Abstract
The calcium ionophore ionomycin cooperates with the S100B protein to rescue a p53-dependent G1 checkpoint control in S100B-expressing mouse embryo fibroblasts and rat embryo fibroblasts (REF cells) which express the temperature-sensitive p53Val135 mutant (C. Scotto, J. C. Deloulme, D. Rousseau, E. Chambaz, and J. Baudier, Mol. Cell. Biol. 18:4272–4281, 1998). We investigated in this study the contributions of S100B and calcium-dependent PKC (cPKC) signalling pathways to the activation of wild-type p53. We first confirmed that S100B expression in mouse embryo fibroblasts enhanced specific nuclear accumulation of wild-type p53. We next demonstrated that wild-type p53 nuclear translocation and accumulation is dependent on cPKC activity. Mutation of the five putative cPKC phosphorylation sites on murine p53 into alanine or aspartic residues had no significant effect on p53 nuclear localization, suggesting that the cPKC effect on p53 nuclear translocation is indirect. A concerted regulation by S100B and cPKC of wild-type p53 nuclear translocation and activation was confirmed with REF cells expressing S100B (S100B-REF cells) overexpressing the temperature-sensitive p53Val135 mutant. Stimulation of S100B-REF cells with the PKC activator phorbol ester phorbol myristate acetate (PMA) promoted specific nuclear translocation of the wild-type p53Val135 species in cells positioned in early G1 phase of the cell cycle. PMA also substituted for ionomycin in the mediating of p53-dependent G1 arrest at the nonpermissive temperature (37.5°C). PMA-dependent growth arrest was linked to the cell apoptosis response to UV irradiation. In contrast, growth arrest mediated by a temperature shift to 32°C protected S100B-REF cells from apoptosis. Our results suggest a model in which calcium signalling, linked with cPKC activation, cooperates with S100B to promote wild-type p53 nuclear translocation in early G1 phase and activation of a p53-dependent G1 checkpoint control.
The tumor suppressor p53 protein has been implicated in cell differentiation (1, 50), cell contact inhibition of growth (65), protection of the cell from the acquisition of genomic abnormalities (32, 33), and cell senescence (53). The mechanisms by which p53 carries out these functions seem to be related to its ability to induce G1 or G2/M cell cycle arrest and/or apoptosis. In some systems, p53-dependent growth arrest may inhibit apoptosis and favor viable cell cycle arrest (46, 49). The diversity of cellular responses to p53 activation indicates that the outcome of p53 activation depends on other signalling pathways upstream and downstream to p53 activation (2, 27). The extremely short half-life of the p53 protein in normal cells suggests that multiple, transient, and probably interdependent control processes regulate cellular p53 at the levels of its synthesis, cytoplasmic anchorage, nuclear translocation, nuclear activities, and degradation. Conformational modulation of p53 between wild-type and mutant-like conformations has also recently emerged as a possible mechanism for regulation of p53 functions (50). Activation of p53 functions following an appropriate stimulus generally initiates a rapid and substantial increase in the total p53 level, achieved at least in part by the stabilization of the normally rapidly degraded wild-type p53 protein in the cell nuclei. On the other hand, stabilization of p53 protein in the absence of stimulus is always a hallmark of loss of function which can occur after gene mutation or interaction with viral oncoprotein (reviewed in reference 7). In tumor cells harboring wild-type and mutant p53 alleles, mutant p53 accumulates in the cell nuclei and acts as a negative dominant fashion and abolishes the functions of the wild-type protein. There is thus considerable interest in understanding the intracellular signalling pathways and mechanisms responsible for conformational stabilization and selective nuclear accumulation of the wild-type p53 conformational species versus those of mutant p53 molecules. We have previously shown that the calcium- and zinc-binding S100B protein (3) can be implicated in activation of wild-type p53 functions (52). The S100B protein is found in astroglial cells in the central nervous system but also in a number of tissues outside the nervous system (42, 43). The synthesis of S100B is tightly regulated. Many cellular stimulations known to activate p53, such as cell contact (65), hypoxia (20), and UV irradiation (56), are also able to stimulate S100B expression (51, 52, 59, 60). In the central nervous system, both S100B and p53 are up regulated in neurodegenerative diseases and might synergize in mechanisms of cell death (9, 26, 52, 54). A functional interaction between S100B and p53 in negative cell growth regulation and cell death was recently demonstrated in p53-negative (p53−/−) mouse embryo fibroblasts (MEF cells) by sequential transfection with the S100B and temperature-sensitive (ts) p53val135 genes and in the rat embryo fibroblast (REF) cell line clone 6, which is transformed by oncogenic Ha-ras and overexpression of p53Val135 (52). Ectopic expression of S100B in clone 6 cells (S100B-REF cells) reverts transformed phenotypes characterized by the rescue of cell density-dependent inhibition of growth (52) and of a G2/M checkpoint in response to double-strand DNA breaks (unpublished data). Moreover, ionomycin stimulation of S100B-MEF and S100B-REF cells was able to rescue a p53-dependent G1 checkpoint control (52). Intracellular calcium elevation mediated by ionomycin not only activates calcium-binding proteins but also contributes to the activation of calcium-dependent protein kinase C (cPKC) isozymes (44, 45). An interdependence is thought to exist between S100B and cPKC-dependent signalling pathways (5, 10, 60). Hence, cPKC activation might also account for the effect of ionomycin on activation of a G1 checkpoint in S100B-MEF and S100B-REF cells. PKC is a family of calcium/diacylglycerol-dependent serine/threonine kinases which play a central role in signal transduction and have been widely implicated in control of cell growth, differentiation, transformation, and apoptosis. The 11 known PKC isoenzymes are classified into three groups: the conventional cPKCs (isoforms α, β, and γ), the novel calcium-independent PKCs (nPKCs; δ, ɛ, and μ), and the atypical PKCs (λ and ζ) (reviewed in reference 23). Initial interest in PKC stemmed from its identification as the major cellular receptor for tumor-promoting phorbol esters (phorbol myristate acetate [PMA]), which act by binding to the diacylglycerol-binding site on the enzymes and subsequently promoting their activation (45). Bryostatin, a macrocyclic lactone with a structure significantly different from that of phorbol ester, was also found to bind and activate PKCs (28). PKC activation can lead to disordered growth, cell transformation, and inhibition of apoptosis (36). On the other hand, PKC activation can also promote cell growth inhibition and induction of apoptosis (18, 39, 66, 67). A key to understanding these diverse responses may be that individual PKC isoenzymes play specific and specialized roles in cell signalling. It is also likely that the genetic background of the cells under investigation and the contribution of other intracellular signalling pathways have a profound effect on the response of the cells to PKC activation. Finally, it is not known whether the long-term effects of PKC activators are due to activation or depletion of PKCs. Because of the importance of PKC isoenzymes in major cellular functions, they have been considered potential targets for therapeutic intervention (23). An important issue is now to characterize the PKC isoenzyme-specific functions and their relationships with other signalling pathways.
We have investigated the contributions of S100B and cPKC signalling pathways to the activation of the wild-type p53. We show that cPKC activation cooperates with S100B in regulating wild-type p53 nuclear translocation and accumulation in early G1 phase of the cell cycle. Moreover, we provide evidence that cPKC-mediated nuclear translocation of the wild-type p53 in early G1 is linked with activation of a p53-dependent G1 checkpoint control.
MATERIALS AND METHODS
Cell cultures.
REF (clone 6) (40) and S100B-REF cells (clones 6β and 9β) (52) cells were grown in RPMI-Glutamax (Gibco) supplemented with 5% fetal calf serum (FCS; Seromed) at 37.5°C. Hygromycin-resistant MEF cells not expressing S100B (clone C) and S100B-MEF cells not expressing (clone J-β) or expressing (clone Jβ2p53) p53Val135 (52) were grown in Dulbecco modified Eagle medium (DMEM)-Glutamax (Gibco) supplemented with 10% FCS (Gibco).
Plasmids and antibodies.
A plasmid encoding mouse wild-type p53 was constructed by inserting full-length p53 cDNA into pUTSV1 (Eurogentech, Seraing, Belgium) under the control of the enhancer and promoter from simian virus 40 (SV40). p53-Ala and p53-Asp mutants were made by PCR using oligonucleotides CGGAATTCCAGTCAGTCTGAGTCAGGCCCCACTTTCTTGACCATTGCTTTTTTATGGCGGGCAGCAGCCTGGCCCTTCTTGGCCTTCAGGTAGCTGGCGTGAGCCCTGCTGTCTCC and CGGAATTCGAGTCAGTCTGAG TCAGGCCCCACTTTCTTGACCATGTCTTTTTTATGGCGGTCATCATC C TGGCCC T TC T TG TCC T TCAGG TAGC TGTCG TGAGCCC TGCTG TC TCC, respectively. The fidelity of PCR synthesis was confirmed by sequencing plasmid constructs. Further details regarding the cloning are available upon request.
