Abstract
Growth factors and cytokines play an important role in supporting cellular viability of various tissues during development due to their ability to suppress the default cell death program in each cell type. To date, neither the triggering molecule nor the transduction pathway of these default apoptosis programs is understood. In this study, we explored the possibility that cytokine receptors are involved in modulating cytokine withdrawal-induced apoptosis (CWIA) in hematopoietic cells. Expression of the exogenous cytokine receptor common β chain (βc), but not the α chains, accelerated CWIA in multiple cytokine-dependent cell lines. Reduction of the expression level of endogenous βc by antisense transcripts resulted in prolonged survival during cytokine deprivation, suggesting a critical role of βc in modulating CWIA. Fine mapping of the βc subunit revealed that a membrane-proximal cytoplasmic sequence, designated the death enhancement region (DER), was critical to the death acceleration effect of βc. Furthermore, DER accelerated cell death either as a chimeric membrane protein or as a cytosolic protein, suggesting that DER functions independently of the cytokine receptor and membrane anchorage. Cross-linking of the chimeric membrane-bound DER molecules by antibody or of the FK506-binding protein–DER fusion protein by a synthetic dimerizing agent, AP1510, did not abrogate the death acceleration effect. Transient transfection assays further indicated that DER promoted cell death in the absence of serum in the nonhematopoietic 293 cell line. In summary, our data suggest that βc plays an important role in modulating CWIA via an anchorage-independent and aggregation-insensitive mechanism. These findings may facilitate further studies on the signaling pathways of CWIA.
During animal development, cells undergo vigorous selection in most tissues where only some will survive. The number of cells that survive within a tissue is thought to be determined by the concentration of its survival factor(s). In a variety of growth factor-dependent cell types, supplementation with exogenous survival growth factors usually results in hypertrophy of a given organ whereas withdrawal of survival factors induces programmed cell death, also called apoptosis (20). As the death-triggering molecule(s) is poorly defined, little is known about the signal transduction pathway of survival factor withdrawal-induced apoptosis. The hematopoietic system is one of the best-characterized examples where the population of each cell lineage is tightly regulated by cytokines (6). In response to an antigenic stimulus, the blood concentration of cytokines increases causing a rapid proliferation and accumulation of certain blood cells. When the antigen is later cleared, massive cell death occurs via apoptosis due to a lack of growth and survival cytokines. This prompt growth and death regulation of primary blood cells by cytokines is well preserved in many cytokine-dependent leukemic cell lines. Therefore, the cytokine-dependent hematopoietic cell line provides an excellent model for exploring the triggering mechanism of cytokine withdrawal-induced apoptosis (CWIA).
Granulocyte-macrophage colony-stimulating factor (GM-CSF), interleukin 5 (IL-5), and IL-3 are potent hematopoietic growth and survival factors that not only promote proliferation but also support survival via their membrane receptor complexes on the cell surface. Functional GM-CSF, IL-5, and IL-3 receptors are composed of a ligand-recruiting α chain, which is specific for each cytokine, and a signal-transducing β subunit (βc), which is shared by all three cytokine receptors (8, 24, 46, 54). Humans possess only one β subunit (hβc) gene, whereas mice have two highly related genes for the signal transducing subunit mβc and mβIL-3. mβc is 56% identical to hβc at the amino acid level and serves as the common subunit for murine IL-3 (mIL-3), GM-CSF, and IL-5 receptors. However, there is an extensive sequence homology between mβc and mβIL-3 (91% identity at the amino acid level). In addition, mβIL-3 forms a high-affinity receptor and transmits a proliferation signal with the mIL-3 receptor α subunit (mIL3Rα) but not with IL5Rα or the GM-CSF receptor α subunit (GMRα) (13, 15, 19). Upon ligand binding, βc becomes heavily tyrosine phosphorylated (46) and associates with many SH2-containing signaling proteins (45, 48). Although the antiapoptotic function of the activated receptor is not clearly elucidated (5), several observations suggest that the receptor α chain and the phosphotyrosine-mediated signals play important roles in activating the antiapoptotic signals. The activation of tyrosine phosphorylation and proliferation function of βc were shown to depend on the presence of the cytoplasmic domain of the receptor α chain (38, 48), which associates with JAK2 kinase. The deletion mutant of βc which lacks a tyrosine residue in the cytoplasmic domain (β590) not only partially lost mitogenic activity but completely lost its antiapoptotic function (4). Furthermore, a cytoplasmic region required for activation of the Ras/Raf/mitogen-activated protein kinase pathway is essential for βc to transduce cytokine-dependent survival activity (21, 23). Expression of activated Ras protein in trans complemented the defect in apoptosis prevention of the mutant βc and supported long-term proliferation in association with GM-CSF (22). Additionally, another serine/threonine kinase, Akt/PKB, was suggested to be involved in the antiapoptotic function of IL-3. Akt/PKB was activated by IL-3 in a phosphatidylinositol 3-kinase-dependent manner in the IL-3-dependent cell line Ba/F3. Active but not inactive forms of Akt/PKB were found to phosphorylate BAD, a distinct member of the Bcl-2 family that promotes cell death, in vivo and in vitro at the same residues that are phosphorylated in response to IL-3 (7, 49).
Like most apoptotic programs, CWIA in the IL-3-dependent cell line requires activation of the caspase-3 like proteases and is sensitive to caspase inhibitors (39). However, to date neither the triggering molecule nor the transduction pathway of this default apoptotic program is well understood. We therefore set out to determine whether the cytokine receptor itself was involved in CWIA. In this report, we show that the βc molecule plays an important role in modulating CWIA. The βc molecule promoted apoptosis via a cytoplasmic sequence, named the death enhancement region (DER), in a membrane anchorage-independent, aggregation-insensitive manner. The novel function of βc in the modulation of apoptosis may shed light on the mechanism of leukemogenesis of hematopoietic cells.
MATERIALS AND METHODS
Cell lines and culture conditions.
Ba/F3, a murine IL-3-dependent pro-B-cell line (41), was a gift from Atsushi Miyajima, Institute of Molecular and Cellular Biosciences, The University of Tokyo, Tokyo, Japan. Ba/F3 cells were maintained in 10% fetal calf serum (FCS)-containing RPMI 1640 medium supplemented with 55 μM β-mercaptoethanol and 10 U of mIL-3 per ml. HT-2, an mIL-2-dependent T-lymphocytic cell line (55) purchased from the American Type Culture Collection (Rockville, Md.), was maintained in a medium similar to that for Ba/F3 cells but supplemented with 10 U of mIL-2 per ml. TF-1, a human GM-CSF- or IL-3-dependent erythroleukemic cell line (25), was a gift from Toshio Kitamura, Department of Hematopoietic Growth Factor, University of Tokyo. TF-1 cells were maintained in a medium similar to that for Ba/F3 but supplemented with 20 U of human GM-CSF per ml. Teo4 is a pTet-Off (14) (Clontech, Palo Alto, Calif.)-transfected TF-1 cell line and was maintained in TF-1 culture medium plus 200 μg of G418 per ml. Antisense hβc transfectants β(AS)1 and β(AS)2 were maintained in the same medium as that used for Teo4, with the inclusion of hygromycin B (200 μg/ml). The ectotropic package cell line BOSC23 (43) and the human embryonic kidney cell line 293 were purchased from the American Type Culture Collection and maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FCS.
Plasmid DNA construction.
Retroviral expression plasmids for wild-type hβc (pBabe- hβc) and hGMRα were constructed by inserting the coding sequences from pKH97 (16) and pKH125 (47) (gifts from A. Miyajima) into the retroviral expression vector pBabeHygro and pBabeNeo (32) (gifts from Hartmut Land, Imperial Cancer Research Fund, London, England), respectively, by standard molecular cloning methods.
The antisense hβc plasmid was constructed by inserting the full-length hβc into the tetracycline-regulatable vector pTRE (Clontech) in a reverse orientation.
Deletion mutants of hβc were created by inserting the Stop linker (top oligonucleotide, 5′ CCCGGGTTAACTTAACTTAAGGATCC 3′; bottom oligonucleotide, 5′ GGATCCTTAAGTTAAGTTAACCCGGG 3′) into the FspI, ApaI, and MunI sites of the hβc cDNA to generate β470, β590, and β772, respectively. To generate mutants β512, β590(Δ473-505), and β560(Δ473-505), the wild-type cytoplasmic domain of hβc was replaced with PCR-amplified DNA fragments at the FspI site. Primers 5′ CGCACGCGTAGAAAGTGGGAGGAGAAG 3′ (5′ primer) and 5′CGGGAATTCTCACCCCTGGTGTGGGGGACT 3′ (3′ primer) were used for the construction of β512, and primers 5′ CGCACGCGTAGTCCCCCACACCAGGGG 3′ (5′ primer) and 5′ CGCGAATTCTCAATCTGAGGCAGCTGGAGT 3′ (3′ primer) were used for the construction of β560(Δ473-505). The 5′ primer of β560(Δ473-505) and the bottom primer of the Stop linker were used to generate β590(Δ473-505), with the mutant β590 used as the template DNA.
