Abstract
Mechanistic insight into protein binding by poly- and perfluoroalkyl substances (PFASs) is critical to understanding how PFASs distribute and accumulate within the body and to developing predictive models within and across classes of PFASs. Fluorine nuclear magnetic resonance spectroscopy (19F NMR) has proven to be a powerful, yet underutilized tool to study PFAS binding; chemical shifts of each fluorine group reflect the local environment along the length of the PFAS molecule. Using bovine serum albumin (BSA), we report dissociation constants, Kd, for four common PFASs well below reported critical micelle concentrations (CMCs) − perfluorooctanoic acid (PFOA), perfluorononanoic acid (PFNA), perfluorohexanesulfonic acid (PFHxS), and perfluorooctanesulfonic acid (PFOS) − as a function of temperature in phosphate buffered saline. Kd values were determined based on the difluoroethyl group adjacent to the anionic headgroups and the terminal trifluoromethyl groups. Our results indicate that the hydrophobic tails exhibit greater binding affinity relative to the headgroup, and that the binding affinities are generally consistent with previous results showing that greater PFAS hydrophobicity leads to greater protein binding. However, the binding mechanism was dominated by entropic hydrophobic interactions attributed to desolvation of the PFAS tails within the hydrophobic cavities of the protein and on the surface of the protein. In addition, PFNA appears to form hemimicelles on the protein surfaces below reported CMC values. This work provides a renewed approach to utilizing 19F NMR for PFAS-protein binding studies and a new perspective on the role of solvent entropy.
Keywords: PFAS, protein binding, albumin, NMR
Graphical Abstract
1. INTRODUCTION
Poly- and perfluoroalkyl substances (PFASs) are a class of highly fluorinated synthetic chemicals used in a variety of industrial and commercial applications, including firefighting foams and non-stick or stain resistant materials.1,2 Due to the strength of the carbon-fluorine bond, these compounds possess unique chemical and physical properties, including high chemical stability, thermal inertness, and ultra-low surface energy.3,4 In addition, PFASs have greater hydrophobicity and acidity than their hydrocarbon analogs,5 resulting in new mechanisms of interaction within environmental and biological systems.6–8 PFASs persist in the environment, withstanding biodegradation, photolysis and hydrolysis. As a result, they may bioaccumulate in the food chain, are transported long distances via air or water and are detected ubiquitously even in remote regions with no history of their production.8–11
Perfluorooctanoic acid (PFOA; C8HF15O2), perfluorononanoic acid (PFNA; C9HF17O2), perfluorohexanesulfonic acid (PFHxS; C7HF15O3S) and perfluorooctanesulfonic acid (PFOS; C8HF17O3S) are four of the most historically used PFASs. Of the estimated thousands of different PFASs, these four compounds have received attention due to their high frequency of detection in the environment and in humans.2,12 Studies have reported widespread exposure in humans, where PFASs were detected in the blood samples of over 94% of the individuals examined in a particular cohort within the United States.13 They have also been detected in cord serum of infants and in breast milk of nursing mothers.14,15 PFAS persistence is demonstrated by their long half-lives in humans, estimated to be 3.8 years for PFOA, 2.5 years for PFNA, 8.5 years for PFHxS, and 5.4 years for PFOS.16,17 From epidemiological studies, the critical effects of PFOA and/or PFOS are an increase in serum total cholesterol in adults,18 a decrease in antibody response for vaccinations,19,20 pregnancy-induced hypertension and preeclampsia,21 and kidney and testicular cancer.22 The mechanisms by which PFASs interact with and transport throughout the human body remain under investigation.19,23 The PFASs listed above are detected primarily in the blood and the liver of humans, highlighting their proteinophilic nature.24
Serum albumin are ligand binding proteins that likely play an important role in the accumulation patterns of PFASs in blood.25–27 For example, human serum albumin (HSA), typically ranging in concentration from 30 to 50 g L−1 (0.45 to 0.75 mM), is the most abundant protein in human blood, transporting natural and exogenous ligands including fatty acids, pharmaceuticals, and small organic anions throughout the body.28 Studies have estimated that over 90% of the total PFASs in the body will be bound to HSA due to their structural similarity with fatty acids – aliphatic tails and anionic head groups.29 HSA contains seven distinct fatty acid binding sites that are asymmetrically distributed around the protein.28 Competition for binding sites between molecules can significantly affect the equilibrium between bound and unbound states of PFASs.26,27 PFAS-albumin binding is important in governing half-lives and determining biodistribution, as protein binding is an essential factor in newly-proposed versions of mechanistic models for PFAS bioaccumulation.6 A recent study has correlated PFAS-HSA association constants (Ka) with placental transfer efficiency (PTE) of PFASs in humans, concluding there is a positive correlation between degree of binding to proteins and PTE.30 PTE is the ratio of PFAS concentration in cord blood to that in maternal blood, an important marker for fetal PFAS burden.31 Each of these studies highlights the importance of accurate PFAS-protein binding parameters as tools for assessing exposure and risk.
Dissociation constants quantify PFAS-HSA binding and have been reported to range from 10−2 M to 10−6 M.25,29,32 The range of Kd over four orders of magnitude can be explained in part by the variety of experimental techniques amenable to different mechanisms of PFAS-albumin binding including van der Waals, hydrophobic, and electrostatic interactions as well as hydrogen bonding. Of these, van der Waals interactions and hydrogen bonding are reportedly dominant.33 There is conflicting literature on the mechanisms of PFAS-albumin binding and the number of binding sites.27,29,32,34,35 For instance, fluorescence quenching indirectly measures PFAS binding based on changes in the chemical environment within the hydrophobic cavities of the protein. Equilibrium dialysis directly measures the amount of bound PFAS within hydrophobic cavities as well as PFAS associated on the protein surface. Once specific hydrophobic sites with high binding affinity are occupied by PFASs, they will continue to be adsorbed non-specifically to albumin surfaces.36 Hence, the number of PFASs bound to a single albumin protein ranges from 1 to 50 in literature.37,38
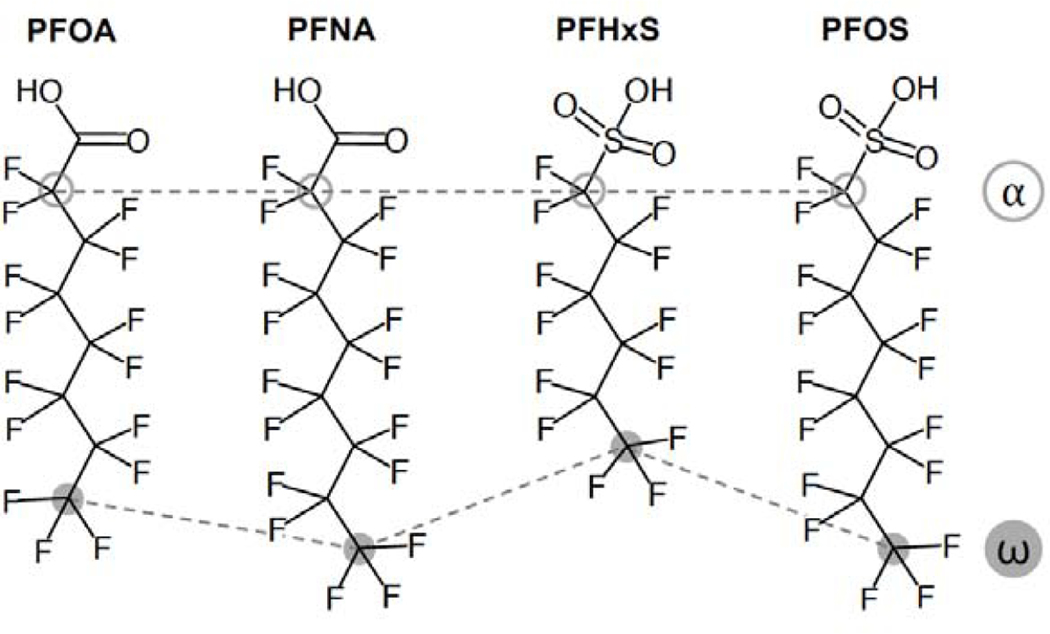
The goal of this study was to further investigate the binding mechanisms of four PFASs (PFOA, PFNA, PFHxS and PFOS) with bovine serum albumin (BSA) at relevant physiological parameters using 19F NMR. PFOA, PFNA, PFHxS and PFOS are the most common PFASs found in the United States population, with average serum concentrations of 1.56, 1.18, 0.577, and 4.72 μg L−1 reported for 2015–2016, respectively.39 BSA was used because it shares a high level of homology with and HSA,40,41 and because it has been used previously for PFAS-albumin protein binding, providing a comparative basis.26,32,33,35 19F NMR spectroscopy is a powerful tool to study PFAS-protein interactions37 because each fluorine atom gives an individual signal in the spectrum that reflects the local chemical environment.24,27,32,42–44 It is therefore uniquely suited to directly examine molecular binding mechanisms. Furthermore, we propose that through synchronous observation of the 19F signals near both the charged head and the fluorinated tail of the PFAS molecules, it is possible to examine the effect of the hydrophilic headgroup and the hydrophobic tail on protein binding for a given PFAS or comparatively across a collection of PFASs (Figure 1).
