Abstract
The major components of the mitogen-activated protein kinase (MAPK) cascades are MAPK, MAPK kinase (MAPKK), and MAPKK kinase (MAPKKK). Recent rapid progress in identifying members of MAPK cascades suggests that a number of such signaling pathways exist in cells. To date, however, how the specificity and efficiency of the MAPK cascades is maintained is poorly understood. Here, we have identified a novel mouse protein, termed Jun N-terminal protein kinase (JNK)/stress-activated protein kinase-associated protein 1 (JSAP1), by a yeast two-hybrid screen, using JNK3 MAPK as the bait. Of the mammalian MAPKs tested (JNK1, JNK2, JNK3, ERK2, and p38α), JSAP1 preferentially coprecipitated with the JNKs in cotransfected COS-7 cells. JNK3 showed a higher binding affinity for JSAP1, compared with JNK1 and JNK2. In similar cotransfection studies, JSAP1 also interacted with SEK1 MAPKK and MEKK1 MAPKKK, which are involved in the JNK cascades. The regions of JSAP1 that bound JNK, SEK1, and MEKK1 were distinct from one another. JNK and MEKK1 also bound JSAP1 in vitro, suggesting that these interactions are direct. In contrast, only the activated form of SEK1 associated with JSAP1 in cotransfected COS-7 cells. The unstimulated SEK1 bound to MEKK1; thus, SEK1 might indirectly associate with JSAP1 through MEKK1. Although JSAP1 coprecipitated with MEK1 MAPKK and Raf-1 MAPKKK, and not MKK6 or MKK7 MAPKK, in cotransfected COS-7 cells, MEK1 and Raf-1 do not interfere with the binding of SEK1 and MEKK1 to JSAP1, respectively. Overexpression of full-length JSAP1 in COS-7 cells led to a considerable enhancement of JNK3 activation, and modest enhancement of JNK1 and JNK2 activation, by the MEKK1-SEK1 pathway. Deletion of the JNK- or MEKK1-binding regions resulted in a significant reduction in the enhancement of the JNK3 activation in COS-7 cells. These results suggest that JSAP1 functions as a scaffold protein in the JNK3 cascade. We also discuss a scaffolding role for JSAP1 in the JNK1 and JNK2 cascades.
The mitogen-activated protein kinase (MAPK) cascades, in which the major components are MAPK, MAPK kinase (MAPKK), and MAPKK kinase (MAPKKK), are conserved eukaryotic signaling pathways (2, 7, 14, 46). The general function of the MAPK cascades is to link a variety of extracellular stimuli to nuclear responses, i.e., the modulation of gene expression (45). MAPK is activated by dual phosphorylation on threonine and tyrosine residues catalyzed by MAPKK, and MAPKK is activated by serine/threonine phosphorylation catalyzed by MAPKKK. In mammals, at least three MAPK cascades have been identified. The MAPKs in each pathway are ERK (extracellular signal-regulated kinase), JNK/SAPK (c-Jun N-terminal kinase/stress-activated protein kinase), and p38. The ERK cascade is mostly responsive to mitogenic and differentiation stimuli, whereas the JNK and p38 cascades are strongly activated by proinflammatory cytokines, such as interleukin 1 (IL-1) and tumor necrosis factor alpha (TNF-α), and extracellular stresses, such as UV irradiation and osmotic shock (4, 23, 29, 36).
In the ERK cascade, Raf (Raf-1, A-Raf, and B-Raf), MEK (MEK1 and MEK2), and ERK (ERK1 and ERK2) correspond to MAPKKK, MAPKK, and MAPK, respectively (36). The p38 cascade contains p38α/CSBP/RK/Mxi2 (12, 25, 37, 56) and p38β (18) as MAPKs and MKK3 (9) and MKK6 (6, 13, 32, 35, 42) as MAPKKs, while in the JNK cascade the MAPKs are JNK1, JNK2, and JNK3 (also known as SAPKγ, SAPKα, and SAPKβ, respectively) (8, 11, 19, 22, 31), and the MAPKKs are SEK1/MKK4/JNKK1 (9, 26, 38) and MKK7/JNKK2 (33, 44, 48). The specificity of the MAPKKKs involved in the JNK and p38 cascades is less clear. For instance, the TAK1 (52), ASK1 (16), and MLK3 (43) MAPKKKs can activate both the JNK and p38 cascades, while the MEKK1 (28, 50, 53) and MEKK4 (10) MAPKKKs selectively activate the JNK cascade.
The identification of numerous components of the MAPK cascades as described above suggests that there are a number of these distinct signaling pathways in cells. Furthermore, studies of JNK3-deficient mice (54) indicate the existence of a JNK3-specific cascade that cannot be complemented by the other JNK family members, even though JNK1, JNK2, and JNK3 exhibit over 80% identity, and these JNKs seem to be similarly regulated by the upstream kinases, at least in transiently transfected cells. The tissue distributions of the JNKs are quite different: JNK3 is specifically expressed in the brain, while JNK1 and JNK2 are widely expressed. How the specificity and efficiency of the MAPK cascades is maintained is largely unknown.
In this paper, we report the molecular cloning and characterization of a novel JNK-binding protein, termed JSAP1 (JNK/SAPK-associated protein 1). Through cotransfection studies of COS-7 cells, we observed preferential interactions between JSAP1 and the JNKs (JNK1, JNK2, and JNK3), SEK1, and MEKK1. The regions of JSAP1 that bind JNK, SEK1, and MEKK1 were distinct from one another. JNK3 exhibited higher binding affinity to JSAP1 compared with JNK1 and JNK2. JNK and MEKK1 also bound JSAP1 in vitro, suggesting that these interactions are direct. In contrast, only the activated form of SEK1 interacted with JSAP1 in cotransfected cells. The unstimulated SEK1 bound to MEKK1; thus, SEK1 might associate with JSAP1 through MEKK1. Overexpressing full-length JSAP1 in COS-7 cells considerably enhanced the JNK3 activation, and modestly enhanced the JNK1 and JNK2 activation, through the MEKK1-SEK1 pathway. JSAP1 mutants lacking the JNK- or MEKK1-binding regions significantly reduced of the enhancement of the JNK3 activation. These results suggest that JSAP1 functions as a scaffold protein in the JNK3 cascade. A scaffolding role of JSAP1 in the JNK1 and JNK2 cascades is also discussed.
MATERIALS AND METHODS
Isolation of cDNAs.
A mouse brain cDNA library (Stratagene) was screened by using a partial JSAP1 cDNA fragment, obtained from a yeast two-hybrid screen, as a probe. A full-length cDNA was isolated, and the open reading frame (ORF) of JSAP1 was sequenced. cDNAs containing the ORFs of JNK1, -2, and -3 were isolated from the same cDNA library. The probes used to screen the library were generated by PCR, with mouse brain cDNA as the template and using the following primers, whose design was based on the rat JNK1, -2, and -3 cDNA sequences (22), respectively: JNK1-S (5′-GCAGATTCTACATTCACAGTCCTA-3′; 5′ end at nucleotide 46), JNK1-A (5′-CATTTCTCCCATAATGCACCCCAC-3′; 5′ end at 654), JNK2-S (5′-GTGGCAGACTCAACTTTCACTGTT-3′; 5′ end at 43), JNK2-A (5′-GTAGCCCATGCCCAGGATGACTTC-3′; 5′ end at 606), JNK3-S (5′-ACAGTTCTAAAGCGCTACCAGAAC-3′; 5′ end at 61), and JNK3-A (5′-GTGACGAACCTATTCTCCCATGAT-3′; 5′ end at 663). Nucleotide numbering starts with +1, which represents the first nucleotide in the initiation codon ATG of the corresponding rat JNK gene.
The partial nucleotide sequence of mouse MEKK1 cDNA has been reported elsewhere (24). MEKK1 residues 656 to 1488 were amplified from mouse spleen cDNA by PCR using the primers 5′-TACACTCCTTGCCACAGTCTGGCA-3′ and 5′-ACTACCACGTGGTACGGAAGACCG-3′.
Mouse MEKK1 cDNA encoding the N-terminal region was isolated from a mouse spleen cDNA library (Stratagene). The probe used to screen the library was generated by two-step PCR using the following primers, whose design was based on the rat MEKK1 cDNA sequence (50): MEKK1-S1 (5′-ACCTGTATGCCTGCCTGGAAGCAC-3′; 5′ end at 544), MEKK1-S2 (5′-TGGTGGTGAAACCAATCCCTATTA-3′; 5′ end at 602), MEKK1-A1 (5′-TTGAGCTACGCCTACTGTGGTATT-3′; 5′ end at 1165), and MEKK1-A2 (5′-TTCCGAGATGGAGCTTTGATTCTT-3′; 5′ end at 1187. Numbering of the nucleotides is as described above.
The first PCR was performed with MEKK1-S1 and -A2 primers with mouse spleen cDNA. An aliquot (1 μl) of the first PCR product was used in the second PCR with the nested MEKK1-S2 and -A1 primers. The full-length human Raf-1 cDNA was obtained from Health Science Research Resources Bank, Osaka, Japan. The ORFs of ERK2, MKK6, p38α, MKK7, MEK1, and SEK1 were amplified by PCR from human lymphocyte (for ERK2 and MKK6), mouse thymus (for p38α, MKK7, and MEK1), or mouse brain (for SEK1) cDNA. The products were inserted into the EcoRV site of pBluescript II KS(+) (Stratagene), and the nucleotide sequences were confirmed by DNA sequencing.
