Abstract
Activation of the c-Jun NH2-terminal kinase (JNK) group of mitogen-activated protein (MAP) kinases is mediated by a protein kinase cascade. This signaling mechanism may be coordinated by the interaction of components of the protein kinase cascade with scaffold proteins. The JNK-interacting protein (JIP) group of scaffold proteins selectively mediates signaling by the mixed-lineage kinase (MLK)→MAP kinase kinase 7 (MKK7)→JNK pathway. The scaffold proteins JIP1 and JIP2 interact to form oligomeric complexes that accumulate in peripheral cytoplasmic projections extended at the cell surface. The JIP proteins function by aggregating components of a MAP kinase module (including MLK, MKK7, and JNK) and facilitate signal transmission by the protein kinase cascade.
The response of cells to extracellular stimuli is mediated by evolutionarily conserved signaling mechanisms that regulate proliferation, differentiation, and survival. These mechanisms include mitogen-activated protein kinase (MAPK) signal transduction pathways. Several groups of MAPKs have been identified in yeast, Drosophila melanogaster, and Caenorhabditis elegans. In mammals, three groups of MAPKs have been described: the extracellular signal-regulated protein kinases (ERK), the c-Jun NH2-terminal kinases (JNK), and the p38 MAPKs. Each of these groups of MAPKs is activated by dual phosphorylation on Thr and Tyr by a MAPK kinase (MAPKK) which, in turn, is activated by a MAPKK kinase (MAPKKK). Comparison of ERK, JNK, and p38 MAPKs indicates that the biological function of each group of MAPKs is different.
The JNK group of MAPKs is activated by treatment of cells with inflammatory cytokines or by exposure to environmental stress (11). Activated JNK phosphorylates many cellular proteins, including components of the AP-1 transcription factor complex (c-Jun and ATF-2). This phosphorylation leads to increased AP-1 transcription activity (11). The role of increased AP-1 transcription activity is unclear. However, JNK signaling and AP-1 activation have been implicated in multiple biological processes (11). For example, genetic analysis of Drosophila demonstrated that JNK is required for early embryonic morphogenesis (20, 26). Genetic epistasis analysis indicated that the effect of JNK on morphogenesis is mediated by activated Drosophila Jun (9, 19, 25). The JNK signaling pathway is also required for mammalian embryogenesis (6, 14, 17, 34).
Three mammalian genes encode JNK protein kinases. JNK1 and JNK2 are expressed ubiquitously, while JNK3 is expressed primarily in the brain (2, 7, 12, 15, 24). A role for JNK in neuronal apoptosis has been demonstrated (31). Recent studies of knockout mice have confirmed this observation. Targeted disruption of the Jnk3 gene causes defects in stress-induced neuronal apoptosis (36), while animals lacking both Jnk1 and Jnk2 genes exhibit defects in developmental neuronal apoptosis (14). JNK is also required for apoptosis of CD4+ CD8+ double-positive thymocytes caused by anti-CD3 in vivo (21, 22). Cellular proliferation, death, and survival may therefore be regulated by the JNK signaling pathway in vivo (11). The JNK signaling pathway also appears to regulate the function of differentiated cells. For example, disruption of the Jnk1 (4) and Jnk2 (22, 35) genes in mice causes defects in T-cell function and immune responses. The JNK signaling pathway therefore contributes to multiple biological processes and represents an important mechanism that is used by cells to respond to extracellular stimulation (11).
JNK is activated by phosphorylation on Thr and Tyr by MKK4 and MKK7 (11). These MAPKKs are activated, in turn, by phosphorylation by MAPKKKs, including ASK1, TPL2, TAK1, and members of the MEKK and mixed-lineage protein kinase (MLK) groups of MAPKKKs (11). Biochemical studies demonstrate that each step in the MAPKKK→MAPKK→JNK signaling pathway can be reconstituted in vitro. However, it is unclear whether these assays faithfully mimic the activation of the JNK pathway in vivo. It is likely that the components of the JNK protein kinase cascade may be organized into defined signaling modules (29). For example, the MAPKKK MEKK1 binds to JNK, MKK4, and the Ste20-related protein kinase NIK (27, 30, 32). These interactions may participate in the transmission of signals from MEKK1 to JNK by the creation of a specific signaling module in vivo (29). A functional signaling module could also be created by the interaction of components of the JNK signaling pathway with other proteins. An example is provided by the scaffold protein JNK-interacting protein 1 (JIP1) (3), which binds JNK, MKK7, MLKs, and the Ste20-related protein kinase HPK1 (28). The JIP1 scaffold mediates signaling to JNK by members of the MLK group of MAPKKKs but does not participate in signaling by the MEKK group of MAPKKKs (28).
The purpose of the study described in this report was to examine the JIP-mediated JNK signaling module. We demonstrate that JIP1 is a member of a group of MAPK scaffold proteins that includes JIP2. Both JIP1 and JIP2 form homo- and hetero-oligomeric complexes with components of the JNK signaling pathway. The JIP scaffolds facilitate JNK activation by MLK protein kinases by aggregating components of the MAPK cascade to form a functional JNK signaling module.
MATERIALS AND METHODS
Molecular cloning of JIP2.
JIP2 cDNA clones were isolated from a human brain λZAPII cDNA library (Stratagene Inc.) by plaque hybridization using a JIP cDNA fragment as a probe. The largest clone (3,355 bp) included the complete open reading frame of human JIP2. The sequence of JIP2 was determined by using an Applied Biosystems 373A machine.
Plasmids.
Expression vectors for JIP2 were constructed by using the plasmids pCDNA3 (Invitrogen Inc.), pEBG (23), and pGEX-4T-1 (Pharmacia-LKB Biotechnology Inc.) by subcloning PCR fragments of JIP2 in the BamHI/NheI, BamHI/NotI, and BamHI/NotI sites, respectively. Expression vectors for MAPKs, MAPKKs, MAPKKKs, JIP1, and JIP1b have been described previously (28).
Antibodies.
The antibodies to the Flag epitope tag (M2; Sigma), the hemagglutinin (HA) epitope tag (12CA5; Boehringer Mannheim), the T7-Tag epitope tag (Novagen Inc.), and glutathione S-transferase (GST) (Pharmacia-LKB Biotechnology Inc.) were purchased from the indicated suppliers. Antibodies to JIP1 and JIP2 were prepared by immunizing mice with purified bacterially expressed proteins by standard techniques (8).
Immunofluorescence analysis.
