Abstract
Many nuclear receptors are capable of recognizing similar DNA elements. The molecular event(s) underlying the functional specificities of these receptors (in regulating the expression of their native target genes) is a very important issue that remains poorly understood. Here we report the cloning and analysis of a novel nuclear receptor coactivator (designated NRIF3) that exhibits a distinct receptor specificity. Fluorescence microscopy shows that NRIF3 localizes to the cell nucleus. The yeast two-hybrid and/or in vitro binding assays indicated that NRIF3 specifically interacts with the thyroid hormone receptor (TR) and retinoid X receptor (RXR) in a ligand-dependent fashion but does not bind to the retinoic acid receptor, vitamin D receptor, progesterone receptor, glucocorticoid receptor, or estrogen receptor. Functional experiments showed that NRIF3 significantly potentiates TR- and RXR-mediated transactivation in vivo but has little effect on other examined nuclear receptors. Domain and mutagenesis analyses indicated that a novel C-terminal domain in NRIF3 plays an essential role in its specific interaction with liganded TR and RXR while the N-terminal LXXLL motif plays a minor role in allowing optimum interaction. Computer modeling and subsequent experimental analysis suggested that the C-terminal domain of NRIF3 directly mediates interaction with liganded receptors through an LXXIL (a variant of the canonical LXXLL) module while the other part of the NRIF3 protein may still play a role in conferring its receptor specificity. Identification of a coactivator with such a unique receptor specificity may provide new insight into the molecular mechanism(s) of receptor-mediated transcriptional activation as well as the functional specificities of nuclear receptors.
Nuclear hormone receptors are ligand-regulated transcription factors that play diverse roles in cell growth, differentiation, development, and homeostasis. The nuclear receptor superfamily has been divided into two subfamilies: the steroid receptor family and the thyroid hormone/retinoid (nonsteroid) receptor family (51). The steroid receptor family includes receptors for glucocorticoids (GR), mineralcorticoids, progestins (PRs), androgens (AR), and estrogens (ERs) (51). The nonsteroid receptor family includes receptors for thyroid hormones (TRs), retinoids (retinoic acid receptors [RARs] and retinoid X receptors [RXRs]), 1,25-(OH)2-vitamin D (VDR), and prostanoids (peroxisome proliferator-activated receptors [PPARs]) as well as many orphan receptors whose ligands (if any) remain to be defined (49, 51). Members of the nuclear receptor superfamily have common structural and functional motifs. Nevertheless, an important difference exists between the two subfamilies. Steroid receptors act primarily as homodimers by binding to their cognate palindromic hormone response elements (HREs) (77, 78). In contrast, members of the nonsteroid receptor family can bind to DNA as monomers, homodimers, and heterodimers (25, 78). Their corresponding HREs are also complex and can be organized as direct repeats, inverted repeats, and everted repeats (49). Therefore, the combination of heterodimerization and HRE complexity provides the potential of generating enormous diversity in receptor-mediated regulation of target gene expression.
Structural and functional studies indicate that the ligand binding domain (LBDs) of many members of the thyroid hormone/retinoid receptor family harbor diverse functions. In addition to binding to ligands, the LBD also plays roles in mediating receptor dimerization, hormone-dependent transactivation, and, with TR and RAR, ligand-relieved gene silencing (54, 61). The carboxyl-terminal helix of the LBD has been implicated in playing an important role in ligand-dependent conformational changes and transactivation (6, 9, 21, 43). Although it has been suggested that an activation function (AF-2) resides in this C-terminal helix, recent studies indicate that AF-2 results from a ligand-induced conformational change involving diverse areas of the LBD (23, 66). Thus, ligand binding serves to switch the receptor from one functional state (e.g., inactive or silencing) to another (e.g., transactivation).
Although much has been learned from studying the structures and functions of these receptors, the detailed molecular mechanism(s) of transcriptional regulation by these receptors is not well understood. Efforts to understand the molecular mechanism of transcriptional repression by unliganded TRs and RARs have led to the description (12) and isolation of the putative corepressor proteins SMRT and N-CoR, which interact with the LBDs of these receptors in the absence of their ligands (15, 36). The recent discovery that both SMRT and N-CoR form complexes with Sin3 and a histone deacetylase suggests that chromatin remodeling by histone deacetylation may play a role in receptor-mediated transcriptional repression (33, 55).
In a somewhat parallel approach, the identification of coactivators has recently received extensive experimental attention in order to elucidate the molecular mechanism(s) of transcriptional activation by nuclear receptors (27). Identified coactivator proteins primarily belong to two groups: the SRC-1 family and the CREB-binding protein (CBP)/p300 family. The SRC-1 family includes SRC-1/NCoA-1 (37, 58, 74) and the related proteins GRIP1/TIF2/NCoA-2 (34, 35, 74, 79) and AIB1/p/CIP/ACTR/RAC3/TRAM-1 (2, 14, 44, 73, 74). The second group of coactivators includes CBP and its homolog p300, which not only influence the activities of nuclear receptors (13, 31, 37) but also functionally interact with many transcription factors such as CREB (3, 16, 40, 46), the Stats (10, 87), AP1 (4, 7), and p53 (28, 45). There are also coactivator proteins that do not belong to these two groups, such as ARA70 (85), PGC-1 (60), and the recently reported RNA coactivator SRA (41). Members of both the SRC-1 family and CBP/p300 family have been shown to possess histone acetyltransferase activities (8, 14, 57, 69), suggesting that chromatin remodeling by histone acetylation is an important mechanism involved in transcriptional activation by ligand-bound nuclear receptors.
Interaction of members of the SRC-1 and CBP/p300 families with nuclear receptors occurs through conserved LXXLL motifs (32), which interact with a hydrophobic cleft in the receptor LBD formed as a result of conformational changes mediated by ligand binding (19, 23, 56). SRC-1/NCoA-1 and GRIP1/TIF2 contain three LXXLL regions or boxes (referred to as LXDs or nuclear receptor boxes) that differentially interact with nuclear receptors so that different nuclear receptors functionally utilize different LXXLL boxes (19, 52). Thus, ER uses the second LXXLL box of SRC-1/NCoA-1 while PR uses both the first and second LXXLL boxes for optimal interaction. In contrast, TR and RAR require both the second and third LXXLL boxes for optimal interaction (52). The specificities of receptor recognition by the different LXXLL boxes of SRC-1/NCoA-1 are primarily mediated by 8 amino acid residues C terminal to the LXXLL motif rather than by the 2 amino acids (XX) within the motif itself. Thus, while members of the SRC-1 family are capable of interacting with many nuclear receptors, the molecular details of such interactions differ for each receptor in the number or combination of LXXLL boxes used as well as in the critical amino acid residues surrounding the LXXLL motifs.
While much has been learned from the study of known coactivators, a number of key mechanistic questions remain to be answered. For example, many nuclear receptors can recognize common DNA elements, (25, 49, 51), while not all are capable of regulating genes containing those elements (20, 47, 65). Thus, how native target genes containing such elements are selectively regulated by specific receptors is a very important but poorly understood problem. Although the various LXXLL boxes of SRC-1 and GRIP1 show differential receptor preference (19, 52), these coactivators are unlikely to play a primary role in mediating effects that are receptor specific since they appear to interact with all ligand-bound nuclear hormone receptors. Thus, the detailed molecular mechanism(s) underlying receptor-specific regulation of gene expression remains to be elucidated. Whether a coactivator(s) contributes to this specificity is currently unknown.
To further our understanding of the molecular events underlying receptor-activated transcription, we sought to identify additional coactivators using a yeast two-hybrid screening strategy (29). In this paper, we report the isolation of a novel coactivator for nuclear receptors, designated NRIF3 (for nuclear receptor-interacting factor 3). Fluorescence microscopy indicates that NRIF3 is a nuclear protein. The yeast two-hybrid and in vitro binding assays revealed that NRIF3 interacts specifically with TR and RXR in a ligand-dependent fashion but does not interact with other examined nuclear receptors. Transfection experiments indicated that NRIF3 selectively potentiates TR- and RXR-mediated transactivation in vivo. The NRIF3 gene encodes a small protein of 177 amino acids and, other than having an N-terminal LXXLL motif, has no homology with known coactivator genes. The results of a combination of computer modeling and domain and mutagenesis analyses suggest that NRIF3 interacts with nuclear receptors through its C-terminal domain that contains a novel LXXIL module while another part of NRIF3 may contribute to its observed receptor specificity. These findings may provide novel insight into the molecular mechanism(s) of receptor-mediated transcriptional activation as well as the functional specificities of nuclear receptors.
MATERIALS AND METHODS
Isolation of NRIFs and the yeast two-hybrid assay.
The Brent two-hybrid system (29) was employed to isolate candidate cDNA clones interacting with LexA-TRα in a ligand-dependent fashion. Full-length chicken TRα (cTRα) was fused in frame to the C terminus of the LexA DNA binding domain (DBD) in pEG202 (29). The LexA-TRα bait, the LacZ reporter (pSH18-34), and a pJG4-5-based HeLa cell cDNA library were transformed into the yeast strain EGY48 (29). The transformants were selected on Gal–Raf–X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) medium in the absence of leucine and were further screened for the expression of LacZ in the presence of 1 μM T3. Blue colonies were picked and reexamined for T3-dependent expression of LacZ. Positive yeast clones were then selected, and plasmids harboring candidate prey cDNAs were isolated. An individual candidate prey plasmid was then amplified in Escherichia coli and retransformed into the original yeast strain to confirm the interaction phenotype. The cDNA inserts were then sequenced with an automatic sequencer. Four novel clones (NRIF1, -2, -3, and -4) were obtained. Among them, NRIF3 was a full-length clone.
