Abstract
An open-source library of implementations for deep-learning based image segmentation and radiotherapy outcomes models is presented in this work. As oncology treatment planning becomes increasingly driven by automation, such a library of model implementations is crucial to (i) validate existing models on datasets collected at different institutions, (ii) automate segmentation, (iii) create ensembles for improving performance and (iv) incorporate validated models in the clinical workflow. The library was developed with the Computational Environment for Radiological Research (CERR) software platform. Centralizing model implementations in CERR builds upon its rich set of radiotherapy and radiomics tools and caters to the world-wide user base. CERR provides well-validated feature extraction pipelines for radiotherapy dosimetry and radiomics with fine control over the calculation settings, allowing users to select the appropriate parameters used in model derivation. Models for automatic image segmentation are distributed via containers, with seamless i/o to and from CERR. Container technology allow segmentation models to be deployed with a variety of scientific computing architectures. The library includes implementations of popular radiotherapy models outlined in the Quantitative Analysis of Normal Tissue Effects in the Clinic effort and recently published literature. Radiomics models include features from the Image Biomarker Standardization Initiative and application-specific features found to be relevant across multiple sites and image modalities. Deep learning-based image segmentation models include state-of-the-art networks such as DeepLab and other problem-specific architectures. The source code is distributed at https://www.github.com/cerr/CERR under the GNU-GPL copyleft with additional restrictions on clinical and commercial use and provision to dual license in future.
Keywords: image segmentation, deep-learning, radiomics, radiotherapy outcomes, normal tissue complication, tumor control, model implementations, library
Introduction:
Oncology is increasingly driven by automation in advanced image analysis and prognostic modeling. In radiotherapy treatment planning automation is crucial for segmenting lesions and organs at risk. Segmentations, along with images and radiotherapy plans, are fed into prediction models to estimate prognosis resulting from the planned treatment. The treatment, in turn, can then be tailored based on the estimated prognosis. Software libraries for deriving radiomics [1] and radiotherapy features as well as outcomes models have grown rapidly. Similarly, automatic image segmentation has seen a great deal of recent progress with deep learning [2]. Segmentation models involve various dependencies related to the choice of the framework, whereas predictive and prognostic models [3] are affected by the choice of feature calculation settings, making it cumbersome to use the published signatures on new datasets. In the present study, we address the issue of variability in model implementations by creating an open-source, validated and comprehensive library of image-segmentation, radiotherapy and radiomics-based outcomes model implementations. The library of model implementations developed in the work herein will be useful to (i) validate existing models on datasets collected at different institutions, (ii) automate segmentation, making it less prone to inter-observer variability, (iii) create ensembles from various models in the library and in-turn improving model performance and (iv) simplify incorporating the models in the clinical workflow.
There have been various efforts to deploy implementations of predictive and prognostic models. The focus has mostly been on developing online tools useful to patients and physicians. These implementations are limited to web interfaces and don’t support batch evaluations on datasets for research. The ability to batch evaluate models is important to validate them on datasets acquired at multiple institutions. For example, DNAmito (https://predictcancer.org) provides online tools and an app that supports 25 cancer models for different sites. These models incorporate patient and radiotherapy treatment characteristics. National Cancer Registration and Analysis Service [4] (https://breast.predict.nhs.uk/) provides an online tool to predict survival rate for early invasive breast cancer treatment. Memorial Sloan Kettering Cancer Center [5, 6] (https://www.mskcc.org/nomograms) provides online tools to predict cancer outcomes based on characteristics of the patient and their disease, for example nomogram to calculate the risk of bladder cancer recurrence after radical cystectomy and pelvic lymph node dissection. There has been a lot of recent progress in the development of toolkits for storing and deploying deep learning segmentation models. These include traditional multi-atlas-based segmentation as well as state of the art deep learning-based models. DeepInfer [7] currently provides segmentation models for prostate gland, biopsy needle trajectory and tip detection in intraoperative MRIs, and brain white matter hyperintensities. DeepInfer can also be invoked from 3DSlicer [8]. NiftyNET [9], a consortium of research organizations, provides a TensorFlow™-based open-source convolutional neural networks (CNN) platform. It provides the implementation of networks such as HighRes3DNet, 3D U-net, V-net in the DeepMedic software [10] which can be used to train new models as well as share pre-trained models. Plastimatch [11] is an open-source software primarily used for image registration and provides a module for multi-atlas segmentation.
In this work, we implemented a research-friendly library of models that is comprehensive in terms of the variety of models, viz. image segmentation, radiotherapy, and radiomics. The library was developed with the Computational Environment for Radiological Research [12] software platform which has been widely used for outcomes modeling and prototyping algorithms useful in radiotherapy treatment planning. The library of model implementations will provide a reproducible pipeline including image segmentation, estimated prediction and prognosis.
Methods:
The library of model implementations was developed as a module within the MATLAB [13] based CERR software platform, allowing users to readily access and plug-in models. The library supports the following three classes of models: (1) Segmentation models based on deep learning, (2) Radiomics models based on imaging biomarkers, and (3) Radiotherapy models based on dose-volume histograms and cell survival. The library builds on CERR’s data import/export, visualization and standardization tools for outcomes modeling. The following sections provide details about the architecture for deploying and using these three classes of models.
