Abstract
Yeast Msh2p forms complexes with Msh3p and Msh6p to repair DNA mispairs that arise during DNA replication. In addition to their role in mismatch repair (MMR), the MSH2 and MSH3 gene products are required to remove 3′ nonhomologous DNA tails during genetic recombination. The mismatch repair genes MSH6, MLH1, and PMS1, whose products interact with Msh2p, are not required in this process. We have identified mutations in MSH2 that do not disrupt genetic recombination but confer a strong defect in mismatch repair. Twenty-four msh2 mutations that conferred a dominant negative phenotype for mismatch repair were isolated. A subset of these mutations mapped to residues in Msh2p that were analogous to mutations identified in human nonpolyposis colorectal cancer msh2 kindreds. Approximately half of the these MMR-defective mutations retained wild-type or nearly wild-type activity for the removal of nonhomologous DNA tails during genetic recombination. The identification of mutations in MSH2 that disrupt mismatch repair without affecting recombination provides a first step in dissecting the Msh-effector protein complexes that are thought to play different roles during DNA repair and genetic recombination.
In bacteria, yeast, and humans, the mismatch repair system (MMR) corrects misincorporation and DNA slippage errors that arise during DNA replication. In Escherichia coli the MutHLS system repairs single-base mismatches and small insertion-deletion mispairs of up to 4 nucleotides (reviewed in reference 46). MMR is thought to be initiated by MutS binding to a mispair. Interactions between MutL and the MutS-mispair complex are thought to result in an ATP-dependent bidirectional translocation of a MutS-MutL complex away from the mispair, resulting in the activation of MutH endonuclease activity at hemimethylated GATC sites. The result of these steps is the removal of the mispair by excising the newly replicated strand (7, 8; reviewed in reference 46).
Three mutS homologues (MSH2, MSH3, and MSH6) and three mutL homologues (MLH1, MLH3, and PMS1) have been identified in Saccharomyces cerevisiae, and a similar number of mutS and mutL homologs that are required in MMR have been identified in humans (reviewed in references 11, 39, and 47). In both yeast and humans, complexes of Msh2p-Msh6p and Msh2p-Msh3p bind to and are required for the repair of base pair and small insertion-deletion mismatches (1, 2, 14, 20, 25, 43, 51, 52). Defects in the bacterial, yeast, and human MutS and MutL homologs lead to a mutator phenotype, an increase in the frequency of repeat-tract instability as a result of DNA slippage, and in humans, to hereditary nonpolyposis colorectal cancer (HNPCC) (reviewed in references 11, 39, and 47).
In addition to their role in MMR, recent studies have shown that the yeast Msh2p-Msh3p complex plays a critical role in removing nonhomologous DNA during genetic recombination (37, 60, 66). In one study, Sugawara et al. (66) identified a role for Msh2p and Msh3p and the excision repair proteins Rad1p and Rad10p in removing nonhomologous DNA that can interfere with the resolution of either gene conversion or single-strand annealing (SSA) events. During recombination, nonhomologous DNA ends of double-strand breaks (DSB) must be removed to enable the invading or annealed 3′ single-stranded DNA end to prime new DNA synthesis from its template. In wild-type strains, DSBs with >30 bp of nonhomology on either end of a DSB can be removed efficiently during gene conversion, but rad1, rad10, msh2, or msh3 strains are severely impaired in this process while other excision repair genes beyond RAD1 and RAD10 are not required (17, 35, 53, 66). Similarly, the removal of long 3′-end single-strand tails during SSA also requires Rad1p, Rad10p, Msh2p, and Msh3p, although here the requirement for Msh2p and Msh3p decreases as the length of the annealed homologous DNA flanking the DSB increases (66). For flanking regions of 205 bp, used in the experiments described below, the requirement for MSH2 and MSH3 was as great as that for RAD1. Interestingly, mutations that disabled other components of the MMR machinery, msh6Δ, mlh1Δ, and pms1Δ, did not affect repair efficiency. These results led to the idea that Msh2p and Msh3p act to facilitate the Rad1p/Rad10p-mediated endonucleolytic cleavage (reviewed in references 17, 35, 53, 60, and 66) of a nonhomologous single-stranded DNA at the junction with duplex DNA. This hypothesis is strongly supported by the observation by Kirkpatrick and Petes (37) that Rad1p and Msh2p are also required to remove heteroduplex single-stranded loops created during meiosis.
The observation that Msh6p, Mlh1p, and Pms1p are not required in recombination events that involve nonhomologous DNA suggests that the Msh2p-Msh3p complex can function in MMR-independent processes. This information also suggests that separation-of-function mutations that affect only MMR can be identified in MSH2 and MSH3. Dominant negative mutations provide an ideal way to test this hypothesis since they have the capacity to disrupt gene function by inhibiting the activity of the wild-type gene product when the mutant proteins are overexpressed (29). Manifestation of a dominant negative phenotype may be due to (i) inactive monomers or multimers that outcompete active monomers or (ii) a mixture of active and inactive monomers producing an inactive multimer (29). The isolation of dominant negative mutations in mutS and its homologues has aided substantially in the identification and characterization of functional domains of these proteins (6, 24, 64, 71). The highly conserved Walker type A nucleotide binding motif was first identified as an important target for mutagenesis in the Salmonella typhimurium and E. coli MutS proteins (24). Substitutions within the phosphate binding loop consensus sequence reduce ATPase activity and result in a dominant negative mutator phenotype in both bacteria and yeast (6, 24, 64, 71).
In this paper we describe a two-part screen yielding 24 msh2 alleles that conferred dominant negativity due to overexpression of the mutant Msh2 proteins by the GAL10 promoter. A subset of these alleles contained mutations analogous to several found in human MSH2 nonpolyposis colorectal cancer (HNPCC) kindreds. As described below, a large number of msh2 alleles that conferred a defect in MMR but were competent in the activities required during genetic recombination were identified. This study also provides important genetic tools that are needed to characterize interactions between Msh proteins and downstream mismatch repair components, only a few of which (i.e., Mlh1p-Pms1p and proliferating-cell nuclear antigen [22, 26, 68]) have been identified in eukaryotic systems.
MATERIALS AND METHODS
Media and chemicals.
E. coli strains were grown in Luria-Bertani broth or on Luria-Bertani agar supplemented with 100 μg of ampicillin per ml when required (45). Yeast strains were grown in either yeast extract-peptone (YP) medium supplemented with 2% glucose, 3% lactate, or 2% galactose or minimal selective medium supplemented with 2% glucose, 2% galactose, or 2% each of galactose and sucrose (57). Selective medium contained 0.7% yeast nitrogen base, 2% agar, and 0.09% drop-out mix that lacks the amino acid used for selection. When required, canavanine was included in minimal selective medium at 60 mg/liter (57). 5-Fluoroorotic acid (5-FOA) and 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) were purchased from U.S. Biologicals. 5-FOA and X-Gal minimal plates were prepared as described previously (57).
E. coli strains.
DH5α [F′ phi80 lacZΔ(lacZYA-argF)U169 recA1 endA1 hsdr17 (rK− mK+) λ− thil gyrA relA1] was used to amplify and manipulate all the plasmids described in this paper.
S. cerevisiae strains.
FY86 (MATα ura3-52 leu2Δ1 his3Δ200), FY23 (MATa ura3-52 leu2Δ1 trp1Δ63), EAY252 (MATa ura3-52 leu2Δ1 trp1Δ63 msh2Δ::TRP1), and EAY281 (MATa ura3-52 leu2Δ1 trp1Δ63 msh2Δ::hisG) are FY-derived strains (69) that were used to identify and characterize the msh2 dominant negative alleles in the mutator and repeat tract instability assays (Table 1).
TABLE 1.
