Abstract
Objectives
Tumor‐associated autoantibodies (AAbs) in individuals with cancer can precede clinical diagnosis by several months to years. The objective of this study was to determine whether the primary immune response in form of IgM and gut mucosa‐associated IgA can aid IgG AAbs in the detection of early‐stage colorectal cancer (CRC).
Methods
We developed a novel protein array comprising 492 antigens seropositive in CRC. The array was used to profile IgG, IgM and IgA antibody signatures in 99 CRC patients and 99 sex‐ and age‐matched non‐cancer controls. A receiver operating curve (ROC), Kaplan–Meier survival analysis and univariate and multivariate Cox regression analyses were conducted.
Results
We identified a panel of 16 multi‐isotype AAbs with a cumulative sensitivity of 91% and specificity of 74% (AUC 0.90, 95% CI: 0.850–0.940) across all CRC stages. IgM and IgG isotypes were conversely associated with disease stage with IgM contributing significantly to improved stage I and II sensitivity of 96% at 78% specificity (AUC 0.928, 95% CI: 0.884–0.973). A single identified IgA AAb reached an overall sensitivity of 5% at 99% specificity (AUC 0.520, 95% CI: 0.440–0.601) balanced across all CRC stages. Kaplan–Meier analysis revealed that se33‐1 (ZNF638) IgG AAbs were associated with reduced 5‐year overall survival (log‐rank test, P = 0.012), whereas cumulative IgM isotype signatures were associated with improved 5‐year overall survival (log‐rank test, P = 0.024).
Conclusion
IgM AAbs are associated with early‐stage colorectal cancer. Combining IgG, IgM and IgA AAbs is a novel strategy to improve early diagnosis of cancers.
Keywords: autoantibodies, biomarker, colorectal cancer, early detection, IgA, IgM
Early‐stage detection of colorectal cancer is critical for patient survival. We have conducted a multi‐isotype autoantibody (AAbs) screen using a 492‐feature CRC protein array. Our study shows that IgM AAbs are associated with early‐stage disease and IgA AAbs, although stage‐independent, encompass humoral responses associated with mucosal surfaces. Combining IgG, IgM and IgA AAbs is a novel strategy for early disease detection and improvement of patient outcomes.

Introduction
Colorectal cancer (CRC) is a major health threat in both men and women. With estimated new cases of more than 1.9 million and 930 000 deaths in 2020, CRC is the third most commonly diagnosed cancer and the second leading cause of cancer deaths worldwide.1 Despite a rapid decline in CRC incidence rates during the 2000s, mostly because of the introduction of population screening modalities, such as colonoscopy and faecal occult blood‐based tests (FOBT), the overall decline in CRC incidence rates and mortality has waned in more recent years.2 The plateauing of mortality rates in CRC in recent years could be attributed to, among others, poor uptake of stool‐based screening3, 4 and the rise in early‐onset CRC incidence by 1–4% among the younger population (age < 50).1 CRC mortality is mainly attributed to nodal and distant metastatic disease often in individuals not diagnosed until the advanced and symptomatic disease has developed.2, 5 The 5‐year CRC survival rates are as high as 90% in early‐stage disease, plummeting down to only 10–15% in metastatic CRC.6 Early disease detection in pre‐symptomatic individuals with broadly acceptable screening modalities is, therefore, a critical factor in reducing CRC mortality rates in the future.7 Despite continuing advances in population screening approaches, a blood‐based biomarker for the early detection of CRC has not yet been developed to meet clinical needs.
Cancers prompt an immunologic response against aberrant self‐antigens resulting in the production of autoantibodies from the onset of disease,8, 9 often preceding clinical manifestation of cancer by several months or years.10, 11 A significant number of reports have documented the diagnostic capability of autoantibodies in various cancers, including ovarian cancer,12 lung cancer,13 prostate cancer,14 breast cancer,15 colorectal cancer,16, 17 gastrointestinal cancer18 and primary cutaneous melanoma.19 However, most of the autoantibody biomarker studies explicitly exploit IgG immunoglobulin‐isotype autoantibodies that postdate the first‐line immune responses. IgM antibodies, in contrast, are considered to be the first immunoglobulin isotype that emerges during a humoral immune response after an initial immunological challenge and have been implicated not only in the early recognition of external invading pathogens, such as bacteria and viruses,20, 21, 22, 23 but also in the recognition and elimination of precancerous and cancerous lesions.24, 25, 26 Consequently, exploring IgM autoantibodies as early cancer detection tools is purposefully more relevant and promising. As cancer progresses, IgM antibody titres decline and IgG responses spike up in the process of isotype switching to provide long‐term protection.27 IgA isotype immunoglobulins have also attracted a lot of attention in recent years, especially because of their presence at mucosal surfaces and their role as mediators of intestinal immunity.28, 29 Although IgA antibodies are known for their antimicrobial properties, their role in cancer has also been explored.30, 31, 32, 33 Serum IgA autoantibodies have been recently found in sera of lung cancer patients.34 Studying the primary (IgM), secondary (IgG) and gut mucosa‐associated (IgA) immune responses points towards unique sets of biomarkers capable of detecting colorectal cancer at its earliest stages with a robust diagnostic performance.
The identification of novel disease‐related autoantibody signatures requires high‐content antigen platforms that allow screening of complex antibody repertoires from patient sera accompanied by ease in data evaluation and interpretation. In the recent decade, protein array technologies have been employed as important platforms to study protein–protein interactions,35 characterisation of binding specificity and cross‐reactivity of antibodies,36 peptide–protein interaction37 and screening of serological markers in cancer.16, 19, 38, 39, 40 The ability to facilitate the characterisation of humoral immune responses to large, annotated collections of proteins in a high‐throughput manner is one of the unique utilities of protein arrays that can be exploited in autoantibody‐based biomarker discovery.
In this study, we utilised a customised colorectal cancer protein array comprising of 492 antigens, to screen sera from a total of 99 colorectal cancer patients and 99 matched non‐cancer controls. We sought to discover autoantibody signatures that can significantly aid in cancer diagnosis, particularly in the early stages of cancer development. This study fundamentally exploited the essence of IgM in early cancer detection and IgA in cancers associated with the gastrointestinal tract as a way of augmenting the more traditional IgG‐based autoantibody signatures.
Results
Autoantibody repertoires in CRC and non‐cancer controls
Serum autoantibodies were characterised using the CRC protein arrays (Figure 1) blinded to the clinicopathological data of 99 CRC patients and 99 non‐cancer controls (Table 1). Distinct IgG, IgM and IgA profiles were established for each subject. Out of the 492 antigens on the CRC protein array, the average numbers of seropositive antigens for IgG, IgM and IgA in CRC patients were 30, 29 and 19, respectively. In non‐cancer controls, the average numbers of seropositive antigens for IgG, IgM and IgA were 25, 27 and 20, respectively (Table 2). We noted higher overall seropositivity in cancer patients compared to non‐cancer controls for all three immunoglobulin isotypes reflected by the higher average number of positives in cancer patients and the overall number of antigens positive at least once in each group (Table 2). In CRC patients, a total of 373, 291 and 228 antigens were seropositive for IgG, IgM and IgA, respectively. On the contrary, the seropositivity in non‐cancer controls was markedly lower with 328, 241 and 168 seropositive antigens for IgG, IgM and IgA, respectively.
Figure 1.
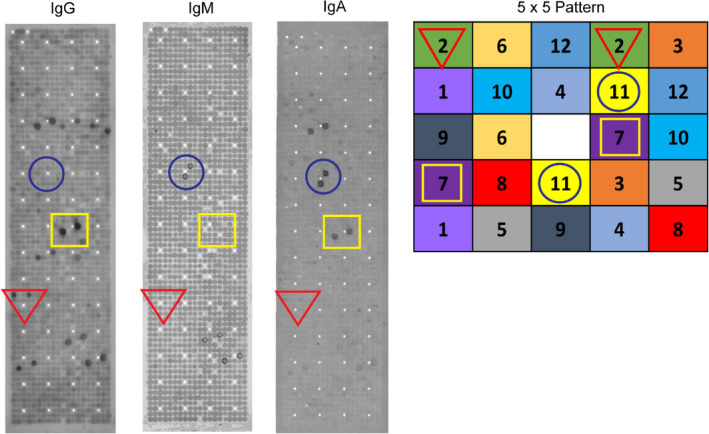
Human recombinant protein array. A total of 720 expression clones corresponding to 492 recombinant human proteins at different cDNA insert lengths were arrayed on a PVDF membrane in duplicate. The depicted array represents IgG, IgM and IgA antibody repertoires from one single patient. The highlighted circle show immunoreactivity for IgM and IgA but not IgG, the square represents seropositivity for IgA and IgG but not IgM, and the triangle represents seropositivity to IgG, but not to IgM and IgA. The enlarged section in the right panel shows the non‐redundant 5 × 5 pattern of protein distribution for rapid and accurate analysis.
