Abstract
We have determined functional elements required for autonomous replication of the Schizosaccharomyces pombe ars2004 that acts as an intrinsic chromosomal replication origin. Internal deletion analysis of a 940-bp fragment (ars2004M) showed three regions, I to III, to be required for autonomously replicating sequence (ARS) activity. Eight-base-pair substitutions in the 40-bp region I, composed of arrays of adenines on a DNA strand, resulted in a great reduction of ARS activity. Substitutions of region I with synthetic sequences showed that no specific sequence but rather repeats of three or more consecutive adenines or thymines, without interruption by guanine or cytosine, are required for the ARS activity. The 65-bp region III contains 11 repeats of the AAAAT sequence, while the 165-bp region II has short adenine or thymine stretches and a guanine- and cytosine-rich region which enhances ARS activity. All three regions in ars2004M can be replaced with 40-bp poly(dA/dT) fragments without reduction of ARS activity. Although spacer regions in the ars2004M enhance ARS activity, all could be deleted when an 40-bp poly(dA/dT) fragment was added in place of region I. Our results suggest that the origin activity of fission yeast replicators depends on the number of adenine/thymine stretches, the extent of their clustering, and presence of certain replication-enhancing elements.
Replication of eukaryotic chromosomes is initiated in the S phase of the cell cycle from a number of distinct loci. The process presumably involves recognition of specific DNA regions by certain protein factors.
In the budding yeast Saccharomyces cerevisiae, distinct chromosome fragments have been shown to replicate autonomously (23, 40). All budding yeast autonomously replicating sequences (ARSs) contain an 11-bp ARS consensus sequence (ACS) that is essential for replication (5, 42). In addition, two or three short elements located in a less than 150-bp region proximal to the ACS are required (28). The ACS is the site for binding of an origin recognition complex (ORC), and this physical interaction is essential for the initiation of replication (4).
Structures of replication origins in other eukaryotes seem to be very different from those in budding yeast. In higher eukaryotic cells, no short chromosome fragments capable of autonomous replication have yet been isolated, although it has been shown that replication of the eukaryotic chromosomes is also initiated from restricted regions, ranging in size from 0.5 to 55 kb (13, 18). The replication origins for the human β-globin gene and the Drosophila chorion gene cluster are located within regions of 2 and 3 kb, respectively (12, 26). Several-kilobase chromosome fragments, including the β-globin origin, are able to initiate replication at another chromosomal location (2). Additional regions apart from the actual initiation sites are required for the origin function (1), and the results suggest that the replication origins in higher eukaryotes are composed of structures more complex than those in the budding yeast.
In the fission yeast Schizosaccharomyces pombe, chromosome fragments capable of autonomous replication are several times larger than the budding yeast ARS fragments (7, 15, 24, 31, 32, 37, 38). Although an 11-bp sequence, similar to the budding yeast ACS, has been found in fission yeast ARSs, it can be deleted without any effect on ARS activity (31). Detailed analyses of three fission yeast ARS elements, ars1, ars3002, and ars3001, have shown that regions containing adenines or thymines asymmetrically on one strand of the DNA duplex are required for ARS activity (11, 15, 25, 43). Although necessary, the adenine- and thymine-rich (AT-rich) regions are different in size and sequence, and it is still not known whether a specific sequence or AT richness is more important. Thus, the nature of functional elements comprising fission yeast replication origins are still not understood.
We have isolated five ARS fragments from fission yeast chromosome II and shown that at least three of them function as chromosomal replication origins (37). The efficiency of utilization of these origins correlates with the efficiency of replication of the corresponding ARS fragments. The most efficient ARS, ars2004, is utilized as a replication origin in almost every cell cycle, with replication initiated from a unique locus within its segment in the chromosome as in the ARS plasmid (37). These results suggest that the ars2004 fragment of 3.2 kb contains sequence elements required for initiation of chromosome replication.
In this study, we show that a 940-bp central fragment of the ars2004 contains three functional regions required for autonomous replication in fission yeast. We have demonstrated that sequences consisting of more than three consecutive adenines or thymines without guanine or cytosine are crucial for ARS function. Fission yeast replication origins appear to be composed of essential adenine or thymine (A/T) stretches and multiple replication-enhancing regions distributed over more than 1 kb.
MATERIALS AND METHODS
Strains and media.
The S. pombe haploid strain used was HM123 (h− leu1), cultured in a complete medium (YPD; 1% yeast extract, 2% polypeptone, 2% glucose) and a minimal medium (EMM [33]). Escherichia coli DH5α (14) was grown in Luria-Bertani medium (LB; 0.5% yeast extract, 1% polypeptone, 1% NaCl [pH 7.5]). For EMM and LB plates, agar was added at 2 and 1.5%, respectively. Plasmid DNA was prepared from E. coli transformants as described previously (30).
Nested deletion derivatives of pARS2004.
Plasmid pYC11 is a derivative of pBluescript KS(+) carrying the S. cerevisiae LEU2 gene (41). pARS2004 (37) contains a 3,207-bp ars2004 fragment (from positions 1 to 3207) and a 27-bp fragment of the BamHI-to-NotI segment of cosmid vector SuperCos1 DNA (37).
For construction of deletions from either end of the 3.2-kb genomic fragment, a double-stranded nested deletion kit (Pharmacia, Piscataway, N.J.) was used as recommended by the manufacturer. The restriction enzymes used were NotI and SacI for N-series derivatives and XbaI and PstI for X-series derivatives. Their designations correspond to positions of the chromosome fragment retained at the deletion boundary. To construct the HD series, N-series deletion derivatives were digested with HindIII and self-ligated. To construct pARS2004M carrying a 940-bp fragment from positions 802 to 1741 of ars2004, PCR products with primers (5′-CAGGCGGCCGCTTACTGCAATTTAAAATGC-3′ and 5′-AGTCAATACGGGTTGGC-3′) were digested with NotI and BamHI and inserted along with the BamHI-HindIII fragment (517 bp) of pARS2004 into the NotI-HindIII sites of pYC11.
Internal-deletion and linker-substitution derivatives.
Internal-deletion derivatives of pARS2004M were generated by PCR. PCR products amplified from pARS2004 with an internal primer containing the Sse8387I recognition site (5′-CCTGCAGG-3′) at its 5′ terminus and the M13-20 primer (5′-GTAAAACGACGGCCAGT-3′) were digested with NotI and Sse8387I. The PCR products with an internal primer and reverse primer (5′-AACAGCTATGACCATG-3′) were digested with Sse8387I and HindIII. A pair of NotI-Sse8387I and Sse8387I-HindIII fragments was inserted into pYC11, resulting in internal-deletion mutants pΔA (lacking the region from positions 802 to 893), pΔB (from 894 to 934), pΔC (from 948 to 990), pΔD (from 991 to 1049), pΔE (from 1042 to 1101), pΔF (from 1102 to 1169), pΔG (from 1157 to 1226), pΔH (from 1227 to 1341), pΔI (from 1342 to 1453), pΔJ (from 1454 to 1486), pΔK (from 1454 to 1552), and pΔL (from 1553 to 1742). Derivatives of pARS2004M, pΔ-II-III lacking region I (from 894 to 934), pI-Δ-III lacking region II (from 1032 to 1146), and pI-II-Δ lacking region III (from 1454 to 1552), and the corresponding derivatives of pARS2004 were made by the same procedures.
