Summary
Small immunoglobulin superfamily (sIGSF) adhesion complexes form a corolla of microdomains around an integrin ring and secretory core during immunological synapse (IS) formation. The corolla recruits and retains major costimulatory/checkpoint complexes, such as CD28, making forces that govern corolla formation of particular interest. Here, we investigated the mechanisms underlying molecular reorganization of CD2, an adhesion and costimulatory molecule of the sIGSF family during IS formation. Computer simulations showed passive distal exclusion of CD2 complexes under weak interactions with the ramified F-actin transport network. Attractive forces between CD2 and CD28 complexes relocate CD28 from the IS center to the corolla. Size-based sorting interactions with large glycocalyx components, such as CD45, or short-range CD2 self-attraction successfully explain the corolla ‘petals.’ This establishes a general simulation framework for complex pattern formation observed in cell-bilayer and cell-cell interfaces, and the suggestion of new therapeutic targets, where boosting or impairing characteristic pattern formation can be pivotal.
Subject areas: Cell biology, Computational bioinformatics, Computational molecular modelling, Immunology, Molecular biology
Graphical abstract
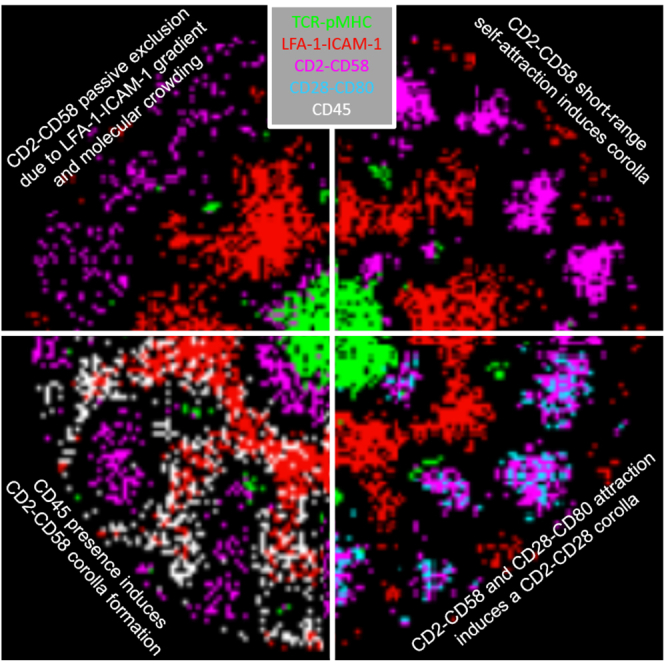
Highlights
-
•
CD2-CD58 were passively excluded due to LFA-1 gradient and molecular crowding
-
•
CD2-CD58 short-range self-attraction induces corolla formation
-
•
CD2-CD58 and CD28-CD80 attraction induces a CD2-CD28 corolla
-
•
CD45 presence induces CD2-CD58 corolla formation
Cell biology; Computational bioinformatics; Computational molecular modelling; Immunology; Molecular biology
Introduction
The organization of membrane receptors at the immunological synapse (IS) interface is important for signal integration and effector function of T cells (Basu et al., 2016; Dustin and Choudhuri, 2016; Hui et al., 2017; Gawden-Bone et al., 2018). The immune synapse is initiated by T cell receptor (TCR) interaction with peptide-major histocompatibility complex (pMHC) to form a specificity complex. The integrin lymphocyte function-associated antigen 1 (LFA-1) forms complexes with intercellular adhesion molecule-1 (ICAM-1) and small immunoglobulin superfamily (sIGSF) member cluster of differentiation 2 (CD2) interacts with CD58 in humans (hCD58) and CD48 in mice (mCD48) in the context of T cell effector function (Monks et al., 1998; Milstein et al., 2008; Van Der Merwe et al., 1994; Milstein et al., 2008). When a live T cell forms an interface with a planar supported lipid bilayer (SLB) containing freely mobile ICAM-1, CD58, and pMHC, the individual complexes are organized into a layered, radially symmetrical pattern with central TCR-pMHC cluster (synaptic cleft and central supramolecular activation cluster [cSMAC]), surrounded by an integrin adhesion ring (peripheral or pSMAC), and fringed by sIGSF microdomains in the distal SMAC (dSMAC), resembling flower petals known as corolla (Monks et al., 1998; Dustin et al., 1998; Grakoui et al., 1999; Kaizuka et al., 2009; Demetriou et al., 2020). cSMAC and pSMAC formation are driven by F-actin-mediated centripetal transport (Kaizuka et al., 2009; Kaizuka et al., 2007; Fritzsche et al., 2017) (Figure S1). Intermittent coupling to the F-actin network (DeMond et al., 2008) with a constant force (Colin-York et al., 2019) is driving TCR-pMHC to the cSMAC. Integrin-mediated adhesion is an active process also regulated by interactions with the F-actin transport network (Nordenfelt et al., 2017). We have previously shown that cSMAC localization of both TCR-pMHC and costimulatory CD28-CD80 complexes can be explained by F-actin-dependent transport (Siokis et al., 2018). sIGSF members operate through highly multivalent microdomains and are less dependent on cellular energy (Milstein et al., 2008), and the contribution of the F-actin network to their localization is poorly characterized. The corolla pattern is completely distinct from that formed by costimulatory and checkpoint complexes, such as CD28-CD80 and PD1-PDL1, which also occupy TCR microclusters and the outer annulus of the cSMAC (Hui et al., 2017; Demetriou et al., 2020; Yokosuka et al., 2008; Tseng et al., 2008; Yokosuka et al., 2012). Strikingly, presence of both CD2-CD58 and CD28-CD80 in the system leads to corolla formation and relocation of the latter from the cSMAC (Yokosuka et al., 2008; Siokis et al., 2018) to the corolla in the dSMAC (Demetriou et al., 2020). Thus, it is important to understand the forces that regulate corolla formation.
To understand the unique localization of sIGSF complexes, we study the positioning of the CD2 receptor. Interacting molecules in the IS display a phase separation behavior based on chemical kinetics coupled to membrane spacing (Hori et al., 2002), depending on the size of the formed complexes. CD2 interaction with mouse CD48 (mCD48) assembles adhesion junctions with an intermembrane gap of 12.8 nm (Milstein et al., 2008; Wang et al., 1999), similar to TCR-pMHC interaction at 13.1 nm (Choudhuri et al., 2005), suggesting limited size-based interaction between them. CD2 interactions with hCD58 or mCD48 are low affinity with fast dissociation kinetics (Van Der Merwe et al., 1994; Dustin et al., 1996; Dustin et al., 1997; Zhu et al., 2006). The long, unstructured, and highly conserved cytoplasmic domain of CD2 is rich in polyproline (PP) motifs that contribute to Src family kinase activation (Kaizuka et al., 2009; Demetriou et al., 2020; Carmo et al., 1993; Bachmann et al., 1999; Skånland et al., 2014; Su et al., 2016). Similar unstructured domains with multiple PP motifs in combination with proteins containing multiple SH3 domains can undergo concentration-dependent liquid-liquid phase separation (Lin et al., 2017). CD2-CD58 complex formation supports T cell polarization (Tibaldi et al., 2002) and also augments and sustains antigen-induced cytoplasmic calcium (Ca2+ increase in T cells (Espagnolle et al., 2007). Blockade of the CD2-CD58 interaction has been shown to impair recruitment of PLCγ1, a key player for downstream signaling in T cells (Espagnolle et al., 2007).
Together with the aforementioned molecules, T cells and antigen-presenting cells (APCs) express numerous other surface glycoproteins that make up the glycocalyx, which plays important roles in T cell interactions, activation, and effector function. A characteristic glycocalyx protein is the transmembrane protein tyrosine phosphatase CD45. No ligand has been described for CD45 on APCs (Figure S1). Nevertheless, it plays an important role for TCR signaling (McNeill et al., 2007), positively regulating T cell activation via dephosphorylation of the inhibitory C-terminal tyrosine of p56lck and p59fyn proteins (Johnson et al., 2000; Thomas and Brown, 1999; Seavitt et al., 1999; Chang et al., 2016), which also interact with CD2 (Carmo et al., 1993). Importantly, CD45 molecules are excluded from the TCR-pMHC microclusters during the initial moments of IS formation and eventually completely relocate to the outer region of the IS, the dSMAC (Johnson et al., 2000; Cordoba et al., 2013; Graf et al., 2007), where the corolla forms (Demetriou et al., 2020). This suggests that CD45 may play a role in the organization of the corolla/dSMAC, although no previous study has addressed this question.
In this study, we focused on understanding the possible mechanisms leading to the CD2 corolla in the dSMAC. We developed a general agent-based model simulating the immunological synapse formation involving all relevant components for corolla pattern emergence, including specificity (TCR-pMHC), intergrin (LFA-1-ICAM-1), sIGSF adhesion (CD2-CD58), and costimulatory (CD28-CD80) complexes as well as glycocalyx molecules (CD45). This complete simulation platform reflects the physical processes of diffusion, chemical kinetics, and agent-agent interactions (Figure S1). In the simulations, the emerging LFA-1 gradient in the pSMAC resulted in two distinct CD2-CD58 populations, one passively following the TCR-pMHC movement toward the cSMAC and the other relocating to the dSMAC and residing in an annular pattern. Attraction between CD2-CD58 and costimulatory CD28-CD80 complexes resulted in the formation of the corolla pattern, where the two classes of interactions can work together. In the absence of costimulatory interactions, self-attraction between CD2-CD58 complexes was sufficient to reproduce the corolla. The origin of such a force could be the interaction of CD2-CD58 complexes with local F-actin asters and focal points but not the centripetal F-actin flow. In simulations with a glycocalyx component (CD45) present, the corolla pattern was reproducible alone by the size-based segregation (SBS) between CD2-CD58 complexes and CD45 molecules. Taken together, these results imply that the corolla pattern formation can be a result of active processes, such as the CD2-CD58 self-attraction and the attraction between CD2-CD58 and CD28-CD80 complexes, or of a physical phase separation process together with competition for space in the presence of glycocalyx components. All these mechanisms can simultaneously contribute during synapse formation, and therefore, additional experiments are required to understand the effect of glycocalyx components and the intermolecular forces.
Results
sIGSF receptor titration fails to reproduce the corolla pattern
In our effort to understand the mechanisms underlying sIGSF adhesion complex localization and interaction with other molecules on the forming synapse, such as costimulatory complexes, we developed a phenomenological agent-based model and validated it by experiments performed on SLBs (Demetriou et al., 2020; Yokosuka et al., 2008). This model takes into account chemical kinetics for the formation of receptor-ligand complexes. SBS of sIGSF versus longer integrin complexes was modeled as a repulsive force and the F-actin-driven centripetal transport as an empirical centrally directed force (Siokis et al., 2018) (Figure S1). As those interactions depend on the biophysical properties of the complexes such as size and on- and off-rates, the following results were generated for a specific example of the involved complexes: the specificity complex TCR-pMHC, the integrin complex LFA-1-ICAM-1, costimulation complexes CD28-CD80 (similar to checkpoint PD1-PDL1), and sIGSF adhesion complex CD2-CD58.
