DNA helicases DHX36 and FANCJ constitute a previously unknown pathway that resolves G-quadruplex structures during DNA replication.
Abstract
G-quadruplex (or G4) structures form in guanine-rich DNA sequences and threaten genome stability when not properly resolved. G4 unwinding occurs during S phase via an unknown mechanism. Using Xenopus egg extracts, we define a three-step G4 unwinding mechanism that acts during DNA replication. First, the replicative helicase composed of Cdc45, MCM2-7 and GINS (CMG) stalls at a leading strand G4 structure. Second, the DEAH-box helicase 36 (DHX36) mediates bypass of the CMG past the intact G4 structure, allowing approach of the leading strand to the G4. Third, G4 structure unwinding by the Fanconi anemia complementation group J helicase (FANCJ) enables DNA polymerase to synthesize past the G4 motif. A G4 on the lagging strand template does not stall CMG but still requires DNA replication for unwinding. DHX36 and FANCJ have partially redundant roles, conferring pathway robustness. This previously unknown genome maintenance pathway promotes faithful G4 replication, thereby avoiding genome instability.
INTRODUCTION
Guanine-rich nucleic acid sequences can adopt four-stranded structures, termed G-quadruplexes (G4s) (1). G4s comprise three or more stacked G-quartets, planar structures formed by four guanines connected via Hoogsteen hydrogen bonding and stabilized by a monovalent cation (2). Vertebrate genomes contain thousands of sequences that can form G4 structures with a wide variety of topologies, arising from parallel and antiparallel G-strand direction and intervening loops (3). Moreover, these motifs preferentially localize to evolutionarily conserved regulatory loci (4). While G4s are involved in a broad range of biological processes such as transcriptional regulation, telomere maintenance, and epigenetic regulation (5–7), these structures have also been linked to genome instability (8).
The eukaryotic genome encodes at least 10 DNA helicases that show G4 unwinding activity in vitro (9). Furthermore, genetic studies have provided important clues that link several helicases to G4 unwinding in vivo. In yeast, the 5′-3′ DNA helicase Petite integration frequency 1 (PIF1) is essential for genome stability at G4s (10). PIF1 is highly conserved in mammals; however, its function appears to predominate in the mitochondria (11). In human cells, the 5′-3′ DNA helicase Fanconi anemia complementation group J (FANCJ) plays a critical role in preventing large deletions near G4 motifs (12), although it seems to be dispensable for this role in mice (13). Biallelic mutations in FANCJ causes Fanconi anemia (FA), a human cancer predisposition syndrome (14–16). While FANCJ acts in the “FA pathway” that repairs DNA interstrand cross-links (ICLs) (17), this role appears to be independent of its poorly characterized function in G4 unwinding (18, 19). Furthermore, a model has been proposed in which FANCJ, together with Werner syndrome ATP-dependent helicase (WRN) and Bloom syndrome protein (BLM), two other G4 resolving helicases that unwind DNA with opposite polarity, promotes DNA replication past G4 structures (5). Another 5′-3′ DNA helicase, Regulator of telomere elongation helicase 1 (RTEL1), specifically suppresses telomeric G4 instability (20). Last, DEAH-box helicase 36 (DHX36) is a 3′-5′ helicase that exhibits an exceptional affinity for DNA G4 structures in vitro (21), but its biological function has been studied mostly in the context of RNA G4 processing (22). This helicase is essential for normal embryogenesis in mice (23), yet its role in DNA G4 unwinding in vivo remains unknown. Notably, depletion of certain helicases, or addition of G4 stabilizing ligands that inhibit G4 unwinding, induces DNA double-strand breaks (DSBs) and can cause chromosomal aberrations (12, 24, 25). Together, this indicates that G4 structures need to be actively unwound by DNA helicases to maintain genome integrity.
Several lines of evidence suggest that G4 structures are predominantly resolved during DNA replication. First, G4 formation is enhanced in S phase but quickly reduced upon progression to G2 phase (26, 27). Second, G4 stabilizing ligands arrest cells in G2 phase with 4N DNA content (28). Third, DNA damage induced by G4 ligands is replication dependent (29). Fourth, several G4 unwinding helicases, such as PIF1, FANCJ, and RTEL1 associate with the replisome, prevent DNA polymerase from stalling at G4s and preserve replication fork speed through G4s (18, 30–32). Fifth, FANCJ-deficient Caenorhabditis elegans shows a G4-specific deletion signature that implies replication fork blockage as a cause of these deletions (8, 33, 34). These insights suggest that G4 unwinding is coupled to DNA replication to prevent replisome arrest at the G4 structure. However, direct evidence that a G4 structure acts as a roadblock for the replisome is hitherto missing, and moreover, virtually nothing is known about the mechanism of G4 unfolding during DNA replication.
Here, we used Xenopus laevis egg extracts to recapitulate unwinding and replication of defined G4 structures. We identify DHX36 and FANCJ as essential helicases involved in this process. Replication past a G4 structure on the leading strand involves stalling of the replicative helicase Cdc45, MCM2-7 and GINS (CMG), followed by DHX36-mediated CMG bypass of the intact G4 structure, allowing the polymerase to approach the G4, and, lastly, the unwinding of the G4 structure by FANCJ enabling G4 motif replication. Although lagging strand G4 resolution does not involve CMG bypass, it still requires active DNA replication and both DHX36 and FANCJ. Our data reveal a robust mechanism that promotes faithful replication of abundant G4 structures and thereby avoids DSBs and chromosomal instability.
RESULTS
FANCJ and DHX36 are required for efficient G4 structure unwinding
Vertebrates contain at least 10 conserved DNA helicases that are able to unwind G4 structures in vitro [FANCJ, BLM, DEAD/H-box helicase 11 (DDX11), DHX9, DHX36, DNA replication helicase/nuclease 2 (DNA2), PIF1, RTEL1, Xeroderma pigmentosum group D (XPD), and WRN] (9), but whether and how these proteins act under physiological conditions are currently unknown. We previously showed that FANCJ promotes the unwinding of G4 structures during replication of single-stranded DNA (ssDNA) templates in Xenopus egg extract (18). However, FANCJ depletion only affected the replication of a subset of these structures, indicating that other G4-unwinding helicases are active in the extract. To identify these helicases, we analyzed the proteins recruited to G4-containing plasmids by mass spectrometry (MS). To this end, we replicated ssDNA plasmids containing a defined parallel G4 or a control sequence (pBS-G4 and pBS-CON; fig. S1A) in a high-speed supernatant (HSS) of total Xenopus egg lysate. Plasmids were pulled down using beads conjugated with LacI, at times when the polymerase is stalled at the G4 (1.5 and 4.5 min) or when the plasmid is fully replicated (50 min) (Fig. 1A and fig. S1B). MS detected eight G4-unwinding helicases on replicating plasmids (Fig. 1B). Some of them were detected on both pBS-G4 and pBS-CON, while others, including FANCJ and DHX36, were specifically enriched on pBS-G4. Moreover, addition of the G4 stabilizing ligand PhenDC3 further increased the enrichment of several helicases. The most strongly enriched helicase was DHX36, which peaked at the earliest time point. To examine the role of DHX36 in DNA G4 unwinding, we immunodepleted it from extract and analyzed replication products of G4 templates by denaturing electrophoresis (Fig. 1C and fig. S1, C and D). Depletion of DHX36 initially enhanced polymerase stalling compared to the mock-depleted extract, but all G4 substrate molecules were eventually replicated (Fig. 1C). In contrast, FANCJ depletion showed extended polymerase stalling on a subset of the template molecules as shown previously (Fig. 1C and fig. S1, C and D) (18). This suggests that DHX36 and FANCJ have different roles in G4 replication. When we depleted both DHX36 and FANCJ, extensive replication stalling at the G4 was observed with almost no replication past the structure for at least an hour (Fig. 1C and fig. S1, C and D). Addition of recombinant wild-type DHX36 or FANCJ (fig. S1, C to F) restored replication to the respective single depletion levels (Fig. 1C). Depletion of DHX36, FANCJ, or both did not affect replication of pBS-CON (fig. S1, G and H). These data indicate that both DHX36 and FANCJ are required for efficient unwinding of G4 structures and that they act partially redundantly.
Fig. 1. G4 unwinding during replication of ssDNA templates requires DHX36 and FANCJ.
(A) Schematic representation of pBS-G4 replication in HSS. Replication is initiated from a primer annealed ~760–nucleotide (nt) upstream of a G4. See also fig. S1 (A and B). (B) pBS-G4 or pBS-CON were replicated in HSS and isolated at various times by LacI pull-down (64, 66). Proteins bound to the plasmids were identified by MS analysis. Relative abundance of proteins is represented by a heatmap showing the mean of the Z scores from four biological replicates. (C) pBS-G4 was replicated in the extracts described in fig. S1C in the presence of 32P-α–2′-deoxycytidine 5′-triphosphate (dCTP). Replication products were extracted, separated by denaturing polyacrylamide gel electrophoresis (PAGE), and visualized by autoradiography. Nascent strands stalled at the G4 sequence (~700 nt) and fully replicated molecules (~3000 nt) are indicated. The G4 stalled and bypassed products of the indicated time points were quantified, and the percentage of stalling versus bypassed products was plotted in a bar graph with SDs (bottom; n = 4).
G4 structure unwinding and replication on double-stranded DNA templates
Replication of ssDNA substrates in Xenopus egg extract does not involve the complete replisome because unwinding of the DNA duplex is not required. To investigate the role of DHX36 and FANCJ at bona fide replication forks, we set out to develop a system that would recapitulate G4 unwinding and replication in the context of a double-stranded DNA (dsDNA) template. Replication of dsDNA in extract involves incubation in HSS to allow the assembly of prereplication complexes, followed by addition of a concentrated nucleoplasmic extract (NPE) to promote initiation and a single round of DNA replication. We constructed dsDNA plasmids that contain a G4 motif or a control sequence similar to the ssDNA templates (pdsG4BOT and pdsCON; fig. S2, A and B). To monitor replication intermediates, we separated replication products on a denaturing agarose gel after linearization with Hinc II. Replication of pdsG4BOT and pdsCON quickly yielded full-length molecules, and no intermediates accumulated (fig. S2C). This suggests that a stable G4 structure does not form in this context, which is consistent with duplex DNA hydrogen bonds being more stable than G4 hydrogen bonds (35).
We then prepared dsDNA plasmids containing a short noncomplementary region in which the G4 structure can be preformed on the top or bottom strand (pG4TOP and pG4BOT), along with non-G4 control plasmids (pCON and pPolyT; fig. S2A). Replication of control plasmids yielded full-length products without accumulation of any intermediates, indicating that the noncomplementary region does not hinder the replication machinery (Fig. 2A). However, replication of pG4BOT and pG4TOP resulted in a transient accumulation of ~2.0- and ~3.6-kb products that were quickly converted to full-length products (Fig. 2A). The accumulation of these fragments was greatly enhanced by PhenDC3 (Fig. 2B) and suppressed when the G4 structure was not preformed by omitting potassium (fig. S2D). These data suggest that replication temporarily halts at the site of the G4 structure and resumes upon G4 unwinding. Notably, the 2.0-kb fragment is less defined and prone to degradation in pG4TOP, similar to the 3.6-kb fragment in pG4BOT, suggesting that these are lagging strand products with free 5′ ends (fig. S2E). This indicates that replication stalling occurs on the G4 structure–containing strand, which was confirmed by digestion with Cla I, which only cleaves the replicated top strand (fig. S2, A, F, and G). In contrast to PhenDC3, the G4 ligand 5,10,15,20-Tetrakis-(N-methyl-4-pyridyl)porphine (TMPyP4) only induced minor replication stalling at the G4 structure (fig. S2H). This is consistent with a previous report that showed that PhenDC3 affects viral replication in human cells much more than TMPyP4 (36).
