Abstract
A subset of male germ line-specific genes, the MAGE-type genes, are activated in many human tumors, where they produce tumor-specific antigens recognized by cytolytic T lymphocytes. Previous studies on gene MAGE-A1 indicated that transcription factors regulating its expression are present in all tumor cell lines whether or not they express the gene. The analysis of two CpG sites located in the promoter showed a strong correlation between expression and demethylation. It was also shown that MAGE-A1 transcription was induced in cell cultures treated with demethylating agent 5′-aza-2′-deoxycytidine. We have now analyzed all of the CpG sites within the 5′ region of MAGE-A1 and show that for all of them, demethylation correlates with the transcription of the gene. We also show that the induction of MAGE-A1 with 5′-aza-2′-deoxycytidine is stable and that in all the cell clones it correlates with demethylation, indicating that demethylation is necessary and sufficient to produce expression. Conversely, transfection experiments with in vitro-methylated MAGE-A1 sequences indicated that heavy methylation suffices to stably repress the gene in cells containing the transcription factors required for expression. Most MAGE-type genes were found to have promoters with a high CpG content. Remarkably, although CpG-rich promoters are classically unmethylated in all normal tissues, those of MAGE-A1 and LAGE-1 were highly methylated in somatic tissues. In contrast, they were largely unmethylated in male germ cells. We conclude that MAGE-type genes belong to a unique subset of germ line-specific genes that use DNA methylation as a primary silencing mechanism.
Most of the mammalian genome is depleted in CpG dinucleotides, which are present at about 20% of their predicted frequency (9). This has been ascribed to the high mutability of 5-methylcytosine, which is present in more than 70% of the CpG nucleotides. In contrast, some regions within the genome, known as CpG islands, have retained a normal amount of CpG sites (9). CpG islands, which account for about 1% of the entire genome, are usually associated with a promoter region. In normal cells, CpG islands are methylated only under two circumstances: on the inactive X chromosome in female somatic cells and on silenced alleles of parentally imprinted genes (42, 45). The preservation of the CpG sites within CpG islands has been ascribed to their lack of methylation (1). In cancer cells, however, CpG islands occasionally undergo aberrant de novo methylation (3). In cultured cell lines, this abnormal de novo methylation may involve as much as half of all CpG islands (2).
There is evidence that in some circumstances DNA methylation can on its own be an efficient mechanism for gene silencing, in cells containing the relevant transcription factors. Methylated silent genes in cultured cell lines have been shown to be activated upon treatment with demethylating drugs (26). In vitro methylation of genes often prevents their transcription in transfection assays (44). In vivo, DNA methylation appears to be responsible for the monoallelic repression of X-linked genes and for that of parentally imprinted genes in cells where the appropriate transcription factors are evidently present (25, 32).
The role of DNA methylation in the differentiation process, i.e., in the programmed expression of tissue-specific genes, has received considerable attention (20). It has been proposed that DNA methylation could be the primary mechanism for the selective expression of tissue-specific genes (41). However, it appears now that DNA methylation often plays a role secondary to the presence or absence of transcription factors. Genes with tissue-specific expression can be divided in two groups according to the content in CpG dinucleotides of their promoter: those with a CpG-rich promoter and those with a CpG-poor promoter. Because CpG islands are unmethylated in all tissues, the selective expression of tissue-specific genes with a CpG-rich promoter must depend solely on the presence of tissue-specific transcription factors (4). On the other hand, tissue-specific genes with CpG-poor promoters often show a higher level of methylation in the tissues where these genes are silent than in those where they are expressed (20). However, it appears that DNA demethylation is not sufficient to activate these genes in nonexpressing cells, presumably because the transcription factors required for their expression are not present (4, 52). Thus, there has been hitherto no clear example of tissue-specific genes that use DNA methylation as a primary mechanism for their regulation.
Moreover, studies using mice carrying a targeted deletion of the gene coding for DNA methyltransferase, the enzyme that maintains cytosine methylation, do not support an important role for DNA methylation in the regulation of tissue-specific genes. Although embryos carrying this deletion died prior to day 11, they displayed significant morphogenesis and tissue differentiation (33). More recently, it was shown that tissue-specific genes are largely demethylated in these mutant embryos but that their expression is not affected (50). Further evidence that DNA methylation does not control the expression of many tissue-specific genes came from the analysis of cancer cells. Although cancer cells often undergo a genomewide demethylation that significantly reduces the overall level of cytosine methylation (18, 22), no massive activation of tissue-specific genes has been observed (3). It therefore appears that differentiation-linked gene methylation may serve only to reinforce an expression arrest that results from a lack of appropriate transcription factors (5). The demethylation of some of these genes in expressing cells is likely to be a consequence of the activation induced by transcription factors (28).
We have shown previously that a subset of male germ line-specific genes are activated in a wide variety of tumors, in which they code for tumor-specific antigens recognized by autologous T lymphocytes (49). The MAGE family contains 18 related genes divided into three clusters (MAGE-A, -B, and -C) located on the X chromosome (11, 34, 35, 37). Several other unrelated families of genes with similar patterns of expression, notably the GAGE and LAGE/ESO-1 families (7, 30, 48), have been identified. These MAGE-type genes are also located on the X chromosome (10, 30). The function of the MAGE, GAGE, and LAGE genes is unknown.
The expression of different MAGE, GAGE, and LAGE genes in tumors is positively correlated, suggesting that these genes are activated by a common mechanism (10, 30). Transfection studies with MAGE-A1 promoter constructs showed that these constructs were transcribed both in tumor cell lines that express the MAGE-A1 gene and in tumor cell lines that do not (13). This finding indicated that in tumor cell lines that do not express MAGE-A1, transcription factors capable of inducing MAGE-A1 promoter activity are present but that the gene is insensitive to their action. Two observations suggested that at least in tumor cell lines, DNA methylation might be involved in this repression mechanism. First, the expression of MAGE-A1 in tumor cell lines correlated closely with demethylation at two CpG sites located within two essential Ets promoter elements (14). Moreover, MAGE-A1 was found to be expressed exclusively in the tumor cell lines that showed a marked decrease in the overall level of CpG methylation (14). Second, MAGE-A1 was shown to be induced in cell cultures treated with 5′-aza-2′-deoxycytidine, a demethylating agent (14, 51).
Our previous analysis of the methylation status of the MAGE-A1 promoter in tumor cell lines involved only two CpG sites (14). We report here a detailed analysis of the methylation status of all CpG sites within the 5′ region of gene MAGE-A1 in tumor cells that either do or do not express the gene. We have made a similar analysis on clones derived from populations treated with 5′-aza-2′-deoxycytidine. We also report that the promoters of MAGE-type genes are characterized by a high content of CpG dinucleotides, which, contrary to all other CpG islands, appear to be methylated in vivo in the tissues where the genes are not expressed.
Our results suggest that methylation is the primary mechanism for the silencing of MAGE-type genes.
MATERIALS AND METHODS
Cell cultures and DNA extraction.