Figure 1.
Chemical structures of perfluorooctanoic acid (PFOA), perfluoronanonoic acid (PFNA), perfluorohexanesulfonic acid (PFHxS), and perfluorooctanesulfonic acid (PFOS). Fluorinated carbons labeled α (open gray circles) and ω (filled gray symbols) represent the -CF2-group near the hydrophilic headgroup and the terminal -CF3 group, respectively.
2. MATERIALS AND METHODS
2.1. Chemicals.
BSA (99% fatty acid free lyophilized powder) and trifluoromethyl acrylic acid (TFMAA) (98%) were obtained from Sigma Aldrich (St. Louis, MO) and deuterium oxide (D2O) (99%) from Fisher Scientific (Agawam, MA). PFOA (99%), PFNA (99%), PFHxS (95%) and PFOS (98%) were obtained from AccuStandard (New Haven, CT). The purities listed are reported by the manufactures. Phosphate buffered saline (PBS) was prepared at pH 7.4 with 2.68 mM KCl, 1.47 mM KH2PO4, 136.89 mM NaCl, and 8.06 mM Na2HPO4•7H20. Chemicals were used as received from the suppliers.
2.2. Nuclear Magnetic Resonance Spectroscopy (19F NMR).
BSA concentrations were held constant at 10 μM in PBS to apply chemical shift perturbation (CSP) analysis, which requires that PFAS be in excess relative to the protein.27,45 BSA solutions were prepared at least one day in advance of 19F NMR measurements and kept at 4 °C overnight. Each PFAS was dissolved in PBS and prepared at least a day in advance of the experiments and stored at room temperature. PFAS stock solutions were stored in polypropylene vials. PFAS solutions were transferred into analytical 5 mm NMR tubes at 90:10 PFAS:D2O ratios summing to 400 μL of solution. D2O was required for NMR lock and calibration. TFMAA (5 μL) was added as a second reference point for the CSP analysis. Spectra were obtained using a Bruker Advance III HD 400 NanoBay spectrometer (Bruker BioSpin, Rheinstetten, Germany) equipped with a 5 mm BBFO z-gradient smart probe using a Bruker Automatic Sample Changer (SampleXpress).
19F NMR spectra were automatically acquired under the control of ICON-NMR (Bruker BioSpin, Rheinstetten, Germany) in the range from +20 to −220 ppm with the Bruker 5 mm auto-band probe tuned to 470 MHz for 19F resonance. Chemical shifts were recorded relative to D2O (0.000 ppm) and TFMAA (−64.866 ppm). A 90° pulse width for 18.0 μs was used for all experiments to provide the maximum signal to noise ratio (S/N) and to minimize the influence of the off-resonance effects on the accuracy of 19F NMR measurements.24 A total of 4096 scans were collected yielding 131072 data points to maximize the S/N. 1D 19F NMR spectra were obtained with a spectral width of 89285.7 Hz, an acquisition time of 0.64 s and a recycle delay of 1 s to ensure full T1 relaxation. For evaluation of the thermodynamic parameters of the PFAS-BSA binding, 19F NMR spectra were recorded at three different temperatures; 298 K, 304 K, and 310 K.
The Bruker pulse program, zgflqn, was used with a receiver gain (RG) of 212. All spectra were automatically phased and baseline corrected for accurate quantitative measurements using the Topspin3.2 software package (Bruker BioSpin, Rheinstetten, Germany), MestReNova software package (Mestrelab Research, Escondido, CA) and Origin Software (Origin Lab, Northampton, MA). Peak shifts were obtained by the electronic Gaussian fit of the expanded regions around diagnostic resonances using Origin Software. All 19F NMR experiments were repeated independently in triplicate.
The following criteria were required to identify PFAS binding mechanisms: (1) the molecular recognition event was sufficiently defined to provide a well-structured binding complex; (2) there were a number of independently varying 19F NMR signals that shift, providing a multidimensional analysis; and (3) the shift of the PFAS 19F resonances was induced by spatial proximity to the protein to access structural information on the whole PFAS molecule.46 By comparing the chemical shifts for both ends of a PFAS molecule, the 1D 19F NMR experiments provided insight into the binding mechanism and structure of the PFAS bound to BSA.32,34,47,48
2.3. Calculated PFAS-BSA Binding Parameters.
The dissociation constant (Kd) is an equilibrium constant that measures the propensity of a complex in the bound state to reversibly dissociate into its constituent parts based on the nature and strength of the intermolecular interactions. In this work, PFAS bound with BSA, [PFAS-BSA], represents the complex while unbound PFAS and BSA represent the free ligands in solution. Kd can be represented from the following reaction and ratio, seen in equation (1) and (2), and is inversely proportional to the association constant, Ka.
| (1) |
| (2) |
The chemical shift of a ligand NMR signal in the presence of a protein is commonly used to monitor the formation of a protein-ligand complex. 1D NMR spectra of small molecules (MW ≤ 500 Da) typically have sharp peaks due to rapid dipole-dipole relaxation. Binding of a ligand to a high molecular weight molecule such as a protein induces peak broadening and a corresponding chemical shift in the NMR signal because the bound ligand experiences the slow relaxation time of the protein compared to the free state of the ligand.49
Kd values were determined based on the resonance chemical shift (δ) of the PFAS bound to the BSA relative to its unbound state in solution.29,34,50 The observed chemical shift is the population-weighted average of free and bound ligands, which allows the determination of Kd from measurement of the peak positions.44,45 The chemical shift of the PFAS resonance peak is sensitive to structural differences of its bound and unbound states, meaning that a genuine binding interaction of PFAS with BSA will produce a perturbation. A change in δ greater than 0.02 ppm indicates that the environmental structure of the ligand experiences some transformation (e.g. change in polarity or electrostatic interactions).45 At a fixed BSA concentration, these perturbations are dependent on the PFAS concentration, reflecting differences in the fraction of PFAS that is bound to BSA. A smaller fraction is bound at high PFAS concentrations, resulting in resonances that more closely resemble those of the free PFAS. These spectral changes are related to the fraction of bound ligand.51
Kd values were determined graphically based on equation (3)
| (3) |
where is the net change in chemical shift of the monitored resonance of the bound ligand, [PFAS]T is the total PFAS concentration, [BSA]T is the total protein concentration, n is the number of binding sites per protein molecule and is the apparent change in the chemical shift for the monitored resonance in the bound state.29,34 The value of Kd is extracted as the negative y-intercept from the plot of the PFAS concentration versus the inverse of the PFAS chemical shift.52 Monitoring the perturbations of the chemical shifts for both the head (difluoroethyl, α) and tail (trifluoromethyl, ω) of PFASs reflects the binding affinity of these two ends of the molecule (Figure 1).
