Abstract
Signal transducers and activators of transcription (STATs) are transcription factors that mediate normal biologic responses to cytokines and growth factors. However, abnormal activation of certain STAT family members, including Stat3, is increasingly associated with oncogenesis. In fibroblasts expressing the Src oncoprotein, activation of Stat3 induces specific gene expression and is required for cell transformation. Although the Src tyrosine kinase induces constitutive Stat3 phosphorylation on tyrosine, activation of Stat3-mediated gene regulation requires both tyrosine and serine phosphorylation of Stat3. We investigated the signaling pathways underlying the constitutive Stat3 activation in Src oncogenesis. Expression of Ras or Rac1 dominant negative protein blocks Stat3-mediated gene regulation induced by Src in a manner consistent with dependence on p38 and c-Jun N-terminal kinase (JNK). Both of these serine/threonine kinases and Stat3 serine phosphorylation are constitutively induced in Src-transformed fibroblasts. Furthermore, inhibition of p38 and JNK activities suppresses constitutive Stat3 serine phosphorylation and Stat3-mediated gene regulation. In vitro kinase assays with purified full-length Stat3 as the substrate show that both JNK and p38 can phosphorylate Stat3 on serine. Moreover, inhibition of p38 activity and thus of Stat3 serine phosphorylation results in suppression of transformation by v-Src but not v-Ras, consistent with a requirement for Stat3 serine phosphorylation in Src transformation. Our results demonstrate that Ras- and Rac1-mediated p38 and JNK signals are required for Stat3 transcriptional activity induced by the Src oncoprotein. These findings delineate a network of tyrosine and serine/threonine kinase signaling pathways that converge on Stat3 in the context of oncogenesis.
Signal transducers and activators of transcription (STATs) were originally discovered as latent cytoplasmic transcription factors that mediate cellular responses to diverse cytokines and growth factors (for reviews, see references 17, 18, and 55). STATs are activated by tyrosine phosphorylation, dimerize, and subsequently translocate to the nucleus, where they regulate the transcription of genes by binding to specific DNA response elements. Studies have implicated normal STAT signaling in controlling fundamental biological processes, including cell differentiation, proliferation, apoptosis, and development (7, 15, 26, 33, 60, 78). Multiple signaling pathways are simultaneously induced in response to cytokine or growth factor stimulation, consistent with complex regulation by signal cross talk. For example, maximum transcriptional activity of certain STATs requires serine phosphorylation mediated by serine/threonine kinases of other signaling pathways (3, 19, 51, 68). The kinases that mediate STAT serine phosphorylation are not fully defined, although evidence implicates multiple serine kinase signals, including mitogen-activated protein kinases (MAPKs)/extracellular signal-regulated kinases (ERKs) (19), an H7-sensitive serine kinase (5), and a MAPK kinase (MKK)-dependent, ERK-independent serine kinase (11).
MAPKs represent a family of serine/threonine protein kinases comprising ERK1/ERK2 (ERKs), p38/HOG1 (p38), and c-Jun N-terminal kinase (JNK)/stress-activated protein kinase (SAPK) (reviewed in references 24, 43, and 59). Ras and Ras-like small G proteins are key regulators in the signaling pathways leading to MAPK activation. For the Ras-ERK branch, sequential protein phosphorylations are mediated by the serine/threonine kinase Raf-1 and the dual-specificity MKKs, which in turn phosphorylate and activate ERKs (24, 48, 49, 72). For the JNK and p38 pathways, the Rac1/Cdc42 subfamily of small G proteins is a key mediator, together with Ras (for reviews, see references 24, 43, and 59). Several serine/threonine protein kinases that are members of the mixed-lineage kinases (MLK), such as dual leucine-zipper bearing kinase (DLK), have been identified as upstream activators of MKKs (23, 24, 38). Activation of JNK is largely induced by MKK4 and MKK7, while MKK3 and MKK6 preferentially activate p38 (22, 24, 62, 75). Activated MAPKs ultimately phosphorylate transcription factors in the nucleus that are responsible for the regulation of immediate-early genes, such as c-fos, whose functional roles include control of cell proliferation (35, 37, 71).
Emerging evidence strongly implicates abnormal activation of STAT signaling in oncogenic transformation. Our laboratory and others have previously reported constitutively active STATs, particularly Stat3 and Stat5, in cells transformed by v-Src, v-Abl, and various other oncoproteins and tumor viruses (4, 9, 13, 16, 29, 45, 46, 76; see reference 28 for a review). Moreover, constitutive activation of Stat3 proteins occurs with high frequency in human tumor cells (10, 12, 29, 31, 61, 66, 77; reviewed in reference 28), suggesting a role for aberrant Stat3 signaling in malignant progression. Recent studies have demonstrated an obligatory requirement for Stat3 signaling in transformation by the Src oncoprotein (6, 63). The mechanisms of subversion of the normal, highly regulated STAT signaling by Src and other oncoproteins are still not fully defined. Understanding how oncoproteins alter STAT signaling should provide further insights into the role of abnormal STAT activation in oncogenesis and may suggest a mechanistic basis for circumventing the oncogenic process. The uniqueness of Stat3 as the sole STAT family member constitutively active in Src-transformed fibroblasts makes this system an excellent model for investigating the regulation of Stat3 signaling in oncogenesis.
Based on this model system, we investigated the signaling pathways involved in aberrant activation of Stat3 in cells expressing the Src oncoprotein. We demonstrate that induction of Stat3-mediated gene regulation by v-Src is strictly Ras dependent in NIH 3T3 cells, since Stat3 function is completely abrogated by the expression of dominant negative Ras. This Ras dependency is reflected in the inhibition of Stat3 transcriptional activity by dominant negative MKK1 or by the MKK1/2 inhibitor PD98059. Similarly, transcriptional regulation by Stat3 is inhibited by dominant negative forms of Rac1, DLK, and MKK4, as well as by the p38 inhibitor SB202190. Constitutive activation of p38 and JNK, together with constitutive Stat3 serine phosphorylation, is observed in Src-transformed cells. Moreover, inhibition of both p38 and JNK is associated with suppression of Stat3 serine phosphorylation and transcriptional activity. Both JNK and p38 phosphorylate Stat3 on serine in vitro. Furthermore, inhibition of p38 activity blocks growth in soft agar of v-Src-transformed cells, consistent with a requirement for p38-mediated Stat3 serine phosphorylation in Src transformation. Thus, we define a network of multiple tyrosine and serine kinase pathways that converge on Stat3 signaling in fibroblasts expressing oncogenic Src and are required for Stat3-mediated gene induction.
MATERIALS AND METHODS
Plasmids.
The Stat3 reporter pLucTKS3, myc-p38mapk, myc-p46sapk, dominant negative DLK (K185A), dominant negative MKK4 (dnMKK4), N17-Ras, and NT-Raf have all been previously described (23, 52, 63). The pLucTKS3 reporter harbors seven copies of a sequence corresponding to the Stat3-specific binding site in the C-reactive protein gene promoter (63). The v-Src expression vector pMvSrc has been described previously (40). Dominant negative forms of ERK2, and MKK1 (34, 69) were generous gifts from M. Weber (University of Virginia) and N. Ahn (University of Colorado), respectively. The Rac1-I115 (activated) and Rac1-17N (dominant negative) vectors were generated by inserting Rac1 cDNA fragments from pZipNeo (41) into pcDNA3 (Invitrogen) at a BamHI site.
Cell culture and transfections.
NIH 3T3, NIH 3T3/v-Src, and NIH 3T3/v-Ras fibroblasts were grown in Dulbecco’s modified Eagle’s medium (DMEM) containing 5% iron-supplemented bovine calf serum. Transient transfections were carried out by the standard calcium phosphate method as previously described (63). NIH 3T3 fibroblasts were seeded at 5 × 105 cells/100-mm plate in DMEM plus 5% bovine calf serum at 18 to 24 h prior to transfection. The total amount of DNA used for transfections was typically 20 μg per plate, including 4 μg of luciferase reporter construct (pLucTKS3), 0.2 μg of β-galactosidase (β-Gal) internal control vector, and the amounts of expression vector indicated in the figure legends. Transfection was terminated 15 h later by aspirating the medium, washing the cells with phosphate-buffered saline (PBS), and adding fresh DMEM. For generation of NIH 3T3/v-Src/TKS3 cell lines stably expressing the Stat3 reporter, NIH 3T3/v-Src cells were transfected with Fugene 6 (Boehringer Mannheim) as specified by the supplier. The transfection mixture contained 5.5 μg of total DNA per 10-cm plate, including 5 μg of the Stat3 reporter pLucTKS3 and 0.5 μg of pcDNA3 that carries the neomycin resistance gene. Individual G418-resistant clones were picked and characterized with regard to Stat3-dependent luciferase activities.
Preparation of cytosolic and nuclear extracts.