Plasmids pCD16/7/β471-590 and pCD16/7/Stop were both derived from plasmid pCD16/7/Syk (30) (provided by Tadatsugu Taniguchi, Institute for Molecular and Cellular Biology, Osaka University, Osaka, Japan). The Stop linker (described above) was inserted into the blunted MluI site at the junction between CD7 and Syk cDNA to generate pCD16/7/Stop. For the construction of pCD16/7/β471-590, the PCR fragment encoding amino acids (aa) 471 to 590 of hβc (amplified by the 5′ primer of β512 and the bottom primer of the Stop linker with β590 as the template DNA) was digested with XbaI and MluI and ligated with the XbaI-MluI DNA fragment containing the CD16/7 extracellular and transmembrane domain to generate the CD16/7/β471-590 chimeric molecule. The resulting plasmid was cut at the XbaI site and blunted and then cut at the EcoRI site to release the entire coding sequence. This fragment was subcloned into the SnaBI and EcoRI sites of pBabeHygro (32) to generate the retroviral expression plasmid pCD16/7/β471-590.
The cDNA sequence encoding aa 471 to 590 of hβc was modified to be flanked by the MluI restriction enzyme site and the stop codon by using standard PCR technique. This PCR product was cloned into the EcoRI site of pcDNA3.1.His-A (Invitrogen) to generate plasmid pcDNA-3/ hβc(471-590). This plasmid encodes a cytoplasmic DER protein that fuses to the hexahistidine tag. An extra IRLRP pentapeptide sequence was created between the expression vector and the hβc coding sequence as a result of the plasmid construction strategy. The entire coding sequence of this chimeric cDNA was then transferred to the retroviral expression vector pBabeNeo by inserting the PmeI fragment into the SnaBI site of pBabeNeo to generate pBabeHisβDER.
Plasmids pBabeF1E and pBabeF1βE were both derived from pCF1E (Ariad Pharmaceuticals, Inc., Cambridge, Mass.). The nucleotide sequence of βc encoding aa 466 to 590 was amplified by PCR and subcloned into the SpeI site of pCF1E to generate pCF1βE. The flanking restriction sites of the entire coding sequences of pCF1E and pCF1βE were modified and subcloned into the SnaBI and BamHI sites of pBabeNeo to generate plasmids pBabeF1E and pBabeF1βE.
Gene transfer by retroviral infection and lipofection.
For retroviral infection, 12 μg of plasmid DNA was transfected into 3 × 106 cells of the packaging cell line BOSC23 (42) with Lipofectamine (Life Science, Gaithersburg, Md.) according to the manufacturer’s protocol, and the 24-h conditioned media from the transfected cells were used to infect mIL-3-dependent cell line Ba/F3, using the standard protocol. The stable infectants were selected in medium containing hygromycin B (500 μg/ml) or G418 (800 μg/ml), and the expression of receptor subunits was confirmed by flow cytometry and Western blot analysis.
TF-1 cells were transfected as previously described (4). Briefly, 2 μg of plasmid DNA was mixed with 2.5 μl of DMRIE-C (Life Science) in 0.5 ml of serum-free DMEM and incubated at room temperature for 30 min; 1.2 × 106 TF-1 cells were washed and resuspended in 50 μl of serum-free DMEM. Cells were mixed with DMRIE-C-bound plasmid DNA and incubated at 37°C for 5 h. The transfected cells were washed twice and resuspended in fresh growth medium for 24 to 48 h before G418 or hygromycin B was added to select for the stable clones.
For each transfection experiment, more than 20 independent subclones were first analyzed by flow cytometry (see below). All candidate clones were further analyzed by Western blotting (see below). More than three positive transfectants were subjected to functional analysis. Since the properties of all positive clones were similar, only the results from one representative clone, unless specifically indicated, are presented and discussed.
Antibodies, immunoprecipitation, Western blot analysis, and antibody cross-linking.
Antibodies were as follows; for immunoprecipitation and flow cytometric analysis of hβc, S-16 (catalog no. sc-457; Santa Cruz Biotechnology, Santa Cruz, Calif.); for Western blot analysis of the carboxyl terminus of hβc, C-20 (catalog no. sc-675; Santa Cruz); for the amino terminus of hβc, N-20 (catalog no. sc-676; Santa Cruz); for immunoprecipitation, flow cytometric analysis, and Western blot analysis of human GMRα (hGMRα), S-50 (catalog no. sc-456; Santa Cruz); for Western blot analysis of phosphotyrosine, PY20 (catalog no. p11120; Transduction Laboratories, Lexington, Ky.); for immunoprecipitation and Western blot analysis of STAT3, C-20 (catalog no. sc-482; Santa Cruz). The monoclonal antibody for immunoprecipitation, flow cytometric analysis, and cross-linking of CD16 was LNK16 (catalog no. MCA1193XZ; Serotec, Oxford, England), and that for Western blot analysis of CD16 was 2H7 (catalog no. NCL-CD16; Novocastra, Newcastle upon Tyne, England).
Cells were lysed in radioimmunoprecipitation assay lysis buffer, and the cell lysates were subjected to immunoprecipitation and Western blot analysis as previously described (4) except that the bands of interest were visualized with an enhanced chemiluminescence (ECL) detection system (Amersham, Little Chalfont, England). When reprobing the same blot with another antibody was necessary, membranes were treated in stripping buffer (100 mM 2-mercaptoethanol, 2% sodium dodecyl sulfate, 62.5 mM Tris-HCl [pH 6.7]) at 55°C for 30 min prior to reprobing.
For antibody cross-linking experiments, cells were initially incubated with 2.5 μg of anti-CD16 monoclonal antibody LNK16 per 107 cells in 0.5 ml of medium at room temperature for 15 min. Cells were then cross-linked with 7.5 μg of goat anti-mouse immunoglobulin G1 antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.) at 37°C for 15 min before harvesting and analysis.
Cell viability, proliferation, and DNA fragmentation assay.
Cell viability was determined by trypan blue exclusion staining. Proliferation activity was measured by a colorimetric assay with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) as a substrate as described by Mosmann et al. (33). For detection of the DNA ladder during cell death, cells were treated as described in the text and subjected to lysis and agarose gel electrophoresis as previously described (59).
Flow cytometric analysis of surface receptor expression.
For analyzing surface receptor expression, 1 million cells were washed and resuspended in 1 ml of staining buffer (2% FCS and 0.1% sodium azide in phosphate-buffered saline). Cells were pelleted after 30 min on ice and mixed with 0.5 μg of primary antibody. After incubation, cells were mixed with fluorescein isothiocyanate-conjugated goat anti-mouse secondary antibody. Fluorescence intensity was analyzed by FACScan (Becton Dickinson, Mountain View, Calif.).
RT-PCR analysis.
A single-tube reverse transcription (RT)-PCR format was used to analyze the expression of the exogenous genes. The cytoplasmic RNA was prepared from the target cells and subjected to RT-PCR analysis as specified for the Access RT-PCR system (Promega, Madison, Wis.).
