Abstract
The four members of the ErbB family of receptor tyrosine kinases are involved in a complex array of combinatorial interactions involving homo- and heterodimers. Since most cell types express more than one member of the ErbB family, it is difficult to distinguish the biological activities of different homo- and heterodimers. Here we describe a method for inducing homo- or heterodimerization of ErbB receptors by using synthetic ligands without interference from the endogenous receptors. ErbB receptor chimeras containing synthetic ligand binding domains (FK506-binding protein [FKBP] or FKBP-rapamycin-binding domain [FRB]) were homodimerized with the bivalent FKBP ligand AP1510 and heterodimerized with the bifunctional FKBP-FRB ligand rapamycin. AP1510 treatment induced tyrosine phosphorylation of ErbB1 and ErbB2 homodimers and recruitment of Src homology 2 domain-containing proteins (Shc and Grb2). In addition, ErbB1 and ErbB2 homodimers activated downstream signaling pathways leading to Erk2 and Akt phosphorylation. However, only ErbB1 homodimers were internalized upon AP1510 stimulation, and only ErbB1 homodimers were able to associate with and induce phosphorylation of c-Cbl. Cells expressing AP1510-induced ErbB1 homodimers were able to associate with and induce phosphorylation of c-Cbl. Cells expressing AP1510-induced ErbB1 homodimers were able to form foci; however, cells expressing ErbB2 homodimers displayed a five- to sevenfold higher focus-forming ability. Using rapamycin-inducible heterodimerization we show that c-Cbl is unable to associate with ErbB1 in a ErbB1-ErbB2 heterodimer most likely because ErbB2 is unable to phosphorylate the c-Cbl binding site on ErbB1. Thus, we demonstrate that ErbB1 and ErbB2 homodimers differ in their abilities to transform fibroblasts and provide evidence for differential signaling by ErbB homodimers and heterodimers. These observations also validate the use of synthetic ligands to study the signaling and biological specificity of selected ErbB dimers in any cell type.
ErbB family receptor tyrosine kinases (RTKs) ErbB1 (also known as human epidermal growth factor [EGF] receptor 1 [HER1] or EGF receptor [EGFR]), ErbB2 (also known as HER2 or Neu), ErbB3, and ErbB4 consist of an extracellular ligand-binding domain, a single transmembrane domain, an uninterrupted tyrosine kinase domain, and a cytoplasmic tail. The ErbB family members play important roles during the growth and development of a number of organs including the heart (9, 17), the mammary gland (9, 27, 77), and the central nervous system (9, 17, 28). In addition, ErbB overexpression is associated with tumorigenesis of the breast, ovaries, brain, and prostate gland (1, 9, 36). Experiments in transgenic mice and cell culture models clearly indicate that ErbB receptors and their ligands can promote the development and progression of mammary tumorigenesis (36, 74).
There are at least 16 different EGF family ligands that bind ErbB receptors (55). The ligands can be grouped into three categories: (i) those that bind to ErbB1 alone (EGF, transforming growth factor α, and amphiregulin), (ii) those that bind to ErbB3 and ErbB4 (neuregulin 1 [NRG1] and NRG2), and (iii) those that bind to ErbB1 and ErbB4 (betacellulin, heparin-binding EGF, NRG3, and epiregulin) (32, 55). The binding of an EGF family ligand to its cognate receptor results in the dimerization and activation of the receptor (76).
ErbB family members partake in a complex process of lateral signaling (also referred to as combinatorial interactions) by forming ligand-induced heterodimers between different family members (1, 18, 55). It is likely that heterodimerization is mediated by ligand bivalency (71). Each ligand has been shown to favor certain dimeric combinations over others, suggesting that in cells expressing all four ErbB receptors a given ligand induces a hierarchical order of ErbB receptor dimerization (72). Among the ErbB RTKs, ErbB2-containing heterodimers are preferred over other ErbB homo- or heterodimers, suggesting that ErbB2 plays a central role in both ligand binding and signal transduction (4, 29, 37, 52, 62, 73). EGF stimulation of cells engineered to lack surface expression of ErbB1 results in defective ErbB2 phosphorylation, and EGF or NRG1 stimulation of cells engineered to lack surface expression of ErbB2 results in impaired ErbB1, ErbB3, and ErbB4 phosphorylation (30). Taken together, these observations highlight the importance of combinatorial interactions in ErbB receptor signaling. Although ErbB2 is recruited into many heterodimers it is likely that different heterodimers have distinct signaling specificities. The evidence that different ligands induce distinct phosphorylation patterns on ErbB1 and ErbB2 is consistent with this possibility (19a, 51).
The activation of ErbB receptors results in the generation of Src homology 2 (SH2)-binding sites for multiple cytoplasmic signaling molecules such as the p85 subunit of phosphoinositide 3′-kinase (PI 3-kinase) (61), phospholipase Cγ1 (16), Src family kinases (5), protein tyrosine phosphatases, SH2 domain-containing tyrosine phosphatases 1 and 2 (25), Shc and Grb2 (13), Grb7 (20), Grb10 (20), c-Cbl (44, 47), Nck (6), Crk (6), Eps8 (22), and Eps15 (23). ErbB receptors also induce tyrosine phosphorylation of proteins involved in cell adhesion signaling such as the focal adhesion kinase (54), Crk-associated substrate (Cas) (50), paxillin (58), cortactin (8), and catenins (35). It is likely that different ErbB dimers recruit or activate different sets of signaling molecules. For example, the p85 subunit of PI 3-kinase is thought to associate only with ErbB3 (24, 39, 53, 64), c-Cbl with ErbB1 (41), and Chk with ErbB2 (80). c-Src associates with both ErbB1 and ErbB2, though it appears that c-Src prefers ErbB2 over ErbB1 (49). Very little is known about how ErbB homodimers and heterodimers differ in their biological properties, and it is also not known whether heterodimers possess unique signaling properties compared to homodimers. Since almost all fibroblasts and mammary epithelial cells express more than one member of the ErbB receptor family, it is not possible to determine the signaling and biological specificities of different ErbB receptor homo- or heterodimers with natural peptide ligands. In this report, we demonstrate that synthetic dimerizing ligands can be used effectively to study the homo- and heterodimerization of chimeric ErbB receptors independent of endogenous receptors and their ligands.
Synthetic dimerizing ligands have been used to induce the dimerization and activation of transcription factors (33, 57), T-cell receptor subunits (70), Src family kinases (69), the guanine nucleotide exchange factor SOS (34), platelet-derived growth factor receptor (78), caspases (45), Fas receptor (68), erythropoetin receptor (7), and integrins (31). Here we demonstrate that synthetic ligand-induced homodimerization of either ErbB1 or ErbB2 in rat fibroblasts results in tyrosine phosphorylation of the receptor, phosphorylation of downstream signaling molecules in a kinase-specific manner, induction of DNA synthesis, ligand-dependent focus formation, and ligand-dependent acquisition of transformed morphology. Our results also indicate that ErbB1 homodimers were five- to sevenfold weaker in their ability to induce focus formation than ErbB2 homodimers. In addition, using a synthetic ligand that selectively induce heterodimers, we demonstrate that c-Cbl prefers ErbB1 in a homodimer over ErbB1 in a heterodimer with ErbB2, suggesting that homo- and heterodimers recruit distinct cytoplasmic signaling proteins.
MATERIALS AND METHODS
DNA constructs.
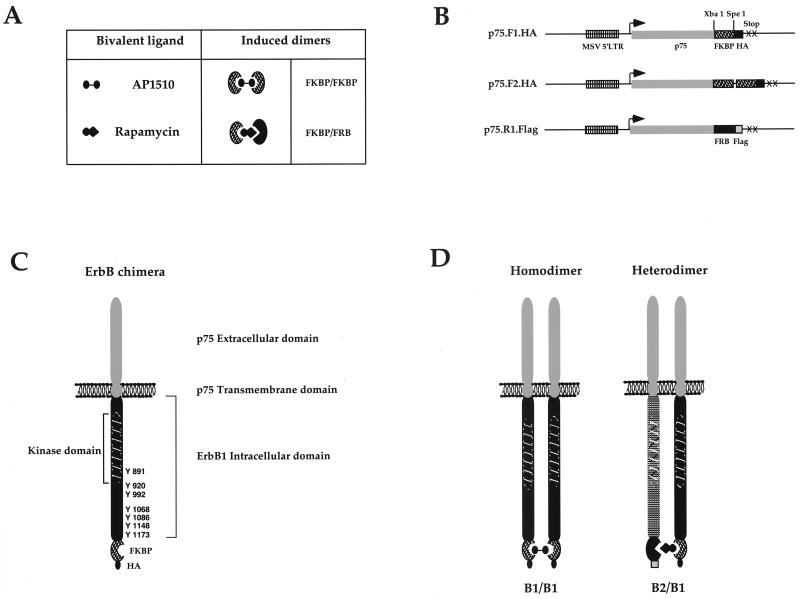
The expression vectors for the ErbB chimeras were constructed as follows: the extracellular and transmembrane domains of low-affinity nerve growth factor receptor (p75) was PCR amplified as an EcoRI/BamHI fragment and subcloned into the retroviral expression vector SRαMSVTKNeo (kindly provided by O. Witte). To generate the EcoRI/BamHI fragment, the 5′ primer was engineered to have an EcoRI site and the 3′ primer was engineered to have in-frame SpeI and XbaI sites followed by either a hemagglutinin (HA) or Flag epitope tag, stop codons, and BamHI restriction site. The resulting vectors were referred to as either p75.HA or p75.Flag. The ligand-binding domains FK506-binding protein (FKBP [one or two copies]) and FKBP-rapamycin-binding domain (FRB) of FKBP-rapamycin-associated protein (FRAP) (57) were subcloned as XbaI/SpeI fragments into p75.HA and p75.Flag to generate p75.F1.HA (F1, one copy of FKBP) or p75.F2.HA (F2, two copies of FKBP) or p75.R1.Flag (R1, one copy of FRB domain) (Fig. 1B). The intracellular domains of ErbB1 (B1) and ErbB2 (B2) were obtained by PCR with Pfu DNA polymerase (Stratagene). The primers were designed such that they contain in-frame XbaI and SpeI restriction sites in the 5′ and 3′ ends, respectively. ErbB1 was amplified with the primers 5′ GCGATCTCTAGACGAAGGCGGCCACATCGTTCGG and 5′ GCATCGACTAGTTGCTCCAATAAATTCACTGCTTTG with a T47D cDNA library (generated by random priming poly[A]-selected mRNA). The kinase-dead ErbB1 (kdB1) was amplified with the Met721Ala mutant human ErbB1 cDNA (kindly provided by Alan Wells). Both ErbB1 and kdB1 PCR fragments were subcloned into a shuttle vector digested with XbaI and SpeI restriction enzymes. The ErbB2 cytoplasmic domain was amplified as two fragments from a rat Neu cDNA (kindly provided by William J. Muller), making use of an internal unique NcoI site. The 5′ fragment was amplified with primers 5′ GCGATCTCTAGAAAACGAAGGAGACAGAAGATCC and 5′ GGAGGTCGGGGTACCTGTCATGG. The 3′ fragment was amplified with primers 5′ CCATCCAGCCCCATGGACAGTACC and 5′ GCATCGACTAGTTACAGGTACATCCAGGCCTAGG. The 5′ and 3′ fragments were subcloned into an XbaI/SpeI cut shuttle vector by a three-way ligation. The ErbB1 and ErbB2 PCR products, in shuttle vectors, were subjected to automated sequencing to verify the nucleotide sequence. The intracellular domains of ErbB1, ErbB2, and kdErbB1 were subcloned as XbaI/SpeI fragments into the XbaI site in p75 fusion vectors (Fig. 1B) to generate p75.B1.F1.HA, p75.B1.F2.HA, p75.B2.F2.HA, and p75.kdB1.R1.Flag.
