor types include luminal breast cancer, Ewing sarcomas, Mantle Cell Lymphomas and others, as well as subgroups within tumor types, such as a portion of lung adenocarcinomas. Among lung adenocarcinoma models, we found a subgroup with a low ratio of CDK6 to CDK4 protein expression that was sensitive to CDK4/6i. The association of low CDK6 expression with increased clinical response to CDK4/6i was further confirmed by analyzing clinical data in NSCLC patients27. Thus, collectively our data indicate that a low ratio of CDK6 to CDK4 expression in the tumor may be used to stratify patients that are likely to benefit from CDK4/6-based therapies. Further, it is becoming increasingly apparent, that in various contexts of intrinsic or acquired resistance, combinatorial targeting of parallel, or convergent growth factor signaling pathways, such as the PI3K/PTEN/AKT55-57 or the RAS/MAPK25,58 pathway may be necessary for effective and durable tumor response. Our findings suggest that selection of patients with low CDK6 relative to CDK4 expression, in combinations of CDK4/6i or CDK4/6 degraders with inhibitors of parallel pathways, such as PI3K, AKT, MEK or ERK inhibitors are likely to be most effective.
We further found that in most tumor cells CDK6 expression confers intrinsic resistance to CDK4/6i. The finding is consistent with CDK6 overexpression being a reported mechanism of acquired resistance to CDK4/6i in luminal breast cancer13,59. However, CDK4/6i have been shown here (Extended Data Fig. 4a) and by others1 to potently bind purified CDK6 in vitro. In addition, a portion of tumors (mostly of certain types of leukemias, such as AML) have been shown to require CDK614 and are sensitive to CDK4/6i, suggesting that CDK6 may or may not confer resistance to CDK4/6i depending on cellular context. We used a potent and selective CDK4/6 degrader that we developed (MS140) as a tool to resolve the discrepancy between the in vitro data and our findings in cells. By employing in-cell binding assays, we found that CDK6 is expressed in different tumors in either of two distinct forms, only one of which is accessible to binding by CDK4/6i (Fig. 7h). We further found that the two CDK6 forms differ dramatically in their thermostability and their dependence on the HSP90/CDC37 chaperone complex. The data are consistent with previous studies showing kinase dependence on HSP90 to be associated with an enrichment of folded, highly active states of the kinase18, which, in the case of CDK6, is structurally predicted to bind strongly to CDK4/6i. The conformation of the active site of the highly active CDK6 in cells likely resembles that of the highly active purified CDK6, commonly used in in vitro kinase and binding assays. Thus, the current clinical CDK4/6i have been developed to strongly bind and inhibit CDK6 selectively in the subgroup of tumor cells that express CDK6 similar to its purified, highly active state.
Recently, two studies have proposed the interaction of CDK4 to either p2760, or p1661 as mechanisms promoting resistance to CDK4/6 inhibitors. In another study, formation of complexes with members of the cyclin D family or with endogenous inhibitors of CDKs was shown to affect the interaction of different members of the CDK family of proteins with inhibitors and HSP90/CDC37 in vitro21. However, we observed p16 to be expressed at undetectable levels in most cancer cell lines tested in our experiments, including the ones that are insensitive to CDK4/6i (Fig. 1e, 7a), and we did not observe a correlation of endogenous expression of members of the cyclin D family with either CDK6 sensitivity to CDK4/6i or HSP90 dependence. Further, p27 is expressed in most CDK4/6i-S and CDK4/6-R cells at similar levels (Fig. 1e, 7a) and knock-down or overexpression of p27 did not affect sensitivity of cell lines to CDK4/6i (not shown). Finally, we found that all cell lines tested expressing CDK4 and low levels of CDK6 are universally sensitive to CDK4/6i, regardless of p16 or p27 status or expression levels. We therefore conclude that although the reported mechanisms may affect target engagement by CDK4/6i or the interaction of CDK4 or CDK6 with HSP90/CDC37 in certain contexts, they are unlikely the predominant mechanism by which large subsets of Rb-proficient tumors diverge in terms of their sensitivity to CDK4/6i.
The duality of CDK6 when expressed in different cells with respect to its dependence on HSP90/CDC37 challenges the current paradigm on the molecular determinants of kinase dependence on HSP90. HSP90/CDC37 dependency of a given kinase has been shown to be altered by genetic alterations, as in the cases of c-Src and v-Src15, wild-type and mutated BRAF19 and wild-type and mutated EGFR20. Previous studies using ectopically expressed proteins, have shown that CDK6 exhibited an intermediate dependency on HSP90 (HSP90 interaction score of 2.23), laying in between the highly dependent CDK4 (3.15) and the entirely HSP90-independent CDK2 (score of 0)15. However, we unexpectedly found that the same kinase (CDK6) can be expressed as a strong or weak HSP90/CDC37 client depending on the cellular context. As regulation of kinase function by the HSP90/CDC37 complex remains incompletely understood15, in the case of CDK6 may be affected by post-translational modification(s), interacting partner(s) or localization, in different cellular environments.
Current clinical CDK4/6i have been optimized to bind and inhibit CDK4 as well as the thermo-unstable, strong HSP90-client form of CDK6. However, based on our data, it is expected that a large number of tumors express thermostable CDK6 that is resistant to these drugs. Drug development efforts directed in potently targeting this newly identified state of CDK6 may bring about more effective therapies for a large number of patients with Rb-proficient tumors.
Methods
Compounds
Commercially available compounds Palbociclib, Ganetespib, Luminespib and Bortezomib were obtained from Selleckchem. Abemaciclib, Ribociclib and MLN4924 were obtained from Medchem Express. Blasticidin S, cycloheximide and doxycycline were purchased from Sigma. Chemical compounds were dissolved either in DMSO or water and stored at −20 °C.
Cell culture
Cell lines HEK293T, HEK293FT, Huh-7, U87MG, A375 and A673 were maintained in DMEM supplemented with 10% fetal bovine serum, 2 mM glutamine, and penicillin/streptomycin, MCF7, T47D, ZR-75-1, CAMA-1, NCI-H358, Colo205, SK-MEL-1, IGROV-1, TC-71, Mino, JeKo-1, Granta 519, Z-138, REC-1, KMS-12-PE, MV4-11, MOLM-14, Pfeiffer, SK-MEL-2, A549, Calu-6, NCI-H1792, NCI-H2087, NCI-H2291, NCI-H1915, NCI-H1666, NCI-H1395, HCC827, PC9, HCT15, HCT116, RKO, LoVo, HT-29, SW620 and BT549 were maintained in RPMI1640 supplemented with 10% fetal bovine serum, 2 mM glutamine, and penicillin/streptomycin (ThermoFisher Scientific).