RESULTS
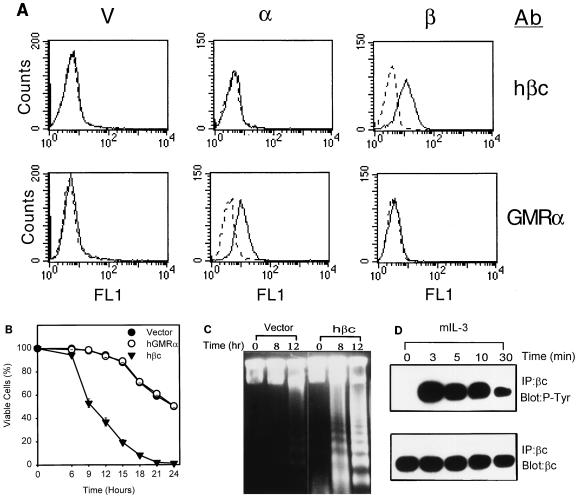
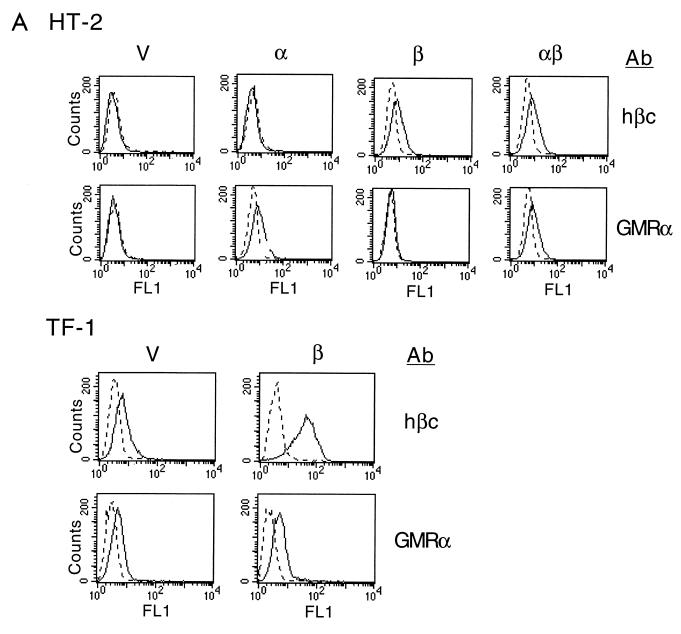
In this study, we explored the potential role of cytokine receptor subunits in the modulation of CWIA. To this end, multiple stable transfectants of mIL-3-dependent cell line Ba/F3 expressing hGMRα (12) or hβc (16) were established and subjected to survival rate measurement by trypan blue staining during mIL-3 deprivation. All transfectants expressing similar amounts of the same type of receptor subunit behaved similarly, and the results of one representative clone of each type are shown in Fig. 1. The surface expression of each receptor subunit in individual stable clones was confirmed by flow cytometric analysis using antibodies specific for hGMRα and hβc (Fig. 1A). While cells expressing hGMRα alone showed death kinetics similar to that of control cells transfected with the retroviral vector alone (half-life [t1/2] ≅ 24 h), cells expressing hβc alone manifested an accelerated death rate after deprivation of mIL-3 (t1/2 ≅ 9 h) (Fig. 1B). Overexpression of the human IL-5 receptor α chain (hIL5Rα) (59) in Ba/F3 cells did not alter the death rate (data not shown). Accelerated death caused by hβc overexpression was mainly due to apoptosis, which was demonstrated by a DNA fragmentation assay (58) (Fig. 1C) and an annexin V binding assay (data not shown). One clone of the hβc transfectant showed an extensive DNA oligonucleosomal ladder at 8 h after deprivation of mIL-3, while control cells did not show apoptotic DNA laddering even up to 12 h (Fig. 1C). To exclude the possibility that hβc accelerates CWIA by interfering with the mIL-3 responsiveness of the host cells, the half-maximum effective dose (ED50) of mIL-3 for an hβc transfectant was determined and shown to be the same as that of the parental Ba/F3 cells (data not shown). The hβc was heavily tyrosine phosphorylated 3 min after stimulation with mIL-3 (Fig. 1D), suggesting that hβc formed a hybrid functional receptor complex with mIL3Rα and was involved in growth signaling of mIL-3 in hβc transfectant cells. To further support the apoptosis-enhancing role of hβc, we expressed hβc in an IL-2-dependent cell line, HT-2 (Fig. 2A), and measured the apoptotic rates of the transfectants. As with Ba/F3 cells, HT-2 subclones expressing the control vector or hGMRα manifested a death rate similar to that of the parental cells (Fig. 2B). Although the expression level of hβc in HT-2 cells was about half of that seen in Ba/F3 cells, HT-2 subclones expressing hβc or hβc plus hGMRα still showed accelerated death and survival reduction by 27% compared to control cells after 24-h depletion of IL-2 (Fig. 2B). Coexpression of hGMRα in hβc-transfected Ba/F3 or HT-2 cells allowed cell growth in human GM-CSF-containing medium (data not shown) but did not alter the accelerated apoptotic rate of hβc transfectants in the absence of cytokines (Fig. 2B). Overexpression of hβc in human GM-CSF-dependent cell line TF-1 (25) (Fig. 2A) also led to an increase in the apoptotic rate by 22% compared to that of vector-expressing TF-1 cells (Fig. 2B). In conclusion, expression of exogenous hβc in both human and murine hematopoietic cell lines accelerates their death rates in the absence of survival factors.
FIG. 1.
Ectopic expression of cytokine receptor hβc, but not hGMRα, accelerates CWIA in Ba/F3 cell line. (A) Surface expression of hβc and hGMRα in Ba/F3 cells. The cells were incubated with receptor specific antibodies (Ab; indicated at the right) and fluorescein isothiocyanate-conjugated secondary antibody. Fluorescence intensity is shown on the x axis, and cell numbers are shown on the y axis. The background fluorescence was detected with a nonimmunized serum and is shown as a dashed line. V, vector-expressing control cells; α, hGMRα-expressing subclone; β, hβc-expressing subclone. (B) Death acceleration by hβc in the mIL-3-dependent Ba/F3 cell line. Ba/F3 cells expressing various exogenous genes were depleted of mIL-3 for various time periods as indicated, and viable cell numbers were determined by trypan blue exclusion assay. Each curve represents the average values of three independent clones expressing vector, hGMRα, or hβc cDNA. (C) Human βc accelerates death via apoptosis. Genomic DNA of hβc- and vector-expressing Ba/F3 cells were prepared from cells treated as described for panel A and analyzed in agarose gel to show the oligonucleosomal DNA ladder. (D) Human βc participates in mIL-3 signaling. Human βc-expressing Ba/F3 cells were cultured in cytokine-free medium for 12 h and restimulated by mIL-3 at 37°C for various time intervals as indicated. Protein lysates were prepared and subjected to immunoprecipitation (IP) with antibody specific to human βc (sc-457) and then to Western blot analysis with antiphosphotyrosine antibody PY20 (P-Tyr) or anti-hβc antibody (βc). The specific bands were visualized by ECL.
FIG. 2.
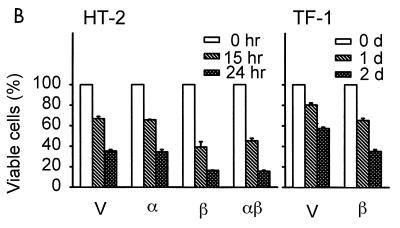
Ectopic expression of cytokine receptor hβc, but not hGMRα, accelerates CWIA in HT-2 and TF-1 cells. (A) Surface expression of receptor molecules in HT-2 and TF-1 cells. Various stable transfectants were stained with antibodies (Ab) and analyzed as described in the legend to Fig. 1. V, vector-expressing control cells; α, hGMRα-expressing subclone; β, hβc-expressing subclone; αβ, hGMRα- and hβc-coexpressing subclone. (B) Death acceleration by hβc in HT-2 and TF-1 cell lines. Stable transfectants of the mIL-2-dependent helper T-cell line HT-2 expressing the indicated genes were depleted of mIL-2 for 0, 15, and 24 h; viable cell numbers were determined and are shown at the left. Stable transfectants of the human GM-CSF-dependent cell line TF-1 expressing the indicated plasmids were depleted of human GM-CSF for 0, 1, and 2 days (d); viable cell numbers were determined and are shown at the right. For most of the genes, each value is the average of two independent duplicated experiments of one stable clone; the value for the hβc gene is the average of two independent stable clones.
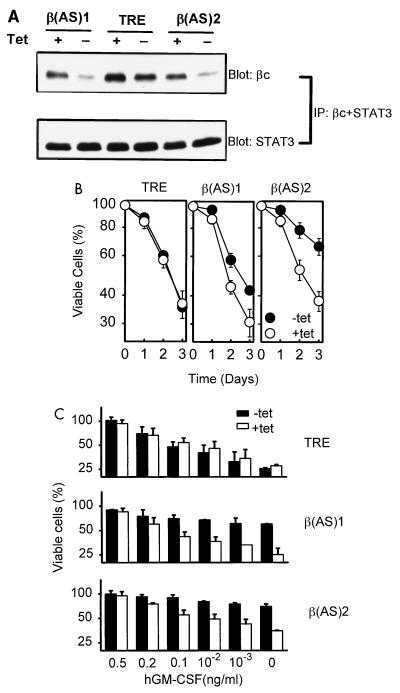
These results suggest that the level of expression of βc in a given cell may determine the rate of apoptosis. To further explore this, we investigated whether down-regulation of the endogenous hβc protein would ameliorate CWIA. We transfected the antisense hβc plasmid into Teo4 cells, a derivative of human TF-1 cell line suitable for inducible expression by tetracycline withdrawal, and selected for hygromycin-resistant clones. Two representative stable clones, β(AS)1 and β(AS)2, were characterized. Upon removal of tetracycline, expression of hβc proteins were reduced to about one-third of that of the control (Fig. 3A) in both cell lines. The antisense hβc transcripts were specific for hβc mRNA and had no effect on the levels of hGMRα (data not shown) or of STAT3 (Fig. 3A). CWIA delay in these cells was demonstrated by two methods. First, a time course study was performed by trypan blue exclusion assay (Fig. 3B). Under conditions used in this study, differences of the survival cell number between cells cultivated in medium with or without Tet [Tet(+) or Tet(−) medium] were discernible 24 h after GM-CSF starvation, and the difference became even more prominent thereafter. Seventy-two hours after cytokine depletion, β(AS)1 cell viability was 30% in Tet(+) medium and 42% in Tet(−) medium; β(AS)2 cell viability was 38 and 66%, respectively. Second, the 48-h time point was chosen to repeat the measurement of increased survival at various concentrations of GM-CSF (below the ED50) by MTT dye reduction assay (33). In these experiments, 2 × 104 cells were seeded per well. After 48 h cultivation, survival percentage was referred to the optical density reading of each sample compared to that of initiating cells. In the absence of tetracyline, the survival cell numbers at all concentrations of GM-CSF tested increased (Fig. 3C). The lower the concentration of GM-CSF, the more profound the protection effects. In GM-CSF-free medium, survival cell numbers increased from 25% in Tet(+) medium to 60% in Tet(−) medium for β(AS)1 cells and from 35 to 70%, respectively, for β(AS)2 cells (Fig. 3C). The antisense construct did not completely prevent CWIA in these experiments, possibly due to incomplete ablation of hβc expression and/or because hβc may not be the only cytokine receptor which can modulate CWIA in TF-1 cells (see below). Nevertheless, CWIA was dramatically inhibited when the expression level of endogenous hβc was reduced. Therefore, not only exogenous hβc but also endogenous hβc regulates the death rate in a seemingly dose-dependent manner.
FIG. 3.
