Abstract
Each language has standard books describing that language’s grammatical rules. Biologists have searched for similar, albeit more complex, principles relating enhancer sequence to gene expression. We introduce dependency grammar, a model where enhancers encode information based on dependencies between enhancer features shaped by mechanistic, evolutionary, and biological constraints. Classifying enhancers based on the types of dependencies may identify unifying principles relating enhancer sequence to gene expression. Such rules would allow us to read the instructions for development within genomes and pinpoint causal enhancer variants underlying disease and evolutionary changes.
Section 1: The promise of enhancer grammar
Enhancers in development, homeostasis, disease, and evolution
Enhancers control the precise patterns of gene expression required for successful development and homeostasis. The human genome appears to contain millions of enhancers that act as switches to regulate the time and location of gene expression (Meuleman et al., 2020) (Figure 1A). As such, enhancers provide the instructions for tissue-specific gene expression, development, homeostasis, and cellular integrity (Levine, 2010). While we can identify putative enhancers using genomic assays, working out which genomic sequences are functional enhancers, the patterns of expression they control, and how enhancers encode these expression patterns are major hurdles in understanding genomes. Sequence changes within enhancers can have significant impact, altering tissue-specific expression and causing phenotypic variation, evolutionary adaptation, and disease. For example, in an enhancer for the membrane protein Duffy, a point mutation results in malarial resistance (Tournamille et al., 1995). In addition, many single base-pair changes in the ZRS enhancer cause aberrant expression of sonic hedgehog in the developing limb bud and changes in digit number (Figure 1B) (Kvon et al., 2020; Lettice et al., 2008). Indeed, the majority of mutations associated with disease lie within enhancers (Maurano et al., 2012; Tak and Farnham, 2015; Visel et al., 2009). Yet enhancers are littered with sequence variation; a critical challenge is pinpointing the causal enhancer variants within a sea of linked inert variants. A set of rules governing how enhancer sequences encode gene expression, known as enhancer grammar, could unlock the instructions for development embedded in our genomes and help us pinpoint causal enhancer variants associated with disease and evolutionary adaptations.
Figure 1: Enhancers encode precise patterns of gene expression; sequence changes in enhancers can alter expression patterns, causing disease and evolutionary adaptations.

(A) Enhancers control the location and timing of gene expression. Ciona heart and neural enhancers are shown (Bertrand et al., 2003; Christiaen et al., 2009). (B) Sequence changes within enhancers can lead to changes in the location and timing of gene expression causing developmental defects such as polydactyly (Kvon et al., 2020; Lettice et al., 2008). The expression of Shh in the developing limb bud is shown in purple, an A to G bp change within the ZRS limb enhancer leads to ectopic expression of Shh and an extra thumb.
Mechanisms governing enhancer function
A set of grammatical rules that define how enhancer sequence encodes tissue-specific expression is an attractive idea first suggested almost 30 years ago (Arnone and Davidson, 1997; Barolo, 2016; Levo and Segal, 2014; Thanos and Maniatis, 1995). Enhancers confer tissue-specific gene expression by the interaction of TFs with enhancer DNA. Generally, it is thought TFs bind to specific motifs known as transcription factor binding sites (TFBSs) within the enhancer sequence; this binding, along with protein-protein interactions, leads to recruitment of transcriptional machinery and activation of gene expression. The hypothesis for grammatical rules is based on the fact that proteins and the enhancer DNA have physical properties. These physical constraints govern the interaction of proteins with DNA and could be read out within the DNA sequence at the level of TFBSs. It is believed that enhancer grammar is composed of constraints on the number, type, and affinity of binding sites within an enhancer and the relative syntax of these sites (orders, orientations, and spacings) (Figure 2).
Figure 2: Enhancer Grammar.
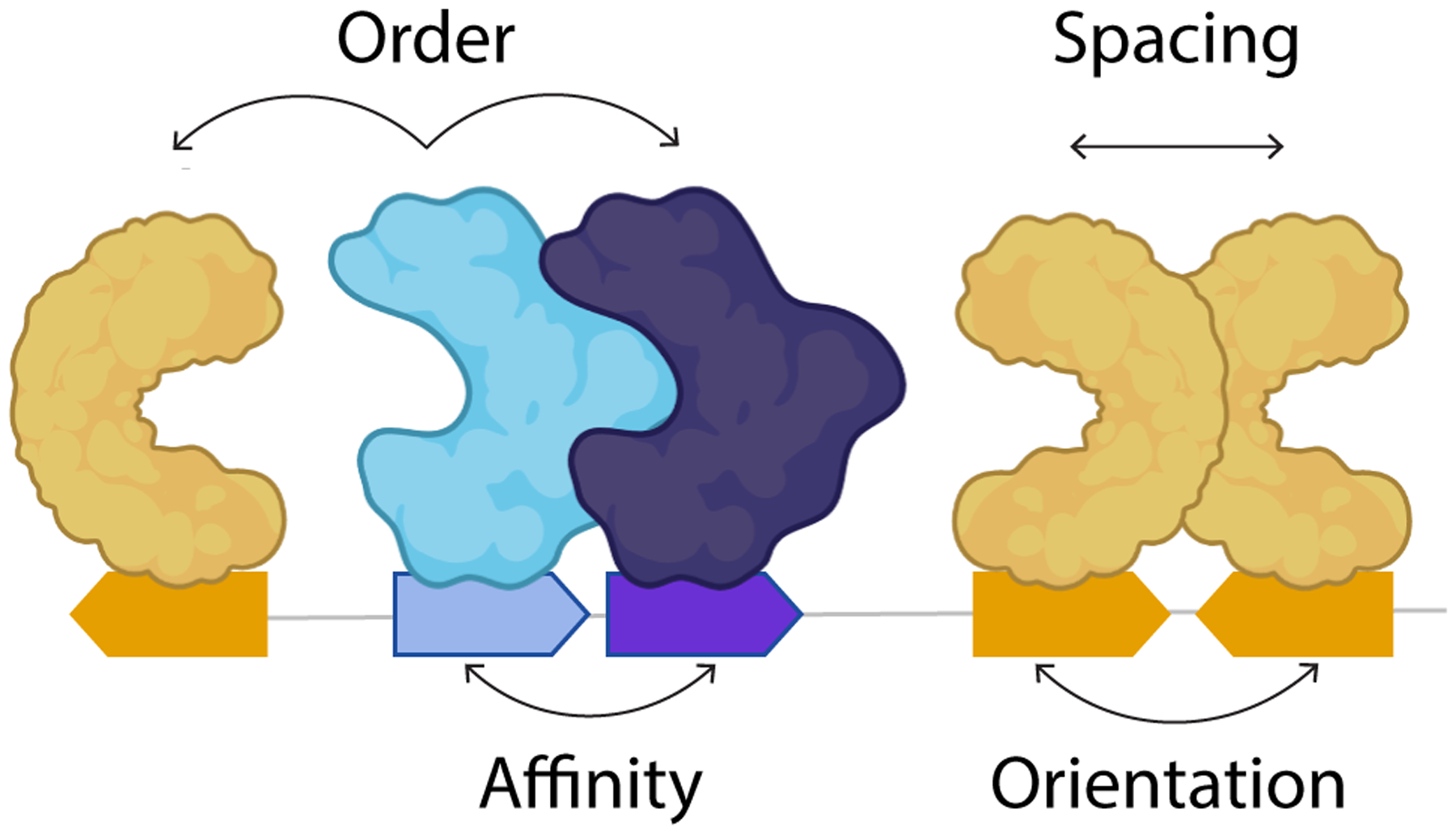
Physical interactions between transcription factors and the DNA along with protein-protein interactions are thought to be important for formation of a functional enhancer complex. Features of enhancer grammar include TFBS type, number, affinity, order, spacing, and orientation.
One of the earliest described examples of enhancer grammar is the IFN-β enhancer, which activates transcription as part of the immune response (Thanos and Maniatis, 1995). Around the same time, several studies of enhancers in the fly embryo suggested that spacing and order of TFBSs are important for encoding developmental expression patterns (Arnosti et al., 1996; Cai et al., 1996; Hanes et al., 1994). In vertebrates, experiments in the developing mouse pituitary demonstrated the importance of spacing between binding sites (Scully et al., 2000). One of the landmark studies often cited as an argument for and against enhancer grammar is the eve stripe 2 enhancer in flies. Enhancers from D.melanogaster, D. yakuba, D.erecta, and D. pseudoobscura were tested for expression in D. melanogaster. Although most of the 17 TFBSs were conserved to some degree across the four species, there are changes in the spacing and sequences between the sites. All four enhancers from the different species drive similar expression patterns during segmentation of the fly embryo despite the differences in the spacing and number of sites; this is often used as an argument against enhancer grammar (Ludwig et al., 1998). This study suggests there is some flexibility in how enhancers can encode gene expression. Interestingly, there are some differences in the levels and uniformity of expression between the four enhancers which could result from the differences in the TFBSs arrangements. A follow-up study showed that chimeric enhancers containing half D. melanogaster and half D. pseudoobscura did not drive the same gene expression patterns as the wild-type enhancers, suggesting that there is less flexibility and more dependency between features within each enhancer than previously appreciated and thus that there are some grammatical constraints within these enhancers (Ludwig et al., 2000).
Section 2. Models of how enhancers encode function
Since the initial hints of enhancer grammar, three models of how enhancers interact with TFs to control gene expression have been proposed.
The enhanceosome model
The enhanceosome model is the most rigid model of enhancer grammar, which suggests that the TFBSs within an enhancer must be precisely arranged to allow the TFs to bind in a functional way and for the enhancer to activate transcription (Bazett-Jones et al., 1994). The most iconic example of an enhanceosome is the IFN-β enhancer (Thanos and Maniatis, 1995). Grammatical constraints were initially found using reporter assays. Since then, the crystal structure of the IFN-β enhancer with TFs bound has been solved and shows a tight arrangement of TFs bound to the enhancer DNA (Panne et al., 2007). Although the crystal structure is the poster child for the enhanceosome, even the IFN-β enhanceosome has some flexibility, with variable spacing of sites leading to the same transcriptional activity, suggesting that there are several ways to encode an enhanceosome (Melnikov et al., 2012; Thanos and Maniatis, 1995). As of yet, no developmental enhancers as rigid as the enhanceosome model have been described.
The billboard model
The billboard model is the polar opposite of the enhanceosome. This model suggests that there are no constraints on how TFBSs are arranged within the enhancer; rather, the TFBSs simply must be present within the enhancer (Kulkarni and Arnosti, 2003). The closest example to a billboard enhancer is the ASE5 enhancer, which mediates autoregulatory expression of Suppressor of Hairless [Su(H)] in the socket cell, a component of sensory bristles in flies (Liu and Posakony, 2012). Within the ASE5 enhancer, 5 Su(H) binding sites are required and sufficient for the expression. However, not every enhancer that contains 5 Su(H) sites drives expression in the socket cell, indicating some other constraints such as spacing or syntax (order, orientation and spacing) of these sites may be important (Liu and Posakony, 2012). The billboard model suggests complete flexibility of arrangement of sites, yet no example of an enhancer having any organization of sites and being functional has been demonstrated. Despite this, the billboard model is often misleadingly invoked when no rules or grammar are found, if there are two or more ways of encoding the same information, or often simply to mean that there is some flexibility in how the sites are arranged.