The tumor cell lines were adapted to culture in our lab from a rhabdomyosarcoma (LB23-SAR), from a melanoma (LB373-MEL), or from a renal carcinoma (LB996-RCC). The cell cultures were maintained in Iscove’s medium supplemented with 10% fetal calf serum, l-arginine (116 mg/ml), l-asparagine (36 mg/ml), and l-glutamine (216 mg/ml). DNA was extracted as previously described (14).
Tissue DNA samples.
Genomic DNA from human lung, colon, peripheral blood lymphocytes, and kidney was kindly provided by Ö. Türeci (Medizinische Klinik I, Homburg, Germany). The testis sample came from a 60-year-old patient with prostate cancer. Histological analysis indicated that the spermatogenesis was slightly diminished but still active (P. Van Cangh, Cliniques Saint-Lue, Brussels, Belgium). The testis and skin tissue fragments were pulverized to a fine powder in liquid nitrogen and extracted as described earlier (14). The sperm sample was obtained from a 33-year-old healthy donor. The sperm sample was rinsed twice in phosphate-buffered saline, and the DNA was extracted as previously described (14) except that 10 mM dithiothreitol was added to the lysis buffer.
Methylation analysis by sodium bisulfite sequencing.
Sodium bisulfite treatment of genomic DNA was performed essentially as described previously (21). Briefly, 5 μg of genomic DNA was digested with EcoRI, denatured in 0.4 M NaOH for 10 min at room temperature, neutralized with 3 M ammonium acetate, and ethanol precipitated. The denatured DNA was then resuspended in 3 μl of water and diluted with 600 μl of a solution of 3.9 M sodium bisulfite and 0.5 mM hydroquinone (pH 5.0). The DNA solution was covered with mineral oil and incubated in a thermal cycler (Trio Block; Biometra) for at least 16 h at 50°C with 5 min at 95°C every 3 h. The DNA was then desalted and concentrated on a silica matrix (Geneclean; Bio 101), incubated in 0.3 M NaOH for 10 min at room temperature, neutralized, and precipitated. The bisulfite-modified genomic DNA was resuspended in 75 μl of water.
For PCR amplification, 10 μl of the bisulfite-modified DNA was added in a final volume of PCR mix containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 0.5 μM each primer, and 2.5 U of TaKaRa Taq (TaKaRa Biomedicals). The primers for PCR amplification of bisulfite-modified DNA were designed to contain either KpnI or HindIII restriction sites at their 5′ ends for subsequent cloning of the PCR products: 5′-CCCAAGCTTTTTTATTTTTATTTAGGTAGGATT-3′ (MAGE-A1; sense, positions −105 to −82 relative to the transcription start site), 5′-CGGGGTACCCCTAATATCTCTCAAAACTTTTAA-3′ (MAGE-A1; antisense, 225 to 202), 5′-CCCAAGCTTTYGGGGTTTATTYGGGGTTTTTTA-3′ (LAGE-1; sense, −112 to −89), 5′-CGGGGTACCAAATACCAAAACCTCCTAAACCAT-3′ (LAGE-1; antisense, 134 to 111), and 5′-CCCAAGCTTGAAGTTGGAATTTTTATTTAGTAA-3′. All primers amplified the upper strand of the corresponding modified promoter sequences. PCR amplifications were performed for 36 cycles of 1 min at 95°C, 1 min at 57°C, and 1 min at 72°C.
PCR products were purified from agarose gels on a silica matrix (Geneclean; Bio 101), and were ligated into the KpnI and HindIII sites of pTZ19R (Pharmacia). DH5αF′IQ bacteria (Life Technologies, GibcoBRL) were transformed by electroporation with the ligation products. Plasmid DNA was prepared from the transformed clones by the boiling miniprep procedure, alkali denatured, and sequenced by using a T7 sequencing kit (Pharmacia Biotech).
5′-Aza-2′-deoxycytidine treatment and cloning of the treated cell line.
Flasks (80 cm2; Nunc) were seeded with 6.25 × 105 LB23-SAR cells or 2.5 × 105 MI665/2-MEL cells in complete medium supplemented 2 μM 5′-aza-2′-deoxycytidine. After 4 days of culture in the presence of the drug, LB23-SAR cells were at 4.6 × 106 cells per flask (about three divisions) and MI665/2-MEL cells were at 1.75 × 106 (about three divisions). We used 4 × 106 cells for RNA extraction as previously described (13) and 107 to 1.5 × 107 cells for DNA extraction. The remaining cells were then rinsed and incubated in fresh, drug-free medium. During the subsequent days, many cells died. However, about 2 weeks after 5′-aza-2′-deoxycytidine removal, enough cells were available for RNA and DNA extractions. At the same time, the cell population was diluted at either 10 or 3 cells/ml, and 100-μl aliquots of these suspensions were inoculated in each well of 96-well plates (Nunc). Twenty-three LB23-SAR clones and 19 MI665/2-MEL clones were amplified, and RNA was extracted from about 105 cells with Trizol (Life Technologies GibcoBRL) between days 28 and 42 after 5′-aza-2′-deoxycytidine removal. Five LB23-SAR clones and five MI665/2-MEL clones were maintained in culture. A first extraction of genomic DNA was performed on these five clones between days 43 and 56 after 5′-aza-2′-deoxycytidine removal. Other extractions of RNA and DNA were performed until day 158 after drug removal.
Reverse transcription-PCR (RT-PCR) analyses.
Reverse transcription was performed as previously described (15). PCR amplification of MAGE-A1, the β-actin gene, LAGE-1, and LAGE-2/ESO-1 cDNAs was performed as described elsewhere (15, 30) except that TaKaRa Taq polymerase was used. β2-Microglobulin gene amplification was carried out for 26 cycles, using 5′-TGAAGCTGACAGCATTCG-3′ as the sense primer and 5′-ATCTTCAAACCTCCATGATG-3′ as the antisense primer with the following conditions: 30 s at 94°C, 1 min at 59°C, and 1 min at 72°C.
Quantitative RT-PCR.
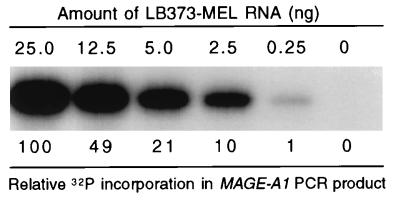
For the evaluation of MAGE-A1 expression levels, we used a quantitative RT-PCR procedure previously described by Lethé et al. (31). Briefly, reverse-transcribed RNA was submitted to 27 cycles of PCR amplification with the MAGE-A1 primers, in the presence of 0.5 μCi of [α-32P]dCTP at 3,000 Ci/mmol. The PCR products were separated in an agarose gel, and the incorporated radioactivity was measured with a PhosphorImager (Molecular Dynamics). Under these conditions, the radioactivity incorporated into the PCR product correlates closely to the amount of MAGE-A1 mRNA in the sample (31) (Fig. 1). To correct variations of integrity, purity, or quantity between different RNA samples, we normalized the value for MAGE-A1 with that for β-actin obtained after 18 cycles of PCR amplification on the same sample, as previously described (31).
FIG. 1.
