Abstract
Summary
Searching for open reading frames is a routine task and a critical step prior to annotating protein coding regions in newly sequenced genomes or de novo transcriptome assemblies. With the tremendous increase in genomic and transcriptomic data, faster tools are needed to handle large input datasets. These tools should be versatile enough to fine-tune search criteria and allow efficient downstream analysis. Here we present a new python based tool, orfipy, which allows the user to flexibly search for open reading frames in genomic and transcriptomic sequences. The search is rapid and is fully customizable, with a choice of FASTA and BED output formats.
Availability and implementation
orfipy is implemented in python and is compatible with python v3.6 and higher. Source code: https://github.com/urmi-21/orfipy. Installation: from the source, or via PyPi (https://pypi.org/project/orfipy) or bioconda (https://anaconda.org/bioconda/orfipy).
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Open reading frames (ORFs) are sequences that have potential to be translated into proteins. They are delineated by start sites, at which translation is initiated by assembly of a ribosome complex, and stop sites, at which translation is terminated and the ribosome complex disassembles (Sieber et al., 2018).
Accurate annotation of the protein coding regions in sequenced genomes remains a challenging task in bioinformatics. For simpler prokaryotic genomes, ORFs correspond to the potential coding sequences (CDS) (Sieber et al., 2018). In eukaryotes, where gene splicing is prevalent, eukaryotic CDS prediction a much more challenging task (Seetharam et al., 2019; Sieber et al., 2018).
Transcriptomic data is critical in addressing this challenge, where presence of an ORF in a mature tranrscript may indicate a potential protein coding gene (Mahmood et al., 2020; Martinez et al., 2020; Seetharam et al., 2019). These data are key to identifying potential orphan genes (Seetharam et al., 2019), young genes unique to a species (Singh and Wurtele, 2020; Tautz and Domazet-Lošo, 2011; Vakirlis et al., 2020); standard ab initio gene-prediction models are trained on canonical gene features and do not work well for identifying orphan genes, which are often sparse in canonical gene features (Heames et al., 2020; Ruiz-Orera et al., 2015; Seetharam et al., 2019).
Depending on data (genomic, transcriptomic or metagenomic) and researcher interest, the computational problem of ORF prediction may be stated in multiple ways (Sieber et al., 2018), yet existing tools lack the flexibility to allow users to fine-tune or customize the search for ORF sequences. Here we present orfipy, an efficient tool for extracting ORFs from nucleotide sequences. orfipy provides rapid, flexible searches in multiple output formats to allow easy downstream analysis of ORFs.
2 Implementation
orfipy is written in python, with the core ORF search algorithm implemented in cython to achieve faster execution times. orfipy uses the pyfastx library (Du et al., 2020) for efficient parsing of input FASTA/FASTQ file. orfipy can leverage multiple cpu-cores to process FASTA sequences in parallel, based on available memory and cpu cores (Supplementary Data).
2.1 Input, flexible search and output
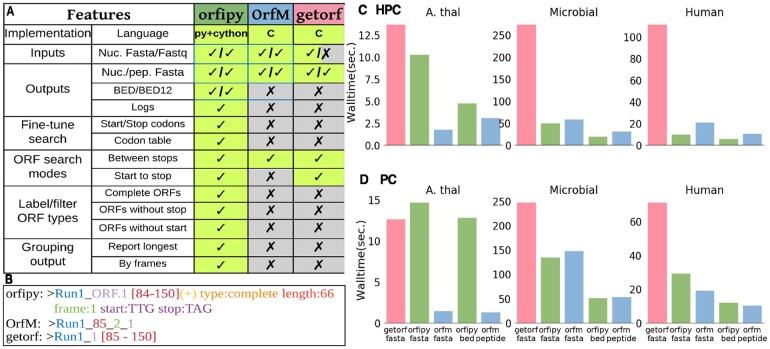
orfipy takes nucleotide sequences in a multi-FASTA/FASTQ, plain or gz-compressed, file as input. Users can provide input parameters that include minimum and maximum size of ORFs, list of start and stop codons and/or a user-defined codon table (Supplementary Data). For efficient and flexible downstream analysis (Fig. 1A, B), orfipy provides multiple output types including BED format. BED files reduce disk space use by storing only the coordinates of the ORFs, and are useful in developing more scalable, flexible downstream analysis pipelines. orfipy also adds relevant information about codon use and ORF types, and can group the output by longest ORF contained in each transcript, or can list each reading frame in each transcript.
Fig. 1.