In the case of stable NIH 3T3/v-Src/TKS3 clones, cells were treated with inhibitors or dimethyl sulfoxide for 6 h before preparation of cytosolic extracts. For transient-expression assays, cytosolic extracts were prepared from cells at 48 h posttransfection as previously described (63). Briefly, after two washes with PBS and equilibration for 5 min with 0.5 ml of PBS–0.5 mM EDTA, the cells were scraped off of the dishes and the cell pellet was obtained by centrifugation (4,500 × g for 2 min at 4°C). The cells were resuspended in 0.4 ml of low-salt HEPES buffer (10 mM HEPES [pH 7.8], 10 mM KCl, 0.1 mM EGTA, 0.1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol) for 15 min, lysed by the addition of 20 μl of 10% Nonidet P-40 (NP-40), and centrifuged (10,000 × g for 30 s at 4°C) to obtain the cytosolic supernatant, which was used for luciferase assays (Promega) with a luminometer and for detection of β-Gal activity by colorimetric assay at an absorbance at 570 nm. As an internal control for transient-transfection efficiency, the results were normalized to β-Gal activity. For electrophoretic mobility shift assay (EMSA), nuclear extracts were prepared from transiently transfected NIH 3T3 cells and volumes containing equal amounts of total protein were incubated with 32P-labeled M67SIE oligonucleotide probe (64), as previously reported (29, 76). Supershift assays were performed with rabbit polyclonal antibodies specific for C-terminal amino acid residues of Stat3 (750 to 769) or Stat1 (688 to 710) proteins (Santa Cruz Biotechnology).
Soft-agar colony formation assay.
Colony formation assays were carried out with six-well dishes. Each well contained 1.5 ml of 1% agarose in DMEM as the bottom layer. The top layer consisted of 1.5 ml of 0.5% agarose in DMEM containing 4,000 or 6,000 NIH 3T3/v-Src or NIH 3T3/v-Ras fibroblasts, respectively. Treatment with inhibitors was initiated 1 day after seeding cells by adding 75 to 100 μl of medium with or without inhibitors and repeated once a week until large colonies were evident. For quantitation, the colonies were stained by adding 20 μl of 1-mg/ml iodonitrotetrazolium violet to each well and incubating at 37°C overnight; stained colonies were counted the next day.
Western blot analysis.
Whole-cell lysates were prepared in boiling sodium dodecyl sulfate (SDS) sample-loading buffer to extract total proteins from the cytoplasm and nucleus as well as preserve the in vivo phosphorylation states. Equivalent amounts of total cellular protein were electrophoresed on an SDS–10% polyacrylamide gel and transferred to nitrocellulose membranes. Probing of nitrocellulose membranes with primary antibodies and detection of horseradish peroxidase-conjugated secondary antibodies by enhanced chemiluminescence (Amersham) were performed as previously described (29, 63, 76). The probes used were rabbit polyclonal antibodies against N-terminal amino acid residues (626 to 640) of Stat3 (Santa Cruz Biotechnology), phosphoserine-727 of Stat3 (25), active (phosphorylated) JNK, p38mapk, or ERKs (New England Biolabs), or total JNK, p38mapk or ERKs (Santa Cruz Biotechnology).
Purification and phosphorylation of Stat3 and recombinant Stat3 proteins.
Stat3 and Stat3β were purified from baculovirus-infected Sf-9 insect cells with biotinylated M67SIE oligonucleotides. Briefly, Sf-9 cells were infected with baculoviruses encoding Stat3 or Stat3β. At 48 h postinfection, the cells were lysed with NP-40 lysis buffer (50 mM HEPES [pH 7.9], 150 mM NaCl, 1% NP-40, 20 mM NaF, 1 mM Na3VO4, 1 mM Na4P2O4, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 2 mM EDTA, 0.1 μM aprotinin, 1 μM leupeptin, 1 μM antipain) and centrifuged (13,000 × g for 15 s at 4°C). The supernatant cell lysates were supplemented with glycerol (to 10%) and 10 μg of poly(dI-dC)-poly(dI-dC) in a final volume of 1 ml and incubated at 4°C for 30 min. Then 2 μg of 5′-biotinylated DNA fragment, containing two copies of the M67SIE sequence (5′-AGCTTCATTTCCCGTAAATCCCTA) (64), was added, and the mixture was further incubated at 4°C for 2 h with slow rotation. Subsequently, 100 μl of avidin-agarose beads (50% slurry) was added to the mixture and incubated for 30 min. The beads were then collected by centrifugation and washed four times with NP-40 lysis buffer and three times with kinase buffer (25 mM HEPES [pH 7.5], 10 mM magnesium acetate). After a final centrifugation (3,000 × g for 2 min), the pellets of Stat3 and Stat3β-bound Sepharose beads were incubated for 5 min at room temperature in 35 μl of kinase buffer containing approximately similar activities of purified p38 (AG Scientific), JNK (BIOMOL), or ERK (BIOMOL) protein kinases. Subsequently, 5 μl of [γ-32P]ATP solution (50 μM ATP; 0.5 μCi/μl) was added, and the mixture was further incubated at 30°C. After 30 min, SDS-polyacrylamide gel electrophoresis loading buffer was added and the samples were electrophoresed on an SDS–8% polyacrylamide gel and exposed for autoradiography.
RESULTS
Ras-mediated signaling is required for Stat3 transcriptional activity.
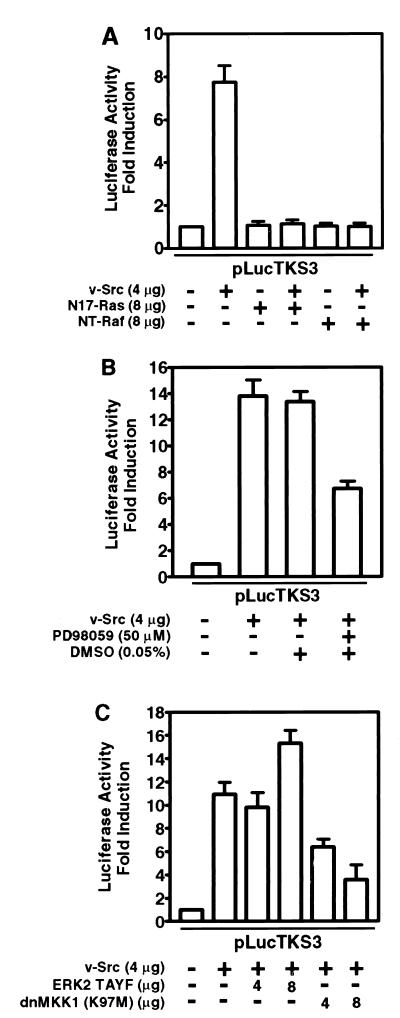
We previously reported that Stat3 is constitutively activated in NIH 3T3 fibroblasts stably transformed by v-Src (76), and we demonstrated its transcriptional potential and its requirement in Src transformation (63). In the present study, we investigated the signaling pathways leading to the induction of Stat3 transcriptional activity by using a Stat3-specific luciferase reporter (pLucTKS3) harboring the Stat3-binding site from the C-reactive protein gene promoter (63). The induction by v-Src of Stat3-specific luciferase reporter was completely abrogated by coexpression of the dominant negative Ras mutant (N17-Ras) or an N-terminal fragment of Raf-1 (NT-Raf) designed to sequester Ras (11, 52) (Fig. 1A). These findings suggest an obligatory requirement of Ras-mediated signaling for Stat3 transcriptional activity in NIH 3T3 fibroblasts expressing v-Src.
FIG. 1.
Ras-MKK1/2-dependent signaling is required for Stat3-mediated gene regulation induced by v-Src. NIH 3T3 cells were transiently transfected with indicated plasmids. Luciferase activities were measured in cytosolic extracts prepared 48 h posttransfection and normalized to β-Gal activity. (A) NIH 3T3 cells were transfected with pLucTKS3 reporter alone or with reporter and v-Src expression vector, pMvSrc, with or without vectors encoding N17-Ras or NT-Raf as indicated. The N17-Ras and NT-Raf proteins inhibit Ras in a dominant negative manner. (B) Cells were transfected with reporter alone or with reporter and pMvSrc and treated with the MKK1/2 inhibitor PD98059 for 6 h or left untreated. (C) Cells were transfected with reporter alone or with reporter and pMvSrc, with or without vectors encoding the dominant negative ERK2 mutant, TAYF, or the MKK1 dominant negative mutant, dnMKK1. Values shown in each panel are means and standard deviations of at least four independent transfections, each performed in triplicate.
We then tested whether the MKK-ERK pathway downstream from Ras is associated with induction of Stat3 transcriptional activity by Src. Results of luciferase reporter assays with the pharmacologic MKK1/2-selective inhibitor PD98059 (21) or the dominant negative MKK1 (dnMKK1) show that inhibition of MKK1/2 activity significantly suppresses transcriptional regulation by Stat3 (Fig. 1B and C). However, expression of a dominant negative form of ERK2, TAYF (19, 34), had no inhibitory effect on Stat3 transcriptional activity (Fig. 1C), suggesting that ERK2 activity is not required for Stat3-mediated gene regulation induced by v-Src. We confirmed the ERK2 dominant negative activity of TAYF by inhibition of another luciferase reporter, pLucSRE, which is not dependent on Stat3 (63) but, rather, is dependent on the activation of the c-fos serum response element by ERKs (36, 73). Because both dnMKK1 and PD98059 block MKK signaling directly, these findings support a role for MKK-mediated signaling in Stat3 transcriptional activity (11). We cannot definitively exclude a possible role for ERKs, since ERKs associate with Stat3 in vivo and in vitro (39) and phosphorylate Stat3 protein in vitro (see below) (14). Together, our results indicate that a Ras-MKK-mediated signaling pathway interacts with Stat3 signaling. The lack of a complete block of Stat3 activity following inhibition of MKK1 or MKK2 (Fig. 1B and C) suggests that other signaling pathways contribute to Stat3 transcriptional activity.
