Abstract
Verticillium dahliae is a particularly notorious vascular wilt pathogen of tomato and poses a reoccurring challenge to crop protection as limited qualitative resistance is available. Therefore, alternative approaches for crop protection are pursued. One such strategy is the impairment of disease susceptibility (S) genes, which are plant genes targeted by pathogens to promote disease development. In Arabidopsis and cotton, the Walls Are Thin 1 (WAT1) gene has shown to be a S gene for V. dahliae. In this study, we identified the tomato WAT1 homolog Solyc04g080940 (SlWAT1). Transient and stable silencing of SlWAT1, based on virus-induced gene silencing (VIGS) and RNAi, respectively, did not consistently lead to reduced V. dahliae susceptibility in tomato. However, CRISPR-Cas9 tomato mutant lines carrying targeted deletions in SlWAT1 showed significantly enhanced resistance to V. dahliae, and furthermore also to Verticillium albo-atrum and Fusarium oxysporum f. sp. lycopersici (Fol). Thus, disabling the tomato WAT1 gene resulted in broad-spectrum resistance to various vascular pathogens in tomato. Unfortunately these tomato CRISPR mutant lines suffered from severe growth defects. In order to overcome the pleiotropic effect caused by the impairment of the tomato WAT1 gene, future efforts should be devoted to identifying tomato SlWAT1 mutant alleles that do not negatively impact tomato growth and development.
Keywords: Verticillium, Fusarium, susceptibility gene, resistance breeding, pleiotropic effect
Introduction
Vascular wilt pathogens cause diseases in many annual and perennial crops (Yadeta and Thomma, 2013). Vascular pathogens of tomato include fungi such as Fusarium (Michielse and Rep, 2009) and Verticillium (Fradin and Thomma, 2006), as well as bacteria such as Clavibacter (Nandi et al., 2018), Ralstonia (Peeters et al., 2013), and Xanthomonas (Potnis et al., 2015). Vascular pathogens are hard to combat once they invaded a plant host (Yadeta and Thomma, 2013). The soil-borne fungus Verticillium dahliae is particularly hard to control due to its wild host range that comprises hundreds of hosts and its persisting resting structures in the soil (Fradin and Thomma, 2006). Crop protection therefore relies on the use of resistant plant varieties. For V. dahliae only one monogenic resistance gene, the Ve1 tomato gene, has been cloned so far (Fradin et al., 2009). This resistance is based on the recognition of the V. dahliae avirulence protein Ave1 by the resistance (R) protein encoded by the Ve1 gene (de Jonge et al., 2012). However, this resistance has been overcome by V. dahliae strains that have purged the Ave1 gene, posing a reoccurring challenge for tomato cultivation (Grogan, 1979; Dobinson et al., 1996; de Jonge et al., 2012; Usami et al., 2017).
To address the recurrent problem of the breakdown of R gene-mediated resistance, alternative approaches can be pursued, such as the impairment of disease susceptibility (S) genes (Pavan et al., 2010; Gawehns et al., 2013). S genes are host genes that play an important role in disease establishment by the pathogen. S genes can function in a multitude of ways, including early recognition of the pathogen, negative regulation of immune responses, or pathogen sustenance (van Schie and Takken, 2014). Nevertheless, S genes also have functions for the host. S gene-mediated resistance, or rather loss-of-susceptibility, is achieved by circumventing the manipulation of these gene products by the pathogen, preferably whilst keeping the intrinsic function for the host intact. In wild germplasm such impaired S gene alleles can occur naturally, for example as loss-of-function mutations or as promoter mutations leading to impaired expression (Chu et al., 2006; Bai et al., 2008; Gao et al., 2015). Alternatively, these impairments can be introduced by random mutagenesis or by targeted genome editing, for example using CRISPR-Cas9 (Zaidi et al., 2018; Dong and Ronald, 2019). Impairment of S genes can be associated with severe pleiotropic effect as a consequence of not only impairment of its function for the pathogen, but also impairment of its intrinsic role for the host. For instance for the defense no death 1 (dnd1) mutant, loss-of-susceptibility to Pseudomonas syringae is accompanied by dwarfism in Arabidopsis and tomato, and spontaneous lesion formation in potato (Sun et al., 2016). An important benefit, however, is that impairment of S genes can lead to non-race specific resistance to different strains of a given pathogen (Jørgensen, 1992), or even to broad-spectrum resistance to multiple pathogens (Wang et al., 2018). This illustrates the potential of S-gene mediated resistance in crop protection.
An example of broad-spectrum resistance to different vascular pathogens is provided by the walls are thin 1 (wat1) mutant (Denancé et al., 2013). This mutant was identified in an Arabidopsis screen for cell wall mutants (Ranocha et al., 2010), and displays resistance to the bacteria Ralstonia solanacearum and Xanthomonas campestris, and the fungi V. dahliae, Verticillium albo-atrum, and Plectoshaerella cucumerina (Denancé et al., 2013). WAT1 encodes a tonoplast-localized auxin transporter (Ranocha et al., 2013), but its exact role in so-called “vascular immunity” is not yet understood. WAT1 orthologs have been characterized in several plant species (Ranocha et al., 2010), and recently its role as susceptibility factor in cotton was investigated, demonstrating that simultaneous transient silencing of three WAT1 homologs resulted in increased resistance to V. dahliae (Tang et al., 2019). In this study, we aimed to identify the tomato WAT1 ortholog and examine its role as a susceptibility factor in tomato for vascular pathogens such as Verticillium spp. and Fusarium oxysporum.
Materials and Methods
Plant and Fungi Growth Conditions
All tomato genotypes were grown in the greenhouse (Unifarm, Wageningen University & Research, Netherlands) at 21/19°C (day/night) at 60% relative humidity and a minimal light intensity of 100 W/m2 in potting soil (Potgrond 4, Horticoop, Katwijk, Netherlands). V. dahliae (strain JR2, race 1), V. albo-atrum (strain CBS385.91, race 1) and F. oxysporum f. sp. lycopersici (stain Bt.01, race 1) were maintained on potato dextrose agar (PDA) at room temperature in the dark.
Virus-Induced Gene Silencing
Virus-induced gene silencing (VIGS) was carried out as described previously using tobacco rattle virus (TRV) (Liu et al., 2002; Fradin et al., 2009; Verlaan et al., 2013). Briefly, a gene-specific 150–300 bp fragment was amplified using Phusion High-Fidelity DNA polymerase (New England Biolabs, Bioké, Leiden, Netherlands) with primers mentioned in Supplementary Table 1. The obtained fragment was cloned into the tobacco rattle virus 2 (TRV2) vector (Liu et al., 2002) using Gateway cloning and subsequently transformed into Agrobacterium tumefaciens strain GV3103. As negative control a TRV2 vector containing a fragment of the β-Glucuronidase (GUS) gene was used (Wu et al., 2011; Senthil-Kumar and Mysore, 2014). Moreover, a TRV2 vector carrying a fragment of the tomato phytoene desaturase (PDS) gene was used as a positive control as it triggers photobleaching upon effective silencing.
Generation of CRISPR-Cas9 and RNAi Lines
To design single guide RNAs (sgRNAs) the ‘‘CCTop -- CRISPR/Cas9 target online predictor’’1 (Stemmer et al., 2015) was used and for target site evaluation the tomato genome (Solanum lycopersicum Solyc2.5) was used as reference. Only sgRNAs with a maximum of one exonic off-target site were selected. All sgRNAs were verified to contain a GC-content2 between 30 and 80% and presence of required secondary structures was evaluated3 (Zuker, 2003) according to Liang et al., 2016. Different scoring tools4,5,6 (Wong et al., 2015; Chari et al., 2017; Sanson et al., 2018) were used to select the best sgRNAs which met most of the criteria. In total, four sgRNAs were designed (Supplementary Table 1).
