Graphical abstract
Keywords: Intrinsically disordered regions, Multi-domain proteins, SMAD, Transcription factor, TGFβ signaling
Abstract
SMAD transcription factors, the main effectors of the TGFβ (transforming growth factor β) network, have a mixed architecture of globular domains and flexible linkers. Such a complicated architecture precluded the description of their full-length (FL) structure for many years. In this study, we unravel the structures of SMAD4 and SMAD2 proteins through an integrative approach combining Small-angle X-ray scattering, Nuclear Magnetic Resonance spectroscopy, X-ray, and computational modeling. We show that both proteins populate ensembles of conformations, with the globular domains tethered by disordered and flexible linkers, which defines a new dimension of regulation. The flexibility of the linkers facilitates DNA and protein binding and modulates the protein structure. Yet, SMAD4FL is monomeric, whereas SMAD2FL is in different monomer–dimer-trimer states, driven by interactions of the MH2 domains. Dimers are present regardless of the SMAD2FL activation state and concentration. Finally, we propose that SMAD2FL dimers are key building blocks for the quaternary structures of SMAD complexes.
1. Introduction
SMAD proteins are transcription factors that play key roles in the transforming growth factor beta (TGFβ) signaling network. TGFβ signaling is initiated upon TGFβ interaction with its specific receptors that in turn activate SMADs to regulate the transcription of specific genes [2], [3]. TGFβ signaling can have contrasting roles in tumor development. It can either promote tumor proliferation, invasion, metastasis, and escape from immune surveillance; or favor tumor suppression through the inhibition of epithelial cell proliferation [4], with SMAD proteins being highly mutated in tumors [5]. These proteins are also affected in rare diseases and several other conditions, including neurological and respiratory diseases [6].
SMAD proteins are conserved in metazoans and they are classified into three functional classes: Receptor-regulated SMADs (R-SMADs, SMAD1/2/3/5/8), Co-mediator SMAD (Co-SMAD, SMAD4), and inhibitors (I-SMADs, SMAD6/7) [7]. R-SMADs and SMAD4 consist of three functional regions: The N-terminal DNA binding domain (MH1); a linker region of about 120 residues; and a C-terminal domain (MH2). Upon receptor activation, the MH2 domain is phosphorylated at two serine residues conserved at the C-terminus [8], [9]. The phosphorylation of the MH2 domain triggers the formation of a hetero-oligomer between R-SMAD and SMAD4 [10], [11], [12]. The linkers are substrates for kinases and phosphatases and act as binding platforms for cofactors and ubiquitin ligases, which label SMAD proteins for activation or degradation [5], [13], [14].
The monomeric and oligomeric nature of FL SMAD proteins have been a matter of debate for many years. Moreover, the observations on the mechanisms of association and on the behaviors in the basal and activated states are contradictory, mostly because of technical limitations [15], [16]. In the 90s, attempts to crystalize FL SMAD4 were unsuccessful [17], most likely because the protein does not adopt a single compact structure. Afterwards, structural studies on SMAD proteins focused on the structured domains bound to DNA and cofactors, providing a fragmented and static view of this system that has prevailed across the last decades [18], [19], [20], [21], [22], [23], [24]. However, SMAD proteins are not static but dynamic, and display different conformations to carry out their function. Indeed, flexibility allows to optimize structures to meet specific functional needs and determines the formation of large multicomponent complexes. At the same time, it adds complexity and requires new strategies to study protein systems in atomic detail. Many signaling proteins and transcription factors like the SMAD proteins, combine disordered linkers and structured modules. Although multidomain proteins represent an evolutionary breakthrough that facilitates molecular diversity and cross-talk, essential to protein function [1], they received little attention in structural biology because of methodological limitations.
We wanted to address how FL SMAD proteins fold and embraced the challenge to provide the first description of their structures. In parallel to this, we define a strategy that could be extended to investigate other transcription factors and modular signaling proteins that share with SMADs the multi-domain architecture of globular domains separated by long flexible linkers. These structures are expected to form ensembles of conformations, because of the intrinsic flexibility of the linker. We also aimed to establish how the linker and the domains -in the FL context- modulate the quaternary structure of SMAD proteins, as well as their association with other partners. For the structural analyses, we selected SMAD4, as it plays a pivotal role in the TGFβ signaling pathway, and SMAD2, as an example of receptor-activated SMADs.
Studying the structures of FL SMAD proteins required streamlined tools to unveil the ensemble of conformations that explain their dynamic behavior. Nuclear Magnetic Resonance (NMR), Small-angle X-ray scattering (SAXS), molecular dynamics simulations, and molecular modeling provided truly complementary data that we have used to study these structures. As a proof of concept of the methodology, we previously analyzed the dynamic behavior of the MH1 domains in solution through an integrative approach [18], [19], [20] based on using the complementary SAXS and NMR techniques. Here, we use this approach to describe flexible regions within the MH2 domains that were not defined in the available structures because of flexibility. In particular, detailed local conformations from NMR were merged with global shape/size fluctuations derived from SAXS. Other hybrid approaches have recently been used for mixed flexible-rigid systems, although none of these systems has linkers as long and flexible as in the SMAD proteins [25], [26], [27], [28], [29]. To analyze the SAXS data corresponding to the complete proteins, we built explicit models of the domains and linkers, comparing the isolated domains and the domains in the FL context. We then selected sub-ensembles that collectively explain the SAXS data, applying the Ensemble Optimization Method (EOM) pipeline.
Our results indicate that SMAD4FL protein is a monomer, whereas SMAD2FL populates ensembles of monomer–dimer-trimers, independently of the activation state. In both FL proteins, the structured MH1 and MH2 domains are flexibly linked and they are not retained in a long and stable compact interaction. Indeed, the proteins populate ensembles of closed and extended conformations, whose equilibria depend on protein concentration and the dynamic behavior of the unstructured linker, which also modulates the quaternary structure of the proteins.
2. Results
2.1. General workflow
Prior to analyzing the full-length protein datasets, we studied the MH2 domains and linkers, to characterize their folding and oligomerization properties in solution and to obtain experimental information to build starting models that accurately represent our systems. Therefore, we obtained high quality SAXS data corresponding to independent fragments and SMAD2FL (S2FL) and SMAD4FL (S4FL) proteins. Then, we applied the ensemble optimization method (EOM) to select those models that fit the experimental SAXS data at different concentrations [30]. In particular, we studied three constructs of S2FL to cover the activated and non-activated states and their role in protein association.
SMAD linkers are inter-domain sequences of approximately 100–120 residues, characteristic of each type of SMAD protein [5], [31]. There is no structural information available, except for short fragments of SMAD1, SMAD2 and SMAD7, which were studied as complexes bound to transcription activators and ubiquitin ligases [14]. Indeed, these interactions involve a portion of the linker, which contains poly-proline rich sequences and small protein modules named WW domains, which are present in the protein partners [32].
The analysis of the sequences indicates that both SMAD2 and SMAD4 linkers (S4L and S2L) have potential flexibility, net charge, and hydrophobicity propensities, similar to Intrinsically Disordered Regions (IDRs) [33] (Supplementary Fig. S1A,B). NMR and SAXS showed that the intrinsically disordered nature was typical of linkers both in isolation and in the context of full-length proteins [34], [35]. Thus, we applied the Flexible-Meccano pipeline, to model these regions as flexible ensembles [36].
Because of the presence of highly flexible linkers, the SAXS data were best described using large ensembles of protein conformations (hundreds of models). Final ensembles were depicted using a schematic representation (Fig. 1B). The schematic experimental workflow and a summary of the constructs and names are shown in Fig. 1A,B and Supplementary Fig. 1C,D.
Fig. 1.
Construct design and general workflow a. S4 and S2 domain composition. Detailed boundaries are shown in Supplementary Fig. S1A,B. b General methodology and the ensemble selection protocol. Models of the full-length proteins were prepared, including monomers, dimers, and trimers. Flexibility of the linkers was verified by NMR. Final models were selected by fitting the generated ensemble to the SAXS data using the EOM pipeline. S4FL models were validated using IM-MS data. Final ensembles were represented using the MH2 as reference (surface representation), with MH1 domains centers-of-mass illustrated as spheres of a diameter that was proportional to the frequency of a given inter-domain position within the ensemble. Linkers have been omitted for clarity.
