Abstract
Drosophila sine oculis and eyes absent genes synergize in compound-eye formation. The murine homologues of these genes, Six and Eya, respectively, show overlapping expression patterns during development. We hypothesized that Six and Eya proteins cooperate to regulate their target genes. Cotransfection assays were performed with various combinations of Six and Eya to assess their effects on a potential natural target, myogenin promoter, and on a synthetic promoter, the thymidine kinase gene promoter fused to multimerized Six4 binding sites. A clear synergistic activation of these promoters was observed in certain combinations of Six and Eya. To investigate the molecular basis for the cooperation, we first examined the intracellular distribution of Six and Eya proteins in transfected COS7 cells. Coexpression of Six2, Six4, or Six5 induced nuclear translocation of Eya1, Eya2, and Eya3, which were otherwise distributed in the cytoplasm. In contrast, coexpression of Six3 did not result in nuclear localization of any Eya proteins. Six and Eya proteins were coimmunoprecipitated from nuclear extracts prepared from cotransfected COS7 cells and from rat liver. Six domain and homeodomain, two evolutionarily conserved domains among various Six proteins, were necessary and sufficient for the nuclear translocation of Eya. In contrast, the Eya domain, a conserved domain among Eya proteins, was not sufficient for the translocation. A specific interaction between the Six domain and homeodomain of Six4 and Eya2 was observed by yeast two-hybrid analysis. Our results suggest that transcription regulation of certain target genes by Six proteins requires cooperative interaction with Eya proteins: complex formation through direct interaction and nuclear translocation of Eya proteins. This implies that the synergistic action of Six and Eya is conserved in the mouse and is mediated through cooperative activation of their target genes.
Six genes are mouse homologues of the Drosophila sine oculis (so) gene, which is essential for compound-eye formation (9, 31). Six members of the Six family of genes have so far been identified in the mouse (17, 18, 27, 28, 35). Each Six gene shows a specific expression pattern during development of the mouse embryo. Six1 and Six2 show expression in mesenchymal cells around E8.5 to E10.5 and in muscles and limb tendons in later stages (28). Six3 is expressed in the rostral forebrain in earlier stages and is confined to the prospective eye region (27). Six4 proteins are distributed in the peripheral region of the mantle layer of the developing brain and spinal cord and in various ganglia between E9.5 and E14.5 (25). Six5 mRNA is expressed as early as E7 and is abundantly expressed in neonatal heart and skeletal muscles (24). Human SIX5 resides downstream of a CTG repeat, whose expansion leads to myotonic dystrophy (DM) (7). Since SIX5 is expressed in several tissues affected by DM and the transcription of SIX5 is repressed by the causative a CTG repeat expansion, it has been proposed that SIX5 is involved in some aspects of DM pathogenesis (7, 12, 20, 34, 37). The expression pattern of Six5 mRNA observed in the mouse suggests a potential link to the DM phenotype observed in humans (24). Optx2/Six9 is first expressed in the most rostral portion of the neural plate at E8.25 and later in the optic vesicle and chiasm (35). These observations imply the important role of Six family genes in vertebrate development. Indeed, ectopic expression of mouse or medaka Six3 leads to lens or retina formation in medaka fish (22, 26), and overexpression of zebrafish six3 induces enlargement of the rostral forebrain and optic stalk (21), suggesting the involvement of Six3 in the formation of rostral forebrain and eye. Six family proteins are characterized by the presence of two evolutionarily conserved regions, the Six domain (110 amino acids) and the Six-type homeodomain (60 amino acids). Both of these domains are required for specific DNA binding (17). Six2, Six4, and Six5 can bind to the same target sequence in the ARE (Atpla1 regulatory element) of the Na,K-ATPase α1 subunit gene (18); however, Six3 did not show specific binding to the element, and the target sequences of Six3 are unknown. Recently, Six1 and Six4 have been shown to bind to the MEF3 site in the myogenin promoter (32). The C-terminal 150-amino-acid region of Six4 has a transactivation activity (17). Thus, it is presumed that Six proteins are transcription factors controlling the expression of multiple target genes by binding to their specific binding sequences.
Eya genes have been identified as homologues of Drosophila eyes absent (eya) gene, which is also essential for the formation of compound eyes in Drosophila (5). Four mouse homologues have been identified (1, 6, 10, 40, 42). Eya1 and Eya2 are highly expressed in cranial placodes, branchial arches, and the central nervous system during organogenesis (40). Eya3 is also expressed in branchial arches and the central nervous system, but not in the cranial placode (40). The recently identified Eya4 is expressed primarily in the craniofacial mesenchyme, the dermomyotome, and the limbs (6). Mutations in human EYA1 have been shown to be responsible for branchio-oto-renal syndrome, with branchial, ear, and renal abnormalities (1). This suggests a role for EYA1 in the development of branchial, otic, and renal organs. However, the molecular function of Eya proteins is not fully understood. The conserved region of Eya is the Eya domain, composed of 271 amino acids (40). The N-terminal portions of mouse Eya1, Eya2, and Eya3 have been shown to possess transactivation activity, and a PST-rich sequence is found in the region of each Eya protein (39). However, no specific DNA binding activity of Eya proteins has been reported.
Ectopic expression of eya in Drosophila embryos induces ectopic eyes (4). Coexpression of so greatly facilitates ectopic-eye formation by eya, suggesting a functional cooperation between so and eya gene products (29). Yeast two-hybrid assays demonstrated that the essential domains for the interaction between So and Eya proteins are the conserved Six and Eya domains (29). Because Eya protein has no specific DNA binding activity, it is thought to function as a coactivator of So. Moreover, eyeless, another Drosophila eye-forming gene, regulates so and eya expression, and eya and eyeless together are more effective in inducing ectopic eye formation than either alone (4). Coexpression of dachshund, a Drosophila gene involved in the formation of the retina, also enhances the ability of eya to induce ectopic eye formation (8). These findings suggest that the complex gene network involving a direct interaction among these gene products and the feedback loops of their gene regulation determines eye identity (8, 29). It is plausible that there are similar functional cooperations among Pax6, Six, Eya, and Dac genes, murine homologues of eyeless, so, eya, and dachshund. As a first step in understanding such gene networks in the mouse, we analyzed the cooperation and interaction between mouse Eya and Six. First, cotransfection assays were performed with various combinations of Six and Eya to assess their effects on a putative natural target, the myogenin gene promoter, as well as on a synthetic promoter. Second, we determined the molecular basis for the cooperation by analyzing the intracellular distribution of Eya and complex formation between Eya and Six. Finally, specific interactions between the Six and Eya proteins were confirmed by yeast two-hybrid analysis. Based on the findings of the present study, we provide a model of the functional cooperation of Six and Eya in transactivation and its implication in various developmental processes.
MATERIALS AND METHODS
Cloning and constructions of expression plasmid for Eya1, Eya2, and Eya3.
Mouse Eya1 and Eya3 cDNAs were obtained by reverse transcription-PCR with poly(A)+ RNA prepared from whole embryos (E10.5; C57B/6) with primers 5′-TTGCAGGTCTATGGAAATGCAGGATC and 5′-ATATGCTGAAATTGGTACATCCTGAAGTCCA for Eya1 and 5′-TCAAGTAAACAACCCAGATGCCAGTGATGAG and 5′-CAGAAAAATTAAAGCACGGTAGCGGCAGC for Eya3. The cDNAs were subcloned into pCR-Script (Stratagene, La Jolla, Calif.). For hemagglutinin (HA)-Eya1 fusion protein expression, the cDNA insert was excised as a NotI-EcoRI fragment and the NotI site was blunt ended to add a HindIII linker and then ligated into the HindIII/EcoRI site of pHM6 (Boehringer Mannheim, Mannheim, Germany) (pHM6Eya1). For the HA-Eya2 fusion protein, to complement the missing 5′ portion of the Eya2 cDNA kindly supplied by N. M. Bonini (42), we performed PCR with primers 5′-CCCAAGCTTGATGTTAGAAGTGGTGACCTCACCCAGCCTCGCAACAAG and 5′-CGCTGTATAGGGTGGTGCC, using the Eya2 cDNA clone as a template. The PCR product was cut with HindIII and NcoI and ligated into pHM6 vector cut with HindIII and KpnI together with an NcoI(164)/KpnI (in the vector) fragment of the Eya2 cDNA (pHM6Eya2). For the HA-Eya3 fusion protein, an HphI fragment (213 to 2232) of Eya3 cDNA from N. M. Bonini (42) was blunt ended and added with a HindIII linker. After ligation into pKS, the BamHI(1006)/NcoI(1064) region in the construct was replaced by a reverse transcription-PCR-derived fragment of Eya3 to remove a point mutation at 1043 in the Eya3 cDNA. The resulting HindIII fragment was subcloned into pHM6 (pHM6Eya3). All regions derived from PCR amplification were verified by sequencing. The HA-Eya domain fusion protein construct was prepared as follows. For pHM6Eya1ED, a HpaII (1100)-EcoRI (3′-terminal) fragment was inserted into the HindIII/EcoRI sites of pHM6 (40). For pHM6Eya2ED, a TaqI (771)-EcoRI (3′-end) fragment was inserted into the HindIII/EcoRI sites of pHM6. For pHM6Eya3ED, an AluI(924)-EcoRI (3′-end) fragment was inserted into the HindIII/EcoRI sites of pHM6.