Fig. 2. DHX36 and FANCJ are essential for G4 unwinding on physiological replication forks.
(A and B) pPolyT, pG4MUT, pG4BOT and pG4TOP were replicated without (A) or with PhenDC3 (B) in the presence of 32P-α-dCTP. Replication intermediates were linearized with Hinc II and analyzed by denaturing agarose gel electrophoresis and autoradiography. Defined nascent stalling products are indicated with an arrowhead, and less defined stalling products are indicated with a bar. (C) Model for pG4BOT replication. Replication of the G4 strand involves polymerase stalling, while the non-G4 strand is replicated without stalling. (D) Mock-depleted, DHX36-depleted, FANCJ-depleted, and DHX36-FANCJ–depleted NPEs were analyzed by Western blot with DHX36 and FANCJ antibodies alongside a dilution series of undepleted extract. (E) pG4BOT was replicated in the indicated extracts in the presence of α-32P-dCTP. Replication intermediates were digested with Hinc II (lower autoradiogram) or Hinc II and Cla I (upper autoradiogram) and analyzed as in (A). Since Cla I specifically digests the replicated top strand, the 5.6-kb product results from the replicated bottom (G4) strand (top). See also fig. S2F. (F) pCON and pG4BOT plasmids were replicated, at various time points (t = 0 is immediately after NPE addition). Plasmids were isolated by LacI pull-down. Bound proteins were analyzed, along with a 2% input sample, by Western blot with the indicated antibodies. Where indicated, HSS was supplemented with Geminin to inhibit DNA replication.
Even in the presence of PhenDC3, when replication stalling was prominent, extension products still readily appeared (Fig. 2B). This suggests that replication of the non-G4 strand precedes G4 structure unwinding. In agreement with this, strand-specific digestion by Cla I showed that replication of non-G4 strand was not affected in the presence of PhenDC3 (fig. S2, F and G). While replication of the G4-containing strand was strongly delayed, at later times, it did reach almost equal intensity compared to the non-G4 strand, indicating that stalling at the G4 is not permanent (fig. S2G). Furthermore, two-dimensional gel electrophoresis demonstrated that the sister chromatids were efficiently resolved upon replication fork stalling at the G4 (fig. S2I). Therefore, we conclude that replication of G4-containing dsDNA templates results in transient replication stalling on the G4 strand, while replication of the opposite strand is not interrupted (Fig. 2C).
DHX36 and FANCJ promote G4 unwinding during DNA replication
We next explored the roles of DHX36 and FANCJ in this system. Similar to what we observed for ssDNA substrates, double depletion of DHX36 and FANCJ greatly increased replication stalling at the G4 structure and severely compromised extension of the G4 strand, while each single depletion had only a minor effect (Fig. 2, D and E). Depletion of DHX36 and FANCJ did not codeplete WRN, BLM, RTEL1, or DNA2; other helicases we found at G4 plasmids during replication; or ssDNA binding protein Replication protein A (RPA) that was reported to unwind G4 structures in vitro (Fig. 1B and fig. S2J) (37). This strongly indicates that replication of the G4 sequence requires G4 unwinding by DHX36 and FANCJ. The extreme G4 replication defect upon DHX36-FANCJ double depletion was also observed in the presence of PhenDC3 and TMPyP4 (fig. S2K), indicating that these proteins are also involved in the resolution of G4 structures stabilized by these ligands.
G4 unwinding could occur before or during DNA replication. To examine this, we pulled down replicating plasmids and monitored DHX36 binding. DHX36 was detected specifically on G4-containing plasmids before replication initiation and was displaced over time, indicative of G4 unwinding during replication (Fig. 2F). When replication initiation was abrogated by addition of Geminin, DHX36 persisted on pG4BOT (Fig. 2F). This indicates that active DNA replication is required for G4 structure resolution. Early accumulation of DHX36 at the G4 was also observed in our MS data in which FANCJ peaked later (Fig. 1B), suggesting that DHX36 and FANCJ act at different stages of G4 unwinding.
G4 structures are resolved during leading and lagging strand synthesis
Since replication forks in our system arrive at the G4 structure either from the left or from the right and fork convergence occurs rapidly on small plasmids, we cannot distinguish whether the G4 is resolved during leading or lagging strand synthesis. To allow this distinction, we made use of an array of lac operator (lacO) repeats situated downstream of the G4 structure (fig. S2A). Upon preincubation with the Lac repressor (LacI), arrival of the leftward fork is efficiently blocked (fig. S3, A and B), and only the rightward fork encounters the G4 on the leading (in pG4BOT) or lagging strand (in pG4TOP) (fig. S3, C and D, cartoons). This also more closely resembles the in vivo situation, since larger interorigin distance will make fork convergence less likely (38). We replicated pG4TOP and pG4BOT in the presence of LacI (and, under these conditions, will refer to them as pG4Lag and pG4Lead, respectively). Analysis of the digested replication products on a sequencing gel showed that the leading and lagging strands were rapidly extended past the G4Lead and G4Lag, respectively (fig. S3, C and D). The extension kinetics were comparable to that of pCON and a convergent fork situation. Therefore, both leading and lagging strand G4s are efficiently resolved, and dual fork collision is not required for G4 structure unwinding.
Leading strand G4 replication is a three-step process
To examine how a G4 structure on the leading strand is resolved, we further analyzed the replication products of pG4Lead. Two faint nascent strand clusters appeared within 15 min and declined to undetectable levels by 30 min (fig. S3C, blue and green brackets). G4 stabilization by PhenDC3 enhanced these clusters that are formed by initial stalling of the leading strand 13– to 26–nucleotide (nt) upstream of G4Lead (“−13 to −26” products), followed by a second stalling event at 1 to 3 nt from the G4Lead (“−1 to −3” products) (Fig. 3, A and B). Quantification of the −13 to −26 and − 1 to −3 clusters indicated that these represent sequential events (Fig. 3C). DNA lesions such as ICLs and DNA-protein cross-links (DPCs) arrest leading strand synthesis roughly 20 to 30 nt from the damage site due to steric hindrance by CMG helicase that translocates along the leading strand template ahead of the polymerase (39, 40). Therefore, the −13 to −26 products likely reflect the CMG footprint upon encountering the G4. Consistent with this, when the G4 was present on the lagging strand template, the −13 to −26 products were hardly detected, while the −1 to −3 products still accumulated (Fig. 3B, lanes 22 to 28). Moreover, the −13 to −26 products reappeared when LacI was omitted and when both lagging and leading strand synthesis occurred (Fig. 3B, lanes 15 to 21).
Fig. 3. Multistep replication bypass of a G4 structure.
(A) Schematic of nascent leading and lagging strand from pG4Lead or pG4Lag replication. Cla I specifically digests the top strand extension products. Brown hexamer, CMG; Green sphere, LacI; T, sequencing primer. (B) pG4Lead, pG4Lag, and pCON were replicated in the presence of PhenDC3 (with or without LacI), digested with Afl III, Bam HI, and Cla I (top gel) or Afl III and Bam HI (bottom gel), separated on denaturing polyacrylamide gel, and visualized by autoradiography. A sequencing ladder was derived from extension of primer T on pG4TOP, and polymerase chain reaction (PCR) products (M; see Materials and Methods) serve as size markers. The −13 to −26 products and the −1 to −3 products are indicated with brackets and bars, respectively. (C) The indicated products from (B) (lanes 8 to 14) were quantified, and the relative intensity compared to the peak signal was plotted. (D) Scheme of the ChIP assay. (E) pG4Lead and pG4Lag were replicated and analyzed by minichromosome maintenance protein 7 (MCM7) ChIP–quantitative PCR (qPCR) with primers for the G4 locus (schematic; left). Where indicated, Geminin was added to inhibit replication. Sonication produces ~600–base pair (bp) fragments; therefore, some fragments will contain both the G4 and lacO loci (420 bp apart). This likely generates the background signal on pG4Lag in which MCM7 is stalled at the LacI-lacO array (dashed line). See Fig. 5A for ChIP signal at the LacO locus for the same experiment.
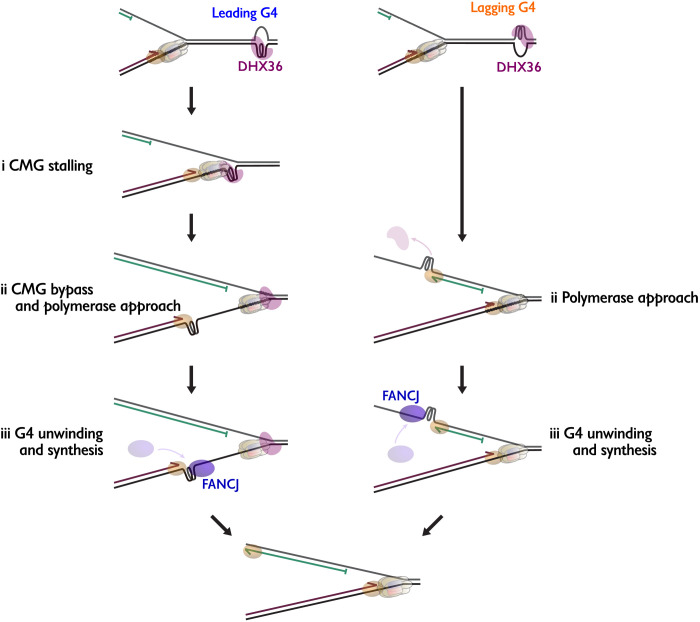
To confirm that CMG stalls at a leading strand G4, we monitored the accumulation of minichromosome maintenance protein 7 (MCM7), a CMG subunit, by plasmid-based chromatin immunoprecipitation (ChIP) (Fig. 3D). During replication of pG4Lag, background levels of MCM7 were detected at the G4 site, most likely due to arrival of CMG at the lacO locus (Fig. 3E). However, during the replication of pG4Lead, MCM7 accumulated at the G4 site, showing a strong peak at 20 min (Fig. 3E) that coincided with the presence of the stalling products (fig. S3C). This indicates that CMG stalling leads to the accumulation of these leading strand products. Together, these data suggest that replication past a G4 structure on the leading strand is a three-step process (Fig. 3B and fig. S3E). First, CMG stalls at a G4 structure resulting in the accumulation of leading strand products 13 to 26 nt from the G4 (CMG stalling products). Subsequently, CMG vacates the G4 site, which enables the polymerase to approach the G4 where it stalls again at 1 to 3 nt from the G4 (polymerase stalling products). Last, the G4 structure is unwound to allow replication of the G4 sequence. Because a G4 on the lagging strand is not encountered by CMG, its replication only involves the latter two steps, polymerase stalling at the G4, followed by G4 unwinding and replication (fig. S3E).