Genetic characterization in mutator and repeat tract instability assays of msh2 alleles that confer a dominant negative phenotype
| mutant residue | Dominance tests (GAL10-msh2 2μm plasmids)a
|
Complementation test (ARS CEN msh2 plasmids)b: relative Canr frequency
|
Plasmid(s) analyzed | ||||
|---|---|---|---|---|---|---|---|
| Relative Canr frequency
|
Relative 5-FOAr frequency
|
||||||
| Mean | Individual results | Mean | Individual results | Mean | Individual results | ||
| Wild-type strain | 1 | 1 | NAc | None | |||
| msh2Δ strain | 45 | 35, 47, 52 | 105 | 103, 107 | 101 | 92, 110 | None |
| MSH2 | 1.0 | 1.6,0.6,0.8 | 1 | 0.7, 1, 1.3 | 1 | p[MSH2] | |
| Δ509–520 | 22 | 23, 21, 22 | 57 | NT | p[msh2–43] | ||
| S517P | 31 | 20, 17, 56 | NTc | NT | p[msh2–29] | ||
| A518P | 13 | 8, 18 | 35 | 32, 37 | 78 | p[msh2–44] | |
| C540R, M691T | 10 | 8, 11, 12 | NT | NT | p[msh2–36] | ||
| R542W | 9 | 14, 5, 8 | NT | NT | p[msh2–10] | ||
| S561P | 11 | 8, 11, 14 | 28 | 19, 36 | 65 | p[msh2–16, p[msh2–20] | |
| K564E | 35 | 31, 38, 36 | 25 | 23, 26 | 96 | p[msh2–5] p[msh2–38] | |
| G566D | 17 | 14, 20 | 27 | 19, 34 | 93 | p[msh2–12], p[msh2–34] | |
| L574S | 9 | 10, 9, 7 | 19 | 19, 19 | 57 | p[msh2–24] | |
| L584P | 18 | 32, 13 | 27 | 25, 29 | 162 | p[msh2–6] | |
| S656P | 39 | 46, 35, 36 | 40 | 24, 56 | 100 | p[msh2–8], p[msh2–41] | |
| G688E | 27 | 16, 18, 47 | NT | NT | p[msh2–4] | ||
| G692E | 28 | 16, 39 | NT | NT | p[msh2–37] | ||
| G693D | 34 | 23, 27, 52 | 73 | 64, 69, 85 | 119 | p[msh2–50] | |
| K694R | 50 | 44, 56 | 25 | 19, 31 | 111 | p[msh2–40] | |
| S695P | 30 | 24, 45, 20 | NT | NT | p[msh2–32] | ||
| R730W | 12 | 6, 17 | 24 | 22, 24, 26 | 69 | p[msh2–25] | |
| R730G, D924V | 14 | 14, 13 | NT | NT | p[msh2–3] | ||
| S736P, G848S | 23 | 25, 20 | NT | NT | p[msh2–26] | ||
| S742P | 28 | 20, 35, 29 | 63 | 51, 58, 81 | 128 | p[msh2–11], p[msh2–19], p[msh2–39] | |
| S742F | 5 | 5, 5, 5 | 44 | 41, 47 | 104 | p[msh2–27] | |
| E768V | 24 | 25, 25, 21 | NT | NT | p[msh2–15], p[msh2–21] | ||
| T773I | 24 | 22, 26 | 32 | 25, 38 | 59 | p[msh2–35] | |
| G855S | 19 | 15, 23 | NT | NT | p[msh2–42] | ||
In the dominance test, wild-type strains (FY86 for the mutator assay, FY23 containing pEAA69 for the repeat tract instability assay) were transformed with the indicated GAL10-msh2 plasmids and examined in a forward mutation assay that tested canavanine resistance and in a repeat tract instability assay that tested 5-FOA resistance (see Materials and Methods). The median frequencies in the mutator and repeat tract instability assays are shown relative to that obtained for a wild-type strain lacking a plasmid (7.33 × 10−7 Canr frequency and 1.6 × 10−4 5-FOAr frequency). Each dominance assay was repeated two to four times.
In the complementation test, EAY281 (msh2Δ) was transformed with the indicated msh2 ARSH4 CEN6 plasmids and tested in the canavanine resistance assay (see Materials and Methods). The median Canr frequencies are presented relative to that obtained for EAY281 bearing MSH2 on a TRP1 ARSH4 CEN6 vector (pEAA54; 3.12 × 10−7).
NT, not tested; NA, not applicable.
tNS1373αu (66) (mata-inc hmrΔ3 HMLα trp1 leu2 ura3-Nco GAL10::HO at THR4) and the derivatives EAY569 (msh2Δ::LEU2), EAY586 (pms1Δ::LEU2), EAY587 (msh6Δ::LEU2), EAY588 (msh3Δ::hisG), and EAY590 (mlh1Δ::hisG) were tested in the DSB-induced recombination assay that measured pFP120 retention (see below). tNS1817 [mat::leu2::hisG hmrΔ3 thr4 leu2 trp1 msh2::kanMX2 THR4-ura3(205 bp)-HOcs-URA3] was the parental strain that was used in the SSA assay (see Fig. 3). tNS1817 contains an SSA substrate with 205-bp URA3 repeats (66). It was derived from tNS62, a strain containing a triplication of the URA3 sequence where the ura3-52 allele was subsequently replaced by the THR4 gene (65). The resulting strain contains an HO cut site flanked by 205-bp repeats derived from the 5′ end of the URA3 gene. tNS1817 was transformed with the following combination of plasmids: pJH727 (GAL::HO LEU2 CEN4) (kindly provided by Anne Plessis) to test SSA in a msh2 null strain, pJH727 and pEAA54 (MSH2 ARSH4 CEN6) (see below) to test SSA in the wild type, and pJH727 and pEAA83 (msh2-R730W ARSH4 CEN6) (see below) to test SSA in a msh2-R730W strain. tNS1357 [mat::leu2::hisG hmr-3Δ ura3 leu2 trp1 thr4 (THR4-ura3-F HOcs-ura3-A)] carrying pFH800 (GAL::HO TRP1) and tNS1826, a msh2::kanMX2 derivative of tNS1357 carrying pEAA83 (msh2-R730W ARSH4 CEN6), were used in the SSA assays involving homeologous URA3 repeats separated by nonhomologous DNA containing an HO-cut site. The 205-bp ura3-FL100 and ura3-A homeologous repeat sequences contain and lack an MspI restriction site, respectively (56, 66).
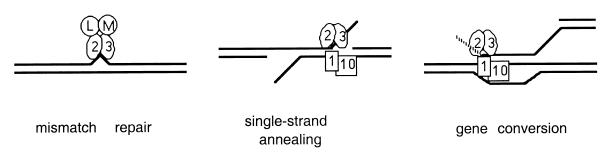
FIG. 3.
Model describing Msh2p-Msh3p interactions with duplex DNA containing nucleotide insertion-deletion mismatches or 3′ tails. In this model, binding of Msh2p-Msh3p (2–3) to a loop mismatch results in recruitment of the Mlh1p-Pms1p (L–M) complex and subsequent MMR steps. During SSA and gene conversion events, binding of Msh2p-Msh3p to duplex DNA containing 3′ tails results in stabilization of the recombination intermediate and allows the Rad1p-Rad10p (1–10) endonuclease to cleave the 3′ tails. We propose that msh2 dominant negative mutations displaying a separation-of-function phenotype do not affect the ability of corresponding Msh2p-Msh3p complexes to bind to DNA mispairs or to duplex DNA containing 3′ tails. However, we hypothesize that these mutant complexes are specifically defective in interactions with downstream MMR factors such as Mlh1p-Pms1p but are capable of stabilizing recombination intermediates for the Rad1p-Rad10p endonuclease.
The msh2Δ::LEU2, pms1Δ::LEU2, mlh1Δ::hisG, msh3Δ::hisG, and msh6Δ::LEU2 alleles contain complete or nearly complete coding-region deletions of their respective genes and were introduced into the FY and tNS strains by single-step transplacement (59). All yeast strains used in this study were transformed with DNA by the lithium acetate method as described by Geitz and Schiestl (19). The primer sequences used to confirm the transplacements by PCR analysis of chromosomal DNA and the plasmids used to make the gene disruptions are available upon request.
Plasmids.
pEAE20 (GAL10-MSH2 URA3 2μm) (2) was used as a template to mutagenize the MSH2 gene by PCR. pK5 (GT14G-LacZ 2μm LEU2) is a derivative of pCIZ1 (62) that was kindly provided by Brett Satterberg and Richard Kolodner. pK5 was transformed into FY86 and was used as a reporter to identify dominant negative msh2 alleles that displayed both a mutator and repeat tract instability phenotype. msh2 dominant negative alleles derived from pEAE20 were subcloned into pEAE86 (GAL10-MSH2 2μm TRP1) and transformed into FY23 so that they could be tested for their effect on repeat tract instability in yeast strains containing plasmid pEAA69 (GT16T-URA3 ARSH4 CEN6 LEU2) (kindly provided by Jayson Bowers).
pFP120 is a URA3-marked vector that contains inverted repeats of LacZ sequences (53, 66). This plasmid was transformed into all strains that were examined for the plasmid retention studies in Table 2. In pFP120, one copy of the LacZ repeat (recipient of information during DSB repair) contains a 40-bp HO cleavage site; the second copy (donor of information) lacks the HO cleavage site and contains an internal 878-bp deletion; the net result is that the LacZ copy containing the HO cleavage site is flanked by LacZ sequences that are deleted in the second “donor” copy.
TABLE 2.