The amount of MAGE-A1 PCR product is proportional to the amount of template RNA. Total RNA from the LB373-MEL cell line, which expresses MAGE-A1, was progressively diluted in total RNA from the LB23-SAR cell line, which does not express MAGE-A1. RNA samples containing the indicated amount of LB373-MEL RNA were converted to cDNA and amplified by PCR for MAGE-A1 (27 cycles) in the presence of [α-32P]dCTP. The PCR products were separated in a 1.7% agarose gel, and the incorporated radioactivity was counted with a PhosphorImager. The relative amount of 32P incorporation is indicated below each band.
In vitro methylation of plasmids.
The plasmid containing the luciferase reporter gene under the control of the MAGE-A1 promoter was obtained by inserting the SacI MAGE-A1 fragment extending between positions −375 and 2636 and the SacI/BbsI MAGE-A1 fragment extending between positions 2636 and 3036 between the SacI and SmaI sites of pXP1. The plasmid containing the 12-kb MAGE-A1 fragment was constructed by inserting into plasmid pTZ18R a SmaI genomic fragment extending from −792 bp to +11 kb relative to the transcription start point of MAGE-A1. A 100-μg aliquot of plasmid DNA was treated in 2 ml with S-adenosylmethionine (160 μM) and SssI methylase (1 U/μg; New England Biolabs), which methylates every CpG site. Complete methylation was verified by digestion with the methylation-sensitive restriction enzyme HpaII. DNAs were purified by phenol and chloform extractions and by ethanol precipitation prior to transfection.
Transfections.
For transfection, we used the calcium phosphate precipitation method as described earlier (12). MZ2-MEL2.2.5 cells (106) were transfected with 25 μg of either unmethylated or methylated MAGE-A1 constructs and with 2.5 μg of pHMR272, which carries the gene for resistance to hygromycin B. The transfectants were selected in medium containing 175 μg of hygromycin B per ml. The luciferase and RT-PCR assays were performed 4 weeks after transfection.
Luciferase assay.
Two cellular extracts were obtained from 1 × 106 and 2 × 106 cells from each group, and their luciferase activities were measured as previously described (13). The reported luciferase activities correspond to the means of the values obtained with the two extracts.
RESULTS
MAGE-A1 demethylation in tumor cell lines that express the gene.
The two Ets binding sites that drive most of the promoter activity of gene MAGE-A1 contain a CpG within their sequences (13). We assessed previously the methylation status of these CpG sites in various tumor cell lines by performing PCR amplification of genomic DNA digested with methyl-sensitive restriction enzyme HpaII (14). The results showed that these sites are demethylated in all the tumor cell lines that express MAGE-A1. In the tumor cell lines that do not express the gene, these sites appeared to be methylated, but the method did not allow us to evaluate the precise extent of this methylation.
We resorted to the sodium bisulfite modification procedure to assess the methylation status of the 17 CpG sites located between positions −81 and 169 in gene MAGE-A1. This method relies on the fact that cytosines but not methylcytosines are changed into uracil by sodium bisulfite (21). The analyzed MAGE-A1 fragment was amplified by PCR applied to genomic DNA treated with sodium bisulfite. The PCR product was then cloned and several clones were sequenced, each sequence representing the methylation profile of an individual DNA molecule.
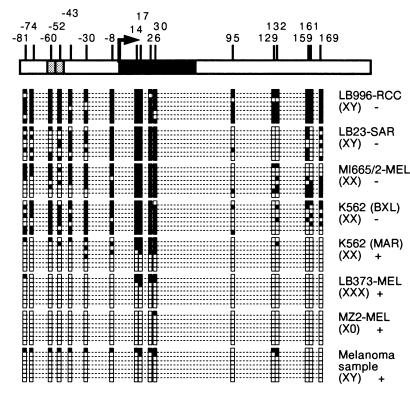
We observed that there was a significant degree of heterogeneity in the methylation of MAGE-A1 within each cell population and that the CpG sites were not all methylated in the cell lines that did not express this gene (Fig. 2). However, in the four tumor cell lines that did not express MAGE-A1, all sequences displayed methylation of at least 7 of the 17 CpG sites analyzed (Fig. 2). In contrast to the tumor cell lines that did not express MAGE-A1, those that expressed the gene contained fully unmethylated MAGE-A1 sequences (Fig. 2). The CpG sites in the essential Ets promoter elements (−60 and −52) were both unmethylated in some of the MAGE-A1 sequences obtained from cell lines LB23-SAR and MI665/2-MEL, which did not express MAGE-A1, indicating that the demethylation of these two sites is not sufficient to allow MAGE-A1 transcription (Fig. 2). A group of five CpG sites surrounding the transcription start site (positions −8, 14, 17, 26, and 30) was heavily methylated in all nonexpressing tumor cell lines, suggesting that methylation of these sites might be required for the arrest of MAGE-A1 transcription.
FIG. 2.
Methylation pattern of MAGE-A1 in tumor cells. The 5′ region of the gene is represented at the top with a broken arrow at the transcription start site, vertical bars at the locations of CpG dinucleotides, and two gray boxes representing the two Ets binding sites of the promoter. For each tumor, the methylation pattern of eight DNA molecules is shown. Black squares correspond to methylated cytosines; empty squares correspond to unmethylated cytosines. For each cell line, the content in X chromosomes and the presence (+) or absence (−) of MAGE-A1 expression are indicated. LB996-RCC is a renal cell carcinoma; LB23-SAR is a sarcoma cell line; MI665/2-MEL, LB373-MEL, and MZ2-MEL are melanoma cell lines; K562 (BXL) and K562 (MAR) are two cultures of the same erythroleukemia cell line that have been maintained separately for a long time and differ with respect to expression of MAGE-A1. The melanoma sample was obtained from a lymph node metastasis.
As each sequence represents the methylation profile of a single DNA molecule, it is possible to detect allele-specific methylation differences linked to X chromosome inactivation. Allele-specific methylation probably accounts for the presence of some methylated sequences in the K562 (MAR) erythroleukemia cell line and the LB373-MEL melanoma cell line, which contain several X chromosomes (Fig. 2). Similar results were obtained with a surgical tumor sample (Fig. 2). Some largely methylated sequences found in this sample probably originated from normal cells present in these lesions.
MAGE expression and demethylation induced by 5′-aza-2′-deoxycytidine.
Previous studies demonstrated that gene MAGE-A1 is induced in cell cultures treated with 5′-aza-2′-deoxycytidine, a demethylating agent (14, 51). Here, we examined whether the induction of individual MAGE-A1 genes by this treatment strictly correlates with the demethylation of their promoter. Because the treatment of a cell line with 5′-aza-2′-deoxycytidine is likely to induce MAGE-A1 in only a fraction of the cells, we analyzed the methylation of the MAGE-A1 promoter in a number of cell clones derived from the treated population.
