The evolutionary history of a common genetic variant is linked to its sex-specific effect on response to nutritional stress.
Abstract
The common deletion of the third exon of the growth hormone receptor gene (GHRd3) in humans is associated with birth weight, growth after birth, and time of puberty. However, its evolutionary history and the molecular mechanisms through which it affects phenotypes remain unresolved. We present evidence that this deletion was nearly fixed in the ancestral population of anatomically modern humans and Neanderthals but underwent a recent adaptive reduction in frequency in East Asia. We documented that GHRd3 is associated with protection from severe malnutrition. Using a novel mouse model, we found that, under calorie restriction, Ghrd3 leads to the female-like gene expression in male livers and the disappearance of sexual dimorphism in weight. The sex- and diet-dependent effects of GHRd3 in our mouse model are consistent with a model in which the allele frequency of GHRd3 varies throughout human evolution as a response to fluctuations in resource availability.
INTRODUCTION
Why does functional genetic variation remain polymorphic? This question is fundamental to evolutionary biology. The complete deletion of the third coding exon of the growth hormone (GH) receptor gene (GHR) in humans provides a compelling model to study this question. This 2.7-kb deletion (GHRd3; esv3604875) is found with allele frequencies between 10 and 80% among extant human populations in the 1000 Genomes phase 3 (1) and Simon Genome Diversity Project (2) datasets. In parallel, GHRd3 was found in Neanderthal and Denisovan genomes (3). This observation is unexpected given that the receptor is highly conserved among mammals and fundamental in various cellular processes, including cell division, immunity, and metabolism (4), and a loss of function mutation of this gene causes recessive Laron syndrome (5, 6). The GHRd3 allele generates a shorter isoform of this critical developmental gene (4). Locus-specific studies consistently and reproducibly associate GHRd3 with altered placental and birth weight (7), time of puberty onset (8), life span (9), and metabolic activity (10). There have been both positive and negative results about the effects of GHRd3 on the response to GH treatments (11, 12). Despite the known relevance of GHRd3 to human phenotypes that are likely crucial for human evolution, the mechanisms through which GHRd3 affects cellular and organismal function and the evolutionary forces that maintain the GHRd3 allele in human populations remain mostly unknown.
RESULTS
GHRd3 has been a common allele since before human-Neanderthal divergence
GHRd3 has a complex evolutionary history in humans. Our previous work determined that GHRd3 is one of 17 exonic deletions polymorphic in extant humans and shared with the Altai Neanderthal or Denisovan genomes (3). This study now extends this observation to include the Vindija (13) and Chagyrskaya (14) Neanderthal genomes, for which coverage is high enough for read depth–based deletion analysis (fig. S1A). The fact that this derived deletion is shared with archaic humans suggests that the GHRd3 allele was formed before the divergence of the human and Neanderthal lineages. Further, while the deletion remains polymorphic in humans, it was likely fixed in the Neanderthal and Denisovan lineages since all of the four archaic human genomes with high coverage we analyzed carry the deletion allele homozygously. We dismissed Neanderthal introgression as a possible explanation for the allele sharing since African populations have the highest allele frequency for this deletion (3). The deleted coding sequence is conserved among mammals (fig. S1B). On a broader scale, exon 3 is significantly more conserved than randomly selected sites on the same chromosome (P = 1.3 × 10−8, Kolmogorov-Smirnov test), suggesting that this exon is not evolving under neutral conditions (fig. S1C). Further, this exon is expressed in all tissues where GHR is expressed unless the individual has the deletion (fig. S2). The effect of GHRd3 at the transcript level was initially thought to be due to alternative splicing but later corrected and described as a “novel mechanism accounting for protein diversity” (15). When these data are observed in conjunction with the existing literature associating GHRd3 with multiple human traits, it raises the possibility that adaptive forces may have shaped the allele distribution of this variant among extant human populations.
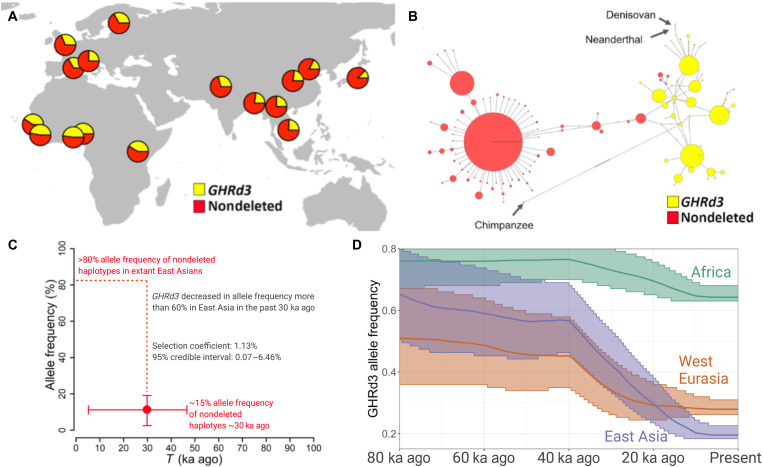
To understand the evolutionary forces that have maintained the GHRd3 allele in human populations, we resolved the linkage disequilibrium (LD) architecture around GHRd3 (fig. S3). We identified 15 nearby single-nucleotide polymorphisms (SNPs) that have r2 > 0.75 with GHRd3 deletions (table S1), a conservative threshold given the genome-wide patterns (16). These variants help construct a short haplotype block (hg19, chr5: 42625443 to 42628325) flanked by known recombination hotspots [recombination rates were retrieved from the 1000 genomes selection browser (http://hsb.upf.edu/)]. This specific haplotype tags the GHRd3 allele. On the basis of these tag SNPs and direct genotyping of the deletion available through the 1000 Genomes dataset, we found that GHRd3 is the major allele in most African populations (Fig. 1A and fig. S4). However, the allele frequency decreases to less than 25% in most Eurasian populations. The single-nucleotide variants with strong LD with the deletion were allowed for the construction of a haplotype network using PopART (version 1.7) (17) (see section S1.1) where one can visualize the clear separation of haplotypes that carry the GHRd3 allele from those that do not (Fig. 1B). One unexpected observation is that the haplotypes that harbor the deletion are more variable and cluster with the chimpanzee haplotype than those that do not (fig. S5). That is, the ancestral nondeleted allele is harbored primarily by recently evolved haplotypes. This observation raises the possibility that GHRd3 was nearly fixed in the ancestral human lineage and that the haplotypes that harbor the ancestral, nondeleted allele rebounded in frequency only recently.
Fig. 1. Evolutionary history of GHRd3.
(A) The geographic distribution of the GHRd3 polymorphism. The deletion status is color-coded for the entire figure. (B) A network of 2504 human haplotypes, the Altai Neanderthal genome, the Denisovan genome, and the chimpanzee reference genome calculated from variations within the high LD region upstream of the GHRd3 location (hg19, chr5: 42624748 to 42628325; see fig. S1). (C) Estimation of the allele frequency of nondeleted haplotypes (and by proxy that of GHRd3) and the date for the onset of selection in the Han Chinese (CHB) population based on the results of the approximate Bayesian computation simulations. The x axis shows time, while the y axis represents allele frequency. This plot shows means of T and ft with 95% credible intervals. The dashed line indicates the difference in allele frequency between the present and t. (D) Inferred allele frequency trajectory using CLUES in three continental populations using Simons Genome Diversity Project data. The ribbons around the lines represent 95% posterior intervals.
Recent population–specific evolutionary trends have shaped the geographic distribution of GHRd3
To test whether recent selection is acting on this locus, we used the single-nucleotide variation data within the GHRd3 upstream haplotype block to calculate Tajima’s D (18) and cross population extended haplotype homozygosity (XP-EHH) (19) values. These statistics measure deviations from expected allele frequency spectra (Tajima’s D) and extended homozygosity differences between populations (XP-EHH) under neutrality. We observed a significantly lower Tajima’s D value for the GHR locus in the East Asian population when compared to randomly selected regions on chromosome 5 [P < 0.05, Mann-Whitney (MW) test; fig. S6A]. Moreover, the nondeleted haplotypes showed lower Tajima’s D values when compared to the deleted haplotypes in the Eurasian populations (P < 0.01, MW test; fig. S6B). Concordant with the Tajima’s D analysis, when the GHRd3 locus of the Han Chinese (CHB) population was compared with that of the West European (CEU) and Yoruba (YRI) populations, we noted XP-EHH values that were higher than genome-wide expectations (fig. S6C). Collectively, these results are parsimonious with a relatively recent sweep of the existing nondeleted haplotypes in the East Asian population.