2.2. The linkers are flexible and behave as intrinsically disordered regions
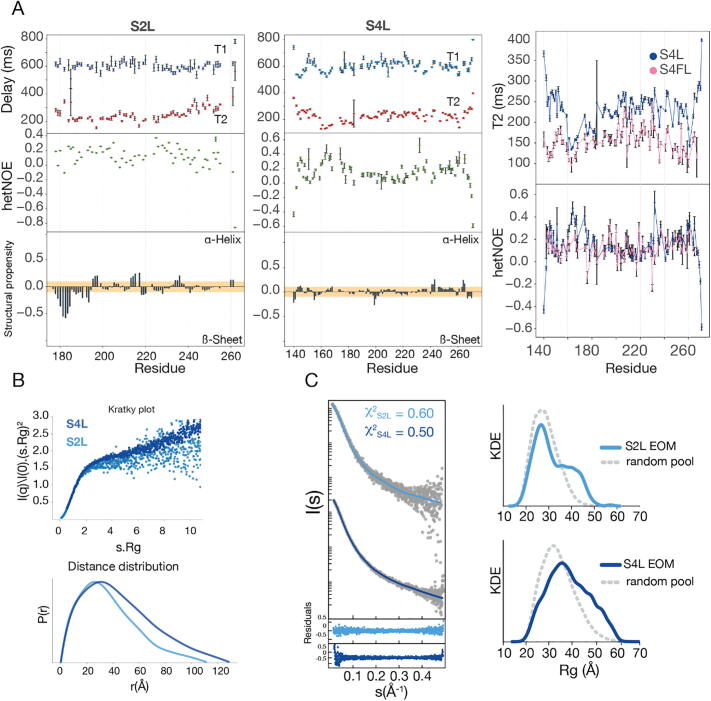
Both S4L and S2L have predicted propensities of Intrinsically Disordered Regions (IDRs) [33] (Supplementary Fig. S1A,B). These propensities are also observed in the 2D 1H-15N Heteronuclear single quantum coherence (HSQC) NMR spectra (Supplementary Fig. S2A). This experiment correlates the amide proton and nitrogen resonances of amino acids, providing a single signal for each of them. The distribution of these resonances indicates if proteins are folded (well-dispersed signals) or unfolded (overlapped signals). For both S4L and S2L, most signals clustered between 7.5 and 8.6 ppm, values that are typical of unfolded proteins [37]. However, the signal overlap was not severe, allowing us to identify most residues in both S2L and S4L constructs using standard 3D backbone experiments. The analysis of the chemical shift confirmed the lack of secondary and tertiary structures characteristic of IDRs. Once the residues were assigned, to decipher their dynamic properties, we started by analyzing the NMR relaxation experiments, that provide information of site-specific internal motions on the subnanosecond timescale [38]. The longitudinal and transverse relaxation experiments (T1, T2), the low heteronuclear Overhauser effect (hetNOE) values (below 0.7), and the absence of propensity to form secondary structure, agreed with values reported in the literature for flexible and disordered regions (Fig. 2A) [39], [40], [41]. Only the N-terminal part of S2L, which is adjacent to the MH1 domain, had a β-sheet/extended propensity, perhaps because of the high Pro content, albeit a flexible nature indicated by the hetNOE values below 0.2.
Fig. 2.
SAXS and NMR data of inter-domain linkers in solution. a S2L and S4L relaxation experiments and secondary structure propensities. The comparison of the spin–spin relaxation time, T2, and the hetNOE for S4FL and S4L. Structural propensities were calculated using ncSPC 50. The colored bar depicts the random-coil threshold and values above or below the bar represent secondary structure propensities (ɑ-helix or β-sheet/extended, respectively). For IDPs, values close to β-sheet propensity imply that these IDPs have an elongated structure. The 1H-15N HSQC spectra showing the narrow 1H chemical shift dispersion characteristic of IDPs is shown as Supplementary Fig. S2B. b Kratky plots for S4L and S2L are shown in dark and light blue, respectively. The high flexibility of both linkers is observed as a monotonic increase along with high s values. Distance distributions derived from the SAXS experimental profiles for S2L and S4L. The color code is the same as for the previous panel. c The experimental SAXS profiles and KDE for Rg distribution for S4L and S2L (in gray) and the solid line simulated curves from each EOM with residuals represented below. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.3. The linker properties are conserved in the FL proteins
We also acquired a 1H-15N HSQC spectrum for S4FL and superimposed it to that of S4L (Supplementary Fig. S2B). Unfortunately, S2FL could not be studied by NMR because of protein oligomerization and precipitation at the concentrations required for these experiments.
Over 75% of the S4FL visible resonances overlapped with the ones of S4L, with low dispersion for the 1H dimension. This indicated that the linker maintained a similar IDP-like behavior both when isolated and in the full-length protein context. Quantitatively, the per-residue distribution of the hetNOE and T2 relaxation values of S4FL resemble those of S4L (Fig. 2A), thus reinforcing the flexible character of the linker within S4FL.
We also assessed the degree of flexibility and/or unfolding of the linker using the SAXS derived Kratky plots, which represent the degree of compactness or flexibility of proteins in solution. The Kratky plots are consistent with an IDR profile (Fig. 2B) since the plot monotonically increased without a clear maximum, indicating conformational heterogeneity and lack of globularity [42]. The asymmetric pair distance distribution functions, P(r), obtained from their scattering data, were also compatible with unfolded particles in solution, having a radius of gyration (Rg) of 30 Å for S2L, and 37 Å for S4L. These Rg values were close to theoretical ones for fully disordered random coils (RgS2LRc = 27 Å, RgS4LRc = 33 Å) (Supplementary Fig. S1B). Their P(r) functions smoothly ended at maximum distances (Dmax) of 111.0 ± 2 Å for S2L, and 128.5 ± 2 Å for S4L (Fig. 2B).
To further characterize the conformational properties of the linkers, we built large ensembles (10,000 conformers) for each isolated linker sequence, using the Flexible-Meccano (FM) algorithm [36]. Ensembles of this size are recommended for highly flexible systems like IDRs [43]. For each pool of conformations, we ran the EOM to yield sub-ensembles that reproduce the scattering profiles. The resulting subsets display Rg distributions enriched in conformations with slightly larger Rg values, in comparison to a theoretical random coil. These distributions fitted with the experimental SAXS curves [44] with χ2 values of 0.6 and 0.5 for S2L and S4L, respectively, with a residual distribution that was better than the one corresponding to theoretical random coils (Fig. 2C). Taken together, these results show that S2L and S4L behave as IDRs and that this behavior is observed in both isolation and full-length protein contexts.
2.4. S4MH2 domain: Visualizing the invisible regions in crystals
In crystals, the S4MH2 fold is defined by a beta-sandwich flanked by a three-helical bundle on one side and by a set of large loops and a helix on the other side (PDB:1YGS) [17]. Undefined areas in the electron density maps covered three regions, the sequence connecting helices H3 and H4 in the three-helical bundle and also the most C-terminal part of the domain that is mostly unobserved (Fig. 3A). This part is partially ordered in crystals corresponding to an extended version of the MH2 domain that contain the SMAD Activation Domain [45] (PDB:1DD1) [46]. In this structure, the H3 is longer and the C-terminal part is folded as two short beta strands, (βt1 and βt2, Fig. 3A). Independently of the MH2 domain boundaries and crystallization conditions, about 30–50 residues located in loops and in the domain, are still undetected in all crystal structures determined so far, probably due to internal flexibility.
Fig. 3.
S4 and S2MH2 domains in solution. a Available crystal structures of S4MH2 domain. Missing residues in the electron density maps are indicated in the structure as dashed lines and as white boxes on the schematic sequence representation (on top). b SAXS scattering curve of S4MH2, corresponding to a merged curve generated from data at several protein concentrations. Residuals showing the agreement between the simulated and experimental profiles are given below the curve. Explicit models satisfying these curves are shown next to the SAXS curve. Green regions in the S4MH2 ensemble depict NMR-supported secondary structures, which were not observed in X-ray structures. c SAXS data corresponding to the S2LMH2 non-phosphorylated domain at two concentrations. In these cases, monomers (M), dimers (D), and trimers (T) (at low concentration), or only trimers (high concentration), contribute to the ensemble of conformations in solution. We provide the Kernel Density Estimation (KDE) calculated for each EOM ensemble compared to that obtained using the starting random pool of models (gray). Explicit models satisfying the curves are shown. d As in c, for S4LMH2. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Given that NMR allows to identify residues in mixed flexible-rigid proteins, we used NMR spectroscopy to clarify the structural characteristics of the S4MH2 core domain in solution. Backbone standard triple resonance experiments facilitate to assign NMR chemical shifts to specific residues and atoms in 2H/13C/15N-enriched proteins. To facilitate the assignment, we acquired the backbone experiments immediately after Deuterium–Hydrogen exchange. In this way, all flexible regions have the amide proton exchanged whereas most residues in the beta-sandwich that participate in hydrogen-bonds remain bound to deuterium, non-observable in the triple resonance experiments. We identified helices, β-strands and flexible loops by comparing the 13C-backbone chemical shifts to random coil values [47]. The chemical shift analysis confirmed the presence and flexibility of the entire H3 as well as the region connecting H3 and H4 that does not adopt a secondary structure in solution (Supplementary Fig. S3A). It also revealed two short extended-regions at the C-terminus, consistent with the βt1 and βt2 strands observed in the S4SADMH2 structure but undetected in crystals of the S4MH2 domain (Fig. 3B).
Overall, our results indicate that in solution, the S4MH2 core domain contains the previously characterized structural elements observed in crystals plus a long H3 and the βt1 and βt2 strands. The complete set of structural elements were included to build the pool of models to analyze all SAXS data related to S4 protein.
2.5. MH2 domains of S2 and S4 display distinctive assembly propensities in solution
The overall structural architecture of both S2MH2 and S4MH2 domains is highly similar. In the case of S2, the helical bundle is more compact, with a shorter helix H3 and an ordered loop connecting helices H3 and H4 (Supplementary Fig. S3B). The second difference is located at the C-terminus that in S2 contains a short motif that is phosphorylated upon activation. In crystals, this 5-residue region becomes well-ordered after phosphorylation and stabilizes the association through homotrimers. SMAD1 MH2 (highly related in sequence to S2) also crystallized as homotrimers with the C-terminal region ordered even in the absence of phosphorylation (PDB:1KHU). Heterotrimers of activated S2 and S4MH2 domains have also been observed in crystals [17], [46], [48].
In solution, there is information on a phosphorylated mimic S2MH2 domain to resemble the activation state that has been studied by SAXS. Due to solubility problems, the domain was tethered to the linker up to the MH1 domain. This phosphorylated mimic variant (S2LMH2EEE) behaves as a trimer in solution [49].
