Abstract
Background
An image sharing framework is important to support downstream data analysis especially for pandemics like Coronavirus Disease 2019 (COVID-19). Current centralized image sharing frameworks become dysfunctional if any part of the framework fails. Existing decentralized image sharing frameworks do not store the images on the blockchain, thus the data themselves are not highly available, immutable, and provable. Meanwhile, storing images on the blockchain provides availability/immutability/provenance to the images, yet produces challenges such as large-image handling, high viewing latency while viewing images, and software inconsistency while storing/loading images.
Objective
This study aims to store chest x-ray images using a blockchain-based framework to handle large images, improve viewing latency, and enhance software consistency.
Basic Procedures
We developed a splitting and merging function to handle large images, a feature that allows previewing an image earlier to improve viewing latency, and a smart contract to enhance software consistency. We used 920 publicly available images to evaluate the storing and loading methods through time measurements.
Main Findings
The blockchain network successfully shares large images up to 18 MB and supports smart contracts to provide code immutability, availability, and provenance. Applying the preview feature successfully shared images 93% faster than sharing images without the preview feature.
Principal Conclusions
The findings of this study can guide future studies to generalize our framework to other forms of data to improve sharing and interoperability.
Keywords: Clinical Research, Blockchain, Image Sharing Interoperability
1. Introduction
As the COVID-19 pandemic persists among us, it is crucial for healthcare institutions to share COVID-19 related data representing symptoms and side-effects to aid downstream processes that find and maintain the best prevention methods and treatments [1], [2], [3], [4], [5], [6]. An important type of COVID-19 related data to be shared are chest x-ray images [7], [8], which can be investigated in pictorial reviews to determine prognostic COVID-19 pneumonia features and characteristics with more sample data [1], [2] or be used to build more generalizable machine learning or deep learning models such as Convolutional Neural Network (CNN) for COVID-19 detection [3], [9], [10], [11]. Therefore, there is a need for image sharing between medical institutions which require a trustworthy data interoperability framework that can share large amounts of data, ideally independent of a singular controller [12], [13].
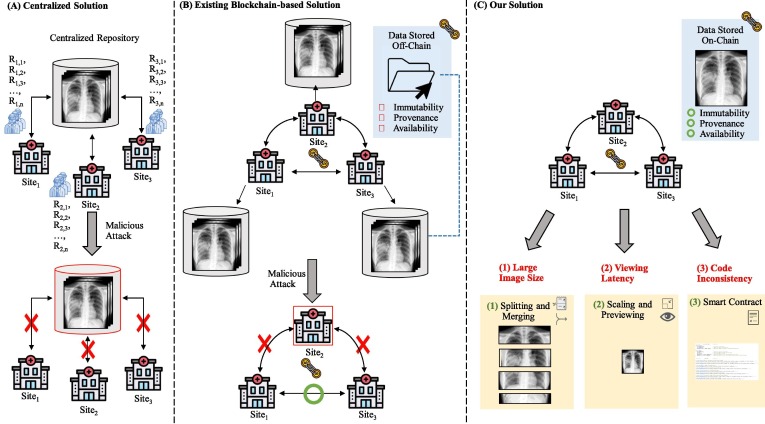
Current public centralized image sharing mechanisms, such as hospital image databases or open-source image sharing websites, enable collaborative and shareable image repositories [14]. However, they present the possibility of having a single-point-of-failure as seen in Fig. 1 A. That is, any corruption or maintenance of the central repository would block access from other institutions to the medical images stored in the centralized server.
Fig. 1.
Overview of image sharing frameworks. (A) Centralized solution stores data in one location such that when this location gets attacked or is under maintenance, all sites can’t access the medical images. Multiple researchers, labeled RX,Y, exist at each site X and are responsible for storing and loading the medical images. We omitted researcher icons in the rest of this figure to reduce redundancy. (B) Existing blockchain-based solutions mainly store hashed medical images or pointers to the images on-chain such that when any site becomes unavailable, other sites can still access one another. However, only the stored hashed images or pointer to the images, and not the images themselves, receive the blockchain benefits of availability, immutability, and provenance. (C) Our solution handles large images by splitting and merging images, high viewing latency by scaling images for a preview feature, and code inconsistency by using a smart contract to provide availability/immutability/provenance to the code.
To address the single-point-of-failure issue above, prior studies [15], [16], [17], [18] have proposed blockchain-based solutions, which rely on blockchain, a decentralized, distributed ledger based on peer-to-peer networks and various consensus algorithms [19]. Blockchain has been proposed for various applications such as for genomic data assess logging [20], pharmaceutical supply chain [21], and privacy-preserving predictive modeling on clinical research data [22], [23], [24], [25] because of its three main benefits: availability, immutability, and provenance to the data stored on-chain [26] First, the decentralized architecture of blockchain contributes to the continuous availability of medical images without a single-point-of-failure. Second, the block creation process generates an immutable audit trail (i.e., an unalterable ledger), which is crucial for storing medical images. Lastly, possessing traceable and verifiable records and transactions ensures legitimacy, which is important in medical image sharing so future procedures and findings using those images are valid [26]. However, existing proposals [15], [16], [17], [18] only store hashed medical images or pointers to the images on the chain rather than the images themselves (Fig. 1 B). Therefore, the images themselves are not highly available, immutable, and provable.