Six protein expression plasmids and reporter gene constructs.
Oligonucleotides containing a Six4 binding site from the ARE sequence of the Na,K-ATPase α1 subunit gene (C3) and its point mutation (C3M) were multimerized (18) and hooked upstream of the herpes simplex virus thymidine kinase (TK) gene promoter (23) (pTKW4FLF and pTKM4FLF) and used for the reporter gene assay. The myogenin promoter-luciferase reporter constructs pGL3MG-185 and its MEF3 site mutation pGL3MG-185M were constructed as follows. An AatI (−182)-HindIII fragment was excised from pMGNLacZ(−4K) (11) and ligated into the SmaI/HindIII sites of pGL3-Basic (Promega, Madison, Wis.) to produce pGL3MG-185. Cassette mutagenesis with the oligonucleotide 5′-TAGAGGGGGGCTGAGTTTTCTGTGGCGT (MEF3 site mutations underlined) and its complementary oligonucleotide was performed for pGL3MG-185M.
Full-length mouse Six2, Six4, and Six5 cDNAs were inserted into pFLAG-CMV-2 (Eastman Kodak, New Haven, Conn.). A BssHII (blunt ended)-Sau3A1 fragment (280 to 1381) of Six2 cDNA (18) was ligated into the blunt-ended ClaI and BamHI sites of pFLAG-CMV-2 (pfSix2). An NcoI (blunt ended)-XmnI fragment (666 to 1616) of Six3α cDNA (18) was added to an XbaI linker (GCTCTAGAGC) and ligated into the XbaI site of pFLAG-CMV-2 (pfSix3). pGSTSMNT was cut with BamHI, blunt ended, and digested with EcoRV (1060). The fragment was ligated into the EcoRV site of pFLAG-CMV-2. The EcoRV (1060)-XbaI (2852) fragment of Six4SM cDNA was cloned into the resulting plasmid digested with EcoRV and XbaI (pfSix4). The missing N-terminal portion of Six5 cDNA was complemented from the genomic clone (18, 24). The most 5′ portion was PCR amplified with primers 5′-TGCTCTAGACATGGCTACCTCGCC (corresponding to a putative initiation codon located between positions 413 and 426) and 5′-TTCCTCCTCCTGCTCCTCGGTCG (496 to 474). After digestion with XbaI (in the primers) and SacII (447), the PCR fragment was ligated into pSK together with the adjacent SacII-XhoI fragment derived from the genomic clone. The 3′ portion downstream of the XhoI site was excised from the Six5 cDNA (18) and inserted into the XhoI site. The resulting full-length Six5 cDNA was excised as a NotI-BglII (2342) fragment and ligated into the NotI/BglII site of pFLAG-CMV-2 (pfSix5). FLAG-Six4 subdomain protein expression vectors were constructed as follows. For pfSix4SDHD, a BsrBI-BsaI (281 to 892) fragment of pfSix4 was blunt ended with Klenow fragment, added to an XbaI linker, and ligated into the XbaI site of pFLAG-CMV-2. For pfSix4SD, the XbaI-Alw26I (717) blunt-ended fragment derived from pfSix4SDHD was ligated into the XbaI/SmaI sites of pFLAG-CMV-2. For pfSix4HD, an RsaI fragment (661 to 924) was added to a SalI linker and ligated into the SalI site of pFLAG-CMV-2. For pfSix5CΔ2, pfSix5 was digested with EcoRI (1119) and BglII (2342), blunt ended with Klenow, and self-ligated.
Cell culture and transfection assays.
COS7 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum with 100 U of penicillin/ml and 100 μg of streptomycin/ml at 37°C under 5% CO2. Transfections were performed by CaCl2 precipitation or with Lipofectamine plus (Gibco, Long Island, N.Y.) as described previously (24) in 3.5-cm-diameter dishes for reporter gene assays and in 6- or 10-cm-diameter dishes for the preparation of nuclear and cytoplasmic extracts.
Antibody production.
Anti-Eya3 sera were prepared by immunizing rabbits with glutathione S-transferase (GST) fusion Eya3 protein expressed in Escherichia coli. A HindIII (219)-Sau3AI (865) fragment was excised from pHM6Eya3, filled by Klenow fragment, and ligated to the SmaI site of pGEX-3X (Amersham Pharmacia Biotech, Uppsala, Sweden). GST fusion protein was purified according to the method recommended by the manufacturer. Rabbits were immunized with 0.1 to 0.3 mg of the antigen six times, separated by intervals of about 2 weeks, and were later sacrificed for antisera.
Preparation of nuclear and cytoplasmic extracts.
Nuclear and cytoplasmic extracts from COS7 cells were prepared as described previously (19). Rat liver nuclear extracts were obtained as described previously (25). The protein concentration of extracts was determined with a protein assay kit (Bio-Rad Laboratories, Hercules, Calif.).
Western blotting.
Nuclear or cytoplasmic extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (9 or 13% acrylamide) and transferred to Hybond-ECL membrane (Amersham). Western analysis was performed with an ECL Kit (Amersham). Anti-FLAG M5 antibody (Eastman Kodak), anti-HA rat antibody (Boehringer Mannheim), anti-Six5 antibody (25), or anti-Eya3 serum was used as the first antibody. Relative quantitation of tagged proteins was performed by measuring exposed X-ray films by using the Discovery Series (Protein Databases, Inc., Huntington, N.Y.).
Immunostaining of tissue culture cells.
Transfected COS7 cells were fixed with 4% paraformaldehyde and analyzed with an LSAB Kit (Dako Corporation, Carpinteria, Calif.) and rat anti-HA antibody.
Coimmunoprecipitation.
Nuclear or cytoplasmic extracts from transfected COS7 cells (5 μg of protein each) were incubated with 0.67 μg of anti-FLAG M5 antibody, or rat liver nuclear extract (100 μg of protein) was incubated with 1.3 μg of anti-Six5 antibody (25) or purified rabbit immunoglobulin G (IgG) (Sigma, St. Louis, Mo.) in a buffer containing 50 mM KCl, 16.7 mM Tris-HCl (pH 7.3 at 25°C), 0.167 mM EDTA, 1.25 mM MgCl2, and 5% (vol/vol) glycerol, as indicated in the text. Immunoprecipitates were recovered by protein G agarose, dissolved in SDS sample buffer, and analyzed by SDS-PAGE followed by Western blotting with anti-HA antibody, anti-Six5 antibody, or anti-Eya3 serum.
Yeast two-hybrid assays.
Saccharomyces cerevisiae strains (EGY48) and interaction assays were described previously (41). Cells harboring a reporter plasmid pSH18-34 were transformed with pEG202Six4SDHD by the lithium acetate method and selected with Ura− His− medium. Each transformant was subsequently transformed by pJG4-5Eya2 constructs and selected with Ura− His− Trp− medium. Each double transformant was placed on Ura− His− Trp− galactose plates supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates for the interaction assay. The interaction was considered positive if the transformant turned blue on X-Gal indicator plates.
For plasmid construction, the SDHD region (see below) from pfSix4SDHD was excised and subcloned into the pEG202 EcoRI/BamHI site (pEG202Six4SDHD). HindIII/NotI fragments were excised from pHM6Eya2 and from pHM6Eya2ED, blunt ended with Klenow fragment, and ligated into the EcoRV site of pSK. EcoRI/XhoI fragments were excised and ligated into pJG4-5 (pJG4-5Eya2 and pJG4-5Eya2ED). pHM6Eya2 was cut with AatII (867), blunt-ended with T4 DNA polymerase, cut with HindIII, and then ligated into the HindIII/EcoRI (blunt-ended) site of pHM6term (a stop codon containing oligonucleotides AATTCTGACTGACTGACGC was inserted in the EcoRI-NotI site of pHM6) to make pHM6Eya2ΔED. The HindIII and NotI fragments were excised and ligated into pSKEcoRV, and then the EcoRI/XhoI fragment was ligated into the pJG4-5 EcoRI and XhoI sites to make pJG4-5Eya2ΔED. For N-terminal deletion proteins of Eya2, the NotI-HindIII fragment of pHM6Eya2 was cut at the BstXI (158), ApaLI(445), or EcoNI(579) site. The digested fragments, termed NΔ1, NΔ2, and NΔ3, were blunt ended with T4 DNA polymerase (for NΔ1) or Klenow fragment (for NΔ2 and NΔ3) and ligated into the pSK EcoRV or pUC119 SmaI site. After digestion with EcoRI and XhoI, fragments from pSKEya2NΔ1, pUC119Eya2NΔ2, and pUC119Eya2NΔ3 were ligated into EcoRI and XhoI sites of pJG4-5 (pJG4-5Eya2NΔ1, pJG4-5Eya2NΔ2, or pJG4-5Eya2NΔ3).
