Abstract
Rsp5 is an E3 ubiquitin-protein ligase of Saccharomyces cerevisiae that belongs to the hect domain family of E3 proteins. We have previously shown that Rsp5 binds and ubiquitinates the largest subunit of RNA polymerase II, Rpb1, in vitro. We show here that Rpb1 ubiquitination and degradation are induced in vivo by UV irradiation and by the UV-mimetic compound 4-nitroquinoline-1-oxide (4-NQO) and that a functional RSP5 gene product is required for this effect. The 26S proteasome is also required; a mutation of SEN3/RPN2 (sen3-1), which encodes an essential regulatory subunit of the 26S proteasome, partially blocks 4-NQO-induced degradation of Rpb1. These results suggest that Rsp5-mediated ubiquitination and degradation of Rpb1 are components of the response to DNA damage. A human WW domain-containing hect (WW-hect) E3 protein closely related to Rsp5, Rpf1/hNedd4, also binds and ubiquitinates both yeast and human Rpb1 in vitro, suggesting that Rpf1 and/or another WW-hect E3 protein mediates UV-induced degradation of the large subunit of polymerase II in human cells.
Ubiquitin-dependent proteolysis involves the covalent ligation of ubiquitin to substrate proteins, which are then recognized and degraded by the 26S proteasome. While many of the components involved in catalyzing protein ubiquitination have been identified and characterized biochemically, we are only beginning to understand how the system specifically recognizes appropriate substrates. At least three classes of activities, known as E1 (ubiquitin-activating), E2 (ubiquitin-conjugating), and E3 (ubiquitin-protein ligase) enzymes, cooperate in catalyzing protein ubiquitination (34). The enzymatic mechanisms and functions of the E1 and E2 proteins have been well characterized. In contrast, the E3 enzymes are a diverse and less-well-characterized group of activities, and many lines of evidence indicate that E3 activities play a major role in determining the substrate specificity of the ubiquitination pathway (14, 28, 34).
The hect (homologous to E6-AP carboxyl terminus) domain defines a family of E3 proteins that were discovered through the characterization of human E6-AP (17). The interaction of E6-AP with the E6 protein of the cervical cancer-associated human papillomavirus types causes E6-AP to associate with and ubiquitinate p53, suggesting that E6 functions in promoting cellular immortalization by, at least in part, stimulating the destruction of this important tumor suppressor protein (16). The hect E3 molecular masses range from 92 to over 500 kDa, with the hect domain comprising the approximately 350 carboxyl-terminal amino acids (17, 34). Exactly five hect E3s are encoded by the Saccharomyces cerevisiae genome, and over 30 have been identified so far in mammalian species. An obligatory intermediate in the ubiquitination reactions catalyzed by hect E3s is a ubiquitin-thioester formed between the thiol group of an absolutely conserved cysteine within the hect domain and the terminal carboxyl group of ubiquitin (33). E3 becomes “charged” with ubiquitin via a cascade of ubiquitin-thioester transfers, in which ubiquitin is transferred from the active-site cysteine of an E1 enzyme to the active-site cysteine of an E2 enzyme and finally to hect E3, which catalyzes isopeptide bond formation between ubiquitin and the substrate. E3 can apparently be recharged with ubiquitin while bound to the substrate and can therefore catalyze ligation of multiple ubiquitin moieties to the substrate, through conjugation either to other lysines on the substrate or to lysine residues on previously conjugated ubiquitin molecules. The resulting multiubiquitinated substrate is then recognized and degraded by the 26S proteasome. Structure-function analyses of human E6-AP and yeast Rsp5 have suggested a model for hect E3 function in which the large and nonconserved amino-terminal domains of these proteins contain determinants for substrate specificity, while the carboxyl-terminal hect domain catalyzes the multiubiquitination of bound substrates (16, 39).
The S. cerevisiae RSP5 gene encodes an essential hect E3 protein, and mutations in the gene have been isolated in multiple genetic screenings, including one for a suppressor of mutations in SPT3 (reference 41; also cited in references 17 and 18). Spt3 is part of the TATA-binding protein recognition component of the SAGA complex, which plays an important role in transcriptional activation in vivo and contains histone acetyltransferase activity (37). Rsp5 has also been identified as being involved in the down-regulation of several plasma membrane-associated permeases, including uracil permease (Fur4), general amino acid permease (Gap1), maltose permease (Mal61), and the plasma membrane H+-ATPase (5, 9, 13, 23). The primary structure of yeast Rsp5 reveals, in addition to its carboxyl-terminal hect domain, two types of domains within the amino-terminal region: C2 (one domain between amino acids 3 and 140) and WW (three domains between amino acids 231 and 418). C2 domains interact with membrane phospholipids, inositol polyphosphates, and proteins, in most cases dependent on or regulated by Ca2+ (31). Although it has not yet been demonstrated, it is possible that the C2 domain of Rsp5 is involved in targeting its membrane-associated substrates either by localizing Rsp5 to the plasma membrane or by directly mediating the interactions with these substrates.
WW domains are protein-protein interaction modules that recognize proline-rich sequences, with the consensus binding site containing either a PPXY (4, 21), PPLP (1a, 7), or PPPGM (2) sequence. WW domains, like SH3 domains, recognize polyproline ligands with high specificity but low affinity (Kd = 1 to 200 μM). The basis of recognition is the N-substituted nature of the proline peptide backbone rather than the proline side chain itself (26). It has been suggested that this explains how WW and SH3 domains can achieve specific but low-affinity recognition of ligands, since proline is the only natural N-substituted amino acid. It has also recently been shown that WW domains can recognize phosphoserine- and phosphothreonine-containing ligands (22), which has important implications for the diversity of substrates that may be recognized by Rsp5 and other WW domain-containing hect E3s. A structure-function analysis of Rsp5 showed that the hect domain and the region spanning WW domains 2 and 3 are necessary and sufficient to support the essential in vivo function of Rsp5, while the C2 domain and WW domain 1 are dispensable, at least under standard growth conditions (39). Together, the results of our structure-function analyses imply that ubiquitination of one or more substrates of Rsp5 is essential for cell viability and that the critical substrate(s) is recognized by the region containing WW domains 2 and 3.