Stat3 transcriptional activity depends on Rac1-mediated signaling.
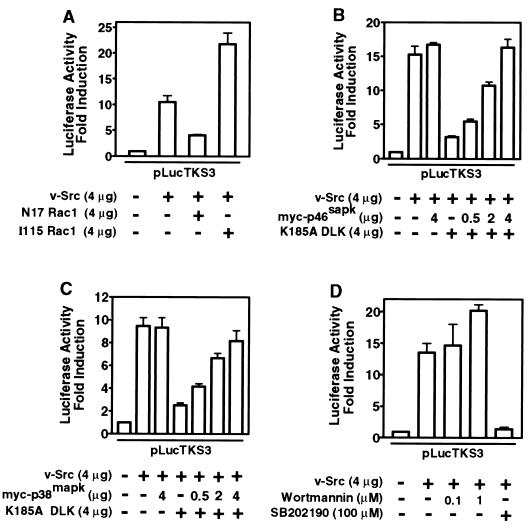
Because the Rac1 subfamily of small G proteins plays a key role in signaling downstream from Ras (24, 43, 59), we investigated the contribution of Rac1-induced signals to Stat3 transcriptional activity. In luciferase reporter assays, the coexpression of dominant negative Rac1 (N17 Rac1) or activated Rac1 (I115 Rac1) mutants significantly inhibited or enhances Stat3 transcriptional activity, respectively (Fig. 2A), suggesting that Rac1 lies in the pathway leading from v-Src to Stat3 activation. To further explore the contribution of Rac1-mediated signals to Stat3 signaling, we examined the role of DLK, a member of the MLK family that participates in activation of the stress pathway by v-Src (23, 47). Dominant negative DLK (K185A) significantly inhibited the induction of Stat3-specific luciferase reporter activity (Fig. 2B). DLK interacts with complexes containing other MLK members (70), and dominant-negative DLK appears to interfere with the function of other members of the MLK family (38a). Therefore, these findings implicate the entire MLK family but do not define which member is required in Stat3 signaling induced by v-Src. Interestingly, the coexpression of JNK1 (myc-p46sapk) or p38 (myc-p38mapk) proteins reversed this inhibitory effect in a concentration-dependent manner (Fig. 2B and C). We infer that the kinase activities of the overexpressed JNK1 or p38 proteins can sustain a level of serine phosphorylation sufficient for maximal Stat3 transcriptional activity even at marginal MLK activity. Together, these findings indicate that Rac1-mediated p38 and JNK activities contribute to Stat3 signaling induced by v-Src.
FIG. 2.
Stat3-mediated gene regulation induced by v-Src requires Rac1- and MLK-dependent p38 and JNK signals. NIH 3T3 cells were transiently transfected with the indicated plasmids, and luciferase activities were assayed as described for Fig. 1. (A) Cells were transfected with the Stat3 reporter pLucTKS3 alone, reporter plus v-Src, or reporter plus v-Src plus dominant negative Rac1 (N17 Rac1), or activated Rac1 (I115 Rac1). (B) Cells were transfected with reporter alone, reporter plus v-Src, or reporter plus v-Src plus dominant negative DLK (K185A) or p46sapk or both. (C) Cells were transfected with reporter alone, reporter plus v-Src, or reporter plus v-Src plus K185A or p38mapk or both. (D) Cells were transfected with reporter alone or reporter plus v-Src and treated or not treated with the p38mapk inhibitor SB202190 or the PI 3-kinase inhibitor wortmannin. Values are means and standard deviations of at least three independent experiments.
That p38 is central to Stat3 signaling is further corroborated by studies showing significant inhibition of Stat3-specific luciferase reporter induction in cells transiently expressing the Stat3-specific reporter and treated with SB202190, a pharmacologic inhibitor selective for p38 (65) (Fig. 2D). Because phosphatidylinositol 3-kinase (PI 3-kinase) involved in Ras-mediated signaling has previously been shown to be activated in Src-transformed cells (27), we investigated any contribution it might make to Stat3 signaling by using wortmannin, a PI 3-kinase inhibitor (74). The results showed no inhibition of Stat3-specific luciferase reporter induction in fibroblasts transiently expressing v-Src and treated with this inhibitor (Fig. 2D), thus excluding a role for PI 3-kinase in transcriptional regulation by Stat3. Our results support the model that induction of Stat3 transcriptional activity by v-Src requires Rac1-mediated p38 and JNK signals.
Evidence of distinct JNK and p38 pathways involved in Stat3 transcriptional activity.
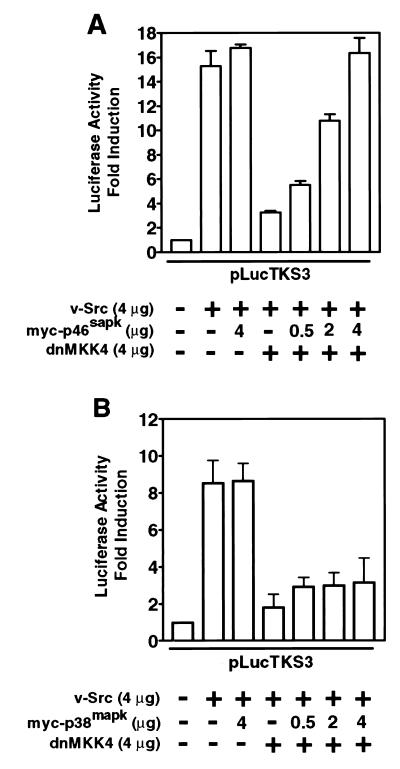
Reports in the literature delineate two distinct pathways leading to the activation of JNK and p38 (see reference 24 for a review). While both pathways utilize a common signal from Rac1, they emerge as separate signals at the level of MKKs. For example, MKK4 and MKK7 largely activate JNK, while MKK3 and MKK6 preferentially activate p38. To test whether this divergence in signaling is relevant to Stat3 function, we first examined the effect of dnMKK4 on transcriptional activation by Stat3. Expression of dnMKK4 significantly blocked Stat3-specific luciferase reporter induction (Fig. 3A), suggesting a requirement for MKK4 in the signaling leading to Stat3 transcriptional activity. The divergence in JNK and p38 signals was evident when only the coexpression of JNK1 (myc-p46sapk), but not p38 (myc-p38mapk), abrogated the inhibitory effect of dnMKK4 and restored Stat3 transcriptional activity (Fig. 3). These results establish that transcriptional activation by Stat3 utilizes the MKK4-JNK pathway and confirm that distinct MKKs mediate the pathways leading to p38 and JNK activation. We infer from these results that Stat3-mediated gene regulation induced by v-Src requires Ras-Rac1-mediated activation of the stress pathway in a manner analogous to normal extracellular stimulus-induced activation of this pathway.
FIG. 3.
v-Src-induced Stat3-mediated gene regulation requires MKK4-dependent JNK signaling. NIH 3T3 cells were transiently transfected with the indicated plasmid vectors, and luciferase activities were assayed as described for Fig. 1. (A) Cells were transfected with pLucTKS3 reporter alone, reporter plus v-Src, or reporter plus v-Src plus dnMKK4 or myc-p46sapk or both. (B) Cells were transfected with reporter alone, reporter plus v-Src, or reporter plus v-Src plus dnMKK4 or myc-p38mapk or both. Values are means and standard deviations of at least three independent transfections.
JNK and p38 kinases mediate the key role of Ras in Stat3 transcriptional activity.
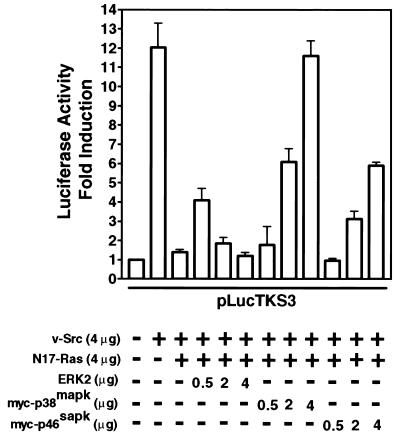
Because inhibition of Ras function (Fig. 1A) is expected to block the activities of downstream MAPKs, we tested whether the abrogation of Stat3 transcriptional activity following dominant negative inhibition of Ras is due to lack of sufficient functional MAPKs. If this is the case, overexpression of the MAPKs would be expected to restore kinase activities and hence Stat3 function. In confirmation of this prediction, coexpression of all three MAPK family proteins brought about recovery of Stat3 transcriptional activity that would otherwise have been blocked by dominant negative inhibition of Ras (Fig. 4). Interestingly, differences emerged in the pattern and extent of restoration of Stat3 transcriptional activity. Similar to p38 or JNK1, the expression of low levels of ERK2 resulted in partial recovery of Stat3 transcriptional activity (Fig. 4). However, as the level of ERK2 expression increases, Stat3 transcriptional activity declines.
