Abstract
E2F transcription factor is subject to stringent regulation by a variety of molecules. We recently observed that prohibitin, a potential tumor suppressor protein, binds to the retinoblastoma (Rb) protein and represses E2F transcriptional activity. Here we demonstrate that prohibitin requires the marked box region of E2F for repression; further, prohibitin can effectively inhibit colony formation induced by overexpression of E2F1 in T47D cells. Prohibitin was also found to interact with the signaling kinase c-Raf-1, and Raf-1 could effectively reverse prohibitin-mediated repression of E2F activity. Agents such as E1A, p38 kinase, and cyclins D and E had no effect on prohibitin-mediated repression of E2F1, but all of these molecules could reverse Rb function. Similarly, stimulation of the immunoglobulin M signaling pathway in Ramos cells could inactivate prohibitin, but this had no effect on Rb function. Serum stimulation of quiescent Ramos cells inactivated Rb and prohibitin with different kinetics; further, while the serum-dependent inactivation of Rb was dependent on cyclin-dependent kinase activity, the inactivation of prohibitin was not. We believe that prohibitin is a novel regulator of E2F function which channels specific signaling cascades to the cell cycle regulatory machinery.
The E2F family of transcription factors plays a significant role in the regulation of mammalian cell cycle progression (33). Studies in recent years have identified E2F as an important downstream target of the retinoblastoma (Rb) family of growth regulatory proteins, and it appears that Rb exerts its growth regulatory function at least in part by regulating E2F activity (15). Since the E2F family of transcription factors is capable of inducing different cell fates such as proliferation, apoptosis, or differentiation, an understanding of their regulation would throw light on the biochemical pathways underlying such phenomena (41).
The term E2F generally refers to a family of six proteins named E2F1 through E2F6 (24). Of these, E2Fs 1 to 5 possess a transcriptional activation domain at the carboxy terminal (50) and can induce transcription from target promoters in association with dimerization partners 1 or 2 (DP1 or DP2) (32). In contrast, E2F6 lacks an activation domain and is repressive in nature; E2F6 has been shown to compete for E2F binding sites on promoters and repress their activity (7, 19, 58). Within the five transcriptionally active E2Fs, there are certain biochemical and functional differences, though it is not yet clear whether they execute distinct functions in normal cells (10, 50). For example, though overexpression of E2F1 can induce S-phase entry in quiescent cells (22), E2F1 can also induce apoptosis under appropriate conditions (1, 18, 28, 45, 67); other E2F family members lack the ability to do so (27). Similarly, E2F1 is capable of transforming primary cells in association with Ras, but it is not yet clear whether other E2F family members are capable of doing so (49); in a recent study, E2F1 was shown to block the differentiation of myeloid cells, but E2F3 could not (53). Thus, despite the fact all five transcriptionally active E2Fs bind to the same DNA recognition site, they may have different functional niches in the cell (13).
E2F family members are also under different levels of control by upstream molecules (15). E2Fs 1, 2, and 3 can all bind to the Rb protein, but E2Fs 4 and 5 preferentially bind to p107 and p130 proteins (3, 10). It has been demonstrated that the Rb family proteins bind to a moiety within the transcriptional activation region of E2Fs, effectively repressing their activity (21, 25, 44). In addition to passive repression of E2F-mediated transcription, Rb has been shown to actively repress transcription from promoters carrying E2F binding sites by recruiting the histone deacetylase HDAC1 (5, 36, 37, 65, 66). Thus, the presence of E2F sites on a promoter does not always indicate that it is induced by E2F but, on the contrary, that it can be repressed through those sites as well (6, 20, 31). A very good example is the E2F1 promoter itself (23). In addition to the preferential interaction with Rb family members, E2Fs 1, 2, and 3 also possess a binding site for cyclins at the amino-terminal region, allowing them to be regulated by the associated cyclin-dependent kinases, mainly cdk2 (26, 69). E2Fs 4 and 5 lack this mode of regulation (10).
Our attempts to characterize additional proteins that bind to Rb family members led to the identification of prohibitin, a potential tumor suppressor protein, as an Rb-binding protein (62). Prohibitin could effectively bind to Rb, p107 and p130 and could repress the transcriptional activity of all of the five E2Fs but had no effect on promoters lacking an E2F binding site. Prohibitin had to interact with Rb to bring about the transcriptional repression of E2F, and this correlated with its ability to suppress colony formation in T47D cells. Interestingly, we also found that adenovirus E1A was unable to reverse prohibitin-mediated repression of E2F1 (62). This led us to believe that prohibitin represses E2F activity through mechanisms different than those used by the Rb protein. The studies described here demonstrate that prohibitin targets a different region of E2F1 than Rb, and prohibitin-mediated repression of E2F can be released by signals which do not target the Rb protein. Our results suggest that prohibitin-mediated repression of E2F is an additional mechanism that allows E2F to function in response to specific signaling pathways.
MATERIALS AND METHODS
Cell lines, vectors, and transfections.
Ramos cells were maintained in Dulbecco modified Eagle medium, T47D and U937 cells grown in RPMI medium, both containing 10% fetal bovine serum (FBS). Transient transfections were conducted on T47D breast carcinoma cells by using the calcium phosphate precipitation method according to standard protocols. Ramos cells were transfected by electroporation with a Bio-Rad Gene Pulser at 250 V, with a 960-μF capacitance. Both Ramos and T47D cells are Rb positive. For serum stimulation experiments, transfected cells were maintained in medium containing 10% FBS for 18 h prior to transfer to medium without serum. After 48 h of starvation, the cells were stimulated by using medium containing 10% FBS for the required periods of time.
A total of 2 μg of plasmids was used in all transfections for reporter analysis, unless noted otherwise; 8 μg of the expression vectors was used when extracts had to be prepared from the transfected cells for biochemical analysis. A 1-μg amount of a pSVβGal vector was included as internal control in all transfections, and the β-galactosidase value varied only slightly within each experiment. In all cases, representative chloramphenicol acetyltransferase (CAT) assay results from multiple experiments are shown. Where data is graphically represented, CAT activity was assessed by scanning the intensity of acetylated chloramphenicol. The total amount of DNA used for transfections was normalized by using salmon sperm DNA in all the lanes.
Expression vectors for GAL4 and VP16 fusions of E2F1 (pCGE2F1 and pCMVE2F1VP16, respectively), as well as the pG5E1BCAT reporter, were a kind gift from David Johnson. E2CAT vector contains the CAT gene driven by the adenovirus E2 promoter carrying two E2F binding sites. Full-length pDCE2F1 vector was used to induce the E2CAT in all transfections except those shown in Fig. 2B, where the wild-type and mutant E2Fs were expressed from the pCR3.1 vector. E2F1 mutants A, B, and C were generated by a PCR-based overlap-extension protocol, and their integrity was confirmed by sequencing. pSVRb, pCDNA3Raf-1 (wild-type as well as Δ28) and pCDNA3-prohibitin have been described before (62). Generation of Raf-1 mutants for mapping of binding domains was described earlier (61). C-terminal deletion mutants of prohibitin were generated in pCR2 vector by using PCR techniques, and the internal deletion mutant Δ185-214 was generated by an overlap-extension strategy. The prohibitin antisense construct was made by cloning the prohibitin cDNA in the reverse orientation in pCR3-1 vector. Other expression vectors used in the study are pRC/CMVCycD1, pCMVCycE, pCMVcdk2D145N (dominant-negative cdk2), pCMVcdk4DN, pCMVcdk6DN, pCMVE1A, and pSRαp38.
FIG. 2.
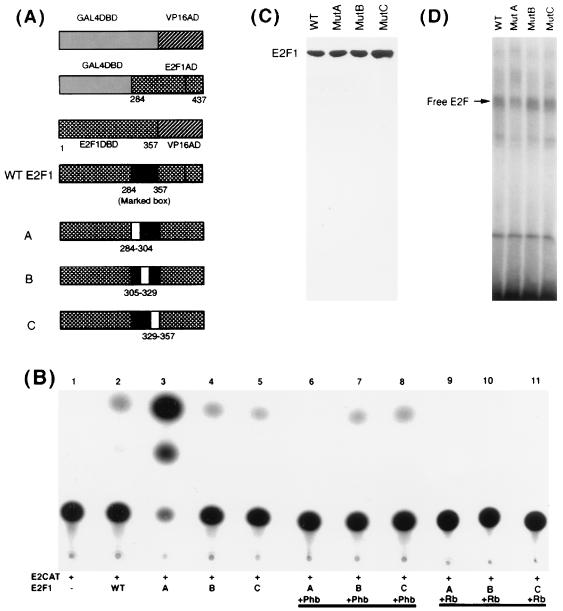
Prohibitin targets the marked box region of E2F1 for repression. (A) Schematic of the different E2F1 fusion proteins, as well as the E2F1 internal deletion mutants, used in the study. Mutants A, B, and C lack a 20-amino-acid sequence between the indicated residues. (B) Repression of E2F1 internal deletion constructs by prohibitin and Rb in T47D cells. Mutant A had approximately 20-fold more transcriptional activity than wild-type (WT) E2F1 (lanes 2 and 3). Prohibitin could fully repress mutant A but had no effect on mutants B and C (lanes 7 and 8). Rb could repress all three (lanes 9 to 11). (C) Western blot analysis of extracts from T47D cells transiently transfected with 8 μg of pCR3.1 E2F1 vectors expressing wild-type or mutant E2F1 proteins. (D) EMSA for E2F in the same extracts. There was no significant difference in the DNA binding activities of the three proteins.
Stable transfections were performed on 35-mm-diameter dishes by using approximately 10,000 cells and subjected to selection in the appropriate antibiotic for 14 days. The total amount of DNA transfected was equalized with salmon sperm DNA in every sample. Cells were fixed and stained with crystal violet, and colonies with more than 20 cells were counted.
In vitro binding assays.
Glutathione S-transferase (GST) fusions of Rb, Raf-1, and prohibitin were prepared as described earlier (61, 62). 35S-labeled Raf-1 and prohibitin proteins were generated by in vitro transcription-translation in rabbit reticulocyte lysates by using standard protocols. First, 8 to 10 μl of synthesized polypeptide was incubated with glutathione beads carrying equal amounts of GST fusion proteins in 200 μl of protein binding buffer (20 mM Tris, pH 7.5; 50 mM KCl; 0.5 mM EDTA; 1 mM dithiothreitol [DTT]; 0.5% NP-40; 3 mg of bovine serum albumin [BSA] per ml) at 4°C for 2 h. The beads were then washed six times with 1 ml of protein binding buffer and eluted with 10 mM glutathione. Eluates were separated in an 8% sodium dodecyl sulfate (SDS)-polyacrylamide gel and visualized by autoradiography. The protein amounts in control input lanes were approximately one-fifth of the total used in binding assay.
