Abstract
Background:
Hepatocellular carcinoma (HCC) is the second leading cause of cancer-related death worldwide. There is an urgent demand for prognostic biomarkers that facilitate early tumor detection, as the incidence of HCC has tripled in the USA in the last three decades. Biomarkers to identify populations at risk would have significant impact on survival. We recently found that expression of Neuronal Calcium Sensor 1 (NCS1), a Ca2+-dependent signaling molecule, predicted disease outcome in breast cancer, but its predictive value in other cancer types is unknown. This protein is potentially useful because increased NCS1 regulates Ca2+-signaling and increased Ca2+-signaling is a hallmark of metastatic cancers, conferring cellular motility and an increasingly aggressive phenotype to tumors.
Methods:
We explored the relationship between NCS1 expression levels and patient survival in two publically available liver cancer cohorts and a tumor micro array using data mining strategies.
Results:
High NCS1 expression levels are significantly associated with worse disease outcome in Asian patients within these cohorts. In addition, a variety of Ca2+-dependent and tumor growth-promoting genes are transcriptionally co-regulated with NCS1 and many of them are involved in cytoskeleton organization suggesting that NCS1 induced dysregulated Ca2+-signaling facilitates cellular motility and metastasis.
Conclusions:
We found NCS1 to be a novel biomarker in HCC. Furthermore, our study identified a pharmacologically targetable signaling complex that can influence tumor progression in HCC.
Impact:
These results lay the foundation for utilizing NCS1 as a prognostic biomarker in prospective cohorts of HCC patients and for further functional assessment of the characterized signaling axis.
Keywords: Ca2+-signaling, cell motility, data mining, LIM Kinase 1, cancer biomarker
Introduction
Hepatocellular carcinoma (HCC) is one of the deadliest cancer types worldwide1, with rapidly increasing rates in the US2,3. Currently, options to cure or palliate HCC are limited to resection, transplantation, and ablative and transarterial therapies, with therapeutic choices dictated by both tumor burden and the extent of hepatic dysfunction. There is only one FDA approved first line, palliative systemic chemotherapeutic agent, an oral tyrosine kinase inhibitor (sorafenib). This treatment is reserved for patients who have failed or progressed after local therapies, offering an overall survival advantage of 3 months with a significant adverse side effect profile4. In order to develop and optimize new treatment modalities for HCC, novel biomarkers are needed that can predict tumor aggressiveness and identify druggable targets that can be exploited to create innovative therapies.
Calcium is long known to be a major regulator of cell cycle progression and cellular proliferation, events promoted by growth factors which often cause oscillatory patterns of Ca2+ release from ER stores and Ca2+ entry from the extracellular space5-7. Furthermore, recent clinical and basic research evidence implicates a Ca2+-driven pro-metastatic phenotype in aggressive tumors with an unfavorable prognosis8-10. To date, only one component of Ca2+-signaling pathways has been proposed as a potential biomarker for HCC. High expression of S100 Ca2+-binding proteins is reported to confer an aggressive phenotype in undifferentiated liver cancers11-14. Although less well characterized, other members of the Ca2+ - signaling cascade have been implicated in cancer progression15.
Neuronal Calcium Sensor 1 (NCS1) is a clinically validated prognostic biomarker in two independent breast cancer cohorts, that is closely related to states of dysregulated Ca2+-signaling10. NCS1, a member of the calmodulin superfamily of EF-hand Ca2+-sensing proteins, is involved in various cellular processes, including exocytosis16, regulation of voltage-gated Ca2+-channels, neuroplasticity17, and calmodulin-mediated signaling18. NCS1 interacts with inositol 1,4,5-triphosphate receptor type 1 (InsP3R1) and leads to an increase in InsP3-dependent Ca2+-release19, especially in an overexpression scenario20,21. NCS1 also is upregulated in injured neurons, acting as a survival factor via activation of PI3K/AKT pathway22. These functional properties and its expression in different extra-neuronal tissues23-26 together with its likely role in breast cancer development make NCS1 a possible contributor to carcinogenesis in other tumor types as well.
To investigate the possible role of NCS1 in Ca2+-mediated tumor progression and metastasis in HCC, we analyzed The Cancer Genome Atlas (TCGA) Liver Hepatocellular Carcinoma (LIHC) dataset to determine if there is an association of NCS1 with patient survival and if NCS1 can distinguish among patients, either by sub-population or etiologic risk factor. In addition, we built a regression model to test for significant co-expression of NCS1 using the approximately 60 000 RNA-sequenced transcripts included in the TCGA LIHC dataset. Using information from the regression model, we identified pathways involved in Ca2+ and NCS1 signaling that influence tumor progression and can be pharmacologically manipulated.
Experimental Procedures
Data Download:
The results shown here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/. TCGA LIHC, breast cancer (BRCA) and lung cancer (LUAD and LUSC) expression data and clinical annotations were download from the NIH’s Genomic Data Commons (GDC) Data Portal (https://portal.gdc.cancer.gov) on 10 October 2017. Plain text files with FPKM values for 60 483 transcripts per patient were joined with corresponding clinical annotations and gene names were added using unique ENSG identifiers (ensembl.org, GRCh38.p11). NCS1 and LIMK1 expression values and clinical annotations related to the International Cancer Genome consortium (ICGC) LIRI-JP liver cancer cohort were downloaded from the ICGC data portal (https://icgcportal.genomics.cn) on 22 February 2018. All data transformation and analyses were performed with R Statistical Programming Language, Version 3.4.0 (2017-04-21)27. Additional information regarding sample processing, patient characteristics, data acquisition and global mutational and expressional analyses are available with The Cancer Genome Atlas Research Network’s publications of these datasets28-31. Data for NCS1, GAPDH, RPS18, HPRT1 and ACTB expression levels in various tissues that lack HCC (assumed to be healthy for the purposes of this study) were accessed through the websites of The Genotype-Tissue Expression (GTEx) Project (https://www.gtexportal.org/home/) as well as The Human Protein Atlas (https://www.proteinatlas.org; both on 1 November 2017). Pre-processed gene expression data of 1019 cancer cell lines are publically available through Broad Institute’s Cancer Cell Line Encyclopedia (CCLE; https://portals.broadinstitute.org/ccle) and were downloaded on 10 January 2018.
Bioinformatics and Statistical Analyses:
The between-group comparisons of overall survival were performed by means of two-sided log-rank tests stratified according to NCS1 (<75% vs. >= 75%), WDR5 (<80% vs. >= 80) and LIMK1 (<60% vs. >60%) expression levels. Cutoff values for dichotomizing continuous expression values to ‘high’ and ‘low’ were derived from receiver operating characteristics (ROC) curves using Youden’s J statistic. A Cox proportional-hazards model that included dichotomized NCS1, LIMK1 and WDR5 expression as covariates was used to estimate hazard ratios and their associated 95% confidence intervals. The Kaplan-Meier method was used to estimate survival curves and a log-rank test to compare survival between distinct expression subgroups. Survival plots and forest plots were generated using the ‘survminer’ R package32. In all analyses, Patient samples were excluded whenever survival data or expression values were missing (<10% per cohort and analysis).
