Abstract
Plasma membrane repair mechanisms are activated within seconds post-injury to promote rapid membrane resealing in eukaryotic cells and prevent cell death. However, less is known about the regeneration phase that follows and how cells respond to injury in the short-term. Here, we provide a genome-wide study into the mRNA expression profile of MCF-7 breast cancer cells exposed to injury by digitonin, a mild non-ionic detergent that permeabilizes the plasma membrane. We focused on the early transcriptional signature and found a time-dependent increase in the number of differentially expressed (> twofold, P < 0.05) genes (34, 114 and 236 genes at 20-, 40- and 60-min post-injury, respectively). Pathway analysis highlighted a robust and gradual three-part transcriptional response: (1) prompt activation of immediate-early response genes, (2) activation of specific MAPK cascades and (3) induction of inflammatory and immune pathways. Therefore, plasma membrane injury triggers a rapid and strong stress and immunogenic response. Our meta-analysis suggests that this is a conserved transcriptome response to plasma membrane injury across different cell and injury types. Taken together, our study shows that injury has profound effects on the transcriptome of wounded cells in the regeneration phase (subsequent to membrane resealing), which is likely to influence cellular status and has been previously overlooked.
Subject terms: Cell biology, Computational biology and bioinformatics, Genetics, Molecular biology
Introduction
It is well established that cells are rapidly able to mount a repair response towards a torn membrane through evolutionary conserved repair mechanisms, thus preventing cell death5,6. However, there is a growing appreciation for what happens after initial membrane resealing, when a continuous membrane is re-established (in the sub-second to second range7–9). The regeneration phase occurs within minutes to hours10–13 and is critical to ensure adequate cellular functioning. In fact, it has been proposed that pathologies may arise not only from defects in membrane resealing but also from the consequences of poor regeneration capacity13. We envision that a better understanding of the underlying mechanisms will pave way for the establishment of strategies that can modulate these processes. These strategies would either promote efficient repair in pathologies caused or worsen by poor membrane integrity11,14 or inhibit repair in cancer cells to counteract their enhanced membrane repair capacity15–19.
It has been reported that the expression profile changes upon plasma membrane (PM) stress2,3,20,21. However, only very few studies have analysed the transcriptome after injury and initial repair of the PM in an unbiased and genome-wide manner1,4,22,23. These studies tend to focus on long-term expression changes (> 6 days post-injury)22,23 and, to our knowledge, no transcriptome analysis has been conducted in the short-term (< 1 h post-injury). Intermediate (few hours) and long-term (24 h) potentiation of membrane resealing at a cellular level, through protein synthesis and gene expression changes respectively, has been suggested as an adaptive response to promote more effective membrane repair3. Therefore, understanding which genes and signalling pathways become activated and inhibited is of interest. Ultimately, such knowledge could help to explain the mechanisms that are key for membrane restructuring after initial resealing.
Models of injury have been mainly neuronal23,24, renal22, cardiac4, smooth muscle21 and skin2,3,20,21, while cancer cells have been largely overlooked. Breast carcinoma cells are of particular interest when investigating cellular changes occurring after wounded cells have resealed their membranes because they are known to be very efficient at recovering from injury, to ensure survival during invasion and metastasis. We have shown that they do this partly by upregulating PM repair proteins17, but other injury-induced responses may also contribute.
The aim of this study was to decipher the short-term transcriptomic response related to membrane injury induced by digitonin, a non-ionic saponin that permeabilizes the PM in a cholesterol dependent manner. The two-part experimental design began with the analysis of the transcriptome profile of MCF-7 breast carcinoma cells early during post-injury (20 min, 40 min and 60 min) by RNA-sequencing (RNA-seq). Firstly, we found that immediate-early response genes (IEGs) were among the most upregulated genes across all timepoints and their mRNA expression increased in a time-dependent manner. IEGs are known to be activated and transcribed within minutes to a wide variety of extrinsic stimuli and have essential functions in cellular stress responses including the immune system25. Secondly, pathway analysis based on the mRNA changes predicted that from 40 min post-injury, but mainly at 60 min, certain kinase-dependent pathways may be activated, including Mitogen-activated protein kinase (MAPK), Protein kinase C (PKC), Phosphoinositide-3 kinase (PI3K) and Extracellular signal-regulated kinase 5 (ERK5). Thirdly, our analysis also predicted a pronounced activation of multiple inflammation and immune-related pathways, including interleukin (IL) signalling, toll-like-receptor (TLR) signalling and cascades mediated by reactive oxygen species (ROS). The second part of our experimental setup investigated the conserved nature of the 1 h transcriptomic response after PM injury or its immediate downstream effect, Ca2+ influx, in other models. Interestingly, our meta-analysis revealed that this three-part transcriptional response to PM injury is conserved1,4. The existence of such early and strong immunogenic response was not previously acknowledged.
Methods
Cell culture
Human breast carcinoma cells, MCF-7, were cultured at 37 °C and 5% CO2 in RPMI 1640 (Gibco) supplemented with 6% heat-inactivated fetal bovine serum (FBS) and 0.25% Penicillin–Streptomycin, which promoted optimal growth conditions.
Detergent-induced PM wounding experiments
Cholesterol-dependent cytolysins are known to create pores in cellular membranes26,27. For example, digitonin is a commonly used membrane pore-forming at cholesterol-rich regions. Because it achieves wide-ranging PM injury, digitonin was used to induce PM wounding. MCF-7 cells were first grown to 70–80% confluency in 10 cm cell culture dishes. Cells were briefly washed with DPBS–Ca2+ and then injured by a 10 min incubation with 10–15 µg/ml digitonin, as indicated, diluted in HBSS ± 1.26 mM Ca2+ (Gibco). The optimal concentration of digitonin was determined prior to its use in the RNA-seq experiment and was one that causes the majority of cells to die when repair mechanisms are compromised (in the absence of Ca2+), while only a minority die when repair mechanisms are competent (under 20% cell death). Therefore, for concentration optimisation experiments, digitonin is incubated in HBSS ± Ca2+, while subsequent experiments use HBSS + Ca2+ and the absence of digitonin as the negative control.
Membrane integrity assay
For cell death measurements, after injury cells were incubated with cell-permeable Hoechst-33342 (5 µg/ml) (Sigma-Aldrich) and cell-impermeable propidium iodide (PI) (0.2 µg/ml) (Sigma-Aldrich) diluted in RPMI media. After 5 min, they were imaged using an IX71 microscope coupled to a DP72 camera and Cell^P software (Olympus), followed by automated particle counting using ImageJ. As part of the quantification pipeline, images were subjected to an arbitrary and fixed intensity threshold and binary images were created. Next, the watershed algorithm in ImageJ ran on these images to resolve “touching objects” (i.e. cells in close proximity). Finally, both Hoechst-positive and PI-positive cells were quantified by the particle analysis plugin.
RNA-seq
At 20-, 40- and 60-min post-injury cells were harvested in TRIzol™ (Invitrogen™) and flash-frozen in liquid nitrogen. Control cells (uninjured) were harvested in the same way. RNA extraction, library construction and transcriptome sequencing were performed by BGI Tech Solutions Hong Kong Co. In total 12 samples (3 samples per condition) were sequenced with 100 bp reads, paired end and at a depth of 20 million reads per sample using Illumina’s HiSeq 4000.
Bioinformatic analysis
The RNA-seq data was pre-processed before being used for downstream analysis28. The pre-processing pipeline included 5 steps: (1) Quality control of the reads was conducted using FastQC, with default settings. (2) Trimmomatic paired end mode was used to trim reads from the 3’ end to remove bases with quality score below 5 and keeping only reads which were longer than 24 bases after trimming. (3) Alignment of the reads to the hg38 reference transcriptome was performed using TopHat2 with the following parameter settings: segment-mismatches = 2, segment-length = 25, and no-coverage-search. (4) Quality control of the aligned reads was conducted using RseQC to get summary statistics about read distribution and transcript coverage distribution. 5) Quantification of reads into integer counts to be used for downstream analysis was performed by using HTSeq with–stranded = no settings.