The TF-collective model
The TF-collective model suggests collective occupancy of an enhancer by TFs via a combination of TFs binding to TFBSs and TF-TF interactions. This co-occupancy occurs in the absence of any specific motif organization, suggesting an alternative mode of gene regulation where cooperative activity of TFs occurs with extensive motif flexibility. It highlights the importance of considering TF-TF interactions along with TF-DNA interactions within enhancers (Junion et al., 2012). Using Chip-seq and reporter assays to study fly heart enhancers, the authors found that some active enhancers must have a critical mass of TFBSs within the enhancer for recruitment of the remaining required factors via TF-DNA interactions. When such a critical mass of TFs are bound to the enhancer, these bound TFs can recruit the remaining TFs via TF-TF interactions. While it is possible that TF-TF interactions are mediating this activity, one cannot rule out the use of very low-affinity motifs within enhancers; such degenerate sites are hard to identify yet prevalent in developmental enhancers (Crocker et al., 2015; Farley et al., 2015, 2016).
In reality, enhancers likely incorporate aspects of all three models. The first two models most likely describe extremes of a spectrum. The TF collective model highlights the layers of complexity when considering enhancer grammar and the importance of protein-protein and protein-DNA interactions. We suggest that considering enhancer grammar as the dependency and interplay between enhancer features (such as TF types, number, spacing, orientation and order) would allow incorporation of all three models into a continuum and may better reflect how developmental enhancers encode gene expression. We call this Dependency Grammar. Dependencies between enhancer features are likely shaped by various forces such as the features within the enhancer, molecular mechanisms, and evolutionary and biological constraints.
Section 3. The role of enhancer grammar in development
Initial studies of the IFN-β enhancer, and eve stripe 2 enhancers, inspired many to search for enhancer grammar to understand how genomes encode development. Evidence in support of and against grammar comes from the manipulation of native enhancers and the creation of synthetic enhancers, both of which are mainly analyzed by reporter assays. Genome-wide approaches such as binding assays and comparative genomics are also commonly employed to look for patterns of enhancer grammar. Similarly, comparisons of many enhancers that give the same expression pattern is another approach that attempts to identify “grammatical” patterns within genomes. So far, very few studies have looked at the impact of perturbing grammar in the endogenous locus to link grammar to phenotype. In Table S1, we summarize studies of grammar within developmental enhancers. While all of these studies are informative, because of the complex nature of enhancers, very few investigate multiple aspects of enhancer grammar in parallel.
Enhancer Grammar - Features rather than precise sequences
The enhancer features important for encoding gene expression include the type and combination of TFBSs, along with the affinity, number, order, orientation, and spacing of TFBSs (Figure 2). Other features such as DNA shape (Li et al., 2017), sequences that occlude nucleosomes (Levo et al., 2017), and yet more currently unknown features may play a role. Initial studies to identify the functional features of enhancers looked for and attempted to perturb conserved sequence motifs; however, it is becoming increasingly clear that the features of enhancers rather than exact sequence conservation are important for encoding gene expression (Farley et al., 2015, 2016; Fuqua et al., 2020). Recently, mutagenesis of the intensively studied fly trichome enhancer E3N, which regulates the gene svb, found that evolutionary sequence conservation was a poor predictor of functional sites highlighting the importance of enhancer features and the challenge in identifying such features (Fuqua et al., 2020). Perturbing the affinities and syntax (order, orientation, and spacing) of these binding sites and seeing if this has any impact on gene expression patterns can then help us understand if there are grammatical constraints on the enhancer. Identifying the functional features that are necessary and sufficient for enhancer activity is critical for such studies as one needs to know all the features before trying to perturb them to understand the relationship between these features (Farley et al., 2015; Johnson et al., 2008). Below, we discuss studies that assess the importance of enhancer features during development.
1. Affinity of TFBSs
Development requires precise patterns of gene expression, and both high- and low-affinity binding sites within enhancers play a role in tissue-specific expression. While high-affinity sites are easy to detect, low-affinity degenerate sites are hard to identify and could explain why TF binding is observed in the absence of TFBS motifs. Low-affinity degenerate sites are known to be important for encoding patterns of gene expression in the developing heart, nervous system, notochord, heart, and eye, in organisms from flies to humans (Catarino and Stark, 2018; Crocker and Ilsley, 2017; Crocker et al., 2016; Datta et al., 2018; Farley et al., 2015, 2016; Gaudet and Mango, 2002; Lorberbaum et al., 2016; Rickels and Shilatifard, 2018; Scardigli et al., 2003; Swanson et al., 2010; Zandvakili et al., 2018). Increasing the affinity of sites within developmental enhancers can lead to ectopic expression. It is thought low-affinity sites confer tissue specificity by ensuring combinatorial control of gene expression (Farley et al., 2015). There is also evidence that low-affinity sites can preferentially bind related transcription factors (Berger et al., 2008). In the Muscle and Heart Enhancer (MHE) active in the fly developing heart and somatic muscle, high-affinity ETS sites preferentially bind the repressive ETS factor, Yan, while the low-affinity sites preferentially bind the ETS activator, Pointed (Figure 3A) (Boisclair Lachance et al., 2018; Halfon et al., 2000). Enhancer grammar may also be key to the functionality of low-affinity sites; several studies have shown that low-affinity binding sites alone cannot explain the expression observed and that incorporating positional information regarding binding sites into computational models improves prediction of functional enhancers (Avsec et al., 2020; Boer et al., 2020; King et al., 2020). Studies in notochord enhancers find other aspects of enhancer grammar, namely spacing and orientation of sites, can compensate for low-affinity sites, presumably stabilizing the interactions to ensure a functional enhancer complex (Farley et al., 2016).
Figure 3: Affinity, Order, Spacing, and Orientation of sites within enhancers are essential for encoding tissue-specific gene expression during development.
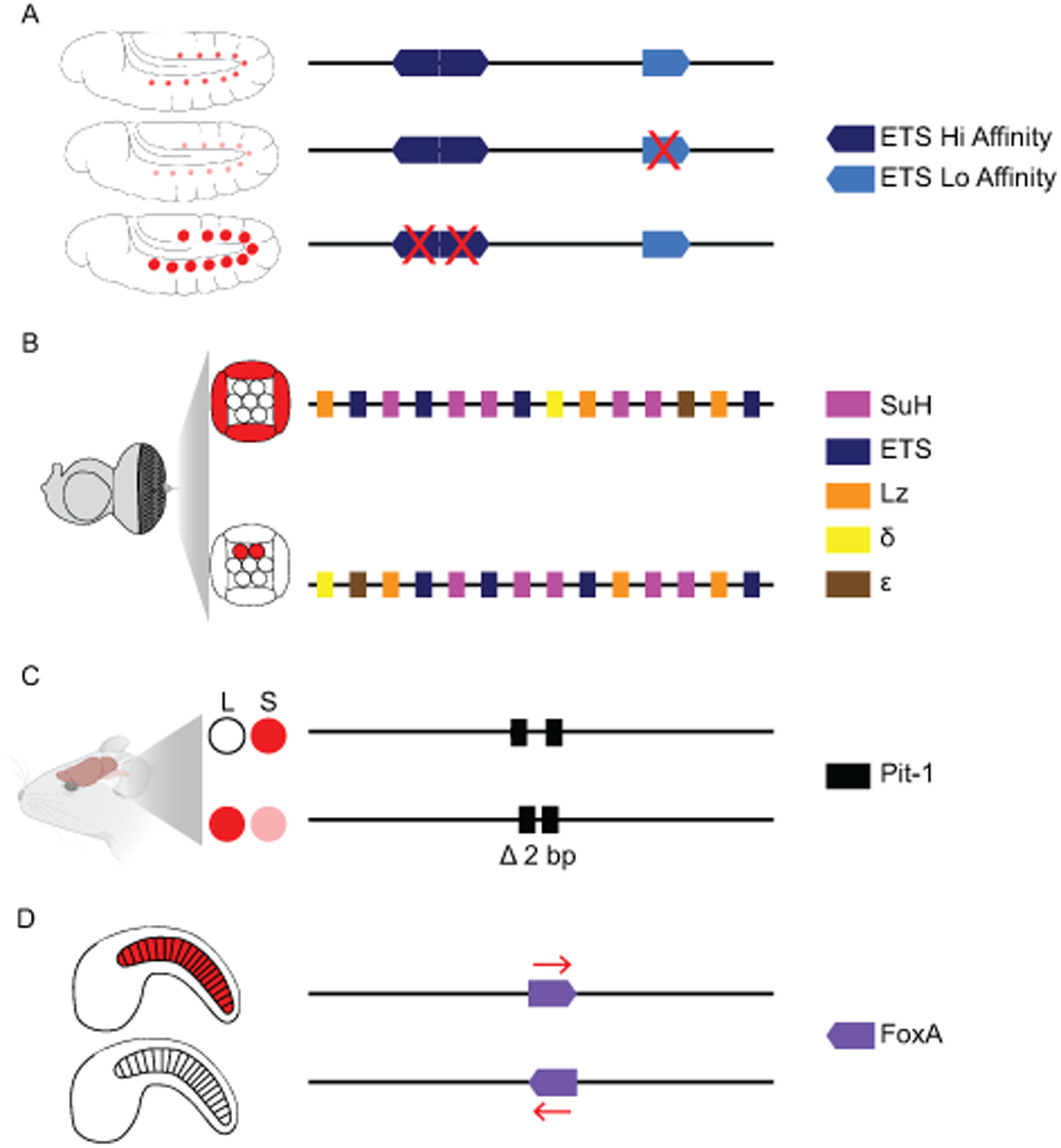
(A) A combination of low-affinity and high-affinity ETS sites within the MHE enhancer restrict expression to the muscle and heart cells of the developing fly. Low-affinity sites bind an activator form of ETS called Pointed, removing low-affinity sites reduces expression. High-affinity sites bind a repressive form of ETS called Yan, loss of high-affinity sites leads to ectopic expression (Boisclair Lachance et al., 2018). (B) In the developing fly eye, specific orders of SuH, ETS, LZ, δ and ε TFBSs activate expression in the cone cells and repress expression in the photoreceptor cells. Changing the order of sites can lead to loss of expression in the cone cells and ectopic expression in the photoreceptor cells (Swanson et al., 2010). (C) In the developing mouse pituitary, a 2-bp spacing within a Pit-1 site allows for expression in the somatotrope (S) cells and repression in lactotrope (L) cells, removing this spacing leads to ectopic expression of GH in lactotrope cells (Scully et al., 2000). (D) In Ciona, the orientation of a FoxA site within the Tune enhancer ensures notochord expression (Passamaneck et al., 2009). Not all sites within each enhancer are shown; for simplicity, we focus on the sites that when manipulated impact gene expression. Expression patterns are shown in red.