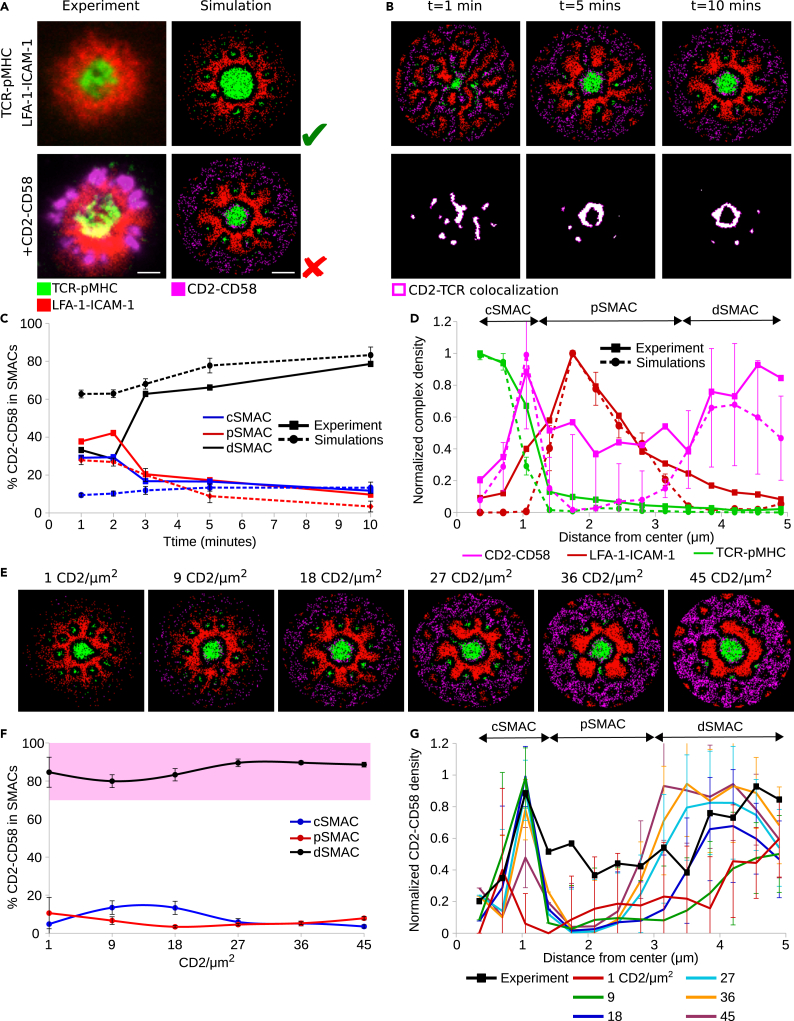
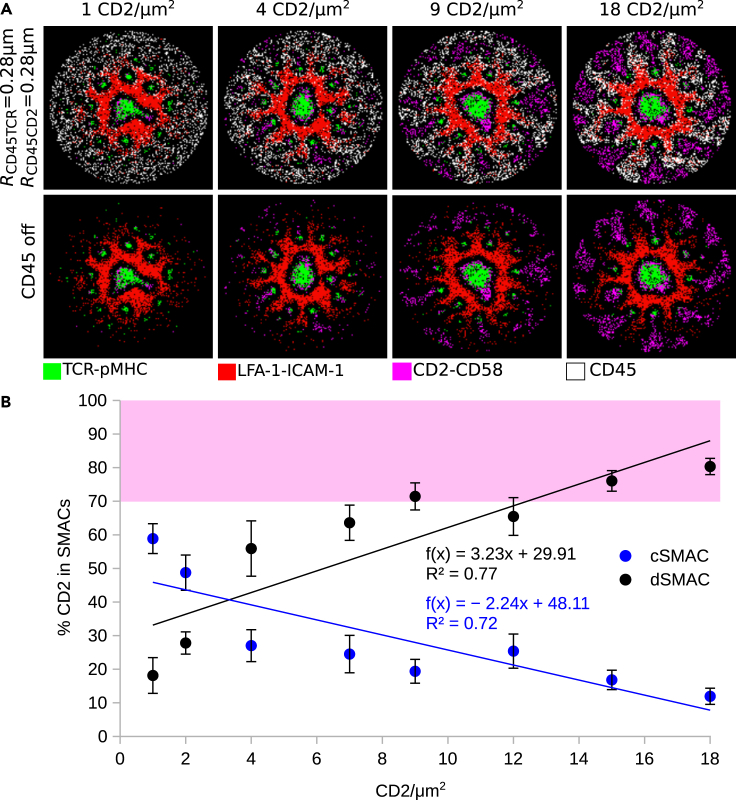
SBS between TCR-pMHC and LFA-1-ICAM-1 complexes was responsible for the formation of TCR-pMHC microclusters, while at the same time, the F-actin-driven centripetal force results in the accumulation of TCR-pMHC in the cSMAC as well as the emergence of an LFA-1-ICAM-1 gradient in the pSMAC (Figure 1A top row). The gradient acted as an exclusion mechanism, altering the localization of free molecules as well as receptor-ligand complexes (Cartwright et al., 2014), which did not interact with the F-actin centripetal flow. SBS between LFA-1-ICAM-1 and CD2-CD58 complexes results in the exclusion of CD2-CD58 complexes toward the dSMAC (Figure 1A bottom row, 1B). The in silico experiments showed that CD2-CD58 and TCR-pMHC complexes initially colocalize in the microclusters (Figure 1B). Measuring the amount of CD2-CD58 in each region of the IS (c, p, dSMAC), we observed that during the first two minutes of IS formation, the in silico and in vitro data did not match (Figure 1C). In vitro, this is the spreading phase of the T cell on the SLB, while in silico the full IS area is already formed. When this phase is over (≥3 min), the in vitro and in silico data are in good agreement (Figure 1C), as can also be seen from the radial density profiles at 10 minutes of IS formation (Figure 1D). Interestingly, a small population of CD2-CD58 complexes passively followed the TCR-pMHC movement toward the center of the IS and localized around the cSMAC (Figures 1A–1C). This population was termed passive followers (Siokis et al., 2018). The majority of CD2-CD58 (≈80%), although excluded to the dSMAC, were correctly positioned but failed to form the corolla pattern (Figures 1A–1D).
Figure 1.
Titration of CD2 in the IS
(A) Experimentally observed IS patterns including different receptor-ligand complexes (left column) and comparison with IS patterns obtained by simulations (right column).
(B) IS formation dynamics between 1 and 10 minutes and colocalization of CD2-CD58 with TCR-pMHC complexes.
(C) Amount of CD2-CD58 complexes in central, peripheral, and distal SMACs during IS formation in simulation versus experiment (Demetriou et al., 2020).
(D) Radial density profiles of TCR, LFA-1, and CD2 complexes along the distance from the center at 10 min.
(E) Titration of the initial CD2 amount from 1 to 45 molecules/μm2.
(F) Amount of CD2-CD58 complexes in central, peripheral, and distal SMACs for different CD2 densities. Pink shaded area denotes the experimental range in the dSMAC.
(G) Radial density profiles of CD2-CD58 complexes along the distance from the center for different CD2 densities. Parameters from Table 1. Varied parameters: 1-45 CD2/μm2; 100 CD58/μm2. Error bars represent SD of N = 10 simulations. TCR-pMHC: green, LFA-1-ICAM-1: red, CD2-CD58: magenta, CD2-TCR colocalization: white.
Demetriou et al. reported that the relative levels of CD2 in different T cell types, including naive and memory CD4+ and CD8+ T cells, and in tumor-infiltrating lymphocytes (TILs) of patients with colorectal cancer (CRC) versus cells from surrounding healthy tissues might play an important role in the emergence of the corolla pattern (Demetriou et al., 2020). TILs of patients with CRC express smaller numbers of CD2 per T cell, resulting in an impaired or completely absent corolla pattern. Therefore, we sought to investigate whether titration of the CD2 amount would result in the corolla pattern in the dSMAC region.
The amount of CD58 in the SLB lattice was abundant (100 CD58/μm2), discounting its role as a limiting factor in CD2-CD58 complex formation. We then studied the localization of CD2-CD58 complexes, starting with population sizes as low as 1 and reaching up to 45 CD2/μm2. With increasing CD2 density, the CD2-CD58 ring in the dSMAC became more prominent (Figure 1E), but the distinct ‘petals’ of the corolla pattern did not emerge. Furthermore, as shown in Figure 1F, increasing CD2 density did not alter the amount of excluded CD2-CD58 (≈90%), reaching the same levels as in Figure 1C. Interestingly, as a result of crowding and SBS between CD2-CD58 and LFA-1-ICAM-1, the passive follower population in the outer cSMAC saturated at a maximum capacity already at 18 CD2/μm2 (Figure 1G). The lack of CD2-CD58 complex clustering in the dSMAC suggests that the amount of CD2 or other similar sIGSF family receptor is not a sufficient driver for corolla formation. Therefore, we investigated different hypotheses and possible mechanisms.
sIGSF adhesion complex self-attraction is sufficient for corolla pattern
In an attempt to recapitulate the clustering of the excluded CD2-CD58 in the corolla, we tested different mechanisms. Both centripetal forces weaker or similar to those acting on LFA-1-ICAM-1 (Figures S2A and S2B) and adhesion between neighboring CD2-CD58 complexes (Figure S2C) failed to reproduce clustering in the dSMAC and thus the corolla.