Image segmentation models
The architecture for implementation of image segmentation models was based on containerizing the models and their dependencies. The containers were designed to accept input images in Hierarchical Data Format (HDF5) file format and return the resulting segmented mask, again in HDF5 file format, thereby allowing models developed with a variety of frameworks to be readily plugged into the library. Currently available implementations of segmentation models use open-source frameworks like Tensorflow™ and PyTorch. Both, Singularity and Docker container technology allow users to securely bundle libraries such that they are compatible with a variety of scientific computing architectures. Singularity containers are well suited for high performance computing (HPC) environments and present various advantages including (i) checksums for making the software stacks reproducible (ii) compatibility with HPC systems and enterprise architectures (iii) software and data controls compliance for HIPAA and (iv) can be run by users without root privileges. Singularity containers are runnable on most flavors of Linux operating system and Mac OS X via “Singularity desktop”. Docker containers, on the other hand, are also runnable on Windows 10 operating system via “Docker desktop”. The implementations of deep learning segmentation models are provided in the form of both, Docker and Singularity containers, such that they can be readily used with different operating systems. The models within the containers can be run with or without using GPU. Figure 1 shows a schematic of invoking deep learning segmentation models from CERR through a Singularity container.
Figure 1:
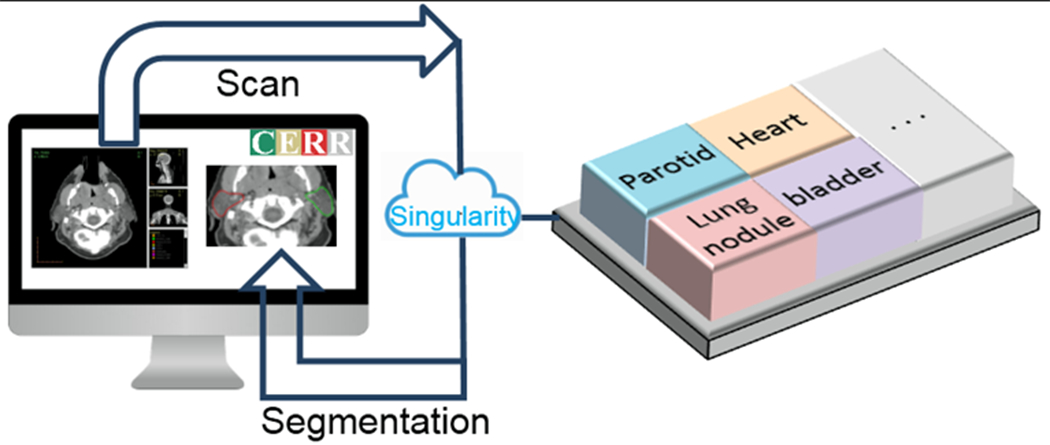
Schematic of invoking the container for deep-learning image segmentation model from CERR. CERR passes images in HDF5 format to the container and displays the resulting segmentation.
Segmentation models often require preprocessing of input images as well as post-processing of output segmentation. These pre and post-processing options are defined through model-specific configuration files. An important pre-processing option is the selection of the region of interest used for segmentation and must correspond to the one used while deriving the model. The options for automatically defining the region of interest include (i) cropping to structure bounds, (ii) cropping by a fixed amount from center, (iii) cropping to the patient’s outline, and (iv) cropping to the shoulder. The cropping operations can also be chained together via union and intersection operators so that the segmentation results can be used as priors for subsequent segmentations. Figure 2 shows examples of selecting regions of interest as inputs to segmentation models. Segmentation models derived on 2-D, 3-D and 2.5-D input images are supported. Various pre-processing filters are supported to populate different channels used by CNNs. Currently, the post-processing options allow filtering segments below a threshold area and limiting the number of connected components.
Figure 2:
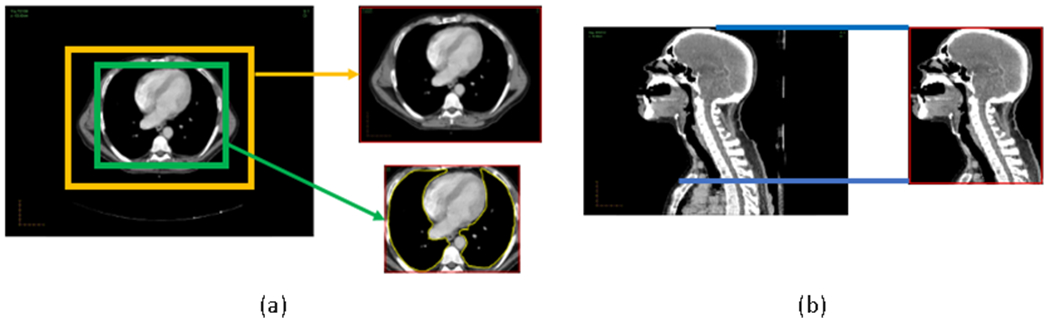
Options to select region of interest as an input to segmentation model. (a) cropping to structure bounds and by a fixed amount. (b) cropping to the shoulder.
