To the Editor:
Fusions of NUTM1 (15q14) define a novel subtype of B-cell precursor acute lymphoblastic leukemia (B-ALL) that is mutually exclusive with sentinel leukemia-driving aberrations based on RNA-sequencing
in children [1–5] and infants [3, 6]. NUTM1 fusions are characterized by increased expression of almost the complete open reading frame of NUTM1 fused to one of several fusion partners. Downstream effects of NUTM1 fusions include upregulation of the proto-oncogene BMI1 [3]. In addition, specific NUTM1 fusions are associated with HOXA gene cluster upregulation [1, 3]. The incidence of NUTM1-rearranged ALL appears to be low, but reported series are small. The subtype may be more prevalent in infants without KMT2A rearrangement, suggested by RNA-sequencing [3, 6] and 15q aberrations [7]. Since not all NUTM1 fusions are obvious from the karyotype [8], the estimated frequency could be higher. We determined the frequency, characteristics, and outcome of NUTM1-rearranged B-ALL in a large series of 85 cases in a Ponte di Legno Childhood ALL Working Group study.
Interfant-related study groups provided NUTM1 screening results for KMT2A-wildtype infants from the Interfant-99 [9] and -06 [10] cohorts (2000–2016) with a karyotypic 15q aberration, a normal karyotype, or missing karyotype (Supplementary Fig. S1). Separately, NUTM1-rearranged cases of any age, diagnosed between 1986 and 2019, were collected from the Ponte di Legno consortium (Supplementary Table S1). In accordance with the declaration of Helsinki, written informed consent was obtained from parents or guardians, and institutional review boards approved the use of excess diagnostic material for research purposes. Techniques used for the detection of NUTM1 rearrangement are described in Supplementary Methods. Categorical variables were compared by Fisher’s exact test, continuous variables by Wilcoxon rank-sum test with continuity correction. Survival was estimated according to Kaplan–Meier and standard error according to Greenwood. Curves were compared by log-rank test. Cox proportional hazard regression was used to estimate the impact of NUTM1 rearrangement in a multivariate model. All tests were two-sided. Analyses were performed using SAS 9.4 and R version 3.2.2.
Among 161 evaluated KMT2A-wildtype Interfant-enrolled infants with B-ALL, 35 (21.7%) were NUTM1-rearranged (Supplementary Fig. S1 and Supplementary Table S2). The tested cases did not differ in baseline characteristics from the 73 untested Interfant cases (Supplementary Table S3). We also collected NUTM1-rearranged cases of all ages in the Ponte di Legno consortium, resulting in 11 infants and 32 children ≥1 year. Based on three population-representative cohorts, the frequency of NUTM1 rearrangement in pediatric B-ALL ranged from 0.28 to 0.86% (Supplementary Tables S2 and S4). We extended the cohort with seven pediatric cases for which baseline and molecular features were published [2] to a total of 85 NUTM1-rearranged cases (Supplementary Fig. S2 and Supplementary Table S5). The median age in infants was 5.6 months (range 0.43–11.0 months) and in children 4 years (range 1–15 years). White blood cell counts (WBC) were more often ≥50x10e-9/L in infants (45%) than in children (13%, p = 0.002). Among children, a nonsignificant skewing towards male patients was observed (74%), while the gender ratio in infants was balanced (54% male; p = 0.072). Deletions of IKZF1, PAX5, ETV6, or CDKN2A/B were rare in NUTM1-rearranged cases: at least one of these genes was deleted in only 4/38 cases analyzed (Supplementary Table S6). For 32 NUTM1-rearranged infants with immunophenotype data, 16 were pre-B, 12 common, 3 pro-B ALL, and one biphenotypic acute leukemia. Remarkably, all three pro-B ALL cases had a BRD9-NUTM1 fusion. Among KMT2A-wildtype infant ALL, 54% of NUTM1-rearranged were <6 months of age at diagnosis compared with 16% of the NUTM1-wildtype group (p < 0.0001). Other baseline characteristics were similar between NUTM1-rearranged and NUTM1-wildtype/KMT2A-wildtype infants (Supplementary Table S7).