To summarize these results, a plausible model is that GHRd3 was nearly fixed in the human-Neanderthal lineage and remained the major allele throughout most of human history but only recently reduced in allele frequency potentially due to a sweep of the existing haplotypes harboring the nondeleted allele. To investigate this model, we used the Fc method that combines site frequency spectrum analyses with LD calculations (20) and found a significant deviation of the Fc value from the simulated neutral expectations in the East Asian population, showing selection starting approximately 27 thousand years (ka) ago (P < 0.01, Kolmogorov-Smirnov test; see section S1.4). This analysis did not detect selection in African or Western European populations. To strengthen this analysis, we used an approximate Bayesian computation method that compares the fit of different evolutionary models to the site frequency spectrum and haplotype structure variation data of a given locus (fig. S8 and section S1.5) (21). This approach selected a model that fits our hypothesis and suggested a recent sweep of the nondeleted haplotypes in East Asia but not in Africa or Europe. Specifically, it dated the onset of this sweep to 29,811 years ago (95% confidence interval between 5103 to 46,710 years ago) while putting the frequency of the nondeleted allele at ~11% before the sweep [log10-scaled approximate Bayes factor, log10(aBF) = 5.250 against neutral; log10(aBF) = 13.257 against hard-sweep] (Fig. 1C).
Collectively, both Fc and Bayesian inferences show similar results, suggesting recent population–specific adaptive forces in shaping the extant allele frequency distribution. Given the ancient origin of the GHRd3 deletion and this recent selection, we then attempted to infer its allele frequency trajectory over time. First, we searched the Allen Ancient DNA Resource (https://reich.hms.harvard.edu/allen-ancient-dna-resource-aadr-downloadable-genotypes-present-day-and-ancient-dna-data; accessed 15 May 2021) to investigate the presence or absence of GHRd3 in ancient samples. We found that rs4590183 (R2 = ~0.9 in extant populations) was genotyped in 1888 ancient samples. Assuming that the LD between this SNP and the GHRd3 deletion is similar in the ancient genomes, we found that the extant allele frequency distribution of GHRd3 is roughly similar to that constructed from ancient genome data (fig. S7). However, virtually, all of these ancient genomes are younger than 30,000 years old (median age of ~10 ka) and do not inform us about the deeper evolutionary history of this allele, which is critical to understand the recent selection in Asia and allele frequencies.
Second, we reconstructed the genealogical history at the GHRd3 locus using RELATE (22). This approach identifies phylogenetic relationships between all modern sequences in the GHRd3 locus and maps the individual mutations onto this phylogeny. To infer allele frequency through time, we calculated the proportion of lineages that carry the GHRd3 allele (as tagged by rs6873545, R2 = 1 in YRI, CEU, and CHB) (see section S1.6). We conducted this analysis using both 1000 Genomes phase 3 data (fig. S9A) (1) and Simons Genome Diversity project data (fig. S9B) (2) with encouragingly similar outcomes. In parallel, we used CLUES (Fig. 1D and fig. S9C) (23), which is an important sampling approach that reweights each sampled genealogical history (obtained from RELATE) according to its likelihood under selection and infers a maximum-likelihood selection coefficient and allele frequency trajectory. We found, in agreement with our earlier predictions, that the allele trajectories of GHRd3 vary among continental populations, most notably due to the marked decrease in allele frequency in Asian populations that started between 20 and 40 ka ago and to a lesser extent a similar, but potentially older, decrease in European populations. The allele frequency in African populations seems to remain stably high (~60 to 70%) in the past 100,000 years. We detected observable (but weak) evidence of selection in East Asians (s ~ 0.002) between 0 and 40 ka ago (log-likelihood ratio = 3), weaker evidence in West Eurasia (log-likelihood ratio = 1.5), and no evidence in Africa (log-likelihood ratio = 0.5) in agreement with our other attempts to detect selection. Overall, our results presented here fit well with a model where GHRd3 has remained at a high allele frequency in human populations until 50 ka ago but reduced in Eurasian populations subsequently, likely under adaptive constraints.
Associations of GHRd3 to metabolic and developmental traits
Dozens of locus-specific studies associated GHRd3 with multiple developmental and metabolic traits (fig. S10), pointing to the pleiotropic effects of this deletion on multiple biological systems at different scales. Further, these studies suggest that the effect of GHRd3 depends on the developmental stage and the metabolic state of the individuals. For example, GHRd3 in the offspring was associated with lower levels of maternal GH levels in the placenta (24), which potentially explains its association with smaller placental and birth size (7, 8). In later developmental stages, the effects of GHRd3 on organismal growth are reversed. It is associated with increased sensitivity to GH (9) and likely as a result with catch-up growth (25) and early onset of puberty in boys (8). In addition, GHRd3 is associated with insulin sensitivity, disposition index in pubescent boys, and higher triglyceride levels (10). In adults, GHRd3 is associated with increased height and longevity (9) and reduced instances of type 2 diabetes but increased rates of metabolic disorder if present in diabetic patients (26). Despite these associations, the exact molecular mechanisms and the genetic model (e.g., recessive, dominant, or additive) through which GHRd3 affects these traits are not known.
Currently, all the associations reported for GHRd3 are from locus-specific studies, which can be prone to type I errors. A recent genome-wide association study (GWAS) was unable to replicate the proposed effect of GHRd3 on GH treatment responsiveness (12), at the very least suggesting that the effect size is smaller than it was reported before (11). The expected genetic model for the effect of this variant is not well established, and the effect of GHRd3 is dependent on the sex (8, 9) and the endocrine/metabolic state (26) of the individual. Therefore, a more focused GWAS design, where sex and metabolic state are controlled and the genetic model is informed by the particular trait being tested, may be needed to produce a strong enough signal to survive stringent multiple hypotheses thresholds of traditional GWAS studies. Regardless, we were able to examine phenotype associations in available GWAS databases, using a single-nucleotide variant that tags the deletion (rs4073476). Assuming an additive genetic model and using a strict nominal P value threshold of P < 10−8 in a phenome-wide association study (PheWAS) of 152 phenotypes (available through https://atlas.ctglab.nl/PheWAS), we found that the derived GHRd3 haplotype is strongly associated with bone mineral density. The next most significant association was height with a nominal P < 10−5 (table S1). We then conducted a similar analysis in the U.K. Biobank dataset phenotypes (available through http://geneatlas.roslin.ed.ac.uk) (27). We found that the GHRd3 haplotype is associated with standing height in this cohort (nominal P < 10−8, in a PheWAS of 742 phenotypes; fig. S11 and table S1). These findings are concordant with previous locus-specific studies that link GHRd3 with developmental and metabolic traits (26). Notably, one such locus-specific study underlined the sex specificity of the phenotypic effect of GHRd3 (9). We found that, among the top 10 traits that are at least nominally associated with GHRd3 in the U.K. Biobank dataset, grip strength has a greater correlation with the deletion in males (left hand, P = 1.29 × 10−5; right hand, P = 1.40 × 10−4) than in females (left hand, P = 3.57 × 10−1; right hand, P = 1.48 × 10−1). This effect was noted independently for both the left and right hands, increasing our confidence in this observed trend.
Response to nutritional deficits may explain the evolutionary history of GHRd3
In their seminal review of ecological literature, Metcalfe and Monaghan (28) have proposed that compensatory growth following a smaller birth due to nutritional deficits may have fitness advantages. However, it may also have multiple adverse consequences later in life, affecting traits such as bone ossification rate, fat deposition rate, growth, age at sexual maturation, insulin regulation, and life span. As described above, the traits associated with GHRd3 show a notable resemblance to the traits that Metcalfe and Monaghan discussed within the context of response to nutritional deficit. On the basis of this insight from the ecological literature, we hypothesize that this deletion may affect fitness within the context of the famine and abundance periods that have been a defining feature of human evolution (Fig. 2A). It was previously speculated that the GHRd3-mediated up-regulation of the GH pathway might be a response to nutritional deprivation (29). Response to starvation is relevant to human evolution because compared to nonhuman great apes, modern and archaic humans generally cope with higher levels of seasonality and unpredictability of resource levels (30). In this ecological context, the fluctuation of resources may have been a major adaptive stressor for humans, especially on traits pertaining to metabolism and reproduction. Studies in other animals suggest that small size at birth and early reproduction, both noted effects of GHRd3, is favored under environmental stress (31).
Fig. 2. Nutritional availability may be a factor in shaping GHRd3 allele frequency.
(A) A plausible model for the evolution of GHRd3. From bottom to top, we indicated GHRd3 allele frequency across time where yellow depicts deleted allele and red depicts nondeleted allele. Next, we show the phylogenetic relationships between extant and archaic humans with the allele frequencies shown with the pie charts at the branch tips. The timeline for ecological variability and the onset of modern human behavior was shown at the top of this panel. LGM, Last Glacial Maximum; Ma ago, million years ago. (B) The association between GHRd3 and edematous severe acute malnutrition (ESAM). GHRd3 is significantly enriched in nonedematous SAM (NESAM) as compared to ESAM, indicating that GHRd3 may provide protection against developing the classically more severe form of acute malnutrition. The graph shows the proportion of nondeleted, heterozygous, and homozygous GHRd3 genotypes in each category. The sample size of each category is indicated on the top of the chart.
