Abstract
The expression of chromosomally integrated transgenes usually varies greatly among independent transfectants. This variability in transgene expression has led to the definition of locus control regions (LCRs) as elements which render expression consistent. Analyses of expression in single cells revealed that the expression of transgenes which lack an LCR is often variegated, i.e., on in some cells and off in others. In many cases, transgenes which show variegated expression were found to have inserted near the centromere. These observations have suggested that the LCR prevents variegation by blocking the inhibitory effect of heterochromatin and other repetitive-DNA-containing structures at the insertion site and have raised the question of whether the LCR plays a similar role in endogenous genes. To address this question, we have examined the effects of deleting the LCR from the immunoglobulin heavy-chain locus of a mouse hybridoma cell line in which expression of the immunoglobulin μ heavy-chain gene is normally highly stable. Our analysis of μ expression in single cells shows that deletion of this LCR resulted in variegated expression of the μ gene. That is, in the absence of the LCR, expression of the μ gene in the recombinant locus could be found in either of two epigenetically maintained, metastable states, in which transcription occurred either at the normal rate or not at all. In the absence of the LCR, the on state had a half-life of ∼100 cell divisions, while the half-life of the off state was ∼40,000 cell divisions. For recombinants with an intact LCR, the half-life of the on state exceeded 50,000 cell divisions. Our results thus indicate that the LCR increased the stability of the on state by at least 500-fold.
Most genes in complex, differentiated organisms, such as metazoa, are expressed in a tissue-specific fashion, on in one subset of cells and off in others. Tissue-specific gene expression is initially established as cells in different environments are subjected to different signals. The signals are each presumed to induce the production of a distinct complement of transcription factors, which are then directed by cis-acting elements to activate or silence adjoining genes. The ontogenetic state of cells is thus considered to reflect their past and present production of trans-acting factors.
Much of our knowledge about gene expression in complex organisms comes from studying the expression of transgenes, DNA segments which have been introduced into animals or cell lines. Early work revealed that the expression of chromosomally integrated transgenes varies greatly, and traditionally this variability has been ascribed to the effects of the neighboring genome. The variability in transgene expression has led to the definition of locus control regions (LCRs) as elements which render expression consistent, at least among cells which are in the same ontogenetic state. Analyses of expression in single cells revealed that the expression of transgenes which lack the LCR is often variegated, i.e., cells which are presumed to be in the same ontogenetic state differ in their expression of the transgene (for a review, see reference 25). The variability in transgene expression thus reflects variations in the fraction of expressing cells. In many cases, transgenes which showed variegated expression were found to have inserted near the centromere (11, 26). The variegated expression of transgenes is thus similar to position effect variegation in Drosophila melanogaster, whereby events such as chromosomal translocations move an endogenous gene into a heterochromatic region. Repeated DNA segments can themselves induce variegation (10, 14), and the variegated expression of transgenes can sometimes result from their insertion as a tandem array. Such observations have suggested that the LCR prevents variegation by blocking the inhibitory effect of heterochromatin and other repetitive-DNA-containing structures (25, 26).
Most genes are located in euchromatin, raising the question of whether the LCR also serves to prevent the repression of endogenous genes. The immunoglobulin heavy chain (IgH) locus of the mouse contains an LCR which includes three distinct elements in the major (VDJ-Cμ) intron: the Eμ core enhancer, the matrix attachment regions (MARs) which flank Eμ, and the switch region (Sμ) (13, 16, 37). A system in which expression of the endogenous μ heavy-chain gene of a mouse hybridoma cell line depends on the integrity of this LCR has been previously described (29, 43). The data in the present analysis indicate that deletion of the LCR resulted in variegated expression of the μ gene. That is, in the absence of the LCR, expression of the μ gene in the recombinant locus could exist in either of two epigenetically maintained, metastable states, in which transcription occurred either at the normal rate or not at all. We have measured the rates at which cells switched between the on and off states. In the absence of the LCR, the on state had a half-life of ∼100 cell divisions, while the half-life of the off state was ∼40,000 cell divisions. Our analysis of recombinants bearing an intact LCR indicated that the LCR increased the stability of the on state by at least 500-fold.
MATERIALS AND METHODS
Cell culture.
Cells were grown in Dulbecco’s modified Eagle’s medium (high glucose; GIBCO H21) supplemented with 13% enriched bovine calf serum (Hyclone) and 3.5 × 10−4% 2-mercaptoethanol (normal medium). MHX medium (27) also contained the additional supplements 10 μg of mycophenolic acid, 15 μg of hypoxanthine, and 25 μg of xanthine per ml.
RNA analysis.
For Northern blot analysis, 10 μg of cytoplasmic RNA (22) was denatured with formamide and size fractionated on 1% agarose gels containing formaldehyde; transfer to nylon membranes and probe preparation by random priming were done according to standard procedures.
Plaque assay for IgM-secreting cells.
We used the plaque assay to detect IgM-secreting cells, as described previously (2). Hybridoma cells were plated in the presence of hapten-coupled sheep erythrocytes in agarose and incubated at 37°C for 2 h. Then, guinea pig complement (Behring) was added, and the cells were incubated further, until plaques formed around IgM-producing hybridoma cells as regions of lysed erythrocytes. Plaque-forming cells were isolated and purified as described previously (2).
Suicide selection for IgM-negative cells.
The method used for the suicide selection of μ-deficient variants was described previously (9). Briefly, hybridoma cells are coupled with trinitrobenzoylsulfate and incubated at limited density in the presence of guinea pig serum as a source of complement. IgM preferentially binds to the same cell that secretes it, thus resulting in the complement-dependent lysis of the IgM-secreting cells. IgM-deficient cells survive this selection.
Flow cytometry.
A total of 106 cells were washed twice in staining buffer (1% bovine calf serum in phosphate-buffered saline) and then fixed in 4% paraformaldehyde (Sigma) for 20 min at 4°C. The cells were then washed twice in permeabilization buffer (1% bovine calf serum in phosphate-buffered saline, 0.1% saponin [Sigma] [pH 7.4 to 7.6]) and resuspended in 100 μl of permeabilization buffer containing 1 μg of fluorescein isothiocyanate-conjugated anti-IgM or isotype-matched anti-IgG2b antibodies (PharMingen; clones R6-60.2 and R12-3, respectively) for 30 min at 4°C. The cells were washed once in permeabilization buffer and once in staining buffer. The cells were then resuspended in staining buffer and analyzed by flow cytometry. Data were analyzed with CellQuest software.
Calculations.
