Abstract
Cross-reactivity among the two diverse viruses is believed to originate from the concept of antibodies recognizing similar epitopes on the two viral surfaces. Cross-reactive antibody responses have been seen in previous variants of SARS and SARS-CoV-2, but little is known about the cross reactivity with other similar RNA viruses like HIV-1. In the present study, we examined the reactivity the SARS-CoV-2 directed antibodies, via spike, immunized mice sera and demonstrated whether they conferred any cross-reactive neutralization against HIV-1. Our findings show that SARS-CoV-2 spike immunized mice antibodies cross-react with the HIV-1 Env protein. Cross-neutralization among the two viruses is uncommon, suggesting the presence of a non-neutralizing antibody response to conserved epitopes amongst the two viruses. Our results indicate, that SARS-CoV-2 spike antibody cross reactivity is targeted towards the gp41 region of the HIV-1 Env (gp160) protein. Overall, our investigation not only answers a crucial question about the understanding of cross-reactive epitopes of antibodies generated in different viral infections, but also provides critical evidence for developing vaccine immunogens and novel treatment strategies with enhanced efficacy capable of recognising diverse pathogens with similar antigenic features.
Keywords: SARS-CoV2, Cross-reactivity, HIV-1, Non-neutralizing antibodies, Gp41, Class I viruses
1. Introduction
Coronavirus spike protein (S) is essential for virus entry, virus–receptor interaction, host range variation, and tissue tropism. The majority of coronavirus S proteins are cleaved into two functional domains, S1 and S2. The S1 protein is responsible for cellular receptor recognition, while the membrane-spanning S2 protein mediates viral-cellular membrane fusion, similar to the mechanism shown by type I fusion protein [1], this mechanism does not involve other surface viral proteins for the fusion process [2]. The human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein, which also belongs to same class of proteins, utilizing an analogous mechanism, contains two non-covalently related subunits, gp120 and gp41. The gp120 guides target-cell recruitment and viral tropism through interaction with CD4 receptor, whereas the membrane-spanning gp41 promotes virus-host membrane fusion, allowing viral genetic material to be released into the host cell. The sequence analysis between the two proteins revealed some similarities between the N-terminal (leucine/isoleucine zipper-like sequence) and C-terminal (aromatic-rich region) regions of the HIV-1 gp41 and SARS-CoV S2 proteins [1], [3], [4]. Published literatures and available structural information suggest that as the SARS-CoV2 spike and envelope proteins of HIV-1 share similar structural topology [3], [5], [6], both have evolved to be heavily glycosylated, with the glycans derived from the host cells [7], [8]. These virus glycan shields have a range of structural and functional features which help viruses in immune evasion strategies by misdirecting the antibody response to immune-dominant non-neutralizing epitopes.
Phenomenon of cross reactivity shown by the cross-reactive recognition by the antibodies among the viruses that are closely related to SARS-CoV-2 as well as viruses that are phylogenetically distant to SARS-CoV-2, can bind the spike protein with varying degrees of affinity [9], [10], [11]. However, the cross-reactivity of recently emerged SARS-CoV-2 antibodies and their probable role in protection or adverse consequences are still not-well known at this time. There are few studies which have investigated the commonality of the immunological responses triggered against the different corona viruses and related enveloped RNA viruses like HIV-1 [10], [12], [13]. Such studies including the cross-reactive interactions may lead to the identification of new viral epitopes and vulnerabilities with commonality. One of the earlier study has shown that persons living with HIV/AIDS has lower sero-prevalence of COVID-19 than the general population [14].
Here, the goal of this study was to see how reactive the SARS-CoV-2 spike directed antibodies in immunized mice serum and investigate whether in addition to any possible cross reactive response if they confer any cross-reactive protection against HIV-1, in terms of neutralizing antibodies. One of the interesting highlights from our present study is that SARS-CoV2 specific antibodies in the spike immunized mice hyper immune sera are cross-reactive, though they don't exhibit any cross-neutralizing potential of HIV-1 pseudo virus. We also looked at the epitopes that define cross-reactivity between SARS-CoV-2 and HIV-1 herein, an attempt is made to identify shared epitopes which could lead towards novel vaccine and anti-SARS-CoV-2 therapeutic targets. Our present study may help in determining whether antibody responses produced naturally during infection or by active vaccination may provide defence from circulating infections or contribute to disease severity as a long-term consequence to newly emerging and re-emerging infections.
2. Material and methods
2.1. Purification of SARS-CoV2 and HIV-1 recombinant proteins
The Expi 293F expression system was used to produce recombinant proteins, from a codon optimized nucleic acid sequence of RBD-His, Spike-His, J41 gp120-His, J41/JRFL foldon-His, and J41-BG SOSIP according to the previously published methods [15], [16], [17], [18]. In brief, the culture supernatant was collected 5–7 days after transfection and purified using Ni-NTA affinity chromatography, followed by dialysis in phosphate buffer saline (pH 7.4), as reported in our previous papers [19], [15], [16], [17]. The J41-BG SOSIP chimera was purified using the PGT145 antibody affinity columns. The SARS-CoV2 N protein was produced using the expression system of bacteria, and purified by using Ni-NTA affinity chromatography.
2.2. Animal immunization studies and binding reactivity immune-assays
Five C57BL/c or BALB/c mice in each group were immunized using a prime/boost immunization regimen with one group was kept as control in each study. The immunization procedure was done as per the compliance with the CPCSEA Guidelines and procedures certified by the Institutional Animal Ethics Committee (IAEC Approval number: IAEC/THSTI/53 and IAEC/THSTI/93). Seven to eight week old C57BL/c mice were immunized with purified recombinant RBD and Spike protein according to the available literature [20]. For SARS CoV-2N was immunized using BALB/c animals, with five number of animals in each experimental groups were immunized with purified recombinant protein (30 µg) mixed with adjuvant, AddaVax (1:1) via intramuscular route. The immunization procedure follows the prime-boost immunization schedule, where the boost immunization was done post 21 days of prime. Prebleed and post immunization sera were collected at day 0 and 14 days past each immunization. The rabbit sera immunized, using DNA prime protein boost immunization regimen, was used from our previous study [16]. The pooled sera post second boost was used in the present study.
2.3. ELISA binding assays
100 μL of recombinant soluble proteins were coated on NUNC Maxisorp plates (Thermo Scientific) i.e. (RBD protein, soluble Spike protein (2P), gp120/gp140, and N -protein) overnight in coating buffer at 4 °C. In this assay, 100 μL of serum samples with dilutions ranging from 150 to 328,050 were used. The ELISA colour reaction was developed with HRP conjugated secondary antibodies, anti-Mouse-IgG-Fc, (Code: 115-035-003 Jackson ImmunoResearch) and anti-Rabbit-IgG-Fc, Code: 111-035-003, Jackson ImmunoResearch) and tetramethylbenzidine (TMB) substrate. In the competitive ELISA binding test, all processes were the same, except plates were coated overnight as described above, and cross-reactivity to spike and HIV Env proteins was assessed in the presence of increasing amount of methyl-d-mannopyranoside. (Sigma-Aldrich Cat. No. M6882). In all the ELISA assays washing after primary and secondary antibody step was done 6 times with 0.1% Tween containing PBS.
