Abstract
Nuclear factor κB (NF-κB) represents a family of dimeric DNA binding proteins, the pleotropic form of which is a heterodimer composed of RelA and p50 subunits. The biological activity of NF-κB is controlled through its subcellular localization. Inactive NF-κB is sequestered in the cytoplasm by physical interaction with an inhibitor, IκBα. Signal-mediated IκBα degradation triggers the release and subsequent nuclear translocation of NF-κB. It remains unknown whether the NF-κB shuttling between the cytoplasm and nucleus is subjected to additional steps of regulation. In this study, we demonstrated that the RelA subunit of NF-κB exhibits strong cytoplasmic localization activity even in the absence of IκBα inhibition. The cytoplasmic distribution of RelA is largely mediated by a leucine-rich sequence homologous to the recently characterized nuclear export signal (NES). This putative NES is both required and sufficient to mediate cytoplasmic localization of RelA as well as that of heterologous proteins. Furthermore, the cytoplasmic distribution of RelA is sensitive to a nuclear export inhibitor, leptomycin B, suggesting that RelA undergoes continuous nuclear export. Interestingly, expression of p50 prevents the cytoplasmic expression of RelA, leading to the nuclear accumulation of both RelA and p50. Together, these results suggest that the nuclear and cytoplasmic shuttling of RelA is regulated by both an intrinsic NES-like sequence and the p50 subunit of NF-κB.
Nuclear factor κB (NF-κB) represents a family of eukaryotic transcription factors participating in the regulation of various cellular genes involved in the immediate early processes of immune, acute-phase, and inflammatory responses as well as genes involved in cell survival (for recent reviews, see references 23, 24, and 59). NF-κB also serves as a key cellular transcriptional activator of a number of human viruses, most notably human immunodeficiency virus type 1 (HIV-1) (30, 34, 35, 48, 53). In mammalian cells, five members of the NF-κB family have been characterized, including p50, p52, RelA (previously termed p65), RelB, and c-Rel. The different NF-κB proteins have significant sequence homology in an N-terminal region (∼300 amino acids), termed the Rel-homology domain (RHD). The RHD contains sequences mediating DNA binding, dimerization, and nuclear translocation functions (47, 56).
In most cell types, the pleotropic-inducible form of NF-κB is a heterodimer composed of p50 and RelA (4). RelA contains a C-terminal transactivation domain in addition to the N-terminal RHD, thus serving as a critical transactivation subunit of NF-κB (6, 42, 45). p50 lacks a transactivation domain, and it is believed to serve as a regulatory subunit modulating the DNA binding affinity of RelA (6, 42, 45). The p50-RelA NF-κB heterodimer is normally sequestered in the cytoplasmic compartment by physical association with inhibitory proteins, including IκBα and related proteins (5). IκBα specifically binds to and masks the nuclear localization signals (NLS) of RelA and p50, thereby preventing the nuclear translocation of the NF-κB heterodimer (7, 21, 25, 61). The latent cytoplasmic NF-κB RelA-p50 complex can be posttranslationally activated by a variety of cellular stimuli (2, 28), which trigger site-specific phosphorylation of IκBα (9, 10, 16, 54) by a multisubunit IκB kinase (IKK) (12, 14, 17, 33, 38, 41, 58, 60, 62). The phosphorylated IκBα becomes rapidly ubiquitinated and degraded by the proteasome complex (11, 16, 40, 44). Following IκBα degradation, the NF-κB heterodimer is rapidly translocated to the nucleus, where it activates the transcription of target genes.
Although the mechanism underlying the inducible degradation of IκBα has been well studied, it has remained unclear whether the cytoplasmic and nuclear shuttling of NF-κB is under the control of additional mechanisms. We report here that the RelA subunit of NF-κB contains a leucine-rich sequence homologous to the recently characterized nuclear export signal (NES) (22). Due to the presence of this NES-like sequence, a large proportion of RelA is localized in the cytoplasm even in the absence of the inhibitory protein IκBα. Interestingly, when coexpressed with p50, the cytoplasmic expression of RelA is completely inhibited, leading to the nuclear accumulation of both RelA and p50. These results strongly suggest that subcellular localization of the RelA subunit of NF-κB is under the regulation of both cis-acting sequences and p50.
MATERIALS AND METHODS
Plasmid constructs.
The cDNA expression vectors encoding wild-type RelA (RelA WT) and its truncation mutants were generated by PCR amplification of human RelA cDNA and subsequent cloning of the PCR products into the pCMV4 mammalian expression plasmid (21). The truncation mutants are named based on the amino acid residues retained in the constructs. For example, RelA(31-551) contains amino acids 31 to 551 of RelA, while RelA(1-450) contains amino acids 1 to 450 of RelA. The RelA(1-450)ΔNES was created by site-directed mutagenesis (Stratagene) to delete four amino acids (L440, L441, Q442, and L443) from the core region of the RelA NES site. The sense oligonucleotide primer sequence used in the site mutagenesis was GGA ACG CTG TCA GAG GCC CAG TTT GAT GAT GAA GAC CTG. To generate RelA(1-420)-NES, a short DNA fragment covering the NES region of RelA was fused to the C terminus of RelA(1-420). RelA-NLS was constructed by fusing a copy of the simian virus 40 large T antigen NLS (PKKKRKV) to the N terminus of RelA by PCR. To generate RelA-NLS-ΔNES, RelA-NLS was subjected to internal deletion to remove the four amino acids (L440, L441, Q442, and L443) from the core region of the RelA NES. p50-GFP was constructed by cloning a HindIII/RsaI fragment of pCMV4-p50 to the pEGFP-N2 plasmid (ClonTech, Inc.) upstream of the green fluorescent protein (GFP) coding region. p50-GFP-NES was generated by inserting the RelA NES to the C terminus of the GFP. pCMV4-IκBα has been reported previously (21), and IκBα-GFP was constructed by inserting GFP to the CMV4-IκBα vector upstream of the IκBα. The κB-TATA-luc reporter plasmid has been reported previously (21).
Immunoblotting and immunofluorescence assays.
