Significance
There is clear and irrefutable evidence that the Notch developmental signaling pathway is an important driver in many cancers, specifically in breast cancer. However, because of the ubiquity of this pathway in normal tissues, therapeutics targeting Notch have been associated with significant deleterious side effects. Herein, we provide evidence that USP9x selectively drives Notch in breast cancer and that therapeutic USP9x inhibition prevents tumor progression without collateral damage to normal tissues.
Keywords: USP9x, Notch, cytokine, breast cancer, TAMs
Abstract
Triple-negative breast cancer (TNBC) is a breast cancer subtype that lacks targeted treatment options. The activation of the Notch developmental signaling pathway, which is a feature of TNBC, results in the secretion of proinflammatory cytokines and the recruitment of protumoral macrophages to the tumor microenvironment. While the Notch pathway is an obvious therapeutic target, its activity is ubiquitous, and predictably, anti-Notch therapies are burdened with significant on-target side effects. Previously, we discovered that, under conditions of cellular stress commonly found in the tumor microenvironment, the deubiquitinase USP9x forms a multiprotein complex with the pseudokinase tribbles homolog 3 (TRB3) that together activate the Notch pathway. Herein, we provide preclinical studies that support the potential of therapeutic USP9x inhibition to deactivate Notch. Using a murine TNBC model, we show that USP9x knockdown abrogates Notch activation, reducing the production of the proinflammatory cytokines, C-C motif chemokine ligand 2 (CCL2) and interleukin-1 beta (IL-1β). Concomitant with these molecular changes, a reduction in tumor inflammation, the augmentation of antitumor immune response, and the suppression of tumor growth were observed. The pharmacological inhibition of USP9x using G9, a partially selective, small-molecule USP9x inhibitor, reduced Notch activity, remodeled the tumor immune landscape, and reduced tumor growth without associated toxicity. Proving the role of Notch, the ectopic expression of the activated Notch1 intracellular domain rescued G9-induced effects. This work supports the potential of USP9x inhibition to target Notch in metabolically vulnerable tissues like TNBC, while sparing normal Notch-dependent tissues.
Triple-negative breast cancer (TNBC), which can be subclassified into basal-like, mesenchymal, immunomodulatory, and luminal androgen subtypes (1), is characterized by the lack of estrogen receptor (ER), progesterone receptor, and the human epidermal growth factor receptor 2. TNBC primarily affects young women in the prime of life, people of African ancestry, and those with BRCA1 mutations. This subtype accounts for only 20% of all breast cancers yet is responsible for a disproportionate number of breast cancer deaths because of its aggressive nature and the lack of effective, targeted treatment (2).
The Notch signaling pathway is an intercellular communication system that tightly regulates tissue patterning during development and adult tissue homeostasis. It plays a central role in normal physiology, controlling a wide range of cellular properties such as stem cell maintenance, cell fate determination, epithelial cell polarity/adhesion, cell division, and apoptosis (3). Aberrantly activated Notch is strongly associated with various malignancies, including breast cancer. The expression of the Notch ligand JAG1 and Notch activation are hallmarks of TNBC and are independent predictors of outcome (4–6).
Compared with less aggressive breast cancer subtypes, TNBCs are heavily infiltrated by immune cells, and the pattern of immune infiltration is strongly associated with outcome. High tumor-associated macrophage (TAM) count is inversely related to survival (7–9), while high tumor-infiltrating lymphocyte (TIL) count is associated with improved survival (10–14). Our recent work shows that Notch shapes the innate immune response in cancer (15). Notch regulates the expression of powerful proinflammatory cytokines in TNBC, including C-C motif chemokine ligand 2 (CCL2) and interleukin-1 beta (IL-1β), which play fundamental roles in TAM recruitment, and predict poor outcome in breast cancer (16–22). Notch also regulates the TGF-β–mediated activation of tumor cells by TAMs, closing a Notch-dependent paracrine signaling loop between these two cell types. This results in a vicious cycle of TAM infiltration and amplified cytokine secretion. TAMs promote cancer by stimulating tumor angiogenesis, tumor cell migration and invasion, and by suppressing tumoricidal cytotoxic–T lymphocytes (CTL) (23–25).
While Notch is considered a potential therapeutic target in TNBC, the benefits of anti-Notch drugs have been outweighed by substantial on-target side effects that reflect the ubiquity of the pathway in normal tissues (26, 27). Obviously, targeting Notch exclusively in cancer cells would have significant, clinical potential. Our previous findings demonstrate that the pseudokinase tribbles homolog 3 (TRB3) and the deubiquitinase (DUB) USP9x are critical to Notch activation in human TNBC cell lines. USP9x forms a protein complex with TRB3 and Mind bomb 1 (MIB1), protecting these proteins from ubiquitin-dependent degradation, and stimulates Notch activity through two independent mechanisms: 1) through TRB3 stabilization and JAG1 up-regulation (28) and 2) through deubiquitination and the activation of MIB1, an E3 ligase required for JAG1 ubiquitination-mediated endocytosis and Notch activation (29). As a sensor of cellular stress, TRB3 is markedly up-regulated during nutrient deprivation, hypoxia, and endoplasmic reticulum stress (ERS) (30, 31), prominent features of TNBC associated with tumor progression and relapse (32). These findings identify USP9x as a putative, therapeutic target, selective to metabolically stressed tumor tissue.
In this study, using a murine TNBC model, namely K14cre;Brca1FL/FL;p53FL/FL (KBP), we demonstrate that USP9x loss of function in mouse mammary tumors results in Notch pathway inactivation, remodeling of the tumor microenvironment (TME) to a “hot” immunoreactive phenotype, and suppression of tumor growth.
Results
Cellular Stress Induces the Notch-TRB3-USP9x Axis in Murine TNBC Cells.
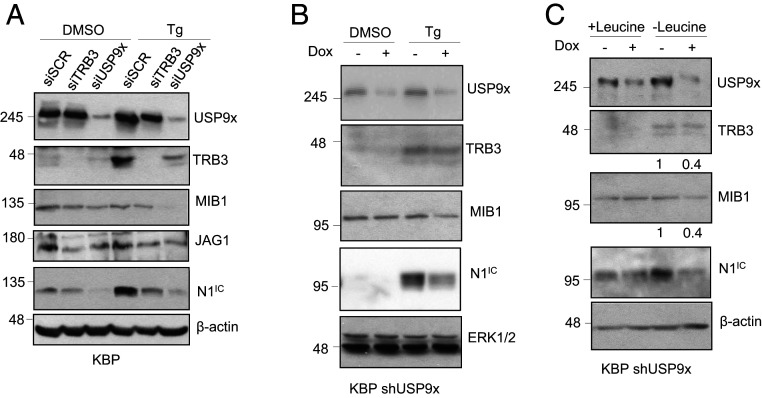
To confirm our previously reported findings in human cells and to prepare for preclinical studies in rodents, we examined the interactions between USP9x and Notch in a mammary carcinoma cell line derived from KBP mice. In these mice, Cre recombinase expressed under the control of the human keratin (K)14 promoter induces specific deletion of BRCA1 and p53 in mammary basal epithelial cells, resulting in the formation of mammary carcinomas reminiscent of human TNBC (33). In KBP cells, TRB3 expression was up-regulated and the Notch pathway activated (as measured by the presence of intracellular NOTCH1, N1IC) upon ERS mediated by thapsigargin, a noncompetitive inhibitor of the sarco/endoplasmic reticulum Ca+2 ATPase (Fig. 1A). Consistent with the role of TRB3 as a transcriptional regulator of JAG1, small interfering (si)RNA–mediated knockdown (KD) of TRB3 reduced both JAG1 protein and Notch activation during ERS. Consistent with our findings in human cells (29), USP9x KD resulted in reduced TRB3 and MIB1 and loss of the Notch response. These effects were reproduced using doxycycline (Dox)-inducible RNA interference, targeting a different sequence within USP9x (KBP short hairpin [sh]USP9x; Fig. 1B) and under nutrient stress mediated by the lack of the essential amino acid, leucine, in the cell growth medium (Fig. 1C). Ets-1 and Itch, two other proteins involved in cancer development and known to be deubiquitinated and stabilized by USP9x (34, 35), were examined in KBP cells, and no significant effect of USP9x KD was seen on either (SI Appendix, Fig. S1).
