Abstract
The type 1 insulin-like growth factor receptor (IGF-1R), activated by its ligands, protects several cell types from a variety of apoptotic injuries. The main signaling pathway for IGF-1R-mediated protection from apoptosis has been previously elucidated and rests on the activation of phosphatidylinositol 3-kinase, Akt/protein kinase B, and the phosphorylation and inactivation of BAD, a member of the Bcl-2 family of proteins. In 32D cells (a murine hemopoietic cell line devoid of insulin receptor substrate 1 [IRS-1]), the IGF-1R activates alternative pathways for protection from apoptosis induced by withdrawal of interleukin-3. One of these pathways leads to the activation of mitogen-activated protein kinase, while a third pathway results in the mitochondrial translocation of Raf and depends on the integrity of a group of serines in the C terminus of the receptor that are known to interact with 14.3.3 proteins. All three pathways, however, result in BAD phosphorylation. The presence of multiple antiapoptotic pathways may explain the remarkable efficacy of the IGF-1R in protecting cells from apoptosis.
The protective effect of the activated type 1 insulin-like growth factor receptor (IGF-1R) on cell survival has been known for some time (see Discussion). The variety of procedures used to induce apoptosis suggests that the wild-type IGF-1R and/or its ligands have a widespread antiapoptotic effect on many death signals. Indeed, McCarthy et al. (50) have suggested that only insulin-like growth factor 1 (IGF-1) and Bcl-2 truly suppress the initiation of the apoptotic program while inhibitors of caspases can arrest the completion of the program but have no effect on its initiation. The mechanism by which the IGF-1R protects cells from apoptosis has been the object of a series of investigations, culminating in a reasonable elucidation of the main pathway used by this receptor for protection against apoptotic injuries. This pathway originates with the interaction of the IGF-1R with one of its major substrates, insulin receptor substrate 1 (IRS-1) (53), which activates phosphatidylinositol 3-kinase (PI3-ki), which in turn activates Akt/protein kinase B (PKB) (17, 34, 36, 38). The concluding step is the phosphorylation, by Akt/PKB, of BAD (13, 14), one of the members of the Bcl-2 family of proteins (see below). This antiapoptotic mechanism is also used by the insulin receptor (IR), at least in mouse embryo fibroblasts (62).
The connection between the IGF-1R and BAD is an important one, since Bcl-2 and the proteins constituting the Bcl-2 family also play an important role in the apoptotic process. There are only a few reports of a link between the IGF-1R and the Bcl-2 family of proteins. For instance, there are two reports (58, 79) indicating that the activated IGF-1R may modulate the level of expression of Bcl-XL by increasing the protein levels. Among other proteins of the Bcl-2 family, BAD is known to be a heterodimeric partner for both Bcl-XL and Bcl-2, neutralizing their protective effect and promoting cell death (91). As mentioned above, in response to survival factors, including IGF-1 (13), the Akt/PKB pathway is activated (21); additionally, BAD is serine phosphorylated by Akt, is no longer capable of forming a heterodimer with Bcl-XL at membrane sites, is sequestered in the cytosol (bound to 14.3.3), and is inactivated as a cell death-promoting protein (94). The various effects of the IGF-1R (mitogenesis, protection from apoptosis, transformation, and differentiation) have been recently mapped to different domains of the receptor (3, 85). It would therefore be relevant, in itself, to investigate the relationship between BAD phosphorylation and the domains of the IGF-1R that are required for this process. This investigation is also justified by a recent report (39) that the IGF-1R may protect cells from apoptosis independently of BAD phosphorylation.
There is another reason to investigate this relationship. While it is clear that the above-mentioned pathway is the main pathway by which the IGF-1R exerts its antiapoptotic effect, some investigators have suggested that this receptor must have alternative pathways. The first clue to alternative pathways has been provided by 32D cells, a murine hemopoietic cell line (26) that is dependent on interleukin-3 (IL-3) for growth and undergoes apoptosis after IL-3 withdrawal. 32D cells lack both IRS-1 and IRS-2 (88, 92, 95); yet, when overexpressing the wild-type IGF-1R, they are protected from apoptosis and actually grow in the absence of IL-3 (16, 61). Interestingly, 32D cells fail to grow without IL-3 when overexpressing the IR, although they do grow without IL-3 when stably transfected with plasmids expressing both the IR and IRS-1 (reference 88 and this paper). Overexpression of IRS-1, by itself, offers only partial protection under the same conditions (93, 95). This alternative pathway is also suggested by the finding that inhibitors of PI3-ki fail to abrogate the protective effect of the IGF-1R against apoptosis (39, 84). Thus, it seems that for mitogenesis and/or survival the IGF-1R can use a pathway that is IRS-1 independent and is not shared with the IR. This is confirmed by the observation that the IGF-1R and the IR have similar antiapoptotic properties when transfected into mouse embryo fibroblasts, which have substantial amounts of IRS proteins (62). The existence of alternative signaling in IGF-1R-mediated survival has also been suggested by other investigators. One suggested that the alternative pathway could involve the mitogen-activated protein kinase (MAPK) pathway (57), which originates, at least in part, from another major substrate of the IGF-1R, the Shc proteins (63, 73, 74).
In this investigation, we have asked two specific questions. First, which domains of the IGF-1R are required for the phosphorylation of BAD? Are they exactly the same as the domains required for protection from apoptosis, or are there differences? Second, which is the alternative pathway(s) used by the IGF-1R to protect 32D cells from apoptosis?
The answer to the first question is that in 32D cells, BAD phosphorylation correlates quite well with IGF-1-dependent survival, and the IGF-1R mutants that sustain survival also maintain high levels of BAD phosphorylation. The answer to the second question is that the IGF-1R, although capable of using the same PI3-ki/Akt/BAD pathway already repeatedly demonstrated in other cells (13, 17, 38), activates at least two alternative pathways that still lead to BAD phosphorylation.
MATERIALS AND METHODS
Plasmids.
pHIT60 and pHIT123 were kindly provided by Alan Kingsman (Oxford University, Oxford, United Kingdom) and are described elsewhere (80). pMSCV neo EB was kindly provided by Robert G. Hawley (University of Toronto, Toronto, Ontario, Canada) and is described by Hawley et al. (29). pMSCV neo IR is described by Prisco et al. (62). The wild-type IGF-1R and all of its mutants are described by Romano et al. (70) (for the retroviral vectors) as well as by D’Ambrosio et al. (11), Hongo et al. (30), and O’Connor et al. (56) (for the original plasmids). pLXSP M-Raf-1 is described by Salomoni et al. (72).
Full-length BAD cDNA was cloned as follows. An aliquot of cDNA obtained after reverse transcription of 2 μg of total RNA from 32D cells was subjected to PCR with the following primers: forward, 5′ ATGGGAACCCCAAAGCAGCC 3′; and reverse, 5′ AGAAGATCACTGGGAGGGGGT 3′. The 620-bp PCR fragment was directly cloned by using a TA cloning kit (Invitrogen), and its identity was confirmed by automatic sequencing, using a Taq Dye Deoxy Terminator Cycle Sequencing kit (Applied Biosystems). Full-length BAD cDNA was then subcloned at the EcoRI site of the pLXSP retroviral vector (59).
Cell lines and retroviral infections.
32D clone 3 cells, described by Valtieri et al. (86), were transduced with a murine leukemia virus-based retroviral vector (80) to express the wild-type IGF-1R, its various mutants, or the IR. 32D/IRS-1 cells, described by Wang et al. (88), were transduced with pMSCV neo IR to generate 32D/IRS-1/IR. All mixed populations were selected in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 10% conditioned medium from the murine myelomonocytic cell line WEHI-3B as a source of IL-3 (61), and 1 mg of G418/ml. 32D cells and derivative cell lines were transduced with pLXSP-BAD, and the selection was carried out with 1.5 μg of puromycin/ml. There were no differences in growth properties or survival between the parental cell lines and those carrying the BAD plasmid.
Survival assay.
Cells growing exponentially were washed three times with Hanks balanced salt solution (HBSS) and seeded (5 × 104/35-mm-diameter plate in 2 ml of medium) in RPMI 1640 supplemented only with 10% FBS; RPMI 1640 supplemented with 10% FBS and 10% WEHI cell-conditioned medium as a source of IL-3, with or without LY294002 (20 μM); or RPMI 1640 supplemented with 10% FBS and 50 ng of IGF-1 or insulin (Gibco BRL)/ml, with or without PD98059 (50 μM) or LY294002 (10 μM). Cell numbers were determined in duplicate after 24 and 48 h; only cells excluding trypan blue were counted as viable.
Evidence of apoptosis.
DNA strand breaks were labeled with biotinylated dUTP (Boehringer Mannheim), using exogenous terminal deoxynucleotidyl transferase (TdT; Boehringer Mannheim). Cells were fixed on ice with buffered 10% methanol-free formaldehyde (Polyscience, Inc.) for 20 min, incubated with 70% ethanol for 1 h at −20°C, and washed three times with phosphate-buffered saline (PBS). DNA strand breaks were detected by in situ labeling of free 3′-OH ends on DNA. Fixed cells were incubated with 50 μl of a reaction mixture (5 U of TdT, 2.5 mM CoCl2, 0.2 M potassium cacodylate, 25 mM Tris-HCl, 0.25% bovine serum albumin, and 0.5 nM biotin–16-dUTP) for 30 min at 37°C and then incubated with staining solution (5 μg of fluorescein isothiocyanate/ml, 4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium acetate], 0.1% Triton X-100, and 5% nonfat dry milk) for 30 min at 37°C. DNA strand breaks were visualized under a fluorescence microscope equipped with a set of excitation-emission filters for fluorescein (excitation, blue; emission, green). To visualize all nuclei, total DNA was additionally stained with 500 μg of bisbenzimide H33258 (Sigma)/ml. Vectashield (Vector Laboratories, Burlingame, Calif.) mounting medium was subsequently applied, and the bromodeoxyuridine labeling index was determined by using a Zeiss microscope in the epifluorescence mode (magnification, ×500).
IGF-1R and IR expression levels.
Cells growing exponentially were lysed in a lysis buffer (20 mM Tris-HCl [pH 8.0], 1% NP-40, 10% glycerol, 137 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 1 mM sodium orthovanadate, 50 mM NaF, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml, 1 mM phenylmethylsulfonyl fluoride [PMSF]), and 20-μg quantities of whole-cell lysates were run on a sodium dodecyl sulfate (SDS) 4 to 15% polyacrylamide minigel (Bio-Rad) and then transferred to a nitrocellulose membrane (Schleicher & Schuell). After being blocked with 5% nonfat milk in TBS-T (10 mM Tri-HCl [pH 7.5], 150 mM NaCl, 0.1% Tween 20), filters were probed with an antibody to the α or β subunit of the IGF-1R or to the β subunit of IR (Santa Cruz Biotechnology, Inc.). Detection was carried out with the ECL system (Amersham Life Science).
BAD detection.