DSB repair efficiency in wild-type and MMR-defective strains containing the recombination substrate pFP120a
| Relevant genotype | msh2 allele in an ARS-CEN vector | % Plasmid retention (mean ± SD) |
|---|---|---|
| Controls | ||
| Wild type | None | 63 ± 8.5 |
| msh2Δ | None | 2.0 ± 0.9 |
| msh3Δ | None | 2.1 ± 0.8 |
| msh3Δ | MSH6, 2μm | 1.8 ± 1.0 |
| msh6Δ | None | 60 ± 4 |
| pms1Δ | None | 60 ± 9 |
| mlh1Δ | None | 55 ± 5 |
| msh2 dominant negative mutations | ||
| msh2Δ | msh2-A518P | 36 ± 11 |
| msh2Δ | msh2-S561P | 64 ± 17 |
| msh2Δ | msh2-K564E | 56 ± 2 |
| msh2Δ | msh2-G566D | 58 ± 0.5 |
| msh2Δ | msh2-L574S | 8.8 ± 3.8 |
| msh2Δ | msh2-L584P | 2.0 ± 1.0 |
| msh2Δ | msh2-S656P | 49 ± 5 |
| msh2Δ | msh2-G693Db | 1.6 ± 0.4 |
| msh2Δ | msh2-K694Rb | 9.2 ± 2.2 |
| msh2Δ | msh2-R730Wb | 58 ± 13 |
| msh2Δ | msh2-S742Pb | 3.5 ± 1.3 |
| msh2Δ | msh2-S742Fb | 30 ± 16 |
| msh2Δ | msh2-T773Ib | 1.4 ± 0.2 |
| msh2Δ | msh2-G855D | 21 ± 3 |
| msh2 interaction domain mutations | ||
| msh2Δ | msh2-A859E | 51 ± 12 |
| msh2Δ | msh2-V862D | 58 ± 8 |
| msh2Δ | msh2-A872D | 27 ± 7 |
| msh2Δ | msh2-Δ863-868 | 1.3 ± 0.3 |
| msh3 ATP binding domain mutation | ||
| msh3Δ | none | 1.0 ± 0.5 |
| msh3Δ | MSH3 | 56 ± 12 |
| msh3Δ | msh3-G825Db | 2.0 ± 0.5 |
Wild-type (tNS1373αu) and MMR-defective derivatives [EAY569 (msh2Δ), EAY588 (msh3Δ), EAY587 (msh6Δ), EAY586 (pms1Δ), and EAY 590 (mlh1Δ)] containing pFP120 and ARSH4 CEN6 plasmids bearing the indicated msh2 and msh3 alleles were examined for the retention of pFP120 following DSB-induced recombination (see Materials and Methods). All measurements were done 3 to 15 times except for msh2-K564E, where only two transformants where examined and the range between the two values is shown.
These mutations map within the highly conserved ATP binding domain identified in mutS homologs (11).
Plasmids pRS414 (ARSH4 CEN6 TRP1) (10), pEAA54 (MSH2 ARSH4 CEN6 TRP1) and the following msh2 derivatives of pEAA54 were transformed into EAY569 strains bearing pFP120 to test the ability of these plasmids to complement a msh2Δ mutation in the pFP120 retention assay: pEAA75 (msh2-A518P), pEAA76 (msh2-S561P), pEAA77 (msh2-K564E), pEAA78 (msh2-G566D), pEAA79 (msh2-L574S), pEAA80 (msh2-L584P), pEAA81 (msh2-S656P), pEAA71 (msh2-G693D), pEAA82 (msh2-K694R), pEAA83 (msh2-R730W), pEAA84 (msh2-S742P), pEAA74 (msh2-S742F), pEAA86 (msh2-T773I), pEAA72 (msh2-G855D), pEAA73 (msh2-A859E), pEAA107 (msh2-V862D), pEAA108(msh2-A872D), and pEAA111(msh2-Δ863–868).
pEAA112 (MSH3 ARSH4 CEN6 TRP1), pEAA115 (msh3-G825D ARSH4 CEN6 TRP1), pEAM49 (MSH6 2μm TRP1), and pRS414 (ARSH4 CEN6 TRP1) were transformed into EAY588 containing pFP120 to test the complementation of these alleles in a msh3Δ mutation in the pFP120 retention assay.
Nucleic acid techniques.
All restriction endonucleases were purchased from New England Biolabs (Beverly, Mass.) and used as specified by the manufacturer. Taq polymerase was purchased from Perkin-Elmer Cetus. Plasmid DNA was isolated by alkali lysis, and all DNA manipulations were performed as described previously (42) (Qiagen Inc. Santa Clarita, Calif.). Yeast chromosomal DNA was prepared as described by Holm et al. (32). Southern blot analysis was performed as described previously (42, 66).
PCR was performed to randomly mutagenize the entire MSH2 open reading frame in pEAE20 as follows. Nine separate 25-μl PCR mixtures were used with 0.5 μg of pEAE20 as a template and standard concentrations of Taq polymerase, primers, and buffer as recommended by the manufacturer (Perkin-Elmer Cetus). Each PCR involved 12 cycles involving a 1-min denaturation step at 94°C, a 1-min annealing step at 50°C, and a 2-min polymerization step at 72°C. In reactions 1 to 5, two primers, AO147 (5′TAAGCGTATTACTGAAAGTTCC) and AO39 (5′GTTAATTTCAGTTAGCGG), that amplified a 2.3-kb 5′ fragment of MSH2 were used; in reactions 6 to 9, two other primers, AO7 (5′GGAAACTTAGAGGATGTC) and AO49 (5′GTCAGAAGTAAGTTGGCCGC) that amplified a 1.9-kb 3′ fragment of MSH2 were used. The amplified fragments were digested with restriction enzymes (XhoI and BglII for the 5′ PCR fragment, BglII and KpnI for the 3′ PCR fragment) and subcloned into the corresponding sites of pEAE20. Each PCR resulted in a pool of ∼3,000 E. coli transformants containing PCR-mutagenized pEAE20.
The msh3-G825D allele was constructed by site-directed mutagenesis by the PCR overlap extension method (30); the amplification conditions and primer sequences that were used to create this allele are available upon request. DNA primer synthesis and DNA sequencing were performed at the Cornell Biotechnology Analytical/Synthesis facility.
Isolation of msh2 dominant negative mutations.
Each of nine PCR pools containing mutagenized pEAE20 were transformed into FY86 containing pK5, and the transformants were grown on minimal selective medium containing 2% galactose and 2% sucrose as carbon sources (these carbon sources were used in all subsequent steps). After growth at 30°C, the transformants were replica plated onto X-Gal minimal plates. Colonies that turned pale blue to dark blue were individually streaked to single colonies on minimal medium. For each candidate, 11 of these single colonies were patched to minimal medium containing canavanine or X-Gal. The PCR-mutagenized pEAE20 plasmid present in each candidate strain was recovered by standard procedures (57). These plasmids were retransformed into FY86 containing pK5 and retested on canavanine and X-Gal plates. A total of 31 plasmids bearing dominant negative alleles were recovered from the 23,000 transformants that were screened.
Determination of mutation frequencies.
Mutation frequencies were determined by measuring the frequency of forward mutation to canavanine resistance (5, 55). Repeat tract instability frequencies were determined by measuring frameshift events that resulted in resistance to 5-FOA in strains containing plasmid pEAA69.
For the dominance mutator tests, FY86 was transformed with each of the 31 GAL10-msh2 plasmids and the transformants were then streaked to form single colonies on selective minimal plates containing galactose plus sucrose. Eleven independent colonies were suspended in water, and appropriate dilutions were plated onto minimal medium containing galactose plus sucrose with or without canavanine. For the dominance repeat tract instability tests, FY23 was cotransformed with pEAA69 and GAL10-msh2 plasmids and transformants were diluted and plated as described above onto minimal medium with or without 5-FOA. The median frequency of canavanine and 5-FOA resistance was determined for each experiment.
For the complementation tests, both quantitative and patch tests were performed. Initially, the canavanine patch assay described above was performed with EAY252 transformed with the 31 GAL10-msh2 plasmids. Of the 31 GAL10-msh2 plasmids, 13 were cotransformed with pK5 into EAY252 so that repeat tract instability phenotypes could be assessed. In the quantitative complementation assay reported in Table 1, 13 of the 31 msh2 alleles present in the pEAE20 plasmid background were subcloned into a ARSH4 CEN6 TRP1 vector so that the mutant alleles were expressed in single copy under the native MSH2 promoter. These plasmids were transformed into EAY281 and tested for their ability to complement the msh2Δ deletion strain in the quantitative canavanine resistance assay described above.
pFP120 retention and SSA assays.