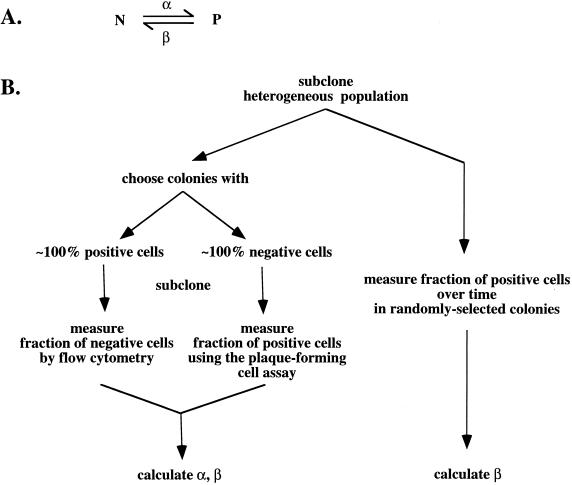
To describe the dynamics of a population of cells which can switch between positive and negative states, we consider that the rate of change in the number of positive cells (P) and the number of negative cells (N) over time (t) can be described in mathematical terms as follows:
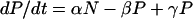 |
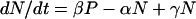 |
where α and β are the transition rates for the conversion of negative to positive cells and positive to negative cells, respectively, and γ is the growth rate of the cells and corresponds to a doubling time of approximately 18 h. Solving this system gives
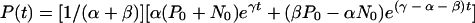 |
1 |
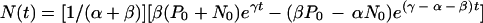 |
2 |
where P0 and N0 are the number of positive and negative cells at time zero.
When cultures are derived from subclones and time is measured from the moment of subcloning, these equations simplify. In this case, a subclone must start from either a positive or a negative cell. Thus,
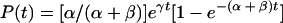 |
for a culture starting from a negative cell, i.e., N0 = 1 and P0 = 0, and
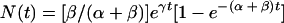 |
for a culture starting from a positive cell, i.e., P0 = 1 and N0 = 0. In these cases, the total number of cells is eγt, so p, the fraction of positive cells, and n, the fraction of negative cells, are given by
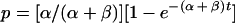 |
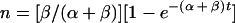 |
The solution of this system for α and β as a function of p, n, and t is
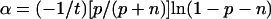 |
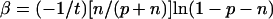 |
For p ≪ n, then α = (−1/t)(p/n)ln(1 − n) and β = (−1/t)ln(1 − n).
As described in the text and shown in Fig. 5, we also calculated β for bulk cultures under circumstances in which βt ≪ 1 and α ≪ β. In this case, equation 2 simplifies to P/P0 = (p/p0) = e−βt, where p0 is the initial fraction of positive cells and, as above, p is the fraction of positive cells at t.
FIG. 5.
Schematic diagram of measurement of transition rates. See text for explanation.
RESULTS
Expression of the μ heavy-chain gene is variegated in recombinants that lack the LCR.
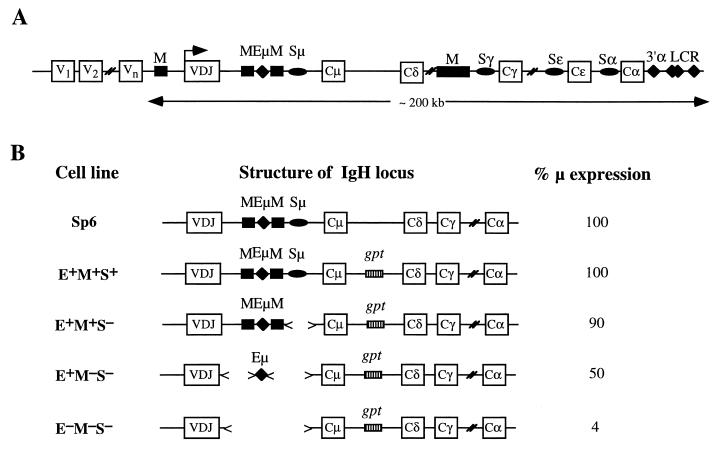
The structure of the functional (rearranged) IgH locus is shown in Fig. 1A. As noted in the introduction, uniform, high-level expression of IgH-derived transgenes in mice requires three elements in the major (VDJ-Cμ) intron: Eμ, the MARs which flank Eμ, and Sμ (13, 16, 37). These elements thus comprise part or all of an LCR. In order to study the molecular requirements for IgH expression in a permanent, cloned cell line, we used a system based on the mouse hybridoma, Sp6, which bears a single copy of the endogenous immunoglobulin μ heavy-chain gene and produces IgM that is specific for the hapten trinitrophenol (TNP). In previous studies (29, 43), various targeted recombinants of the Sp6 cell line that lacked one or more of the elements Eμ (E), MARs (M), or Sμ (S) (Fig. 1B) were constructed and analyzed. To facilitate recovery of the targeted recombinants, the targeting vector included the gpt gene of Escherichia coli, which allows cells to utilize xanthine and thus to grow in medium containing xanthine and mycophenolic acid (such as MHX medium) (27). Expression of the μ gene in these targeted recombinants depends on elements in the VDJ-Cμ intron. That is, while expression of the μ gene was normal in the E+M+S+ recombinants, μ expression in the E−M−S− recombinant hybridoma cell lines was greatly reduced, and this expression varied significantly among independent E−M−S− recombinants (29, 43).
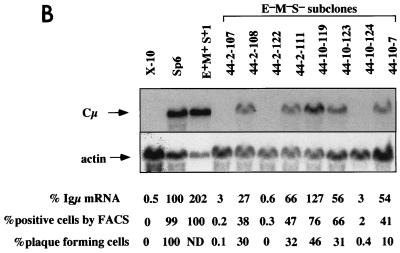
FIG. 1.
μ expression in targeted recombinants. (A) Structure of the IgH locus. V, exons encoding the variable regions; C, exons encoding the constant regions; M, MARs; S, Sμ. The promoter of the VDJ-Cμ transcription unit is indicated by the arrow. (B) Targeted recombinants used in this study. These recombinants were described previously (29, 43), and their structure and average level of μ mRNA are indicated.
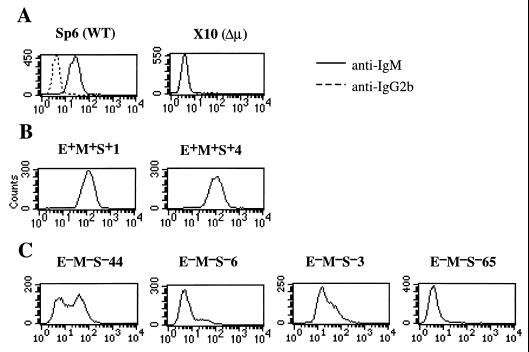
We initially compared expression of the μ gene in E+M+S+ and E−M−S− recombinants. In each case, we examined multiple independent recombinants. Independent recombinants are designated by the isolate number, e.g., E−M−S−3 and E−M−S−44 denote two independently generated recombinants, 3 and 44, lacking the MAR-Eμ-MAR-Sμ segment; specific subclones of these recombinants are then designated by appended numbers, e.g., E−M−S−44-10-119. To test whether deletion of the MAR-Eμ-MAR-Sμ segment resulted in the variegated expression of the μ gene, we examined μ expression in single cells by using a fluorescent μ-specific antibody to label intracellular μ chains and then analyzing the cells by flow cytometry (Fig. 2). To standardize this analysis, we used two cell lines, the parental hybridoma (Sp6) and a mutant derivative of Sp6 that has the μ gene deleted (X10). The mean fluorescence intensity of the Sp6 cells with the μ-specific antibody was ∼20-fold higher than with an isotype-matched control (Fig. 2A). The μ-deleted cell line, X10, gave the same low, background-level signal with the μ-specific antibody as with the isotype-matched control (Fig. 2A). In the case of the wild-type E+M+S+ recombinants, the populations were homogeneous and contained the normal level of IgM (Fig. 2B). By contrast, the E−M−S− recombinants had heterogeneous populations: some cells stained at the normal level and some stained at the level of the μ-deleted mutant (Fig. 2C).