Fig. 1.
Cellular stress induces TRB3/USP9x-dependent Notch activation in KBP cells. (A) Immunoblot of USP9x, TRB3, MIB1, JAG1, and N1IC in murine KBP cells transfected with either scrambled control (siSCR), TRB3, or USP9x siRNA and treated with either DMSO or 1 μM thapsigargin (Tg). (B) Immunoblot of USP9x, TRB3, MIB1, and N1IC in KBP cells stably expressing a Dox-inducible, shRNA–targeting USP9x, without (−) or with (+) Dox and treated with either DMSO or Tg. (C) Immunoblot of USP9x, TRB3, MIB1, and N1IC in KBP shUSP9x cells cultured in complete (+Leucine) or leucine-deprived (−Leucine) medium without and with Dox. Densitometry of the TRB3 and MIB1 protein bands are presented, relative to β-actin. The expression of β-actin or ERK1/2 are included as loading controls. Molecular weight markers are shown in kilodaltons.
To further demonstrate the importance of USP9x as a regulator of Notch, KBP shUSP9x cells were transduced with a retrovirus-expressing, activated, and cleaved Notch1 receptor (KBP shUSP9x-N1IC) and tested for the rescued expression of Notch target genes Il1b (15) and Plau (36) after USP9x KD (SI Appendix, Fig. S2). Indeed, the thapsigargin-induced expression of the Il1b and Plau genes was USP9x dependent, and this effect was lost in cells that constitutively expressed N1IC. Interestingly, in the absence of cellular stress, N1IC alone was insufficient to up-regulate Il1b and Plau, suggesting the requirement of additional stress-induced factors. These data demonstrate the importance of USP9x as a mediator of Notch activation during conditions of cellular stress in murine TNBC cells.
Murine TNBC Progression Is USP9x Dependent.
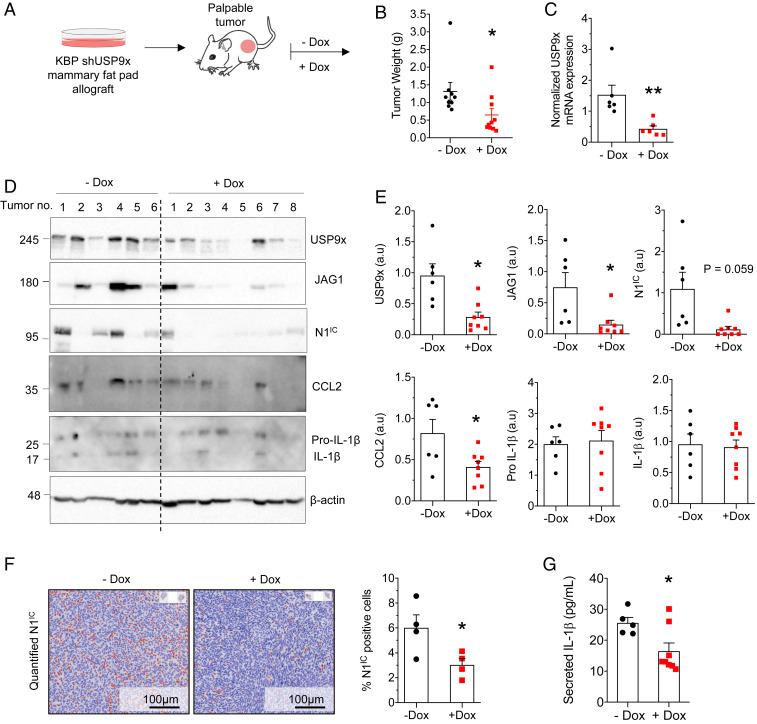
To test whether USP9x is required for tumor progression, we modeled USP9x loss of function in a murine TNBC allograft (Fig. 2A). KBP shUSP9x cells were engrafted into the mammary fat pads of syngeneic, immune-competent mice. Compared with untreated animals, treatment with Dox resulted in smaller tumors (Fig. 2B) with reduced expression of USP9x, JAG1, and N1IC (Fig. 2 C–F).
Fig. 2.
The USP9x-TRB3-Notch axis is required for the progression of murine TNBC tumors. (A) Schematic diagram of the experimental design (see main text for details). (B) Weight of KBP shUSP9x allografts in mice given regular (−Dox, n = 9 tumors) or Dox-containing diet (n = 10 tumors) at tumor onset. (C) qPCR analysis of USP9x in KBP shUSP9x tumors 4 wk after tumor onset. Expression levels were normalized to RNA expression of GAPDH (n = 6 tumors per group). (D) Immunoblot of USP9x, JAG1, N1IC, CCL2, Pro-IL-1β (precursor), and IL-1β (mature) in lysates from KBP shUSP9x tumors. The expression of β-actin is included as the loading control. Molecular weight markers are shown in kilodaltons. (E) Expression of USP9x, JAG1, N1IC, CCL2, pro-IL-1β, and IL-1β from D, determined by densitometry and presented relative to β-actin (a.u = arbitrary units). (F) N1IC expression determined by immunohistochemistry (representative images from KBP shUSP9x tumors treated without and with Dox; Left) and quantified according to N1IC-positive cell density (Right) (n = 4 tumors per group). (Scale Bars, 100 µm.) (G) Enzyme-linked immunosorbent assay of secreted IL-1β in the conditioned media of tumor cells from KBP shUSP9x tumors (−Dox, n = 5 tumors and +Dox, n = 8 tumors). Error bars represent SEM. *P < 0.05 and **P < 0.01. P values are determined by the two-tailed Student’s t test with Welch’s correction for unequal variances.
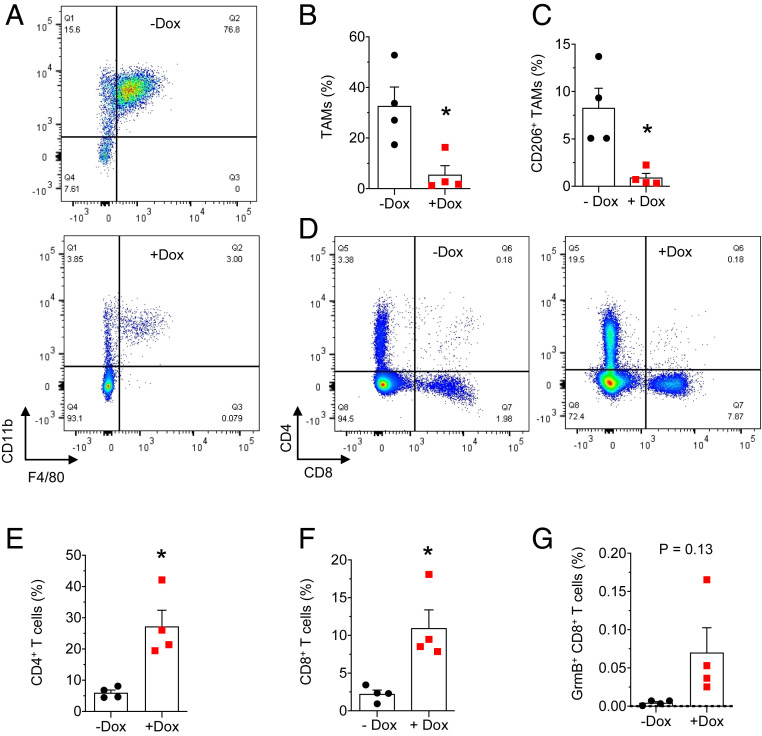
Since inflammation and immune evasion are Notch-regulated hallmarks of tumor progression, we examined the effect of USP9x depletion on cytokine secretion and immune infiltration. USP9x KD resulted in reduced intracellular CCL2, while the intracellular precursor and mature forms of IL-1β were unchanged (Fig. 2 D and E). Since basal-like breast cancer cells express elevated levels of inflammasome components (15), which facilitate the maturation and secretion of IL-1β (37), we tested for secreted IL-1β; predictably, USP9x KD tumors secreted decreased amounts compared with the untreated group (Fig. 2G). Accordingly, TAM recruitment was reduced by sixfold (Fig. 3 A and B) after USP9x KD. This was accompanied by a ninefold decrease in the immunosuppressive, M2-polarized TAM subtype identified by the CD206 marker (38) (Fig. 3C) and a fivefold increase in both CD4+ and CD8+ T cells (Fig. 3 D–F), with granzyme B–positive, cytotoxic CD8+ T cells demonstrating a trend toward increased infiltration (Fig. 3G). To rule out the effects of Dox treatment alone, mice with KBP tumors were treated with or without drugs, and no difference in tumor weight or immune infiltration was observed (SI Appendix, Fig. S3). Collectively, these findings support a model in which USP9x-mediated Notch signaling drives a protumoral inflammatory response.
