Abstract
Cellular immunotherapy has recently emerged as a fourth pillar in cancer treatment co-joining surgery, chemotherapy and radiotherapy. Where, the discovery of immune checkpoint blockage or inhibition (ICB/ICI), anti-PD-1/PD-L1 and anti-CTLA4-based, therapy has revolutionized the class of cancer treatment at a different level. However, some cancer patients escape this immune surveillance mechanism and become resistant to ICB-therapy. Therefore, a more advanced or an alternative treatment is required urgently. Despite the functional importance of epitranscriptomics in diverse clinico-biological practices, its role in improving the efficacy of ICB therapeutics has been limited. Consequently, our study encapsulates the evidence, as a possible strategy, to improve the efficacy of ICB-therapy by co-targeting molecular checkpoints especially N6A-modification machineries which can be reformed into RNA modifying drugs (RMD). Here, we have explained the mechanism of individual RNA-modifiers (editor/writer, eraser/remover, and effector/reader) in overcoming the issues associated with high-dose antibody toxicities and drug-resistance. Moreover, we have shed light on the importance of suppressor of cytokine signaling (SOCS/CISH) and microRNAs in improving the efficacy of ICB-therapy, with brief insight on the current monoclonal antibodies undergoing clinical trials or already approved against several solid tumor and metastatic cancers. We anticipate our investigation will encourage researchers and clinicians to further strengthen the efficacy of ICB-therapeutics by considering the importance of epitranscriptomics as a personalized medicine.
Keywords: epitranscriptomics, immune checkpoint blockage (ICB) therapy, anti-PD-1/PD-L1 drug resistance, personalized medicine, CISH, microRNAs
1. Introduction
The advent of immunotherapy given in-combination with standard chemotherapeutic drugs has greatly control the cancer-spread over decades. However, still some cancer patients develop resistance against these therapeutic approaches, alarming the discovery of further advanced medicines. The invention of programmed cell death protein-1 and its ligand-1 (PD-1/PD-L1) was breakthrough in the history of cancer treatment, but still some tumors escape these immune surveillance mechanisms and relapse to grow continuously. Therefore, a more creative and advanced treatment is required instantly to overcome the issues largely associated with high-dose antibody/drug toxicities and drug-resistances, conceivably in the form of personalized medicines. In this study, we have summarized epitranscriptomic mechanisms to improve the efficacy of ICB-therapy by targeting N6A-modification machineries especially m6A-modifiers. Moreover, we have also emphasized co-targeting immune checkpoint proteins (PD-1 and PD-L1) along with intracellular checkpoint molecules (CISH, SOCS-1 and microRNAs) in enhancing the efficacy of ICB-therapeutics by combining immunotherapy. A recent human clinical trial NCT04426669, NCT03538613 [1,2] evidenced the success of targeting CISH/SOCS-1 in NK-cells [3,4], T-cells [5,6], and DCs [7] in further strengthening the efficacy of ICB-therapy against broad range of solid tumors and metastatic gastrointestinal cancers (Table 1, Table 2 and Table 3) and (Figure 1).
Table 1.
Epigenetic modifiers and microRNAs in improving the efficacy of ICB-therapy.
| RNA (m6A)-Modifiers (Editors/Erasers/Effectors) | |||||
|---|---|---|---|---|---|
| RNA Modifiers | Disease Condition | Target | Disease Mechanism | Therapeutic Strategies | Ref. |
| Writers Mettl3/14 | up-regulated in colorectal cancer and melanoma |
IFNγ, STAT1, IRF1, Cxcl-9 and Cxcl-10 | By reducing CD8+T-cells infiltrations in TME | CRISPR/cas9 silencing of Mettl3/14 via YTHDF2 | [8] |
| Mettl-3 | down-regulated in M1/M2-med. lung metastasis | Spred-2 | By recruiting immunosuppresive T-reg and MDSCs | Overexpressing Mettl3 via polarizing M1/M2-macrophages | [9] |
| m6A | m6A-mediated regulation of PD-L1 in HNSCC | G2M checkpoint and PI3K/AKT/ mTOR signaling | Analysed via cancer genome atlas TCGA and GSE65858 cohort | By targeting m6A regulatated signature genes | [10] |
|
Erasers FTO |
up-regulated in melanoma | PD-1, CXCR4 and SOX10 | Impairs anti-PD1 effect by reducing target gene expressions | Selective inhibition of FTO to enhance anti-PD1 effects | [11] |
| FTO | up-regulated in colon cancer | PD-L1 | Up-regulates PD-L1 expression in IFNγ signaling-independent manner | Selective inhibition of FTO inhibits PD-L1 to control colon cancer | [12] |
| ALKBH5 | up-regulated in melanoma | Mct4/Slc16a3 | By recruiting immunosuppresive T-reg and MDSCs | Anti-ALKBH5 enhances the effect of anti-PD1 therapy. | [13] |
|
Readers YTHDF1 |
up-regulated in solid tumors | Lysosomal cathepsins | Degrade neo-antigen and impair dendritic cell presentation | Anti-YTHDF1 suppress cathepsins and enhance DC cross-presentation | [14] |
| YTHDF2 | up-regulated in LGG (brain tumor) and several other immune cells | PD-1, CTLA4, TIM3 | Impair immune checkpoint signalling | Anti-YTHDF2 in combination with immunecheckpoint immunotherapy | [15,16] |
| DNA and Histone Modifiers in ICB-Therapeutics | |||||
| Epigenetic Regulators | Disease Condition | Target | Mechanism | Therapeutic Strategies | Ref. |
| DNA methylation | down-ragulates CTLA4 in HNSCC | CTLA4, CD28, CD80/86, ICOS | DNA methylation affects HNSCC | Selective DNA (DNMTs) inhibitors | [17] |
| DNA methylation | down-regulates PD-L1 in melanoma | Interfron signalling | cpG DNA methylation regulate melanoma | [18] | |
| DNA methylation | up-regulates PD-1 & CTLA4 in NSCLC | PD-1 (PDCD-1) CTLA4 |
Hypo-methylation increases PD-1, CTLA4 expression in NSCLC | Selective DNA (5hmC) inhibitors | [19] |
| DNA methylation | up-regulates PD-L1 & PD-L2 in HNSCC | PD-L1 (CD274) PD-L2 (PDCD1LG2) |
Hypo-methylation increases PD-L1 & PD-L2 expression | Combining DNA inh. with Nivolumab and Pembrolizumab | [20] |
| DNA methylation | up-regulates PD-L1 in CRC | PD-L1 (CD274) | DNA-methylation control PD-L1 exp. | Selective DNA (TETs) inhibitors | [21] |
| HDAC | up-ragulates CTLA4 in B-cell associated function | CTLA4 and LAG3 | Tcf1 regulate CTLA4 expression in TFH-cells | HDACi control CTLA4-mediated B-cell help | [22] |
| HDAC6 | up-regulates PD-L1 in melanoma | PD-L1 (CD274) STAT3 |
HDAC6 increase PD-L1 expression by recruiting STAT3 | HDAC6-inhibitor decreases PD-L1 by de-activating STAT3 | [23] |
| Active H3K4me3 | up-regulates PD-L1 in breast cancer | EMT-induced PD-L1 expression | Active H3K4me3 modifications in Breast cancer |
Selective histone inhi. enhance the efficacy of ICB-Abs | [24] |
| Active H3K4me3 | up-regulates PD-L1 in pancreatic cancer |
PD-L1 (CD274) | MLL1 catalyzed H3K4me3 to bind with PD-L1 promoter and increase its expression | MLL1 inhibitor in combination with anti-PD-L1,anti-PD-1 improves efficacy | [25] |
| Repressive H3K27me3 | down-regulates PD-L1 in HCC | PD-L1, IRF1 | EZH2 negatively regulate PD-L1 exp. by recruiting repressive H3K27me3 in HCC | Selective H3K27me3 inhibitor could enhance ICB efficacy | [26] |
| HDACi (Belinostat) |
up-regulates PD-L1 & CTLA4 in HCC | Increase IFN-γ & reduce T-reg populations |
Belinostat treatment increase anti-tumor immunity against HCC | Combining belinostat enhances the efficacy of ICB therapy | [27] |
| SAHA | Increases CTLA4 and Foxp3 exp. cardiac transplant | Foxp3 CTLA4 |
SAHA increases suppressive function of T-reg to prolong allograft survival | SAHA (HDACi) couls be a promissing immunosuppressive agent with CNI drug | [28] |
| H3Ac | up-regulates PD-L1 in drug resistant cancer cell | H3Ac enhance PD-L1 exp. | drug resistant issues in cancer cells | HDACi in combination with anti-PD-L1 | [29] |
| MicroRNAs in ICB-Therapeutics | |||||
| miRNAs | Disease Condition | Target | Mechanism | Therapeutic Strategies | Ref. |
| miR-15a,b miR-16, miR-193a-3p | down-regulated in MPM | Direct target of PD-L1 | miR-15a, miR-16 and miR-193a-3p (−)vely regulates PD-L1 | Respective miRNA mimics combined ICB-therapeutics | [30] |
| miR-17-5p | down-regulated in melanoma | Directly binds 3′-UTR PD-L1 |
miR-17-5p (−)vely regulates PD-L1 | miR-17-5p mimics with anti-PD-L1 Abs | [31] |
| miR-18a (miR-140, 142, 340, 383) | up-regulated in cervical cancer | PI3K/AKT, WNK2, SOX6, p53 PTEN, MEK | miR-18a (+)vely and miR-140, 142, 340, 383 (−)ly regulates PD-L1 | Respective miRNA antagomiR & mimics with ICB-therapy | [32] |
| miR-20b-21-130b | up-regulated in colorectal cancer | PTEN, B7-H1 (PD-1) |
miRs (+)vely regulates B7-H1 (PD-1) exp. | Respective miRNAs AntagomiRs in combination with ICB-therapeutics | [33] |
| miR-21 (CD4+T-cells) | up-regulated in arthritis and GC | PDCD4, Th17, STAT5, T-reg | miR-21 (−)vely regulates PDCD4, PD-1 | [34,35] | |
| miR-23a-3p | up-regulated in (MΦ) liver cancer | PTEN, AKT pathways | miR-23a-3p (+)vely regulates PD-L1 exp. | Anti-miR-23a-3p (antagomiR therapy) with anti-PD-L1 Abs | [36] |
| miR-25-93- 106b cluster | down-regulated in pancreatic cancer | CXCL12, PD-L1 | miR-25-93- 106b−/− mice increases PD-L1 | miR-93, miR-106b mimics with BET inh. | [37] |
| miR-28 | melanoma | PD-1 | miR-28 (−)vely regulates PD-1 | miR-28 mimics | [38] |
| miR-33a | down-regulated in Lung A. carcinoma | PD-L1,CTLA4, PD-1, CAND1 | miR-33a (−)vely regulates PD-1/PD-L1 | miR-33a mimics with combined ICB-Abs | [39] |
| miR-34a | down-regulated in AML, lymphoma | EBF-1 and 3′-UTR PD-L1 | miR-34a (−)vely regulates PD-L1 exp. | ICB therapy combined miRNA | [40,41,42,43,44] |
| miR-138-5p | down-regulated in CRC | Target 3′-UTR PD-L1 | miR-138-5p (−)vely regulatesPD-L1 exp. | miR-138-5p mimics combined ICB-Abs | [45] |
| miR-140 | down-regulated in NSCLC | miR-140/ PD-L1/cyclinE pathways | miR-140 target 3′-UTR PD-L1 (−)vely regulates its exp. |
miR-140 mimics with anti-PD-L1 therapy | [46] |
| miR-142-5p | down-regulated in pancreatic cancer | miR-142-5p target 3′-UTR PD-L1 | miR-142-5p (−)vely regulates PD-L1 exp. | miR-142-5p mimics + anti-PD-L1 therapy | [47] |
| miR-145 | down-regulated in ovarian carcinoma | Cisplatin cMYc (TcF) |
miR-145 (−)vely regulates PD-L1 exp. | miR-145 mimic (restoration therapy) with anti-PD-L1 Abs | [48] |
| miR-146a | up-regulated in melanoma | STAT1-IFNγ axis | miR-146a (+)vely regulates PD-L1 exp. | miR-146a antagomiR with anti-PD-L1 Abs | [49] |
| miR-148a -3p | down-regulated in dMMR/MSI-H CRC | miR-148a-3p binds to 3′-UTR PD-L1 | miR-148a-3p (−)vely regulates PD-L1 exp. | Respective miRNA mimics with anti-PD-L1 therapy | [50] |
| miR-155 | up-regulated in B-cell lymphoma | AKT and ERK | miR-155 (+)vely regulates PD-L1 exp. | miR-155 antagomiR + PD-L1 antagonists | [51] |
| miR-191-5p | down-regulated in colon-adenocarcinoma | PD-L1 | miR-191-5p (−)vely regulates PD-L1 exp. | miR-191-5p mimics | [52] |
| miR-195 | down-regulated in PC and DLBCL | PD-L1 | miR-191-5p (−)vely regulates PD-L1 exp. | miR-191 mimics | [53,54] |
| miR-197 | down-regulated in NSCLC | CKS1B/STAT3 (Bcl-2, c-Myc, CyclinD1) |
miR-197 (−)vely regulates PD-L1 exp. | miR-193 mimics (replacement therapy) + ICB-therapeutics | [55] |
| miR-200b, miR-152 | down-regulated in gastric cancer (GC) | B7-H1 (PD-1) | miR-200b and miR-152 (−)vely regulates B7-H1 | Respective miRNA mimics combined PD-L1 antagonists | [43,56,57] |
| miR-214 | down-regulated in B-cell lymphoma (DLBCL) | miR-214 atrget 3′-UTR PD-L1 | miR-214 (−)vely regulates PD-L1 exp. | miR-214 mimic in combination with anti-PD-L1 Abs | [58] |
| miR-217 | down-regulated in laryngeal cancer | AEG-1 and PD-L1 | miR-217 (−)vely regulates PD-L1 exp | miR-217 mimics with anti-PD-L1 therapy | [59] |
| miR-324-5p miR-338-5p | downregulated in Mycobateria-responsive hedgehog sign | PD-L1, SHH signaling |
(−)vely regulate PD-L1 |
miRNA mimics | [60] |
| miR-340 | down-regulated in Cervical cancer | PD-L1 | miR-340 (−)vely regulates PD-L1 exp. | miR-340 mimics | [61] |
| miR-375 | down-regulated in HNSCC | JAK2 | Inhibits JAK2-STAT1 axis suppressing PD-L1 exp. | miR-375 mimics | [62] |
| miR-424 (322) | down-regulated in ovarian cancer | PD-1/PD-L1, CD80/CTLA4 | miR-424 (322) (−)vely regulates PD-1/PD-L1, CD80/CTLA4 exp. | miR-424 (322) mimics (restoration therapy) + ICB-therapeutics | [63] |
| up-regulated in Colon cancer | CD28, CD80 and CD86 | up regulated miR-424 impairs anti-tumor immunity | modified tumor-secreted EVs with miR-424 knocked down |
[64] | |
| miR-497-5p | down-regulated in RCC (ccRCC) | Cell proliferation | miR-497-5p (−)vely regulates PD-L1 exp. | miR-497-5p mimic with anti-PD-L1 Abs | [65] |
| miR-513 | cholangiocytes in response to C. parvum infection | B7-H1 (PD-1) | miR-513 (−)vely regulates PD-1 exp. | miR-513 mimics | [66] |
| miR-570 | down-regulated in gastric cancer | B7-H1 (PD-1) | SNP (polymorphism) disrupts miR-570- B7-H1 interactions | Restoration therapy combined ICB-Abs | [43,67] |
| miR-873 | down-regulated in breast cancer | PI3K/Akt, ERK1/2 pathways | miR-873 (−)vely regulates PD-L1 by binding to 3′-UTR | miR-873 mimics with PD-1/PD-L1 inhibitor | [68] |
| miR-3127-5p | up-regulated in NSCLC | pSTAT3 | Upregulates PD-L1 by suppressing p-STAT3 | Anti-miR-3127-5p (antagomiR therapy) | [69] |
| miR-3609 | down-regulated in breast cancer | PD-L1 | miR-3609 (−)vely regulates PD-L1 exp. | miR-3609 mimics | [70] |
| miR-4717 | down-regulated in HBV | PD-1 | miR-4717 (−)vely regulates PD-1 exp. | miR-4717 mimics | [71] |
Table 2.
Biopharmaceutical companies developing immune checkpoint blockade (ICB)-antibodies.
| Immune Cell Targeted Antibodies (Anti-PD-1 Therapy) | ||||||
|---|---|---|---|---|---|---|
| Company | Antibody FDA Approval |
Brand/ Other Name |
Combination | Disease | Clinical Trial | Ref. |
| Bristol-Meyers Squibb | Nivolumab (Human IgG4) 2014 |
Opdivo®, BMS-936558, MDX-1106 ONO-4538 |
LAG3 (BMS-986016), B7-H3 (Enoblituzumab), KIR (Lirilumab), 4-1BB (Urelumab), ICOS (JTX-2011), CD27 (Varlilumab), GM.CD40L (vaccine for lung NSCLC) |
Broad range of tumor types and Lymphomas |
NCT01968109 NCT02817633 NCT01714739 NCT02253992 NCT02904226 NCT02335918 NCT02466568 NCT01673867 |
[83,84] |
| Medimmune | MEDI0680 (AMP-514) | - |
NCT02118337 Phase I |
[85,86] | ||
| Regeneron/ Sanofi | REGN2810 | - | Phase I/II NCT02383212 NCT02760498 |
|||
| Novartis | PDR001 | GITR (GWN323) | NCT02740270 | [87] | ||
| Merck | Pembrolizumab (Humanized IgG4k) 2014 |
Keytruda® MK-3475, lambrolizumab |
B7-H3 (Enoblituzumab), Multi-kinase inhibitor (Sunitinib) |
Melanoma, Lung, NSCLC, HNC, cervical, thyroid cancer |
NCT02475213 NCT02599779 NCT01295827 |
[83,88] |
| Cure Tech | Pidilizumab (Humanized IgG1k) |
CT-011 |
Pidilizumab (formerly CT-011), anti-delta like-1 (DLL1), anti-PD-1 | Malignant gliomas | Phase I/ II NCT01952769 |
|
| Sanofi | Cemiplimab 2018 |
Libtayo® | Cervical cancer CSCC |
Phase III | [83] | |
| Immune Cell Targeted Antibodies (Anti-CTLA4 Therapy) | ||||||
| Medarex/ Bristol-Meyers Squibb | Ipilimumab (IgG1 isotype) 2011 |
Yervoy® (BMS-734016, MDX-010, MDX-101) |
Nivolumab, Gemcitabine, Cisplatin |
Melanoma, SCLC, Bladder, prostate cancer |
NCT00527735 NCT01524991 NCT00323882 |
[83,89,90,91,92] |
| Pfizer/ AstraZeneca | Tremolimumab (IgG2 isotype) 2015 |
Orphan drug approval, CP-675, 206 |
Metastatic melanoma, Solid Tumor | Phase III NCT02527434 NCT03703297 |
[93,94,95,96,97] | |
| Tumor Cell/APC-Targeted Antibodies (Anti-PD-L1/L2 Therapy) | ||||||
| Roche/ Genentech |
Anti-PD-L1 Atezolizumab (Humanized IgG1k), 2016 |
Tecentriq®, MPDL3280A, RG7446, RO5541267 |
CD27 (Varlilumab), VEGF inhibitors (Bevacizumab cediranib) |
Ovarian, Urothelial, Lung Cancer, HNCLC |
NCT02543645 NCT02659384 |
[83] |
| Merck, EMD, Serono/Pfizer | Avelumab 2017 |
Bavencio® MSB0010718C | Metastatic MCC | Urothelial, RCC, Merkel | NCT02603432 | [83,98] |
| Medimmune/ AstraZeneca |
Anti-PD-L1 Durvalumab (Human IgG1k), 2017 |
Imfinzi® MEDI4736 | Osimertinib, Olaparib and Sunitinib |
NSCLC, Solid Tumor, urothelial carcinoma |
Reference [70] NCT02221960 NCT02484404 |
[99,100,101] |
| Bristol-Meyers Squibb | Anti-PD-L1 (Human IgG4) |
BMS-936559 (MDX1105) |
- | HIV-1, Sepsis, NSCLC |
Phase I NCT02028403 |
[102,103,104] |
| Amplimmune/ Glaxo Smith Klein |
Anti-PD-L2 | AMP-224 | - | MCC | NCT02298946 | [105] |
| Anti-PD-L2 AMP-514 (fusion protein) |
MEDI0680 | - | kidney cancer, melanoma |
Phase I NCT02013804 |
[86] | |
Table 3.
