ABSTRACT
Microbial DNA, shed from human skin, can be distinctive to its host and, thus, help individualize donors of forensic biological evidence. Previous studies have utilized single-locus microbial DNA markers (e.g., 16S rRNA) to assess the presence/absence of personal microbiota to profile human hosts. However, since the taxonomic composition of the microbiome is in constant fluctuation, this approach may not be sufficiently robust for human identification (HID). Multimarker approaches may be more powerful. Additionally, genetic differentiation, rather than taxonomic distinction, may be more individualizing. To this end, the nondominant hands of 51 individuals were sampled in triplicate (n = 153). They were analyzed for markers in the hidSkinPlex, a multiplex panel comprising candidate markers for skin microbiome profiling. Single-nucleotide polymorphisms (SNPs) with the highest Wright’s fixation index (FST) estimates were then selected for predicting donor identity using a support vector machine (SVM) learning model. FST is an estimate of the genetic differences within and between populations. Three different SNP selection criteria were employed: SNPs with the highest-ranking FST estimates (i) common between any two samples regardless of markers present (termed overall); (ii) each marker common between samples (termed per marker); and (iii) common to all samples used to train the SVM algorithm for HID (termed selected). The SNPs chosen based on criteria for overall, per marker, and selected methods resulted in an accuracy of 92.00%, 94.77%, and 88.00%, respectively. The results support that estimates of FST, combined with SVM, can notably improve forensic HID via skin microbiome profiling.
IMPORTANCE There is a need for additional genetic information to help identify the source of biological evidence found at a crime scene. The human skin microbiome is a potentially abundant source of DNA that can enable the identification of a donor of biological evidence. With microbial profiling for human identification, there will be an additional source of DNA to identify individuals as well as to exclude individuals wrongly associated with biological evidence, thereby improving the utility of forensic DNA profiling to support criminal investigations.
KEYWORDS: hidSkinPlex, skin microbiome, microbial forensics, human identification, massive parallel sequencing
INTRODUCTION
Determining the source of DNA evidence from a crime scene is the primary goal of forensic genetics. Identifying the molecular profile of a donor typically involves comparing short tandem repeat (STR) markers from an unknown sample(s) with a reference sample from a person(s) of interest. STRs are highly polymorphic, thereby providing high powers of discrimination. However, forensic genetic evidence can often be degraded and contain small amounts of human DNA, making it difficult to obtain even a partial STR profile for human identification (HID). When an incomplete (or partial) STR profile is obtained, the discrimination power is reduced substantially. In such cases, there is a need to consider alternative approaches to assist in criminal investigations.
The human microbiome provides a promising alternative source of DNA that could supplement forensic human DNA analyses. The number of microbes in and on the human body is estimated to be 1:1 compared to human cells, but when only human nucleated cells are considered, the ratio is estimated to be 10:1 (1, 2). The ratio may be higher for skin swab samples. Indeed, the skin microbiome is an abundant source of microbes, with an estimated ∼10,000 bacteria/cm2 (3). In contrast, human nuclear DNA (nDNA) is far less abundant on a per-copy basis. For example, Schmedes et al. (4), swabbing a similar area of the skin, obtained a quantity of human DNA equivalent to four diploid cells. In contrast, the DNA of the human skin microbiome from the same extract provided sufficient information for identifying the donor of the sample (4).
The 16S rRNA marker has traditionally been used in the context of human microbiome profiling. The human skin microbiome has been characterized for multiple individuals and multiple body sites using 16S rRNA sequencing, demonstrating that the human skin microbiome is a potential source of trace evidence (5–14), but there still is a need for improvement. These studies have focused on the taxonomic diversity of specific microbial species to determine the relationship between an unknown sample and its potential donor. However, previous studies have had various success rates for HID and were typically based on a small number of samples (i.e., <15 individuals) (15–18). The limited success of these investigations could be attributed to their reliance on the presence/absence (or quantitation) of specific microbes as evidence for a “match” between an unknown sample and a reference sample. Environmental interactions and temporal shifts are common phenomena in microbiomes (19). Specifically, microbes from the skin can also be shared and exchanged between cohabiting and noncohabitating individuals when they come in contact with each other or with items (9, 20–22). Moreover, several studies have also claimed that 16S rRNA lacks the necessary phylogenetic resolution for HID (6, 9, 16, 23–27). All of the above suggests that the taxonomic and phylogenetic constitution of the microbiome is in constant fluctuation. Using the presence/absence of specific microbial taxa as evidence of a match could be limiting or possibly misleading.
However, a better system possibly consists of identifying discriminatory skin microbial features in which stability decays minimally over time. Consequently, recent work has focused on targeting a number of stable taxon-specific markers to improve the accuracy of HID (4, 28, 29). Oh et al. (28) completed one of the first whole-genome sequence studies of the human skin microbiome for multiple body sites, providing detailed information about abundant and stable microorganisms. The hidSkinPlex (4), for example, is a multiplex panel based on the data of Oh et al. (28) and includes 286 markers, ranging from the level of the genus to subspecies of 22 different microbial clades. The markers were selected based on their abundance and temporal stability (up to 3 years) as well as their prevalence across body sites (4, 28). Using specific stable markers with a wide phylogenetic range allows for the selection of specific features from the skin microbiome that may improve HID. For example, the markers chosen by Schmedes et al. (4) were able to achieve accuracies with a range of 54.00% to 100.00% using presence/absence and nucleotide diversity with two machine learning methods, albeit with a limited sample size.
A promising approach to identify human hosts could be to use measures of genetic differentiation, specifically the F-statistics (for example, the fixation index, or FST) (30) for assessing microbial populations. Ancestry informative markers (AIM) regularly used in human bioancestry studies commonly have high FST (31, 32). A few high-FST markers are first mined from genomes and then used to predict population groups. FST can be estimated by evaluating orthologous SNPs in two different skin microbiome populations (i.e., skin microbiome samples from different individuals). FST estimates could provide insight into whether the alleles of a marker observed between microbial populations are identical by descent, allowing for better discrimination between microbial populations, which in turn may improve the accuracy of associating a skin microbiome sample with its respective human host.