The green fluorescent protein (GFP) expression plasmid pEGFP-N1 was purchased from Clontech. The luciferase expression plasmid PGL3-control, encoding luciferase under the control of the SV40 enhancer and promoter, was from Promega. p53-specific monoclonal antibodies PAb240, PAb246, PAb242, and PAb421 were purified from hybridoma supernatants by protein A-agarose chromatography. Affinity-purified polyclonal rabbit anti-p21 and anti-p27 antibodies were from Santa Cruz Biotechnology. Anti-Rb (clone PMG3-245) was from Pharmingen. Anti-PKCs and rat brain extracts used as controls were from Transduction Laboratory. Anti-β-tubulin was a gift from L. Paturle and D. Job.
Transfections.
MEF cells and S100B-MEF cells grown in DMEM-Glutamax supplemented with 10% FCS in 60-mm-diameter dishes at 37.5°C were transiently transfected with Fugene-6 (Boehringer); 1 to 4 μg of wild-type p53 plasmid was cotransfected with 0.5 μg of pEGFP-N1 or PGL3-control in the presence of 12 μl of Fugene-6 as recommended by the manufacturer. Cells were harvested 36 h after transfection. We found that 30 to 50% of cells were efficiently transfected in these experimental conditions.
Protein concentration and Western blot analysis.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer, and protein concentration was estimated by the bicinchoninic acid method (Pierce), using bovine serum albumin as a standard. Western blot analysis used cell extracts in RIPA buffer mixed with an equal volume of 1% sodium dodecyl sulfate (SDS) containing 20% glycerol, 50 mM dithiothreitol (DTT), and a trace of bromophenol blue. Samples were boiled, and 50 μg of protein was loaded on SDS-containing 12% (p21, p27, tubulin, and Mdm2) or 7.5% (Rb) polyacrylamide gels. Proteins were transferred to nylon membrane.
Flow cytometry.
Cell cycle and cell sorting analysis by flow cytometry was performed on a FACStar+ (Becton Dickinson). For cell cycle parameter analysis, cells were collected in phosphate-buffered saline (PBS), rapidly vortexed with 0.2% Triton X-100, and fixed with 4% formaldehyde. DNA was stained with Hoechst 33258 (2 μg/ml) just prior to flow cytometry analysis.
Mitotic detachment.
Cells were grown at 37.5°C to 80% confluence. The plates were gently shaken for 5 s. The media containing mitotic cells were pooled and centrifuged at low speed. Cell pellets were resuspended in culture medium. This procedure yielded populations that consisted of early G1 and mitotic cells (37). Mitotic cells were seeded on polylysine (0.1 mg/ml)-coated Permanox slides from Nunc, Inc.
Immunofluorescence cell staining.
Cells were fixed with 4% paraformaldehyde for 30 min and permeabilized for 3 min with 0.2% Triton X-100. After washing with PBS, cells were incubated at 4°C overnight in PBS containing 5% goat serum with either purified wild-type specific monoclonal antibody PAb246 or purified mutant specific monoclonal antibody PAb240 (1 μg/ml). The cells were then washed five times with PBS and incubated for 1 h with fluorescein isothiocyanate or cyanin 3-conjugated secondary antibodies. Coverslips were mounted in Aquamount and observed on a Bio-Rad confocal microscope or a Zeiss Axioplan microscope (×40) equipped with an exposure command system MC80; 400 ASA color slides were used.
Nuclear extracts.
Nuclear extracts were prepared as previously described (11), with minor modifications. S100B-REF cells (clone 6β) were grown to 80% confluence in 100-mm-diameter dishes and labeled in methionine-free medium supplemented with [35S]Met-Cys mix (50 μCi/ml) for 3 h. Cells were or were not stimulated with PMA, washed once with PBS, and frozen by putting the culture dishes on a layer of liquid nitrogen. Cells were immediately thawed in 1 ml of buffer A (20 mM Tris-HCl [pH 7.6], 0.2% Triton X-100, 12% sucrose, 2 mM EGTA, 1 mM Pefabloc, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml, 1 μM microcystin, 1 mM vanadate, 1 mM NaF). Nuclei were pelleted by low-speed centrifugation and lysed in 200 μl of buffer B (10 mM Tris-HCl [pH 7.5], 0.5 M NaCl, 1 mM Pefabloc, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml, 1 μM microcystin, 1 mM vanadate, 1 mM NaF). After centrifugation at 200,000 × g for 20 min to remove DNA, the supernatant (200 μl) was diluted in 600 μl of buffer C (20 mM Tris-HCl [pH 7.4], 5 mM MgCl2, 5% glycerol, 0.5% NP-40, 1 mM Pefabloc, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml, 1 μM microcystin, 1 mM vanadate, 1 mM NaF). Diluted nuclear extracts were then centrifuged for 15 min at 20,000 × g. The supernatants were used either for immunoprecipitation or for DNA-binding studies.
S100B-MEF cells were labeled with [35S]Met-Cys mix (100 μCi/ml) for 5 h. Nuclear extracts were prepared as described above except that the freezing step was omitted.
Immunoprecipitation.
Nuclear extracts were incubated with purified p53 monoclonal antibodies (5 μg/ml) and protein G-agarose for 30 min. The immunoprecipitates were washed three times with 1 ml of buffer C. 35S-labeled immunoprecipitated proteins were resuspended in 1% SDS–10 mM DTT and analyzed on SDS–12% polyacrylamide gels.
DNA-binding studies.
A biotinylated double-stranded oligonucleotide corresponding to the sequence 5′-biotin-TTTTTTTGCAGGAATTCGATAGGCATGTCTAGGCATGTCTATCAAGCTTATCGAT-3′ was synthesized; it comprises the consensus binding site for p53 (16). Nuclear extracts were incubated for 30 min with biotin-DNA probe (0.6 μg/ml), salmon sperm DNA (20 μg/ml), and streptavidin-agarose. The streptavidine-agarose was then washed three times with 1 ml of buffer C. 35S-labeled proteins bound to biotin-target DNA were resuspended in 1% SDS–10 mM DTT and analyzed on SDS–12% polyacrylamide gels.
RESULTS
Contribution of S100B to wild-type p53 accumulation.
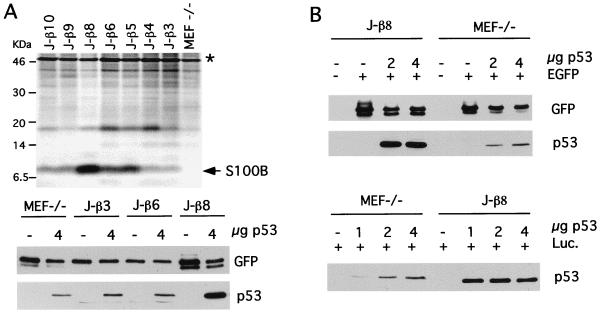
To investigate the contribution of S100B to wild-type p53 accumulation, we used hygromycin-resistant p53−/−MEF cells either expressing or not expressing S100B (52). After a few passages in culture, p53−/−MEF cells stably transfected with the S100B gene drastically lost S100B expression, suggesting that S100B synthesis is detrimental for cell growth. S100B-producing clone J-β cells (52) were therefore subcloned by limiting dilution, and seven subclones were selected and analyzed for S100B production. Only one clone (J-β8) still expressed a significant amount of S100B (Fig. 1A, upper panel). Other selected clones were characterized by drastic down regulation of S100B synthesis. We next compared p53 levels in MEF cells not expressing (MEF−/−) or expressing different S100B levels (clone J-β3, J-β6, and J-β8 cells). Cells were transfected with a plasmid encoding wild-type murine p53 under the control of the SV40 enhancer and promoter, which allow low protein expression. Transfection efficiencies were evaluated with a plasmid encoding enhanced GFP (EGFP) under the control of cytomegalovirus (CMV) promoter. Stronger accumulation of p53 was observed in S100B-producing J-β8 cells than in parental MEF cells or clone J-β3 and clone J-β6 cells that down regulated S100B expression (Fig. 1A, lower panel). This observation was confirmed with experiments utilizing different p53 plasmid concentrations and transfection efficiencies evaluated either with the plasmid encoding EGFP under the control of CMV promoter (Fig. 1B, upper panel) or a plasmid encoding luciferase under the control of the enhancer and promoter from SV40 (Fig. 1B, lower panel). We controlled so that S100B has no effect on CMV and SV40 promoters.
FIG. 1.