Golden Gate Cloning (Engler et al., 2008) was used to clone the constructs, and plasmids were obtained from Addgene7 : pICH86966 (level 0 plasmid for amplification), pICSL01009 (level 0 plasmid containing AtU6), pICH47751 (level 1 sgRNA1), pICH47761 (level 1 sgRNA2), pICH47772 (level 1 sgRNA3), pICH47781 (level 1 sgRNA4), pICH47732 (level 1 containing NPTII), pICH47742 (level 1 containing Cas9), pICH41822 (linker), and pAGM4723 (level 2 binary vector) (Weber et al., 2011). Phusion High-Fidelity DNA Polymerase (Thermo Scientific, Bleiswijk, Netherlands) was used to amplify sgRNAs, and PCR products were purified with QIAquick PCR Purification Kit (Qiagen Benelux B.V., Venlo, Netherlands). Level 1 plasmids were digested using BsaI/Eco31I and ligated using T4 DNA ligase (Thermo Scientific, Bleiswijk, Netherlands) and cloned into Escherichia coli strain DH5α (Thermo Scientific, Bleiswijk, Netherlands). Plasmids were purified using QIAprep Spin Miniprep Kit (Qiagen Benelux B.V., Venlo, Netherlands). Level 2 plasmids were digested using BpiI/BpsI and ligated using T4 DNA ligase (Thermo Scientific, Bleiswijk, Netherlands), cloned into E. coli strain DH5α, purified and sequenced. All plasmids were cloned into A. tumefaciens strain AGL1+virG. Transformation of tomato cultivar Moneymaker (MM) was carried out as described previously (Huibers et al., 2013).
To generate the WAT1 RNAi construct, the same fragment that was used for VIGS was cloned from the TRV2 vector into the pHellsgate8 vector (Helliwell and Waterhouse, 2003) using Gateway cloning (Katzen, 2007). Subsequently the construct was transformed into A. tumefaciens strain AGL1+virG. Tomato transformation of cultivar MM was carried out as described previously (Huibers et al., 2013).
Pathogen Inoculations, Phenotyping, and Fungal Biomass Quantification
Verticillium dahliae, V. albo-atrum, and F. oxysporum (Fol) inoculations were carried out with root dipping as described previously (Fradin et al., 2009; Boshoven, 2017). For phenotyping, stunting (%) between inoculated and mock-inoculated plants was calculated based on plant canopy area at 21 days post inoculation (dpi) using ImageJ (Abramoff et al., 2004) as follows:
To quantify fungal biomass, stems sections (∼ 2 cm around the cotyledons) were harvested at 21 dpi and freeze-dried for 48 h. Subsequently, material was ground, and DNA was isolated using CTAB buffer (200 mM Tris–HCl pH 7.5, 50 mM EDTA pH 8.0, 2 M NaCl, 2% CTAB). Fungal biomass was determined on genomic DNA targeting the ITS gene (V. dahliae, V. albo-atrum, and F. oxysporum) relative to the reference gene SlRUB (Supplementary Table 1) with the CFX96 Real-time System (Bio-Rad, Veenendaal, Netherlands) and SYBR Green Master Mix (Bio-Rad, Veenendaal, Netherlands). Data were normalized to MM with the 2–ΔΔCt method (Livak and Schmittgen, 2001).
DNA Isolation and Genotyping
To genotype RNAi and CRISPR plants, DNA was isolated from young leaves using CTAB buffer (1 M Tris–HCl pH 7.5, 0.5 M EDTA pH 8.0, 5 M NaCl, 2% CTAB). PCR was performed with DreamTaq DNA polymerase (Thermo Scientific, Bleiswijk, Netherlands) and corresponding primers (Supplementary Table 1). PCR products were sequenced by Marcrogen Europe (Amsterdam, Netherlands).
RNA Isolation and Quantitative Reverse Transcription PCR
To quantify silencing levels in RNAi plants, root material was harvested at 21 dpi and snap-frozen in liquid nitrogen. Total RNA was isolated with the MagMAX-96 Total RNA Isolation Kit (Invitrogen, Bleiswijk, Netherlands) using a KingFisher Flex System (Thermo Scientific, Bleiswijk, Netherlands) and synthesis of cDNA was performed with the iScript cDNA Synthesis Kit (Bio-Rad, Veenendaal, Netherlands) according to the manufacturer’s instructions. Quantitative reverse transcription PCR (qRT-PCR) was carried out with the CFX96 Real-time System (Bio-Rad, Veenendaal, Netherlands) and SYBR Green Master Mix (Bio-Rad, Veenendaal, Netherlands) according to the manufacturer’s instructions. Gene expression was determined using the 2–ΔΔCt method (Livak and Schmittgen, 2001) relative to the tomato elongation factor 1 α (SlEF1α) (Supplementary Table 2). Data were normalized to transformants devoid of the silencing construct or, when not available, to MM plants.
Results
Transient Silencing of WAT1 in Tomato Might Reduce Susceptibility to V. dahliae
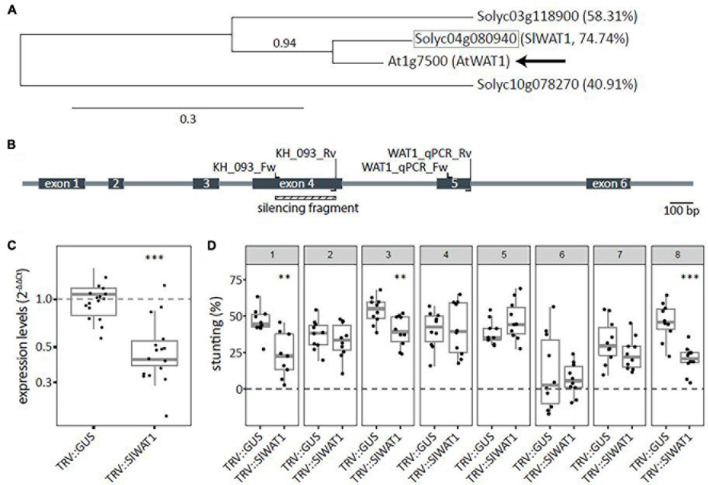
To identify tomato WAT1 orthologs, the amino acid sequence of the Arabidopsis WAT1 gene (At1g75500) was obtained from TAIR8 and used as query in a blastP search against the Sol genomics database (ITAG release 4.09). Two close homologs were identified and phylogenetic trees were constructed using Phylogeny.fr (Dereeper et al., 2008). Results showed that the tomato gene Solyc04g080940 (hereafter SlWAT1) has the highest homology of 74.7% to AtWAT1 (Figure 1A). To functionally test the SlWAT1 gene for a role as S gene in tomato, A. tumefaciens-mediated VIGS was used for transient silencing of SlWAT1. One TRV construct (TRV::SlWAT1) was made (Figure 1B), which resulted in a significant reduction in relative expression of SlWAT1 to approximately 49.6% in plants treated with TRV::SlWAT1 when compared with TRV::GUS-treated plants (Figure 1C). To screen for reduced susceptibility to V. dahliae resulted from silencing SlWAT1, stunting based on canopy area was calculated between mock- and V. dahliae-inoculated plants at 21 dpi. Compared with V. dahliae-inoculated TRV::GUS plants, V. dahliae-inoculated plants treated with the TRV::SlWAT1 construct showed significantly less stunting in three out of the eight performed experiments (Figure 1D, panels 1, 3, and 8).
FIGURE 1.
Transient silencing of WAT1 in tomato resulted in reduced susceptibility to Verticillium dahliae. (A) Phylogenetic tree based on amino acid sequences for WAT1 including potential ortholog from tomato (highlighted in gray square). Percentages indicate protein sequence similarity to AtWAT1 marked with an arrow. Number above node indicate branch support values. (B) Schematic overview of SlWAT1 (Solyc04g080940) with intron/exon locations as well as primers for cloning the VIGS fragment and for monitoring the SlWAT1 expression. (C) Expression levels (2–ΔΔCt) of plants treated with TRV::SlWAT1 in stems normalized to TRV::GUS 2 weeks after Agrobacterium tumefaciens treatment on a log10 scale. Data of two independent experiments with n ≥ 6 per experiment per genotype (t-test when compared with TRV::GUS with ∗∗∗p < 0.001). (D) Stunting (%) in Verticillium dahliae-inoculated plants when compared with the average of mock-inoculated plants at 21 dpi after transient silencing of SlWAT in eight independent experiments with n ≥ 8 per experiment per genotype (n ≥ 8, t-test when compared with TRV::GUS with ∗∗p < 0.01 and ∗∗∗p < 0.001).
RNAi Knock-Down of SlWAT1 Did Not Confirm Loss of Susceptibility to V. dahliae
Since the silencing effect via VIGS was transient and patchy, an RNAi approach was taken to further verify the role of SlWAT1 in V. dahliae susceptibility. An RNAi construct was made with the VIGS fragment (Figure 1A) and used to transform the tomato cultivar MM. Several primary transformants (T1) were evaluated by testing for stable integration of the silencing construct and by determining residual WAT1 expression levels. In the T1 transformants relative WAT1 expression varied greatly, from 11 to 270%, when compared with the expression levels found in leaves of control plants (Supplementary Table 2). Five T1 transformants with reduced SlWAT1 expression were transferred to the greenhouse for T2 seed production. However, seeds were only obtained from three transformants, TV181034, TV181036, and TV181037, which were used for further study (Supplementary Table 2).