DHX36 and FANCJ collaborate to unwind a leading strand G4
We next examined the roles of FANCJ and DHX36 in these steps. During pG4Lead replication, depletion of DHX36 resulted in a transient accumulation of the CMG stalling products, whereas depletion of FANCJ enhanced the polymerase stalling products (Fig. 4A, arrowheads). Double depletion of DHX36 and FANCJ strongly enhanced both stalling clusters and severely impaired the extension past the G4 structure (Fig. 4A, lanes 16 to 21). Notably, although similar polymerase stalling bands are observed on DHX36-FANCJ double depletion compared to incubation with PhenDC3, the most intense band is 1 nt closer to the G4 sequence, which is consistent with our previous observation on ssDNA templates (fig. S4A, arrowheads) (18). The replication deficiency was restored to single depletion levels by addition of wild-type DHX36 and FANCJ proteins, but not their adenosine triphosphatase (ATPase) mutants (figs. S1, E and F, and S4, B to E) (41, 42). Therefore, the major role of DHX36 during leading strand G4 replication is likely to promote polymerase approach by facilitating CMG clearance from the G4 site, while FANCJ mostly acts in G4 unwinding, although they also act in part redundantly in both processes. In agreement with this, depletion of DHX36 had no effect on G4Lag replication that does not involve CMG stalling (fig. S4F). FANCJ depletion caused polymerase stalling on the lagging strand on pG4Lag that was further enhanced by DHX36-FANCJ double depletion (fig. S4, F and G), indicating that DHX36 and FANCJ also act redundantly in G4 unwinding on the lagging strand. In contrast, depletion of the other G4 unwinding helicases BLM, WRN, and RTEL1 did not affect G4 replication (fig. S5), suggesting that it predominantly relies on DHX36 and FANCJ in our system.
Fig. 4. DHX36 and FANCJ cooperate in unwinding a leading strand G4 structure.
(A) pGLead was replicated in the indicated extracts, digested with Afl III, Bam HI, and Cla I (top gel) or with Afl III and Bam HI (bottom gel), separated on denaturing polyacrylamide gel, and visualized by autoradiography. The CMG stalling products and the polymerase stalling products are indicated with a bracket and bars, respectively. The schematic representation of the generated fragments is shown on the left. Brown hexamer, CMG; green sphere, LacI. (B) pG4Lead and pCON were replicated and analyzed by ChIP-qPCR with DHX36 (i) and FANCJ (ii) antibodies using a primer pair for the G4 locus (schematic representation in left panel). Where indicated, Geminin was added to inhibit replication initiation. To compare the timing of accumulation for both proteins, DHX36 and FANCJ ChIP data in the absence of Geminin were plotted together in one graph (iii).
To gain insight into how FANCJ and DHX36 collaborate to replicate a G4 on the leading strand template, we monitored their recruitment to pG4Lead by ChIP. Consistent with our plasmid pull-down assay, DHX36 was present at pG4Lead before replication initiation and mostly disappeared by 30 min (Fig. 4B, i). In contrast, while some FANCJ was present before replication, it further accumulated at pG4Lead simultaneously with the disappearance of DHX36, and FANCJ only vacated the G4 plasmid at later times (Fig. 4B, ii and iii). The initial FANCJ binding before replication is independent of the G4 structure and likely mediated by the ssDNA of the non-G4 strand, while the accumulation at later times during replication is dependent on the G4 structure (Fig. 4B, ii, and fig. S4H). Inhibition of replication initiation by Geminin caused persistent DHX36 binding at pG4Lead and prevented FANCJ from accumulating (Fig. 4B). Similar results were obtained with pG4Lag (fig. S4I). These data indicate that DHX36 and FANCJ are sequentially bound to both leading and lagging strand G4s during replication. Since DHX36 binds to the G4 structure before FANCJ and its depletion induced CMG stalling at the G4, we envision that it facilitates CMG clearance to allow polymerase approach to the G4 site.
DHX36 promotes CMG bypass by generating ssDNA downstream of the G4
The CMG helicase could vacate the G4 site by two distinct mechanisms, unloading or bypass (43–45). To distinguish between these possibilities, we replicated pG4Lag and pG4Lead and analyzed the accumulation of MCM7 at the region flanking the lacO array by ChIP (Fig. 5A). If CMG is unloaded at the leading strand G4, then we would expect no MCM7 to be detected at the LacI-bound lacO locus for pG4Lead, while it would accumulate at the locus in pG4Lag. As anticipated, MCM7 was readily detected at the lacO locus on pG4Lag in the presence of LacI (Fig. 5A). This accumulation was due to the LacI-bound lacO array as it was abolished in the absence of LacI. On pG4Lead, we observed MCM7 accumulation to similar levels but with a delay of about 20 min (Fig. 5A). This delay is consistent with the initial accumulation of MCM7 at the leading strand G4, followed by the loss of MCM7 at the G4 that coincides with accumulation at the lacO locus (Figs. 3E and 5A). In addition, double depletion of DHX36 and FANCJ enhanced MCM7 accumulation at the G4Lead site and diminished it at the lacO locus (fig. S6A). Therefore, we conclude that DHX36 facilitates CMG bypass of the G4Lead, rather than extracting it from the DNA. Consistent with this, inhibition of the CMG unloader p97 (43, 46) did not enhance the CMG footprint at the leading strand G4 (fig. S6, B to D).
Fig. 5. DHX36 promotes CMG bypass by generating ssDNA downstream of the G4 structure.
(A) pG4Lead and pG4Lag were replicated and analyzed by ChIP-qPCR with the MCM7 antibody using a primer pair for the lacO locus (295- to 388-bp downstream from the G4) or the G4 locus. Where indicated, LacI was omitted, or Geminin was added to inhibit replication initiation. The MCM7 levels on the G4 locus (from Fig. 3D) were added to the graph for comparison. If CMG bypasses the G4, then it will be detected on the lacO locus as depicted in the left panel. (B) pG4UP and pG4DOWN were replicated, digested with Afl III and Bam HI, separated on denaturing polyacrylamide gel, and visualized by autoradiography. The CMG stalling products and the polymerase stalling products are indicated with brackets and bars, respectively.
Since a G4 recognition mutant of DHX36 (DHX36Y53A) did not promote CMG bypass (fig. S4, B and C), G4 binding of DHX36 is critical for efficient bypass. In addition, DHX36 is reported to translocate and unwind duplex DNA with a 3′-5′ directionality (47) and could therefore generate ssDNA downstream of the G4 structure. Consistently, our purified Xenopus laevis (xl) DHX36 unwound duplex DNA downstream of the G4 structure in an ATPase-dependent manner (fig. S6E). Generation of ssDNA by helicase-mediated unwinding has recently been shown to promote CMG bypass of a DPC (45). We therefore examined whether downstream ssDNA is sufficient for the CMG to bypass the G4. To this end, we inserted a noncomplementary 25-nt polythimine (PolyT) sequence downstream of the leading strand G4 (pG4DOWN; Fig. 5B). As a control, we also prepared a plasmid in which the PolyT stretch was inserted upstream of the G4 (pG4UP). While replication of pG4UP resulted in the accumulation of CMG stalling products upon DHX36 and DHX36-FANCJ depletion, these were largely absent when pG4DOWN was replicated (Fig. 5B). This indicates that ssDNA downstream of the G4 structure is sufficient for CMG bypass. Consistent with this, CMG stalling at a G4 in a DHX36-FANCJ–depleted extract was rescued by fork convergence (fig. S6F). However, while G4 bypass was facilitated in pG4DOWN, DNA synthesis past the G4 was still potently blocked, indicating that the G4 structure was stable even in the presence of downstream ssDNA (Fig. 5B and fig. S6G). Therefore, arrival of the second fork or downstream ssDNA likely facilitate CMG bypass of the intact G4 structure (fig. S6H). In support of a model in which DHX36 does not promote G4 unwinding but generates ssDNA beyond the G4, we found that G4 unwinding by DHX36 requires a long stretch of 3′ ssDNA [see also (47)], while duplex unwinding does not show this absolute requirement (fig. S6, E and I).
CMG bypass is essential for FANCJ-mediated G4 unwinding on the leading strand
Given the role of DHX36 in CMG bypass and our observation that FANCJ replaces DHX36 at the G4 structure during G4 resolution, we hypothesized that CMG bypass might be a prerequisite for this helicase switch. To address this, we first examined the localization of the ATPase mutant of DHX36 to the G4 during replication by ChIP. Consistent with a defect in translocation, we found this DHX36E327A mutant to be retained at the G4 site in contrast to the wildtype protein that readily vacates the site during replication (Fig. 6, A and B, i). The DHX36E327A mutant also impaired CMG bypass as seen by the enhanced levels of MCM7 at G4Lead locus and the reduced MCM7 levels at the lacO locus (Fig. 6B, ii and iv). Moreover, the DHX36E327A mutant prevented accumulation of FANCJ at the G4Lead locus during replication (Fig. 6B, iii). Consistent with a lack of FANCJ accumulation, the mutant enhanced the CMG stalling signature and extended it to at least 2.5 hours (Fig. 6C), indicating that the G4 structure is hardly unwound. CMG stalling at the G4 was more extensive in the presence of the DHX36 mutant than in the absence of DHX36 [Fig. 6, B (ii) and C], indicating that persistence of DHX36 at the G4 site acts in a dominant negative fashion. Conversely, when CMG bypass was facilitated by allowing replication fork convergence, FANCJ accumulation was restored (fig. S7A), and the replication defect of G4Lead in the presence of the DHX36 mutant was rescued (Fig. 6D). Together, these data indicate that CMG bypass is required for G4 unwinding most likely because it promotes FANCJ accumulation at the leading strand G4. Consistent with this sequence of events, G4 unwinding does not seem to be required for CMG bypass since an ATPase inactive mutant of FANCJ persisted on the G4 and induced polymerase stalling but did not affect DHX36 dissociation (fig. S7, B and C).
Fig. 6. CMG bypass is a prerequisite for unwinding of a leading strand G4.
(A) DHX36-depleted NPE supplemented with buffer, wild-type (WT) DHX36, or DHX36E327A (EA) were analyzed by Western blot with the DHX36 antibody alongside a dilution series of undepleted extract. (B) pG4Lead was replicated in the indicated extracts and analyzed by ChIP-qPCR with DHX36 (i), MCM7 (ii and iv), or FANCJ (iii) antibodies using a primer pair for the G4 locus (i to iii) or for the lacO locus (iv). Schematic representation of qPCR loci on top. Brown hexamer, CMG; green sphere, LacI. (C) pG4Lead was replicated in the indicated extracts, digested with Afl III, Bam HI, and Cla I (top gel) or Afl III and Bam HI (bottom gel), separated on a denaturing polyacrylamide gel, and visualized by autoradiography. The CMG stalling and polymerase stalling products are indicated with brackets. (D) pG4UP (G4 on bottom strand and T25 upstream of G4) was replicated in the indicated extracts and analyzed as in (C) but only digested with Afl III and Bam HI. Extension product of the nascent leading strand past the G4 is shown in the top panel. Where indicated, LacI was released by addition of isopropyl-β-d-1-thiogalactopyranoside (IPTG) at 15 min (lanes 25 to 48). The CMG stalling and polymerase stalling products are indicated with brackets. To monitor replication fork block by LacI, extracted samples were digested with Hinc II, separated on native agarose gel, and visualized by autoradiography (bottom gels).
DHX36 and FANCJ have partially redundant roles
Since the CMG stalling signature is more extensive in the DHX36-FANCJ double-depleted extract compared to the DHX36-depleted extract (Fig. 4A), it seems that FANCJ can partially replace the function of DHX36 in CMG bypass. Consistent with this, FANCJ levels at the G4 region are enhanced roughly twofold before replication initiation in a DHX36-depleted extract, suggesting that FANCJ can readily bind the vacated G4 (Fig. 6B, iii, and fig. S7A, ii). Since FANCJ is a 5′-3′ DNA helicase, it could translocate along the non-G4 strand to generate ssDNA downstream of the G4 structure to facilitate CMG bypass. To test this possibility, we introduced a stable DPC on the non-G4 strand of pG4Lead to prevent FANCJ translocation (pG4Lead-DPC; fig. S7B) (40). During pG4Lead-DPC replication, the leading strand mostly extended past the G4 as observed for a control plasmid that did not contain DPC (pG4Lead-CON; fig. S7, C and D), indicating that the DPC does not affect the G4Lead replication. Notably, the CMG stalling footprint in the DHX36-depleted extract was enhanced by the presence of the DPC to the same extend as observed in a DHX36-FANCJ double-depleted extract (fig. S7D). These data indicate that, in absence of DHX36, FANCJ facilitates CMG bypass by translocating on the non-G4 strand. DHX36 can also partially replace FANCJ’s function in G4 unwinding (Figs. 1C and 4A). Consistent with this, DHX36 appears to persist longer on both leading and lagging strand G4s in the absence of FANCJ (fig. S7E). Collectively, DHX36 and FANCJ have partially redundant roles during both CMG bypass and G4 unwinding processes.