Table 1.
Clinicopathological features of the cohort
| Colorectal cancer patients | Non‐cancer controls | |
|---|---|---|
| n = 99 | n = 99 | |
| Gender, n (%) | ||
| Male | 51 | 49 |
| Female | 48 | 50 |
| Age (years) | ||
| Mean | 63 | 56 |
| Median | 65 | 57 |
| Range (age) | 35–87 | 19–84 |
| > 55 | 77 | 53 |
| < 55 | 22 | 46 |
| Tumor site | ||
| Colon | 95 | |
| Rectum | 4 | |
| Tumor (T) stage | ||
| T1/PT1 | 2 | |
| T2/PT2 | 18 | |
| T3/PT3 | 59 | |
| T4/PT4 | 20 | |
| Node (N) stage | ||
| N0 | 57 | |
| N1 | 19 | |
| N2 | 23 | |
| Metastasis (M) stage | ||
| M0 | 47 | |
| M1 | 19 | |
| Mx | 33 | |
| Differentiation | ||
| Well | 16 | |
| Moderately | 70 | |
| Poorly | 13 | |
Table 2.
Autoantibody seroprevalence in cancer and controls analysed against 492 antigens
| Immunoglobulin isotype | Subjects | Average seropositive per subject (mean) | Overall seropositive antigens (at least once) |
|---|---|---|---|
| IgG | Cancer (n = 99) | 30 | 373 (76%) |
| Control (n = 99) | 25 | 328 (67%) | |
| IgM | Cancer (n = 99) | 29 | 291 (59%) |
| Control (n = 99) | 27 | 241 (49%) | |
| IgA | Cancer (n = 99) | 19 | 228 (46%) |
| Control (n = 99) | 20 | 168 (33%) |
Autoantibody signatures in CRC detection
All seropositive antigens were ranked for each immunoglobulin isotype in order to discriminate between CRC and non‐cancer controls. Data analysis revealed that CRC sensitivity of the best‐performing markers ranging from 5% to 21%, with specificities of 93–100%. A combination of the individual markers led to a multi‐marker panel of 16 antigens comprising 6 IgG, 9 IgM and 1 IgA antigens showing the highest combined seropositivity for CRC (Table 3). Criteria for patient assignment to the CRC group required at least one positive marker evaluation for the 16‐marker panel, with 43 patients seropositive to one marker, 29 seropositive to two markers and 18 seropositive to three or more markers.
Table 3.
A panel of 16 autoantibody biomarkers and their frequency of occurrence in CRC and the non‐cancer control group
| Marker Uniprot ID | UniProt KB | Cellular location | Antibody isotype | CRC (n = 99) | Non‐cancer control (n = 99) |
|---|---|---|---|---|---|
| P53 | P04637 | Nucleus | IgG | 21 | 1 |
| CERS5 | Q9D6K9 | ER | IgG | 21 | 2 |
| CIRBP | Q14011 | Nucleus | IgG | 18 | 7 |
| ZNF787 | Q6DD87 | Nucleus | IgM | 16 | 5 |
| FAM13A | O94988 | Cytosol | IgG | 12 | 3 |
| TBR1 | Q16650 | Nucleus | IgM | 11 | 0 |
| CAMSAP3 | Q9P1Y5 | Cytoskeleton | IgM | 11 | 1 |
| VCL | P18206 | Plasma membrane | IgM | 10 | 2 |
| TCP4 | P53999 | Nucleus | IgM | 10 | 4 |
| MBD3 | O95983 | Nucleus | IgM | 10 | 5 |
| GOSR2 | O14653 | ER, Golgi | IgM | 8 | 0 |
| FARP1 | Q9Y4F1 | Plasma membrane | IgM | 7 | 0 |
| ZNF638 | Q14966 | Nucleus | IgG | 6 | 1 |
| CCDC50 | Q8IVM0 | Cytosol | IgG | 6 | 1 |
| PSMD10 | O75832 | Nucleus | IgM | 5 | 0 |
| MON1A | Q86VX9 | Cytosol | IgA | 5 | 1 |
In order to assess the diagnostic performance of the 16‐marker panel, a receiver operating characteristics (ROC) curve analysis was constructed. The analysis resulted in an optimal sensitivity of 91% at 74% specificity (AUC 0.90, 95% CI: 0.850–0.940, Figure 2a). We further evaluated the diagnostic values of the multi‐marker panel in different stages of CRC in our cohort. To achieve a balanced analysis, we grouped stages I and II together because of the relatively small number of stage I patients in the cohort. Interestingly, the sensitivity and specificity of the panel increased to 96% and 78%, respectively, for early‐stage CRC (stage I and II) with high accuracy (AUC 0.928, 95% CI: 0.884–0.973, Figure 2b). The sensitivity and specificity decreased in more advanced CRC. The sensitivity of the panel was 86% at 80% specificity for CRC stage III (AUC 0.884, 95% CI: 0.800–0.968, Figure 2c) and 84% at 82% specificity for CRC stage IV (AUC 0.885, 95% CI: 0.781–0.898, Figure 2d).
Figure 2.
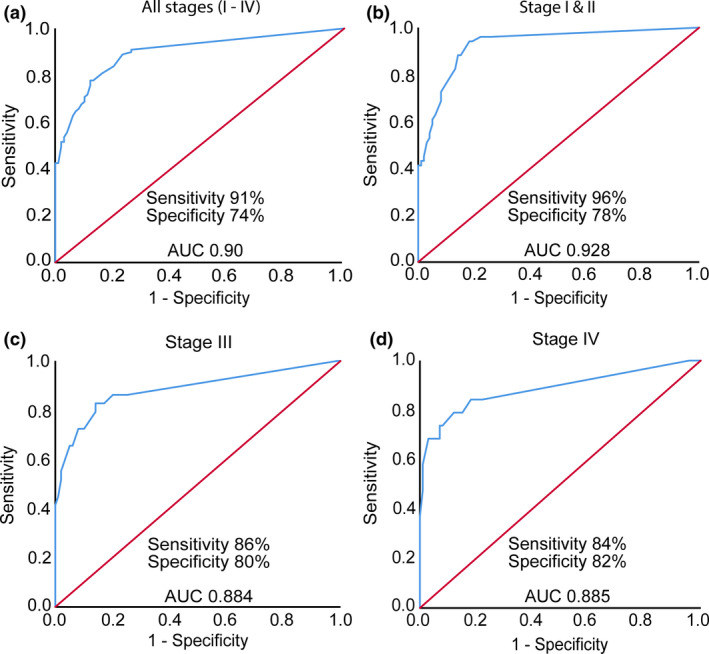
Diagnostic performance of 16‐antigen multi‐marker panel at different stages of CRC. ROC curve analysis of the multi‐marker panel showed (a) overall sensitivity of 91% at 74% specificity with high accuracy (AUC 0.90, 95% CI: 0.850–0.940) in discriminating CRC patients (n = 99) from non‐cancer controls (n = 99), (b) sensitivity of 96% at 78% specificity (AUC 0.928, 95% CI: 0.884–0.973) in detecting early‐stage (stages I and II) CRC (CRC n = 51, controls n = 99), (c) sensitivity and specificity of 86% and 80% (AUC 0.884, 95% CI: 0.800–0.968), respectively, for CRC that had nodal spreads (stage III) (CRC n = 29, controls n = 99) and (d) overall diagnostic sensitivity of 84% at 82% specificity (AUC 0.885, 95% CI: 0.781–0.898) for advanced‐stage (stage IV) CRC that had metastatic disease (CRC n = 19, controls n = 99).
