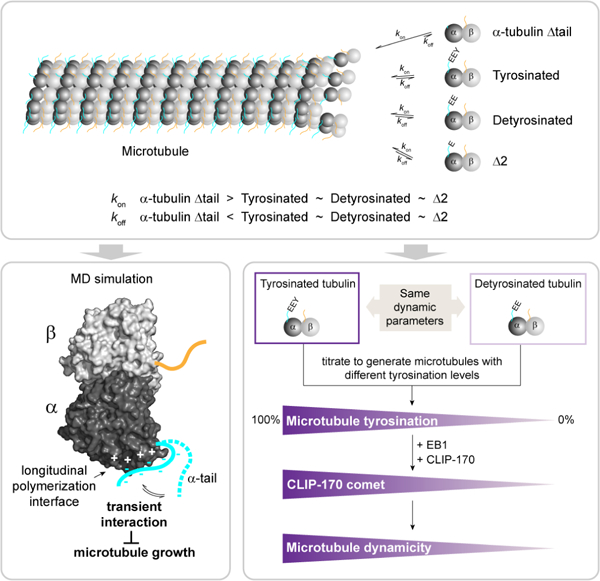
Summary
Microtubules are non-covalent polymers of αβ-tubulin dimers. Posttranslational processing of the intrinsically disordered C-terminal α-tubulin tail produces detyrosinated and ∆2-tubulin. Although these are widely employed as proxies for stable cellular microtubules, their effect (and of the α-tail) on microtubule dynamics remains uncharacterized. Using recombinant, engineered human tubulins, we now find that neither detyrosinated nor ∆2-tubulin affect microtubule dynamics, while the α-tubulin tail is an inhibitor of microtubule growth. Consistent with the latter, molecular dynamics simulations show the α-tubulin tail transiently occluding the longitudinal microtubule polymerization interface. The marked differential in vivo stabilities of the modified microtubule subpopulations, therefore, must result exclusively from selective effector recruitment. We find that tyrosination quantitatively tunes CLIP-170 density at the growing plus-end, and that CLIP170 and EB1 synergize to selectively up-regulate the dynamicity of tyrosinated microtubules. Modification-dependent recruitment of regulators thereby results in microtubule subpopulations with distinct dynamics, a tenet of the tubulin code hypothesis.
Graphical Abstract
eToc
Microtubules functionalized with different posttranslational modifications have different stabilities in cells. Chen et al. show that tyrosinated, detyrosinated and ∆2 microtubules have the same dynamic parameters and that their different dynamicity in cells results from the tyrosination-dependent recruitment of CLIP-170.
Introduction
Microtubules are mesoscopic non-covalent polymers built from αβ-tubulin heterodimers. They switch stochastically between polymerization and depolymerization, a phenomenon known as dynamic instability (Walker, 1988; Horio, 1986; Mitchison and Kirschner, 1984). The transition from growth to depolymerization is called catastrophe and the transition from depolymerization state to growth is called rescue. Dynamic instability is essential for chromosome segregation, cellular motility and differentiation (Kirschner and Mitchison, 1986). Many cellular factors modulate microtubule polymerization and depolymerization. The last two decades have seen an explosion in our understanding of their mechanism of action (reviewed in Alfaro-Aco and Petry, 2015; Akhmanova and Steinmetz, 2010b). However, our understanding of the relationship between the diversity of tubulin itself and its dynamic properties lags behind. Microtubules in cells are functionalized through chemically diverse and evolutionarily conserved posttranslational modifications (Janke and Magiera, 2020; Roll-Mecak, 2020; Verhey and Gaertig, 2007). The majority of these modifications concentrate on the C-terminal tails of the αβ-tubulin heterodimer. These are intrinsically disordered elements that project from the compact, folded globular core of the tubulin dimer and form a negatively-charged dense brush on the microtubule surface. The effects of tubulin tail modifications on microtubule dynamics are unknown, even though these modifications have been widely used for decades as surrogates for microtubule stability in cellular studies where the dynamics of microtubule subpopulations are inferred from their posttranslational modification status detected with specific antibodies.
Some of the most widely used proxies for microtubule stability in cells are detyrosination and tyrosination, for stable and dynamic microtubules, respectively. Detyrosination and tyrosination involve the removal and addition of the terminal tyrosine on α-tubulin, respectively. Tyrosination was the first tubulin posttranslational modification discovered (Barra, et al., 1973). The terminal tyrosine is removed by carboxypeptidases vasohibins/SVBP (Aillaud, et al., 2017; Nieuwenhuis, et al., 2017) and added back by tubulin tyrosine ligase (Flavin and Murofushi, 1984; Argarana, et al., 1978; Raybin and Flavin, 1977; Barra, et al., 1973) as part of this detyrosination/tyrosination cycle. In certain cell types, most prominently in neurons, a large portion of the detyrosinated α-tubulin pool proceeds to a second modification step which involves the removal of the penultimate glutamate in the α-tail resulting in ∆2-tubulin (Paturle-Lafanechere, et al., 1994; Paturle-Lafanechere, et al., 1991). Approximately 35% of brain tubulin is in this state (Paturle-Lafanechere, et al., 1994). This modification is also associated with stable microtubule populations (Paturle-Lafanechere, et al., 1994). Unlike detyrosination, which can be reversed by tubulin tyrosine ligase, the ∆2 modification is irreversible and thus removes this tubulin variant from the detyrosination/tyrosination cycle (Paturle-Lafanechere, et al., 1991). These posttranslational modifications are spatially and temporally regulated and are important in a wide array of biological processes ranging from cell division (Akera, et al., 2017; Iwata-Otsubo, et al., 2017; Barisic, et al., 2015; Peris, et al., 2006) to neurogenesis (Aillaud, et al., 2017; Erck, et al., 2005), myogenesis (Chang, et al., 2002; Gundersen, et al., 1989) and cardiomyocyte function (Robison, et al., 2016); reviewed in Roll-Mecak, 2019). For example, microtubules that orient in the direction of cell migration are enriched in detyrosinated tubulin (Gundersen and Bulinski, 1988); kinetochore microtubules are also enriched in detyrosinated tubulin, while astral microtubules are enriched in tyrosinated tubulin (Gundersen and Bulinski, 1986a). Stable microtubules in axons and cilia are enriched in ∆2 (Paturle-Lafanechere, et al., 1994) and detyrosinated tubulin (Gundersen and Bulinski, 1986b), while the dynamic microtubules in growth cones are mostly tyrosinated (Marcos, et al., 2009; Witte, et al., 2008; Robson and Burgoyne, 1989). The presence or absence of the C-terminal tyrosine on α-tubulin recruits microtubule effectors. For example, the microtubule tip binding protein CLIP-170 (Bieling, et al., 2008; Peris, et al., 2006), the depolymerizing kinesin-13 MCAK (Sirajuddin, et al., 2014; Peris, et al., 2009) and dynactin (McKenney, et al., 2016; Nirschl, et al., 2016; Peris, et al., 2006) associate preferentially with tyrosinated microtubules, while the CENP-E motor prefers detyrosinated microtubules (Barisic, et al., 2015) and detyrosination mediates the interaction between microtubules and intermediate filaments (Kreitzer, et al., 1999).
Pioneering work by several laboratories showed that, in cells, microtubules with slow turnover (t1/2 ~ tens of minutes to hours) and those resistant to cold or pharmacologically induced depolymerization are enriched in detyrosinated and ∆2-tubulin while dynamic microtubules that turn over rapidly (t1/2 ~ 2–5 min) are enriched in tyrosination (Webster and Borisy, 1989; Kreis, 1987; Schulze and Kirschner, 1987; Webster, et al., 1987b). Since then, cell biologists use antibodies against these modifications to distinguish between “stable” and “dynamic” microtubules in cells (Xu, et al., 2011; Whipple, et al., 2010; Shintani, et al., 2009; Infante, et al., 2000; Webster and Borisy, 1989; Kreis, 1987; Schulze and Kirschner, 1987; Webster, et al., 1987a). In fact, this has become almost the “de facto” tool for distinguishing between these two subpopulations of microtubules in cells. However, we still do not know the answer to the fundamental question: how do these modifications affect microtubule dynamics? Do they impact microtubule dynamics directly or do they recruit regulators that impact their dynamics in trans? Kirschner and colleagues speculated that the selective recruitment of hypothetical factors to the detyrosinated microtubule could further stabilize it as part of a positive feedback loop (Schulze and Kirschner, 1987). The effect of posttranslational modifications on polymer dynamics has remained an unsolved problem in the field for more than three decades because of the inability to make well-defined preparations of unmodified and differentially modified tubulin for in vitro reconstitution assays.
An early clue to the effects of the detyrosination/tyrosination cycle came from cellular work that examined the stability of microtubules in detergent-extracted cells that were treated with carboxypeptidase, which non-specifically removes C-terminal amino acids (Khawaja, et al., 1988). These experiments showed that tyrosinated and detyrosinated microtubules respond similarly to nocadozole treatment as well as dilution. However, these studies did not examine microtubule dynamics. Microtubule dynamics parameters of pure tyrosinated, detyrosinated or ∆2-tubulin microtubules have yet to be reported. This gap in our knowledge of the dynamic parameters of different types of tubulins stems from the fact that biochemical investigations into microtubule functions have relied in the last four decades on tubulin purified from brain tissue. While tubulin from this source is easily isolated and abundant (Weisenberg, 1972), it is highly heterogeneous as it is abundantly and diversely posttranslationally modified. Thus, this tubulin is not appropriate for understanding the individual contributions of posttranslational modifications to microtubule functions. Recent successes with the expression and purification of single tubulin isotypes recombinantly (Ti, et al., 2018; Vemu, et al., 2016; Minoura, 2013) enable us to explore the relationship between tubulin sequence, structure, and dynamics and establish whether and how tubulin modifications regulate intrinsic polymer properties.
Here, we use recombinant homogenous and isotypically pure α1A/βIII tubulin to understand the role of the α-tubulin tail and its modifications in regulating microtubule dynamics. Combining in vitro microtubule dynamics assays and molecular dynamics simulations we show that the intrinsically disordered α-tubulin tail negatively regulates microtubule polymerization by transiently interacting with the α-tubulin longitudinal polymerization interface. In contrast, loss of only the ultimate tyrosine (detyrosination) and penultimate glutamate (∆2 modification) in the α-tubulin tail does not affect microtubule dynamic parameters. However, we show that α-tubulin tyrosination quantitatively tunes the density of CLIP-170 at the microtubule plus end and that CLIP-170 synergizes with EB1 to selectively increase the dynamicity of tyrosinated microtubules. Thus, our work establishes that tyrosinated, detyrosinated and the ∆2 modified microtubules have similar intrinsic dynamic parameters and demonstrates that tyrosination-dependent recruitment of microtubule regulators can generate microtubule subpopulations with distinct dynamic properties, laying the foundation for the regulation of microtubule dynamics by the tubulin code in cells.
Results
The Intrinsically Disordered α-tubulin Tail Inhibits Microtubule Growth and Dynamicity
The last twelve residues in the α-tubulin tail are intrinsically disordered. The C-terminal tails of tubulin are not resolved in all cryo-electron microscopy microtubule reconstructions to date, including reconstructions of homogenous single-isotype microtubules (Ti, et al., 2018; Vemu, et al., 2016). Cryo-EM studies revealed that the C-terminal tails are ordered only when bound to microtubule effectors such as the Ndc80 complex (Alushin, et al., 2010) or the tubulin tyrosine ligase-like 7 (TTLL7) glutamylase (Garnham, et al., 2015). The proteolytic removal of both α and β‒tails lowers the critical concentration for tubulin polymerization, as measured by sedimentation assays, and results in the formation of heterogenous polymeric structures such as ribbons, sheets and hooks (Knipling, et al., 1999; Serrano, et al., 1988; White, et al., 1987). However, these experiments relied on the non-specific proteolytic cleavage of both C-terminal tails that yields a heterogenous sample with varying degrees of tubulin tail removal. Moreover, all these studies used proteases that cleave the β-tail before the α-tail (Bhattacharyya, et al., 1985) and thus could not establish the effects of the α-tail alone on microtubule properties. To study the contribution of the α-tubulin tail to microtubule dynamics, we recombinantly expressed and purified wild-type human α1A/βIII tubulin with a native carboxy-terminus (Vemu, et al., 2016), and an α1A∆-tail/βIII tubulin which misses the last twelve residues of α-tubulin (Figures 1A, S1A, B and S2A; STAR Methods). We then reconstituted microtubule dynamics in vitro by nucleating microtubules from GMPCPP-stabilized microtubule seeds and imaged the microtubules by label-free interference reflection microscopy (IRM) (Mahamdeh, et al., 2018; Figure 1B). Label-free imaging avoids the confounding effects of fluorescently labeled tubulin on microtubule dynamics (Vigers, et al., 1988) and allows data collection with high temporal resolution and minimal photodamage.
Figure 1: The α-tubulin tail inhibits microtubule polymerization and catastrophe.