Monkey kidney COS cells were cultured in Iscove’s medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and antibiotics. For immunoblotting assays, the cells were transfected in six-well plates as previously described (21). Whole-cell extracts were subjected to Western blot assays (21). For immunofluorescence assays, the cells were seeded on four-well chamber slides and transfected by the DEAE-dextran method (26). In the single transfections, 0.25 μg of the indicating cDNA expression vectors was used. For the double transfections, 0.25 μg of RelA and 0.5 μg of p50-GFP (or p50-GFP-NES) were used. After 48 h, recipient cells were lysed in ELB cell lysis buffer (21) and then subjected to immunoblotting with the indicated antibodies. For immunofluorescence assays, COS cells were seeded onto coverslips and transfected by the DEAE-dextran method. After 48 h, the cells were fixed, permeabilized, and sequentially incubated with the indicated primary antibodies, followed by donkey anti-rabbit immunoglobulin (Ig) covalently coupled to Texas red dye (21). The subcellular localization of transfected proteins was detected by fluorescence microscopy with a rhodamine filter by using an Olympus BH-2 fluorescence microscope. Digital images were collected by Optimas 6.2 and then transferred to Adobe Photoshop 4.0. Expression of the GFP fusion proteins were visualized directly with a fluorescein isothiocyanate (FITC) filter. Cells were also counterstained with 1 μg of Hoechst 33258 (added together with the secondary antibody; Sigma) per ml and visualized with a UV filter. For inhibition of protein nuclear export, transfected cells were incubated with 40 ng of leptomycin B (LMB) per ml for 6 h prior to immunofluorescence staining.
Luciferase reporter gene assays.
COS cells were transfected, in 24-well plates, with 50 ng of the κB-TATA-luc reporter (21) together with 100 ng of cDNA expression vectors encoding wild-type RelA or the indicated RelA truncation mutants. After 40 to 48 h of transfection, the recipient cells were lysed in a reporter lysis buffer (Promega). Luciferase activity was detected by mixing 5 μl of extract with 25 μl of luciferase substrate (Promega) and measured with a single photon channel of a scintillation counter (Beckman).
RESULTS
Cytoplasmic expression of RelA is mediated by a C-terminal region adjacent to its transactivation domain.
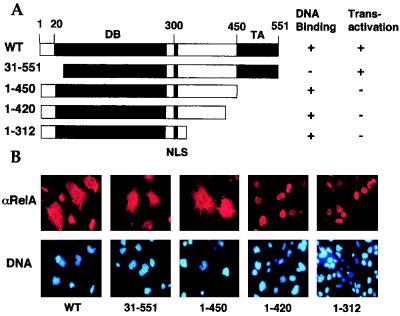
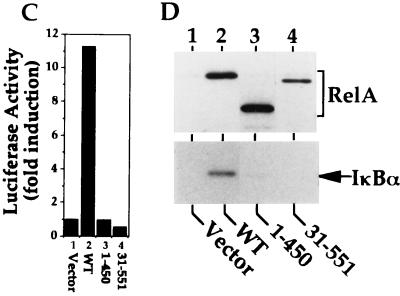
Previous studies demonstrate that when expressed in COS cells, a large proportion of RelA is retained in the cytoplasm, while the p50 subunit is exclusively expressed in the nucleus (21). Since RelA is a transactivator of the gene encoding its inhibitor IκBα (13, 31, 46, 51), one potential mechanism mediating the cytoplasmic expression of RelA may be the induction of endogenous IκBα. To examine this possibility, we first determined whether the cytoplasmic expression of RelA was dependent on its transactivation activity. RelA WT and its truncation mutants (Fig. 1A) were subjected to indirect immunofluorescence and parallel transactivation assays. As expected, RelA WT was predominantly located in the cytoplasm (Fig. 1B), and the cytoplasmic localization of RelA was correlated with its transactivation function (Fig. 1C and D, lanes 2). However, an N-terminally truncated form of RelA [RelA(31-551)] defective in DNA binding (21) still exhibited a strong cytoplasmic expression activity, generating a whole-cell distribution pattern (Fig. 1B). As previously demonstrated (21), this RelA mutant failed to transactivate the κB-TATA-luc reporter (Fig. 1C) and to induce the expression of endogenous IκBα (Fig. 1D, lane 4). Thus, IκBα inhibition may not be the only mechanism mediating the cytoplasmic expression of RelA. In further support of this notion, removal of the C-terminal transactivation domain of RelA [RelA(1-450)] (Fig. 1A) did not significantly alter its subcellular localization pattern (Fig. 1B), although this truncated form of RelA completely lost its transactivation activity (Fig. 1C and D, lanes 3). To further examine the mechanism regulating the subcellular localization of RelA, additional truncation mutants of RelA were subjected to the immunofluorescence assays. Interestingly, a deletion of 30 amino acids or more beyond the transactivation domain generated RelA mutants [RelA(1-420) and RelA(1-312)] exhibiting predominantly nuclear expression (Fig. 1B). Together, these results strongly suggest that the cytoplasmic expression of RelA involves not only IκBα inhibition but also a regulatory mechanism mediated by a C-terminal sequence element located adjacent to its transactivation domain.
FIG. 1.
Cytoplasmic expression of RelA is mediated by its C-terminal sequences located beyond the transactivation domain. (A) Primary domain structure of RelA WT and its truncation mutants. The locations of the DNA binding domain (DB) and the transactivation domain (TA) are presented according to previous studies (6, 21, 45). The DNA binding and transactivation activities of the mutants were as determined in a previous study (21) and for Fig. 1C and D. (B) Immunofluorescence assays to determine the subcellular localization of RelA WT and truncation mutants. COS cells were transfected with the indicated cDNA expression vectors. After 48 h, the transfected cells were subjected to indirect immunofluorescence assays with antisera recognizing the C terminus (WT and 31-551) or N terminus (1-450, 1-420, and 1-312) of RelA (21) and Texas red-conjugated anti-rabbit Ig secondary antibody (upper panels). To localize the nuclei of the transfected cells, the cells were counterstained with Hoechst 33258 and visualized with a UV filter (lower panels). (C) Luciferase reporter gene assay determining the transactivation activity of RelA and its truncation mutants. COS cells were transfected with the κB-TATA-luc reporter plasmid together with either an empty vector, cDNA expression vectors encoding the wild type (WT), or the indicated truncation mutants of RelA. Luciferase activity is presented as the fold induction relative to the basal level measured in cells transfected with the empty vector. (D) Immunoblotting analysis of the whole-cell extracts isolated from COS cells transfected with either an empty vector or cDNA expression vectors encoding RelA WT or its truncation mutants [RelA(1-450) and RelA(31-551)]. Immunoblotting was performed with antisera that reacted with the N terminus (lanes 1 to 3) or C terminus (lane 4) of RelA or IκBα. The RelA and its truncation mutants are labeled with a bracket, and IκBα is indicated by an arrow. Only RelA WT induces expression of endogenous IκBα.
