Abstract
Analysis of global gene expression in Saccharomyces cerevisiae by the serial analysis of gene expression technique has permitted the identification of at least 302 previously unidentified transcripts from nonannotated open reading frames (NORFs). Transcription of one of these, NORF5/HUG1 (hydroxyurea and UV and gamma radiation induced), is induced by DNA damage, and this induction requires MEC1, a homolog of the ataxia telangiectasia mutated (ATM) gene. DNA damage-specific induction of HUG1, which is independent of the cell cycle stage, is due to the alleviation of repression by the Crt1p-Ssn6p-Tup1p complex. Overexpression of HUG1 is lethal in combination with a mec1 mutation in the presence of DNA damage or replication arrest, whereas a deletion of HUG1 rescues the lethality due to a mec1 null allele. HUG1 is the first example of a NORF with important biological functional properties and defines a novel component of the MEC1 checkpoint pathway.
A major accomplishment of genome-era research was the complete elucidation of the genomic sequence of the eukaryote Saccharomyces cerevisiae. As a direct result of this effort, 6,275 open reading frames (ORFs) representing all ORFs larger than 100 contiguous amino acids were identified (10, 14). However, identification of genes encoded by small ORFs (<100 amino acids) based on sequence analysis alone has been severely limited by high false-positive rates, and traditional functional screens have been similarly hampered by the small target size for mutagenesis (4). Evidence from several microorganisms suggests that a significant fraction of genomes are encoded by small genes. For example, the Escherichia coli genome encodes 381 proteins of less than 100 amino acids in length from a total of 4,288 annotated ORFs (8.9% [37a]), and random protein sequencing in the fully sequenced cyanobacterium Synechocystis revealed that 11.8% of the total proteins were encoded by ORFs of <100 codons (8a). Extrapolation of such studies to yeast would suggest that there may be as many as 800 small ORFs in the entire yeast genome, of which only 177 have been identified (20a). The subset of small ORFs will likely encode important proteins in all organisms, including humans. In S. cerevisiae, these small proteins include mating pheromones, proteins involved in energy metabolism, proteolipids, chaperonins, stress proteins, transporters, transcriptional regulators, nucleases, ribosomal proteins, thioredoxins, and metal ion chelators. In multicellular organisms, there is a rich diversity of short peptides, including many hormones, antibacterial defensins, cecroporins, and magainins (3). There are also small ORFs encoding transporter proteins, homeobox proteins, transcription factors, and kinase regulatory subunits reported in the nematode Caenorhabditis elegans (29a).
Analysis of global gene expression in S. cerevisiae by the serial analysis of gene expression (SAGE) technique (39, 40) has permitted the identification of at least 302 previously unidentified transcripts from nonannotated ORFs (NORFs). Whether any of these NORFs are important for the growth and biology of yeast is unclear. We report herein the first systematic analysis of NORFs in the yeast genome and the characterization of NORF5/HUG1. Our analysis of the 30 most highly transcribed NORFs has shown that 12 of the 30 NORF genes are evolutionarily conserved with mammalian homologs (28a). NORF5/HUG1 was chosen for further analysis because its dramatic expression in hydroxyurea (HU)-treated cells suggested a potential role in transcriptional response after replication arrest and DNA damage.
Several checkpoint genes in S. cerevisiae are required for transcriptional induction of a large regulon of genes that facilitate DNA repair, cause cell cycle arrest, and mediate recovery from DNA damage (12, 41). A central component of these checkpoints is MEC1, the budding yeast homolog of the hereditary ataxia telangiectasia ATM gene and a member of the phoshatidylinositol-3-kinase family (32, 45). Signals of DNA damage normally pass from sensor genes such as RAD9, RAD17, RAD24, MEC3, and DDC1 to MEC1, leading to phosphorylation of Rad53p, replication protein A, and potentially other targets, causing cell cycle arrest and transcriptional response (2, 8, 12, 19, 29, 36). We found that genes in the MEC1 checkpoint pathway are required for the transcriptional induction of NORF5/HUG1 in response to replication arrest and DNA damage. Additional experiments have shown that NORF5/HUG1 has distinct genetic interactions with MEC1. These findings highlight the importance of the development and application of new technologies in the total-genome sequence era to fully understand the genetic complexity of an organism.
MATERIALS AND METHODS
Analysis of NORF data.
Yeast genome intergenic regions were defined as regions outside annotated ORFs or the 500-bp region downstream of annotated ORFs (yeast genome sequence and tables of annotated ORFs were obtained from the Stanford Genome Database (35a). Based on sequence analysis, a total of 9,524 putative ORFs of 25 to 99 amino acids were present in the intergenic regions. Of the 60,633 SAGE tags analyzed, there were 302 unique SAGE tags that matched the genome uniquely, were in the correct orientation, and were expressed at levels greater than 0.3 transcript copies per cell. The 302 unique SAGE tags were either within or adjacent to intergenic ORFs (100 bp upstream or 500 bp downstream of the ORF). Homology searches for 30 highly transcribed NORFs can be obtained from reference 28a.
Strains and plasmids used.
The strains used included YPH499 (MATa ura3-52 lys2-801 ade2-101 his3-Δ200 trp1-Δ63 leu2-Δ1), YPH987 (MATa/α ura3-52/ura3-52 lys2-801/lys2-801 ade2-101/ade2-101 trp1-Δ63/trp1-Δ63 leu2-Δ1/leu2-Δ1 his3-Δ200/his3-Δ200 CFIII CEN3L. YPH983TRP1SUP11), YMB711 (MATα ura3-52 lys2-801 ade2-101 his3-Δ200 trp1Δ63 leu2-Δ1 hug1Δ1::HIS3), and YMB847 (MATa ura3-52 lys2-801 ade2-101 his3-Δ200 trp1-Δ63 leu2-Δ1 hug1Δ2::HIS3) (our collection); Y203 (MATa ade2-1 his3 leu2-3,112 lys2 trp1 ura3-Δ100 rnr3::RNR3-URA3-TRP1), Y203-dun1 (dun1 in Y203), Y217(MATa ade2-1 his3 leu2-3, 112 lys2 trp1 ura3-Δ100 rnr3::RNR3-URA3-TRP1 crt4-2/tup1), Y231 (same as Y217, except with crt8-91/ssn6 instead of crt4-2/tup1), Y300 (MATa can1-100 ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1), and Y577 (crt1-Δ1::LEU2 in Y300) from S. Elledge (16); W1588-4A (MATa leu2,3-112 ade2-1 can1-100 his3-11, 15 ura3-1 trp1 RAD5), U952-3C (sml1Δ::HIS3 in W1588-4A), U953-61D (mec1Δ::TRP1 sml1Δ::HIS3 in W1688-4A), and U971 (MATα leu2,3-112 ade2-1 can1-100 his3-11, 15 ura3-1 trp1 RAD5 dun1Δ::URA3) from R. Rothstein (46); TWY312 (MATa ura3 trp1 his7 rad53/mec2-1), TWY316 (MATa ura3 trp1 his3 mec3-1), TWY397 (MATa ura3 his7 trp1 leu2), DLY62 (MATa ura3 leu2 his3 trp1 ade2), and DLY258 (MATa ura3 leu2 his3 trp1 ade2 mec1-1 sml1) from T. Weinert (44); and YMP10381 (MATa ade2 ade3-130 ura3 leu2 trp1 cyh2 SCR::URA3), YMP10535 (rad9Δ::LEU2 in YMP10381), YMP11108 (rad17Δ::LEU2 in YMP10381), YMP10533 (rad24Δ::TRP1 in YMP10381), YEF610 (MATa ade2 ade3 leu2 ura3 trp1 mec1Δ::TRP1 sml1-1 [pEF208=URA3 ADE3 MEC1 CEN] lacking pEF208 by loss on 5-fluoroorotic acid (5-FOA) medium, YEF630 (MATa leu2 ura3 his3 sml1-1), and yPP8 (MATα ade2 ade3 leu2 ura3 trp1 mec1Δ::TRP1 his3 [pEF208=URA3 ADE3 MEC1 CEN] from the L. Hartwell laboratory. Plasmid pMB363 (HUG1 LEU2 CEN) contains the HUG1 ORF and sequences 272 bp upstream of the start codon and 191 bp downstream of the stop codon of HUG1 in pRS315 (35). Plasmid pMB366 (3HA-HUG1 LEU2 CEN) contains three copies of the hemagglutinin (HA) epitope after the second amino acid in Hug1p and was derived by ligation of 3HA from plasmid pSM937, a gift from S. Michaelis. Plasmid pMB379 (GAL1-HUG1 URA3-2μ) contains the HUG1 ORF and sequences 36 bp upstream of the start codon and 66 bp downstream of the stop codon of HUG1 in pRS426GAL1 (GAL1-URA3-2μ) (22). Plasmid pMB386 (HUG1∫sLEU2CEN) contains a frameshift in the HUG1 ORF at codon 14 of HUG1.