S100B stabilizes wild-type p53. (A) The upper panel shows a comparison of S100B contents in MEF−/− cells and J-β subclones selected by limiting dilution. Cells were metabolically labeled with [35S]Met-Cys mix, and S100B was immunoprecipitate with rabbit polyclonal anti-S100B antibody as previously described (52). The asterisk indicates nonspecific binding. The lower panel shows levels of p53 in MEF−/− cells and J-β3, J-β6, and J-β8 subclones following transient transfection as determined by Western blotting. Transfection efficiencies were determined by analysis of GFP expression. (B) Levels of p53 in S100B-MEF (clone J-β8) cells and MEF−/− cells in transient transfection assays using different p53 plasmid concentrations. Upper panel, transfection efficiencies determined by Western blot analysis of GFP expression in parallel with that of p53; lower panel, transfection efficiencies determined by analyzing luciferase activities for each cell line, using a plasmid encoding luciferase (Luc.) under the control of the SV40 enhancer and promoter. Note that p53 expression in S100B-producing J-β8 cells resulted in cell growth arrest with a high incidence of cell death and decrease in luciferase activities. Hence, loading samples for Western blot analysis of p53 were adjusted to equal luciferase activities determined in the absence of p53 plasmids.
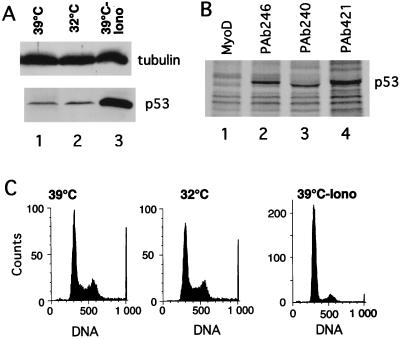
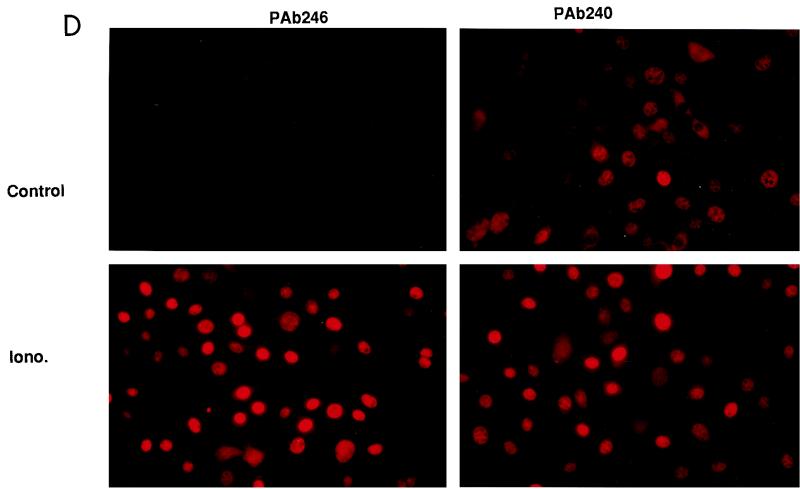
The S100B-dependent accumulation of p53 in MEF cells is reminiscent of that observed with the ts p53Val135 mutant (52). Expression of S100B in MEF cells cooperates with the calcium ionophore ionomycin to induce specific accumulation of the wild-type p53Val135 conformational species and to rescue wild-type p53 functions at the nonpermissive temperature 37.5°C (52). At 37.5°C, a significant amount of the p53Val135 can be folded under a wild-type conformation (37). Hence, one cannot exclude the possibility that ionomycin activates such a minor wild-type p53 population independently of S100B. To investigate whether S100B contributes to the nuclear accumulation and activation of p53Val135 under a wild-type conformation, we reproduced the experiment at 39°C, a temperature far above the nonpermissive temperature. Ionomycin stimulation produced a drastic accumulation of the p53Val135 protein (Fig. 2A). Accumulation of p53Val135 correlated with accumulation of the cells in the G1 phase of the cell cycle (Fig. 2C). Both immunoprecipitation (Fig. 2B) and indirect immunofluorescence analysis (Fig. 2D) revealed that at 39°C, ionomycin caused specific nuclear accumulation of p53Val135 with a wild-type conformation (PAb246+), resulting in high wild-type/mutant ratios in the cell nuclei. Note that in control S100B-MEF cells grown at 39°C, the level of wild-type p53Val135 was below the detection limit (Fig. 2D). Note also that in control cells, the PAb240 immunoreactivity accumulated in the cell nuclei. Only a small number of cells with small rounded nuclei showed cytoplasmic staining; these cells were most likely in early G1 phase of the cell cycle (see also Fig. 4).
FIG. 2.
Ionomycin stimulation, but not permissive temperature, promotes specific activation of wild-type p53 in S100B-MEF cells. (A) Comparison of p53Val135 contents in clone J-β2p53 cells grown at 39°C (lane 1), kept at 32°C for 22 h (lane 2), or stimulated with ionomycin (Iono) for 22 h at 39°C (lane 3). (B) Ionomycin stimulation promotes nuclear accumulation of the wild-type p53Val135 species in S100B-MEF cells grown at 39°C. Clone J-β2p53 cells grown at 39°C were stimulated for 12 h with 1 μM ionomycin prior to labeling with [35S]Met-Cys mix (100 μCi/ml) for 5 h. Nuclear extracts were prepared, and 35S-labeled p53Val135 was immunoprecipitated with anti-MyoD immunoglobulin G used as a control (lane 1), the wild-type-specific PAb246 (lane 2), the mutant-specific PAb240 (lane 3), or the pan-specific PAb421 (lane 4). (C) Flow cytometry analysis of the DNA content of clone J-β2P53 cells grown at 39°C (39°C), shifted to 32°C for 24 h (32°C), or grown at 39°C and stimulated with ionomycin for 36 h (39°C-Iono). (D) Immunofluorescence analysis of PAb246 and PAb240 immunoreactivities in subconfluent clone J-β2p53 cells grown at 39°C not stimulated (control) or stimulated with 1 μM ionomycin (Iono.) for 20 h.
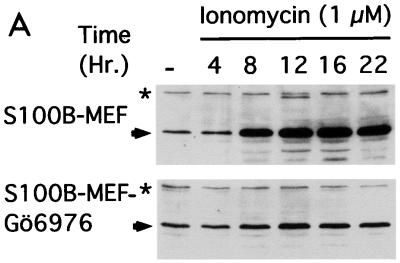
FIG. 4.
Down regulation of cPKC by Gö6976 counteracts nuclear accumulation of the wild-type p53Val135 conformational species in S100B-MEF cells. (A) Western blot analysis of p53Val135 protein accumulation in clone J-β2p53 cells stimulated with 1 μM ionomycin in the absence (S100B-MEF) or in the presence of 1 μM Gö6976 (S100B-MEF-Gö6976). p53 was detected by a mixture of monoclonal antibodies PAb421 and PAb240. The asterisk indicates a cross-reacting protein that serves as internal loading control. (B) Microscopic analysis of PAb246 immunoreactivities in S100B-MEF clone J-β2p53 cells grown at 37.5°C and stimulated for 18 h with 1 μM ionomycin in the absence (Iono.) or in the presence (Iono. Gö6976) of 1 μM Gö6976. Left panels show DNA staining with Hoechst 33258.
Shifting S100B-MEF cells to the permissive temperature (32°C) had no significant effect on p53Val135 accumulation and cell cycle parameters (Fig. 2A and C). These observations confirm that calcium signalling and S100B are linked to specific activation of wild-type p53. They also suggest that temperature-dependent conformational shift at 32°C is not sufficient to activate wild-type p53Val135 function when the protein is present at a low concentration.
cPKC activation mediates nuclear translocation and accumulation of the wild-type p53 in S100B-MEF cells.
Previous studies pointed to a possible role of PKC in the regulation of p53 nuclear translocation (11). In vitro, p53 is a cPKC substrate that is phosphorylated on at least five serine and threonine residues (4, 12). To investigate a possible contribution of direct p53 phosphorylation by cPKC on p53 nuclear translocation, the five putative cPKC phosphorylation sites on wild-type murine p53 (Ser360, Thr365, Ser370, Ser372, and Thr377) were mutated to either an Ala residue to prevent phosphorylation or an Asp residue to mimic phosphorylation. The accumulations and subcellular localizations of the p53 mutants were compared in transient transfection assays using S100B-MEF cells. Mutations had no significant effect on p53 nuclear accumulation (Fig. 3). However, incubation of S100B-MEF cells with the cPKC-specific inhibitor Gö6976 (38) prevented nuclear accumulation of both wild-type p53 and the p53-Asp mutant (Fig. 3). Together, these observations suggest that cPKC indirectly regulates wild-type p53 nuclear translocation (see also Discussion).