FIG. 1.
Synthetic ligands for ErbB dimerization. The ligand-binding domains FKBP12 and FRB (A) were subcloned as single or double copies to generate the expression vectors shown in panel B. The intracellular domains of ErbB receptors were PCR amplified and subcloned into the expression vectors as described in Materials and Methods. The addition of AP1510 to cells expressing FKBP-fused ErbB receptors (C) will result in the generation of homodimers (D), while the addition of rapamycin to cells coexpressing the ErbB1-FKBP chimera (p75.B1.F1.HA) and ErbB2-FRB chimera (p75.B2.R1.Flag) will result in a p75.B1.F1.HA-p75.B2.R1.Flag heterodimer (D).
Retroviral stocks.
Retroviral stocks were prepared by using the Phoenix packaging cells and following a previously described protocol (49a). The viral stocks were stored at −80°C.
Cell culture and stable cell lines.
Rat1 cells (kindly provided by Peter Siegel and William J. Muller) were grown in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum (FBS) and antibiotics. Stable cell lines expressing p75.B1.F1.HA, p75.B1.F2.HA, and p75.B2.F2.HA were derived by infecting Rat1 fibroblasts with retrovirus expressing the respective RTK chimera and selecting infected cells with 500-μg/ml G418-containing media. Clones were screened by either anti-HA blots or by fluorescence-activated cell sorting (FACS) of cells stained with anti-p75 antibodies and fluorescein isothiocyanate-conjugated anti-mouse secondary antibodies. Clones that showed comparable levels of p75 surface staining were used for the experiments (p75.B1.F1.HA, clone 9; p75.B1.F2.HA, clone 3 or 6; and p75.B2.F2.HA, clone 4). Cells coexpressing FKBP and FRB (p75.kd.B1.R1.Flag) in fusion were derived by transfecting the p75.B1.F2 (clone 6)- and p75.B2.F2 (clone 4)-expressing cells with the p75.kdB1.R1.Flag and pBabe Hygro (48) and selected in media containing 200 μg of hygromycin (Boehringer Mannheim) per ml. Hygromycin-resistant colonies were pooled, and early passages (between 3 and 10) were used for heterodimerization experiments (see Fig. 9). Transient assays in COS7 cells (see Fig. 10) were carried out by plating 1.2 × 106 cells per 10-cm-diameter plate, and lipofection was carried out 16 to 18 h after plating. The lipofection mix was prepared with 30 μl of Lipofectamine, 3.0 μg of p75.kdB1.R1.Flag, and 1.0 μg of either p75.B1.F2.HA or p75.B2.F2.HA following the manufacturer’s protocol (Gibco-BRL). Lipofection was carried out for 5 h, and the cells were analyzed 48 h after transfection.
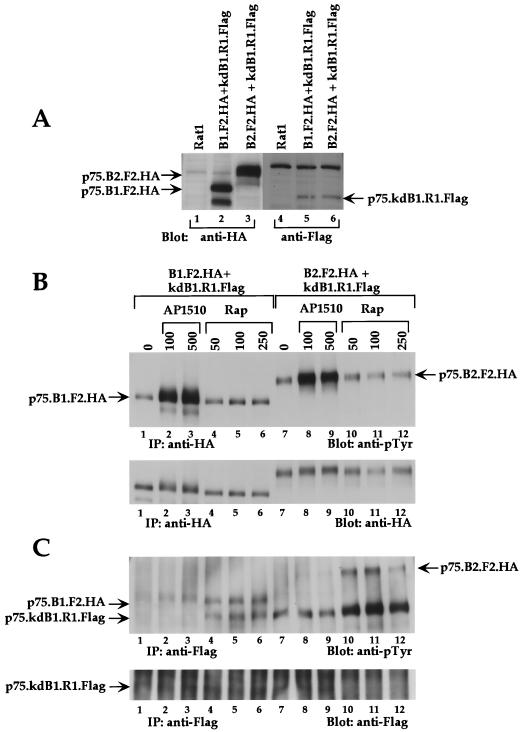
FIG. 9.
Synthetic ligand-induced heterodimerization between ErbB receptors. (A) P75.B1.F2.HA- and p75.B2.F2.HA-expressing cells were transfected with p75.kdB1.R1.Flag, and stable pools containing B1.F2.HA plus kdB1.R1.Flag and B2.F2.HA plus kdB1.R1.Flag were selected. The relative expression levels of each chimera in both pools were examined by immunoblotting cell lysates with either anti-HA or anti-Flag antibodies. The parental Rat1 cell lysate was used as a negative control. The pools were stimulated with either AP1510 (concentrations shown are nanomolar) (lanes 2, 3, 8, and 9) or with rapamycin (nanomolar) (lanes 4 to 6 and 10 to 12), immunoprecipitated (IP) with either anti-HA (B) or anti-Flag (C) antibodies, and immunoblotted with anti-pTyr antibodies. The blots in the upper panels (B and C) were stripped and reprobed with anti-HA and anti-Flag antibodies, respectively (lower panels).
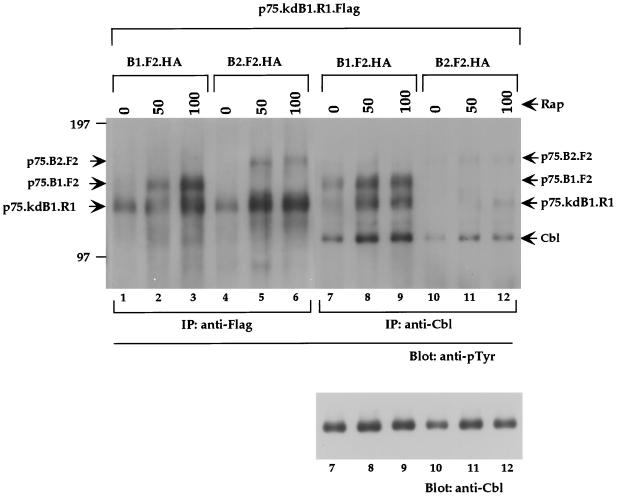
FIG. 10.
c-Cbl can differentiate ErbB1 in homodimers from ErbB1 in ErbB1-ErbB2 heterodimers. COS7 cells were cotransfected with p75.kdB1.R1.Flag and p75.B1.F2.HA (lanes 1 to 3 and 7 to 9) or p75.B2.F2.HA (lanes 4 to 6 and 10 to 12). The cells were stimulated with indicated amounts of rapamycin (nanomolar), and 1.5 mg of lysate was used for immunoprecipitation (IP) with anti-Flag antibodies (lanes 1 to 6) or anti-Cbl antibodies (lanes 7 to 12) and immunoblotted with anti-pTyr antibodies (upper panel). The c-Cbl portion of the blot (lanes 7 to 12) was stripped and reprobed with anti-Cbl antibodies (lower panel).
Cell lysis and immunoprecipitation.
Subconfluent or confluent cultures were stimulated with indicated amounts of AP1510 or rapamycin for 15 min at 37°C. The ligands were stored as a x2,000 stock in 100% ethanol at −20°C. After stimulation, the cells were rinsed once with ice-cold phosphate-buffered saline (PBS) containing 1 mM sodium orthovanadate and lysed in Triton X-100 lysis buffer (150 mM NaCl, 50 mM Tris · Cl [pH 8.0], 5 mM NaF, 1% Triton X-100, 1 mM sodium orthovanadate, 5 μg of aprotinin per ml, and 5 μg of leupeptin per ml) for 25 to 35 min. The lysates were cleared by centrifugation at 16,000 × g for 15 min at 4°C. Protein concentrations were measured by using the Bradford assay (Bio-Rad). Cell lysates were incubated with anti-HA (HA.11; BabCo) or anti-flag (anti-Flag M2 beads; Sigma) or anti-Cbl (SC 14; Santacruz) antibodies in a 500-μl total volume. Protein G-Sepharose beads (Pharmacia) were added to the lysate-antibody mix and incubated on a rotating platform for 2.5 to 3.5 h at 4°C and washed three to four times with a lysis buffer. The immunoprecipitates or total cell lysates were resolved on a sodium dodecyl phosphate (SDS)–8.0 or 9.0% polyacrylamide gel and transferred onto polyvinylidene difluoride membranes (NEN). The blots were blocked for 1.5 to 3.0 h in 3% bovine serum albumin in TBS-T (20 mM Tris · Cl [pH 7.5], 150 mM NaCl, 0.1% Tween 20) and immunoblotted for 1.5 to 3.0 h with either anti-pTyr (PY20-HRP; Transduction Labs; 1:3,000), anti-EGFR (1:1,000; Transduction Labs), anti-Erk2 (1:1,000; Upstate Biotechnology Inc.) antiphospho-473 Akt (1:1,000; New England Biolabs), anti-Akt (1:1,000; New England Biolabs), anti-Flag (1:1,000; M2; BabCo), anti-Shc (1:1,000; Transduction Labs), anti-Grb2 (1:1,000; Transduction Labs), or anti-beta1 integrin (1:1,000) antibodies. The immunoblots were washed five to seven times, incubated with appropriate horseradish peroxidase-conjugated secondary antibody subsequently, washed five to seven times, reacted with enhanced chemiluminescence (NEN), and subjected to autoradiography. For stripping the blots were incubated in a strip buffer (62.5 mM Tris · Cl [pH 6.8], 2% SDS, 0.7% mercaptoethanol) at 50°C for 30 min.
Receptor internalization experiments were carried out as follows: cells were stimulated with AP1510 or the ethanol alone for the indicated lengths of time. The cells were rinsed three times with ice-cold PBS (pH 8.0) and incubated with PBS containing 0.5 mg of NHS-S-S-Biotin (Pierce) at 4°C for 1 h. The cells were subsequently rinsed three times with ice-cold PBS and lysed in 1× modified radioimmunoprecipitation assay buffer (150 mM NaCl, 20 mM Tris · HCl [pH 7.5], 0.1% SDS, 1.0% sodium deoxycholate, 1.0% Triton X-100, 2 μg of aprotinin per ml, and 2 μg of leupeptin per ml). Equal amounts of lysates (300 μg) were incubated with 75 μl of Sepharose beads coupled to NeutrAvidin (Pierce) on a rotating platform for 1 to 1.5 h. The immunoprecipitates were washed three times with modified radioimmunoprecipitation assay buffer and resuspended in 1× sample buffer.
Cell cycle analysis.