Wild-type NRIF3, the β3-endonexin long form (EnL) and short form (EnS), and the L9A NRIF3 mutant protein were examined for their interaction with various nuclear receptors in a yeast two-hybrid assay. The following receptor baits were used: the LexA-cTRα LBD, LexA-human TRβ (hTRβ) LBD, LexA-hRARα LBD, LexA-hRXRα LBD, and LexA-hGR LBD. The NRIF3 C-terminal domain (NCD) was fused in frame with the LexA DBD and examined for interaction with receptor LBDs with the following preys: the B42-cTRα LBD, B42-hRARα LBD, and B42-hRXRα LBD expressed from pJG4-5. Yeast cells harboring appropriate plasmids were grown in selective media with Gal-Raf in the presence or absence of cognate ligand (1 μM T3 for TR, all trans or 9-cis RA for RAR, 9-cis RA for RXR, and 10 μM deoxycorticosterone for GR) overnight before β-galactosidase activity was assayed with o-nitrophenyl β-d-galactopyranoside as the substrate. β-Galactosidase units were calculated with the formula (OD420 × 1,000)/(minutes of incubation × OD600 of yeast suspension), where OD420 and OD600 are the optical densities at 420 and 600 nm, respectively.
Fluorescence microscopy.
Full-length NRIF3 was cloned into the green fluorescent protein (GFP) fusion protein expression vector pEGFP (Clontech). The resulting GFP-NRIF3 vector and the control plasmid pEGFP were transfected into HeLa cells by calcium phosphate coprecipitation. Cells were incubated at 37°C for 24 h before the examination with a fluorescence microscope to determine the subcellular location of GFP-NRIF3 or the GFP control.
In vitro binding assay.
Full-length NRIF3 was cloned into pGEX2T, a bacterial glutathione S-transferase (GST) fusion protein expression vector (Pharmacia). The GST-NRIF3 fusion protein was expressed in E. coli and affinity purified with glutathione-agarose beads (30). 35S-labeled full-length cTRα, hRARα, hRXRα, hVDR, hGR, hPR, and hER were generated by in vitro transcription and translation with a reticulocyte lysate system (Promega). Binding was performed as previously described (30) with the following buffer: 20 mM HEPES (pH 7.9)–1 mM MgCl2–1 mM dithiothreitol–10% glycerol–0.05% Triton X-100–1 μM ZnCl2–150 mM KCl. Appropriate ligands were added into the binding reaction mixture where indicated in the figures in the following concentrations: 1 μM T3 for TR, 1 μM all-trans RA or 9-cis RA for RAR, 1 μM 9-cis RA for RXR, and 150 nM 1,25-(OH)2-vitamin D3, dexamethasone, progesterone, or estradiol for VDR, GR, PR, or ER, respectively. After the binding reaction, the beads were washed three times and the labeled receptors bound to the beads were examined by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis followed by autoradiography. Five percent of the 35S-labeled receptor input was also electrophoresed in the same gel.
Transfection studies.
Most reporters used in this study, including IR-ΔMTV-CAT, DR4-ΔMTV-CAT, GH-TRE-tk-CAT, and IR+3 (ERE)-ΔMTV-CAT, have been described previously (5, 25, 78). A DR1-ΔMTV-CAT reporter responsive to RXR was obtained from Ron Evans. A GRE/PRE-tk-CAT reporter was obtained from Gunther Schutz. (IR)2-TATA-CAT was constructed in our laboratory by cloning two copies of the inverted-repeat (IR) sequence (AGGTCA TGACCT) upstream of a TATA element derived from the thymidine kinase (tk) promoter. An hVDR expression vector and VDRE-ΔMTV-CAT containing the VDRE from the osteocalcin promoter were obtained from J. Wesley Pike. Vectors expressing cTRα, hRARα, hRXRα, rat GR (rGR), hPR, and hER have been described previously (17, 25, 26, 50, 53, 81). The NRIF3 expression vector was constructed by cloning full-length NRIF3 into a pExpress vector (25). Appropriate plasmids were transfected into HeLa cells by calcium phosphate coprecipitation with 25 to 100 ng of the receptors, 250 to 500 ng of the chloramphenicol acetytransferase (CAT) reporters, and 750 ng of the NRIF3 or control pExpress vector. After transfection, cells were incubated at 37°C (with or without cognate ligands) for 42 h before being harvested. CAT assays were carried out as previously described (30). Relative CAT activity was determined as the percent acetylation of the substrate per 30 μg of cell protein in a 15-h incubation at 37°C. The results were calculated from duplicate or quadruplicate samples, and the variation among samples was less than 10%.
Domain and mutagenesis analyses.
To construct pJG4-5-derived vectors expressing EnL or EnS, the pJG4-5/NRIF3 plasmid was digested with NcoI and XhoI and the resulting vector fragment was gel purified. This fragment was then ligated to an EnL or EnS insert generated from pExpress-EnL or pExpress-EnS by NcoI/SalI double digestion. The resulting pJG4-5/EnL or pJG4-5/EnS plasmid was confirmed by sequence analysis. The L9A mutant form of NRIF3 was generated by site-directed mutagenesis by a PCR-based method, and the mutation was confirmed by sequence analysis. pJG4-5-derived vectors expressing EnL, EnS, or the L9A NRIF3 mutant form were transformed into yeast strains harboring the LacZ reporter (pSH18-34) and appropriate bait plasmids (LexA-TR, LexA-RAR, LexA-RXR, and LexA-GR). Transformants were subjected to quantitative assays of β-galactosidase activity as described above.
To construct the bait plasmid expressing LexA-NCD, a derivative of pEG202 (which contains a new polylinker) was digested with NcoI and XhoI and ligated to synthetic oligonucleotides that encode the last 16 amino acids of NRIF3 (residues 162 to 177). Similarly, the mutant NCD was generated by using oligonucleotides that contain the designed mutations in the ligation reaction. Both constructs were confirmed by sequence analysis. Bait plasmids expressing LexA-NCD or LexA-mutant NCD were transformed together with one of the following prey plasmids, B42-TR LBD, B42-RXR LBD, or B42-RAR LBD, into the yeast strain that harbors the LacZ reporter (pSH18-34). Subsequent two-hybrid assays were carried out as described above.
Docking of coactivator peptides to receptors.
We built a model of the interaction between the 17-residue C-terminal peptide of NRIF3 (KASRHLDSYEFLKAILN) and the LBDs of several receptors (TRα was used as an example in the experiment reflected in Fig. 10). An LXXIL motif within the NRIF3 peptide is underlined. A similar modeling procedure was carried out on a 20-residue peptide (SLTERHKILHRLLQEGSPSD) of the second LXXLL box of SRC-1 (52). We hypothesized that the LXXIL motif of the C terminus of NRIF3 contacts the coactivator binding site of the nuclear receptors, and the automatic docking procedure was carried out towards this site (71, 75, 76). Two critical features of the interaction between the LBDs of nuclear hormone receptors and their coactivators were used to build the models. (i) One was the “charge clamp,” initially observed in the complex between SRC-1 and PPARγ (56), where a conserved glutamate and lysine at opposite ends of the hydrophobic cavity of the receptors contact the backbone of the coactivator’s LXXLL box. This feature enabled us to orient the NRIF3 helical peptide. (ii) The other feature was that the leucines of the LXXLL motif of SRC-1 are buried in the hydrophobic cavity of the receptor. This feature allowed us to predict the side of the NRIF3 peptide which faces the receptor.
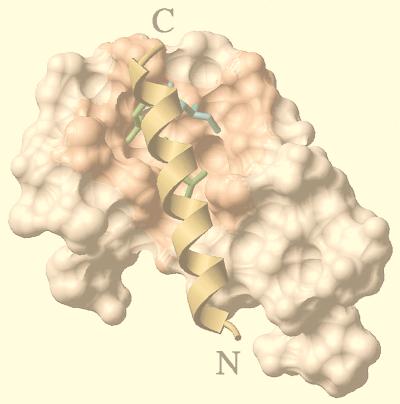
FIG. 10.
Hypothetical model of the interaction of the NCD and the liganded LBD. The docking of the C-terminal helix of NRIF3, which contains an LXXIL module, to the ligand-bound LBDs was carried out as described in Materials and Methods. The NCD-TR LBD model is shown here as an example. The side chains of the two leucines (green) and one isoleucine (cyan) of the LXXIL core fit within a hydrophobic groove (salmon) on the surface of the liganded LBD (80). A similar modeling procedure was carried out with an LXXLL box of SRC-1 (result not shown). Putative binding energies (−21 kcal/mol for the NCD and −18 kcal/mol for the LXXLL box of SRC-1) were calculated as described in Materials and Methods. See the text for details.
The coactivator peptides were assigned a helical secondary structure, the backbone φ and ψ angles being −62 and −41 degrees, respectively. The ω angle was set to 180 degrees. Loose distance restraints were set between the charge clamp of the receptors (56) and Cα atoms of the peptide. The energy of the complex was minimized in the internal coordinate space by using the modified ECEPP/3 potentials. The subset of the variables minimized by the ICM method (1, 71, 76) included the side chains of the receptor, six positional variables of the helix, and the side chain torsion angles of the helix.
Binding energy calculation.