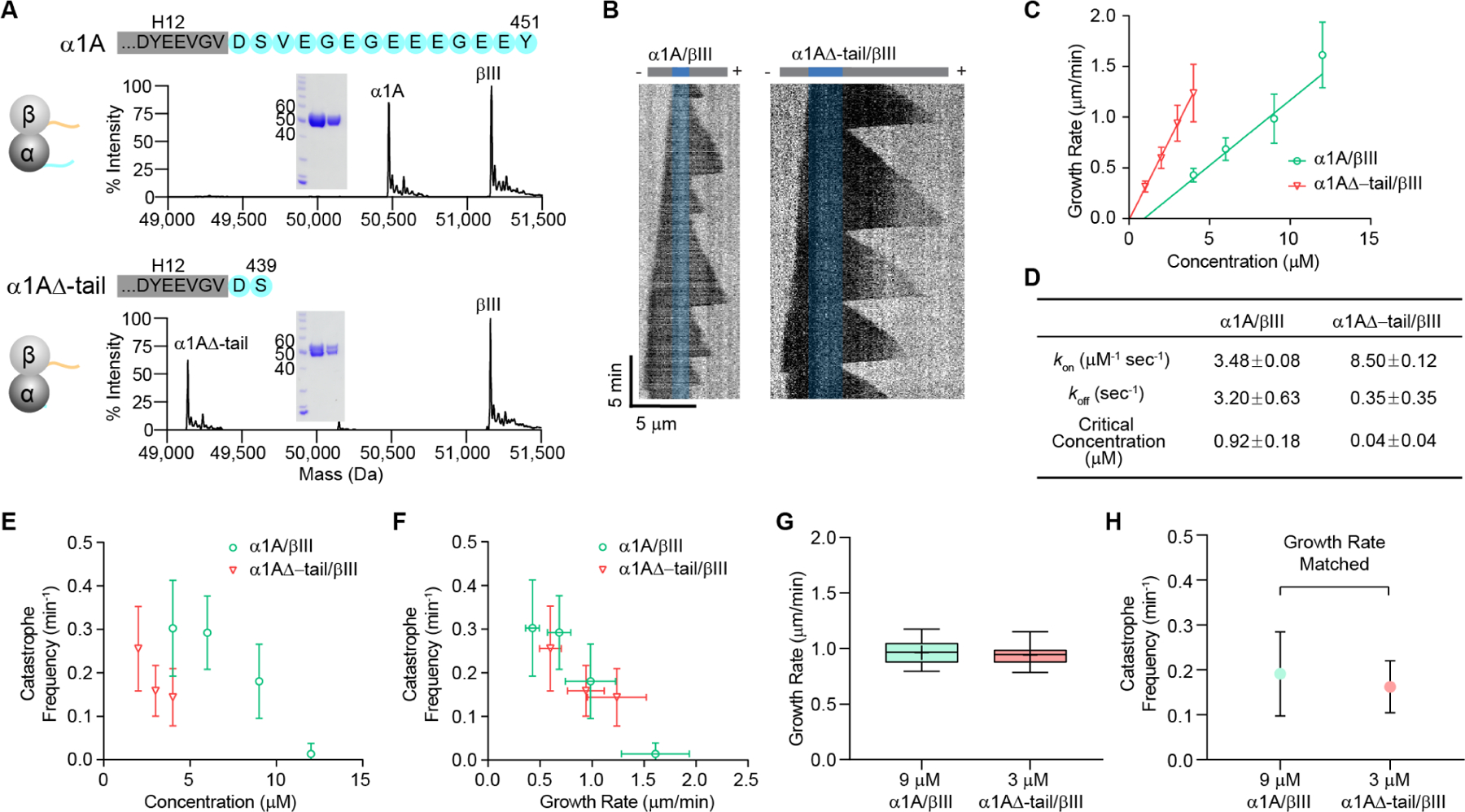
(A) Mass spectra and SDS-polyacrylamide gel (inset: 5µgs and 2µgs of tubulin) of recombinant human α1A/βIII and α1A∆-tail/βIII tubulin purified to > 99% homogeneity (see Figures S1A, B and STAR Methods). The C-terminal sequences of human α1A and α1A∆-tail tubulin are shown in cyan. The sequence in the grey rectangle denotes the H12 helix, based on the cryo-EM structure of α1A/βIII microtubules (PDB ID: 5JCO; (Vemu, et al., 2016)).
(B) Representative kymographs showing microtubule growth at 3 µM for α1A/βIII (left) and α1A∆-tail/βIII (right) microtubules. GMPCPP-stabilized seeds are pseudo-colored in blue.
(C) Plus-end growth rates as a function of tubulin concentration for α1A/βIII and α1A∆-tail/βIII tubulin. Error bars represent S.D. (n= 181, 256, 608, 82 growth phases for α1A/βIII at 4, 6, 9, 12 µM, respectively; n= 214, 518, 529, 424 growth phases for α1A∆-tail/βIII at 1, 2, 3, 4 µM, respectively).
(D) Tubulin apparent on- and off-rates at the plus-end, and critical concentration determined by fitting a linear regression of the mean growth rates versus tubulin concentration shown in (C); p < 0.0001 as determined by the Tukey range test for the differences in kon and koff rates between α1A/βIII and α1A∆-tail/βIII tubulin.
(E) Microtubule plus-end catastrophe frequencies as a function of tubulin concentration. Error bars represent S.D. (n= 27, 29, 86, 59 microtubules for α1A/βIII at 4, 6, 9, 12 µM, respectively; n= 59, 63, 73, 70, 23 microtubules for α1A∆-tail/βIII at 2, 3, 4, 6 µM, respectively).
(F) Microtubule plus-end catastrophe frequencies plotted against growth rates. Error bars represent S.D. (n values as in (C) and (E)).
(G) Box-whisker plot of α1A/βIII and α1A∆-tail/βIII microtubules with matched growth rates within 10% S.D. (0.96 ± 0.10 μm/min, n= 61 α1A/βIII microtubules at 9 µM; 0.94 ± 0.09 μm/min, n= 59 α1A∆-tail/βIII microtubules at 3 µM). Whiskers indicate minimum and maximum.
(H) Catastrophe frequencies for growth-rate matched α1A/βIII and α1A∆-tail/βIII microtubules shown in (G); n= 61 and 59 microtubules for α1A/βIII and α1A∆-tail/βIII, respectively; error bars represent S.D.
We find that tailless α1A∆-tail/βIII microtubules nucleate from seeds robustly at tubulin concentrations as low as 1 μM. Consistent with this, the critical concentration for the α-tailless tubulin is 0.04 ± 0.04 μM for α1A∆-tail/βIII versus 0.94 ± 0.18 μM for the α1A/βIII wild-type construct, as determined from a linear regression of the mean growth rates as a function of tubulin concentration (Figure 1C; (Oosawa, 1970); STAR Methods). This analysis also shows that the absence of the α-tail increases the apparent tubulin on-rate at the plus end by 2.4-fold (8.50 ± 0.12 versus 3.48 ± 0.08 μM−1 s−1 for α1A∆-tail/βIII and α1A/βIII microtubules, respectively, Figure 1D) while also decreasing the apparent off-rate by almost ten-fold (0.35 ± 0.35 versus 3.20 ± 0.63 s−1 for α1A∆-tail/βIII and α1A/βIII microtubules, respectively, Figure 1D). Thus, α-tailless microtubules grow considerably faster. For example, at 4 μM tubulin, α1A∆-tail/βIII microtubules grow 2.9-fold faster than wild-type microtubules. This increase in growth rate is accompanied by a decrease in catastrophe rates (Figure 1E). As a result, at 4 µM tubulin, in the absence of the α-tail, microtubules grow almost 6-fold longer before they depolymerize [(7.82 ± 0.35 µm versus 1.32 ± 0.07 µm for α1A∆-tail/βIII and α1A/βIII microtubules, respectively (Figure S1C)]. Rescues are very rare both for α1A/βIII and α1A∆-tail/βIII microtubules and were thus not quantified, similar to the low rescue rates observed for heterogenous brain microtubules (Vemu, et al., 2016; Walker, 1988). Thus, our in vitro dynamics microtubule assays reveal that the α-tubulin tail inhibits microtubule growth and promotes catastrophe.
A growing microtubule is protected from depolymerization by a GTP-tubulin cap generated by the faster rate of GTP-tubulin addition to the microtubule end compared to that of GTP hydrolysis after tubulin lattice incorporation (Carlier, 1982; Carlier and Pantaloni, 1981). The microtubule undergoes catastrophe when its GTP cap erodes (Duellberg, et al., 2016). Plotting plus-end catastrophe frequencies against growth rate shows that microtubules catastrophe less frequently when growth rates increase and that they are similar between the wild-type and tailless microtubules at similar growth rates, i.e., at similar rates of GTP-tubulin addition to the growing plus end (Figure 1F). We further refined our investigation into whether catastrophe frequencies are due to differences in the size of the stabilizing GTP-tubulin cap by determining catastrophe frequencies of α1A/βIII and α1A∆-tail/βIII microtubules with growth rates that matched within less than 10% standard deviation (Figure 1G). This analysis also shows that catastrophe frequencies are statistically indistinguishable (Figure 1H). Thus, the decrease in catastrophe frequency seen with the α-tailless tubulin is due primarily to the differences in the kinetics of tubulin addition at the microtubule end.
The α-tubulin Tail Interacts Transiently with the Longitudinal Polymerization Interface
We hypothesized that the strong attenuation of microtubule growth rates by the α-tubulin tail may be explained by a transient interaction with surfaces on the tubulin dimer involved in polymerization (Figure 2A). Thus, we used all-atom explicit solvent MD simulations of free GTP-tubulins to investigate the dynamics of the intrinsically disordered tubulin tails (Figure S3A). These simulations revealed that the α-tubulin tail interacts transiently with the α-tubulin longitudinal polymerization interface, thus likely directly interfering with its addition to a protofilament in the microtubule (Figure 2B, Movie S1). We observe this interaction in all of our five independent 1 µs simulations. The α-tail associates with the longitudinal interface for 83 ± 20% of the simulation time, with as little as 52% in one simulation, but in some 100% of the time. Engagement in relatively long-lasting interactions with the tubulin body explains the variable occlusion durations in our simulations. A metadynamics simulation, in which an extra energy potential is applied to the C-terminus of the tail in order to accelerate its dynamics and ensure better sampling of configuration space also revealed an association of the α-tubulin tail with the longitudinal polymerization interface (Figure S4A, B, C). Association of the α-tubulin tail occludes up to 33% of the solvent accessible area of the longitudinal interface (Figure 2C). We mapped the regions on the tubulin body surface with which the α-tail prefers to interact by computing the probabilities of contact between the α-tubulin tail and residues in the tubulin body (Figure 2D). The residues with highest contact probabilities map to the α-tubulin longitudinal polymerization interface (Figures 2D, E, F and Figure S4C). The simulations indicate that the interaction with the α-tail is facilitated by multiple contacts with conserved positively charged residues at the interface through the negatively charged glutamates in the α-tail (Figures 2G and S3B). The individual tubulin tail residues have similar contact probabilities with the α-tubulin longitudinal interface (Figure S3C). This suggests that there isn’t a single, specific residue on the α-tail that nucleates this interaction. The high-density of glutamates in the tail is conserved across all α-tubulin isotypes (Figure S2), despite the higher sequence variability in this region compared to the tubulin core. Moreover, the length of the α-tubulin tails is more stringently conserved compared to that of β-tubulin tails (Roll-Mecak, 2015; Luduena, 2013 and Figure S2), consistent with an evolutionary pressure to maintain important functional interactions.
Figure 2: The α-tubulin tail occludes the longitudinal polymerization interface transiently.
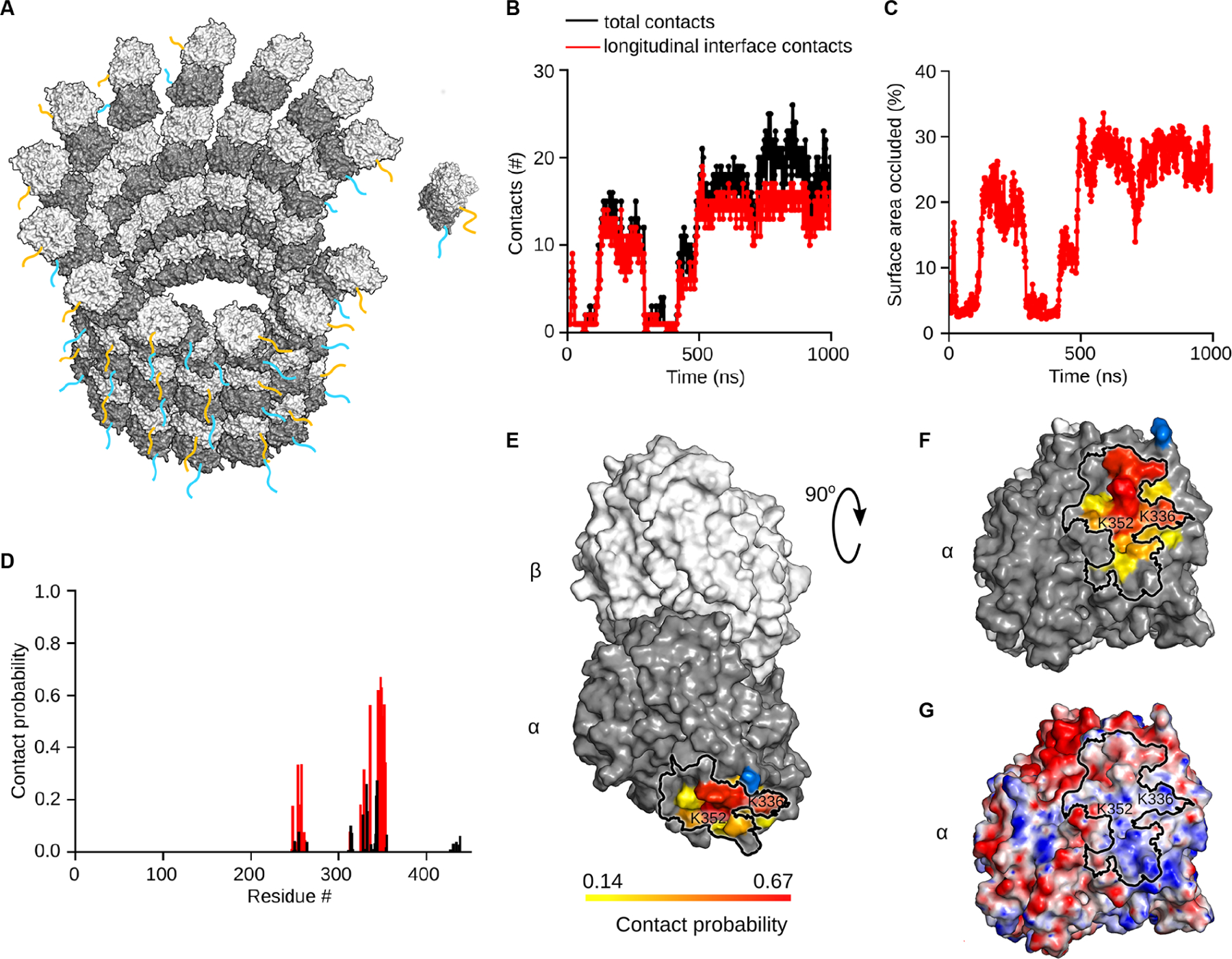
(A) Schematic of tubulin dimer incorporation into a protofilament at the growing microtubule plus-end. α-tubulin tail, cyan, β-tubulin tail, orange in cartoon representation.