Parental Rat1 cells or p75.B1.F1.HA (clone 9), p75.B1.F2.HA (clone 6), or p75.B2.F2.HA (clone 4) were plated at a density of 8 × 104 cells per well in a six-well plate. After 48 h the cells were switched to serum-free media for 24 h and subsequently stimulated with indicated amounts of AP1510 or 10 ng of EGF per ml for 18 to 20 h. Cells were trypsinized, resuspended in 1.0 ml of 10% serum-containing medium, and transferred to a tube containing 10.0 ml of 1× PBS. The cells were pelleted by centrifugation at 3,000 × g for 4 min. Pellets were washed again with 10.0 ml of 1× PBS and then resuspended in 0.5 ml of 1× PBS. The resuspended cells were fixed overnight in 5.0 ml of ice-cold 100% ethanol. Ethanol was added very slowly, while vortexing, to avoid cell clumping. The fixed cells were pelleted and washed twice in 5.0 ml of 1× PBS containing 2.0% FBS. After the second wash the cells were resuspended in 0.5 ml of 1× PBS containing 2% FBS, 0.1% Tween 20; 20 μg of RNase A per ml, and 10 μg of propidium iodide per ml, incubated at 37°C for 2 to 3 h, and analyzed by FACS. The data were analyzed by the ModFit program (Becton Dickinson) to calculate the percentage of cells in the G0-G1, S, and G2-M stages of the cell cycle. The fold increase in the percentage of cells that are in S and G2-M phases was plotted.
Focus-forming assay.
Rat1 fibroblasts were plated at 2 × 104 cells per well in a 12-well plate. The cells were infected with retroviruses expressing the appropriate ErbB fusion at the rate of 150 to 250 CFU per well. Infected cells (24 h after infection) were trypsinized and replated on 10-cm-diameter plates containing different concentrations of AP1510. Cells from one infected well were plated onto 10-cm-diameter plates in media containing 500 μg of G418 per ml for 10 to 12 days. The focus assay was carried out for 14 days by changing the drug-containing media once every 3 days. The plates were fixed with 4% formalin and stained with 4% Giemsa. Previous experiments have shown that AP1510 is active for at least 4 days as dilute aqueous solutions at 37°C (data not shown). For another experiment (see Fig. 7), Rat1 cells were plated at 3 × 105 cells per 10-cm-diameter plate and infected with a larger amount of virus stock and the exact number of CFU per plate was estimated by trypsinizing and plating one infected plate under 750 μg of G418 per ml.
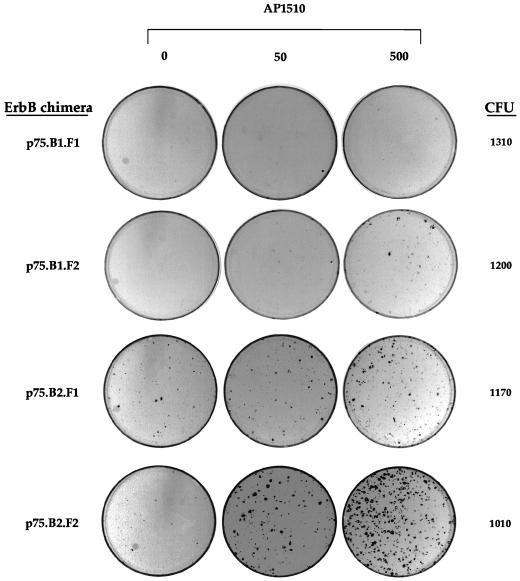
FIG. 7.
ErbB1 and ErbB2 homodimers differ in their abilities to induce focus formation in rat fibroblasts. Rat1 cells were infected with retroviruses containing different ErbB fusions. The cells were maintained in media containing the indicated amounts of AP1510 (nanomolar) for 14 days, fixed, and stained with Giemsa stain. One set of infected cells were trypsinized, and 1/10 of the cells were replated in media containing G418 to ascertain the number of CFU infected per plate.
RESULTS
Synthetic ligand-mediated ErbB dimerization.
To study the signaling and biological specificities of different ErbB dimers, we have designed a dimerization strategy that employs synthetic bivalent dimerizing ligands (also called chemical inducers of dimerization) and their binding proteins (Fig. 1; for a review, see reference 67). For homodimerization, we employed ligands that bind to the FK506-binding protein, FKBP12. The first reported homodimerizing compound, FK1012, was derived by chemical coupling of the monomeric FKBP ligand FK506 (70). Each molecule of FK1012 or a synthetic analog, AP1510 (2), binds to two copies of the ligand binding domain FKBP (Fig. 1A) and can induce the homodimerization of proteins fused to FKBP. To induce heterodimerization, we used the natural product rapamycin, which binds to one copy of FKBP and one copy of the FRB domain of FRAP (Fig. 1A).
To create dimerizable ErbB receptors, the FKBP or FRB domain and an epitope tag were fused to the C-terminal end of the ErbB cytoplasmic domain (Fig. 1C). To prevent the binding of EGF family ligands released by autocrine secretion, the extracellular and transmembrane domains of the low-affinity NGF receptor p75 were substituted for the analogous domains of ErbB1 and ErbB2. Using FKBP- or FRB-containing ErbB receptor chimeras it is possible to generate homodimers with AP1510 or heterodimers with rapamycin (Fig. 1D).
Dimerization of ErbB1 cytoplasmic domain with synthetic ligands results in a dose-dependent stimulation of receptor and substrate phosphorylation.
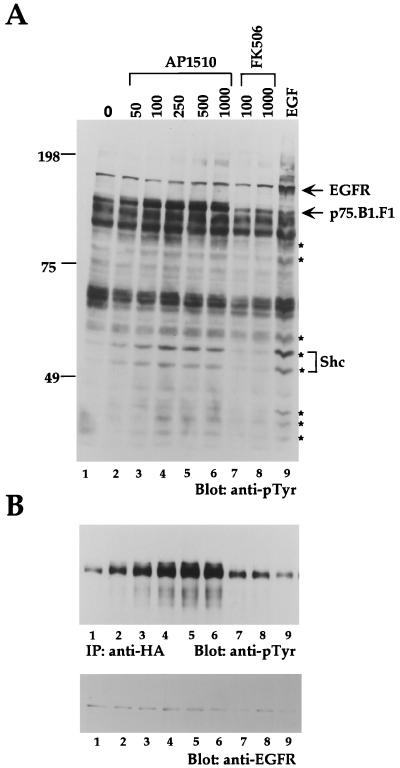
To establish whether synthetic ligands can be used to dimerize ErbB receptors, we examined the dimerization and activation of the ErbB1 cytoplasmic domain (B1) fused to one copy of FKBP (F1) (denoted as p75.B1.F1.HA) (Fig. 1C). Stable cell lines expressing the p75.B1.F1.HA chimera were derived by using Rat1 fibroblasts. AP1510, but not FK506, treatment resulted in a dose-dependent increase in tyrosine phosphorylation of cellular proteins (Fig. 2A; compare lanes 1 to 6 and 7 to 8). As expected, the cells were still sensitive to EGF stimulation (Fig. 2A, lane 9). Interestingly, the pattern of tyrosine phosphorylation after AP1510 stimulation was comparable to the pattern obtained after EGF stimulation (Fig. 2; compare lanes 6 and 9). For example, the adapter protein Shc and proteins with approximate molecular masses of 35, 42, 44, 60, 79, and 100 kDa (marked with asterisks) were phosphorylated by both EGF and AP1510 stimulation (Fig. 2A). As expected, EGF stimulation resulted in phosphorylation of endogenous EGFR, while AP1510 stimulation did not result in phosphorylation of any protein in the mobility range of EGFR (Fig. 1A; compare lanes 2 to 6 and 9). Similar results were obtained in three independent Rat1 clones expressing p75.B1.F1.HA (data not shown). AP1510 stimulation of the parental Rat1 fibroblasts did not have any effect on the tyrosine phosphorylation of cellular proteins (data not shown). To specifically examine the tyrosine phosphorylation status of the chimeric receptor, anti-HA immunoprecipitates were immunoblotted with anti-pTyr antibodies. The p75.B1.F1 chimera was inducibly tyrosine phosphorylated with maximal stimulation at 500 nM AP1510 (Fig. 2B, lanes 1 to 6). As expected, neither the monomeric ligand nor EGF induced tyrosine phosphorylation of the ErbB1 chimera (Fig. 2B, lanes 7 to 9). Synthetic ligand-mediated activation of the p75.B1.F1.HA receptor did not show any increase in tyrosine phosphorylation levels of the endogenous ErbB2 receptor (data not shown). Thus, the activation of the chimeric receptor appears to be independent of both EGF ligands and endogenous ErbB receptors.
FIG. 2.
Dimerization of ErbB1 cytoplasmic domain with synthetic ligands results in a dose-dependent stimulation of receptor and substrate phosphorylation. (A) Rat1 fibroblasts expressing ErbB1 fused to one copy of FKBP (p75.B1.F1.HA) were stimulated with increasing amounts of AP1510 (nanomolar) (lanes 2 to 6) or FK506 (lanes 7 and 8) for 15 min or stimulated with 50 ng of EGF per ml for 5 min. Cell lysates were collected, and 45 μg of protein was resolved and immunoblotted with anti-phosphotyrosine (anti-pTyr) antibodies. The blot was stripped and reprobed with anti-Shc antibodies, and the p46 and p52 isoforms of Shc and other cellular proteins that were tyrosine phosphorylated by ligand stimulation are indicated by asterisks. (B) Seven hundred micrograms of lysate was used for immunoprecipitation with anti-HA antibodies and immunoblotted with anti-pTyr (upper panel). The anti-pTyr blot was subsequently stripped and reprobed with anti-EGFR antibodies (lower panel).
Synthetic ligand-activated receptors are competent in recruiting signaling molecules and activating a downstream target.
Stimulation of the wild-type ErbB1 receptor by EGF results in a recruitment of multiple cytoplasmic signaling molecules, including Grb2 and Shc (1). To examine whether AP1510-activated chimeric ErbB1 receptors can recruit SH2-containing proteins, we immunoprecipitated the chimeric receptors and immunoblotted them with antibodies to Grb2 and Shc (Fig. 3A). Both Grb2 and Shc coimmunoprecipitated with tyrosine phosphorylated p75.B1.F1.HA in AP1510-treated cells (Fig. 3A, lanes 4 to 7), suggesting that synthetic ligand-activated ErbB1 receptors were competent to recruit known cytoplasmic signaling molecules.
FIG. 3.
Synthetic ligand-activated receptors are competent in recruiting signaling molecules and activating a downstream target. (A) HA epitope-containing proteins were immunoprecipitated (IP) (lanes 4 to 7) from 500 μg of cell lysate, and the membrane was probed with antibodies against anti-pTyr (first panel), anti-Grb2 (second panel), or anti-Shc (third panel). The blot from the first panel was stripped and reprobed with anti-EGFR (fourth panel). Normal mouse serum (NMS; lanes 1 to 3) was used as a nonspecific control. (B) Total cell lysates from p75.B1.F1.HA or Rat1 cells stimulated with AP1510 (nanomolar) were immunoblotted with anti-Erk2 antibodies.
Activation of the EGFR is known to activate a signal transduction pathway, leading to the activation of extracellular signal-regulated kinase 2 (Erk2 or MAPK). To test whether activation of the chimeric ErbB1 receptor results in activation of Erk2, p75.B1.F1.HA-expressing cells or the parental Rat1 cells were stimulated with AP1510 and total cell lysates were immunoblotted with anti-Erk2 antibodies. Interestingly, AP1510 stimulation induced a characteristic mobility shift of Erk2, suggesting that synthetic ligand-mediated dimerization of p75.B1.F1.HA leads to activation of downstream signaling targets.
