Abstract
Seventeen new carbazole alkaloid derivatives, including a trimeric carbazole racemate, (±)-microphyltrine A (1), 15 dimeric carbazole racemates, (±)-microphyldines A–O (2–16), and a C-6–C-3″-methyl-linked dimeric carbazole, microphyldine P (17), were isolated from the leaves and stems of Murraya microphylla (Merr. et Chun) Swingle. The structures of the new compounds were elucidated on the basis of HRESIMS and NMR data analysis. The optically pure isomers of these isolated carbazole alkaloids were obtained by chiral HPLC separation and their absolute configurations were determined by electronic circular dichroism (ECD) data analysis.
Keywords: Murraya microphylla, trimeric carbazole, dimeric carbazole, racemates, absolute configurations, ECD
1. Introduction
Carbazole alkaloids, one type of bioactive constituents from the Murraya genus, have been demonstrated to possess anti-inflammatory, antitumor, antimicrobial, antioxidant, and antidiabetic properties [1]. Many of the biologically active carbazole alkaloids have been isolated from four closely related genera, Clausena, Glycosmis, Murraya, and Micromelum of the family Rutaceae [2,3,4]. Murraya microphylla (Merr. et Chun) Swing (M. microphylla) is a shrub distributed in the thickets of sandy areas or coastal regions in the Hainan Province of China [5]. Previous chemical investigations have confirmed that M. microphylla contains abundant carbazole alkaloids [6,7].
In the course of the search for bioactive carbazole alkaloids from Murraya species, the 95% aqueous ethanol extract of the leaves and stems of M. microphylla was investigated and 17 new carbazole alkaloid derivatives, namely (±)-microphyltrine A (1), (±)-microphyldines A–O (2–16), and microphyldine P (17) (Figure 1) were obtained. (±)-Microphyltrine A (1) is an unprecedented racemate of trimeric carbazole atropisomers. Microphyldines A–P (2–17) are new carbazole dimers, among which, (±)-microphyldines A–J (2–11) are 10 biphenyl-type carbazole dimeric racemates, while (±)-microphyldines K–O (12–16) are five pyranocarbazole dimeric racemates, and microphyldine P (17) is a C-6–C-3″-methyl-linked dimeric carbazole alkaloid. The optically pure isomers of these isolated carbazole racemates were obtained by chiral HPLC separation. Herein, we describe the isolation and structural characterization of compounds 1–17, as well as their activity screening on lipopolysaccharide (LPS)-induced nitric oxide (NO) production in BV-2 microglial cells and murine monocytic RAW 264.7 macrophages and cytotoxicities on HepG2, Du145, HCT116, and HeLa cells.
Figure 1.
Structures of compounds 1–17.
2. Results
2.1. Structural Elucidation
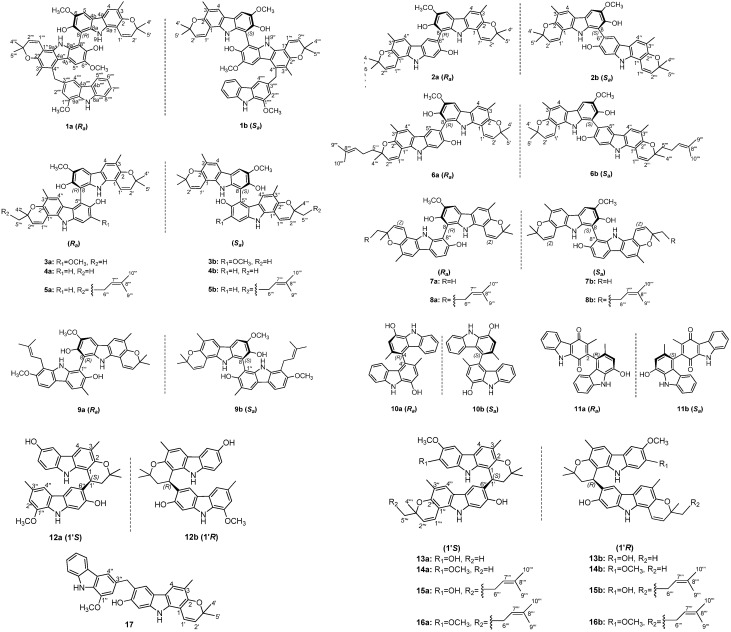
(±)-Microphyltrine A (1) was obtained as a white amorphous powder. Its molecular formula was determined as C52H47N3O7 based on the HRESIMS (m/z 824.3330 [M − H]−, calcd. for C52H46N3O7, 824.3336) and 13C NMR data. The UV spectrum showed absorptions at 227, 252, 293, and 332 nm, suggesting the presence of a pyranocarbazole skeleton in the molecule [8,9,10]. Analysis of the 1H NMR data (Table 1) revealed the presence of five labile proton signals (δH 7.05 (1H, br s, 7″-OH), 7.16 (1H, br s, 7-OH), 9.55 (1H, br s, 9″-NH), 9.58 (1H, br s, 9-NH), 10.25 (1H, br s, 9⁗-NH)), five aromatic doublets and singlets (δH 7.15 (1H, d, J = 2.0 Hz, H-2⁗), 7.56 (1H, d, J = 2.0 Hz, H-4⁗), 7.59 (1H, s, H-5″), 7.60 (1H, s, H-5), 7.62 (1H, s, H-4)), an ortho-substituted phenyl moiety (δH 7.10 (1H, t, J = 8.0 Hz, H-6⁗), 7.34 (1H, t, J = 8.0 Hz, H-7⁗), 7.55 (1H, d, J = 8.0 Hz, H-8⁗), 7.91 (1H, d, J = 8.0 Hz, H-5⁗)), three methoxy groups (δH 3.82 (3H, s, 6″-OCH3), 3.98 (3H, s, 1⁗-OCH3), 4.01 (3H, s, 6-OCH3)), two methyl groups (δH 2.28 (3H, s, 3-CH3) 2.42 (3H, s, 3″-CH3)), a methylene singlet (δH 4.84 (2H, s, 3⁗-CH2)), and two 2,2-dimethyl-2H-pyran moieties (δH 1.37/1.45 (3H, s, H-5‴/H-5′), 1.38/1.49 (3H, s, H-4‴/H-4′), 5.48/5.59 (1H, d, J = 10 Hz, H-2‴/H-2′), 6.66/6.81 (1H, d, J = 10 Hz, H-1‴/H-1′)). In the 13C NMR data of 1, there were 52 carbon resonances, comprising 42 olefinic, six methyl, three methoxy, and one methylene carbons. These data suggested 1 to be a carbazole trimer, consisting of three pyranocarbazole units similar to two koenigine [11,12] and a murrayafoline A moiety [13], in the combination of the HSQC and HMBC spectral analysis. The murrayafoline A unit was deduced to be connected to the middle koenigine unit via a C-4″–C-3⁗-methyl-linked mode, on the basis of HMBC correlations from 3⁗-CH2 to C-3″, C-4″, C-4a″, C-2⁗, C-3⁗, and C-4⁗, and the reverse correlations of H-2⁗/4⁗ to C-3⁗. The deficiency of H-8 and H-8″, together with the HMBC correlations from 7-OH to C-8 and from 7″-OH to C-8″, implied the two koenigine units were linked by a C-8–C-8″ bond (Figure 2). Thus, the planar structure of microphyltrine A was proposed as shown (Figure 1).
Table 1.
1H NMR and 13C NMR data of compound 1 in acetone-d6 (δ in ppm, J in Hz, 1H NMR: 400 MHz; 13C NMR: 100 MHz) a.
| Position | δ H | δ C | Position | δ H | δ C |
|---|---|---|---|---|---|
| 1 | 105.1, C | 1‴ | 6.66, d (10.0) | 119.9, CH | |
| 2 | 149.5, C | 2‴ | 5.48, d (10.0) | 129.6, CH | |
| 3 | 117.9, C | 3‴ | 76.5, C | ||
| 4 | 7.62, s | 120.9, CH | 4‴ | 1.38, s | 28.1, CH3 |
| 4a | 119.3, C | 5‴ | 1.37, s | 28.4, CH3 | |
| 4b | 115.9, C | 1⁗ | 147.6, C | ||
| 5 | 7.60, s | 105.9, CH | 2⁗ | 7.15, d (2.0) | 108.4, CH |
| 6 | 144.1, C | 3⁗ | 132.9, C | ||
| 7 | 144.5, C | 4⁗ | 7.56, d (2.0) | 112.9, CH | |
| 8 | 105.1, C | 4a⁗ | 125.7, C | ||
| 8a | 136.6, C | 4b⁗ | 124.6, C | ||
| 9a | 136.5, C | 5⁗ | 7.91, d (8.0) | 121.3, CH | |
| 1′ | 6.81, d (10.0) | 119.8, CH | 6⁗ | 7.10, t (8.0) | 120, CH |
| 2′ | 5.59, d (10.0) | 129.3, CH | 7⁗ | 7.34, t (8.0) | 126.6, CH |
| 3′ | 76.7, C | 8⁗ | 7.55, d (8.0) | 112.6, CH | |
| 4′ | 1.49, s | 27.9, CH3 | 8a⁗ | 141.6, C | |
| 5′ | 1.45, s | 28.5, CH3 | 9a⁗ | 130.1, C | |
| 1″ | 106.2, C | 3-CH3 | 2.28, s | 16.8, CH3 | |
| 2″ | 149.5, C | 6-OCH3 | 4.01, s | 57.7, CH3 | |
| 3″ | 117.7, C | 7-OH | 7.16, br s | ||
| 4″ | 134, C | 9-NH | 9.58, br s | ||
| 4a″ | 118.6, C | 3″-CH3 | 2.42, s | 12, CH3 | |
| 4b″ | 115.9, C | 6″-OCH3 | 3.82, s | 57.4, CH3 | |
| 5″ | 7.59, s | 103, CH | 7″-OH | 7.05, br s | |
| 6″ | 143.5, C | 9″-NH | 9.55, br s | ||
| 7″ | 145, C | 1⁗-OCH3 | 3.98, s | 56.4, CH3 | |
| 8″ | 105.4, C | 3⁗-CH2 | 4.84, s | 37.3, CH2 | |
| 8a″ | 136.6, C | 9⁗-NH | 10.25, br s | ||
| 9a″ | 136.6, C |
a Assignments were based on HSQC and HMBC experiments.
Figure 2.
Key HMBC correlations of compounds 1, 5, 15, and 17.