FIG. 2.
μ expression in recombinant cell lines as analyzed by flow cytometry. For each of the indicated cell lines, the cells were fixed, stained with fluorescein isothiocyanate-labeled rat monoclonal IgG2b specific for the Cμ2 domain of mouse IgM or another IgG2b as an isotype-matched control, and analyzed by flow cytometry. Fluorescence intensity is plotted on the horizontal axis, while the number of cells is plotted on the vertical axis.
Positive and negative cells can interconvert.
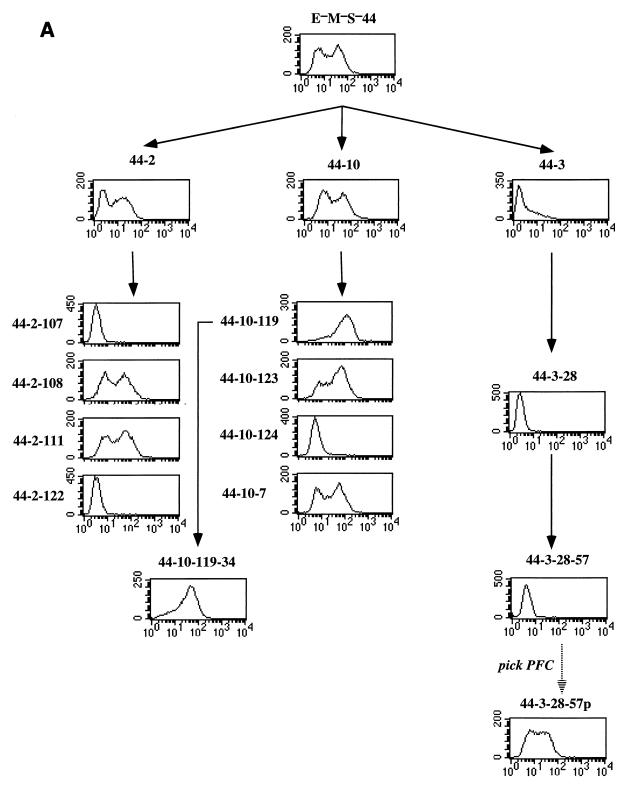
Our finding that the E−M−S− recombinant cells occurred in two populations with the staining intensity of the wild-type and μ-deleted controls suggested that transcription in these cells was bimodal, with cells positive or negative for μ expression. The fact that these cells had been subcloned several times also suggested that positive and negative cells could interconvert. Considering that μ mRNA is stable for >15 h (15) and that the recombinant hybridoma cells have a doubling time of ∼18 h, the presence of two distinct peaks in the flow cytometry analysis suggested that the positive and negative transcriptional states were stable for at least several cell doublings. We tested these interpretations by examining the properties of subclones of the E−M−S− recombinants. We envisaged that any particular subclone must start from either a positive or a negative cell. If positive and negative transcriptional states persisted over several generations, independent subclones should vary greatly in the fractions of positive and negative cells, if the subclones are observed long before reaching equilibrium. As illustrated in Fig. 3A, we readily obtained subclones which differed greatly in the fraction of positive cells.
FIG. 3.
Persistent variegated μ expression in the E−M−S− recombinant. (A) Flow cytometry of subclones. The E−M−S−44 recombinant was subcloned, yielding subclones 44-2, 44-10, and 44-3, which were analyzed by flow cytometry. These subclones were in turn resubcloned to obtain the indicated resubclones, which were analyzed by flow cytometry and for μ mRNA (B). These resubclones were also used to obtain further resubclones as noted in the text and Fig. 4 and 6. (B) Analysis of μ mRNA by Northern blot. Aliquots of the same culture were used to extract cytoplasmic RNA and for analysis by flow cytometry and the plaque-forming cell assay. RNA was isolated from the indicated subclones of the E−M−S−44-2 and E−M−S−44-10 recombinants and analyzed by Northern blot with a probe specific for the Cμ3 and Cμ4 domains and actin (43). The amount of μ- and actin-specific RNA was measured by PhosphorImager analysis. The relative percentages of mRNA given below the blot were calculated by subtracting a background value from each band intensity and dividing by the intensity of the Sp6 band. Cells were analyzed by flow cytometry as described in the legend to Fig. 2 and in Materials and Methods. For the plaque-forming cell assay, cells were plated in the presence of hapten-coupled sheep erythrocytes and complement, as described in Materials and Methods. The frequency of plaque-forming cells was corrected for the efficiency of plaque-forming cells (∼40%), which was determined by plating a defined number (∼200) of wild-type Sp6 cells. FACS, fluorescence-activated cell sorter.
Among the subclones obtained were some with mostly positive (88%) or mostly negative (>99%) cells (e.g., subclones E−M−S−44-10-119 and E−M−S−44-3-28, respectively) (Fig. 3A). We have also used these highly biased subclones to test whether cells could switch between positive and negative states. First, we found that cultures of mostly positive cells and cultures of mostly negative cells grew at the same rate and that resubcloning these cultures yielded colonies at the same high rates of efficiency (data not shown). These observations indicated that growth rate and cloning efficiency were the same for positive and negative cells. We resubcloned these nearly homogeneous colonies with the view that the resubclones derived from E−M−S−44-10-119 were very likely to have arisen from a positive cell, while the resubclones derived from E−M−S−44-3-28 were very likely to have originated from a negative cell. We then examined 10 resubclones of each of these two subclones to measure the frequency of switched cells (Table 1 and Fig. 4A). In the case of resubclones arising from positive cells (Fig. 4A), negative cells were sufficiently numerous to be detected by flow cytometry. In the case of resubclones arising from negative cells, positive cells were too rare to detect in this way. In this case, we took advantage of the fact that positive cells secrete IgM, and we assayed positive cells by their capacity to produce plaques on TNP-coupled erythrocytes (Table 1). Each of the 10 resubclones derived from the highly positive subclone E−M−S−44-10-119 contained negative cells (Fig. 4A; Table 1), and likewise each of the 10 resubclones derived from the negative subclone E−M−S−44-3-28 contained positive cells (Table 1). Taken together, these results indicate that cells can switch between the positive and negative states.
TABLE 1.