RESULTS
Cooperation of Six and Eya proteins in transactivation.
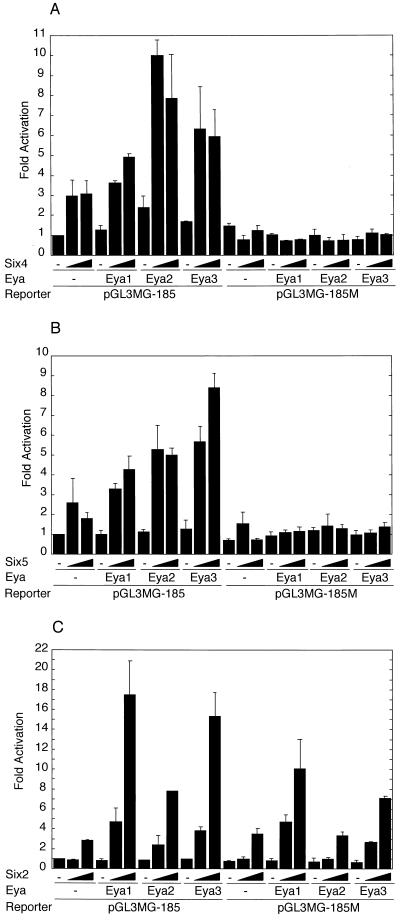
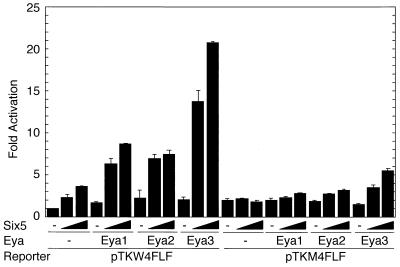
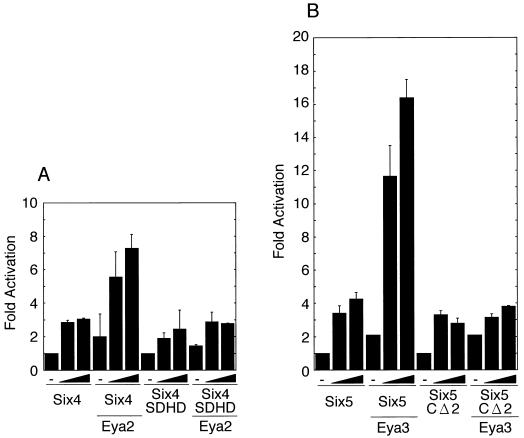
The functional synergy between the so and eya genes of Drosophila in the formation of ectopic eyes (29) suggests similar functional cooperation between the mouse Six and Eya gene families. To test whether the synergistic action is mediated by cooperative transactivation of their target genes, we analyzed the effects of Six and Eya proteins on target gene expression with transient transfection assays. Plasmids expressing various Six proteins as FLAG fusion proteins were used to monitor the expression level separately from endogenous proteins (Fig. 1A). Likewise, Eya proteins were expressed as HA fusion proteins (Fig. 1C). We tested the effects of Six and Eya proteins on the proximal promoter (−185 to +49) of the myogenin gene, a putative natural target (32), fused to the luciferase reporter gene (pGL3MG-185) (Fig. 2). Transfection of pfSix4 resulted in an approximately threefold increase in luciferase activity. The activation level was increased to approximately 10-fold by the coexpression of Eya2, 6-fold by Eya3, and 5-fold by Eya1 (Fig. 2A, left). In contrast, virtually no activation by Six4 and Eya proteins was observed with the mutation reporter pGL3MG-185M in which the MEF3 site (to which Six1 and Six4 can bind) was mutated (Fig. 2A, right); thus, the Six4 binding activity was reduced by more than 25-fold (data not shown). This indicates that the transcriptional response to Eya is dependent on a functional Six4 binding site. Transfection of pfSix5 resulted in a 2.5-fold increase in the luciferase activity of pGL3MG-185. Coexpression of Eya1 showed fourfold activation, Eya2 showed fivefold activation, and Eya3 showed eightfold activation (Fig. 2B, left). A weak activation by pfSix5 was observed for pGL3MG-185M in any combination with Eya (Fig. 2B, right). This indicates that the transcriptional response to Eya is again dependent on the functional Six4 binding site, to which Six5 can bind (data not shown). When we transfected pfSix2, a threefold increase in luciferase activity was observed for pGL3MG-185 (Fig. 2C). Coexpression of Eya1 led to an 18-fold increase, while Eya2 showed an 8-fold increase and Eya3 showed 16-fold activation. A similar but less efficient activation was also observed for pGL3MG-185M by Six2 in combination with Eya. This was probably due to the presence of other binding sequences for Six2 in the myogenin promoter fragment, although the mutated MEF3 site greatly diminished the specific binding of Six2 (data not shown). We also tested the effects on another reporter gene of the TK promoter fused to multimerized Six4 binding sites derived from the Na,K-ATPase α1 subunit gene (pTKW4FLF). Transfection of pfSix5 activated luciferase activity of pTKW4FLF 3.5-fold. Coexpression of Eya1 or Eya2 resulted in 8- to 9-fold activation, while coexpression of Eya3 led to 21-fold activation, but only a marginal activation was observed for pTKM4FLF, a reporter with mutated Six4 binding sites (Fig. 3). In contrast, Six4 or Six2 showed moderate or marginal transactivation, respectively, with this particular reporter construct (data not shown). These results clearly indicate that Six and Eya proteins act cooperatively to transactivate natural and synthetic target promoters containing Six4 binding sites and that the magnitude of cooperation among various Six and Eya proteins varies depending on their combinations.
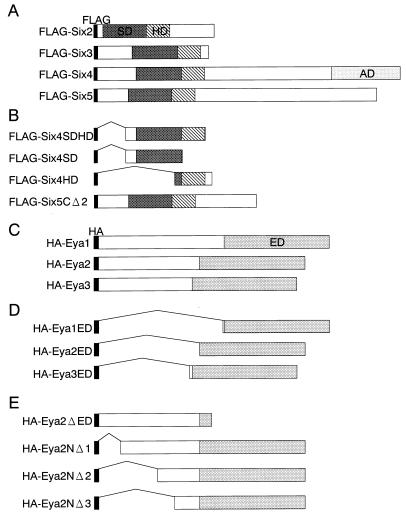
FIG. 1.
Six and Eya expression constructs used in this study. (A) FLAG fusion Six2, Six3, Six4, and Six5 proteins were expressed from pFLAG-CMV-2 constructs. Conserved Six domain (SD) and homeodomain (HD) are indicated by shaded and hatched boxes, respectively. The activation domain of Six4 is indicated (AD). (B) Subdomains of Six4 and Six5 proteins were fused to FLAG. (C) HA fusion Eya1, Eya2, and Eya3 proteins were expressed from pHM6 constructs. The conserved Eya domains are indicated by the lightly shaded box (ED). (D) Eya domains of Eya1, Eya2, and Eya3 were fused to HA. (E) Various deletion mutations of Eya2 were constructed for the yeast two-hybrid assays. The indicated regions were expressed as a fusion protein to B42 activation domain. Open boxes indicate protein region, and bent lines indicate deleted region.
FIG. 2.
Activation of myogenin promoter by Six2, Six4, and Six5 proteins and effects of Eya proteins on the activation. One microgram of the myogenin luciferase reporters pGL3MG-185 or pGL3MG-185M was cotransfected with pfSix and/or pHM6Eya plasmid. Luciferase activity in the cell lysate was normalized with β-galactosidase activity of pEFBOSβ-gal as an internal control. (A) Increasing amounts (0, 0.2, or 0.4 μg) of pfSix4 and 1.5 μg of pHM6Eya1 or 0.5 μg of pHM6Eya2 or pHM6Eya3 were used for cotransfection. (B) Increasing amounts (0, 0.1, or 0.2 μg) of pfSix5 and 1.5 μg of pHM6Eya1 and 0.5 μg of pHM6Eya2 and pHM6Eya3 were used for cotransfection. (C) Increasing amounts (0, 0.2, or 0.4 μg) of pfSix2 and 1.5 μg of pHM6Eya1 and 0.5 μg of pHM6Eya2 and pHM6Eya3 were used for transfection. The activity of each datum point is relative to that obtained by the control pFLAG-CMV-2 vector (−). The mean fold activation from three independent experiments (each performed in duplicate or triplicate) is shown with the standard deviation.
FIG. 3.