2. Order of TFBSs
Developmental enhancers typically contain combinations of TFs that work together to ensure precise patterns of gene expression. These can be homotypic clusters (Liu and Posakony, 2012), but more commonly, enhancers contain several types of TFBSs. Therefore, one aspect of enhancer grammar is the order of TFBSs within the enhancer. TFs interact both with the enhancer DNA but also with each other via protein-protein interactions. These interactions can be hetero or homodimers and higher-order interactions. Switching the order of binding sites can disrupt such interactions. Indeed, the order of sites is critical to encode developmental patterning. In the developing fly eye, the sparkling enhancer drives expression in cone cells, changing the order of TFBSs switches expression from cone cell-specific to photoreceptor cell-specific (Figure 3B) (Swanson et al., 2010). One caveat of this study is that in the process of changing the order of sites, a significant amount of sequence is also altered. This highlights a major hurdle in studies of enhancer grammar, the need to know the necessary and sufficient sites before manipulating other features, and the need to avoid the creation of new sites. Within the mɑ enhancer, which drives expression in the non-sensory organ precursor (SOP) cells of fly proneural clusters, changing the order of sites leads to loss of expression in the non-SOP cells of fly proneural cells and ectopic expression in other parts of the developing fly. This hints that TFBS order is not only necessary for enhancer activity in the tissue of interest, but can also prevent activity in ectopic tissues; this is known as preventative grammar (Liu and Posakony, 2012). Similar results have been found in mouse heart developmental enhancers (Luna-Zurita et al., 2016). Order of binding sites has been shown to be important in the mouse liver and stem cells (Fiore and Cohen, 2016; King et al., 2020; Smith et al., 2013). Order of binding sites is an understudied aspect of enhancer grammar that needs to be included in future studies. The prevalent use of degenerate binding sites in vivo makes studying the order of sites a real challenge, as the functional features must first be identified, and then any changes in order must be checked to ensure the new arrangement is not creating or ablating other functional sites. This is further complicated by the fact that there may indeed be more than one encoding of gene expression within the same enhancer region. Nonetheless, these studies demonstrate that the order of TFBSs within an enhancer is important in encoding developmental expression patterns.
3. Spacing between binding sites
Constraints on spacing between TFBSs may ensure all TFs within an enhancer can access their sites, or certain spacings could be essential to allow both TF-DNA interactions along with protein-protein interactions such as binding of dimers (e.g., (Nam et al., 2007)). In some enhancers, including the IFN-β enhancer and several developmental enhancers, helical phasing of sites within enhancers is important for expression (Cai et al., 1996; Hanes et al., 1994; Melnikov et al., 2012; Passamaneck et al., 2009; Thanos and Maniatis, 1995). Studies in the fly neural ectoderm and Ciona nervous system and notochord show that even single base-pair changes in spacing affect levels of expression (Crocker et al., 2008; Farley et al., 2016). Spacing between binding sites can also dictate the location of expression (Figure 3C). In the developing mouse pituitary gland, a two bp spacing between Pitx-1 half-sites within an enhancer is vital for activation of growth hormone in the somatotrope cells (Scully et al., 2000). This two bp spacing also prevents growth hormone expression in another cell type of the developing pituitary, the lactotrope. This is likely not an isolated example. Future studies should also consider the interplay of spacing and affinity of sites, as most transcription factor interaction pairs recognize composite sites that are markedly different from the motif of each TF alone (Jolma et al., 2015).
4. Orientation of TFBSs
Orientation of binding sites refers to the relative direction of TFBSs. Changing the orientation of a TFBS causes the TF to bind the other strand of DNA in a flipped orientation. This may affect how that transcription factor interacts with other TFs bound to the enhancer and the creation of a functional complex. The first demonstration of this phenomenon was in a Drosophila immunity enhancer; here, the orientation of Gata and Rel are important for enhancer activity (Senger et al., 2004). Soon after, the orientation of paired SuH sites were shown to be important for Notch signaling-dependent activity in Drosophila wing and eye imaginal discs (Cave et al., 2005). In Ciona development, certain orientations of Brachyury and FoxA binding sites, and ETS and Zic sites, are necessary for notochord specific gene expression (Figure 3D) (Farley et al., 2016; Passamaneck et al., 2009). In stem cells, four TFs (Oct4, Sox2, Klf4/5, and Esrrb) work together to encode pluripotency. These are known collectively as OSKE. Studies of synthetic enhancers with different OSKE grammars found that the orientation of sites is crucial for encoding expression in mouse ES cells (Fiore and Cohen, 2016; King et al., 2020).
Section 4. Signatures of enhancer grammar from genome-wide assays and comparative genomics
Our discussion of the features important for enhancer grammar has so far focused on examples where changes in enhancer grammar are measured by reporter assays, which can functionally demonstrate the impact of changing grammatical features on transcriptional output. Genome-wide studies and comparative genomic approaches are also powerful tools to find patterns within the genome; however, without functional validation, these can only provide hints about the link between enhancer grammar and gene expression (Halfon, 2019). In this section, we look at the putative signatures of grammar found using genome-wide and comparative genomic approaches.
Searching for grammar using binding data:
Millions of putative enhancers have been identified using Chip-seq in combination with chromatin accessibility data (e.g. (Meuleman et al., 2020; Zinzen et al., 2009)). This technique provides a list of potential enhancers, but the true signal in this type of data can be clouded by the fact that not all these regions that show binding or open chromatin are functional enhancers (Grossman et al., 2017; Halfon, 2019; King et al., 2020; Ryan and Farley, 2020). Even so, many studies attempt to look for grammar patterns in such data (Cheng et al., 2013; Guo et al., 2012; Nandi et al., 2013; Ng et al., 2014; Wang et al., 2012). Other studies functionally test a subset of the resulting genomic regions to validate their findings (Junion et al., 2012; Menoret et al., 2013; Sorge et al., 2012; Zinzen et al., 2009).
From flies to humans, orthologs of Tbox5, Nkx2-5, and GATA4 are thought to work together to pattern the developing heart. Two genome-wide studies have looked for signatures of enhancer grammar in the developing fly and mouse heart, respectively, each comes to different conclusions. These differences could be due to species-specific differences or limitations of the studies. The first in the developing fly combined Chip-Seq with transgenic reporter assays and found that TF co-binding could occur via a combination of TF-DNA and TF-TF interactions but did not find any grammatical constraints (Junion et al., 2012). The second study in mouse cardiac cells using Chip-exo and structural studies also noted the importance of TF-TF along with TF-DNA interactions and found a complex but well-defined motif grammar involving the spacing and orientation of TFBSs. Furthermore, in the mouse studies, knocking out one TF caused the other members to bind to different locations, providing some evidence that enhancer grammar is important not only to recruit TFs to the correct region, but also to prevent recruitment of these TFs to other regions in order to avoid inappropriate gene expression (Luna-Zurita et al., 2016). Another study in mouse blood cells saw patterns of spacing between TFs at bound loci and functionally validated that such spacing was important for levels of expression using reporter assays (Ng et al., 2014). In human cells, 92 pairs of TFs show position and orientation preferences between co-binding TFs (Wang et al., 2012). While these signatures and patterns provide hints, the true signal is likely clouded by non-functional binding, which is further compounded by the challenges of identifying functional binding sites, such as the large footprint of standard Chip-seq, and the use of degenerate motifs. A combination of eRNA data along with Chip-seq data could help detect functional enhancers (Perez-Cervantes et al., 2020), while methods such as Chip-exo allow more precise location of the TFBSs (Avsec et al., 2020; Luna-Zurita et al., 2016). It is also important to note that the lack of patterns or even presence of patterns within genomic data does not mean there is a grammar; ultimately, to harness the most power from any signatures seen in genomic regions, functional validation at scale is needed.
Searching for grammar using comparative genomics
Conservation of sequences or TFBSs across species is often used to look for grammatical patterns or signatures within genomes. Approaches range from looking at the same enhancer across multiple species, to looking at many enhancers with similar expression or comparing genomic regions with similar binding patterns within an organism or across organisms (Erives and Levine, 2004; Ilsley et al., 2013; Li and Wunderlich, 2017; Lusk and Eisen, 2010; Markstein et al., 2004; Papatsenko et al., 2009). These attempts have had mixed results. In some cases, common signatures pop out, which could be signatures of dimers or common interacting partners. There are also rare examples of highly conserved order, spacing, and orientation of motifs across bilateria (Rebeiz et al., 2012), or examples where the entire enhancer sequence is conserved (Kvon et al., 2016; Lettice et al., 2003). However, in the majority of cases, comparative genomic approaches find no patterns and often conclude that no grammar exists within genomes. Before such conclusions can be drawn, we need to shift our focus from looking for exact sequence motifs to looking for features, even degenerate ones. We also need to move away from looking for exact patterns towards a more nuanced understanding of the dependency between features and functionally validate any hypothesis.
Comparative approaches combined with functional dissection are rare but have been the most informative. A common misconception is that lack of sequence conservation, or rapid evolution of an enhancer, is inconsistent with the idea of enhancer grammar. Two classical studies demonstrate this point, the eve stripe 2 enhancer, and the sparkling enhancer (Ludwig et al., 1998, 2000; Swanson et al., 2010, 2011). Careful dissection of the D. melanogaster sparkling enhancer elegantly shows the importance of enhancer grammar (Swanson et al., 2010). Yet, searching for this grammar in the orthologous enhancer in different fly species found no common signatures (Swanson et al., 2011). Indeed, in other species, the sparkling enhancer has rapid turnover of motifs, yet the expression pattern is maintained. These findings are consistent with the idea that shuffling of specific binding sites in orthologous regions by compensatory motif turnover may make the enhancer unrecognizable by comparative genomics but keep enhancer tissue-specificity (Evans et al., 2012; Ludwig et al., 1998, 2000; Swanson et al., 2010, 2011; Taher et al., 2011; Weirauch and Hughes, 2010). In such cases, one cannot rule out that there are simply several ways to encode the same outcome or that grammatical rules can be conserved whilst encoding the information in seemingly different ways. As more studies investigate enhancer grammar using massively parallel reporter assays, it will be crucial to identify if any underlying grammatical constraints exist that can explain how these different solutions, in different organisms, can encode the same expression pattern. Another exciting future direction will be using comparative genomics, not only to look for signatures or patterns of grammar that stay constant, but to understand how tweaking enhancer grammar could contribute to evolutionary adaptations. We are far from pinpointing grammatical changes associated with evolutionary changes, but there are hints that such a goal is possible. In the developing fly neural ectoderm, changes in spacing between TFBSs correlates with changes in morphogen gradients and expression patterns between species (Crocker et al., 2008).