Since we plan to determine the S2FL ensemble of conformations in both phosphorylated and non-phosphorylated states, we studied the non-phosphorylated S2LMH2 using the same boundaries as in S2LMH2EEE. For comparison, we also generated two extended constructs of S4MH2 domain, one construct containing the SAD region (S4SADMH2), and the other containing the full linker up to the MH1 domain (S4LMH2) (Supplementary Fig. S1C,D).
For the non-phosphorylated S2LMH2 domain, we observed that the size distribution of the particles in solution varies with the concentration, covering distances that agree with compact monomeric structures and larger ones corresponding to different association states. To determine the nature of these ensembles we built a pool of conformations containing monomers as well as trimers and dimers, the latter built after removing one monomer of the trimer. We have included dimers in the pool of conformations because they have been previously detected by size exclusion chromatography using S2 FL proteins purified from endogenous expression in cells [16]. Our molecular dynamics simulations confirmed that the network of van der Waals interactions between two monomers is stable during the simulation (Supplementary Fig. S3C). In addition, one of the two non-phosphorylated C-terminal tails remains bound to the second monomer whereas the other tail is exposed to the solvent and flexible.
The EOM-selected ensemble analysis indicated that the S2LMH2 domain coexisted as a monomer–dimer-trimer equilibrium in solution. Remarkably, in this equilibrium, 25% of the total population is a dimer at low concentration whereas 50% of the population was assembled as a trimer already at concentrations of 15 and 30 μM. The trimer was the main species at 60 μM (χ2 = 0.69). We observed that the freedom displayed by the linker is larger in monomers and decreases concomitantly with the formation of dimers and trimers (Fig. 3C).
In the case of S4MH2 domains, independently of the linker length and concentrations, the selected models that fitted the data only contained monomeric species (S4MH2: χ2 = 0.77, Rg = 22 Å, S4SADMH2: χ2 = 0.87, Rg = 25 Å, S4LMH2: χ2 = 0.63, Rg = 37 Å) (Fig. 3B,D and Supplementary Fig. S3D). In fact, the fitting of the SAXS curve to purely trimeric ensembles yields a χ2 = 21, which is not compatible with the experimental data (Supplementary Fig. S3E). Furthermore, the presence or absence of the SAD domain, or of the linker preceding the MH2 domain, did not affect the quaternary structure of the fragments at different concentrations. Indeed, the fragments were always monomeric, and not trimeric as in crystals (Supplementary Fig. S3E,F).
The mechanisms that govern the association propensities of the MH2 domains have been a matter of debate for many years. Our data reveal that, besides the sequence and fold conservation between the SMAD4 and SMAD2 MH2 domains (39% identity and 51% similarity), their MH2 domains display distinctive assembly propensities in solution. Whereas S4MH2 and S4LMH2 domains are monomeric, the S2LMH2 domain populates dimeric and trimeric states, even without a phosphorylation-dependent trigger. In the trimer, almost all linkers are exclusively positioned on one side of the trimer plane because of steric hindrance. This restraint is key to define the relative orientation of the MH1 and MH2 domains in the FL context.
2.6. S4FL is a monomer and populates multiple conformational states in solution
To gain insights into the conformational landscape of full-length SMAD proteins, we acquired SAXS data for SMAD4 and SMAD2 proteins at different concentrations (Fig. 4A).
Fig. 4.
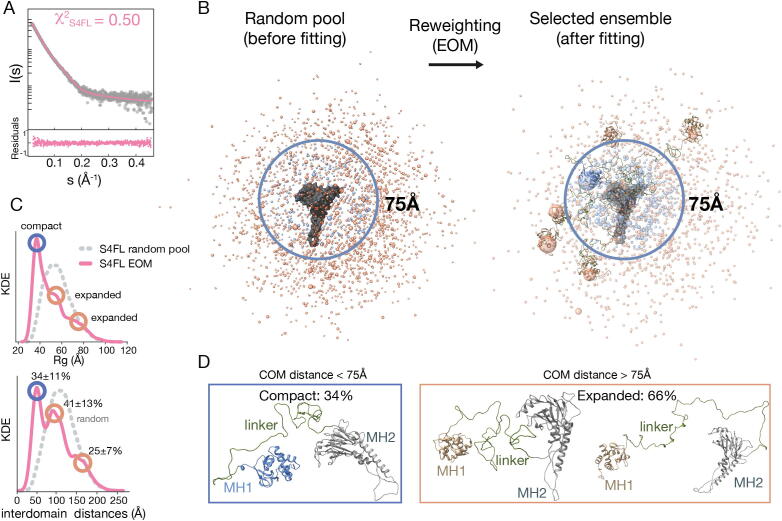
S4FL conformational landscape in solution. a The SAXS EOM simulated profile corresponding to S4FL is shown in pink. It is overlaid with the experimental profile in gray and the respective residuals are represented at the bottom panel. b EOM and random pool S4FL conformational landscapes containing 10 000 conformations. To visualize the domains in the ensemble, the MH2 domain is depicted as a surface (gray) and was used as the reference to fit all conformers. On the contrary, the MH1 domain is simplified as a sphere whose radius is proportional to the probability of occurrence for a given conformation. Large spheres that represent highly populated distributions have the MH1 domain represented as a cartoon. Blue spheres represent distributions up to a distance of 75 Å between domains, and tan spheres indicate expanded conformations. Linkers have been hidden from the representation for clarity, c Size distribution of the ensemble of conformations in the random pool and after EOM selection. The distribution shows compact (34%) and expanded (66 %) conformations, respectively. d Most representative conformations are indicated as explicit models. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In solution, S4FL shows a moderate flexibility, as reflected by the asymmetry of the SAXS-derived P(r) distribution (Dmax = 171.4 Å and Rg = 47 Å) intermediate values between IDR and globular structures, as expected from the mixed architecture of the protein (Supplementary Fig. S1B and 4A). The Kratky plot also shows a plateau characteristic of modular proteins, in which domains are separated by flexible and non-structured linkers [50].
To describe the ensemble of conformers that satisfy the experimental SAXS restraints, we generated explicit models using structured MH1 and MH2 domains tethered by a linker that was modeled as an IDR. Although the fragments containing the MH2 domains indicated a monomeric behavior, we built models to include monomer, dimer, and trimer populations through MH2 domain contacts to explore all possible associations in the FL context. Since the linker behaves as an IDR, we generated a large ensemble pool of 10,000 models with inter-domain distances covering the broad distribution range of the linkers experimentally characterized.
Sub-ensembles that collectively fit the SAXS curve were selected using EOM. The minimal sub-ensemble size (Nse) of 50 models was empirically determined by searching for the smallest size at which the increase of the Nse did not lead to a significant improvement in χ2, to avoid overfitting (Supplementary Fig. S4B). As for the S4 constructs without the MH1 domain, only monomers were compatible with the experimental data.
We simultaneously fitted a single S4FL EOM pool (truncated as required for each SAXS experimental profile) to data acquired on three linker constructs, in isolation, attached to the MH2 domain, and in the context of the full-length protein. This way, we confirmed that linker conformations had similar overall flexibility profiles in the three scenarios, unaffected by the presence of the structured MH1 and MH2 domains in the full-length protein. Also, we observed that in the three cases, the linkers adopted slightly more compact conformations than those expected for a theoretical random coil distribution. This multi-curve fitting approach yielded excellent χ2 statistics of 0.78, 0.95, and 0.76, for S4L, S4LMH2, and S4FL, respectively (Supplementary Fig. S4C).
Since the linker can adopt a variety of conformations, we grouped the EOM-selected models on the basis of the inter-domain center-of-mass (COM) distances. This approach resulted in three major clusters with distances of approximately 50 Å, 100 Å, and 160 Å (Fig. 4B,C). These clusters correspond to Rg values of around 35 Å, 50 Å, and 70 Å, respectively, resulting in a compact to expanded conformations ratio of about 1:2. When compared to the random ensemble, which followed a Gaussian distribution centered at inter-domain distances of about 115 Å and Rg of about 55 Å, these experimental clusters were more compact (Fig. 4C,D). The three clusters satisfying the experimental SAXS data displayed inter-domain distances too big for MH1 and MH2 to directly interact, even in the most compact cluster, which has an inter-domain COM distance of 50 Å. The other two clusters show very large values with an average separation of approximately 93 and 155 Å. The ratio between compact and expanded conformations was validated by Ion mobility followed by Mass Spectrometry (IM-MS). Using this approach, we clearly observed monomeric proteins and similar ratios for various m/z (Supplementary Fig. S4D).
To visualize the ensemble of models (pool and EOM-selected), we generated a representation overlaying the ensemble with respect to the MH2 domain (surface representation). We represented the MH1 domains as spheres condensed at the COM, as indicated in the general workflow description. In the pool, all spheres representing the MH1 domain have a similar diameter and are uniformly distributed. In contrast, in the ensembles that fit the experimental data, the spheres have different diameters. More populated regions are represented by larger spheres (Fig. 4B) since volumes are proportional to how often a given domain populates one region in the conformational space around the MH2 domain. Regions with an inter-domain distance up to 75 Å are shown in blue and the rest in tan.
The final ensemble agreed with the experimental data for models corresponding to monomeric proteins with a χ2 = 0.50 and a random dispersion of the residuals. Some representative models of compact and extended conformations are indicated in Fig. 4D. As expected, the pool ensembles are unable to explain the experimental SAXS data (χ2 = 1.98) (Supplementary Fig. S4E).