Plasmid construction.
The ORFs of JNK1, JNK2, JNK3, and ERK2 were amplified by PCR. The products (each of which contains a NcoI site at the 5′ end and a BamHI site at the 3′ end of the sense strand) were first digested with NcoI, filled in, ligated with a phosphorylated NotI linker (5′-pGCGGCCGC-3′), and then digested with NotI and BamHI. The NotI-BamHI fragments were subcloned into NotI/BamHI-digested pFlag-CMV2 (Kodak) to generate pFlag-CMV2-JNK1, -JNK2, -JNK3, and -ERK2. The ORF of p38α was amplified by PCR. The product (containing a NotI site at the 5′ end and a BamHI site at the 3′ end of the sense strand) was inserted into the EcoRV site of pBluescript II KS(+). The NotI-BamHI fragment of the plasmid was subcloned into NotI/BamHI-digested pFlag-CMV2 to generate pFlag-CMV2-p38α.
A double-stranded Flag linker consisting of annealed and phosphorylated oligonucleotides 5′-pAGCTACCATGGACTACAAAGACGATGACGACA-3′ and 5′-pAGCTTGTCGTCATCGTCTTTGTAGTCCATGGT-3′ was inserted at the HindIII site of pcDNA3 (Invitrogen) to generate pcDNA3-Flag. The ORFs of SEK1, MEK1, MKK6, and MKK7 were amplified by PCR. The products (each of which contains a HindIII site at the 5′ end and an XbaI site at the 3′ end of the sense strand) were inserted into the EcoRV site of pBluescript II KS(+). The HindIII-XbaI fragments of the plasmids were subcloned into HindIII/XbaI-digested pcDNA3-Flag to generate pcDNA3-Flag-SEK1, -MEK1, -MKK6, and -MKK7. The N-terminal region (residues 1 to 327) of Raf-1 was amplified by PCR. The product (containing an EcoRI site at the 5′ end and a stop codon at the 3′ end of the sense strand) was digested with EcoRI and subcloned into EcoRI/EcoRV-digested pcDNA3-Flag to generate pcDNA3-Raf-N. Similarly, the C-terminal region (residues 316 to 648) of Raf-1 was amplified by PCR. The product (containing a stop codon followed by an XhoI site at the 3′ end of the sense strand) was digested with XhoI and subcloned into EcoRV/XhoI-digested pcDNA3-Flag to generate pcDNA3-Flag-Raf-C. pcDNA3-Flag-ΔRaf is identical to pcDNA3-Flag-Raf-C. The region encoding residues 1169 to 1488 of MEKK1 was amplified by PCR. The product (containing a HindIII site at the 5′ end and a BamHI site at the 3′ end of the sense strand) was digested with HindIII and BamHI and subcloned into HindIII/BamHI-digested pcDNA3-Flag to generate pcDNA3-Flag-ΔMEKK. The BamHI-NcoI fragment (containing a 15-bp 5′ flanking sequence and 1,653-bp coding sequence [residues 1 to 551]) was excised from MEKK1 N-terminal cDNA. The region encoding residues 551 to 640 of MEKK1 was amplified by PCR. The product (containing an NcoI site at the 5′ end and a stop codon followed by an EcoRI site at the 3′ end of the sense strand) was digested with NcoI and EcoRI. The BamHI-NcoI and NcoI-EcoRI fragments were subcloned together into BamHI/EcoRI-digested pcDNA3-Flag to generate pcDNA3-Flag-MEKK-N. The NcoI-EcoRV fragment (containing an 2,814-bp coding sequence [residues 551 to 1488], a stop codon, an 8-bp 3′ flanking sequence, and a HindIII site followed by an EcoRV site at the 3′ end of the sense strand) was excised from a mouse MEKK1 cDNA. The BamHI-NcoI and NcoI-EcoRV fragments were subcloned together into BamHI/EcoRV-digested pcDNA3-Flag to generate pcDNA3-Flag-MEKK1. pcDNA3-Flag-MEKK1 was used for in vitro transcription-translation.
A 0.2-kb BalI-XhoI fragment of pET32b (Novagen) containing multiple cloning sites, a His-tag coding sequence, and an S-tag coding sequence was inserted into HindIII (filled in)/XhoI-digested pcDNA3 to generate pcDNA3-His-S. The N-terminal region (residues 1 to 343) of JSAP1 was amplified by PCR. The product (containing an EcoRI site at the 5′ end and a HindIII site at the 3′ end of the sense strand) was digested with EcoRI and HindIII and subcloned into EcoRI/HindIII-digested pcDNA3-His-S to generate pcDNA3-His-S-JSAP1-N. The C-terminal region (residues 1233 to 1305) of JSAP1 was amplified by PCR. The product (containing an ApaI site at the 5′ end and a stop codon followed by an XhoI site at the 3′ end of the sense strand) was digested with ApaI and XhoI. The HindIII-ApaI fragment (encoding residues 343 to 1233) was excised from the JSAP1 cDNA. The HindIII-ApaI and ApaI-XhoI fragments were subcloned together into HindIII/XhoI-digested pcDNA3-His-S-JSAP1-N to generate pcDNA3-His-S-JSAP1. To generate deletion mutants of JSAP1, portions of JSAP1 were amplified by PCR, and the products were inserted into pET32b or pcDNA3-His-S. The ORF of SEK1 was amplified by PCR. The products (containing a BamHI site at the 5′ end and an XhoI site at 3′ end of the sense strand) was digested with BamHI and XhoI. The BamHI-XhoI fragment was subcloned into BamHI/XhoI-digested pET32a to generate pET32-SEK1. The deletion mutants pcDNA3-His-S-JSAP1-Δ1, -Δ2, -Δ3, -Δ4, and -Δ5 encode residues 1 to 1053, 744 to 1305, 1054 to 1305, 343 to 1053, and 1 to 343, respectively. The deletion mutants pcDNA3-His-S-JSAP1-ΔJ, -ΔS, and -ΔM include deletions of the JNK-binding region (residues 115 to 233), the SEK1-binding region (residues 1054 to 1305), and the MEKK1-binding region (residues 343 to 744), respectively. pcDNA3-His-S-JSAP1-ΔS is identical to pcDNA3-His-S-JSAP1-Δ1. The deletion in pcDNA3-His-S-JSAP1-ΔJ was generated by overlapping PCR (1). The HindIII site at nucleotide 1025 in the JSAP1 cDNA was changed to a BamHI site by PCR. The BamHI site and the other BamHI site at nucleotides 2228 were ligated to generate the deletion in the pcDNA3-His-S-JSAP1-ΔM. pcDNA3-Flag-MEKK was first digested with BamHI, filled in, ligated with a phosphorylated HindIII linker (5′-CCAAGCTTGG-3′), and then digested with HindIII. The HindIII fragment containing the ORF of MEKK1 was subcloned into HindIII-digested pcDNA3-His-S to generate pcDNA3-His-S-MEKK1. The HindIII-XbaI fragments of pcDNA3-Flag-SEK1 and -MEK1 were subcloned into HindIII/XbaI-digested pcDNA3-His-S to generate pcDNA3-His-S-SEK1 and -MEK1, respectively.
To generate expression vectors for Trx (thioredoxin)-His-S-JSAP1 (T-0, T-1, T-2, and T-3), site-directed mutagenesis was carried out by using overlapping PCR (1). Mutated sequences were confirmed by DNA sequencing. The glutathione S-transferase (GST)–c-Jun (residues 1 to 79) expression vector was described previously (15).
The primers 5′-AAATCTAGATAAACGCTCAACTTTGGCC-3′ and 5′-TTTACTGTCACCCATGGCGTA-3′ were used in a PCR with SRα-3XHA (20) (a gift from T. Deng) as the template. The PCR product (which contains an XbaI site at the 5′ end and an NcoI site at the 3′ end of the sense strand and HA3 [hemagglutinin] coding sequence) was inserted into the EcoRV site of pBluescript II KS(+). The resulting plasmid (termed pKS-3XHA) has two XbaI sites outside the HA3 coding sequence. The region encoding residues 1169 to 1488 of MEKK1 was amplified by PCR. The product (containing an NcoI site at the 5′ end and a BamHI site at the 3′ end of the sense strand) was digested with NcoI and BamHI and subcloned into NcoI/BamHI-digested pKS-3XHA to generate pKS-3XHA-ΔMEKK. The XbaI filler fragment of pEF-BOS (30) was replaced with the XbaI fragment of pKS-3XHA-ΔMEKK to generate pEF-3XHA-ΔMEKK. A 0.28-kb BamHI-XhoI fragment of pCS3+MT (a gift from D. Turner and R. Rupp) containing a Myc-tag coding sequence was filled in and inserted into HindIII (filled in)-digested pcDNA3 to generate pcDNA3-Myc. pcDNA3-Flag-MEK1 was digested with HindIII, filled in, and then digested with XbaI. The HindIII (filled in)-XbaI fragment containing MEK1 ORF was subcloned into NotI (filled in)/XbaI-digested pcDNA3-Myc to generate pcDNA3-Myc-MEK1.
pGEM-3Zf(+) (Promega) was digested with EcoRI, filled in, ligated with phosphorylated NcoI linker (5′-pGCCATGGC-3′), digested with NcoI, and self-ligated to generate pGEM-NCO. The ORF of JNK3 was amplified by PCR. The product (containing an NcoI site at the 5′ end a BamHI site at the 3′ end of the sense strand) was digested with NcoI and BamHI and subcloned into NcoI/BamHI-digested pGEM-NCO and pAS2 (Clontech) to generate pGEM-JNK3 and pAS2-JNK3, respectively. pGEM-JNK3 and pAS2-JNK3 were used for in vitro transcription-translation and the yeast two-hybrid screen, respectively.