Synthetic ligands activate other members of the ErbB family, and the activated receptors retain their kinase specificity.
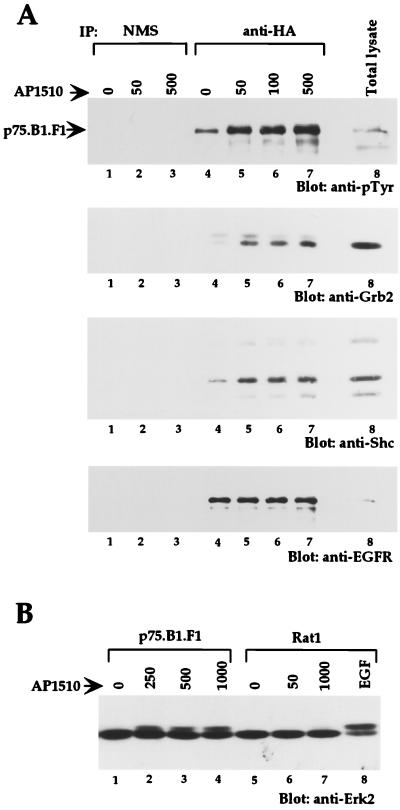
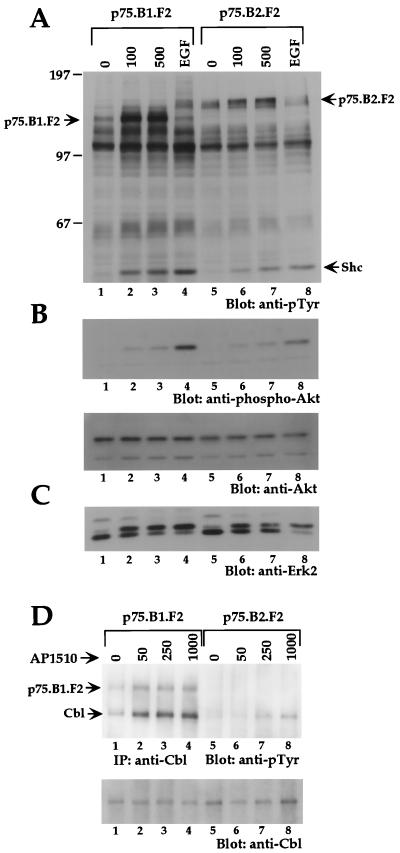
In order to examine whether dimerization of cytoplasmic domains can activate other ErbB family members and whether synthetic ligand-activated ErbB kinases retain their kinase specificity, we derived Rat1-based stable cell lines expressing AP1510-inducible ErbB2 chimeras. We found that cell lines expressing ErbB2 chimeras with one copy of FKBP (p75.B2.F1.HA) had very high levels of basal tyrosine phosphorylation (data not shown); however, cells expressing p75.B2.F2.HA, which contains two copies of FKBP (F2), had low levels of basal tyrosine phosphorylation and showed AP1510-inducible phosphorylation. To make an objective comparison between ErbB1 and ErbB2 homodimers, we derived cell lines expressing two FKBP variants of either ErbB1 or ErbB2 (p75.B1.F2.HA or p75.B2.F2.HA). As observed with cells expressing the single FKBP ErbB1 chimera (p75.B1.F1.HA), addition of synthetic ligand to cells expressing the double FKBP ErbB1 chimera (p75.B1.F2.HA) resulted in increased phosphorylation of the chimeric receptor and selected proteins (Fig. 4A, lanes 1 to 3). AP1510 stimulation also resulted in tyrosine phosphorylation of the ErbB2 chimera, p75.B2.F2.HA, and other protein substrates (Fig. 4A, lanes 5 to 7).
FIG. 4.
Synthetic ligands can activate other members of the ErbB family, and the activated receptors retain their kinase specificity. (A) Total cell lysates from cells stimulated with AP1510 (nanomolar) or EGF (50 ng/ml) were resolved and blotted with anti-pTyr antibodies. The positions of p75.B1.F2.HA, p75.B2.F2.HA, and Shc are indicated. (B) The top two-thirds of the blot in panel A was stripped and reprobed with anti-phospho-473 Akt antibody (upper panel), and the blot was restripped and blotted with anti-Akt antibody (lower panel). (C) The lower third of the blot in panel A was stripped and reprobed with anti-Erk2. (D) c-Cbl was immunoprecipitated (IP) from 750 μg of lysate and immunoblotted with anti-pTyr antibodies (upper panel), and the blot was subsequently stripped and reprobed with anti-Cbl antibodies (lower panel). The position of the coimmunoprecipitated p75.B1.F2.HA is indicated.
In order to examine whether downstream signaling pathways are activated by the ErbB2 chimera, we examined the activation of Erk2 and Akt, a serine or threonine protein kinase that is activated by ErbB receptors (10). Homodimerization of both ErbB1 and ErbB2 resulted in the characteristic mobility shift of Erk2 (Fig. 4C). To examine activation of Akt, total cell lysates were immunoblotted with anti-Akt antibodies that specifically recognize Akt phosphorylated at serine 473 (Fig. 4B). Phosphorylation of both Thr 308 and Ser 473 on Akt is required for full activation (19). AP1510 stimulation induced Ser 473 phosphorylation in cells expressing either ErbB1 or ErbB2 chimeras (Fig. 4B). The difference in signal strength between lanes 1 to 4 and 5 to 8 (Fig. 4A) is likely due to a difference in the levels of protein loaded (compare lanes 1 to 4 and 5 to 8 in Fig. 4B, lower panel). It should be noted that both cell lines used in this experiment express comparable levels of the ErbB chimera as determined by FACS analysis (see Materials and Methods).
It is known that c-Cbl is tyrosine phosphorylated only by ErbB1 and not by other ErbB family members (41). We examined c-Cbl tyrosine phosphorylation status upon synthetic ligand-mediated dimerization of either ErbB1 or ErbB2. Homodimerization of ErbB1 resulted in c-Cbl tyrosine phosphorylation (Fig. 4D), whereas phosphorylation of c-Cbl was barely detectable following ErbB2 homodimerization. This differential phosphorylation of c-Cbl was also observed in COS7 cells transiently transfected with either p75.B1.F2.HA or p75.B2.F2.HA chimeras (data not shown). These observations suggest that dimerization of ErbB receptors activated by synthetic ligands can retain their kinase-specific functions as determined by their ability to phosphorylate c-Cbl.
ErbB1 but not ErbB2 chimeras are internalized after AP1510 stimulation.
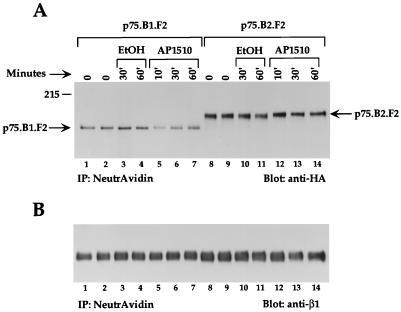
EGF activation has been shown to induce the endocytosis of activated ErbB1 receptors (65). It has also been shown that ErbB chimeras containing the extracellular domain of ErbB1 and the cytoplasmic domains of ErbB2, ErbB3, or ErbB4 do not undergo EGF-induced receptor endocytosis (3). In order to determine whether synthetic ligand-activated ErbB receptors undergo internalization, we stimulated p75.B1.F2.HA- or p75.B2.F2.HA-expressing cells with AP1510 for different lengths of time, and the cell surface proteins were subsequently labeled with biotin at 4°C. The cell lysates were then subjected to precipitation with NeutrAvidin-coupled beads, and the bound proteins were immunoblotted with HA tag antibodies to determine the amount of chimeric receptor present at the cell surface. AP1510 treatment of ErbB1-expressing (Fig. 5A, lanes 5 to 7) but not ErbB2-expressing (Fig. 5A, lanes 12 to 14) cells resulted in a significant decrease in the amount of chimeric receptors present at the cell surface. This observation suggests that AP1510 stimulation results in the internalization of activated ErbB1 receptors. The significant decrease in the levels of ErbB1 receptors at the cell surface after 10 min of AP1510 stimulation is consistent with the internalization rates observed for the peptide ligand EGF (65). Stimulation of p75.B1.F2.HA-expressing cells with the carrier alone did not result in any change in the levels of the ErbB1 chimera (Fig. 5A, lanes 1 to 4). Since NeutrAvidin would precipitate all biotin-labeled cell surface proteins, the same blot was reprobed with anti-beta1 integrin antibodies (Fig. 5B) to demonstrate that the decrease in the p75.B1.F2 levels was receptor specific.
FIG. 5.
ErbB1 but not ErbB2 chimeras are internalized after AP1510 stimulation. Cell lines expressing either ErbB1 or ErbB2 chimeras were stimulated with carrier alone (ethanol [EtOH]) or with 500 nM AP1510 for the indicated lengths of time. The cell surface proteins were subsequently biotinylated by incubating with NHS-S-S-Biotin at 4°C for 1 h. The biotinylated proteins were immunoprecipitated (IP) with Sepharose-conjugated NeutrAvidin beads and immunoblotted with either anti-HA (A) or anti-beta1 integrin (B) antibodies.
Immunofluorescent labeling of ErbB1-expressing cells with anti-p75 antibodies also showed a ligand activation-dependent internalization of the chimeric p75.B1.F2.HA receptor (data not shown). These observations suggest that ErbB1 chimeras undergo synthetic ligand-dependent endocytosis and also demonstrate that the ErbB chimeras retain their differential regulation of receptor internalization.
Activation of ErbB homodimers results in induction of cell cycle progression.
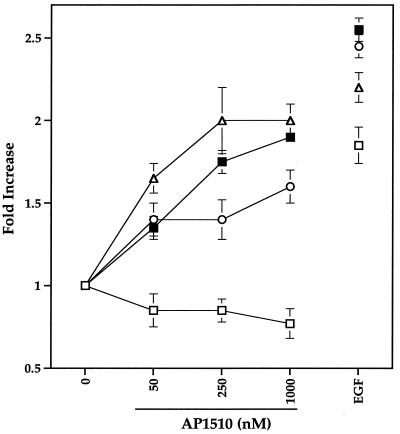
To establish whether activation of ErbB receptors by synthetic ligands can induce cell cycle progression, cells expressing either ErbB1 fused to one copy of FKBP (p75.B1.F1.HA), ErbB1 fused to two copies of FKBP (p75.B1.F2.HA), or ErbB2 fused to two copies of FKBP (p75.B2.F2.HA) were starved in serum-free media for 24 h. The cells were then stimulated with indicated amounts of AP1510 for 16 to 18 h, and the DNA content was measured by FACS of propidium iodide-labeled cells (Fig. 6). The fold increase in the percentage of cells that have left the G0-G1 stage of the cell cycle was calculated (see Materials and Methods). AP1510 stimulation of p75.B1.F1.HA-, p75.B1.F2.HA-, and p75.B2.F2.HA-expressing cells resulted in a 1.5- to 2.0-fold increase, and EGF stimulation resulted in a 1.8- to 2.5-fold increase in the percentages of cells that have left the G0-G1 stage of the cell cycle (Fig. 6). These results suggest that ErbB1 and ErbB2 chimeras activated by synthetic ligands were able to promote cell cycle progression.