The retention of pFP120 in yeast strains following DSB-induced recombination was measured as follows. Two to 15 colonies from each strain transformed with both pFP120 and an ARSH4 CEN6 TRP1 vector containing a particular msh2 or msh3 allele were suspended in distilled water and plated at an equivalent concentration onto both YP-glucose and YP-galactose plates. Colonies from each plate were then replica plated onto minimal glucose plates lacking uracil and tryptophan, and the number of Ura+ Trp+ colonies was scored. The percent plasmid retention was calculated by determining the ratio of Ura+ Trp+ colonies obtained after cell growth on YP-galactose medium to Ura+ Trp+ colonies obtained after growth on YP-glucose medium. The SSA assays in Fig. 2C were carried out as described by Sugawara et al. (66). To measure whether the msh2-R730W allele was defective in the repair of base pair mismatches created during SSA, cultures of tNS1357 and the derivative tNS1826 were grown from single colonies in minimal selective medium. The overnight cultures were diluted into YP-lactate medium and grown to a cell density of 106 to 107 cells/ml. Expression of HO endonuclease in these strains was induced by adding galactose to 2% (wt/vol) final concentration. Unbudded cells were isolated by micromanipulation on YP-dextrose (YPD) plates within 30 min following a 1-h HO induction and grown to individual colonies. DNA was prepared from these colonies, digested with MspI, and analyzed by Southern blotting with the URA3 HindIII sequence.
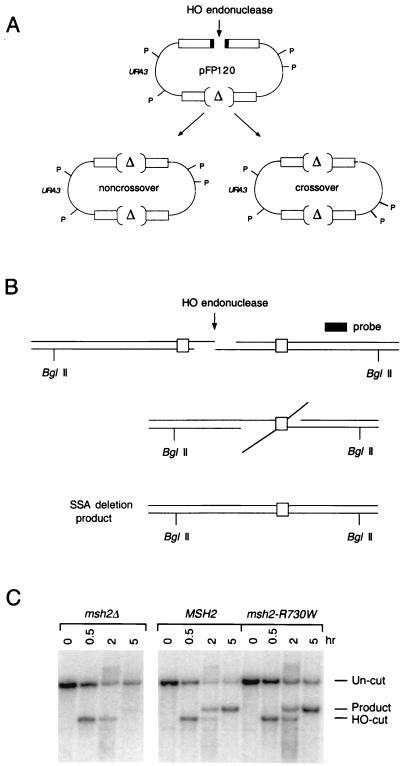
FIG. 2.
(A) Structure of pFP120 and predicted gene conversion events, with and without crossing over, following HO cleavage. P denotes PstI sites that distinguish crossover from noncrossover recombinants. A detailed description of the structure of pFP120 can be found in reference 53. (B) SSA between identical 205-bp URA3 gene segments that flank an HO-induced DSB. Following HO cleavage, 5′-3′ exonuclease digestion produces 3′ single-stranded tails that can anneal with complementary sequences. Removal of 3′ nonhomologous tails results in the formation of the deletion product. BglII sites that distinguish uncut (7.4 kb) and HO-cut (4.8 kb) chromosomal DNA from the deletion product (5.5 kb) are indicated. (C) Southern blot analysis showing the kinetics of HO-induced SSA in wild-type (tNS1817 containing pJH727 and pEAA54), msh2Δ (tNS1817 containing pJH727), and msh2-R730W (tNS1817 containing pJH727 and pEAA83) strains that contain repeats of the 205-bp URA3 gene segment illustrated in panel B. Chromosomal DNA from the indicated strains was isolated at the indicated time points after HO induction, digested with BglII, and probed with a DNA fragment centromere proximal to the URA3 gene (66). The ratios of plasmid-bearing colonies after and before induction were 0.09, 1.8, and 1.2 for msh2Δ, MSH2, and msh2-R730W strains, respectively. These values reflect the value of product formation, the extent of induction, and the growth of the culture during the 5-h induction (66).
Computer analysis.
Yeast and human Msh2p amino acid sequences were aligned with DNASTAR MegAlign (v.3.16) by the CLUSTAL method with a PAM250 residue weight table (DNASTAR Inc). The alignment shown in Fig. 1 was shaded in MacBoxshade v.2.15 (Baron and Felciano, Mercutio 1.52). The human Msh2p sequence used in the alignment was obtained from the Protein Sequence Database, Protein Identification Resource entry I37550 (53a). The human HNPCC mutations were obtained from The Human Genome Mutation Database Cardiff (32a) and the HNPCC mutation database (29a).
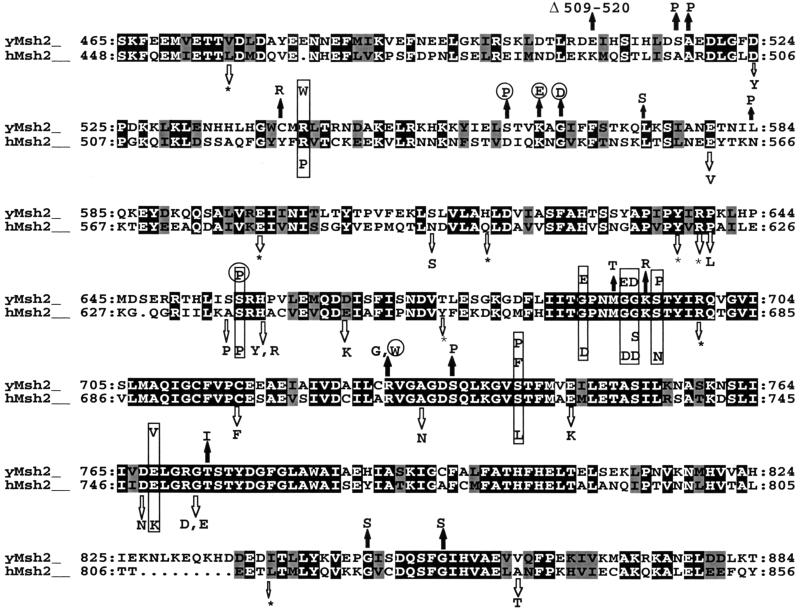
FIG. 1.
Map of mutated Msh2p residues in S. cerevisiae and in human HNPCC kindreds. Residues corresponding to the carboxyl termini of the two homologs are shown. Identical and similar residues are shaded black and gray, respectively. The dominant negative yeast mutations are indicated by solid arrows above the yeast Msh2 (yMsh2) sequence; mutations identified in the human kindreds are indicated by open arrows below the human Msh2 (hMsh2) sequence. Of the 59 human MSH2 mutations that were examined, 31 mapped to the region shown (see Materials and Methods). Yeast mutations in residues identical to HNPCC mutated residues are indicated by the boxes. Mutations in yeast MSH2 that confer wild-type activity in the removal of nonhomologous DNA during DSB repair are indicated by circles. The ATP binding domain is located between residues 686 and 700 (region I), 739 and 745 (region II), 762 and 773 (region III), and 798 and 803 (region IV) with respect to the yeast Msh2p sequence (14, 48). The location of the msh2-D924V mutation in the double-mutant allele p[msh2-3] is not shown. ∗, nonsense mutation.
RESULTS
Isolation of dominant negative msh2 alleles.
The two-part screen used to identify dominant negative alleles in MSH2 is based on the observation that msh2Δ strains display repeat tract instability and mutator phenotypes (see Materials and Methods) 63; reviewed in reference 39). These phenotypes were measured in the wild-type strain FY86 containing pK5, a 2μm plasmid that contains an out-of-frame GT repeat sequence inserted into the LACZ open reading frame (GT14G-LACZ) (28, 62). pK5 was used to assess repeat tract instability because it can monitor defects in MMR without the use of a fluctuation assay (62). On X-Gal plates, nearly 100% of msh2Δ strains containing pK5 display a blue colony phenotype; in contrast, less than 0.5% of wild-type colonies are blue. The mutator phenotype was measured in FY86 containing pK5 by using patch assays to identify canavanine-resistant colonies (see Materials and Methods). In the canavanine assay, msh2Δ strains papillate to canavanine resistance at a 50-fold higher frequency than the wild type does. To identify dominant negative msh2 alleles, the MSH2 gene expressed in pEAE20 under the GAL10 promoter was PCR mutagenized (see Materials and Methods). Pools of mutagenized plasmids were transformed into FY86 bearing pK5, and 31 independently derived plasmids were obtained that conferred a dominant negative phenotype in both assays when cells were grown on galactose.
DNA sequencing of msh2 alleles.
DNA sequencing of the 31 msh2 plasmids revealed that 3 plasmids each contained two mutations, 1 contained a 36-bp deletion, and the remaining 27 each contained single mutations, six of which were identified more than once from independent pools (Table 1). Of the 20 unique single mutations, 10 were located within the highly conserved ATP binding domain (Fig. 1). Interestingly, a mutation in the phosphate binding loop (msh2-G693D) was isolated that was identical to a dominant negative allele that had been created previously by site-directed mutagenesis (6). We were surprised to find no substitutions within the N-terminal 509 amino acids of Msh2p, even though both the amino-terminal end and the carboxyl-terminal ends of the protein were mutagenized in the same procedure (see Materials and Methods).