A putative NES sequence promotes cytoplasmic expression of RelA.
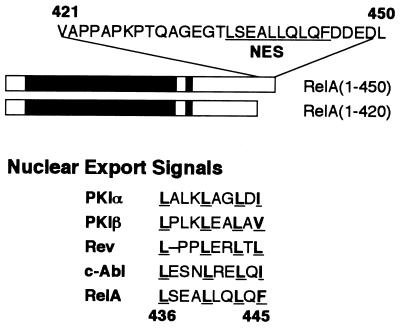
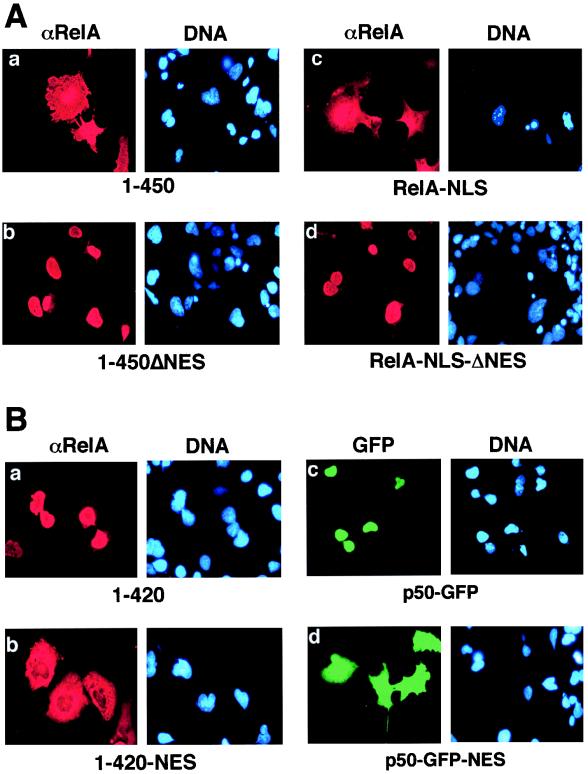
After analyzing the sequence located between amino acids 420 and 450 of RelA, we observed a leucine-rich sequence element (amino acids 436 to 445) homologous to the NES motifs identified from a number of proteins, such as the protein kinase inhibitor alpha and beta subunits (57), the HIV-1 Rev protein (18), and the proto-oncogene product c-Abl (52) (Fig. 2). An important structural feature of NES is the presence of four conserved hydrophobic amino acid residues (22). The NES-like sequence of RelA contains all these conserved residues (Fig. 2). To determine the potential role of this putative NES in the cytoplasmic expression of RelA, a RelA mutant [RelA(1-450)ΔNES] was generated by deleting four amino acid residues (L440, L441, Q442, and L443) from this leucine-rich segment of RelA(1-450). As shown in Fig. 3A, this modification completely altered the subcellular localization pattern of RelA(1-450) (compare panels a and b). While the unmodified RelA(1-450) mutant exhibited a whole-cell expression pattern, the NES-deficient mutant, RelA(1-450)ΔNES, was located predominantly in the nucleus (Fig. 3). Thus, the NES-like sequence is responsible for the whole-cell expression of RelA(1-450). To examine whether this NES also functions in the full-length RelA, we first generated a RelA containing an N-terminal-tagged NLS derived from the simian virus 40 large T antigen NLS (RelA-NLS). Since the N terminus of RelA is not masked by IκBα (3, 27, 29), the nuclear import of this N-terminal-tagged RelA should not be affected by IκBα. Interestingly, a significant amount of RelA-NLS was still located in the cytoplasm even though it was fused with a strong NLS (Fig. 3A). Furthermore, the cytoplasmic localization pattern of RelA-NLS was completely abolished when the NES of RelA was deleted (Fig. 3A). These results suggest that the NES of RelA functions in both the truncated and full-length forms of RelA.
FIG. 2.
Identification of a NES-like sequence in RelA. The upper panel shows the amino acid sequence of the C-terminal region of RelA mediating its cytoplasmic distribution. The NES-like sequence is indicated. The lower panel shows the sequence homology of the RelA NES with the NES characterized from various other proteins, including the alpha and beta subunits of the protein kinase inhibitor (PKI) (57), the HIV Rev protein (18), and c-Abl (52). The four hydrophobic amino acids, most of which are leucines, are bold and underlined. The NES of RelA is located between amino acids 436 and 445.
FIG. 3.
The NES-like sequence of RelA promotes cytoplasmic localization of RelA as well as p50. (A) COS cells were transfected with cDNA expression vectors encoding the RelA proteins indicated below each of the panels. The cells were stained with anti-RelA plus Texas red-conjugated rabbit IgG (αRelA), counterstained with Hoechst, and visualized as described in the legend for Fig. 1B. (B) COS cells were transfected with cDNA expression vectors encoding the proteins indicated below the panels. The cells were stained with an anti-RelA antiserum and Texas red-conjugated rabbit IgG. The expression of the RelA mutants (a and b) was visualized with a rhodamine filter (αRelA), whereas the expression of p50-GFP fusion proteins (c and d) was visualized via the autofluorescence of GFP by using an FITC filter (GFP). The nuclei of the cells were visualized by DNA staining with Hoechst (DNA).