Biotech companies/Universities entering into personalized medicine targeting intracellular immune checkpoints in combination with ICB-therapeutics.
| Biopharmaceutical Company/University | Target | Combined Therapeutic Approach |
Clinical Trial | Indication | Ref. |
|---|---|---|---|---|---|
| Natural Killer Cells (NK-cells) Clinicaltrials.gov, accessed on 15 July 2021 | |||||
| ONK therapeutics (Ireland) 2015 www.onktherapeutics.com | CISH−/− NK-cells NK-cells |
CISH−/− NK-cells in combination with ICB-antibodies |
ONK102 ONK103 ONK104 |
M. Myeloma NSCLC AML |
[135] |
| Fate Therapeutics San Diego, USA |
iPSC-derived NK Cells (FT500) | Nivolumab (anti-PD-1) Pembrolizumab (anti-PD-1) Atezolizumab (anti-PD-L1) Interleukin-2 (IL-2) |
NCT03841110 NCT04106167 (Phase-I) |
Advanced solid tumors and lymphoma |
[136,137,138,139,140] |
| Innate Pharma S. A |
NK cell (NKG2A) | Durvalumab (Phase-I/II) Nivolumab (Phase-I) Ipilimumab (Phase-I) Nivolumab + 5-Aza (Ph-I) |
NCT02671435 NCT01592370 NCT01750580 NCT02599649 |
Metastatic Cancer | [141,142] |
| Altor Biosciences corporation |
IL-15 super agonist mediated NK-cells | Nivolumab (anti-PD-1) |
NCT02523469 (Phase-I/II) |
NSCLC | [142] |
| ImmunityBio, Inc. | High-affinity Natural Killer (haNK) Cell | Avelumab (Bavencio®) (anti-PD-L1) |
NCT03387085 (Phase-I/II) |
Triple Negative Breast Cancer | - |
| SignalRX Pharmaceuticals, Inc. | SF1126 (dual inhibitor of PI3K and BRD4) |
Nivolumab (anti-PD-1) | NCT03059147 | Advanced HCC | [83] |
| Effector Therapeutics | Tomivosertib (eFT-508) | Pembrolizumab (anti-PD-1) |
NCT03616834 Phase-II Completed 2021 |
Solid tumors and NSCLC | [83] |
| NantKwest Inc., and Chan Soon-Shiong Institute for Medicine, USA | CD16-targeted NK-cell (haNKTM) with N-803 (IL-15 superagonist) | Avelumab (Bavencio®) (anti-PD-L1) |
NCT03853317 (Phase-II) |
Merkel cell carcinoma | [139,143] |
| National Cancer Institute, Naples | NK-cells (Tregs and NKs) |
Nivolumab (anti-PD-1) | NCT03891485 | Renal cell carcinoma | [144] |
| Gachon University & Severance hospital, Republic of Korea | Allogeneic NK-Cells (SMT-NK) | Pembrolizumab (anti-PD-1) Keytruda |
NCT03937895 (Phase-I/II) |
Biliary tract cancer | [139] |
| Fox Chase Cancer Center, USA | NK-cells and T-cells | Pembrolizumab (anti-PD-1) |
NCT02535247 (Phase-I/II) |
Lymphoma | [144,145,146] |
| Jilin University Hospital, China | NK-cells | PD-1 Ab |
NCT03958097 (Phase-II) |
Non-small cell lung cancer | [139] |
| MD Anderson Cancer Center, USA | DF1001 (a new molecule targeting NK-cell activations) |
Drug: DF1001 Pembrolizumab (anti-PD-1) |
NCT04143711 (Phase-I/II) |
Advanced Solid Tumors | [139,144] |
| T-Cells: Tumor-Infiltrating Lymphocytes (TILs) | |||||
| Intima Bioscience, Inc. with University of Minnesota |
CISH-deleted Tumor-Infiltrating Lymphocytes (TIL) | CISH checkpoint-deleted TILs combined with Cyclophosphamide, Fludarabine, Aldesleukin and ICB-therapeutics |
NCT04426669 (Phase-I/II) |
Solid tumors & gastro-intestinal cancers | [1,147] |
| CISH−/− T-cells (TILs) | NCT03538613 (Phase-I/II) |
Gastro-intestinal cancers | [2,5] | ||
| Hangzhou Cancer Hospital in collabration with Anhui Kedgene Biotechnology Co.,Ltd | PD-1 Knockout T-Cells | CRISPR/Cas9-deleted PD-1 in T-Cells with hydrocortisone |
NCT03081715 (Phase-I) Completed, 2018 |
Advanced Esophageal Squamous Cell Carcinoma | [2,144] |
| Sichuan University in collabration with Chengdu MedGenCell |
PD-1 Knockout T-Cells | CRISPR/Cas9-deleted PD-1 in T-Cells with Cyclophosphamide |
NCT02793856 (Phase-I) Completed, 2020 |
Metastatic Non-small Cell Lung Cancer | [2,144,148] |
| Peking University and (Cell Biotech) |
PD-1 Knockout Engineered T Cells | PD-1-KO-T-cells with IL-2 and Cyclophosphamide |
NCT02863913 NCT02867345 NCT02867332 (Phase-I) |
Bladder, Prostate and Renal Cell Carcinoma | [2,5] |
| University of Pennsylvania, with Tmunity Therapeutics |
NY-ESO-1 redirected autologous T cells | TCR-deleted and PD-1-deleted T cells |
NCT03399448 | Myeloma, melanoma and several cancers | [2,5,149] |
| Nanjing University Medical School | PD-1 Knockout EBV-CTLs | PD-1-KO-EBV-CTL with IL-2, Fludarabine and Cyclophosphamide |
NCT03044743 (Phase-I/II) |
EBV associated Malignancies | [2,5] |
| Dendritic Cells (DCs) | |||||
| H. Lee Moffitt Cancer Center, BMS and MultiVir, Inc. | DC-based p53 Vaccine | Ipilimumab (anti-CTLA4) Nivolumab (anti-PD-1) |
NCT03406715 (Phase-II) |
Small Cell Lung Cancer | [137] |
| Allife Medical Sc. and Technology Co., Ltd. | DC-NK YNYY-01 (DC-NK Cells) |
Pembrolizumab (anti-PD-1) Keytruda |
NCT03815084 (Phase-I) |
Solid tumors | [144,150] |
| Bristol-Myers Squibb and Duke Cancer Inst. |
DC Vaccines | Nivolumab (anti-PD-1) |
NCT02529072 NCT02775292 (Phase-I) |
Recurrent Brain Tumors | |
| Northwest Biotherapeutics, BMS and JCCC | Autologous DCs pulsed with tumor lysate | Nivolumab (anti-PD-1) |
NCT03014804 (Phase-II) |
Recurrent Glioblastoma | |
| University of Pennsylvania | Autologous DC pulsed peptide | Pembrolizumab (anti-PD-1) |
NCT03092453 (Phase-I) |
Advanced Melanoma | |
| Mayo Clinic in collabration with National Cancer Inst. | Autologous DC pulsed tumor Ags | Pembrolizumab (anti-PD-1) |
NCT03035331 (Phase-I/II) |
Aggressive Non-Hodgkin Lymphoma | |
| Oslo University Hospital in collabration with NCS and MSDC | Autologous DC | Pembrolizumab (anti-PD-1) Rituximab, GM-CSF and anti-TNF-alpha therapy |
NCT02677155 (Phase-II) |
Follicular Lymphoma | |
| Capital Medical Univ. in collabration with Duke Univ. | Autologous DC-CIK cell | Pembrolizumab Anti-PD-1 + DC-CIK (Ph-I) Anti-PD-1 alone (Ph-II) |
NCT03190811 NCT03360630 |
Advanced Solid Tumors and NSCLC |
|
| Sun Yat-sen University | DC-CIK cell (Cytokine-induced Killer Cell) |
Anti-PD-1 antibody |
NCT02886897 (Phase-I) Completed, 2019 |
Refractory Solid Tumors | |
| Beth Israel Deaconess Medical Center | Dendritic Cell Fusion Vaccine | Pidilizumab (anti-PD-1) |
NCT01067287 (Phase-I) |
Multiple Myeloma | |
| Cancer Insight in collabration with Elios Therapeutics, LLC | Autologous DC (TLPLDC Vaccine) | Checkpoint Inhibitor |
NCT02678741 (Phase-I/II) |
Metastatic Melanoma | |
| Grupo Espanol Multidisciplinario del Cancer Digestivo | Autologous DC Vaccine (AVEVAC) | Avelumab (Bavencio®) (anti-PD-L1) |
NCT03152565 (Phase-I/II) Completed, 2020 |
Metastatic Colorectal Carcinoma | |
| Dana-Farber Cancer Institute in collabration with Celgene | DC/AML Fusion Vaccine | Durvalumab (Imfinzi®) (anti-PD-L1) |
NCT03059485 (Phase-II) |
Acute Myelogenous Leukemia | |
| Radboud University in collabration with Dutch Cancer Society |
MiHA-loaded PD-L1/L2 silenced DC Vaccination | PD-L1/L2-silenced DC (siRNA silenced) |
NCT02528682 (Phase-I/II) Completed, 2021 |
Hematological Malignancies | |
| Johns Hopkins University, USA | TLR3 agonist enhace DC activation | Anti-PD-1 in combination with DCs | - | Glioblastoma | [16] |
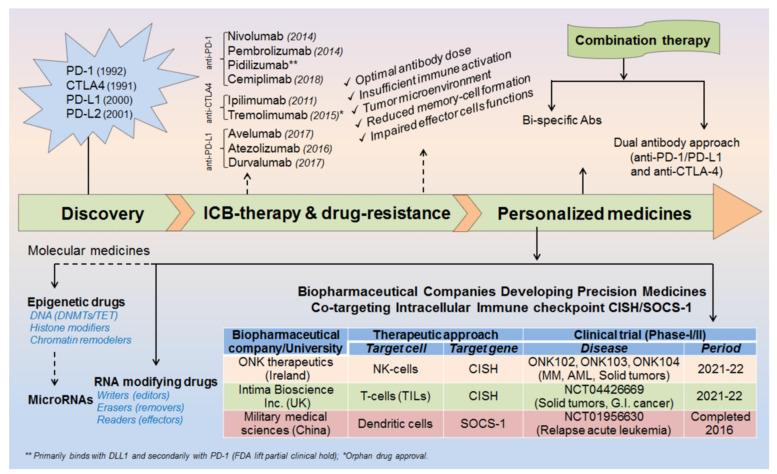
Figure 1.
Milestones in the development of ICB-therapeutics. Discovery of immune checkpoint markers. Year of FDA-approved ICB-antibodies. Factor affecting antibody/drug-resistance. Recent strategies to improve ICB-efficacy by combining molecular medicines. Biopharmaceutical companies developing personalized medicines co-targeting epitranscriptomics and intracellular immune checkpoint (CISH/SOCS-1) in NK-cells, TILs and DCs with relevant clinical trial were summarized.
Before coming to the main stream of this review, a very logical question arises: (i) why even after so strong therapeutic approaches still some cancer cells escape these immune surveillance mechanisms? (ii) What could be the best possible combinations to overcome the issues associated with drug-resistance and high-dose antibody toxicities [72,73,74,75] and (iii) what would be the best diagnostic biomarkers or alternative strategies to completely eliminate these cancerous cells? Such questions provoked the scientist to further understand in-depth of the molecular mechanism of immune cell regulation and ICB-drug resistance. This revealed that; immune cells contains both inhibitory (break) as well as activator (acceleratory) marker to maintain immune homeostasis, or to avoid a situation called autoimmunity and self-tolerance phenomena. This understanding led to the discovery of (i) first immune checkpoint marker PD-1 or PDCD1 (CD279) in 1992 [76] and (ii) immune cell inhibitory marker cytotoxic T-lymphocyte-associated protein-4 (CTLA-4 or CD152) in 1991 [77] or 1995 [78,79]. However the first anti-CTLA4-based therapy ‘Ipilimumab’ was approved in 2011 by (James P. Allison, Nobel laureate, physiology or medicine, 2018) Medarex and Bristol-Myers Squibb for the treatment of melanoma, and the first anti-PD-1 therapy was approved in 2014 for melanoma and in 2015 for non-small-cell lung carcinoma (NSCLC) treatment (Table 2). Later, the tumor-cell inhibitory marker PD-L1 (CD274, previously known as B7-H1) was discovered in 1999-2000 [80] and PD-L2 (CD273, previously known as B7-DC) in 2001 [81] and was considered even much better control over immune cell checkpoint-based therapeutic targets [82] (Figure 2).
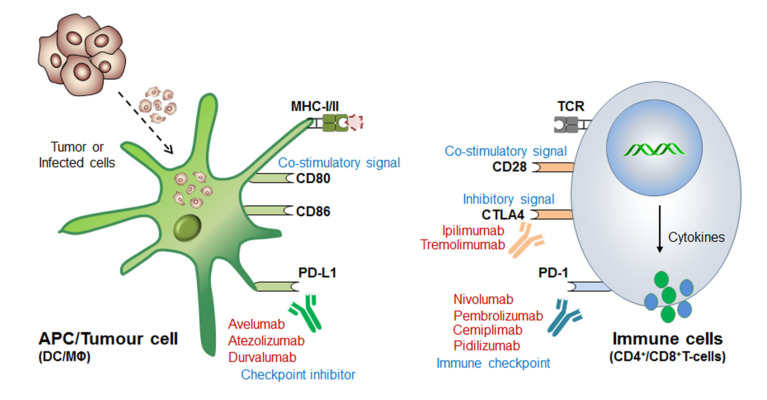
Figure 2.
Mechanism of immune checkpoint blockage or inhibition (ICB/ICI) therapy. The antigen presenting cells (APCs), especially dendritic cells and macrophages recognize and engulf the virus-infected or cancerous cells. The immune cells now processed and present the antigen to the naive T-cells in conjugation with MHC-I/II. The T-cell receptor (TCR) present on the immune cells recognizes this processed antigen and activates humoral as well as cell-mediated immune response. However, interestingly, immune cells, like CD8+T-cells also express PD-1 marker which function as “immune checkpoint” before cytolytic activation. On the other hand, tumor-engulfed DCs also expresses PD-L1 and PD-L2 (ligand for PD-1) and inhibitor bypass the function of immune activation called “immune checkpoint inhibitor” and thus T-cells filed to recognize it and considered as ‘self’ rather than ‘foreign’. Therefore tumor cell escapes this immune-surveillance mechanism and proliferates rapidly. Blocking these immune checkpoint markers by means of specific antibodies endorsed the discovery of ICB-therapeutics, for example, (i) Anti-PD-1 therapy (or Immune cell targeted therapy): Nivolumab (Opdivo®), Pembrolizumab (Keytruda®), Pedilizumab (CT-011) and Cemiplimab (Libtayo®) block PD-1 receptor and bypass the ‘self-recognition’ mechanism of T-cells, and thereby allowing rapid recognition and cytolytic activation to kill tumor cells. (ii) Anti-CTLA4 therapy: Immune cell (T-cells) expresses CTLA-4 to maintain normal homeostasis by regulating the hyper activation of other immune cells and also to avoid autoimmunity, just like ‘speed breaker’. But due to its impairments under the TME it is required to be constantly activated, and so anti-CTLA4 antibodies, like Ipilimumab (Yervoy®) and Tremolimumab efficiently block the inhibitory effect of CTLA-4. Moreover, since it is highly homologous to CD28-receptor functions, thereby further activating CD8+T effector function to enhance anti-tumor immunity. (iii) Tumor targeted therapy (or, immune checkpoint inhibitor): The anti-PD-L1 antibodies, like Atezolizumab (Tecentriq®), Avelumab (Bavencio®) and Durvalumab (Imfinzi®) blocks the inhibitory signal generated by tumor expressing PD-L1 (ligand for PD-1) to stop its self-defense mechanism, resulting in rapid tumor killing by T-cell attack. The detail mechanism of antigen presentation, ICB-therapy and strategies to overcome drug-resistance is well discribed in these articles [64,75].
1.1. Connotation of Immune Checkpoint Markers
The significance of these immune checkpoint markers (PD-1, PD-L1/L2) as a ‘remarkable discoveries’ was initially ratified after the experiments in mouse models, suggesting the requirement of these markers are equally vital in maintaining immune homeostasis by regulating a balance between ‘immune response’ and ‘immune tolerance’ via its acceleratory/co-stimulatory (CD28) as well as inhibitory (PD-1, CTLA4) receptors. For example, (i) the immune inhibitory function of PD-1 was demonstrated by characterizing autoimmune phenotype in PD1-deficient (PD-1−/−) mice, suggesting the loss of peripheral tolerance [106]. (ii) lupus like arthritis and glomerulus-nephritis in PD-1−/− C57BL/6 mice [107]. (iii) fatal myocarditis in PD-1−/− Balb/c and MRL mice [108,109]. (iv) Type-I diabetes in PD-1−/− NOD-mice [110,111]. (v) host vs graft disease in PD-1−/− mice crossed with H-2LD-specific 2C-TCR transgenic mice [107], and (vi) hydronephrosis associated abnormalities in PD-1−/− Balb/c mice [112]. Similarly, the first report on CTLA-4 blockade (negative regulator of T-cell activation [113]) in anti-tumor immunity was demonstrated in 1996 [114] and the first clinical report of CTLA-4 against melanoma in 2003 [115,116]. These discoveries were sufficient enough to encourage scientists to investigate its human relevance and further clinical trial (CT) studies.
1.2. ICB Drug-Resistance and Toxicities
Beside patient‘s age, cancer stage (I–IV) and various environmental factors; there might be several other factors for increased drug-resistance and reduced efficacy of ICB therapeutics [75]. For example, sub-optimal antibody dose, insufficient immune cell activation, intra-tumoral microenvironment, reduced memory cell formation and impaired effector cell functions after first course of treatment schedule. Sometimes, high-dose antibody toxicity also becomes a major concern for its adverse consequences (Figure 1). Therefore, a more advanced and unique therapy is required promptly to overcome this major issues.
Conclusively, our study devotes to improve the efficacy of ICB-therapy by co-targeting (i) epitranscriptomics (ii) intracellular immune checkpoints and (iii) microRNAs. More importantly, our investigation would help to design a specialized approach or custom-made strategies to improve the efficacy of ICB-therapy [7,117,118].
2. Milestones in ICB therapeutics
2.1. Discovery of ICB Therapy
The invention of ICB-therapy was started after the discovery of PD-1, PD-L1 and CTLA-4 like immune checkpoint markers. Write after that, several other immune-based markers were tested against broad range of tumor types, covered extensively in these articles [119,120,121,122]. However, this section diagrammatically simplifies the milestone in the development of ICB-therapy and recent strategies to improve the efficacy of ICB-antibodies by combining personalized medicines (Figure 1).