Measurement of transition rates by resubclone analysis
| Subclone type and recombinant (medium) | No. of subclones analyzed | No. of days in culture after plating | Fraction of cells
|
Transition rate (per cell-day)
|
|||
|---|---|---|---|---|---|---|---|
| Highest | Lowest | Median | α | β | |||
| Mostly negativea | Positive | ||||||
| E−M−S−44 (MHX) | 10 | 21 | 3.1 × 10−3 | 3.1 × 10−5 | 2.4 × 10−4 | 1.2 × 10−5 | |
| E−M−S−6 (MHX) | 5 | 26 | 6.0 × 10−4 | <1.4 × 10−4 | 3.5 × 10−4 | 1.5 × 10−5 | |
| Mostly positiveb | Negative | ||||||
| E−M−S−44 (MHX) | 10 | 28 | 4.4 × 10−1 | 2.6 × 10−2 | 8.1 × 10−2 | 3.0 × 10−3 | |
| E−M−S−44 (NM) | 8 | 25 | 3.5 × 10−1 | <1.0 × 10−2 | 1.0 × 10−1 | 4.2 × 10−3 | |
| E−M−S−6 (MHX) | 7 | 27 | 4.6 × 10−1 | 2.8 × 10−2 | 1.9 × 10−1 | 7.8 × 10−3 | |
| E+M+S+1 (MHX) | 10 | 28 | <1.0 × 10−2 | <2.9 × 10−3 | <1.0 × 10−2 | <3.6 × 10−4 | |
| E+M+S+1 (NM) | 10 | 23 | <1.0 × 10−2 | <1.0 × 10−2 | <1.0 × 10−2 | <4.4 × 10−4 | |
Mostly negative subclones of E−M−S−44 and E−M−S−6 were resubcloned in MHX medium and allowed to grow for the indicated time in culture. The resubclones were then assayed for plaque-forming cells to determine the fraction of positive cells. The plaque-forming efficiency of positive cells (usually ∼40%) was determined by plating ∼200 Sp6 cells, which constituted a homogeneously positive population. The median values were used to calculate transition rate, α, the rate at which negative cells become positive, as described in Materials and Methods.
Mostly positive subclones of the E−M−S−44 and E−M−S−6 recombinants were resubcloned in either normal medium (NM) or MHX-containing medium and allowed to grow for the indicated number of days. The resubclones were then assayed for negative cells by flow cytometry. The median values were used to calculate the transition rate, β, the rate at which positive cells become negative, as described in Materials and Methods.
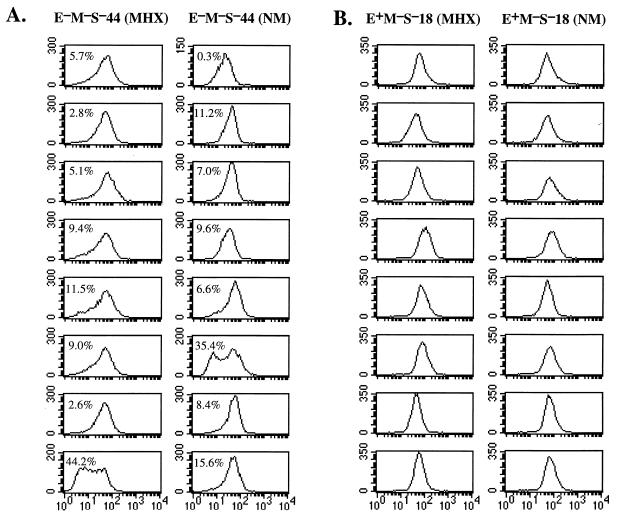
FIG. 4.
Measurement of transition rates for E−M−S− and E+M−S− recombinants by subcloning method. (A) The E−M−S−44-10-119-34 subclone of the E−M−S−44 recombinant was resubcloned and cultured for 3 to 4 weeks in either normal (NM) or MHX-containing medium. These cultures were then analyzed by flow cytometry. The fraction of negative cells is indicated in each panel. As described in the text, the asymmetry of the labeled peaks suggested that many transitions had occurred recently and should be included in the population of negative cells. In these cases, to estimate the negative population we assumed that the peak of positive cells was symmetrical and set a gate to count the right half of the peak. This fraction was multiplied by 2 to obtain the fraction of positive cells. This value was then sub-tracted from 100% to estimate the fraction of negative cells. (B) Subclones of the E+M−S−18 recombinant were analyzed by flow cytometry as described above (A).
Positive and negative staining corresponds to positive and negative transcriptional states.
The high variability among independent subclones allowed us to ascertain the level of μ mRNA in positive and negative cells. Thus, for some of the E−M−S−44 subclones illustrated in Fig. 3A, we measured the steady-state μ mRNA levels and the fraction of positive cells, as assessed either by flow cytometry or by their capacity to form plaques in an IgM-complement-dependent lysis assay (Fig. 3B). For all clones analyzed, the relative fraction of μ mRNA produced by the culture correlated closely with the fraction of positive cells (Fig. 3B). This correlation is consistent with a model in which positive cells contained the same amount of μ mRNA as wild-type recombinants, while negative cells had none (<2% of normal μ mRNA). It has previously been shown with nuclear run-on experiments that the reduced level of μ mRNA in the E−M−S− recombinants reflects reduced transcription (29). Our observation that the cells were bimodal in their μ mRNA content thus indicates that the cells were bimodal for transcription.
Rate of switching between the positive and negative states in E−M−S− recombinants.
Our finding that cells could switch between positive and negative states suggested that these transitions might occur at characteristic rates and that the relative number of positive and negative cells at equilibrium would correspond to the ratio of these rates (Fig. 5A). In analyzing these experiments, we assumed that the probability of a cell switching from one state to the other is a constant throughout the lifetime of the cultures. As noted above, positive and negative cells grew at the same rate. On this basis, we derived the formulas (see “Calculations” in Materials and Methods) relating the fraction of positive and negative cells and the transition rates. In these formulations, time is expressed in days but could as well be expressed in cell divisions (1 cell division = ∼0.7 day).
To measure α, the rate at which cells switch from the negative to the positive state, we used subclones which were composed of nearly all negative cells and resubcloned them (Fig. 5; Table 1). Under these circumstances, nearly all resubclones arose from negative cells, and the frequency of positive cells in these resubclones was then a measure of switching from the negative to the positive state. In this case, the frequency of positive cells was uniformly very low, so we measured their frequency by the number of plaque-forming cells (Table 1), using the median value as the best simple measurement to estimate the number of transitions (23). Our results for two independently generated recombinants, E−M−S−44 and E−M−S−6, indicated values for α of 1.2 × 10−5 and 1.3 × 10−5/day, respectively.
Using a similar approach to measure β, the rate of switching from the positive to the negative state, we resubcloned a mostly positive subclone and then measured the frequency of negative cells in the resubclones by flow cytometry (Fig. 4). We again measured this rate for two independently generated recombinants, E−M−S−44 and E−M−S−6. However, in the case of the E−M−S−65 recombinant, extensive subcloning did not yield a subclone with a sufficiently high fraction of positive cells for this type of analysis. As illustrated for the E−M−S−44 recombinant (Fig. 4A), we found that some resubclones were clearly divided into positive and negative populations, while others appeared to contain some cells with an intermediate level of IgM, as judged by the asymmetry of the peak. Intermediate levels of expression might correspond to cells which had only recently extinguished expression of the μ gene and therefore contained an intermediate level of μ mRNA, reflecting the effects of dilution and decay. Accordingly, we counted such cells as negative; the significance of the seemingly large fraction of recently extinguished cells is considered further in the Discussion. As summarized in Table 1, our measurements for two recombinants, E−M−S−44 and E−M−S−6, yielded estimates of β which ranged from ∼3 × 10−3 to ∼8 × 10−3/day, with an average of ∼5 × 10−3/day.