Cell extracts from strains simultaneously overexpressing Msh6p and one of eight mutant Msh2 proteins (msh2-Δ509–520p, msh2-R542Wp, msh2-G566Dp, msh2-L574Sp, msh2-G688Ep, msh2-G693Dp, msh2-R730Wp, and msh2-S742Fp) were examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Expression levels of both Msh6p and the corresponding mutant Msh2 proteins were comparable to those of strains overexpressing Msh6p and Msh2p, suggesting that the mutant Msh2 proteins are stable in vivo (references 2 and 6 and data not shown).
The yeast and human Msh2p amino acid sequences were aligned to determine if any of the yeast mutations corresponded to mutations identified in HNPCC kindreds. A search of online databases (see Materials and Methods) revealed that of the 20 msh2 alleles that contained unique single mutations, 9 had mutations that corresponded to substitution mutations identified in hMSH2 kindreds (Fig. 1). In addition, 8 of the remaining 11 yeast single-substitution mutations were located within seven of fewer residues of the HNPCC mutations (Fig. 1).
msh2 dominant negative mutations fail to complement a msh2Δ strain.
The 24 unique (20 single, 1 deletion, and 3 double mutations) msh2 alleles identified in pEAE20 derived plasmids were tested in canavanine patch assays for their ability to complement a msh2Δ strain (EAY252 [see Materials and Methods). As expected, the wild-type MSH2 gene complemented the msh2Δ mutator phenotype; however, all of the mutant plasmids conferred a high papillation phenotype that was indistinguishable from that observed in msh2Δ strains. Thirteen of these plasmids were also cotransformed with pK5 into EAY252 to test the repeat tract instability phenotype in patch assays on X-Gal plates. While pEAE20 conferred a white colony phenotype, all 13 plasmids containing msh2 mutations conferred a blue colony phenotype that was indistinguishable from that observed in msh2Δ strains (data not shown).
We also tested whether the failure of the dominant negative msh2 alleles to complement the msh2Δ strain was due to overexpression of the alleles on the GAL10 2μm vector. Thirteen msh2 dominant negative alleles (Table 1) were subcloned into an ARSH4 CEN6 TRP1 vector so that expression of MSH2 was driven by the native MSH2 promoter and expressed in single copy. These plasmids were transformed into msh2Δ strain EAY281 and tested for complementation of the msh2Δ phenotype in quantitative canavanine assays. All of these msh2 alleles conferred a strong mutator phenotype that was indistinguishable from that observed in a msh2Δ strain. Taken together, these data indicate that none of the msh2 dominant negative mutations could complement the msh2Δ MMR defect (Table 1).
Assessment of the dominant negative phenotype in quantitative mutator and repeat tract instability assays.
The dominant negative phenotype conferred by overexpressing the Msh2 mutant proteins described above was assessed in quantitative canavanine and 5-FOA resistance assays. The Canr frequencies in strains overexpressing these mutant proteins were 5- to 50-fold higher than in a wild-type strain that lacked a MSH2 overexpression plasmid (Table 1). Fourteen GAL10-msh2 alleles were subcloned into 2μm TRP1 vectors and tested for a dominant negative phenotype in an assay that measures simple repeat tract alterations in a (GT)16G-URA3 fusion construct. Repeat tract instability events were measured by determining the frequency of cells resistant to 5-FOA (28). The relative rates of DNA slippage events conferred by these plasmids varied from 19 to 73 with respect to the wild-type strain bearing wild-type MSH2 (Table 1). Overall, the strongest mutators also showed the strongest repeat tract instability phenotypes. The one exception was the msh2-S742F allele, which conferred a weak dominant negative phenotype in the canavanine resistance assay but a strong dominant negative phenotype in the tract alteration assay (Table 1).
Eight (msh2-A518P, msh2-S561P, msh2-K564E, msh2-G566D, msh2-S656P, msh2-G693D, msh2-R730W, and msh2-S742F) of the above msh2 alleles were subcloned into ARSH4 CEN6 TRP1 vectors to express the mutant proteins in single copy under the native MSH2 promoter (see Materials and Methods). These plasmids were transformed into the wild-type strain FY23 and tested in quantitative canavanine assays. None of the mutations conferred a canavanine resistance frequency above the wild-type level, indicating that overexpression of these alleles was required to observe a mutator phenotype (data not shown).
A subset of dominant negative msh2 mutants are proficient in removing nonhomologous DNA during gene conversion and SSA.
We tested whether the dominant negative msh2 mutations described above would confer wild-type function in the DSB-induced gene conversion and SSA annealing assays in Fig. 2. Gene conversion events were examined in tNS1373αu-derived yeast strains containing pFP120 (Fig. 2) (see Materials and Methods). Gene conversion events in strains bearing pFP120 were induced by creating a DSB in pFP120 through HO endonuclease cleavage, and the formation of successful gene conversion events was assessed in a plasmid retention assay (see Materials and Methods) (53, 66).
As shown in control experiments presented in Table 2, plasmid retention, as measured by the formation of Ura+ colonies, was indistinguishable in wild-type, msh6Δ (EAY587), pms1Δ (EAY586), and mlh1Δ (EAY590) strains and ranged from 55 to 63%. In msh2Δ and msh3Δ strains, plasmid retention was reduced 30-fold. The values for wild-type, msh2Δ (EAY569), and msh3Δ (EAY588) strains closely agree with those reported previously (66). Expression of MSH6 on a high-copy-number 2μm vector did not improve plasmid retention levels in msh3Δ mutants. This was tested because previous studies had shown that Msh3p and Msh6p display overlapping functions during MMR (36, 43).
Thirteen of the dominant negative msh2 alleles described in Table 1 and Fig. 2, as well as the previously described msh2-G855D dominant allele (6), were subcloned into ARSH4 CEN6 TRP1 plasmids and transformed into EAY569. These subcloning steps were performed to examine pFP120 retention in strains containing the msh2 alleles expressed in single copy from the native MSH2 promoter. Two other sets of alleles were also expressed in similar vectors and tested: msh2 mutations A859E, V862D, and Δ863–868, which were shown previously to disrupt Msh2p-Msh6p interactions (6), and msh3-G825D, which is analogous to the phosphate binding loop mutation present in msh2-G693D (48). The msh2-A859E, msh2-V862D, and msh2-Δ863–868 mutations were shown to confer a MMR defect identical to that found in msh2Δ strains (6). In repeat tract instability assays, the msh3-G825D mutation conferred a tract instability frequency (9.4 × 10−3) that was 50-fold higher than that in the wild type and was similar to that observed in msh3Δ strains (2.8 × 10−3).
As shown in Table 2, four of the six mutations that mapped to the Msh2p ATP binding domain (msh2-G693D, msh2-K694R, msh2-S742P, and msh2-T773I) and the msh3-G825D mutation conferred a defect in plasmid retention that was similar to that observed in msh2Δ or msh3Δ mutants. However, the msh2-G855D mutation (21% retention), which was shown previously to disrupt Msh2p-Msh6p ATPase activity to a level similar to that observed for the msh2-G693D mutation (6), and two mutations that map to residues in the ATP binding domain consensus sequence that are conserved among mutS homologs (msh2-R730W [58% retention] and msh2-S742F, [30% retention]), displayed relatively high levels of plasmid retention. Plasmid retention was conferred at wild-type or nearly wild-type levels for five mutations that map to locations in MSH2 that have not been previously implicated in function (msh2-A518P, msh2-S561P, msh2-K564E, msh2-G566D, and msh2-S656P). In contrast, only two other mutations that map to regions of unknown function, msh2-L574S and msh2-L584P, conferred low plasmid retention levels.
Site-directed mutations in MSH2 that were created previously and were shown to cause both an MMR and a Msh2p-Msh6p interaction defect were also tested in the pFP120 retention assay (6). Two mutations that conferred a moderate defect in Msh2p-Msh6p interactions (msh2-A859E and msh2-A862D) (6) conferred wild-type levels of plasmid retention. However, msh2 mutants that displayed a more severe Msh2p-Msh6p interaction-defective phenotype also displayed lower levels of plasmid retention. As shown in Table 2, the msh2-A872D mutation, which conferred a stronger Msh2p-Msh6p interaction defect than the msh2-A859E or msh2-V862D mutation, conferred a lower level of plasmid retention. The msh2-Δ863–868 mutation, which conferred the strongest Msh2p-Msh6p interaction defect, conferred a plasmid retention defect similar to that observed for the msh2Δ mutation (6).
