Abstract
Understanding mechanisms of protective antibody recognition can inform vaccine and therapeutic strategies against SARS-CoV-2. We report a monoclonal antibody, 910-30, targeting the SARS-CoV-2 receptor-binding site for ACE2 as a member of a public antibody response encoded by IGHV3-53/IGHV3-66 genes. Sequence and structural analyses of 910-30 and related antibodies explore how class recognition features correlate with SARS-CoV-2 neutralization. Cryo-EM structures of 910-30 bound to the SARS-CoV-2 spike trimer reveal binding interactions and its ability to disassemble spike. Despite heavy-chain sequence similarity, biophysical analyses of IGHV3-53/3-66-encoded antibodies highlight the importance of native heavy:light pairings for ACE2-binding competition and SARS-CoV-2 neutralization. We develop paired heavy:light class sequence signatures and determine antibody precursor prevalence to be ∼1 in 44,000 human B cells, consistent with public antibody identification in several convalescent COVID-19 patients. These class signatures reveal genetic, structural, and functional immune features that are helpful in accelerating antibody-based medical interventions for SARS-CoV-2.
Keywords: SARS-CoV-2, public antibody, neutralization, yeast display, B-cell, biotechnology, virology, immunity
Graphical abstract
Banach et al. report a SARS-CoV-2 neutralizing antibody along with genetic, structural, and functional features of public antibody responses targeting SARS-CoV-2. These data reveal how structural interactions with the SARS-CoV-2 receptor-binding domain correlate with viral neutralization and demonstrate the importance of native antibody heavy:light pairings in convergent antibody responses.
Introduction
The highly infectious nature of severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) and significant prevalence of severe disease has caused immense global, social, and economic disruption (Cucinotta and Vanelli, 2020; Liu et al., 2020b). SARS-CoV-2 marks the third known emergence of a novel beta-coronavirus in the past 2 decades, following its closest documented human pathogen SARS-CoV in 2002 and Middle East respiratory syndrome (MERS)-CoV in 2012 (Cui et al., 2019; Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020; Graham and Baric, 2010; Ksiazek et al., 2003; de Wit et al., 2016; Zaki et al., 2012). Both SARS and SARS-CoV-2 infect human cells by binding the angiotensin convertase 2 receptor (ACE2) via the trimeric spike (S) class I fusion protein (Hoffmann et al., 2020; Wrapp et al., 2020a). S protein comprises two subunits, S1 and S2. The S1 subunit contains a receptor-binding domain (RBD), which binds to ACE2. To enter cells, S undergoes a protease cleavage event that allows S1 to shed and expose the hydrophobic fusion peptide of the S2 subunit. SARS-CoV predominantly enters cells via endosomes, assisted by cathepsin cleavage in the low pH (5.5–4.5) endosomal environment. SARS-CoV-2 acquired a new protease cleavage site that enables entry either at the cell surface after cleavage with TMPRSS2 or inside endosomes via protease cleavage similar to SARS, and the route of SARS-CoV-2 entry is likely dependent on the protease expression profile in target cells (Ou et al., 2020; Tang et al., 2020). ACE2 interactions appear to play a role in pre-fusion S1 shedding (Benton et al., 2020; Cai et al., 2020).
A detailed understanding of SARS-CoV-2 antibody neutralization should help to accelerate progress in medical interventions. Antibodies from several coronavirus disease 2019 (COVID-19) patients revealed public antibody responses against SARS-CoV-2 via shared genetic and structural elements in IGHV3-53 and IGHV3-66 heavy-chain V-genes. This public antibody class targets a conserved RBD epitope in the S1 subunit that overlaps with the ACE2 binding site (Barnes et al., 2020; Brouwer et al., 2020; Cao et al., 2020; Chi et al., 2020; Du et al., 2020; Hansen et al., 2020; Hurlburt et al., 2020; Liu et al., 2020a; Rogers et al., 2020; Seydoux et al., 2020; Shi et al., 2020; Wu et al., 2020b; Yuan et al., 2020a). IGHV3-53/3-66 public class antibodies share common genetic features, including IGHV-gene-encoded motifs NY in the CDR-H1, SGGS in the CDR-H2, a relatively short CDR-H3 length, and comparatively low levels of antibody somatic hypermutation (SHM) (Barnes et al., 2020; Du et al., 2020; Wu et al., 2020a; Yuan et al., 2020a). Both kappa and lambda light chains are represented in antibodies of this class (Catalan-Dibene, 2020; Du et al., 2020; Wang et al., 2020a; Wrapp et al., 2020b; Wu et al., 2020b, 2020b; Yuan et al., 2020a). Despite heavy-chain gene similarities, IGHV3-53/3-66 anti-RBD antibodies show a broad range of neutralization potencies (half-maximal inhibitory concentrations [IC50s] from 0.003 to 2.547 μg/mL) (Brouwer et al., 2020; Cao et al., 2020; Ju et al., 2020; Liu et al., 2020a; Robbiani et al., 2020; Rogers et al., 2020; Shi et al., 2020; Wu et al., 2020b; Yuan et al., 2020a; Zost et al., 2020). Given the low SHM observed and the importance of germline-encoded recognition motifs, it remains unclear what unique molecular features lead to the diverse range of SARS-CoV-2 neutralization potencies among class members.
SARS-CoV-2 S displays a pH-dependent conformational switch that causes the “up” position of the RBD to rotate to a “down” position (Walls et al., 2020; Zhou et al., 2020c). The RBD-up position is required for ACE2 engagement and for IGHV3-53/3-66 class antibody binding (Du et al., 2020; Walls et al., 2020; Wrapp et al., 2020b). A mutational variant, D614G, also emerged that influences the RBD-up versus -down state, and has constituted >97% of isolates worldwide (Korber et al., 2020; Long et al., 2020; Volz et al., 2021; Yurkovetskiy et al., 2020; Zhang et al., 2020). D614G is proximal to the RBD, and D614G appears to favor more RBD-up at both serological and endosomal pHs (Grubaugh et al., 2020; Yurkovetskiy et al., 2020; Zhou et al., 2020c). The D614G substitution enhances viral infectivity, competitive fitness, and transmission (Hou et al., 2020; Mansbach et al., 2021; Yurkovetskiy et al., 2020; Zhang et al., 2020); further information related to the effects of D614G on IGHV3-53/3-66 class recognition and neutralization is needed (Grubaugh et al., 2020).
Here, we discover a new member of the IGHV3-53/3-66 antibody class, mAb 910-30, with moderate neutralization capacity. To understand the features of potent IGHV3-53/3-66 class neutralization, we explore molecular and genetic features of 910-30 and other related antibodies, including heavy- and light-chain structural recognition motifs, biophysical correlates of neutralization, and the influence of D614 versus D614G variants on IGHV3-53/3-66 class member interactions. Our study provides a detailed molecular understanding of how the public IGHV3-53/3-66 class leverages native heavy- and light-chain binding interactions, providing important data to accelerate medical interventions against the SARS-CoV-2 RBD epitope.
Results
Isolation and structural characterization of a novel neutralizing class member
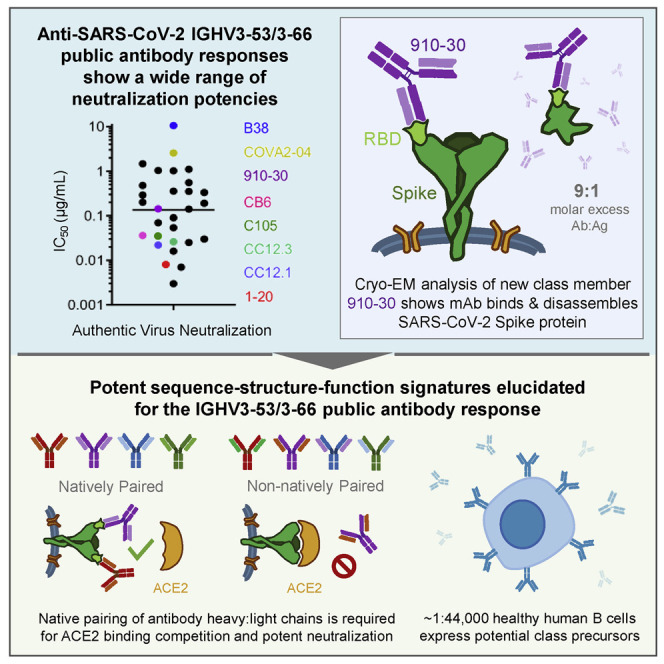
We screened the immune repertoire of a COVID-19 convalescent patient, Donor 910 (To et al., 2020), to identify a new member of the public IGHV3-53/3-66 antibody class. Donor 910 serum ELISA showed potent S trimer recognition, and pseudovirus neutralization assays confirmed potent SARS-CoV-2 serum neutralization (Figure S1) (Wang et al., 2020b). Donor 910 cryopreserved peripheral blood mononuclear cells (PBMCs) were selected for analysis using a recently described method to clone natively paired heavy- and light-chain antibody-variable regions into yeast display for functional screening (Wang et al., 2018). Yeast antibody display libraries were screened for binding against SARS-CoV-2 S protein probes by fluorescence-activated cell sorting (FACS) (Figure S1). Bioinformatic interrogation of yeast display next-generation sequencing (NGS) data revealed that monoclonal antibody (mAb) 910-30 enriched 90-fold in the round 2 sorted library against biotinylated S trimer, and 2,296-fold in round 3 sorted libraries against His-labeled S trimer. Based on its strong enrichment, mAb 910-30 was expressed as immunoglobulin G (IgG) in HEK293 cells for neutralization assays. 910-30 showed an IC50 of 0.071 μg/mL against a vesicular stomatitis virus (VSV) SARS-CoV-2 pseudovirus (Liu et al., 2020a) and 0.142 μg/mL against authentic SARS-CoV-2 (Figure 1 A) (Huo et al., 2020; ter Meulen et al., 2006; Yuan et al., 2020b).
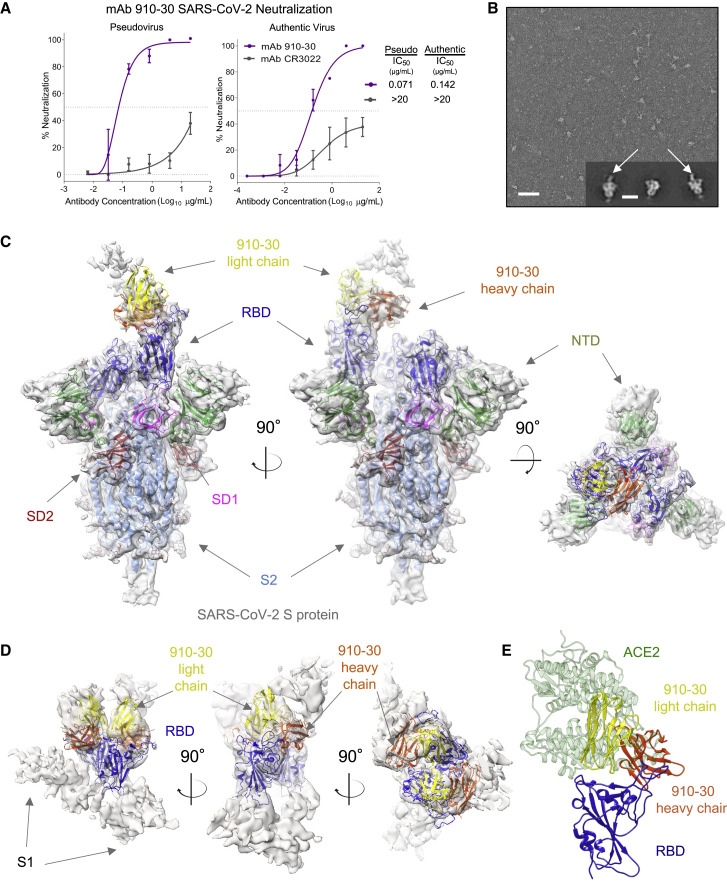
Figure 1.