Two-hybrid screen.
A mouse brain cDNA library in pGAD10 (Clontech) was transfected into Saccharomyces cerevisiae CG1945 harboring pAS2-JNK3. The Clontech yeast two-hybrid system was used according to the manufacturer’s instructions.
Northern blotting analysis.
Northern blotting analysis was performed as described previously (55), using 32P-labeled JNK3, JSAP1, and β-actin cDNA probes.
Analyses of protein-protein interactions in vitro and in vivo.
Trx-His-S-JSAP1 proteins were expressed in Escherichia coli and purified with S-protein–agarose or nickel-nitrilotriacetic acid-agarose according to the manufacturers’ instructions. The TNT T7 Quick Coupled transcription-translation system (Promega) was used for in vitro translation. The Trx-His-S-JSAP1 proteins bound to S-protein–agarose were mixed with in vitro-translated 35S-labeled JNK3 or MEKK1 in buffer A (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% NP-40), rotated for 2 h at 4°C, spun, and washed three times with buffer A. The precipitates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and subjected to autoradiography. The phosphorylated form of Trx-His-S-JSAP1 (residues 115 to 274) was prepared as follows. Flag-JNK3 that was activated by ΔMEKK in COS-7 cells was immunoprecipitated with anti-Flag monoclonal antibody M5 (Kodak), resuspended in buffer B (50 mM HEPES [pH 7.5], 150 mM NaCl, 1% NP-40, 10% glycerol, 2 mM MgCl2, 1 mM EGTA, 20 mM β-glycerophosphate, 2 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 0.2 mM dithiothreitol) containing unphosphorylated Trx-His-S-JSAP1 (residues 115 to 274), incubated for 30 min at 30°C in the presence of 1 mM ATP, and purified with S-protein–agarose. COS-7 cells transfected with Flag-tagged expression vectors were lysed in buffer C (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% NP-40, 0.5 mM EDTA, 0.5 mM dithiothreitol), mixed with immobilized Trx-His-S protein, Trx-His-S-JSAP1, or Trx-His-S-SEK1, rotated for 2 h, spun, and washed three times with buffer C. The precipitates were examined by immunoblotting with anti-Flag monoclonal antibody M5. For the analysis of protein-protein interactions in intact cells, the expression vectors were cotransfected into COS-7 cells with TransIT-LT1 (Mirus) according to the manufacturer’s instructions. After 34 h, cells were lysed in buffer B and precipitated by S-protein–agarose. The recovered fractions were separated by SDS-PAGE and transferred to Immobilon-P (Millipore). The membranes were probed with anti-Flag monoclonal antibody M5 or anti-c-Myc monoclonal antibody 9E10 (Boehringer Mannheim) and visualized with the Amersham enhanced chemiluminescence detection system. The phosphorylated forms of JNK3 and SEK1 were detected by phospho-specific JNK/SAPK and SEK1/MKK4 antibodies, respectively (New England Biolabs).
Protein kinase assay.
COS-7 cells were transfected with pFlag-CMV2-JNK3 with expression vectors, using TransIT-LT1. After 34 h, the cells were lysed in buffer B and precipitated with anti-Flag monoclonal antibody M5 bound to protein G-agarose. The immunocomplex kinase assay was performed as described previously (8).
Nucleotide sequence accession numbers.
The nucleotide sequences of mouse JNK1, -2, and -3 cDNAs have been deposited in DDBJ/EMBL/GenBank with accession no. AB005663, AB005664, and AB005665, respectively. The nucleotide sequence of the mouse MEKK1 cDNA encoding the N-terminal region has been deposited in DDBJ/EMBL/GenBank with accession no. AB014614. The nucleotide sequence of JSAP1 cDNA has been deposited in DDBJ/EMBL/GenBank with accession no. AB005662.
RESULTS
Molecular cloning of JSAP1, a novel JNK-binding protein.
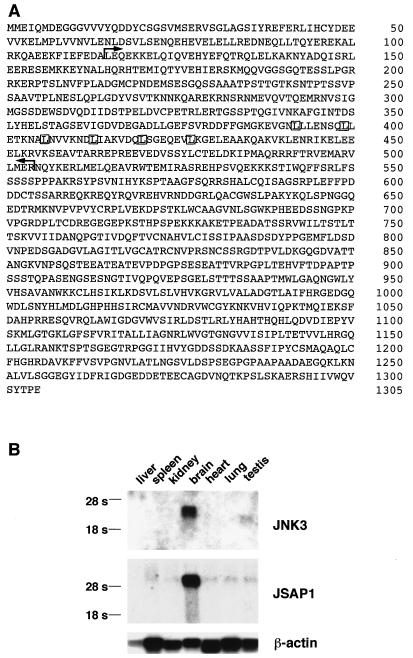
We used a yeast two-hybrid system to search for proteins that directly interact with JNK3. A group of positive clones encoding a protein, termed JSAP1, was identified by screening approximately 2 × 106 transformants. To identify a full-length clone, we screened a mouse brain cDNA library with a partial JSAP1 cDNA insert obtained from the yeast two-hybrid screen. The full-length JSAP1 cDNA was found to encode a protein of 1,305 amino acids with a calculated relative molecular weight of 144,131 (Fig. 1A). Databank searches indicated that JSAP1 represents a novel protein. However, significant amino acid sequence homology to a human sperm-specific protein (accession no. X91879) (40) was found. No function has yet been ascribed to this human gene. JSAP1 contains a leucine zipper motif with a periodic repeat of leucines every seven residues (residues 392 to 427 [Fig. 1A]). We analyzed the tissue distributions of mouse JNK3 and JSAP1 by Northern blotting analysis (Fig. 1B). As in humans (31), mouse JNK3 mRNA was expressed almost exclusively in the brain. Of the other tissues tested, only the testis showed JNK3 expression, which was at a level markedly lower than that seen in the brain. The approximately 6-kb JSAP1 transcript was also particularly abundant in the brain, and very weak signals of JSAP1 mRNA were detected in other tissues. JSAP1 protein was localized to the cytoplasm in P19 cells that were induced to differentiate by treatment with retinoic acid (data not shown).
FIG. 1.
(A) Deduced amino acid sequence of JSAP1. The region (residues 115 to 504) isolated in the yeast two-hybrid system is shown by L-shaped arrows. The leucine residues of the leucine zipper are boxed. (B) Expression of JNK3 and JSAP1 mRNAs in mouse tissues. Poly(A)+ RNA samples isolated from various mouse tissues were examined by Northern blotting analysis. The positions of 28S and 18S rRNAs are indicated on the left. β-Actin mRNA was included as a loading control.
JSAP1 specifically interacts with JNK and not with other mammalian MAPKs.
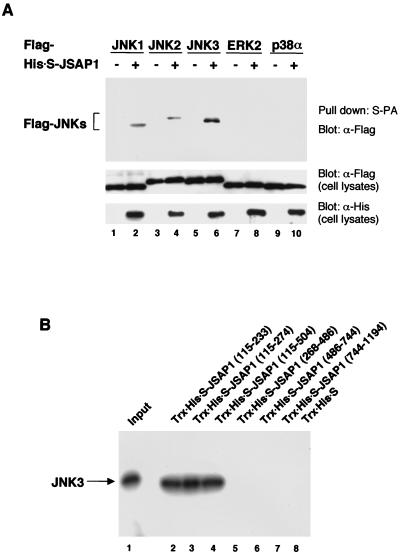
The binding specificity of JSAP1 for the various MAPKs was studied in cotransfection experiments (Fig. 2A). We transiently expressed His-S-tagged full-length JSAP1 with Flag epitope-tagged JNK1, JNK2, JNK3, ERK2, or p38α in COS-7 cells. The His-S-JSAP1 proteins were recovered from the cell extracts by affinity binding to S-protein–agarose, and the precipitates were examined for the presence of the MAPKs by immunoblotting with an anti-Flag antibody. JNK1, JNK2, and JNK3 interacted with JSAP1, while no or very low binding of ERK2 and p38α to JSAP1 was observed. JNK3 showed higher binding affinity to JSAP1 than did JNK1 or JNK2.
FIG. 2.