2.2. Mechanism of ICB/ICI-Therapeutics
The detail mechanism of ICB-therapy including current drug-resistance issues was already well described by Wei and Allison et al., 2018 [123], Jenkins et al., 2018 [73], Kalbasi et al., 2020 [75] and Barrueto et al., 2020 [124]. However, this section briefly simplifies the understanding of immune checkpoint markers and its implications in developing therapeutic antibodies. Although, our main focus is to resolve the issues associated with ICB drug-resistances by promoting personalized therapy (Figure 2).
2.3. Strategies to Overcome ICB Drug-Resistance
This section describes the strategies to overcome the issues mainly associated with drug-resistance and high-dose antibody toxicities. For example, (i) Epitranscriptomic approach: by targeting N6A modifiers: editor/writer, eraser/remover and effector/reader [125,126]. (ii) Bi-specific antibody approach: by co-targeting PD-1 and CD47 markers enlightened by ImmuneOncia therapeutics Inc. Korea [127,128] and AstraZeneca [129]. (iii) Antibody combination: by combining two antibodies targeting PD-1, PD-L1/L2 and CTLA-4 targets [123]. (iv) Precision medicines/personalized therapy: combining immunotherapy targeting intracellular immune checkpoints (CISH/SOCS-1) in specific immune cells [130,131]. (v) Molecular medicine: epigenetic modifiers targeting DNA, histone proteins and chromatin remodelers [22,132] and (vi) microRNAs [41,133,134] (Table 1 and Table 3, Figure 1). The detail of ICB-therapy and strategies to overcome ICB-drug resistance is well described in this review [75], however covering all is out of scope of this review.
3. Epitranscriptomics in ICB-Therapeutics
Epitranscriptomics has contributed greatly to the clinico-biological practices due to its diverse role in regulating at post-transcriptional and translational level. Epitranscriptomics generally referred to chemical modifications in the RNA molecule without changing the nucleotide sequence. So far more than 160 chemical modifications have been identified [151] playing a crucial role in regulating various biological processes, for example, in acute myeloid leukemia treatment [125], lung adenocarcinoma [152] gastric cancer [153] and broad range tumor types [151,154,155]. The major epitranscriptomic machineries (writer/editor, eraser/remover and readers/effector [156] not only regulate RNAs by specific regulatory mechanism [157,158] but also decide the fate of the cells and its associated immune disorders in cellular context-dependent manner. In this section, we have described the clinical application of epitranscriptomics in overcoming the issues associated with ICB drug-resistance by combining personalized approach.
3.1. Editors (Writers):
3.1.1. Mettl-3/14 in Anti-PD-1 Resistance (Colorectal Cancer)
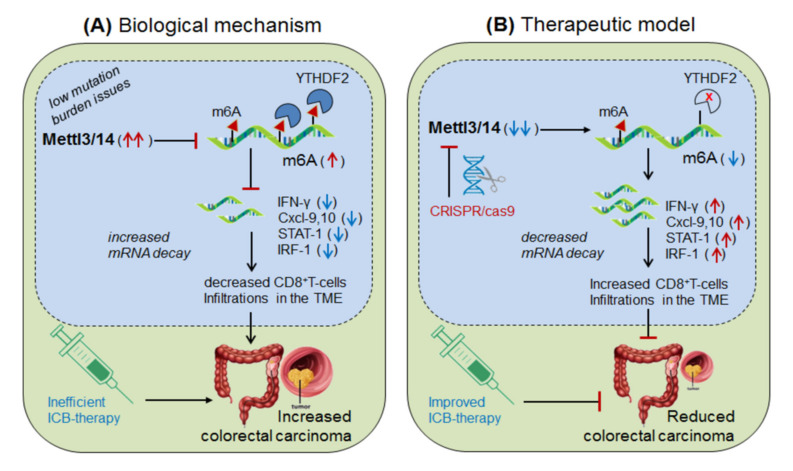
Wang, et al., 2020 [8] demonstrated the role of Mettl-3/14 (m6A-writer enzyme) in improving the efficacy of anti-PD-1 therapy. They found that even after standard anti-PD-1 treatment, still some patients with colorectal cancer and melanoma develop resistance, because of insufficient immune response generated by the tumors with low mutation burden issues (mismatch-repair-proficient or microsatellite instability-low ‘pMMR-MSI-L’) constituting ~85% of the patients [159]. They found that, these patients have significantly increased level of Mettl-3/14, which has impaired the function of certain crucial genes under the tumor microenvironment (TME). Interestingly, CRISPR/cas9-mediated deletion of Mettl-3/14 in colorectal cancer cell line (CT26) and murine melanoma cell line (B16) has not only increased cytotoxic CD8+T-cell (CTL) infiltrations in the TME but also provided durable adoptive immune response. Mechanistically, they justified that, the loss of Mettl-3/14 augmented mRNA-stability of IFNγ, STAT-1 and IRF-1 by promoting IFNγ-STAT1-IRF1-signalling through YTHDF2 reader proteins [157,158], leading to prolong secretion of these cytokines in the TME, resulting in strong immune response. These investigations suggest the key role of Mettl-3/14 in inhibiting the efficacy of anti-PD-1 therapy by decreasing IFNγ, Cxcl-9 and Cxcl10-mediated immune response. Conclusively, this study endorsed the immunotherapeutic potential of m6A-writer in improving the efficacy of anti-PD-1 antibody by silencing Mettl-3/14 in the TME [8]. Moreover, overexpressing FTO (m6A-demethylase) or by targeting intracellular YTHDF2 (m6A-reader protein) in decreasing Mettl-3/14 methylation could be considered as an alternative strategy to improve anti-PD-1 therapeutics (Figure 3).
Figure 3.
Therapeutic model targeting ‘Mettl-3/14’ in colorectal cancer. (A) Biological mechanism: Mettl-3/14 is up-regulated in colorectal cancer and melanoma, and inhibits the expression of IFNγ-STAT1-IRF1 signaling via YTHDF2-mediated (decreased mRNA decay) mechanism and thereby decreases the efficacy of anti-PD-1 effect by lowering CD8+T-cell infiltrations in the TME, and thus facilitated disease progression. (B) Therapeutic model: Anti-Mettl-3/14 therapy: CRISPR/cas9-silencing of Mettl-3/14 increases the expression of its target IFNγ-STAT1-IRF1 genes/signaling by reducing the recruitment of YTHDF2-mediated decay mechanism, and thus enhances the efficacy of anti-PD-1 antibody by increasing infiltrations of CD8+T-cell in the TME. Moreover, FTO overexpression might decrease Mettl-3/14 level via balancing mechanisms, and ‘anti-YTHDF2 therapy’ by directly augmenting target gene expressions, via its mRNA stability mechanisms, might have therapeutic benefits.
3.1.2. Mettl-3 in Anti-PD-1 Resistance (Lung Metastasis)
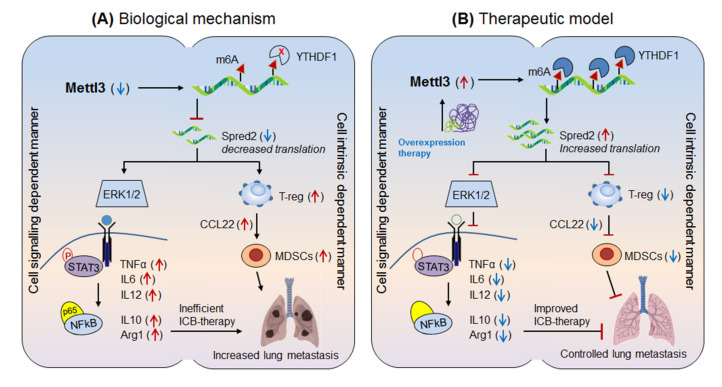
Yin et al., 2021 [9] demonstrated the molecular mechanism of anti-PD-1 resistance by Mettl3-mediated macrophage polarization, and enlightened the significance of decreased Mettl3-level in lung metastasis. Yin and collogues identified that the in vitro co-culture of bone marrow derived macrophages (BMDM) with B16 (skin melanoma) or LLC (lewis lung carcinoma) cell lines decreases the expression of Mettl-3. Moreover, the in vivo implantation of B16 and LLC cell lines into syngeneic mice also decreases Mettl-3 expression in tumor associated macrophages (TAM: CD11b+F4/80+), suggesting the loss of Mettl-3 in promoting tumor growth and thus survival defect. To investigate the underlying mechanism, they used specific mouse model selectively depleted with Mettl-3 by crossing Mettl3f1/f1 and Lyz2-cre mice. Interestingly, B16/LLC injected mice showed rapid tumor progression as well as lung metastasis in Mettl3-deficient (Mettl3fl/flLyz2cre/+ or Mettl3cKO) mice as compared to the wild type (Mettl3fl/flLyz2+/+ or Mettl3WT) mice. In addition, abnormal macrophage polarization characterized by increased M1-pro-inflammatory/anti-tumor (CD11b+F4/80+NOS2highIL-12high) and decreased M2-anti-inflammatory/pro-tumor (CD11b+F4/80+ARG1highIL-10high) were also noted in Mettl3cKO mice, along with impaired response to effector T-cell functions. More importantly, the flow cytometry analysis of tumor bearing mice (TBM) revealed increased infiltration of immunosuppressive cells like, regulatory T-cells (T-reg: CD4+CD25+Foxp3+) and myeloid derived suppressor cells (MDSCs: CD11b+Gr1+) in the TME evidenced by increased expression of CCL22-migratory marker in Mettl3cKO mice. Reciprocally, selective depletion of T-reg (anti-CD25) and macrophages (clodronate liposomes) significantly decreased both tumor growth and lung metastasis. This result clearly suggests that anti-PD-1 resistance has occurred due to (i) increased abundance of immunosuppressive populations. (ii) impaired CD8+ T-cell effector function and (iii) hyper-polarization of M1/M2-macrophage in the TME of Mettl3cKO mice mimicking diseased model. Mechanistically, m6A-methylated RNA-immunoprecipitation followed by high throughput sequencing (MeRIP-Seq) of the RNA isolated from BMDMs from Mettl3WT and Mettl3cKO mice revealed ‘spred2’ as a potential downregulated target of Mettl-3 overlapping MAPK/ERK pathways. This suggests that spred2 is an upstream target of NFκB and STAT3 pathway in polarizing M1/M2-macrophage, as well as negative regulator of ERK/MAPK-signaling [160]. The above findings were further validated by reverting the M1/M2-polarizations by selective inhibition of STAT3 (S3I-201) and NFκB (BAY-11-7082) pathways, justified by chromatin immunoprecipitation (ChIP) for increased STAT3 binding to Arg1 promoter (M2-polarization marker) in Mettl3cKO. Next, with regard to epigenetic regulation, the overexpression of Mettl-3 increases the translation of ‘spred2’ by YTHDF1-mediated mechanism, confirmed by increased binding of YTHDF1 to spred2 via RNA-IP. Conversely, knockdown of YTHDF1 (siRNA) diminishes spred2 level. This result further supports ‘spred2’ as a target of Mettl-3 and is regulated by YTHDF1-mediated mechanism [126] rather than by targeting mRNA-stability or promoter-dependent translation mechanisms [161], and thereby activated ERK-mediated (being spred2 as a negative regulator of ERK signaling) other downstream signaling pathways in polarizing M1/M2-macrophages. Additionally, polysome profiling for translation-active (>80S) regulatory site and m6A-conserved motif ‘GGAC’ analysis further authenticate spred2 regulation by Mettl3-methylation mechanisms, validated by decreased spred2 expression in mutant (GCTC) as compared to the wild-type (GGAC) motif. Lastly, the link between Mettl3-driven spred2 and ERK1/2-NFκB-STAT3 signaling confirms the polarization of M1/M2-macrophage by aggravating TNFα and IL-6 (M1: pro-inflammatory) and IL-10, Arg1 (M2: anti-inflammatory) cytokines, validated by diminished expression of the same by selective signaling inhibitors. Taken together, these results suggest the crucial role of Mettl-3 in impairing anti-PD-1 efficacy by (i) polarizing M1/M2-macrophage via activating spred2-mediated ERK1/2-NFκB-STAT3 signaling cascade through cytokine milieu and (ii) by recruiting immunosuppressive cell populations in the TME. Conclusively, Mettl-3 is key player in reducing the efficacy of anti-PD-1 therapy, and therefore targeting (overexpressing) Mettl-3 could be a promising approach to control cancer metastasis by enhancing the efficacy of anti-PD-1 antibodies (Figure 4) [9]. This hypothesis was further supported by Yi, et al., 2020 in regulating PD-L1 mediated HNSCC control by implicating m6A-modifiers, and thus potentiating its therapeutic value by targeting G2M checkpoint, mTORC1 and PI3K/AKT/mTOR signaling analyzed via cancer genome atlas TCGA (n = 499) and GSE65858 (n = 270) cohorts [10].
Figure 4.
Therapeutic model targeting ‘Mettl-3’ in lung metastasis. (A) Biological mechanism: Mettl-3 is significantly down-regulated in tumor associated macrophage (TAM) and thereby alters M1/M2-macrophage-polarization and thus increases the infiltration of immunosuppressive populations (T-reg and MDSCs) in the tumor microenvironment, resulting in increased tumor growth and lung metastasis. (B) Therapeutic model: overexpression therapy: overexpression of Mettl-3 recruited YTHDF1-reader protein which increases the expression of its target ‘spred2’ gene, resulting in decreased infiltration of immunosuppressive cells by reducing ERK1/2 signaling, that finely reduces lung metastasis by improving the efficacy of anti-PD-1 therapeutics.
3.2. Erasers (Removers):
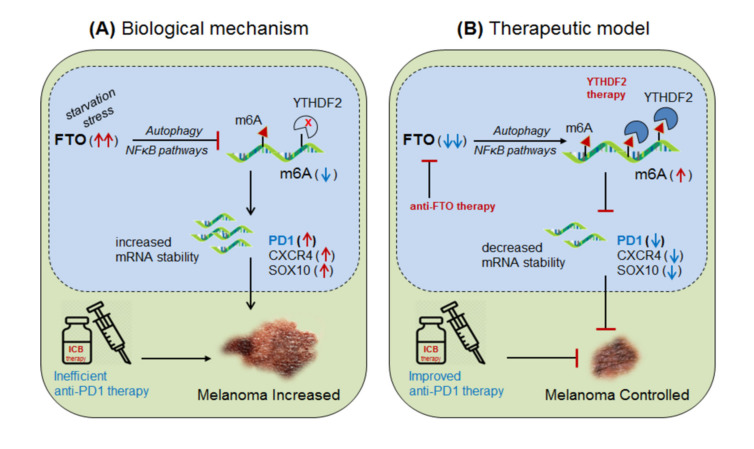
3.2.1. FTO in Anti-PD1 Resistance (Melanoma):
Yang et al., 2019 [11] demonstrated the role of m6A-eraser protein ‘FTO’ in melanoma progression, a type of skin cancer, and enlightened the intrinsic mechanism to improve the efficacy of anti-PD-1 therapy by targeting FTO. Yang and colleagues found that FTO is significantly up-regulated in human melanoma patients (metastatic skin samples n = 65) including human (Mel624) and mouse (B16F10) cell lines, and facilitated rapid tumorigenesis, caused by metabolic starvation stress in mice requiring autophagy and NFκB pathway [162]. However, selective depletion of ‘FTO’ not only increases sensitivity to anti-PD-1 therapy but also increases m6A methylation-inhibition of critical pro-tumorigenic (tumor-promoting) genes. Mechanistically, they proved that FTO-deficiency increases m6A-methylation at 5′UTR and 3′UTR of target genes; PD-1 (PDCD1), CXCR4 and SOX10, and thereby causing rapid mRNA-degradation by recruiting YTHDF2-reader proteins [155,157,158], confirmed by YTHDF2-knockdown in ‘increasing’ and YTHDF2-overexpression in ‘decreasing’ melanoma growth. Moreover, FTO-deficiency enhances the sensitivity of anti-PD-1 treatment by IFNγ-mediated cytokine response. These results clearly suggest that FTO plays a crucial role in melanoma tumorigenesis by regulating mTOR signalling through limiting the nutrient supply to the tumours [162]. Therefore, co-targeting FTO in combination with ICB-antibodies would be a promising approach to control melanoma progression [11]. This hypothesis was also supported by Singh et al., 2016 in controlling triple-negative inflammatory breast cancer cells using FTO (MO-I-500) inhibitor [125,163,164]. Theoretically, targeted overexpression of Mettl-3 might also control melanoma progression by decreasing FTO via balancing mechanism, and also by directly inhibiting the expression of pro-tumorigenic genes via recruiting YTHDF2 reader proteins (Figure 5).
Figure 5.
Therapeutic model targeting ‘FTO’ in melanoma. (A) Biological mechanism: FTO is highly up-regulated in melanoma (due to starvation stress through NFκB-pathways and autophagy) leading to increased mRNA transcript of the critical pro-tumorigenic genes (PD-1, CXCR4 and SOX10) by decreasing m6A-methylation mark, resulting in increased melanoma progression. (B) Therapeutic model: (i) Anti-FTO therapy: selective inhibition of FTO (FTO inhibitor [125]) or intracellular silencing of ‘FTO’ controls melanoma progression by selectively increasing the methylation-inhibition of its pro-tumorigenic genes, including PD-1 immune checkpoint markers by increasing the efficacy of anti-PD-1 antibody [163]. (ii) YTHDF2 therapy: YTHDF2 overexpression would control melanoma progression by accelerating the mRNA-decay of critical pro-tumorigenic genes. Moreover, targeted overexpression of Mettl-3 might control melanoma progression by destabilizing critical tumor-promoting genes by recruiting YTHDF2-reader proteins. Furthermore, targeting NFκB/mTOR signaling might also control melanoma progression by limiting nutrient supply to the tumors.
3.2.2. FTO in Anti-PD-1 Resistance (Colon Cancer)
Tsuruta et al., 2020 [12] demonstrated the role of FTO in colon cancer progression and enlightened the molecular mechanism to control cancer carcinogenesis by targeting FTO. They found that FTO is aberrantly expressed in colon cancer cell line (HCT-116). Moreover, immune checkpoint molecule ‘PD-L1’ expression was also highly up-regulated. Therefore, targeting FTO by selective depletion (siRNA) not only reduced FTO-level but also significantly decreased PD-L1 expression in IFNγ signaling-independent manner at both mRNA and protein levels. This result clearly suggests that FTO facilitates colon cancer progression by promoting the expression of PD-L1 markers. Mechanistically, they proved (via RNA immunoprecipitation) that FTO binds to m6A-marked PD-L1 mRNA and elevates its expression probably by decreasing mRNA-decay mechanism. Taken together, this study reveals the critical role of FTO in facilitating colon carcinoma by increasing PD-L1 expression, and therefore targeting FTO by means of either selective FTO inhibitor [125] or CRISPR/Cas9-based methods could hold the potential to control colon cancer in combination with anti-PD-L1 therapeutics (Table 1) [12].