The downstream analyses included differential expression analysis, gene ontology analysis, pathway enrichment analysis and analysis of upstream pathway regulators. The differential expression analysis was carried out using edgeR29 and the quasi-likelihood generalized linear model, after removal of genes expressed at low levels and normalization for library size. We defined significantly expressed genes as genes having FDR < 0.05 and absolute log2 fold change > 1. GO analysis was carried out on both the significantly upregulated and downregulated genes (separated analysis) from the expression analysis using the TopGO R package30 (default parameters). The following parameters are calculated: the total number of genes that are annotated with that specific term is denoted (“Annotated”), the number of significantly differentially expressed genes associated with that term (“Significant”), and the number of genes that would be expected to be significantly associated to each term by random chance (“Expected”). The analysis uses the Fisher’s Exact Test to test the association, and the P-values and FDR are calculated.
Pathway enrichment analysis was conducted using ReactomePA31 and Ingenuity Pathway Analysis, IPA (Qiagen). Both map different functional pathways to the network of proteins produced by the input genes (related to the mRNA transcripts) based on reference datasets, which for the former is the Ingenuity Knowledge Base32 and the latter is the Reactome Knowledge Base33. For the analyses using ReactomePA, only DE genes (related to the mRNA transcripts) were considered. This R-package had the p-value and FDR cutoffs set to 0.05. For the analysis using IPA, all genes (related to the mRNA transcripts) were considered along with their relative expression changes and p-values. The cut-off used for the predictive pathway analysis was of − log(P-value) > 1.3, which corresponds to P-value < 0.05. IPA was not only used for predicting the canonical pathways related to the transcriptomic changes, but also for upstream regulator analyses. The calculated z-scores reflect the predicted activation state of the pathway or potential upstream regulator (a positive/negative z-score indicates activation/inhibition, respectively). While its directionality reflects the predicted activation state, its absolute value predicts the degree of activation/inhibition in a relative manner. This is achieved by comparing transcriptomic changes to molecular networks and literature-derived regulation direction (activation/inhibition)32.
All the scripts and processed data related to the bioinformatic analyses are reported in the GitHub repository https://github.com/ELELAB.
Results
Plasma membrane injury by digitonin in MCF-7 breast carcinoma cells
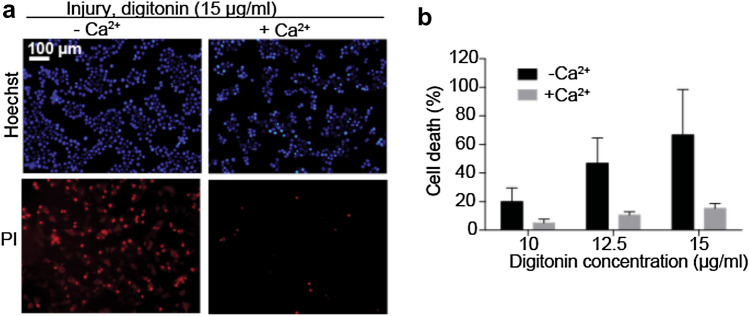
Digitonin was used to induce PM injury in MCF-7 breast cancer cells and activates an efficient calcium (Ca2+)-dependent repair mechanism to prevent propidium iodide (PI) cellular inclusion (Fig. 1a), in accordance with our earlier findings6,17,34,35. To determine a concentration of digitonin that would led to PM damage to a degree that could be repaired, MCF-7 cells were injured in presence or absence of Ca2+ for 10 min followed by a 5-min repair period (Fig. 1b). In these experiments PI is not only being used as an indicator of necrotic death, but also a measure of increased membrane permeability (that of the PM and nuclear envelope). All future experiments were conducted using 15 µg/ml of digitonin.
Figure 1.
Cells repair from digitonin-induced PM injury in a Ca2+-dependent manner. (a) Representative images of MCF-7 cells injured by 10 min incubation in 15 µg/ml digitonin in the absence (left panel) or presence (right panel) of Ca2+ and then assayed using impermeable PI staining and Hoechst 33,342 (permeable). (b) Percentages of PI-positive cells after injury with 10, 12.5 or 15 µg/ml digitonin. Mean and SD of biological triplicates.
Transcriptome analysis of MCF-7 cells post-injury
Differentially expressed genes
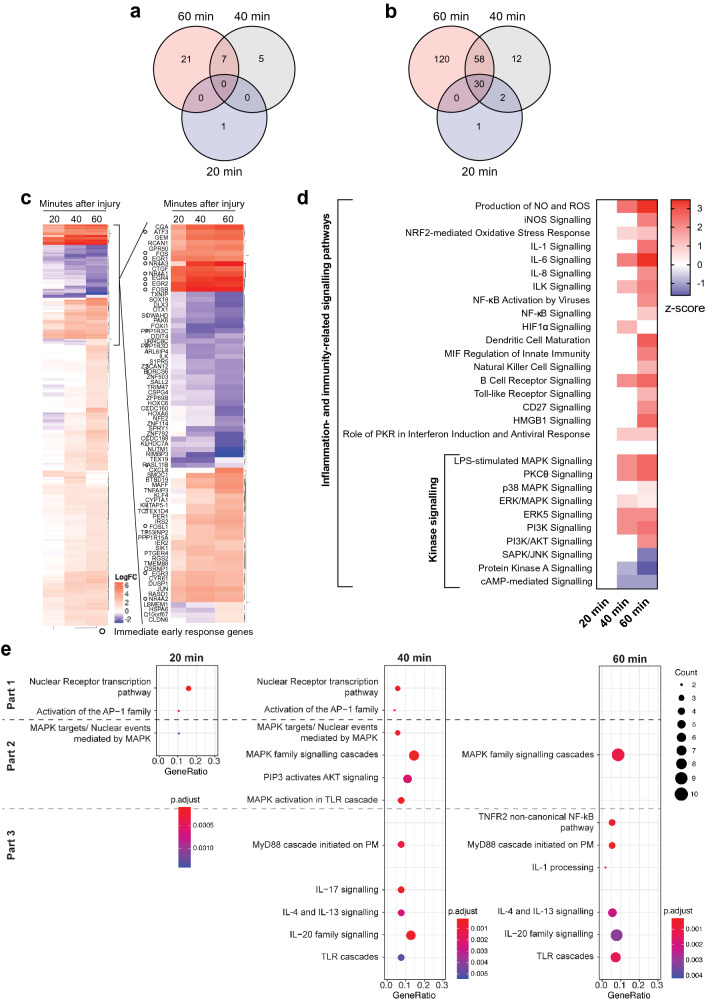
To assess the effect of PM injury at the transcriptional level, MCF-7 cells were treated with a sub-lethal digitonin concentration and the transcriptome was analysed by RNA-seq at 20-, 40- and 60-min post-injury. The number of differentially expressed (DE) mRNA transcripts (fold change ≥ 2 and ≤ -2; P-value < 0.05) compared to uninjured cells increased over time, with 33, 102 and 208 mRNA transcripts being significantly upregulated (see Supplementary Table S1 online) and 1, 12 and 28 mRNA transcripts being significantly downregulated (see Supplementary Table S2 online) at 20-, 40- and 60-min, respectively. Although there was no overlap in downregulated DE transcripts across all the timepoints assessed (Fig. 2a), several of the upregulated transcripts were shared (Fig. 2b). The majority of these encode for IEGs, such as early growth response (EGR) 2–4, nuclear receptor subfamily members (NR4A) 1–3, FOS and JUN transcription factors25 (Fig. 2c). There is a high correlation between the expression of DE IEGs and post-injury time (Multiple linear regression: R2 = 0.9872, P-value < 0.05). EGR4, ATF3 and NR4A3 show a positive trend across the timepoints assessed (i.e. its relative mRNA expression increases over time). FOS, on the other hand, despite being also upregulated throughout the post-injury window assessed, peaks at 20 min. The other upregulated IEGs remain largely unchanged across the timepoints assessed. Even when IEGs are excluded, overall the significant transcriptomic changes correlate with post-injury time (Multiple linear regression: R2 = 0.9618, P-value < 0.05).
Figure 2.
Early injury-induced transcriptional response in MCF-7 breast cancer cells. Overlap of significantly downregulated (a) and upregulated (b) genes (> twofold, P < 0.05) across the post-injury timepoints assessed (20-, 40- and 60-min post-injury). (c) Heatmap depicting the log2 fold change of differentially expressed genes (DE) (> twofold, P < 0.05) across the analysed timepoints. The most significantly changed genes are listed and the majority of these show a positive correlation between expression and post-injury time. Round open circles mark immediate-early response genes (IEGs). (d) Heatmap showing the signalling cascades predicted by the Ingenuity Pathway Analysis (IPA) to be the most significantly changed at different timepoints post-injury, based on the differentially expressed transcripts (RNA-seq data). These pathways are significantly associated to the dataset (-log(P-value) > 1.3) and have a z-score that reflects their predicted activation state (a positive/negative z-score indicates activation/inhibition). Pathways enriched are related to inflammation and immunity and a subset are kinase-dependent. (e) Dot-plot showing enriched pathways at 20-, 40- and 60-min post-injury as a function of Gene Ratio (i.e. percentage of total differentially expressed genes in the given GO term), Count (i.e. number of input genes associated to a specific pathway) and P-adjusted values. For each timepoint, enriched pathways were grouped into the three parts of the transcriptomic response (Part 1: induction of immediate-early response genes; Part 2: activation of specific kinase-dependent pathways; Part 3: activation of inflammatory and immune responses). Pathway enrichment analysis was conducted by the ReactomePA.
