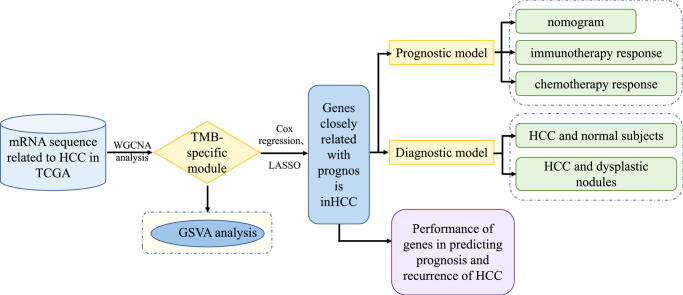
Graphical abstract
Keywords: TMB, Hepatocellular carcinoma (HCC), Prognosis, Diagnosis, Immune checkpoint
Abstract
Introduction
The development and prognosis of HCC involve complex molecular mechanisms, which affect the effectiveness of its treatment strategies. Tumor mutational burden (TMB) is related to the efficacy of immunotherapy, but the prognostic role of TMB-related genes in HCC has not yet been determined clearly.
Objectives
In this study, we identified TMB-specific genes with good prognostic value to build diagnostic and prognostic models and provide guidance for the treatment of HCC patients.
Methods
Weighted gene co-expression network analysis (WGCNA) was applied to identify the TMB-specific genes. And LASSO method and Cox regression were used in establishing the prognostic model.
Results
The prognostic model based on SMG5 and MRPL9 showed patients with higher prognostic risk had a remarkedly poorer survival probability than their counterparts with lower prognostic risk in both a TCGA cohort (P < 0.001, HR = 1.93) and an ICGC cohort (P < 0.001, HR = 3.58). In addition, higher infiltrating fractions of memory B cells, M0 macrophages, neutrophils, activated memory CD4 + T cells, follicular helper T cells and regulatory T cells and higher expression of B7H3, CTLA4, PD1, and TIM3 were present in the high-risk group than in the low-risk group (P < 0.05). Patients with high prognostic risk had higher resistance to some chemotherapy and targeted drugs, such as methotrexate, vinblastine and erlotinib, than those with low prognostic risk (P < 0.05). And a diagnostic model considering two genes was able to accurately distinguish patients with HCC from normal samples and those with dysplastic nodules. In addition, knockdown of SMG5 and MRPL9 was determined to significantly inhibit cell proliferation and migration in HCC.
Conclusion
Our study helps to select patients suitable for chemotherapy, targeted drugs and immunotherapy and provide new ideas for developing treatment strategies to improve disease outcomes in HCC patients.
Introduction
As the most common type of primary liver cancer, hepatocellular carcinoma (HCC) is listed as the sixth most common tumor and the fourth leading cause of cancer-related death worldwide, bringing a heavy health burden to society [1]. Since HCC is usually diagnosed at an advanced stage, it is not suitable for surgical treatment, such as radical liver resection or liver transplantation; as such, the prognosis of patients is usually poor, and the probability of recurrence is high [2]. The molecular mechanisms leading to the evolution of HCC tumors are extremely complex, involving multiple genetic abnormalities, including genomic instability, single nucleotide polymorphisms (SNPs), somatic mutations, and dysregulation of signaling pathways related to HCC occurrence and development [3], [4]. Analysis of the complex relationship between genes, proteins and other molecules aids the understanding of the underlying oncogenic molecular mechanisms of HCC; in addition, specific gene expression characteristics obtained from such analysis may help accurately predict the risk of HCC and be translated into novel diagnostic, prognostic and treatment indicators in clinical interventions, providing new options for treatment [5], [6].
In recent years, immunotherapy, including immune checkpoint inhibitors, has attracted wide attention in the field of anticancer therapy as a new treatment option [7]. Many studies have confirmed that multiple types of tumor cells, including HCC cells, carry new antigens induced by gene mutations on the surface that may be recognized by the immune system, making them targets for activated immune cells [8], [9], [10]. However, there is currently no reliable molecular predictive marker that can clarify which HCC patients respond to immunotherapy. Tumor mutation burden (TMB) has been proven to be related with the efficacy of immunotherapy; the higher the TMB is, the better the tumor remission effect and clinical benefits obtained from immunotherapy [11], [12]. TMB is interpreted as the total number of nonsynonymous mutations occurring in each coding region in the tumor genome [13]. Some studies have shown that some types of cancer (such as non-small-cell lung cancer and colorectal cancer) with high TMB values are likely to have a poor prognosis [14], [15], but the prognostic value of the TMB in HCC has not yet been determined clearly.
The rise of high-throughput sequencing technology creates an opportunity to find specific gene expression characteristics that can effectively predict the risk and prognosis in HCC [16]. In this study, we conducted comprehensive analysis on the genomic sequence in HCC and identified genes bearing important mutations contributing to TMB and closely related with the prognosis in HCC patients to establish diagnostic and prognostic models for HCC. We assessed the capability of the prognostic model in predicting the response to chemotherapy, targeted drugs and immune checkpoint inhibitors in HCC patients. Our findings will help develop individualized therapeutic plans for HCC patients and improve their disease outcomes.
Materials and Methods
Determination of differentially expressed hub genes in HCC
The mRNA sequence and corresponding clinical information of HCC patients were obtained from the TCGA database, including 19,676 annotated mRNA sequences and data from 370 HCC tissue samples and 50 normal tissue samples. The limma R package was performed to determine differentially expressed genes (DEGs) with cut-off values of absolute log 2-fold change (FC) > 1 and adjusted P value < 0.05.
Calculation of TMB and definition of high- and low-TMB groups
Somatic mutation profiles were downloaded from the General Genomic Data (GDC) website. Maftools were adopted to evaluate somatic mutations. The formula used to calculate the TMB was as follows: TMBi = 1.0 * NTMi + 1.5 * TMi [17].
TMBi represents the TMB of sample i. NTMi represents the total number of nontruncated mutations, including missense, in-frame deletion, in-frame insertion and nonstop deletions. TMi represents the total number of truncated mutations, including nonsense, frame-shift deletion, frame-shift insertion, and splice-site deletions. Silent mutations were not integrated into the calculation of TMB because they do not cause any downstream changes. According to Foundation Medicine official reports, TMB levels were divided into three groups: low (1–5 mutations/mb), intermediate (6–19 mutations/mb), and high (≥20 mutations/mb) [18]. In this study, we classified samples with TMB < 6 mutations/mb into the low TMB group and samples with TMB ≥ 20 mutations/mb into the high TMB group.
WGCNA for identifying the TMB-specific module
WGCNA was applied in identifying the HCC TMB-specific module using the R package “WGCNA” [19], [20]. The expression profiles of DEGs between HCC patients and normal samples in the TCGA database were used as input for the WGCNA, and TMB was calculated and defined as the phenotype of the sample. A signed scale-free co-expression gene network was ensured through setting the power of β = 10 and scale-free R2 of 0.90 as the soft threshold parameters. A co-expression matrix was established based on the β value and the input gene expression matrix to classify genes with similar expression patterns into the same gene module, thereby generating co-expression modules. The correlation between the module Eigengenes (ME) and TMB was calculated using the Eigengenes function. A heat map was adopted to visualize the correlation between each module and TMB. And the dark green module depicted the highest correlation with TMB among these modules was identified as the TMB-specific module. Then, the correlation matrix between the TMB and the genes in the TMB-specific module was calculated.
Gene set variation analysis (GSVA) of the genes in the TMB-specific module
GSVA was accomplished with the R package “GSVA” [21] to evaluate differentially enriched metabolic pathways in which genes in the TMB-specific module were involved. Pathways used for GSVA were obtained from the Molecular Signatures Database (MSigDB) (http://software.broadinstitute.org/gsea/msigdb).
Determination of genes in the TMB-specific module closely related to the prognosis in HCC
Cox regression and the LASSO method were adopted to assess the prognostic value of the genes in the HCC TMB-specific module. We first used univariate Cox regression to choose genes associated with the prognosis of HCC. Genes with P < 0.05 represents statistical significance. The LASSO method with an L1 penalty was performed to determine the genes with the greatest impact on the prognosis of HCC. LASSO applies an L1 penalty to reduce the regression coefficient and reduce the number of indicators with a final weight of nonzero, thus determining the indicators with the highest contribution [22]. In this study, LASSO to cut down the number of genes was accomplished by the glmnet package in R using 10-fold cross validations and 1000 iterations. Related parameters were set as follows: max_iter = 1000 and cv = 10. After 1000 iterations of LASSO, the higher the nonzero coefficient, the stronger the capability of the corresponding gene in predicting prognosis was. The selected genes were then incorporated in a multivariate Cox regression model and the gene set with the best prognostic value of HCC was determined through forward selection and backward elimination.