FIG. 3.
Nuclear translocation of p53 is down regulated by Gö6976, a specific cPKC inhibitor. S100B-MEF clone J-β8 cells were cotransfected with EGFP and plasmids encoding wild-type p53, mutant p53-Ala, or mutant p53-Asp as indicated. After 12 h, cells culture medium was changed to medium without or with 1 μM Gö6976 as indicated. After 20 h cells were fixed. Immunofluorescence of PAb246 immunoreactivity was analyzed in parallel with DNA staining with Hoechst 33258 and EGFP autofluorescence.
The contribution of cPKC activity to wild-type p53 nuclear translocation in S100B-MEF cells was also shown with S100B-MEF cells stably expressing the ts p53Val135 mutant. With these cells, incubation with Gö6976 drastically decreased ionomycin-mediated p53 accumulation (Fig. 4A) and totally inhibited ionomycin-mediated wild-type p53Val135 nuclear translocation (Fig. 4B). The strong correlation that exists between nuclear translocation and accumulation indicates that nuclear localization contributes to wild-type p53 stabilization.
cPKC activation promotes nuclear translocation of the wild-type p53Val135 conformational species in S100B-REF cells in early G1.
S100B-REF cells are derived from transformed REF cells expressing oncogenic Ha-ras and overexpressing p53Val135 (clone 6 cells) (40) transfected with the S100B gene (52). The amount of p53Val135 in S100B-REF cells is about 500-fold higher than the amount in S100B-MEF cells, and overexpression of p53Val135 in S100B-REF cells results in the presence of a significant amount of wild-type p53Val135 conformation species at the nonpermissive temperature (52). The loss of growth-suppressive function of the wild-type p53Val135 species in exponentially growing S100B-REF cells at 37.5°C is probably due to nuclear exclusion of the wild-type p53Val135 conformational species during the early G1 phase of the cell cycle (11, 37) through interaction with a cytoplasmic anchor protein (20). To confirm a role for cPKC in regulation of wild-type p53 nuclear translocation, we analyzed the effect of PMA stimulation on the subcellular localization of the p53Val135 protein in S100B-REF cells synchronized in early G1 by mitotic detachment (Fig. 5). PMA stimulation resulted in a rapid nuclear translocation of the wild-type p53Val135 conformational species (PAb246+), whereas the mutant species (PAb240+) remained cytoplasmic. Such conformational specificity was restricted to cells in early G1. As the cells progressed through the cell cycle, the mutant p53Val135 conformational species also translocated to the nucleus (not shown; see also references 11 and 37). Immunoprecipitation studies with 35S-labeled nuclear extracts confirmed that PMA stimulation of S100B-REF cells produced rapid nuclear translocation of the wild-type p53Val135 species, with a maximum effect after 5 min of stimulation (Fig. 6A, lanes 1 and 2; Fig. 6B). PMA stimulation was also linked with p53 DNA binding activation. The DNA binding activity of the nuclear p53Val135 was tested in a DNA-binding assay based on the interaction of 35S-labeled p53 with a biotinylated consensus p53 (p53-CON) target DNA (Fig. 6A, lanes 3 and 4; and Fig. 6C). The specificity of the interaction between p53 and the biotinylated target DNA was demonstrated by competition with nonbiotinylated p53-CON target DNA (Fig. 6A, lanes 3 and 4). PMA stimulation induced a rapid increase in 35S-labeled p53Val135 which bound to biotinylated p53-CON target DNA (Fig. 6C, lanes 1 to 3). Depletion of the wild-type p53Val135 species from nuclear extracts by immunoprecipitation with PAb246 suppressed the PMA-dependent increase in DNA binding activity (Fig. 6C, lanes 4 and 5).
FIG. 5.
Effect of PMA on cellular localization of wild-type and mutant p53Val135 in S100B-REF cells synchronized in the early G1 phase of the cell cycle. Shown are the results of confocal microscope analysis of PAb246 and PAb240 immunoreactivities in clone 6β cells synchronized by mitotic detachment and grown for 1 h on polylysine-coated coverslips allowing cells to pass through mitosis. Cells were either untreated (control) or stimulated with 8 nM PMA for 5 min (PMA).
FIG. 6.
PMA stimulates nuclear translocation and DNA binding of wild-type p53Val135 in S100B-REF cells. (A) Interaction of wild-type p53 with biotinylated p53-CON target DNA. Clone 6β cell nuclear extracts were prepared, and 35S-labeled p53 was immunoprecipitated with anti-MyoD immunoglobulin G used as a control (lane 1) or wild-type-specific PAb246 (lane 2). Nuclear extracts were also incubated with streptavidin-agarose and biotinylated p53-CON DNA target in the absence (lane 3) or presence (lane 4) of 20 μg of nonbiotinylated p53-CON DNA used as a specific competitor. (B) PMA stimulates wild-type p53 (PAb246+) nuclear translocation. Clone 6β cells were not stimulated (−) or were stimulated with 15 nM PMA for 2, 5, 10, and 15 min as indicated. Wild-type p53 was immunoprecipitated with PAb246. (C) PMA stimulates wild-type p53 binding to biotinylated target DNA. Clone 6β cells were not stimulated (−) or stimulated with 15 nM PMA for 2 or 5 min as indicated. Lanes 1 to 3, nuclear extracts were incubated with biotinylated p53-CON DNA target and streptavidin (Strep.)-agarose. Lanes 4 and 5, nuclear extracts were first incubated with PAb246 and protein G-agarose; the remaining supernatants were then incubated with biotinylated p53-DNA target and streptavidin-agarose. Arrows indicate positions of 35S-labeled p53 which bound to PAb246 or to a biotinylated DNA probe that was visualized by gel electrophoresis and autoradiography.
Dynamic nuclear translocation of the wild-type p53 mediated by PMA stimulation was followed by long-term cytostatic effects. With parental REF cells, not expressing S100B, single stimulation with PMA (5 to 50 nM) decreased the rate of DNA synthesis by 50% (Fig. 7A), but cell cycle parameters were not significantly modified (Fig. 7B). With S100B-REF cells, single stimulation with 5 nM PMA induced a total inhibition of DNA synthesis (Fig. 7A). Fluorescence-activated cell sorting analysis showed that this inhibition corresponds to a G1 phase growth arrest (Fig. 7B).
FIG. 7.
PMA induces G1 phase growth arrest of S100B-REF cells. (A) Effect of PMA concentration on the rate of DNA synthesis of clone 6 (⧫), clone 6β (○), and clone 9β (□) cells grown at 37.5°C. Subconfluent cells were treated for 20 h with a single dose of PMA as indicated, and [3H]thymidine uptake was measured during the last 2 h. (B) Flow cytometry analysis of the DNA content of clone 6 and clone 6β cells stimulated with 8 nM PMA for 8 and 20 h as indicated. (C) Effect of PMA on cell cycle regulatory proteins in S100B-REF cells. Shown is a time course of p21 protein induction and Rb dephosphorylation in clone 6β after PMA stimulation. Cells grown at 37.5°C were not stimulated (lane 1) or stimulated with 8 nM PMA for 1 h (lane 2), 2 h (lane 3), 3 h (lane 4), 4 h (lane 5), 8 h (lane 6), and 16 h (lane 7). Total cell extracts were analyzed by Western blotting using anti-β-tubulin, anti-p27, anti-p21, and anti-Rb antibodies as indicated. (D) Time course of B99 protein induction in clone 6β cells after PMA stimulation (lanes 1 to 7) and comparison with growth-arrested cells at 32°C (lane 8). The asterisk indicates a cross-reacting protein that serves as internal loading control.
PMA-dependent growth arrest of S100B-REF cells was phenotypically indistinguishable from ionomycin-mediated G1 arrest (52). The arrest was characterized by induction of the p21WAF1 cyclin-dependent kinase inhibitor protein but not the p27 inhibitor protein, and with dephosphorylation of the Rb protein (Fig. 7C). Time course analysis of Rb phosphorylation status revealed a biphasic dephosphorylation of Rb protein, with a first maximum effect between 2 and 3 h poststimulation, when the cell cycle parameters were not yet affected by the treatment. A trivial explanation for this phenomenon is that PMA activates a phosphatase which dephosphorylates Rb. Alternatively, it is also possible that PMA mediates p53-dependent transactivation of a phosphatase, like Wip1 (17), which dephosphorylates Rb protein independently of the position of the cells within the cell cycle.