The segmentation models within the library can also be invoked from CERR’ user interface. Figure 3 shows the selection of model and its parameters through the graphical user interface.
Figure 3:
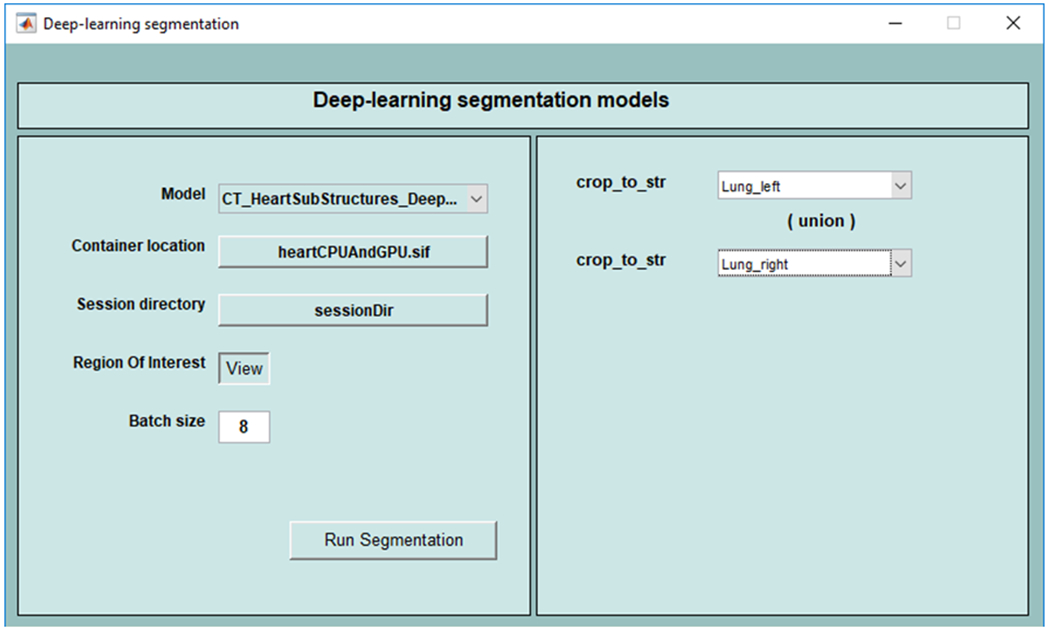
Graphical User Interface to invoke deep learning segmentation models from CERR Viewer. In this example, the union of left and right lungs is used as a prior to select the region of interest.
Parameters such as structure names to define the region of interest are selected using the interface.
In addition to deep-learning-based segmentation models, multi-atlas segmentation is supported. Atlas fusion methods such as majority-vote and STAPLE [14] are supported. Plastimatch is used for deformable image registration allowing users to choose from a variety of cost functions, optimizers, and registration algorithms. It allows users to easily compare state-of-the-art with traditional segmentation models. CERR provides metrics to evaluate the performance of auto-segmentation methods such as Dice coefficient, Hausdorff distance, and deviance of the resulting segmentation from the ground truth.
Radiotherapy models
Radiotherapy models involve modeling Normal Tissue Complication (NTCP) and Tumor Control probabilities (TCP). NTCP models are usually based on dose-volume histogram (DVH) characteristics whereas TCP models involve estimating cell survival from radiobiology. QUANTEC series of papers provides a comprehensive description of models for various treatment sites. CERR provides utility functions to import DICOM RT-DOSE and RT-PLAN objects as well as tools to apply linear-quadratic (LQ) corrections [15] for fraction size and to sum dose distributions from multiple courses of radiation. The library of radiotherapy models has access to the wide variety of DVH metrics in CERR such as minimum dose to hottest x percent volume (Dx), volume receiving at least x Gy of dose (Vx), mean of hottest x% volume (MOHx), mean of coldest x% volume (MOCx) and generalized equivalent dose (gEUD). Various types of NTCP models include Lyman-Kutcher-Burman (LKB) [16], linear, logistic, bi-exponential and the ability to correct for risk factors [17, 18]. TCP models include cell survival model based on the dominant lesion for the prostate [19] and Jeong et al’s model for lung based on dose and treatment schedule [20]. The models are defined via JavaScript Object Notation (JSON) files as shown in figure 4, allowing users to readily adjust the model specific parameters.