The binding energy was calculated by the partitioning method as described elsewhere (64). Briefly, the binding energy function is partitioned into three terms: the surface (or hydrophobic) term, determined as the product of the solvent-accessible surface by a surface tension of 30 cal/mol/Å2; the electrostatic term, calculated by a boundary element algorithm, with a dielectric constant of 8; and the entropic term, which results from the decrease in conformational freedom of residue side chains partially or completely buried upon complexation.
RESULTS
Cloning of NRIF3 cDNA.
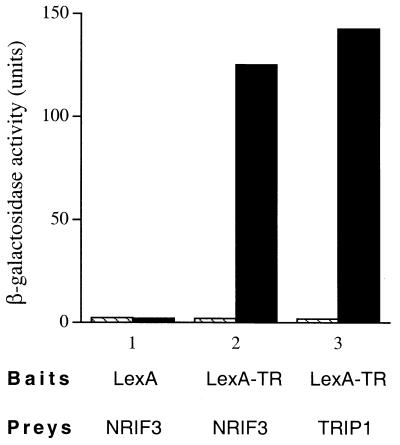
To isolate potential coactivators mediating the transcriptional activation functions of nuclear receptors, we employed a yeast two-hybrid screening strategy (29). A bait expressing a full-length TRα fused to the C terminus of the LexA DBD was used to screen a HeLa cell cDNA library cloned into pJG4-5 (29). Candidate clones that exhibited a thyroid hormone (T3)-dependent interaction with LexA-TRα were selected and further examined and sequenced. Four novel clones were identified, and all were found to exhibit levels of interaction with the LBD of TRα similar to the levels they exhibited with the full-length receptor (data not shown). These clones were designated NRIF1, -2, -3, and -4. Not surprisingly, the LBD of TRβ was also found to interact with these NRIFs in a T3-dependent manner (data not shown). Among these four isolated NRIFs, NRIF3 was a full-length clone. As shown in Fig. 1, LexA alone (negative control) did not interact with NRIF3 (as indicated by the low β-galactosidase activity) and incubation with T3 had no effect. Similarly, no interaction was detected between the LexA-TR LBD and B42 alone with or without T3 (data not shown). The LexA-TR LBD also showed little interaction with NRIF3 in the absence of T3. However, incubation with T3 resulted in strong stimulation of the NRIF3-TR LBD interaction (Fig. 1). The extent of T3-dependent interaction between NRIF3 and the LexA-TR LBD was similar to that of Trip1 (Fig. 1), one of the first TR-interacting factors cloned in a two-hybrid screen (42).
FIG. 1.
Hormone-dependent interaction of NRIF3 with the LBD of TR. Induction of β-galactosidase activity by thyroid hormone (T3) was measured in the yeast strain EGY48 transformed with a bait vector expressing the LexA-cTRα LBD and a prey plasmid expressing NRIF3 fused to the B42 activation domain (29). The bait LexA alone was used as the negative control. The prey B42–Trip1 was used as the positive control. Hatched bars, without T3; filled bars, with 1 μM T3.
Sequence analysis of NRIF3.
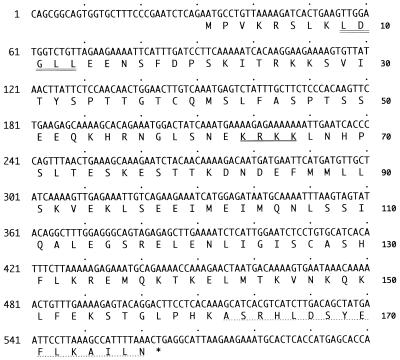
Sequence analysis of the NRIF3 cDNA revealed a single open reading frame encoding a polypeptide of 177 amino acids (Fig. 2). NRIF3 has no homology with members of the SRC-1 and CBP/p300 families. The size of NRIF3 is in sharp contrast to the size of CBP/p300 (around 300 kDa) or of SRC-1 family members (around 160 kDa). NRIF3 contains a putative nuclear localization signal (KRKK), as well as one copy of an LXXLL motif (amino acids 9 to 13) that was recently identified as being essential for the interaction of a number of putative coactivators with nuclear receptors (32).
FIG. 2.
Nucleotide and deduced amino acid sequences of NRIF3. Only part of the cDNA sequence is shown. A putative nuclear localization signal (KRKK) is underlined. The putative LXXLL motif is shown with a double underline. NRIF3 and EnL have 95% identity. They differ only in their C termini, where the last 16 amino acids (dotted underline) in NRIF3 are replaced with 9 different amino acids (GQPQMSQPL) in EnL. EnS consists of 111 amino acids and is 100% identical to the first 111 amino acids of NRIF3 or EnL.
A database search identified two highly related homologs of NRIF3, which were previously designated β3-endonexin short and long forms (67). The endonexin short form (EnS) was originally isolated from a two-hybrid screen intended to clone factors that interact with the cytoplasmic tail of integrin β3 (67). The long form (EnL) was then identified as an alternatively spliced product of the same gene. However, the long form does not bind to integrin β3 (67). Nucleotide sequence comparisons between cDNAs of NRIF3 and EnS or EnL indicate that NRIF3 is a third alternatively spliced product of the same gene (alignment not shown). The precise function(s) of the two endonexin proteins is under investigation (reference 66a and see Discussion).
NRIF3 localizes to the cell nucleus.
Although a putative nuclear localization signal was found in NRIF3, we considered it important to identify the subcellular location of the NRIF3 protein since extensive homology was found between NRIF3 and the two endonexins. The entire NRIF3 open reading frame was fused to the C terminus of GFP (18). The resulting GFP-NRIF3 fusion protein was expressed in HeLa cells by transient transfection, and the subcellular location of the fusion protein was visualized by fluorescence microscopy. As shown in Fig. 3, the control GFP protein distributed throughout the cell while GFP-NRIF3 localized exclusively to the nucleus. This result suggests that NRIF3 is a nuclear protein, which is compatible with its putative role as a nuclear receptor coactivator.
FIG. 3.
NRIF3 is a nuclear protein. HeLa cells were transfected with an expression vector for GFP (left panel) or GFP-NRIF3 (right panel). The cellular location of the expressed proteins was visualized by fluorescence microscopy.
Selective interaction of NRIF3 with liganded nuclear receptors in yeast.
Although NRIF3 was originally cloned with full-length TRα as the bait, we later identified that the region of the receptor responsible for NRIF3 binding is its LBD (Fig. 1). A common feature among most of the known coactivators that show ligand-dependent interaction with nuclear receptors is the presence of the LXXLL motif(s) in their receptor interaction domains. The LXXLL motif appears to be involved in direct contact with a structurally conserved surface in the ligand-bound LBDs of the receptors (23), which may provide the molecular basis for the broad spectrum of receptor binding by coactivators such as SRC-1 and GRIP1. Since a putative LXXLL motif is also present in NRIF3 (amino acids 9 to 13), we asked whether NRIF3 also interacts with the LBDs of other nuclear receptors.
The LBDs of several nuclear receptors were examined for interaction with NRIF3 in a yeast two-hybrid assay. As shown in Table 1, NRIF3 does not interact with LexA alone (negative control) with or without ligand. LexA-TR and LexA-RXR showed little (if any) interaction with NRIF3 in the absence of their cognate ligands. However, the presence of T3 (for TR) or 9-cis RA (for RXR) resulted in a strong stimulation of their interaction with NRIF3, as indicated by the induction of β-galactosidase activity (Table 1). Interestingly, when LexA-RAR or LexA-GR was used as the bait, no interaction was detected with NRIF3 in the presence or absence of their cognate ligands (Table 1). The finding that NRIF3 interacts with TR but not RAR was surprising in light of a recent study which showed that TR and RAR functionally interact with the same LXXLL boxes (boxes 2 and 3) of SRC-1/NCoA-1 (52). As positive controls, we confirmed that both LexA-RAR and LexA-GR exhibited ligand-dependent interaction with other coactivators that are not receptor specific (data not shown). Taken together, these results suggest that NRIF3 exhibits differential specificities in its interactions with different nuclear receptors.
TABLE 1.
Interaction of NRIF3 with nuclear receptors in yeasta
| Bait | Prey | β-Galactosidase activity
|
||
|---|---|---|---|---|
| Without ligand | With ligand | Fold stimulation | ||
| LexA | NRIF3-B42 | 2.3 | 1.9 | 0.8 |
| LexA-TR | NRIF3-B42 | 1.8 | 125 | 69 |
| LexA-RAR | NRIF3-B42 | 0.1 | 0.1 | 1 |
| LexA-RXR | NRIF3-B42 | 0.2 | 63 | 315 |
| LexA-GR | NRIF3-B42 | 0.8 | 0.6 | 0.8 |
The LacZ reporter activities were determined for yeast strains harboring the indicated bait and prey plasmids in the presence or absence of cognate ligands as described in Materials and Methods. See the text for detailed explanations.
NRIF3 specifically binds to TR and RXR but not to other nuclear receptors in vitro.