We then examined whether adding back the NES would alter the subcellular localization pattern of the nuclear forms of RelA. For these studies, the RelA NES was fused to the C terminus of RelA(1-420), a RelA truncation mutant predominantly located in the nucleus (Fig. 1B). Interestingly, attachment of the NES-like sequence to this short form of RelA generated a derivative protein [RelA(1-420)-NES] exhibiting a whole-cell expression pattern (Fig. 3B). To examine whether the NES-like sequence of RelA is sufficient to mediate cytoplasmic expression of heterologous proteins, the effect of this NES sequence on the subcellular localization of p50 was investigated. The RelA NES-like sequence was tagged to the C terminus of a fusion protein composed of p50 and GFP. Like p50, the p50-GFP fusion protein was located in the nucleus (Fig. 3B). However, when tagged with the NES-like sequence, the p50-GFP protein exhibited a whole-cell expression pattern reminiscent of that observed with RelA (Fig. 3B). These results clearly demonstrated that the NES-like sequence of RelA is both required and sufficient to mediate cytoplasmic expression of RelA as well as the heterologous protein p50-GFP.
LMB inhibits the cytoplasmic distribution of RelA.
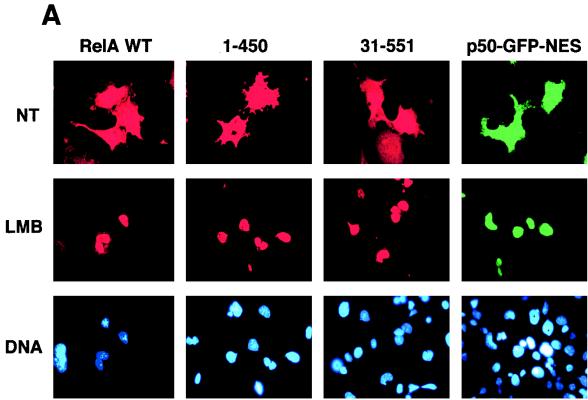
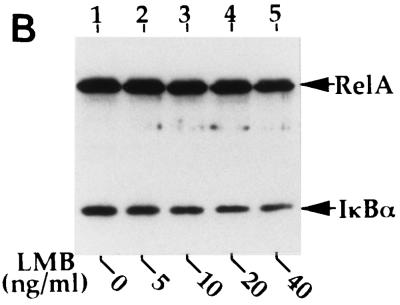
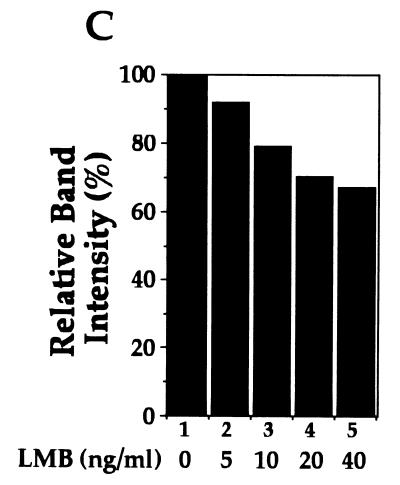
Recent studies demonstrate that the nuclear export of NES-containing proteins is mediated by a receptor protein termed CRM1 (19, 20, 36, 37, 49). A drug, LMB, has been shown to specifically bind CRM1, thereby inhibiting NES-mediated nuclear protein export (20). To further determine whether the cytoplasmic expression of RelA is mediated through its continuous nuclear export, the effect of LMB on the subcellular localization of RelA was examined. Incubation of the RelA-transfected cells with LMB for 6 h markedly inhibited the cytoplasmic distribution of RelA, leading to its nuclear accumulation (Fig. 4A). A shorter period (1 to 2 h) of LMB treatment also yielded a partial effect on RelA subcellular localization, but a maximal effect was detected at 6 h (data not shown), which was also the time period used in other studies (52). Parallel immunoblotting assays revealed that LMB moderately inhibited the inducible expression of IκBα in cells transfected with RelA WT (Fig. 4B and C). However, the IκBα inhibition does not seem to be a major mechanism by which LMB blocks the cytoplasmic expression of RelA. As shown in Fig. 4A, LMB also efficiently blocked the cytoplasmic expression of two truncated forms of RelA [RelA(1-450) and RelA(31-551)], which were defective in IκBα induction (Fig. 1D), as well as p50-GFP-NES. Thus, it is likely that LMB inhibited the nuclear export of these proteins containing the RelA NES.
FIG. 4.
Cytoplasmic expression of RelA is sensitive to a nuclear export inhibitor, LMB. (A) COS cells were transfected with cDNA expression vectors encoding the indicated RelA proteins or p50-GFP-NES. After 48 h, the cells were either not treated (NT) or treated for 6 h with 40 ng of LMB per ml, followed by immunofluorescence staining. The RelA and its mutants were stained with anti-RelA and Texas red-conjugated rabbit IgG and were visualized with a rhodamine filter, while the p50-GFP-NES proteins were directly visualized with an FITC filter. Nuclei of the cells were stained with Hoechst, and the images are shown in the lower panels. (B and C) COS cells were transfected with RelA WT, and 48 h posttransfection, the cells were incubated for 6 h with the indicated amounts of LMB. The expression of the transfected RelA and the induced endogenous IκBα was detected by a Western blot assay with anti-RelA and anti-IκBα (B). The intensity of the protein bands was quantitated by densitometry and presented as a percentage of that from the untreated cells (C).
p50 prevents the cytoplasmic expression of RelA mediated by the putative NES.