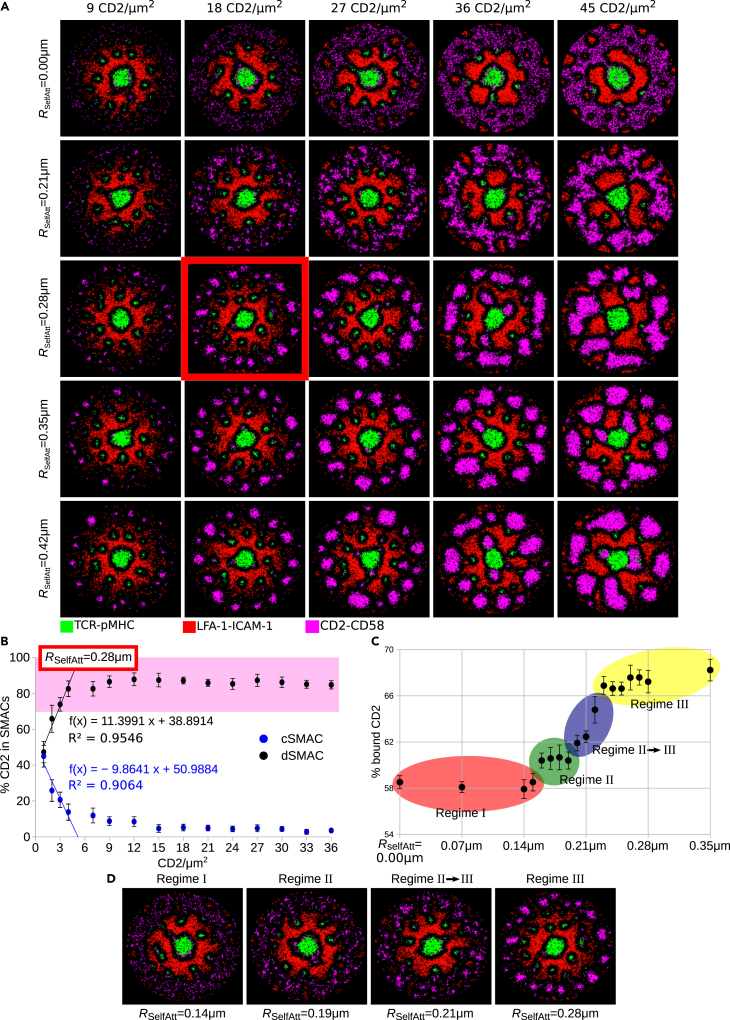
We then hypothesized an attractive interaction between neighboring CD2-CD58 complexes. Such an attraction could be the result of CD2 interacting with F-actin asters that are not part of the ramified transport network (Gowrishankar et al., 2012). This self-attraction was implemented as an attractive force between CD2-CD58 complexes over a distance described by a varying radius RSelfAtt = 0.00–0.42μm and with strength WSelfAtt ∈ (0, 1]. For attraction radius RSelfAtt = 0.28 μm and CD2 concentrations ≥18/μm2, distinct clustering appeared, emulating the corolla pattern (Figure 2A). Increased CD2 density on the T cell surface led to a linear increase of the number of cells, developing a corolla pattern in vitro (Demetriou et al., 2020). Titration of the CD2 amount with fixed attraction radius RSelfAtt = 0.28 μm divided CD2-CD58 into two populations, one localized around the cSMAC and the other in the corolla (Figure 2B). With the increasing amount of CD2, the sIGSF adhesion complex population in the corolla increased in a linear fashion (see linear regression line for CD2 density ≤4/μm2 in Figure 2B) before saturation at ≈90%, as before (Figures 1C and 1F). Assuming ergodicity, the linear increase of the number of cells with developed corolla in vitro (Demetriou et al., 2020) corroborates the linear increase of the number of CD2-CD58 complexes in the corolla with increasing CD2 density in silico.
Figure 2.
CD2 self-attraction results in the corolla pattern
(A) Rows: Different radii of self-attraction, RSelfAtt, between CD2-CD58 complexes. Columns: Different densities of CD2 molecules.
(B) Amount of CD2-CD58 complexes in the central and distal SMAC. Pink shaded area denotes the experimental range in the dSMAC. The function f(x) is a linear regression based on the four lowest CD2 densities, i.e., ≤ 4/μm2.
(C and D) Amount of CD2 that formed complexes with CD58 and the emergence of four regimes based on the amount of CD2-CD58 complex formation and the clustering in the IS considering 18 CD2/μm2. Parameters from Table 1. Varied parameters: 1-45 CD2/μm2; 100 CD58/μm2. Error bars represent SD of N = 10 simulations. TCR-pMHC: green, LFA-1-ICAM-1: red, CD2-CD58: magenta.
Interestingly, stronger self-attraction increased the amount of CD2 molecules that formed complexes with CD58 (Figure 2C). Comparing the amount of bound CD2 with the state of clustering in the dSMAC, we identified four regimes (Figures 2C and 2D). In Regime I, RSelfAtt≤0.15 μm, CD2-CD58 were located in the dSMAC but not clustered. In Regime II, 0.16 ≤ RSelfAtt≤0.19 μm, some individual CD2-CD58 stuck together, while in Regime II→III, a transition regime, 0.20 ≤ RSelfAtt≤0.22 μm, denser CD2-CD58 areas appeared. Finally, in Regime III, RSelfAt≥0.23 μm, the CD2-CD58 clustering was more pronounced and the corolla pattern became apparent. It is remarkable that the simulations suggest a sharp transition from regime II to III.
For the case of self-attraction with radius RSelfAtt = 0.28 μm, we observed that CD2-CD58 complexes initially (t < 3 min) colocalized with TCR-pMHC in the microclusters (Figure S3 [top and middle row] and Video S1). As TCR-pMHC accumulated in the cSMAC (t > 3 min), some of the CD2-CD58 complexes passively followed while the majority were relocated in the dSMAC, where the corolla pattern eventually formed (t = 10 min). This recapitulates the path described experimentally (Demetriou et al., 2020). Plotting the tracks of twenty random CD2-CD58 complexes (Figure S3 [bottom row] and Video S2), we saw that they initially moved inward, and when the LFA-1-ICAM-1 complexes formed in the pSMAC, they were excluded toward the dSMAC. The strength of the self-attraction, WSelfAtt ∈ (0, 1], did not alter these results (Figure S4): The transition from the CD2-CD58 ring in the dSMAC to the corolla occurred for RSelfAtt>0.21 μm (Figure S4A), while their localization in the IS was not substantially affected (Figures S4B–S4D).
These results together showed that a phenomenological attractive force between sIGSF adhesion complexes, possibly originating from their interaction with local F-actin asters or networks of signaling proteins such as Lck, can reproduce in silico the experimentally observed corolla pattern.
sIGSF adhesion complexes alter CD28 localization
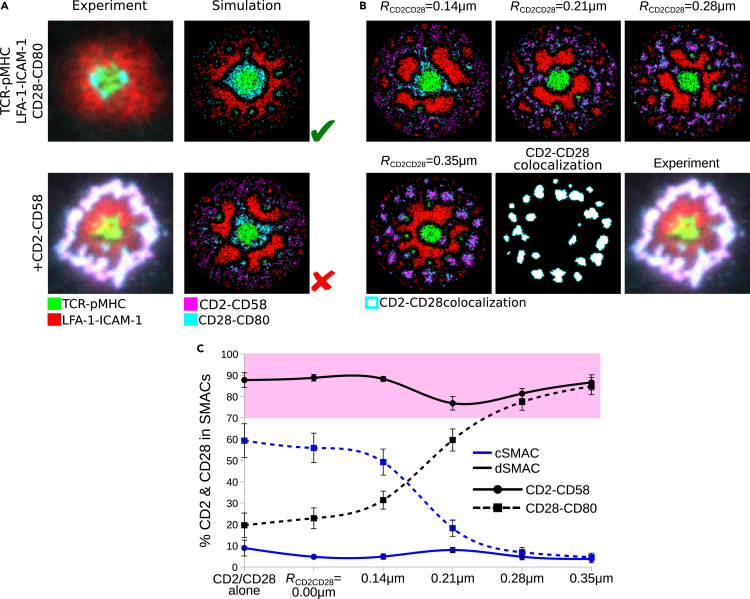
Costimulatory CD28-CD80 complexes (Yokosuka et al., 2008) are structurally similar to the sIGSF adhesion complexes CD2-CD58, but their localization around the cSMAC was completely different in a minimal IS in which LFA-1-ICAM-1 and TCR-pMHC complexes were involved (Figure 3A-top row). Their accumulation around the core of TCR-pMHC complexes in the cSMAC was a combined result of SBS with LFA-1-ICAM-1, passively following TCR-pMHC, and being actively transported toward the center owing to interaction with the F-actin centripetal flow (Figure 3A top row).
Figure 3.
CD28-CD80 complexes relocate to the CD2-CD58 dSMAC based on an attractive force between both complexes that results in the corolla formation
(A) Experimentally observed IS patterns including different receptor-ligand complexes (left column) and comparison with IS patterns obtained by simulations (right column).
(B) Attraction between CD2-CD58 and CD28-CD80 complexes within radius RCD2CD28 = 0.14–0.35 μm and weight WCD2CD28 = 1.0, colocalization of CD2-CD58 and CD28-CD80 complexes for RCD2CD28 = 0.35 μm and experimental data.
(C) Amount of CD2-CD58 (solid) and CD28-CD80 (dashed) complexes in the central and distal SMACs for various CD2-CD28 attractive force radii. Pink shaded area denotes the experimental range of CD2-CD58 and CD28-CD80 in the dSMAC. Parameters from Table 1. Error bars represent SD of N = 10 simulations. Scale bar: 2 μm. TCR-pMHC: green, LFA-1-ICAM-1: red, CD2-CD58: magenta, CD28-CD80: cyan, CD2-CD28 colocalization: white.
Interestingly, it was recently shown experimentally that the sIGSF adhesion complex can relocate costimulatory/checkpoint complexes to the corolla (Figure 3A bottom row) (Demetriou et al., 2020). Considering the aforementioned experimental and theoretical results for CD28 (Yokosuka et al., 2008; Siokis et al., 2018), we run simulations with both CD28/CD80 and CD2/CD58. Owing to the similar sizes of CD2-CD58, CD28-CD80, and TCR-pMHC complexes (≈12.8–13.1 nm), we did not assume any interaction between them, but only introduced SBS between each of these and LFA-1-ICAM-1 complexes.
Introduction of CD2-CD58 complexes into the synapse did not affect the annular localization of CD28-CD80 costimulatory complexes, in contrast to the experimental observation (Figure 3A bottom row). This was also confirmed by measuring the amount of CD2-CD58 and CD28-CD80 complexes in the cSMAC and dSMAC of the IS (Figure 3C). The majority of CD28-CD80 (≈60%) still resided in the cSMAC, while almost 90% of CD2-CD58 complexes were excluded to the dSMAC, where they resided in a ring-like structure. This set of simulations showed that to (i) relocate costimulatory complexes from the cSMAC to the dSMAC and (ii) form the CD2-CD58 corolla in the dSMAC, as also seen earlier (Figures 1 and 2), additional mechanisms are required.
For the relocation of costimulatory complexes to the dSMAC, the similarities of sIGSF adhesion and costimulatory complexes in protein interaction network and size could suggest that the two kinds of complexes prefer to stay together during IS formation, as found experimentally (Demetriou et al., 2020). This was modeled as an attractive interaction defined by interaction radius (RCD2CD28) and strength (WCD2CD28). The attractive force was able to overcome the centripetal force on CD28-CD80 and pull them into the dSMAC region (Figures 3B and 3C, Video S3). The greater the attraction radius, the more the costimulatory complexes relocated in the dSMAC. The amount of CD28-CD80 in the dSMAC increased from ≈20% to ≈80%. The attraction was also sufficient to cluster CD2-CD58 and CD28-CD80 complexes (Figure 3B), recapitulating the experimentally observed corolla pattern (Demetriou et al., 2020).