MCF7, T47D, ZR-75-1, CAMA-1, NCI-H358, Colo205, A375, A549, SK-MEL-2, Calu-6, NCI-H1792, NCI-H2087, NCI-H2291, NCI-H1915, NCI-H1666, NCI-H1395, HCC827, RKO, HCT15, HCT116, SW620, LoVo, HT-29 and HEK293T were purchased from ATCC. PC9 cells were purchased from Sigma-Aldrich. U87MG, IGROV-1 and BT549 were kindly provided by Dr. Ramon Parsons. Huh-7 cells were provided by Dr. Amaia Lujambio. Z-138 cells were kindly provided by Dr. E. Premkumar Reddy. REC-1, KMS-12-PE and Pfeiffer were kind gifts from Dr. Samir Parekh. MV-4-11 and MOLM-14 cells were provided by Dr. Iannis Aifantis (New York Univeristy). Mino, JeKo-1, Granta 519 were kind gifts from Dr. Shannon M. Buckley (University of Nebraska). A673 and TC-71 were provided by Dr. Christine A. Pratilas (Johns Hopkins). HEK293FT cells were a kind gift from Dr. William Kaelin Jr (Harvard Medical School).
Plasmids
Tet-pLKO-puro (item #21915) and pcDNA3-FLAG-CRBN (item #107380) was purchased from Addgene. shCDK4 (TRCN0000018364 and TRCN0000010520), shCDK6 (TRCN0000010081: and TRCN0000000488) and shCDC37 (TRCN0000116633 and TRCN0000116632) (Supplementary Table 1) were subcloned into Tet-pLKO plasmid. Human CDK6 coding sequences were amplified from HEK293H cDNA using a SuperScript III First-Strand kit (Thermo Fisher). C-terminal V5-tagged CDK6 were subcloned using pcDNA3 as a backbone. shRNA-resistant form of V5-CDK6 and S178P V5-CDK6 were generated with a QuikChange II XL Site-Directed Mutagenesis Kit (Agilent).
siRNA knockdown
A673 , TC-71 and NSCLC cell lines were transfected with SMARTpool ON-TARGETplus Non-targeting pool, or SMARTpool ON-TARGETplus CDK4-siRNA (L-003238-00-0005), or SMARTpool ON-TARGETplus CDK6-siRNA (L-003240-00-0005) (Dharmacon) using Lipofectamine RNAiMAX (Thermo Fisher).
Western Blot and immunoprecipitation
Cells were washed with PBS and lysed on ice for 10 min in NP40 buffer (50mM Tris pH 7.5, 1% NP40, 150mM NaCl, 10% Glycerol, 1mM EDTA) supplemented with protease and phosphatase inhibitors (Roche). Lysates were centrifuged at 15,000 rpm for 10 min and the protein concentration was quantified using BCA (Thermo Fisher). Protein G agarose (Thermo Fisher) was used for immunoprecipitations. The following antibodies were used: phospho-Rb (Ser807/811, 1:1000), Rb (4H1, 1:1000), PLK1 (208G4, 1:1000), cyclin D2 (D52F9, 1:1000), cyclin E2 (1:1000), Akt1 (2H10, 1:1000), JAK2 (D2E12, 1:1000), DHFR, ß-Actin (13E5, 1:1000), anti-rabbit, HRP (1:1000) and anti-mouse, HRP (1:1000) (Cell Signaling), CDK4 (H-22), CDK6 (C-21; DCS-83, 1:1000), CDK2 (D-12, 1:1000), CDK7 (C-19, 1:2000), CDK9 (D-7, 1:3000), p15/16 (C-7, 1:400), p16 (C-20, 1:500), p18 (118.2, 1:250), p57 (KP39, 1:500), cyclin A (H-432, 1:2000; B-8, 1:1000), cyclin D1 (M-20, 1:400; A-12, 1:250), cyclin D3 (D-7, 1:500), cyclin E1 (HE12, 1:2000), CDC37 (H-271; E-4, 1:2000), Hsp70 (W27, 1:8000) and Hsp90α/ß (F-8, 1:8000) (Santa Cruz), p27 (1:1000) and c-Raf (1:2000) (BD Transduction Laboratories), p21 (1:1000) and phospho-CDK4-T172 (1:1000) (ABclonal), CRBN (1:500) (Novus Biologicals) and V5 (1:5000) (Thermo Fisher), FLAG M2 (1:3000) (Sigma).
In vitro CDK4 and CDK6 kinase assay
Palbociclib, MS140 and ribociclib were assayed by Reaction Biology Corporation for in vitro kinase activity. Briefly, kinase substrates were added to base reaction buffer [20 mM Hepes (pH 7.5), 10 mM MgCl2, 1 mM EGTA, 0.02% Brij35, 0.02 mg/ml BSA, 0.1 mM Na3VO4, 2 mM DTT, 1% DMSO]. Differential CDK4/6 complexed with cyclin D1 were then diluted in the substrate solution. A range concentrations of compounds were incubated with the kinase reaction mixture by acoustic technology (Echo550; nanoliter range) and incubated for 20 minutes at room temperature followed by the addition of 10 μ Ci/μL 33P-ATP. Reactions were incubated for 2 hours at room temperature and radioactivity was detected by filter binding methods. Kinase activity data were presented as % kinase activity in samples relative to DMSO control. IC50 values were calculated using GraphPad Prism 5.
Lentivirus transduction and stable cell lines
Lentivirus was produced by transfecting HEK293FT cells with a lentiviral transfer vector, psPAX2 and pMD.G at a 5:4:1 ratio using Lipofectamine 2000 (Thermo Fisher). The viral supernatant was collected 72 h after transfection and filtered through a 0.45μm filter unit (Millipore). For Dox-inducible stable CDK4/6 knockdown cell lines, cells were transduced with shCDK4 or shCDK6 lentivirus in the presence of 8 μg/ml polybrene (EMD Millipore) and selected with 2 μg/ml puromycin (Thermo Fisher) for 5-7 days.
Cellular thermal shift assay
For basal CDK6 thermal stability assay, cells resuspended in 1 ml PBS containing protease inhibitor cocktail (Roche) were aliquoted with 100 μl cell suspension each into nine PCR tubes (~ 3 x 106 cells). Heat the PCR tubes at different temperature endpoints (35-59 °C) for 3 min in the Veriti 96-well thermal cycler (Thermo Fisher). Remove and incubate the tubes at room temperature for 3 min. Snap-freeze the samples immediately in liquid nitrogen. Freeze and thaw the cells three times using liquid nitrogen and a thermal cycler at 25 °C. Cell lysates were centrifuged at 20,000 x g for 15 min at 4 °C. Supernatants were collected for Western blots.
For PB or MS140-ve-induced thermal shift assay, cells were treated with 1 μM PB or 15 μM MS140-Ve for 2 hr. Cells resuspended in PBS containing protease inhibitor cocktail (Roche) were aliquoted with 100 μl cell suspension each into nine PCR tubes. Samples were heated at different temperature endpoints for 3 min using the Veriti 96-well thermal cycler (Thermo Fisher), followed by incubation at room temperature for 3 min and snap-freezing in liquid nitrogen. After three freeze and thaw cycles, samples were centrifuged at 20,000 x g for 15 min at 4 °C. Supernatants were collected for Western blot analysis.
