Caveolae were originally identified as flask-shaped invaginations of the plasma membrane in endothelial and epithelial cells (14). Prior to the development of biochemical methods for their purification, caveolae were thought to principally mediate the transcellular movement of molecules (101, 145). Recently, the development of novel purification procedures has greatly expanded our knowledge regarding the putative functions of caveolae in vivo. In this review, we seek to update the working definition of caveolae, describe the functional roles of the caveolin gene family, and summarize the evidence that supports a role for caveolae as mediators of a number of cellular signaling processes.
OVERVIEW: CAVEOLAE AND CAVEOLA-RELATED DOMAINS ARE LIQUID-ORDERED MICRODOMAINS
Although caveolae were classically defined as plasma membrane invaginations with a characteristic diameter of ∼50 to 100 nm, this morphological description is inadequate. Caveolae can be invaginated, flat within the plane of the plasma membrane, or detached vesicles. In addition, caveolae can fuse to form grape-like structures (132) and tubules (116) with sizes significantly larger than 100 nm. Morphologically, they are abundant in endothelia, muscle cell types, adipocytes, and lung epithelial cells (34, 112). Recent investigations have also revealed that caveola-like structures are present within the nervous system (15, 50, 71).
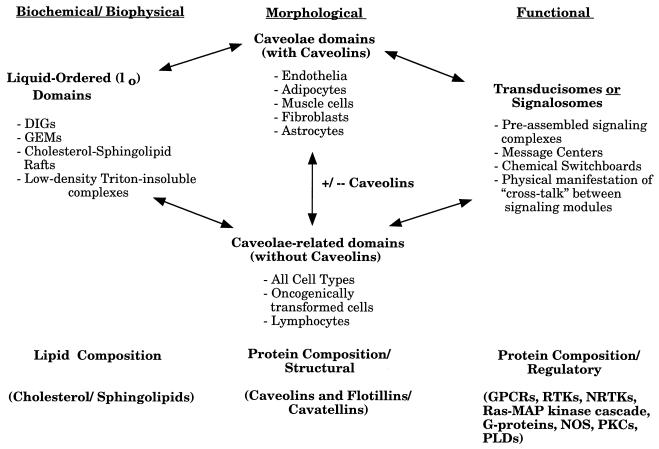
Caveolae have a unique lipid composition. They are mainly composed of cholesterol and sphingolipids. In contrast, noncaveolar regions of the plasma membrane are composed mainly of phospholipids. Cholesterol and sphingolipids can form a liquid-ordered (lo) phase, which is resistant to detergent solubilization (13). These detergent-resistant liquid-ordered domains purified from mammalian cells and tissues are currently referred to as detergent-insoluble glycolipid-rich membranes, cholesterol-sphingolipid rafts, glycolipid-enriched membranes, detergent-resistant membranes, caveolin-enriched membranes, low-density Triton-insoluble domains, caveola-like domains, and caveola-related domains. Here, we will refer to liquid-ordered domains that contain caveolins as caveolae and liquid-ordered domains lacking caveolins as caveola-related domains (Fig. 1). In addition, experiments with liposomes in vitro have provided evidence that cholesterol and sphingolipids alone can form liquid-ordered lipid domains which are resistant to detergent solubilization (13). The idea that caveolae and caveola-related domains are liquid-ordered membranous structures is not new and has been proposed by other investigators as well (1, 12, 13, 120, 142). For a more complete definition of liquid-ordered domains, see the work of Brown and London (12, 13). Furthermore, by using multiple independent approaches, several laboratories have now provided evidence that these microdomains exist in living cells in vivo (12, 13, 46, 65, 72, 120).
FIG. 1.
Caveolae and caveola-related domains. A diagram summarizing the various biochemical, morphological, and functional features of these plasma membrane-associated microdomains is shown. See text for details. DIGs, detergent-insoluble glycolipid-rich membranes; GEMs, glycolipid-enriched membranes; GPCRs, G-protein-coupled receptors; RTKs, receptor tyrosine kinases; NRTKs, nonreceptor tyrosine kinases; PLDs, phospholipase D.
Caveolins are the defining protein components of caveolae. Interestingly, caveolins bind cholesterol directly. In addition, cholesterol binding may stabilize the formation of caveolin homo-oligomeric complexes. Since liquid-ordered domains are dramatically enriched in cholesterol, caveolins may be attracted or partition into liquid-ordered domains through a direct interaction with cholesterol. Thus, caveolin homo-oligomers may interact with each other within liquid-ordered domains to drive the formation of the invaginated flask-shaped membrane domains that we see as caveolae. Since liquid-ordered domains can also form within the Golgi apparatus, the partitioning of caveolins into these liquid-ordered domains may begin to occur at the level of the Golgi apparatus, initiating the biogenesis of caveolae. Thus, both caveolae and caveola-related liquid-ordered domains may be transported to the cell surface as vesicular organelles. In conclusion, caveola-related domains or rafts can exist independently of caveolae, but they must exist prior to formation of caveolae for proper insertion of caveolin into membranes. In this regard, in cells that express caveolins, these structures would function as precursors to caveolae (12, 65, 112).
Caveolae and caveola-related domains are enriched in molecules that play pivotal roles in intracellular signal transduction. These molecules include G-protein-coupled receptors, heterotrimeric G proteins, receptor tyrosine kinases, components of the Ras–mitogen-activated protein (MAP) kinase pathway, Src family tyrosine kinases, protein kinase C’s (PKCs), and nitric oxide synthase (NOS). As a consequence, caveolae and caveola-related domains function as preassembled signaling complexes, message centers, or chemical switchboards for integrating signal transduction. As such, these liquid-ordered structures may represent a physical manifestation of cross talk between distinct signaling modules. However, it must be cautioned that the localization of signaling molecules to caveolae may not be absolute (for possible exceptions, see references 69 and 173). This may also reflect the dynamic nature of these structures.
This type of organization is best illustrated by signal transduction initiated by cholera toxin. The B subunit of cholera toxin, a bacterial toxin, first binds to GM1, a glycosphingolipid that is highly concentrated in caveolae and caveola-related domains. Next, the A subunit of cholera toxin pierces the membrane and ADP-ribosylates the heterotrimeric G protein, GαS, that is concentrated on the cytoplasmic surface of caveolae. Chemically modified GαS now activates adenylyl cyclase and produces cyclic AMP. Efficient cholera toxin-mediated adenylyl cyclase activation is thus made possible by concentrating not only GM1 (cholera toxin receptor) but also GS (signal transducer) and adenylyl cyclase (effector) within caveolae and caveola-related domains. Thus, long before we knew of the existence of liquid-ordered domains, bacteria developed specific toxins that are tailor-made to take advantage of the organized capacity of liquid-ordered domains for signal transduction. In addition, since cholera toxin is internalized via caveolae, the actions and processing of cholera toxin nicely illustrate the dual role that caveolae play in both signal transduction and vesicular transport processes. Thus, these plasma membrane domains may be thought of as signalosomes or transducisomes, i.e., organelles specialized for signal transduction (Fig. 1).
PURIFICATION OF CAVEOLAR MEMBRANES
The purification of caveolar membranes has been recently reviewed (1, 90, 91, 112, 139). Each biochemical method for the purification of caveolae generates a membrane fraction that is highly enriched in caveolin (128), and most generate a fraction enriched in cholesterol, sphingomyelin, and the ganglioside GM1 (11, 136, 151). In general, caveolae purified by density centrifugation have similar protein compositions. These methodologies have been devised by utilizing the biochemical characteristics that caveolae are (i) resistant to detergent solubilization and (ii) have light buoyant density, which is commonly seen in all liquid-ordered domains.
LIPID CONSTITUENTS OF CAVEOLAE
The precise lipid composition of caveolae is not known. However, caveolae are enriched in a variety of specific lipids (Table 1). The principal structural lipids of caveolae are cholesterol and sphingolipids (sphingomyelin and glycosphingolipids). Depending on the cell type, caveolae reportedly contain ∼4 to ∼30% of plasma membrane cholesterol and up to ∼95% of cellular sphingomyelin, indicating that these lipids are highly enriched within caveolar microdomains of the plasma membrane. Brown and Rose (11) isolated detergent-resistant membrane domains from MDCK cells with a number of properties of caveolae. These vesicles contain 30 mol% cholesterol, 14 mol% sphingomyelin, and 19 mol% phosphatidylethanolamine. The precise lipid composition of caveolae is likely to be different among distinct cell types (Table 1). Altering caveolar lipid composition may be a means of regulating caveolar function. Caveolae are also enriched in a number of signaling lipids in addition to sphingomyelin (Table 1).
TABLE 1.
Caveolar lipid composition
| Lipid | % of lipid in caveolaea | Source (reference) |
|---|---|---|
| Cholesterol | 26 | MDCK cells (11) |
| 7 | Fibroblasts (149) | |
| Sphingomyelin | 96 | MDCK cells (11) |
| 50–70 | Fibroblasts (95) | |
| Gangliosides | 67 | MDCK cells (11) |
| Phosphatidylethanolamine | 6 | MDCK cells (11) |
| Phosphatidylserine | 10 | MDCK cells (11) |
| Phosphatidylcholine | 5 | MDCK cells (11) |
| Phosphatidylinositol | 5 | MDCK cells (11) |
| Phosphatidylinositol P2 | 50 | A431 (117) |
| Ceramide | 50 | Fibroblasts (95) |
| Diacyl-glycerol | 50 | Fibroblasts (95) |
Note that the percentage of component in caveolae is relative to the whole-cell lipid content.
The compartmentalized production of lipid mediators of signal transduction has been reported to occur within caveolae (18, 95, 117). Nerve growth factor (NGF) induces ceramide production by binding to the p75 NGF receptor within caveolae in oligodendrocytes, where ceramide may cause cell apoptosis by activating stress-activated protein kinase or Jun kinase (7, 18, 180). The substrate for ceramide production is sphingomyelin, which is a major lipid component of caveolae and caveola-related domains. The presence of p75 NGF receptors within caveolae is further substantiated by evidence that the p75 NGF receptor forms a physical complex with caveolin-1 in heterologous expression systems (7). The relative amount of caveolin-1 expression may also determine the direction of NGF signaling involving either p75 NGF or TrkA, since caveolin-1 expression enhances p75 NGF receptor-mediated ceramide production and, in turn, attenuates TrkA receptor-mediated neuronal differentiation in PC12 cells (6).
INTERNALIZATION AND VESICULATION OF PLASMALEMMAL CAVEOLAE
Vesicles that are the same size as caveolae have been identified within the cytosol. The molecular machinery for caveolar internalization and vesiculation has been studied extensively by using GM1 as a marker. GM1 gangliosides are enriched within purified detergent-resistant caveolar membranes (11, 30, 42, 80, 115, 136) and can bind and concentrate the B subunit of cholera toxin (137, 163). Utilizing the cholera toxin-GM1 interaction as a caveolar marker, Parton et al. (115) convincingly showed that the toxin is internalized into the cell’s interior via a caveola-mediated process largely distinct from that of clathrin-coated pits.
In addition, it has been demonstrated that this distinct caveola-based endocytic pathway differs from clathrin-mediated endocytosis in several ways. First, it is believed that ligand internalization by caveolae is significantly slower by approximately two- to fourfold (45, 163). Second, the protein phosphatase inhibitor okadaic acid stimulates caveola-mediated endocytosis but inhibits the formation of clathrin-coated vesicles (115). Third, the sterol binding agent filipin has little or no effect on clathrin-mediated endocytosis, yet inhibits the internalization of caveolae (138). Finally, activators of PKC, which do not inhibit clathrin-mediated endocytosis, prevent the formation of caveolar invaginations (150, 152).
From the wealth of observations described above, it appeared likely that additional proteins associated with caveolae could sustain the liberation of these plasma membrane invaginations into discrete vesicles in a regulated manner. A likely candidate for this scission process is the large GTPase dynamin that has been implicated in the formation of clathrin-coated vesicles. Dynamin was first identified from mammalian brain based on its ability to bind microtubules in a nucleotide-dependent manner (144). The GTPase activity of dynamin can be stimulated in vitro through interaction with effector molecules at its proline-rich C-terminal domain. Insights into dynamin function emerged by demonstrating substantial identity with the Drosophila shibire gene product (19, 167) which, at the restrictive temperature, is deficient at an early step in endocytosis, mainly, the ability to form coated vesicles at the plasma membrane (76, 79). In support of these studies, transient overexpression of dominant-negative GTP binding mutants of dynamin blocked clathrin-mediated endocytosis (24, 25, 68, 168). Dynamin has been localized to clathrin-coated pits at the plasma membrane in cultured cells (25) and to the necks of membrane invaginations and clathrin-coated pits in an isolated synaptosomal preparation (160) most recently. For a recent review on dynamin, see reference 99.
From the original observations on dynamin function, it was attractive to predict that in addition to clathrin-mediated endocytosis, this mechano-enzyme may participate in multiple endocytic processes such as caveolar internalization and fluid phase uptake. In contrast to this model, one study had demonstrated that cultured cells, expressing a mutant neuronal dynamin form, were unable to perform clathrin-mediated endocytosis while other endocytic processes were actually increased (24). From this single criterion, the authors concluded that dynamin is specifically required for clathrin-dependent endocytosis (82).
Subsequent to these observations, two distinct laboratories accumulated independent results implicating dynamin in the scission of plasma membrane-attached caveolae. Initially, Schnitzer and colleagues (137) convincingly demonstrated that fission of caveolae from the plasma membrane in a cell-free assay was stimulated through the addition of GTP but prevented by the nonhydrolyzable analog guanosine 5′-O-(3-thiotriphosphate). Subsequent to these observations, Henley and McNiven (67) tested the function of dynamin in vivo by microinjecting specific inhibitory antibodies into cultured murine hepatocytes. Using a functional assay and ultrastructural analysis, they found that cells injected with antidynamin antibodies did not internalize fluorescent conjugates of transferrin and accumulated long plasmalemmal invaginations with attached clathrin-coated pits. In addition to these structures, a marked accumulation of numerous plasmalemmal specializations resembling caveolae was observed. These plasmalemmal vesicles were distinct from clathrin-coated pits in that they lacked bristle-like coats, were ∼65 to 75 nm in diameter, and had a characteristic omega or flask shape.