Previously, Woerner et al. (29) estimated FST values between two sample populations: a sample incorrectly associated with another host. Their work showed that even though the central value (i.e., mean) FST would also lead to an incorrect classification, the use of high FST SNPs would lead to the correct classification. However, the Woerner et al. study was only a proof of concept because only two samples were analyzed, and classification of the hosts based on the FST estimations was not performed. In the current study, a novel approach to accurately associate skin microbiota with their respective hosts is described. The nondominant hands of 51 individuals were sampled in triplicate, and the DNA was analyzed using the hidSkinPlex panel. FST estimates were then computed using SNPs across the sequenced markers to assess genetic differentiation between inter- and intra-individual microbiome populations. A select number of SNPs displaying the highest FST estimates were chosen, applying three different approaches: those with the highest-ranking FST estimates (i) common between any two samples regardless of taxonomy (termed overall); (ii) per common marker between samples (forcing a more uniform distribution on taxonomy, termed per marker); and (iii) markers common to all samples that are used to train the subsequent machine learning algorithm (termed selected). Each approach focused on a specific hypothesis to determine if using the overall highest-ranking SNPs, maximizing taxa, or a common selected panel could increase the classification accuracy of unknown skin microbiome samples. These SNPs were used as data points for classification by a support vector machine (SVM) learning approach. The predictive capabilities of the SVM to match samples to their human hosts were compared across all three methods of SNP selection.
RESULTS
FST estimations for skin microbiome samples.
As previously described in Woerner et al. (29), 51 individual's nondominant hands were sampled in triplicate and analyzed for the markers in the hidSkinPlex panel. The samples were split into training (n = 26 individuals in triplicate) and test data (n = 25 individuals in triplicate) sets. A total of ∼69 million quality-controlled reads with a mean of 893,355 (standard deviation [SD] = 362,436) per sample remained after read preprocessing for the training set. The test data set had ∼72 million mapped reads with a mean of 964,161 (SD = 418,058) mapped reads per sample. FST was estimated over all pairs of individuals for every orthologous nucleotide in the hidSkinPlex within the training and test data sets.
After estimating FST for all pairwise comparisons with at least 1× read coverage, the mean number of nucleotides with an FST estimate greater than zero for each pairwise comparison in the training data set was 24,809 (SD = 8,502; 2,590 minimum to 52,459 maximum) (see Table S1 in the supplemental material). The test data set had a mean of 22,789 (SD = 9,657) for single-nucleotide positions with an FST estimate greater than zero. Typically, 1× read coverage is not sufficient to call SNPs. However, it was initially used here to identify potential variants and their nucleotide positions. Subsequently, various read depths were tested in the optimization of the machine learning approaches. When analyzing FST estimates for all pairs, 236 markers of the 286 markers in the hidSkinPlex were seen in at least two samples being compared from the data set. As a reminder, each marker in the hidSkinPlex is associated with some level of microbial taxonomy (e.g., stably present in Cutibacterium acnes at the species level). The reduced number of markers was only from eight species and one family (Table 1). Corynebacterium pseudogenitalium was only seen in one comparison of two samples, from the training data set, with both samples collected from the same individual.
TABLE 1.
Number of markers present for each species seen in the data analyzed by the hidSkinPlex
| Family | Genus | Species or phage | No. of markers |
|---|---|---|---|
| Propionibacteriaceae | — a | —a | 3 |
| Cutibacterium | acnes | 197 | |
| Cutibacterium | humerusii | 23 | |
| Cutibacterium | namnetense | 4 | |
| Cutibacterium | granulosum | 4 | |
| Corynebacteriaceae | Corynebacterium | tuberculostearicum | 4 |
| Corynebacterium | pseudogenitalium | 2 | |
| Micrococcaceae | Rothia | mucilaginosa | 1 |
| Siphoviridae | Pahexavirus | Propionibacterium phage P1. 1 | 1 |
—, genus/species not determined.
SVM analysis of training data set.
SVMs are natural binary classifiers, and, for the purposes of this study, each person is considered a separate class. SVMs can be extended to multiclass classification by using one-versus-one (OvO) decomposition, wherein a classifier is built for each pair of classes (individuals). OvO classifiers were created using SNPs, selected based on high-ranking FST estimates, specific to the pair of individuals. The multiclass classification was estimated by using a simple tally of votes (see Materials and Methods). Parameter optimization included varying the number of SNPs and the minimum number of reads and the SVM cost (C), and the best combination of parameters was identified for each SNP selection method. The best combination was selected from the training data based on classification accuracy with a tie-breaking rule using the mean prediction accuracy.
The three methods of selecting SNPs with the highest-ranking FST estimates were termed overall, per marker, and selected. While all three methods focused on the SNPs with the highest-ranking FST estimations, each method varied on the number of markers and SNPs used to classify an unknown sample. The variation in the three methods was developed to answer distinct hypotheses about how SNP selection methods affect HID and to determine which method had the highest accuracy, as assessed in the test data set. The overall method tested whether accuracies can be increased by selecting the highest-ranking SNPs, regardless of the markers present. The overall method selected SNPs with the highest-ranking FST estimates in each pair of samples (although it could lead to less diverse distribution of taxa). The per marker method tested whether maximizing the number of taxa used for classification could increase classification accuracy, even if doing so relied on SNPs with lower FST estimates. The per marker method selected the SNPs with the highest-ranking FST in each orthologous marker in a pair of samples. The selected method tested whether using SNPs that were common to all samples in the training data, used to train the SVM, could be used to increase accuracy of identification. The selected method relied on a predetermined number of common SNPs, which had high-ranking FST estimations for all comparisons in the training data set. Each selection method was then compared under different parameter values (i.e., the number of SNPs, minimum sequence reads, and SVM cost) using a customized SVM approach designed specifically for HID (see Materials and Methods).