FIG. 4.
Stat3-mediated gene regulation induced by v-Src requires Ras-dependent p38mapk and JNK activities. NIH 3T3 cells were transiently transfected with pLucTKS3 reporter alone, reporter plus v-Src, or reporter plus v-Src plus N17-Ras with or without vectors encoding ERK2, myc-p38mapk or myc-p46sapk. Luciferase activities were assayed as described for Fig. 1. Values are means and standard deviations of three independent experiments.
While there is not yet an explanation for this phenomenon, it is consistent with previous reports that ERK kinase activity can down-regulate Stat3 function via a number of mechanisms. These include inhibition of upstream kinases, such as JAK family members (56), dephosphorylation of phosphotyrosine in Stat3 (14), and formation of an ERK-Stat3 complex (39). While we do not exclude any of these events in our system, it is also relevant that constitutive activation of ERKs is not detected in many fibroblast cell lines stably transformed by v-Src (32, 58) (see Fig. 6). As compelling evidence that p38 and, to a lesser extent, JNK mediate the role of Ras in Stat3 transcriptional activity, coexpression of either of these MAPKs caused a complete or partial rescue of Stat3 function from inhibition by dominant negative Ras (Fig. 4). The extent of this restoration was dependent on the level of p38 or JNK expression. We deduce that the overexpressed p38 or JNK proteins compensate for the loss of kinase activities. Together, our findings provide strong evidence of cooperation of Ras-mediated p38 and JNK pathways with v-Src for the induction of Stat3 transcriptional activity.
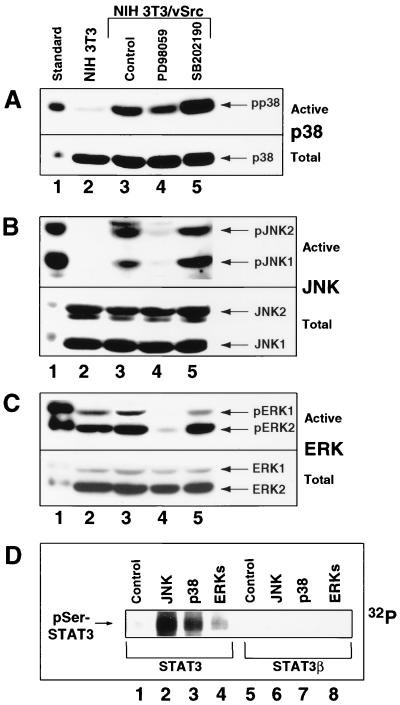
FIG. 6.
p38 and JNK are constitutively induced in Src-transformed cells and phosphorylate Stat3 in vitro. (A) Western blot analysis of whole-cell lysates prepared from normal NIH 3T3 fibroblasts (lane 2) or Src-transformed counterparts treated with or without PD98059 or SB202190 (lanes 3 to 5). Samples were probed with antibody specific to phospho-p38 (top) or total p38 (bottom). (B) The whole-cell extracts described in panel A were analyzed by Western blotting. Samples were probed with antibody specific to phospho-JNK1/2 (top) or total JNK1/2 (bottom). (C) The whole-cell extracts described in panel A were analyzed by Western blotting. Samples were probed with antibody specific to phospho-ERK1/2 (top) or total ERK1/2 (bottom). (D) In vitro serine phosphorylation of Stat3 by JNK, p38, and ERKs. Purified baculovirus-expressed Stat3 (lanes 1 to 4) and Stat3β (lanes 5 to 8) were incubated with [γ-32P]ATP together with or without purified JNK, p38, or ERKs for 30 min and subjected to SDS-polyacrylamide gel electrophoresis and autoradiography. For positive identification, cell lysates from anisomycin-treated C6 glioma cells with highly induced ERKs, p38, and JNK, served as standards (lane 1).
Serine phosphorylation and DNA-binding activity of Stat3 in fibroblasts expressing v-Src.
In the context of transformation by v-Src, our results suggest a cross-communication of signals involving the p38 and JNK serine/threonine kinases and Stat3. The prediction is that in addition to tyrosine phosphorylation, Stat3 undergoes constitutive serine phosphorylation in Src-transformed cells for induction of transcriptionally functional Stat3. To test this assumption, we first assayed for Stat3 serine phosphorylation levels by Western blot analysis with phosphoserine-727-specific anti-Stat3 antibodies (25, 30). Strikingly, our results showed that Stat3 was constitutively phosphorylated on serine 727 in Src-transformed fibroblasts compared to their normal counterparts (Fig. 5A, lanes 1 and 2). To determine if MAPK members are required for this event, we treated Src-transformed fibroblasts with PD98059 or SB202190 and prepared cell lysates for phosphoserine-Stat3 Western blot analysis. Treatment with either PD98059 or SB202190 blocked serine phosphorylation of Stat3 (lanes 2 to 4). These results establish that Stat3 serine phosphorylation is constitutive in NIH 3T3 fibroblasts stably transformed by Src and provide evidence that MAPK family members are major mediators of this effect.
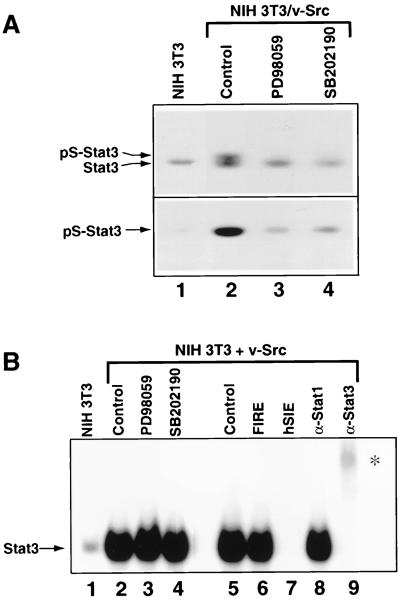
FIG. 5.
Analyses of constitutive Stat3 serine phosphorylation and SIE-binding activity induced by v-Src. (A) Western blot analysis of whole-cell lysates prepared from normal NIH 3T3 fibroblasts and Src-transformed counterparts treated with PD98059 or SB202190 for 6 h or left untreated (lanes 2 to 4). Samples were probed with antibodies specific to phosphoserine-727 (bottom) or the N-terminal portion (top) of Stat3. (B) Nuclear extracts were prepared from NIH 3T3 cells transfected with v-Src. Equal amounts of total protein were incubated with 32P-labeled M67SIE and subjected to EMSA. Cells were transfected with empty vector alone (NIH 3T3) or v-Src vector and treated with PD98059 or SB202190 for 6 h or left untreated (lanes 2 to 5). Competitions of radiolabeled M67SIE-binding activity present in nuclear extracts of NIH 3T3 cells transfected with v-Src alone (lanes 6 and 7) were performed with a 100-fold molar excess of unlabeled M67SIE or the unrelated FIRE oligonucleotides. Supershifts (lanes 8 and 9) were performed with antibodies specifically recognizing either Stat1 or Stat3 (α-Stat1, α-Stat3). The asterisk indicates positions of supershifted complexes.
We next explored whether PD98059 and SB202190 have an influence on the Stat3 DNA-binding activity induced by v-Src. Nuclear extracts were prepared from fibroblasts expressing v-Src that have been treated with inhibitors or left untreated. STAT DNA-binding activities in extracts containing equal amounts of total proteins were analyzed by EMSA with an oligonucleotide probe corresponding to the M67 variant of the c-fos gene sis-inducible element (SIE), which binds both activated Stat1 and Stat3 (64). As previously reported (29, 63, 76), expression of v-Src induced Stat3 tyrosine phosphorylation and DNA-binding activity (Fig. 5B, lanes 1 and 2). Moreover, treatment of v-Src-expressing cells with PD98059 or SB202190 had no effect on Stat3 DNA-binding activity induced by v-Src (lanes 2 to 4). For controls, the binding of Stat3 to M67SIE was competitively inhibited by a molar excess of cold, unlabelled M67SIE but not by the unrelated c-fos intragenic regulatory element (FIRE) oligonucleotide, showing the specificity of DNA binding. Furthermore, Stat3 binding was blocked and supershifted by anti-Stat3 antibodies but not by anti-Stat1 antibodies, demonstrating that the DNA-binding complex in this case contained Stat3. We conclude from these results that inhibition of Stat3 serine phosphorylation has no effect on the constitutive Stat3 DNA-binding activity in cells expressing v-Src, consistent with earlier findings that Stat3 DNA-binding activity is independent of serine phosphorylation (67). Taken together, our findings demonstrate that constitutive Stat3 serine phosphorylation in Src-transformed cells is dependent on signaling through MAPK family members.
p38 and JNK are activated in Src-transformed fibroblasts.