Exponentially growing cells were washed three times with HBSS and seeded in RPMI 1640 supplemented with 10% FBS only, with 10% FBS and 10% IL-3 (see above), or with 10% FBS and 50 ng of IGF-1 or insulin (Gibco BRL)/ml, with or without PD98059 (50 μM) or LY294002 (50 μM). Cells were harvested at various hours after IL-3 withdrawal and lysed as described before; 80-μg quantities of the whole-cell lysates were resolved on an SDS–15% polyacrylamide gel (30% acrylamide-bisacrylamide solution [29:1] and TEMED were from Bio-Rad; ammonium persulfate was from Sigma) and transferred to a nitrocellulose membrane. The anti-BAD antibody was from Transduction Laboratories. When required, the filters were stripped at 50°C for 30 min with a stripping buffer (100 mM 2-mercaptoethanol, 2% SDS, 62.5 mM Tris-HCl [pH 6.7]) and incubated with anti-Grb2 (Transduction Laboratories).
AKT assay.
Cells were washed three times with HBSS and seeded (3 × 105/ml) in RPMI 1640 supplemented with 10% FBS. After 5 h, the cells were stimulated with (i) 10% WEHI-conditioned medium for 10 min; (ii) 50 μM LY 294002 for 15 min, followed by 10% WEHI-conditioned medium for 10 min; (iii) 50 ng of IGF-1 or insulin/ml for 10 min; or (iv) 50 μM LY 294002 for 15 min, followed by 50 ng of IGF-1 or insulin for 10 min. Cells were lysed after stimulation. The lysis was performed as described above, and 50-μg quantities of the whole-cell lysate was loaded on an SDS–4 to 15% polyacrylamide minigel, subjected to electrophoresis, and transferred to a nitrocellulose filter. Phosphorylated AKT and total AKT were detected by using a Phosphoplus AKT (Ser473) antibody kit (New England Biolabs), which provides both phosphospecific AKT and AKT protein antibodies.
MAPK activation.
32D cell lines were incubated in serum-free medium for 3 h, stimulated with 50 ng of growth factor (IGF-1, IGF-2, or insulin)/ml for various periods of time, collected by centrifugation, and rinsed in ice-cold PBS. Cells were lysed in 100 μl of lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10% glycerol, 1% Triton X-100, 0.1 mM PMSF, 0.5 μg of aprotinin/ml, 0.5 μg of leupeptin/ml, and 0.1 mM Na3 VO4). Lysates (100 μg/lane) were resolved on 10% polyacrylamide gels, transferred to nitrocellulose, and immunoblotted with a phosphospecific antibody against ERK1/ERK2 as recommended by the manufacturer (Promega, Madison, Wis.). To confirm equal loading of proteins, filters were stripped and reblotted with an anti-ERK1 antibody which recognizes both ERK1 and ERK2 (Santa Cruz).
Immunoprecipitation.
Cells were washed three times with HBSS and seeded in RPMI 1640 supplemented with 10% FBS only, with 10% FBS and 10% IL-3 (WEHI cell-conditioned medium), or with 10% FBS and 50 ng of IGF-1 (Gibco BRL)/ml, with or without PD98059 (50 μM). Cells were harvested after 16 h and lysed as described above. Cell lysate (350 μg) was immunoprecipitated with an anti-BAD antibody (Santa Cruz). After resolution on an SDS–15% polyacrylamide gel and transfer to a nitrocellulose membrane, the immunoprecipitate was probed with an anti-14-3-3-β antibody (Santa Cruz). The filter was stripped and probed with the anti-BAD antibody (Transduction Laboratories).
Detection of mitochondrial Raf-1.
Exponentially growing cells were washed five times with HBSS and seeded in RPMI 1640 medium supplemented with 10% FBS only, with 10% FBS and 10% IL-3 (WEHI cell-conditioned medium), or with 10% FBS and 50 ng of IGF-1/ml. Cells were harvested after 30 h, washed twice with cold PBS, and lysed in buffer A (20 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 250 mM sucrose, 1 mM dithiothreitol, 1 mM PMSF, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml, 10 mM benzamidine, 0.2 mM sodium orthovanadate) at 4°C for 30 min. After homogenization (25 strokes with a Dounce homogenizer B pestle), samples were centrifuged two times (2,500 × g for 5 min) to remove the nuclei and centrifuged again at 13,000 × g for 30 min to obtain the heavy membrane pellet. This fraction was resuspended in buffer A and centrifuged again at 13,000 × g for 30 min. Finally, the pellet was resuspended in lysis buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 0.5% Triton X-100, 10 μg of aprotinin/ml, 1 mM PMSF, 1 mM sodium orthovanadate). Samples were then centrifuged at 13,000 × g for 20 min and measured for protein content. After SDS–4 to 15% polyacrylamide gel electrophoresis and blotting to membranes were performed, the membranes were probed with a Raf-1 antibody (Transduction Laboratories). The membranes were then stripped and reprobed with a cytochrome oxidase subunit IV antibody (Molecular Probes).
Inhibitors.
LY294002 was from BioMol. PD98059 was from New England Biolabs.
RESULTS
In previous experiments with 32D and other hemopoietic cell lines, the cells were stably transfected with plasmids expressing the wild-type IGF-1R or one of its mutants (16, 56, 61). In all of the following experiments, 32D cells were transduced with retroviral vectors (see Materials and Methods), which have two advantages: (i) the efficiency of transduction is high (very useful in 32D cells, which are notoriously difficult to transfect) and allows the selection of mixed populations, thus avoiding clonal variations; and (ii) the level of expression of the insert is usually reasonably uniform and high enough to ensure that most of the cells have a sufficient number of receptors to carry out adequately the various functions of the IGF-1R. Table 1 gives a list of the retroviral vectors used in these experiments; the 32D cell lines generated by transduction carry the same names as the retroviral vectors.
TABLE 1.
Retroviral vectors expressing the IGF-1 receptors used in these experimentsa
| Vector | Mutation(s) |
|---|---|
| GR15 | None (wild-type receptor) |
| GR18 | Tyrosines 950, 1131, 1135, and 1136 to phenylalanines |
| GR25 | Residues 1289, 1290, 1293, and 1294 to alanines |
| GR35 | Serines at positions 1280 to 1283 to alanines |
| GR36 | Truncated at residue 1245 |
| GR46 | Tyrosine 950 to phenylalanine |
| GR47 | Residues 1293 and 1294 to alanines |
| GR48 | Tyrosines 1131, 1135, and 1136 to alanines |
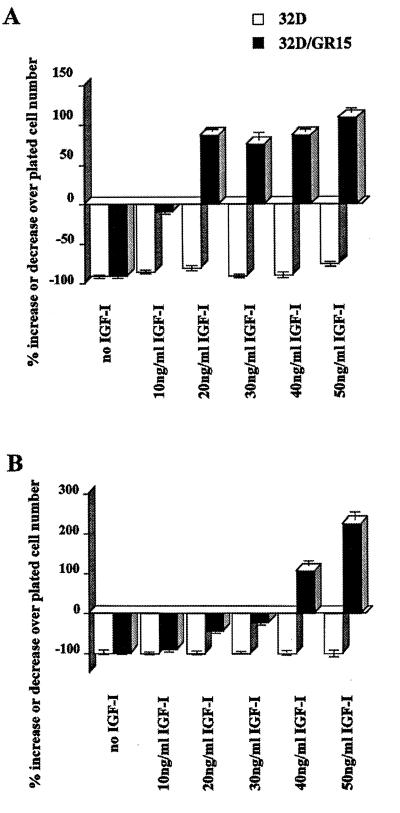
As a preliminary study, we determined the optimal levels of IGF-1 to be added to the FBS for protection of cells from apoptosis. Figure 1A shows that the parental 32D cells die very rapidly after IL-3 withdrawal, with 80 to 90% of the cells already dead by 24 h; only at 50 ng of IGF-1/ml does one notice a slight improvement in survival. With 32D/GR15 cells (wild-type receptor), survival at 24 h is already almost complete at 10 ng of IGF-1/ml; at higher concentrations, the cell numbers actually increase. However, if the cells are counted at 48 h, it is evident (Fig. 1B) that at least a 40-ng/ml concentration of IGF-1 is required to maintain the cell number above the number of cells plated. At lower concentrations, the cells begin to die. In all subsequent experiments, the FBS was supplemented with 50 ng of IGF-1/ml.
FIG. 1.
Effect of IGF-1 concentrations on survival of 32D cells expressing the wild-type IGF-1R. Parental 32D cells (open bars) and 32D cells expressing the wild-type IGF-1R (closed bars) were cultured in medium containing 10% FBS and the indicated concentrations of IGF-1. The percentages of surviving cells were determined at 24 h (A) and 48 h (B) after removal of IL-3 (WEHI-conditioned medium).
Survival of 32D cells expressing mutant IGF-1 receptors.
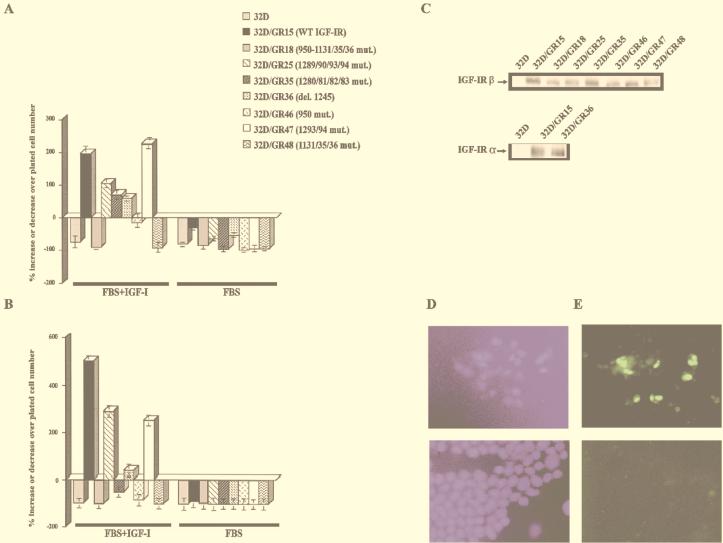
Figure 2 shows the survival of the various 32D cell lines (Table 1) at 24 h (Fig. 2A) and 48 h (Fig. 2B) after IL-3 withdrawal. With the same clone of 32D cells (clone 3), we had already shown that an overexpressed IGF-1R promotes survival and stimulates cell growth (14, 57, 90). This is again confirmed in Fig. 2A. All cell lines grew very well in the presence of IL-3 (data not shown), and all cell lines died in FBS without IGF-1 (Fig. 2). At 24 h after IL-3 withdrawal, the addition of IGF-1 had little or no effect on the survival of parental 32D cells, 32D/GR18 (Y950F/3YF), or 32D/GR48 (3YF). The number of 32D/GR46 (Y950F) cells was about 10% lower than the number of cells plated, but the cells responded to IGF-1 (compare with the same cells in FBS only). All of the other cell lines survived and grew with IGF-1. Clearly, the tyrosine kinase domain is essential for protection from apoptosis while the Y950F mutant receptor offers diminished protection.
FIG. 2.
Survival of 32D cells expressing wild-type (WT) and mutant IGF-1R. 32D cells were stably transduced with retroviral vectors expressing the indicated receptors (see also Table 1), and survival was determined at 24 h (A) and 48 h (B) after IL-3 withdrawal. FBS+IGF-1, 10% FBS plus IGF-1 (50 ng/ml), no IL-3; FBS, 10% fetal bovine serum, no IGF-1 or IL-3. The behavior in FBS supplemented with IL-3 was omitted since all cell lines grew vigorously in this medium. mut., mutation; del., deletion. (C) Western blots for the IGF-1R of the cell lines used in panels A and B. In all instances, we used an antibody to the C terminus of the IGF-1R (see Materials and Methods), except for GR36, that is truncated at residue 1245. For this receptor, we used an antibody to the α subunit. (D) Staining for DNA. (E) TUNEL staining. The upper figures are for the GR18 mutant; the lower figures are for the GR15 cells (wild-type receptor).