For construction of linker-substitution derivatives carrying the Sse8387I sequence in region I or region II, NotI-Sse8387I and Sse8387I-BamHI fragments made as described above were ligated with the NotI-BamHI fragment of pARS2004M. The name of each resulting plasmid reflects the first position of substitution. Plasmids pI906S, pI916S and pI926S, carrying 10-bp insertions at positions 906, 916, and 926, respectively, were constructed by the same procedures.
Replacement of essential regions in pARS2004M.
For construction of pI-I-I, carrying three copies of region I at the sites of regions I, II, and III, oligonucleotides 5′-CCGGGTTAAAAAAAATTAAAAATTAACAAAAAAAAAAAAAAAAAAAC-3′ and 5′-CCGGGTTTTTTTTTTTTTTTTTTTGTTAATTTTTAATTTTTTTTAAC-3′ were annealed and inserted into the AvaI site of pYC11-Sse carrying a Sse8387I site in place of the BamHI site of pYC11, resulting in pYC-I and pYC-Ix2, containing one and two copies of the region I fragment, respectively. The region I fragment excised by PstI digestion was inserted into the Sse8387I sites of pI-Δ-III and pI-II-Δ, resulting in pI-I-III and pI-II-I. Insertion of two copies of region I resulted in pI-Ix2-III and pI-II-Ix2. The NotI-BamHI fragment of pI-I-III and BamHI-HindIII fragment of pI-II-I were ligated with NotI-HindIII-digested pYC11 to construct pI-I-I. Similarly, pI-Δ-Δ was made from pI-Δ-III and pI-II-Δ, pI-I-Δ was made from pI-I-III and pI-II-Δ, pI-Δ-Ix2 was made from pI-Δ-III and pI-II-Ix2, and pI-Ix2-Δ was made from pI-Ix2-III and pI-II-Δ. For construction of pIx3-Δ-Δ, the Sse8387I fragment pYC-Ix2 was inserted into the Sse8387I site of pS963, resulting in pIx3-II-III. Then its NotI-HinfI fragment and the HinfI-HindIII fragment of pI-Δ-Δ were inserted into pYC11.
The region II fragment (positions 1030 to 1146) was PCR amplified with a set of primers, 5′-GAGCCTGCAGGTAATTTTAATTGTTTTA-3′ and 5′-GAGCCTGCAGGGAATAAAAAAATTAAG-3′, and digested with Sse8387I. The Sse8387I fragment was inserted into the Sse8387I sites of pΔ-II-III and pI-II-Δ, resulting in pII-II-III and pI-II-II. The NotI-BamHI fragment of pII-II-III and BamHI-HindIII fragment pI-II-II were ligated with NotI-HindIII-digested pYC11 to construct pII-II-II.
Region III (1454 to 1562) amplified with primers 5′-GCGCCTGCAGGGAAACTTGTATATTATTTC-3′ and 5′-AGACCTGCAGGTTCCAGAAGACCTACG-3′ was inserted into pΔ-II-III and pI-Δ-III, resulting in pIII-II-III and pI-III-III. The NotI-HinfI fragment of pIII-II-III and the HinfI-HindIII fragment of pI-III-III were inserted into pYC11, resulting in pIII-III-III.
Derivatives carrying artificial sequences.
A pair of complementary oligonucleotides, 5′-GGA40CTGCA-3′ with 5′-GT40CCTGCA-3′, 5′-GG(AAAT)10CTGCA-3′ with 5′-G(ATTT)10CCTGCA-3′, 5′-GG(AAT)13CTGCA-3′ with 5′-G(ATT)13CCTGCA-3′, or 5′-GG(AAAC)10CTGCA-3′ with 5′-G(TTTG)10CCTGCA-3′, and a self-complementary oligonucleotide, 5′-GG(AT)20CCTGCA-3′, were annealed and inserted into the Sse8387I sites of pΔ-II-III, pI-Δ-III, and pI-II-Δ.
For construction of minimum ARS plasmids without spacer regions, the region I fragment or the (A/T)40 fragment was inserted into the PstI site of pYC11, resulting in pM-I, pM-A40, pM-T80, and pM-A120. Then, the region II fragment amplified by PCR with primers 5′-AAAGGATCCTAATTTTAATTGTTTTAAAATGAG-3′ and 5′-AAAGGATCCGAATAAAAAAATTAAGTTAG-3′ was inserted into the BamHI site, and the region III fragment amplified with 5′-AAATCTAGATTTATTTTTATTTTAATTTTATTTTTTAC-3′ and 5′-AAATCTAGACCTACGAAAAAATAAAATAA-3′ was inserted into the XbaI site, resulting in pMI-II-III, pMA40-II-III, pMT80x2-II-III, and pMA120-II-III. Their nucleotide sequences were confirmed.
Transformation of S. pombe cells.
The electroporation method was used to transform S. pombe cells (22). HM123 cells (107 cells/ml) were washed three times and suspended in cold 1.2 M sorbitol at a concentration of 109 cells/ml. To this cell suspension (0.1 ml), 0.2 μg of plasmid DNA was added with 5 μg of sonicated salmon testis DNA. After electroporation at 2,000 V, 200 Ω, and 25 μF, 1/20 of the suspension was spread on an EMM plate and incubated for 3 or 4 days at 30°C. Relative transformation efficiency was calculated as the ratio of the number of transformants to that of the parental plasmid.
Stability of ARS plasmids.
The stability of ARS plasmids was determined by the method described by Heyer et al. (21). Transformants grown in EMM to 107 cells/ml were diluted and plated onto YPD plates. Colonies which formed after 2 days at 30°C were replica plated onto both EMM and YPD plates to determine the percentage of plasmid-containing cells under the selective conditions (A). Cells in the EMM culture were then diluted to 103 per ml with YPD and grown at 30°C for about 10 generations without selection. After scoring the cell number (n), diluted cells were plated onto YPD plates. The colonies formed were then replicated on EMM and YPD plates to determine the percentage of plasmid-containing cells under the nonselective conditions (B). Plasmid loss rate per generation was calculated with the equation 1 − (B/A)1/N, where N = 3.3 log10n − 10.
RESULTS
Unidirectional deletion analysis of the ars2004 fragment.
To examine the regions required for autonomous replication of the ars2004 fragment, we first made a series of unidirectional nested deletions from either end of the 3.2-kb ARS fragment. ARS activity was examined by measuring the efficiency of transformation of a haploid S. pombe leu1 strain to Leu+ as described in the Materials and Methods.