Members of our group previously reported the results of a biochemical approach for identifying substrates of Rsp5, which led to the identification of Rpb1, the largest subunit (LS) of RNA polymerase II (Pol II), as a substrate of Rsp5 (18). Rpb1 is very efficiently ubiquitinated by Rsp5 in vitro, and the WW domain region mediates binding to Rpb1, with WW domain 2 being most critical. Since the requirements for Rpb1 binding and ubiquitination parallel those for the essential function of Rsp5, Rpb1 is a candidate for being at least one of the substrates related to the essential function of Rsp5. The biological relevance of Rpb1 ubiquitination was not initially clear, however, since Rpb1 is an abundant, long-lived protein in vivo. Interestingly, another study showed that the Pol II LS is subject to ubiquitination and degradation in response to UV irradiation (3, 30); however, the enzymatic components of the ubiquitin system responsible for this phenomenon were not identified or characterized. We show here that UV irradiation or treatment with a UV-mimetic chemical induces the degradation of Rpb1 in yeast cells and that Rsp5 and the 26S proteasome mediate this effect. Furthermore, we show that human Rpf1, a WW domain-containing hect (WW-hect) E3 protein, binds and ubiquitinates Rpb1 in vitro, suggesting that this may be the E3 protein that mediates UV-induced degradation of the Pol II LS in human cells.
MATERIALS AND METHODS
Yeast strains and plasmids.
FY56 (RSP5), FW1808 (rsp5-1), and the Gal-RSP5 strain were described previously (18, 39). The sen3-1 (MHY811) and SEN3 (MHY810) strains (6) were kindly provided by Mark Hochstrasser (University of Chicago). The tom1 null mutant strain was made by single-step gene disruption in the diploid strain W303, and haploid tom1Δ colonies were isolated by the sporulation and dissection of the heterozygous TOM1/tom1Δ diploid. All plasmids that promote the expression of Rsp5 and Rpb1 were described previously (18, 39). Plasmids that promote the bacterial expression of glutathione S-transferase (GST)–Rpf1 fusion proteins were generated by PCR amplification of regions of the Rpf1 open reading frame in plasmid pBKC-hRPF1 (19). The GST-Rpf1 N protein contains amino acids 13 to 192 of Rpf1, the GST-WW protein contains amino acids 193 to 506, the GST-C protein contains amino acids 506 to 901, and the GST–WW-hect protein contains amino acids 193 to 901. This numbering is based on the assumption that amino acid 29 of the protein sequence given in GenBank (accession no. D42055) is the initiating methionine. pGEX-5x-1 (Pharmacia, Piscataway, N.J.) was the cloning vector for the expression of all the GST fusion proteins except for GST–WW-hect, which was expressed by pGEX-6p-1.
Protein purification and biochemical assays.
GST fusion proteins for ubiquitination assays and protein binding assays were expressed in Escherichia coli by standard methods and affinity purified on glutathione-Sepharose (Pharmacia). Ubiquitination assays utilized hect E3 proteins (Rsp5, the Rsp5 C-A mutant, human E6-AP, and Rpf1 WW-hect) that were cleaved from the GST portion of the molecule with PreScission protease (Pharmacia). These proteins were then used in ubiquitination assays with 35S-labeled yeast Rpb1 that had been translated in vitro with a TNT rabbit reticulocyte lysate system (Promega, Madison, Wis.), as described previously (18).
Rpb1 binding assays were performed by mixing 100 ng of GST-E3 fusion protein bound to 10 μl of glutathione-Sepharose with 80 μg of total HeLa cell lysate (cell lysis buffer: 0.1 M Tris [pH 8.0], 0.1 M NaCl, and 1% NP-40), with the remainder of the 125-μl volume consisting of 25 mM Tris (pH 8.0) and 125 mM NaCl. Reaction mixtures were rotated for 2 h at 4°C, and the beads were washed three times with 500 μl of cell lysis buffer. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer was added directly to the Sepharose and heated at 95°C for 5 min, and proteins were analyzed by SDS-PAGE and Western blotting with either anti-carboxyl-terminal domain (anti-CTD) antibody (generously provided by Danny Reinberg, University of Medicine and Dentistry of New Jersey, Piscataway) or anti-Pol II antibody N-20 from Santa Cruz Biotechnology (Santa Cruz, Calif.).
Analysis of UV- and 4-NQO-treated cells.
HeLa cells were maintained in Dulbecco’s modified Eagle medium with 10% fetal bovine serum, and UV irradiation was performed on tissue culture dishes after the removal of the medium. A germicidal lamp emitting light at 254 nm with an incident dose rate of 1.5 J per m2 per s was used, and the time of irradiation was generally 15 s, for a total dose of 22.5 J per m2. Fresh medium was then added to the cells, which were then allowed to recover for various times at 37°C. 4-Nitroquinoline-1-oxide (4-NQO) (Sigma), prepared as a 0.5-mg/ml stock solution in ethanol, was added directly to the medium at various concentrations and times. Extracts were made by lysing cells directly in SDS-PAGE loading buffer.
Yeast cells were irradiated as follows. Log-phase liquid cultures (5 optical density [OD] units) were concentrated by centrifugation to 0.5 ml, and then the cells were spread onto 10-cm agar plates. The liquid was allowed to absorb into the plates for 30 min at 30°C, and then the plates were irradiated for 15 s, as described above for HeLa cells. The cells were then collected from the plates and extracts were prepared as described below. Log-phase liquid yeast cultures (5 OD units) were treated with 4-NQO by adding a 0.5-mg/ml stock solution in ethanol directly to the culture medium for either 30 or 60 min. Yeast cell extracts were prepared by the method of Silver et al. (36). Briefly, 5 OD units of cells were resuspended in 1 ml of 0.25 M NaOH–1% β-mercaptoethanol and incubated on ice for 10 min. A volume of 0.16 ml of 50% trichloroacetic acid was added, and incubation on ice was continued for 10 min. The precipitate was collected by microcentrifugation at 4°C for 10 min, and then the pellet was washed with cold acetone, dried, and resuspended in 200 μl of SDS-PAGE sample buffer. Samples were heated at 95°C for 10 min prior to being loaded onto SDS-PAGE gels. Protein from the equivalent of 0.1 to 0.25 OD unit of cells was analyzed on SDS–7% PAGE gels for Western analyses of Rpb1. Immunoprecipitation and Western blotting (see Fig. 4) were performed by diluting 40 μl of yeast extract with 1.4 ml of 25 mM Tris (pH 7.9)–125 mM NaCl, followed by the addition of antibody and 20 μl of protein A-Sepharose (Pharmacia). The mixture was rotated at 4°C for 4 h; the Sepharose beads were collected, washed, and boiled in sample buffer; and then the proteins were analyzed by SDS-PAGE followed by immunoblotting.
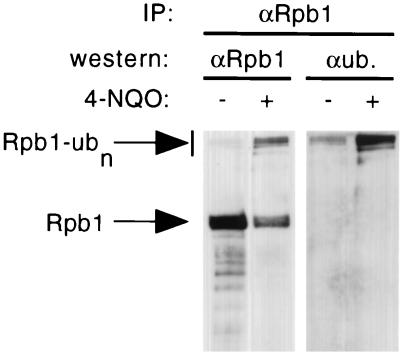
FIG. 4.