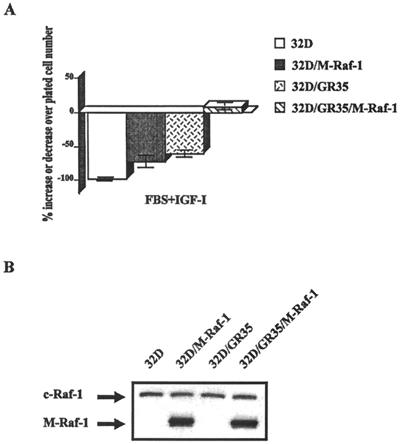
The results were slightly different at 48 h after IL-3 withdrawal (Fig. 2B; note the different scale of the ordinate above the baseline). The wild-type receptor and three of the mutant receptors were still effective in protection of cells from apoptosis. However, 32D/GR46 cells were now dying almost to the same degree as 32D/GR48 cells, and one other mutant joined the group of antiapoptosis-defective receptors: 32D/GR35, the 4-serine mutant. Note that both the GR35 and the GR46 receptors were still functional receptors, since addition of IGF-1 increases cell survival (compare cells in IGF-1 with cells in FBS only in Fig. 2A). Furthermore, these same mutant receptors were found to protect mouse embryo fibroblasts (which have IRS-1) from a form of apoptosis called anoikis (70).
The levels of expression of the wild-type and mutant IGF-1R, as evidenced by Western blotting, are shown in Fig. 2C. Parental 32D cells actually do have low levels of IGF-1R (16, 61), but it was not detectable under the conditions used in this experiment. Although there were variations, all transduced cell lines overexpressed the IGF-1R. By comparing the 32D-derived cells and mouse embryo fibroblast cell lines having a known number of IGF-1R (71) in Western blot experiments, we estimated the average number of IGF-1R in the 32D cell lines as ranging from 1 × 105 to 2 × 105 receptors/cell, considerably above the number required for full IGF-1R activity, which is 3 × 103 receptors/cell (71). Note that the GR36 receptor (truncated at residue 1245) can only be detected with an antibody to the α subunit.
Although it has been repeatedly shown that IL-3 withdrawal causes apoptosis of 32D cells (16, 69, 93, 95), we have monitored our cells by the TdT-mediated dUTP-biotin nick end labeling (TUNEL) method. Representative results are given in Fig. 2D and E, where two cell lines are shown: GR15 and the tyrosine kinase mutant, stained both for DNA and for apoptotic cells. In all instances in which the cell number decreased, the cells were shown to undergo apoptosis. One word of caution is necessary. With some mutants (and in the parental cells), cell death is so rapid after IL-3 withdrawal that one has to examine them within 14 h. By 24 h, most cells have disintegrated.
The IR and IRS-1 in 32D cells.
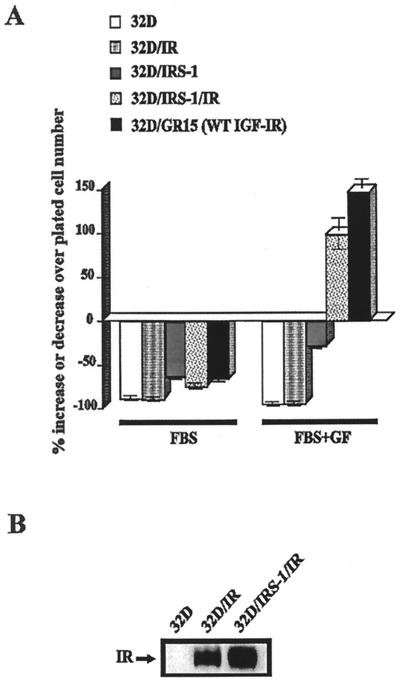
As mentioned in the introduction, 32D cells are completely devoid of both IRS-1 and IRS-2 (88, 93, 95). The experiments just described and previous reports from the literature (93, 95) indicate that IRS-1 offers partial protection from apoptosis. It is not, however, as effective as the wild-type IGF-1R. In a previous report, Wang et al. (88) had reported that overexpression of the IR in 32D cells was not sufficient for insulin stimulation of thymidine incorporation; however, when 32D cells overexpressed both the IR and IRS-1, they responded to insulin by undergoing DNA synthesis (Fig. 4 of their paper). On the contrary, overexpression of the IGF-1R is known to completely protect 32D cells from apoptosis caused by IL-3 withdrawal and to actually stimulate their growth (16, 61, 95). We confirm these reports in Fig. 3A. The IR offered no protection at all; the 32D/IR cells behaved essentially as the parental cells. 32D cells expressing IRS-1 did better when insulin was added to the medium. When both the IR and IRS-1 were overexpressed, the cells survived and grew, almost as well as 32D/GR15 cells. In these experiments, the IR and IR/IRS-1 cells were stimulated with insulin while the 32D/GR15 cells were stimulated with IGF-1.
FIG. 3.
Survival of 32D cells expressing the IR and/or IRS-1. The cell lines used were 32D cells expressing either IRS-1 only (32D/IRS-1), the insulin receptor only (32D/IR), or both (32D/IRS-1/IR). (A) Survival was determined at 24 h after IL-3 withdrawal. 32D/GR15 cells and parental 32D cells are also shown for comparison. FBS+GF, FBS supplemented with insulin (50 ng/ml), with the exception of the 32D/GR15 cells, which were supplemented with IGF-1 (50 ng/ml). WT, wild type. (B) Levels of expression of the IR in the two cell lines overexpressing it.
Figure 3B shows that the levels of expression of the IR were essentially similar in the cell lines overexpressing this receptor; a comparison with cells expressing the IGF-1R indicates that at least 2 × 105 IR/cell were expressed (data not shown). As in the case of the Western blot for the IGF-1R, the IR of parental 32D cells was not detectable under the conditions used, although 32D cells did have a small number of IR (88). The levels of IRS-1 in these cells have already been published in several reports from this and other laboratories (88, 95). Our results therefore clearly establish that in 32D cells, under the same conditions, the IGF-1R (but not the IR) can dispense with IRS-1 (or IRS-2) for protection from apoptosis caused by IL-3 withdrawal.
Phosphorylation of BAD.
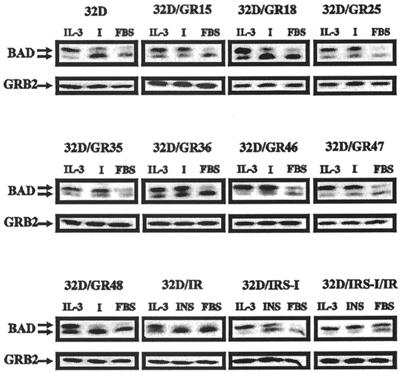
BAD can be detected under appropriate conditions as two separate bands, the upper band being the phosphorylated form of BAD and the lower band the hypophosphorylated or dephosphorylated form of BAD (94). For simplicity, we will refer to the lower band as the dephosphorylated form of BAD. Figure 4 shows BAD phosphorylation in the various 32D cells tested for survival in Fig. 2 and 3. Although experiments were carried out at several intervals after IL-3 withdrawal, for clarity we are presenting only the results obtained at 3 h (Fig. 4). The experiments have been repeated several times, especially with certain cell lines. The variations in BAD levels were largely due to our efforts to show the best exposure, which will give the most information on the state of BAD phosphorylation. In the presence of IL-3, all cell lines showed two forms of BAD, with the phosphorylated form predominating over the dephosphorylated form, as expected (94). In some cases, only the phosphorylated form was visible; in other cases, both forms were detectable, but the phosphorylated form was in excess over the dephosphorylated form. In the presence of FBS (no IGF-1 added), all cell lines promptly lost, within 3 h, the phosphorylated form of BAD, which was barely or no longer detectable. The only exception was the 32D/IRS1/IR, for which the two forms seemed to be in equilibrium. When IGF-1 was added to the medium, BAD was still predominantly in the phosphorylated form in 32D cells expressing the wild-type receptor (GR15), the 1290 region mutant (GR25), the 4-serine mutant (GR35), the δ1245 mutant (GR36), the GR46 mutant (Y950F), and the 1293/1294 mutant (GR47). These are the same cell lines that survived at 24 h when IL-3 was withdrawn (Fig. 2). The only exception was the GR46 cells (Y950F), which were only partially protected after IL-3 withdrawal. With the cell lines that did not survive (parental cells, GR18, and GR48), the phosphorylated form of BAD was already markedly decreased after 3 h in the absence of IL-3, despite the presence of IGF-1. Thus, there is an almost perfect correlation between survival at 24 h and BAD phosphorylation at 3 h. The results were essentially the same at 16 h after IL-3 withdrawal, with two notable differences. In surviving cell lines, the level of phosphorylated BAD remained high, while in the cells expressing the Y950F (GR46) mutant or the 4-serine mutant (GR35), the dephosphorylated form became predominant. We will return to these two mutant receptors later. The same correlation was seen with the 32D cells expressing either IRS-1, the IR, or both. Cell survival correlated with BAD phosphorylation.
FIG. 4.
BAD phosphorylation in 32D cells expressing the wild-type and mutant IGF-1 receptors. BAD phosphorylation was determined, as described in Materials and Methods, under three different sets of conditions, all in medium with 10% FBS: with IL-3, with IGF-1 (I), or with no additions. BAD phosphorylation was determined at 3 h after IL-3 withdrawal. The cell lines and the treatments are indicated above the BAD blots. In the case of the cell lines expressing IRS-1, the IR, or both, the FBS was supplemented with insulin (50 ng/ml). In all other cases, IGF-1 (50 ng/ml) was used. Under the BAD blots are Western blots of Grb2, used as indicators of protein amounts in each lane.
An objection may be raised that 32D cells that were not protected from apoptosis might have been losing phosphorylated BAD because protein synthesis or degradation in general might have been altered. As a control, we used Grb2, an adapter protein of the IGF-1R signaling pathway. We have previously reported that in 32D cells, even at 16 h and even in cells programmed for cell death, many proteins are still present in normal amounts—for instance, Grb2, exogenous IRS-1, and the simian virus 40 T antigen (95). We have confirmed these results in Fig. 4, where we show the levels of Grb2 in the various cell lines at 3 h after IL-3 withdrawal. Interestingly, Grb2 was still detectable at normal or only slightly decreased amounts even at 16 h after IL-3 withdrawal, even in cells programmed to die (data not shown), confirming our previous findings (95).
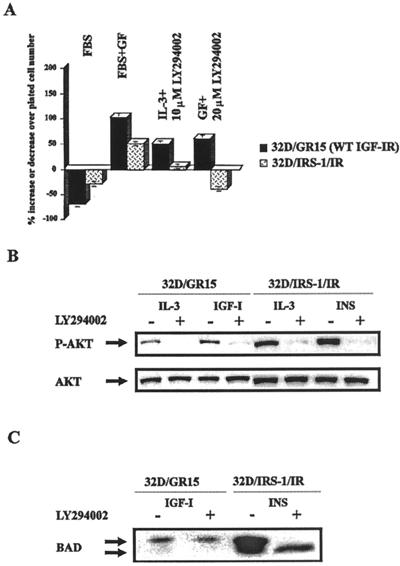
Effect of PI3-ki inhibitors on survival of 32D-derived cells.