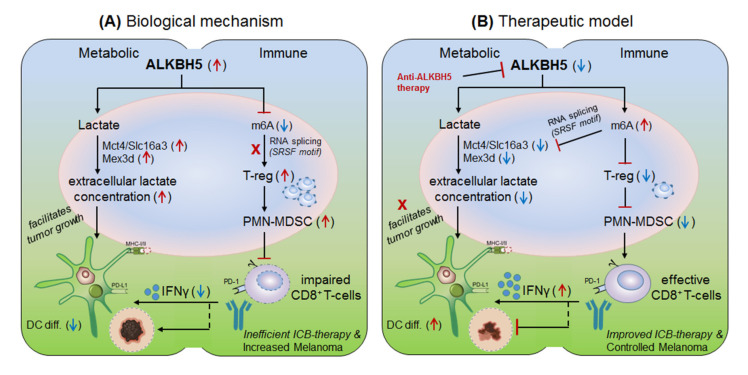
3.2.3. ALKBH5 in Anti-PD-1 Resistance (Melanoma)
Li et al., 2020 [13] explained the role of another m6A-eraser protein ‘ALKBH5’ in progression of melanoma-associated metastatic cancer, and enlightened the molecular mechanism to overcome anti-PD-1 resistance by targeting ALKBH5. Based on their previous studies [8] for the role of Mettl-3/14 in melanoma progression, the authors hypothesized that ALKBH5 might also have significant role in regulating the efficacy of anti-PD-1 therapeutics. To this end, Li and colleagues used B16 (mouse melanoma) and CT26 (colorectal carcinoma)-induced TBM model, and selectively depleted ALKBH5 and/or FTO (CRISPR/Cas9-mediated silencing) in B16 and CT26 cell lines respectively, and injected subcutaneously into wild-type C57BL/6 and BALB/c mice to create tumor, followed by 1-day prior vaccination with irradiated B16 cells secreting GM-CSF ‘GVAX’ to induce sufficient antitumor T-cell response, and finally anti-PD-1 antibody treatment was given to check its efficacy. Interestingly, ALKBH5−/− TBM showed prolonged survival and slower tumor growth as compared to the non-transfected (NTC) control mice, suggesting the direct involvement of ‘ALKBH5’ in interfering with the efficacy of anti-PD-1 antibody. To further elucidate the role of ALKBH5 in modulating GVAX/anti-PD-1 treatment, they analysed tumor infiltrating lymphocytes (TILs) by FACS and found that among total CD45+CD4+CD8+ gated populations, ALKBH5−/− mice have elevated granzyme-B (GZMB)+CD8+, GZMB+CD4+ T-cell, NK-cell (CD56+) and dendritic cell (DCs: CD45+Ly6C-MHC-II+CD24hiF4/80lo) numbers, but more importantly, T-reg (CD4+Foxp3+) and polymorphonuclear myeloid derived suppressor cell (PMN-MDSCs: CD45+CD11b+Ly6G+Ly6CloF4/80−MHC-II−) populations were drastically reduced as compared to the control mice. This was further validated by immunohistochemistry staining (IHC) of the MDSC-mLy6G, however, no differences in other immune cell populations (MDSC and macrophage) were noted. This suggests that ALKBH5 has the potential to recruit immunosuppressive (T-reg and PMN-MDSCs) populations in the TME during ICB therapy. Again, to stamp the selective function of immunosuppressive cells in inhibiting anti-PD-1 effect, they specifically depleted T-regs (anti-CD25) and PMN-MDSCs cells in the NTC control mice, resulting in delayed tumor progression as compared to the ALKBH5−/− model (due to already fewer T-reg numbers), confirming the immunosuppressive function of T-regs (induced by MDSCs) in impairing the efficacy of anti-PD-1 antibody by inhibiting CD8+T-cells effector functions through decreasing DC-differentiation (CD45+Ly6C-MHC-II+CD24hiF4/80lo) markers [165]. These observations clearly suggest that ALKBH5 recruits immunosuppressive populations in the TME and thereby interfering with the efficacy of anti-PD-1 therapy. Next, to identify the molecular targets, they sequenced RNA isolated from ALKBH5/FTO−/− B16 tumors and compared it with the NTC-control TBM on day-12 after GVAX/anti-PD1 treatment. Interestingly, the gene ontology (GO) analysis of the differentially expressed genes (DEG) revealed ALKBH5 is associated with metabolic genes especially ‘Mct4/Slc16a3’ involved in lactate metabolism, whereas, FTO is associated with IFNγ and chemokine signalling pathways. This was validated by increased IFNγ intermediates (qRT-PCR expression) upon in-vitro stimulation of IFNγ to the FTO−/− B16 cells. Moreover, the comparison of mouse DEGs with human melanoma patients (n = 21 anti-PD1 therapy responder) and (n = 17 non-responder) reveals eight common genes associated with ALKBH5-deficiency and eleven common genes with FTO-deficiency, indicating ‘conserved’ and potential targets of ALKBH5 and FTO in mouse as well as human receiving anti-PD1 therapy. This suggests that ALKBH5 modulates anti-PD-1 resistance by recruiting immunosuppressive T-reg cells and by modulating metabolic genes whereas FTO works by targeting IFNγ and by modulating inflammatory chemokine-mediated signalling pathways in the TME. Next, epigenetic analysis via LC-MS/MS reveals higher m6A-abundance in ALKBH5-deficient as compared to FTO-deficient B16 tumours, which meaningfully suppresses the expression of m6A-mediated ‘Mct4/Slc16a3’ in ALKBH5 alone and ‘Mex3d’ in ALKBH5 and FTO both. Moreover, MeRIP-seq reveals enriched SRSF motif (a subunit of SAG core involved in RNA splicing [166]) in ALKBH5-deficient tumors as compared to FTO, suggesting different mechanisms of action of these two de-methylases in modulating anti-PD1 efficacies. Collectively, these results suggest that ALKBH5 and FTO target metabolic genes and increase the expression of Mct4/Slc16a3 and Mex3d (supplementing lactate to the tumour) by inhibiting m6A methylation-mediated RNA-splicing mechanisms, supported by Zaho et al., 2014 [167], (Figure 6, therapeutic model). Furthermore, to dig out the m6A-modulated genes via RNA-splicing mechanism, they identified m6A-enriched transcripts around 5′-3′ splice sites by m6-CLIP and found the involvement of three immunotherapeutic resistance genes Eif4a2, Arid4b and USP15 affecting the response of anti-PD-1 therapeutics by regulating transcription, translation and T-reg activation via TGF-β signalling in the TME. (Figure 6) Taken together, ALKBH5 is playing a crucial role in promoting tumour metastasis, and therefore intracellular silencing of ALKBH5 in the TME would hold the potential to control tumor metastasis via increasing the efficacy to anti-PD-1 therapeutics [13].
Figure 6.
Therapeutic model targeting intracellular checkpoint ‘ALKBH5’ in melanoma. (A) Biological mechanism: The ALKBH5 abnormally expressed in melanoma and colorectal cell carcinomas, and impairs the efficacy of anti-PD-1 therapy by (i) recruiting immunosuppressive; regulatory T-cell (T-reg) and polymorphonuclear myeloid derived suppressor cells (PMN-MDSC) abundances in the TME. (ii) by impairing DC-differentiation resulting in decreased CD8+T-cell effector functions. (iii) by increasing extracellular lactate availability to the tumors by up-regulating the expression of Mct4/Slc16a3 genes due to decreased m6A-methylation mark associated mechanism. (B) Therapeutic model: (i) Anti-ALKBH5 therapy: selective inhibition of ALKBH5 [168] by increasing m6A methylation-mediated inhibition of crucial genes essential to increase the efficacy of CD8+T-effector cells. (ii) T-reg/PMN-MDSCs depletion therapy: could also show the therapeutic propensity by rescuing the immunosuppressive environment. (iii) Increasing DC-differentiation: could be also a promising approach to enhance DC-mediated CD8+T-cell effector function. (iv) Targeting metabolic genes: could be an alternative approach to control melanoma tumorigenesis by limiting extracellular lactate accumulation in the TME. Collectively, all these approach seems promising in overcoming the issues associated with ICB drug-resistance.
3.3. Effectors (Readers):
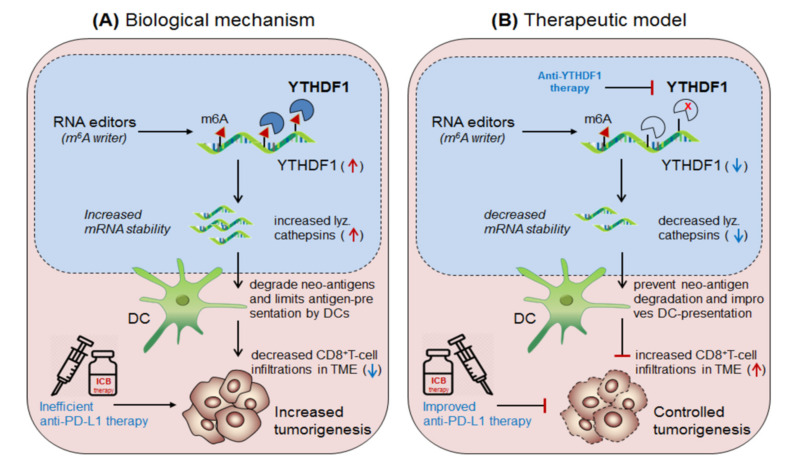
3.3.1. YTHDF1 in Anti-PD1 Resistance (Solid Tumors)
Han et al., 2019 [14] demonstrated the synergistic role of dendritic cells expressing ‘m6A-writer’ and ‘YTHDF1-readers’ proteins in anti-tumor immunity. They found that despite the presence of numerous neo-antigens, some patients still failed to generate sufficient anti-tumor response. To this end, in discovering the intrinsic molecular mechanism, they generated dendritic cell-specific conditional knockout mice depleted with YTHDF1 (YTHDF1cKO) gene. Surprisingly, the loss of YTHDF1 enhances antigen-recognition and cross-presentation ability of DCs in-vivo, resulting in elevated CD8+ T-cell infiltration in the TME as compared to the control wild type (YTHDF1WT) mice. Moreover YTHDF1cKO mice showed enhanced response to anti-PD1 therapy [159]. Mechanistically, they proved that the wild type mice, in the presence of m6A mRNA-methylation machineries recruited YTHDF1 reader proteins at the lysosomal-cathepsins mRNA axis, resulting in increased mRNA-stability, and thereby increased the abundance of cathepsin proteins in the phagosomal compartments of the DCs, causing severe degradation of the neo-antigens and thus limiting the antigen availability to the DCs for antigen-recognition and further cross-presentation to CD8+T-cells in the cytosol. This result suggests that YTHDF1 is playing a crucial role in suppressing anti-tumor immunity [14], and therefore intracellular silencing of YTHDF1 in DCs designates its potential to enhance anti-tumor immunity. Collectively, this discovery reveals two important mechanisms to enhance anti-tumor immunity by co-targeting (i) anti-YTHDF1 therapy: where, YTHDF1-deficiency protect ‘antigen-degradation’ and allows efficient recognition and presentation by DCs, in-turn, further increases the abundance of DC-mediated effector CD8+T-cells by cross-presentation mechanism, supported by Ding et al., 2021 [169] and (ii) by enhancing anti-PD-1/PD-L1 efficacy: which further potentiates the efficacy of anti-PD-1 immunotherapy by enhancing the effector function of CD8+T-cells in the TME [159] Figure 7.
Figure 7.
Therapeutic model targeting intracellular checkpoint ‘YTHDF1’ in enhancing DC-mediated anti-tumor immunity. (A) Biological mechanism: The ‘YTHDF1’ reader protein recognizes m6A-marked cathepsin transcript and increases it’s mRNA and protein level, which translocate into the phagosome and degrades neo-antigens, and thus limiting its recognition and cross-presentation by the DCs, and thereby the impaired DCs decreases CD8+T-cell effector function, leading to decreased efficacy of anti-PD-1 therapy, resulting in increased tumor growth. (B) Therapeutic model: Anti-YTHDF1 therapy: inhibits cathepsin level and thus unable to degrade neo-antigens, resulting in effective antigen recognition and cross-presentation by DCs and thereby enhanced CD8+T-cell effector function which improves the efficacy of anti-PD-1 therapy.
3.3.2. YTHDF2 in Anti-PD1 Resistance (Brain Tumors)
Lin et al., 2020 [15] demonstrated the role of YTHDF2 in progression of lower-grade glioma (LGG) also called ‘pilocytic astrocytoma’, a type of early stage brain tumor. They showed that YTHDF2 is abnormally expressed in various types of cancers and reduces overall longevity and survival. The higher expression of YTHDF2 has been positively correlated with immune cell (B-cells, T-cells, DCs, MΦ and neutrophils) expressing PD-1, TIM-3 and CTLA-4 markers. Therefore, targeting YTHDF2 in DCs would hold the potential to enhance anti-tumor immunity in combination with ICB-therapeutics [14,15]. A similar pre-clinical trial was proposed by jubilant-therapeutics targeting PD-1 inhibitor (with brain penetrant PRMT5) in controlling LGG, potentiating the scope to utilize in combinations with immune cells targeting intracellular checkpoints as targeted therapy. Moreover, Garzon-Muvdi et al., 2018, have supported the prominence of DC activation in enhancing the efficacy of anti-PD-1 immunotherapy against glioblastoma [16]. Taken together, this study reveals the importance of targeting YTHDF2 in combination with DC-immunotherapy [7] to enhance the efficacy of ICB therapy against early stage brain tumors [15], (Table 3).
4. Immune Cells: Targeting Intracellular Checkpoint ‘CISH’ in Combination with ICB-Therapeutics and Recent Clinical Trials
Cytokine-inducible SH2-domain containing protein (CISH or CIS) is one of the eighth members of SOCS family of proteins, recently gaining high attention due to its widespread regulatory role in cytokine signalling [170,171] and its involvement in more than 349-diseased (https://platform.opentargets.org/target/ENSG00000114737/associations; accessed on: 25 July 2021) [172,173,174] phenotypes. The therapeutic significance of ‘CISH’ can be evidenced by a recent clinical trial (NCT04426669, NCT03538613 by Intima Bioscience, UK; and ONKT102, ONKT103 and ONKT104 by ONK therapeutics, Ireland) targeting NK-cells, TILs and DCs for the treatment of broad range of metastatic cancers [1,2]. The so-called personalized medicine targeting ‘CISH’ in immune cells has shown promising effect in improving the efficacy of ICB-therapeutics [3,5]. Therefore, this section highlights another layer of strengthening ICB-therapy by targeting intracellular immune checkpoint ‘CISH’ in different immune cells. A few important links/references are also provided in (Box-2) supporting ‘CISH/SOCS’ to be used as potential markers in developing personalized medicine [7,175,176,177,178] (Table 3, Box-1).
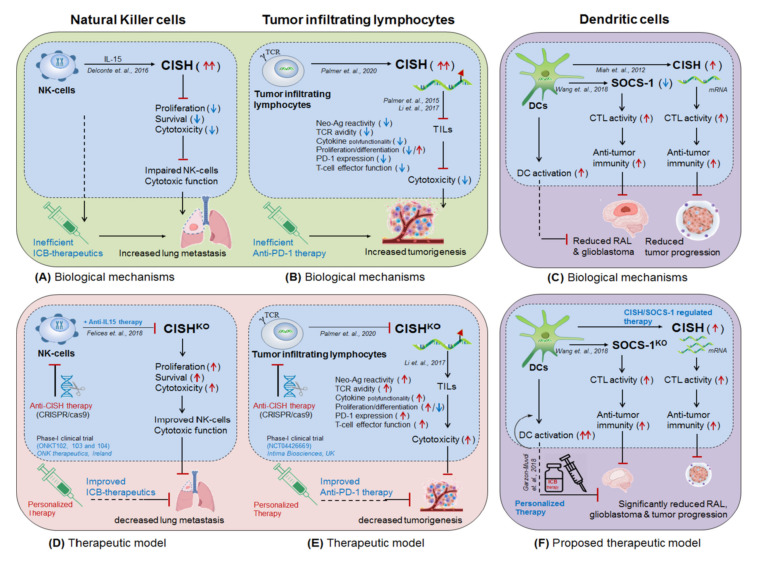
4.1. NK-Cells Targeting CISH in ICB Therapeutics
Delconte et al., 2016 [3] demonstrated the therapeutic benefit of anti-PD-1 and anti-CTLA-4 antibodies in combination with intracellular checkpoint targeting CISH (CISH-deletion) in NK-cells for the treatment of lung metastasis and melanomas in murine model. They showed that intravenous (i.v.) administration of melanoma cell line (B16/F10) and prostate cancer cell line (RM-1) into CISH-deficient (CISH−/−) NK-cells have significantly reduced melanoma growth and metastatic nodule formation as compared to the wild-type (CISH+/+) mice, indicating the critical role of CISH in NK-cell cytotoxicity. The specificity of NK-cell function was confirmed by selective depletion of NK-cells (anti-asiolo GM1) in rendering susceptibility to B16F10 metastasis in CISH−/− mice. Moreover, the adoptive transfer of CISH−/− NK-cells into NK-cell−/− recipient mice (Mcl1f/f Ncr1-iCre) showed fewer B16F10 metastases as compared to the mice receiving CISH+/+ NK cells. These results clearly suggest that (i) CISH is playing a crucial role in NK-cell activation. (ii) CISH is a negative regulator of NK-cell cytotoxicity and (iii) CISH−/− NK cells are intrinsically more active. Moreover, combining anti-PD-1 and anti-CTLA-4 antibody treatment with CISH−/− NK-cells drastically reduced lung metastasis as compared to the IgG control and CISH+/+ NK-cells alone in the adoptive transfer model, highlighting the potential therapeutic benefit that could be achieved when anti-PD-1 and anti-CTLA-4 therapy was combined with loss of CISH function. A similar result targeting intracellular checkpoint ‘CISH in NK-cells’ in combination with ICB-antibodies in increasing anti-tumor immunity was described by Putz et al., 2017 [4], Bernard et al., 2021 [179], Felices et al., 2018 [180] and Andre et al., 2018 [181]. Furthermore, a recent phase-I clinical trial (ONKT102, ONKT103 and ONKT104) targeting CISH-deletion in NK-cells was proposed by ONK therapeutics, Ireland, until 2021𠄲2022 against hematological malignancies (multiple myeloma and acute myeloid leukemia) and solid tumors (ovarian, NSCLC and breast cancers) [127,135,142] (Figure 8A,D). In addition to ‘CISH’, other immune checkpoint markers in NK-cells were nicely described by Chiossone et al., 2018 [121].
Figure 8.
Immunotherapeutic model co-targeting intracellular checkpoint (CISH/SOCS-1) in combination with ICB-therapeutics. (A) NK-cell biological mechanism: CISH expression in natural killer cell impairs its cytolytic function by reducing NK-cell proliferation, fitness and survival inside the TME [190] and thus insufficient in protecting lung metastasis [3]. (B) Tumor infiltrating lymphocytes (TILs) biological mechanism: CISH expression in TILs also impairs T-cells function by reducing TCR avidity, cytokine poly-functionality and CD8+T-cells effector functions and thereby facilitating tumorigenesis [6]. (C) Dendritic cell biological mechanism: SOCS1-deficiency and CISH-expression in DCs has been proven in regulating DC-mediated anti-tumor immunity against broad range of solid tumors by increasing DC-activation and CTL-activity [7,184]. (D) Therapeutic model targeting CISH in NK-cells: (i) anti-CISH therapy: CISH deletion (CRISPR/Cas9) in NK-cells improves overall NK-cell survival, proliferation, fitness and effector functions, and thereby potentiating its cytotoxic activity. (ii) Personalized therapy: co-targeting CISH−/− NK-cells with ICB-antibodies significantly improves ICB-efficacy in controlling lung metastasis [190]. A relevant human clinical trial (ONKT102, ONKT103 and ONKT104) targeting CISH-deficient NK-cells has been proposed by ONK therapeutics, Ireland, against hematological malignancies and solid tumors [135] (Box-1). (E) Therapeutic model targeting CISH in TILs: (i) anti-CISH therapy: CISH deletion (CRISPR/Cas9) in TILs improves T-cell effector functions by increasing TCR avidity and cytokine poly-functionality, and thereby potentiating its effector function. (ii) Personalized therapy: co-targeting CISH−/− TILs with ICB-antibodies significantly improves ICB-efficacy in increasing anti-tumor immunity. A relevant human clinical trial (NCT04426669, NCT03538613) targeting CISH−/− TILs has been proposed by Intima Bioscience, Inc. UK, against solid tumors and metastatic gastrointestinal cancers [1,2] (Box-1). (F) Proposed therapeutic model targeting SOCS-1/CISH in DCs: (i) SOCS-1/CISH regulated therapy: Regulation of SOCS-1/CISH in DC-mediated anti-tumor immunity has been already shown in increasing CTL-activity against broad range of solid tumors [7,182,183,184], however, (ii) Personalized therapy: Co-targeting SOCS-1/CISH in DCs in combination with ICB-antibodies would further potentiates the efficacy of ICB-therapy, and therefore would be efficient in solving the issues associated with high-dose antibody toxicity and drug-resistance. A relevant human clinical trial (NCT01956630) targeting SOCS1−/− in DCs has been conducted by academy of military medical sciences, China against leukemia [7] (Box-1) Moreover, a future research targeting epitranscriptomic machineries (m6A-modifiers) and microRNAs might further potentiate DCs by regulating CISH diversity [191], (Table 1).