Antiubiquitin antibody recognizes high-molecular-weight forms of Rpb1 from 4-NQO-treated cells. Yeast cells were treated with 4-NQO at 4 μg/ml for 30 min, and whole-cell extracts were prepared. Rpb1 was immunoprecipitated (IP) in duplicate from each sample with anti-CTD antibody. The immunoprecipitates were then analyzed by SDS-PAGE followed by immunoblotting with either anti-CTD (αRpb1) or antiubiquitin (αub.) antibody.
Antibodies utilized in this study were either anti-CTD rabbit polyclonal antibody (used for yeast Rpb1 Western analyses and Rpb1 immunoprecipitations; generously provided by Danny Reinberg), anti-human Pol II rabbit polyclonal antibody N-20 (Santa Cruz Biotechnology), antiubiquitin mouse monoclonal antibody (Santa Cruz Biotechnology), antihemagglutinin rabbit polyclonal antibody (Santa Cruz Biotechnology), anti-Rsp5 mouse monoclonal antibody (39), or anti-Rfa1 rabbit polyclonal antibody (generously provided by Steve Brill, Rutgers University). Horseradish peroxidase-linked secondary antibodies and chemiluminescent reagents were obtained from DuPont NEN.
RESULTS
UV irradiation and 4-NQO induce the degradation of Rpb1 in both human and yeast cells.
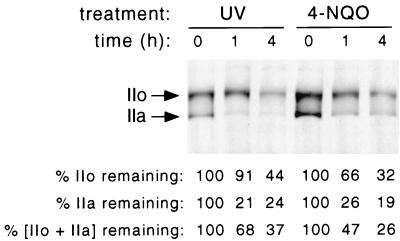
4-NQO is considered a UV mimetic because it is metabolized to yield a compound that reacts with purine nucleotides of DNA, and these adducts are processed by the nucleotide excision repair (NER) system in a manner similar to that of dipyrimidine photoproducts induced by 254-nm UV light (15, 29). It was previously shown that UV irradiation of human cells induces the ubiquitination and degradation of human Rpb1 (hRpb1) (3, 30). Figure 1 demonstrates this effect in HeLa cells. Cells were irradiated with 254-nm UV light at a dose of 22.5 J per m2, and cell extracts were made at various times, up to 4 h after irradiation. Extracts were analyzed by SDS-PAGE, followed by immunoblotting with an antibody that recognizes the amino-terminal region of hRpb1 and therefore detects both hypophosphorylated (IIa) and hyperphosphorylated (IIo) forms of the protein. The degradation of the IIa form was more rapid and more complete than the degradation of the IIo form, with the IIa form reaching a minimum degradation of 20 to 25% of the initial amount after 1 h, while the IIa form reached a minimum degradation of 40 to 50% of the initial amount after 4 h. 4-NQO treatment stimulated the degradation of hRpb1 over a similar time course, again with the IIa form disappearing more rapidly and more completely than the IIo form. Lactacystin, a highly specific inhibitor of the proteasome, inhibited both UV- and 4-NQO-induced degradation of hRpb1 (not shown), which is consistent with previous reports that this effect is mediated by the 26S proteasome of the ubiquitin system (30).
FIG. 1.
hRpb1 levels following UV irradiation and 4-NQO treatment of HeLa cells. HeLa cells were irradiated with 254-nm UV light at 22.5 J per m2 as described in Materials and Methods, and cell extracts were prepared immediately or 1 or 4 h postirradiation. For 4-NQO treatment, the chemical was added directly to the culture medium at a final concentration of 0.5 μg/ml, and cell extracts were prepared immediately or 1 or 4 h later. Relative hRpb1 levels were determined by SDS-PAGE and immunoblotting and quantitated by densitometry. Levels are expressed as the percentage of Rpb1 remaining relative to the level in untreated cells.
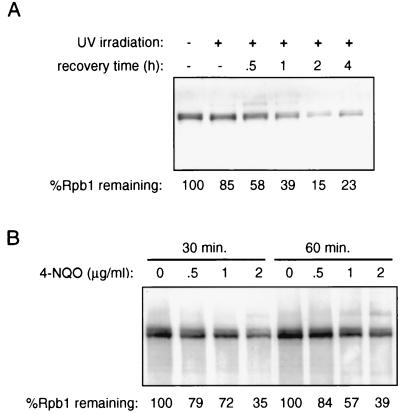
Figure 2 shows that the degradation of Rpb1 was also induced in S. cerevisiae by both UV irradiation and 4-NQO treatment. UV irradiation of intact yeast cells on agar plates led to a dose- and time-dependent decrease in the steady-state level of Rpb1 (Fig. 2A). Rpb1 levels reached a minimum of 15 to 20% of the initial amount between 1 and 2 h after irradiation and began to return to normal after 4 h. 4-NQO also elicited a dose-dependent decrease in Rpb1 levels, reaching a minimum 30 to 60 min after the addition of 4-NQO (Fig. 2B). The amount of Rpb1 remaining in the experiment whose results are shown was 35 to 40% of the initial amount; however, in other experiments, the minimum was generally 25 to 30% (Fig. 3). Neither UV irradiation nor 4-NQO treatment resulted in a significant loss of viability at doses necessary to elicit maximal Rpb1 degradation. Unlike hRpb1, the hypo- and hyperphosphorylated forms of yeast Rpb1 migrate as a very closely spaced doublet and are not easily distinguished by SDS-PAGE. Therefore, it is difficult to conclude whether there is an apparent preferential disappearance of one form over the other, as there is with hRpb1.
FIG. 2.
(A) Rpb1 levels following UV irradiation of yeast. Yeast cells (strain FY56) were irradiated at 22.5 J per m2 as described in Materials and Methods, and whole-cell extracts were made at the indicated times postirradiation. Rpb1 was detected by SDS-PAGE followed by immunoblotting with anti-CTD antibody. Rpb1 levels were quantitated by densitometry and are expressed as the percentage of Rpb1 remaining relative to the level in untreated cells. (B) Rpb1 levels following 4-NQO treatment. 4-NQO was added to liquid cultures of log-phase yeast at the indicated concentrations, and cells were collected at the indicated times following addition. Whole-cell extracts were prepared, and Rpb1 was detected by SDS-PAGE and immunoblotting.