ATP-biotin competition assay
Cell lysates were prepared and labeled according to the manual instructions for the Pierce Kinase Enrichment Kits and ActivX Probes (Thermo Scientific). Briefly, cells were pre-treated with PB for 2 hr, lysed and centrifuged at 16,000 x g for 10 min at 4 °C. The supernatant was desalted through Zeba Spin Desalting Columns. 1 mg of total cell lysates in 500 μl were used for ATP competition reaction with a final concentration of 5 μM desthiobiotin-ADP probe for 10 min at room temperature. Samples were mixed with 500 μl 8 M urea and 50 μl streptavidin agarose for 1 hr at room temperature on a rotator. Beads were washed with 4 M Urea/lysis buffer and collected by centrifugation at 1000 x g for 1 min. Proteins were eluted with 2 x sample buffer at 95 °C for 5 min. Samples were analyzed by immunoblotting.
Generation of CRBN−/− ZR-75-1 cell line by CRISPR/Cas9
Human CRBN gRNA (5’-CACCGTAAACAGACATGGCCGGCGA-3’) was designed using the following website: http://crispr.mit.edu/ and cloned into BsmBI-digested lentiCRISPRv2 (Addgene, #52961). ZR-75-1 cells were transduced with sgCRBN lentivirus in the presence of 8 μg/ml polybrene followed by selecting with 2 μg/ml puromycin for 5 days. Cells were plated at 0.3 cells/well in a 96-well plate. After 2-3 weeks, individual clones were expanded. CRBN homozygous knockout clones were validated by genotyping with primers (F : 5’- AAG TCA TGC TAA GGG CTG GAA C −3’, R: 5’- GGA TGG GTT TCC TGT TCT TAA TAG −3’) and Western Blotting.
Sample preparation for quantitative mass spectrometry analysis
Colo205 cells were treated with 0.3 μM MS140 or MS140-ve for 5 hr in duplicate. Cell pellets were lysed in lysis buffer containing 8 M urea, 50 mM Tris, pH 8.0, 75 mM NaCl, 1 mM MgCl2, and 500 units Benzonase. Proteins were reduced with DTT and alkylated with iodoacetamide. After precipitation, proteins were first digested with LysC for 4 hr at 37 °C. The solution was diluted 4-fold with 25 mM Tris, pH 8.0, 1 mM CaCl2 and further digested with trypsin (Promega) for 12 hr at 37 °C. Peptides were desalted on Sep-Pak Light C18 cartridges (Waters, Milford, MA) and dissolved in 30% acetonitrile, 0.1% TFA before loading on a 300-m Source 15S (GE Healthcare, Pittsburgh, PA) column for basic reversed phase chromatography (bRPLC). A linear LC gradient was performed by increasing buffer B from 0 to 70% within 60 min, where buffer A was aqueous 10 mM ammonium formate, and buffer B was 90% AcCN (Acetonitrile) in 10 mM ammonium formate. A total of 30 fractions were collected for each of the basal (WHIM2) and luminal (WHIM16) samples and non-contiguously recombined to five fractions per sample. The fractions were dried and desalted using a stop-and-go-extraction tip (StageTip) protocol containing 4 1-mm C18 extraction disk (3 M).
Liquid Chromatography-Tandem Mass Spectrometry
Samples were desalted using PepClean C18 spin columns (Pierce) according to the manufacturer’s directions and resuspended in aqueous 0.1% formic acid. Sample analysis was performed via reversed phase LC-MS/MS using a Proxeon 1000 nano-LC system coupled to a Q Exactive mass spectrometer (Thermo Scientific, San Jose, CA). The Proxeon system was configured to trap peptides using a 3-cm long, 100-m inner diameter C18 column at 5 l/min liquid flow that was diverted from the analytical column via a vent valve, whereas elution was performed by switching the valve to place the trap column in line with a 15-cm long, 75-m inner diameter, 3.5-m, 300-Å particle C18 analytical column. Analytical separation of all the tryptic peptides was achieved with a linear gradient of 2–30% buffer B over 240 min at a 250 nl/min flow rate, where buffer A was aqueous 0.1% formic acid, and buffer B was acetonitrile in 0.1% formic acid. LC-MS experiments were also performed in a data-dependent mode with full MS (externally calibrated to a mass accuracy of 5 ppm and a resolution of 70,000 at m/z 200) followed by high energy collision-activated dissociation-MS/MS of the top 20 most intense ions. High energy collision-activated dissociation-MS/MS was used to dissociate peptides at a normalized collision energy of 27 eV in the presence of nitrogen bath gas atoms. All five bRPLC fractions were derived from three process technical replicates of each tumor sample and were subjected to two independent LC-MS runs resulting in the production of 20 LC-MS runs for global peptide analysis. Mass spectra were processed, and peptide identification was performed using the Andromeda search engine found in MaxQuant software version 1.3.0.5 (Max Planck Institute, Germany). All protein database searches were performed against the UniProt human and mouse protein sequence database downloaded from the Clinical Proteomic Tumor Analysis Consortium Data Portal. This database contains 105,001 annotated proteins, and the sequences were derived from the UniProt December 2012 assembly. Peptides were identified with a target-decoy approach using a combined database consisting of reverse protein sequences of the UniProt human, mouse, and common repository of adventitious proteins. The common repository of adventitious proteins database was obtained from the Global Proteome Machine. Peptide inference was made with a false discovery rate (FDR) of 1%, and peptides were assigned to proteins with a protein FDR of 5%. A precursor ion mass tolerance of 20 ppm was used for the first search that allowed for m/z retention time recalibration of precursor ions that were then subjected to a main search using a precursor ion mass tolerance of 6 ppm and a product ion mass tolerance 0.5 Da. Search parameters included up to two missed cleavages at Lys/Arg on the sequence, oxidation of methionine, and protein N-terminal acetylation as a dynamic modification. Carbamidomethylation of cysteine residues was considered as a static modification. Peptide identifications are reported by filtering of reverse and contaminant entries and assigning to their leading razor protein. All of the mass spectrometry data on PDX tumor samples were deposited at the CPTAC Data Coordinating Center as raw and mzML files for public access.
Peptide and Protein Quantitation
LFQ was performed based on peak area. The measured area under the curve of m/z and the retention time-aligned extracted ion chromatogram of a peptide were performed via the label-free quantitation module found in MaxQuant version 1.3.0.5 (30). All replicates for each PDX were included in the LFQ experimental design with peptide-level quantitation performed using unique and razor peptide features corresponding to identifications filtered with a posterior error probability of 0.06, peptide FDR of 0.01, and protein FDR of 0.05. The MaxQuant peptide and protein groups files were processed and stored in an Oracle database, and statistical analysis, model building, and visualization of a majority of data were performed based on Statistical Analysis Software code and R script that was developed in-house.