To further test this notion, we genotyped GHRd3 in 176 Malawian children with severe acute malnutrition (SAM) (see section S1.8) (32). We found that GHRd3 is depleted among children who suffer from the more severe, edematous form of SAM (kwashiorkor) (P = 0.0066, chi-square test) (Fig. 2B). Thus, individuals carrying GHRd3 may fare better under nutritional stress, supporting the hypothesis that GHRd3 may confer a fitness advantage under these conditions. Note that previous work showed that children who suffer from edematous SAM (ESAM) have significantly higher birth weight than those who suffer from less severe nonedematous SAM (NESAM) (33). Thus, it is plausible that lower birth weight at birth, which GHRd3 is significantly associated in (7), may lead to a more favorable metabolic response to malnutrition and may increase fitness under nutritional stress.
It is notable that GHRd3 emerged 1 million to 2 million years ago, yet it is still segregating today. This emergence time coincides with the spread of the Homo genus within and outside Africa (34), likely carrying the deletion across the world. A strongly deleterious effect is therefore highly unlikely, and given the pleiotropic effects and evidence of recent adaptive effects on modern humans, it appears plausible that GHRd3 increased the fitness of these archaic populations who were facing new ecologies and potential resource fluctuations. This scenario also fits with the observation that GHRd3 is potentially fixed in ancient Eurasian hominins (i.e., Neanderthals and Denisovans), who may have been exposed to even greater seasonal environmental stress than most African hominins (35).
Diet- and sex-dependent effect of GHRd3 on size
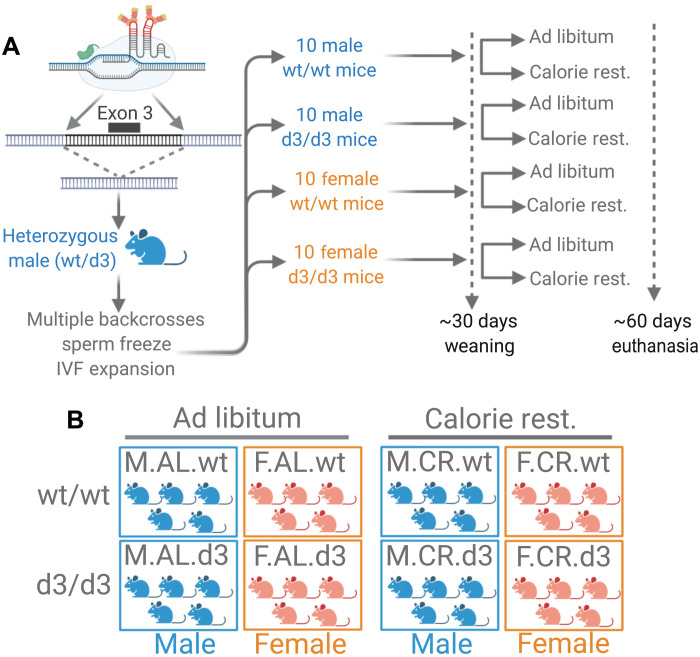
On the basis of our evolutionary model, we expected GHRd3 to have pronounced phenotypic effects when resources are limited. Further, given the previous data (9) suggesting sex-specific effects of GHRd3, we expected the phenotypic effects of GHRd3 to be confined to males. To test these expectations and further investigate the biological effects of GHRd3, we developed a mouse model by generating a ~2.5-kb deletion using CRISPR-Cas9. This deletion mimicked the human variant and removed the otherwise conserved exon 3 ortholog from the C57BL/6 genome. This procedure involved editing the genome of mouse embryos using a pair of single guide RNAs that complement the sequences flanking the third exon of the GH receptor gene (Ghr) in the mouse genome (see Fig. 3A and section S2.1). The resulting founder mouse was a heterozygous (wt/d3) male, which was then used to generate offspring with wild-type (wt/wt) mice. We backcrossed the initial wt/d3 mice with wt/wt mice for at least five generations before conducting the experiments to prevent off-target effects. No deviations from Mendelian expectations were observed in the initial colony or subsequently established colonies of this mouse line. Further, we sequenced the genomes of three Ghrd3 homozygous mice and found no evidence for off-target effects (see section S2.3). We confirmed mouse genotypes using gel electrophoresis (fig. S12A). Further, we showed that wt/wt mice abundantly express exon 3, while Ghrd3 mice show no expression of exon 3 as expected (fig. S13). To test the specific expectations outlined above, we established eight cohorts of five mice (table S2) based on all possible combinations of sex (female or male), diet [constantly available, i.e., ad libitum (AL) or 40% calorie-restricted (CR)], and genotype (wt/wt or d3/d3) under controlled settings (Fig. 2B). We raised all of the cohorts under identical conditions until weaning at 30 days old and provided the differential diets for the next 30 days.
Fig. 3. Experimental design.
(A) Generation of the mouse model using a CRISPR-Cas9 approach with single guide RNAs targeting each side of the deletion. After multiple backcrosses, we raised the male and female mice with or without the deletion under AL and 40% CR dietary conditions. IVF, in vitro fertilization. (B) On the basis of the experimental design, we used eight different cohorts, each having five mice.
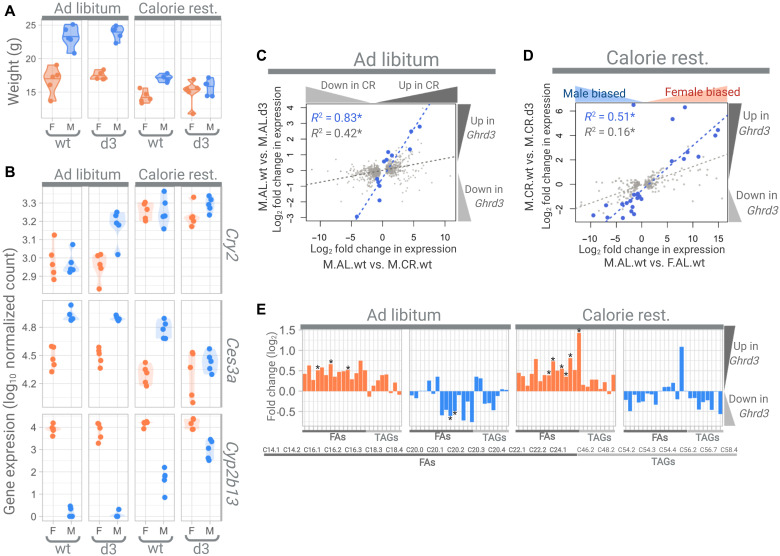
At ~60 days, we observed no significant effect of Ghrd3 on the weight of mice that were fed the AL diet (Fig. 4A and table S2). However, under CR, we observed that Ghrd3 leads to a −8.4% (P = 0.016, MW test) and a +4.75% change in mean weight in males and females, respectively. As a result, sexual differentiation in weight completely disappears under CR among d3/d3 mice. This is a remarkable finding considering that male mice are ~39 and 20% larger in wt/wt mouse cohorts grown under AL and CR, respectively (P < 0.01, MW test). Overall, our results are consistent with the notion that the phenotypic effect of Ghrd3 is strong but manifests in a sex- and environment-dependent manner.
Fig. 4. The phenotypic effects of Ghrd3.
(A) Weights of mice. Female and male mice are labeled by orange and blue, respectively. (B) Examples of genes that show significant differences between wt/wt and d3/d3 male mice. Cry2 is an example of a circadian gene, while Cyp2b13 and Ces3a are examples of genes with sex-specific expression. (C) Correlation between the effect of calorie restriction (x axis) and Ghrd3 under an AL diet (y axis). The light gray dots show the genes that have significantly (Padj < 0.01) different expression levels between wt/wt male mice that were fed AL and CR diets. The blue dots show the genes that have significantly (Padj < 0.01) different expression levels between wt/wt and d3/d3 male mice that were grown under AL diets. (D) Correlation between the effect of sex (x axis) and Ghrd3 under calorie restriction (y axis). The light gray dots show the genes that have significantly (Padj < 0.01) different expression levels between males and females, while blue dots show the genes that have significantly (Padj < 0.01) different expression levels between wt/wt and d3/d3 male mice that were grown under a CR diet. (E) Log2 fold changes between wt/wt and d3/d3 mice for FA (fatty acid) and TAG (triacylglyceride) levels in the blood serum. Asterisk indicates significant changes (nominal P < 0.05, t test).