To assess transcriptional co-regulation of the 60 482 transcripts with NCS1 expression in LIHC, BRCA, LUAD and LUSC datasets, simple linear regression analysis was performed. P-values derived from these analyses were corrected using Bonferroni’s method and top hits were identified by means of corrected p-values. We pre-specified a threshold for Pearson’s correlation coefficient of 0.6 or higher to indicate a strong correlation, mainly to reduce the false positive rate33. Again, a univariate linear regression model was used to compute the relationship between NCS1 and LIMK1 or WDR5 in the TCGA, ICGC and CCLE datasets.
Exploratory Gene Association Networks (EGAN)34 software was used to combine NCS1-correlated genes and database-derived gene and pathway annotations into meaningful network plots. A list of 40 genes, comprising the top 10 hits from all 4 datasets, was examined for overlaps with pathways from the Molecular Signature Database (MSigDB, http://software.broadinstitute.org/gsea/msigdb) and by means of EGAN software. Results with an FDR q-value < 0.05 were called significant and listed as relevant. Welch’s two sample t-test was used to test for statistical differences of sample means if not otherwise specified and Fisher’s exact test was used to analyze contingency tables.
Liver Cancer Tissue Array:
A commercially available hepatocellular carcinoma microarray with 49 cases (2 cores per case, 41 tumors with matched non-neoplastic adjacent tissue, 6 tumors without matched non-neoplastic tissue, and 2 normal liver tissues without corresponding tumors) was purchased from US Biomax, Inc. (OD-CT-DgLivT10-024, HLiv-HCC180Bch-01). Immunofluorescence staining for NCS1 and arginase was performed and automated quantitative analysis (AQUA) with arginase as a marker for tumor tissue compared to stromal tissue was used to quantify NCS1 protein levels in tumors and adjacent non-neoplastic tissues, as previously described10. Biochemical indicators (AFP, CA19-9, CEA, hepatitis B/C markers, Creatinine, BUN) and tumor stage data are available on the supplier’s website.
Results
Tissue NCS1 expression increases during HCC carcinogenesis.
To assess physiological NCS1 expression in normal liver tissue from healthy individuals, we explored two databases; The Genotype-Tissue Expression (GTEx) Project and The Human Protein Atlas (HPA). Analysis of both of these datasets suggests very low overall NCS1 levels in healthy human liver (mean NCS1 expression levels derived from RNA-sequencing specified as 0.9 TPM and 0.7 RPKM in 10 and 119 specimens in the HPA and GTEx datasets, respectively). Further, when compared to other organs, liver expression levels rank last and second to last among 31 and 36 analyzed tissues in the HPA and GTEx datasets, respectively. Because inter-experimental comparison of expression units is usually not possible, we calculated the ratio of NCS1 to the housekeeping gene β-actin (ACTB) in HPA, GTEx and TCGA LIHC datasets (Fig. 1a). The ratio of NCS1 to ACTB was 1.7- and 1.4-fold higher in cancerous tissue (p < 0.01 for the comparison of TCGA vs. GTEx and TCGA vs. HPA, respectively). To validate the choice of ACTB for normalization, three additional housekeeping genes (HPRT1, GAPDH and RPS1) were used to normalize the NCS1 levels (Supp. Fig. 1). An increase in NCS1 expression levels in the HCC samples was observed with all housekeeping genes. These results suggest NCS1 levels increase in liver tissue during HCC development.
Fig. 1: NCS1 Expression in Non-Neoplastic and Cancerous Tissue.
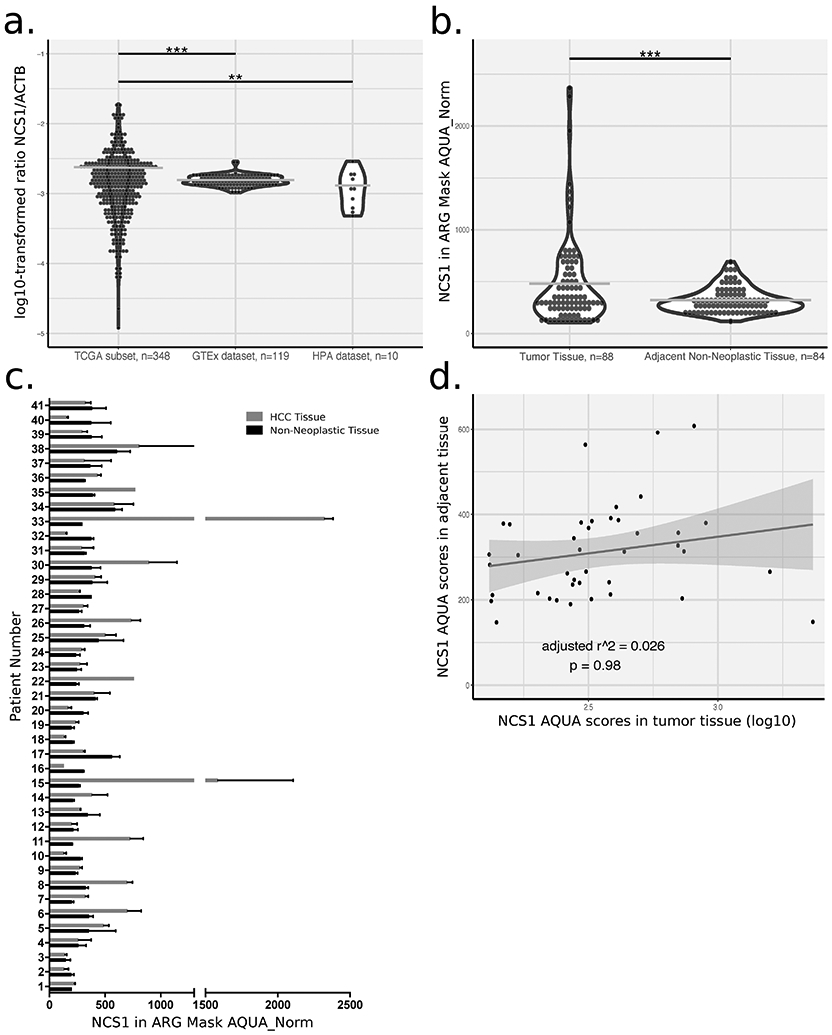
a. Dot plot showing the ratio of NCS1 to the housekeeping gene ACTB on a log10-transformed scale for 3 different datasets. *** denotes p-value <0.0001, ** denotes p-value <0.001 and horizontal lines indicate sample means. Violin plots visualize the overall data distribution per sample. Data on NCS1 and ACTB expression are only available for a subset of TCGA samples. b. Dot plot showing AQUA scores for NCS1 in tumor tissue compared to adjacent non-neoplastic tissue. Data is derived from a tumor micro array and a “tumor mask” was generated using of arginase (ARG) signal to discriminate tumorous from stromal tissue compartments. A paired t-test was used to assess group differences. c. Bar plot showing AQUA scores for tumors and adjacent normals of 41 patients. Error bars represent standard deviations of n = 2 cores per sample and bars represent sample means. d. Scatter plot for NCS1 AQUA scores in tumor tissue vs. matched adjacent non-neoplastic tissues. Trend line with 95% confidence interval (CI) is based on a simple linear regression model to visualize random distribution of the data and summary statistics of that regression model are added to the plot.