IgM autoantibodies improve sensitivity for early‐stage cancer
Our multi‐marker panel included 9 IgM autoantibodies. We sought to specifically understand whether IgM autoantibodies could aid in the early recognition of cancer. Intriguingly, the diagnostic value of the IgM autoantibody panel was consistently higher for early‐stage CRC than for advanced disease. Although the overall sensitivity and specificity for all stages were 51% and 85% (AUC 0.701, 95% CI 0.628–0.775, Figure 3a and e), the sensitivity of cumulative IgM autoantibodies increased to 57% at 86% specificity for cancer stages I and II (AUC 0.738, 95% CI 0.644–0.831, Figure 3b and e). The sensitivity decreased for advanced stages with a minor increase in specificity. The cumulative sensitivity and specificity of the IgM panel were 48% and 89% for stage III cancer (AUC 0.689, 95% CI 0.564–0.814, Figure 3c and e) and 37% and 92% for stage IV (AUC 0.677, 95% CI 0.531–0.822, Figure 3d and e), respectively.
Figure 3.
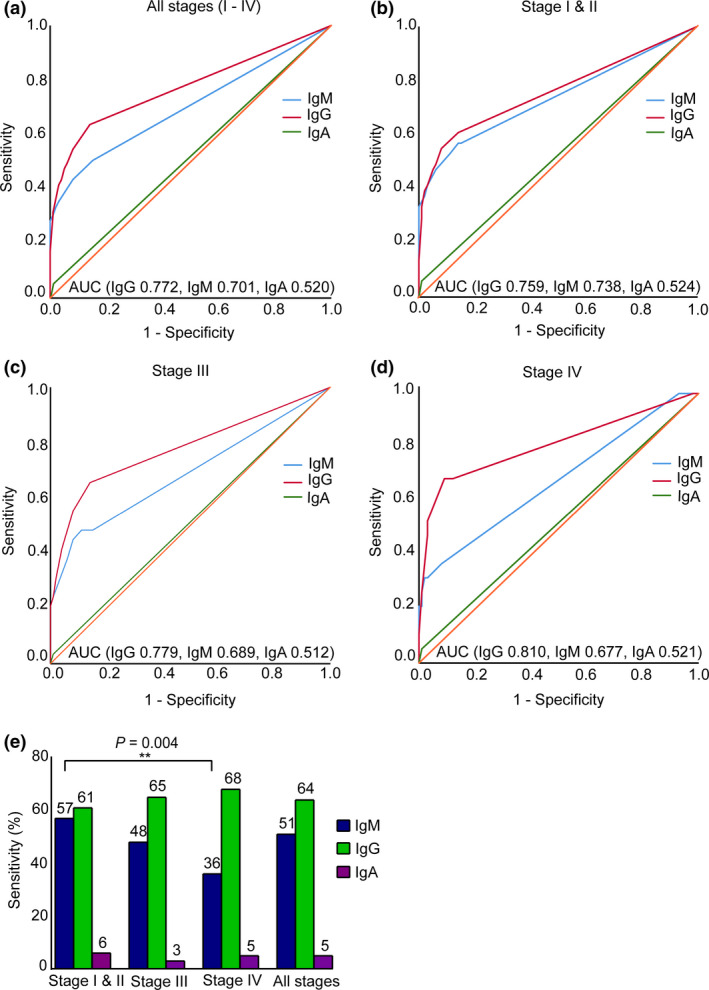
Autoantibody isotypes differentiate between clinical stages of CRC. ROC curve analysis of IgM, IgG and IgA‐specific autoantibody markers revealed (a) overall sensitivity of the panel of IgG, IgM and IgA autoantibodies in the cohort (CRC n = 99, controls n = 99) (b) sensitivities of the markers in detecting early‐stage (stages I and II) CRC (CRC n = 51, controls n = 99), (c) sensitivities of the markers in detecting stage III CRC (CRC n = 29, controls n = 99), (d) sensitivities in detecting metastatic CRC (stage IV) (CRC n = 19, controls n = 99) and (e) sensitivities of IgG, IgM and IgA autoantibody markers in different CRC stages. IgM autoantibody panel showed significantly higher sensitivity in the early cancer stages and then gradually declined in the advanced stages (Fisher exact test, P = 0.004), whereas IgG showed a relatively lower sensitivity and specificity profile for early stages than for the advanced stages.
In contrast, the IgG autoantibody subset of the panel showed relatively lower sensitivity for early stages and higher for advanced stages. The overall sensitivity of the cumulative IgG autoantibodies was 64% at 86% specificity for all stages (AUC 0.772, 95% CI 0.705–0.839, Figure 3a and e). For early stage (stages I and II), the sensitivity of the IgG panel was 61% at 86% specificity (AUC 0.759, 95% CI 0.669–0.850, Figure 3b and e). The sensitivity and specificity for stage III were 65% and 86% (AUC 0.779, 95% CI 0.667–0.891, Figure 3c and e) and for stage IV, 68% and 91% (AUC 0.870, 95% CI 0.680–0.940, Figure 3d and e), respectively.
Our panel included a single IgA autoantibody signature. The sensitivity and specificity of the IgA marker were consistent throughout different cancer stages. The overall sensitivity and specificity were 5% and 99% (AUC 0.520, 95% CI 0.440–0.601, Figure 3a and e). Early‐stage sensitivity of IgA was 6% at 99% specificity (AUC 0.524, 95% CI 0.425–0.623, Figure 3b and e). At 99% specificity, the sensitivity for stage III was 3.4% (AUC 0.522, 95% CI 0.391–0.634, Figure 3c and e) and for stage IV was 5.3% (AUC 0.521, 95% CI 0.375–0.667, Figure 3d and e).
Autoantibodies and CRC prognosis
Since the 9‐marker IgM autoantibody panel was predominantly associated with early‐stage CRC and IgG autoantibodies were found more frequently in advanced cancers, we investigated whether patient groups always positive for IgM or IgG showed different clinical outcomes. In order to investigate patient survival in those two groups, we conducted a 5‐year Kaplan–Meier analysis (Figure 4, Supplementary figures 1 and 2). The IgM autoantibodies were significantly associated with improved patient survival (log‐rank test, P = 0.024, Figure 4a). A significant association was observed between a number of clinicopathological factors and overall survival in univariate Cox regression analysis. Univariate Cox analysis showed that IgM autoantibodies were significantly correlated with better patient survival (P = 0.039, Supplementary table 2). However, the prognostic value of IgM autoantibodies was not independent of patients' clinicopathological variables as observed in a multivariate Cox regression analysis (P = 0.069, Supplementary table 2). We next examined whether the presence of autoantibodies to the six identified IgG autoantigens showed any association with clinical outcomes. Kaplan–Meier analysis revealed that the group of 63 CRC patients positive for IgG autoantibodies did not differ in clinical outcome when compared to patients without IgG antibodies (log‐rank test, P = 0.353, Figure 4b).
Figure 4.
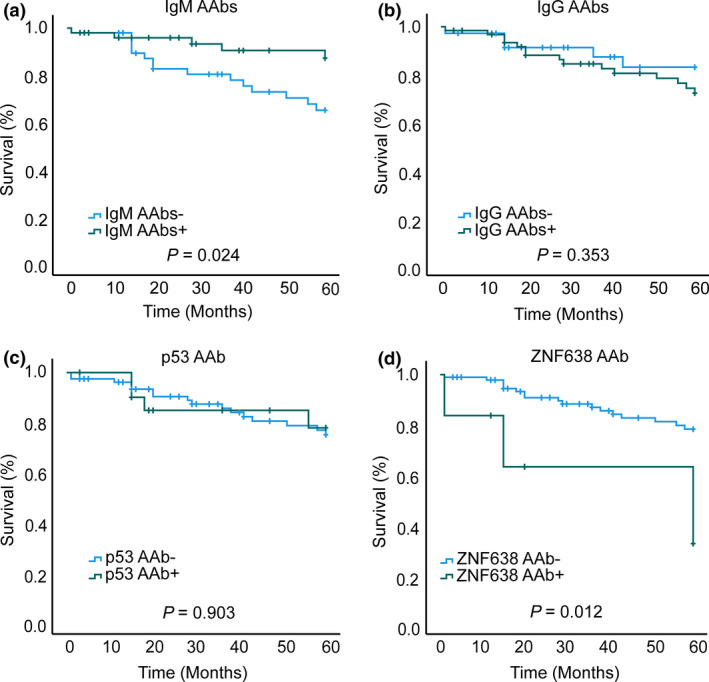
Autoantibodies and patient survival. Kaplan–Meier survival analysis of autoantibody panels revealed (a) cumulative presence of IgM autoantibodies (+ seropositive, − seronegative) was significantly associated with improved survival (IgM+ n = 50, IgM− n = 49) (log‐rank test, P = 0.024), (b) cumulative presence (+) and absence (−) of IgG autoantibodies had no association with overall patient survival (n = 63, IgG− n = 36) (log‐rank test, P = 0.353), (c) presence of anti‐p53 IgG autoantibodies showed no association with overall patient survival (p53+ n = 21, p53− n = 78) (log‐rank test, P = 0.903) and (d) presence of anti‐CTLC‐associated antigen se33‐1 (ZNF638) IgG autoantibodies was associated with poor 5‐year survival in CRC patients (ZNF638+ n = 6, ZNF638− n = 93) (log‐rank test, P = 0.012).