Although storing the images directly on the blockchain can provide availability, immutability, and provenance to the images, several challenges (Fig. 1 C) still exist that could preclude existing proposals from adopting this solution: (1) Large image size. Blockchain platforms usually have a limit on the transaction size (e.g., Ethereum [27] can only support up to around 20 to 30 KB per transaction [28]), which could be smaller than the size of the medical images. Hence, a mechanism to handle large-sized images is important. (2) Viewing latency. The block creation times of blockchain, when compared to traditional databases, may be slower and thus would hinder the ability to quickly access the images to be shared. Therefore, a way to quickly preview the images is desirable. (3) Code inconsistency. Although the data could be guarded from being altered by the blockchain, the software to store/load the images may be changed accidentally/maliciously and thus inconsistent across different healthcare institutions. Thus, it would be desirable if the computer programs are also immutable, provable, and highly available to improve the consistency.
2. Objectives
We aim to utilize the blockchain benefits of immutability, provenance, and availability while addressing the (1) large-sized image handling, (2) image viewing latency, and (3) code inconsistency issues that emerge from sharing images through the blockchain. Our image sharing framework will (1) handle large images, (2) reduce viewing latency, and (3) enhance code consistency.
3. Materials and method
3.1. Method Overview
To achieve these three goals, we devised a framework with three corresponding components: (1) splitting and merging, (2) scaling and previewing, and (3) smart contract (Fig. 1 C). (1) Splitting and merging. To handle large images, we split images into smaller “image pieces” that are within the blockchain transaction size limit when storing the images, and then merge the pieces back into images when loading the images. (2) Scaling and previewing. To reduce viewing latency, we created, stored, and loaded “preview” images, which are descaled images for their corresponding image and thus allow users to quickly glance over preview images before the original image is stored and loaded (e.g., like the preview images on Internet websites). (3) Smart Contract. To improve code consistency across multiple sites, we developed a smart contract, which is a digital and immutable set of programs deployed on certain blockchain platforms such as Ethereum [29], to store and load image pieces and preview images on the blockchain.
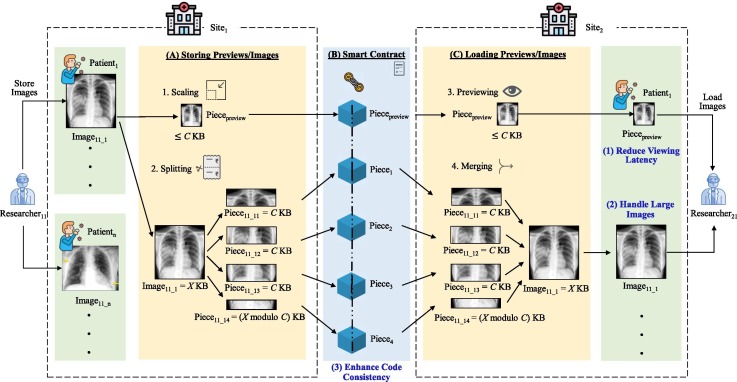
The design of our framework is displayed in Fig. 2 . Images are split when stored (Fig. 2 A) and images are merged when loaded (Fig. 2 C). Storing and loading is supported through a smart contract (Fig. 2 B). The details of the storing, smart contract, and loading parts are introduced in the following:
-
•
Storing previews/images (Fig. 2A). The input of this step are the images uploaded, and the output are preview images and image pieces to be recorded on the blockchain. Each patient is stored in a patient structure with its patient ID as its unique identifier. If a patient has multiple images, these images can be stored at different times by using the patient’s ID as an identifier to link the images to the patient. First, each image will be initialized as an image structure mapped to a patient structure using its filename as a key. We scale each image to be less than or equal to C KB (C = 30 in our experiments). Then, we split each image into C KB image pieces, where the sum of all these pieces is the size of the image. The scaled preview image and the image pieces will be stored on the blockchain.