Immunoprecipitation and Western blots.
Polyclonal antibodies to prohibitin were a kind gift of J. Keith McClung, and monoclonal antiprohibitin antibodies were obtained from NeoMarkers. Anti-cRaf-1 monoclonal antibody was obtained from Transduction Laboratories; anti-Rb and anti-c-Myc antibodies were purchased from Oncogene Science-Calbiochem. Antibodies to p16, p107, p130, and p38 were obtained from Santa Cruz Biotechnologies; anti-human immunoglobulin M (IgM) antibody was from Southern Biotechnologies, and the anti-pTyr antibodies were from UBI.
Whole-cell extracts were prepared by hypotonic shock followed by salt extraction, as described previously (8). Portions (50 to 200 μg) of whole-cell extracts were treated with 5 μl of the appropriate primary antibody in a volume of 100 μl at 4°C for 1 h. Then, 3 mg of protein A-Sepharose or protein G-Sepharose in a 100-μl volume was added and incubated for an additional hour. The binding was performed in a buffer containing 20 mM HEPES (pH 7.9), 40 mM KCl, 1 mM MgCl2, 0.1 mM EGTA, 0.1 mM EDTA, 0.1 mM DTT, 0.1 mM NaF, 0.1 mM Na3VO4, 0.5% NP-40, and 3 mg of BSA per ml. The beads were washed six times with 600 μl of the same buffer, boiled in 20 μl of SDS sample buffer, and separated on 8 or 10% polyacrylamide gels. After semidry transfer to supported nitrocellulose membrane, the blots were probed with the appropriate antibody. The proteins were detected by using an enhanced chemiluminescence assay system from Amersham.
EMSA.
Electrophoretic mobility shift assay (EMSA) after immunoprecipitation was performed as previously described (8, 71). An EcoRI-HindIII fragment of the adenovirus E2 promoter containing two E2F binding sites (TTTCGCGC) was end labeled by using Klenow fragment and was used as the probe in all of the assays. Briefly, 8 μg of whole-cell extracts prepared as described above was incubated with approximately 0.2 ng of 32P-labeled E2F probe in a buffer containing 20 mM HEPES (pH 7.9); 40 mM KCl; 0.1 mM concentrations each of MgCl2, EGTA, EDTA, DTT, NaF, and Na3VO4; 1% NP-40, 1 μg of salmon sperm DNA per ml, and 10 μg of BSA per ml. After incubation at room temperature for 20 min, the reactions were separated on a 4% polyacrylamide gel in 0.25× TBE at 300 V for 3 h. The gel was dried, and the bands were detected by autoradiography.
RESULTS
Prohibitin and Rb target different regions of E2F1 for repression.
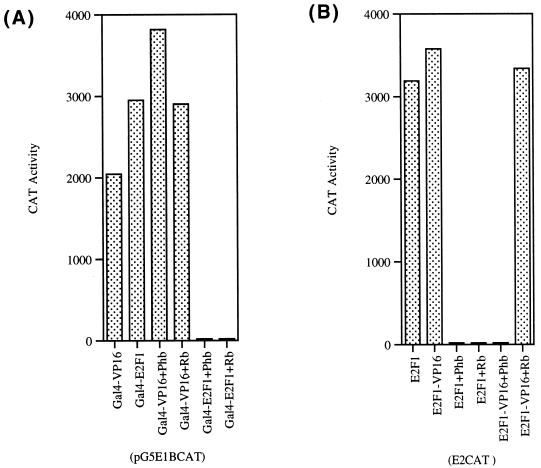
Since we found that prohibitin can specifically repress all transcriptionally active members of the E2F family (62), we attempted to identify the domain of E2F that is targeted by prohibitin. Studies were conducted on E2F1 to this end. As a first step, we examined the effect of prohibitin on two chimeric E2F1 proteins by transient-transfection experiments in T47D cells. In the first experiment, a chimeric E2F protein with the carboxy-terminal 153 amino acids of E2F1 (which contains the transcriptional activation domain) fused to a GAL4 DNA-binding domain (11) was tested for its ability to respond to prohibitin. As shown in Fig. 1A, the GAL4-E2F1 construct, as well as a control GAL4VP16 vector, could induce transcription from a pG5E1BCAT reporter; cotransfection of prohibitin or Rb had no effect on the transcription induced by the GAL4-VP16 but specifically abolished the transcriptional activity of the GAL4-E2F1 protein. This result suggests that prohibitin can target E2F1 specifically and that the carboxy-terminal domain containing the transcriptional activation domain can respond to prohibitin.
FIG. 1.
Repression of E2F1 fusion proteins by prohibitin and Rb. (A) A pG5E1BCAT reporter was induced by transfecting the indicated GAL4 fusion proteins. Cotransfection of 2 μg of prohibitin or Rb specifically represses GAL4E2F1 but not GAL4VP16 protein. (B) An E2CAT reporter was cotransfected with expression vectors for E2F1 or E2F1-VP16AD fusion protein. Prohibitin could repress both the constructs, but Rb does not repress E2F1-VP16 (unlike prohibitin).
In a second experiment, we examined whether prohibitin can target the amino-terminal region of E2F1 that contains the DNA-binding domain. An E2F1-VP16 construct that contains the region from residues 1 to 357 of E2F1 fused to a VP16 activation domain (11) was tested for its ability to respond to prohibitin. As shown in Fig. 1B, the E2F1-VP16 construct could activate transcription from an E2CAT vector as effectively as E2F1. It was found that cotransfection of prohibitin could effectively repress both wild-type E2F1 and the E2F1-VP16 fusion protein. This raised the possibility that prohibitin either targets multiple regions of E2F1 for repression or targets a region of E2F1 shared between the E2F1-VP16 and GAL4-E2F1 constructs.
In contrast to repression by prohibitin, Rb could repress the activity of full-length E2F1 but had no effect on the E2F1-VP16 construct (Fig. 1B). The observation that Rb can repress GAL4-E2F1 as well as full-length E2F1, but not E2F1-VP16 (which lacks an E2F1 activation domain), is consistent with the fact that Rb specifically targets the activation domain of E2F1 for repression.
Prohibitin targets the marked box region of E2F1.
Experiments were designed to further delineate the region of E2F1 targeted by prohibitin. As shown in Fig. 2A, the region from residues 284 to 357 of E2F1 is present in GAL4-E2F1 and in the E2F1-VP16 constructs, both of which were repressed by prohibitin. This region corresponds to the highly conserved marked box region of E2F (50). Attempts were made to create internal deletions of this region and to assess whether such mutant E2Fs could respond to prohibitin. A deletion of the entire region from residues 284 to 357 of E2F1 totally abolished its transcriptional activity (data not shown). Hence we divided this region into three parts (regions from residues 284 to 304, 304 to 329, and 329 to 357) and generated internal deletions of each segment by using a PCR-based overlap-extension protocol. A schematic of the different E2F mutants, as well as of the E2F1 fusion proteins, is shown in Fig. 2A.
The ability of each internal deletion to induce transcription from an E2CAT vector was tested (Fig. 2B, lanes 2 to 5). Mutants B and C (which had internal deletions of regions from residues 304 to 329 and 329 to 357) had amounts of transcriptional activity comparable to that of wild-type E2F1; surprisingly, deletion mutant A, which lacked residues 284 to 304, had 20-fold more transcriptional activity compared to the wild-type E2F1 (lane 2). The ability of Rb and prohibitin to repress the three mutant E2Fs was next examined by a cotransfection experiment. As shown in lane 6, prohibitin could effectively repress mutant A; in contrast, prohibitin had no effect on the transcriptional activity of mutants B and C (lanes 7 and 8), suggesting that the region of E2F1 spanning residues 304 to 357 is essential for prohibitin-mediated repression. It was found that Rb could effectively repress all three mutants (Fig. 2B, lanes 9 to 11), since all had an intact activation domain. This result suggests that prohibitin targets a region of E2F1 at the carboxy terminal of the marked box domain and not the activation domain itself.
A Western blot analysis of T47D cells transiently transfected with the mutant E2F constructs showed that all are expressed equally well and produce stable proteins (Fig. 2C). In addition, there was no significant difference in the DNA-binding activity of the mutants (Fig. 2D), and it is not clear at this moment why mutant A has such a high transcriptional activity.
Prohibitin can bind to E2F in vitro and in vivo.
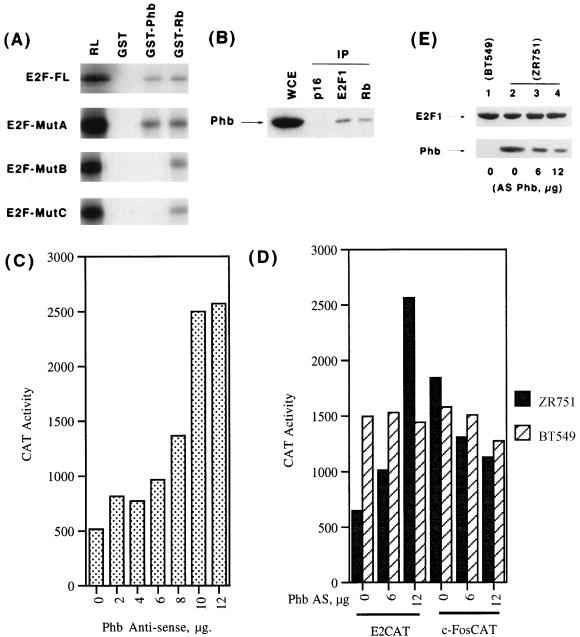
Since we found that prohibitin can efficiently repress E2F activity through the marked box domain, we examined whether prohibitin can physically interact with E2F1. As a first step, we examined whether E2F1 can bind to prohibitin in an in vitro GST binding assay. E2F1 was synthesized by in vitro transcription-translation in rabbit reticulocyte lysates, and its binding to GST fusions of prohibitin or Rb was tested. As shown in Fig. 3A, top panel, the full-length E2F1 could efficiently bind to both the fusion proteins; there was no binding to control unprimed GST beads. This suggested that prohibitin might be interacting with E2F1 to bring about the repression. Since we found that the marked box of E2F1 responds to prohibitin, we tested the mutants of this region for their ability to bind to prohibitin. As shown in Fig. 3A, E2F1 mutant A could bind efficiently to prohibitin as well as to Rb; in contrast, mutants B and C could bind only to GST-Rb and not to prohibitin. This correlates with their ability to respond to prohibitin and Rb, since we find that mutant A responds to Rb, as well as prohibitin, but that mutants B and C can be repressed only by Rb.