The Kd values were measured for PFAS concentrations ranging from 10 μM to 1 mM for PFOA, PFNA, PFHxS and PFOS. This concentration range of PFAS is above the ~1 μM detection limit of the 19F NMR method and can be found in a body of a highly exposed individual.43
Dissociation of PFAS with BSA is accompanied by a change of the standard Gibb’s free energy, , determined as
| (4) |
where R is the ideal gas constant and T is absolute temperature. can be further related to the changes in the standard enthalpy, , and standard entropy, , of the binding.
| (5) |
and , were determined from the slope () and the y-intercept () based on the van’t Hoff equation.
| (6) |
The signs of and (+ or −) can be used to determine the dominant intermolecular forces for PFAS-BSA binding: hydrophobic interactions when and , electrostatic interactions when and , or van der Waals interactions and hydrogen bonding when and .33,35,53,54
3. RESULTS AND DISCUSSION
3.1. Carboxylic acids (PFCAs).
PFOA and PFNA exhibited 1D 19F NMR spectra consistent with linear n-PFCAs free of impurities or branched isomers detected (Figure S1), consistent with prior results.24,29,34 The highest PFCA concentrations examined (1000 μM) were below reported critical micelle concentrations (CMCs) and no evidence of micellization, reported as an upfield shift in the −CF3 group due to shielding,55 was observed. It should be noted that reported CMC values vary significantly and have not be examined as functions of pH, salt composition or concentration, or temperature.
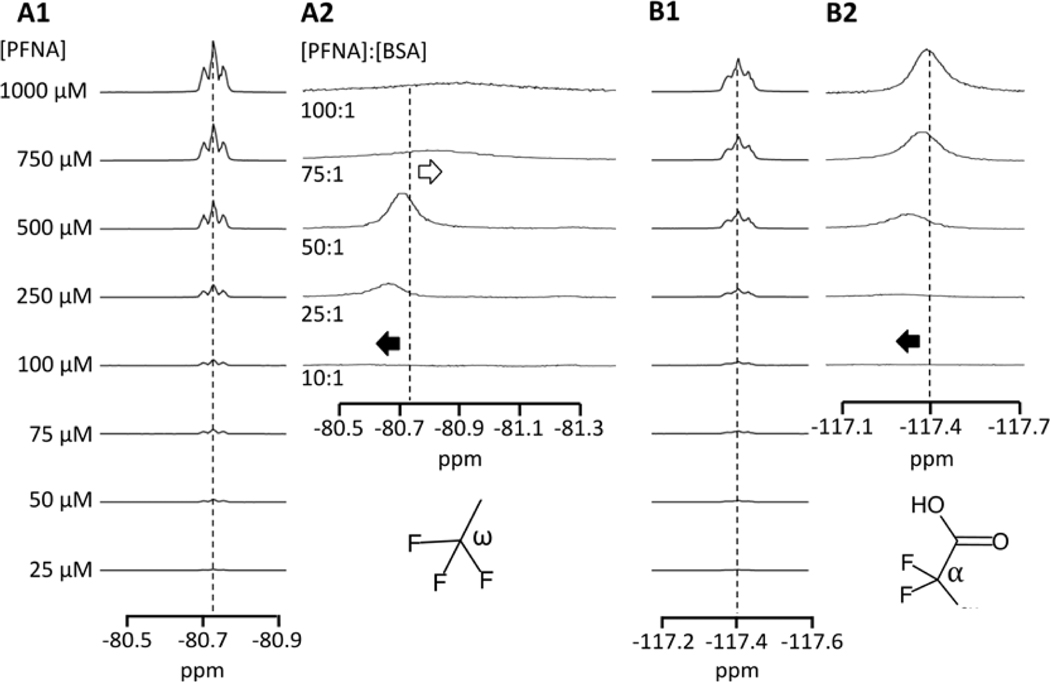
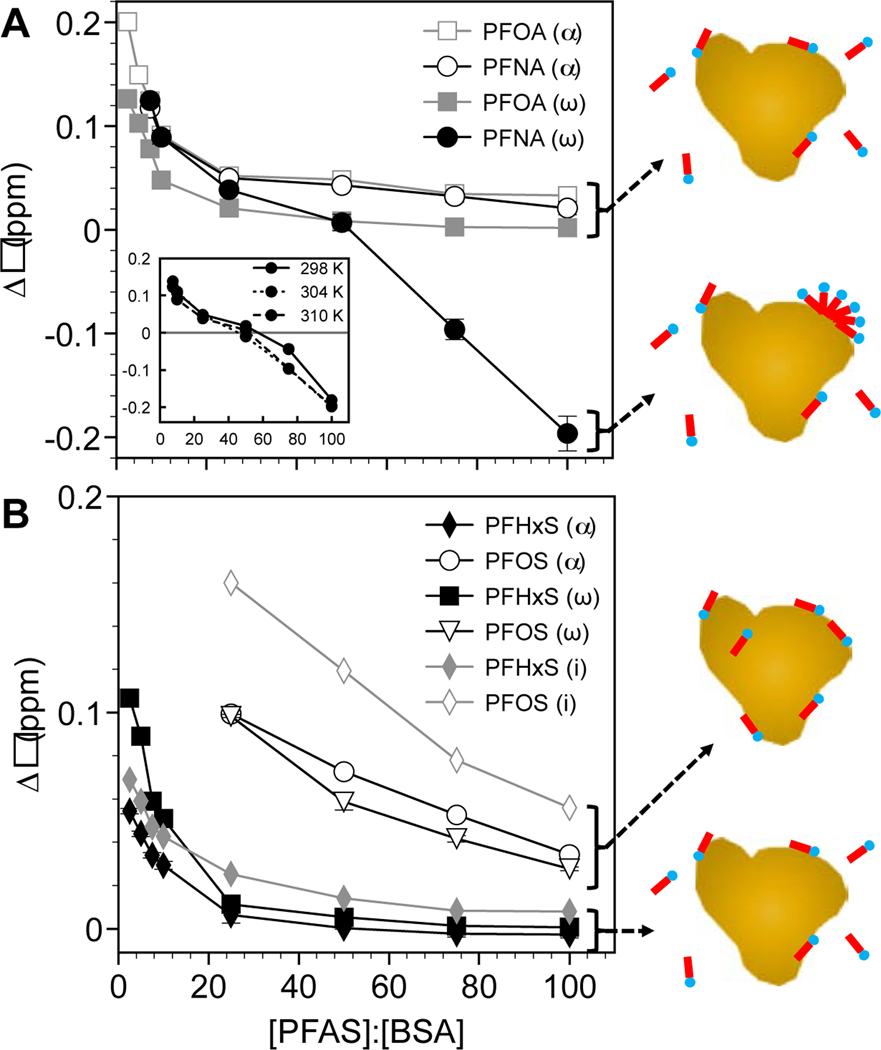
Significant 19F chemical shifts, peak broadening, and reductions in peak intensity were observed for both the α and ω groups of PFOA and PFNA upon binding to BSA, reflecting the formation of a PFAS-BSA complex. Exemplary 1D 19F NMR spectra are shown for PFNA in Figure 2 in the absence and presence of BSA. At low PFCA concentrations corresponding to [PFOA]:[BSA] < 2.5:1 and [PFNA]:[BSA] < 7.5:1 nearly all measurable PFOA was protein-bound, and the greatest deshielding (downfield shift) was observed for the α and ω resonances. As [PFCA]:[BSA] was increased the α and ω resonances approached their native chemical shift positions in the absence of BSA. This is demonstrated with the −CF2− α peak of PFNA in Figures 2B1-B2 (310 K, pH 7.4 PBS). Also shown in Figure 2A2 is a shielding of the −CF3 ω at [PFNA]:[BSA] > 50:1, which will be discussed later in more detail. The fluorine α resonance on the carbon adjacent to the headgroup is the most sensitive to protein binding and does not fully return to its original position, similar to results from prior studies.34
Figure 2.
19F resonance peaks for the (A) −CF3 (ω, tail) and −CF2− (α, head) groups of PFNA in pH 7.4 PBS at 310 K in the absence (A1, B1) and presence (A2, B2) of 10 μM BSA.