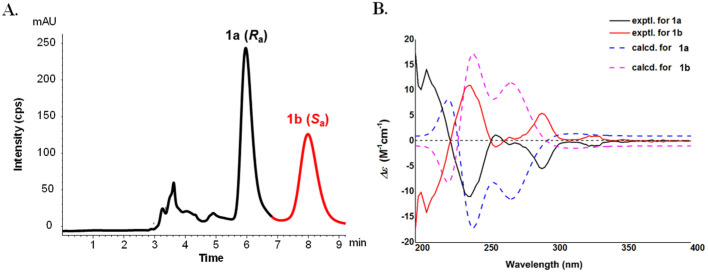
Considering the high steric hindrance at the central biaryl axis of C-8–C-8″, 1 was supposed to be with an axial chirality. However, its specific rotation value approached zero and no Cotton effects were observed in its ECD spectrum, suggesting 1 to be a pair of atropisomers coexisting as a racemic mixture. Compound 1 was isolated by a chiral HPLC with n-hexane-isopropanol (85:15, v/v, 1 mL/min) as the mobile phase to give the enantiomers 1a and 1b in an approximate ratio of 1:1. Their specific rotations were detected as −74 (c 0.1, MeOH) for 1a and +84 (c 0.1, MeOH) for 1b, respectively. The ECD spectrum of 1a exhibited a sequential negative and positive Cotton effect at 245 nm and 218 nm, i.e., a negative couplet derived from the electronic transition of the two carbazole monomers, indicating an (Ra) configuration for (−)-microphyltrine A (1a) and, accordingly, the configuration of (+)-microphyltrine A (1b) was defined as (Sa) [14]. Furthermore, the ECD spectra of (Ra)- and (Sa)-1 were calculated using the TDDFT method at the B3LYP/6-311G(d) level to confirm the results of ECD exciton coupling. The computed ECD spectrum of (Ra)-1 matched well with the experimental curve for 1a and the computed ECD spectrum of (Sa)-1 matched well with the experimental curve for 1b (Figure 3). This is the second report for trimeric carbazole alkaloids from a natural source, for the first was murratrines A and B from M. tetramera [4], whose units are three simple carbazoles linked with a bismethylene ether and a C-3-methyl-linked mode.
Figure 3.
The chiral HPLC separation (A) and experimental and calculated ECD data (B) of compounds 1a and 1b.
(±)-Microphyldine A (2) was obtained as a brown amorphous powder, and its molecular formula was determined as C37H34N2O5 from the 13C NMR spectroscopic data and the HRESIMS ion at m/z 585.2387 [M − H]− (calcd. for C37H33N2O5, 585.2389). Four labile proton signals (δH 7.43 (1H, br s, 7-OH), 7.43 (1H, br s, 7″-OH), 9.58 (1H, br s, 9-NH), 10.14 (1H, br s, 9″-NH)), five aromatic singlet protons (δH 7.04 (1H, s, H-8″), 7.61 (1H, s, H-5), 7.63 (1H, s, H-4″), 7.65 (1H, s, H-4), 7.87 (1H, s, H-5″)), a methoxy group (δH 4.02 (3H, s, 6-OCH3)), two methyl groups (δH 2.29 (3H, s, 3″-CH3) 2.31 (3H, s, 3-CH3)), and two 2,2-dimethyl-2H-pyran moieties (δH 1.42/1.49 (each 3H, s, H-5′/H-5‴), 1.43/1.50 (each 3H, s, H-4′/H-4‴), 5.58/5.80 (each 1H, d, J = 10 Hz, H-2′/H-2‴), 6.87/6.94 (each 1H, d, J = 10 Hz, H-1′/H-1‴)) were observed in the 1H NMR data (Table 2).
Table 2.
1H NMR data of compounds 2–11 and 17 in acetone-d6 (δ in ppm, J in Hz) a.
| Position | 2 | 3 | 4 | 5 b | 6 b | 7 | 8 | 9 c | 10 b | 11 b | 17 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 7.03, s | ||||||||||
| 4 | 7.65, s | 7.66, s | 7.66, s | 7.67, s | 7.62, s | 7.66, s | 7.65, s | 7.64, s | 7.49, s | ||
| 5 | 7.61, s | 7.71, s | 7.72, s | 7.73, s | 7.60, s | 7.63, s | 7.63, s | 7.58, s | 7.44, d (8.0) | 8.31, d (8.0) | 7.68, s |
| 6 | 7.12, t (8.0) | 7.43, t (8.0) | |||||||||
| 7 | 6.59, t (8.0) | 7.49, t (8.0) | |||||||||
| 8 | 6.52, d (8.0) | 7.71, d (8.0) | 6.69, s | ||||||||
| 1′ | 6.87, d (10.0) | 6.66, d (10.0) | 6.64, d (10.0) | 6.64, d (10.0) | 6.86, d (10.0) | 6.72, d (10.0) | 6.74, d (10.0) | 6.37, d (10.0) | 6.87, d (10.0) | ||
| 2′ | 5.58, d (10.0) | 5.51, d (10.0) | 5.49, d (10.0) | 5.50, d (10.0) | 5.58, d (10.0) | 5.55, d (10.0) | 5.53, d (10.0) | 5.57, d (10.0) | 7.03, s | 6.92, s | 5.74, d (10.0) |
| 4′ | 1.43, s | 1.43, s | 1.38, s | 1.39, s | 1.43, s | 1.41, s | 1.41, s | 1.47, s | 1.45, s | ||
| 5′ | 1.42, s | 1.42, s | 1.35, s | 1.36, s | 1.43, s | 1.41, s | 1.41, s | 1.47, s | 7.44, d (8.0) | 7.78, d (8.0) | 1.45, s |
| 6′ | 7.12, t (8.0) | 6.94, t (8.0) | |||||||||
| 7′ | 6.59, t (8.0) | 7.29, t (8.0) | |||||||||
| 8′ | 6.52, d (8.0) | 7.56, d (8.0) | |||||||||
| 9′ | |||||||||||
| 10′ | |||||||||||
| 2″ | 7.03, s | ||||||||||
| 4″ | 7.63, s | 6.44, s | 6.42, s | 6.43, s | 7.65, s | 7.67, s | 7.67, s | 7.86, s | 7.65, s | ||
| 5″ | 7.87, s | 7.86, s | 7.82, d (8.0) | 7.81, d (8.0) | 7.81, d (8.0) | 8.01, d (8.0) | |||||
| 6″ | 6.87, d (8.0) | 6.86, d (8.0) | 6.87, d (8.0) | 7.13, t (8.0) | |||||||
| 7″ | 7.00, d (8.0) | 7.00, d (8.0) | 7.35, t (8.0) | ||||||||
| 8″ | 7.04, s | 7.09, s | 7.31, d (8.0) | 7.32, d (8.0) | 7.03, s | 7.55, d (8.0) | |||||
| 1‴ | 6.94, d (10.0) | 6.87, d (10.0) | 6.87, d (10.0) | 6.92, d (10.0) | 6.98, d (10.0) | 6.75, d (10.0) | 6.75, d (10.0) | 3.44, d (7.2) | |||
| 2‴ | 5.80, d (10.0) | 5.58, d (10.0) | 5.69, d (10.0) | 5.69, d (10.0) | 5.78, d (10.0) | 5.56, d (10.0) | 5.57, d (10.0) | 5.12, t (7.2) | |||
| 4‴ | 1.50, s | 1.39, s | 1.38, s | 1.50, s | 1.47, s | 1.41, s | 1.39, s | 1.26, s | |||
| 5‴ | 1.49, s | 1.36, s | 1.38, s | 1.70, m | 1.80, m | 1.41, s | 1.71, m | 1.26, s | |||
| 6‴ | 2.11, m | 2.23, m | 2.15, m | ||||||||
| 7‴ | 5.08, t (6.0) | 5.16, t (6.0) | 5.11, t (6.0) | ||||||||
| 9‴ | 1.61, s | 1.66, s | 1.63, s | ||||||||
| 10‴ | 1.52, s | 1.59, s | 1.55, s | ||||||||
| 1-OH | 8.65, br s | ||||||||||
| 3-CH3 | 2.31, s | 2.29, s | 2.28, s | 2.28, s | 2.31, s | 2.31, s | 1.81, s | 2.37, s | 2.06, s | 1.89, s | 2.23, s |
| 6-OCH3 | 4.02, s | 4.05, s | 4.04, s | 4.04, s | 4.08, s | 4.04, s | 4.03, s | 4.13, s | |||
| 7-OH | 7.43, br s | 7.08, br s | 7.23, br s | 7.23, br s | 7.62, br s | 7.66, br s | 7.00, br s | 6.21, br s | 8.20, br s | ||
| 9-NH | 9.58, br s | 9.40, br s | 9.40, br s | 9.40, br s | 9.57, br s | 9.58, br s | 10.02, br s | 7.47, br s | 10.11, br s | 11.70, br s | 9.92, br s |
| 1′-OH | 8.65, br s | 8.80, br s | |||||||||
| 3′-CH3 | 2.06, s | 2.25, s | |||||||||
| 9′-NH | 10.11, br s | 10.26, br s | |||||||||
| 1″-OCH3 | 3.96, s | ||||||||||
| 2″-OH | 5.56, br s | ||||||||||
| 3″-CH2 | 4.28, s | ||||||||||
| 3″-CH3 | 2.29, s | 1.85, s | 1.81, s | 1.81, s | 2.30, s | 2.32, s | 2.33, s | 2.54, s | |||
| 6″-OH | 7.02, br s | ||||||||||
| 6″-OCH3 | 3.99, s | ||||||||||
| 7″-OH | 7.43, br s | 6.71, br s | 7.00, br s | 7.42, br s | 7.67, br s | 7.67, br s | |||||
| 7″-OCH3 | 3.92, s | ||||||||||
| 9″-NH | 10.14, br s | 9.95, br s | 10.02, br s | 10.02, br s | 10.15, br s | 9.66, br s | 9.67, br s | 7.66, br s | 10.20, br s |
a Assignments were based on HSQC and HMBC experiments.b Measured in 500 MHz, and others in 400 MHz. c 9 was measured in CDCl3 and others were measured in acetone-d6.
The 13C NMR data (Table 3) showed 37 carbon resonances, comprising 30 olefinic, six methyl, and one methoxy carbons. The above data, coupled with information from the literature [4,14,15] and 2D NMR analysis, indicated a dimeric carbazole skeleton of 2, and the two carbazole units were deduced as koenigine [12] and murrayamine A [16] moieties, respectively. The HMBC correlations from H-5″ to C-8 indicated that the two units were linked by a C-8–C-6″ bond (Figure S12, Supporting Information). Therefore, the planar structure of 2 was assigned as shown (Figure 1).
Table 3.
13C NMR data of compounds 2–11 and 17 (Measured in acetone-d6, δ in ppm) a.