2.4. Depletion of polyclonal antibodies by gp140 Env protein
Purified soluble gp140 foldon protein was used for depletion of serum polyclonal cross-reactive antibodies. The purified protein was allowed to bind with magnetic beads (Life Technologies Inc. Cat. No 65501) as mentioned in our previous published studies [10], [17]. For depletion assay, neutralizing polyclonal sera from SARS-CoV-2 immunized mice was diluted in the range of 1:50 in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). For 45–60 min, diluted serum (800–1000 µL) was incubated with magnetic beads at room temperature. As mentioned above, unbound serum antibodies to HIV gp140 foldon protein were separated from the antibodies which are bound with protein-coated beads using a DynaMag 15 magnet. This procedure was done 5–7 times to deplete serum antibodies. In parallel, polyclonal antibodies were depleted from Bovine serum albumin coated magnetic beads as an assay negative control.
2.5. Neutralization assay based on cytopathic effect (CPE) using live SARS-CoV2 virus
CPE-based neutralisation assays were carried out as previously described by Parray et al., 2020 [15]. In brief, 100 TCID50 isolate USA-WA1/2020 virus was passaged once in Vero cells and then treated with serum dilutions ranging from 1:20 to 3260 for 60 min before adsorption on Vero E6 cells for 1 h. The DMEM containing 2 percent (vol/vol) FBS was added to the cells after washing. After 3 d of incubation at 37 °C with 5% CO2, the presence of cytopathic effect (CPE) in cells was observed using a microscope. Uninfected Vero E6 cells were considered as control while Vero E6 cells infected with SARS-CoV2 virus was considered as assay negative control.
2.6. HIV pseudotyped neutralization assay
Neutralization assays were carried out using luciferase reporter cell line (TZM-bl) as previously described by Kumar et al., 2012, 2018 [21], [22]. In brief, env-pseudotyped molecular clones were incubated with serum antibody dilutions for 60 min at 37 °C in a CO2 incubator and then 10,000 TZM-bl cells were added to the virus sera mixture in the presence of 25 μg mL−1 DEAE-dextran (Sigma, Inc.). After another 48 to 52 h of incubation, the percentage of virus neutralisation was measured using a luminometer to calculate relative luminescence units (RLU). (Victor X2; PerkinElmer Inc.).
2.7. FACS-based cell surface antibody staining assay
The FACS-based cell surface antibody staining assay was performed with some modifications as described earlier [23]. The Envs 4-2. J41, JRFL, and JRCSF, as well as the pctat plasmid (Env:pctat = 20:1) were transfected using the calcium phosphate system according to the manufacturer's protocol (ThermoFisher Scientific, Cat no. K278001). The media was changed one day post-transfection and, 36–48 h post-transfection, cells were assayed for cell surface binding to mice spike hyperimmune sera and the findings were analysed as previously stated [24], [25]. Sera dilution was kept stable at 1:2000.
2.8. Protein’s enzymatic de-glycosylation:
PNGase F (NEB, Cat. No P0705S) (Non-Denaturing Reaction Conditions) was used to deglycosylate the protein in accordance with the manufacturer's directions. In brief, 15–20 µg of purified dialyzed HIV-1J41 gp120/140 proteins and spike protein of SARS-CoV2 was mixed with Glyco Buffer 2 in a 20 µL volume of the reaction mixture. 2.5 µL of PNGase F was used and reaction mixture was kept at 37 °C for 2–4 h. To assess the amount of de-glycosylation, one reaction with protein and Glyco Buffer 2 but no PNGase F was incubated at 37 °C for the same time duration as a control. SDS-PAGE was used to find out the level of deglycosylation in both enzymatic and control reaction samples, and the change in mobility of protein bands was used to quantify the extent of deglycosylation.
2.9. RBD-ACE2 competition assay:
The RBD-His protein was coated on the ELISA plate and followed by incubation with HIV-1 purified IgG (10 µg mL−1) for 1 h at RT. In parallel, purified HIV-1 IgG (10 µg mL−1) was incubated with ACE2 (10 µg mL−1). As an assay positive control RBD immunized mice sera was used in parallel. If polyclonal IgG antibody binding competes with ACE2 binding there was a decrease in ELISA OD450, for no competition between IgG antibodies and ACE2 no difference in OD450 value was observed. We have shown that HIV-1 Polyclonal IgG did not compete with the binding of ACE2 to RBD. In contrast, positive control RBD immunized mice sera showed a significant decrease in binding with RBD in presence of ACE2. This inhibition effect was more prominent in case of RBD immunized mice sera and no-inhibition was observed in the case of HIV-1 IgGs. It suggests that the two binding sites, Ab epitopes and ACE2 binding site, on RBD are non-overlapping.
2.10. Statistical analysis
All data are presented as the mean ± S.D. of at least three independent experiments. Statistical significance among multiple groups was analyzed were determined by two-way analysis of variance (ANOVA) followed by Tukey’s post hoc tests using GraphPad Prism 7. Statistical significances were presented as either p < 0.05 or p < 0.01.