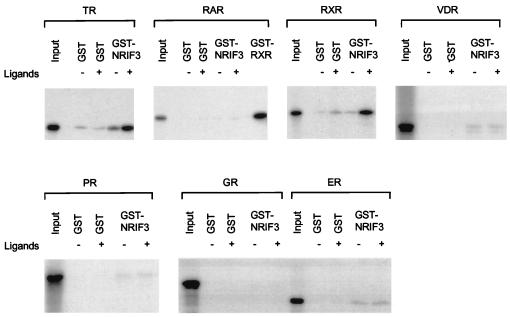
To further examine the interaction between NRIF3 and various nuclear receptors as well as to confirm the potential receptor specificity of NRIF3, in vitro GST binding assays were performed (30). 35S-labeled nuclear receptor, generated by in vitro transcription and translation, was incubated with purified GST-NRIF3 or the GST control bound to glutathione-agarose beads. All binding assays were carried out with or without the cognate ligand of the examined receptor. As shown in Fig. 4 (top left), TR and NRIF3 interact poorly in the absence of T3. Addition of T3 resulted in a strong increase in TR binding to GST-NRIF3, confirming that NRIF3 associates with TR in a T3-dependent manner. Using similar binding assays, we also studied the interaction of NRIF3 with six other nuclear receptors. Consistent with our findings from the yeast two-hybrid experiments (Table 1), NRIF3 interacted with RXR in vitro in a ligand-dependent manner (Fig. 4) but showed little or no binding to other nuclear receptors (RAR, VDR, GR, PR, and ER) in the presence or absence of their cognate ligands (Fig. 4). Taken together, the results of the yeast two-hybrid (Table 1) and the in vitro binding (Fig. 4) assays suggest that NRIF3 possesses a distinct receptor specificity.
FIG. 4.
Characterization of the NRIF3 interaction with nuclear receptors in vitro. A 35S-labeled full-length receptor (cTRα, hRARα, hRXRα, hVDR, hPR, hGR, or hER) was incubated with an affinity-purified GST control or GST-NRIF3 linked to glutathione-agarose beads. The binding was performed in the absence (−) or presence (+) of cognate ligands as described in Materials and Methods. After incubation and washing, the bound receptors were analyzed by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis and detected by autoradiography. The input lane in each binding assay represents 5% of the total 35S-labeled receptor used in each incubation. GST-RXR was used as a positive control for RAR binding.
NRIF3 selectively potentiates TR- and RXR-mediated transactivation in vivo.
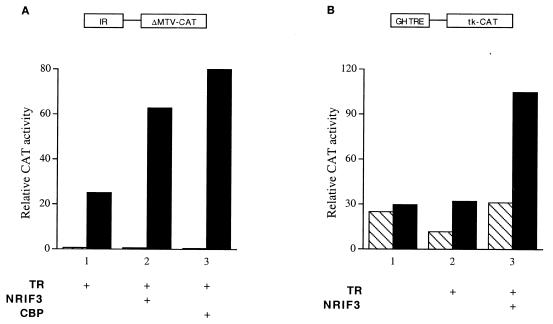
To examine the potential role of NRIF3 in TR-mediated transactivation, transfection studies were carried out. HeLa cells, which lack endogenous TR (25), were transfected with a vector expressing TR and a CAT reporter under the control of the ΔMTV basal promoter linked to an idealized IR (AGGTCATGACCT) TRE sequence (IR-ΔMTV-CAT) (25), along with either a control plasmid or a vector expressing NRIF3. As shown in Fig. 5A, NRIF3 significantly enhances TR-mediated activation of the CAT reporter (typically 2.5- to 3-fold). As a control, we also examined the effect of CBP, a reported coactivator for nuclear receptors (13, 37), and found that its expression results in a degree of enhancement similar to that with NRIF3 (around threefold) (Fig. 5A).
FIG. 5.
NRIF3 enhances TR-mediated transactivation in vivo. HeLa cells were transfected with a vector expressing cTRα and the IR-ΔMTV-CAT reporter (A) or the GH-TRE-tk-CAT reporter (B) in the presence (filled bars) or the absence (hatched bars) of 1 μM T3. The vector expressing NRIF3 or the empty control vector was cotransfected to examine the effect of NRIF3 on TR-mediated activation. In panel A, the effect of CBP was compared to that of NRIF3. GH, growth hormone.
We also examined another CAT reporter controlled by the herpesvirus tk promoter linked to native rat growth hormone TRE sequences (5). NRIF3 was found to also enhance TR-mediated activation of this reporter (about 3.5-fold) (Fig. 5B). In addition, using similar transfection assays, we found that NRIF3 enhances TR-mediated activation of two other reporters, (IR)2-TATA-CAT and DR4-ΔMTV-CAT (data not shown). Therefore, NRIF3 potentiates TR-mediated transactivation in a variety of different TRE and promoter contexts. Taken together, the results of these transfection studies suggest that NRIF3 can function as a coactivator of TR.
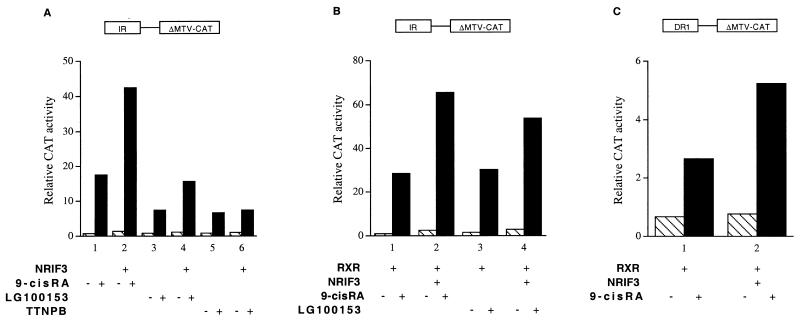
To examine whether NRIF3 can also act as a coactivator for RXR, HeLa cells were transfected with the IR-ΔMTV-CAT reporter, whose IR sequence can also function as a strong response element for the RXR(s) and RAR(s) (25, 49, 61). HeLa cells express the endogenous RXR(s) and RAR(s), as the activity of the IR-ΔMTV-CAT reporter was strongly stimulated by their cognate ligands, even without cotransfection of any receptor expression plasmid (Fig. 6A, bars 1, 3, and 5). Cotransfection of NRIF3 enhanced the activation of this reporter by either 9-cis RA, or LG100153 (72), an RXR-specific ligand (Fig. 6A, bars 1 and 2 and bars 3 and 4). In contrast, although the RAR-specific ligand TTNPB (68) also activated the IR-ΔMTV-CAT reporter, cotransfection of NRIF3 had no effect (Fig. 6A, bars 5 and 6). These results indicate that NRIF3 potentiates the activity of the endogenous RXR(s) but not the RAR(s), which is consistent with the distinct receptor specificity of NRIF3 revealed from the yeast two-hybrid assay (Table 1) and in vitro binding experiments (Fig. 4).
FIG. 6.
NRIF3 functions as a coactivator for RXR but not RAR. (A) NRIF3 potentiates the activity of the endogenous RXR(s) but not the RAR(s). HeLa cells were transfected with the IR-ΔMTV-CAT reporter (without any receptor expression vector) to examine the activation by endogenous retinoid receptors. The NRIF3 expression vector or the empty control vector was cotransfected to examine the effect of NRIF3 on the activity of the endogenous RXR(s) or RAR(s). Relative CAT activity was determined in the presence (filled bars) or absence (hatched bars) of the indicated ligands (1 μM). (B and C) NRIF3 potentiates the activity of the exogenously expressed RXR. A vector expressing hRXRα was cotransfected into HeLa cells with the IR-ΔMTV-CAT reporter (B) or the DR1-ΔMTV-CAT reporter (C) in the presence (filled bars) or absence (hatched bars) of the indicated ligands (1 μM). The effect of NRIF3 on RXR-mediated transactivation was examined as described for panel A. TTNPB, a synthetic ligand for RAR.
To further document that NRIF3 can function as a coactivator for RXR, a vector expressing exogenous RXR was cotransfected with IR-ΔMTV-CAT. Exogenous RXR expression enhanced the activation of this CAT reporter by either 9-cis RA or LG100153 (compare Fig. 6A and B, bars 1 and 3). This RXR-mediated activation of reporter expression was further stimulated by NRIF3 (Fig. 6B). Finally, we also examined the activation of a DR1-ΔMTV-CAT reporter. This DR1 (AGGTCANAGGTCA [where N is any nucleotide]) sequence is thought to be a specific response element for RXR (39, 51). Although we found that this DR1 is a weaker response element than the IR sequence, cotransfection of an RXR expression vector led to ligand-induced activation of this DR1 reporter, which was also further enhanced by NRIF3 (Fig. 6C).
NRIF3 does not potentiate the activities of GR, PR, ER, and VDR in vivo.
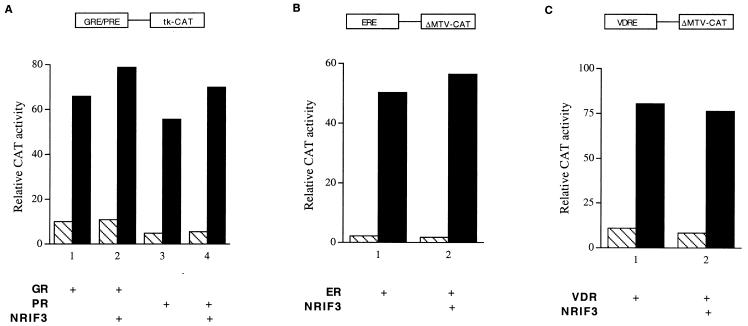
The selective coactivation of TR and RXR (but not RAR) by NRIF3 is consistent with its distinct binding specificities to these receptors. To further establish that NRIF3 acts as a receptor-specific coactivator, we next examined the effect of NRIF3 on the activities of four additional nuclear receptors, including GR, PR, ER, and VDR, by transfection experiments. HeLa cells were transfected with a GRE/PRE-tk-CAT reporter along with a vector expressing either GR or PR. As shown in Fig. 7A, cognate hormone treatment results in activation of the CAT reporter. However, expression of NRIF3 has little effect (Fig. 7A). Similar experiments were carried out with ER and ERE-ΔMTV-CAT or VDR and VDRE-ΔMTV-CAT. As shown in Fig. 7B and C, NRIF3 was found to have little or no effect on the activities of these receptors as well. Taken together, the combined results of our transfection studies support the notion that NRIF3 is a coactivator with a unique receptor specificity.