Section 5. Dependency grammar
Since not all clusters of binding sites drive expression and changes to orientation, order, and spacing along with affinity of binding sites impact enhancer function, it is clear that some sort of grammatical constraints on gene regulation exist; however, the extent of any constraints appears to differ in different enhancers. The majority of studies of grammar within developmental enhancers find that enhancers fall between the two extreme models, the billboard, and the enhanceosome, respectively. We suggest that Dependency Grammar - a model that considers the dependency and interplay between enhancer features and how these are shaped by evolutionary, biological, and mechanistic constraints may allow us to us classify enhancers into different bins on the spectrum between billboard and enhanceosome. Such classification may help us get to unifying principles that relate enhancer sequence to expression and, ultimately, which variants cause grammatical errors leading to evolutionary and disease phenotypes (Figure 4).
Figure 4: Dependency Grammar.
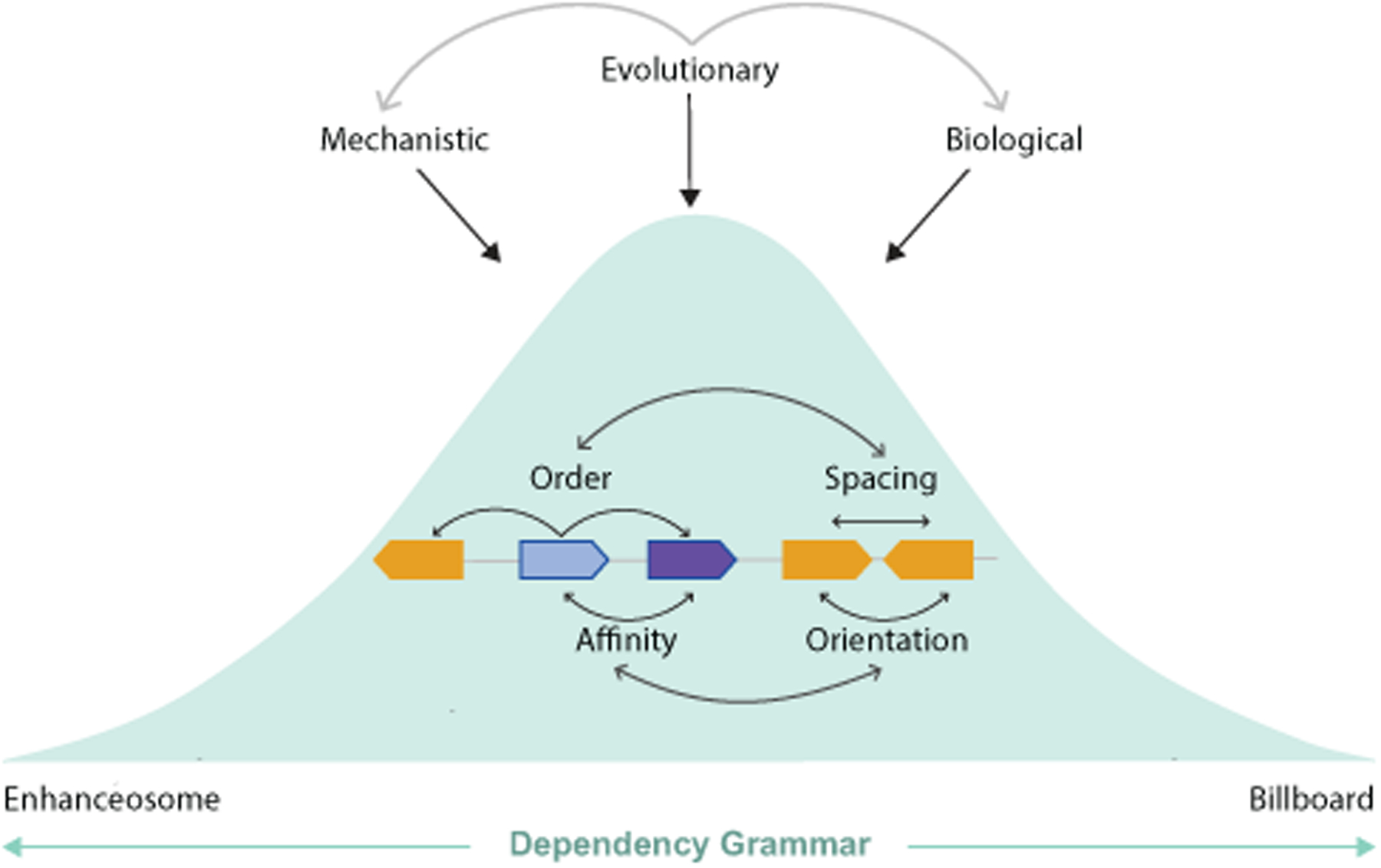
Enhanceosome and Billboard models are extremes of a spectrum. TF-collective model suggests motif flexibility and highlights the importance of both TF-DNA and protein-protein interactions. Enhancers fall at various positions on this spectrum based on the dependency between enhancer features which are shaped by mechanistic, evolutionary, and biological constraints.
Dependency Grammar - Interplay between spacing and affinity affects enhancer specificity
In ascidians, ETS and Zic bind to enhancers and activate notochord-specific expression of genes, including Brachyury and Mnx (Farley et al., 2016; Matsumoto et al., 2007). While Zic and ETS sites are necessary for notochord expression, a synthetic screen found not all enhancers containing these TFBSs were functional. Functional notochord enhancers had particular spacing, number, and orientation of TFBSs. Based on screens of synthetic enhancers, a set of crude rules were defined. These were used to turn non-functional enhancer variants into notochord specific enhancers. Rather than a binary and rigid organization of sites, an interplay between affinity and spacing of Zic and ETS sites is important for notochord expression (Figure 5). Furthermore, because of the interplay between features, there were several ways to adhere to the grammatical rules and encode the same expression pattern. Functional enhancers could be made less functional by increasing the spacing between sites, but the activity of the enhancer could be recovered by increasing the affinity of the binding sites to compensate for the worse spacing (Farley et al., 2016). This interplay is an example of how affinity and syntax (spacing, order, orientation) of binding sites in functional enhancers can be dependent on each other and how grammar may be encoded in a seemingly flexible yet grammatical way (Figure 5). A similar interdependence is seen in the regulation of pluripotency in ES cells. Certain combinations of Oct4, Sox2, Klf, and Esrrb are equally functional, but there are also combinations that are inert, and thermodynamic models are more accurate when TF interdependencies are considered (Fiore and Cohen, 2016; King et al., 2020). If features are interdependent, then tweaking some features and not others could explain the continuum from billboard to enhanceosome, and how enhancers can encode the same information with seemingly different sequences and organization of TFBSs.
Figure 5: Notochord activity depends on the interplay between affinity and spacing of Zic and Ets sites.
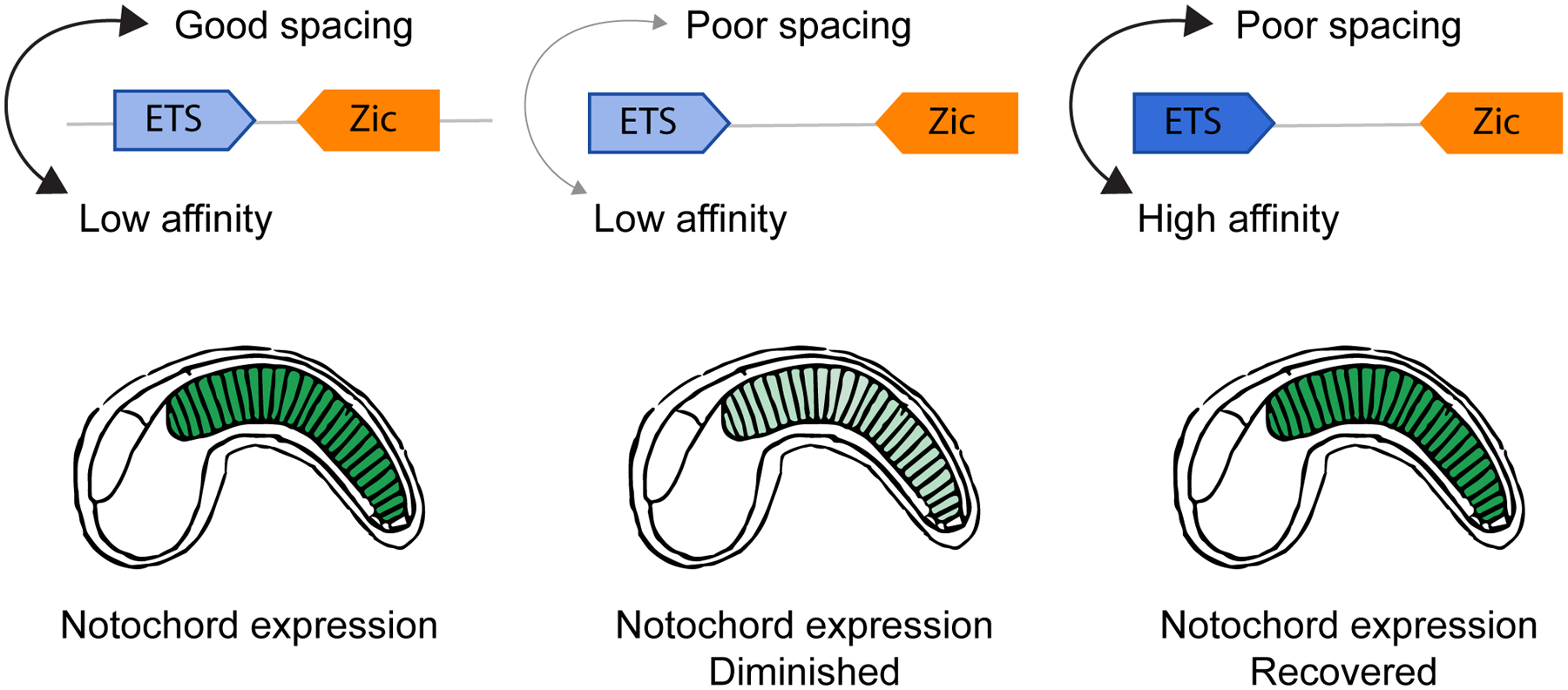
Compensation between affinity and spacing encodes notochord expression. When both spacing and affinity are poor, notochord expression is lost; when one is good and the other poor, notochord expression is recovered. (Farley et al., 2016).