Activation of TK promoter fused to multimerized Six4-binding sites. Increasing amounts (0, 0.1, or 0.2 μg) of pfSix5 were transfected with 1.5 μg of pHM6 and pHM6Eya1 and 0.5 μg of pHM6Eya2 or pHM6Eya3. One microgram of pTKW4FLF or pTKM4FLF was used as a reporter gene. pEFBOSβ-gal was included as an internal control. The relative luciferase activity normalized by β-galactosidase activity is indicated. The activity of each datum point is relative to that obtained by the control pFLAG-CMV-2 vector (−). A result typical of three independent experiments (each performed in duplicate), which yielded essentially the same results, is shown with the standard deviation.
Distribution of Six and Eya proteins between nucleus and cytoplasm.
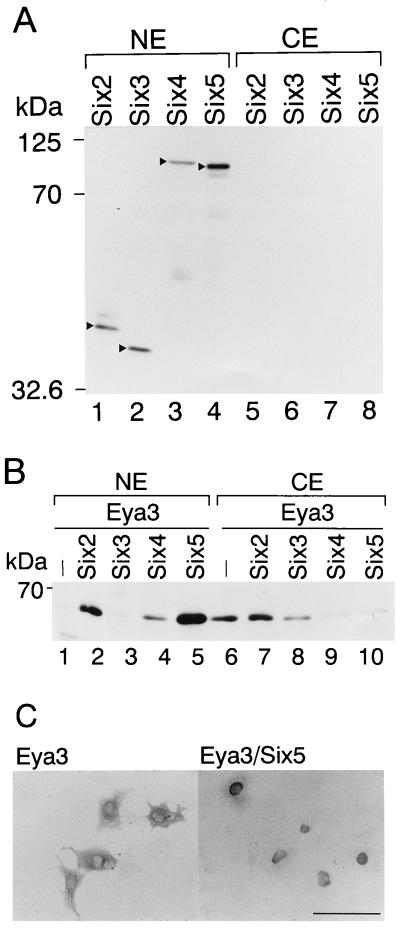
To gain insight into the molecular mechanism of the cooperation of Six and Eya in target gene activation, we analyzed the intracellular distribution of FLAG-Six and HA-Eya fusion proteins expressed in COS7 cells. Nuclear and cytoplasmic extracts from COS7 cells transfected with various combinations of pfSix and pHM6Eya were prepared and analyzed by Western blotting. The distribution of Six2, Six3, Six4, and Six5 proteins was detected by anti-FLAG M5 antibody. Most Six proteins were found in the nucleus but not in the cytoplasm (Fig. 4A), and the distribution remained unchanged with coexpression of Eya proteins (data not shown). Subsequently, the distribution of Eya proteins was analyzed with anti-HA antibody. Eya3 protein was found mostly in the cytoplasm in the absence of Six coexpression (Fig. 4B, lanes 1 and 6), while a significant increase in nuclear Eya3 was detected with coexpression of Six2, Six4, or Six5 (Fig. 4B, lanes 2, 4 to 5 and 7, and 9 to 10). Interestingly, the intracellular distribution of Eya3 remained unchanged with the coexpression of Six3 (Fig. 4B, lanes 3 and 8). The nuclear and cytoplasmic distribution of Eya3 protein in the presence or absence of Six5 coexpression in COS7 cells was confirmed by immunostaining with anti-HA antibody (Fig. 4C). Apparent cytoplasmic staining was observed in COS7 cells transfected with pHM6Eya3 alone (Fig. 4C, left), while intense nuclear staining was observed in COS7 cells transfected with both pHM6Eya3 and pfSix5 (Fig. 4C, right). The nuclear distribution of Eya3 protein was also observed with the coexpression of Six2 or Six4 by immunostaining (data not shown). The distribution of Eya1 and Eya2 between nucleus and cytoplasm with or without coexpression of Six2, Six3, Six4, or Six5 was analyzed in a manner similar to that of Eya3 (data not shown). The quantitative results are summarized in Table 1. The relative amount of Eya1 protein in the nucleus was 3.7% in the absence of Six coexpression. The amount of nuclear Eya1 markedly increased to 92.3% with coexpression of Six2, to 60.5% with Six4, and to 38.2% with Six5. A certain amount (19.8%) of Eya2 resided in the nucleus without coexpression of Six. However, the nuclear amount significantly increased to 94.9% with coexpression of Six2, to 70.8% with Six4, and to 87.0% with Six5. The nuclear amount of Eya3 was 3.3% in the absence of Six coexpression. It increased to 29.9% by coexpression of Six2, to 66.8% with Six4, and to 87.4% with Six5. Of note, coexpression of Six3 did not increase the nuclear amount of any Eya proteins (Table 1). These results suggest that Six2, Six4, and Six5, but not Six3, induce nuclear translocation of Eya proteins. The efficiency of translocation varied depending on the combination of Eya and Six proteins. Specifically, Eya1 was translocated most efficiently by Six2, moderately by Six4, and weakly by Six5. Furthermore, Eya2 was translocated efficiently by Six2 and Six5 and moderately by Six4, whereas Eya3 was most efficiently translocated by Six5, moderately by Six4, and weakly by Six2.
FIG. 4.
Nuclear translocation of Eya proteins by coexpression of Six proteins. (A) FLAG-Six proteins were expressed in COS7 cells. Nuclear (NE; lanes 1 to 4) and cytoplasmic (CE; lanes 5 to 8) extracts were analyzed by Western blotting with anti-FLAG antibody. The amount of nuclear protein analyzed was 1.5, 3.0, 4.0, and 0.75 μg for lanes 1, 2, 3, and 4, respectively. The amount of cytoplasmic protein used was 2.9, 1.8, 7.7, and 0.6 μg for lanes 5, 6, 7, and 8, respectively. The positions of detected FLAG-Six fusion proteins are indicated by arrowheads. The positions of molecular mass markers are shown on the left. Small amounts of proteins were detected in cytoplasmic extracts. (B) HA-Eya3 fusion protein was expressed with Six2, Six3, Six4, or Six5 or without (−) Six in COS7 cells. Nuclear (NE; lanes 1 to 5) and cytoplasmic (CE; lanes 6 to 10) extracts were analyzed by Western blotting with anti-HA antibody, and HA-Eya3 protein was detected. A total of 3.5 μg of nuclear extract was used for each of lanes 1 to 5, and 6.2, 3.0, 3.3, 4.9, and 3.6 μg of cytoplasmic protein was used for lanes 6, 7, 8, 9, and 10, respectively. (C) COS7 cells were transfected with pHM6Eya3 (left) or pHM6Eya3 and pfSix5 (right). The cells were fixed by 4% paraformaldehyde followed by immunostaining with anti-HA antibody. Bar, 100 μm.
TABLE 1.
Intracellular distribution of Eya proteinsa
| Six protein | Distribution (%)
|
|||||
|---|---|---|---|---|---|---|
| Eya1
|
Eya2
|
Eya3
|
||||
| Nucleus | Cytoplasm | Nucleus | Cytoplasm | Nucleus | Cytoplasm | |
| None | 3.7 | 96.3 | 19.8 | 80.2 | 3.3 | 96.7 |
| Six2 | 92.3 | 7.7 | 94.9 | 5.1 | 29.9 | 70.1 |
| Six3 | 1.8 | 98.2 | 22.5 | 77.5 | 5.2 | 94.8 |
| Six4 | 60.5 | 39.5 | 70.8 | 30.0 | 66.8 | 33.2 |
| Six5 | 38.2 | 61.8 | 87.0 | 13.0 | 87.4 | 12.6 |
Nuclear and cytoplasmic extracts from COS7 cells coexpressed with a combination of Six and Eya were analyzed by Western blotting with anti-HA. The relative amount of each HA-Eya fusion protein detected was calculated as a percentage in a set of nuclear and cytoplasmic extracts from COS7 cells. The fraction loaded on the gel was taken into consideration to calculate the total amount of Eya protein and the percentage of nuclear and cytoplasmic distribution. The amount of DNA used for transfections was adjusted to produce roughly the same amount of protein among Eya proteins and among Six proteins, i.e., 1.0 μg for pHM6Eya1 and pHM6Eya2; 0.2 μg for pHM6Eya3; and 0.06, 0.1, 3.0, and 0.48 μg for pfSix2, pfSix3, pfSix4, and pfSix5, respectively. A representative result of three similar independent experiments is shown.
Complex formation by Eya and Six.