4.2. T-cells Targeting CISH in ICB Therapeutics
Palmer et al., 2020 [5] demonstrated the improved efficacy ICB-antibodies when combined with CISH-depleted (CISH−/−) TILs. Palmer and colleagues showed that the adoptive transfer of neoantigen specific TILs, derived from antigen expressing tumors, was failed in constantly eliciting durable tumor regression. Moreover, an altered expression of CD39, Tox and PD-1 marker was observed, suggesting the impaired function of effector CD8+T-cells in the TME. Interestingly, depletion of CISH (CRISPR/Cas9) in TILs significantly improved neoantigen recognition, TCR avidity, T-cell activation/expansion and tumor cytolysis, resulting in rapid-control over tumorigenesis. However an increased expression of PD-1 marker was also observed. Thus, co-targeting CISH−/− TILs in combination with anti-PD1 antibody has proficiently controlled the tumor progression. This result clearly suggests the negative regulatory role of CISH in impairing T-cell effector functions, supported previously by Palmer et al., 2015 [6] and Periasamy et al., 2011 [147]. Therefore, co-targeting CISH−/− TILs in combination with ICB-therapy would hold the potential to control tumor progression by improving the efficacy of ICB-antibodies as well as CD8+T-cell effector function. A relevant human Phase-I/II clinical trial (NCT04426669, NCT03538613) targeting CISH−/− TILs in combination with ICB-therapy was proposed by Intima Bioscience, UK, until 2021𠄲2022 against wide range of tumor types and gastrointestinal cancer [1,2,115] (Figure 8B,E, Table 3, Box-1).
4.3. Dendritic Cells Targeting SOCS-1/CISH in ICB-Therapeutics
Wang et al., 2018 [7] demonstrated the therapeutic benefit of targeting intracellular checkpoint SOCS-1, one of the members of CISH family, in DCs (SOCS-1−/− DCs [182]) in controlling relapsed acute leukemia (RAL). They showed that the adoptive transfer of genetically modified DCs plus CIK cells is safe & effective in prolonging the survival of RAL patients (n = 48), by increasing DC activation, DC-maturation and TAA-induced CTL response. A relevant human phase-I/II clinical trial (NCT01956630) was conducted by the academy of military medical sciences, China and recommends it safe in-use [7]. A similar result was observed by Shen et al., 2004 in increasing anti-tumor immunity by silencing SOCS-1 in DCs [183]. More relevantly, Miah et al., 2012, demonstrated the importance of CISH-expressing DCs in increasing anti-tumor immunity by enhancing CTL activity in CISH−/− CD11c mouse model [184], however it would be interesting to further investigate the role by combining ICB-antibodies. These findings suggest that targeting intracellular checkpoints ‘CISH/SOCS-1 in DCs’ would hold the potential to treat several cancers even-in-combination with ICB-therapeutics [7,185] (Figure 8C,F and Table 1 and Table 3).
In addition, several researches support the improved efficacy of DC-immunotherapy when combined with ICB-antibodies. For example; Zhang et al., 2019 [186] demonstrated the role of PD-1 blockade in increasing anti-tumor activity of specific DCs called DC-stimulated cytokine-induced killer cells (DC-CIK) generated in presence of anti-CD3 antibody, IFNγ, poly-hydroxyalkanoates and IL-2; characterized by co-expression of CD56 and CD3 or CD3 and CD8 markers. The authors have shown that the adoptive transfer of pre-treated DC-CIK with PD-1 inhibitor (Pembrolizumab) block PD-1/PD-L1 axis and therefore increased its cytotoxic activity as compared to the null-DCs. Moreover, an increased infiltration of effector CD8+T-cells was noted in a nude mouse xenograft model with hepatocellular carcinoma (HCC), resulting in reduced tumor growth. This study suggests the improved efficacy of pre-treated ‘PD-1 inhibitor DC-CIK’ in controlling HCC recurrence [186]. Similarly, Lim et al., 2016 supported the above hypothesis and further emphasized the anti-tumor activity of PD-1−/− DCs in controlling HCC [187]. They showed that PD-1 expression on DCs reduces T-cell proliferation and suppresses CD8+T-cells effector function, resulting in decreased anti-tumor immunity. The adoptive transfer of PD-1−/− DCs increases CD8+T-cells infiltrations along with IFNγ, IL-2, perforin and GZMB secretions in the TME and thereby causing rapid tumor control. This result suggests the improved efficacy of PD-1−/− DCs in controlling HCC [187]. Next, Garzon-Muvdi et al., 2018 showed improved efficacy of anti-PD-1 when given in combination with DC-immunotherapy in controlling glioblastoma [16]. They showed that DC-activation through TLR3 agonist increases anti-tumor immunity in vitro. Moreover, TLR3 agonist poly (I:C)-injected mice showed increased DC-activation, antigen presentation and T-cell proliferation and thus enhancing the efficacy of ICB- therapy against glioblastoma [16].
Furthermore, Peng et al., 2020 [188] demonstrated the role of PD-L1 (ligand for PD-1) in the impairment of DCs. They showed that in response to antigenic exposure and IFN-II, type-I DCs (cDC1) increases the expression of PD-L1 and suppress CTL activity. Interestingly, the blocking of PD-L1 significantly improves DC-mediated T-cell infiltration and killing abilities in vitro. This result clearly suggests that PD-L1 expression is playing a crucial role in cDC1-impairment, and therefore targeting PD-L1 would hold the potential to enhance therapeutic benefits [188]. Similarly, Go et al., 2021 underlined the role of PD-L1-expressing DCs in reducing helicobacter-induced gastritis [189]. Go and colleagues showed that the treatment of anti-PD-L1 or PD-L1−/− in bone marrow transplantation enhances gastritis. Upon a closer look, the loss of Ftl3 (Flt3−/−) or Zbtb46-diphtheria toxin receptor (DTR) mice showed decreased DC-abundances causing severe mucosal metaplasia, and suggesting the protective role of PD-L1 expressing DCs in controlling gastritis [189]. Furthermore, miRNA-200b and miRNA-152 have been found to be downregulated in HP-induced gastric cancer tissues, suggesting a negative correlation of miRNA-200b and miRNA-152 in B7-H1 (PD-1) expression. Therefore, targeting miRNA (miRNA-restoration or miRNA-mimics) would have therapeutic benefit against gastric cancer [57] (Table 1). Collectively, these investigations suggest the potential therapeutic benefit of targeting intracellular immune checkpoint ‘CISH/SOCS-1’ in enhancing the efficacy of ICB-therapy if combined with DC-immunotherapy (Table 3, Figure 8C,F).
5. MicroRNAs and Epigenetic Modifiers (DNA and Histone Proteins) in ICB-Therapy
5.1. MicroRNAs in ICB-Therapeutics
In addition to co-targeting epitranscriptomics, intracellular immune checkpoints (m6A-modifiers, CISH/SOCS-1) and microRNA can be another potential targets to overcome the issues associated with ICB-drug resistances and high-dose antibody toxicities. The significance of ‘microRNAs’ also cannot be ignored because of their versatile roles in regulating numerous genes associated with immune checkpoint inhibitors. Kumar and colleagues have extensively described the therapeutic potential of microRNAs in treating DC-mediated Th1/Th2-associated immune disorders [41,192]. However, this section highlights some of the recent advancements in the utilization of ‘microRNAs’ in improving the efficacy of ICB-therapeutics. For example, miR-21, miR-34, miR-146a, miR-155 including many others miRNAs (Table 3) [193,194] have shown astonishing results by targeting PD-1, PD-L1 and CTLA4 immune markers summarized well in these references [132,133].
5.2. Epigenetic Modifiers (DNA and Histone Proteins) in ICB-Therapeutics
Like microRNAs, other epigenetic modifiers such as DNA and histone modifiers also hold great potential to increase the efficacy of ICB-therapeutics. Therefore, this section summarizes in brief about the systemic utilization of DNA-modifiers alone or in combination with other immunotherapeutic procedures. The DNA modification machineries, also known as writers/editors: DNMTs; removers/erasers: TET-proteins; readers/effectors: histone proteins HATs (acetylases) & HDACs (de-acetylases) [195] and chromatin remodelers: SWI/SNF chromatin remodelling complexes [196,197] have significant role in modulating the genes associated with immune checkpoint markers. Collectively, section-5 nurtures the potential of DNA-epigenetic modifiers and microRNAs in developing efficient molecular medicines in resolving the issues associated with ICB-drug resistance and toxicities [198,199,200,201] (Table 3).
6. Biopharmaceutical Companies Developing Personalized Medicines: Targeting Intracellular Checkpoint ‘CISH’ in Combination with ICB-Therapeutics and Recent Clinical Trials
In this section, we have described some of the biopharmaceutical/cell-therapy companies entering into developing personalized medicines by targeting immune cells expressing ‘intracellular checkpoint CISH’ in combination with ICB-antibodies to overcome the issues associated with ICB drug-resistance and high-dose antibody toxicities in several caners.
6.1. ONK Therapeutics Limited
ONK therapeutics is an Ireland-based cell therapy company, founded in 2015, conducting phase-I clinical trial against multiple myeloma, NSCLC and AML by targeting intracellular checkpoint ‘CISH’ in NK-cells. It was disclosed that the deletion of CISH improves the cytotoxic activity of NK-cells and therefore can be used efficiently to enhance the efficacy in combination with ICB-therapeutics. The respective phase-I clinical trials (ONKT102, ONKT103 and ONKT104) are estimated to complete until 2021𠄲2022 (https://www.onktherapeutics.com/pipeline; accessed on: 25 July 2021). A relevant patent (US10034925B2 and EP3434762A1) was also filed for securing global license to use CISH knockout NK-cells from Australia’s WEHI” on 28 May 2021 (www.onktherapeutics.com; accessed on: 25 July 2021) [135].
6.2. Intima Bioscience, Inc.
Intima Bioscience is a UK-based Biotechnology Company, founded in 2021, conducting phase-I/II clinical trials (NCT04426669) against metastatic gastrointestinal (GIT) cancer patients by administering CISH-inactivated TILs by CRISPR/Cas9 system [1]. It is estimated to complete the trial by 31 October 2022 in collaboration with Masonic Cancer Center, University of Minnesota, USA [5].
In addition to the above companies some other biopharmaceutical companies are also involved in encouraging personalized medicines by targeting immune cells are: AstraZeneca, Acepodia, Affimed, Avid Biotics, Bristol-Myers Squibb, Celgene, Cellular Therapeutics, Celularity, Crispr Therapeutics, Dragonfly Therapeutics, Effector Therapeutics, Fate Therapeutics Inc., Fortress Biotech Inc., Genentech, Glycostem Therapeutics, Green Cross Lab Cell Korea, Gamida Cell, GT Biopharma, ImmuneOncia therapeutics, Korea [127,128], Innate Pharma, ImmunityBio, Inc. (NCT03387085), Intima Bioscience, Inc. (NCT04426669 and NCT03538613), Juno Therapeutics Inc., Kyowa Hakko Kirin, Kiadis Pharma, Mentrik Biotech, Multimmune GmbH, NantKwest Inc., Nektar Therapeutics, Nkarta Therapeutics, NOXXON Pharma, Northwest Biotherapeutics, ONK therapeutics, Roche Glycart, Rubius Therapeutics, Sanofi, Senti Biosciences, SignalRX Pharmaceuticals Inc., Sorrento Therapeutics Inc., XNK Therapeutics in collaboration with Sanofi’s and NextGenNK competence center coordinated by Karolinska institute conducting (EudraCT No: 2010-0223330-83 phase-I/II and NCT04558853) clinical trial, and Ziopharm Oncology Inc. [83,139,142,143,175], (Table 1 and Table 3).
Box 1. Immune cells targeting intracellular checkpoint ‘CISH/SOCS-1’ in improving ICB-efficacy.
-
■
NK-cells targeting intracellular checkpoint CISH: ONK therapeutics, Ireland, estimated to conduct Phase-I clinical trial (ONKT102, ONKT103 and ONKT104) by 2021𠄲2022 for the treatment of haematological malignancies (multiple myeloma and AML) and solid tumors (ovarian, NSCLC and breast cancers) [135].
-
■
TILs targeting intracellular checkpoint CISH: Intima Bioscience, UK, estimated to conduct Phase-I/II clinical trial (NCT04426669, NCT03538613) by 2021𠄲2022 for the treatment of wide range of tumor types and gastrointestinal cancer [1,2,5].
-
■
DCs targeting intracellular checkpoint SOCS-1: Military medical sciences, China, conducted Phase-I clinical trial (NCT01956630) in 2018 for the treatment of RAL [7,182].
Box 2. Important links targeting intracellular checkpoint ‘CISH’ in developing personalized medicine (accessed on: 28 July 2021).
https://clinicaltrials.gov/ct2/show/NCT04426669
https://www.onktherapeutics.com/pipeline/
https://acir.org/weekly-digests/2020/october/a-new-internal-t-cell-checkpoint-cish
7. Conclusions
In this review, we have summarized the strategies to improve the efficacy of immune checkpoint blockade therapy by combining personalized medicines. In our opinion, we have put forwarded the strategies that are worth-considering regarding the importance of epitranscriptomics, in improving the efficacy of ICB-therapy. Moreover, combining immunotherapy by targeting intracellular immune checkpoints ‘CISH/SOCS-1’ in NK-cells, TILs and DCs would further potentiates the efficacy of ICB-therapy. We anticipate our investigation would boost clinicians and researchers in further strengthening the efficacy of ICB-antibodies by considering the significance of personalized medicines towards solving the issues largely associated with high-dose antibody toxicity and drug-resistance. Further investigation is warranted targeting CISH in DCs to check its immunotherapeutic competency in controlling Th1/Th2-associated immune disorders.
8. Future Prospective
In addition to NK-cells and T-cells; clinical trial co-targeting CISH-expressing DCs in combination with ICB-antibodies has not been scheduled yet providing an opportunity into exploring the unattended avenues of CISH as a new intracellular checkpoint. Several evidences (epitranscriptomics [202,203,204], microRNAs or CRISPR therapeutics [184,191]) certainly signpost the potential of targeting CISH-expressing DCs not only to overcome the issues associated with high-dose antibody toxicities and drug-resistances but also holds the potential to improve body‘s own defence mechanism by enhancing cellular immunity [14,16] as well as humoral immunities [205].
Acknowledgments
Thanks to all the lab members for their support.
Abbreviations
| AIA | Ag-induced arthritis |
| ALKBH5 | alpha-ketoglutarate-dependent hydroxylase |
| CAND1 | Cullin-associated NEDD8-dissociated protein 1 |
| EZH2 | Enhancer of zeste 2 polycomb repressive complex-2 subunit |
| EBF-1 | Early B-cell factor-1 |
| FTO | Fat Mass and Obesity Associated Protein |
| MPM | Malignant pleural mesothelioma |
| Spred2 | Sprouty related EVH1 domain containing protein-2 |
Author Contributions
Conceptualization, S.K.; formal analysis, S.K.; investigation, S.K.; data curation, S.K., P.S., V.K. and I.M.; writing-original draft preparation, S.K.; writing-review and editing, S.K., Y.-S.B. and M.U.A.; visualization, Y.-S.B., S.K., M.U.A. and M.-H.K.; supervision, Y.-S.B. and S.K.; project administration, Y.-S.B. and S.K.; funding acquisition, S.K. and Y.-S.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the grant of Individual Basic Science and Engineering Research Program (2018R1D1A1B07048567) funded by the Korean National Research Foundation. This work was also supported by a National Research Foundation (NRF) grant funded by the Korea Ministry of Science and ICT (SRC-2017R1A5A1014560) and in part by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, Republic of Korea (HI16C1074).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Palmer D., Webber B., Patel Y., Johnson M., Kariya C., Lahr W., Parkhurst M., Gartner J., Prickett T., Lowery F., et al. 333 Targeting the apical intracellular checkpoint CISH unleashes T cell neoantigen reactivity and effector program. J. Immunol. Ther. Cancer. 2020;8:A204. doi: 10.1136/jitc-2020-SITC2020.0333. [DOI] [Google Scholar]
- 2.Plieth J. Crispr: Nice Valuation, but Where’s the Clinical Trial? [(accessed on 20 July 2021)]. Available online: https://www.evaluate.com/node/13152/pdf.