FIG. 6.
Activation of ErbB homodimers results in induction of cell cycle progression. The parental Rat1 cells or cell lines expressing different ErbB chimeras were serum starved, stimulated, and analyzed by FACS as described in Materials and Methods. The graph shows the fold increase ± standard deviations (error bars) in the percentages of cells that have left the G0-G1 stage of the cell cycle. The parental Rat1 cells (open squares), cells expressing ErbB1 with one copy FKBP (p75.B1.F1) (closed squares), cells expressing ErbB1 with two copies of FKBP (p75.B1.F2) (open circles), and cells expressing ErbB2 with two copies of FKBP (p75.B2.F2) (open triangles) were used for the analysis. EGF was used at a 10-ng/ml concentration.
ErbB1 and ErbB2 homodimers differ in their abilities to induce focus formation in Rat1 fibroblasts.
Since ErbB family members are known to be potent oncogenes, we examined whether ErbB1 or ErbB2 homodimers can induce focus formation in fibroblasts. Rat1 fibroblasts were infected with retroviruses expressing either ErbB1 or ErbB2 fused to one or two copies of FKBP (p75.B1.F1.HA, p75.B1.F2.HA, p75.B2.F1.HA, or p75.B2.F2.HA). The infected cells were maintained in the presence of different doses of AP1510 for 14 days. The number of infected cells was determined by G418 selection, since the virus carried the gene coding for neomycin (see Materials and Methods). Fifty-five to sixty-five percent of cells infected with p75.B2.F2.HA retrovirus formed foci in the presence of AP1510 (Fig. 7 and Table 1). In contrast, the expression of ErbB1 chimera fused to two FKBP (p75.B1.F2.HA) showed weak focus-forming activity (Fig. 7), with only 10% of the infected cells forming foci (Table 1). In addition, the foci induced by the ErbB1 chimera had a diffuse morphology and showed faint Giemsa staining relative to the dense, intensely stained foci induced by the ErbB2 chimera (Fig. 7). The expression of a ErbB2 chimera with one copy of FKBP (p75.B2.F1.HA) resulted in a low level of ligand-independent focus formation and nevertheless showed a six- to sevenfold ligand inducibility (Fig. 7 and Table 1). Surprisingly, the ErbB1 chimera with one copy of FKBP (p75.B1.F1.HA) failed to induce any detectable focus formation (Fig. 7 and Table 1). The difference in the focus-inducing ability was not due to differences in expression levels of the chimeric proteins, since we detected comparable levels of expression by both anti-HA immunoblots and FACS analyses of the infected Rat1 cells stained with anti-p75 antibodies (data not shown).
TABLE 1.
Transformation of Rat1 fibroblasts by ErbB1 or ErbB2 homodimersa
| ErbB chimera | No. of foci (± SD) per 100 CFU with AP1510 (nM)
|
|||||
|---|---|---|---|---|---|---|
| 0 | 50 | 100 | 250 | 500 | 1,000 | |
| p75.B1.F1 | 0 | 0 | 0 | 0 | 0 | 1 |
| p75.B1.F2 | 0 | 2 ± 1 | 7 ± 3 | 10 ± 4 | 8 ± 1 | 7 ± 1 |
| p75.B2.F1 | 6 ± 1 | 6 ± 2 | 8 ± 1 | 19 ± 2 | 47 ± 6 | 34 ± 10 |
| p75.B2.F2 | 1 | 16 ± 3 | 38 ± 7 | 55 ± 3 | 62 ± 9 | 58 ± 6 |
Each value represents an average of at least three independent experiments.
Both ErbB1 and ErbB2 homodimers can induce a reversible morphological transformation of fibroblasts.
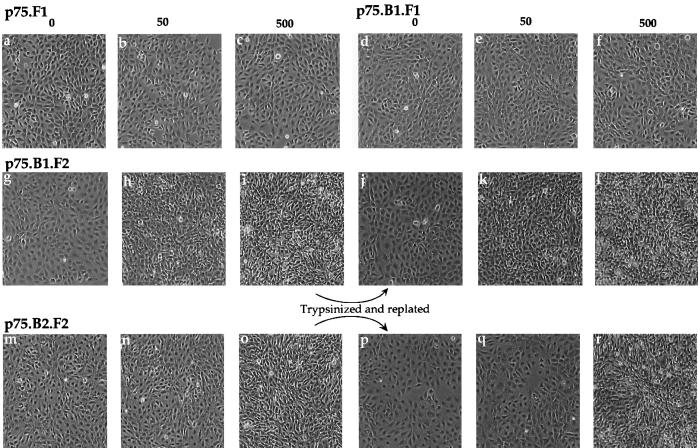
Transformed fibroblasts are known to display a refractile morphology and lose contact inhibition. We examined whether dimerization of the cytoplasmic domains of ErbB1 or ErbB2 induces morphological changes and whether these changes are reversible after ligand withdrawal. Stable cell lines expressing comparable levels of ErbB1 with one copy of FKBP (p75.B1.F1.HA), ErbB1 with two copies of FKBP (p75.B1.F2.HA), or ErbB2 with two copies of FKBP (p75.B2.F2.HA) (see Materials and Methods) were grown in the presence of AP1510 for 48 h, and the changes in morphology were recorded (Fig. 8). Cells expressing p75.B1.F2.HA (Fig. 8g to i) or p75.B2.F2.HA (Fig. 8m to o), but not cells expressing p75.F1.HA (a p75 chimera without ErbB cytoplasmic domain; Fig. 1B) (Fig. 8a to c), lost their contact-inhibited flat morphology and assumed a transformed, refractile morphology in the presence of AP1510. Consistent with a lack of focus-forming ability, AP1510 did not induce morphological changes in cells expressing p75.B1.F1.HA (Fig. 8d to f).
FIG. 8.
Both ErbB1 and ErbB2 homodimers are able to induce reversible morphological transformation of fibroblasts. The cells were plated in the presence of AP1510 (concentrations shown are nanomolar) and allowed to grow for 48 h. The morphologies of the cells were recorded (a to i and m to o). The cells in panels i and o were trypsinized and replated either in media without (j and p) or with AP1510 (k, l, q, and r). P75.F1 (Fig. 1B) corresponds to cells expressing the chimera without the ErbB cytoplasmic domain.
After trypsinization and replating of the AP1510-treated cells in media without the ligand, the cells reverted to a nontransformed state, displaying a well-spread morphology and contact inhibition (Fig. 8j and p). This observation suggests that the morphological changes require the continuous presence of the dimerizing ligand.
Synthetic ligand-induced heterodimerization between ErbB receptors.
It has not been possible to study the signaling specificities of different ErbB heterodimers in isolation, since most cell types express more than one ErbB family member. In order to establish whether synthetic ligands can be used to form heterodimers of selected ErbB receptors, we generated a chimera containing the kinase-dead variant of ErbB1 (kdB1) fused to the FRB domain and a Flag epitope tag (p75.kdB1.R1.Flag; see Materials and Methods). The p75.kdB1.R1.Flag chimera was not phosphorylated on tyrosine when expressed transiently in COS7 cells (data not shown). Stable pools of Rat1 cells coexpressing the kinase-dead ErbB1-FRB chimera (p75.kdB1.R1.Flag) and either the wild-type ErbB1-FKBP chimera (p75.B1.F2.HA) or the wild-type ErbB2-FKBP chimera (p75.B2.F2.HA) were generated (Fig. 9A). As illustrated in Fig. 1D, addition of the FKBP-binding ligand AP1510 to these cells should result in the formation of either B1-B1 or B2-B2 homodimers, while treatment with the heterodimerizing ligand rapamycin should result in either B1-kdB1 or B2-kdB1 heterodimers. As expected, AP1510 induced homodimerization and tyrosine phosphorylation of both ErbB1-FKBP (p75.B1.F2.HA; Fig. 9B, lanes 1 to 3) and ErbB2-FKBP (p75.B2.F2.HA; Fig. 9B, lanes 7 to 9) chimeras. Interestingly, the kinase-dead ErbB1 chimera fused to the FRB domain, expressed in the same cell, was not tyrosine phosphorylated by AP1510 stimulation (Fig. 9C, lanes 1 to 3 and 7 to 9), likely due to the inability of AP1510 to bind FRB-fused chimeras.
However, rapamycin stimulation resulted in tyrosine phosphorylation of the kinase-dead ErbB1-FRB chimera (p75.kdB1.R1.Flag) (Fig. 9C, lanes 4 to 6 and 10 to 12) by both kinase-active ErbB1-FKBP (lanes 4 to 6) and kinase-active ErbB2-FKBP (lanes 10 to 12). Since rapamycin is known to dimerize an FKBP domain and an FRB domain (Fig. 1), this observation suggests that rapamycin can induce heterodimers between the kinase-dead ErbB1-FRB and kinase-active ErbB-FKBP receptors. Rapamycin stimulation did not change the phosphorylation status of either kinase-active ErbB1-FKBP (Fig. 9B, lanes 4 to 6) or kinase-active ErbB2-FKBP (Fig. 9B, lanes 10 to 12), possibly because the dimer comprises one kinase-active and one kinase-dead receptor. This is consistent with the notion that the tyrosine phosphorylation of ErbB receptors occurs primarily by trans-phosphorylation within a dimer (76). The kinase-active receptors observed in the anti-Flag immunoprecipitates from rapamycin-stimulated cell lysates (Fig. 9C, lanes 4 to 6 and lanes 10 to 12) were due to the ability of rapamycin to induce a stable heterodimeric complex between the Flag-tagged kinase-dead and weakly phosphorylated kinase-active ErbB receptors (Fig. 9B, lanes 1 and 7). Since the tyrosine phosphorylation levels of neither ErbB1 (Fig. 9B; compare lane 1 and lanes 4 to 6) nor ErbB2 (Fig. 9B; compare lane 7 and lanes 10 to 12) change in response to rapamycin stimulation, it is unlikely that rapamycin stimulation affects the tyrosine phosphorylation status of FKBP-fused kinase-active ErbB chimeras. These results demonstrate that rapamycin can be used to form heterodimers in the absence of homodimers and AP1510 can be used to form homodimers in the absence of heterodimers.
c-Cbl can differentiate ErbB1 in homodimers from ErbB1 in ErbB1-ErbB2 heterodimers.