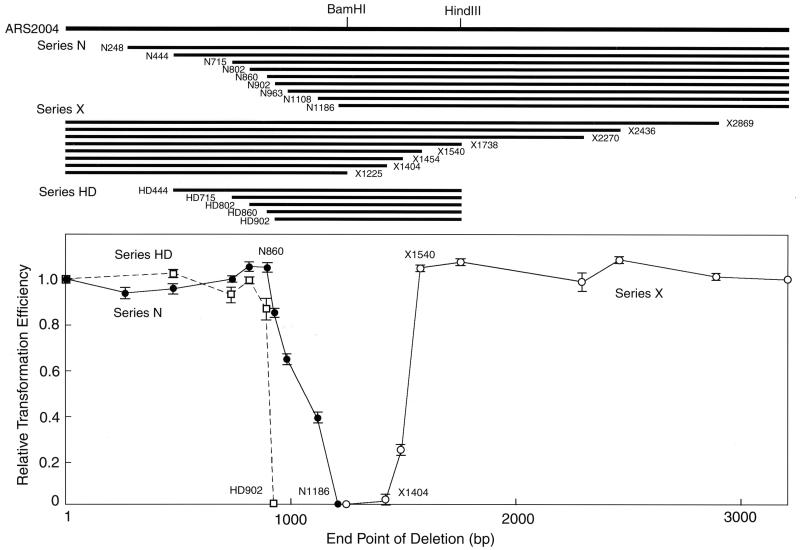
As shown in Fig. 1, derivatives with deletions differing in length from the left end (N series) to position 860 formed transformants at the same efficiency as the parental plasmid, pARS2004. Further deletion derivatives (pN902, pN963, and pN1018) exhibited gradually reduced transformation efficiency and pN1186 gave no transformants. Deletions from the right end (X series) up to about a 1.7-kb segment did not affect transformation efficiency (pX1540). However, deletion of a further 150-bp segment (pX1404) completely abolished activity. These results pointed to the existence of an element required for ARS activity in a distinct region between positions 860 and 1540.
FIG. 1.
Effects of deletions on autonomous replication of ars2004. Derivatives of pARS2004 with nested unidirectional deletions were constructed as described in Materials and Methods. Plasmid DNA was introduced into S. pombe HM123 cells, and the number of Leu+ colonies was scored after incubation for 4 days at 30°C. The transformation efficiency relative to the value for the parental pARS2004 was determined. Regions retaining the natural sequence in the deletion derivatives are shown by lines on the top. Series N and X constructs contain deletions from the left and right ends of the insert, respectively. Series HD constructs are series N derivatives lacking the region right of position 1741. Transformation efficiencies of deletion derivatives in series N (filled circles), series X (open circles), and series HD (open squares) relative to that of the parental plasmid pARS2004 are shown with standard deviations.
To determine the minimum region sufficient for autonomous replication, the region right of position 1741 was removed from the N-series derivatives (HD series). pHD444, pHD714, pHD802, and pHD860 showed almost the same transformation efficiencies as pARS2004, although the pHD802 and pHD860 transformants grew slightly slower than the pARS2004 transformants. In contrast, pHD902 yielded no transformants. These results confirmed the presence of a crucial sequence element right of position 860. The difference between pHD902 and pN902 in replication efficiency suggested the region right of position 1741 to contain an element(s) compensating for lack of the region between 860 and 902. For more detailed analysis of the region required for ARS function, we used a 940-bp fragment from positions 802 to 1741, designated ars2004M.
Identification of regions essential for replication.
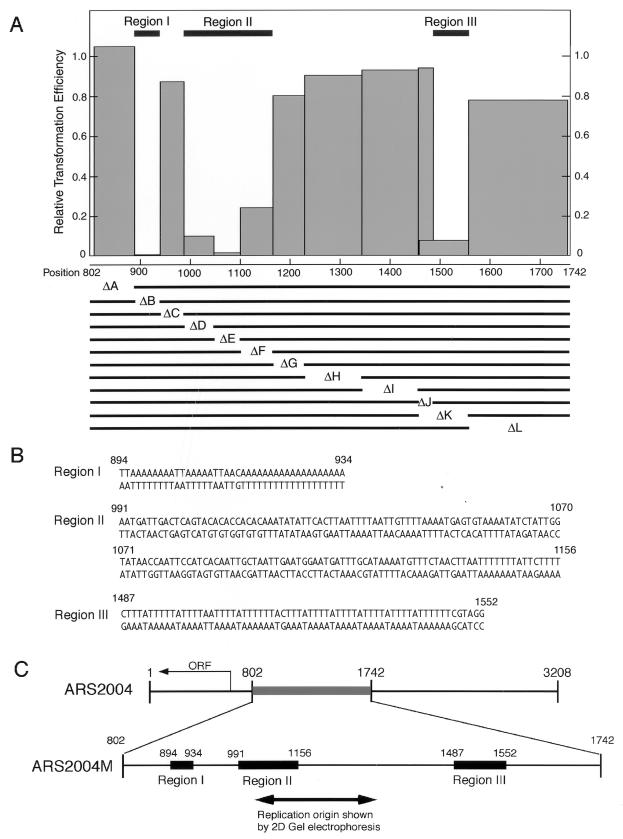
To identify functional regions required for autonomous replication of ars2004M, we deleted 50- to 200-bp internal segments and examined the effects on ARS activity (Fig. 2A). A derivative lacking segment B yielded no transformants. Deletion of segment D, E, F, or K greatly reduced transformation efficiency, and the resultant transformants grew very slowly. In contrast, deletion of segment A, C, G, H, I, J, or L had little effect on transformation efficiency. These results showed at least three distinct regions, I (from positions 894 to 934), corresponding to segment B, II (from 991 to 1156), containing segments D, E, and F, and region III (from 1487 to 1552), corresponding to segment K excluding segment J, to be important for autonomous replication of ars2004M. It should be noted that the boundaries for ARS activity detected by analyses of nested deletions from either end of the 3.2-kb ars2004 fragment are colocalized at regions I and III.
FIG. 2.
Regions required for autonomous replication of the 940-bp ars2004 fragment. (A) Internal segments of 50 to 200 bp (A to L) were deleted from the 940-bp ars2004M fragment, and effects on transformation of HM123 cells were examined. Transformation efficiencies relative to the pARS2004M value are shown by columns. Regions retained in the deletion derivatives are shown by lines at the bottom. Three regions that abolished or greatly reduced ARS activity are indicated by bars at the top. (B) Nucleotide sequences of the three regions required for autonomous replication of ars2004M. (C) Primary structures of ars2004 and ars2004M. The ars2004M targeted for internal deletion analysis is shown by the thick gray line. Three regions required for autonomous replication of ars2004M are indicated by thick black lines. The line with arrowheads shows the region in which the replication origin on the chromosome and on the ARS plasmid was mapped by neutral/neutral two-dimensional (2D) gel electrophoresis (37).