There is an ongoing debate on whether autoantibodies to p53 are associated with patient outcomes in cancer.24, 41, 42, 43 In order to investigate the association between patient outcome and autoantigens, we performed a 5‐year Kaplan–Meier survival analysis for all 16 autoantibody markers identified in our study. Notably, the presence of autoantibodies to p53 showed no significant association with patient survival (log‐rank test, P = 0.903, Figure 4c). No association with patient survival was seen in 14 further antigens (Supplementary figures 1–3), apart from CTLC‐associated antigen se33‐1 (ZNF638). Patients seropositive for se33‐1 (ZNF638) IgG autoantibodies had a significantly poorer 5‐year survival (Figure 4d) than those who were negative for the marker (log‐rank test, P = 0.012). Univariate Cox regression analysis revealed that se33‐1 (ZNF638) autoantibodies were associated with poor patient survival (HR = 4.139, 95% CI 1.260–14.800, P = 0.02, Supplementary table 3). In the univariate analysis, other significant prognostic factors for patient survival included age, cancer stages such as tumor (T) and node (N) stage. Subsequent multivariate Cox regression analysis revealed that se33‐1 (ZNF638) was not an independent predictor of 5‐year survival (HR = 2.473, 95% CI 0.685–8.930, P = 0.167, Supplementary table 3). However, as expected, there was a significant association between metastasis and patient survival (HR = 5.812, 95% CI 2.168–15.581, P = 0.000) as well as with age (HR = 1.077, 95% CI 1.02–1.138, P = 0.008, Supplementary table 3).
Discussion
This study aimed to investigate whether IgM and IgA immune responses specific to selected tumor antigens are associated with cancer and whether these, in combination with IgG autoantibodies, may improve detection of early‐stage CRC. To achieve this, we characterised IgG, IgM and IgA antibody repertoires in CRC patients and matched controls using a customised array of 492 proteins previously shown to be immunoreactive in CRC.16 Our rationale was that, as several antigens identified in an earlier IgG autoantibody screening were also immunoreactive with IgM in the same patients, a wider screen encompassing several hundred CRC antigens would likely identify IgM autoantibodies potentially detectable prior to measurable IgG response.24 We also sought to extend the study to IgA antibody responses, to take into account the potential activation of intestinal mucosal immunity in CRC as demonstrated in other cancers including recently in lung cancer.34
Serum analysis conducted in this study revealed a novel set of 16‐biomarkers comprising a combination of IgG, IgM and IgA autoantibodies with a robust performance in detecting early‐stage CRC. We report for the first time to our knowledge specifically the potential of IgM autoantibodies in detecting early‐stage cancer. The IgM autoantibody signatures identified in this study were more sensitive in detecting early‐stage cancer, with 57% for stages I and II, decreasing to 48% for stage III and 37% for stage IV. Antigen‐specific IgG sensitivity, on the other hand, was initially 61% for stages I and II and rose gradually to 65% for stage III and 68% for stage IV. These data highlight the substantial contribution of the IgM autoantibody subset to improving the overall sensitivity for early‐stage disease. As the sensitivity and specificity of the identified single IgA autoantibody were similar across all stages, the stage‐dependent IgM and IgG autoantibody conversion potentially reflects isotype switching in antigen‐activated B cells while the anti‐tumor immune responses are building up during cancer progression.27 IgM autoantibodies present at advanced stages of CRC are likely attributed to new antigens evolving during tumor progression.
Autoantibodies to tumor antigens are understood to rise in patients at very early disease stages because of possible changes in antigen expression and structural conformity within the tumor, loss of tolerance mechanisms, presence of mutations or disease‐associated post‐translation modifications.9 It is further conceivable that proteins that incite autoantibody production may have a plethora of biological functions in humans. Overall, we have identified eight proteins that are located in the nucleus, three cytosolic proteins, two proteins located in the endoplasmic reticulum, two plasma membrane proteins and one cytoskeleton protein. Among the identified antigens, we have previously reported autoantibodies to p53, se33‐1 (ZNF638) and CerS5.16, 44, 45 Antibodies to the tumor suppressor protein p53 have been frequently reported in CRC and other types of cancer, such as oesophageal cancer, lung cancer, ovarian cancer, breast cancer, lymphoma, bladder cancer, hepatocellular carcinoma and melanoma.19, 46 Recently, p53 has been exploited as a target to stimulate T‐cell‐mediated killing of cancer cells in an antibody‐based therapy, indicating that antigen‐targeted antibody‐based therapies might present a route to targeted cancer treatment.47 Novel antigens associated with the humoral immune response could therefore be exploited similarly. Zinc finger protein 638 (ZNF638) for instance, also known as cutaneous T‐cell lymphoma‐associated antigen (CTCL) Se33‐1 or NP220, is a DNA‐binding nuclear protein,48 which has been previously identified as a marker for cutaneous T‐cell lymphoma (CTCL).49 While most studies have focused on anti‐p53 autoantibodies,41, 42, 43, 50, 51, 52 the prognostic utility of other autoantibodies has also been explored.24, 51, 53, 54, 55, 56, 57 In this current study, we have reported that patients with autoantibodies to the ZNF638 protein had poorer 5‐year survival. However, autoantibodies to p53 and the remaining 14 markers in our panel showed no prognostic ability, which is consistent with our previous study.24 Ceramide synthase 5 (CerS5), one of the members of ceramide synthases (CerS1‐6), is not only found to be involved in apoptosis, autophagy and cell proliferation58, 59, 60 but also in tumor suppression.61 Although CerS5 is expressed ubiquitously in mammalian tissue,62, 63 we have previously shown that high CerS5 expression in colorectal cancer tissue is correlated with poor patient survival.44 In addition, CerS5 has also been found to be associated with other cancers, such as endometrial cancer,60 breast cancer64 and large‐cell lung carcinoma.65 Autoantibodies to CIRBP were previously reported to be elevated in breast cancer patients, predicting a transition of ductal carcinoma in situ into invasive breast cancer.66
There were several limitations to our study. First, the 492 proteins constituting the protein array were selected mainly because of their ability to induce immunoreactivity with IgG antibodies in cancer patients.16 Although we showed that IgG‐specific antigens can be immunoreactive with IgM antibodies,24 we are potentially missing subsets of IgM antigens that did not yet develop IgG responses. Second, the antigens in our study are derived from a eukaryotic Escherichia coli expression system; hence, they do not incorporate any posttranslational modifications excluding those from detectable antibody repertoires. Furthermore, the protein array is developed under denaturing conditions, thereby omitting the possibility to identify structural epitopes. Nevertheless, our results demonstrate the capability of this protein array platform to identify dozens of novel antigens, which is further reinforced by the identification of established antigens such as p53, which contains predominantly linear epitopes.19, 46 Finally, a critical aspect of this study is its moderate cohort size of 99 CRC patients and 99 controls. However, prior characterisation of all 492 antigens as immunoreactive in a cohort of 43 CRC patients in our previous study makes this set substantially more robust.16 The robustness of the screening is yet again highlighted by the identification of autoantibodies previously characterised by other groups and in our earlier studies.