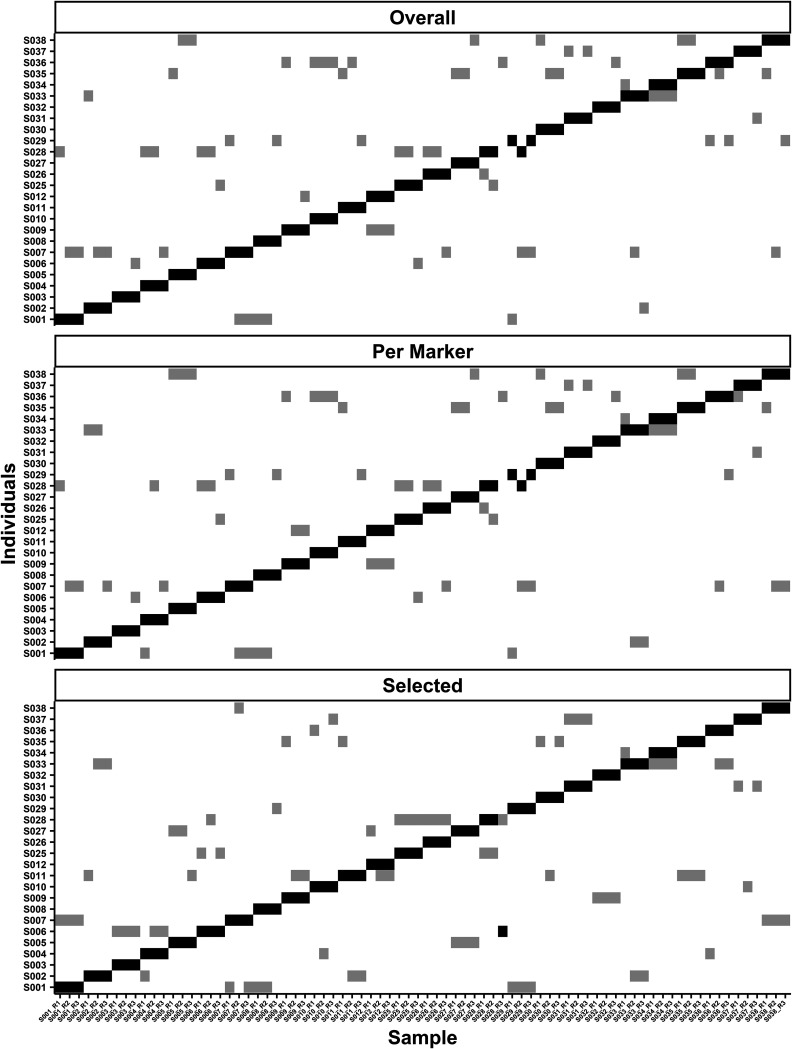
(i) Overall method. The overall FST selection focused on choosing the highest FST SNPs for each pairwise comparison (i.e., 500, 1,000, or 2,000; note with this method that some of the highest-ranking SNPs had FST estimates close to zero). The number of selected high-ranking SNPs was tested with all possible combinations of minimum reads and SVM cost (i.e., the C hyperparameter). The data training set compared the accuracy of 75 parameter combinations, and 12 combinations performed best, classifying 76 out of 78 samples correctly, yielding a 97.44% accuracy (Table 2). Using the highest prediction probability to break the tie of the 12 options, the 500 SNPs with the highest FST estimations, minimum read depth of 250, and SVM cost of 1 were the optimal parameters. The two incorrectly classified samples were S028_R3 and S029_R2 (Fig. 1), which had some of the lowest numbers of markers (mean ± SD) (S028_R3, 120.80 ± 0.47; S029_R2, 152.00 ± 11.91) and SNPs (S028_R3, 823.50 ± 131.28; S029_R2, 1,059.00 ± 144.45) for analysis among the training set samples. The mean number of markers for the overall method with the training data set was 146.20 (SD = 15.72), and the mean number of SNPs was 1,036.00 (SD = 160.25). The mean number of taxa seen was only 3.89 (SD = 0.86).
TABLE 2.
Number of SNPs, read minimum, and cost for SVM modeling with the highest accuracy for all three-nucleotide selection methods optimized with the training dataa
| No. of SNPs | Read minimum | Cost | Mean confidence prediction |
|---|---|---|---|
| Overall | |||
| 500 | 250 | 1 | 0.7651584 |
| 500 | 250 | 10 | 0.7651583 |
| 500 | 250 | 1,000 | 0.7651581 |
| 500 | 250 | 100 | 0.7651579 |
| 1000 | 250 | 100 | 0.7643158 |
| 1000 | 250 | 10 | 0.7643155 |
| 1000 | 250 | 1,000 | 0.7643153 |
| 1000 | 250 | 1 | 0.7643141 |
| 2000 | 250 | 10 | 0.7642741 |
| 2000 | 250 | 1 | 0.7642740 |
| 2000 | 250 | 100 | 0.7642738 |
| 2000 | 250 | 1,000 | 0.7642734 |
| Per marker | |||
| 5 | 250 | 10 | 0.7660739 |
| 5 | 250 | 1,000 | 0.7660738 |
| 5 | 250 | 100 | 0.7660735 |
| 10 | 250 | 1,000 | 0.7651763 |
| 10 | 250 | 100 | 0.7651763 |
| 10 | 250 | 1 | 0.7651759 |
| 10 | 250 | 10 | 0.7651747 |
| 25 | 250 | 100 | 0.7644139 |
| 25 | 250 | 10 | 0.7644138 |
| 25 | 250 | 1 | 0.7644134 |
| 25 | 250 | 1,000 | 0.7644130 |
| Selected | |||
| 150 | 500 | 1,000 | 0.7626328 |
| 150 | 1,000 | 1,000 | 0.7626328 |
| 150 | 250 | 1,000 | 0.7626327 |
| 150 | 100 | 1,000 | 0.7626327 |
| 150 | 250 | 100 | 0.7626327 |
| 150 | 10 | 1,000 | 0.7626326 |
| 150 | 500 | 100 | 0.7626326 |
| 150 | 10 | 100 | 0.7626325 |
| 150 | 100 | 100 | 0.7626325 |
| 150 | 1,000 | 100 | 0.7626323 |
Parameters are in descending order by the mean confidence prediction produced by the SVM model. The overall selection method analyzed 75 different parameter combinations, and 12 parameters had a 97.40% accuracy. The per marker method analyzed 75 different parameter combinations, and 11 parameters had an accuracy of 97.40%. The selected method analyzed 175 total parameter combinations, and 10 parameters had an accuracy of 98.70%. The column headings indicate the optimized parameters used on the test data set for each nucleotide position selection method.
FIG 1.
Training data set matrices showing rank numbers 1 (black) and 2 (gray) for classification. The three matrices are labeled with the nucleotide selection method (i.e., per marker, overall, or selected) used at the top of the individual graphs. The three selection methods chose SNPs with the highest-ranking FST estimates. The overall method optimized 250 SNPs for the pairwise comparison, per marker method optimized 5 SNPs per marker, and selected had a set of 150 SNPs that were common in the training data set. The x axis lists all samples with the individual number and replicates (S0## = individual number, R# = replicate number). The y axis lists the possible groups, i.e., individuals, a sample could be classified.