IRS-1 is a major player in the activation of PI3-ki (2, 53, 54). Since IRS-1 is absent from parental 32D cells, we tested the effect of PI3-ki inhibitors on the survival of selected 32D cell lines. We tested two inhibitors: LY294002 and wortmannin. The results of several experiments with LY294002 are summarized in Fig. 5A. LY294002 (10 μM) completely abrogated the survival of 32D/IRS1/IR cells, thus confirming the necessity of PI3-ki for the antiapoptotic effect of the IR–IRS-1 combination. However, 32D/GR15 cells survived even this high concentration of inhibitor, indicating that the antiapoptotic effect of the IGF-1R has an alternative pathway which is not shared by the IR. Cells in IL-3 were also resistant to LY294002, even at a concentration of 20 μM. Essentially the same results were obtained with wortmannin (data not shown). These experiments confirm and extend the findings of Kulik and Weber (39).
FIG. 5.
Effect of PI3-ki inhibitors on survival of 32D cells. (A) Cell survival. 32D/GR15 and 32D/IRS1/IR cells were incubated in medium with FBS only or with FBS supplemented with the respective growth factors (GF) (IGF-1 for 32D/GR15, insulin for 32D/IRS1/IR cells, or IL-3), plus or minus the indicated concentrations of LY294002, an inhibitor of PI3-ki. The numbers of surviving cells were determined at 24 h after IL-3 withdrawal and addition of the growth factors and the inhibitor. WT, wild type. (B) Phosphorylation (upper panel) and amounts (lower panel) of Akt/PKB in the same cell lines and with different treatments. The presence (+) or absence (−) of LY294002 (10 μM) is indicated. P-AKT, phosphorylated Akt. (C) BAD phosphorylation under the same conditions.
Phosphorylation of Akt in 32D-derived cells.
We have experienced some problems in obtaining reproducible results for PI3-ki activity in 32D cells, and for this reason we elected to go directly to the activation of Akt/PKB, which is achieved with PI3-ki (21) (see above). In addition, Akt/PKB activation is important for BAD phosphorylation (13). We tested Akt activation by using an antibody that detects an activated Akt in cell lysates (see Materials and Methods). All cells in IL-3 had activated Akt (Fig. 5B), as expected (81). Akt was also activated in 32D/GR15 cells after supplementation with IGF-1, but not as markedly as in 32D/IRS1/IR cells following stimulation with insulin (Fig. 5B). It is true that there was more Akt protein in the lanes of the 32D/IRS1/IR cells, but even so, IGF-1 stimulation of 32D/GR15 cells seemed to be less effective than insulin stimulation of 32D/IRS1/IR cells. The modest activation by IGF-1 of Akt in 32D/GR15 cells can be attributed to the reported binding of the p85 subunit of PI3-ki to tyrosine 1316 of the receptor (76). Addition of LY294002 abrogates the activation of Akt, even in 32D/GR15 cells, which survive under these conditions. These results indicate that the IRS-1 pathway is the major activating pathway of Akt in 32D/IRS1/IR cells and that the PI3-ki inhibitors effectively block this pathway. Akt activation alone, however, does not explain the protective effect of the IGF-1R, since Akt/PKB activation was abrogated by LY294002 in 32D/GR15 cells and yet these cells survived. The lower panel of Fig. 5B shows that the amounts of Akt/PKB protein in untreated cells and in cells treated with the PI3-ki inhibitors were essentially the same.
Under the same conditions, BAD remained phosphorylated in 32D/GR15 cells, even in the presence of LY294002 (Fig. 5C). However, in 32D/IRS1/IR cells, the addition of LY294002 caused the complete disappearance of the phosphorylated form while the dephosphorylated form was clearly detectable. These experiments confirm the importance of the PI3-ki/Akt pathway in survival mediated by IRS-1, while the IGF-1R maintains survival and BAD phosphorylation even in the presence of PI3-ki inhibitors.
Role of MAPK in the antiapoptotic function of the IGF-1R.
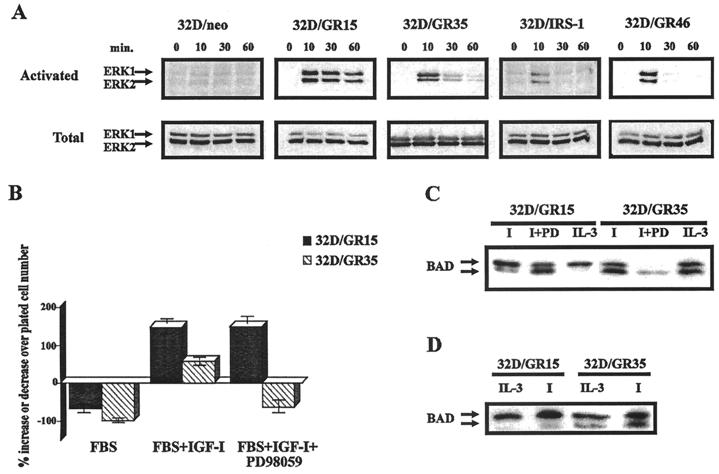
Activation of MAPK has often been implicated in protection from apoptosis by a variety of agents (45), including IGF-1 (57). Activation of MAPK by the IR or the IGF-1R is also very well established (see, for instance, reference 32). The work of Morris White and collaborators has indicated that IRS-1 plays a major role in MAPK activation by the IR (89); in its absence, MAPK activation is strongly reduced. Figure 6A confirms that MAPKs (both ERK-1 and ERK-2) were phosphorylated in 32D/GR15 cells stimulated with IGF-1. The activation was sustained for at least 1 h. In the absence of an overexpressed IGF-1R (32Dneo), we could not detect activation of MAPK by IGF-1. Activation of MAPK was also detectable in 32D/GR35 cells, although after an initial burst the level of activation decreased; it was still detectable at 1 h. Not so with the Y950F mutant (GR46), in which stimulation by IGF-1 resulted in a sharp increase in MAPK activation at 10 min, with a return to basal levels immediately afterward. These experiments have been repeated several times, and we have also tested other 32D cell lines expressing mutant receptors. All of the cell lines that survived after addition of IGF-1 had a sustained activation of MAPK, but for simplicity we are showing only the results for the cell lines that are relevant in subsequent experiments. The inability of the Y950F mutant receptor to induce a sustained activation of MAPK suggests that the 32D cells expressing this receptor are defective in this signaling pathway (see Discussion). This may explain why (in the absence of IRS-1) they began to die even in the first 24 h. 32D/GR35 cells behaved differently: MAPK was not activated as strongly as with the wild-type receptor, but the activation was sustained and the cells did not begin to die until 24 h after IL-3 withdrawal. We therefore hypothesized that 32D/GR46 cells died because they failed to activate the MAPK pathway (57) while 32D/GR35 cells were resistant for the first 24 h but eventually died because they could not activate a third pathway.
FIG. 6.
Role of MAPK in IGF-1R-mediated survival of 32D cells. (A) MAPK activation in selected cell lines. MAPK phosphorylation and protein amounts were determined as described in Materials and Methods. Stimulation was with IGF-1 (50 ng/ml), and lysates were prepared at the times (in minutes) indicated above the blots. The cell lines are also indicated above the blots. (B) Survival of 32D/GR15 and 32D/GR35 cells in the presence of PD98059, an inhibitor of MEK. The indicated cell lines were incubated in FBS (no IL-3) for 24 h, in the presence of only IGF-1 or both IGF-1 and PD98059. Survival is expressed as a percentage of cell number increase or decrease over the number of cells plated. (C) BAD phosphorylation in the same cell lines, with or without PD98059 (I, IGF-1; PD, the inhibitor). (D) Same procedures as in Fig. 4, except that 32D/GR15 and 32D/GR35 cells were examined at 16 h after IL-3 withdrawal. Lysates were prepared from cells in FBS supplemented with either IL-3 or IGF-1 (I).
The 4-serine mutant.
In the case of mutant IGF-1 receptors that do not protect 32D cells from apoptosis (GR18 and GR48) and in which receptor function is impaired, the respective cell lines died massively during the first 24 h. As shown in Fig. 2 and 3, 32D/GR35 cells (4-serine mutant) were protected from apoptosis caused by IL-3 withdrawal in the first 24 h but succumbed between 24 and 48 h. 32D/GR35 cells also responded to IGF-1 with a sustained activation of both ERK1 and ERK2 (Fig. 6A), albeit not as strongly as the cells with the wild-type receptor. To test whether the activation of MAPK is critical for protection from apoptosis, we treated 32D/GR15 and 32D/GR35 cells with an inhibitor of MEK, PD98059 (Fig. 6B). In the absence of the inhibitor, both 32D/GR15 and 32D/GR35 survived and even grew in the first 24 h (Fig. 2). In the presence of the inhibitor, 32D/GR35 died very rapidly in the first 24 h while 32D/GR15 cells still survived (30% increase over the number of cells plated).
In Fig. 6C, we show the effect of the MEK inhibitor (PD98059) on BAD phosphorylation in the same two cell lines. The inhibitor had an effect on BAD phosphorylation; even in 32D/GR15 cells (which survived) there was an increase in the amount of dephosphorylated BAD, which was then about half of the total BAD protein. However, in 32D/GR35 cells, only dephosphorylated BAD could be detected in the presence of the MEK inhibitor. This experiment was repeated with an antibody specific to the phosphorylated BAD (Phospholus Bad S136 antibody; New England Biolabs) with the same results (data not shown).
As shown in Fig. 4, 32D/GR35 cells maintained a phosphorylated form of BAD 3 h after IL-3 withdrawal. However, when these cells were examined at 16 h after IL-3 withdrawal (no inhibitors present, but supplementation with IGF-1), a considerable amount of dephosphorylated BAD was present in these cells, almost to the same level as the phosphorylated BAD (Fig. 6D; compare with Fig. 4). This is in contrast to the 32D/GR15 cells (wild-type receptor), in which, even at 16 h, BAD was mostly in the phosphorylated form. These results indicate that under the experimental conditions used, 32D/GR35 cells slowly lost their ability to maintain BAD in the phosphorylated form, which could explain why they began to die at between 24 and 48 h.
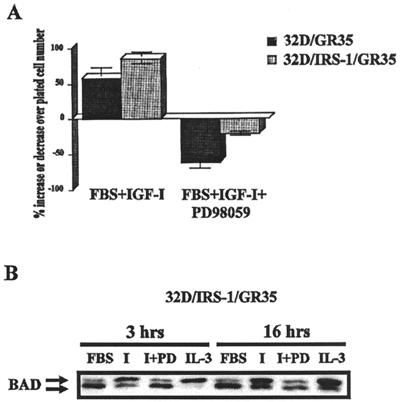
A corollary of these findings is that a double-mutant receptor (mutated at Y950 and at the 4 serines 1280 to 1283) should have little or no survival value. Indeed, 32D cells expressing this double-mutant receptor died very rapidly, with less than 1% of the plated cells being viable 24 h after IL-3 withdrawal (data not shown). Another corollary is that even in the presence of the MEK inhibitor the introduction of IRS-1 into 32D/GR35 cells should result in survival by restoring the PI3-ki/Akt pathway. This was, indeed, the case (Fig. 7A). In the presence of PD98059, 32D/GR35 cells died, as previously shown, while 32D/IRS1/GR35 cells were at least partially protected. At the same time, BAD phosphorylation was partially maintained in 32D/IRS1/GR35 cells, although not as well as when the inhibitor was omitted (Fig. 7B). 32D/GR35 cells completely lost phosphorylated BAD in the presence of the MEK inhibitor (Fig. 6C).