Success in cancer treatment largely depends on the stage at diagnosis, with earlier diagnosis offering better outcomes of curative treatment and long‐term patient survival.67, 68, 69, 70 Although many studies have focused on cancer diagnosis, early cancer detection remains challenging, particularly because of the difficulties in finding biomarkers with high sensitivity and specificity.71, 72 A significant number of reports to date have documented the diagnostic capability of serum autoantibodies in various cancers, with sensitivity and specificity ranging from 55% to 84% and from 80% to 98%, respectively.12, 13, 14, 15, 16, 18, 19 However, studies particularly focused on early‐stage cancer detection with autoantibodies are scarce and predominantly exploit IgG responses. Many of these study outcomes exhibit low sensitivity and specificity, ranging from 22% to 65% only for early‐stage CRC, which is far from reaching the current clinical expectations.73 We argue that studies conducted in early‐stage cancer research have not focused enough on the early events in a growing tumor. Thus, our study specifically aimed at early immune responses, such as the IgM response, to create an opportunity for improved early cancer detection. Previously, IgM autoantibodies against recombinant scFv have been exploited in early‐stage non‐small‐cell lung cancer (NSCLC) detection, which reported a better sensitivity and specificity of 80% and 87%.74 In our earlier studies, we have shown that a combination of IgG and IgM autoantibodies can reach high predictive values for the presence of colorectal cancer, albeit with a moderate sensitivity of 77.3% at 82.1% specificity.24 Several other studies have investigated the diagnostic value of IgM, IgG and IgA autoantibodies, with most of these studies reporting single or two protein marker assays.18, 75 In contrast, this study has identified a comprehensive 16‐marker multi‐isotype panel comprising IgM, IgG and IgA autoantibodies.
Currently, colonoscopy is the gold standard screening tool with a sensitivity and specificity of over 98%. However, colonoscopy is an expensive procedure, invasive in nature, shows increased risk of perforation and requires frequent repetitions usually every 3–5 years once pathologies were identified.7 Concurrently, there is a growing popularity of faecal occult blood testing (FOBT) for population screening, but adherence to FOBT is as low as 40% and the test, especially immunochemical FOBT (FIT), has lower sensitivity in detecting early‐stage cancer (stages I and II) than advanced stages (stages III and IV)76, 77, 78 and may also lead to overdiagnosis of benign polyps and therefore to overtreatment.79 Moreover, screening uptake rates among indigenous and ethnic minority groups worldwide are even lower because of multiple socio‐psychological and cultural factors, such as the feeling of shame, lack of confidence in self‐screening test procedures.80 A significant preference for blood‐based CRC screening tests over primary colonoscopy, sigmoidoscopy and FOBT is observed among a wide range of ethnicity and race.81
Moreover, the incidence rate of interval cancers (I‐CRC) is on the rise, which could be as high as 9%.82, 83 I‐CRC occurs because of missed lesions by any screening means or a new cancer growth between screening appointments.84 There has been a growing concern about the monitoring of interval cancers because of a lack of available tools. The development of autoantibody‐based tools to complement the existing diagnostic procedures and patient follow‐ups has been recently attempted in breast cancer recently.85 Similar developments in other cancers, including CRC, would further bolster the ongoing efforts. From a clinical perspective, autoantibody‐based biomarker panels with a robust sensitivity for all cancer stages could play an important role in the monitoring of occult and minimal residual disease. Nonetheless, very few autoantibodies have been examined as biomarkers in this manner, and in this aspect, our study may present an exciting new approach in the future.
In conclusion, we show for the first time that screening serum cancer‐associated IgM autoantibodies may enhance the detection of early‐stage cancer. By showing that IgM autoantibodies are found more frequently in early‐stage CRC and combined with IgG autoantibodies, which are more sensitive in advanced CRC, we demonstrate a crucial complementarity of multi‐isotope antibody responses in patients with CRC. Although IgA autoantibodies were less prevalent in CRC, the identified IgA antibody did improve the overall performance by incorporating antigens potentially associated with mucosal membranes in the gut. In summary, combining IgG, IgM and IgA represents a novel isotype‐augmented strategy for detecting cancer in the very early stages of disease, thereby improving patient outcomes.
Methods
Patient cohort and clinical samples
This study was approved by Mater Misericordiae Ltd, Australia, Human Ethics Committee HREA Study Reference Number: HREC/18/MHS/58. Informed consent was obtained from all patients by the Victorian Cancer Biobank (VCB), Australia (VCB number: 18020), and clinical samples were collected between 2000 and 2013. Clinicopathological information and 5‐year follow‐up data were recorded. In total, serum samples from 99 confirmed CRC patients collected before any therapeutic interventions and 99 non‐cancer controls were included in this study. CRC patients and non‐cancer controls had no previous history of cancer, systemic inflammatory or autoimmune disease and were not on immunosuppressive medication. The serum samples were kept at −80°C until used.
The cohort comprised 51 male and 48 female CRC patient samples, and 49 male and 50 female non‐cancer controls (Table 1). Ninety‐five CRC patients had the tumor in their colon and there were four patients with rectal tumors. The average age for CRC patients was 63 and 56 for non‐cancer control subjects. Tumors were staged according to the 6th edition of the UICC TNM classification of malignant tumors.86 Mx cases with uncertain or not evaluated metastatic spread were assigned the respective lower (i.e. less advanced) stage categories in accordance with the UICC TNM system rules. In total, 52% of the (51/99) patients were in early‐stage cancer (T1–4, N0, M0), 29% in (29/99) had nodal spread with no distant metastases (T1–4, N1–2, M0), and 19% (19/99) had metastatic disease (T1–4, N1–2, M1). A total of 70 patients had moderately differentiated, 16 well‐differentiated and 13 poorly differentiated tumors.
Customised CRC protein arrays
Colorectal cancer protein arrays comprising 492 proteins were used to screen serum samples for autoantibodies. Protein array composition was selected based on protein immunoreactivity in CRC patients from data analysis collated in an earlier CRC autoantibody study published by this group.16 The IgG antibody repertoires from 43 CRC patients and 40 non‐cancer controls identified using large 37 830‐clone recombinant human protein arrays were reanalysed. Expression clones immunoreactive with CRC patients were identified and sequenced (Engine, Berlin, Germany). Annotated proteins predominantly or exclusively immunoreactive with autoantibodies from CRC patients were selected for the CRC protein array. Tumor‐associated antigens identified by IgG autoantibodies were shown to also elicit IgM responses in cancer patients by ELISA.24 In addition, naturally occurring antigens immunoreactive in most of the CRC patients and non‐cancer controls were chosen as positive control antigens and added to the CRC protein array. The CRC protein arrays were custom‐made and QC‐certified by Engine GmbH, Germany, by availing of the hEx1 human recombinant protein expression library.87 A total of 720 E. coli expression clones encoding the total of 492 proteins were spotted in duplicates and induced on PVDF membranes in the presence of isopropyl β‐d‐1‐thiogalactopyranoside (IPTG) allowing overexpression of the His6‐tagged fusion proteins. The CRC protein array contains full‐length as well as shorter‐length cDNA clones representing full‐length as well as partial proteins. Clones expressing identical proteins at different cDNA inserts were spotted on the array separately.