Antisense hβc transcript delays CWIA of GM-CSF-dependent cells. (A) Suppression of endogenous hβc protein expression by the antisense hβc plasmid in stable cell lines. Two antisense hβc-expressing TF-1 subclones [β(AS)1 and β(AS)2] and control cells (TRE) were cultured with (+) or without (−) tetracycline (2 μg/ml) for 48 h. Cell lysates were then prepared and subjected to immunoprecipitation (IP) and Western blot analysis (Blot) with anti-hβc antibody (βc) and anti-STAT3 antibody (STAT3), respectively. (B) Time course study of retarded CWIA of stable cell lines expressing hβc antisense RNA. Viable cell numbers of each cell line were determined by trypan blue staining and are relative to that of initial seeded cell number during cytokine depletion in Tet(+) or Tet(−) medium. The values are averages of two independent experiments performed in duplicate. (C) Survival effect of antisense hβc in low-GM-CSF medium. Experiments were conducted as for panel B except that various low concentrations of human GM-CSF were used in the presence (open bar) or absence (solid bar) of tetracycline and samples were analyzed only at the 48-h time point by MTT dye reduction assay. Percentage of survival is relative to that of cells initially seeded. The values are averages of two independent experiments performed in triplicate.
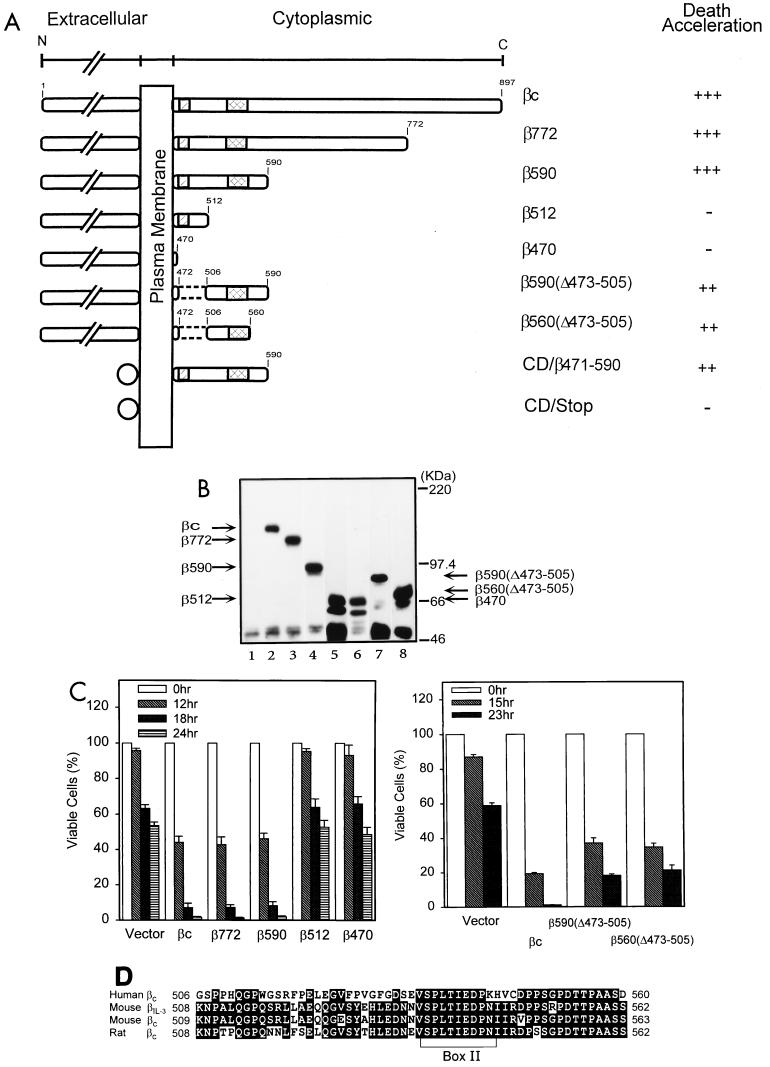
The structural domains essential for transducing proliferation signals of the hβc molecule have been mapped (45, 47). To determine whether the domains needed for proliferation overlap with that for apoptosis modulation, we constructed a series of deletion mutants to delineate sequence domains of hβc critical for the death acceleration effect (Fig. 4A). These constructs were introduced into Ba/F3 cells by retroviral infection, and the hygromycin-resistant clones were analyzed and verified for the expression of hβc mutants by flow cytometry (data not shown) and Western blot analysis (Fig. 4B). At least three independent clones for each mutation were subjected to survival kinetics study after depletion of mIL-3. The death acceleration ability of each mutation is summarized in Fig. 4A. The hβc mutations with C-terminal deletions up to aa 773 (β772) or 591 (β590) retained full death acceleration ability. However, with further deletions to aa 513 (β512) and 471 (β470), death-promoting activity was completely lost (Fig. 4C). Internal deletion of the DNA sequence containing the conserved box I (31, 34, 40) from both β590 [i.e., β590(Δ473-505)] and β560 [i.e., β560(Δ473-505)] did not abolish, but resulted in a slight reduction in, death acceleration ability (Fig. 4C). These data suggest that the box I sequence is not essential for the apoptosis-enhancing activity of hβc per se but may be required for optimizing its death activity. The minimum sequence essential for accelerating apoptosis, designated the DER, is mapped by this deletion analysis to a 54-aa peptide fragment from aa 506 to 560 (Fig. 4D). In contrast to the dependence on the presence of hGMRα for transducing proliferation signals of GM-CSF (24), coexpression of hGMRα with all hβc mutants tested did not alter the survival kinetics in the absence of cytokines (data not shown).
FIG. 4.
Mapping of the DER of hβc. (A) Schematic depiction of cytoplasmic deletion mutants of cytokine receptor hβc. Position 1 is the first methionine of the open reading frame βc, full-length wild-type molecule of hβc; hatched and double-hatched boxes, conserved cytoplasmic box I and box II sequences, respectively; dashed lines, the internal deletion; circles, extracellular domains of the chimeric CD16/7/β471-590 and control CD16/7/Stop molecules. Death acceleration is indicated as +++ (strong enhancement [decrease of 50 to 60% of survival cells compared to the control]), ++ (moderate enhancement [decrease of 20 to 40%]), and − (no enhancement). (B) Expression of the mutant hβc proteins in Ba/F3 cells. Stable Ba/F3 subclones infected with vector pBabeHygro (lane 1) or with hβc (lane 2), β772 (lane 3), β590 (lane 4), β512 (lane 5), β470 (lane 6), β590(Δ473-505) (lane 7), and β560(Δ473-505) (lane 8) were lysed and subjected to Western blot analysis using a polyclonal antibody specific to the extracellular domain of the hβc protein. The predicted position of each mutant hβc in the blot is indicated by arrows. (C) Survival of cells expressing various hβc mutants during the course of cytokine deprivation. Viability of cells expressing various hβc mutants at various time points were measured as described in legend to Fig. 1A. Each number is the average of two experiments of two to three independent subclones of each mutation. (D) Alignment of DER sequences of different species of βc. The amino acid sequence from 506 to 560 of human βc was compared to equivalent regions from mβIL-3, mβc, and rat βc. Identical amino acids are shown as white letters; the conserved box II sequence is indicated by a bracket.
These observations question whether DER can accelerate apoptosis via a mechanism independent of the cytokine receptor complex. To address this question, we fused the cDNA sequence encoding aa 471 to 590 of hβc, which contains DER, to a heterologous transmembrane protein, the chimeric CD16-CD7 molecule (30) (Fig. 4A, CD/β471-590), to test the ability of this chimeric hβc molecule to affect CWIA. The construct CD16/7/Stop (Fig. 4A, CD/Stop), which contains no cytoplasmic domain, was introduced to serve as a negative control. Surface expression of these chimeric CD16 antigens in all transfectants was confirmed by flow cytometry (Fig. 5A) and Western blot analysis (Fig. 5B). Ba/F3 cells expressing CD16/7/Stop did not show an altered rate of CWIA compared to parental cells (Fig. 5D, CD/S), whereas Ba/F3 cells expressing CD16/7/β471-590 showed moderately accelerated CWIA compared to parental cells, with the decrease of viable cells from 80 to 50% at the 12-h time point (Fig. 5D, CD/β). These data suggest that promotion of CWIA is independent of the GM-CSF receptor complex and is mediated via cis element(s) distinct from those required for proliferation.
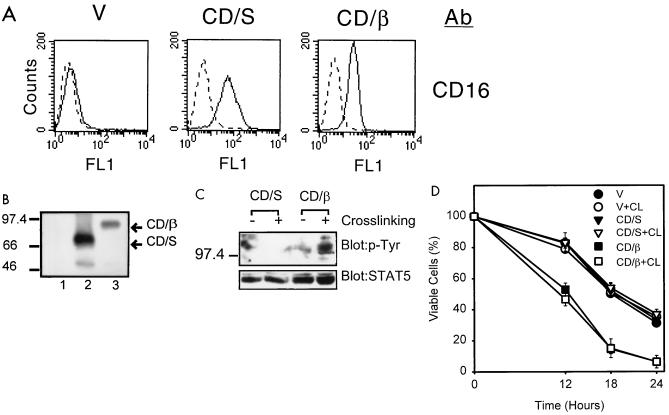
FIG. 5.
