Abstract
BACKGROUND AND AIMS:
Circular RNAs (circRNAs) are a class of endogenous noncoding RNAs that form covalently closed circles. Although circRNAs influence many biological processes, little is known about their role in intestinal epithelium homeostasis. We surveyed circRNAs required to maintain intestinal epithelial integrity and identified circHIPK3 as a major regulator of intestinal epithelial repair after acute injury.
METHODS:
Intestinal mucosal tissues were collected from mice exposed to cecal ligation and puncture (CLP) for 48 h and patients with inflammatory bowel diseases (IBD) and sepsis. We isolated primary enterocytes from the small intestine of mice and derived intestinal organoids. The levels of circHIPK3 were silenced in intestinal epithelial cells (IECs) by transfection with siRNAs targeting the circularization junction of circHIPK3 or elevated using a plasmid vector that overexpressed circHIPK3. Intestinal epithelial repair was examined in an in vitro injury model by removing part of the monolayer. The association of circHIPK3 with miR-29b was determined by biotinylated RNA pulldown assays.
RESULTS:
Genome-wide profile analyses identified ~300 circRNAs, including circHIPK3, differentially expressed in the intestinal mucosa of CLP-mice relative to sham mice. Intestinal mucosa from patients with IBD and sepsis had reduced levels of circHIPK3. Increasing the levels of circHIPK3 enhanced intestinal epithelium repair after wounding, whereas circHIPK3 silencing repressed epithelial recovery. CircHIPK3 silencing also inhibited growth of IECs and intestinal organoids, and circHIPK3 overexpression promoted intestinal epithelium renewal in mice. Mechanistic studies revealed that circHIPK3 directly bound to miR-29b and inhibited miR-29 activity, thus increasing expression of Rac1, Cdc42, and cyclin B1 in IECs after wounding.
CONCLUSIONS:
In studies of mice, IECs, and human tissues, our results indicate that circHIPK3 improves repair of the intestinal epithelium at least in part by reducing miR-29b availability.
Keywords: circRNAs, IEC, microRNAs, mucosal injury, IBD
Lay Summary
Circular RNA circHIPK3 enhances intestinal epithelial repair and promotes mucosal growth by reducing microRNA 29b activity, but its levels decrease in IBD patients.
Graphical Abstract
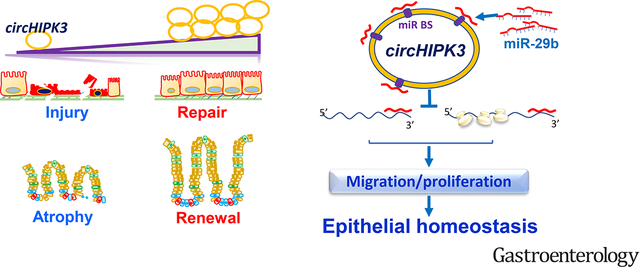
INTRODUCTION
The intestinal epithelium is a single layer of columnar cells lining the luminal surface of the intestinal mucosa and functions as a dynamic physical barrier between sterile mucosal tissues and a wide array of luminal noxious substances and microbiota.1 Homeostasis of the intestinal epithelium is preserved via tightly controlled mechanisms that rely on intestinal epithelial cells (IECs) to quickly change gene expression patterns to regulate cell survival, migration, proliferation, differentiation, and cell-to-cell interaction.2,3 Early epithelial restitution is a primary repair modality in the intestinal mucosa and rapidly reseals superficial wounds by migrating visible remaining epithelial cells from areas adjacent to the injured surface to cover the wounded area, followed by proliferation and epithelial remodeling.4,5 Nonetheless, intestinal epithelium injury occurs commonly in patients with various critical disorders, including inflammatory bowel disease (IBD), sepsis, trauma, severe thermal injury, and emergent surgical laparotomies.5,6 Disrupted integrity of the intestinal epithelium by severe and diffusive mucosal injury/erosions leads to the translocation of luminal toxic substances and bacteria to the bloodstream and, in some instances, might result in multiple organ dysfunction syndrome and death in critically ill patients.6,7
The majority of the mammalian genome is transcribed into a vast transcriptome of noncoding RNAs (ncRNAs), whereas protein-coding sequences only account for a minority (< 4%) of the cellular transcriptional output.8 Circular RNAs (circRNAs) represent a class of widespread and diverse endogenous ncRNAs that are often expressed in a tissue- and developmental stage-specific manner.9 CircRNAs are mainly synthesized from precursor RNAs undergoing splicing, but the 5’ to 3’ termini of splicing byproducts become covalently religated.9,10 Most circRNAs are believed to have noncoding functions, although some circRNAs encode peptides.9 Several circRNAs have been reported to harbor one or several binding sites for a single microRNA (miRNA), and some harbor binding sites for multiple miRNAs.9,11,12 Many circRNAs have been proposed to function as decoys or sponges that reduce the number of freely available miRNAs, but binding of a miRNA to a circRNA might not always result in suppression of the miRNA.13 CircRNAs also interact with RNA-binding proteins (RBPs) to jointly regulate gene expression and possess multiple cellular functions.14,15 The differential expression of circRNAs during disease progression suggests a significant role for circRNAs in human pathologies.16,17
CircRNA circHIPK3 (circHIPK3) is derived from the exon 2 of the HIPK3 gene and formed by direct backsplicing supported by intronic RNA pairings.9,18 CircHIPK3 is expressed in various mammalian tissues including the intestine and confers cell-type specific regulation of cell functions by interacting with different miRNAs.19 CircHIPK3 expression levels change dramatically in disease conditions such as cancer, although it can be tumor-repressive or pro-ncogenic depending on its cellular context, tumor type, and target miRNAs.18,19 However, no reports so far have addressed the role of circHIPK3 in regulating intestinal epithelium homeostasis. In this study, we provided evidence that the expression patterns of circRNAs in the intestinal mucosa changed in mice exposed to septic stress, and that human intestinal mucosa with injury/erosions and inflammation from patients with IBD and sepsis exhibited reduced circHIPK3 levels. We further discovered that circHIPK3 enhanced intestinal epithelial repair after wounding and promoted epithelial renewal partially by acting as a sponge for miR-29b. These findings revealed that circHIPK3 is essential for sustaining homeostasis of the intestinal epithelium and point to circHIPK3/miR-29b axis as a novel therapeutic target for interventions to protect the epithelial integrity in patients with critical illnesses.
MATERIALS AND METHODS
Murine studies
C57BL/6J mice (male and female, 6–9 weeks old) were purchased from The Jackson Laboratory and housed in a pathogen-free animal facility at the Baltimore VA Medical Center. All animal experiments were conducted in accordance with NIH guidelines and were approved by the Institutional Animal Care and Use Committee of University Maryland School of Medicine and Baltimore VA hospital. Animals were deprived of food but allowed free access to tap water for 24 h before experiments. To generate the model of cecal ligation and puncture (CLP)-induced injury, mice were anesthetized by Nembutal, and CLP was performed as described.20 Forty-eight hours after CLP, two 4-cm segments taken from the middle of the small intestine were removed in each animal as described previously.21,22
Histology and Immunohistochemistry
Human tissue samples were obtained from surplus discarded tissue from Department of Surgery, University of Maryland Health Science Center and commercial tissue banks. The study was approved by the University Maryland Institutional Review Board. Dissected and opened intestines were mounted onto a solid surface and fixed in formalin and paraffin. The immunofluorescence staining procedure was carried out according to the method described.23,24 All slides were incubated with the primary antibody against F-actin and then incubated with secondary antibody conjugated with Alexa Fluor-594, and images were processed using Photoshop software.
Procedures of several methods, including cell culture,25 intestinal organoid culture,23,26 plasmid construction,27,28 reverse transcription followed by quantitative real-time PCR (Q-PCR) or droplet digital Q-PCR analyses,36 circRNA microarray analysis,22,29 biotin-labeled RNA pull-down assays,25 RNA fluorescence in situ hybridization (RNA-FISH) assay,30,31 lentivirus infection,31 and measurements of epithelial repair and barrier function,3,31,32 are described in the Supplementary Materials for more detail.
Statistical analysis
All values were expressed as the means ± SEM. Unpaired, two-tailed Student’s t test was used when indicated with P < 0.05 considered significant. When assessing multiple groups, one-way ANOVA was utilized with Tukey’s post hoc test.33 The statistical software used was GraphPad Instat Prism 9.0 (San Diego, CA). For non-parametric analysis rank comparison, the Kruskal-Wallis test was conducted.
RESULTS
Changes in circRNA expression profiles in intestinal epithelium responding to critical stress
To determine the involvement of circRNAs in intestinal epithelium homeostasis, a mouse CLP model20 was used in this study. As we and other reported previously,21,30,34,35 exposure of mice to CLP for 48 h induced mucosal lesions in the small intestine, as shown by severely sloughed cells, denuded villi with dilated capillaries, and by frank hemorrhage microscopically, and bleeding, edematous and swollen mucosa with areas of red streaks macroscopically. CLP for 48 h also led to an acute gut barrier dysfunction, as indicated by an increased mucosal permeability to fluorescein isothiocyanate (FITC) dextran, similar to the observations reported previously.21,30 The small intestinal mucosal tissues, containing >91% intestinal epithelial cells,31 were scraped from the underlying smooth muscle as described.22 Total RNA was isolated from the mucosa and then digested with RNase R to remove all linear RNAs. The remaining circRNAs were amplified and transcribed into fluorescent cRNA utilizing a random priming method,14 and microarray-based interrogation of global circRNAs expression profiles was performed. In total, ~9360 circRNAs were detectable in the small intestinal mucosa of mouse (Supplementary Table 1).
A comparison of the circRNA expression profiles in control mice relative to mice exposed to CLP for 48 h showed that ~300 circRNAs, including circHIPK3, were differentially expressed in the damaged mucosa (Figure 1A; Supplementary Figure 1A). Figure 1B summarizes the ten most increased or decreased circRNAs in the mucosa of mice exposed to CLP when compared with sham mice. Although a sizeable subset of circRNAs showed altered abundance in the damaged mucosa, experiments in this study were highly focused on circHIPK3, since it is highly conserved in mammals18,36 and because there were potential interactions of circHIPK3 with several miRNAs including miR-29b, based on computational prediction algorithms and previous publications.18,37 Moreover, some circRNAs that showed significant changes in CLP-mice were undetectable in human IECs, whereas the basal level of circHIPK3 was higher than many other circRNAs detected in the small intestinal mucosal tissue and in cultured human and rat IECs. Reverse transcription (RT) followed by Q-PCR analysis using primer pairs that spanned the circularization junction (Supplementary Fig. 1B) confirmed these changes and revealed that exposure to CLP for 48 h significantly increased circHIPK3 levels in the small intestinal mucosa (Figure 1C), while it did not change the levels of circRMDN2, circSTIM1, and circPABPN1.