Fig. 3.
USP9x regulates the inflammatory response in KBP allografts. (A and B) Representative flow cytometry plots (A) and the quantification of TAM infiltration (Gr-1− F4/80+ CD11b+) (B) in KBP shUSP9x tumors from mice treated without and with Dox (n = 4 tumors per group). (C) Quantification of M2-polarized TAMs (Gr-1− F4/80+ CD11b+ CD206+). (D–G) Representative flow cytometry plots (D) and quantification of CD4+ (E), CD8+ (F), and cytotoxic (CD8+ GrmB+) (G) T cell infiltrates in KBP shUSP9x tumors. All values are expressed as a percentage of CD45+ cells. Error bars represent SEM. * = P < 0.05. P values are determined by the two-tailed Student’s t test with Welch’s correction for unequal variances.
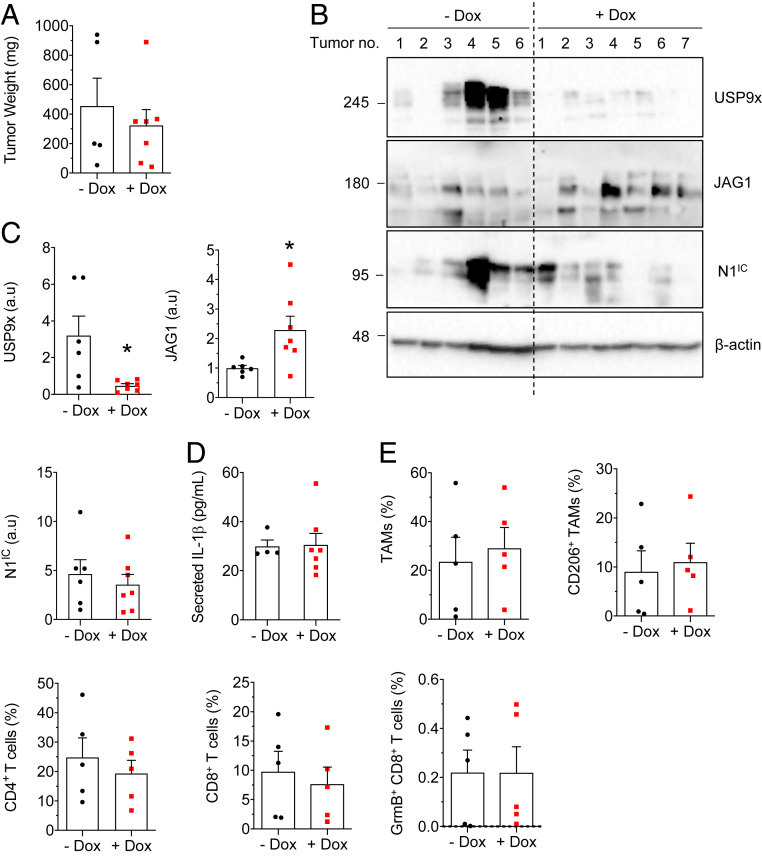
To prove that Notch is a mediator of USP9x in mammary tumors, allografts were created using KBP shUSP9x in which Notch was constitutively activated (KBPshUSP9x-N1IC). In these tumors, weight was unaffected by USP9x KD (Fig. 4A). JAG1 levels were increased upon USP9x KD (Fig. 4 B and C), likely owing to the ectopic, N1IC-mediated up-regulation of JAG1 in cells incapable of undergoing USP9x-MIB1–mediated JAG1 recycling (29, 39–41). N1IC expression (Fig. 4 B and C), IL-1β secretion (Fig. 4D), and immune infiltration (Fig. 4E) were unaffected by USP9x KD in KBPshUSP9x-N1IC tumors. Supporting the cell culture data (SI Appendix, Fig. S2), these data demonstrate that ectopic N1IC can rescue the effects of USP9x KD and they implicate Notch as the downstream effector of USP9x.
Fig. 4.
Ectopic N1IC rescues USP9x knockdown in KBP allografts. (A) Estimated weight of KBP shUSP9x; hN1IC allografts given regular (n = 6 tumors) or Dox diet (n = 7 tumors). (B) Immunoblot of USP9x, JAG1, and N1IC in KBP shUSP9x; hN1IC tumors 3 wk after tumor onset. The expression of β-actin is included as the loading control. Molecular weight markers are shown in kilodaltons. (C) Expression of USP9x, JAG1, and N1IC from B, determined by densitometry and presented, relative to β-actin. (D) Enzyme-linked immunosorbent assay analyses of secreted IL-1β in conditioned media from overnight cultures of tumor cells from KBP shUSP9x; hN1IC tumors (−Dox, n = 4 tumors and +Dox, n = 7 tumors). (E) Quantification (by flow cytometry) of TAMs, M2 TAMs, CD4+ T cells, CD8+ T cells, and cytotoxic T cells in KBP shUSP9x; hN1IC tumors 3 wk after tumor onset (n = 5 tumors per group). All values are expressed as a percentage of CD45+ cells. Error bars represent SEM. *P < 0.05. P values are determined by the two-tailed Student’s t test with Welch’s correction for unequal variances. a.u = arbitrary units.
Previous studies have described a link between hypoxia and Notch pathway activation in both physiological (42–45) and pathological settings (46, 47). KBP tumors underwent immunostaining for the hypoxia marker pimonidazole and N1IC. Predictably, hypoxic regions were enriched for N1IC-positive cells (SI Appendix, Fig. S4 A and B). This relationship persisted in untreated mice harboring KBP shUSP9x allografts (SI Appendix, Fig. S4C) but was lost after USP9x KD in Dox-treated mice (SI Appendix, Fig. S4D). These data confirm the relationship between cellular stress and USP9x-mediated Notch activation in mouse TNBC.