- 3.Delconte R.B., Kolesnik T.B., Dagley L.F., Rautela J., Shi W., Putz E.M., Stannard K., Zhang J.G., Teh C., Firth M., et al. CIS is a potent checkpoint in NK cell-mediated tumor immunity. Nat. Immunol. 2016;17:816–824. doi: 10.1038/ni.3470. [DOI] [PubMed] [Google Scholar]
- 4.Putz E.M., Guillerey C., Kos K., Stannard K., Miles K., Delconte R.B., Takeda K., Nicholson S.E., Huntington N.D., Smyth M.J. Targeting cytokine signaling checkpoint CIS activates NK cells to protect from tumor initiation and metastasis. Oncoimmunology. 2017;6:e1267892. doi: 10.1080/2162402X.2016.1267892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer D.C., Webber B.R., Patel Y., Johnson M.J., Kariya C.M., Lahr W.S., Parkhurst M.R., Gartner J.J., Prickett T.D., Lowery F.J., et al. Internal checkpoint regulates T cellneoantigen reactivity and susceptibility to PD1 blockade. bioRxiv. 2020 doi: 10.1101/2020.09.24.306571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmer D.C., Guittard G.C., Franco Z., Crompton J.G., Eil R.L., Patel S.J., Ji Y., Van Panhuys N., Klebanoff C.A., Sukumar M., et al. Cish actively silences TCR signaling in CD8+ T cells to maintain tumor tolerance. J. Exp. Med. 2015;212:2095–2113. doi: 10.1084/jem.20150304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D., Huang X.F., Hong B., Song X.T., Hu L., Jiang M., Zhang B., Ning H., Li Y., Xu C., et al. Efficacy of intracellular immune checkpoint-silenced DC vaccine. JCI Insight. 2018;3 doi: 10.1172/jci.insight.98368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L., Hui H., Agrawal K., Kang Y., Li N., Tang R., Yuan J., Rana T.M. m(6) A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J. 2020;39:e104514. doi: 10.15252/embj.2020104514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin H., Zhang X., Yang P., Zhang X., Peng Y., Li D., Yu Y., Wu Y., Wang Y., Zhang J., et al. RNA m6A methylation orchestrates cancer growth and metastasis via macrophage reprogramming. Nat. Commun. 2021;12:1394. doi: 10.1038/s41467-021-21514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi L., Wu G., Guo L., Zou X., Huang P. Comprehensive Analysis of the PD-L1 and Immune Infiltrates of m(6)A RNA Methylation Regulators in Head and Neck Squamous Cell Carcinoma. Mol. Ther. Nucleic Acids. 2020;21:299–314. doi: 10.1016/j.omtn.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang S., Wei J., Cui Y.H., Park G., Shah P., Deng Y., Aplin A.E., Lu Z., Hwang S., He C., et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 2019;10:2782. doi: 10.1038/s41467-019-10669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuruta N., Tsuchihashi K., Ohmura H., Yamaguchi K., Ito M., Ariyama H., Kusaba H., Akashi K., Baba E. RNA N6-methyladenosine demethylase FTO regulates PD-L1 expression in colon cancer cells. Biochem. Biophys. Res. Commun. 2020;530:235–239. doi: 10.1016/j.bbrc.2020.06.153. [DOI] [PubMed] [Google Scholar]
- 13.Li N., Kang Y., Wang L., Huff S., Tang R., Hui H., Agrawal K., Gonzalez G.M., Wang Y., Patel S.P., et al. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc. Natl. Acad. Sci. USA. 2020;117:20159–20170. doi: 10.1073/pnas.1918986117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han D., Liu J., Chen C., Dong L., Liu Y., Chang R., Huang X., Liu Y., Wang J., Dougherty U., et al. Anti-tumour immunity controlled through mRNA m(6)A methylation and YTHDF1 in dendritic cells. Nature. 2019;566:270–274. doi: 10.1038/s41586-019-0916-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin X., Wang Z., Yang G., Wen G., Zhang H. YTHDF2 correlates with tumor immune infiltrates in lower-grade glioma. Aging (Albany NY) 2020;12:18476–18500. doi: 10.18632/aging.103812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garzon-Muvdi T., Theodros D., Luksik A.S., Maxwell R., Kim E., Jackson C.M., Belcaid Z., Ganguly S., Tyler B., Brem H., et al. Dendritic cell activation enhances anti-PD-1 mediated immunotherapy against glioblastoma. Oncotarget. 2018;9:20681–20697. doi: 10.18632/oncotarget.25061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Vos L., Grunwald I., Bawden E.G., Dietrich J., Scheckenbach K., Wiek C., Zarbl R., Bootz F., Landsberg J., Dietrich D. The landscape of CD28, CD80, CD86, CTLA4, and ICOS DNA methylation in head and neck squamous cell carcinomas. Epigenetics. 2020;15:1195–1212. doi: 10.1080/15592294.2020.1754675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Micevic G., Thakral D., McGeary M., Bosenberg M.W. PD-L1 methylation regulates PD-L1 expression and is associated with melanoma survival. Pigment Cell Melanoma Res. 2019;32:435–440. doi: 10.1111/pcmr.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marwitz S., Scheufele S., Perner S., Reck M., Ammerpohl O., Goldmann T. Epigenetic modifications of the immune-checkpoint genes CTLA4 and PDCD1 in non-small cell lung cancer results in increased expression. Clin. Epigenet. 2017;9:51. doi: 10.1186/s13148-017-0354-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franzen A., Vogt T.J., Muller T., Dietrich J., Schrock A., Golletz C., Brossart P., Bootz F., Landsberg J., Kristiansen G., et al. PD-L1 (CD274) and PD-L2 (PDCD1LG2) promoter methylation is associated with HPV infection and transcriptional repression in head and neck squamous cell carcinomas. Oncotarget. 2018;9:641–650. doi: 10.18632/oncotarget.23080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goltz D., Gevensleben H., Dietrich J., Dietrich D. PD-L1 (CD274) promoter methylation predicts survival in colorectal cancer patients. Oncoimmunology. 2017;6:e1257454. doi: 10.1080/2162402X.2016.1257454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li F., Zhao X., Zhang Y., Shao P., Ma X., Paradee W.J., Liu C., Wang J., Xue H.H. TFH cells depend on Tcf1-intrinsic HDAC activity to suppress CTLA4 and guard B-cell help function. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2014562118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lienlaf M., Perez-Villarroel P., Knox T., Pabon M., Sahakian E., Powers J., Woan K.V., Lee C., Cheng F., Deng S., et al. Essential role of HDAC6 in the regulation of PD-L1 in melanoma. Mol. Oncol. 2016;10:735–750. doi: 10.1016/j.molonc.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Darvin P., Sasidharan Nair V., Elkord E. PD-L1 Expression in Human Breast Cancer Stem Cells Is Epigenetically Regulated through Posttranslational Histone Modifications. J. Oncol. 2019;2019:3958908. doi: 10.1155/2019/3958908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu C., Paschall A.V., Shi H., Savage N., Waller J.L., Sabbatini M.E., Oberlies N.H., Pearce C., Liu K. The MLL1-H3K4me3 Axis-Mediated PD-L1 Expression and Pancreatic Cancer Immune Evasion. J. Natl. Cancer Inst. 2017;109 doi: 10.1093/jnci/djw283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao G., Jin L.L., Liu C.Q., Wang Y.C., Meng Y.M., Zhou Z.G., Chen J., Yu X.J., Zhang Y.J., Xu J., et al. EZH2 negatively regulates PD-L1 expression in hepatocellular carcinoma. J. Immunother. Cancer. 2019;7:300. doi: 10.1186/s40425-019-0784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Llopiz D., Ruiz M., Villanueva L., Iglesias T., Silva L., Egea J., Lasarte J.J., Pivette P., Trochon-Joseph V., Vasseur B., et al. Enhanced anti-tumor efficacy of checkpoint inhibitors in combination with the histone deacetylase inhibitor Belinostat in a murine hepatocellular carcinoma model. Cancer Immunol. Immunother. 2019;68:379–393. doi: 10.1007/s00262-018-2283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X., Han S., Kang Y., Guo M., Hong S., Liu F., Fu S., Wang L., Wang Q.X. SAHA, an HDAC inhibitor, synergizes with tacrolimus to prevent murine cardiac allograft rejection. Cell. Mol. Immunol. 2012;9:390–398. doi: 10.1038/cmi.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H., Fu C., Du J., Wang H., He R., Yin X., Li H., Li X., Wang H., Li K., et al. Enhanced histone H3 acetylation of the PD-L1 promoter via the COP1/c-Jun/HDAC3 axis is required for PD-L1 expression in drug-resistant cancer cells. J. Exp. Clin. Cancer Res. 2020;39:29. doi: 10.1186/s13046-020-1536-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kao S.C., Cheng Y.Y., Williams M., Kirschner M.B., Madore J., Lum T., Sarun K.H., Linton A., McCaughan B., Klebe S., et al. Tumor Suppressor microRNAs Contribute to the Regulation of PD-L1 Expression in Malignant Pleural Mesothelioma. J. Thorac. Oncol. 2017;12:1421–1433. doi: 10.1016/j.jtho.2017.05.024. [DOI] [PubMed] [Google Scholar]
- 31.Audrito V., Serra S., Stingi A., Orso F., Gaudino F., Bologna C., Neri F., Garaffo G., Nassini R., Baroni G., et al. PD-L1 up-regulation in melanoma increases disease aggressiveness and is mediated through miR-17-5p. Oncotarget. 2017;8:15894–15911. doi: 10.18632/oncotarget.15213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong P., Xiong Y., Yu J., Chen L., Tao T., Yi S., Hanley S.J.B., Yue J., Watari H., Sakuragi N. Correction: Control of PD-L1 expression by miR-140/142/340/383 and oncogenic activation of the OCT4-miR-18a pathway in cervical cancer. Oncogene. 2019;38:3972. doi: 10.1038/s41388-019-0677-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu J., Chen L., Zou L., Yang P., Wu R., Mao Y., Zhou H., Li R., Wang K., Wang W., et al. MiR-20b, -21, and -130b inhibit PTEN expression resulting in B7-H1 over-expression in advanced colorectal cancer. Hum. Immunol. 2014;75:348–353. doi: 10.1016/j.humimm.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 34.Iliopoulos D., Kavousanaki M., Ioannou M., Boumpas D., Verginis P. The negative costimulatory molecule PD-1 modulates the balance between immunity and tolerance via miR-21. Eur. J. Immunol. 2011;41:1754–1763. doi: 10.1002/eji.201040646. [DOI] [PubMed] [Google Scholar]
- 35.Zheng X., Dong L., Wang K., Zou H., Zhao S., Wang Y., Wang G. MiR-21 Participates in the PD-1/PD-L1 Pathway-Mediated Imbalance of Th17/Treg Cells in Patients After Gastric Cancer Resection. Ann. Surg. Oncol. 2019;26:884–893. doi: 10.1245/s10434-018-07117-6. [DOI] [PubMed] [Google Scholar]
- 36.Liu J., Fan L., Yu H., Zhang J., He Y., Feng D., Wang F., Li X., Liu Q., Li Y., et al. Endoplasmic Reticulum Stress Causes Liver Cancer Cells to Release Exosomal miR-23a-3p and Up-regulate Programmed Death Ligand 1 Expression in Macrophages. Hepatology. 2019;70:241–258. doi: 10.1002/hep.30607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cioffi M., Trabulo S.M., Vallespinos M., Raj D., Kheir T.B., Lin M.L., Begum J., Baker A.M., Amgheib A., Saif J., et al. The miR-25-93-106b cluster regulates tumor metastasis and immune evasion via modulation of CXCL12 and PD-L1. Oncotarget. 2017;8:21609–21625. doi: 10.18632/oncotarget.15450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Q., Johnston N., Zheng X., Wang H., Zhang X., Gao D., Min W. miR-28 modulates exhaustive differentiation of T cells through silencing programmed cell death-1 and regulating cytokine secretion. Oncotarget. 2016;7:53735–53750. doi: 10.18632/oncotarget.10731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boldrini L., Giordano M., Niccoli C., Melfi F., Lucchi M., Mussi A., Fontanini G. Role of microRNA-33a in regulating the expression of PD-1 in lung adenocarcinoma. Cancer Cell. Int. 2017;17:105. doi: 10.1186/s12935-017-0474-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anastasiadou E., Stroopinsky D., Alimperti S., Jiao A.L., Pyzer A.R., Cippitelli C., Pepe G., Severa M., Rosenblatt J., Etna M.P., et al. Epstein-Barr virus-encoded EBNA2 alters immune checkpoint PD-L1 expression by downregulating miR-34a in B-cell lymphomas. Leukemia. 2019;33:132–147. doi: 10.1038/s41375-018-0178-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar S., Ashraf M.U., Kumar A., Bae Y.S. Therapeutic Potential of microRNA Against Th2-associated Immune Disorders. Curr. Top. Med. Chem. 2021;21:753–766. doi: 10.2174/1568026621666210303150235. [DOI] [PubMed] [Google Scholar]
- 42.Wang X., Li J., Dong K., Lin F., Long M., Ouyang Y., Wei J., Chen X., Weng Y., He T., et al. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell. Signal. 2015;27:443–452. doi: 10.1016/j.cellsig.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 43.Boussiotis V.A. Molecular and Biochemical Aspects of the PD-1 Checkpoint Pathway. N. Engl. J. Med. 2016;375:1767–1778. doi: 10.1056/NEJMra1514296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cortez M.A., Ivan C., Valdecanas D., Wang X., Peltier H.J., Ye Y., Araujo L., Carbone D.P., Shilo K., Giri D.K., et al. PDL1 Regulation by p53 via miR-34. J. Natl. Cancer Inst. 2016;108 doi: 10.1093/jnci/djv303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao L., Yu H., Yi S., Peng X., Su P., Xiao Z., Liu R., Tang A., Li X., Liu F., et al. The tumor suppressor miR-138-5p targets PD-L1 in colorectal cancer. Oncotarget. 2016;7:45370–45384. doi: 10.18632/oncotarget.9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie W.B., Liang L.H., Wu K.G., Wang L.X., He X., Song C., Wang Y.Q., Li Y.H. MiR-140 Expression Regulates Cell Proliferation and Targets PD-L1 in NSCLC. Cell. Physiol. Biochem. 2018;46:654–663. doi: 10.1159/000488634. [DOI] [PubMed] [Google Scholar]
- 47.Jia L., Xi Q., Wang H., Zhang Z., Liu H., Cheng Y., Guo X., Zhang J., Zhang Q., Zhang L., et al. miR-142-5p regulates tumor cell PD-L1 expression and enhances anti-tumor immunity. Biochem. Biophys. Res. Commun. 2017;488:425–431. doi: 10.1016/j.bbrc.2017.05.074. [DOI] [PubMed] [Google Scholar]
- 48.Sheng Q., Zhang Y., Wang Z., Ding J., Song Y., Zhao W. Cisplatin-mediated down-regulation of miR-145 contributes to up-regulation of PD-L1 via the c-Myc transcription factor in cisplatin-resistant ovarian carcinoma cells. Clin. Exp. Immunol. 2020;200:45–52. doi: 10.1111/cei.13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mastroianni J., Stickel N., Andrlova H., Hanke K., Melchinger W., Duquesne S., Schmidt D., Falk M., Andrieux G., Pfeifer D., et al. miR-146a Controls Immune Response in the Melanoma Microenvironment. Cancer Res. 2019;79:183–195. doi: 10.1158/0008-5472.CAN-18-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashizawa M., Okayama H., Ishigame T., Thar Min A.K., Saito K., Ujiie D., Murakami Y., Kikuchi T., Nakayama Y., Noda M., et al. miRNA-148a-3p Regulates Immunosuppression in DNA Mismatch Repair-Deficient Colorectal Cancer by Targeting PD-L1. Mol. Cancer Res. 2019;17:1403–1413. doi: 10.1158/1541-7786.MCR-18-0831. [DOI] [PubMed] [Google Scholar]
- 51.Zheng Z., Sun R., Zhao H.J., Fu D., Zhong H.J., Weng X.Q., Qu B., Zhao Y., Wang L., Zhao W.L. MiR155 sensitized B-lymphoma cells to anti-PD-L1 antibody via PD-1/PD-L1-mediated lymphoma cell interaction with CD8+T cells. Mol. Cancer. 2019;18:54. doi: 10.1186/s12943-019-0977-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X.Y., Zhang J., Hou L.D., Zhang R., Chen W., Fan H.N., Huang Y.X., Liu H., Zhu J.S. Upregulation of PD-L1 predicts poor prognosis and is associated with miR-191-5p dysregulation in colon adenocarcinoma. Int. J. Immunopathol. Pharmacol. 2018;32:2058738418790318. doi: 10.1177/2058738418790318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tao Z., Xu S., Ruan H., Wang T., Song W., Qian L., Chen K. MiR-195/-16 Family Enhances Radiotherapy via T Cell Activation in the Tumor Microenvironment by Blocking the PD-L1 Immune Checkpoint. Cell. Physiol. Biochem. 2018;48:801–814. doi: 10.1159/000491909. [DOI] [PubMed] [Google Scholar]
- 54.He B., Yan F., Wu C. Overexpressed miR-195 attenuated immune escape of diffuse large B-cell lymphoma by targeting PD-L1. Biomed. Pharmacother. 2018;98:95–101. doi: 10.1016/j.biopha.2017.11.146. [DOI] [PubMed] [Google Scholar]
- 55.Fujita Y., Yagishita S., Hagiwara K., Yoshioka Y., Kosaka N., Takeshita F., Fujiwara T., Tsuta K., Nokihara H., Tamura T., et al. The clinical relevance of the miR-197/CKS1B/STAT3-mediated PD-L1 network in chemoresistant non-small-cell lung cancer. Mol. Ther. 2015;23:717–727. doi: 10.1038/mt.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen L., Gibbons D.L., Goswami S., Cortez M.A., Ahn Y.H., Byers L.A., Zhang X., Yi X., Dwyer D., Lin W., et al. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie G., Li W., Li R., Wu K., Zhao E., Zhang Y., Zhang P., Shi L., Wang D., Yin Y., et al. Helicobacter Pylori Promote B7-H1 Expression by Suppressing miR-152 and miR-200b in Gastric Cancer Cells. PLoS ONE. 2017;12:e0168822. doi: 10.1371/journal.pone.0168822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun J.R., Zhang X., Zhang Y. MiR-214 prevents the progression of diffuse large B-cell lymphoma by targeting PD-L1. Cell Mol Biol Lett. 2019;24:68. doi: 10.1186/s11658-019-0190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miao S., Mao X., Zhao S., Song K., Xiang C., Lv Y., Jiang H., Wang L., Li B., Yang X., et al. miR-217 inhibits laryngeal cancer metastasis by repressing AEG-1 and PD-L1 expression. Oncotarget. 2017;8:62143–62153. doi: 10.18632/oncotarget.19121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holla S., Stephen-Victor E., Prakhar P., Sharma M., Saha C., Udupa V., Kaveri S.V., Bayry J., Balaji K.N. Mycobacteria-responsive sonic hedgehog signaling mediates programmed death-ligand 1- and prostaglandin E2-induced regulatory T cell expansion. Sci. Rep. 2016;6:24193. doi: 10.1038/srep24193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong P., Xiong Y., Yu J., Chen L., Tao T., Yi S., Hanley S.J.B., Yue J., Watari H., Sakuragi N. Control of PD-L1 expression by miR-140/142/340/383 and oncogenic activation of the OCT4-miR-18a pathway in cervical cancer. Oncogene. 2018;37:5257–5268. doi: 10.1038/s41388-018-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Q., Zhao Y., Sun Y., Yan X., Wang P. miR-375 inhibits IFN-gamma-induced programmed death 1 ligand 1 surface expression in head and neck squamous cell carcinoma cells by blocking JAK2/STAT1 signaling. Oncol. Rep. 2018;39:1461–1468. doi: 10.3892/or.2018.6177. [DOI] [PubMed] [Google Scholar]
- 63.Xu S., Tao Z., Hai B., Liang H., Shi Y., Wang T., Song W., Chen Y., OuYang J., Chen J., et al. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat. Commun. 2016;7:11406. doi: 10.1038/ncomms11406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao X., Yuan C., Wangmo D., Subramanian S. Tumor-Secreted Extracellular Vesicles Regulate T-Cell Costimulation and Can Be Manipulated To Induce Tumor-Specific T-Cell Responses. Gastroenterology. 2021;161:560–574. doi: 10.1053/j.gastro.2021.04.036. [DOI] [PubMed] [Google Scholar]