FIG. 3.
Prohibitin binds to the marked box region of E2F1. (A) In vitro binding assay showing the interaction of full-length E2F1 or mutants A, B, or C to GST fusions of prohibitin or Rb. RL indicates one-fifth of the loading material, and GST indicates unprimed beads. (B) Association of prohibitin with E2F1 in Ramos cells. Ramos whole-cell extracts were immunoprecipitated with the indicated antibodies, and the presence of prohibitin was examined by Western blotting. WCE indicates an equivalent amount of whole-cell extract. (C) Endogenous prohibitin levels affect E2F activity. ZR751 cells were transfected with 12 μg of E2CAT vector alone or with increasing amounts of an antisense prohibitin construct. Transcriptional activity from the E2CAT reporter increases with the amount of antisense prohibitin construct used. (D) The effect of antisense prohibitin construct on 12 μg of E2CAT or 4 μg of c-FosCAT vectors in ZR751 and BT549 cells. Transcription from the E2CAT vector is increased in response to the antisense construct in ZR751 cells but not in BT549; there was no increase in c-FosCAT in either cell line. (E) Western blot analysis of E2F1 and prohibitin in BT549 cells, as well as ZR751 cells, transiently transfected with the indicated amounts of antisense prohibitin construct.
It was next examined whether the prohibitin-E2F1 interaction can be detected in vivo; an immunoprecipitation-Western blot analysis was used for this purpose. Whole-cell extract prepared from the human B-cell line Ramos was immunoprecipitated with antibodies to p16, E2F1, and Rb; the presence of prohibitin was examined by Western blotting by using a monoclonal antiprohibitin antibody. As shown in Fig. 3B, there was no prohibitin detected in the p16 immunoprecipitate, but prohibitin was found to be associated with E2F1, as well as Rb. This suggests that prohibitin and E2F1 can associate in vivo, and this association can be detected without overexpressing any component.
We previously reported that the level of endogenous prohibitin in human breast cancer cells could affect the activity of an E2CAT reporter, but not a c-fos promoter or AP1CAT reporter. To assess whether endogenous E2F activity was affected by altering the amounts of prohibitin already present in cells, we used an antisense strategy. The cell line ZR751, which has abundant levels of endogenous prohibitin, was used for this purpose. Transfection of 12 μg of an E2CAT reporter resulted in a low level of CAT activity in ZR751 cells, but cotransfection of increasing levels of an antisense prohibitin construct led to an increase in the observed CAT conversion. This reflects an increase in the transcriptional activity of the endogenous E2F present in the cell when prohibitin level is reduced (Fig. 3C).
Two separate control experiments were performed to verify the above result. First, cotransfection of 6 or 12 μg of antisense prohibitin did not lead to an increase in E2F activity in BT549 cells, which have no detectable amount of prohibitin (Fig. 3D, striped bars). Second, to rule out that the repression of E2F by prohibitin is through a nonspecific effect on components of the general transcriptional machinery, a similar antisense experiment was conducted with a c-FosCAT reporter. As shown in Fig. 3D, whereas increasing amounts of antisense prohibitin construct elevated the transcriptional activity of the E2CAT in ZR751 cells, it did not induce c-FosCAT activity, suggesting that the effect on E2F is a specific event. The antisense prohibitin had no significant effect on either reporter in BT549 cells, which lack endogenous prohibitin (Fig. 3D, striped bars).
A Western blot experiment was conducted to examine whether transfection of antisense prohibitin did reduce the levels of prohibitin protein. As shown in Fig. 3E, there is a comparable amount of E2F1 present in BT549 and ZR751 cells, and the levels of E2F1 do not change by transfecting antisense prohibitin. In contrast, BT549 cells have no detectable prohibitin (Fig. 3E, lane 1); further, the level of prohibitin in ZR751 cells is reduced upon transfecting the antisense construct, correlating with the increase in E2F activity. Taken together, these results strongly suggest that the level of prohibitin in cells has a direct bearing on E2F mediated transcription.
Prohibitin can repress E2F1-mediated induction of cell proliferation.
Overexpression of E2F1 has been demonstrated to induce cell proliferation in many mammalian cell lines, and the Rb protein could repress this efficiently (1, 14, 22). Since prohibitin has strong antiproliferative activity that correlated with its ability to bind to the Rb protein, we designed experiments to examine whether prohibitin could repress E2F1-mediated induction of cell proliferation. Toward this purpose, a colony formation assay was performed on T47D cells by using standard protocols as described previously (62).
As shown in Table 1, transfection of a pSV-NEO vector gave rise to ca. 119 colonies after 14 days of selection; in contrast, transfection of 2 μg of a prohibitin expression vector reduced the number of colonies to ca. 43. All the E2F1 constructs could stimulate the proliferation of T47D cells at the levels transfected (3 μg); the induction was most pronounced in the case of mutant A, which doubled the number of colonies. Prohibitin could effectively suppress the colonies induced by the transfection of wild-type E2F1 and mutant A; surprisingly, prohibitin could not suppress the colony formation when mutants B and C were transfected. This experiment suggests that prohibitin and E2F1 have antagonistic effects on cell proliferation, and there is a correlation between the ability of prohibitin to repress the transcriptional activity of E2F1 and its ability to reverse E2F1-mediated cell proliferation.
TABLE 1.
Modulation of E2F1-induced colony formation by prohibitina
| Vector transfected | No. of colonies
|
|
|---|---|---|
| Expt 1 | Expt 2 | |
| pSVNeo | 119 | 115 |
| pCDNA3Prohibitin | 43 | 46 |
| pCR3.1E2F1WT | 195 | 199 |
| pCR3.1E2F1MutA | 231 | 241 |
| pCR3.1E2F1MutB | 146 | 144 |
| pCR3.1E2F1MutC | 175 | 181 |
| pCR3.1E2F1WT+Phb | 51 | 55 |
| pCR3.1E2F1MutA+Phb | 49 | 45 |
| pCR3.1E2F1MutB+Phb | 149 | 142 |
| pCR3.1E2F1MutC+Phb | 141 | 145 |
Approximately 10,000 T47D cells were transfected with 2 μg of the indicated vectors; 3 μg of the E2F constructs was used. Colonies with 20 or more cells were counted after 14 days of selection in 40 μg of neomycin per ml. Phb, prohibitin.
Raf-1 can bind to prohibitin and regulate its function.
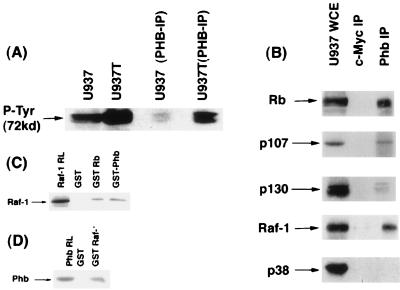
It has been reported that prohibitin is associated with the IgM receptor (55), and recently it was shown to associate with the mixed lineage kinase-2 (MLK2) (46). Since these observations indicated that prohibitin could be involved in mediating signal transduction cascades and since prohibitin had a potential tyrosine phosphorylation site, we felt it was prudent to examine whether prohibitin was tyrosine phosphorylated. To address this, we immunoprecipitated whole-cell extracts from proliferating or tetradecanoyl phorbol acetate (TPA)-treated U937 cells with an antiprohibitin antibody and probed the immunoprecipitate with an anti-phosphotyrosine antibody in a Western blot. There was no detectable tyrosine phosphorylated band corresponding to the size of prohibitin; surprisingly, the major band recognized by the phosphotyrosine antibody in the prohibitin immuneprecipitate was at ca. 72 to 76 kDa (Fig. 4A). The 76-kDa band was more pronounced in the U937 extracts that were treated with TPA and the prohibitin immuneprecipitate of this TPA-treated extract. It is known that Raf-1 is ca. 72 to 76 kDa, that it is tyrosine phosphorylated (16, 40), and that Raf-1 tyrosine phosphorylation increases after TPA treatment (2). Further, Raf-1 is known to be involved in IgM-mediated signaling cascades (57). Hence, we decided to examine whether the protein associated with prohibitin was Raf-1. This was verified by a coimmunoprecipitation-Western blot experiment, with whole-cell extracts from dividing U937 cells. As shown in Fig. 4B, all the three Rb family members could be detected in association with prohibitin; we observed that in two other cell lines as well: Ramos and Daudi (62). Interestingly, Raf-1 could be detected in the prohibitin immune precipitate but not in an unrelated kinase (p38). These interactions were specific, since neither Raf-1 nor Rb family proteins could be detected in a c-Myc immune precipitate.
FIG. 4.
Prohibitin associates with Raf-1 in vivo and in vitro. (A) Western blot analysis with an anti-P-Tyr antibody on whole-cell extracts or prohibitin immunoprecipitates from dividing or TPA-treated U937 cells. The major 72-kDa protein band comigrates with Raf-1. (B) Association of Rb family proteins and Raf-1 with prohibitin in U937 cells. Extracts from dividing U937 cells were immunoprecipitated with an anti-c-Myc antibody as a control or an antiprohibitin antibody. The immune precipitates were tested for the presence of the indicated proteins by Western blotting. Raf-1 can be detected in prohibitin immune precipitates but not p38 kinase. (C) GST-binding assay with in vitro-synthesized Raf-1 and GST fusions of Rb and prohibitin. RL indicates one-fifth of the amount of rabbit reticulocyte lysate used in the binding reaction. (D) In a similar experiment, in vitro-synthesized prohibitin was tested for binding to GST–Raf-1 beads or beads primed with GST protein. Prohibitin specifically binds to GST–Raf-1.
Additional experiments were conducted to verify whether Raf-1 and prohibitin could interact directly. GST binding assays were conducted toward this purpose, and the results are shown in Fig. 4C and D. In the first experiment, Raf-1 was synthesized in vitro in rabbit reticulocyte lysates, and its binding to beads carrying GST as control, or to GST fusions of Rb or prohibitin, was assessed. As shown in Fig. 4C, Raf-1 could bind to Rb and prohibitin very efficiently. Under identical conditions, Raf-1 had no binding to a variety of other GST fusion proteins, indicating that its binding to Rb and prohibitin is a specific interaction.
Similarly, the binding of in vitro-synthesized prohibitin to GST–Raf-1 was assessed. As shown in Fig. 4D, prohibitin could bind efficiently to GST–Raf-1 beads but not to control GST beads. These experiments suggest that prohibitin and Raf-1 can bind to each other, and this interaction is probably direct.
Different regions of Raf-1 are involved in binding to Rb and prohibitin.