Because the results presented above suggest that p38 and JNK are key components of the signaling leading to Stat3 transcriptional activity induced by v-Src, we determined whether these kinases are constitutively activated in cell lines stably transformed by Src. The activity levels of p38, JNK, and ERKs were assayed by Western blot analysis with antibodies specific to the phosphorylated, activated forms. Significantly, we observed that both p38 and JNK1/2 were highly activated in v-Src-transformed compared to normal NIH 3T3 fibroblasts (Fig. 6A and B, lanes 2 and 3). In contrast, no substantial induction of ERK1/2 was observed in Src-transformed over normal NIH 3T3 cells, consistent with previous reports (32, 58) (Fig. 6C, lanes 2 and 3).
We next investigated the effects of PD98059 and SB202190 on the activation of these MAPKs. As expected, treatment of Src-transformed fibroblasts with PD98059 caused a complete block of basal ERKs activity (Fig. 6C, lanes 3 and 4). Surprisingly, however, treatment with the same MKK1/2 inhibitor caused complete or partial suppression of JNK1/2 and p38 induction, respectively (Fig. 6A and B, lanes 3 and 4). These results suggest either that the MKK1/2 inhibitor has a nonspecific effect on other MKKs upstream of JNK and p38 or that MKK1/2 are involved in JNK and p38 activation, as previously reported (50). Combined with the results in Fig. 1B, these findings indicate that the suppression of Stat3 transcriptional activity by PD98059 is the sum of the effects of this inhibitor on MKK1/2, JNK, and p38 activities. While the block by PD98059 of basal ERK1/2 and induced JNK1/2 activities is complete, there is only a partial inhibition of Stat3-mediated gene regulation (compare Fig. 1B with Fig. 6B and C, lanes 3 and 4). We speculate that the limited effect of PD98059 on Stat3 function is at least in part due to its incomplete inhibition of p38 activity, thus pointing to this MAPK member as the major serine/threonine kinase required for Stat3 signaling. Because SB202190 directly blocks p38 kinase activity, treatment of Src-transformed cells with this inhibitor did not significantly alter the phosphorylation of MAPK members, including p38 (Fig. 6A to C, lanes 3 and 5). The apparent high induction of p38 phosphorylation when SB202190 was present may be due to a positive feedback response by MKK3 or MKK6 to the diminished p38 kinase activity. Although the activation of overexpressed exogenous JNK by v-Src has been previously reported (23, 47), this is the first evidence of constitutive activation of both endogenous p38 and JNK in stable Src-transformed fibroblasts and supports the model that the activated stress pathway cooperates with Stat3 signaling induced by Src. Altogether, our findings provide evidence of cross talk between Ras-Rac1-mediated activities of p38/JNK and Stat3 signaling in Src-transformed cells.
Because our results implicate p38 and, to a lesser extent, JNK1/2 as the key serine/threonine kinases involved in Stat3 signaling in Src-transformed cells, we tested whether Stat3 can be a direct substrate for these MAPKs in vitro. The results shown in Fig. 6D indicate that p38 and JNK can effectively phosphorylate Stat3 in vitro, partly consistent with a previous report (14). The notable difference between our study and this previous report was our use of full-length Stat3 as the substrate and purified proteins of the MAPK family. Stat3 phosphorylation by ERK, however, was minimal compared to the levels achieved for JNK and p38. For a control, we used a splice variant of full-length Stat3 with a C-terminal deletion, Stat3β, which lacks serine 727 and therefore cannot transactivate in many cell types (8, 63). The Stat3 and Stat3β proteins used as substrates in this assay maintained correct protein folding as they were purified by virtue of their DNA-binding activity to a Stat3-specific site. The results showed that Stat3β did not undergo serine phosphorylation by any of the MAPKs (Fig. 6D), consistent with serine 727 being the site of phosphorylation (14, 67). These results suggest that all three MAPKs are capable of using Stat3 as substrate in vivo, although the actual contributions of the individual MAPK family members in vivo would be expected to depend on the extent of their activation by v-Src.
Inhibition of p38 activity blocks constitutive Stat3 signaling and Src transformation.
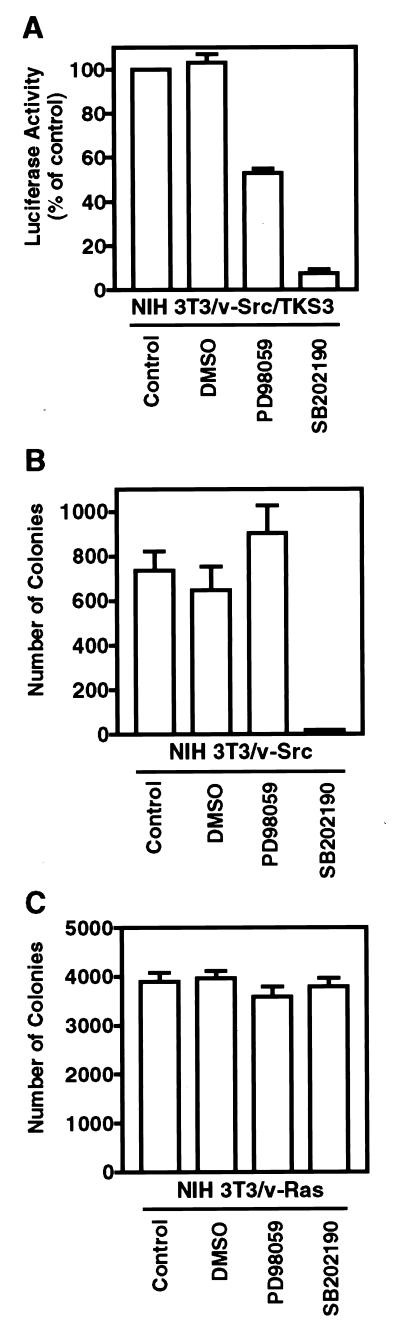
The above results of transient-transfection assays with reporter constructs suggest that MKK-mediated p38 and, to a lesser extent, JNK activities are required for constitutive Stat3 signaling in Src-transformed cells. To confirm this conclusion, we tested the effects of inhibition of MKKs or p38 on the induction of the Stat3-dependent luciferase reporter, pLucTKS3, in v-Src-transformed fibroblasts that stably express this reporter. Because Stat3 is constitutively activated in Src-transformed cells (76), NIH 3T3/v-Src/TKS3 cells stably expressing the Stat3 reporter exhibit very high luciferase activity, reflecting constitutive Stat3-dependent induction of this reporter. As seen in transient transfections, treatment of NIH 3T3/v-Src/TKS3 cells with PD98059 or SB202190 partially or completely suppressed constitutive induction of the Stat3-dependent luciferase reporter, respectively (Fig. 7A), consistent with an obligatory requirement for p38 in constitutive Stat3 signaling in Src-transformed cells. The present studies, combined with previous reports (6, 63), raise the possibility that p38-mediated Stat3 serine phosphorylation is required for v-Src transformation. We tested this hypothesis by investigating the effects of inhibition of p38 on anchorage-independent growth of Src-transformed fibroblasts in soft-agar suspension. Treatment of cells in agar with SB202190 completely blocked colony formation of Src-transformed cells (Fig. 7B). In contrast, treatment with the same inhibitor had no significant effects on colony formation by Ras-transformed fibroblasts, which do not require Stat3 activation (Fig. 7C) (6, 29, 63). Thus, the inhibition by SB202190 of Src transformation is not the outcome of gross cytotoxicity. These studies suggest that p38 activity and Stat3 serine phosphorylation are required for transformation by Src but not by Ras. We also tested the effect of inhibition of MKK1/2 by PD98059 on anchorage-independent growth of fibroblasts transformed by v-Src or v-Ras. The results showed a lack of significant effect of this inhibitor on transformation by either oncoprotein (Fig. 7B and C), suggesting that inhibition of MKK1/2 is not sufficient to block Src or Ras transformation. Together, these results demonstrate that p38 activity is required for Stat3-mediated gene regulation and v-Src transformation.
FIG. 7.
Inhibition of p38 activity blocks cell transformation by v-Src and not by v-Ras. (A) NIH 3T3/v-Src/TKS3 cells stably transfected with the Stat3-dependent luciferase reporter, pLucTKS3, were treated with the indicated inhibitors for 6 h prior to cytosolic extract preparation and luciferase assays as described for Fig. 1. Values are the means and standard deviations of six independent assays. (B) NIH 3T3/v-Src fibroblasts seeded in soft-agar suspension were treated once weekly with the indicated inhibitors until large-colony formation was evident. Values are the means and standard deviations of 12 independent assays. (C) NIH 3T3/v-Ras fibroblasts seeded in soft-agar suspension were treated once weekly with the indicated inhibitors, and colonies were counted as in panel B. Values are the means and standard deviations of nine independent assays. DMSO, dimethyl sulfoxide.