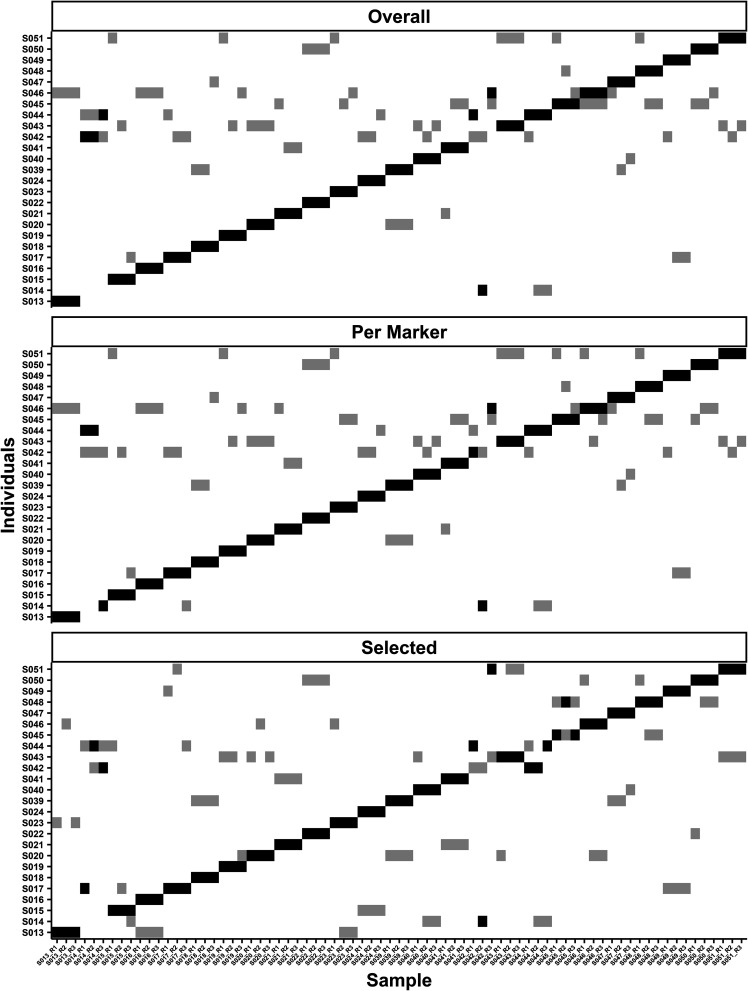
Applying the optimized parameters to the test data set (n = 25 samples in triplicate) yielded a classification accuracy of 92.00% (69/75) with classification error of the model likely between 2.99% and 16.60% with 95% confidence (R package exactci [33]) (Fig. 2). The test data set for the overall method assayed a larger number of markers (152.10 ± 14.16) but had fewer SNPs (1,026.00 ± 166.89) on mean compared to the training set. While six samples were incorrectly classified, four of the incorrect classifications involved S014 and S042. The other two incorrectly classified samples S044 and S046 ranked as number 1 (Fig. 2).
FIG 2.
Test data set matrices showing rank numbers 1 (black) and 2 (gray) for classification of samples for the three methods of selecting the highest-ranking SNPs based on their FST estimation. The top matrix is the overall method, which chose the 250 highest SNPs in any given pairwise comparison. The second matrix shows the per marker method using training set optimized parameters of 5 SNPs per marker in a pairwise comparison. The bottom matrix shows the selected method that had 150 prechosen SNPs that were common and had the highest-ranking FST estimates in the training data set.
(ii) Per marker method. The per marker method focused on the highest-ranking FST estimates within each marker to achieve the largest taxonomic diversity possible. With the per marker approach, up to a specified number of SNPs with the highest-ranking FST estimates (i.e., 5, 10, or 25) were selected per orthologous marker in a pair of samples. The per marker approach allowed for the widest variety of taxa (5.11 ± 1.15) and the largest number of markers (151.40 ± 27.98) to be used for classification with the training data. There were a total of 75 parameter combinations, and 11 parameter combinations provided the same highest prediction accuracy (Table 2). Using 5 SNPs per marker with a minimum read depth threshold of 250 and an SVM cost of 10 yielded the highest accuracy and the highest mean confidence prediction. Each SVM analysis for the optimized parameters for the per marker method had a mean of 1,650.00 SNPs per SVM classification (SD = 309.15). The per marker training set generated a 97.44% accuracy with only two misclassifications out of 78 samples. S028_R3 and S029_R2 were also incorrectly classified samples with the overall method (Fig. 1).
Using the optimized parameters, five SNPs with the highest-ranking FST estimates per marker, 250 read minimum, and an SVM cost of 10, the test data set produced an accuracy of 94.70%, with classification error between 1.47% to 13.11% (binomial 95% confidence interval). Four samples were classified incorrectly out of 75 (Fig. 2). All four comparisons were from two samples, S014_R1/R2 and S042_R2/R3. One sample in the test data set, S014_R3, had a three-way tie, based on votes, with three potential candidates, S014, S042, and S044. S014 was ranked number 1 of potential candidates because it had the highest mean prediction accuracy out of the three possible choices. All three replicates for S014 had S044 ranked number 1 or 2. Additionally, S044 was classified correctly, but it had a close association with S014 and S042, with those two classes ranked numbers 2 and 3 for S044. Having the same classes ranked highly for S014, S042, and S044 is of particular interest because S042_R2 was classified as S014, indicating that they could have arisen from the same host, a potential sample mix-up, or close relationship of the hosts.
(iii) Selected method. The selected method used predetermined SNPs for analysis (i.e., 50 to 2,000). The number of SNPs was selected based on the number of the markers in the hidSkinPlex and their base pair length. The different number of SNPs chosen for the selected method was determined based on the maximum number of SNPs (∼2,000) used in the previous two methods that had the highest classification accuracy. Out of 175 parameter combinations, 10 combinations yielded the highest accuracy of 98.70%. The optimized parameters for selected FST were 150 high FST SNPs, a minimum of 500 reads, and an SVM cost of 1,000 (Table 2). The 150 common SNPs represented 22 markers, one family and two species (Propionibacteriaceae, Cutibacterium acnes, and Cutibacterium humerusii) from the hidSkinPlex. The selected method had a training accuracy of 98.70%, with only one sample incorrectly classified. The incorrectly classified sample, S028_R3, was also incorrectly identified with the other two selection methods. The difference in the selected method was that S028_R3 ranked number 2 based on its votes, and S006 was ranked number 1 by votes (Fig. 1) and was a notable change in the rank of the correct group classification for S028_R3, which changed from rank 10 (in per marker and overall methods) to rank 2. In the training data set, S028 R3 had a mean of 43.94 markers (SD = 0.42) for all possible pairs.
When the test data set was evaluated with the parameters of 150 SNPs with the highest-ranking FST estimates from the training data set, 500 minimum reads, and a cost of 1,000 for the selected FST method, the accuracy decreased to 88.00% with a classification error of 5.63% to 21.56%. Only 66 out of 75 samples were correctly classified. Of the 11 incorrectly classified samples, three belonged to S014, three to S042, two to S017, two to S044, and one to S045 (Fig. 2). Individuals S042 and S045 did not have as many SNPs in their replicates, 146.90 ± 4.33 and 140.10 ± 1.85 across all replicates, respectively, compared to other individuals, which may have impacted classification. However, missing data alone cannot explain the decreased accuracy with the selected method, as other replicate sample pairs did not contain all 150 specified SNPs and were classified correctly.