To further examine this hypothesis, a commercially available microarray composed of hepatocellular carcinoma samples and adjacent non-neoplastic tissues was analyzed using immunofluorescence to calculate AQUA scores. Matched adjacent non-neoplastic tissues were available for most tumor samples (41 / 47) on the array. A significant (p <0.0001) upregulation of NCS1 was observed in tumors (Fig. 1b), with 59% (24/41) of them showing increased NCS1 AQUA scores compared to matched non-neoplastic samples (Fig. 1c). However, in all of the samples, the levels of NCS1 in the non-neoplastic tissue is low and there is a very small range, suggesting that baseline NCS1 will not be a predictor for developement of HCC and that high baseline NCS1 expression cannot be a prerequisite for the delevopment of HCC. This is more clearly shown by the relationship between AQUA scores in tumors and non-neoplastic samples (Fig. 1d). Each data point is the expression of NCS1 in tumors and non-neoplastic samples in the same patient and the scatter of the data points show that there is no correlation. This relationship suggests that baseline NCS1 levels do not predict the expression level of NCS1 in the tumor samples and provides further evidence that increased NCS1 expression is associated with tumor development. An analysis of the limited clinical annotations available for the array showed no association of NCS1 AQUA scores in tumor tissue with either tumor stage (Supp. Fig. 2b), hepatitis B/C status, tumor markers (CEA, CA19-9, AFP) or any of the patient characteristics (age, gender; data not shown). No additional information on the spatial relationship between tumor samples and non-neoplastic adjacent tissues were available for analysis.
NCS1 levels predict survival status and time to death in a subgroup of Asian patients.
To test whether NCS1 levels correlate with tumor stage and outcome in the TCGA cohort of liver cancer patients, we examined the relationship between NCS1 expression measured in FPKM units and clinical tumor stage for 377 patients. Although advanced tumors did not show increased NCS1 expression compared to early stage tumors in this cohort (Supp. Fig. 2a), in the subgroup of Asian patients who died, average NCS1 expression was significantly increased compared to the survivors (t-test corrected p < 0.05 for male Asian patients, Fig. 1a). In addition, Asian patients had a higher mean NCS1 expression than white patients (t-test p = 0.056) and a significantly (p < 0.01) smaller proportion of NCS1low Asian patients died during the course of their disease (21.4% vs. 39.0% for Asian and white patients respectively, Fig. 1b), irrespective of tumor stage. This comparison suggests a protective effect of low NCS1 expression in NCS1low Asian patients. Differences in the expression profiles of proteins associated with HCC among races has recently been described35. With respect to Ca2+-signaling pathways, indel mutations in ryanodine receptors (RYRs; intracellular Ca2+ channels) were identified in Thai patients in this study35. Because the number of subjects from races other than Asians and whites constitute only a small part of the overall sample (Supp. Tab. 1), they were excluded from further analyses.
In the next analysis, we examined the prognostic value of dichotomized NCS1 expression levels (to NCS1high and NCS1low) with regard to patient survival at the last follow-up and over time. To explore the optimum cutoff for dichotomization of continuous FPKM values, we created a receiver operating characteristics (ROC) curve by calculating sensitivity and specificity for a range of different cutoffs and calculated Youden’s J statistic to capture the performance of our binary test (Fig. 1c). As a result, we categorized 25% of patients as NCS1high and 75% as NCS1low. When applying this cutoff to the survival status of Asian patients (i.e., patients being either dead or alive at the time of last follow-up), the estimated odds ratio of 0.26 shows a significant (Fisher’s exact test p < 0.001) association of high NCS1 expression levels with an unfavorable disease outcome (Fig. 1e). However, the relationship between NCS1 expression and prognosis is not true for white patients (Fig. 1d), confirming the result of our prior analysis.
We next estimated survival curves via the Kaplan-Meier method and applied a Cox proportional-hazards model with NCS1 as a single covariate to calculate hazard ratios for all patients in the TCGA LIHC cohort (Fig. 3a), as well as race-stratified for the subgroup of Asian (Fig. 3b) and for the subgroup of white patients (Fig. 3c). With this approach, Asians showed a significantly higher probability of death if categorized as NCS1high (Hazard ratio (HR) 3.5, p < 0.0001), whereas the time to death in white patients was independent of their NCS1 status (HR 0.89, p = 0.68). NCS1 status was also significantly associated with worse outcome in the Kaplan-Meier analysis of all patients (HR 1.7, p = 0.006), although with a hazard ratio closer to 1 than the ratio calculated for Asian patients only.
Fig. 3: NCS1 Status and Patient Survival.