The effect of GHRd3 on liver transcriptome in male mice is compensated through circadian regulation
To understand the mechanistic underpinnings of the biological effects of Ghrd3, we conducted a comparative transcriptomics analysis of the liver tissues of mice from the eight cohorts at ~60 days (see table S3 and section S2.4). At this age range, the mice have reached a point of sexual maturity but were still in a period of growth and development (36). The liver is a natural tissue choice for this analysis because GHR is expressed abundantly in the liver and its specific functions in this tissue have been extensively studied (37). Moreover, the overall expression trends of mouse livers for both sexes are well studied (38). Since GH secretion patterns are circadian and sex specific (39, 40), we expected the downstream effects of Ghrd3 to be highly variable because they are intrinsically dependent on GH availability. Thus, only the strongest effects would likely be significant when comparing wt/wt and d3/d3 liver transcriptomes. We found no genes with significant expression differences when we compared F.AL.wt and F.AL.d3 or F.CR.wt and F.CR.d3 cohorts (see Fig. 3B for cohort descriptions). These results are consistent with the notion that Ghrd3 has a minimal effect in females, independent of diet, at least at the 2-month developmental snapshot. In contrast, we found 13 differentially expressed genes when we compared M.AL.wt and M.AL.d3 and 28 differentially expressed genes when we compared M.CR.wt and M.CR.d3 cohorts (Padj < 0.01, Wald test with Benjamini-Hochberg multiple testing correction; Table 1). Unexpectedly, these sets of genes do not overlap, indicating that the effect of Ghrd3 on gene expression in male mouse livers differs based on dietary conditions.
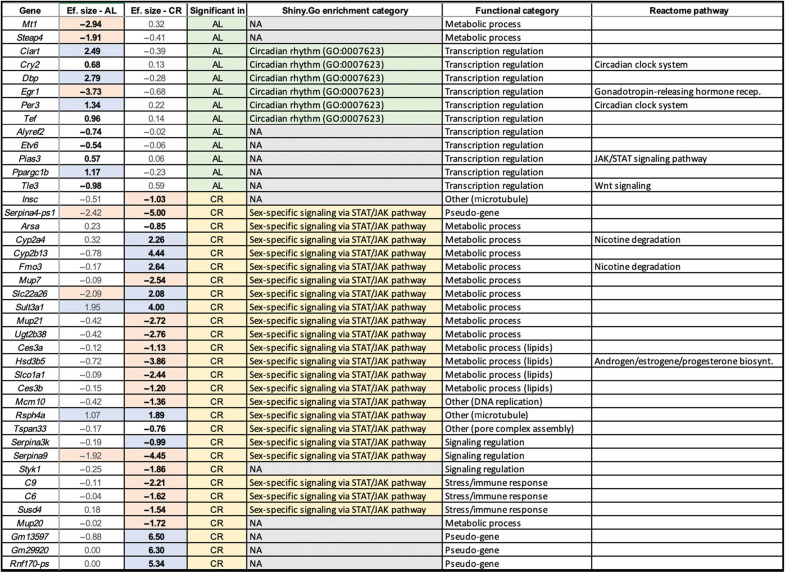
Table 1. The effect sizes of genes that show significant expression level differences between wt/wt and d3/d3 male mice under AL and CR conditions.
The orange and blue colors indicate genes that are up-regulated or down-regulated more than twofold in d3/d3 mice compared to wt/wt mice, respectively. The fold changes that are shown to be significant (Padj < 0.01) are bolded. The top enrichment classifications in Shiny.Go (more detailed information can be found in table S4), manually curated Panther classifications, and Reactome pathway classifications, respectively, are in the last three columns. NA, not available.
To further understand this phenomenon, we first conducted an enrichment analysis of functional categories for the genes that are differentially expressed between M.AL.wt and M.AL.d3 livers (see table S4 and section S2.4). We found that 5 of the 13 genes (~38%) are related to circadian rhythm processes, indicating a significant enrichment from stochastic expectations [false discovery rate (FDR) = 2.33 × 10−6]. Notably, these include the up-regulation of three primary circadian rhythm regulators, Per3, Cry2, and Ciart (Table 1). A closer inspection of the expression trends in all the mouse cohorts revealed that the same circadian rhythm genes are also up-regulated in wt/wt mice as a response to calorie restriction (e.g., Cry2; Fig. 4B). Circadian response to dietary change directly interacts with the GH pathway and involves the regulation of Per and Cry2 expression (41, 42). The expression levels of circadian rhythm regulators in M.AL.d3 mice resemble those of wt/wt mice under CR.
Calorie restriction dampens GH cyclicity in mice, leading to the flattened pulsation of its downstream signal (43). This flattening is partially compensated by the activation of the circadian rhythm pathway, involving Per and Cry (41, 42). Since Ghrd3 transduces GH signaling ~30% faster than the full-length isoform (11), we hypothesize that it has a similar dampening effect, reducing the pulsating downstream signaling in the GH pathway even under an AL diet. Supporting this hypothesis, we found a significant correlation between the gene expression response to calorie restriction in wt/wt mice and the effect of GHRd3. Specifically, the expression change from M.AL.wt to M.AL.d3 significantly correlates with the expression change from M.AL.wt to M.CR.wt (P < 10−5; Fig. 4C). Overall, our results support the notion that the effect of GHRd3 is similar to calorie restriction in male mice under normal dietary conditions.
Under calorie restriction, GHRd3 leads to female-like gene expression in male livers
Next, we conducted an enrichment analysis on the 28 genes that are differentially expressed in M.CR.d3 mice as compared to M.CR.wt mice. We found that 13 of these 28 genes are involved in “HNF4A-dependent sex-specific differences in mice liver” (44) and were highly enriched (~46%; FDR = 1.62 × 10−12). A closer inspection revealed that multiple well-established male-biased genes [e.g., major urinary protein (Mup) genes and carboxylesterases (Ces3a and Ces3b)] were down-regulated in M.CR.d3 livers when compared to M.CR.wt livers, while notable female-biased genes (e.g., Cypb13, Cyp2a4, Sult3a1, and Fmo3) were up-regulated (Table 1). When we investigated the expression of those genes in all eight cohorts, we found a remarkable similarity between the expression of these genes in M.CR.d3 mice and female mice in general, regardless of the diet and genotype (e.g., Cyp2b13 and Ces3a; Fig. 4B). In that regard, the expression of these genes mimics the weight variation. To recap, M.CR.d3 mice are significantly lighter than M.CR.wt mice, making them virtually identical in weight to F.CR.d3 mice (Fig. 4A).
On the basis of these observations, we hypothesize that Ghrd3 leads to the female-like gene expression in male livers under calorie restriction, which potentially leads to the observed size reduction. Note that all of these genes were shown to be downstream gene targets of the Hnf4α and Stat5b pathways and that their concerted signaling regulates several sexually dimorphic genes in the liver (45). We also found a significant correlation between the effect of Ghrd3 in males under calorie restriction (measured by comparing M.CR.wt and M.CR.d3 cohorts) and sex-specific differences in gene expression under AL dietary conditions (measured by comparing F.AL.wt and M.AL.wt cohorts) (P < 10−4; Fig. 4D).
These results are consistent with a model where Ghrd3 leads to a female-like gene expression pattern in male mice livers under calorie restriction. It was suggested that the pulsatile secretion of GH promotes male-biased gene expression while suppressing female-biased gene expression (45). Conversely, persistent GH treatment disrupts the intermittent nature of signal transducers and activators of transcription (Stat)/Janus kinase (Jak) activity, which leads to the promotion of female-biased gene expression while suppressing male-biased expression. As mentioned before, GHRd3 was shown to transduce GH signaling faster than the ancestral type of the protein (11). It is plausible that this stronger binding leads to a more persistent downstream signaling. This effect is likely compensated under an AL diet through the up-regulation of the expression of circadian rhythm genes as described above. However, this compensation mechanism may not be strong enough to overcome the combined dampening effect of Ghrd3 and calorie restriction. Therefore, we predict that Ghrd3 under calorie restriction dampens the GH signaling cycle, leading to the female-like gene expression in the male liver. These results also suggest that the smaller size observed in male d3/d3 mice under calorie restriction is at least partially the result of these transcriptome-level effects we document in the male liver.
Sex-specific effects of GHRd3 on serum lipidome
Given previous reports on the effect of GHR on lipid metabolism (37), as well as our own results that identify multiple lipid metabolism–related genes (e.g., multiple Mup and Ces genes), we investigated the downstream effects of Ghrd3 on the blood serum lipid composition using liquid chromatography–mass spectrometry (LC-MS) (see section S2.5). We chose to focus on serum lipids because we wanted to understand the global metabolic effect of Ghrd3 at the organismal level. Furthermore, serum lipids are routinely measured for diagnostic purposes, and we wanted to construct a comparable dataset for future studies. We documented a notable, opposite effect of Ghrd3 on fatty acid and triglyceride composition in the serum of males and females, independent of diet (Fig. 4E and table S5). The opposing effect is particularly prominent for fatty acids. Specifically, we found that all 15 fatty acid species that we analyzed were up-regulated in female Ghrd3 mice compared to their wt/wt counterparts, whereas most of these lipids were down-regulated in male Ghrd3 mice. The effects of Ghrd3 on serum lipids seem to be similar in mice that are fed with AL and CR diets and thus differ from the observations in the liver. This observation is consistent with the previously proposed notion that the effect of the GH receptor varies between different tissues (46).