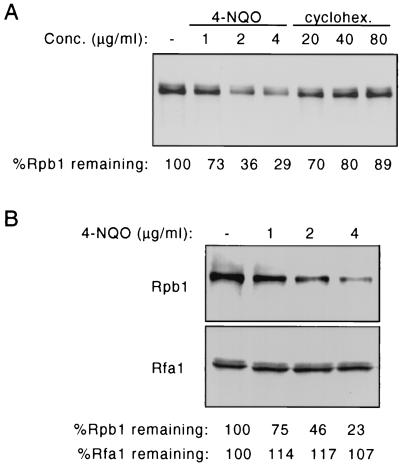
FIG. 3.
(A) Yeast cells (FY56 [RSP5]) were treated with the indicated doses of 4-NQO or cycloheximide (cyclohex.) for 30 min, cell extracts were prepared, and Rpb1 levels were examined by SDS-PAGE and immunoblotting with anti-CTD antibody. (B) Yeast cells (FY56 [RSP5]) were treated with the indicated doses of 4-NQO for 30 min, cell extracts were prepared, and Rpb1 levels and Rfa1 levels were examined by SDS-PAGE and immunoblotting.
To rule out the possibility that the decrease in yeast Rpb1 levels accompanying UV or 4-NQO treatment was simply the result of inhibition of the synthesis of Rpb1, 4-NQO-treated cells were compared to cells treated with cycloheximide. As shown in Fig. 3A, cycloheximide treatment led to only a slight decrease in Rpb1 levels after 45 min, whereas 4-NQO treatment resulted in the reduction in Rpb1 levels as described above. Total cellular protein levels were not affected by 4-NQO treatment, and Coomassie blue staining of SDS-PAGE gels indicated that the effect of 4-NQO was specific for Rpb1. This was confirmed by immunoblotting for an unrelated nuclear protein, Rfa1, a component of replication protein A. Figure 3B shows that levels of Rfa1 were not affected by 4-NQO treatment under conditions in which Rpb1 degradation was induced.
The appearance of slower-migrating forms of Rpb1, suggestive of ubiquitinated intermediates, was evident in some experiments at higher concentrations of 4-NQO and on longer film exposures. These slower-migrating bands were shown to be ubiquitinated forms of Rpb1 by immunoprecipitating them with anti-Rpb1 antibody, followed by immunoblotting with either anti-CTD or antiubiquitin antibody (Fig. 4). While the accumulation of ubiquitinated forms of Rpb1 was clearly stimulated by 4-NQO, there was some reaction of the Rpb1 immunoprecipitate with the antiubiquitin antibody even in untreated cells. This may reflect a basal level of Rpb1 ubiquitination in normally growing cells, as suggested previously (18).
4-NQO-induced degradation of Rpb1 is dependent on RSP5 and SEN3/RPN2.
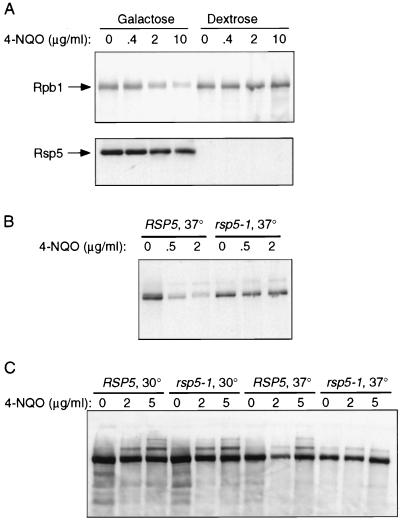
Members of our group previously showed that Rsp5 ubiquitinates Rpb1 in vitro (18). To determine if the in vivo-induced degradation of Rpb1 was dependent on Rsp5, we first took advantage of a yeast strain that contains a single copy of a conditionally expressed wild-type RSP5 gene. The Gal-RSP5 yeast strain contains an epitope-tagged RSP5 gene under the control of the GAL1 promoter, which is integrated at the RSP5 chromosomal locus (18). This strain was grown to early log phase in galactose-containing medium, and then it was switched to dextrose-containing medium for 48 h. Figure 5A shows that Rsp5 protein levels were dramatically reduced after 48 h in dextrose. The cells were still fully viable at this point and resumed growth when shifted back to galactose-containing medium. The dextrose-shifted cells were treated with 4-NQO and compared to log-phase cells that had been maintained in galactose-containing medium. 4-NQO-induced Rpb1 degradation occurred in the cells maintained in galactose, but not in Rsp5-depleted cells. These results suggest that 4-NQO-induced degradation of Rpb1 is dependent on RSP5.
FIG. 5.
(A) 4-NQO treatment of the Gal-RSP5 strain maintained in galactose or shifted to dextrose. The Gal-RSP5 strain was grown to early log phase in galactose-containing medium, and then the cells were either shifted to dextrose-containing medium for 48 h or maintained in galactose-containing medium. The cultures were then treated with 4-NQO at the indicated concentrations for 30 min, and whole-cell extracts were prepared and analyzed by SDS-PAGE and immunoblotting with either an anti-Rsp5 monoclonal antibody (bottom) or anti-CTD antibody (top). (B) 4-NQO treatment of the rsp5-1 temperature-sensitive mutant. Strains FY56 (RSP5) and FW1808 (rsp5-1) were grown to mid-log phase at 30°C and then shifted to 37°C for 1 h. 4-NQO was then added at the indicated concentrations for 30 min. Whole-cell extracts were prepared, and Rpb1 was detected by SDS-PAGE and immunoblotting. (C) Experiment similar to that in panel B, except that cells were treated with 4-NQO at both 30 and 37°C.
To independently confirm the importance of Rsp5 in the induced degradation of Rpb1, we examined the effect of 4-NQO on the temperature-sensitive rsp5-1 mutant. Temperature sensitivity is conferred by a single amino acid change (amino acid 733) within the hect domain that directly affects the catalytic activity of the protein (39). The rsp5-1 strain grows with a slightly longer doubling time than an isogenic RSP5 strain at 30°C but arrests within 30 to 60 min after a shift to 37°C. Figure 5B shows that Rpb1 degradation was induced by 4-NQO in an isogenic wild-type RSP5 strain at 37°C, while little or no loss of Rpb1 was seen in the rsp5-1 strain at 37°C. Figure 5C shows the results of an experiment in which multiubiquitinated forms of Rpb1 were evident following 4-NQO treatment. The accumulation of these forms was seen in the wild-type RSP5 strain at both 30 and 37°C and in the rsp5-1 strain at 30°C, but not at 37°C. These results again indicate that 4-NQO-induced ubiquitination and degradation of Rpb1 are RSP5 dependent.