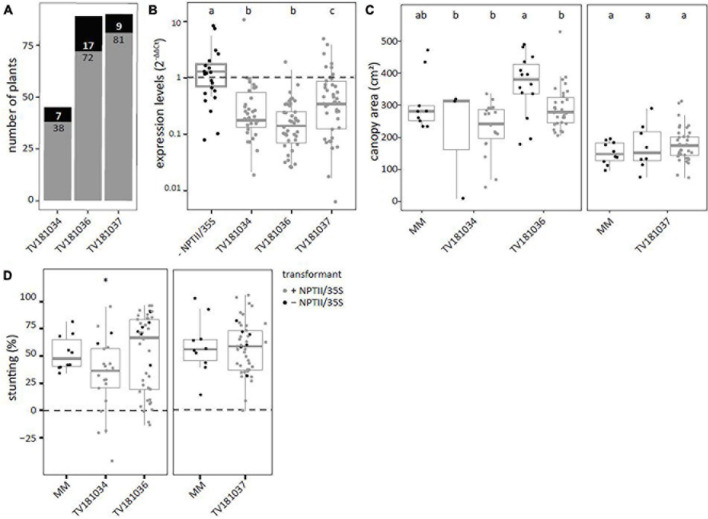
T2 plants derived from the three transformants, TV181034, TV181036, and TV181037, were tested for presence of the silencing construct with a NPTII- and 35S specific-PCR (Supplementary Table 1). This revealed that 7 out of 45 plants (15.5%), 17 out of 89 plants (19.1%), and 9 out of 90 plants (10.0%) of the plants of the T2 families TV181034, TV181036, and TV181037, respectively, did not carry the silencing construct (Figure 2A). When compared with plants lacking the construct (− NPTII/35S) of all families, the expression of SlWAT1 in the roots of plants carrying the silencing construct (+ NPTII/35S) was significantly reduced, with the residual expression of on average 20.2, 54.4, and 65.5% for the family TV181036, TV181034, and TV181037, respectively (Figure 2B). We also determined whether the presence of the silencing construct affected plant growth in the absence of V. dahliae inoculation. No significant difference in canopy area of mock-inoculated plants was found for any of the T2 families compared with mock-inoculated MM plants (Figure 2C).
FIGURE 2.
Reduced susceptibility to V. dahliae was found in SlWAT1 T2 RNAi family TV181034, but not for TV181036 and TV181037. (A) Total number of plants with (gray) and without (black) the silencing construct for each of the obtained T2 RNAi families. (B) Expression levels of SlWAT1 in roots collected at 21 dpi of plants with the silencing construct (+ NPTII/35S, gray dots) relatively to plants without the silencing construct (– NPTII/35S, black dots) for all families. Data were normalized using 2–ΔΔCt to SlEF1α with n ≥ 4 per family (ANOVA with Fisher’s unprotected LSD with p = 0.05 on ΔCt values). (C) Canopy area (cm2) of mock-inoculated plants at 21 dpi for T2 RNAi families with (gray) and without (black) the silencing construct (n ≥ 3, ANOVA with Fisher’s unprotected LSD, p = 0.05). (D) Stunting of V. dahliae-inoculated plants when compared with the average stunting of mock-inoculated plants per genotype at 21 dpi (n ≥ 5, t-test with ∗p < 0.05). a–c: Genotypes having average values with a letter in common are not statistically significant different (at P = 0.05).
To test for loss of susceptibility to V. dahliae, plants from all three T2 families were challenged with V. dahliae. To this end, stunting based on canopy area was calculated between mock- and V. dahliae-inoculated plants for each of the genotypes at 21 dpi and compared with V. dahliae-induced stunting in MM plants (Figure 2D). Of the three tested families, a significant reduction in stunting of V. dahliae-inoculated plants was found only in the family TV181034 with a value of on average 32.2 vs 52.6% in V. dahliae-inoculated MM plants.
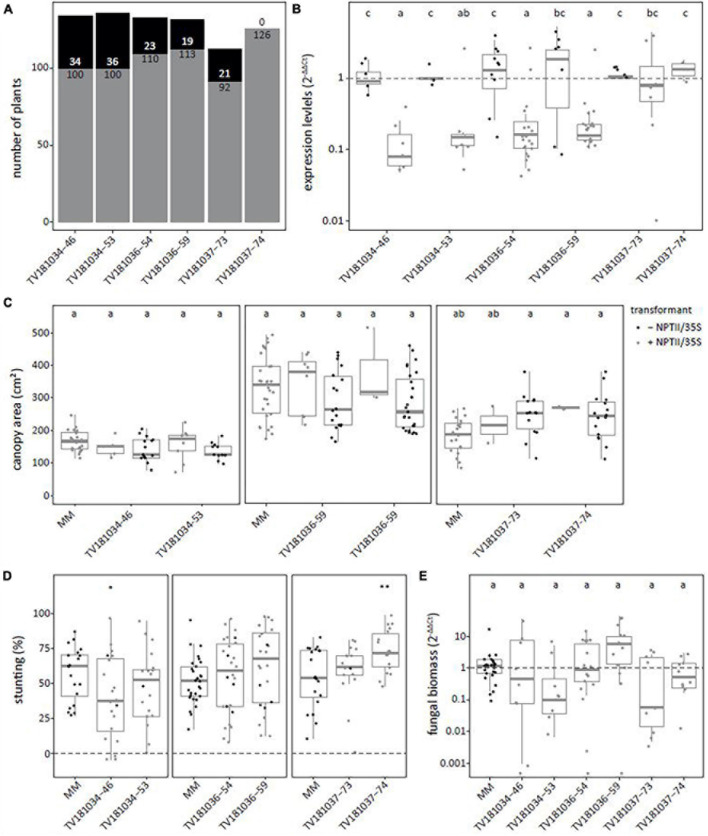
To confirm the results of the T2 generation, two plants per T2 family were kept for T3 seed production. T3 plants with and without the RNAi construct were identified (Figure 3A). In the T3 families derived from T2 plants of TV181034-46 and –53 as well as TV181036-54 and –59, a significant reduction of SlWAT1 expression to 12.0, 41.4, 35.2 and 30.6%, respectively, were found in the roots of plants carrying the RNAi constructs (Figure 3B). Unfortunately, SlWAT1 was not silenced at all in plants carrying the RNAi construct for families TV181037-73 and –74, as SlWAT1 expression was on average 39.4 and 36.5% higher than in control plants lacking the RNAi construct, respectively (Figure 3B). No statistically significant difference in canopy area was found for plants of any of the T3 RNAi families when compared with the canopy area of MM plants (Figure 3C). This is in agreement with the results obtained in the T2 families (Figure 2C), indicating that the presence of the silencing construct did not significantly affect plant growth at this developmental stage.
FIGURE 3.
Loss of susceptibility to V. dahliae was not confirmed in all SlWAT1 T3 RNAi families. (A) Total number of plants with (gray) and without (black) the silencing construct for six T3 RNAi families which were obtained from three different T2 RNAi families. (B) Expression levels of T3 SlWAT1 RNAi families relative to plants without the silencing construct (– NPTII/35S, gray) and normalized using 2–ΔΔCt to SlEF1α with n ≥ 4 per family with (gray) and without (black) the silencing construct (ANOVA with Fisher’s unprotected LDS with p = 0.05 on ΔCt values). (C) Canopy area (cm2) of mock-inoculated plants at 21 days post inoculation (dpi) for T3 SlWAT1 RNAi families with (gray) and without (black) the silencing construct. Data from two independent experiments with n ≥ 2 (ANOVA with Fisher’s unprotected LSD, p = 0.05). (D) Stunting (%) of V. dahliae-inoculated plants when compared with the average stunting of mock-inoculated plants per genotype at 21 dpi. Box plots represent data with n ≥ 8 plants per experimental repeat (t-test when compared with MM with *p < 0.05 and **p < 0.01). (E) Relative fungal biomass in the stems of RNAi plants at 21 dpi with V. dahliae. This was calculated as the ratio of the V. dahliae ITS gene amplification in comparison with the tomato SlRUB gene (Supplementary Table 1) and normalized the V. dahliae-inoculated MM plants using 2–ΔΔCt on a log10 scale with n ≥ 7 per family (ANOVA with Fisher’s unprotected LSD, p = 0.05 on ΔCt). a–c: Genotypes having average values with a letter in common are not statistically significant different (at P = 0.05).