Rin5F insulinoma cells were grown on glass coverslips, washed, and then fixed and permeabilized with methanol at −20°C for 8 min. Indirect immunofluorescence was performed by incubation with 10 μg of a primary monoclonal antibody to JIP1 or JIP2/ml in 50 mM Tris-Cl (pH 7.4)–150 mM NaCl–3% (wt/vol) bovine serum albumin for 12 h at 4°C. The secondary antibody was ALEXA 594-conjugated anti-mouse immunoglobulin (Ig) antibody (1:200; Molecular Probes), and nuclei were visualized by using SYTOX Green (Molecular Probes). Confocal images were prepared with a Leica microscope. Double-labeled immunofluorescence images were prepared with a conventional Zeiss Axiophot microscope by using cells stained with a fluorescein isothiocyanate-conjugated monoclonal antibody to JIP1 (2.5 μg/ml) and a monoclonal antibody to JIP2 (2.5 μg/ml) together with a Texas red-conjugated anti-mouse Ig secondary antibody (1:100; Jackson Immunoresearch). Nuclei were detected with 4,6-diamidino-2-phenylindole.
Biochemical assays.
Rin5F insulinoma cells and COS-7 cells were cultured in RPMI medium and Dulbecco modified Eagle medium, respectively, supplemented with 10% fetal calf serum (Life Technologies Inc.). Transfection assays were performed with COS-7 cells by the Lipofectamine method (Life Technologies Inc.). The cells were solubilized in lysis buffer (20 mM Tris-Cl [pH 7.4], 137 mM NaCl, 2 mM EDTA, 25 mM β-glycerophosphate, 2 mM pyrophosphate, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin/ml, 10% glycerol, 1% Triton X-100). GST fusion proteins were isolated by incubation with glutathione-agarose (Pharmacia-LKB Biotechnology Inc.) beads (20 μl) for 3 h at 4°C. Proteins were immunoprecipitated by incubation for 3 h at 4°C with antibodies bound to protein G-Sepharose (Pharmacia-LKB Biotechnology Inc.). Immunoblot analysis was performed by using enhanced-chemiluminescence detection (Kirkegaard & Perry). Binding assays using purified bacterially expressed proteins were performed by methods described previously (28). Protein kinase activity was measured by using the in-gel method with 0.25 mg of substrate (GST–c-Jun)/ml polymerized in the gel (28).
Nucleotide sequence accession number.
The sequence of JIP2 has been deposited in GenBank under accession no. AF136382.
RESULTS
Molecular cloning of JIP2.
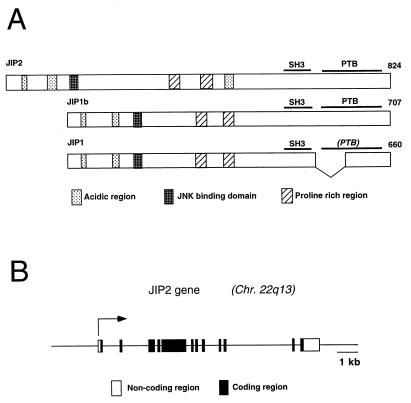
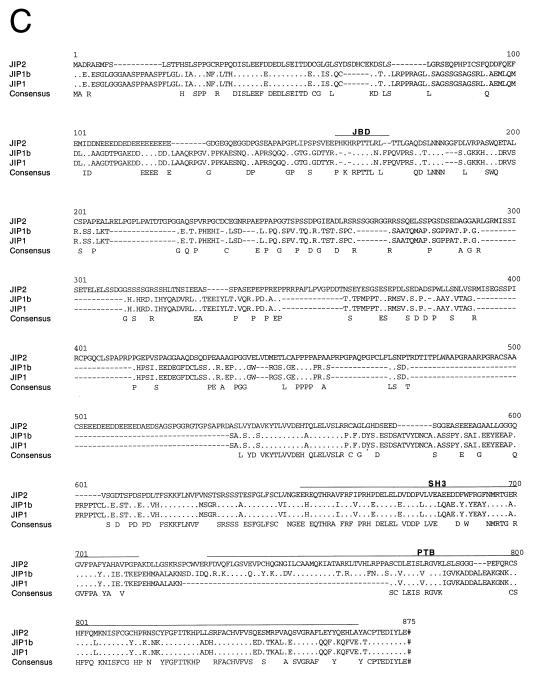
The JIP1 protein has been identified as a putative scaffold that binds components of the JNK signaling pathway (28). We investigated whether additional members of the JIP group are expressed by mammalian cells. These studies led to the molecular cloning of human JIP2 (Fig. 1A). The JIP2 gene is composed of 12 exons and is located on human chromosome 22q13 (Fig. 1B). Sequence analysis of JIP2 cDNA clones demonstrated the presence of a JNK binding domain (JBD) in the NH2-terminal region and both an SH3 domain and a PTB domain in the COOH-terminal region (Fig. 1C). This domain structure is similar to that of JIP1. However, unlike JIP1, we obtained no evidence for the expression of alternatively spliced variants of JIP2 with a deletion in the PTB domain.
FIG. 1.
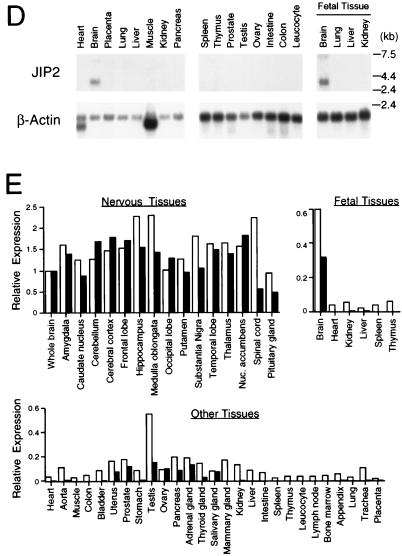
JIP2 is a member of the JIP group of scaffold proteins. (A) The structure of the JIP group of scaffold proteins is illustrated schematically. The JBD, the SH3 domain, and the PTB domain are indicated. Comparison of the NH2-terminal region of JIP2 (residues 1 to 603) with the corresponding region of JIP1 indicates 41% identity and 46% similarity (Line-up Program, Wisconsin package, version 9.1; Genetics Computer Group). The SH3 domains of JIP2 (residues 604 to 665) and JIP1 are 66% identical and 80% similar. The PTB domains of JIP2 (residues 683 to 812) and JIP1b are 64% identical and 74% similar. (B) The human JIP2 gene consists of 12 exons and is located on chromosome 22q13 (GenBank accession no. U62317). The structure of the gene is illustrated schematically. Solid boxes, coding regions; open boxes, noncoding regions of exons 1 and 12. (C) The primary sequence of human JIP2 deduced from the sequence of cDNA clones is presented in single-letter code and compared to the sequences of JIP1 and JIP1b. Periods, residues that are identical to JIP2 residues; dashes, deletions; pound signs (#), termination codons. The JBD, the SH3 domain, and the PTB domain are indicated. The sequences of JIP1, JIP1b, and JIP2 have been deposited in GenBank under accession no. AF003115, AF054611, and AF136382, respectively. (D) The expression of JIP2 mRNA was examined by Northern blot analysis of different human tissues (Clontech Inc.) using JIP2 cDNA as a probe. (E) Comparison of the tissue distribution of the expression of the human JIP1 and JIP2 genes. Immobilized human mRNA was hybridized with human JIP1 and JIP2 cDNA probes labeled with 32P by random priming. The data were quantitated by PhosphorImager analysis and are presented graphically. The data are normalized to the expression in the whole brain (arbitrarily set at 1.0). JIP1 and JIP2 are represented by open and filled bars, respectively.