Mass spectrometry to identify CDK6-interacting proteins
Preparation of Samples for Mass Spectrometry
The affinity purified proteins were reduced, alkylated, and digested with trypsin directly on the beads. Briefly, the beads were resuspended in 100uL 100mM ammonium bicarbonate. Proteins were reduced with 2μl of 0.2M dithiothreitol (Sigma) for one hour at 57 °C at pH 7.5, alkylated with 2μl of 0.5M iodoacetamide (Sigma) for 45 minutes at room temperature in the dark, and digested using 200ng sequencing grade trypsin (Promega) overnight at room temperature with gentle shaking. The solution was transferred to a new tube and the digestion stopped by adding 100 ul of a 5% formic acid and 0.2% trifluoroacetic acid (TFA) R2 50 μm Poros (Applied Biosystems) beads slurry in water. The samples were allowed to shake at 4°C for 3 hour. The beads were loaded onto C18 ziptips (Millipore), equilibrated with 0.1% TFA, using a microcentrifuge for 30 seconds at 6,000 rpm. The beads were washed with 0.5% acetic acid. Peptides were eluted with 40% acetonitrile in 0.5% acetic acid followed by 80% acetonitrile in 0.5% acetic acid. The organic solvent was removed using a SpeedVac concentrator and the sample reconstituted in 0.5% acetic acid.
Mass Spectrometry Analysis
1/10th of each sample was loaded onto an Acclaim PepMap trap column (2 cm x 75 μm) in line with an EASY-Spray analytical column (50 cm x 75 μm ID PepMap C18, 2 μm bead size) using the auto sampler of an EASY-nLC 1200 HPLC (Thermo Fisher Scientific) with solvent A consisting of 2% acetonitrile in 0.5% acetic acid and solvent B consisting of 80% acetonitrile in 0.5% acetic acid. The peptides were gradient eluted into a Thermo Fisher Scientific Q Exactive HF-X Mass Spectrometer using the following gradient: 5 - 35% in 60 min, 35 - 45% in 10 min, followed by 45 - 100% in 10 min. High resolution full MS spectra were recorded with a resolution of 45,000, an AGC target of 3e6, with a maximum ion time of 45ms, and a scan range from 400 to 1500m/z. The MS/MS spectra were collected using a resolution of 15,000, an AGC target of 1e5, maximum ion time of 120ms, one microscan, 2 m/z isolation window, and Normalized Collision Energy (NCE) of 27.
Data Processing
The MS/MS spectra were searched against the Uniprot human reference proteome database containing common contaminant proteins using Sequest within Proteome Discoverer 2.3. The search parameters were as follows: precursor mass tolerance ±10 ppm, fragment mass tolerance ±0.02 Da, digestion parameters trypsin allowing two missed cleavages, fixed modification of carbamidomethyl on cysteine, variable modification of oxidation on methionine, variable modification of deamidation on glutamine and asparagine, and a 1% peptide and protein FDR searched against a decoy database. The results were filtered to only include proteins identified by at least two unique peptides.
Cell viability assay
Cells were seeded at 3-10 x 103 cells/well in 96-well plates. 24 hr after seeding, cells were treated with palbociclib and MS140 at a range of concentrations for 72 hr. 10 μl of 0.1 mg/ml resazurin (Sigma-Aldrich) or 20 μl of CellTiter 96® AQueous One Solution Reagent (Promega) was added to cells and incubated for 2-3 hr at 37°C. Cell viability was determined by measuring the fluorescence at 560 nm excitation wavelength and 590 nm emission wavelength (resazurin) or 490 nm (CellTiter 96® AQueous One Solution Reagent) using a Molecular Devices Spectramax M5 plate reader. IC50 values were calculated using log-transformed, normalized data in GraphPad Prism 5.0.
Crystal violet cell growth assay
Cells were seeded at 1-10 x 103 cells/well in six-well plates. The next day, cells were treated with the increasing concentrations of palbociclib and MS140 for 10-15 days. Cell culture medium was replaced every 2 days in the presence or absence of inhibitors. Cells were fixed with 10% formalin solution (Sigma-Aldrich) for 5 min at RT followed by 0.05% crystal violet for 25 min. cells were de-stained with tap water and air-dried.
Quantitative Real-time PCR
Total RNA was extracted using Trizol Reagent (Thermo Fisher). Complementary DNA was synthesized with a SuperScript IV First-Strand kit (Thermo Fisher). Quantitative real-time PCR was performed using a Fast SYBR Green Master Mix kit (Thermo Fisher) with a 7500 Fast real-time PCR system (Applied Biosystems). PCR primers are listed in Supplementary Table 1. Differences in expression were calculated by the ΔΔCt method.
Genetic dependency data and genomics data
CRISPR dependency data were obtained from the 20Q1 public Avana dataset containing genome-scale CRISPR knockout screens for 18,333 genes in 739 cell lines. The gene dependencies were estimated for each gene and cell lines by the CERES algorithm31. RNA interference (RNAi) dependency data were derived from combination of the Broad Institute Project Achilles, Novartis Project Drive, and Marcotte et al. database32-35. The genetic dependencies were estimated using the DEMETER2 model across 712 unique cancer cell lines36. Cancer cell line mRNA expression were taken from the DepMap 20Q1 data release. Cancer cell line encyclopedia (CCLE) proteomics data were obtained by quantitative profiling of proteins by mass spectrometry across 375 cell lines1. The normalized protein quantification data and cell line omics data can be downloaded at DepMap (depmap.org/protal/). All cell line omics data can be downloaded at DepMap depmap.org/portal/.
Animal experiments
To determine in vivo degradation efficacy, 5-7 week-old female athymic Nude-Foxn1nu mice obtained from Envigo Laboratories were injected subcutaneously with 1 x 107 JeKo-1 cells in 1:1 PBS/ Matrigel GFR membrane Matrix (Corning) or 5 x 106 Colo205 cells in PBS. Mice were treated with vehicle (5% DMSO and 95% PEG 300) or MS140 (25 or 30 mg/kg) intraperitoneally, twice daily, or palbociclib (50 or 60 mg/kg) orally once daily for 3 days when tumors reach around 100 mm3. 5 h after the last dose, tumors were collected for further analysis. Liver and kidney were collected for qPCR analysis.
For efficacy of MS140 on tumor xenograft, 5 x 106 Colo205 cells in PBS or 10 x 106 JeKo-1 cells in 1:1 PBS/ Matrigel GFR membrane Matrix were injected subcutaneously on the left flank in 6-week old female athymic Nude-Foxn1nu mice. Tumors were allowed to reach 100 mm3 in size before the animals were randomized in two groups of 5-8 mice per group. Mice were treated with vehicle (5% DMSO and 95% PEG 300) or MS140 (25 or 30 mg/kg) intraperitoneally, twice daily, or palbociclib (50 or 60 mg/kg) orally once daily for 3 weeks. Tumor size was measured using caliper every 3 days and tumor volume was calculated as the following formula: (Length x Width2)/2. Tumor samples and organs were collected at the end of treatment for further analysis. All experiments were conducted under a protocol approved by Mount Sinai School of Medicine Institutional Animal Care and Use Committee (IACUC-2016-0066). All mice were maintained in specific pathogen-free barrier conditions under 12 hr light/12 hr dark cycle and with temperature and humidity set points at 20-25% and 30-70%, respectively.