Figure 4:
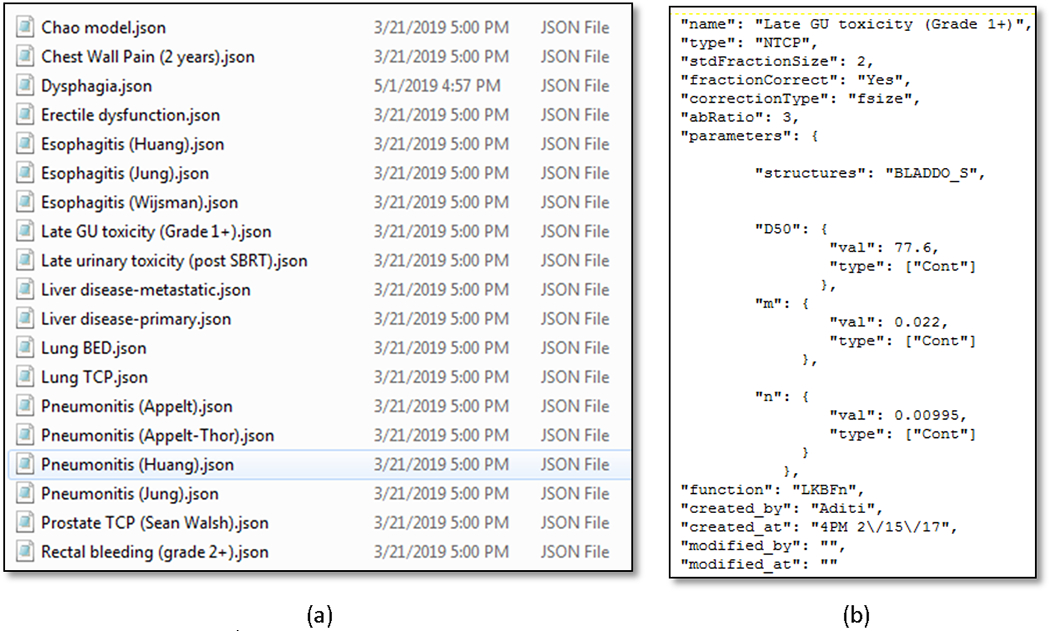
(a) JSON files to define models. (b) Parameters for a model are defined in the corresponding JSON file.
Radiomics models
Radiomics models require the derivation of image and morphological features. These include image histogram-based features as well as texture derived from gray-level co-occurrences, run-lengths, size-zones, gray level dependence, and gray tone difference matrices. CERR supports the calculation of a variety of flavors of these features using the radiomics extensions [21]. CERR’s data structure supports longitudinal images as well as multiple modalities such as CT, MR, PET, SPECT, and US. This allows for the deployment of models derived from multiple modalities and various time points, for example, Hypoxia prediction model from Crispen-Ortuzar et al [22] which used features from PET and CT scans. In addition to the Image Biomarker Standardization Initiative (IBSI) [23] defined radiomics features, CERR supports computation of modality-specific features such as Tofts model [24] parameters and non-model-based parameters such as time to half peak (TTHP) for Dynamic Contrast-Enhanced (DCE) MRI [25] and Intravoxel Incoherent Motion (IVIM) parameters for Diffusion Weighted Imaging (DWI). Parameter extraction is driven via configuration files in JSON format.
Quality assurance
Implemented models were validated by comparing results with those provided by the developers of these models. The results from the developers were available in the form of publications, test datasets or open source code. CERR’s DVH-based features used in radiotherapy models have been independently validated by comparing with commercial treatment planning software. Radiotherapy models were thoroughly validated by creating test sets with single-voxel structures and then comparing the results with manual calculations. Radiomics features in CERR are compliant with IBSI and validated with other popular libraries such as ITK (https://itk.org/) and PyRadiomics [26]. A test suite was developed, including test datasets for the implemented models and is run following each software update using Jenkins [27] continuous integration platform. This ensures accurate implementation, stability, and reproducibility with respect to software updates. The test suite along with test datasets is distributed along with CERR.
License
The codebase for implementations of models uses the GNU-GPL copyleft license (https://www.gnu.org/licenses/lgpl-3.0.en.html) to allow open-source distribution with additional restrictions. The license retains the ability to propagate any changes to the codebase back to the open-source community along with the following restrictions (i) No Clinical Use, (ii) No Commercial Use, and (iii) Dual Licensing which reserve the right to diverge and/or modify and/or expand the model implementations library to have a closed source/proprietary version along with the open source version in future. We would like to highlight that the library of implementations presented in this work is not approved by the U.S. Food and Drug Administration and should not be used to make clinical decisions for treating patients. The library merely provides implementations of the developed models, whereas the creators of models retain the copyright to their work.
Results:
CERR’s Model Implementation Library is distributed as an open-source, GNU-copyrighted software with additional restrictions along with the CERR platform. Documentation for the available models and their usage is available at https://github.com/cerr/CERR/wiki. Tables 1–3 list various models implemented for image segmentation, radiotherapy, and radiomics based outcomes prediction. Image segmentation models include the popular DeepLab [28] network for segmentation of prostate, heart sub structures, chewing and swallowing structures as well as specialized architectures such as MRRN [29] for segmentation of lung nodules and Self-attention networks for segmentation of head & neck organs at risk [30]. Radiotherapy and radiomics models from the published literature demonstrating satisfactory performance were included. Radiotherapy based models include the prediction of normal tissue complication resulting from lung, prostate and head & neck radiotherapy. Radiomics models include features that have been shown to be predictive in a variety of published studies. For example, the image features used in overall survival prediction model for lung cancer patients [31] have been shown to be important across multiple treatment sites and image types. Similarly, CoLIAGe [32] features have been shown to predict brain necrosis as well as breast cancer sub-types.