Gene ontology (GO) analysis and functional pathway analysis
When focusing on significantly associated biological process GO terms, the most enriched were “response to cytokine”, “response to cAMP”, “negative regulation of ERK1 and ERK2 cascades”, “response to lipopolysaccharide”, “cytokine-mediated signalling pathway” and “inactivation of MAPK activity” (see Supplementary Fig. S1a online). As expected, in addition to the “response to cAMP”, other GO terms induced by Ca2+- and mechanical-stimulation were also associated (see Supplementary Fig. S1b online). Interestingly, other significantly associated GO terms were related to cellular stress and survival (see Supplementary Fig. S1c online) and both inflammatory and immune responses (see Supplementary Fig. S1d online).
To obtain insight into the functional significance of the GO terms associated to the post-injury transcriptome, we conducted pathway analyses using both the Ingenuity Pathway Analysis (IPA) (Fig. 2d) and ReactomePA (Fig. 2e). The use of multiple platforms is an attempt to counteract knowledge biases (an inherent limitation of this knowledge-guided analysis that has been previously reviewed36), achieving a more comprehensive pathway analysis. Together, the GO analysis and pathway analyses performed support a dynamic and gradual three-part transcriptomic response.
The first part of the response, occurring predominantly at 20 min, consists of the activation of transcription factors, such as those that form the AP-1 complex, which influence the expression of stress-related genes (Fig. 2e, part 1). At 40 min post-injury, the predominant transcriptomic changes are associated to kinase-dependent cascades (Fig. 2e, part 2, and Fig. 2d). These include multiple branches of the MAPK signalling cascade (the canonical ERK, p38 MAPK and the atypical ERK5 – see Supplementary Fig. S2 online37–39) and the PKC and PI3K cascades (Fig. 2d).
Overall, the activation of kinase-dependent pathways increases as a function of post-injury time, which is reflected by the increasing z-score (Fig. 2d) and the number of DE genes associated to the pathway (Fig. 2e, part 2). Importantly, the induction of kinase-mediated pathways occurred in a specific rather than global manner – evident from the inhibition of the Stress-activated protein kinases (SAPK)/Jun amino-terminal kinases (JNK) branch of MAPK, PKA and cAMP-mediated signalling pathways (Fig. 2d).
The final part of the transcriptomic response is associated to the induction of inflammation- and immunity-related pathways (Fig. 2d,e, part 3). Both pathway analyses predicted the activation of IL signalling, TLR cascades and NF-kB-mediated pathways (Fig. 2d,e, part 3). In addition, IPA predicted a pronounced activation of redox-mediated pathways, as well as key processes that regulate innate immunity (Fig. 2d).
Although transcriptomic changes at 60-min post-injury can be best characterised by changes in inflammation- and immunity-related pathways, these are activated throughout the response (Fig. 2d). At 40 min particular IL family cascades are significantly associated, namely IL-6 (Fig. 2d), IL-4, IL-13, IL-17 and IL-20 (Fig. 2e, 40 min), while at 60 min post-injury IL-1, IL-6, IL-8 (Fig. 2d), IL-4, IL-13 and IL-20 (Fig. 2e, 60 min) pathways are active. Likewise, ROS-mediated pathways (Fig. 2d) and TLR cascades (Fig. 2d, 40 min and 60 min) are significantly associated to the transcriptome at 40 min and 60 min post-injury, but have an increased z-score or count, respectively, at 60 min.
Taken together, the cellular consequences of digitonin-induced PM injury activate a rapid (from 20 min) and sustained (for at least 1 h post-injury) transcriptional response in MCF-7 breast cancer cells that is very dynamic. Initially, the mRNA expression of IEGs is induced and MAPK pathways are predicted to become activated. Later, processes related to cellular stress, inflammation and immunity are activated.
Predicted upstream regulators of the transcriptomic response
Our data support a dynamic transcriptomic response to PM injury, both in the nature of the genes expressed and the pathways altered, as well as the magnitude of the transcriptomic changes over time. As a preliminary attempt to understand how these changes could be regulated, we performed an upstream regulator analysis using IPA (see Supplementary Fig. S3 online). Four key transcriptional regulators were found to be activated in a post-injury time-dependent manner (Platelet-derived growth factor (PDGF-BB), Nuclear protein 1 (NUPR1), Tumour necrosis factor (TNF) and ERK) and one inhibited (Histone deacetylases (HDAC)). Of relevance are also G-protein coupled estrogen receptor 1 (GPER1), Triggering receptor expressed on myeloid cells 1 (TREM1), IL1A, IL1B, Signal transducer and activator of transcription 3 (STAT3), Epidermal Growth Factor (EGF) and JUN, which have smaller absolute z-scores but also increase in a time-dependent manner.
Only JUN and some members of the TNF family were found to be upregulated in our RNA-seq data. Specifically, JUN recorded log2 fold changes of 3.069, 3.614, 3.469 at 20-, 40- and 60-min post-injury, while the transcription of TNF superfamily member 9 (TNFSF9) and TNF alpha induced protein 3 (TNFAIP3) was associated with log2 fold changes of 1.365 and 1.590 and 1.661 and 3.878, respectively, at 40- and 60-min, and the TNF receptor superfamily member 11b (TNFRSF11B) of 2.086 at 60-min. Therefore, JUN and TNF family proteins are putative upstream regulators that could explain the observed transcriptomic changes.
Meta-analysis of the post-injury transcriptome
We next performed a small meta-analysis to better elucidate the transcriptomic response directly or indirectly associated to PM injury. To our knowledge this is the first one in the field and included the only two published studies addressing genome-wide transcriptomic changes following PM injury or PM damage-related events in the short-term (1 h post-injury), along with our dataset. One of the studies analysed was conducted by Wales et al.1 and assessed the exposure of MCF-7 breast cancer cells to ionomycin, which induces increased cytosolic Ca2+ and cytoskeletal remodelling. These are direct cellular consequences of PM injury. The other study was perfomed by Rysä et al.4 and investigated the response of cardiomyocytes to mechanical stretch, which is known to cause Ca2+ influx40,41 due to compromised membrane integrity42–45. The nature of the meta-analysis not only allows for greater power to detect differential expression and associated pathways46, but also questions the conserved nature of the transcriptomic response across different cell and injury types. Furthermore, because Ca2+ influx is the common signalling cascade stimulus across all injury types (namely digitonin- and stretch-induced) and “pseudo-injury” (i.e. ionomycin) considered, similarities in transcriptome profile suggest that Ca2+ is the driver of the transcriptomic response to PM injury.
Differentially expressed genes
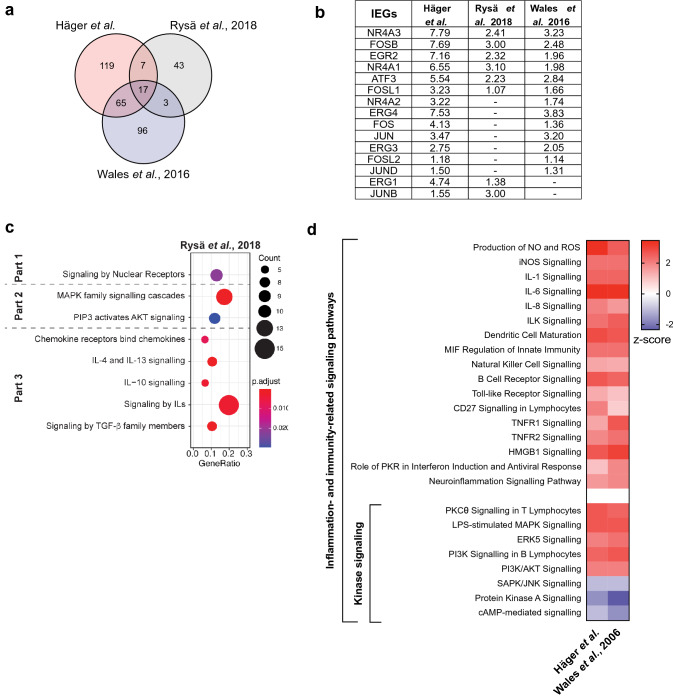
Consistent with our RNA-seq data, the majority of gene expression changes recorded represent upregulation (Fig. 3a) rather than downregulation (see Supplementary Fig. S4a online), when the same cut-off criteria are applied (fold change ≥ 2 and ≤ -2; P-value < 0.05). Interestingly, 17 DE mRNA transcripts were consistently upregulated in all studies, including the IEGs EGR2, NR4A 1–3 and FOS (FOSB and FOSL1) (Fig. 3b).