It is important to note that the dominant negative msh2 alleles that were subcloned into ARSH4 CEN6 vectors were also tested for complementation of the msh2Δ mutator phenotype in EAY569. In patch assays, the msh2 dominant negative mutations all conferred an increase in Canr papillation frequency indistinguishable from that observed in the msh2Δ strain, and this increase was ∼40-fold higher than that observed in the wild type (data not shown). These data indicate that the msh2 dominant negative mutations conferred a similar MMR defect in both the tNS1373αu and FY strain backgrounds (Table 1 and data not shown).
Based on the studies presented in Table 2 and the finding that the MSH2 and MSH3 gene products are required for the removal of nonhomologous DNA tails during both DSB-induced gene conversion and SSA events (66), we tested whether a msh2 mutant (msh2-R730W) that was functional in the pFP120 plasmid retention assay would show a similar activity in the SSA assay (see Materials and Methods). As shown in the Southern blot in Fig. 2C, the msh2-R730W strain displayed cell viability and SSA product formation levels that were indistinguishable from those of the wild type. As shown previously, a strain bearing a msh2 null mutation showed a dramatic decrease in cell viability and SSA product formation (Fig. 2C and data not shown).
Because strains carrying the msh2-R730W allele were proficient for SSA but displayed a mutator phenotype, we examined whether the msh2-R730W allele conferred a defect in the repair of base pair mismatches created during SSA. To test this, an SSA substrate containing 205-bp homeologous repeats (3% divergence) was constructed in wild-type and msh2-R730W strains (tNS1357 and tNS1826, respectively [see Materials and Methods). The URA3 repeats were previously described and are referred to as ura3-FL100 (56) and ura3-A (66). After inducing DSB formation for 1 h, unbudded cells were isolated by micromanipulation and allowed to grow into single colonies. DNA was isolated from each colony and analyzed by Southern blot hybridization (data not shown) to score for an MspI site that is present in ura3-FL100 but absent in ura3-A; this polymorphism allows the identification of “sectored” colonies that have failed to undergo MMR in heteroduplex DNA created by SSA. For the wild-type strain, only 11% of the colonies were sectored (2 of 19 colonies). In contrast, 68% of the colonies containing the msh2-R730W allele were sectored (17 of 25 colonies), indicating that the msh2-R730W allele is defective for MMR during SSA.
DISCUSSION
Using a two-part genetic screen, we identified 24 unique msh2 alleles that confer an MMR defect and cause a dominant negative phenotype in both mutator and repeat tract instability assays. About half of the alleles contain mutations that map within the highly conserved ATP binding domain. A screen for dominant negative mutS mutations in E. coli also yielded a high percentage of mutations that mapped to the ATP binding domain (71). In vitro studies of bacterial, yeast, and human MutS homolog proteins that display ATP hydrolysis-related defects as the result of mutations in the P-loop motif provide a biochemical explanation for the dominant negative effect. In all three cases, the wild-type proteins bind specifically to mismatches in the absence of ATP but lose this specificity in the presence of ATP. In contrast, ATPase-defective complexes (i.e., yeast msh2-G693Dp-Msh6p and Msh2p-msh6-G987Dp) retain mismatch binding specificity similar to the wild type in both the absence and presence of ATP (6, 24, 34, 64, 71). This analysis suggested that the ATPase-defective complexes can compete with the wild-type protein for mismatch binding. The retention of mismatch binding in these mutant proteins is hypothesized to result in a dominant negative phenotype because the mutant Msh proteins are unable to function in ATP-dependent translocation steps that are thought to occur after a mismatch has been identified (7, 64).
In the recombination assay described in Fig. 2A and Table 2, seven msh2 alleles that displayed at least 50% of wild-type activity were identified. Interestingly, two of these alleles contained mutations in the ATP binding domain (Fig. 1) while five contained mutations in residues that are conserved among Msh proteins but map to regions for which no function has been described. We hypothesize that these seven mutants with separation-of-function mutations in MSH2 are disrupted for interactions between Msh2p and downstream MMR factors such as Mlh1p and Pms1p because the latter are not required for the removal of nonhomologous DNA during recombination (66). It is likely that these mutations confer a dominant negative phenotype for MMR but a proficiency in recombination because their gene products can function in steps that involve binding to a mispair or to a branched structure containing a nonhomologous 3′ tail (Fig. 3) (see below). Thus, we expect that they would retain domains in Msh2p that are required for both DNA binding (i.e., the putative N-terminal mismatch binding-domain amino acids 27 to 52) (41) and subunit interactions (6). Consistent with this idea, we did not identify any dominant negative mutations that mapped to the putative DNA binding domain. In addition, a mutation in MSH2 that disrupts Msh2p-Msh6p interactions (msh2-Δ863–868) and is hypothesized to disrupt Msh2p-Msh3p interactions (based on human protein studies) resulted in a strong defect in both MMR and in recombination (Table 2) (6, 23). It is important to note that none of the dominant negative msh2 alleles identified in this study mapped within this region.
The requirement for Msh2p and Msh3p in nonhomologous DNA tail removal adds to the growing list of recombination activities that involve MutS homolog proteins. Previous studies have shown that Msh2p and Msh3p act to suppress recombination between divergent DNA sequences (12, 33, 61) and that Msh2p also plays an important role in establishing gene conversion gradients at recombination hotspots (5, 55). Interestingly, mutations in the MutS homologs MSH4 and MSH5 do not confer an MMR defect but, instead, confer a defect in crossing over that results in an increase in meiosis I nondisjunction events (31, 58). Biochemical studies of the Msh proteins also support a role for these proteins in genetic recombination. Different associations of these proteins (i.e., Msh2p, Msh2p-Msh3p, and Msh2p-Msh6p) bind to a variety of DNA structures such as base and palindromic insertion mismatches, Y junctions, and synthetic Holliday junctions (1, 3, 4, 6, 16, 20, 25, 44, 52). The requirement for MSH2 and MSH3 in the removal of nonhomologous DNA and the DNA binding properties of Msh proteins suggests that the Msh2p-Msh3p complex binds to branched DNA structures containing a nonhomologous 3′ end in the same way that it binds to DNA containing mispairs (Fig. 3).
Studies with E. coli and yeast have identified several proteins that function in MMR and also interact with MutS or Msh2p. Gel retardation studies and affinity chromatography studies have shown that E. coli MutS can physically interact with MutL in steps that require both mismatch substrates and ATP (21, 70). A similar analysis has shown that yeast Msh2p-Msh3p and Msh2p-Msh6p interact with Pms1p-Mlh1p in steps that require mismatched substrates (26, 27). Two-hybrid and coimmunoprecipitation studies have shown that S. cerevisiae Msh2p interacts with the POL30, EXO1, and RAD2, RAD10, RAD14, and RAD25 gene products (9, 67, 68). The yeast 5′-3′ exonuclease Exo1p is thought to be active in DNA replication, recombination, and MMR (15, 54, 67), the POL30 gene encodes the DNA replication clamp PCNA that is required in both mismatch and nucleotide excision repair (22, 49, 68), and the RAD2, RAD10, RAD14, and RAD25 genes encode products that participate in nucleotide excision repair (18).
A biochemical analysis of Msh2p-Msh3p complexes containing the dominant negative msh2 mutations and downstream MMR factors should provide a direct way to test for specific interaction defects. The msh2-R730W and msh2-S742F alleles, which map to the Msh2p ATP binding domain, are good choices to test for Mlh1p-Pms1p interaction defects. Based on the observation that MutS-MutL interactions require ATP, we hypothesize that the msh2-R730W and msh2-S742F mutations disrupt ATP-dependent conformational changes in Msh2p that are required for Mlh1p-Pms1p interactions but do not affect the ability of ATP to modulate mismatch recognition functions that are required for interactions with Rad1p-Rad10p (Fig. 3) (8, 21, 64) (see below). These proposed studies will require new overexpression methods to purify sufficient quantities of prospective partners to examine interactions with a large number of mutant Msh2p-Msh3p and/or Msh2p-Msh6p complexes. In yeast, for example, overproduction of the Msh2p-Msh3p and Mlh1p-Pms1p complexes is not straightforward, since only microgram quantities of these complexes have been obtained from several hundred grams of yeast cells induced for overexpression of the relevant gene products (25, 26). New genetic approaches and biochemical assays will also be required to characterize interactions between wild-type and mutant Msh2 proteins and to identify downstream MMR factors such as helicases and exonucleases.