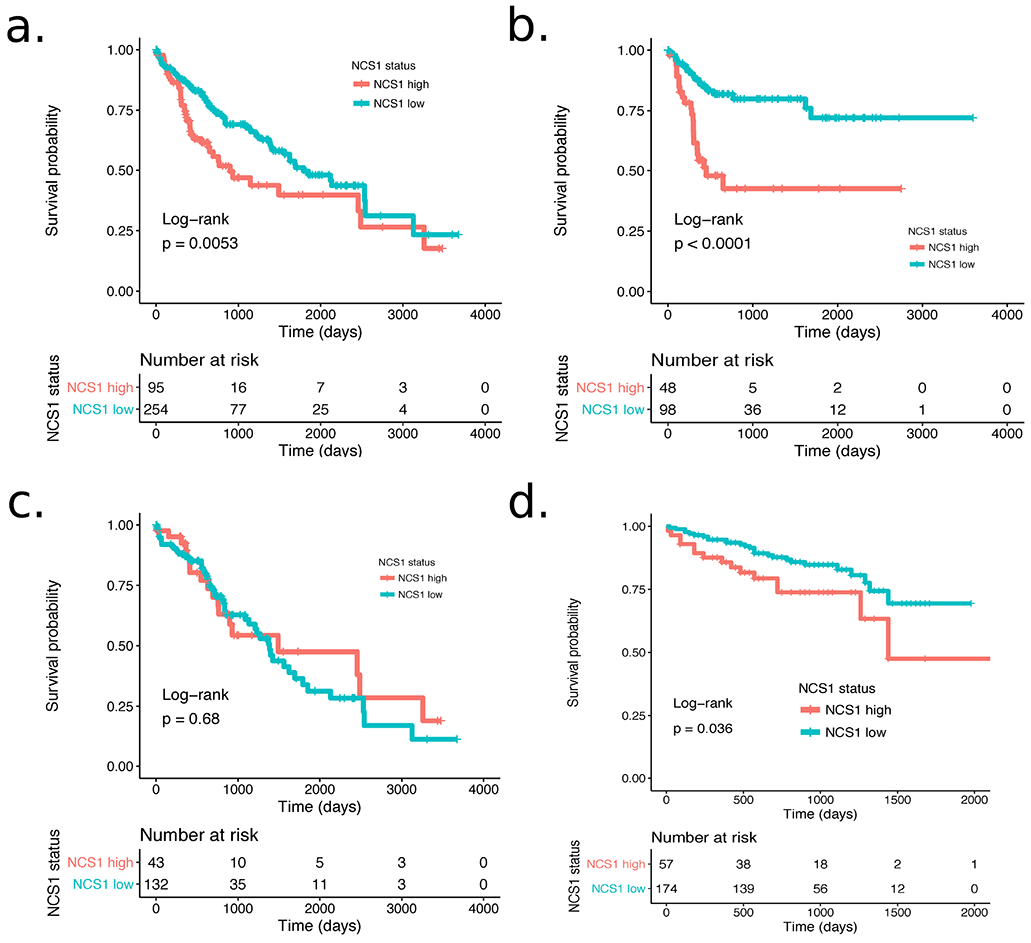
a. Kaplan-Meier plot showing survival probability for 349 patients from the TCGA LIHC cohort. Hazard ratio = 1.7 (p = 0.006, 95% CI 1.2 to 2.4). b. Kaplan-Meier plot showing survival probability for n = 146 Asian patients from the TCGA LIHC cohort. Hazard ratio = 3.5 (p < 0.0001, 95% CI 1.9 to 6.5). c. Kaplan-Meier plot showing survival probability for n = 175 white patients from the TCGA LIHC cohort. Hazard ratio = 0.89 (p = 0.68, 95% CI 0.52 to 1.5). d. Kaplan-Meier plot showing survival probability for 231 patients from the ICGC LIRI-JP cohort. Hazard ratio = 1.96 (p = 0.039, 95% CI 1.03 to 3.7).
NCS1 status predicts survival in an independent cohort of Japanese HCC patients.
To validate the strong relationship between high NCS1 expression and survival of Asian patients in the TCGA LIHC cohort, we then examined the International Cancer Genome Consortium (ICGC) LIRI-JP cohort36 that comprises 231 HCC samples from Japanese patients. After dichotomizing continuous NCS1 expression levels in NCS1high and NCSlow as described above, we again found that high expression was significantly associated with worse patient survival over time (Fig. 3d; with an unadjusted HR 1.96, p = 0.039).
LIMK1 and WDR5 are transcriptionally co-regulated with NCS1.
Because the exact signaling pathway that is influenced by NCS1 expression in tumor cells remains to be elucidated, we performed simple linear regression analyses of NCS1 with 60 483 RNA-sequenced transcripts from the TCGA LIHC dataset to assess transcriptional co-regulation of certain transcripts with NCS1. After correcting for multiple hypothesis testing, a total of 54 genes were significantly (p < 10−30 and Pearson’s correlation coefficient above the pre-specified threshold of 0.6) and positively correlated with NCS1 expression (Supp. Fig. 3 and Supp. Tab. 2). LIM Domain Kinase 1 (LIMK1) was the most significantly co-expressed gene and remarkably, LIMK1 stands out, separate from all the other genes that are associated with NCS1 (Supp. Fig. 3 and Supp. Fig. 4a, NCS1 vs. LIMK1 Pearson’s r = 0.76, p < 0.0001). We further validated the strong co-expression by exploring the relationship of NCS1 and LIMK1 in the ICGC LIRI-JP cohort (Supp. Fig. 4b, Pearson’s r = 0.72, p < 0.0001).
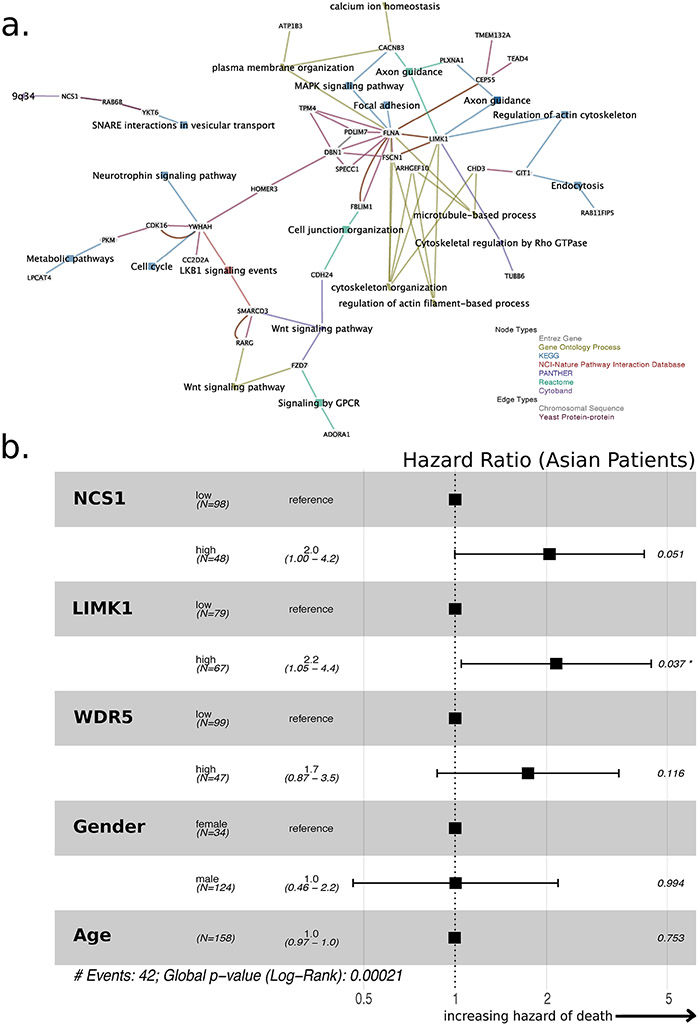
With this information, we constructed a co-expression network37 using Exploratory Gene Association Networks (EGAN, Fig. 4a)34 software. We found cytoskeleton organization to be the dominant pathway in the derived network. To further minimize bias and improve robustness of the approach, we analyzed three additional TCGA cohorts (LUAD, LUSC and BRCA; Supp. Tab. 2 for top hits and Supp. Fig. 5 for gene association networks) using the same methods and included only the 10 most significant results from each cohort in our pathway analysis.