Figure 3.
Meta-analysis of the injury-induced transcriptional response (at 1 h post-injury) shows that the transcriptional response is conserved. (a) Overlap of significantly upregulated genes (> twofold, P < 0.05) between our RNA-seq dataset derived from digitonin-injured MCF-7 breast cancer cells and two others from ionomycin-treated MCF7 breast cancer cells (Wales et al., 20161) and mechanically stretched cardiomyoctes (Rysä et al., 20184). (b) log2 fold changes of immediate-early response genes (IEGs) (with P < 0.05) across all datasets included in the meta-analysis. (c) Dot-plot showing the predicted enriched pathways from the transcriptomic dataset by Rysä et al.4 as a function of Gene Ratio (i.e. percentage of total differentially expressed genes in the given GO term), Count (i.e. number of input genes associated to a specific pathway) and P-adjusted values. The enriched pathways were grouped into the three parts of the transcriptomic response (Part 1: induction of immediate-early response genes; Part 2: activation of specific kinase-dependent pathways; Part 3: activation of inflammatory and immune responses). Pathway enrichment analysis was conducted by the ReactomePA. (d) Heatmap showing the signalling cascades predicted by the Ingenuity Pathway Analysis to be the most significantly changed in our RNA-seq data and the transcriptomic data reported by Wales et al., 20161. These pathways are significantly associated to the dataset (-log(P-value) > 1.3) and have a z-score that reflects their predicted activation state (a positive/negative z-score indicates activation/inhibition).
GO analysis and functional pathway analysis
Next, we questioned whether the same GO terms would be significantly associated to all three studies (see Supplementary Table S3 online). Indeed, five GO terms were in common. The transcriptome profile of our model was also associated to the same eight GO terms as the model from Wales et al.1, including GO terms related to calcium signalling, stress responses, kinase signalling and inflammation.
To identify key changes in signalling cascades that could drive the association of these biological processes (GO terms) to the transcriptomic response of all three models in question, we performed pathway analysis using ReactomePA and IPA. As with our digitonin-induced PM injury model, analysis of the transcriptome published by Rysä et al.4 reveals a response that can be characterised in three parts. Firstly, along with the upregulation of IEGs (Fig. 3b), nuclear-based pathways are upregulated (Fig. 3c, part 1). This is expected given that most IEGs are transcription factors. Secondly, specific kinase-dependent cascades are upregulated, namely MAPK (Fig. 3c, part 2). Thirdly, multiple inflammatory and immune-related pathways are induced, such as chemokine and cytokine signalling (in particular IL signalling). The heatmap shows that there is great overlap (i.e. similarity) between the transcriptome profile in our model and Rysä et al.4 (Fig. 3c). Likewise, the transcriptomic dataset presented by Wales et al.1 also supports the activation of specific kinase-dependent signalling and enrichment of multiple inflammation- and immunity-related signalling pathways (Fig. 3d). Taken together, the activation of IEGs, certain kinase-dependent cascades and inflammation- and immunity-related pathways are common transcriptional events observed in our model and others1,4.
Predicted upstream regulators of the transcriptomic response
Our meta-analysis identified that all datasets shared largely the same predicted upstream transcriptional regulators (see Supplementary Fig. S4b online). Some of these were predicted to drive the time-dependent transcriptomic changes in our model of injury, namely JUN and TNF (see Supplementary Fig. S3 online). Other predicted upstream transcriptional regulators include transcription factors (e.g. NF-kB47), transcription activators (e.g. AP-1 complex48), cytokines49,50 and TLRs51.
Of the predicted upstream regulators in common across all three studies some are upregulated in the RNA-seq of digitonin-injured MCF-7 cells (see Supplementary Fig. S4c online). Some of these regulate in inflammatory and immune responses, by modulating the expression of chemokine receptors52, cytokines53 and the NF-kB signalling54,55. In addition, some of the putative regulators, such as EGR1, are themselves IEGs and/or modulate the expression of IEGs53,56, as well as MAPK components56. Interestingly, AP1 was the only predicted upstream regulator upregulated in all three studies (see Supplementary Fig. S4c online), thus may be a relevant regulator of the conserved transcriptomic response following the PM injury or its associated downstream effects.
Discussion
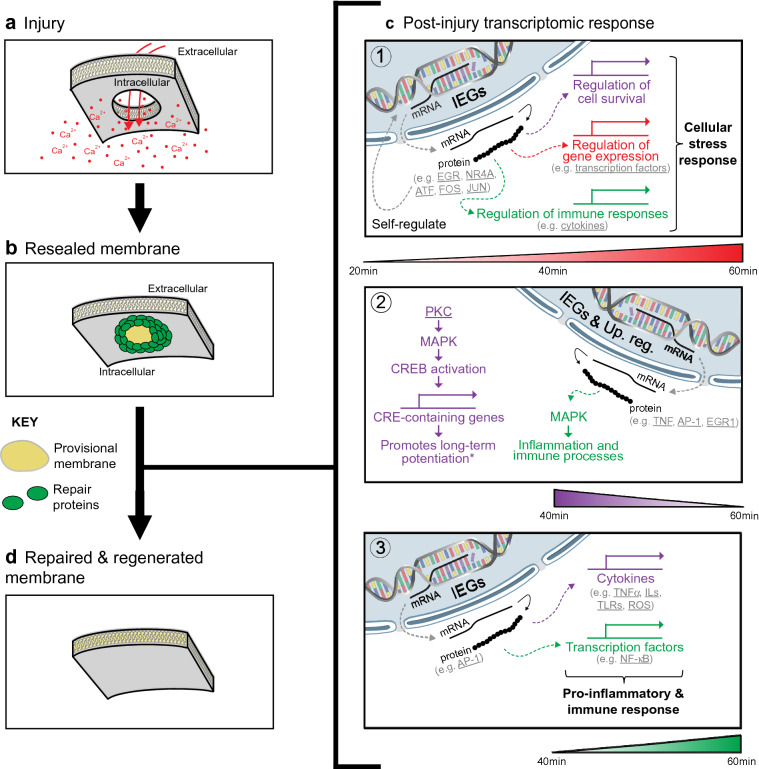
Disruptions to the PM trigger a robust repair response that is predominantly driven by the abrupt influx of Ca2+. While the first phase of the response aims to physically re-seal the PM, less is known about the secondary regeneration phase that follows. Here, we show that PM injury induced a strong transcriptional response in MCF-7 breast cancer cells (summarised in Fig. 4). This response is gradual and dynamic, involving (1) the activation of IEGs, (2) the activation of specific MAPK cascades and (3) the induction of inflammatory and immune pathways.
Figure 4.
Short-term transcriptomic response following PM injury. (a) Injury at the PM results in Ca2+ influx. (b) Membrane repair mechanisms are rapidly activated and result in immediate PM repair to halt the influx of Ca2+ and outflux of intracellular components (e.g. by forming a provisional patch membrane). (c) Injury triggers a three-part transcriptomic response: 1. Activation of immediate-early response genes (IEGs): Increased expression of IEGs regulates the expression of genes involved in the regulation of cell survival, gene expression and immune responses, mounting a cellular stress response. IEGs can self-regulation by inducing their own expression. This initial response dominates the transcriptomic profile at 20 min, although the expression of the majority of IEGs increased as a function of time. 2. Activation of specific MAPK signalling cascades: Induction of CRE-containing genes may promote long-term potentiation of the membrane for increased resilience to repeated wounding (indicated by the asterisk)2, 3. In parallel, increased expression of IEGs and upstream regulators (abbreviated as “Up. reg.”) drives MAPK-mediated signalling, which induces inflammation- and immunity-related processes. 3. Induction of pro-inflammatory and immune responses: IEGs induce the expression of cytokines and transcription factors that result in a pro-inflammatory milieu and drive a strong immune response. (d) Ultimately, the PM fully repairs and is regenerated to adopt its original composition and properties. We hypothesise that this post-injury transcriptional response is key for adequate membrane repair and regeneration, restoring plasma membrane functionality. Genes, proteins and protein families underlined were found to be changed in the RNA-seq data, while pathways and transcriptional events mentioned that are not underlined were predicted from the RNA-seq data (except for that marked with an asterisk).