Establishment and validation of a prognostic model
A prognostic model was constructed from the gene set identified by the multivariate Cox regression. The prognostic score formula was organized as follows: prognostic index (PI) = (β1 * expression level of SMG5) + (β2 * expression level of MRPL9). The median was set as a threshold to divide HCC patients with survival data into high-risk and low-risk groups. The predictive performance of the prognostic model was examined by K-M curves and ROC curve analysis.
Construction and assessment of a predictive nomogram
Independence analysis of the prognosis-predicting ability of the prognostic model compared to traditional clinical prognostic variables (containing age, AFP, weight, vascular tumor cell invasion, sex, pathologic grade and TNM stage) was carried out using univariate and multivariate Cox regression analyses. The confirmed independent predictive factors were integrated to construct a nomogram and corresponding calibration map via “rms” R software. The calibration curves and consistency index (C-index), which was calculated via a bootstrap method with 1000 resamples, were applied in measuring the consistency between the predicted results of the nomogram and the actual results. ROC curves were generated to compare the specificity and sensitivity of the prediction ability of the nomogram with single independent predictors. Decision curve analysis (DCA) was employed to analyze the clinical benefits achieved by the predictive results of the nomogram compared to those of single independent predictors.
Validation of the prognostic value of gene expression for HCC
Further validation of the prognostic value of SMG5 and MRPL9 was performed in the HCC cohort from the UALCAN database. The Wilcoxon signed-rank test was employed to analyze the expression differences of SMG5 and MRPL9 between tumor tissues and nontumor tissues and examine the effect of the expression of the two genes on the survival time and recurrence time of HCC patients.
Determination of immune cell infiltration
CIBERSORT analysis was adopted to quantitatively convert transcriptome data into the absolute abundance of immune cell and matrix cell types in heterogeneous tissues to evaluate the proportions of 22 human immune cell subsets [23]. Standard annotation files were used to organize gene expression profiles. mRNA data converting into infiltration levels of nontumor cells in tumor microenvironments was realized using the R package “CIBERSORT”.
Cell culture and siRNA treatment
Human hepatocellular carcinoma cell lines (including SK-HEP1 and LM3) were obtained from the American Type Culture Collection (ATCC) (Manassas, VA, USA). The cells were cultured in DMEM containing 10% fetal bovine serum (FBS), 2 mM L-glutamine and 100 U/ml penicillin–streptomycin solution at 37 °C in 5% CO2 in an incubator.
SMG5 and MRPL9 siRNAs were synthesized by GeneChem (Shanghai, China) and transfected into cells using Lipofectamine 2000 (Invitrogen, California, USA) according to the manufacturer’s instructions. The cells were cultured in basic DMEM media for 6–8 h before the media was for DMEM media containing FBS and penicillin–streptomycin.
Western blot analysis
RIPA lysis buffer (Invitrogen) containing PMSF (Bio-Rad, Shanghai, China) was used to collect proteins from SK-HEP1 and LM3 cells. 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was applied in separating protein samples and a polyvinylidene fluoride membrane (PVDF) membrane (Invitrogen, Carlsbad, USA) was used to transfer the separated protein. The membrane was blocked in 5% skim milk at room temperature for 2 h in a shaker, and then incubated with the primary antibody at 4 °C overnight and subsequent incubation with the secondary antibody for 2 h. The blots were detected and imaged using the iBright FL1500 intelligent imaging system (Invitrogen, Carlsbad, USA). The details of antibodies in this study are listed in the supporting information.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cultured SK-HEP1 cells and LM3 cells using TRIzol® reagent (Invitrogen, Carlsbad, USA) following the manufacturer's protocol. Then, the RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, USA) was used for reverse transcribing total RNA to cDNA, and qRT-PCR was performed using Real-time PCR Master Mix (SYBR Green; TOYOBO, Japan). The relative expression (fold change) of the target genes was determined using the 2-ΔΔCT methodology. β-Actin served as the internal control. Each experiment was repeated at least three times. The primers used for qRT-PCR were purchased from RiboBio Co.Ltd. The primers used for qRT-PCR are listed in the supporting information.
Cell proliferation assay
After transfection with SMG5 and MRPL9 siRNA for 48 h, SK-HEP1 and LM3 cells were cultured in 96-well plates (3000 cells/plate in 200 µl DMEM). The proliferative capacity of treated cells was detected at 0, 24, 48 and 72 h. Cell Counting Kit-8 (CCK8) reagent (Yeasen, Shanghai, China) was added to each plate according to the kit instructions, and the OD450 value was analyzed by a microplate spectrophotometer (Thermo Fisher Scientific, MA, USA). Cell proliferation was also measured using 5-ethynyl-2′-deoxyuridine (EdU) agent (Ruibo, Guangzhou, China), and the EdU assay was conducted following the manufacturer's protocol.
Transwell migration assay
SK-HEP1 and LM3 cells were transfected with SMG5 and MRPL9 siRNA for 48 h and cultured in 24-well culture plates with 8 mm pore-containing membrane inserts to measure cell migration capacity. Serum-free DMEM was added to the upper chamber, and DMEM (Gibco, NY, USA) supplemented with 10% fetal bovine serum was added to the lower chamber. Cells were incubated at 37 °C for three days. Cells in the lower chamber (below the membrane) were stained with 0.4% trypan blue (Invitrogen) and counted under a light microscope. Each experiment was repeated three times.
Statistical analyses
R language (version 3.5.2) and GraphPad Prism 7 software (version 7.0) were used to perform statistical analysis and generate figures. Differences in data with a normal distribution between paired groups were compared using Student’s t-tests, and the results are exhibited as the mean ± standard deviation (SD). Differences in data with a nonnormal distribution between paired groups were compared using the Wilcox rank sum test, and the results are expressed as the median (interquartile range). P < 0.05 was considered to represent statistical significance.
Results
Acquisition of differentially expressed hub genes in TMB in HCC
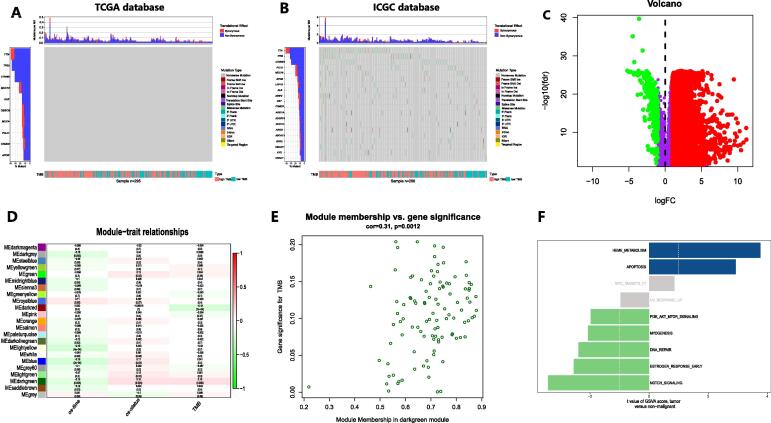
The TMB characteristics for the TCGA HCC cohort and ICGC HCC cohort are presented in Fig. 1A,B. We identified 6,800 HCC-related DEGs from TCGA (Fig. 1C). Through WGCNA, we grouped these genes into modules to aggregate genes with similar traits (Figure S1). Then, the “dark green” module with the highest correlation with TMB (Cor = 0.15, P < 0.01) was identified as the HCC TMB-specific module (Fig. 1D). Fig. 1E indicates that there is a close correlation between the expression characteristics of genes in the TMB-specific module and TMB level. Then, GSVA was performed on the genes in this module. A total of 50 hallmark pathways obtained from the Molecular Signatures Database (MSigDB) (http://software.broadinstitute.org/gsea/msigdb) were incorporated in the analysis (Table S1). We found that the gene set was mainly enriched in the heme metabolism and apoptosis signaling pathways in HCC patients (Fig. 1F).
Fig. 1.