Complete blood count
5-7 week-old female C57BL/6 mice (Envigo Laboratories) were dosed intraperitoneally with 25 mg/kg MS140 twice daily or 50 mg/kg PB once daily via oral gavage for 3 weeks. Mouse blood samples were collected in K3EDTA tubes before treatment and last treatment. Complete blood counts were performed with Coulter Ac·T 5diff Hematology Analyzer (BECKMAN).
Trial design
JUNIPER Study: A randomized phase III study of Abemaciclib with best supportive care versus Erlotinib with best supportive care in patients with stage IV non-small-cell lung cancer with a detectable KRAS mutation whose disease has progressed after platinum-based chemotherapy.
RNA sequence
148 patients have evaluable FFPE tumor sections for CDK6 RNA sequencing and further statistical analysis. RNA Exome sequencing was performed by Almac with the Illumina TruSeq RNA Sample Preparation Kit v2. The CDK6 sequencing data were quantile-normalized across samples.
Statistical method and cut point selection
The Kaplan-Meier product limit method were used to estimate the survival curve (OS or PFS) for treatment arms in both CDK6 expression low and high subgroup. Unstratified Cox proportional hazard model were performed to compare treatment effect in both CDK6 expression low and high subgroup, Hazard ratio (HR) estimates and it’s 95% CIs from the Cox model were reported on the Kaplan-Meier plot. Cutoff to define CDK6 expression low and high subgroup was derived from unstratified Cox proportional hazard interaction model by minimizing the treatment by CDK6 interaction p-value in terms of overall survival.
Software
R programming language
Chemical synthesis
Chemistry General Procedures.
HPLC spectra for all compounds were acquired using an Agilent 1200 Series system with DAD detector. Chromatography was performed on a 2.1×150 mm Zorbax 300SB-C18 5 μm column with water containing 0.1% formic acid as solvent A and acetonitrile containing 0.1% formic acid as solvent B at a flow rate of 0.4 mL/min. The gradient program was as follows: 1% B (0–1 min), 1–99% B (1–4 min), and 99% B (4–8 min). High-resolution mass spectra (HRMS) data were acquired in positive ion mode using an Agilent G1969A API-TOF with an electrospray ionization (ESI) source. Nuclear Magnetic Resonance (NMR) spectra were acquired on a Bruker DRX-600 spectrometer (600 MHz 1H, 150 MHz 13C) or a Varian Mercury spectrometer (400 MHz 1H, 100 MHz 13C). Chemical shifts are reported in ppm (δ). Preparative HPLC was performed on Agilent Prep 1200 series with UV detector set to 254 nm. Samples were injected into a Phenomenex Luna 75 x 30 mm, 5 μm, C18 column at room temperature. The flow rate was 40 mL/min. A linear gradient was used with 10% (or 50%) of MeOH (A) in H2O (with 0.1 % TFA) (B) to 100% of MeOH (A). HPLC was used to establish the purity of target compounds. All final compounds had > 95% purity using the HPLC methods described above.
(2-(2,6-Dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)glycine.
A solution of 2-(2,6-dioxopiperidin-3-yl)-4-fluoroisoindoline-1,3-dione (1.38 g, 5.0 mmol), tert-butyl glycinate (0.66 g, 5.0 mmol), and N,N-diisopropylethylamine (1.31 mL, 7.5 mmol) in DMF (10 mL) was heated to 85 °C in a microwave reactor for 40 min. After cooling to RT, the reaction was diluted with water and extracted with ethyl acetate (3x). Combined organic phase was dried over anhydrous sodium sulfate and concentrated under reduced pressure. The resulting residue was purified by silica gel chromatography to give the desired tBu ester intermediate as oil. This intermediate was treated with a solution of hydrogen chloride in dioxane (10 mL, 4.0 M) for 16 h. The reaction was concentrated under reduced pressure to give desired acid product (0.24 g, 14%). 1H NMR (600 MHz, CD3OD) δ 7.57 (dd, J = 8.5, 7.1 Hz, 1H), 7.11 (d, J = 7.1 Hz, 1H), 6.95 (d, J = 8.5 Hz, 1H), 5.07 (dd, J = 12.6, 5.5 Hz, 1H), 4.12 (s, 2H), 2.86 (ddd, J = 18.0, 14.4, 5.4 Hz, 1H), 2.74 – 2.67 (m, 2H), 2.15 – 2.08 (m, 1H). MS (ESI) m/z 332.1 [M+H]+.
4-((2-(4-(6-((6-Acetyl-8-cyclopentyl-5-methyl-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidin-2-yl)amino)pyridin-3-yl)piperazin-1-yl)-2-oxoethyl)amino)-2-(2,6-dioxopiperidin-3-yl)isoindoline-1,3-dione (MS140).
To a solution of (2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)glycine (0.017 g, 0.051 mmol) in DCM (10 mL) and DMSO (2 mL) were added palbociclib (0.023 g, 0.051 mmol), 4-methylmorpholine (0.020 g, 0.20 mmol), 1-hydroxy-7-azabenzotriazole (0.0090 g, 0.066 mmol) and N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride (0.013 g, 0.066 mmol). The reaction mixture was allowed to stir at rt for 16 h, before being concentrated under reduce pressure. The resulting residue was purified by reverse-phase prep-HPLC to yield the product (0.028 g, 72%) as yellow solid. 1H NMR (600 MHz, CD3OD) δ 9.08 (s, 1H), 8.21 (dd, J = 9.6, 2.8 Hz, 1H), 7.86 (d, J = 2.8 Hz, 1H), 7.55 – 7.50 (m, 2H), 7.03 (d, J = 7.0 Hz, 1H), 6.99 (d, J = 8.5 Hz, 1H), 6.04 – 5.97 (m, 1H), 5.08 (dd, J = 12.7, 5.5 Hz, 1H), 4.24 (s, 2H), 3.88 – 3.73 (m, 4H), 3.42 – 3.31 (m, 4H), 2.92 – 2.82 (m, 1H), 2.80 – 2.67 (m, 2H), 2.50 (s, 3H), 2.43 (s, 3H), 2.35 – 2.27 (m, 2H), 2.15 – 2.06 (m, 3H), 1.94 – 1.87 (m, 2H), 1.73 – 1.66 (m, 2H). HRMS (ESI-TOF) m/z: [M+H]+ calcd for C39H41N10O7, 761.3160; found: 761.3150.
Statistics and Reproducibility
No data were excluded in our analysis. All data represent single experiments. In animal experiments, the tumors were randomized across treatments, the investigators were not blinded to allocation during experiments and outcome assessment. Statistical analysis was performed using a two-tailed, paired or unpaired Student’s t-tests between two groups. Variation is indicated with S.D., presented as mean ± S.D. unless otherwise specified. GraphPad Prism software (v5) and R v.3.6.3 were used for statistical calculations.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data Availability Statement
The mass spectrometry raw files for the CDK6 complex analysis are available at the Mass Spectrometry Interactive Virtual Environment (MassIVE) (https://massive.ucsd.edu) under MassIVE ID: MSV000086571.