Using dependency grammar to find enhancers in the genome
The true test of any model of enhancer grammar is using it to find enhancers in the genome from sequence alone (Phillips, 2015; Sayal et al., 2016; Vincent et al., 2016). To test if the dependency grammar identified for Zic and ETS is broadly applicable, we used what we learned about constraints on Zic and ETS TFBSs, affinity, and arrangement, along with the interplay between features to search for notochord enhancers in the Ciona genome. We previously showed that having all optimal features such as highest affinity and best spacing for transcriptional output gave ectopic expression (Farley et al., 2015). This appears to be due to the loss of combinatorial control of the enhancer and aberrant activation of the enhancer even at low levels of either activating TF. Therefore, specificity of gene expression may require enhancers that contain degenerate low-affinity motifs, and/or suboptimal organization of sites to ensure tissue-specific expression (Farley et al., 2015, 2016). To rigorously test the idea of dependency grammar, whilst also considering our findings regarding suboptimization, we searched for putative enhancers with very low affinity but with good organization. We identified two enhancers with incredibly low affinity but good orientation and spacing of sites. Both of these enhancers activate expression in the notochord and one lies close to the notochord-specific genes Mnx, the other is close to Brachyury (Farley et al., 2016). This demonstrates that dependency grammar can identify enhancers in the genome, that there is indeed an interplay between features, and that organization can compensate for low affinity. The extreme compensation of organization for low affinity could explain the functionality of low-affinity sites. Indeed, there could be an entire class of developmental enhancers with poor affinity but good organization of TFBSs. It is still unclear why the notochord enhancers we’ve studied and the pluripotency enhancers within stem cells appear to use low-affinity sites and grammar, while some enhancers such as the ASE5 have much less constraint and the IFN-β enhancer has a very rigid grammar. It could simply be that evolution has come up with more than one solution to the same problem. Regardless of the number of solutions, understanding what shapes the dependencies between features and understanding why different enhancers use different degrees of grammar will be vital in cracking the enhancer code.
Dependency grammar parallels to Natural Language Processing
We came to the term Dependency grammar because of our research on notochord and neural enhancers that exemplify the dependency and interplay between enhancer features. We later discovered that Dependency grammar is also studied in linguistics and considered in Natural Language Processing (NLP) methods to describe how languages encode meaning (de Marneffe and Nivre, 2019). In linguistics, dependency grammar aims to understand the meaning of a sentence based on how words depend on each other. Another approach in linguistics is constituency grammar, which is focused on looking for phrases within languages. These phrases, or common signatures, are akin to dimers that have been identified in enhancers such as Sox and Oct working together in mouse stem cells or many of the signatures identified from genome-wide chip data (Kuroda et al., 2005; Maurano et al., 2012; Rodda et al., 2005). In NLP applications, constituency grammar was the preferred mode for understanding how language is encoded; however, focus has recently shifted to dependency grammar and neural network methods. This parallels the search for enhancer grammar, as we have found many dimer interactions that appear to be important and are now moving towards understanding the dependency between features. Machine learning approaches are starting to provide insight into enhancer grammar yet mainly focus on finding commonly co-occurring combinations of motifs or “motif phrases” akin to constituency grammar (Avsec et al., 2020; Boer et al., 2020; Chen and Capra, 2020). As we amass more functional data from massively parallel reporter assays in combination with epigenomic, comparative genomic data, and computational approaches, it will be interesting to see how far the parallels between dependency grammar in language and enhancers extend. While the metaphor of language is attractive, we want to emphasize that there is a complexity and multidimensionality to enhancers far beyond the linearity of language.
Section 6: Organizing principles of enhancer grammar
The use of grammatical rules to identify enhancers in the genome from sequence alone is in its infancy with very few successful examples (Farley et al., 2016; Khoueiry et al., 2010; King et al., 2020). If we are able to predict enhancers from sequence alone, will there be a different set of grammatical rules for each enhancer, or will there be unifying principles relating grammatical principles to enhancer function? Classifying enhancers based on the type of dependency grammar they use may help us get to organizing principles, and elucidate how enhancer sequence encodes function. We may even ultimately be able to pinpoint which variants impact evolutionary and disease phenotypes. Key to this will be working out what shapes the dependencies between features within different types of enhancers and how we classify enhancers based on the dependencies between features. Below we speculate on the mechanistic, biological, and evolutionary constraints that could shape the dependencies between enhancer features.
Transcriptional mechanisms could shape enhancer grammar
One crucial step in gene regulation is how enhancers communicate with promoters. There are many modes of enhancer-promoter communication, and these different modes may shape grammatical constraints on enhancers. Enhancers extremely proximal to the promoter or indeed enhancers intertwined with the promoter may have a more rigid grammar as the proximity to the pre-initiation complex puts further constraint on assembly of a functional complex. There is some evidence for rigid grammar signatures in regions close to promoters; however, these are yet to be functionally validated (Minnoye et al., 2020; Rebeiz et al., 2012). Most enhancers lie far from their target promoter and are thought to activate transcription via various mechanisms, including looping, TADs, chromatin decompaction, and even eRNA mediated enhancer-promoter interactions (Benabdallah et al., 2019; Cajigas et al., 2018; Furlong and Levine, 2018; Isoda et al., 2017; Tsai et al., 2018). These different mechanisms of enhancer-promoter interactions could shape the grammatical constraints and the organization of features within enhancers. It will be interesting to see if enhancers that use these different mechanisms of enhancer-promoter communication depend on different types of enhancer grammar.
The micro-environment surrounding enhancers before and upon activation of transcription is a poorly understood yet active area of research. Different enhancers may well experience different types of microenvironments, which could shape the dependencies between enhancer features – and thus enhancer grammar. At some genomic regions, high concentrations of TFs, co-factors, and transcriptional machinery appear to aggregate with enhancers and promoters before and upon transcriptional activation. These are sometimes called TF hubs. The high concentration of TFs in these “hubs” may enable binding of TFs to low-affinity sites (Lim et al., 2018; Tsai et al., 2017, 2019). Although controversial, some suggest these hubs are phase-separated condensates and that the entropy of TFs binding to enhancers is required for phase separation and transcriptional activation (Hnisz et al., 2017; Shrinivas et al., 2019). If phase separation is occurring and driven by energy derived from binding of TFs to the enhancer, the requirement to phase separate may shape enhancer grammar. As the field advances, no doubt, we will work out if different enhancers experience different microenvironments, the properties of these, and how these impact enhancer grammar and gene regulation.
Grammatical constraints could be skewed by other mechanistic constraints, such as chromatin dynamics (Avsec et al., 2020; Luna-Zurita et al., 2016). A recent study found evidence for FoxA functioning as a pioneer factor at some enhancers, but not at other enhancers, with both types of enhancers showing putative evidence for different grammars (Geusz et al., 2020). TFs with chromatin remodeling capability such as PU.1, Sox2, and Zelda may have similar modes of action, where different grammars exist depending on whether the TF is functioning in a pioneer or non-pioneer role. Temporal dynamics of TF binding, which may be influenced by numerous factors, could also shape the dependency between features. Other aspects of transcriptional regulation that may affect enhancer grammar include enhancer-enhancer interactions, additive or synergistic activity of enhancers, and transvection (Fukaya and Levine, 2017; Hay et al., 2016; Maekawa et al., 1989).
Biological constraints on enhancer grammar
The most obvious example of the biological constraints on enhancer grammar is the need for tissue-specific gene expression. In Section 3, we discuss many examples where changes in enhancer grammar change gene expression (Farley et al., 2016; Liu and Posakony, 2012; Swanson et al., 2010). It is also possible that the organization of sites is essential, not only for activation in one cell type but for preventing expression in another cell type (Scully et al., 2000). Tissue specificity can be achieved using tissue-specific TFs, and these may be shaped by grammar differently from enhancers that are responding to pleiotropic factors or enhancers that are controlled by activators and repressors. The concentration and type of TFs binding to the enhancers in different cell types also likely shapes the grammatical constraints. Certain grammars could even ensure the coupling of two developmental processes. In chick motor neuron development, a combination of direct binding at the enhancer and protein-protein interactions are thought to be important for coupling of neurogenesis and motor neuron specification (Lee and Pfaff, 2003). The amount of constraint may also be linked to the developmental time point, cell type, and the role of the enhancer. ASE5 is the most billboard-like developmental enhancer identified so far and seems only to require 5 Su(H) sites, although the spacing and possibly orientation of these sites may be important. The flexibility of this enhancer may reflect the simplicity of the biological need and cell type, as this enhancer only needs to maintain autoregulated Su(H) expression in a terminally differentiated cell type (Liu and Posakony, 2012). Other aspects of biological constraint that could shape grammar include temporal dynamics of gene expression, for example, oscillating genes in development (Kageyama et al., 2018; Rebeiz et al., 2012).
Evolutionary constraints may shape enhancer grammar
The hourglass model suggests evolutionary constraint on gene regulation changes across developmental time and tissue (von Baer, 1828; Duboule, 1994; Raff, 1996). Although this model was first coined to describe the similarity in morphology seen within developing embryos, gene expression patterns and gene regulation also follow this hourglass pattern (Domazet-Lošo and Tautz, 2010; Kalinka et al., 2010; Liu et al., 2020; Stergachis et al., 2013). Indeed, a study of developing mouse forebrain, liver, and heart using epigenomic data and reporter assays, found the level of evolutionary conservation and constraint on putative enhancers followed the hourglass pattern with greatest constraint seen during mid-embryogenesis; similar results are seen in fly enhancers (Liu et al., 2020; Nord et al., 2013). Thus, the grammatical constraints on enhancers may be more rigid during the bottleneck of the hourglass than at early and later developmental stages.
Conversely, it is also possible that the grammatical constraints within some enhancers in the bottleneck of the hourglass may have more flexibility. A common mechanism for robustness through this bottleneck is the use of multiple redundant enhancers, known as shadow, or redundant enhancers (Barolo, 2012; Bomblies et al., 1999; Cannavò et al., 2016; Garnett et al., 2012; Hong et al., 2008; Osterwalder et al., 2018; Perry et al., 2010). The use of multiple seemingly redundant enhancers ensures transcriptional robustness in the face of environmental and genetic perturbations (Frankel et al., 2010; Osterwalder et al., 2018; Perry et al., 2010). The number of shadow enhancers involved in a process could shape the extent and flavor of grammar seen within the enhancer, allowing flexibility in regulatory logic. Although we sometimes call these redundant enhancers, studies of Drosophila mesodermal enhancers suggest many of the redundant enhancers are in reality actually only partially redundant, encoding function in one tissue at one time point and another at another, or encoding similar but not identical patterns of expression (Cannavò et al., 2016; El-Sherif and Levine, 2016). In enhancers that encode one or more expression patterns, the enhancer logic for both expression patterns may be intertwined within the enhancer sequence. This could be another layer of potential constraint on the dependency between enhancer features shaped by both evolutionary and biological pressures. As well as having multiple enhancers to ensure robustness, there may well be redundancy encoded within enhancers. This second type of redundancy could be yet another mechanism that shapes the grammatical constraints and could confound studies of enhancers, as apparent flexibility in the grammatical constraints could indeed be a mirage of redundancy.