Identification of DEGs affecting TMB in HCC. A and B Characteristics of TMB for the TCGA HCC cohort (A) and ICGC HCC cohort (B). C DEGs in the TCGA HCC cohort. D Modules with different traits identified via WGCNA. E Determination of genes in the HCC TMB-specific module. F GSVA for genes in the HCC TMB-specific module.
Comprehensive analysis to determine the association of genes and the prognosis of HCC
Cox regression and the LASSO method were performed on 370 HCC samples with survival data from TCGA to determine the prognostic relevance of the genes in the HCC TMB-specific module. The univariate Cox regression results indicated that the expression of 41 genes was clearly correlated with the prognosis of HCC (P < 0.05) (Table S2). Then, 5 of these genes were further selected through the LASSO method with 10-fold cross validation in 1000 substitution samplings (Figure S2). The subsequent multivariate Cox regression analysis finally determined that 2 genes (SMG5 and MRPL9) could serve as markers for the prediction of prognosis in HCC (P < 0.05) (Table S3).
Construction of the prognostic model and validation of its predictive performance
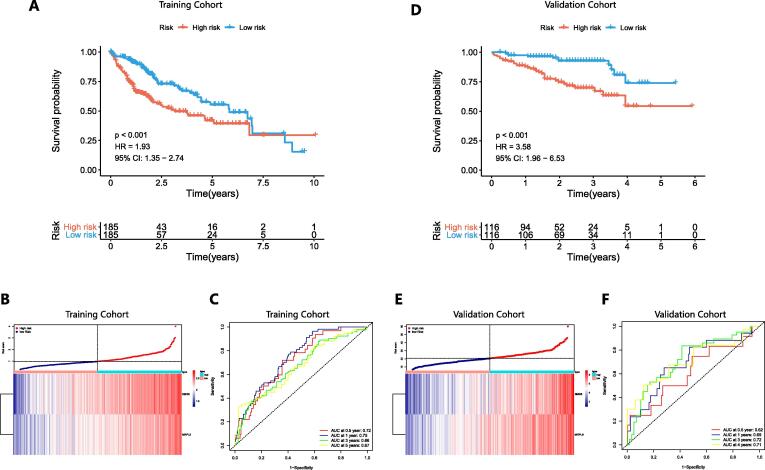
A prognostic model based on the 2 genes (SMG5 and MRPL9) was constructed. The prediction score of the model was calculated as follows: PI = (0.369 * expression level of SMG5) + (0.404 * expression level of MRPL9). A total of 370 HCC samples with survival data from TCGA were employed as a training set, and 243 HCC samples with survival information from ICGC were adopted as a validation set to assess the predictive performance of the prognostic model. HCC samples were divided into high-risk and low-risk groups using the median as a threshold. K-M curves indicated that patients with high risk had a significantly lower overall survival (OS) probability than their counterparts in both the training set (P < 0.001, HR = 1.93) (Fig. 2A) and validation set (P < 0.001, HR = 3.58) (Fig. 2D). Fig. 2B and 2E show the expression profiles of SMG5 and MRPL9 and the prognostic risk of HCC patients. The AUC values of ROC curves at 0.5, 1, 3, and 5 years reached 0.72, 0.75, 0.66, and 0.67, respectively, in the training set (Fig. 2C) and 0.62, 0.69, 0.72 and 0.71, respectively, in the validation set (Fig. 2F), confirming the high specificity and sensitivity of the prognostic model for predicting the prognosis of HCC. We then further explored the correlation between the prognostic model and TMB in the HCC cohort from TCGA and confirmed that the prognostic score of HCC patients was positively correlated with TMB (Figure S3).
Fig. 2.
Survival analysis results, distribution of risk scores, and predictive performance in the training set (A-C) and validation set (D-F). A and D K-M curves showing the prognosis differences between the high-risk and low-risk groups. B and E Distribution of prognostic risk and expression of SMG5 and MRPL9 in patients with HCC. C and F ROC curves for validating the specificity and sensitivity of the prognosis model.
Construction and validation of a nomogram integrating independent predictive factors
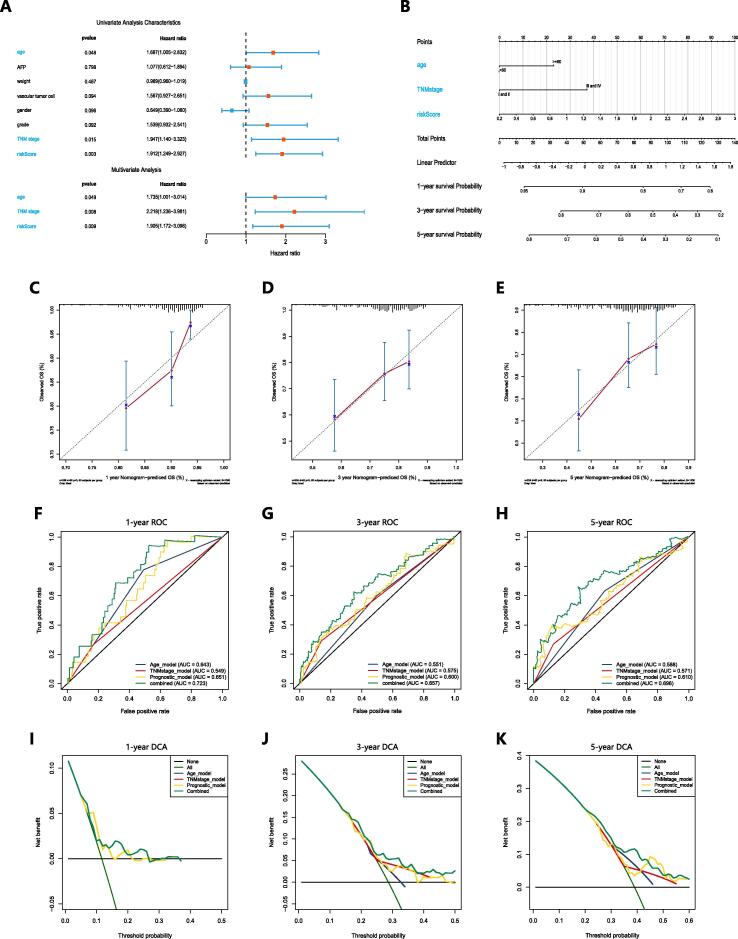
370 HCC samples with clinical information from TCGA was adopted to evaluate the independent performance of the prognostic score in predicting prognosis compared with that of clinical characteristics (including age, AFP, weight, vascular tumor cell invasion, sex, pathologic grade and TNM stage) using univariate and multivariate Cox regression analyses. The results determined that age (P < 0.05, HR = 1.735), TNM stage (P < 0.05, HR = 2.218) and the prognostic score (P < 0.05, HR = 1.905) were independent predictive factors of prognosis in HCC (Fig. 3A) (Table S4). A nomogram based on these independent predictive factors was established to quantify survival probability in HCC patients at 1, 3 and 5 years (Fig. 3B). The calibration curves of the nomogram were close to the 45° line (Fig. 3C–E), and the C-index was 0.65, indicating high consistency between the nomogram's prediction and actual results. ROC curves confirmed that the specificity and sensitivity of the nomogram for predicting prognosis were superior to those of any single independent predictive factor (Fig. 3F–H). DCA also demonstrated that prognosis prediction through the nomogram provided the best clinical benefits, suggesting that the nomogram has operational value in clinical practice (Fig. 3I–K).
Fig. 3.
The independence of the prognosis model its correlation with clinical pathological features in prognosis prediction. A Forest map showing independent predictive factors for prognosis in HCC. B Nomogram for predicting the survival probability in HCC patients at 1, 3 and 5 years. C-E Calibration charts for validating the predictive accuracy of the 1-year, 3-year, and 5-year survival probabilities of the nomogram. F-H ROC curves comparing the predicted performance of the nomogram and single independent predictive factors. I-K Evaluation of the clinical benefits that the nomogram can achieve.