Figure 1.
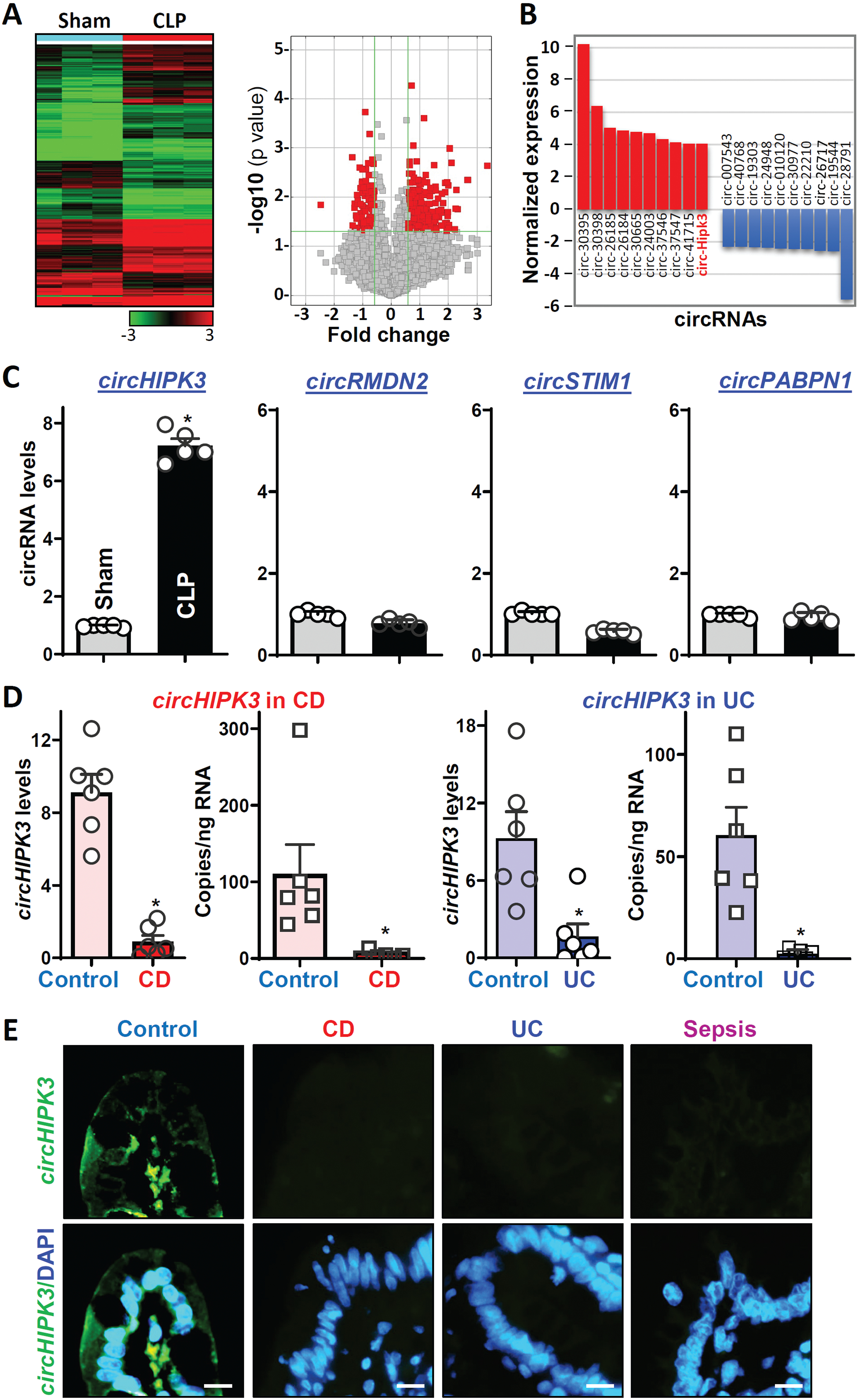
Changes in expression of circRNAs in the intestinal mucosa in septic mice and patients with critical illnesses. (A) Heat map (a) and scatter plot (b) depictions of circRNAs differentially expressed in the small intestinal mucosa in mice exposed to CLP for 48 h as measured by circRNA microarray. (B) Differential expression analysis of circRNAs in results described in A. Values are the means from three animals. (C) Levels of mucosal circHIPK3 and other circRNAs in the small intestine as examined by Q-PCR analysis. Values are the means ± SEM (n = 5). * P < 0.05 compared with sham mice. (D) Total levels and quantification of copy numbers of circHIPK3 in the intestinal mucosa from patients with active CD and UC as measured by Q-PCR and droplet digital Q-PCR analyses, respectively. Values are the means ± SEM (n = 6). * P < 0.05 compared with controls. (E) RNA-FISH analysis of circHIPK3 with fluorescent LNA-RNA detection probe in the mucosa from patients with CD, UC, and sepsis, as shown in yellow-green. Scale bars: 25 μm. All these experiments were repeated in tissue samples obtained from 4 control individuals, 4 patients with CD, 4 patients with UC 4 or 4 patients with sepsis and showed similar results.
To explore the implications of altered circHIPK3 clinically, we examined changes in the levels of circHIPK3 in human intestinal mucosal tissues from patients with critical illnesses. Small intestine and colonic mucosal tissues from patients with Crohn’s disease (CD), ulcerative colitis (UC), and sepsis were collected for measurements of circHIPK3 levels, whereas tissue samples from patients without gut mucosal injury/erosions, inflammation, or disrupted barrier served as controls. Interestingly, intestinal mucosal tissues from patients with active CD and UC exhibited decreased levels of circHIPK3, as measured by Q-PCR analysis (Figure 1D, left). Droplet digital Q-PCR analysis further showed that copy numbers of circHIPK3 in the mucosa decreased dramatically in IBD patients relative to controls (Figure 1D, right). In this study, circHIPK3 was read from at least 10,000 droplets in each well of the mucosal samples for absolute quantification. RNA-FISH detection in the mucosa of the intestine showed that circHIPK3 was predominantly distributed in the cytoplasm of epithelial cells and there were no significant differences in the circHIPK3 expression levels between villi and crypts in control individuals (Supplementary Figure 2A). Accordingly, the intestinal mucosal tissues obtained from all patients with CD, UC, and sepsis displayed a significant decrease in the levels of circHIPK3-FITC intensity when compared with those observed in controls (Figure 1E, and Supplementary Figure 2B). The decreased levels of the mucosal circHIPK3 in patients with CD, UC, and sepsis were associated with severe mucosal injury/erosions, inflammation, delayed healing (Supplementary Figure 2C), and gut barrier dysfunction; similar results were published in our previous studies.22,23
Analysis of cultured Caco-2 cells further showed that inhibition of cell proliferation and disrupted barrier by depleting cellular polyamines with D,L-α-difluoromethylornithine (DFMO; a specific inhibitor of polyamine biosynthesis)7 or by silencing RBP HuR were also associated with a decrease in the levels of cellular circHIPK3. Consistent with previous findings,38 treatment with DFMO for 6 days almost completely depleted cellular polyamines putrescine and spermidine. Decreasing the levels of cellular polyamines by DFMO also significantly decreased circHIPK3 abundance without affecting circPABPN1 levels (Supplementary Figure 2D, left). Exogenous putrescine given together with DFMO rescued circHIPK3 expression, which was associated with restoration of cell proliferation and epithelial barrier function.38,39 In addition, HuR silencing by transfection with HuR-directed siRNA (siHuR) also decreased the levels of cellular circHIPK3 (Supplementary Figure 2E, left), which was associated with G1 phase growth arrest and delayed epithelial repair after wounding as reported previously.29 Together, these results obtained from mice, human tissues, and cultured IECs indicate that expression levels of circHIPK3 in the intestinal mucosa are remarkably altered in response to various critical stresses and strongly suggest that circHIPK3 is widely implicated in the intestinal epithelium homeostasis and plays a role in gut mucosal pathologies.
CircHIPK3 regulates intestinal epithelial repair by altering multiple gene expression
To define the function of circHIPK3 in the regulation of intestinal epithelium repair after acute injury, we employed an in vitro epithelial injury model described previously.3,32 To determine if elevation of circHIPK3 abundance enhanced intestinal epithelial repair after wounding, the levels of cellular circHIPK3 were increased by transfecting Caco-2 cells with an expression vector. By 48 h after transfection with the circHIPK3 expression vector, circHIPK3 levels increased dramatically and specifically (Figure 2A, left), since there were no differences in the levels of circPABPN1, HIPK3 mRNA or HIPK3 protein (Figure 2A, right) between circHIPK3-transfected cells and cells transfected with empty control vector. As shown in Figure 2B, epithelial repair occurred quickly after wounding, as demonstrated by a significant increase in cells migrating over the denuded (wounded) area at 6 h; the wounded areas were further healed at 16 and 24 h thereafter. Ectopically expressed circHIPK3 stimulated epithelial repair after wounding, and the numbers of cells over the denuded area were significantly higher in cells overexpressing circHIPK3 than in cells transfected with control vector. In contrast, decreasing the levels of endogenous circHIPK3 by transfection with siRNA specifically targeting the circularization junction of circHIPK3 (si-cHIPK3) inhibited epithelial repair after wounding. Transient transfection with si-cHIPK3 remarkably decreased the levels of cellular circHIPK3 (Figure 2C, left), although it did not alter the levels of HIPK3 mRNA or HIPK3 protein (Figure 2C, right). CircHIPK3 silencing significantly inhibited epithelial repair after wounding (Figure 2D), and the epithelial repair decreased by ~52% at 6 h and ~64% at 16 h in si-cHIPK3-transfected cells, respectively, when compared with cells transfected with control siRNA (C-siRNA). On the other hand, neither circHIPK3 overexpression nor its silencing by transfection with si-cHIPK3 affected cell viability (Supplementary Figure 3A, bottom). In fact, cell viability increased by ~2-fold 48 hours after transfection to overexpress circHIPK3 but decreased significantly after circHIPK3 silencing by transfection with si-cHIPK3 (Supplementary Figure 3A, top) as measured by MTT assays. These results indicate that circHIPK3 contributes to normal intestinal epithelial repair after wounding.