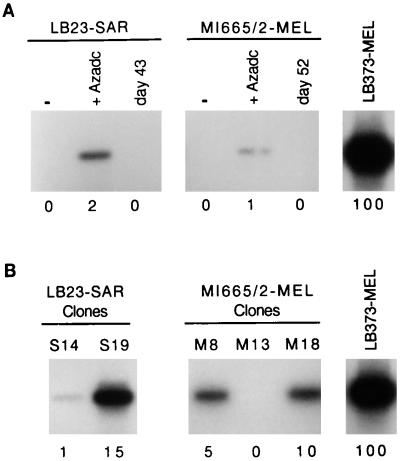
Sarcoma cell line LB23-SAR and melanoma cell line MI665/2-MEL, which do not express MAGE-A1, were incubated for 4 days with 2 μM 5′-aza-2′-deoxycytidine. This treatment induced MAGE-A1 in both cell lines (Fig. 3A). However, using a quantitative RT-PCR procedure previously described by Lethé et al. (31) and providing accurate evaluation of the amount of MAGE-A1 template in RNA samples (Fig. 1), we found that the level of MAGE-A1 expression induced in the LB23-SAR and MI665/2-MEL cell lines represented only a small percentage of the level observed in many cell lines that express the gene spontaneously, such as LB373-MEL (Fig. 3A). These low levels of expression supported the notion that MAGE-A1 was induced in only a fraction of the treated LB23-SAR and MI665/2-MEL cells. Moreover, the expression of MAGE-A1 in both treated cell lines decreased progressively after withdrawal of the demethylating agent and disappeared almost completely after about 6 weeks of culture in the absence of further drug treatment (Fig. 3A).
FIG. 3.
MAGE-A1 expression levels in 5′-aza-2′-deoxycytidine-treated cell lines and clones. (A) RNA was extracted from cell lines LB23-SAR and MI665/2-MEL before treatment (−), at the end of 5′-aza-2′-deoxycytidine treatment (+Azadc), or at the number of days indicated after the end of treatment. RT-PCR amplifications were performed with primers specific to MAGE-A1 and in the presence of [α-32P]dCTP. Band intensities were quantified with a PhosphorImager. The expression level (below each PCR band) was normalized with the β-actin messenger level of the same sample and expressed relative to the expression level of melanoma cell line LB373-MEL. Values are the mean of two independent PCR experiments. (B) The same analysis was carried out with cell clones derived from the 5′-aza-2′-deoxycytidine-treated LB23-SAR and MI665/2-MEL cell lines.
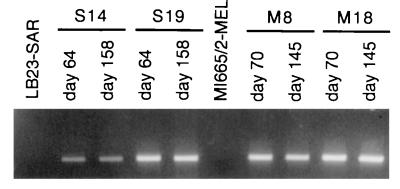
The gradual disappearance of MAGE-A1 expression in cell population after the end of 5′-aza-2′-deoxycytidine treatment might result either from a gradual remethylation of the demethylated cells or from a selection against the most demethylated cells. To decide between these possibilities, the treated LB23-SAR and MI665/2-MEL populations were cloned by limiting dilutions 11 and 6 days, respectively, after drug removal, at a time when MAGE-A1 expression was still present. The clones were grown in the absence of the demethylating agent, and RNA was extracted from individual clones as soon as enough cells were obtained (between day 28 and 56 after 5′-aza-2′-deoxycytidine removal). Expression of MAGE-A1 was observed in three of the 23 LB23-SAR clones and in three of 19 MI665/2-MEL clones. Quantitative RT-PCR carried out on four positive clones showed levels of expression of MAGE-A1 ranging from 1 to 15% of that of LB373-MEL (Fig. 3B). These four clones were maintained in normal medium for more than 20 weeks in the absence of the demethylating agent. Unlike the corresponding cell lines, these clones continued to express MAGE-A1 after withdrawal of the 5′-aza-2′-deoxycytidine (Fig. 4). We conclude that the activation of MAGE-A1 expression following 5′-aza-2′-deoxycytidine treatment is stable but that the heavily demethylated cells, where MAGE-A1 induction occurs, have a growth impairment resulting in their counterselection in the treated populations.
FIG. 4.
Stable expression of gene MAGE-A1 in clones derived from cell lines LB23-SAR and MI665/2-MEL treated with 5′-aza-2′-deoxycytidine. Cell clones derived from the treated LB23-SAR cell line (S14 and S19) and from the treated MI665/2-MEL cell line (M8 and M18) were maintained in culture in the absence of the demethylating agent. The expression of MAGE-A1 was assessed by RT-PCR on RNA extracted from these clones at two different times. Indicated days correspond to the days after drug withdrawal from the two cell lines.
The methylation status of the MAGE-A1 promoter region was analyzed in clones that expressed MAGE-A1 and in clones that did not. Sequencing of bisulfite-treated DNA indicated that the 5′ region of MAGE-A1 had a lower level of methylation in the clones where the gene was active than in the clones where it was not (Fig. 5), or in the untreated LB23-SAR and MI665/2-MEL cell lines (Fig. 2). The residual MAGE-A1 methylation observed in clones S14, S19, and M8 may explain why the expression level of MAGE-A1 is lower in these clones than in the LB373-MEL cell line, where most DNA molecules appear to have a completely demethylated 5′ end of MAGE-A1 (Fig. 2). These differences in expression levels may also be partly due to differences in the abundance of transcription activators, as transfection experiments showed that the activity of the MAGE-A1 promoter varies among tumor cell lines regardless of whether they express the gene (13).
FIG. 5.
Methylation status of the MAGE-A1 5′ end in cell clones derived from the LB23-SAR and MI665/2-MEL cell lines initially treated with 5′-aza-2′-deoxycytidine. The sodium bisulfite DNA modification procedure was used to analyze the methylation pattern of clones that do not (−) and that do (+) express the gene. The methylation patterns of eight DNA molecules are given. Black squares correspond to methylated cytosines; empty squares correspond to unmethylated cytosines. DNAs of the LB23-SAR and MI665/2-MEL cell line clones were extracted between 43 and 56 days after the end of treatment.
In vitro methylation before transfection of gene MAGE-A1 prevents expression.
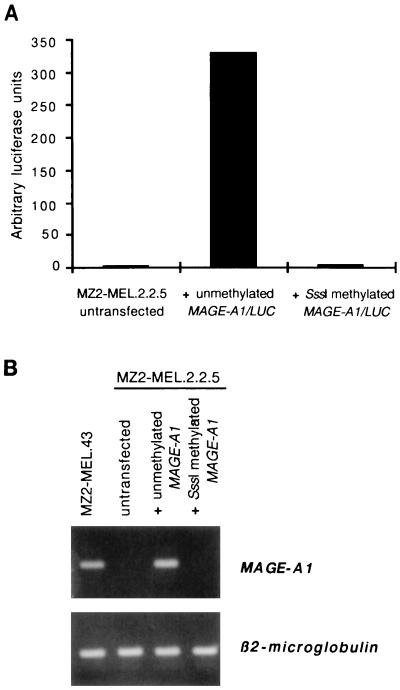
A construct with a luciferase reporter gene controlled by the MAGE-A1 promoter was made and methylated in vitro with SssI, an enzyme which methylates every CpG. The methylated and unmethylated constructs were both cotransfected with a plasmid conferring resistance to hygromycin into MZ2-MEL.2.2.5. This subclone of the MZ2-MEL cell line has been previously selected for its resistance to anti-MAGE-A1 cytolytic T lymphocytes and had been found to have lost MAGE-A1 expression as a result of a deletion of the gene (47). Four weeks after transfection, we tested the luciferase activity in transfectants selected with hygromycin (Fig. 6A). The cells transfected with the methylated construct showed no luciferase expression. In contrast, those transfected with the unmethylated construct showed expression.