FIG. 7.
IRS-1 protects GR35 cells from apoptosis. The two cell lines tested were 32D/GR35 (4-serine mutant) and 32D/IRS1/GR35 (expressing IRS-1). (A) Survival was determined after 24 h as in previous experiments. The cells were tested in medium containing serum supplemented with IGF-1, plus or minus the MEK inhibitor PD98059. (B) Lysates from 32D/IRS1/GR35 cells, under the same conditions, were examined for BAD phosphorylation. I, IGF-1; PD, PD98059.
Emergence of a third pathway for IGF-1R-mediated protection from apoptosis.
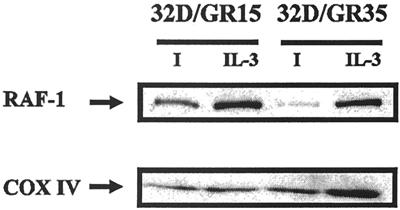
The previous results suggest that the IGF-1R, in the absence of IRS-1, uses the MAPK pathway to exert its antiapoptotic function but that in addition to this second pathway the wild-type IGF-1R (but not the 4-serine mutant) has a third pathway which also results in BAD phosphorylation. This third pathway is clearly dependent on the integrity of the serine quartet at positions 1280 to 1283. Serine 1283 of the IGF-1R is known to bind the 14.3.3 family of proteins (9, 24). Interestingly, 14.3.3 binds to IRS-1 even better than it binds to the IGF-1R (37), but it does not bind to the IR (9, 24). 14.3.3 proteins have been implicated in many cellular functions, among which are the stabilization of phosphorylated BAD (94), the enhancement of Raf kinase activity (22, 44), and the stabilization of activated Raf-1 (15). We first determined whether Raf-1 activity in 32D/GR15 cells was significantly different from that in 32D/GR35 cells. We first determined Raf activity (data not shown). The results of three separate experiments indicated that IL-3 induced an increase in Raf activity (from 51 to 80% over that in serum-free medium), and so did IGF-1 in cells expressing the wild-type IGF-1R (from 50 to 93%). In cells expressing the 4-serine mutant receptor, the increase in Raf activity never exceeded 35%, and in some experiments it was not increased at all over that in serum-free medium. However, the variability of the assay precluded an extension of these results to other mutants and to the IR. We therefore explored another possibility.
It has been reported that Bcl-2 targets the protein kinase Raf-1 to mitochondria (87), allowing this kinase to phosphorylate BAD. We tested for the presence of mitochondrial Raf (mitRaf) in 32D/GR15 cells and in 32D/GR35 cells after IL-3 withdrawal. The results of such an experiment, repeated several times, are shown in Fig. 8. When IL-3 was present, mitRaf-1 could be detected in both cell lines. However, when IL-3 was withdrawn, mitRaf was still clearly detectable in 32D/GR15 cells but not in 32D/GR35 cells. The difference was not due to differences in amounts of mitochondrial proteins, as shown by the amounts of cytochrome oxidase in the lysates. The signal was higher in IL-3-containing cultures, probably because these cells were growing exponentially whereas 32D/GR15 cells are programmed for differentiation (85). These results are compatible with the previous findings that BAD is still phosphorylated in 32D/GR15 cells while in 32D/GR35 cells, BAD is partially dephosphorylated (Fig. 6).
FIG. 8.
Presence of mitRaf in 32D/GR15 and 32D/GR35 cells. Mitochondrial lysates were prepared as described in Materials and Methods, and the same amounts of protein were blotted for Raf-1 (upper panel). The amounts of mitochondrial proteins in each lane were monitored with an antibody to cytochrome oxidase (COX IV) (lower panel). I, IGF-1.
To confirm that mitRaf has value with regard to survival of these cells, we transfected a constitutively active mutant Raf which localizes exclusively to the mitochondria (72, 87) into 32D and 32D/GR15 cells. It was previously shown by one of our laboratories (72), and also by Wang et al. (87), that this Raf mutant localizes exclusively to the mitochondria. The results on cell survival are shown in Fig. 9A, where we compared the survival of 32D and 32D/GR35 cells versus that of the same cells expressing the mutant mitRaf. At 48 h after IL-3 withdrawal, in the presence of serum supplemented with IGF-1, 32D cells expressing the mitRaf still die. On the contrary, 32D/GR35 cells with the mitRaf survive (10% increase over the number of plated cells). The expression of the mutant Raf in both cell lines was confirmed by Western blotting (Fig. 9B).
FIG. 9.
Effect of mitRaf on cell survival. (A) 32D and 32D/GR35 cells were stably transfected with constitutively activated mutant Raf that localizes to the mitochondria. Transduction and selection of cell lines are described in Materials and Methods. Survival was determined at 48 h after IL-3 withdrawal, under the usual conditions. (B) Detection of mitRaf in the various cell lines. Detection was carried out with an anti-Raf1 antibody (Santa Cruz). The size of the mutant Raf is different from that of wild-type Raf-1, which is visible above the mutant Raf.
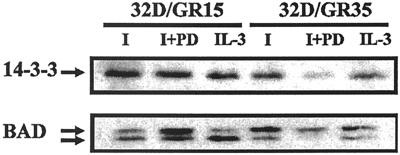
Finally, we tested the ability of phosphorylated BAD to coprecipitate 14.3.3 (13, 94). Lysates from 32D/GR15 cells were immunoprecipitated with an antibody to BAD, and the blot was stained with an antibody to the β isoform of 14.3.3. The results are shown in Fig. 10: 14.3.3 could be detected in the BAD immunoprecipitates, even when the cells had been treated with the MEK inhibitor PD98059. Some 14.3.3 was visible also in the immunoprecipitates from 32D/GR35 cells, but not when the cells were treated with the MEK inhibitor. The blot was stripped and reprobed with an antibody to BAD, which was visible in the immunoprecipitates of both cell lines.
FIG. 10.
Coprecipitation of 14.3.3 and BAD. BAD immunoprecipitation, detection of 14.3.3, and detection of BAD itself on the stripped blot were carried out as described in Materials and Methods. The cell lines used were 32D/GR15 and 32D/GR35. PD, MEK inhibitor PD98059 at the same concentration as that used for Fig. 7; I, IGF-1 (50 ng/ml).
DISCUSSION
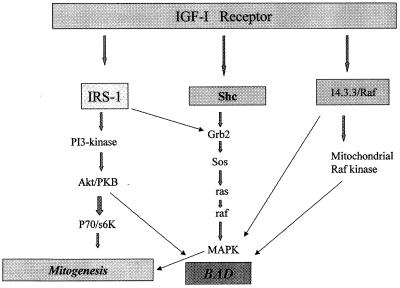
The main pathway for the antiapoptotic effect of the IGF-1R is the well-established pathway that through IRS-1 activates PI3-ki (53) and through Akt/PKB (17, 34, 38) induces phosphorylation of BAD (13). None of the experiments described in this paper contradicted this statement. However, our experiments also indicated that the IGF-1R (but not the IR) has alternative pathways for protection from apoptosis that are independent of the IRS-1 pathways. In favor of such a conclusion are the findings that the IGF-1R protects 32D cells from apoptosis (i) in the absence of IRS-1; (ii) in the presence of inhibitors of PI3-ki, at concentrations that abrogate the protection of the combined IR–IRS-1 overexpression; and (iii) upon complete inhibition of Akt/PKB phosphorylation. Our results suggest that the IGF-1R actually has two alternative pathways, one through MAPK activation and another through the activation of Raf-1 and its translocation to the mitochondria (Fig. 11). All three pathways result in the maintenance of BAD phosphorylation. We discuss these points separately below.
FIG. 11.
Diagram of the multiple antiapoptotic pathways of the IGF-1R. The diagram is based on the results presented in this paper and previous papers from this and other laboratories.
The IGF-1R, activated by its ligands, is a powerful inhibitor of apoptosis caused by a variety of injuries. Addition of IGF-1 inhibits apoptosis induced by IL-3 withdrawal in hemopoietic cells (51, 69) as well as by c-myc overexpression (28), IL-1β-converting enzyme protease expression (33), serum withdrawal (57), anticancer drugs (18), and transforming growth factor β1 (31) in fibroblasts. An overexpressed and activated IGF-1R protects cells from apoptosis induced by a variety of agents and conditions, including etoposide (77), IL-3 withdrawal (51, 56, 61), osmotic shock (79), tumor necrosis factor alpha (90), p53 (61), ionizing and nonionizing radiation (38, 55, 83), and okadaic acid (11).
Conversely, a down-regulation of IGF-1R function, either by antisense strategies (6, 40, 47, 66, 67, 78), by dominant-negative mutations (12, 60, 65), or by triple-helix formation (68), causes apoptosis of tumor cells in vivo and/or abrogation of tumorigenesis and metastases (6, 19, 48). Our finding in the present study that the IGF-1R has multiple antiapoptotic pathways may help to explain why it is such a powerful inhibitor of apoptosis.
In 32D cells, the protective effect of the activated IGF-1R is dependent on the availability of IGF-1 (Fig. 1). Although serum contains both IGF-1 and IGF-2, 32D cells, even when overexpressing the IGF-1R, still require the addition of IGF-1 for survival. This requirement was not as apparent in cells that have IRS-1 (56). While it is true that in our 32D cells the IGF-1R is overexpressed, it has often been reported that IGF-1 can protect cells from apoptosis even when the receptor is at normal levels (51, 46), although some cooperative factors may be required in this case.
A series of experiments by several laboratories has firmly established that the main antiapoptotic pathway of activated IGF-1R is through IRS-1, PI3-ki, and Akt/PKB (see the introduction). Datta et al. (13) made the connection with BAD by showing that Akt/PKB phosphorylates BAD; in its phosphorylated form, BAD cannot bind to antiapoptotic proteins of the Bcl-2 family and therefore cannot induce cell death (91). Furthermore, it has been shown that phosphorylated BAD is associated with 14.3.3 (94). Several investigators, however, had already noticed that the antiapoptotic effect of the activated IGF-1R was insensitive or almost insensitive to inhibitors of PI3-ki (39). It was suggested that the IGF-1R might be using an alternative pathway when the PI3-ki pathway was unavailable or inhibited (57). Valentinis et al. (84) examined the situation in mouse embryo fibroblasts plated on polyHEMA (Sigma) plates, in which the cells are denied attachment to the substratum and undergo a form of apoptosis that has been called anoikis (23). This system was chosen because cells in a state of anchorage independence have a strict requirement for a functional IGF-1R for survival, a requirement that is not as strict when the cells are in monolayer cultures (3, 4). In addition, IGF-1 rescues mouse embryo fibroblasts from anoikis without inducing DNA synthesis, which could obscure the antiapoptotic signaling. In this tightly regulated system, Valentinis et al. (84) showed that the PI3-ki/Akt/PKB pathway was not required for IGF-1-mediated protection from anoikis while Ras activation had a clearly protective effect.