Fig. 4: NCS1 Associated Signaling Pathways in HCC.
a. Co-expression network constructed from 54 NCS1 associated genes using EGAN software. 34 genes are actually part of the network. A legend in the lower right corner explains node and edge types. To not introduce bias, all pathways that comprise at least 2 of the 54 genes are included in the network. b. Forest plot showing hazard ratios derived from a Cox proportional-hazards model that includes dichotomized NCS1, LIMK1 and WDR5 expression, patient age, and gender as covariates to predict survival of Asian TCGA LIHC patients. Dichotomization cutoffs are chosen according to ROC curves. Number of events = 42, p = 0.0002 for all covariates together.
In this expanded cohort, two genes occurred among the top 10 NCS1-associated genes in at least two of four samples, namely LIMK1 (overlapped between two datasets) and WD Repeat Domain 5 (WDR5, overlapped between three datasets). We calculated a Cox proportional-hazards model that includes dichotomized NCS1, LIMK1 and WDR5 expression as covariates and adjusts for patient age and gender to investigate the prognostic significance of these three NCS1 associated genes in the TCGA LIHC cohort. Although not all of the calculated hazard ratios become significant due to the relatively small number of events, all analyses predicted an increased risk of death in patients with high expression of the respective genes compared to patients with low expression (Fig. 4b, Supp. Fig. 6a shows the respective forest plot for white patients). These effects were stable and independent of patient age or gender. In addition, we examined the genes that overlap with pathways from the Molecular Signature Database (MSigDB). Again, we found that the genes identified in our analysis are related to either Ca2+-dependent signaling pathways (cell cycle progression, cytoskeleton organization), neurogenesis or cancer-related pathways (Supp. Fig. 6b).
We also examined Broad Institute’s Cancer Cell Line Encyclopedia (CCLE) dataset that comprises RNA sequencing derived expression data of 1019 cell lines to further test and potentially validate the positive correlation between NCS1 and LIMK1 or WDR5. Consistent with our pathway analysis, the relationship between NCS1 and LIMK1 is observed in breast and liver cancers, but not in cell lines derived from hematological malignancies that do not require increased motility and cytoskeleton remodeling for metastasis. In contrast, NCS1 and WDR5 did not show a significant correlation in the CCLE dataset, independent of cancer type (Supp. Fig. 7a, b).
Discussion
In this study, we systematically examined the prognostic role of NCS1 and transcriptionally associated genes in a liver cancer cohort from a publically available database. Interestingly, healthy liver tissue exhibited lower overall NCS1 levels compared to HCC, suggesting an upregulation of NCS1 during tumorigenesis. We were able to validate this finding by using a tissue micro array where 59% of tumors harbored NCS1 levels (as assessed by a quantitative immunofluorescence technique) that were increased compared to the matched non-neoplastic tissues. We did not see a correlation between NCS1 levels in these matched adjacent non-neoplastic tissues and NCS1 levels in the respective tumor samples. This result supports our hypothesis that NCS1 is upregulated during carcinogenesis, in a manner independent of basal levels in the healthy organ or tumor-adjacent parenchyma. We suggest that NCS1 expression is a fixed, tumor dependent phenomenon that is associated with risk factors or concomitant liver diseases, irrespective of stage at diagnosis.
By correlating RNA-sequencing based expression to clinical outcome measures, specifically survival status of patients at the end of follow-up and time to death, we demonstrated a significantly worse prognosis for Asian patients with high NCS1 expression compared to Asians with low NCS1 expression. We validated this finding by investigating an additional, independent cohort of Asian HCC patients. Although we found NCS1 to be predictive of survival in an analysis of all TCGA samples, we hypothesize that the observed effect is mainly driven by the subpopulation of Asian patients because we did not see an association of NCS1 with survival for white patients. In addition to these analyses, we identified an associated signaling network whose central components further improved survival predictions. The expression pattern of these genes suggests a path for the development of prognostic biomarkers and potential targeted therapies.
One possible cause for the observed race-specific differences regarding survival might be the overall higher expression of NCS1 among Asian cancer patients. This differential expression may be caused by hepatocellular carcinoma-associated risk factors, including hepatitis or alcohol exposure, although a mechanistic explanation is beyond the scope of this study. Nevertheless, it is known that HCCs differ between Asian and white patients in terms of genomic properties35 and with respect to pro-carcinogenic risk factors38. In this regard, our study adds to an increasing body of literature investigating possible contributors to these phenomena. Importantly, our study suggests a specific cellular mechanism, dysregulated Ca2+-signaling, as a hallmark of HCC in a subgroup of patients.
To facilitate a better understanding of the biology of tumors expressing high levels of NCS1, we investigated additional cancer datasets, two lung cancer and one breast cancer datasets, for significant co-regulation of NCS1 with other transcripts. We found a range of pro-oncogenic genes to be highly expressed in NCS1high patients and identified the associated pathways to be cytoskeleton organization, neurogenesis and cell cycle progression. Furthermore, most of the identified genes are involved in cancer- and metastasis-related pathways, underscoring the validity of the analysis.
Two genes appeared to be NCS1-associated in at least two of the four examined TCGA datasets (WDR5 and LIMK1). WDR5 is a protein shown to be Ca2+-sensitive in amphibians39 and it has a critical role in recruiting the highly oncogenic transcription factor MYC40 and epigenetic modifiers MLL1 – 441,42 to chromatin43. This study is the first time WDR5 has been proposed to be associated with NCS1 as a crucial (dys)regulator of intracellular Ca2+-signaling. The WDR5/MLL complex has been suggested as a contributor to an unfavorable disease outcome in HCC44 and MLL4 is a known target of HBV integration, especially in Chinese patients45. However, WDR5 ranks only among the top 500 genes when analyzing the TCGA LIHC dataset. To include WDR5 as a critical component of tumorigenesis would require a robust validation of a functional role of WDR5 beyond mere association, especially as it is located in close chromosomal proximity to NCS1 and was not correlated in an analysis of RNA sequencing data from the CCLE.