A novel SARS-CoV-2 neutralizer in the reproducible IGHV3-53/3-66 antibody class targets the ACE2 binding site of both ordered and disassembled spike
(A) Antibody 910-30 shows moderately potent neutralization capacity compared in both VSV-pseudo-type virus and authentic virus assays. Data are represented as means ± SDs.
(B) Representative micrograph of pH 5.5 negative-staining electron microscopy showing 910-30 Fab bound to SARS-CoV-2 S2P protein. Inset shows representative 2D class averages; arrows point to bound Fab fragments. Scale bars: 50 nm (micrographs) 20 nm (2D class averages).
(C) Cryo-EM map and molecular model of 910-30 Fab in complex with SARS-CoV-2 spike at 4.75 Å resolution.
(D) Cryo-EM map obtained of 9:1 molar ratio 910-30 Fab:spike at pH 5.5.
(E) The structural superposition of ACE2 (PDB: 6M0J) and 910-30 (PDB: 7KS9) in complex with SARS-CoV-2 RBD shows a representative ACE2 competition mechanism defining IGHV3-53/3-66 class neutralization.
See also Figures S1 and S2 and Table S1.
910-30 structural recognition was next characterized by cryoelectron microscopy (cryo-EM). Negative-staining EM revealed particles of 910-30 fragment antigen-binding (Fab) bound to SARS-CoV-2 S2P at pH 5.5 (Figure 1B) (Zhou et al., 2020c) and also across a broader pH range of 4.0–7.4 (Figure S1). Subsequent cryo-EM mapping and molecular modeling of 910-30 Fab in complex with SARS-CoV-2 S2P protein at pH 5.5 showed 1 Fab bound to 1 RBD in the up position when mixing Fab and spike in a 1:1 molar ratio (Figures 1C and S2; Table S1), whereas a 9:1 Fab:spike molar ratio revealed mostly disordered spike (Figures 1D and S2; Table S1), with an RBD that still fit the cryo-EM map consistent with Figure 1C. Structural modeling of ACE2 (PDB: 6M0J) and 910-30 (PDB: 7KS9) in complex with SARS-CoV-2 RBD confirmed ACE2 binding site recognition (Figure 1E). Binding analysis against yeast-displayed aglycosylated N343Q RBD (333-537) confirmed that 910-30 recognized a glycan-independent epitope (Figure S3A) (Starr et al., 2020). Antibody titrations showed a 910-30 IgG KD to RBD of 230 pM (191–268 pM, 95% confidence interval) (Figure S3B), and that 910-30 competes with human ACE2 (hACE2) for binding RBD, like other IGHV3-53/3-66 class members (Figure S3C).
Potent antibodies of the IGHV3-53/3-66 class compete strongly with ACE2 for binding to spike
Structural analysis of IGHV3-53/3-66 antibody recognition shows substantial overlap between the ACE2 binding site and the shared class epitope (Figure 2 A) (Barnes et al., 2020; Cao et al., 2020; Hansen et al., 2020; Ju et al., 2020; Liu et al., 2020a; Shi et al., 2020; Walls et al., 2020; Wu et al., 2020b). Despite low reported SHM and common epitopes, IGHV3-53/3-66 antibody class members still show a broad range of neutralization potencies (Figure 2B; Table S2). To better understand the molecular features of potent antibody neutralization in this class, we assessed the biophysical performance of a panel of weak, moderate, and potent IGHV3-53/3-66 class members. We selected 1-20 (a potent neutralizer), 910-30 (a moderate neutralizer), and B38 (a weak neutralizer), along with a VH-gene matched control (mAb 4-3) that neutralizes poorly and likely targets a different site on RBD (Table S3) (Wu et al., 2020a; Zhou et al., 2020a). Preliminary IgG ELISA revealed that the more potent class members (1-20, 910-30) bound more tightly to full-length spike but had similar binding to RBD (Figure 2C). Pseudovirus and authentic virus neutralization assays showed two orders of magnitude across IC50 neutralization potencies for the panel (Figure 2D), confirming that antibody neutralization within the class is driven by more than simple recognition of the ACE2 binding site on RBD.
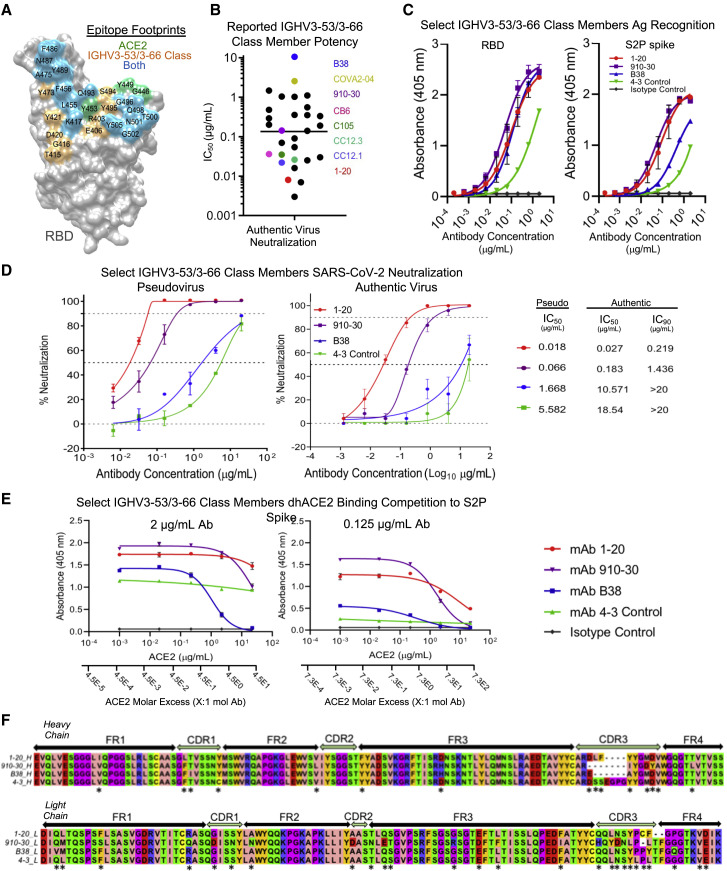
Figure 2.
IGHV3-53/3-66 class neutralization potency is driven by strong competition with ACE2 for spike S2P recognition
(A) Comparison between ACE2 binding site and the IGHV3-53/3-66 class epitope on RBD show substantial overlap.
(B) Dot chart of reported wild-type authentic virus neutralization IC50 titers for previously published IGHV3-53/3-66 anti-RBD antibodies. Data were plotted without correcting for any differences in neutralization assay protocols. The line indicates the mean of IC50 values. A list of antibodies, IC50 values, and citations are provided in Table S2.
(C) IgG ELISA binding titrations for select IGHV3-53/3-66 class members against S2P spike and RBD antigens, with an IGHV gene-matched control (mAb 4-3) and an isotype control. Data are represented as means ± SDs.
(D) Pseudovirus and authentic virus neutralization show that IC50 neutralization potency ranges 2 orders of magnitude between the selected IGHV3-53/3-66 class members, along with an IGHV gene-matched control (mAb 4-3). Data are represented as means ± SDs.
(E) dhACE2 competition ELISA against SARS-CoV-2 S2P spike showing IgG binding response to increasing dhACE2 (ACE2) concentrations. dhACE2 concentration is provided as both μg/mL and as ACE2 molar excess units. Data are represented as means ± SDs.
(F) Sequence alignment of heavy chain (upper) and light chain (lower) genes selected for detailed investigation.
We next assessed the ability of potent and weak neutralizers to compete with dimerized human ACE2 (dhACE2) for binding to spike (S2P). In a competition ELISA with pre-mixed antibody and dhACE2, we found that more potently neutralizing class members competed more strongly with dhACE2 than less potent Abs (Figure 2E). The most potent mAb (1-20) required 73 dhACE2 molecules for 50% binding inhibition of 1 IgG molecule. 1-20 was 6-fold more competitive with dhACE2 than the moderate neutralizer 910-30 (dhACE2 molar excess IC50 = 12) and 150-fold more competitive than B38 (dhACE2 molar excess IC50 = 0.48). Sequence analysis confirmed high sequence similarity and low levels of SHM for the panel, as previously reported for the IGHV3-53/3-66 antibody class (Figure 2F) (Hurlburt et al., 2020; Yuan et al., 2020a).
Unique heavy- and light-chain interactions drive potent neutralization for the IGHV3-53/3-66 neutralizing antibody class
Given the broad variations in potency despite high sequence similarity, we next sought to understand the features associated with potent SARS-CoV-2 neutralization. IGHV3-53/IGHV3-66 anti-SARS-CoV-2 antibodies show diverse light chains, with the two defining heavy-chain genes (IGHV3-53 and IGHV3-66) pairing with at least 14 light-chain genes (Table S2). We constructed a panel of 12 non-native heavy-light swapped antibody variants to explore light-chain pairing features using 4 IGHV3-53/3-66-encoded mAbs (1-20, 910-30, B38, and 4-3 included as an IGHV gene control). Of the 12 non-native antibodies, 11 expressed successfully and were assayed for SARS-CoV-2 pseudovirus neutralization (B38 heavy:1-20 light failed to express). Heavy-light swaps revealed substantially reduced neutralization for nearly all non-native heavy-light combinations, with only the most potent antibody heavy chain (1-20) achieving significant neutralization with a non-native light chain (Figure 3 A). More potent neutralization was correlated with strong dhACE2 competition (Figure 3B), consistent with natively paired antibody performance (Figure 2E), as all four heavy-chain genes have low SHM and high sequence similarity (Figure 2F; Table S3). Figure 3A shows that native light chains make substantial contributions to neutralization performance, and that non-native pairings reduced antibody performance, despite conserved heavy and light sequence signatures. To explore structural features, we generated in silico models of 910-30/B38 heavy:light swaps by superimposing the light chains and replacing the native light chain. Structural alignment of the 910-30/B38 swaps on RBD showed significant complementary determining region-L3 (CDR-L3) displacement, especially at the heavy:light interface. Changes in the heavy:light-chain contact region can affect the orientation of all CDRs, resulting in suboptimal binding geometry for non-natively paired heavy:light swaps. These in silico data suggested that native heavy:light-chain assembly was important for proper CDR orientation and protective RBD recognition in this antibody class.