The mass spectrometry files for global protein degradation have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD023137.
The normalized protein quantification data and cell line omics data can be downloaded at DepMap (depmap.org/protal/).
Reagents generated in this study will be made available on request, but we may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
Code Availability
Data analysis was performed in R v.3.6.3 using custom-made or publicly available R packages. The code is available from GitHub (https://github.com/lijin0303/CDK46_expression_dependency). Figure 3e and Extended Data Figure 3b were generated with R 3.6.3 (Packages taigr, dplyr, tidyr, ggplot2, ggpubr), the corresponding code can be found in https://github.com/lijin0303/CDK46_expression_dependency.
Extended Data
Extended Data Fig. 1. Intrinsic resistance to CDK4/6i is associated with incomplete inhibition of Rb/E2F and expression of CDK6.
a, Cell growth crystal violet assay for the indicated cell lines treated with increasing concentrations of PB for 10-16 days and stained with crystal violet. CDK4/6i-sensitive cell lines were highlighted in blue, CDK4/6i-resistant cell lines were in red. b, MCF7 and HCT116 were treated with 1 μM PB for 24, 48 and 72 hr and lysates were immunoblotted with the indicated antibodies. c, Colo205 cells were treated with 1 μM PB at the indicated time points. Cell lysates were immunoblotted with the indicated antibodies.
Extended Data Fig. 2. Low expression of CDK6 predicts for sensitivity to CDK4/6i in NSCLC.
a, The indicated cell lines were treated with increasing concentrations of PB for 24 hr. Lysates were immunoblotted with the indicated antibodies. b, GI90 values of PB and CDK4/6 dependency in NSCLC cell lines.
Extended Data Fig. 3. Tumors expressing both CDK4 and CDK6 depend selectively on CDK6.
a, A673 and TC-71 cells were transfected with non-targeting control or siCDK4 or siCDK6 for 72 hr. Lysates were immunoblotted with the indicated antibodies. b, Relationship between CDK4 and CDK6 expression (CCLE RNA-seq) and DepMap CRISPR–Cas9 single-gene knockout scores (CERES; 20Q1 public dataset). All expression values are in log2(TPM +1). Cell lines harboring COSMIC hotspot mutations to RB1 are annotated in orange. P-values were calculated based on linear regression analysis.
Extended Data Fig. 4. Development of MS140, a potent and selective CDK4/6-degrader (PROTAC).
a. IC50 of in vitro kinase activity assays for PB and MS140 against CDK4/cyclin D1 and CDK6/cyclin D1. b, T47D cells were pretreated with either the proteasome inhibitor 100 nM bortezomib (BOR), 10 μM PB, 10 μM pomalidomide (POM) or 1 μM MLN4924 (MLN) for 4 hr, followed by treatment with MS140 (100 nM/3 hr). Lysates were subjected to immunoblotting with the indicated antibodies. c, Chemical structure of the MS140 negative control (MS140-ve) that does not bind CRBN.
Extended Data Fig. 5. CDK4/6-directed degradation is more effective than CDK4/6i in CDK4/6i-S tumor cells.
a, MCL cell lines were treated with 0.1 μM PB or MS140 at different time points. Lysates were immunoblotted with the indicated antibodies. b, Colo205 cells expressing Dox-inducible shCDK4 or shCDK6 were treated with or without 0.1 μg/ml doxycycline for 72 hr and cell lysates were subjected to immunoblotting with the indicated antibodies. c, Colo205 cells expressing Dox-inducible shCDK4 or shCDK6 were treated with or without 0.1 μg/ml doxycycline for 10 days followed by crystal violet staining. d, Dependency score of CDK4 and CDK6 from cancer cell line encyclopedia (CCLE) and Depmap portal database. e, GI50 values of PB and MS140 in hematologic cancer cell lines. f, Growth curve for an efficacy assay in JeKo-1 tumor xenografts in nude mice treated with vehicle or MS140 (25 mg/kg, b.i.d) or PB (50 mg/kg, q.d.) for 21 days. Each treatment contained 8 animals (n=8). Data represent mean ± SEM. g, Body weight in mice bearing JeKo-1 tumors treated with vehicle (n=8) or PB (50 mg/kg, q.d., n=8) or MS140 (25 mg/kg, b.i.d., n=8) in the course of the experiment (21 days). Data are presented as mean ± S.D. h, White blood cell, lymphocytes and red blood cell counts in C57BL/6 mice before treatment and post treatment with PB (50 mg/kg, q.d., n=8) or MS140 (25 mg/kg, b.i.d., n=7) for 21 days. Data are presented as mean ± S.D. Statistical significance was determined by paired two-tailed Student’s t-test.
Extended Data Fig. 6. In CDK4/6-R cells, CDK4/6 degraders fail to degrade CDK6 due to weak binding of compound.
a, Calu6 cells transiently expressing pcDNA3 (Ev) or pcDNA3-Flag-CRBN were treated with increasing concentrations of MS140 for 24 hr. Lysates were subjected to immunoblotting with the indicated antibodies. b, MV4-11 and A375 were treated MS140 (3 nM) or YKL-06-102 (3 nM) or BSJ-02-162 (3 nM) at different time points. Lysates were subjected to immunoblotting with the indicated antibodies. c, KMS-12-PE and Calu6 were treated MS140 (3 nM) or YKL-06-102 (3 nM) or BSJ-02-162 (3 nM) at different time points. Lysates were subjected to immunoblotting with the indicated antibodies.
Extended Data Fig. 7. CDK4/6i-resistant cells express CDK6 as a thermostable, weak HSP90 client protein.
a, Comparison of total peptide-spectrum match (PSM) for CDK6-interacting proteins by mass spectrometry in KMS-12-PE and Calu6. b, Cell lysates from Colo205 and Calu6 were either subjected to Co-IP with a CDK6 antibody followed by immunoblotting with HSP90, CDC37 and CDK6, or immunoblotted with the indicated antibodies. c, The indicated cell lines were treated with increasing concentrations of Ganetespib (GAN) for 24 hr. Lysates were subjected to immunoblotting with the indicated antibodies. d, The indicated cell lines were treated with 40 nM Luminespib (LUM) at the indicated time points. Lysates were subjected to immunoblotting with the indicated antibodies. e. Calu6 cell line expressing Dox-inducible shCDC37 were treated with or without 0.1 μg/ml doxycycline for 72 hr and cell lysates were subjected to immunoblotting with the indicated antibodies. f, CDK4-dependent cell lines were treated with 30 nM GAN for the indicated time points. Lysates were subjected to immunoblotting with the indicated antibodies. g, KMS-12-PE and Calu6 were treated with increasing concentrations of GAN for 24 hr. Lysates were subjected to immunoblotting with the indicated antibodies. h, Lysates from A375 cells ectopically expressing V5-CDK6 or V5-CDK6 S178p were immunoprecipited with a V5 antibody. The immunoprecipitates were subjected to kinase assay with recombinant Rb protein as substrate.