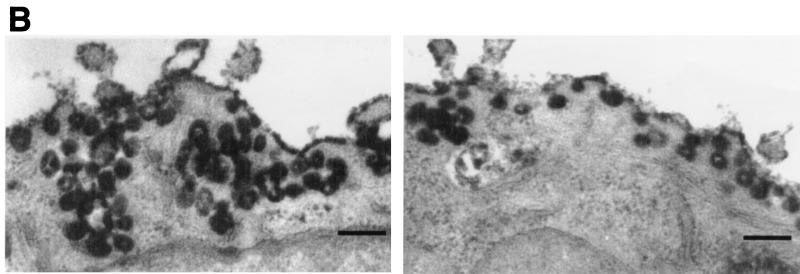
These initial findings suggested that caveolae may accumulate at the plasma membrane through the inhibition of either GTP hydrolysis or dynamin function and provided an incentive for both laboratories to pursue a direct study on the participation of dynamin in the scission of caveolae (66, 110). Concomitantly, these groups conducted functional studies to determine if caveola-mediated internalization of cholera toxin B subunit could be inhibited in intact or permeabilized cells by the addition of antidynamin antibodies. As for previous studies on caveolar internalization, cholera toxin B subunit was employed as a marker. Control experiments demonstrated that toxin uptake was largely independent of clathrin-mediated endocytosis in cultured epithelial cells, since depletion of cytoplasmic potassium ions did not inhibit the accumulation of the labeled toxin within perinuclear compartments but did block the internalization of fluorophore-labeled transferrin. When permeabilized or intact cells were incubated or microinjected with irrelevant antibodies and then incubated with fluorescent or horseradish peroxidase-conjugated toxin and viewed by fluorescence or electron microscopy, it was found that the toxin was readily internalized by control cells to a perinuclear compartment with little toxin labeling detected at the plasma membrane. Instead, toxin was concentrated within cytoplasmic organelles, including endocytic vesicles and, notably, elements of the rough endoplasmic reticulum (ER) and the nuclear envelope. In contrast, cells exposed to antidynamin antibodies did not transport toxin to intracellular compartments but instead concentrated toxin within numerous complex caveolar invaginations that had accumulated at the cell surface.
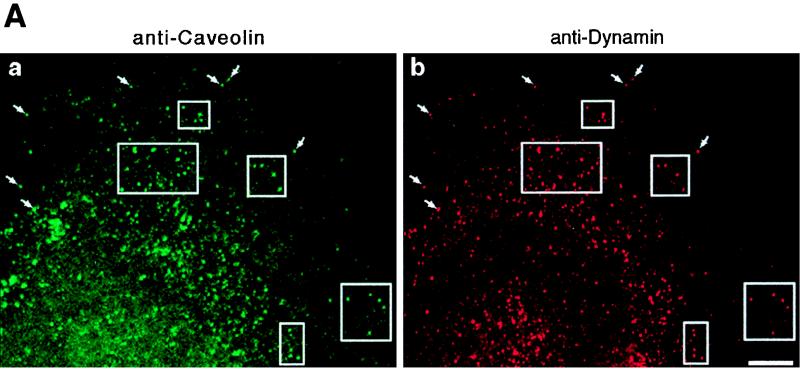
To support these functional observations, both laboratories tested for colocalization between dynamin and caveolae by using morphological and biochemical methods. Pan-dynamin antibodies coupled to magnetic beads were used to immunoisolate caveolar membranes from a hepatocyte postnuclear membrane fraction of cultured cells. By immunoblot analysis, most of the caveolin detected in the starting fraction was associated with the beads, while very little could be detected in the remaining nonbound fraction. To provide morphological support for this association, double-label immunofluorescence microscopy was used to show a significant colocalization of dynamin and caveolin. By electron microscopy, double immunogold labeling of ultrathin cryosections also showed a significant colocalization of these two proteins on plasmalemmal caveolae in lung endothelial cells. Thus, functional assays in combination with fluorescence microscopy and ultrastructural and biochemical analyses from two independent laboratories provided strong evidence that dynamin II participates in the scission of caveolae from the plasma membrane in addition to a role in clathrin-mediated endocytosis (Fig. 2).
FIG. 2.
Dynamin localizes to caveolae in cultured epithelial cells. (A) Fluorescence micrographs representing laser scanning confocal microscopy of cultured hepatocytes that were double-labeled with a monoclonal anti-caveolin-1 antibody as a marker for caveolae (a) and a polyclonal antibody to dynamin to label the endogenous dynamin (b). A significant number of vesicular structures are labeled with both antibodies (arrows and outlined areas), indicating colocalization of dynamin and caveolin-1. Bar, 8.0 μm. (B) Horseradish peroxidase-cholera toxin B is sequestered within caveolae in dynamin-inhibited cells. Electron micrographs showing hepatocytes that were injected with an inhibitory dynamin antibody and incubated with peroxidase-conjugated cholera toxin are shown. Most of the toxin, represented by the peroxidase reaction product, has not been internalized but instead resides on the plasma membrane grape-like caveolar clusters. Bars, 0.2 μm.
The observations described above raise several interesting questions that need to be addressed. These include defining the specifics of the dynamin-caveola interactions. Does dynamin bind directly or indirectly to caveolin? If this is a direct binding, what regions of the two proteins interact with each other? Is this binding regulated by the cell? It is attractive to predict that okadaic acid, which, as described above, stimulates caveolar internalization, may do so through an activation of dynamin function. Finally, which dynamin forms may interact with caveolae? Do only specific dynamin proteins interact with caveolae? Three distinct dynamin genes have been identified in mammals and include dynamin I (Dyn1), which appears to be expressed exclusively in neurons (109); dynamin II (Dyn2), found in all tissues (20, 158); and dynamin III (Dyn3), which is restricted to the testis, brain, lung, and muscle tissues (16). Each dynamin gene encodes four or more alternatively spliced isoforms (122, 166). Thus, while some epithelial tissues appear to express only the four different Dyn2 spliced variants, other tissues such as muscle, brain, lung, or testis may express over 20 different dynamin forms. It is interesting to note that some of these tissues, such as lung and muscle, which express the most dynamin forms, are also extremely rich in caveolae. Whether this coenrichment is of biological importance remains to be tested.
CAVEOLAE AND POTOCYTOSIS
One of the most recently proposed functions for caveolae is potocytosis (2). Potocytosis is a mechanism for the uptake of small molecules or solutes independent of an endocytic process. A molecule binds to a receptor in a flat or open caveola. The caveola then invaginates and may transiently form a sealed compartment independent of the extracellular space but still contiguous with the plasma membrane. The formation of a sealed microenvironment then facilitates the uptake of the molecules across the plasma membrane. The invaginated caveola then flattens or opens, and the cycle is repeated. Although potocytosis has only been clearly demonstrated for the uptake of folic acid (2), the concept is entirely compatible with the selective uptake of cholesterol esters from high-density lipoprotein (HDL) particles. HDL receptors are clustered in caveolae (see below) (3). Caveolae that are depleted of cholesterol are flat within the plasma membrane and maximally accessible to extracellular material (124). Invaginated or closed caveolae are highly enriched in cholesterol and are inaccessible to extracellular material.
THE CAVEOLIN GENE FAMILY
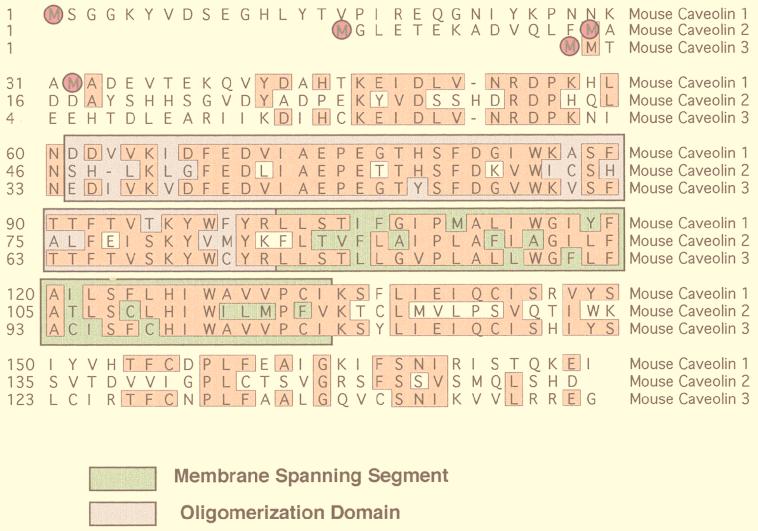
Caveolins are a family of 21- to 25-kDa integral membrane proteins that have been implicated in a variety of cellular functions (34, 112) (Fig. 3). Currently, three caveolin genes are known to exist. Caveolin-1 and caveolin-2 are ubiquitously expressed (130, 132–134), while the expression of caveolin-3 is muscle specific (116, 156, 161). In addition, recent evidence indicates that caveolin-1 and -2 are also present in neuronal and glial cell populations (15, 50) and caveolin-3 is present within astrocytes (71). Caveolin-1 undergoes alternate initiation during translation to generate two isoforms of caveolin-1, caveolin-1α and caveolin-1β (134).
FIG. 3.
The caveolin gene family. An alignment of the protein sequences of murine caveolin-1, -2, and -3 is shown. Identical residues are boxed and highlighted. Note that caveolin-1 and -3 are most closely related, while caveolin-2 is divergent. Translation initiation sites are circled. In addition, the positions of the membrane-spanning segment (green) and the oligomerization domain (purple) are indicated.
Caveolins are thought to associate with membranes via a central 33-amino-acid hydrophobic domain (81). This allows both the N- and C-terminal domains to remain entirely cytosolic. Recently, a cytosolic pool of caveolin-1 has also been identified (165). In addition to localization of caveolin-1 within plasma membrane caveolae (>90% under normal steady-state conditions) (123), a small but significant amount of caveolin-1 has been localized to intracellular compartments, such as the ER (105, 149) and the trans-Golgi network (80). This subcellular distribution of caveolin-1 is thought to reflect the dynamic trafficking of caveolins or caveolae between these intracellular compartments and the plasma membrane. Thus far, three important related functional roles have been described for caveolins: (i) the principle structural protein components of caveolae membranes; (ii) shuttle proteins in the biosynthetic trafficking of cholesterol from the ER to the plasma membrane, and (iii) scaffolding proteins to organize and inactivate signaling molecules that are concentrated on the cytoplasmic surface of caveolar membranes.
Caveolin-1 was first identified by Glenney and co-workers as a major v-Src substrate in Rous sarcoma virus-transformed chicken embryo fibroblasts (58, 60, 61). Thus, caveolin-1 was first discovered in the context of tyrosine phosphorylation, signal transduction, and cell transformation. Only later was caveolin-1 implicated as a component of the caveolar coat by Rothberg et al. (123). Caveolin-1 has since been shown to induce the formation of caveola-sized vesicles (88) and morphologically invaginated caveolae (33, 48). Caveolin-1 forms a homo-oligomeric complex in vitro and in vivo (105, 157), and caveolins 1 and 2 can also form stable hetero-oligomeric complexes with each other (129, 130). Additionally, evidence suggests that these caveolin homo- and hetero-oligomers may represent the functional assembly units of the striated coat associated with the cytoplasmic face of caveolae (123, 126). This may provide a cytoplasmic scaffold onto which signaling molecules may assemble (126). Caveolins are thought to facilitate the formation of invaginated caveolae through their interactions with cholesterol.
The first evidence supporting a role for caveolin-1 in cholesterol trafficking was the discovery that caveolin-1 can functionally bind cholesterol (89, 107). Later, recombinant expression of caveolin-1 was shown to facilitate the transport of newly synthesized cholesterol from the ER to the plasma membrane, where cholesterol rapidly diffuses out of caveolae (153). Caveolin-1 appears to mediate the intracellular movement of cholesterol as a part of an immunophilin complex that includes HSP-56, cyclophilin 40, and cyclophilin A (165). The relative contribution of other caveolins (i.e., caveolin-2 and caveolin-3) to the cellular trafficking of cholesterol has yet to be determined.
Caveolins have been functionally implicated in a wide variety of signal transduction processes (Table 2). In general, caveolin-1 and caveolin-3 repress the enzymatic activity of a wide variety of signal transducing molecules, while caveolin-2 has little or no effect. One notable exception is the insulin receptor tyrosine kinase; caveolin-1 and -3 augment the ability of the insulin receptor to phosphorylate one of its major substrates, IRS-1 (178).
TABLE 2.
Caveolin-interacting signaling molecules
| Caveolin-interacting proteins | Reference(s) |
|---|---|
| GαS | 85 |
| G-protein-coupled receptor kinase | 17 |
| Adenylyl cyclase | 162 |
| Gαi1 | 52 |
| Gαi2 | 85 |
| Go | 85 |
| Ha-Ras | 155 |
| c-Src/Fyn | 84 |
| EGF receptor | 22 |
| cNeu | 32 |
| PDGF receptor | 177 |
| eNOS | 38, 55 |
| nNOS | 169 |
| p75 NGF receptor | 6 |
| TrkA | 6 |
| PKCα | 22, 111 |
| Insulin receptor | 178 |
| Phospholipase D1 | 23, 77 |
| Fyn | 172, 176 |
| Integrins | 172, 176 |
| MEK/ERK | 31 |
| PKA | 118 |
THE CAVEOLIN SCAFFOLDING DOMAIN, SIGNAL INTEGRATION, AND ONCOGENESIS
It is thought that caveolin family members function as scaffolding proteins (126) to organize and concentrate specific lipids (cholesterol and glycosphingolipids) and lipid-modified signaling molecules (Src-like kinases, H-Ras, endothelial NOS (eNOS), and G proteins [84, 85, 87, 155]) within caveolae. In support of this idea, caveolin-1 binding can functionally suppress the GTPase activity of heterotrimeric G proteins and inhibit the kinase activity of Src family tyrosine kinases through a common caveolin domain, termed the caveolin scaffolding domain (155).
Since the identification of the caveolin scaffolding domain and caveolin-binding sequence motifs, these observations have been extended to other caveolin-interacting proteins. Functional caveolin-binding motifs have been deduced in both tyrosine and serine/threonine kinases, as well as eNOS. In all cases examined, the caveolin-binding motif is located within the enzymatically active catalytic domain of a given signaling molecule. For example, in the case of tyrosine and serine/threonine kinases, a kinase domain consists of 11 conserved subdomains (I to XI) (22, 31, 111). The caveolin-binding motif is located within conserved kinase subdomain number IX, suggesting that caveolin could function as a general kinase inhibitor (112). This hypothesis has been substantiated by the observation that the caveolin scaffolding domain inhibits Src family tyrosine kinases (c-Src/Fyn), extracellular growth factor R (EGF-R), Neu, PKC, and protein kinase A (PKA) with similar potencies (see reference 118 and references cited within).