LIMK1 is a promising therapeutic target that was previously studied with regard to metastasis and appeared prominently in our analyses of TCGA and CCLE datasets. Cellular motility, a hallmark of metastatic cancers, is mainly facilitated via actin polymerization46 and LIMK1 is a key regulator of this process47. In addition, Ca2+-dependent LIMK1 activation was observed in neuronal outgrowth48. The second most significantly co-expressed gene in the analyzed liver cancer cohort is ARHGEF10, a RhoA guanine nucleotide exchange factor that acts upstream of LIMK149. From the potential functional combination of these proteins along with components of our co-expression network (Fig. 4a), we hypothesize an increased activation of the RhoA – Rho-associated protein kinase (ROCK) – LIMK1 signaling axis in tumor cells exhibiting increased NCS1 levels50. This functional complex confers motility and allows tumor cells to interact with the stroma and microenvironment, and expressional upregulation would ultimately lead to decreased patient survival. Because potent inhibitors of LIMK1 have been developed that can reduce cellular motility in vitro51, LIMK1 is a potential clinical target in advanced HCC, possibly in combination with inhibitors that address additional components of a larger signaling network. Furthermore, our results suggest that the NCS1/LIMK1 signaling network can be modelled using cancer cell lines.
Other Ca2+-dependent pathways that have been causally related to HCC development (e.g. the lysophosphatidic acid pathway52) may be directly or indirectly associated with NCS1 expression. However, these pathways were not identified in the cohort included in this study.
The main limitations of this study are the relatively small number of Asian patients in the TCGA LIHC cohort, the focus on RNA-sequencing as the primary measure of protein expression, and the limited availability of patients’ past medical histories. Our hypothesis that NCS1 increases during hepatocarcinogenesis is based on a TMA with HCC samples and non-neoplastic adjacent tissues. To test this hypothesis, a longitudinal studies is required to explore the role of NCS1 during the evolution of HCC in greater depth. This is especially import with regard to recent evidence suggesting that liver tissue adjacent to a primary liver tumor might have expression patterns or phenotypes different from normal hepatocytes53.
Moreover, the excellent overall survival rate of about 65% after 10 years in the TCGA study suggests a bias in this cohort towards patients that were treated with a curative intention and thus had on average a longer survival than usually observed with HCC. Nevertheless, our study points to cytoskeleton remodeling, possibly facilitated by an interaction of NCS1 and the RhoA/ROCK/LIMK1 pathway, as an important determinant of aggressive HCC. The question whether disruption of this pathway applies only to a distinct subgroup of patients or to all HCC patients with NCS1 levels beyond a certain cutoff, will be addressed in subsequent studies.
In summary, using a database-focused proof-of-concept study to analyze the predictive capacity of the multifunctional Ca2+-binding protein NCS1 in liver cancer, we found that high expression of NCS1 and transcriptionally co-regulated proteins is associated with an unfavorable clinical outcome in HCC. This study lays the foundation for utilizing NCS1 as a prognostic biomarker in prospective cohorts of HCC patients. Furthermore, our work can serve as a starting point to assess the identified signaling complex and demonstrate a functional contribution to tumorigenesis and metastasis.
Supplementary Material
Fig. 2: NCS1 Expression and Patient Survival.
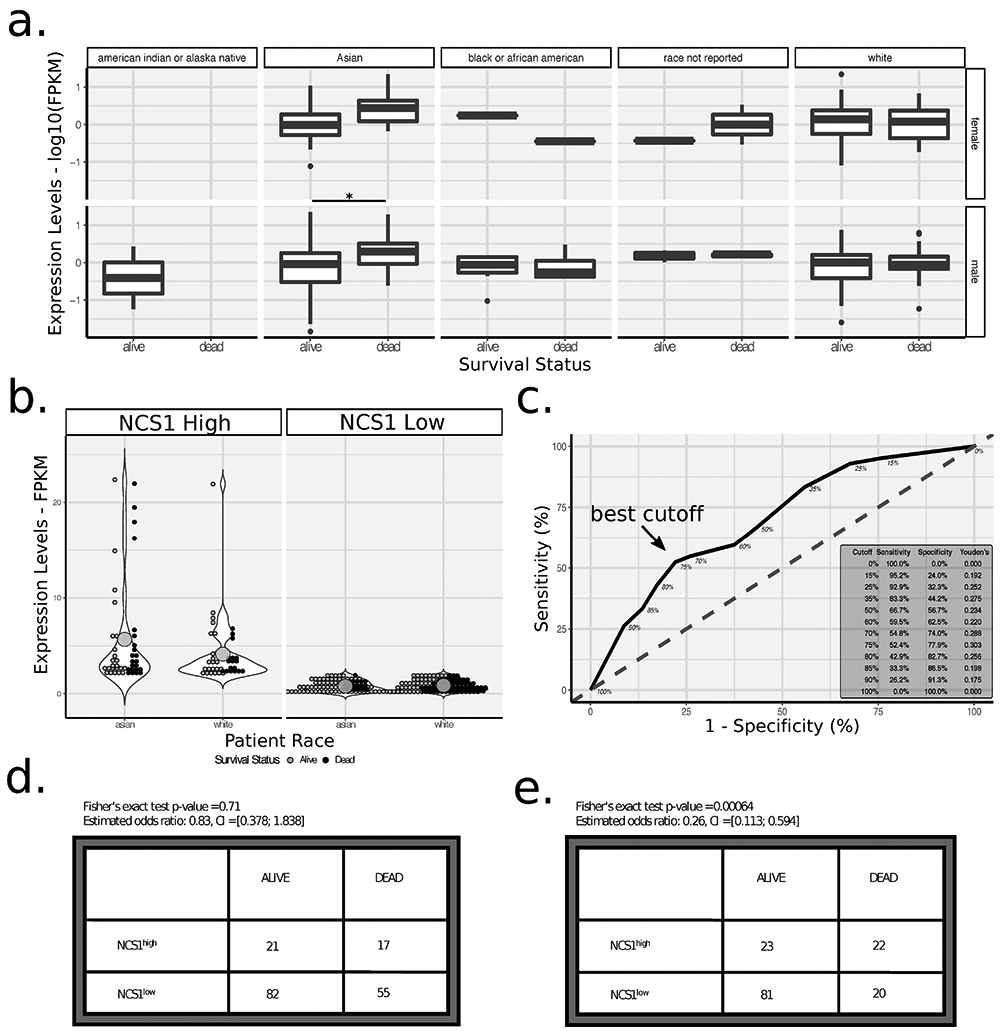
a. Boxplots showing log10-transformed, continuous NCS1 expression levels for male and female patients in rows and different races in columns. Adjacent boxplots partition respective patient populations by survival status. See Supp. Tab. 1 for additional information on sample size. * indicates corrected p-value < 0.05 in a two-sided t-test. b. Dot plots comparing mean (represented by large dots) NCS1 expression levels for NCS1high and NCS1low Asian and white patients. Violin plots visualize overall data distribution. Colors represent survival status of patients. Two-sided t-test p-value = 0.056 for comparison of NCS1 expression levels in Asian vs. white patients (estimated means 2.28 vs. 1.59, respectively). c. Receiver operating characteristic curve showing the optimum cutoff for dichotomizing NCS1 expression in Asian patients to ‘high’ and ‘low’ to be 75% (75% of patients categorized as ‘low’, 25% as ‘high’; indicated by an arrow). Inset in lower right corner lists sensitivity, specificity and Youden’s J statistic for different cutoffs.
d., e. Contingency tables showing NCS1 and survival status for white and Asian patients, respectively. P-values were calculated using Fisher’s exact test, odds ratios and 95% CIs are listed above tables.