To test whether passage through the negative state detectably altered the positive state, we isolated a plaque-forming cell from a mostly negative subclone and purified it by repeated plaque picking and limiting-dilution subcloning (44-3-28-57p in Fig. 3A). The resulting colony had approximately 50% negative cells, a fraction similar to that of a subclone, such as E−M−S−44-10-119, that was grown in culture for a comparable amount of time (63 days).
The apparent rate at which cells switched from positive to negative was not affected by selection for the expression of the adjoining gpt gene, i.e., the apparent rate was the same for cells which were grown in normal medium and in MHX-containing medium (Table 1; Fig. 4). In fact, direct measurement of the frequency of thioxanthine-resistant colonies indicated that the rate at which cells extinguish the gpt gene is <10−6/cell generation. The gpt gene thus appears to be >104-fold more stable than the adjoining μ gene, although both lie within the IgH locus. These results are consistent with earlier findings that a population of cells which had only ∼4% of the normal level of μ mRNA had normal or even higher-than-normal levels of gpt mRNA (29).
We used a related method to estimate the rate of positive-to-negative switches in bulk cultures. In this case, we took subclones with mostly positive cells and measured how this fraction decreased over time (Fig. 6). As presented in Table 2, these results show that β ranged from 4 × 10−3 to 3 × 10−2/day for the E−M−S−3, E−M−S−6, and E−M−S−44 recombinants and was thus somewhat higher than the estimate of 5 × 10−3/day derived from measuring switches during the outgrowth of individual subclones.
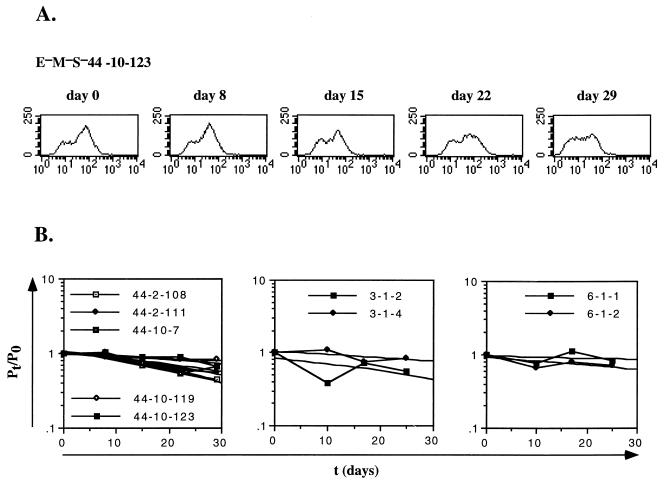
FIG. 6.
Alternative measurement of transition rates for E−M−S− recombinant. Various subclones of independent E−M−S− recombinants were analyzed by flow cytometry at successive times during a 20- to 30-day interval and the fraction of positive cells, p, was determined for each time point. (A) The results for one particular recombinant, E−M−S−44-10-123, are shown. (B) The results of measuring subclones of the indicated recombinants are shown. The values for p, divided by the value for p0 (the value for time zero) were plotted and fitted to a straight line. The calculated slopes were then used to estimate the rate of switching from the positive to negative state, as described in Table 2.
TABLE 2.
Measurement of transition rates in bulk culturesa
| Recombinant and subclone | β (per cell-day) |
|---|---|
| E−M−S−44 | |
| 2-108 | 3.1 × 10−2 |
| 2-111 | 2.3 × 10−2 |
| 10-7 | 1.9 × 10−2 |
| 10-119 | 6.7 × 10−3 |
| 10-123 | 1.2 × 10−2 |
| 3-28-57-4p | 1.1 × 10−2 |
| E−M−S−3 | |
| 1-2 | 1.6 × 10−2 |
| 1-4 | 9.6 × 10−3 |
| E−M−S−6 | |
| 1-1 | 4.1 × 10−3 |
| 1-2 | 1.1 × 10−2 |
The data shown in Fig. 6B, which plots p/p0 as a function of time in culture for the subclones of the E−M−S−44, E−M−S−3, and E−M−S−6 recombinants, were used to calculate β, the rate at which positive cells switched to the negative state. For each of the indicated subclones, the best-fitting straight line was approximated to the data. β was then calculated according to the formula βt = ln(p/p0).
μ expression is stable in recombinants which bear the core enhancer.
Several results indicate that the Eμ core enhancer alone is sufficient to render μ expression stably positive in the recombinant IgH locus. First, as shown for the two E+M+S+ recombinants in Fig. 2, the three E+M+S− and three E+M−S− recombinants which we examined were each homogeneous by flow cytometry (data not shown). To further assess the homogeneity of Eμ-containing recombinants, we examined 20 subclones of both the E+M−S−18 and the E−M−S−44 recombinants; in each case, 10 were grown in normal medium and 10 were grown in MHX medium. Whereas substantial populations of negative cells were evident in nearly all subclones of the E−M−S−44 recombinant after ∼30 days in culture (Fig. 4A), all subclones of the E+M−S−18 recombinant were homogeneously positive (Fig. 4B). Reconstruction experiments indicated that we would have detected negative cells, if >1% of the population had corresponded to fully negative cells. These results thus indicate that for the E+M−S− recombinant, β was <4 × 10−4/day.
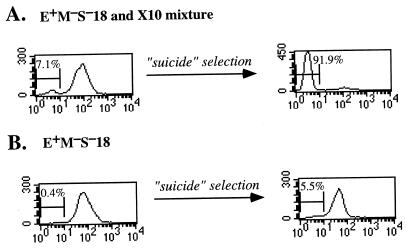
In order to increase the sensitivity of detecting negative cells, we used the suicide selection procedure to selectively kill positive cells (9). In this procedure, cells are coupled with the hapten TNP and then allowed to secrete IgM in the presence of complement. Under conditions in which cells are incubated at low density, IgM binds preferentially to the same cell from which it was secreted and not to other cells, thus rendering that cell sensitive to complement-mediated lysis. We used reconstruction experiments to assess the power of this procedure to enrich for negative cells, i.e., we prepared mixtures of the E+M−S− recombinant with different amounts of the μ-deleted mutant, X10, and compared the fraction of negative cells before and after the suicide selection. As shown in Fig. 7, this procedure enriched the mixture ∼100 fold for μ-negative cells. After application of this enrichment protocol to a pure culture of subclones of the E+M−S−18 and E+M−S−66 recombinants, the frequency of negative cells was 5 and 4%, respectively. Assuming that this procedure enriched the mixture ∼100 fold as in the reconstruction experiment, these values imply that negative cells comprised ∼0.05 and ∼0.04% of the cells in the E+M−S−18 and E+M−S−66 recombinants, respectively. Because the population of μ-negative cells is expected to include some cells with a mutation in the μ gene (9), this fraction defines an upper limit to the frequency of epigenetically μ-negative cells. Considering that these cultures had grown in normal medium for ∼40 days prior to enrichment, these results imply that β was <10−5/day. Thus, the inclusion of Eμ increased the stability of the positive state by >500-fold. A similar analysis applied to a E+M+S+ recombinant yielded a comparable estimate of stability, i.e., β was <4 × 10−6/day.