For the training data set, the difference in methods and parameters only resulted in 1.26% (or one sample) difference in classification accuracy, but the goal of the training data set is to find the parameter combinations that result in the highest accuracy. Testing the optimized parameters on the test data set provides a better indication of how the method and optimized parameters perform on unknown data. The classification accuracy results of the three methods ranged from 88.00% to 94.00%, but accuracy rates are not significantly different (McNemar's chi-square test) (Table 3). All three methods had issues determining the correct classification for samples from S014 and S042, but the selected method also had difficulty correctly associating S017, S044, and S045. While the selected method performed better than the other methods on the training data, it was the method predicted to most likely be overfit due to SNPs being chosen based on their presence in the training data set.
TABLE 3.
Results for McNemar’s chi-square test for accuracies from the test data seta
| Comparison | Chi-squared | P value |
|---|---|---|
| Overall vs selected | 1.33 | 0.25 |
| Per marker vs overall | 0.5 | 0.48 |
| Per marker vs selected | 3.2 | 0.07 |
The SNP selection method accuracies were compared for the test data set, and no significant differences were observed between the accuracies of the methods.
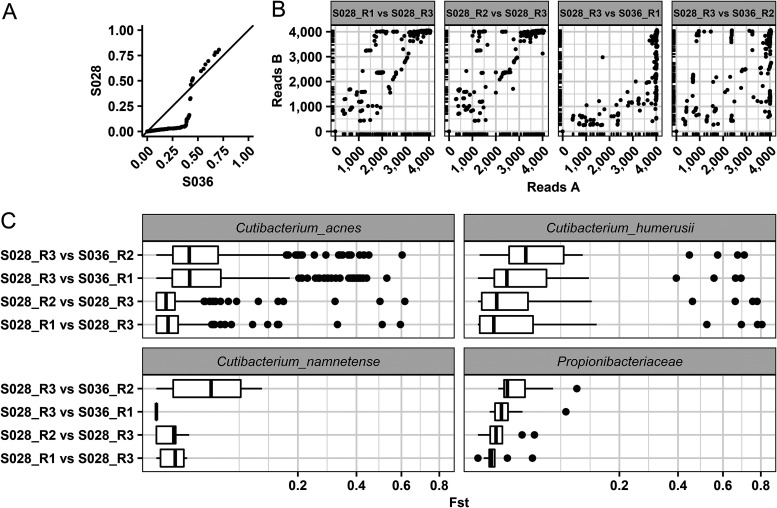
(iv) Study of misclassified sample. In this study, a misclassification was considered any unknown sample that was assigned to an incorrect individual. In essence, this error assumes the analysis achieves uniqueness, which may not be realistic with these data. Thus, a misclassification may not be a true error. More studies with refined markers/SNPs and larger sample sizes are needed to determine the host resolution of the system. Plausible explanations were sought as to why one sample was consistently misclassified in the training data set before the optimized parameters were used with the test data set. In the overall and selected methods, S028_R3 was classified as S036 (full rankings are in Table S2). S028_R3, which only had a total of 16 votes (per marker and overall methods) out of the potential 25 votes, ranking it number 10 on the list of potential donors, was the only sample in the training data set that did not have the actual contributor ranked in the top three potential candidates. For the selected method, S028_R3 was ranked 2 and had 24 votes, while S006 was ranked 1. Compared to S036, S028_R3 had much lower read coverage for the markers that are orthologous between samples. S028_R3 had the fewest markers in common when estimating FST compared to any other sample that was analyzed. The reduced amount of data available for classification may be associated with individual S028_R3 having low read depth coverage or no reads for the SNPs of interest (Fig. 3B).
FIG 3.
Comparisons of S028_R3 that were incorrectly classified as S036. (A) A quantile-quantile plot of FST estimates for sample S028_R3 compared to individual S036. The distribution of FST estimates between S028 (y axis) and S036 (x axis) and from comparing S028_R3 to other technical replicates. The FST estimates were computed for SNPs that were orthologous in at least two samples. The main diagonal represents S028 and S036, having equal values of FST estimates. Points below the main diagonal represent a greater differentiation between S036 and S028, while points above the diagonal show greater differentiation within S028. (B) First sample in the graph labeled on the x axis and the second sample on the y axis with the number of reads plotted for the SNPs. The ticks on the x and y axis show the density of the corresponding area on the graph to provide clarity about the density of plotted points. Overall, S028_R3 had less read coverage for SNPs in common with S036 than with S028. (C) A boxplot of the FST estimates for each pairwise comparison. The distribution of FST estimates for the 36 markers S036 and S028 had in common tend to have higher FST for sample comparisons within S028 than between S036.
The test data also had samples that were incorrectly classified by all three methods. Specifically, the samples from S014 and S042 were often classified as S044, S046, or each other. While some of the highest-ranking FST estimates are higher between S014 and S042, overall, there were more SNPs with high FST estimates within the individual than between individuals. For all replicates of S014, there did not tend to be any notable differences in the reads between selected SNPs. For S042_R2 and R3, the incorrect classifications may be due to S042_R3 having low read coverage for selected SNPs.
DISCUSSION
This study investigated the potential of selecting high FST markers to improve HID using the skin microbiome. Previous work with the hidSkinPlex using presence/absence or nucleotide diversity with nearest neighbor or normalized logistic regression achieved accuracy rates between 54.20% and 100.00% when classifying eight individuals with samples from three body sites collected in triplicate (4). Woerner et al. (29) expanded the number of individuals to 51 and sampled from the nondominant hand in triplicate; using the same panel they achieved accuracies of 78.00% and 83.70% using phylogenetic distance or nucleotide diversity, respectively, for classification with nearest neighbor machine learning approaches. There was a decrease in classification accuracy classification when the sample size was increased to 51 individuals compared to the eight samples in Schmedes et al. (4). The study here reanalyzed the same sequence data as those in Woerner et al. (29) with a novel method for SNP selection based on FST estimates and SVM and achieved higher accuracies (P = 0.03, chi-squared test, comparing the most accurate approaches in both studies). The accuracies of the three FST SNP selection methods also have increased accuracies compared to any of the previous studies using the targeted hidSkinPlex sequence data.