Northern blot analysis of the expression of JIP2 in human tissues demonstrated that transcripts were detected in the brain but not in the other tissues examined (Fig. 1D). This selective expression of JIP2 in the brain differs from JIP1, which is expressed more widely throughout the body (3). To directly compare the distribution of these JIP proteins, we examined the amounts of JIP1 and JIP2 mRNAs in different human tissues. This analysis demonstrated that both JIP1 and JIP2 are expressed throughout the nervous system (Fig. 1E). JIP1 was also expressed in many additional tissues. A low level of JIP2 expression was also detected in a limited number of human tissues, including the uterus, prostate, colon, testis, ovary, pancreas, adrenal gland, thyroid gland, and salivary gland.
The JIP2 scaffold binds components of the JNK cascade.
JIP1 selectively binds components of the JNK signaling pathway (28). We therefore examined the interaction of JIP2 with MAPKKKs, MAPKKs, and MAPKs.
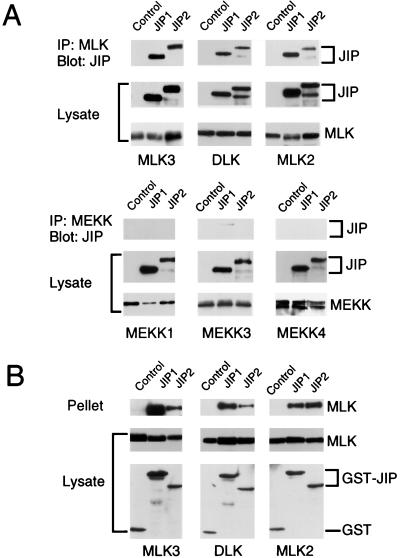
The interaction of JIP1 and JIP2 with MAPKKKs was examined in coimmunoprecipitation assays (Fig. 2). The JIP proteins did not bind members of the MEKK group, for example, MEKK1 and MEKK4 (Fig. 2A). However, a very small amount of coimmunoprecipitation of MEKK3 was detected in experiments using JIP1 but not JIP2 (Fig. 2A). Furthermore, a very small amount of coimmunoprecipitation of the MAPKKK ASK1 with both JIP1 and JIP2 was detected (data not shown). In contrast, three different MLKs (DLK, MLK2, and MLK3) coimmunoprecipitated with JIP1 and JIP2 (Fig. 2A). Reciprocal precipitation-immunoblot analysis confirmed the interaction between the JIP proteins and MLKs (Fig. 2B). These data demonstrated that both JIP1 and JIP2 selectively interact with the MLK group of MAPKKKs. The COOH-terminal region of JIP1 (28) or JIP2 (Fig. 2B) was sufficient for the observed interaction with MLK3.
FIG. 2.
Selective binding of JIP1 and JIP2 to the mixed-lineage group of MAPKKKs. (A) Epitope-tagged JIP1 or JIP2 was expressed in cells together with epitope-tagged MAPKKKs. Control experiments were performed by transfection of the empty expression vector instead of the JIP expression vector. The expression of JIP and MAPKKK proteins was examined by immunoblot analysis of the cell lysates. The MAPKKK proteins were immunoprecipitated with the M2 monoclonal antibody to the Flag epitope. The presence of JIP proteins in the immunoprecipitates (IP) was examined by immunoblot analysis using a monoclonal antibody that binds the T7-Tag epitope. Coimmunoprecipitation was not observed in experiments using MEKK1, MEKK3, or MEKK4. In contrast, JIP1 and JIP2 coimmunoprecipitated with the MLKs DLK, MLK2, and MLK3. (B) The COOH-terminal regions of JIP1 (residues 283 to 660) and JIP2 (residues 499 to 824) were expressed in cells as GST fusion proteins together with epitope-tagged MLK. Control experiments were performed by transfection with the GST expression vector pEBG. The amounts of the GST proteins and MLK in the cell lysates were examined by protein immunoblot analysis. The GST fusion proteins were precipitated from cell lysates with glutathione-agarose, and MLK present in the pellet was detected by protein immunoblot analysis.
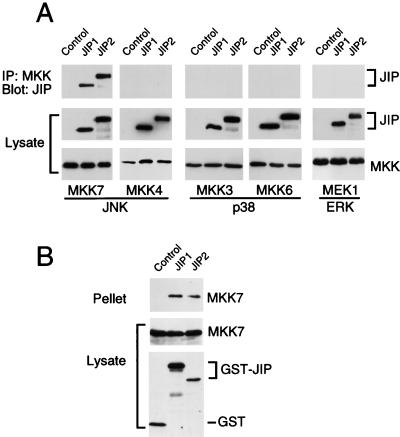
Coimmunoprecipitation assays were performed to investigate the interaction of JIP proteins with MAPKKs (Fig. 3). No evidence for interaction with JIP1 or JIP2 was obtained in experiments using activators of the ERK (MEK1) or p38 MAPK (MKK3 and MKK6) pathways (Fig. 3A). Similarly, MKK4, an activator of JNK, did not coimmunoprecipitate with the JIP proteins. However, both JIP1 and JIP2 were found to coimmunoprecipitate with the JNK activator MKK7 (Fig. 3A). The interaction of MKK7 with the JIP proteins was confirmed by reciprocal precipitation-immunoblot analysis (Fig. 3B). Thus, both JIP1 and JIP2 selectively bind the MAPKK isoform MKK7. The COOH-terminal region of JIP1 (28) or JIP2 (Fig. 3B) was sufficient for the observed interaction with MKK7.
FIG. 3.
Selective binding of JIP1 and JIP2 to the MAPKK MKK7. (A) Epitope-tagged JIP1 and JIP2 were expressed in cells together with epitope-tagged MEK1, MKK3, MKK4, MKK6, or MKK7. Control experiments were performed by transfection of the empty expression vector instead of the JIP expression vector. The expression of JIP and MAPKK proteins was examined by immunoblot analysis of the cell lysates. The MAPKK proteins were immunoprecipitated, and the presence of JIP1 and JIP2 in the immunoprecipitates (IP) was examined by immunoblot analysis using a monoclonal antibody that binds the T7-Tag epitope. (B) The COOH-terminal regions of JIP1 (residues 283 to 660) and JIP2 (residues 499 to 824) were expressed in cells as GST fusion proteins together with epitope-tagged MKK7. Control experiments were performed by transfection with the GST expression vector pEBG. The amounts of the GST proteins and MKK7 in the cell lysates were examined by protein immunoblot analysis. The GST fusion proteins were precipitated from cell lysates with glutathione-agarose, and MKK7 present in the pellet was detected by protein immunoblot analysis using an antibody to the Flag epitope.