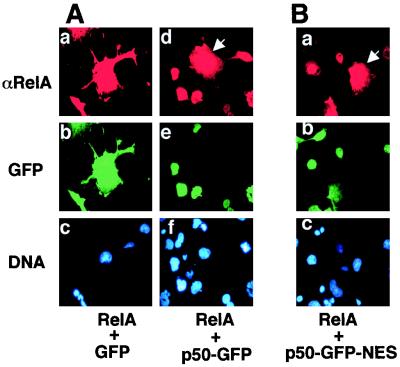
Under physiological conditions, nuclear RelA is present predominantly as a heterodimer with p50. The p50 protein is known to serve as a regulatory subunit modulating the DNA binding affinity of RelA (47). To examine whether p50 also regulates the cytoplasmic and nuclear shuttling of RelA, immunofluorescence assays were performed to examine the effect of p50 on RelA subcellular localization. For these studies, COS cells were transfected with RelA together with cDNA expression vectors encoding either GFP or the p50-GFP fusion protein (Fig. 5A). Expression of RelA in the cells was detected by immunostaining with anti-RelA followed by Texas red-conjugated anti-rabbit IgG, whereas the expression of GFP and p50-GFP was directly visualized via the autofluorescence of GFP (Fig. 5A). As expected, coexpression with GFP did not change the subcellular localization of RelA, which was still largely located in the cytoplasm (Fig. 5A). Interestingly, when RelA was expressed together with p50-GFP, these two NF-κB subunits were colocalized to the nucleus (Fig. 5A). Similar results were obtained with a p50 lacking GFP (data not shown). The nuclear localization of RelA was specifically triggered by expression of p50-GFP in the same cells, since RelA was still retained in the cytoplasm in cells lacking p50-GFP expression (Fig. 5A). Similarly, the cytoplasmic distribution of RelA(1-450) was also inhibited when this truncated form of RelA was coexpressed with p50 (data not shown). Thus, the p50 subunit of NF-κB promotes nuclear accumulation of RelA. To determine whether this specific property of p50 is due to its lack of nuclear export activity, studies were performed to examine the effect of p50-GFP-NES on the subcellular localization of RelA. Unexpectedly, attachment of the RelA NES to p50 did not affect its ability to induce the nuclear accumulation of RelA (Fig. 5B). Furthermore, the NES-mediated cytoplasmic expression of p50-GFP-NES was also inhibited when this fusion protein was coexpressed with RelA (Fig. 5B). These results suggest that lack of a NES is unlikely the molecular basis of p50-mediated stimulation of RelA nuclear accumulation. Further studies showed that the C-terminal truncation mutants RelA(1-312) and RelA(1-420) also induced the nuclear expression of RelA (data not shown). Thus, it seems likely that the lack of the long RelA C-terminal tail sequence in p50 contributes to its specific function in promoting RelA nuclear localization.
FIG. 5.
p50 induces the nuclear accumulation of RelA. (A) COS cells were transfected with RelA together with cDNA expression vectors encoding either GFP (a to c) or p50-GFP (d to f). The cells were stained with the C-terminal specific anti-RelA antibody plus Texas red-conjugated rabbit IgG. The expression of RelA and GFP or p50-GFP in the same cells was visualized with rhodamine (upper panels) and FITC (middle panels) filters, respectively. The nuclei of the cells were visualized by Hoechst staining (lower panels). Note that most of the cells were cotransfected. A cell expressing only RelA is indicated by the arrow. (B) COS cells were transfected with RelA and p50-GFP-NES, and the transfected cells were subjected to immunofluorescence as described above. A cell expressing only RelA is indicated by the arrow, which shows cytoplasmic expression of RelA.
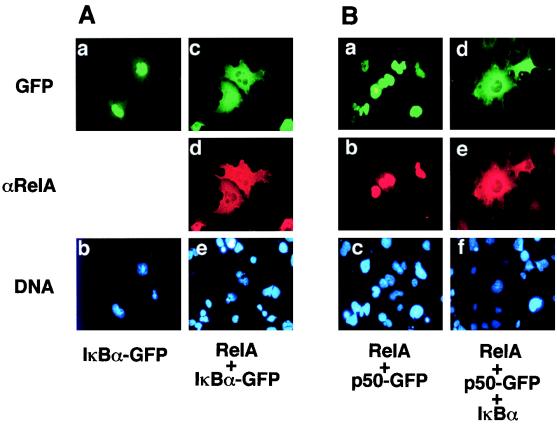
Free IκBα accumulates in the nucleus.
A recent study suggested that IκBα contains a NES which mediates active nuclear export in the Xenopus oocyte (1). To examine whether the nuclear export of IκBα is a dominant event in its subcellular distribution in mammalian somatic cells, immunofluorescence assays were performed with an IκBα-GFP fusion protein. Interestingly, the expressed IκBα fusion protein was predominantly detected in the nucleus (Fig. 6A). As previously demonstrated (21, 61), when IκBα was coexpressed with RelA, the nuclear expression of both proteins was largely blocked (Fig. 6A). Furthermore, when IκBα was coexpressed with RelA in the presence of p50, all three proteins were completely excluded from the nucleus (Fig. 6B and data not shown). Thus, although p50 promotes the nuclear expression of free RelA (Fig. 5A and 6A), it is unable to override the inhibitory function of IκBα. Therefore, regulation of RelA subcellular localization by p50 likely occurs after the degradation of IκBα during a cellular stimulation.
FIG. 6.
Free IκBα accumulates in the nucleus but is excluded from the nucleus when coexpressed with RelA and p50. (A) COS cells were transfected with an expression vector encoding the IκBα-GFP fusion protein either alone (left panels) or together with RelA (right panels). The transfected cells were subjected to immunostaining with anti-RelA, and the subcellular localization of IκBα-GFP and RelA was visualized by a fluorescence microscope with FITC (upper panels) and rhodamine (middle panels) filters, respectively. DNA staining is shown in the lower panels. (B) COS cells were transfected with p50-GFP together with RelA (left panels) or p50-GFP together with RelA and IκBα (right panels). The cells were subjected to immunofluorescence analyses as described above, and the subcellular localization of p50-GFP and RelA was visualized with FITC (upper panels) and rhodamine (middle panels) filters, respectively. DNA staining is shown in the lower panels.
DISCUSSION
The biological activity of NF-κB is regulated at the level of subcellular localization. NF-κB is normally sequestered in the cytoplasmic compartment by physical association with IκBα, which specifically binds to and masks the NLS of both RelA and p50 subunits of NF-κB (7, 21, 61). Upon cellular stimulation, IκBα is rapidly degraded, and the liberated NF-κB heterodimer concomitantly moves to the nucleus. Since both RelA and p50 contain an NLS, it is generally believed that both of these proteins will localize to the nucleus in the absence of IκBα. When expressed in various cell types, p50 is indeed predominantly nuclear (8, 21, 25). Unexpectedly, however, a large proportion of RelA is retained in the cytoplasm even when it is expressed in the absence of IκBα (Fig. 1) (21). This finding prompted us to explore additional mechanisms regulating the subcellular localization of RelA. Our studies demonstrate that RelA contains a leucine-rich sequence homologous to the recently characterized NES (22). This NES-like sequence is both required and sufficient for maintaining the whole-cell expression pattern of RelA. When fused to p50, the RelA NES is able to alter the subcellular localization pattern of p50 from nuclear to whole cell, suggesting that this NES also functions on heterologous proteins. Unlike IκBα and the C-terminal sequences of p105 and p100 (7, 32, 39, 61), the RelA NES-like sequence does not inhibit the nuclear import of RelA or heterologous proteins since these proteins accumulate in the nucleus when nuclear export is blocked by LMB. The LMB-induced nuclear accumulation of RelA and its derivatives is insensitive to the protein synthesis inhibitor cycloheximide (data not shown), suggesting that these NES-containing proteins are continuously shuttling rather than just passing through the nucleus after being newly synthesized.