To further investigate how the corolla was affected by the LFA-1-ICAM-1 gradient (Siokis et al., 2018), in silico simulations were performed with different strengths of the centripetal force on LFA-1-ICAM-1 complexes (CLFA = 0.00–0.20). The attraction of the CD2-CD58 and CD28-CD80 complexes was kept constant at RCD2CD28 = 0.35 μm with strength WCD2CD28 = 1.0 (Figure S5). In the absence of centripetal forces (CLFA = 0.00), no LFA-1-ICAM-1 gradient developed and the CD2-CD28 clusters localized to the pSMAC region rather than being excluded to the dSMAC. As the centripetal force increased in strength (CLFA≥0.03), the CD2-CD28 clusters were excluded to the dSMAC. This result shows the importance of the LFA-1-ICAM-1 gradient for exclusion of sIGSF adhesion and costimulatory complexes to the dSMAC during IS formation.
Glycocalyx proteins residing in the dSMAC promote corolla pattern
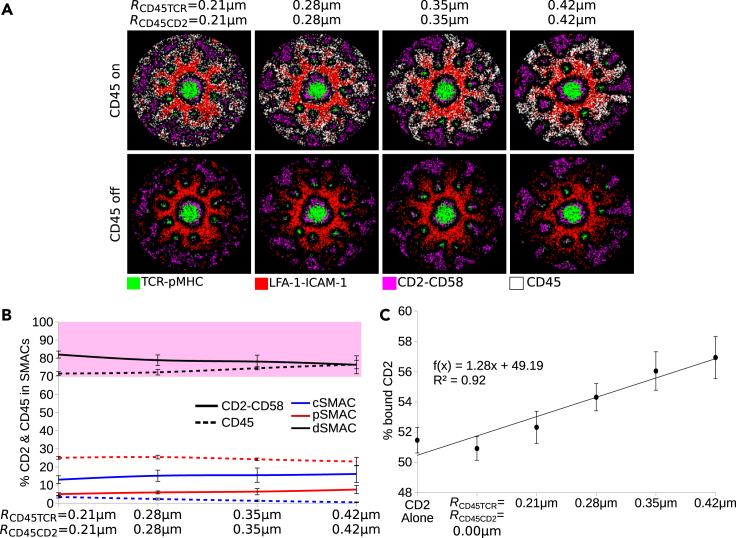
In reality, many more molecules are present in the IS region contributing to a dense glycocalyx. Next, we asked whether such molecules affect the sIGSF adhesion complex localization and the transition from the ring in the dSMAC to the corolla pattern. An example is the transmembrane protein tyrosine phosphatase CD45. CD45 molecules are important for T cell activation (McNeill et al., 2007; Johnson et al., 2000; Thomas and Brown, 1999; Seavitt et al., 1999; Chang et al., 2016), and their concentration in the dSMAC (Johnson et al., 2000; Cordoba et al., 2013; Graf et al., 2007) positions them to regulate sIGSF adhesion complexes in this compartment.
At first, we asked how CD45 molecules are excluded from the IS. Their size, ≈20 nm, is smaller than the maximal reach of LFA-1-ICAM-1 complexes, and the exclusion from the TCR-pMHC microclusters (Johnson et al., 2000; Graf et al., 2007) suggested SBS between CD45 and TCR-pMHC. For simplicity, we first investigated the IS dynamics only with TCR, LFA-1, and CD45 on the T cell and pMHC and ICAM-1 on the SLB. The repulsive force between TCR-pMHC and CD45 with radius RCD45TCR led to initial exclusion of CD45 molecules from TCR microclusters. Together with the LFA-1-ICAM-1 gradient, CD45 was eventually excluded from the cSMAC and inner pSMAC, restricting CD45 to the outer pSMAC and dSMAC (Figures S6A and S6C). Accordingly, we investigated how additional SBS between LFA-1-ICAM-1 complexes and CD45 affects the IS dynamics and pattern. A small distance repulsion (RCD45LFA = 0.14 μm) between the LFA-1-ICAM-1 complexes and CD45 resulted in complete exclusion of CD45 from the pSMAC. Interestingly, for increasing radii, RCD45LFA>0.21 μm, clusters of CD45 appeared in the pSMAC and around the cSMAC (Figure S6B). This resulted in a slightly decreasing amount of CD45 in the dSMAC, but still more than 80% remained there (Figure S6D). Because CD45 is experimentally shown to form a ring in the dSMAC (Johnson et al., 2000; Graf et al., 2007), we decided to include SBS only between TCR-pMHC and CD45.
TCR-pMHC and CD2-CD58 complexes have similar sizes, and therefore, we included the same SBS between both of these and CD45. No self-attraction between CD2-CD58 was included in these in silico experiments. For a repulsion radius of RCD45TCR = RCD45CD2 = 0.21 μm, CD2-CD58 complexes colocalized with CD45 in the dSMAC but did not form a distinct corolla (Figure 4A). The corolla emerged for RCD45TCR = RCD45CD2≥0.28 μm (Video S4). The mechanisms leading to the corolla and the final characteristic pattern per se were not affected by different lattice resolutions, ranging between 35 and 100 nm (Figure S7). Around 80% of the total CD2-CD58 participated in the formation of the corolla in the dSMAC (Figure 4B), a fraction in accordance with the experimental data (Demetriou et al., 2020). Interestingly, with increasing repulsion strength, the amount of CD2 molecules that formed complexes with CD58 increased in a linear fashion (Figure 4C). These results suggested that the corolla pattern formation can also be driven by a phase separation process with competition for space between a percolating phase enriched in CD45 and microdomains enriched in CD2-CD58. Other glycocalyx components could contribute to corolla formation (Shurer et al., 2019).
Figure 4.
Interaction of CD45 with CD2-CD58 and TCR-pMHC complexes
(A) IS pattern at 10 minutes for different radii of repulsion between CD45 and CD2-CD58 as well as CD45 and TCR-pMHC. The same plots are shown with and without resolving CD45.
(B) Amount of CD45 and CD2-CD58 in the central, peripheral, and distal SMACs for different radii of repulsion. Pink shaded area denotes the experimental range of CD2-CD58 in the dSMAC.
(C) Percent of CD2 molecules that formed complexes with CD58. The function f(x) is a linear regression. Parameters from Table 1. Error bars represent SD of N = 10 simulations. TCR-pMHC: green, LFA-1-ICAM-1: red, CD2-CD58: magenta, CD45: white.
Next, we performed CD2 titration while keeping the repulsion at RCD45TCR = RCD45CD2=0.28 μm. For CD2 densities between 1 and 4/μm2, no clustering appeared in the corolla and a large fraction of CD2-CD58 complexes resided in the central IS region (Figures 5A and 5B). As the CD2 amount increased, not only the majority of CD2 was excluded, reaching more than 70% in the dSMAC, in accordance with experiment (Demetriou et al., 2020), but also clustering in the corolla was induced (Figures 5A and 5B). These results together showed that the amount of CD2 can affect corolla formation (Demetriou et al., 2020), provided the presence of large, SBS inducing glycocalyx molecules in the dSMAC. Therefore, reduced CD2 numbers on T cells of CRC TILs, indeed, can result in the absence of CD2-CD58 corolla formation, with implications for the balance of costimulatory and checkpoint function, as suggested before (Demetriou et al., 2020).
Figure 5.
CD2 titration in the presence of CD45
(A) Titration of CD2 amount, with constant repulsion between CD45 and CD2-CD58 and TCR-pMHC (RCD45TCR = RCD45CD2 = 0.28 μm) with CD45 shown (top row) or not (bottom row).
(B) Amount of CD2-CD58 in cSMAC and dSMAC, during CD2 amount titration. Pink shaded area denotes the experimental range of CD2-CD58 in the dSMAC. The function f(x) is a linear regression. Parameters from Table 1. Varied parameters: 1-18 CD2/μm2. Error bars represent SD of N = 10 simulations. TCR-pMHC: green, LFA-1-ICAM-1: red, CD2-CD58: magenta, CD45: white.
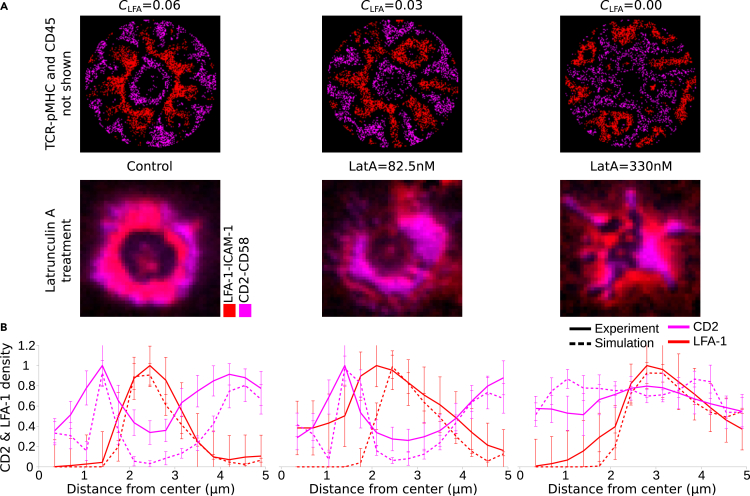
Finally, to confirm the presence and also to show the importance of the LFA-1 gradient, we performed in vitro experiments where after allowing the synapse to form for 20 min, the cells were treated with Latrunculin A (LatA). LatA is known to disrupt the ramified F-actin transport network and therefore the centripetal flow of LFA-1 complexes. In accordance with the experiments, we performed simulations where the centripetal flow of LFA-1 (CLFA) was altered after 20 minutes of synapse formation. Interestingly, we observed both in vitro and in silico that depending on the LatA dose, and accordingly depending on strength of the centripetal flow CLFA, the corolla pattern is disrupted and eventually ceases to exist. CD2-CD58 complexes then move to the center of the IS (Figure 6). For LatA = 330 nM, the corolla completely disappeared in vitro, as was also observed for CLFA = 0.00 in silico (Figure 6A). The radial density profiles of CD2 and LFA-1 complexes were similar between experiments and simulations (Figure 6B). These data provide an additional qualitative validation of the IS simulation model. These experiments could be also performed with acto-myosin inhibitors targeting Arp2/3 such as CK666 (Murugesan et al., 2016). Acto-myosin leads to F-actin arc formation and the consequent centripetal transport of complexes, and therefore, partial or complete inhibition of it will lead to impaired synapse formation.
Figure 6.
LatA treatment and alteration of the LFA-1 centripetal transport strength (CLFA), 20 minutes after synapse formation
(A) IS simulations of altered CLFA strength (top) and cells treated with different doses of LatA.
(B) Radial density profiles of CD2-CD58 and LFA-1-ICAM-1 complexes along the distance from the center for different CLFA strengths (dashed lines) and LatA doses (solid lines). Parameters from Table 1. Varied parameters: 14 TCR, pMHC/μm2; 23 CD2, CD58/μm2; CLFA = 0.00–0.06. Error bars represent SD of N = 10 simulations and at least N = 15 treated cells. LFA-1-ICAM-1: red, CD2-CD58: magenta.