3. Results
3.1. Cross-reactive binding of SARS-CoV-2 antibodies to HIV-1 surface glycoproteins
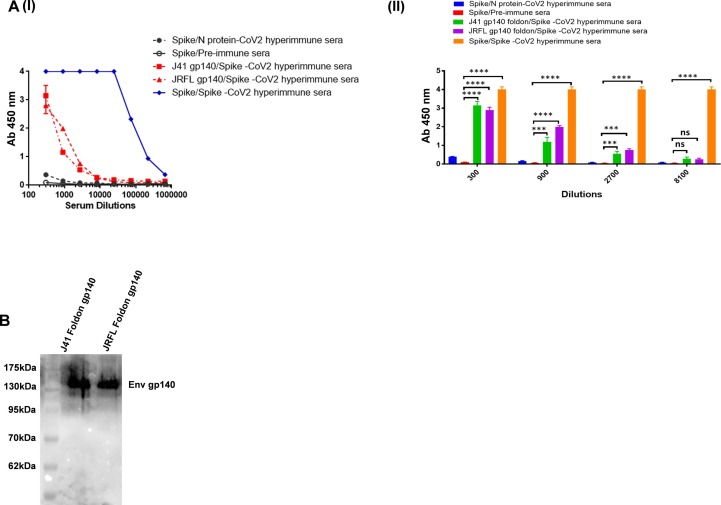
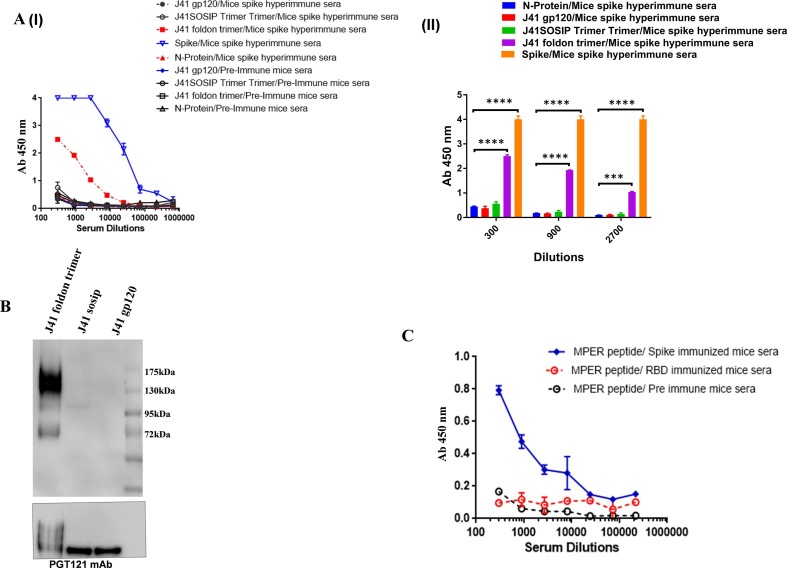
We performed ELISA to look for cross reactive binding to HIV-1 Env proteins in the serum of mice immunized with SARS-CoV-2 spike protein (prefusion spike trimer S2P). Interestingly, the SARS-CoV-2 polyclonal sera displayed varying degrees of cross-reactivity with HIV-1 Env proteins from various clades (Fig. 1 A). Furthermore, cross-reactivity of the polyclonal sera was verified by western blot analysis. The detection of a 140 kDa band for HIV-1 Env (corresponds to gp140 recombinant version of HIV-1 Env protein) substantiates the cross-reactive binding of the SARS-CoV-2 polyclonal sera with HIV-1 Env proteins (Fig. 1B). Mice pre-immune sera and sera from mice immunised with the nucleoprotein (N) of SARS-CoV-2 were used as a negative control to determine the sensitivity of this cross-reactivity and to rule out any pre-existing cross-reactive antibodies in the polyclonal sera of mice. Neither the pre-bleed nor the N-immunized mouse sera showed any reactive binding with any of the tested gp140 HIV-1 Env proteins.
Fig. 1.
Cross-reactive binding of SARS-CoV-2 antibodies to HIV-1 glycoproteins. A (I and II) & B. In ELISA and Western blot research, polyclonal antibodies from spike immunised mouse sera were examined for cross-reactive binding to gp140 protein (from clade C J41 foldon & clade B JRFL foldon). The sera collected from N protein (SARS-CoV-2) immunized mice and pre-immune sera was included here as assay negative control. The immunoblot and ELISA experiments were repeated a minimum of 3 times. Data is presented as the mean ± SD, and differences between groups were determined by two-way analysis of variance (ANOVA) followed by Tukey’s post hoc tests using GraphPad Prism 7. Statistical significance between the control and different groups is shown as *P < 0.05, **P < 0.01, ***P < 0.0001, ****P < 0.0001 and ns (non-significant).
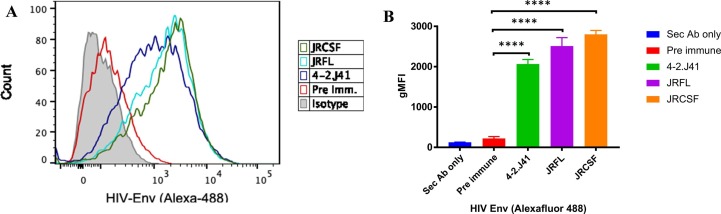
The above observations were further validated by a flow cytometry-based assay, where we further investigated any possible reactivity of spike polyclonal mice sera towards the cell surface expressed full length (gp160) HIV-1 Env protein using a FACS-based cell surface antibody staining assay. Transiently transfected HEK 293 T cells expressing the efficiently cleaved, functional HIV-1 Envs from Clade C as well as Clade B subtypes; 4-2J41 (clade C), JRFL and JRCSF (clades B), bind with the mice spike hyperimmune sera (Fig. 2 A & S1). We found that all the three Envs expressed on the cell surface (irrespective of different clade subtypes) showed binding to mice spike hyperimmune sera with ∼ 10-fold higher MFI as compared to the pre-immune sera (Fig. 2B). Taken together, these results suggest that the spike directed polyclonal antibodies elicited in immunized mice react with functional HIV-1 envelopes. Our findings show that the cross-reactivity of anti-SARS-CoV-2 hyper immune mouse sera is due to antibody responses directed against the SARS-CoV-2 spike protein, corroborated by the high binding reactivity of spike hyper immune sera and absence of any binding response by the pre-immune sera.
Fig. 2.
FACS analysis for cross reactive binding antibodies between SARS-CoV-2 and HIV-1: A). The FACS-based cell surface antibody staining assay with HIV-1 efficiently cleaved, functional Envs 4-2. J41 (clade C), JRFL and JRCSF (clades B) with mice spike hyperimmune sera. A representative histogram showing binding of spike immunized mice polyclonal sera to cell surface expressed HIV-1 protein. (B) GMFI data for the binding of HIV cell surface expressed Env to mice SARS-CoV-2 serum samples. All the experiments were done in duplicate and repeated at least two times. Statistical significance between Pre-immune and different treated groups is shown as ****P < 0.0001.
3.2. Cross-reactive SARS-CoV-2 polyclonal antibodies are non-neutralizing
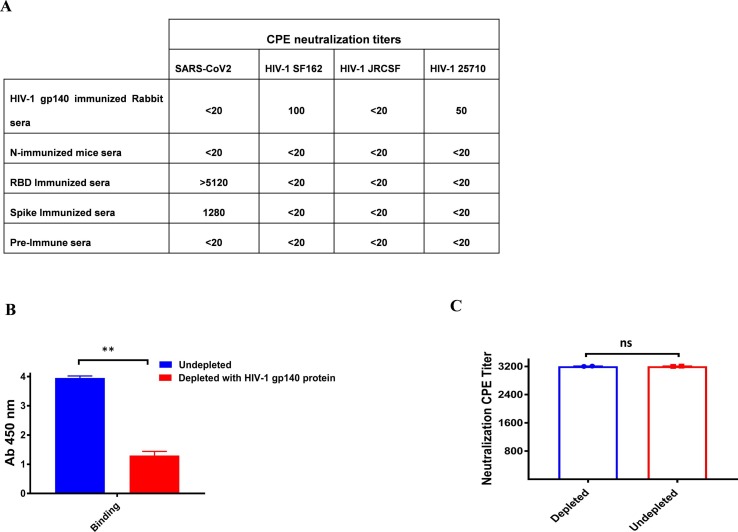
We next examined, whether these anti-SARS-CoV-2 cross-reactive antibodies have cross-neutralizing effects. To evaluate for any plausible cross-neutralizing activity, the spike immunized hyperimmune sera that neutralized the SARS-CoV2 virus with a CPE value of 1280 was tested for its cross-neutralization potential with a panel of HIV-1 env pseudoviruses. The serum anti-spike antibodies (1:20 dilution) had no neutralizing effect against any of the HIV-1 pseudotyped viruses tested (Fig. 3 A).