By analogy to their role in MMR, it appears likely that Msh protein interactions with the repair and recombination machinery allow them to play key roles in both the processing and resolution of DNA intermediates. During MMR, ATP-mediated conformational changes in the MutS-mispair complex are thought to allow interactions with MutL (7, 8, 64; reviewed in reference 47). Is a similar mechanism used to facilitate physical interactions between Msh2p-Msh3p and Rad1p-Rad10p when Msh2p-Msh3p is bound to branched DNA structures? Our data suggest that ATP-dependent conformational changes in Msh2p-Msh3p may not be required for interaction with Rad1p-Rad10p. First, we observed that dominant negative mutations in highly conserved residues of the Msh2p ATP binding domain that are expected to severely affect ATP binding, hydrolysis, or both do not affect pFP120 retention to the same extent (i.e. msh2-G693D versus msh2-K694R [Table 2] [6, 34, 64]). Second, we found that the msh2-G855D mutation, which was shown previously to disrupt the ATPase activity of Msh2p to a level similar to that observed for the msh2-G693D mutation, conferred a relatively high level of pFP120 retention (21% for msh2-G855D versus 1.6% for msh2-G693D [Table 2] [6]).
As described in Fig. 3, we propose that Msh2p-Msh3p functions to remove nonhomologous DNA during recombination by binding and stabilizing branched DNA structures. Msh2p-Msh3p is then displaced in steps that require ATP, thus allowing the Rad1p-Rad10p endonuclease to cleave the 3′ single-stranded DNA tails. Based on this model, the strong defect in pFP120 retention observed for some ATP binding domain msh2 mutants is due to the inability of the mutant complexes to be displaced in an ATP-dependent fashion from a branched DNA substrate. This defect in turn prevents cleavage of the 3′ nonhomologous tail by Rad1p-Rad10p. In support of this idea, we found previously that the msh2-G855Dp-Msh6p complex (21% pFP120 retention) was displaced more easily from a mismatch oligonucleotide substrate by ATP than was the msh2-G693Dp-Msh6p complex (1.6% pFP120 retention [6]). Genetic support for this model is provided by the fact that SSA reactions analogous to those described in Fig. 2C become independent of MSH2 and MSH3 functions but not RAD1 functions when the duplicated regions approach 1 kb in size (66). These data suggest that Rad1p-Rad10p does not require Msh2p-Msh3p to remove nonhomologous DNA when stable branched structures are formed (66). The data also suggest that Msh2p-Msh3p stabilizes branched intermediates that might otherwise fall apart prior to the removal of 3′ tails by the Rad1p-Rad10p endonuclease (66). With this logic, the recruitment of Rad1p-Rad10p to the branched structures does not require a protein-protein interaction with Msh2p-Msh3p that is analogous to the interaction required to form a Msh2p-Msh3p-Mlh1p-Pms1p complex on a mismatch substrate.
Many of the yeast dominant negative mutations have human HNPCC counterparts.
A large number of the dominant negative yeast msh2 alleles that resulted from single point mutations (9 of 20) were associated with corresponding mutations in human MSH2 that have been identified from HNPCC kindreds. Four of these alleles (msh2-S656P, msh2-G693D, msh2-S742P, and msh2-S742F) were tested in the nonhomologous tail removal assay (Fig. 2), and two (msh2-S656P and msh2-S742F) were shown to display at least 50% of the wild-type activity. The progression to HNPCC in MSH2 kindreds is thought to result from the loss of heterozygosity in cells from individuals heterozygous for a mutated copy of MSH2 (reviewed in references 38 and 40). One possible explanation for this progression, based on the data presented in this report, may be that some of the human HNPCC msh2 alleles are not null alleles but encode polypeptides that can function in a subset of MMR steps when expressed in either heterozygous or homozygous mutant cells (13, 50). Further studies are required to test this idea and explore whether the expression of non-null alleles can be related to the manifestation of the HNPCC disease phenotype.
ACKNOWLEDGMENTS
We thank Elizabeth Evans for extensive comments on the manuscript and helpful discussions and Tanya Sokolsky for providing reagents and unpublished data.
E.A. and B.S. were supported by National Institutes of Health grant GM53085 and USDA Hatch grant NYC-1867424, B.S. was also supported by a New York State Fellowship and a Cornell University Anonymous Donor Fellowship, G.P. was supported by an undergraduate summer research fellowship from the Howard Hughes Medical Institute awarded to Cornell University, and J.E.H. and N.S. were supported by NIH grant GM20056.
REFERENCES
- 1.Acharya S, Wilson T, Gradia S, Kane M F, Guerrette S, Marsischky G T, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani E. The Saccharomyces cerevisiae Msh2p and Msh6p form a complex that specifically binds to duplex oligonucleotides containing mismatched DNA base pairs. Mol Cell Biol. 1996;16:5604–5615. doi: 10.1128/mcb.16.10.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alani E, Chi N-W, Kolodner R D. The Saccharomyces cerevisiae Msh2 protein specifically binds to duplex oligonucleotides containing mismatched DNA base pairs and loop insertions. Genes Dev. 1995;9:234–247. doi: 10.1101/gad.9.2.234. [DOI] [PubMed] [Google Scholar]
- 4.Alani E, Lee S, Kane M F, Griffith J, Kolodner R D. Saccharomyces cerevisiae MSH2, a mispaired base recognition protein, also recognizes Holliday junctions in DNA. J Mol Biol. 1997;265:289–301. doi: 10.1006/jmbi.1996.0743. [DOI] [PubMed] [Google Scholar]
- 5.Alani E, Reenan R A G, Kolodner R D. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics. 1994;137:19–39. doi: 10.1093/genetics/137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alani E, Sokolsky T, Studamire B, Miret J J, Lahue R S. Genetic and biochemical analysis of Msh2p-Msh6p: role of ATP hydrolysis and Msh2p-Msh6p subunit interactions in mismatch base pair recognition. Mol Cell Biol. 1997;17:2436–2447. doi: 10.1128/mcb.17.5.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen D J, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, Griffith J D. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ban C, Yang W. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell. 1998;95:541–552. doi: 10.1016/s0092-8674(00)81621-9. [DOI] [PubMed] [Google Scholar]
- 9.Bertrand P, Tishkoff D X, Filosi N, Dasgupta R, Kolodner R D. Physical interaction between components of DNA mismatch repair and nucleotide excision repair. Proc Natl Acad Sci USA. 1998;95:14278–14283. doi: 10.1073/pnas.95.24.14278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 11.Crouse G F. Mismatch repair systems in Saccharomyces cerevisiae. In: Nickoloff J, Hoekstra M, editors. DNA damage and repair—biochemistry, genetics and cell biology. Clifton, N.J: Humana Press; 1996. pp. 411–448. [Google Scholar]
- 12.Datta A, Adjiri A, New L, Crouse G F, Jinks-Robertson S. Crossovers between diverged sequences are regulated by mismatch repair proteins in yeast. Mol Cell Biol. 1996;16:1085–1093. doi: 10.1128/mcb.16.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drotschmann K, Clark A B, Tran H T, Resnick M A, Gordenin D A, Kunkel T A. Mutator phenotypes of yeast strains heterozygous for mutations in the MSH2 gene. Proc Natl Acad Sci USA. 1999;96:2970–2975. doi: 10.1073/pnas.96.6.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drummond J T, Li G-M, Longley M J, Modrich P. Mismatch recognition by an hMSH2-GTBP heterodimer and differential repair defects in tumor cells. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 15.Fiorentini P, Huang K N, Tishkoff D X, Kolodner R D, Symington L S. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fishel R A, Ewel A, Lee S, Lescoe M K, Griffith J. Binding of mismatched microsatellite DNA sequences by the human MSH2 protein. Science. 1994;266:1403–1405. doi: 10.1126/science.7973733. [DOI] [PubMed] [Google Scholar]
- 17.Fishman-Lobell J, Haber J E. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 18.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C: American Society for Microbiology; 1995. pp. 233–281. [Google Scholar]
- 19.Geitz R D, Schiestl R H. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast. 1991;7:253–263. doi: 10.1002/yea.320070307. [DOI] [PubMed] [Google Scholar]
- 20.Genschel J, Littman S J, Drummond J T, Modrich P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 21.Grilley M, Welsh K M, Su S-S, Modrich P. Isolation and characterization of the Escherichia coli mutL gene product. J Biol Chem. 1989;264:1000–1004. [PubMed] [Google Scholar]
- 22.Gu L, Hong Y, McCulloch S, Watanabe H, Li G M. ATP-dependent interaction of human mismatch repair proteins and dual role of PCNA in mismatch repair. Nucleic Acids Res. 1998;26:1173–1178. doi: 10.1093/nar/26.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerrette S, Wilson T, Gradia S, Fishel R. Interactions of human hMSH2 with hMSH3 and hMSH2 with hMSH6 examination of mutations found in hereditary nonpolyposis colorectal cancer. Mol Cell Biol. 1998;18:6616–6623. doi: 10.1128/mcb.18.11.6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haber L T, Walker G C. Altering the conserved nucleotide binding motif in the Salmonella typhimurium MutS mismatch repair protein affects both its ATPase and mismatch binding activities. EMBO J. 1991;10:2707–2715. doi: 10.1002/j.1460-2075.1991.tb07815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Habraken Y, Sung P, Prakash L, Prakash S. Binding of insertion/deletion DNA mismatches by the heterodimer of yeast mismatch repair proteins MSH2 and MSH3. Curr Biol. 1996;6:1185–1187. doi: 10.1016/s0960-9822(02)70686-6. [DOI] [PubMed] [Google Scholar]
- 26.Habraken Y, Sung P, Prakash L, Prakash S. Enhancement of MSH2-MSH3-mediated mismatch recognition by the yeast MLH1-PMS1 complex. Curr Biol. 1997;7:790–793. doi: 10.1016/s0960-9822(06)00337-x. [DOI] [PubMed] [Google Scholar]
- 27.Habraken Y, Sung P, Prakash L, Prakash S. ATP-dependent assembly of a ternary complex consisting of a DNA mismatch and the yeast MSH2-MSH6 and MLH1-PMS1 protein complexes. J Biol Chem. 1998;273:9837–9841. doi: 10.1074/jbc.273.16.9837. [DOI] [PubMed] [Google Scholar]
- 28.Henderson S T, Petes T D. Instability of simple sequence DNA in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2749–2757. doi: 10.1128/mcb.12.6.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herskowitz I. Functional inactivation of genes by dominant negative mutations. Nature. 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- 29a.HNPCC Mutation Database. HNPCC mutation sequences. [Online.] http://www.nfdht.nl/database/msh2.htm. [10 April 1999, last date accessed.]