Garcia-Cardena et al. have performed site-directed mutagenesis to modify the predicted caveolin-binding motif (from FSAAPFSGW to ASAAPASGA) within eNOS (55). It is known from in vitro studies that aromatic residues (W, F, or Y) are required for the proper recognition of the caveolin-binding motif (21). In their work, they show that mutation of the caveolin-binding motif within eNOS blocks the ability of caveolin-1 to inhibit eNOS activity in vivo (55). These findings provide the first demonstration that a caveolin-binding motif is relevant and functional in vivo.
The direct interaction of caveolin with signaling molecules leads to their inactivation (112). Since many of these signaling molecules can cause cellular transformation when constitutively activated, it is reasonable to speculate that caveolin itself may possess transformation suppressor activity. Consistent with this hypothesis, both caveolae and caveolins are most abundantly expressed in terminally differentiated cells: adipocytes, endothelial cells, and muscle cells (10). In addition, caveolin-1 mRNA and protein expression are lost or reduced during cell transformation by activated oncogene products such as v-Abl and H-Ras (G12V); caveolae are absent from these cell lines (78). The potential transformation suppressor activity of caveolin-1 has recently been evaluated by using an inducible expression system to upregulate caveolin-1 expression in oncogenically transformed cells. Induction of caveolin-1 expression in v-Abl- and H-Ras (G12V)-transformed NIH 3T3 cells abrogated the anchorage-independent growth of these cells in soft agar and resulted in the de novo formation of caveolae (33). Thus, downregulation of caveolin-1 expression and caveolar organelles may be critical for maintaining the transformed phenotype.
Caveolae have also been implicated in signaling through the p42/44 MAP kinase (MAPK) pathway. Morphological studies have directly shown that extracellular signal-regulated kinase 1/2 (ERK-1/2) is concentrated in plasma membrane caveolae in vivo by using immunoelectron microscopy (96). Evidence suggesting that other components of the p42/44 MAPK cascade are localized within caveolae has been presented. These components include receptor tyrosine kinases (EGF-R, platelet-derived growth factor R, Ins-R) (93, 97, 102, 151), H-Ras (102, 155), Raf kinase (102), 14-3-3 proteins (97), ERK (97), Shc (97), Grb-2 (97), mSos-1 (97), and Nck (97).
Recently, we examined the functional role of caveolins in regulating signaling along the MAPK cascade (31). Coexpression with caveolin-1 dramatically inhibited signaling from EGF-R, Raf, MEK-1, and ERK-2 to the nucleus in vivo (31). By using a variety of caveolin-1 deletion mutants, this in vivo inhibitory activity was mapped to caveolin-1 residues 32 to 95. In addition, peptides derived from this region of caveolin-1 (i.e., the caveolin scaffolding domain) also inhibited the in vitro kinase activity of purified MEK-1 and ERK-2 (31). Thus, caveolin-1 can inhibit signal transduction from the p42/44 MAPK cascade both in vitro and in vivo by acting as a natural endogenous inhibitor of both MEK and ERK.
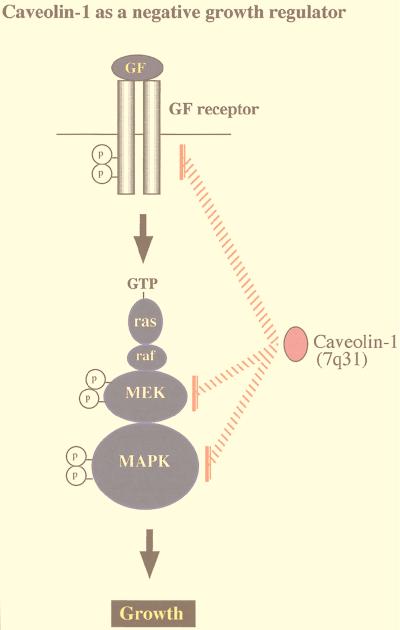
Conversely, a prediction of these findings would be that downregulation of caveolin-1 should lead to constitutive activation of the p42/44 MAPK pathway. Since constitutive activation of the p42/44 MAPK cascade is sufficient to mediate cell transformation, another predicted consequence of caveolin-1 downregulation would be cell transformation. This hypothesis was directly tested by employing an antisense approach to derive stable NIH 3T3 cell lines that express dramatically reduced levels of caveolin-1 but contain normal amounts of caveolin-2 (49). Antisense-mediated reductions in caveolin-1 protein expression were sufficient to drive oncogenic transformation and constitutively activate the p42/44 MAPK cascade (49). In normal NIH 3T3 cells, caveolin-1 expression levels were downregulated in rapidly dividing cells and dramatically upregulated at confluency. Thus, upregulation of caveolin-1 expression levels may be important in mediating normal contact inhibition and in negatively regulating the activation state of the p42/44 MAPK cascade (Fig. 4). This is the first demonstration that a loss of caveolin-1 expression is sufficient to mediate cellular transformation. In accordance with these current findings, the caveolin-1 gene is localized to a suspected tumor suppressor locus in humans (7q31.1/D7S522) that is deleted in many forms of cancer (Fig. 5) (34–36).
FIG. 4.
Caveolins negatively regulate signaling along the p42/44 MAPK cascade. Caveolae have been implicated in signaling through the p42/44 MAPK pathway. Caveolin-1 can inhibit signal transduction from the p42/44 MAPK cascade both in vitro and in vivo by acting as a natural endogenous inhibitor of EGF-R, MEK, and ERK (31). Conversely, when NIH 3T3 cells are used, antisense-mediated reductions in caveolin-1 protein expression are sufficient to constitutively activate the p42/44 MAPK cascade and drive oncogenic transformation (49). In normal NIH 3T3 cells, caveolin-1 expression levels are downregulated in rapidly dividing cells and dramatically upregulated at confluency. Thus, upregulation of caveolin-1 expression levels may be important in mediating normal contact inhibition and in negatively regulating the activation state of the p42/44 MAPK cascade. In accordance with these findings, the caveolin-1 gene is localized to a suspected tumor suppressor locus that is deleted in many forms of human cancer (7q31.1/D7S 522 locus) (34, 36).
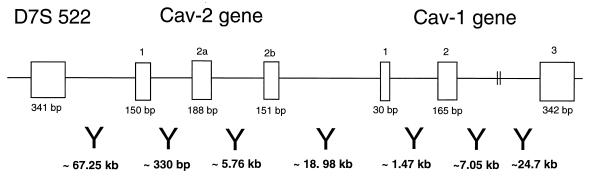
FIG. 5.
Detailed organization of the human caveolin-1 and -2 locus and its relationship to D7S 522, a microsatellite marker that is deleted in many forms of human cancer. The sizes of the exons and the distances between them are indicated. Note that the marker D7S 522 is located ∼67 kb upstream of the caveolin-2 gene and that the caveolin-2 gene is located ∼19 kb upstream of the caveolin-1 gene.
In one case, Huang and colleagues failed to detect an interaction between caveolin-1 and G proteins (69). These authors indicated that this may have been due to technical difficulties in maintaining the solubility of their caveolin-1 fusion protein (69). However, this appears to represent an isolated negative result, since many other groups have now shown that the caveolin scaffolding domain specifically recognizes different classes of signaling molecules (17, 112). In addition, after the paper by Huang et al. was published, other investigators were able to independently confirm a specific interaction between caveolin-1 and G-protein alpha subunits see Fig. 2C in reference (154).
SIGNALING PATHWAYS THAT REGULATE CAVEOLIN-1 GENE EXPRESSION
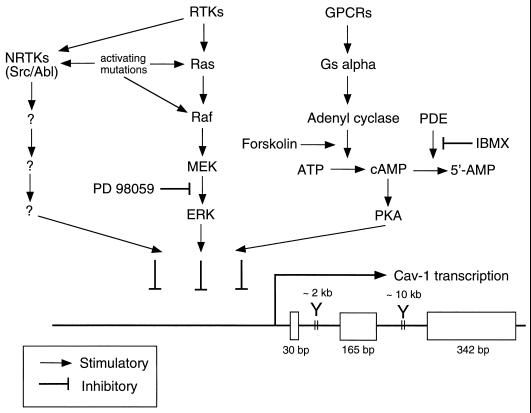
The signaling pathways that govern caveolin-1 gene expression have recently been examined (37). Evidence suggesting that caveolin-1 gene expression is directly regulated by activation of the p42/44 MAPK cascade has been presented. For example, treatment of Ras (H-, K-, and N-Ras)- or v-Raf-transformed NIH 3T3 cells with a well-characterized MEK inhibitor (PD 98059) restores the expression of the caveolin-1 protein (37). In contrast, treatment of v-Src- and v-Abl-transformed NIH 3T3 cells with PD 98059 has no effect on caveolin-1 expression (37). Thus, there are at least two pathways for downregulating caveolin-1 expression, one that is p42/44 MAPK dependent and others that are p42/44 MAPK independent and depend on the activation of nonreceptor tyrosine kinases (such as Src or Abl).
The activity of caveolin-1 promoter constructs was also evaluated by using expression in H-Ras (G12V)-transformed NIH 3T3 cells (37). Caveolin-1 promoter activity was upregulated approximately fivefold through inhibition of the p42/44 MAPK cascade with PD 98059. In addition, transient transfection of CHO cells with ERK-2 dramatically downregulates caveolin-1 promoter activity. To determine if different complexes form on the caveolin-1 promoter in normal and Ras (G12V)-transformed NIH 3T3 cells, electromobility shift assays were performed. The data suggest that the caveolin-1 promoter from −156 to −561 is differentially bound by transcription factors in normal and H-Ras (G12V)-transformed cells.
The effects of the PKA pathway on caveolin-1 gene expression have also been evaluated (37). Activation of the PKA pathway by pharmacological agents or by overexpression of the PKA catalytic subunit is sufficient to downregulate caveolin-1 promoter activity and caveolin-1 protein expression. Thus, there may be three independent signaling pathways (Ras-p42/44 MAPK, nonreceptor tyrosine kinases, and PKA) that can transcriptionally downregulate caveolin-1 gene expression. These results are summarized schematically in Fig. 6.
FIG. 6.
Schematic diagram summarizing the signaling pathways that down-regulate caveolin-1 gene expression via transcriptional control. The points of control that are affected by oncogenic activating mutations (Ras, Raf, Src, and Abl) and pharmacological agents (PD 98059, forskolin, and IBMX) are as indicated. RTKs, receptor tyrosine kinases; cAMP, cyclic AMP; NRTKs, nonreceptor tyrosine kinases; GPCRs, G-protein-coupled receptors; PDE, cyclic nucleotide phosphodiesterase. The overall structure of the murine caveolin-1 gene is as we described previously (34–36).
Interestingly, the caveolin-1 protein product can act as an inhibitor of many elements of these signaling cascades, such as Src (84), EGF-R (22), c-Neu (32), vascular endothelial growth factor receptor (94), Raf (31), MEK (31, 49), ERK (31, 49), G-protein alpha subunits (86, 133, 161), adenyl cyclase (162), and PKA (118), by the recognition of a common caveolin-binding motif (21, 22), and many of these proteins have been localized to caveolar membranes (34, 112). These observations suggest a general pattern of negative reciprocal regulation. In this sense, caveolin-1 is both upstream and downstream of these signaling pathways.
CAVEOLIN-MEDIATED REGULATION OF ENOS ACTIVATION
eNOS is targeted by acylation to caveolae, where it interacts with caveolin-1 (56, 143). Mutations of eNOS that prevent caveolar targeting result in less agonist-stimulated NO release (75, 92, 141), suggesting that movement into caveolae is important for temporal and spatial activation of the enzyme. While in caveolae, both caveolin-1 and caveolin-3 appear to bind eNOS through two scaffolding domains present in caveolin-1 and -3, but not caveolin-2 (38, 55), and in the case of caveolin-1 can interact with the tyrosine-phosphorylated form of eNOS (54). In vitro, these caveolin scaffolding domain peptides and native caveolin-1 inhibit eNOS activity by interfering with electron flux from the reductase to the oxygenase domain of the enzyme (55, 57, 73). Interestingly, this inhibition can be antagonized by the addition of calmodulin in a calcium-dependent manner (73, 100). This discovery has led to the proposal of a caveolin-calmodulin regulatory cycle of eNOS inactivation and activation.
More recent studies have demonstrated that calmodulin promotes the dissociation of caveolin from eNOS but does not influence the caveolar localization of the enzyme. Interestingly, the membrane-associated, depalmitoylated form of eNOS interacts more weakly with caveolin, suggesting that movement within domains of the caveolae and/or plasma membrane may be regulated by the cycle of palmitoylation and depalmitoylation (41). The significance of the inhibitory caveolin scaffolding domain in regulating eNOS activity in vivo has also been highlighted by a recent report which has shown that caveolin and caveolin peptides can inhibit eNOS activity in cultured ventricular myocytes (40). In addition, the inhibition of eNOS altered the spontaneous contraction frequency of these cells, suggesting that the effects of caveolin on eNOS activity are relevant in vivo. Negative functional regulation of eNOS by caveolin is biologically important, since (i) hypercholesterol-induced impairment of NO production is likely induced by the hypercholesterol-mediated upregulation of caveolin-1 expression (39) and (ii) vascular flow- and pressure-dependent eNOS activation is associated with eNOS dissociation from caveolin-1 and association with calmodulin (121). Neuronal NOS (nNOS) is also functionally regulated directly by caveolin-3 binding (55, 169).
PHYSICAL AND FUNCTIONAL INTERACTIONS BETWEEN CAVEOLIN-1 AND -2
Caveolin-1 and -2 are coexpressed, and they form a hetero-oligomeric complex in many cell types, with particularly high levels in adipocytes, endothelial cells, and fibroblasts. These caveolin hetero-oligomers are thought to represent the functional assembly units that drive caveolar formation in vivo.
Recently, we investigated the mechanism by which caveolin-1 and -2 form hetero-oligomers (26). This reciprocal interaction was reconstituted in vivo and in vitro by using a variety of complementary approaches, including the generation of glutathione S-transferase (GST) fusion proteins and synthetic peptides. Taken together, our results indicate that the membrane-spanning domains of both caveolin-1 and -2 play a critical role in mediating their ability to interact with each other (26). This is the first demonstration that these unusual membrane-spanning regions found in the caveolin family play a specific role in protein-protein interactions. However, the functional significance of the interaction between caveolin-1 and -2 remains unknown.