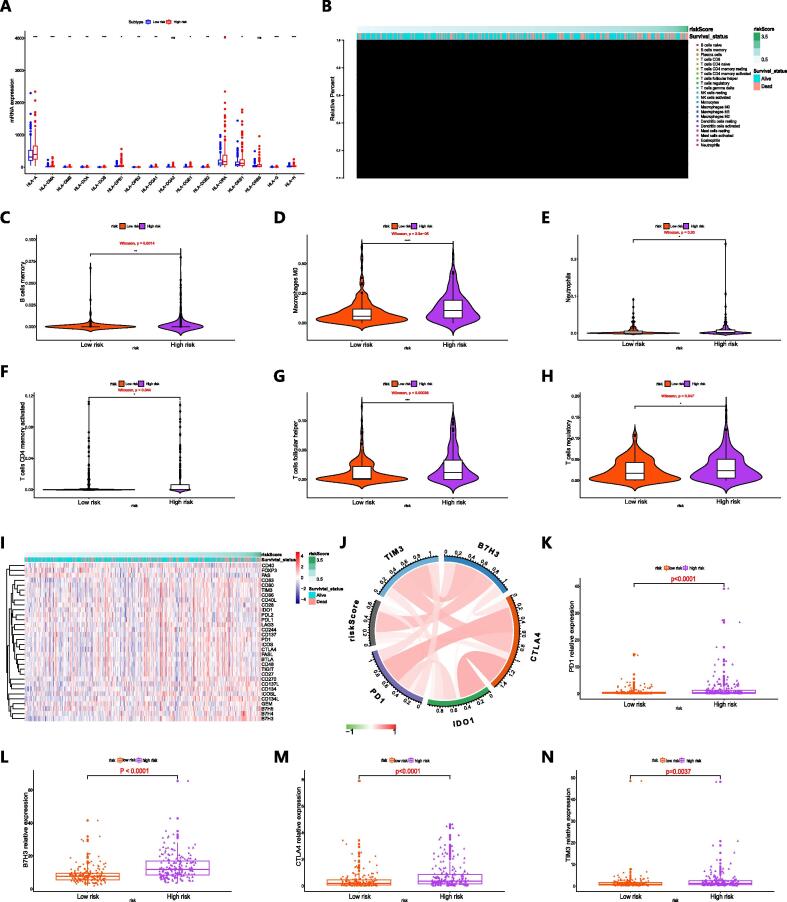
Determination of differences in immune cell infiltration and immune checkpoint expression in the high-risk and low-risk groups
We determined the level of immune cell infiltration in the TCGA cohort, and the median was used to divide HCC patients with prognostic data into high-risk and low-risk groups. Fig. 4A shows the expression of multiple subtypes of human leukocyte antigen (HLA) in the high-risk and low-risk groups. The distributions of prognosis risk and the 22 infiltrating immune cell fractions in tumor tissues are shown in Fig. 4B. Comparing with the low-risk group, the high-risk group exhibited a higher proportion of infiltrating memory B cells, M0 macrophages, neutrophils, activated memory CD4 + T cells, follicular helper T cells and regulatory T cells within tumor tissues (Fig. 4C–H). We found that the T cell receptor signaling pathway enrichment score was obviously higher in high-risk patients comparing with low-risk patients (Figure S4).
Fig. 4.
The correlation between immune cell infiltration and the expression of immune checkpoints and prognostic model components. A HLA subtype expression in high-risk and low-risk patients. B Distribution of prognostic risk and immune cell infiltration within tumor tissues in patients with HCC. C-H Violin charts revealing the relationship between the fraction of immune cells and prognostic risk score (C memory B cells; D M0 macrophages; E neutrophils; F activated memory CD4 T cells; G follicular helper T cells; H regulatory T cells). I Distribution of prognostic risk score and immune checkpoint expression in patients with HCC. J The association between prognosis risk and immune checkpoints. K-N Column charts showing the expression of immune checkpoints in high-risk and low-risk patients (K PD1; L B7H3; M CTLA4; N TIM3).
As an innovative therapy, immunotherapy based on immune checkpoint inhibitors is gradually being used in the treatment of various types of advanced cancer. Tumors usually upregulate immune checkpoints to avoid detection and killing by the immune system, while immune checkpoint inhibitors can rejuvenate host immune cells and target the adaptive immune system to tumor cells [24]. Fig. 4I shows expression profiles of immune checkpoint molecules and prognostic risk for 370 HCC samples from TCGA. The correlation between prognosis risk score and the expression of immune checkpoints is visualized in Fig. 4J. We found that a positive correlation was exhibited between prognosis risk score and immune checkpoints such as B7H3 and CTLA4 (Table S5). Then, we further revealed that higher relative expression of PD1, B7H3, CTLA4 and TIM3 was presented in patients with higher prognostic risk comparing with patients with lower prognostic risk (Fig. 4K–N), suggesting that patients with high risk may obtain more clinical benefits from immunotherapy.
Prediction of therapeutic response to chemotherapy drugs and targeted drugs according to prognostic risk in patients with HCC
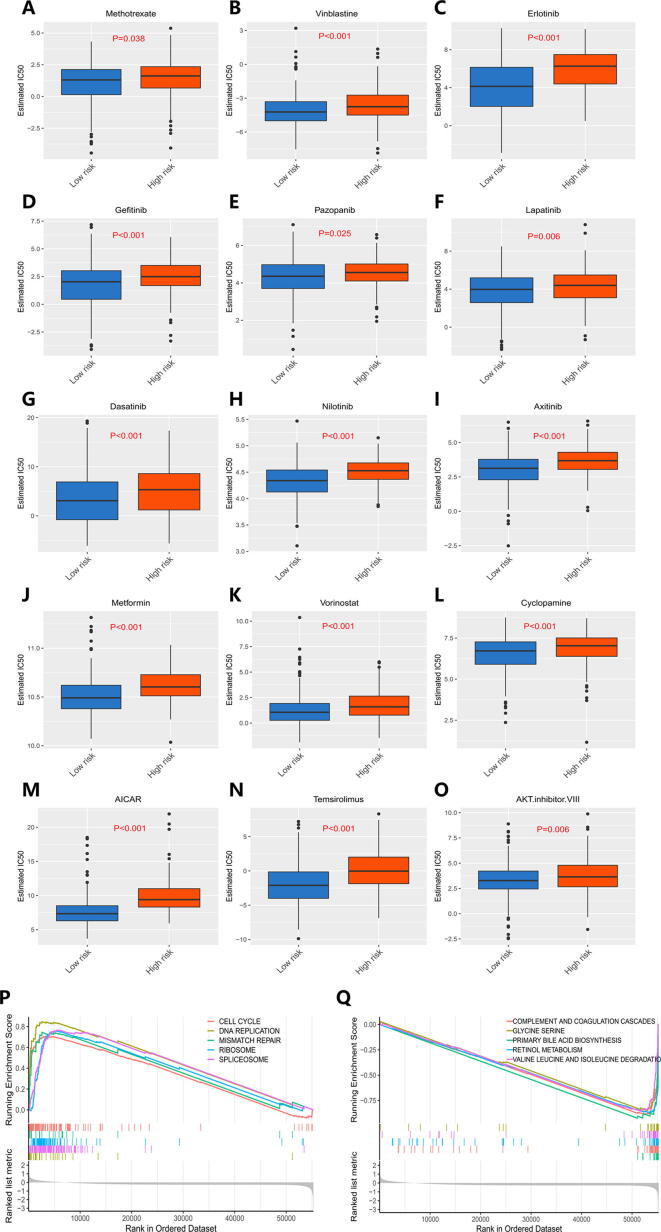
Chemotherapy and targeted drugs are common treatments for patients with HCC, especially suitable for patients with HCC who cannot undergo surgery to reduce tumor volume and prolong their survival as is postoperative adjuvant chemotherapy to inhibit the recurrence and progression of tumor [25], but drug resistance is a critical issue influencing the effectiveness of anticancer drugs. We evaluated the therapeutic response of HCC patients to 266 anticancer drugs on the Genomics of Drug Sensitivity in Cancer (GDSC) website with reference to half the maximum inhibitory concentration (IC50). The responses of HCC patients to traditional chemotherapeutic drugs and molecularly targeted chemotherapeutic drugs such as methotrexate, vinblastine, erlotinib and gefitinib are presented in Fig. 5A–O. We found that more resistance to chemotherapy was exhibited in high-risk patients were than in low-risk patients (P < 0.05). The top 5 positively regulated pathways are presented in Fig. 5P, and the top 5 negatively regulated pathways are presented in Fig. 5Q. Through GSEA, we found that the prognosis model was related to signaling pathways involved in physiological processes including the cell cycle, mismatch repair and spliceosome signaling, and biological processes including the cell cycle [26], mismatch repair [27] and spliceosome signalling [28] were shown to have a regulatory effect on cancer cell chemoresistance in previous studies, and differences in these pathways in patients may lead to different prognosis risks and different resistance to chemotherapy.