DISCUSSION
Our results provide one of the very few human examples (47) where the effects of a common genetic variant are sex and environment dependent. In that regard, we suggest that GHRd3 has important ramifications for metabolic disorders, such as obesity and diabetes, but only for males within particular environmental contexts. For example, it was reported that GHRd3 has a preventative impact on type 2 diabetes. However, in the small number of diabetes patients who are homozygous for GHRd3, a significant increase in further metabolic complications was observed (26). This parallels our observation that a significant size difference was observed between wt/wt and d3/d3 mice only under calorie restriction for males. Similarly, we expect the reported effects on birth and placental size, time to menarche, and longevity to vary considerable for different dietary and endocrinological contexts. Following this thread, we expect that GHRd3 may have other important roles that would be visible only when specific sexes, environmental conditions, and developmental stages are investigated. Our study has characterized the effect of homozygous Ghrd3. Given that this gene codes for a protein that self-dimerizes for it to function properly, the functional impact of Ghrd3 in heterozygous individuals remains a fascinating question. Previous studies suggest that the heterozygous GHRd3 may contribute to different traits in an additive, recessive, or dominant manner. Our mouse model makes it possible to test all these different perspectives in future studies.
The sex-specific effects of GHRd3 raise additional evolutionary considerations, especially because the GH pathway is a major driver of sexual dimorphism in mammals (48). Moreover, multiple genes in this pathway were recently implicated in sexually antagonistic transmission distortions in humans (49). In this regard, GHRd3 provides an interesting case. The traditional understanding of the evolution of sex-specific effects of genetic variation first considers sexual conflict where the functional effect of the variant leads to increased fitness in one sex but decreased fitness in the other (50). This conflict leads to maintenance of the variation in the population through balancing selection until the conflict is resolved by additional genetic variation that moderates the effect of the variant in a sex-specific manner. GHRd3 does not fit this scenario. Instead, the sex-specific effect of GHRd3 is instantaneous because the GH pathway is already operating in a sex-specific manner. This observation raises questions about the extent of the evolutionary effects of genetic variation in pathways that already operate in a sex-specific manner.
The evolutionary history of GHRd3 is complex. Our model suggests that geography-specific adaptive pressures over time have shaped the allele frequency of GHRd3 across human evolutionary history, effectively maintaining this variation in human populations over 700 ka. In this regard, GHRd3 evolutionary history fits a very broad definition of balancing selection, i.e., maintenance of advantageous variation over long periods of time, overcoming fixation due to varying strengths of selective advantage through time, including drift and potentially negative selection. However, the signatures of GHRd3’s evolutionary history do not fit the expectations of more narrowly defined balancing selection models, such as heterozygous advantage or frequency-dependent selection (51). Instead, GHRd3 evidences the existence of loci under selective pressures that do not appear to conform with classical sweep or balancing selection models, and future work should investigate the prevalence of such loci across the genome (52–57).
Our insights into this genetic variation at the GHR locus bring forth additional questions concerning recent human evolution. The initial allele frequency increase in GHRd3 across evolutionary time coincides with unstable environments that mark recent hominin evolution (58). Given its enhanced effect under calorie restriction, it is plausible that GHRd3 provided a fitness advantage to early hominins, perhaps facilitating adaptation to new environments during Homo migrations out of Africa starting ~1 million years ago. Our finding that Ghrd3 is protective against the severe consequences of malnutrition in male children supports this hypothesis. The sex-specific nature of this adaptation may be better understood by considering times of nutritional stress and assuming that the increased size in males is a derived trait. During these periods, survival may be a stronger adaptive force than the fitness benefits of increased size, causing smaller sizes in males to be favored (28). Thus, GHRd3 may be favored under environmental stress because it increases survival although it reduces sexual dimorphism and thus reduces competitiveness for mate choice among males.
The marked reduction in GHRd3 allele frequencies coincides with the emergence of technologically advanced material culture, such as bone tools, fish hooks, and composite weapons, along with a concurrent population expansion, seemingly occurring independently in different parts of the world between 90 and 30 ka ago (59). Emerging technological innovations that may have allowed these populations to adapt to diverse environments (60) may also have acted as a buffer against the effects of fluctuating resource levels, thereby changing the selective pressures acting on the GHR locus. Note that the reduction in GHRd3 allele frequency coincides with major demographic and ecological changes including the Last Glacial Maximum (61). Thus, the combined effect of cultural and climate change could explain the rapid adaptive decrease in GHRd3 allele frequencies in all human populations, the most marked of which was observed in East Asia.
MATERIALS AND METHODS
We used multiple population genetics approaches to elucidating the evolutionary history of GHRd3. These include analysis of haplotypic variation, conservation across mammalian species, and calculation of empirical measures such as Tajima’s D (62) and XP-EHH (19), and 2DSFS (two-dimensional site frequency spectrum) (20). In parallel, we used simulation-based inferences, including a Bayesian approach to estimating the mode and tempo of potential natural selection (21) and an MC-EM (Monte Carlo expectation maximum) approach (CLUES) to estimating allele frequency trajectories (23). For all these population genetics inferences, we primarily used 1000 Genomes phase 3 (1) and Simons Genome Diversity datasets (2). We searched for phenotype associations in GWAS Atlas (http://geneatlas.roslin.ed.ac.uk/phewas/) and GeneATLAS (http://geneatlas.roslin.ed.ac.uk/) databases. We used the curated datasets from U.K. Biobank (www.nealelab.is/uk-biobank/) to search for sex-specific associations. We tested the association between GHRd3 and outcomes from severe malnutrition in a cohort of Malawi children with ESAM and NESAM (32) by directly genotyping GHRd3 in these individuals.
To understand the mechanisms through which GHRd3 may affect biological traits, we constructed a mouse model containing a deletion of the orthologous exon using CRISPR-Cas9. Using this model, we generated several mice cohorts (Fig. 3B), which we comparatively analyzed for variation in size, liver transcriptome, and serum lipidome. For RNA sequencing analysis, we used a standard Kallisto (63)/DeSeq (64) pipeline to comparatively analyze the transcript abundances. We used LC-MS to measure the abundance of individual lipid species in serum. Detailed descriptions of all these approaches are provided in the Supplementary Materials.
Acknowledgments
We are grateful to the Roswell Park Cancer Institute Gene Targeting and Transgenic Resource, especially to A. Stablewski, for help in establishing the Ghrd3 mouse line. We are grateful for the help of G. (PJ) Perry and A. Arner regarding the navigation of the U.K. Biobank dataset. We would like to thank E. Lucotte for discussion on potential sex-specific selection. We would like to thank Laura R. Parisi for her contributions to the lipidomics analysis. We would like to acknowledge K. Dean and V. Gellatly for help in genotyping. We thank our colleagues M. Xu-Friedman, D. J. Taylor, V. J. Lynch, M. S. Halfon, and V. A. Albert for constructive criticism throughout this study. Funding: This work was financially supported by the National Science Foundation (no. 1714867 to O.G.). Work in the MuLab was also supported by grants from the BrightFocus Foundation (G2016024) and the National Eye Institute (R01EY020545 and R01EY029705). The mouse work in the Jackson Laboratory was supported by an NIH grant (#P30AG038070). S.R. received funding during his first 2 years of research from the Collaborative Learning and Integrated Mentoring in the Biosciences (CLIMB) program. The work in the laboratory of C.L. is supported in part by the operational funds from the First Affiliated Hospital of Xi’an Jiaotong University. Human genetic studies of severe malnutrition were supported by the Doris Duke Charitable Foundation (#2013096) and the USDA, ARS cooperative agreement (58-3092-5-001). L.S. acknowledges support provided by the Sir Henry Wellcome fellowship (220457/Z/20/Z). Author contributions: O.G. designed, oversaw, and managed the study. O.G. contributed to analysis in each step. X.M. helped design the experiments and oversaw the study. M.S. conducted transcriptomics and population genetics analysis. S.R. conducted the majority of the mouse work and genotyping. A.J.P. and G.E.A.-G. conducted lipidomics analysis. S.N. and Y.S. conducted simulation-based selection analysis. L.S. ran the RELATE and CLUES analyses. F.W. helped in tissue collection in mice. L.R. and G.C. designed and oversaw mice experiments in the Jackson Laboratory. N.A.H., N.J.H., and N.C.L. contributed analysis of malnutrition outcome association study. C.L. and Q.Z. contributed to transcriptomics and genomic sequencing for looking for potential off-target effects from CRISPR-Cas9 experiments. O.G., S.R., and M.S. wrote the manuscript and prepared the figures. Competing interests: The authors declare that they have no competing interests. Data and materials availability: The RNA-seq fastq files are available through GEO (www.ncbi.nlm.nih.gov/geo/; accession no. GSE171716). The whole-genome sequencing data for three mice that we used to ensure that no off-target effects are present are available through SRA (BioSample accession nos. SAMN19643319, SAMN19643320, and SAMN19643321). All other data are available in the Supplementary Materials. The Ghrd3 mouse line can be provided through The Mutant Mouse Resource and Research Center at The Jackson Laboratory through its standard conditions of use. Requests should be submitted to O.G. (gokcumen@gmail.com) or L.R. (Laura.reinholdt@jax.org).