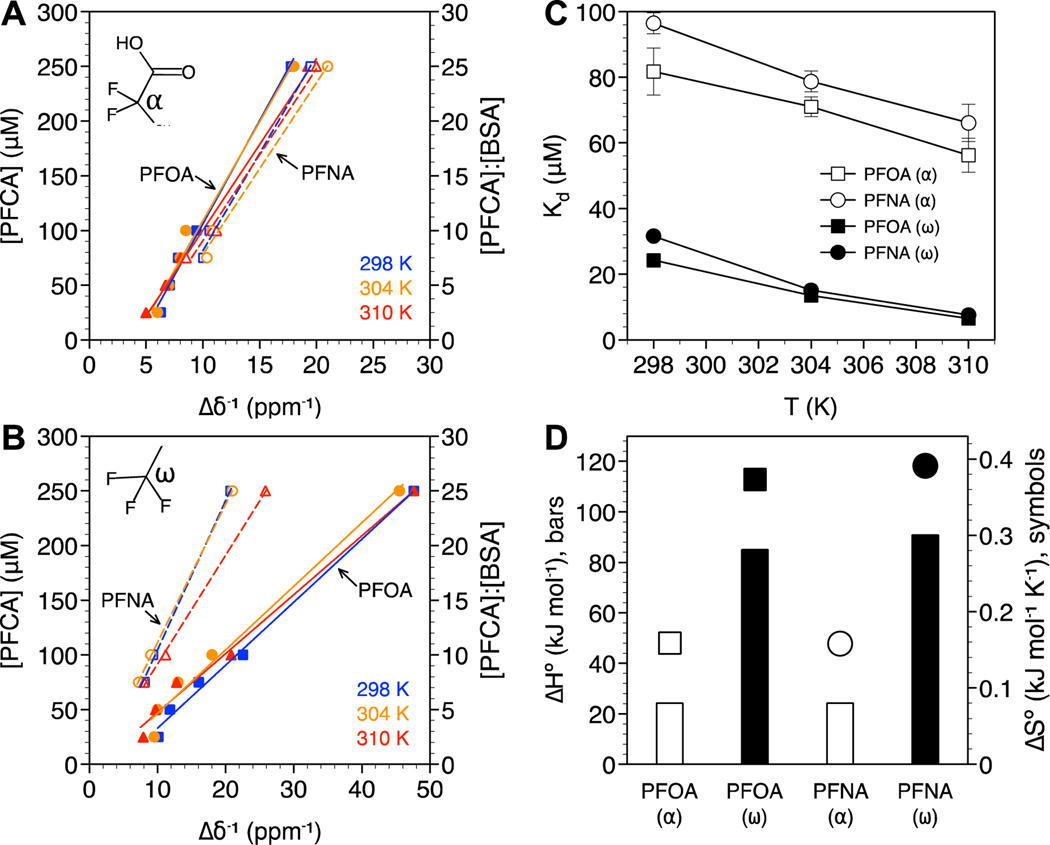
PFCA Kd values were determined based on changes in the chemical shift of the α (head) and ω (tail) positions as a function of temperature, from room to physiological (Figures 3A and 3B, respectively). Only the linear range of [PFCA] vs. up to [PFCA]:[BSA] ratios of 25:1 were considered for this analysis. All calculated Kd values were on the order of 10−6 to 10−5 M and decreased with increasing temperature, consistent with binding driven by hydrophobic interactions (Figure 3C). Values for Kd of the same order of magnitude have been calculated from fluorescence quenching experiments33,47 and electrospray ionization mass spectrometry.56 This was confirmed by the positive values for enthalpy () and entropy () of binding (Figure 3D). The lowest Kd values (highest protein affinity) were also observed for the −CF3 ω position, which is more hydrophobic itself, compared to the −CF2− α position, and expected to preferentially bind to hydrophobic regions on the protein. PFOA exhibited slightly lower Kd values than PFNA despite PFNA being more hydrophobic (PFNA with C8 fluorinated carbons vs. PFOA with C7). While these results differ from prior 19F NMR work reporting higher binding affinity of PFNA to BSA compared to PFOA,29 they are in agreement with BSA-water distribution coefficients determined by equilibrium dialysis.32 The higher Kd observed for PFNA may be attributed to steric hindrance. PFASs with long carbon tails have been shown to adopt helical conformations that hinder binding to the hydrophobic pockets of BSA.57
Figure 3.
PFCA binding analysis based on the (A) −CF2− α and (B) −CF3 ω positions. PFOA (solid symbols, solid lines) or PFNA (open symbols, dashed lines) concentration are plotted against the inverse change in chemical shift (Δδ−1) with and without BSA at 298 K (blue squares), 304 K (orange circles), and 310 K (red triangles). The molar ratio of PFCA:BSA are also shown. R2 values for the linear fits ranged from 0.983 to 0.994. (C) Dissociation constants (Kd) as a function of temperature and (D) standard enthalpy and entropy of BSA binding for the −CF3 ω (closed symbols) and −CF2− α (open symbols) positions. Standard error bars in (C) are based on n = 3. Error bars not visible are smaller than the symbols.
3.2. Sulfonic acids (PFSAs).
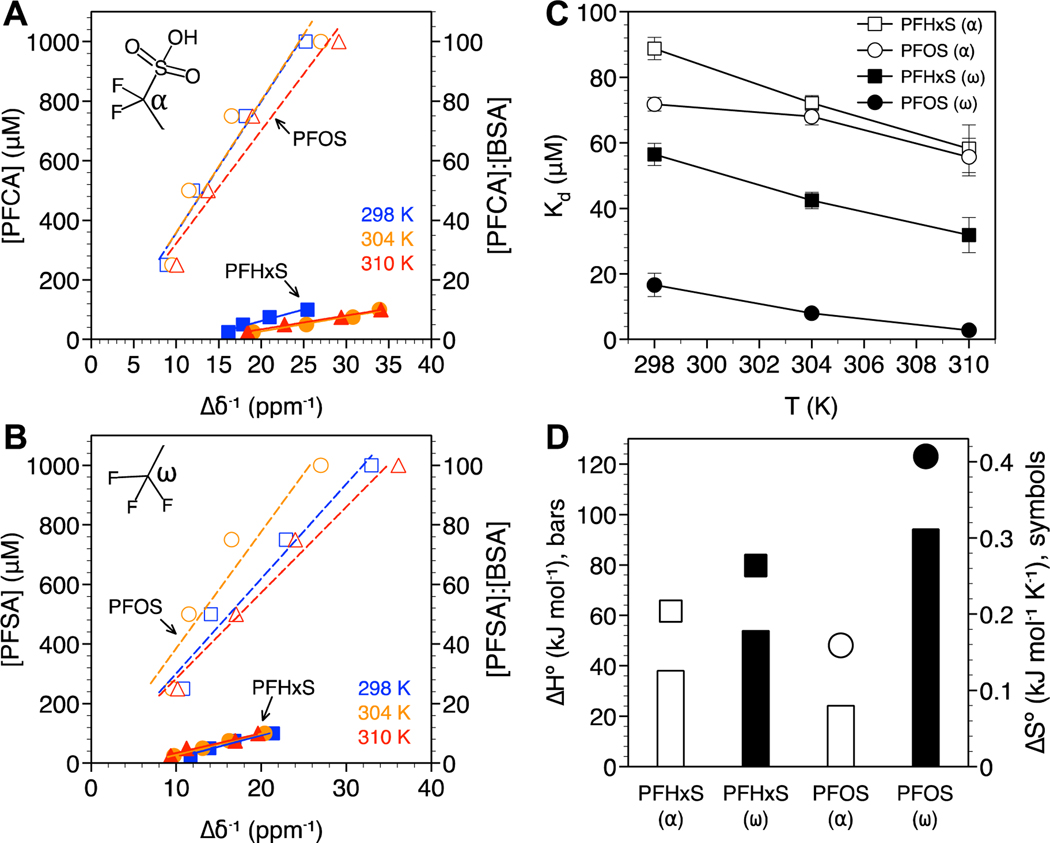
In addition to the α and ω groups for PFHxS and PFOS 19F NMR spectra, both contained branched isomer content as commonly observed for PFSAs (Figure S2).42,52,58 Understanding how the protein binding of isomers differs from linear PFSAs has not been studied in detail by 19F NMR due to challenges connected with the coalescing and splitting of isomer peaks, and the isolation of PFSA isomers.58 In this work, the binding properties of PFSAs were determined based on the α and ω groups (Figure 4A, B), and the additional resonance peak observed at −71.868 ppm (labeled i; Figure 5A). This resonance peak corresponded to the most abundant branched isomer found in both PFHxS and PFOS and is consistent with the CF3 isopropyl group previously identified.58 The carbon chain lengths of the PFHxS and PFOS isomers are reduced by one and the presence of the branched CF3 isopropyl groups on the carbon adjacent to the sulfonic head group is expected to modify the interaction with BSA compared to with linear structures.