Interestingly, the expression of caveolin-1 appears to be required for the transport of caveolin-2 from the Golgi complex to the plasma membrane (114). A human erythroleukemic cell line, K562, that expresses caveolin-2 but fails to express detectable levels of caveolin-1 was identified. Expression of caveolin-1 in K562 cells reconstituted the de novo formation of caveolae in these cells. In addition, recombinant expression of caveolin-1 allowed caveolin-2 to form high-molecular-mass oligomers that were targeted to caveolae (114). In striking contrast, in the absence of caveolin-1 expression, caveolin-2 formed low-molecular-mass oligomers that were retained at the level of the Golgi complex. In addition, expression of caveolin-1 in K562 cells dramatically upregulated the expression of endogenous caveolin-2 (114). Thus, the formation of a hetero-oligomeric complex between caveolin-1 and -2 stabilizes the caveolin-2 protein product and allows caveolin-2 to be transported from the Golgi complex to the plasma membrane. Similar results were obtained by Mora et al. (106).
CAVEOLIN-1 MEMBRANE TOPOLOGY AND MEMBRANE ATTACHMENT
Several independent lines of evidence indicate that caveolin-1 is an integral membrane protein with a cytoplasmic N-terminal domain and a cytoplasmic C-terminal domain. First, wild-type caveolin-1 contains a central 33-amino-acid hydrophobic stretch of amino acids that encodes a putative membrane-spanning segment, and biochemically caveolin-1 is carbonate inextractable (5, 30, 127), behaving as expected of an integral membrane protein. Second, both the N- and C-terminal domains face the cytoplasm, as shown by using specific N- and C-terminus-directed antipeptide antibodies and by employing N- and C-terminal epitope tags (27, 30, 104, 134). In addition, both the N- and C-terminal domains undergo cytoplasmic modifications: the extreme N terminus is tyrosine phosphorylated by v-Src on caveolin-1 (Y14) and several cysteines within the C-terminal domain are S-acylated by palmitoylation (27, 59, 87). Third, caveolin-1 is inaccessible to cell surface biotinylation, indicating that no detectable portion of the protein is extracellular (125). Fourth, consistent with the idea that caveolin-1 is an integral membrane protein, caveolin-1 in native plasma membranes can be photoaffinity labeled with a variety of lipids, including a glycosphingolipid (GM1) and a free fatty acid (47, 164); additional evidence that caveolin-1 is a cholesterol binding protein has been presented (89, 107, 153). Taken together, these data have been used to construct a model of caveolin-1 in which the putative membrane-spanning domain forms a hairpin loop within the membrane, allowing both the N- and C-terminal domains of caveolin-1 to remain entirely cytoplasmic (the hairpin loop model). However, no experimental evidence demonstrating that this central hydrophobic stretch (residues 102 to 134) is required for membrane attachment of caveolin-1 has yet been presented.
Recently, a series of caveolin-1 deletion mutants were created to examine whether the membrane-spanning segment is required for membrane attachment of caveolin-1 in vivo (135). One mutant, Cav-1(1-101), contained only the cytoplasmic N-terminal domain and lacked the membrane-spanning domain and the C-terminal domain. Interestingly, Cav-1(1-101) still behaved as an integral membrane protein after transient expression in 293T cells (135). However, it lacked any known signals for lipid modification. Consistent with its behavior as an integral membrane protein, Cav-1(1-101) was targeted to the plasma membrane, as demonstrated by immunofluorescence microscopy. In striking contrast, another deletion mutant, Cav-1(1-81) behaved as a soluble protein. These results implicate caveolin-1 residues 82 to 101 (also known as the caveolin scaffolding domain) in membrane attachment. In accordance with the postulated role of the caveolin-1 scaffolding domain as an inhibitor of signal transduction, Cav-1(1-101) retained the ability to functionally inhibit signaling along the p42/44 MAPK cascade, while Cav-1(1-81) was completely ineffective (135).
To rule out the possibility that membrane attachment mediated by the caveolin scaffolding domain was indirect (i.e., mediated by its interaction with another protein), the membrane binding of caveolin-1 was reconstituted in vitro. When purified GST–caveolin-1 fusion proteins and reconstituted lipid vesicles were used, the caveolin-1 scaffolding domain appeared to be both necessary and sufficient for membrane attachment in vitro (135).
In addition, GST–Cav-1(135-178), encoding the entire caveolin-1 C-terminal domain, also showed membrane binding activity (135). Interestingly, the C-terminal domain of caveolin-1 is lipid modified by palmitoylation on cysteine residues in vivo (27); however, bacterially expressed GST fusion proteins are not lipid modified. This suggests that independent of lipid modification by palmitoylation, the C-terminal domain of caveolin-1 may play a role in its membrane attachment. However, the putative membrane-spanning domain (residues 102 to 134) did not show any physical association with membranes in this in vitro system (135). Taken together, these results provide strong evidence that the caveolin scaffolding domain and the C-terminal domain contribute to the membrane attachment of caveolin-1.
CAVEOLIN AND THE PATHOGENESIS OF NEURODEGENERATIVE DISEASES
Senile plaques are the characteristic brain pathology of Alzheimer’s disease (AD) (140). The β-amyloid peptide, a major protein component of the senile plaque, is generated from its precursor protein, termed amyloid precursor protein (APP), by enzymatic digestion involving β- and γ-secretase activities. Another enzymatic cleavage activity, α-secretase, cuts APP in the middle of the amyloid region (63, 147, 148), thereby precluding amyloid production. In nonneuronal cells, α-secretase-mediated APP shedding plays a major role in APP metabolism, whereas in neurons, β- and γ-secretase-mediated processing is the main metabolic pathway for APP processing.
Recent findings provide clear evidence that APP is enriched within caveolae, where caveolin-1 provides a direct means for APP to be concentrated in this microdomain of the plasma membrane. Caveolin-1 expression also regulates APP processing by promoting α-secretase activity in cultured kidney epithelial cell lines (70). It has been recently reported that caveola-related domains or detergent-insoluble glycolipid-rich membranes isolated from whole brain contain not only APP (9) but also presenilins 1 and 2 together with the Aβ amyloid peptide (83). Therefore, it has been suggested that brain caveola-related domains are the site where amyloid biogenesis or transport takes place.
In support of this notion, cholesterol depletion, which leads to the loss of caveola-related domain integrity, efficiently inhibits Aβ-amyloid peptide secretion in cultured hippocampal neurons (146). The role of caveolin in the brain pathology of AD has recently been clarified (108). In brain tissue sections from authentic AD patients and an established transgenic mouse model of AD, dramatic upregulation of caveolin-3 immunoreactivity in astroglial cells surrounding senile plaques has been observed. In addition, caveolin-3 physically interacts and biochemically colocalizes with APP both in vivo and in vitro. Recombinant overexpression of caveolin-3 in cultured cells stimulated β-secretase-mediated processing of APP. Immunoreactivities of APP and presenilins were concomitantly increased in caveolin-3-positive astrocytes. Since the presenilins also form a physical complex with caveolin-3, caveolin-3 may provide a common platform for APP and the presenilins to coassociate in astrocytes. In AD, augmented expression of caveolin-3 and presenilins in reactive astrocytes may alter APP processing, leading to the overproduction of its toxic amyloid metabolites.
Recent evidence also suggests the involvement of caveola-related domains in prion protein processing (74). Efficient formation of the scrapie isoform of prion protein [PrP(Sc)] requires targeting of PrP(Sc) by glycosylphosphatidyl inositol (GPI) anchors to caveola-related domains. Redirecting the cellular isoform of prion protein [PrP(C)] to clathrin-coated pits by creating chimeric PrP molecules with four different COOH-terminal transmembrane domains prevented the formation of PrP(Sc). PrP(Sc) was formed only from GPI-anchored PrP(C), which was targeted to caveola-related domains, and not from transmembrane PrP(C), which was targeted to clathrin-coated pits. Therefore, PrP(Sc) formation is restricted to a specific subcellular compartment and is likely to involve auxiliary macromolecules found within caveola-related domains in the brain.
CAVEOLIN-3, A MUSCLE-SPECIFIC CAVEOLIN FAMILY MEMBER
Caveolin-3 is localized to the sarcolemma (muscle cell plasma membrane) and coincides with the distribution of another muscle-specific plasma membrane marker protein, dystrophin (156). Caveolin-3 cofractionates with cytoplasmic signaling molecules (G proteins and Src-like kinases) and members of the dystrophin complex (dystrophin, alpha-sarcoglycan, and beta-dystroglycan).
It was also shown that caveolin-3 forms a physical complex with dystrophin, suggesting an essential role for caveolin-3 in muscle biology (156). Caveolin-3 protein expression is dramatically induced during the differentiation of C2C12 skeletal myoblasts in culture (156). In addition, caveolin-3 is transiently associated with T-tubules during muscle development and may be involved in the early development of the T-tubule system (116).
Caveolin-3 also interacts with phosphofructokinase-M in muscle. This interaction is (i) highly regulated by the extracellular concentration of glucose and (ii) can be stabilized by a number of relevant intracellular metabolites, such as fructose 1,6-bisphosphate and fructose 2,6-bisphosphate, which are known allosteric activators of phosphofructokinase-M (131). Glucose-dependent plasma membrane recruitment of activated phosphofructokinase-M by caveolin-3 may play a key role in regulating energy metabolism in skeletal muscle fibers (131).
Genetic evidence highlights the importance of caveolin-3 in muscle functioning (98, 103, 119, 159). A new form of autosomal dominant limb girdle muscular dystrophy (LGMD) associated with a severe deficiency of caveolin-3 in muscle fibers has been reported (103). The human caveolin-3 gene was mapped to chromosome 3p25 (159), and two mutations in the gene were identified, a missense mutation in the membrane-spanning region (P→L) and a microdeletion in the scaffolding domain (ΔTFT) (103). These mutations may interfere with caveolin-3 oligomerization and possibly disrupt caveolar formation at the muscle cell plasma membrane. However, the molecular mechanisms by which these two mutations cause muscular dystrophy remain unknown.
Recently, the phenotypic behavior of these caveolin-3 mutations was investigated by using heterologous expression (53). Wild-type caveolin-3 or caveolin-3 mutants were transiently expressed in NIH 3T3 cells. LGMD-1C mutations led to formation of unstable high-molecular-mass aggregates of caveolin-3 that were retained within the Golgi complex and not targeted to the plasma membrane (53). Consistent with its autosomal dominant form of genetic transmission (103), LGMD-1C mutants of caveolin-3 behaved in a dominant-negative fashion, causing the retention of wild-type caveolin-3 at the level of the Golgi complex (53). These data provide a molecular explanation for why caveolin-3 levels are downregulated in patients with LGMD-1C.
CAVEOLINS AND CHOLESTEROL HOMEOSTASIS
Depleting cells of cholesterol reduces caveolin expression and the number of morphologically invaginated caveolae at the cell surface (64, 124). In contrast, elevating free cellular cholesterol by addition of low-density lipoproteins to the culture media increases the expression of caveolin-1 (43). Cholesterol appears to modulate caveolin-1 expression through a steroid regulatory binding element (SRE) present in the caveolin-1 promoter and SRE binding protein 1 (SREBP-1) (8). This observation is of particular interest since SREBP-1 is released from the ER and activates transcription of the low-density lipoprotein receptor when cells are depleted of cholesterol (179). In contrast, the SRE present in the caveolin-1 promoter suppresses the expression of caveolin-1 (8).
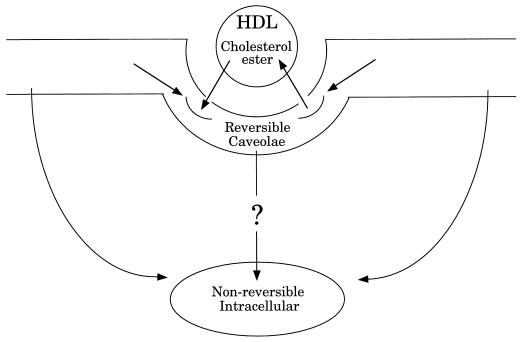
The relationship between cholesterol and caveolar function has been the subject of a number of studies. In contrast, the role of caveolae in regulating cellular cholesterol levels has only recently gained attention. Studies by Oram and Yokoyama indicate that cholesterol is constantly exiting the cell (113), while caveolin-1 appears to constitutively transport cholesterol to the cell surface (153). Caveolae have been shown to efflux cellular cholesterol to HDLs, the principal acceptor of cellular cholesterol in the reverse cholesterol transport pathway (44). More recently, the putative HDL receptor, SR-BI, has been localized to caveolar membranes (3, 174). Further, SR-BI mediates the uptake of HDL cholesterol esters into caveolae (62). Within the plasma membrane, caveolae constitute a reversible pool of cholesterol esters (Fig. 7). Caveolar cholesterol esters are then trafficked to nonreversible pools within both the plasma membrane and an intracellular membrane compartment, presumably the ER. The mechanism for the internalization of caveolar cholesterol esters remains unknown. Therefore, caveolae appear to be a principal site for cholesterol exchange between cells and HDL, where they are involved in both efflux of free cholesterol and uptake of esterified cholesterol. Collectively, these data suggest that cellular cholesterol levels and caveolae are intimately related and may display reciprocal regulation.
FIG. 7.
Caveolae mediate SR-BI-dependent uptake of cholesterol esters from HDL. HDL cholesterol esters are initially associated with plasma membrane caveolae. While in caveolae, cholesterol esters may either efflux back to HDL or translocate to nonreversible pools within the plasma membrane or an intracellular membrane compartment. The mechanism for internalization of caveolar cholesterol esters remains unknown.
CAVEOLINS AND CELL ADHESION
Integrins are heterodimeric adhesion molecules implicated in many biological processes characterized by cell movement, including inflammation, tissue remodeling, growth, and tumorigenesis. Integrin-dependent cellular adhesiveness depends not only on specific integrin classes expressed by cells but also on ligand-induced integrin redistribution and signal transduction leading to cytoskeletal reorganization. Signaling through the integrin family of adhesion receptors requires their association with cytoplasmic elements capable of signal transduction because integrins lack intrinsic signaling capacity. Possible signal mediators of integrins are integrin-associated protein, CD98, and a GPI-linked nonintegrin receptor, the urokinase receptor (uPAR1).
Recent evidence suggests that caveolin-1 is also an important component of integrin-mediated signaling. Caveolin-1 was found to coimmunoprecipitate with integrins and uPAR in uPAR-transfected 293 cells (175). Wary et al. (171) also observed that caveolin-1 coprecipitates with integrins in A431 cells. A fraction of cellular caveolin-1 associates with integrins and promotes Fyn-dependent Shc phosphorylation in response to integrin ligation, leading to MAPK activation and cellular growth (172, 176). Thus, caveolins may contribute to the general regulation of integrin functioning.