The plants of the six T3 RNAi families were challenged with V. dahliae, and similar levels of stunting of V. dahliae-inoculated plants were found for most plants of the T3 families when compared with V. dahliae-inoculated MM plants (Figure 2D). Only in one T3 family, TV181034-46, stunting of V. dahliae-inoculated plants was significantly reduced to (on average) 39.5% when compared with V. dahliae-inoculated MM plants that displayed an average stunting of 56.6%.
To quantify V. dahliae colonization in the T3 RNAi families, fungal biomass was quantified on genomic DNA by targeting the ITS gene of V. dahliae relative to the tomato reference gene SlRUB (Supplementary Table 1) in stems of V. dahliae-inoculated plants at 21 dpi for each genotype. No significant reduction in fungal biomass in plants of all six T3 RNAi families was found when compared with V. dahliae-inoculated MM plants (Figure 3E).
Targeted Deletion in SlWAT1 Leads to Loss-of-Susceptibility to V. dahliae Despite Severe Growth Defects
In order to circumvent interference of residual SlWAT1 expression as shown for the RNAi families, we explored approaches for targeted knock-out. To this end, stable transformants using CRISPR-Cas9 were generated. The CRISPR-Cas9 construct was designed with four sgRNAs that targeted sequences in exons 3, 4, and 5 of the SlWAT1 gene (Figure 4A). The use of multiple sgRNAs increased the possibility of creating large deletions due to the occurrence of double stranded breaks at multiple sgRNAs locations simultaneously (Do et al., 2019). T1 transformants were evaluated for the occurrence of mutations in SlWAT1 with a gene-specific PCR and gel electrophoresis to detect aberrantly sized PCR products (Supplementary Table 1). By focusing on large deletions, small deletions, small insertions, and single nucleotide polymorphisms might have been missed. Three transformants (#10, #19, and #28) showed a relatively large deletion, while for a fourth transformant (#23) an additional band appeared above the wild-type band (Supplementary Figure 1). However, T2 seeds from only one of these mutants, TV181046 (#19), were obtained as the other transformants were either not successfully transferred from in vitro conditions to soil, did not set fruits, or did not produce seeds.
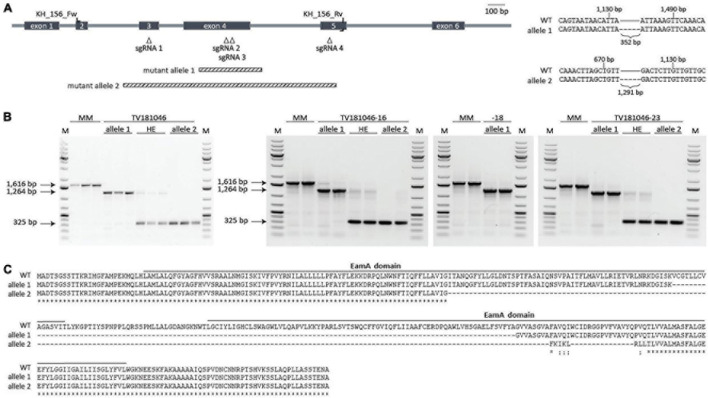
FIGURE 4.
CRISPR T2 family TV181046 and its T3 progeny carry a bi-allelic mutation in SlWAT1. (A) Schematic overview of SlWAT1 (Solyc04g080940) indicating location of the sgRNAs, primers used for genotyping and the two mutant alleles (right). Sequencing revealed a 352 and 1,291 bp deletion for mutant alleles 1 and 2, respectively, compared with SlWAT1 wild-type (WT) (right). (B) Gel electrophoresis (1% TAE, ethidium bromide) of gene-specific PCR with (wild-type band at 1,616 bp) showing the two mutant alleles as well as heterozygous (HE) plants for the T2 (left) and the T3 (right) plants with a 1 kb ladder (M). (C) Protein alignments of SlWAT1 wild-type (WT) with mutant alleles 1 and 2 showing a 121 and 197 amino acid deletion, respectively. Mutant alleles were translated into protein using http://www.softberry.com/berry.phtml. CLUSTAL multiple sequence alignment was done using https://www.ebi.ac.uk/Tools/msa/muscle/. Solid bars indicate predicted protein domains annotated as EamA domain (https://www.ebi.ac.uk/interpro).
First, plants from the T2 CRISPR family TV181046 were genotyped to confirm the presence of a mutation by sequencing. In fact, seedlings of TV181046 were found to carry bi-allelic mutations with either a smaller deletion (allele 1), or a larger deletion (allele 2), or heterozygous plants that carry both types of deletions (Figures 4A,B). The smaller 352 bp deletion (allele 1), located in exon 4, led to a 121 amino acid deletion, while the larger 1,291 bp deletion (allele 2), spanning exons 3, 4, and partly 5, resulted in a 197 amino acid deletion. As only one T2 CRISPR line was obtained, we propagated plants with the heterozygous deletions as well as homozygous plants for each mutant allele to obtain a larger panel of genotypes (T3) for testing. Seeds from three T3 CRISPR lines were obtained, TV181046-16, –18, and –23, genotyped and found to be heterozygous for the deletions (TV181046-16 and –23) and homozygous for allele 1 (TV181046-18) (Figure 4B).
As for neither of the two deletions (allele 1 and allele 2, Figure 4C) a premature stop codon was predicted,10 we subsequently investigated whether these deletions affected any known domain within SlWAT1. To this end, protein domains were predicted using InterPro.11 For wild-type SlWAT1 two EamA domains were found. Most EamA domain-containing proteins are classified as metabolite transporters that usually carry two copies of this domain (Jack et al., 2001). For the SlWAT1 mutant alleles both predicted EamA domains were affected (Supplementary Figure 2). As AtWAT1 is located in the tonoplast (Ranocha et al., 2010), and also because many EamA domain-containing proteins carry multiple transmembrane domains, we further predicted the transmembrane domains for wild-type SlWAT1, mutant allele 1 and mutant allele 2. Wild-type SlWAT1 was predicted to contain ten transmembrane domains which was described before for WAT1 in Arabidopsis and cotton as well (Ranocha et al., 2010; Tang et al., 2019). For mutant allele 1 only seven out of ten transmembrane domains were found and for mutant allele 2 only four (Supplementary Figure 2). Collectively, our data suggested that both mutant alleles carry a deletion which affects known domains in SlWAT1, presumably leading to loss of its biological function. This allows us to further study these knock-out CRISPR lines for loss of susceptibility to V. dahliae.
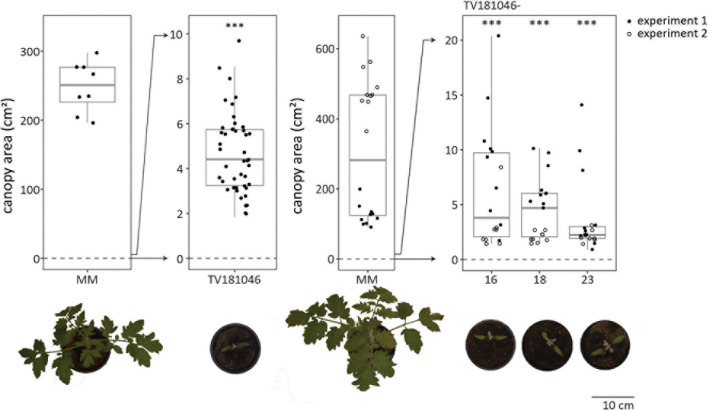
Irrespective of the type of mutation, all plants of the T2 and the T3 generation displayed severe growth and development defects (in absence of V. dahliae inoculation); the germination rate was low, seedlings were small and light in color, and overall plant growth remained severely compromised (Figure 5 and Supplementary Figure 3). To quantify the size difference, we determined canopy area of mock-inoculated plants for all genotypes in the absence of V. dahliae inoculation. For MM Plants, canopy area of mock-inoculated plants was heavily reduced when compared with mock-inoculated plants measured at 21 dpi. While the canopy area of mock-inoculated MM plants was 300 cm2 on average, the canopy area of most mock-inoculated CRISPR T2 and 3 plants was less than 10 cm2. However, the observed aberrations alleviated slowly during further plant development and even though the CRISPR plants remained smaller than MM plants, they developed flowers and set fruits (Supplementary Figure 3).