FIG. 6.
Lack of MAGE-A1 expression after in vitro methylation. (A) A fragment of the MAGE-A1 gene extending from −375 to +3036 was cloned upstream to the luciferase reporter gene. This construct, either unmethylated or methylated with SssI methylase, was stably transfected into MZ2-MEL.2.2.5 cells. Transcription directed by the MAGE-A1 promoter was assessed by measuring the amount of luciferase activity in the transfectants. (B) A construct with a 12-kb genomic fragment containing MAGE-A1 was either unmethylated or methylated with SssI and was stably transfected in MZ2-MEL.2.2.5 cells. RT-PCR were performed to test the expression of the transfected MAGE-A1 gene.
A similar experiment was carried out with a vector containing a 12-kb genomic fragment containing the entire sequence of gene MAGE-A1. The methylated construct was not expressed in stable transfectants, whereas the unmethylated construct was expressed (Fig. 6B).
We conclude that the complete methylation produced by SssI suffices to block expression of MAGE-A1 even in cells that contain transcription factors capable of inducing a high level of expression of this gene.
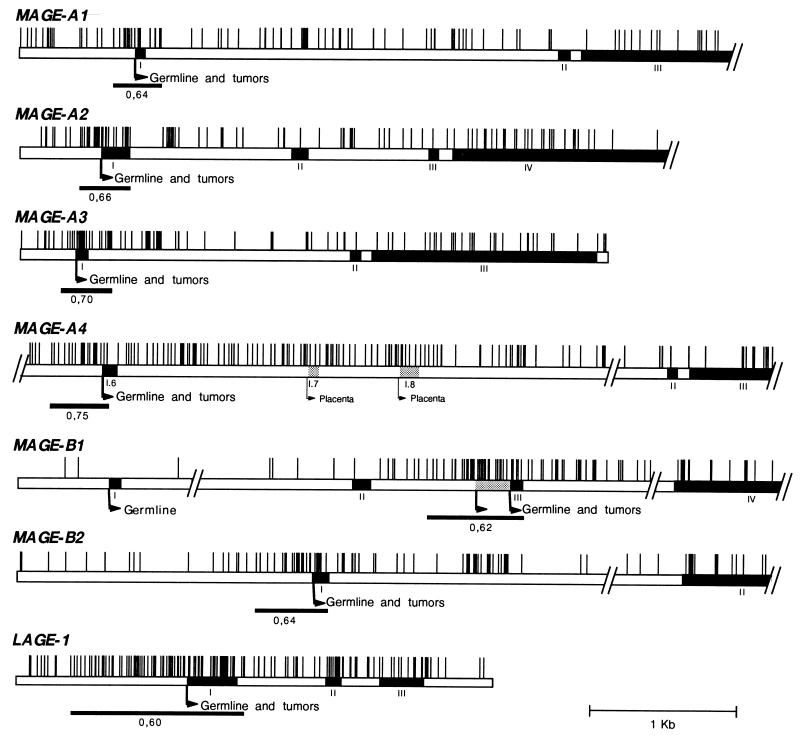
CpG islands associated with the promoter of MAGE-type genes.
A region of about 300 bp which covers a part of the promoter of gene MAGE-A1, the first exon, and a part of the first intron is rich in CpG sites. It matches generally accepted CpG island criteria, which are a G+C content over 50% and an observed/expected CpG ratio of at least 0.6 (23). The sequences of the promoter region and of the first intron are not available for all the genes of the MAGE family. However, those that could be examined were also found to have a CpG island at their 5′ end (Fig. 7). Gene MAGE-B1 has two alternative promoters. The first, located before exon 1, is active only in male germ line cells, whereas the second, located before exon 3, directs a MAGE-type pattern of expression, being active in both germ line cells and tumor cells (35). Interestingly, only the latter promoter of MAGE-B1 is included in a CpG-rich region. Another MAGE-type gene, LAGE-1, was also found to have a CpG island at its 5′ end.
FIG. 7.
Distribution of CpG dinucleotides in MAGE-type genes that are expressed in tumors. The vertical bars correspond to locations of CpG dinucleotides. Promoter regions that fit the CpG island criteria, as previously defined (23), are underlined, and their observed/expected CpG ratios are indicated. The diagram gives a condensed view of the density of the CpG dinucleotides. Exons are indicated by black boxes; alternatively used exons are indicated by shaded boxes. Broken arrows correspond to the transcription start sites. “Germline” or “Germline and tumors” indicates that transcription from the start site occurs only in male germ cells or in male germ cells and in some tumors, respectively.
Methylation of CpG-rich promoters in somatic tissues.
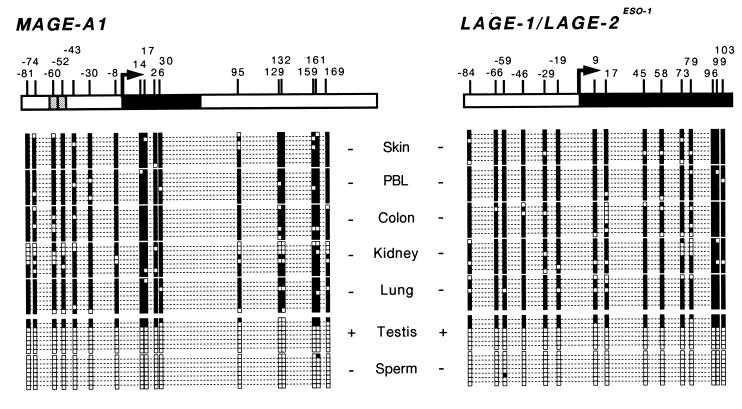
Since CpG-rich promoters are classically unmethylated in all normal cells, it was important to find out whether the methylation of MAGE-A1 observed in tumor cell lines that do not express this gene also occurred in normal somatic tissues, which never express the gene. The analysis of sequences derived from DNA of five normal human tissues treated with sodium bisulfite showed that the promoter and the first exon of MAGE-A1 were heavily methylated (Fig. 8). The methylation of MAGE-1 was not restricted to the inactivated X chromosome, as methylation was observed in tissues of male origin and on all copies in tissues of female origin.
FIG. 8.
Methylation pattern of MAGE-A1 and LAGE promoters in normal tissues. The gene fragments that were amplified from sodium bisulfite-modified DNA are represented. LAGE-1 and LAGE-2/ESO-1 promoter sequences are identical and could therefore not be distinguished. Black boxes correspond to the first exon. The broken arrow indicates the transcription start site, and the vertical bars correspond to the location of CpG dinucleotides. The methylation pattern deduced from eight sequences is represented for each DNA sample. Black squares correspond to methylated cytosines; empty squares correspond to unmethylated cytosines. + or − indicates whether the gene is expressed or not, respectively, in each tissue sample. The colon sample is of female origin. The other tissue samples are of male origin.