Physical aggregation did not abrogate the function of DER. (A) Surface expression of the chimeric CD16-DER molecules in Ba/F3 cells. The stable cell lines were treated with anti-CD16 antibody (Ab) and analyzed by flow cytometry as described for Fig. 1. CD/S, pCD16/7/Stop-expressing cell line; CD/β, pCD16/7/β471-590-expressing cell line. (B) Protein expression of the CD16 chimeric genes. Cell lysates were prepared from control cells (lane 1), CD/S cells (lane 2), and CD/β cells (lane 3) and were subjected to Western blot analysis with the anti-CD16 antibody. Sizes are indicated in kilodaltons. (C) Cross-linking of the DER sequence of hβc by anti-CD16 antibody. Ba/F3 subclones as described for panel A were cross-linked with (+) or without (−) anti-CD16 antibody. The total cell lysates were prepared and analyzed for the protein tyrosine phosphorylation by Western blot analysis using antiphosphotyrosine (anti-p-Tyr) antibody. The STAT5 protein levels are shown as loading controls for each lane. (D) The survival rate of cells expressing various CD16/7 chimera with or without antibody cross-linking (CL). Cells as indicated were treated as described for panel B and were depleted of cytokine. Viable cell numbers were determined at various time points after cytokine depletion; the values are averages of two independent experiments performed in duplication. V, vector-expressing cells.
By taking advantage of the presence of the CD16 tag in the chimeric CD16/7/β471-590 protein, we further tested whether dimerization or aggregation of the DER sequence would abrogate death acceleration activity. Since both box I and box II motifs of hβc are present in the chimeric CD16/7/β471-590 molecule and the JAK kinases were reported to be preassociated with the box I sequence (38, 43), cross-linking of this molecule is likely to activate the associated tyrosine kinases and result in tyrosine phosphorylation of certain cellular proteins. As indicated by the appearance of tyrosine-phosphorylated protein signals in Western blot analysis (Fig. 5C, rightmost lane), we successfully cross-linked the surface CD16/7/β471-590 molecules. Several tyrosine-phosphorylated cellular proteins (ranging from 97 to 150 kDa) were detectable in CD16/7/β471-590-expressing cells when the surface CD16 was cross-linked. Upon aggregation, the CD16/7/β471-590 molecules retained their ability to accelerate apoptosis (Fig. 5D, CD/β + CL). At 18 h, the surviving cells decreased from 55% for all control cells to 15% for CD16/7/β471-590-expressing cells regardless of cross-linking with the antibody. Our data strongly suggest that physical aggregation caused by antibody cross-linking does not down-regulate the death acceleration activity of the DER sequence. Given the fact that subclones expressing CD16/7/β471-590 grew well in mIL-3-containing medium and that hβc-expressing HT-2 cells grew satisfactorily in mIL-2, cytokines obviously abrogate in trans the apoptosis enhancing activity of DER.
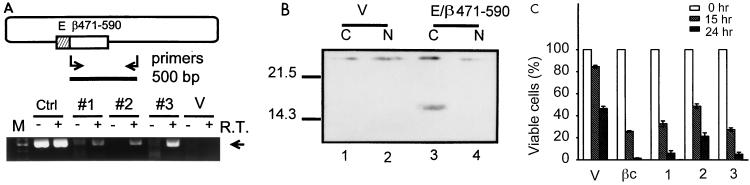
Many membrane proteins function in an anchorage-dependent manner, due to the special membrane localization of their signaling components. To further understand the mechanism of death promotion by hβc and to explore whether this apoptosis acceleration activity is anchorage dependent, we constructed the retroviral expression plasmid pBabeHisβDER, encoding a hexahistidine-tagged cytoplasmic DER of hβc, and established several pBabeHisβDER-expressing cell lines by retroviral infection. Although the protein product of pBabeHisβDER was readily detectable by Western blot analysis in a transient transfection assay with HeLa cells (data not shown), the expression level of the cytoplasmic chimeric hβc protein in these stable lines was very low and undetectable by a conventional Western blot analysis. Instead, we demonstrated the presence of the RNA transcript of pBabeHisβDER by RT-PCR using a set of transgene specific oligonucleotide primers as depicted in Fig. 6A. Three pBabeHisβDER-expressing clones, but not the control vector transformant cells, showed a reverse transcriptase-dependent 500-bp specific PCR product (Fig. 6A, lanes 4, 6, and 8 versus lane 10). Additionally, we enriched the histidine tag-containing proteins with Ni-nitrilotriacetic acid beads from about 2 mg of the cytoplasmic and nuclear proteins and analyzed these protein samples as for conventional Western blotting except that the histidine tag-containing proteins were probed with horseradish peroxidase-labeled nickel (Kirkegaard & Perry Laboratories, Gaithersburg, Md.). As shown in Fig. 6B, a 14-kDa protein was detected in the cytoplasmic fraction of pBabeHisβDER-expressing cells (lane 3) but not in the nuclear fraction or in vector-expressing cells (lanes 1, 2, and 4). In the death rate determination experiment (Fig. 6C), clones 1 and 3 showed death kinetics comparable to that of the wild-type hβc transformant (columns 1 and 3 versus column V). Clone 2 showed a death rate slightly lower than the wild-type hβc control rate but still much higher than the control rate (column 2). Therefore, the DER sequence is capable of accelerating CWIA in an anchorage-independent manner.
FIG. 6.
The DER functions via an anchorage-independent mechanism. (A) Expression of the cytoplasmic hexahistidine-tagged DER in Ba/F3 cells. In the upper portion, the plasmid pBabeHisβDER is shown as an oblong. The hatched and open bars represent the histidine epitope tag from pcDNA3.1 (E box) and the hβc cytoplasmic domain (aa 471 to 590; containing the DER sequence), respectively. The pair of arrows and solid bar indicate the oligonucleotide primers used for RT-PCR and the 500-bp PCR product, respectively. Cytoplasmic RNA prepared from pBabeHisβDER-expressing Ba/F3 cells (clones 1 to 3) and from vector-expressing cells (clone V) were subjected to RT-PCR. Reactions performed without RT served as controls for genomic DNA contamination. A plasmid DNA control (lane Ctr1) was included to indicate the position of the PCR product (arrow). M, 1-kb DNA marker. (B) Expression of cytoplasmic histidine-tagged DER protein. The cytosolic and nuclear (N) extracts were prepared as described by Dignam et al. (10). The histidine tag-containing proteins were then enriched by passing 2 mg of extracts through Ni-nitrilotriacetic acid beads and were analyzed as for conventional Western blot analysis except that the proteins were probed with horseradish peroxidase-labeled nickel. The signals were visualized with the ECL system. (C) Moderate acceleration of CWIA by cytosolic DER sequence. Cells expressing indicated plasmids were subjected to cytokine deprivation and viable cell measurement as described for Fig. 1A. A cell line expressing wild-type hβc (βc) was included as a positive control. The pBabe vector-containing cell line (V) was included as a negative control. Sizes are indicated in kilodaltons.
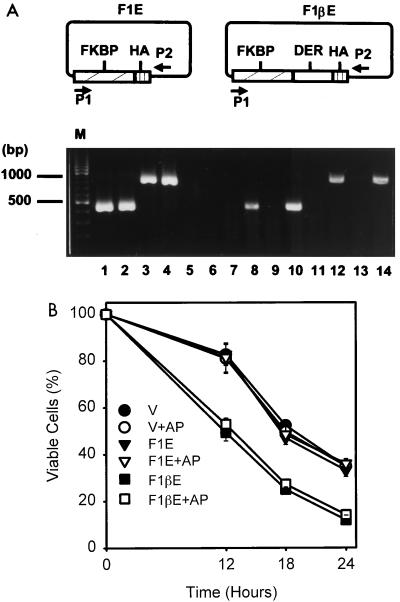
To further elucidate the mode of DER’s function, we expressed cytoplasmic DER as an FK506-binding protein (FKBP) fusion protein to explore the effect of dimerization on the deleterious function of DER. Intriguingly, the cytoplasmic FKBP-DER fusion protein was again undetectable in stable transfectants with anti-FKBP or hemagglutinin antigen antibody even though it was highly expressed in a transient transfection experiment (data not shown). An RT-PCR analysis was carried out instead to demonstrate the expression of the fusion gene. Using the pair of primers depicted in Fig. 7A, DNA fragments the size of ∼500 and ∼1,000 bp were specifically amplified from the plasmids pBabeF1E and pBabeF1βE, respectively (Fig. 7A, lanes 1 to 4). The stable clone expressing pBabe vector showed no PCR product (lanes 5 and 6), and clones expressing pBabeF1E and pBabeF1βE showed a RT-dependent PCR product of predicted size (lanes 8, 10, 12, and 14). Apoptotic kinetics was then studied in the presence of the dimerizing agent AP1510. Following the course of cytokine deprivation, pBabe-F1βE-expressing cells manifested accelerated death faster than that of controls regardless of the presence of AP1510 (Fig. 7B). These results further supported the notion that DER-enhanced apoptosis is independent of membrane anchorage and insensitive to physical dimerization.
FIG. 7.