Ras activation is known to be an important step in the activation of MAPK (74). Since MAPKs are activated by IGF-1 (32), we hypothesized, as already suggested by Parrizas et al. (57), that MAPK activation might be the alternative pathway to IGF-1R-mediated protection from apoptosis. Our experiments suggest that MAPK activation is one of the alternative pathways: 32D/GR46 cells (Y950F mutant) fail to achieve a sustained activation of MAPK after IGF-1 stimulation and begin to die within the first 24 h. The fact that this receptor gives partial protection suggested, however, that another pathway (also IRS-1 independent) was operative. The failure of the Y950F mutant to induce a sustained activation of MAPK is compatible with results from the literature. The Y950 residue of the IGF-1R is known to interact with the Shc proteins (10, 82), which in turn are involved in the activation of the MAPK pathway (64, 73, 74). The cell lines that survive show a sustained activation (at least 1 h) of MAPK. Marshall (49) has already made an eloquent case for the need for sustained MAPK activation for their biological effect. Only when MAPK activation is sustained is there nuclear translocation and activation of the genes that initiate cell cycle progression and/or survival (35, 41).
MAPK activation cannot be, however, the sole alternative pathway for IGF-1R-mediated protection from apoptosis, since an MEK inhibitor, PD98059, failed to abrogate the protective effect of the IGF-1R in 32D/GR15 cells (Fig. 6B). A clue to a third pathway used by the IGF-1R came from the 4-serine mutant receptor, expressed by 32D/GR35 cells. These cells are protected in the first 24 h but then die rapidly between 24 and 48 h. When 32D/GR35 cells are exposed to the MEK inhibitor, they die very rapidly in the first 24 h. These data suggested that the IGF-1R, in the absence of IRS-1, has two alternative pathways, one leading to the activation of MAPK and another that depends on the integrity of the serines in the 1280 to 1283 positions. It is known that 14.3.3 proteins bind to the IGF-1R (9) at serine residues located in the C terminus, between positions 1272 and 1284, and that this interaction depends on the phosphorylation of the appropriate serines. According to Furlanetto et al. (24), the 14.3.3 β isoform binds to serine 1283 and possibly to tyrosine 1316 in the C terminus. In addition, Craparo et al. (9) and other laboratories (37) have shown that 14.3.3 proteins also bind to IRS-1; in fact, they may bind to IRS-1 more effectively than to the IGF-1R (37). Interestingly, the IR fails to interact with 14.3.3 proteins (9, 24). The inability of the 4-serine mutant to protect 32D cells in the presence of an MEK inhibitor is suggestive of an intervention by 14.3.3 in IGF-1R-mediated protection from apoptosis in the absence of IRS-1. This suggestion is strengthened by our finding that the wild-type receptor and the 4-serine mutant have similar antiapoptotic activities in mouse embryo fibroblasts that express IRS-1 (70). Furthermore, the fact that the IR (which does not interact with 14.3.3 proteins) cannot protect 32D cells in the absence of IRS-1 (which instead binds to 14.3.3 proteins) also indicates that 14.3.3 proteins may be involved in IGF-1R-mediated protection from apoptosis. It would have been desirable to demonstrate a coprecipitation of 14.3.3 proteins with the wild-type IGF-1R but not with the 4-serine mutant receptor. Unfortunately, no one has yet shown such a coprecipitation in mammalian cells; the evidence for interaction is based on the yeast two-hybrid system. Our attempts to coprecipitate 14.3.3 proteins with the IGF-1R were also unsatisfactory (data not shown). The increases in Raf-1 activity were reproducible when the wild-type receptor was compared to the 4-serine mutant, but in other instances the results were inconclusive. Alternatively, the 32D/GR35 cells may activate MAPK by a Raf-independent mechanism; protein kinase C, the proto-oncogene c-mos, and G protein-coupled receptors have been reported to activate MAPK independently of Raf (8). Both protein kinase C and G protein-coupled receptors have been shown to have signaling pathways that cross talk with the IR and IGF-1R pathways (1).
In a recent paper, Kulik and Weber (39) reported that BAD phosphorylation is not necessarily involved in IGF-1-mediated protection from apoptosis. Our results show that regardless of the pathway used, protection from apoptosis correlates strictly with a phosphorylated BAD. BAD is predominantly phosphorylated when the cells express receptors that protect them from apoptosis. BAD remains predominantly phosphorylated even in 32D/GR15 cells that are treated with inhibitors of PI3-ki or an MEK inhibitor, or even both (data not shown). When protection is partially decreased (see, for instance, 32D/GR15 cells incubated with the MEK inhibitor [Fig. 6]), BAD phosphorylation is also partially decreased. We have no explanation for the discrepancy between the results of Kulik and Weber (39) and our own, except the obvious ones that the cell lines used were different and the methods used to induce apoptosis were also different.
BAD phosphorylation through the Akt/PKB pathway is well documented (see above), but there is also evidence in the literature that BAD can be phosphorylated, or at least stabilized in its phosphorylated form, through the activation of MAPK (75) or a mitochondrion-anchored protein kinase A (92). In addition, there are alternative explanations for the antiapoptotic effect of Akt, including its ability to inactivate caspase-9 (93) or the forkhead genes (91). This mechanism may be valid in the protection or partial protection of 32D cells by mutant receptors like the Y950F and the 4-serine mutant in the first 24 h after IL-3 withdrawal. Especially in the latter case, this pathway must play a critical role, since an MEK inhibitor causes massive cell death in the first 24 h, as well as a decrease in BAD phosphorylation. This conclusion is supported by the finding that a double-mutant receptor (Y950F and S1280-1283A) has no survival value.
Confirmation that BAD phosphorylation plays a role in these alternative pathways comes from the experiments with mitRaf. mitRaf is detectable in 32D cells in medium with IL-3 and in 32D/GR15 cells in medium supplemented with IGF-1. It is not detectable in 32D/GR35 cells, which have dephosphorylated BAD after 16 h following IL-3 withdrawal. mitRaf has been thought to phosphorylate BAD, which causes its dissociation from antiapoptotic proteins and its release into the cytosol (25, 87), where it remains phosphorylated (94). Although it may be coincidental, the isoform of 14.3.3 that preferentially binds to the IGF-1R, the β isoform (24), is also one of the 14.3.3 isoforms that bind to Raf-1 (20). In support of this view are also findings that in cells with the wild-type receptor, 14.3.3 coprecipitates with BAD. No 14.3.3 is detectable in immunoprecipitates of BAD from lysates of 32D/GR35 cells in the presence of the MEK inhibitor. Finally, the introduction of the mitRaf mutant, which localizes exclusively to the mitochondria (72, 87), into 32D/GR35 cells yields protection from apoptosis, at least partially. The parental 32D cells, on the contrary, are not protected, which indicates that IGF-1R signaling is still required for survival.
We summarize our interpretation of these experiments as follows: (i) the main antiapoptotic pathway of the IGF-1R is the well-established IRS-1/PI3-ki–Akt/PKB pathway; (ii) in the absence of IRS-1, the IGF-1R has alternative pathways that are not shared with the IR or possibly other growth factor receptors; (iii) one of these alternative pathways is the sustained activation of the MAPK pathway, which is largely (but not exclusively) dependent on the integrity of Y950; (iv) another alternative pathway originates from the serines at positions 1280 to 1283, probably through the intervention of 14.3.3 proteins; (v) this last pathway results in translocation of Raf-1 to mitochondria; and, finally, (vi) all of these pathways, at least in these cells, exhibit the stabilization of BAD in its phosphorylated form. The results presented here are compelling enough to justify our suggestion that the IGF-1R has not one but two alternative pathways for protection from apoptosis and that BAD phosphorylation is an important end point of all three pathways. The multiplicity of signaling pathways used by the IGF-1R may explain why this receptor has such a powerful and widespread antiapoptotic activity (3, 50).
ACKNOWLEDGMENTS
This work was supported by grants GM 33694 and PO 1 CA 56309 from the National Institutes of Health.
REFERENCES
- 1.Avruch J. Insulin signal transduction through protein kinase cascades. Mol Cell Biochem. 1998;182:31–48. [PubMed] [Google Scholar]
- 2.Backer J M, Myers M G, Jr, Sun X J, Chi D J, Shoelson S E, Miralpeix M, White M F. Association of IRS-1 with insulin receptor and the phosphatidylinositol 3′-kinase. J Biol Chem. 1993;268:8204–8212. [PubMed] [Google Scholar]
- 3.Baserga R, Hongo A, Rubini M, Prisco M, Valentinis B. The IGF-I receptor in cell growth, transformation and apoptosis. Biochim Biophys Acta. 1997;1332:105–126. doi: 10.1016/s0304-419x(97)00007-3. [DOI] [PubMed] [Google Scholar]
- 4.Baserga R. The price of independence. Exp Cell Res. 1997;236:1–3. doi: 10.1006/excr.1997.3732. [DOI] [PubMed] [Google Scholar]
- 5.Brunet A, Bonni A, Zigmond M J, Lin M Z, Jun P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Akt promotes cell survival by phosphorylating and inhibiting a forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 6.Burfeind P, Chernicky C L, Rininsland F, Ilan J, Ilan J. Antisense RNA to the type I insulin-like growth factor receptor suppresses tumor growth and prevents invasion by rat prostate cancer cells in vivo. Proc Natl Acad Sci USA. 1996;93:7263–7268. doi: 10.1073/pnas.93.14.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardone M H, Roy N, Stennicke H R, Salvesen G S, Franke T F, Stanbridge E, Frisch S, Reed J C. Regulation of cell death protease caspase-9 by phosphorylation. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 8.Cobb M H, Goldsmith E J. How MAP kinases are regulated. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- 9.Craparo A, Freund R, Gustafson T A. 14.3.3 interacts with the insulin-like growth factor I receptor and insulin receptor substrate 1 in a phosphoserine-dependent manner. J Biol Chem. 1997;272:11663–11669. doi: 10.1074/jbc.272.17.11663. [DOI] [PubMed] [Google Scholar]