PMA stimulation of S100B-REF cells was also accompanied by increased expression of B99 (Fig. 7D), another cellular protein that is induced by wild-type p53 activity in REF cells (61). In NIH 3T3 cells, B99 is selectively induced in G2/M phase of the cell cycle when p53 is activated (61). As expected, PMA-dependent B99 induction in S100B-REF cells correlates with progression of the cells from S to G2/M phase, and B99 synthesis decreases when cells accumulate in G1 phase. It is noteworthy that in contrast to PMA-mediated G1 arrest, B99 synthesis was sustained in S100B-REF cells growth arrested at 32°C (Fig. 7D; compare lanes 7 and 8), suggesting that temperature shift-mediated growth arrest is distinct from PMA-dependent G1 arrest (see also below and Discussion).
PMA-mediated G1 arrest of S100B-REF cells was totally abolished by the cPKC-specific inhibitor Gö6976 (Fig. 8A and B), suggesting that the PMA effect is mediated via activation of a cPKC isoform. The implication of cPKC isoforms in the long-term cytostatic effect of PMA was confirmed by the use of bryostatin, a structurally distinct PKC activator. In many systems, bryostatin induces only a subset of the responses to PMA and blocks those which it does not induce (22, 28, 57). The mechanisms by which bryostatin and PMA induce divergent long-term responses are linked with differential activation and down regulation of PKC isoforms (22, 57). With S100B-REF cells, bryostatin induced only partial inhibition of DNA synthesis at low concentrations (0.5 to 5 nM) (Fig. 8C) and antagonized the long-term cytostatic effect of PMA at higher concentrations (5 to 20 nM) (Fig. 8C). The divergent long-term responses of the cells to PMA or bryostatin stimulation correlated with specific down regulation of the calcium-dependent PKCα and PKCγ isoforms by bryostatin but not by PMA (Fig. 8D). In contrast to cPKC, the nPKCɛ was down regulated by both PMA and bryostatin (Fig. 8D).
FIG. 8.
cPKC inhibition suppresses the long-term cytostatic effect of PMA. (A) Subconfluent S100B-REF clone 6β cells were treated for a total of 20 h with 15 nM PMA in the absence or in the presence of the PKC inhibitor Gö6976 (1 μM) as indicated, and [3H]thymidine uptake was measured during the last 2 h. Results are the averages of values from three experiments performed in duplicate. C, control. (B) Effect of Gö6976 (1 μM) on the cell cycle parameters of clone 6β cells that were not stimulated or stimulated with 15 nM PMA for 20 h as indicated. (C) Upper panel, effect of bryostatin (Bryo.) concentration on the rate of DNA synthesis of clone 6β cells grown at 37.5°C; lower panel, effect of bryostatin concentration on PMA-mediated inhibition of DNA synthesis of clone 6β cells. Subconfluent cells were treated for 20 h with a single dose of bryostatin without (upper panel) or with (lower panel) 15 nM PMA as indicated, and [3H]thymidine uptake was measured during the last 2 h. (D) Comparison of the effects of PMA (PMA) and bryostatin (Bryo.) on down regulation of PKC isoenzymes in clone 6β cells. Cells grown at 37.5°C were not stimulated (lanes 1 and 5) or stimulated with PMA (15 nM) (lanes 2 to 4) or bryostatin (20 nM) (lanes 6 to 8) for 2 h (lanes 2 and 6), 8 h (lanes 3 and 7), and 22 h (lanes 4 and 8). Lane 9 is bovine brain extract, which was used as control. Total cell extracts (50 μg) were analyzed by Western blotting using anti-PKCα, anti-PKCγ, and anti-PKCɛ antibodies as indicated.
cPKC-dependent G1 arrest but not permissive temperature restores full G1 checkpoint control in S100B-REF cells.
Parental REF cells showed only moderate apoptosis upon UV irradiation (52), and apoptosis was not significantly increased if cells were stimulated with PMA prior to UV irradiation (Fig. 9A). Exponentially growing S100B-REF cells are also insensitive to UV irradiation (52) but showed full apoptotic response if they were first arrested in G1 upon PMA stimulation (Fig. 9B). Internucleosomal DNA cleavage in apoptotic cells was confirmed by agarose gel electrophoresis (Fig. 9C, lanes 1 to 3). Apoptosis of S100B-REF cells was rapid and maximal 24 h after UV irradiation if PMA was removed from the culture media after irradiation (Fig. 9B). It is important to note that if irradiated cells were left in the presence of PMA, the kinetics of apoptosis were considerably retarded, indicating that cell cycle progression is probably required for this apoptosis. Apoptosis still occurred when the transcriptional inhibitor actinomycin D was added 1 h before UV irradiation and maintained after irradiation (Fig. 9C, lane 4). We controlled so that the cells treated with actinomycin D alone did not undergo apoptosis upon UV irradiation (not shown). p53Val135-dependent apoptosis in the absence of de novo transcription and translation in response to UV irradiation has also been observed in immortalized somatotropic progenitor cells expressing p53Val135 (8). We also compared the apoptotic responses to UV irradiation of S100B-REF cells whose growth was arrested by shifting the temperature to 32°C and which were or were not stimulated with PMA (Fig. 9D). No apoptosis was observed in cells growth arrested at 32°C. Moreover, apoptosis could not be restored if the growth-arrested cells at 32°C were subsequently stimulated with PMA. At 32°C, apoptosis could be observed only when S100B-REF cells were first growth arrested with PMA prior to UV irradiation (Fig. 9D; Fig. 9C, lanes 5 and 6). Together these data confirm that S100B and PMA act in concert to specifically rescue a p53 apoptosis pathway in REF cells. They also show that temperature shift-mediated G1 arrest at 32°C protects cells from UV-mediated apoptosis and is more likely associated with enhanced cell survival (see also Discussion).
FIG. 9.
S100B cooperates with PMA in triggering apoptosis of clone 6 cells upon UV irradiation. (A and B) Time course of induction of apoptosis in clone 6 (A) and clone 6β (B) cells by UV irradiation. Clone 6 and clone 6β cells were stimulated for 24 h with 4 nM PMA (PMA/24h). Cells were then irradiated (10 J/m2), changed to fresh medium without PMA, and collected 8 h (UV/8h) or 24 h (UV/24h) after irradiation. (C) Agarose gel analysis of DNA fragmentation in apoptotic clone 6β cells. Lane 1, control cells. Lanes 2 and 3, cells were first stimulated with 4 nM PMA, UV irradiated (10J/m2), and analyzed after 8 h (lane 2) or 24 h (lane 3). Lane 4, cells were as in lane 3 but incubated with actinomycin D (2.5 mg ml−1) for 1 h prior to irradiation. After irradiation, actinomycin D was kept in the medium for another 24 h prior to cell analysis. Lanes 5 and 6 correspond to cells in panel D (32°C, UV and 32°C-PMA, UV). (D) PMA stimulation but not temperature shift restores full G1 checkpoint control. Clone 6β cells were growth arrested by shifting the temperature to 32°C. After 24 h at 32°C, cells were UV irradiated (32°C, UV). Cells were first growth arrested at 32°C; after 24 h, cells were stimulated with PMA for another 20 h and UV irradiated (32°C, PMA-UV). Cells were stimulated with PMA at the time of the temperature shift to 32°C. After 24 h, cells were UV irradiated (32°C-PMA, UV). In all experiments, PMA was used at an 8 nM final concentration, the irradiation dose was 10 J/m2, and the culture media were replaced by media without PMA after irradiation. Cells were analyzed 24 h postirradiation.