- 65.Qu F., Ye J., Pan X., Wang J., Gan S., Chu C., Chu J., Zhang X., Liu M., He H., et al. MicroRNA-497-5p down-regulation increases PD-L1 expression in clear cell renal cell carcinoma. J. Drug Target. 2019;27:67–74. doi: 10.1080/1061186X.2018.1479755. [DOI] [PubMed] [Google Scholar]
- 66.Gong A.Y., Zhou R., Hu G., Liu J., Sosnowska D., Drescher K.M., Dong H., Chen X.M. Cryptosporidium parvum induces B7-H1 expression in cholangiocytes by down-regulating microRNA-513. J. Infect. Dis. 2010;201:160–169. doi: 10.1086/648589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang W., Li F., Mao Y., Zhou H., Sun J., Li R., Liu C., Chen W., Hua D., Zhang X. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum. Genet. 2013;132:641–648. doi: 10.1007/s00439-013-1275-6. [DOI] [PubMed] [Google Scholar]
- 68.Gao L., Guo Q., Li X., Yang X., Ni H., Wang T., Zhao Q., Liu H., Xing Y., Xi T., et al. MiR-873/PD-L1 axis regulates the stemness of breast cancer cells. EBioMedicine. 2019;41:395–407. doi: 10.1016/j.ebiom.2019.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang D., Zhao D., Wu Y., Yao R., Zhou L., Lu L., Gao W., Sun Y. The miR-3127-5p/p-STAT3 axis up-regulates PD-L1 inducing chemoresistance in non-small-cell lung cancer. J. Cell. Mol. Med. 2018 doi: 10.1111/jcmm.13657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li D., Wang X., Yang M., Kan Q., Duan Z. miR3609 sensitizes breast cancer cells to adriamycin by blocking the programmed death-ligand 1 immune checkpoint. Exp. Cell. Res. 2019;380:20–28. doi: 10.1016/j.yexcr.2019.03.025. [DOI] [PubMed] [Google Scholar]
- 71.Zhang G., Li N., Li Z., Zhu Q., Li F., Yang C., Han Q., Lv Y., Zhou Z., Liu Z. microRNA-4717 differentially interacts with its polymorphic target in the PD1 3′ untranslated region: A mechanism for regulating PD-1 expression and function in HBV-associated liver diseases. Oncotarget. 2015;6:18933–18944. doi: 10.18632/oncotarget.3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang D.Y., Johnson D.B., Davis E.J. Toxicities Associated With PD-1/PD-L1 Blockade. Cancer J. 2018;24:36–40. doi: 10.1097/PPO.0000000000000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jenkins R.W., Barbie D.A., Flaherty K.T. Mechanisms of resistance to immune checkpoint inhibitors. Br. J. Cancer. 2018;118:9–16. doi: 10.1038/bjc.2017.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fares C.M., Van Allen E.M., Drake C.G., Allison J.P., Hu-Lieskovan S. Mechanisms of Resistance to Immune Checkpoint Blockade: Why Does Checkpoint Inhibitor Immunotherapy Not Work for All Patients? Am. Soc. Clin. Oncol. Educ. Book. 2019;39:147–164. doi: 10.1200/EDBK_240837. [DOI] [PubMed] [Google Scholar]
- 75.Kalbasi A., Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 2020;20:25–39. doi: 10.1038/s41577-019-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ishida Y., Agata Y., Shibahara K., Honjo T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992;11:3887–3895. doi: 10.1002/j.1460-2075.1992.tb05481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bashyam H. CTLA-4: From conflict to clinic. J. Exp. Med. 2007;204:1243. doi: 10.1084/jem.2046fta. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waterhouse P., Penninger J.M., Timms E., Wakeham A., Shahinian A., Lee K.P., Thompson C.B., Griesser H., Mak T.W. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science. 1995;270:985–988. doi: 10.1126/science.270.5238.985. [DOI] [PubMed] [Google Scholar]
- 79.Tivol E.A., Borriello F., Schweitzer A.N., Lynch W.P., Bluestone J.A., Sharpe A.H. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity. 1995;3:541–547. doi: 10.1016/1074-7613(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 80.Dong H., Zhu G., Tamada K., Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat. Med. 1999;5:1365–1369. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- 81.Latchman Y., Wood C.R., Chernova T., Chaudhary D., Borde M., Chernova I., Iwai Y., Long A.J., Brown J.A., Nunes R., et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat. Immunol. 2001;2:261–268. doi: 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- 82.De Sousa Linhares A., Battin C., Jutz S., Leitner J., Hafner C., Tobias J., Wiedermann U., Kundi M., Zlabinger G.J., Grabmeier-Pfistershammer K., et al. Therapeutic PD-L1 antibodies are more effective than PD-1 antibodies in blocking PD-1/PD-L1 signaling. Sci. Rep. 2019;9:11472. doi: 10.1038/s41598-019-47910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lucibello G., Mograbi B., Milano G., Hofman P., Brest P. PD-L1 regulation revisited: Impact on immunotherapeutic strategies. Trends. Mol. Med. 2021 doi: 10.1016/j.molmed.2021.06.005. [DOI] [PubMed] [Google Scholar]
- 84.Borghaei H., Paz-Ares L., Horn L., Spigel D.R., Steins M., Ready N.E., Chow L.Q., Vokes E.E., Felip E., Holgado E., et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015;373:1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu Z., Man S., Sun R., Li Z., Wu Y., Zuo D. Recent advances and challenges of immune checkpoint inhibitors in immunotherapy of non-small cell lung cancer. Int. Immunopharmacol. 2020;85:106613. doi: 10.1016/j.intimp.2020.106613. [DOI] [PubMed] [Google Scholar]
- 86.Naing A., Infante J., Goel S., Burris H., Black C., Marshall S., Achour I., Barbee S., May R., Morehouse C., et al. Anti-PD-1 monoclonal antibody MEDI0680 in a phase I study of patients with advanced solid malignancies. J. Immunother. Cancer. 2019;7:225. doi: 10.1186/s40425-019-0665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Naing A., Gainor J.F., Gelderblom H., Forde P.M., Butler M.O., Lin C.C., Sharma S., Ochoa de Olza M., Varga A., Taylor M., et al. A first-in-human phase 1 dose escalation study of spartalizumab (PDR001), an anti-PD-1 antibody, in patients with advanced solid tumors. J. Immunother. Cancer. 2020;8 doi: 10.1136/jitc-2020-000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Garon E.B., Rizvi N.A., Hui R., Leighl N., Balmanoukian A.S., Eder J.P., Patnaik A., Aggarwal C., Gubens M., Horn L., et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 89.Galsky M.D., Wang H., Hahn N.M., Twardowski P., Pal S.K., Albany C., Fleming M.T., Starodub A., Hauke R.J., Yu M., et al. Phase 2 Trial of Gemcitabine, Cisplatin, plus Ipilimumab in Patients with Metastatic Urothelial Cancer and Impact of DNA Damage Response Gene Mutations on Outcomes. Eur. Urol. 2018;73:751–759. doi: 10.1016/j.eururo.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 90.Slovin S.F., Higano C.S., Hamid O., Tejwani S., Harzstark A., Alumkal J.J., Scher H.I., Chin K., Gagnier P., McHenry M.B., et al. Ipilimumab alone or in combination with radiotherapy in metastatic castration-resistant prostate cancer: Results from an open-label, multicenter phase I/II study. Ann. Oncol. 2013;24:1813–1821. doi: 10.1093/annonc/mdt107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Iwama S., De Remigis A., Callahan M.K., Slovin S.F., Wolchok J.D., Caturegli P. Pituitary expression of CTLA-4 mediates hypophysitis secondary to administration of CTLA-4 blocking antibody. Sci. Transl. Med. 2014;6:230ra245. doi: 10.1126/scitranslmed.3008002. [DOI] [PubMed] [Google Scholar]
- 92.Hodi F.S., O’Day S.J., McDermott D.F., Weber R.W., Sosman J.A., Haanen J.B., Gonzalez R., Robert C., Schadendorf D., Hassel J.C., et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tomillero A., Moral M.A. Gateways to clinical trials. Methods Find Exp. Clin. Pharmacol. 2008;30:643–672. doi: 10.1358/mf.2008.30.5.1236622. [DOI] [PubMed] [Google Scholar]
- 94.Poust J. Targeting metastatic melanoma. Am. J. Health Syst. Pharm. 2008;65:S9–S15. doi: 10.2146/ajhp080461. [DOI] [PubMed] [Google Scholar]
- 95.Reuben J.M., Lee B.N., Li C., Gomez-Navarro J., Bozon V.A., Parker C.A., Hernandez I.M., Gutierrez C., Lopez-Berestein G., Camacho L.H. Biologic and immunomodulatory events after CTLA-4 blockade with ticilimumab in patients with advanced malignant melanoma. Cancer. 2006;106:2437–2444. doi: 10.1002/cncr.21854. [DOI] [PubMed] [Google Scholar]
- 96.Senan S., Okamoto I., Lee G.W., Chen Y., Niho S., Mak G., Yao W., Shire N., Jiang H., Cho B.C. Design and Rationale for a Phase III, Randomized, Placebo-controlled Trial of Durvalumab With or Without Tremelimumab After Concurrent Chemoradiotherapy for Patients With Limited-stage Small-cell Lung Cancer: The ADRIATIC Study. Clin. Lung. Cancer. 2020;21:e84–e88. doi: 10.1016/j.cllc.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 97.Ribas A., Kefford R., Marshall M.A., Punt C.J., Haanen J.B., Marmol M., Garbe C., Gogas H., Schachter J., Linette G., et al. Phase III randomized clinical trial comparing tremelimumab with standard-of-care chemotherapy in patients with advanced melanoma. J. Clin. Oncol. 2013;31:616–622. doi: 10.1200/JCO.2012.44.6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Powles T., Park S.H., Voog E., Caserta C., Valderrama B.P., Gurney H., Kalofonos H., Radulovic S., Demey W., Ullen A., et al. Avelumab Maintenance Therapy for Advanced or Metastatic Urothelial Carcinoma. N. Engl. J. Med. 2020;383:1218–1230. doi: 10.1056/NEJMoa2002788. [DOI] [PubMed] [Google Scholar]
- 99.Paz-Ares L., Dvorkin M., Chen Y., Reinmuth N., Hotta K., Trukhin D., Statsenko G., Hochmair M.J., Ozguroglu M., Ji J.H., et al. Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): A randomised, controlled, open-label, phase 3 trial. Lancet. 2019;394:1929–1939. doi: 10.1016/S0140-6736(19)32222-6. [DOI] [PubMed] [Google Scholar]
- 100.Antonia S.J., Villegas A., Daniel D., Vicente D., Murakami S., Hui R., Yokoi T., Chiappori A., Lee K.H., de Wit M., et al. Durvalumab after Chemoradiotherapy in Stage III Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017;377:1919–1929. doi: 10.1056/NEJMoa1709937. [DOI] [PubMed] [Google Scholar]
- 101.Antonia S.J., Villegas A., Daniel D., Vicente D., Murakami S., Hui R., Kurata T., Chiappori A., Lee K.H., de Wit M., et al. Overall Survival with Durvalumab after Chemoradiotherapy in Stage III NSCLC. N. Engl. J. Med. 2018;379:2342–2350. doi: 10.1056/NEJMoa1809697. [DOI] [PubMed] [Google Scholar]
- 102.Gay C.L., Bosch R.J., Ritz J., Hataye J.M., Aga E., Tressler R.L., Mason S.W., Hwang C.K., Grasela D.M., Ray N., et al. Clinical Trial of the Anti-PD-L1 Antibody BMS-936559 in HIV-1 Infected Participants on Suppressive Antiretroviral Therapy. J. Infect. Dis. 2017;215:1725–1733. doi: 10.1093/infdis/jix191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hotchkiss R.S., Colston E., Yende S., Angus D.C., Moldawer L.L., Crouser E.D., Martin G.S., Coopersmith C.M., Brakenridge S., Mayr F.B., et al. Immune Checkpoint Inhibition in Sepsis: A Phase 1b Randomized, Placebo-Controlled, Single Ascending Dose Study of Antiprogrammed Cell Death-Ligand 1 Antibody (BMS-936559) Crit. Care Med. 2019;47:632–642. doi: 10.1097/CCM.0000000000003685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Corrales L., Scilla K., Caglevic C., Miller K., Oliveira J., Rolfo C. Immunotherapy in Lung Cancer: A New Age in Cancer Treatment. Adv. Exp. Med. Biol. 2018;995:65–95. doi: 10.1007/978-3-030-02505-2_3. [DOI] [PubMed] [Google Scholar]
- 105.Floudas C.S., Brar G., Mabry-Hrones D., Duffy A.G., Wood B., Levy E., Krishnasamy V., Fioravanti S., Bonilla C.M., Walker M., et al. A Pilot Study of the PD-1 Targeting Agent AMP-224 Used With Low-Dose Cyclophosphamide and Stereotactic Body Radiation Therapy in Patients With Metastatic Colorectal Cancer. Clin. Colorectal. Cancer. 2019;18:e349–e360. doi: 10.1016/j.clcc.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nishimura H., Okazaki T., Tanaka Y., Nakatani K., Hara M., Matsumori A., Sasayama S., Mizoguchi A., Hiai H., Minato N., et al. Autoimmune dilated cardiomyopathy in PD-1 receptor-deficient mice. Science. 2001;291:319–322. doi: 10.1126/science.291.5502.319. [DOI] [PubMed] [Google Scholar]
- 107.Nishimura H., Nose M., Hiai H., Minato N., Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11:141–151. doi: 10.1016/S1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- 108.Okazaki T., Tanaka Y., Nishio R., Mitsuiye T., Mizoguchi A., Wang J., Ishida M., Hiai H., Matsumori A., Minato N., et al. Autoantibodies against cardiac troponin I are responsible for dilated cardiomyopathy in PD-1-deficient mice. Nat. Med. 2003;9:1477–1483. doi: 10.1038/nm955. [DOI] [PubMed] [Google Scholar]
- 109.Wang J., Okazaki I.M., Yoshida T., Chikuma S., Kato Y., Nakaki F., Hiai H., Honjo T., Okazaki T. PD-1 deficiency results in the development of fatal myocarditis in MRL mice. Int. Immunol. 2010;22:443–452. doi: 10.1093/intimm/dxq026. [DOI] [PubMed] [Google Scholar]
- 110.Wang J., Yoshida T., Nakaki F., Hiai H., Okazaki T., Honjo T. Establishment of NOD-Pdcd1-/- mice as an efficient animal model of type I diabetes. Proc. Natl. Acad. Sci. USA. 2005;102:11823–11828. doi: 10.1073/pnas.0505497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yoshida T., Jiang F., Honjo T., Okazaki T. PD-1 deficiency reveals various tissue-specific autoimmunity by H-2b and dose-dependent requirement of H-2g7 for diabetes in NOD mice. Proc. Natl. Acad. Sci. USA. 2008;105:3533–3538. doi: 10.1073/pnas.0710951105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Okazaki T., Otaka Y., Wang J., Hiai H., Takai T., Ravetch J.V., Honjo T. Hydronephrosis associated with antiurothelial and antinuclear autoantibodies in BALB/c-Fcgr2b-/-Pdcd1-/- mice. J. Exp. Med. 2005;202:1643–1648. doi: 10.1084/jem.20051984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Grosso J.F., Jure-Kunkel M.N. CTLA-4 blockade in tumor models: An overview of preclinical and translational research. Cancer Immunol. 2013;13:5. [PMC free article] [PubMed] [Google Scholar]
- 114.Leach D.R., Krummel M.F., Allison J.P. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271:1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 115.Hodi F.S., Mihm M.C., Soiffer R.J., Haluska F.G., Butler M., Seiden M.V., Davis T., Henry-Spires R., MacRae S., Willman A., et al. Biologic activity of cytotoxic T lymphocyte-associated antigen 4 antibody blockade in previously vaccinated metastatic melanoma and ovarian carcinoma patients. Proc. Natl. Acad. Sci. USA. 2003;100:4712–4717. doi: 10.1073/pnas.0830997100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Phan G.Q., Yang J.C., Sherry R.M., Hwu P., Topalian S.L., Schwartzentruber D.J., Restifo N.P., Haworth L.R., Seipp C.A., Freezer L.J., et al. Cancer regression and autoimmunity induced by cytotoxic T lymphocyte-associated antigen 4 blockade in patients with metastatic melanoma. Proc. Natl. Acad. Sci. USA. 2003;100:8372–8377. doi: 10.1073/pnas.1533209100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sharma P., Allison J.P. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 118.Rosenberg S.A., Restifo N.P. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62–68. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pardoll D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Iwai Y., Hamanishi J., Chamoto K., Honjo T. Cancer immunotherapies targeting the PD-1 signaling pathway. J. Biomed. Sci. 2017;24:26. doi: 10.1186/s12929-017-0329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chiossone L., Dumas P.Y., Vienne M., Vivier E. Natural killer cells and other innate lymphoid cells in cancer. Nat. Rev. Immunol. 2018;18:671–688. doi: 10.1038/s41577-018-0061-z. [DOI] [PubMed] [Google Scholar]
- 122.Waldman A.D., Fritz J.M., Lenardo M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020;20:651–668. doi: 10.1038/s41577-020-0306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wei S.C., Duffy C.R., Allison J.P. Fundamental Mechanisms of Immune Checkpoint Blockade Therapy. Cancer Discov. 2018;8:1069–1086. doi: 10.1158/2159-8290.CD-18-0367. [DOI] [PubMed] [Google Scholar]
- 124.Barrueto L., Caminero F., Cash L., Makris C., Lamichhane P., Deshmukh R.R. Resistance to Checkpoint Inhibition in Cancer Immunotherapy. Transl. Oncol. 2020;13:100738. doi: 10.1016/j.tranon.2019.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kumar S., Nagpal R., Kumar A., Ashraf M.U., Bae Y.S. Immunotherapeutic Potential of m6A-Modifiers and MicroRNAs in Controlling Acute Myeloid Leukaemia. Biomedicines. 2021;9:690. doi: 10.3390/biomedicines9060690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell. Biol. 2017;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gordon S.R., Maute R.L., Dulken B.W., Hutter G., George B.M., McCracken M.N., Gupta R., Tsai J.M., Sinha R., Corey D., et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature. 2017;545:495–499. doi: 10.1038/nature22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cheong K.H. Novel Immunotherapies to Combine with PD-1/PD-L1 Treatment. [(accessed on 25 July 2021)]. Available online: https://media.nature.com/original/magazine-assets/d43747-020-00338-3/d43747-020-00338-3.pdf.
- 129.Dovedi S.J., Elder M.J., Yang C., Sitnikova S.I., Irving L., Hansen A., Hair J., Jones D.C., Hasani S., Wang B., et al. Design and Efficacy of a Monovalent Bispecific PD-1/CTLA4 Antibody That Enhances CTLA4 Blockade on PD-1(+) Activated T Cells. Cancer Discov. 2021;11:1100–1117. doi: 10.1158/2159-8290.CD-20-1445. [DOI] [PubMed] [Google Scholar]
- 130.Kakimi K., Karasaki T., Matsushita H., Sugie T. Advances in personalized cancer immunotherapy. Breast Cancer. 2017;24:16–24. doi: 10.1007/s12282-016-0688-1. [DOI] [PubMed] [Google Scholar]
- 131.Sahin U., Tureci O. Personalized vaccines for cancer immunotherapy. Science. 2018;359:1355–1360. doi: 10.1126/science.aar7112. [DOI] [PubMed] [Google Scholar]
- 132.Zhang H., Dai Z., Wu W., Wang Z., Zhang N., Zhang L., Zeng W.J., Liu Z., Cheng Q. Regulatory mechanisms of immune checkpoints PD-L1 and CTLA-4 in cancer. J. Exp. Clin. Cancer Res. 2021;40:184. doi: 10.1186/s13046-021-01987-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Han Y., Liu D., Li L. PD-1/PD-L1 pathway: Current researches in cancer. Am. J. Cancer Res. 2020;10:727–742. [PMC free article] [PubMed] [Google Scholar]
- 134.Catela Ivkovic T., Voss G., Cornella H., Ceder Y. microRNAs as cancer therapeutics: A step closer to clinical application. Cancer Lett. 2017;407:113–122. doi: 10.1016/j.canlet.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 135.Nowers C. Maximizing Synergy and Mitigating Resistance: Novel Dual-Targeted Natural Killer Cell Therapies for Cancer. [(accessed on 20 July 2021)]. Available online: https://www.onktherapeutics.com/wp/wp-content/uploads/2021/03/Nature-Biotech-Dealmakers-ONK-Therapeutics-March-2021.pdf.