We then examined whether replication of the 3.2-kb ars2004 was dependent on regions I, II, and III. Lack of the 940-bp ars2004M segment from ars2004 abolished ARS activity (Table 1). In contrast, deletion of any one of regions I, II, and III did not abolish ARS activity, although resultant transformants lost plasmids at frequencies of 8.4, 3.2, and 5.1% per generation, respectively, in all cases higher than the 2.2% for pARS2004 transformants. However, the derivative lacking both regions I and III yielded no transformants (Table 1). Although derivatives lacking regions I and II or regions II and III yielded transformants, the transformants grew very slowly and the plasmid loss rates increased to 14 or 8.5% per generation, respectively (Table 1). Thus, regions I and III are required for replication of ars2004 but lack of regions I and II or regions II and III can be partly compensated for by certain elements existing outside the ars2004M.
TABLE 1.
Role of regions I, II, and III in replication of ars2004
| Plasmid | Region
|
Colony formationa | Colony sizeb | Mitotic stabilityc (%) | ||
|---|---|---|---|---|---|---|
| I | II | III | ||||
| 2004 | + | + | + | + | Large | 2.2 ± 0.8 |
| 2004Δ940 | − | − | − | − | NT | |
| 2004ΔI | − | + | + | + | Medium | 8.4 ± 1.7 |
| 2004ΔII | + | − | + | + | Large | 3.2 ± 1.8 |
| 2004ΔIII | + | + | − | + | Medium | 5.1 ± 2.0 |
| 2004ΔIΔII | − | − | + | +/− | Small | 14.0 ± 1.6 |
| 2004ΔIΔIII | − | + | − | − | NT | |
| 2004ΔIIΔIII | − | − | + | + | Medium | 8.5 ± 3.2 |
The number of transformants was counted after incubation for 3 days at 30°C. +, efficiency similar to that of ars2004; +/−, about 20% of the efficiency of ars2004; −, no transformant formed.
Transformants formed after a 3-day incubation were classified into three groups based on colony size.
The mitotic loss rate of plasmid per generation was determined as described in Materials and Methods. NT, not tested.
The nucleotide sequences of regions I, II, and III together with a schematic illustration of the ars2004 structure are shown in Fig. 2B and C. Region I is extremely rich in adenines and thymines, containing 8, 5, and 19 consecutive adenine residues in the upper strand. Region III consists of 11 repeats of TTTTA or variants. On the other hand, region II does not contain any long characteristic sequence but has short AT-rich sequences with intervening guanine- and cytosine-rich (GC-rich) sequences.
Requirement of A stretches in region I.
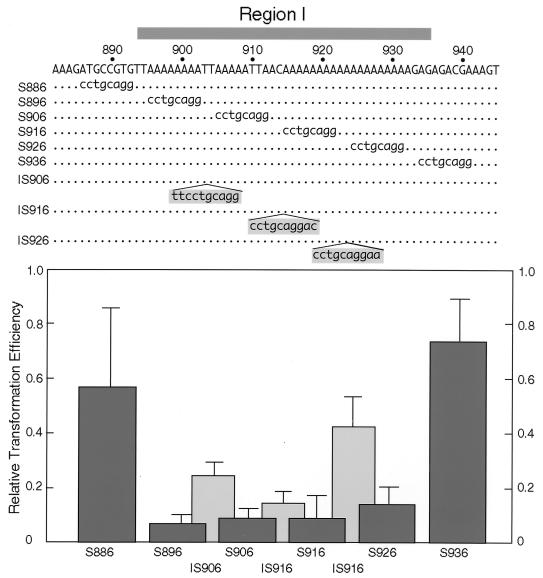
To evaluate the importance of adenine (A) stretches in region I, an 8-bp segment was serially replaced with the Sse8387I recognition sequence (CCTGCAGG). Although the number of transformants after 4 days was not reduced by substitution, they grew significantly more slowly than with pARS2004M transformants. Since the growth rate of ARS plasmid transformants correlates with ARS activity (37), the results indicated partial loss with the base substitutions. To detect reduction in growth rates of transformants quantitatively, the transformants were counted at 3 days instead of 4 days after transformation.
As shown in Fig. 3, all substitutions in region I (S896, S906, S916, and S926) reduced the transformation efficiency at 3 days to about 1/10 the parental value, while substitutions outside region I (S886 and S936) exerted only slight effects, indicating that all the A stretches in region I are required for efficient replication. The fact that none of the substitutions completely abolished the ARS activity suggested that the remaining A-stretch array retained substantial function.
FIG. 3.
Effects of Sse8387I linker substitution in region I on autonomous replication. Various 8-bp sequences from positions 886 to 939 in ars2004M were replaced with an Sse8387I sequence. Derivatives carrying 10-bp insertions were made by recombining linker-substitution derivatives at the Sse8387I site. ARS activity of plasmids was examined as described for Fig. 1 except that incubation was for 3 instead of 4 days to more sensitively detect reduction of ARS activity. Altered bases are indicated by lowercase letters, and the natural sequences are shown by dots. Transformation efficiencies of linker-substitution and insertion derivatives relative to the value of pARS2004M are shown by dark and light gray columns, respectively.
We next tested whether continuity of the A stretches in region I was required for the ARS activity, by inserting the Sse8387I site within or between A stretches. With a 10-bp insertion at position 906 (IS906), the transformation efficiency was reduced to one-fourth of the parental value (Fig. 3). Insertion at position 916 (IS916) or 926 (IS926) also reduced ARS activity (Fig. 3). These results suggested continuity of A stretches to be important for ARS activity. However, the transformation efficiency with IS906 was three times higher than that with S896, in which the left-most 8-bp A stretch was substituted (Fig. 3). The value with IS926 was also three times higher than that with S926, suggesting that the sequestered 8-bp A stretches contribute to ARS activity.
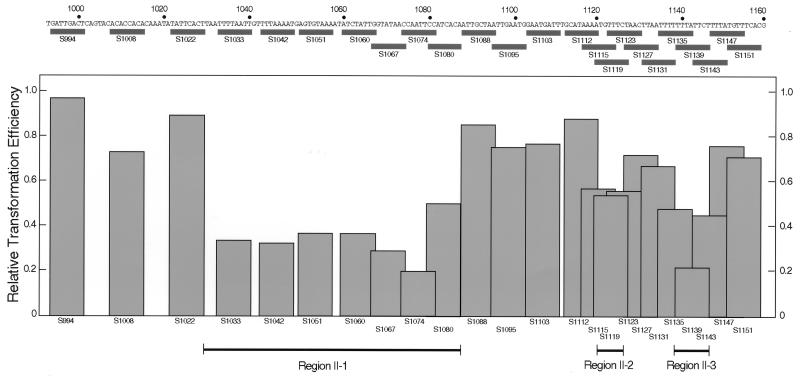
Multiple elements in region II.