FIGURE 5.
CRISPR T2 family TV181046 and its T3 progeny display severe growth and development defects. Canopy area of mock-inoculated plants at 21 dpi for T2 CRISPR line TV181046 (left) and T3 CRISPR lines TV181046-16, –18, and –23 (right). Data from one or two independent experiments with n ≥ 8 (t-test compared with MM with ∗∗∗p = 0.001).
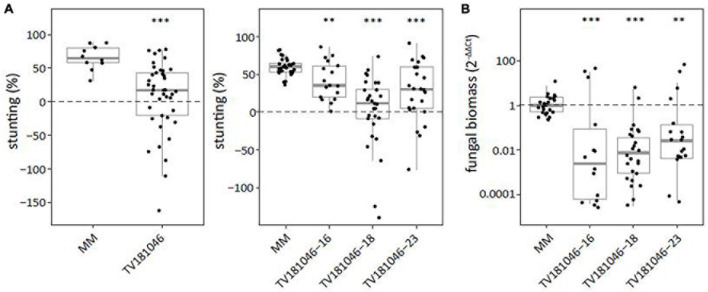
To test for loss of susceptibility, plants of the T2 and the T3 generation were inoculated with V. dahliae. Stunting based on canopy area was calculated between mock- and V. dahliae-inoculated plants for each of the genotypes. Stunting of V. dahliae-inoculated T2 plants was significantly reduced to on average 7.1% for line TV181046 when compared with V. dahliae-inoculated MM plants with on average 65.5% stunting (Figure 6A and Supplementary Figure 4). In the T3 families, stunting of TV181046- 16–, –18, and –23 was significantly reduced to 41.7, 1.4, and 26.2% on average, respectively, compared with V. dahliae-induced stunting of 60.0% on average in MM plants. Due to the stunting calculations being based on the average of mock-inoculated plants and due to variation in plant size observed in the mutant lines, the differences in stunting of V. dahliae-inoculated plants were pronounced in the mutant lines when compared with the MM plants. To quantify the effect on V. dahliae proliferation, fungal biomass was determined in stems of V. dahliae-inoculated plants at 21 dpi for each genotype. In the V. dahliae-inoculated plants of all CRISPR T3 families, fungal biomass was significantly reduced to around 1% of the biomass in the V. dahliae-inoculated MM plants (Figure 6B).
FIGURE 6.
Targeted knockout of SlWAT1 lead to loss of susceptibility to V. dahliae. (A) Stunting (%) of V. dahliae inoculated T2 (left) and T3 (right) plants when compared with the average stunting of mock-inoculated plants at 21 days post inoculation (dpi). Box plots represent data with n ≥ 9 plants per experimental repeat (t-test when compared with MM with ∗∗p < 0.01 and ∗∗∗p < 0.001). (B) Fungal biomass of V. dahliae-inoculated T3 CRISPR plants of all three lines in stems at 21dpi. This was calculated as the ratio of the V. dahliae ITS gene amplification in comparison with the tomato reference SlRUB gene (Supplementary Table 1) and normalized to V. dahliae-inoculated MM plants using 2–ΔΔCt on a log10 scale with n ≥ 9 repeat per experimental repeat (t-test on ΔCt when compared with MM with ∗∗p < 0.01 and ∗∗∗p < 0.001).
Targeted Deletion in SlWAT1 Leads to Loss-of-Susceptibility to V. albo-atrum and Fusarium oxysporum f. sp. Lycopersici
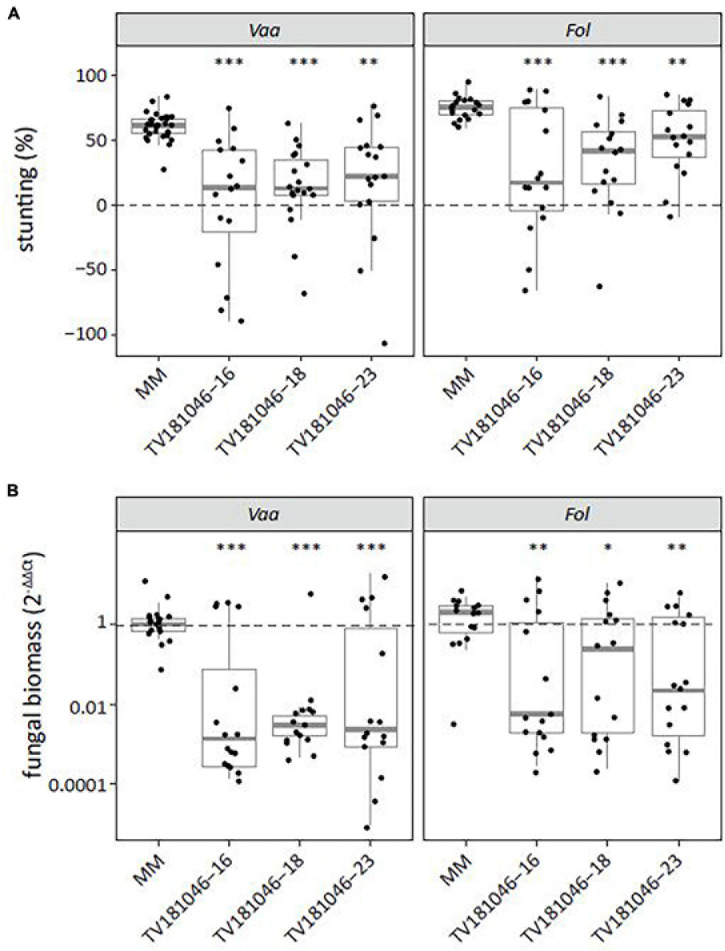
As S gene-mediated resistance can lead to broad-spectrum resistance to multiple pathogens (Wang et al., 2018), we also challenged T3 CRISPR plants with V. albo-atrum and Fol. For both pathogens, inoculated T3 CRISPR plants showed significantly reduced stunting when compared with inoculated MM plants (Figure 7A). Moreover, fungal biomass was significantly reduced in V. albo-atrum- and Fol-inoculated T3 CRISPR plants of all three lines when compared with inoculated MM plants (Figure 7B).
FIGURE 7.
Knockout of SlWAT1 lead to loss-of-susceptibility to V. albo-atrum and Fusarium oxysporum f. sp. lycopersici. (A) Stunting (%) of V. albo-atrum (Vaa) and F. oxysporum f. sp. lycopersici (Fol) inoculated T3 plants when compared with the average stunting of mock-inoculated plants at 21 days post inoculation (dpi). Box plots represent data with n ≥ 7 plants per experimental repeat (t-test when compared with MM with ∗∗p < 0.01 and ∗∗∗p < 0.001). (B) Fungal biomass of Vaa- and Fol-inoculated T3 CRISPR plants at 21 dpi. This was determined on genomic DNA targeting the ITS gene (Vaa and Fol) relative to the tomato reference gene SlRUB (Supplementary Table 1) and then normalized to Vaa and Fol inoculated MM plants using 2–ΔΔCt on a log10 scale with n ≥ 7 per experimental repeat (t-test on ΔCt when compared with MM with ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001).
Discussion
For vascular pathogens such as V. dahliae, for which only few sources of monogenic resistance are known, crop protection mainly relies on alternative strategies. The impairment of S genes has gained increasing attention in resistance breeding over the last years (Pavan et al., 2010; Gawehns et al., 2013; van Schie and Takken, 2014), particularly in the light of recent advances in genome editing in plants (Andolfo et al., 2016; Langner et al., 2018; Zaidi et al., 2018; Yin and Qiu, 2019). Here, we show that targeted deletion of SlWAT1 using CRISPR-Cas9 led to loss of susceptibility to V. dahliae in tomato. Plants of T3 CRISPR lines showed reduced disease symptoms upon challenge with V. dahliae as well as reduced fungal biomass when compared with susceptible MM plants (Figure 6). The loss of susceptibility to V. dahliae, as observed in plants of the CRISPR lines, could not be demonstrated consistently in plants carrying the RNAi silencing construct (Figures 1, 2). This can likely be attributed to the relatively high degree of residual WAT1 expression in most plants of the T2 and T3 RNAi lines, which likely compromised the efficacy of silencing too much to monitor effects on V. dahliae infection.