Figure 4.
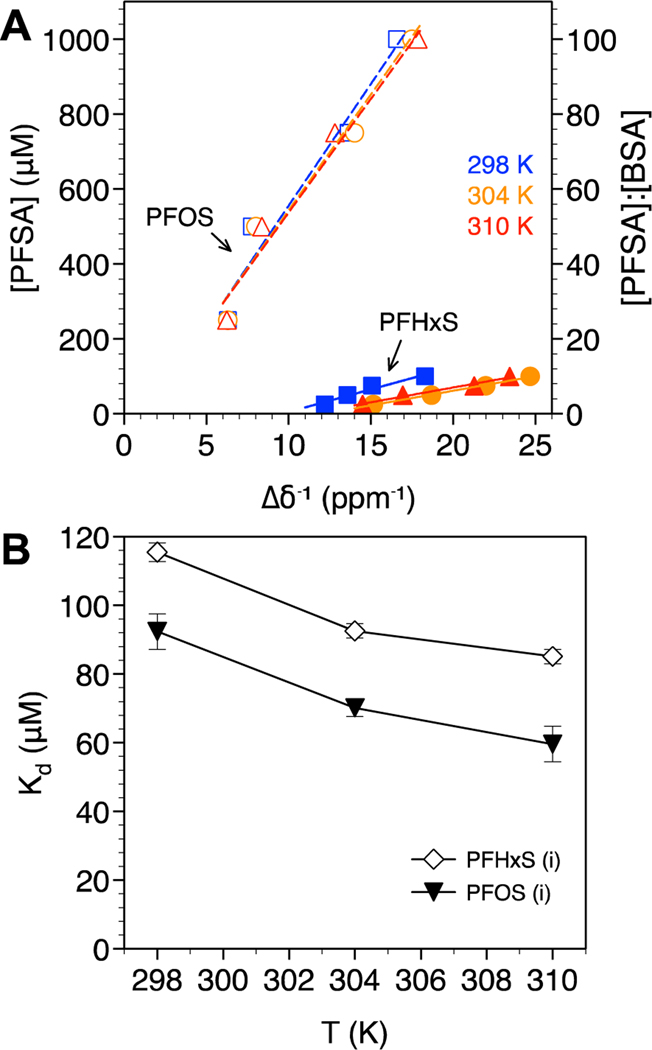
PFSA binding analysis based on the (A) −CF2− α and (B) −CF3 ω positions. PFHxS (solid symbols, solid lines) or PFOS (open symbols, dashed lines) concentration are plotted against the inverse change in chemical shift (Δδ−1) with and without BSA at 298 K (blue squares), 304 K (orange circles), and 310 K (red triangles). The molar ratio of PFCA:BSA are also shown. R2 values for the linear fits ranged from 0.9 to 0.996. (C) Dissociation constants (Kd) as a function of temperature and (D) standard enthalpy and entropy of BSA binding for the −CF3 ω (closed symbols) and −CF2− α (open symbols) positions. Standard error bars in (C) are based on n = 3. Error bars not visible are smaller than the symbols.
Figure 5.
PFSA isomer binding analysis based on (A) the branched CF3 isopropyl group for PFHxS (solid symbols, solid lines) and PFOS (open symbols, dashed lines) at 298 K (blue squares), 304 K (orange circles), and 310 K (red triangles). The molar ratio of PFCA:BSA are also shown. R2 values for the linear fits ranged from 0.953 to 0.983. (B) Dissociation constants (Kd) as a function of temperature. Standard error bars in (B) are based on n = 3.
PFSA Kd values based on the α and ω positions are shown in Figure 4C. High Kd values (low binding affinity) were determined for PFHxS compared to PFOS, consistent with the dominant role of hydrophobic interactions in PFAS-BSA binding, as observed for PFCAs. Based on the positive values for binding enthalpy and entropy, PFSA binding was driven by hydrophobic interactions. The binding of PFHxS and PFOS isomers (i) was also hydrophobic, though the presence of the branched CF3 isopropyl group near the headgroup coupled with a shorter perfluoroalkyl tail led to these isomers exhibiting the lowest binding affinity (highest Kd) of all PFASs examined (Figure 5), consistent with previous work using ultrafiltration to measure albumin binding.59
4. DISCUSSION
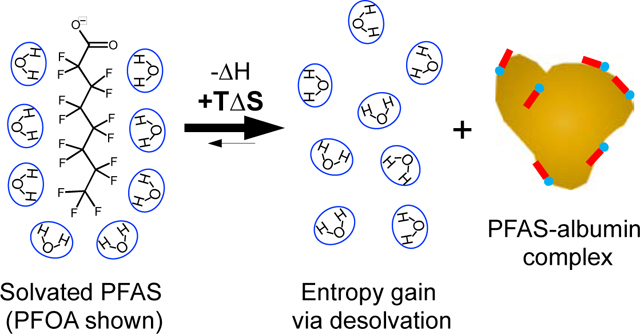
Association constants () for fatty acid binding to albumin are reported to decrease with temperature, characterized by a favorable exothermic process () with a modest gain in entropy due to desolvation of the alkyl tail.60 PFAS binding is also reported as being a favorable exothermic process driven by attractive van der Waals interactions and H-bonding ( and ).33,35,53,54 For the PFASs studied herein BSA binding is entropic, with significant gains in entropy that compensate for an unfavorable endothermic process (positive ). This observation, that PFAS binding is driven by hydrophobic interactions, can be partially explained by fluoroalkyls exhibiting strong hydrophobic interactions, stronger than their hydrocarbon analogs, due to the large structured water cavity required to solvate the fluorine groups.5 The observed pattern of PFAS association with BSA at 310 K follows the van der Waals volume (shown in parentheses; calculated using MarvinSketch) of the compounds examined with the largest PFAS (PFOS) exhibiting the highest value: PFOS (275 Å3) > PFOA (231 Å3) ≈ PFNA (258 Å3) > PFHxS (221 Å3). To our knowledge the dominance of entropic hydrophobic interactions have not been reported for PFASs despite the general observation that protein binding increases with PFAS hydrophobicity (excluding steric hinderance). This may be attributed to the use of 19F NMR, which is specific to the PFAS molecules and changes in their chemical environment.
Ka values for fatty acid binding to HSA at 310 K have been reported as 15 mM−1 for hexanoic acid, 34 mM−1 for octanoic acid, and 100 mM−1 for decanoic acid.61 Comparatively, we report Ka values of 17 mM−1 (α) and 31 mM−1 (ω) for PFHxS; 18 mM−1 (α) and 155 mM−1 (ω) for PFOA; and 18 mM−1 (α) and 357 mM−1 (ω) for PFOS. Values corresponding to the terminal −CF3 (ω) group for PFOA and PFOS, and greater association of PFOS than PFOA, are in good agreement with recent results based on mass spectrometry.56 In this prior work, Ka values for HSA binding were approximately half that compared to BSA. Hence, PFHxS, PFOA, and PFOS are expected to competitively bind with albumin proteins compared to their hydrocarbon analogs.25
Studies have shown that PFAS binding to BSA follows a two-step Langmuir sequence; the first and most favorable being PFAS binding within the protein hydrophobic binding sites, followed by surface adsorption.62 This explains in part why up to approximately 50 apparent albumin binding sites have been reported for PFOA.25,34,48 Changes in the chemical shift with increasing ratio of [PFAS]:[BSA] observed by 19F NMR support a two-step binding process (Figure 6). PFASs preferentially bind to hydrophobic sites as depicted by a steep decrease in with increasing [PFAS]:[BSA]. At a critical [PFAS]:[BSA] ratio, Δδ begins to plateau and approach their original positions for each PFAS. For PFOA, PFNA, and PFHxS this ratio was [PFAS]:[BSA] ≈ 12–14, denoting the shift from binding site to surface adsorption. Beyond this ratio, up to [PFAS]:[BSA] ≈ 25, additional adsorption was likely driven by hydrophobic interactions with hydrophobic patches comprised of apolar amino acids on the BSA surface.63,64
Figure 6.