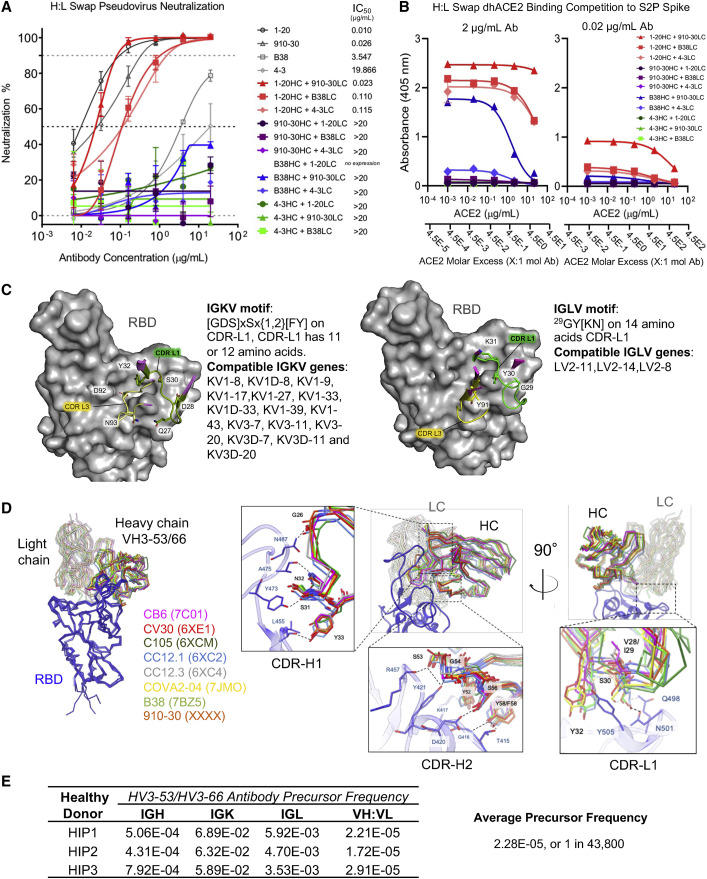
Figure 3.
Heavy- and light-chain analyses reveal critical contributions of both VH and VL for potent antibody neutralization in the IGHV3-53/3-66 antibody class
(A) SARS-CoV-2 pseudovirus neutralization results for heavy-light swapped IgG panel. Data are represented as means ± SDs.
(B) dhACE2 competition ELISA against SARS-CoV-2 S2P protein showing heavy-light swapped IgG binding response to increasing dhACE2 (ACE2) concentrations. dhACE2 concentration is provided in μg/mL and also as ACE2 molar excess units. Data are represented as means ± SDs.
(C) Conserved IGHV3-53/3-66 class light-chain kappa (left panel) and lambda (right panel) genetic elements associated with contacting the RBD interface. CDR-L1 residues are not specific to IGKV1-33 (910-30) and IGLV2-8 (C105) (Table S4).
(D) Left panel: structural superposition of IGHV3-53/3-66 Fab variable domains in complex with RBD shows the same binding orientation for 8 different class antibodies aligned on RBD. Right panel: close-up views of the Fab:RBD interface for the 8 IGHV3-53/3-66 antibodies superimposed on RBD. Conserved interactions of CDR-H1, -H2, and -L1 define the structural signatures responsible for viral neutralization by the IGHV3-53/3-66 antibody class.
(E) Estimated probability of IGHV3-53/3-66 class pre-cursor antibodies derived from healthy donor (HIP1, HIP2, HIP3) immune repertories.
To better understand the determinants of antibody recognition, we next performed a structure-based alignment to analyze interactions and identify light-chain signatures of class membership (Lefranc et al., 2003; Zhu et al., 2013). We followed numerous anti-RBD IGHV3-53/3-66 antibody lineages natively paired with different light chain V-genes, including KV1-33, KV1-9, KV1-39, KV3-20, and LV2-8 (Barnes et al., 2020; Hurlburt et al., 2020; Shi et al., 2020; Wu et al., 2020b; Yuan et al., 2020a). Structural analyses of antibody contact sites revealed that conserved residues in VH and VL genes contributed 56%–75% of the binding surface area (BSA) (Figures 3C and S3D). Figure S3D shows the IGHV3-53/3-66 heavy-chain projected surface with germline-encoded amino acids, including interaction residues 31SNY on CDR-H1 and 52YSGxSxY on CDR-H2 (x indicates any residue) that provide multiple hydrogen bonds interactions with Thr415, Gly416, Lys417, Asp420, Tyr421, Leu455, Tyr473, Ala475, and Asn487 in RBD. We verified previous reports that class members have shorter 6–11 amino acid CDR-H3s, and that key CDR-H1 and CDR-H2 motifs are only present in IGHV3-53/3-66 genes. Sequence-structure alignments revealed that kappa chain class members use a conserved [DGS]xSx{1,2}[FY] motif of 11 or 12 amino acids starting at residue 27a or 28 in the CDR-L1 to form hydrogen bonds with RBD residues Gln498 and Asn501 (Figure 3C). In contrast, lambda chains in the class use a different 29GY[KN] motif with 14 amino acids in CDR-L1 that interacts with RBD residues Gly502 and Tyr505 (Figure 3C). We defined the 27a/28[DGS]xSx{1,2}[FY] motif of 11 or 12 amino acids in the CDR-L1 as a signature of kappa chain class members and the 29GY[KN] motif on 14 amino acids CDR-L1 as a signature of lambda class members. We did not observe conserved residues in the CDR-L3 targeting RBD, and we also note that the CDR-L1 motifs defined here can be encoded by multiple light-chain V-genes (Figure 3C; Table S4).
Structural comparison of eight natively paired IGHV3-53/3-66 class members showed that variable domains bound RBD with the same orientation, reflecting conserved heavy-chain recognition and defining conserved light-chain residues (Figure 3D). Structural alignment revealed a conserved hydrogen bond network responsible for RBD recognition by CDR-H1. The backbone carbonyl of Gly26HC interacts with the amide group of Asn487, the backbone CO of Ser31HC contacts the hydroxyl of Tyr473, the side-chain amide of Asn32HC contacts the carbonyl of Ala475, and the hydroxyl of Tyr33HC acts as hydrogen bond donor to Leu455 backbone CO within CDR-H1. For the CDR-H2, the hydroxyl group of Ser53HC targets both the backbone CO of R457 and the hydroxyl of Tyr421, the latter being involved in a hydrogen bond with the NH of Gly54HC as well; and the hydroxyl of Ser56HC interacts with the carboxyl of Asp420. Overall, CDR-H2 interactions are less conserved among IGHV3-53/3-66 members compared to CDR-H1: the hydrogen bond between the Tyr52HC hydroxyl and the Lys417 amine is observed only in B38 and CV30, while the hydroxyl of Thr415 and the NH of Gly416 are targeted by the hydroxyl of Tyr/Phe58HC only in B38, CV30, and 910-30. Structural comparison of light-chain residues shows a strongly conserved tyrosine (Tyr32LC) in CDR-L1 at the heavy:light interface, which provides a stabilizing hydrophobic environment to the aromatic ring of Tyr505, together with Val28/29LC (in CV30, CC12.3, COVA2-04), Ile29LC (in 910-30, B38, CB6, CC12.1) or Tyr30LC (in C105). For CC12.1, B38, and 910-30, Ser30LC interacts with the side chains of Gln498 and Asn501.
We used published antibody repertoire data and these new defined sequence signatures to estimate the prevalence of antibody class precursors in healthy human immune repertoires (Bräuninger et al., 2001; Sethna et al., 2019; Soto et al., 2019). Antibody lineages with anti-SARS-CoV-2 IGHV3-53/3-66 signatures were identified in ∼1 in 44,000 reported human antibody sequences (Figure 3E) (Soto et al., 2019), which was high compared to previously studied anti-HIV-1 VRC01-class antibody precursors that occur in ∼1 in 1–4 million human antibodies (Zhou et al., 2013). This frequency estimate reflects the prevalence of precursor B cells with the potential to develop into protective mAbs of this antibody class; however, we emphasize that not all 1:44,000 potential precursor B cells are anticipated to develop into protective mAbs. The comparatively high frequency of IGHV3-53/3-66 anti-RBD precursors in human B cells supports the recovery of antibodies from this class in multiple convalescent COVID-19 patients.