(A) Binding of JSAP1 to mammalian MAPKs. COS-7 cells were transiently cotransfected with 0.4 μg of pFlag-CMV2-JNK1, -JNK2, -JNK3, -p38α or -ERK2 along with 1.1 μg of either pcDNA3-His-S empty vector or pcDNA3-His-S-JSAP1. Cell lysates were precipitated with S-protein-linked agarose (S-PA) and analyzed by immunoblotting with anti-Flag antibody. Expression of Flag-MAPKs and His-S-JSAP1 was examined by immunoblotting 1/10 of the cell lysates used in the binding reactions. (B) Mapping of the JNK3-binding region on JSAP1. In vitro-translated 35S-labeled JNK3 was incubated with 0.5 μg of either immobilized Trx-His-S protein or Trx-His-S-tagged segments of JSAP1. The protein complexes were extensively washed, and the bound JNK3 was detected by SDS-PAGE and autoradiography. One-tenth of the 35S-labeled protein used in the binding reactions was loaded as a positive control.
To define the region of JSAP1 responsible for its interaction with JNK, a series of Trx-His-S-JSAP1 fusion proteins containing various portions of JSAP1 were expressed in bacteria, purified by using S-protein–agarose, and assayed for the ability to bind JNK. The agarose-bound fusion proteins were mixed with in vitro-translated 35S-labeled JNK3, recovered, and analyzed by SDS-PAGE and autoradiography (Fig. 2B). Essentially the same results were obtained when JNK1 and JNK2 were used instead of JNK3 (data not shown). The results indicate that the JNK-binding region is located between residues 115 and 233 in JSAP1.
JSAP1 is phosphorylated by JNK.
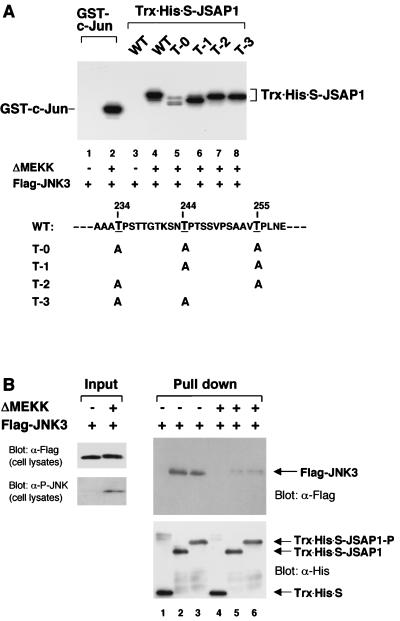
We then examined whether JSAP1 could serve as a substrate for JNK, because three potential sites for phosphorylation by proline-directed serine/threonine kinases, such as JNK, were located at residues 234, 244, and 255 (Fig. 3A), adjacent to the JNK-binding region (residues 115 to 233). Flag-JNK3 was transiently expressed in COS-7 cells with or without truncated MEKK1 (ΔMEKK), a strong activator of JNKs in vivo (28, 53), immunoprecipitated with an anti-Flag antibody, and examined for the ability to phosphorylate Trx-His-S-JSAP1 (residues 115 to 274). GST–c-Jun (residues 1 to 79) was used as a positive control (8, 11, 22). As shown in Fig. 3A, JSAP1 was phosphorylated by the activated JNK3 as efficiently as c-Jun (lanes 2 and 4). Several bands were seen, which probably reflected the phosphorylation of several sites in JSAP1. In fact, three mutant forms of JSAP1, in which each potential phosphorylation site was intact and the other two sites were changed to alanine, were all efficiently phosphorylated (lanes 6 to 8). Essentially the same results were obtained when JNK1 and JNK2 were used instead of JNK3 (data not shown). Although residues outside the region (residues 115 to 274) may also be phosphorylation sites for JNK, these results indicate that JSAP1 is an in vitro substrate for JNK and that all three sites at residues 234, 244, and 255 in JSAP1 can be phosphorylated by JNK.
FIG. 3.
Phosphorylation of JSAP1 by JNK3 (A) and its effect on the interaction with JNK3 (B). (A) COS-7 cells were transiently transfected with 1.5 μg of pFlag-CMV2-JNK3 with or without 0.05 μg of pEF-3XHA-ΔMEKK. Cell lysates were immunoprecipitated with anti-Flag antibody, and the in vitro kinase assay was carried out as described in Materials and Methods. GST–c-Jun (residues 1 to 79) and Trx-His-S-JSAP1 (residues 115 to 274) were used as substrates. The mutants of JSAP1 (T-0, T-1, T-2, and T-3) were as indicated. WT, wild type. The concentration of each substrate was 1 pmol/μl in 30 μl of total volume. (B) COS-7 cells were transiently transfected with 1.5 μg of pFlag-CMV2-JNK3 with or without 0.05 μg of pEF-3XHA-ΔMEKK. The unstimulated (lanes 1 to 3) and stimulated (lanes 4 to 6) Flag-JNK3 were precipitated from cell lysates with 0.5 μg of immobilized Trx-His-S protein (lanes 1 and 4) or the unphosphorylated (lanes 2 and 5) or phosphorylated (lanes 3 and 6) Trx-His-S-JSAP1 and analyzed by immunoblotting with anti-Flag or anti-His antibody. Expression of Flag-JNK3 was examined by immunoblotting 1/10 of the cell lysates used in the binding reactions. The phosphorylated form of Flag-JNK3 was detected with a phospho-specific JNK (P-JNK) antibody.
We performed an in vitro association experiment to examine whether the phosphorylation of JSAP1 has an effect on its interaction with JNK. Flag-JNK3 was first transiently expressed with or without ΔMEKK in COS-7 cells. Unstimulated and stimulated Flag-JNK3s were precipitated with the bacterially expressed, either unphosphorylated Trx-His-S-JSAP1 (residues 115 to 274) or the corresponding phosphorylated Trx-His-S-JSAP1 (at residues 234, 244, and 255), respectively. The precipitates were analyzed for the presence of JNK3 by Western blotting using an anti-Flag antibody (Fig. 3B). The phosphorylation state of Trx-His-S-JSAP1 was confirmed by its mobility in the gel (Fig. 3B). The results indicated that stimulated JNK3 had a much lower binding affinity for both the unphosphorylated and phosphorylated JSAP1 than unstimulated JNK3 did.
JSAP1 binds SEK1 MAPKK and MEKK1 MAPKKK.
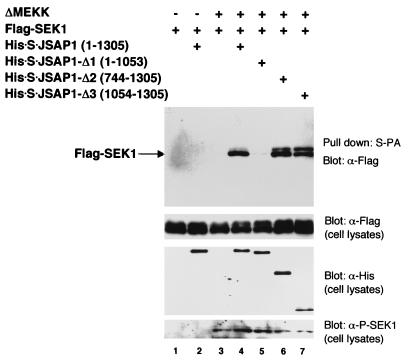
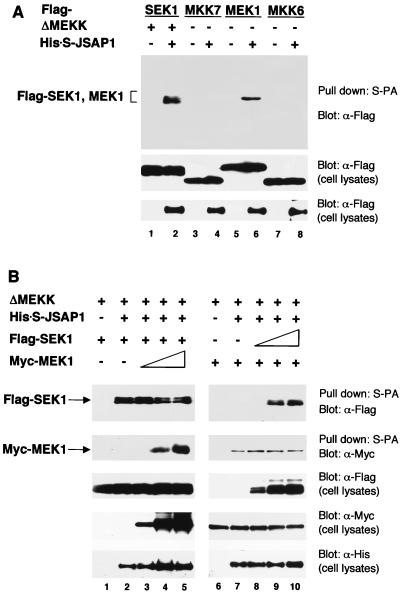
We next examined whether SEK1, a MAPKK in the JNK cascade, could bind to JSAP1 (Fig. 4). Flag-SEK1 and the full-length His-S-JSAP1 expression vectors were cotransfected into COS-7 cells with or without ΔMEKK expression vector and analyzed as in the MAPK/JSAP1 binding experiment. Although the unstimulated Flag-SEK1 and the His-S-JSAP1 were not copurified from the cells, ΔMEKK, an activator of SEK1 (53), significantly enhanced the binding affinity of Flag-SEK1 for His-S-JSAP1 (lanes 2 and 3). The presence of the phosphorylated, activated form of Flag-SEK1 was confirmed by Western blotting using a phospho-specific SEK1 antibody (Fig. 4, bottom). Furthermore, the SEK1-binding region in JSAP1 was mapped using a series of His-S-tagged deletion mutants of JSAP1 (Fig. 4, lanes 5 to 7). The results indicate that the C-terminal region (residues 1054 to 1305) of JSAP1 is responsible for the interaction with SEK1. Thus, the SEK1-binding region is distinct from the JNK-binding region (residues 115 to 233) in JSAP1. MKK7, MEK1, and MKK6, which are MAPKKs in the JNK, ERK, and p38 cascades, respectively, were also examined for the ability to bind JSAP1. Interestingly, MEK1, and not MKK7 or MKK6, bound to JSAP1 as efficiently as SEK1 (Fig. 5A). When MKK7, MEK1, and MKK6 MAPKKs were activated by UV irradiation, truncated Raf-1 (ΔRaf, a constitutively active Raf-1 [41]), and osmotic shock, respectively, they showed similar binding profiles for JSAP1 with the corresponding unstimulated MAPKKs under our assay conditions (data not shown). Increasing the amount of MEK1 had essentially no effect on the JSAP1-SEK1 interaction; similarly, increasing the amount of SEK1 did not affect the JSAP1-MEK1 interaction (Fig. 5B), indicating that the MEK1-binding site is distinct from the SEK1-binding site in JSAP1.