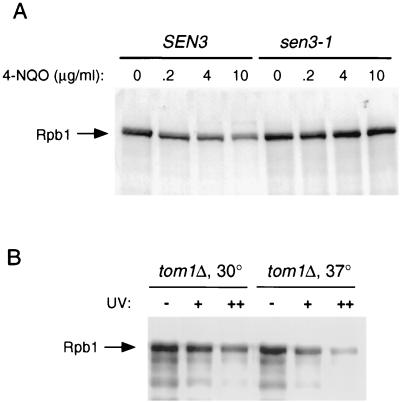
A strain containing a mutation in a subunit of the 26S proteasome was used to determine if the 4-NQO-induced degradation of Rpb1 was proteasome dependent. SEN3/RPN2 encodes an essential non-ATPase regulatory subunit of the 26S proteasome (6). The sen3-1 mutant shows a growth defect at 30°C (doubling time of 4.5 h) and a more severe growth defect at higher temperatures. The MATα2 transcription factor and certain artificial substrates of the ubiquitin system (Ub-Pro-β-galactosidase and Ub-Leu-β-galactosidase) have been shown to be stabilized in this mutant at 30°C. We compared the sen3-1 mutant to an isogenic wild-type SEN3 strain for its ability to support 4-NQO-induced degradation of Rpb1. As shown in Fig. 6A, the sen3-1 mutant was defective in 4-NQO-induced degradation of Rpb1 compared to the SEN3 strain. This result indicates that UV-induced degradation of Rpb1 is proteasome dependent, consistent with the observation that proteasome inhibitors blocked the degradation of the human Pol II LS in response to UV irradiation (30).
FIG. 6.
(A) 4-NQO treatment of SEN3 and sen3-1 strains at 37°C. 4-NQO was added at the indicated concentrations for 30 min. Whole-cell extracts were prepared, and Rpb1 was detected by SDS-PAGE and immunoblotting. (B) The tom1Δ mutant was grown at 30°C and then either maintained at 30°C or shifted for 4 h to 37°C. Cells were then irradiated at either 25 (+) or 50 (++) J/m2, followed by a 1-h recovery period at their respective temperatures. Whole-cell extracts were then prepared, and Rpb1 was detected by SDS-PAGE and immunoblotting.
A caveat to the experiments utilizing the GAL-RSP5, rsp5-1, and sen3-1 strains is that both RSP5 and SEN3/RPN2 are essential genes, and their inactivation results in growth inhibition. Therefore, indirect effects cannot be ruled out as being responsible for the block in 4-NQO-induced Rpb1 degradation seen in these mutants. To rule out the possibility that the block in Rpb1 degradation is due to a general growth arrest, we examined a temperature-sensitive mutation in a gene not predicted to affect either Rsp5 or Rpb1. We examined a tom1 null mutant, since TOM1 encodes a hect E3 protein that does not interact with Rpb1. Interestingly, Tom1 appears to influence transcription through effects on ADA coactivators, possibly by targeting the Spt7 protein for ubiquitination (32). The tom1 null mutant has a near-normal doubling time at 30°C but exhibits a strong growth arrest within 2 h after a shift to 37°C. The tom1 mutant was UV irradiated either at 30°C or 4 h after a shift to 37°C, and Rpb1 levels were examined. Figure 6B shows that the degradation of Rpb1 was induced at both temperatures. Therefore, the lack of induced degradation in the rsp5 and sen3 mutants is unlikely to be due to general growth arrest or cell stress.
Rpf1/hNedd4, a human hect E3 protein related to Rsp5, binds and ubiquitinates Rpb1 in vitro.
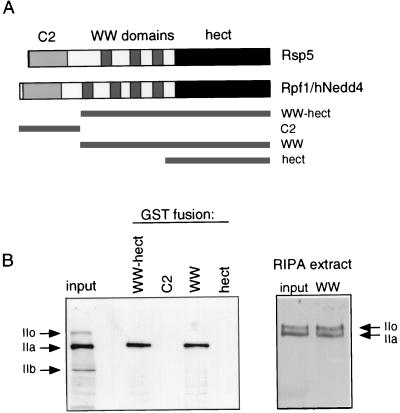
Rpf1, also known as human Nedd4 (hNedd4), has a C2 domain at its extreme amino terminus, four WW domains in the central portion of the molecule, and a carboxyl-terminal hect domain (Fig. 7A). Rpf1 is one of at least seven human hect E3s that have this general organization, with a variable number of WW domains (two to four). GST-Rpf1 proteins were expressed as indicated in Fig. 7A, and equivalents amounts (100 ng) of each protein were assayed for the ability to bind to hRpb1. The full-length Rpf1 protein was not used in this analysis because it was produced in small amounts in bacteria and, furthermore, was not catalytically active, as judged by ubiquitin-thioester assays (not shown). Rpf1 WW-hect and the isolated WW domain region stably bound the hRpb1 present in the HeLa cell extract (Fig. 7B, left panel), whereas neither the isolated C2 domain nor the hect domain bound to hRpb1. These results are consistent with previous results showing that the WW domain region of Rsp5 is necessary and sufficient for binding to yeast Rpb1 (18, 39). In addition, a well-characterized proteolyzed form of hRpb1 (form IIb) that lacks the CTD did not bind to Rpf1, also consistent with previous results showing that the CTD is the binding site for Rsp5 (18, 39). There was an apparent preferential binding of Rpf1 to the hypophosphorylated (IIa) form of hRpb1 in this experiment; however, the degree to which the phosphorylated (IIo) form of hRpb1 associated with Rpf1 was dependent on the cell extraction buffer. When the cell lysis buffer conditions were harsher (radioimmunoprecipitation assay buffer instead of NP-40 lysis buffer [11]), an equivalent portion of hyperphosphorylated hRpb1 bound to Rpf1 (Fig. 7B, right panel). This suggests that the interaction of the hyperphosphorylated CTD with other proteins might preclude binding to Rpf1 and that Rsp5 and Rpf1 have an inherent ability to bind to both forms of the protein. This interpretation is consistent with previous results showing that Rsp5 could bind to both the IIo and IIa forms of purified Pol II holoenzyme in vitro (18).
FIG. 7.
(A) Schematic representation of yeast Rsp5 and human Rpf1/Nedd4. GST-Rpf1 fusions to the regions of Rpf1 indicated by the solid bars were made. (B) (Left) HeLa cell extract was prepared in NP-40 lysis buffer (see Materials and Methods). The binding of hRpb1 to GST-Rpf1 fusion proteins immobilized on glutathione-Sepharose was analyzed by SDS-PAGE and immunoblotting. The “input” shows hRpb1 in the extract with forms IIo, IIa, and IIb. (Right) Similar experiment, with HeLa cell extract prepared in radioimmunoprecipitation assay (RIPA) buffer. The input and binding to GST-WW are shown.