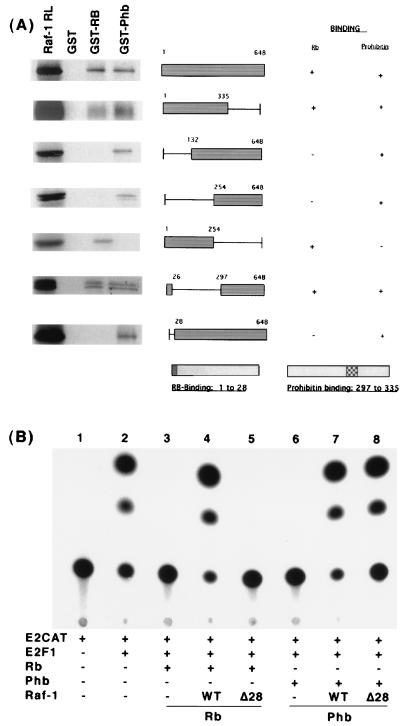
Attempts were made to identify the region of Raf-1 involved in binding to Rb and prohibitin. GST-binding assays using a panel of Raf-1 deletion mutants had shown that the extreme amino-terminal 28 amino acids of Raf-1 are involved in binding to Rb (61). The ability of the same set of Raf-1 deletion mutants to bind to prohibitin was tested similarly. As shown in Fig. 5A, full-length Raf-1 protein, as well as one spanning residues 1 to 335, could bind to both Rb and prohibitin. Deletion of the amino-terminal 132 amino acids abolished the ability of Raf-1 to bind to Rb, but it could bind to prohibitin very well. A similar binding pattern was observed with a Raf-1 molecule lacking the amino-terminal 254 amino acids. In contrast, the amino-terminal 254 amino acids could bind to Rb but not to prohibitin. A constitutively active form of Raf-1, Raf-1 BXB, whose amino-terminal 26 amino acids were fused to the carboxy-terminal residues 297 to 648, could bind to both Rb and prohibitin. This indicated that the prohibitin-binding region of Raf-1 spans residues 297 to 335. Interestingly, the Raf-1Δ28 construct, which could not bind to Rb, was very efficient in binding to prohibitin.
FIG. 5.
Raf-1 binds to Rb and prohibitin through distinct domains. (A) Binding assay results with different deletion mutants of Raf-1 synthesized in vitro and GST-Rb and GST-prohibitin. Filled box represents the region used for binding, and the line indicates the deleted region. RL represents one-fifth of the lysate used for binding. The regions of Raf-1 involved in binding to Rb and prohibitin are also shown. (B) Reversal of prohibitin-mediated repression of E2F1 by Raf-1. T47D cells were transfected with an E2CAT reporter and E2F1. Transfection of Rb (lanes 3 to 5) or prohibitin (lanes 6 to 8) could repress transcription. Cotransfection of wild-type Raf-1 could reverse both Rb-mediated (lane 4) and prohibitin-mediated (lane 7) repression; Raf-1Δ28 had no effect on Rb (lane 5) but could fully reverse prohibitin-mediated repression (lane 8).
Since our earlier experiments had shown that Raf-1 could effectively reverse Rb-mediated repression of E2F activity, we next examined whether Raf-1 could reverse prohibitin-mediated repression of E2F1 activity as well. A transient-transfection experiment in T47D cells showed that wild-type Raf-1 could reverse Rb-mediated repression of E2F1 activity, (Fig. 5B, lane 4), whereas Raf-1Δ28 had no effect. In contrast, prohibitin-mediated repression of E2F1 could be effectively reversed by Raf-1 wild type (lane 7), as well as Raf-1Δ28. This experiment suggests that Raf-1 can reverse prohibitin-mediated repression of E2F activity and that residues 1 to 28 of Raf-1 are essential only for targeting Rb function.
A colony formation assay was performed on T47D cells to verify whether Raf-1 could affect prohibitin-mediated suppression of cell proliferation. As shown in Table 2, transfection of prohibitin reduced the number of colonies about threefold, whereas transfection of Raf-1 could increase the number of colonies formed. Cotransfection of Raf-1 could effectively reverse prohibitin-mediated repression of cell proliferation, but the effect was not as pronounced as that on Rb.
TABLE 2.
Raf-1 can reverse prohibitin-mediated repression of colony formationa
| Vector transfected | No. of colonies
|
|
|---|---|---|
| Expt 1 | Expt 2 | |
| pSVNeo | 141 | 149 |
| pBABE-Puro | 135 | 158 |
| Puro+Neo | 133 | 146 |
| Raf-1–Neo, WT | 208 | 208 |
| Puro-Rb | 36 | 51 |
| Prohibitin-Neo | 41 | 86 |
| Rb+Raf-1, WT | 231 | 225 |
| Prohibitin+Raf-1, WT | 135 | 129 |
Suppression of colony formation by Rb and prohibitin is reversed by Raf-1. T47D cells grown in 35-mm-diameter dishes were stably transfected with the indicated vectors and selected in antibiotic for 14 days. Colonies with 20 or more cells were counted. pCMV-Rb was cotransfected with pBABE-Puro; pCDNA3–Raf-1 vector carried neomycin resistance marker. Selection was done in 40 μg of neomycin or 1 μg of puromycin per ml; a combination of both antibiotics was used when Rb and Raf-1 were cotransfected. WT, wild type.
Raf-1 targets the carboxy-terminal region of prohibitin.
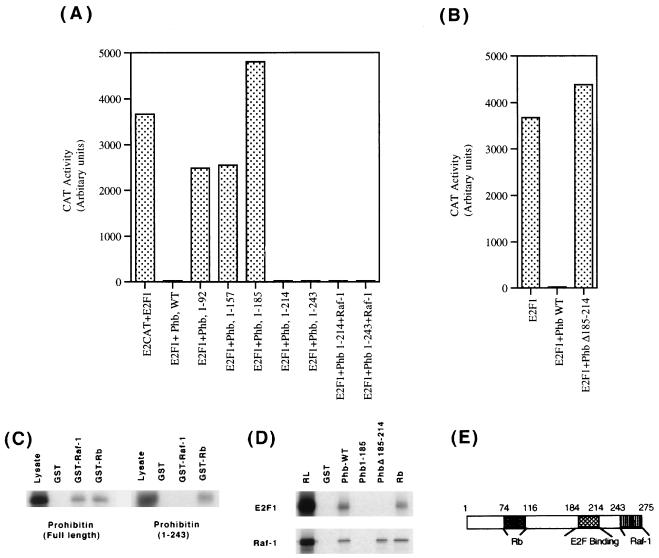
We had earlier found that the Rb-binding region (residues 74 to 116) is necessary for prohibitin to repress the transcriptional activity of E2F1. Experiments were designed to examine whether additional regions of prohibitin were necessary for the repressive activity of prohibitin. As a first step, we made deletion mutants of prohibitin that lacked various numbers of residues from the C-terminal end and examined their ability to repress E2F activity. A prohibitin construct expressing residues 1 to 157 was unable to repress E2F activity (Fig. 6A); similarly, a construct spanning residues 1 to 185 had no repressive effects on E2F1. Prohibitin constructs having residues 1 to 214, as well as residues 1 to 243, could repress E2F activity as effectively as the full-length prohibitin, suggesting that the region spanning residues 185 to 214 is required for repression of E2F activity, in addition to the Rb-binding domain (residues 74 to 116). This was confirmed by making a prohibitin construct with an internal deletion of residues 185 to 214 (as shown in Fig. 6B), whereas wild-type prohibitin could repress E2F1 activity, the deletion construct was unable to do so.
FIG. 6.
Raf-1 targets the carboxy-terminal end of prohibitin. (A) Mapping of the regions of prohibitin required for repression of E2F activity. C-terminal deletion mutants of prohibitin were tested for their ability to repress E2F1-mediated transcription in T47D cells. Prohibitins 1-157 and 1-185 were unable to repress transcription, but prohibitin 1-214 and 1-243 could. While both the 1-214 and 1-243 mutants of prohibitin could repress E2F1, the repression was not affected by cotransfected Raf-1. (B) The involvement of residues 185 to 214 in transcriptional repression was tested by using an internal deletion mutant of prohibitin. Deletion of the residues 185 to 214 impaired the ability of prohibitin to repress E2F1 activity. (C) The carboxy-terminal end of prohibitin interacts with Raf-1. A GST-binding assay showing the binding of prohibitin to GST fusions of Raf-1 and Rb; while full-length prohibitin could bind to both the beads, prohibitin 1-243 could bind only to Rb. (D) Residues 185 to 214 of prohibitin is involved in binding to E2F1. E2F1 synthesized in vitro was checked for binding to GST fusions of full-length prohibitin or prohibitin Δ185-214, or Rb. E2F1 could bind only to full-length prohibitin and Rb (upper panel), whereas Raf-1 could bind to all the three GST fusion proteins. (E) Schematic showing the different regions of prohibitin involved in binding to Rb, E2F1, and Raf-1.
The ability of Raf-1 to reverse the repression mediated by the C-terminal truncated prohibitin molecules was examined by a cotransfection experiment in T47D cells. As shown in Fig. 6A, Raf-1 was unable to reverse the repression mediated by prohibitin 1-243; this result suggested that Raf-1 targets the C-terminal region of prohibitin spanning residues 243 to 275. It was examined by using a GST-binding assay whether the region of prohibitin residues 243 to 275 that was targeted by Raf-1 was involved in the binding to Raf-1 also. As shown in Fig. 6C, left panel, in vitro-synthesized full-length prohibitin was able to bind to GST-Rb as well as GST–Raf-1 but not to control GST beads. In contrast, prohibitin 1-243 could bind to GST-Rb but not to GST–Raf-1 (right panel). This suggests that the extreme C-terminal region of prohibitin is involved in binding to Raf-1, as well as responding to Raf-1-mediated signaling events. Similar experiments were conducted to examine whether there was any correlation between the abilities of prohibitin to bind E2F and to repress transcription. E2F1 made in rabbit reticulocyte lysates could bind very well to GST fusions of full-length prohibitin and Rb, but it could not bind to the truncated prohibitin 1-185 or the internal deletion mutant Δ185-214 (Fig. 6D, top panel). This suggests that prohibitin has to associate with E2F1 to repress its transcription. The binding of Raf-1 synthesized in vitro to the same beads was also tested; it was found that Raf-1 could not bind to the truncated prohibitin 1-185, but it could bind to the internal deletion mutant Δ185-214 (Fig. 6D, bottom panel). This result confirms the earlier findings that Raf-1 targets the C-terminal region of prohibitin. These experiments help delineate three distinct functional domains of prohibitin: the region from residues 74 to 116, which is involved in Rb binding; the region from residues 185 to 214, which is necessary for transcriptional repression; and the carboxy-terminal 243-275 region involved in responding to Raf-1 (Fig. 6E).