Figure 2.
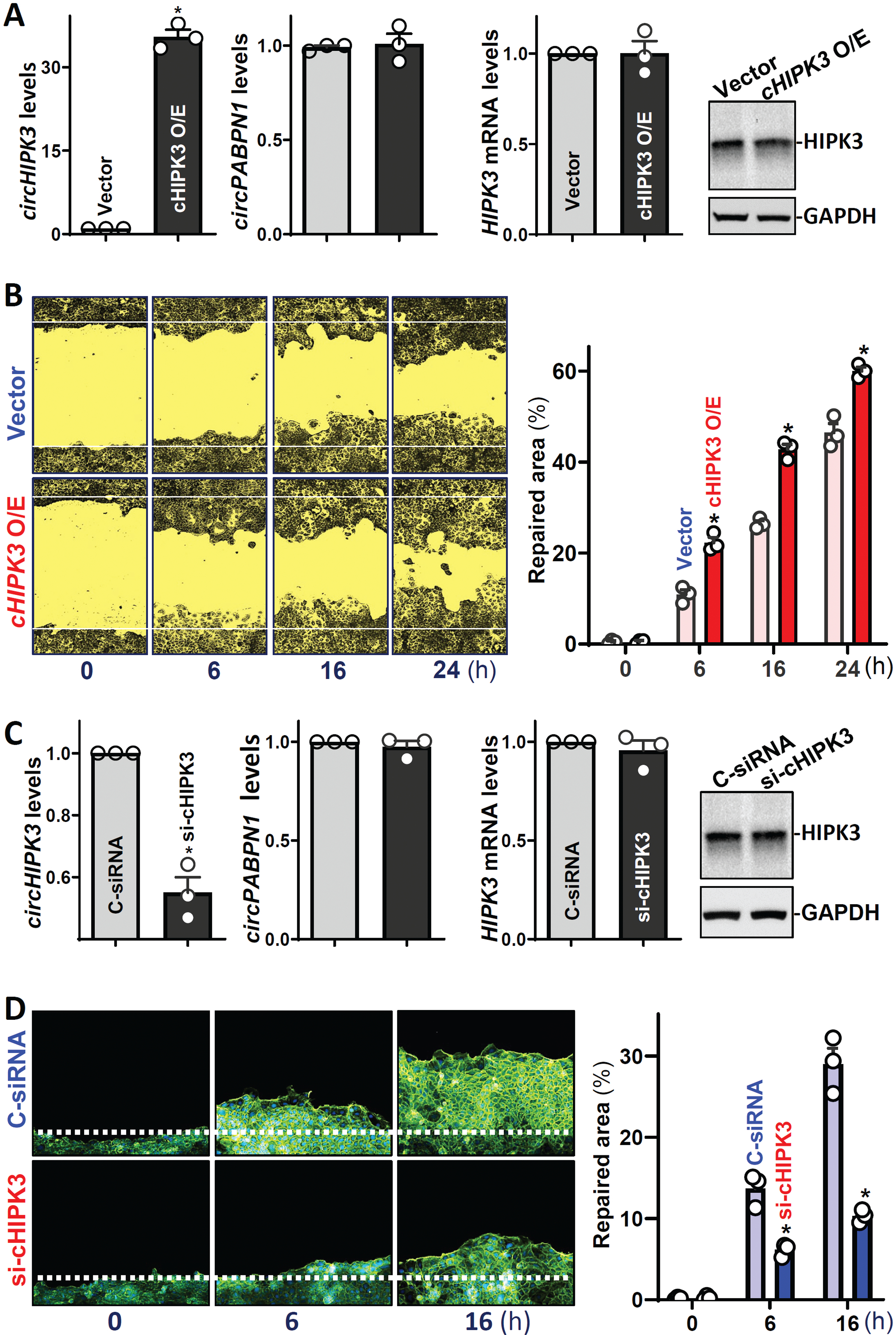
CircHIPK3 enhances intestinal epithelial repair after wounding. (A) Levels of circHIPK3 (left) and HIPK3 mRNA and HIPK3 protein (right) in Caco-2 cells 48 h after transfection with the circHIPK3 expression vector. Values are the means ± SEM (n = 3). * P < 0.05 compared with control. (B) Epithelial repair after wounding in cells described in A. Left: images of epithelial repair; and right: summarized data. Values are the means ± SEM (n = 6). (C) Levels of circHIPK3 (left) and the mRNA and protein (right) of HIPK3 48 h after transfection. Values are the means ± SEM (n = 3). (D) Epithelial repair after wounding in cells described in C. Values are the means ± SEM (n = 6).
To define the mechanism underlying circHIPK3 in the regulation of intestinal epithelial repair, we examined expression patterns of small Rho GTPases including Rac1, Cdc42, and RhoA (well-known regulators of cell migration)40 and proliferation-associated proteins in the presence or absence of cellular circHIPK3 after wounding. As shown in Figure 3A (top panel-left) and Supplementary Figure 3B, the levels of cellular Rac1, Cdc42, and RhoA increased rapidly in the control group after wounding, peaked at 6 h, and then began to decrease. By 24 h after wounding, abundance of cellular Rac1, Cdc42, and RhoA were returned to basal levels. Interestingly, circHIPK3 silencing by transfection with si-cHIPK3 prevented the increased levels of Rac1, Cdc42, and RhoA during the process of epithelial repair after wounding (Figure 3A, top panel-right). In circHIPK3-deficient cells, the levels of cellular Rac1, Cdc42, and RhoA after wounding were indistinguishable from their basal levels (unwounded monolayer). As expected, the levels of cyclin B1 (CCNB1) increased at 16 and 24 h after wounding, whereas the levels of p21 (CDNK1A) gradually decreased during the epithelial repair in control group (Figure 3A, low panel-left). CircHIPK3 silencing by si-cHIPK3 transfection abolished the increased cyclin B1 and decreased p21 after wounding (Figure 3A, low panel-right). Although there were no significant changes in the levels of p27 (CDKN1B) after wounding in cells transfected with C-siRNA, its abundance increased dramatically during the repair process in circHIPK3-deficient cells. On the other hand, wounding did not alter cellular levels of cyclin-dependent kinase 4 (CDK4) in both controls and circHIPK3-deficient cells.
Figure 3.
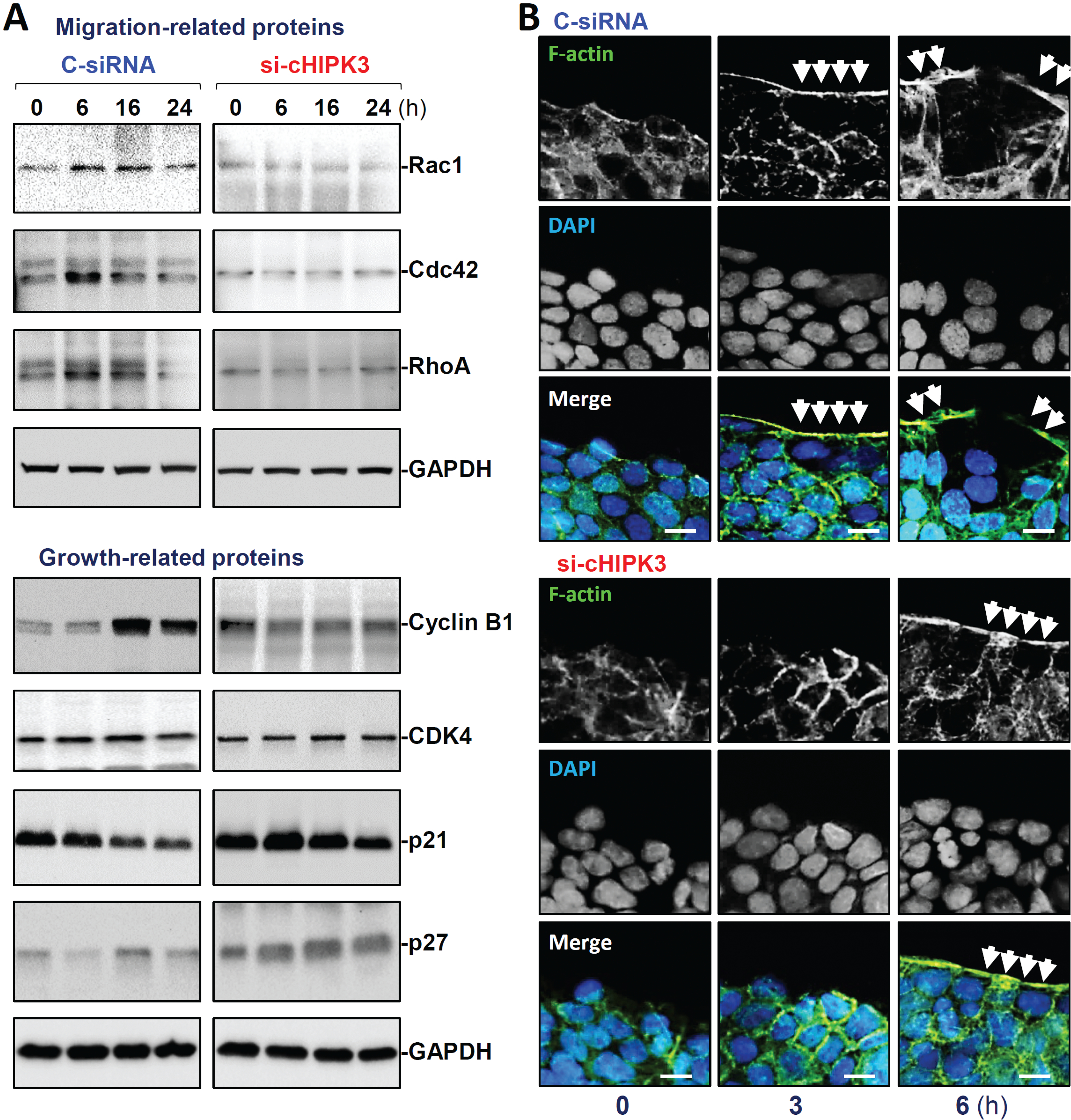
CircHIPK3 silencing prevents increase in the levels of migration- and growth-associated proteins and alters actin dynamics. (A) Immunoblots of various proteins relevant to cell migration (top) and growth (bottom) after wounding. (B) Immunostaining of F-actin in cells described in A. F-actin: green; nuclei: blue. Scale bar, 25 μm. Three separate experiments were performed and showed similar results.