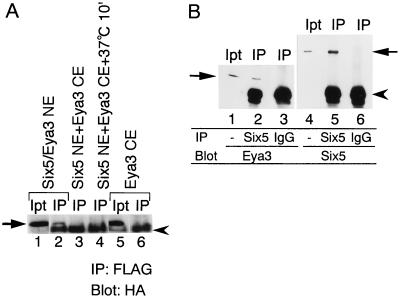
Translocation of Eya into the nucleus by coexpression of Six suggests a specific interaction between Six and Eya leading to complex formation. To investigate whether the translocated Eya in the nucleus forms a complex with Six, we performed immunoprecipitation analysis. Nuclear extracts from COS7 cells transfected with both pfSix5 and pHM6Eya3 were incubated with anti-FLAG antibody, recovered by protein G agarose, and then developed by SDS-PAGE followed by Western blotting with anti-HA antibody. Figure 5A shows that the HA-Eya3 protein was coimmunoprecipitated with FLAG-Six5 by anti-FLAG antibody (lane 2). In contrast, Eya3 was not immunoprecipitated from the cytoplasmic extract by anti-FLAG antibody (Fig. 5A, lane 6). Even when the cytoplasmic extract containing Eya3 and nuclear extract containing Six5 were mixed and incubated for 1.5 h at 4°C or for 10 min at 37°C before 1.5-h incubation at 4°C, Eya3 was not coimmunoprecipitated (Fig. 5A, lanes 3 and 4). HA-Eya2 was coimmunoprecipitated with FLAG-Six4 from nuclear extract of transfected COS7 cells (data not shown). These results indicate that translocated Eya proteins in the nucleus form a complex with Six proteins and suggest that the cotranslation or cotranslocation of Eya with Six is essential for Six-Eya complex formation.
FIG. 5.
Six5 and Eya3 complex formation. (A) Nuclear extracts from COS7 cells transfected with pfSix5 and pHM6Eya3 (lanes 1 and 2), a mixture of a nuclear extract from COS7 cells transfected with pfSix5 and a cytoplasmic extract from COS7 cells transfected with pHM6Eya3 (lanes 3 and 4), and a cytoplasmic extract from COS7 cells transfected with pHM6Eya3 (lanes 5 and 6) were incubated with anti-FLAG antibody and then precipitated by protein G agarose beads (lanes 2 to 4 and 6). The precipitates were dissolved in SDS sample buffer followed by Western blotting analysis with anti-HA antibody. For lanes 1 and 5, 60% of the amount of protein used for lanes 2 and 6, respectively, was dissolved in SDS sample buffer and loaded. The position of HA-Eya3 fusion protein is indicated by the arrow. The position of IgG polypeptide, detected with the second antibody, is indicated by the arrowhead. Ipt, input; IP, immunoprecipitate. (B) Rat liver nuclear extract was incubated with anti-Six5 antibody (lanes 2 and 5) or purified rabbit IgG (lanes 3 and 6) and then precipitated by protein G agarose beads. The precipitates were dissolved in SDS sample buffer followed by Western blotting analysis with anti-Eya3 (lanes 2 and 3) or anti-Six5 (lanes 5 and 6) antibody. For lanes 1 and 4, 6% of the amount of protein used for lanes 2 and 3 and 5 and 6, respectively, was dissolved in SDS sample buffer and loaded. The positions of Eya3 (lanes 1 to 3) and Six5 (lane 4 to 6) are indicated by arrows. The position of IgG polypeptide, detected with the second antibody is indicated by the arrowhead. Ipt, input; IP, immunoprecipitate; Six5, Six5 antibody; IgG, purified rabbit IgG.
To test whether endogenous Six and Eya proteins form a complex in the nucleus in vivo, we performed immunoprecipitation analysis with a nuclear extract from rat liver, in which Six5 is known to be abundantly produced (25). The extract was incubated with anti-Six5 antibody or purified rabbit IgG as a control for 1.5 h at 4°C followed by Western blotting with anti-Eya3 serum or anti-Six5 antibody. Eya3 was coimmunoprecipitated with Six5 (Fig. 5B, lane 2) but not with rabbit IgG (Fig. 5B, lane 3). In addition, Six5 and Eya3 were coimmunoprecipitated from nuclear extracts prepared from P19 and HeLa cells (data not shown). These results indicate that complex formation between Six and Eya is relevant in vivo.
Domains necessary for nuclear translocation of Eya.
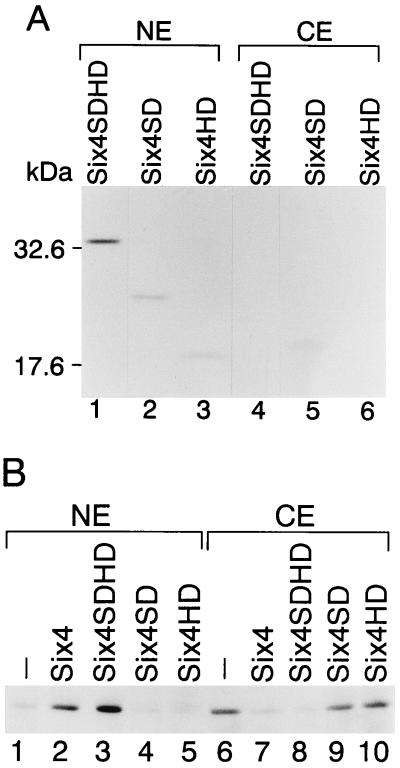
Nuclear translocation of three different Eya proteins was induced by either Six2, Six4, or Six5, suggesting that the conserved domains of Six and Eya are critical for the translocation. To analyze the involvement of conserved domains of Six in Eya translocation, we constructed expression plasmids for FLAG-Six4 fusion proteins containing Six domain only (SD), homeodomain alone (HD), and both Six domain and homeodomain (SDHD) (Fig. 1B). These fusion proteins were distributed in the nucleus when expressed in COS7 cells, indicating that each Six domain and homeodomain has an intrinsic nuclear localization signal (Fig. 6A). The distribution of Eya2 protein was analyzed in nuclear and cytoplasmic extracts from COS7 cells cotransfected with pfSix4SD, pfSix4HD, or pfSix4SDHD. Nuclear Eya2 was significantly increased by coexpression of full-length Six4 or Six4SDHD compared with that in the absence of Six4 coexpression (Fig. 6B, compare lanes 1 to 3 and 6 to 8). In contrast, coexpression of Six4SD or Six4HD did not increase nuclear Eya2 (Fig. 6B, lanes 4 to 5 and 9 to 10). These observations were confirmed by immunostaining (data not shown). The distribution of Eya3 was analyzed in a similar fashion (data not shown). The quantitative results of protein distribution analysis of Eya2 and Eya3 with coexpression of Six4 subdomain proteins are summarized in Table 2. Nuclear Eya2 markedly increased from 18.7 to 84.7% by coexpression of full-length Six4 or to 40.9% with Six4SDHD. In contrast, nuclear Eya2 did not increase with coexpression of Six4SD or Six4HD. Nuclear Eya3 significantly increased from 1.2 to 28.7% by coexpression of Six4 or to 14.3% with Six4SDHD and only marginally increased to 5.3 or 3.7% by coexpression of Six4SD and Six4HD, respectively. These results suggest that both the Six domain and homeodomain are necessary and sufficient for the nuclear translocation of Eya.
FIG. 6.
Domains essential for the nuclear translocation of Eya proteins. (A) FLAG-Six4SDHD (lanes 1 and 4), FLAG-Six4SD (lanes 2 and 5), and FLAG-Six4HD (lanes 3 and 6) were expressed in COS7 cells with HA-Eya2, and the nuclear (NE; lanes 1 to 3) and cytoplasmic (CE; lanes 4 to 6) extracts were prepared and analyzed by Western blotting with anti-FLAG antibody. Ten micrograms of protein from nuclear extracts for each of lanes 1 to 3 and 6.0, 7.5, and 6.7 μg of cytoplasmic extracts for lanes 4, 5, and 6, respectively, were analyzed. (B) HA-Eya3 was expressed alone (lanes 1 and 6) or with FLAG-Six4 (lanes 2 and 7), FLAG-Six4SDHD (lanes 3 and 8), FLAG-Six4SD (lanes 4 and 9), or FLAG-Six4HD (lanes 5 and 10). Nuclear (NE; lanes 1 to 5) and cytoplasmic (CE; lanes 6 to 10) extracts were analyzed by Western blotting with anti-HA antibody. Four micrograms of protein from nuclear extracts for each of lanes 1 to 5 and 10.8, 11.0, 7.4, 12.2, and 9.8 μg of protein from cytoplasmic extracts for lanes 6, 7, 8, 9, and 10, respectively, were used. The detected HA-Eya3 protein is shown.
TABLE 2.
Domains of Six4 protein required for Eya nuclear translocationa
| Six protein | Distribution (%)
|
|||
|---|---|---|---|---|
| Eya2
|
Eya3
|
|||
| Nucleus | Cytoplasm | Nucleus | Cytoplasm | |
| None | 18.7 | 81.3 | 1.2 | 98.8 |
| Six4 | 84.7 | 15.3 | 28.7 | 71.3 |
| Six4SDHD | 40.9 | 59.1 | 14.3 | 85.7 |
| Six4SD | 8.2 | 91.8 | 5.3 | 94.7 |
| Six4HD | 2.4 | 97.6 | 3.7 | 96.3 |
The relative amounts of nuclear and cytoplasmic Eya proteins from COS7 cells coexpressed with Six4 subdomain proteins were calculated as described for Table 1. The amount of DNA used for transfections was 10 μg each except for pfSix4SDHD (1 μg) to adjust the amounts of the expressed proteins. Two independent sets of experiments yielded essentially the same results.