Our meta-analysis investigating transcriptional changes in response to PM injury and/or its associated cellular consequences (such as Ca2+ influx) also supported this three-part response. In this comparative approach, the DE genes identified in our dataset (derived from MCF-7 cells injured with digitonin) were compared with those identified from MCF-7 cells treated with ionomycin (Wales et al.1) and cardiomyocytes injured by mechanical stretching (Rysä et al.4). To our knowledge, these are the only unbiased, genome-wide study looking at short-term transcriptomic changes. When analysed in parallel to our transcriptomic data, they aid in the understanding of what changes could be driven by the directly related calcium influx, which is an inevitable cellular consequence of PM injury57. This is because Wales and colleagues1 used a pharmacological approach to increased cytosolic Ca2+ (acting as a “pseudo” injury), while Rysä et al.4 exposed a different cell line to mechanical stretching, which also results in increased Ca2+ influx40,41 due to compromised membrane integrity42–45.
Our findings add PM injury to the established list of broad stimuli25 that activate IEGs and our time-course analysis suggests that this transcriptional response occurs within minutes, being significantly detected at 20-min post-injury. Specifically, the mRNA expression of the nuclear receptors NR4A 1–3 and the transcription factors EGR 2–4, FOS and JUN are induced (Fig. 2b). Most of these were also induced upon membrane stress4 and cytosolic Ca2+ influx1 (Fig. 3b). Although this upregulation was previously reported by Rysä and colleagues4, our time-course analysis of wounded MCF-7 cells and our meta-analysis validate and support the conserved nature of the transcriptomic response across different cell and injury types.
IEGs regulate the expression of target genes involved in the regulation of gene expression24,25,48,58,59, cell survival58,59 and immune responses24,25,48. These activities place the AP-1 protein complex (encoded by FOS and JUN), as well as other IEGs25, at a key position in the regulation of cellular stress responses. It is therefore not surprising that the induction of IEG expression is a prevailing part of the transcriptomic response to PM injury and its associated cellular consequences. Likewise, IEGs have been found by others to be induced at 1 h post-injury20, as well as days after injury23. Moreover, it has been proposed that c-fos gene expression (and possibly other IEGs too) is proportional to the degree of PM disruption (due to its Ca2+-responsive element), influencing the activation of its target genes21. Wales et al.1 also found IEGs to be part of a cohort of genes directly regulated by Ca2+, through the phenomenon of Ca2+-mediated actin reset (i.e. the sudden shift of actin distribution in response to increased intracellular Ca2+)1. Given that PM injury also results in pronounced Ca2+ influx, this phenomenon is likely to modulate, at least in part, the post-injury transcriptomic response.
Beyond functioning as a notable activator of signalling cascades (e.g. activating PM repair proteins60), abrupt Ca2+ influx imposes some degree of cytotoxicity61. The AP-1 complex is also known to directly influence the balance between cell death and survival58, although the end-effect is influenced by cell type, cellular status, expression levels and the composition of the complexes62,63. The literature is controversial and c-jun has been associated with both cell death64,65 and cell survival59 in tumours. In MCF-7, specifically, the literature favours its pro-survival activity66, as does transient induction of IEGs compared to chronic58. Indeed, “cell stress and survival” was one of the most associated GO terms (see Supplementary Fig. S1c online) and JNK signalling, which is believed to be important in initiating apoptosis67, was predicted to be downregulated (Fig. 2d). Also, within the post-injury window investigated, we did not observe the induction of pro-apoptotic genes in digitonin-induced MCF-7 cells subsequent to the early expression of IEGs. Therefore, the induction of IEGs may function to promote cell survival in response to PM injury (in cells that were able to reseal their membranes), which is hypothesised to occur frequently during cancer metastasis and invasion17.
Beyond dictating cellular fate, IEGs might ensure that an adequate cellular response is mounted towards injury. This would be advantageous for cells for multiple reasons. Firstly, by being regulated by Ca2+, its expression/activity can be proportional to the degree of injury. Secondly, IEG induction is hypothesised to occur early in the repair response (detected at 20 min post-injury) and IEGs are capable of driving the transcriptomic response that follows. In fact, IEGs were predicted to be key upstream regulators of the transcriptomic response (see Supplementary Fig. S3, S4b and S4c online). Moreover, other predicted upstream regulators may also regulate IEG expression, namely PDGF-BB68, NUPR169, TNF53, ERK70, HDAC71, TREM172, IL1B73, STAT374, EGF75 and JUN76. Two other factors that would favour an advantage regulatory role of IEGs in the response to injury is the fact that they self-regulate the expression of each other77 and seem to be conserved across different cell and injury types. Therefore, IEGs are likely to govern major aspects of the injury response—a concept that was previously proposed24,25 but not linked to PM injury.
A predominant part of the transcriptomic response that we characterize here is the robust activation of cascades involved in inflammation and immunity. Both IEGs48,77 and kinase-dependent pathways78–85 have been suggested to play a key role in inflammatory. The AP-1 complex can, for example, directly regulate the expression of cytokines (including TNF-α and ILs) and transcription factors (e.g. NF-κB)48,77,86. Other predicted upstream regulators that could regulate inflammatory and immune responses are NUPR169, TNF53, ERK87, TREM188, IL1A89, IL1B73, STAT374 and EGF90. Some of these exert this effect through MAPK-mediated signalling (e.g. TNF53) or through their intrinsic kinase activity91. Interestingly, as with the expression of IEGs, Ca2+-mediated actin reset may also regulate the expression of inflammatory genes1.
We predict that the transcriptional response mounted upon PM injury is mainly pro-inflammatory, as the majority of the activated pathways are pro-inflammatory47,92 (involving TLR, IL-1, IL-8, TNF, NF-kB, iNOS and ROS). TLRs are known to mediate innate immunity in response to a wide variety of pathogenic threats through recognition of conserved pathogen-associated molecular motifs93, although PM injury has not been previously added to that list. TLRs and proinflammatory cytokines activate the canonical pathway of NF-kB signalling, which regulates the expression of proinflammatory and cell survival genes. Furthermore, through its canonical and alternative pathways (identified in Fig. 2d,e 60 min respectively), NF-kB signalling mediates both innate and adaptive immunity47,94. This pro-inflammatory and immunogenic profile arise at 40 min post-injury (Fig. 2d,e) and becomes gradually activated throughout the response. Likewise, transcriptomic data from Rysä et al.4 and Wales et al.1 also predicted a strong immunogenic response using two independent pathway analysis software (Fig. 3c,d).
An injury-induced inflammatory and immunogenic response has not been given much focus in the literature. However, there is some evidence to suggest its existence, namely the activation of NF-kB upon PM disruption21 and the upregulation of cytokines (including TGF-β) due to PM stress22. These events are believed to trigger the expression of stress-induced genes and promote wound-healing21,22. This supports our hypothesis that the immediate-early response and the inflammatory and immune responses are key in the transcriptomic response post-injury. However, it remains to be determined the functional significance of the pro-inflammation and immunogenic response post-injury. Whether it contributes the phenomenon of long-term potentiation3 to improve repair, or is an intrinsic part of membrane regeneration following injury (resembling inflammation and remodelling in wound healing95), remains to be answered.