We also analyzed the methylation status of LAGE-1. Due to the high sequence similarity of LAGE-1 and LAGE-2/ESO-1, we were unable to distinguish between the promoter sequences of these two genes. However, as both genes display a MAGE-type expression, this should not confound the methylation analysis. Like the promoter of MAGE-A1, the LAGE promoters were heavily methylated in the normal somatic tissues that were analyzed.
Demethylation of MAGE-A1 and LAGE promoters in testis and sperm.
Among the MAGE-A1 and LAGE sequences obtained from testis, the vast majority was almost completely unmethylated, whereas a small minority was heavily methylated (Fig. 8). As the adult testicular tissue contains a majority of germ line cells and a minority (<15%) of somatic cells, these observations are consistent with the notion that these genes, which are expressed only in germ line cells (46), are also unmethylated only in these cells. Remarkably, both genes were also found to be demethylated in sperm, even though they are not expressed. This is presumably due to the general arrest of transcription in these cells.
DISCUSSION
We find that the 5′ region of gene MAGE-A1 is much less methylated in male germ line cells, which express MAGE-A1, than in all normal somatic cells, which do not express the gene. For tumor cells also, the 5′ end of MAGE-A1 is invariably much less methylated in those that express the gene than in those that do not. After treatment with 5′-aza-2′-deoxycytidine, stable expression of the gene is found in all the cells where the 5′ end of MAGE-A1 has been demethylated and only in those cells. Finally, transfection of the unmethylated MAGE-A1 5′ end region invariably results in transcriptional activity, even when applied to recipient cells that do not express the gene. In contrast, no transcription is observed when a heavily methylated MAGE-A1 sequence is transfected. All of these results are compatible with the notion that all normal and tumor cells carry the transcription factors necessary for MAGE-A1 expression, whether or not they express the gene, and that demethylation of the 5′ end of the gene is a necessary and sufficient condition for its expression.
The pattern of demethylation of the 5′ end of MAGE-A1 did not reveal individual CpG sites that were invariably methylated in silent cells and demethylated in cells that express the gene. However, the methylation of the five CpG located near the transcription initiation site appeared to correlate strongly with gene silencing. A complete correlation between gene silencing and the methylation of CpG sites around the transcription start site has been reported for genes on the inactive X chromosome (39, 40). Like MAGE-A1, these genes, namely, the HPRT and the PGK1 genes, contain a CpG-rich TATA-less promoter. It is tempting to propose that the methylation of CpG sites near the transcription start site inhibits the setting of the basal transcription machinery on these genes and therefore strongly represses their expression. This, of course, does not exclude that the methylation of CpG sites located further from the transcription initiation site contributes to the silencing of MAGE-A1, either by inhibiting the binding of Ets transcription factors (14) or by attracting methyl-binding proteins that induce a regional repressive chromatin structure (27, 38).
MAGE-A1 was found previously to be induced in cell cultures treated with 5′-aza-2′-deoxycytidine, a demethylating agent (14, 51). We now show that the expression of MAGE-A1 is maintained indefinitely after withdrawal of the demethylating agent in positive cell clones that were derived from the treated cell lines. Consistently, we showed that the 5′ region of MAGE-A1 is hypomethylated in positive cell clones that were cultured for more than 6 weeks after drug withdrawal. Hypomethylation of the MAGE-A1 promoter was still observed in these clones at day 101 after drug removal (data not shown). The maintenance of the MAGE-A1 promoter hypomethylation contrasts with the considerable variability of methylation that we observed at specific sites within each cell population. Similar variations in methylation patterns have been observed in other genes and probably reflect a dynamic process of methylation maintenance (39).
Other genes of the MAGE family, and genes of the GAGE and LAGE families were also shown to be activated by 5′-aza-2′-deoxycytidine treatment (10, 28, 32), and we found that their expression is stably maintained in clones derived from the treated tumor cell lines (data not shown). This suggests that the expression of most MAGE-type genes is regulated by DNA methylation.
The MAGE-type genes appear to be characterized by the presence of a CpG-rich region at their 5′ end. These CpG-rich regions match the commonly accepted CpG island criteria, as their percentage of G+C is over 50% and their ratio of observed to expected CpG is higher than or equal to 0.6. Whereas the CpG island in LAGE-1 has a size that is close to the average size of CpG islands in vertebrates (1,000 bp), the CpG islands in MAGE genes are smaller (300 to 650 bp). However, a survey of CpG islands in vertebrates shows that they have no typical size and range between 200 and 3,000 bp (29). Moreover, a lack of methylation in sperm DNA was proposed as a criterion for the designation of promoter regions as CpG islands (50). The 5′ regions of MAGE-type genes such as MAGE-A1 and LAGE-1 were found to be mostly unmethylated in sperm DNA.
Unlike all other CpG islands that are not subject to X inactivation or imprinting, the MAGE-type CpG islands are methylated in all normal somatic tissues. CpG-rich sequences showing high methylation in somatic tissues have been detected in repetitive sequences, like Alu or long interspersed nuclear elements (24, 53). MAGE-type 5′ regions do not contain such elements. The MAGE-type genes are demethylated in male germ line cells. The precise methylation pattern of MAGE-A1 and LAGE-1 promoters throughout the male germ lineage is not known. The MAGE-A1 protein has been detected in spermatogonia and in young spermatocytes (46). It is therefore likely that the promoter of MAGE-A1 is demethylated in these early stages of spermatogenesis. Our results suggest that this unmethylated status is maintained in the subsequent stages of spermatogenesis and in mature sperm, although MAGE-A1 is silent at these stages of sperm differentiation. The silencing of MAGE-A1 in these later stages of spermatogenesis coincides with the inactivation of the X chromosome, which is not associated with DNA methylation in male germ cells (19, 36). There is at present no evidence in favor of or against demethylation of MAGE-type genes in female germ line cells.
The methylation of CpG was proposed to be the origin of their relative scarcity in the mammalian genome, because of the high mutability of methylcytosine (1). The high density of CpG in CpG islands would therefore result from their general demethylation. The same explanation may apply to the MAGE-type CpG islands, notwithstanding their general methylation, because they are demethylated in germ line cells and may therefore be transmitted to the offspring without being subjected to the high mutation rate associated with cytosine methylation.
Remarkably, two previous suggestive examples of genes controlled by DNA methylation concerned germ line-specific genes, namely, the histone tH2B gene in rats (8) and the lactate dehydrogenase LDH-C gene in humans (6). DNA methylation appears therefore to be particularly suitable for the regulation of germ line-specific genes. This is probably related to the fact that these genes retain a high CpG content because their tissue-specific demethylation occurs in germ cells. On the other hand, the global demethylation process that occurs during the development of spermatogenic cells may provide the mechanism by which these germ line-specific genes are demethylated (17, 43), thus eliminating the need for specific demethylation factors. Although DNA methylation is appropriate for controlling germ line-specific expression, it seems that most testis-specific genes use other regulation mechanisms. This was suggested by the observation that, unlike MAGE genes, 14 randomly selected testis-specific genes were not induced in cell cultures treated with 5′-aza-2′-deoxycytidine (16).