Consistent with a reduction in the small Rho GTPases, circHIPK3 silencing decreased actin dynamics in migrating cells at early epithelial restitution after wounding. As shown in Figure 3B (top), significant actin-based plasma membrane protrusions such as membrane ruffles, blebs, and lamellipodia at the wounding edge were observed at 3 and 6 h after epithelial injury in control cells. However, the decreased levels of Rac1, Cdc42, and RhoA by circHIPK3 silencing were accompanied by a remarkable inhibition of the membrane protrusions (Figure 3B, low panel and Supplementary Figure 3C); 6 h after wounding, no typical actin-based membrane blebs and lamellipodia were observed in cells transfected with si-cHIPK3. In addition, circHIPK3 silencing also decreased the formation of F-actin stress fibers in the cytoplasm, similar to the observations identified in cells after silencing Rac1, Cdc42, or RhoA.3,7 These results indicate that circHIPK3 regulates intestinal epithelial repair after wounding at least in part by altering the activity of Rac1, Cdc42, and RhoA and modulating the expression levels of cyclin B1, p21, and p27.
CircHIPK3 enhances renewal of the intestinal epithelium
To define the role of circHIPK3 in the regulation of intestinal epithelium renewal, we examined the effect of decreasing the levels of circHIPK3 on the growth of Caco-2 cells and primary cultured intestinal organoids isolated from the small intestine in mice. In an in vitro model, the decrease in circHIPK3 levels by transfection of si-cHIPK3 (Figure 2C) lowered proliferation as indicated by a significant decrease in cell numbers (Figure 4A). We also examined the effect of circHIPK3 on proliferation of other cultured IECs and demonstrated that circHIPK3 silencing inhibited growth of HCT116 cells (Supplementary Figure 3D, left). The numbers of HCT116 cells decreased by ~20% on day 3 and by >60% on days 4, 5, 6 after transfection with si-cHIPK3, respectively, when compared with cells transfected with control siRNA. Similarly, circHIPK3 silencing inhibited proliferation of cultured IEC-6 cells (Supplementary Figure 3D, right). In an ex vivo model, decreasing circHIPK3 levels by transfection with si-cHIPK3 also inhibited the growth of intestinal organoids. As shown in Figure 4B, an intestinal organoid was initiated from a tiny proliferating crypt, but by 8 days after culture, the structures of organoids consisted of multiple buds and cells in both control and si-cHIPK3-transfected organoids. However, decreasing the levels of circHIPK3 by transfecting organoids with si-cHIPK3 (Supplementary Figure 4A) inhibited DNA synthesis in multiple cells markedly, as determined by a decrease in BrdU incorporation (Figure 4B, left panel and Supplementary Fig 4B), and it also decreased the sizes of intestinal organoids as shown by a reduction in their surface areas (Figure 4C, top) and the numbers of BrdU-positive cells in organoids (Figure 4C, low panel). In agreement with these results, circHIPK3 silencing in intestinal organoids also decreased the levels of Cdc42, Rac1, RhoA, and cyclin B1 proteins (Supplementary Figure 4C). On the other hand, circHIPK3 silencing failed to alter IEC differentiation, because there were no significant differences in Paneth cells (lysozyme-positive) and enterocytes (marked by villin staining) between circHIPK3-deficient organoids and organoids transfected with control siRNA (Figure 4D). Moreover, decreasing the levels of circHIPK3 by transfection with si-cHIPK3 did not affect the epithelial barrier function when examined in 2-dimensional (2D) intestinal organoid culture model (Supplementary Figure 4D). To exclude off-target effects of si-cHIPK3, another siRNA targeting circHIPK3 (si-cHIPK3b), was tested and showed similar results (Supplementary 5A).
Figure 4.
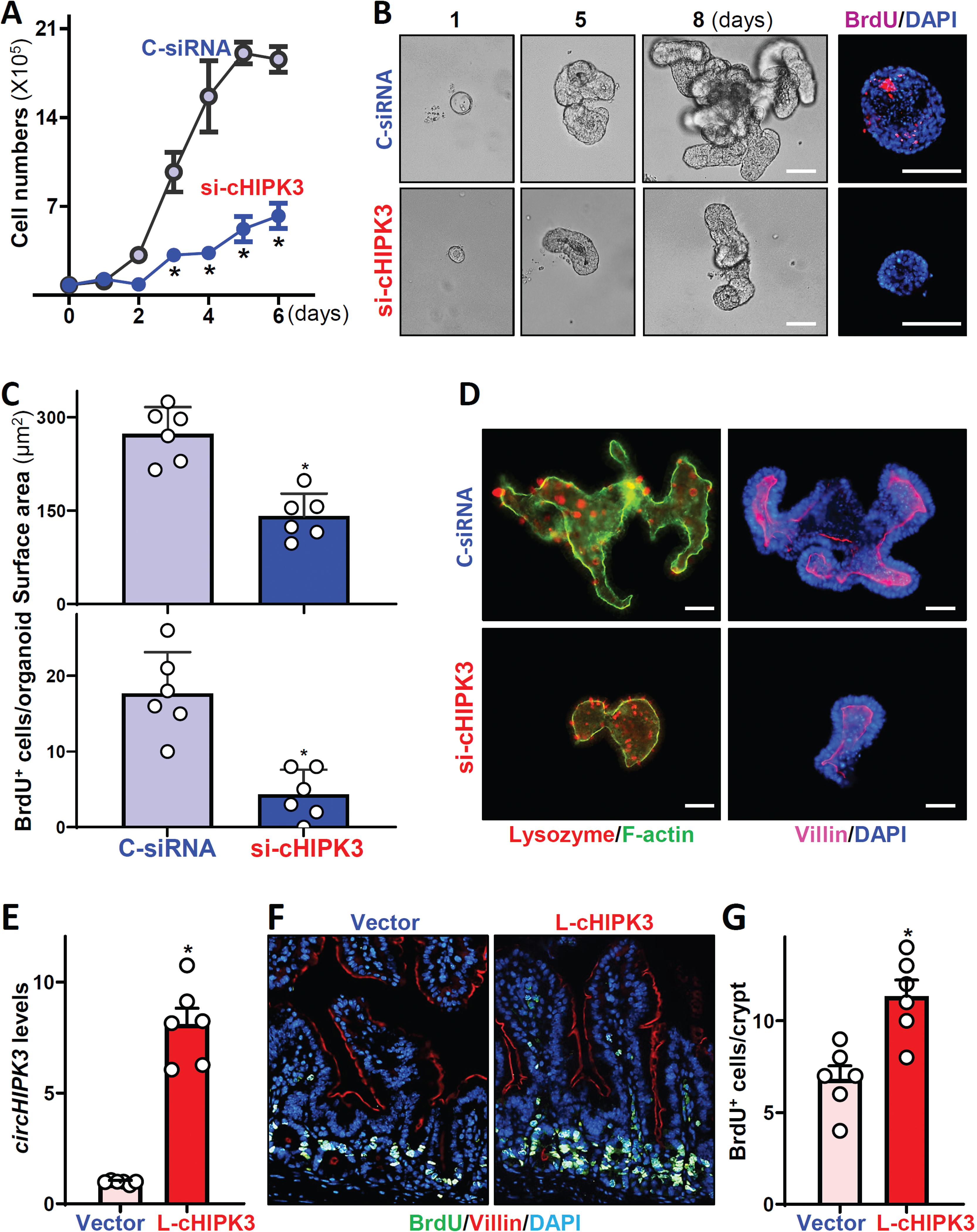
CircHIPK3 is essential for renewal of the intestinal epithelium. (A) Cell growth after circHIPK3 silencing in vitro. Values are means ± SEM (n = 3). * P < 0.05 compared with C-siRNA. (B) Growth of small intestinal organoids after circHIPK3 silencing ex vivo. Left: bright field microscopy analysis of growth of organoids on day 8; and right: confocal analysis of BrdU (red) and DAPI (blue) on day 3 after culture. Scale bars: 100 μm. (C) Quantification of surface area (top) and BrdU positive cells (bottom) of the organoids described in B (n = 6). (D) Immunostainings of lysozyme- (left) and villin-positive (right) cells in intestinal organoids on day 5 after transfection with si-cHIPK3 or C-siRNA. Red, lysozyme; purple, villin. Scale bars: 100 μm. (E) Levels of circHIPK3 in the small intestinal mucosa of mice on day 5 after intraperitoneal injection with a recombinant circHIPK3 lentiviral expression vector (L-cHIPK3) or empty control lentiviral vector (Vector). * P < 0.05 compared with control vector (n = 6). (F) Proliferating cells in small intestinal crypts as measured by BrdU labeling (1 h post-injection, green) in mice described in E. (G) Quantification of BrdU-positive cells in the mucosa described in F. * P < 0.05 compared with control vector (n = 6).
In an effort to define the in vivo importance of circHIPK3 in regulating intestinal mucosa growth, we increased the levels of circHIPK3 by infecting mice with a recombinant circHIPK3 lentiviral expression vector (L-cHIPK3) as described.31 L-cHIPK3 containing intron-HIPK3(exon2)-intron/GFP was under the control of the suCMV-promoter. By 5 days after infection with L-cHIPK3, there was a sustained increase (~8-fold) in the levels of circHIPK3 in the intestinal mucosa (Figure 4E) compared with those from animals infected with the control empty lentiviral vector. Consistent with findings obtained from cultured IECs and intestinal organoids, ectopic expression of circHIPK3 promoted growth of the small intestinal mucosa, as indicated by a significant increase in the proliferating crypt cell population (marked by BrdU) in L-cHIPK3-infected mice compared with animals infected with the control vector (Figure 4F). The numbers of BrdU-positive cells in crypts of the intestinal mucosa increased by ~2-fold in L-cHIPK3-infected mice relative to controls (Figure 4G). In contrast, overexpression of circHIPK3 in mice did not alter enterocyte migration along the crypt-villus axis as examined by BrdU pulse-chase assays (Supplementary Figure 5B). Together, these results indicate that circHIPK3 is essential for renewal of the intestinal mucosa but plays a smaller role in the regulation of IEC differentiation.