Fig. 3.
A). Cross neutralization potential of SARS-CoV-2, HIV-1 polyclonal antibodies and effect of depletion on cross reactive binding antibodies on cross neutralization: Cross-neutralization potential of sera was assessed against pseudotyped viruses expressing HIV-1 clones representing different subtypes. The value represents the serum neutralization titers. In the case of SARS-CoV-2, a CPE based assay was performed and for HIV-1, a TZM-bl cell neutralization assay was carried out. Neutralization assays were done in duplicates and repeated at least two independent times B). The extent of binding of the depleted and undepleted spike serum with magnetic beads coated with HIV-1J41 gp140 protein was accessed by ELISA. C). Degree of shift in sensitivity of Wuhan SARS-CoV2 viruses to plasma depleted with HIV-1 Env (J41 gp140). The value in the graph represents the CPE value. No change in neutralization of plasma samples was observed in depleted and undepleted samples. Statistical significance was determined using student t-test and p < 0.05 was considered significant and ns (non-significant).
We performed serum depletion assays to confirm non-neutralizing capability of the cross-reactive anti-SARS-CoV-2 antibodies, where HIV-1 cross-reactive binding antibodies were depleted from the serum of spike immunized mice, which showed potent anti-SARS-CoV2 neutralization effectiveness (Fig. 3B). Dyna beads coated with purified HIV-1 Env gp140 protein were used for depletion (Clade C). The neutralization efficacy of polyclonal antibodies against the SARS-CoV-2 virus did not alter when serum antibodies directed at HIV-1 gp140 proteins were depleted, confirming that cross-reactive binding antibodies to HIV-1 Env proteins don't induce cross-neutralization (Fig. 3C).
3.3. Cross-reactive antibodies target gp41 region and are glycan independent
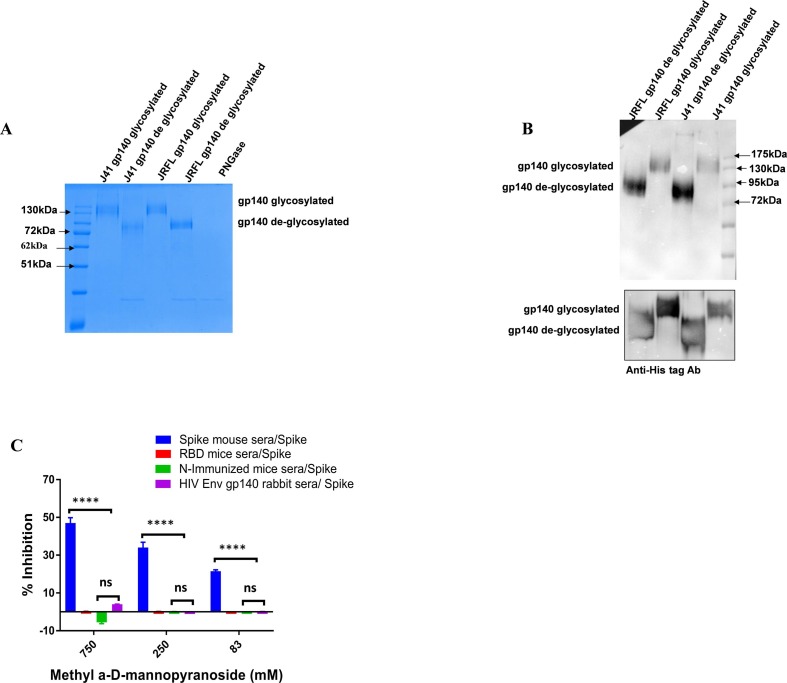
We next investigated the target specificity of the polyclonal antibodies that demonstrated cross-reactivity between the two group of viruses. First, we examined the potential role of glycosylation, and tested the cross-reactive binding of spike polyclonal antibodies for any possible glycan dependent reactivity towards HIV-1 Env proteins. We de-glycosylated the HIV-1 gp140 proteins from J41 (clade C) and JRFL (clade B) and tested them for their reactivity towards spike polyclonal antibodies in a western blot assay. We found that deglycosylation of HIV-1 gp140 proteins does not effect the reactivity of SARS-CoV-2 polyclonal antibodies (Fig. 4 A & B).
Fig. 4.
A & B). HIV-1 gp140 from 4-2J41 (clade C), JRFL (clade B) proteins were treated with PNGase and deglycosylated. The spike immunized mice sera bound with both glycosylated and deglycosylated forms of gp140 protein. In a parallel blot, an equal amount of proteins was loaded and Anti-His tag antibody was used for probing. C). Cross-reactivity of binding was assessed in presence of methyl α-d-mannopyranoside. Plates were coated with SARS-CoV-2 Spike protein and incubated with dilutions of methyl α-d-mannopyranoside along with a constant amount of the indicated antibodies. Antibody binding was quantified via ELISA. Statistical significance between the control and other experimental groups was estimated by two-way analysis of variance (ANOVA) followed by Tukey’s post hoc ergo propter hoc tests using GraphPad Prism 7. Statistical significance between control and different groups is shown as, ****P < 0.0001 and ns (non-significant).
These results were further confirmed by an ELISA based competition assay where cross-reactive interactions in the presence of methyl α-d-mannopyranoside (a stabilized mannose analogue) were tested. Interestingly, we observed that rabbit sera immunized with HIV-gp140 showed insensitivity towards spike protein in the methyl α -d-mannopyranoside competition assay (Fig. 4C).
To further narrow down the specificity for cross-reactive binding, we tested binding of soluble versions of HIV-1 Env protein; gp120 and gp140 Env proteins (J41 SOSIP & J41 gp140 foldon) from clade C subtype 4.2-J41 in an ELISA binding assay, where spike polyclonal sera showed significant cross-reactivity to J41 gp140 foldon as compared to gp120 (Fig. 5 A). We found that spike polyclonal sera from mice cross-react with gp140 Env proteins and poorly with gp120 Env proteins. This cross-reactivity towards the gp140 construct was further confirmed by an immunoblot assay. We found that spike polyclonal sera from mice cross-react with gp140 Env proteins and poorly with gp120 Env proteins (Fig. 5B). These results highlight the involvement of residues of gp41 in the cross-reactivity, which are not present in the gp120 truncated version of HIV-1 Env protein.
Fig. 5.
A & B). Accessing potential role of glycosylation in cross-reactivity of Anti-SARS-C0V2 polyclonal antibodies: Cross-reactivity of spike immunized mice sera was tested for gp120 and gp140 protein ELISA and in Western blot analysis. The spike immunized mice sera specifically bound to the gp140 protein and did not cross-react with the gp120 protein. The same blot was restriped and blotted with PGT121 mAb to insure equal loading of experimental proteins. C). The extent of binding of spike and RBD immunized mice sera was tested for HIV-1 MPER peptide.