Discussion
A limitation of previous immune synapse simulation models was a failure to incorporate two major classes of molecules. Our studies and others have simulated basic features of IS formation related to the cSMAC and pSMAC only (Qi et al., 2001; Weikl and Lipowsky, 2004; Figge and Meyer-Hermann, 2006; Carlson and Mahadevan, 2015). Recently, we refined the model to incorporate costimulation complexes and a more accurate simulation of the pSMAC (Siokis et al., 2018). We also now recognize that rules developed for costimulatory interactions likely also apply to checkpoint molecules that counteract costimulatory signals (Hui et al., 2017). In this study, we have generalized this agent-based model to incorporate sIGSF adhesion complexes such as CD2-CD58 and large glycocalyx molecules such as CD45 to simulate a newly described feature of human T cell IS (Demetriou et al., 2020). The in silico experiments pointed to two possible mechanisms contributing to CD2 corolla formation. One active, energy-consuming mechanism, where self-attraction between sIGSF adhesion complexes as well as attraction between sIGSF adhesion and costimulatory complexes led to their clustering and together with exclusion by the integrin adhesion complex gradient, to the formation of the corolla pattern (Figures 2, 3, S3, and S5). The CD2 corolla can also be formed by an extracellular size-dependent phase separation mechanism in combination with competition for space. In this case, the presence of other, SBS-inducing molecules in the dSMAC, such as glycocalyx component CD45, led to sIGSF adhesion complex clustering and consequently the corolla pattern (Figures 4 and 5). These molecular and mechanistic features establish a general model and simulation platform that can recapitulate complex pattern formation processes observed in cell-bilayer and cell-cell interfaces.
We have categorized many potential interactions into larger groups based on specificity (TCR-pMHC), integrin adhesion complexes (LFA-1-ICAM-1), sIGSF adhesion complexes (CD2-CD58), and costimulatory/checkpoint complexes (CD28-CD80 and PD-1-PDL1). We expected that methods we have used to simulate sIGSF adhesion also apply to hemophilic SLAM family members and heterophilic but promiscuous adhesion molecules such as CR-TAM-CADM1 (Takeuchi et al., 2015). Costimulatory interactions may also include ICOS-ICOSL interactions and tumor necrosis factor (TNF) receptor superfamily members that can interact with sIGSF or TNF superfamily ligands (Demetriou et al., 2020; Mintz et al., 2019). The similarities in size and interaction networks of CD2-CD58 sIGSF adhesion complex and CD28-CD80 costimulatory complex together with the indirect links with F-actin (Hutchings et al., 2003) supported the notion that sIGSF adhesion complexes bind to the centripetal flow of F-actin, just like CD28-CD80 complexes (Siokis et al., 2018). Interactions of individual engaged CD2 with F-actin would be transient owing to the brief lifetime of CD2-CD58 interactions. These interactions may not result in domain transport as high lateral mobility within the microdomains might instead generate convective mixing within the microdomains and deformation away from perfect circles dictated by self-attraction or size-dependent phase separation mechanisms. Although, in some cases, a weak centripetal force seemed compatible with corolla formation, the amount of CD2-CD58 complexes residing around the cSMAC contradicted the experimental data (Figures S2A and S2B) (Demetriou et al., 2020). The localization of sIGSF adhesion complexes then more resembled that of costimulatory complexes (Yokosuka et al., 2008; Siokis et al., 2018), rather than the corolla (Demetriou et al., 2020). In general, adhesion molecules are expressed at 10-fold greater abundance than costimulatory complexes, but our results suggest there are additional differences.
The attractive force between sIGSF adhesion complexes and costimulatory/checkpoint complexes or among sIGSF adhesion complexes that are sufficient for corolla formation in simulation may arise from different types of lateral interactions. There are two classes of interactions to consider: active solids such as F-actin asters (Hutchings et al., 2003) and active liquids such as cytoplasmic condensates implication in TCR signaling (Su et al., 2016). Linking to an active solid such as F-actin cytoskeleton requires adapter proteins. For example, the cytoplasmic domain of CD2 interacts with CapZ through CD2AP (CD2-associated protein) (Hutchings et al., 2003). This interaction is believed to orchestrate protein patterning in the IS (Dustin et al., 1998). The multiple PP motifs in the long, unstructured cytoplasmic domain of CD2 are a signature of proteins that can participate in condensate formation (Hyman et al., 2014). There are several possibilities for the implementation of such an interaction including hybrids in which liquid condensates interact with F-actin filaments. The necessary range of self-attraction suggests that the organizing structure would be on the order of >200-350 nm. Alternately, short-range interactions of neighboring complexes, similar to TCR cross-linking (Dixon et al., 1989; Minguet et al., 2007), were unable to reproduce the CD2-CD58 corolla pattern (Figure S2C).
The second mechanism able to recapitulate the corolla was a passive mechanism based on SBS and local crowding. CD45 is abundantly expressed on T cells and more generally on all leukocytes and is important for signaling (McNeill et al., 2007; Zamoyska, 2007). CD45 localization in the dSMAC together with the strong SBS (Johnson et al., 2000; Graf et al., 2007) suggested a repulsive interaction between the two. The confining effects of this force on sIGSF adhesion complexes may account for corolla formation. In general, the model suggests that crowding in the IS and specifically in the dSMAC can induce pattern formation without self-attraction between the sIGSF adhesion complexes. Additional experiments are required for an improved understanding of whether CD45 or any other kind of molecule or complex present in the dSMAC is responsible for the CD2 corolla formation. A combination of knockouts targeting particular core proteins and glycosylation pathways might be able to sufficiently modulate the glycocalyx of T cells to test this mechanism.
The incorporation of the sIGSF adhesion complex corolla and alternative models for how the structure can form greatly increases the fundamental and practical utility of IS simulations. Genetic variants of CD2 and its ligand CD58 are important in rheumatoid arthritis (Raychaudhuri et al., 2009). Individuals with variants of CD58 that decreased expression are at a greater risk of developing multiple sclerosis (De Jager et al., 2009). In cancer, loss of CD58 is associated with relapse in Hodgkins disease (Razak et al., 2016) and low CD2 expression is associated with poor outcome on melanoma (Patel et al., 2017). Moreover, exhausted T cells with low CD2 are observed in patients with CRC and have a similar impact on TCR signaling to checkpoint PD-1 engagement (Demetriou et al., 2020). This general model is now also likely to be predictive for patterns of checkpoint complex forming including interactions of PD-1 and its ligands. Additional features such as cis interaction of CD80 and PD-L1, and PD-1 and PD-L1 between adjacent molecules in the same membrane (Zhao et al., 2018; Chaudhri et al., 2018), or introduction of another major class of adhesion molecules such as cadherins (Brasch et al., 2012; Wu et al., 2010) could be rapidly implemented.
It has to be noted that Demetriou et al., 2020 showed that the corolla pattern emerges and is similar when formed on T cell-APC and T cell-SLB conjugates. The greater imaging convenience of the latter leads to the wider use of SLBs. Similar to T cells, the cytoskeleton of APCs is also dynamically changing during synapse formation, and these changes may not only affect (by assisting or even resisting) the movement of certain molecules but also signaling that can be altered which consequently may affect synapse formation. Nonetheless, the benefits of SLB use are widely acceptable in the biological community despite being a simplification of the real biological system. Our model is validated on data from SLBs (Demetriou et al., 2020; Yokosuka et al., 2008) because data from three-dimensional (3D) T cell-APC conjugates are scarce. Our model has to be considered as a phenomenological model in an attempt to understand possible mechanisms leading to characteristic pattern formation. Our predictions need to be validated by additional experiments either on SLBs or with real APCs, allowing us to further validate and improve the current model in an iterative process. A 3D representation of T cells and APCs together with signaling in the forming synapse will allow the in silico experiments to better represent physiological synapse dynamics. The physics of the membranes of the two interacting cells as well as the signaling cascades affecting pattern formation have to be considered as future developments of the current agent-based model.
The current model ignored additional complexity emerging from endocytosis, exocytosis/recycling, as well as molecule influx into and outflux out of the T cell-APC/SLB interface (Liu et al., 2000; Martín-Cófreces et al., 2014), but these mechanisms are not expected to alter the emerging characteristic patterns (Figure S8). Although during synapse formation the number of participating molecules seems to remain constant and speak for flow equilibrium, effects of such dynamics on the pattern merit further investigation.
The presented agent-based simulation platform can be easily extended to study spatial reorganization and complex pattern formation of transmembrane molecules in the plasma membrane of cells composed of lipids. Data obtained by cell-cell (Comrie et al., 2015; Duchez et al., 2011), cell-SLB, or cell-glass coverslip synapses of epithelial cells, cancer cells, or even neurons (Salaita et al., 2010; Yu et al., 2015; Pautot et al., 2005) can further inform the simulation platform. Commonly acknowledged mechanisms derived from the experimental data can assist the computer simulation and lead to predictions and suggestions of possible mechanisms for the molecular organization on the different cell-cell/SLB/glass interfaces.
Limitations of the study
In this study, we presented a phenomenological model validated on T cell-SLB conjugates. The predictions made in this study should be validated by new experiments. To better describe physiological synapse dynamics, a 3D representation of T cells and APCs including complex membrane physics is required. In addition, signaling should also be considered as it may also affect the dynamics of synapse formation. Further complexity arising from endocytosis, recycling, influx, and outflux of molecules and complexes during synapse formation, although briefly discussed in this study, should be examined in detail in the future.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Software and algorithms | ||
| C++ ISO/IEC 14882:2011 | ISO/IEC 14882:2011 Information technology—Programming languages—C | https://www.iso.org/standard/50372.html |
| Qt creator | https://download.qt.io/archive/qt/5.2/5.2.0/ | |
| Microsoft Office | https://www.microsoft.com/de-de/download/office.aspx | |
Resource availability
Lead contact
Further information and requests can be addressed to Michael Meyer-Hermann (mmh@theoretical-biology.de) or Anastasios Siokis (anastasiossio@gmail.com).
Materials availability
This study did not generate new unique reagents.
Method details
Agent-based model
The agent-based model used in this article was formulated in order to replicate experiments performed on supported lipid bilayers (SLBs) instead of APCs (Siokis et al., 2018; Siokis et al., 2017). The model consists of two square lattices, with each node occupied by only one freely diffusing agent, namely the in silico molecules. Each agent can diffuse to one of its eight neighboring nodes (Moore neighborhood). Diffusion is implemented as a random walk with a probability defined by the speed of diffusion (Table 1) and the simulation timestep. Binding and unbinding of molecules can happen only with ligands on the exact same position on the opposite lattices, with probabilities defined by (Figge and Meyer-Hermann, 2009):
where τ is the simulation timestep, kon and koff are the on- and off-rates, respectively, V the volume of the complex about to form and NA the Avogadro's number. The kon and koff for each specific complex forming in the IS are shown in Table 1.
Table 1.