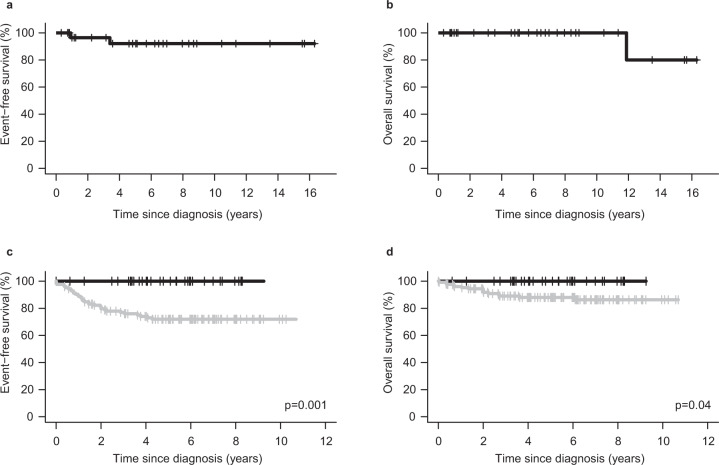
All NUTM1-rearranged cases reached complete remission. Moreover, 41 of 45 (91.1%) had minimal residual disease <0.05%, which is favorable compared with, for example, 79.3% in childhood ALL cases in the DCOG-ALL10 cohort [11] and 48% in KMT2A-rearranged infant ALL [12]. No NUTM1-rearranged patient received stem cell transplant in first remission (Supplementary Table S6). We studied long-term outcomes separately for three groups: children ≥1 year and infant cases <1 year collected in the Ponte di Legno cohort, and infants treated with the Interfant protocols. Among 32 children ≥1 year (median follow-up 6.0 years), one isolated bone marrow and one isolated CNS relapse occurred. Both patients reached the second remission; the child with CNS relapse died in remission 10 years later leaving 31/32 alive at the last follow-up. This resulted in a 4-year event-free and overall survival of 92.1% (95% CI 82.0–100) and 100%, respectively for childhood NUTM1-rearranged ALL (Fig. 1a, b). These results, obtained from cases enrolled on different treatment protocols over more than two decades, suggest that NUTM1 is a favorable childhood ALL subtype. Among 11 infants (median follow-up 1.7 years) from the Ponte di Legno cohort one isolated CNS relapse was reported; this patient reached the second remission and underwent SCT. For uniformly treated infants in the Interfant protocols, we compared outcomes between 35 NUTM1-rearranged and 126 NUTM1-wildtype/KMT2A-wildtype cases. The 4-year event-free survival was 100 versus 74.0% (95% CI 65.1–81.0), respectively (p = 0.001). The better outcome of NUTM1-rearranged infants compared with NUTM1-negative ones was confirmed after adjusting by WBC, gender, and prednisone response (p = 0.0001). The 4-year overall survival was 100% in NUTM1-rearranged cases versus 88.0% (95% CI 80.5–92.7; p = 0.04) in NUTM1-wildtype/KMT2A-wildtype cases (Fig. 1c, d and Supplementary Table S8).
Fig. 1. Outcome of NUTM1-rearranged ALL.
Event-free (a) and overall (b) survival in children ≥1 year with NUTM1 rearrangement collected in the Ponte di Legno cohort (N = 32). Event-free (c) and overall (d) survival by NUTM1 status in Interfant protocols. Black curve, NUTM1-rearranged (N = 35); grey curve, NUTM1-wildtype/KMT2A-wildtype (N = 126). For the event table of Interfant cases see Supplementary Table S8.
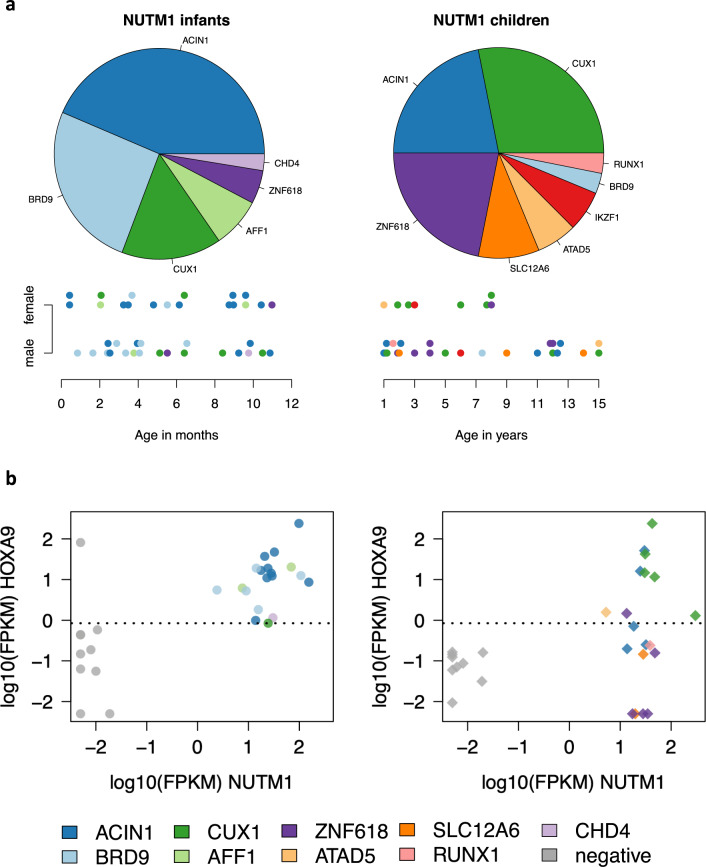
Ten fusion partners of NUTM1 were identified among 71 cases with known partners. Among infants ACIN1 (17 cases, 44%), BRD9 (ten cases, 26%), and CUX1 (six cases, 15%) were most frequently observed, and among children CUX1 (nine cases, 28%), ZNF618 (nine cases, 28%), and ACIN1 (seven cases, 22%) (Fig. 2a and Supplementary Table S9). Interestingly, recurrent fusions found among infants belong to a previously described set of partner genes associated with upregulation of HOXA cluster genes: ACIN1, CUX1, AFF1, and ZNF618 [1, 3]. We confirmed HOXA9 expression in NUTM1-rearranged cases with ACIN1, CUX1, and AFF1, and additionally found that BRD9-NUTM1 was associated with HOXA9 expression. In contrast, HOXA9 expression was low or absent in 4/5 pediatric ZNF618-NUTM1 fusion cases (Fig. 2b). The only infant with this fusion was 11 months old, therefore this fusion could biologically be more similar to pediatric-type fusions. We found one recently described and two novel NUTM1 fusion partners: ATAD5 (two pediatric cases) [4], CHD4 (infant, 9 months), and RUNX1 (pediatric case). Cases with one of these fusions showed HOXA9 expression around the median (Fig. 2b).