CircHIPK3 directly binds to miR-29b and inhibits miR-29b activity
Given the fact that circHIPK3 was mainly distributed in the cytoplasm of IECs (Figure 1E), we tested the possibility that circHIPK3 regulated the intestinal epithelium homeostasis at the posttranscriptional level by interacting with specific miRNAs. Through searching for circHIPK3-interacting miRNAs using the TargetScan algorithm, miR-29b was found to be a potential target of circHIPK3, as there were several potential miR-29b binding sites on circHIPK3 (Figure 5A). Since miR-29b-1–3p (miR-29b) is highly expressed in IECs and plays an important role in the control of intestinal epithelial renewal and gut barrier function,25,27 we investigated the direct interaction between these two ncRNAs by using biotinylated miR-29b, as described previously.22 miR-29b levels increased significantly 24 h after transfection with biotin-labeled miR-29b compared with cells transfected with control scramble oligomer (Figure 5B), although there were no changes in the levels of U6 RNA between these two groups. As expected, the levels of circHIPK3 in samples pulled down using biotin-labeled miR-29b were much higher than those in samples pulled down by using biotin-labeled scramble oligomer (Figure 5C, left). Biotin-labeled miR-29b did not pull down other circRNAs such as Cdr1as or circPABPN1; transfection with biotin-labeled miR-29b also failed to alter the levels of total input circRNAs (Figure 5C, right).
Figure 5.
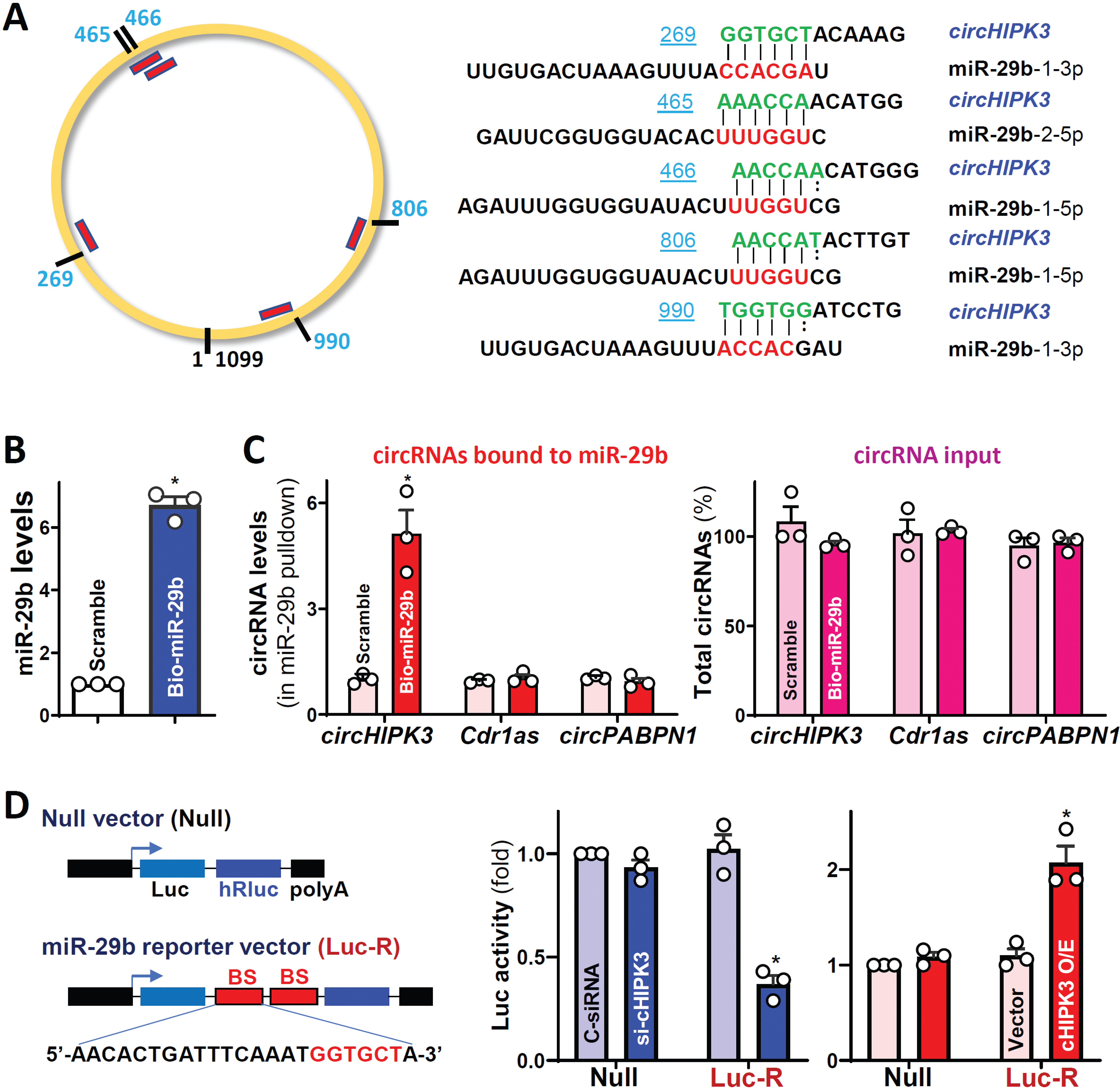
CircHIPK3 acts as a sponge of miR-29b in vitro. (A) Schematic of the putative binding sites of miR-29b on circHIPK3 transcript. (B) Levels of miR-29b 24 h after transfection with biotinylated miR-29b. Values are the means ± SEM (n = 3). * P <0.05 compared with control scramble oligomer. (C) Binding of biotinylated miR-29b to various circRNAs. Left, levels of circHIPK3, Cdr1as, and circPABPN1 in the materials pulled down by biotin-miR-29b; and right, levels of total input circRNAs. (D) Levels of luciferase reporter activity after circHIPK3 silencing or its overexpression. Left: Schematic of the chimeric mRNA vector bearing two perfect binding sites (BS) of miR-29b. * P <0.05 compared with cells transfected with C-siRNA or control vector.
To further examine if circHIPK3 could interact with miR-29b in IECs, the miRNA-luciferase reporter assay was employed, as reported.28 A vector expressing chimeric RNA that contained the luciferase coding region and a 3’-UTR bearing two perfect target sequences of miR-29b were constructed (Figure 5D, left). As shown in Figure 5D (middle), luciferase activity decreased in circHIPK3-deficient cells when transfected with the miR-29b reporter vector (Luc-R), primarily resulting from an increasing availability of miR-29b by circHIPK3 silencing. In contrast, luciferase activity increased in cells overexpressing circHIPK3 (Figure 5D, right), due to the decreased activity of miR-29b. Luciferase activity was not affected by circHIPK3 silencing or its overexpression when cells were transfected with null vector (without miR-29b binding sequences). These results suggest that circHIPK3 sequesters and inhibits miR-29b activity in cultured IECs.
Interaction between circHIPK3 and miR-29b regulates epithelial repair
To define the role of circHIPK3/miR-29b interaction in regulating intestinal epithelial repair after wounding, the following three sets of experiments were carried out. First, we examined the association of miR-29b with mRNAs encoding Rac1, Cdc42, RhoA, cyclin B1, CDK4, and p21 by RNA pulldown assays using biotin-labeled miR-29b.25 Twenty-four hours after transfecting biotin-labeled miR-29b (Figure 6A), Rac1, Cdc42, and cyclin B1 mRNAs were enriched in the materials pulled down by biotin-miR-29b but not in materials from cells transfected with a biotin-labeled control scramble oligomer (Figure 6B). However, biotin-miR-29b did not pull down the mRNAs encoding RhoA, CDK4, and p21. Additionally, transfection with biotin-labeled miR-29b did not affect whole cell input mRNA levels (data not shown).
Figure 6.