DISCUSSION
This work provides direct evidence that a G4 structure efficiently blocks progression of the vertebrate replisome and establishes the first comprehensive mechanism of how G4 structures are resolved during DNA replication. On the leading strand template, this requires three steps including CMG bypass of the intact G4 structure, while on the lagging strand, only the final two steps are required, polymerase stalling at the G4 and unwinding of the G4 structure (Fig. 7). Duplex unwinding ahead of the G4 structure by DHX36 facilitates CMG bypass. This mechanism ensures that G4 unwinding and DNA synthesis occur in close temporal proximity and could avoid unscheduled ssDNA generation that could induce DSBs. Partial redundancy between DHX36 and FANCJ makes this mechanism further robust (fig. S8A).
Fig. 7. Model for replication-coupled G4 structure unwinding.
Models for replication of a leading strand G4 (left) and a lagging strand G4 (right). See Discussion for details. Brown hexamer, CMG; magenta oval, DHX36; blue oval, FANCJ; brown oval, the leading strand DNA polymerase; green oval, the lagging strand DNA polymerase.
CMG stalling at a leading strand G4
During leading strand synthesis, the CMG helicase is the first replisome component that encounters the G4 structure. This causes CMG to stall and induces leading strand stalling at 13- to 26-nt upstream of the G4 structure (Fig. 3B and fig. S3C). Within this CMG footprint, there are roughly two clusters, one at −21 to −26, and another at −13 to −17 nt from the ICL (Fig. 3B). In the absence of PhenDC3, the first cluster is relatively faint and quickly converted to the second cluster (fig. S3C), suggesting that CMG stalls at the G4 in two steps. Since the G4 structure is too large to fit into the CMG channel (48, 49), the initial cluster likely reflects stalling of the CMG at the G4 structure (fig. S8B, i). Consistently, a similar footprint was observed at an ICL and a DPC (39, 40). However, the second cluster of CMG-arrested products can only arise if the G4 can be accommodated within the N-tier channel of the CMG that first encounters the structure (fig. S8B, ii). We envision at least two mechanisms that could support this. First, the N-tier channel changes its conformation upon stalling to create space for the G4 structure. In support of this model, purified MCM complex was shown to accommodate a ~5-kDa DPC adduct (50). Alternatively, CMG or another DNA helicase might remodel the G4 structure to allow it to fit within the channel. Our observation that the G4 ligand PhenDC3 inhibits the transition to the second CMG stalling cluster (Fig. 3B) favors the latter possibility. The channel of the CMG C-terminal tier contains several protruding loops (49), which could prevent further passage of the G4 and accumulation of the second stalling cluster. Similar stalling products (−11 to −24 products) are also observed during DPC repair, only when the DPC is proteolyzed to a peptide (40). This suggests that the “jammed state” might ubiquitously occur, when an impediment fits the N-tier but not the C-tier of the CMG.
Mechanism of CMG bypass
After initial stalling, CMG readily bypasses a G4 structure on the leading strand template through the action of DHX36, but after that, the polymerase stalls at the G4 (as seen under FANCJ depletion condition; Fig. 4A). This raises two possibilities; CMG bypass proceeds without unfolding of the structure, or the G4 structure is unwound but quickly refolds after bypass. We favor the first option because biophysical experiments showed that parallel G4 structures require ~200 s to fold (51). If the G4 structure is not unwound, then CMG bypass most likely involves opening of the CMG ring. Such a model is consistent with recent evidence that the MCM2-7 ring opens during replication initiation (52) and possibly also during DPC repair (45). Notably, the requirement of DHX36 for CMG bypass is abolished by a converging fork or when ssDNA is placed downstream of the G4 (Figs. 5B and 6D and fig. S6F). These observations indicate that the primary function of DHX36 is to generate ssDNA downstream of the structure, as shown for RTEL1 in DPC repair (45), and that the ssDNA triggers CMG bypass via ring opening by a currently unknown mechanism (Fig. 7, i and ii). Given the directionality and requirements of their ATPase activities (fig. S4, B to E), DHX36 likely translocates along the leading strand template, and when it is absent, FANCJ translocates along lagging strand template for ssDNA generation (fig. S8A). This model is consistent with our observation that before replication initiation, DHX36 directly binds the G4 structure, while FANCJ binds the opposite displaced strand (Figs. 2F and 4B and fig. S4H).
The principle of CMG bypass appears similar between G4 replication and DPC repair, but the mechanism for ssDNA generation past the obstruction differs in important aspects. During DPC repair, CMG bypass requires the 5′-3′ DNA helicase RTEL1 that accumulates at stalled replication forks through proliferating cell nuclear antigen (PCNA) binding (30, 45). Unlike DPC repair, DHX36 and FANCJ helicases accumulate at a G4-forming region before replication (Fig. 4B and fig. S6A). DHX36 appears to directly bind the G4 via its extremely high affinity for G4s (KD of <10 pM) (21), because a G4 recognition mutant DHX36Y53A does not promote CMG bypass (fig. S4B). In contrast, FANCJ initially binds to the displaced ssDNA. Given the direct interaction and cellular colocalization between FANCJ and RPA, the ssDNA binding protein RPA might facilitate FANCJ accumulation (53). While both helicases can promote ssDNA generation past the G4, DHX36 predominates in this process, since depletion of DHX36 enhances the CMG stalling signature, while depletion of FANCJ does not (Figs. 4A and 6C). Moreover, addition of the DHX36 ATPase mutant to a DHX36-depleted extract further enhanced CMG stalling probably because its persistence at the G4 site inhibits FANCJ from unwinding DNA past the G4. However, while we assume that DHX36 normally generates the ssDNA past the G4, DHX36 is likely inactive before replisome arrival at the G4 structure, as it persists on the G4 in the absence of DNA replication (Fig. 4B). We therefore speculate that the replication fork plays a direct role in activating the DHX36 helicase activity through a currently unknown mechanism. A backup helicase on the non-G4 strand, such as FANCJ, would be advantageous, for example, when DHX36 fails to recognize the structure.
It should be noted that, although delayed, the CMG is still able to bypass the G4 when both DHX36 and FANCJ are depleted (Fig. 4A) or the G4 is stabilized (Fig. 3B), resulting in leading strand extension to the G4. We therefore assume the existence of yet another bypass mechanism.
CMG bypass is required for G4 unwinding
While we observed some DHX36/FANCJ-independent CMG bypass, G4 unwinding and DNA synthesis beyond the G4 strictly depend on these two helicases (Fig. 4A and fig. S5). Furthermore, CMG bypass is essential for G4 unwinding, based on the following evidence. First, the ATPase mutant DHX36E327A that blocks CMG bypass also abrogates G4-dependent FANCJ recruitment and DNA synthesis past the G4. This defect is rescued by a converging fork, suggesting that CMG bypass is a prerequisite for G4 unwinding (Fig. 6D and fig. S7A). Second, under a single fork condition, not only CMG bypass but also DNA synthesis past a stabilized G4 is slower compared to a converging fork condition (Fig. 3B), strongly indicating that G4 unwinding depends on CMG bypass. However, why is CMG bypass required for FANCJ accumulation? On a lagging strand G4 template, which does not involve CMG bypass, FANCJ replaces DHX36 with similar kinetics compared to a leading strand G4 (fig. S4I, iii), indicating that CMG bypass per se does not promote the helicase switch. An event that occurs during replication of both strands but requires CMG bypass on the leading strand is the collision of the polymerase with the G4 structure (fig. S8). Stalling of the DNA polymerase could lower the affinity for DHX36 that requires a free 3′ end to efficiently bind the G4, while FANCJ does not (54, 55). Therefore, the DNA polymerase could facilitate FANCJ accumulation by preventing reaccumulation of DHX36 that binds to G4s with ~100-fold higher affinity than FANCJ (21, 54). This model for the helicase switch is consistent with our observation that DNA replication is also required for this switch on a lagging strand G4 (fig. S4I). Future biochemical work is required to establish further details of the helicase switch mechanism.
G4 structure unwinding in the presence of G4 stabilizing ligands
Both in the presence and absence of PhenDC3 or TMPyP4, double depletion of DHX36 and FANCJ markedly inhibited G4 replication (Fig. 2E and fig. S2K), indicating that these proteins are also involved in the resolution of stabilized G4 structures. However, in agreement with our previous study (18), we observed slight differences in the polymerase stalling products in the presence and absence of PhenDC3. The PhenDC3-treated samples showed additional stalling at −1, −5, and −6 from the G4, and the most intense stalling band was 1 nt closer to the G4 sequence (fig. S4, A and G). These small differences in polymerase stalling position in the presence of PhenDC3 could be caused by minor structural rearrangements in the G4 structure that allow the polymerase to approach the structure more efficiently or could point to additional differences in the G4 resolution mechanism.
G4 unwinding during DNA replication in vivo
G4 structures have been suggested to affect DNA replication in two ways. First, they can regulate replication origins (56). However, on our G4-containing DNA templates, we do not observe preferred origin activation at the G4 structure (fig. S2I). Second, G4 structures are thought to hinder the DNA replication machinery. Our work establishes an important role for DHX36 and FANCJ in the progression of DNA replication past these structures. Such a role was implicated for FANCJ previously by experiments in C. elegans (8, 33, 34) that showed increased deletions, likely caused by replication blockage, at G4 sites in the absence of deletion of guanine rich DNA, the FANCJ ortholog in the worm. Notably, the deletion start sites found in this study fit very well with our polymerase stalling position, suggesting that polymerase stalling, but not CMG stalling, at G4s could be a source of replication-associated breaks in vivo. However, under unperturbed conditions, the presence of a large number of G4 structures in S phase does not directly lead to an accumulation of DNA damage (25, 26), suggesting that efficient G4 unwinding mechanisms such as described here are active. Moreover, even in dog-1–deficient C. elegans, deletions at G4 sites are rare (33, 34), indicating that redundant helicases such as DHX36 could act in an evolutionary conserved pathway of replication-coupled G4 unwinding. In line with this, DSB formation in FANCJ-depleted human cells is severely exacerbated by G4 ligand treatment that inhibits G4 unwinding by other helicases including DHX36 (19). Nevertheless, on the basis of the large variety of G4 conformations that can be formed and the preference of DHX36 to interact with parallel G4 structures, we envision other helicases, besides DHX36 and FANCJ, to be involved in the replication of other G4 structures. Consistently, a recent study suggested that the replication machinery can detect and unwind a G4 structure ahead of the replication fork through a Timeless and DDX11-mediated mechanism in a specific avian locus (57). Moreover, G4 structure resolution is important not only during DNA replication but also during other cellular processes (58). This implies DNA replication–independent G4 unwinding mechanism(s) might exist. Consistent with this, we observed a slow release of DHX36 from the G4 templates, indicating G4 unwinding, even in the absence of DNA replication (Fig. 4B).