DISCUSSION
The first observation reported in this study is the synchronized and concerted regulation of wild-type p53 nuclear translocation, accumulation, and activation by S100B and cPKC. A direct implication of S100B in wild-type p53 activation was first suggested by the fact that MEF cells expressing S100B tolerate only very low expression levels of the p53Val135 protein (52). Inversely, REF cells expressing a very high level of p53Val135 tolerate only low S100B expression levels (52). Moreover, calcium-dependent accumulation and activation of the wild-type p53Val135 functions in MEF and REF cells is strictly conditioned by the presence of S100B (52). Although S100B expression in p53−/−MEF cells was found to be sufficient for the accumulation of wild-type p53 in transient transfection assays (Fig. 1), in stable transfected MEF clones expressing both S100B and a low level of the ts p53Val135 mutant, ionomycin stimulation was required to promote nuclear accumulation of p53Val135 under a wild-type conformation (Fig. 2). These apparent discrepancies for calcium requirement in mediating S100B-dependent wild-type p53 accumulation are not clearly understood. It could be that in transient transfection assays, cellular stress associated with transfection conditions causes changes in intracellular calcium homeostasis similar to that produced by the calcium ionophore. It is also possible that calcium binding to S100B is not absolutely required for S100B to function in regulating wild-type p53. Other ions such as Zn2+, which bind to S100B with much higher affinity (KD, 10 nM) and which induce Ca2+-like conformational changes (3), could also be implicated in regulating S100B functions. Strict calcium-dependent regulation of the wild-type p53Val135 in S100B-MEF cells could also be due to the heat sensitivity of this peculiar p53 mutant and its low expression level. In vitro, S100B interacts in a calcium-dependent manner with the C-terminal regulatory domain of p53 (KD, 20 ± 10 nM) so as to protect p53 from thermal denaturation (13). The high-affinity and specific S100B-binding site on the C-terminal domain of p53 overlaps the multifunctional domain on p53 implicated in stabilization against Mdm2-directed degradation (29) and cytoplasmic anchorage (34) and is a critical determinant for the thermostability of p53 (13). Hence, in its calcium-bound state S100B could transiently interact with the p53Val135 to favor its nuclear translocation and subsequent accumulation. A correlation has also been observed between expression of S100A4 with enhanced levels of p53 in melanoma cells (55). Moreover, S100A2, which is 55% homologous to S100B, is a putative transcriptional target for p53 (58). This raises the possibility that p53 activation is a property shared by other S100 proteins. Finally, like calmodulin, S100B is likely to regulate multiple target proteins. Hence, other potential effector proteins of S100B, including cytoskeletal proteins and cytoskeleton-associated kinases (41) or other signalling pathways, may additionally synergize with S100B in p53 activation. The complexity of the calcium- and S100B-dependent regulation of wild-type p53 is further illustrated with the finding that cPKC is also implicated in the transduction of the calcium signal linked with p53 nuclear translocation, accumulation, and activation in the G1 phase of the cell cycle. In the case of S100B-MEF cells expressing the ts p53Val135 mutant, the cPKC inhibitor Gö6967 inhibited nuclear translocation of the wild-type p53Val135 conformational species and severely decreased p53 accumulation mediated by ionomycin (Fig. 4). This finding indicates that wild-type p53 degradation is probably linked with cytoplasmic sequestration and that nuclear localization contributes to p53 stabilization. Cytoplasmic sequestration and degradation of wild-type p53 would allow cells to maintain a low level of p53 in the absence of p53 activation signals. Activation of cPKC by ionomycin or PMA could either induce phosphorylation of the cytoplasmic p53 or induce phosphorylation of cytoplasmic anchoring proteins (19). To resolve these issues, we performed studies to evaluate the phosphorylation status of the nuclear wild-type p53 in S100B-REF cells before and after PMA stimulation. The results showed that short-term (5-min) PMA stimulation enhanced nuclear accumulation of wild-type p53Val135 without affecting the phosphorylation status of the nuclear protein (data not shown). Moreover, mutations within the five putative PKC phosphorylation sites on murine wild-type p53 were found to have no significant effect on the nuclear translocation of the protein in transient transfection assays (Fig. 3A), suggesting that cPKC activity on p53 nuclear translocation is indirect. We consider that it is more likely that cPKC activity affects the phosphorylation status of cytoplasmic anchoring proteins (19) or of proteins implicated in p53 nuclear transport. It is noteworthy that the high-affinity S100B-binding domain within p53 (13) corresponds to a cytoplasmic sequestration domain on p53 (34). Hence, S100B might cooperate with cPKC by modulating interactions between p53 and cytoplasmic anchoring proteins to favor p53 nuclear translocation. Whether or not direct cPKC phosphorylation of p53 regulates other functions of p53 is also under investigation. In support of a more general role for calcium signalling in the regulation of p53 nuclear translocation in the G1 phase of the cell cycle, a correlation can be established between the inhibition of nuclear translocation of p53 in G1 phase by the antiapoptotic protein Bcl2 (6, 48) and the capacity of Bcl2 to negatively regulate free intracellular calcium elevation by modulation of mitochondria and endoplasmic reticulum calcium pumps (30, 31, 63). The S100B and calcium-dependent activation of wild-type p53 which we have observed in MEF and REF cells could also be of more general occurrence. Many cellular stimulations known to activate p53, such as cell contact (65), hypoxia (20), and UV irradiation (57), are also able to stimulate S100B expression (51, 52, 59, 60). Moreover, like p53, S100B has been implicated in cell differentiation, cellular scenescence (21, 64), neurodegenerative disorders (26, 54), and negative tumor growth (24, 25). It is likely that in both normal cells and tumor cells, in neurodegenerative diseases and senescence, S100B induction could be associated with p53 nuclear translocation and activation.
The second major observation reported in this study is that cPKC-mediated p53 nuclear translocation rescues apoptotic response in S100B-REF cells at both permissive (37.5°C) and nonpermissive (32°C) temperatures (Fig. 9). In contrast, temperature shift-mediated growth arrest at 32°C is linked with enhanced cell survival. Much of the existing data implicates p53 in G1 checkpoint control. However, in some systems, p53-dependent growth arrest may inhibit apoptosis and favor viable cell cycle arrest (46, 49). A central question related to p53 and optimization of anti-cancer therapies is how a cell decides whether to undergo either a viable growth arrest and/or apoptosis when p53 is activated (56). It has been suggested that the outcome of wild-type p53 activation could be dependent on the timing of p53 nuclear translocation. p53-dependent apoptosis requires nuclear translocation of the protein during a critical period in early G1 (14, 48). Similarly, we would like to propose that in S100B-REF cells, S100B and cPKC act in concert to activate a p53-dependent G1 checkpoint by synchronizing nuclear translocation and transcriptional activation of the wild-type p53 in early G1. At this stage, p53 is probably capable of the transactivation of genes linked with both growth arrest and engagement of cells into a preapoptotic program (47). This model fits with the fact that UV-irradiated S100B-REF cells proceeding from G1 enter apoptosis in the absence of de novo transcription (Fig. 9C, lane 4). When shifted to 32°C, REF cells and S100B-REF cells overexpressing the p53Val135 stop growing, and this growth arrest protects cells from UV-mediated apoptosis (Fig. 9D). It is noteworthy that growth arrest at 32°C also protects cells from apoptosis mediated by the DNA-damaging agent doxorubicin (unpublished data). The antiapoptotic activity of wt p53Val135 at 32°C could be linked to the fact that p53Val135 accumulates passively in the nucleus in mid or late G1 subphases, triggering cell growth arrest after the G1 restriction point. The ability of overexpressed p53 to bind to and inhibit the function of the cellular DNA replication factors such as RP-A could be one mechanism by which p53Val135 functions to suppress cell growth at 32°C beyond the G1 restriction point at the G1/S boundary (15). That arrest can also involve p53-mediated transactivation of a target gene like p21. p53-mediated transactivation of p21 after the G1 restriction point is indeed possible (35). The ability of p21 to inhibit proliferating cell nuclear antigen, a factor which is involved in DNA replication, could also be one other mechanism by which p53Val135 functions in late G1 by preventing cells progression to S phase (62). That model would also explain why, in contrast to PMA-mediated G1 arrest, cell growth arrest at 32°C is associated with enhanced synthesis of the B99 protein (Fig. 7D). B99, which is normally down regulated in G1, is thought to play a role in mediating specific activities of wild-type p53 in later phases of the cell cycle (61).
Conclusion.
p53 does not appear to play an essential function in normal cell growth and development but plays a critical role in protection from neoplasia (33). One of the principal mechanisms by which wild-type p53 becomes activated is through its stabilization. One avenue of anticancer therapy might be to exploit mechanisms involved in the normal regulation of p53 conformation and stabilization. We have shown here that forced S100B expression coupled to cPKC activation can promote wild-type p53 nuclear translocation and accumulation. Moreover, we have demonstrated that the dominant negative activity of mutant p53 can be suppressed by forcing the wild-type p53 to translocate into the cell nuclei prior to mutant protein. Our results may constitute a basis for the rational design of p53 coactivators in the development of p53 gene therapy to restore wild-type p53 function in tumor cells harboring wild-type and mutant p53 alleles.
ACKNOWLEDGMENTS
This work was supported by grants from the Association pour la Recherche sur le Cancer, Ligue National Contre le Cancer, to J.B.
We thank Licio Collavin and Claudio Schneider for the generous gift of B99 antibodies.