In WAT studies in Arabidopsis and cotton, reduced Verticillium wilt symptoms and reduced fungal proliferation were observed in knock-out mutants or upon transient silencing of WAT1, respectively (Denancé et al., 2013; Tang et al., 2019). Remarkably, the loss of susceptibility in Arabidopsis wat1 mutants was further extended to other vascular pathogens including bacteria and fungi (Denancé et al., 2013). S gene-mediated broad-spectrum resistance to multiple pathogens was described before (Wang et al., 2018), and highlights the potential of using impaired S genes for the control of multiple pathogens simultaneously. In fact, we also demonstrated loss of susceptibility of knock-out slwat1 mutants to another pathogenic Verticillium species, V. albo-atrum, as well to another vascular pathogen, Fol (Figure 7). Also for these pathogens, disease symptoms and fungal biomass were significantly reduced when compared with susceptible MM plants. Together, this indicates that the function of WAT1 in susceptibility to different vascular pathogens seems to be conserved across plant species, and therefore impairment of WAT1 might offers an approach to combat different vascular pathogens in multiple crops.
To date, the function of WAT1 in so called “vascular immunity” remains to be elucidated. WAT1 was originally identified in a cell wall mutant screening in zinnia (Zinnia elegans) (Pesquet et al., 2005; Ranocha et al., 2010) and the homolog of Arabidopsis was shown to be a tonoplast-localized auxin transporter (Ranocha et al., 2010). In Arabidopsis wat1 mutants, cell wall-related phenotypes in stems were described with altered cell elongation and reduced secondary cell walls of fiber cells, hence its name Walls Are Thin 1. Furthermore, wat1 mutants showed altered contents of auxin (indole-3-acetic acid, IAA), tryptophan and salicylic acid (SA) (Ranocha et al., 2010; Denancé et al., 2013). The IAA content in roots was reduced in wat1 mutants while the SA content was found to be elevated when compared with wild-type plants (Ranocha et al., 2010), which is in line with the previously described antagonism of auxin and SA in plant immunity and development (Wang et al., 2007; Robert-Seilaniantz et al., 2011). SA does not seem to play a role in basal plant defense against V. dahliae as different Arabidopsis mutants with a deficiency in SA signaling, such as enhanced disease susceptibility (eds1-2 and eds5-1), nonexpresser of PR genes (npr1-1 and npr1-3), and phytoalexin deficient 4 (pad4-1), show similar symptoms and levels of fungal biomass as control plants upon V. dahliae infection (Pantelides et al., 2010; Fradin et al., 2011). In contrast, a role was assigned to auxin in V. dahliae susceptibility as two auxin receptor mutants, auxin signaling F-box 1 and 3 (afb1 and afb3), as well as auxin transporter mutant auxin resistant 4 (axr4) display reduced symptoms and less fungal biomass upon challenge with V. dahliae (Fousia et al., 2018). For another vascular wilt pathogen, F. oxysporum, two transcription factor mutants, auxin response factor 1 and 2 (arf1 and arf2), showed significantly less disease levels although fungal biomass was not quantified (Lyons et al., 2015). Collectively, auxin seems to play a crucial role in V. dahliae susceptibility to vascular wilt fungi, and therefore auxin-related genes may be further studied to test their potential as susceptibility factors for V. dahliae.
Even though SlWAT1 CRISPR plants of tomato showed loss of susceptibility to V. dahliae, V. albo-atrum, and F. oxysporum, the targeted deletion in SlWAT1 was accompanied by severe growth defects. It may be argued that the significantly reduced stunting and reduced fungal biomass accumulation in the SlWAT1 CRISPR plants is an indirect consequence of the dramatically impaired growth and development of these mutants. In our experiments, we have performed fungal inoculations on 10-day-old seedlings. Since the observed developmental aberrations alleviated slowly during further plant development, it may be worthwhile to inoculate tomato SlWAT1 CRISPR plants at a later time point than the control plants, when both plants have a similar overall appearance, for example by inoculating 1-month-old SlWAT1 CRISPR plants. However, this approach obviously has the downside that mutant and control plants will not be tested at the same age.
Impairment of S genes is known to cause pleiotropic side effects in some cases (Clough et al., 2000; Sun et al., 2016), and also for WAT1 such effects were described in other plant species. For Arabidopsis wat1 mutants, no abnormalities were found in early stages of development, but older plants were stunted when compared with wild-type plants (Ranocha et al., 2010). Transient silencing of WAT1 in cotton resulted in reduced root length and shorter first internodes (Tang et al., 2019). Such growth defects can certainly be attributed to the imbalance between auxin and SA. Firstly, it is well known that auxin plays an essential role in many aspects of plant development (Korver et al., 2018) and its downregulation, as shown in Arabidopsis wat1 mutants, might negatively affect growth. Secondly, Arabidopsis wat1 mutants also showed higher SA levels, which is known to affect plant growth as observed in the constitutive expressor of PR genes 5 (cpr5) mutant which shows high SA levels accompanied by severe dwarfism (Bowling et al., 1997). Evidently, pleiotropic effects of impaired S genes are not desirable for breeding purposes, as it might affect yield but also overall development (Hückelhoven et al., 2013; Engelhardt et al., 2018). Additionally, special attention should also be given to resistance against other pathogens as an altered hormone balance, as observed in WAT1-mediated resistance (Denancé et al., 2013), can influence resistance to other pathogens (Thomma et al., 1998). Therefore, alternatives for obtaining mutants without such pleiotropic effects need to be explored. For example, potential natural allelic variants of WAT1 in wild germplasm that can no longer be exploited by the pathogen, but that do not display pleiotropic effects, could be used for breeding. Alternatively, mutant populations can be used to identify wat1 mutants omitting the severe growth defects. Certainly, mutants with smaller targeted deletions or even single base pair changes could also be studied, to find essential domains that are only required by Verticillium spp. for disease development, but that are not involved in tomato growth and development. Lastly, targeted modification in the promoter region of a S gene could circumvent pleiotropy by preventing binding the pathogen effector, as it was demonstrated by the xa13-mediated resistance against bacterial blight (Chu et al., 2006; Zaka et al., 2018).
In the case of WAT1, two Arabidopsis wat1 mutant alleles that each carry a T-DNA insertion have been described (Ranocha et al., 2010). The T-DNA insertion located 55 bp upstream of the ATG translation start codon leads to lack of WAT1 expression in the wat1-1 mutant line. In contrast, the wat1-2 mutant line that carries the T-DNA insertion 28 bp downstream of the stop codon has about 50% residual WAT1 expression when compared with wild-type plants. Compared with wat1-1, a less strong phenotype was displayed by wat1-2 plants (Ranocha et al., 2010). In our study, the tomato WAT1 RNAi plants had a similar canopy area to the control MM plants until 6 weeks after sowing (Figure 2C). This finding may indicate that the residual WAT1 expression in the tomato RNAi lines is sufficient to prevent negative effects on plant growth. Unfortunately, the WAT1 expression levels were not sufficiently reduced to hamper Verticillium infection in most RNAi tomato plants, since a significant reduction in stunting upon V. dahliae inoculation was only observed in the T2 family TV181034 (Figure 2) and the derived T3 family TV181034-46 (Figure 3). Variation in stunting was shown between individual T2 and T3 plants, with some plants showing a very low level of stunting (Figures 2, 3), which may be associated with variation in residual WAT1 expression. Therefore, in further studies, it could be worthwhile to determine the residual level of WAT1 expression and correlate that with stunting levels of individual RNAi plants in order to assess a direct correlation between these characteristics. Such direct correlation justifies the further search for a natural wat1 alleles or the generation of mutations in the promoter of the WAT1 gene that can associate reduced WAT1 expression with reduced Verticillium-induced stunting.
Summarizing, it remains challenging to identify WAT1 alleles in tomato, as well as in other crops, that cannot be exploited by Verticillium spp. for disease development, yet that do not negatively impact plant growth and development.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
KH, HS, BT, and YB conceived the study and wrote the manuscript. KH designed and performed the experiments and analyzed the data. DS, SC, and MO helped in collecting data. All authors read and approved the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Bert Essenstam and Andre Maassen at Unifarm of WUR for excellent plant care and to Eleni Koseoglou for discussions.