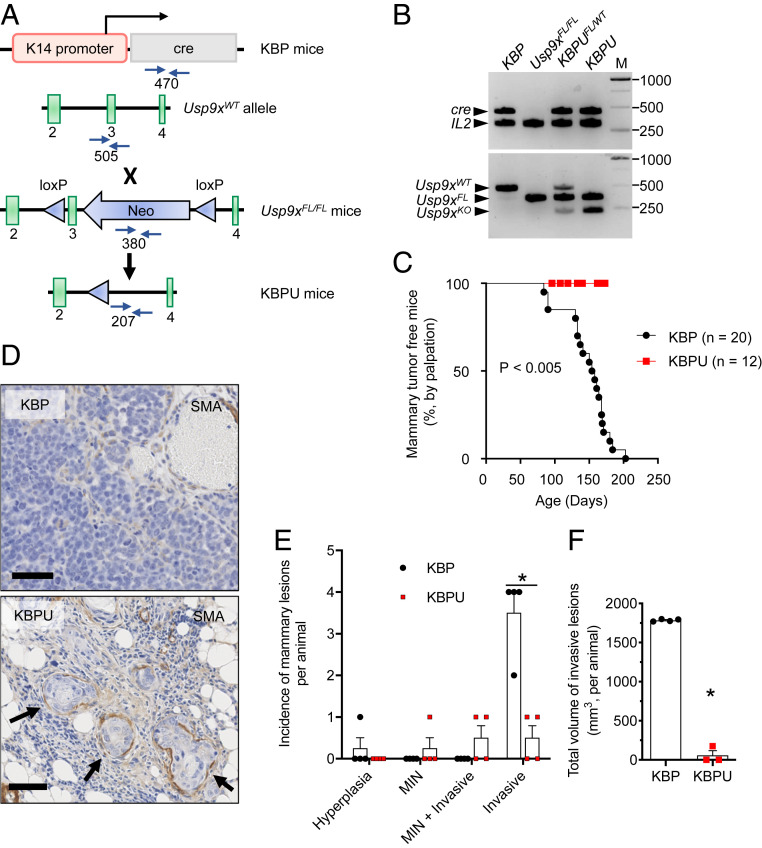
To further evaluate the importance of USP9x in mammary cancer, USP9x loss was explored in a model of spontaneous tumorigenesis. Usp9xFL/FL mice were interbred with KBP mice to generate KBP;Usp9xFL/FL (KBPU) mice. We confirmed K14 cre-dependent knockout (KO) of Usp9x in squamous epithelium of the tail (Fig. 5 A and B). As expected, all KBP control animals developed spontaneous mammary carcinomas by 200 d of life (Fig. 5 C and D). KBPU mice began to lose weight by 90 to 120 d but remained free of palpable mammary carcinomas until 175 d, at which point they had to be euthanized because of the emergence of aggressive oral papillomas, infection, septicemia, or other undiagnosed causes for failure to thrive. At euthanasia, KBPU mammary glands contained primarily mammary intraepithelial neoplasms (including ductal carcinomas in situ) (Fig. 5 D and E). KBPU animals also contained invasive lesions, but these were less numerous and smaller than those found in age-matched KBP mice (Fig. 5 E and F). These findings suggest a requirement for USP9x during the progression of premalignant mammary neoplasms to invasive carcinoma.
Fig. 5.
USP9x loss of function suppresses tumor progression in a model of spontaneous murine TNBC. (A) Schematic of the generation of KBPU mice via KBP × Usp9xFL/FL cross-breeding. In the conditional allele, loxP sites (blue triangles) flank exon 3 of the endogenous Usp9x locus and the neomycinR cassette (Neo). Primer sites used for genotyping (blue arrows) and the expected PCR product sizes are shown. (B) Confirmation of conditional KO of Usp9x in genomic DNA isolated from tail tissue obtained from KBP × Usp9xFL/FL crosses. Shown is the PCR using primers to detect cre and Il2 (internal control, 324 bp) genes (Top) and the Usp9x wild-type, floxed, and KO alleles (Bottom). KBP and Usp9xFL/FL mice are shown as controls. Note that tail DNA from the KBP × Usp9xFL/FL crosses contains both the Usp9xFL and Usp9xKO alleles resulting from heterogenous K14 expression in tail tissue. M = molecular weight markers in base pairs. (C) Kaplan–Meier analysis showing the latency of palpable mammary tumors in KBP (n = 20 mice) and KBPU mice (n = 12 mice). The P value is determined by the log-rank test. (D) Representative images of anti-SMA–stained sections of mammary glands from KBP and KBPU mice (euthanized at 175 and 173 d of life, respectively). Note that the KBP mammary gland contains a typical invasive mammary carcinoma; the KBPU mammary gland demonstrates mammary intraepithelial neoplasia (MIN) contained within anti-SMA–stained basement membrane (arrows). (Scale bars, 50 µm.) (E) Incidence of mammary lesions per animal at euthanasia (age ∼165 to 180 d, n = 4 mice per group). (F) Total volume of invasive lesions in mammary glands of KBP and KBPU mice from C. Error bars represent SEM. P value is determined by the Student’s t test and corrected for multiple comparisons using the Holm–Sidak method.
Pharmacologic USP9x Inhibition Reduces Notch Activity and Tumor Growth and Remodels the Tumor Immune Microenvironment.
The emerging role of USP9x in multiple malignancies has prompted the development of small-molecule DUB inhibitors. G9 is a partially selective DUB inhibitor with activity against USP9x in mouse and human cells (48). In ubiquitination assays, a minimum of 0.5 μM G9 was required to prevent the deubiquitination of immunoprecipitated TRB3 (SI Appendix, Fig. S5A) and MIB1 (SI Appendix, Fig. S5B), with the appearance of dominant bands migrating at 62 and 121 kDa, the predicted molecular weights of these monoubiquitinated fusion proteins, respectively. These findings are consistent with a previous report that identified 0.312 to 1.25 μM G9 as the range for in vitro antitumor cell activity (48). Next, it was confirmed that G9 treatment resulted in reduced MIB1, TRB3, and N1IC under ERS (SI Appendix, Fig. S5C).
Progress toward therapeutically inhibiting Notch in various cancer types has been slow, as the pathway is critical for tissue homeostasis. The most prominent side effects induced by Notch antagonists are gastrointestinal (GI) toxicities, which occur because of a resulting imbalance between intestinal epithelial cell types maintained by Notch (49, 50). Our in vitro findings suggest the theoretical potential of USP9x inhibition as an antagonist of Notch in stressed conditions only. To explore this possibility, G9 was tested in mice at 12 mg/kg once every other day (SI Appendix, Fig. S6A). After 3 wk of treatment, compared with vehicle-treated mice, body weight was unaffected by G9, indicating no major toxicity (SI Appendix, Fig. S6B). Interestingly, small bowel secretory cells were slightly more numerous in G9-treated animals (SI Appendix, Fig. S6C), confirmed with Alcian blue–periodic acid–Schiff (AB-PAS) staining (SI Appendix, Fig. S6D) and consistent with the known role of Notch in promoting the differentiation to absorptive cells at the expense of cells of the secretory cell lineage (51–53). Thus, G9 treatment at the dose used results in subtle GI tract changes without the apparent GI toxicity associated with γ-secretase inhibitors. We also noted that short-term G9 treatment did not phenocopy the deleterious effects of genetic USP9x KO seen in KBPU animals.