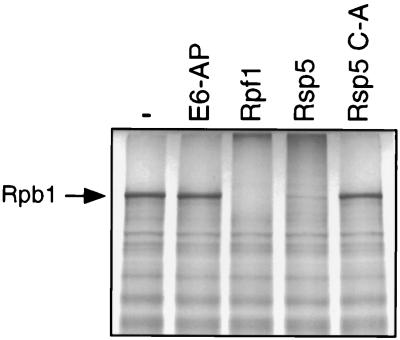
To determine if Rpf1 can ubiquitinate Rpb1, the Rpf1 WW-hect protein was cleaved from the purified GST fusion protein and assayed for its ability to ubiquitinate in vitro-translated yeast Rpb1. Rpf1 was as efficient in stimulating multiubiquitination of Rpb1 as yeast Rsp5 (Fig. 8). Neither the mutant of Rsp5 with a change of the active-site cysteine to alanine nor human E6-AP ubiquitinated Rpb1. Together, the binding and ubiquitination results suggest that Rpf1 may mediate the DNA damage-induced degradation of the Pol II LS in human cells.
FIG. 8.
Ubiquitination of Rpb1 by Rpf1 in vitro. Rpb1 was translated in vitro in rabbit reticulocyte lysate in the presence of [35S]-methionine. Purified hect E3 proteins (human E6-AP, human Rpf1 [WW-hect; amino acids 193 to 901], yeast Rsp5, and the mutant of Rsp5 with a change of the active-site Cys to Ala [C-A]) were incubated as indicated with Rpb1 in the presence of ATP, ubiquitin, E1 enzyme, and E2 enzyme (Arabidopsis thaliana Ubc8) as previously described (18).
DISCUSSION
Rpb1 was initially identified as a substrate of Rsp5 based on a biochemical screening for proteins that were bound and ubiquitinated by Rsp5 in vitro (18). While Rsp5 was found to efficiently multiubiquitinate Rpb1 in vitro, the biological function of this was unclear, since Rpb1 is an abundant and stable protein in vivo. The steady-state level of Rpb1 was found to increase modestly (approximately three- to fivefold) on prolonged transcriptional repression of RSP5, providing evidence that Rpb1 may be a bona fide substrate of Rsp5 in vivo, even if the half-life of Rpb1 under normal growth conditions is relatively long. Other studies have shown that the inhibition of transcription caused by the exposure of mammalian cells to DNA-damaging agents or treatment, including α-amanitin, actinomycin D, cisplatin, and UV irradiation, leads to the degradation of the Pol II LS (3, 27). Ratner et al. further demonstrated that the degradation of the Pol II LS induced by UV irradiation was ubiquitin and proteasome dependent (30). Together, these results suggested that the recognition of Rpb1 by Rsp5 might be enhanced in response to DNA damage. The experiments described here showed that, as in human cells, DNA damage induces the ubiquitination and degradation of Rpb1 in S. cerevisiae and that this is dependent on the Rsp5 ubiquitin-protein ligase. In addition, a human hect E3 protein closely related to Rsp5, Rpf1/hNedd4, is shown to bind and ubiquitinate Rpb1 in vitro, suggesting that this hect E3 protein might mediate UV-induced degradation of Rpb1 in human cells.
It has long been recognized that RNA synthesis is down-regulated in response to DNA damage and that stalled RNA polymerase at sites of DNA damage might serve as a signal for the recruitment of the NER machinery (10, 24). This is thought to be the basis of a specialized form of NER, transcription-coupled repair (TCR), in which lesions within the transcribed strand of genes are repaired more rapidly than lesions on the nontranscribed strand or outside of the transcription units. TCR also occurs in E. coli, where the transcription repair coupling factor binds to and releases RNA polymerase stalled at a lesion and then stimulates the recruitment of the repair machinery (35). Several lines of evidence suggest that the mechanism of TCR is more complex in eukaryotes, and it is generally thought that a stalled RNA polymerase can resume transcript synthesis following repair. This is based in part on the stability of stalled RNA polymerase-template-RNA complexes in vitro and the idea that it would be energetically wasteful to abort transcript synthesis entirely. The finding that a fraction of the Pol II LS is ubiquitinated and degraded in response to DNA damage suggests an alternative mechanism for the down-regulation of transcription in response to DNA damage: irreversible disassembly of transcription complexes by the degradation of the major catalytic subunit of Pol II.
It is not yet clear which form of Pol II is targeted for ubiquitin-mediated degradation following DNA damage. The CTD, which is necessary and sufficient for Rsp5 binding, is subject to phosphorylation and dephosphorylation events during the transcription cycle and is also the site of interaction of many components of the transcription machinery (25). The CTD is hypophosphorylated (IIa) in Pol II transcription initiation complexes and undergoes phosphorylation upon promoter clearance to yield a hyperphosphorylated (IIo) form that persists throughout transcription elongation. Ratner et al. (30) reported that ubiquitinated forms of hRpb1 detected after UV irradiation reacted with an antibody that is specific for the hyperphosphorylated form of hRpb1, suggesting that Pol II complexes arrested at intragenic damage sites might be the preferential substrate for ubiquitination. This is not consistent, however, with the observation that the hypophosphorylated form of hRpb1 preferentially disappears in response to either UV irradiation or 4-NQO treatment. In order to explain this discrepancy, Ratner et al. suggested that the apparent loss of hypophosphorylated hRpb1 upon UV irradiation might reflect a rapid conversion of hypo- to hyperphosphorylated Rpb1 in order to compensate for the loss of hyperphosphorylated Rpb1. While we cannot exclude this possibility, the data are also consistent with a model in which the hypophosphorylated form of Pol II is actually the preferential substrate for ubiquitination but that the kinetics of its ubiquitination and degradation are too rapid to allow the detection of ubiquitinated intermediates.
While further studies are clearly necessary to determine which form of Pol II is targeted for ubiquitin-mediated degradation in response to DNA damage in vivo, our in vitro results suggest that there is not a specific requirement for the recognition of Rpb1 by Rsp5 in terms of the phosphorylation state of the CTD. Phosphorylation of the CTD is not a prerequisite for Rsp5 recognition, since in vitro-translated Rpb1 and GST-CTD produced in bacteria are both efficiently recognized by Rsp5. We also showed previously that the hypo- and hyperphosphorylated forms of purified human Pol II holoenzyme bind equally well to GST-Rsp5 (18). In addition, both Rsp5 and Rpf1 bind to the hypophosphorylated form of hRpb1 present in human cell extracts; however, the degree to which Rsp5 and Rpf1 can bind to hyperphosphorylated hRpb1 is a function of the cell extraction buffer, with more stringent extraction buffers resulting in more binding of the hyperphosphorylated forms. Together, these results suggest that the association of other transcription factors with Pol II, and specifically with the CTD, might block recognition by Rsp5 in vivo. Changes in Pol II transcription complexes in response to DNA damage, such as the dissociation of specific CTD-associated proteins or the dissociation of the elongated polymerase complex from the template, might then allow Rsp5 to bind and ubiquitinate Rpb1.