Exploiting G4-induced DNA damage
Impaired G4 unwinding leads to excessive DSBs. Homologous recombination is important for repair of the G4-induced DSB, as G4 ligands induce synthetic lethality in BRCA1/2-deficient cancer cells (59). We showed that the mechanism of replication-dependent G4 unwinding does not change in the presence of G4 ligands, indicating that this pathway can be a therapeutic target for homologous recombination-deficient cancers. Similar to BRCA1 and BRCA2, FANCJ is the third most common cancer susceptibility gene in ovarian cancer (60) and also plays an important role in homologous recombination. Therefore, DHX36 could be a potential target for therapy of FANCJ-deficient cancers.
MATERIALS AND METHODS
Xenopus laevis
Egg extracts were prepared using eggs from adult X. laevis female frogs (aged >2 years; Nasco, catalog no. LM00535), and demembranated sperm chromatin was prepared from the testes of adult X. laevis male frogs (purchased from the European Xenopus Resource Centre). All animal procedures and experiments were performed in accordance with national animal welfare laws and were reviewed by the Animal Ethics Committee of the Royal Netherlands Academy of Arts and Sciences (KNAW). All animal experiments were conducted under a project license granted by the Central Committee Animal Experimentation (CCD) of the Dutch government and approved by the Hubrecht Institute Animal Welfare Body (IvD), with project license number AVD80100201711044. Sample sizes were chosen on the basis of previous experience, and randomization and blinding are not relevant to this study.
Insect cell line
Sf9 cells (Thermo Fisher Scientific, catalog no. B82501) were cultured at 27°C for overexpression of xlFANCJ. Insect cells were cultured in Sf-900 III SFM medium (Thermo Fisher Scientific, catalog no. 12658019).
Bacterial strains
Escherichia coli LOBSTR (DE3) strain (Kerafast, catalog no. EC1001) was used for overexpression of xlDHX36. E. coli BL21 (DE3) strain (New England Biolabs, catalog no. C2527) was used for overexpression of xlGeminin and the N-terminal residues (1 to 170) of xlDHX36. E. coli T7 Express strain (New England Biolabs, catalog no. C2566) was used for overexpression of LacI and HpaII Methyltransferase (M.HpaII). E. coli XL1-Blue strain (Agilent Technologies, catalog no. 200249) was used for ssDNA plasmid preparation. E. coli cells were cultured in LB medium.
Preparation of plasmid substrates
ssDNA plasmids containing a site-specific G4 motif (5′-GGGAGGGTGGGAGGG-3′) or a control motif (5′-GGGACCCTGGGAGGG-3′) were prepared by viral replication using a helper phage M13K07 (New England Biolabs, catalog no. N0315S). E. coli XL1-Blue cells were transformed with pBluescript SK(−) plasmid containing the motif and cultured in 50 ml of LB medium supplemented with ampicillin (100 μg/ml final concentration) at 37°C until the optical density at 600 nm reaches ~0.05. Fifty microliters of M13K07 (1 × 1011 plaque-forming units/ml) phage was then added, and the cells were cultured for further 18 hours in the presence of kanamycin (70 μg/ml final concentration). The medium was collected after centrifugation (4000g) for 10 min and mixed with 0.2 volumes of polyethylene glycol (PEG) solution (2.5 M NaCl and 20% PEG-8000) for an hour at 4°C. After centrifugation (12,000g) for 10 min at 23°C, the pellet was resuspended in 1.6 μl of TE [10 mM tris-HCl (pH 8) and 1 mM EDTA] and centrifuged by 17,000g for a minute at 23°C to remove any remaining cells. The supernatant was collected after which 0.2 volumes of PEG solution was added and the solution was centrifuged by 17,000g for 10 min at 23°C. The pellet was resuspended with 300 μl of TE and purified by phenol/chloroform extraction. Purified DNA was then ethanol-precipitated with 20 μg of glycogen, resuspended with 25 μl of TE, and stored at −80°C.
The double-stranded plasmids containing a DNA ICL derived from cisplatin (pICLPt) was prepared as described (61). dsDNA plasmids containing a G4 motif were prepared by a similar method. First, short DNA duplexes were generated by heating a pair of oligonucleotides (table S1) to 80°C for 5 min and slowly cooling down to room temperature in ~2 hours in annealing buffer [10 mM tris-HCl (pH 8) and 25 mM KCl]. To generate pdsG4BOT and pdsCON, oligonucleotides A and B or C and D were used, respectively. For pPolyT, pCON, pG4BOT, pG4DOWN, and pG4UP, oligonucleotides E and F, G, H, I, or J were used, respectively. For pG4TOP and pG4DOUBLE, oligonucleotides K and either L or H were used, respectively. The resulting duplexes were ligated into the Bbs I sites of the pSVRlacO vector (62). After ligation, the closed circular plasmid was purified using a cesium chloride gradient ultracentrifugation, followed by butanol extraction, concentration, and buffer exchange to TE with an Amicon Ultra-4 centrifuge filter unit (30-kDa cutoff; Merck Millipore, catalog no. UFC803024).
To make a pG4Lead-DPC, pKS was created by replacing the Bbs I fragment of pSVRlacO with a duplex, consisting of oligonucleotides M and N, containing two Nt.BbvCI nicking sites. pKS was nicked with Nt.BbvCI and purified using Wizard SV Gel and PCR (polymerase chain reaction) Clean-Up System (Promega, catalog no. A9281). The resulting short ssDNA fragment was then replaced with a 5-fluoro-2′-deoxycytidine (FdC)–modified oligonucleotide (400 nM) by heating the mixture (containing 50 nM nicked pKS) to 80°C for 5 min and cooled down to room temperature in ~2 hours. To avoid reannealing of the original fragment, an excess (2.4 μM) of oligonucleotide O (complementary to the original fragment) was added. The annealed fragment was ligated with T4 DNA ligase (66.7 U/μl at a final concentration; New England Biolabs, catalog no. M0202M) by incubating in reaction buffer [50 mM tris-HCl (pH 7.5), 10 mM MgCl2, 4 mM adenosine 5′-triphosphate (ATP), and 10 mM dithiothreitol (DTT)] for 18 hours at 16°C. The modified plasmid was purified and concentrated to ~150 ng/μl with Wizard SV Gel and PCR Clean-Up System using elution buffer [10 mM tris-HCl (pH 7.5), 10 mM KCl, and 0.1 mM EDTA]. To induce a covalent cross-link between FdC and the catalytic cysteine of DNA methyltransferase M.HpaII (fig. S7C) (63), the purified plasmid was mixed with His6-tagged M.HpaII, which was methylated on lysine residues to prevent proteolysis in Xenopus egg extract (a gift from J. C. Walter) (64), for 18 hours at 37°C in cross-link buffer [50 mM potassium acetate, 20 mM tris-acetate, 10 mM magnesium acetate, bovine serum albumin (BSA; 0.1 mg/ml), 100 μM S-adenosylmethionine, 30 mM KCl, 9% glycerol, and 0.3 mM DTT (pH 7.9)].
Xenopus egg extracts and DNA replication
X. laevis female frogs were used as a source of eggs. Egg production, preparation of Xenopus HSS, demembranated sperm chromatin, NPE, and DNA replication were performed as previously described (65).
For replication of ssDNA templates, ssDNA plasmids (20 ng/μl final concentration) were incubated with primer A (1.0 μM final concentration) in annealing buffer for 5 min at 80°C and cooled down to room temperature in ~2 hours to allow the G4 structure to form and the primer to anneal. Where indicated, the G4 stabilizing compound PhenDC3 (5 μM final concentration) and/or TMPyP4 (20 μM final concentration) was added to the mixture at 50°C and incubated for 30 min after which the mixture was further cooled down to room temperature. To prevent 5′-3′ DNA resection in egg extract, the primer was synthesized with the 12 most 5′ nucleotides connected by phosphorothioate bonds. To initiate replication, the primed template (2.7 ng/μl final concentration) was added to HSS supplemented with nocodazole (3 ng/μl), 18.9 mM phosphocreatine, 1.9 mM ATP, and creatine phosphokinase (4.7 ng/μl) at room temperature. For nascent strand labeling, HSS was supplemented with 32P-α–2′-deoxycytidine 5′-triphosphate (dCTP). For extraction of replicated samples, aliquots of the reaction (5 μl) were stopped with 45 μl of stop solution II [50 mM tris (pH 7.5), 0.5% SDS, and 10 mM EDTA (pH 8.0)] at the indicated time points. Samples were then treated with ribonuclease A (0.15 mg/ml) for 30 min at 37°C, followed by Proteinase K (0.5 mg/ml) treatment overnight at room temperature. DNA was phenol/chloroform-extracted, ethanol-precipitated with glycogen (20 μg), and resuspended in 5 μl of TE.
For replication of dsDNA templates, the G4 structure was induced on dsDNA plasmids (75 ng/μl final concentration), and where indicated, G4 ligands PhenDC3 and TMPyP4 were introduced before replication by the same method as for the ssDNA template. For replication of pG4BOT-CON and pG4Lead-DPC, replication was conducted in a SprT-like N-terminal domain–depleted background to prevent M.HpaII degradation in Xenopus egg extracts (64). The plasmids (15 ng/μl for plasmid pull-down assay and 9 ng/μl for all other assays) were then incubated with HSS for 20 min at room temperature to assemble prereplication complexes. Two volumes of NPE [diluted to 40% with egg lysis buffer-sucrose buffer containing 10 mM Hepes-KOH (pH 7.7), 50 mM KCl, 2.5 mM MgCl2, and 250 mM sucrose] supplemented with 10 mM DTT, 15.5 mM phosphocreatine, 1.5 mM ATP, and creatine phosphokinase (3.8 ng/μl) were then added to fire a single round of DNA replication. For nascent strand labeling, NPE was supplemented with 32P-α-dCTP. To inhibit replication initiation, recombinant xlGeminin (400 nM final concentration) was added to HSS and incubated for 10 min at room temperature before DNA addition. To block CMG unloading, an inhibitor of the p97 segregase, NMS-873 (Sigma-Aldrich, catalog no. SML1128-5MG; 200 μM final concentration), was added to NPE and incubated for 10 min at room temperature before mixing with HSS. For replication in the presence of LacI, plasmids were incubated with 1.33 volumes of 30 μM biotinylated LacI for 30 min at room temperature before HSS addition. Where indicated, LacI was released from DNA by adding isopropyl-β-d-1-thiogalactopyranoside (IPTG; 9 mM final concentration) to replication reaction. The 0-min time point was taken immediately after NPE addition. DNA was extracted from replication reactions by the same method as described for the ssDNA templates. To monitor the replication fork blockage by LacI bound to the lacO locus, extracted replication products were digested with Hinc II for 3 hours at 37°C and separated on a 0.8% agarose gel in 1× TBE buffer (89 mM tris-borate and 2 mM EDTA). DNA was visualized by autoradiography using Typhoon TRIO+ (GE Healthcare).