MAPK signalling (the canonical ERK, p38 MAPK and the atypical ERK5) is known to response to a diverse array of stress stimuli96. Moreover, the activation of specific MAPK cascades following injury has also been previously given some degree of attention2–4,20,22,23. Likewise, our findings support the gradual activation of specific MAPK cascades clearly from 40 min post-injury (Fig. 2d,e), but possibly as early as 20 min (Fig. 2e, part 1), and across different models (Fig. 3d). PKA activation, for example, has been implicated in membrane resealing kinetics97 and early potentiation of Ca2+-regulated exocytosis to facilitate subsequent wound resealing3. However, our time-course analysis and meta-analysis predicted that PKA and cAMP-mediated signalling pathways are inhibited (Figs. 2d, 3d). This discrepancy may be due to cell type differences (the studies referenced were performed in neurons and fibroblasts) or, more likely, differences in post-injury times assayed. PKA activation was detected early post-injury in the study (< 5 min)97, thus the predicted inactivation of PKA and cAMP-mediated signalling pathways observed in our study may reflect a regulatory negative feedback loop, which would be expected following acute activation. At a later stage post-injury, PKC was reported to mediate cAMP response element-binding protein (CREB) activation via a p38 MAPK pathway2 to induce expression of cAMP-response element (CRE)-containing genes and promote long-term potentiation2,3. Likewise, our predictive pathway analyses predicted the activation of MAPK- and PKC-mediated signalling cascades (Figs. 2d, 3d) and CREB1 and p38 MAPK are predicted activated upstream regulators (see Supplementary Fig. S3 and S4 online). Therefore, these specific kinase-dependent cascades may also become activated in our study to promote long-term potentiation, improving the regenerative ability of cells. Although this requires further investigation, modulating kinase-dependent pathways to facilitate regeneration could have clinical significance.
Although the existence of injury-induced changes in gene expression has been previously hypothesised3,21 and investigated in a gene-specific manner20,21, the field lacked evidence regarding the temporal-specificity of global transcriptomic changes. In particular, short-term (< 1 h) transcriptomic responses were not previously investigated, as opposed to long-term changes (> 6 h)22–24. Our study is therefore a step towards deciphering these, although the findings presented would benefit from further validation. This should include evaluation of the transcriptome profile using an additional and independent method and assessment of whether transcriptional changes translate to protein levels and changes in the predicted pathways. Once validated, functional studies should assess the individual and cumulative roles of the three distinct parts of the transcriptomic response (i.e. activation of IEGs, activation of MAPK cascades and induction of inflammation and immune-related processes) on post-injury cellular dynamics. Furthermore, it would be also interesting to understand whether the injury-induced transcriptome is driven as a defensive response to acute trauma and as an adaptive response to potentiate membrane resealing for subsequent injuries. This would not only be significant within the field of membrane repair, but also within cell death and survival, as injury and repair processes are likely to be intimately related.
We show that MCF-7 cancer cells exposed to digitonin-induced membrane injury respond, at a transcriptional level, by expressing IEGs, activating specific kinase-dependent cascades and mounting a robust pro-inflammatory and immunogenic response. Our meta-analysis demonstrated that this three-part response may be conserved across different cell types and injuries, although this should be addressed further.
Supplementary Information
Acknowledgements
This study was supported by the Danish Council for Independent Research, Natural Sciences (6108-00378A, 9040-00252B) (JN, SCH, SLS), the Novo Nordisk Foundation (NNF18OC0034936) (JN, CD, ASH), the Scientific Committee Danish Cancer Society (R90-A5847-14-S2) (JN, SLS), and the Danmarks Grundforskningsfond (DNRF125).
Abbreviations
- PM
Plasma membrane
- IEGs
Immediate-early response genes
- MAPK
Mitogen-activated protein kinase
- PKC
Protein kinase C
- PI3K
Phosphoinositide-3 kinase
- ERK5
Extracellular signal-regulated kinase 5
- PKA
Protein kinase A
- IL
Interleukin
- TLR
Toll-like-receptor
- ROS
Reactive oxygen species
- FBS
Fetal bovine serum
- PI
Propidium iodide
- EGR
Early growth response
- NR4A
Nuclear receptor subfamily members
- GO
Gene ontology
- IPA
Ingenuity Pathway Analysis
- SAPK
Stress-activated protein kinases
- JNK
Jun amino-terminal kinases
- PDGF-BB
Platelet-derived growth factor
- NUPR1
Nuclear protein 1
- TNF
Tumour necrosis factor
- HDAC
Histone deacetylases
- GPER1
G-protein coupled estrogen receptor 1
- TREM1
Triggering receptor expressed on myeloid cells 1
- STAT3
Signal transducer and activator of transcription 3
- EGF
Epidermal Growth Factor
- TNFSF9
TNF superfamily member 9
- TNFAIP3
TNF alpha induced protein 3
- TNFRSF11B
TNF receptor superfamily member 11b
- TIAF1
TGFB1-induced anti-apoptotic factor 1
- CREB
CAMP response element-binding protein
- CREBR
CREB3 regulator factor
- CRE
CAMP-response element
Author contributions
Cell biological studies were performed by S.C.H and S.L.S. RNA-sequencing was conducted by A.V.O and I.P. Bioinformatic analyses were undertaken by C.D., A.V.O and I.P. J.N conceived and designed the study, acquired funding and supervised the work. J.N. and C.D. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Swantje Christin Häger and Catarina Dias.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-98420-y.
References
- 1.Wales P, et al. Calcium-mediated actin reset (CaAR) mediates acute cell adaptations. Elife. 2016 doi: 10.7554/eLife.19850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Togo T. Long-term potentiation of wound-induced exocytosis and plasma membrane repair is dependent on cAMP-response element-mediated transcription via a protein kinase C- and p38 MAPK-dependent pathway. J. Biol. Chem. 2004;279:44996–45003. doi: 10.1074/jbc.M406327200. [DOI] [PubMed] [Google Scholar]
- 3.Togo T, Alderton JM, Steinhardt RA. Long-term potentiation of exocytosis and cell membrane repair in fibroblasts. Mol. Biol. Cell. 2003;14:93–106. doi: 10.1091/mbc.e02-01-0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rysä J, Tokola H, Ruskoaho H. Mechanical stretch induced transcriptomic profiles in cardiac myocytes. Sci. Rep. 2018;8:4733. doi: 10.1038/s41598-018-23042-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bement WM, Mandato CA, Kirsch MN. Wound-induced assembly and closure of an actomyosin purse string in Xenopus oocytes. Curr. Biol. 1999;9:579–587. doi: 10.1016/s0960-9822(99)80261-9. [DOI] [PubMed] [Google Scholar]
- 6.Boye TL, et al. Annexin A4 and A6 induce membrane curvature and constriction during cell membrane repair. Nat. Commun. 2017;8:1623. doi: 10.1038/s41467-017-01743-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plattner H, Hentschel J. Sub-second cellular dynamics: time-resolved electron microscopy and functional correlation. Int. Rev. Cytol. 2006;255:133–176. doi: 10.1016/S0074-7696(06)55003-X. [DOI] [PubMed] [Google Scholar]
- 8.He H, Chang DC, Lee YK. Using a micro electroporation chip to determine the optimal physical parameters in the uptake of biomolecules in HeLa cells. Bioelectrochemistry. 2007;70:363–368. doi: 10.1016/j.bioelechem.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 9.Hibino M, Itoh H, Kinosita KJ. Time courses of cell electroporation as revealed by submicrosecond imaging of transmembrane potential. Biophys. Soc. 1993;64:1789–1800. doi: 10.1016/S0006-3495(93)81550-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrews NW, Almeida PE, Corrotte M. Damage control: cellular mechanisms of plasma membrane repair. Trends Cell Biol. 2014;24:734–742. doi: 10.1016/j.tcb.2014.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper ST, McNeil PL. Membrane repair: mechanisms and pathophysiology. Physiol. Rev. 2015;95:1205–1240. doi: 10.1152/physrev.00037.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Häger SC, Nylandsted J. Annexins: players of single cell wound healing and regeneration. Commun. Integr. Biol. 2019;12:162–165. doi: 10.1080/19420889.2019.1676139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moe AM, Golding AE, Bement WM. Cell healing: calcium, repair and regeneration. Semin. Cell Dev. Biol. 2015;45:18–23. doi: 10.1016/j.semcdb.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guiraud S, et al. The pathogenesis and therapy of muscular dystrophies. Annu. Rev. Genom. Hum. Genet. 2015;16:281–308. doi: 10.1146/annurev-genom-090314-025003. [DOI] [PubMed] [Google Scholar]