FIG. 7.
NRIF3 does not potentiate the activity of GR, PR, ER, or VDR. HeLa cells were transfected with the following CAT reporters and appropriate receptor expression vectors: GRE/PRE-tk-CAT and rGR or hPR (A), ERE-ΔMTV-CAT and hER (B), and VDRE-ΔMTV-CAT and hVDR (C). Cells were incubated in the presence (filled bars) or absence (hatched bars) of 100 nM dexamethasone for GR, progesterone for PR, estradiol for ER, and 1,25-(OH)2-vitamin D3 for VDR. Cotransfection of NRIF3 was found to have little effect on the activities of these receptors.
A novel C-terminal domain in NRIF3 is essential for ligand-dependent interactions with TR and RXR.
The LXXLL signature motif has been found to be present in the receptor-interacting domains of many identified coactivators, such as SRC-1/NCoA-1 and GRIP1/TIF-2 (32). The broad spectrum of receptor binding by coactivators such as SRC-1 suggests that the LXXLL-containing interacting domain may recognize structurally similar surfaces of these LBDs. Indeed, recent structural and functional studies revealed that the LXXLL motif and its nearby flanking amino acids are involved in direct contact with a hydrophobic cleft of the target surfaces presented by the ligand-bound LBDs of nuclear receptors (19, 23, 52, 56). The facts that NRIF3 also contains an LXXLL motif (amino acids 9 to 13) (Fig. 2 and 8A) and exhibits a distinct receptor specificity raise the possibility that (i) the motif and surrounding amino acids are involved in mediating receptor-specific interaction of NRIF3 or (ii) another region of NRIF3 (alone or in concert with the LXXLL motif region) plays an important role in mediating such an interaction.
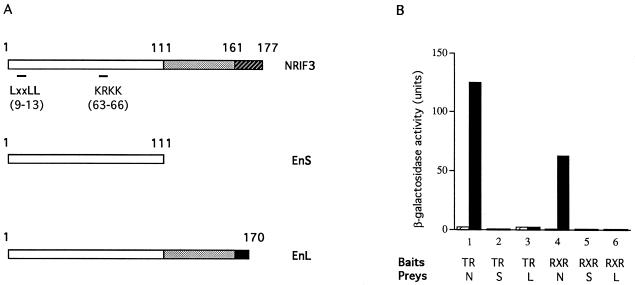
FIG. 8.
The NCD is essential for the interaction with liganded TR or RXR. (A) Schematic comparison of NRIF3 with EnS and EnL. EnS is 100% identical to the first 111 amino acids of NRIF3 and EnL (open boxes). The regions from amino acids 112 to 161 in NRIF3 and EnL (stippled boxes) are 100% identical. NRIF3 and EnL differ in their C termini (16 amino acids in NRIF3 [hatched box] and 9 amino acids in EnL [filled box]). The positions of the LXXLL motif and a putative nuclear localization signal (KRKK) are also indicated. (B) NRIF3 (N), EnS (S), or EnL (L) was examined for interaction with LexA-TR or LexA-RXR in a yeast two-hybrid assay as described in Materials and Methods. The assays were performed in the absence (hatched bars) or the presence (filled bars) of 1 μM T3 (for TR) or 9-cis RA (for RXR).
To explore these issues, we examined whether EnS and EnL, which contain the same LXXLL motif and flanking amino acids as NRIF3, can interact with nuclear receptors in a yeast two-hybrid assay (Fig. 8). EnS consists of 111 amino acids and is 100% identical to the first 111 residues of NRIF3, while EnL consists of 170 amino acids, the first 161 of which are also 100% identical to the same region in NRIF3 (Fig. 2 legend and Fig. 8A). Thus, NRIF3 and EnL differ only in their C termini, with NRIF3 having a unique region of 16 amino acids and EnL having a unique region of 9 amino acids (Fig. 8A). Interestingly, despite their extensive identity with NRIF3, the interaction with liganded TR or RXR is completely abolished in EnS and EnL (Fig. 8B). We also examined other nuclear receptors that do not interact with NRIF3 and found that they also do not interact with EnS or EnL (data not shown). These results indicate that the unique C-terminal domain in NRIF3 (NCD) (residues 162 to 177) is essential for its specific interaction with liganded TR and RXR while the N-terminal LXXLL motif (amino acids 9 to 13) and its flanking sequences are not sufficient to allow for detectable receptor interactions.
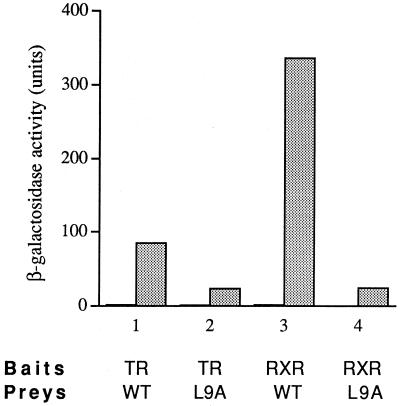
Although the LXXLL motif was found to be insufficient for interaction, we examined whether this N-terminal motif of NRIF3 contributes to the NRIF3-receptor interaction by mutating the first leucine of the LXXLL motif into alanine (L9A) by site-directed mutagenesis. Previous experiments have shown that the three leucine residues are essential for an LXXLL module to interact with receptor LBDs and that the replacement of any of them with alanine abolishes the interaction (32). We examined the L9A NRIF3 mutant form for its interaction with TR and RXR in a yeast two-hybrid assay. As shown in Fig. 9, the L9A mutant was still capable of ligand-dependent interaction with TR and RXR (∼25-fold induction by ligand). However, the introduced mutation reduced the interaction by about 4-fold (for TR) or 14-fold (for RXR). These results suggest that although the LXXLL motif is not absolutely essential for NRIF3 interaction with liganded receptors, it plays a role in allowing an optimum interaction to occur.
FIG. 9.
The LXXLL motif of NRIF3 is required for optimum interaction with TR and RXR. Wild-type NRIF3 (WT) or the L9A NRIF3 mutant (L9A) was examined for interaction with LexA-TR or LexA-RXR in a yeast two-hybrid assay as described in Materials and Methods. β-Galactosidase activities were determined in the absence (filled bars) or presence (stippled bars) of cognate ligands (1 μM T3 for TR; 1 μM 9-cis RA for RXR).
Computer modeling suggests that the NCD docks into the hydrophobic cleft of the liganded LBDs.
Secondary-structure analysis of the C-terminal domain of NRIF3 predicted the formation of an α-helix. Moreover, inspection of the putative C-terminal helix revealed an LXXIL motif (amino acids 172 to 176), which is reminiscent of the canonical LXXLL. Although the ultimate elucidation of the molecular basis of the NRIF3-receptor interaction awaits future studies such as X-ray crystallography, the putative helix structure of the NCD and its LXXIL motif suggest that the NCD may interact with the liganded LBDs in a fashion similar to that of the receptor-interacting domains that employ the canonical LXXLL motif(s). To explore this possibility, we modeled the interaction of the C terminus of NRIF3 with the liganded LBDs, using algorithms developed mainly by the staff of the laboratory of one of the authors (R. Abagyan and coworkers) (1, 63, 70, 74, 75). The background information and procedures used for constructing these models are described in Materials and Methods. The results of our modeling suggest that the NCD fits well into the hydrophobic cleft formed on the LBDs as a result of ligand binding. An example of such a model (NCD-TR LBD) is shown in Fig. 10. In this model, the two leucines and one isoleucine of the LXXIL motif are predicted to be deeply buried in the central cavity of the hydrophobic groove formed by the liganded LBD of the receptor. We also calculated the putative binding energy for the modeled NCD-TR complex, using an improved partitioning binding energy function, with continuum representation of the electrostatics of the system (64). The calculated binding energy for the modeled NCD-TR complex is about −21 kcal/mol. As a control, we carried out a similar modeling procedure using the second LXXLL box within the receptor-interacting domain of SRC-1. This LXXLL box has been shown to be required for interaction with TR (52). Our calculated binding energy for this LXXLL box with liganded TR LBD is −18 kcal/mol, a value that is very close to the one calculated for the NCD. Altogether, our modeling and calculations suggest a mechanism in which the NCD directly mediates interaction with liganded LBDs through an LXXIL motif.
Functional interaction of the NCD with liganded LBDs and the essential role of its LXXIL motif.
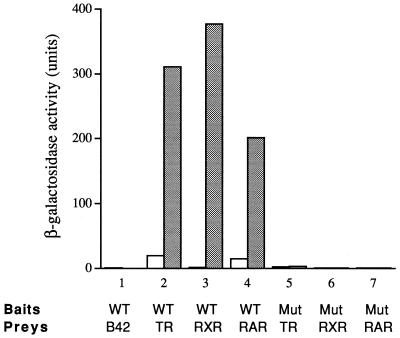
To explore the possibility suggested from our computer modeling, the NCD (amino acids 162 to 177) was fused to the LexA DNA binding domain and was examined for interaction with the receptor LBDs in a yeast two-hybrid assay. The LexA-NCD fusion protein alone does not activate the LacZ reporter in yeast (data not shown). As a negative control, we also found that LexA-NCD does not interact with the B42 activation domain itself (Fig. 11) and that LexA alone does not interact with the receptor LBDs (data not shown). However, when the LexA-NCD and the LBD of TR or RXR (fused with B42) were used in the two-hybrid assay, a strong ligand-dependent interaction was observed, as indicated by the induction of β-galactosidase activity by their cognate ligands (Fig. 11). These results suggest that the NCD can directly interact with the LBDs of TR and RXR in a ligand-dependent manner.