It is possible that heterodimers have different signaling specificities than homodimers. We tested whether the kinase-dead ErbB1 receptor, phosphorylated by either a kinase-active ErbB1 (homodimer) or a kinase-active ErbB2 (heterodimer), differed in its ability to associate with cytoplasmic signaling molecules. Since c-Cbl has been shown to bind selectively to ErbB1 but not to other ErbB receptors (41), we asked whether c-Cbl can differentiate between the kinase-dead ErbB1 phosphorylated by a kinase-active ErbB1 receptor (homodimer) from a kinase-dead ErbB1 phosphorylated by a kinase-active ErbB2 receptor (heterodimer). The kinase-dead ErbB1 receptor fused to the FRB domain (p75.kdB1.R1.Flag) was cotransfected with either kinase-active ErbB1-FKBP chimera (p75.B1.F2.HA) or kinase-active ErbB2-FKBP chimera (p75.B2.F2.HA) in COS7 cells. In the presence of rapamycin the kinase-active FKBP-fused ErbB1 and ErbB2 receptors coimmunoprecipitate with the kinase-dead ErbB1-FRB.Flag chimera (Fig. 9 and 10). Rapamycin stimulation resulted in increased phosphorylation of the kinase-dead ErbB1-FRB chimera (p75.kdB1.R1.Flag) by both ErbB1-FKBP (p75.B1.F2.HA) (Fig. 10, lanes 1 to 3) and ErbB2-FKBP (p75.B2.F2.HA) (Fig. 10, lanes 4 to 6) receptors, determined by anti-Flag immunoprecipitation and anti-pTyr immunoblotting. Endogenous c-Cbl was also immunoprecipitated from the same cell lysates, and the precipitates were blotted with anti-pTyr antibodies (Fig. 10, lanes 7 to 12) to identify the ErbB chimeras that bind to c-Cbl. c-Cbl was able to coimmunoprecipitate the tyrosine phosphorylated, kinase-dead, ErbB1-FRB chimera (p75.kdB1.R1.Flag) only when the kinase-dead ErbB1 was phosphorylated by kinase-active ErbB1 chimera (p75.B1.F2.HA) (Fig. 10, lanes 7 to 9) but not when kinase-dead ErbB1 was phosphorylated by kinase-active ErbB2 chimera (p75.B2.F2.HA) (Fig. 10, lanes 10 to 12). These observations suggest that c-Cbl can differentiate ErbB1 molecules in a homodimer (p75.B1.F2-p75.kdB1.R1) from ErbB1 molecules in a heterodimer with ErbB2 (p75.B2.F2-p75.kdB1.R1).
DISCUSSION
We demonstrate that synthetic dimerizing ligands can selectively activate homo- and heterodimers of the ErbB family of receptors and result in the activation of signal transduction pathways. Synthetic ligand-mediated dimerization and activation also induce a dose-dependent stimulation of phenotypic alterations known to be regulated by ErbB receptors (e.g., stimulation of cell proliferation, morphological transformation, and focus formation). In addition, using a heterodimerizing ligand we demonstrate that c-Cbl can associate with ErbB1 in an ErbB1-ErbB1 homodimer but does not associate with ErbB1 in an ErbB1-ErbB2 heterodimer. These observations suggest that this approach can be used to study the signaling and biological specificities of ErbB homodimers and heterodimers and provide the first clear evidence for differential signaling by ErbB homo- and heterodimers.
The extracellular cysteine-rich domains of ErbB receptors play important roles in ligand binding and receptor dimerization (11, 12, 14, 59, 60, 66). The mutation or addition of Cys residues at the juxtamembrane region and generation of unpaired Cys residues result in dimerization of ErbB2 (12, 60). Interestingly, a number of such dimers fail to induce transformation, suggesting that forced dimerization is not sufficient for a functional activation of ErbB2 (11). The transmembrane and the juxtamembrane regions of ErbB2 form a helical structure, and receptor dimerization is thought to promote a helix-helix interaction (12). It is proposed that some of the Cys modification-induced dimerization may result in packing the helices in an unfavorable or nonpermissive orientation for signaling (12). Results presented in this report suggest that dimerization of the cytoplasmic domain was sufficient to activate both ErbB1 and ErbB2 receptors. This is consistent with an earlier report which suggests that membrane localization of the cytoplasmic domain of ErbB2 was sufficient to induce kinase activation and transformation (26). It will be interesting to progressively change the orientation or location of the synthetic ligand-binding domains within the chimera to ask whether the ErbB cytoplasmic domains require a specific dimerization interface.
Synthetic ligand-inducible dimerization was able to activate both biochemical and biological processes that are known to be stimulated by ErbB receptors. Both ErbB1 and ErbB2 homodimers were able to induce activation of the serine threonine kinase Akt (Fig. 4). Akt is a known downstream target of activated PI 3-kinase. It is unclear how either of these homodimers activates the PI 3-kinase pathway, since neither ErbB1 nor ErbB2 possesses the binding site for the SH2 domain of p85 subunit of PI 3-kinase (24, 39, 53, 64). It is possible that c-Cbl mediates the ErbB1 homodimer-induced activation of PI 3-kinase (63), whereas c-Cbl is unlikely to play a role in ErbB2 homodimer-induced activation of the PI 3-kinase pathway since ErbB2 does not show strong association with c-Cbl (Fig. 4 and 8). Further experiments will be necessary to understand the underlying mechanism leading to the activation of PI 3-kinase by ErbB1 and ErbB2 homodimers.
The activation of ErbB1 or ErbB2 dimers with synthetic ligands resulted in a ligand-dependent acquisition of transformed-cell morphology, while the removal of AP1510 caused a reversion to a normal morphology. These observations demonstrate the continuous requirement of a dimerization signal for maintenance of the transformed morphology. It will be of interest to determine whether the ligand-dependent transformation and reversion can be induced in animal models where other mutations are involved during tumorigenesis. The development of such a model with inducible activation of specific homo- and heterodimers of different ErbB receptors would be very useful in understanding the early events of tumor progression in adult animals.
The biological effects of ErbB1 homodimerization in fibroblasts or other cell types are unclear, since almost all cell types express more than one member of the ErbB receptor family. Previous reports have shown that ErbB1 is able to induce focus formation in a ligand-dependent manner in NIH 3T3 clones which either lack ErbB1 (21) or lack expression of all ErbB family members (79) and that EGF-activated ErbB1 possesses weaker focus-forming activity than activated ErbB2 (21). It is possible that these cell lines are not devoid of ErbB receptors, and the observed phenotype may be a result of heterodimers involving ErbB1. Studies using ErbB receptor-deficient hemopoietic cells that require interleukin 3 for survival and growth have suggested that ErbB1 homodimers are not effective in inducing proliferation but can induce interleukin 3-independent survival (56). The approach presented here enables us to study the effect of homodimers in the absence of lateral or combinatorial interactions with other ErbB family members in cells that naturally express these receptors; hence, we believe that our observations provide a clear demonstration of the biological differences between ErbB1 and ErbB2 homodimers in fibroblasts (see below).
It is unclear why synthetic ligand-induced ErbB1 homodimers possess a five- to sevenfold weaker focus-forming activity than ErbB2 homodimers. It is possible that ErbB1 homodimers do not activate signaling molecules that mediate transformation as effectively as ErbB2 (58). Alternatively, the homodimers may couple to negative regulators of signaling. The heterodimerization of ErbB1 with other ErbB receptors either can generate novel autophosphorylation sites for activating cytoplasmic signaling molecules or may fail to generate certain autophosphorylation sites to preclude interactions with a negative regulator(s). We present evidence which suggests that ErbB1 homodimers and ErbB1-ErbB2 heterodimers differ in their abilities to recruit a cytoplasmic signaling protein, c-Cbl. It is possible that such differences may play a role in determining the biological specificity of homo- and heterodimers.
c-Cbl has been implicated both as a positive and negative regulator of cell signaling (44, 47). The mechanism by which Cbl functions is not known. Recent observations suggest that c-Cbl promotes the ubiquitination and degradation of activated EGF and platelet-derived growth factor receptors (42, 47). Interestingly, only ErbB1, and not ErbB3, is ubiquitinated and downregulated by c-Cbl, and this is dependent on the presence of the ErbB1 cytoplasmic tail (42). It is possible that ErbB1 homodimers and ErbB1-ErbB2 heterodimers are differentially ubiquitinated and downregulated. Such a possibility is consistent with the observation that ErbB1 homodimers are endocytosed (Fig. 5) and degraded, while ErbB1-ErbB2 heterodimers are recycled to the membrane after EGF stimulation (40). However, the differential association with c-Cbl may also regulate multiple downstream signaling pathways that play a role in signaling by ErbB1 and ErbB2 homo- and heterodimers.
It is unclear why ErbB1 chimeras with one copy of FKBP (p75.B1.F1) did not induce morphological changes or focus formation (Fig. 7 and 8). One copy of FKBP was sufficient to activate the receptor since AP1510 induced tyrosine phosphorylation of multiple cellular proteins including Erk2 (Fig. 2), as well as stimulation of DNA synthesis in cells expressing the p75.B1.F1.HA chimera (Fig. 6). In addition, AP1510 activation of the single FKBP version of the ErbB2 chimera, p75.B2.F1, results in induction of focus formation (Fig. 7 and Table 1). It is possible that a simple dimerization of ErbB1 is not sufficient for morphological transformation whereas dimerization of ErbB2 is sufficient. Consistent with that possibility, the Val664-Glu mutation in ErbB2 promotes homodimerization, activation of the kinase, and transformation of fibroblasts (75). Interestingly, the insertion of a similar mutation into ErbB1 does not result in the ligand-independent transformation of fibroblasts (15, 38, 46), suggesting that homodimerization of ErbB1 may not be sufficient for transformation. Our results suggest that the p75.B1.F2.HA chimera, which contains two copies of FKBP, can induce only a weak transformation. It should be noted that two FKBP-containing chimeras can form higher-order complexes; however, sucrose gradient centrifugation of p75.B1.F2.HA-expressing cell lysate showed a ligand-dependent formation of a dimeric complex (data not shown). Further experiments are required to better understand the difference between the ErbB1 chimeras consisting of one or two copies of FKBP.
We will not be able to use the heterodimerizing ligand rapamycin in biological studies since rapamycin is a known immunosuppressive drug that can negatively regulate cellular kinases FRAP and p70S6K. However, synthetic versions of rapamycin (“rapalogs”) have been generated which do not bind endogenous FRAP and instead can only bind to a FRAP molecule that has been appropriately engineered to fit the modification on rapamycin (18a, 43). These rapalogs possess no immunosuppressive functions (18a, 43). We are in the process of constructing ErbB chimeras with the modified FRB domain which will enable us to study the biological effects of distinct ErbB heterodimers.
To our knowledge the results presented here provide the first direct evidence for differential signaling by ErbB1 homodimers and ErbB1-ErbB2 heterodimers. It will be of interest to apply this strategy to understand the signaling specificities of different ErbB receptor homo- and heterodimers. Since the synthetic ligand-mediated activation of chimeric ErbB receptors is independent of the endogenous levels of ErbB receptor expression, this approach could be well suited to study the biological effects of different ErbB receptor dimers in the cell type of choice.
ACKNOWLEDGMENTS
We thank Stuart Schreiber and Jerry Crabtree for the FKBP12 plasmid, Bill Muller and Peter Siegel for the Rat1 fibroblasts and rat Neu cDNA, Alan Wells for the M721A mutant of ErbB1, Owen Witte for the SRαMSVTKNeo retroviral plasmid, Richard Hynes for anti-beta1 integrin antibody, Kermit Carraway for technical assistance with sucrose gradient centrifugation, and Jane Amara, Victor Rivera, Sridar Natesan, Roy Pollock, and Tim Clackson for plasmids containing combinations of FKBP and FRB domains and the p75 receptor extracellular and transmembrane domain. We also thank members of the Brugge laboratory for their helpful suggestions.
This work was supported by a grant from the National Institutes of Health (CA78773 to J.S.B) and by a grant from the U.S. Army Research and Materiel Command (DAMD17-97-1-7237 to S.K.M).