Supplementary Materials
This PDF file includes:
Supplementary Methods
Figs. S1 to S13
Tables S6 to S8, and S10
References
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S5, S9 and S11
REFERENCES AND NOTES
- 1.Sudmant P. H., Rausch T., Gardner E. J., Handsaker R. E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M. H.-Y., Konkel M. K., Malhotra A., Stütz A. M., Shi X., Casale F. P., Chen J., Hormozdiari F., Dayama G., Chen K., Malig M., Chaisson M. J. P., Walter K., Meiers S., Kashin S., Garrison E., Auton A., Lam H. Y. K., Mu X. J., Alkan C., Antaki D., Bae T., Cerveira E., Chines P., Chong Z., Clarke L., Dal E., Ding L., Emery S., Fan X., Gujral M., Kahveci F., Kidd J. M., Kong Y., Lameijer E.-W., McCarthy S., Flicek P., Gibbs R. A., Marth G., Mason C. E., Menelaou A., Muzny D. M., Nelson B. J., Noor A., Parrish N. F., Pendleton M., Quitadamo A., Raeder B., Schadt E. E., Romanovitch M., Schlattl A., Sebra R., Shabalin A. A., Untergasser A., Walker J. A., Wang M., Yu F., Zhang C., Zhang J., Zheng-Bradley X., Zhou W., Zichner T., Sebat J., Batzer M. A., McCarroll S. A.; 1000 Genomes Project Consortium, Mills R. E., Gerstein M. B., Bashir A., Stegle O., Devine S. E., Lee C., Eichler E. E., Korbel J. O., An integrated map of structural variation in 2,504 human genomes. Nature 526, 75–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mallick S., Li H., Lipson M., Mathieson I., Gymrek M., Racimo F., Zhao M., Chennagiri N., Nordenfelt S., Tandon A., Skoglund P., Lazaridis I., Sankararaman S., Fu Q., Rohland N., Renaud G., Erlich Y., Willems T., Gallo C., Spence J. P., Song Y. S., Poletti G., Balloux F., van Driem G., de Knijff P., Romero I. G., Jha A. R., Behar D. M., Bravi C. M., Capelli C., Hervig T., Moreno-Estrada A., Posukh O. L., Balanovska E., Balanovsky O., Karachanak-Yankova S., Sahakyan H., Toncheva D., Yepiskoposyan L., Tyler-Smith C., Xue Y., Abdullah M. S., Ruiz-Linares A., Beall C. M., Di Rienzo A., Jeong C., Starikovskaya E. B., Metspalu E., Parik J., Villems R., Henn B. M., Hodoglugil U., Mahley R., Sajantila A., Stamatoyannopoulos G., Wee J. T. S., Khusainova R., Khusnutdinova E., Litvinov S., Ayodo G., Comas D., Hammer M. F., Kivisild T., Klitz W., Winkler C. A., Labuda D., Bamshad M., Jorde L. B., Tishkoff S. A., Watkins W. S., Metspalu M., Dryomov S., Sukernik R., Singh L., Thangaraj K., Pääbo S., Kelso J., Patterson N., Reich D., The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538, 201–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin Y.-L., Pavlidis P., Karakoc E., Ajay J., Gokcumen O., The evolution and functional impact of human deletion variants shared with archaic hominin genomes. Mol. Biol. Evol. 32, 1008–1019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks A. J., Waters M. J., The growth hormone receptor: Mechanism of activation and clinical implications. Nat. Rev. Endocrinol. 6, 515–525 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Zoledziewska M., Sidore C., Chiang C. W. K., Sanna S., Mulas A., Steri M., Busonero F., Marcus J. H., Marongiu M., Maschio A., Del Vecchyo D. O., Floris M., Meloni A., Delitala A., Concas M. P., Murgia F., Biino G., Vaccargiu S., Nagaraja R., Lohmueller K. E.; UK10K consortium, Timpson N. J., Soranzo N., Tachmazidou I., Dedoussis G., Zeggini E.; Understanding Society Scientific Group, Uzzau S., Jones C., Lyons R., Angius A., Abecasis G. R., Novembre J., Schlessinger D., Cucca F., Height-reducing variants and selection for short stature in Sardinia. Nat. Genet. 47, 1352–1356 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amselem S., Duquesnoy P., Duriez B., Dastot F., Sobrier M.-L., Valleix S., Goossens M., Spectrum of growth hormone receptor mutations and associated haplotypes in Laron syndrome. Hum. Mol. Genet. 2, 355–359 (1993). [DOI] [PubMed] [Google Scholar]
- 7.Padidela R., Bryan S. M., Abu-Amero S., Hudson-Davies R. E., Achermann J. C., Moore G. E., Hindmarsh P. C., The growth hormone receptor gene deleted for exon three (GHRd3) polymorphism is associated with birth and placental weight. Clin. Endocrinol. 76, 236–240 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sørensen K., Aksglaede L., Petersen J. H., Leffers H., Juul A., The exon 3 deleted growth hormone receptor gene is associated with small birth size and early pubertal onset in healthy boys. J. Clin. Endocrinol. Metab. 95, 2819–2826 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Ben-Avraham D., Govindaraju D. R., Budagov T., Fradin D., Durda P., Liu B., Ott S., Gutman D., Sharvit L., Kaplan R., Bougnères P., Reiner A., Shuldiner A. R., Cohen P., Barzilai N., Atzmon G., The GH receptor exon 3 deletion is a marker of male-specific exceptional longevity associated with increased GH sensitivity and taller stature. Sci. Adv. 3, e1602025 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sørensen K., Aksglaede L., Munch-Andersen T., Aachmann-Andersen N. J., Leffers H., Helge J. W., Hilsted L., Juul A., Impact of the growth hormone receptor exon 3 deletion gene polymorphism on glucose metabolism, lipids, and insulin-like growth factor-I levels during puberty. J. Clin. Endocrinol. Metab. 94, 2966–2969 (2009). [DOI] [PubMed] [Google Scholar]
- 11.Dos Santos C., Essioux L., Teinturier C., Tauber M., Goffin V., Bougnères P., A common polymorphism of the growth hormone receptor is associated with increased responsiveness to growth hormone. Nat. Genet. 36, 720–724 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Dauber A., Meng Y., Audi L., Vedantam S., Weaver B., Carrascosa A., Albertsson-Wikland K., Ranke M. B., Jorge A. A. L., Cara J., Wajnrajch M. P., Lindberg A., Camacho-Hübner C., Hirschhorn J. N., A genome-wide pharmacogenetic study of growth hormone responsiveness. J. Clin. Endocrinol. Metab. 105, 3203–3214 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prüfer K., de Filippo C., Grote S., Mafessoni F., Korlević P., Hajdinjak M., Vernot B., Skov L., Hsieh P., Peyrégne S., Reher D., Hopfe C., Nagel S., Maricic T., Fu Q., Theunert C., Rogers R., Skoglund P., Chintalapati M., Dannemann M., Nelson B. J., Key F. M., Rudan P., Kućan Ž., Gušić I., Golovanova L. V., Doronichev V. B., Patterson N., Reich D., Eichler E. E., Slatkin M., Schierup M. H., Andrés A. M., Kelso J., Meyer M., Pääbo S., A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358, 655–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mafessoni F., Grote S., de Filippo C., Slon V., Kolobova K. A., Viola B., Markin S. V., Chintalapati M., Peyrégne S., Skov L., Skoglund P., Krivoshapkin A. I., Derevianko A. P., Meyer M., Kelso J., Peter B., Prüfer K., Pääbo S., A high-coverage Neandertal genome from Chagyrskaya Cave. Proc. Natl. Acad. Sci. U.S.A. 117, 15132–15136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pantel J., Machinis K., Sobrier M.-L., Duquesnoy P., Goossens M., Amselem S., Species-specific alternative splice mimicry at the growth hormone receptor locus revealed by the lineage of retroelements during primate evolution. J. Biol. Chem. 275, 18664–18669 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Gabriel S. B., Schaffner S. F., Nguyen H., Moore J. M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., Liu-Cordero S. N., Rotimi C., Adeyemo A., Cooper R., Ward R., Lander E. S., Daly M. J., Altshuler D., The structure of haplotype blocks in the human genome. Science 296, 2225–2229 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Leigh J. W., Bryant D., POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116 (2015). [Google Scholar]