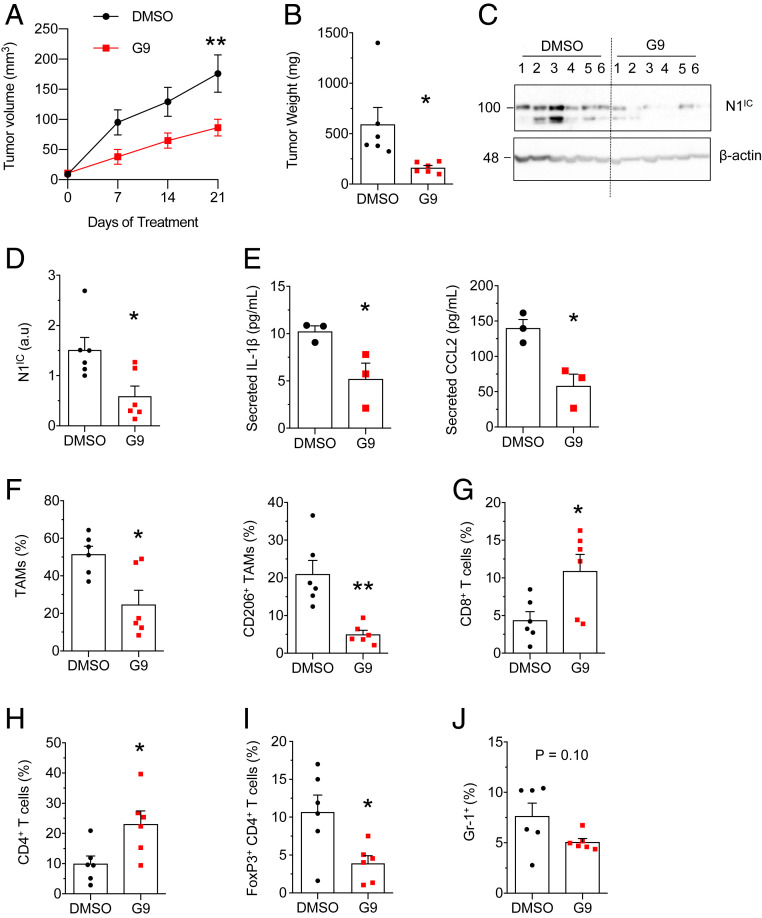
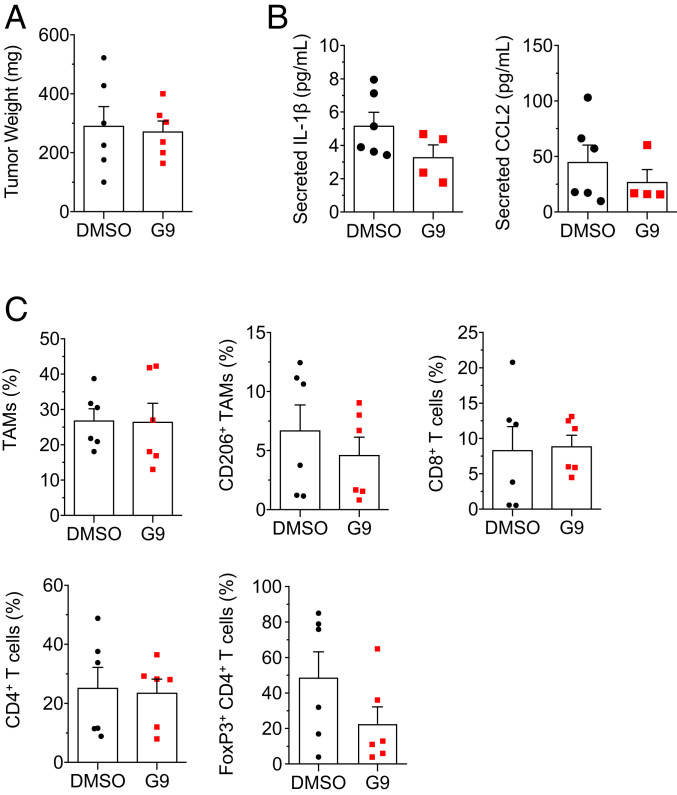
Next, the effect of G9 on tumorigenesis was tested in the KBP allograft model. Consistent with genetic USP9x KO, G9 impeded tumor growth (Fig. 6 A and B), decreased Notch activation (Fig. 6 C and D), and decreased the secretion of IL-1β and CCL2 (Fig. 6E). With G9, TAM infiltration, specifically the CD206+ subtype, was reduced (Fig. 6F), and CD8+ (Fig. 6G) and CD4+ T (Fig. 6H) lymphocytes were increased. Within the CD4+ population, there was a proportional decrease in the FoxP3+ immunosuppressive (regulatory T cell) subtype (Fig. 6I). In addition, there was a trend toward decreased myeloid-derived suppressor cells (Fig. 6J), a population of activated, immature myeloid cells, involved in inhibiting adaptive immune response against tumors (54). Again, implicating Notch, ectopic N1IC reversed the effect of G9 on tumor weight (Fig. 7A), secreted IL-1β and CCL2 (Fig. 7B), and immune infiltrates (Fig. 7C). These findings indicate that G9 transforms the tumor immune landscape, from a protumor to an antitumor phenotype, through Notch inhibition.
Fig. 6.
G9 suppresses tumor growth and cytokine expression and remodels the tumor immune microenvironment. (A) Estimated volume of KBP allografts treated with G9 or vehicle (n = 6 tumors per group). (B) Weight of KBP allografts treated with G9 or vehicle. (C) Immunoblot of N1IC in tumor lysates of KBP allografts. Molecular weight markers are shown in kilodaltons. (D) Expression of N1IC from C, determined by densitometry and presented relative to the loading control, β-actin. (E) Enzyme-linked immunosorbent assay of secreted IL-1β and CCL2 in conditioned media from overnight cultures of tumor cells from KBP tumors (n = 3 tumors per group). Cytokine levels are expressed per 106 cells. (F–J) Quantification (by flow cytometry) of TAMs (Gr-1− F4/80+ CD11b+), M2 TAMs (Gr-1− F4/80+ CD11b+ CD206+) (F), cytotoxic T cells (CD8+) (G), helper T cells (CD4+) (H), regulatory T cells (CD4+ FoxP3+) (I), and myeloid-derived suppressor cells (Gr-1+) (J) in KBP allografts after 3 wk of treatment (n = 6 tumors per group). Values are expressed as percentage of CD45+ cells (F–H and J) or as percentage of CD4+ T cells (I). Error bars represent SEM. *P < 0.05 and **P < 0.01. P values are determined by two-way ANOVA (A) and Student’s t test (B and D–J). a.u = arbitrary units.
Fig. 7.
Ectopic N1IC rescues the effect of G9. (A) Weight of KBP; hN1IC allografts treated with G9 or vehicle (n = 6 tumors per group). (B) Enzyme-linked immunosorbent assay of secreted IL-1β and CCL2 in conditioned media from overnight cultures of tumor cells from KBP; hN1IC tumors. Cytokine levels are expressed per 106 cells. (C) Quantification (by flow cytometry) of TAMs (Gr-1− F4/80+ CD11b+), M2 TAMs (Gr-1− F4/80+ CD11b+ CD206+), cytotoxic T cells (CD8+), helper T cells (CD4+), and regulatory T cells (CD4+ FoxP3+) in KBP; hN1IC 3 wk after treatment. Values for immune infiltrates are expressed as percentage of CD45+ cells except for FoxP3+ CD4+ T cells, which are expressed as percentage of CD4+ T cells. Error bars represent SEM. P values are determined by the two-tailed Student’s t test with Welch’s correction for unequal variances.
To simulate the treatment of human breast cancer, mice that generated spontaneous KBP tumors were treated with G9. N1IC was down-regulated in the tumors of G9-treated mice (SI Appendix, Fig. S7 A and B), but this failed to meet statistical significance because of the paradoxical increase of N1IC in one out of five treated mice [G9 (3)]. Tumor growth rate correlated with N1IC expression, demonstrating the dependence of KBP tumors on activated Notch (SI Appendix, Fig. S7C). Predictably, G9 (3) had an elevated growth rate and likely represented a case of treatment resistance due to G9-refractory Notch activation. Overall, G9 treatment was associated with reduced tumor growth (SI Appendix, Fig. S7D), demonstrating the therapeutic potential of USP9x inhibition in TNBC.
Discussion
Herein, using a preclinical mouse model of TNBC, we demonstrate that USP9x promotes Notch pathway activation, the secretion of CCL2 and IL-1β, and the recruitment of immune infiltrates consistent with an immunosuppressive TME. The genetic or pharmaceutical blockade of USP9x deactivates Notch, reduces proinflammatory cytokine secretion, and induces a switch to an immunologically hot tumor phenotype characterized by a decrease in immunosuppressive FoxP3+ regulatory T cells and M2-polarized TAMs, the emergence of cytotoxic and helper T cells, and a reduction in tumor growth.
The last decade has witnessed a revolution in cancer treatment toward precision medicine. This includes therapies that precisely modify the immune system, such as chimeric antigen receptor T cell transplantation and immune checkpoint blockade (ICB). The overarching aim of these immunotherapies is to boost cancer-specific T cell responses. Despite these advances, durable, clinical responses have been appreciated in a minority of cancer patients. This underscores the need for a deeper understanding of differing tumor immune microenvironment configurations and how they may predict outcome and therapeutic response.