Cell cycle arrest and Northern and Western blot analyses.
For cell cycle arrest and exposure to DNA damage, we used early-logarithmic-phase cultures. For replication arrest, cells were incubated in the presence of HU (0.1 M) for 3.5 h; for G1 arrest, cells were incubated with alpha factor (Sigma; T-6901) (3 × 10−2 M) for 2 h; for G2/M arrest, cells were incubated with nocodazole (Sigma; M-1404) (15 μg/ml) for 90 min at 30°C. For each arrest (>90%), we examined cell morphology and determined DNA content by flow cytometry (5). For exposure to UV radiation, cells were spread on the surface of yeast extract-peptone-dextrose (YPD) plates and irradiated (Stratagene; UV Stratalinker 2400) at 60 J/m2. For exposure to gamma radiation, liquid cultures were irradiated with a dose of 2 Gy with a Shepherd Mark 137I-Cs irradiator. After irradiation with UV and gamma radiation, cells were incubated at 30°C for 1 h. For thermal stress, cells were shifted to 37°C for 2 h. Cells from each treatment were washed, and cell pellets were frozen at −70°C for RNA preparation. Total RNA was made by the hot phenol method as described previously (3) from cell pellets (−70°C) of treated or untreated cultures, and Northern blot analysis was performed as described previously (11). Quantitation was done with a Fuji Phosphoimager, model BAS1500. We have previously determined that the SAGE tag abundance for TUB2 is 10:7:8 and that of ACT1 is 81:38:84 in log-phase–S phase–G2/M-phase cells (40). Hence, we used TUB2 as the loading control for RNA. For most of the blots, we detected very low levels of HUG1 transcript in the no treatment (control) lane. For example, we determined that the ratio of the intensity of the HUG1 signal to the TUB2 signal (HUG1/TUB2) in the control lane ranges from 0.04 to a maximum of 1.2 in one case. The ratio of HUG1/TUB2 was set to 1.0 for the control lane, and the value of the ratio of HUG1/TUB2 in the treated lanes was divided by the value of the ratio in the control lane. The result of this ratio is presented at the bottom of each panel as HUG1/TUB2. Background values were subtracted from the values obtained for each observation. Exceptions are in Fig. 1D and 2D, lane 5 (see figure legends).
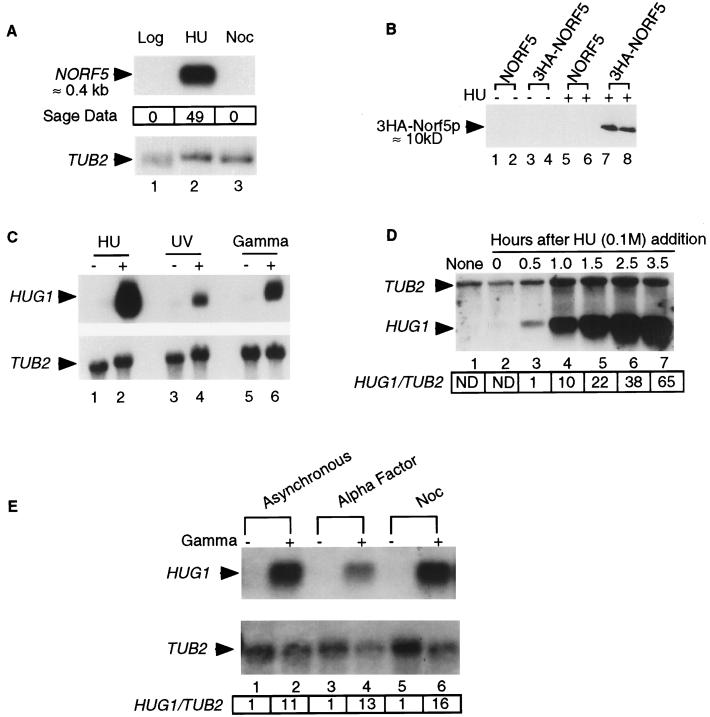
FIG. 1.