Three methods for selecting the highest-ranking FST estimations were used to assess how SNPs from the skin microbiome may be chosen for HID. All three methods of selecting informative SNPs had high classification accuracies. The per marker method achieved the highest accuracy (94.70%), which indicates that inclusion of more taxa could increase classification accuracies. The per marker method allowed for the broadest selection of markers and SNPs in common in each single pairwise comparison, resulting in the method's higher accuracy. The overall method performed well, with a 92.00% accuracy, even though the number of SNPs used for analysis was less than the per marker method. While the selected method had the lowest accuracy of 88.00%, even though it initially had the highest training accuracy at 98.70%, the method still showed that a predetermined panel of chosen SNPs could include or exclude a particular individual as the donor of a sample. An additional increase in accuracy might be achieved if minimum requirements were implemented to remove samples that have low read coverage causing a drop of informative SNPs. The results of this study provide support that using high-ranking FST estimates to select SNPs with SVM increased accuracies of classification to 94.70% and can be used in a fashion similar to that for AIM in human populations analyses.
The investigation into S028_R3 in the training data and S042_R3 in the test data set suggested that low read coverage and low diversity of a sample impact classification accuracy. If one of the three replicates from an individual has low read coverage and/or low diversity, the ability to correctly classify other replicates from the same individual may be impacted. Perhaps implementing minimum thresholds for analyzing a sample eliminates poor-quality samples from being searched. Additional research on potential minimum requirements, such as overall read coverage and depth and the number of total SNPs, may reduce the number of false positives (or, for now, better stated as adventitious hits). For the test data set, individuals S014 and S042 were incorrectly classified in all three methods of SNP selection. Individuals S014 and S042 were also incorrectly classified to some degree by Woerner et al. (29) for both classification methods tested in their study. This observation suggests that replicates of individuals S014 and S042 have been switched, contamination occurred during handling or processing, and/or these individuals share a genetically and taxonomically similar microbiome. It is also possible that the SNPs selected for distinct individuals still need refinement and/or that thresholds for minimum data requirements need to be considered further. Additionally, studies need to be performed to determine why a few high-FST SNPs could impact incorrect classification when the data as a whole support the correct classification.
Although the performance decreased with the test set, the selected method is of particular interest in that it provides a predetermined set of SNPs to be used in every classification of the unknown samples. For the optimized parameter of 150 SNPs there were only two species and one family level marker represented, which were Cutibacterium acnes, Cutibacterium humerusii, and Propionibacteriaceae. These two species and one family-level marker are common and abundant on the human skin and often have multiple subspecies or strains within individuals (28). The decrease in accuracy from 98.70% in the training data to 88.00% in the test data is most likely due to overfitting, both in the SNP ascertainment and in the SVM model itself. With more data for training, it may be possible to adjust the predetermined SNPs, but some level of overfitting will likely persist. A predetermined panel would allow for the redesign of the hidSkinPlex to reduce the number and size of the markers in the panel with a potential increase in assay robustness.
Using FST estimates permitted selection of SNPs to be input into an SVM model. With a refined MPS targeted skin microbiome panel, it will also be possible to further investigate how the SNPs of specific microorganisms change due to environment, health status, and other external factors. With a set of informative SNPs, studies can begin on determining the stability of markers over time, which is an important criterion for forensic applications. Refinement of informative SNPs may provide an increase in the accuracy to include or exclude an individual as a potential contributor of a microbiome sample when time has passed. The human skin microbiome has the potential to be supportive evidence to more traditional DNA evidence for law enforcement. In the study here, the selection criteria for SNPs may be considered ad hoc. Future work shall optimize methods using other machine learning approaches to determine informative features in the current data set and additional novel data sets.
MATERIALS AND METHODS
Samples.
Targeted sequence data from samples originally described in Woerner et al. (29) were used in this study. Briefly, skin swabs from 51 individuals were collected in triplicate from the nondominant hand (Hp) of each individual (n = 153, replicates R1, R2, and R3). These samples were then analyzed using the hidSkinPlex, a targeted genome sequencing panel (4) drawn from the MetaPhlAn2 database (34). This panel targets 22 clades, with genus to subspecies level information, comprising 286 markers that were determined to be abundant and relatively stable on human skin (35). The University of North Texas Health Science Center Institutional Review Board approved the collection and analyses of these samples.
Sequence data and analysis.
All fastq files from the MiSeq were trimmed with cutadapt (36) to remove bases with a quality score less than 20 and reads less than 50 bases long, as described in Woerner et al. (29). MetaPhlAn2 (34) was used to align sequence reads to the MetaPhlAn2 reference database. SAMtools (37) was used to calculate read depth and coverage and to generate base pileups for each aligned marker in the hidSkinPlex panel.
Computation, statistics, and FST estimation.
All statistics were performed in the R (v. 3.4.2) (38) or Python (v. 2.7.17; Python Software Foundation, https://www.python.org/) programming language with plots created by ggplot2 (39). Welch two-sample t tests and McNemar's chi-squared test were performed using the stats package (38). Hudson et al. (40) proposed estimating FST as FST = 1 − (Hw/Hb), where Hw is the mean number of pairwise differences within a population and Hb is the mean number of pairwise differences between two populations (40). FST was estimated for all relevant nucleotide positions with a read depth minimum of one. It is worth noting that FST is only defined when Hb > 0 and that a minimum read depth parameter was optimized in the machine learning approach. When estimating FST, the two samples (i.e., two populations) must each have at least one orthologous SNP being compared and have >1× read depth for the analysis (for example, sample A at SNP position 25 has 2 reads of A, and sample B has 2 reads of C). An additional read depth parameter was optimized during the analysis of the training data set. A 3-fold cross validation holding out one of the technical replicates then was performed.
Machine learning strategy.
A training set was used to optimize the linear support vector machine (SVM) C hyperparameter as well as a threshold on a maximum number of SNPs and minimum read depth. The test data set was used to determine how the SVM performed on unseen data. The training data set comprises 26 samples in triplicate (S001 to S012 and S025 to S037, where S0## represents an individual), and the test data set consists of 25 samples (S013 to S024 and S038 to S051).
The SVM approach embeds the distance (FST) between two individuals relative to a single query point into the Cartesian coordinate system. The embedding begins by considering four samples, two samples for each class (a class represents two samples from the same individual), and selecting the highest-ranking SNPs for each sample compared to the unknown sample. While embedding distances in the Cartesian coordinate system generally is not possible without error or loss, it is possible to use distance with a binary classifier when the distance is constrained to a single (query) point. A further benefit of the approach is it can be trained only on comparable data, in this case SNPs, between just the two samples and the query, in contrast to requiring the presence of each SNP in all samples. This allows the SVM to handle dropout in a way that avoids imputation and uses the variants to separate two individuals based on their common microbes.