Rsp5 is the only hect E3 protein in yeast that has a C2 domain and WW domains, while at least seven human hect E3s with C2 and WW domains have been identified. The WW domains, as well characterized protein-protein interaction modules, are likely to mediate the interaction with at least some of the substrates of Rsp5, including Rpb1 (39). WW domains bind proline-rich ligands, with the best-characterized ligand being the PY motif (containing a PPXY sequence). In addition, it has recently been shown that WW domains can also recognize phosphoserine- and phosphothreonine-containing ligands (22), suggesting that there are two disparate types of WW domain ligands. The CTD heptapeptide consensus (YSPTSPS) may be a nonconsensus PY motif in the context of the repeating heptapeptide (YXPXXPXYXPXXPX). Alternatively, if the phosphorylated form of Rpb1 is the in vivo substrate of Rsp5, phosphorylation at the serine and/or threonine residues may contribute to recognition, although as mentioned above, phosphorylation is not required for the binding of Rsp5 to the CTD in vitro. Our finding that Rpf1/hNedd4 can bind and ubiquitinate hRpb1 in vitro suggests that this may be the E3 enzyme responsible for this effect in human cells. Preliminary results, however, indicate that other WW-hect E3s can also bind to Rpb1 in vitro (1). It is possible that while several of the WW-hect E3s can bind and ubiquitinate Rpb1 in vitro, intracellular localization is the key determinant of which E3 can target Rpb1 in vivo. Mouse Nedd4 and yeast Rsp5 are primarily cytoplasmic (12, 40); however, there is now a precedent for the ubiquitin-mediated degradation of nuclear proteins being linked to their export from the nucleus to the cytoplasm (8, 38).
While it is now established that DNA damage induces the degradation of Rpb1 in both yeast and human cells, the relevance of this to DNA repair is not yet clear. Rsp5 mutants do not show any apparent UV sensitivity, although we cannot yet rule out more subtle effects of Rsp5 on the efficiency of DNA repair. The fact that both CSA and CSB Cockayne syndrome cells were found to be defective in UV-induced Rpb1 degradation in human cells suggested that this is related to the process of TCR. However, a rad26 null mutant (Rad26 is the yeast CSB homolog and the only yeast protein known to be required for TCR but not for NER) exhibited no defect in 4-NQO-induced Rpb1 degradation (data not shown). This suggests that TCR may not be directly linked to DNA damage-induced degradation of Rpb1, at least in yeast, and again raises the question of which form of Pol II is the in vivo substrate of Rsp5. The expression of Rpf1/hNedd4 in yeast cannot functionally substitute for expression by RSP5 in terms of either cell viability or the UV-induced effect on Rpb1 (data not shown). The basis of this noncomplementation is not known but could be related to an inability of Rpf1 to productively interact with other components of the ubiquitin system in yeast.
Several examples of regulated substrate ubiquitination have now been characterized. In many cases, modification of the substrate, often by phosphorylation, can serve as a signal for recognition by specific E3 ubiquitin-protein ligases, as in the recognition of phosphorylated Sic1 by SCFCdc4 (28). In other cases, the unmasking of ubiquitination signals can occur when a substrate dissociates from an interacting protein, as in the case of the mutual destruction of the MATα2 and MATa1 transcription factors upon the dissociation of the heterodimer (20). An unmasking of the recognition signals on Rpb1 in response to DNA damage may account for the observations that Rpb1 is freely and efficiently recognized by Rsp5 under several different experimental conditions in vitro yet is normally a stable and long-lived protein in vivo. It seems likely that the nature of the Rpb1 CTD, as an organizational center for many components of the basal transcription machinery, might preclude Rsp5 from interacting with Rpb1 during the normal transcription cycle. DNA damage may signal alterations in Pol II complexes in a manner that allows Rsp5 to recognize and ubiquitinate Rpb1. Further studies on the effects of DNA damage of Pol II holoenzyme complexes will aid in addressing this hypothesis.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (CA72943, J.M.H., and DK50495, D.P.D.), an award to J.M.H. from the Charles and Johanna Busch Foundation, and a U.S. Army Medical Research and Materiel Command Predoctoral Fellowship to M.R.H.
We thank Mark Hochstrasser for yeast strains and Keven Sweder and Kiran Madura for helpful discussions.
REFERENCES
- 1.Beaudenon, S. L., and J. M. Huibregtse. Unpublished results.