Compared with hormone receptor–positive breast cancers, TNBCs contain higher levels of TILs (12, 55). In TNBC, TIL number is inversely correlated with the rates of relapse and death (10–14, 56, 57) and is predictive of the increased response to neoadjuvant and adjuvant chemotherapy (55, 58). These findings make evident the immunogenicity of some TNBC and an opportunity to treat these cancers through immune modulation, specifically by boosting cytotoxic CD8+ T cell activity. That being said, attempts to boost CTL activity through ICB have only demonstrated modest activity in TNBC, with objective response rates (ORRs) in phase 1 studies for single-agent treatment of around 10% and 40% when in combination with nab-paclitaxel (59, 60). Recent phase 3 trials in early [KEYNOTE-522 (61)] and advanced [IMpassion130 (62)] TNBC, that compared standard chemotherapy plus ICB to chemotherapy alone, demonstrated improvements for the ICB regimen in complete pathologic response (64.8 versus 51.2%, respectively—KEYNOTE-522) and ORR (56.0 versus 45.9%, respectively—IMpassion130). These promising, yet less-than-satisfactory, results support the continued pursuit of immune-based technologies to treat these cancers. TAMs can function as gatekeepers within the TNBC immune microenvironment by suppressing CTL function and number through ICB-resistant mechanisms (63, 64), and our findings raise the possibility that anti-Notch therapies can reduce the load of immunosuppressive TAMs in TNBC, promoting immunoconversion to ICB sensitivity.
Dysregulated Notch signaling has long been recognized as an important driver in multiple cancer types (3). Notch steers cancer progression by controlling epithelial-to-mesenchymal transition, angiogenesis, inflammation, immune evasion, and metabolism (65–68) and clinically confers resistance to radiation and chemotherapy (65). However, Notch is ubiquitously expressed in normal tissues and so achieving antitumor levels of small-molecule Notch inhibitors without debilitating side effects has been a challenge (69, 70). This highlights the importance of precisely targeting Notch in cancer cells. TRB3, which is dispensable during normal cellular physiology (71) but plays a vital role in Notch activation in cancer cells (29), is a potential therapeutic target. However, it lacks enzymatic activity, making it technically elusive in this capacity. Here, we have shown in a mouse model of TNBC, and previously in human TNBC (29), that USP9x is required to maintain elevated TRB3 levels, stabilize MIB1, and to activate Notch under cellular stress. The key role of USP9x in mediating Notch activity in this respect is demonstrated classically through the rescue of the USP9x null phenotype by ectopic Notch1. This identifies USP9x as a potential anti-Notch therapeutic target, and it predicts that drugs that target USP9x will be selective for cancer cells while displaying minimal collateral damage to normal tissues. Indeed, while the K14 cre-mediated permanent loss of USP9x was incompatible with a normal lifespan, the short-term G9 treatment of adult mice suppressed tumor growth without inducing weight loss or other deleterious side effects. These findings strengthen the concept of leveraging TME stress-induced pathways in malignant cells to devise targeted and safe anticancer therapeutics.
The growing interest in understanding the role of USP9x in development and disease has identified several proteins as their interactors and/or substrates. The context-dependent interactions of USP9x control a wide range of cell signaling pathways, as well as cellular processes (72). Indeed, the opposing roles of USP9x have been reported; for instance, it can deubiquitinate and stabilize both oncoproteins, such as MCL-1 (73), as well as tumor-suppressors, such as FBW7 (74). Inferring which USP9x–substrate interactions are dominant in a given context is pertinent to understand the net consequence of USP9x activity. In cancer, in which TME stress is a major driver of cellular adaptations, the activation of the unfolded protein response (UPR) pathway is an early step in tumor development (66). The UPR pathway potentially directs the activity of USP9x to either promote or suppress tumor progression. While our data indicate a tumor-promoting role for USP9x in TNBC, we lack an exhaustive list of other signaling pathways that may be influenced by USP9x in this context. Itch and Ets-1, known for their broad effects in cell signaling pathways, were not primarily regulated by USP9x in the KBP system. Nevertheless, further investigation will be needed to comprehensively elucidate the input of USP9x on the vast signaling network in TNBC and other hypoxic cancer types.
In summary, both USP9x and the Notch pathway confer pleiotropic effects that can be tumor promoting or tumor suppressing, dependent on the cell type. Our study indicates that in TNBC the activities of USP9x and Notch converge to promote tumor growth by programming the TME stress response, inflammation, and immune evasion. This work provides preclinical evidence that the therapeutic targeting of USP9x in TNBC is feasible and provides a rationale for the continued development of anti-USP9x precision therapeutics to be used in combination with ICB.
Materials and Methods
Cell Culture, Transfection, and Chemical Reagents.
KBP cells were a kind gift from Chiara Gorrini, Cancer Therapeutic Unit, The Institute of Cancer Research, London, United Kingdom and were maintained as described (75). Human embryonic kidney 293T (HEK293T) cells were from ATCC. To dissociate adherent cells, cells were washed with phosphate-buffered saline (PBS)-MgCl2-CaCl2 and trypsinized with 0.05% Trypsin-ethylenediaminetetraacetic acid (EDTA) (Gibco). For siRNA transfection, KBP cells were seeded at 3 × 105 per well in a 6-well format and treated with 25 pmol siRNAs (SI Appendix, Table S1) using lipofectamine RNAiMAX (Invitrogen). Transfected cells were cultured for 72 h before experiments. To induce ER stress, KBP cells were treated with thapsigargin (Sigma) at 1 μM for either 18 h (protein analysis) or 5 h (RNA analysis). To induce leucine deprivation condition, KBP cells were grown in Dulbecco’s Modified Eagle Medium (DMEM)/nutrient F-12 ham base (Sigma) supplemented with 0.1545 gm/L CaCl2, 0.365 gm/L glutamine, 0.0612 gm/L MgCl2, 0.04884 gm/L MgSO4, 1.2 gm/L NaHCO3, 0.09125 gm/L lysine, 0.0172 gm/L methionine, and lacking 0.05905 gm/L leucine. MG132 (Sigma) was used at 10 μM and stored frozen as a 10 mM stock solution. EOAI3402143 (referred to as G9) was obtained from Aobious and stored frozen as a 6 mg/mL stock solution. Pimonidazole was obtained from Hypoxyprobe and stored in saline as 12 mg/mL.
Stable Expression of Dox-Inducible shUSP9x and N1IC.
For inducible USP9x KD, a specific shRNA sequence for murine USP9x (76) (SI Appendix, Table S1) was cloned into the lentiviral vector Tet-pLKO-puro (Addgene plasmid No. 21915). Lentiviral shUSP9x particles were produced using pMD2.G (Addgene plasmid No. 12259) and psPAX2 (Addgene plasmid No. 12260) in lenti-X 293T cells and concentrated using lenti-X concentrator (Clontech). Concentrated lentivirus was used to infect KBP cells in the presence of 8 μg/mL polybrene (Sigma). For ectopic N1IC expression, complementary DNA (cDNA) containing N1IC as described in ref. 15 was cloned into the retroviral vector pMIG (Addgene plasmid No. 9044). Retroviral N1IC particles were produced in Phoenix-packaging cells and concentrated using a retro-X concentrator (Clontech). Concentrated retrovirus was used to infect KBP cells and KBP shUSP9x cells in the presence of 8 μg/mL polybrene.
Quantitative Real-Time PCR.
Primer sequences were designed using the IDT PrimerQuest Tool and are listed in SI Appendix, Table S1. cDNA was prepared from 1 μg RNA using the iScript cDNA synthesis kit and subjected to quantitative real-time PCR using the default PCR cycle on a 7900HT Fast Real-Time PCR System (Applied Biosystems). Amplified DNA products were detected and quantified by SYBR green using power SYBR green PCR master mix. Each sample was tested in triplicate for each primer set. Dissociation curve analyses were also performed to ensure the absence of nonspecific amplification. Normalization was done using murine GAPDH expression levels.
Western Blotting and Antibodies.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris pH 7.6, 150 mM NaCl, 1% Nonidet P-40, 1% DOC, and 0.1% SDS). Tumor tissue samples were lysed in a modified RIPA buffer (50 mM Hepes pH 7.86, 150 mM NaCl, 1.5 mM EDTA, 10% glycerol, 1% Nonidet P-40, 1% DOC, and 0.1% SDS). Lysates were sonicated and centrifuged at 13,000× g for 10 min. The concentration of proteins in the supernatant was measured and equal amounts of protein samples were then resolved by reducing SDS/polyacrylamide gel electrophoresis and transferred to PVDF membranes (Bio-Rad). Blots were incubated overnight in corresponding primary antibodies (listed in SI Appendix, Table S2), followed by detection with HRP-conjugated secondary antibodies. Proteins of interest were visualized using enhanced, luminol-based chemiluminescent (GE Healthcare). When required, immunoblots were quantified by ImageJ software.