To determine the sequence element of region II required for ARS activity, effects of serial 8-bp substitutions were examined. With substitutions in a region from positions 1033 to 1087, the transformation efficiency after 3 days of incubation was reduced to about one-third of the parental value (Fig. 4). Substitutions from positions 1139 to 1146 (S135, S1139, and S1143), and to a lesser extent 1120 to 1126, also caused reduction. Other substitutions did not significantly affect transformation efficiency. These results suggested that region II contains three subdomains extending from positions 1033 to 1087 (region II-1), 1120 to 1126 (region II-2), and 1139 to 1146 (region II-3). All substitution derivatives yielded almost the same number of transformants as pARS2004M after 4 days of incubation, showing that the 8-bp substitution in region II did not abolish but rather diminished its function.
FIG. 4.
Effects of Sse8387I-linker substitution in region II on autonomous replication. Locations of linker substitutions are shown by rectangles below the sequence of region II. Transformation efficiencies relative to pARS2004M after 3 days of incubation are shown by columns. Three regions that reduced the ARS activity are indicated at the bottom.
Replacement of essential regions.
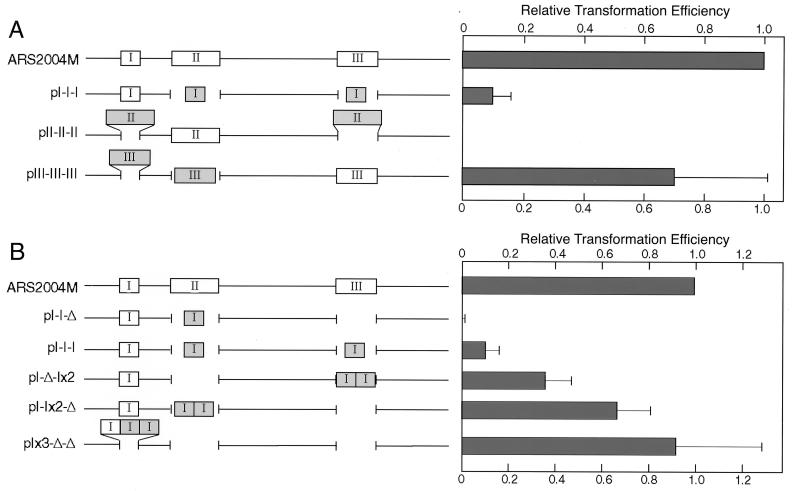
To evaluate the relative importance of regions I, II, and III in autonomous replication, we constructed derivatives carrying three copies of only one of these, replacing the other two. The derivative with two additional copies of region I at the positions of regions II and III gave about one-tenth as many transformants as the parental construct (pI-I-I in Fig. 5A). That with two additional copies of region III (pIII-III-III) yielded transformants as efficiently as pARS2004M. In contrast, the derivative with three copies of region II (pII-II-II) yielded no transformants (Fig. 5A).
FIG. 5.
Effects of substitutions of region I, II, or III for the other two regions of ars2004M on ARS activity. (A) Two of three regions required for the ARS activity of ars2004M were replaced with copies of the other region. Transformation efficiencies relative to the pARS2004M value after 3 days of incubation are presented. The fragments inserted in tandem orientation are shown by shaded boxes. (B) Effects of clustering of region I fragments on ARS activity. Transformation efficiencies with derivatives of ars2004M lacking regions II and III but carrying three tandem copies of the region I fragment either at three or two separate places or at a single location are presented. ARS activity was enhanced as the region I fragments were placed closer together.
Although the transformation efficiency with pI-I-I was much lower than that with pARS2004M itself, the ARS activity was greatly affected by clustering of region I fragments. A derivative carrying two tandem copies of the region I fragment at the region III site without region II (pI-Δ-Ix2) exhibited transformation efficiency three times higher than that of pI-I-I (Fig. 5C). Further elevation was observed with a derivative carrying two copies of the region I fragment at the region II site without region III (pI-Ix2-Δ). Moreover, the plasmid with three copies of the region I fragment at the region I site without regions II and III (pIx3-Δ-Δ) yielded transformants as efficiently as pARS2004M. These results showed closely located region I fragments to be much more effective than when separated.
Specific sequences required for autonomous replication.
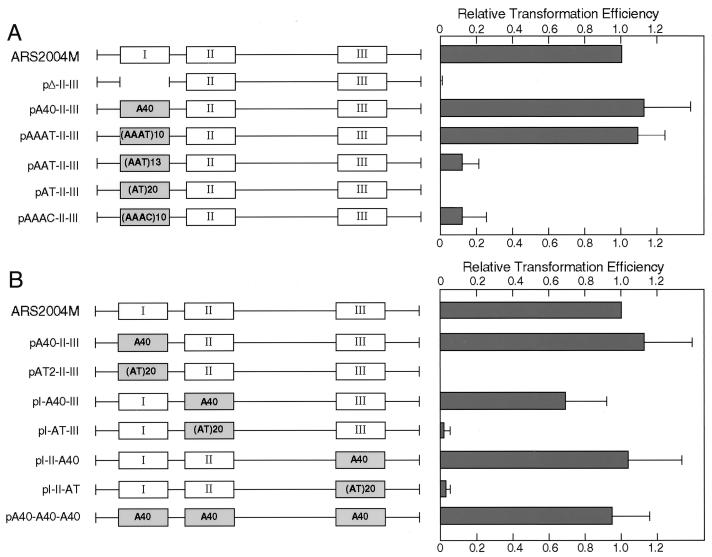
To examine whether specific sequences or merely AT richness in region I has importance for autonomous replication, region I of pARS2004M was replaced with a synthetic (A/T)40, (AAAT/TTTA)10, (AAT/TTA)13, or (AT/TA)20 fragment, with almost the same numbers of adenines and thymines. As shown in Fig. 6A, the derivative carrying (A/T)40 or (AAAT/TTTA)10 transformed as efficiently as pARS2004M. That with (A/T)40 in the opposite orientation had similar ARS activity (37a). In contrast, the derivative with (AAT/TTA)13 yielded about 1/10 as many transformants (pAAT-II-III in Fig. 6A), and none were obtained with (AT/TA)20 (pAT-II-III in Fig. 6A). These results demonstrated that specific sequences rather than mere AT richness are required for ARS activity. Furthermore, replacement of region I with (AAAC/TTTG)10 reduced the transformation efficiency to about 1/10 of the parental value, showing that the presence of cytosine or guanine impairs the function of region I. From these results, we concluded that three or more consecutive A/T stretches without intervening guanine or cytosine are required for ARS function.
FIG. 6.
Substitutions of artificial sequences for regions required for autonomous replication of ars2004M. (A) Region I of ars2004M was replaced with a synthetic (A/T)40, (AAAT/TTTA)10, (AAT/TTA)13, (AT/TA)20, or (AAAC/TTTG)10 fragment. Transformation efficiencies relative to that of pARS2004M after 3 days of incubation are presented. (B) Each region or all three regions were replaced with (A/T)40 or (AT/TA)20. A/T stretches, but not AT alternates, function like the natural essential regions.
As shown in Fig. 6B, regions II and III could also be functionally replaced with (A/T)40 but not (AT/TA)20. Moreover, the derivative with (A/T)40 fragments at positions of regions I, II, and III yielded the same number of transformants as pARS2004M.
Minimum ARS fragments.