The finding that LMB causes the nuclear accumulation of RelA WT is somewhat surprising since RelA WT induces expression of endogenous IκBα (Fig. 4A). One possible explanation is that a significant proportion of RelA may be present as free forms in overexpressed cells, which would translocate to the nucleus and accumulate there in the presence of LMB. Another possibility is that the dynamic nature of the RelA-IκBα interaction may allow the occasional release and nuclear translocation of RelA. Of course, the moderate inhibition of IκBα synthesis observed in cells treated with LMB (Fig. 4B) may also contribute in part to the nuclear expression of RelA WT. In any case, our LMB dose-responsive assays show that RelA WT appears to be less sensitive to LMB than p65(1-450) and p50-GFP-NES. At a low concentration (10 ng/ml), LMB induced the complete nuclear expression of the truncated RelA and p50-GFP-NES but only caused a partial effect on RelA WT (data not shown). The lower LMB sensitivity of RelA WT may reflect the effect of endogenous IκBα on RelA nuclear import. Nevertheless, given the strong inhibitory effect of LMB on the cytoplasmic expression of various RelA mutants and p50-GFP-NES, which do not induce IκBα expression, it is likely that the cytoplasmic expression of free RelA is at least partially contributed to by its active nucleus export.
What biological roles can the RelA NES play? First, the NES sequence may function to restrict the nuclear translocation of RelA in the absence of a partner like p50. Indeed, the RelA homodimer is rare in most cell types, whereas the p50-RelA heterodimer is predominant. Second, the NES of RelA may facilitate the nuclear export of RelA and p50 when bound to IκBα in the nucleus. In this regard, a recent study suggests that IκBα contains a NES, which mediates nuclear export of IκBα in the Xenopus oocyte (1). Surprisingly, we have shown that an IκBα-GFP fusion protein accumulates in the nucleus when expressed in COS cells (Fig. 6). Nuclear accumulation of transfected IκBα has also been observed in other studies (15, 61), although a whole-cell expression pattern can be detected under different transfection conditions (43, 55). These studies suggest that a high efficiency of IκBα nuclear export may require its binding to RelA, which provides a second NES. These findings support the notion that newly synthesized free IκBα efficiently enters the nucleus. Binding of IκBα to RelA and p50 may not only block the nuclear translocation of these proteins but also promote the nuclear export of the inactive NF-κB–IκBα complex. In this regard, the NLS of IκBα is located in its second ankyrin repeat (43). Notably, recent structural studies of the IκBα-RelA complex suggest that the first and second ankyrin repeats of IκBα are involved in extensive interaction with the NLS region of RelA (3, 27, 29). IκBα forces the otherwise unstructured RelA NLS and flanking sequences into an α-helical conformation that may not be recognized by nuclear import proteins (3). Thus, formation of the NF-κB–IκBα complex will mask the NLS of both IκBα and the NF-κB subunits. In contrast to the NLS, the putative NES of RelA is located outside of the region involved in binding to IκBα (3, 27, 29). It is likely that the NES-like sequence of RelA is exposed when RelA is bound by IκBα. However, this notion needs to be confirmed by further biochemical or structural analysis since the RelA NES region is not included in the crystal structure of the RelA-IκBα complex. The NES of IκBα is located in its sixth ankyrin repeat (1), which is involved in its physical interaction with RelA and p50 (27, 29). Indeed, the NES (previously named the QL-rich region) of IκBα is essential for its physical association with RelA (50). It remains to be determined whether the NES of IκBα or that of RelA or both are required for the nuclear export of the NF-κB–IκBα complex.
An interesting finding of this study is that p50 promotes the nuclear accumulation of RelA. When coexpressed in cells, RelA and p50 are colocalized in the nucleus, while free RelA exhibits a whole-cell expression pattern. The mechanism mediating this function of p50 remains unclear. Since p50 contains an NLS but lacks a NES, the RelA-p50 heterodimer possesses a higher NLS-to-NES ratio (2:1) than the RelA-RelA homodimer (2:2). However, this structural difference does not seem to contribute to the strong nuclear distribution activity of the RelA-p50 heterodimer. As shown in Fig. 5B, fusion of the RelA NES to p50 did not affect the activity of p50 to stimulate the nuclear accumulation of RelA, although the NES did cause cytoplasmic expression of free p50 (Fig. 3B). Additionally, the subcellular localization of p50 and RelA appear to be mutually regulated, since both p50-NES and RelA are located in the nucleus when they are coexpressed (Fig. 5B). Thus, the mechanism by which p50 promotes RelA nuclear localization appears to be complex. One possibility is that heterodimer formation induces conformational changes in RelA and p50, which may result in the masking of the NES and better exposure of the NLS of these proteins. Alternatively, the combination of a p50 NLS with a RelA NLS may favor the nuclear localization of the NF-κB heterodimer. Nevertheless, our finding suggests that heterodimer formation serves as a step in the regulation of NF-κB nuclear expression.
ACKNOWLEDGMENTS
We greatly acknowledge M. Yoshida for providing the LMB, W. C. Greene for the RelA expression vectors, and Dave Antonetti and the Penn State Retina Research Group for the use of their microscope.
E.W.H. is supported by NIH predoctoral training grant 5 T32 CA 6039-5. This study was supported by Public Health Service grant 1 R01 CA68471 to S.-C.S.