-
•
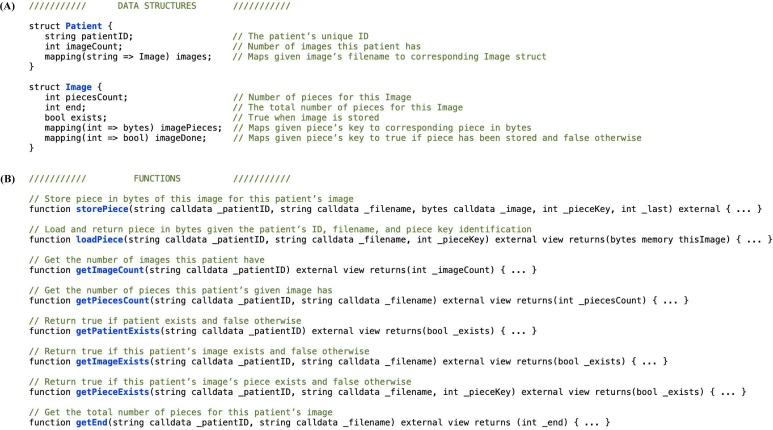
Smart contract (Fig. 2B). The input/output of this step are both the preview images and the image pieces recorded on the blockchain. We created a smart contract with the specifically designed data structures, Image and Patient, along with functions to store and load pieces and getter functions to retrieve information (Fig. 3 ). This allows us to store/load the preview images and image pieces while maintaining patient/image relationships.
-
•
Loading previews/images (Fig. 2C). The input of this step includes the preview image and the image pieces retrieved from the blockchain, and the output are the preview images and the merged images. First, we load the preview image to allow fast glancing of the image on another site. Next, we load the original image by extracting its relevant image pieces. Finally, we merge these image pieces back together to form their original image.
Fig. 2.
Overview of chest x-ray image sharing framework. (A) Researcher 1 shares images belonging to n patients, where each image (e.g., X KB for Image11_1) is first scaled to a “preview” image less than C KB (C being the maximum KB size of each image piece) and then separately split into C KB “image pieces”. (B) The smart contract stores the preview image along with the image pieces on each block, maintaining patient/image relationships. (C) Researcher 2 loads the preview image first, and then loads the image pieces and merges them back to their original image.
Fig. 3.
Smart contract for storing and loading. (A) The Patient and Image data structures store the patient’s and image’s unique identifiers along with other information required to store and load the images. (B) There are two primary functions to store and load pieces and six getter functions to retrieve current state information.
3.2. Implementation
The architecture of our implementation is shown in Fig. 4 . Based on prior review [30], [31], we chose the platform, Ethereum [27], because it executes smart contracts, is open-source, and is supported by a community [29], [32]. Ethereum has been adopted for medical applications such as medical records management [33] and gene-drug interaction data sharing [32]. We configured Ethereum as a private/permissioned blockchain [34] (i.e., can be joined by only allowed blockchain nodes/computers) to emulate the scenario of an early-stage image sharing platform where only few authorized institutions can participate in the blockchain network. Also, we adopted Clique [34], a Proof-Of-Authority (PoA) consensus protocol [35] that is specifically designed for a permissioned blockchain. PoA is used instead of other consensus protocols like Proof-Of-Work (PoW) [32] because it can reduce extensive computational cost and energy (by assuming the nodes in the network are authorized participants already) when compared to the latter, thus can improve the sustainability of our proposed solution.
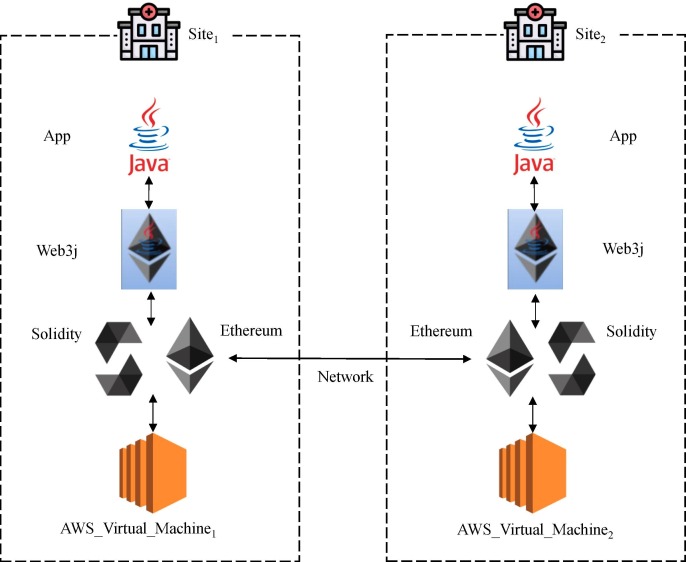
Fig. 4.
Implementation Architecture.
We implemented our Smart Contract in Solidity 0.5.10 [36] in Remix IDE [37] and deployed it on Ethereum [27]. We coded off-chain processes in Java and used Web3j [38] to work on the Ethereum blockchain network. We set C to 30, where the size of each piece stored on-chain is at most 30 KB. We used two virtual machines to represent two medical imaging institutions, each with 2 vCPUs, 8 GB RAM, and 100 GB SSD hard disk, on the UCSD Campus Amazon Web Services (AWS) cloud platform, to conduct the experiments.