(B) Number of contacts between residues in the tubulin body and the α-tubulin tail as a function of time identified from the MD simulation. Black dots indicate total number of contacts, red dots indicate contacts with the α-tubulin longitudinal polymerization interface.
(C) Percentage of solvent-accessible surface area of the α-tubulin longitudinal interface occluded by the α-tubulin tail in the simulation shown in (B).
(D) Contact probability for residues in the α-tubulin core with the α-tubulin tail based on five independent one-microsecond-long simulation runs (STAR Methods). Red bars correspond to contacts with the α-tubulin polymerization interface, black bars, contacts with the tubulin surface elsewhere.
(E) Molecular surface of the αβ-tubulin dimer colored on a gradient from yellow to red according to the probability of its contact with the α-tubulin tail. Residue S439 where the tail originates is shown in blue.
(F) View of the longitudinal polymerization interface colored as in (E). Black outline delineates the longitudinal polymerization interface.
(G) Electrostatic potential distribution mapped on the surface of the tubulin dimer in the same orientation as in panel (F). Red and blue colors correspond to negative and positive electrostatic potential values, respectively. Black outline delineates the longitudinal polymerization interface. See also Figures S3, S4.
Tyrosinated, detyrosinated and ∆2 microtubules have similar dynamics in vitro
Cells posttranslationally remove the ultimate and penultimate residues in the α-tubulin tail to generate detyrosinated and ∆2 microtubules, respectively. Detyrosinated and ∆2 microtubules are associated with stable microtubule subpopulations in cells (Paturle-Lafanechere, et al., 1994; Webster and Borisy, 1989; Kreis, 1987; Schulze and Kirschner, 1987; Webster, et al., 1987b). Since removing the entire intrinsically disordered α-tubulin tail has dramatic effects on microtubule stability, we next examined whether detyrosination and the ∆2 modification also affect intrinsic polymer properties. We performed in vitro dynamics assays with recombinantly expressed, homogenous and isotypically pure human tyrosinated (α1A/βIII), detyrosinated (α1A-Y/βIII) and ∆2 (α1A-∆2/βIII) tubulin (Figures 3A, 3B). Our tagging scheme produces an α-tubulin with a native carboxy-terminus (Vemu, et al., 2016; STAR Methods) and thus suitable for examining the effects of the α-tail modification cycle without any confounding effects from a tag. Using our IRM-based in vitro dynamics assays we find that detyrosinated (α1A-Y/βIII) and ∆2 (α1A-∆2/βIII) microtubules have similar growth rates across a range of concentrations (Figure 3C). A linear regression analysis of the growth rates as a function of tubulin concentration yields similar tubulin apparent on and off-rates at the plus-end for all three tubulin variants (Figure 3D). Both detyrosinated and ∆2-tubulin show a statistically significant but very small increase (11% and 9%, respectively) in their apparent on-rates compared to tyrosinated tubulin, consistent with our molecular dynamics simulations which show that the two terminal tubulin tail residues do not have a dominant contribution to the interaction with the α-tubulin longitudinal interface, having similar contact probabilities as other tail residues (Figure S3C). Our analysis also reveals that tyrosinated, detyrosinated and ∆2 tubulin have similar critical concentrations for nucleation. Lastly, the three microtubule types also have similar catastrophe frequencies across a range of concentrations (Figure 3E). The speeds at which the three different types of microtubule depolymerize are also statistically indistinguishable (Figures 3F, G). In conclusion, our in vitro results demonstrate quantitatively that the in vivo differences in dynamics between detyrosinated, ∆2 and tyrosinated microtubule subpopulations are exclusively due to the differential recruitment of effectors and not to intrinsic differences in polymer dynamics.
Figure 3: Tyrosinated, detyrosinated and ∆2 microtubules have similar dynamic parameters.
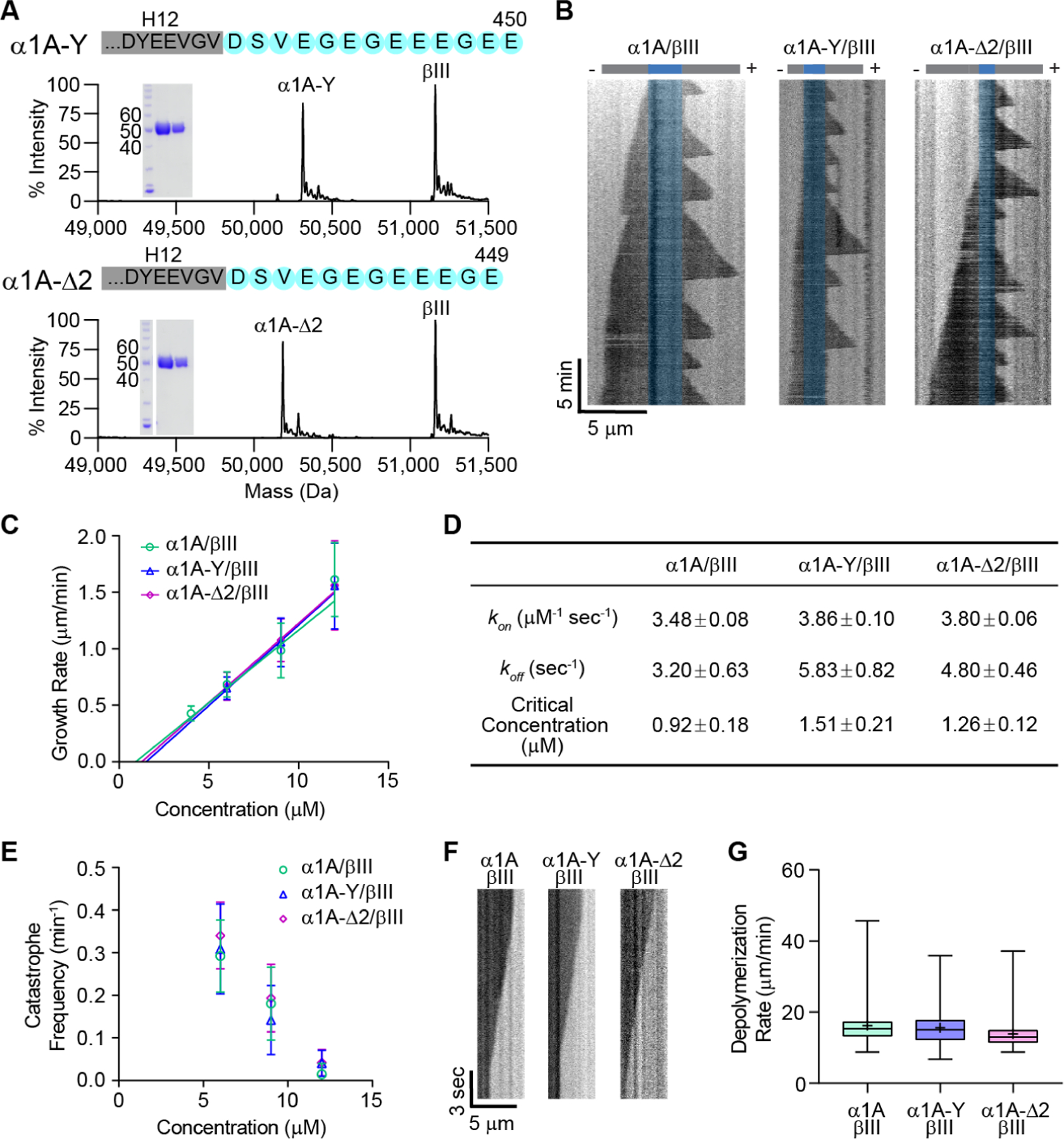
(A) Mass spectra and SDS-polyacrylamide gel (inset: 5µgs and 2µgs of tubulin) of recombinant human α1A-Y/βIII (top) and α1A-∆2/βIII (bottom) tubulin purified to > 99% homogeneity.
(B) Representative kymographs showing microtubule growth at 6µM for α1A/βIII (left), α1A-Y/βIII (middle), and α1A-∆2/βIII (right) microtubules. GMPCPP-stabilized seeds are pseudo-colored in blue.
(C) Plus-end growth rates as a function of tubulin concentration for α1A/βIII, α1A-Y/βIII and α1A-∆2/βIII microtubules. Error bars represent S.D. (n= 181, 256, 608, 82 growth phases for α1A/βIII at 4, 6, 9, 12 µM, respectively; n= 357, 638, 63 growth phases for α1A-Y/βIII at 6, 9, 12 µM, respectively; n= 1139, 729, 67 growth phases for α1A-∆2/βIII at 6, 9, 12 µM, respectively).
(D) Tubulin apparent on- and off-rates at the plus-end and critical concentration determined by fitting a linear regression of the mean growth rates versus tubulin concentration shown in (C); p = 0.003 for α1A /βIII vs. either α1A-Y/βIII or α1A-∆2/βIII on-rates ; p = 0.85 for α1A-Y/βIII vs α1A-∆2/βIII on-rates. Differences in intercepts (off-rate and nucleation concentration) are not statistically significant. Statistical analysis performed using a Tukey range test.
(E) Microtubule plus-end catastrophe frequencies as a function of tubulin concentration. Error bars represent S.D.; n= 29, 86, 59 microtubules for α1A/βIII at 6, 9, 12 µM, respectively; n= 37, 116, 21 microtubules for α1A-Y/βIII at 6, 9, 12 µM, respectively; n= 96, 97, 22 microtubules for α1A-∆2/βIII at 6, 9, 12 µM, respectively.
(F) Representative kymographs showing depolymerization at 6µM for α1A/βIII (left), α1A-Y/βIII (middle), and α1A-∆2/βIII (right) microtubules.
(G) Box-whisker plot (whiskers indicate minimum and maximum) of depolymerization rates; n= 26, 50, and 48 growth phases for α1A/βIII, α1A-Y/βIII, and α1A-∆2/βIII tubulin, respectively.
Selective recruitment of CLIP-170 to tyrosinated microtubules increases their dynamicity
One of the tenets of the tubulin code hypothesis is that posttranslational modifications tune microtubule dynamics either directly or indirectly, through the recruitment of cellular factors, thus creating microtubule subpopulations with different dynamic properties in cells. We thus wanted to test whether the tyrosination specific recognition of a cellular effector can selectively regulate microtubule stability. The dynamic behavior of microtubules in cells is modulated by a large array of factors. Most notable among them are what are known as “tip-tracking” factors because they preferentially associate with the growing end of the microtubule (reviewed in (Akhmanova and Steinmetz, 2008)). The end-binding (EB) family of proteins are the master regulators at the growing microtubule end where they recruit an entire network of proteins that synergize to regulate microtubule dynamics (reviewed in Akhmanova and Steinmetz, 2010a). Among them, CLIP-170 is expressed in mammals, invertebrates and yeast (Akhmanova, et al., 2005; reviewed in Gouveia and Akhmanova, 2010) and its association with microtubules is enhanced by tubulin tyrosination (Bieling, et al., 2008). Biochemical and structural studies showed that this specific recognition is mediated by the N-terminal cytoskeleton-associated protein glycine-rich (CAP-Gly) domains of CLIP-170 that recognize the terminal tyrosine in the- EEY motif present both in the C-terminal tail of α-tubulin and EB1 (Bjelic, et al., 2012; Bieling, et al., 2008; Mishima, et al., 2007; Weisbrich, et al., 2007). The association of CLIP170 with EB1 is stronger than that with the tyrosinated α-tubulin tail (Chen, et al., 2019) and CLIP-170 requires EB1 for tip-tracking (Dixit, et al., 2009; Bieling, et al., 2008), while detyrosination reduces CLIP-170 end tracking activity both in vitro (Bieling, et al., 2008) and in vivo (Peris, et al., 2006). Thus, differential tyrosination of the microtubule lattice can potentially function as a quantitative tuner of CLIP-170 recruitment to the microtubule tip and microtubule dynamics.
We used our recombinant tubulin to reconstitute a minimal tip-tracking complex with EB1 and CLIP-170 (Figure 4). We used a CLIP-170 construct, H2, which contains the two N-terminal CAP-Gly domains and part of the coiled-coil domain needed for dimerization. This construct localizes to the growing plus-end both in vitro and in cells (Bieling, et al., 2008; Perez, et al., 1999; Scheel, et al., 1999). We titrated α1A/βIII with α1A-Y/βIII to generate microtubules with different percentages of tyrosination (0%, 25%, 50%, 75%, and 100%). We then reconstituted GFP-H2 tip-tracking on the differentially tyrosinated microtubules (Figure 4A) and characterized the comet profiles (Figure 4B). The fluorescence intensity of GFP-H2 comets increases with tyrosination and is highest on the 100% tyrosinated microtubules (Figures 4B, C). This difference is due exclusively to the selective recognition of the tyrosinated tubulin by CLIP-170 as EB1 comets exhibit no distinguishable differences between α1A/βIII with α1A-Y/βIII microtubules (Figure S5A, B, C). This is consistent with previous observations which showed that treatment of brain tubulin with carboxypeptidase A, which removes residues from the C-terminus of a protein, does not affect EB1 microtubule association, but affects the recruitment of CLIP-170 to microtubule ends (Bieling, et al., 2008). Moreover, growth rate-matched porcine brain microtubules which are ~ 25–50% detyrosinated (Paturle, et al., 1989) show similar GFP-H2 comet profiles to those with the 50/50 tyrosinated/detyrosinated recombinant tubulin mixture, while the GFP-H2 comet intensity on growth rate-matched tyrosinated tubulin purified from tsA201 cells (Vemu, et al., 2017) is similar to that of 100% tyrosinated α1A/βIII recombinant tubulin (Figure 4B, C). These results indicate that tyrosination is the dominant factor recruiting CLIP-170 to microtubules of diverse tubulin isotype composition and that it quantitatively tunes the density of CLIP-170 at the growing microtubule ends.