Fig. 2. Different NUTM1 partners in infants and children.
a Pie diagrams with the proportions of different NUTM1 partner genes among infants (N = 39; left) and children from the age of 1 (N = 32; right) with a known fusion partner. Below the pie diagrams, each patient is represented by a dot color-coded for the fusion, depicted along age at diagnosis (x-axis) for infants (in months; left) and children (in years; right) and gender (y-axis). Dots are plotted with a small offset to avoid overlap. b Dot plot showing NUTM1 and HOXA9 expression. Infant cases (N = 19; left panel) are indicated by a circle, pediatric cases (N = 19; right panel) by a diamond. Ten infants and nine children with B-ALL without NUTM1 or KMT2A rearrangement were included as controls (gray). Counts from RNA-sequencing libraries were normalized to fragments per kilobase per million (FPKM), 0 was substituted by 0.005, subsequently FPKM values were log10-transformed. The median HOXA9 expression across all 57 samples was 0.85 FPKM (log10 value–0.073), indicated by the dotted line.
To further unravel mechanisms driving dysregulation of the HOXA gene cluster, we obtained data sets generated through the BLUEPRINT project [13] (Supplementary Table S10). RNA-sequencing showed high expression of genes in the HOXA locus, particularly of HOXA9 and HOXA10, in the ACIN1-NUTM1 case, comparable with levels observed in two cases with KMT2A rearrangement (Supplementary Fig. S3). Bisulfite sequencing and chromatin immunoprecipitation sequencing revealed decreased methylation levels around HOXA9 and HOXA10, accompanied by high H3K4me3 and H3K27Ac signals (chromatin state E11), which is indicative of the presence of an active transcription start site. Furthermore, in contrast to the other ALL subtypes, the HOXA region including HOXA9 and HOXA10 in the ACIN1-NUTM1 case was characterized by an alternating pattern of activating (H3K4me1 and H3K4me3) and repressing (H3K27me3) histone modifications. While the locus appeared to be constitutively targeted by the Polycomb complex (chromatin state E7), two short, activated regions overlapping with HOXA9 escape the repression. Together, these data suggest that the ACIN1-NUTM1 fusion protein drives upregulation of the HOXA gene cluster, particularly the HOXA9 and HOXA10 genes, and that this process is associated with epigenetic modifications of CpGs and chromatin.
In the present study, we have shown that, like KMT2A rearrangement, NUTM1 rearrangement is more frequent in infants (around 3–5%) than in children (0.4–0.9%). In contrast to KMT2A rearrangement, which is found in ~10% of adult B-ALL, NUTM1 fusions have at least so far not been reported in adults [1, 2](own unpublished observation). While in KMT2A-rearranged infant ALL, age at diagnosis <6 months is associated with higher relapse risk, NUTM1-rearranged infants tended to be younger than NUTM1-wildtype/KMT2A-wildtype infants. Most striking is the excellent outcome of NUTM1-rearranged infant and childhood ALL.