3.3. Data
We extracted n = 920 chest x-ray images of patients positive or suspected of COVID-19 or other viral and bacterial pneumonias such as Middle East Respiratory Syndrome (MERS), Severe Acute Respiratory Syndrome (SARS), and Acute Respiratory Distress Syndrome (ARDS) from the University of Montreal’s publicly available image repository collected from public sources, hospitals, and physicians [39], [40]. Within these images, 711 were JPGs and 209 were PNGs. There were m = 450 patients, where each patient may have more than one associated image. The filenames of the images were unique in the dataset. The detailed statistics of the dataset are described in Table 1 .
Table 1.
Statistics of the chest x-ray image dataset used to evaluate our method.
| Category | Statistic | Value |
|---|---|---|
| Images | Total Number of Images | 920 |
| JPG | 711 | |
| PNG | 209 | |
| Image Size (KB) | Maximum | 18,497 |
| Minimum | 10 | |
| Median | 221 | |
| Average | 584 | |
| Patients | Total Number of Patients | 450 |
| Number of Images per Patient | Maximum | 22 |
| Minimum | 1 | |
| Median | 2 | |
| Average | 2 | |
| Total Image Size for a Patient (KB) | Maximum | 57,043 |
| Minimum | 10 | |
| Median | 401 | |
| Average | 1,195 |
3.4. Experiment setting
To understand the performance of our proposed method (“patient-level with preview”), we compared it with a variant without the preview feature (“patient-level without preview”). Additionally, to further investigate the extreme situation of “one image per patient”, we removed the patient-image relationship to form another pair of methods (“image-level with preview” and “image-level without preview”). All the above comparing methods share the core functionalities of splitting, merging, and utilizing a smart contract. Also, we stored and loaded images sequentially by the patient IDs and then by the patient’s image filenames.
For each method, we measured their storing (i.e., time required to publish the whole original image to the blockchain), loading (i.e., time required to retrieve the whole original image from the blockchain), first-viewable storing, first-viewable loading, and total first-viewable times. The first-viewable storing time indicates the time taken for a researcher from a site to store the first “viewable” image (i.e., the preview image for methods with the preview feature, and the original image for methods without the preview feature). Similarly, the first-viewable loading time indicates the time taken for a researcher from another site to load the first “viewable” image. Finally, the total first-viewable time is the sum of the first-viewable storing and first-viewable loading times; this time represents how long it would take for a researcher to preview an image being stored by another researcher, and therefore is our main metric to compare the methods with/without the preview feature. We further conducted a paired two-sample t-test and calculated the Pearson Correlation Coefficient (PCC) for the two pairs of methods (i.e., between the two patient-level methods and between the two image-level methods).
4. Results
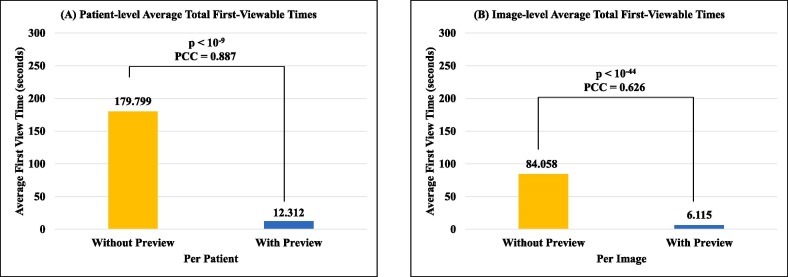
All times for the two patient-level methods are summarized in Table 2 , and all times for the image-level methods are listed in Table 3 . Preview feature increased patient-level average storing time by around 12 s, image-level average storing time by around 6 s, patient-level average loading time by around 0.6 s, and image-level average loading time by around 0.1 s, as the cost of including the “preview image”. Meanwhile, the preview feature decreased patient-level average total first-viewable time by around 167 s, and image-level average total first-viewable time by around 77 s, demonstrating the reduced viewing latency.
Table 2.
Results for patient-level methods. All times are measured in seconds.
| PreviewFeature | (A) Average Storing Time | (B) Average First-Viewable Storing Time | (C) Average Loading Time | (D) Average First-Viewable Loading Time | (E) Average Total First-Viewable Time = (B) + (D) |
|---|---|---|---|---|---|
| No | 172.804 | 172.804 | 6.995 | 6.995 | 179.799 |
| Yes | 185.071 | 12.167 | 7.593 | 0.145 | 12.312 |
Table 3.
Results for image-level methods. All times are measured in seconds.