Transcription of NORF5/HUG1 is induced by replication arrest and DNA damage. (A) NORF5 transcription is upregulated in cells arrested with HU. Results are from Northern blot analysis with wild-type cells (YPH499) grown logarithmically (lane 1) and arrested with HU (lane 2) or nocodazole (Noc) (lane 3). The expression pattern observed by SAGE is indicated at the bottom (0:49:0) (40). (B) NORF5 is translated in cells arrested with HU. Western blot analysis was done with protein extracts from transformants (YMB711) containing pMB366 (3HA-NORF5 LEU2 CEN) or plasmid pMB363 (NORF5 LEU2 CEN) grown logarithmically (lanes 1 to 4) or arrested with HU (lanes 5 to 8) and probed with HA antibody as described previously. (C) Transcription of NORF5/HUG1 is HU and UV and gamma radiation induced. Results are from Northern blot analysis with wild-type cells (YPH499) grown logarithmically (lanes 1, 3, and 5), arrested with HU (lane 2), exposed to UV radiation (lane 4), or exposed to gamma radiation (lane 6). (D) HUG1 transcription is delayed upon replication arrest with HU. Results are from Northern blot analysis using logarithmically grown wild-type cells (YPH499) (lane 1) or after addition of HU (0.1 M) and incubation for 0 h (lane 2), 0.5 h (lane 3), 1.0 h (lane 4), 1.5 h (lane 5), 2.5 h (lane 6), or 3.5 h (lane 7) at 30°C. The levels of HUG1 in lanes 1 and 2 were below the background level and hence are denoted as ND (not detected). HUG1/TUB2 indicates the ratio of the intensity of the HUG1 signal to the TUB2 signal normalized to the HUG1/TUB2 ratio in lane 3 (0.5 h) set to 1.0 as described in Materials and Methods. (E) HUG1 transcription is independent of the cell cycle stage. Northern blot analysis was done with wild-type cells (YPH499) grown logarithmically (lanes 1 and 2), arrested in G1 phase by treatment with alpha factor (lanes 3 and 4), and arrested in G2/M with nocodazole (lanes 5 and 6), either before (lanes 1, 3, and 5) or after exposure to gamma radiation (lanes 2, 4, and 6). The arresting agents were present throughout the incubations. HUG1/TUB2 for lanes 2, 4, and 6 indicates the ratio of the intensity of the HUG1 signal to the TUB2 signal normalized to the HUG1/TUB2 ratio in control lanes 1, 3, and 5, respectively, as described in Materials and Methods.
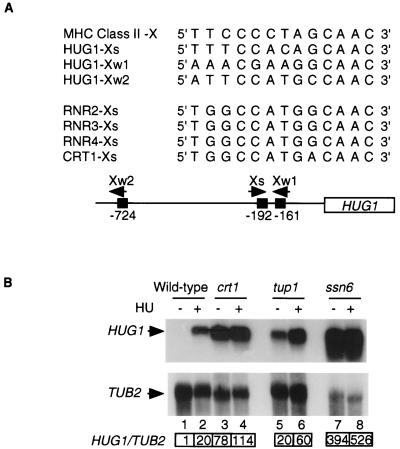
FIG. 2.
Crt1p, Ssn6p, and Tup1p are negative regulators of HUG1 transcription in the absence of DNA damage or replication arrest. (A) The promoter of HUG1 contains X-box-related sequences Xs and Xw, with strong and weak homology, respectively, to the consensus sequence in mammalian MHC class II and S. cerevisiae RNR and CRT1 genes (16, 26, 27). (B) Transcription of HUG1 in the absence of DNA damage is repressed by the Crt1p-Ssn6p-Tup1p complex. Northern blot analysis was performed with the wild-type strain (Y300) and the crt1-Δ1::LEU2 (Y577), crt4-2/tup1 (Y217), and crt8-91/ssn6 (Y231) strains, grown logarithmically (lanes 1, 3, 5, and 7) or arrested with HU (lanes 2, 4, 6, and 8). HUG1/TUB2 for lanes 2 to 8 indicates the ratio of the intensity of the HUG1 signal to the TUB2 signal normalized to the HUG1/TUB2 ratio in control lane 1 as described in Materials and Methods.
Sensitivity to HU, UV radiation, ionizing radiation, and methyl methanesulfonate was determined as described previously (21). Western blot analysis was done as described previously (18) by using whole-cell extracts from hug1Δ1::HIS3 (YMB711) transformants containing plasmid pMB363 or pMB366. Filters were incubated with the primary HA antibody (1:5,000 dilution) followed by secondary antibody GAMHRP (goat anti-mouse horseradish peroxidase) (dilution of 1:10,000) and then with the enhanced chemiluminescence reagent (Amersham) and exposed to film.
Genetic analysis.
The HUG1 ORF was replaced by HIS3 by a PCR-based method (6). hug1Δ1::HIS3 (YMB711) replaces the HUG1 ORF, including sequences 153 bp upstream of the start codon (ATG) and 65 bp downstream of the stop codon (TAA). hug1Δ2::HIS3 (YMB847) replaces the HUG1 ORF, including sequences 153 bp upstream of the start codon (ATG) and 53 bp upstream of the stop codon (TAA). Deletions were made in diploid strain YPH987. Deletion of HUG1 was verified by PCR and Southern blot analysis, the diploid was sporulated, and tetrad analysis showed 2:2 segregation of the hug1Δ::HIS3 in each of the tetrads. For genetic interactions with MEC1, tetrad analyses of two independent matings were done. In the first case, YMB711 was mated to YEF610 lacking pEF208. From a total of 14 tetrads dissected, we obtained 3, 8, and 3 tetrads containing 4, 3, and 2 viable spores, respectively. Among these were 14 hug1Δ1::HIS3 and 15 mec1Δ::TRP1 spores, and from these, 7 were hug1Δ1::HIS3 mec1Δ::TRP1. In a second experiment we analyzed tetrads from a mating between YMB847 and yPP8. The strain yPP8 is inviable without the pMEC1 plasmid (pEF208). From a total of 34 tetrads, we obtained 14, 9, and 11 tetrads containing 4, 3, and 2, viable spores, respectively. Among these were 40 hug1Δ2::HIS3 and 32 mec1Δ::TRP1 spores, and from these, 15 were hug1Δ2::HIS3 mec1Δ::TRP. The latter spores were viable without pMEC1, as evidenced by growth on 5-FOA (7). Genetic interactions between DUN1 and HUG1 were determined by tetrad analysis of a diploid derived by mating strains YMB847 and U971. From a total of 22 tetrads, we obtained 41 hug1Δ2::HIS3 and 37 dun1Δ::URA3 spores: 19 of these were dun1Δhug1Δ2, and all of the double mutants were resistant to HU.
RESULTS
SAGE analysis reveals transcription from NORFs that are evolutionarily conserved.
As previously reported (40), SAGE has identified transcripts that correspond to NORFs in the intergenic regions of S. cerevisiae. We performed a systematic analysis of the SAGE tags that correspond to the NORFs (see Materials and Methods). Of the 60,633 SAGE tags analyzed, there were 302 unique SAGE tags that were either within or adjacent to intergenic ORFs of <100 amino acids. The 302 SAGE tags were expressed at levels ranging from 0.6 to 94 transcript copies per cell. The 30 most abundant of the transcripts detected by SAGE were observed at least nine times. We found that 12 of the 30 highly expressed NORF genes are evolutionarily conserved with mammalian homologs (28a). Northern blot analysis of four of the NORFs (NORF1, NORF5, NORF14, and NORF17) has confirmed their transcription in S. cerevisiae (data not shown). In addition, the SAGE data facilitated the addition of 27 new ORFs (<100 amino acids) to the S. cerevisiae genome database (35b).
Transcription of NORF5/HUG1 is induced by replication arrest and DNA damage.