We used NMR titrations to further explore the potential interaction between the MH1 and MH2 domains in solution using the isolated domains (up to 3 equivalents, Supplementary Fig. S4F). The analysis did not reveal significant differences in the resonances (or changes in the intensity) of the MH1 domain. Therefore, MH1 and MH2 did not form stable complexes in solution, both in the context of the full-length protein and of isolated domains.
Overall, these results indicate that, in solution, S4FL is a monomer and presents a ratio of compact to expanded conformations of 1:2 (Fig. 4A,B).
2.7. S2FL exists as a monomer–dimer-trimer equilibrium, shaped by phosphomimetic mutations
To explore the conformational equilibrium of S2FL in solution and its potential dependence on the activation of the MH2 domain, we acquired SAXS data on wild type SMAD2FL (S2FLWT) and on a phosphomimetic SMAD2 (S2FLEEE). We also prepared a mutant that contains a premature stop codon at position 460, which produces a protein without the phosphorytable region (S2FL460* Supplementary Fig. S1D). This variant is listed in the COSMIC database as being present in some tumors [51].
Data were acquired at different concentrations: 13.6, 24.3, 46.8, and 56.1 μM, for S2FLWT; and 9.4, 18.7, and 28.1 μM for S2FLEEE (Fig. 5A,B and Supplementary Fig. S5A,B). Then, these datasets were analyzed using explicit ensemble models including monomer, dimer, and trimer populations, based on MH2 domain contacts. All χ2 values for the EOM-selected ensembles agreed with the SAXS data for both S2FLWT and S2FLEEE. In the case of S2FLWT, the trimeric state ranged from 1.0% at 13.8 μM, to 16.4% at 56.1 μM. For the S2FLEEE variant instead, the trimer population was 44.8% at 28.1 μM. In both SMAD proteins (Fig. 6 and Fig. 7), trimers were enriched at higher concentrations and their formation was enhanced by the phosphomimetic mutations. The EOM-selected models are shown using similar representations as for S4FL.
Fig. 5.
S2FL equilibrium distribution in solution. a and b SAXS curves for S2FLWT or S2FLEEE at two different concentrations in gray and EOM fittings in blue and red. Next to each SAXS curve are the Kernel density contour plots for Dmax and Volume, calculated from the EOM ensembles. M, D, and T are abbreviations for monomer, dimer, and trimer species, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 6.
S2FLWT conformational landscape in solution. Left: distribution of MH1-MH2 inter-domain distances in the pool ensembles, in comparison to those obtained after the EOM-selection. Compact structures were classified as those with inter-domain distances less than 75 Å. Right: Models derived from SAXS data at 56.1 µM are shown following a similar approximation as that used for S4FL proteins. Representative conformations (indicated with arrows) are depicted as explicit models. To facilitate the identification of monomers, dimers, and trimers, the MH2 domains are colored in purple, green, and yellow, whereas the MH1 domains are shown in red. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
Conformational landscape of S2FL phosphomimetic variant (S2FLEEE) in solution. Inter-domain distance distribution and models derived from SAXS data corresponding to this variant. The representations are prepared following the same representations as in Fig. 6 and derived from SAXS data at 28.1 µM.
Remarkably, for S2FLWT, the dimer formation reached a maximum of 18% at 56 μM. For S2FLEEE instead, dimer populations were almost invariable with concentration, with a constant fraction of approximately 36% (Fig. 6 and Fig. 7). This concentration independence suggests that formation of dimers is on one hand, an intermediate step in the monomer-trimer equilibrium. On the other hand, dimers seem to exist as stable entities during long periods that permit their identification. Since trimers, especially heterotrimers of S4 and R-SMAD proteins, are believed to define the functional unit of SMAD proteins, the presence of S2 dimers will facilitate the formation of heterotrimers after binding to monomeric S4. They will also define the stoichiometry of the trimeric form, to be 2S2-1S4, an open question in the field for decades.
For S2FL460*, almost no trimer (4%) was observed at high concentrations (74 μM), thus confirming the essential role of the C-terminal residues for trimer formation even in the absence of activation (Supplementary Fig. S5C). Abolishing TGFβ activation or the possibility to associate with other SMAD proteins in the basal state could indicate that tumors harboring S2FL460* drastically reduce the tumor-suppressor capacity [52], [53].
We also explored the inter-domain distances, using the EOM-derived ensembles. We found that an increase in protein concentration shifted these distances towards more compact arrangements, especially in S2FLWT (Fig. 6 and Supplementary Fig. S5D). Remarkably, the ≈50Å inter-domain distance resembled that of S4FL (Fig. 4A-C).
3. Discussion
Conventional structural biology approaches are confronted with the challenge of studying mixed rigid and flexible proteins that populate ensembles of conformations instead of compact models. Protein dynamics covers structural fluctuations from minor local movements in the structured domains up to large-scale changes modulated by linkers and inter-domain contacts. Moreover, the dynamic behavior is inherent to modular proteins, which combine structured domains and linkers of variable length and flexibility, to enhance their functional versatility. Describing the global structure and flexibility has proved challenging in multidomain proteins containing long linkers, like the SMAD transcription factors. Because of the technical limitation imposed by flexibility, we only have access to structures of folded domains, which have been dissected from the full-length (FL) proteins. We also have several studies of FL proteins carried out using biochemical approaches and using exogenous and endogenous SMAD proteins purified from cell lysates. In these cases, the drawback is related to the characterization of the SMAD complexes, which is based on protein retention time in columns and in the use of specific antibodies that detect individual proteins. These experiments have yielded results that have been interpreted contradictorily [15], [16].
We tackled the SMAD complex system through an integrated structural biology approach that allowed us to study the conformational landscape and self-assembly properties of SMAD4 and SMAD2 FL proteins and provide new hypotheses on how SMAD proteins associate. These hypotheses provide a view where past experiments and current experiments performed in cell lines and with native proteins fit together with the results obtained here using recombinant SMAD proteins.
The analysis of the SAXS data revealed how SMAD proteins undergo large conformational changes that affect their overall shape, exposing or covering the functional regions of the proteins depending on the open or extended state. Without external cofactors, the MH1 and MH2 domains are flexibly linked. In solution, the linker between the two domains acts like a mechanical element. Indeed, it allows to approximate and separate the MH1 and MH2 domains, without retaining them through a long and stable compact interaction, but allowing them to participate in direct contacts. These closed conformations have been previously described in the literature and are proposed to play essential inhibitory roles in tumor mutated SMAD4 proteins [54]. However, since these interactions are transient in the wild type proteins, the absence of stable inter domain interactions allows the domains to explore different relative orientations, without assuming compact and globular architectures.
Our results indicated that the linker, although very long and unstructured, restricts the relative orientation of the domains, with some distances and orientations preferred with respect to others (Fig. 4, Fig. 6, Fig. 7). This is particularly remarkable in S2 and in the dimers and trimers, which condition the orientation of the linkers to cluster on one side of the MH2 trimeric plane. Short linker distances are observed in the compact states whereas the expanded states illustrate how linker modification and cofactor association can occur. Indeed, from a biological perspective, linker compactness could act as a protective mechanism to prevent SMAD2 from non-specific interactions with other proteins, modulating its conformational landscape in a concentration-dependent way. In fact, spatial and temporal variations of protein concentration regulate the function of other transcription factors, both in vivo and in vitro [55], [56], [57], [58].
Expanded structures exposed linker accessible regions separated by almost no energy barriers. Remarkably, these extended conformations also exposed the domains to interact with DNA and other protein partners, as linker kinases and phosphatases or cofactors. This observation solves open questions related to SMAD’s association that had been in the field for decades. Remarkably, in the case of SMAD2 (WT and phosphomimetic variant), we also observed an increase in compact conformations dominated by shorter interdomain distances upon dimer-trimer formation.
One of the excruciating questions in the field has been related to how SMAD proteins form heteromeric complexes of variable composition (homo or hetero dimers and trimers [16], [17], [59], [60]). We observed that S4 is mostly monomeric (and not a trimer, as observed in crystals of MH2 domains). On the contrary, S2 populates an equilibrium containing monomers, dimers, and trimers, with trimers being less abundant among FL proteins (activated and inactivated) than in the context of isolated MH2 domains. Thus, the presence of the MH1 domain and the linker seems to modulate the association propensity of S2 MH2 domains, a feature overlooked when working with isolated MH2 domain fragments.
The inter-domain association, however, was reported in the past, using purified S2 protein endogenously expressed in cells, and it was hypothesized that MH1 and MH2 stable contacts might play a role in tuning the homotrimerization propensities [15], [16]. Nevertheless, our data favor a different explanation, where transient interactions between either the MH1 or the linker with the MH2 domain suffice to modulate the oligomerization properties of the MH2 domain in the FL protein context. Moreover, the presence of S2 dimers—and not only trimers—offers a plausible explanation for the heterotrimeric association of SMAD proteins in cells. In this scenario, a S4 (monomer) and a dimer of S2 can yield a 1S4-2S2 hetero-trimer (Fig. 8) defining for the first time the mechanism of heterotrimer assembly. The S2 dimer can also associate with a monomeric S2 (or S3) to form trimers without the presence of S4, as observed in experiments in cells [61], [62], [63].
Fig. 8.