FIG. 7.
Use of the suicide selection to estimate numbers of negative cells in the E+M−S− recombinant. (A) To estimate how much the suicide selection enriches for negative cells, a culture was prepared by mixing a small amount of the μ-deleted cell line (X10) with a subclone of the E+M−S−18 recombinant. This culture was divided in two parts. One part was untreated (left) while the other (right) was subjected to the suicide selection. The cells were then cultured for several days and analyzed by flow cytometry. The fraction of negative cells is indicated. Enrichment was calculated as the ratio (1/nb) × (1/pa), where nb is the fraction of negative cells before enrichment and pa is the fraction of positive cells after enrichment. (B) A subclone of the E+M−S−18 recombinant was subjected to the suicide selection. The fraction of negative cells is indicated.
DISCUSSION
Switching between states involves an epigenetic change.
The MAR-Eμ-MAR-Sμ segment of the mouse IgH locus is part of an LCR, in that inclusion of this segment is required for the uniform high-level expression of IgH-derived transgenes (13, 16, 37). As shown here, the effect of deleting these elements from the endogenous IgH locus of a hybridoma cell line is to render μ expression metastable. That is, E+M+S+ recombinants expressed the μ gene uniformly and stably, while E−M−S− recombinants which lacked this segment switched between states in which μ expression was fully on (positive) and fully off (negative). This dynamic state implies that expression cannot be characterized simply by measuring the rate of transcription. For this reason, we sought to describe expression by measuring the rates at which cells switched between the positive and negative states. In the simplest case of this type, cells would be of only two types, positive and negative, with a characteristic and unvarying rate of switching. Our analysis indicated that the E−M−S− recombinants switch from the positive to the negative state at a rate of ∼5 × 10−3/day, while the reverse switch, from negative to positive, occurred at a rate of ∼1.2 × 10−5/day. Although we do not know whether the switches occur as a function of time or cell division, it is interesting that the value of β, 5 × 10−3/day, corresponds to a half-life for the positive state of ∼100 cell divisions and that the value of α, 1.2 × 10−5/day, corresponds to a half-life for the negative state of 40,000 cell divisions.
In this metastable system, the fraction of positive cells at equilibrium should correspond to the ratio α/β, or 0.2%. This fraction is consistent with previous reports that E−M−S− recombinants generally have very little μ mRNA (29, 43). Nevertheless, these rates might not be a sufficient descriptor, i.e., states with higher transition rates might also occur. As noted in Results, the flow cytometry profiles suggest that, for some subclones, many cells have an intermediate level of μ mRNA, as if they had recently extinguished μ expression. This observation suggests that there might be two types of positive cells, a relatively unstable type for which the half-life is ∼1 month and another more stable type which yields negative cells only after first converting to the former, unstable type of positive cell. It is possible that the repeated subcloning, which we used to derive the predominantly positive cultures for measuring transition rates, selected relatively stable variants. Consequently, our analysis might have underestimated the transition rates of the unstable positive cells.
Even though this repetitive subcloning method tends to underestimate transition rates, the rates of switching for the E−M−S− recombinants from positive to negative and from negative to positive were much higher than normal mutation rates. In particular, the corresponding mutation rates for the μ gene in this cell line were 10−6 per cell generation from μ+ to μ− and <10−9 per cell generation for reversion from μ− to μ+ (9). The high rates argue that the switch between positive and negative states in the E−M−S− recombinants has an epigenetic cause. The interconverting positive and negative states are thus similar to epiphenotypes of Schizosaccharomyces pombe (17) and reminiscent of the phenomenon of phenotypic switching in another yeast, Candida albicans (38).
Previous analyses have indicated that the MAR-Eμ-MAR segment is required to maintain the expression of IgH-derived transgenes in pre-B-cell lines (19, 34). In one case, this segment was excised only a few weeks before expression was measured, but nevertheless excision resulted in <1% residual expression (34). By comparison, >50% of the E−M−S− recombinant hybridoma cells were usually still positive after the same interval. Several explanations might account for this difference. (i) The pre-B and hybridoma cells might differ in crucial trans-acting factors. (ii) The endogenous IgH locus of our recombinant hybridoma cells might contain an element which helps maintain IgH expression but was missing from the transgenes tested in the pre-B-cell lines. (iii) Transgene expression might have been extinguished by an extraneous component of the transgene, e.g., the neo cassette, which has been implicated in other cases of gene silencing (1, 8, 12, 21, 31, 33).
Role of the LCR in transcription of the μ gene.
The rate at which the E−M−S− recombinant switched from the positive to the negative state was at least 500-fold higher than in the case of the E+M+S+ and E+M−S− recombinants, indicating that at least one role of the core enhancer in this LCR is to stabilize μ expression in the on state. The presence of Eμ might impede transitions from the positive to negative state, or it might be that transitions occur but the presence of Eμ allows transcription to reinitiate efficiently. Although the MARs and Sμ are also components of the LCR, these elements were not needed to maintain μ expression in the recombinant hybridoma cells. This discrepancy also invites two explanations. One explanation postulates that the requirements for initiating transcription are different from the requirements for maintaining transcription. Thus, Eμ, the MARs, and Sμ might all be needed to initiate expression, but only Eμ is needed to maintain it. Alternatively, the IgH locus might include other functionally equivalent elements which collaborate with Eμ to generate a functional LCR in the absence of the MARs and Sμ (29, 30). Analyses of knockout mice lacking components of this LCR provide independent evidence that the MARs are redundant in the natural locus (8, 36).
Two hypotheses suggesting how LCRs and other distal elements might activate expression have been set forth. One hypothesis, which was proposed to explain how the enhancer of the SV40 virus (simian vaculating virus) increased the frequency of expressing cells, is that gene expression occurs when chromatin is in an active, accessible state and that transcriptional activators increase the probability of forming and/or maintaining this state (42). Enhancers derived from the β-globin and metallothionine loci and MARs from the human interferon γ locus also appear to act in this fashion (5, 6, 24, 39). As well, this mode of action is supported by in vitro studies of enhancers from the T-cell receptor α- and β-globin genes (3, 4). Transcriptional activators of various types, operating in conjunction with RNA polymerases I and III as well as RNA polymerase II, have been found to increase the probability that a gene is active without greatly altering the rate of transcription of those active genes (for a review, see references 32 and 40). Our finding, that the active and inactive states of μ expression are metastable in the absence of the LCR, correlates well with this model. The stability of the positive state which we have found for the E−M−S− recombinants is generally similar to that observed for enhancer-deficient transgenes, i.e., the median rate for the positive to negative switch in transgene expression ranged from 0.1 to 0.01/day, although much higher and lower rates were also observed for these transgenes (24, 40). In contrast to our findings for the μ gene in the recombinant IgH locus, the silenced transgenes were not generally capable of re-expression.