Changes in PFAS chemical shifts at 310 K as a function of the [PFAS]:[BSA] molar ratio for (A) PFCAs and (B) PFSAs. The inset in (A) is for PFNA at 298 K, 304 K, and 310 K. Standard error bars are based on n = 3. Error bars not visible are smaller than the symbols.
PFNA exhibited unique behavior at high concentrations – shielding of the −CF3 (ω) group leading to negative values for Δδ observed at each of the three temperatures examined (Figure 6A). Shielding and broadening of the line width (Figure 2A2) reflect self-assembly processes, likely micelle formation as observed for PFOS using 19F NMR.55 In the presence of a charged surface, such as a polymer or activated carbon, PFAS hemimicelles are reported to form at values well below the CMC.65–69 The CMC of PFNA has been reported to be roughly 2 to 3 mM in deionized water at 298 K. Based on our results and this prior work, we propose that PFNA at concentrations greater than 0.5 mM formed partial micelles on the surface of BSA from 298 to 310 K driven again by entropic hydrophobic interactions.70 While the biological implications of hemimicelle formation are unclear, this would represent an additional mechanism by which an albumin protein could binding and transport a PFAS.
Finally, the 19F NMR signal for PFOS (⍺ and ω) was particularly weak and could not be observed in the presence of BSA below a [PFAS]:[BSA] ratio of 25:1. This is reflected in the high Δδ values at high [PFAS]:[BSA] ratios relative to the other PFASs. The weakened signal may be partially due to the presence of significant isomeric impurities in the commercial samples, consistent with prior work.42 Despite the weak PFOS signal, the Kd values and how they compare to the other PFASs are in agreement with literature values and trends, respectively. While PFOS exhibited the greatest binding of all PFASs examined, the values reported only correspond to the linear and isopropyl isomers tracked by 19F NMR.
5. CONCLUSIONS
By using 19F NMR, which directly probed the PFASs examined, entropic hydrophobic interactions were determined to be dominant in PFCA and PFSA binding to BSA. These interactions, which reflect the size of the structured water cage required to solvate a fluoroalkyl tail and the entropy gained from desolvating upon protein binding, should be characteristic for PFASs based on the number of fluorinated carbons and structure of the fluoroalkyl tail. This provides new insight to modeling PFAS-protein binding based on the local solvent structure. The importance of water interactions and solvation structure is gaining attention in protein-ligand binding and drug discovery.71
Results for Kd values are consistent with previous reports using methods such as equilibrium dialysis and mass spectrometry as these account for PFASs bound in hydrophobic cavities and on protein surfaces. However, advantages of 19F NMR include the ability to directly determine the nature of binding, and also to directly examine isomer impurities and PFAS self-assembly. This was demonstrated for PFOS and PFHxS isopropyl isomers, which exhibited lower binding affinity compared to the linear PFSAs, and for PFNA where protein-bound hemimicelles appear to have formed below the CMC, respectively.
Supplementary Material
Highlights.
PFAS-albumin binding measured for four common PFASs found in the U.S. population.
Entropic gains through desolvation revealed by 19F NMR proportional to PFAS volume.
Association constants suggesting that PFASs may compete for fatty acid binding sites.
PFAS bound per protein ranged from 12–14 via binding site and surface adsorption.
Acknowledgements
This work was funded by the National Institute of Environmental Health Science Sources, Transport, Exposure & Effects of PFASs (STEEP) Superfund Research Program under grant P42ES027706. STEEP is a partnership of the University of Rhode Island, the Harvard T.H. Chan School of Public Health Department of Environmental Health, and the Silent Spring Institute. We are grateful to Rainer Lohmann, Bongsup Cho, and Michael Greenfield for their valuable insight.
Footnotes
Michael Fedorenko: Conceptualization, methodology, formal analysis, investigation, writing - original draft preparation. Jessica Alesio: Validation, writing - review & editing. Anatoliy Fedorenko: Conceptualization, Methodology. Angela Slitt: Conceptualization, Project Administration. Geoffrey D. Bothun: Conceptualization, formal analysis, visualization, supervision.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Lindstrom AB, Strynar MJ, Libelo EL. Polyfluorinated compounds: Past, present, and future. Environ Sci Technol. 2011;45(19):7954–7961. doi: 10.1021/es2011622 [DOI] [PubMed] [Google Scholar]
- 2.Hu XC, Andrews DQ, Lindstrom AB, et al. Detection of Poly- and Perfluoroalkyl Substances (PFASs) in U.S. Drinking Water Linked to Industrial Sites, Military Fire Training Areas, and Wastewater Treatment Plants. Environ Sci Technol Lett. 2016;3(10):344–350. doi: 10.1021/acs.estlett.6b00260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henry BJ, Carlin JP, Hammerschmidt JA, et al. A critical review of the application of polymer of low concern and regulatory criteria to fluoropolymers. Integr Environ Assess Manag. 2018;14(3):316–334. doi: 10.1002/ieam.4035 [DOI] [PubMed] [Google Scholar]
- 4.Dorrance LR, Kellogg S, Love AH. What you should know about per- and polyfluoroalkyl substances (PFAS) for environmental claims. Environ Claims J. 2017;29(4):290–304. doi: 10.1080/10406026.2017.1377015 [DOI] [Google Scholar]
- 5.Dalvi VH, Rossky PJ. Molecular origins of fluorocarbon hydrophobicity. Proc Natl Acad Sci. 2010;107(31):13603–13607. doi: 10.1073/pnas.0915169107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng CA, Hungerbühler K. Bioconcentration of perfluorinated alkyl acids: How important is specific binding? Environ Sci Technol. 2013;47(13):7214–7223. doi: 10.1021/es400981a [DOI] [PubMed] [Google Scholar]
- 7.Dassuncao C, Hu XC, Nielsen F, Weihe P, Grandjean P, Sunderland EM. Shifting Global Exposures to Poly- and Perfluoroalkyl Substances (PFASs) Evident in Longitudinal Birth Cohorts from a Seafood-Consuming Population. Environ Sci Technol. 2018;52(6):3738–3747. doi: 10.1021/acs.est.7b06044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ahrens L, Bundschuh M. Fate and effects of poly- and perfluoroalkyl substances in the aquatic environment: A review. Environ Toxicol Chem. 2014;33(9):1921–1929. doi: 10.1002/etc.2663 [DOI] [PubMed] [Google Scholar]
- 9.Ng CA, Hungerbühler K. Bioaccumulation of perfluorinated alkyl acids: Observations and models. Environ Sci Technol. 2014;48(9):4637–4648. doi: 10.1021/es404008g [DOI] [PubMed] [Google Scholar]
- 10.Boisvert G, Sonne C, Rigét FF, Dietz R, Letcher RJ. Bioaccumulation and biomagnification of perfluoroalkyl acids and precursors in East Greenland polar bears and their ringed seal prey. Environ Pollut. 2019;252:1335–1343. doi: 10.1016/j.envpol.2019.06.035 [DOI] [PubMed] [Google Scholar]