Serum screening
To characterise IgG, IgM and IgA antibodies in patient sera, CRC proteins arrays were prepared with minor modifications as previously described.16 Briefly, all serum samples and detection antibodies were preabsorbed for 4 h in blocking solution on PVDF sheets coated with E. coli containing an empty expression vector to remove cross‐reactive background antibodies. Each CRC protein array was assigned to a particular patient or control sample and incubated with preabsorbed serum at 1:200 dilution overnight at room temperature. The arrays were washed in Tris buffer and incubated with rabbit anti‐human IgM antibody (SAB3701400; Sigma‐Aldrich, St Louis, MO, USA) at 1:1000 dilution followed by a secondary goat anti‐rabbit‐680 antibody (5366; Cell Signaling Technology, Danvers, MA, USA) at 1:25 000 dilution. The arrays were scanned at 700 nm using an Odyssey CLx imaging system (LI‐COR Biosciences, Lincoln, NE, USA) to identify IgM binding. The protein arrays were stripped with pre‐warmed stripping solution (500 mm Tris–HCl, SDS solution 20% (w/v) 100 mL, 0.07% (v/v) 2‐mercaptoethanol) and re‐probed for IgA detection with a goat anti‐human IgA antibody (I0884; Sigma‐Aldrich) at 1:1000 dilution followed by a secondary donkey IRDye680 anti‐goat antibody (925‐68074; LI‐COR Biosciences) at 1:25 000 dilution and scanned using Odyssey CLx at 700 nm. At this point, protein arrays were stripped again and re‐probed for IgG detection. To identify IgG antibodies in sera, the stripped protein arrays were incubated with mouse anti‐human IgG (I6260, Clone GG‐7; Sigma‐Aldrich) at 1:5000 dilution followed by alkaline phosphatase (AP)‐conjugated goat anti‐mouse IgG‐AP antibody (A1418; Sigma‐Aldrich) at 1:5000 dilution and AttoPhos substrate (S1000; Promega, Madison, WI, USA). The arrays were scanned using GE Typhoon FLA 9000 Gel Imaging Scanner (GE Healthcare, Chicago, IL, USA).
Protein array analysis
High‐resolution images of CRC protein arrays were analysed using Visual Grid (GPC Biotech, Martinsried, Germany) as previously described.16 Briefly, each protein is present in duplicate in a specific pattern on the array; only proteins showing distinct signals in both positions were defined as positive (Figure 1). Unique IgG, IgM and IgA antibody signatures were generated on the CRC protein arrays for each subject and were then compared between cancer and non‐cancer control groups. Non‐specific secondary antibody binding was identified in serum‐free experiments and excluded as previously described.88
Statistical analyses
We applied a two‐step approach to select a differential autoantibody marker combination. To find biomarkers with high diagnostic potentials, IgG, IgM and IgA positives identified on the protein array were ranked according to their frequency of occurrence in cancer patients and non‐cancer controls. Thus, biomarkers with a high frequency of occurrence in cancer and low frequency in non‐cancer controls were compiled. Markers with the most significant association with the cancer group compared to the non‐cancer control group were then selected. Next, we applied binary logistic regression to the selected IgG, IgM and IgA autoantibody markers to create predicted probability values. A receiver operating characteristics (ROC) curve was constructed based on the predicted probability scores to evaluate the performance of the combinatorial biomarker panel. At least one positive marker evaluation was required to assign an individual to the CRC group. The predicted probability values were evaluated to determine optimal cut‐off points for prediction, in this case, maximum sensitivity and specificity. Five‐year survival analysis was done using the Kaplan–Meier method. Patients with follow‐up information over 5 years were censored at year 5 post‐diagnosis. Univariate and multivariate Cox regression analyses for age, sex, differentiation and tumor, node, metastasis (TNM) stages were conducted using the Cox proportional hazard regression model to investigate the impact of the variables on overall patient survival. All statistical analyses were conducted in SPSS 27 (SPSS, Chicago, IL, USA), GraphPad Prism 8.3.1 (GraphPad Software, San Diego, CA, USA) and MS Excel. All the findings were considered statistically significant at *P < 0.05, **P < 0.01 and ***P < 0.001.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
Md Saiful Islam Roney: Data curation; Formal analysis; Investigation; Methodology; Software; Writing‐original draft; Writing‐review & editing. Catharine Lanagan: Data curation; Investigation; Writing‐review & editing. Yong Hua Sheng: Investigation; Methodology. Karen Lawler: Formal analysis; Writing‐review & editing. Christopher Schmidt: Data curation. Nam‐Trung Nguyen: Writing‐review & editing. Jakob Begun: Resources. Gregor Stefan Kijanka: Conceptualization; Data curation; Formal analysis; Funding acquisition; Investigation; Methodology; Project administration; Resources; Software; Supervision; Writing‐original draft; Writing‐review & editing.
Supporting information
Acknowledgments
This study was supported by the Mater Foundation Research Fellowship and Griffith University Retention of High Performing Research Fellows Research Grant to GSK. MSR received the Research Training Program (RTP) Scholarship from the University of Queensland (UQ) and Frank Clair Scholarship from Mater Research Institute‐UQ. Odyssey CLx was obtained with the support of The Lions Prostate Cancer Research Project Grant, Lions Club to GSK. The Translational Research Institute (TRI) is supported by a grant from the Australian Government. The Victorian Cancer Biobank through the Cancer Council Victoria as Lead Agency is supported by the Victorian Government through the Victorian Cancer Agency—a business unit of the Department of Health and Human Services.
References
- 1.Sung H, Ferlay J, Siegel RLet al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021; 71: 209–249. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020; 70: 7–30. [DOI] [PubMed] [Google Scholar]
- 3.Mozdiak E, Wicaksono AN, Covington JA, Arasaradnam RP. Colorectal cancer and adenoma screening using urinary volatile organic compound (VOC) detection: early results from a single‐centre bowel screening population (UK BCSP). Tech Coloproctol 2019; 23: 343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chido‐Amajuoyi OG, Sharma A, Talluri R, Tami‐Maury I, Shete S. Physician‐office vs home uptake of colorectal cancer screening using FOBT/FIT among screening‐eligible US adults. Cancer Med 2019; 8: 7408–7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahlquist DA. Universal cancer screening: revolutionary, rational, and realizable. NPJ Precis Oncol 2018; 2: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siegel RL, Miller KD, Goding Sauer Aet al. Colorectal cancer statistics, 2020. CA Cancer J Clin 2020; 70: 145–164. [DOI] [PubMed] [Google Scholar]
- 7.Shah R, Jones E, Vidart V, Kuppen PJ, Conti JA, Francis NK. Biomarkers for early detection of colorectal cancer and polyps: systematic review. Cancer Epidemiol Biomarkers Prev 2014; 23: 1712–1728. [DOI] [PubMed] [Google Scholar]
- 8.Anderson KS, LaBaer J. The sentinel within: exploiting the immune system for cancer biomarkers. J Proteome Res 2005; 4: 1123–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaenker P, Gray ES, Ziman MR. Autoantibody production in cancer‐the humoral immune response toward autologous antigens in cancer patients. Autoimmun Rev 2016; 15: 477–483. [DOI] [PubMed] [Google Scholar]
- 10.Caron M, Choquet‐Kastylevsky G, Joubert‐Caron R. Cancer immunomics using autoantibody signatures for biomarker discovery. Mol Cell Proteomics 2007; 6: 1115–1122. [DOI] [PubMed] [Google Scholar]