RBD-up/-down conformation influences S protein recognition for the IGHV3-53/3-66 antibody class
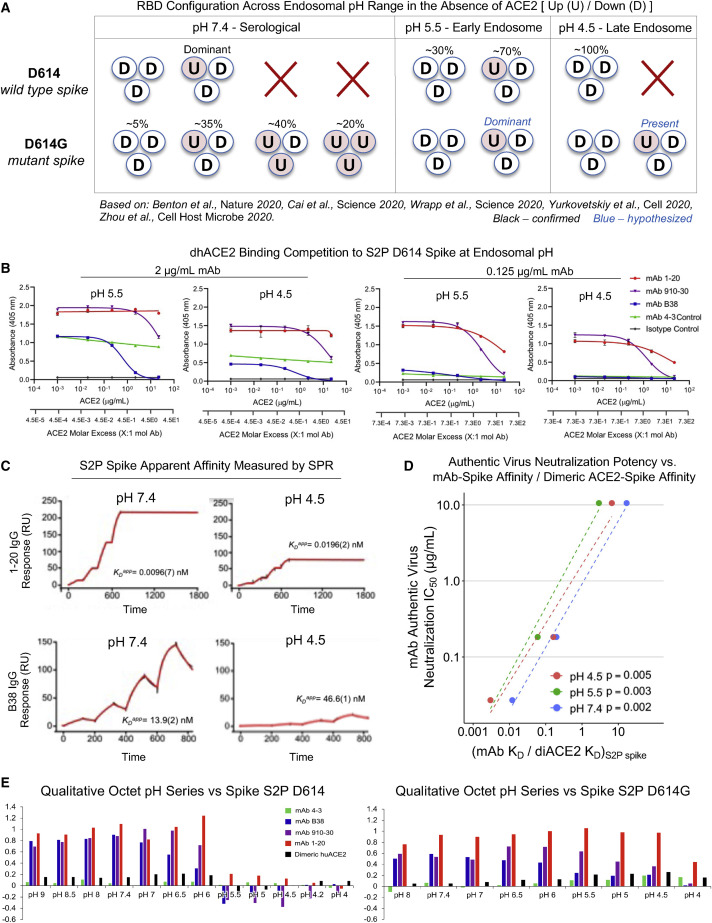
The RBD-up position is required for ACE2 engagement, as well as IGHV3-53/3-66 class antibody binding (Du et al., 2020; Walls et al., 2020; Wrapp et al., 2020b). Cryo-EM analysis at endosomal pH has revealed a pH-mediated conformational switch that rotates RBD domains down at pH 5.5–4.5 (Zhou et al., 2020b), and the D614G mutation also alters RBD-up versus -down dynamics. To understand how pH-based alteration of up versus down RBD prevalence influence IGHV3-53/3-66 class recognition, we investigated antibody binding at three pH values related to known RBD-up versus -down states (Figure 4 A) (Benton et al., 2020; Cai et al., 2020; Walls et al., 2020; Wrapp et al., 2020b; Yurkovetskiy et al., 2020; Zhou et al., 2020b, 2020c). dhACE2 competition ELISA at pH 5.5 and 4.5 showed that IGHV3-53/3-66 class members compete in a concentration-dependent manner with dimeric human ACE2 for binding to SARS-CoV-2 S2P spike and to D614G S2P spike (Figures 4B and S3). Single-cycle surface plasmon resonance showed that the extremely potent mAb 1-20 recognized S protein and RBD with no loss in affinity at endosomal pH, whereas less potent antibodies 910-30 and B38 showed reduced affinity in the endosomal pH range (Figures 4C and S4). We compared authentic virus neutralization IC50 potencies (from Figure 2D) to the ratio of mAb-spike affinity (Figure S4) divided by reported dhACE2-spike affinity (Zhou et al., 2020b), which suggested that potent mAb neutralization was correlated with mAb affinity across all tested pH values (Figure 4D). Finally, a qualitative Octet pH series analysis using D614 S2P spike showed that as pH reduces (and RBDs preferentially rotate down), the potent neutralizer mAb 1-20 exhibited strong recognition of D614 S2P spike for pH 6.0, whereas 910-30 showed reduced binding below pH 6.5, and the least potent B38 binding showed reduced binding below pH 7.0 (Figure 2E, left panel). In contrast, all class members maintained strong binding to mutant D614G S2P spike in the endosomal pH range (where 1 RBD likely remains up), and the potent mAb 1-20 recognized D614G spike down to pH 4.0 (Figure 2E, right panel). Thus, the most potent antibodies maintained a bound state at endosomal pH conditions for D614 S2P spike. We also observed that antibody class members effectively recognized the native RBD-up conformation for D614G across a broad pH range. Our data support ACE2 competition as a functional signature of IGHV3-53/3-66 public antibody class neutralization, and we show that the RBD-up versus RBD-down conformation substantially influenced the ability of IGHV3-53/3-66 class antibodies to recognize spike trimer.
Figure 4.
Up/down conformational changes of RBD influence IGHV3-53/3-56 antibody class recognition of spike protein across the serological to endosomal pH range
(A) Schematic of RBD conformational states inferred by cryo-EM and experimental analysis for un-ligated D614 and D614G spike. U and D denote up and down RBD configurations, respectively. Percentages denote observed particle populations.
(B) dhACE2 competition ELISA at endosomal pH against SARS-CoV-2 S2P D614 protein. dhACE2 concentration is provided as both μg/mL and ACE2 molar excess units. Data are represented as means ± SDs.
(C) Single-cycle SPR kinetic assays for 1-20 and B38 IgG at serological and endosomal pH against biotinylated spike. Black traces represent experimental data and red traces represent the fit to a 1:1 interaction model. The number in parentheses represents the error of the fit in the last digit.
(D) Correlations between authentic virus neutralization potency (Figure 2D) versus the ratio of antibody IgG affinity to spike S2P (Figure S4A) divided by dhACE2 affinity to spike S2P (reported from Zhou et al., 2020b).
(E) Qualitative octet pH series for wild-type S2P spike and escape mutant D614G S2P spike across a range of pH values.
See also Figures S3 and S4.
Discussion
Enhanced understanding of IGHV3-53/3-66 class-based spike recognition can provide insight into immune monitoring, antibody discovery, and vaccine design against SARS-CoV-2. Structural analysis of a novel class member mAb 910-30 revealed previously undescribed spike disassembly at high occupancy, and our antibody class comparative studies showed that native heavy:light pairing was essential for potent neutralization, despite high similarities in heavy-chain sequences. We note that non-native heavy:light swaps still encoded the general structural signatures features required for effective SARS-CoV-2 neutralization by antibodies of this class, and we anticipate that directed evolution could restore effective neutralization of non-native heavy-light antibody pairings.
Comparative sequence-structure analyses enabled the identification of conserved light-chain class signatures, defined as 27a/28[GDS]xSx{1,2}[FY] (kappa) and 29GY[KN] (lambda) residues in CDR-L1 that made important contributions to RBD recognition. We also note that class member light chains use common aromatic/hydrophobic residues 28Val, 29Ile/Val, or 30/32Tyr30/32 to achieve similar interactions with 505Tyr in the RBD, which is part of the shared ACE2 and IGHV3-53/3-66 class binding epitope. These shared light-chain features illuminate the structural rationale for broader light chain diversity among IGHV3-53/3-66 class members. Interestingly, conserved residues in both the heavy and light chain of IGHV3-53/3-66 antibodies targeted RBD residues that have been mutated in emerging SARS-CoV-2 variants. The effect of single mutations observed in B.1.1.7 and B.1.351 on 910-30 neutralization potency has been reported recently (Wang et al., 2021). A decrease in 910-30 efficacy is observed for the K417N and the N501Y mutants, while only a minor change is observed for the E484K mutant. These results are consistent with the structural data, as Lys417 and Asn501 are part of the epitope of 910-30, while Glu484 is not.
The frequency of anti-SARS-CoV-2 IGHV3-53/3-66 precursor antibodies in healthy donors (∼1 in 44,000) was more common than the previously studied anti-HIV-1 VRC01-class antibody precursors observed in 1/1–4 million antibodies (Zhou et al., 2013). In addition, it has been shown that anti-HIV-1 VRC01-class antibodies require much higher levels of SHM to achieve potent neutralization (Zhou et al., 2013). The comparably limited SHM required for anti-SARS-CoV-2 IGHV3-53/3-66 class antibodies appears to be a feature of IGHV germline gene-neutralizing interactions and the need to recognize more similar viral variants, as compared to HIV-1 broadly neutralizing antibodies that must recognize broadly diverse viral variants and show limited germline gene neutralization. These findings help explain the observed reproducibility of public IGHV3-53/3-66 anti-RBD antibodies in convalescent COVID-19 patients.
D614G S2P spike shows a greater prevalence of RBD-up than D614G, which may enhance spike and the ACE2 host receptor recognition to confer higher D614G viral infectivity (Hou et al., 2020; Mansbach et al., 2021; Yurkovetskiy et al., 2020; Zhang et al., 2020). Conversely, a sustained RBD-up also could make the virus more sensitive to neutralization, as the exposed up RBD enhances exposure of vulnerable epitopes (Mansbach et al., 2021; Zhou et al., 2020c). We outlined differences in RBD display caused by the D614G mutation that enhance antibody class recognition of spike across a broad pH range, and we show that D614G had no detrimental impact on IGHV3-53/3-66 antibody class neutralization, which agrees with prior reports (Plante et al., 2021; Weisblum et al., 2020; Weissman et al., 2021). Interestingly, only the most potent antibodies could bind to the D614 variant at endosomal pH, demonstrating that high-affinity antibody recognition can prevent D614 RBD from rotating down at pH 5.5–4.5. These data imply that screening for antibody recognition of D614 S2P at endosomal pH could be an effective method to identify potent anti-SARS-CoV-2 antibodies, and other studies have reported potent antibodies recognizing S trimer even in the context of RBD-down conformations (Tortorici et al., 2020). We also found that ACE2 competition at pH 7.4 was correlated with potent antibody protection, consistent with cell surface attachment via ACE2 at serological pH.
In summary, here, we report the discovery of a new public IGHV3-53/3-66 antibody class member and outline the unique heavy- and light-chain interactions that lead to potent immune recognition of both D614G and D614G spike variants. These data enhance our understanding of the public IGHV3-53/3-66 antibody class and highlight its convergent neutralization features to accelerate anti-SARS-CoV-2 antibody mapping and inform future efforts to identify and elicit neutralizing antibody responses against COVID-19.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal 910-30 | This paper | N/A |
| Monoclonal 1-20 | Ho Lab | Liu et al. 2020a |
| Monoclonal B38 | Ho Lab | Wu et al., 2020b |
| Monoclonal C105 | Bjorkman Lab | Barnes et al., 2020 |
| Monoclonal CR3022 | Neil King and David Veesler at the University of Washington | RRID: AB_2848080 |
| Goat anti-Human IgG Fc PE | Invitrogen | Cat #12-4998-82; RRID: AB_465926 |
| Monoclonal Anti-MYC-FITC | Sigma-Aldrich | Cat #SAB4700448; RRID: AB_10896411 |
| Monoclonal Anti-FLAG-FITC | Sigma-Aldrich | Cat #F4049; RRID: AB_439701 |
| Bacterial and virus strains | ||
| High-efficiency electrocompetent E. coli | New England Biolabs | Cat #C2987H |
| Authentic SARS-CoV-2 | Ho Lab | Ho et al., 2020 |
| AWY101 (MATα AGA1::GAL1-AGA1::URA3 PDI1::GAPDH-PDI1::LEU2 ura3-52 trp1 leu2Δ1 his3Δ200 pep4::HIS3 prb1Δ1.6R can1 GAL) | Eric Shusta Lab, University of Wisconsin-Madison | N/A |
| Biological samples | ||
| Plasma from SARS-CoV-2 patients | Hong Kong University | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 RBD | Vaccine Research Center | Zhou et al., 2020c |
| SARS-CoV-2 S2P | Vaccine Research Center | Zhou et al., 2020c |
| Dna Clean & Concentrator Kits, Zymo Research | Zymo research | Cat #D4005 |
| Expi293 Expression Medium | GIBCO | Cat #A1435102 |
| EasySep Human B cell enrichment kit w/o CD43 depletion | STEMCELL Technologies | Cat #17963 |
| Monarch DNA Gel Extraction Kit | NEB | Cat #T1020 |
| NEBuilder HiFi DNA | NEB | Cat #E5520 |
| SDCAA medium | Teknova | Cat#2S0540 |
| FuGENE 6 | Promega | Cat #E2693 |
| ACE2-Fc | Neil King and David Veesler at the University of Washington. | N/A |
| NHS-Ester biotin | ThermoFisher EZLink Biotin | Cat #20217 |
| YPD medium | ThermoFisher | Cat #242820 |
| Frozen-EZ Yeast Transformation II kit | Zymo Research | Cat #T2001 |
| Human CD27 Microbeads | Miltenyi Biotec | Cat #130-051-601 |
| SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase | Thermo Fisher Scientific | Cat #12574026 |
| Deposited data | ||
| Sequence data | GenBank | MY291105 |
| Sequence data | GenBank | MY291106 |
| Structures | PDB | 7KS9 |
| Electron microscopy data | EMDB | EMD-23016 |
| Electron microscopy data | EMDB | EMD-23039 |
| Supplemental tables | Mendeley | https://doi.org/10.17632/6rtg8t48zk.1 |
| Experimental models: cell lines | ||
| Expi293F human cells | GIBCO | Cat #A14527 |
| Irradiated 3T3-CD40L fibroblast | Gift from John Mascola, Vaccine Research Center, NIAID | N/A |
| Vero cells | ATCC | Cat #CRL-1586 |
| HEK293T | Ho Lab | Ho et al., 2020 |
| Oligonucleotides | ||
| VH:VL amplification and cloning primers | IDT | McDaniel et al., 2016 |
| VH:VL amplification and cloning primers | IDT | Wang et al., 2018 |
| Recombinant DNA | ||
| pCT-VHVL-K1 | NIAID | Wang et al., 2018 |
| BRS-hIgG1 | NIAID | Wang et al., 2018 |
| EVB 114 kc | NIAID | Wang et al., 2018 |
| EVB100_Lc | NIAID | Wang et al., 2018 |
| pCMV3-SARS-CoV-2-spike | Sino Biological | N/A |
| Software and algorithms | ||
| Usearch | USEARCH | https://www.drive5.com/usearch/ |
| MiXCR | MiLaboratory, LLC | https://mixcr.readthedocs.io/en/master/ |
| Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/; RRID: SCR_002798 |
| R | The R Foundation | https://cran.r-project.org |
| tidyverse | R studio | https://www.tidyverse.org/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Brandon J. DeKosky (dekosky@ku.edu).