Table 1.
Image segmentation models
| Site | Modality | Organ/s | Model / Framework |
|---|---|---|---|
| Lung [34] | CT | Heart, heart sub-structures, pericardium, atria, ventricles | DeepLabV3+ / Pytorch |
| Lung [29] | CT | Tumor nodules | Incremental MRRN / Keras, Tensorflow™ |
| Prostate (Elguindi et al, under review) | MRI | Bladder, prostate and seminal vesicles (CTV), penile bulb, rectum, urethra and rectal spacer | DeepLabV3+, Tensorflow™-GPU |
| Head & Neck [36] | CT | Masseters (left, right), medial pterygoids (left, right), constrictor muscles, and larynx | DeepLabV3+ / Pytorch |
| Head & Neck [30] | CT | Parotid glands (left, right), Sub-mandibular glands (left, right), mandible and brain stem | Self-attention UNet / Pytorch |
Table 3.
Radiomics models
| Site | Endpoint | Features |
|---|---|---|
| Ovarian [48] | Survival for high grade Ovarian cancer | CluDiss (inter-tumor heterogeneity) |
| Breast, Brain [32] | (i) Radiation necrosis in T1-w MRI Brain, (ii) Subtype classification in DCE-MRI Breast | CoLlAGe (gradient orientations) |
| Head & Neck [22] | Hypoxia | P90 of FDG PET SUV, LRHGLE of CT |
| Lung, H&N [31] | Survival for NSCLC and HNSCC | Statistics Energy, Shape Compactness, Grey Level Nonuniformity, wavelet Grey Level Nonuniformity HLH |
One of the advantages of the library of implementations developed in this work is to be able to readily apply models to datasets at different institutions. It is demonstrated by applying the segmentation model for cardiac sub-structures [33, 34], derived at Memorial Sloan Kettering Cancer Center, to segment 490 scans from the RTOG 0617 [35] dataset at the University of Pennsylvania. Features extracted from these sub-structures will further be used for outcomes modeling.
Discussion:
The library of model implementations can be used to create ensembles for improving performance in addition to inter-institutional validation as well as collaboration. It can be used in developing tools to personalize treatment planning and to drive clinical segmentation. The following describes applications that make use of the library of model implementations.
Radiotherapy Outcomes Estimator
Radiotherapy Outcomes Estimator (ROE) is a tool for exploring the impact of dose scaling on TCP and NTCP in radiotherapy. Users define outcomes models and clinical constraints, following a simple pre-defined syntax. Inputs are specified in easily edited JSON files containing details of treatment protocols and outcomes models. Model files specify tumor and critical structures; associated outcomes models; model parameters including clinical risk factors (age, gender, stage etc.); and clinical constraints on outcomes. Predictive models supplied with ROE include Lyman-Kutcher-Burman [16], linear, biexponential, logistic, and TCP models for lung and prostate cancers. ROE supports multiple display modes including (1) NTCP vs. BED, (2) NTCP vs. TCP, and (3) TCP/NTCP dose responses as functions of the prescription dose (varied by fraction size or number) and indicates where the clinical limits are first violated. Users can modify the clinical and model variables to visualize the simultaneous impact on TCP/NTCP. Figure 5 shows the ROE graphical user interface.
Figure 5:
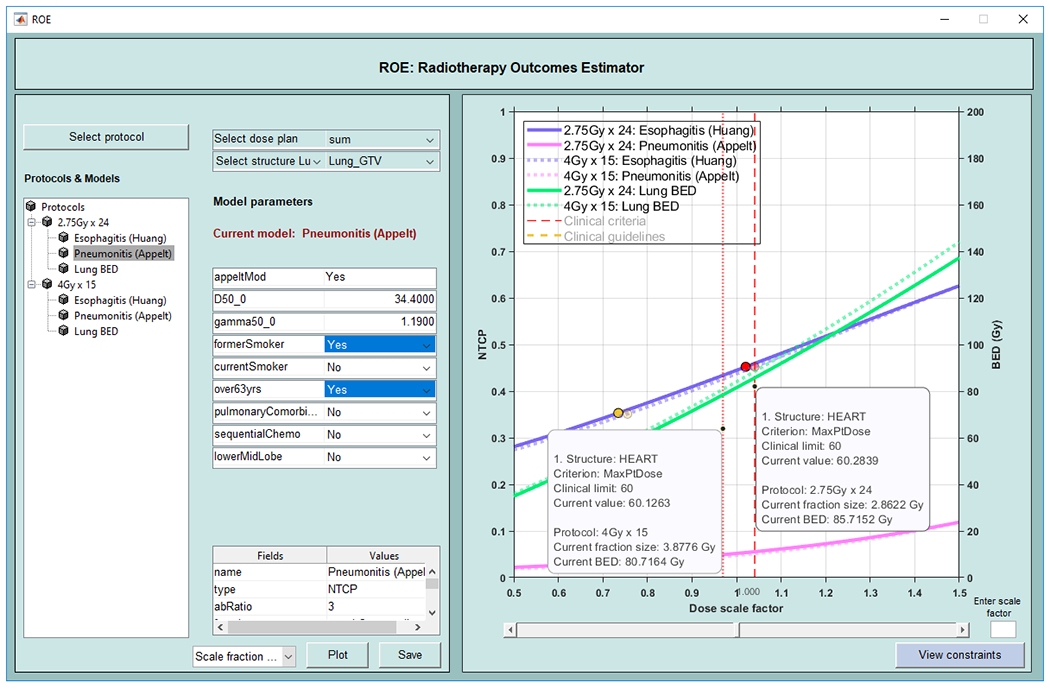
Radiotherapy Outcomes Explorer (ROE). Graphical user interface allows the selection of protocols which includes models and constraints. Additionally, users can input risk factors for the patient. NTCP curves for esophagitis and pneumonitis models and TCP curve for BED are shown in this example along with the points on the NTCP curves where the clinical limits are reached.