- 10.Craparo A, O’Neill T J, Gustafson T A. Non-SH2 domains within insulin receptor substrate-1 and Shc mediate their phosphotyrosine-dependent interaction with the NPEY motif of the insulin-like growth factor 1 receptor. J Biol Chem. 1995;270:15639–15643. doi: 10.1074/jbc.270.26.15639. [DOI] [PubMed] [Google Scholar]
- 11.D’Ambrosio C, Valentinis B, Prisco M, Reiss K, Rubini M, Baserga R. Protective effect of the insulin-like growth factor I receptor on apoptosis induced by okadaic acid. Cancer Res. 1997;57:3264–3271. [PubMed] [Google Scholar]
- 12.D’Ambrosio C, Ferber A, Resnicoff M, Baserga R. A soluble insulin-like growth factor I receptor that induces apoptosis of tumor cells in vivo and inhibits tumorigenesis. Cancer Res. 1996;56:4013–4020. [PubMed] [Google Scholar]
- 13.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 14.Del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Interleukin-3 phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 15.Dent P, Jelinek T, Morrison D K, Weber M J, Sturgill T W. Reversal of raf-1 activation by purified and membrane-associated protein phosphatases. Science. 1995;268:1902–1906. doi: 10.1126/science.7604263. [DOI] [PubMed] [Google Scholar]
- 16.Dews M, Nishimoto I, Baserga R. IGF-I receptor protection from apoptosis in cells lacking the IRS proteins. Recept Signal Transduct. 1997;7:231–239. [PubMed] [Google Scholar]
- 17.Dudek H, Datta S R, Franke T F, Birnbaum M J, Yao R, Cooper G M, Segal R A, Kaplan D R, Greenberg M E. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 18.Dunn S E, Hardman R A, Kari F W, Barrett C J. Insulin-like growth factor 1 (IGF-I) alters drug sensitivity of HBL100 human breast cancer cells by inhibition of apoptosis induced by diverse anticancer drugs. Cancer Res. 1997;57:2687–2693. [PubMed] [Google Scholar]
- 19.Dunn S E, Wehrlich M, Sharp N J H, Reiss K, Solomon G, Hawkins R, Baserga R, Barrett J C. A dominant negative mutant of the insulin-like growth factor 1 receptor inhibits the adhesion, invasion and metastasis of breast cancer. Cancer Res. 1998;58:3353–3361. [PubMed] [Google Scholar]
- 20.Fantl W J, Muslin A J, Kikuchi A, Martin J A, MacNicol A M, Gross R W, Williams L T. Activation of Raf-1 by 14-3-3 proteins. Nature. 1994;371:612–614. doi: 10.1038/371612a0. [DOI] [PubMed] [Google Scholar]
- 21.Franke T F, Kaplan D R, Cantley L C, Toker A. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-biphosphate. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- 22.Freed E, Symons M, McDonald S G, McCormick F, Ruggieri R. Binding of 14.3.3 proteins to the protein kinase Raf and effects on its activation. Science. 1994;265:1713–1716. doi: 10.1126/science.8085158. [DOI] [PubMed] [Google Scholar]
- 23.Frisch S M, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furlanetto R W, Dey B R, Lopaczynski W, Nissley S P. 14.3.3 proteins interact with the insulin-like growth factor receptor but not with the insulin receptor. Biochem J. 1997;327:765–771. doi: 10.1042/bj3270765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gajewski T F, Thompson C B. Apoptosis meets signal transduction: elimination of a BAD influence. Cell. 1996;87:589–592. doi: 10.1016/s0092-8674(00)81377-x. [DOI] [PubMed] [Google Scholar]
- 26.Greenberger J S, Sakakeeny M A, Humphries R K, Eaves C J, Eckner R J. Demonstration of permanent factor-dependent multipotential (erythroid, neutrophil, basophil) hematopoietic progenitor cell lines. Proc Natl Acad Sci USA. 1983;80:2931–2935. doi: 10.1073/pnas.80.10.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harada H, Becknell B, Wilm M, Mann M, Huang L J, Taylor S S, Scott J D, Korsmeyer S J. Phosphorylation and inactivation of BAD by mitochondria-anchored protein kinase A. Mol Cell. 1999;3:413–422. doi: 10.1016/s1097-2765(00)80469-4. [DOI] [PubMed] [Google Scholar]
- 28.Harrington E A, Bennett M R, Fanidi A, Evan G I. c-myc-induced apoptosis in fibroblasts is inhibited by specific cytokines. EMBO J. 1994;13:3286–3295. doi: 10.1002/j.1460-2075.1994.tb06630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawley R G, Lieu F H, Fong A Z, Hawley T S. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- 30.Hongo A, D’Ambrosio C, Miura M, Morrione A, Baserga R. Mutational analysis of the mitogenic and transforming activities of the insulin-like growth factor I receptor. Oncogene. 1996;12:1231–1238. [PubMed] [Google Scholar]
- 31.Hsing A Y, Kadomatsu K, Bonham N J, Danielpour D. Regulation of apoptosis induced by transforming growth factor β1 in nontumorigenic and tumorigenic rat prostatic epithelial cell lines. Cancer Res. 1996;56:5146–5149. [PubMed] [Google Scholar]
- 32.Hugl S R, White M F, Rhodes C J. Insulin-like growth factor I (IGF-I)-stimulated pancreatic β-cell growth is glucose-dependent. J Biol Chem. 1998;273:17771–17779. doi: 10.1074/jbc.273.28.17771. [DOI] [PubMed] [Google Scholar]
- 33.Jung Y K, Miura M, Yuan J. Suppression of interleukin-1 beta converting enzyme-mediated cell death by insulin-like growth factor. J Biol Chem. 1996;271:5112–5117. doi: 10.1074/jbc.271.9.5112. [DOI] [PubMed] [Google Scholar]
- 34.Kennedy S G, Wagner A J, Conzen S D, Jordan J, Bellacosa A, Tsichlis P N, Hay N. The PI 3-kinase/Akt signaling pathway delivers an anti-apoptotic signal. Genes Dev. 1997;11:701–713. doi: 10.1101/gad.11.6.701. [DOI] [PubMed] [Google Scholar]
- 35.Khokhlatchev A V, Canagarajah B, Wilsbacher J, Robinson M, Atkinson M, Goldsmith E, Cobb M H. Phosphorylation of the MAP kinase ERK2 promotes its homodimerization and nuclear translocation. Cell. 1998;93:605–615. doi: 10.1016/s0092-8674(00)81189-7. [DOI] [PubMed] [Google Scholar]
- 36.Khwaja A, Rodriguez-Viciana P, Wennstrom S, Warne P H, Downward J. Matrix adhesion and ras transformation both activate a phosphoinositide 3-OH kinase and protein kinase B/AAkt cellular survival pathway. EMBO J. 1997;16:2783–2793. doi: 10.1093/emboj/16.10.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kosaki A, Yamada K, Suga J, Otaka A, Kuzuya H. 14.3.3β protein associates with insulin receptor substrate-1 and decreases insulin-stimulated phosphatidylinositol 3′-kinase activity in 3T3L1 adipocytes. J Biol Chem. 1998;273:940–944. doi: 10.1074/jbc.273.2.940. [DOI] [PubMed] [Google Scholar]
- 38.Kulik G, Klippel A, Weber M J. Antiapoptotic signaling by the insulin-like growth factor I receptor, phosphatidylinositol 3-kinase, and Akt. Mol Cell Biol. 1997;17:1595–1606. doi: 10.1128/mcb.17.3.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulik G, Weber M J. Akt-dependent and -independent survival signaling pathways utilized by insulin-like growth factor I. Mol Cell Biol. 1998;18:6711–6718. doi: 10.1128/mcb.18.11.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee C T, Wu S, Gabrivolich D, Chen H, Nadaf-Rahrov S, Ciernick I F, Carbone D P. Antitumor effect of an adenovirus expressing antisense insulin-like growth factor I receptor on human lung cancer cell lines. Cancer Res. 1996;56:3038–3041. [PubMed] [Google Scholar]
- 41.Lenormand P, Sardet C, Pages G, L’Allemain G, Brunet A, Pousseygur J. Growth factors induce nuclear translocation of MAP kinases (p42mpk and p44mpk) but not of their activator MAP kinase kinase (p45mpkk) in fibroblasts. J Cell Biol. 1993;122:1079–1088. doi: 10.1083/jcb.122.5.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li S, Ferber A, Miura M, Baserga R. Mitogenicity and transforming activity of the insulin-like growth factor I receptor with mutations in the tyrosine kinase domain. J Biol Chem. 1994;269:32558–32564. [PubMed] [Google Scholar]
- 43.Li S, Resnicoff M, Baserga R. Effects of mutations at serines 1280–1283 on the mitogenic and transforming activities of the insulin-like growth factor I receptor. J Biol Chem. 1996;271:12254–12260. doi: 10.1074/jbc.271.21.12254. [DOI] [PubMed] [Google Scholar]
- 44.Li S, Janosch P, Tanji M, Rosenfeld G C, Waymire J C, Mischak H, Kolch W, Sedivy J M. Regulation of Raf-1 kinase activity by the 14.3.3 family of proteins. EMBO J. 1995;14:685–696. doi: 10.1002/j.1460-2075.1995.tb07047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin T H, Chen Q, Howe A, Juliano H L. Cell anchorage permits efficient signal transduction between ras and its downstream kinases. J Biol Chem. 1997;272:8849–8852. [PubMed] [Google Scholar]
- 46.Liu Q, Ning W, Dantzer R, Freund G G, Kelley K W. Activation of protein kinase C-zeta and phosphatidylinositol 3′-kinase and promotion of macrophage differentiation by insulin-like growth factor 1. J Immunol. 1998;160:1393–1401. [PubMed] [Google Scholar]
- 47.Liu X, Turbyville T, Fritz A, Whitesell L. Inhibition of insulin-like growth factor 1 receptor expression in neuroblastoma cells induces the regression of established tumors in mice. Cancer Res. 1998;58:5432–5438. [PubMed] [Google Scholar]
- 48.Long L, Rubin R, Baserga R, Brodt P. Loss of the metastatic phenotype in murine carcinoma cells expressing an antisense RNA to the insulin-like growth factor I receptor. Cancer Res. 1995;55:1006–1009. [PubMed] [Google Scholar]
- 49.Marshall C J. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 50.McCarthy N J, Whyte M K B, Gilbert C S, Evan G I. Inhibition of Ced3/ICE-related proteases does not prevent cell death induced by oncogenes, DNA damage, or the Bcl-2 homologue Bak. J Cell Biol. 1997;136:215–227. doi: 10.1083/jcb.136.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCubrey J A, Stillman L S, Mayhew M W, Algate P A, Dellow R A, Kaleko M. Growth promoting effects of insulin-like growth factor 1 (IGF-1) on hematopoietic cells. Overexpression of introduced IGF-1 receptor abrogates interleukin-3 dependency of murine factor dependent cells by ligand dependent mechanism. Blood. 1991;78:921–929. [PubMed] [Google Scholar]
- 52.Miura M, Li S, Baserga R. Effect of a mutation at tyrosine 950 of the insulin-like growth factor I receptor on the growth and transformation of cells. Cancer Res. 1995;55:663–667. [PubMed] [Google Scholar]
- 53.Myers M G, Jr, Grammer T C, Wang L M, Sun X J, Pierce J H, Blenis J, White M F. Insulin receptor substrate-1 mediates phosphatidylinositol 3′-kinase and p70S6K signaling during insulin, insulin-like growth factor-1, and interleukin-4 stimulation. J Biol Chem. 1994;269:28783–28789. [PubMed] [Google Scholar]
- 54.Myers M G, Jr, Sun X J, Cheatham B, Jachna B R, Glasheen E M, Backer J M, White M F. IRS-1 is a common element in insulin and insulin-like growth factor-I signaling to the phosphatidylinositol 3′-kinase. Endocrinology. 1993;132:1421–1430. doi: 10.1210/endo.132.4.8384986. [DOI] [PubMed] [Google Scholar]