Plasmid pull-down MS
Replicating ssDNA plasmids were pulled down as previously described (64, 66). Streptavidin magnetic beads (Dynabeads M-280, Thermo Fisher Scientific, catalog no. DB11205) were washed three times with 1 volume of wash buffer I [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, and 0.02% Tween 20], resuspended with 1 volume of wash buffer I, and incubated with biotinylated LacI (2 μM final concentration) for 45 min with mixing every 10 min at room temperature. The beads were washed four times with 1 volume of IP buffer [ELBS buffer supplemented with BSA (0.25 mg/ml) and 0.02% Tween 20], resuspended with 6.64 volume of IP buffer, and stored at 4°C. To capture replication intermediates via LacI that nonspecifically binds to ssDNA and dsDNA (67), 150 μl of the replication reaction was mixed with 750 μl of bead solution (containing 113 μl of biotin-LacI-bound streptavidin magnetic beads) at the indicated times and incubated for 30 min at 0° to 2°C with mixing every 10 min. The beads were washed three times with 750 μl of IP buffer containing 0.03% Tween 20 and resuspended in 40 μl of 1× SDS sample buffer [75 mM tris-HCl (pH 6.8), 10% glycerol, 2.5% SDS, 10% (v/v) Bond-Breaker TCEP Solution (Thermo Fisher Scientific, catalog no. 7772), and 0.02% (w/v) bromophenol blue]. The samples were heated at 95°C for 5 min and separated on a 12% bis-tris SDS–polyacrylamide gel electrophoresis (PAGE) gel (Bio-Rad).
Mass spectrometry
Data collection
The gel was run for 2 to 3 cm and stained with colloidal coomassie dye G-250 (Gel Code Blue Stain Reagent, Thermo Fisher Scientific) after which each lane was cut out. Gel pieces were reduced, alkylated, and digested overnight with trypsin at 37°C. The peptides were extracted with 100% acetonitrile and dried in a vacuum concentrator. Samples were resuspended in 10% (v/v) formic acid for ultrahigh-performance liquid chromatography (UHPLC)–MS/MS. The data were acquired using an UHPLC 1290 system coupled to an Orbitrap Q Exactive Biopharma HF mass spectrometer (Thermo Fisher Scientific). Samples were first trapped (Dr. Maisch ReproSil C18, 3 μm, 2 cm by 100 μm) before being separated on an analytical column (Agilent Poroshell EC-C18, 278 μm, 40 cm by 75 μm), using a gradient of 100 min at a column flow of 300 nl/min. Trapping was performed at 5 μl/min for 5 min in solvent A (0.1% formic acid in water), and the gradient was as follows: 13 to 44% solvent B (0.1% formic acid in 80% acetonitrile) in 95 min, 44 to 100% in 3 min, 100% solvent B for 1 min, and 100 to 0% in 1 min. Full-scan MS spectra from mass/charge ratio (m/z) 375 to 1600 were acquired at a resolution of 60,000 at m/z 400 after accumulation to a target value of 3 × 106. Up to 10 most intense precursor ions were selected for fragmentation. Higher-energy C-trap dissociation fragmentation was performed at a normalized collision energy of 27% after the accumulation to a target value of 1 × 105. MS/MS was acquired at a resolution of 30,000. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (68) partner repository with the dataset identifier PXD027623.
Data analysis
Raw data were analyzed with the MaxQuant software (version 1.5.0.17) using label-free quantification (69). A false discovery rate of 0.01 and a minimum peptide length of seven amino acids were used. MS/MS spectra were searched against a nonredundant Xenopus database (70). For the Andromeda search, the enzyme trypsin was chosen allowing for cleavage N-terminal to proline. Cysteine carbamidomethylation was selected as a fixed modification, and protein N-terminal acetylation and methionine oxidation were selected as variable modifications. Two missed cleavages were allowed maximally. Initial mass deviation of precursor ion was up to 7 parts per million, and mass deviation for fragment ions was 0.05 Da. Protein identification required one unique peptide to the protein group, and “Match between run” was enabled.
Statistical analysis
All bioinformatics analysis was carried out with the Perseus software version 1.6.10.0. For each comparison, the processed data were filtered to contain at least three valid values in at least one of the replicate group (four repeats per condition).
Antibodies and immunodepletion
Antibodies against xlFANCJ (18), xlMCM7 (71), xlPCNA (72), xlRPA (71), xlRTEL1-N (45), xlSPRTN (64), and histone H3 (Abcam, catalog no. ab1791) were previously described. The xlDHX36 antibody was raised against N-terminal residues (1 to 170). The complementary DNA (cDNA) encoding the fragment was codon-optimized for E. coli, synthesized (gBlocks Gene Fragments, Integrated DNA Technologies), and ligated into the Bam HI–Xho I sites of the pETDuet-1 vector (Novagen, catalog no. 71146-3). The fragment was overexpressed in E. coli BL21 (DE3) cells with a N-terminal His6-tag and purified by the method described previously (73). The purified antigen was used for immunization of rabbits (Pocono rabbit farm and laboratory, Canadensis, USA). Antibodies used for ChIP experiments were purified with rProtein A Sepharose (PAS) beads (GE Healthcare, catalog no. 171279-01). The affinity-purified xlBLM antibody was raised against C-terminal residues of two isoforms (1354 to 1367: CQTNRHFLKPSYSLF; and 1356 to 1369: CQPNRRFLKPSYSMF) by New England Peptide. The affinity-purified xlDNA2 and xlWRN antibodies were raised against N-terminal residues (1 to 16: MTSLQRKLPEWMSVKC) and C-terminal residues (1037 to 1053: CENQIYDLPEGAHEHFPV), respectively by New England Peptide. Specificity of the antisera or purified antibodies was confirmed by immunoblotting.
For depletion, 1 volume of Dynabeads Protein A beads (Thermo Fisher Scientific, catalog no. 10008D) was preincubated with half volumes of antisera for 30 min at room temperature. When affinity-purified antibodies were used, the Protein A beads were saturated with the antibody [beads (0.45 μg/μl)] by incubating for 30 min at room temperature. For mock depletion, preimmunized rabbit serum was used. To deplete HSS and NPE, 1 volume of each antibody-bound beads was incubated with 1.5 volume of extract for 30 min at room temperature for two rounds.
Protein purification
Recombinant C-terminal FLAG-tagged xlFANCJ (18), xlGeminin (74), biotinylated LacI (75), and methylated M.HpaII (64) were prepared as previously described. The xlFANCJK52R mutant was created by a site-directed mutagenesis using oligonucleotides K52R for and K52R rev (table S1) and purified by the same method as wild-type xlFANCJ.
For purification of xlDHX36, the cDNA encoding full-length xlDHX36 (gBlocks Gene Fragments, Integrated DNA Technologies) were ligated into the Bam HI–Xho I sites of the pETDuet-1 vector, and the protein were overexpressed as a N-terminal His6-tagged protein in the E. coli LOBSTR (DE3) cells cultured in 4 liters of LB medium, as previously described (76). The cells were collected, resuspended in buffer A [50 mM tris-HCl (pH 8.0), 10% glycerol, 0.5 M NaCl, 1 mM phenylmethylsulfonyl fluoride, 20 mM imidazole, 0.1% Tween 20, and 5 mM DTT], and disrupted by sonication. The supernatant was then separated from the cell debris by centrifugation (39,191g) for 25 min at 4°C and treated with polyethyleneimine [0.05% (v/v)]. After centrifugation (16,639g) for 10 min at 4°C, the supernatant was mixed with nickel–nitrilotriacetic acid (Ni-NTA) agarose resin (1.2 ml; Thermo Fisher Scientific, catalog no. R90115) at 4°C for an hour. The Ni-NTA beads were packed into a disposable chromatography column (Bio-Rad, catalog no. 731-1550) and were washed with 60 ml of buffer A, followed by 30 ml of buffer B [50 mM tris-HCl (pH 8.0), 10% glycerol, 250 mM NaCl, 20 mM imidazole, and 5 mM DTT]. His6-tagged xlDHX36 was eluted with 8 ml of buffer B containing 400 mM imidazole and loaded on 1 ml of HiTrap Heparin HP (GE Healthcare, catalog no. 17040601) equilibrated with buffer C [20 mM tris-HCl (pH 8.0), 10% glycerol, 250 mM NaCl, and 5 mM DTT]. The column was subsequently washed with buffer C, and the protein was eluted with 60 ml of linear gradient of 250 to 1000 mM NaCl in buffer C. Peak fractions were collected, concentrated with Amicon Ultra-4 Centrifugal Filter Unit (100-kDa cutoff; Merck Millipore, catalog no. UFC810024) to ~1 mg/ml, snap-frozen, and stored at −80°C. The xlDHX36 mutants were created by a site-directed mutagenesis using oligonucleotides Y53A for and Y53A rev (for DHX36Y53A) or E327A for and E327A rev (for DHX36E327A) and purified by the same method as the wild-type protein. For rescue experiments, depleted NPE (for dsDNA template replication) or HSS (for ssDNA template replication) was supplemented with 35 nM recombinant xlFANCJ or 250 nM recombinant xlDHX36. The concentration of the recombinant proteins was determined by SDS-PAGE with Coomassie Brilliant Blue staining, using BSA as a standard protein.
G4 unwinding assay
Oligonucleotides G4 15-nt and G4 3-nt (2 μM final concentration) were radiolabeled with T4 polynucleotide kinase (New England Biolabs, catalog no. M0201S) and 32P-γ-dATP, diluted by 10 times with annealing buffer, incubated 5 min at 95°C, and cooled down to room temperature in ~2 hours to inactivate T4 polynucleotide kinase and allow the G4 structure. The substrate (5 nM final concentration) was incubated with either wild-type xlDHX36 or the DHX36E327A mutant in 10 μl of reaction solution [24 mM tris (pH 8.0), 2 mM DTT, 100 mM NaCl, acetylated BSA (0.05 mg/ml; Promega, catalog no. R3961), 2 mM MgCl2, 2 mM ATP, 2% glycerol, and 20 nM oligonucleotide Q] for 30 min at room temperature. The reaction was stopped by Proteinase K treatment [2 μl of 1.67% SDS and Proteinase K (8.3 mg/ml)] and was further incubated for 15 min at room temperature. The sample was then analyzed by 10% native PAGE in 1× TBE buffer at 4°C. The gel was dried with cellophane (Sigma-Aldrich, catalog no. Z377570-1PAK) and exposed to a phosphor screen. The band intensity was measured with Typhoon TRIO+ and quantified using ImageQuant TL software.
Duplex unwinding assay
Oligonucleotides R (2 μM final concentration) was radiolabeled by the same method as for G4 unwinding assay, heated for 5 min at 95°C, and cooled down on ice to inactivate T4 polynucleotide kinase. The labeled oligonucleotide (0.2 μM final concentration) was incubated with either oligonucleotide G4 15-nt, G4 3-nt, or PolyT 15-nt (3 μM final concentration) in annealing buffer for 5 min at 95°C and cooled down to room temperature in ~2 hours to allow the G4 structure to form and the oligonucleotides to anneal. The labeled substrate (4 nM final concentration) was incubated with either wild-type xlDHX36 or the DHX36E327A mutant in 10 μl of reaction solution [24 mM tris (pH 8.0), 2 mM DTT, 100 mM NaCl, acetylated BSA (0.05 mg/ml), 2 mM MgCl2, 2 mM ATP, 2% glycerol, and 200 nM nonlabeled oligonucleotide R] for an hour at room temperature. The reaction was stopped, and the samples were analyzed by the same method as for G4 unwinding assay.
Nascent strand analysis
Nascent strands on ssDNA templates were analyzed as previously described (18). Extracted replication products were mixed with the same volume of Gel Loading Buffer II (Thermo Fisher Scientific, catalog no. AM8547), heated for 4 min at 98°C, snap-cooled on ice, and separated on 6% urea-PAGE gels, which were subsequently dried and exposed to a phosphor screen. DNA was visualized using a Typhoon TRIO+.
Nascent strands on dsDNA templates were analyzed as previously described (77). Extracted replication products were digested with Hinc II or Hinc II and Cla I for 3 hours at 37°C, ethanol-precipitated, and resuspended in 12 μl of alkaline loading buffer (50 mM NaOH, 2.5% Ficoll-400, and 1 mM EDTA). Fragments were then separated on a 0.8% agarose gel in alkaline buffer (50 mM NaOH and 1 mM EDTA), after which the gel was dried on Amersham Hybond-XL membrane (GE Healthcare, catalog no. RPN203S) and exposed to a phosphor screen.