| Position | 2 | 3 | 4 | 5 b | 6 b | 7 | 8 | 9 c | 10 b | 11 b | 17 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 104.9, C | 104.7, C | 104.9, C | 104.3, C | 104.9, C | 104.9, C | 104.7, C | 104.9, C | 141.8, C | 179.1, C | 104.5, C |
| 2 | 148.3, C | 148.1, C | 149.1, C | 147.7, C | 148.3, C | 148.2, C | 148.2, C | 149.1, C | 112.6, CH | 142.2, C | 148.2, C |
| 3 | 116.8, C | 116.5, C | 118.1, C | 116.2, C | 116.9, C | 116.8, C | 118.0, C | 118.7, C | 126.8, C | 145.6, C | 116.8, C |
| 4 | 119.7, CH | 119.8, CH | 120.7, CH | 119.3, CH | 120.4, CH | 119.6, CH | 119.7, CH | 120.0, CH | 125.0, C | 183.1, C | 120.0, CH |
| 4a | 117.9, C | 118.0, C | 118.8, C | 117.4, C | 117.9, C | 114.8, C | 116.7, C | 116.2, C | 123.9, C | 116.5, C | 117.3, C |
| 4b | 115.0, C | 114.7, C | 115.8, C | 114.4, C | 115.0, C | 118.0, C | 114.8, C | 117.3, C | 123.4, C | 124.3, C | 116.8, C |
| 5 | 101.0, CH | 101.7, CH | 102.9, CH | 101.5, CH | 101.0, CH | 101.8, CH | 101.7, CH | 101.8, CH | 110.6, CH | 122.4, CH | 120.5, CH |
| 6 | 142.3, C | 142.8, C | 143.8, C | 142.4, C | 142.7, C | 142.8, C | 142.8, C | 142.1, C | 124.6, CH | 123.8, CH | 121.4, C |
| 7 | 142.7, C | 143.4, C | 144.5, C | 143.1, C | 142.8, C | 143.8, C | 143.8, C | 141.6, C | 118.0, CH | 126.5, CH | 153.2, C |
| 8 | 109.1, C | 106.6, C | 106.7, C | 109.5, C | 109.0, C | 103.5, C | 103.5, C | 101.7, C | 121.2, CH | 113.6, CH | 96.5, CH |
| 8a | 135.1, C | 134.9, C | 135.9, C | 134.5, C | 135.1, C | 135.3, C | 135.5, C | 132.7, C | 140.4, C | 138.0, C | 140.1, C |
| 9a | 135.3, C | 135.3, C | 136.2, C | 134.8, C | 135.3, C | 135.7, C | 135.1, C | 134.7, C | 128.5, C | 136.2, C | 135.2, C |
| 1′ | 118.4, CH | 118.3, CH | 119.2, CH | 117.8, CH | 118.4, CH | 118.3, CH | 118.3, CH | 117.3, CH | 141.8, C | 142.6, C | 117.9, CH |
| 2′ | 128.3, CH | 128.2, CH | 129.2, CH | 127.7, CH | 128.3, CH | 128.4, CH | 128.4, CH | 129.4, CH | 112.6, CH | 112.3, CH | 128.9, CH |
| 3′ | 75.2, C | 75.1, C | 76, C | 74.6, C | 75.2, C | 75.2, C | 75.1, C | 75.7, C | 126.8, C | 118.6, C | 75.3, C |
| 4′ | 27, CH3 | 27, CH3 | 27.9, CH3 | 26.4, CH3 | 27, CH3 | 26.8, CH3 | 27, CH3 | 27.4, CH3 | 125, C | 126.8, C | 26.9, CH3 |
| 4a′ | 123.9, C | 122.4, C | |||||||||
| 4b′ | 123.4, C | 123.0, C | |||||||||
| 5′ | 26.9, CH3 | 26.8, CH3 | 27.8, CH3 | 26.4, CH3 | 26.9, CH3 | 26.8, CH3 | 27, CH3 | 27.5, CH3 | 110.6, CH | 120.7, CH | 26.9, CH3 |
| 6′ | 124.6, CH | 118.8, CH | |||||||||
| 7′ | 118.0, CH | 125.1, CH | |||||||||
| 8′ | 121.2, CH | 111.3, CH | |||||||||
| 8a′ | 140.4, C | 140.4, C | |||||||||
| 9′ | |||||||||||
| 9a′ | 128.5, C | 128.2, C | |||||||||
| 10′ | |||||||||||
| 1″ | 104.6, C | 104, C | 105.7, C | 103.2, C | 104.5, C | 104.9, C | 104.9, C | 102.1, C | 145.7, C | ||
| 2″ | 148.6, C | 148.1, C | 150, C | 148.7, C | 148.7, C | 148.3, C | 148.4, C | 150.0, C | 107.8, CH | ||
| 3″ | 117.1, C | 116, C | 117.1, C | 115.4, C | 116.8, C | 117.0, C | 117.7, C | 118.1, C | 133.7, C | ||
| 4″ | 120.3, CH | 121.8, CH | 123.6, CH | 122.3, CH | 119.7, CH | 119.7, CH | 119.8, CH | 121.6, CH | 112.4, CH | ||
| 4a″ | 117.3, C | 117.7, C | 118.7, C | 116.7, C | 117.2, C | 117.7, C | 116.7, C | 118.5, C | 124, C | ||
| 4b″ | 117.5, C | 116, C | 124.3, C | 122.8, C | 117.5, C | 116.8, C | 116.8, C | 118.7, C | 123.4, C | ||
| 5″ | 122.1, CH | 112.8, C | 113.6, C | 112.2, C | 122.0, CH | 119.2, CH | 119.6, CH | 117.3, CH | 119.9, CH | ||
| 6″ | 114.2, C | 139.2, C | 149.6, C | 148.2, C | 114.2, CH | 108.9, CH | 108.8, C | 104.7, CH | 118.5, CH | ||
| 7″ | 153.4, C | 146.5, C | 114.5, CH | 113.1, CH | 153.4, C | 153.2, C | 153.2, C | 154.7, C | 125.1, CH | ||
| 8″ | 97.9, CH | 93.9, CH | 111.5, CH | 110.1, CH | 97.8, CH | 103.7, C | 103.7, C | 111.0, C | 111.2, CH | ||
| 8a″ | 141.5, C | 133.9, C | 135.6, C | 134.3, C | 141.5, C | 141.0, C | 141.0, C | 140.4, C | 140.2, C | ||
| 9a″ | 135.5, C | 135.4, C | 137.3, C | 136.0, C | 135.5, C | 135.4, C | 135.3, C | 137.7, C | 128.6, C | ||
| 1‴ | 117.9, CH | 117.9, CH | 118.7, CH | 117.7, CH | 118.2, CH | 118.3, CH | 118.7, CH | 24, CH2 | |||
| 2‴ | 129.1, CH | 128.5, CH | 129.8, CH | 127, CH | 128.2, CH | 128.5, CH | 127.6, CH | 122.1, CH | |||
| 3‴ | 75.4, C | 75.2, C | 76.3, C | 77.3, C | 77.7, C | 75.2, C | 77.5, C | 132.7, C | |||
| 4‴ | 26.9, CH3 | 26.9, CH3 | 27.9, CH3 | 24.7, CH3 | 25.3, CH3 | 27, CH3 | 26.8, CH3 | 25, CH3 | |||
| 5‴ | 26.9, CH3 | 26.8, CH3 | 27.8, CH3 | 40.2, CH2 | 40.7, CH2 | 26.8, CH3 | 40.6, CH2 | 16.9, CH3 | |||
| 6‴ | 22.1, CH2 | 22.6, CH2 | 22.6, CH2 | ||||||||
| 7‴ | 123.8, CH | 124.3, CH | 124.3, CH | ||||||||
| 8‴ | 130.4, C | 131.0, C | 130.9, C | ||||||||
| 9‴ | 24.7, CH3 | 24.9, CH3 | 24.9, CH3 | ||||||||
| 10‴ | 16.2, CH3 | 16.7, CH3 | 16.7, CH3 | ||||||||
| 3-CH3 | 15.4, CH3 | 15.4, CH3 | 16.4, CH3 | 14.9, CH3 | 15.4, CH3 | 15.4, CH3 | 16.3, CH3 | 16.1, CH3 | 18.2, CH3 | 12.7, CH3 | 15.2, CH3 |
| 6-OCH3 | 56.3, CH3 | 56.6, CH3 | 57.5, CH3 | 56.1, CH3 | 56.3, CH3 | 56.3, CH3 | 56.3, CH3 | 56.9, CH3 | |||
| 3″-CH3 | 15.3, CH3 | 15.5, CH3 | 16.3, CH3 | 15, CH3 | 15.3, CH3 | 15.5, CH3 | 15.4, CH3 | 16.9, CH3 | 18.2, CH3 | 18.4, CH3 | |
| 3″-CH2 | 36.4, CH3 | ||||||||||
| 1″-OCH3 | 54.9, CH3 | ||||||||||
| 6″-OCH3 | |||||||||||
| 7″-OCH3 | 55.7, CH3 | 56.7, CH3 |
a Assignments were based on HSQC and HMBC experiments.b Measured in 500 MHz, and others in 400 MHz. c 9 was measured in CDCl3 and others were measured in acetone-d6.
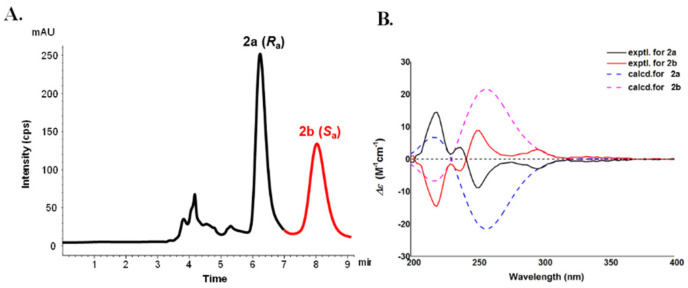
Compound 2 could also be a racemate owing to the disappeared specific rotation and Cotton effects in the ECD spectrum, thus, it was then separated by a chiral HPLC to give the enantiomers 2a and 2b for almost equal quantity, which possess the opposite ECD curves and specific rotations. Similar to 1, the ECD spectrum of 2a exhibited sequential negative and positive Cotton effects at 252 nm and 224 nm, indicating an (Ra) configuration for (−)-microphyldine A (2a) [14] and, accordingly, the configuration of (+)-microphyldine A (2b) was defined as (Sa). Furthermore, the ECD spectra of (Ra)- and (Sa)-2 were calculated and compared with the experimental spectra to support the results of ECD exciton coupling (Figure 4).
Figure 4.
The chiral HPLC separation (A) and experimental and calculated ECD data (B) of compounds 2a and 2b.
(±)-Microphyldine B (3) gave a molecular formula of C38H36N2O6, as established by 13C NMR data and an [M − H]− ion at m/z 615.2486 [M − H]−(calcd for C38H35N2O6, 615.2495) in the HRESIMS. The 1H and 13C NMR data (Table 2 and Table 3) of 3 showed close resemblance to those of 2, except that one of the phenyl singlets in 3 was missing, but an additional methoxy signal was observed at δH 3.99 (3H, s). After 2D NMR analysis (Figure S19, Supporting Information), the two units were deduced to be both koenigine units [12]. The deficiency of H-8 and H-5″ signals suggested 3 to be a C-8–C-5″-linked dimeric carbazole. Thus, the planar structure of 3 was assigned as shown (Figure 1).
Similar to 2, compound 3 is also an atropisomeric racemate, and the following chiral HPLC resolution afforded 3a and 3b in a ratio of 1:1, and their absolute configurations were established as (Ra) and (Sa), respectively, by comparison of their ECD (Figure S18, Supporting Information) and specific rotations with those of 2a and 2b [14].
(±)-Microphyldine C (4) exhibited an [M − H]− ion at m/z 585.2389 in the HRESIMS, which, in conjunction with the 13C NMR data, suggested a molecular formula of C37H34N2O5 (calcd. for C37H33N2O5, 585.2389). Analysis of 1D and 2D NMR data (Table 2 and Table 3) suggested that the structure of 4 resembled that of 3, except for the disappearance of one methoxy group in 4, and the replacement of an aromatic singlet in 3 by two aromatic doublets (δH 7.00 (1H, d, J = 8.0 Hz, H-7″), 7.31 (1H, d, J = 8.0 Hz, H-8″)). This suggested that 4 is the demethoxy derivative of 3. Further 2D NMR analysis (Figure S26, Supporting Information) deduced the structure of 4 as shown (Figure 1). The isolation of individual enantiomers (4a and 4b) was accomplished by a chiral HPLC separation and their absolute configurations were established as (Ra) and (Sa), respectively, by comparison of their ECD (Figure S25, Supporting Information) and specific rotations with those of 2a and 2b [14].