Materials availability
Plasmids for antibody 910-30 generated in this study are available upon request for non-commercial research purposes.
Experimental model and subject details
Patient samples in the form of peripheral blood mononuclear cells (PBMCs) for B cell sorting were obtained from a convalescent SARS-CoV-2 patient, Donor 910. Cell line Expi293F cell was purchased from Thermo Fisher Scientific. Cell line HEK293T cell was purchased from Sino Biological. Cell line Vero C1008 (Vero-E6 cell) was purchased from ATCC. The cells were maintained and used following manufacturer suggestions and as described in detail below.
Method details
Human sample collection
Informed consent was obtained for all study participants under IRB-AAAS9010 (Hong Kong University). Donor 910 (To et al., 2020) was a 73 year-old male. Patient serum was collected for ELISA and neutralization assays, and PBMCs were cryopreserved for subsequent B cell receptor gene capture and antibody screening.
Expression and purification of SARS-CoV-2 antigens
The antigen probes used for sorting yeast surface displayed libraries were prepared as previously described (Liu et al., 2020a). Briefly, expression vectors encoding the ectodomain of the SARS-CoV-2 S protein was transiently transfected into Expi293 cells and then purified five days post transfection using on-column purification methods.
Production of SARS-CoV-2 pseudovirus
SARS-CoV-2 pseudovirus was generated using recombinant Indiana vesicular stomatitis virus (rVSV) as previously described (Han et al., 2021; Liu et al., 2020a; Nie et al., 2020; Whitt, 2010). HEK293T cells were grown to 80% confluency then used for transfection of pCMV3-SARS-CoV-2-spike (kindly provided by Dr. Peihui Wang, Shandong University, China) using FuGENE 6 (Promega). Cells were cultured to grow overnight at 37 °C with 5% CO2. Then medium was removed and VSV-G pseudotyped ΔG-luciferase (G∗ΔG-luciferase, Kerafast) was harvested to infect the cells in DMEM at a MOI of 3 for 1 h. Then cells were washed three times with 1 × DPBS. DMEM supplemented with anti-VSV-G antibody (I1, mouse hybridoma supernatant from CRL-2700; ATCC) and was added to the inoculated cells. The cells were then cultured overnight. The supernatant was removed the following day and clarified by centrifugation at 300 g for 10 mins before storing at −80 °C.
Emulsion overlap extension RT-PCR and yeast display library generation
B cells were isolated from Donor 910 cryopreserved PBMCs. Non-B cells were depleted by magnetic bead separation, and CD27+ antigen-experienced B cells were isolated by positive magnetic bead separation (EasySep Human B cell enrichment kit w/o CD43 depletion, STEMCELL Technologies, Vancouver, Canada, and CD27 Human Microbeads, Miltenyi Biotec, Auburn, CA, USA). Antigen-experienced B cells (memory B cells) were stimulated in vitro for 5 days to enhance antibody gene transcription. For stimulation, cells were incubated 5 days in the presence of Iscove’s Modified Dulbecco’s Medium (IMDM) (Thermo Fisher Scientific) supplemented with 10% FBS, 1x GlutaMAX, 1x non-essential amino acids, 1x sodium pyruvate and 1x penicillin/streptomycin (Life Technologies) along with 100 units/mL IL-2 and 50 ng/mL IL-21 (PeproTech, Rocky Hill, NJ, USA). B cells were co-cultured with irradiated 3T3-CD40L fibroblast cells that secrete CD40L (kind gift of John Mascola, Vaccine Research Center, NIAID) to aid B cell expansion. Single B cells were captured in emulsion droplets via a flow focusing device with concentric nozzles flowing suspended cells, lysis buffer with mRNA capture magnetic oligo (dT)-coated magnetic beads, and a viscous oil solution to form stable droplets compartmentalizing single B cells with lysis buffer and the mRNA capture beads (McDaniel et al., 2016). Captured beads loaded with single-cell mRNA were re-emulsified and the captured RNA product was reverse transcribed using a SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase (Thermo Fisher Scientific). The specific immunoglobulin VH and VL genes were then processed with an overlap-extension RT-PCR to link native heavy and light chains into a single amplicon, introducing two restriction sites: NheI and NcoI between the VH and VL genes for downstream yeast library generation (Wang et al., 2018). Natively paired antibody heavy and light chain sequencing and yeast surface display library generation were performed as described previously (DeKosky et al., 2013, 2015; Lagerman et al., 2019; McDaniel et al., 2016; Wang et al., 2018).
For yeast library generation, cDNA libraries were amplified with primers containing the yeast display vector restriction sites: AscI and NotI, used for subcloning into the yeast display vector. PCR amplified products were purified by agarose gel extraction and digested with AscI and NotI restriction enzymes followed by subsequent ligation into the yeast display vector backbone. This step was performed in duplicate for each library with separate Kappa- or Lambda- gene-specific primers and a corresponding Kappa or Lambda display vector to generate Kappa and Lambda libraries. Ligated plasmid libraries were transformed into high-efficiency electrocompetent E. coli, expanded overnight, and maxiprepped to isolate the plasmid library DNA product. Maxiprepped plasmid libraries were digested with NheI and NcoI restriction enzymes to remove the native linker from VH:VL pairing. Digested product was purified by agarose gel extraction, and then ligated with a pre-digested DNA gene encoding a bidirectional Gal1/Gal10 promoter inserted between the VH and VL sequences. The resulting ligated product was again transformed into high-efficiency electrocompetent E. coli, expanded overnight, and maxiprepped to isolate the plasmid library DNA product now containing the bidirectional promoter. A final PCR amplification was performed to amplify the VH:bidirectional promoter:VL amplicon with overhanging homologous ends to the pCT backbone for high-efficiency yeast transformation into AWY101 using an homologous recombination method previously described (Benatuil et al., 2010). Transformed libraries were passaged twice in SD-CAA to ensure a 1:1 ratio of plasmid DNA to yeast colony (Benatuil et al., 2010).
FACS screening of yeast libraries
To induce Fab surface expression yeast libraries were incubated in SGD-CAA media at 20°C, 225 rpm for 36 hr. For the first round of sorting, 3x107 presorted cells were washed twice with staining buffer (1x PBS with 0.5% BSA and 2 mM EDTA). Washed yeast display libraries were stained with 20 nM of trimer antigen and a monoclonal anti-FLAG-FITC marker to measure Fab expression (Monoclonal ANTI-FLAG M2-FITC antibody, Sigma-Aldrich, St. Louis, MO, USA). For staining with the NHS-Biotin S-Trimer Protein probe, cells were mixed with 20 nM un-labeled antigen and a monoclonal anti-FLAG-FITC marker (Monoclonal ANTI-FLAG M2-FITC antibody, Sigma-Aldrich, St. Louis, MO, USA) used to measure VL surface expression. This mix was incubated for 15 minutes at 4°C with gentle agitation on a platform shaker. Following incubation, a Streptavidin PE conjugate (Streptavidin, R-Phycoerythrin Conjugate Premium Grade, Thermo Fisher Scientific, Waltham, MA, USA) was added to the re-suspended mix to fluorescently label the biotinylated antigen protein and the sample was again incubated for 15 minutes at 4°C with gentle agitation on a platform shaker. These NHS-Biotin S-Trimer Protein samples were then washed 3x and re-suspended in a final volume of 1 mL in staining buffer before being filtered through a 35 micron-filter cap FACS tube. For staining the with His-Strep-II S-Trimer Protein probe, cells were incubated with un-labeled antigen for 15 minutes at 4°C with gentle agitation on a platform shaker. Samples were then washed 3x with staining buffer, and resuspended in a common mix containing the monoclonal anti-FLAG-FITC marker and a monoclonal anti-His-PE antibody (PE anti-His Tag Antibody, BioLegend, San Diego, CA, USA) to label surface expressed, antigen bound Fab. These cells were again incubated for 15 minutes in the fluorophore mix at 4°C with gentle agitation on a platform shaker. The fluorescently labeled samples were then washed 3x and resuspended in a final volume of 1 mL staining buffer before being filtered through a 35 micron-filter cap FACS tube. Samples were kept in the dark on ice until sorting. Subsequent rounds of enrichment sorting were performed using the same staining procedure, but for only 5x106 input cells and 250 μL final resuspension volume.
A SONY Multi-Application 900 cell sorter running SONY LE-MA900FP Cell Sorter Software was used to detect all FITC+/PE+ cells from each sample and sort them into low pH SD-CAA media. The gating strategy used was previously described (Wang et al., 2018). Sorted yeast were expanded for 24-48 hr at 30°C, 225 rpm and then passaged into SGD-CAA media to induce Fab expression for the next round of sorting. This process was repeated for 3-4 rounds of sorting to enrich for Fab-expressing antigen-binding library populations. In addition to the antigen-positive sorts, an aliquot of each yeast library was washed and stained with only the anti-FLAG-FITC marker, and all FITC+ (i.e., VL+) cells were sorted and sequenced for use as a reference database for NGS enrichment ratio calculations. Analysis of flow cytometry data was conducted using Flowjo10.4 (Flowjo, LLC, Oregon, USA).