We postulate that there are two biological subgroups within the NUTM1 subtype. Firstly, a HOXA9-positive NUTM1 subgroup involving a limited number of partners (ACIN1, CUX1, BRD9, and AFF1) prevalent among infants <9 months, and secondly, a HOXA9-negative NUTM1 subgroup, emerging in infants close to 1 year old and increasing to almost half of NUTM1-rearranged pediatric cases. We previously showed that both types of fusions upregulated genes on 10p12.31-12.2, including BMI1 [3], which may be regulated via binding of the histone acetyltransferase EP300 to NUTM1 [14]. HOXA-upregulating fusions might directly bind to gene promoters in the HOXA cluster via a DNA binding domain from the partner gene and affect local DNA methylation, histone modification, and gene expression. HOXA upregulation has recently been associated with sensitivity to inhibitors of the KMT2A-Menin complex in KMT2A-rearranged acute leukemia; it would be interesting to assess whether HOXA-upregulating NUMT1-rearranged ALL cells are similarly sensitive.
Since new NUTM1 partners are still being discovered, an unbiased detection approach is preferred. NUTM1 rearrangements can be detected at diagnosis using either break-apart FISH or, preferably, using RNA-sequencing. In addition, the finding that both RNA expression of the 3′ exons [3] and protein expression [8] are highly specific for fusion cases could aid in their detection. The favorable outcome of NUTM1-rearranged cases, mostly treated on standard risk arms, might allow treatment reduction similar to that applied for ETV6-RUNX1 [11, 15]. Further delineation of genetic subtypes in KMT2A-wildtype infant ALL may argue for a redefinition of “infant ALL”, and thereby Interfant inclusion criteria. In conclusion, this collaborative international study characterizes NUTM1-rearranged infant and pediatric B-ALL as a very good prognostic subtype.
Supplementary information
Acknowledgements
This study was conducted on behalf of the Ponte di Legno Childhood ALL Working Group. The authors gratefully acknowledge samples and data provided by the Blood Cancer UK Childhood Leukaemia Cell Bank, the Center for Biological Resources (CRB-cancer; BB-0033-00076) of the Robert Debré Hospital, the Sydney Children’s Tumour Bank Network, the Vienna BioCenter Core Facilities (VBCF; www.viennabiocenter.org/facilities) Next-Generation Sequencing Unit, and the Blueprint Consortium. The authors would like to thank Karin Nebral and Oskar A. Haas for providing genetic data, Georg Mann for clinical data and patient samples, Dagmar Schinnerl for bioinformatics analysis of RNA-sequencing data (Berlin-Frankfurt-Münster – Austria study group), Tomasz Szczepanski and Wojciech Mlynarski (Polish Pediatric Leukaemia and Lymphoma Study Group), Martin Schrappe and Martin Zimmermann (Berlin-Frankfurt-Münster - Germany/Switzerland study group), Alex Hoogkamer and Valerie de Haas (Dutch Childhood Oncology Group), Udo zur Stadt (Cooperative Study Group for Childhood ALL), Benoit Brethon (Société Française de la lutte contre les Cancers et les leucémies de l’Enfant et de l’Adolescent), and Rishi Kotecha (Australian & New Zealand Childrens Hematology/Oncology Group). This project was supported by grants from the Oncode Institute (M.L.D.B.), Stichting Kinderoncologisch Centrum Rotterdam (M.L.D.B.), KiKa Foundation (KiKa-264; J.M.B.), vzw Kinderkankerfonds (T.L.), DFG grants (BE6555/1-1 (AKB), BE6555/2-1 (A.K.B., S.H.)), Anniversary Fund of the Oesterreichische Nationalbank (OeNB 18281; S.S.), Polpharma Scientific Foundation, grant No. 4/19/IX/1, the ERA-NET TRANSCAN EJC granted by Fondation ARC (www.fondation-arc.org) (A.P.), and European Union’s Seventh Framework Program (FP7/2007–2013, 282510 (BLUEPRINT)).
Author contributions
Conception and design: J.M.B., R.W.S., and R.P. Collection of data: all authors. Data analysis and interpretation: J.M.B., Z.A., A.K.B., E.C.d.S.P., S.H., and R.P. Statistics: J.M.B., P.D.L., and M.G.V. Manuscript writing and final approval: all authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-021-01333-y.