To examine whether the conserved Eya domain is sufficient for translocation, we constructed expression plasmids containing only Eya domains (ED) from Eya1, Eya2, and Eya3 (Fig. 1D). We measured the distribution of Eya1ED, Eya2ED, and Eya3ED in nuclear and cytoplasmic extracts from transfected COS7 cells (Table 3). More than 90% of Eya1ED, Eya2ED, and Eya3ED were distributed in the cytoplasm without Six coexpression. Nuclear Eya1ED showed only a marginal increase from 4.6 to 8.4% by coexpression of Six5. Nuclear Eya2ED also increased, from 1.3 to 2.8%, by coexpression of Six4, and nuclear Eya3ED exhibited no increase (6.6 to 6.3%) by coexpression of Six5. The proportions of nuclear EyaED proteins were apparently lower than those of full-length Eya proteins coexpressed with Six4 or Six5 (Table 1). These results indicate that the Eya domain is not adequate for efficient nuclear translocation.
TABLE 3.
Eya domain is not sufficient for nuclear translocationa
| Six protein | Distribution (%)
|
|||||
|---|---|---|---|---|---|---|
| Eya1ED
|
Eya2ED
|
Eya3ED
|
||||
| Nucleus | Cytoplasm | Nucleus | Cytoplasm | Nucleus | Cytoplasm | |
| None | 4.6 | 95.4 | 1.3 | 98.7 | 6.6 | 93.4 |
| Six4 | NDb | ND | 2.8 | 97.2 | ND | ND |
| Six5 | 8.4 | 91.6 | ND | ND | 6.3 | 93.7 |
The relative amounts of nuclear and cytoplasmic Eya domain proteins from COS7 cells coexpressed with Six4 or Six5 were calculated as described for Table 1. The amount of DNA used for transfections was 10 μg each. Three independent sets of experiments yielded essentially the same results.
ND, not determined.
The results of these domain analyses suggest that Six proteins interact with Eya proteins through the conserved Six domain and homeodomain and recruit Eya proteins into the nucleus. Eya domain is not adequate for the interaction, but rather an additional domain is required, as discussed below.
Interaction between Six domain and homeodomain and Eya in yeast two-hybrid assay.
Nuclear translocation of Eya was observed by coexpression of Six4SDHD. This predicts a specific interaction between the conserved Six domain-homeodomain and Eya protein. To test whether the interaction is direct, we analyzed SixSDHD and Eya2 interaction in various assays. We did not observe binding of bacterially expressed Eya to immobilized GST-Six4SDHD or GST-Six4HD and could not detect Eya-Six4SDHD ternary complex formation on DNA containing a Six4 binding site by gel mobility shift assay (data not shown). Therefore, we used the more sensitive yeast two-hybrid assay and examined the binding of Six4SDHD to Eya2. Six4SDHD showed binding to full-length Eya2 but not to Eya2ED, consistent with the results of the nuclear translocation assay (Table 4). Eya2ΔED containing the N-terminal half of Eya2 but not the Eya domain did not show specific binding to the Six domain-homeodomain. To map the region required for the specific interaction of Eya2 with Six4SDHD, a series of N-terminal deletion proteins (Fig. 1E) were expressed in yeast, in addition to an examination of the interaction with Six4SDHD. Eya2NΔ1 and Eya2NΔ2 showed comparable binding to Six4SDHD with full-length Eya2. Eya2NΔ3 showed diminished binding based on X-Gal color development but still retained interaction (Table 4). These results indicate that the presence of the adjacent 62-amino-acid region in addition to Eya2ED is necessary for specific interaction between Six4SDHD and Eya2. Specific interaction between Six4 and Eya2 was also observed by yeast two-hybrid analysis (data not shown).
TABLE 4.
Interaction between various domains of Eya2 and Six4SDHDa
| Eya2 subdomain protein | Interaction with Six4SDHD |
|---|---|
| Eya2 | ++ |
| Eya2ED | − |
| Eya2ΔED | − |
| Eya2NΔ1 | ++ |
| Eya2NΔ2 | ++ |
| Eya2NΔ3 | + |
Yeast strain EGY48 was cotransformed with pJG4-5 constructs expressing various Eya2 subdomain proteins and pEG202Six4SDHD. An interaction was scored as strongly positive (++) or positive (+) if the transformant turned blue on X-Gal indicator plates containing galactose and negative (−) if it did not. The regions contained in pJG4-5 Eya2 constructs are shown in Fig. 1E.
Activation domain required for myogenin promoter activation.
To gain insight into the cooperative activation mechanism of Six and Eya proteins on the myogenin promoter, we tested the effect of the C-terminal domain of Six4 by comparing transactivation by full-length Six4 and Eya2 with that by Six4SDHD and Eya2. Transfection of pfSix4 resulted in threefold activation of the promoter, while cotransfection of pHM6Eya2 led to sevenfold activation. However, transfection of pfSix4SDHD resulted in twofold activation of the promoter, while cotransfection of pHM6Eya2 resulted in no further activation (Fig. 7A). These results suggest that although coexpression of Six4SDHD is sufficient for the nuclear translocation of Eya2, domains other than the Six domain-homeodomain of Six4 are required for activation and that the N-terminal region of Eya, which exhibits transactivation activity (39), could not simply act as a transactivation domain by tethering to a specific DNA site. We also compared transactivation by Six5 and Eya3 to that by Six5CΔ2, which lacks the potential C-terminal activation domain of Six5 (unpublished observation) (Fig. 1B) and Eya3. Compared to the 16-fold cooperative activation by Six5 and Eya3, Six5CΔ2 showed only a marginal activation of the myogenin promoter with Eya3 (Fig. 7B). Six5CΔ2 caused nuclear translocation of Eya3 as efficiently as Six5 (data not shown). This result suggests a vital role for the C-terminal region of Six5 in the cooperative activation with Eya3.
FIG. 7.
Six domain-homeodomain is not sufficient for transactivation of the myogenin promoter. The myogenin luciferase reporters pGL3MG-185 and pGL3MG-185M were cotransfected with pfSix and pHM6Eya plasmids. Luciferase activity in the cell lysate was normalized by β-galactosidase activity of pEFBOSβ-gal as an internal control. (A) Increasing amounts of pfSix4 (0, 0.1, or 0.2 μg) or pfSix4SDHD (0, 0.1, or 0.2 μg) was cotransfected with 0.5 μg of pHM6Eya2 and pHM6Eya2ED. (B) Increasing amounts of pfSix5 (0, 0.1, or 0.2 μg) or pfSix5CΔ2 (0, 0.02, and 0.04 μg to adjust the expressed protein amount in COS7 cells as Six5) was cotransfected with 0.5 μg of pHM6Eya3 or pHM6Eya3ED. The activity of each datum point is expressed relative to that of the control pFLAG-CMV-2 vector (−). The mean fold activation from three independent experiments (each performed in duplicate or triplicate) is shown with the standard deviation.
DISCUSSION
Cooperative activation of myogenin promoter by Six and Eya.
The Six4 protein was originally purified as a binding factor to the transcriptional regulatory region of the Na,K-ATPase α1 subunit gene, which is essential for the maintenance of the Na+ and K+ ion gradient across the cell membrane (17). More than eight factors can interact with the most important regulatory region of the gene, termed ARE (19, 33). Coexpression of Six and Eya proteins showed only a marginal effect on promoter activity in transient transfection assays (unpublished observation), probably because other factors had already activated the promoter through the ARE. However, cooperative activation was observed when a synthetic promoter containing multimerized Six4 binding sites derived from the ARE was used (Fig. 3). A recent finding that the myogenin promoter is controlled by the Six1 and Six4 proteins through a conserved MEF3 binding site (32) prompted us to test the cooperative effect of Six and Eya on the promoter. A combination of Six2-Eya1, Six4-Eya2, and Six5-Eya3 exhibited the most prominent activation of the myogenin promoter in COS7 cells compared with other combinations of Six and Eya (Fig. 2). These results are the first indication of cooperation between Six and Eya proteins in the transcriptional activation of their target genes. Myogenin is a key regulator for skeletal muscle development, and we observed a five- to eightfold increase in promoter activity by coexpression of Six5 with Eya2 or Eya3. In patients with DM, the CTG-repeat expansion results in a reduction in the expression level of SIX5 mRNA (20, 34), which may in turn reduces the expression of its target genes and causes muscle immaturity (30). Our observation that Six5 can activate the myogenin promoter with Eya protein is consistent with the notion that the reduced expression of SIX5 causes immaturity of skeletal muscles through reduced myogenin expression in some patients with DM. However, considering the fact that Six2 and Six4 can also activate the myogenin promoter, the Six and Eya proteins that are genuinely involved in the regulation of myogenin during muscle development must be carefully identified.
Nuclear translocation of Eya proteins by Six.