CAVEOLINS, P75 NEUROTROPHIN RECEPTOR, AND APOPTOSIS
In neural cell lines and glial cells, neurotrophin binding to p75 neurotrophin stimulates the generation of ceramide (18, 28, 29) and enhancement of Jun kinase activity (18). NGF-mediated activation of p75 neurotrophin receptor signaling in cultured oligodendrocytes leads to apoptosis (18).
The role of p75 neurotrophin receptor in cellular apoptosis has been critically reevaluated by using genetically engineered mice which lack either p75 neurotrophin receptor or neurotrophin brain-derived neurotrophic factor or both (4). The results provided firm evidence that the p75 neurotrophin receptor mediates apoptosis in vivo.
Recent studies have revealed that the p75 neurotrophin receptor is located within caveolae and caveola-related domains and forms a physical complex with caveolin-1 in a heterologous expression system (6, 7). Upon ligand binding, it is likely that p75 neurotrophin receptor utilizes caveolar sphingomyelin as a substrate to generate ceramide and induces apoptosis. Interleukin 1 receptor seems to adopt a similar apoptotic signaling via caveolae (95). Thus, it is possible that ligand-mediated enzymatic processing of caveolar sphingomyelin is a common mechanism whereby death receptors induce cell apoptosis.
FUTURE DIRECTIONS: CAVATELLINS
We have identified another family of integral membrane proteins that may contribute to the structural organization of caveolae membranes (5, 51). Microsequence analysis of purified caveolin-rich membrane domains isolated from lung tissue revealed a novel ∼45-kDa component of caveola membranes, termed flotillin-1 (5, 51). Molecular cloning of flotillin and analysis of the cDNA for this protein have provided new avenues by which to explore the structure and function of caveola organelles. Interestingly, flotillin is a close homologue of ESA (epidermal surface antigen), and together they define a new flotillin family of caveolar integral membrane proteins (flotillin-1 and flotillin-2/ESA) (5, 51).
Recently, we examined the cell type- and tissue-specific expression of the flotillin gene family (170). Taken together, our data suggest that the expression levels of flotillin-1 and -2 are independently regulated and do not strictly correlate with the known expression patterns of caveolin family members. However, when caveolins and flotillins are coexpressed within the same cell, they form a stable hetero-oligomeric caveolar complex. In support of these observations, we demonstrated that heterologous expression of murine flotillin-1 in Sf21 insect cells using baculovirus-based vectors is sufficient to drive the formation of caveola-like vesicles (170). These results suggest that flotillins may participate functionally in the formation of caveolae or caveola-like vesicles in vivo. As a consequence of these findings, we proposed that the new functional term cavatellins be used (instead of flotillins) to describe this gene family (170). Thus, cavatellins represents new integral membrane protein markers for the slightly larger caveola-related domains (50 to 200 nm) that are observed in certain cell types that fail to express caveolins.
ACKNOWLEDGMENTS
This work is supported by an NIH grant from the National Cancer Institute (CA-80250; to M.P.L.) and grants from the Charles E. Culpeper Foundation (to M.P.L.), G. Harold and Leila Y. Mathers Charitable Foundation (to M.P.L. and P.E.S.), and the Sidney Kimmel Foundation for Cancer Research (to M.P.L.). P.E.S. is supported by a pilot grant from the AECOM DRTC, a grant from Pfizer Corp., and a grant from the American Diabetes Association. E.J.S. and G.A.G. are supported by grants from the NIH (HL-58475 and HL-62844; to E.J.S.) and the American Heart Association (KY-97-GS-28 and 9950038N; to E.J.S.). T.O. is supported by an NIH FIRST Award (MH-56036). M.A.M. is supported by an NIH grant (DK-44560). J.A.E. is supported by NIH Medical Scientist Training Program grant T32-GM-07288. W.C.S. is an Established Investigator of the American Heart Association and is supported by grants from the National Institutes of Health (HL-51948 and HL-50974) and a grant-in-aid from the American Heart Association.
REFERENCES
- 1.Anderson R G W. The caveolae membrane system. Annu Rev Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- 2.Anderson R G W, Kamen B A, Rothberg K G, Lacey S W. Potocytosis: sequestration and transport of small molecules by caveolae. Science. 1992;255:410–411. doi: 10.1126/science.1310359. [DOI] [PubMed] [Google Scholar]
- 3.Babitt J, Trigatti B, Rigotti A, Smart E J, Anderson R G, Xu S, Krieger M. Murine SR-BI, a high density lipoprotein receptor that mediates selective lipid uptake, is N-glycosylated and fatty acylated and colocalizes with plasma membrane caveolae. J Biol Chem. 1997;272:13242–13249. doi: 10.1074/jbc.272.20.13242. [DOI] [PubMed] [Google Scholar]
- 4.Bamji S X, Majdan M, Pozniak C D, Belliveau D J, Aloyz R, Kohn J, Causing C G, Miller F D. The p75 neurotrophin receptor mediates neuronal apoptosis and is essential for naturally occurring sympathetic neuron death. J Cell Biol. 1998;140:911–923. doi: 10.1083/jcb.140.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bickel P E, Scherer P E, Schnitzer J E, Oh P, Lisanti M P, Lodish H F. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J Biol Chem. 1997;272:13793–13802. doi: 10.1074/jbc.272.21.13793. [DOI] [PubMed] [Google Scholar]
- 6.Bilderback T R, Gazula V R, Lisanti M P, Dobrowsky R T. Caveolin interacts with Trk A and p75(NTR) and regulates neurotrophin signaling pathways. J Biol Chem. 1999;274:257–263. doi: 10.1074/jbc.274.1.257. [DOI] [PubMed] [Google Scholar]
- 7.Bilderback T R, Grigsby R J, Dobrowsky R T. Association of p75(NTR) with caveolin and localization of neurotrophin-induced sphingomyelin hydrolysis to caveolae. J Biol Chem. 1997;272:10922–10927. doi: 10.1074/jbc.272.16.10922. [DOI] [PubMed] [Google Scholar]
- 8.Bist A, Fielding P E, Fielding C J. Two sterol regulatory element-like sequences mediate up-regulation of caveolin gene transcription in response to low density lipoprotein free cholesterol. Proc Natl Acad Sci USA. 1997;94:10693–10698. doi: 10.1073/pnas.94.20.10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouillot C, Prochiantz A, Rougon G, Allinquant B. Axonal amyloid precursor protein expressed by neurons in vitro is present in a membrane fraction with caveolae-like properties. J Biol Chem. 1996;271:7640–7644. doi: 10.1074/jbc.271.13.7640. [DOI] [PubMed] [Google Scholar]
- 10.Bretscher M S, Whytock S. Membrane-associated vesicles in fibroblasts. J Ultrastruct Res. 1977;61:215–217. doi: 10.1016/s0022-5320(77)80088-9. [DOI] [PubMed] [Google Scholar]
- 11.Brown D, Rose J K. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 12.Brown D A, London E. Structure and origin of ordered lipid domains in biological membranes. J Membr Biol. 1998;164:103–114. doi: 10.1007/s002329900397. [DOI] [PubMed] [Google Scholar]
- 13.Brown D A, London E. Structure of detergent-resistant membrane domains: does phase separation occur in biological membranes? Biochem Biophys Res Commun. 1997;240:1–7. doi: 10.1006/bbrc.1997.7575. [DOI] [PubMed] [Google Scholar]
- 14.Bruns R R, Palade G E. Studies on blood capillaries. I. General organization of blood capillaries in muscle. J Cell Biol. 1968;37:244–276. doi: 10.1083/jcb.37.2.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cameron P L, Ruffin J W, Bollag R, Rasmussen H, Cameron R S. Identification of caveolin and caveolin-related proteins in the brain. J Neurosci. 1997;17:9520–9535. doi: 10.1523/JNEUROSCI.17-24-09520.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao H, Garcia F, McNiven M A. Differential distribution of dynamin isoforms in mammalian cells. Mol Biol Cell. 1998;9:2595–2609. doi: 10.1091/mbc.9.9.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carman C V, Lisanti M P, Benovic J L. Regulation of G protein-coupled receptor kinases by caveolin. J Biol Chem. 1999;274:8858–8864. doi: 10.1074/jbc.274.13.8858. [DOI] [PubMed] [Google Scholar]
- 18.Casaccia-Bonnefil P, Carte R B D, Dobrowsky R T, Chao M V. Death of oligodendrocytes mediated by the interaction of nerve growth factor with its receptor p75. Nature. 1996;383:716–719. doi: 10.1038/383716a0. [DOI] [PubMed] [Google Scholar]
- 19.Chen M S, Obar R A, Schroeder C C, Austin T W, Poodry C A, Wadsworth S C, Vallee R B. Multiple forms of dynamin are encoded by shibire, a Drosophila gene involved in endocytosis. Nature. 1991;351:583–586. doi: 10.1038/351583a0. [DOI] [PubMed] [Google Scholar]
- 20.Cook T A, Urrutia R, McNiven M A. Identification of dynamin 2, an isoform ubiquitously expressed in rat tissues. Proc Natl Acad Sci USA. 1994;91:644–648. doi: 10.1073/pnas.91.2.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Couet J, Li S, Okamoto T, Ikezu T, Lisanti M P. Identification of peptide and protein ligands for the caveolin-scaffolding domain. Implications for the interaction of caveolin with caveolae-associated proteins. J Biol Chem. 1997;272:6525–6533. doi: 10.1074/jbc.272.10.6525. [DOI] [PubMed] [Google Scholar]
- 22.Couet J, Sargiacomo M, Lisanti M P. Interaction of a receptor tyrosine kinase, EGF-R, with caveolins: caveolin-binding negatively regulates tyrosine and serine/threonine kinase activities. J Biol Chem. 1997;272:30429–30438. doi: 10.1074/jbc.272.48.30429. [DOI] [PubMed] [Google Scholar]
- 23.Czarny M, Lavie Y, Fiucci G, Liscovitch M. Localization of phospholipase D in detergent-insoluble, caveolin-rich membrane domains. Modulation by caveolin-1 expression and caveolin-1 82-101. J Biol Chem. 1999;274:2717–2724. doi: 10.1074/jbc.274.5.2717. [DOI] [PubMed] [Google Scholar]
- 24.Damke H, Baba T, van der Bliek A M, Schmid S L. Clathrin-independent pinocytosis is induced in cells overexpressing a temperature-sensitive mutant of dynamin. J Cell Biol. 1995;131:69–80. doi: 10.1083/jcb.131.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Damke H, Baba T, Warnock D E, Schmid S L. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das K, Lewis R Y, Scherer P E, Lisanti M P. The membrane spanning domains of caveolins 1 and 2 mediate the formation of caveolin hetero-oligomers. Implications for the assembly of caveolae membranes in vivo. J Biol Chem. 1999;274:18721–18726. doi: 10.1074/jbc.274.26.18721. [DOI] [PubMed] [Google Scholar]
- 27.Dietzen D J, Hastings W R, Lublin D M. Caveolin is palmitoylated on multiple cysteine residues: palmitoylation is not necessary for localization of caveolin to caveolae. J Biol Chem. 1995;270:6838–6842. doi: 10.1074/jbc.270.12.6838. [DOI] [PubMed] [Google Scholar]
- 28.Dobrowsky R T, Jenkins G M, Hannun Y A. Neurotrophins induce sphingomyelin hydrolysis. Modulation by co-expression of p75NTR with Trk receptors. J Biol Chem. 1995;270:22135–22142. doi: 10.1074/jbc.270.38.22135. [DOI] [PubMed] [Google Scholar]
- 29.Dobrowsky R T, Werner M H, Castellino A M, Chao M V, Hannun Y A. Activation of the sphingomyelin cycle through the low-affinity neurotrophin receptor. Science. 1994;265:1596–1599. doi: 10.1126/science.8079174. [DOI] [PubMed] [Google Scholar]
- 30.Dupree P, Parton R G, Raposo G, Kurzchalia T V, Simons K. Caveolae and sorting of the trans-Golgi network of epithelial cells. EMBO J. 1993;12:1597–1605. doi: 10.1002/j.1460-2075.1993.tb05804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engelman J A, Chu C, Lin A, Jo H, Ikezu T, Okamoto T, Kohtz D S, Lisanti M P. Caveolin-mediated regulation of signaling along the p42/44 MAP kinase cascade in vivo. A role for the caveolin-scaffolding domain. FEBS Lett. 1998;428:205–211. doi: 10.1016/s0014-5793(98)00470-0. [DOI] [PubMed] [Google Scholar]
- 32.Engelman J A, Lee R J, Karnezis A, Bearss D J, Webster M, Siegel P, Muller W J, Windle J J, Pestell R G, Lisanti M P. Reciprocal regulation of neu tyrosine kinase activity and caveolin-1 protein expression in vitro and in vivo. Implications for human breast cancer. J Biol Chem. 1998;273:20448–20455. doi: 10.1074/jbc.273.32.20448. [DOI] [PubMed] [Google Scholar]
- 33.Engelman J A, Wykoff C C, Yasuhara S, Song K S, Okamoto T, Lisanti M P. Recombinant expression of caveolin-1 in oncogenically transformed cells abrogates anchorage-independent growth. J Biol Chem. 1997;272:16374–16381. doi: 10.1074/jbc.272.26.16374. [DOI] [PubMed] [Google Scholar]
- 34.Engelman J A, Zhang X, Galbiati F, Volonte D, Sotgia F, Pestell R G, Minetti C, Scherer P E, Okamoto T, Lisanti M P. Molecular genetics of the caveolin gene family: implications for human cancers, diabetes, Alzheimer disease, and muscular dystrophy. Am J Hum Genet. 1998;63:1578–1587. doi: 10.1086/302172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engelman J A, Zhang X L, Galbiati F, Lisanti M P. Chromosomal localization, genomic organization, and developmental expression of the murine caveolin gene family (Cav-1, -2, and -3). Cav-1 and Cav-2 genes map to a known tumor suppressor locus (6-A2/7q31) FEBS Lett. 1998;429:330–336. doi: 10.1016/s0014-5793(98)00619-x. [DOI] [PubMed] [Google Scholar]