DISCUSSION
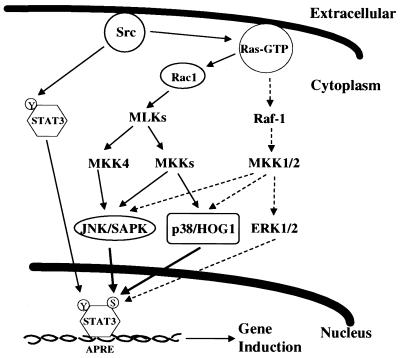
Recent studies have established that constitutive activation of Stat3 signaling participates in cell transformation by the oncogenic Src tyrosine kinase (6, 63). Here we demonstrate that, in parallel to the constitutive DNA-binding activity and tyrosine phosphorylation of Stat3, the Src oncoprotein recruits additional signaling pathways crucial for Stat3 function (Fig. 8). Upstream of these signals is Ras, which functions to coordinately integrate serine/threonine kinase activities necessary for efficient Stat3 transcriptional activity. As one of the Ras-mediated pathways, MKK-ERK signaling interacts with that of Stat3 (14, 19, 39, 51, 56). The interaction between ERKs and Stat3 signaling, however, is complicated by results which indicate that ERKs can down-regulate (14, 39, 56) as well as enhance (19, 51) Stat3 tyrosine phosphorylation and transcriptional activity. This disparity may be explained by our finding that low levels of ERK2 induce while higher levels inhibit Stat3-mediated gene regulation (Fig. 4). At the same time, the evidence indicates a role for MKK1/2-mediated, ERK-independent signals in Stat3 transcriptional activity (11), consistent with transformation of NIH 3T3 fibroblasts by activated MKK1 mutants independently of ERKs (2) and raising the possibility that MKK1/2 recruits p38 and JNK serine/threonine kinases (50) for Stat3 signaling induced by v-Src.
FIG. 8.
Model of Stat3 phosphorylation by tyrosine and serine/threonine kinase signaling pathways in Src oncogenesis. APRE, acute-phase response element.
Positioned downstream from Ras, the Rac1 family of small G proteins is key to signals that induce p38 and JNK serine/threonine kinases (37, 71). We confirm that Rac1 signaling is recruited by v-Src (23, 47) and extend these studies to demonstrate that Stat3 signaling induced by v-Src requires components of Rac1 signaling, including MLK family members and MKK4. The rescue of Stat3 function by p38 and JNK proteins from inhibition induced by dominant negative Ras provides compelling evidence that these serine/threonine kinases are key in Src-induced Stat3 signaling. Thus, the essential role of Ras in this Stat3 signaling is the recruitment of Rac1-mediated p38 and JNK activities. It is also highly significant that both p38 and JNK activities are constitutively induced in cells stably transformed by Src. The aberrant constitutive activation of these two kinases may be essential to maintain the observed elevated Stat3 serine phosphorylation and transcriptional activity in Src-transformed cells. This is the first demonstration of constitutive induction of p38, JNK, and Stat3 serine phosphorylation in cells stably transformed by Src and provides evidence that these events are associated. Whether these events are unique to Src transformation or are common to cells transformed by other oncogenic tyrosine kinases remains to be determined. Our results are consistent with the finding that transformation by the nonreceptor tyrosine kinase, v-Fps/Fes, requires Ras- and Rac1-mediated activation of MAPK family members (42). These findings set precedents for investigating possible augmentation by Ras, Rac1, p38, and JNK pathways of aberrant STAT signaling in human malignancies that harbor constitutively activated STAT proteins (reviewed in reference 28).
We do not exclude the possibility that other serine/threonine kinases contribute to Stat3 transcriptional activity in Src-transformed fibroblasts. Notably, studies show that v-Src activates various isoforms of protein kinase C (53), suggesting that the latter may play a role in Src transformation. Others have also noted a role for H7-sensitive serine kinases in Stat3 transcriptional activity (5). On the other hand, our observations suggest that the PI 3-kinase pathway is unlikely to contribute to Stat3 signaling, although it has previously been shown to be activated in Src-transformed cells (27). Furthermore, we do not anticipate any role for the serine/threonine kinase AKT2, which is a downstream target of PI 3-kinase that is also activated by v-Src (20, 44). That PI 3-kinase and AKT2 may not be required for Stat3 signaling is supported by the report that dominant negative inhibition of PI 3-kinase inhibits transformation by V12 Ras but not by v-Src (54).
Our findings presented here define signal transduction networks from v-Src to Stat3 in NIH 3T3 fibroblasts that integrate tyrosine and serine/threonine kinase pathways (Fig. 8). This model predicts a key role for Ras, which regulates the contributions of the MKK1/2 cascade and the Rac1-mediated stress-activated pathways involving p38 and JNK. While Ras plays an essential role in transformation of NIH 3T3 cells by v-Src (57), more recent studies have demonstrated that Ras is not required for Src transformation of chicken embryo fibroblasts or Rat-2 fibroblasts (1). Thus, the requirement for Ras-mediated signaling in Src transformation is cell type specific, raising the possibility that Ras-independent pathways activate p38 and JNK signaling leading to Stat3 transcriptional activity in different cell types. On the other hand, NIH 3T3 cells harboring activated Ras do not exhibit activated Stat3 (29), suggesting that Ras signaling is necessary but not sufficient for transformation of these cells by Src. Our results also indicate that downstream events, such as p38 and JNK signaling, are not sufficient to induce Stat3 transcriptional activity in the absence of Src. Nevertheless, activation of the stress signaling pathways involving p38 and JNK is obligatory for Stat3 function.
Because serine phosphorylation of Stat3 is required for its maximal transcriptional activity (68) and because Stat3 signaling is obligatory for Src transformation (6, 63), we infer from the present study that p38- and JNK-mediated Stat3 serine phosphorylation is necessary for Src oncogenesis. Consistent with this conclusion, earlier studies have demonstrated that a Stat3 mutant with a Ser-727-to-Ala mutation blocks Src-mediated transformation in a dominant negative manner (6). Thus, it is highly significant that inhibition of p38-mediated Stat3 serine phosphorylation blocks transformation by v-Src and not other oncoproteins like Ras, which do not induce Stat3 signaling. These findings underscore the functional importance of p38 in mediating Stat3 serine phosphorylation in Src oncogenesis. In addition, the pathways delineated here are relevant to normal Stat3 signaling because recent studies demonstrated that p38 induces Stat3 serine phosphorylation in T cells in response to interleukin-12 and interleukin-2 (30). Our findings provide the first evidence detailing cross talk between the Ras/Rac1-mediated p38/JNK pathways and Stat3 signaling leading to serine phosphorylation of Stat3 in the context of oncogenesis. While there are likely to be other pathways essential for Stat3 function and Src transformation, our study demonstrates a convergence at the level of Stat3 of multiple signaling pathways activated by Src. These novel observations provide new insight into some of the signaling pathways induced by the Src oncoprotein that potentially play critical roles in cell transformation and human cancer.
ACKNOWLEDGMENTS
We thank M. Weber and N. Ahn for generously providing dominant negative ERK2 (TAYF) and MKK1, respectively, and members of the laboratory for stimulating discussions.
This work was supported by the Molecular Biology and Molecular Imaging Core Facilities of the Moffitt Cancer Center and Research Institute and by NCI grants CA55652 (to R.J.) and CA76661 (to S.S.).