REFERENCES
- 1.Aloni-Grinstein R, Schwartz D, Rotter V. Accumulation of wt p53 protein upon γ-irradiation induces a G2 arrest-dependent immunoglobulin κ light chain gene expression. EMBO J. 1995;14:1392–1401. doi: 10.1002/j.1460-2075.1995.tb07125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates S, Vousden K. p53 in signalling checkpoint arrest or apoptosis. Curr Opin Genet Dev. 1996;6:12–19. doi: 10.1016/s0959-437x(96)90004-0. [DOI] [PubMed] [Google Scholar]
- 3.Baudier J, Glasser N, Gérard D. Ions binding to S100 protein. Calcium and zinc-binding properties of bovine brain S100αα, S100a (αβ), and S100B (ββ) protein: Zn2+ regulates Ca2+ binding on S100B protein. J Biol Chem. 1986;261:8192–8203. [PubMed] [Google Scholar]
- 4.Baudier J, Delphin C, Grunwald D, Khoshbin S, Lawrence J J. Characterization of the tumor suppressor protein p53 as a protein kinase C substrate and a S100B-binding protein. Proc Natl Acad Sci USA. 1992;89:11627–11631. doi: 10.1073/pnas.89.23.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudier J, Bergeret E, Bertacchi N, Weintraub H, Gagnon J, Garin J. Interactions of myogenic bHLH transcription factors with calcium-binding calmodulin and S100a (αα) proteins. Biochemistry. 1995;34:7834–7846. doi: 10.1021/bi00024a007. [DOI] [PubMed] [Google Scholar]
- 6.Beham A, Marin M C, Fernandez A, Herrmann J, Brisbay S, Tari A M, Lopez-Berestein G, Lozano G, Sarkiss M, McDonnell T J. Bcl-2 inhibits p53 nuclear import following DNA damage. Oncogene. 1997;15:2767–2772. doi: 10.1038/sj.onc.1201464. [DOI] [PubMed] [Google Scholar]
- 7.Blagosklonny M V. Loss of function and p53 protein stabilization. Oncogene. 1997;15:1889–1893. doi: 10.1038/sj.onc.1201374. [DOI] [PubMed] [Google Scholar]
- 8.Caelles C, Heimberg A, Karim M. p53 dependent apoptosis in the absence of transcriptional activation of p53-target genes. Nature. 1994;370:220–224. doi: 10.1038/370220a0. [DOI] [PubMed] [Google Scholar]
- 9.De la Monte S M, Sohn Y K, Ganju N, Wands J R. p53- and CD95-associated apoptosis in neurodegenerative diseases. Lab Investig. 1998;78:401–411. [PubMed] [Google Scholar]
- 10.Deloulme J C, Sensenbrenner M, Baudier J. Interactions of S100 proteins with protein kinase substrates, biological implication. In: Pochet R, Lawson E M, Heizmann K, editors. Calcium binding proteins in normal and transformed cells. New York, N.Y: Plenum Press; 1990. pp. 153–157. [DOI] [PubMed] [Google Scholar]
- 11.Delphin C, Baudier J. The protein kinase C activator, phorbol ester, cooperates with the wt p53 species of ras-transformed embryo fibroblasts growth arrest. J Biol Chem. 1994;269:29579–29587. [PubMed] [Google Scholar]
- 12.Delphin C, Huang K P, Scotto C, Chapel A, Vincon M, Chambaz E, Garin J, Baudier J. The in vitro phosphorylation of p53 by calcium-dependent protein kinase C. Characterization of a protein kinase C-binding site on p53. Eur J Biochem. 1997;245:684–692. doi: 10.1111/j.1432-1033.1997.t01-1-00684.x. [DOI] [PubMed] [Google Scholar]
- 13.Delphin C, Ronjat M, Deloulme J C, Garin G, Debussche L, Higashimoto Y, Sakaguchi K, Baudier J. Calcium-dependent interaction of S100B with the C-terminal domain of the tumour suppressor p53. J Biol Chem. 1999;274:10539–10544. doi: 10.1074/jbc.274.15.10539. [DOI] [PubMed] [Google Scholar]
- 14.Di Leonardo A, Linke S P, Clarkin K, Wahl G M. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 15.Dutta A, Ruppert S M, Aster J C, Winchester E. Inhibition of DNA replication factor RPA by p53. Nature. 1993;365:79–82. doi: 10.1038/365079a0. [DOI] [PubMed] [Google Scholar]
- 16.El-Deiry W S, Kern S E, Pietenpol J A, Kinzler K W, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 17.Fiscella M, Zhang H L, Fan S, Sakaguchi K, Shen S, Mercer W E, Vande Woude G F, O’Connor P M, Appella E. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci USA. 1997;94:6048–6053. doi: 10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frey M R, Saxon M L, Zhao X, Rollins A, Evan S S, Black J D. Protein kinase C isozyme-mediated cell cycle arrest involves induction of p21waf1/cip1 and p27kip1 and hypophosphorylation of the retinoblastoma protein in intestinal epithelial cells. J Biol Chem. 1997;272:9424–9435. doi: 10.1074/jbc.272.14.9424. [DOI] [PubMed] [Google Scholar]
- 19.Gannon J V, Lane D P. Protein synthesis required to anchor a mutant p53 protein which is temperature-sensitive for nuclear transport. Nature. 1991;349:802–805. doi: 10.1038/349802a0. [DOI] [PubMed] [Google Scholar]
- 20.Graeber T G, Peterson J F, Tsai M, Monica K, Fornace A J, Giaccia A. Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol Cell Biol. 1994;14:6264–6277. doi: 10.1128/mcb.14.9.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffin W S T, Sheng J G, Mrak R E. Senescence-accelerated overexpression of S100b in brain of SAMP6 mice. Neurobiol Aging. 1998;19:71–76. doi: 10.1016/s0197-4580(97)00167-x. [DOI] [PubMed] [Google Scholar]
- 22.Hocevar B A, Fields A P. Selective translocation of βII-protein kinase C to the nucleus of human promyelocytic (HL60) leukemia cells. J Biol Chem. 1991;266:28–33. [PubMed] [Google Scholar]
- 23.Hofmann J. The potential for isoenzyme-selective modulation of protein kinase C. FASEB J. 1997;650:649–669. doi: 10.1096/fasebj.11.8.9240967. [DOI] [PubMed] [Google Scholar]
- 24.Karnell R, Von Schoultz E, Hansson L O, Nilsson B, Arstrand K, Kagedal B. S100B protein, 5-S-cysteinyldopa and 6-hydroxy-5-methoxyndole-2-carboxylic acid as biochemical markers for survival prognosis in patients with malignant melanoma. Melanoma Res. 1997;7:393–399. doi: 10.1097/00008390-199710000-00005. [DOI] [PubMed] [Google Scholar]
- 25.Kernohan N M, Rankin R. S100 protein: a prognostic indicator in cutaneous malignant melanoma? Histopathology. 1987;11:1285–1293. doi: 10.1111/j.1365-2559.1987.tb01873.x. [DOI] [PubMed] [Google Scholar]
- 26.Kitamura Y, Shimohama S, Kamoshima W, Matsuoka Y, Nomura Y, Taniguchi T. Changes of p53 in the brains of patients with Alzheimer’s disease. Biochem Biophys Res Commun. 1997;232:418–421. doi: 10.1006/bbrc.1997.6301. [DOI] [PubMed] [Google Scholar]
- 27.Ko L J, Prive C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 28.Kraft A S, Smith J B, Berkow R L. Bryostatin, an activator of the calcium phospholipid-dependent protein kinase, blocks phorbol ester-induced differentiation of human promyelocytic leukemia cells HL-60. Proc Natl Acad Sci USA. 1986;83:1334–1338. doi: 10.1073/pnas.83.5.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubbutat M H, Ludwig R L, Ashcroft M, Vousden K. Regulation of Mdm2-directed degradation by C terminus of p53. Mol Cell Biol. 1998;18:5690–5698. doi: 10.1128/mcb.18.10.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo T H, Kim H-R C, Zhu L, Lin H-M, Tsang W. Modulation of endoplasmic reticulum calcium pump by Bcl2. Oncogene. 1998;17:1903–1910. doi: 10.1038/sj.onc.1202110. [DOI] [PubMed] [Google Scholar]
- 31.Lam M, Dubyak G, Chen L, Nunez G, Miesfeld R L, Distelhorst C W. Evidence that bcl-2 represses apoptosis by regulating endoplasmic reticulum-associated Ca2+ fluxes. Proc Natl Acad Sci USA. 1994;91:6569–6573. doi: 10.1073/pnas.91.14.6569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lane D P. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 33.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 34.Liang S H, Hong D, Clarke M F. Cooperation of a single lysine mutation and a C-terminal domain in the cytoplasmic sequestration of the p53 protein. J Biol Chem. 1998;273:19817–19821. doi: 10.1074/jbc.273.31.19817. [DOI] [PubMed] [Google Scholar]