- 36.Engelman J A, Zhang X L, Lisanti M P. Sequence and detailed organization of the human caveolin-1 and -2 genes located near the D7S522 locus (7q31.1). Methylation of a CpG island in the 5′ promoter region of the caveolin-1 gene in human breast cancer cell lines. FEBS Lett. 1999;448:221–230. doi: 10.1016/s0014-5793(99)00365-8. [DOI] [PubMed] [Google Scholar]
- 37.Engelman, J. A., X. L. Zhang, R. G. Pestell, and M. P. Lisanti. p42/44 MAP kinase-dependent and -independent signaling pathways regulate caveolin-1 gene expression. Activation of Ras-MAP kinase and PKA signaling cascades transcriptionally down-regulates caveolin-1 promoter activity. J. Biol. Chem., in press. [DOI] [PubMed]
- 38.Feron O, Belhassen L, Kobzik L, Smith T W, Kelly R A, Michel T. Endothelial nitric oxide synthase targeting to caveolae. J Biol Chem. 1996;271:22810–22814. doi: 10.1074/jbc.271.37.22810. [DOI] [PubMed] [Google Scholar]
- 39.Feron O, Dessy C, Moniotte S, Desager J P, Balligand J L. Hypercholesterolemia decreases nitric oxide production by promoting the interaction of caveolin and endothelial nitric oxide synthase. J Clin Invest. 1999;103:897–905. doi: 10.1172/JCI4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feron O, Dessy C, Opel D J, Arstall M A, Kelly R A, Michel T. Modulation of the endothelial nitric-oxide synthase-caveolin interaction in cardiac myocytes. Implications for the autonomic regulation of heart rate. J Biol Chem. 1998;273:30249–30254. doi: 10.1074/jbc.273.46.30249. [DOI] [PubMed] [Google Scholar]
- 41.Feron O, Michel J B, Sase K, Michel T. Dynamic regulation of endothelial nitric oxide synthase: complementary roles of dual acylation and caveolin interactions. Biochemistry. 1998;37:193–200. doi: 10.1021/bi972307p. [DOI] [PubMed] [Google Scholar]
- 42.Fiedler K, Kobayashi T, Kurzchalia T, Simons K. Glycosphingolipid-enriched, detergent-insoluble complexes in protein sorting in epithelial cells. Biochemistry. 1993;32:6365–6373. doi: 10.1021/bi00076a009. [DOI] [PubMed] [Google Scholar]
- 43.Fielding C, Bist A, Fielding P. Caveolin mRNA levels are up-regulated by free cholesterol and down-regulated by oxysterols in fibroblast monolayers. Proc Natl Acad Sci USA. 1997;94:3753–3758. doi: 10.1073/pnas.94.8.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fielding P E, Fielding C J. Plasma membrane caveolae mediate the efflux of cellular free cholesterol. Biochemistry. 1995;34:14288–14292. doi: 10.1021/bi00044a004. [DOI] [PubMed] [Google Scholar]
- 45.Fishman P H. Internalization and degradation of cholera toxin by cultured cells: relationship to toxin action. J Cell Biol. 1982;93:860–865. doi: 10.1083/jcb.93.3.860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fivaz M, Abrami L, van der Goot F G. Landing on lipid rafts. Trends Cell Biol. 1999;9:212–213. doi: 10.1016/s0962-8924(99)01567-6. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 47.Fra A M, Masserini M, Palestini P, Sonnino S, Simons K. A photo-reactive derivative of ganglioside GM1 specifically cross-links VIP21-caveolin on the cell surface. FEBS Lett. 1995;375:11–14. doi: 10.1016/0014-5793(95)95228-o. [DOI] [PubMed] [Google Scholar]
- 48.Fra A M, Williamson E, Simons K, Parton R G. De novo formation of caveolae in lymphocytes by expression of VIP21-caveolin. Proc Natl Acad Sci USA. 1995;92:8655–8659. doi: 10.1073/pnas.92.19.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Galbiati F, Volonte D, Engelman J A, Watanabe G, Burk R, Pestell R G, Lisanti M P. Targeted downregulation of caveolin-1 is sufficient to drive cell transformation and hyperactivate the p42/44 MAP kinase cascade. EMBO J. 1998;17:6633–6648. doi: 10.1093/emboj/17.22.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Galbiati F, Volonte D, Gil O, Zanazzi G, Salzer J L, Sargiacomo M, Scherer P E, Engelman J A, Schlegel A, Parenti M, Okamoto T, Lisanti M P. Expression of caveolins 1 and 2 in differentiating PC12 cells and dorsal root ganglion cells. Proc Natl Acad Sci USA. 1998;95:10257–10262. doi: 10.1073/pnas.95.17.10257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galbiati F, Volonte D, Goltz J S, Steele Z, Sen J, Jurcsak J, Stein D, Stevens L, Lisanti M P. Identification, sequence and developmental expression of invertebrate flotillins from Drosophila melanogaster. Gene. 1998;210:229–237. doi: 10.1016/s0378-1119(98)00064-x. [DOI] [PubMed] [Google Scholar]
- 52.Galbiati F, Volonte D, Meani D, Milligan G, Lublin D M, Lisanti M P, Parenti M. The dually acylated NH2-terminal domain of gi1alpha is sufficient to target a green fluorescent protein reporter to caveolin-enriched plasma membrane domains. Palmitoylation of caveolin-1 is required for the recognition of dually acylated G-protein alpha subunits in vivo. J Biol Chem. 1999;274:5843–5850. doi: 10.1074/jbc.274.9.5843. [DOI] [PubMed] [Google Scholar]
- 53.Galbiati F, Volonte V, Minetti C, Chu J B, Lisanti M P. Phenotypic behavior of caveolin-3 mutations that cause autosomal dominant limb girdle muscular dystrophy (LGMD-1C). Retention of LGMD-1C caveolin-3 mutants within the Golgi complex. J Biol Chem. 1999;274:25632–25641. doi: 10.1074/jbc.274.36.25632. [DOI] [PubMed] [Google Scholar]
- 54.Garcia-Cardena G, Fan R, Stern D F, Liu J, Sessa W C. Endothelial nitric oxide synthase is regulated by tyrosine phosphorylation and interacts with caveolin-1. J Biol Chem. 1996;271:27237–27240. doi: 10.1074/jbc.271.44.27237. [DOI] [PubMed] [Google Scholar]
- 55.Garcia-Cardena G, Martasek P, Siler-Masters B S, Skidd P M, Couet J C, Li S, Lisanti M P, Sessa W C. Dissecting the interaction between nitric oxide synthase (NOS) and caveolin: functional significance of the NOS caveolin binding domain in vivo. J Biol Chem. 1997;272:25437–25440. doi: 10.1074/jbc.272.41.25437. [DOI] [PubMed] [Google Scholar]
- 56.Garcia-Cardena G, Oh P, Liu J, Schnitzer J E, Sessa W C. Targeting of nitric oxide synthase to endothelial cell caveolae via palmitoylation: implications for nitric oxide signaling. Proc Natl Acad Sci USA. 1996;93:6448–6453. doi: 10.1073/pnas.93.13.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghosh S, Gachhui R, Crooks C, Wu C, Lisanti M P, Stuehr D J. Interaction between caveolin-1 and the reductase domain of endothelial nitric-oxide synthase. Consequences for catalysis. J Biol Chem. 1998;273:22267–22271. doi: 10.1074/jbc.273.35.22267. [DOI] [PubMed] [Google Scholar]
- 58.Glenney J R. The sequence of human caveolin reveals identity with VIP 21, a component of transport vesicles. FEBS Lett. 1992;314:45–48. doi: 10.1016/0014-5793(92)81458-x. [DOI] [PubMed] [Google Scholar]
- 59.Glenney J R. Tyrosine phosphorylation of a 22 kD protein is correlated with transformation with Rous sarcoma virus. J Biol Chem. 1989;264:20163–20166. [PubMed] [Google Scholar]
- 60.Glenney J R, Soppet D. Sequence and expression of caveolin, a protein component of caveolae plasma membrane domains phosphorylated on tyrosine in RSV-transformed fibroblasts. Proc Natl Acad Sci USA. 1992;89:10517–10521. doi: 10.1073/pnas.89.21.10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Glenney J R, Zokas L. Novel tyrosine kinase substrates from Rous sarcoma virus transformed cells are present in the membrane cytoskeleton. J Cell Biol. 1989;108:2401–2408. doi: 10.1083/jcb.108.6.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Graf G A, Connell P M, van der Westhuyzen D R, Smart E J. The class B, type I scavenger receptor promotes the selective uptake of high density lipoprotein cholesterol ethers into caveolae. J Biol Chem. 1999;274:12043–12048. doi: 10.1074/jbc.274.17.12043. [DOI] [PubMed] [Google Scholar]
- 63.Haass C, Koo E H, Mellon A, Hung A Y, Selkoe D J. Targeting of cell-surface beta-amyloid precursor protein to lysosomes: alternative processing into amyloid-bearing fragments. Nature. 1992;357:500–503. doi: 10.1038/357500a0. [DOI] [PubMed] [Google Scholar]
- 64.Hailstones D, Sleer L S, Parton R G, Stanley K K. Regulation of caveolin and caveolae by cholesterol in MDCK cells. J Lipid Res. 1998;39:369–379. [PubMed] [Google Scholar]
- 65.Harder T, Scheiffele P, Verkade P, Simons K. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J Cell Biol. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Henley J R, Krueger E W, Oswald B J, McNiven M A. Dynamin-mediated internalization of caveolae. J Cell Biol. 1998;141:85–99. doi: 10.1083/jcb.141.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Henley J R, McNiven M A. Association of a dynamin-like protein with the Golgi apparatus in mammalian cells. J Cell Biol. 1996;133:761–775. doi: 10.1083/jcb.133.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Herskovits J S, Burgess C C, Obar R A, Vallee R B. Effects of mutant rat dynamin on endocytosis. J Cell Biol. 1993;122:565–578. doi: 10.1083/jcb.122.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang C, Hepler J R, Chen L T, Gilman A G, Anderson R G W, Mumby S M. Organization of G proteins and adenylyl cyclase at the plasma membrane. Mol Biol Cell. 1997;8:2365–2378. doi: 10.1091/mbc.8.12.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ikezu T, Trapp B D, Song K S, Schlegel A, Lisanti M P, Okamoto T. Caveolae, plasma membrane microdomains for alpha-secretase-mediated processing of the amyloid protein precursor. J Biol Chem. 1998;273:10485–10495. doi: 10.1074/jbc.273.17.10485. [DOI] [PubMed] [Google Scholar]
- 71.Ikezu T, Ueda H, Trapp B D, Nishiyama K, Sha J F, Volonte D, Galbiati F, Byrd A L, Bassell G, Serizawa H, Lane W S, Lisanti M P, Okamoto T. Affinity-purification and characterization of caveolins from the brain. Differential expression of caveolin-1, -2, and -3 in brain endothelial and astroglial cell types. Brain Res. 1998;804:177–192. doi: 10.1016/s0006-8993(98)00498-3. [DOI] [PubMed] [Google Scholar]
- 72.Jacobson K, Dietrich C. Looking at lipid rafts? Trends Cell Biol. 1999;9:87–91. doi: 10.1016/s0962-8924(98)01495-0. . (Forum/comment.) [DOI] [PubMed] [Google Scholar]
- 73.Ju H, Zou R, Venema V J, Venema R C. Direct interaction of endothelial nitric-oxide synthase and caveolin-1 inhibits synthase activity. J Biol Chem. 1997;272:18522–18525. doi: 10.1074/jbc.272.30.18522. [DOI] [PubMed] [Google Scholar]
- 74.Kaneko K, Vey M, Scott M, Pilkuhn S, Cohen F E, Prusiner S B. COOH-terminal sequence of the cellular prion protein directs subcellular trafficking and controls conversion into the scrapie isoform. Proc Natl Acad Sci USA. 1997;94:2333–2338. doi: 10.1073/pnas.94.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kantor D B, Lanzrein M, Stary S J, Sandoval G M, Smith W B, Sullivan B M, Davidson N, Schuman E M. A role for endothelial NO synthase in LTP revealed by adenovirus-mediated inhibition and rescue. Science. 1996;274:1744–1748. doi: 10.1126/science.274.5293.1744. [DOI] [PubMed] [Google Scholar]
- 76.Kessell I, Holst B D, Roth T F. Membranous intermediates in endocytosis are labile, as shown in a temperature-sensitive mutant. Proc Natl Acad Sci USA. 1989;86:4968–4972. doi: 10.1073/pnas.86.13.4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim J H, Han J M, Lee S, Kim Y, Lee T G, Park J B, Lee S D, Suh P G, Ryu S H. Phospholipase D1 in caveolae: regulation by protein kinase C alpha and caveolin-1. Biochemistry. 1999;38:3763–3769. doi: 10.1021/bi982478+. [DOI] [PubMed] [Google Scholar]
- 78.Koleske A J, Baltimore D, Lisanti M P. Reduction of caveolin and caveolae in oncogenically transformed cells. Proc Natl Acad Sci USA. 1995;92:1381–1385. doi: 10.1073/pnas.92.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kosaka T, Ikeda K. Reversible blockage of membrane retrieval and endocytosis in the garland cell of the temperature-sensitive mutant of Drosophila melanogaster, shibirets1. J Cell Biol. 1983;97:499–507. doi: 10.1083/jcb.97.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kurzchalia T, Dupree P, Parton R G, Kellner R, Virta H, Lehnert M, Simons K. VIP 21, a 21-kD membrane protein, is an integral component of trans-Golgi-network-derived transport vesicles. J Cell Biol. 1992;118:1003–1014. doi: 10.1083/jcb.118.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kurzchalia T V, Dupree P, Monier S. VIP21-caveolin, a protein of the trans-Golgi network and caveolae. FEBS Lett. 1994;346:88–91. doi: 10.1016/0014-5793(94)00466-8. [DOI] [PubMed] [Google Scholar]
- 82.Lamaze C, Schmid S L. The emergence of clathrin-independent pinocytic pathways. Curr Opin Cell Biol. 1995;7:573–580. doi: 10.1016/0955-0674(95)80015-8. [DOI] [PubMed] [Google Scholar]
- 83.Lee S-J, Liyanage U, Bickel P E, Xia W, Lansbury P T, Kosik K S. A detergent-insoluble membrane compartment contains Abeta in vivo. Nat Med. 1998;4:730–734. doi: 10.1038/nm0698-730. [DOI] [PubMed] [Google Scholar]