The cytoplasmic FKBP-DER fusion protein accelerates apoptosis. (A) Expression of the FKBP-DER chimeric gene in Ba/F3 stable transfectants. The cytoplasmic RNA samples were prepared from cells expressing pBabe vector (lanes 5 and 6), pBabeF1E (lanes 7 to 10), and pBabeF1βE (lanes 11 to 14) and subjected to RT-PCR analysis with a pair of primers depicted as P1 and P2 in the scheme. Two plasmid DNA controls (lanes 1 to 4) are included, and the PCR products of 500 bp for pBabeF1E (lanes 1 and 2) and of 1,000 bp for pBabeF1βE (lanes 3 and 4) are indicated at the left. Samples in odd numbered lanes are products without RT, and those in even numbered lanes are products with RT. (B) Dimerization-insensitive death acceleration effect of the cytosolic FKBP-DER fusion protein. Cells expressing the indicated plasmids were subjected to cytokine deprivation and were treated with or without 10 μM AP1510 simultaneously. At various time points, the viable cells were measured as described for Fig. 1A. The values are averages of two independent determinations performed in duplicate. The pBabe vector-containing cell line (V) was included as a negative control. F1E, pBabeF1E-expressing cells; F1βE, pBabeF1βE-expressing cells; AP, dimerization with AP1510.
Finally, we examined whether hβc can promote death in a nonhematopoietic cell line. This issue was explored in the human kidney cell line 293 by a transient assay measuring loss of green fluorescence protein (GFP) expression in apoptotic cells as previously described (4). As observed in the control group, neither pBabe vector nor pcDNA3 vector caused any significant loss of GFP expression in the absence of serum (Table 1). However, the expression of wild-type hβc led to a decrease of 18.95% GFP+ cells upon serum starvation compared to cells in serum medium (Table 1). The expression of cytosolic DER-containing sequence, i.e., pcDNA-hβc(471-590), also caused a decrease of 11.25% GFP+ cells upon serum starvation (Table 1). These data confirm the anchorage-independent nature of the death acceleration activity of DER and further suggest that serum starvation sensitizes the 293 cells to the death-promoting effect of hβc.
TABLE 1.
Expression of both hβc and cytoplasmic DER of hβc affected the viability of human renal epithelial cell line 293
| Construct | Serum addition | Expt 1
|
Expt 2
|
Avg gain or loss ± SE | ||
|---|---|---|---|---|---|---|
| No. of GFP-positive cellsa | % Gain or lossb | No. of GFP-positive cells | % Gain or loss | |||
| pBabe | + | 6,476 | 5,121 | |||
| − | 6,321 | −2.4 | 5,001 | −2.3 | −2.35 ± 0.05 | |
| pBabe-hβc | + | 4,993 | 4,721 | |||
| − | 3,981 | −20.3 | 3,891 | −17.6 | −18.95 ± 1.35 | |
| pcDNA3 | + | 7,007 | 4,751 | |||
| − | 7,387 | +5.4 | 4,876 | +2.6 | +4.00 ± 1.40 | |
| pcDNA-hβc(471-590) | + | 5,181 | 4,520 | |||
| − | 4,595 | −11.3 | 4,012 | −11.2 | −11.25 ± 0.05 | |
Number of cells expressing GFP detected by flow cytometry from 5 × 104 transfected cells.
{[(Number of GFP-positive cells in medium without serum) − (number of GFP-positive cells in medium with serum)]/(number of GFP-positive cells in medium with serum)} × 100.
DISCUSSION
Hematopoietic cells differ from most other cell types in that their survival in vitro has an absolute requirement for specific growth factors. Growth factors are required continuously throughout the developmental program, and removal of growth factors at any stage during differentiation into the mature cells leads to apoptosis. Therefore, programmed cell death was suggested to have a physiological significance in hematopoiesis (9). In many factor-dependent hematopoietic cell lines, apoptosis was observed when the relevant cytokines were removed from culture media. However, cell viability could be maintained in these cell lines by the addition of cycloheximide (56), suggesting that new protein synthesis is essential and a positive control mechanism is hypothesized to CWIA. In this report, we provide evidence that the βc molecule is involved in modulating CWIA. We further demonstrate that the βc molecule modulates apoptosis with the following distinct features. First, its expression levels closely correlated with the rate of apoptosis when cytokines were depleted (Fig. 1 to 3). Second, the sequence essential for death acceleration was distinct from that required for proliferation (Fig. 4). Third, the death accelerated by βc was sensitive to cycloheximide treatment (data not shown). Fourth, its enhancement of cell death did not depend on membrane anchorage (Fig. 6 and 7; Table 1). Fifth, physical aggregation of βc did not abrogate its death-enhancing effect (Fig. 5 and 7). Finally, βc itself was sufficient to promote cell death in the nonhematopoietic 293 cell line under serum-free conditions (Table 1).
Although the expression level of hβc in a given cell line strongly correlated with the death rate of this cell line in the absence of cytokine, it did not seem to have a good correlation between cell lines from different origins. We noticed that TF-1 cells were less sensitive to CWIA than HT-2 and Ba/F3 cells. Moreover, TF-1 cells were also less sensitive to the death-enhancing effect of hβc overexpression. Low-level expression of hβc in HT-2 cells resulted in a 20% increase in cell death within 24 h. However, an eightfold increase in hβc expression in TF-1 cells exacerbated cell death by 21% compared to control cells at 48 h (compare Fig. 1B and 2B). Although there are many explanations for these results, the most likely is the distinct genetic background of these cells.
Amino acid sequence alignment of the DER sequence of human βc and those of mβc (13), mβIL-3 (13), and rat βc (1) revealed a highly conserved 26-aa motif (Fig. 4D) which is rich in proline (6 residues of 26), serine/threonine (6 of 26), and acidic amino acids (4 of 26). The DER sequence also includes a short sequence motif known as box II that is highly conserved among several type I cytokine receptors. This observation is consistent with our data that both hGMRα (Fig. 1A) and hIL5Rα (data not shown) contain no box II sequence in their cytoplasmic domains and lack cell death-enhancing ability. An intriguing observation is that the cytokine receptors containing the box II sequence, including gp130, βc, the erythropoietin and G-CSF receptors, and IL-2Rβ (31, 34, 40), are key signal transducers and are expressed with limited overlap in terms of distribution in hematopoietic cell lineages. It would be of interest to investigate whether these receptors also regulate apoptosis.
The dual role of βc in growth and death regulation may confer an IL-3-, IL-5, or GM-CSF dependence on βc-expressing cells when these receptor α chains are coexpressed. During hematopoiesis, IL3Rα was highly expressed in the totipotent stem cell population and mature cell lineages. The expression of βc and GMRα was not detectable in the stem cell populations and was induced in the multilineage progenitors and along the erythroid and myeloid differentiation pathways (2, 28, 57). IL5Rα was expressed even later during the differentiation of eosinophil lineage (51). This stage-specific expression pattern was concomitant with the responsiveness and the dependence of myeloid cells on IL-3, GM-CSF, and IL-5 during differentiation (for a review, see reference 29). For those cell lineages expressing βc molecules, the continuous presence of IL-3, GM-CSF, or IL-5 in vitro and in vivo will allow the activation of survival signals via the αβc heterodimeric receptor complex and ensure further differentiation. Mice lacking the receptor βc have recently been generated. Except with the development of a pulmonary alveolar proteinosis-like disease and having reduced numbers of peripheral eosinophils, these mβc-null mice seem to have normal hematopoiesis (36, 37, 50). The expression of another highly homologous protein, mβIL-3, in these mβc-null mice was suggested to be responsible for transducing appropriate proliferation and differentiation signals during hematopoiesis. Similarly, since the DER sequence of mβIL-3 is almost identical to that of mβc, it is possible that in these mβc-null mice, mβIL-3 can substitute for mβc to regulate apoptosis in a subset of hematopoietic cells. If this scenario is the case and given the fact that the receptor βc levels affect the death rate of a given cell type, it would then be interesting to examine whether the expression level of mβIL-3 in these mβc-null mice is elevated, a compensatory effect that is frequently observed in mice lacking one member of an important gene family (18, 26).
The functional and structural properties of DER are distinct from those of the death domains identified in the tumor necrosis factor receptor family and their adaptor signaling molecules (53). These death domains can override the survival effects of growth factors and drive the target cell to undergo apoptosis. Functionally, DER is more closely related to the addiction/dependence domains (ADD) in the nerve growth factor receptor (NGFR) (44) and the androgen receptor (AR) (3) wherein the expression of these receptors establishes a ligand-dependent cellular status. The ADD-containing receptors confer host cells with proliferation activity in the presence of ligands and will drive cells to undergo apoptosis only when ligands are removed from the culture medium. However, these three functionally related peptide motifs are structurally very distinct. DER of the receptor βc is a proline-rich peptide, ADD of NGFR is an α-helical peptide containing two critical basic residues (17), whereas ADD of AR is a stretch of glutamines that results from the trinucleotide repeats in the AR coding sequence (3). It remains to be determined how these structurally distinct motifs can manifest similar biological functions.
Although our results strongly suggest that the receptor βc is involved in modulating growth factor withdrawal-induced apoptosis, the underlying mechanism is still not clear. On the basis of current knowledge on the signaling molecules involved in growth and death control, we propose the following three likely explanations. (i) Forced expression of βc might result in sequestering some common signaling components which interact with the box II sequence, and consequently these cells die faster upon deprivation of their dependent growth factors. (ii) The CIS/SOCS/SSI/Jab gene family could be induced by the receptor βc through the JAK/STAT pathway (11, 27, 35, 52, 60). Induction of these negative regulators may lead to premature termination of the residual survival signal of the receptor after cytokine removal, and the CWIA rate is subsequently enhanced. (iii) In the absence of cytokines, the receptor βc can trigger a death cascade that eventually kill cells. While these three mechanisms are distinct from each other, they are not necessarily mutually exclusive in the context of βc function. More experiments are required to unravel this issue.