- 18.Tajima F., Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sabeti P. C., Varilly P., Fry B., Lohmueller J., Hostetter E., Cotsapas C., Xie X., Byrne E. H., McCarroll S. A., Gaudet R., Schaffner S. F., Lander E. S.; International HapMap Consortium , Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujito N. T., Satta Y., Hayakawa T., Takahata N., A new inference method for detecting an ongoing selective sweep. Genes Genet. Syst. 93, 149–161 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Nakagome S., Hudson R. R., Di Rienzo A., Inferring the model and onset of natural selection under varying population size from the site frequency spectrum and haplotype structure. Proc. R. Soc. B 286, 20182541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Speidel L., Forest M., Shi S., Myers S. R., A method for genome-wide genealogy estimation for thousands of samples. Nat. Genet. 51, 1321–1329 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stern A. J., Wilton P. R., Nielsen R., An approximate full-likelihood method for inferring selection and allele frequency trajectories from DNA sequence data. PLOS Genet. 15, e1008384 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen R. B., Boas M., Nielsen J. E., Maroun L. L., Jørgensen A., Larsen T., Main K. M., Juul A., A common deletion in the growth hormone receptor gene (d3-GHR) in the offspring is related to maternal placental GH levels during pregnancy. Growth Horm. IGF Res. 55, 101360 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Schreiner F., Stutte S., Bartmann P., Gohlke B., Woelfle J., Association of the growth hormone receptor d3-variant and catch-up growth of preterm infants with birth weight of less than 1500 grams. J. Clin. Endocrinol. Metab. 92, 4489–4493 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Strawbridge R. J., Kärvestedt L., Li C., Efendic S., Ostenson C. G., Gu H. F., Brismar K., GHR exon 3 polymorphism: Association with type 2 diabetes mellitus and metabolic disorder. Growth Horm. IGF Res. 17, 392–398 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Canela-Xandri O., Rawlik K., Tenesa A., An atlas of genetic associations in UK Biobank. Nat. Genet. 50, 1593–1599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metcalfe N. B., Monaghan P., Compensation for a bad start: Grow now, pay later? Trends Ecol. Evol. 16, 254–260 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Millar D. S., Lewis M. D., Horan M., Newsway V., Rees D. A., Easter T. E., Pepe G., Rickards O., Norin M., Scanlon M. F., Krawczak M., Cooper D. N., Growth hormone (GH1) gene variation and the growth hormone receptor (GHR) exon 3 deletion polymorphism in a West-African population. Mol. Cell. Endocrinol. 296, 18–25 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Antón S. C., Potts R., Aiello L. C., Evolution of early Homo: An integrated biological perspective. Science 345, 1236828 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Czorlich Y., Aykanat T., Erkinaro J., Orell P., Primmer C. R., Rapid sex-specific evolution of age at maturity is shaped by genetic architecture in Atlantic salmon. Nat. Ecol. Evol. 2, 1800–1807 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulze K. V., Swaminathan S., Howell S., Jajoo A., Lie N. C., Brown O., Sadat R., Hall N., Zhao L., Marshall K., May T., Reid M. E., Taylor-Bryan C., Wang X., Belmont J. W., Guan Y., Manary M. J., Trehan I., McKenzie C. A., Hanchard N. A., Edematous severe acute malnutrition is characterized by hypomethylation of DNA. Nat. Commun. 10, 5791 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forrester T. E., Badaloo A. V., Boyne M. S., Osmond C., Thompson D., Green C., Taylor-Bryan C., Barnett A., Soares-Wynter S., Hanson M. A., Beedle A. S., Gluckman P. D., Prenatal factors contribute to the emergence of kwashiorkor or marasmus in severe undernutrition: Evidence for the predictive adaptation model. PLOS ONE 7, e35907 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergström A., Stringer C., Hajdinjak M., Scerri E. M. L., Skoglund P., Origins of modern human ancestry. Nature 590, 229–237 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Smith T. M., Austin C., Green D. R., Joannes-Boyau R., Bailey S., Dumitriu D., Fallon S., Grün R., James H. F., Moncel M.-H., Williams I. S., Wood R., Arora M., Wintertime stress, nursing, and lead exposure in Neanderthal children. Sci. Adv. 4, eaau9483 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.P. J. Danneman, M. A. Suckow, C. Brayton, The Laboratory Mouse (CRC Press, 2012). [Google Scholar]
- 37.Fan Y., Menon R. K., Cohen P., Hwang D., Clemens T., DiGirolamo D. J., Kopchick J. J., Le Roith D., Trucco M., Sperling M. A., Liver-specific deletion of the growth hormone receptor reveals essential role of growth hormone signaling in hepatic lipid metabolism. J. Biol. Chem. 284, 19937–19944 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang X., Schadt E. E., Wang S., Wang H., Arnold A. P., Ingram-Drake L., Drake T. A., Lusis A. J., Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 16, 995–1004 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sassin J. F., Parker D. C., Mace J. W., Gotlin R. W., Johnson L. C., Rossman L. G., Human growth hormone release: Relation to slow-wave sleep and sleep-walking cycles. Science 165, 513–515 (1969). [DOI] [PubMed] [Google Scholar]
- 40.Jansson J. O., Edén S., Isaksson O., Sexual dimorphism in the control of growth hormone secretion. Endocr. Rev. 6, 128–150 (1985). [DOI] [PubMed] [Google Scholar]
- 41.Vakili H., Jin Y., Cattini P. A., Evidence for a circadian effect on the reduction of human growth hormone gene expression in response to excess caloric intake. J. Biol. Chem. 291, 13823–13833 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaudhari A., Gupta R., Patel S., Velingkaar N., Kondratov R., Cryptochromes regulate IGF-1 production and signaling through control of JAK2-dependent STAT5B phosphorylation. Mol. Biol. Cell 28, 834–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Steyn F. J., Huang L., Ngo S. T., Leong J. W., Tan H. Y., Xie T. Y., Parlow A. F., Veldhuis J. D., Waters M. J., Chen C., Development of a method for the determination of pulsatile growth hormone secretion in mice. Endocrinology 152, 3165–3171 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Holloway M. G., Miles G. D., Dombkowski A. A., Waxman D. J., Liver-specific hepatocyte nuclear factor-4α deficiency: Greater impact on gene expression in male than in female mouse liver. Mol. Endocrinol. 22, 1274–1286 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lau-Corona D., Suvorov A., Waxman D. J., Feminization of male mouse liver by persistent growth hormone stimulation: Activation of sex-biased transcriptional networks and dynamic changes in chromatin states. Mol. Cell. Biol. 37, e00301-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duran-Ortiz S., Noboa V., Kopchick J. J., Tissue-specific disruption of the growth hormone receptor (GHR) in mice: An update. Growth Horm. IGF Res. 51, 1–5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ober C., Loisel D. A., Gilad Y., Sex-specific genetic architecture of human disease. Nat. Rev. Genet. 9, 911–922 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lichanska A. M., Waters M. J., How growth hormone controls growth, obesity and sexual dimorphism. Trends Genet. 24, 41–47 (2008). [DOI] [PubMed] [Google Scholar]
- 49.E. A. Lucotte, C. Albiñana, R. Laurent, C. Bhérer, Genome of the Netherland Consortium, T. Bataillon, B. Toupance, Detection of sexually antagonistic transmission distortions in trio datasets. bioRxiv 2020.09.11.293191 [Preprint]. 11 September 2020. 10.1101/2020.09.11.293191. [DOI] [PMC free article] [PubMed]
- 50.Mank J. E., Population genetics of sexual conflict in the genomic era. Nat. Rev. Genet. 18, 721–730 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Charlesworth D., Balancing selection and its effects on sequences in nearby genome regions. PLOS Genet. 2, e64 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andrés A. M., Dennis M. Y., Kretzschmar W. W., Cannons J. L., Lee-Lin S.-Q., Hurle B.; NISC Comparative Sequencing Program, Schwartzberg P. L., Williamson S. H., Bustamante C. D., Nielsen R., Clark A. G., Green E. D., Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLOS Genet. 6, e1001157 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pajic P., Lin Y.-L., Xu D., Gokcumen O., The psoriasis-associated deletion of late cornified envelope genes LCE3B and LCE3C has been maintained under balancing selection since Human Denisovan divergence. BMC Evol. Biol. 16, 265 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hollox E. J., Armour J. A. L., Directional and balancing selection in human beta-defensins. BMC Evol. Biol. 8, 113 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun C., Huo D., Southard C., Nemesure B., Hennis A., Cristina Leske M., Wu S.-Y., Witonsky D. B., Olopade O. I., Di Rienzo A., A signature of balancing selection in the region upstream to the human UGT2B4 gene and implications for breast cancer risk. Hum. Genet. 130, 767–775 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pasvol G., Weatherall D. J., Wilson R. J., Cellular mechanism for the protective effect of haemoglobin S against P. falciparum malaria. Nature 274, 701–703 (1978). [DOI] [PubMed] [Google Scholar]