The contribution of the various domains of prohibitin to growth suppression was assessed by a colony formation assay on T47D cells (Table 3). It was found that full-length prohibitin could effectively inhibit colony formation after 14 days of antibiotic selection; interestingly, prohibitin mutants 1-157 and 1-185 had little effect on the number of colonies. In addition, prohibitin constructs possessing an internal deletion of the region from residues 74 to 116 or 185 to 214 were not able to suppress colony formation. Thus, it appears that the ability of prohibitin to exert growth regulation coincides with its ability to repress transcription mediated by E2F, as in the case of Rb.
TABLE 3.
Regions of prohibitin involved in growth suppression and Raf-1 responsea
| Vector transfected | No. of colonies
|
|
|---|---|---|
| Expt 1 | Expt 2 | |
| pSVNeo | 165 | 175 |
| Raf-1–Neo, WT | 221 | 235 |
| Prohibitin-Neo, WT | 46 | 48 |
| Prohibitin 1-157 | 171 | 158 |
| Prohibitin 1-185 | 139 | 149 |
| Prohibitin 1-214 | 41 | 44 |
| Prohibitin 1-243 | 58 | 61 |
| Prohibitin Δ74-116 | 158 | 155 |
| Prohibitin Δ185-214 | 154 | 169 |
| Prohibitin+Raf-1 | 245 | 233 |
| Prohibitin 1-157+Raf-1 | 217 | 225 |
| Prohibitin 1-185+Raf-1 | 219 | 211 |
| Prohibitin 1-214+Raf-1 | 52 | 55 |
| Prohibitin 1-243+Raf-1 | 58 | 54 |
Approximately 10,000 T47D cells were transfected with 2 μg each of the indicated vectors. PCDNA3Raf-1 and pCDNA3 prohibitin vectors carried the neomycin resistance marker; colonies with 20 or more cells were counted after 14 days of selection in 40 μg of neomycin per ml. WT, wild type.
In contrast, the constructs lacking the carboxy-terminal end (constructs 1-214 and 1-243) were quite efficient in repressing proliferation. Since these two mutants of prohibitin could not respond to Raf-1 in transient-transfection assays, we examined whether Raf-1 could affect the growth suppression mediated by these constructs. It was found that Raf-1 could reverse the suppression of colony formation mediated by wild-type prohibitin; similarly, the presence of Raf-1 increased the number of colonies when the truncation mutants 1-157 and 1-185 were present. Raf-1 had no effect, however, on the suppression of colony formation mediated by the prohibitin mutants 1-214 and 1-243; this again suggests that the carboxy terminus of prohibitin is the functional target of Raf-1. Further, it appears that the ability of prohibitin to suppress cell proliferation correlates with its ability to repress transcription and that agents that can reverse the transcriptional repression can also affect the growth suppression mediated by prohibitin.
E2F regulation by Rb and prohibitin respond to different signals.
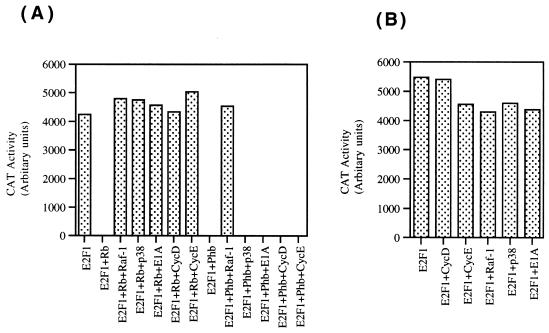
We had observed that E1A was unable to reverse prohibitin-mediated repression of E2F1. Since the results described above demonstrate that Raf-1 was quite effective in releasing prohibitin-mediated repression of E2F1, we performed a series of transient-transfection experiments to assess the effect of different agents on Rb- and prohibitin-mediated repression of E2F1. First, we compared the effects of E1A, Raf-1, and p38 kinase on Rb and prohibitin (Fig. 7A). As we have observed earlier, E1A, Raf-1, and p38 all could effectively inactivate Rb, reversing the repression of E2F transcriptional activity (63). In contrast, only Raf-1 was able to reverse prohibitin-mediated inhibition of E2F activity (Fig. 7A), indicating that Rb and prohibitin probably respond to different signal transduction cascades. A summary of these results is presented in Table 4.
FIG. 7.
Differential regulation of Rb and prohibitin by upstream molecules. (A) Transient-transfection experiment in T47D cells with an E2CAT reporter, which is induced by E2F1. Both Rb and prohibitin could repress E2F1 activity. Whereas Raf-1 could reverse both Rb- and prohibitin-mediated repression of E2F1, p38 and E1A could only reverse Rb but not prohibitin repression. Similarly, cotransfection of cyclins D and E can reverse Rb-mediated repression of E2F1 but had no effect on prohibitin. (B) Control experiment showing that the different agents used do not directly affect E2F1 activity in T47D cells.
TABLE 4.
Differential response of Rb and prohibitin to upstream regulatorsa
| Signal | Presence (+) or absence (−) of response
|
|
|---|---|---|
| Rb | Prohibitin | |
| E1A | + | − |
| Raf-1 | + | + |
| p38 | + | − |
| CycD | + | − |
| CycE | + | − |
| Serum stimulation | + | +b |
| IgM | − | + |
A summary of the effect of different upstream regulators on Rb and prohibitin. All of the agents tested except IgM could reverse Rb-mediated inhibition of E2F activity. In contrast, only Raf-1 and IgM could affect prohibitin. Serum stimulation was also able to overcome prohibitin-mediated inhibition of E2F but followed different kinetics. CycD, cyclin D; CycE, cyclin E.
Delayed.
Since cyclins D and E, along with their dependent kinases, are known to be the major regulators of Rb function during the course of G1 progression (35, 64), we decided to examine whether these molecules had any effect on prohibitin function. In a similar transient transfection experiment on T47D cells, cotransfection of either cyclin D or cyclin E could effectively repress Rb-mediated repression of E2F transcriptional activity (Fig. 7A). Both of these cyclins had no effect on prohibitin-mediated repression of E2F1 activity, suggesting that prohibitin is not regulated by the same cyclin dependent kinases that regulate Rb. Cotransfection of E1A, p38, Raf-1, or cyclins D and E had only minimal effect on E2F1 directly in T47D cells (Fig. 7B).
Prohibitin-mediated repression of E2F is reversed by IgM receptor stimulation.
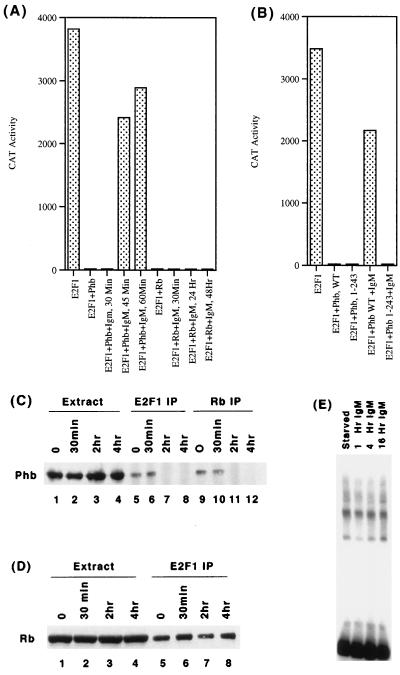
Prohibitin and a prohibitin-related protein have been reported to be associated with IgM receptors in B-cell lines (55), and stimulation of IgM receptors has been shown to activate Raf-1 kinase (57). Since prohibitin can reverse E2F activity and since it associates with IgM receptors as well as Raf-1, we examined whether prohibitin-mediated repression of E2F activity is affected by stimulating the IgM receptors. The human B-cell line Ramos was transiently transfected with E2CAT reporter whose activity was stimulated by cotransfecting E2F-1. Interestingly, treating the transfected cells with 1 μg of an anti-IgM antibody per ml could reverse prohibitin-mediated repression of E2F (Fig. 8A). The reversal was reflected in an increase in CAT activity, which could be observed as early as 45 min after treatment; treatment of the cells for additional periods of time (up to 48 h) did not result in any further increase in E2F activity. Identical results were obtained in two other B-cell lines, Daudi and Raji (data not shown).
FIG. 8.
IgM stimulation can modulate prohibitin function. (A) Ramos cells were transiently transfected with E2CAT and E2F1, along with prohibitin or Rb. Cells were stimulated with 1 μg of an anti-IgM antibody per ml for the indicated periods of time; prohibitin-mediated repression of E2F1 is reversed within 45 min of stimulation. IgM stimulation, even for prolonged periods of time, had no effect on Rb. (B) The carboxy-terminal region of prohibitin is necessary for responding to IgM. In a cotransfection experiment, full-length prohibitin or prohibitin 1-243 was used to repress E2F1. IgM stimulation for 2 h reversed the repression mediated by the full-length prohibitin but not prohibitin 1-243. (C) Association of E2F1 and Rb with prohibitin is disrupted upon IgM signaling. Extracts from Ramos cells stimulated with an anti-IgM antibody for the indicated period of time were immunoprecipitated with an anti-E2F1 antibody (lanes 5 to 8) or anti Rb antibody (lanes 9 to 12). Prohibitin was detected by Western blotting. Lanes 1 to 4 show a prohibitin level in an equivalent amount of the extracts. (D) Similar experiment showing the association of Rb with E2F1 (lanes 5 to 8). Rb remains associated with E2F1 during the course of IgM stimulation. (E) EMSA showing the E2F binding activity in Ramos cells stimulated with anti-IgM antibody for the indicated periods of time.
It was next examined whether the effect of IgM receptor stimulation was restricted to prohibitin. In a similar cotransfection experiment on Ramos cells, Rb was used to repress E2F1 activity instead of prohibitin. Stimulation of the IgM receptor with 1 μg of an anti-IgM antibody per ml had no effect on Rb-mediated repression of E2F1 activity even after 48 h (Fig. 8A). This experiment suggests that the IgM receptor stimulation specifically targets prohibitin and that prohibitin can respond to signals from this receptor. It was next examined whether the C-terminal deletion mutant of prohibitin that could not respond to Raf-1 was inactivated by IgM-mediated signaling. As shown in Fig. 8B, IgM stimulation could reverse repression mediated by full-length but not the C-terminal truncated prohibitin, indicating that Raf-1 and IgM target the same domain of prohibitin.