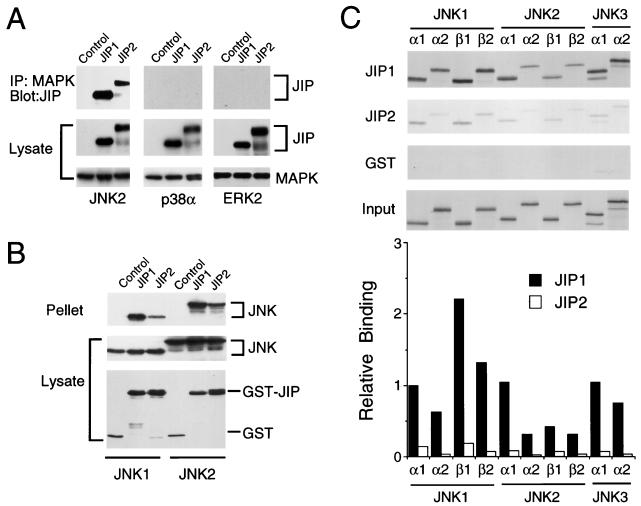
To test the interaction of the JIP proteins with MAPKs, we performed coimmunoprecipitation assays with JNK, ERK, and p38 MAPK (Fig. 4). These experiments demonstrated that ERK and p38 MAPK did not interact with JIP1 or JIP2 proteins. In contrast, both JIP proteins coimmunoprecipitated with JNK (Fig. 4A). Reciprocal precipitation-immunoblot analysis confirmed that JNK interacts with JIP proteins (Fig. 4B). These data demonstrate that JIP1 and JIP2 selectively bind the JNK group of MAPKs. The NH2-terminal region of JIP1 (28) or JIP2 (Fig. 4B) was sufficient for the observed interaction with JNK.
FIG. 4.
Selective binding of JIP1 and JIP2 to the JNK group of MAPKs. (A) Epitope-tagged (T7-Tag) JIP1 and JIP2 were expressed in cells with the HA-tagged MAPKs ERK2, p38α, and JNK2α2. Control experiments were performed by transfection of the empty expression vector instead of the JIP expression vectors. The MAPKs were immunoprecipitated with an antibody to HA. The presence of JIP in the immunoprecipitates (IP) was detected on immunoblots probed with an antibody to T7-Tag. The amounts of JIP and MAPKs in the cell lysates were examined by protein immunoblot analysis. (B) The NH2-terminal regions of JIP1 (residues 1 to 282) and JIP2 (residues 1 to 229) were expressed in cells as GST fusion proteins together with epitope-tagged JNK1α1 or JNK2α2. Control experiments were performed by transfection with the GST expression vector pEBG instead of the JIP expression vector. The amounts of the GST fusion proteins and JNK in the cell lysates were examined by protein immunoblot analysis. The GST fusion proteins were precipitated from cell lysates with glutathione-agarose, and JNK present in the pellet was detected by protein immunoblot analysis using an antibody to the HA epitope. (C) Comparison of the binding of 10 human JNK isoforms to JIP1 and JIP2. The JNK protein kinases were prepared by in vitro translation in the presence of [35S]methionine. The JNK proteins were incubated with GST-JIP1, GST-JIP2, or GST immobilized on glutathione-agarose beads. The beads were washed with lysis buffer. Bound JNK was detected by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and autoradiography. The amount of JNK bound was quantitated by PhosphorImager analysis. The data are presented graphically as relative binding by defining the amount of JNK1α1 binding to JIP1 as 1.0.
Comparison of JIP1 and JIP2 indicates that these proteins exhibit similar binding properties in assays using MAPKKKs and MAPKKs (Fig. 2 and 3). In contrast, the interaction with MAPKs appears to differ between JIP1 and JIP2. While both JIP proteins selectively bind JNK, a greater amount of JNK binding to JIP1 than to JIP2 was detected (Fig. 4A and B). To quantitate this difference and to examine the relative binding to different JNK isoforms, we performed assays using JNK prepared by in vitro translation in the presence of [35S]methionine. The binding of JNK isoforms to immobilized recombinant JIP1 and JIP2 was examined (Fig. 4C). These experiments demonstrated that 10 different JNK isoforms bound to both JIP proteins. However, differences in binding were detected. First, the amount of JNK binding to JIP1 was markedly greater than the binding to JIP2. Second, some JNK isoforms (e.g., JNK1α1) were found to bind JIP more strongly than other JNK isoforms (e.g., JNK2α2). These data indicate that JIP proteins interact with JNK and can distinguish between JNK isoforms.
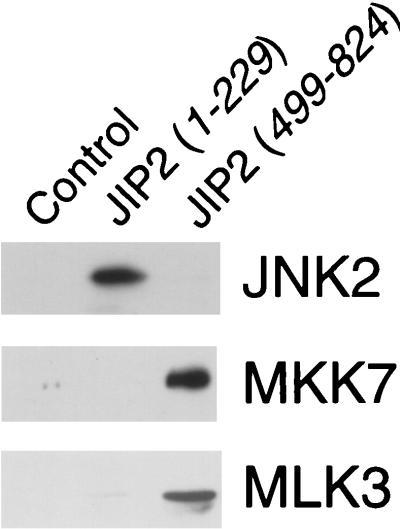
To test whether the interaction of JIP2 with JNK, MKK7, and MLK3 might be direct, we investigated the binding of JIP2 to these protein kinases using purified bacterially expressed recombinant proteins. This analysis demonstrated that JIP2 binds to JNK, MKK7, and MLK3 (Fig. 5). Similarly, purified recombinant JIP1 binds to purified recombinant JNK, MKK7, and MLK3 (28).
FIG. 5.
JIP2 binds JNK, MKK7, and MLK3 in vitro. Purified bacterially expressed GST and GST-JIP2 fusion proteins were immobilized on glutathione-agarose and incubated with purified bacterially expressed JNK2, MKK7, or MLK3 (residues 1 to 204). Bound JNK2, MKK7, and MLK3 were detected by immunoblot analysis.
JIP2 regulates signal transduction by the JNK signaling pathway.
The binding to MLKs, MKK7, and JNK suggests that JIP2 may act as a scaffold protein. We therefore examined the effect of JIP2 overexpression on the JNK signal transduction pathway. Initial studies of the effect of JIP2 overexpression indicated that JIP2, like JIP1, acted as a powerful inhibitor of JNK signaling. For example, studies of the chimeric transcription factor GAL4–ATF-2 demonstrated that both JIP1 and JIP2 inhibited JNK-dependent reporter gene expression by more than 90% (data not shown). This inhibition of JNK signaling by JIP proteins (3) is probably mediated by the sequestration of limiting components of the JNK signaling pathway into separate complexes by the overexpressed scaffold protein (28).