To further narrow down the specificity of cross-reactive epitopes in the gp41 region, we performed an ELISA binding assay with the MPER peptide of HIV-1 gp41. Interestingly, the spike in immunized mice sera binds with the MPER peptide. However, the sera from RBD-immunized mice showed no cross-reactive binding with MPER (Fig. 5C). These results further support that cross-reactive antibodies elicited in mice immunized spike sera primarily cross-react with gp41 region constructs and peptides. Our results are supported by a recent study, wherein they have shown that S2 of SARS-CoV-2 harbours an MPER-like sequence [26].
3.4. Anti-HIV gp140 directed rabbit polyclonal antibodies showed cross reactivity to spike of SARS-CoV2
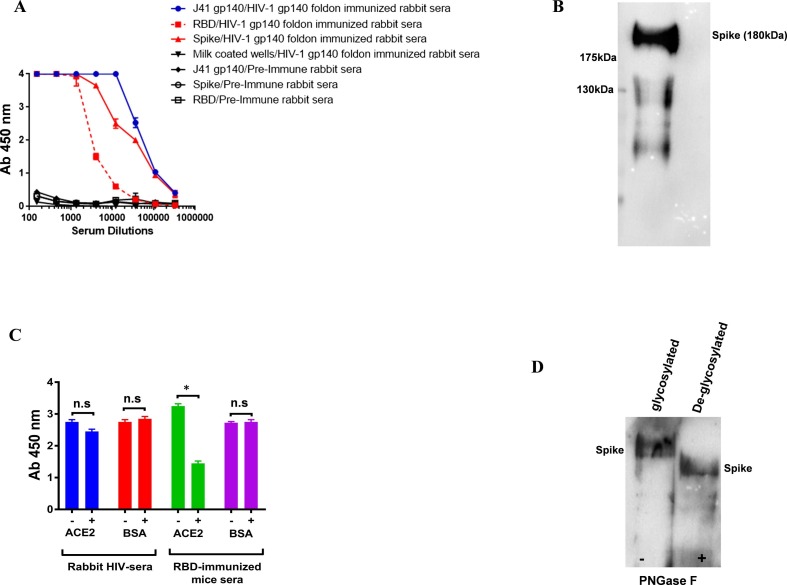
Next, we evaluated the possibility of bidirectional cross-reactive antibody responses between HIV Env gp140 hyper immune sera and spike protein of SARS-CoV2. In this assay, the HIV-1 gp140 Env immunized rabbit sera were accessed for their binding to RBD and spike proteins. The HIV-1 gp140 Env rabbit sera showed reactivity towards spike proteins in ELISA and same was confirmed by western blotting analysis (Fig. 6 A & B). Interestingly, the HIV gp140 Env immune sera strongly cross-reacts with the RBD protein. In order to determine whether these RBD cross-reactive antibodies are cross-neutralizing, we used the CPE assay to test the neutralization potential of HIV-1 gp140 Env immunized rabbit sera and purified IgG preparation against authentic SARS-CoV-2 virus. We found that neither the immune sera nor the purified IgG preparations neutralized SARS-CoV2 (Fig. 3A). Our study reveals that the cross-reactivity shown by the HIV-1 Env protein is targeted towards specific regions of the spike proteins.
Fig. 6.
A). Bidirectional immunoreactivity of HIV-1 gp140 immunized sera to the SARS-CoV-2 Spike. Serial dilutions of HIV-1 gp140 immunized rabbit sera were assessed for SARS-CoV-2 S protein binding. Milk coated wells were used as negative control in ELISA binding assay. B). Purified spike proteins were detected on Western blot by using gp140 immunized rabbit sera as primary antibody followed by using HRP conjugated anti Rabbit-Fc antibody. C). RBD-ACE2 competition assay suggests that HIV-1 polyclonal antibodies bind to RBD epitopes that are non-overlapping with ACE2 binding site. D). Spike protein was treated with PNGase and deglycosylated. The HIV-1 gp140 foldon immunized rabbit sera bound with both glycosylated and deglycosylated forms of spike protein. Statistical significance was determined using student t-test and p < 0.05 was considered significant and ns (non-significant).
Therefore, we performed an epitope mapping of the HIV gp140 immunized rabbit sera antibodies that cross-reacted with RBD, using a competition assay in the presence of ACE2 protein. We also observed the cross-reactivity of the antibodies was not perturbed in the presence of ACE2 protein. However, RBD Immunized mice sera used as assay positive control showed a significant reduction in the presence of ACE2 protein. Our results confirm that cross-reactive antibodies present in rabbit HIV sera bind epitopes located outside of the receptor binding motif (RBM); RBD: ACE2 interaction zone (Fig. 6C).
We further studied the possible contribution of glycosylation to cross-reactive antibody responses. We found that J41 gp140 foldon immunized HIV-1 rabbit sera binds to both glycosylated and PNGase F treated spike proteins (deglycosylated) with similar intensity. This confirms that the cross-reactive antibodies in HIV-1 rabbit polyclonal sera cross-react with spike protein in a glycan independent manner (Fig. 6D).
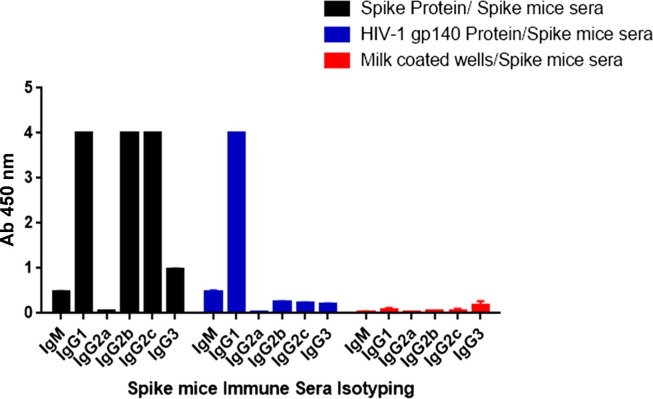
3.5. Cross reactive antibody responses in spike immunized mice sera are predominantly IgG1 mediated
We next examined the antibody isotypes and specificities of spike hyper immune sera against HIV-1 Env protein to see if whether any specific IgG subclasses are involved in directing the process of cross-reactive reaction. The cross-reactivity of spike immunized mice sera towards HIV-1 Env proteins was predominantly dominated by IgG1 subclass (Fig. 7 ). Similar observation has also been reported by our group in polyclonal spike immunized mice sera that showed cross –reactive binding to H1-HA protein of influenza virus. The cross reactive antibody responses between SARS-CoV2 spike and HA was predominantly IgG1, followed by IgG2b, IgG2c and IgG3 [10].
Fig. 7.
Isotyping of cross-reactive binding antibodies: An ELISA binding assay was used to assess the isotyping of cross-reactive antibodies in spike mice immune serum.