Ensemble of Voxel-wise Attributors
Ensemble of Voxel-wise Attributors (EVA) is an extensible pipeline developed at Memorial Sloan Kettering Cancer Center to deploy deep learning-based image segmentation algorithms into the clinic. It works in conjunction with MIM software (https://www.mimsoftware.com/) and uses segmentation models from the library presented in this work. EVA has been deployed in clinical use for segmentation of prostate structures [Elguindi et al, under review] and is currently undergoing testing for other sites. Figure 6 shows the workflow for EVA. Models from the library are invoked on the processing server and the resulting segmentation is archived in MIM Software.
Figure 6:
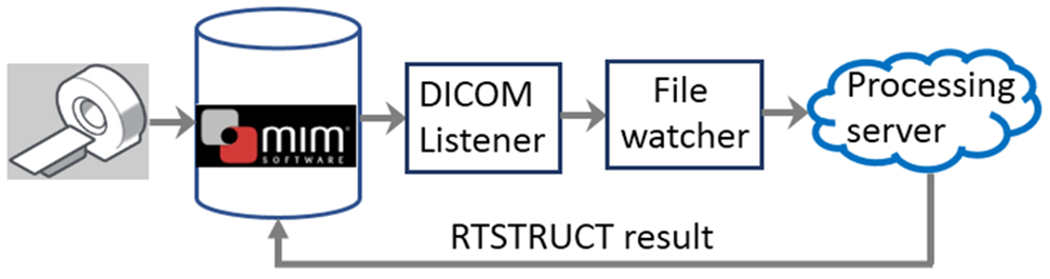
Schematic of EVA pipeline. Image transfer to MIM software triggers segmentation on the processing server and the resulting segmentation gets archived in MIM.
One of the limitations of using Singularity containers is the requirement of Linux operating system. For Mac OS, there is a beta version of “Singularity Desktop” which allows users to natively run Singularity containers. “Docker Desktop” runs as a native application by enabling virtualization only in Windows 10 operating system. Users using versions below Windows 10 are required to run containers in a virtual Linux environment for Windows. MATLAB users also have the option to purchase the Deep Learning toolbox which supports exchanging models with TensorFlow™ and PyTorch through the ONNX format. We plan to make implementations of segmentation models available in ONNX format and running them natively in MATLAB.
Conclusion:
A library of model implementations for segmentation of organs and risk and tumors and outcomes models was developed using the CERR platform. The library makes it efficient to use and deploy segmentation, radiotherapy and radiomics based models. It facilitates reproducible application of models and the development of automated pipelines starting from imaging and radiotherapy inputs to applying the final model. In the present study, it was demonstrated by sharing the segmentation model for cardiac sub-structures between the two institutions.
Table 2.
Radiotherapy models
| Site | Endpoint | Model |
|---|---|---|
| Prostate [37–40] | Rectal bleeding | LKB |
| Late urinary toxicity | ||
| Late urinary toxicity post SBRT | ||
| Lung[41] | Radiation-induced liver disease | LKB |
| Prostate[42] | Erectile dysfunction | Linear regression |
| Lung [43] | Esophagitis | Logistic regression |
| Lung[44] | Dysphagia | Logistic regression |
| Head & Neck[45] | Xerostomia | Bi-exponential |
| Lung[46] | Chest wall pain | Non-standard |
| Lung[17] | Pneumonitis | Logistic with correction for clinical risk factors |
| Lung[19] | Tumor control probability | Radiobiological |
| Prostate[47] | Tumor control probability | Logistic |
Acknowledgements
This research was partially funded by NIH grant 1R01CA198121 and NIH/NCI Cancer Center Support grant P30 CA008748.