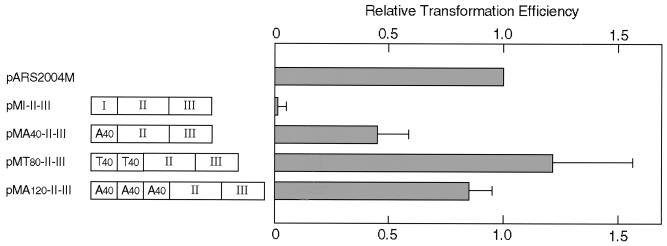
Since serial deletions of a 50- to 200-bp segment except for regions I, II, and III had little effect on ARS activity (Fig. 2), we tested whether segments other than regions I, II, and III were dispensable for ARS activity. A derivative carrying a set of region I, region II, and region III fragments in the native order and direction yielded no significant transformants (pMI-II-III [Fig. 7]), showing that the spacer regions are required. However, pMA40-II-III carrying (A/T)40 instead of the region I fragment yielded transformants at about one-third the level for pARS2004M. Insertion of additional copies of (A/T)40 increased transformation to almost the same efficiency as pARS2004M (pMT80-II-III and pMA120-II-III [Fig. 7]), indicating that the functions of all spacer regions could be replaced by the presence of additional A stretches. A derivative carrying three copies of (A/T)40 alone did not transform efficiently (data not shown). Since we failed to construct a derivative with longer A stretches, it has not been determined whether an A stretch alone, if long enough, functions as an ARS element.
FIG. 7.
Autonomous replication of ars2004M without spacer regions. Transformation efficiencies with derivatives of pYC11 carrying combinations of regions I, II, and III and (A/T)40 fragments without spacer regions of ars2004M were measured after 4 days of incubation. Fragments inserted into the vector are schematically shown. Regions I, II, and III were joined in the natural order and direction. The boxes labeled T40 represent (A/T)40 fragments inserted in the opposite orientation.
Our previous study demonstrated that efficient ARS fragments are maintained as monomeric plasmids in the transformants, while less efficient ARS fragments are present as multimeric plasmid forms (37). We examined the minimum ARS plasmids lacking spacer regions for their form maintained as extrachromosomal elements. Plasmid DNA from the Leu+ transformants grown under selective conditions was separated by agarose gel electrophoresis and analyzed by Southern hybridization. The parental plasmid pARS2004M was maintained as monomers (37a). Plasmids pMA40-II-III, pMT80-II-III, and pMA120-II-III were maintained as monomers, although transformants that grew faster than others also contained dimer and trimer forms (38b). These results confirmed that minimum ARS fragments lacking spacer regions are maintained as extrachromosomal elements.
DISCUSSION
We have previously shown that the ars2004 fragment contains genetic information necessary for efficient initiation of replication from a distinct region (37). In the present study, we identified three functional regions in a 940-bp fragment that is sufficient for ARS activity. The regions were not found to contain short essential sequences like the ACS in the budding yeast replication origins. Instead, ARS activity appears to depend on multiple arrays of A (or T) stretches.
A stretches are essential for replication in fission yeast.
Deletion analysis of the 940-bp internal segment of the ars2004 revealed that three distinct regions are required for the ARS activity, with region I or III being essential. Both regions are exclusively composed of adenine and thymine residues which are asymmetrically present in a strand of DNA. Moreover, the fact that regions I, II, and III in ars2004M could all be replaced by (A/T)40 without significant reduction in ARS activity (Fig. 6B) demonstrated that A stretches are sufficient for the functions of three regions in autonomous replication of ars2004M.
It has been reported that some fission yeast ARS fragments contain a match to an 11-bp sequence, (A/T)(A/G)TTTATTTA(A/T), which is similar to the budding yeast ACS (31). However, deletion of this sequence does not affect the ARS activity (31), and ars2004 does not contain any match. Zhu et al. have proposed a consensus sequence motif, AA(A/T)AA(A/T)A(A/T)AA(A/T)(A/T), that is critical for replication of ars3002 (43). The 19 consecutive adenines in region I of ars2004 match this sequence motif. However, region I can be replaced without significant reduction in ARS activity by region III, which does not contain a match, suggesting that the motif is not essential. Extensive studies of three ARS elements, ars3002 (16), ars1 (11), and ars3001 (25), have shown that the regions required for ARS activity contain adenines (or thymine) clustered on a strand. However, the A-rich regions differ in size and sequence, and it is not clear whether a specific sequence or merely AT-rich sequence is required for fission yeast ARS activity. From the finding that region I can be functionally replaced with synthetic (A/T)40 and (AAAT/TTTA)10 but not with (AT/TA)20, (AAT/TTA)13, or (AAAC/TTTG)10, we conclude that clustered sequences composed of three or more consecutive adenines or thymines without an intervening guanine or cytosine play critical roles in replication of ars2004M. Thus, no highly specific sequence but a certain DNA structure made by A stretches might be required for fission yeast ARS activity. The effects of base substitutions in region I (Fig. 3) suggest that ARS activity depends on the numbers of clustered adenine or thymine residues. Moreover, the fact that insertion of a GC-rich sequence in region I diminished ARS activity suggests that continuity of A/T stretches is important. Importance of clustering of A stretches was clearly shown by the elevation of ARS activity in the order pI-I-I, pI-Δ-Ix2, pI-Ix2-Δ, and pIx3-Δ-Δ (Fig. 6B), suggesting cooperation of A stretches in autonomous replication.
It has been shown that the ACS in budding yeast replication origins is recognized by ORC and their interaction is necessary for initiation of replication (3). Finding of counterparts to ORC components in many eukaryotes (8, 9, 17, 19, 27, 34) has raised the possibility that ORC has an essential role in eukaryotic DNA replication. The fission yeast orp1+ and orp2+ genes, counterparts of budding yeast ORC1 and ORC2, respectively, are required for cell growth (17, 27, 34). Analysis of the temperature-sensitive orp1-4 mutant has shown that Orp1 protein functions in an early step of DNA replication (20). We have shown that Orp1 protein is specifically associated with chromosomal replication origins, such as the ars2004 and ars3002 loci (36a). It is possible that the fission yeast ORC complex binds to the essential A/T stretches in the replication origins. Recently, the fission yeast Orp4, a homologue of budding yeast Orc4, has been shown to contain AT hook motifs that are involved in interaction with minor groove of AT tracts in DNA (10). The N-terminal domain of Orp4p with nine AT hook motifs specifically binds to the fission yeast ars1 fragment that contain A/T stretches. The DNA binding activity of Orp4p might participate in recognition of fission yeast replication origins. Further genetic and biochemical studies are required for an understanding of the nature of the fission yeast ORC complex.
Stimulation of ARS activity by region II.