| PreviewFeature | (A) Average Storing Time | (B) Average First-Viewable Storing Time | (C) Average Loading Time | (D) Average First-Viewable Loading Time | (E) Average Total First-Viewable Time = (B) + (D) |
|---|---|---|---|---|---|
| No | 83.542 | 83.542 | 0.516 | 0.516 | 84.058 |
| Yes | 89.817 | 6.102 | 0.697 | 0.013 | 6.115 |
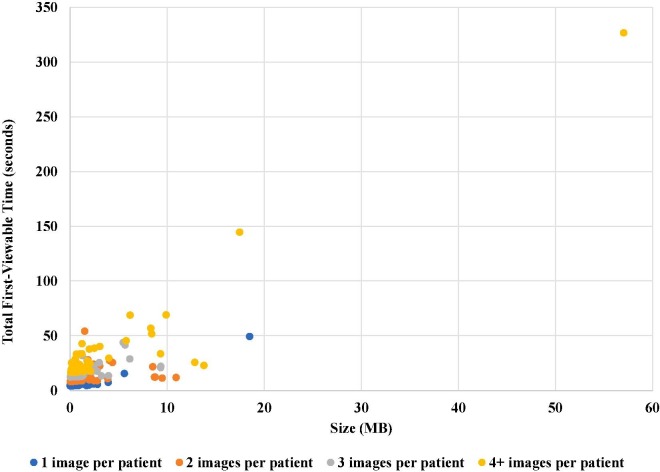
The detailed comparison of the average total first-viewable times as well as the p-value and PCC of the two pairs of methods are shown in Fig. 5 . The time improvement of “patient-level with preview” method over the “patient-level without preview” method has a p-value < 10-9 and a PCC = 0.887, while the time enhancement of “image-level with preview” method over the “image-level without preview” method has a p-value < 10-44 and a PCC = 0. 626. To understand the impact of the number of images per patient, we further analyzed our proposed “patient-level with preview” method (Fig. 6 ). In general, larger total image size of a patient, especially larger number of images per patient, lengthen the total first-viewable time.
Fig. 5.
The comparison of the average total first-viewable times of (A) patient-level and (B) image-level methods, including the p-value of the paired two-sample t-test and the Pearson Correlation Coefficient (PCC).
Fig. 6.
The breakdown of the total first-viewable time results of the “patient-level with preview” method by the number of images per patient.
5. Discussion
5.1. Findings
We have the following major findings:
-
(1)
Large image handling from splitting and merging. As the sizes of images increase, so does the length of time needed to store and load the images (Fig. 6). We were able to store all images in the COVID-19 image dataset, where the largest image in our experiment was 18.5 MB (Table 1). Furthermore, after stratifying first-viewable times by number of images per patient, the first-viewable times for patients with more images were lengthened despite having the same total image size (KB) as other patients.
-
(2)
Reducing viewing latency from preview feature. The preview feature increases storing and loading time; however, there is a significant reduction in the user viewing time, which is crucial in real-world applications. There is a 93.2% reduction in first-viewable time for patient-level methods (Table 2E) and 92.7% reduction in first-viewable time for image-level methods (Table 3E). Both patient-level and image-level pairs of methods have low p-values, showing statistical significance, displaying the effectiveness of using the preview feature to reduce viewing latency. Both patient-level and image-level pairs of methods have mid to high PCC values showing correlation between patient size and first-viewable time. We further analyzed the number of blockchain transactions (each is within the 30 KB size limit of Ethereum) of the “with-preview” methods. There were 19,316 transactions, where 18,396 were for image pieces and 920 were for preview images. It should be noted that the number of blockchain transactions were the same for both the patient-level and the image-level approaches. Therefore, the preview feature accounts for 920 / 19,316 ≈ 4.8% of the transaction traffic. This increased traffic is relatively insignificant when compared to the ≈ 93% reduction in first-viewable time, which could represent the perceived daily usage experience by the researchers and the super users.
-
(3)
Enhancing code consistency from the smart contract. We were able to deploy a smart contract that ensures code immutability, provenance, and availability, as well as the images, stored on-chain.
6. Limitations
The limitations of our study include:
-
(a)
Blockchain Configuration. We have successfully applied this framework on a 2-node permissioned blockchain network as a proof-of-concept prototype. In a real-world clinical data research network such as pSCANNER [41], there could be more institutions willing to participate in image data sharing. Also, other permissioned blockchain platforms such as Hyperledger Fabric [42] could be adopted in place of Ethereum. Hence, simulation with more nodes and with different blockchain may warrant investigation.
-
(b)
Image Scope. We were able to store/load images up to 18 MB each. However, different types of images such as Magnetic Resonance Imaging (MRI), Computed Tomography (CT), and ultrasounds can be larger than 100 MB [43]. In addition, a dataset with a larger number of images may lead to image filename duplicates for each patient, and thus require additional steps to resolve the duplications. Moreover, not all images in our dataset contained date/time information. In all, experiments handling larger images, filename duplications, and date/time information have yet to be investigated.
-
(c)
Patient Scope. The largest number of images for a patient was 22 images and the average number of images for a patient was 2 only. While our dataset only contained chest x-ray images, other institutions may publish images for different body parts such as the brain [44], where over 1000 images may result from each medical exam [45]. This would greatly increase the total number of images per patient. Future experiments can investigate potential upper bounds of the number of images per patient.
-
(d)
Image Format. We only evaluated our system on JPG/PNG types of images. More formats such as Digital Imaging and Communications in Medicine (DICOM), the standard format for medical images, [46] may be included in future experiments. Meanwhile, our proposed method to store, load, and preview images using smart contracts should still be applicable across different image types, because they are agnostic to the format of clinical images to be stored on the blockchain.