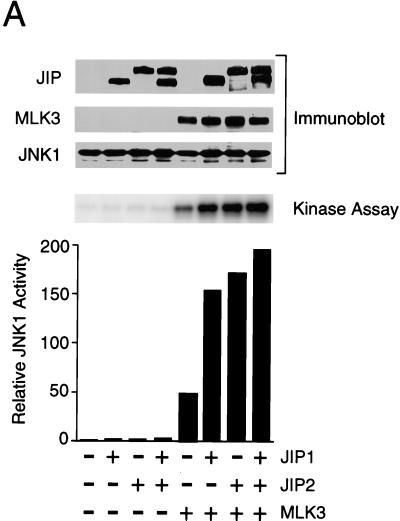
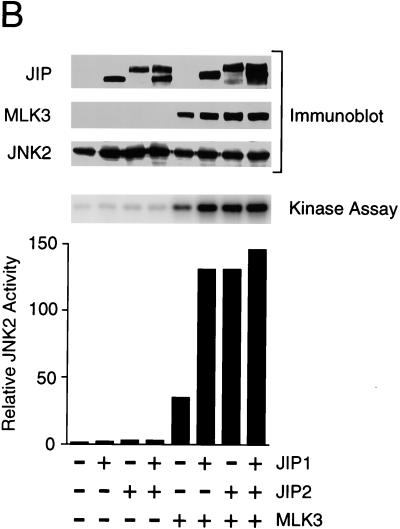
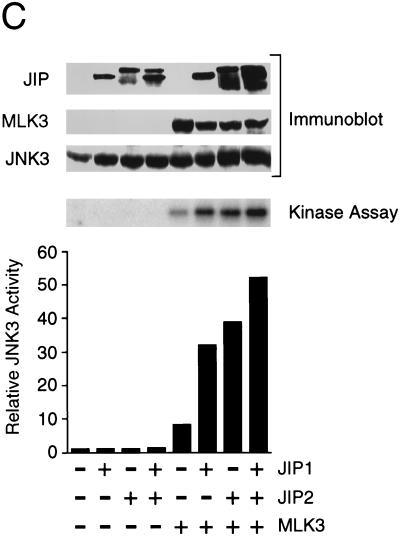
To test the effect of JIP2 under conditions where sequestration of limiting components of the JNK signaling pathway does not occur, we coexpressed JIP proteins with components of the JNK signaling pathway (Fig. 6). JNK activity was measured in an in vitro protein kinase assay using recombinant c-Jun as the substrate. This analysis demonstrated that MLK3 caused JNK activation. Expression of JIP1 and JIP2 enhanced the activation of JNK caused by MLK3. Similar data were obtained in experiments using JNK1, JNK2, or JNK3. Together, these data demonstrated that JIP2 increases JNK activation by the MLK signaling pathway. The effect of JIP2 in increasing JNK activation was similar to that caused by JIP1.
FIG. 6.
JIP1 and JIP2 enhance JNK activation by MLK3. Epitope-tagged (HA) JNK1α1 (A), JNK2α2 (B), and JNK3α2 (C) were each expressed alone or coexpressed with JIP1 and/or JIP2. JNK activity was examined by using c-Jun as a substrate. The effect of coexpression with MLK3 was examined. The amounts of JIP, MLK, and JNK expressed were examined by immunoblot analysis.
Coexpression of JIP1 with JIP2 did not cause further enhancement of JNK activation beyond that caused by either JIP1 or JIP2 alone (Fig. 6). Since the binding of JNK to JIP1 was markedly greater than the binding of JNK to JIP2 (Fig. 4), the similar effects of JIP1 and JIP2 on JNK activation (Fig. 6) suggest that the function of JIP proteins to potentiate MLK-stimulated JNK activation may be catalytic and may not require the formation of stable complexes with JNK.
JNK scaffold complexes are formed by JIP1 and JIP2.
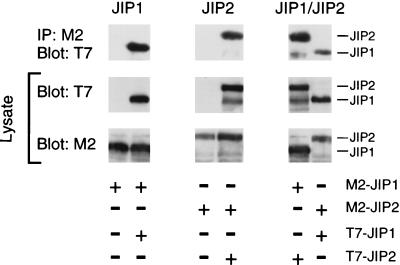
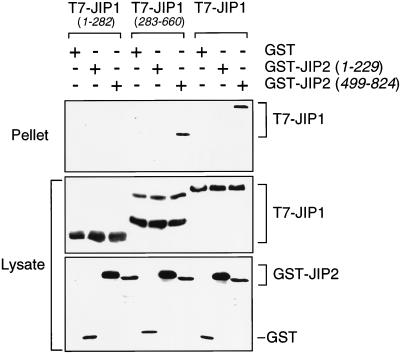
JIP1 and JIP2 are individually capable of forming scaffold complexes for the MLK→MKK7→JNK signaling pathway. However, both JIP1 and JIP2 are expressed in the brain (Fig. 1). This coexpression of JIP1 and JIP2 raises questions about whether the JIP1 and JIP2 scaffold complexes are independent or whether these complexes may interact. To examine this question, we performed coimmunoprecipitation analysis of JIP1 and JIP2 (Fig. 7). These experiments demonstrated that JIP1 interacts with JIP2. Furthermore, both JIP1 and JIP2 were observed to self-associate. These data indicated that JIP1 and JIP2 can form both homo-oligomers and hetero-oligomers. Deletion analysis demonstrated that the COOH-terminal regions of JIP1 and JIP2 were sufficient for the formation of hetero-oligomeric complexes (Fig. 8).
FIG. 7.
JIP proteins form homo- and hetero-oligomeric scaffold complexes. Epitope-tagged JIP1 and JIP2 proteins (Flag and T7-Tag) were expressed in COS-7 cells. The expression of epitope-tagged JIP proteins was examined by immunoblot analysis of cell lysates with antibodies to Flag and T7-Tag. Coimmunoprecipitation analysis was performed by immunoblot analysis (T7-Tag antibody) of immunoprecipitates (IP) prepared by using the M2 monoclonal antibody to the Flag epitope.
FIG. 8.
The interaction of JIP1 and JIP2 is mediated by the COOH-terminal region. Epitope-tagged JIP1 was expressed in COS-7 cells together with GST or GST-JIP2 fusion proteins. The expression of JIP1 was detected by immunoblot analysis using an antibody to the T7 epitope tag. GST and GST-JIP2 fusion proteins were detected by immunoblot analysis using an antibody that binds GST. The interaction of JIP1 with JIP2 was examined by isolation of GST and GST-JIP2 complexes using glutathione-agarose and immunoblot analysis of the bound T7-tagged JIP1. Deletion analysis demonstrated that the COOH-terminal regions, but not the NH2-terminal regions, of JIP1 and JIP2 were sufficient for the formation of hetero-oligomeric complexes.