NGS analysis of sorted yeast libraries
After each round of FACS enrichment, yeast libraries were expanded via incubation at 30°C for 24-48 hr. An aliquot of this culture was used for high-efficiency yeast plasmid DNA extraction (Whitehead et al., 2012). A high-fidelity polymerase (Kapa Hifi HotStart Mastermix, Kapa Biosystems, Massachusetts, USA) and primers targeting the yeast display vector backbone were used to amplify HC and LC genes from each library (Wang et al., 2018). A second round of primer-extension PCR with barcoded primers added a unique identifier to all HC and LC from a particular library (McDaniel et al., 2016). Sorted libraries were sequenced on the Illumina 2x300 MiSeq platform and sequencing was performed for each library after each round of FACS enrichment. Data processing of Illumina Raw FASTQ data was performed as reported previously (McDaniel et al., 2016; Wang et al., 2018). Briefly, Illumina sequences were quality-filtered to improve read quality, followed by V(D)J gene identification and annotation of CDR3 regions using IgBLAST (Ye et al., 2013). Antibody clonal lineages were tracked across yeast sort rounds by their CDR-H3 amino acid sequence and enrichment ratio. Enrichment ratios were calculated by comparing sequence prevalence in each sorted libraries to the unsorted, Fab-expressing (VL+) antibody library.
Antibody production and purification
The 910-30 antibody was codon-optimized, cloned into mammalian expression plasmid, and expressed as full human antibody IgG1s by co-transfection into Expi293 cells. Heavy and light chain plasmids were co-transfected into Expi293F (ThermoFisher) mammalian cells using the ExpiFectamineTM 293 Transfection Kit (Thermo Fisher Scientific, Massachusetts, USA) and culture in 37°C shaker at 125 rpm and 8% CO2. On day 6 post transfection, the supernatant from transient transfection culture were purified with Protein G or A resin (GenScript, New Jersey, USA) and concentrated using an Amicon Ultra-4 Centrifugal 30K Filter Unit (MilliporeSigma, Maryland, USA), then stored at 4°C.
ELISA binding assays to S trimer and RBD
S trimer and RBD enzyme-linked immunosorbent assays (ELISAs) (Figure 2C) were performed in triplicate. 175 ng of antigen per well was coated onto 96-well ELISA plates at 4 °C overnight. Plates were washed and then blocked with 100 μL of blocking buffer at 37 °C for 2 hr. Purified antibodies were serial diluted using dilution buffer, added to the antigen-coated blocked plates, and then incubated at 4 °C for 1 hr. Plates were washed and 50uL of a secondary anti-human kappa light chain detection antibody (A18853, Invitrogen, Carlsbad, CA) was added to each well and incubated at room temperature for 1 hr. After the final wash, 50 μL TMB substrate (00-4203-56, ThermoFisher Scientific, Waltham, MA) was used to detect antibody binding to antigen measuring absorbance at 405 nm.
Pseudovirus SARS-CoV-2 viral neutralization assay
SARS-CoV-2 pseudovirus neutralization assays were performed as previously described (Liu et al., 2020a). Briefly, pseudovirus particles were generated from recombinant Indiana VSV (rVSV) expressing SARS-CoV-2 S protein. Neutralization was assessed by incubating pseudoviruses with serial dilutions of purified antibody, and scored by the reduction in luciferase gene expression.
Authentic SARS-CoV-2 viral neutralization assay
Authentic virus neutralization assays were performed as previously described (Liu et al., 2020a). Briefly, to measure the neutralizing activity of purified mAbs an end-point dilution assay in a 96-well plate format was performed. Each mAb was 5-fold serially diluted starting at 20 μg/mL in triplicate. Dilutions were incubated with live SARS-CoV-2 for 1 hr at 37°C, and post-incubation the virus-antibody mixture was transferred onto a monolayer of Vero-E6 cells and incubated for 70 hr. CPE from the resulting cell incubations were visually scored for each well in a blinded fashion by two independent observers.
dhACE2 competition ELISA
Antibodies were assayed for dhACE2 competition by enzyme-linked immunosorbent assays (ELISAs) (Figures 2E, 3C, and S3I) in triplicate. ELISA experiments were performed in parallel at three pH values 7.4, 5.5, and 4.5. ELISA 96-well plates were coated with 175 ng per well of antigen in pH-adjusted PBS and incubated at room temperature for 1 hr. Ag-coated ELISA plates were washed and blocked with 100 μL of blocking buffer and incubated at room temperature for 1 hr. Purified antibodies were serial diluted and pre-mixed with dhACE2 using pH-adjusted dilution buffer. Ab:dhACE2 premixes were added to the pre-blocked, antigen-coated plates and incubated at room temperature for 2 hr. Plates were washed and 50 μL of 1:2000 diluted, pH-adjusted secondary anti-human kappa light chain detection antibody (A18853, Invitrogen, Carlsbad, CA) solution was added to each well and incubated at room temperature for 1 hr. After the final wash, 50 μL Super AquaBlue substrate was used to detect antibody binding to antigen measuring absorbance at 405 nm.
RBD glycan recognition analysis via yeast display
For plasmid construction, pJS699 (S-RBD (333-537)-N343Q for fusion to the C terminus of AGA2) was synthesized by PCR amplifying pUC19-S-ecto with primers PJS-P2196/PJS-P2197 (2.9kb) and PJS-P2198/PJS-P2199 (0.65kb). The resulting products were fractionated by agarose gel electrophoresis and the bands corresponding to the desired products were excised from the gel and purified using a Monarch DNA Gel Extraction Kit (NEB). The fragments were assembled using NEBuilder HiFi DNA assembly master mix (NEB) according to the manufacturer’s instructions and 5 μL of the reaction was transformed into chemically competent E. coli Mach1 (Invitrogen) and selected on LB agar supplemented with 50 μg/mL kanamycin.
To create the display construct of S-RBD (333-537)-N343Q fused to the C terminus of Aga2p, pJS697 was digested with BsaI-HFv2 (NEB) and purified using a Monarch PCR & DNA Cleanup Kit (NEB). pJS699 was digested with NotI-HF (NEB), the reaction fractionated by agarose gel electrophoresis, and the band corresponding to S-RBD (0.83kb) excised and purified using a Monarch DNA Gel Extraction Kit (NEB). The two fragments were co-transformed (in a 2.4:1 molar ratio of S-RBD to backbone) into chemically competent S. cerevisiae EBY100 (Boder and Wittrup, 1997) and selected on M19D agar. M19D contained 5 g/L casamino acids, 40 g/L dextrose, 80 mM 2-(N-morpholino) ethanesulfonic acid (MES free acid), 50 mM citric acid, 50 mM phosphoric acid, 6.7 g/L Yeast Nitrogen Base (Sigma), and was adjusted to pH 7 with 9M NaOH, 1M KOH.
Recombinant human ACE2-Fc and CR3022 were received as a gift from Neil King and David Veesler at the University of Washington. Human ACE2-Fc was produced and purified as described (Walls et al., 2020). CR3022 (ter Meulen et al., 2006) was expressed by transient transfection in Expi293F cells and purified by protein A affinity chromatography and SEC using a Superdex 200 10/300 GL. Specificity was verified by measuring binding to SARS-CoV-2 RBD and irrelevant antigen.
For yeast display screening, EBY100 harboring the RBD display plasmid was grown in 1 mL M19D overnight at 30°C. Expression was induced by resuspending the M19D culture to OD600 = 1 in M19G (5 g/L casamino acids, 40 g/L galactose, 80 mM MES free acid, 50 mM citric acid, 50 mM phosphoric acid, 6.7 g/L yeast nitrogen base, adjusted to pH 7 with 9 M NaOH, 1 M KOH) and growing 22 hr at 22°C with shaking at 300 rpm. Yeast surface display titrations were performed as described (Chao et al., 2006) with an incubation time for 910-30 of 4 hr and using secondary labels anti-c-myc-FITC (Miltenyi Biotec) and Goat anti-Human IgG Fc PE conjugate (Invitrogen Cat. No. 12-4998-82). Titrations were performed in biological replicate.
Glycosylation-independent binding for antibody 910-30
EBY100 harboring the RBD display plasmid was grown in 1 mL M19D overnight at 30°C. Expression was induced by resuspending the M19D culture to OD600 = 1 in M19G (5 g/L casamino acids, 40 g/L galactose, 80 mM MES free acid, 50 mM citric acid, 50 mM phosphoric acid, 6.7 g/L yeast nitrogen base, adjusted to pH 7 with 9 M NaOH, 1 M KOH) and growing 22 h at 22°C with shaking at 300 rpm. Yeast surface display titrations were performed as described (Chao et al., 2006) with an incubation time for 910-30 of 4 hr at room temperature and the secondary labels anti-c-myc-FITC (Miltenyi Biotec) and Goat anti-Human IgG Fc PE conjugate (Invitrogen Catalog # 12-4998-82). Titrations were performed in biological replicate (n = 2) with three technical replicates.
910-30 IgG was chemically biotinylated using NHS-Ester biotin (ThermoFisher EZ-Link Biotin Cat. No. 20217) at a 20:1 molar ratio of biotin:IgG according to manufacturer’s instructions. 1x105 yeast cells were labeled with no protein or 100 nM non-biotinylated CR3022, hACE2 or 910-30 for 30 min at room temperature in PBSF (PBS containing 1 g/L BSA). The same cells were then labeled with 1 nM chemically biotinylated 910-30, in the same tube without washing, for 30 min at room temperature in PBSF. The cells were centrifuged and washed with 200 μL PBSF. They were labeled with 0.6 μL FITC, 0.25 μL SAPE and 49.15 μL PBSF for 10 min at 4°C. Cells were then centrifuged, washed with PBSF, and analyzed on a flow cytometer. Experiments were performed with three technical replicates and two biological replicates.
Delineation of IGHV3-53/3-66 sequence signatures
A structure-based method was applied to define sequence signatures for the HV3-53/3-66 class COVID neutralizing antibody (Zhu et al., 2013). Briefly, protein structures of IGHV3-53/3-66 antibodies complexed with RBD or spike were selected for analysis, and the buried surface area (BSA) between antibody and RBD was calculated by the PDBePISA server (https://www.ebi.ac.uk/pdbe/pisa/). We examined the BSA larger than 20 Å2, and residues making contacting with the RBD projected surface that were encoded by the conserved germline sequence were selected as initial class sequence signatures, and amino acids from somatic hypermutations were used to refine the signature of the class antibody. For germline sequence alignments, heavy and light chain germline sequences were downloaded from IMGT (Lefranc et al., 2003) and the sequences of CDR1 and CDR2 were extracted and aligned based on Kabat numbering. ANARCI server was used to number amino acid sequences of antibody (http://opig.stats.ox.ac.uk/webapps/newsabdab/sabpred/anarci).