Cartoon describing our hypothesis on the mechanism for the hetero-trimer association of SMAD proteins. Schematic representation of the S2FL and S4FL proteins and their quaternary structures. The MH1 and MH2 domains are represented as silhouettes generated from 3D structures. Flexible connectors have been simplified and sketched as lines. The S2FL dimer can associate either with monomeric S4FL forming a hetero-trimer, or with another monomeric R-SMAD to generate homo-trimeric R-SMAD assemblies. Each monomer of S2FL is colored with a shade of blue and monomeric S4FL is shown in orange. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.) (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In both cases, the formation of heterotrimers starting from an intermediate dimer seems to be more favorable than competitive displacement of an already formed homotrimer triggered by SMAD4, as was previously thought [60]. Although experimental proof is needed, it is tempting to speculate that a similar mechanism of hetero-trimer formation starting from homodimers also holds true for SMAD1/5/8 proteins. Indeed, in these proteins, their MH1 domains are prone to define dimers, perhaps enhancing the dimerization propensity of the entire protein [20].
Another conclusion derived from the conformational fluctuations of the linkers relates to the recognition of specific DNA sites in cis-regulatory elements. Linkers, regardless of how flexible or rigid they could be, limit the overall freedom of the MH1 domains by tethering them to the MH2 domain trimer. Our hypothesis is that this restriction has a positive impact in DNA binding, by speeding up the process of identifying optimal binding sites through an adapted “Monkey Bar” mechanism [64]. Through such mechanism and thanks to the flexibility provided by the linkers, the interaction of one MH1 domain with the DNA suffices to approximate the entire complex to a given promoter. This leads to explore the possibilities to produce a second and third interaction of all MH1 domains present in the trimeric SMAD complex. Optimal binding sites will correspond to those where two or three MH1 domains can interact effectively.
Overall, these results on the conformational landscape of SMAD full length proteins start unveiling how these transcription factors associate and work in native contexts. They also pave the way to study the interactions with cofactors and the effects of SMAD modifications (ubiquitination, phosphorylation, or disease-associated mutations) on the dynamics, in the context of FL proteins and outside conserved domains.
Finally, the methodology and computational tools that we optimized could find broad application to study other multi-domain proteins with long and disordered linkers. Indeed, these linkers are abundant among transcription factors and yet poorly represented in structural studies.
4. Methods
The general experimental workflow is indicated in Fig. 1 A,B and explained at the beginning of the Results section.
4.1. Recombinant protein production and cloning
S2FL, S2FL460*, S2LMH2, S2FLEEE, and S2LMH2EEE constructs were cloned in the pETM10 vector with an N-terminal His-tag, whereas S2L, S2MH1-E3, S2MH1E3, S4LMH2, S4SADMH2, S4MH2, and S4L were cloned in the pETM11 vector. S4FL was cloned in the pCOOFY34 vector with an N-terminal streptavidin tag and a 3C protease cleavage site. Successful cloning was confirmed by DNA sequencing (GATC Biotech). All constructs are shown in Supplementary Fig. S1C,D.
Cloning was performed using standard protocols as described [18]. For protein production, all protein constructs were expressed in the E. coli BL21 (DE3) strain. Cells were cultured at 37 °C in Luria-Bertani (LB) medium until reaching an OD600 of 0.6–0.8. After induction with IPTG (Isopropyl β-D-1-thiogalactopyranoside) at a final concentration of 0.5 mM, and overnight expression at 20 °C, bacterial cultures were centrifuged at 4000xg for 20 min and resuspended in lysis buffer (40 mM Tris, pH 7.5, 400 mM NaCl, 0.1% Tween-20, 40 mM imidazole, 1 mM TCEP (tris(2-carboxyethyl) phosphine) and a protease inhibitor cocktail (S8820 SIGMAFAST™). Cells were lysed using a refrigerated EmulsiFlex-C5 (Avestin) at 20,000 psi and the lysed solution was centrifuged at 35000xg for 45 min at 4 °C to discard insoluble material. Soluble supernatants were purified by affinity chromatography (StepTag or HiTrap Chelating HP columns, GE Healthcare Life Science) using an NGC Quest 10 Plus Chromatography System (BIO-RAD) and a buffer gradient starting at 0% buffer A (lysis buffer) up to 100% buffer B in 15 column volumes. Buffer B was 40 mM Tris, pH 7.5, 200 mM NaCl, 2.5 mM desthiobiotin for S4FL and 40 mM Tris, pH 7.5, 400 mM NaCl, 400 mM imidazole for the rest. Eluted proteins were digested at 4 °C with specific proteases and further purified by ion-exchange chromatography using a HiTrap SP HP or monoQ (GE Healthcare) columns and a gradient running from 0% buffer A (40 mM TRIS, pH 7.2) to 100% buffer B (40 mM TRIS, 1 M NaCl, pH 7.2). As a final purification, step size-exclusion chromatography was performed using 40 mM TRIS, 150 mM NaCl, pH 7.2 buffer.
For the purification of the inter-domain linkers S2L and S4L, the proteins were expressed as described above but the resulting proteins were insoluble. In these cases, the lysis and protein elution were performed in denaturing conditions (40 mM TRIS, 400 mM NaCl, 8 M Urea, 40 mM imidazole, 1 mM TCEP, 0.1% tween20, pH 7.5). Proteins were refolded bound to the resin, using four washing steps and increasing the ratio of refolding/lysis buffers from zero to four (refolding buffer: 40 mM TRIS, 400 mM NaCl, 40 mM imidazole, 1 mM TCEP, 0.1% tween20, pH 7.5). After elution, proteins were cleaved and further purified by gel filtration chromatography and stored in 40 mM TRIS, 150 mM NaCl, pH 7.2. 15N-and 13C-labeled proteins were prepared as previously described [19], [20] and purified as above. Aliquots were kept frozen at -80 °C. Protein purity was assessed by SDS-PAGE (sodium dodecyl sulfate–polyacrylamide gel electrophoresis) and mass spectrometry. DNA binding capacity was compared and equal to the capacity reported in the literature to ensure that the Full-length proteins were functional.
4.2. Nuclear magnetic resonance spectroscopy
NMR data were acquired on a Bruker Avance III 600-MHz spectrometer (IRB Barcelona) or Bruker Avance IIIHD 850-MHz (IBS-ISBG, Grenoble), both equipped with a Cryo TCI (1H, 13C, 15N, 2H), 5 mm, with z-gradients. S4L and S2L samples were studied in 40 mM TRIS, 150 mM NaCl, 10% D2O, pH 6.6 at 278 K. The HSQC experiments were processed using TOPSPIN v3.5 (Bruker). All other experiments were processed using NMRPipe [65] and analyzed with the CcpNmr Analysis [66] software suite or with CARA. The NMR backbone assignment followed established protocols employing CBCANH, CBCA(CO)NH, HN(COCA)NH, HN(CA)NH, HN(CA)CO, HNCO and N HCACB and NHCACOCB experiments for deuterated proteins using Non-Uniform Sampling (NUS) and BEST-TROSY backbone experiments [67], [68], [69], [70], [71]. Proline residues were connected using a set of specific experiments [72]. T1 and T2 relaxation measurements were acquired using standard pulse sequences at 278 K [67], essentially as described in [20]. T1 relaxation experiments used inversion recovery delays of 20, 110, 160, 270, 430, 540, 700, 860, 1080, 1400, 1720 and 2000 ms. The delays used for the T2 experiment were 0, 20, 40, 60, 80, 120, 160, 200, 280 and 400 ms. The size of the fid for all experiments was (1H)1024 × (15N)256 points and the interscan delay was set to 3 s. Relaxation rates were retrieved by fitting peak intensities to an exponential function implemented in CcpNnmr analysis [66].
The NMR titration of the S4MH2 into the S4MH1 construct was performed in the same buffer described above, with 1 equivalent corresponding to a final concentration of 130 μM.
Chemical shifts were quantified using equation (1),
| (1) |
where δH and δN are the 1H and 15N chemical shift differences, respectively.
Secondary structure propensities were calculated using the ncSPC (Neighbor Corrected Structural Propensity Calculator) method [41] using 13C, 15N and 1HN backbone chemical shifts. Values between -0.1 and 0.1 are random coils, with values above 0.1 and below -0.1 corresponding to ɑ-helix and β-sheet propensity, respectively.
4.3. Protein disorder propensities
The protein disorder propensity was calculated with MetaDisorder [73]. Uversky plot was calculated using CIDER [74].
4.4. Small-angle X-ray scattering data acquisition
SAXS data were acquired on Beamline 29 (BM29) at the European Synchrotron Radiation Facility (ESRF, Grenoble, France). Protein samples were centrifuged for 10 min at 10000xg prior to data acquisition. Experiments on BM29 were collected on 45 μL samples with the following settings: 12.5 keV, 100% transmission, low viscosity and 0 s wait time. Data were recorded on a Pilatus 1 M detector, at 10 ˚C. Ten frames per sample were collected for 1 s each. Solvent from each sample elution was collected and their scattering data were acquired to account for buffer contribution. Image conversion to the 1D profile, scaling, buffer subtraction and radiation damage accession was done using the in-house software pipeline available at BM29. Further processing was done by the ATSAS software suite [75] and the ScÅtter package (http://www.bioisis.net/). For SMAD4 constructs, the reported SAXS profiles were concentration-independent and were merged in ATSAS2.8 and used for subsequent analysis. For SMAD2, the S2L construct was merged at the reported concentrations, and all other constructs were analyzed for each concentration individually, due to their concentration-dependence (Supplementary Table 1). Fitting to the experimental data was calculated using the reduced χ2 metric in equation (2).
| (2) |
where K is the number of data points for each SAXS profile (), are the standard deviations of the scattering intensities and is a scaling factor. are the theoretical scattering intensities for each model.