- 57.Xu S., Nielsen R., Huerta-Sanchez E., The history and evolution of the Denisovan-EPAS1 haplotype in Tibetans. Proc. Natl. Acad. Sci. U.S.A. e2020803118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Potts R., Dommain R., Moerman J. W., Behrensmeyer A. K., Deino A. L., Riedl S., Beverly E. J., Brown E. T., Deocampo D., Kinyanjui R., Lupien R., Owen R. B., Rabideaux N., Russell J. M., Stockhecke M., deMenocal P., Faith J. T., Garcin Y., Noren A., Scott J. J., Western D., Bright J., Clark J. B., Cohen A. S., Keller C. B., King J., Levin N. E., Shannon K. B., Muiruri V., Renaut R. W., Rucina S. M., Uno K., Increased ecological resource variability during a critical transition in hominin evolution. Sci. Adv. 6, eabc8975 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Powell A., Shennan S., Thomas M. G., Late Pleistocene demography and the appearance of modern human behavior. Science 324, 1298–1301 (2009). [DOI] [PubMed] [Google Scholar]
- 60.Roberts P., Stewart B. A., Defining the ‘generalist specialist’ niche for Pleistocene Homo sapiens. Nat. Hum. Behav. 2, 542–550 (2018). [DOI] [PubMed] [Google Scholar]
- 61.Clark P. U., Dyke A. S., Shakun J. D., Carlson A. E., Clark J., Wohlfarth B., Mitrovica J. X., Hostetler S. W., McCabe A. M., The Last Glacial Maximum. Science 325, 710–714 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Tajima F., Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 135, 599–607 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wegmann M. G., Thankamony A., Roche E., Hoey H., Kirk J., Shaikh G., Ivarsson S.-A., Söder O., Dunger D. B., Juul A., Jensen R. B., The exon3-deleted growth hormone receptor gene polymorphism (d3-GHR) is associated with insulin and spontaneous growth in short SGA children (NESGAS). Growth Horm. IGF Res. 35, 45–51 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., Handsaker R. E., Lunter G., Marth G. T., Sherry S. T., McVean G., Durbin R.; 1000 Genomes Project Analysis Group , The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.The Chimpanzee Sequencing Consortium , Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437, 69–87 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P. H., de Filippo C., Li H., Mallick S., Dannemann M., Fu Q., Kircher M., Kuhlwilm M., Lachmann M., Meyer M., Ongyerth M., Siebauer M., Theunert C., Tandon A., Moorjani P., Pickrell J., Mullikin J. C., Vohr S. H., Green R. E., Hellmann I., Johnson P. L. F., Blanche H., Cann H., Kitzman J. O., Shendure J., Eichler E. E., Lein E. S., Bakken T. E., Golovanova L. V., Doronichev V. B., Shunkov M. V., Derevianko A. P., Viola B., Slatkin M., Reich D., Kelso J., Pääbo S., The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reich D., Green R. E., Kircher M., Krause J., Patterson N., Durand E. Y., Viola B., Briggs A. W., Stenzel U., Johnson P. L. F., Maricic T., Good J. M., Marques-Bonet T., Alkan C., Fu Q., Mallick S., Li H., Meyer M., Eichler E. E., Stoneking M., Richards M., Talamo S., Shunkov M. V., Derevianko A. P., Hublin J.-J., Kelso J., Slatkin M., Pääbo S., Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468, 1053–1060 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu D., Jaber Y., Pavlidis P., Gokcumen O., VCFtoTree: A user-friendly tool to construct locus-specific alignments and phylogenies from thousands of anthropologically relevant genome sequences. BMC Bioinformatics 18, 426 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bandelt H. J., Forster P., Röhl A., Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 16, 37–48 (1999). [DOI] [PubMed] [Google Scholar]
- 72.South A., rworldmap: A new R package for mapping global data. R J. 3, 35–43 (2011). [Google Scholar]
- 73.Kuhn R. M., Haussler D., Kent W. J., The UCSC genome browser and associated tools. Brief. Bioinform. 14, 144–161 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kumar S., Stecher G., Li M., Knyaz C., Tamura K., MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pollard K. S., Hubisz M. J., Rosenbloom K. R., Siepel A., Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pybus M., Dall’Olio G. M., Luisi P., Uzkudun M., Carreño-Torres A., Pavlidis P., Laayouni H., Bertranpetit J., Engelken J., 1000 Genomes Selection Browser 1.0: A genome browser dedicated to signatures of natural selection in modern humans. Nucleic Acids Res. 42, D903–D909 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.H. Wickham, Ggplot2: Elegant Graphics for Data Analysis (Springer Publishing Company Incorporated, ed. 2, 2009). [Google Scholar]
- 79.Satta Y., Zheng W., Nishiyama K. V., Iwasaki R. L., Hayakawa T., Fujito N. T., Takahata N., Two-dimensional site frequency spectrum for detecting, classifying and dating incomplete selective sweeps. Genes Genet. Syst. 94, 283–300 (2020). [DOI] [PubMed] [Google Scholar]
- 80.Hudson R. R., Generating samples under a Wright–Fisher neutral model of genetic variation. Bioinformatics 18, 337–338 (2002). [DOI] [PubMed] [Google Scholar]
- 81.Osada N., Nakagome S., Mano S., Kameoka Y., Takahashi I., Terao K., Finding the factors of reduced genetic diversity on X chromosomes of Macaca fascicularis: Male-driven evolution, demography, and natural selection. Genetics 195, 1027–1035 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakagome S., Fukumizu K., Mano S., Kernel approximate Bayesian computation in population genetic inferences. Stat. Appl. Genet. Mol. Biol. 12, 667–678 (2013). [DOI] [PubMed] [Google Scholar]
- 83.L. Speidel, L. Cassidy, R. W. Davies, G. Hellenthal, Inferring population histories for ancient genomes using genome-wide genealogies. bioRxiv 2021.02.17.431573 [Preprint]. 17 February 2021. 10.1101/2021.02.17.431573. [DOI] [PMC free article] [PubMed]
- 84.Mashal R. D., Koontz J., Sklar J., Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nat. Genet. 9, 177–183 (1995). [DOI] [PubMed] [Google Scholar]
- 85.Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S. L., TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li H., Durbin R., Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ewels P., Magnusson M., Lundin S., Käller M., MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zerbino D. R., Achuthan P., Akanni W., Amode M. R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Girón C. G., Gil L., Gordon L., Haggerty L., Haskell E., Hourlier T., Izuogu O. G., Janacek S. H., Juettemann T., To J. K., Laird M. R., Lavidas I., Liu Z., Loveland J. E., Maurel T., McLaren W., Moore B., Mudge J., Murphy D. N., Newman V., Nuhn M., Ogeh D., Ong C. K., Parker A., Patricio M., Riat H. S., Schuilenburg H., Sheppard D., Sparrow H., Taylor K., Thormann A., Vullo A., Walts B., Zadissa A., Frankish A., Hunt S. E., Kostadima M., Langridge N., Martin F. J., Muffato M., Perry E., Ruffier M., Staines D. M., Trevanion S. J., Aken B. L., Cunningham F., Yates A., Flicek P., Ensembl 2018. Nucleic Acids Res. 46, D754–D761 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W., BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440 (2005). [DOI] [PubMed] [Google Scholar]
- 94.S. Ge, D. Jung, ShinyGO: A graphical enrichment tool for animals and plants. bioRxiv 315150 [Preprint]. 4 May 2018. 10.1101/315150. [DOI]
- 95.del Solar V., Lizardo D. Y., Li N., Hurst J. J., Brais C. J., Atilla-Gokcumen G. E., Differential regulation of specific sphingolipids in colon cancer cells during staurosporine-induced apoptosis. Chem. Biol. 22, 1662–1670 (2015). [DOI] [PubMed] [Google Scholar]
- 96.Saghatelian A., Trauger S. A., Want E. J., Hawkins E. G., Siuzdak G., Cravatt B. F., Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry 43, 14332–14339 (2004). [DOI] [PubMed] [Google Scholar]
- 97.Tuck M. K., Chan D. W., Chia D., Godwin A. K., Grizzle W. E., Krueger K. E., Rom W., Sanda M., Sorbara L., Stass S., Wang W., Brenner D. E., Standard operating procedures for serum and plasma collection: Early detection research network consensus statement standard operating procedure integration working group. J. Proteome Res. 8, 113–117 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Figs. S1 to S13
Tables S6 to S8, and S10
References
Tables S1 to S5, S9 and S11