Footnotes
Funding
This project was financially supported by the Foundation Topconsortium voor Kennis en Innovatie (TKI) Starting Materials, project number KV 1409-026.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.721674/full#supplementary-material
Gel electrophoresis (1% TBE, Gelred, image colors inverted) of gene-specific PCR with primers KH_156 on primary transformants (T1) of plants transformed with SlWAT1 CRISPR-Cas9 construct. Wild-type PCR product (1,616 bp) indicated with an arrow and highlighted are mutants #10, #19, #23, and #28 that were transferred to the greenhouse, and of which seeds were only obtained from #19 (TV181046).
Predicted transmembrane domains for wild-type SlWAT1 (left), mutant allele 1 (right), and mutant allele 2 (bottom). Graphs were generated with TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
CRISPR T2 family TV181046 and its T3 progeny TV181046-16, –18, and –23 display severe growth and development defects. Pictures of MM and T3 CRISPR WAT1 plants at different time points.
Canopy area pictures mock- and V. dahliae-inoculated (strain JR2) plants for CRISPR T2 family TV181046 and T3 families TV181046-16, –18, and –23 at 21 dpi compared to MM plants of respective experiment.
Primers used in this study.
Overview of primary transformants (T1) with presence/absence of silencing construct (NPTII/35S), relative SlWAT1 expression normalized to control plants at 1 (2–ΔΔCt) and plants from which T2 seeds were obtained.
References
- Abramoff M. D., Magalhaes P. J., Ram S. J. (2004). Image processing with ImageJ. Biophotonics Int. 11 36–42. [Google Scholar]
- Andolfo G., Iovieno P., Frusciante L., Ercolano M. R. (2016). Genome-editing technologies for enhancing plant disease resistance. Front. Plant Sci. 7:1813. 10.3389/fpls.2016.01813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Pavan S., Zheng Z., Zappel N. F., Reinstädler A., Lotti C., et al. (2008). Naturally occurring broad-spectrum powdery mildew resistance in a entral American tomato accession is caused by loss of MLO function. Mol. Plant Microbe Interact. 21 30–39. 10.1094/mpmi-21-1-0030 [DOI] [PubMed] [Google Scholar]
- Boshoven J. C. (2017). Virulence Contribution and Recognition of Homologs of the Verticillium Dahliae Effector Ave1. Ph.D. Thesis. Wageningen: Wageningen University. [Google Scholar]
- Bowling S. A., Clarke J. D., Liu Y., Klessig D. F., Dong X. (1997). The cpr5 mutant of arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 9 1573–1584. 10.2307/3870444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari R., Yeo N. C., Chavez A., Church G. M. (2017). sgRNA scorer 2.0 - a species independent model to predict CRISPR/Cas9 activity. ACS Synth. Biol. 19 902–904. 10.1021/acssynbio.6b00343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Z., Yuan M., Yao J., Ge X., Yuan B., Xu C., et al. (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev. 20 1250–1255. 10.1101/gad.1416306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Fengler K. A., Yu I. C., Lippok B., Smith R. K., Bent A. F. (2000). The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. U.S.A. 97 9323–9328. 10.1073/pnas.150005697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge R., Peter van Esse H., Maruthachalam K., Bolton M. D., Santhanam P., Saber M. K., et al. (2012). Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 109 5110–5115. 10.1073/pnas.1119623109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denancé N., Ranocha P., Oria N., Barlet X., Rivière M. P., Yadeta K. A., et al. (2013). Arabidopsis wat1 (walls are thin1)-mediated resistance to the bacterial vascular pathogen, Ralstonia solanacearum, is accompanied by cross-regulation of salicylic acid and tryptophan metabolism. Plant J. 73 225–239. 10.1111/tpj.12027 [DOI] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36 465–469. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do P. T., Nguyen C. X., Bui H. T., Tran L. T. N., Stacey G., Gillman J. D., et al. (2019). Demonstration of highly efficient dual gRNA CRISPR/Cas9 editing of the homeologous GmFAD2-1A and GmFAD2-1B genes to yield a high oleic, low linoleic and α-linolenic acid phenotype in soybean. BMC Plant Biol. 19:311. 10.1186/s12870-019-1906-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobinson K. F., Tenuta G. K., Lazarovits G. (1996). Occurrence of race 2 of Verticillium dahliae in processing tomato fields in southwestern Ontario. Can. J. Plant Pathol. 18 55–58. 10.1080/07060669609500655 [DOI] [Google Scholar]
- Dong O. X., Ronald P. C. (2019). Genetic engineering for disease resistance in plants: recent progress and future perspectives. Plant Physiol. 180 26–38. 10.1104/pp.18.01224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt S., Stam R., Hückelhoven R. (2018). Good riddance? Breaking disease susceptibility in the era of new breeding technologies. Agronomy 8:114. 10.3390/agronomy8070114 [DOI] [Google Scholar]
- Engler C., Kandzia R., Marillonnet S. (2008). A one pot, one step, precision cloning method with high throughput capability. PLoS One 3:e3647. 10.1371/journal.pone.0003647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fousia S., Tsafouros A., Roussos P. A., Tjamos S. E. (2018). Increased resistance to Verticillium dahliae in Arabidopsis plants defective in auxin signalling. Plant Pathol. 67 1749–1757. 10.1111/ppa.12881 [DOI] [Google Scholar]
- Fradin E. F., Abd-El-Haliem A., Masini L., van den Berg G. C. M., Joosten M. H. A. J., Thomma B. P. H. J. (2011). Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 156 2255–2265. 10.1104/pp.111.180067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin E. F., Thomma B. P. H. J. (2006). Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 7 71–86. 10.1111/j.1364-3703.2006.00323.x [DOI] [PubMed] [Google Scholar]
- Fradin E. F., Zhang Z., Juarez Ayala J. C., Castroverde C. D. M., Nazar R. N., Robb J., et al. (2009). Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150 320–332. 10.1104/pp.109.136762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao D., Appiano M., Huibers R. P., Loonen A. E. H. M., Visser R. G. F., Wolters A. M. A., et al. (2015). Natural loss-of-function mutation of EDR1 conferring resistance to tomato powdery mildew in Arabidopsis thaliana accession C24. Mol. Plant Pathol. 16 71–82. 10.1111/mpp.12165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawehns F., Cornelissen B. J. C., Takken F. L. W. (2013). The potential of effector-target genes in breeding for plant innate immunity. Microb. Biotechnol. 6 223–229. 10.1111/1751-7915.12023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grogan R. G. (1979). Verticillium wilt on resistant tomato cultivars in California: virulence of isolates from plants and soil and relationship of inoculum density to disease incidence. Phytopathology 69 1176–1180. 10.1094/phyto-69-1176 [DOI] [Google Scholar]
- Helliwell C., Waterhouse P. (2003). Constructs and methods for high-throughput gene silencing in plants. Methods 30 289–295. 10.1016/s1046-2023(03)00036-7 [DOI] [PubMed] [Google Scholar]
- Hückelhoven R., Eichmann R., Weis C., Hoefle C., Proels R. K. (2013). Genetic loss of susceptibility: a costly route to disease resistance? Plant Pathol. 62 56–62. 10.1111/ppa.12103 [DOI] [Google Scholar]
- Huibers R. P., Loonen A. E. H. M., Gao D., Van den Ackerveken G., Visser R. G. F., Bai Y. (2013). Powdery mildew resistance in tomato by impairment of SlPMR4 and SlDMR1. PLoS One 8:e67467. 10.1371/journal.pone.0067467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack D. L., Yang N. M., Saier M. H. (2001). The drug/metabolite transporter superfamily. Eur. J. Biochem. 268 3620–3639. 10.1046/j.1432-1327.2001.02265.x [DOI] [PubMed] [Google Scholar]
- Jørgensen J. H. (1992). Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 63 141–152. 10.1007/bf00023919 [DOI] [Google Scholar]
- Katzen F. (2007). Gateway® recombinational cloning: a biological operating system. Expert. Opin. Drug Discov. 2 571–589. 10.1517/17460441.2.4.571 [DOI] [PubMed] [Google Scholar]
- Korver R. A., Koevoets I. T., Testerink C. (2018). Out of shape during stress: a key role for auxin. Trends Plant Sci. 23 783–793. 10.1016/j.tplants.2018.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langner T., Kamoun S., Belhaj K. (2018). CRISPR crops: crops: plant genome editing toward disease resistance. Annu. Rev. Phytopathol. 56 479–512. 10.1146/annurev-phyto-080417-050158 [DOI] [PubMed] [Google Scholar]