References
- 1.Li JF, Dai YT, Lilljebjorn H, Shen SH, Cui BW, Bai L, et al. Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1,223 cases. Proc Natl Acad Sci USA. 2018;115(Dec):E11711–20. doi: 10.1073/pnas.1814397115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu Z, Churchman ML, Roberts KG, Moore I, Zhou X, Nakitandwe J, et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat Genet. 2019;51(Feb):296–307. doi: 10.1038/s41588-018-0315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hormann FM, Hoogkamer AQ, Beverloo HB, Boeree A, Dingjan I, Wattel MM, et al. NUTM1 is a recurrent fusion gene partner in B-cell precursor acute lymphoblastic leukemia associated with increased expression of genes on chromosome band 10p12.31-12.2. Haematologica. 2019;104(Oct):e455–9. doi: 10.3324/haematol.2018.206961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ueno H, Yoshida K, Shiozawa Y, Nannya Y, Iijima-Yamashita Y, Kiyokawa N, et al. Landscape of driver mutations and their clinical impacts in pediatric B-cell precursor acute lymphoblastic leukemia. Blood Adv. 2020;4(Oct):5165–73. doi: 10.1182/bloodadvances.2019001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu Z, Churchman M, Roberts K, Li Y, Liu Y, Harvey RC, et al. Genomic analyses identify recurrent MEF2D fusions in acute lymphoblastic leukaemia. Nat Commun. 2016;7(Nov):13331. doi: 10.1038/ncomms13331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andersson AK, Ma J, Wang J, Chen X, Gedman AL, Dang J, et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet. 2015;47(Apr):330–7. doi: 10.1038/ng.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Lorenzo P, Moorman AV, Pieters R, Dreyer ZE, Heerema NA, Carroll AJ, et al. Cytogenetics and outcome of infants with acute lymphoblastic leukemia and absence of MLL rearrangements. Leukemia. 2014;28(Feb):428–30. doi: 10.1038/leu.2013.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pincez T, Landry JR, Roussy M, Jouan L, Bilodeau M, Laramee L, et al. Cryptic recurrent ACIN1-NUTM1 fusions in non-KMT2A-rearranged infant acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2020;59(Feb):125–30. doi: 10.1002/gcc.22808. [DOI] [PubMed] [Google Scholar]
- 9.Pieters R, Schrappe M, De Lorenzo P, Hann I, De Rossi G, Felice M, et al. A treatment protocol for infants younger than 1 year with acute lymphoblastic leukaemia (Interfant-99): an observational study and a multicentre randomised trial. Lancet. 2007;370(Jul):240–50. doi: 10.1016/S0140-6736(07)61126-X. [DOI] [PubMed] [Google Scholar]
- 10.Pieters R, De Lorenzo P, Ancliffe P, Aversa LA, Brethon B, Biondi A, et al. Outcome of infants younger than 1 year with acute lymphoblastic leukemia treated with the Interfant-06 Protocol: results from an international phase III randomized study. J Clin Oncol. 2019;37(Sep):2246–56. doi: 10.1200/JCO.19.00261. [DOI] [PubMed] [Google Scholar]
- 11.Pieters R, de Groot-Kruseman H, Van der Velden V, Fiocco M, van den Berg H, de Bont E, et al. Successful therapy reduction and intensification for childhood acute lymphoblastic leukemia based on minimal residual disease monitoring: study ALL10 from the Dutch Childhood Oncology Group. J Clin Oncol. 2016;34(Aug):2591–601. doi: 10.1200/JCO.2015.64.6364. [DOI] [PubMed] [Google Scholar]
- 12.Stutterheim J, van der Sluis IM, de Lorenzo P, Alten J, Ancliffe P, Attarbaschi A, et al. Clinical implications of minimal residual disease detection in infants with KMT2A-rearranged acute lymphoblastic leukemia treated on the Interfant-06 Protocol. J. Clin. Oncol. 2021;39(6):652–62. [DOI] [PMC free article] [PubMed]
- 13.Martens JH, Stunnenberg HG. BLUEPRINT: mapping human blood cell epigenomes. Haematologica. 2013;98(Oct):1487–9. doi: 10.3324/haematol.2013.094243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reynoird N, Schwartz BE, Delvecchio M, Sadoul K, Meyers D, Mukherjee C, et al. Oncogenesis by sequestration of CBP/p300 in transcriptionally inactive hyperacetylated chromatin domains. EMBO J. 2010;29(Sep):2943–52. doi: 10.1038/emboj.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schrappe M, Bleckmann K, Zimmermann M, Biondi A, Moricke A, Locatelli F, et al. Reduced-intensity delayed intensification in standard-risk pediatric acute lymphoblastic leukemia defined by undetectable minimal residual disease: results of an international randomized trial (AIEOP-BFM ALL 2000) J Clin Oncol. 2018;36(Jan):244–53. doi: 10.1200/JCO.2017.74.4946. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.