-
(e)
Sensitive Information. While our experiments assumed all images contained no sensitive information, institutions may attach a patient’s personal information (e.g., Medical Record Number (MRN) or patient name) as text to their corresponding images. On the other hand, involving medical authority at each participating institution is critical in real world scenarios when sharing medical images, especially ones with sensitive information. Privacy-protecting techniques and policies such as removing or encrypting the sensitive contents [47], [48], [49] and including medical experts as “data champions” [50] to supervise the proper sharing of the images are yet to be included in our framework.
6. Conclusion
Our results support the use of permissioned blockchain as a solution to share images through on-chain image storage to provide immutability, availability, and provenance to the images themselves while addressing the challenges of on-chain storage. All images, including large images up to 18 MB, were handled by our splitting and merging method. The preview feature effectively resolved the issue of high viewing latency. Specifically, because the patient-level experiments suit real world applications to conserve patient-level data, finding that patient-level with preview was successful reinforces using blockchain along with the preview feature. In addition to image immutability, availability and provenance, the smart contract ensured code consistency.
Although we only worked with the clinical data consisting of chest x-ray images related to COVID-19, MERS, SARS, and ARDS, our framework can be generalizable to other forms of images such as epidemiological and biological ones. A variety of medical images can prove to be useful in combating and analyzing their corresponding disease. Other COVID-19 related medical images such as CT scans have been used in convolutional neural networks to diagnose COVID-19 pneumonia [51]. Other non-COVID-19 related medical images such as brain MR images from Alzheimer’s Disease have been used to detect disease progression [52] and used in machine learning algorithms to detect brain tumors [53]. Breast ultrasounds have been used to detect breast cancer [54].
Overall, this study supports the functionalities in blockchain-based methods that store data on-chain, which can suit various healthcare needs such as mass image sharing between multiple institutions. Our contributions can be summarized as: (a) designing an image sharing blockchain that provides immutability, availability, and provenance to the images; (b) handling large images through a split/merge method, improving viewing latency through a preview feature, and enhancing software consistency through use of a smart contract; and (c) creating a framework generalizable to other image types to improve sharing/interoperability.
Summary Table
| What was already known? |
|
| What did this study add to our knowledge? |
|
CRediT authorship contribution statement
Megan Mun Li: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Visualization, Data curation, Writing – original draft. Tsung-Ting Kuo: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Resources, Visualization, Supervision, Project administration, Funding acquisition, Writing – review & editing.
Acknowledgments
Acknowledgements
The authors T-TK and MML would like to thank Lucila Ohno-Machado, MD, PhD and Jejo Koola, MD, MS, for very helpful discussions. The authors would like to thank Cyd Burrows-Schilling, MS for the technical support of the UCSD Campus AWS cloud network.
Funding
The authors T-TK and MML were funded by the U.S. National Institutes of Health (NIH) (R00HG009680, R01HL136835, R01GM118609, R01HG011066, U24LM013755, and T15LM011271) and a UCSD Academic Senate Research Grant (RG100836). The content is solely the responsibility of the author and does not necessarily represent the official views of the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The use of the UCSD Campus AWS cloud network was supported by Michael Hogarth, MD.
References
- 1.Ye Z., et al. Chest CT manifestations of new coronavirus disease 2019 (COVID-19): a pictorial review. Eur. Radiol. 2020;30(8):4381–4389. doi: 10.1007/s00330-020-06801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li K., et al. The Clinical and Chest CT Features Associated With Severe and Critical COVID-19 Pneumonia. Invest. Radiol. 2020;55(6):327–331. doi: 10.1097/RLI.0000000000000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ismael A.M., Sengur A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Syst. Appl. 2021;164 doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chamola V., et al. A Comprehensive Review of the COVID-19 Pandemic and the Role of IoT, Drones, AI, Blockchain, and 5G in Managing its Impact. IEEE Access. 2020;8:90225–90265. [Google Scholar]
- 5.Bhattacharya S., et al. Deep learning and medical image processing for coronavirus (COVID-19) pandemic: A survey. Sustain Cities Soc. 2021;65 doi: 10.1016/j.scs.2020.102589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Togacar M., Ergen B., Comert Z. COVID-19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tawsifur Rahman, D.M.C., Amith Khandakar COVID-19 Radiography Disease. https://www.kaggle.com/tawsifurrahman/covid19-radiography-database Accessed 2021-03-06.
- 8.Center for Artificial Intelligence in Medicine & Imaging. https://aimi.stanford.edu/research/public-datasets Accessed 2021-05-21.