- 1a.Bedford M T, Chan D C, Leder P. FBP WW domains and the Abl SH3 domain bind to a specific class of proline-rich ligands. EMBO J. 1997;16:2376–2383. doi: 10.1093/emboj/16.9.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedford M T, Reed R, Leder P. WW domain-mediated interactions reveal a spliceosome-associated protein that binds a third class of proline-rich motif: the proline glycine and methionine-rich motif. Proc Natl Acad Sci USA. 1998;95:10602–10607. doi: 10.1073/pnas.95.18.10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bregman D B, Halaban R, van Gool A J, Henning K A, Friedberg E C, Warren S L. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc Natl Acad Sci USA. 1996;93:11586–11590. doi: 10.1073/pnas.93.21.11586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H I, Sudol M. The WW domain of YES-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci USA. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de la Fuente N, Maldonado A M, Portillo F. Glucose activation of the yeast plasma membrane H+-ATPase requires the ubiquitin-proteasome proteolytic pathway. FEBS Lett. 1997;411:308–312. doi: 10.1016/s0014-5793(97)00721-7. [DOI] [PubMed] [Google Scholar]
- 6.DeMarini D J, Papa F R, Swaminathan S, Ursic D, Rasmussen T P, Culbertson M R, Hochstrasser M. The yeast SEN3 gene encodes a regulatory subunit of the 26S proteasome complex required for ubiquitin-dependent protein degradation in vivo. Mol Cell Biol. 1995;15:6311–6321. doi: 10.1128/mcb.15.11.6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ermekova K S, Zambrano N, Linn H, Minopoli G, Gertler F, Russo T, Sudol M. The WW domain of neural protein FE65 interacts with proline-rich motifs in Mena, the mammalian homolog of Drosophila enabled. J Biol Chem. 1997;272:32869–32877. doi: 10.1074/jbc.272.52.32869. [DOI] [PubMed] [Google Scholar]
- 8.Freedman D A, Levine A J. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galan J M, Moreau V, Andre B, Volland C, Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- 10.Hanawalt P C. Transcription-coupled repair and human disease. Science. 1994;266:1957–1958. doi: 10.1126/science.7801121. [DOI] [PubMed] [Google Scholar]
- 11.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 12.Hatakeyama S, Jensen J P, Weissman A M. Subcellular localization and ubiquitin-conjugating enzyme (E2) interactions of mammalian HECT family ubiquitin protein ligases. J Biol Chem. 1997;272:15085–15092. doi: 10.1074/jbc.272.24.15085. [DOI] [PubMed] [Google Scholar]
- 13.Hein C, Springael J-Y, Volland C, Haguenauer-Tsapis R, André B. NPI1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- 14.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 15.Hoeijmakers J H. Nucleotide excision repair. II. From yeast to mammals. Trends Genet. 1993;9:211–217. doi: 10.1016/0168-9525(93)90121-w. [DOI] [PubMed] [Google Scholar]
- 16.Huibregtse J M, Maki C G, Howley P M. Ubiquitination of the p53 tumor suppressor. In: Peters J-M, Harris J R, Finley D, editors. Ubiquitin and the biology of the cell. New York, N.Y: Plenum; 1997. pp. 323–343. [Google Scholar]
- 17.Huibregtse J M, Scheffner M, Beaudenon S, Howley P M. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huibregtse J M, Yang J C, Beaudenon S L. The large subunit of RNA polymerase II is a substrate of the Rsp5 ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imhof M O, McDonnell D P. Yeast RSP5 and its human homolog hRPF1 potentiate hormone-dependent activation of transcription by human progesterone and glucocorticoid receptors. Mol Cell Biol. 1996;16:2594–2605. doi: 10.1128/mcb.16.6.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson P R, Swanson R, Rakhilina L, Hochstrasser M. Degradation signal masking by heterodimerization of MATalpha2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell. 1998;94:217–227. doi: 10.1016/s0092-8674(00)81421-x. [DOI] [PubMed] [Google Scholar]
- 21.Linn H, Ermekova K S, Rentschler S, Sparks A B, Kay B K, Sudol M. Using molecular repertoires to identify high-affinity peptide ligands of the WW domain of human and mouse YAP. Biol Chem. 1997;378:531–537. doi: 10.1515/bchm.1997.378.6.531. [DOI] [PubMed] [Google Scholar]
- 22.Lu P-J, Zhou X Z, Shen M, Lu K P. Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science. 1999;283:1325–1328. doi: 10.1126/science.283.5406.1325. [DOI] [PubMed] [Google Scholar]
- 23.Medintz I, Jiang H, Michels C A. The role of ubiquitin conjugation in glucose-induced proteolysis of Saccharomyces maltose permease. J Biol Chem. 1998;273:34454–34462. doi: 10.1074/jbc.273.51.34454. [DOI] [PubMed] [Google Scholar]
- 24.Mellon I, Spivak G, Hanawalt P C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 25.Myers L C, Gustafsson C M, Bushnell D A, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen J T, Turck C W, Cohen F E, Zuckermann R N, Lim W A. Exploiting the basis of proline recognition by SH3 and WW domains: design of N-substituted inhibitors. Science. 1998;282:2088–2092. doi: 10.1126/science.282.5396.2088. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen V T, Giannoni F, Dubois M-F, Seo S-J, Vigneron M, Kedinger C, Bensaude O. In vivo degradation of RNA polymerase II largest subunit triggered by α-amanitin. Nucleic Acids Res. 1996;24:2924–2929. doi: 10.1093/nar/24.15.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peters J-M. SCF and APC: the yin and yang of cell cycle regulated proteolysis. Curr Opin Cell Biol. 1998;10:759–768. doi: 10.1016/s0955-0674(98)80119-1. [DOI] [PubMed] [Google Scholar]
- 29.Prakash S, Sung P, Prakash L. DNA repair genes and proteins of Saccharomyces cerevisiae. Annu Rev Genet. 1993;27:33–70. doi: 10.1146/annurev.ge.27.120193.000341. [DOI] [PubMed] [Google Scholar]
- 30.Ratner J N, Balasubramanian B, Corden J, Warren S L, Bregman D B. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. Implications for transcription-coupled DNA repair. J Biol Chem. 1998;273:5184–5189. doi: 10.1074/jbc.273.9.5184. [DOI] [PubMed] [Google Scholar]
- 31.Rizo J, Sudhof T C. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- 32.Saleh A, Collart M, Martens J A, Genereaux J, Allard S, Cote J, Brandl C J. TOM1p, a yeast hect-domain protein which mediates transcriptional regulation through the ADA/SAGA coactivator complexes. J Mol Biol. 1998;282:933–946. doi: 10.1006/jmbi.1998.2036. [DOI] [PubMed] [Google Scholar]
- 33.Scheffner M, Nuber U, Huibregtse J M. Protein ubiquitination involving an E1-E2-E3 enzyme thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 34.Scheffner M, Smith S, Jentsch S. The ubiquitin-conjugation system. In: Peters J-M, Harris J R, Finley D, editors. Ubiquitin and the biology of the cell. New York, N.Y: Plenum; 1997. pp. 65–98. [Google Scholar]
- 35.Selby C P, Sancar A. Structure and function of transcription-repair coupling factor. I. Structural domains and binding properties. J Biol Chem. 1995;270:4882–4889. doi: 10.1074/jbc.270.9.4882. [DOI] [PubMed] [Google Scholar]
- 36.Silver P A, Chiang A, Sadler I. Mutations that alter both localization and production of a yeast nuclear protein. Genes Dev. 1988;2:707–717. doi: 10.1101/gad.2.6.707. [DOI] [PubMed] [Google Scholar]
- 37.Sterner D E, Grant P A, Roberts S M, Duggan L J, Belotserkovskaya R, Pacella L A, Winston F, Workman J L, Berger S L. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol Cell Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomoda K, Kubota Y, Kato J. Degradation of the cyclin-dependent-kinase inhibitor p27kip1 is instigated by Jab1. Nature. 1999;398:160–165. doi: 10.1038/18230. [DOI] [PubMed] [Google Scholar]
- 39.Wang G, Yang J, Huibregtse J M. Functional domains of the Rsp5 ubiquitin-protein ligase. Mol Cell Biol. 1999;19:342–352. doi: 10.1128/mcb.19.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, G., and J. M. Huibregtse. Unpublished results.
- 41.Winston, F., et al. Unpublished results.