In contrast to the highly clustered A stretches in regions I and III, region II was found to contain short A stretches scattered in II-1, II-2, and II-3. Since base substitutions in these segments reduced ARS activity, they must contribute to stimulation of autonomous replication. However, this was also the case for the 35-bp GC-rich segment of II-1. It has been shown that specific sequence elements enhance autonomous replication of budding yeast and human chromosome fragments. In the budding yeast ars1, the element B3 that stimulates ARS activity is a site for binding of the transcription factor ABF1 (6, 28, 39). An 18-bp REE1 sequence that enhances autonomous replication of human chromosome fragments also interacts with the human transcription factor YY1 (35, 36). These findings suggest that autonomous replication can be stimulated through interaction of certain transcription factors with specific sequence elements. It should be noted that the ars2004 is located upstream of an open reading frame for an unknown product. Another well-characterized replication origin, ars3002, is similarly located upstream of an open reading frame (15). Furthermore, we have found that the region upstream of fission yeast homologue of nucleosome assembly protein (nap1) gene exhibits autonomous replication activity (38a). These results suggest that certain transcription factors that bind to specific elements in the origins may participate in stimulation of replication in fission yeast. Further investigations are necessary to identify functional relations of the elements required for replication and those involved in regulation of transcription.
We have previously shown that replication of the ars2004 plasmid is initiated from a distinct region the same as in its native chromosomal location (37). The initiation site was mapped by neutral/neutral two-dimensional gel electrophoresis to an approximately 200-bp region, close to region II but rather distant from the essential regions I and III. The replication enhancing activity of region II might facilitate assembly of replication machinery. The relationship between essential regions and the initiation site is under investigation.
Functional domain structures of ars2004.
Although deletion of any 100-bp segment in the spacer regions between regions I, II, and III hardly affected ARS activity, region I, II, and III fragments joined without spacer regions proved insufficient for autonomous replication. This and the fact that ARS function was restored by supplementation with (A/T)40 fragments suggest that the 940-bp ars2004M is composed of two types of functional regions, essential A/T stretches in regions I and III and stimulatory elements in region II and the spacer regions.
The 940-bp ars2004M that contains elements sufficient for autonomous replication is essential for replication of ars2004, because its deletion abolishes ARS activity. Deletion of both regions I and III from ars2004 completely abolishes ARS activity, indicating that their A/T stretches are essential for ARS activity. However, regions I, II, and III can individually be deleted from the 3.2-kb ars2004 without significant effect on transformation efficiency. The region outside ars2004M thus contains an element(s) that compensates for the lack of a functional region of ars2004M. Therefore, ars2004 is composed of essential A/T stretches and multiple enhancing elements scattered over a region larger than 1 kb.
Essential A/T stretches were found to be highly clustered in regions I and III in ars2004. ars3001 and ars3002 also contain regions consisting of A/T clusters (16, 25), while they are not extensively clustered in the ars1 (11) or ars-nap1. The ars2004 appears to be more efficient for replication than ars1 or ars-nap1, judging from the growth rates of transformants (37a). Our studies suggest that the origin activity of fission yeast replicators may depend on the number of A/T stretches, the extent of their clustering, and the presence of certain enhancing elements.
Replication of higher eukaryotic chromosomes is initiated from restricted regions (13). Recently, Aladjem et al. have shown that a several-kilobase fragment containing a highly AT-rich sequence derived from the β-globin origin can promote initiation of replication when translocated to a different chromosomal location (2). Functional elements of higher eukaryotic replicators have not been as extensively studied as those in yeasts, because of the lack of ARS fragments that can replicate as efficiently as chromosome DNA. We have previously shown that a human chromosome fragment replicating autonomously at several-times-higher efficiency than random fragments contains a replication-enhancing element and a several-kilobase-pair region rich in adenine residues asymmetrically in one strand (29, 36). The characteristics of fission yeast replicators, such as the absence of a short essential sequence and the presence of clustered A/T stretches, are thus similar to those observed for human ARS fragments. A/T stretches are more dispersed in larger regions in human ARS than in fission yeast replicators and are not characteristic for budding yeast replicators. Fission yeast replicators might thus be a prototype for higher eukaryotic replicators. Therefore, it is important to elucidate the roles of functional elements in fission yeast replicators for an understanding of the more complex replication origins in higher eukaryotes.
ACKNOWLEDGMENTS
We thank Y. Sakakibara, J. Tomizawa, T. Tsurimoto, D. Gilbert, and T. Yonesaki for critical reading of the manuscript and helpful discussions.
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture of Japan.
Footnotes
Present address: Department of Biochemistry and Molecular Biology, SUNY Health Science Center, Syracuse, NY 13210.
REFERENCES
- 1.Aladjem M I, Groudine M, Brody L L, Dieken E S, Fournier R E K, Wahl G M, Epner E M. Participation of the human β-globin locus control region in initiation of DNA replication. Science. 1995;270:815–819. doi: 10.1126/science.270.5237.815. [DOI] [PubMed] [Google Scholar]
- 2.Aladjem M I, Rodewald L W, Kolman J L, Wahl G M. Genetic dissection of a mammalian replicator in the human β-globin locus. Science. 1998;281:1005–1009. doi: 10.1126/science.281.5379.1005. [DOI] [PubMed] [Google Scholar]
- 3.Bell S P, Kobayashi R, Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993;262:1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- 4.Bell S P, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 5.Broach J R, Li Y-Y, Feldman J, Jayaram M, Abraham J, Nasmyth K A, Hicks J B. Localization and sequence analysis of yeast origins of DNA replication. Cold Spring Harbor Symp Quant Biol. 1983;47:1165–1173. doi: 10.1101/sqb.1983.047.01.132. [DOI] [PubMed] [Google Scholar]