Reference parameter values, used throughout the article
| Name | Parameter | Value | References |
|---|---|---|---|
| Radius of contact region | R (μm) | 4.9 | (Siokis et al., 2018; Figge and Meyer-Hermann, 2009) |
| Lattice constant | a (μm) | 0.07 | (Siokis et al., 2018; Figge and Meyer-Hermann, 2009) |
| Free molecule diffusion constant | Dm (μm2/s) | 0.10 | (Siokis et al., 2018; Figge and Meyer-Hermann, 2009) |
| Complex diffusion constant | Dc (μm2/s) | 0.06 | (Siokis et al., 2018; Figge and Meyer-Hermann, 2009) |
| Radius of SBS between TCR-pMHC and LFA-1-ICAM-1 | RSBS,TCR-LFA (μm) | 0.42 | (Siokis et al., 2018; Figge and Meyer-Hermann, 2009) |
| Weight of SBS between TCR-pMHC and LFA-1-ICAM-1 | WSBS,TCR-LFA | −1.0 | (Siokis et al., 2018; Figge and Meyer-Hermann, 2009) |
| Radius of SBS between CD28-CD80 and LFA-1-ICAM-1 | RSBS,CD28-LFA (μm) | 0.42 | (Siokis et al., 2018) |
| Weight of SBS between CD28-CD80 and LFA-1-ICAM-1 | WSBS,CD28-LFA | −1.0 | (Siokis et al., 2018) |
| Radius of SBS between CD2-CD58 and LFA-1-ICAM-1 | RSBS,CD2-LFA (μm) | 0.42 | fixed |
| Weight of SBS between CD2-CD58 and LFA-1-ICAM-1 | WSBS,CD2-LFA | −1.0 | fixed |
| Radius of SBS between CD45 and TCR-pMHC | RCD45TCR (μm) | 0.28 | varied: 0.00–0.42 |
| Weight of SBS between CD45 and TCR-pMHC | WCD45TCR | −1.0 | fixed |
| Radius of SBS between CD45 and LFA-1-ICAM-1 | RCD45LFA (μm) | 0.00 | varied: 0.00–0.28 |
| Weight of SBS between CD45 and LFA-1-ICAM-1 | WCD45LFA | 0.0 | varied: −1.00-0.00 |
| Radius of SBS between CD45 and CD2-CD58 | RCD45CD2 (μm) | 0.28 | varied: 0.21–0.42 |
| Weight of SBS between CD45 and CD2-CD58 | WCD45CD2 | −1.0 | fixed |
| Radius of attraction between CD2-CD58 and CD28-CD80 | RCD2CD28 (μm) | 0.35 | varied: 0.00–0.42 |
| Weight of attraction between CD2-CD58 and CD28-CD80 | WCD2CD28 | 1.0 | fixed |
| Radius of self-attraction between CD2-CD58 and CD2-CD58 | RSelfAtt (μm) | 0.28 | varied: 0.00–0.42 |
| Weight of self-attraction between CD2-CD58 and CD2-CD58 | WSelfAtt | 1.0 | varied: 0.2–1.0 |
| Dissociation rate TCR-pMHC | koff,TCR (1/s) | 0.1 | (Siokis et al., 2018; Figge and Meyer-Hermann, 2009) |
| Association rate TCR-pMHC | kon,TCR (1/Ms) | 2∗104 | (Siokis et al., 2018; Figge and Meyer-Hermann, 2009) |
| Dissociation rate LFA-1-ICAM-1 | koff,LFA (1/s) | 0.03 | (Siokis et al., 2018; Figge and Meyer-Hermann, 2009) |
| Association rate LFA-1-ICAM-1 | kon,LFA (1/Ms) | 3∗105 | (Siokis et al., 2018; Figge and Meyer-Hermann, 2009) |
| Dissociation rate CD28-CD80 | koff,CD28 (1/s) | 1.6 | (Siokis et al., 2018) |
| Association rate CD28-CD80 | koff,CD28 (1/Ms) | 4∗105 | (Siokis et al., 2018) |
| Dissociation rate CD2-CD58 | kon,CD2 (1/s) | 4 | (Van Der Merwe et al., 1994; Dustin et al., 1997; Dustin et al., 1996) |
| Association rate CD2-CD58 | koff,CD2 (1/Ms) | 6∗105 | (Van Der Merwe et al., 1994; Dustin et al., 1997; Dustin et al., 1996) |
| TCR-pMHC centrally directed force | CTCR | 1.00 | varied: 0.00–1.00 |
| LFA-1-ICAM-1 centrally directed force | CLFA | 0.06 | varied: 0.00–0.10 |
| CD28-CD80 centrally directed force | CCD28 | 0.20 | (Siokis et al., 2018) |
| CD2-CD58 centrally directed force | CCD2 | 0.00 | varied: 0.00–0.10 |
| Density of TCR, pMHC | (1/μm2) | 9 | varied: 9-14 |
| Density of LFA-1, ICAM-1 | (1/μm2) | 23 | fixed |
| Density of CD28, CD80 | (1/μm2) | 11 | fixed |
| Density of CD2, CD58 | (1/μm2) | 18 | varied: 1-100 |
| Density of CD45 | (1/μm2) | 21 | fixed |
The forces between interacting complexes are modeled as weighted vectors from one complex towards all its interacting neighbors. The forces are exerted within a radius Rforce and all the vectors are summed. The strength of the force is represented as a weight, Wforce, where negative weight represents repulsion (SBS) and positive attraction (Siokis et al., 2018). In a similar manner, interaction of the IS molecules with the centripetal flow of F-actin is modeled as an empirical centrally directed force added to the vector of the interaction forces discussed above. The strength of this force, CX > 0, where X represents each specific complex, shows the coupling strength to F-actin (Siokis et al., 2018).
Adhesion between neighboring CD2 pairs was implemented as a phenomenological factor, g(N), reducing the movement probability, pmove → pmoveg(N), with g(N) given by (Figge and Meyer-Hermann, 2009):
where β ε [1, 2] is the strength and N is the number of neighbors considered. Different hypotheses were tested for different number of adhering neighbors, N, where N ≤ γ, with γ = 2, 4, 6 and 8 (Figure S2).
In addition, a variety of the model robustness tests has been previously performed (Siokis et al., 2018). Different lattice constants, ranging from 35-100 nm (Figure S7), different densities of molecules either participating in the synapse formation process or not, barriers that block the movement of complexes but not free molecules, similar to (DeMond et al., 2008) and finally an exchange algorithm where complexes trying to move to each other's position can swap places have all resulted in unchanged characteristic pattern formation dynamics (Siokis et al., 2018). Additional mechanisms such as endocytosis, recycling of molecules back to the T cell surface as well as influx and outflux of molecules from the synapse interface (Figure S8) did not alter the presented results. A random position (x, y) on the T cell lattice is selected and the molecule/complex present is recognized. Based on (Liu et al., 2000) we have assumed internalization rate of TCR-pMHC complexes to be approximately 0.6%/minute. Due to data scarcity, we have assumed the same internalization rate for CD2-CD58 and LFA-1-ICAM-1 complexes. When a complex (TCR-pMHC, CD2-CD58 or LFA-1-ICAM-1) is selected for internalization the 8 nearest neighbors are also internalized representing the way the membrane engulfs a region of the surface molecules. In addition, we have assumed a total rate of recycling and influx of molecules equal to the internalization rate mentioned above. This assumption followed experimental evidence suggesting that the number of molecules in the synapse region during synapse formation tends to remain constant despite all the influxes and outfluxes of molecules, in a flow-equilibrium state. The molecular trafficking dynamics require additional investigations which will increase the complexity of the current article and therefore were not considered.
Cell culture
Donor specific CD4 T cells were plated at 1x106 cells/ml with CD3/CD28 activation beads at a 1:1 ratio with 50 U/ml IL-2 at 2 ml per well in a 24 well plate and expanded for three days at 37°C in a CO2 incubator. On day three, cells were resuspended, counted and replenished with fresh media to reach the original density of 1x106 cells/ml. The cells were expanded for four more days replenished with 50 U/ml IL-2 and fresh culture media every second day. The cells were left without IL-2 the day before they were plated on planar supported lipid bilayers (PSLBs).
Preparation of PSLBs for high throughput imaging
96-well glass bottom plates (Brooks Life Science Systems) were incubated with 1% Hellmanex III in 50% isopropanol overnight at room temperature (RT). The wells were then rinsed with a plate washer (Wellwash Versa, Thermo Scientific). To remove residual water, the plates were centrifuged upside down for 1 min at 2000 rpm and dried with nitrogen gas. The wells were then cleaned with 3M NaOH for 1 hour at RT and rinsed as before. 50 μl 12.5% NTA lipids in DOPC were then loaded per well and the plate incubated for 20 min at RT. The wells were subsequently washed with 2 ml 0.1% BSA in HBS (20 mM HEPES, 137 mM NaCl, 1.7 mM KCl, 0.7 mM Na2PO4, 5 mM glucose, 2 mM MgCl2, 1 mM CaCl2, pH 7.2), blocked with 400 μl blocking solution (2% BSA in HBS) and mixed for 3 min at 300 rpm. The plate was then incubated for 20 min at RT before it was washed with 2 ml 0.1% BSA in HBS.
Each well was loaded with 100 μl of a protein mix containing ICAM-1-AF647, UCHT1 (unlabeled) and CD58-AF488. The protein concentrations were calibrated by flow cytometry to yield a density of 200 molecules/μm2 ICAM-1, 100 molecules/μm2 UCHT1, and 200 molecules/μm2 CD58. Proteins and lipids were then mixed for 3 min at 300 rpm and incubated for 30 min at RT. Each well was subsequently washed with 2 ml 0.1% BSA in HBS. Cells were incubated on the bilayers for 20 min. at 37°C in a CO2 incubator. They were then treated with either DMSO, 82.5 nM latrunculin A or 330 nM latrunculin A for another 15 min. The cells were then fixed with 100 μl 4% PFA for 10 min at RT. Following fixation, the cells were imaged with an IN Cell Analyzer 6000 (GE Healthcare) with a 40x (0.75 NA) objective.
Quantification and statistical analysis
The in silico radial density plots are calculated inside equally spaced concentric rings as the fraction of occupied grid points by each type of complex and is normalized by the maximum value. The presented simulation curves and error bars represent the mean values and standard deviation (SD), respectively, of at least ten computer simulations. The central, peripheral and distal SMACs are defined at the end of the in silico experiment, which also determines the amount of each complex in each SMAC in the simulations.
The experimental images were analyzed using the Matlab based TIAM software (https://github.com/uvmayya/TIAM_HT) described previously (Valvo et al., 2017). Following this analysis, individual cells were chosen at random and their radial averages analysed using the Fiji macro RadAv (https://github.com/donFellus/radAv). This macro is specifically designed to analyze the average position of subcellular structures within the immunological synapse. It rotates a selected image 360 times 1 degree and creates a copy following each rotation. It then merges all the copies to create a radial positional average of the fluorescence signals in the original image. Further details can be found in the respective Figure legends.
Acknowledgments
This work was supported by the Human Frontier Science Program (RGP0033-2015), the Wellcome Trust (100262Z/12/Z), the Kennedy Trust for Rheumatology Research, and the European Commission (ERC-2014-AdG_670930). A.S. was also funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) - Project-ID 97850925 - SFB 854. A.S. was part of the project ZT-I-0010 funded by the Helmholtz Association. A.K. was supported by The Research Council of Norway in conjunction with Marie Sklodowska-Curie Actions (Project number 275466).