- 11.Pedersen JW, Gentry‐Maharaj A, Fourkala E‐Oet al. Early detection of cancer in the general population: a blinded case‐control study of p53 autoantibodies in colorectal cancer. Br J Cancer 2013; 108: 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatterjee M, Mohapatra S, Ionan Aet al. Diagnostic markers of ovarian cancer by high‐throughput antigen cloning and detection on arrays. Cancer Res 2006; 66: 1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyle P, Chapman CJ, Holdenrieder Set al. Clinical validation of an autoantibody test for lung cancer. Ann Oncol 2011; 22: 383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Yu J, Sreekumar Aet al. Autoantibody signatures in prostate cancer. N Engl J Med 2005; 353: 1224–1235. [DOI] [PubMed] [Google Scholar]
- 15.Chapman C, Murray A, Chakrabarti Jet al. Autoantibodies in breast cancer: their use as an aid to early diagnosis. Ann Oncol 2007; 18: 868–873. [DOI] [PubMed] [Google Scholar]
- 16.Kijanka G, Hector S, Kay EWet al. Human IgG antibody profiles differentiate between symptomatic patients with and without colorectal cancer. Gut 2010; 59: 69–78. [DOI] [PubMed] [Google Scholar]
- 17.Ushigome M, Nabeya Y, Soda Het al. Multi‐panel assay of serum autoantibodies in colorectal cancer. Int J Clin Oncol 2018; 23: 917–923. [DOI] [PubMed] [Google Scholar]
- 18.Zayakin P, Ancāns G, Siliņa Ket al. Tumor‐associated autoantibody signature for the early detection of gastric cancer. Int J Cancer 2013; 132: 137–147. [DOI] [PubMed] [Google Scholar]
- 19.Zaenker P, Lo J, Pearce Ret al. A diagnostic autoantibody signature for primary cutaneous melanoma. Oncotarget 2018; 9: 30539–30551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janeway CA Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol 1989; 54: 1–13. [DOI] [PubMed] [Google Scholar]
- 21.Ben‐Aissa‐Fennira F, Ben Ammar‐El Gaaied A, Bouguerra A, Dellagi K. IgM antibodies to P1 cytoadhesin of Mycoplasma pneumoniae are part of the natural antibody repertoire expressed early in life. Immunol Lett 1998; 63: 59–62. [DOI] [PubMed] [Google Scholar]
- 22.Ochsenbein AF, Fehr T, Lutz Cet al. Control of early viral and bacterial distribution and disease by natural antibodies. Science 1999; 286: 2156–2159. [DOI] [PubMed] [Google Scholar]
- 23.Vollmers HP, Brandlein S. The, “early birds”: natural IgM antibodies and immune surveillance. Histol Histopathol 2005; 20: 927–937. [DOI] [PubMed] [Google Scholar]
- 24.Fitzgerald S, O'Reilly J‐A, Wilson Eet al. Measurement of the IgM and IgG autoantibody immune responses in human serum has high predictive value for the presence of colorectal cancer. Clin Colorectal Cancer 2019; 18: e53–e60. [DOI] [PubMed] [Google Scholar]
- 25.Vollmers HP, Brandlein S. Natural antibodies and cancer. N Biotechnol 2009; 25: 294–298. [DOI] [PubMed] [Google Scholar]
- 26.Brandlein S, Pohle T, Ruoff N, Wozniak E, Muller‐Hermelink HK, Vollmers HP. Natural IgM antibodies and immunosurveillance mechanisms against epithelial cancer cells in humans. Cancer Res 2003; 63: 7995–8005. [PubMed] [Google Scholar]
- 27.Racine R, McLaughlin M, Jones DDet al. IgM production by bone marrow plasmablasts contributes to long‐term protection against intracellular bacterial infection. J Immunol 2011; 186: 1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pabst O. New concepts in the generation and functions of IgA. Nat Rev Immunol 2012; 12: 821–832. [DOI] [PubMed] [Google Scholar]
- 29.Palm N, de Zoete M, Cullen Tet al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014; 158: 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baseler MW, Maxim PE, Veltri RW. Circulating IgA immune complexes in head and neck cancer, nasopharyngeal carcinoma, lung cancer, and colon cancer. Cancer 1987; 59: 1727–1731. [DOI] [PubMed] [Google Scholar]
- 31.Staff C, Magnusson CGM, Hojjat‐Farsangi Met al. Induction of IgM, IgA and IgE antibodies in colorectal cancer patients vaccinated with a recombinant CEA protein. J Clin Immunol 2012; 32: 855–865. [DOI] [PubMed] [Google Scholar]
- 32.Yoneyama K, Shibata R, Igarashi Aet al. Proteomic identification of dihydrolipoamide dehydrogenase as a target of autoantibodies in patients with endometrial cancer. Anticancer Res 2014; 34: 5021–5027. [PubMed] [Google Scholar]
- 33.Erić‐Nikolić A, Milovanović Z, Sánchez Det al. Overexpression of calreticulin in malignant and benign breast tumors: relationship with humoral immunity. Oncology 2012; 82: 48–55. [DOI] [PubMed] [Google Scholar]
- 34.Pan J, Yu L, Wu Qet al. Integration of IgA and IgG autoantigens improves performance of biomarker panels for early diagnosis of lung cancer. Mol Cell Proteomics 2020; 19: 490–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grelle G, Kostka S, Otto Aet al. Identification of VCP/p97, carboxyl terminus of Hsp70‐interacting protein (CHIP), and amphiphysin II interaction partners using membrane‐based human proteome arrays. Mol Cell Proteomics 2006; 5: 234–244. [DOI] [PubMed] [Google Scholar]
- 36.Kijanka G, IpCho S, Baars Set al. Rapid characterization of binding specificity and cross‐reactivity of antibodies using recombinant human protein arrays. J Immunol Methods 2009; 340: 132–137. [DOI] [PubMed] [Google Scholar]
- 37.Larkin D, Murphy D, Reilly DFet al. ICln, a novel Integrin αIIbβ3‐associated protein, functionally regulates platelet activation. J Biol Chem 2004; 279: 27286–27293. [DOI] [PubMed] [Google Scholar]
- 38.Kijanka G, Murphy D. Protein arrays as tools for serum autoantibody marker discovery in cancer. J Proteomics 2009; 72: 936–944. [DOI] [PubMed] [Google Scholar]
- 39.Casiano CA, Mediavilla‐Varela M, Tan EM. Tumor‐associated antigen arrays for the serological diagnosis of cancer. Mol Cell Proteomics 2006; 5: 1745–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hudson ME, Pozdnyakova I, Haines K, Mor G, Snyder M. Identification of differentially expressed proteins in ovarian cancer using high‐density protein microarrays. Proc Natl Acad Sci USA 2007; 104: 17494–17499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lenner P, Wiklund F, Emdin SOet al. Serum antibodies against p53 in relation to cancer risk and prognosis in breast cancer: a population‐based epidemiological study. Br J Cancer 1999; 79: 927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodell V, Salazar LG, Urban Net al. Antibody immunity to the p53 oncogenic protein is a prognostic indicator in ovarian cancer. J Clin Oncol 2006; 24: 762–768. [DOI] [PubMed] [Google Scholar]
- 43.Lai CL, Tsai CM, Tsai TTet al. Presence of serum anti‐p53 antibodies is associated with pleural effusion and poor prognosis in lung cancer patients. Clin Cancer Res 1998; 4: 3025–3030. [PubMed] [Google Scholar]
- 44.Fitzgerald S, Sheehan KM, Espina Vet al. High CerS5 expression levels associate with reduced patient survival and transition from apoptotic to autophagy signalling pathways in colorectal cancer. J Pathol Clin Res 2015; 1: 54–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Reilly J‐A, Fitzgerald J, Fitzgerald Set al. Diagnostic potential of zinc finger protein‐specific autoantibodies and associated linear B‐cell epitopes in colorectal cancer. PLoS One 2015; 10: e0123469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soussi T. p53 Antibodies in the sera of patients with various types of cancer: a review. Cancer Res 2000; 60: 1777–1788. [PubMed] [Google Scholar]
- 47.Hsiue EH, Wright KM, Douglass Jet al. Targeting a neoantigen derived from a common TP53 mutation. Science 2021; 371: eabc8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inagaki H, Matsushima Y, Nakamura K, Ohshima M, Kadowaki T, Kitagawa Y. A large DNA‐binding nuclear protein with RNA recognition motif and serine/arginine‐rich domain. J Biol Chem 1996; 271: 12525–12531. [DOI] [PubMed] [Google Scholar]
- 49.Eichmuller S, Usener D, Dummer R, Stein A, Thiel D, Schadendorf D. Serological detection of cutaneous T‐cell lymphoma‐associated antigens. Proc Natl Acad Sci USA 2001; 98: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeda A, Shimada H, Nakajima Ket al. Serum p53 antibody as a useful marker for monitoring of treatment of superficial colorectal adenocarcinoma after endoscopic resection. Int J Clin Oncol 2001; 6: 45–49. [DOI] [PubMed] [Google Scholar]