- 15.Boye TL, Nylandsted J. Annexins in plasma membrane repair. Biol. Chem. 2016;397:961–969. doi: 10.1515/hsz-2016-0171. [DOI] [PubMed] [Google Scholar]
- 16.Draeger A, Monastyrskaya K, Babiychuk EB. Plasma membrane repair and cellular damage control: the annexin survival kit. Biochem. Pharmacol. 2011;81:703–712. doi: 10.1016/j.bcp.2010.12.027. [DOI] [PubMed] [Google Scholar]
- 17.Jaiswal JK, et al. S100A11 is required for efficient plasma membrane repair and survival of invasive cancer cells. Nat. Commun. 2014;5:3795. doi: 10.1038/ncomms4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng S, et al. Annexin A1, A2, A4 and A5 play important roles in breast cancer, pancreatic cancer and laryngeal carcinoma, alone and/or synergistically. Oncol. Lett. 2013;5:107–112. doi: 10.3892/ol.2012.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duncan R, Carpenter B, Main LC, Telfer C, Murray GI. Characterisation and protein expression profiling of annexins in colorectal cancer. Br. J. Cancer. 2008;98:426–433. doi: 10.1038/sj.bjc.6604128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenfeldt H, Lee DJ, Grinnell F. Increased c-fos mRNA expression by human fibroblasts contracting stressed collagen matrices. Mol. Cell Biol. 1998;18:2659–2667. doi: 10.1128/mcb.18.5.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grembowicz KP, Sprague D, McNeil PL. Temporary disruption of the plasma membrane is required for c-fos expression in response to mechanical stress. Mol. Biol. Cell. 1999;10:1247–1257. doi: 10.1091/mbc.10.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunnen SJ, Malas TB, Semeins CM, Bakker AD, Peters DJM. Comprehensive transcriptome analysis of fluid shear stress altered gene expression in renal epithelial cells. J. Cell Physiol. 2018;233:3615–3628. doi: 10.1002/jcp.26222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arthur-Farraj PJ, et al. Changes in the coding and non-coding transcriptome and DNA methylome that define the Schwann cell repair phenotype after nerve injury. Cell Rep. 2017;20:2719–2734. doi: 10.1016/j.celrep.2017.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arthur-Farraj PJ, et al. c-Jun reprograms Schwann cells of injured nerves to generate a repair cell essential for regeneration. Neuron. 2012;75:633–647. doi: 10.1016/j.neuron.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bahrami S, Drabløs F. Gene regulation in the immediate-early response process. Adv. Biol. Regul. 2016;62:37–49. doi: 10.1016/j.jbior.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 26.Brito C, Cabanes D, Sarmento MF, Sousa S. Mechanisms protecting host cells against bacterial pore-forming toxins. Cell Mol. Life Sci. 2019;76:1319–1339. doi: 10.1007/s00018-018-2992-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolfmeier H, et al. Ca2+-dependent repair of pneumolysin pores: a new paradigm for host cellular defense against bacterial pore-forming toxins. Biochim. Biophys. Acta. 1853;2045–2054:2015. doi: 10.1016/j.bbamcr.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Korpelainen, E. & Lehtivaara, M. RNA-seq hands-on tutorial. https://research.csc.fi/rnaseq-tutorial.
- 29.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.topGO: Enrichment Analysis for Gene Ontology (2020).
- 31.Yu G, He QY. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst. 2016;12:477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- 32.Krämer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jassal B, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bendix PM, et al. Interdisciplinary synergy to reveal mechanisms of annexin-mediated plasma membrane shaping and repair. Cells. 2020 doi: 10.3390/cells9041029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sønder SL, et al. Annexin A7 is required for ESCRT III-mediated plasma membrane repair. Sci. Rep. 2019;9:6726. doi: 10.1038/s41598-019-43143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin L, et al. Pathway-based analysis tools for complex diseases: a review. Genom. Proteom. Bioinf. 2014;12:210–220. doi: 10.1016/j.gpb.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tatsukawa Y, Kiyosue T, Arita M. Mechanical stretch increases intracellular calcium concentration in cultured ventricular cells from neonatal rats. Heart Vessels. 1997;12:128–135. doi: 10.1007/BF02767130. [DOI] [PubMed] [Google Scholar]
- 41.Kaur S, Shen X, Power A, Ward ML. Stretch modulation of cardiac contractility: importance of myocyte calcium during the slow force response. Biophys. Rev. 2020;12:135–142. doi: 10.1007/s12551-020-00615-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geddes DM, Cargill RS, LaPlaca MC. Mechanical stretch to neurons results in a strain rate and magnitude-dependent increase in plasma membrane permeability. J. Neurotrauma. 2003;20:1039–1049. doi: 10.1089/089771503770195885. [DOI] [PubMed] [Google Scholar]
- 43.Kim TJ, Sun J, Lu S, Qi YX, Wang Y. Prolonged mechanical stretch initiates intracellular calcium oscillations in human mesenchymal stem cells. PLoS ONE. 2014;9:e109378. doi: 10.1371/journal.pone.0109378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lovering RM, Roche JA, Bloch RJ, De Deyne PG. Recovery of function in skeletal muscle following 2 different contraction-induced injuries. Arch. Phys. Med. Rehabil. 2007;88:617–625. doi: 10.1016/j.apmr.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 45.Michielin, F., Serena, E., Pavan, P. & Elvassore, N. Microfluidic-assisted cyclic mechanical stimulation affects cellular membrane integrity in a human muscle dystrophy in vitro model. RSC Adv. (2015).
- 46.Rau A, Marot G, Jaffrézic F. Differential meta-analysis of RNA-seq data from multiple studies. BMC Bioinf. 2014;15:91. doi: 10.1186/1471-2105-15-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu, T., Zhang, L., Joo, D. & Sun, S. C. NF-κB signaling in inflammation. Signal Transduct. Target Ther.2, 10.1038/sigtrans.2017.23 (2017). [DOI] [PMC free article] [PubMed]
- 48.Ye N, Ding Y, Wild C, Shen Q, Zhou J. Small molecule inhibitors targeting activator protein 1 (AP-1) J. Med. Chem. 2014;57:6930–6948. doi: 10.1021/jm5004733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rauch I, Müller M, Decker T. The regulation of inflammation by interferons and their STATs. JAKSTAT. 2013;2:e23820. doi: 10.4161/jkst.23820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Howland KC, Ausubel LJ, London CA, Abbas AK. The roles of CD28 and CD40 ligand in T cell activation and tolerance. J. Immunol. 2000;164:4465–4470. doi: 10.4049/jimmunol.164.9.4465. [DOI] [PubMed] [Google Scholar]
- 51.Jezierska A, Kolosova IA, Verin AD. Toll like receptors signaling pathways as a target for therapeutic interventions. Curr. Signal Transduct. Ther. 2011;6:428–440. doi: 10.2174/157436211797483930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sung HJ, Kim YS, Kang H, Ko J. Human LZIP induces monocyte CC chemokine receptor 2 expression leading to enhancement of monocyte chemoattractant protein 1/CCL2-induced cell migration. Exp. Mol. Med. 2008;40:332–338. doi: 10.3858/emm.2008.40.3.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loupasakis K, et al. Tumor Necrosis Factor dynamically regulates the mRNA stabilome in rheumatoid arthritis fibroblast-like synoviocytes. PLoS ONE. 2017;12:e0179762. doi: 10.1371/journal.pone.0179762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miraghazadeh B, Cook MC. Nuclear Factor-kappaB in autoimmunity: man and mouse. Front. Immunol. 2018;9:613. doi: 10.3389/fimmu.2018.00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chang NS, et al. Cloning and characterization of a novel transforming growth factor-beta1-induced TIAF1 protein that inhibits tumor necrosis factor cytotoxicity. Biochem. Biophys. Res. Commun. 1998;253:743–749. doi: 10.1006/bbrc.1998.9846. [DOI] [PubMed] [Google Scholar]