Each comparison between two samples (one of them being the unknown data point) selected the highest-ranking FST estimates (i.e., SNPs) based on the selection method (i.e., overall, per marker, or selected). After SNP selection, a matrix with the four samples (rows) and the selected SNPs (columns) was formed. If any SNP was not present in the other (up to three) comparisons, because it was not present as a high-ranking SNP, it was filled in with the FST estimate from the original data that met the minimum read requirement. Missing data were filled in with zero. FST values for common markers for all four comparisons were input into an in-house SVM code that used LibSVM (v. 1.7-3) (41) (R package e1071) as a feature vector with two labeled classes and a single unlabeled sample. The unknown sample was then provided as a vector of zeros as an additional feature vector to represent FST estimates of the unknown sample compared to itself. The SVM provided a prediction about which of the two potential classes the unknown sample belonged and provided a percentage representing the SVM's confidence in its prediction. Each time the binary SVM made a prediction, the corresponding class was given a vote. The votes were tallied, and the maximum number of votes determined the classification of the unknown sample.
Three approaches to select SNPs for analysis were developed to determine which method would provide the highest accuracy. Each method of high FST selection focused on a distinct approach to provide insight into whether the number of highest-ranking FST estimates increases classification accuracy or a common set of markers would more effectively improve accuracy of unknown sample prediction. The first approach, overall, selected either up to 500, 1,000, or 2,000 SNPs with the highest-ranked FST estimates across all markers, but not from any specific marker, to determine the minimum number of SNPs that could be used and still provide accurate classification. The second method, FST per marker, selected either 5, 10, or 25 SNPs contained within a marker with the highest-ranking FST per marker common between the two populations that were compared. The third method, called selected FST, used all FST estimates with reads greater than 10 from the training samples to select SNPs that were seen most often in pairwise comparisons and had the highest-ranking FST estimates. The number of SNPs selected with the highest-ranking FST estimates was set at 50 to 2,000. The selected method chose SNPs by arranging FST estimates in descending order for each marker seen in all pairwise comparison in the training data set. All three selection methods were optimized under the objective of maximizing classification accuracy.
Parameter optimization.
Three parameters were varied for all SVM models. The three parameters were the number of SNPs with the highest-ranking FST estimates in a pairwise comparison, the minimum reads at each SNP compared, and the cost (C parameter) for the linear SVM. The number of SNPs selected with the highest-ranking FST estimates depended on which method was used, i.e., FST per marker, overall FST, and selected FST. A minimum read depth threshold was assessed with each approach, and the thresholds were 10, 100, 250, 500, or 1,000. Cost, the degree of misclassification allowed in the SVM, was set at 0.1, 1, 10, 100, or 1,000. The selection of optimal parameters for each FST selection method was evaluated by looking at the number of times each possible combination of all three parameters was used to predict 78 unknown samples with SVM. The accuracy was determined by the number of times that the unknown sample was predicted correctly (i.e., the highest rank).
Data availability.
Custom R and Python scripts can be accessed at https://github.com/CardiShire/PopulationInformativeMarkers.
ACKNOWLEDGMENTS
We thank Sarah Schmedes for the design of the hidSkinPlex and the initial development of sample processing. Additionally, we thank Angie Ambers, Rachel Kieser, Frank Wendt, Nicole Novroski, and Jonathan King for the countless hours they contributed to collecting/processing samples and providing feedback on the next steps for HID using the skin microbiome. Last but not least, we thank Utpal Smart, Sammed Mandape, Ben Crysup, and Jonathan King for all the time they spent advising on code and debugging support.
This study was supported in part by the National Institute of Justice, award numbers 2015-NE-BX-K006 and 2020-R2-CX-0046.
Footnotes
Supplemental material is available online only.
Contributor Information
Allison J. Sherier, Email: allisonsherier@my.unthsc.edu.
Andrew J. McBain, University of Manchester
REFERENCES
- 1.Sender R, Fuchs S, Milo R. 2016. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol 14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sender R, Fuchs S, Milo R. 2016. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 164:337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 3.Grice EA, Kong HH, Renaud G, Young AC, Program NCS, Bouffard GG, Blakesley RW, Wolfsberg TG, Turner ML, Segre JA, NISC Comparative Sequencing Program . 2008. A diversity profile of the human skin microbiota. Genome Res 18:1043–1050. doi: 10.1101/gr.075549.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmedes SE, Woerner AE, Novroski NMM, Wendt FR, King JL, Stephens KM, Budowle B. 2018. Targeted sequencing of clade-specific markers from skin microbiomes for forensic human identification. Forensic Sci Int Genet 32:50–61. doi: 10.1016/j.fsigen.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Hampton-Marcell JT, Larsen P, Anton T, Cralle L, Sangwan N, Lax S, Gottel N, Salas-Garcia M, Young C, Duncan G, Lopez JV, Gilbert JA. 2020. Detecting personal microbiota signatures at artificial crime scenes. Forensic Sci Int 313:110351. doi: 10.1016/j.forsciint.2020.110351. [DOI] [PubMed] [Google Scholar]
- 6.Lax S, Hampton-Marcell JT, Gibbons SM, Colares GB, Smith D, Eisen JA, Gilbert JA. 2015. Forensic analysis of the microbiome of phones and shoes. Microbiome 3:21. doi: 10.1186/s40168-015-0082-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lax S, Smith DP, Hampton-Marcell J, Owens SM, Handley KM, Scott NM, Gibbons SM, Larsen P, Shogan BD, Weiss S, Metcalf JL, Ursell LK, Vazquez-Baeza Y, Van Treuren W, Hasan NA, Gibson MK, Colwell R, Dantas G, Knight R, Gilbert JA. 2014. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345:1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lax S, N C, Gilbert JA. 2015. Our interface with the built environment: immunity and the indoor microbiota. Trends Immunol 36:121–123. doi: 10.1016/j.it.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richardson M, Gottel N, Gilbert JA, Lax S. 2019. Microbial similarity between students in a common dormitory environment reveals the forensic potential of individual microbial signatures. mBio 10:e01054-19. doi: 10.1128/mBio.01054-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luongo JC, Barberán A, Hacker-Cary R, Morgan EE, Miller SL, Fierer N. 2017. Microbial analyses of airborne dust collected from dormitory rooms predict the sex of occupants. Indoor Air 27:338–344. doi: 10.1111/ina.12302. [DOI] [PubMed] [Google Scholar]