Figure 4. CLIP-170 and EB1 synergize to selectively increase the growth and catastrophe rates of tyrosinated microtubules.
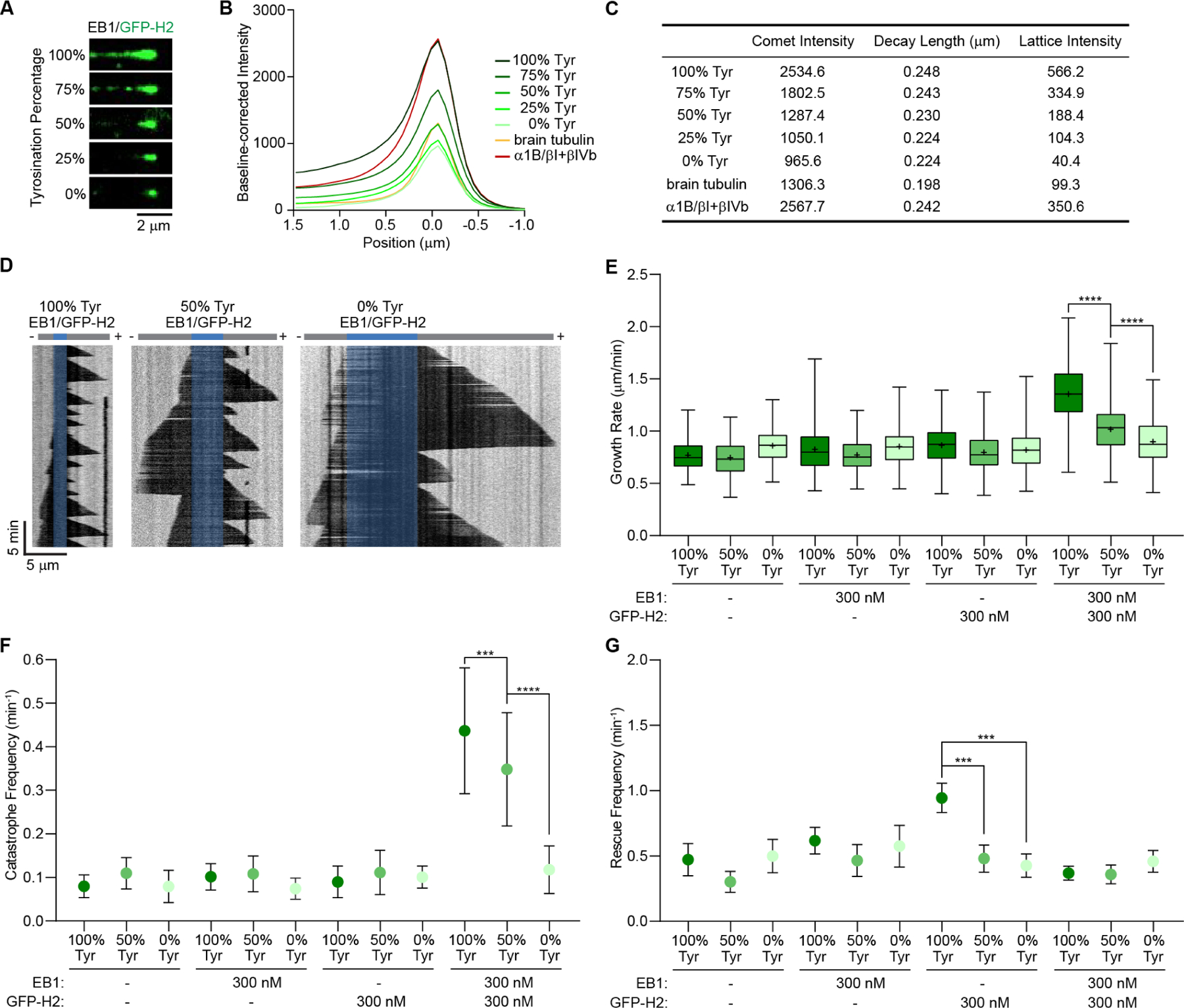
(A) TIRF microscopy images of GFP-H2 (75 nM, green) on recombinant microtubules with different percentages of tyrosination in the presence of 300 nM unlabeled EB1. Recombinant α1A/βIII and α1A-Y/βIII were mixed proportionally to generate a gradient of tyrosination with a total tubulin concentration of 15 μM.
(B) Averaged fluorescence intensity profiles of GFP-H2 comets on microtubules with different tyrosination content (15 μM recombinant tubulin), growth rate-matched heterogenous brain microtubules (10 μM), and growth rate-matched α1B/βI+βIV microtubules purified from tsA201 cells (12 μM) in the presence of 300 nM unlabeled EB1. (C) Comet profile parameters. Comet profiles were fitted using Gaussian (−1.5 to 0 μm) and single exponential functions (0 to 1.5 μm; STAR Methods); n= 1305, 1676, 1890, 1163, and 442 for recombinant tubulin with 100%, 75%, 50%, 25%, and 0% tyrosination, respectively; n= 2500 and 1618 for brain and α1B/βI+βIV tubulin, respectively.
(D) Representative kymographs of tyrosinated (100% Tyr; left), 50/50 tyrosinated/detyrosinated (50% Tyr; middle), and detyrosinated (0% Tyr; right) microtubules (6 μM recombinant tubulin) in the presence of 300 nM EB1 and 300 nM GFP-H2. GMPCPP-stabilized seeds are pseudo-colored in blue.
(E, F, G) Plus-end growth rates (E), catastrophe frequencies (F) and rescue frequencies (G) of 6 μM α1A/βIII and α1A-Y/βIII microtubules with or without 300 nM EB1 and 300 nM GFP-H2. (E) Box-whisker plot (whiskers represent minimum and maximum) of growth rates; n= 86, 202, 105 growth phases for 100%, 50%, 0% tyrosinated microtubules, respectively; n= 223, 247, 80 growth phases for 100%, 50%, 0% tyrosinated microtubules with EB1, respectively; n= 223, 246, 204 growth phases for 100%, 50%, 0% tyrosinated microtubules with GFP-H2, respectively; n= 934, 587, 106 growth phases for 100%, 50%, 0% tyrosinated microtubules with EB1 and GFP-H2, respectively; **** denotes p < 0.0001 as determined by an unpaired t-test. (F) Catastrophe frequencies; n= 18, 33, 22 microtubules for 100%, 50%, 0% tyrosinated microtubules, respectively; n= 43, 39, 17 microtubules for 100%, 50%, 0% tyrosinated microtubules with EB1, respectively; n= 41, 43, 36 microtubules for 100%, 50%, 0% tyrosinated microtubules with GFP-H2, respectively; n= 63, 48, 21 microtubules for 100%, 50%, 0% tyrosinated microtubules with EB1 and GFP-H2, respectively. Error bars indicate S.D. (G) Rescue frequencies; n values as in (F). Error bars represent S.E.M. **** denotes p < 0.0001 as determined by a Mann-Whitney test. See also Figure S5, S6.
But does this tyrosination-dependent increase in CLIP-170 recruitment to the microtubule tip change microtubule dynamics? Quantitation of dynamic parameters on tyrosinated and detyrosinated microtubules reveals that addition of both EB1 and H2 increases the polymerization speed of tyrosinated α1A/βIII microtubules by ~50% compared to detyrosinated α1A-Y/βIII (Figure 4D, E). Neither EB1 nor H2 increase growth speeds individually (Figure 4E), indicating that the two factors synergize at the microtubule tip in a tyrosination-dependent manner to selectively increase the polymerization speed of tyrosinated microtubules. This effect is proportional, since having a 50/50 tyrosinated/detyrosinated tubulin mixture results in an intermediate effect on microtubule growth. The effect on growth rates depends on the amount of CLIP-170 recruited at the tip and is more modest at lower H2 concentrations (300 versus 75 nM H2; Figure S5D). Surprisingly, the two factors also synergize to increase the catastrophe frequency of tyrosinated microtubules by ~3.7 fold (Figure 4F). Again, the effect is proportional to the tyrosination levels as microtubules containing 50% tyrosinated tubulin show an intermediate effect on catastrophe rates. Neither EB1 nor H2 increase catastrophe rates individually under these conditions. The rescue frequencies of α1A/βIII and α1A-Y/βIII microtubules showed no statistically distinguishable differences when EB1 and CLIP-170 are present together (Figure 4G). However, in the absence of EB1, CLIP-170 increases selectively the rescue frequency of tyrosinated microtubules and not detyrosinated ones (Figure 4G, S6A) by binding with higher affinity along the tyrosinated microtubule lattice (Figure S6B). Taken together, our in vitro experiments demonstrate that tyrosination quantitatively tunes CLIP-170 recruitment to the microtubule end and that this tyrosination-dependent recruitment increases microtubule dynamicity proportionately.
Discussion
In this study we used the power of recombinant well-defined tubulin variants to interrogate the effects of the α-tubulin tail and its three abundant posttranslational modifications on microtubule dynamics. We find that the intrinsically disordered α-tubulin tail is a potent inhibitor of microtubule growth and catastrophe by regulating both the apparent on-rate and off-rates of tubulin at the growing microtubule end (Figure 1D). We speculate that the marked effect of the α-tail on the apparent off-rate at the growing plus-end is due to the fact that at the higher rates of tubulin addition of the α-tailless construct, microtubule growth is dominated by the addition of new GTP-tubulin to the GTP-cap, while in the wild-type tubulin, the rate of addition is closer matched to that of GTP hydrolysis and the tips have a mixture of GDP and GTP tubulins, resulting in a higher apparent tubulin off-rate at the growing tip. Consistent with the effect on growth, our molecular dynamics simulations reveal that the α-tubulin tail interacts with the α-tubulin body and occludes the longitudinal polymerization interface (Figure 2B). Recent NMR studies of Tetrahymena thermophila tubulin demonstrated that α-tubulin tail peptides have different conformations in isolation versus in the context of the αβ-tubulin dimer (Wall, et al., 2016), suggesting transient interactions between the tail and the folded tubulin body that could modulate tubulin polymerization; however, the identity of the tubulin surfaces involved in these interactions was not assigned, nor the effects of the α-tubulin tail loss on microtubule dynamics characterized. The transient occlusion of the longitudinal polymerization interface by the α-tail is facilitated through charge-charge interactions by the large glutamate density in the α-tubulin tail. A positive effect on microtubule growth speed was recently reported after removal of the β-tubulin C-terminal tail using controlled proteolysis (Fees and Moore, 2018). However, in contrast to the α-tail, removal, of the β-tail leads to an increase and not a decrease in catastrophe rates (Fees and Moore, 2018). Thus, the two tubulin tails have evolved to have complex, distinct effects on microtubule dynamics. Since their evolution is not as tightly constrained as that of the folded polymerization interfaces, they likely evolved to provide tunable properties to the microtubule polymer.
The inhibitory role of the α-tubulin tail has interesting parallels to that recently described for a bacterial tubulin homolog from Bacillus subtilis, FtsZ which also uses its intrinsically disordered C-terminal tail to attenuate its incorporation into filaments (Cohan, et al., 2020). More generally, intrinsically disordered regions are frequently employed in proteins to tune the equilibrium between active and inactive states (Trudeau, et al., 2013). In the case of tubulin, it is important to note that the vast majority of microtubule effectors interact with the tubulin tails (Tan, et al., 2019; Yadav, et al., 2014; Knipling and Wolff, 2006; Westermann, et al., 2005; Skiniotis, et al., 2004; Wang and Sheetz, 2000; Marya, et al., 1994; Serrano, et al., 1985; Serrano, et al., 1984a; reviewed in Roll-Mecak, 2015) and some of them are completely dependent on the tails for high-affinity microtubule binding (Zehr, et al., 2020; Roll-Mecak and Vale, 2005). Early studies demonstrated that removal of both α- and β-tubulin tails by subtilisin lowers tubulin critical concentration to that found in the presence of microtubule associated proteins (MAPs) (Sackett, et al., 1985; Serrano, et al., 1984b). Thus, the occlusion of the longitudinal polymerization interface by the α-tubulin tail raises the interesting possibility that MAPs increase the tubulin on-rate by sequestering and changing the dynamics of the α-tubulin tail.
In contrast to the marked effect of the α-tail on microtubule dynamics, we find that microtubules composed of detyrosinated and ∆2-tubulin have similar dynamics to tyrosinated microtubules (Figure 3D), even though these two modifications correlate strongly with increased microtubule stability in cells and are used widely as reporters for microtubule stability. However, we show that tyrosination quantitatively recruits CLIP-170 to the plus-ends and facilitates a synergy between CLIP-170 and EB1 to increase growth and catastrophe rates (Figure 4), providing proof in a reconstituted system that a microtubule posttranslational modification can selectively regulate microtubule dynamic parameters through the specific recruitment of effectors. In contrast, a modification on the tubulin body also frequently used as a marker for microtubule stability in cells, Lys40 acetylation, has a direct effect on microtubule properties by protecting them from breakage (Portran, et al., 2017; Xu, et al., 2017).