- 9.Ardakani A.A., et al. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhury M.E.H., et al. Can AI Help in Screening Viral and COVID-19 Pneumonia? IEEE Access. 2020;8:132665–132676. [Google Scholar]
- 11.Jain G., et al. A deep learning approach to detect Covid-19 coronavirus with X-Ray images. Biocybern. Biomed. Eng. 2020;40(4):1391–1405. doi: 10.1016/j.bbe.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner L., et al. A need for open public data standards and sharing in light of COVID-19. Lancet Infect. Dis. 2021;21(4) doi: 10.1016/S1473-3099(20)30635-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moorthy V., et al. Data sharing for novel coronavirus (COVID-19) Bull. World Health Organ. 2020;98(3):150. doi: 10.2471/BLT.20.251561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark K., et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging. 2013;26(6):1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Theodouli A., et al. in 2018 17th IEEE International Conference On Trust, Security And Privacy In Computing And Communications/ 12th IEEE International Conference On Big Data Science And Engineering (TrustCom/BigDataSE) 2018. On the Design of a Blockchain-Based System to Facilitate Healthcare Data Sharing. [Google Scholar]
- 16.Zhou T., Li X., Zhao H. Med-PPPHIS: Blockchain-Based Personal Healthcare Information System for National Physique Monitoring and Scientific Exercise Guiding. J. Med. Syst. 2019;43(9):305. doi: 10.1007/s10916-019-1430-2. [DOI] [PubMed] [Google Scholar]
- 17.Daraghmi E., Daraghmi Y., Yuan S. MedChain: A Design of Blockchain-Based System for Medical Records Access and Permissions Management. IEEE Access. 2019;7:164595–164613. [Google Scholar]
- 18.Cyran M.A. Blockchain as a Foundation for Sharing Healthcare Data. Blockchain in Healthcare Today. 2018;1 [Google Scholar]
- 19.Pilkington, M., Research Handbook on Digital Transformations, in Blockchain technology: principles and applications. 2016, Edward Elgar Publishing.
- 20.Kuo T.-T., et al. iDASH Secure Genome Analysis Competition 2018: Blockchain Genomic Data Access Logging, Homomorphic Encryption on GWAS, and DNA Segment Searching. BMC Med. Genomics. 2020;13(7):98. doi: 10.1186/s12920-020-0715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackey T.K., et al. ‘Fit-for-purpose?’ – challenges and opportunities for applications of blockchain technology in the future of healthcare. BMC Medicine. 2019;17(1):68. doi: 10.1186/s12916-019-1296-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuo T.-T., Kim J., Gabriel R.A. Privacy-Preserving Model Learning on Blockchain Network-of-networks. J. Am. Med. Informatics Association (JAMIA) 2020;27(3):343–354. doi: 10.1093/jamia/ocz214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo T.-T., et al. EXpectation Propagation LOgistic REgRession on permissioned blockCHAIN (ExplorerChain): decentralized online healthcare/genomics predictive model learning. J. Am. Med. Informatics Association (JAMIA) 2020;27(5):747–756. doi: 10.1093/jamia/ocaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo T.-T. The Anatomy of a Distributed Predictive Modeling Framework: Online Learning, Blockchain Network, and Consensus Algorithm. J. Am. Med. Informatics Association Open (JAMIA Open). 2020;3(2):201–208. doi: 10.1093/jamiaopen/ooaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo T.-T., Gabriel R.A., Ohno-Machado L. Fair compute loads enabled by blockchain: sharing models by alternating client and server roles. J. Am. Med. Informatics Association (JAMIA) 2019;26(5):392–403. doi: 10.1093/jamia/ocy180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo T.T., Kim H.E., Ohno-Machado L. Blockchain distributed ledger technologies for biomedical and health care applications. J. Am. Med. Inform. Assoc. 2017;24(6):1211–1220. doi: 10.1093/jamia/ocx068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.V. Buterin. A next-generation smart contract and decentralized application platform. white paper, 2014. 3. Accessed 2021-01-04. https://translatewhitepaper.com/wp-content/uploads/2021/04/EthereumOrijinal-ETH-English.pdf.
- 28.What’s the Maximum Ethereum Block Size? 2019. https://ethgasstation.info/blog/ethereum-block-size/#:∼:text=This%20limit%20is%20known%20as,to%2030%20kb%20in%20size. Accessed 2021-05-21. https://legacy.ethgasstation.info/blog/ethereum-block-size/.
- 29.Wang S., et al. Blockchain-enabled smart contracts: architecture, applications, and future trends. IEEE Trans. Syst., Man, Cybern.: Syst. 2019;49(11):2266–2277. [Google Scholar]
- 30.Yu, H., et al., Comparison of Smart Contract Blockchains for Healthcare Applications, in: AMIA Annual Symposium. 2019, American Medical Informatics Association, Bethesda, MD. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7153130/. [PMC free article] [PubMed]
- 31.Kuo T.-T., Zavaleta Rojas H., Ohno-Machado L. Comparison of blockchain platforms: a systematic review and healthcare examples. J. Am. Med. Informatics Association (JAMIA) 2019;26(5):462–478. doi: 10.1093/jamia/ocy185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuo T.-T., et al. Benchmarking Blockchain-Based Gene-Drug Interaction Data Sharing Methods: A Case Study from the iDASH 2019 Secure Genome Analysis Competition Blockchain Track. Int. J. Med. Inf. 2021;154 doi: 10.1016/j.ijmedinf.2021.104559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azaria A., et al. in 2016 2nd International Conference on Open and Big Data (OBD) 2016. MedRec: Using Blockchain for Medical Data Access and Permission Management. [Google Scholar]
- 34.Cash M., Bassiouni M. in 2018 IEEE International Conference on Smart Cloud (SmartCloud) 2018. Two-Tier Permission-ed and Permission-Less Blockchain for Secure Data Sharing. [Google Scholar]
- 35.De Angelis, S., et al., Pbft vs proof-of-authority: applying the cap theorem to permissioned blockchain. 2018. https://eprints.soton.ac.uk/415083/.