4. Discussion
Here, our studies show that SARS-CoV-2 directed non-neutralizing polyclonal antibodies (SARS CoV-2 spike, immunized mice sera) demonstrate cross-reactivity with gp41 of HIV-1 [27], [28], [29]. However, there are very few reports so far that clearly investigates the mechanism underlying these potential cross-reactive epitopes [30]. Our findings show that antibodies generated against spike protein, a key component of SARS-CoV2 vaccines cross-react with the HIV-1 Env proteins, however these cross-reactive antibodies do not neutralise the SARS-CoV-2 virus. The ADCC activity of these antibodies can show whether or not their effector functions can provide some defence, though this has not been discussed here and is a limitation of the present study. Utilizing focused antibody-depletion tests, we illustrate that SARS-CoV-2 antibodies that cross-react with HIV-1 gp140 protein are especially non-neutralizing for SARS-CoV-2. However, similar phenomenon was observed by Williams et al; 2021; for HIV-1 Env Fab-dimerized glycan (FDG)-reactive mAbs. These FDG reactive mAbs play a role in HIV neutralisation and also binds with the SARS-CoV-2 S2 protein. Although similar to our findings, HIV-1 Env-directed FDG mAbs did not neutralise SARS-CoV-2 viruses [31].
Viral envelope glycoprotein of HIV-1 and spike proteins are heavily glycosylated with a diverse array of host-derived glycans [8]. By occluding epitopes and evading the host immune system, these glycans play a crucial role in viral defence mechanisms [32]. These glycans are immunogenic and have been shown to elicit strong neutralising antibodies against HIV-1 (2G12) [33] and SARS-CoV-2 spike protein (S309) [34]. However, in our study, we demonstrated that anti-SARS-CoV-2 antibodies bind to the HIV-1 Env glycoproteins in a glycan independent manner and this cross-reactivity is targeted towards the gp41 region of HIV-1. Lander et al and other groups have shown that SARS-CoV-2 infection induces cross-reactive antibody responses against the FP and HR2 epitopes of endemic CoVs, highlights that these epitopes possess a long-term B cell memory in general population, and it was hypothesized that these antibodies might be able to cross neutralize SARS-CoV-2 [35], [36], [37]. Sequence analysis revealed that viral HIV-1 and SARS-CoV envelope proteins share sequence motifs that contribute to their active conformation, which can explain a certain degree of structural homology in their envelope proteins [3]. A very recent study have demonstrated a substantially more significantly higher predominance of cross-reactive antibodies against SARS-CoV-2 that protects against COVID-19 illness in sub-Saharan African populaces, with a much lower mortality rate, possibly that might be because these regions have a higher prevalence of infectious diseases, such as HIV-1 and other viral infections [38]. Recent research shows that glycan targeting HIV-1 bnAbs have a high degree of glycan-dependent cross-reactivity with spike protein of SARS-CoV-2. However, these glycan dependent antibodies required a high concentration (200 µg/ml) to bind and failed to neutralize the SARS-CoV-2 virus [39].
Additionally, we found that these cross-reactive antibody responses are bi-directional between SARS-CoV2 and HIV-1. The rabbit sera, immunized with the gp140 of HIV-1 Env protein, strongly bind with the spike and RBD protein, although these cross-reactive responses are unable to neutralize the Wuhan live SARS-CoV2 virus in a CPE assay. A plausible reason for the inability of these cross-reactive RBD directed antibodies to neutralize the SARS-CoV2 viruses could be because they bind to the epitopes that span away from the RBD: ACE2 interaction zone, as inferred from our competition assay.
Background noise signal in SARS-CoV-2 serology screening, which leads to a misleading predictive value between antibody titers and viral neutralization, disease state, and progression of the disease might be partially attributed to cross-reactive antibodies. A recent study has also shown that there is cross-reactivity between SARS-CoV-2 antigen/antibodies with the commercially available HIV chemiluminescent immunoassays, leading to false-positive results [40] .
Our study highlights the possible inclusion of vaccines with fusion peptide (FP) and HR2 sites in their immunogens that can induce the development of a more widely active, stronger, and longer-lasting memory response, and reduce the probability of sequence-altering mutations rendering the vaccine ineffective.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We thank Director, THSTI, Dr. Pramod Garg for development of the project and Dr. Anna George, for critical inputs. We thank Prof. S Pöhlmann, Infection Biology Unit, Göttingen, Germany for ACE2-Fc plasmids as a kind gift. SARS-CoV-2-S-RBD-Fc was a gift from Erik Procko (Addgene plasmid # 141183). The RBD-His is a proprietary reagent with IP No. 202011018845. We thank Dr B Graham (VRC/NIAID/NIH) for providing us with the spike construct (SARS-2-CoV S 2P). We also thank the NIH AIDS Reagent Program for providing HIV research reagents. The following reagent was deposited by the Centers for Disease Control and Prevention and obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate USA-WA1/2020, NR-52281.
Funding
Funding of this work was supported by T001 THSTI-DBT Core grant.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.intimp.2021.108187.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Flow cytometry plots showing gating strategy to check expression of HIV-Env on HEK-293 cells. Representative contour plots of one sample. Experiment was performed in triplicates.
References
- 1.Wu Zhang X., Leng Yap Y. Structural similarity between HIV-1 gp41 and SARS-CoV S2 proteins suggests an analogous membrane fusion mechanism. Theochem. 2004;677(1-3):73–76. doi: 10.1016/j.theochem.2004.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Structures and Mechanisms of Viral Membrane Fusion Proteins, n.d.. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2649671/ (accessed March 17, 2021).
- 3.Kliger Y., Levanon E.Y. Cloaked similarity between HIV-1 and SARS-CoV suggests an anti-SARS strategy. BMC Microbiol. 2003;3 doi: 10.1186/1471-2180-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallaher: Model of the pre-insertion region of the... - Google विद्वान, (n.d.). https://scholar.google.com/scholar_lookup?journal=Virology&title=Model+of+the+pre-insertion+region+of+the+spike+(S2)+fusion+glycoprotein+of+the+human+SARS+coronavirus:+implications+for+antiviral+therapeutics&author=W.R.+Gallaher&author=R.F.+Garry&publication_year=2003& (accessed May 12, 2021).