The synergy between EB1 and CLIP-170 in promoting catastrophe on tyrosinated microtubules is surprising because CLIP-170 is largely thought of as a catastrophe suppressing factor (Martin-Garcia and Mulvihill, 2009; Brunner and Nurse, 2000) and previous work has also shown that CLIP-170 can synergize with EB1 to elicit a modest decrease in catastrophe rates on brain microtubules (Lopus, et al., 2012). EB1 on the other hand has been shown to increase catastrophe rates (Zanic, et al., 2013; Vitre, et al., 2008; Bieling, et al., 2007), presumably by accelerating a conformational transition at the plus-end of the microtubule (Zhang, et al., 2018; Zhang, et al., 2015). While EB1 alters the interaction of CLIP-170 with the microtubule by recruiting it to the tip, shown in this work and many others (Dixit, et al., 2009; Bieling, et al., 2008; Peris, et al., 2006), our data also support the model where CLIP-170 itself alters the association of EB1 at the plus end and thus possibly accelerates the kinetics of the structural transitions that lead to catastrophe. Such an interaction between EB1 and CLIP-170 is in line with previous work showing that CLIP-170 can recruit to the microtubule a truncated EB1 construct that lacks microtubule binding (Gupta, et al., 2010). When not recruited to the tip by EB1, CLIP-170 promotes rescues preferentially of tyrosinated microtubules (Figure 4G), consistent with previous studies identifying it as a rescue factor (Henrie, et al., 2020; Arnal, et al., 2004; Komarova, et al., 2002). Previous work has shown that MCAK13 selectively depolymerizes tyrosinated microtubules and this was also proposed to increase their dynamicity (Peris, et al., 2009). Thus, tyrosination can act through a combination of effectors to increase microtubule turnover in cells.
We note that the α-tubulin tail -EEY motif recognized by CLIP-170 (Bjelic, et al., 2012; Mishima, et al., 2007; Weisbrich, et al., 2007) is invariant in six of the eight α-tubulin isotypes except for α4A, and α8, (Figure S2A). α4A, which lacks the C-terminal tyrosine, is upregulated in megakaryocytes as they differentiate into platelets and is responsible for the high proportion of detyrosinated tubulin found in these cells (Strassel, et al., 2019). Its loss leads to hypertyrosination and defects in establishing the stable bundle of microtubules, known as the marginal band, that is essential for differentiation into platelets (Strassel, et al., 2019). We speculate that the failure to transition the cytoskeleton to this stable microtubule structure could be in part due to the increased dynamicity of the microtubules which are now hypertyrosinated compared to wild-type. Tubulin α8, which has a unique C-terminal -EEF motif, plays a critical role in cortical progenitor differentiation by not being subject to detyrosination and the subsequent terminal processing into Δ2-tubulin (Ramos, et al., 2020). Since an α-tubulin tail with a terminal phenylalanine is also recognized by the CAP-Gly domain of CLIP-170 (Mishima, et al., 2007; Honnappa, et al., 2006), we propose that expression of α8 generates a permanent subpopulation of microtubules with increased turnover because of their persistent recruitment of CLIP-170 and possibly other factors. Thus, in addition to the dynamic tyrosination/detyrosination cycle, variations in the genetically encoded terminal residue of the α-tubulin tail itself can be used to regulate microtubule dynamics through the differential interaction with plus-end effectors.
Our experiments underscore the importance of the microtubule substrate itself in mediating the effects of microtubule regulators on microtubule dynamics and demonstrate how detyrosination or the ∆2 modification of transiently stabilized microtubules can give rise to persistent subpopulations of microtubules in cells that are less dynamic. A newly born microtubule is tyrosinated because tubulin is synthesized with a terminal tyrosine and the tubulin tyrosine ligase acts preferentially on the soluble tubulin dimer (Szyk, et al., 2011; Wehland and Weber, 1987; Raybin and Flavin, 1975). The detyrosinating enzymes act preferentially on the microtubule polymer (Li, et al., 2020; Wang, et al., 2019; Aillaud, et al., 2017; Nieuwenhuis, et al., 2017; Gundersen, et al., 1987; Kumar and Flavin, 1981). Thus, a transient stabilization of a microtubule can result in progressive detyrosination, which leads, as we demonstrate, to the gradual concomitant loss of tip factors that then decrease its dynamicity, thus further stabilizing the microtubule through positive-feedback. The detyrosinated tubulin can further be converted to ∆2-tubulin, a modification that is irreversible and not reset through tyrosination upon depolymerization, thus permanently committing this tubulin subpopulation to slower growth and turnover. Our work shows how the tubulin code can constitute the chemical memory of the initial stabilization, allowing it to affect cytoskeleton fate in the future.
Limitations of the study
We note that while our simulations reveal that the α-tubulin tail interacts transiently with the longitudinal polymerization interface, we could not explicitly model tubulin incorporation into the microtubule lattice or protofilaments at the microtubule tip. Thus, our conclusion that the observed marked increase in microtubule growth rates is due to this transient interaction with the polymerization interface, is a plausible extrapolation. The α1A tail is highly conserved in mammals, but shows more sequence divergence in lower vertebrates, invertebrates, algae and plants, both in length and in the number of negatively charged residues (Figure S2B). We predict that loss of the α-tail will potentiate microtubule growth, regardless of α-tubulin species or isotype; however, future biochemical and molecular dynamics studies will be needed to establish whether loss of the α-tail leads to similar magnitude effects on microtubule growth rates in these cases as we observe here for the human isotype. More recent work also revealed the existence of ∆3-tubulin in neuronal tissue, which misses the last three residues (EEY) of the α-tubulin tail (Aillaud, et al., 2016). It is unclear whether the loss of one more glutamate in the ∆3-tubulin will have a pronounced effect on growth rates compared to ∆2-tubulin due to a reduced interaction of the tail with the polymerization interface. Our simulations predict that the effect would be modest because of the cumulative small contributions that each tail residue has out of the total of 12, but this will only be established with certainty through future experimental work.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Antonina Roll-Mecak (antonina@mail.nih.gov).
Material availability
All plasmids and cell lines used in this study are available upon request from the authors.
Data and code availability
MATLAB scripts used in this study for microtubule dynamics analysis and CLIP-170 comet analysis are available on GitHub: https://github.com/RollmecakLab.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All recombinant human tubulin were expressed in Sf9 insect cells. The cells were cultured in Sf-900™ III SFM media at 27 °C, 125 rpm for 48 hrs after infection. GFP-H2 was expressed in E. coli BL21(DE3) CodonPlus RIL cells. GFP-EB1 and unlabeled EB1 were expressed in E. coli BL21(DE3) pLysS cells. Unmodified human α1B/βI+βIVb tubulin was purified from tsA201 cells. The tsA201 cells were cultured in Freestyle293 supplemented with 2% FBS and 1xPenicillin/Streptomycin at 37 °C, 125 rpm, 8% CO2, and 70% humidity.
METHOD DETAILS
Purification of recombinant tubulin isotypes
All tubulin variants were expressed using baculovirus and purified as described previously (Vemu, et al., 2016). Briefly, codon optimized α1A with an internal His-tag in the acetylation loop and PreScission protease cleaveable FLAG-tag on βIII tubulin were custom synthesized by IDT™ and cloned into a pFAST™-Dual vector. Tubulin variants, α1A-Y/βIII, α1A∆2/βIII and α1A∆-tail /βIII, were made using Quick change mutagenesis. To ensure no insect tubulin contamination, α1A with an internal His-tag and βIII with a C-terminal cleavable Flag tag was purified using a Ni-NTA column (Qiagen) and anti-flag G1 affinity resin (Gen Script). The tubulin was then purified by ion exchange chromatography using a Resource Q anion exchange column (GE Healthcare). Peak fractions were combined and buffer exchanged into BRB80 (80 mM PIPES, pH 6.8, 1 mM EGTA, 1 mM MgCl2) supplemented with 20 µM GTP using a PD10 column (GE Healthcare). All tubulin variants were subjected to liquid chromatography electrospray ionization time of flight mass spectrometry (LC-ESI-TOF MS) analysis and no posttranslational modifications or endogenous insect tubulin contaminants were detected. The dynamic parameters are consistent between different tubulin growths and preparations (Figure S1A, B).
Purification of GFP-H2 and EB1
The GFP-H2 construct was a gift from the Surrey lab. Expression and purification of GFP-H2 was carried out as described previously (Bieling, et al., 2008) with slight modifications. Briefly, His-tagged GFP-H2 was expressed in E. coli BL21(DE3) CodonPlus-RIL. Cells were induced with 1 mM IPTG for 5 h at 30 °C and lysed by using high-pressure homogenizer in ice-cold lysis buffer (50 mM KPi, pH 7.5, 500 mM NaCl, 1 mM MgCl2, 1 mM β-mercaptoethanol) with Roche protease inhibitors. Precleared lysate was loaded onto a Ni2+ -NTA column and eluted with lysis buffer containing 300 mM imidazole. GFP-H2 was then purified using GE Q Sepharose Fast Flow with a gradient elution (50 mM KPi, pH 7.5, 0.1–1 M NaCl, 1 mM MgCl2, 1 mM β-mercaptoethanol). Overnight TEV digestion at 4 °C removed the His-tag. GFP-H2 was then loaded onto a Ni2+ -NTA column again. The flow-through was collected and further purified on a GE HiLoad Superdex 200 column. Peak fractions were pooled, supplemented with glycerol to a final concentration of 16%, flash frozen in liquid nitrogen and stored at −80 °C.
The EB1-GFP and EB1 constructs were expressed and purified as described previously (Petry, et al., 2011; Dixit, et al., 2009). Briefly, the His-tagged GFP-EB1 was expressed in E.coli BL21(DE3) pLysS, purified with affinity chromatography (Ni-NTA), ion exchange chromatography (GE Q Sepharose FF) and followed by size-exclusion chromatography in 80 mM PIPES, pH6.8, 100 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.1% β-mercaptoethanol (GE HiLoad Superdex 200). The unlabeled EB1 has a N-terminal GST-tag followed by a PreScission cleavage site. The protein was expressed in E.coli BL21(DE3) pLysS and purified using glutathione resin. The GST-tag was removed by PreScission protease and the protein was further purified by Q Sepharose ion exchange and buffer exchanged into 80 mM PIPES, pH6.8, 100 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.1% β-mercaptoethanol.
In vitro microtubule dynamics assays
GMPCPP-stabilized microtubule seeds were prepared as described (Gell, et al., 2010) and immobilized in flow chambers using neutravidin. The final imaging buffer is BRB80 (80 mM PIPES, pH 6.8, 1 mM EGTA, 1 mM MgCl2) supplemented with 1mM GTP, 0.1% methylcellulose 4,000 cP, 1% pluronic F-127 and 0.1 mg/ml casein. An objective heater (Bioptechs) was used to warm the chamber to 30 °C. All chambers were sealed and allowed to equilibrate on the microscope stage for 5 min prior to imaging. IRM images were acquired on a Nikon Eclipse Ti-E equipped with a Hamamatsu ORCA Flash4.0 V2 sCMOS camera every 5 s for 30 or 60 min. For depolymerization rate measurements, the frame rate used was 20 frames/s.
EB1 and CLIP170 in vitro tip-tracking
Biotin-labeled GMPCPP stabilized microtubule seeds were double cycled from porcine brain tubulin and immobilized using neutravidin. The final assay buffer contains BRB80 (80 mM PIPES, pH 6.8, 1 mM EGTA, 1 mM MgCl2) supplemented with 95 mM KCl, 85 mM KAc, 0.1% methylcellulose 4000 mPa.S, 0.5% pluronic F-127, 100 μg/ml κ-casein, 1 mM β-mercaptoethanol, and oxygen scavengers (Bieling, et al., 2008). All assays were carried out at 30 °C. All microtubule CLIP-170 end tracking assays were carried out at 30 °C, in presence of 6 or 15 μM recombinant tubulin, 300 nM EB1 and 75 nM GFP-H2 unless otherwise specified. Synchronous TIRF/IRM imaging was performed on a Nikon Eclipse Ti-E equipped with a LED light source, 488-laser and EMCCD cameras (Andor) with an exposure time of 100 ms per frame. Images were collected at 1 frame per second for 10 min. To obtain microtubule dynamic parameters in the presence of EB1 or CLIP-170, IRM images were recorded every 5 second for 1 h on a sCMOS camera (Hamamatsu).
MD simulations
A detailed description of MD simulations is provided in (Fedorov, et al., 2019). Computational architecture and performance of the simulations was described in (Fedorov, et al., 2018). In order to investigate the intradimer interaction of the α-tubulin tail we generated molecular models of tubulin dimers or tetramers (two longitudinally attached dimers; Figure S3A) based on microtubule cryo-EM structures (PDB IDs 3j6f and 3j6e (Alushin, et al., 2014)). The nonhydrolyzable GTP analog (GMPCPP) in the 3j6e structures was converted into GTP as described previously (Fedorov, et al., 2019). Unresolved amino acid chains, including C-terminal α- and β-tubulin tails were added in all the models, using the Modeller software (Webb and Sali, 2014). α-tubulin tails have the following sequence: VEGEGEEEGEEY, β-tubulin tails have the ATADEQGEFEEEGEEDEA sequence. Ionizable amino acid residues were protonated with Propka software (Olsson, et al., 2011). Intrinsic cavities in tubulins were identified and solvated with Dowser software (Morozenko and Stuchebrukhov, 2016). Tubulin dynamics were simulated using the GROMACS 5 software package with the CHARMM27 force field (MacKerell, et al., 2004; MacKerell, et al., 1998) in a virtual cubic reaction volume with periodic boundary conditions. The reaction volume size was selected so that the distance from the protein surface to the nearest box boundary was at least 2 nm. The virtual reaction volume was filled with TIP3P water and K+ and Cl− ions of 100 mM total ionic strength and zero total system charge. Each tubulin system was subjected to an energy-minimization, employing the steepest descent algorithm and then to a two-step equilibration, described in (Fedorov, et al., 2019). The production simulation runs were carried out in the NPT ensemble at 300K, using the Parrinello-Rahman algorithm (Parrinello and Rahman, 1981) and the V-rescale thermostat. The particle mesh Ewald method was used to treat the long-range electrostatics. All-bond P-LINCS constraints and mass rescaling (partial transfer of mass from heavy atoms to bound hydrogens (Feenstra, et al., 1999)) allowed molecular dynamics simulations with a 4 fs time step. All simulations of whole tubulins were one-microsecond-long and sampled with 1 ns intervals for analysis. Pymol (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC) was used for visualization.