- 11.Adams RI, Bateman AC, Bik HM, Meadow JF. 2015. Microbiota of the indoor environment: a meta-analysis. Microbiome 3:49. doi: 10.1186/s40168-015-0108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujiyoshi S, Tanaka D, Maruyama F. 2017. Transmission of airborne bacteria across built environments and its measurement standards: a review. Front Microbiol 8:2336. doi: 10.3389/fmicb.2017.02336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meadow JF, Altrichter AE, Green JL. 2014. Mobile phones carry the personal microbiome of their owners. PeerJ 2:e447. doi: 10.7717/peerj.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meadow JF, Altrichter AE, Kembel SW, Moriyama M, O'Connor TK, Womack AM, Brown GZ, Green JL, Bohannan BJ. 2014. Bacterial communities on classroom surfaces vary with human contact. Microbiome 2:7. doi: 10.1186/2049-2618-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fierer N, Lauber CL, Zhou N, McDonald D, Costello EK, Knight R. 2010. Forensic identification using skin bacterial communities. Proc Natl Acad Sci USA 107:6477–6481. doi: 10.1073/pnas.1000162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park J, Kim SJ, Lee J-A, Kim JW, Kim SB. 2017. Microbial forensic analysis of human-associated bacteria inhabiting hand surface. Forensic Sci Int 6:e510–e512. doi: 10.1016/j.fsigss.2017.09.210. [DOI] [Google Scholar]
- 17.Watanabe H, Nakamura I, Mizutani S, Kurokawa Y, Mori H, Kurokawa K, Yamada T. 2018. Minor taxa in human skin microbiome contribute to the personal identification. PLoS One 13:e0199947. doi: 10.1371/journal.pone.0199947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J, Tsukimi T, Yoshikawa M, Suzuki K, Takeda T, Tomita M, Fukuda S. 2019. Cutibacterium acnes (Propionibacterium acnes) 16S rRNA genotyping of microbial samples from possessions contributes to owner identification. mSystems 4:e00594-19. doi: 10.1128/mSystems.00594-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doleckova I, Capova A, Machkova L, Moravcikova S, Maresova M, Velebny V. 2020. Seasonal variations in the skin parameters of Caucasian women from Central Europe. Skin Res Technol 27:358–369. doi: 10.1111/srt.12951. [DOI] [PubMed] [Google Scholar]
- 20.Ross AA, Doxey AC, Neufeld JD. 2017. The skin microbiome of cohabiting couples. mSystems 2:e00043-17. doi: 10.1128/mSystems.00043-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, Caporaso JG, Knights D, Clemente JC, Nakielny S, Gordon JI, Fierer N, Knight R. 2013. Cohabiting family members share microbiota with one another and with their dogs. Elife 2:e00458. doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neckovic A, van Oorschot RAH, Szkuta B, Durdle A. 2020. Investigation of direct and indirect transfer of microbiomes between individuals. Forensic Sci Int Genet 45:102212. doi: 10.1016/j.fsigen.2019.102212. [DOI] [PubMed] [Google Scholar]
- 23.Bosshard PP, Zbinden R, Abels S, Boddinghaus B, Altwegg M, Bottger EC. 2006. 16S rRNA gene sequencing versus the API 20 NE system and the VITEK 2 ID-GNB card for identification of nonfermenting Gram-negative bacteria in the clinical laboratory. J Clin Microbiol 44:1359–1366. doi: 10.1128/JCM.44.4.1359-1366.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mignard S, Flandrois JP. 2006. 16S rRNA sequencing in routine bacterial identification: a 30-month experiment. J Microbiol Methods 67:574–581. doi: 10.1016/j.mimet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Fox GE, Wisotzkey JD, JurtshukP, Jr.. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol 42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 26.Lee S-Y, Woo S-K, Lee S-M, Eom Y-B. 2016. Forensic analysis using microbial community between skin bacteria and fabrics. Toxicol Environ Health Sci 8:263–270. doi: 10.1007/s13530-016-0284-y. [DOI] [Google Scholar]
- 27.Gu Y, Zha L, Yun L. 2017. Potential usefulness of SNP in the 16S rRNA gene serving as informative microbial marker for forensic attribution. Forensic Sci Int Genet 6:e451–e452. doi: 10.1016/j.fsigss.2017.09.176. [DOI] [Google Scholar]
- 28.Oh J, Byrd AL, Park M, Program NCS, Kong HH, Segre JA, NISC Comparative Sequencing Program . 2016. Temporal stability of the human skin microbiome. Cell 165:854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woerner AE, Novroski NMM, Wendt FR, Ambers A, Wiley R, Schmedes SE, Budowle B. 2019. Forensic human identification with targeted microbiome markers using nearest neighbor classification. Forensic Sci Int Genet 38:130–139. doi: 10.1016/j.fsigen.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Wright S. 1951. The genetical structure of populations. Ann Eugen 15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 31.Kidd KK, Speed WC, Pakstis AJ, Furtado MR, Fang R, Madbouly A, Maiers M, Middha M, Friedlaender FR, Kidd JR. 2014. Progress toward an efficient panel of SNPs for ancestry inference. Forensic Sci Int Genet 10:23–32. doi: 10.1016/j.fsigen.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 32.Phillips C, Parson W, Lundsberg B, Santos C, Freire-Aradas A, Torres M, Eduardoff M, Borsting C, Johansen P, Fondevila M, Morling N, Schneider P, Consortium EU-N, Carracedo A, Lareu MV, EUROFORGEN-NoE Consortium . 2014. Building a forensic ancestry panel from the ground up: the EUROFORGEN global AIM-SNP set. Forensic Sci Int Genet 11:13–25. doi: 10.1016/j.fsigen.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 33.Fay MP. 2010. Two-sided exact tests and matching confidence intervals for discrete data. J R Project 2:53–58. doi: 10.32614/RJ-2010-008. [DOI] [Google Scholar]
- 34.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. 2015. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 35.Schmedes SE, Woerner AE, Budowle B. 2017. Forensic human identification using skin microbiomes. Appl Environ Microbiol 83:e01672-17. doi: 10.1128/AEM.01672-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17:3. [Google Scholar]
- 37.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/. [Google Scholar]
- 39.Wickham H, Chang W, Henry L, Pedersen TL, Takahashi K, Wilke C, Woo K. 2016. ggplot2: elegant graphics for data analysis, vol 2018. Springer-Verlag, New York, NY. [Google Scholar]
- 40.Hudson RR, Slatkin M, Maddison WP. 1992. Estimation of levels of gene flow from DNA sequence data. Genetics 132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer DE, Dimitriadou Hornik K, Weingessel A, Leisch F. 2019. e1071: misc functions of the Department of Statistics, Probability Theory Group (formerly: E1071), TU Wien. R package version 1.7–3. https://CRAN.R-project.org/package=e1071.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 legend and Table S2. Download AEM.01208-21-s0001.pdf, PDF file, 0.01 MB (13.8KB, pdf)
Table S2. Download AEM.01208-21-s0002.csv, CSV file, 0.2 MB (250.8KB, csv)
Data Availability Statement
Custom R and Python scripts can be accessed at https://github.com/CardiShire/PopulationInformativeMarkers.