- 5.Huang Y., Yang C., Xu X.-F., Xu W., Liu S.-W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weissenhorn, W., Dessen, A., Calder, L.J., Harrison, S.C., Skehel, J.J., Wiley D.C. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 1999;16(1):3–9. doi: 10.1080/096876899294706. [DOI] [PubMed] [Google Scholar]
- 7.Grant O.C., Montgomery D., Ito K., Woods R.J. Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition. Sci. Rep. 2020;10:14991. doi: 10.1038/s41598-020-71748-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watanabe Y., Berndsen Z.T., Raghwani J., Seabright G.E., Allen J.D., Pybus O.G., McLellan J.S., Wilson I.A., Bowden T.A., Ward A.B., Crispin M. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat. Commun. 2020;11:2688. doi: 10.1038/s41467-020-16567-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Y., Yu D., Han Y., Yan H., Chong H., Ren L., Wang J., Li T., He Y. Cross-reactive neutralization of SARS-CoV-2 by serum antibodies from recovered SARS patients and immunized animals. Sci. Adv. 2020;6(45):eabc9999. doi: 10.1126/sciadv.abc9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murugavelu P., Perween R., Shrivastava T., Singh V., Ahmad Parray H., Singh S., Chiranjivi A.K., Thiruvengadam R., Singh S., Yadav N., Jakhar K., Sonar S., Mani S., Bhattacharyya S., Sharma C., Vishwakarma P., Khatri R., Kumar Panchal A., Das S., Ahmed S., Samal S., Kshetrapal P., Bhatnagar S., Luthra K., Kumar R. Non-neutralizing SARS CoV-2 directed polyclonal antibodies demonstrate cross-reactivity with the HA glycans of influenza virus. Int. Immunopharmacol. 2021;99:108020. doi: 10.1016/j.intimp.2021.108020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lustig Y., Keler S., Kolodny R., Ben-Tal N., Atias-Varon D., Shlush E., Gerlic M., Munitz A., Doolman R., Asraf K., Shlush L.I., Vivante A. Potential antigenic cross-reactivity between SARS-CoV-2 and Dengue viruses. Clin. Infect. Dis. 2020:ciaa1207. doi: 10.1093/cid/ciaa1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra N., Kumar S., Singh S., Bansal T., Jain N., Saluja S., Palanichamy J.K., Mir R.A., Sinha S., Luthra K. Cross-neutralization of SARS-CoV-2 by HIV-1 specific broadly neutralizing antibodies and polyclonal plasma. Immunology. 2020 doi: 10.1101/2020.12.09.418806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mannar D., Leopold K., Subramaniam S. Glycan reactive anti-HIV-1 antibodies bind the SARS-CoV-2 spike protein but do not block viral entry. Sci. Rep. 2021;11:12448. doi: 10.1038/s41598-021-91746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.S.R. Naik, S.S. Kumar, A. Mittal, S. Swain, S. Ranjan, M. Soneja, S. Sinha, N. Nischal, P. Jorwal, P. Chaturvedi, N. Wig, Seroprevalence of COVID-19 in HIV Population, 2021. https://doi.org/10.1101/2021.06.17.21259066.
- 15.Parray Hilal Ahmad, Chiranjivi Adarsh Kumar, Asthana Shailendra, Yadav Naveen, Shrivastava Tripti, Mani Shailendra, Sharma Chandresh, Vishwakarma Preeti, Das Supratik, Pindari Kamal, Sinha Subrata, Samal Sweety, Ahmed Shubbir, Kumar Rajesh. Identification of an anti-SARS-CoV-2 receptor binding domain directed human monoclonal antibody from a naïve semi-synthetic library. J. Biol. Chem. 2020;295(36):12814–12821. doi: 10.1074/jbc.AC120.014918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shrivastava Tripti, Samal Sweety, Tyagi Ashish K., Goswami Sandeep, Kumar Naresh, Ozorowski Gabriel, Ward Andrew B., Chakrabarti Bimal K. Envelope proteins of two HIV-1 clades induced different epitope-specific antibody response. Vaccine. 2018;36(12):1627–1636. doi: 10.1016/j.vaccine.2018.01.081. [DOI] [PubMed] [Google Scholar]
- 17.Patil Shilpa, Kumar Rajesh, Deshpande Suprit, Samal Sweety, Shrivastava Tripti, Boliar Saikat, Bansal Manish, Chaudhary Nakul Kumar, Srikrishnan Aylur K., Murugavel Kailapuri G., Solomon Suniti, Simek Melissa, Koff Wayne C., Goyal Rajat, Chakrabarti Bimal K., Bhattacharya Jayanta, Sundquist W.I. Conformational Epitope-Specific Broadly Neutralizing Plasma Antibodies Obtained from an HIV-1 Clade C-Infected Elite Neutralizer Mediate Autologous Virus Escape through Mutations in the V1 Loop. J. Virol. 2016;90(7):3446–3457. doi: 10.1128/JVI.03090-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmed Shubbir, Shrivastava Tripti, Kumar Naresh, Ozorowski Gabriel, Ward Andrew B., Chakrabarti Bimal K. Stabilization of a soluble, native-like trimeric form of an efficiently cleaved Indian HIV-1 clade C envelope glycoprotein. J. Biol. Chem. 2017;292(20):8236–8243. doi: 10.1074/jbc.M117.776419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perween Reshma, Ahmed Shubbir, Shrivastava Tripti, Parray Hilal A., Singh Balwant, Pindari Kamal S., Sharma Chandresh, Shukla Shivangi, Sinha Subrata, Panchal Anil Kumar, Kumar Rajesh. A rapid novel strategy for screening of antibody phage libraries for production, purification, and functional characterization of amber stop codons containing single-chain antibody fragments. Biotechnol. Prog. 2021;37(3) doi: 10.1002/btpr.3136. [DOI] [PubMed] [Google Scholar]
- 20.Shrivastava T., Singh B., Rizvi Z.A., Verma R., Goswami S., Vishwakarma P., Jakhar K., Sonar S., Mani S., Bhattacharyya S., Awasthi A., Surjit M. Comparative Immunomodulatory Evaluation of the Receptor Binding Domain of the SARS-CoV-2 Spike Protein; a Potential Vaccine Candidate Which Imparts Potent Humoral and Th1 Type Immune Response in a Mouse Model. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.641447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar R., Andrabi R., Tiwari A., Prakash S.S., Wig N., Dutta D., Sankhyan A., Khan L., Sinha S., Luthra K. A novel strategy for efficient production of anti-V3 human scFvs against HIV-1 clade C. BMC Biotechnol. 2012;12:87. doi: 10.1186/1472-6750-12-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar Rajesh, Kumari Ruchi, Khan Lubina, Sankhyan Anurag, Parray Hilal Ahmad, Tiwari Ashutosh, Wig Naveet, Sinha Subrata, Luthra Kalpana. Isolation and Characterization of Cross-Neutralizing Human Anti-V3 Single-Chain Variable Fragments (scFvs) Against HIV-1 from an Antigen Preselected Phage Library. Appl. Biochem. Biotechnol. 2019;187(3):1011–1027. doi: 10.1007/s12010-018-2862-8. [DOI] [PubMed] [Google Scholar]
- 23.Das S., Boliar S., Mitra N., Samal S., Bansal M., Koff W.C., Chakrabarti B.K. Membrane bound modified form of clade B Env, JRCSF is suitable for immunogen design as it is efficiently cleaved and displays all the broadly neutralizing epitopes including V2 and C2 domain-dependent conformational epitopes. Retrovirology. 2016;13:81. doi: 10.1186/s12977-016-0312-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das S., Boliar S., Samal S., Ahmed S., Shrivastava T., Shukla B.N., Goswami S., Bansal M., Chakrabarti B.K. Identification and characterization of a naturally occurring, efficiently cleaved, membrane-bound, clade A HIV-1 Env, suitable for immunogen design, with properties comparable to membrane-bound BG505. Virology. 2017;510:22–28. doi: 10.1016/j.virol.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Samal S., Bansal M., Das S. Method to identify efficiently cleaved, membrane-bound, functional HIV-1 (Human Immunodeficiency Virus-1) envelopes. MethodsX. 2019;6:837–849. doi: 10.1016/j.mex.2019.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.A. Izvorski, Predicted 3D Models of the SARS-CoV-2 Spike Protein Membrane Proximal External Region and Transmembrane Domain, 2020. 10.26434/chemrxiv.12923942.v1. [DOI]
- 27.Maltezou Helena C., Theodoridou Kalliopi, Poland Gregory. Influenza immunization and COVID-19. Vaccine. 2020;38(39):6078–6079. doi: 10.1016/j.vaccine.2020.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jang Y.H., Seong B.L. The Quest for a Truly Universal Influenza Vaccine. Front. Cell Infect. Microbiol. 2019;9:344. doi: 10.3389/fcimb.2019.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fink Günther, Orlova-Fink Nina, Schindler Tobias, Grisi Sandra, Ferrer Ana Paula S, Daubenberger Claudia, Brentani Alexandra. Inactivated trivalent influenza vaccination is associated with lower mortality among patients with COVID-19 in Brazil. BMJ Evid. Based Med. 2021;26(4):192–193. doi: 10.1136/bmjebm-2020-111549. [DOI] [PubMed] [Google Scholar]
- 30.The effect of influenza vaccination on trained immunity: impact on COVID-19 | medRxiv, (n.d.). https://www.medrxiv.org/content/10.1101/2020.10.14.20212498v1 (accessed March 17, 2021).