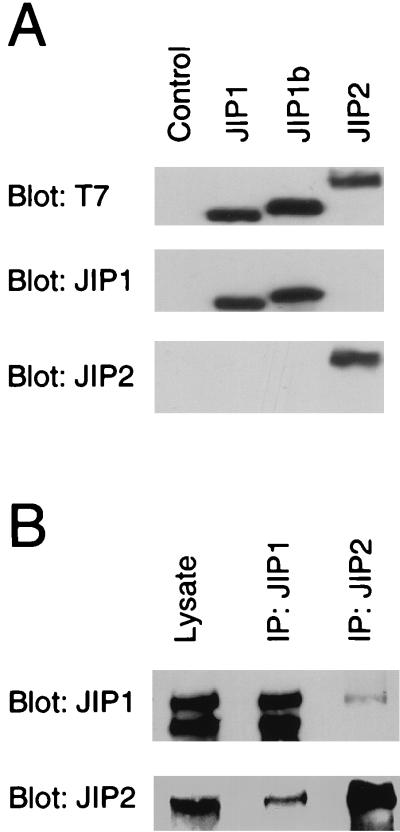
To examine whether JIP1 and JIP2 form oligomeric complexes in vivo, we prepared antibodies to JIP1 and JIP2 (Fig. 9A). These antibodies detect endogenous JIP1 and JIP2 proteins expressed by cultured cells (Fig. 9B). Reciprocal immunoprecipitation-immunoblot analysis of cell extracts demonstrated coprecipitation of JIP1 and JIP2 (Fig. 9B). These data confirm the observation made with transfected recombinant JIP proteins in cultured cells by demonstrating an interaction between the endogenous JIP proteins in vivo.
FIG. 9.
Identification of endogenous oligomeric JIP complexes. (A) Characterization of monoclonal antibodies to JIP1 and JIP2. COS-7 cell lysates containing T7-Tag-labeled JIP proteins were examined by protein immunoblot analysis using antibodies to the T7-Tag and monoclonal antibodies to JIP1 and JIP2. (B) JIP1 and JIP2 interact in vivo. Extracts were prepared from Rin5F insulinoma cells and examined by protein immunoblot analysis by probing with murine antibodies to JIP1 and JIP2. The endogenous JIP1 and JIP2 proteins were detected in the cell lysates and in immunoprecipitates (IP) prepared by using antibodies to JIP1 and JIP2.
Subcellular localization of JIP scaffold complexes.
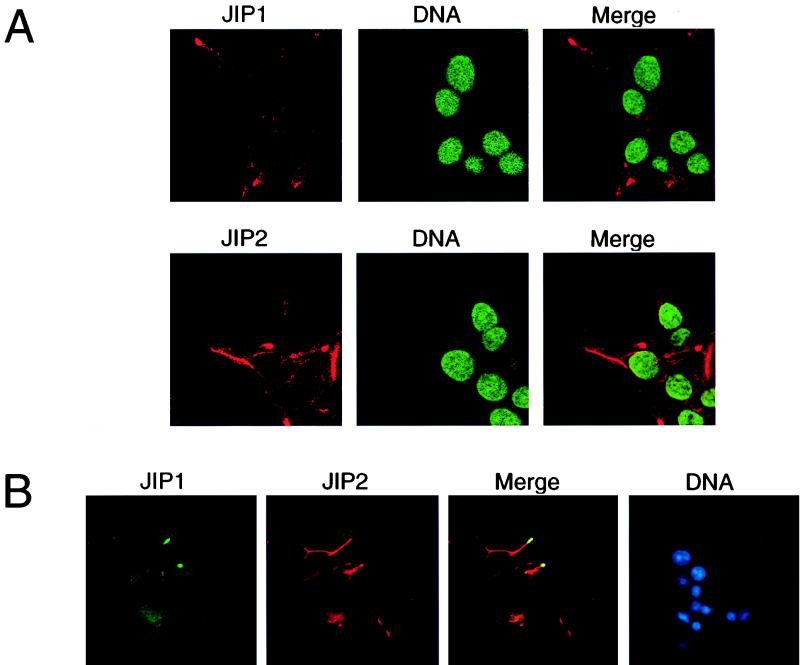
We investigated the subcellular localization of JIP proteins using monoclonal antibodies to JIP1 and JIP2. Confocal immunofluorescence microscopy demonstrated that both JIP1 and JIP2 were located in the cytoplasm and were largely excluded from the nucleus (Fig. 10A). This distribution of JIP proteins was unaffected when JNK was activated following treatment of the cells with interleukin-1 or exposure to UV-C radiation (data not shown). Comparison of JIP1 and JIP2 indicated that the patterns of expression of these proteins differed. Immunofluorescence analysis demonstrated a punctate appearance of JIP1 in the cytoplasm and an accumulation of JIP1 in cell surface projections (Fig. 10A). A similar punctate expression of JIP2 was observed, but more extensive cytoplasmic accumulation of JIP2 than JIP1 was detected.
FIG. 10.
Cytoplasmic location of endogenous JIP proteins. (A) The endogenous JIP1 and JIP2 proteins expressed by Rin5F insulinoma cells were detected by confocal immunofluorescence analysis using monoclonal antibodies to JIP1 and JIP2 (red). The nucleus was detected by staining DNA with SYTOX green. (B) Double-label immunofluorescence analysis of JIP1 (green) and JIP2 (red) was performed by conventional microscopy. The nucleus was detected by staining DNA with 4,6-diamidino-2-phenylindole (blue).
To compare the distribution of JIP1 and JIP2, we performed double-label immunofluorescence analysis (Fig. 10B). JIP1 and JIP2 colocalized in the peripheral cell surface projections where most of the JIP1 protein was detected. In addition, the punctate cytoplasmic distributions of JIP1 and JIP2 were similar. However, the majority of JIP2 was found in cytoplasmic structures that did not appear to contain JIP1. Thus, some JIP2 molecules colocalize with JIP1 (Fig. 10B). This conclusion is consistent with the observation that a fraction of the total JIP2 molecules coimmunoprecipitated with JIP1 (Fig. 9B). Together, these data indicate that the JIP1 and JIP2 scaffolds exist in vivo as both homo- and hetero-oligomeric complexes.
DISCUSSION
Two members of the JIP group have been identified: JIP1 (1, 3, 13) and JIP2 (Fig. 1). These JIP proteins are related in structure (Fig. 1), but the genes that encode these proteins are not closely linked in the genome. The human JIP1 gene is located on human chromosome 11 (11p11.2-p12), while a JIP1 pseudogene is located on human chromosome 17 (17q21) (16). In contrast, the JIP2 gene is located on human chromosome 22 (22q13) (Fig. 1). Comparison of the expression of the JIP1 and JIP2 genes indicates that both JIP proteins are widely expressed in many regions of the brain (Fig. 1). In addition, expression of JIP1 and JIP2 was detected in other tissues.
The JIP1 and JIP2 proteins interact with multiple components of the JNK signaling pathway, including the JNK group of MAPKs, the MAPKK isoform MKK7, and members of the MLK group of MAPKKKs (Fig. 2 to 4). Functional analysis demonstrated that these complexes cause marked potentiation of JNK activation (Fig. 6). These data suggest that the JIP1 and JIP2 proteins act as molecular scaffolds that mediate the activation of the JNK signaling pathway in vivo.