FIG. 4.
Binding of JSAP1 to SEK1 MAPKK and mapping of the SEK1-binding region on JSAP1. COS-7 cells were transiently cotransfected with 0.4 μg of pcDNA3-Flag-SEK1 and 1.1 μg of pcDNA3-His-S-JSAP1, -Δ1, -Δ2, or -Δ3 with or without 0.04 μg of pEF-3XHA-ΔMEKK, as indicated. Cell lysates were precipitated with S-protein-linked agarose (S-PA) and analyzed by immunoblotting with anti-Flag antibody. Expression of Flag-SEK1 and His-S-JSAP1, -Δ1, -Δ2, and -Δ3 was examined by immunoblotting 1/10 of the cell lysates used in the binding reactions. The phosphorylated form of Flag-SEK1 was detected using a phospho-specific SEK1 (P-SEK1) antibody.
FIG. 5.
(A) Binding of JSAP1 to mammalian MAPKKs. COS-7 cells were transiently cotransfected with 0.4 μg of pcDNA3-Flag-SEK1, -MKK7, -MEK1, or -MKK6 with 1.1 μg of either pcDNA3-His-S empty vector or pcDNA3-His-S-JSAP1 in the absence or presence of 0.04 μg of pEF-3XHA-ΔMEKK, as indicated. Cell lysates were precipitated with S-protein-linked agarose (S-PA) and analyzed as for Fig. 2A. Expression of Flag-MAPKKs and His-S-JSAP1 was examined by immunoblotting 1/10 of the cell lysates used in the binding reactions. (B) Competition analysis of SEK1 and MEK1 in the interaction with JSAP1. COS-7 cells were transiently cotransfected with 0.2 μg of pcDNA3-Flag-SEK1, 0.02 μg of pEF-3XHA-ΔMEKK, and 0.2 μg of either pcDNA3-His-S empty vector or pcDNA3-His-S-JSAP1 with different amounts of pcDNA3-Myc-MEK1 (0, 0, 0.05, 0.2, and 0.5 μg in lanes 1 to 5, respectively). COS-7 cells were transiently cotransfected with 0.1 μg of pcDNA3-Myc-MEK1, 0.02 μg of pEF-3XHA-ΔMEKK, and 0.2 μg of either pcDNA3-His-S empty vector or pcDNA3-His-S-JSAP1 with different amounts of pcDNA3-Flag-SEK1 (0, 0, 0.1, 0.3, and 0.6 μg in lanes 6 to 10, respectively). Total DNA was kept at 1.5 μg per transfection with pcDNA3-His-S empty vector. Cell lysates were precipitated with S-protein-linked agarose (S-PA) and analyzed as for Fig. 2A. Expression of Flag-SEK1, Myc-MEK1, and His-S-JSAP1 was examined by immunoblotting 1/10 of the cell lysates used in the binding reactions.
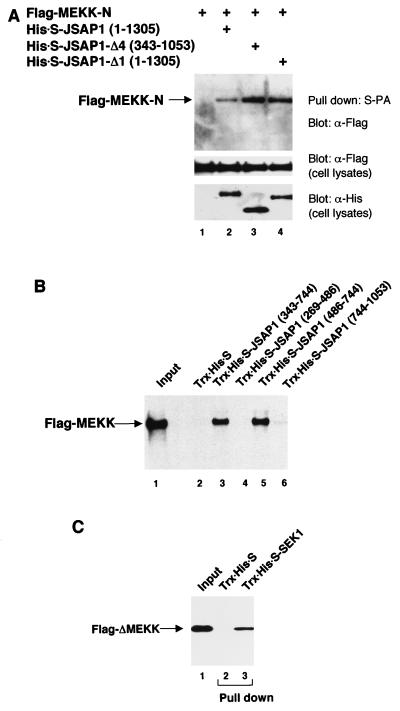
We next analyzed whether JSAP1 could bind to MEKK1, a MAPKKK in the JNK cascade, in cotransfected COS-7 cells. When full-length MEKK1 was expressed in these cells, smaller species of MEKK1 were detected, as was also observed by Xu et al. (50), and the expression level of the full-length protein was very low. We therefore expressed Flag-tagged portions of MEKK1 in these cells and examined their ability to bind to His-S-JSAP1. As shown in Fig. 6A, the N-terminal region (residues 1 to 640) of MEKK1 (MEKK-N) was found to interact with full-length JSAP1 (lane 2). None of the other regions of MEKK1 interacted with full-length JSAP1 under our assay conditions (data not shown). Moreover, the MEKK1-binding region of JSAP1 was mapped by using a series of His-S-tagged deletion mutants of JSAP1 (Fig. 6A, lanes 2 to 4). The results indicate that the internal region (residues 343 to 1053) of JSAP1 is responsible for the interaction with MEKK1. The C-terminal SEK1-binding region of JSAP1 could work as an inhibitor of the JSAP1-MEKK1 interaction, because MEKK1 bound to the JSAP1 mutants that lacked the C-terminal region with higher affinity than to full-length JSAP1 (lanes 2 to 4). Since the expression level of a JSAP1 fragment containing residues 745 to 1053 was very low, we used an in vitro pull-down method, as in the JNK3- and JSAP1-binding experiments, for more precise mapping of the MEKK1-binding region on JSAP1. Portions of JSAP1 were expressed as Trx-His-S-tagged fusion proteins in bacteria, purified by using S-protein–agarose, and assayed for the ability to bind MEKK1. The agarose-bound fusion proteins were mixed with in vitro-translated 35S-labeled full-length MEKK1, recovered, and analyzed by SDS-PAGE and autoradiography (Fig. 6B). The results indicate that the MEKK1-binding region is located between residues 486 and 744. Thus, the MEKK1-binding region is distinct from both the JNK3- and SEK1-binding regions (residues 115 to 233 and 1054 to 1305, respectively) in JSAP1.
FIG. 6.
Binding of JSAP1 to MEKK1 MAPKKK, mapping of the MEKK1-binding region on JSAP1, and binding of MEKK1 MAPKKK to SEK1 MAPKK. (A) COS-7 cells were transiently cotransfected with 0.4 μg of pcDNA3-Flag-MEKK-N with 1.1 μg of pcDNA3-His-S empty vector or pcDNA3-His-S-JSAP1, -Δ1, or -Δ4. Cell lysates were precipitated with S-protein-linked agarose (S-PA) and analyzed as for Fig. 2A. Expression of Flag-MEKK-N and His-S-JSAP1, -Δ1, and -Δ4 was examined by immunoblotting 1/10 of the cell lysates used in the binding reactions. (B) In vitro-translated 35S-labeled full-length MEKK was incubated with 0.5 μg of either immobilized Trx-His-S protein or Trx-His-S-tagged segments of JSAP1 and analyzed as for Fig. 2B. One-tenth of the 35S-labeled protein used in the binding reactions was loaded as a positive control. (C) COS-7 cells were transiently transfected with 1.5 μg of pcDNA3-Flag-ΔMEKK. Cell lysate was precipitated with 0.5 μg of either immobilized Trx-His-S protein (lane 2) or Trx-His-S-SEK1 (lane 3) and analyzed by immunoblotting with anti-Flag antibody. One-tenth of the cell lysate used in the binding reactions was loaded as a control (lane 1).
As only the stimulated SEK1 interacted with JSAP1 (Fig. 4), we further examined whether MEKK1 could bind to the unstimulated SEK1 as noted by Xia et al. (49). Flag-ΔMEKK was transiently expressed in COS-7 cells and pulled down with bacterially expressed, S-protein-agarose-bound Trx-His-S or Trx-His-S-SEK1. The precipitates were examined for the presence of Flag-ΔMEKK by immunoblotting with an anti-Flag antibody (Fig. 7C). The results indicates that ΔMEKK binds to the unstimulated SEK1.
FIG. 7.
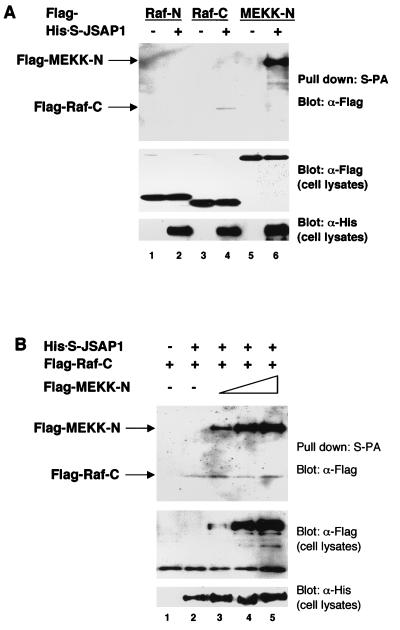
(A) Binding of JSAP1 to mammalian MAPKKKs. COS-7 cells were transiently cotransfected with 0.4 μg of pcDNA3-Flag-Raf-N, -Raf-C, or -MEKK-N with 1.1 μg of either pcDNA3-His-S empty vector or pcDNA3-His-S-JSAP1. (B) Competition analysis of MEKK1 and Raf-1 in the interaction with JSAP1. COS-7 cells were transiently cotransfected with 0.7 μg of pcDNA3-Flag-Raf-C and 0.2 μg of either pcDNA3-His-S empty vector or pcDNA3-His-S-JSAP1 with different amounts of pcDNA3-Flag-MEKK-N (0, 0, 0.05, 0.2 and 0.6 μg in lanes 1 to 5, respectively). Total DNA was kept at 1.5 μg per transfection with pcDNA3-His-S empty vector. Cell lysates were precipitated with S-protein-linked agarose (S-PA) and analyzed as for Fig. 2A. Expression of Flag-MAPKKKs and His-S-JSAP1 was examined by immunoblotting 1/10 of the cell lysates used in the binding reactions.