NORF5, a putative 68-amino-acid protein, corresponds to a previously unidentified ORF transcribed in HU-arrested cells (40) HU, a potent inhibitor of ribonucleotide reductase (RNR), which is required for deoxynucleoside triphosphate (dNTP) synthesis, leads to replication arrest in S phase (13, 15). The transcript abundance for NORF5 in logarithmically grown yeast cells was <1 copy/cell, whereas in HU-arrested cells, it was 37 copies/cell, exhibiting a higher level of differential gene expression in HU-arrested cells than any other S. cerevisiae gene (40). Northern blot analysis supported SAGE data, because a transcript of approximately 400 bp, corresponding to NORF5, is present in RNA prepared from HU-arrested cells (Fig. 1A). Consistent with these results, Western blot analysis of the candidate epitope-tagged 68-amino-acid ORF (chromosome 13, coordinates 158760 to 158966) confirmed a protein of about 10 kDa in HU-arrested cells (Fig. 1B). Transcription of NORF5 is also induced in cells exposed to UV or gamma radiation (Fig. 1C). The transcriptional induction of NORF5 appears to be specific to replication arrest and DNA damage, since there was no induction of NORF5 in cells subjected to heat shock (data not shown) or nocodazole-induced G2/M arrest (Fig. 1A). On the basis of its transcription pattern, we named the NORF5 gene HUG1. We found that following addition of HU, low levels of HUG1 transcription are detected at earlier time periods of 0.5 and 1.0 h, followed by an almost linear increase until 3.5 h post HU addition (Fig. 1D). The DNA damage-dependent transcription of HUG1 is not restricted to any particular stage of the cell cycle. Cells arrested in G1 with alpha factor or G2/M with nocodazole show similar patterns of transcription of HUG1 compared to asynchronous populations upon exposure to gamma radiation (Fig. 1E), and, therefore, DNA damage-induced transcription of HUG1 can occur in the G1 and G2/M phases.
Crt1p, Ssn6p, and Tup1p are negative regulators of HUG1 transcription in the absence of DNA damage or replication arrest.
Promoters of DNA damage- or replication arrest-inducible genes, such as RNR2, RNR3, RNR4, and CRT1, often contain X-box sequences (16). In S. cerevisiae, the X box mediates Crt1p-dependent repression of the RNR genes by recruitment of the general repressors Ssn6p and Tup1p (37) to the promoters of damage-inducible genes (16). X-box sequences sharing a high degree of identity to those found in the promoters of mammalian major histocompatibility complex (MHC) class II genes (26) and S. cerevisiae genes were found in the promoter of HUG1 (Fig. 2A), suggesting that HUG1 may also be repressed by Crt1p. Accordingly, Northern blot analysis showed that HUG1 is constitutively transcribed in crt1, ssn6, and tup1 mutants that are deficient for Crt1p-mediated repression. In the absence of DNA damage, HUG1 is transcribed at levels 78-, 394-, and 20-fold higher in the crt1, ssn6, and tup1 mutants than wild-type cells (Fig. 2B). Thus, Crt1p, Ssn6p, and Tup1p are negative regulators of HUG1 transcription in the absence of DNA damage or replication arrest.
Checkpoint genes in the MEC1 pathway are required for the transcriptional induction of HUG1.
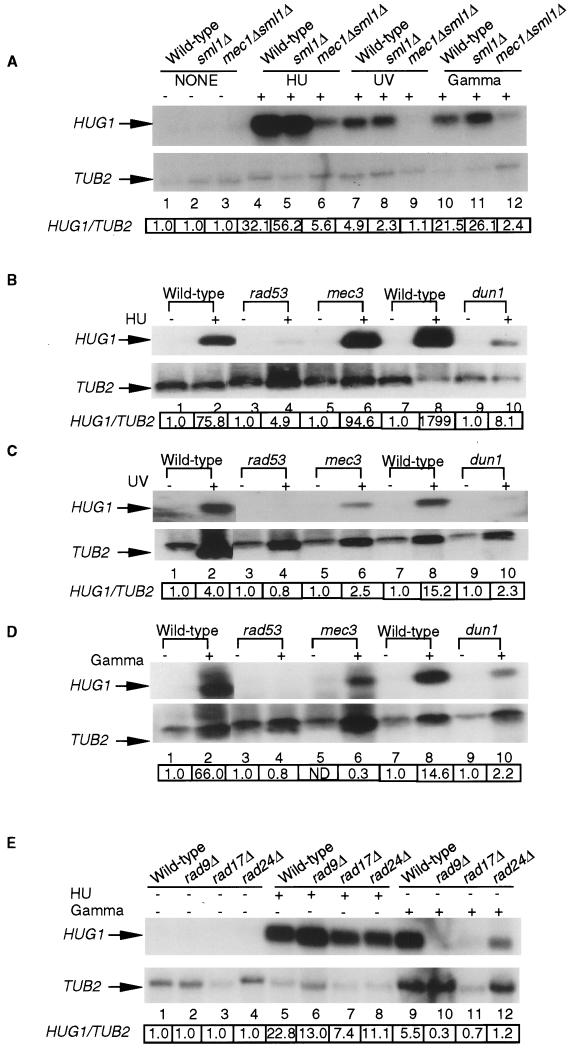
Checkpoint genes in the MEC1 pathway are required for the alleviation of DNA damage-dependent repression of RNR genes by the Crt1p-Ssn6p-Tup1p complex (16). The checkpoint genes mediate multiple responses following damage to DNA or the spindle apparatus including cell cycle arrest, transcriptional induction of damage-inducible genes, and repair of DNA damage (12). Unlike most other checkpoint genes, null alleles of MEC1 (mec1Δ) are lethal (17, 47), but mutations in SML1 (sml1-1 or sml1Δ) (46), CRT1 (16), or CLN1 and CLN2 (38) can suppress this lethality. Since the sml1Δ mutation does not affect the transcription of HUG1 (Fig. 3A), we decided to use a mec1Δ sml1Δ strain for evaluation of the role of MEC1 in the transcriptional induction of HUG1. Northern blot analysis showed that MEC1 is required for the transcriptional induction of HUG1 in response to replication arrest with HU and DNA damage from UV or gamma radiation (Fig. 3A). In contrast, TEL1, a functional homolog of MEC1 (21), is not required for the HU-induced transcription of HUG1 (data not shown). These results prompted us to determine if other genes in the MEC1 pathway (see Fig. 7) were required for the transcriptional induction of HUG1. Our results showed that the HU (Fig. 3B)-, UV (Fig. 3C), and gamma (Fig. 3D) radiation-induced transcription of HUG1 is dependent on RAD53 and partially dependent on DUN1. Additionally, transcriptional induction of HUG1 is dependent on MEC3 (Fig. 3A, B, and C), RAD9, RAD17, and RAD24 (Fig. 3E) upon exposure to UV and gamma radiation, but independent of these genes in the presence of HU. These effects do not appear to be simply due to delayed induction, since no HUG1 induction is detected in the mutants after 3.5 h in 0.1 M HU, whereas marked induction of HUG1 is observed as early as 1 h in wild-type cells (Fig. 1D). We conclude that the transcriptional induction of HUG1 is dependent on MEC1 and other genes in the checkpoint pathway (Fig. 3 and 7).
FIG. 3.