The JIP scaffold proteins were found to self-associate to form intermolecular complexes (Fig. 7). Coimmunoprecipitation studies demonstrated the presence of both homo- and hetero-oligomeric complexes. These oligomeric JIP complexes may contribute to the scaffold function of JIP proteins by aggregating components of the JNK signaling pathway to create a functional protein kinase cascade. This oligomeric structure of JIP is analogous to the Ste5p scaffold protein in yeast, which also forms oligomeric complexes containing more than one Ste5p molecule (5, 10, 33).
Previous studies of overexpressed epitope-tagged JIP1 indicate that it is located exclusively in the cytoplasm (3). Similarly, overexpressed epitope-tagged JIP2 was also detected in the cytoplasm (data not shown). Together, these observations suggest that the endogenous JIP proteins expressed by cells may also be located in the cytoplasm. To examine this question, we prepared monoclonal antibodies to JIP1 and JIP2 and examined the distribution of the endogenous JIP proteins in cultured cells (Fig. 10). These data confirmed that both JIP proteins were detected in the cytoplasm. Interestingly, the majority of the JIP1 protein accumulated in peripheral cytoplasmic projections that extend from the cell surface. The JIP2 protein colocalized in these cell surface projections but was also found to accumulate in a cytoplasmic compartment that did not include JIP1. It is tempting to speculate that the observed cell surface location of JIP proteins may represent a site of action of these scaffold proteins in vivo, as described for the yeast scaffold protein Ste5p (18).
In a recent study, JIP1 was implicated in the expression of the Glut2 and insulin genes (1). Further studies are required to determine whether there is a similar role for JIP2. The JIP1 function of regulating Glut2 and insulin gene expression was proposed to be mediated by nuclear JIP1 (1). Our immunofluorescence analysis of endogenous JIP proteins did not detect significant amounts of nuclear JIP1 or JIP2. We cannot eliminate the possibility that JIP proteins might accumulate in the nucleus under some experimental conditions. However, it is possible that the effect of JIP1 on Glut2 and insulin gene expression may be indirectly mediated by changes in JNK activity (or some other JIP-regulated activity).
The JIP group of MAPK scaffold proteins includes at least two members. Both JIP1 and JIP2 strongly potentiate JNK activation by the MLK/MKK7 signaling pathway. The function of the JIP scaffold proteins appears similar to that of the Ste5p scaffold, which mediates activation of the Ste11p (MAPKKK), Ste7p (MAPKK), and Fus3p (MAPK) signaling pathway in yeast (for a review, see reference 29). The results of the present study demonstrate that JIP proteins exist as oligomers in vivo and are detected in peripheral cell surface projections. These properties are similar to those of the yeast Ste5p scaffold, which also functions as an oligomer (5, 10, 33) and is present in pheromone-induced cell projections (18). Together, these data suggest that mammalian JIP and yeast Ste5p may function by analagous mechanisms that involve the selective aggregation of MAPK pathway components in a discrete cellular compartment.
The JIP scaffold proteins interact with MLKs, MKK7, and JNK. Interactions with other components of the JNK signaling pathway, including MKK4 and members of the MEKK group of MAPKKKs, were not observed. This selectivity indicates that JIP scaffold complexes represent only one mechanism of JNK activation. Signals mediated by MEKK protein kinases and MKK4 function independently of the JIP scaffold complexes (29). These signals may be mediated by other scaffold complexes. Alternatively, JNK may be activated by a mechanism involving intermolecular associations between signaling components, as described recently for signaling mediated by MEKK1 (27, 30, 32). The concept that scaffold-dependent and scaffold-independent signaling pathways can coexist has been established by studies of MAPK signaling in yeast (for a review, see reference 29). An important goal for future research will be to define the relative roles of these alternative mechanisms that mediate JNK activation. The analysis of mice with targeted disruptions of genes that encode components of the JNK signaling pathway will facilitate the genetic dissection of these processes.
ACKNOWLEDGMENTS
We thank our colleagues for providing essential reagents, L. Gangwani for assistance with monoclonal antibody production, W. E. Theurkauf for assistance with confocal microscopy, T. Barrett and A. Quail for DNA sequence analysis, and K. Gemme for administrative assistance.
J.Y. was supported, in part, by the Foundation for Promotion of Cancer Research in Japan. R.J.D. is an investigator of the Howard Hughes Medical Institute. This study was supported by a grant from the National Cancer Institute.
REFERENCES
- 1.Bonny C, Nicod P, Waeber G. IB1, a JIP-1-related nuclear protein present in insulin-secreting cells. J Biol Chem. 1998;273:1843–1846. doi: 10.1074/jbc.273.4.1843. [DOI] [PubMed] [Google Scholar]
- 2.Dérijard B, Hibi M, Wu I-H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 3.Dickens M, Rogers J S, Cavanagh J, Raitano A, Xia Z, Halpern J R, Greenberg M E, Sawyers C L, Davis R J. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science. 1997;277:693–696. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 4.Dong C, Yang D D, Wysk M, Whitmarsh A J, Davis R J, Flavell R A. Defective T cell differentiation in the absence of Jnk1. Science. 1998;282:2092–2095. doi: 10.1126/science.282.5396.2092. [DOI] [PubMed] [Google Scholar]
- 5.Feng Y, Song L Y, Kincaid E, Mahanty S K, Elion E A. Functional binding between Gβ and the LIM domain of Ste5 is required to activate the MEKK Ste11. Curr Biol. 1998;8:267–278. doi: 10.1016/s0960-9822(98)70108-3. [DOI] [PubMed] [Google Scholar]
- 6.Ganiatsas S, Kwee L, Fujiwara Y, Perkins A, Ikeda T, Labow M A, Zon L I. SEK1 deficiency reveals mitogen-activated protein kinase cascade crossregulation and leads to abnormal hepatogenesis. Proc Natl Acad Sci USA. 1998;95:6881–6886. doi: 10.1073/pnas.95.12.6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta S, Barrett T, Whitmarsh A J, Cavanagh J, Sluss H K, Derijard B, Davis R J. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 8.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 9.Hou X S, Goldstein E S, Perrimon N. Drosophila Jun relays the Jun amino-terminal kinase signal transduction pathway to the Decapentaplegic signal transduction pathway in regulating epithelial cell sheet movement. Genes Dev. 1997;11:1728–1737. doi: 10.1101/gad.11.13.1728. [DOI] [PubMed] [Google Scholar]