- 11.Abercrombie SA, de Perre C, Choi YJ, et al. Larval amphibians rapidly bioaccumulate poly- and perfluoroalkyl substances. Ecotoxicol Environ Saf. 2019;178(March):137–145. doi: 10.1016/j.ecoenv.2019.04.022 [DOI] [PubMed] [Google Scholar]
- 12.Wang Z, Dewitt JC, Higgins CP, Cousins IT. A Never-Ending Story of Per- and Polyfluoroalkyl Substances (PFASs)? Environ Sci Technol. 2017;51(5):2508–2518. doi: 10.1021/acs.est.6b04806 [DOI] [PubMed] [Google Scholar]
- 13.Daly ER, Chan BP, Talbot EA, et al. Per- and polyfluoroalkyl substance (PFAS) exposure assessment in a community exposed to contaminated drinking water, New Hampshire, 2015. Int J Hyg Environ Health. 2018;221(3):569–577. doi: 10.1016/j.ijheh.2018.02.007 [DOI] [PubMed] [Google Scholar]
- 14.Mogensen UB, Grandjean P, Nielsen F, Weihe P, Budtz-Jørgensen E. Breastfeeding as an Exposure Pathway for Perfluorinated Alkylates. Environ Sci Technol. 2015;49(17):10466–10473. doi: 10.1021/acs.est.5b02237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karrman A, Harada KH, Inoue K, Takasuga T, Ohi E, Koizumi A. Analysis of Perfluorochemicals (PFCs) in diet duplicates and serum from Japan. Env Int. 2009. [DOI] [PubMed]
- 16.Olsen GW, Burris JM, Ehresman DJ, et al. Half-Life of Serum Elimination of Perfluorooctanesulfonate,Perfluorohexanesulfonate, and Perfluorooctanoate in Retired Fluorochemical Production Workers. Environ Health Perspect. 2007;115(9):1298–1305. doi: 10.1289/ehp.10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Beesoon S, Zhu L, Martin JW. Biomonitoring of perfluoroalkyl acids in human urine and estimates of biological half-life. Environ Sci Technol. 2013;47(18):10619–10627. doi: 10.1021/es401905e [DOI] [PubMed] [Google Scholar]
- 18.MacNeil J, Steenland NK, Shankar A, Ducatman A. A cross-sectional analysis of type II diabetes in a community with exposure to perfluorooctanoic acid (PFOA). Environ Res. 2009;109(8):997–1003. doi: 10.1016/j.envres.2009.08.002 [DOI] [PubMed] [Google Scholar]
- 19.Grandjean P, Andersen EW, Budtz-Jørgensen E, et al. Serum Vaccine Antibody Concentrations in Children Exposed to Perfluorinated Compounds. JAMA. 2012;307(4). doi: 10.1001/jama.2011.2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grandjean P, Heilmann C, Weihe P, Nielsen F, Mogensen UB, Budtz-Jørgensen E. Serum vaccine antibody concentrations in adolescents exposed to perfluorinated compounds. Environ Health Perspect. 2017;125(7). doi: 10.1289/EHP275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darrow LA, Stein CR, Steenland K. Serum Perfluorooctanoic Acid and Perfluorooctane Sulfonate Concentrations in Relation to Birth Outcomes in the Mid-Ohio Valley, 2005–2010. Environ Health Perspect. 2013;121(10):1207–1213. doi: 10.1289/ehp.1206372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barry V, Winquist A, Steenland K. Perfluorooctanoic Acid (PFOA) Exposures and Incident Cancers among Adults Living Near a Chemical Plant. Environ Health Perspect. 2013;121(11–12):1313–1318. doi: 10.1289/ehp.1306615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun M, Arevalo E, Strynar M, et al. Legacy and Emerging Perfluoroalkyl Substances Are Important Drinking Water Contaminants in the Cape Fear River Watershed of North Carolina. Environ Sci Technol Lett. 2016;3(12):415–419. doi: 10.1021/acs.estlett.6b00398 [DOI] [Google Scholar]
- 24.Goecke-Flora CM, Reo N V. Influence of Carbon Chain Length on the Hepatic Effects of Perfluorinated Fatty Acids. A 19 F- and 31 P-NMR Investigation. Chem Res Toxicol. 1996;9(4):689–695. doi: 10.1021/tx950217k [DOI] [PubMed] [Google Scholar]
- 25.Nordby GL, Luck JM. Perfluorooctanoic acid interactions with human serum albumin. J Biol Chem. 1956;219(1):399–404. [PubMed] [Google Scholar]
- 26.Jones PD, Hu W, De Coen W, Newsted JL, Giesy JP. BINDING OF PERFLUORINATED FATTY ACIDS TO SERUM PROTEINS. Environ Toxicol Chem. 2003;22(11):2639. doi: 10.1897/02-553 [DOI] [PubMed] [Google Scholar]
- 27.D’eon JC, Simpson AJ, Kumar R, Baer AJ, Mabury SA. Determining the molecular interactions of perfluorinated carboxylic acids with human sera and isolated human serum albumin using nuclear magnetic resonance spectroscopy. Environ Toxicol Chem. April2010:n/a-n/a. doi: 10.1002/etc.204 [DOI] [PubMed]
- 28.Peters T. Advances in Protein Chemistry. Volume 37. New York: Academic Press; 1985. doi: 10.1016/S0065-3233(08)60065-0 [DOI] [Google Scholar]
- 29.MacManus-Spencer LA, Tse ML, Hebert PC, Bischel HN, Luthy RG. Binding of Perfluorocarboxylates to Serum Albumin: A Comparison of Analytical Methods. Anal Chem. 2010;82(3):974–981. doi: 10.1021/ac902238u [DOI] [PubMed] [Google Scholar]
- 30.Gao K, Zhuang T, Liu X, et al. Prenatal Exposure to Per- and Polyfluoroalkyl Substances (PFASs) and Association between the Placental Transfer Efficiencies and Dissociation Constant of Serum Proteins-PFAS Complexes. Environ Sci Technol. 2019;53(11):6529–6538. doi: 10.1021/acs.est.9b00715 [DOI] [PubMed] [Google Scholar]
- 31.Gao K, Zhuang T, Liu X, et al. Prenatal Exposure to Per- and Polyfluoroalkyl Substances (PFASs) and Association between the Placental Transfer Efficiencies and Dissociation Constant of Serum Proteins–PFAS Complexes. Environ Sci Technol. 2019;53(11):6529–6538. doi: 10.1021/acs.est.9b00715 [DOI] [PubMed] [Google Scholar]
- 32.Bischel HN, Macmanus-Spencer LA, Zhang C, Luthy RG. Strong associations of short-chain perfluoroalkyl acids with serum albumin and investigation of binding mechanisms. Environ Toxicol Chem. 2011;30(11):2423–2430. doi: 10.1002/etc.647 [DOI] [PubMed] [Google Scholar]
- 33.Qin P, Liu R, Pan X, Fang X, Mou Y. Impact of Carbon Chain Length on Binding of Perfluoroalkyl Acids to Bovine Serum Albumin Determined by Spectroscopic Methods. J Agric Food Chem. 2010;58(9):5561–5567. doi: 10.1021/jf100412q [DOI] [PubMed] [Google Scholar]
- 34.Han X, Snow TA, Kemper RA, Jepson GW. Binding of Perfluorooctanoic Acid to Rat and Human Plasma Proteins. Chem Res Toxicol. 2003;16(6):775–781. doi: 10.1021/tx034005w [DOI] [PubMed] [Google Scholar]
- 35.Chen H, He P, Rao H, Wang F, Liu H, Yao J. Systematic investigation of the toxic mechanism of PFOA and PFOS on bovine serum albumin by spectroscopic and molecular modeling. Chemosphere. 2015;129:217–224. doi: 10.1016/j.chemosphere.2014.11.040 [DOI] [PubMed] [Google Scholar]
- 36.Ren X-M, Qin W-P, Cao L-Y, et al. Binding interactions of perfluoroalkyl substances with thyroid hormone transport proteins and potential toxicological implications. Toxicology. 2016;366–367:32–42. doi: 10.1016/j.tox.2016.08.011 [DOI] [PubMed]
- 37.Liu X, Fang M, Xu F, Chen D. Characterization of the binding of per- and poly-fluorinated substances to proteins: A methodological review. TrAC - Trends Anal Chem. 2019;116:177–185. doi: 10.1016/j.trac.2019.05.017 [DOI] [Google Scholar]
- 38.Luebker DJ, Hansen KJ, Bass NM, Butenhoff JL, Seacat AM. Interactions of fluorochemicals with rat liver fatty acid-binding protein. Toxicology. 2002;176(3):175–185. doi: 10.1016/S0300-483X(02)00081-1 [DOI] [PubMed] [Google Scholar]
- 39.Fourth National Report on Human Exposure to Environmental Chemicals.