Comparison of orfipy features and performance with getorf and OrfM. We compared attributes of orfipy with two commonly used tools for ORF identification. (A) Comparison of orfipy features with getorf and OrfM. orfipy provides a number of options to fine-tune ORF search, this includes labeling the ORF type, reporting only the longest ORF and reporting ORFs by translation frame. To allow reproducible analysis, orfipy logs the commands. (B) Example of FASTA headers written to output files by each tool. orfipy output provides information about each ORF that can be readily used in downstream analyses. (C and D) Runtimes, using plain FASTA input, on HPC (128 GB RAM; 28 cores) (C) and PC (16 GB RAM; 8 cores) (D) environments (Supplementary Data). Each analysis was run three times, via pyrpipe (Singh et al., 2020), and the mean runtime is reported. orfipy runtimes are comparable to OrfM for the large microbial and human transcriptome data. orfipy is fastest when ORFs are saved to a BED file; OrfM is fastest when ORFs are saved to peptide FASTA. Data sizes: A.thaliana genome 120 MB; microbial sequences 1.5 GB; human transcriptome 370 MB. fasta, output ORFs to nucleotide and peptide FASTA; bed, output ORFs to BED file; peptide, output ORFs to peptide-only FASTA
orfipy enables researchers to fully fine-tune ORF searches using a variety of options (Fig. 1A). For example, users can limit ORF searching to a specific start codon or choose to output ORFs without an inframe start codon. orfipy labels each ORF for users to easily comprehend results (Supplementary Data).
2.2 Comparison with existing tools
We compared orfipy with two popular ORF searching tools, getorf (Rice et al., 2000) and OrfM (Woodcroft et al., 2016). What sets orfipy apart is its flexibility and the options to fine-tune ORF searches and output (Fig. 1A, B). Runtimes (Fig. 1C, D) depend on software, environment, input (FASTA input is shown) and output-type. In all scenarios except using a PC to analyze the A.thaliana genome, orfipy is much faster than getorf, and comparable to OrfM, with OrfM being faster for FASTQ input (Supplementary Data).
Supplementary Material
Acknowledgements
The authors were grateful to all reviewers for careful reviews and helpful suggestions. They thank the developer of OrfM, Ben Woodcroft, for suggestions regarding extending orfipy functionality. They also thank all the members of Wurtele lab.
Funding
This work was funded in part by National Science Foundation [IOS 1546858], Orphan Genes: An Untapped Genetic Reservoir of Novel Traits, and by the Center for Metabolic Biology, Iowa State University. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation [ACI-1548562], through allocations TG-MCB190098 and TG-MCB200123 awarded from XSEDE.
Conflict of Interest: none declared.
Contributor Information
Urminder Singh, Bioinformatics and Computational Biology Program, Iowa State University, Ames, IA 50011, USA; Center for Metabolic Biology, Iowa State University, Ames, IA 50011, USA; Department of Genetics Development and Cell Biology, Iowa State University, Ames, IA 50011, USA.
Eve Syrkin Wurtele, Bioinformatics and Computational Biology Program, Iowa State University, Ames, IA 50011, USA; Center for Metabolic Biology, Iowa State University, Ames, IA 50011, USA; Department of Genetics Development and Cell Biology, Iowa State University, Ames, IA 50011, USA.
References
- Du L. et al. (2020) Pyfastx: a robust python package for fast random access to sequences from plain and gzipped fasta/q files. Brief. Bioinf. doi: 10.1093/bib/bbaa368. [DOI] [PubMed] [Google Scholar]
- Heames B. et al. (2020) A continuum of evolving de novo genes drives protein-coding novelty in drosophila. J. Mol. Evol., 88, 382–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood K. et al. (2020) De novo transcriptome assembly, functional annotation, and expression profiling of rye (Secale cereale l.) hybrids inoculated with ergot (Claviceps purpurea). Sci. Rep., 10, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez T.F. et al. (2020) Accurate annotation of human protein-coding small open reading frames. Nat. Chem. Biol., 16, 458–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice,P. et al. (2000) EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet., 16, 276–277. [DOI] [PubMed] [Google Scholar]
- Ruiz-Orera J. et al. (2015) Origins of de novo genes in human and chimpanzee. PLoS Genet., 11, e1005721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharam A.S. et al. (2019) Maximizing prediction of orphan genes in assembled genomes. BioRxiv. doi: 10.1101/2019.12.17.880294. [DOI] [PMC free article] [PubMed]
- Sieber P. et al. (2018) The definition of open reading frame revisited. Trends Genet., 34, 167–170. [DOI] [PubMed] [Google Scholar]
- Singh U., Wurtele E.S. (2020) Genetic novelty: how new genes are born. Elife, 9, e55136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh U. et al. (2020) pyrpipe: a python package for rna-seq workflows. bioRxiv. doi: 10.1101/2020.03.04.925818. [DOI] [PMC free article] [PubMed]
- Tautz D., Domazet-Lošo T. (2011) The evolutionary origin of orphan genes. Nat. Rev. Genet., 12, 692–702. [DOI] [PubMed] [Google Scholar]
- Vakirlis N. et al. (2020) Synteny-based analyses indicate that sequence divergence is not the main source of orphan genes. Elife, 9, e53500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodcroft B.J. et al. (2016) OrfM: a fast open reading frame predictor for metagenomic data. Bioinformatics, 32, 2702–2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.