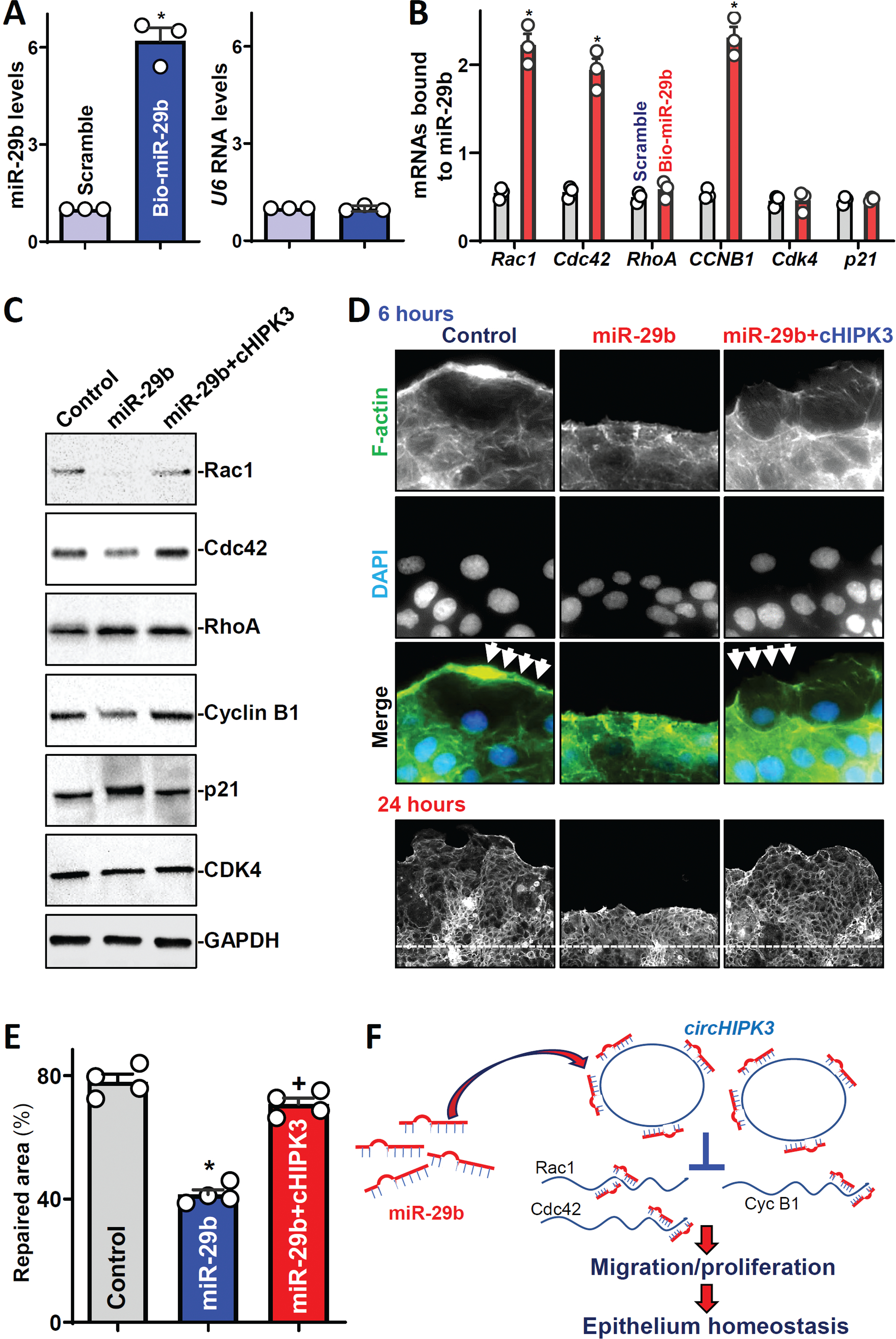
CircHIPK3 regulates intestinal epithelial repair by interacting with miR-29b. (A) Levels of miR-29b (left) and U6 RNA (right) 24 h after transfection with biotinylated miR-29b. Values are the means ± SEM (n = 3). * P <0.05 compared with control scramble. (B) Levels of mRNAs encoding Rac1, Cdc42, RhoA, cyclin B1 (CycB1), CDK4, and p21 in the materials pulled down by biotin-miR-29b. * P <0.05 compared with control scramble. (C) Immunoblots of various proteins. (D) Immunostaining of F-actin at 6 (top) and 24 (bottom) h after wounding in cells described in C. (E) Summarized data in cells described D (bottom). *, +P < 0.05 compared with control and miR-29b expression alone, respectively. (F) Model proposed to explain the influence of circHIPK3 upon intestinal epithelial repair after wounding.
Second, we investigated whether increasing the levels of miR-29b through transfection with its precursor (pre-miR-29b) inhibited the expression of Rac1, Cdc42, and cyclin B1 and if this inhibition was prevented by overexpressing circHIPK3. As shown in our previous studies,41,42 transient transfection with pre-miR-29b increased the levels of cellular miR-29b remarkably without altering the abundance of other miRNAs such as miR-222 and miR-503. Increasing miR-29b levels specifically inhibited the expression of Rac1, Cdc42, and cyclin B1, slightly elevated p21 abundance, and did not affect the levels of RhoA or CDK4 (Figure 6C, middle; Supplementary Figure 6A). On the other hand, the ectopic rise in circHIPK3 levels rescued expression of Rac1, Cdc42, and cyclin B1 in cells overexpressing miR-29b (Figure 6C, right). Elevation of cellular circHIPK3 also abolished miR-29-mediated induction in p21 levels, although biotin-miR-29b did not pull down p21 mRNA (Figure 6B). As Rac1 mRNA is a novel target of miR-29b identified in the current study, further analysis revealed that miR-29b repressed Rac1 expression by interacting with the coding region of Rac1 mRNA rather than with its 3’-UTR (Supplementary Figure 6B).
Third, we tested if increasing the levels of circHIPK3 prevented the miR-29b-induced inhibition of intestinal epithelial repair after wounding. Similar to observations in circHIPK-deficient cells (Figure 3), ectopically expressed miR-29b also inhibited actin-based plasma membrane protrusions at early epithelial restitution (Figure 6D; Supplementary Figure 6C) and delayed intestinal epithelial repair after wounding (Figure 6E). Consistent with its stimulatory effect on the expression of Rac1, Cdc42, and cyclin B1, elevation of cellular circHIPK3 levels rescued the formation of membrane protrusions in cells overexpressing miR-29b and restored the epithelial repair to near normal level. Interestingly, the levels of circHIPK3 increased rapidly after wounding, which was associated with a decrease in the levels of miR-29b (Supplementary Figure 6D). Taken together, our findings suggest a model whereby circHIPK3 regulates the intestinal epithelium homeostasis at least in part by interacting with miR-29b (Figure 6F). We propose that circHIPK3 acts by neutralizing miR-29b and preventing its repressive effect on the expression of Rac1, Cdc42, and cyclin B1 after wounding. CircHIPK3 also indirectly regulates RhoA, p21, and p27 via unknown mechanisms. By modulating the expression of these specific target transcripts, at least in part through miR-29b, elevation of circHIPK3 enhances IEC migration and proliferation, thereby contributing to the maintenance of the intestinal epithelial integrity under various stressful environments.
DISCUSSION
With the recent progress in high-throughput RNA-sequencing and bioinformatic analysis, tens of thousands of circRNAs have been identified from various animal genomes. Many circRNAs are highly stable and display tissue-specific expression patterns and evolutionary conservation,9,11 but the functions of most circRNAs in physiology and human diseases remain unknown. In the present study, we identified hundreds of circRNAs expressed differentially in the small intestinal epithelium after exposure to CLP-induced septic stress in mice, and found that circHIPK3 acts as a biological regulator of the intestinal epithelium homeostasis by enhancing epithelial repair after acute injury and by promoting constant renewal of the epithelium. Molecular characterization of circHIPK3 revealed that it may function as a molecular decoy for miRNAs in cultured IECs and increases expression of Rac1, Cdc42, and cyclin B1 by interacting with miR-29b. These findings link circRNA with the maintenance of the intestinal epithelial integrity and suggest that disruptions of the circHIPK3/miR-29b axis may underlie the pathogenesis of mucosal injury, delayed repair, and gut barrier dysfunction in patients with various critical illnesses.
The results reported here indicate that the epithelium of the intestinal mucosa expressed high levels of circHIPK3 and its levels were dramatically altered in various pathologies. CircHIPK3 levels increased in acute mucosal injury induced CLP in mice but decreased dramatically in the mucosal tissues from patients with active IBD and sepsis. The discrepancy in the expression patterns of circHIPK3 most likely stems from differences in the nature of the mucosal damage in CLP-mice versus critically ill patients.20–23 Intestinal mucosal injury induced by CLP in mice is acute and rapidly repaired, but mucosal injury in critically ill patients is chronic with unhealed wounds, persistently disrupted epithelial renewal, and massive mucosal inflammation. Injury and repair of the intestinal mucosa are separate, but closely linked biological processes. Repair occurs immediately after acute mucosal injury and can be coupled and even co-exist with mucosal injury. Changes in the levels of circHIPK3 and other factors observed in the damaged mucosa could be related either to new injury or to rapid repair of damaged tissues. In studies of mice and cultured IECs and intestinal organoids, we found that circHIPK3 is required for rapid mucosal repair and constant intestinal epithelial renewal. Thus, it is perhaps expected that chronic and unhealed injury and disrupted renewal of the intestinal mucosa observed in critically ill patients was associated with significantly reduced tissue levels of circHIPK3. Although the exact role of the altered levels of circHIPK3 in the pathogenesis of impaired repair and mucosal growth inhibition in patients with IBD and sepsis remains to be fully investigated, these findings expand our knowledge of the biological function of circHIPK3 in sustaining intestinal epithelial homeostasis.
The results presented here also show that circHIPK3 promotes early epithelial restitution after wounding by altering F-actin dynamics via activation of small GTPase signals. IECs rapidly migrate over the wounded area after superficial injury and this coordinated process is tightly regulated by numerous factors including small GTPases.3,45 Rho-family GTPases function as molecular switches by cycling between inactive GDP and active GTP-bound states of GTPases to quickly alter the subcellular organization of actin cytoskeleton.43 In humans, there are >20 members of the Rho family, but the relative contribution of each Rho GTPase is dependent on the environment and cell type.40 The roles of three GTPases, including Rac1, Cdc42, and RhoA, in the regulation of actin/myosin activity and subsequent cell migration have been extensively investigated in various cell types.43,44 Decreasing the levels of cellular GTPases or inactivation of their enzyme activity inhibits IEC migration over the wounded area and delays early epithelial restitution.45,46 In this study, expression levels of Rac1, Cdc42, and RhoA decreased significantly in circHIPK3-deficient cells after wounding, and this reduction in the levels of small GTPases was associated with a decrease in F-actin-containing protrusions and an inhibition of IEC migration, indicating the importance of small GTPase signaling in circHIPK3-mediated stimulation of intestinal epithelial restitution after wounding.
Another important finding from this study is that circHIPK3 acts as a molecular decoy for miR-29b. An increasing body of evidence indicates that circRNAs usually play their regulatory roles by acting as natural binding RNAs for miRNAs,11,12 although there also are studies suggesting that circRNAs do not necessarily function as miRNA sponges in human and mouse cells.47,48,49 CircHIPK3 harbors multiple binding sites for several miRNAs, but only a few of such binding sites are identified to trap particular miRNAs, based on experimental validations. It has been reported that circHIPK3 has potential binding sites for miR-29b,18 but the exact role of the circHIPK3/miR-29b interaction in the regulation of miR-29b activity remains to be elucidated. Using sequence-specific affinity assay and functional studies, our results revealed that circHIPK3 directly bound to miR-29b and inhibited miR-29b activity in cultured IECs. Moreover, the mRNAs encoding Rac1, Cdc42, and cyclin B1 are the targets of miR-29b, and their expression levels decline by miR-29b overexpression. However, ectopically expressed circHIPK3 prevented miR-29b-induced inhibition of Rac1, Cdc42, and cyclin B1 and restored epithelial repair to near normal level in cells overexpressing miR-29b, suggesting that there was ‘functional stoichiometry’ between circHIPK3 and miR-29b. In support of this notion, Cdc42 is known to be a direct target of miR-29 family members (miR-29a, miR-29b and miR-29c) in HeLa cells.50 On the other hand, although there were significant changes in the levels of RhoA, p21, and p27 in circHIPK3-deficent cells, miR-29b did not bind to the respective mRNAs and failed to reduce the levels of the encoded proteins. Together, these results indicate that circHIPK3 regulates the expression of Rac1, Cdc42, and cyclin B1 by directly interacting with miR-29b, but it modulates the production of RhoA, p21, and p27 via distinct processes, independent of miR-29b.