- 30.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 31.Hollingsworth N M, Ponte L, Halsey C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 1995;9:1728–1739. doi: 10.1101/gad.9.14.1728. [DOI] [PubMed] [Google Scholar]
- 32.Holm C, Meeks-Wagner D W, Fangman W L, Botstein D. A rapid, efficient method for isolating DNA from yeast. Gene. 1986;42:169–173. doi: 10.1016/0378-1119(86)90293-3. [DOI] [PubMed] [Google Scholar]
- 32a.Human Genome Mutation Database Cardiff. HNPCC mutation sequences. [Online.] http://www.uwcm.ac.uk/uwcm/mg/ns/1/203983.html. [10 April 1999, last date accessed.]
- 33.Hunter N, Chambers S R, Louis E J, Borts R H. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 1996;15:1726–1733. [PMC free article] [PubMed] [Google Scholar]
- 34.Iaccarino I, Marra G, Palombo F, Jiricny J. hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSα. EMBO J. 1998;17:2677–2686. doi: 10.1093/emboj/17.9.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ivanov E L, Haber J E. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:2245–2251. doi: 10.1128/mcb.15.4.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson R E, Kovvali G K, Prakash L, Prakash S. Requirement of the yeast MSH3 and MSH6 genes for MSH2 dependent genomic stability. J Biol Chem. 1996;271:7285–7288. doi: 10.1074/jbc.271.13.7285. [DOI] [PubMed] [Google Scholar]
- 37.Kirkpatrick D T, Petes T D. Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature. 1997;387:929–931. doi: 10.1038/43225. [DOI] [PubMed] [Google Scholar]
- 38.Knudson A G. Hereditary cancer, oncogene and anti-oncogene. Cancer Res. 1985;45:1437–1443. [PubMed] [Google Scholar]
- 39.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 40.Lynch H T, Smyrk T. An update on Lynch syndrome. Curr Opin Oncol. 1998;10:349–356. doi: 10.1097/00001622-199807000-00012. [DOI] [PubMed] [Google Scholar]
- 41.Malkov V A, Biswas I, Camerini-Otero R D, Hsieh P. Photocross-linking of the NH2-terminal region of Taq MutS protein to the major groove of a heteroduplex DNA. J Biol Chem. 1997;272:23811–23817. doi: 10.1074/jbc.272.38.23811. [DOI] [PubMed] [Google Scholar]
- 42.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 43.Marsischky G T, Filosi N, Kane M F, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 44.Marsischky G T, Lee S, Griffith J, Kolodner R D. Saccharomyces cerevisiae MSH2/6 complex interacts with Holliday junctions and facilitates their cleavage by phage resolution enzymes. J Biol Chem. 1999;274:7200–7206. doi: 10.1074/jbc.274.11.7200. [DOI] [PubMed] [Google Scholar]
- 45.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 46.Modrich P. Strand-specific mismatch repair in mammalian cells. J Biol Chem. 1997;272:24727–24730. doi: 10.1074/jbc.272.40.24727. [DOI] [PubMed] [Google Scholar]
- 47.Modrich P, Lahue R S. Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 48.New L, Liu K, Crouse G F. The yeast gene MSH3 defines a new class of eukaryotic MutS homologues. Mol Gen Genet. 1993;239:97–108. doi: 10.1007/BF00281607. [DOI] [PubMed] [Google Scholar]
- 49.Nichols A F, Sancar A. Purification of PCNA as a nucleotide excision-repair protein. Nucleic Acids Res. 1992;20:3559–3564. doi: 10.1093/nar/20.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicolaides N C, Littman S, Modrich P, Kinzler K W, Vogelstein B. A naturally occurring hPMS2 mutation can confer a dominant negative phenotype. Mol Cell Biol. 1998;18:1635–1641. doi: 10.1128/mcb.18.3.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D’Arrigo A, Truong O, Hsuan J J, Jiricny J. GTBP, a 160 kD protein essential for mismatch binding activity in human cells. Science. 1995;268:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 52.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSβ, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 53.Pâques F, Haber J E. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53a.Protein Sequence Database. Protein Identification Resource entry I37550. [Online.] http://www.nbrf.georgetown.edu/pir. [10 April 1999, last date accessed.]
- 54.Qiu J, Qian Y, Chen V, Guan M-X, Shen B. Human exonuclease 1 functionally complements its yeast homologues in DNA recombination, RNA primer removal, and mutation avoidance. J Biol Chem. 1999;274:17893–17900. doi: 10.1074/jbc.274.25.17893. [DOI] [PubMed] [Google Scholar]
- 55.Reenan R A G, Kolodner R D. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics. 1992;132:975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rose M, Grisafi P, Botstein D. Structure and function of the yeast URA3 gene: expression in Escherichia coli. Gene. 1984;29:113–124. doi: 10.1016/0378-1119(84)90172-0. [DOI] [PubMed] [Google Scholar]
- 57.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- 58.Ross-Macdonald P, Roeder G S. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell. 1994;79:1069–1080. doi: 10.1016/0092-8674(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 59.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 60.Saparbaev M, Prakash L, Prakash S. Requirement of mismatch repair genes MSH2 and MSH3 in the RAD1-RAD10 pathway of mitotic recombination in Saccharomyces cerevisiae. Genetics. 1996;142:727–736. doi: 10.1093/genetics/142.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Selva E M, New L, Crouse G F, Lahue R S. Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:1175–1188. doi: 10.1093/genetics/139.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shimodaira H, Filosi N, Shibata H, Suzuki T, Radice P, Kanamaru R, Friend S H, Kolodner R D, Ishioka C. Functional analysis of human MLH1 mutations in Saccharomyces cerevisiae. Nat Genet. 1998;19:384–389. doi: 10.1038/1277. [DOI] [PubMed] [Google Scholar]
- 63.Strand M, Prolla T A, Liskay R M, Petes T D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 64.Studamire B, Quach T, Alani E. The Saccharomyces cerevisiae Msh2p and Msh6p ATPase activities are both required during mismatch repair. Mol Cell Biol. 1998;18:7590–7601. doi: 10.1128/mcb.18.12.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sugawara N, Haber J E. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sugawara N, Pâques F, Colaiácovo M, Haber J E. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc Natl Acad Sci USA. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tishkoff D X, Boerger A L, Bertrand P, Filosi N, Gaida G M, Kane M F, Kolodner R D. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci USA. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Umar A, Buermeyer A B, Simon J A, Thomas D C, Clark A B, Liskay R M, Kunkel T A. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 69.Winston F, Dollard C, Ricupero-Hovasse S L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 70.Wu T, Marinus M G. Deletion mutation analysis of the mutS gene in Escherichia coli. J Biol Chem. 1999;274:5948–5942. doi: 10.1074/jbc.274.9.5948. [DOI] [PubMed] [Google Scholar]
- 71.Wu T-H, Marinus M G. Dominant negative mutator mutations in the mutS gene of Escherichia coli. J Bacteriol. 1994;176:5393–5400. doi: 10.1128/jb.176.17.5393-5400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]