Metadynamics simulations of α-tail interactions
For enhanced sampling, we carried out a 2 µs-long metadynamics simulation using the GROMACS 2019.4 augmented with the PLUMED 2.5 package (Tribello, et al., 2014). Molecular model of tubulin was constructed as described previously (Fedorov, et al., 2019), using the structure of undecorated GMPCPP microtubule (PDB ID 6dpu (Zhang, et al., 2018). The simulated system contained α-tubulin body and α-tubulin tail, surrounded by a 10.4 × 12.8 × 9.7 nm box, centered around the origin of the α-tubulin tail. Cα - atoms of the tubulin core were position restrained. MD simulations were carried out as described in the section above, with the exception that an additional Gaussian energy potential with 0.1 kT depth and 0.1 nm width was applied to the center of mass of Y451 residue (Figure S4A). The position of the potential was updated every 0.4 ps (100 MD computation steps). Simulation results were processed with a custom Python script to plot the volumes around α-tubulin core, where the maximal numbers of additional energy potentials were applied to the tip of the C-terminal tail throughout the simulation (Figure S4B, C).
QUANTIFICATION AND STATISTICAL ANALYSIS
Microtubule dynamics analysis
Microtubule dynamics parameters were measured in ImageJ as previously described (Vemu, et al., 2016). Briefly, kymographs were generated from IRM images and hand traced to obtain growth and depolymerization speeds from the slopes of the growing and depolymerizing microtubules. Catastrophe frequency is defined as the number of catastrophes observed divided by the total time the microtubule spent in the growth phase. Rescue frequency is defined as the number of rescues observed divided by the total time the microtubule spent in the depolymerization phase. Tubulin association rate (kon), the tubulin dissociation rate (koff) and critical concentration were calculated using the slope, X and Y intercepts, respectively, of a simple linear regression fit in Prism using the mean growth rates at different tubulin concentrations and assuming 1625 dimers per micron of microtubule (Walker, 1988; Oosawa, 1970). The error reported is the standard error of the fit.
EB1 and CLIP-170 plus-end tracking analysis
The fluorescence signal of microtubule plus-end comets was analyzed as previously described (Vemu, et al., 2017; Bieling, et al., 2007). First, kymographs of the growing microtubules were drawn in ImageJ. Next, a custom-written MATLAB script was used to identify the maximum intensity in each line of the kymograph. Then an exponential fit to the line profile covering 1.5 μm of the lattice and a Gaussian fit to the line profile extending 1.5 μm beyond the microtubule tip was performed. For all exponential and Gaussian fitting, R2 > 0.99. All fitted comet profiles were aligned, binned and averaged. The averaged comet profile was then fitted to a single exponential fit from the comet peak to the end of the comet with an offset that represents the lattice intensity. These comet profiles were then plotted in Prism and background corrected by subtracting the minimum background value. The reported comet intensity is the averaged peak value with background correction, and the lattice intensity is the background-corrected offset value, determined by the exponential fitting of comet tail.
Analysis of α-tubulin tail dynamics in MD simulations
Residues belonging to the tubulin polymerization interface were defined as those located closer than 4.5 Å to any of the residues of the adjacent longitudinally attached tubulin subunit. The fraction of polymerization interface occluded by the α-tubulin tail was calculated as follows. First, we artificially cut out the α-tubulin tail from the simulated MD trajectory of the tubulin dimer and determined the total solvent accessible surface area (SASA) for all amino acids belonging to the polymerization interface, every 1 ns of the simulation. SASA was computed using the shrake_rupley function of the MDTraj open library (McGibbon, et al., 2015). Then we repeated the calculation of the total SASA for the amino acids belonging to the polymerization interface without removing the tail from the MD trajectory. The fraction of occluded SASA was then calculated as the ratio of the difference between SASA in the absence and in the presence of the α-tubulin tail to the total SASA for the polymerization interface. Contacts between the α-tubulin tail and tubulin body were characterized with a custom Python script which identifies and counts residues of the tubulin body that are within 4.5 Å from residues of the α-tubulin tail. Probabilities of contacts between the tail of α-tubulin and its body were calculated as the average fraction of contacts over the whole simulation time, based on two simulations with one tubulin dimer and three simulations with the longitudinally assembled tubulin dimers (only the minus-end proximal α-tubulins were considered for tetramers) (Figure S3A). The distribution of electrostatic potential on the tubulin surface was mapped in Pymol based on Poisson-Boltzmann calculations performed using the ProKSim software (Khruschev, et al., 2015).
Supplementary Material
Movie S1 (related to Figure 2).
Visualization of the interaction between the α-tubulin tail and body showing prolonged interactions with the longitudinal polymerization interface.
α-tubulin and β-tubulin are colored dark and light grey, respectively. Areas with which the α-tubulin tail interacts most frequently in 5 independent GTP-tubulin simulations, are colored according to contact probability as in Figures 2E and F. Residues in the α-tubulin tail are shown as cyan van der Waals spheres and the αβ-tubulin body is shown as molecular surface.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli BL21(DE3) CodonPlus-RIL competent cells | Agilent | Cat#230245 |
| E. coli BL21(DE3) pLysS competent cells | ThermoFisher | Cat#606010 |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant human tubulin α1A/βIII | (Vemu, et al., 2016) | N/A |
| Recombinant human tubulin α1A-Y/βIII | This study | N/A |
| Recombinant human tubulin α1A-Δ2/βIII | This study | N/A |
| Recombinant human tubulin α1A-Δtail/βIII | This study | N/A |
| Recombinant human CLIP-170 H2 fragment with C-terminal GFP tag | (Bieling, et al., 2008) | N/A |
| Recombinant full-length human EB1 with C-terminal GFP-His tag | (Petry, et al., 2011) | N/A |
| Recombinant full-length human EB1, non-tagged | (Dixit, et al., 2009) | N/A |
| Porcine brain tubulin | Cytoskeleton, Inc. | Cat#T238P |
| Biotin-labeled porcine brain tubulin | Cytoskeleton, Inc. | Cat#T333P |
| α1B/βI+βIVb tubulin purified from tsA201 cells | (Vemu, et al., 2017) | N/A |
| Experimental models: cell lines | ||
| Sf9 insect cells | ThermoFisher | Cat#11496015 |
| tsA201 cells | Millipore, Sigma | 96121229–1VL |
| Oligonucleotides | ||
| α1A/βIII with PreScission site before FLAG-tag: CCGAGGCCCAGGGACCCAAACTGGAGGTGCTGTTCCAGGGTCCCGGTGGTAGCGGAGGAGACTAC; GTAGTCTCCTCCGCTACCACCGGGACCCTGGAACAGCACCTCCAGTTTGGGTCCCTGGGCCTCGG |
This study | N/A |
| α1A-Y/βIII generated by QuikChange mutagenesis: GAAGAGGAGGGCGAAGAATGAAAGCTGTCGAGAAG; CTTCTCGACAGCTTTCATTCTTCGCCCTCCTCTTC |
This study | N/A |
| α1A-Δ2/βIII generated by QuikChange mutagenesis: GGCGAAGAGGAGGGCGAATGAAAGCTGTCGAGAAGTACTAGAGG; CCTCTAGTACTTCTCGACAGCTTTCATTCGCCCTCCTCTTCGCC |
This study | N/A |
| α1A-Δtail/βIII generated by insertion of a stop codon: GGCGTCGATAGCTAAGTGGAGGGCGAG; CTCGCCCTCCACTTAGCTATCGACGCC |
This study | N/A |
| Recombinant DNA | ||
| pFast™-Dual vector | ThermoFisher | Cat#10712024 |
| H. sapiens tubulin α1A/βIII in pFast™-Dual | (Vemu, et al., 2016) | N/A |
| H. sapiens tubulin α1A-Y/βIII in pFast™-Dual | This study | N/A |
| H. sapiens tubulin α1A-Δ2/βIII in pFast™-Dual | This study | N/A |
| H. sapiens tubulin α1A-Δtail/βIII in pFast™-Dual | This study | N/A |
| H. sapiens CLIP-170 H2 construct in pETEM | Dr. Thomas Surrey, Francis Crick Institute | N/A |
| H. sapiens GFP-tagged full-length EB1 in pET28 | Dr. Kevin Slep, UNC | N/A |
| H. sapiens full-length EB1 in pGEX6p2 | Dr. Sabine Petry, Princeton | N/A |
| Software and algorithms | ||
| Fiji | (Schindelin, et al., 2012) | https://imagej.net/Fiji |
| MATLAB 2019b | MathWorks | N/A |
| Prism | Graphpad | N/A |
| Jalview | (Waterhouse, et al., 2009) | https://www.jalview.org/ |
| ESPript3.0 | (Robert and Gouet, 2014) | https://espript.ibcp.fr/ESPript/ESPript/ |
| Modeller | (Webb and Sali, 2014) | https://salilab.org/modeller/ |
| Propka | (Olsson, et al., 2011) | https://pypi.org |
| Dowser | (Morozenko and Stuchebrukhov, 2016) | N/A |
| GROMACS 5 | (MacKerell, et al., 2004; MacKerell, et al., 1998) | https://www.gromacs.org/ |
| Parrinello-Rahman algorithm | (Parrinello and Rahman, 1981) | N/A |
| PyMOL v.2.0 | PyMOL by Schrodinger | https://pymol.org/2/ |
| PLUMED 2.5 plugin for GROMACS | (Tribello, et al., 2014) | https://www.plumed.org/doc-v2.5/user-doc/html/index.html |
| MDTraj open library | (McGibbon, et al., 2015) | https://www.mdtraj.org/1.9.5/index.html |
| ProKSim | (Khruschev, et al., 2015) | N/A |
| MATLAB scripts for microtubule dynamic analysis and comet analysis | (Vemu, et al., 2017) This study |
https://github.com/RollmecakLab |
Highlights.
The tubulin α-tail inhibits polymerization by interacting with a polymerization interface
The dynamics of tyrosinated, detyrosinated and ∆2 microtubules are similar
Tyrosination quantitatively tunes the recruitment of CLIP-170 to the microtubule
Tyrosination-dependent factor recruitment creates dynamic microtubules subpopulations
Acknowledgements
We thank Jeff Spector (National Institute of Neurological Disorders and Stroke) for advice with comet analysis, Annapurna Vemu (National Institute of Neurological Disorders and Stroke) for advice on microtubule dynamics assays and initial efforts on the project, D-Y. Lee from the Biophysics Core (National Heart, Lung and Blood Institute) for mass spectrometry access and advice, and Thomas Surrey (Center for Genomic Regulation) for the GFP-H2 plasmid. A.R-M. is supported by the intramural programs of the National Institute of Neurological Disorders and Stroke (NINDS) and the National Heart, Lung and Blood Institute (NHLBI). MD simulations were done with support from Russian Foundation for Basic Research (research project #20-34-70159) and RF President’s grant MK-1869.2020.4 to N.B.G., using the equipment of the shared research facilities of the high-performance computational resources at Lomonosov Moscow State University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing financial interests.