4.5. Ion mobility mass spectrometry data acquisition
Ion mobility mass spectrometry experiments were performed using a Synapt G1-HDMS mass spectrometer (Waters, Manchester, UK). Samples were buffer-exchanged to a 200 mM ammonium acetate buffer and infused by an automated chip-based nanoelectrospray using a Triversa Nanomate system (Advion BioSciences, Ithaca, NY, USA). The ionization was performed in positive mode using a spray voltage and a gas pressure of 1.70 kV and 0.5 psi, respectively. Cone voltage, extraction cone and source temperature were set to 40 V, 2 V and 20 °C, respectively. Trap and transfer collision energies were set to 10 V and 10 V, respectively. The pressure in the trap and transfer T-Wave regions were 5.8410-2 mbar of Ar and the pressure in the IMS T-Wave was 0.460 mbar of N2. Trap gas and IMS gas flows were 8 and 24 mL/sec, respectively. The travelling wave used in the IMS T-Wave for mobility separation was operated at 300 m/sec. The wave amplitude was fixed to 10 V. The bias voltage for entering in the T-wave cell was 15 V. The instrument was calibrated over the m/z range 500–8000 Da using a solution of cesium iodide. MassLynx (v4.1) and Driftscope (v2.4) were used for data processing and analysis. Drift time calibration of the T-Wave cell was performed using the following calibrants: β-Lactoglobulin (bovine milk), transthyretin (human plasma), avidin (egg white), serum albumin (bovine) and concanavalin A (Canavalia ensiformis) in 200 mM ammonium acetate at 20 μM. All measurements followed the same experimental protocol stated above. The reduced cross-sections (Ω’) were retrieved from previous results [76] and plotted against corrected drift times (tD). A power law fit of Ω’ vs. tD was performed to extract the calibration coefficients (Prism v6, GraphPad Software Inc.). Finally, Gaussian curves were fitted to the drift time distributions used to extract the experimental CCS.
4.6. Structural modeling of SMAD2 and SMAD4 linkers
Random coil ensemble models of S2L and S4L containing 10,000 conformations each [43] were generated using Flexible-Meccano (FM) [36], where torsion angle pairs were selected randomly from a database of amino acid-specific conformations in loop regions of high-resolution X-ray structures. Side-chains modeling with SCCOMP [77], and energy-minimization in explicit solvent using GROMACS 5.1.1 were then carried out [78]. We used the force field AMBER99sb-ILDN [79] and the TIP3P water model [80]. We used CRYSOL [81] to compute the theoretical SAXS profiles from conformational ensembles of S2L and S4L. All theoretical curves were obtained with 101 points and a maximum scattering vector of 0.5 Å-1 using 25 harmonics. Using the ensemble optimization method (EOM) [30], we select from the S2L and S4L structural pools the linker structures whose theoretical SAXS profiles collectively fit their experimental SAXS profiles, using the reduced χ2 metric. The theoretical SAXS profile for each generated conformation was computed and then averaged over the selected sub-ensembles.
4.7. Reconstruction of the missing fragments of SMAD2 and SMAD4 MH2 domains
Ensembles of missing terminal disordered fragments were re-built using FM and attached to the X-ray templates of SMADs MH1 and MH2 using in-house scripts as in [82], [83]. We used the following PDB structures as templates: SMAD4 MH1 (PDB:3QSV) and MH2 (PDB:1DD1) domains, and SMAD2 MH1 (PDB:6H3R) and MH2 (PDB:1KHX) domains. For each built segment, side-chains were added using SCCOMP and then pre-processed with Rosetta 3.5 fixbb-module to alleviate steric clashes. Internal segments and disordered loops were built using the RosettaCM application as previously described for S2MH1 [19], outputting 5,000 structures per domain. We assessed the quality of the ensembles by examining their averaged SAXS curves against the respective experimental data, using the reduced χ2 metric. We subsequently used these conformers to build LS-MH2, full-length (i.e., MH1-LS-MH2), and oligomeric constructs. All models were energy-minimized in water as above.
4.8. Modeling and fittings of full-length SMAD proteins and mutants
We modeled the full-length proteins (i.e, S4FL, S2FLWT, S2FLEEE and S2FL460* variants) using the pools created for each region (i.e., MH1, MH2, MH2460*, and LS), explained above. To create S2FLEEE, we in-silico mutated the phospho-serine sites (pSer) of MH2 located at the C-terminal residues Cys-Ser-Ser-Met-Ser (SSXSS motif) by phosphomimetic glutamic acid residues. To generate ensemble models for the C-terminally-truncated SMAD2FL variant (S2FL460*), we removed the seven last residues from the MH2 crystal structure. Different conformers were randomly selected and added to new unique explicit models without steric clashes. The final models were energy-minimized with GROMACS 5.1.1 [78].
Following this workflow, we created an ensemble of 10,000 unique combinations for each system/scenario, including different oligomeric forms, i.e., their monomeric, dimer, and trimer representations. S4 and S2 MH2 domains form crystallographic homo-trimers. We used the trimers as starting models to generate monomeric and dimeric versions by removing one or two chains. Then, to probe the oligomeric preferences of SMAD4 and SMAD2 FL and variants, identical monomer, dimer, and trimer populations were defined in the initial shared pool and analyzed with EOM. To this end, we used CRYSOL [81] to compute the theoretical SAXS profiles from each conformational ensemble, and with EOM, we selected those structures reproducing the experimental SAXS data. A minimal sub-ensemble size (Nse) was empirically determined by searching for the smallest Nse = 50 with the global lowest SAXS discrepancy (reduced χ2), checking for over-fitting biases. To further detail the SMAD4 conformational landscape and add robustness to the modeling, we also used multiple SAXS curves from the deletion mutant S4LMH2, S4L, and full-length protein as a strategy to enhance the structural content of SAXS data and improve model discrimination. The ensemble multi-curve fitting with a single pool successfully improved the structural analysis of disordered tau protein [84]. With the assumption that S4L remains disordered in the full-length context, we selected those conformers that fitted the SAXS profiles of S4L, S4LMH2, and S4FL simultaneously, by minimizing the sum of χ2 between the experimental () and average theoretical () SAXS intensities:
| (3) |
| (4) |
| (5) |
where K is the number of data points of each SAXS profile (), () are the standard deviations of the scattering intensities, and is a scaling factor. was obtained by averaging the scattering of 50 explicit models (Nse) per variant (i.e., = S4L, S4LMH2 or S4FL). Subsequent ensemble analysis obtained was done using MDanalysis [85].
4.9. Molecular dynamics simulations
To assess the stability of MH2 dimers, we ran a molecular dynamics trajectory of 500 ns. Molecular dynamics simulations were performed for the dimer with GROMACS 5.1.1 using the Amber99sb force field [78]. The system was solvated in a dodecahedron box with TIP3P water. It was minimized for a maximum of 50,000 steps or until the force constant was less than 1000 kJ/mol·nm, using the steepest descent algorithm implemented in GROMACS. The cutoff distance used for the non-bonded interactions, using the Particle mesh Ewald (PME) method, was 10 Å. Before the final production simulation, the system was equilibrated using the NPT ensemble for 500 ps, followed by 50 ps in the NVT ensemble. Finally, the system was simulated for 0.5 μs with a 2 fs integration step. The first 100 ns were discarded assuming system equilibration. Temperature coupling was done with the Nose–Hoover algorithm at 300 K. Pressure coupling was done with the Parrinello–Rahman algorithm at 1 bar. Root-mean-squared deviation (RMSD) was calculated using built-in GROMACS analysis routines and plotted using Xmgrace.
5. Data availability
Protein ensembles are deposited at the Protein Ensemble Database (PED) [86], PED00193-PED00202. NMR data are available at the BMRB with accession codes 50738 (SMAD2) and 50737 (SMAD4). SAXS data are available at SASBDB, accession numbers are detailed in Supplementary Table 1. All remaining data are available in the main text or the supplementary materials.
CRediT authorship contribution statement
Tiago Gomes: Methodology, Investigation, Writing - review & editing. Pau Martin-Malpartida: Methodology, Writing - review & editing. Lidia Ruiz: Investigation. Eric Aragón: Investigation. Tiago N. Cordeiro: Conceptualization, Methodology, Investigation, Supervision, Writing - review & editing. Maria J. Macias: Conceptualization, Investigation, Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We thank the ESRF group for the help and access to the synchrotron Bio-SAXS BM29 beamline, and the Mass Spectrometry and Protein Expression Core Facilities (IRB Barcelona) for help and reagents. We also thank Dr. B. Brutscher (Institut de Biologie Structurale, Biomolecular NMR Spectroscopy Group, Grenoble, France) for the help with the acquisition of the NMR data at 850 MHz; A. Vea and C. Torner for support with protein purification; Dr. P. Bernardó for the support and suggestions on the methodology; and Dr. J. Massagué for insightful discussions. Access to Bio-SAXS BM29 ESRF was part of the MX-1941 BAG proposal, access to ALBA synchrotron was through the BAG proposal 2018092972.
Funding
This work has been financed through the Spanish MINECO program (BFU2014-53787-P and BFU2017-82675-P, M.J.M); the IRB Barcelona; The BBVA Foundation; the Horizon 2020 Programme; the iNEXT program (grant SMAD4 MH2 domain PID:7192, the FCT – Fundação para a Ciência e a Tecnologia, I.P., Project MOSTMICRO-ITQB with references UIDB/04612/2020 and UIDP/04612/2020 (to ITQB-NOVA); FEDER Funds through COMPETE 2020 (0145-FEDER-007660). We gratefully acknowledge institutional funding from the CERCA Programme of the Catalan Government and from the Spanish Ministry of Economy, Industry and Competitiveness (MINECO) through the Centres of Excellence Severo Ochoa award.