Constraints shaping enhancer sequence and function are not solely due to things that directly affect the enhancer activity. Several studies in embryonic stem cells have found that promoters can function as enhancers (Diao et al., 2017). Another example of dual encoding are duons, elements that encode both an enhancer and part of an exon (Agoglia and Fraser, 2016; Stergachis et al., 2013; Xing and He, 2015). In duons, functional codon usage and enhancer sequence constrain the dependency between features within the enhancer. Similarly, it has been suggested that some enhancers also serve as origins of replication (Pherson et al., 2019). Polycomb response elements (PREs) mediate gene silencing; some of these elements also encode enhancer activity in other cell types (Erceg et al., 2017). The linking of both silencing and spatiotemporal gene expression may ensure precise expression and maintenance of cell identity. Thus, these dual-use genomic regions and the enhancer features themselves could also be constrained by the need to maintain other functions such as replication or correct encoding of a promoter, a protein, or a PRE. This dual use of genomic regions will likely shape dependencies between features in ways that aren’t obvious or transcription-centric.
While we have discussed the mechanistic, evolutionary, and biological constraints on enhancers separately here, the three are intertwined and together likely shape the dependencies between enhancer features (Figure 4). Gaining insight into the modes of transcriptional regulation, and biological and evolutionary constraints on enhancers at all levels will help us further understand the role of enhancer grammar in gene regulation and development and may ultimately allow us to classify enhancers based on the grammatical rules that govern them.
Section 7: Reading genomes and pinpointing grammatical changes that alter phenotype
As we find more and more enhancer variants associated with disease and evolutionary adaptations, it is becoming increasingly important to work out which sequence variants are causal within a sea of inert variants associated via linkage. Our lack of understanding of how an enhancer encodes expression patterns, the lack of sequence conservation within most enhancers, and the use of low-affinity sites, combined with linkage disequilibrium, make it hard to identify causal SNPs. Grammar could provide mechanistic insight to overlay on top of statistical approaches such as GWAS or eQTL analysis. A combined mechanistic and statistical approach could provide a powerful way to pinpoint causal variants. So far, there have been no attempts to do this, and it is an exciting future direction. There are many steps to this, and we’ve addressed defining grammatical constraints, classifying enhancers, and the dependencies between features, but another critical area is understanding what types of grammar changes will impact phenotype. Enhancer redundancy makes this a challenge. Gain-of-function mutations that lead to aberrant expression could be an obvious starting point and may be successful in some enhancers. Identifying causal variants in other enhancers is likely more nuanced. In some enhancers, extensive genetic epistasis within developmental enhancers may mean some SNPs can buffer the functional impact of large-effect variants in TFBSs making it harder to identify the causal variants (Cannavò et al., 2016). Furthermore, there are examples where causal SNPs are found in regions that are not thought to harbor TFBSs. Why such SNPs have an impact remains to be seen, this could be due to our inability to identify degenerate or non-canonical TFBSs. To our knowledge, only one study has experimentally investigated the link between enhancer grammar and phenotype. In the developing fly, manipulating the grammar of the MHE enhancer active in the somatic muscle and heart affects fitness, but only in homozygote animals exposed to stress (Boisclair Lachance et al., 2018; Halfon et al., 2000). Many more studies are needed to understand the link between enhancer grammar and phenotype.
Conclusion:
The promise of enhancer grammar to unlock the instructions for development encoded in our genomes is tantalizing yet elusive. Here we review the literature on enhancer grammar within development. The two most common models of enhancer grammar have been around for decades, and, in reality, these likely represent extremes of a spectrum. The third model highlights the importance of protein-protein interactions in addition to TF-enhancer interactions and motif flexibility. We propose dependency grammar as a conceptual model that unifies the three current models and more accurately reflects the range of enhancer grammars discovered so far. Dependency grammar considers the interplay and dependency between the type, affinity, and syntax of sites along with the biological, evolutionary, and mechanistic constraints that shape these dependencies (Figure 5). Understanding what shapes these dependencies and classifying enhancers based on where they lie on this spectrum of grammatical constraints may help us reach the holy grail of unifying principles of enhancer grammar – A Strunk and White for enhancers. The true test of such a model uses grammar to predict enhancers in the genome and design synthetic tissue-specific enhancers at scale. Beyond this, as most variants associated with disease lie within enhancers, finding ways to effectively pinpoint causal variants within a sea of linked inert variants is paramount. If grammatical rules are found, violations in these rules may hold the key to pinpointing such causal variants. Towards this goal, the relationship between enhancer grammar and phenotype must be interrogated. Ultimately one day, we may be able to read the instructions for development encoded in our genomes and pinpoint causal enhancer variants underlying disease and evolutionary adaptation. It is an exciting time, full of potential. Nevertheless, the complexity of enhancer sequence space, as well as the diversity of TFs, co-factors and transcriptional machinery, and the complexity of organisms means the study of enhancer grammar is highly complex, and much work remains in order to discover the “Strunk and White” of enhancer grammar in living organisms.
Supplementary Material
Acknowledgments
We thank the Farley lab, Jim Posakony, and Marc Halfon for helpful discussions. Figures created with BioRender.com. We apologize to colleagues whose work we could not cite due to space constraints. G.A.J. is supported by a Hartwell Fellowship and has past support from American Heart Association Grant 18POST34030077, NIH T32HL007444, and UC San Diego Chancellor’s Research Excellence Scholars Program. E.K.F. is supported by DP2HG010013.
References:
- Agoglia RM, and Fraser HB (2016). Disentangling Sources of Selection on Exonic Transcriptional Enhancers. Mol. Biol. Evol 33, 585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnone MI, and Davidson EH (1997). The hardwiring of development: organization and function of genomic regulatory systems. Development 124, 1851–1864. [DOI] [PubMed] [Google Scholar]
- Arnosti DN, Barolo S, Levine M, and Small S (1996). The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development 122, 205–214. [DOI] [PubMed] [Google Scholar]
- Avsec Ž, Weilert M, Shrikumar A, Krueger S, Alexandari A, Dalal K, Fropf R, McAnany C, Gagneur J, Kundaje A, et al. (2020). Deep learning at base-resolution reveals cis-regulatory motif syntax. BioRxiv 737981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Baer KE (1828). Entwicklungsgeschichte der Thiere: Beobachtung und Reflexion. (Developmental history of animals: Observations and reflections) (Konigsberg: Borntrager; ). [Google Scholar]
- Barolo S (2012). Shadow enhancers: Frequently asked questions about distributed cis-regulatory information and enhancer redundancy. Bioessays 34, 135–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S (2016). How to tune an enhancer. Proc. Natl. Acad. Sci 113, 6330–6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazett-Jones D, Leblanc B, Herfort M, and Moss T (1994). Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science 264, 1134–1137. [DOI] [PubMed] [Google Scholar]
- Benabdallah NS, Williamson I, Illingworth RS, Kane L, Boyle S, Sengupta D, Grimes GR, Therizols P, and Bickmore WA (2019). Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Mol. Cell 76, 473–484.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Peña-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. (2008). Variation in Homeodomain DNA Binding Revealed by High-Resolution Analysis of Sequence Preferences. Cell 133, 1266–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand V, Hudson C, Caillol D, Popovici C, and Lemaire P (2003). Neural Tissue in Ascidian Embryos Is Induced by FGF9/16/20, Acting via a Combination of Maternal GATA and Ets Transcription Factors. Cell 115, 615–627. [DOI] [PubMed] [Google Scholar]
- Boer C.G. de, Vaishnav ED, Sadeh R, Abeyta EL, Friedman N, and Regev A (2020). Deciphering eukaryotic gene-regulatory logic with 100 million random promoters. Nat. Biotechnol 38, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisclair Lachance J-F, Webber JL, Hong L, Dinner AR, and Rebay I (2018). Cooperative recruitment of Yan via a high-affinity ETS supersite organizes repression to confer specificity and robustness to cardiac cell fate specification. Genes Dev. 32, 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Dagenais N, and Weigel D (1999). Redundant Enhancers Mediate Transcriptional Repression of AGAMOUS by APETALA2. Dev. Biol 216, 260–264. [DOI] [PubMed] [Google Scholar]
- Cai HN, Arnosti DN, and Levine M (1996). Long-range repression in the Drosophila embryo. Proc. Natl. Acad. Sci 93, 9309–9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajigas I, Chakraborty A, Swyter KR, Luo H, Bastidas M, Nigro M, Morris ER, Chen S, VanGompel MJW, Leib D, et al. (2018). The Evf2 Ultraconserved Enhancer lncRNA Functionally and Spatially Organizes Megabase Distant Genes in the Developing Forebrain. Mol. Cell 71, 956–972.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavò E, Khoueiry P, Garfield DA, Geeleher P, Zichner T, Gustafson EH, Ciglar L, Korbel JO, and Furlong EE (2016). Shadow enhancers are pervasive features of developmental regulatory networks. Curr. Biol 26, 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catarino RR, and Stark A (2018). Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation. Genes Dev. 32, 202–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cave JW, Loh F, Surpris JW, Xia L, and Caudy MA (2005). A DNA Transcription Code for Cell-Specific Gene Activation by Notch Signaling. Curr. Biol 15, 94–104. [DOI] [PubMed] [Google Scholar]
- Chen L, and Capra JA (2020). Learning and interpreting the gene regulatory grammar in a deep learning framework. PLOS Comput. Biol 16, e1008334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Kazemian M, Pham H, Blatti C, Celniker SE, Wolfe SA, Brodsky MH, and Sinha S (2013). Computational Identification of Diverse Mechanisms Underlying Transcription Factor-DNA Occupancy. PLOS Genet. 9, e1003571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiaen L, Stolfi A, Davidson B, and Levine M (2009). Spatio-temporal intersection of Lhx3 and Tbx6 defines the cardiac field through synergistic activation of Mesp. Dev. Biol 328, 552–560. [DOI] [PubMed] [Google Scholar]
- Crocker J, and Ilsley GR (2017). Using synthetic biology to study gene regulatory evolution. Curr. Opin. Genet. Dev 47, 91–101. [DOI] [PubMed] [Google Scholar]