Another view of LCR function, developed in part to account for competition between genes of the β-globin locus, is that the LCR interacts intimately with the transcription unit and that the level of expression reflects the frequency of this interaction (for a review, see reference 18). Because expression of the individual genes in the β-globin locus persists for only ∼8 min, this model implies that the effects of the LCR are short lived. In the simplest case, our finding that μ expression in the IgH locus continues for many weeks in the absence of the LCR is incompatible with this model. However, as noted above, one possibility is that the IgH locus contains another element which is independent of the intronic LCR and can maintain and occasionally reinitiate transcription of the μ gene.
Regulation of transcription in the absence of the enhancer.
The intronic IgH enhancer was originally detected because it activates the expression of transgenes in B cells. However, this enhancer can also extinguish expression in non-B cells (41, 45). The function of the enhancer is therefore determined by the available trans-acting factors and by the epigenetic state of the genes for these factors. In the case of the enhancer-deficient (E−M−S−) recombinants, we do not know whether the primary epigenetic determinant of the positive and negative states of μ expression lies in the IgH locus itself or in some other locus encoding a factor which acts on μ in trans. To give an example of the latter possibility, transcription of the E−M−S− μ gene might require an additional factor which is not needed to transcribe the intact μ gene, and transitions between the positive and negative states of μ transcription might then reflect changes in the expression of this factor. In the case of homozygous mice bearing diploid transgenes, each transgene was expressed independently of the other, thus showing that variegation was not due to changes in the expression of a trans-acting factor (11, 26). We have undertaken related experiments to generate hybridoma cells with two copies of the E−M−S− recombinant IgH loci to test whether the two μ genes are expressed independently or coordinately and so indicate whether the primary epigenetic determinant lies in the IgH or another locus.
Several mechanisms have been proposed to explain how epigenetic states might be propagated. One class of mechanisms is based on the possibility that the methylation of cytosine (in CpG) can extinguish expression. In this case, the epigenetic determinant corresponds to methylation or demethylation which is introduced de novo, either spontaneously or following externally derived signals. Inheritance is then achieved by the strong preference of maintenance methylases for hemimethylated sites (35). Other models propose that the epigenetic determinant corresponds to deacetylation or acetylation of histones, which decreases or increases the accessibility of genes in chromatin, thus regulating transcription. In this case, epigenetic inheritance might be achieved by a mechanism which restricts the availability of newly replicated DNA and deacetylated histones to the same time or place (7, 44). The recent finding that transcriptional repression by a methyl-CpG binding protein involves a histone deacetylase offers a specific vision of how control by cytosine methylation and control by nucleosome structure might be related (20, 28). Any of these models could potentially account for the metastable states of μ expression. As noted above, the epigenetic determinant which distinguishes the positive and negative states of expression might be in the IgH locus itself, and if so, biochemical comparisons of the IgH locus in positive and negative cells might directly reveal the primary basis of this form of epigenetic inheritance.
ACKNOWLEDGMENTS
We thank C. Collins for excellent technical assistance; J. Ellis, P. Sadowski, and F. Tsui for their critical reading of the manuscript; and A. Igelfeld for help in the mathematical formulations.
This work was supported by grants from Ciba-Geigy/Novartis and from the Medical Research Council of Canada.
REFERENCES
- 1.Artelt P, Grannemann R, Stocking C, Friel J, Bartsch J, Hauser H. The prokaryotic neomycin-resistance-encoding gene acts as a transcriptional silencer in eukaryotic cells. Gene. 1991;99:249–254. doi: 10.1016/0378-1119(91)90134-w. [DOI] [PubMed] [Google Scholar]
- 2.Baar J, Shulman M J. The Ig heavy chain switch region is a hotspot for insertion of transfected DNA. J Immunol. 1995;155:1911–1920. [PubMed] [Google Scholar]
- 3.Bagga R, Emerson B E. An HMG I/Y-containing repressor complex and supercoiled DNA topology are critical for long-range enhancer-dependent transcription in vitro. Genes Dev. 1997;11:629–639. doi: 10.1101/gad.11.5.629. [DOI] [PubMed] [Google Scholar]
- 4.Barton M C, Madani N, Emerson B M. Distal enhancer regulation by promoter derepression in topologically constrained DNA in vitro. Proc Natl Acad Sci USA. 1997;94:7257–7262. doi: 10.1073/pnas.94.14.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bode J, Schlake T, Rios-Ramirez M, Mielke C, Stengert M, Kay V, Klehr-Wirth D. Scaffold/matrix-attached regions: structural properties creating transcriptionally active loci. Int Rev Cytol. 1995;162A:389–454. doi: 10.1016/s0074-7696(08)61235-8. [DOI] [PubMed] [Google Scholar]
- 6.Boyes J, Felsenfeld G. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J. 1996;15:2496–2507. [PMC free article] [PubMed] [Google Scholar]
- 7.Brown K E, Baxter J, Graf D, Merkenschlager M, Fisher A G. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Young F, Bottaro A, Stewart V, Smith R K, Alt F W. Mutations of the intronic IgH enhancer and its flanking sequences differentially affect accessibility of the JH locus. EMBO J. 1993;12:4635–4645. doi: 10.1002/j.1460-2075.1993.tb06152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connor A, Collins C, Jiang L, McMaster M, Shulman M J. Isolation of new nonsense and frameshift mutations in the immunoglobulin mu heavy chain gene of hybridoma cells. Somatic Cell Mol Genet. 1993;19:313–320. doi: 10.1007/BF01232744. [DOI] [PubMed] [Google Scholar]
- 10.Dorer D R, Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77:993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- 11.Festenstein R, Tolaini M, Corbella P, Mamalaki C, Parrington J, Fox M, Miliou A, Jones M, Kioussis D. Locus control region function and heterochromatin-induced position effect variegation. Science. 1996;271:1123–1125. doi: 10.1126/science.271.5252.1123. [DOI] [PubMed] [Google Scholar]
- 12.Fiering S, Kim C G, Epner E E, Groudine M G. An “in-out” strategy using gene targeting and FLP recombinase for the functional dissection of complex DNA regulatory elements: analysis of the β-globin locus control region. Proc Natl Acad Sci USA. 1993;90:8469–8473. doi: 10.1073/pnas.90.18.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forrester W C, van-Genderen C, Jenuwein T, Grosschedl R. Dependence of enhancer-mediated transcription of the immunoglobulin mu gene on nuclear matrix attachment regions. Science. 1994;265:1221–1225. doi: 10.1126/science.8066460. [DOI] [PubMed] [Google Scholar]