TNC is a recipient of Stimulus of Scientific Employment, Individual Support from Fundação para a Ciência e a Tecnologia, Ministério da Ciência, Tecnologia e Ensino Superior, CEECIND/01443/2017 and M.J.M is an ICREA Programme Investigator.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.09.009.
Contributor Information
Tiago N. Cordeiro, Email: tiago.lopes@irbbarcelona.org.
Maria J. Macias, Email: maria.macias@irbbarcelona.org.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Bornberg-Bauer E., Albà M.M. Dynamics and adaptive benefits of modular protein evolution. Curr Opin Struct Biol. 2013;23(3):459–466. doi: 10.1016/j.sbi.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 2.Heldin C.H., Miyazono K., ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 3.Massague J. The transforming growth factor-beta family. Annu Rev Cell Biol. 1990;6(1):597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 4.Massagué J. TGFβ signalling in context. Nat Rev Mol Cell Biol. 2012;13(10):616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macias M.J., Martin-Malpartida P., Massagué J. Structural determinants of SMAD function in TGF-β signaling. Trends Biochem Sci. 2015;40(6):296–308. doi: 10.1016/j.tibs.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kashima R., Hata A. The role of TGF-beta superfamily signaling in neurological disorders. Acta Biochim Biophys Sin (Shanghai) 2018;50:106–120. doi: 10.1093/abbs/gmx124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Massagué J., Seoane J., Wotton D. SMAD transcription factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- 8.Kretzschmar M., Liu F., Hata A., Doody J., Massague J. The TGF-beta family mediator SMAD1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes Dev. 1997;11(8):984–995. doi: 10.1101/gad.11.8.984. [DOI] [PubMed] [Google Scholar]
- 9.Macias-Silva M., Abdollah S., Hoodless P.A., Pirone R., Attisano L., Wrana J.L. MADR2 is a substrate of the TGFbeta receptor and its phosphorylation is required for nuclear accumulation and signaling. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 10.Lagna G., Hata A., Hemmati-Brivanlou A., Massagué J. Partnership between DPC4 and SMAD proteins in TGF-beta signalling pathways. Nature. 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 11.Candia AF, et al. Cellular interpretation of multiple TGF-beta signals: intracellular antagonism between activin/BVg1 and BMP-2/4 signaling mediated by SMADs. Development 124, 4467-4480 (1997). [DOI] [PubMed]
- 12.Wu R.Y., Zhang Y., Feng X.H., Derynck R. Heteromeric and homomeric interactions correlate with signaling activity and functional cooperativity of SMAD3 and SMAD4/DPC4. Mol Cell Biol. 1997;17(5):2521–2528. doi: 10.1128/mcb.17.5.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aragón E., Goerner N., Xi Q., Gomes T., Gao S., Massagué J. Structural basis for the versatile interactions of SMAD7 with regulator WW domains in TGF-β Pathways. Structure. 2012;20(10):1726–1736. doi: 10.1016/j.str.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aragon E., Goerner N., Zaromytidou A.-I., Xi Q., Escobedo A., Massague J. A SMAD action turnover switch operated by WW domain readers of a phosphoserine code. Genes Dev. 2011;25(12):1275–1288. doi: 10.1101/gad.2060811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawabata M., Inoue H., Hanyu A., Imamura T., Miyazono K. SMAD proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J. 1998;17(14):4056–4065. doi: 10.1093/emboj/17.14.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayaraman L., Massagué J. Distinct oligomeric states of SMAD proteins in the transforming growth factor-beta pathway. J Biolog Chem. 2000;275:40710–40717. doi: 10.1074/jbc.M005799200. [DOI] [PubMed] [Google Scholar]
- 17.Shi Y., Hata A., Lo R.S., Massagué J., Pavletich N.P. A structural basis for mutational inactivation of the tumour suppressor SMAD4. Nature. 1997;388(6637):87–93. doi: 10.1038/40431. [DOI] [PubMed] [Google Scholar]
- 18.Martin-Malpartida P., Batet M., Kaczmarska Z., Freier R., Gomes T., Aragón E. Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors. Nat Commun. 2017;8(1) doi: 10.1038/s41467-017-02054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aragón E., Wang Q., Zou Y., Morgani S.M., Ruiz L., Kaczmarska Z. Structural basis for distinct roles of SMAD2 and SMAD3 in FOXH1 pioneer-directed TGF-β signaling. Genes Dev. 2019;33(21-22):1506–1524. doi: 10.1101/gad.330837.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz L., Kaczmarska Z., Gomes T., Aragon E., Torner C., Freier R. Unveiling the dimer/monomer propensities of SMAD MH1-DNA complexes. Comput Struct. Biotechnol J. 2021;19:632–646. doi: 10.1016/j.csbj.2020.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyazono K.-I., Ito T., Fukatsu Y., Wada H., Kurisaki A., Tanokura M. Structural basis for transcriptional coactivator recognition by SMAD2 in TGF-β signaling. Sci Signal. 2020;13(662):eabb9043. doi: 10.1126/scisignal.abb9043. [DOI] [PubMed] [Google Scholar]
- 22.Guca E, et al. TGIF1 homeodomain interacts with SMAD MH1 domain and represses TGF-β signaling. Nucleic Acids Research 46, 9220-9235 (2018). [DOI] [PMC free article] [PubMed]
- 23.Miyazono K.-I., Moriwaki S., Ito T., Kurisaki A., Asashima M., Tanokura M. Hydrophobic patches on SMAD2 and SMAD3 determine selective binding to cofactors. Sci Signal. 2018;11(523):eaao7227. doi: 10.1126/scisignal.aao7227. [DOI] [PubMed] [Google Scholar]
- 24.Murayama K., Kato-Murayama M., Itoh Y., Miyazono K., Miyazawa K., Shirouzu M. Structural basis for inhibitory effects of SMAD7 on TGF-beta family signaling. J Struct Biol. 2020;212 doi: 10.1016/j.jsb.2020.107661. [DOI] [PubMed] [Google Scholar]
- 25.Jussupow A., Messias A.C., Stehle R., Geerlof A., Solbak S.M.Ø., Paissoni C. The dynamics of linear polyubiquitin. Sci Adv. 2020;6(42):eabc3786. doi: 10.1126/sciadv.abc3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen A.H., Wang Y., Bottaro S., Grudinin S., Arleth L., Lindorff-Larsen K. Combining molecular dynamics simulations with small-angle X-ray and neutron scattering data to study multi-domain proteins in solution. PLoS Comput Biol. 2020;16(4):e1007870. doi: 10.1371/journal.pcbi.1007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brosey C.A., Tainer J.A. Evolving SAXS versatility: solution X-ray scattering for macromolecular architecture, functional landscapes, and integrative structural biology. Curr Opin Struct Biol. 2019;58:197–213. doi: 10.1016/j.sbi.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Q., Li M., Lai L., Liu Z. Allostery of multidomain proteins with disordered linkers. Curr Opin Struct Biol. 2020;62:175–182. doi: 10.1016/j.sbi.2020.01.017. [DOI] [PubMed] [Google Scholar]
- 29.Cordeiro T.N., Herranz-Trillo F., Urbanek A., Estaña A., Cortés J., Sibille N. Small-angle scattering studies of intrinsically disordered proteins and their complexes. Curr Opin Struct Biol. 2017;42:15–23. doi: 10.1016/j.sbi.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 30.Bernadó P., Mylonas E., Petoukhov M.V., Blackledge M., Svergun D.I. Structural characterization of flexible proteins using small-angle X-ray scattering. J Am Chem Soc. 2007;129(17):5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 31.Sorrentino G.M., Gillis W.Q., Oomen-Hajagos J., Thomsen G.H. Conservation and evolutionary divergence in the activity of receptor-regulated SMADs. Evodevo. 2012;3:22. doi: 10.1186/2041-9139-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Macias M.J., Wiesner S., Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513:30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- 33.Uversky V.N., Gillespie J.R., Fink A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins Struct Funct Bioinf. 2000 doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 34.Babu M.M. The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem Soc Trans. 2016;44:1185–1200. doi: 10.1042/BST20160172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lieutaud P., Ferron F., Uversky A.V., Kurgan L., Uversky V.N., Longhi S. How disordered is my protein and what is its disorder for? A guide through the “dark side” of the protein universe. Intrinsically Disord Proteins. 2016;4(1):e1259708. doi: 10.1080/21690707.2016.1259708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozenne V., Bauer F., Salmon L., Huang J.-r., Jensen M.R., Segard S. Flexible-meccano: a tool for the generation of explicit ensemble descriptions of intrinsically disordered proteins and their associated experimental observables. Bioinformatics. 2012;28(11):1463–1470. doi: 10.1093/bioinformatics/bts172. [DOI] [PubMed] [Google Scholar]
- 37.Yao J., Dyson H.J., Wright P.E. Chemical shift dispersion and secondary structure prediction in unfolded and partly folded proteins. FEBS Lett. 1997;419:285–289. doi: 10.1016/s0014-5793(97)01474-9. [DOI] [PubMed] [Google Scholar]
- 38.Ishima R., Torchia D.A. Protein dynamics from NMR. Nat Struct Biol. 2000;7:740–743. doi: 10.1038/78963. [DOI] [PubMed] [Google Scholar]
- 39.Brutscher B. NMR methods for the study of instrinsically disordered proteins structure, dynamics, and interactions: general overview and practical guidelines. Adv Exp Med Biol. 2015;870:49–122. doi: 10.1007/978-3-319-20164-1_3. [DOI] [PubMed] [Google Scholar]