REFERENCES
- 1.Aftab D T, Kwan J, Martin G S. Ras-independent transformation by v-Src. Proc Natl Acad Sci USA. 1997;94:3028–3033. doi: 10.1073/pnas.94.7.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alessandrini A, Greulich H, Huang W, Erikson R L. Mek1 phosphorylation site mutants activate Raf-1 in NIH 3T3 cells. J Biol Chem. 1996;271:31612–31618. doi: 10.1074/jbc.271.49.31612. [DOI] [PubMed] [Google Scholar]
- 3.Beadling C, Ng J, Babbage J W, Cantrell D A. Interleukin-2 activation of STAT5 requires the convergent action of tyrosine kinases and a serine/threonine kinase pathway distinct from the Raf-1/ERK2 MAP kinase pathway. EMBO J. 1996;15:1902–1913. [PMC free article] [PubMed] [Google Scholar]
- 4.Besser D, Bromberg J F, Darnell J E, Jr, Hanafusa H. A single amino acid substitution in the v-Eyk intracellular domain results in activation of Stat3 and enhances cellular transformation. Mol Cell Biol. 1999;19:1401–1409. doi: 10.1128/mcb.19.2.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boulton T G, Zhong Z, Wen Z, Darnell J E, Jr, Stahl N, Yancopoulos G D. STAT3 activation by cytokines utilizing gp130 and related transducers involves a secondary modification requiring H7-sensitive kinase. Proc Natl Acad Sci USA. 1995;92:6915–6919. doi: 10.1073/pnas.92.15.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bromberg J F, Horvath C M, Besser D, Lathem W W, Darnell J E., Jr Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bromberg J F, Horvath C M, Wen Z, Schreiber R D, Darnell J E., Jr Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc Natl Acad Sci USA. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldenhoven E, van Dijk T B, Solari R, Armstrong J, Raaijmakers J A M, Lammers J W J, Koenderman L, de Groot R P. STAT3beta, a splice variant of transcription factor STAT3, is a dominant negative regulator of transcription. J Biol Chem. 1996;271:13221–13227. doi: 10.1074/jbc.271.22.13221. [DOI] [PubMed] [Google Scholar]
- 9.Cao X, Tay X, Guy G R, Tan Y H. Activation and association of Stat3 with Src in v-Src-transformed cell lines. Mol Cell Biol. 1996;16:1595–1603. doi: 10.1128/mcb.16.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Catlett-Falcone R, Landowski T H, Oshiro M M, Turkson J, Levitzki A, Savino R, Ciliberto G, Moscinski L, Fernandez-Luna J L, Nunez G, Dalton W S, Jove R. Constitutive activation of Stat3 signaling confers resistance to apoptosis in human U266 myeloma cells. Immunity. 1999;10:105–115. doi: 10.1016/s1074-7613(00)80011-4. [DOI] [PubMed] [Google Scholar]
- 11.Ceresa B P, Horvath C M, Pessin J E. Signal transducer and activator of transcription-3 serine phosphorylation by insulin is mediated by a Ras/Raf/MEK-dependent pathway. Endocrinology. 1997;138:4131–4137. doi: 10.1210/endo.138.10.5266. [DOI] [PubMed] [Google Scholar]
- 12.Chai S K, Nichols G L, Rothman P. Constitutive activation of JAKs and STATs in BCL-Abl-expressing cell lines and peripheral blood cells derived from leukemic patients. J Immunol. 1997;159:4720–4728. [PubMed] [Google Scholar]
- 13.Chaturvedi P, Sharma S, Reddy E P. Abrogation of interleukin-3 dependence of myeloid cells by the v-src oncogene requires SH2 and SH3 domains which specify activation of STATs. Mol Cell Biol. 1997;17:3295–3304. doi: 10.1128/mcb.17.6.3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung J, Uchida E, Grammer T C, Blenis J. STAT3 serine phosphorylation by ERK-dependent and -independent pathways negatively modulates its tyrosine phosphorylation. Mol Cell Biol. 1997;17:6508–6515. doi: 10.1128/mcb.17.11.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cressman D E, Greenbaum L E, DeAngelis R A, Ciliberto G, Furth E E, Poli V, Taub R. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 16.Danial N N, Pernis A, Rothman P B. Jak-STAT signaling induced by the v-abl oncogene. Science. 1995;269:1875–1877. doi: 10.1126/science.7569929. [DOI] [PubMed] [Google Scholar]
- 17.Darnell J E., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 18.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 19.David M, Petricoin III E, Benjamin C, Pine R, Weber M J, Larner A C. Requirement for MAP kinase (ERK2) activity in interferon alpha- and interferon beta-stimulated gene expression through STAT proteins. Science. 1995;269:1721–1723. doi: 10.1126/science.7569900. [DOI] [PubMed] [Google Scholar]
- 20.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Interleukin 3-induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 21.Dudley D T, Pang L, Decker S J, Bridges A J, Saltiel A R. A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc Natl Acad Sci USA. 1995;92:7686–7689. doi: 10.1073/pnas.92.17.7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enslen H, Raingeaud J, Davis R J. Selective activation of p38 mitogen-activated protein (MAP) kinase isoforms by MAP kinase kinases MKK3 and MKK6. J Biol Chem. 1998;273:1741–1748. doi: 10.1074/jbc.273.3.1741. [DOI] [PubMed] [Google Scholar]
- 23.Fan G, Merritt S E, Kortenjann M, Shaw P E, Holzman L B. Dual leucine zipper-bearing kinase (DLK) activates p46sapk and p38mapk but not ERK2. J Biol Chem. 1996;271:24788–24793. doi: 10.1074/jbc.271.40.24788. [DOI] [PubMed] [Google Scholar]
- 24.Fanger G R, Gerwins P, Widmann C, Jarpe M B, Johnson G L. MEKKs, GCKs, MLKs, PAKs, TAKs, and tpls: upstream regulators of the c-Jun amino-terminal kinases? Curr Opin Genet Dev. 1997;7:67–74. doi: 10.1016/s0959-437x(97)80111-6. [DOI] [PubMed] [Google Scholar]
- 25.Frank D A, Mahajan S, Ritz J. B lymphocytes from patients with chronic lymphocytic leukemia contain signal transducer and activator of transcription (STAT) 1 and STAT3 constitutively phosphorylated on serine residues. J Clin Investig. 1997;100:3140–3148. doi: 10.1172/JCI119869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fukada T, Hibi M, Yamanaka Y, Takahashi-Tezuka M, Fujitani Y, Yamaguchi T, Nakajima K, Hirano T. Two signals are necessary for cell proliferation induced by a cytokine receptor gp130: involvement of STAT3 in anti-apoptosis. Immunity. 1996;5:449–460. doi: 10.1016/s1074-7613(00)80501-4. [DOI] [PubMed] [Google Scholar]
- 27.Fukui Y, Saltiel A R, Hanafusa H. Phosphatidylinositol-3 kinase is activated in v-src, v-yes, and v-fps transformed chicken embryo fibroblasts. Oncogene. 1991;6:407–411. [PubMed] [Google Scholar]
- 28.Garcia R, Jove R. Activation of STAT transcription factors in oncogenic tyrosine kinase signaling. J Biomed Sci. 1998;5:79–85. doi: 10.1007/BF02258360. [DOI] [PubMed] [Google Scholar]
- 29.Garcia R, Yu C-L, Hudnall A, Catlett R, Nelson K L, Smithgall T, Fujita D J, Ethier S P, Jove R. Constitutive activation of Stat3 in fibroblasts transformed by diverse oncoproteins and in breast carcinoma cells. Cell Growth Differ. 1997;8:1267–1276. [PubMed] [Google Scholar]
- 30.Gollob J A, Schnipper C P, Murphy E A, Ritz J, Frank D A. The functional synergy between IL-12 and IL-2 involves p38 MAP kinase and is associated with the augmentation of STAT serine phosphorylation. J Immunol. 1999;162:4472–4481. [PubMed] [Google Scholar]
- 31.Grandis J R, Drenning S D, Chakraborty A, Zhou M-Y, Zeng Q, Pitt A S, Tweardy D J. Requirement of Stat3 but not Stat1 activation for epidermal growth factor receptor-mediated cell growth in vitro. J Clin Investig. 1998;102:1385–1392. doi: 10.1172/JCI3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greulich H, Reichman C, Hanafusa H. Delay in serum stimulation of Erk activity caused by oncogenic transformation. Oncogene. 1996;12:1689–1695. [PubMed] [Google Scholar]
- 33.Hauser P J, Agrawal D, Hackney J, Pledger W J. STAT3 activation accompanies keratinocyte differentiation. Cell Growth Differ. 1998;10:847–855. [PubMed] [Google Scholar]
- 34.Her J-H, Lakhani S, Zu K, Vila J, Dent P, Sturgill T W, Weber M J. Dual phosphorylation and autophosphorylation in mitogen-activated protein (MAP) kinase activation. Biochem J. 1993;296:25–31. doi: 10.1042/bj2960025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hill C S, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- 36.Hill C S, Treisman R. Differential activation of c-fos promoter elements by serum, lysophosphatidic acid, G proteins and polypeptide growth factors. EMBO J. 1995;14:5037–5047. doi: 10.1002/j.1460-2075.1995.tb00186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hill C S, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 38.Holland P M, Suzanne M, Campbell J S, Noselli S, Cooper J A. MKK7 is a stress-activated mitogen-activated protein kinase kinase functionally related to hemipterous. J Biol Chem. 1997;272:24994–24998. doi: 10.1074/jbc.272.40.24994. [DOI] [PubMed] [Google Scholar]
- 38a.Holzman, L. Unpublished results.