References
- Aillaud C, Bosc C, Peris L, Bosson A, Heemeryck P, Van Dijk J, Le Friec J, Boulan B, Vossier F, Sanman LE, et al. (2017). Vasohibins/SVBP are tubulin carboxypeptidases (TCPs) that regulate neuron differentiation. Science 358, 1448–1453. [DOI] [PubMed] [Google Scholar]
- Aillaud C, Bosc C, Saoudi Y, Denarier E, Peris L, Sago L, Taulet N, Cieren A, Tort O, Magiera MM, et al. (2016). Evidence for new C-terminally truncated variants of alpha- and beta-tubulins. Molecular biology of the cell 27, 640–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akera T, Chmatal L, Trimm E, Yang K, Aonbangkhen C, Chenoweth DM, Janke C, Schultz RM, and Lampson MA (2017). Spindle asymmetry drives non-Mendelian chromosome segregation. Science 358, 668–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmanova A, Mausset-Bonnefont AL, van Cappellen W, Keijzer N, Hoogenraad CC, Stepanova T, Drabek K, van der Wees J, Mommaas M, Onderwater J, et al. (2005). The microtubule plus-end-tracking protein CLIP-170 associates with the spermatid manchette and is essential for spermatogenesis. Genes Dev 19, 2501–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmanova A, and Steinmetz MO (2008). Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol 9, 309–22. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, and Steinmetz MO (2010a). Microtubule +TIPs at a glance. Journal of cell science 123, 3415–9. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, and Steinmetz MO (2010b). Microtubule+ TIPs at a glance. Journal of cell science 123, 3415–3419. [DOI] [PubMed] [Google Scholar]
- Alfaro-Aco R, and Petry S (2015). Building the Microtubule Cytoskeleton Piece by Piece. The Journal of biological chemistry 290, 17154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Lander GC, Kellogg EH, Zhang R, Baker D, and Nogales E (2014). High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell 157, 1117–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, and Nogales E (2010). The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature 467, 805–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argarana CE, Barra HS, and Caputto R (1978). Release of [14C]tyrosine from tubulinyl-[14C]tyrosine by brain extract. Separation of a carboxypeptidase from tubulin-tyrosine ligase. Mol Cell Biochem 19, 17–21. [DOI] [PubMed] [Google Scholar]
- Arnal I, Heichette C, Diamantopoulos GS, and Chretien D (2004). CLIP-170/tubulin-curved oligomers coassemble at microtubule ends and promote rescues. Curr Biol 14, 2086–95. [DOI] [PubMed] [Google Scholar]
- Barisic M, Silva e Sousa R, Tripathy SK, Magiera MM, Zaytsev AV, Pereira AL, Janke C, Grishchuk EL, and Maiato H (2015). Mitosis. Microtubule detyrosination guides chromosomes during mitosis. Science 348, 799–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barra HS, Rodriguez JA, Arce CA, and Caputto R (1973). A soluble preparation from rat brain that incorporates into its own proteins ( 14 C)arginine by a ribonuclease-sensitive system and ( 14 C)tyrosine by a ribonuclease-insensitive system. J Neurochem 20, 97–108. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya B, Sackett DL, and Wolff J (1985). Tubulin, hybrid dimers, and tubulin S. Stepwise charge reduction and polymerization. The Journal of biological chemistry 260, 10208–16. [PubMed] [Google Scholar]
- Bieling P, Kandels-Lewis S, Telley IA, van Dijk J, Janke C, and Surrey T (2008). CLIP-170 tracks growing microtubule ends by dynamically recognizing composite EB1/tubulin-binding sites. J Cell Biol 183, 1223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Laan L, Schek H, Munteanu EL, Sandblad L, Dogterom M, Brunner D, and Surrey T (2007). Reconstitution of a microtubule plus-end tracking system in vitro. Nature 450, 1100–5. [DOI] [PubMed] [Google Scholar]
- Bjelic S, De Groot CO, Scharer MA, Jaussi R, Bargsten K, Salzmann M, Frey D, Capitani G, Kammerer RA, and Steinmetz MO (2012). Interaction of mammalian end binding proteins with CAP-Gly domains of CLIP-170 and p150(glued). Journal of structural biology 177, 160–7. [DOI] [PubMed] [Google Scholar]
- Brunner D, and Nurse P (2000). CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell 102, 695–704. [DOI] [PubMed] [Google Scholar]
- Carlier M-F (1982). Guanosine-5′-triphosphate hydrolysis and tubulin polymerization. Molecular and cellular biochemistry 47, 97–113. [DOI] [PubMed] [Google Scholar]
- Carlier MF, and Pantaloni D (1981). Kinetic analysis of guanosine 5’-triphosphate hydrolysis associated with tubulin polymerization. Biochemistry 20, 1918–24. [DOI] [PubMed] [Google Scholar]
- Chang W, Webster DR, Salam AA, Gruber D, Prasad A, Eiserich JP, and Bulinski JC (2002). Alteration of the C-terminal amino acid of tubulin specifically inhibits myogenic differentiation. The Journal of biological chemistry 277, 30690–8. [DOI] [PubMed] [Google Scholar]
- Chen Y, Wang P, and Slep KC (2019). Mapping multivalency in the CLIP-170-EB1 microtubule plus-end complex. The Journal of biological chemistry 294, 918–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan MC, Eddelbuettel AMP, Levin PA, and Pappu RV (2020). Dissecting the Functional Contributions of the Intrinsically Disordered C-terminal Tail of Bacillus subtilis FtsZ. J Mol Biol 432, 3205–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R, Barnett B, Lazarus JE, Tokito M, Goldman YE, and Holzbaur EL (2009). Microtubule plus-end tracking by CLIP-170 requires EB1. Proceedings of the National Academy of Sciences of the United States of America 106, 492–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duellberg C, Cade NI, Holmes D, and Surrey T (2016). The size of the EB cap determines instantaneous microtubule stability. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erck C, Peris L, Andrieux A, Meissirel C, Gruber AD, Vernet M, Schweitzer A, Saoudi Y, Pointu H, Bosc C, et al. (2005). A vital role of tubulin-tyrosine-ligase for neuronal organization. Proceedings of the National Academy of Sciences of the United States of America 102, 7853–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov VA, Kholina EG, Kovalenko IB, and Gudimchuk N (2018). Performance Analysis of Different Computational Architectures: Molecular Dynamics in Application to Protein Assemblies, Illustrated by Microtubule and Electron Transfer Proteins. Journal of Supercomputing Frontiers and Innovations 5, 111–114. [Google Scholar]
- Fedorov VA, Orekhov PS, Kholina EG, Zhmurov AA, Ataullakhanov FI, Kovalenko IB, and Gudimchuk NB (2019). Mechanical properties of tubulin intra- and inter-dimer interfaces and their implications for microtubule dynamic instability. PLoS Comput Biol 15, e1007327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feenstra KA, Hess B, and Berendsen HJC (1999). Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. Journal of Computational Chemistry. 20, 786–798. [DOI] [PubMed] [Google Scholar]
- Fees CP, and Moore JK (2018). Regulation of microtubule dynamic instability by the carboxy-terminal tail of beta-tubulin. Life science alliance 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavin M, and Murofushi H (1984). Tyrosine incorporation in tubulin. Methods in enzymology 106, 223–37. [DOI] [PubMed] [Google Scholar]
- Garnham CP, Vemu A, Wilson-Kubalek EM, Yu I, Szyk A, Lander GC, Milligan RA, and Roll-Mecak A (2015). Multivalent Microtubule Recognition by Tubulin Tyrosine Ligase-like Family Glutamylases. Cell 161, 1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gell C, Bormuth V, Brouhard GJ, Cohen DN, Diez S, Friel CT, Helenius J, Nitzsche B, Petzold H, Ribbe J, et al. (2010). Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol 95, 221–45. [DOI] [PubMed] [Google Scholar]
- Gouveia SM, and Akhmanova A (2010). Cell and molecular biology of microtubule plus end tracking proteins: end binding proteins and their partners. Int Rev Cell Mol Biol 285, 1–74. [DOI] [PubMed] [Google Scholar]
- Gundersen GG, and Bulinski JC (1986a). Distribution of tyrosinated and nontyrosinated alpha-tubulin during mitosis. J Cell Biol 102, 1118–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen GG, and Bulinski JC (1986b). Microtubule arrays in differentiated cells contain elevated levels of a post-translationally modified form of tubulin. Eur J Cell Biol 42, 288–94. [PubMed] [Google Scholar]
- Gundersen GG, and Bulinski JC (1988). Selective stabilization of microtubules oriented toward the direction of cell migration. Proceedings of the National Academy of Sciences of the United States of America 85, 5946–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen GG, Khawaja S, and Bulinski JC (1987). Postpolymerization detyrosination of alpha-tubulin: a mechanism for subcellular differentiation of microtubules. J Cell Biol 105, 251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen GG, Khawaja S, and Bulinski JC (1989). Generation of a stable, posttranslationally modified microtubule array is an early event in myogenic differentiation. J Cell Biol 109, 2275–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta KK, Joyce MV, Slabbekoorn AR, Zhu ZC, Paulson BA, Boggess B, and Goodson HV (2010). Probing interactions between CLIP-170, EB1, and microtubules. J Mol Biol 395, 1049–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrie H, Bakhos-Douaihy D, Cantaloube I, Pilon A, Talantikite M, Stoppin-Mellet V, Baillet A, Pous C, and Benoit B (2020). Stress-induced phosphorylation of CLIP-170 by JNK promotes microtubule rescue. J Cell Biol 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S, Okhrimenko O, Jaussi R, Jawhari H, Jelesarov I, Winkler FK, and Steinmetz MO (2006). Key interaction modes of dynamic +TIP networks. Molecular cell 23, 663–71. [DOI] [PubMed] [Google Scholar]
- Horio TH, H. (1986). Visualization of the dynamic instability of individual microtubules by dark-field microscopy. Nature 321, 605–607. [DOI] [PubMed] [Google Scholar]
- Infante AS, Stein MS, Zhai Y, Borisy GG, and Gundersen GG (2000). Detyrosinated (Glu) microtubules are stabilized by an ATP-sensitive plus-end cap. Journal of cell science 113 (Pt 22), 3907–19. [DOI] [PubMed] [Google Scholar]
- Iwata-Otsubo A, Dawicki-McKenna JM, Akera T, Falk SJ, Chmatal L, Yang K, Sullivan BA, Schultz RM, Lampson MA, and Black BE (2017). Expanded Satellite Repeats Amplify a Discrete CENP-A Nucleosome Assembly Site on Chromosomes that Drive in Female Meiosis. Curr Biol 27, 2365–2373 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, and Magiera MM (2020). The tubulin code and its role in controlling microtubule properties and functions. Nat Rev Mol Cell Biol 21, 307–326. [DOI] [PubMed] [Google Scholar]
- Khawaja S, Gundersen GG, and Bulinski JC (1988). Enhanced stability of microtubules enriched in detyrosinated tubulin is not a direct function of detyrosination level. J Cell Biol 106, 141–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khruschev SS, Abaturova AM, Diakonova AN, Fedorov VA, Ustinin DM, Kovalenko IB, Riznichenko GY, and Rubin AB (2015). [Brownian dynamics simulations of protein-protein interactions in photosynthetic electron transport chain]. Biofizika 60, 270–92. [PubMed] [Google Scholar]
- Kirschner M, and Mitchison T (1986). Beyond self-assembly: from microtubules to morphogenesis. Cell 45, 329–42. [DOI] [PubMed] [Google Scholar]
- Knipling L, Hwang J, and Wolff J (1999). Preparation and properties of pure tubulin S. Cell motility and the cytoskeleton 43, 63–71. [DOI] [PubMed] [Google Scholar]
- Knipling L, and Wolff J (2006). Direct interaction of Bcl-2 proteins with tubulin. Biochem Biophys Res Commun 341, 433–9. [DOI] [PubMed] [Google Scholar]
- Komarova YA, Akhmanova AS, Kojima S, Galjart N, and Borisy GG (2002). Cytoplasmic linker proteins promote microtubule rescue in vivo. J Cell Biol 159, 589–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis TE (1987). Microtubules containing detyrosinated tubulin are less dynamic. The EMBO journal 6, 2597–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitzer G, Liao G, and Gundersen GG (1999). Detyrosination of tubulin regulates the interaction of intermediate filaments with microtubules in vivo via a kinesin-dependent mechanism. Molecular biology of the cell 10, 1105–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, and Flavin M (1981). Preferential action of a brain detyrosinolating carboxypeptidase on polymerized tubulin. The Journal of biological chemistry 256, 7678–86. [PubMed] [Google Scholar]
- Li F, Li Y, Ye X, Gao H, Shi Z, Luo X, Rice LM, and Yu H (2020). Cryo-EM structure of VASH1-SVBP bound to microtubules. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopus M, Manatschal C, Buey RM, Bjelic S, Miller HP, Steinmetz MO, and Wilson L (2012). Cooperative stabilization of microtubule dynamics by EB1 and CLIP-170 involves displacement of stably bound P(i) at microtubule ends. Biochemistry 51, 3021–30. [DOI] [PubMed] [Google Scholar]
- Luduena RF (2013). A hypothesis on the origin and evolution of tubulin. Int Rev Cell Mol Biol 302, 41–185. [DOI] [PubMed] [Google Scholar]
- MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. (1998). All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102, 3586–616. [DOI] [PubMed] [Google Scholar]
- MacKerell AD Jr., Feig M, and Brooks CL 3rd (2004). Improved treatment of the protein backbone in empirical force fields. J Am Chem Soc 126, 698–9. [DOI] [PubMed] [Google Scholar]
- Mahamdeh M, Simmert S, Luchniak A, Schaffer E, and Howard J (2018). Label-free high-speed wide-field imaging of single microtubules using interference reflection microscopy. Journal of microscopy 272, 60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos S, Moreau J, Backer S, Job D, Andrieux A, and Bloch-Gallego E (2009). Tubulin tyrosination is required for the proper organization and pathfinding of the growth cone. PLoS One 4, e5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Garcia R, and Mulvihill DP (2009). Myosin V spatially regulates microtubule dynamics and promotes the ubiquitin-dependent degradation of the fission yeast CLIP-170 homologue, Tip1. Journal of cell science 122, 3862–72. [DOI] [PubMed] [Google Scholar]
- Marya PK, Syed Z, Fraylich PE, and Eagles PA (1994). Kinesin and tau bind to distinct sites on microtubules. Journal of cell science 107 (Pt 1), 339–44. [DOI] [PubMed] [Google Scholar]
- McGibbon RT, Beauchamp KA, Harrigan MP, Klein C, Swails JM, Hernandez CX, Schwantes CR, Wang LP, Lane TJ, and Pande VS (2015). MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys J 109, 1528–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenney RJ, Huynh W, Vale RD, and Sirajuddin M (2016). Tyrosination of alpha-tubulin controls the initiation of processive dynein-dynactin motility. The EMBO journal 35, 1175–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoura I, Hachikubo Y, Yamakita Y, Takazaki H, Ayukawa R, Uchimura S & Muto E (2013). Overexpression, purification, and functional analysis of recombinant human tubulin dimer. FEBS Lett. 587, 3450–3455. [DOI] [PubMed] [Google Scholar]
- Mishima M, Maesaki R, Kasa M, Watanabe T, Fukata M, Kaibuchi K, and Hakoshima T (2007). Structural basis for tubulin recognition by cytoplasmic linker protein 170 and its autoinhibition. Proceedings of the National Academy of Sciences of the United States of America 104, 10346–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, and Kirschner M (1984). Dynamic instability of microtubule growth. Nature 312, 237–42. [DOI] [PubMed] [Google Scholar]