(±)-Microphyldine D (5) was isolated as an amorphous powder. Its 13C NMR and negative-ion HRESIMS data at m/z 653.3023 [M − H]− (calcd for C42H41N2O5, 653.3015) established a molecular formula of C42H42N2O5. The 1H and 13C NMR data (Table 2 and Table 3) of 5 were comparable to those of 4, except for the presence of a set of additional resonances for isopentenyl group (δH 1.52 (3H, s, H-10‴), 1.61 (3H, s, H-9‴), 2.11 (2H, m, H-6‴), 5.08 (1H, t, J = 6.0 Hz, H-7‴), δC 16.2 (C-10‴), 22.1 (C-6‴), 24.7 (C-9‴), 123.8 (C-7‴), 130.4 (C-8‴)) in 5. The HMBC correlations from H-5‴ to C-2‴ and C-7‴ and from H-6‴ to C-3‴, C-7‴, and C-8‴ suggested that the isopentenyl moiety is located at C-5‴ (Figure 2). Compound 5 is also a pair of atropisomers, and the following chiral HPLC resolution afforded 5a and 5b in a ratio of 1:1, and their absolute configurations were established as (Ra) and (Sa), respectively, by comparison of their ECD (Figure S32, Supporting Information) and specific rotations with those of 4a and 4b [14]. The experimental ECD spectra of 5a/5b are almost similar to those of 4a/4b, indicating that the C-3″′configuration did not have much influence on the ECD curves, which was proofed by the calculated ECD data of the different configurations of C-3‴. Thus, the C-3‴configuration was undetermined in this paper.
(±)-Microphyldine E (6) was obtained as an amorphous powder with a molecular formula of C42H42N2O5, as deduced from the 13C NMR and HRESIMS (m/z 653.3023 [M − H]− (calcd for C42H41N2O5, 653.3015) data. Its 1H and 13C NMR (Table 2 and Table 3) data showed many similarities to those of 2, except for the presence of a set of additional resonances for isopentenyl group (δH 1.59 (3H, s, H-10‴), 1.66 (3H, s, H-9‴), 2.23 (2H, m, H-6‴), 5.16 (1H, t, J = 6.0 Hz, H-7‴), δC 16.7 (C-10‴), 22.6 (C-6‴), 24.9 (C-9‴), 124.3 (C-7‴), 131.0 (C-8‴)) in 6. The HMBC correlations from H-5‴ to C-2‴ and C-7‴ and from H-6‴ to C-3‴, C-7‴, and C-8‴ suggested that the isopentenyl moiety is located at C-5‴ (Figure S39, Supporting Information). Compound 6 was separated by a chiral HPLC to afford the enantiomers 6a and 6b. The ECD spectra of 6a/6b were calculated using the TDDFT method at the B3LYP/6-311G(d) level to determine the absolute configuration. The computed ECD spectrum of (Ra)-6 matched the experimental curve for 6a. and the computed ECD spectrum of (Sa)-6 matched the experimental curve for 6b. Thus, the absolute configuration of 6a was defined as (Ra) and, accordingly, 6b was defined as (Sa) (Figure S38, Supporting Information).
(±)-Microphyldine F (7) was shown to have the same molecular formula as 2, according to its 13C NMR data and the [M − H]− ion at m/z 585.2390 in the HRESIMS (calcd. for C37H33N2O5, 585.2389). Analysis of 1H and 13C NMR data (Table 2 and Table 3) of 7 showed a close structural resemblance to 2, a dimeric carbazole formed by koenigine [12] and murrayamine A [16] units. The difference between them is that the linkage mode is shifted from C-8–C-6″ in 2 to C-8–C-8″ in 7, as deduced from the deficiency of H-8 and H-8″ signals in 7. Accordingly, the structure of 7 was determined as shown (Figure 1). A subsequent chiral HPLC isolation was performed to obtain the pure enantiomers of 7a and 7b in a ratio of 1:1. After ECD and specific rotation determination, the absolute configurations of 7a and 7b were established as (Ra) and (Sa), respectively, by comparison of their ECD (Figure S45, Supporting Information) and specific rotations with those of 2a and 2b [14].
(±)-Microphyldine G (8) was isolated as an amorphous powder. The HRESIMS gave a deprotonated molecular ion at m/z 653.3007 [M − H]− (calcd for C42H41N2O5, 653.3015), corresponding to a molecular formula of C42H42N2O5. The 1H and 13C NMR data (Table 2 and Table 3) of 8 were found to be similar to those of 7. Their apparent difference was the presence of a set of resonances for isopentenyl group (δH 1.55 (3H, s, H-10‴), 1.63 (3H, s, H-9‴), 2.15 (2H, m, H-6‴), 5.11 (1H, t, J = 6.0 Hz, H-7‴), δC 16.7 (C-10‴), 22.6 (C-6‴), 24.9 (C-9‴), 124.3 (C-7‴), 130.9 (C-8‴)) in 8. The HMBC correlations from H-5‴ to C-2‴ and C-7‴ and from H-6‴ to C-3‴, C-7‴, and C-8‴ suggested that the isopentenyl moiety is located at C-5‴ (Figure S53, Supporting Information). Thus, the planar structure of 8 was assigned as shown (Figure 1). Compound 8 was also a pair of atropisomer mixtures and was then separated by a chiral HPLC to give the enantiomers 8a and 8b. The ECD spectra of 8a/8b were calculated using the TDDFT method at the B3LYP/6-311G(d) level to determine the absolute configuration. The computed ECD spectrum of (Ra)-8 matched the experimental curve for 8a. and the computed ECD spectrum of (Sa)-8 matched the experimental curve for 8b. Thus, the absolute configuration of 8a was defined as (Ra) and accordingly, 8b was defined as (Sa). (Figure S52, Supporting Information).
(±)-Microphyldine H (9) was obtained as a brown amorphous powder with a molecular formula of C38H38N2O5, as deduced from the 13C NMR and HRESIMS data (m/z 601.2692 [M − H]−, calcd for C38H37N2O5, 601.2702). The 1H and 13C NMR data (Table 2 and Table 3) analysis revealed that the structure of 9 is a dimeric carbazole formed by a koenigine [12] and an isomurrayafoline B [17,18] unit. The 2D NMR data (Figure S60, Supporting Information) analysis indicated the linkage mode of 9 was C-8–C-1″ due to the absence of H-8 and H-1″ protons. Accordingly, the structure of 9 was determined as shown (Figure 1). A subsequent chiral HPLC isolation was performed to obtain the pure enantiomers of 9a and 9b in a ratio of 1:1. After ECD and specific rotation determination, the absolute configurations of 9a and 9b were established as (Ra) and (Sa), respectively, by comparison of their ECD (Figure S59, Supporting Information) and specific rotations with those of 2a and 2b [14].
(±)-Microphyldine I (10) was isolated as a brown amorphous powder. Its molecular formula was defined as C26H20N2O2 via its 13C NMR and HRESIMS data (m/z 391.1440 [M − H]−, calcd for C26H19N2O2, 391.1447). The 13C NMR data of 10 exhibited only 13 carbon signals, suggesting that it is a symmetrical carbazole dimer. The NMR data (Table 2 and Table 3) of the monomeric unit of 10 resembled those of 1-hydroxy-3-methylcarbazole [19], except for the absence of H-4 proton and a shift of the C-4 signal downfield to δC 125.0, indicating that the two units are linked through C-4–C-4′. Compound 10 was a pair of atropisomer mixtures inferred from its almost zero specific rotation and weak ECD Cotton effects. The pure enantiomers of 10a and 10b were obtained by a chiral HPLC separation. The absolute configurations of 10a and 10b were defined as (Ra) and (Sa), respectively, from their experimental ECD spectra and computed ECD spectra using the TDDFT method at the B3LYP/6-311 + G(d) level (Figure S66, Supporting Information).
(±)-Microphyldine J (11) was obtained as an amorphous powder. It has a molecular formula of C26H18N2O3 determined by the HRESIMS data showing a deprotonated molecular ion at m/z 405.1232 [M − H]− (calcd for C26H17N2O3, 405.1239) and its 13C NMR data. Its 1D and 2D NMR data (Table 2 and Table 3) showed many similarities to those of murrayaquinone A [20], except for the replacement of a methoxy singlet in murrayaquinone A by a hydroxy singlet (δH 8.80 (1H, br s)) in 11. The 2D NMR analysis, especially of HMBC correlation from OH-1′ to C-9a, from CH3-3 to C-2, and from CH3-3′ to C-4′ proved the above deduction (Figure S74, Supporting Information). Compound 11 was separated by a chiral HPLC to give the enantiomers of 11a and 11b. The absolute configurations of 11a and 11b were defined as (Ra) and (Sa), respectively, from their experimental ECD and computed ECD spectra (Figure S73, Supporting Information).
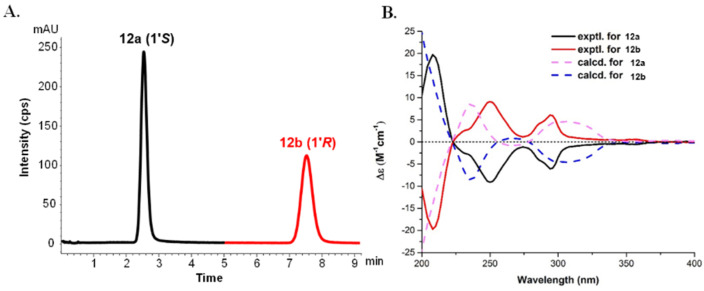
(±)-Microphyldine K (12) was obtained as a brown amorphous powder with a molecular formula of C32H30N2O4 based on the HRESIMS (m/z 505.2125 [M − H]−, calcd. for C32H29N2O4, 505.2127) and 13C NMR data. The 1H NMR data (Table 4) of 12 were found to be similar to those of murrafoline D [21]. The apparent differences were the replacement of two aromatic signals in murrafoline D by two active hydrogen signals in 12, suggesting that 12 is a dihydroxy derivative of murrafoline D [21]. The two hydroxy groups were deduced to be located at C-6 and C-7″, respectively, via the HMBC correlation of one hydroxy proton (δH 7.66 (1H, br s)) with C-5/C-7, and the other hydroxy proton (δH 8.23 (1H, br s) with C-6″ and C-8″ (Figure S80, Supporting Information). The optical inactivity of 12 indicated that it is a pair of enantiomer mixture, thus a chiral HPLC isolation was performed to obtain the pure enantiomers of 12a and 12b. The ECD spectra of 12a/12b were calculated using the TDDFT method at the B3LYP/6-311G(d) level to determine the absolute configuration. The computed ECD spectrum of (1′S)-12a matched the experimental curve for 12a (Figure 5). Thus, the absolute configuration of 12a was defined as (1′S) and, accordingly, 12b was defined as (1′R).
Table 4.