- 39.Jain N, Zhang T, Fong S L, Lim C P, Cao X. Repression of Stat3 activity by mitogen-activated protein kinase (MAPK) Oncogene. 1998;17:3157–3167. doi: 10.1038/sj.onc.1202238. [DOI] [PubMed] [Google Scholar]
- 40.Johnson P J, Coussens P M, Danko A V, Shalloway D. Overexpressed pp60c-src can induce focus formation without complete transformation of NIH 3T3 cells. Mol Cell Biol. 1985;5:1073–1083. doi: 10.1128/mcb.5.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khosravi-Far R, Solski P A, Clark G J, Kinch M S, Der C J. Activation of Rac1, RhoA, and mitogen-activated protein kinases is required for Ras transformation. Mol Cell Biol. 1995;15:6443–6453. doi: 10.1128/mcb.15.11.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Smithgall T E. Fibroblasts transformation by Fps/Fes tyrosine kinases requires Ras, Rac, and Cdc42 and induces extracellular signal-regulated and c-Jun N-terminal kinase activation. J Biol Chem. 1998;273:13828–13834. doi: 10.1074/jbc.273.22.13828. [DOI] [PubMed] [Google Scholar]
- 43.Lim L, Manser E, Leung T, Hall C. Regulation of phosphorylation pathways by p21 GTPases. The p21 Ras-related Rho subfamily and its role in phosphorylation signaling pathways. Eur J Biochem. 1996;242:171–185. doi: 10.1111/j.1432-1033.1996.0171r.x. [DOI] [PubMed] [Google Scholar]
- 44.Liu A X, Testa J R, Hamilton T C, Jove R, Nicosia S V, Cheng J Q. AKT2, a member of the protein kinase B family, is activated by growth factors, v-Ha-ras, and v-src through phosphatidylinositol 3-kinase in human ovarian epithelial cancer cells. Cancer Res. 1998;58:2973–2977. [PubMed] [Google Scholar]
- 45.Lund T C, Garcia R, Medveczky M M, Jove R, Medveczky P G. Activation of STAT transcription factors by herpesvirus saimiri Tip-484 requires p56lck. J Virol. 1997;71:6677–6682. doi: 10.1128/jvi.71.9.6677-6682.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Migone T S, Lin J X, Cereseto A, Mulloy J C, O’Shea J J, Franchini G, Leonard W J. Constitutively activated Jak-STAT pathway in T cells transformed with HTLV-I. Science. 1995;269:79–81. doi: 10.1126/science.7604283. [DOI] [PubMed] [Google Scholar]
- 47.Minden A, Lin A, Claret F-X, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPase Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 48.Minden A, Lin A, McMahon M, Lange-Carter C, Derijard B, Davis R J, Johnson G L, Karin M. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 49.Moodie S A, Willumsen B M, Weber M J, Wolfman A. Complexes of Ras-GTP with Raf-1 and mitogen-activated protein kinase kinase. Science. 1993;260:1658–1661. doi: 10.1126/science.8503013. [DOI] [PubMed] [Google Scholar]
- 50.Morooka T, Nishida E. Requirement of p38 mitogen-activated protein kinase-activated protein kinase for neuronal differentiation in PC12 cells. J Biol Chem. 1998;273:24285–24288. doi: 10.1074/jbc.273.38.24285. [DOI] [PubMed] [Google Scholar]
- 51.Ng J, Cantrell D. STAT3 is a serine kinase target in T lymphocytes. J Biol Chem. 1997;272:24542–24549. doi: 10.1074/jbc.272.39.24542. [DOI] [PubMed] [Google Scholar]
- 52.Pumiglia K, Chow Y-H, Fabian J, Morrison D, Decker S, Jove R. Raf-1 N-terminal sequences necessary for Ras-Raf interaction and signal transduction. Mol Cell Biol. 1995;15:398–406. doi: 10.1128/mcb.15.1.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qureshi S A, Alexandropoulos K, Rim M, Joseph C K, Bruder J T, Rapp U R, Foster D A. Evidence that Ha-Ras mediates two distinguishable intracellular signals activated by v-Src. J Biol Chem. 1992;267:17635–17639. [PubMed] [Google Scholar]
- 54.Rodriguez-Viciana P, Warne P H, Khwaja A, Marte B M, Pappin D, Das P, Waterfield M D, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 55.Schindler C, Darnell J E., Jr Transcriptional responses to polypeptide ligands: the Jak-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 56.Sengupta T K, Talbot E S, Scherle P A, Ivashkiv L B. Rapid inhibition of interleukin-6 signaling and Stat3 activation mediated by mitogen-activated protein kinases. Proc Natl Acad Sci USA. 1998;95:11107–11112. doi: 10.1073/pnas.95.19.11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stacey D W, Roudebush M, Day R, Mosser S D, Gibbs J B, Feig L A. Dominant inhibitory Ras mutants demonstrate the requirement for Ras activity in the action of tyrosine kinase oncogenes. Oncogene. 1991;6:2297–2304. [PubMed] [Google Scholar]
- 58.Stofega M R, Yu C-L, Wu J, Jove R. Activation of extracellular signal-regulated kinase (ERK) by mitogenic stimuli is repressed in v-Src transformed cells. Cell Growth Differ. 1997;8:113–119. [PubMed] [Google Scholar]
- 59.Symons M. Rho family of GTPases: the cytoskeleton and beyond. Trends Biochem Sci. 1996;21:178–181. [PubMed] [Google Scholar]
- 60.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci USA. 1997;94:3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takemoto S, Mulloy J C, Cereseto A, Migone T S, Patel B K, Matsuoka M, Yamaguchi K, Takatsuki K, Kamihira S, White J D, Leonard W J, Waldmann T, Franchini G. Proliferation of adult T cell leukemia/lymphoma cells is associated with the constitutive activation of JAK/STAT proteins. Proc Natl Acad Sci USA. 1997;94:13897–13902. doi: 10.1073/pnas.94.25.13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tibbles L A, Ing Y L, Kiefer F, Chan J, Iscove N, Woodgett J R, Lassam N J. MLK-3 activates the SAPK/JNK and p38 pathways via SEK1 and MKK3/6. EMBO J. 1996;15:7026–7035. [PMC free article] [PubMed] [Google Scholar]
- 63.Turkson J, Bowman T, Garcia R, Caldenhoven E, De Groot R P, Jove R. Stat3 activation by Src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wagner B J, Hayes T E, Hoban C J, Cochran B H. The SIF binding elements confers sis/PDGF inducibility onto the c-fos promoter. EMBO J. 1990;9:4477–4484. doi: 10.1002/j.1460-2075.1990.tb07898.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Z, Canagarajah B J, Boehm J C, Kassisa S, Cobb M H, Young P R, Abdel-Meguid S, Adams J L, Goldsmith E J. Structural basis of inhibitor selectivity in MAP kinases. Structure. 1998;6:1117–1128. doi: 10.1016/s0969-2126(98)00113-0. [DOI] [PubMed] [Google Scholar]
- 66.Weber-Nordt R M, Egen C, Wehinger J, Ludwig W, Gouilleux-Gruart V, Mertelsmann R, Finke J. Constitutive activation of STAT proteins in primary lymphoid and myeloid leukemia cells and in Epstein-Barr virus (EBV)-related lymphoma cell lines. Blood. 1996;88:809–816. [PubMed] [Google Scholar]
- 67.Wen Z, Darnell J E., Jr Mapping of Stat3 serine phosphorylation to a single residue (727) and evidence that serine phosphorylation has no influence on DNA binding of Stat1 and Stat3. Nucleic Acids Res. 1997;25:2062–2067. doi: 10.1093/nar/25.11.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wen Z, Zhong Z, Darnell J E., Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 69.Whalen A M, Galasinski S C, Shapiro P S, Nahreini T S, Ahn N G. Megakaryocytic differentiation induced by constitutive activation of mitogen-activated protein kinase kinase. Mol Cell Biol. 1997;17:1947–1958. doi: 10.1128/mcb.17.4.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whitmarsh A J, Cavanagh J, Tournier C, Yasuda J, Davis R J. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 71.Whitmarsh A J, Yang S-H, Su M S-S, Sharrocks A D, Davis R J. Role of p38 and JNK mitogen-activated protein kinases in the activation of ternary complex factors. Mol Cell Biol. 1997;17:2360–2371. doi: 10.1128/mcb.17.5.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu J, Harrison J K, Dent P, Lynch K R, Weber M J, Sturgill T W. Identification and characterization of a new mammalian mitogen-activated protein kinase kinase, MKK2. Mol Cell Biol. 1993;13:4539–4548. doi: 10.1128/mcb.13.8.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yamauchi K, Holt K, Pessin J E. Phosphatidylinositol 3-kinase functions upstream of Ras and Raf in mediating insulin stimulation of c-fos transcription. J Biol Chem. 1993;268:14597–14600. [PubMed] [Google Scholar]
- 74.Yano H, Nakanishi S, Kimura K, Hanai N, Saitoh Y, Fukui Y, Nonomura Y, Matsuda Y. Inhibition of histamine secretion by wortmannin through the blockade of phosphatidylinositol 3-kinase in RBL-2H3 cells. J Biol Chem. 1993;268:25846–25856. [PubMed] [Google Scholar]
- 75.Yao Z, Diener K, Wang X S, Zukowski M, Matsumoto G, Zhou G, Mo R, Sasaki T, Nishina H, Hui C C, Tan T H, Woodgett J P, Penninger J M. Activation of stress-activated protein kinases/c-Jun N-terminal protein kinases (SAPK/JNKs) by a novel mitogen-activated protein kinase kinase (MKK7) J Biol Chem. 1997;272:32378–32383. doi: 10.1074/jbc.272.51.32378. [DOI] [PubMed] [Google Scholar]
- 76.Yu C-L, Meyer D J, Campbell G S, Larner A C, Carter-Su C, Schwartz J, Jove R. Enhanced DNA-binding activity of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 77.Zhang Q, Nowak I, Vonderheid E C, Rook A H, Kadin M E, Nowell P C, Shaw L M, Wasik M A. Activation of Jak/STAT proteins involved in signal transduction pathway mediated by receptor for interleukin 2 in malignant T lymphocytes derived from cutaneous anaplastic large T-cell lymphoma and Sezary syndrome. Proc Natl Acad Sci USA. 1996;93:9148–9153. doi: 10.1073/pnas.93.17.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhong Z, Wen Z, Darnell J E., Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]