REFERENCES
- 1.Alroy I, Yarden Y. The ErbB signaling network in embyrogensis and oncogenesis: signal diversification through combinatorial ligand-receptor interactions. FEBS Lett. 1997;410:83–86. doi: 10.1016/s0014-5793(97)00412-2. [DOI] [PubMed] [Google Scholar]
- 2.Amara J F, Clackson T, Rivera V M, Guo T, Keenan T, Natesan S, Pollock R, Yang W, Courage N L, Holt D A, Gilman M. A versatile synthetic dimerizer for the regulation of protein-protein interactions. Proc Natl Acad Sci USA. 1997;94:10618–10623. doi: 10.1073/pnas.94.20.10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baulida J, Kraus M H, Alimandi M, Di Fiore P P, Carpenter G. All ErbB receptors other than the epidermal growth factor receptor are endocytosis impaired. J Biol Chem. 1996;271:5251–5257. doi: 10.1074/jbc.271.9.5251. [DOI] [PubMed] [Google Scholar]
- 4.Beerli R R, Graus-Porta D, Woods-Cook K, Chen X, Yarden Y, Hynes N E. Neu differentiation factor activation of ErbB-3 and ErbB-4 is cell specific and displays a differential requirement for ErbB-2. Mol Cell Biol. 1995;15:6496–6505. doi: 10.1128/mcb.15.12.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belsches, A. P., M. D. Haskell, and S. J. Parsons. 1997. Role of c-Src tyrosine kinase in EGF-induced mitogenesis. Front. Biosci. 2:d501–d518. [DOI] [PubMed]
- 6.Birge R B, Knudsen B S, Besser D, Hanafusa H. SH2 and SH3-containing adaptor proteins: redundant or independent mediators of intracellular signal transduction. Genes Cells. 1996;1:595–613. doi: 10.1046/j.1365-2443.1996.00258.x. [DOI] [PubMed] [Google Scholar]
- 7.Blau C A, Peterson K R, Drachman J G, Spencer D M. A proliferation switch for genetically modified cells. Proc Natl Acad Sci USA. 1997;94:3076–3081. doi: 10.1073/pnas.94.7.3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer B, Roche S, Denoyelle M, Thiery J P. Src and Ras are involved in separate pathways in epithelial cell scattering. EMBO J. 1997;16:5904–5913. doi: 10.1093/emboj/16.19.5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burden S, Yarden Y. Neuregulins and their receptors: a versatile signaling module in organogenesis and oncogenesis. Neuron. 1997;18:847–855. doi: 10.1016/s0896-6273(00)80324-4. [DOI] [PubMed] [Google Scholar]
- 10.Burgering B M, Coffer P J. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. . (Comment, 376:553–554.) [DOI] [PubMed] [Google Scholar]
- 11.Burke C L, Lemmon M A, Coren B A, Engelman D M, Stern D F. Dimerization of the p185neu transmembrane domain is necessary but not sufficient for transformation. Oncogene. 1997;14:687–696. doi: 10.1038/sj.onc.1200873. [DOI] [PubMed] [Google Scholar]
- 12.Burke C L, Stern D F. Activation of Neu (ErbB-2) mediated by disulfide bond-induced dimerization reveals a receptor tyrosine kinase dimer interface. Mol Cell Biol. 1998;18:5371–5379. doi: 10.1128/mcb.18.9.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell S L, Khosravi-Far R, Rossman K L, Clark G J, Der C J. Increasing complexity of Ras signaling. Oncogene. 1998;17:1395–1413. doi: 10.1038/sj.onc.1202174. [DOI] [PubMed] [Google Scholar]
- 14.Cao H, Bangalore L, Dompe C, Bormann B J, Stern D F. An extra cysteine proximal to the transmembrane domain induces differential cross-linking of p185neu and p185neu. J Biol Chem. 1992;267:20489–20492. [PubMed] [Google Scholar]
- 15.Carpenter C D, Ingram H A, Cochet C, Walton G M, Lazar C S, Sowadski J M, Rosenfeld M G, Gill G N. Structural analysis of the transmembrane domain of the epidermal growth factor receptor. J Biol Chem. 1991;266:5750–5755. [PubMed] [Google Scholar]
- 16.Carpenter G. Receptor tyrosine kinase substrates: src homology domains and signal transduction. FASEB J. 1992;6:3283–3289. doi: 10.1096/fasebj.6.14.1385243. [DOI] [PubMed] [Google Scholar]
- 17.Carraway K L., III Involvement of the neuregulins and their receptors in cardiac and neural development. Bioessays. 1996;18:263–266. doi: 10.1002/bies.950180403. [DOI] [PubMed] [Google Scholar]
- 18.Carraway K L, III, Cantley L C. A neu acquaintance for erbB3 and erbB4: a role for receptor heterodimerization in growth signaling. Cell. 1994;78:5–8. doi: 10.1016/0092-8674(94)90564-9. [DOI] [PubMed] [Google Scholar]
- 18a.Clackson, T. Personal communication.
- 19.Coffer P J, Jin J, Woodgett J R. Protein kinase B (c-Akt): a multifunctional mediator of phosphatidylinositol 3-kinase activation. Biochem J. 1998;335:1–13. doi: 10.1042/bj3350001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Crovello, C. S., and K. L. Carraway III. Personal communication.
- 20.Daly R J. The Grb7 family of signalling proteins. Cell Signal. 1998;10:613–618. doi: 10.1016/s0898-6568(98)00022-9. [DOI] [PubMed] [Google Scholar]
- 21.Di Fiore P P, Segatto O, Taylor W G, Aaronson S A, Pierce J H. EGF receptor and erbB-2 tyrosine kinase domains confer cell specificity for mitogenic signaling. Science. 1990;248:79–83. doi: 10.1126/science.2181668. [DOI] [PubMed] [Google Scholar]
- 22.Fazioli F, Minichiello L, Matoska V, Castagnino P, Miki T, Wong W T, Di Fiore P P. Eps8, a substrate for the epidermal growth factor receptor kinase, enhances EGF-dependent mitogenic signals. EMBO J. 1993;12:3799–3808. doi: 10.1002/j.1460-2075.1993.tb06058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fazioli F, Minichiello L, Mat̂oŝkovā B, Wong W T, Di Fiore P P. eps15, a novel tyrosine kinase substrate, exhibits transforming activity. Mol Cell Biol. 1993;13:5814–5828. doi: 10.1128/mcb.13.9.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fedi P, Pierce J H, di Fiore P P, Kraus M H. Efficient coupling with phosphatidylinositol 3-kinase, but not phospholipase Cγ or GTPase-activating protein, distinguishes ErbB-3 signaling from that of other ErbB/EGFR family members. Mol Cell Biol. 1994;14:492–500. doi: 10.1128/mcb.14.1.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng G S, Pawson T. Phosphotyrosine phosphatases with SH2 domains: regulators of signal transduction. Trends Genet. 1994;10:54–58. doi: 10.1016/0168-9525(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 26.Flanagan J G, Leder P. neu protooncogene fused to an immunoglobulin heavy chain gene requires immunoglobulin light chain for cell surface expression and oncogenic transformation. Proc Natl Acad Sci USA. 1988;85:8057–8061. doi: 10.1073/pnas.85.21.8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fowler K J, Walker F, Alexander W, Hibbs M L, Nice E C, Bohmer R M, Mann G B, Thumwood C, Maglitto R, Danks J A, et al. A mutation in the epidermal growth factor receptor in waved-2 mice has a profound effect on receptor biochemistry that results in impaired lactation. Proc Natl Acad Sci USA. 1995;92:1465–1469. doi: 10.1073/pnas.92.5.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gassmann M, Lemke G. Neuregulins and neuregulin receptors in neural development. Curr Opin Neurobiol. 1997;7:87–92. doi: 10.1016/s0959-4388(97)80125-0. [DOI] [PubMed] [Google Scholar]
- 29.Graus-Porta D, Beerli R R, Hynes N E. Single-chain antibody-mediated intracellular retention of ErbB-2 impairs Neu differentiation factor and epidermal growth factor signaling. Mol Cell Biol. 1995;15:1182–1191. doi: 10.1128/mcb.15.3.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graus-Porta D, Beerli R R, Daly J M, Hynes N E. ErbB-2, the preferred heterodimerization partner of all ErbB receptors, is a mediator of lateral signaling. EMBO J. 1997;16:1647–1655. doi: 10.1093/emboj/16.7.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hato T, Pampori N, Shattil S J. Complementary roles for receptor clustering and conformational change in the adhesive and signaling functions of integrin alphaIIb beta3. J Cell Biol. 1998;141:1685–1695. doi: 10.1083/jcb.141.7.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hijazi M M, Young P E, Dougherty M K, Bressette D S, Cao T T, Pierce J H, Wong L M, Alimandi M, King C R. NRG-3 in human breast cancers: activation of multiple erbB family proteins. Int J Oncol. 1998;13:1061–1067. doi: 10.3892/ijo.13.5.1061. [DOI] [PubMed] [Google Scholar]
- 33.Ho S N, Biggar S R, Spencer D M, Schreiber S L, Crabtree G R. Dimeric ligands define a role for transcriptional activation domains in reinitiation. Nature. 1996;382:822–826. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- 34.Holsinger L J, Spencer D M, Austin D J, Schreiber S L, Crabtree G R. Signal transduction in T lymphocytes using a conditional allele of Sos. Proc Natl Acad Sci USA. 1995;92:9810–9814. doi: 10.1073/pnas.92.21.9810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoschuetzky H, Aberle H, Kemler R. Beta-catenin mediates the interaction of the cadherin-catenin complex with epidermal growth factor receptor. J Cell Biol. 1994;127:1375–1380. doi: 10.1083/jcb.127.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hynes N, Stern D F. The biology of erb-2/neu/HER-2 and its role in cancer. Biochim Biophys Acta. 1994;1198:165–184. doi: 10.1016/0304-419x(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 37.Karunagaran D, Tzahar E, Beerli R R, Chen X, Graus-Porta D, Ratzkin B J, Seger R, Hynes N E, Yarden Y. ErbB-2 is a common auxiliary subunit of NDF and EGF receptors: implications for breast cancer. EMBO J. 1996;15:254–264. [PMC free article] [PubMed] [Google Scholar]
- 38.Kashles O, Szapary D, Bellot F, Ullrich A, Schlessinger J, Schmidt A. Ligand-induced stimulation of epidermal growth factor receptor mutants with altered transmembrane regions. Proc Natl Acad Sci USA. 1988;85:9567–9571. doi: 10.1073/pnas.85.24.9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim H H, Sierke S L, Koland J G. Epidermal growth factor-dependent association of phosphatidylinositol 3-kinase with the erbB3 gene product. J Biol Chem. 1994;269:24747–24755. [PubMed] [Google Scholar]
- 40.Lenferink A E, Pinkas-Kramarski R, van de Poll M L, van Vugt M J, Klapper L N, Tzahar E, Waterman H, Sela M, van Zoelen E J, Yarden Y. Differential endocytic routing of homo- and hetero-dimeric ErbB tyrosine kinases confers signaling superiority to receptor heterodimers. EMBO J. 1998;17:3385–3397. doi: 10.1093/emboj/17.12.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levkowitz G, Klapper L N, Tzahar E, Freywald A, Sela M, Yarden Y. Coupling of the c-Cbl protooncogene product to ErbB-1/EGF-receptor but not to other ErbB proteins. Oncogene. 1996;12:1117–1125. [PubMed] [Google Scholar]
- 42.Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon W Y, Beguinot L, Geiger B, Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liberles S D, Diver S T, Austin D J, Schreiber S L. Inducible gene expression and protein translocation using nontoxic ligands identified by a mammalian three-hybrid screen. Proc Natl Acad Sci USA. 1997;94:7825–7830. doi: 10.1073/pnas.94.15.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y, Altman A. Cbl: complex formation and functional implications. Cell Signal. 1998;10:377–385. doi: 10.1016/s0898-6568(97)00179-4. [DOI] [PubMed] [Google Scholar]
- 45.MacCorkle R A, Freeman K W, Spencer D M. Synthetic activation of caspases: artificial death switches. Proc Natl Acad Sci USA. 1998;95:3655–3660. doi: 10.1073/pnas.95.7.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miloso M, Mazzotti M, Vass W C, Beguinot L. SHC and GRB-2 are constitutively activated by an epidermal growth factor receptor with a point mutation in the transmembrane domain. J Biol Chem. 1995;270:19557–19562. doi: 10.1074/jbc.270.33.19557. [DOI] [PubMed] [Google Scholar]
- 47.Miyake S, Lupher M L, Jr, Andoniou C E, Lill N L, Ota S, Douillard P, Rao N, Band H. The Cbl protooncogene product: from an enigmatic oncogene to center stage of signal transduction. Crit Rev Oncog. 1997;8:189–218. doi: 10.1615/critrevoncog.v8.i2-3.30. [DOI] [PubMed] [Google Scholar]
- 48.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muthuswamy S K, Muller W J. Direct and specific interaction of c-Src with Neu is involved in signaling by the epidermal growth factor receptor. Oncogene. 1995;11:271–279. [PubMed] [Google Scholar]
- 49a.Nolan, G. P. 1997, posting date. Protocol. [Online.] http://www.stanford.edu/group/nolan/NL-retropage.html. [11 August 1999, last date accessed.]