REFERENCES
- 1.Arenzana-Seisdedos F, Turpin P, Rodriguez M, Thomas D, Hay R T, Virelizier J-L, Dargemont C. Nuclear localization of IκBα promotes active transport of NF-κB from the nucleus to the cytoplasm. J Cell Sci. 1997;110:369–378. doi: 10.1242/jcs.110.3.369. [DOI] [PubMed] [Google Scholar]
- 2.Baeuerle P, Baichwal V R. NF-κB as a frequent target for immunosuppressive and anti-inflammatory molecules. Adv Immunol. 1997;65:111–137. [PubMed] [Google Scholar]
- 3.Baeuerle P A. IκB-NF-κB structures: at the interface of inflammation control. Cell. 1998;95:729–731. doi: 10.1016/s0092-8674(00)81694-3. [DOI] [PubMed] [Google Scholar]
- 4.Baeuerle P A, Baltimore D. A 65-kD subunit of active NF-κB is required for inhibition of NF-κB by IκB. Genes Dev. 1989;3:1689–1698. doi: 10.1101/gad.3.11.1689. [DOI] [PubMed] [Google Scholar]
- 5.Baldwin A S., Jr The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 6.Ballard D W, Dixon E P, Peffer N J, Bogerd H, Doerre S, Stein B, Greene W C. The 65-kDa subunit of human NF-κB functions as a potent transcriptional activator and a target for v-Rel-mediated repression. Proc Natl Acad Sci USA. 1992;89:1875–1879. doi: 10.1073/pnas.89.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beg A A, Ruben S M, Scheinman R I, Haskill S, Rosen C A, Baldwin A S., Jr IκB interacts with the nuclear localization sequence of the subunits of NF-κB: a mechanism for cytoplasmic retention. Genes Dev. 1992;6:1899–1913. doi: 10.1101/gad.6.10.1899. [DOI] [PubMed] [Google Scholar]
- 8.Blank V, Kourilsky P, Isräel A. Cytoplasmic retention, DNA-binding and processing of the NF-κB p50 precursor are controlled by a small region in its C-terminus. EMBO J. 1991;10:4159–4167. doi: 10.1002/j.1460-2075.1991.tb04994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D A. Coupling of a signal response domain in IκBα to multiple pathways for NF-κB activation. Mol Cell Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U. Control of IκB-α proteolysis by site-specific, signal-induced phosphorylation. Science. 1995;267:1485–1488. doi: 10.1126/science.7878466. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z, Hagler J, Palombella V, Melandri F, Scherer D, Ballard D, Maniatis T. Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z J, Parent L, Maniatis T. Site specific phosphorylation of IκBα by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 13.Chiao P J, Miyamoto S, Verma I M. Autoregulation of IκBα activity. Proc Natl Acad Sci USA. 1994;91:28–32. doi: 10.1073/pnas.91.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen L, Henzel W J, Baeuerle P A. IKAP is a scaffold protein of the IkappaB kinase complex. Nature. 1998;17:292–296. doi: 10.1038/26254. [DOI] [PubMed] [Google Scholar]
- 15.Cressman D E, Taub R. IκBα can localize in the nucleus but shows no direct transactivation potential. Oncogene. 1993;8:2567–2573. [PubMed] [Google Scholar]
- 16.DiDonato J, Mercurio F, Rosette C, Wu-Li J, Suyang H, Ghosh S, Karin M. Mapping of the inducible IκB phosphorylation sites that signal its ubiquitination and degradation. Mol Cell Biol. 1996;16:1295–1304. doi: 10.1128/mcb.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DiDonato J A, Hayakawa M, Rothwarf D M, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- 18.Fischer U, Huber J, Boelens W C, Mattaj I W, Lührmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 19.Fornerod M, Ohno M, Yoshida M, Mattaj I W. Crm1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 20.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 21.Ganchi P A, Sun S-C, Greene W C, Ballard D W. IκB/MAD-3 masks the nuclear localization signal of NF-κB p65 and acts with the C-terminal activation domain to inhibit NF-κB p65 DNA binding. Mol Biol Cell. 1992;3:1339–1352. doi: 10.1091/mbc.3.12.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerace L. Nuclear export signals and the fast track to the cytoplasm. Cell. 1995;82:341–344. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- 23.Gerondakis S, Grumont R, Rourke I, Grossmann M. The regulation and roles of Rel/NF-κB transcription factors during lymphocyte activation. Curr Opin Immunol. 1998;10:353–359. doi: 10.1016/s0952-7915(98)80175-1. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh S, May M J, Kopp E B. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 25.Henkel T, Zabel U, van Zee K, Müller J M, Fanning E, Baeuerle P A. Intramolecular masking of the nuclear location signal and dimerization domain in the precursor for the p50 NF-κB subunit. Cell. 1992;68:1121–1133. doi: 10.1016/0092-8674(92)90083-o. [DOI] [PubMed] [Google Scholar]
- 26.Holbrook N, Gulino A, Ruscetti F. Cis-acting transcriptional regulatory sequences in the gibbon ape leukemia virus (GALV) long terminal repeat. Virology. 1987;157:211–219. doi: 10.1016/0042-6822(87)90330-8. [DOI] [PubMed] [Google Scholar]
- 27.Huxford T, Huang D-B, Malek S, Ghosh G. The crystal structure of the IκBα/NF-κB complex reveals mechanisms of NF-κB inactivation. Cell. 1998;95:759–770. doi: 10.1016/s0092-8674(00)81699-2. [DOI] [PubMed] [Google Scholar]
- 28.Israël A. A role for phosphorylation and degradation in the control of NF-κB activity. Trends Genet. 1995;11:203–205. doi: 10.1016/s0168-9525(00)89045-9. [DOI] [PubMed] [Google Scholar]
- 29.Jacobs M D, Harrison S C. Structure of an IκBα/NF-κB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- 30.Kaufman J D, Valanora G, Rodriguez G, Bushar G, Giri C, Norcross M D. Phorbol ester enhances human immunodeficiency virus-promoted gene expression and acts on a repeated 10-base-pair functional enhancer element. Mol Cell Biol. 1987;7:3759–3766. doi: 10.1128/mcb.7.10.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LeBail O, Schmidt-Ullrich R, Israël A. Promoter analysis of the gene encoding the IκB-α/MAD3 inhibitor of NF-κB: positive regulation by members of the rel/NF-κB family. EMBO J. 1993;12:5043–5049. doi: 10.1002/j.1460-2075.1993.tb06197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercurio F, DiDonato J A, Rosette C, Karin M. p105 and p98 precursor proteins play an active role in NF-κB-mediated signal transduction. Genes Dev. 1993;7:705–718. doi: 10.1101/gad.7.4.705. [DOI] [PubMed] [Google Scholar]