Acknowledgements:
Helpful discussions with Michael H. Nathanson (Yale University) and Joseph K. Lim (Yale University) are acknowledged.
Financial Support: DS received a scholarship from the German National Merit Foundation. NIH grant 5P01DK057751 (BEE and MER) supported this work.
List of Abbreviations:
- HCC
Hepatocellular carcinoma
- NCS1
Neuronal Calcium Sensor 1
- TCGA
The Cancer Genome Atlas
- LIHC
Liver Hepatocellular Carcinoma
- BRCA
Breast Cancer
- LUAD
Lung Adenocarcinoma
- LUSC
Lung Squamous Cell Carcinoma
- ICGC
International Cancer Genome Consortium
- InsP3R1
Inositol 1,4,5-triphosphate receptor type 1
- EGAN
Exploratory Gene Association Networks
- AQUA
Automated quantitative analysis
- ACTB
β-actin
- GTEx
The Genotype-Tissue Expression
- HPA
The Human Protein Atlas
- ROC
Receiver operating characteristics
- RYR
Ryanodine receptor
- LIMK1
LIM Domain Kinase 1
- WDR5
WD Repeat Domain 5
Footnotes
Conflict of interest: DS, LMM, and THT state that they have no competing interests. MER is a member of the Merck Speaker’s Bureau. BEE is a co-founder of Osmol Therapeutics.
Supplementary Information accompanies the paper.
References:
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015; 136: E359–E386. [DOI] [PubMed] [Google Scholar]
- 2.Ryerson AB, Eheman CR, Altekruse SF, Ward JW, Jemal A, Sherman RL et al. Annual Report to the Nation on the Status of Cancer, 1975–2012, featuring the increasing incidence of liver cancer. Cancer 2016; 122: 1312–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong CR, Nguyen MH, Lim JK. Hepatocellular carcinoma in patients with non-alcoholic fatty liver disease. World J Gastroenterol 2016; 22: 8294–8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc J et al. Sorafenib in Advanced Hepatocellular Carcinoma. N Engl J Med 2008; 359: 378–390. [DOI] [PubMed] [Google Scholar]
- 5.Berridge MJ. Calcium signalling and cell proliferation. BioEssays 1995; 17: 491–500. [DOI] [PubMed] [Google Scholar]
- 6.Berridge MJ. Inositol triphosphate and calcium signalling. Nature 1993. [DOI] [PubMed] [Google Scholar]
- 7.Berridge MJ. Neuronal Calcium Signaling. Neuron 1998; 21: 13–26. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Chen Y, Chiu W, Shen M. Remodeling of calcium signaling in tumor progression. J Biomed Sci 2013; 20: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prevarskaya N, Skryma R, Shuba Y. Calcium in tumour metastasis: new roles for known actors. Nat Rev Cancer 2011; 11: 609–618. [DOI] [PubMed] [Google Scholar]
- 10.Moore LM, England A, Ehrlich BE, Rimm DL. Neuronal Calcium Sensor 1 (NCS-1) Promotes Tumor Aggressiveness and Predicts Patient Survival. Mol Cancer Res 2017; : 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hass HG, Vogel U, Scheurlen M, Jobst J. Gene-expression Analysis Identifies Specific Patterns of Dysregulated Molecular Pathways and Genetic Subgroups of Human Hepatocellular Carcinoma. Anticancer Res 2016; 5096: 5087–5095. [DOI] [PubMed] [Google Scholar]
- 12.Wu R, Duan L, Cui F, Cao J, Xiang Y, Tang Y et al. S100A9 promotes human hepatocellular carcinoma cell growth and invasion through RAGE-mediated ERK1 / 2 and p38 MAPK pathways. Exp Cell Res 2015; 334: 228–238. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Zhang K, Jiang X. S100A6 as a Potential Serum Prognostic Biomarker and Therapeutic Target in Gastric Cancer. 2014; : 2136–2144. [DOI] [PubMed] [Google Scholar]
- 14.Luo X, Xie H, Long X, Zhou M, Xu Z, Shi B et al. EGFRvIII Mediates Hepatocellular Carcinoma Cell Invasion by Promoting S100 Calcium Binding Protein A11 Expression. PLoS One 2013; 8: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y, Liu Y, Duan J, Yan H, Zhang J, Zhang H et al. Hippocalcin-like 1 suppresses hepatocellular carcinoma progression by promoting p21Waf/Cip1stabilization by activating the ERK1/2-MAPK pathway. Hepatology 2016; 63: 880–897. [DOI] [PubMed] [Google Scholar]
- 16.Koizumi S, Rosa P, Willars GB, Challiss RAJ, Taverna E, Francolini M et al. Mechanisms Underlying the Neuronal Calcium Sensor-1-evoked Enhancement of Exocytosis in PC12 Cells. J Biol Chem 2002; 277: 30315–30324. [DOI] [PubMed] [Google Scholar]
- 17.Weiss JL, Hui H, Burgoyne RD. Neuronal Calcium Sensor-1 Regulation of Calcium Channels, Secretion, and Neuronal Outgrowth. Cell Mol Neurobiol 2010; : 1283–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaad NC, Castrot EDE, Neft S, Hegit S, Hinrichsent R, Martone ME et al. Direct modulation of calmodulin targets by the neuronal calcium sensor NCS-1. Proc Natl Acad Sci U S A 1996; 93: 9253–9258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Choe C, Ehrlich BE. The Inositol 1,4,5-Trisphosphate Receptor (IP3R) and Its Regulators: Sometimes Good and Sometimes Bad Teamwork. Sci STKE 2006; : 1–12. [DOI] [PubMed] [Google Scholar]
- 20.Schlecker C, Boehmerle W, Jeromin A, Degray B, Varshney A, Sharma Y et al. Neuronal calcium sensor-1 enhancement of InsP 3 receptor activity is inhibited by therapeutic levels of lithium. J Clin Invest 2006; 116: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boehmerle W, Splittgerber U, Lazarus MB, McKenzie KM, Johnston DG, Austin DJ et al. Paclitaxel induces calcium oscillations via an inositol 1,4,5-trisphosphate receptor and neuronal calcium sensor 1-dependent mechanism. Proc Natl Acad Sci 2006; 103: 18356–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura TY, Jeromin A, Smith G, Kurushima H, Koga H, Nakabeppu Y et al. Novel role of neuronal Ca2+ sensor-1 as a survival factor up-regulated in injured neurons. J Cell Biol 2006; 172: 1081–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gromada J, Bark C, Smidt K, Efanov AM, Janson J, Mandic SA et al. Neuronal calcium sensor-1 potentiates glucose-dependent exocytosis in pancreatic islet cells through activation of phosphatidylinositol 4-kinase beta. Proc Natl Acad Sci 2005; 102: 10303–10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blasiole B, Kabbani N, Boehmler W, Thisse B, Thisse C, Canfield V et al. Neuronal Calcium Sensor-1 Gene ncs-1a Is Essential for Semicircular Canal Formation in Zebrafish Inner Ear. Wiley Period 2005. doi: 10.1002/neu.20138. [DOI] [PubMed] [Google Scholar]