Genes in the MEC1 checkpoint pathway are required for the DNA damage- and replication arrest-induced transcription of HUG1. (A) Northern blot analysis was done with logarithmically grown, HU-arrested, UV or gamma radiation-exposed cells. The strains used were isogenic to the wild-type strain (W1588-3A) and the sml1Δ::HIS3 (U952-3C) and mec1Δ::TRP1 sml1Δ::HIS3 (U953-61D) strains. HUG1/TUB2 indicates the ratio of the intensity of the HUG1 signal to the TUB2 signal normalized to the HUG1/TUB2 ratio in control lanes 1, 2, and 3 (lanes 4, 7, and 10 normalized to lane 1, lanes 5, 8, and 11 to lane 2, and lanes 6, 9, and 12 to lane 3) as described in Materials and Methods (B, C and D) Northern blot analysis was done with strains grown logarithmically, arrested with HU (B), or exposed to UV (C) or gamma (D) radiation. The strains used were wild type (TWY397), rad53/mec2-1 (TWY312), mec3-1 (TWY316), wild type (Y203), and dun1 (Y203-dun1). Two lanes between lanes 2 and 3 in panels B, C, and D were deleted because they represented data not relevant to the experiment. HUG1/TUB2 indicates the ratio of the intensity of the HUG1 signal to the TUB2 signal in cells treated with HU or UV or gamma radiation and normalized to the HUG1/TUB2 ratio in control lanes without treatment (lane 1 normalized to lane 2, lane 3 to lane 4, lane 5 to lane 6, lane 7 to lane 8, lane 9 to lane 10, and lane 11 to lane 12). Transcription of TUB2 is not induced by UV or gamma radiation; the data reflect unequal loading of the lanes as evidenced by ethidium bromide staining of the gels (data not shown). (For Fig. 2D, lane 5, the level of HUG1 was below the background level and hence was denoted as not detected [ND].) The wild-type strain isogenic to the rad53 and mec3 mutants is represented in lanes 1 and 2. The wild-type strain isogenic to the dun1 mutant is represented in lanes 9 and 10. (E) Northern blot analysis using logarithmically grown cells, arrested with HU or exposed to gamma radiation. The strains used were isogenic to the wild-type strain (YMP10381), rad9Δ::LEU2 (YMP10535), rad17Δ::LEU2 (YMP11108), and rad24Δ::TRP1 (YMP10533). HUG1/TUB2 indicates the ratio of the intensity of the HUG1 signal to the TUB2 signal in cells treated with HU or gamma radiation and normalized to the HUG1/TUB2 ratio in control lanes without treatment (lanes 5 and 9 normalized to lane 1, lanes 6 and 10 to lane 2, lanes 7 and 11 to lane 3, and lanes 8 and 12 to lane 4).
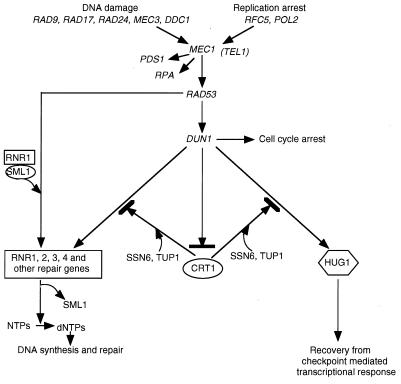
FIG. 7.
HUG1 is a critical component of the checkpoint response. Signals received from the sensors for DNA damage and replication arrest are transduced through the kinases MEC1 and TEL1, leading to phosphorylation and activation of RAD53 and DUN1, causing cell cycle arrest and transcriptional induction, which can be DUN1 independent or dependent (12). SML1 (46) and CRT1 (16) function to negatively regulate the MEC1 effectors RNR1 and RNR1 to 4, respectively. Transcription of HUG1 is induced in response to replication arrest and DNA damage in a checkpoint-dependent manner. Deletion of HUG1 rescues the lethality of mec1Δ and the HU sensitivity of dun1Δ strains; overexpression of HUG1 is lethal in combination with a mec1 mutation in the presence of replication arrest or DNA damage. These observations, along with the delayed induction of HUG1 in response to HU, suggest that HUG1 may function, in part, through the negative regulation of MEC1 effectors, perhaps facilitating recovery from the transcriptional response after DNA damage and replication arrest.
Deletion of HUG1 suppresses mec1 lethality, and overexpression of HUG1 increases the sensitivity of the mec1 sml1-1 strain to HU.
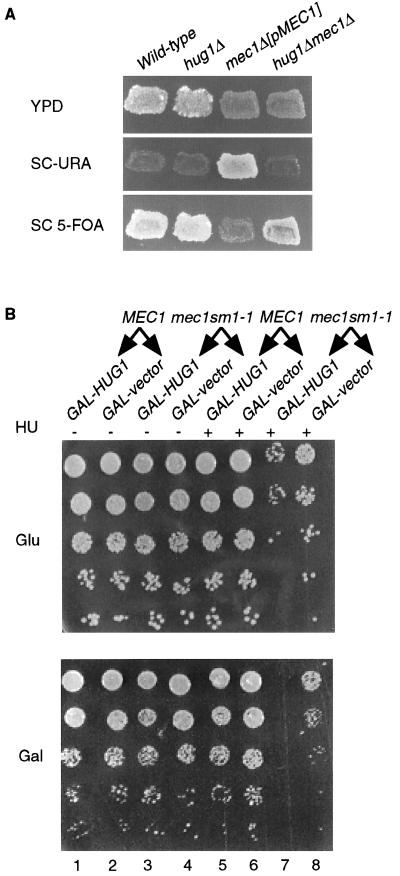
To elucidate the role of Hug1p in DNA damage and replication arrest, we deleted the HUG1 ORF and examined several phenotypes. Deletion of HUG1 in a haploid strain does not affect growth, sensitivity to DNA-damaging agents, or HU (data not shown). Given the transcriptional dependence of HUG1 on MEC1, we examined the effect of hug1Δ on the essential and checkpoint functions of MEC1. The mec1Δ sml1-1 strain is viable due to the sml1-1 mutation (46). We mated a hug1Δ SML1 strain to a mec1Δ sml1-1 strain, sporulated the heterozygous diploid, and analyzed the tetrads. Genetic analysis showed that hug1Δ suppresses the lethality due to mec1Δ, because we obtained hug1Δ mec1Δ spores at the expected frequency (see Materials and Methods). The hug1Δ mec1Δ strain is as sensitive to DNA damage and replication arrest as the parent mec1Δ sml1-1 strain (data not shown). These results were confirmed by tetrad analysis of a mating between the hug1Δ and mec1Δ[pMEC1 CEN URA3] strains. We obtained hug1Δ mec1Δ[pMEC1 CEN URA3] spores that were viable without the pMEC1 plasmid, thus confirming the suppression of mec1Δ lethality by deletion of HUG1 (Fig. 4A). Therefore, hug1Δ suppresses mec1Δ lethality, but not sensitivity to DNA damage or replication arrest. These results also suggest that HUG1 may be transcribed either at low levels or in a small fraction of the cells in the absence of DNA damage or replication arrest.
FIG. 4.