- Morozenko A, and Stuchebrukhov AA (2016). Dowser++, a new method of hydrating protein structures. Proteins 84, 1347–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis J, Adamopoulos A, Bleijerveld OB, Mazouzi A, Stickel E, Celie P, Altelaar M, Knipscheer P, Perrakis A, Blomen VA, et al. (2017). Vasohibins encode tubulin detyrosinating activity. Science 358, 1453–1456. [DOI] [PubMed] [Google Scholar]
- Nirschl JJ, Magiera MM, Lazarus JE, Janke C, and Holzbaur EL (2016). alpha-Tubulin Tyrosination and CLIP-170 Phosphorylation Regulate the Initiation of Dynein-Driven Transport in Neurons. Cell Rep 14, 2637–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson MH, Sondergaard CR, Rostkowski M, and Jensen JH (2011). PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J Chem Theory Comput 7, 525–37. [DOI] [PubMed] [Google Scholar]
- Oosawa F (1970). Size distribution of protein polymers. J Theor Biol 27, 69–86. [DOI] [PubMed] [Google Scholar]
- Parrinello M, and Rahman A (1981). Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied Physics 52, 7182–7190. [Google Scholar]
- Paturle-Lafanechere L, Edde B, Denoulet P, Van Dorsselaer A, Mazarguil H, Le Caer JP, Wehland J, and Job D (1991). Characterization of a major brain tubulin variant which cannot be tyrosinated. Biochemistry 30, 10523–8. [DOI] [PubMed] [Google Scholar]
- Paturle-Lafanechere L, Manier M, Trigault N, Pirollet F, Mazarguil H, and Job D (1994). Accumulation of delta 2-tubulin, a major tubulin variant that cannot be tyrosinated, in neuronal tissues and in stable microtubule assemblies. Journal of cell science 107 (Pt 6), 1529–43. [DOI] [PubMed] [Google Scholar]
- Paturle L, Wehland J, Margolis RL, and Job D (1989). Complete separation of tyrosinated, detyrosinated, and nontyrosinatable brain tubulin subpopulations using affinity chromatography. Biochemistry 28, 2698–704. [DOI] [PubMed] [Google Scholar]
- Perez F, Diamantopoulos GS, Stalder R, and Kreis TE (1999). CLIP-170 highlights growing microtubule ends in vivo. Cell 96, 517–27. [DOI] [PubMed] [Google Scholar]
- Peris L, Thery M, Faure J, Saoudi Y, Lafanechere L, Chilton JK, Gordon-Weeks P, Galjart N, Bornens M, Wordeman L, et al. (2006). Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. J Cell Biol 174, 839–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris L, Wagenbach M, Lafanechere L, Brocard J, Moore AT, Kozielski F, Job D, Wordeman L, and Andrieux A (2009). Motor-dependent microtubule disassembly driven by tubulin tyrosination. J Cell Biol 185, 1159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S, Pugieux C, Nedelec FJ, and Vale RD (2011). Augmin promotes meiotic spindle formation and bipolarity in Xenopus egg extracts. Proceedings of the National Academy of Sciences of the United States of America 108, 14473–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portran D, Schaedel L, Xu Z, Thery M, and Nachury MV (2017). Tubulin acetylation protects long-lived microtubules against mechanical ageing. Nature cell biology 19, 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos SI, Makeyev EV, Salierno M, Kodama T, Kawakami Y, and Sahara S (2020). Tuba8 Drives Differentiation of Cortical Radial Glia into Apical Intermediate Progenitors by Tuning Modifications of Tubulin C Termini. Developmental cell 52, 477–491 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raybin D, and Flavin M (1975). An enzyme tyrosylating alpha-tubulin and its role in microtubule assembly. Biochem Biophys Res Commun 65, 1088–95. [DOI] [PubMed] [Google Scholar]
- Raybin D, and Flavin M (1977). Enzyme which specifically adds tyrosine to the alpha chain of tubulin. Biochemistry 16, 2189–94. [DOI] [PubMed] [Google Scholar]
- Robert X, and Gouet P (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic acids research 42, W320–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison P, Caporizzo MA, Ahmadzadeh H, Bogush AI, Chen CY, Margulies KB, Shenoy VB, and Prosser BL (2016). Detyrosinated microtubules buckle and bear load in contracting cardiomyocytes. Science 352, aaf0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson SJ, and Burgoyne RD (1989). Differential localisation of tyrosinated, detyrosinated, and acetylated alpha-tubulins in neurites and growth cones of dorsal root ganglion neurons. Cell motility and the cytoskeleton 12, 273–82. [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A (2015). Intrinsically disordered tubulin tails: complex tuners of microtubule functions? Seminars in cell & developmental biology 37, 11–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A (2019). How cells exploit tubulin diversity to build functional cellular microtubule mosaics. Current opinion in cell biology 56, 102–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A (2020). The Tubulin Code in Microtubule Dynamics and Information Encoding. Developmental cell 54, 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A, and Vale RD (2005). The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr Biol 15, 650–5. [DOI] [PubMed] [Google Scholar]
- Sackett DL, Bhattacharyya B, and Wolff J (1985). Tubulin subunit carboxyl termini determine polymerization efficiency. The Journal of biological chemistry 260, 43–5. [PubMed] [Google Scholar]
- Scheel J, Pierre P, Rickard JE, Diamantopoulos GS, Valetti C, van der Goot FG, Haner M, Aebi U, and Kreis TE (1999). Purification and analysis of authentic CLIP-170 and recombinant fragments. The Journal of biological chemistry 274, 25883–91. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nature methods 9, 676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze E, and Kirschner M (1987). Dynamic and stable populations of microtubules in cells. J Cell Biol 104, 277–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano L, Avila J, and Maccioni RB (1984a). Controlled proteolysis of tubulin by subtilisin: localization of the site for MAP2 interaction. Biochemistry 23, 4675–81. [DOI] [PubMed] [Google Scholar]
- Serrano L, de la Torre J, Maccioni RB, and Avila J (1984b). Involvement of the carboxyl-terminal domain of tubulin in the regulation of its assembly. Proceedings of the National Academy of Sciences of the United States of America 81, 5989–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano L, Montejo de Garcini E, Hernandez MA, and Avila J (1985). Localization of the tubulin binding site for tau protein. Eur J Biochem 153, 595–600. [DOI] [PubMed] [Google Scholar]
- Serrano L, Wandosell F, de la Torre J, and Avila J (1988). Effect of specific proteolytic cleavages on tubulin polymer formation. Biochem J 252, 683–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Ihara M, Tani S, Sakuraba J, Sakuta H, and Noda M (2009). APC2 plays an essential role in axonal projections through the regulation of microtubule stability. J Neurosci 29, 11628–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirajuddin M, Rice LM, and Vale RD (2014). Regulation of microtubule motors by tubulin isotypes and post-translational modifications. Nature cell biology 16, 335–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiniotis G, Cochran JC, Muller J, Mandelkow E, Gilbert SP, and Hoenger A (2004). Modulation of kinesin binding by the C-termini of tubulin. The EMBO journal 23, 989–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassel C, Magiera MM, Dupuis A, Batzenschlager M, Hovasse A, Pleines I, Gueguen P, Eckly A, Moog S, Mallo L, et al. (2019). An essential role for alpha4A-tubulin in platelet biogenesis. Life science alliance 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyk A, Deaconescu AM, Piszczek G, and Roll-Mecak A (2011). Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nature structural & molecular biology 18, 1250–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan R, Lam AJ, Tan T, Han J, Nowakowski DW, Vershinin M, Simo S, Ori-McKenney KM, and McKenney RJ (2019). Microtubules gate tau condensation to spatially regulate microtubule functions. Nature cell biology 21, 1078–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ti SC, Alushin GM, and Kapoor TM (2018). Human beta-Tubulin Isotypes Can Regulate Microtubule Protofilament Number and Stability. Developmental cell 47, 175–190 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribello GA, Bonomi M, Branduardi D, Camilloni C, and Bussi G (2014). PLUMED 2: New feathers for an old bird. Computer Physics Communications 185, 604–613. [Google Scholar]
- Trudeau T, Nassar R, Cumberworth A, Wong ET, Woollard G, and Gsponer J (2013). Structure and intrinsic disorder in protein autoinhibition. Structure 21, 332–41. [DOI] [PubMed] [Google Scholar]
- Vemu A, Atherton J, Spector JO, Moores CA, and Roll-Mecak A (2017). Tubulin isoform composition tunes microtubule dynamics. Molecular biology of the cell 28, 3564–3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemu A, Atherton J, Spector JO, Szyk A, Moores CA, and Roll-Mecak A (2016). Structure and Dynamics of Single-isoform Recombinant Neuronal Human Tubulin. The Journal of biological chemistry 291, 12907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey KJ, and Gaertig J (2007). The tubulin code. Cell cycle 6, 2152–2160. [DOI] [PubMed] [Google Scholar]
- Vigers GP, Coue M, and McIntosh JR (1988). Fluorescent microtubules break up under illumination. J Cell Biol 107, 1011–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitre B, Coquelle FM, Heichette C, Garnier C, Chretien D, and Arnal I (2008). EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nature cell biology 10, 415–21. [DOI] [PubMed] [Google Scholar]
- Walker RA, O’Brien ET, Pryer NK, Soboeiro MF, Voter WA, Erickson HP & Salmon ED (1988). Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J. Cell Biol. 107, 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall KP, Pagratis M, Armstrong G, Balsbaugh JL, Verbeke E, Pearson CG, and Hough LE (2016). Molecular Determinants of Tubulin’s C-Terminal Tail Conformational Ensemble. ACS Chem Biol 11, 2981–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N, Bosc C, Ryul Choi S, Boulan B, Peris L, Olieric N, Bao H, Krichen F, Chen L, Andrieux A, et al. (2019). Structural basis of tubulin detyrosination by the vasohibin-SVBP enzyme complex. Nature structural & molecular biology 26, 571–582. [DOI] [PubMed] [Google Scholar]
- Wang Z, and Sheetz MP (2000). The C-terminus of tubulin increases cytoplasmic dynein and kinesin processivity. Biophys J 78, 1955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, and Barton GJ (2009). Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B, and Sali A (2014). Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Bioinformatics 47, 5 6 1–32. [DOI] [PubMed] [Google Scholar]
- Webster DR, and Borisy GG (1989). Microtubules are acetylated in domains that turn over slowly. Journal of cell science 92 (Pt 1), 57–65. [DOI] [PubMed] [Google Scholar]
- Webster DR, Gundersen GG, Bulinski JC, and Borisy GG (1987a). Assembly and turnover of detyrosinated tubulin in vivo. J Cell Biol 105, 265–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster DR, Gundersen GG, Bulinski JC, and Borisy GG (1987b). Differential turnover of tyrosinated and detyrosinated microtubules. Proceedings of the National Academy of Sciences of the United States of America 84, 9040–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehland J, and Weber K (1987). Turnover of the carboxy-terminal tyrosine of alpha-tubulin and means of reaching elevated levels of detyrosination in living cells. Journal of cell science 88 (Pt 2), 185–203. [DOI] [PubMed] [Google Scholar]
- Weisbrich A, Honnappa S, Jaussi R, Okhrimenko O, Frey D, Jelesarov I, Akhmanova A, and Steinmetz MO (2007). Structure-function relationship of CAP-Gly domains. Nature structural & molecular biology 14, 959–67. [DOI] [PubMed] [Google Scholar]
- Weisenberg RC (1972). Microtubule formation in vitro in solutions containing low calcium concentrations. Science 177, 1104–5. [DOI] [PubMed] [Google Scholar]
- Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, and Barnes G (2005). Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Molecular cell 17, 277–90. [DOI] [PubMed] [Google Scholar]
- Whipple RA, Matrone MA, Cho EH, Balzer EM, Vitolo MI, Yoon JR, Ioffe OB, Tuttle KC, Yang J, and Martin SS (2010). Epithelial-to-mesenchymal transition promotes tubulin detyrosination and microtentacles that enhance endothelial engagement. Cancer research 70, 8127–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White EA, Burton PR, and Himes RH (1987). Polymorphic assembly of subtilisin-cleaved tubulin. Cell motility and the cytoskeleton 7, 31–8. [DOI] [PubMed] [Google Scholar]
- Witte H, Neukirchen D, and Bradke F (2008). Microtubule stabilization specifies initial neuronal polarization. J Cell Biol 180, 619–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Das M, Reilly J, and Davis RJ (2011). JNK regulates FoxO-dependent autophagy in neurons. Genes Dev 25, 310–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Schaedel L, Portran D, Aguilar A, Gaillard J, Marinkovich MP, Thery M, and Nachury MV (2017). Microtubules acquire resistance from mechanical breakage through intralumenal acetylation. Science 356, 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S, Verma PJ, and Panda D (2014). C-terminal region of MAP7 domain containing protein 3 (MAP7D3) promotes microtubule polymerization by binding at the C-terminal tail of tubulin. PLoS One 9, e99539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanic M, Widlund PO, Hyman AA, and Howard J (2013). Synergy between XMAP215 and EB1 increases microtubule growth rates to physiological levels. Nature cell biology 15, 688–93. [DOI] [PubMed] [Google Scholar]
- Zehr EA, Szyk A, Szczesna E, and Roll-Mecak A (2020). Katanin Grips the beta-Tubulin Tail through an Electropositive Double Spiral to Sever Microtubules. Developmental cell 52, 118–131 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Alushin GM, Brown A, and Nogales E (2015). Mechanistic Origin of Microtubule Dynamic Instability and Its Modulation by EB Proteins. Cell 162, 849–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, LaFrance B, and Nogales E (2018). Separating the effects of nucleotide and EB binding on microtubule structure. Proceedings of the National Academy of Sciences of the United States of America 115, E6191–E6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1 (related to Figure 2).
Visualization of the interaction between the α-tubulin tail and body showing prolonged interactions with the longitudinal polymerization interface.
α-tubulin and β-tubulin are colored dark and light grey, respectively. Areas with which the α-tubulin tail interacts most frequently in 5 independent GTP-tubulin simulations, are colored according to contact probability as in Figures 2E and F. Residues in the α-tubulin tail are shown as cyan van der Waals spheres and the αβ-tubulin body is shown as molecular surface.
Data Availability Statement
MATLAB scripts used in this study for microtubule dynamics analysis and CLIP-170 comet analysis are available on GitHub: https://github.com/RollmecakLab.