- 6.Buchman A R, Kimmerly W J, Rine J, Kornberg R D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caddle S, Calos M P. Specific initiation at an origin of replication from Schizosaccharomyces pombe. Mol Cell Biol. 1994;14:1796–1805. doi: 10.1128/mcb.14.3.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calza R E, Eckhardt L A, DelGiudice T, Schildkraut C L. Changes in gene position are accompanied by a change in time of replication. Cell. 1984;36:689–696. doi: 10.1016/0092-8674(84)90349-0. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter P B, Mueller P R, Dunphy W G. Role for a Xenopus Orc-2-related protein in controlling DNA replication. Nature. 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- 10.Chuang R-Y, Kelly T J. The fission yeast homologue of Orc4 binds to replication origin DNA via multiple AT-hooks. Proc Natl Acad Sci USA. 1999;96:2656–2661. doi: 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clyne R K, Kelly T J. Genetic analysis of an ARS element from the fission yeast Schizosaccharomyces pombe. EMBO J. 1995;14:6348–6357. doi: 10.1002/j.1460-2075.1995.tb00326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delidakis C, Kafatos F C. Amplification enhancers and replication origins in the autosomal chorion gene cluster of Drosophila. EMBO J. 1989;8:891–901. doi: 10.1002/j.1460-2075.1989.tb03450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DePamphilis M L. Eukaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- 14.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubey D D, Zhu J, Carlson D, Sharma K, Huberman J A. Three ARS elements contribute to the ura4 replication origin in the fission yeast, Schizosaccharomyces pombe. EMBO J. 1994;13:3638–3647. doi: 10.1002/j.1460-2075.1994.tb06671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubey D D, Kim S-M, Todorov I T, Huberman J A. Large, complex modular structure of a fission yeast DNA replication origin. Curr Biol. 1996;6:467–473. doi: 10.1016/s0960-9822(02)00514-6. [DOI] [PubMed] [Google Scholar]
- 17.Gavin K A, Hidaka M, Stillman B. Conserved initiator proteins in eukaryotes. Science. 1996;270:1667–1671. doi: 10.1126/science.270.5242.1667. [DOI] [PubMed] [Google Scholar]
- 18.Gilbert D M. Replication origins in yeast versus metazoa: separation of the haves and the have nots. Curr Opin Gen Dev. 1998;8:194–199. doi: 10.1016/s0959-437x(98)80141-x. [DOI] [PubMed] [Google Scholar]
- 19.Gossen M, Pak D T S, Hansen S K, Acharya J K, Botchan M R. A Drosophila homolog of the yeast origin recognition complex. Science. 1995;270:1674–1677. doi: 10.1126/science.270.5242.1674. [DOI] [PubMed] [Google Scholar]
- 20.Grallert B, Nurse P. The ORC1 homolog orp1 in fission yeast plays a key role in regulating onset of S phase. Genes Dev. 1996;10:2644–2654. doi: 10.1101/gad.10.20.2644. [DOI] [PubMed] [Google Scholar]
- 21.Heyer W-D, Sipiczki M, Kohli J. Replicating plasmids in Schizosaccharomyces pombe: improvement of symmetric segregation by a new genetic element. Mol Cell Biol. 1986;6:80–89. doi: 10.1128/mcb.6.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hood M T, Stachow C. Transformation of Schizosaccharomyces pombe by electroporation. Nucleic Acids Res. 1990;18:688–692. doi: 10.1093/nar/18.3.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsiao C-L, Carbon J. High-frequency transformation of yeast by plasmids containing the cloned yeast ARG4 gene. Proc Natl Acad Sci USA. 1979;76:3829–3833. doi: 10.1073/pnas.76.8.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnston L H, Baker D G. Characterization of an autonomously replicating sequence from the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1987;207:161–164. doi: 10.1007/BF00331504. [DOI] [PubMed] [Google Scholar]
- 25.Kim S, Huberman J A. Multiple orientation-dependent, synergistically interacting similar domains in the ribosomal DNA replication origin of the fission yeast, Schizosaccharomyces pombe. Mol Cell Biol. 1998;18:7294–7303. doi: 10.1128/mcb.18.12.7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kitsberg D, Selig S, Keshet I, Cedar H. Replication structure of the human β-globin gene domain. Nature. 1993;366:588–590. doi: 10.1038/366588a0. [DOI] [PubMed] [Google Scholar]
- 27.Leatherwood J, Lopez-Girona A, Russel P. Interaction of Cdc2 and Cdc18 with a fission yeast ORC2-like protein. Nature. 1996;379:360–363. doi: 10.1038/379360a0. [DOI] [PubMed] [Google Scholar]
- 28.Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 29.Masukata H, Satoh H, Obuse C, Okazaki T. Autonomous replication of human chromosomal DNA fragments in human cells. Mol Biol Cell. 1993;4:1121–1132. doi: 10.1091/mbc.4.11.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masukata H, Tomizawa J. Control of primer formation for ColE1 plasmid replication: conformational change of the primer transcript. Cell. 1986;44:125–136. doi: 10.1016/0092-8674(86)90491-5. [DOI] [PubMed] [Google Scholar]
- 31.Maundrell K, Hutchison A, Shall S. Sequence analysis of ARS elements in fission yeast. EMBO J. 1988;7:2203–2209. doi: 10.1002/j.1460-2075.1988.tb03059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maundrell K, Wright A P H, Piper M, Shall S. Evaluation of heterologous ARS activity in S. cerevisiae using cloned DNA from S. pombe. Nucleic Acids Res. 1985;13:3711–3722. doi: 10.1093/nar/13.10.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchison J M. Physiological and cytological methods for Schizosaccharomyces pombe. Methods Cell Physiol. 1970;4:131–165. [Google Scholar]
- 34.Muzi-Falconi M, Kelly T. Orp1, a member of the Cdc18/Cdc6 family of S-phase regulators, is homologous to a component of the origin recognition complex. Proc Natl Acad Sci USA. 1995;92:12475–12479. doi: 10.1073/pnas.92.26.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obuse C, Okazaki T, Masukata H. Interaction of transcription factor YY1 with a replication-enhancing element, REE1, in an autonomously replicating human chromosome fragment. Nucleic Acids Res. 1998;26:2392–2397. doi: 10.1093/nar/26.10.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Obuse C, Okuno Y, Okazaki T, Masukata H. A replication-enhancing element with transcriptional silencer activity in autonomously replicating human chromosomal fragments. Mol Biol Cell. 1996;7:43–55. doi: 10.1091/mbc.7.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Ogawa Y, Takahashi T, Masukata H. Association of fission yeast Orp1 and Mem6 proteins with chromosomal replication origins. Mol Cell Biol. 1999;19:7228–7236. doi: 10.1128/mcb.19.10.7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okuno Y, Okazaki T, Masukata H. Identification of a predominant replication origin in fission yeast. Nucleic Acids Res. 1997;25:530–536. doi: 10.1093/nar/25.3.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37a.Okuno, Y. Unpublished data.
- 38.Olsson T, Ekwall K, Ruusala T. The silent P mating type locus in fission yeast contains two autonomously replicating sequences. Nucleic Acids Res. 1993;21:855–861. doi: 10.1093/nar/21.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.Nakata, T., and H. Masukata. Unpublished data.
- 38b.Sekiguchi, M. Unpublished data.
- 39.Shore D, Stillman D J, Brand A H, Nasmyth K A. Identification of silencer binding proteins from yeast: possible roles in SIR control and DNA replication. EMBO J. 1987;6:461–467. doi: 10.1002/j.1460-2075.1987.tb04776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Struhl K, Stinchcomb D T, Scherer S, Davis R W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci USA. 1979;76:1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takahashi K, Murakami S, Chikashige Y, Funabiki H, Niwa O, Yanagida M. A low copy number central sequence with strict symmetry and unusual chromatin structure in fission yeast centromere. Mol Biol Cell. 1992;3:819–835. doi: 10.1091/mbc.3.7.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Houten J V, Newlon C S. Mutational analysis of the consensus sequence of a replication origin from yeast chromosome III. Mol Cell Biol. 1990;10:3917–3925. doi: 10.1128/mcb.10.8.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu J, Carlson D L, Dubey D D, Sharma K, Huberman J A. Comparison of the two major ARS elements of the ura4 replication origin region with other ARS elements in the fission yeast, Schizosaccharomyces pombe. Chromosoma. 1994;103:414–422. doi: 10.1007/BF00362286. [DOI] [PubMed] [Google Scholar]