- 10.Inouye C, Dhillon N, Thorner J. Ste5 RING-H2 domain: role in Ste4-promoted oligomerization for yeast pheromone signaling. Science. 1997;278:103–106. doi: 10.1126/science.278.5335.103. [DOI] [PubMed] [Google Scholar]
- 11.Ip Y T, Davis R J. Signal transduction by the c-Jun NH2-terminal kinase (JNK)—from inflammation to development. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 12.Kallunki T, Su B, Tsigelny I, Sluss H K, Derijard B, Moore G, Davis R J, Karin M. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- 13.Kim I-J, Lee K-W, Park B Y, Lee J-K, Park J, Choi I Y, Eom S-J, Chang T-S, Kim M J, Yeom Y I, Chang S K, Lee Y D, Choi E-J, Han P-L. Molecular cloning of multiple splice variants of JIP-1 preferentially expressed in brain. J Neurochem. 1999;72:1335–1343. doi: 10.1046/j.1471-4159.1999.721335.x. [DOI] [PubMed] [Google Scholar]
- 14.Kuan C-Y, Yang D D, Roy D R S, Davis R J, Rakic P, Flavell R A. The Jnk1 and Jnk2 protein kinases are required for regional specific apoptosis during early brain development. Neuron. 1999;22:667–676. doi: 10.1016/s0896-6273(00)80727-8. [DOI] [PubMed] [Google Scholar]
- 15.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 16.Mooser V, Maillard A, Bonny C, Steinmann M, Shaw P, Yarnall D P, Burns D K, Schorderet D F, Nicod P, Waeber G. Genomic organization, fine-mapping, and expression of the human islet-brain 1 (IB1)/c-Jun-amino-terminal kinase interacting protein-1 (JIP-1) gene. Genomics. 1999;55:202–208. doi: 10.1006/geno.1998.5641. [DOI] [PubMed] [Google Scholar]
- 17.Nishina H, Vaz C, Billia P, Nghiem M, Sasaki T, la Pompa J L, Furlonger K, Paige C, Hui C, Fischer K D, Kishimoto H, Iwatsubo T, Katada T, Woodgett J R, Penninger J M. Defective liver formation and liver cell apoptosis in mice lacking the stress signaling kinase SEK1/MKK4. Development. 1999;126:505–516. doi: 10.1242/dev.126.3.505. [DOI] [PubMed] [Google Scholar]
- 18.Pryciak P M, Huntress F A. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riesgo-Escovar J R, Hafen E. Drosophila Jun kinase regulates expression of decapentaplegic via the ETS-domain protein Aop and the AP-1 transcription factor DJun during dorsal closure. Genes Dev. 1997;11:1717–1727. doi: 10.1101/gad.11.13.1717. [DOI] [PubMed] [Google Scholar]
- 20.Riesgo-Escovar J R, Jenni M, Fritz A, Hafen E. The Drosophila Jun-N-terminal kinase is required for cell morphogenesis but not for DJun-dependent cell fate specification in the eye. Genes Dev. 1996;10:2759–2768. doi: 10.1101/gad.10.21.2759. [DOI] [PubMed] [Google Scholar]
- 21.Rincon M, Whitmarsh A, Yang D D, Weiss L, Derijard B, Jayaraj P, Davis R J, Flavell R A. The JNK pathway regulates the in vivo deletion of immature CD4+ CD8+ thymocytes. J Exp Med. 1998;188:1817–1830. doi: 10.1084/jem.188.10.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabapathy K, Hu Y, Kallunki T, Schreiber M, David J P, Jochum W, Wagner E F, Karin M. JNK2 is required for efficient T-cell activation and apoptosis but not for normal lymphocyte development. Curr Biol. 1999;9:116–125. doi: 10.1016/s0960-9822(99)80065-7. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez I, Hughes R T, Mayer B J, Yee K, Woodgett J R, Avruch J, Kyriakis J M, Zon L I. Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature. 1994;372:794–798. doi: 10.1038/372794a0. [DOI] [PubMed] [Google Scholar]
- 24.Sluss H K, Barrett T, Dérijard B, Davis R J. Signal transduction by tumor necrosis factor mediated by JNK protein kinases. Mol Cell Biol. 1994;14:8376–8384. doi: 10.1128/mcb.14.12.8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sluss H K, Davis R J. Embryonic morphogenesis signaling pathway mediated by JNK targets the transcription factor JUN and the TGF-beta homologue decapentaplegic. J Cell Biochem. 1997;67:1–12. [PubMed] [Google Scholar]
- 26.Sluss H K, Han Z, Barrett T, Davis R J, Ip Y T. A JNK signal transduction pathway that mediates morphogenesis and an immune response in Drosophila. Genes Dev. 1996;10:2745–2758. doi: 10.1101/gad.10.21.2745. [DOI] [PubMed] [Google Scholar]
- 27.Su Y C, Han J, Xu S, Cobb M, Skolnik E Y. NIK is a new Ste20-related kinase that binds NCK and MEKK1 and activates the SAPK/JNK cascade via a conserved regulatory domain. EMBO J. 1997;16:1279–1290. doi: 10.1093/emboj/16.6.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitmarsh A J, Cavanagh J, Tournier C, Yasuda J, Davis R J. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 29.Whitmarsh A J, Davis R J. Structural organization of MAP kinase signaling modules in yeast and mammals. Trends Biochem Sci. 1998;23:481–485. doi: 10.1016/s0968-0004(98)01309-7. [DOI] [PubMed] [Google Scholar]
- 30.Xia Y, Wu Z, Su B, Murray B, Karin M. JNKK1 organizes a MAP kinase module through specific and sequential interactions with upstream and downstream components mediated by its amino-terminal extension. Genes Dev. 1998;12:3369–3381. doi: 10.1101/gad.12.21.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Opposing effects of ERK and JNK-p38 MAP kinases on apoptosis. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 32.Xu S, Cobb M H. MEKK1 binds directly to the c-Jun N-terminal kinases/stress-activated protein kinases. J Biol Chem. 1997;272:32056–32060. doi: 10.1074/jbc.272.51.32056. [DOI] [PubMed] [Google Scholar]
- 33.Yablonski D, Marbach I, Levitzki A. Dimerization of Ste5, a mitogen-activated protein kinase cascade scaffold protein, is required for signal transduction. Proc Natl Acad Sci USA. 1996;93:13864–13869. doi: 10.1073/pnas.93.24.13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang D, Tournier C, Wysk M, Lu H-T, Xu J, Davis R J, Flavell R A. Targeted disruption of the MKK4 gene causes embryonic death, inhibition of c-Jun NH2-terminal kinase activation and defects in AP-1 transcriptional activity. Proc Natl Acad Sci USA. 1997;94:3004–3009. doi: 10.1073/pnas.94.7.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang D D, Conze D, Whitmarsh A J, Barrett T, Davis R J, Rincon M, Flavell R A. Differentiation of CD4+ T cells to Th1 cells requires MAP kinase JNK2. Immunity. 1998;9:575–585. doi: 10.1016/s1074-7613(00)80640-8. [DOI] [PubMed] [Google Scholar]
- 36.Yang D D, Kuan C-Y, Whitmarsh A J, Rincon M, Zheng T S, Davis R J, Rakic P, Flavell R A. Absence of excitotoxicity-induced apoptosis in the hippocampus of mice lacking the JNK3 gene. Nature. 1997;389:865–870. doi: 10.1038/39899. [DOI] [PubMed] [Google Scholar]