- Crocker J, Tamori Y, and Erives A (2008). Evolution Acts on Enhancer Organization to Fine-Tune Gradient Threshold Readouts. PLOS Biol. 6, e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J, Abe N, Rinaldi L, McGregor AP, Frankel N, Wang S, Alsawadi A, Valenti P, Plaza S, Payre F, et al. (2015). Low Affinity Binding Site Clusters Confer Hox Specificity and Regulatory Robustness. Cell 160, 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker J, Noon EP-B, and Stern DL (2016). The soft touch: low-affinity transcription factor binding sites in development and evolution. In Current Topics in Developmental Biology, (Elsevier; ), pp. 455–469. [DOI] [PubMed] [Google Scholar]
- Datta RR, Ling J, Kurland J, Ren X, Xu Z, Yucel G, Moore J, Shokri L, Baker I, and Bishop T (2018). A feed-forward relay integrates the regulatory activities of Bicoid and Orthodenticle via sequential binding to suboptimal sites. Genes Dev. 32, 723–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao Y, Fang R, Li B, Meng Z, Yu J, Qiu Y, Lin KC, Huang H, Liu T, Marina RJ, et al. (2017). A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat. Methods 14, 629–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Lošo T, and Tautz D (2010). A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature 468, 815–818. [DOI] [PubMed] [Google Scholar]
- Duboule D (1994). Temporal colinearity and the phylotypic progression: a basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Development 135–142. [PubMed] [Google Scholar]
- El-Sherif E, and Levine M (2016). Shadow Enhancers Mediate Dynamic Shifts of Gap Gene Expression in the Drosophila Embryo. Curr. Biol 26, 1164–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erceg J, Pakozdi T, Marco-Ferreres R, Ghavi-Helm Y, Girardot C, Bracken AP, and Furlong EEM (2017). Dual functionality of cis-regulatory elements as developmental enhancers and Polycomb response elements. Genes Dev. 31, 590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erives A, and Levine M (2004). Coordinate enhancers share common organizational features in the Drosophila genome. Proc. Natl. Acad. Sci 101, 3851–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans NC, Swanson CI, and Barolo S (2012). Sparkling Insights into Enhancer Structure, Function, and Evolution. In Current Topics in Developmental Biology, (Elsevier; ), pp. 97–120. [DOI] [PubMed] [Google Scholar]
- Farley EK, Olson KM, Zhang W, Brandt AJ, Rokhsar DS, and Levine MS (2015). Suboptimization of developmental enhancers. Science 350, 325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley EK, Olson KM, Zhang W, Rokhsar DS, and Levine MS (2016). Syntax compensates for poor binding sites to encode tissue specificity of developmental enhancers. Proc. Natl. Acad. Sci 113, 6508–6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore C, and Cohen BA (2016). Interactions between pluripotency factors specify cis-regulation in embryonic stem cells. Genome Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Davis GK, Vargas D, Wang S, Payre F, and Stern DL (2010). Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466, 490–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T, and Levine M (2017). Transvection. Curr. Biol 27, R1047–R1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuqua T, Jordan J, van Breugel ME, Halavatyi A, Tischer C, Polidoro P, Abe N, Tsai A, Mann RS, Stern DL, et al. (2020). Dense and pleiotropic regulatory information in a developmental enhancer. Nature 587, 235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong EEM, and Levine M (2018). Developmental enhancers and chromosome topology. Science 361, 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett AT, Square TA, and Medeiros DM (2012). BMP, Wnt and FGF signals are integrated through evolutionarily conserved enhancers to achieve robust expression of Pax3 and Zic genes at the zebrafish neural plate border. Development 139, 4220–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet J, and Mango SE (2002). Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science 295, 821–825. [DOI] [PubMed] [Google Scholar]
- Geusz RJ, Wang A, Chiou J, Lancman JJ, Wetton N, Kefalopoulou S, Wang J, Qiu Y, Yan J, Aylward A, et al. (2020). Pancreatic progenitor epigenome maps prioritize type 2 diabetes risk genes with roles in development. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Zhang X, Wang L, Engreitz J, Melnikov A, Rogov P, Tewhey R, Isakova A, Deplancke B, Bernstein BE, et al. (2017). Systematic dissection of genomic features determining transcription factor binding and enhancer function. Proc. Natl. Acad. Sci 114, E1291–E1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Mahony S, and Gifford DK (2012). High Resolution Genome Wide Binding Event Finding and Motif Discovery Reveals Transcription Factor Spatial Binding Constraints. PLOS Comput. Biol 8, e1002638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon MS (2019). Studying Transcriptional Enhancers: The Founder Fallacy, Validation Creep, and Other Biases. Trends Genet. 35, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfon MS, Carmena A, Gisselbrecht S, Sackerson CM, Jiménez F, Baylies MK, and Michelson AM (2000). Ras Pathway Specificity Is Determined by the Integration of Multiple Signal-Activated and Tissue-Restricted Transcription Factors. Cell 103, 63–74. [DOI] [PubMed] [Google Scholar]
- Hanes SD, Riddihough G, Ish-Horowicz D, and Brent R (1994). Specific DNA Recognition and Intersite Spacing Are Critical for Action of the Bicoid Morphogent. Mol Cell Biol 14, 3364–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay D, Hughes JR, Babbs C, Davies JOJ, Graham BJ, Hanssen LLP, Kassouf MT, Oudelaar AM, Sharpe JA, Suciu MC, et al. (2016). Genetic dissection of the α-globin super-enhancer in vivo. Nat. Genet 48, 895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, and Sharp PA (2017). A Phase Separation Model for Transcriptional Control. Cell 169, 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J-W, Hendrix DA, and Levine MS (2008). Shadow Enhancers as a Source of Evolutionary Novelty. Science 321, 1314–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilsley GR, Fisher J, Apweiler R, DePace AH, and Luscombe NM (2013). Cellular resolution models for even skipped regulation in the entire Drosophila embryo. Elife 2, e00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoda T, Moore AJ, He Z, Chandra V, Aida M, Denholtz M, Piet van Hamburg J, Fisch KM, Chang AN, Fahl SP, et al. (2017). Non-coding Transcription Instructs Chromatin Folding and Compartmentalization to Dictate Enhancer-Promoter Communication and T Cell Fate. Cell 171, 103–119.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LA, Zhao Y, Golden K, and Barolo S (2008). Reverse-Engineering a Transcriptional Enhancer: A Case Study in Drosophila. Tissue Eng. Part A 14, 1549–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, and Taipale J (2015). DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527, 384–388. [DOI] [PubMed] [Google Scholar]
- Junion G, Spivakov M, Girardot C, Braun M, Gustafson EH, Birney E, and Furlong EEM (2012). A Transcription Factor Collective Defines Cardiac Cell Fate and Reflects Lineage History. Cell 148, 473–486. [DOI] [PubMed] [Google Scholar]
- Kageyama R, Shimojo H, and Isomura A (2018). Oscillatory Control of Notch Signaling in Development. In Molecular Mechanisms of Notch Signaling, Borggrefe T, and Giaimo BD, eds. (Cham: Springer International Publishing; ), pp. 265–277. [DOI] [PubMed] [Google Scholar]
- Kalinka AT, Varga KM, Gerrard DT, Preibisch S, Corcoran DL, Jarrells J, Ohler U, Bergman CM, and Tomancak P (2010). Gene expression divergence recapitulates the developmental hourglass model. Nature 468, 811–814. [DOI] [PubMed] [Google Scholar]
- Khoueiry P, Rothbächer U, Ohtsuka Y, Daian F, Frangulian E, Roure A, Dubchak I, and Lemaire P (2010). A cis-Regulatory Signature in Ascidians and Flies, Independent of Transcription Factor Binding Sites. Curr. Biol 20, 792–802. [DOI] [PubMed] [Google Scholar]
- King DM, Hong CKY, Shepherdson JL, Granas DM, Maricque BB, and Cohen BA (2020). Synthetic and genomic regulatory elements reveal aspects of cis-regulatory grammar in mouse embryonic stem cells. ELife 9, e41279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni MM, and Arnosti DN (2003). Information display by transcriptional enhancers. Development 130, 6569–6575. [DOI] [PubMed] [Google Scholar]
- Kuroda T, Tada M, Kubota H, Kimura H, Hatano S, Suemori H, Nakatsuji N, and Tada T (2005). Octamer and Sox Elements Are Required for Transcriptional cis Regulation of Nanog Gene Expression. Mol. Cell. Biol 25, 2475–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Kamneva OK, Melo US, Barozzi I, Osterwalder M, Mannion BJ, Tissières V, Pickle CS, Plajzer-Frick I, and Lee EA (2016). Progressive loss of function in a limb enhancer during snake evolution. Cell 167, 633–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Zhu Y, Kelman G, Novak CS, Plajzer-Frick I, Kato M, Garvin TH, Pham Q, Harrington AN, Hunter RD, et al. (2020). Comprehensive In Vivo Interrogation Reveals Phenotypic Impact of Human Enhancer Variants. Cell 180, 1262–1271.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-K, and Pfaff SL (2003). Synchronization of Neurogenesis and Motor Neuron Specification by Direct Coupling of bHLH and Homeodomain Transcription Factors. Neuron 38, 731–745. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, and de Graaff E (2003). A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet 12, 1725–1735. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Hill AE, Devenney PS, and Hill RE (2008). Point mutations in a distant sonic hedgehog cis-regulator generate a variable regulatory output responsible for preaxial polydactyly. Hum. Mol. Genet 17, 978–985. [DOI] [PubMed] [Google Scholar]
- Levine M (2010). Transcriptional enhancers in animal development and evolution. Curr. Biol. CB 20, R754–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levo M, and Segal E (2014). In pursuit of design principles of regulatory sequences. Nat. Rev. Genet 15, 453–468. [DOI] [PubMed] [Google Scholar]
- Levo M, Avnit-Sagi T, Lotan-Pompan M, Kalma Y, Weinberger A, Yakhini Z, and Segal E (2017). Systematic Investigation of Transcription Factor Activity in the Context of Chromatin Using Massively Parallel Binding and Expression Assays. Mol. Cell 65, 604–617.e6. [DOI] [PubMed] [Google Scholar]
- Li L, and Wunderlich Z (2017). An Enhancer’s length and composition are shaped by its regulatory task. Front. Genet 8, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Sagendorf JM, Chiu T-P, Pasi M, Perez A, and Rohs R (2017). Expanding the repertoire of DNA shape features for genome-scale studies of transcription factor binding. Nucleic Acids Res. 45, 12877–12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Heist T, Levine M, and Fukaya T (2018). Visualization of Transvection in Living Drosophila Embryos. Mol. Cell 70, 287–296.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, and Posakony JW (2012). Role of Architecture in the Function and Specificity of Two Notch-Regulated Transcriptional Enhancer Modules. PLOS Genet. 8, e1002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Viales RR, Khoueiry P, Reddington JP, Girardot C, Furlong EEM, and Robinson-Rechavi M (2020). The hourglass model of evolutionary conservation during embryogenesis extends to developmental enhancers with signatures of positive selection. BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorberbaum DS, Ramos AI, Peterson KA, Carpenter BS, Parker DS, De S, Hillers LE, Blake VM, Nishi Y, and McFarlane MR (2016). An ancient yet flexible cis-regulatory architecture allows localized Hedgehog tuning by patched/Ptch1. Elife 5, e13550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Patel NH, and Kreitman M (1998). Functional analysis of eve stripe 2 enhancer evolution in Drosophila: rules governing conservation and change. Development 125, 949–958. [DOI] [PubMed] [Google Scholar]