- 84.Li S, Couet J, Lisanti M P. Src tyrosine kinases, G alpha subunits and H-Ras share a common membrane-anchored scaffolding protein, caveolin. Caveolin binding negatively regulates the auto-activation of Src tyrosine kinases. J Biol Chem. 1996;271:29182–29190. doi: 10.1074/jbc.271.46.29182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li S, Okamoto T, Chun M, Sargiacomo M, Casanova J E, Hansen S H, Nishimoto I, Lisanti M P. Evidence for a regulated interaction between hetero-trimeric G proteins and caveolin. J Biol Chem. 1995;270:15693–15701. doi: 10.1074/jbc.270.26.15693. [DOI] [PubMed] [Google Scholar]
- 86.Li S, Okamoto T, Chun M, Sargiacomo M, Casanova J E, Hansen S H, Nishimoto I, Lisanti M P. Evidence for a regulated interaction between heterotrimeric G proteins and caveolin. J Biol Chem. 1995;270:15693–15701. doi: 10.1074/jbc.270.26.15693. [DOI] [PubMed] [Google Scholar]
- 87.Li S, Seitz R, Lisanti M P. Phosphorylation of caveolin by Src tyrosine kinases: the alpha-isoform of caveolin is selectively phosphorylated by v-Src in vivo. J Biol Chem. 1996;271:3863–3868. [PubMed] [Google Scholar]
- 88.Li S, Song K S, Koh S S, Kikuchi A, Lisanti M P. Baculovirus-based expression of mammalian caveolin in Sf21 insect cells. A model system for the biochemical and morphological study of caveolae biogenesis. J Biol Chem. 1996;271:28647–28654. doi: 10.1074/jbc.271.45.28647. [DOI] [PubMed] [Google Scholar]
- 89.Li S, Song K S, Lisanti M P. Expression and characterization of recombinant caveolin. Purification by polyhistidine tagging and cholesterol-dependent incorporation into defined lipid membranes. J Biol Chem. 1996;271:568–573. [PubMed] [Google Scholar]
- 90.Lisanti M P, Sargiacomo M, Scherer P E. Purification of caveolae-derived membrane microdomains containing lipid-anchored signaling molecules, such as GPI-anchored proteins, H-Ras, Src-family tyrosine kinases, eNOS and G-protein alpha-, beta-, and gamma-subunits. Methods Mol Biol. 1999;116:51–60. doi: 10.1385/1-59259-264-3:51. [DOI] [PubMed] [Google Scholar]
- 91.Lisanti M P, Tang Z-T, Scherer P, Sargiacomo M. Caveolae purification and GPI-linked protein sorting in polarized epithelia. Methods Enzymol. 1995;250:655–668. doi: 10.1016/0076-6879(95)50103-7. [DOI] [PubMed] [Google Scholar]
- 92.Liu J, Garcia-Cardena G, Sessa W C. Palmitoylation of endothelial nitric oxide synthase is necessary for optimal stimulated release of nitric oxide: implications for caveolae localization. Biochemistry. 1996;35:13277–13281. doi: 10.1021/bi961720e. [DOI] [PubMed] [Google Scholar]
- 93.Liu J, Oh P, Horner T, Rogers R A, Schnitzer J E. Organized endothelial cell surface signal transduction in caveolae. J Biol Chem. 1997;272:7211–7222. doi: 10.1074/jbc.272.11.7211. [DOI] [PubMed] [Google Scholar]
- 94.Liu J, Razani B, Tang S, Terman B I, Ware J A, Lisanti M P. Angiogenesis activators and inhibitors differentially regulate caveolin-1 expression and caveolae formation in vascular endothelial cells. Angiogenesis inhibitors block VEGF-induced down-regulation of caveolin-1. J Biol Chem. 1999;274:15781–15785. doi: 10.1074/jbc.274.22.15781. [DOI] [PubMed] [Google Scholar]
- 95.Liu P, Anderson R G W. Compartmentalized production of ceramide at the cell surface. J Biol Chem. 1995;270:27179–27185. doi: 10.1074/jbc.270.45.27179. [DOI] [PubMed] [Google Scholar]
- 96.Liu P, Ying Y, Anderson R G W. Platelet-derived growth factor activates mitogen-activated protein kinase in isolated caveolae. Proc Natl Acad Sci USA. 1997;94:13666–13670. doi: 10.1073/pnas.94.25.13666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu P, Ying Y, Ko Y G, Anderson R G W. Localization of platelet-derived growth factor-stimulated phosphorylation cascade to caveolae. J Biol Chem. 1996;271:10299–10303. doi: 10.1074/jbc.271.17.10299. [DOI] [PubMed] [Google Scholar]
- 98.McNally E M, de Sa Moreira E, Duggan D J, Bonnemann C G, Lisanti M P, Lidov H G W, Vainzof M, Passos-Bueno M R, Hoffman E P, Zatz M, Kunkel L M. Caveolin-3 in muscular dystrophy. Hum Mol Genet. 1998;7:871–877. doi: 10.1093/hmg/7.5.871. [DOI] [PubMed] [Google Scholar]
- 99.McNiven M A. Dynamin: a molecular motor with pinchase action. Cell. 1998;94:151–154. doi: 10.1016/s0092-8674(00)81414-2. [DOI] [PubMed] [Google Scholar]
- 100.Michel J B, Feron O, Sase K, Prabhakar P, Michel T. Caveolin versus calmodulin. Counterbalancing allosteric modulators of endothelial nitric oxide synthase. J Biol Chem. 1997;272:25907–25912. doi: 10.1074/jbc.272.41.25907. [DOI] [PubMed] [Google Scholar]
- 101.Milici A J, Watrous N E, Stukenbrok H, Palade G E. Transcytosis of albumin in capillary endothelium. J Cell Biol. 1987;105:2603–2612. doi: 10.1083/jcb.105.6.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mineo C, James G L, Smart E J, Anderson R G W. Localization of the EGF-stimulated Ras/Raf-1 interaction to caveolae membrane. J Biol Chem. 1996;271:11930–11935. doi: 10.1074/jbc.271.20.11930. [DOI] [PubMed] [Google Scholar]
- 103.Minetti C, Sotgia F, Bruno C, Scartezzini P, Broda P, Bado M, Masetti E, Mazzocco M, Egeo A, Donati M A, Volonte’ D, Galbiati F, Cordone G, Bricarelli F D, Lisanti M P, Zara F. Mutations in the caveolin-3 gene cause autosomal dominant limb-girdle muscular dystrophy. Nat Genet. 1998;18:365–368. doi: 10.1038/ng0498-365. [DOI] [PubMed] [Google Scholar]
- 104.Monier S, Parton R G, Vogel F, Behlke J, Henske A, Kurzchalia T. VIP21-caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol Biol Cell. 1995;6:911–927. doi: 10.1091/mbc.6.7.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Monier S, Parton R G, Vogel F, Behlke J, Henske A, Kurzchalia T V. VIP21-caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol Biol Cell. 1995;6:911–927. doi: 10.1091/mbc.6.7.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mora R, Bonilha V, Marmostein A, Brown D, Scherer P E, Lisanti M P, Rodriguez-Boulan E. Caveolin-2 localizes to the Golgi complex but redistributes to plasma membrane, caveolae, and rafts when co-expressed with caveolin-1. J Biol Chem. 1999;274:25708–25717. doi: 10.1074/jbc.274.36.25708. [DOI] [PubMed] [Google Scholar]
- 107.Murata M, Peranen J, Schreiner R, Weiland F, Kurzchalia T, Simons K. VIP21/caveolin is a cholesterol-binding protein. Proc Natl Acad Sci USA. 1995;92:10339–10343. doi: 10.1073/pnas.92.22.10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nishiyama K, Trapp B D, Ikezu T, Ransohoff R M, Tomita T, Iwatsubo T, Kanazawa I, Hsiao K K, Lisanti M P, Okamoto T. Caveolin-3 up-regulation activates beta-secretase mediated cleavage of the amyloid precursor protein (APP) in Alzheimer’s disease. J Neurosci. 1999;19:6538–6548. doi: 10.1523/JNEUROSCI.19-15-06538.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Obar R A, Collins C A, Hammarback J A, Shpetner H S, Vallee R B. Molecular cloning of the microtubule-associated mechanochemical enzyme dynamin reveals homology with a new family of GTP-binding proteins. Nature. 1990;347:256–261. doi: 10.1038/347256a0. [DOI] [PubMed] [Google Scholar]
- 110.Oh P, McIntosh D P, Schnitzer J E. Dynamin at the neck of caveolae mediates their budding to form transport vesicles by GTP-driven fission from the plasma membrane of endothelium. J Cell Biol. 1998;141:101–114. doi: 10.1083/jcb.141.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Oka N, Yamamoto M, Schwencke C, Kawabe J, Ebina T, Ohno S, Couet J, Lisanti M P, Ishikawa Y. Caveolin interaction with protein kinase C. Isoenzyme-dependent regulation of kinase activity by the caveolin scaffolding domain peptide. J Biol Chem. 1997;272:33416–33421. doi: 10.1074/jbc.272.52.33416. [DOI] [PubMed] [Google Scholar]
- 112.Okamoto T, Schlegel A, Scherer P E, Lisanti M P. Caveolins, a family of scaffolding proteins for organizing “preassembled signaling complexes” at the plasma membrane. J Biol Chem. 1998;273:5419–5422. doi: 10.1074/jbc.273.10.5419. [DOI] [PubMed] [Google Scholar]
- 113.Oram J F, Yokoyama S. Apolipoprotein-mediated removal of cellular cholesterol and phospholipids. J Lipid Res. 1996;37:2473–2491. [PubMed] [Google Scholar]
- 114.Parolini I, Sargiacomo M, Galbiati F, Rizzo G, Grignani F, Engelman J A, Okamoto T, Ikezu T, Scherer P E, Peschle C, Lisanti M P. Expression of caveolin-1 is required for the transport of caveolin-2 to the plasma membrane. Retention of caveolin-2 at the level of the Golgi complex. J Biol Chem. 1999;274:25718–25725. doi: 10.1074/jbc.274.36.25718. [DOI] [PubMed] [Google Scholar]
- 115.Parton R G, Joggerst B, Simons K. Regulated internalization of caveolae. J Cell Biol. 1994;127:1199–1215. doi: 10.1083/jcb.127.5.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Parton R G, Way M, Zorzi N, Stang E. Caveolin-3 associates with developing T-tubules during muscle differentiation. J Cell Biol. 1997;136:137–154. doi: 10.1083/jcb.136.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pike L J, Casey L. Localization and turnover of phosphatidylinositol 4,5-bisphosphate in caveolin-enriched membrane domains. J Biol Chem. 1996;271:26453–26456. doi: 10.1074/jbc.271.43.26453. [DOI] [PubMed] [Google Scholar]
- 118.Razani B, Rubin C S, Lisanti M P. Regulation of cAMP-mediated signal transduction via interaction of caveolins with the catalytic subunit of protein kinase A. J Biol Chem. 1999;274:26353–26360. doi: 10.1074/jbc.274.37.26353. [DOI] [PubMed] [Google Scholar]
- 119.Repetto S, Bado M, Broda P, Lucania G, Masetti E, Sotgia F, Carbone I, Pavan A, Bonilla E, Cordone G, Lisanti M P, Minetti C. Increased number of caveolae and caveolin-3 over-expression in Duchenne muscular dystrophy. Biochem Biophys Res Commun. 1999;261:547–550. doi: 10.1006/bbrc.1999.1055. [DOI] [PubMed] [Google Scholar]
- 120.Rietveld A, Simons K. The differential miscibility of lipids as the basis for the formation of functional membrane rafts. Biochim Biophys Acta. 1998;1376:467–479. doi: 10.1016/s0304-4157(98)00019-7. [DOI] [PubMed] [Google Scholar]
- 121.Rizzo V, McIntosh D P, Oh P, Schnitzer J E. In situ flow activates endothelial nitric oxide synthase in luminal caveolae of endothelium with rapid caveolin dissociation and calmodulin association. J Biol Chem. 1998;273:34724–34729. doi: 10.1074/jbc.273.52.34724. [DOI] [PubMed] [Google Scholar]
- 122.Robinson M S. The role of clathrin, adaptors and dynamin in endocytosis. Curr Opin Cell Biol. 1994;6:538–544. doi: 10.1016/0955-0674(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 123.Rothberg K G, Heuser J E, Donzell W C, Ying Y, Glenney J R, Anderson R G W. Caveolin, a protein component of caveolae membrane coats. Cell. 1992;68:673–682. doi: 10.1016/0092-8674(92)90143-z. [DOI] [PubMed] [Google Scholar]
- 124.Rothberg K G, Ying Y, Kamen B A, Anderson R G W. Cholesterol controls the clustering of the glycophopholipid-anchored membrane receptor for 5-methyltetrahydrofolate. J Cell Biol. 1990;111:2931–2938. doi: 10.1083/jcb.111.6.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sargiacomo M, Scherer P E, Tang Z, Kubler E, Song K S, Sanders M C, Lisanti M P. Oligomeric structure of caveolin: implications for caveolae membrane organization. Proc Natl Acad Sci USA. 1995;92:9407–9411. doi: 10.1073/pnas.92.20.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sargiacomo M, Scherer P E, Tang Z-L, Kubler E, Song K S, Sanders M C, Lisanti M P. Oligomeric structure of caveolin: implications for caveolae membrane organization. Proc Natl Acad Sci USA. 1995;92:9407–9411. doi: 10.1073/pnas.92.20.9407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sargiacomo M, Sudol M, Tang Z, Lisanti M P. Signal transducing molecules and glycosyl-phosphatidylinositol-linked proteins form a caveolin-rich insoluble complex in MDCK cells. J Cell Biol. 1993;122:789–807. doi: 10.1083/jcb.122.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sargiacomo M, Sudol M, Tang Z-L, Lisanti M P. Signal transducing molecules and GPI-linked proteins form a caveolin-rich insoluble complex in MDCK cells. J Cell Biol. 1993;122:789–807. doi: 10.1083/jcb.122.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Scheiffele P, Verkade P, Fra A M, Virta H, Simons K, Ikonen E. Caveolin-1 and -2 in the exocytic pathway of MDCK cells. J Cell Biol. 1998;140:795–806. doi: 10.1083/jcb.140.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Scherer P E, Lewis R Y, Volonté D, Engelman J A, Galbiati F, Couet J, Kohtz D S, van Donselaar E, Peters P, Lisanti M P. Cell-type and tissue-specific expression of caveolin-2. Caveolins 1 and 2 co-localize and form a stable hetero-oligomeric complex in vivo. J Biol Chem. 1997;272:29337–29346. doi: 10.1074/jbc.272.46.29337. [DOI] [PubMed] [Google Scholar]
- 131.Scherer P E, Lisanti M P. Association of phosphofructokinase-M with caveolin-3 in differentiated skeletal myotubes. Dynamic regulation by extracellular glucose and intracellular metabolites. J Biol Chem. 1997;272:20698–20705. doi: 10.1074/jbc.272.33.20698. [DOI] [PubMed] [Google Scholar]