Antibody class frequency estimation
The frequency of antibody class was estimated using OLGA software based on defined motif (Sethna et al., 2019). NGS samples of three healthy donors (NCBI Short Read Archive accession code: PRJNA511481) were used to analyze heavy and light chain lineage precursor frequencies (Soto et al., 2019). The ratio of human kappa and light chain (60:40) was obtained from Bräuninger et al. (2001). Antibody class precursor frequency was calculated as:
(Frequency of heavy chain × Frequency of kappa chain × kappa chain ratio) + (Frequency of heavy chain × Frequency of lambda chain × lambda chain ratio)
Negative stain cryo-EM
Samples were diluted to a spike concentration of about 20 μg/ml. A 4.7-μl drop of the diluted sample was applied to a glow-discharged carbon-coated copper grid. The grid was washed with a buffer with the same pH as the sample buffer (10 mM HEPES with 150 mM NaCl for pH 7.4; 10 mM acetate with 150 mM NaCl for the lower pH values). Protein molecules adsorbed to the carbon were negatively stained with 0.75% uranyl formate. Datasets were collected using a ThermoFisher Talos F200C electron microscope equipped with a Ceta CCD camera. The microscope was operated at 200 kV, the pixel size was 2.53 Å (nominal magnification: 57,000), and the defocus was set at −1.2 μm. Particles were picked and extracted automatically using in-house written software (data not shown). 2D classification was performed using Relion 1.4 (Scheres, 2012).
Cryo-EM sample preparation
SARS-CoV-2 S2P spike was expressed and purified as described in Wrapp et al. (2020a). 910-30 Fab was prepared by incubating the full 910-30 IgG with immobilized papain for 3 hr at 37°C in 50 mM phosphate buffer, 120 mM NaCl, 30 mM cysteine, 1 mM EDTA, pH 7. Purified SARS-CoV-2 spike was diluted to a final trimer concentration of 0.33 mg/mL and mixed with 910-30 Fab in a 1:1 molar ratio (sample 1) or 1:9 molar ratio (sample 2). The final buffer for both samples was 10 mM sodium acetate, 150 mM NaCl, pH 5.5; 0.005% w/v n-Dodecyl β-D-maltoside (DDM) was added to the mixture to prevent aggregation during vitrification. After incubation for 1 hour on ice, a volume of 2 μL was applied to a glow-discharged carbon-coated copper grid (CF 1.2/1.3 300 mesh) and vitrified using a Vitrobot Mark IV with a wait time of 30 s and a blot time of 3 s.
Cryo-EM data collection, processing and model fitting
Cryo-EM data were collected using the Leginon software (Suloway et al., 2005) installed on a Titan Krios electron microscope operating at 300 kV, equipped with a Gatan K3-BioQuantum direct detection device. The total dose was fractionated for 3 s over 60 raw frames. Data processing including motion correction, CTF estimation, particle picking and extraction, 2D classification, ab initio model generation, 3D refinements and local resolution estimation for both sample 1 and sample 2 datasets were carried out in cryoSPARC 2.15 (Punjani et al., 2017). The coordinates of SARS CoV-2 spike with 1 RBD up, (PDB: 6VYB) (Walls et al., 2020), were employed as initial template to model the cryo-EM map of 910-30 Fab in complex with SARS-CoV-2 spike (sample 1). The RBDs were modeled using the crystallographic structure of RBD in complex with B38 Fab (PDB: 7BZ5) (Wu et al., 2020b) as a template. The variable region of 910-30 Fab was modeled separately (PDB: 7BZ5, 5SX4 (Sickmier et al., 2016) for the heavy and light chain. The residues at the Fab:RBD interface were modeled by structural comparison of 910-30 Fab with 7 different antibodies belonging to the IGHV3-53/3-66 class. Automated and manual model building were iteratively performed using real space refinement in Phenix (Adams et al., 2004) and Coot (Emsley and Cowtan, 2004) respectively. EMRinger (Barad et al., 2015) and Molprobity (Davis et al., 2004) were used to validate geometry and check structure quality at each iteration step. UCSF Chimera (Pettersen et al., 2004) and Chimera X (Pettersen et al., 2021) were used to calculate map-fitting cross correlation (Fit-in-Map tool) and to prepare figures.
Octet binding experiments
Binding of mAbs 4-3, B38, 910-30, and 1-20 to SAR-CoV-2 S2P D614 and D614G variants was assessed an a FortéBio Octet HTX instrument (FortéBio). Experiments were run in tilted black 384-well plates (Geiger Bio-One) at 30°C and 1,000 rpm agitation. Running buffer was comprised of 10mM of the corresponding pH buffer plus 150mM NaCl, 0.02% Tween20, 0.1% BSA and 0.05% sodium azide. The following buffers were used to achieve the range of pH: pH 9 (borate), pH 8.5 (Tris), pH 8 (Tris), pH 7.4 (PBS), pH 7 (HEPES), pH 6.5 (MES), pH 6 (MES), pH 5.5 (NaAc), pH 5 (NaAc), pH 4.5 (NaAc), pH 4.2 (NaAc), pH 4.0 (NaAc). 300 nM IgG solution was used for immobilization at pH 7.4 on anti-human IgG Fc capture biosensors (FortéBio) that were pre-hydrated for 30 minutes. Sensors were then equilibrated in pH 7.4 buffer for 30 s followed by 180 s in the altered pH buffer. Binding was assessed at 200nM S2P D614 or D614G and response recorded for 180 s. Dissociation in the respective buffer was measured for 300 s. The Data Analysis Software HT v12.0 (Fortebio) was used to subtract reference well signal from loaded sensor dipped into buffer without spike protein. The maximum association response (nm) is reported at each pH.
SPR binding experiments
SPR binding experiments were performed using a Biacore T200 biosensor, equipped with a Series S SA chip. The running buffer varied depending on the pH of the binding reaction; experiments at pH 7.4 were performed in a running buffer of 10 mM HEPES pH 7.4, 150 mM NaCl, 0.1% (v/v) Tween-20; at pH 5.5 experiments were performed in 10mM sodium acetate pH 5.5, 150 mM NaCl, 0.1% (v/v) Tween-20; and at pH 4.5 in 10 mM sodium acetate pH 4.5, 150 mM NaCl, 0.1% (v/v) Tween-20. All measurements were performed at 25°C.
Biotinylated S2P was captured over independent flow cells at 750-900 RU. 910-30 and 1-20 IgGs were tested over the biotinylated S2P surfaces at four concentrations ranging from 1-27nM, while B38 and 4-3 were tested at four concentrations ranging from 3-81 nM, to account for higher binding KDs. Biotinylated RBD was captured over independent flow cells at 250-500 RU and B38 was tested at four concentrations ranging from 3-81 nM, 910-30 and 4-3 were tested at four concentrations of 1-27 nM and 1-20 at four concentrations ranging from 0.333-9 nM, to account for differences in their binding affinities. To avoid the need for surface regeneration that arises with the slowly dissociating interactions, we used single-cycle kinetics binding experiments. The four concentrations for each IgG were prepared in running buffers at each of pH, using a three-fold dilution series.
Binding of 910-30, 4-3 and B38 over the S2P or RBD surface as well as over a streptavidin reference surface was monitored for 120 s, followed by a dissociation phase of 120-1080 s depending on the interaction at 50 μL/min. For the interaction of 1-20 with the RBD, which showed an unusually slow dissociation rate, an extended dissociation phase of 4500 s was necessary to extrapolate accurate apparent dissociation constants. Four blank buffer single cycles were performed by injecting running buffer instead of Fab to remove systematic noise from the binding signal. The data was processed and fit to 1:1 single cycle model using the Scrubber 2.0 (BioLogic Software). The results from these assays, are reported in terms of apparent kinetic parameters and KDs to account for potential avidity effects arising from the binding of bivalent IgGs to trivalent S2P.
Quantification and statistical analysis
IC50 calculations were reported using GraphPad Prism software (version 8.4.3). Briefly, experimental data was imported and modeled using a least-squares regression method to fit the data to a variable slope (four parameter) inhibitor versus response curve with bottom parameters constrained to zero. Flow cytometry analysis was carried out using FlowJo software (version 10.4). The Spearman rank order correlation was calculated using cor.test function in base R. Spearman ρ and the p values for the test were used to determine the strength of the correlation between tested variables.
Acknowledgments
We thank Jennifer Hackett for help with Illumina sequencing, and R. Grassucci, Y.-C. Chi, and Z. Zhang from the Columbia University Cryo-EM Center for assistance with data collection. This work was supported by the University of Kansas Departments of Pharmaceutical Chemistry and Chemical Engineering, COVID-19 Fast Grants, the Jack Ma Foundation, the American Lung Association, the Madison and Lila Self Graduate Fellowship Program, the Balsells Fellowship program, the Intramural Research Program of the Vaccine Research Center, the National Institute of Allergy and Infectious Diseases, the National Institutes of Health (NIH), and by NIH grants DP5OD023118, R01AI141452, R21AI143407, and R21AI144408. This work was supported in part with federal funds from the Frederick National Laboratory for Cancer Research, NIH, under contract no. HHSN261200800001.
Author contributions
B.B.B., G.C., A.S.F., C.-H.S., S.N.L.A., K.-Y.Y., T.A.W., D.D.H., P.D.K., L.S., and B.J.D. designed the experiments. B.B.B., G.C., A.S.F., C.-H.S., M.O., P.S.K., Y.T., P.W., M.S.N., Y.H., I.M.F.U., P.J.S., L.L., S.N.L.A., A.F.N., J.R.W., Y.L., X.P., B.M., A.D.L., and R.N. performed the experiments. A.S.O., I.-T.T., J.Y., T.Z., E.R.R., and J.B. provided reagents for the experiments. B.B.B., G.C., A.S.F., C.-H.S., P.S.K., I.M.F.U., P.J.S., M.G.-G., B.M., S.N.L.A., X.P., and B.J.D. analyzed the data. B.B.B., G.C., A.S.F., C.-H.S., P.D.K., L.S., and B.J.D. wrote the manuscript, with feedback from all of the authors.
Declaration of interests
B.B.B., A.S.F., M.O., P.W., L.L., S.N.L.A., J.R.W., X.P., B.M., D.D.H., and B.J.D. declare competing financial interests in the form of a provisional patent application filed by the University of Kansas.
Published: October 5, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109771.
Supplemental information
Data and code availability
Cryo-EM coordinates and maps are deposited in the Protein Data Bank with accession code 7KS9 and in the Electron Microscopy Data Bank with accession code EMD-23016 (trimeric spike) and EMD-23039 (disrupted spike). The 910-30 neutralizing antibody variable heavy and variable light chain sequences have been deposited in GenBank with accession numbers MY291105 and MY291106, respectively. This study did not generate any unique code.