Since prohibitin was found to bind to E2F1 in vivo, we attempted to assess whether the physical interaction between prohibitin and E2F1 is disrupted upon IgM signaling. Whole-cell extracts prepared from Ramos cells stimulated with an anti-IgM antibody for various periods of time were immunoprecipitated with antibodies to E2F1 or Rb (Fig. 8C). The presence of prohibitin in the immunoprecipitates was examined by Western blot analysis. The level of prohibitin in the extracts did not change upon IgM stimulation. There was a considerable amount of prohibitin associated with E2F1 and Rb in unstimulated cells and cells stimulated for 30 min with the IgM antibody but, interestingly, there was no detectable amount of prohibitin associated with E2F1 or Rb after 2 or 4 h of stimulation. Thus, it appears that the reversal of prohibitin-mediated repression of E2F1 by IgM correlates with a disruption of its interaction with Rb and/or E2F1. In a parallel experiment, we tested whether there is any change in the amount of Rb associated with E2F1 when cells are stimulated with IgM. As shown in Fig. 8D, the amount of Rb associated with E2F1 remains constant during the course of stimulation. It appears that IgM receptor stimulation leads to a specific dissociation of prohibitin from Rb and E2F.
There was no significant change in the DNA binding activity of E2F and E2F-containing complexes in whole-cell extracts of Ramos cells treated with IgM (Fig. 8E). This might suggest that the release of repression from prohibitin may not involve changes in the DNA binding activity of E2F1 but might be occurring through the inactivation of additional repressors tethered by prohibitin onto E2F1. Though it has been reported that p107-E2F complexes are altered in response to IgM stimulation, such changes occur only after prolonged periods of stimulation (30).
IgM-mediated inactivation of prohibitin does not require cyclin-dependent kinase activity.
Attempts were made to assess which signaling cascades are involved in mediating the effects of IgM receptor stimulation or prohibition. As a first step, IgM stimulation was conducted in the presence of chemical inhibitors of different kinases, and the results are shown in Fig. 9A. IgM stimulation for 2 h in the presence of PD98059, a specific inhibitor of MEK1 kinase (39), totally blocked IgM-mediated reversal of prohibitin function. Olomoucine, which inhibits cdk2 and cdc2, had no effect on prohibitin function, ruling out a role for these kinases in the process. Further, a p38 kinase inhibitor, SB203580 (56, 70), could also not block IgM-mediated inactivation of prohibitin. This experiment suggested that IgM-mediated reversal of prohibitin occurs mainly through the mediation of the mitogen-activated protein (MAP) kinase cascade; the direct role of Raf-1 in this process is not clear.
FIG. 9.
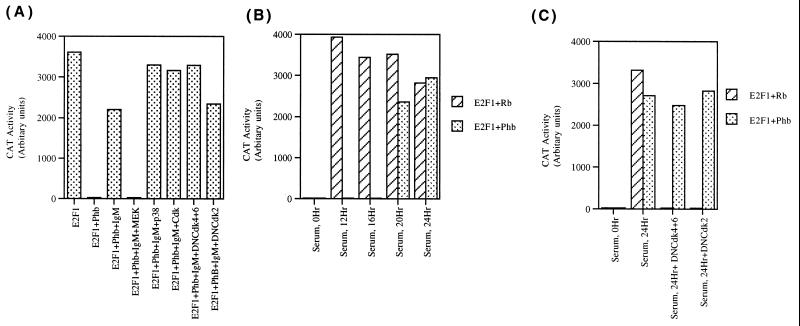
Cyclin-dependent kinases are not involved in the regulation of prohibitin function. (A) Ramos cells were transiently transfected with E2CAT, E2F1, and prohibitin as in Fig. 8A. Stimulation with anti-IgM antibody for 2 h released prohibitin-mediated repression of E2F1; MEK inhibitor (PD98059, 100 μM) could block IgM-mediated reversal of prohibitin function, but inhibitors of p38 kinase (SB203580, 10 μM) or cdk2 (Olomoucine, 200 μM) had no effect. A similar experiment was conducted to check whether dominant-negative cyclin-dependent kinases (4 μg each of dominant-negative cdk4 and cdk6 and 4 μg of dominant-negative cdk2) could block IgM-mediated inactivation of prohibitin. Stimulation with anti-IgM could still reverse prohibitin-mediated repression of E2F1. (B) Serum stimulation of quiescent Ramos cells reverses prohibitin and Rb function. Ramos cells were transiently transfected with E2CAT and E2F1, along with Rb and prohibitin; serum stimulation reversed Rb-mediated repression of E2F1 within 12 h; repression by prohibitin was released only after 20 h. (C) Cyclin–cyclin-dependent kinase activity is required for reversing Rb, but not prohibitin, function. Ramos cells were transiently transfected with E2CAT, E2F1, Rb, or prohibitin as in panel B. Serum stimulation for 24 h reversed both Rb and prohibitin, but cotransfected dominant-negative cdk4 and cdk6 blocked inactivation of Rb not prohibitin. Similar results were obtained with dominant-negative cdk2.
The possibility that cyclin-dependent kinases are not involved in IgM-mediated regulation of prohibitin was examined by a direct experiment. A transient-transfection experiment was conducted, where a dominant-negative cdk4 and cdk6, or cdk2 was included. As shown in Fig. 9A, cotransfection of a combination of dominant-negative cdk4 and cdk6 or a dominant-negative cdk2 had no significant effect on IgM-mediated reversal of prohibitin function. This finding is consistent with the observation that cyclins D and E had no direct effect on prohibitin-mediated repression of E2F activity and supports the results obtained with Olomoucine.
Since it was found that cyclins and cyclin-dependent kinases had no significant effect on prohibitin function, it was examined whether prohibitin-mediated repression of E2F1 activity responds to serum stimulation (Fig. 9B). Ramos cells were transiently transfected with E2CAT and E2F1, and E2F activity was repressed by cotransfecting Rb or prohibitin. Serum stimulation of the transfected cells showed that Rb-mediated repression of E2F1 activity was completely reversed within 12 h; there was no change in prohibitin-mediated repression of E2F1 at the same time point. It was found that prohibitin-mediated inhibition of E2F1 activity was reversed only after 20 h of serum stimulation. Similarly, serum stimulation resulted in a delayed release of prohibitin-mediated repression of E2F1 activity in T47D cells also. This experiment suggests that Rb- as well as prohibitin-mediated repression of E2F1 activity can respond to serum stimulation but that they follow different kinetics.
G1 progression after serum stimulation of quiescent cells is known to involve the inactivation of the Rb protein by cyclins and cyclin-dependent kinases (35, 54, 64). Since Rb and prohibitin responded differently to IgM stimulation and serum stimulation, attempts were made to evaluate whether dominant-negative cyclin-dependent kinases had any differential effects on Rb and prohibitin. Ramos cells were transiently transfected with E2CAT and E2F1, along with Rb or prohibitin. The transfected cells were serum stimulated for 24 h, and both Rb- and prohibitin-mediated repression of E2F1 were released by this time (Fig. 9C). Cotransfection of a combination of cdk4 and cdk6 totally prevented the release of Rb-mediated repression of E2F1 in response to serum; a dominant-negative cdk2 construct also had a similar effect. Interestingly, neither cdk4 and cdk6 nor cdk2 dominant-negative constructs could block serum-mediated inactivation of prohibitin. This experiment shows that though prohibitin and Rb can respond to serum stimulation, they are inactivated by different signaling pathways.
DISCUSSION
The transcriptional activity of E2F1 is regulated at multiple levels, mainly through its interaction with a variety of cellular proteins (15, 41). The interaction of E2F1 with Rb has attracted considerable attention since E2F appears to be the major downstream target of Rb in modulating G1/S transition. In addition, E2F1 can interact with positive modulators such as DP1 and DP2, as well as the adenovirus E4 protein (11, 12, 33). Further, the transcriptional coactivator p300/CBP has been suggested to be a positive modulator of E2F1 activity (34, 59, 60). E2F1 activity has been shown to be modulated by cyclin A and cdk2 after direct binding to its amino-terminal domain (69). This results in the phosphorylation of E2F1 or DP1, which affects the transcriptional activity of the former (29). Further, proteins like Mdm2 and p53 have been suggested to bind to E2F1 or to affect its transcriptional activity indirectly (43, 51, 68). Our results suggest that prohibitin is a novel regulator of E2F function and that prohibitin is probably acting as a link between certain specific signal transduction pathways and the cell cycle machinery.
We have previously shown that the repression mediated by Rb and prohibitin showed qualitative differences, mainly based on the observation that adenovirus E1A protein could not affect prohibitin-mediated repression of E2F activity. Further, Rb could repress only E2Fs 1, 2, and 3, whereas prohibitin could inhibit all five transcriptionally active E2Fs. We show here that Rb and prohibitin target different regions of the E2F1 protein for repression, since prohibitin requires the marked box region for exerting its effects. The amino-terminal end of the marked box region is involved in binding to DP1 (50), but the region targeted by prohibitin has no known function. Interestingly, this region is fairly conserved between the different E2F family members, probably enabling prohibitin to repress all of them, unlike Rb family members. Our experiments show that prohibitin can physically interact with E2F1 through this region of the marked box. Nevertheless, the finding that the Rb binding region is also necessary for repressing E2F activity raises the possibility that prohibitin has to tether to Rb or an Rb family member to effectively bind E2F and repress its activity. The precise mechanism by which prohibitin represses the transcriptional activity of E2F is not yet clear, and in preliminary experiments we do not find prohibitin affecting the DNA binding activity of E2F1 or its interaction with DP1 (data not shown). Hence, we believe that prohibitin could be recruiting additional corepressor molecules onto E2F (62); a good candidate appears to be HDAC1, which has been shown to be involved in mediating Rb-mediated transcriptional repression (5, 36, 37). As we had proposed earlier, it may be imagined that prohibitin is acting as an adaptor between such repressors and Rb/E2F. The identity of the repressors, as well as the stoichiometry of the interactions, remains to be elucidated.
One common feature of the repression of E2F by Rb and prohibitin is the correlation between their ability to inhibit E2F activity and their ability to suppress cell proliferation. It has been established in the case of Rb that E2F can antagonize Rb-mediated growth suppression and that an E2F molecule that could not bind to Rb could induce cell proliferation irrespective of the presence of Rb (48). We find that a similar situation exists in the case of prohibitin as well, since prohibitin could block induction of colony formation by wild-type E2F, as well as E2F1 mutant A, both of which it could repress. Prohibitin had no effect on the induction of colony formation by mutants B or C, which it could not repress. This is analogous to our finding that a prohibitin mutant that could not bind to Rb and repress E2F activity could not suppress colony formation in T47D cells. Though growth-suppressive properties of Rb correlated with its ability to repress E2F, other phenomena, such as induction of a senescent phenotype in Saos-2 osteosarcoma cells, did not require the binding or repression of E2F activity (47). It would be interesting to see whether prohibitin has similar functions related to growth control which are separable from its E2F regulation.