To determine exact replication stalling positions, extracted replication products were analyzed by sequencing gel as previously described (39). The samples were digested either with Afl III and Bam HI or with Afl III, Bam HI, and Cla I for 3 hours at 37°C, mixed with the same volume of Gel Loading Buffer II, heated at 98°C for 5 min, snap-cooled on ice for 5 min, and separated on a 7% urea–polyacrylamide sequencing gel prepared in 0.8× TTE buffer [71 mM tris, 23 mM taurine, and 0.4 mM EDTA (pH 8.9)]. After gel drying, the products were visualized by autoradiography. The size of the stalling products was determined using sequencing ladders and PCR-amplified “−1” and “−2” fragments. The ladders were generated using either primer T or primer S and, as a template, either pdsG4BOT or pG4BOT by the Thermo Sequenase Cycle Sequencing Kit (Thermo Fisher Scientific, catalog no. 785001KT) according to the manufacturer’s protocol. −1 and −2 products on bottom strand were prepared by PCR using pdsG4BOT and primer T labeled with 32P at the 5′ end and either primer T-1 or primer T-2, respectively.
Plasmid pull-down assay
Replicating dsDNA plasmids were pulled down as previously described (66). At the indicated times, 10 μl of replication samples was mixed with 7.5 μl of biotin-LacI–bound streptavidin magnetic beads suspended in 50 μl of IP buffer containing 0.03% Tween 20 and incubated for 30 min at 0° to 2°C without pipeting. The beads were washed three times with 75 μl of IP buffer and suspended in 20 μl of 1× SDS sample buffer. Plasmid-bound proteins were then separated by SDS-PAGE and visualized by Western blot using the indicated antibodies.
Two-dimensional gel electrophoresis
Two-dimensional gel electrophoresis was performed as previously described (78). Extracted replication samples were digested with Hinc II for 3 hours at 37°C and analyzed by two consecutive electrophoreses. For the first dimension, the Hinc II–digested samples were separated with 0.4% agarose gel in 0.5× TBE buffer at 0.86 V/cm for 24 hours at room temperature. The lanes of interest were cut out, casted across the top of the second-dimension gel consisting of 1% agarose with ethidium bromide (0.3 μg/ml), and run in 0.5× TBE buffer containing ethidium bromide (0.3 μg/ml) with buffer circulation at 3.5 V/cm for 14.5 hours at room temperature. The gel was dried on Amersham Hybond-XL membrane and exposed to a phosphor screen. DNA was visualized using a Typhoon TRIO+.
Chromatin immunoprecipitation
ChIP was performed similar to described previously (79). At the indicated times, replication samples (3 μl) were cross-linked with 47 μl of ELBS buffer containing 1% formaldehyde for 10 min at room temperature. An unrelated nondamaged control plasmid (pQuant; 0.5 ng/μl) was coincubated with HSS to be used as an internal control for quantifications. After quenching the formaldehyde by addition of 5 μl of 1.25 M glycine, the samples were passed through a Micro Bio-Spin 6 Chromatography column (Bio-Rad, catalog no. 7326222), sonicated, and immunoprecipitated with the indicated antibodies (5 μg) bound to PAS beads. The protein-bound DNA fragments were eluted with ChIP elution buffer [50 mM tris (pH 7.5), 10 mM EDTA, and 1% SDS], and the cross-links were reversed by consecutive incubation for 6 hours at 42°C and then for 9 hours at 70°C. DNA was phenol/chloroform-extracted, followed by quantitative PCR in 10 μl of reaction buffer [6 mM tris (pH 8.3), 25 mM KCl, 2.5 mM MgCl2, 0.3 mM deoxynucleotide triphosphates, 0.1% Tween 20, BSA (0.1 mg/ml), 1:66,500 SYBR Green I (Sigma-Aldrich, catalog no. S9430), and Hot Start Taq DNA polymerase], using 0.25 μM of following primer pairs (table S1): G4 for and G4 rev [for G4 locus, 37– to 136–base pair (bp) upstream from G4s], lacO for and lacO rev (for lacO locus, 295- to 388-bp downstream from G4s), and pQuant for and pQuant rev (for assessment of background binding of the proteins on pQuant). The values from pQuant primers were subtracted from the values for G4 and lacO primers. ChIP data are plotted as the percentage of peak value with the highest value set to 1.
Acknowledgments
We thank J. C. Walter, M. Tijsterman, and F. Mattirolli for feedback on the manuscript; J. C. Walter and J. L Sparks for pSVRlacO, anti-SPRTN antibody, and methylated M.HpaII; the Hubrecht animal caretakers for animal support; and the other members of the Knipscheer laboratory for discussions. We are grateful to M. Räschle for providing us with the X. laevis protein database. Funding: K.S. was supported by the Uehara Memorial Foundation, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, and the Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship for Research Abroad. N.M.-P. was supported by the European Commission, and the plasmid pull-downs for MS, the data analysis, and the production of DHX36 antibodies were funded by the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement 750035 (ReXeG) to N.M.-P. This work was supported by a project grant from the Dutch Cancer Society (KWF HUBR 2015-7736) to P.K. and the Gravitation program CancerGenomiCs.nl from the Netherlands Organisation for Scientific Research (NWO), part of the Oncode Institute, which is partly financed by the Dutch Cancer Society. This research was part of the Netherlands X-omics Initiative and partially funded by NWO (project 184.034.019). Author contributions: K.S. and P.K. designed the experiments and wrote the manuscript. K.S. performed the experiments with Xenopus egg extracts. N.M.-P. prepared the samples for MS. H.P. performed the MS under supervision of M.A. N.M.-P. and H.P. analyzed the MS data. All of the authors read and approved the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Table S1
REFERENCES AND NOTES
- 1.Gellert M., Lipsett M. N., Davies D. R., Helix formation by guanylic acid. Proc. Natl. Acad. Sci. U.S.A. 48, 2013–2018 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sen D., Gilbert W., Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 334, 364–366 (1988). [DOI] [PubMed] [Google Scholar]
- 3.Burge S., Parkinson G. N., Hazel P., Todd A. K., Neidle S., Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 34, 5402–5415 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huppert J. L., Balasubramanian S., Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 33, 2908–2916 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarkies P., Reams C., Simpson L. J., Sale J. E., Epigenetic instability due to defective replication of structured DNA. Mol. Cell 40, 703–713 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siddiqui-Jain A., Grand C. L., Bearss D. J., Hurley L. H., Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. U.S.A. 99, 11593–11598 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zahler A. M., Williamson J. R., Cech T. R., Prescott D. M., Inhibition of telomerase by G-quartet DNA structures. Nature 350, 718–720 (1991). [DOI] [PubMed] [Google Scholar]
- 8.Cheung I., Schertzer M., Rose A., Lansdorp P. M., Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 31, 405–409 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Lerner L. K., Sale J. E., Replication of G Quadruplex DNA. Genes 10, 95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myung K., Chen C., Kolodner R. D., Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature 411, 1073–1076 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Snow B. E., Mateyak M., Paderova J., Wakeham A., Iorio C., Zakian V., Squire J., Harrington L., Murine Pif1 interacts with telomerase and is dispensable for telomere function in vivo. Mol. Cell. Biol. 27, 1017–1026 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.London T. B., Barber L. J., Mosedale G., Kelly G. P., Balasubramanian S., Hickson I. D., Boulton S. J., Hiom K., FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 283, 36132–36139 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuzaki K., Borel V., Adelman C. A., Schindler D., Boulton S. J., FANCJ suppresses microsatellite instability and lymphomagenesis independent of the Fanconi anemia pathway. Genes Dev. 29, 2532–2546 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Litman R., Peng M., Jin Z., Zhang F., Zhang J., Powell S., Andreassen P. R., Cantor S. B., BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell 8, 255–265 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Levitus M., Waisfisz Q., Godthelp B. C., de Vries Y., Hussain S., Wiegant W. W., Elghalbzouri-Maghrani E., Steltenpool J., Rooimans M. A., Pals G., Arwert F., Mathew C. G., Zdzienicka M. Z., Hiom K., De Winter J. P., Joenje H., The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet 37, 934–935 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Levran O., Attwooll C., Henry R. T., Milton K. L., Neveling K., Rio P., Batish S. D., Kalb R., Velleuer E., Barral S., Ott J., Petrini J., Schindler D., Hanenberg H., Auerbach A. D., The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat. Genet. 37, 931–933 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Ishiai M., Sato K., Tomida J., Kitao H., Kurumizaka H., Takata M., Activation of the FA pathway mediated by phosphorylation and ubiquitination. Mutat. Res. 803-805, 89–95 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Castillo Bosch P., Segura-Bayona S., Koole W., van Heteren J. T., Dewar J. M., Tijsterman M., Knipscheer P., FANCJ promotes DNA synthesis through G-quadruplex structures. EMBO J. 33, 2521–2533 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y., Shin-ya K., Brosh R. M. Jr., FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 28, 4116–4128 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding H., Schertzer M., Wu X., Gertsenstein M., Selig S., Kammori M., Pourvali R., Poon S., Vulto I., Chavez E., Tam P. P., Nagy A., Lansdorp P. M., Regulation of murine telomere length by Rtel: An essential gene encoding a helicase-like protein. Cell 117, 873–886 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Giri B., Smaldino P. J., Thys R. G., Creacy S. D., Routh E. D., Hantgan R. R., Lattmann S., Nagamine Y., Akman S. A., Vaughn J. P., G4 resolvase 1 tightly binds and unwinds unimolecular G4-DNA. Nucleic Acids Res. 39, 7161–7178 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schult P., Paeschke K., The DEAH helicase DHX36 and its role in G-quadruplex-dependent processes. Biol. Chem. 402, 581–591 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Lai J. C., Ponti S., Pan D., Kohler H., Skoda R. C., Matthias P., Nagamine Y., The DEAH-box helicase RHAU is an essential gene and critical for mouse hematopoiesis. Blood 119, 4291–4300 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Bharti S. K., Sommers J. A., George F., Kuper J., Hamon F., Shin-ya K., Teulade-Fichou M. P., Kisker C., Brosh R. M. Jr., Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability. J. Biol. Chem. 288, 28217–28229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez R., Miller K. M., Forment J. V., Bradshaw C. R., Nikan M., Britton S., Oelschlaegel T., Xhemalce B., Balasubramanian S., Jackson S. P., Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 8, 301–310 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biffi G., Tannahill D., McCafferty J., Balasubramanian S., Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 5, 182–186 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu H.-Y., Zhao Q., Zhang T.-P., Wu Y., Xiong Y.-X., Wang S.-K., Ge Y.-L., He J.-H., Lv P., Ou T.-M., Tan J.-H., Li D., Gu L.-Q., Ren J., Zhao Y., Huang Z.-S., Conformation selective antibody enables genome profiling and leads to discovery of parallel G-quadruplex in human telomeres. Cell Chem. Biol. 23, 1261–1270 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Izbicka E., Wheelhouse R. T., Raymond E., Davidson K. K., Lawrence R. A., Sun D., Windle B. E., Hurley L. H., Von Hoff D. D., Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res. 59, 639–644 (1999). [PubMed] [Google Scholar]