1H NMR and 13C NMR data of compounds 12–16 in acetone-d6 (δ in ppm, J in Hz) a.
| Position | 12 | 13 | 14 b | 15 b | 16 b | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| δ H | δ C | δ H | δ C | δ H | δ C | δ H | δ C | δ H | δ C | |
| 1 | 106.6, C | 106.8, C | 108.0, C | 108.4, C | 106.3, C | |||||
| 2 | 151.9, C | 150.7, C | 152.0, C | 152.0, C | 150.3, C | |||||
| 3 | 118.1, C | 117.9, C | 117.6, C | 120.1, C | 117.7, C | |||||
| 4 | 7.65, s | 119.4, CH | 7.62, s | 118.5, CH | 7.65, s | 120.0, CH | 7.63, s | 119.5, CH | 7.66, s | 118.2, CH |
| 4a | 115.8, C | 115.4, C | 117.5, C | 117.9, C | 115.9, C | |||||
| 4b | 124.4, C | 116.5, C | 117.0, C | 116.8, C | 115.3, C | |||||
| 5 | 7.32, d (2.0) | 103.9, CH | 7.47, s | 101.8, CH | 7.47, s | 104.3, CH | 7.48, s | 103.3, CH | 7.48, s | 102.6, CH |
| 6 | 150.6, C | 145.0, C | 145.7, C | 143.8, C | 143.9, C | |||||
| 7 | 6.61, dd (2.0, 8.0) | 112.4, CH | 142.4, C | 149.6, C | 146.4, C | 147.9, C | ||||
| 8 | 6.87, d (8.0) | 110.8, CH | 6.55, d (8.0) | 97.0, CH | 6.69, s | 96.7, CH | 6.56, s | 98.4, CH | 6.70, s | 94.9, CH |
| 8a | 134.0, C | 134.8, C | 135.7, C | 136.2, C | 133.9, C | |||||
| 9a | 138.7, C | 137.6, C | 138.9, C | 139.0, C | 137.1, C | |||||
| 1′ | 4.94, dd (7.2, 10.0) | 30.1, CH | 4.93, dd (7.2, 10.0) | 30.5, CH | 4.93, dd (7.2, 10.0) | 31.9, CH | 4.94, dd (7.2, 10.0) | 30.2, CH | 4.95, dd (7.2, 10.0) | 30.1, CH |
| 2′a | 2.07, dd (10.0, 14.0) | 42.8, CH2 | 2.06, dd (10.0, 14.0) | 43, CH2 | 2.08, dd (10.0, 14.0) | 44.2, CH2 | 2.13, dd (10.0, 14.0) | 44.4, CH2 | 2.05, dd (10.0, 14.0) | 42.4, CH2 |
| 2′b | 2.42, dd (7.2, 14.0) | 2.39, dd (7.2, 14.0) | 2.40, dd (7.2, 14.0) | 2.41, dd (7.2, 14.0) | 2.37, dd (7.2, 14.0) | |||||
| 3′ | 74.4, C | 74.2, C | 75.4, C | 75.6, C | 73.8, C | |||||
| 4′ | 1.37, s | 23.9, CH3 | 1.35, s | 23.8, CH3 | 1.36, s | 25.1, CH3 | 1.37, s | 25.2, CH3 | 1.38, s | 23.3, CH3 |
| 5′ | 1.45, s | 29.1, CH3 | 1.44, s | 29.3, CH3 | 1.45, s | 30.4, CH3 | 1.46, s | 30.4, CH3 | 1.47, s | 28, CH3 |
| 1″ | 145.3, C | 104.5, C | 105.7, C | 105.7, C | 103.8, C | |||||
| 2″ | 6.63, d (2.0) | 106.4, CH | 148.4, C | 149.6, C | 149.9, C | 148.0, C | ||||
| 3″ | 128.5, C | 117.0, C | 118.3, C | 118.1, C | 116.3, C | |||||
| 4″ | 7.01, d (2.0) | 111.6, CH | 7.25, s | 120, CH | 7.27, s | 121.3, CH | 7.27, s | 121.5, CH | 7.28, s | 119.6, CH |
| 4a″ | 124.5, C | 117.0, C | 119.4, C | 118.3, C | 117.6, C | |||||
| 4b″ | 117.1, C | 117.4, C | 118.2, C | 118.8, C | 116.9, C | |||||
| 5″ | 7.46, s | 119.6, CH | 7.39, s | 118.7, CH | 7.42, s | 119.9, CH | 7.41, s | 119.9, CH | 7.42, s | 118.2, CH |
| 6″ | 122.2, C | 122.2, C | 123.4, C | 123.6, C | 121.7, C | |||||
| 7″ | 153.9, C | 153.0, C | 154.2, C | 154.3, C | 152.5, C | |||||
| 8″ | 7.13, s | 97.1, CH | 7.02, s | 97.0, CH | 7.04, s | 98.3, CH | 7.04, s | 98.4, CH | 7.05, s | 96.6, CH |
| 8a″ | 140.2, C | 140.2, C | 141.4, C | 141.5, C | 139.7, C | |||||
| 9a″ | 127.9, C | 135.2, C | 136.5, C | 136.2, C | 134.8, C | |||||
| 1‴ | 6.83, d (10.0) | 118, CH | 6.84, d (10.0) | 119, CH | 6.88, d (10.0) | 119.3, CH | 6.89, d (10.0) | 117.6, CH | ||
| 2‴ | 5.72, d (10.0) | 128.9, CH | 5.72, d (10.0) | 130.2, CH | 5.71, d (10.0) | 129.4, CH | 5.72, d (10.0) | 127.6, CH | ||
| 3‴ | 75.3, C | 76.6, C | 79.1, C | 77.2, C | ||||||
| 4‴ | 1.41, s | 27, CH3 | 1.41, s | 28.1, CH3 | 1.40, s | 26.2, CH3 | 1.40, s | 24.7, CH3 | ||
| 5‴ | 1.41, s | 27, CH3 | 1.40, s | 28.1, CH3 | 1.73, m | 42.0, CH2 | 1.73, m | 40.1, CH2 | ||
| 6‴ | 2.15, m | 23.9, CH2 | 2.15, m | 22.1, CH2 | ||||||
| 7‴ | 5.11, t (6.0) | 125.7, CH | 5.11, t (6.0) | 123.8, CH | ||||||
| 8‴ | 132.3, C | 130.4, C | ||||||||
| 9‴ | 1.62, s | 26.2, CH3 | 1.61, s | 24.7, CH3 | ||||||
| 10‴ | 1.54, s | 18.1, CH3 | 1.54, s | 16.2, CH3 | ||||||
| 3-CH3 | 2.36, s | 16.3, CH3 | 2.36, s | 16.3, CH3 | 2.37, s | 17.6, CH3 | 2.37, s | 17.7, CH3 | 2.38, s | 15.8, CH3 |
| 6-OH | 7.66, br s | |||||||||
| 6-OCH3 | 3.88, s | 56.2, CH3 | 3.82, s | 57.4, CH3 | 3.89, s | 57.6, CH3 | 3.83, s | 55.7, CH3 | ||
| 7-OH | 7.14, br s | 7.12, br s | ||||||||
| 7-OCH3 | 3.64, s | 56.5, CH3 | 3.66, s | 54.8, CH3 | ||||||
| 9-NH | 8.55, br s | 8.43, br s | 8.40, br s | 8.43, br s | 8.42, br s | |||||
| 1″-OCH3 | 3.91, s | 54.8, CH3 | ||||||||
| 3″-CH3 | 2.30, s | 20.8, CH3 | 2.12, s | 15.0, CH3 | 2.13, s | 16.3, CH3 | 2.16, s | 16.4, CH3 | 2.15, s | 14.6, CH3 |
| 7″-OH | 8.23, br s | 8.18, br s | 8.14, br s | 8.16, br s | 8.15, br s | |||||
| 9″-NH | 9.88, br s | 9.97, br s | 9.97, br s | 9.96, br s | 9.98, br s | |||||
a Assignments were based on HSQC and HMBC experiments. b1H NMR: 500 MHz; 13C NMR: 125 MHz. Others: 1H NMR: 400 MHz; 13C NMR: 100 MHz.
Figure 5.
The chiral HPLC separation (A) and experimental and calculated ECD data (B) of compounds 12a and 12b.
(±)-Microphyldine L (13) gave a molecular formula of C37H36N2O5 on the basis of its 13C NMR and HRESIMS (m/z 587.2543 [M − H]−, calcd. for C37H35N2O5, 587.2546). The UV and NMR data (Table 4) of 13 were closely comparable to those of microphyldine K (12), indicating that it also has a biscarbazole skeleton like 12. The difference between them is that 13 is a dimeric carbazole formed by koenigine [12] and murrayamine A [16] units, which were further deduced from the 2D NMR data. The linkage of these two units was determined to be via the C-1′-C-6″ bond, supported by the HMBC correlations of H-2′ and C-6″, and H-1′ and C-5″/C-7″ (Figure S87, Supporting Information). Accordingly, the structure of 13 was determined as shown (Figure 1). A subsequent chiral HPLC isolation was performed to obtain the pure enantiomers of 13a and 13b in a ratio of 1:1. The absolute configurations of 13a and 13b were defined as (1′S) and (1′R), respectively, by comparison of the experimental and calculated ECD spectra (Figure S86, Supporting Information).
(±)-Microphyldine M (14) gave a molecular formula of C38H38N2O5 based on the 13C NMR and a deprotonated ion at m/z 601.2712 [M − H]− (calcd. for C38H37N2O5, 601.2702) in the negative-ion HRESIMS. Its 1D (Table 4) and 2D NMR data showed many similarities to those of 13, except for the replacement of a hydroxy singlet (δH 7.14 (1H, br s)) in 13 by a methoxy singlet (δH 3.64 (3H, s)) in 14. This suggested that 14 is a 7-methoxy derivative of 13, as deduced from the HMBC correlation of the methoxy protons with C-7 (δC 149.6) (Figure S94, Supporting Information). Hence, the structure of 14 was defined as shown (Figure 1). The pure enantiomers of 14a and 14b were obtained by a chiral HPLC separation. A comparison of the experimental and calculated ECD spectra facilitated the assignment of the absolute configurations of 14a and 14b as (1′S) and (1′R), respectively (Figure S93, Supporting Information).
(±)-Microphyldine N (15) gave a molecular formula of C42H44N2O5, as determined from its 13C NMR and HRESIMS data (m/z 655.3156 [M − H]−, calcd. for C42H43N2O5, 655.3172). Its 1D (Table 4) and 2D NMR data showed many similarities to those of 13 and the apparent difference was the presence of a set of resonances for the isopentenyl group (δH 1.54 (3H, s, H-10‴), 1.62 (3H, s, H-9‴), 2.15 (2H, m, H-6‴), 5.11 (1H, t, J = 6.0 Hz, H-7‴), δC 18.1 (C-10‴), 23.9 (C-6‴), 26.2 (C-9‴), 125.7 (C-7‴), 132.3 (C-8‴)) in 15. The HMBC correlations from H-5‴ to C-2‴, C-6‴, and C-7‴ and from H-6‴ to C-3‴ and C-5‴ suggested that the isopentenyl moiety is located at C-5‴(Figure 2). Hence, the planar structure of 15 was assigned as shown (Figure 1). The following chiral HPLC resolution and ECD determination defined the absolute configurations of 15a and 15b as (1′S) and (1′R), respectively (Figure S100, Supporting Information).
(±)-Microphyldine O (16) was isolated as an amorphous powder with a molecular formula of C43H46N2O5, as deduced from the 13C NMR and HRESIMS data (m/z 669.3328 [M − H]−, calcd for C43H45N2O5, 669.3328). The 1H and 13C NMR data (Table 4) of 16 were found to be similar to those of 14. The obvious difference was the presence of a set of resonances for the isopentenyl group (δH 1.54 (3H, s, H-10‴), 1.61 (3H, s, H-9‴), 2.15 (2H, m, H-6‴), 5.11 (1H, t, J = 6.0 Hz, H-7‴), δC 16.2 (C-10‴), 22.1 (C-6‴), 24.7 (C-9‴), 123.8 (C-7‴), 130.4 (C-8‴)) in 16. The HMBC correlations from H-5‴ to C-6‴ and C-7‴ and from H-6‴ to C-3‴ and C-5‴ suggested that the isopentenyl moiety is located at C-5‴ (Figure S107, Supporting Information). Thus, the planar structure of 16 was assigned as shown (Figure 1). Compound 16 is also a racemate and was then separated by a chiral HPLC to give the enantiomers of 16a and 16b. By comparison of the experimental and calculated ECD spectra, the absolute configurations of 16a and 16b were established as (1′S) and (1′R), respectively (Figure S106, Supporting Information).