- Ludwig MZ, Bergman C, Patel NH, and Kreitman M (2000). Evidence for stabilizing selection in a eukaryotic enhancer element. Nature 403, 564–567. [DOI] [PubMed] [Google Scholar]
- Luna-Zurita L, Stirnimann CU, Glatt S, Kaynak BL, Thomas S, Baudin F, Samee MAH, He D, Small EM, Mileikovsky M, et al. (2016). Complex Interdependence Regulates Heterotypic Transcription Factor Distribution and Coordinates Cardiogenesis. Cell 164, 999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk RW, and Eisen MB (2010). Evolutionary Mirages: Selection on Binding Site Composition Creates the Illusion of Conserved Grammars in Drosophila Enhancers. PLoS Genet. 6, e1000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T, Imamoto F, Merlino GT, Pastan I, and Ishii S (1989). Cooperative Function of Two Separate Enhancers of the Human Epidermal Growth Factor Receptor Proto-oncogene. J. Biol. Chem 264, 5488–5494. [PubMed] [Google Scholar]
- Markstein M, Zinzen R, Markstein P, Yee K-P, Erives A, Stathopoulos A, and Levine M (2004). A regulatory code for neurogenic gene expression in the Drosophila embryo. Development 131, 2387–2394. [DOI] [PubMed] [Google Scholar]
- de Marneffe M-C, and Nivre J (2019). Dependency Grammar. Annu. Rev. Linguist 197–218. [Google Scholar]
- Matsumoto J, Kumano G, and Nishida H (2007). Direct activation by Ets and Zic is required for initial expression of the Brachyury gene in the ascidian notochord. Dev. Biol 306, 870–882. [DOI] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. (2012). Systematic Localization of Common Disease-Associated Variation in Regulatory DNA. Science 337, 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, Feizi S, Gnirke A, Callan CG, Kinney JB, et al. (2012). Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat. Biotechnol 30, 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menoret D, Santolini M, Fernandes I, Spokony R, Zanet J, Gonzalez I, Latapie Y, Ferrer P, Rouault H, White KP, et al. (2013). Genome-wide analyses of Shavenbaby target genes reveals distinct features of enhancer organization. Genome Biol. 14, R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuleman W, Muratov A, Rynes E, Halow J, Lee K, Bates D, Diegel M, Dunn D, Neri F, Teodosiadis A, et al. (2020). Index and biological spectrum of human DNase I hypersensitive sites. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnoye L, Taskiran II, Mauduit D, Fazio M, Van Aerschot L, Hulselmans G, Christiaens V, Makhzami S, Seltenhammer M, Karras P, et al. (2020). Cross-species analysis of enhancer logic using deep learning. Genome Res. 30, 1815–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y, Sliz P, Pear WS, Aster JC, and Blacklow SC (2007). Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc. Natl. Acad. Sci 104, 2103–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi S, Blais A, and Ioshikhes I (2013). Identification of cis-regulatory modules in promoters of human genes exploiting mutual positioning of transcription factors. Nucleic Acids Res. 41, 8822–8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng FS, Schütte J, Ruau D, Diamanti E, Hannah R, Kinston SJ, and Göttgens B (2014). Constrained transcription factor spacing is prevalent and important for transcriptional control of mouse blood cells. Nucleic Acids Res. 42, 13513–13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, et al. (2013). Rapid and Pervasive Changes in Genome-wide Enhancer Usage during Mammalian Development. Cell 155, 1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder M, Barozzi I, Tissières V, Fukuda-Yuzawa Y, Mannion BJ, Afzal SY, Lee EA, Zhu Y, Plajzer-Frick I, and Pickle CS (2018). Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panne D, Maniatis T, and Harrison SC (2007). An Atomic Model of the Interferon-β Enhanceosome. Cell 129, 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatsenko D, Goltsev Y, and Levine M (2009). Organization of developmental enhancers in the Drosophila embryo. Nucleic Acids Res. 37, 5665–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passamaneck YJ, Katikala L, Perrone L, Dunn MP, Oda-Ishii I, and Gregorio AD (2009). Direct activation of a notochord cis-regulatory module by Brachyury and FoxA in the ascidian Ciona intestinalis. Development 136, 3679–3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Cervantes C, Smith LA, Nadadur RD, Hughes AEO, Wang S, Corbo JC, Cepko C, Lonfat N, and Moskowitz IP (2020). Enhancer transcription identifies cis -regulatory elements for photoreceptor cell types. Development 147, dev184432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, and Levine M (2010). Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol 20, 1562–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pherson M, Misulovin Z, Gause M, and Dorsett D (2019). Cohesin occupancy and composition at enhancers and promoters are linked to DNA replication origin proximity in Drosophila. Genome Res. 29, 602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips R (2015). Theory in Biology: Figure 1 or Figure 7? Trends Cell Biol. 25, 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff RA (1996). The Shape of Life: Genes, Development, and the Evolution of Animal Form (Chicago: University of Chicago Press; ). [Google Scholar]
- Rebeiz M, Castro B, Liu F, Yue F, and Posakony JW (2012). Ancestral and conserved cis-regulatory architectures in developmental control genes. Dev. Biol 362, 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickels R, and Shilatifard A (2018). Enhancer logic and mechanics in development and disease. Trends Cell Biol. 28, 608–630. [DOI] [PubMed] [Google Scholar]
- Rodda DJ, Chew J-L, Lim L-H, Loh Y-H, Wang B, Ng H-H, and Robson P (2005). Transcriptional Regulation of Nanog by OCT4 and SOX2. J. Biol. Chem 280, 24731–24737. [DOI] [PubMed] [Google Scholar]
- Ryan GE, and Farley EK (2020). Functional genomic approaches to elucidate the role of enhancers during development. WIREs Syst. Biol. Med 12, e1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayal R, Dresch JM, Pushel I, Taylor BR, and Arnosti DN (2016). Quantitative perturbation-based analysis of gene expression predicts enhancer activity in early Drosophila embryo. ELife 5, e08445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scardigli R, Bäumer N, Gruss P, Guillemot F, and Le Roux I (2003). Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development 130, 3269–3281. [DOI] [PubMed] [Google Scholar]
- Scully KM, Jacobson EM, Jepsen K, Lunyak V, Viadiu H, Carriere C, Rose DW, Hooshmand F, Aggarwal AK, and Rosenfeld MG (2000). Allosteric Effects of Pit-1 DNA Sites on Long-Term Repression in Cell Type Specification. Science 290, 1127–1131. [DOI] [PubMed] [Google Scholar]
- Senger K, Armstrong GW, Rowell WJ, Kwan JM, Markstein M, and Levine M (2004). Immunity Regulatory DNAs Share Common Organizational Features in Drosophila. Mol. Cell 13, 19–32. [DOI] [PubMed] [Google Scholar]
- Shrinivas K, Sabari BR, Coffey EL, Klein IA, Boija A, Zamudio AV, Schuijers J, Hannett NM, Sharp PA, Young RA, et al. (2019). Enhancer Features that Drive Formation of Transcriptional Condensates. Mol. Cell 75, 549–561.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RP, Taher L, Patwardhan RP, Kim MJ, Inoue F, Shendure J, Ovcharenko I, and Ahituv N (2013). Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat. Genet 45, 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorge S, Ha N, Polychronidou M, Friedrich J, Bezdan D, Kaspar P, Schaefer MH, Ossowski S, Henz SR, and Mundorf J (2012). The cis-regulatory code of Hox function in Drosophila. EMBO J. 31, 3323–3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, et al. (2013). Developmental Fate and Cellular Maturity Encoded in Human Regulatory DNA Landscapes. Cell 154, 888–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson CI, Evans NC, and Barolo S (2010). Structural Rules and Complex Regulatory Circuitry Constrain Expression of a Notch- and EGFR-Regulated Eye Enhancer. Dev. Cell 18, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson CI, Schwimmer DB, and Barolo S (2011). Rapid Evolutionary Rewiring of a Structurally Constrained Eye Enhancer. Curr. Biol 21, 1186–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taher L, McGaughey DM, Maragh S, Aneas I, Bessling SL, Miller W, Nobrega MA, McCallion AS, and Ovcharenko I (2011). Genome-wide identification of conserved regulatory function in diverged sequences. Genome Res. 21, 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak YG, and Farnham PJ (2015). Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin 8, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanos D, and Maniatis T (1995). Virus Induction of Human IFNP Gene Expression Requires the Assembly of an Enhanceosome. Cell 10. [DOI] [PubMed] [Google Scholar]
- Tournamille C, Colin Y, Cartron JP, and Kim CLV (1995). Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy–negative individuals. Nat. Genet 10, 224–228. [DOI] [PubMed] [Google Scholar]
- Tsai A, Muthusamy AK, Alves MR, Lavis LD, Singer RH, Stern DL, and Crocker J (2017). Nuclear microenvironments modulate transcription from low-affinity enhancers. ELife 6, e28975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai A, Alves MR, and Crocker J (2019). Multi-enhancer transcriptional hubs confer phenotypic robustness. ELife 8, e45325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai P-F, Dell’Orso S, Rodriguez J, Vivanco KO, Ko K-D, Jiang K, Juan AH, Sarshad AA, Vian L, Tran M, et al. (2018). A Muscle-Specific Enhancer RNA Mediates Cohesin Recruitment and Regulates Transcription In trans. Mol. Cell 71, 129–141.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent BJ, Estrada J, and DePace AH (2016). The appeasement of Doug: a synthetic approach to enhancer biology. Integr. Biol 8, 475–484. [DOI] [PubMed] [Google Scholar]
- Visel A, Rubin EM, and Pennacchio LA (2009). Genomic views of distant-acting enhancers. Nature 461, 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhuang J, Iyer S, Lin X, Whitfield TW, Greven MC, Pierce BG, Dong X, Kundaje A, Cheng Y, et al. (2012). Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 22, 1798–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, and Hughes TR (2010). Conserved expression without conserved regulatory sequence: the more things change, the more they stay the same. Trends Genet. 26, 66–74. [DOI] [PubMed] [Google Scholar]
- Xing K, and He X (2015). Reassessing the “Duon” Hypothesis of Protein Evolution. Mol. Biol. Evol 32, 1056–1062. [DOI] [PubMed] [Google Scholar]
- Zandvakili A, Campbell I, Gutzwiller LM, Weirauch MT, and Gebelein B (2018). Degenerate Pax2 and Senseless binding motifs improve detection of low-affinity sites required for enhancer specificity. PLoS Genet. 14, e1007289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen RP, Girardot C, Gagneur J, Braun M, and Furlong EEM (2009). Combinatorial binding predicts spatio-temporal cis -regulatory activity. Nature 462, 65–70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