Fig. 5.
Response of HCC patients to chemotherapy drugs. A-O Differences in response to chemotherapy drugs between high-risk and low-risk patients. P Top 5 signaling pathways positively regulated by the TMB-specific prognostic model. Q Top 5 signaling pathways negatively regulated by the TMB-specific prognostic model.
Construction and validation of diagnostic models to accurately distinguish patients with HCC from normal subjects and those with dysplastic nodules
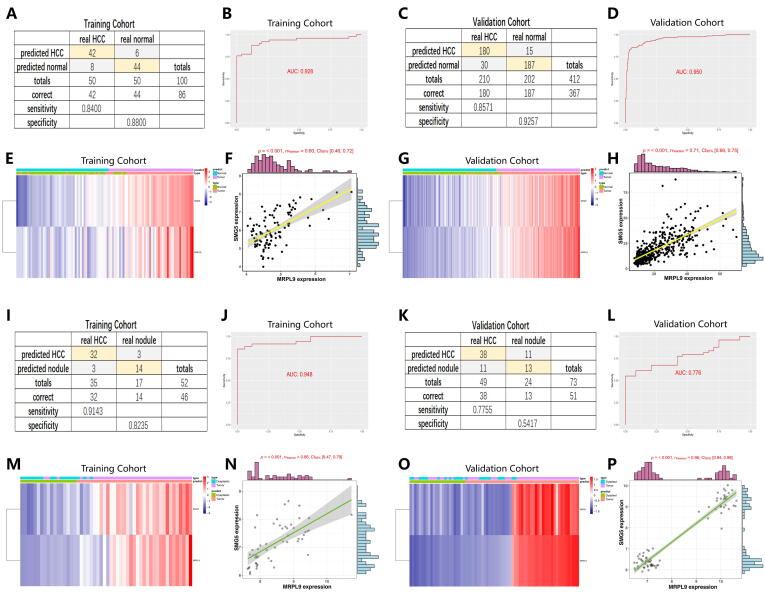
For improving the probability of early detection in HCC, a stepwise logistic regression method was performed to build a diagnostic model. The diagnostic formula is determined as follows: logit (P-HCC) = -7.819 + (0.160 × SMG5 expression level) + (1.014 × MRPL9 expression level). 50 normal samples and 50 paired HCC samples from TCGA were used as a training set, and 202 normal samples and 210 paired tumor samples from ICGC were adopted as a validation set to determine the performance of the diagnostic model. The results suggested that the diagnostic model achieved 88.00% specificity and 84.00% sensitivity in the training set (Fig. 6A) and 92.57% specificity and 85.71% sensitivity in the validation set (Fig. 6C). The AUCs of the training set and validation set were 0.928 (Fig. 6B) and 0.950 (Fig. 6D), respectively, confirming that the diagnostic model made a accurate distinction between HCC and normal subjects. Fig. 6C and 6G show the distribution of SMG5 and MRPL9 expression in HCC samples, as well as the predicted disease status and actual status. High expression of SMG5 and MRPL9 was exhibited in HCC patients. Next, we explored the relationship between SMG5 and MRPL9 and revealed that there is a positive correlation between the expression levels of the two genes (P < 0.001) (Fig. 6F and H).
Fig. 6.
A diagnostic model for differentiating HCC from normal (A-H) and dysplastic nodule (I-P) samples. A and C Confusion matrix of the binary results in the diagnostic model for distinguishing HCC and normal subjects. B and D ROC curves confirming the predictive accuracy of the diagnostic model. E and G Expression levels of SMG5 and MRPL9 in patients with HCC: distribution of the predicted results and actual results. F and H The correlation between the expression of SMG5 and MRPL9. I and K The specificity and sensitivity of the diagnostic model for distinguishing HCC lesions from dysplastic nodules. J and L ROC curves validating the predictive performance of the diagnostic model. M and O Expression characteristics of SMG5 and MRPL9 in patients with HCC: distribution of the predicted results and actual results. N and P Positive correlation of SMG5 and MRPL9 expression levels.
Currently, early diagnosis of HCC mainly relies on radiology examination combined with pathological diagnosis, but small nodules<2 cm have an increased likelihood of being missed because they are difficult to characterize via pathological or radiology examination [29]. We built a diagnostic model integrating SMG5 and MRPL9 in a training set (GSE6467) with 35 HCC samples and 17 dysplastic nodule samples to detect whether HCC and dysplastic nodules could be accurately distinguished. The diagnostic formula was identified as follows: logit (P-HCC) = -51.308 +(2.967 × SMG5 expression level) + (3.966 × MRPL9 expression level). GSE98620, which contains data from 49 HCC samples and 24 dysplastic nodule samples, was adopted as a validation set to further validate the diagnostic capability of the diagnostic model. The results determined that the diagnostic model could correctly distinguish HCC from dysplastic nodules, with specificity of 82.35% and sensitivity of 91.43% in the training set (Fig. 6I) and specificity of 54.17% and sensitivity of 77.55% in the validation set (Fig. 6K). The AUC reached 0.948 in the training set (Fig. 6J) and 0.776 in the validation set (Fig. 6L), indicating high consistency between predicted diagnostic status and actual status. The expression of SMG5 and MRPL9 in patients, predicted disease status and actual status are shown in Fig. 6M and P. Consistent with the above finding, the expression levels of SMG5 and MRPL9 were positively correlated (P < 0.001) (Fig. 6N and 6P).
Determination of the correlation of SMG5 and MRPL9 with prognosis and recurrence of HCC
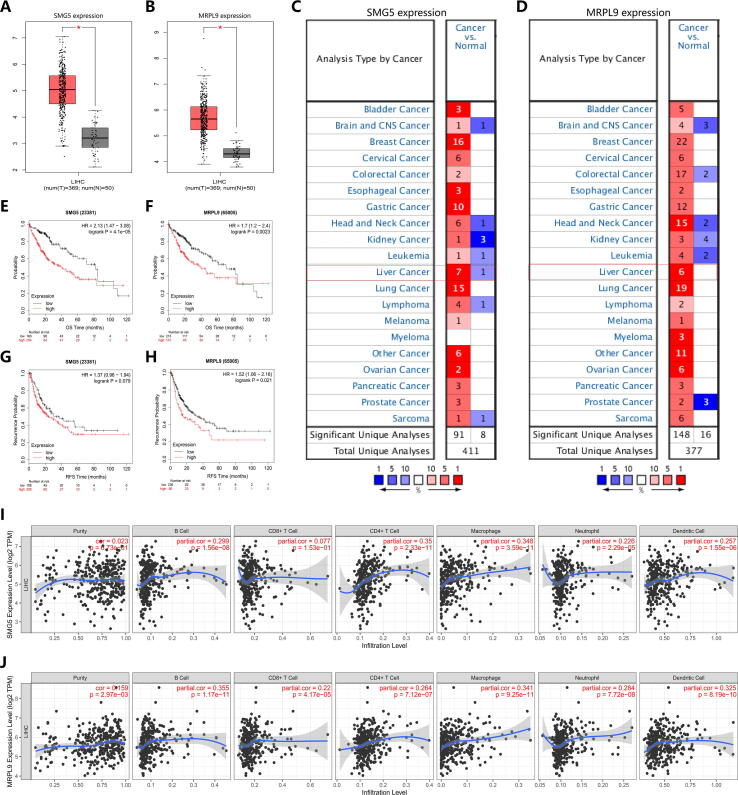
We explored the expression characteristics of SMG5 and MRPL9 through the online database Oncomine and the data visualization platform GEPIA for external validation. We found that in the Oncomine database, higher expression levels of SMG5 and MRPL9 was exhibited in tumor tissues compared to normal tissues (Fig. 7A,B), and the results of analysis in GEPIA were similar (Fig. 7C,D). A log-rank test and K-M curve analysis were conducted to further assess the predictive value of SMG5 and MRPL9 expression for prognosis and recurrence in HCC. The results determined that patients with HCC and higher expression levels of SMG5 and MRPL9 had remarkedly shorter survival time (Fig. 7E,F) and lower relapse-free survival (RFS) time (Fig. 7G,H) than those with lower expression levels of SMG5 and MRPL9. The above findings confirmed that the prognostic model based on SMG5 and MRPL9 is reasonable and holds value in clinical practice. Besides, we used the data of HCC patients in the TCGA-LIHC cohort to evaluate the correlation between SMG5 and MRPL9 and immune cells in the TIMBER database. Fig. 7I,J shows that the expression of these two genes conducted correlation with the infiltration level of immune cells, such as B cells, CD4 + T cells, macrophages, and neutrophils.