- 31.Williams Wilton B., Meyerhoff R. Ryan, Edwards R.J., Li Hui, Manne Kartik, Nicely Nathan I., Henderson Rory, Zhou Ye, Janowska Katarzyna, Mansouri Katayoun, Gobeil Sophie, Evangelous Tyler, Hora Bhavna, Berry Madison, Abuahmad A. Yousef, Sprenz Jordan, Deyton Margaret, Stalls Victoria, Kopp Megan, Hsu Allen L., Borgnia Mario J., Stewart-Jones Guillaume B.E., Lee Matthew S., Bronkema Naomi, Moody M. Anthony, Wiehe Kevin, Bradley Todd, Alam S. Munir, Parks Robert J., Foulger Andrew, Oguin Thomas, Sempowski Gregory D., Bonsignori Mattia, LaBranche Celia C., Montefiori David C., Seaman Michael, Santra Sampa, Perfect John, Francica Joseph R., Lynn Geoffrey M., Aussedat Baptiste, Walkowicz William E., Laga Richard, Kelsoe Garnett, Saunders Kevin O., Fera Daniela, Kwong Peter D., Seder Robert A., Bartesaghi Alberto, Shaw George M., Acharya Priyamvada, Haynes Barton F. Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies. Cell. 2021;184(11):2955–2972.e25. doi: 10.1016/j.cell.2021.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balzarini Jan. Targeting the glycans of glycoproteins: a novel paradigm for antiviral therapy. Nat. Rev. Microbiol. 2007;5(8):583–597. doi: 10.1038/nrmicro1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Structure of 2G12 Fab2 in Complex with Soluble and Fully Glycosylated HIV-1 Env by Negative-Stain Single-Particle Electron Microscopy, (n.d.). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4136306/ (accessed March 17, 2021). [DOI] [PMC free article] [PubMed]
- 34.Pinto Dora, Park Young-Jun, Beltramello Martina, Walls Alexandra C., Tortorici M. Alejandra, Bianchi Siro, Jaconi Stefano, Culap Katja, Zatta Fabrizia, De Marco Anna, Peter Alessia, Guarino Barbara, Spreafico Roberto, Cameroni Elisabetta, Case James Brett, Chen Rita E., Havenar-Daughton Colin, Snell Gyorgy, Telenti Amalio, Virgin Herbert W., Lanzavecchia Antonio, Diamond Michael S., Fink Katja, Veesler David, Corti Davide. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583(7815):290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 35.Ladner J.T., Henson S.N., Boyle A.S., Engelbrektson A.L., Fink Z.W., Rahee F., D’ambrozio J., Schaecher K.E., Stone M., Dong W., Dadwal S., Yu J., Caligiuri M.A., Cieplak P., Bjørås M., Fenstad M.H., Nordbø S.A., Kainov D.E., Muranaka N., Chee M.S., Shiryaev S.A., Altin J.A. Epitope-resolved profiling of the SARS-CoV-2 antibody response identifies cross-reactivity with an endemic human CoV. BioRxiv. 2020 doi: 10.1101/2020.07.27.222943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorse Geoffrey J., Patel Gira B., Vitale Joseph N., O'Connor Theresa Z. Prevalence of antibodies to four human coronaviruses is lower in nasal secretions than in serum. Clin Vaccine Immunol. 2010;17(12):1875–1880. doi: 10.1128/CVI.00278-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan K.-H., Chan J.F.-W., Tse H., Chen H., Lau C.C.-Y., Cai J.-P., Tsang A.K.-L., Xiao X., To K.K.-W., Lau S.K.-P., Woo P.C.-Y., Zheng B.-J., Wang M., Yuen K.-Y. Cross-reactive antibodies in convalescent SARS patients’ sera against the emerging novel human coronavirus EMC by both immunofluorescent and neutralizing antibody tests. J. Infect. 2012;67(2013):130–140. doi: 10.1016/j.jinf.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tso F.Y., Lidenge S.J., Peña P.B., Clegg A.A., Ngowi J.R., Mwaiselage J., Ngalamika O., Julius P., West J.T., Wood C. High prevalence of pre-existing serological cross-reactivity against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) in sub-Saharan Africa. Int. J. Infect. Dis. 2021;102:577–583. doi: 10.1016/j.ijid.2020.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glycan reactive anti-HIV-1 antibodies bind the SARS-CoV-2 spike protein but do not block viral entry | bioRxiv, (n.d.). https://www.biorxiv.org/content/10.1101/2021.01.03.425141v1 (accessed March 17, 2021). [DOI] [PMC free article] [PubMed]
- 40.Tan Shaun S, Chew Ka Lip, Saw Sharon, Jureen Roland, Sethi Sunil. Cross-reactivity of SARS-CoV-2 with HIV chemiluminescent assay leading to false-positive results. J. Clin. Pathol. 2021;74(9) doi: 10.1136/jclinpath-2020-206942. 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flow cytometry plots showing gating strategy to check expression of HIV-Env on HEK-293 cells. Representative contour plots of one sample. Experiment was performed in triplicates.