References
- Adams P.D., Gopal K., Grosse-Kunstleve R.W., Hung L.-W., Ioerger T.R., McCoy A.J., Moriarty N.W., Pai R.K., Read R.J., Romo T.D., et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J. Synchrotron Radiat. 2004;11:53–55. doi: 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]
- Barad B.A., Echols N., Wang R.Y.-R., Cheng Y., DiMaio F., Adams P.D., Fraser J.S. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods. 2015;12:943–946. doi: 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benatuil L., Perez J.M., Belk J., Hsieh C.-M. An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng. Des. Sel. 2010;23:155–159. doi: 10.1093/protein/gzq002. [DOI] [PubMed] [Google Scholar]
- Benton D.J., Wrobel A.G., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588:327–330. doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boder E.T., Wittrup K.D. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- Bräuninger A., Goossens T., Rajewsky K., Küppers R. Regulation of immunoglobulin light chain gene rearrangements during early B cell development in the human. Eur. J. Immunol. 2001;31:3631–3637. doi: 10.1002/1521-4141(200112)31:12<3631::aid-immu3631>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y., Zhang J., Xiao T., Peng H., Sterling S.M., Walsh R.M., Jr., Rawson S., Rits-Volloch S., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 2020;584:450–456. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalan-Dibene J. Human antibodies can neutralize SARS-CoV-2. Nat. Rev. Immunol. 2020;20:350. doi: 10.1038/s41577-020-0313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao G., Lau W.L., Hackel B.J., Sazinsky S.L., Lippow S.M., Wittrup K.D. Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO Declares COVID-19 a Pandemic. Acta Biomed. 2020;91:157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis I.W., Murray L.W., Richardson J.S., Richardson D.C. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKosky B.J., Ippolito G.C., Deschner R.P., Lavinder J.J., Wine Y., Rawlings B.M., Varadarajan N., Giesecke C., Dörner T., Andrews S.F., et al. High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire. Nat. Biotechnol. 2013;31:166–169. doi: 10.1038/nbt.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKosky B.J., Kojima T., Rodin A., Charab W., Ippolito G.C., Ellington A.D., Georgiou G. In-depth determination and analysis of the human paired heavy- and light-chain antibody repertoire. Nat. Med. 2015;21:86–91. doi: 10.1038/nm.3743. [DOI] [PubMed] [Google Scholar]
- Du S., Cao Y., Zhu Q., Wang G., Du X., He R., Xu H., Zheng Y., Wang B., Bai Y., et al. Structures of potent and convergent neutralizing antibodies bound to the SARS-CoV-2 spike unveil a unique epitope responsible for exceptional potency. Cell. 2020;183:1013–1023.e13. [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Graham R.L., Baric R.S. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J. Virol. 2010;84:3134–3146. doi: 10.1128/JVI.01394-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh N.D., Hanage W.P., Rasmussen A.L. Making Sense of Mutation: What D614G Means for the COVID-19 Pandemic Remains Unclear. Cell. 2020;182:794–795. doi: 10.1016/j.cell.2020.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Duan X., Yang L., Nilsson-Payant B.E., Wang P., Duan F., Tang X., Yaron T.M., Zhang T., Uhl S., et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature. 2021;589:270–275. doi: 10.1038/s41586-020-2901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schäfer A., Nakajima N., Takahashi K., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo J., Zhao Y., Ren J., Zhou D., Duyvesteyn H.M.E., Ginn H.M., Carrique L., Malinauskas T., Ruza R.R., Shah P.N.M., et al. Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike. Cell Host Microbe. 2020;28:445–454.e6. doi: 10.1016/j.chom.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlburt N.K., Seydoux E., Wan Y.-H., Edara V.V., Stuart A.B., Feng J., Suthar M.S., McGuire A.T., Stamatatos L., Pancera M. Structural basis for potent neutralization of SARS-CoV-2 and role of antibody affinity maturation. Nat. Commun. 2020;11:5413. doi: 10.1038/s41467-020-19231-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Sheffield COVID-19 Genomics Group Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., et al. SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Lagerman C.E., López Acevedo S.N., Fahad A.S., Hailemariam A.T., Madan B., DeKosky B.J. Ultrasonically-guided flow focusing generates precise emulsion droplets for high-throughput single cell analyses. J. Biosci. Bioeng. 2019;128:226–233. doi: 10.1016/j.jbiosc.2019.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc M.-P., Pommié C., Ruiz M., Giudicelli V., Foulquier E., Truong L., Thouvenin-Contet V., Lefranc G. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev. Comp. Immunol. 2003;27:55–77. doi: 10.1016/s0145-305x(02)00039-3. [DOI] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.-W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Liu Y., Ning Z., Chen Y., Guo M., Liu Y., Gali N.K., Sun L., Duan Y., Cai J., Westerdahl D., et al. Aerodynamic analysis of SARS-CoV-2 in two Wuhan hospitals. Nature. 2020;582:557–560. doi: 10.1038/s41586-020-2271-3. [DOI] [PubMed] [Google Scholar]
- Long S.W., Olsen R.J., Christensen P.A., Bernard D.W., Davis J.J., Shukla M., Nguyen M., Saavedra M.O., Yerramilli P., Pruitt L., et al. Molecular Architecture of Early Dissemination and Massive Second Wave of the SARS-CoV-2 Virus in a Major Metropolitan Area. mBio. 2020;11:e02707-20. doi: 10.1128/mBio.02707-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansbach R.A., Chakraborty S., Nguyen K., Montefiori D.C., Korber B., Gnanakaran S. The SARS-CoV-2 Spike Variant D614G Favors an Open Conformational State. Sci. Adv. 2021;7:eabf3671. doi: 10.1126/sciadv.abf3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel J.R., DeKosky B.J., Tanno H., Ellington A.D., Georgiou G. Ultra-high-throughput sequencing of the immune receptor repertoire from millions of lymphocytes. Nat. Protoc. 2016;11:429–442. doi: 10.1038/nprot.2016.024. [DOI] [PubMed] [Google Scholar]
- Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L., Nie L., Qin H., Wang M., et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg. Microbes Infect. 2020;9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., et al. Spike mutation D614G alters SARS-CoV-2 fitness and neutralization susceptibility. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H.W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethna Z., Elhanati Y., Callan C.G., Walczak A.M., Mora T. OLGA: fast computation of generation probabilities of B- and T-cell receptor amino acid sequences and motifs. Bioinformatics. 2019;35:2974–2981. doi: 10.1093/bioinformatics/btz035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux E., Homad L.J., MacCamy A.J., Parks K.R., Hurlburt N.K., Jennewein M.F., Akins N.R., Stuart A.B., Wan Y.-H., Feng J., et al. Analysis of a SARS-CoV-2-Infected Individual Reveals Development of Potent Neutralizing Antibodies with Limited Somatic Mutation. Immunity. 2020;53:98–105.e5. doi: 10.1016/j.immuni.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Sickmier E.A., Kurzeja R.J.M., Michelsen K., Vazir M., Yang E., Tasker A.S. The Panitumumab EGFR Complex Reveals a Binding Mechanism That Overcomes Cetuximab Induced Resistance. PLoS ONE. 2016;11:e0163366. doi: 10.1371/journal.pone.0163366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C., Bombardi R.G., Branchizio A., Kose N., Matta P., Sevy A.M., Sinkovits R.S., Gilchuk P., Finn J.A., Crowe J.E., Jr. High frequency of shared clonotypes in human B cell receptor repertoires. Nature. 2019;566:398–402. doi: 10.1038/s41586-019-0934-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Crawford K.H., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., Veesler D., Bloom J.D. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C.S., Carragher B. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L.M., Marissen W.E., Leung C.S.W., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E., et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., Yip C.C.-Y., Cai J.-P., Chan J.M.-C., Chik T.S.-H., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E.M., Hill V., McCrone J.T., Price A., Jorgensen D., O’Toole A., Southgate J.A., Johnson R., Jackson B., Nascimento F.F., et al. Evaluating the effects of SARS-CoV-2 Spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184:64–75.e11. doi: 10.1016/j.cell.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., DeKosky B.J., Timm M.R., Lee J., Normandin E., Misasi J., Kong R., McDaniel J.R., Delidakis G., Leigh K.E., et al. Functional interrogation and mining of natively paired human VH:VL antibody repertoires. Nat. Biotechnol. 2018;36:152–155. doi: 10.1038/nbt.4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Liu L., Nair M.S., Yin M.T., Luo Y., Wang Q., Yuan T., Mori K., Solis A.G., Yamashita M. SARS-CoV-2 Neutralizing Antibody Responses Are More Robust in Patients with Severe Disease. Emerg. Microbes Infect. 2020;9:2091–2093. doi: 10.1080/22221751.2020.1823890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C.C., Muecksch F., Rutkowska M., Hoffmann H.-H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman D., Alameh M.-G., de Silva T., Collini P., Hornsby H., Brown R., LaBranche C.C., Edwards R.J., Sutherland L., Santra S., et al. D614G Spike Mutation Increases SARS CoV-2 Susceptibility to Neutralization. Cell Host Microbe. 2021;29:23–31.e4. doi: 10.1016/j.chom.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead T.A., Chevalier A., Song Y., Dreyfus C., Fleishman S.J., De Mattos C., Myers C.A., Kamisetty H., Blair P., Wilson I.A., Baker D. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat. Biotechnol. 2012;30:543–548. doi: 10.1038/nbt.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitt M.A. Generation of VSV pseudotypes using recombinant ΔG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J. Virol. Methods. 2010;169:365–374. doi: 10.1016/j.jviromet.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Hoffmann M., Pöhlmann S., et al. VIB-CMB COVID-19 Response Team Structural Basis for Potent Neutralization of Betacoronaviruses by Single-Domain Camelid Antibodies. Cell. 2020;181:1004–1015.e15. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N.C., Yuan M., Liu H., Lee C.D., Zhu X., Bangaru S., Torres J.L., Caniels T.G., Brouwer P.J.M., van Gils M.J., et al. An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain. Cell Rep. 2020;33:108274. doi: 10.1016/j.celrep.2020.108274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Ma N., Madden T.L., Ostell J.M. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell. 2020;183:739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., Ojha A., Rangarajan E.S., Izard T., Farzan M., Choe H. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. 2020 doi: 10.1101/2020.06.12.148726. [DOI] [Google Scholar]
- Zhou T., Zhu J., Wu X., Moquin S., Zhang B., Acharya P., Georgiev I.S., Altae-Tran H.R., Chuang G.-Y., Joyce M.G., et al. NISC Comparative Sequencing Program Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Duyvesteyn H.M.E., Chen C.-P., Huang C.-G., Chen T.-H., Shih S.-R., Lin Y.-C., Cheng C.-Y., Cheng S.-H., Huang Y.-C., et al. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 2020;27:950–958. doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Tsybovsky Y., Olia A.S., Gorman J., Rapp M.A., Cerutti G., Katsamba P.S., Nazzari A., Schön A., Wang P., et al. Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains. Cell Host Microbe. 2020;28:867–879.e5. doi: 10.1016/j.chom.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Wu X., Zhang B., McKee K., O’Dell S., Soto C., Zhou T., Casazza J.P., Mullikin J.C., Kwong P.D., et al. NISC Comparative Sequencing Program De novo identification of VRC01 class HIV-1-neutralizing antibodies by next-generation sequencing of B-cell transcripts. Proc. Natl. Acad. Sci. USA. 2013;110:E4088–E4097. doi: 10.1073/pnas.1306262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C., et al. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM coordinates and maps are deposited in the Protein Data Bank with accession code 7KS9 and in the Electron Microscopy Data Bank with accession code EMD-23016 (trimeric spike) and EMD-23039 (disrupted spike). The 910-30 neutralizing antibody variable heavy and variable light chain sequences have been deposited in GenBank with accession numbers MY291105 and MY291106, respectively. This study did not generate any unique code.