Cellular Ubiquitination Assay.
TRB3-Flag, mMIB1-Flag, and HA-UB constructs were prepared as described (29). HEK293T cells were seeded at 4 × 105 per well in a 6-well format and transfected with the indicated plasmids using Lipofectamine 2000. Around 48 h after transfection, cells were treated with MG132 (Sigma) for 30 min prior to G9 treatment. Around 7 h after G9 treatment, cells were lysed in ionic lysis buffer as described (29), followed by immunoprecipitation with anti-Flag M2 beads and Western blotting with anti-HA.
Mouse KBP Mammary Tumor Model.
KBP cell lines (parental, shUSP9x, shUSP9x-N1IC, or hN1IC) (6 × 105) were prepared in 25 μL PBS for injection into the mammary fat pads of wild-type FVB mice (The Jackson Laboratory). Once tumors were palpable, mice were matched by tumor size and assigned to the indicated treatment groups. Dox was administered through the rodent diet (Teklad) at 625 mg/kg diet. Dox-containing diet was changed twice every week. G9 was administered in DMSO: PEG300 (1:1) by intraperitoneal injection every other day at 12 mg/kg. After 3 wk, tumor-engrafted mice were euthanized, followed by excision of tumors. K14cre;Brca1FL/FL;p53FL/FL mice, developed by Liu et al. (33), were a gift from T.W.M. The genotype of all animals was confirmed by PCR analyses of the tail tips of 3-wk-old female mice, as previously described (33, 77, 78). All mouse protocols were approved by the Animal Care and Use Committee of the University Health Network.
Flow Cytometry.
Tumors were minced and then digested overnight using digestion buffer (5% calf serum, 1 mg/mL collagenase, 30 μg/mL DNase I, and 1% penicillin–streptomycin in DMEM/F12). The digested samples were filtered through 70-μm Falcon cell strainers, stained with a fixable viability dye and fluorophore-conjugated antibodies (listed in SI Appendix, Table S2), and analyzed with a BD Biosciences LSRFortessa Analyzer using FlowJo software (TreeStar).
Enzyme-Linked Immunosorbent Assay.
To determine IL-1β and CCL2 expression levels in tumor cells, freshly dissociated tumors were enriched for tumor cells using a mouse epithelial cell enrichment kit (StemCell Technologies). After overnight culture of enriched tumor cells, conditioned media was centrifuged at 500× g for 10 min at 4 °C, and supernatant was collected to determine cytokine levels using Quantikine enzyme-linked immunosorbent assay kits (listed in SI Appendix, Table S2).
Histological Analysis and Quantification.
Formalin-fixed, paraffin-embedded mouse tissues were cut into 4-μm tissue sections, and antigen retrieval was performed in 10 mM, pH 6.0 citrate or 10 mM, pH 9.0 Tris-EDTA (for N1IC). Sections were incubated with the primary antibody (listed in SI Appendix, Table S2) overnight at 4 °C. Secondary antibody and ABC reagent (Vector Laboratories) were added sequentially, and sections were developed with DAB reagent (Vector Laboratories) and counterstained with hematoxylin. Stained slides were scanned on a whole-slide scanner (Nanozoomer 2.0-HT) to acquire the whole-slide image. The quantification of immunohistochemistry stained whole-slide image was done using QuPath software (79).
The small intestines were dissected and cut longitudinally, opened and washed first with PBS and later with fixative (50% ethanol and 5% acetic acid in distilled water), roll processed, fixed overnight in 10% neutral buffered formalin, and embedded into paraffin for hematoxylin and eosin staining and AB-PAS staining.
Statistics.
Student’s t test (two tailed) was used for the statistical analysis of all experiments unless otherwise specified. Mammary tumor latency was analyzed using the Kaplan–Meier method, and the difference in mean latency was tested using the log-rank test. The correlation coefficient was measured using Pearson correlation. P values < 0.05 were considered significant. All statistical analysis was done using GraphPad Prism software.
Supplementary Material
Acknowledgments
This study was supported by funds to M.R. and P.S.O. from the Canadian Cancer Society (Award No. 705788). This research was funded in part by the Ontario Ministry of Health and Long-Term Care.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101592118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Lehmann B. D., et al., Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Invest. 121, 2750–2767 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foulkes W. D., Smith I. E., Reis-Filho J. S., Triple-negative breast cancer. N. Engl. J. Med. 363, 1938–1948 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Siebel C., Lendahl U., Notch signaling in development, tissue homeostasis, and disease. Physiol. Rev. 97, 1235–1294 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Dickson B. C., et al., High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod. Pathol. 20, 685–693 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Reedijk M., et al., JAG1 expression is associated with a basal phenotype and recurrence in lymph node-negative breast cancer. Breast Cancer Res. Treat. 111, 439–448 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Stylianou S., Clarke R. B., Brennan K., Aberrant activation of notch signaling in human breast cancer. Cancer Res. 66, 1517–1525 (2006). [DOI] [PubMed] [Google Scholar]
- 7.DeNardo D. G., et al., CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 16, 91–102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahmoud S. M. A., et al., Tumour-infiltrating macrophages and clinical outcome in breast cancer. J. Clin. Pathol. 65, 159–163 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Schedin P., O’Brien J., Rudolph M., Stein T., Borges V., Microenvironment of the involuting mammary gland mediates mammary cancer progression. J. Mammary Gland Biol. Neoplasia 12, 71–82 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Adams S., et al., Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J. Clin. Oncol. 32, 2959–2966 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García-Teijido P., Cabal M. L., Fernández I. P., Pérez Y. F., Tumor-infiltrating lymphocytes in triple negative breast cancer: The future of immune targeting. Clin. Med. Insights Oncol. 10 (suppl. 1), 31–39 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Savas P., et al., Clinical relevance of host immunity in breast cancer: From TILs to the clinic. Nat. Rev. Clin. Oncol. 13, 228–241 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Denkert C., et al., Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. 28, 105–113 (2010). Correction in: J. Clin. Oncol. 28, 708 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Loi S., et al., Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J. Clin. Oncol. 31, 860–867 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Shen Q., et al., Notch shapes the innate immunophenotype in breast cancer. Cancer Discov. 7, 1320–1335 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Jin L., et al., Expression of interleukin-1beta in human breast carcinoma. Cancer 80, 421–434 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Kolb R., Liu G.-H., Janowski A. M., Sutterwala F. S., Zhang W., Inflammasomes in cancer: A double-edged sword. Protein Cell 5, 12–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian B.-Z., et al., CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 475, 222–225 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saji H., et al., Significant correlation of monocyte chemoattractant protein-1 expression with neovascularization and progression of breast carcinoma. Cancer 92, 1085–1091 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Ueno T., et al., Significance of macrophage chemoattractant protein-1 in macrophage recruitment, angiogenesis, and survival in human breast cancer. Clin. Cancer Res. 6, 3282–3289 (2000). [PubMed] [Google Scholar]
- 21.Valković T., Lucin K., Krstulja M., Dobi-Babić R., Jonjić N., Expression of monocyte chemotactic protein-1 in human invasive ductal breast cancer. Pathol. Res. Pract. 194, 335–340 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Pollard J. W., Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer 4, 71–78 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Coussens L. M., Zitvogel L., Palucka A. K., Neutralizing tumor-promoting chronic inflammation: A magic bullet? Science 339, 286–291 (2013). Correction in: Science 339, 1522 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gajewski T. F., Schreiber H., Fu Y.-X., Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 14, 1014–1022 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmid M. C., Varner J. A., Myeloid cells in the tumor microenvironment: Modulation of tumor angiogenesis and tumor inflammation. J. Oncol. 2010, 201026 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersson E. R., Lendahl U., Therapeutic modulation of Notch signalling–Are we there yet? Nat. Rev. Drug Discov. 13, 357–378 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Majumder S., et al., Targeting Notch in oncology: The path forward. Nat. Rev. Drug Discov. 