- Liang G., Zhang H., Lou D., Yu D. (2016). Selection of highly efficient sgRNAs for CRISPR/Cas9-based plant genome editing. Sci. Rep. 6:21451. 10.1038/srep21451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S. P. (2002). Virus-induced gene silencing in tomato. Plant J. 31 777–786. 10.1046/j.1365-313x.2002.01394.x [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lyons R., Stiller J., Powell J., Rusu A., Manners J. M. (2015). Fusarium oxysporum triggers tissue-specific transcriptional reprogramming in Arabidopsis thaliana. PLoS One 10:e0121902. 10.1371/journal.pone.0121902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielse C. B., Rep M. (2009). Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 10 311–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi M., Macdonald J., Liu P., Weselowski B., Yuan Z. C. (2018). Clavibacter michiganensis ssp. michiganensis: bacterial canker of tomato, molecular interactions and disease management. Mol. Plant Pathol. 19 2036–2050. 10.1111/mpp.12678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantelides I. S., Tjamos S. E., Paplomatas E. J. (2010). Ethylene perception via ETR1 is required in Arabidopsis infection by Verticillium dahliae. Mol. Plant Pathol. 11 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan S., Jacobsen E., Visser R. G. F., Bai Y. (2010). Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. 25 1–12. 10.1007/s11032-009-9323-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters N., Guidot A., Vailleau F., Valls M. (2013). Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol. Plant Pathol. 14 651–662. 10.1111/mpp.12038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesquet E., Ranocha P., Legay S., Digonnet C., Barbier O., Pichon M., et al. (2005). Novel markers of xylogenesis in zinnia are differentially regulated by auxin and cytokinin. Plant Physiol. 139 1821–1839. 10.1104/pp.105.064337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potnis N., Timilsina S., Strayer A., Shantharaj D., Barak J. D., Paret M. L., et al. (2015). Bacterial spot of tomato and pepper: diverse Xanthomonas species with a wide variety of virulence factors posing a worldwide challenge. Mol. Plant Pathol. 16 907–920. 10.1111/mpp.12244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranocha P., Denancé N., Vanholme R., Freydier A., Martinez Y., Hoffmann L., et al. (2010). Walls are thin 1 (WAT1), an Arabidopsis homolog of Medicago truncatula NODULIN21, is a tonoplast-localized protein required for secondary wall formation in fibers. Plant J. 63 469–483. 10.1111/j.1365-313x.2010.04256.x [DOI] [PubMed] [Google Scholar]
- Ranocha P., Dima O., Nagy R., Felten J., Corratgé-Faillie C., Novák O., et al. (2013). Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat. Commun. 4:2625. 10.1038/ncomms3625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J. D. G. (2011). Hormone crosstalk in plant disease and defense: more than just JASMONATE-SALICYLATE antagonism. Annu. Rev. Phytopathol. 49 317–343. 10.1146/annurev-phyto-073009-114447 [DOI] [PubMed] [Google Scholar]
- Sanson K. R., Hanna R. E., Hegde M., Donovan K. F., Strand C., Sullender M. E., et al. (2018). Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun. 9:5416. 10.1038/s41467-018-07901-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil-Kumar M., Mysore K. S. (2014). Tobacco rattle virus-based virus-induced gene silencing in Nicotiana benthamiana. Nat. Protoc. 9 1549–1562. 10.1038/nprot.2014.092 [DOI] [PubMed] [Google Scholar]
- Stemmer M., Thumberger T., Del Sol Keyer M., Wittbrodt J., Mateo J. L. (2015). CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One 10:e0124633. 10.1371/journal.pone.0124633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K., Wolters A. A., Loonen A. E. H. M., Huibers R. P., van der Vlugt R., Goverse A., et al. (2016). Down-regulation of Arabidopsis DND1 orthologs in potato and tomato leads to broad-spectrum resistance to late blight and powdery mildew. Transgenic. Res. 25 123–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Zhang Z., Lei Y., Hu G., Liu J., Hao M., et al. (2019). Cotton WATs modulate SA biosynthesis and local lignin deposition participating in plant resistance against Verticillium dahliae. Front. Plant Sci. 10:526. 10.3389/fpls.2019.00526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B. P. H. J., Eggermont K., Penninckx I. A. M. A., Mauch-Mani B., Vogelsang R., Cammue B. P. A., et al. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. U.S.A. 95 15107–15111. 10.1073/pnas.95.25.15107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami T., Momma N., Kikuchi S., Watanabe H., Hayashi A., Mizukawa M., et al. (2017). Race 2 of Verticillium dahliae infecting tomato in Japan can be split into two races with differential pathogenicity on resistant rootstocks. Plant Pathol. 66 230–238. 10.1111/ppa.12576 [DOI] [Google Scholar]
- van Schie C. C. N., Takken F. L. W. (2014). Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52 551–581. 10.1146/annurev-phyto-102313-045854 [DOI] [PubMed] [Google Scholar]
- Verlaan M. G., Hutton S. F., Ibrahem R. M., Kormelink R., Visser R. G. F., Scott J. W., et al. (2013). The tomato yellow leaf curl virus resistance genes Ty-1 and Ty-3 are allelic and code for DFDGD-class RNA-dependent RNA polymerases. PLoS Genet. 9:e1003399. 10.1371/journal.pgen.1003399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Pajerowska-Mukhtar K., Culler A. H., Dong X. (2007). Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17 1784–1790. 10.1016/j.cub.2007.09.025 [DOI] [PubMed] [Google Scholar]
- Wang Y., Tan J., Wu Z., VandenLangenberg K., Wehner T. C., Wen C., et al. (2018). Staygreen, Stay healthy: a loss-of-susceptibility mutation in the staygreen gene provides durable, broad-spectrum disease resistances for over 50 years of US cucumber production. New Phytol. 221 415–430. 10.1111/nph.15353 [DOI] [PubMed] [Google Scholar]
- Weber E., Engler C., Gruetzner R., Werner S., Marillonnet S. (2011). A modular cloning system for standardized assembly of multigene constructs. PLoS One 6:e16765. 10.1371/journal.pone.0016765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong N., Liu W., Wang X. (2015). WU-CRISPR: characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 16:218. 10.1186/s13059-015-0784-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Jia L., Goggin F. (2011). The reliability of virus-induced gene silencing experiments using tobacco rattle virus in tomato is influenced by the size of the vector control. Mol. Plant Pathol. 12 299–305. 10.1111/j.1364-3703.2010.00669.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadeta K. A., Thomma B. P. H. J. (2013). The xylem as battleground for plant hosts and vascular wilt pathogens. Front. Plant Sci. 4:97. 10.3389/fpls.2013.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K., Qiu J.-L. (2019). Genome editing for plant disease resistance: applications and perspectives. Philos. Trans. 374:20180322. 10.1098/rstb.2018.0322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi S. S., Mukhtar M. S., Mansoor S. (2018). Genome editing: targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 36 898–906. 10.1016/j.tibtech.2018.04.005 [DOI] [PubMed] [Google Scholar]
- Zaka A., Grande G., Coronejo T., Quibod I. L., Chen C. W., Chang S. J., et al. (2018). Natural variations in the promoter of OsSWEET13 and OsSWEET14 expand the range of resistance against Xanthomonas oryzae pv. Oryzae. PLoS One 13:e0203711. 10.1371/journal.pone.0203711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gel electrophoresis (1% TBE, Gelred, image colors inverted) of gene-specific PCR with primers KH_156 on primary transformants (T1) of plants transformed with SlWAT1 CRISPR-Cas9 construct. Wild-type PCR product (1,616 bp) indicated with an arrow and highlighted are mutants #10, #19, #23, and #28 that were transferred to the greenhouse, and of which seeds were only obtained from #19 (TV181046).
Predicted transmembrane domains for wild-type SlWAT1 (left), mutant allele 1 (right), and mutant allele 2 (bottom). Graphs were generated with TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/).
CRISPR T2 family TV181046 and its T3 progeny TV181046-16, –18, and –23 display severe growth and development defects. Pictures of MM and T3 CRISPR WAT1 plants at different time points.
Canopy area pictures mock- and V. dahliae-inoculated (strain JR2) plants for CRISPR T2 family TV181046 and T3 families TV181046-16, –18, and –23 at 21 dpi compared to MM plants of respective experiment.
Primers used in this study.
Overview of primary transformants (T1) with presence/absence of silencing construct (NPTII/35S), relative SlWAT1 expression normalized to control plants at 1 (2–ΔΔCt) and plants from which T2 seeds were obtained.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.