Fig. 7.
The value of SMG5 and MRPL9 in predicting the prognosis and recurrence of HCC. A-B The expression of SMG5 (A) and MRPL9 (B) as assessed in Oncomine. C-D The expression of SMG5 (C) and MRPL9 (D) as assessed in GEPIA. E-F Survival analysis for SMG5 (E) and MRPL9 (F). G-H Recurrence analysis for SMG5 (G) and MRPL9 (H). I-J The correlation of the expression of SMG5 (I) and MRPL9 (J) with the infiltration of different immune cells.
Measurement of the oncogenic effect of SMG5 and MRPL9 in HCC cells
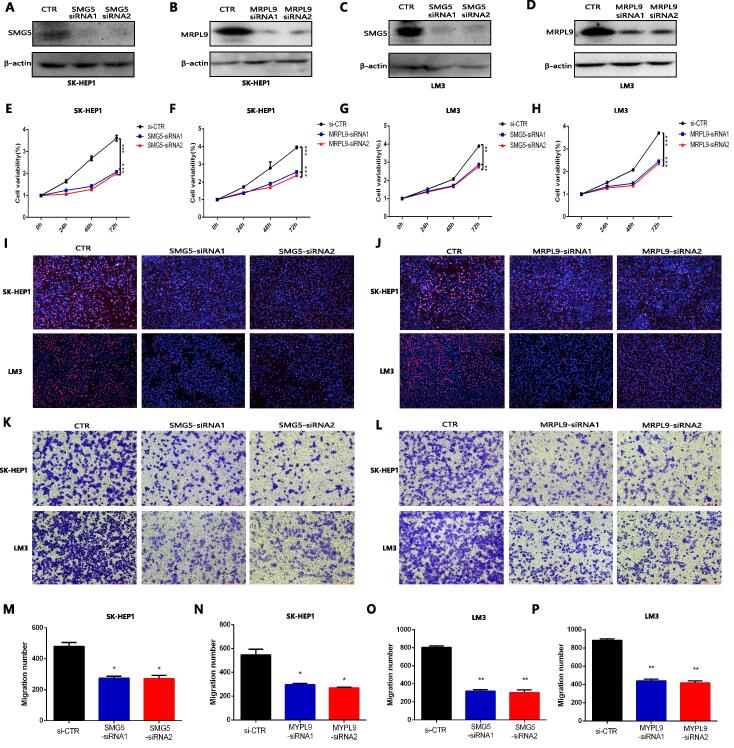
To further explore the role of SMG5 and MRPL9 on the proliferation and migration of HCC cells, the expression of SMG5 and MRPL9 in human LO2 hepatocytes and different HCC cell lines was detected using qPCR. The results showed that SMG5 and MRPL9 exhibited the highest expression in SK-HEP1 and LM3 cells (Figure S5A and S5B), so we used SKHEP1 and LM3 to perform follow-up experiments. We knocked down the levels of SMG5 and MRPL9 in HCC cell lines (including SK-HEP1 and LM3 cells). Western blotting showed that the administration of SMG5 siRNA and MRPL9 siRNA significantly inhibited the expression of SMG5 and MRPL9 in cells (Fig. 8A–D). The CCK assay indicated that both SMG5 inhibition (Fig. 8E and G) and MRPL9 inhibition (Fig. 8F and 8H) significantly inhibited the proliferation of HCC cells. The EdU assay also determined that SMG5 and MRPL9 inhibition showed a significant inhibitory effect on the proliferation of SK-HEP1 cells and LM3 cells. (Fig. 8I,J). In addition, the results of transwell analysis revealed that SMG5 and MRPL9 inhibition obviously inhibited the migration of SK-HEP1 cells and LM3 cells (Fig. 8K–L). The quantitative statistics are presented in Fig. 8M–P. The findings above confirmed that the expression of SMG5 and MRPL9 had a close association with the proliferation and migration of HCC cells and further confirmed the value of SMG5 and MRPL9 in predicting the prognosis of HCC.
Fig. 8.
The effect of SMG5 and MRPL9 on the progression of HCC. A–D Western blot analysis confirmed that the expression of SMG5 and MRPL9 was inhibited by SMG5 and MRPL9 siRNA administration. E-H CCK8 assay indicated that SMG5 and MRPL9 inhibition significantly suppressed the proliferation of SK-HEP1 cells (E-F) and LM3 cells (G-H). I-J EdU assay revealed that SMG5 and MRPL9 inhibition showed a significant inhibitory effect on the proliferation of SK-HEP1 cells and LM3 cells, respectively. K-L Transwell migration assays confirmed that SMG5 and MRPL9 inhibition obviously inhibited the migration of SK-HEP1 cells and LM3 cells. M−P Quantitative statistical results of the effects of SMG5 and MRPL9 expression on the migration of SK-HEP1 cells (M−N) and LM3 cells (O-P). Data are shown as the mean ± SD of at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
HCC is a major health problem worldwide with its increasing morbidity and mortality [30]. HCC exhibits obvious molecular heterogeneity, including various somatic genome mutations in addition to epigenetic modifications [2]. However, to date, no proposed molecular markers can accurately predict the prognosis or recurrence of HCC. TMB is a characteristic representing the total number of somatic coding mutations in tumors [31]. There is increasing evidence that TMB has value as a potential biomarker in non-small-cell lung cancer, rectal cancer and other cancer types [18], [32], [33]. Therefore, identifying and utilizing genes that carry mutations that provide important contributions to TMB may help optimize the early diagnosis of HCC and predict the prognostic risk of patients.
The increased ease of use of high-throughput sequencing technology opens up opportunities for us to better understand the pathological process of HCC and identify key genes involved in the carcinogenesis of HCC [34]. In this study, we focused our research on those mutated genes mostly intricately involved in TMB and tried to clarify the prognostic and diagnostic value of these key genes in HCC through a series of comprehensive analyses to accurately stratify the disease risk of patients with HCC and develop individualized treatments. We used integrated analysis, Cox regression, and the LASSO method to determine that SMG5 and MRPL9 have strong prognostic value in HCC. Knockdown of SMG5 and MRPL9 was determined to significantly inhibit cell proliferation and migration in HCC, suggesting that the expression of these two genes has a close association with the progression of HCC. A prognostic model based on the two genes could independently predict the prognosis of HCC patients with high specificity and sensitivity. SMG5 is a regulator of the nonsense-mediated messenger RNA (mRNA) decay (NMD) pathway in cells, which is a cell quality control and post-transcriptional gene regulation mechanism. SMG5 is thought to have important regulatory effects on the viability of most multicellular organisms [35]. MRPL9 is a nuclear gene encoding protein components in the ribosome in mitochondria, which is essential for mitochondrial function [36].
HCC has an insidious onset and progresses rapidly, and it is usually diagnosed in advanced stages (BCLC stage B and C). Chemotherapy is one of the most important treatments for patients with advanced HCC, especially for those who cannot undergo surgical resection, have a poor response to TACE, and have extrahepatic metastases or vascular invasion [37]. However, tumor cells may develop intrinsic or acquired resistance during the process of chemotherapy, resulting in poor chemotherapy effects and affecting the prognosis of patients [38]. Changes in gene expression profiles associated with somatic mutations in HCC play a significant role in regulating the sensitivity of tumor cells to chemotherapy [39]. In addition to drug-resistant regulatory factors and carcinogenic drivers, somatic mutations can also produce uniquely altered proteins, including new antigens to which specific antitumor immunity can be raised. These mutation-derived antigens can be recognized by the immune system and targeted by immunotherapy [40], [41]. The more somatic mutations a tumor has, the more neoantigens it may form. Clinical studies have confirmed a significant correlation between the production of neoantigens and immune-mediated clinical responses [42]. TMB is thus also regarded as a potential marker for immunotherapy and can provide a useful estimate of tumor neoantigen load [43]. Through comprehensive analysis of genes that play an important role in TMB, potential markers may be found to identify patients sensitive to chemotherapy and suitable for immunotherapy.