Microphyldine P (17) was obtained as an amorphous powder. Its molecular formula was determined as C32H28N2O3 base on its 13C NMR and HRESIMS data (m/z 487.2016 [M − H]−, calcd for C32H27N2O3, 487.2022). The 1H and 13C NMR data (Table 2 and Table 3) analysis revealed that the structure of 17 is a dimeric carbazole formed by murrayamine A [16] and murrayafoline A [22,23] units. Additionally, the HMBC correlations from the 3″-CH2 protons to C-5/C-6/C-7/C-2″/C-3″/C-4″, and H-5/H-2″/H-4″ to 3″-CH2 revealed a C-6–C-3″-methyl linkage between the two carbazole moieties (Figure 2). Thus, the structure of microphyldine P (17) was defined as shown (Figure 1).
2.2. Bioactivity
All of these isolates (1–17) were subjected to an evaluation of their inhibition on nitric oxide (NO) production stimulated by lipopolysaccharide (LPS) in BV-2 microglial cells and RAW 264.7 macrophages in reference to the traditional anti-inflammation use of Murraya genera [24,25]. Moreover, we tested their cytotoxic activities on HepG2, Du145, HCT116, and HeLa cells. However, none of these isolates displayed significant NO inhibitory or cytotoxic activities at 50 μM. Further testing of the biological activities of these isolates is under progress.
3. Discussion
Seventeen previously undescribed carbazole alkaloids were isolated and identified from the 95% aqueous EtOH extract of M. microphylla. (±)-Microphyltrine A (1) is a racemate of a pair of novel trimeric carbazole atropisomers from a natural source. (±)-Microphyldines A–P (2–17) are new carbazole dimeric racemates, among which, (±)-microphyldines A–J (2–11) are 10 pairs of biphenyl-type carbazole atropisomers, and (±)-microphyldines K–O (12–16) are five pairs of C1′−C6″-linked pyranocarbazole dimeric enantiomers. The chirally pure isomers of carbazole alkaloids (1a/1b–16a/16b) were obtained by chiral HPLC separation, and their absolute configurations were determined by electronic circular dichroism (ECD) data analysis. Unfortunately, none of the isolated alkaloids demonstrated significant NO inhibition or cytotoxicity at 50 μM.
4. Materials and Methods
4.1. General Experimental Procedures
UV spectra were recorded on a Shimadzu UV-2450 spectrophotometer (Shimadzu Co., Tokyo, Japan). Optical rotations were measured on a Rudolph Autopol IV automatic polarimeter (NJ, USA). ECD data were acquired on a J-810 CD spectrophotometer (JASCO, Japan). IR spectra were recorded on a Thermo Nicolet Nexus 470 FT-IR spectrometer (MA, USA). The NMR spectra were measured with a Bruker Plus-400 NMR spectrometer (Bruker Co., Switzerland) or a Varian INOVA-500 NMR spectrometer (Varian Co., USA), using acetone-d6 or CDCl3 as solvent, and the chemical shifts were referenced to the solvent residual peak. HRESIMS experiments were measured on a Waters Xevo G2 Q-TOF mass spectrometer (Waters Co., Milford, MA, USA). Column chromatography (CC) was performed on silica gel (100−200 mesh or 200−300 mesh, Qingdao Marine Chemical Co. Ltd., Qingdao, China). Semipreparative HPLC was carried out using a ZORBAX Eclipse XDB-C18 column (10 mm × 250 mm, 5 μm) on an Agilent 1200 series LC instrument with a DAD detector (Agilent Technologies, Palo Alto, CA, USA). Preparative TLC and TLC analyses were carried out on the pre-coated silica gel GF254 plates (Qingdao Marine Chemical Co. Ltd., Qingdao, China). Spots were visualized under the UV lights (254 and 365 nm) or by heating after spraying with 2% vanillin-H2SO4 solution. All the solvents used for isolation were of analytical grade and the solvents used for HPLC were of HPLC grade.
4.2. Plant Material
The dry leaves and stems of Murraya microphylla were collected from the Hainan Province, People’s Republic of China, in July 2015. The plant material was identified by one of the authors (P.F. Tu). A voucher specimen (no. MM201507) has been deposited at the Herbarium of the Peking University Modern Research Center for Traditional Chinese Medicine.
4.3. Extraction and Isolation
Air-dried and finely powdered leaves and stems of M. microphylla (16 kg) were refluxed three times with 95% aqueous EtOH (160 L × 2 h) and concentrated under reduced pressure to obtain a dry extract (730 g). The extract was suspended in H2O and extracted with CH2Cl2 and n-BuOH, successively. The CH2Cl2 extract (410 g) was subjected to a silica gel column and eluted with a stepwise gradient of petroleum ether–acetone (98:2, 96:4, 90:10, 80:20, 60:40, and 40:60, v/v) to obtain eight fractions (A−H). Fraction F (8.0 g) was subjected to a Sephadex LH-20 column eluted with MeOH–CH2Cl2 (1:1, v/v) and produced three subfractions (F1–F3). Subfraction F2 (2.2 g) was separated into three fractions (F2a–F2c) using an ODS column eluted with a stepwise gradient of MeOH–H2O (40:60, 50:50, 70:30, and 100:0, v/v). Fraction F2b (230 mg) was purified by semipreparative HPLC (3.0 mL/min, 0–25 min MeCN/H2O (75:25)) to yield 1 (8.0 mg, tR 11.8 min), 5 (11.0 mg, tR 14.4 min), 7 (5.0 mg, tR 17.9 min), and 12 (4.0 mg, tR 20.1 min). Fraction G (12.0 g) was subjected to a Sephadex LH-20 column eluted with MeOH–CH2Cl2 (1:1, v/v) and produced three subfractions (G1–G3). Fraction G2 (132 mg) was purified by semipreparative HPLC (3.0 mL/min, 0–25 min MeCN/H2O (60:40)) to yield 2 (6.0 mg, tR 13.4 min), 3 (5.0 mg, tR 15.4 min), and 4 (4.0 mg, tR 19.1 min). Subfraction G3 (4.2 g) was separated into five fractions (G3a–G3e) using an ODS column eluted with a stepwise gradient of MeOH–H2O (40:60, 60:40, and 80:20, v/v). Fraction G3a (230 mg) was purified by semipreparative HPLC (3.0 mL/min, 0–25 min MeCN/H2O (60:40)) to yield 6 (9.0 mg, tR 9.8 min), 8 (8.0 mg, tR 13.4 min), 9 (5.0 mg, tR 16.9 min), and 14 (7.0 mg, tR 22.1 min). Fraction G3e (60 mg) was purified by semipreparative HPLC (3.0 mL/min, 0–25 min MeCN/H2O (60:40)) to yield 10 (8.0 mg, tR 12.8 min), 13 (7.0 mg, tR 15.4 min), 15 (9.0 mg, tR 18.9 min), and 17 (5.0 mg, tR 21.1 min). Fraction H (5.0 g) was subjected to a Sephadex LH-20 column eluted with MeOH–CH2Cl2 (1:1, v/v) and produced three subfractions (H1–H3). Fraction H2 (132 mg) was purified by semipreparative HPLC (3.0 mL/min, 0–25 min MeCN/H2O (60:40)) to yield 16 (7.0 mg, tR 15.4 min) and 11 (9.0 mg, tR 19.1 min).
Chiral separations of 1–16 were performed on a semipreparative NP-HPLC using a Chiralpak AD-H column (4.6 mm × 250 mm, 5 mm, Daicel, Nanning, China), eluting with n-hexane-isopropanol in a ratio of 85:15 (v/v), 75:25 (v/v), 75:25 (v/v), 75:25 (v/v), 75:25 (v/v), 75:25 (v/v), 75:25 (v/v), 70:30 (v/v), 75:25 (v/v), 75:25 (v/v), 78:22 (v/v), 60:40 (v/v), 60:40 (v/v), 60:40 (v/v), 60:40 (v/v), and 60:40 (v/v), respectively. The detection wavelength was 238 nm and the flow rate was 1 mL/min. Finally, compounds 1a (3.8 mg, tR 6.0 min), 1b (3.8 mg, tR 8.0 min), 2a (2.6 mg, tR 6.2 min), 2b (2.9 mg, tR 8.0 min), 3a (2.4 mg, tR 4.4 min), 3b (2.3mg, tR 5.8 min), 4a (1.8 mg, tR 4.3 min), 4b (1.9 mg, tR 6.8 min), 5a (4.7 mg, tR 6.3 min), 5b (5.0 mg, tR 7.8 min), 6a (3.9 mg, tR 6.1 min), 6b (4.2 mg, tR 7.6 min), 7a (2.3 mg, tR 3.4 min), 7b (2.5 mg, tR 5.4 min), 8a (3.6 mg, tR 4.8 min), 8b (3.8 mg, tR 7.1 min), 9a (2.4 mg, tR 4.0 min), 9b (2.4 mg, tR 6.8 min), 10a (3.8 mg, tR 3.9 min), 10b (3.8 mg, tR 6.3 min), 11a (4.3 mg, tR 5.4 min), 11b (4.5 mg, tR 7.5 min), 12a (1.9 mg, tR 2.6 min), 12b (2.0 mg, tR 7.5 min), 13a (3.3 mg, tR 4.2 min), 13b (3.5 mg, tR 8.3 min), 14a (3.4 mg, tR 2.6 min), 14b (3.4 mg, tR 7.8 min), 15a (4.3 mg, tR 5.4 min), 15b (4.5 mg, tR 7.6 min), 16a (3.3 mg, tR 4.4 min), and 16b (3.5 mg, tR 7.9 min) were yielded, respectively.
4.3.1. (±)-. Microphyltrine A (1)
White amorphous powder; UV (MeOH) λmax (log ε) 227 (4.64), 252 (4.63), 293 (2.60), 332 (2.06) nm; IR (KBr) νmax 3421, 1633, 1454, 1259, 1129, 1025, 670 cm−1; 1H and 13C NMR data, see Table 1; HRESIMS m/z 824.3330 [M − H]− (calcd. for C52H46N3O7, 824.3336).
(−)-Microphyltrine A (1a): White amorphous powder, −74 (c 0.1, MeOH); ECD (MeOH) λmax (Δε) 218 (+13.79), 245 (−10.01) nm.
(+)-Microphyltrine A (1b): White amorphous powder, +84 (c 0.1, MeOH); ECD (MeOH) λmax (Δε) 218 (−13.68), 245 (+10.30) nm.
4.3.2. (±)-. Microphyldine A (2)
Brown amorphous powder; UV (MeOH) λmax (log ε) 224 (3.85), 247 (3.86), 305 (3.33), 340 (2.73) nm; IR (KBr) νmax 3413, 2968, 1720, 1628, 1462, 1301, 1128, 1025, 890, 838, 728 cm−1; 1H and 13C NMR data, see Table 2 and Table 3; HRESIMS m/z 585.2387 [M − H]− (calcd for C37H33N2O5, 585.2389).