In addition, Raf-1 MAPKKK, which is involved in the ERK cascade, was examined for its ability to interact with JSAP1 (Fig. 7A). Since the expression level of full-length Raf-1 was very low, as was observed for MEKK1 in transfected COS-7 cells, the N-terminal half (residues 1 to 327) and the C-terminal half (residues 316 to 648) of Raf-1 (Raf-N and Raf-C, respectively) were expressed separately in the cells, and their interactions with JSAP1 were examined. Raf-C, and not Raf-N, bound to JSAP1; however, the binding affinity of Raf-C for JSAP1 was very low compared with that of MEKK1 with JSAP1 (lanes 4 and 6). Increasing the amount of MEKK-N had essentially no effect on the JSAP1–Raf-C interaction (Fig. 7B), and increasing the amount of Raf-C did not affect the JSAP1–MEKK-N interaction (data not shown), indicating that the MEKK1-binding site is distinct from the Raf-1-binding site in JSAP1.
Full-length JSAP1 enhances JNK activation.
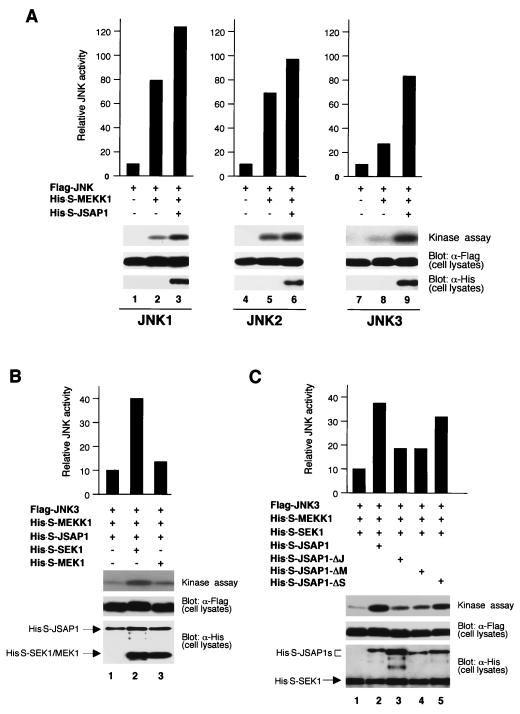
The results of our JSAP1-binding experiments suggest that JSAP1 acts as a scaffold protein in the JNK cascades. To examine this possibility, we analyzed the effect of overexpressing full-length JSAP1 on JNK activation by full-length MEKK1 (Fig. 8A). Flag-JNK1, -JNK2, or -JNK3 was transiently expressed alone or with His-S-MEKK1 in the absence or presence of His-S-JSAP1 in COS-7 cells. Cell lysates were immunoprecipitated with an anti-Flag antibody, and the JNK activity was measured by using GST–c-Jun (residues 1 to 79) as the substrate. MEKK1 activated JNK1, JNK2, and JNK3 8-, 6.8-, and 2.6-fold, respectively, and the activation was further enhanced 1.6-, 1.4-, and 3-fold, respectively, in the presence of JSAP1. JSAP1 alone without MEKK1 had little effect on the activity of JNK1, JNK2, and JNK3 in COS-7 cells (data not shown).
FIG. 8.
JSAP1 enhances the activation of JNK3 by MEKK1 and SEK1. (A) Effect of overexpressing JSAP1 on JNK activation by MEKK1. COS-7 cells were transiently transfected with 0.2 μg of pFlag-CMV2-JNK1 (lane 1), -JNK2 (lane 4), or -JNK3 (lane 7) alone or with 0.02 μg of pcDNA3-His-S-MEKK1 in the absence or presence of 1 μg of pcDNA3-His-S-JSAP1, as indicated. (B) Effect of overexpressing SEK1 and MEK1 on JNK3 activation by JSAP1 and MEKK1. COS-7 cells were transiently cotransfected with 0.2 μg of pFlag-CMV2-JNK3, 0.06 μg of pcDNA3-His-S-MEKK1, and 1 μg of pcDNA3-His-S-JSAP1 in the absence or presence of 0.2 μg of either pcDNA3-His-S-SEK1 or pcDNA3-His-S-MEK1, as indicated. (C) Effect of overexpressing JSAP1 mutants on JNK3 activation by MEKK1 and SEK1. COS-7 cells were transiently cotransfected with 0.2 μg of pFlag-CMV2-JNK3, 0.06 μg of pcDNA3-His-S-MEKK1, and 0.2 μg of pcDNA3-His-S-SEK1 with 1 μg of pcDNA3-His-S empty vector and pcDNA3-His-S-JSAP1, -ΔJ, -ΔM, or -ΔS, as indicated. Total DNA was kept at 1.5 μg per transfection with pcDNA3-His-S empty vector. Cell lysates were immunoprecipitated with anti-Flag antibody, and kinase activity was measured by using GST–c-Jun (residues 1 to 79) as the substrate. Expression of Flag-JNKs, His-S-JSAP1, His-S-SEK1, and His-S-MEK1 was examined by immunoblotting 1/10 of the cell lysates used in the kinase assays.
We next examined whether SEK1 or MEK1 could activate JNK in the presence of JSAP1 and MEKK1 (Fig. 8B), since they both interacted with JSAP1 (Fig. 5A). Flag-JNK3, His-S-JSAP1, and His-S-MEKK1 were transiently coexpressed in the absence or presence of either His-S-SEK1 or His-S-MEK1 in COS-7 cells. The JNK3 activity was enhanced fourfold by SEK1 in the presence of JSAP1 and MEKK1. When JNK1 (or JNK2) was used instead of JNK3, a modest enhancement of the JNK activation was observed (data not shown). MEK1 had little effect on the JNK3 activation, suggesting that MEK1 could not activate JNK3 in the presence of JSAP1 and MEKK1. We further studied whether JSAP1 mutants lacking the JNK-, SEK1-, and MEKK1-binding regions could enhance the JNK3 activation by MEKK1 and SEK1 as full-length JSAP1 did in COS-7 cells (Fig. 8C). JSAP1 mutants lacking the JNK- or MEKK1-binding regions resulted in significant reduction of the enhancement of the JNK3 activation. In contrast, a JSAP1 mutant lacking the SEK1-binding region acted similarly to full-length JSAP1. Taken together, these results suggest that JSAP1 functions as a scaffold protein in the JNK3 cascade and that stimulated SEK1 could activate JNK3 without direct interaction with JSAP1.
DISCUSSION
In this study we have identified a novel mouse JNK-binding protein, JSAP1, and examined its interactions with a variety of mammalian MAPKs, MAPKKs, and MAPKKKs through cotransfection studies of COS-7 cells. JSAP1 coprecipitated with JNK1, JNK2, and JNK3 MAPKs but not with ERK2 or p38α MAPKs. JNK3 showed higher binding affinity to JSAP1 than did JNK1 or JNK2. Furthermore, JSAP1 interacted with SEK1 MAPKK and MEKK1 MAPKKK, which are involved in the JNK cascades. Importantly, the regions of JSAP1 that bound JNK, SEK1, and MEKK1 were distinct from one another. JNK and MEKK1 also interacted with JSAP1 in vitro; thus, these interactions are likely to be direct. In contrast, only the stimulated SEK1 interacted with JSAP1, and the unstimulated SEK1 interacted with MEKK1. The amino- and carboxy-terminal regions of MEKK1 interacted with JSAP1 and SEK1, respectively. Thus, SEK1 could indirectly associate with JSAP1 through MEKK1. Although there is no direct evidence, the binding properties of JSAP1 and MEKK1 as described above indicate that JSAP1 may tether JNK MAPK, SEK1 MAPKK, and MEKK1 MAPKKK in a complex, a role similar to that the yeast scaffold protein Ste5. Ste5 forms a multicomponent complex with Fus3 (or Kss1) MAPK, Ste7 MAPKK, and Ste11 MAPKKK to facilitate the specific and efficient activation of the mating pheromone pathway (3, 21, 27, 34).