FIG. 11.
Interaction of the NCD with the receptor LBDs and the role of the LXXIL motif. The wild-type NCD (WT) or the NCD mutant form (Mut) in which the three core hydrophobic residues of the LXXIL motif (two leucines and one isoleucine) are changed into alanines was examined for interaction with the LBDs of TR, RXR, and RAR in a yeast two-hybrid assay as described in Materials and Methods. β-Galactosidase activities were determined in the absence (open bars) or presence (stippled bars) of cognate ligands. The prey expressing B42 alone was used as a negative control.
Since NRIF3 harbors a distinct receptor specificity in interacting only with TR and RXR and not other receptors (e.g., RAR), we next asked whether the NCD also harbors a receptor specificity. To our surprise, the NCD was found to interact efficiently with the LBD of RAR in a ligand-dependent manner (Fig. 11). Therefore, while our results clearly suggest that the NCD is an important surface for receptor interactions, as the NCD is found to be both essential for (Fig. 8) and sufficient to mediate (Fig. 11) such interactions, it nevertheless does not appear to be (solely) responsible for the receptor specificity of NRIF3. It is possible that another region of the NRIF3 molecule contributes to the observed receptor specificity of NRIF3 and/or that the specificity is determined by the overall three-dimensional structure of NRIF3.
Since our model predicts the importance of the LXXIL motif in the NCD-receptor interaction (Fig. 10), we tested this by changing the three core residues of the motif (two leucines and one isoleucine) into alanines. As expected, interaction with the LBDs is completely abolished in the resulting mutant NCD (Fig. 11), confirming that the LXXIL motif is essential for the interaction.
DISCUSSION
Recent efforts in understanding receptor-mediated transcription have led to the identification of a number of coactivators for nuclear hormone receptors, which can be categorized into two main groups based on overall homology, the SRC-1 family (including SRC-1/NCoA-1, TIF2/GRIP1/NCoA-2, and AIB1/p/CIP/ACTR/RAC3/TRAM-1) (2, 14, 34, 35, 37, 44, 58, 73, 74, 79) and the CBP/p300 family (13, 31, 37). Other putative coactivators (e.g., ARA70 and PGC-1) that do not belong to the SRC-1 or CBP/p300 family have also been identified (60, 85). In addition, p/CAF may also be involved in receptor action through its association with nuclear receptors as well as with other coactivators (11, 14, 38, 83). Among these known coactivators, CBP/p300, members of the SRC-1 group, and p/CAF all possess histone acetyltransferase activities (8, 14, 57, 69, 83).
In this study we report the identification of a novel nuclear protein (NRIF3) which exhibits specific ligand-dependent interactions with TR and RXR but not with RAR, VDR, GR, PR, or ER. Functional experiments indicated that NRIF3 potentiates TR- and RXR-mediated transactivation in vivo but exhibits little or no effect on the activities of other examined receptors. Therefore, NRIF3 represents a novel coactivator with a distinct receptor specificity and, thus, may shed light on clarifying the molecular mechanism(s) underlying receptor-specific regulation of gene expression.
A database search indicated that NRIF3 has no homology with any known coactivators except in a single LXXLL motif. An unusual feature of NRIF3 is its relatively small size, which is in sharp contrast to the sizes of SRC-1 and CBP/p300. A homology search identified two alternatively spliced isoforms of NRIF3 which were previously designated β3-endonexin short and long forms (67). Preliminary studies with these two endonexins indicate that, like NRIF3, they localize to the cell nucleus (43a, 66a). Interestingly, despite their extensive identities with NRIF3, both EnS and EnL fail to exhibit interaction with liganded nuclear receptors (Fig. 8). Consistent with this finding, we found that EnS and EnL have little effect on receptor-mediated transcription in transfection experiments (data not shown). Therefore, the precise roles of these two endonexins remain to be elucidated. We suggest two not mutually exclusive possibilities. First, since both EnL and EnS appear to localize to the nucleus, it is possible that they act as cofactors for other transcriptional regulators. Second, since the EnS can interact with the cytoplasmic tail of β3-integrin (22, 67), it may communicate signals generated at the plasma membrane to the cell nucleus. An example of a protein which is involved in both cell adhesion and transcriptional regulation is β-catenin (82).
Previous study of the endonexins identified the presence of NRIF3-related mRNAs (by Northern blotting) in a wide range of human tissues (67). Because NRIF3 and EnL contain almost identical nucleotide sequences and differ only by an alternative splice site which results in the removal of a small exon in NRIF3, it is difficult to specifically identify NRIF3 mRNA by Northern blotting. A search of the expressed sequence tag database indicates that NRIF3 mRNA, as well as both EnL and EnS mRNAs, is expressed. However, the precise determination of cell and tissue distribution of NRIF3, EnS, and EnL will require the development of highly selective antibodies. Nevertheless, the wide expression pattern of NRIF3-related mRNAs is consistent with the role of NRIF3 as a coactivator of the TRs, which are also widely expressed (70), or the RXRs, which are ubiquitously expressed (48).
A key goal concerning the action of nuclear hormone receptors is to understand the molecular events underlying the functional specificities of different receptors in regulating the expression of their target genes. Determinants of specificity include specific ligand binding and selective binding of the receptors to their cognate response elements, as well as specific expression patterns of different receptors. These determinants alone, however, are not always sufficient to explain the extents of specificity observed for members of the nuclear receptor family. For example, several members of the thyroid hormone/retinoid receptor subfamily may bind similarly to common DNA elements while target genes containing those elements are only selectively activated by certain receptors (20, 47). Therefore, it is likely that additional factors (determined by cell and promoter contexts) are involved in determining receptor functional specificity. In this respect, most known coactivators do not appear to be receptor specific. For example, members of the SRC-1 and CBP/p300 families interact with and appear to be involved in the actions of many nuclear receptors (13, 14, 34, 37). Two known coactivators that may be involved in receptor-specific functions are ARA70 and PGC-1. The AR coactivator ARA70 has been reported to potentiate the activity of AR more efficiently than it does the activities of other nuclear receptors (85). However, whether ARA70 can associate with other receptors remains to be thoroughly examined. The expression of PGC-1 is restricted mainly to the brown fat tissue and is thought to be directly involved in the regulation of thermogenesis by PPARγ (60). Nevertheless, PGC-1 exhibits a relatively broad spectrum of binding to different nuclear receptors. Therefore, the identification of NRIF3 represents the first example of a coactivator with such a clearly defined receptor specificity.
The receptor specificity of NRIF3 raises an interesting question about its molecular mechanism. Domain analysis suggests that the LXXLL motif (amino acids 9 to 13) and its flanking sequences in NRIF3 are not sufficient for interaction with liganded nuclear receptors. In fact, such interaction is completely abolished in EnL, an alternatively spliced product which has the same LXXLL motif and contains the first 161 amino acids (of a total of 177 amino acids) of NRIF3. This result suggests that a putative domain consisting of the last 16 amino acids of NRIF3 (residues 162 to 177) is essential for its interaction with liganded receptors. Inspection of this NCD indicates that it contains an LXXIL motif (amino acids 172 to 176), and secondary-structure analysis predicts the formation of an α-helix. The predicted helix structure and the similarity of LXXIL to the canonical LXXLL raise the possibility that this LXXIL-containing region plays a direct role in NRIF3-receptor interactions.
Our modeling of the NCD-LBD interaction (Fig. 10) suggests that the same hydrophobic groove in the ligand-bound LBD, which has been shown by previous studies to be the binding site for coactivators such as SRC-1/NCoA-1 and GRIP1 (19, 23, 56), may also be a suitable site for the docking of the C-terminal helix of NRIF3. Thus, the utilization of the complementary pair of an α-helix (in the coactivator) and a hydrophobic groove (in the receptor) for interaction seems to be a general scheme that also applies to NRIF3. The binding energy estimated for the NCD and the TR LBD (−21 kcal/mol) is similar to the value calculated for the second LXXLL box of SRC-1/NCoA-1 and the TR LBD (−18 kcal/mol). To explore the mechanisms suggested by the modeling, we found that the NCD can directly mediate interaction with the LBDs in a ligand-dependent manner (Fig. 11). Moreover, the LXXIL motif contained in the NCD was found to be essential for such interactions (Fig. 11). In summary, the results of a combination of a computer modeling and domain and mutagenesis analyses clearly suggest that the NCD is an important surface that is directly involved in interaction with the LBDs of the receptors, where the LXXIL motif of the NCD mimics the function of a canonical LXXLL. The AF-2 helix (which is a critical constituent of the hydrophobic groove formed upon ligand binding) of the LBD has been shown to be important for interaction with the LXXLL boxes of the coactivators (23). Interestingly, we have examined two TR AF-2 mutants (66) and found that in both cases, ligand-dependent interaction with NRIF3 was abolished (43a).