ACKNOWLEDGMENTS
We thank Young-Sun Lin, Yu-Chung Yang, and Atsushi Miyajima for critical comments on the manuscript, David Baltimore for the BOSC23 packaging cell line, Hartmut Land for retroviral expression vectors, and Ariad Pharmaceuticals for plasmid pCF1E and the synthetic dimerizing agent AP1510. We are also grateful to Derek W. Gilroy for preparation of the manuscript.
This work was supported in part by Academia Sinica, Taiwan (J.J.-Y.Y.) and the National Science Council of Taiwan (J.J.-Y.Y.). S.-F.L. was supported by a postdoctoral fellowship from Academia Sinica, Taiwan. H.-M.H. and J.-R.C. were supported by the postdoctoral fellowships of the National Science Council of Taiwan.
REFERENCES
- 1.Appel K, Buttini M, Sauter A, Gebicke-Haerter P J. Cloning of rat interleukin-3 receptor β-subunit from cultured microglia and its mRNA expression in vivo. J Neurosci. 1995;15:5800–5809. doi: 10.1523/JNEUROSCI.15-08-05800.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashihara E, Vannucchi A M, Migliaccio G, Migliaccio A R. Growth factor receptor expression during in vitro differentiation of partially purified populations containing murine stem cells. J Cell Physiol. 1997;171:343–356. doi: 10.1002/(SICI)1097-4652(199706)171:3<343::AID-JCP13>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 3.Bredesen D, Ye X, Tasinato A, Sperandio S, Wang J J, Assa-Munt N, Rabizadeh S. p75NTR and the concept of cellular dependence: seeing how the other half die. Cell Death Differ. 1998;5:365–371. doi: 10.1038/sj.cdd.4400378. [DOI] [PubMed] [Google Scholar]
- 4.Chao J R, Wang J M, Lee S F, Peng H W, Lin Y H, Chou C H, Li J C, Huang H M, Chou C K, Kuo M L, Yen J J Y, Yang-Yen H F. mcl-1 is an immediate-early gene activated by the granulocyte-macrophage colony-stimulating factor (GM-CSF) signaling pathway and is one component of the GM-CSF viability response. Mol Cell Biol. 1998;18:4883–4898. doi: 10.1128/mcb.18.8.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cleveland J L, Dean M, Rosenberg N, Wang J Y, Rapp U R. Tyrosine kinase oncogenes abrogate interleukin-3 dependence of murine myeloid cells through signaling pathways involving c-myc: conditional regulation of c-myc transcription by temperature-sensitive v-abl. Mol Cell Biol. 1989;9:5685–5695. doi: 10.1128/mcb.9.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowling G J, Dexter T M. Apoptosis in the haemopoietic system. Philos Trans R Soc Lond Ser B. 1994;345:257–263. doi: 10.1098/rstb.1994.0103. [DOI] [PubMed] [Google Scholar]
- 7.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 8.Devos R, Plaetinck G, Van-der-Heyden J, Cornelis S, Vandekerckhove J, Fiers W, Tavernier J. Molecular basis of a high affinity murine interleukin-5 receptor. EMBO J. 1991;10:2133–2137. doi: 10.1002/j.1460-2075.1991.tb07747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dexter T M, Whetton A D, Heyworth C M. The relevance of protein kinase C activation, glucose transport and ATP generation in the response of haemopoietic cells to growth factors. In: Kahn P, Graf T, editors. Oncogenes and growth control. Berlin, Germany: Springer-Verlag Press; 1986. pp. 163–169. [Google Scholar]
- 10.Dignam J, Lebovitz R, Roeder R. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endo T A, Masuhara M, Yokouchi M, Suzuki R, Sakamoto H, Mitsui K, Matsumoto A, Tanimura S, Ohtsubo M, Misawa H, Miyazaki T, Leonor N, Taniguchi T, Fujita T, Kanajura Y, Komiya S, Yoshimura A. A new protein containing an SH2 domain that inhibits JAK kinases. Nature. 1997;387:921–924. doi: 10.1038/43213. [DOI] [PubMed] [Google Scholar]
- 12.Gearing D P, King J A, Gough N M, Nicola N A. Expression cloning of a receptor for human granulocyte-macrophage colony-stimulating factor. EMBO J. 1989;8:3667–3676. doi: 10.1002/j.1460-2075.1989.tb08541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorman D M, Itoh N, Kitamura T, Schreurs J, Yonehara S, Yahara I, Arai K, Miyajima A. Cloning and expression of a gene encoding an interleukin 3 receptor-like protein: identification of another member of the cytokine receptor gene family. Proc Natl Acad Sci USA. 1990;87:5459–5463. doi: 10.1073/pnas.87.14.5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hara T, Miyajima A. Two distinct functional high affinity receptors for mouse interleukin-3 (IL-3) EMBO J. 1992;11:1875–1884. doi: 10.1002/j.1460-2075.1992.tb05239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashida K, Kitamura T, Gorman D M, Arai K, Yokota T, Miyajima A. Molecular cloning of a second subunit of the receptor for human granulocyte-macrophage colony-stimulating factor (GM-CSF): reconstitution of a high-affinity GM-CSF receptor. Proc Natl Acad Sci USA. 1990;87:9655–9659. doi: 10.1073/pnas.87.24.9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hileman M R, Chapman B S, Rabizadeh S, Krishnan V V, Bredesen D, Assa-Munt N, Plesniak L A. A cytoplasmic peptide of the neurotrophin receptor p75NTR: induction of apoptosis and NMR determined helical conformation. FEBS Lett. 1997;415:145–154. doi: 10.1016/s0014-5793(97)01113-7. [DOI] [PubMed] [Google Scholar]
- 18.Hummler E, Cole T J, Blendy J A, Ganss R, Aguzzi A, Schmid W, Beermann F, Schutz G. Targeted mutation of the CREB gene: compensation within the CREB/ATF family of transcription factors. Proc Natl Acad Sci USA. 1994;91:5647–5651. doi: 10.1073/pnas.91.12.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itoh N, Yonehara S, Schreurs J, Gorman D M, Maruyama K, Ishii A, Yahara I, Arai K, Miyajima A. Cloning of an interleukin-3 receptor: a member of a distinct receptor gene family. Science. 1990;247:324–327. doi: 10.1126/science.2404337. [DOI] [PubMed] [Google Scholar]
- 20.Jacobson M D, Weil M, Raff M C. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 21.Kinoshita T, Shirouzu M, Kamiya A, Hashimoto K, Yokoyama S, Miyajima A. Raf/MAPK and rapamycin-sensitive pathways mediate the anti-apoptotic function of p21Ras in IL-3-dependent hematopoietic cells. Oncogene. 1997;15:619–627. doi: 10.1038/sj.onc.1201234. [DOI] [PubMed] [Google Scholar]
- 22.Kinoshita T, Yokota T, Arai K, Miyajima A. Regulation of Bcl-2 expression by oncogenic Ras protein in hematopoietic cells. Oncogene. 1995;10:2207–2212. [PubMed] [Google Scholar]
- 23.Kinoshita T, Yokota T, Arai K, Miyajima A. Suppression of apoptosis death in hematopoietic cells by signalling through the IL-3/GM-CSF receptors. EMBO J. 1995;14:266–275. doi: 10.1002/j.1460-2075.1995.tb07000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kitamura T, Sato N, Arai K, Miyajima A. Expression cloning of the human IL-3 receptor cDNA reveals a shared beta subunit for the human IL-3 and GM-CSF receptors. Cell. 1991;66:1165–1174. doi: 10.1016/0092-8674(91)90039-2. [DOI] [PubMed] [Google Scholar]
- 25.Kitamura T, Tange T, Terasawa T, Chiba S, Kuwaki T, Miyagawa K, Piao Y F, Miyazono K, Urabe A, Takaku F. Establishment and characterization of a unique human cell line that proliferates dependently on GM-CSF, IL-3, or erythropoietin. J Cell Physiol. 1989;140:323–334. doi: 10.1002/jcp.1041400219. [DOI] [PubMed] [Google Scholar]
- 26.LeCouter J E, Kablar B, Hardy W R, Ying C, Megeney L A, May L L, Rudnicki M A. Strain-dependent myeloid hyperplasia, growth deficiency, and accelerated cell cycle in mice lacking the Rb-related p107 gene. Mol Cell Biol. 1998;18:7455–7465. doi: 10.1128/mcb.18.12.7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto A, Masuhara M, Mitsui K, Yokouchi M, Ohtsubo M, Misawa H, Miyajima A, Yoshimura A. CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood. 1997;89:3148–3154. [PubMed] [Google Scholar]
- 28.McKinstry W J, Li C L, Rasko J E, Nicola N A, Johnson G R, Metcalf D. Cytokine receptor expression on hematopoietic stem and progenitor cells. Blood. 1997;89:65–71. [PubMed] [Google Scholar]
- 29.Metcalf D. Hematopoietic regulators: redundancy or subtlety? Blood. 1993;82:3515–3523. [PubMed] [Google Scholar]