- 25.Zhang K, Heidrich FM, Degray B, Boehmerle W, Ehrlich BE. Paclitaxel accelerates spontaneous calcium oscillations in cardiomyocytes by interacting with NCS-1 and the InsP 3 R. J Mol Cell Cardiol 2010; 49: 829–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weiss JL, Archer DA, Burgoyne RD. Neuronal Ca Sensor-1 / Frequenin Functions in an Autocrine Pathway Regulating Ca Channels in Bovine Adrenal Chromaffin Cells. J Biol Chem 2000; 275: 40082–40087. [DOI] [PubMed] [Google Scholar]
- 27.R Team. R: A language and environment for statistical computing. 2017.https://www.r-project.org/. [Google Scholar]
- 28.The Cancer Genome Atlas Network. Comprehensive and Integrative Genomic Characterization of Hepatocellular Carcinoma. Cell 2017; 169: 1327–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The Cancer Genome Atlas Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012; 489: 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The Cancer Genome Atlas Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014; 511: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kassambara ANT, Kosinski M. survminer: Drawing Survival Curves using ‘ggplot2’. 2017. [Google Scholar]
- 33.Benjamin DJ, Berger JO, Johannesson M, Nosek BA, Wagenmakers E, Berk R et al. Redefine statistical significance. Nat Hum Behav 2017. doi: 10.1038/s41562-017-0189-z. [DOI] [PubMed] [Google Scholar]
- 34.Paquette J, Tokuyasu T. EGAN : exploratory gene association networks. 2010; 26: 285–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaisaingmongkol J, Budhu A, Dang H, Ruchirawat M, Wang XW. Common Molecular Subtypes Among Asian Hepatocellular Carcinoma and Cholangiocarcinoma. Cancer Cell 2017; 32: 57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujimoto A, Furuta M, Totoki Y, Tsunoda T, Kato M, Shiraishi Y et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat Genet 2016; 48: 500–509. [DOI] [PubMed] [Google Scholar]
- 37.Saha A, Kim Y, Gewirtz ADH, Jo B, Gao C, Ian C et al. Co-expression networks reveal the tissue-specific regulation of transcription and splicing. Genome Res 2017; 27: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kutsenko A, Ladenheim MR, Kim N, Nguyen P, Chen V, Jayasekera C et al. Increased Prevalence of Metabolic Risk Factors in Asian Americans With Hepatocellular Carcinoma. J Clin Gastroenterol 2017; 51: 384–390. [DOI] [PubMed] [Google Scholar]
- 39.Bibonne A, Néant I, Batut J, Leclerc C, Moreau M, Gilbert T. Three calcium-sensitive genes, fus, brd3 and wdr5, are highly expressed in neural and renal territories during amphibian development. BBA - Mol Cell Res 2013; 1833: 1665–1671. [DOI] [PubMed] [Google Scholar]
- 40.Rahl PB, Lin CY, Seila AC, Flynn RA, Mccuine S, Burge CB et al. c-Myc Regulates Transcriptional Pause Release. Cell 2010; 141: 432–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karatas H, Li Y, Liu L, Ji J, Lee S, Chen Y et al. Discovery of a Highly Potent, Cell-Permeable Macrocyclic Peptidomimetic (MM-589) Targeting the WD Repeat Domain 5 Protein (WDR5)–Mixed Lineage Leukemia (MLL) Protein–Protein Interaction. J Med Chem 2017; : 4818–4839. [DOI] [PubMed] [Google Scholar]
- 42.Schapira M, Tyers M, Torrent M, Arrowsmith CH. WD40 repeat domain proteins: a novel target class? Nat Rev Drug Discov 2017; 16: 773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas LR, Wang Q, Grieb BC, Phan J, Foshage AM, Olejniczak ET et al. Interaction with WDR5 Promotes Target Gene Recognition and Tumorigenesis by MYC. Mol Cell 2015; 58: 440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quagliata L, Matter MS, Piscuoglio S, Arabi L, Ruiz C, Procino A et al. Long Noncoding RNA HOTTIP/HOXA13 Expression is Associated With Disease Progression and Predicts Outcome in Hepatocellular Carcinoma Patients. Hepatology 2013; : 911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong H, Zhang L, Qian Z, Zhu X, Zhu G, Chen Y. Identification of HBV-MLL4 Integration and Its Molecular Basis in Chinese Hepatocellular Carcinoma. PLoS One 2015; 10. doi: 10.1371/journal.pone.0123175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olson MF, Sahai E. The actin cytoskeleton in cancer cell motility. Clin Exp Metastasis 2009; 26: 273–287. [DOI] [PubMed] [Google Scholar]
- 47.Li R, Doherty J, Antonipillai J, Chen S, Devlin M, Visser K et al. LIM kinase inhibition reduces breast cancer growth and invasiveness but systemic inhibition does not reduce metastasis in mice. Clin Exp Metastasis 2013; 30: 483–495. [DOI] [PubMed] [Google Scholar]
- 48.Takemura M, Mishima T, Wang Y, Kasahara J, Fukunaga K, Ohashi K et al. Ca 2+/Calmodulin-dependent Protein Kinase IV-mediated LIM Kinase Activation Is Critical for Calcium Signal-induced Neurite Outgrowth. J Biol Chem 2009; 284: 28554–28562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aoki T, Ueda S, Kataoka T, Satoh T. Regulation of mitotic spindle formation by the RhoA guanine nucleotide exchange factor ARHGEF10. BMC Cell Biol 2009; 10: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanna S, El-Sibai M. Signaling networks of Rho GTPases in cell motility. Cell Signal 2013; 25: 1955–1961. [DOI] [PubMed] [Google Scholar]
- 51.Prunier C, Prudent R, Kapur R, Sadoul K. LIM kinases : cofilin and beyond. Oncotarget 2017; 8: 41749–41763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakagawa S, Wei L, Song WM, Zhang B, Fuchs BC, Hoshida Y. Liver Cancer Prevention in Cirrhosis by Organ Transcriptome Analysis and Lysophosphatidic Acid Pathway Inhibition. Cancer Cell 2016; 30: 879–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aran D, Camarda R, Odegaard J, Paik H, Oskotsky B, Krings G et al. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat Commun 2017; 8: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.