However, the NCD alone does not appear to harbor the same specificity as NRIF3 (Fig. 11). Thus, it seems likely that another part of the NRIF3 molecule contributes to the observed specificity and/or that the specificity is determined by the overall three-dimensional structure of NRIF3. In this regard, the potential role of the N-terminal LXXLL motif is intriguing. Although the N-terminal LXXLL motif (amino acids 9 to 13) is insufficient alone to mediate an interaction with TR or RXR (Fig. 8), it can influence the interaction of NRIF3 with these receptors, as the L9A NRIF3 mutant retains a significant but nevertheless reduced level of association with liganded TR or RXR (Fig. 9). Thus, NRIF3 appears to employ at least two regions in interacting with liganded TR or RXR, with the NCD playing an essential role and the N-terminal LXXLL motif playing a secondary role. A simplified explanation for these findings is that the NCD provides a major surface for receptor binding while the N-terminal LXXLL motif makes some minor contact with either the same receptor molecule or, more likely, with the other partner of a homodimer or heterodimer to further stabilize the NRIF3-receptor interaction. An example of a coactivator molecule employing two separate regions to interact with the two partners of a receptor dimer has been documented for the recently solved crystal structure of liganded PPARγ complexed with SRC-1/NCoA-1 (56). If NRIF3 indeed employs both its NCD and its N-terminal LXXLL motif in receptor interactions, the specificity may result from the intramolecular dialog between the two regions as well as the intermolecular dialog among NRIF3 and the receptors. However, it remains possible that the N-terminal LXXLL plays only a more indirect role and that the overall three-dimensional structure of NRIF3 is responsible for its observed specificity.
Accumulating evidence suggests that the actions of transcriptional activating proteins are (usually) mediated by multiprotein complexes (59), and such a scheme is also likely for nuclear receptors. For example, biochemical evidence suggests that multiprotein complexes associate with liganded TR and VDR (24, 62, 86). Interestingly, many of the proteins identified in these studies are not known coactivators. While the study of known coactivators such as CBP/p300 and members of the SRC-1 family has suggested that histone acetylation may play an important role in receptor-mediated transactivation (8, 14, 57, 69), detailed elucidation of the transactivation mechanism(s) used by these receptors awaits the identification and study of additional cofactors involved in the transactivation process.
Our results with NRIF3 suggest that transcriptional activation by nuclear receptors may employ a receptor-specific coactivator(s) in addition to the generally used coactivators such as CBP and SRC-1. Therefore, coactivators with NRIF3-like properties may contribute to the functional specificities of nuclear receptors in vivo. Based on our results with NRIF3 and the results of previous studies of nuclear receptor action, we suggest a combinatorial specificity model where a coactivation complex is likely composed of two kinds of factors: general factors that interact with and are involved in the action of many nuclear receptors (such as CBP and SRC-1) and specific factors that exhibit receptor specificity (such as NRIF3). In addition to interacting with the liganded receptor, coactivators may also communicate with each other within the coactivation complex through protein-protein interactions (e.g., CBP/p300 can interact with SRC-1/NCoA-1 or p/CIP) (37, 74, 84). An intriguing possibility is that the combinatorial actions of specific factors and other partners involved in the transactivation process facilitate the recruitment of specific coactivation complexes for different receptors (under different cell, promoter, and HRE contexts), which would provide an important mechanistic layer for receptor functional specificity. An advantage of employing such a combinatorial strategy is that a broad array of diversity can be generated from a relatively small number of involved factors. Further study of NRIF3 with known and possibly other yet to be identified coactivators, as well as analysis of the interplay among these coactivators, should provide important insights into the molecular mechanism(s) underlying the specificity of receptor-mediated regulation of target gene expression.
ACKNOWLEDGMENTS
We are grateful to Richard Goodman, Bert O’Malley, Ming-Jer Tsai, Michael Garabedian, J. Wesley Pike, Gunther Schutz, Ron Evans, and David Moore for plasmids and Richard Heyman of Ligand, Inc., for providing the retinoids. We thank Sanford Shattil for GFP-EnL and GFP-EnS; Gordon Fishell for help with fluorescence microscopy; Chun Wong, Ula Huang, Sidney Guo, and Paul Modlinger for experimental assistance; and Bruce Raaka and Fred Stanley for advice with graphic preparations.
This research was supported by NIH grant DK16636 (to H.H.S.), NRSA postdoctoral fellowship award DK09581 (to D.L.), DOD grant DAMD179818133, NIH grant GM5541801, DOE grant DEFG0296ER62268 (to R.A. and M.S.), and an NIH short-term training grant for students in health professional schools (DK07421) (to E.L.). V.D.-Y. was supported in part by The Aaron Diamond Foundation (grant HRI817-5332F). H.H.S., V.D.-Y., and R.A. are members of the NYUMC Cancer Center (grant CA16087). Sequence analysis and database searches were through the NYUMC Research Computing Resource, which received support from the National Science Foundation (grant DIR-8908095).
REFERENCES
- 1.Abagyan R A, Totrov M M, Kuznetsov D A. ICM: a new method for structure modeling and design: applications to docking and structure prediction from the distorted native conformation. J Comp Chem. 1994;15:488–506. [Google Scholar]
- 2.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 3.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature. 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 4.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 5.Au-Fliegner M, Helmer E, Casanova J, Raaka B M, Samuels H H. The conserved ninth C-terminal heptad in thyroid hormone and retinoic acid receptors mediates diverse responses by affecting heterodimer but not homodimer formation. Mol Cell Biol. 1993;13:5725–5737. doi: 10.1128/mcb.13.9.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baniahmad A, Leng X, Burris T P, Tsai S Y, Tsai M-J, O’Malley B W. The τ4 activation domain of the thyroid hormone receptor is required for release of a putative corepressor(s) necessary for transcriptional silencing. Mol Cell Biol. 1995;15:76–86. doi: 10.1128/mcb.15.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bannister A J, Kouzarides T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 9.Barettino D, Ruiz M D M V, Stunnenberg H G. Characterization of the ligand-dependent transactivation domain of thyroid hormone receptor. EMBO J. 1994;13:3039–3049. doi: 10.1002/j.1460-2075.1994.tb06603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston D M. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 11.Blanco J C G, Minucci S, Lu J, Yang X J, Walker K K, Chen H, Evans R M, Nakatani V, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Casanova J, Helmer E, Selmi-Ruby S, Qi J-S, Au-Fliegner M, Desai-Yajnik V, Koudinova N, Yarm F, Raaka B M, Samuels H H. Functional evidence for ligand-dependent dissociation of thyroid hormone and retinoic acid receptors from an inhibitory cellular factor. Mol Cell Biol. 1994;14:5756–5765. doi: 10.1128/mcb.14.9.5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 14.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 15.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 16.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 17.Conneely O M, Sullivan W P, Toft D O, Birnbaumer M, Cook R G, Maxwell B L, Zaraucko-Schulz T, Greene G L, Schraeder W T, O’Malley B W. Molecular cloning of the chicken progesterone receptor. Science. 1986;233:767–770. doi: 10.1126/science.2426779. [DOI] [PubMed] [Google Scholar]
- 18.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 19.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desai-Yajnik V, Samuels H H. The NF-κB and Sp1 DNA motifs of the human immunodeficiency virus type 1 long terminal repeat function as novel thyroid hormone response elements. Mol Cell Biol. 1993;13:5057–5069. doi: 10.1128/mcb.13.8.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durand B, Saunders M, Gausdon C, Roy B, Losson R, Chambon P. Activation function 2 (AF-2) of retinoic acid receptor and 9-cis retinoic acid receptor: presence of a conserved autonomous constitutive activating domain and influence of the nature of the response element on AF-2 activity. EMBO J. 1994;13:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eigenthaler M, Hofferer L, Shattil S J, Ginsberg M H. A conserved sequence motif in the integrin β3 cytoplasmic domain is required for its specific interaction with β3-endonexin. J Biol Chem. 1997;272:7693–7698. doi: 10.1074/jbc.272.12.7693. [DOI] [PubMed] [Google Scholar]
- 23.Feng W, Ribeiro R C, Wagner R L, Nguyen H, Apriletti J W, Fletterick R J, Baxter J D, Kushner P J, West B L. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 24.Fondell J D, Ge H, Roeder R G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forman B M, Casanova J, Raaka B M, Ghysdael J, Samuels H H. Half-site spacing and orientation determines whether thyroid hormone and retinoic acid receptors and related factors bind to DNA response elements as monomers, homodimers, or heterodimers. Mol Endocrinol. 1992;6:429–442. doi: 10.1210/mend.6.3.1316541. [DOI] [PubMed] [Google Scholar]
- 26.Giguere V, Ong E S, Segui P, Evans R M. Identification of a receptor for the morphogen retinoic acid. Nature. 1987;330:624–629. doi: 10.1038/330624a0. [DOI] [PubMed] [Google Scholar]
- 27.Glass C K, Rose D W, Rosenfeld M G. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 28.Gu W, Shi X L, Roeder R G. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 29.Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 30.Hadzic E, Desai-Yajnik V, Helmer E, Guo S, Wu S, Koudinova N, Casanova J, Raaka B M, Samuels H H. A 10-amino-acid sequence in the N-terminal A/B domain of thyroid hormone receptor α is essential for transcriptional activation and interaction with the general transcription factor TFIIB. Mol Cell Biol. 1995;15:4507–4517. doi: 10.1128/mcb.15.8.4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 33.Heinzel T, Lavinsky R M, Mullen T M, Soderstrom M, Laherty C D, Torchia J, Yang W M, Brard G, Ngo S D, Davie J R, Seto E, Eisenman R N, Rose D W, Glass C K, Rosenfeld M G. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 34.Hong H, Kohli K, Garabedian M, Stallcup M R. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamil Y, Soderstrom M, Glass C K, Rosenfeld M G. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 37.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 38.Korzus E, Torchia J, Rose D W, Xu L, Kurokawa R, McInerney E M, Mullen T M, Glass C K, Rosenfeld M G. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 39.Kurokawa R, DiRenzo J, Boehm M, Sugarman J, Glass B, Rosenfeld M G, Heyman R A, Glass C K. Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature. 1994;371:528–531. doi: 10.1038/371528a0. [DOI] [PubMed] [Google Scholar]
- 40.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 41.Lanz R B, McKenna N J, Onate S A, Albrecht U, Wong J, Tsai S Y, Tsai M J, O’Malley B W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 42.Lee J W, Choi H-S, Gyuris J, Brent R, Moore D D. Two classes of proteins dependent on either the presence or absence of thyroid hormone for interaction with the thyroid hormone receptor. Mol Endocrinol. 1995;9:243–254. doi: 10.1210/mend.9.2.7776974. [DOI] [PubMed] [Google Scholar]
- 43.Leng X, Blanco J, Tsai S Y, Ozato K, O’Malley B W, Tsai M-J. Mouse retinoid X receptor contains a separable ligand-binding and transactivation domain in its E region. Mol Cell Biol. 1995;15:255–263. doi: 10.1128/mcb.15.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.Li, D., and H. H. Samuels. Unpublished data.