- 50.Ojaniemi M, Vuori K. Epidermal growth factor modulates tyrosine phosphorylation of p130Cas. Involvement of phosphatidylinositol 3′-kinase and actin cytoskeleton. J Biol Chem. 1997;272:25993–25998. doi: 10.1074/jbc.272.41.25993. [DOI] [PubMed] [Google Scholar]
- 51.Olayioye M A, Graus-Porta D, Beerli R R, Rohrer J, Gay B, Hynes N E. ErbB-1 and ErbB-2 acquire distinct signaling properties dependent upon their dimerization partner. Mol Cell Biol. 1998;18:5042–5051. doi: 10.1128/mcb.18.9.5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pinkas-Kramarski R, Soussan L, Waterman H, Levkowitz G, Alroy I, Klapper L, Lavi S, Seger R, Ratzkin B J, Sela M, Yarden Y. Diversification of Neu differentiation factor and epidermal growth factor signaling by combinatorial receptor interactions. EMBO J. 1996;15:2452–2467. [PMC free article] [PubMed] [Google Scholar]
- 53.Prigent S A, Gullick W. Identification of c-erbB-3 binding sites for phosphotidyl 3′-kinase and SHC using an EGF receptor/c-erbB-3 chimera. EMBO J. 1994;13:2831–2841. doi: 10.1002/j.1460-2075.1994.tb06577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rankin S, Hooshmand-Rad R, Claesson-Welsh L, Rozengurt E. Requirement for phosphatidylinositol 3′-kinase activity in platelet-derived growth factor-stimulated tyrosine phosphorylation of p125 focal adhesion kinase and paxillin. J Biol Chem. 1996;271:7829–7834. doi: 10.1074/jbc.271.13.7829. [DOI] [PubMed] [Google Scholar]
- 55.Riese D J, II, Stern D F. Specificity within the EGF family/ErbB receptor family signaling network. Bioessays. 1998;20:41–48. doi: 10.1002/(SICI)1521-1878(199801)20:1<41::AID-BIES7>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 56.Riese D J, II, van Raaji T M, Plowman G D, Andrews G C, Stern D F. The cellular response to neuregulins is governed by complex interactions of the erbB receptor family. Mol Cell Biol. 1995;15:5770–5776. doi: 10.1128/mcb.15.10.5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rivera V M, Clackson T, Natesan S, Pollock R, Amara J F, Keenan T, Magari S R, Phillips T, Courage N L, Cerasoli F, Jr, Holt D A, Gilman M. A humanized system for pharmacologic control of gene expression. Nat Med. 1996;2:1028–1032. doi: 10.1038/nm0996-1028. . (Comment, 2:977–978.) [DOI] [PubMed] [Google Scholar]
- 58.Romano A, Wong W T, Santoro M, Wirth P J, Thorgeirsson S S, Di Fiore P P. The high transforming potency of erbB-2 and ret is associated with phosphorylation of paxillin and a 23 kDa protein. Oncogene. 1994;9:2923–2933. [PubMed] [Google Scholar]
- 59.Siegel P M, Dankort D L, Hardy W R, Muller W J. Novel activating mutations in the neu proto-oncogene involved in induction of mammary tumors. Mol Cell Biol. 1994;14:7068–7077. doi: 10.1128/mcb.14.11.7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Siegel P M, Muller W J. Mutations affecting conserved cysteine residues within the extracellular domain of Neu promote receptor dimerization and activation. Proc Natl Acad Sci USA. 1996;93:8878–8883. doi: 10.1073/pnas.93.17.8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Skolnik E Y, Margolis B, Mohammadi M, Lowenstein E, Fischer R, Drepps A, Ullrich A, Schlessinger J. Cloning of PI3 kinase-associated p85 utilizing a novel method for expression/cloning of target proteins for receptor tyrosine kinases. Cell. 1991;65:83–90. doi: 10.1016/0092-8674(91)90410-z. [DOI] [PubMed] [Google Scholar]
- 62.Sliwkowski M X, Schaefer G, Akita R W, Lofgren J A, Fitzpatrick V D, Nuijens A, Fendly B M, Cerione R A, Vandlen R L, Carraway K L., III Coexpression of erbB2 and erbB3 proteins reconstitutes a high affinity receptor for heregulin. J Biol Chem. 1994;269:14661–14665. [PubMed] [Google Scholar]
- 63.Soltoff S P, Cantley L C. p120cbl is a cytosolic adapter protein that associates with phosphoinositide 3-kinase in response to epidermal growth factor in PC12 and other cells. J Biol Chem. 1996;271:563–567. doi: 10.1074/jbc.271.1.563. [DOI] [PubMed] [Google Scholar]
- 64.Soltoff S P, Carraway III K L, Prigent S A, Gullick W G, Cantley L C. ErbB3 is involved in activation of phosphatidylinositol 3-kinase by epidermal growth factor. Mol Cell Biol. 1994;14:3550–3558. doi: 10.1128/mcb.14.6.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sorkin A, Waters C M. Endocytosis of growth factor receptors. Bioessays. 1993;15:375–382. doi: 10.1002/bies.950150603. [DOI] [PubMed] [Google Scholar]
- 66.Sorokin A, Lemmon M A, Ullrich A, Schlessinger J. Stabilization of an active dimeric form of the epidermal growth factor receptor by introduction of an inter-receptor disulfide bond. J Biol Chem. 1994;269:9752–9759. [PubMed] [Google Scholar]
- 67.Spencer D M. Creating conditional mutations in mammals. Trends Genet. 1996;12:181–187. doi: 10.1016/0168-9525(96)10013-5. [DOI] [PubMed] [Google Scholar]
- 68.Spencer D M, Belshaw P J, Chen L, Ho S N, Randazzo F, Crabtree G R, Schreiber S L. Functional analysis of Fas signaling in vivo using synthetic inducers of dimerization. Curr Biol. 1996;6:839–847. doi: 10.1016/s0960-9822(02)00607-3. [DOI] [PubMed] [Google Scholar]
- 69.Spencer D M, Graef I, Austin D J, Schreiber S L, Crabtree G R. A general strategy for producing conditional alleles of Src-like tyrosine kinases. Proc Natl Acad Sci USA. 1995;92:9805–9809. doi: 10.1073/pnas.92.21.9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spencer D M, Wandless T J, Schreiber S L, Crabtree G R. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. . (Comment, 262:989.) [DOI] [PubMed] [Google Scholar]
- 71.Tzahar E, Pinkas-Kramarski R, Moyer J D, Klapper L N, Alroy I, Levkowitz G, Shelly M, Henis S, Eisenstein M, Ratzkin B J, Sela M, Andrews G C, Yarden Y. Bivalence of EGF-like ligands drives the ErbB signaling network. EMBO J. 1997;16:4938–4950. doi: 10.1093/emboj/16.16.4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tzahar E, Waterman H, Chen X, Levkowitz G, Karunagaran D, Lavi S, Ratzkin B J, Yarden Y. A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol Cell Biol. 1996;16:5276–5287. doi: 10.1128/mcb.16.10.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang L M, Kuo A, Almandi M, Veri M C, Lee C C, Kapoor V, Ellmore N, Chen X H, Pierce J H. ErbB2 expression increases the spectrum and potency of ligand-mediated signal transduction through ErbB4. Proc Natl Acad Sci USA. 1998;95:6809–6814. doi: 10.1073/pnas.95.12.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Webster M A, Muller W J. Mammary tumorigenesis and metastasis in transgenic mice. Semin Cancer Biol. 1994;5:69–76. [PubMed] [Google Scholar]
- 75.Weiner D B, Liu J, Cohen J A, Williams W V, Greene M I. A point mutation in the neu oncogene mimics ligand induction of receptor aggregation. Nature. 1989;339:230–231. doi: 10.1038/339230a0. [DOI] [PubMed] [Google Scholar]
- 76.Weiss A, Schlessinger J. Switching signals on or off by receptor dimerization. Cell. 1998;94:277–280. doi: 10.1016/s0092-8674(00)81469-5. [DOI] [PubMed] [Google Scholar]
- 77.Xie W, Paterson A J, Chin E, Nabell L M, Kudlow J E. Targeted expression of a dominant negative epidermal growth factor receptor in the mammary gland of transgenic mice inhibits pubertal mammary duct development. Mol Endocrinol. 1997;11:1766–1781. doi: 10.1210/mend.11.12.0019. [DOI] [PubMed] [Google Scholar]
- 78.Yang J, Symes K, Mercola M, Schreiber S L. Small-molecule control of insulin and PDGF receptor signaling and the role of membrane attachment. Curr Biol. 1998;8:11–18. doi: 10.1016/s0960-9822(98)70015-6. [DOI] [PubMed] [Google Scholar]
- 79.Zhang K, Sun J, Liu N, Wen D, Chang D, Thomason A, Yoshinaga S K. Transformation of NIH 3T3 cells by HER3 or HER4 receptors requires the presence of HER1 or HER2. J Biol Chem. 1996;271:3884–3890. [PubMed] [Google Scholar]
- 80.Zrihan-Licht S, Lim J, Keydar I, Sliwkowski M X, Groopman J E, Avraham H. Association of csk-homologous kinase (CHK) (formerly MATK) with HER- 2/ErbB-2 in breast cancer cells. J Biol Chem. 1997;272:1856–1863. doi: 10.1074/jbc.272.3.1856. [DOI] [PubMed] [Google Scholar]