Six proteins reside in the nucleus, while most Eya proteins are located in the cytoplasm when they are expressed separately in COS7 cells. Coexpression of Six induced a nuclear translocation of Eya (Fig. 4 and Table 1). Although there were some differences in the efficiency of nuclear translocation among various combinations of Six and Eya, Six2, Six4, and Six5 could translocate any of the Eya proteins examined. Nuclear translocation of Eya is a prerequisite for the cooperative activation of their target genes. In contrast, Six3 never induced translocation of any Eya protein. Phylogenetic analysis of various Six family genes based on amino acid sequence similarity revealed that there are three major classes of Six genes (unpublished observation): a group including Six1 and Six2, another group containing Six3 and Optx2/Six9, and a third group containing Six4 and Six5. Considering the evolutionary aspects of Six family genes, Six3 may interact with Eya gene family products other than Eya1, Eya2, and Eya3 or other as-yet-unidentified comolecules distinct from Eya proteins.
β-Catenin, known as a coactivator of the high-mobility-group protein LEF-1, directly interacts with LEF-1 and translocates into the nucleus (2, 14, 36). Such translocation is regulated by Wnt signaling (13). It is possible that nuclear translocation of Eya by Six might be regulated by an as-yet-unidentified signaling pathway.
Direct interaction of Six and Eya.
Translocation of Eya by Six suggests a direct interaction between Six and Eya. In fact, Six4-Eya2 and Six5-Eya3 complexes were immunoprecipitated from nuclear extracts of coexpressed COS7 cells and of rat liver (Fig. 5). The interaction between the Six domain-homeodomain of Six4 and full-length Eya2 was observed in yeast two-hybrid analysis (Table 4). Because Drosophila So and Eya interact through their conserved Six and Eya domains by yeast two-hybrid analysis (29), the Six domain of mouse Six was expected to be sufficient for interaction with mouse Eya. Surprisingly, both Six4SD and Six4HD were necessary for the translocation and Eya2ED alone was not sufficient for the translocation (Fig. 6B and Tables 2 and 3). Therefore, we also tested the interaction of Eya2 with Six4SD and Six4HD separately. A similar degree of interaction was observed between Eya2 and Six4SD but not between Eya2 and Six4HD in yeast two-hybrid analysis (data not shown). However, nuclear translocation did not occur when we coexpressed Six4SD and Eya2 in COS7 cells (Fig. 6B). Thus, the interaction between the Six domain alone and the Eya protein does not seem to be sufficient for nuclear translocation (Fig. 8). This suggests that involvement of another factor or conformational changes of the Six-Eya complex induced by the homeodomain might be necessary for efficient translocation in addition to the specific interaction between the Six domain and the Eya protein.
FIG. 8.
Summary of domain analyses of Six4 and Eya2. The regions necessary for the interaction were analyzed by yeast two-hybrid analysis, those for nuclear translocation were analyzed by Western blotting of nuclear and cytoplasmic fractions of transfected COS7 cells, and those for transcriptional activation were analyzed by reporter gene assay with myogenin promoter.
Temporal and spatial overlapping expression of Six and Eya genes.
Cooperation between Six and Eya was manifested by the reporter gene assays and nuclear translocation assays described above (Fig. 2 to 4 and Table 1). If these cooperative interactions are relevant in vivo, both Six and Eya proteins should exist at the same location in similar developmental stages. In addition to our observation of the colocalization of Six5 and Eya3 in the nuclei of adult rat livers, analyses of the expression patterns of Six and Eya genes in mouse development also indicated colocalization of mRNAs or proteins of both genes. For example, Eya1 and Eya2 mRNAs are expressed in the head mesenchyme and in the presomitic mesoderm at E8.5 and in brain, pharyngeal pouch, nephrogenic cord, and branchial arches at E9.5 to E10.5 (40). Furthermore, Six2 mRNA is distributed in the head mesoderm and paraxial mesoderm, including somites, at E8.5 and is expressed in the otic vesicle, presomitic mesoderm, and nephrogenic cord at E9.5 (28). Six4 protein is detected in the brain at E9.5 to E11.5, and Six2 protein is also found in the nephrogenic cord at E10.5 to E12.5 (25). We also observed a strong expression of the lacZ gene in branchial arches of mice harboring a Six4-lacZ fusion gene (unpublished observation). Eya1 and Eya2 mRNAs are expressed in various ganglia, such as facioacoustic ganglia (VII to VIII) and glossopharyngeal (IX) and vagus (X) ganglia, and Eya2 is expressed in trigeminal (V) ganglia (40), where Six4 proteins are produced at E10.5 to E11.5 (25). Moreover, Eya3 mRNA is expressed in the head and branchial arch mesenchyme and the limbs at E9.5 to E10.5 (40), while the Six2 gene is expressed in the head mesenchyme at E9.5 to E10.5 or in the limbs at E12.5 (28). The precise expression pattern of the Six5 gene has not been determined; however, the expression of Six5 mRNA is observed as early as E7 through E17 by Northern analysis (24). Eya3 expression is also detected from E7 through the embryonic period (42). Abundant expression of Eya1 mRNA in the heart and skeletal muscles of adult mice resembles the expression pattern of Six5 mRNA in adult mice (1, 24). Thus, coexpression of various combinations of the Eya gene and Six genes occurs in mouse embryos and also in adults.
Coactivation mechanism of Six and Eya.
Mouse Eya genes show specific temporal and spatial expression patterns and are thought to be involved in differentiation and morphogenesis (6, 15, 40). The results of several experiments in the present study indicated that Eya can function as a coactivator of Six. Thus, Eya is considered to be a tissue-specific coactivator involved in differentiation and morphogenesis.
Six4 is known to have an intrinsic activation domain in its C-terminal domain (17). The N-terminal portions of Eya1, Eya2, and Eya3 exhibit transactivation activities (39). A simple model for cooperative activation is that the association of Eya with Six could simply serve to tether the activation domain of Eya to specific sites in DNA. Alternatively, the activation domains of Eya could collaborate with other regions of Six to activate target gene transcription. In the case of Six4, it was clearly seen that the Six domain-homeodomain was not sufficient for cooperative activation with Eya2 (Fig. 7A). In the case of Six5, it was demonstrated that the C-terminal region deleted from Six5CΔ2 was essential for cooperative activation with Eya3. Thus, the potential activation domain of Eya2 or Eya3 did not work properly, at least with the Six domain-homeodomain alone, which is sufficient to cause nuclear translocation of Eya (Fig. 6 and 8). This might suggest that the potential activation domain situated in the C-terminal region of Six4 or Six5 is unmasked by interaction with Eya or that both activation domains of Six and Eya give a composite activation domain surface. Detailed domain analyses are necessary to identify the precise cooperative activation mechanism. Furthermore, we cannot rule out the possibility that this mechanism of action might vary, depending on the promoters. Such analysis is currently under way in our laboratory.
Eya has been reported to be associated with Dac and Msx (8, 38), suggesting that Eya acts as a coactivator of several transcription factors and integrates their effects. CBP/p300 coactivator interacts with many transcription factors and has been shown to act as an integrator of diverse signal transduction pathways (3, 16). The Eya gene family is also thought to have similar functional properties, in the sense that they form a complex gene network with various types of transcription factors and integrate their diverse regulatory pathways.
ACKNOWLEDGMENTS
We thank N. Bonini for providing Eya2 and Eya3 cDNAs, A. Fujisawa-Sehara for providing myogenin promoter, and R. Brent for allowing us to use the yeast two-hybrid LexA system. We also thank M. Kobayashi for preparing rat liver nuclear extract, P. Xu for communicating unpublished results, and K. Ikeda for useful discussion.
This work was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan and from the Ministry of Health and Welfare of Japan.