- 132.Scherer P E, Lisanti M P, Baldini G, Sargiacomo M, Mastick C C, Lodish H F. Induction of caveolin during adipogenesis and association of GLUT4 with caveolin-rich vesicles. J Cell Biol. 1994;127:1233–1243. doi: 10.1083/jcb.127.5.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Scherer P E, Okamoto T, Chun M, Nishimoto I, Lodish H F, Lisanti M P. Identification, sequence, and expression of caveolin-2 defines a caveolin gene family. Proc Natl Acad Sci USA. 1996;93:131–135. doi: 10.1073/pnas.93.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Scherer P E, Tang Z, Chun M, Sargiacomo M, Lodish H F, Lisanti M P. Caveolin isoforms differ in their N-terminal protein sequence and subcellular distribution. Identification and epitope mapping of an isoform-specific monoclonal antibody probe. J Biol Chem. 1995;270:16395–16401. doi: 10.1074/jbc.270.27.16395. [DOI] [PubMed] [Google Scholar]
- 135.Schlegel A, Schwab R B, Scherer P E, Lisanti M P. A role for the caveolin-scaffolding domain in mediating the membrane attachment of caveolin-1. The caveolin-scaffolding domain is both necessary and sufficient for membrane binding in vitro. J Biol Chem. 1999;274:22660–22667. doi: 10.1074/jbc.274.32.22660. [DOI] [PubMed] [Google Scholar]
- 136.Schnitzer J, McIntosh D, Dvorak A M, Liu J, Oh P. Separation of caveolae from associated microdomains of GPI-anchored proteins. Science. 1995;269:1435–1439. doi: 10.1126/science.7660128. [DOI] [PubMed] [Google Scholar]
- 137.Schnitzer J E, Oh P, McIntosh D P. Role of GTP hydrolysis in fission of caveolae directly from plasma membranes. Science. 1996;274:239–242. doi: 10.1126/science.274.5285.239. [DOI] [PubMed] [Google Scholar]
- 138.Schnitzer J E, Oh P, Pinney E, Allard J. Filipin-sensitive caveolae-mediated transport in endothelium: reduced transcytosis, scavenger endocytosis, and capillary permeability of select macromolecules. J Cell Biol. 1994;127:1217–1232. doi: 10.1083/jcb.127.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schwab R, Okamoto T, Scherer P E, Lisanti M P. Analysis of the association of proteins with membranes. In: Bonifacino J S, Dasso M, Harford J B, Lippincott-Schwartz J, Yamada K, editors. Current protocols in cell biology. New York, N.Y: John Wiley & Sons, Inc.; 1999. pp. 1–17. [DOI] [PubMed] [Google Scholar]
- 140.Selkoe D J. Cell biology of the amyloid beta-protein precursor and the mechanism of Alzheimer’s disease. Annu Rev Cell Biol. 1994;10:373–403. doi: 10.1146/annurev.cb.10.110194.002105. [DOI] [PubMed] [Google Scholar]
- 141.Sessa W C, Garcia-Cardena G, Liu J, Keh A, Pollock J S, Bradley J, Thiru S, Braverman I M, Desai K M. The Golgi association of endothelial nitric oxide synthase is necessary for the efficient synthesis of nitric oxide. J Biol Chem. 1995;270:17641–17644. doi: 10.1074/jbc.270.30.17641. [DOI] [PubMed] [Google Scholar]
- 142.Shaul P W, Anderson R G. Role of plasmalemmal caveolae in signal transduction. Am J Physiol. 1998;275:L843–L851. doi: 10.1152/ajplung.1998.275.5.L843. [DOI] [PubMed] [Google Scholar]
- 143.Shaul P W, Smart E J, Robinson L J, German Z, Yuhanna I S, Ying Y, Anderson R G W, Michel T. Acylation targets endothelial nitric-oxide synthase to plasmalemmal caveolae. J Biol Chem. 1996;271:6518–6522. doi: 10.1074/jbc.271.11.6518. [DOI] [PubMed] [Google Scholar]
- 144.Shpetner H S, Vallee R B. Identification of dynamin, a novel mechanochemical enzyme that mediates interactions between microtubules. Cell. 1989;59:421–432. doi: 10.1016/0092-8674(89)90027-5. [DOI] [PubMed] [Google Scholar]
- 145.Simionescu N, Lupu F, Simionescu M. Rings of membrane sterols surround the openings of vesicles and fenestrae, in capillary endothelium. J Cell Biol. 1983;97:1592–600. doi: 10.1083/jcb.97.5.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Simons M, Keller P, De Strooper B, Beyreuther K, Dotti C G, Simons K. Cholesterol depletion inhibits the generation of beta-amyloid in hippocampal neurons. Proc Natl Acad Sci USA. 1998;95:6460–6464. doi: 10.1073/pnas.95.11.6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sisodia S S. Beta-amyloid precursor protein cleavage by a membrane-bound protease. Proc Natl Acad Sci USA. 1992;89:6075–6079. doi: 10.1073/pnas.89.13.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sisodia S S, Koo E H, Beyreuther K, Unterbeck A, Price D L. Evidence that beta-amyloid protein in Alzheimer’s disease is not derived by normal processing. Science. 1990;248:492–495. doi: 10.1126/science.1691865. [DOI] [PubMed] [Google Scholar]
- 149.Smart E, Ying Y-S, Conrad P, Anderson R G W. Caveolin moves from caveolae to the Golgi apparatus in response to cholesterol oxidation. J Cell Biol. 1994;127:1185–1197. doi: 10.1083/jcb.127.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Smart E J, Foster D C, Ying Y S, Kamen B A, Anderson R G W. Protein kinase C activators inhibit receptor-mediated potocytosis by preventing internalization of caveolae. J Cell Biol. 1994;124:307–313. doi: 10.1083/jcb.124.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Smart E J, Ying Y, Mineo C, Anderson R G W. A detergent free method for purifying caveolae membrane from tissue cultured cells. Proc Natl Acad Sci USA. 1995;92:10104–10108. doi: 10.1073/pnas.92.22.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Smart E J, Ying Y-S, Anderson R G W. Hormonal regulation of caveolae internalization. J Cell Biol. 1995;131:929–938. doi: 10.1083/jcb.131.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Smart E J, Ying Y S, Donzell W C, Anderson R G W. A role for caveolin in transport of cholesterol from endoplasmic reticulum to plasma membrane. J Biol Chem. 1996;271:29427–29435. doi: 10.1074/jbc.271.46.29427. [DOI] [PubMed] [Google Scholar]
- 154.Smine A, Xu X, Nishiyama K, Katada T, Gambetti P, Yadav S P, Wu X, Shi Y-C, Yasuhara S, Homburger V, Okamoto T. Regulation of brain G-protein, Go, by Alzheimer disease gene presenilin-1. J Biol Chem. 1998;273:16281–16288. doi: 10.1074/jbc.273.26.16281. [DOI] [PubMed] [Google Scholar]
- 155.Song K S, Li S, Okamoto T, Quilliam L, Sargiacomo M, Lisanti M P. Copurification and direct interaction of Ras with caveolin, an integral membrane protein of caveolae microdomains. Detergent free purification of caveolae membranes. J Biol Chem. 1996;271:9690–9697. doi: 10.1074/jbc.271.16.9690. [DOI] [PubMed] [Google Scholar]
- 156.Song K S, Scherer P E, Tang Z-L, Okamoto T, Li S, Chafel M, Chu C, Kohtz D S, Lisanti M P. Expression of caveolin-3 in skeletal, cardiac, and smooth muscle cells. Caveolin-3 is a component of the sarcolemma and co-fractionates with dystrophin and dystrophin-associated glycoproteins. J Biol Chem. 1996;271:15160–15165. doi: 10.1074/jbc.271.25.15160. [DOI] [PubMed] [Google Scholar]
- 157.Song K S, Tang Z, Li S, Lisanti M P. Mutational analysis of the properties of caveolin-1. A novel role for the C-terminal domain in mediating homo-typic caveolin-caveolin interactions. J Biol Chem. 1997;272:4398–4403. doi: 10.1074/jbc.272.7.4398. [DOI] [PubMed] [Google Scholar]
- 158.Sontag J M, Fykse E M, Ushkaryov Y, Liu J P, Robinson P J, Sudhof T C. Differential expression and regulation of multiple dynamins. J Biol Chem. 1994;269:4547–4554. [PubMed] [Google Scholar]
- 159.Sotgia F, Minetti C, Lisanti M P. Localization of the human caveolin-3 gene to the D3S18/D3S4163/D3S4539 locus (3p25), in close proximity to the human oxytocin receptor gene. Identification of the caveolin-3 gene as a candidate for deletion in 3p-syndrome. FEBS Lett. 1999;452:177–180. doi: 10.1016/s0014-5793(99)00658-4. [DOI] [PubMed] [Google Scholar]
- 160.Takei K, Haucke V, Slepnev V, Farsad K, Salazar M, Chen H, De Camilli P. Generation of coated intermediates of clathrin-mediated endocytosis on protein-free liposomes. Cell. 1998;94:131–141. doi: 10.1016/s0092-8674(00)81228-3. [DOI] [PubMed] [Google Scholar]
- 161.Tang Z, Scherer P E, Okamoto T, Song K, Chu C, Kohtz D S, Nishimoto I, Lodish H F, Lisanti M P. Molecular cloning of caveolin-3, a novel member of the caveolin gene family expressed predominantly in muscle. J Biol Chem. 1996;271:2255–2261. doi: 10.1074/jbc.271.4.2255. [DOI] [PubMed] [Google Scholar]
- 162.Toya Y, Schwencke C, Couet J, Lisanti M P, Ishikawa Y. Inhibition of adenylyl cyclase by caveolin peptides. Endocrinology. 1998;139:2025–2031. doi: 10.1210/endo.139.4.5957. [DOI] [PubMed] [Google Scholar]
- 163.Tran D, Carpentier J L, Sawano F, Gorden P, Orci L. Ligands internalized through coated or noncoated invaginations follow a common intracellular pathway. Proc Natl Acad Sci USA. 1987;84:7957–7961. doi: 10.1073/pnas.84.22.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Trigatti B L, Anderson R G, Gerber G E. Identification of caveolin-1 as a fatty acid binding protein. Biochem Biophys Res Commun. 1999;255:34–39. doi: 10.1006/bbrc.1998.0123. [DOI] [PubMed] [Google Scholar]
- 165.Uittenbogaard A, Ying Y, Smart E J. Characterization of a cytosolic heat-shock protein-caveolin chaperone complex. Involvement in cholesterol trafficking. J Biol Chem. 1998;273:6525–6532. doi: 10.1074/jbc.273.11.6525. [DOI] [PubMed] [Google Scholar]
- 166.Urrutia R, Henley J R, Cook T, McNiven M A. The dynamins: redundant or distinct functions for an expanding family of related GTPases? Proc Natl Acad Sci USA. 1997;94:377–384. doi: 10.1073/pnas.94.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.van der Bliek A M, Meyerowitz E M. Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature. 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- 168.van der Bliek A M, Redelmeier T E, Damke H, Tisdale E J, Meyerowitz E M, Schmid S L. Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Venema V J, Ju H, Zou R, Venema R C. Interaction of neuronal nitric-oxide synthase with caveolin-3 in skeletal muscle. Identification of a novel caveolin scaffolding/inhibitory domain. J Biol Chem. 1997;272:28187–28190. doi: 10.1074/jbc.272.45.28187. [DOI] [PubMed] [Google Scholar]
- 170.Volonté D, Galbiati F, Li S, Nishiyama K, Okamoto T, Lisanti M P. Flotillins/cavatellins are differentially expressed in cells and tissues and form a hetero-oligomeric complex with caveolins in vivo. Characterization and epitope-mapping of a novel flotillin-1 monoclonal antibody probe. J Biol Chem. 1999;274:12702–12709. doi: 10.1074/jbc.274.18.12702. [DOI] [PubMed] [Google Scholar]
- 171.Wary K K, Mainiero F, Isakoff S J, Marcantonio E E, Giancotti F G. The adaptor protein Shc couples a class of integrins to the control of cell cycle progression. Cell. 1996;87:733–743. doi: 10.1016/s0092-8674(00)81392-6. [DOI] [PubMed] [Google Scholar]
- 172.Wary K K, Mariotti A, Zurzolo C, Giancotti F G. A requirement for caveolin-1 and associated kinase Fyn in integrin signaling and anchorage-dependent cell growth. Cell. 1998;94:625–634. doi: 10.1016/s0092-8674(00)81604-9. [DOI] [PubMed] [Google Scholar]
- 173.Waugh M G, Lawson D, Tan S K, Hsuan J J. Phosphatidylinositol 4-phosphate synthesis in immunoisolated caveolae-like vesicles and low buoyant density non-caveolar membranes. J Biol Chem. 1998;273:17115–17121. doi: 10.1074/jbc.273.27.17115. [DOI] [PubMed] [Google Scholar]
- 174.Webb N R, Connell P M, Graf G A, Smart E J, de Villiers W J, de Beer F C, van der Westhuyzen D R. SR-BII, an isoform of the scavenger receptor BI containing an alternate cytoplasmic tail, mediates lipid transfer between high density lipoprotein and cells. J Biol Chem. 1998;273:15241–15248. doi: 10.1074/jbc.273.24.15241. [DOI] [PubMed] [Google Scholar]
- 175.Wei Y, Lukashev M E, Simon D I, Bodary S C, Rosenberg S, Doyle M V, Chapman H A. Regulation of integrin function by the urokinase receptor. Science. 1996;273:1551–1555. doi: 10.1126/science.273.5281.1551. [DOI] [PubMed] [Google Scholar]
- 176.Wei Y, Yang X, Liu Q, Wilkins J A, Chapman H A. A role for caveolin and the urokinase receptor in integrin-mediated adhesion and signaling. J Cell Biol. 1999;144:1285–1294. doi: 10.1083/jcb.144.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Yamamoto M, Toya Y, Jensen R A, Ishikawa Y. Caveolin is an inhibitor of platelet-derived growth factor receptor signaling. Exp Cell Res. 1999;247:380–388. doi: 10.1006/excr.1998.4379. [DOI] [PubMed] [Google Scholar]
- 178.Yamamoto M, Toya Y, Schwencke C, Lisanti M P, Myers M G, Ishikawa Y. Caveolin is an activator of insulin receptor signaling. J Biol Chem. 1998;273:26962–26968. doi: 10.1074/jbc.273.41.26962. [DOI] [PubMed] [Google Scholar]
- 179.Yokoyama C, Wang X, Briggs M R, Admon A, Wu J, Hua X, Goldstein J L, Brown M S. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- 180.Yoon S O, Casaccia-Bonnefil P, Carter B, Chao M V. Competitive signaling between TrkA and p75 nerve growth factor receptors determines cell survival. J Neurosci. 1998;18:3273–3281. doi: 10.1523/JNEUROSCI.18-09-03273.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]