Genetic interactions between HUG1 and MEC1. (A) Deletion of HUG1 suppresses the lethality of mec1Δ. Strains derived from a mating between the hug1Δ (YMB847) and mec1ΔSML1 (pMEC1) (yPP8) strains were plated on control medium YPD and then replica plated to SC-Ura and SC with 5-FOA. The mec1Δ SML1 strain is inviable without the pMEC1 plasmid (pEF208) (growth on SC-Ura, 5-FOA sensitive). The wild-type, hug1Δ and hug1Δ mec1Δ strains can lose the pMEC1 plasmid (no growth on SC-Ura, 5-FOA resistant). (B) Overexpression of HUG1 (pMB379) increases the sensitivity of mec1 sml1-1 (DLY258) mutants to replication arrest, with no effect in the wild-type strain (DLY62). Strains were grown logarithmically in either the absence or presence of HU (0.1 M) for 3.5 h, and 5 μl of a fivefold serial dilution series was plated on SC-Ura with glucose (Glu) or SC-Ura with raffinose plus galactose (Gal).
Consistent with the ability of a HUG1 deletion to suppress mec1Δ lethality, we found that overexpression of HUG1 (GAL1-HUG1) increased the sensitivity of the mec1 sml1-1 strain (DLY258) to HU (Fig. 4B) and UV radiation (data not shown) and had no phenotype in wild-type cells (DLY62) (Fig. 4B). Almost identical results were obtained with another mec1Δ sml1-1 strain (YEF610 lacking pEF208), suggesting that the dosage lethality phenotype is not strain specific (data not shown). The phenotype was specifically due to the HUG1 protein, because a frameshift mutation in the HUG1 ORF abolished the dosage lethality phenotype (data not shown).
HUG1 and SML1 are adjacent to each other and are transcribed independently.
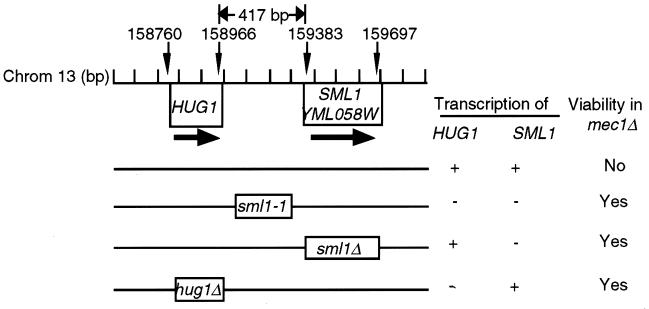
Similar to the phenotype of a hug1Δ, mutations in SML1 (sml1-1 or sml1Δ) also suppress mec1Δ lethality (46). The start codon of SML1 is 417 bp downstream of the stop codon of HUG1, and both genes are transcribed from the same strand of DNA (Fig. 5). Hence, we examined whether SML1 played a role in the phenotype of suppression of mec1Δ lethality by a deletion of HUG1. We determined that HUG1 and SML1 are transcribed and regulated independently (Fig. 5). For example, unlike HUG1, the transcription of SML1 is not induced by replication arrest or DNA damage and is unaffected by mutations in checkpoint genes (data not shown and reference 46). Additionally, HUG1 transcription is not affected by a deletion of SML1 or vice versa (Fig. 5). We also determined that SML1 is transcribed in a hug1Δ mec1Δ strain (data not shown). It is interesting to note that in contrast to sml1Δ, the sml1-1 mutation present in most laboratory mec1 strains (46) overlaps with the 3′ untranslated region of HUG1 and abolishes the transcription of HUG1 (Fig. 5).
FIG. 5.
HUG1 and SML1 are transcribed independently, and deletions of either gene suppress mec1Δ lethality. The strains used were the wild type (W1588-4A) and the sml1-1 (YEF630), sml1Δ(U952-3C), and hug1Δ2 (YMB847) mutants. Transcription of SML1 was detected in logarithmically grown cells, whereas that of HUG1 was only detected in cells arrested with HU. The sml1-1 mutation (46) deletes a 290-bp region between two direct repeats of 11 bp; the first repeat is 7 bp downstream of the HUG1 stop codon. The sml1-1 mutation is present in most laboratory mec1 strains (25, 46).
Deletion of HUG1 suppresses the HU sensitivity of the dun1Δ strain.
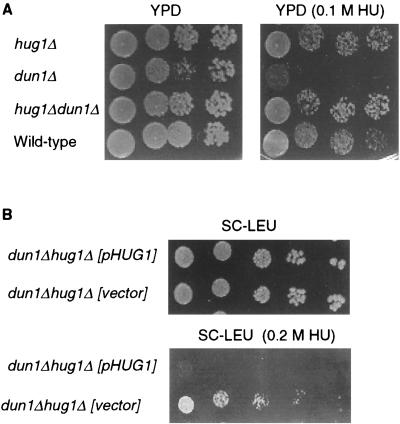
The protein kinase DUN1 gene acts downstream of MEC1 (1, 12, 24, 48) and is required for the efficient induction of HUG1 following replication arrest and DNA damage (Fig. 3 and 7). The dun1 mutants exhibit an HU sensitivity that can be suppressed by overexpression of RNR1 (46). Given the ability of HUG1 overexpression to increase the sensitivity of the mec1 sml1-1 strain to replication arrest and DNA damage, we determined whether HUG1 expression might modulate the HU sensitivity of the dun1Δ strain. Genetic analysis demonstrated that a deletion of HUG1 (hug1Δ) suppressed the HU sensitivity of a dun1Δ strain (Fig. 6A). As expected, this HU sensitivity was restored in the dun1Δ hug1Δ strain by a HUG1-containing plasmid (Fig. 6B). These findings further support the role of HUG1 as a critical downstream mediator of the MEC1 pathway.
FIG. 6.
Deletion of HUG1 suppresses the HU sensitivity of the dun1Δ strain. (A) hug1Δ dun1Δ strains are resistant to HU. Spores from tetrad analysis of a mating between the hug1Δ2 (YMB847) and dun1Δ (U971) strains were plated on YPD medium with or without HU (0.1 M). (B) HUG1 restores HU sensitivity in a dun1Δ hug1Δ strain. The hug1Δ dun1Δ spores from panel A were transformed with pMB363 (CEN HUG1 LEU2) or vector alone (pRS315) and plated on SC-Leu with or without HU (0.2 M).
DISCUSSION
Here, we show that the SAGE technique (39, 40) used to determine global gene expression can identify transcripts corresponding to NORFs. Systematic analysis of SAGE tags corresponding to intergenic regions suggests the presence of at least 302 NORFs. These NORFs may correspond to small ORFs (<99 amino acids) or large ORFs (>99 amino acids) that may have been overlooked due to possible sequencing errors. Homology searches have shown that 12 of the 30 most highly transcribed NORFs are evolutionarily conserved. One of the NORFs, NORF5/HUG1, encodes a novel DNA damage and replication arrest-induced gene that is transcriptionally regulated by the genes in the MEC1 pathway. Our results validate the idea that the NORFs are biologically relevant and highlight the importance of global approaches such as SAGE to identify a significant number of genes in yeast and other organisms that may be missed by sequence analysis alone.