20, 125–144 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Izrailit J., Berman H. K., Datti A., Wrana J. L., Reedijk M., High throughput kinase inhibitor screens reveal TRB3 and MAPK-ERK/TGFβ pathways as fundamental Notch regulators in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 110, 1714–1719 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Izrailit J., Jaiswal A., Zheng W., Moran M. F., Reedijk M., Cellular stress induces TRB3/USP9x-dependent Notch activation in cancer. Oncogene 36, 1048–1057 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Wennemers M., et al., Tribbles homolog 3 denotes a poor prognosis in breast cancer and is involved in hypoxia response. Breast Cancer Res. 13, R82 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corcoran C. A., et al., Genotoxic and endoplasmic reticulum stresses differentially regulate TRB3 expression. Cancer Biol. Ther. 4, 1063–1067 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Chen X., et al., XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature 508, 103–107 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X., et al., Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc. Natl. Acad. Sci. U.S.A. 104, 12111–12116 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Potu H., et al., Usp9x regulates Ets-1 ubiquitination and stability to control NRAS expression and tumorigenicity in melanoma. Nat. Commun. 8, 14449 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mouchantaf R., et al., The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J. Biol. Chem. 281, 38738–38747 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Shimizu M., et al., Plasminogen activator uPA is a direct transcriptional target of the JAG1-Notch receptor signaling pathway in breast cancer. Cancer Res. 71, 277–286 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Franchi L., Eigenbrod T., Muñoz-Planillo R., Nuñez G., The inflammasome: A caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat. Immunol. 10, 241–247 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Movahedi K., et al., Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 70, 5728–5739 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Chen X., et al., Jagged1 expression regulated by Notch3 and Wnt/β-catenin signaling pathways in ovarian cancer. Oncotarget 1, 210–218 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manderfield L. J., et al., Notch activation of Jagged1 contributes to the assembly of the arterial wall. Circulation 125, 314–323 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Foldi J., et al., Autoamplification of Notch signaling in macrophages by TLR-induced and RBP-J-dependent induction of Jagged1. J. Immunol. 185, 5023–5031 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hiyama A., et al., Hypoxia activates the notch signaling pathway in cells of the intervertebral disc: Implications in degenerative disc disease. Arthritis Rheum. 63, 1355–1364 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gustafsson M. V., et al., Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev. Cell 9, 617–628 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Ciria M., et al., Mesenchymal stem cell migration and proliferation are mediated by hypoxia-inducible factor-1α upstream of Notch and SUMO pathways. Stem Cells Dev. 26, 973–985 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diez H., et al., Hypoxia-mediated activation of Dll4-Notch-Hey2 signaling in endothelial progenitor cells and adoption of arterial cell fate. Exp. Cell Res. 313, 1–9 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Bertout J. A., et al., Heterozygosity for hypoxia inducible factor 1α decreases the incidence of thymic lymphomas in a p53 mutant mouse model. Cancer Res. 69, 3213–3220 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X., et al., Notch3 signaling promotes the development of pulmonary arterial hypertension. Nat. Med. 15, 1289–1297 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peterson L. F., et al., Targeting deubiquitinase activity with a novel small-molecule inhibitor as therapy for B-cell malignancies. Blood 125, 3588–3597 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Takebe N., Nguyen D., Yang S. X., Targeting notch signaling pathway in cancer: Clinical development advances and challenges. Pharmacol. Ther. 141, 140–149 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Es J. H., et al., Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435, 959–963 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Fre S., et al., Notch signals control the fate of immature progenitor cells in the intestine. Nature 435, 964–968 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Stanger B. Z., Datar R., Murtaugh L. C., Melton D. A., Direct regulation of intestinal fate by Notch. Proc. Natl. Acad. Sci. U.S.A. 102, 12443–12448 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pellegrinet L., et al., Dll1- and dll4-mediated notch signaling are required for homeostasis of intestinal stem cells. Gastroenterology 140, 1230–1240.e1-7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gabrilovich D. I., Nagaraj S., Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 9, 162–174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stanton S. E., Adams S., Disis M. L., Variation in the incidence and magnitude of tumor-infiltrating lymphocytes in breast cancer subtypes: A systematic review. JAMA Oncol. 2, 1354–1360 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Ali H. R., et al., Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann. Oncol. 25, 1536–1543 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Mahmoud S. M. A., et al., Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J. Clin. Oncol. 29, 1949–1955 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Pruneri G., Vingiani A., Denkert C., Tumor infiltrating lymphocytes in early breast cancer. Breast 37, 207–214 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Emens L. A., et al., Long-term clinical outcomes and biomarker analyses of atezolizumab therapy for patients with metastatic triple-negative breast cancer: A phase 1 study. JAMA Oncol. 5, 74–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adams S., et al., Atezolizumab plus nab-paclitaxel in the treatment of metastatic triple-negative breast cancer with 2-year survival follow-up: A phase 1b clinical trial. JAMA Oncol. 5, 334–342 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmid P.et al.; KEYNOTE-522 Investigators , Pembrolizumab for early triple-negative breast cancer. N. Engl. J. Med. 382, 810–821 (2020). [DOI] [PubMed] [Google Scholar]
- 62.Schmid P.et al.; IMpassion130 Trial Investigators , Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N. Engl. J. Med. 379, 2108–2121 (2018). [DOI] [PubMed] [Google Scholar]
- 63.Noy R., Pollard J. W., Tumor-associated macrophages: From mechanisms to therapy. Immunity 41, 49–61 (2014). Correction in: Immunity 41, 866 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katakura T., Miyazaki M., Kobayashi M., Herndon D. N., Suzuki F., CCL17 and IL-10 as effectors that enable alternatively activated macrophages to inhibit the generation of classically activated macrophages. J. Immunol. 172, 1407–1413 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Ranganathan P., Weaver K. L., Capobianco A. J., Notch signalling in solid tumours: A little bit of everything but not all the time. Nat. Rev. Cancer 11, 338–351 (2011). [DOI] [PubMed] [Google Scholar]
- 66.Yadav R. K., Chae S.-W., Kim H.-R., Chae H. J., Endoplasmic reticulum stress and cancer. J. Cancer Prev. 19, 75–88 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Timmerman L. A., et al., Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 18, 99–115 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garcia A., Kandel J. J., Notch: A key regulator of tumor angiogenesis and metastasis. Histol. Histopathol. 27, 151–156 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Kloe G. E., De Strooper B., Small molecules that inhibit Notch signaling. Methods Mol. Biol. 1187, 311–322 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Rizzo P., et al., The role of notch in the cardiovascular system: Potential adverse effects of investigational notch inhibitors. Front. Oncol. 4, 384 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Okamoto H., et al., Genetic deletion of Trb3, the mammalian Drosophila tribbles homolog, displays normal hepatic insulin signaling and glucose homeostasis. Diabetes 56, 1350–1356 (2007). [DOI] [PubMed] [Google Scholar]
- 72.Murtaza M., Jolly L. A., Gecz J., Wood S. A., La FAM fatale: USP9X in development and disease. Cell. Mol. Life Sci. 72, 2075–2089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schwickart M., et al., Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature 463, 103–107 (2010). [DOI] [PubMed] [Google Scholar]
- 74.Khan O. M., et al., The deubiquitinase USP9X regulates FBW7 stability and suppresses colorectal cancer. J. Clin. Invest. 128, 1326–1337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gorrini C., et al., Estrogen controls the survival of BRCA1-deficient cells via a PI3K-NRF2-regulated pathway. Proc. Natl. Acad. Sci. U.S.A. 111, 4472–4477 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pérez-Mancera P. A.et al.; Australian Pancreatic Cancer Genome Initiative , The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature 486, 266–270 (2012). Correction in: Nature 494, 390 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jonkers J., et al., Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29, 418–425 (2001). [DOI] [PubMed] [Google Scholar]
- 78.Stegeman S., et al., Loss of Usp9x disrupts cortical architecture, hippocampal development and TGFβ-mediated axonogenesis. PLoS One 8, e68287 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bankhead P., et al., QuPath: Open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.