- 55.Nakamura S, Watanabe H, Miura M, Sasaki T. Effect of the insulin-like growth factor I receptor on ionizing radiation-induced cell death in mouse embryo fibroblasts. Exp Cell Res. 1997;235:287–294. doi: 10.1006/excr.1997.3683. [DOI] [PubMed] [Google Scholar]
- 56.O’Connor R, Kauffmann-Zeh A, Liu Y, Lehar S, Evan G I, Baserga R, Blättler W A. Identification of domains of the insulin-like growth factor I receptor that are required for protection from apoptosis. Mol Cell Biol. 1997;17:427–435. doi: 10.1128/mcb.17.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Parrizas M, Saltiel A R, LeRoith D. Insulin-like growth factor I inhibits apoptosis using the phosphatidylinositol 3′-kinase and mitogen-activated protein kinase pathways. J Biol Chem. 1997;272:154–161. doi: 10.1074/jbc.272.1.154. [DOI] [PubMed] [Google Scholar]
- 58.Parrizas M, LeRoith D. Insulin-like growth factor 1 inhibition of apoptosis is associated with increased expression of the bcl-xL gene product. Endocrinology. 1997;138:1355–1358. doi: 10.1210/endo.138.3.5103. [DOI] [PubMed] [Google Scholar]
- 59.Perrotti D, Bonatti S, Trotta R, Martinez R, Skorski T, Salomoni P, Grassilli E, Iozzo R, Cooper D R, Calabretta B. TLS/FUS, a proto-oncogene involved in multiple chromosomal translocations, is a novel regulator of BCR/ABL-mediated leukemogenesis. EMBO J. 1998;17:4442–4445. doi: 10.1093/emboj/17.15.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prager D, Li H L, Asa S, Melmed S. Dominant negative inhibition of tumorigenesis in vivo by human insulin-like growth factor receptor mutant. Proc Natl Acad Sci USA. 1994;91:2181–2185. doi: 10.1073/pnas.91.6.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prisco M, Hongo A, Rizzo M G, Sacchi A, Baserga R. The insulin-like growth factor I receptor as a physiological relevant target of p53 in apoptosis caused by interleukin-3 withdrawal. Mol Cell Biol. 1997;17:1084–1092. doi: 10.1128/mcb.17.3.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prisco M, Romano G, Peruzzi F, Valentinis B, Baserga R. Insulin and IGF-I receptors signaling in protection from apoptosis. Horm Metab Res. 1999;31:80–89. doi: 10.1055/s-2007-978703. [DOI] [PubMed] [Google Scholar]
- 63.Pronk G J, McGlade J, Pelicci G, Pawson T, Bos J L. Insulin-induced phosphorylation of the 46- and 52-kDa Shc proteins. J Biol Chem. 1993;268:5748–5753. [PubMed] [Google Scholar]
- 64.Pruett W, Yuan Y, Rose E, Batzer A G, Harada N, Skolnik E Y. Association between GRB2/Sos and insulin receptor substrate 1 is not sufficient for activation of extracellular signal-regulated kinases by interleukin-4: implications for Ras activation by insulin. Mol Cell Biol. 1995;15:1778–1785. doi: 10.1128/mcb.15.3.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reiss K, D’Ambrosio C, Tu X, Tu C, Baserga R. Inhibition of tumor growth by a dominant negative mutant of the insulin-like growth factor I receptor with a bystander effect. Clin Cancer Res. 1998;4:2647–2655. [PubMed] [Google Scholar]
- 66.Resnicoff M, Abraham D, Yutanawiboonchai W, Rotman H, Kajstura J, Rubin R, Zoltick P, Baserga R. The insulin-like growth factor I receptor protects tumor cells from apoptosis in vivo. Cancer Res. 1995;55:2463–2469. [PubMed] [Google Scholar]
- 67.Resnicoff M, Burgaud J-L, Rotman H L, Abraham D, Baserga R. Correlation between apoptosis, tumorigenesis and levels of insulin-like growth factor I receptors. Cancer Res. 1995;55:3739–3741. [PubMed] [Google Scholar]
- 68.Rininsland F, Johnson T R, Chernicky C L, Schulze E, Berfeind B, Ilan J, Ilan J. Suppression of insulin-like growth factor I receptor by a triple-helix strategy inhibits IGF-I transcription and tumorigenic potential of rat C6 glioblastoma cells. Proc Natl Acad Sci USA. 1997;94:5854–5859. doi: 10.1073/pnas.94.11.5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodriguez-Tarduchy G, Collins M K L, Garcia I, Lopez-Rivas A. Insulin-like growth factor I inhibits apoptosis in IL-3 dependent hemopoietic cells. J Immunol. 1992;149:535–540. [PubMed] [Google Scholar]
- 70.Romano G, Prisco M, Zanocco-Marani T, Peruzzi F, Valentinis B, Baserga R. Dissociation between resistance to apoptosis and the transformed phenotype in IGF-I receptor signaling. J Cell Biochem. 1999;72:294–310. doi: 10.1002/(sici)1097-4644(19990201)72:2<294::aid-jcb14>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 71.Rubini M, Hongo A, D’Ambrosio C, Baserga R. The IGF-I receptor in mitogenesis and transformation of mouse embryo cells: role of receptor number. Exp Cell Res. 1997;230:284–292. doi: 10.1006/excr.1996.3430. [DOI] [PubMed] [Google Scholar]
- 72.Salomoni P, Wasik M A, Riedel R F, Reiss K, Choi J K, Skorski T, Calabretta B. Expression of constitutively active Raf-1 in the mitochondria restores antiapoptotic and leukemogenic potential of a transformation-deficient BCR/ABL mutant. J Exp Med. 1998;187:1995–2007. doi: 10.1084/jem.187.12.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sasaoka T, Draznin B, Leitner J W, Langlois W J, Olefsky J M. Shc is the predominant signaling molecule coupling insulin receptors to activation of guanine nucleotide releasing factor and p21ras-GTP formation. J Biol Chem. 1994;269:10734–10738. [PubMed] [Google Scholar]
- 74.Sasaoka T, Rose D W, Jhun B H, Saltiel A R, Draznin B, Olefsky J M. Evidence for a functional role of Shc proteins in mitogenic signaling induced by insulin, insulin-like growth factor-1, and epidermal growth factor. J Biol Chem. 1994;269:13689–13694. [PubMed] [Google Scholar]
- 75.Scheid M P, Duronio V. Dissociation of cytokine-induced phosphorylation of Bad and activation of PKB/akt: involvement of MEK upstream of Bad phosphorylation. Proc Natl Acad Sci USA. 1998;95:7439–7444. doi: 10.1073/pnas.95.13.7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seely B L, Reichart D R, Staubs P A, Jhun B H, Hsu D, Maegawa H, Milarski K L, Saltiel L R, Olefsky J M. Localization of the insulin-like growth factor 1 receptor binding sites for the SH2 domain proteins p85, Syp and GTPase activating protein. J Biol Chem. 1995;270:19151–19157. doi: 10.1074/jbc.270.32.19151. [DOI] [PubMed] [Google Scholar]
- 77.Sell C, Baserga R, Rubin R. Insulin-like growth factor I (IGF-I) and the IGF-I receptor prevent etoposide-induced apoptosis. Cancer Res. 1995;55:303–306. [PubMed] [Google Scholar]
- 78.Shapiro D N, Jones B G, Shapiro L H, Dias P, Houghton P J. Antisense-mediated reduction in insulin-like growth factor 1 receptor expression suppresses the malignant phenotype of a human alveolar rhabdomyosarcoma. J Clin Investig. 1994;94:1235–1242. doi: 10.1172/JCI117441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Singleton J R, Dixit V M, Feldman E L. Type 1 insulin-like growth factor receptor activation regulates apoptotic proteins. J Biol Chem. 1996;271:31791–31794. doi: 10.1074/jbc.271.50.31791. [DOI] [PubMed] [Google Scholar]
- 80.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Songyang Z, Baltimore D, Cantley L C, Kaplan D R, Franke T F. Interleukin-3-dependent survival by the Akt protein kinase. Proc Natl Acad Sci USA. 1997;94:11345–11350. doi: 10.1073/pnas.94.21.11345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tartare-Deckert S, Sawka-Verhelle D, Murdaca J, van Obberghen E. Evidence for a differential interaction of Shc and the insulin receptor substrate-1 (IRS-1) with the insulin-like growth factor-I (IGF-I) receptor in the yeast two-hybrid system. J Biol Chem. 1995;270:23456–23460. doi: 10.1074/jbc.270.40.23456. [DOI] [PubMed] [Google Scholar]
- 83.Turner B C, Haffty B G, Narayanan L, Yuan J, Havre P A, Gumbs A A, Kaplan L, Burgaud J L, Carter D, Baserga R, Glazer P M. Insulin-like growth factor 1 receptor overexpression mediates cellular resistance and local breast cancer recurrence after lumpectomy and radiation. Cancer Res. 1997;57:3079–3083. [PubMed] [Google Scholar]
- 84.Valentinis B, Morrione A, Peruzzi F, Prisco M, Reiss K, Baserga R. Anti-apoptotic signaling of the IGF-I receptor in fibroblasts following loss of matrix adhesion. Oncogene. 1999;18:1827–1836. doi: 10.1038/sj.onc.1202471. [DOI] [PubMed] [Google Scholar]
- 85.Valentinis B, Romano G, Peruzzi F, Morrione A, Prisco M, Soddu S, Cristofanelli B, Sacchi A, Baserga R. Growth and differentiation signals by the insulin-like growth factor 1 receptor in hemopoietic cells are mediated through different pathways. J Biol Chem. 1999;274:12423–12430. doi: 10.1074/jbc.274.18.12423. [DOI] [PubMed] [Google Scholar]
- 86.Valtieri M, Tweardy D J, Caracciolo D, Johnson K, Mavilio F, Altmann S, Santoli D, Rovera G. Cytokine dependent granulocytic differentiation. J Immunol. 1987;138:3829–3835. [PubMed] [Google Scholar]
- 87.Wang H-G, Rapp U R, Reed J C. Bcl-2 targets the protein kinase Raf-1 to mitochondria. Cell. 1996;87:629–638. doi: 10.1016/s0092-8674(00)81383-5. [DOI] [PubMed] [Google Scholar]
- 88.Wang L M, Myers M G, Jr, Sun X J, Aaronson S A, White M, Pierce J H. IRS-1: essential for insulin- and IL-4-stimulated mitogenesis in hemopoietic cells. Science. 1993;261:1591–1594. doi: 10.1126/science.8372354. [DOI] [PubMed] [Google Scholar]
- 89.White M F. The IRS-signalling system: a network of docking proteins that mediate insulin action. Mol Cell Biochem. 1998;182:3–11. [PubMed] [Google Scholar]
- 90.Wu Y, Tewari M, Cui S, Rubin R. Activation of the insulin-like growth factor I receptor inhibits tumor necrosis factor-induced cell death. J Cell Physiol. 1996;168:499–509. doi: 10.1002/(SICI)1097-4652(199609)168:3<499::AID-JCP2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 91.Yang E, Zha J, Jockel J, Boise L H, Thompson C B, Korsmeyer S J. Bad, a heterodimeric partner for Bcl-xL and Bcl-2, displaces Bax and promotes cell death. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 92.Yenush L, Makati K J, Smith-Hall J, Ishibashi O, Myers M G, Jr, White M F. The pleckstrin homology domain is the principal link between the insulin receptor and IRS-1. J Biol Chem. 1996;271:24300–24306. doi: 10.1074/jbc.271.39.24300. [DOI] [PubMed] [Google Scholar]
- 93.Zamorano J, Wang H Y, Wang L-M, Pierce J H, Keegan A D. IL-4 protects cells from apoptosis via the insulin receptor substrate pathway and a second independent signaling pathway. J Immunol. 1996;157:4926–4934. [PubMed] [Google Scholar]
- 94.Zha J, Harada H, Yang E, Jockel J, Korsmeyer S J. Serine phosphorylation of death agonist Bad in response to survival factor results in binding to 14.3.3 not Bcl-xL. Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 95.Zhou-Li F, Xu S Q, Dews M, Baserga R. Co-operation of simian virus 40 T antigen and insulin receptor substrate-1 in protection from apoptosis induced by interleukin-3 withdrawal. Oncogene. 1997;15:961–970. doi: 10.1038/sj.onc.1201265. [DOI] [PubMed] [Google Scholar]