Further characterization of the transcriptional regulation of HUG1 showed that the promoter of HUG1 contains three X-box-related sequences (16, 26): one strongly conserved X box (Xs) and two weakly conserved X boxes (Xw). X-box sequences (13 bp in length) were originally identified in the promoters of all MHC class II genes (26) and subsequently found in the promoters of RNR2, RNR3, RNR4, and CRT1 (16). There is a high degree of conservation between the X boxes; for example, 10 of the 13 bases of Xs in HUG1 are identical to the Xs of the MHC class II X box (26). Also, the location and orientation of the X boxes in HUG1 are similar to those of the other X-box-containing genes in S. cerevisiae; Xs and Xw in HUG1 are in opposite orientations located 30 bp apart (16). It has been shown that Crt1p binds specifically to X-box sequences in the promoters of RNR genes and mediates repression of these genes by recruitment of the Tup1p-Ssn6p corepressor complex to the promoters of these genes. DNA damage leads to hyperphosphorylation of Crt1p with loss of DNA binding and loss of repression (16). Northern blot analysis showed that HUG1 is constitutively transcribed in crt1, ssn6, and tup1 mutants that are deficient for Crt1p-mediated repression. The degree of derepression of HUG1 transcription was as follows: ssn6>crt1>tup1 mutants. Similar results were reported for the derepression of the RNR2 promoter in the ssn6, crt1, and tup1 mutants (16).
In S. cerevisiae, there is a large regulon of genes that show increased transcription in response to DNA damage and replication arrest (1, 12, 19, 20, 24, 28). The checkpoints that are sensitive to DNA damage or replication arrest act in multiple phases of the cell cycle (G1, S, or G2 phases) (2, 12, 23, 34, 41–43). The checkpoint genes regulate transcription, facilitate the repair of DNA, and mediate cell cycle arrest and recovery from DNA damage-induced responses (12, 41). The results presented in this paper show that the DNA replication arrest and damage-induced transcription of HUG1 are dependent on the signal transduction pathway involving the checkpoint genes RAD9, RAD17, RAD24, MEC3, MEC1, RAD53, and DUN1.
Despite the major advances in the delineation of the MEC1 checkpoint pathway, the full complexity of this pathway is just beginning to be addressed (16, 30, 33, 38, 41, 46). The current findings suggest that the small protein Hug1p, the product of a NORF, is a critical mediator of the MEC1 pathway. Induction of HUG1 by DNA damage and replication arrest requires an intact MEC1 pathway, and a deletion of HUG1 can rescue phenotypes associated with defects in the MEC1 pathway. Although the precise mechanism of action of HUG1 remains unclear, several observations suggest that HUG1 may function, in part, through the negative regulation of MEC1 pathway effectors, perhaps facilitating the recovery from the transcriptional response after DNA damage and replication arrest. First, mutations in the other two genes (SML1 and CRT1) besides HUG1 that can rescue mec1Δ lethality function to negatively regulate effectors of the MEC1 pathway (16, 46). Second, overexpression of HUG1 is lethal in combination with a mec1 mutation in the presence of DNA damage or replication arrest; this is in contrast to the MEC1 effectors RNR1 and RNR3, whose overexpression rescues mec1Δ lethality (9). Third, transcription of HUG1 is delayed in response to replication arrest (Fig. 1D), unlike the rapid induction of RNR3 (16). This delay in HUG1 induction may allow time for DNA synthesis and repair before recovery. Taken together, these results suggest that HUG1 is a critical component of the checkpoint response (Fig. 7).
Consistent with the importance of the coordinated response to DNA damage, several key features of these pathways are conserved in human, yeast, and other systems. The S. cerevisiae MEC1 gene, for example, is homologous to the Schizosaccharomyces pombe rad3+ gene, the Drosophila melanogaster mei-41 gene, and the human ATM gene (31). By analogy, a HUG1 homolog regulated by ATM or p53 may be present in humans. It is not surprising that database searches have failed to detect a homolog of HUG1, because it has only been detected in DNA-damaged or replication-arrested cells. Identification and characterization of homologs of HUG1 from other organisms, including humans, may further our understanding of the role of MEC1 in budding yeast and may allow greater insight into the ATM- and p53-mediated checkpoint pathway in humans.
ACKNOWLEDGMENTS
We gratefully acknowledge the gifts of strains and plasmids, suggestions, or support from A. Basrai, D. Bassett, J. Boeke, G. Brush, C. Connelly, S. Elledge, K. Gupta, the L. Hartwell laboratory, M. Huang, K. Hyland, M. Johnston, M. Kenna, I. Kirsch, R. Kitagawa, D. Koshland, R. Krishnan, S. Michaelis, D. Morrow, P. Paddison, R. Rothstein, R. Skibbens, F. Spencer, B. Vogelstein, T. Weinert, J. Zhang, X. Zhao, and our laboratory members. We thank J. Vogelstein for analysis of NORF data; S. Dwight, T. Roe, C. Ball, A. Malekian, and M. Cherry of the Stanford Genome Database for annotation of new ORFs based on SAGE analysis; J. Flook for assistance with flow cytometry; and L. Dillehay for assistance with gamma irradiation. M.A.B. thanks C. Greider for constant support and M. Huang and P. Paddison for valuable suggestions.
K.W.K. received research funding from Genzyme Molecular Oncology (Genzyme). P.H. was supported by grants NIH CA16519 and NIH HD24605.
REFERENCES
- 1.Aboussekhra A, Vialard J E, Morrison D E, de la Torre-Ruiz M A, Cernakova L, Fabre F, Lowndes N F. A novel role for the budding yeast RAD9 checkpoint gene in DNA damage-dependent transcription. EMBO J. 1996;15:3912–3922. [PMC free article] [PubMed] [Google Scholar]
- 2.Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, editor. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 4.Basrai M A, Hieter P, Boeke J D. Small open reading frames: beautiful needles in the haystack. Genome Res. 1997;7:768–771. doi: 10.1101/gr.7.8.768. [DOI] [PubMed] [Google Scholar]
- 5.Basrai M A, Kingsbury J, Koshland D, Spencer F, Hieter P. Faithful chromosome transmission requires Spt4p, a putative regulator of chromatin structure in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2838–2847. doi: 10.1128/mcb.16.6.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 8.Brush G S, Morrow D M, Hieter P, Kelly T J. The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc Natl Acad Sci USA. 1996;93:15075–15080. doi: 10.1073/pnas.93.26.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a.Cyano2Dbase. 14 January 1999, revision date. ORFs. [Online.] http://www.kazusa.or.jp/tech/sazuka/cyano/proteome.html. [23 August 1999, last date accessed.]