Overexpression of full-length JSAP1 in COS-7 cells led to a considerable enhancement of the JNK3 activation, and modest enhancement of the JNK1 and JNK2 activation, by the MEKK1-SEK1 pathway. Deletion of the JNK- and MEKK1-binding regions significantly reduced of the enhancement of the JNK3 activation in COS-7 cells. However, a JSAP1 mutant lacking the SEK1-binding region acted similarly to wild-type full-length JSAP1. Taken together, these results suggest that JSAP1 functions as a scaffold protein in the JNK3 cascade and that stimulated SEK1 could activate JNK3 without direct interaction with JSAP1. What is the role of the binding of the stimulated SEK1 to JSAP1? Although SEK1 can activate both JNK and p38 (9, 26), the SEK1-JSAP1 interaction may prohibit the stimulated SEK1 by JSAP1-associated MEKK1 from activating p38. It is also possible that a JSAP1 complex binding stimulated SEK1 is involved in amplification of JNK3 activation. At present, a scaffolding role of JSAP1 in the JNK1 and JNK2 cascades is not clear. While JNK3 and JSAP1 are expressed predominantly in the brain, both JNK1 and JNK2 are widely expressed. Thus, COS-7 cells, for example, may include multicomponent complexes containing JNK1 and JNK2, organized by a scaffold protein(s) other than JSAP1. The higher activation of JNK1 and JNK2 than of JNK3 by MEKK1 alone (Fig. 8A) might support this idea. The formation of a complex by the transiently coexpressed JNK1 (or JNK2), JSAP1, and MEKK1 in COS-7 cells could be interfered with by the presence of other scaffold proteins. Taking this into account, we cannot rule out the possibility that JSAP1 can also function as a scaffold protein in the JNK1 and JNK2 cascades. To clarify this issue, it should be useful to examine the effect of JSAP1 on the JNK1 (and JNK2) activation in JNK-free system, such as yeast. Recently, Xia et al. (49) have proposed a sequential interaction model for the organization of the MEKK1-SEK1-JNK module. JSAP1 may stabilize this signaling module further.
JSAP1 is an in vitro substrate for JNK, and three sites at residues 234, 244, and 255, adjacent to the JNK-binding region (residues 115 to 233), can be phosphorylated by JNK (Fig. 3A). Although we have not examined whether the other sites in JSAP1 can also be phosphorylated by JNK, our preliminary results showed that these three sites, and not others, are the major phosphorylation sites for JNK (17). Phosphorylated JSAP1 had little binding affinity for the activated form of JNK3 (Fig. 3B). Taken together, these data suggest that once activated, JNK dissociates from JSAP1 by phosphorylating it. Thus, a mutant JSAP1 containing substitutions at residues 234, 244, and 255 could be developed as a specific inhibitor for the JNK cascades.
JSAP1 contains a leucine zipper motif with six leucine repeats (residues 392 to 427 [Fig. 1A]), which may mediate the homo- and/or heterodimerization of JSAP1. In fact, two different epitope-tagged JSAP1 fragments encompassing the leucine zipper motif associated with each other in cotransfected cells (17). The yeast scaffold protein Ste5 can also self-associate, and importantly, dimerization of Ste5 is essential for the mating pheromone pathway (51). Thus, it should be interesting to examine whether dimerization of JSAP1 is required for its function.
JSAP1 exhibited different binding affinities for the signaling components of the ERK cascade in cotransfected COS-7 cells. JSAP1 coprecipitated with MEK1 MAPKK and Raf-1 MAPKKK but not ERK2 MAPK. Thus, even though MEK1 and Raf-1 did not interfere with the binding of JSAP1 to SEK1 and MEKK1, respectively, JSAP1 may affect the ERK cascade. In fact, overexpression of full-length JSAP1 inhibited the activation of the ERK cascade in COS-7 cells (17). Highly expressed JSAP1 could absorb MEK1 and/or Raf-1 from an ERK signaling complex tethered by an unidentified scaffold protein or an adapter protein such as MP1 (39), resulting in the inhibition of ERK activation. At present, how JSAP1 is involved in vivo in signaling pathways other than the JNK cascades is not clear. However, an interesting possibility is that JSAP1 expression is positively autoregulated by the JNK cascades, so that the activation of the JNK cascades leads to the increased expression of JSAP1. Inhibition of the other cascades by JSAP1 would ensure the specific activation of the JNK cascades. In support of this idea, our preliminary experiments showed that a 5.2-kb genomic fragment containing the JSAP1 promoter sequence increases its transcriptional activity in response to overexpression of full-length JSAP1 (17). Further study is required to clarify this issue.
Recently, Whitmarsh et al. (47) reported that JIP-1 works as a scaffold factor in the JNK cascades. JSAP1 is clearly distinct from JIP-1, because JSAP1 contains a leucine zipper motif but not an SH3 domain; JIP-1 contains an SH3 domain but not a leucine zipper motif. More importantly, JSAP1 and JIP-1 bind distinct sets of kinases. JSAP1 organizes the MEKK1-SEK1-JNK signaling module; JIP-1 organizes the MLK-MKK7-JNK signaling module. In spite of these differences, JSAP1 and JIP-1 appear to function similarly in cells, selectively enhancing the activation of signaling pathways. Furthermore, Schaeffer et al. (39) and Cohen et al. (5) have identified an adapter protein, MP1, for MEK1 and ERK1, and a scaffold protein, IKAP, in the IκB kinase complex. In addition, current work in our laboratory has revealed the existence of other JSAP1 family members. These scaffold/adapter proteins, together with unidentified related proteins, could contribute to the specificity determination of numerous distinct signaling pathways in cells.
ACKNOWLEDGMENTS
M. Ito and K. Yoshioka contributed equally to this work.
We thank Rikiro Fukunaga for helpful discussion, Atsushi Yamashita for RNA samples, and Yoshiyuki Sakaki for encouragement.
This work was supported in part by grants from the Kitasato Research Foundation (M.I.), the Waksman Foundation of Japan Inc. (T.S.), grants-in-aid from the Ministry of Education, Science, Sports and Culture in Japan (K. Yoshioka), and the Kato Memorial Foundation (K. Yoshioka).
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1990. [Google Scholar]
- 2.Cano E, Mahadevan L C. Parallel signal processing among mammalian MAPKs. Trends Biochem Sci. 1995;20:117–122. doi: 10.1016/s0968-0004(00)88978-1. [DOI] [PubMed] [Google Scholar]
- 3.Choi K-Y, Satterberg B, Lyons D M, Elion E A. Ste5 tethers multiple protein kinases in the MAP kinase cascade required for mating in S. cerevisiae. Cell. 1994;78:499–512. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 4.Cobb M H, Goldsmith E J. How MAP kinases are regulated. J Biol Chem. 1995;270:14843–14846. doi: 10.1074/jbc.270.25.14843. [DOI] [PubMed] [Google Scholar]
- 5.Cohen L, Henzel W J, Baeuerle P A. IKAP is a scaffold protein of the IκB kinase complex. Nature. 1998;395:292–296. doi: 10.1038/26254. [DOI] [PubMed] [Google Scholar]
- 6.Cuenda A, Alonso G, Morrice N, Jones M, Meier R, Cohen P, Nebreda A R. Purification and cDNA cloning of SAPKK3, the major activator of RK/p38 in stress- and cytokine-stimulated monocytes and epithelial cells. EMBO J. 1996;15:4156–4164. [PMC free article] [PubMed] [Google Scholar]
- 7.Davis R J. MAPKs: new JNK expands the group. Trends Biochem Sci. 1994;19:470–473. doi: 10.1016/0968-0004(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 8.Dérijard B, Hibi M, Wu I-H, Barrett T, Su B, Deng T, Karin M, Davis R J. JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 9.Dérijard B, Raingeaud J, Barrett T, Wu I-H, Han J, Ulevitch R J, Davis R J. Independent human MAP kinase signal transduction pathway defined by MEK and MKK isoforms. Science. 1995;267:682–685. doi: 10.1126/science.7839144. [DOI] [PubMed] [Google Scholar]
- 10.Gerwins P, Blank J L, Johnson G L. Cloning of a novel mitogen-activated protein kinase kinase kinase, MEKK4 that selectively regulates the c-Jun amino terminal kinase pathway. J Biol Chem. 1997;272:8288–8295. doi: 10.1074/jbc.272.13.8288. [DOI] [PubMed] [Google Scholar]
- 11.Gupta S, Barrett T, Whitmarsh A J, Cavanagh J, Sluss H K, Dérijard B, Davis R J. Selective interaction of JNK protein isoforms with transcription factors. EMBO J. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 12.Han J, Lee J-D, Bibbs L, Ulevitch R J. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cell. Science. 1994;265:808–811. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 13.Han J, Lee J-D, Jiang Y, Li Z, Feng L, Ulevitch R J. Characterization of the structure and function of a novel MAP kinase kinase (MKK6) J Biol Chem. 1996;271:2886–2891. doi: 10.1074/jbc.271.6.2886. [DOI] [PubMed] [Google Scholar]
- 14.Herskowitz I. MAP kinase pathway in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 15.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 16.Ichijo H, Nishida E, Irie K, Dijke P, Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K, Gotoh Y. Induction of Apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 17.Ito, M., and K. Yoshioka. Unpublished data.