REFERENCES
- 1.Abdelhak S, Kalatzis V, Heilig R, Compain S, Samson D, Vincent C, Weil D, Cruaud C, Sahly I, Leibovici M, Bitner-Glindzicz M, Francis M, Lacombe D, Vigneron J, Charachon R, Boven K, Bedbeder P, Van Regemorter N, Weissenbach J, Petit C. A human homologue of the Drosophila eyes absent gene underlies branchio-oto-renal (BOR) syndrome and identifies a novel gene family. Nat Genet. 1997;15:157–164. doi: 10.1038/ng0297-157. [DOI] [PubMed] [Google Scholar]
- 2.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya S, Michels C L, Leung M K, Arany Z P, Kung A L, Livingston D M. Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev. 1999;13:64–75. doi: 10.1101/gad.13.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonini N M, Bui Q T, Gray-Board G L, Warrick J M. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development. 1997;124:4819–4826. doi: 10.1242/dev.124.23.4819. [DOI] [PubMed] [Google Scholar]
- 5.Bonini N M, Leiserson W M, Benzer S. The eyes absent gene: genetic control of cell survival and differentiation in the developing Drosophila eye. Cell. 1993;72:379–395. doi: 10.1016/0092-8674(93)90115-7. [DOI] [PubMed] [Google Scholar]
- 6.Borsani G, DeGrandi A, Ballabio A, Bulfone A, Bernard L, Banfi S, Gattuso C, Mariani M, Dixon M, Donnai D, Metcalfe K, Winter R, Robertson M, Axton R, Brown A, van Heyningen V, Hanson I. EYA4, a novel vertebrate gene related to drosophila eyes absent. Hum Mol Genet. 1999;8:11–23. doi: 10.1093/hmg/8.1.11. [DOI] [PubMed] [Google Scholar]
- 7.Boucher C A, King S K, Carey N, Krahe R, Winchester C L, Rahman S, Creavin T, Meghji P, Bailey M E, Chartier F L, et al. A novel homeodomain-encoding gene is associated with a large CpG island interrupted by the myotonic dystrophy unstable (CTG)n repeat. Hum Mol Genet. 1995;4:1919–1925. doi: 10.1093/hmg/4.10.1919. [DOI] [PubMed] [Google Scholar]
- 8.Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- 9.Cheyette B N, Green P J, Martin K, Garren H, Hartenstein V, Zipursky S L. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–996. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 10.Duncan M K, Kos L, Jenkins N A, Gilbert D J, Copeland N G, Tomarev S I. Eyes absent: a gene family found in several metazoan phyla. Mamm Genome. 1997;8:479–485. doi: 10.1007/s003359900480. [DOI] [PubMed] [Google Scholar]
- 11.Fujisawa-Sehara A, Hanaoka K, Hayasaka M, Hiromasa-Yagami T, Nabeshima Y. Upstream region of the myogenin gene confers transcriptional activation in muscle cell lineages during mouse embryogenesis. Biochem Biophys Res Commun. 1993;191:351–356. doi: 10.1006/bbrc.1993.1224. [DOI] [PubMed] [Google Scholar]
- 12.Heath S K, Carne S, Hoyle C, Johnson K J, Wells D J. Characterisation of expression of mDMAHP, a homeodomain-encoding gene at the murine DM locus. Hum Mol Genet. 1997;6:651–657. doi: 10.1093/hmg/6.5.651. [DOI] [PubMed] [Google Scholar]
- 13.Hsu S C, Galceran J, Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol Cell Biol. 1998;18:4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann B G, Kemler R. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech Dev. 1996;59:3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- 15.Kalatzis V, Sahly I, El-Amraoui A, Petit C. Eya1 expression in the developing ear and kidney: towards the understanding of the pathogenesis of Branchio-Oto-Renal (BOR) syndrome. Dev Dyn. 1998;213:486–499. doi: 10.1002/(SICI)1097-0177(199812)213:4<486::AID-AJA13>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 16.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 17.Kawakami K, Ohto H, Ikeda K, Roeder R G. Structure, function and expression of a murine homeobox protein AREC3, a homologue of Drosophila sine oculis gene product, and implication in development. Nucleic Acids Res. 1996;24:303–310. doi: 10.1093/nar/24.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawakami K, Ohto H, Takizawa T, Saito T. Identification and expression of six family genes in mouse retina. FEBS Lett. 1996;393:259–263. doi: 10.1016/0014-5793(96)00899-x. [DOI] [PubMed] [Google Scholar]
- 19.Kawakami K, Yanagisawa K, Watanabe Y, Tominaga S, Nagano K. Different factors bind to the regulatory region of the Na+,K+-ATPase alpha 1-subunit gene during the cell cycle. FEBS Lett. 1993;335:251–254. doi: 10.1016/0014-5793(93)80740-l. [DOI] [PubMed] [Google Scholar]
- 20.Klesert T R, Otten A D, Bird T D, Tapscott S J. Trinucleotide repeat expansion at the myotonic dystrophy locus reduces expression of DMAHP. Nat Genet. 1997;16:402–406. doi: 10.1038/ng0897-402. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi M, Toyama R, Takeda H, Dawid I B, Kawakami K. Overexpression of the forebrain-specific homeobox gene six3 induces rostral forebrain enlargement in zebrafish. Development. 1998;125:2973–2982. doi: 10.1242/dev.125.15.2973. [DOI] [PubMed] [Google Scholar]
- 22.Loosli F, Winkler S, Wittbrodt J. Six3 overexpression initiates the formation of ectopic retina. Genes Dev. 1999;13:649–654. doi: 10.1101/gad.13.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luckow B, Schutz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murakami Y, Ohto H, Ikeda U, Shimada K, Momoi T, Kawakami K. Promoter of mDMAHP/Six5: differential utilization of multiple transcription initiation sites and positive/negative regulatory elements. Hum Mol Genet. 1998;7:2103–2112. doi: 10.1093/hmg/7.13.2103. [DOI] [PubMed] [Google Scholar]
- 25.Ohto H, Takizawa T, Saito T, Kobayashi M, Ikeda K, Kawakami K. Tissue and developmental distribution of Six family gene products. Int J Dev Biol. 1998;42:141–148. [PubMed] [Google Scholar]
- 26.Oliver G, Loosli F, Koster R, Wittbrodt J, Gruss P. Ectopic lens induction in fish in response to the murine homeobox gene Six3. Mech Dev. 1996;60:233–239. doi: 10.1016/s0925-4773(96)00632-6. [DOI] [PubMed] [Google Scholar]
- 27.Oliver G, Mailhos A, Wehr R, Copeland N G, Jenkins N A, Gruss P. Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development. 1995;121:4045–4055. doi: 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- 28.Oliver G, Wehr R, Jenkins N A, Copeland N G, Cheyette B N, Hartenstein V, Zipursky S L, Gruss P. Homeobox genes and connective tissue patterning. Development. 1995;121:693–705. doi: 10.1242/dev.121.3.693. [DOI] [PubMed] [Google Scholar]
- 29.Pignoni F, Hu B, Zavitz K H, Xiao J, Garrity P A, Zipursky S L. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- 30.Sarnat H B. New insights into the pathogenesis of congenital myopathies. J Child Neurol. 1994;9:193–201. doi: 10.1177/088307389400900218. [DOI] [PubMed] [Google Scholar]
- 31.Serikaku M A, O’Tousa J E. sine oculis is a homeobox gene required for Drosophila visual system development. Genetics. 1994;138:1137–1150. doi: 10.1093/genetics/138.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spitz F, Demignon J, Porteu A, Kahn A, Concordet J P, Daegelen D, Maire P. Expression of myogenin during embryogenesis is controlled by Six/sine oculis homeoproteins through a conserved MEF3 binding site. Proc Natl Acad Sci USA. 1998;95:14220–14225. doi: 10.1073/pnas.95.24.14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki-Yagawa Y, Kawakami K, Nagano K. Housekeeping Na,K-ATPase alpha 1 subunit gene promoter is composed of multiple cis elements to which common and cell-type-specific factors bind. Mol Cell Biol. 1992;12:4046–4055. doi: 10.1128/mcb.12.9.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thornton C A, Wymer J P, Simmons Z, McClain C, Moxley R T., III Expansion of the myotonic dystrophy CTG repeat reduces expression of the flanking DMAHP gene. Nat Genet. 1997;16:407–409. doi: 10.1038/ng0897-407. [DOI] [PubMed] [Google Scholar]
- 35.Toy J, Sundin O H. Expression of the Optx2 homeobox gene during mouse development. Mech Dev. 1999;83:183–186. doi: 10.1016/s0925-4773(99)00049-0. [DOI] [PubMed] [Google Scholar]
- 36.van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, Peifer M, Mortin M, Clevers H. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 37.Winchester C L, Ferrier R K, Sermoni A, Clark B J, Johnson K J. Characterization of the expression of DMPK and SIX5 in the human eye and implications for pathogenesis in myotonic dystrophy. Hum Mol Genet. 1999;8:481–492. doi: 10.1093/hmg/8.3.481. [DOI] [PubMed] [Google Scholar]
- 38.Xu, P. X. Personal communication.
- 39.Xu P X, Cheng J, Epstein J A, Maas R L. Mouse Eya genes are expressed during limb tendon development and encode a transcriptional activation function. Proc Natl Acad Sci USA. 1997;94:11974–11979. doi: 10.1073/pnas.94.22.11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu P X, Woo I, Her H, Beier D R, Maas R L. Mouse Eya homologues of the Drosophila eyes absent gene require Pax6 for expression in lens and nasal placode. Development. 1997;124:219–231. doi: 10.1242/dev.124.1.219. [DOI] [PubMed] [Google Scholar]
- 41.Zervos A S, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 42.Zimmerman J E, Bui Q T, Steingrimsson E, Nagle D L, Fu W, Genin A, Spinner N B, Copeland N G, Jenkins N A, Bucan M, Bonini N M. Cloning and characterization of two vertebrate homologs of the Drosophila eyes absent gene. Genome Res. 1997;7:128–141. doi: 10.1101/gr.7.2.128. [DOI] [PubMed] [Google Scholar]