References
- [1].Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44. [DOI] [PubMed] [Google Scholar]
- [3].Lughezzani G, Briganti A, Karakiewicz PI, Kattan MW, Montorsi F, Shariat SF, et al. Predictive and prognostic models in radical prostatectomy candidates: a critical analysis of the literature. Eur Urol. 2010;58:687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wishart GC, Azzato EM, Greenberg DC, Rashbass J, Kearins O, Lawrence G, et al. PREDICT: a new UK prognostic model that predicts survival following surgery for invasive breast cancer. Breast Cancer Res. 2010;12:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Van Zee KJ, Manasseh D-ME, Bevilacqua JLB, Boolbol SK, Fey JV, Tan LK, et al. A Nomogram for Predicting the Likelihood of Additional Nodal Metastases in Breast Cancer Patients With a Positive Sentinel Node Biopsy. Annals of Surgical Oncology. 2003;10:1140–51. [DOI] [PubMed] [Google Scholar]
- [6].Kattan MW, Vickers AJ, Yu C, Bianco FJ, Cronin AM, Eastham JA, et al. Preoperative and postoperative nomograms incorporating surgeon experience for clinically localized prostate cancer. Cancer. 2009;115:1005–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mehrtash A, Pesteie M, Hetherington J, Behringer PA, Kapur T, Wells WM 3rd, et al. DeepInfer: Open-Source Deep Learning Deployment Toolkit for Image-Guided Therapy. Proc SPIE Int Soc Opt Eng. 2017;10135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging. 2012;30:1323–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gibson E, Li W, Sudre C, Fidon L, Shakir DI, Wang G, et al. NiftyNet: a deep-learning platform for medical imaging. Comput Methods Programs Biomed. 2018;158:113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kamnitsas K, Ledig C, Newcombe VFJ, Simpson JP, Kane AD, Menon DK, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal. 2017;36:61–78. [DOI] [PubMed] [Google Scholar]
- [11].Zaffino P, Raudaschl P, Fritscher K, Sharp GC, Spadea MF. Technical Note: plastimatch mabs, an open source tool for automatic image segmentation. Med Phys. 2016;43:5155. [DOI] [PubMed] [Google Scholar]
- [12].Deasy JO, Blanco AI, Clark VH. CERR: a computational environment for radiotherapy research. Med Phys. 2003;30:979–85. [DOI] [PubMed] [Google Scholar]
- [13].MATLAB and Deep Learning Toolbox, The MathWorks, Inc., Natick, Massachusetts, United States. [Google Scholar]
- [14].Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging. 2004;23:903–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fowler JF. The linear-quadratic formula and progress in fractionated radiotherapy. Br J Radiol. 1989;62:679–94. [DOI] [PubMed] [Google Scholar]
- [16].Burman C, Kutcher GJ, Emami B, Goitein M. Fitting of normal tissue tolerance data to an analytic function. Int J Radiat Oncol Biol Phys. 1991;21:123–35. [DOI] [PubMed] [Google Scholar]
- [17].Appelt AL, Vogelius IR, Farr KP, Khalil AA, Bentzen SM. Towards individualized dose constraints: Adjusting the QUANTEC radiation pneumonitis model for clinical risk factors. Acta Oncologica. 2014;53:605–12. [DOI] [PubMed] [Google Scholar]
- [18].Thor M, Deasy J, Iyer A, Bendau E, Fontanella A, Apte A, et al. Toward personalized dose-prescription in locally advanced non-small cell lung cancer: Validation of published normal tissue complication probability models. Radiother Oncol. 2019;138:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Walsh S, van der Putten W. A TCP model for external beam treatment of intermediate-risk prostate cancer. Med Phys. 2013;40:031709. [DOI] [PubMed] [Google Scholar]
- [20].Jeong J, Oh JH, Sonke JJ, Belderbos J, Bradley JD, Fontanella AN, et al. Modeling the Cellular Response of Lung Cancer to Radiation Therapy for a Broad Range of Fractionation Schedules. Clin Cancer Res. 2017;23:5469–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Apte AP, Iyer A, Crispin-Ortuzar M, Pandya R, van Dijk LV, Spezi E, et al. Technical Note: Extension of CERR for computational radiomics: A comprehensive MATLAB platform for reproducible radiomics research. Med Phys. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Crispin-Ortuzar M, Apte A, Grkovski M, Oh JH, Lee NY, Schoder H, et al. Predicting hypoxia status using a combination of contrast-enhanced computed tomography and [(18)F]-Fluorodeoxyglucose positron emission tomography radiomics features. Radiother Oncol. 2018;127:36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zwanenburg A, Leger S, Vallières M, Lock S. Image biomarker standardisation initiative. ArXiv. 2016;1612.07003. [Google Scholar]
- [24].Tofts PS, Berkowitz B, Schnall MD. Quantitative analysis of dynamic Gd-DTPA enhancement in breast tumors using a permeability model. Magn Reson Med. 1995;33:564–8. [DOI] [PubMed] [Google Scholar]
- [25].Lee SH, Rimner A, Gelb E, Deasy JO, Hunt MA, Humm JL, et al. Correlation Between Tumor Metabolism and Semiquantitative Perfusion Magnetic Resonance Imaging Metrics in Non-Small Cell Lung Cancer. Int J Radiat Oncol Biol Phys. 2018;102:718–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017;77:e104–e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].https://jenkins.io/.2019.