We had reported previously that Raf-1 could interact with the Rb protein and reverse its growth-suppressive properties (61). We find that Raf-1 is capable of interacting with prohibitin as well; in fact, our studies on the Rb–Raf-1 interaction stemmed from the observation that Raf-1 can bind to prohibitin, which in turn could bind to Rb. So far, we have no evidence to suggest that prohibitin, Rb, and Raf-1 exist in a ternary complex, since all three proteins can bind to each other in a GST binding assay. The domains of Raf-1 and prohibitin involved in interacting with each other are distinct from the domains involved in Rb binding. Unlike the interaction of Raf-1 with Rb, which is serum inducible, we do not find any change in the status of the prohibitin–Raf-1 interaction. Prohibitin protein has a certain degree of similarity to the 14-3-3 class of adapter proteins and may be functioning as an adapter itself. This raises the possibility that prohibitin functions as a specific adapter that can stably interact with a variety of molecules, facilitating their modulation by signaling cascades. Interestingly, it has been reported recently that prohibitin physically interacts with mixed lineage kinase-2 (MLK2), which is a MAP kinase-kinase-kinase like Raf-1, that functions in the p38/JNK signaling pathway (17, 46).
A role for prohibitin in replicative senescence, as well as mitochondrial functions, has been reported (4, 9, 52), and a putative amino-terminal domain is believed to facilitate these functions. Deletion of the amino-terminal 34 amino acids did not affect prohibitin-mediated repression of E2F activity, suggesting that this region plays a distinct role in growth regulation separate from the potential mitochondrial functions. Further, prohibitin, as well as a prohibitin-related protein, has been reported to be associated with the IgM receptor in B cells, but the functional consequence of this interaction is not known. We find that IgM stimulation can specifically release prohibitin-mediated repression of E2F activity, which corresponds with a dissociation of prohibitin from E2F1 as well as Rb. It is possible that the release of prohibitin causes the inactivation or dissociation of a corepressor leading to the transcriptional activity. So far, we do not find any change in the DNA-binding activity of E2F in response to IgM stimulation. It has been reported that IgM stimulation can affect the Rb-related p107 protein and its regulation of E2F activity, but this does not occur until 48 h after stimulation (30). Hence, we believe that the regulation of prohibitin by the IgM receptor is a distinct event which occurs early in the signaling cascade. It may be that prohibitin-mediated repression of E2F is released first in response to IgM signaling and, after a significant lapse of time, the p107-E2F complex is disrupted by secondary events taking place in the cell.
The release of prohibitin-mediated repression of E2F activity apparently involves the MAP kinase pathway, since the chemical inhibitor of MEK1 could inhibit this; we are not sure whether the concentration of the inhibitor used can affect Raf-1 activity. It has been reported that IgM receptor stimulation leads to an activation of Raf-1 kinase. Interestingly, IgM stimulation cannot affect a truncated prohibitin protein that could not respond to Raf-1. Based on these observations, it is tempting to speculate that IgM-mediated inactivation of prohibitin occurs through the recruitment of the Raf-1 protein, but so far we have not found an increased binding of Raf-1 to prohibitin upon IgM stimulation. We had observed earlier that serum stimulation of human diploid fibroblasts leads to the nuclear translocation of a small fraction of Raf-1, allowing it to colocalize with Rb (59). Hence, although it may be that a similar event is taking place upon IgM stimulation, it is not yet clear whether Raf-1 has to physically interact with prohibitin to bring about this effect. This has been difficult to determine, since the prohibitin-binding domain of Raf-1 is immediately adjacent to its kinase domain, and mutations of this region might alter the kinase activity of Raf-1. The actual mechanisms of prohibitin inactivation remain unclear, since in the preliminary in vitro kinase reaction prohibitin could not be phosphorylated either by Raf-1 or ERK2.
One of the intriguing aspects of prohibitin-mediated repression of E2F is that the inhibitory effects of prohibitin do not respond to many agents that reverse Rb-mediated repression of E2F. A case in point that is notable is the adenovirus E1A protein. Similarly, cyclins D and E, which inactivate the Rb protein by phosphorylation, had no effect on prohibitin-mediated repression of E2F1. This was observed in direct overexpression experiments, where cyclins D and E could reverse Rb, but not prohibitin, function. Further, we find that though prohibitin-mediated repression of E2F1 is negated upon prolonged serum stimulation of quiescent T47D cells, this reversal could not be blocked by dominant-negative cyclin-dependent kinases. This observation implies that the inactivation of prohibitin during cell cycle progression is mediated by a different set of kinases or by other means that are independent of phosphorylation. Similarly, while we find that overexpression of Raf-1 can inactivate prohibitin, p38 kinase has no effect; in contrast, both Raf-1 and p38 can target the Rb protein. This scenario further supports the notion that prohibitin-mediated regulation of E2F responds to a different set of signals than those regulating Rb.
Prohibitin was originally cloned based on its ability to bring about a very potent G1/S arrest (38, 42), and our earlier results suggest that prohibitin brings about the growth arrest by repressing E2F activity. The results presented here suggest that prohibitin is a novel regulator of E2F function and that prohibitin facilitates the cell cycle machinery to respond to extracellular signals that cannot target Rb and its family members. These findings, we believe, will help us further understand the molecular mechanisms by which prohibitin regulates cell proliferation, as well as how extracellular signals contact the cell cycle machinery.
ACKNOWLEDGMENTS
We thank David Johnson for the kind gift of E2F1 fusion proteins and for helpful suggestions and J. Keith McClung for anti-prohibitin antibodies.
This work was supported by a grant from the National Cancer Institute (RO-1 CA63136). S.P.C. is a recipient of the Irma-Hirschl Trust Research Award.
REFERENCES
- 1.Adams P D, Kaelin W J. The cellular effects of E2F overexpression. Curr Top Microbiol Immunol. 1996;208:79–93. doi: 10.1007/978-3-642-79910-5_4. [DOI] [PubMed] [Google Scholar]
- 2.Barnard D, Diaz B, Clawson D, Marshall M. Oncogenes, growth factors and phorbol esters regulate Raf-1 through common mechanisms. Oncogene. 1998;17:1539–1547. doi: 10.1038/sj.onc.1202061. [DOI] [PubMed] [Google Scholar]
- 3.Beijersbergen R L, Bernards R. Cell cycle regulation by the retinoblastoma family of growth inhibitory proteins. Biochim Biophys Acta. 1996;1287:103–120. doi: 10.1016/0304-419x(96)00002-9. [DOI] [PubMed] [Google Scholar]
- 4.Berger K H, Yaffe M P. Prohibitin family members interact genetically with mitochondrial inheritance components in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:4043–4052. doi: 10.1128/mcb.18.7.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 6.Bremner R, Cohen B L, Sopta M, Hamel P A, Ingles C J, Gallie B L, Phillips R A. Direct transcriptional repression by pRB and its reversal by specific cyclins. Mol Cell Biol. 1995;15:3256–3265. doi: 10.1128/mcb.15.6.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cartwright P, Muller H, Wagener C, Holm K, Helin K. E2F-6: a novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene. 1998;17:611–623. doi: 10.1038/sj.onc.1201975. [DOI] [PubMed] [Google Scholar]
- 8.Chellappan S P, Hiebert S, Mudryj M, Horowitz J M, Nevins J R. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 9.Coates P J, Jamieson D J, Smart K, Prescott A R, Hall P A. The prohibitin family of mitochondrial proteins regulate replicative lifespan. Curr Biol. 1997;7:607–610. doi: 10.1016/s0960-9822(06)00261-2. [DOI] [PubMed] [Google Scholar]
- 10.Cobrinik D. Regulatory interactions among E2Fs and cell cycle control proteins. Curr Top Microbiol Immunol. 1996;208:32–63. doi: 10.1007/978-3-642-79910-5_2. [DOI] [PubMed] [Google Scholar]
- 11.Cress W D, Nevins J R. Interacting domains of E2F1, DP1, and the adenovirus E4 protein. J Virol. 1994;68:4213–4219. doi: 10.1128/jvi.68.7.4213-4219.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cress W D, Nevins J R. Use of the E2F transcription factor by DNA tumor virus regulatory proteins. Curr Top Microbiol Immunol. 1996;208:63–78. doi: 10.1007/978-3-642-79910-5_3. [DOI] [PubMed] [Google Scholar]
- 13.DeGregori J, Leone G, Miron A, Jakoi L, Nevins J R. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc Natl Acad Sci USA. 1997;94:7245–7250. doi: 10.1073/pnas.94.14.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeGregori J, Leone G, Ohtani K, Miron A, Nevins J R. E2F-1 accumulation bypasses a G1 arrest resulting from the inhibition of G1 cyclin-dependent kinase activity. Genes Dev. 1995;9:2873–2887. doi: 10.1101/gad.9.23.2873. [DOI] [PubMed] [Google Scholar]
- 15.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 16.Fabian J R, Daar I O, Morrison D K. Critical tyrosine residues regulate the enzymatic and biological activity of Raf-1 kinase. Mol Cell Biol. 1993;13:7170–7179. doi: 10.1128/mcb.13.11.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fanger G R, Gerwins P, Widmann C, Jarpe M B, Johnson G L. MEKKs, GCKs, MLKs, PAKs, TAKs, and tpls: upstream regulators of the c-Jun amino-terminal kinases? Curr Opin Genet Dev. 1997;7:67–74. doi: 10.1016/s0959-437x(97)80111-6. [DOI] [PubMed] [Google Scholar]