- 14.Garrick D, Fiering S, Martin D I K, Whitelaw E. Repeat-induced gene silencing in mammals. Nat Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 15.Genovese C, Milcarek C. Increased half-life of μ immunoglobulin mRNA during mouse B-cell development increases its abundancy. Mol Immunol. 1990;27:733–743. doi: 10.1016/0161-5890(90)90082-b. [DOI] [PubMed] [Google Scholar]
- 16.Gram H, Zenke G, Geisse S, Kleuser B, Burki K. High level expression of a human immunoglobulin transgene depends on switch region sequences. Eur J Immunol. 1992;22:1185–1191. doi: 10.1002/eji.1830220512. [DOI] [PubMed] [Google Scholar]
- 17.Grewal S I V, Klar A J S. Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell. 1996;86:95–101. doi: 10.1016/s0092-8674(00)80080-x. [DOI] [PubMed] [Google Scholar]
- 18.Gribnau J, deBoer E, Trimborn T, Wijgerde M, Milot E, Grosveld F, Fraser P. Chromatin interaction mechanism of transcriptional control in vivo. EMBO J. 1998;17:6020–6027. doi: 10.1093/emboj/17.20.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grosschedl R, Marx M. Stable propagation of the active transcriptional state of an immunoglobulin mu gene requires continuous enhancer function. Cell. 1988;55:645–654. doi: 10.1016/0092-8674(88)90223-1. [DOI] [PubMed] [Google Scholar]
- 20.Jones P L, Veenstra G J C, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 21.Kim C G, Epner E M, Forrester W C, Groudine M. Inactivation of the human β-globin gene by targeted insertion into the β-globin locus control region. Genes Dev. 1992;6:928–938. doi: 10.1101/gad.6.6.928. [DOI] [PubMed] [Google Scholar]
- 22.Kowalski J, Denhardt D T. Regulation of the mRNA for monocyte-derived neutrophil-activating peptide in differentiating HL60 promyelocytes. Mol Cell Biol. 1989;9:1946–1957. doi: 10.1128/mcb.9.5.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lea D E, Coulson C A. The distribution of the numbers of mutants in bacterial populations. J Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 24.Magis W, Fiering S, Groudine M, Martin D I K. An upstream activator of transcription coordinately increases the level and epigenetic stability of gene expression. Proc Natl Acad Sci USA. 1996;93:13914–13918. doi: 10.1073/pnas.93.24.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin D I K, Whitelaw E. The vagaries of variegating transgenes. Bioessays. 1996;18:919–923. doi: 10.1002/bies.950181111. [DOI] [PubMed] [Google Scholar]
- 26.Milot E, Strouboulis J, Trimborn T, Wijgerde M, deBoer E, Langeveld A, Grosveld F, Fraser P. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell. 1996;87:105–114. doi: 10.1016/s0092-8674(00)81327-6. [DOI] [PubMed] [Google Scholar]
- 27.Mulligen R C, Berg P. Expression of a bacterial gene in mammalian cells. Science. 1980;209:1422–1427. doi: 10.1126/science.6251549. [DOI] [PubMed] [Google Scholar]
- 28.Nan X, Ng H-H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the lethyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 29.Oancea A E, Berru M, Shulman M J. Expression of the (recombinant) endogenous immunoglobulin heavy-chain locus requires the intronic matrix attachment regions. Mol Cell Biol. 1997;17:2658–2668. doi: 10.1128/mcb.17.5.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oancea A E, Tsui F W L, Shulman M J. Targeted removal of the mu switch region from mouse hybridoma cells: a test of its role in gene expression in the endogenous IgH locus. J Immunol. 1995;155:5678–5683. [PubMed] [Google Scholar]
- 31.Olson E N, Arnold H H, Rigby P W J, Wold B J. Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell. 1996;85:1–4. doi: 10.1016/s0092-8674(00)81073-9. [DOI] [PubMed] [Google Scholar]
- 32.Osheim Y N, Mougey E B, Windle J, Anderson M, O’Reilly M, Miller O L, Jr, Beyer A, Sollner-Webb B. Metazoan rDNA enhancer acts by making more genes transcriptionally active. J Cell Biol. 1996;133:943–954. doi: 10.1083/jcb.133.5.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pham C T N, MacIvor D M, Hug B A, Heusel J W, Ley T J. Long-range disruption of gene expression by a selectable marker cassette. Proc Natl Acad Sci USA. 1996;93:13090–13095. doi: 10.1073/pnas.93.23.13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porton B, Zaller D M, Lieberson R, Eckhardt L A. Immunoglobulin heavy-chain enhancer is required to maintain transfected γ-2A gene expression in a pre-B-cell line. Mol Cell Biol. 1990;10:1076–1083. doi: 10.1128/mcb.10.3.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Razin A. CpG methylation, chromatin structure and gene silencing—a three-way connection. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakai E, Bottaro A, Davidson L, Sleckman B P, Alt F W. Recombination and transcription of the endogenous Ig heavy chain locus is effected by the Ig heavy chain intronic enhancer core region in the absence of the matrix attachment regions. Proc Natl Acad Sci USA. 1999;96:1526–1531. doi: 10.1073/pnas.96.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sigurdardottir D, Sohn J, Kass J, Selsing E. Regulatory regions 3′ of the immunoglobulin heavy chain intronic enhancer differentially affect expression of a heavy chain transgene in resting and activated B cells. J Immunol. 1995;154:2217–2225. [PubMed] [Google Scholar]
- 38.Slutsky B, Buffo J, Soll D R. High-frequency switching of colony morphology in Candida albicans. Science. 1985;230:666–669. doi: 10.1126/science.3901258. [DOI] [PubMed] [Google Scholar]
- 39.Walters M C, Fiering S, Eidemiller J, Magis W, Groudine M, Martin D I K. Enhancers increase the probability but not the level of gene expression. Proc Natl Acad Sci USA. 1995;92:7125–7129. doi: 10.1073/pnas.92.15.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walters M C, Magis W, Fiering S, Eidemiller J, Scalzo D, Groudine M, Martin D I K. Transcriptional enhancers act in cis to suppress position-effect variegation. Genes Dev. 1996;10:185–195. doi: 10.1101/gad.10.2.185. [DOI] [PubMed] [Google Scholar]
- 41.Wasylyk C, Wasylyk B. The immunoglobulin heavy-chain B-lymphocyte enhancer efficiently stimulates transcription in non-lymphoid cells. EMBO J. 1986;5:553–560. doi: 10.1002/j.1460-2075.1986.tb04246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weintraub H. Formation of stable transcription complexes as assayed by analysis of individual templates. Proc Natl Acad Sci USA. 1988;85:5819–5823. doi: 10.1073/pnas.85.16.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiersma E J, Ronai D, Berru M, Tsui F W L, Shulman M J. Role of the intronic elements in the endogenous immunoglobulin heavy chain locus. J Biol Chem. 1999;274:4858–4862. doi: 10.1074/jbc.274.8.4858. [DOI] [PubMed] [Google Scholar]
- 44.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 45.Yu H, Porton B, Shen L Y, Eckhardt L A. Role of the octamer motif in hybrid cell extinction of immunoglobulin gene expression: extinction is dominant in a two enhancer system. Cell. 1989;58:441–448. doi: 10.1016/0092-8674(89)90425-x. [DOI] [PubMed] [Google Scholar]