- 40.Konrat R. NMR contributions to structural dynamics studies of intrinsically disordered proteins. J Magn Reson. 2014;241:74–85. doi: 10.1016/j.jmr.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamiola K., Mulder F.A.A. Using NMR chemical shifts to calculate the propensity for structural order and disorder in proteins. Biochem Soc Trans. 2012;40:1014–1020. doi: 10.1042/BST20120171. [DOI] [PubMed] [Google Scholar]
- 42.Rambo R.P., Tainer J.A. Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers. 2011;95:559–571. doi: 10.1002/bip.21638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tria G., Mertens H.D.T., Kachala M., Svergun D.I. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCrJ. 2015;2(2):207–217. doi: 10.1107/S205225251500202X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rambo R.P., Tainer J.A. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496(7446):477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Caestecker M.P., Hemmati P., Larisch-Bloch S., Ajmera R., Roberts A.B., Lechleider R.J. Characterization of functional domains within SMAD4/DPC4. J Biol Chem. 1997;272(21):13690–13696. doi: 10.1074/jbc.272.21.13690. [DOI] [PubMed] [Google Scholar]
- 46.Qin B., Lam S.SW., Lin K. Crystal structure of a transcriptionally active SMAD4 fragment. Structure. 1999;7(12):1493–1503. doi: 10.1016/s0969-2126(00)88340-9. [DOI] [PubMed] [Google Scholar]
- 47.Wishart D.S., Sykes B.D., Richards F.M. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31(6):1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 48.Chacko B.M. Structural basis of heteromeric SMAD protein assembly in TGF-beta signaling. Mol Cell. 2004;15:813–823. doi: 10.1016/j.molcel.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 49.Chacko B.M., Qin B., Correia J.J., Lam S.S., de Caestecker M.P., Lin K. The L3 loop and C-terminal phosphorylation jointly define SMAD protein trimerization. Nat Struct Biol. 2001;8:248–253. doi: 10.1038/84995. [DOI] [PubMed] [Google Scholar]
- 50.Receveur-Brechot V., Durand D. How random are intrinsically disordered proteins? A small angle scattering perspective. Curr Protein Pept Sci. 2012;13:55–75. doi: 10.2174/138920312799277901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forbes SA, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic acids research 43, D805-811 (2015). [DOI] [PMC free article] [PubMed]
- 52.Shi Y, Massagué J. Mechanisms of TGFB Signaling from Cell Membrane to the Nucleus. Cell 113, 16-16 (2003). [DOI] [PubMed]
- 53.David C.J., Massagué J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat Rev Mol Cell Biol. 2018;19(7):419–435. doi: 10.1038/s41580-018-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hata A., Lo R.S., Wotton D., Lagna G., Massagué J. Mutations increasing autoinhibition inactivate tumour suppressors SMAD2 and SMAD4. Nature. 1997;388(6637):82–87. doi: 10.1038/40424. [DOI] [PubMed] [Google Scholar]
- 55.Wang J., Choi J.-M., Holehouse A.S., Lee H.O., Zhang X., Jahnel M. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell. 2018;174(3):688–699.e16. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hnisz D., Shrinivas K., Young R.A., Chakraborty A.K., Sharp P.A. A phase separation model for transcriptional control. Cell. 2017;169(1):13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sabari B.R. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361 doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du M., Chen Z.J. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018;361(6403):704–709. doi: 10.1126/science.aat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakao A. TGF-beta receptor-mediated signalling through SMAD2, SMAD3 and SMAD4. EMBO J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu J.-W., Fairman R., Penry J., Shi Y. Formation of a stable heterodimer between SMAD2 and SMAD4. J Biolog Chem. 2001;276(23):20688–20694. doi: 10.1074/jbc.M100174200. [DOI] [PubMed] [Google Scholar]
- 61.Lucarelli P., Schilling M., Kreutz C., Vlasov A., Boehm M.E., Iwamoto N. Resolving the combinatorial complexity of SMAD protein complex formation and its link to gene expression. Cell Syst. 2018;6(1):75–89.e11. doi: 10.1016/j.cels.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 62.Inman G.J., Hill C.S. Stoichiometry of active SMAD-transcription factor complexes on DNA. J Biolog Chem. 2002;277(52):51008–51016. doi: 10.1074/jbc.M208532200. [DOI] [PubMed] [Google Scholar]
- 63.Hill C.S. Transcriptional control by the SMADs. Cold Spring Harb Perspect Biol. 2016;8(10):a022079. doi: 10.1101/cshperspect.a022079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vuzman D., Azia A., Levy Y. Searching DNA via a “Monkey Bar” mechanism: the significance of disordered tails. J Mol Biol. 2010;396(3):674–684. doi: 10.1016/j.jmb.2009.11.056. [DOI] [PubMed] [Google Scholar]
- 65.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 66.Vranken W.F., Boucher W., Stevens T.J., Fogh R.H., Pajon A., Llinas M. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59(4):687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 67.Barbato G., Ikura M., Kay L.E., Pastor R.W., Bax A. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: the central helix is flexible. Biochemistry. 1992;31:5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- 68.Lescop E., Schanda P., Brutscher B. A set of BEST triple-resonance experiments for time-optimized protein resonance assignment. J Magn Reson. 2007;187(1):163–169. doi: 10.1016/j.jmr.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 69.Orekhov V.Y., Jaravine V.A. Analysis of non-uniformly sampled spectra with multi-dimensional decomposition. Prog nucl magn reson spectrosc. 2011;59(3):271–292. doi: 10.1016/j.pnmrs.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 70.Pervushin K., Riek R., Wider G., Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94(23):12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Solyom Z., Schwarten M., Geist L., Konrat R., Willbold D., Brutscher B. BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J Biomol NMR. 2013;55(4):311–321. doi: 10.1007/s10858-013-9715-0. [DOI] [PubMed] [Google Scholar]
- 72.Bottomley M.J., Macias M.J., Liu Z., Sattler M. A novel NMR experiment for the sequential assignment of proline residues and proline stretches in 13C/15N-labeled proteins. J Biomol NMR. 1999;13:381–385. doi: 10.1023/a:1008393903034. [DOI] [PubMed] [Google Scholar]
- 73.Kozlowski L.P., Bujnicki J.M. MetaDisorder: a meta-server for the prediction of intrinsic disorder in proteins. BMC Bioinf. 2012;13:111. doi: 10.1186/1471-2105-13-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holehouse A.S., Das R.K., Ahad J.N., Richardson M.O.G., Pappu R.V. CIDER: Resources to Analyze Sequence-Ensemble Relationships of Intrinsically Disordered Proteins. Biophys J. 2017;112(1):16–21. doi: 10.1016/j.bpj.2016.11.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Petoukhov M.V., Franke D., Shkumatov A.V., Tria G., Kikhney A.G., Gajda M. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr. 2012;45(2):342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bush M.F., Hall Z., Giles K., Hoyes J., Robinson C.V., Ruotolo B.T. Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology. Anal Chem. 2010;82(22):9557–9565. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- 77.Eyal E., Najmanovich R., Mcconkey B.J., Edelman M., Sobolev V. Importance of solvent accessibility and contact surfaces in modeling side-chain conformations in proteins. J Comput Chem. 2004;25(5):712–724. doi: 10.1002/jcc.10420. [DOI] [PubMed] [Google Scholar]
- 78.Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1-2:19–25. [Google Scholar]
- 79.Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78(8):1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- 81.Svergun D., Barberato C., Koch M.H.J. CRYSOL – a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J Appl Crystallogr. 1995;28(6):768–773. [Google Scholar]
- 82.De Biasio A., de Opakua A.I., Mortuza G.B., Molina R., Cordeiro T.N., Castillo F. Structure of p15(PAF)-PCNA complex and implications for clamp sliding during DNA replication and repair. Nat Commun. 2015;6(1) doi: 10.1038/ncomms7439. [DOI] [PubMed] [Google Scholar]
- 83.Cordeiro T.N., Sibille N., Germain P., Barthe P., Boulahtouf A., Allemand F. Interplay of protein disorder in retinoic acid receptor heterodimer and its corepressor regulates gene expression. Structure. 2019;27(8):1270–1285.e6. doi: 10.1016/j.str.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 84.Mylonas E., Hascher A., Bernadó P., Blackledge M., Mandelkow E., Svergun D.I. Domain conformation of tau protein studied by solution small-angle X-ray scattering. Biochemistry. 2008;47(39):10345–10353. doi: 10.1021/bi800900d. [DOI] [PubMed] [Google Scholar]
- 85.Michaud-Agrawal N., Denning E.J., Woolf T.B., Beckstein O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J Comput Chem. 2011;32(10):2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lazar T., Martínez-Perez E., Quaglia F., Hatos A., Chemes L.B., Iserte J.A. PED in 2021: a major update of the protein ensemble database for intrinsically disordered proteins. Nucleic Acids Research. 2021;49(D1):D404–D411. doi: 10.1093/nar/gkaa1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Protein ensembles are deposited at the Protein Ensemble Database (PED) [86], PED00193-PED00202. NMR data are available at the BMRB with accession codes 50738 (SMAD2) and 50737 (SMAD4). SAXS data are available at SASBDB, accession numbers are detailed in Supplementary Table 1. All remaining data are available in the main text or the supplementary materials.