Cancer cells often undergo a genomewide demethylation process, which increases during tumor progression (18, 22). Although this overall hypomethylation was initially expected to induce the aberrant expression of many genes in cancer cells, this appears not to be a common association (3). This observation probably reflects the fact that although many genes may be included in this demethylation process, most of them are not activated by this modification because of lack of transcription factors. The gene encoding testis-specific phosphoglycerate kinase 2 (PGK2) appears to provide a good example of such demethylation insensitive genes. PGK2 belongs to the testis-specific genes that were not induced in tumor cells upon 5′-aza-2′-deoxycytidine treatment (16). We have observed that the PGK2 promoter, which is largely methylated in most normal somatic tissues, is completely demethylated in some tumor cells where the gene is silent (data not shown). Hence, it seems likely that only genes with ubiquitous transcription factors, such as MAGE-A1 and LAGE-1, constitute targets for gene activation consequently to this genomewide hypomethylation in cancer cells.
DNA methylation does not seem to be the primary control mechanism regulating the programmed expression of most tissue-specific genes resulting in tissue differentiation. But our analysis of MAGE-type genes indicates that DNA methylation can serve as the primary control mechanism for the expression of a number of germ line-specific genes. Whether this mechanism is also applicable to some genes with specific expression in other tissues remains an open question.
ACKNOWLEDGMENTS
We are grateful to Adrian Bird for advice and critical review of the manuscript. We also thank Etienne De Plaen for comments on the manuscript. We thank Özlem Tureci, who kindly provided human DNA samples. The excellent technical assistance of M.-C. Letellier is gratefully acknowledged.
V.M. was supported by the Fonds pour la Recherche Scientifique dans l’Industrie et l’Agriculture (Belgium). This work was partially supported by the Belgian program on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister’s Office, Office for Science, Technology and Culture, by the Fonds Maisin, by the Association contre le Cancer (Belgium), by the CGER-Assurances (Belgium), and by the European Community program BIO-MED for Research and Technological Development.
REFERENCES
- 1.Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antequera F, Boyes J, Bird A. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;62:503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- 3.Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J-P. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 4.Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 5.Bird A P. Gene number, noise reduction and biological complexity. Trends Genet. 1995;11:94–100. doi: 10.1016/S0168-9525(00)89009-5. [DOI] [PubMed] [Google Scholar]
- 6.Bonny C, Goldberg E. The CpG-rich promoter of human LDH-C is differentially methylated in expressing and nonexpressing tissues. Dev Genet. 1995;16:210–217. doi: 10.1002/dvg.1020160213. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Scanlan M, Sahin U, Türeci Ö, Gure A, Tsang S, Williamson B, Stockert E, Pfreundschuh M, Old L. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc Natl Acad Sci USA. 1997;94:1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi Y-C, Chae C-B. DNA hypomethylation and germ cell-specific expression of testis-specific H2B histone gene. J Biol Chem. 1991;266:20504–20511. [PubMed] [Google Scholar]
- 9.Cross S H, Bird A P. CpG islands and genes. Curr Opin Genet Dev. 1995;5:309–314. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- 10.De Backer O, Arden K C, Boretti M, Vantomme V, De Smet C, Czekay S, Viars C S, De Plaen E, Brasseur F, Chomez P, Van den Eynde B, Boon T, van der Bruggen P. Characterization of the GAGE genes that are expressed in various human cancers and in normal testis. Cancer Res. 1999;59:3157–3165. [PubMed] [Google Scholar]
- 11.De Plaen E, Arden K, Traversari C, Gaforio J J, Szikora J-P, De Smet C, Brasseur F, van der Bruggen P, Lethé B, Lurquin C, Brasseur R, Chomez P, De Backer O, Cavenee W, Boon T. Structure, chromosomal localization and expression of twelve genes of the MAGE family. Immunogenetics. 1994;40:360–369. doi: 10.1007/BF01246677. [DOI] [PubMed] [Google Scholar]
- 12.De Plaen E, Lurquin C, Van Pel A, Mariamé B, Szikora J-P, Wölfel T, Sibille C, Chomez P, Boon T. Immunogenic (tum−) variants of mouse tumor P815: cloning of the gene of tum− antigen P91A and identification of the tum− mutation. Proc Natl Acad Sci USA. 1988;85:2274–2278. doi: 10.1073/pnas.85.7.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Smet C, Courtois S J, Faraoni I, Lurquin C, Szikora J-P, De Backer O, Boon T. Involvement of two Ets binding sites in the transcriptional activation of the MAGE1 gene. Immunogenetics. 1995;42:282–290. doi: 10.1007/BF00176446. [DOI] [PubMed] [Google Scholar]
- 14.De Smet C, De Backer O, Faraoni I, Lurquin C, Brasseur F, Boon T. The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proc Natl Acad Sci USA. 1996;93:7149–7153. doi: 10.1073/pnas.93.14.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Smet C, Lurquin C, van der Bruggen P, De Plaen E, Brasseur F, Boon T. Sequence and expression pattern of the human MAGE2 gene. Immunogenetics. 1994;39:121–129. doi: 10.1007/BF00188615. [DOI] [PubMed] [Google Scholar]
- 16.De Smet C, Martelange V, Lucas S, Brasseur F, Lurquin C, Boon T. Identification of testis-specific transcripts and analysis of their expression in tumor cells. Biochem Biophys Res Commun. 1997;241:653–657. doi: 10.1006/bbrc.1997.7868. [DOI] [PubMed] [Google Scholar]
- 17.del Mazo J, Prantera G, Miguel T, Ferraro M. DNA methylation changes during mouse spermatogenesis. Chromosome Res. 1994;2:147–152. doi: 10.1007/BF01553493. [DOI] [PubMed] [Google Scholar]
- 18.Diala E S, Cheah M S C, Rotwich D, Hoffman R M. Extent of methylation in human tumor cells. J Natl Cancer Inst. 1983;71:755–764. [PubMed] [Google Scholar]
- 19.Driscoll D J, Migeon B R. Sex differences in methylation of single-copy genes in human meiotic germ cells: implication for X chromosome inactivation, parental imprinting, and origin of CpG mutations. Somatic Cell Mol Genet. 1990;16:267–282. doi: 10.1007/BF01233363. [DOI] [PubMed] [Google Scholar]
- 20.Eden S, Cedar H. Role of DNA methylation in the regulation of transcription. Curr Opin Genet Dev. 1994;4:255–259. doi: 10.1016/s0959-437x(05)80052-8. [DOI] [PubMed] [Google Scholar]