Supplementary Material
Acknowledgments
We thank A. Lujambio, S. Parekh, R. Parsons, E.P. Reddy, R. Sachidanandam, (Icahn School of Medicine at Mount Sinai), I. Aifantis (New York University), S.M. Buckley (University of Nebraska), C.A. Pratilas (Johns Hopkins University) and W.G. Kaelin Jr. (Harvard Medical School) for sharing cell lines and reagents. P.I.P. is supported by the NIH/NCI (R01CA204314, R01 CA240362 and R01CA238229), the Irma T. Hirschl Trust, the Manhasset Women’s Coalition against Breast Cancer, the Breast Cancer Alliance, the Melanoma Research Foundation, the Melanoma Research Alliance and Tisch Cancer Institute developmental awards. T.A.A. has been supported by grant T32CA078207, and Z.K. would like to acknowledge the 2017 Robin Chemers Neustein Postdoctoral Fellowship. W.R.S acknowledges support from the NIH (1R01CA233626) and the Ludwig Center at Harvard, T.I. is supported by the Department of Defense Peer Reviewed Cancer Research Program Horizon Award (W81XWH-19-1-0271). S.A.A. acknowledges support from a grant from the Breast Cancer Research Foundation. This work was supported in part by the P30CA196521 grant (to J.J.) from the US National Institutes of Health and an endowed professorship from the Icahn School of Medicine at Mount Sinai (to J.J.). This work utilized the AVANCE NEO 600 MHz NMR Spectrometer System that was upgraded with funding from a National Institutes of Health SIG grant 1S10OD025132-01A1.
Footnotes
Competing interests
X.W., X.Y., Y.X., T.A., Z.K., J.L., J.J and P.I.P hold a patent on CDK4/6-directed degradation (WO/2018/106870). The Jian Jin laboratory received research funds from Celgene Corporation, Levo Therapeutics, and Cullgen, Inc. J.J. is an equity shareholder and consultant of Cullgen, Inc. H.W. and S.G.B. are employees of Eli Lilly. All other authors declare no competing interests.
References
- 1.Sherr CJ, Beach D & Shapiro GI Targeting CDK4 and CDK6: From Discovery to Therapy. Cancer Discov 6, 353–367, doi: 10.1158/2159-8290.CD-15-0894 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Leary B, Finn RS & Turner NC Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol 13, 417–430, doi: 10.1038/nrclinonc.2016.26 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Otto T & Sicinski P Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer 17, 93–115, doi: 10.1038/nrc.2016.138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finn RS et al. Palbociclib and Letrozole in Advanced Breast Cancer. N Engl J Med 375, 1925–1936, doi: 10.1056/NEJMoa1607303 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Hortobagyi GN et al. Ribociclib as First-Line Therapy for HR-Positive, Advanced Breast Cancer. N Engl J Med 375, 1738–1748, doi: 10.1056/NEJMoa1609709 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Ingham M & Schwartz GK Cell-Cycle Therapeutics Come of Age. J Clin Oncol, JCO2016690032, doi: 10.1200/JCO.2016.69.0032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goetz MP et al. MONARCH 3: Abemaciclib As Initial Therapy for Advanced Breast Cancer. J Clin Oncol 35, 3638–3646, doi: 10.1200/JCO.2017.75.6155 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Im SA et al. Overall Survival with Ribociclib plus Endocrine Therapy in Breast Cancer. N Engl J Med 381, 307–316, doi: 10.1056/NEJMoa1903765 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Knudsen ES & Witkiewicz AK The Strange Case of CDK4/6 Inhibitors: Mechanisms, Resistance, and Combination Strategies. Trends Cancer 3, 39–55, doi: 10.1016/j.trecan.2016.11.006 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamilton E & Infante JR Targeting CDK4/6 in patients with cancer. Cancer Treat Rev 45, 129–138, doi: 10.1016/j.ctrv.2016.03.002 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Gong X et al. Genomic Aberrations that Activate D-type Cyclins Are Associated with Enhanced Sensitivity to the CDK4 and CDK6 Inhibitor Abemaciclib. Cancer Cell 32, 761–776 e766, doi: 10.1016/j.ccell.2017.11.006 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Kim S et al. The potent and selective cyclin-dependent kinases 4 and 6 inhibitor ribociclib (LEE011) is a versatile combination partner in preclinical cancer models. Oncotarget 9, 35226–35240, doi: 10.18632/oncotarget.26215 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang C et al. Acquired CDK6 amplification promotes breast cancer resistance to CDK4/6 inhibitors and loss of ER signaling and dependence. Oncogene, doi: 10.1038/onc.2016.379 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Placke T et al. Requirement for CDK6 in MLL-rearranged acute myeloid leukemia. Blood 124, 13–23, doi: 10.1182/blood-2014-02-558114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taipale M et al. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 150, 987–1001, doi: 10.1016/j.cell.2012.06.047 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaughan CK et al. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol Cell 23, 697–707, doi: 10.1016/j.molcel.2006.07.016 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stepanova L, Leng X, Parker SB & Harper JW Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev 10, 1491–1502, doi: 10.1101/gad.10.12.1491 (1996). [DOI] [PubMed] [Google Scholar]
- 18.Taipale M et al. Chaperones as thermodynamic sensors of drug-target interactions reveal kinase inhibitor specificities in living cells. Nat Biotechnol 31, 630–637, doi: 10.1038/nbt.2620 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grbovic OM et al. V600E B-Raf requires the Hsp90 chaperone for stability and is degraded in response to Hsp90 inhibitors. Proc Natl Acad Sci U S A 103, 57–62, doi: 10.1073/pnas.0609973103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawai A et al. Inhibition of Hsp90 down-regulates mutant epidermal growth factor receptor (EGFR) expression and sensitizes EGFR mutant tumors to paclitaxel. Cancer Res 68, 589–596, doi: 10.1158/0008-5472.CAN-07-1570 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hallett ST et al. Differential Regulation of G1 CDK Complexes by the Hsp90-Cdc37 Chaperone System. Cell Rep 21, 1386–1398, doi: 10.1016/j.celrep.2017.10.042 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai AC & Crews CM Induced protein degradation: an emerging drug discovery paradigm. Nat Rev Drug Discov 16, 101–114, doi: 10.1038/nrd.2016.211 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brand M et al. Homolog-Selective Degradation as a Strategy to Probe the Function of CDK6 in AML. Cell Chem Biol 26, 300–306 e309, doi: 10.1016/j.chembiol.2018.11.006 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang B et al. Development of Dual and Selective Degraders of Cyclin-Dependent Kinases 4 and 6. Angew Chem Int Ed Engl 58, 6321–6326, doi: 10.1002/anie.201901336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruscetti M et al. NK cell-mediated cytotoxicity contributes to tumor control by a cytostatic drug combination. Science 362, 1416–1422, doi: 10.1126/science.aas9090 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finn RS et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res 11, R77, doi: 10.1186/bcr2419 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldman JW et al. A randomized phase 3 study of abemaciclib versus erlotinib in previously treated patients with stage IV NSCLC with KRAS mutation: JUNIPER. Journal of Clinical Oncology 36, 9025–9025, doi: 10.1200/JCO.2018.36.15_suppl.9025 (2018). [DOI] [Google Scholar]