In our study, we used a TMB-based prognostic model to stratify HCC risk and assess immune cell infiltration and immune checkpoint expression in patients with different risks and test patient response to different chemotherapy drugs; by so doing, we further explored the effect of the expression of SMG5 and MRPL9 on the response of HCC patients to immunosuppressive therapy and chemotherapy. We found that compared with low-risk patients, high-risk patients showed higher levels of immune cell infiltration and higher expression levels of immune checkpoints in tumor tissues, and high-risk patients were more resistant to chemotherapy drugs. Subsequent GSEA suggested that the genes involved in the prognostic model were related to the signaling pathways related to physiological processes such as the cell cycle [26], mismatch repair [27] and the spliceosome [28], which have been found to have a regulatory effect on the chemoresistance of cells in previous studies. It was reported that SMG5 is involved in regulating the expression of a series of genes with a wide range of cellular activity functions, including cell cycle progression [44], which may partly explain the differences in chemoresistance among patients with different prognostic risks. The findings above indicate that in clinical management, high-risk patients may be the beneficiaries of targeted immunotherapy, while low-risk patients can benefit more from chemotherapy.
The early diagnosis of HCC mainly relies on radiological examination and pathological examination. The development of imaging technology has improved the recognition rate of HCC, enabling small HCC tumors (<2 cm) to be diagnosed at an early stage for further histological identification [45], [46]. However, increasing evidence suggests that small nodules are difficult to characterize. There is no clear boundary in morphology and histology between high-grade proliferative nodules and well-differentiated HCC lesions, and it is difficult to accurately distinguish them [47], [48]. Therefore, it is urgent to find molecular markers to objectively and accurately identify HCC. Two diagnostic models including SMG5 and MRPL9 expression were confirmed to accurately distinguish HCC from normal and dysplastic nodule samples (<2 cm) in this study, suggesting that the model may be beneficial to improve the early detection rate of HCC and is conducive to early clinical intervention in patients with HCC, thus improving the prognosis and likelihood of relapse of patients. Our findings lay the foundation for using SMG5 and MRPL9 as biomarkers for the early diagnosis of HCC patients.
Inevitably, there is some limitations in our study. First, when using the LASSO method to filter genes in the early analysis, the limitations of the LASSO method itself led to the potential for missing some genes with similarly important contributions when the regression coefficients were adjusted. The clinical characteristics considered in the independence analysis of the prognostic model and the construction of the nomogram are traditionally recognized as important factors affecting the carcinogenesis of HCC, while some clinical variables with similar contributions, such as geographical factors, dietary patterns, and exposure to endogenous or exogenous chemicals, were not included in the study due to insufficient sample information, which may affect the results. When identifying HCC lesions and nodules, the small sample size also affected the evaluation of the performance of the diagnostic model. Besides, we will further confirm the performance of the prognostic model for evaluating immunotherapy and chemotherapy response in follow-up clinical trials.
Conclusion
The prognostic model based on SMG5 and MRPL9 expression was proven to have accurate prediction capability, and it helped to identify patients suitable for immunotherapy. Patients with higher prognostic risks may obtain better clinical benefits through immunotherapy and achieve greater tumor remission. Prognostic risk was confirmed to have a close correlation with chemotherapeutic response. Low-risk patients had a significant survival advantage compared to high-risk patients receiving chemotherapy. Diagnostic models constructed using SMG5 and MRPL9 can make an accurate distinction between HCC and normal and dysplastic nodule samples. The comprehensive analysis of multidimensional genomic data opens up new ideas for determining patient treatment strategies and improving disease outcomes.
Data and materials availability
The data and materials applied in supporting the findings in this study are available from the corresponding author upon request.
Author Contributions
Jiansong Ji and Risheng Yu conceived and designed the experiments. Bufu Tang and Jinyu Zhu performed the experiments. Zhongwei Zhao, Chenying Lu, Siyu Liu and Shiji Fang analyzed the data. Liyun Zheng, Nannan Zhang, Minjiang Chen and Min Xu contributed analysis tools. Jinyu Zhu and Bufu Tang wrote the paper. Jiansong Ji and Risheng Yu edited the paper. All authors read and approved the final manuscript.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Declaration of Competing Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Acknowledgments
The authors of the present work are truly grateful to the TCGA, ICGC and GEO databases for the availability of their data.
Funding
This study was supported by National Natural Science Foundation of China (Nos. 81803778), and The Key Research and development Project of Zhejiang Province (No. 2018C03024), and The Public Welfare Research Program of Zhejiang Province (Nos. LQ20H160056 and LGD19h160002), The Natural Science Foundation of Zhejiang Province (No. LYQ20H280003).
Glossary
- HCC
hepatocellular carcinoma
- SMG5
SMG5 nonsense-mediated mRNA decay factor
- MRPL9
mitochondrial ribosomal protein L9
- LASSO
the least absolute shrinkage and selection operator
- HR
hazard ratio
- SNPs
single-nucleotide polymorphisms
- TMB
tumor mutation burden
- WGCNA
weighted correlation network analysis
- PI
prognostic index
- ROC curve
receiver operating characteristic curve
- AUC
area under the curve
- DCA
decision curve analysis
- OS
overall survival
- RFS
relapse-free survival
- HLA
human leukocyte antigen
- IC50
half the maximum inhibitory concentration
- GSEA analysis
gene set enrichment analysis
- GSVA
gene set variation analysis
- GDC
General Genomic Data
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2021.01.018.
Contributor Information
Risheng Yu, Email: risheng-yu@zju.edu.cn.
Jiansong Ji, Email: lschrjjs@163.com.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Yang J.D., Hainaut P., Gores G.J., Amadou A., Plymoth A., Roberts L.R. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589–604. doi: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forner A., Reig M., Bruix J. Hepatocellular carcinoma. Lancet (London, England) 2018;391:1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- 3.Aravalli R.N., Steer C.J., Cressman E.N. Molecular mechanisms of hepatocellular carcinoma. Hepatology (Baltimore, MD) 2008;48:2047–2063. doi: 10.1002/hep.22580. [DOI] [PubMed] [Google Scholar]
- 4.Farazi P.A., DePinho R.A. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674–687. doi: 10.1038/nrc1934. [DOI] [PubMed] [Google Scholar]
- 5.Weston A.D., Hood L. Systems biology, proteomics, and the future of health care: toward predictive, preventative, and personalized medicine. J Proteome Res. 2004;3:179–196. doi: 10.1021/pr0499693. [DOI] [PubMed] [Google Scholar]
- 6.Thorgeirsson S.S., Lee J.S., Grisham J.W. Functional genomics of hepatocellular carcinoma. Hepatology (Baltimore, MD) 2006;43:S145–S150. doi: 10.1002/hep.21063. [DOI] [PubMed] [Google Scholar]
- 7.S.L. Topalian, J.D. Wolchok, T.A. Chan, I. Mellman, and K.D.J.C. Wittrup, Immunotherapy: The Path to Win the War on Cancer? 161 (2015) 185-6. [DOI] [PMC free article] [PubMed]
- 8.Brown Z.J., Heinrich B., Greten T.F. Mouse models of hepatocellular carcinoma: an overview and highlights for immunotherapy research. Nat Rev Gastroenterol Hepatol. 2018;15:536–554. doi: 10.1038/s41575-018-0033-6. [DOI] [PubMed] [Google Scholar]