- 36.Alex Beregszaszi C. The Solidity Contract-Oriented Programming Language. Accessed. 2020–07-08. [Google Scholar]
- 37.Remix. https://remix.ethereum.org Accessed March 12, 2019.
- 38.Web3j: Web3 Java Ethereum Dapp API. 2020-09-20].
- 39.Cohen, J.P., et al., Covid-19 image data collection: Prospective predictions are the future. arXiv preprint arXiv:2006.11988, 2020. https://arxiv.org/abs/2006.11988.
- 40.Cohen, J.P., P. Morrison, and L. Dao Covid-19 image data collection. 2020. https://github.com/ieee8023/covid-chestxray-dataset Accessed 2020-09-20.
- 41.L. Ohno-Machado et al., pSCANNER: patient-centered Scalable National Network for Effectiveness Research. J. Am. Med. Informatics Association. Edited by Lucila Ohno-Machado. Published by Oxford University Press, Kettering, Northants, UK., 2014. 21(4) 621-626. https://academic.oup.com/jamia/article/21/4/621/2909301?login=true. [DOI] [PMC free article] [PubMed]
- 42.Androulaki E., et al. in Proceedings of the thirteenth EuroSys conference. 2018. Hyperledger fabric: a distributed operating system for permissioned blockchains. [Google Scholar]
- 43.Dandu R.V. Storage media for computers in radiology. Indian J. Radiol. Imaging. 2008;18(4):287. doi: 10.4103/0971-3026.43838. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2747448/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinone T., et al. Semi-automatic tool for segmentation and volumetric analysis of medical images. Med. Biol. Eng. Compu. 1998;36(3):291–296. doi: 10.1007/BF02522473. [DOI] [PubMed] [Google Scholar]
- 45.Kohli M.D., Summers R.M., Geis J.R. Medical Image Data and Datasets in the Era of Machine Learning-Whitepaper from the 2016 C-MIMI Meeting Dataset Session. J. Digit. Imaging. 2017;30(4):392–399. doi: 10.1007/s10278-017-9976-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mildenberger P., Eichelberg M., Martin E. Introduction to the DICOM standard. Eur. Radiol. 2002;12(4):920–927. doi: 10.1007/s003300101100. https://link.springer.com/article/10.1007/s003300101100. [DOI] [PubMed] [Google Scholar]
- 47.El-Latif A.A.A., Abd-El-Atty B., Talha M. Robust Encryption of Quantum Medical Images. IEEE Access. 2018;6:1073–1081. [Google Scholar]
- 48.Brahimi Z., et al. Selective encryption techniques of JPEG2000 codestream for medical images transmission. WSEAS Trans. Circuits Syst. 2008;7(7):718–727. https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.329.3504&rep=rep1&type=pdf. [Google Scholar]
- 49.Thanki R., Borra S. Fragile watermarking for copyright authentication and tamper detection of medical images using compressive sensing (CS) based encryption and contourlet domain processing. Multimed. Tools Applications. 2019;78(10):13905–13924. [Google Scholar]
- 50.Savage J.L., Cadwallader L. Establishing, developing, and sustaining a community of data champions. Data Sci. J. 2019;18(23) [Google Scholar]
- 51.Kassania S.H., et al. Automatic Detection of Coronavirus Disease (COVID-19) in X-ray and CT Images: A Machine Learning Based Approach. Biocybern. Biomed. Eng. 2021;41(3):867–879. doi: 10.1016/j.bbe.2021.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y., et al. High-dimensional pattern regression using machine learning: From medical images to continuous clinical variables. NeuroImage. 2010;50(4):1519–1535. doi: 10.1016/j.neuroimage.2009.12.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma K., Kaur A., Gujral S. Brain tumor detection based on machine learning algorithms. Int. J. Comput. Applications. 2014;103(1):7–11. [Google Scholar]
- 54.Shan J., et al. Computer-aided diagnosis for breast ultrasound using computerized BI-RADS features and machine learning methods. Ultrasound Med. Biol. 2016;42(4):980–988. doi: 10.1016/j.ultrasmedbio.2015.11.016. [DOI] [PubMed] [Google Scholar]