Our results have potential clinical relevance, because human intestinal mucosa with injury/erosions and inflammation from patients with IBD and sepsis exhibited decreased levels of circHIPK3. As reported,11,12,18,19 deregulated expression of circHIPK3 commonly occurs in patients with various diseases including different cancers, osteoarthritis, and age-related cataracts. Abnormalities in tissue circHIPK3 abundance can alter many biological processes including proliferation, migration, and apoptosis and also affect cancer growth, metastasis, and angiogenesis. In this study, we identified a novel role for circHIPK3 in maintaining intestinal epithelium integrity. Increasing the levels of cellular circHIPK3 enhanced epithelial repair after acute injury and sustained constant renewal of the intestinal epithelium. We also found evidence that circHIPK3 is required for increasing expression of Rac1, Cdc42, and cyclin B1 after wounding via its direct interaction with miR-29b. In sum, our results indicate that circHIPK3 plays an important role in intestinal epithelium homeostasis mainly by acting as a sponge for miR-29b. These findings help us to understand how circRNA/miRNA interaction controls intestinal mucosal adaptation under various stressful environments and uncover novel therapeutic targets to protect the intestinal epithelium integrity in pathologic conditions.
Supplementary Material
Supplementary Figure 1. Changes in cicrRNA expression profiles in the small intestinal mucosa of CLP-mice. (A) Summarized data showing changes in circRNA expression profiles in mice exposed to CLP for 48 as measured by circRNA microarray. (B) Methods used to confirm results obtained from microarray assays: a) RT-PCR primers; and b) PCR results. Green arrows indicates the location and direction of the divergent primers used for PCR analysis. The PCR product was analyzed in 1% agarose gel.
Supplementary Figure 2. (A) Distribution of circHIPK3 in the mucosa of small intestine and colon as measured by RNA-FISH assay. (B) Quantification of circHIPK3-FITC intensity in the mucosa from patients with CD, UC, or sepsis as described in Fig. 1E. * P < 0.05 compared with controls (n = 4). (C) H&E staining of intestinal mucosa from control individuals and patients with UC, CD or sepsis. (D) Levels of circHipk3 (left) and circPabpn1 (right) in Caco-2 cells treated with DFMO (5 mmol/L) with or without exogenous putrescine (Put, 10 μmol/L) for 6 days. Values are the means ± SEM (n = 3). * P < 0.05 compared with control or DFMO plus Put. (E) Levels of circHipk3 (left) and circPabpn1 (right) in cells transfected with siRNA targeting HuR (siHuR) or control siRNA (C-siRNA) for 48 h. * P < 0.05 compared with C-siRNA (n = 3).
Supplementary Figure 3. (A) Cell viability 48h after transfection with si-cHIPK3 or circHIPK3 expression vector, as measured by MTT assay. * P < 0.05 compared with C-siRNA or control vector (n = 3). (B) Semi-quantitative analysis of immunoblots of various proteins after wounding as described in Fig. 3A. * P < 0.05 compared with C-siRNA. (C) Quantification of cell plasma protrusions 6 h after wounding in cells treated as described in Fig. 3B. * P < 0.05 compared with C-siRNA. (D) circHIPK3 silencing by transfection with si-cHIPK3 inhibits growth of HCT-116 and IEC-6 cells. * P < 0.05 compared with C-siRNA (n = 3).
Supplementary Figure 4. (A) Levels of circHIPK3 in primarily cultured intestinal organoids on day 3 after transfection with si-cHIPK3 or C-siRNA. Values are the means ± SEM (n = 3). * P < 0.05 compared with C-siRNA. (B) BrdU intensity of the intestinal organoids treated as described in A. * P < 0.05 compared with C-siRNA (n = 6). (C) Levels of various proteins in intestinal organoids treated as described in A. Left, representative immunoblots; right, semi-quantitative analysis of immunoblots. * P < 0.05 compared with C-siRNA (n = 3). (D) Epithelial barrier function in 2D organoid culture model (left) as indicated by transepithelial electrical resistance (TEER) (middle) and FITC-dextran paracellular permeability (right) after circHIPK3 silencing as described in A (n = 6)
Supplementary Figure 5. (A) Effect of transfection with second siRNA targeting circHIPK3, si-cHIPK3b (a), on levels of circHIPK3 (b) and cell growth (c). Levels of cellular circHIPK3 were examined 48 h after the transfection. * P < 0.05 compared with C-siRNA (n = 3). (B) Intestinal epithelial cell migration in the small intestinal mucosa after circHIPK3 overexpression. Mice were intraperitoneally injected with the recombined circHIPK3 lentiviral vector (Lenti-cHIPK3) or control lentiviral vector (Vector), and BrdU was given on day 5 after the injection. Cell migration was examined 24 h after administration of BrdU (n = 6).
Supplementary Figure 6. (A) Quantitative results of immunoblots from densitometry analysis in cells transfected with miR-29b alone or co-transfected with miR-29b and cHIPK3 as described in Fig. 6C. *P < 0.05 compared with control or miR-29b+cHIPK3 (n = 3). (B) Ectopically expressed miR-29b inhibits activity of the Rac1 luciferase reporter in Caco-2 cells. Left, schematic of firefly luciferase reporter constructs containing fragments of the Rac1 5’-untranslated region (UTR), coding region (CR), and 3’-UTR. Right, levels of Rac1 luciferase reporter activity in cells transfected with the miR-29b expression vector for 48 h. Values are the means ± SEM (n = 3). * P < 0.05 compared with control. (C) Quantification of cell plasma protrusions 6 h after wounding in cells treated as described in Fig. 6D. *,+ P < 0.05 compared with control and miR-29b alone, respectively (n = 6). (D) Levels of circHIPK3 and miR-29 after wounding (6 h) in Caoco-2 cells. * P < 0.05 compared with C-siRNA.
BACKGROUND AND CONTEXT.
Circular RNAs (circRNAs) are a novel class of endogenous noncoding RNAs with closed loop circles. This study surveyed circRNAs essential for intestinal epithelium homeostasis.
NEW FINDINGS
Three hundred of circRNAs, including circHIPK3, were differentially expressed in the intestinal mucosa of septic-mice relative to sham mice. circHIPK3 levels decreased in the mucosa of patients with IBD and sepsis. circHIPK3 enhanced epithelial repair after wounding and promoted constant mucosal renewal partially by reducing miR-29b function.
LIMITATIONS
There are only limited results showing the exact role of circHIPK3 in the pathogenesis of IBD and sepsis-induced gut mucosal pathologies.
IMPACT
These findings point to circHIPK3/miR-29b axis as a novel therapeutic target for interventions to protect the epithelial integrity in patients with critical illnesses.
Funding
This work was supported by Merit Review Awards (to J.Y.W.; J.N.R.) from US Department of Veterans Affairs; grants from National Institutes of Health (DK57819, DK61972, DK68491 to J.Y.W.); and funding from the National Institute on Aging-Intramural Research Program, NIH (to M.G.).