- 136.Ryan B.S.M., Gaidarova S., Abujarour R., Clarke R., Stokely L., Rogers P., Ge M., Robinson M., Rezner B., Lee T.T., et al. Abstract 3576: FT500, an off-the-shelf NK cell cancer immunotherapy derived from a master pluripotent cell line, enhances T-cell activation and recruitment to overcome checkpoint blockade resistance. Cancer Res. Immunol. 2018 doi: 10.1158/1538-7445.AM2018-3576. [DOI] [Google Scholar]
- 137.Guo H., He Y., Chen P., Wang L., Li W., Chen B., Liu Y., Wang H., Zhao S., Zhou C. Combinational immunotherapy based on immune checkpoints inhibitors in small cell lung cancer: Is this the beginning to reverse the refractory situation? J. Thorac. Dis. 2020;12:6070–6089. doi: 10.21037/jtd-20-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wang F., Lau J.K.C., Yu J. The role of natural killer cell in gastrointestinal cancer: Killer or helper. Oncogene. 2021;40:717–730. doi: 10.1038/s41388-020-01561-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shin M.H., Kim J., Lim S.A., Kim J., Kim S.J., Lee K.M. NK Cell-Based Immunotherapies in Cancer. Immune Netw. 2020;20:e14. doi: 10.4110/in.2020.20.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Goodridge J.P., Mahmood S., Zhu H., Gaidarova S., Blum R., Bjordahl R., Cichocki F., Chu H.-y., Bonello G., Lee T., et al. FT596: Translation of First-of-Kind Multi-Antigen Targeted Off-the-Shelf CAR-NK Cell with Engineered Persistence for the Treatment of B Cell Malignancies. Blood. 2019;134:301. doi: 10.1182/blood-2019-129319. [DOI] [Google Scholar]
- 141.Bjordahl R., Gaidarova S., Woan K., Cichocki F., Bonello G., Robinson M., Ruller C., Pribadi M., Dinella J., Fong L., et al. FT538: Preclinical Development of an Off-the-Shelf Adoptive NK Cell Immunotherapy with Targeted Disruption of CD38 to Prevent Anti-CD38 Antibody-Mediated Fratricide and Enhance ADCC in Multiple Myeloma When Combined with Daratumumab. Blood. 2019;134:133. doi: 10.1182/blood-2019-131138. [DOI] [Google Scholar]
- 142.Veluchamy J.P., Kok N., van der Vliet H.J., Verheul H.M.W., de Gruijl T.D., Spanholtz J. The Rise of Allogeneic Natural Killer Cells As a Platform for Cancer Immunotherapy: Recent Innovations and Future Developments. Front. Immunol. 2017;8:631. doi: 10.3389/fimmu.2017.00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang C., Hu Y., Shi C. Targeting Natural Killer Cells for Tumor Immunotherapy. Front. Immunol. 2020;11:60. doi: 10.3389/fimmu.2020.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.U.S. National Library of Medicine. [(accessed on 7 July 2021)]; Available online: https://clinicaltrials.gov/
- 145.Barta S.K., Zain J., MacFarlane A.W.t., Smith S.M., Ruan J., Fung H.C., Tan C.R., Yang Y., Alpaugh R.K., Dulaimi E., et al. Phase II Study of the PD-1 Inhibitor Pembrolizumab for the Treatment of Relapsed or Refractory Mature T-cell Lymphoma. Clin. Lymphoma Myeloma Leuk. 2019;19:356–364. doi: 10.1016/j.clml.2019.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Barta S.K., Fowler N.H., Zain J., Ruan J., Smith S.M., Schuster S.J., Nasta S.D., Svoboda J., Gerson J.N., Landsburg D.J., et al. Pembrolizumab and Copanlisib for the Treatment of Relapsed or Refractory Mature T-Cell Lymphomas. Blood. 2019;134:4031. doi: 10.1182/blood-2019-125385. [DOI] [Google Scholar]
- 147.Periasamy S., Dhiman R., Barnes P.F., Paidipally P., Tvinnereim A., Bandaru A., Valluri V.L., Vankayalapati R. Programmed death 1 and cytokine inducible SH2-containing protein dependent expansion of regulatory T cells upon stimulation With Mycobacterium tuberculosis. J. Infect. Dis. 2011;203:1256–1263. doi: 10.1093/infdis/jir011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lu Y., Xue J., Deng T., Zhou X., Yu K., Deng L., Huang M., Yi X., Liang M., Wang Y., et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 2020;26:732–740. doi: 10.1038/s41591-020-0840-5. [DOI] [PubMed] [Google Scholar]
- 149.Stadtmauer E.A., Fraietta J.A., Davis M.M., Cohen A.D., Weber K.L., Lancaster E., Mangan P.A., Kulikovskaya I., Gupta M., Chen F., et al. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367 doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Versteven M., Van den Bergh J.M.J., Marcq E., Smits E.L.J., Van Tendeloo V.F.I., Hobo W., Lion E. Dendritic Cells and Programmed Death-1 Blockade: A Joint Venture to Combat Cancer. Front. Immunol. 2018;9:394. doi: 10.3389/fimmu.2018.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang T., Kong S., Tao M., Ju S. The potential role of RNA N6-methyladenosine in Cancer progression. Mol. Cancer. 2020;19:88. doi: 10.1186/s12943-020-01204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li Y., Gu J., Xu F., Zhu Q., Chen Y., Ge D., Lu C. Molecular characterization, biological function, tumor microenvironment association and clinical significance of m6A regulators in lung adenocarcinoma. Brief. Bioinform. 2020 doi: 10.1093/bib/bbaa225. [DOI] [PubMed] [Google Scholar]
- 153.Zhang B., Wu Q., Li B., Wang D., Wang L., Zhou Y.L. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer. 2020;19:53. doi: 10.1186/s12943-020-01170-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Han S.H., Choe J. Diverse molecular functions of m(6)A mRNA modification in cancer. Exp. Mol. Med. 2020;52:738–749. doi: 10.1038/s12276-020-0432-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dai X.Y., Shi L., Li Z., Yang H.Y., Wei J.F., Ding Q. Main N6-Methyladenosine Readers: YTH Family Proteins in Cancers. Front. Oncol. 2021;11:635329. doi: 10.3389/fonc.2021.635329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Elcheva I.A., Spiegelman V.S. Targeting RNA-binding proteins in acute and chronic leukemia. Leukemia. 2021;35:360–376. doi: 10.1038/s41375-020-01066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hou G., Zhao X., Li L., Yang Q., Liu X., Huang C., Lu R., Chen R., Wang Y., Jiang B., et al. SUMOylation of YTHDF2 promotes mRNA degradation and cancer progression by increasing its binding affinity with m6A-modified mRNAs. Nucleic Acids Res. 2021;49:2859–2877. doi: 10.1093/nar/gkab065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G., et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kim D.J., Iwasaki A. YTHDF1 Control of Dendritic Cell Cross-Priming as a Possible Target of Cancer Immunotherapy. Biochemistry. 2019;58:1945–1946. doi: 10.1021/acs.biochem.9b00200. [DOI] [PubMed] [Google Scholar]
- 160.Kachroo N., Valencia T., Warren A.Y., Gnanapragasam V.J. Evidence for downregulation of the negative regulator SPRED2 in clinical prostate cancer. Br. J. Cancer. 2013;108:597–601. doi: 10.1038/bjc.2012.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Barbieri I., Tzelepis K., Pandolfini L., Shi J., Millan-Zambrano G., Robson S.C., Aspris D., Migliori V., Bannister A.J., Han N., et al. Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature. 2017;552:126–131. doi: 10.1038/nature24678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gulati P., Cheung M.K., Antrobus R., Church C.D., Harding H.P., Tung Y.C., Rimmington D., Ma M., Ron D., Lehner P.J., et al. Role for the obesity-related FTO gene in the cellular sensing of amino acids. Proc. Natl. Acad. Sci. USA. 2013;110:2557–2562. doi: 10.1073/pnas.1222796110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Singh B., Kinne H.E., Milligan R.D., Washburn L.J., Olsen M., Lucci A. Important Role of FTO in the Survival of Rare Panresistant Triple-Negative Inflammatory Breast Cancer Cells Facing a Severe Metabolic Challenge. PLoS ONE. 2016;11:e0159072. doi: 10.1371/journal.pone.0159072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Su R., Dong L., Li Y., Gao M., Han L., Wunderlich M., Deng X., Li H., Huang Y., Gao L., et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell. 2020;38:79–96. doi: 10.1016/j.ccell.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Fujimura T., Kambayashi Y., Aiba S. Crosstalk between regulatory T cells (Tregs) and myeloid derived suppressor cells (MDSCs) during melanoma growth. Oncoimmunology. 2012;1:1433–1434. doi: 10.4161/onci.21176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Adhikari S., Xiao W., Zhao Y.L., Yang Y.G. m(6)A: Signaling for mRNA splicing. RNA Biol. 2016;13:756–759. doi: 10.1080/15476286.2016.1201628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhao X., Yang Y., Sun B.F., Shi Y., Yang X., Xiao W., Hao Y.J., Ping X.L., Chen Y.S., Wang W.J., et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Selberg S., Seli N., Kankuri E., Karelson M. Rational Design of Novel Anticancer Small-Molecule RNA m6A Demethylase ALKBH5 Inhibitors. ACS Omega. 2021;6:13310–13320. doi: 10.1021/acsomega.1c01289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ding Z., Li Q., Zhang R., Xie L., Shu Y., Gao S., Wang P., Su X., Qin Y., Wang Y., et al. Personalized neoantigen pulsed dendritic cell vaccine for advanced lung cancer. Signal. Transduct. Target. Ther. 2021;6:26. doi: 10.1038/s41392-020-00448-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yoshimura A., Ito M., Chikuma S., Akanuma T., Nakatsukasa H. Negative Regulation of Cytokine Signaling in Immunity. Cold Spring Harb. Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a028571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yoshimura A., Nishinakamura H., Matsumura Y., Hanada T. Negative regulation of cytokine signaling and immune responses by SOCS proteins. Arthritis Res. Ther. 2005;7:100–110. doi: 10.1186/ar1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shouda T., Yoshida T., Hanada T., Wakioka T., Oishi M., Miyoshi K., Komiya S., Kosai K., Hanakawa Y., Hashimoto K., et al. Induction of the cytokine signal regulator SOCS3/CIS3 as a therapeutic strategy for treating inflammatory arthritis. J. Clin. Investig. 2001;108:1781–1788. doi: 10.1172/JCI13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hunter M.G., Jacob A., O’Donnell L.C., Agler A., Druhan L.J., Coggeshall K.M., Avalos B.R. Loss of SHIP and CIS recruitment to the granulocyte colony-stimulating factor receptor contribute to hyperproliferative responses in severe congenital neutropenia/acute myelogenous leukemia. J. Immunol. 2004;173:5036–5045. doi: 10.4049/jimmunol.173.8.5036. [DOI] [PubMed] [Google Scholar]
- 174.Ochoa D., Hercules A., Carmona M., Suveges D., Gonzalez-Uriarte A., Malangone C., Miranda A., Fumis L., Carvalho-Silva D., Spitzer M., et al. Open Targets Platform: Supporting systematic drug-target identification and prioritisation. Nucleic Acids Res. 2021;49:D1302–D1310. doi: 10.1093/nar/gkaa1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Martz L. Innate harmony. [(accessed on 21 July 2021)]. Available online: https://www.innate-pharma.com/sites/default/files/072816in_coverstory_innateharmony.pdf.
- 176.Trengove M.C., Ward A.C. SOCS proteins in development and disease. Am. J. Clin. Exp. Immunol. 2013;2:1–29. [PMC free article] [PubMed] [Google Scholar]
- 177.Hernandez C., Bogdanov P., Gomez-Guerrero C., Sampedro J., Sola-Adell C., Espejo C., Garcia-Ramirez M., Prieto I., Egido J., Simo R. SOCS1-Derived Peptide Administered by Eye Drops Prevents Retinal Neuroinflammation and Vascular Leakage in Experimental Diabetes. Int. J. Mol. Sci. 2019;20:3615. doi: 10.3390/ijms20153615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Chikuma S., Kanamori M., Mise-Omata S., Yoshimura A. Suppressors of cytokine signaling: Potential immune checkpoint molecules for cancer immunotherapy. Cancer Sci. 2017;108:574–580. doi: 10.1111/cas.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bernard P.-L., Delconte R.B., Pastor S., Laletin V., Goubard A., Josselin E., Castellano R., Vernerey J., Vivier E., Huntington N.D., et al. CISH targeting in NK cells activates natural cytotoxicity receptor signaling and reduce cell exhaustion to unsilence primary anti-tumor response. bioRxiv. 2021 doi: 10.1101/2021.03.16.435571. [DOI] [Google Scholar]
- 180.Felices M., Lenvik A.J., McElmurry R., Chu S., Hinderlie P., Bendzick L., Geller M.A., Tolar J., Blazar B.R., Miller J.S. Continuous treatment with IL-15 exhausts human NK cells via a metabolic defect. JCI Insight. 2018;3 doi: 10.1172/jci.insight.96219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Andre P., Denis C., Soulas C., Bourbon-Caillet C., Lopez J., Arnoux T., Blery M., Bonnafous C., Gauthier L., Morel A., et al. Anti-NKG2A mAb Is a Checkpoint Inhibitor that Promotes Anti-tumor Immunity by Unleashing Both T and NK Cells. Cell. 2018;175:1731–1743. doi: 10.1016/j.cell.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gilboa E. Knocking the SOCS1 off dendritic cells. Nat. Biotechnol. 2004;22:1521–1522. doi: 10.1038/nbt1204-1521. [DOI] [PubMed] [Google Scholar]
- 183.Shen L., Evel-Kabler K., Strube R., Chen S.Y. Silencing of SOCS1 enhances antigen presentation by dendritic cells and antigen-specific anti-tumor immunity. Nat. Biotechnol. 2004;22:1546–1553. doi: 10.1038/nbt1035. [DOI] [PubMed] [Google Scholar]
- 184.Miah M.A., Yoon C.H., Kim J., Jang J., Seong Y.R., Bae Y.S. CISH is induced during DC development and regulates DC-mediated CTL activation. Eur. J. Immunol. 2012;42:58–68. doi: 10.1002/eji.201141846. [DOI] [PubMed] [Google Scholar]
- 185.Kobayashi T., Yoshimura A. Keeping DCs awake by putting SOCS1 to sleep. Trends Immunol. 2005;26:177–179. doi: 10.1016/j.it.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 186.Zhang W., Song Z., Xiao J., Liu X., Luo Y., Yang Z., Luo R., Li A. Blocking the PD-1/PD-L1 axis in dendritic cell-stimulated Cytokine-Induced Killer Cells with pembrolizumab enhances their therapeutic effects against hepatocellular carcinoma. J. Cancer. 2019;10:2578–2587. doi: 10.7150/jca.26961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lim T.S., Chew V., Sieow J.L., Goh S., Yeong J.P., Soon A.L., Ricciardi-Castagnoli P. PD-1 expression on dendritic cells suppresses CD8(+) T cell function and antitumor immunity. Oncoimmunology. 2016;5:e1085146. doi: 10.1080/2162402X.2015.1085146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Peng Q., Qiu X., Zhang Z., Zhang S., Zhang Y., Liang Y., Guo J., Peng H., Chen M., Fu Y.X., et al. PD-L1 on dendritic cells attenuates T cell activation and regulates response to immune checkpoint blockade. Nat. Commun. 2020;11:4835. doi: 10.1038/s41467-020-18570-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Go D.M., Lee S.H., Lee S.H., Woo S.H., Kim K., Kim K., Park K.S., Park J.H., Ha S.J., Kim W.H., et al. Programmed Death Ligand 1-Expressing Classical Dendritic Cells MitigateHelicobacter-Induced Gastritis. Cell. Mol. Gastroenterol. Hepatol. 2021;12:715–739. doi: 10.1016/j.jcmgh.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zhu H., Blum R.H., Bernareggi D., Ask E.H., Wu Z., Hoel H.J., Meng Z., Wu C., Guan K.L., Malmberg K.J., et al. Metabolic Reprograming via Deletion of CISH in Human iPSC-Derived NK Cells Promotes In Vivo Persistence and Enhances Anti-tumor Activity. Cell Stem Cell. 2020;27:224–237. doi: 10.1016/j.stem.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Li H.B., Tong J., Zhu S., Batista P.J., Duffy E.E., Zhao J., Bailis W., Cao G., Kroehling L., Chen Y., et al. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548:338–342. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kumar S., Jeong Y., Ashraf M.U., Bae Y.S. Dendritic Cell-Mediated Th2 Immunity and Immune Disorders. Int. J. Mol. Sci. 2019;20:2159. doi: 10.3390/ijms20092159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Huemer F., Leisch M., Geisberger R., Zaborsky N., Greil R. miRNA-Based Therapeutics in the Era of Immune-Checkpoint Inhibitors. Pharmaceuticals (Basel) 2021;14:89. doi: 10.3390/ph14020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Skafi N., Fayyad-Kazan M., Badran B. Immunomodulatory role for MicroRNAs: Regulation of PD-1/PD-L1 and CTLA-4 immune checkpoints expression. Gene. 2020;754:144888. doi: 10.1016/j.gene.2020.144888. [DOI] [PubMed] [Google Scholar]
- 195.Kumar S., Kim Y. An endoparasitoid wasp influences host DNA methylation. Sci. Rep. 2017;7:43287. doi: 10.1038/srep43287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kumar S., Venkata P., Kim Y. Suppressive activity of a viral histone H4 against two host chromatin remodelling factors: Lysine demethylase and SWI/SNF. J. Gen. Virol. 2016;97:2780–2796. doi: 10.1099/jgv.0.000560. [DOI] [PubMed] [Google Scholar]
- 197.He C., Lan F. RNA m(6)A meets transposable elements and chromatin. Protein Cell. 2021 doi: 10.1007/s13238-021-00859-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Sharma P., Allison J.P. Immune checkpoint targeting in cancer therapy: Toward combination strategies with curative potential. Cell. 2015;161:205–214. doi: 10.1016/j.cell.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hu Z., Ott P.A., Wu C.J. Towards personalized, tumour-specific, therapeutic vaccines for cancer. Nat. Rev. Immunol. 2018;18:168–182. doi: 10.1038/nri.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Blass E., Ott P.A. Advances in the development of personalized neoantigen-based therapeutic cancer vaccines. Nat. Rev. Clin. Oncol. 2021;18:215–229. doi: 10.1038/s41571-020-00460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Perrier A., Didelot A., Laurent-Puig P., Blons H., Garinet S. Epigenetic Mechanisms of Resistance to Immune Checkpoint Inhibitors. Biomolecules. 2020;10:1061. doi: 10.3390/biom10071061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wang H., Hu X., Huang M., Liu J., Gu Y., Ma L., Zhou Q., Cao X. Mettl3-mediated mRNA m(6)A methylation promotes dendritic cell activation. Nat. Commun. 2019;10:1898. doi: 10.1038/s41467-019-09903-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wu H., Xu Z., Wang Z., Ren Z., Li L., Ruan Y. Exosomes from dendritic cells with Mettl3 gene knockdown prevent immune rejection in a mouse cardiac allograft model. Immunogenetics. 2020;72:423–430. doi: 10.1007/s00251-020-01180-8. [DOI] [PubMed] [Google Scholar]
- 204.Feng Y., Dong H., Sun B., Hu Y., Yang Y., Jia Y., Jia L., Zhong X., Zhao R. METTL3/METTL14 Transactivation and m(6)A-Dependent TGF-beta1 Translation in Activated Kupffer Cells. Cell Mol. Gastroenterol. Hepatol. 2021;12:839–856. doi: 10.1016/j.jcmgh.2021.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yao Y., Yang Y., Guo W., Xu L., You M., Zhang Y.C., Sun Z., Cui X., Yu G., Qi Z., et al. METTL3-dependent m(6)A modification programs T follicular helper cell differentiation. Nat. Commun. 2021;12:1333. doi: 10.1038/s41467-021-21594-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.