(−)-Microphyldine A (2a): Brown amorphous powder, −20 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 224 (+14.81), 252 (−7.66) nm.
(+)-Microphyldine A (2b): Brown amorphous powder, +20 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 224 (−14.73), 252 (+7.68) nm.
4.3.3. (±)-. Microphyldine B (3)
Brown amorphous powder; UV (MeOH) λmax (log ε) 229 (4.37), 301 (2.16) nm; IR (KBr) νmax 3552, 2968, 2919, 1642, 1453, 1375, 1168, 1128, 1023, 891, 577, 451 cm−1; 1H and 13C NMR data, see Table 2 and Table 3; HRESIMS m/z 615.2486 [M − H]− (calcd for C38H35N2O6, 615.2495).
(−)-Microphyldine B (3a): Brown amorphous powder, −100 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 226 (+13.87), 250 (−7.95) nm.
(+)-Microphyldine B (3b): Brown amorphous powder, +100 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 226 (−14.23), 250 (+8.39) nm.
4.3.4. (±)-. Microphyldine C (4)
Brown amorphous powder; UV (MeOH) λmax (log ε) 228 (4.86), 303 (3.26) nm; IR (KBr) νmax 3418, 2969, 2922, 1715, 1640, 1444, 1423, 1377, 1186, 1128, 1024, 663 cm−1; 1H and 13C NMR data, see Table 2 and Table 3; HRESIMS m/z 585.2389 [M − H]− (calcd for C37H33N2O5, 585.2389).
(−)-Microphyldine C (4a): Brown amorphous powder, −80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 226 (+81.61), 252 (−32.80) nm.
(+)-Microphyldine C (4b): Brown amorphous powder, +80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 226 (−79.45), 254 (+32.11) nm.
4.3.5. (±)-. Microphyldine D (5)
Brown amorphous powder; UV (MeOH) λmax (log ε) 227 (4.86), 248 (4.83), 307 (3.26) nm; IR (KBr) νmax 3444, 2922, 1746, 1644, 1454, 1428, 1185, 1020, 578 cm−1; 1H and 13C NMR data, see Table 2 and Table 3; HRESIMS m/z 653.3023 [M − H]− (calcd for C42H41N2O5, 653.3015).
(−)-Microphyldine D (5a): Brown amorphous powder, −80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 222 (+9.61), 252 (−6.80), 296 (−2.78) nm.
(+)-Microphyldine D (5b): Brown amorphous powder, +80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 222 (−9.45), 254 (+7.11), 298 (+2.63) nm.
4.3.6. (±)-. Microphyldine E (6)
Brown amorphous powder; UV (MeOH) λmax (log ε) 217 (4.86), 224 (4.43), 300 (3.26) nm; IR (KBr) νmax 3445, 2979, 2901, 2123, 1646, 1454, 1383, 1160, 1044, 577 cm−1; 1H and 13C NMR data, see Table 2 and Table 3; HRESIMS m/z 653.3023 [M − H]− (calcd for C42H41N2O5, 653.3015).
(−)-Microphyldine E (6a): Brown amorphous powder, −80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 218 (+3.31), 272 (−0.50) nm.
(+)-Microphyldine E (6b): Brown amorphous powder, +80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 218 (−3.15), 272 (+0.50) nm.
4.3.7. (±)-. Microphyldine F (7)
Brown amorphous powder; UV (MeOH) λmax (log ε) 227 (4.86), 248.8 (4.83), 296.6 (2.80) nm; IR (KBr) νmax 3646, 3612, 3361, 2921, 2851, 1713, 1642, 1503, 1458, 1379, 1143, 1019, 576 cm−1; 1H and 13C NMR data, see Table 2 and Table 3; HRESIMS m/z 585.2390 [M − H]− (calcd for C37H33N2O5 585.2389).
(−)-Microphyldine F (7a): Brown amorphous powder, −80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 218 (+7.11), 248 (−3.10) nm.
(+)-Microphyldine F (7b): Brown amorphous powder, +80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 218 (−7.45), 248 (+2.91) nm.
4.3.8. (±)-. Microphyldine G (8)
Brown amorphous powder; UV (MeOH) λmax (log ε) 227 (4.86), 296.6 (4.80), 334 (4.26) nm; IR (KBr) νmax 3647, 3420, 2966, 2920, 2851, 1728, 1608, 1448, 1428, 1185, 1018, 578 cm−1; 1H and 13C NMR data, see Table 2 and Table 3; HRESIMS m/z 653.3007 [M − H]− (calcd for C42H41N2O5, 653.3015).
(−)-Microphyldine G (8a): Brown amorphous powder, −80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 200 (+8.61), 242 (−1.80) nm.
(+)-Microphyldine G (8b): Brown amorphous powder, +80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 200 (−8.45), 244 (+2.11) nm.
4.3.9. (±)-. Microphyldine H (9)
Brown amorphous powder; UV (MeOH) λmax (log ε) 238.8 (4.83), 309 (4.80), 334 (4.26) nm; IR (KBr) νmax 3712, 3444, 2920, 1729, 1610, 1454, 1419, 1175, 1019, 575 cm−1; 1H and 13C NMR data, see Table 2 and Table 3; HRESIMS m/z 601.2692 [M − H]− (calcd for C38H37N2O5, 601.2702).
(−)-Microphyldine H (9a): Brown amorphous powder, −80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 218 (+23.61), 254 (−13.80) nm.
(+)-Microphyldine H (9b): Brown amorphous powder, +80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 218 (−23.45), 254 (+13.11) nm.
4.3.10. (±)-. Microphyldine I (10)
Brown amorphous powder; UV (MeOH) λmax (log ε) 234 (4.86), 248 (4.83), 291 (4.80), 334 (4.26) nm; IR (KBr) νmax 3419, 2969, 2920, 1714, 1613, 1453, 1387, 1174, 1019, 582 cm−1; 1H and 13C NMR data, see Table 2 and Table 3; HRESIMS m/z 391.1440 [M − H]− (calcd for C26H19N2O2, 391.1447).
(−)-Microphyldine I (10a): Brown amorphous powder, −80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 203 (+11.61), 275 (−24.80) nm.
(+)-Microphyldine I (10b): Brown amorphous powder, +80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 204 (−19.45), 275 (+25.11) nm.
4.3.11. (±)-. Microphyldine J (11)
Brown amorphous powder; UV (MeOH) λmax (log ε) 226 (4.85), 243 (4.81), 294 (4.76), 334 (4.26) nm; IR (KBr) νmax 3404, 1746, 1633, 1470, 1453, 1111, 1019, 579 cm−1; 1H and 13C NMR data, see Table 2 and Table 3; HRESIMS m/z 405.1232 [M − H]− (calcd for C26H17N2O3, 405.1239).
(−)-Microphyldine J (11a): Brown amorphous powder, −80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 218 (−12.61), 272 (+25.20), 266 (−16.28) nm.
(+)-Microphyldine J (11b): Brown amorphous powder, +80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 218 (+19.45), 274 (−24.11), 268 (+16.63) nm.
4.3.12. (±)-. Microphyldine K (12)
Brown amorphous powder; UV (MeOH) λmax (log ε) 238 (4.83), 306 (4.80), 324 (4.26) nm; IR (KBr) νmax 3601, 3425, 2969, 2917, 2855, 1720, 1631, 1464, 1135, 1025, 584 cm−1; 1H and 13C NMR data, see Table 4; HRESIMS m/z 505.2125 [M − H]− (calcd for C32H29N2O4, 505.2127).
(−)-Microphyldine K (12a): Brown amorphous powder, −80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 214 (+19.61), 252 (−8.80), 296 (−5.78) nm.
(+)-Microphyldine K (12b): Brown amorphous powder, +80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 213 (−19.45), 254 (+9.11), 298 (+5.63) nm.
4.3.13. (±)-. Microphyldine L (13)
Brown amorphous powder; UV (MeOH) λmax (log ε) 243 (4.76), 300 (4.30), 334 (4.26) nm; IR (KBr) νmax 3525, 3459, 3363, 2969, 2923, 2855, 1706, 1622, 1450, 1420, 1186, 1047, 580 cm−1; 1H and 13C NMR data, see Table 4; HRESIMS m/z 587.2543 [M − H]− (calcd for C37H35N2O5, 587.2546).
(−)-Microphyldine L (13a): Brown amorphous powder, −80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 222 (−4.61), 296 (−3.78) nm.
(+)-Microphyldine L (13b): Brown amorphous powder, +80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 222 (+4.45), 298 (+3.63) nm.
4.3.14. (±)-. Microphyldine M (14)
Brown amorphous powder; UV (MeOH) λmax (log ε) 224 (4.86), 238.8 (4.83), 296.6 (4.80), 334 (4.26) nm; IR (KBr) νmax 3525, 3459, 3363, 2969, 2923, 2855, 1706, 1622, 1450, 1420, 1186, 1047, 580 cm−1; 1H and 13C NMR data, see Table 4; HRESIMS m/z 601.2712 [M − H]− (calcd for C38H37N2O5, 601.2702).
(−)-Microphyldine M (14a): Brown amorphous powder, −80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 208 (+6.81), 242 (−7.20), 296 (−2.78) nm.
(+)-Microphyldine M (14b): Brown amorphous powder, +80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 208 (−6.45), 244 (+7.11), 296 (+2.63) nm.
4.3.15. (±)-. Microphyldine N (15)
Brown amorphous powder; UV (MeOH) λmax (log ε) 224 (4.86), 245 (4.83), 301 (4.80), 334 (4.26) nm; IR (KBr) νmax 3725, 3600, 3383, 2964, 2917, 2850, 1721, 1617, 1494, 1475, 1259, 1025, 580 cm−1; 1H and 13C NMR data, see Table 4; HRESIMS m/z 655.3156 [M − H]− (calcd for C42H43N2O5, 655.3172).
(−)-Microphyldine N (15a): Brown amorphous powder, −80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 214 (+1.21), 248 (−1.10), 326 (−0.78) nm.
(+)-Microphyldine N (15b): Brown amorphous powder, +80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 213 (−1.45), 247 (+1.11), 328 (+0.63) nm.
4.3.16. (±)-. Microphyldine O (16)
Brown amorphous powder; UV (MeOH) λmax (log ε) 226 (4.86), 238.8 (4.83), 302 (4.80), 334 (4.26) nm; IR (KBr) νmax 3725, 3624, 3383, 2920, 2849, 1719, 1617, 1476, 1438, 1162, 1028, 580 cm−1; 1H and 13C NMR data, see Table 4; HRESIMS m/z 669.3328 [M − H]− (calcd for C43H45N2O5, 669.3328).
(−)-Microphyldine O (16a): Brown amorphous powder, −80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 218 (+5.91), 252 (−5.80), 296 (−0.78) nm.
(+)-Microphyldine O (16b): Brown amorphous powder, +80 (c 0.01, MeOH); ECD (MeOH) λmax (Δε) 218 (−5.85), 254 (+5.71), 298 (+0.93) nm.
4.3.17. Microphyldine P (17)
Brown amorphous powder; UV (MeOH) λmax (log ε) 245 (4.83), 294 (4.80), 334 (4.26) nm; IR (KBr) νmax 3442, 2954, 2920, 2850, 1734, 1656, 1494, 1458, 1214, 1024, 578 cm−1; 1H and 13C NMR data see, Table 2 and Table 3; HRESIMS m/z 487.2016 [M − H]− (calcd for C32H27N2O3, 487.2022).