- 56.Arora S, et al. Egr1 regulates the coordinated expression of numerous EGF receptor target genes as identified by ChIP-on-chip. Genome Biol. 2008;9:R166. doi: 10.1186/gb-2008-9-11-r166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dias C, Nylandsted J. Plasma membrane integrity in health and disease: significance and therapeutic potential. Cell Discov. 2021;7:4. doi: 10.1038/s41421-020-00233-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 2002;4:E131–136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 59.Shaulian E. AP-1–The Jun proteins: oncogenes or tumor suppressors in disguise? Cell Signal. 2010;22:894–899. doi: 10.1016/j.cellsig.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 60.Bittel, D. C. et al. Annexin A2 Mediates Dysferlin Accumulation and Muscle Cell Membrane Repair. Cells9, 10.3390/cells9091919 (2020). [DOI] [PMC free article] [PubMed]
- 61.Farber JL. The role of calcium ions in toxic cell injury. Environ. Health Perspect. 1990;84:107–111. doi: 10.1289/ehp.9084107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jacobs-Helber SM, Wickrema A, Birrer MJ, Sawyer ST. AP1 regulation of proliferation and initiation of apoptosis in erythropoietin-dependent erythroid cells. Mol. Cell Biol. 1998;18:3699–3707. doi: 10.1128/mcb.18.7.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hess J, Angel P, Schorpp-Kistner M. AP-1 subunits: quarrel and harmony among siblings. J. Cell Sci. 2004;117:5965–5973. doi: 10.1242/jcs.01589. [DOI] [PubMed] [Google Scholar]
- 64.Gee JM, Barroso AF, Ellis IO, Robertson JF, Nicholson RI. Biological and clinical associations of c-jun activation in human breast cancer. Int. J. Cancer. 2000;89:177–186. doi: 10.1002/(sici)1097-0215(20000320)89:2<177::aid-ijc13>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 65.Lerdrup M, et al. Depletion of the AP-1 repressor JDP2 induces cell death similar to apoptosis. Biochim. Biophys. Acta. 2005;1745:29–37. doi: 10.1016/j.bbamcr.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 66.Duan L, et al. Inducible overexpression of c-Jun in MCF7 cells causes resistance to vinblastine via inhibition of drug-induced apoptosis and senescence at a step subsequent to mitotic arrest. Biochem. Pharmacol. 2007;73:481–490. doi: 10.1016/j.bcp.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Davis RJ. Signal transduction by the JNK group of MAP kinases. Cell. 2000;103:239–252. doi: 10.1016/s0092-8674(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 68.Chen J, et al. Multi-omics analysis reveals regulators of the response to PDGF-BB treatment in pulmonary artery smooth muscle cells. BMC Genom. 2016;17:781. doi: 10.1186/s12864-016-3122-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hamidi T, et al. Nuclear protein 1 promotes pancreatic cancer development and protects cells from stress by inhibiting apoptosis. J. Clin. Invest. 2012;122:2092–2103. doi: 10.1172/JCI60144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Uhlitz F, et al. An immediate-late gene expression module decodes ERK signal duration. Mol. Syst. Biol. 2017;13:928. doi: 10.15252/msb.20177554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miotto B, et al. Chameau HAT and DRpd3 HDAC function as antagonistic cofactors of JNK/AP-1-dependent transcription during Drosophila metamorphosis. Genes. Dev. 2006;20:101–112. doi: 10.1101/gad.359506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dower K, Ellis DK, Saraf K, Jelinsky SA, Lin LL. Innate immune responses to TREM-1 activation: overlap, divergence, and positive and negative cross-talk with bacterial lipopolysaccharide. J. Immunol. 2008;180:3520–3534. doi: 10.4049/jimmunol.180.5.3520. [DOI] [PubMed] [Google Scholar]
- 73.Swindell WR, et al. RNA-Seq analysis of IL-1B and IL-36 responses in epidermal keratinocytes identifies a shared MyD88-dependent gene signature. Front. Immunol. 2018;9:80. doi: 10.3389/fimmu.2018.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carpenter RL, Lo HW. STAT3 Target Genes Relevant to Human Cancers. Cancers (Basel) 2014;6:897–925. doi: 10.3390/cancers6020897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gregg J, Fraizer G. Transcriptional regulation of EGR1 by EGF and the ERK signaling pathway in prostate cancer cells. Genes. Cancer. 2011;2:900–909. doi: 10.1177/1947601911431885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mechta-Grigoriou F, Gerald D, Yaniv M. The mammalian Jun proteins: redundancy and specificity. Oncogene. 2001;20:2378–2389. doi: 10.1038/sj.onc.1204381. [DOI] [PubMed] [Google Scholar]
- 77.Ji Z, He L, Regev A, Struhl K. Inflammatory regulatory network mediated by the joint action of NF-kB, STAT3, and AP-1 factors is involved in many human cancers. Proc. Natl. Acad. Sci. U S A. 2019;116:9453–9462. doi: 10.1073/pnas.1821068116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frazier WJ, Xue J, Luce WA, Liu Y. MAPK signaling drives inflammation in LPS-stimulated cardiomyocytes: the route of crosstalk to G-protein-coupled receptors. PLoS ONE. 2012;7:e50071. doi: 10.1371/journal.pone.0050071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chand S, Mehta N, Bahia MS, Dixit A, Silakari O. Protein kinase C-theta inhibitors: a novel therapy for inflammatory disorders. Curr. Pharm. Des. 2012;18:4725–4746. doi: 10.2174/138161212802651625. [DOI] [PubMed] [Google Scholar]
- 80.Pfeifhofer-Obermair C, et al. Role of PKCtheta in macrophage-mediated immune response to Salmonella typhimurium infection in mice. Cell Commun. Signal. 2016;14:14. doi: 10.1186/s12964-016-0137-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Herlaar E, Brown Z. p38 MAPK signalling cascades in inflammatory disease. Mol. Med. Today. 1999;5:439–447. doi: 10.1016/s1357-4310(99)01544-0. [DOI] [PubMed] [Google Scholar]
- 82.Lu, N. & Malemud, C. J. Extracellular signal-regulated kinase: a regulator of cell growth, inflammation, chondrocyte and bone cell receptor-mediated gene expression. Int. J. Mol. Sci.20, 10.3390/ijms20153792 (2019). [DOI] [PMC free article] [PubMed]
- 83.Arthur JS, Ley SC. Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 2013;13:679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- 84.Hawkins PT, Stephens LR. PI3K signalling in inflammation. Biochim. Biophys. Acta. 1851;882–897:2015. doi: 10.1016/j.bbalip.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 85.Oldenburger A, et al. Anti-inflammatory role of the cAMP effectors Epac and PKA: implications in chronic obstructive pulmonary disease. PLoS ONE. 2012;7:e31574. doi: 10.1371/journal.pone.0031574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Atsaves, V., Leventaki, V., Rassidakis, G. Z. & Claret, F. X. AP-1 Transcription factors as regulators of immune responses in cancer. Cancers (Basel)11, 10.3390/cancers11071037 (2019). [DOI] [PMC free article] [PubMed]
- 87.Carter AB, Monick MM, Hunninghake GW. Both Erk and p38 kinases are necessary for cytokine gene transcription. Am. J. Respir. Cell Mol. Biol. 1999;20:751–758. doi: 10.1165/ajrcmb.20.4.3420. [DOI] [PubMed] [Google Scholar]
- 88.Ornatowska M, et al. Functional genomics of silencing TREM-1 on TLR4 signaling in macrophages. Am. J. Physiol. Lung Cell Mol. Physiol. 2007;293:L1377–1384. doi: 10.1152/ajplung.00140.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Werman A, et al. The precursor form of IL-1alpha is an intracrine proinflammatory activator of transcription. Proc. Natl. Acad. Sci. U S A. 2004;101:2434–2439. doi: 10.1073/pnas.0308705101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim JM, Choo JE, Lee HJ, Kim KN, Chang SE. Epidermal growth factor attenuated the expression of inflammatory cytokines in human epidermal keratinocyte exposed to. Ann. Dermatol. 2018;30:54–63. doi: 10.5021/ad.2018.30.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Karin, M. Inflammation-activated protein kinases as targets for drug development. Proc. Am. Thorac. Soc.2, 386–390; discussion 394–385, 10.1513/pats.200504-034SR (2005). [DOI] [PMC free article] [PubMed]
- 92.Yazdi AS, Ghoreschi K. The Interleukin-1 Family. Adv. Exp. Med. Biol. 2016;941:21–29. doi: 10.1007/978-94-024-0921-5_2. [DOI] [PubMed] [Google Scholar]
- 93.Leulier F, Lemaitre B. Toll-like receptors–taking an evolutionary approach. Nat. Rev. Genet. 2008;9:165–178. doi: 10.1038/nrg2303. [DOI] [PubMed] [Google Scholar]
- 94.Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guo S, Dipietro LA. Factors affecting wound healing. J. Dent. Res. 2010;89:219–229. doi: 10.1177/0022034509359125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Obata T, Brown GE, Yaffe MB. MAP kinase pathways activated by stress: the p38 MAPK pathway. Crit. Care Med. 2000;28:N67–77. doi: 10.1097/00003246-200004001-00008. [DOI] [PubMed] [Google Scholar]
- 97.Spaeth CS, Boydston EA, Figard LR, Zuzek A, Bittner GD. A model for sealing plasmalemmal damage in neurons and other eukaryotic cells. J. Neurosci. 2010;30:15790–15800. doi: 10.1523/JNEUROSCI.4155-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.