- 21.Frommer M, MacDonald L E, Millar D S, Collis C M, Watt F, Grigg G W, Molloy P L, Paul C L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gama-Sosa M A, Slagel V A, Trewyn R W, Oxenhandler R, Kuo K C, Gehrke C W, Ehrlich M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 24.Hellmann-Blumberg U, Hintz M F, Gatewood J M, Schmid C W. Developmental differences in methylation of human Alu repeats. Mol Cell Biol. 1993;13:4523–4530. doi: 10.1128/mcb.13.8.4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones P A, Taylor S M, Mohandas T, Shapiro L J. Cell cycle-specific reactivation of an inactive X-chromosome locus by 5-azadeoxycytidine. Proc Natl Acad Sci USA. 1982;79:1215–1219. doi: 10.1073/pnas.79.4.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones P A, Wolkowicz M J, Harrington M A, Gonzales F. Methylation and expression of the Myo D1 determination gene. Philos Trans R Soc Lond Ser B. 1990;326:277–284. doi: 10.1098/rstb.1990.0011. [DOI] [PubMed] [Google Scholar]
- 27.Kass S U, Landsberger N, Wolffe A P. DNA methylation directs a time-dependent repression of transcription initiation. Curr Biol. 1997;7:157–65. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- 28.Kirillov A, Kistler B, Mostoslavsky R, Cedar H, Wirth T, Bergman Y. A role for nuclear NF-kB in B-cell-specific demethylation of the Igk locus. Nat Genet. 1996;13:435–441. doi: 10.1038/ng0895-435. [DOI] [PubMed] [Google Scholar]
- 29.Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- 30.Lethé B, Lucas S, Michaux L, De Smet C, Godelaine D, Serrano A, De Plaen E, Boon T. LAGE-1, a new gene with tumor specificity. Int J Cancer. 1998;76:903–908. doi: 10.1002/(sici)1097-0215(19980610)76:6<903::aid-ijc22>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 31.Lethé B, van der Bruggen P, Brasseur F, Boon T. MAGE-1 expression threshold for the lysis of melanoma cell lines by a specific cytotoxic T lymphocyte. Melanoma Res. 1997;7(Suppl. 2):S83–S88. [PubMed] [Google Scholar]
- 32.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;25:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 33.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 34.Lucas S, De Smet C, Arden K C, Viars C S, Lethé B, Lurquin C, Boon T. Identification of a new MAGE gene with tumor-specific expression by representational difference analysis. Cancer Res. 1998;58:743–752. [PubMed] [Google Scholar]
- 35.Lurquin C, De Smet C, Brasseur F, Muscatelli F, Martelange V, De Plaen E, Brasseur R, Monaco A P, Boon T. Two members of the human MAGE-B gene family located in Xp21.3 are expressed in tumors of various histological origins. Genomics. 1997;46:397–408. doi: 10.1006/geno.1997.5052. [DOI] [PubMed] [Google Scholar]
- 36.McCarrey J R, Berg W M, Paragioudakis S J, Zhang P L, Dilworth D D, Arnold B L, Rossi J J. Differential transcription of Pgk genes during spermatogenesis in the mouse. Dev Biol. 1992;154:160–168. doi: 10.1016/0012-1606(92)90056-m. [DOI] [PubMed] [Google Scholar]
- 37.Muscatelli F, Walker A P, De Plaen E, Stafford A N, Monaco A P. Isolation and characterization of a new MAGE gene family in the Xp21.3 region. Proc Natl Acad Sci USA. 1995;92:4987–4991. doi: 10.1073/pnas.92.11.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 39.Park J G, Chapman V M. CpG island promoter region methylation patterns of the inactive-X-chromosome hypoxanthine phosphoribosyltransferase (Hprt) gene. Mol Cell Biol. 1994;14:7975–7883. doi: 10.1128/mcb.14.12.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfeifer G P, Steigerwald S D, Hansen R S, Gartler S M, Riggs A D. Polymerase chain reaction-aided genomic sequencing of an X chromosome-linked CpG island: methylation patterns suggest clonal inheritance, CpG site autonomy, and an explanation of activity state stability. Proc Natl Acad Sci USA. 1990;87:8252–8256. doi: 10.1073/pnas.87.21.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riggs A D, Jones P A. 5-methylcytosine, gene regulation, and cancer. Adv Cancer Res. 1983;40:1–30. doi: 10.1016/s0065-230x(08)60678-8. [DOI] [PubMed] [Google Scholar]
- 42.Riggs A D, Pfeifer G P. X-chromosome inactivation and cell memory. Trends Genet. 1992;8:169. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- 43.Rocamora N, Mezquita C. Chicken spermatogenesis is accompanied by a genome-wide loss of DNA methylation. FEBS Lett. 1989;247:415–418. doi: 10.1016/0014-5793(89)81382-1. [DOI] [PubMed] [Google Scholar]
- 44.Stein R, Razin A, Cedar H. In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proc Natl Acad Sci USA. 1982;79:3418–3422. doi: 10.1073/pnas.79.11.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stögert R, Kubicka P, Liu C-G, Kafri T, Razin A, Cedar H, Barlow D P. Maternal specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- 46.Takahashi K, Shichijo S, Noguchi M, Hirohata M, Itoh K. Identification of MAGE-1 and MAGE-4 proteins in spermatogonia and primary spermatocytes of testis. Cancer Res. 1995;55:3478–3482. [PubMed] [Google Scholar]
- 47.Traversari C, van der Bruggen P, Van den Eynde B, Hainaut P, Lemoine C, Ohta N, Old L, Boon T. Transfection and expression of a gene coding for a human melanoma antigen recognized by autologous cytolytic T lymphocytes. Immunogenetics. 1992;35:145–152. doi: 10.1007/BF00185107. [DOI] [PubMed] [Google Scholar]
- 48.Van den Eynde B, Peeters O, De Backer O, Gaugler B, Lucas S, Boon T. A new family of genes coding for an antigen recognized by autologous cytolytic T lymphocytes on a human melanoma. J Exp Med. 1995;182:689–698. doi: 10.1084/jem.182.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van den Eynde B, van der Bruggen P. T cell defined tumor antigens. Curr Opin Immunol. 1997;9:684–693. doi: 10.1016/s0952-7915(97)80050-7. [DOI] [PubMed] [Google Scholar]
- 50.Walsh C P, Bestor T H. Cytosine methylation and mammalian development. Genes Dev. 1999;13:26–34. doi: 10.1101/gad.13.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber J, Salgaller M, Samid D, Johnson B, Herlyn M, Lassam N, Treisman J, Rosenberg S A. Expression of the MAGE1 tumor antigen is upregulated by the demethylating agent 5-aza-2′-deoxycytidine. Cancer Res. 1994;54:1766–1771. [PubMed] [Google Scholar]
- 52.Weih F, Nitsch D, Schütz G, Becker P B. Analysis of CpG methylation and genomic footprinting at the tyrosine aminotransferase gene: DNA methylation alone is not sufficient to prevent protein binding in vivo. EMBO J. 1991;10:2556–2567. doi: 10.1002/j.1460-2075.1991.tb07796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woodcock D M, Lawler C B, Linsenmeyer M E, Doherty J P, Warren W D. Asymmetric methylation in the hypermethylated CpG promoter region of the human L1 retrotransposon. J Biol Chem. 1997;272:7810–7816. doi: 10.1074/jbc.272.12.7810. [DOI] [PubMed] [Google Scholar]