4.4. Computational Methods
The Ra/Sa configurations of compounds 1, 2, 6, 8, 10, 11 and the (1′S)/(1′R) configurations of compounds 12–16 were submitted to random conformational analysis, respectively, with the MMFF94s force field using the Sybyl-X 2.0 software package. The conformers were further optimized by using the TDDFT method at the B3LYP/6-31G(d) level, and the frequency was calculated at the same level of theory. The stable conformers without imaginary frequencies were subjected to ECD calculation by the TDDFT method at the B3LYP/6-31+G(d) level with the CPCM model in MeOH. ECD spectra of different conformers were simulated using SpecDis v1.51 with a half-band width of 0.3 eV, and the final ECD spectra were computed according to the Boltzmann-calculated contribution of each conformer. The calculated ECD spectra were compared with the experimental data. All calculations were performed with the Gaussian 09 program package [26,27].
4.5. Anti-Inflammatory Activity Assay
The murine BV-2 microglial cells or the monocytic RAW 264.7 macrophages were purchased from Peking Union Medical College (PUMC) Cell Bank (Beijing, China). Cell maintenance, experimental procedures, and data determination for the inhibition of NO production are the same as previously described [15,28]. Cell viability was evaluated by MTT assay. Dexamethasone was used as a positive control.
4.6. Cytotoxicity Assay
HepG2, Du145, HCT116, and HeLa cells (PUMC Cell Bank, Beijing, China) were used for the cytotoxicity assays. Cytotoxic activities were determined using the MTT method. Cell culture, experimental procedures, and data processing were performed according to the literature report [29], with taxol serving as a positive control.
Supplementary Materials
The following are available online, Figures S1, the HRESIMS, 1H NMR, 13C NMR, HSQC, and HMBC spectra of compounds 1–17, along with the chiral HPLC separation and ECD data of compounds 3–11 and 13–16.
Author Contributions
Conceptualization, P.T. and Y.J.; methodology, X.M. and Y.J.; validation, X.M., H.C., and S.Z.; investigation, X.M.; writing, review, and editing, H.C., X.M., and Y.J.; supervision, project administration and funding acquisition, Y.J. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by National Natural Science Foundation of China (NSFC; Nos. 81973199, 81773864, and 81473106), and National Key R&D Program of China (No. 2019YFC1711000).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of the compounds are not available from the authors.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nandy B.C., Gupta A.K., Mittal A., Vyas V. Carbazole: It’s biological activity. J. Biomed. Pharm. Res. 2014;3:42–48. [Google Scholar]
- 2.Greger H. Phytocarbazoles: Alkaloids with great structural diversity and pronounced biological activities. Phytochem. Rev. 2017;16:1095–1153. doi: 10.1007/s11101-017-9521-5. [DOI] [Google Scholar]
- 3.Lv H., Zhou Y., Wen R., Shi M., Zeng K., Xia F., Tu P., Jiang Y. Murradiate and murradiol, two structurally unique heterodimers of carbazole-monoterpene and carbazole-phenylethanol from Murraya tetramera. Phytochem. Lett. 2016;15:113–115. doi: 10.1016/j.phytol.2015.12.002. [DOI] [Google Scholar]
- 4.Lv H., Wen R., Zhou Y., Zeng K., Li J., Guo X., Tu P., Jiang Y. Nitrogen oxide inhibitory trimeric and dimeric carbazole alkaloids from Murraya tetramera. J. Nat. Prod. 2015;78:2432–2439. doi: 10.1021/acs.jnatprod.5b00527. [DOI] [PubMed] [Google Scholar]
- 5.Editorial Committee of Flora of China . Flora of China. Science Press; Beijing, China: 1997. p. 146. [Google Scholar]
- 6.Ma X., Cao N., Zhang C., Guo X., Zhao M., Tu P., Jiang Y. Cytotoxic carbazole alkaloid derivatives from the leaves and stems of Murraya microphylla. Fitoterapia. 2018;127:334–340. doi: 10.1016/j.fitote.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Zou L., Yang C., Zhang H. Study on carbazole alkaloids of Murraya microphylla. J. Chin. Med. Mater. 1999;22:458–460. [PubMed] [Google Scholar]
- 8.Gassner C., Hesse R., Schmidt A.W., Knoelker H. Total synthesis of the cyclic monoterpenoid pyrano[3,2-a]carbazole alkaloids derived from 2-hydroxy-6-methylcarbazole. Org. Biomol. Chem. 2014;12:6490–6499. doi: 10.1039/C4OB01151A. [DOI] [PubMed] [Google Scholar]
- 9.Ito C., Wu T.S., Furukawa H. New carbazole alkaloids from Murraya euchrestifolia. Chem. Pharm. Bull. 1988;36:2377–2380. doi: 10.1248/cpb.36.2377. [DOI] [Google Scholar]
- 10.Ya Q., Lu W., Chen J., Tan X. Study on the chemical constituent from Murraya tetramera Huang. Guangxi Sci. 2010;17:347–348. [Google Scholar]
- 11.Ma Q., Tian J., Yang J., Wang A., Ji T., Wang Y., Su Y. Bioactive carbazole alkaloids from Murraya koenigii (L.) Spreng. Fitoterapia. 2013;87:1–6. doi: 10.1016/j.fitote.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Joshi T., Jain T., Mahar R., Singh S.K., Srivastava P., Shukla S.K., Mishra D.K., Bhatta R.S., Banerjee D., Kanojiya S. Pyranocarbazoles from Murraya koenigii (L.) Spreng. as antimicrobial agents. Nat. Prod. Res. 2018;32:430–434. doi: 10.1080/14786419.2017.1308363. [DOI] [PubMed] [Google Scholar]
- 13.Furukawa H., Wu T.S., Ohta T., Kuoh C.S. Chemical constituents of Murraya euchrestifolia Hayata. Structures of novel carbazolequinones and other new carbazole alkaloids. Chem. Pharm. Bull. 1985;33:4132–4138. doi: 10.1248/cpb.33.4132. [DOI] [Google Scholar]
- 14.Chen Y., Cao N., Lv H., Zeng K., Yuan J., Guo X., Zhao M., Tu P., Jiang Y. Anti-inflammatory and cytotoxic carbazole alkaloids from Murraya kwangsiensis. Phytochemistry. 2020;170:112186. doi: 10.1016/j.phytochem.2019.112186. [DOI] [PubMed] [Google Scholar]
- 15.Cao N., Chen Y., Ma X., Zeng K., Zhao M., Tu P., Li J., Jiang Y. Bioactive carbazole and quinoline alkaloids from Clausena dunniana. Phytochemistry. 2018;151:1–8. doi: 10.1016/j.phytochem.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Schuster C., Julich-Gruner K.K., Schnitzler H., Hesse R., Jaeger A., Schmidt A.W., Knoelker H. Total syntheses of murrayamines E, I, and K. J. Org. Chem. 2015;80:5666–5673. doi: 10.1021/acs.joc.5b00630. [DOI] [PubMed] [Google Scholar]
- 17.Hesse R., Kataeva O., Schmidt A.W., Knoelker H. Synthesis of prenyl- and geranyl-substituted carbazole alkaloids by DIBAL-H promoted reductive pyran ring opening of dialkylpyrano[3,2-a]carbazoles. Chem. Eur. J. 2014;20:9504–9509. doi: 10.1002/chem.201403645. [DOI] [PubMed] [Google Scholar]
- 18.Rehman F., Khan M.F., Khan I., Shareef H., Marwat S.K. Analgesic activity of carbazolealkaloid from Murraya paniculata Linn. (Rutaceae) World Appl. Sci. J. 2014;32:1631–1636. [Google Scholar]
- 19.Humne V., Dangat Y., Vanka K., Lokhande P. Iodine-catalyzed aromatization of tetrahydrocarbazoles and its utility in the synthesis of glycozoline and murrayafoline A: A combined experimental and computational investigation. Org. Biomol. Chem. 2014;12:4832–4836. doi: 10.1039/C4OB00635F. [DOI] [PubMed] [Google Scholar]
- 20.Bedford R.B., Bowen J.G., Weeks A.L. Synthesis of murrayaquinone A and analogues via ring-closing C-H arylation. Tetrahedron. 2013;69:4389–4394. doi: 10.1016/j.tet.2013.02.055. [DOI] [Google Scholar]
- 21.Furukawa H., Ito C., Wu T.S., McPhail A.T. Structural elucidation of murrafolines, six novel binary carbazole alkaloids isolated from Murraya euchrestifolia. Chem. Pharm. Bull. 1993;41:1249–1254. doi: 10.1248/cpb.41.1249. [DOI] [Google Scholar]
- 22.Chakthong S., Bindulem N., Raknai S., Yodwaree S., Kaewsanee S., Kanjana-Opas A. Carbazole-pyranocoumarin conjugate and two carbazole alkaloids from the stems of Clausena excavata. Nat. Prod. Res. 2016;30:1690–1697. doi: 10.1080/14786419.2015.1135143. [DOI] [PubMed] [Google Scholar]
- 23.Boerger C., Kataeva O., Knoelker H. Novel approach to biscarbazole alkaloids via Ullmann coupling–synthesis of murrastifoline-A and bismurrayafoline-A. Org. Biomol. Chem. 2012;10:7269–7273. doi: 10.1039/c2ob26229k. [DOI] [PubMed] [Google Scholar]
- 24.Bhattacharya K., Samanta S.K., Tripathi R., Mallick A., Chandra S., Pal B.C., Shaha C., Mandal C. Apoptotic effects of mahanine on human leukemic cells are mediated through crosstalk between Apo-1/Fas signaling and the Bid protein and via mitochondrial pathways. Biochem. Pharmacol. 2010;79:361–372. doi: 10.1016/j.bcp.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 25.Ito C., Itoigawa M., Nakao K., Murata T., Tsuboi M., Kaneda N., Furukawa H. Induction of apoptosis by carbazole alkaloids isolated from Murraya koenigii. Phytomedicine. 2006;13:359–365. doi: 10.1016/j.phymed.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Zhang C., Wang S., Zeng K., Li J., Ferreira D., Zjawiony J.K., Liu B., Guo X., Jin H., Jiang Y., et al. Nitric oxide inhibitory dimeric sesquiterpenoids from Artemisia rupestris. J. Nat. Prod. 2016;79:213–223. doi: 10.1021/acs.jnatprod.5b00894. [DOI] [PubMed] [Google Scholar]
- 27.Sun J., Zhu Z., Song Y., Dong D., Zheng J., Liu T., Zhao Y., Ferreira D., Zjawiony J.K., Tu P., et al. Nitric oxide inhibitory meroterpenoids from the fungus Penicillium purpurogenum MHZ 111. J. Nat. Prod. 2016;79:1415–1422. doi: 10.1021/acs.jnatprod.6b00160. [DOI] [PubMed] [Google Scholar]
- 28.Lv H., Wang S., Zeng K., Li J., Guo X., Ferreira D., Zjawiony J.K., Tu P., Jiang Y. Anti-inflammatory coumarin and benzocoumarin derivatives from Murraya alata. J. Nat. Prod. 2015;78:279–285. doi: 10.1021/np500861u. [DOI] [PubMed] [Google Scholar]
- 29.Ma K., Wang J., Luo J., Yang M., Kong L. Tabercarpamines A-J, apoptosis-inducing indole alkaloids from the leaves of tabernaemontana corymbosa. J. Nat. Prod. 2014;77:1156–1163. doi: 10.1021/np401098y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.