- 35.Linke S P, Harris M P, Neugebauer S E, Clarkin K C, Shepard H P, Maneval D C, Wahl G M. P53-mediated accumulation of hypophosphorylated pRb after the G1 restriction point fails to halt cell cycle progression. Oncogene. 1997;15:337–345. doi: 10.1038/sj.onc.1201200. [DOI] [PubMed] [Google Scholar]
- 36.Lucas M, Sanchez-Margalet V. Protein kinase C involvement in apoptosis. Gen Pharmacol. 1995;26:881–887. doi: 10.1016/0306-3623(94)00295-x. [DOI] [PubMed] [Google Scholar]
- 37.Martinez J, Georgoff I, Martinez J, Levine A J. Cellular localization and cell cycle regulation by a temperature-sensitive p53 protein. Genes Dev. 1991;5:151–159. doi: 10.1101/gad.5.2.151. [DOI] [PubMed] [Google Scholar]
- 38.Martiny-Baron G, Kazanietz M G, Mischak H, Blumberg P M, Kochs G, Hug H, Dieter M, Schachtele C. Selective inhibition of protein kinase isozymes by the indolocarbazole Gö6976. J Biol Chem. 1993;268:9194–9197. [PubMed] [Google Scholar]
- 39.McConkey D J, Orrenius S. Signal transduction pathways to apoptosis. Trends Cell Biol. 1994;4:370–375. doi: 10.1016/0962-8924(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 40.Michalovitz D, Halevy O, Oren M. Conditional inhibition of transformation and of cell proliferation by a temperature-sensitive mutant of p53. Cell. 1990;62:671–680. doi: 10.1016/0092-8674(90)90113-s. [DOI] [PubMed] [Google Scholar]
- 41.Millward T A, Heizmann C W, Schäfer B W, Hemmings B A. Calcium regulation of Ndr protein kinase mediated by S100 calcium-binding proteins. EMBO J. 1998;17:5913–5922. doi: 10.1093/emboj/17.20.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morii K, Tanaka R, Takahashi Y, Kuwano R. Cloning of cDNA encoding human S100 a and b subunits and their differential expression in human tumor cell lines. J Neurosci Res. 1992;32:27–33. doi: 10.1002/jnr.490320104. [DOI] [PubMed] [Google Scholar]
- 43.Nakajima T, Watanabe S, Sato Y, Kameya T, Hirota T, Shimosato Y. An immunoperoxidase study of S100 protein distribution in normal and neoplastic tissues. Am J Surg Pathol. 1982;6:715–727. doi: 10.1097/00000478-198212000-00003. [DOI] [PubMed] [Google Scholar]
- 44.Nishizuka Y. Intracellular signalling by hydrolysis of phospholipids and activation of protein kinase C. Science. 1992;258:607–614. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- 45.Nishizuka Y. Protein kinase C and lipid signaling for sustained cellular responses. FASEB J. 1995;9:484–496. [PubMed] [Google Scholar]
- 46.Polyak K, Waldman T, He T C, Kinzler K, Vogelstein B. Genetic determinants of p53-induced apoptosis and growth arrest. Genes Dev. 1996;10:1945–1952. doi: 10.1101/gad.10.15.1945. [DOI] [PubMed] [Google Scholar]
- 47.Polyak K, Xia Y, Zweier J L, Kinzler K W, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 48.Ryan J J, Prochownik E, Gottlieb C A, Apel I J, Merino R, Nunez G, Clarke M. c-myc and bcl-2 modulate p53 function by altering p53 subcellular trafficking during the cell cycle. Proc Natl Acad Sci USA. 1994;9:5878–5882. doi: 10.1073/pnas.91.13.5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryan K M, Vousden K H. Characterization of structural p53 mutants which show selective defects in apoptosis but not cell cycle arrest. Mol Cell Biol. 1998;18:3692–3698. doi: 10.1128/mcb.18.7.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sabapathy K, Klemm M, Jaenisch R, Wagner E F. Regulation of ES cell differentiation by functional and conformational modulation of p53. EMBO J. 1997;16:6217–6229. doi: 10.1093/emboj/16.20.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider S A, Fukuyama K, Maceira J, Epstein W L. Effect of ultraviolet B radiation on S100 protein antigen in epidermal Langerhans cells. J Investig Dermatol. 1985;84:146–148. doi: 10.1111/1523-1747.ep12275395. [DOI] [PubMed] [Google Scholar]
- 52.Scotto C, Deloulme J C, Rousseau D, Chambaz E, Baudier J. Calcium and S100B regulation of p53-dependent cell growth arrest and apoptosis. Mol Cell Biol. 1998;18:4272–4281. doi: 10.1128/mcb.18.7.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Serrano M, Lin A W, McCurrach M E, Beach D, Lowe S W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 54.Sheng J G, Mrak R E, Griffin W S T. S100b protein expression in Alzheimer’s disease: potential role in the pathogenesis of neuritic plaques. J Neurosci Res. 1994;39:398–404. doi: 10.1002/jnr.490390406. [DOI] [PubMed] [Google Scholar]
- 55.Sherbet G V, Lakshmi M S. S100A4 (MTS1) calcium binding protein in cancer growth, invasion and metastasis. Anticancer Res. 1998;18:2415–2422. [PubMed] [Google Scholar]
- 56.Smith M L, Fornace A J. p53-mediated protective responses to UV irradiation. Proc Natl Acad Sci USA. 1997;94:12255–12257. doi: 10.1073/pnas.94.23.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Szallasi Z, Smith C B, Pettit G R, Blumberg P M. Differential regulation of protein kinase C isozymes by bryostatin 1 and phorbol 12-myristate 13-acetate in NIH 3T3 fibroblasts. J Biol Chem. 1994;269:2118–2124. [PubMed] [Google Scholar]
- 58.Tan M, Heizmann C W, Guan K, Schafer B W, Sun Y. Transcriptional activation of the human S100A2 promoter by wild-type p53. FEBS Lett. 1999;445:265–268. doi: 10.1016/s0014-5793(99)00135-0. [DOI] [PubMed] [Google Scholar]
- 59.Tronnier M, Missler U, Grotrian K, Kock N. Does ultraviolet radiation exposure influence S100b protein plasma level? Br J Dermatol. 1998;138:1098–1100. doi: 10.1046/j.1365-2133.1998.02294.x. [DOI] [PubMed] [Google Scholar]
- 60.Tsoporis J N, Marks A, Kahn H J, Butany J, Liu P P, O’Hanlon D, Parker T G. S100β inhibits α1-adrenergic induction of the hypertrophic phenotype in cardiac myocytes. J Biol Chem. 1997;272:31915–31921. doi: 10.1074/jbc.272.50.31915. [DOI] [PubMed] [Google Scholar]
- 61.Utrera R, Collavin L, Lazarevic D, Delia D, Schneider C. A novel p53-inducible gene coding for a microtubule-localized protein with G2-phase-specific expression. EMBO J. 1998;17:5015–5025. doi: 10.1093/emboj/17.17.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waga S, Hannon G J, Beach D, Stillman B. The p21 inhibitor of cyclin-dependent kinases controls DNA replication by interaction with PCNA. Nature. 1994;369:574–578. doi: 10.1038/369574a0. [DOI] [PubMed] [Google Scholar]
- 63.Wang E, Murphy A N, Bredesen D E, Cortopassi G, Fiskum G. Bcl-2 potentiates the maximal calcium uptake capacity of neural cell mitochondria. Proc Natl Acad Sci USA. 1996;93:9893–9898. doi: 10.1073/pnas.93.18.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Whitaker-Amitia P, Wingate M, Borella A, Gerlai R, Roder J, Azmitia E. Transgenic mice overexpressing the neurotrophic factor S100b show neuronal cytoskeletal and behavioral signs of altered aging processes: implication for Alzheimer’s disease and Down’s syndrome. Brain Res. 1997;776:51–60. doi: 10.1016/s0006-8993(97)01002-0. [DOI] [PubMed] [Google Scholar]
- 65.Yahanda A M, Bruner J M, Donehower L A, Morrison R S. Astrocytes derived from p53-deficient mice provide a multistep in vitro model for development of malignant gliomas. Mol Cell Biol. 1995;15:4249–4259. doi: 10.1128/mcb.15.8.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zezula J, Sexl V, Hutter C, Karel A, Schutz W, Freissmuth M. The cyclin-dependent kinase inhibitor p21cip1 mediates the growth inhibitory effect of phorbol esters in human venous endothelial cells. J Biol Chem. 1997;272:29967–29974. doi: 10.1074/jbc.272.47.29967. [DOI] [PubMed] [Google Scholar]
- 67.Zhao X, Gschwend J E, Powell T, Foster R G, Day K C, Day M L. Retinoblastoma protein-dependent growth signal conflict and caspase activity are required for protein kinase C-signaled apoptosis of prostate epithelial cells. J Biol Chem. 1997;272:22751–22757. doi: 10.1074/jbc.272.36.22751. [DOI] [PubMed] [Google Scholar]