- 18.Field S J, Tsai F Y, Kuo F, Zubiaga A M, Kaelin W J, Livingston D M, Orkin S H, Greenberg M E. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 19.Gaubatz S, Wood J G, Livingston D M. Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F-6. Proc Natl Acad Sci USA. 1998;95:9190–9195. doi: 10.1073/pnas.95.16.9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamel P A, Gill R M, Phillips R A, Gallie B L. Transcriptional repression of the E2-containing promoters EIIaE, c-myc, and RB1 by the product of the RB1 gene. Mol Cell Biol. 1992;12:3431–3438. doi: 10.1128/mcb.12.8.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helin K, Lees J A, Vidal M, Dyson N, Harlow H, Fattaey A. A cDNA encoding a pRB-binding protein with properties of the transcription factor E2F. Cell. 1992;70:337–350. doi: 10.1016/0092-8674(92)90107-n. [DOI] [PubMed] [Google Scholar]
- 22.Johnson D G, Schwarz J K, Cress W D, Nevins J R. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 23.Johnson D G, Ohtani K, Nevins J R. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 24.Johnson D G, Schneider-Broussard R. Role of E2F in cell cycle control and cancer. Front Biosci. 1998;3:447–448. doi: 10.2741/a291. [DOI] [PubMed] [Google Scholar]
- 25.Kaelin W G, Krek W, Sellers W R, DeCaprio J A, Ajchenbaum F, Fuchs C S, Chittenden T, Li Y, Farnham P J, Blanar M A E A. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell. 1992;70:351–364. doi: 10.1016/0092-8674(92)90108-o. [DOI] [PubMed] [Google Scholar]
- 26.Kitagawa M, Higashi H, Suzuki T I, Segawa K, Hanks S K, Taya Y, Nishimura S, Okuyama A. Phosphorylation of E2F-1 by cyclin A-cdk2. Oncogene. 1995;10:229–236. [PubMed] [Google Scholar]
- 27.Kowalik T F, DeGregori J, Leone G, Jakoi L, Nevins J R. E2F1-specific induction of apoptosis and p53 accumulation, which is blocked by Mdm2. Cell Growth Differ. 1998;9:113–118. [PubMed] [Google Scholar]
- 28.Kowalik T F, DeGregori J, Schwarz J K, Nevins J R. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krek W, Ewen M, Shirodkar S, Arany Z, Kaelin W J, Livingston D M. Negative regulation of the growth-promoting transcription factor E2F-1 by a stably bound cyclin A-dependent protein kinase. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 30.Lam E W, Choi M S, van der Sman J, Burbidge S A, Klaus G G. Modulation of E2F activity via signaling through surface IgM and CD40 receptors in WEHI-231 B lymphoma cells. J Biol Chem. 1998;273:10051–10057. doi: 10.1074/jbc.273.16.10051. [DOI] [PubMed] [Google Scholar]
- 31.Lam E W, Watson R J. An E2F-binding site mediates cell-cycle regulated repression of mouse B-myb transcription. EMBO J. 1993;12:2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.La Thangue N B. Dp and E2F proteins: an expanding family of heterodimeric transcription factors implicated in cell cycle control. Curr Opin Cell Biol. 1994;6:443–450. doi: 10.1016/0955-0674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 33.La Thangue N B. E2F and the molecular mechanisms of early cell-cycle control. Biochem Soc Trans. 1996;24:54–59. doi: 10.1042/bst0240054. [DOI] [PubMed] [Google Scholar]
- 34.Lee C W, Sorensen T S, Shikama N, La Thangue N B. Functional interplay between p53 and E2F through co-activator p300. Oncogene. 1998;16:2695–2710. doi: 10.1038/sj.onc.1201818. [DOI] [PubMed] [Google Scholar]
- 35.Lundberg A S, Weinberg R A. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/mcb.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 37.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 38.McClung J K, Jupe E R, Liu X-T, Dell’Orco R T. Prohibitin: potential role in senescence, development and tumor suppression. Exp Gerontol. 1995;30:99–124. doi: 10.1016/0531-5565(94)00069-7. [DOI] [PubMed] [Google Scholar]
- 39.Morooka T, Nishida E. Requirement of p38 mitogen-activated protein kinase for neuronal differentiation in PC12 cells. J Biol Chem. 1998;273:24285–24288. doi: 10.1074/jbc.273.38.24285. [DOI] [PubMed] [Google Scholar]
- 40.Morrison D K, Kaplan D R, Escobedo J A, Rapp U R, Roberts T M, Williams L T. Direct activation of the serine/threonine kinase activity of Raf-1 through tyrosine phosphorylation by the PDGF beta-receptor. Cell. 1989;58:649–657. doi: 10.1016/0092-8674(89)90100-1. [DOI] [PubMed] [Google Scholar]
- 41.Nevins J R. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 1998;9:585–593. [PubMed] [Google Scholar]
- 42.Nuell M J, Stewart D A, Walker L, Friedman V, Wood C M, Owens G A, Smith J R, Schneider E L, Dell O R, Lumpkin C K, et al. Prohibitin, an evolutionarily conserved intracellular protein that blocks DNA synthesis in normal fibroblasts and HeLa cells. Mol Cell Biol. 1991;11:1372–1381. doi: 10.1128/mcb.11.3.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Connor D J, Lam E W, Griffin S, Zhong S, Leighton L C, Burbidge S A, Lu X. Physical and functional interactions between p53 and cell cycle co-operating transcription factors, E2F1 and DP1. EMBO J. 1995;14:6184–6192. doi: 10.1002/j.1460-2075.1995.tb00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin X Q, Livingston D M, Ewen M, Sellers W R, Arany Z, Kaelin W G., Jr The transcription factor E2F-1 is a downstream target of RB action. Mol Cell Biol. 1995;15:742–755. doi: 10.1128/mcb.15.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin X Q, Livingston D M, Kaelin W J, Adams P D. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rasmussen R K, Ji H, Eddes J S, Moritz R L, Reid G E, Simpson R J, Dorow D S. Two-dimensional electrophoretic analysis of mixed lineage kinase 2 N-terminal domain binding proteins. Electrophoresis. 1998;19:809–817. doi: 10.1002/elps.1150190535. [DOI] [PubMed] [Google Scholar]
- 47.Sellers W R, Novitch B G, Miyake S, Heith A, Otterson G A, Kaye F J, Lassar A B, Kaelin W G., Jr Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shan B, Durfee T, Lee W H. Disruption of RB/E2F-1 interaction by single point mutations in E2F-1 enhances S-phase entry and apoptosis. Proc Natl Acad Sci USA. 1996;93:679–684. doi: 10.1073/pnas.93.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh P, Wong S H, Hong W. Overexpression of E2F-1 in rat embryo fibroblasts leads to neoplastic transformation. EMBO J. 1994;13:3329–3338. doi: 10.1002/j.1460-2075.1994.tb06635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slansky J E, Farnham P J. Introduction of the E2F family: protein structure and gene regulation. Curr Top Microbiol Immunol. 1996;208:1–30. doi: 10.1007/978-3-642-79910-5_1. [DOI] [PubMed] [Google Scholar]
- 51.Sorensen R S, Girling R, Lee C-W, Gannon J, Bandara L, La Thangue N B. Functional interaction between DP-1 and p53. Mol Cell Biol. 1996;16:5888–5895. doi: 10.1128/mcb.16.10.5888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steglich G, Neupert W, Langer T. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol Cell Biol. 1999;19:3435–3442. doi: 10.1128/mcb.19.5.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strom D K, Cleveland J L, Chellappan S, Nip J, Hiebert S W. E2F-1 and E2F-3 are functionally distinct in their ability to promote myeloid cell cycle progression and block granulocyte differentiation. Cell Growth Differ. 1998;9:59–69. [PubMed] [Google Scholar]
- 54.Taya Y. RB kinases and RB-binding proteins: new points of view. Trends Biochem Sci. 1997;22:14–17. doi: 10.1016/s0968-0004(96)10070-0. [DOI] [PubMed] [Google Scholar]
- 55.Terashima M, Kim K M, Adachi T, Nielsen P J, Reth M, Kohler G, Lamers M C. The IgM antigen receptor of B lymphocytes is associated with prohibitin and a prohibitin-related protein. EMBO J. 1994;13:3782–3792. doi: 10.1002/j.1460-2075.1994.tb06689.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tong L, Pav S, White D M, Rogers S, Crane K M, Cywin C L, Brown M L, Pargellis C A. A highly specific inhibitor of human p38 MAP kinase binds in the ATP pocket. Nat Struct Biol. 1997;4:311–316. doi: 10.1038/nsb0497-311. [DOI] [PubMed] [Google Scholar]
- 57.Tordai A, Franklin R A, Patel H, Gardner A M, Johnson G L, Gelfand E W. Cross-linking of surface IgM stimulates the Ras/Raf-1/MEK/MAPK cascade in human B lymphocytes. J Biol Chem. 1994;269:7538–7543. [PubMed] [Google Scholar]
- 58.Trimarchi J M, Fairchild B, Verona R, Moberg K, Andon N, Lees J A. E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proc Natl Acad Sci USA. 1998;95:2850–2855. doi: 10.1073/pnas.95.6.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trouche D, Cook A, Kouzarides T. The CBP co-activator stimulates E2F1/DP1 activity. Nucleic Acids Res. 1996;24:4139–4145. doi: 10.1093/nar/24.21.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trouche D, Kouzarides T. E2F1 and E1A(12S) have a homologous activation domain regulated by RB and CBP. Proc Natl Acad Sci USA. 1996;93:1439–1442. doi: 10.1073/pnas.93.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang S, Ghosh R, Chellappan S. Raf-1 physically interacts with Rb and regulates its function: a link between mitogenic signaling and cell cycle regulation. Mol Cell Biol. 1998;18:7487–7498. doi: 10.1128/mcb.18.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang S, Nath N, Adlam M, Chellappan S. Prohibitin, a potential tumor suppressor, interacts with Rb and regulates E2F function. Oncogene. 1999;18:3501–3510. doi: 10.1038/sj.onc.1202684. [DOI] [PubMed] [Google Scholar]
- 63.Wang S, Nath N, Minden A, Chellappan S. Regulation of Rb and E2F by signal transduction cascades: divergent effects of JNK1 and p38 kinases. EMBO J. 1999;18:1559–1570. doi: 10.1093/emboj/18.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 65.Weintraub S J, Chow K N, Luo R X, Zhang S H, He S, Dean D C. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 66.Weintraub S J, Prater C A, Dean D C. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 67.Wu X, Levine A J. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci USA. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiao Z X, Chen J, Levine A J, Modjtahedi N, Xing J, Sellers W R, Livingston D M. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375:694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 69.Xu M, Sheppard K A, Peng C Y, Yee A S, Piwnica W H. Cyclin A/CDK2 binds directly to E2F-1 and inhibits the DNA-binding activity of E2F-1/DP-1 by phosphorylation. Mol Cell Biol. 1994;14:8420–8431. doi: 10.1128/mcb.14.12.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Young P R, McLaughlin M M, Kumar S, Kassis S, et al. Pyridinyl imidazole inhibitors of p38 mitogen-activated protein kinase bind in the ATP site. J Biol Chem. 1997;272:12116–12121. doi: 10.1074/jbc.272.18.12116. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y, Chellappan S P. Cloning and characterization of human DP2, a novel dimerization partner of E2F. Oncogene. 1995;10:2085–2093. [PubMed] [Google Scholar]