- 9.Segal N.H., Parsons D.W., Peggs K.S., Velculescu V., Kinzler K.W., Vogelstein B. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68:889–892. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- 10.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snyder A., Makarov V., Merghoub T., Yuan J., Zaretsky J.M., Desrichard A. Genetic basis for clinical response to CTLA-4 blockade in melanoma. New England J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melendez B., Van Campenhout C., Rorive S., Remmelink M., Salmon I., D'Haene N. Methods of measurement for tumor mutational burden in tumor tissue. Translat Lung Cancer Res. 2018;7:661–667. doi: 10.21037/tlcr.2018.08.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chalmers Z.R., Connelly C.F., Fabrizio D., Gay L., Ali S.M., Ennis R. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Owada-Ozaki Y., Muto S., Takagi H., Inoue T., Watanabe Y., Fukuhara M. Prognostic Impact of Tumor Mutation Burden in Patients With Completely Resected Non-Small Cell Lung Cancer: Brief Report. J Thoracic Oncol Off Publicat Int Assoc Study Lung Cancer. 2018;13:1217–1221. doi: 10.1016/j.jtho.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Innocenti F., Ou F.S., Qu X., Zemla T.J., Niedzwiecki D., Tam R. Mutational Analysis of Patients With Colorectal Cancer in CALGB/SWOG 80405 Identifies New Roles of Microsatellite Instability and Tumor Mutational Burden for Patient Outcome. J Clin Oncol Off J American Soc Clin Oncol. 2019;37:1217–1227. doi: 10.1200/JCO.18.01798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyd S.D. Diagnostic applications of high-throughput DNA sequencing. Annual Rev Pathol. 2013;8:381–410. doi: 10.1146/annurev-pathol-020712-164026. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z., Li M., Jiang Z., Wang X. A Comprehensive Immunologic Portrait of Triple-Negative Breast Cancer. Transl Oncol. 2018;11:311–329. doi: 10.1016/j.tranon.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodman A.M., Kato S., Bazhenova L., Patel S.P., Frampton G.M., Miller V. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther. 2017;16:2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niemira M., Collin F., Szalkowska A., Bielska A., Chwialkowska K., Reszec J. Molecular Signature of Subtypes of Non-Small-Cell Lung Cancer by Large-Scale Transcriptional Profiling: Identification of Key Modules and Genes by Weighted Gene Co-Expression Network Analysis (WGCNA) Cancers. 2019;12 doi: 10.3390/cancers12010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tibshirani R. Regression shrinkage selection via the LASSO. J Roy Stat Soc B. 2011;73:273–282. [Google Scholar]
- 23.Gentles A.J., Newman A.M., Liu C.L., Bratman S.V., Feng W., Kim D. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.K.M. Olthoff, M.H. Rosove, C.R. Shackleton, D.K. Imagawa, D.G. Farmer, P. Northcross, A.L. Pakrasi, P. Martin, L.I. Goldstein, A. Shaked, and et al., Adjuvant chemotherapy improves survival after liver transplantation for hepatocellular carcinoma. Annals of surgery 221 (1995) 734-41; discussion 731-43. [DOI] [PMC free article] [PubMed]
- 26.Cheng J., Cashman J.R. PAWI-2 overcomes tumor stemness and drug resistance via cell cycle arrest in integrin β(3)-KRAS-dependent pancreatic cancer stem cells. Sci Rep. 2020;10:9162. doi: 10.1038/s41598-020-65804-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higuchi F., Nagashima H., Ning J., Koerner M.V.A., Wakimoto H., Cahill D.P. Restoration of Temozolomide Sensitivity by PARP Inhibitors in Mismatch Repair Deficient Glioblastoma is Independent of Base Excision Repair. Clinical Cancer Res Off J American Assoc Cancer Res. 2020;26:1690–1699. doi: 10.1158/1078-0432.CCR-19-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wojtuszkiewicz A., Assaraf Y.G., Maas M.J., Kaspers G.J., Jansen G., Cloos J. Pre-mRNA splicing in cancer: the relevance in oncogenesis, treatment and drug resistance. Expert Opin Drug Metab Toxicol. 2015;11:673–689. doi: 10.1517/17425255.2015.993316. [DOI] [PubMed] [Google Scholar]
- 29.Kojiro M. Liver transplantation : official publication of the American Association for the Study of Liver Diseases and the International Liver Transplantation Society. 2004. Focus on dysplastic nodules and early hepatocellular carcinoma: an Eastern point of view; pp. S3–S8. [DOI] [PubMed] [Google Scholar]
- 30.Ogunwobi O.O., Harricharran T., Huaman J., Galuza A., Odumuwagun O., Tan Y. Mechanisms of hepatocellular carcinoma progression. World J Gastroenterol. 2019;25:2279–2293. doi: 10.3748/wjg.v25.i19.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fancello L., Gandini S., Pelicci P.G., Mazzarella L. Tumor mutational burden quantification from targeted gene panels: major advancements and challenges. J ImmunoTher Cancer. 2019;7:183. doi: 10.1186/s40425-019-0647-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rizvi N.A., Hellmann M.D., Snyder A., Kvistborg P., Makarov V., Havel J.J. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science (New York, N.Y.) 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balar A.V., Galsky M.D., Rosenberg J.E., Powles T., Petrylak D.P., Bellmunt J. Atezolizumab as first-line treatment in cisplatin-ineligible patients with locally advanced and metastatic urothelial carcinoma: a single-arm, multicentre, phase 2 trial. Lancet (London, England) 2017;389:67–76. doi: 10.1016/S0140-6736(16)32455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt B., Hildebrandt A. Next-generation sequencing: big data meets high performance computing. Drug Discovery Today. 2017;22:712–717. doi: 10.1016/j.drudis.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 35.Nicholson P., Gkratsou A., Josi C., Colombo M., Muhlemann O. Dissecting the functions of SMG5, SMG7, and PNRC2 in nonsense-mediated mRNA decay of human cells. RNA (New York, N.Y.) 2018;24:557–573. doi: 10.1261/rna.063719.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graack H.R., Grohmann L., Kitakawa M., Schafer K.L., Kruft V. YmL9, a nucleus-encoded mitochondrial ribosomal protein of yeast, is homologous to L3 ribosomal proteins from all natural kingdoms and photosynthetic organelles. Eur J Biochem. 1992;206:373–380. doi: 10.1111/j.1432-1033.1992.tb16937.x. [DOI] [PubMed] [Google Scholar]
- 37.Kudo M., Trevisani F., Abou-Alfa G.K., Rimassa L. Hepatocellular Carcinoma: Therapeutic Guidelines and Medical Treatment. Liver cancer. 2016;6:16–26. doi: 10.1159/000449343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ikeda M., Mitsunaga S., Ohno I., Hashimoto Y., Takahashi H., Watanabe K. Systemic Chemotherapy for Advanced Hepatocellular Carcinoma: Past, Present, and Future. Diseases (Basel, Switzerland) 2015;3:360–381. doi: 10.3390/diseases3040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma J., Zeng S., Zhang Y., Deng G., Qu Y., Guo C. BMP4 promotes oxaliplatin resistance by an induction of epithelial-mesenchymal transition via MEK1/ERK/ELK1 signaling in hepatocellular carcinoma. Cancer Lett. 2017;411:117–129. doi: 10.1016/j.canlet.2017.09.041. [DOI] [PubMed] [Google Scholar]
- 40.Riaz N., Morris L., Havel J.J., Makarov V., Desrichard A., Chan T.A. The role of neoantigens in response to immune checkpoint blockade. Int Immunol. 2016;28:411–419. doi: 10.1093/intimm/dxw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ladanyi A., Timar J. Immunologic and immunogenomic aspects of tumor progression. Semin Cancer Biol. 2020;60:249–261. doi: 10.1016/j.semcancer.2019.08.011. [DOI] [PubMed] [Google Scholar]
- 42.Van Allen E.M., Miao D., Schilling B., Shukla S.A., Blank C., Zimmer L. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science (New York N.Y.) 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samstein R.M., Lee C.H., Shoushtari A.N., Hellmann M.D., Shen R., Janjigian Y.Y. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rehwinkel J., Letunic I., Raes J., Bork P., Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA (New York, N.Y.) 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barreiros A.P., Piscaglia F., Dietrich C.F. Contrast enhanced ultrasound for the diagnosis of hepatocellular carcinoma (HCC): comments on AASLD guidelines. J Hepatol. 2012;57:930–932. doi: 10.1016/j.jhep.2012.04.018. [DOI] [PubMed] [Google Scholar]
- 46.Parisi G. Should a radiological diagnosis of hepatocellular carcinoma be routinely confirmed by a biopsy? Yes. Eur J Intern Med. 2012;23:34–36. doi: 10.1016/j.ejim.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 47.Sakamoto M., Hirohashi S., Shimosato Y. Early stages of multistep hepatocarcinogenesis: adenomatous hyperplasia and early hepatocellular carcinoma. Hum Pathol. 1991;22:172–178. doi: 10.1016/0046-8177(91)90039-r. [DOI] [PubMed] [Google Scholar]
- 48.Terminology of nodular hepatocellular lesions. Hepatology (Baltimore, Md.) 22 (1995) 983-93. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.