Author contributions
A.S. conceived and designed the study. A.S. designed the code and performed and analyzed the simulations. P.A.R. and M.M.-H. supervised the work. A.S., M.L.D., and M.M.-H. wrote and edited the manuscript. P.D., A.K., S.V., V.M., and M.L.D. provided the experimental figures and made intellectual contributions.
Declaration of interests
The authors have no competing interests to declare.
Published: October 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103100.
Contributor Information
Michael L. Dustin, Email: michael.dustin@kennedy.ox.ac.uk.
Michael Meyer-Hermann, Email: mmh@theoretical-biology.de.
Supplemental information
Data and code availability
The software used for this study is developed in C++ according to all the algorithmic details explained in (Siokis et al., 2017; Siokis et al., 2018) and is available on https://github.com/AnastasiosSiokis/IS_Cell.
Any additional information required to reanalyze and resimulate the data reported in this paper is available from the lead contact upon request.
References
- Bachmann M.F., Barner M., Kopf M. CD2 sets quantitative thresholds in T cell activation. J. Exp. Med. 1999;190:1383–1392. doi: 10.1084/jem.190.10.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R., Whitlock B.M., Husson J., Le Floc’h A., Weiyang J., Oyler-Yaniv A., Farokh D., Giannone G., Hivroz C., Biais N. Cytotoxic T cells use mechanical force to potentiate target cell killing. Cell. 2016;165:100–110. doi: 10.1016/j.cell.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasch J., Harrison O.J., Honig B., Shapiro L. Thinking outside the cell: how cadherins drive adhesion. Trends Cell Biol. 2012;22:299–310. doi: 10.1016/j.tcb.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson A., Mahadevan L. Elastohydrodynamics and kinetics of protein patterning in the immunological synapse. Plos Comput. Biol. 2015;11:e1004481. doi: 10.1371/journal.pcbi.1004481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo A.M., Mason D.W., Beyers A.D. Physical association of the cytoplasmic domain of CD2 with the tyrosine kinases p56lck and p59fyn. Eur. J. Immunol. 1993;23:2196–2201. doi: 10.1002/eji.1830230922. [DOI] [PubMed] [Google Scholar]
- Cartwright A.N., Griggs J., Davis D.M. The immune synapse clears and excludes molecules above a size threshold. Nat. Commun. 2014;5:1–13. doi: 10.1038/ncomms6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang V.T., Fernandes R.A., Ganzinger K.A., Lee S.F., Siebold C., McColl J., Jönsson P., Palayret M., Harlos K., Coles C. Initiation of T cell signaling by CD45 segregation at’close contacts. Nat. Immunol. 2016;17:574–582. doi: 10.1038/ni.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhri A., Xiao Y., Klee A.N., Wang X., Zhu B., Freeman G.J. PD-L1 binds to B7-1 only in cis on the same cell surface. Cancer Immunol. Res. 2018;6:921–929. doi: 10.1158/2326-6066.CIR-17-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhuri K., Wiseman D., Brown M.H., Brown K., van der Merwe P.A. T-cell receptor triggering is critically dependent on the dimensions of its peptide-MHC ligand. Nature. 2005;436:578. doi: 10.1038/nature03843. [DOI] [PubMed] [Google Scholar]
- Colin-York H., Javanmardi Y., Skamrahl M., Kumari S., Chang V.T., Khuon S., Taylor A., Chew T.-L., Betzig E., Moeendarbary E. Cytoskeletal control of antigen-dependent T cell activation. Cell Rep. 2019;26:3369–3379. doi: 10.1016/j.celrep.2019.02.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comrie W.A., Li S., Boyle S., Burkhardt J.K. The dendritic cell cytoskeleton promotes T cell adhesion and activation by constraining ICAM-1 mobility. J. Cell Biol. 2015;208:457–473. doi: 10.1083/jcb.201406120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordoba S.-P., Choudhuri K., Zhang H., Bridge M., Basat A.B., Dustin M.L., van der Merwe P.A. The large ectodomains of CD45 and CD148 regulate their segregation from and inhibition of ligated T-cell receptor. Blood. 2013;121:4295–4302. doi: 10.1182/blood-2012-07-442251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager P.L., Baecher-Allan C., Maier L.M., Arthur A.T., Ottoboni L., Barcellos L., McCauley J.L., McCauley S., Goris A., Saarela J. The role of the CD58 locus in multiple sclerosis. Proc. Natl. Acad. Sci. U S A. 2009;106:5264–5269. doi: 10.1073/pnas.0813310106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetriou P., Abu-Shah E., Valvo S., McCuaig S., Mayya V., Kvalvaag A., Starkey T., Korobchevskaya K., Lee L.Y.W., Friedrich M. A dynamic CD2-rich compartment at the outer edge of the immunological synapse boosts and integrates signals. Nat. Immunol. 2020:1–12. doi: 10.1038/s41590-020-0770-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMond A.L., Mossman K.D., Starr T., Dustin M.L., Groves J.T. T cell receptor microcluster transport through molecular mazes reveals mechanism of translocation. Biophysical J. 2008;94:3286–3292. doi: 10.1529/biophysj.107.119099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J.F.P., Law J.L., Favero J.J. Activation of human T lymphocytes by crosslinking of anti-CD3 monoclonal antibodies. J. Leukoc. Biol. 1989;46:214–220. doi: 10.1002/jlb.46.3.214. [DOI] [PubMed] [Google Scholar]
- Duchez S., Rodrigues M., Bertrand F., Valitutti S. Reciprocal polarization of T and B cells at the immunological synapse. J. Immunol. 2011;187:4571–4580. doi: 10.4049/jimmunol.1100600. [DOI] [PubMed] [Google Scholar]
- Dustin M.L., Choudhuri K. Signaling and polarized communication across the T cell immunological synapse. Annu. Rev. Cell Dev. Biol. 2016;32:303–325. doi: 10.1146/annurev-cellbio-100814-125330. [DOI] [PubMed] [Google Scholar]
- Dustin M.L., Ferguson L.M., Chan P.-Y., Springer T.A., Golan D.E. Visualization of CD2 interaction with LFA-3 and determination of the two-dimensional dissociation constant for adhesion receptors in a contact area. J. Cell Biol. 1996;132:465–474. doi: 10.1083/jcb.132.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dustin M.L., Golan D.E., Zhu D.-M., Miller J.M., Meier W., Davies E.A., van der Merwe P.A. Low affinity interaction of human or rat T cell adhesion molecule CD2 with its ligand aligns adhering membranes to achieve high physiological affinity. J. Biol. Chem. 1997;272:30889–30898. doi: 10.1074/jbc.272.49.30889. [DOI] [PubMed] [Google Scholar]
- Dustin M.L., Olszowy M.W., Holdorf A.D., Li J., Bromley S., Desai N., Widder P., Rosenberger F., van der Merwe P.A., Allen P.M. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell. 1998;94:667–677. doi: 10.1016/s0092-8674(00)81608-6. [DOI] [PubMed] [Google Scholar]
- Espagnolle N., Depoil D., Zaru R., Demeur C., Champagne E., Guiraud M., Valitutti S. CD2 and TCR synergize for the activation of phospholipase Cγ1/calcium pathway at the immunological synapse. Int. Immunol. 2007;19:239–248. doi: 10.1093/intimm/dxl141. [DOI] [PubMed] [Google Scholar]
- Figge M.T., Meyer-Hermann M. Geometrically repatterned immunological synapses uncover formation mechanisms. PLoS Comput. Biol. 2006;2:e171. doi: 10.1371/journal.pcbi.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge M.T., Meyer-Hermann M. Modeling receptor-ligand binding kinetics in immunological synapse formation. Eur. Phys. J. D Atomic. 2009;51:153–160. [Google Scholar]
- Fritzsche M., Fernandes R.A., Chang V.T., Colin-York H., Clausen M.P., Felce J.H., Galiani S., Erlenkämper C., Santos A.M., Heddleston J.M. Cytoskeletal actin dynamics shape a ramifying actin network underpinning immunological synapse formation. Sci. Adv. 2017;3:e1603032. doi: 10.1126/sciadv.1603032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawden-Bone C.M., Frazer G.L., Richard A.C., Ma C.Y., Strege K., Griffiths G.M. PIP5 kinases regulate membrane phosphoinositide and actin composition for targeted granule secretion by cytotoxic lymphocytes. Immunity. 2018;49:427–437. doi: 10.1016/j.immuni.2018.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar K., Ghosh S., Saha S., Rumamol C., Mayor S., Rao M. Active remodeling of cortical actin regulates spatiotemporal organization of cell surface molecules. Cell. 2012;149:1353–1367. doi: 10.1016/j.cell.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Graf B., Bushnell T., Miller J. LFA-1-mediated T cell costimulation through increased localization of TCR/class II complexes to the central supramolecular activation cluster and exclusion of CD45 from the immunological synapse. J. Immunol. 2007;179:1616–1624. doi: 10.4049/jimmunol.179.3.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grakoui A., Bromley S.K., Sumen C., Davis M.M., Shaw A.S., Allen P.M., Dustin M.L. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- Hori Y., Raychaudhuri S., Chakraborty A.K. Analysis of pattern formation and phase separation in the immunological synapse. J. Chem. Phys. 2002;117:9491–9501. [Google Scholar]
- Hui E., Cheung J., Zhu J., Su X., Taylor M.J., Wallweber H.A., Sasmal D.K., Huang J., Kim J.M., Mellman I. T cell costimulatory receptor CD28 is a primary target for PD-1--mediated inhibition. Science. 2017;355:1428–1433. doi: 10.1126/science.aaf1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings N.J., Clarkson N., Chalkley R., Barclay A.N., Brown M.H. Linking the T cell surface protein CD2 to the actin-capping protein CAPZ via CMS and CIN85. J. Biol. Chem. 2003;278:22396–22403. doi: 10.1074/jbc.M302540200. [DOI] [PubMed] [Google Scholar]
- Hyman A.A., Weber C.A., Jülicher F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- Johnson K.G., Bromley S.K., Dustin M.L., Thomas M.L. A supramolecular basis for CD45 tyrosine phosphatase regulation in sustained T cell activation. Proc. Natl. Acad. Sci. U S A. 2000;97:10138–10143. doi: 10.1073/pnas.97.18.10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizuka Y., Douglass A.D., Vardhana S., Dustin M.L., Vale R.D. The coreceptor CD2 uses plasma membrane microdomains to transduce signals in T cells. J. Cell Biol. 2009;185:521–534. doi: 10.1083/jcb.200809136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizuka Y., Douglass A.D., Varma R., Dustin M.L., Vale R.D. Mechanisms for segregating T cell receptor and adhesion molecules during immunological synapse formation in Jurkat T cells. Proc. Natl. Acad. Sci. U S A. 2007;104:20296–20301. doi: 10.1073/pnas.0710258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Currie S.L., Rosen M.K. Intrinsically disordered sequences enable modulation of protein phase separation through distributed tyrosine motifs. J. Biol. Chem. 2017;292:19110–19120. doi: 10.1074/jbc.M117.800466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Rhodes M., Wiest D.L., Vignali D.A. On the dynamics of TCR: CD3 complex cell surface expression and downmodulation. Immunity. 2000;13:665–675. doi: 10.1016/s1074-7613(00)00066-2. [DOI] [PubMed] [Google Scholar]