- 29.Xu H., Di Antonio M., McKinney S., Mathew V., Ho B., O’Neil N. J., Santos N. D., Silvester J., Wei V., Garcia J., Kabeer F., Lai D., Soriano P., Banath J., Chiu D. S., Yap D., Le D. D., Ye F. B., Zhang A., Thu K., Soong J., Lin S. C., Tsai A. H., Osako T., Algara T., Saunders D. N., Wong J., Xian J., Bally M. B., Brenton J. D., Brown G. W., Shah S. P., Cescon D., Mak T. W., Caldas C., Stirling P. C., Hieter P., Balasubramanian S., Aparicio S., CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 8, 14432 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vannier J. B., Sandhu S., Petalcorin M. I., Wu X., Nabi Z., Ding H., Boulton S. J., RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science 342, 239–242 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Alabert C., Bukowski-Wills J. C., Lee S. B., Kustatscher G., Nakamura K., de Lima Alves F., Menard P., Mejlvang J., Rappsilber J., Groth A., Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat. Cell Biol. 16, 281–291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwab R. A., Nieminuszczy J., Shin-ya K., Niedzwiedz W., FANCJ couples replication past natural fork barriers with maintenance of chromatin structure. J. Cell Biol. 201, 33–48 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemmens B., van Schendel R., Tijsterman M., Mutagenic consequences of a single G-quadruplex demonstrate mitotic inheritance of DNA replication fork barriers. Nat. Commun. 6, 8909 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kruisselbrink E., Guryev V., Brouwer K., Pontier D. B., Cuppen E., Tijsterman M., Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr. Biol. 18, 900–905 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Phan A. T., Mergny J. L., Human telomeric DNA: G-quadruplex, i-motif and Watson-Crick double helix. Nucleic Acids Res. 30, 4618–4625 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madireddy A., Purushothaman P., Loosbroock C. P., Robertson E. S., Schildkraut C. L., Verma S. C., G-quadruplex-interacting compounds alter latent DNA replication and episomal persistence of KSHV. Nucleic Acids Res. 44, 3675–3694 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salas T. R., Petruseva I., Lavrik O., Bourdoncle A., Mergny J. L., Favre A., Saintome C., Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 34, 4857–4865 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berezney R., Dubey D. D., Huberman J. A., Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108, 471–484 (2000). [DOI] [PubMed] [Google Scholar]
- 39.Räschle M., Knipscheer P., Enoiu M., Angelov T., Sun J., Griffith J. D., Ellenberger T. E., Scharer O. D., Walter J. C., Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134, 969–980 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duxin J. P., Dewar J. M., Yardimci H., Walter J. C., Repair of a DNA-protein crosslink by replication-coupled proteolysis. Cell 159, 346–357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cantor S. B., Bell D. W., Ganesan S., Kass E. M., Drapkin R., Grossman S., Wahrer D. C., Sgroi D. C., Lane W. S., Haber D. A., Livingston D. M., BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 105, 149–160 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Tran H., Schilling M., Wirbelauer C., Hess D., Nagamine Y., Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol. Cell 13, 101–111 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Fullbright G., Rycenga H. B., Gruber J. D., Long D. T., p97 promotes a conserved mechanism of helicase unloading during DNA cross-link repair. Mol. Cell. Biol. 36, 2983–2994 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu R. A., Semlow D. R., Kamimae-Lanning A. N., Kochenova O. V., Chistol G., Hodskinson M. R., Amunugama R., Sparks J. L., Wang M., Deng L., Mimoso C. A., Low E., Patel K. J., Walter J. C., TRAIP is a master regulator of DNA interstrand crosslink repair. Nature 567, 267–272 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sparks J. L., Chistol G., Gao A. O., Räschle M., Larsen N. B., Mann M., Duxin J. P., Walter J. C., The CMG helicase bypasses DNA-protein cross-links to facilitate their repair. Cell 176, 167–181.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Semlow D. R., Zhang J., Budzowska M., Drohat A. C., Walter J. C., Replication-dependent unhooking of DNA interstrand cross-links by the NEIL3 glycosylase. Cell 167, 498–511.e14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yangyuoru P. M., Bradburn D. A., Liu Z., Xiao T. S., Russell R., The G-quadruplex (G4) resolvase DHX36 efficiently and specifically disrupts DNA G4s via a translocation-based helicase mechanism. J. Biol. Chem. 293, 1924–1932 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Do N. Q., Phan A. T., Monomer-dimer equilibrium for the 5′-5′ stacking of propeller-type parallel-stranded G-quadruplexes: NMR structural study. Chemistry 18, 14752–14759 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Yuan Z., Bai L., Sun J., Georgescu R., Liu J., O’Donnell M. E., Li H., Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat. Struct. Mol. Biol. 23, 217–224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakano T., Miyamoto-Matsubara M., Shoulkamy M. I., Salem A. M., Pack S. P., Ishimi Y., Ide H., Translocation and stability of replicative DNA helicases upon encountering DNA-protein cross-links. J. Biol. Chem. 288, 4649–4658 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gray R. D., Trent J. O., Arumugam S., Chaires J. B., Folding landscape of a parallel G-Quadruplex. J. Phys. Chem. Lett. 10, 1146–1151 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bochman M. L., Schwacha A., The Mcm2-7 complex has in vitro helicase activity. Mol. Cell 31, 287–293 (2008). [DOI] [PubMed] [Google Scholar]
- 53.Gupta R., Sharma S., Sommers J. A., Kenny M. K., Cantor S. B., Brosh R. M. Jr., FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood 110, 2390–2398 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu C. G., Spies M., G-quadruplex recognition and remodeling by the FANCJ helicase. Nucleic Acids Res. 44, 8742–8753 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen M. C., Tippana R., Demeshkina N. A., Murat P., Balasubramanian S., Myong S., Ferre-D’Amare A. R., Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature 558, 465–469 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cayrou C., Coulombe P., Vigneron A., Stanojcic S., Ganier O., Peiffer I., Rivals E., Puy A., Laurent-Chabalier S., Desprat R., Mechali M., Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 21, 1438–1449 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lerner L. K., Holzer S., Kilkenny M. L., Svikovic S., Murat P., Schiavone D., Eldridge C. B., Bittleston A., Maman J. D., Branzei D., Stott K., Pellegrini L., Sale J. E., Timeless couples G-quadruplex detection with processing by DDX11 helicase during DNA replication. EMBO J., e104185 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Varshney D., Spiegel J., Zyner K., Tannahill D., Balasubramanian S., The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 21, 459–474 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McLuckie K. I., Di Antonio M., Zecchini H., Xian J., Caldas C., Krippendorff B. F., Tannahill D., Lowe C., Balasubramanian S., G-quadruplex DNA as a molecular target for induced synthetic lethality in cancer cells. J. Am. Chem. Soc. 135, 9640–9643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rafnar T., Gudbjartsson D. F., Sulem P., Jonasdottir A., Sigurdsson A., Jonasdottir A., Besenbacher S., Lundin P., Stacey S. N., Gudmundsson J., Magnusson O. T., le Roux L., Orlygsdottir G., Helgadottir H. T., Johannsdottir H., Gylfason A., Tryggvadottir L., Jonasson J. G., de Juan A., Ortega E., Ramon-Cajal J. M., Garcia-Prats M. D., Mayordomo C., Panadero A., Rivera F., Aben K. K., van Altena A. M., Massuger L. F., Aavikko M., Kujala P. M., Staff S., Aaltonen L. A., Olafsdottir K., Bjornsson J., Kong A., Salvarsdottir A., Saemundsson H., Olafsson K., Benediktsdottir K. R., Gulcher J., Masson G., Kiemeney L. A., Mayordomo J. I., Thorsteinsdottir U., Stefansson K., Mutations in BRIP1 confer high risk of ovarian cancer. Nat. Genet. 43, 1104–1107 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Enoiu M., Ho T. V., Long D. T., Walter J. C., Schärer O. D., Construction of plasmids containing site-specific DNA interstrand cross-links for biochemical and cell biological studies. Methods Mol. Biol. 920, 203–219 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J., Dewar J. M., Budzowska M., Motnenko A., Cohn M. A., Walter J. C., DNA interstrand cross-link repair requires replication-fork convergence. Nat. Struct. Mol. Biol. 22, 242–247 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen L., MacMillan A. M., Chang W., Ezaz-Nikpay K., Lane W. S., Verdine G. L., Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry 30, 11018–11025 (1991). [DOI] [PubMed] [Google Scholar]
- 64.Larsen N. B., Gao A. O., Sparks J. L., Gallina I., Wu R. A., Mann M., Räschle M., Walter J. C., Duxin J. P., Replication-coupled DNA-protein crosslink repair by SPRTN and the proteasome in xenopus egg extracts. Mol. Cell 73, 574–588 e577 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sparks J., Walter J. C., Extracts for analysis of DNA replication in a nucleus-free system. Cold Spring Harb. Protoc. 2019, pdb.prot097154 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Budzowska M., Graham T. G., Sobeck A., Waga S., Walter J. C., Regulation of the Rev1-pol ζ complex during bypass of a DNA interstrand cross-link. EMBO J. 34, 1971–1985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Riggs A. D., Lin S., Wells R. D., Lac repressor binding to synthetic DNAs of defined nucleotide sequence. Proc. Natl. Acad. Sci. U.S.A. 69, 761–764 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., Tiwary S., Cox J., Audain E., Walzer M., Jarnuczak A. F., Ternent T., Brazma A., Vizcaino J. A., The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cox J., Mann M., MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Temu T., Mann M., Raschle M., Cox J., Homology-driven assembly of NOn-redundant protEin sequence sets (NOmESS) for mass spectrometry. Bioinformatics 32, 1417–1419 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Walter J., Newport J., Initiation of eukaryotic DNA replication: Origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell 5, 617–627 (2000). [DOI] [PubMed] [Google Scholar]
- 72.Kochaniak A. B., Habuchi S., Loparo J. J., Chang D. J., Cimprich K. A., Walter J. C., van Oijen A. M., Proliferating cell nuclear antigen uses two distinct modes to move along DNA. J. Biol. Chem. 284, 17700–17710 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brandsma I., Sato K., van Rossum-Fikkert S. E., van Vliet N., Sleddens E., Reuter M., Odijk H., van den Tempel N., Dekkers D. H. W., Bezstarosti K., Demmers J. A. A., Maas A., Lebbink J., Wyman C., Essers J., van Gent D. C., Baarends W. M., Knipscheer P., Kanaar R., Zelensky A. N., HSF2BP interacts with a conserved domain of BRCA2 and is required for mouse spermatogenesis. Cell Rep. 27, 3790–3798 e3797 (2019). [DOI] [PubMed] [Google Scholar]
- 74.McGarry T. J., Kirschner M. W., Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93, 1043–1053 (1998). [DOI] [PubMed] [Google Scholar]
- 75.Dewar J. M., Budzowska M., Walter J. C., The mechanism of DNA replication termination in vertebrates. Nature 525, 345–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takahashi D., Sato K., Shimomuki M., Takata M., Kurumizaka H., Expression and purification of human FANCI and FANCD2 using Escherichia coli cells. Protein Expr. Purif. 103, 8–15 (2014). [DOI] [PubMed] [Google Scholar]
- 77.Sato K., Brandsma I., van Rossum-Fikkert S. E., Verkaik N., Oostra A. B., Dorsman J. C., van Gent D. C., Knipscheer P., Kanaar R., Zelensky A. N., HSF2BP negatively regulates homologous recombination in DNA interstrand crosslink repair. Nucleic Acids Res. 48, 2442–2456 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Long D. T., Räschle M., Joukov V., Walter J. C., Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 333, 84–87 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pacek M., Tutter A. V., Kubota Y., Takisawa H., Walter J. C., Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol. Cell 21, 581–587 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Table S1