- 28.Pacheco J & Schenk E CDK4/6 inhibition alone and in combination for non-small cell lung cancer. Oncotarget 10, 618–619, doi: 10.18632/oncotarget.26545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghandi M et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508, doi: 10.1038/s41586-019-1186-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato JY, Matsuoka M, Strom DK & Sherr CJ Regulation of cyclin D-dependent kinase 4 (cdk4) by cdk4-activating kinase. Mol Cell Biol 14, 2713–2721, doi: 10.1128/mcb.14.4.2713 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McFarland JM et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat Commun 9, 4610, doi: 10.1038/s41467-018-06916-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyers RM et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet 49, 1779–1784, doi: 10.1038/ng.3984 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsherniak A et al. Defining a Cancer Dependency Map. Cell 170, 564–576 e516, doi: 10.1016/j.cell.2017.06.010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McDonald ER 3rd et al. Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell 170, 577–592 e510, doi: 10.1016/j.cell.2017.07.005 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Nusinow DP et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 180, 387–402 e316, doi: 10.1016/j.cell.2019.12.023 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marcotte R et al. Functional Genomic Landscape of Human Breast Cancer Drivers, Vulnerabilities, and Resistance. Cell 164, 293–309, doi: 10.1016/j.cell.2015.11.062 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gadd MS et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat Chem Biol 13, 514–521, doi: 10.1038/nchembio.2329 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen P et al. Spectrum and Degree of CDK Drug Interactions Predicts Clinical Performance. Mol Cancer Ther 15, 2273–2281, doi: 10.1158/1535-7163.MCT-16-0300 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Bondeson DP et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol 11, 611–617, doi: 10.1038/nchembio.1858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winter GE et al. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348, 1376–1381, doi: 10.1126/science.aab1433 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chamberlain PP et al. Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nat Struct Mol Biol 21, 803–809, doi: 10.1038/nsmb.2874 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Zhang C et al. Proteolysis Targeting Chimeras (PROTACs) of Anaplastic Lymphoma Kinase (ALK). Eur J Med Chem 151, 304–314, doi: 10.1016/j.ejmech.2018.03.071 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fischer ES et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512, 49–53, doi: 10.1038/nature13527 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lebraud H, Wright DJ, Johnson CN & Heightman TD Protein Degradation by In-Cell Self-Assembly of Proteolysis Targeting Chimeras. ACS Cent Sci 2, 927–934, doi: 10.1021/acscentsci.6b00280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sumi NJ, Kuenzi BM, Knezevic CE, Remsing Rix LL & Rix U Chemoproteomics Reveals Novel Protein and Lipid Kinase Targets of Clinical CDK4/6 Inhibitors in Lung Cancer. ACS Chem Biol 10, 2680–2686, doi: 10.1021/acschembio.5b00368 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bisi JE et al. Preclinical development of G1T38: A novel, potent and selective inhibitor of cyclin dependent kinases 4/6 for use as an oral antineoplastic in patients with CDK4/6 sensitive tumors. Oncotarget 8, 42343–42358, doi: 10.18632/oncotarget.16216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinez Molina D et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87, doi: 10.1126/science.1233606 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Jafari R et al. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat Protoc 9, 2100–2122, doi: 10.1038/nprot.2014.138 (2014). [DOI] [PubMed] [Google Scholar]
- 49.Patricelli MP et al. In situ kinase profiling reveals functionally relevant properties of native kinases. Chem Biol 18, 699–710, doi: 10.1016/j.chembiol.2011.04.011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verba KA et al. Atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science 352, 1542–1547, doi: 10.1126/science.aaf5023 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boczek EE et al. Conformational processing of oncogenic v-Src kinase by the molecular chaperone Hsp90. Proc Natl Acad Sci U S A 112, E3189–3198, doi: 10.1073/pnas.1424342112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J, Yang PL & Gray NS Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer 9, 28–39, doi: 10.1038/nrc2559 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bockstaele L, Bisteau X, Paternot S & Roger PP Differential regulation of cyclin-dependent kinase 4 (CDK4) and CDK6, evidence that CDK4 might not be activated by CDK7, and design of a CDK6 activating mutation. Mol Cell Biol 29, 4188–4200, doi: 10.1128/MCB.01823-08 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nomanbhoy TK et al. Chemoproteomic Evaluation of Target Engagement by the Cyclin-Dependent Kinase 4 and 6 Inhibitor Palbociclib Correlates with Cancer Cell Response. Biochemistry 55, 5434–5441, doi: 10.1021/acs.biochem.6b00629 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Romano G et al. A Preexisting Rare PIK3CA(E545K) Subpopulation Confers Clinical Resistance to MEK plus CDK4/6 Inhibition in NRAS Melanoma and Is Dependent on S6K1 Signaling. Cancer Discov 8, 556–567, doi: 10.1158/2159-8290.CD-17-0745 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Herrera-Abreu MT et al. Early Adaptation and Acquired Resistance to CDK4/6 Inhibition in Estrogen Receptor-Positive Breast Cancer. Cancer Res 76, 2301–2313, doi: 10.1158/0008-5472.CAN-15-0728 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Costa C et al. PTEN Loss Mediates Clinical Cross-Resistance to CDK4/6 and PI3Kalpha Inhibitors in Breast Cancer. Cancer Discov 10, 72–85, doi: 10.1158/2159-8290.CD-18-0830 (2020). [DOI] [PubMed] [Google Scholar]
- 58.de Leeuw R et al. MAPK Reliance via Acquired CDK4/6 Inhibitor Resistance in Cancer. Clin Cancer Res 24, 4201–4214, doi: 10.1158/1078-0432.CCR-18-0410 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Z et al. Loss of the FAT1 Tumor Suppressor Promotes Resistance to CDK4/6 Inhibitors via the Hippo Pathway. Cancer Cell 34, 893–905 e898, doi: 10.1016/j.ccell.2018.11.006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guiley KZ et al. p27 allosterically activates cyclin-dependent kinase 4 and antagonizes palbociclib inhibition. Science 366, doi: 10.1126/science.aaw2106 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Green JL et al. Direct CDKN2 Modulation of CDK4 Alters Target Engagement of CDK4 Inhibitor Drugs. Mol Cancer Ther 18, 771–779, doi: 10.1158/1535-7163.MCT-18-0755 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry raw files for the CDK6 complex analysis are available at the Mass Spectrometry Interactive Virtual Environment (MassIVE) (https://massive.ucsd.edu) under MassIVE ID: MSV000086571.
The mass spectrometry files for global protein degradation have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD023137.
The normalized protein quantification data and cell line omics data can be downloaded at DepMap (depmap.org/protal/).
Reagents generated in this study will be made available on request, but we may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.