- 9.Desany B A, Alcasabas A A, Bachant J B, Elledge S J. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dujon B, Alexandraki D, Andre B, Ansorge W, Baladron V, Ballesta J P, Banrevi A, Bolle P A, Bolotin-Fukuhara M, Bossier P, et al. Complete DNA sequence of yeast chromosome XI. Nature. 1994;369:371–378. doi: 10.1038/369371a0. [DOI] [PubMed] [Google Scholar]
- 10a.E. coli Genome Center. 5 September 1997, revision date. ORFs. [Online.] E. coli Genome Center, University of Wisconsin, Madison. http://genetics/wisc.edu/. [23 August 1999, last date accessed.]
- 11.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 12.Elledge S J. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 13.Elledge S J, Zhou Z, Allen J B, Navas T A. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays. 1993;15:333–339. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- 14.Goffeau A, Barrell B G, Bussey H, Davis R W, Dujon B, Feldmann H, Galibert F, Hoheisel J D, Jacq C, Johnston M, Louis E J, Mewes H W, Murakami Y, Philippsen P, Tettelin H, Oliver S G. Life with 6000 genes. Science. 1996;274:546. doi: 10.1126/science.274.5287.546. , 563–567. [DOI] [PubMed] [Google Scholar]
- 15.Huang M, Elledge S J. Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6105–6113. doi: 10.1128/mcb.17.10.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang M, Zhou Z, Elledge S J. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- 17.Kato R, Ogawa H. An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22:3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenna M A, Brachmann C B, Devine S E, Boeke J D. Invading the yeast nucleus: a nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol Cell Biol. 1998;18:1115–1124. doi: 10.1128/mcb.18.2.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiser G L, Weinert T A. Distinct roles of yeast MEC and RAD checkpoint genes in transcriptional induction after DNA damage and implications for function. Mol Biol Cell. 1996;7:703–718. doi: 10.1091/mbc.7.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClanahan T, McEntee K. DNA damage and heat shock dually regulate genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:90–96. doi: 10.1128/mcb.6.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.MIPS. 9 August 1999, revision date. ORFs. [Online.] Munich Information Centre for Protein Sequences, Martinsfried, Germany. http://www.mips.biochem.mpg.de/proj/yeast/tables/small_orfs.html. [23 August 1999, last date accessed.]
- 21.Morrow D M, Tagle D A, Shiloh Y, Collins F S, Hieter P. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 22.Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navas T A, Zhou Z, Elledge S J. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 24.Pati D, Keller C, Groudine M, Plon S E. Reconstitution of a MEC1-independent checkpoint in yeast by expression of a novel human fork head cDNA. Mol Cell Biol. 1997;17:3037–3046. doi: 10.1128/mcb.17.6.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulovich A G, Margulies R U, Garvik B M, Hartwell L H. RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics. 1997;145:45–62. doi: 10.1093/genetics/145.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reith W, Herrero-Sanchez C, Kobr M, Silacci P, Berte C, Barras E, Fey S, Mach B. MHC class II regulatory factor RFX has a novel DNA-binding domain and a functionally independent dimerization domain. Genes Dev. 1990;4:1528–1540. doi: 10.1101/gad.4.9.1528. [DOI] [PubMed] [Google Scholar]
- 27.Reith W, Ucla C, Barras E, Gaud A, Durand B, Herrero-Sanchez C, Kobr M, Mach B. RFX1, a transactivator of hepatitis B virus enhancer I, belongs to a novel family of homodimeric and heterodimeric DNA-binding proteins. Mol Cell Biol. 1994;14:1230–1244. doi: 10.1128/mcb.14.2.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruby S W, Szostak J W. Specific Saccharomyces cerevisiae genes are expressed in response to DNA-damaging agents. Mol Cell Biol. 1985;5:75–84. doi: 10.1128/mcb.5.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Sagenet. 7 March 1999, revision date. ORFs. [Online.] http://www.sagenet.org/NORF/NORF.html. [23 August 1999, last date accessed.]
- 29.Sanchez Y, Desany B A, Jones W J, Liu Q, Wang B, Elledge S J. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 29a.Sanger Website. 13 August 1999, revision date. ORFs. [Online.] http://www.songer.ac.uk/Projects/C_elegans. [23 August 1999, last date accessed.]
- 30.Santocanale C, Diffley J F. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 31.Savitsky K, Sfez S, Tagle D A, Ziv Y, Sartiel A, Collins F S, Shiloh Y, Rotman G. The complete sequence of the coding region of the ATM gene reveals similarity to cell cycle regulators in different species. Hum Mol Genet. 1995;4:2025–2032. doi: 10.1093/hmg/4.11.2025. [DOI] [PubMed] [Google Scholar]
- 32.Shiloh Y. Ataxia-telangiectasia and the Nijmegen breakage syndrome: related disorders but genes apart. Annu Rev Genet. 1997;31:635–662. doi: 10.1146/annurev.genet.31.1.635. [DOI] [PubMed] [Google Scholar]
- 33.Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 34.Siede W, Friedberg A S, Friedberg E C. RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:7985–7989. doi: 10.1073/pnas.90.17.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Stanford Genome Database. 17 July 1999, revision date. ORFs. [Online.] Department of Genetics, Stanford University School of Medicine, Stanford, Calif. http://genome-www.stanford.edu/Saccharomyces/. [23 August 1999, last date accessed.]
- 35b.Stanford Genome Database. 17 July 1999, revision date. ORFs. [Online.] http://genome-www.stanford.edu/Saccharomyces/newORF.html. [23 August 1999, last date accessed.]
- 36.Sun Z, Fay D S, Marini F, Foiani M, Stern D F. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 37.Tzamarias D, Struhl K. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev. 1995;9:821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- 38.Vallen E A, Cross F R. Interaction between the MEC1-dependent DNA synthesis checkpoint and G1 cyclin function in Saccharomyces cerevisiae. Genetics. 1999;151:459–471. doi: 10.1093/genetics/151.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 40.Velculescu V E, Zhang L, Zhou W, Vogelstein J, Basrai M A, Bassett D E, Jr, Hieter P, Vogelstein B, Kinzler K W. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 41.Weinert T. DNA damage checkpoints update: getting molecular. Curr Opin Genet Dev. 1998;8:185–193. doi: 10.1016/s0959-437x(98)80140-8. [DOI] [PubMed] [Google Scholar]
- 42.Weinert T A, Hartwell L H. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinert T A, Hartwell L H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 44.Weinert T A, Kiser G L, Hartwell L H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 45.Zakian V A. ATM-related genes: what do they tell us about functions of the human gene? Cell. 1995;82:685–687. doi: 10.1016/0092-8674(95)90463-8. [DOI] [PubMed] [Google Scholar]
- 46.Zhao X, Muller E G, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 47.Zheng P, Fay D S, Burton J, Xiao H, Pinkham J L, Stern D F. SPK1 is an essential S-phase-specific gene of Saccharomyces cerevisiae that encodes a nuclear serine/threonine/tyrosine kinase. Mol Cell Biol. 1993;13:5829–5842. doi: 10.1128/mcb.13.9.5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Z, Elledge S J. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell. 1993;75:1119–1127. doi: 10.1016/0092-8674(93)90321-g. [DOI] [PubMed] [Google Scholar]