- 33.Mercurio F, Zhu H, Murray B W, Shevchenko A, Bennett B L, Li J W, Young D B, Barbose M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science. 1997;278:860–865. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 34.Muesing M A, Smith D H, Capon D J. Regulation of mRNA accumulation by human immunodeficiency virus transactivator protein. Cell. 1987;48:691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- 35.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 36.Neville M S, Stutz F, Lee L, Davis L I, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 37.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of Crm1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 38.Regnier C H, Song H Y, Cao Z, Rothe M. Identification and characterization of an IkB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- 39.Rice N R, MacKichan M L, Israel A. The precursor of NF-κB p50 has IκB-like functions. Cell. 1992;71:243–253. doi: 10.1016/0092-8674(92)90353-e. [DOI] [PubMed] [Google Scholar]
- 40.Roff M, Thompson J, Rodriguez M S, Jacque J-M, Baleux F, Arenzana-Seisdedos F, Hay R T. Role of IκBα ubiquitination in signal-induced activation of NF-κB in vivo. J Biol Chem. 1996;271:7844–7850. doi: 10.1074/jbc.271.13.7844. [DOI] [PubMed] [Google Scholar]
- 41.Rothwarf D M, Zandi E, Natoli G, Karin M. IKK-gamma is an essential regulatory subunit of the IkappaB kinase complex. Nature. 1998;17:297–300. doi: 10.1038/26261. [DOI] [PubMed] [Google Scholar]
- 42.Ruben S M, Narayanan R, Klement J F, Chen C-H, Rosen C A. Functional characterization of the NF-κB p65 transcriptional activator and an alternatively spliced derivative. Mol Cell Biol. 1992;12:444–454. doi: 10.1128/mcb.12.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sachdev S, Hoffmann A, Hannink M. Nuclear localization of IκBα is mediated by the second ankyrin repeat: the IκBα ankyrin repeats define a novel class of cis-acting nuclear import sequences. Mol Cell Biol. 1998;18:2524–2534. doi: 10.1128/mcb.18.5.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scherer D C, Brockman J A, Chen A, Maniatis T, Ballard D W. Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc Natl Acad Sci USA. 1995;92:11259–11263. doi: 10.1073/pnas.92.24.11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitz M L, Baeuerle P. The p65 subunit is responsible for the strong transcription activating potential of NF-κB. EMBO J. 1991;10:3805–3817. doi: 10.1002/j.1460-2075.1991.tb04950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scott M L, Fujita T, Liou H-C, Nolan G P, Baltimore D. The p65 subunit of NF-κB regulates IκB by two distinct mechanisms. Genes Dev. 1993;7:1266–1276. doi: 10.1101/gad.7.7a.1266. [DOI] [PubMed] [Google Scholar]
- 47.Siebenlist U, Franzoso G, Brown K. Structure, regulation and function of NF-κB. Annu Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 48.Siekevitz M, Josephs S F, Dukovich M, Peffer N J, Wong-Staal F, Greene W C. Activation of the HIV-1 LTR by T-cell mitogens and the tat-I protein of HTLV-I. Science. 1987;238:1575–1578. doi: 10.1126/science.2825351. [DOI] [PubMed] [Google Scholar]
- 49.Stade K, Ford C S, Guthrie C, Weiss K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 50.Sun S-C, Elwood J, Greene W C. Both amino- and carboxyl-terminal sequences within IκBα regulate its inducible degradation. Mol Cell Biol. 1996;16:1058–1065. doi: 10.1128/mcb.16.3.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun S-C, Ganchi P A, Ballard D W, Greene W C. NF-κB controls expression of inhibitor IκBα: evidence for an inducible autoregulatory pathway. Science. 1993;259:1912–1915. doi: 10.1126/science.8096091. [DOI] [PubMed] [Google Scholar]
- 52.Taagepera S, McDonald D, Loeb J E, Whitaker L L, McElroy A K, Wang J Y J, Hope T J. Nuclear-cytoplasmic shuttling of c-ABL tyrosine kinase. Proc Natl Acad Sci USA. 1998;95:7457–7462. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tong-Starksen S E, Luciw P A, Peterin B M. Human immunodeficiency virus long terminal repeat responds to T cell activation signals. Proc Natl Acad Sci USA. 1987;84:6845–6849. doi: 10.1073/pnas.84.19.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Traenckner E B-M, Pahl H L, Henkel T, Schmidt K N, Wilk S, Baeuerle P A. Phosphorylation of human IκB-α on serine 32 and 36 controls IκB-α proteolysis and NF-kB activation in response to diverse stimuli. EMBO J. 1995;14:2876–2883. doi: 10.1002/j.1460-2075.1995.tb07287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turpin P, Hay R T, Dargemont C. Characterization of IκBα nuclear import pathway. J Biol Chem. 1999;274:6804–6812. doi: 10.1074/jbc.274.10.6804. [DOI] [PubMed] [Google Scholar]
- 56.Verma I M, Stevenson J K, Schwarz E M, Antwerp D V, Miyamoto S. Rel/NF-κB/IκB family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 57.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 58.Woronicz J D, Gao X, Cao Z, Rothe M, Goeddel D V. IκB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science. 1997;278:866–869. doi: 10.1126/science.278.5339.866. [DOI] [PubMed] [Google Scholar]
- 59.Wulczyn F G, Krappmann D, Scheidereit C. The NF-kappa B/Rel and I kappa B gene families: mediators of immune response and inflammation. J Mol Med. 1996;74:749–769. doi: 10.1007/s001090050078. [DOI] [PubMed] [Google Scholar]
- 60.Yamaoka S, Courtois G, Bessia C, Whiteside S T, Weil R, Agou F, Kirk H E, Kay R J, Israel A. Complementation cloning of NEMO, a component of the IkappaB kinase complex essential for NF-kappaB activation. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 61.Zabel U, Henkel T, dos Santos Silva M, Baeuerle P. Nuclear uptake control of NF-κB by MAD-3, an IκB protein present in the nucleus. EMBO J. 1993;12:201–211. doi: 10.1002/j.1460-2075.1993.tb05646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zandi E, Rothwarf D M, Delhase M, Hayakawa M, Karin M. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell. 1997;91:243–252. doi: 10.1016/s0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]