- [28].Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans Pattern Anal Mach Intell. 2018;40:834–48. [DOI] [PubMed] [Google Scholar]
- [29].Jiang J, Hu YC, Liu CJ, Halpenny D, Hellmann MD, Deasy JO, et al. Multiple Resolution Residually Connected Feature Streams for Automatic Lung Tumor Segmentation From CT Images. IEEE Trans Med Imaging. 2019;38:134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jiang Jue ES, Um Hyemin, Berry Sean L., Veeraraghavan Harini. Local block-wise self attention for normal organ segmentation. ArXiv. 2019;abs/1909.05054. [Google Scholar]
- [31].Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Prasanna P, Tiwari P, Madabhushi A. Co-occurrence of Local Anisotropic Gradient Orientations (CoLlAGe): A new radiomics descriptor. Sci Rep. 2016;6:37241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hotca A, Thor M, Deasy JO, Rimner A. Dose to the cardio-pulmonary system and treatment-induced electrocardiogram abnormalities in locally advanced non-small cell lung cancer. Clin Transl Radiat Oncol. 2019;19:96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Haq R, Hotca A, Apte A, Rimner A, Deasy JO, Thor M. Cardio-Pulmonary Substructure Segmentation of CT Images Using Convolutional Neural Networks. Cham: Springer International Publishing; 2019. p. 162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Bradley JD, Paulus R, Komaki R, Masters G, Blumenschein G, Schild S, et al. Standard-dose versus high-dose conformal radiotherapy with concurrent and consolidation carboplatin plus paclitaxel with or without cetuximab for patients with stage IIIA or IIIB non-small-cell lung cancer (RTOG 0617): a randomised, two-by-two factorial phase 3 study. Lancet Oncol. 2015;16:187–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Iyer A, Thor M, Haq R, Deasy JO, Apte AP. Deep learning-based auto-segmentation of swallowing and chewing structures in CT. bioRxiv. 2019. [Google Scholar]
- [37].Michalski JM, Gay H, Jackson A, Tucker SL, Deasy JO. Radiation dose-volume effects in radiation-induced rectal injury. Int J Radiat Oncol Biol Phys. 2010;76:S123–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Cheung MR, Tucker SL, Dong L, de Crevoisier R, Lee AK, Frank S, et al. Investigation of bladder dose and volume factors influencing late urinary toxicity after external beam radiotherapy for prostate cancer. Int J Radiat Oncol Biol Phys. 2007;67:1059–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kole TP, Tong M, Wu B, Lei S, Obayomi-Davies O, Chen LN, et al. Late urinary toxicity modeling after stereotactic body radiotherapy (SBRT) in the definitive treatment of localized prostate cancer. Acta Oncol. 2016;55:52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Pan CC, Kavanagh BD, Dawson LA, Li XA, Das SK, Miften M, et al. Radiation-associated liver injury. Int J Radiat Oncol Biol Phys. 2010;76:S94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Roach M 3rd, Nam J, Gagliardi G, El Naqa I, Deasy JO, Marks LB. Radiation dose-volume effects and the penile bulb. Int J Radiat Oncol Biol Phys. 2010;76:S130–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Roach M 3rd, Chinn DM, Holland J, Clarke M. A pilot survey of sexual function and quality of life following 3D conformal radiotherapy for clinically localized prostate cancer. Int J Radiat Oncol Biol Phys. 1996;35:869–74. [DOI] [PubMed] [Google Scholar]
- [43].Huang EX, Bradley JD, El Naqa I, Hope AJ, Lindsay PE, Bosch WR, et al. Modeling the risk of radiation-induced acute esophagitis for combined Washington University and RTOG trial 93-11 lung cancer patients. Int J Radiat Oncol Biol Phys. 2012;82:1674–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Rancati T, Schwarz M, Allen AM, Feng F, Popovtzer A, Mittal B, et al. Radiation dose-volume effects in the larynx and pharynx. Int J Radiat Oncol Biol Phys. 2010;76:S64–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chao KS, Deasy JO, Markman J, Haynie J, Perez CA, Purdy JA, et al. A prospective study of salivary function sparing in patients with head-and-neck cancers receiving intensity-modulated or three-dimensional radiation therapy: initial results. Int J Radiat Oncol Biol Phys. 2001;49:907–16. [DOI] [PubMed] [Google Scholar]
- [46].Din SU, Williams EL, Jackson A, Rosenzweig KE, Wu AJ, Foster A, et al. Impact of Fractionation and Dose in a Multivariate Model for Radiation-Induced Chest Wall Pain. International Journal of Radiation Oncology Biology Physics. 2015;93:418–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Fontanella A, Robinson C, Zuniga A, Apte A, Thorstad W, Bradley J, et al. SU-E-T-312: Test of the Generalized Tumor Dose (gTD) Model with An Independent Lung Tumor Dataset. Medical Physics. 2014;41:296-. [Google Scholar]
- [48].Veeraraghavan H, Vargas HA, Sanchez AJ, Micco M, Mema E, Capanu M, et al. Computed Tomography Measures of Inter-site tumor Heterogeneity for Classifying Outcomes in High-Grade Serous Ovarian Carcinoma: a Retrospective Study. 2019. [Google Scholar]