- 18.Jiang Y, Chen C, Li Z, Guo W, Gegner J A, Lin S, Han J. Characterization of the structure and function of a new mitogen-activated protein kinase (p38β) J Biol Chem. 1996;271:17920–17926. doi: 10.1074/jbc.271.30.17920. [DOI] [PubMed] [Google Scholar]
- 19.Kallunki T, Su B, Tsigelny I, Sluss H K, Dérijard B, Moore G, Davis R J, Karin M. JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev. 1994;8:2996–3007. doi: 10.1101/gad.8.24.2996. [DOI] [PubMed] [Google Scholar]
- 20.Kallunki T, Deng T, Hibi M, Karin M. c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- 21.Kranz J E, Satterberg B, Elion E A. The MAP kinase Fus3 associates with and phosphorylates the upstream signaling component Ste5. Genes Dev. 1994;8:313–327. doi: 10.1101/gad.8.3.313. [DOI] [PubMed] [Google Scholar]
- 22.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. The stress-activated protein kinase subfamily of c-Jun kinases. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 23.Kyriakis J M, Avruch J. Sounding the alarm: protein kinase cascades activated by stress and inflammation. J Biol Chem. 1996;271:24313–24316. doi: 10.1074/jbc.271.40.24313. [DOI] [PubMed] [Google Scholar]
- 24.Lange-Carter C A, Pleiman C M, Gardner A M, Blumer K J, Johnson G L. A divergence in the MAP kinase regulatory network defined by MEK kinase and Raf. Science. 1993;260:315–319. doi: 10.1126/science.8385802. [DOI] [PubMed] [Google Scholar]
- 25.Lee J C, Laydon J T, McDonnell P C, Gallagher T F, Kumar S, Green D, McNulty D, Blumenthal M J, Heys J R, Landvatter S W, Strickler J E, McLaughlin M M, Siemens I R, Fisher S M, Livi G P, White J R, Adams J L, Young P R. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 26.Lin A, Minden A, Martinetto H, Claret F-X, Lange-Carter C, Mercurio F, Johnson G L, Karin M. Identification of a dual specificity kinase that activates the Jun kinases and p38-Mpk2. Science. 1995;268:286–290. doi: 10.1126/science.7716521. [DOI] [PubMed] [Google Scholar]
- 27.Marcus S, Polverino A, Barr M, Wigler M. Complex between STE5 and components of the pheromone-responsive mitogen-activated protein kinase module. Proc Natl Acad Sci USA. 1994;91:7762–7766. doi: 10.1073/pnas.91.16.7762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minden A, Lin A, McMahon M, Lange-Carter C, Dérijard B, Davis R J, Johnson G L, Karin M. Differential activation of ERK and JNK mitogen-activated protein kinases by Raf-1 and MEKK. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 29.Minden A, Karin M. Regulation and function of the JNK subgroup of MAP kinases. Biochim Biophy Acta. 1997;1333:F85–F104. doi: 10.1016/s0304-419x(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 30.Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohit A A, Martin J H, Miller C A. p493F12 kinase: a novel MAP kinase expressed in a subset of neurons in the human nervous system. Neuron. 1995;14:67–78. doi: 10.1016/0896-6273(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 32.Moriguchi T, Kuroyanagi N, Yamaguchi K, Gotoh Y, Irie K, Kano T, Shirakabe K, Muro Y, Shibuya H, Matsumoto K, Nishida E, Hagiwara M. A novel kinase cascade mediated by mitogen-activated protein kinase kinase 6 and MKK3. J Biol Chem. 1996;271:13675–13679. doi: 10.1074/jbc.271.23.13675. [DOI] [PubMed] [Google Scholar]
- 33.Moriguchi T, Toyoshima F, Masuyama N, Hanafusa H, Gotoh Y, Nishida E. A novel SAPK/JNK kinase, MKK7, stimulated by TNFα and cellular stresses. EMBO J. 1997;16:7045–7053. doi: 10.1093/emboj/16.23.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Printen J A, Sprague G F., Jr Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics. 1994;138:609–619. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raingeaud J, Whitmarsh A J, Barrett T, Dérijard B, Davis R J. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol Cell Biol. 1996;16:1247–1255. doi: 10.1128/mcb.16.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson M J, Cobb M H. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 37.Rouse J, Cohen P, Trigon S, Morange M, Alonso-Llamazares A, Zamanillo D, Hunt T, Nebreda A R. A novel kinase cascade triggered by stress and heat shock that stimulates MAPKAP kinase-2 and phosphorylation of the small heat shock proteins. Cell. 1994;78:1027–1037. doi: 10.1016/0092-8674(94)90277-1. [DOI] [PubMed] [Google Scholar]
- 38.Sánchez I, Hughes R T, Mayer B J, Yee K, Woodgett J R, Avruch J, Kyriakis J M, Zon L I. Role of SAPK/ERK kinase-1 in the stress-activated pathway regulating transcription factor c-Jun. Nature. 1994;372:794–798. doi: 10.1038/372794a0. [DOI] [PubMed] [Google Scholar]
- 39.Schaeffer H J, Catling A D, Eblen S T, Collier L S, Krauss A, Weber M J. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science. 1998;281:1668–1671. doi: 10.1126/science.281.5383.1668. [DOI] [PubMed] [Google Scholar]
- 40.Shankar S, Mohapatra B, Suri A. Cloning of a novel human testis mRNA specifically expressed in testicular haploid germ cells, having unique palindromic sequences and encoding a leucine zipper dimerization motif. Biochem Biophy Res Commun. 1998;243:561–565. doi: 10.1006/bbrc.1997.7943. [DOI] [PubMed] [Google Scholar]
- 41.Stanton V P, Jr, Nichols D W, Laudano A P, Cooper G M. Definition of the human raf amino-terminal regulatory region by deletion mutagenesis. Mol Cell Biol. 1989;9:639–647. doi: 10.1128/mcb.9.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stein B, Brady H, Yang M X, Young D B, Barbosa M S. Cloning and characterization of MEK6, a novel members of the mitogen-activated protein kinase kinase cascade. J Biol Chem. 1996;271:11427–11433. doi: 10.1074/jbc.271.19.11427. [DOI] [PubMed] [Google Scholar]
- 43.Tibbles L A, Ing Y L, Kiefer F, Chan J, Iscove N, Woodgett J R, Lassam N J. MLK-3 activates the SAPK/JNK and p38/RK pathways via SEK1 and MKK3/6. EMBO J. 1996;15:7026–7035. [PMC free article] [PubMed] [Google Scholar]
- 44.Tournier C, Whitmarsh A J, Cavanagh J, Barrett T, Davis R J. Mitogen-activated protein kinase kinase 7 is an activator of the c-Jun NH2-terminal kinase. Proc Natl Acad Sci USA. 1997;94:7337–7342. doi: 10.1073/pnas.94.14.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Treisman R. Regulation of transcription by MAP kinase cascades. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 46.Waskiewicz A J, Cooper J A. Mitogen and stress response pathway: MAP kinase cascades and phosphatase regulation in mammals and yeast. Curr Opin Cell Biol. 1995;7:798–805. doi: 10.1016/0955-0674(95)80063-8. [DOI] [PubMed] [Google Scholar]
- 47.Whitmarsh A J, Cavanagh J, Tournier C, Yasuda J, Davis R J. A mammalian scaffold complex that selectively mediates MAP kinase activation. Science. 1998;281:1671–1674. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 48.Wu Z, Wu J, Jacinto E, Karin M. Molecular cloning and characterization of human JNKK2, a novel Jun NH2-terminal kinase-specific kinase. Mol Cell Biol. 1997;17:7407–7416. doi: 10.1128/mcb.17.12.7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xia Y, Wu Z, Su B, Murray B, Karin M. JNKK1 organizes a MAP kinase module through specific and sequential interactions with upstream and downstream components mediated by its amino-terminal extension. Genes Dev. 1998;12:3369–3381. doi: 10.1101/gad.12.21.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu S, Robbins D J, Christerson L B, English J M, Vanderbilt C A, Cobb M H. Cloning of Rat MEK kinase 1 cDNA reveals an endogenous membrane-associated 195-kDa protein with a large regulatory domain. Proc Natl Acad Sci USA. 1996;93:5291–5295. doi: 10.1073/pnas.93.11.5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yablonski D, Marbach I, Levitzki A. Dimerization of Ste5, a mitogen-activated protein kinase cascade scaffold protein, is required for signal transduction. Proc Natl Acad Sci USA. 1996;93:13864–13869. doi: 10.1073/pnas.93.24.13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K. Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science. 1995;270:2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- 53.Yan M, Dai T, Deak J C, Kyriakis J M, Zon L I, Woodgett J R, Templeton D J. Activation of stress-activated protein kinase by MEKK1 phosphorylation of its activator SEK1. Nature. 1994;372:798–800. doi: 10.1038/372798a0. [DOI] [PubMed] [Google Scholar]
- 54.Yang D D, Kuan C-Y, Whitmarsh A J, Rincón M, Zheng T S, Davis R J, Rakic P, Flavell R A. Absence of excitotoxicity-induced apoptosis in the hippocampus of mice lacking the Jnk3 gene. Nature. 1997;389:865–870. doi: 10.1038/39899. [DOI] [PubMed] [Google Scholar]
- 55.Yoshioka K, Deng T, Cavigelli M, Karin M. Antitumor promotion by phenolic antioxidants: inhibition of AP-1 activity through induction of Fra expression. Proc Natl Acad Sci USA. 1995;92:4972–4976. doi: 10.1073/pnas.92.11.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zervos A S, Faccio L, Gatto J P, Kyriakis J M, Brent R. Mxi2, a mitogen-activated protein kinase that recognizes and phosphorylates Max protein. Proc Natl Acad Sci USA. 1995;92:10531–10534. doi: 10.1073/pnas.92.23.10531. [DOI] [PMC free article] [PubMed] [Google Scholar]