- Martín-Cófreces N.B., Baixauli F., Sánchez-Madrid F. Immune synapse: conductor of orchestrated organelle movement. Trends Cell Biol. 2014;24:61–72. doi: 10.1016/j.tcb.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill L., Salmond R.J., Cooper J.C., Carret C.K., Cassady-Cain R.L., Roche-Molina M., Tandon P., Holmes N., Alexander D.R. The differential regulation of Lck kinase phosphorylation sites by CD45 is critical for T cell receptor signaling responses. Immunity. 2007;27:425–437. doi: 10.1016/j.immuni.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Milstein O., Tseng S.-Y., Starr T., Llodra J., Nans A., Liu M., Wild M.K., van der Merwe P.A., Stokes D.L., Reisner Y. Nanoscale increases in CD2-CD48-mediated intermembrane spacing decrease adhesion and reorganize the immunological synapse. J. Biol. Chem. 2008;283:34414–34422. doi: 10.1074/jbc.M804756200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minguet S., Swamy M., Alarcón B., Luescher I.F., Schamel W.W.A. Full activation of the T cell receptor requires both clustering and conformational changes at CD3. Immunity. 2007;26:43–54. doi: 10.1016/j.immuni.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Mintz M.A., Felce J.H., Chou M.Y., Mayya V., Xu Y., Shui J.-W., An J., Li Z., Marson A., Okada T. The HVEM-BTLA Axis restrains T cell help to germinal center B cells and functions as a cell-extrinsic suppressor in lymphomagenesis. Immunity. 2019 doi: 10.1016/j.immuni.2019.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks C.R.F., Freiberg B.A., Kupfer H., Sciaky N., Kupfer A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 1998;395:82. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- Murugesan S., Hong J., Yi J., Li D., Beach J.R., Shao L., Meinhardt J., Madison G., Wu X., Eric B. Formin-generated actomyosin arcs propel T cell receptor microcluster movement at the immune synapse. J. Cell Biol. 2016;215:383–399. doi: 10.1083/jcb.201603080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordenfelt P., Moore T.I., Mehta S.B., Kalappurakkal J.M., Swaminathan V., Koga N., Lambert T.J., Baker B., Waters J.C., Oldenbourg R. Direction of actin flow dictates integrin LFA-1 orientation during leukocyte migration. Nat. Commun. 2017;8:2047. doi: 10.1038/s41467-017-01848-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S.J., Sanjana N.E., Kishton R.J., Eidizadeh A., Vodnala S.K., Cam M., Gartner J.J., Jia L., Steinberg S.M., Yamamoto T.N. Identification of essential genes for cancer immunotherapy. Nature. 2017;548:537. doi: 10.1038/nature23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautot S., Lee H., Isacoff E.Y., Groves J.T. Neuronal synapse interaction reconstituted between live cells and supported lipid bilayers. Nat. Chem. Biol. 2005;1:283–289. doi: 10.1038/nchembio737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S.Y., Groves J.T., Chakraborty A.K. Synaptic pattern formation during cellular recognition. Proc. Natl. Acad. Sci. U S A. 2001;98:6548–6553. doi: 10.1073/pnas.111536798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S., Thomson B.P., Remmers E.F., Eyre S., Hinks A., Guiducci C., Catanese J.J., Xie G., Stahl E.A., Chen R. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat. Genet. 2009;41:1313. doi: 10.1038/ng.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razak F.R.A., Diepstra A., Visser L., Berg A. CD58 mutations are common in Hodgkin lymphoma cell lines and loss of CD58 expression in tumor cells occurs in Hodgkin lymphoma patients who relapse. Genes Immun. 2016;17:363. doi: 10.1038/gene.2016.30. [DOI] [PubMed] [Google Scholar]
- Salaita K., Nair P.M., Petit R.S., Neve R.M., Das D., Gray J.W., Groves J.T. Restriction of receptor movement alters cellular response: physical force sensing by EphA2. Science. 2010;327:1380–1385. doi: 10.1126/science.1181729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seavitt J.R., White L.S., Murphy K.M., Loh D.Y., Perlmutter R.M., Thomas M.L. Expression of the p56 lck Y505F mutation in CD45-deficient mice rescues thymocyte development. Mol. Cell Biol. 1999;19:4200–4208. doi: 10.1128/mcb.19.6.4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurer C.R., Kuo J.C.-H., Roberts L.M., Gandhi J.G., Colville M.J., Enoki T.A., Pan H., Su J., Noble J.M., Hollander M.J. Physical principles of membrane shape regulation by the glycocalyx. Cell. 2019;177:1757–1770.e21. doi: 10.1016/j.cell.2019.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siokis A., Robert P.A., Demetriou D., Dustin M.L., Meyer-Hermann M. F-Actin-Driven CD28-CD80 localization in the immune synapse. Cell Rep. 2018;24:1151–1162. doi: 10.1016/j.celrep.2018.06.114. [DOI] [PubMed] [Google Scholar]
- Siokis A., Robert P.A., Meyer-Hermann M. In: Baldari C.T., Dustin M.L., editors. vol 1584. Humana Press; 2017. Mathematical modeling of synaptic patterns; pp. 171–182. (Hrsg. The Immune Synapse. Methods in Molecular Biology). [DOI] [PubMed] [Google Scholar]
- Skånland S.S., Moltu K., Berge T., Aandahl E.M., Taskén K. T-cell co-stimulation through the CD2 and CD28 co-receptors induces distinct signalling responses. Biochem. J. 2014;460:399–410. doi: 10.1042/BJ20140040. [DOI] [PubMed] [Google Scholar]
- Su X., Ditlev J.A., Hui E., Xing W., Banjade S., Okrut J., King D.S., Taunton J., Rosen M.K., Vale R.D. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science. 2016;352:595–599. doi: 10.1126/science.aad9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi A., Badr M.E.S.G., Miyauchi K., Ishihara C., Onishi R., Guo Z., Sasaki Y., Ike H., Takumi A., Tsuji N.M. CRTAM determines the CD4+ cytotoxic T lymphocyte lineage. J. Exp. Med. 2015;213:123–138. doi: 10.1084/jem.20150519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M.L., Brown E.J. Positive and negative regulation of Src-family membrane kinases by CD45. Immunol. Today. 1999;20:406–411. doi: 10.1016/s0167-5699(99)01506-6. [DOI] [PubMed] [Google Scholar]
- Tibaldi E.V., Salgia R., Reinherz E.L. CD2 molecules redistribute to the uropod during T cell scanning: implications for cellular activation and immune surveillance. Proc. Natl. Acad. Sci. U S A. 2002;99:7582–7587. doi: 10.1073/pnas.112212699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng S.-Y., Waite J.C., Liu M., Vardhana S., Dustin M.L. T cell-dendritic cell immunological synapses contain TCR-dependent CD28-CD80 clusters that recruit protein kinase Cθ. J. Immunol. 2008;181:4852–4863. doi: 10.4049/jimmunol.181.7.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvo S., Mayya V., Seraia E., Afrose J., Novak-Kotzer H., Ebner D., Dustin M.L. In: Baldari C.T., Dustin M.L., editors. vol 1584. Humana Press; 2017. Comprehensive analysis of immunological synapse phenotypes using supported lipid bilayers; pp. 423–441. (Hrsg. The Immune Synapse. Methods in Molecular Biology). [DOI] [PubMed] [Google Scholar]
- Van Der Merwe P.A., Barclay A.N., Mason D.W., Davies E.A., Morgan B.P., Tone M., Krishnam A.K., Ianelli C., Davis S.J. Human cell-adhesion molecule CD2 binds CD58 (LFA-3) with a very low affinity and an extremely fast dissociation rate but does not bind CD48 or CD59. Biochemistry. 1994;33:10149–10160. doi: 10.1021/bi00199a043. [DOI] [PubMed] [Google Scholar]
- Wang J.-H., Smolyar A., Tan K., Liu J.-H., Kim M., Zhen-yu J.S., Wagner G., Reinherz E.L. Structure of a heterophilic adhesion complex between the human CD2 and CD58 (LFA-3) counterreceptors. Cell. 1999;97:791–803. doi: 10.1016/s0092-8674(00)80790-4. [DOI] [PubMed] [Google Scholar]
- Weikl T.R., Lipowsky R. Pattern formation during T-cell adhesion. Biophys. J. 2004;87:3665–3678. doi: 10.1529/biophysj.104.045609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Jin X., Harrison O., Shapiro L., Honig B.H., Ben-Shaul A. Cooperativity between trans and cis interactions in cadherin-mediated junction formation. Proc. Natl. Acad. Sci. U S A. 2010;107:17592–17597. doi: 10.1073/pnas.1011247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosuka T., Kobayashi W., Sakata-Sogawa K., Takamatsu M., Hashimoto-Tane A., Dustin M.L., Tokunaga M., Saito T. Spatiotemporal regulation of T cell costimulation by TCR-CD28 microclusters and protein kinase C θ translocation. Immunity. 2008;29:589–601. doi: 10.1016/j.immuni.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosuka T., Takamatsu M., Kobayashi-Imanishi W., Hashimoto-Tane A., Azuma M., Saito T. Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J. Exp. Med. 2012;209:1201–1217. doi: 10.1084/jem.20112741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.-H., Rafiq N.B.M., Cao F., Zhou Y., Krishnasamy A., Biswas K.H., Ravasio A., Chen Z., Wang Y.-H., Kawauchi K. Integrin-beta3 clusters recruit clathrin-mediated endocytic machinery in the absence of traction force. Nat. Commun. 2015;6:1–12. doi: 10.1038/ncomms9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamoyska R. Why is there so much CD45 on T cells? Immunity. 2007;27:421–423. doi: 10.1016/j.immuni.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Harrison D.L., Song Y., Ji J., Huang J., Hui E. Antigen-presenting cell-intrinsic PD-1 neutralizes PD-L1 in cis to attenuate PD-1 signaling in T cells. Cell Rep. 2018;24:379–390. doi: 10.1016/j.celrep.2018.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D.-M., Dustin M.L., Cairo C.W., Thatte H.S., Golan D.E. Mechanisms of cellular avidity regulation in CD2--CD58-mediated T cell adhesion. ACS Chem. Biol. 2006;1:649–658. doi: 10.1021/cb6002515. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The software used for this study is developed in C++ according to all the algorithmic details explained in (Siokis et al., 2017; Siokis et al., 2018) and is available on https://github.com/AnastasiosSiokis/IS_Cell.
Any additional information required to reanalyze and resimulate the data reported in this paper is available from the lead contact upon request.