- 51.Litvak DA, Gupta RK, Yee R, Wanek LA, Ye W, Morton DL. Endogenous immune response to early‐ and intermediate‐stage melanoma is correlated with outcomes and is independent of locoregional relapse and standard prognostic factors. J Am Coll Surg 2004; 198: 27–35. [DOI] [PubMed] [Google Scholar]
- 52.Ushigome M, Shimada H, Miura Yet al. Changing pattern of tumor markers in recurrent colorectal cancer patients before surgery to recurrence: serum p53 antibodies, CA19‐9 and CEA. Int J Clin Oncol 2020; 25: 622–632. [DOI] [PubMed] [Google Scholar]
- 53.Fässler M, Diem S, Mangana Jet al. Antibodies as biomarker candidates for response and survival to checkpoint inhibitors in melanoma patients. J Immunother Cancer 2019; 7: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pallasch CP, Struss A‐K, Munnia Aet al. Autoantibodies against GLEA2 and PHF3 in glioblastoma: tumor‐associated autoantibodies correlated with prolonged survival. Int J Cancer 2005; 117: 456–459. [DOI] [PubMed] [Google Scholar]
- 55.Kurtenkov O, Klaamas K, Rittenhouse‐Olson Ket al. IgG immune response to tumor‐associated carbohydrate antigens (TF, Tn, αGal) in patients with breast cancer: impact of neoadjuvant chemotherapy and relation to the survival. Exp Oncol 2005; 27: 136–140. [PubMed] [Google Scholar]
- 56.Dong J, Zeng B‐H, Xu L‐Het al. Anti‐CDC25B autoantibody predicts poor prognosis in patients with advanced esophageal squamous cell carcinoma. J Transl Med 2010; 8: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Albanopoulos K, Armakolas A, Konstadoulakis MMet al. Prognostic significance of circulating antibodies against carcinoembryonic antigen (anti‐CEA) in patients with colon cancer. Am J Gastroenterol 2000; 95: 1056–1061. [DOI] [PubMed] [Google Scholar]
- 58.Morad SA, Cabot MC. Ceramide‐orchestrated signalling in cancer cells. Nat Rev Cancer 2013; 13: 51–65. [DOI] [PubMed] [Google Scholar]
- 59.Brachtendorf S, Wanger RA, Birod Ket al. Chemosensitivity of human colon cancer cells is influenced by a p53‐dependent enhancement of ceramide synthase 5 and induction of autophagy. Biochim Biophys Acta Mol Cell Biol Lipids 2018; 1863: 1214–1227. [DOI] [PubMed] [Google Scholar]
- 60.Mojakgomo R, Mbita Z, Dlamini Z. Linking the ceramide synthases (CerSs) 4 and 5 with apoptosis, endometrial and colon cancers. Exp Mol Pathol 2015; 98: 585–592. [DOI] [PubMed] [Google Scholar]
- 61.Novgorodov SA, Szulc ZM, Luberto Cet al. Positively charged ceramide is a potent inducer of mitochondrial permeabilization. J Biol Chem 2005; 280: 16096–16105. [DOI] [PubMed] [Google Scholar]
- 62.Levy M, Futerman AH. Mammalian ceramide synthases. IUBMB Life 2010; 62: 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mullen TD, Hannun YA, Obeid LM. Ceramide synthases at the centre of sphingolipid metabolism and biology. Biochem J 2012; 441: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wegner M‐S, Wanger RA, Oertel Set al. Ceramide synthases CerS4 and CerS5 are upregulated by 17β‐estradiol and GPER1 via AP‐1 in human breast cancer cells. Biochem Pharmacol 2014; 92: 577–589. [DOI] [PubMed] [Google Scholar]
- 65.Qian H, Deng J, Lu Cet al. Ceramide synthases: insights into the expression and prognosis of lung cancer. Exp Lung Res 2021; 47: 37–53. [DOI] [PubMed] [Google Scholar]
- 66.Mange A, Lacombe J, Bascoul‐Mollevi Cet al. Serum autoantibody signature of ductal carcinoma in situ progression to invasive breast cancer. Clin Cancer Res 2012; 18: 1992–2000. [DOI] [PubMed] [Google Scholar]
- 67.Cohen JD, Javed AA, Thoburn Cet al. Combined circulating tumor DNA and protein biomarker‐based liquid biopsy for the earlier detection of pancreatic cancers. Proc Natl Acad Sci USA 2017; 114: 10202–10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cohen JD, Li LU, Wang Yet al. Detection and localization of surgically resectable cancers with a multi‐analyte blood test. Science 2018; 359: 926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet 2014; 383: 1490–1502. [DOI] [PubMed] [Google Scholar]
- 70.Crosby D, Lyons N, Greenwood Eet al. A roadmap for the early detection and diagnosis of cancer. Lancet Oncol 2020; 21: 1397–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Benson AB, Desch CE, Flynn PJet al. 2000 update of American Society of Clinical Oncology colorectal cancer surveillance guidelines. J Clin Oncol 2000; 18: 3586–3588. [DOI] [PubMed] [Google Scholar]
- 72.Chao M, Gibbs P. Caution is required before recommending routine carcinoembryonic antigen and imaging follow‐up for patients with early‐stage colon cancer. J Clin Oncol 2009; 27: e279–e280. [DOI] [PubMed] [Google Scholar]
- 73.Wang H, Li X, Zhou D, Huang J. Autoantibodies as biomarkers for colorectal cancer: a systematic review, meta‐analysis, and bioinformatics analysis. Int J Biol Markers 2019; 34: 334–347. [DOI] [PubMed] [Google Scholar]
- 74.Pedchenko T, Mernaugh R, Parekh D, Li M, Massion PP. Early detection of NSCLC with scFv selected against IgM autoantibody. PLoS One 2013; 8: e60934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.De Chiara L, Páez de la Cadena M, Rodríguez‐Berrocal Jet al. CD26‐related serum biomarkers: sCD26 protein, DPP4 activity, and anti‐CD26 isotype levels in a colorectal cancer‐screening context. Dis Markers 2020; 2020: 4347936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vernon SW. Participation in colorectal cancer screening: a review. J Natl Cancer Inst 1997; 89: 1406–1422. [DOI] [PubMed] [Google Scholar]
- 77.Bond JH. Fecal occult blood test screening for colorectal cancer. Gastrointest Endosc Clin N Am 2002; 12: 11–21. [DOI] [PubMed] [Google Scholar]
- 78.Elsafi SH, Alqahtani NI, Zakary NY, Al Zahrani EM. The sensitivity, specificity, predictive values, and likelihood ratios of fecal occult blood test for the detection of colorectal cancer in hospital settings. Clin Exp Gastroenterol 2015; 8: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kalager M, Wieszczy P, Lansdorp‐Vogelaar I, Corley DA, Bretthauer M, Kaminski MF. Overdiagnosis in colorectal cancer screening: time to acknowledge a blind spot. Gastroenterology 2018; 155: 592–595. [DOI] [PubMed] [Google Scholar]
- 80.Christou A, Thompson SC. Colorectal cancer screening knowledge, attitudes and behavioural intention among Indigenous Western Australians. BMC Public Health 2012; 12: 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Taber JM, Aspinwall LG, Heichman KA, Kinney AY. Preferences for blood‐based colon cancer screening differ by race/ethnicity. Am J Health Behav 2014; 38: 351–361. [DOI] [PubMed] [Google Scholar]
- 82.Sanduleanu S, Masclee AM, Meijer GA. Interval cancers after colonoscopy‐insights and recommendations. Nat Rev Gastroenterol Hepatol 2012; 9: 550–554. [DOI] [PubMed] [Google Scholar]
- 83.Singh S, Singh PP, Murad MH, Singh H, Samadder NJ. Prevalence, risk factors, and outcomes of interval colorectal cancers: a systematic review and meta‐analysis. Am J Gastroenterol 2014; 109: 1375–1389. [DOI] [PubMed] [Google Scholar]
- 84.Samadder NJ, Curtin K, Tuohy TMFet al. Characteristics of missed or interval colorectal cancer and patient survival: a population‐based study. Gastroenterology 2014; 146: 950–960. [DOI] [PubMed] [Google Scholar]
- 85.Henderson MC, Silver M, Tran Qet al. A noninvasive blood‐based combinatorial proteomic biomarker assay to detect breast cancer in women over age 50 with BI‐RADS 3, 4, or 5 assessment. Clin Cancer Res 2019; 25: 142–149. [DOI] [PubMed] [Google Scholar]
- 86.Sobin LH, Wittekind CL (eds). TNM Classification of Malignant Tumors, 6th edn. New York, NY: John Wiley & Sons Inc, 2002. [Google Scholar]
- 87.Bussow K, Cahill D, Nietfeld Wet al. A method for global protein expression and antibody screening on high‐density filters of an arrayed cDNA library. Nucleic Acids Res 1998; 26: 5007–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lemass D, O'Kennedy R, Kijanka GS. Referencing cross‐reactivity of detection antibodies for protein array experiments. F1000Res 2016; 5: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