- 30.Minami Y, Nakagawa Y, Kawahara A, Miyazaki T, Sada K, Yamamura H, Taniguchi T. Protein tyrosine kinase Syk is associated with and activated by the IL-2 receptor: possible link with the c-myc induction pathway. Immunity. 1995;2:89–100. doi: 10.1016/1074-7613(95)90081-0. [DOI] [PubMed] [Google Scholar]
- 31.Miura O, Cleveland J L, Ihle J N. Inactivation of erythropoietin receptor function by point mutations in a region having homology with other cytokine receptors. Mol Cell Biol. 1993;13:1788–1795. doi: 10.1128/mcb.13.3.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morgensten J P, Land H. Advanced mammalian gene transfer: high titer retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 34.Murakami M, Narazaki M, Hibi M, Yawata H, Yasukawa K, Hamaguchi M, Taga T, Kishimoto T. Critical cytoplasmic region of the interleukin 6 signal transducer gp130 is conserved in the cytokine receptor family. Proc Natl Acad Sci USA. 1991;88:11349–11353. doi: 10.1073/pnas.88.24.11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naka T, Narazaki M, Hirata M, Matsumoto T, Minamoto S, Aono A, Nishimoto N, Kajita T, Taga T, Yoshizaki K, Akira S, Kishimoto T. Structure and function of a new STAT-induced STAT inhibitor. Nature. 1997;387:924–929. doi: 10.1038/43219. [DOI] [PubMed] [Google Scholar]
- 36.Nishinakamura R, Nakayama N, Hirabayashi Y, Inoue T, Aud D, McNeil T, Azuma S, Yoshida S, Toyoda Y, Arai K, Miyajima A, Murray R. Mice deficient for the IL-3/GM-CSF/IL-5 βc receptor exhibit lung pathology and impaired immune response, while βIL-3 receptor-deficient mice are normal. Immunity. 1995;2:211–222. doi: 10.1016/1074-7613(95)90046-2. [DOI] [PubMed] [Google Scholar]
- 37.Nishinakamura R, Miyajima A, Mee P J, Tybulewixz V L J, Murray R. Hematopoiesis in mice lacking the entire granulocyte-macrophage colony-stimulating factor/interleukin-5 functions. Blood. 1996;88:2458–2464. [PubMed] [Google Scholar]
- 38.Ogata N, Kouro T, Yamada A, Koike M, Hanai N, Ishikawa T, Takatsu K. JAK2 and JAK1 constitutively associate with an interleukin-5 (IL-5) receptor α and βc subunit, respectively, and are activated upon IL-5 stimulation. Blood. 1998;91:2264–2271. [PubMed] [Google Scholar]
- 39.Ohta T, Kinoshita T, Naito M, Nozaki T, Masutani M, Tsuruo T, Miyajima A. Requirement of the caspase-3/CPP32 protease cascade for apoptotic death following cytokine deprivation in hematopoietic cells. J Biol Chem. 1997;272:23111–23116. doi: 10.1074/jbc.272.37.23111. [DOI] [PubMed] [Google Scholar]
- 40.O’Neal K D, Yu-Lee L Y. The proline-rich motif (PRM): a novel feature of the cytokine/hematopoietin receptor superfamily. Lymphokine Cytotokine Res. 1993;12:309–312. [PubMed] [Google Scholar]
- 41.Palacios R, Steinmetz M. IL-3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell. 1985;41:727–734. doi: 10.1016/s0092-8674(85)80053-2. [DOI] [PubMed] [Google Scholar]
- 42.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;88:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quelle F W, Sato N, Witthuhn B A, Inhorn R C, Eder M, Miyajima A, Griffin J D, Ihle J N. JAK2 associates with the βc chain of the receptor for granulocyte-macrophage colony-stimulating factor, and its activation requires the membrane-proximal region. Mol Cell Biol. 1994;14:4335–4341. doi: 10.1128/mcb.14.7.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rabizadeh S, Oh J, Zhong L T, Yang J, Bitler C M, Butcher L L, Bredesen D E. Induction of Apoptosis by the low-affinity NGF receptor. Science. 1993;261:345–348. doi: 10.1126/science.8332899. [DOI] [PubMed] [Google Scholar]
- 45.Rao P, Mufson R A. A membrane proximal domain of the human interleukin-3 receptor βc subunit that signals DNA synthesis in NIH 3T3 cells specifically binds a complex of Src and Janus family tyrosine kinases and phosphatidylinositol 3-kinase. J Biol Chem. 1995;270:6886–6893. doi: 10.1074/jbc.270.12.6886. [DOI] [PubMed] [Google Scholar]
- 46.Sakamaki K, Miyajima I, Kitamura T, Miyajima A. Critical cytoplasmic domains of the common β subunit of the human GM-CSF, IL-3 and IL-5 receptors for growth signal transduction and tyrosine phosphorylation. EMBO J. 1992;11:3541–3549. doi: 10.1002/j.1460-2075.1992.tb05437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato N, Sakamaki K, Terada N, Arai K, Miyajima A. Signal transduction by the high-affinity GM-CSF receptor: two distinct cytoplasmic regions of the common β subunit responsible for different signaling. EMBO J. 1993;12:4181–4189. doi: 10.1002/j.1460-2075.1993.tb06102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato S, Katagiri T, Takaki S, Kikuchi Y, Hitoshi Y, Yonehara S, Tsukada S, Kitamura D, Watanabe T, Witte O, Takatsu K. IL-5 receptor-mediated tyrosine phosphorylation of SH2/SH3-containing proteins and activation of Bruton’s tyrosine and Janus 2 kinases. J Exp Med. 1994;180:2101–2111. doi: 10.1084/jem.180.6.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheid M P, Duronio V. Dissociation of cytokine-induced phosphorylation of Bad and activation of PKB/akt: involvement of MEK upstream of Bad phosphorylation. Proc Natl Acad Sci USA. 1998;95:7439–7444. doi: 10.1073/pnas.95.13.7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scott C L, Hughes D A, Cary D, Nicola N A, Begley C G, Robb L. Functional analysis of mature hematopoietic cells from mice lacking the βc chain of the granulocyte-macrophage colony-stimulating factor receptor. Blood. 1998;92:4119–4127. [PubMed] [Google Scholar]
- 51.Shalit M, Sekhsaria S, Malech H L. Modulation of growth and differentiation of eosinophils from human peripheral blood CD34+ cells by IL5 and other growth factors. Cell Immunol. 1995;160:50–57. doi: 10.1016/0008-8749(95)80008-7. [DOI] [PubMed] [Google Scholar]
- 52.Starr R, Willson T A, Viney E M, Murray L J L, Rayner J R, Jenkins B J, Gonda T J, Alexander W S, Metcalf D, Nicola N A, Hilton D J. A family of cytokine-inducible inhibitors of signalling. Nature. 1997;387:917–921. doi: 10.1038/43206. [DOI] [PubMed] [Google Scholar]
- 53.Tartaglia L A, Ayres T M, Wong G H W, Goeddel D V. A novel domain within the 55 kd TNF receptor signals cell death. Cell. 1993;74:845–853. doi: 10.1016/0092-8674(93)90464-2. [DOI] [PubMed] [Google Scholar]
- 54.Tavernier J, Devos R, Cornelis S, Tuypens T, Van-der-Heyden J, Fiers W, Plaetinck G. A human high affinity interleukin-5 receptor (IL5R) is composed of an IL5-specific α chain and a β chain shared with the receptor for GM-CSF. Cell. 1991;66:1175–1184. doi: 10.1016/0092-8674(91)90040-6. [DOI] [PubMed] [Google Scholar]
- 55.Watson J. Continuous proliferation of murine antigen-specific helper T lymphocytes in culture. J Exp Med. 1979;150:1510–1519. doi: 10.1084/jem.150.6.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams G T, Smith C A, Spooncer E, Dexter T M, Taylor D R. Haemopoietic colony stimulating factors promote cell survival by suppressing apoptosis. Nature. 1990;343:76–79. doi: 10.1038/343076a0. [DOI] [PubMed] [Google Scholar]
- 57.Wognum A W, de-Jong M O, Wagemaker G. Differential expression of receptors for hemopoietic growth factors on subsets of CD34+ hemopoietic cells. Leuk Lymphoma. 1996;24:11–25. doi: 10.3109/10428199609045710. [DOI] [PubMed] [Google Scholar]
- 58.Wyllie A H. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature. 1980;284:555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- 59.Yen J J Y, Hsieh Y C, Yen C L, Chang C C, Lin S, Yang-Yen H F. Restoring the apoptosis suppression response to IL-5 confers on erythroleukemic cells a phenotype of IL-5-dependent growth. J Immunol. 1995;154:2144–2152. [PubMed] [Google Scholar]
- 60.Yoshimura A, Ohkubo T, Kigushi T, Jenkins N A, Gilbert D J, Copeland N G, Hara T, Miyajima A. A novel cytokine-inducible gene CIS encodes an SH2-containing protein that binds to tyrosine-phosphorylated interleukin 3 and erythropoietin receptors. EMBO J. 1995;14:2816–2826. doi: 10.1002/j.1460-2075.1995.tb07281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]