- 44.Li H, Gomes P J, Chen J D. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 46.Lundblad J R, Kwok R P, Laurance M E, Harter M L, Goodman R H. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature. 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 47.MacDonald P N, Dowd D R, Nakajima S, Galligan M A, Reeder M C, Haussler C A, Ozato K, Haussler M R. Retinoid X receptors stimulate and 9-cis retinoic acid inhibits 1,25-dihydroxyvitamin D3-activated expression of the rat osteocalcin gene. Mol Cell Biol. 1993;13:5907–5917. doi: 10.1128/mcb.13.9.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mangelsdorf D J, Borgmeyer U, Heyman R A, Zhou J Y, Ong E S, Oro A E, Kakizuka A, Evans R M. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev. 1992;6:329–344. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- 49.Mangelsdorf D J, Evans R M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 50.Mangelsdorf D J, Ong E S, Dyck J A, Evans R M. Nuclear receptor that identifies a novel retinoic acid response pathway. Nature. 1990;345:224–229. doi: 10.1038/345224a0. [DOI] [PubMed] [Google Scholar]
- 51.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McInerney E M, Rose D W, Flynn S E, Westin S, Mullen T M, Krones A, Inostroza J, Torchia J, Nolte R T, Assa-Munt N, Milburn M V, Glass C K, Rosenfeld M G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miesfeld R, Rusconi S, Godowski P J, Maler B A, Okret S, Wikstrom A-C, Gustafsson J-A, Yamamoto K R. Genetic complementation of a glucocorticoid receptor deficiency by expression of cloned receptor cDNA. Cell. 1986;46:389–399. doi: 10.1016/0092-8674(86)90659-8. [DOI] [PubMed] [Google Scholar]
- 54.Nagpal S, Friant S, Nakshatri H, Chambon P. RARs and RXRs: evidence for two autonomous transactivation functions (AF-1 and AF-2) and heterodimerization in vivo. EMBO J. 1993;12:2349–2360. doi: 10.1002/j.1460-2075.1993.tb05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nagy L, Kao H V, Chakravarti D, Lin R J, Hassig C A, Ayer D E, Schreiber S L, Evans R M. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 56.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 57.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 58.Onate S A, Tsai S Y, Tsai M-J, O’Malley B W. Sequence and characterization of a coactivator of the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 59.Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- 60.Puigserver P, Wu Z, Park C W, Graves R, Wright M, Spiegelman B M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 61.Qi J-S, Desai-Yajnik V, Greene M E, Raaka B M, Samuels H H. The ligand binding domains of the thyroid hormone/retinoid receptor gene subfamily function in vivo to mediate heterodimerization, gene silencing, and transactivation. Mol Cell Biol. 1995;15:1817–1825. doi: 10.1128/mcb.15.3.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rachez C, Suldan Z, Ward J, Chang C P, Burakov D, Erdjument-Bromage H, Tempst P, Freedman L P. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Renaud J-P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-γ ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 64.Schapira M, Totrov M, Abagyan R A. Prediction of the binding energy for small molecules, peptides and proteins. J Mol Recognit. 1999;12:177–190. doi: 10.1002/(SICI)1099-1352(199905/06)12:3<177::AID-JMR451>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 65.Schule R, Umesono K, Mangelsdorf D J, Bolado J, Pike J W, Evans R M. Jun-Fos and receptors for vitamins A and D recognize a common response element in the human osteocalcin gene. Cell. 1990;61:497–504. doi: 10.1016/0092-8674(90)90531-i. [DOI] [PubMed] [Google Scholar]
- 66.Selmi-Ruby S, Casanova J, Malhotra S, Roussett B, Raaka B M, Samuels H H. Role of the conserved C-terminal region of thyroid hormone receptor-α in ligand-dependent transcriptional activation. Mol Cell Endocrinol. 1998;138:105–114. doi: 10.1016/s0303-7207(98)00016-1. [DOI] [PubMed] [Google Scholar]
- 66a.Shattil, S. J. Personal communication.
- 67.Shattil S J, O’Toole T, Eigenthaler M, Thon V, Williams M, Babior B M, Ginsberg M H. β3-Endonexin, a novel polypeptide that interacts specifically with the cytoplasmic tail of the integrin β3 subunit. J Cell Biol. 1995;131:807–816. doi: 10.1083/jcb.131.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sheikh M S, Shao Z-M, Li X-S, Dawson M, Jetten A M, Wu S, Conley B A, Garcia M, Rochefort H, Fontana J A. Retinoid-resistant estrogen receptor-negative human breast carcinoma cells transfected with retinoic acid receptor-α acquire sensitivity to growth inhibition by retinoids. J Biol Chem. 1994;269:21440–21447. [PubMed] [Google Scholar]
- 69.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O’Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 70.Strait K, Schwartz H L, Perez-Castillo A, Oppenheimer J H. Relationship of c-erbA mRNA content to tissue triiodothyronine nuclear binding capacity and function in developing and adult rats. J Biol Chem. 1990;265:10514–10521. [PubMed] [Google Scholar]
- 71.Strynadka N C, Eisenstein M, Katchalski-Katzir E, Shoichet B K, Kuntz I D, Abagyan R, Totrov M, Janin J, Cherfils J, Zimmerman F, Olson A, Duncan B, Rao M, Jackson R, Sternberg M, James M N. Molecular docking programs successfully predict the binding of a beta-lactamase inhibitory protein to TEM-1 beta-lactamase. Nat Struct Biol. 1996;3:233–239. doi: 10.1038/nsb0396-233. [DOI] [PubMed] [Google Scholar]
- 72.Sugawara A, Sanno N, Takahashi N, Osamura R Y, Abe K. Retinoid X receptors in the kidney: their protein expression and functional significance. Endocrinology. 1997;138:3175–3180. doi: 10.1210/endo.138.8.5351. [DOI] [PubMed] [Google Scholar]
- 73.Takeshita A, Cardona G R, Koibuchi N, Suen C S, Chin W W. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 74.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 75.Totrov M, Abagyan R. Detailed ab initio prediction of lysozyme-antibody complex with 1.6 Å accuracy. Nat Struct Biol. 1994;1:259–263. doi: 10.1038/nsb0494-259. [DOI] [PubMed] [Google Scholar]
- 76.Totrov M, Abagyan R. Flexible protein-ligand docking by global energy optimization in internal coordinates. Proteins. 1997;1997(Suppl. 1):215–220. doi: 10.1002/(sici)1097-0134(1997)1+<215::aid-prot29>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
- 77.Umesono K, Evans R M. Determinants of target gene specificity for steroid/thyroid hormone receptors. Cell. 1989;57:1139–1146. doi: 10.1016/0092-8674(89)90051-2. [DOI] [PubMed] [Google Scholar]
- 78.Umesono K, Murakami K K, Thompson C C, Evans R M. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Voegel J J, Heine M J S, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 80.Wagner R L, Apriletti J W, McGrath M E, West B L, Baxter J D, Fletterick R J. A structural role for hormone in the thyroid hormone receptor. Nature. 1995;378:690–697. doi: 10.1038/378690a0. [DOI] [PubMed] [Google Scholar]
- 81.Waterman M L, Adler S, Nelson C, Greene G L, Evans R M, Rosenfeld M G. A single domain of the estrogen receptor confers deoxyribonucleic acid binding and transcriptional activation of the rat prolactin gene. Mol Endocrinol. 1988;2:14–21. doi: 10.1210/mend-2-1-14. [DOI] [PubMed] [Google Scholar]
- 82.Willert K, Nusse R. Beta-catenin: a key mediator of Wnt signaling. Curr Opin Genet Dev. 1998;8:95–102. doi: 10.1016/s0959-437x(98)80068-3. [DOI] [PubMed] [Google Scholar]
- 83.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 84.Yao T P, Ku G, Zhou N, Scully R, Livingston D M. The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yeh S, Chang C. Cloning and characterization of a specific coactivator, ARA70, for the androgen receptor in human prostate cells. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yuan C X, Ito M, Fondell J D, Fu Z Y, Roeder R G. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci USA. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath C M, Darnell J E., Jr Two contact regions between Stat1 and CBP/p300 in interferon gamma signaling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]