Abbreviations used:
- CLP
cecal ligation and puncture
- IECs
intestinal epithelial cells
- LncRNAs
long noncoding RNAs
- miRNAs
microRNAs
- circRNAs
circular RNAs
- RBPs
RNA-binding proteins
- Q
quantitative
- RT
reverse transcription
Footnotes
Conflicts of interest
The authors disclose no conflicts.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Bankaitis ED, Ha A, Kuo CJ, et al. Reserve stem cells in intestinal homeostasis and injury. Gastroenterology 2018; 155:1348–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson-Rolf A, Zilbauer M, Koo BK, et al. Stem cells in repair of gastrointestinal epithelia. Physiology 2017; 32:278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu L, Zhuang R, Xiao L, et al. HuR enhances early restitution of the intestinal epithelium by increasing Cdc42 translation. Mol Cell Biol 2017; 37:e00574–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aihara E, Montrose MH. Importance of Ca2+ in gastric epithelial restitution-new views revealed by real-time in vivo measurements. Curr Opin Pharmacol 2014; 19:76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Assimakopoulos SF, Triantos C, Thomopoulos K, et al. Gut-origin sepsis in the critically ill patient: pathophysiology and treatment. Infection 2018; 46:751–760. [DOI] [PubMed] [Google Scholar]
- 6.Assimakopoulos SF, Triantos C, Maroulis M, et al.The role of the gut barrier function in health and disease. Gastroenterology Res 2018; 11:261–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao JN, Xiao L, Wang JY. Polyamines in gut epithelial renewal and barrier function. Physiology 2020; 35:328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell 2013; 152:1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Yang L, Chen LL. The Biogenesis, functions, and challenges of circular RNAs. Mol Cell 2018; 71:428–442. [DOI] [PubMed] [Google Scholar]
- 10.Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. EMBO J 2013;32:923–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kristensen LS, Hansen TB, Venø MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 2018; 37:555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384–388. [DOI] [PubMed] [Google Scholar]
- 13.Piwecka M, Glažar P, Hernandez-Miranda LR, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017;357:eaam8526. [DOI] [PubMed] [Google Scholar]
- 14.Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by circPABPN1. RNA Biol 2017;14:361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li XX, Xiao L, Chung HK, et al. Interaction between HuR and circPABPN1 Modulates autophagy in the intestinal epithelium by altering ATG16L1 translation. Mol Cell Biol 2020; 40:e00492–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P, He X. Current research on circular RNAs associated with colorectal cancer. Scand J Gastroenterol 2017; 52:1203–1210. [DOI] [PubMed] [Google Scholar]
- 17.Chen L, Zhang S, Wu J, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene 2017; 36:4551–4561. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016; 7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Zheng F, Xiao X, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep 2017; 18:1646–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubbard WJ, Choudhry M, Schwacha MG, et al. Cecal ligation and puncture. Shock 2005;24 Suppl 1:52–57. [DOI] [PubMed] [Google Scholar]
- 21.Yu TX, Chung HK, Xiao L, et al. Long Noncoding RNA H19 impairs the intestinal barrier by suppressing autophagy and lowering paneth and goblet cell function. Cell Mol Gastroenterol Hepatol 2020; 9:611–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao L, Wu J, Wang JY, et al. Long noncoding RNA uc.173 promotes renewal of the intestinal mucosa by inducing degradation of microRNA 195. Gastroenterology 2018; 154:599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao L, Li XX, Chung HK, et al. RNA-binding protein HuR regulates Paneth cell function by altering membrane localization of TLR2 via post-transcriptional control of CNPY3. Gastroenterology 2019; 157:731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L, Xiao L, Chung HK, et al. RNA-binding protein HuR regulates Rac1 nucleocytoplasmic shuttling through nucleophosmin in the intestinal epithelium. Cell Mol Gastroenterol Hepatol 2019; 8:475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao L, Rao JN, Zou T, et al. miR-29b represses intestinal mucosal growth by inhibiting translation of cyclin-dependent kinase 2. Mol Biol Cell 2013; 24:3038–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindemans CA, Calafiore M, Mertelsmann AM, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature 2015; 528:560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang JY, Cui YH, Xiao L, et al. Regulation of intestinal epithelial barrier function by long noncoding RNA uc.173 through interaction with microRNA 29b. Mol Cell Biol 2018; 38:e00010–e00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo R, Abdelmohsen K, Morin PJ, et al. Novel microRNA reporter uncovers repression of let-7 by GSK-3β. PLoS One 2013;8:e66330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L, Christodoulou-Vafeiadou E, Rao JN, et al. RNA-binding protein HuR promotes growth of small intestinal mucosa by activating the Wnt signaling pathway. Mol Biol Cell 2014; 25:3308–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu TX, Wang PY, Rao JN, et al. Chk2-dependent HuR phosphorylation regulates occludin mRNA translation and epithelial barrier function. Nucleic Acids Res 2011;39:8472–8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao L, Rao JN, Cao S, et al. Long noncoding RNA SPRY4-IT1 regulates intestinal epithelial barrier function by modulating the expression levels of tight junction proteins. Mol Biol Cell 2016; 27:617–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhuang R, Rao JN, Zou T, et al. miR-195 competes with HuR to modulate stim1 mRNA stability and regulate cell migration. Nucleic Acids Res 2013;41:7905–7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harter JL. Critical values for Duncan’s new multiple range tests. Biometric 1960; 16:671–685. [Google Scholar]
- 34.Li GX, Wang XM, Jiang T, et al. Berberine prevents damage to the intestinal mucosal barrier during early phase of sepsis in rat through mechanisms independent of the NOD-like receptors signaling pathway. Eur J Pharmacol 2014; 730:1–7. [DOI] [PubMed] [Google Scholar]
- 35.Chen S, He Y, Hu Z, et al. Heparanase mediates intestinal inflammation and injury in a mouse model of sepsis. J Histochem Cytochem 2017;65:241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leng Q, Lin Y, Zhan M, et al. An integromic signature for lung cancer early detection. Oncotarget 2018; 9:24684–24692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereira AL, Magalhães L, Pantoja RP, et al. The biological role of sponge circular RNAs in gastric cancer: Main players or coadjuvants? Cancers (Basel) 2020; 12:E1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu L, Li L, Rao JN, et al. Polyamine-modulated expression of c-myc plays a critical role in stimulation of normal intestinal epithelial cell proliferation. Am J Physiol 2005; 288:C89–C99. [DOI] [PubMed] [Google Scholar]
- 39.Wang JY, McCormack SA, Viar MJ, et al. Decreased expression of protooncogenes c-fos, c-myc, and c-jun following polyamine depletion in IEC-6 cells. Am J Physiol 1993; 265:G331–G338. [DOI] [PubMed] [Google Scholar]
- 40.Guan X, Guan X, Dong C, et al. Rho GTPases and related signaling complexes in cell migration and invasion. Exp Cell Res 2020; 388:111824. [DOI] [PubMed] [Google Scholar]
- 41.Ouyang M, Su W, Xiao L, et al. Modulation by miR-29b of intestinal epithelium homoeostasis through the repression of menin translation. Biochem J 2015; 465:315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Y, Chen G, Wang JY, et al. Post-transcriptional regulation of Wnt co-receptor LRP6 and RNA-binding protein HuR by miR-29b in intestinal epithelial cells. Biochem J 2016; 473:1641–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tybulewicz VLJ, Henderson RB. Rho family GTPases and their regulators in lymphocytes. Nat Rev Immunol 2009; 9:630–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Machesky LM, Hall A. Role of actin polymerization and adhesion to extracellular matrix in Rac- and Rho-induced cytoskeletal reorganization. J Cell Biol 1997; 138:913–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao JN, Li L, Golovina VA, et al. Ca2+-RhoA signaling pathway required for polyaminedependent intestinal epithelial cell migration Am J Physiol 2001;280:C993–C1007. [DOI] [PubMed] [Google Scholar]
- 46.Santos MF, McCormack SA, Guo Z, et al. Rho proteins play a critical role in cell migration during the early phase of mucosal restitution. J Clin Invest 1997; 100:216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014; 15:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 2015; 58:870–885. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Zhao R, Shen C, et al. Exosomal circHIPK3 released from hypoxia-induced cardiomyocytes regulates cardiac angiogenesis after myocardial infarction. Oxid Med Cell Longev 2020; 2020:8418407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park SY, Lee JH, Ha M, et al. MiR-29 miRNAs activate p53 by targeting p85a and CDC42. Nat Struct Mol Biol 2009; 16:23–29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Changes in cicrRNA expression profiles in the small intestinal mucosa of CLP-mice. (A) Summarized data showing changes in circRNA expression profiles in mice exposed to CLP for 48 as measured by circRNA microarray. (B) Methods used to confirm results obtained from microarray assays: a) RT-PCR primers; and b) PCR results. Green arrows indicates the location and direction of the divergent primers used for PCR analysis. The PCR product was analyzed in 1% agarose gel.
Supplementary Figure 2. (A) Distribution of circHIPK3 in the mucosa of small intestine and colon as measured by RNA-FISH assay. (B) Quantification of circHIPK3-FITC intensity in the mucosa from patients with CD, UC, or sepsis as described in Fig. 1E. * P < 0.05 compared with controls (n = 4). (C) H&E staining of intestinal mucosa from control individuals and patients with UC, CD or sepsis. (D) Levels of circHipk3 (left) and circPabpn1 (right) in Caco-2 cells treated with DFMO (5 mmol/L) with or without exogenous putrescine (Put, 10 μmol/L) for 6 days. Values are the means ± SEM (n = 3). * P < 0.05 compared with control or DFMO plus Put. (E) Levels of circHipk3 (left) and circPabpn1 (right) in cells transfected with siRNA targeting HuR (siHuR) or control siRNA (C-siRNA) for 48 h. * P < 0.05 compared with C-siRNA (n = 3).
Supplementary Figure 3. (A) Cell viability 48h after transfection with si-cHIPK3 or circHIPK3 expression vector, as measured by MTT assay. * P < 0.05 compared with C-siRNA or control vector (n = 3). (B) Semi-quantitative analysis of immunoblots of various proteins after wounding as described in Fig. 3A. * P < 0.05 compared with C-siRNA. (C) Quantification of cell plasma protrusions 6 h after wounding in cells treated as described in Fig. 3B. * P < 0.05 compared with C-siRNA. (D) circHIPK3 silencing by transfection with si-cHIPK3 inhibits growth of HCT-116 and IEC-6 cells. * P < 0.05 compared with C-siRNA (n = 3).
Supplementary Figure 4. (A) Levels of circHIPK3 in primarily cultured intestinal organoids on day 3 after transfection with si-cHIPK3 or C-siRNA. Values are the means ± SEM (n = 3). * P < 0.05 compared with C-siRNA. (B) BrdU intensity of the intestinal organoids treated as described in A. * P < 0.05 compared with C-siRNA (n = 6). (C) Levels of various proteins in intestinal organoids treated as described in A. Left, representative immunoblots; right, semi-quantitative analysis of immunoblots. * P < 0.05 compared with C-siRNA (n = 3). (D) Epithelial barrier function in 2D organoid culture model (left) as indicated by transepithelial electrical resistance (TEER) (middle) and FITC-dextran paracellular permeability (right) after circHIPK3 silencing as described in A (n = 6)
Supplementary Figure 5. (A) Effect of transfection with second siRNA targeting circHIPK3, si-cHIPK3b (a), on levels of circHIPK3 (b) and cell growth (c). Levels of cellular circHIPK3 were examined 48 h after the transfection. * P < 0.05 compared with C-siRNA (n = 3). (B) Intestinal epithelial cell migration in the small intestinal mucosa after circHIPK3 overexpression. Mice were intraperitoneally injected with the recombined circHIPK3 lentiviral vector (Lenti-cHIPK3) or control lentiviral vector (Vector), and BrdU was given on day 5 after the injection. Cell migration was examined 24 h after administration of BrdU (n = 6).
Supplementary Figure 6. (A) Quantitative results of immunoblots from densitometry analysis in cells transfected with miR-29b alone or co-transfected with miR-29b and cHIPK3 as described in Fig. 6C. *P < 0.05 compared with control or miR-29b+cHIPK3 (n = 3). (B) Ectopically expressed miR-29b inhibits activity of the Rac1 luciferase reporter in Caco-2 cells. Left, schematic of firefly luciferase reporter constructs containing fragments of the Rac1 5’-untranslated region (UTR), coding region (CR), and 3’-UTR. Right, levels of Rac1 luciferase reporter activity in cells transfected with the miR-29b expression vector for 48 h. Values are the means ± SEM (n = 3). * P < 0.05 compared with control. (C) Quantification of cell plasma protrusions 6 h after wounding in cells treated as described in Fig. 6D. *,+ P < 0.05 compared with control and miR-29b alone, respectively (n = 6). (D) Levels of circHIPK3 and miR-29 after wounding (6 h) in Caoco-2 cells. * P < 0.05 compared with C-siRNA.