- 40.Huang BX, Kim H-Y, Dass C. Probing three-dimensional structure of bovine serum albumin by chemical cross-linking and mass spectrometry. J Am Soc Mass Spectrom. 2004;15(8):1237–1247. doi: 10.1016/j.jasms.2004.05.004 [DOI] [PubMed] [Google Scholar]
- 41.Majorek KA, Porebski PJ, Dayal A, et al. Structural and immunologic characterization of bovine, horse, and rabbit serum albumins. Mol Immunol. 2012;52(3–4):174–182. doi: 10.1016/j.molimm.2012.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vyas SM, Kania-Korwel I, Lehmler H-J. Differences in the isomer composition of perfluoroctanesulfonyl (PFOS) derivatives. J Environ Sci Heal Part A. 2007;42(3):249–255. doi: 10.1080/10934520601134031 [DOI] [PubMed] [Google Scholar]
- 43.Moody CA, Kwan WC, Martin JW, Muir DCG, Mabury SA. Determination of Perfluorinated Surfactants in Surface Water Samples by Two Independent Analytical Techniques: Liquid Chromatography/Tandem Mass Spectrometry and 19 F NMR. Anal Chem. 2001;73(10):2200–2206. doi: 10.1021/ac0100648 [DOI] [PubMed] [Google Scholar]
- 44.Williamson MP. Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 45.FIELDING L. NMR methods for the determination of protein–ligand dissociation constants. Prog Nucl Magn Reson Spectrosc. 2007;51(4):219–242. doi: 10.1016/j.pnmrs.2007.04.001 [DOI] [Google Scholar]
- 46.Zhao Y, Markopoulos G, Swager TM. 19 F NMR Fingerprints: Identification of Neutral Organic Compounds in a Molecular Container. J Am Chem Soc. 2014;136(30):10683–10690. doi: 10.1021/ja504110f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macmanus-spencer L a Tse ML, Hebert PC Bischel HN, Luthy RG. Binding of Perfluorocarboxylates to Serum Albumin : A Comparison of Analytical Methods quantitative information about PFCA - albumin interac-. Anal Chem. 2010;82(3):974–981. [DOI] [PubMed] [Google Scholar]
- 48.Okaru AO, Brunner TS, Ackermann SM, et al. Application of 19 F NMR Spectroscopy for Content Determination of Fluorinated Pharmaceuticals. J Anal Methods Chem. 2017;2017:1–7. doi: 10.1155/2017/9206297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shortridge MD, Hage DS, Harbison GS, Powers R. Estimating Protein−Ligand Binding Affinity Using High-Throughput Screening by NMR. J Comb Chem. 2008;10(6):948–958. doi: 10.1021/cc800122m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugiki T, Furuita K, Fujiwara T, Kojima C. Current NMR Techniques for Structure-Based Drug Discovery. Molecules. 2018;23(1):148. doi: 10.3390/molecules23010148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swift TJ, Connick RE. NMR-Relaxation Mechanisms of O 17 in Aqueous Solutions of Paramagnetic Cations and the Lifetime of Water Molecules in the First Coordination Sphere. J Chem Phys. 1962;37(2):307–320. doi: 10.1063/1.1701321 [DOI] [Google Scholar]
- 52.Arsenault G, Chittim B, Gu J, McAlees A, McCrindle R, Robertson V. Separation and fluorine nuclear magnetic resonance spectroscopic (19F NMR) analysis of individual branched isomers present in technical perfluorooctanesulfonic acid (PFOS). Chemosphere. 2008;73(1):S53–S59. doi: 10.1016/j.chemosphere.2007.06.096 [DOI] [PubMed] [Google Scholar]
- 53.Zhang X, Chen L, Fei X-C, Ma Y-S, Gao H-W. Binding of PFOS to serum albumin and DNA: insight into the molecular toxicity of perfluorochemicals. BMC Mol Biol. 2009;10(1):16. doi: 10.1186/1471-2199-10-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao S, Liu R. Comprehensive insights into the interaction mechanism between perfluorodecanoic acid and human serum albumin. New J Chem. 2018;42(11):9065–9073. doi: 10.1039/c8nj00124c [DOI] [Google Scholar]
- 55.Wang X, Chen J, Wang D, Dong S, Hao J, Hoffmann H. Monitoring the different micelle species and the slow kinetics of tetraethylammonium perfluorooctane-sulfonate by 19F NMR spectroscopy. Adv Colloid Interface Sci. 2017;246:153–164. doi: 10.1016/j.cis.2017.05.016 [DOI] [PubMed] [Google Scholar]
- 56.Chi Q, Li Z, Huang J, Ma J, Wang X. Interactions of perfluorooctanoic acid and perfluorooctanesulfonic acid with serum albumins by native mass spectrometry, fluorescence and molecular docking. Chemosphere. 2018;198:442–449. doi: 10.1016/j.chemosphere.2018.01.152 [DOI] [PubMed] [Google Scholar]
- 57.Ellis DA, Denkenberger KA, Burrow TE, Mabury SA. The Use of 19 F NMR to Interpret the Structural Properties of Perfluorocarboxylate Acids: A Possible Correlation with Their Environmental Disposition. J Phys Chem A. 2004;108(46):10099–10106. doi: 10.1021/jp049372a [DOI] [Google Scholar]
- 58.Liu P, Goddard JD, Arsenault G, et al. Theoretical studies of the conformations and 19F NMR spectra of linear and a branched perfluorooctanesulfonamide (PFOSAmide). Chemosphere. 2007;69(8):1213–1220. doi: 10.1016/j.chemosphere.2007.06.006 [DOI] [PubMed] [Google Scholar]
- 59.Beesoon S, Martin JW. Isomer-Specific Binding Affinity of Perfluorooctanesulfonate (PFOS) and Perfluorooctanoate (PFOA) to Serum Proteins. Environ Sci Technol. 2015;49(9):5722–5731. doi: 10.1021/es505399w [DOI] [PubMed] [Google Scholar]
- 60.Bojesen E, Bojesen IN. Albumin Binding of Long-Chain Fatty Acids: Thermodynamics and Kinetics. J Phys Chem. 1996;100(45):17981–17985. doi: 10.1021/jp962141m [DOI] [Google Scholar]
- 61.Spector AA. Fatty acid binding to plasma albumin. J Lipid Res. 1975;16(3):165–179. http://www.ncbi.nlm.nih.gov/pubmed/236351. [PubMed] [Google Scholar]
- 62.Salvalaglio M, Muscionico I, Cavallotti C. Determination of Energies and Sites of Binding of PFOA and PFOS to Human Serum Albumin. J Phys Chem B. 2010;114(46):14860–14874. doi: 10.1021/jp106584b [DOI] [PubMed] [Google Scholar]
- 63.Jr Nicolau D V., Paszek E, Fulga F, Nicolau DV Mapping Hydrophobicity on the Protein Molecular Surface at Atom-Level Resolution. Carloni P, ed. PLoS One. 2014;9(12):e114042. doi: 10.1371/journal.pone.0114042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dorh N, Zhu S, Dhungana KB, et al. BODIPY-Based Fluorescent Probes for Sensing Protein Surface-Hydrophobicity. Sci Rep. 2015;5(1):18337. doi: 10.1038/srep18337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu Q, Zhang R, Deng S, Huang J, Yu G. Sorption of perfluorooctane sulfonate and perfluorooctanoate on activated carbons and resin: Kinetic and isotherm study. Water Res. 2009;43(4):1150–1158. doi: 10.1016/j.watres.2008.12.001 [DOI] [PubMed] [Google Scholar]
- 66.Ning C, Ma H, Pedersen CM, Chang H, Wang Y, Qiao Y. Interaction between environmental contaminant PFOA and PAMAM in water: 19F and 1H NMR studies. J Mol Liq. 2019;283:45–50. doi: 10.1016/j.molliq.2019.03.057 [DOI] [Google Scholar]
- 67.Lath S, Knight ER, Navarro DA, Kookana RS, McLaughlin MJ. Sorption of PFOA onto different laboratory materials: Filter membranes and centrifuge tubes. Chemosphere. 2019;222:671–678. doi: 10.1016/j.chemosphere.2019.01.096 [DOI] [PubMed] [Google Scholar]
- 68.Johnson RL, Anschutz AJ, Smolen JM, Simcik MF, Penn RL. The Adsorption of Perfluorooctane Sulfonate onto Sand, Clay, and Iron Oxide Surfaces. J Chem Eng Data. 2007;52(4):1165–1170. doi: 10.1021/je060285g [DOI] [Google Scholar]
- 69.Brusseau ML. Assessing the potential contributions of additional retention processes to PFAS retardation in the subsurface. Sci Total Environ. 2018;613–614:176–185. doi: 10.1016/j.scitotenv.2017.09.065 [DOI] [PMC free article] [PubMed]
- 70.Torres MF, de Rossi RH, Fernández MA. Aggregation Behavior of Perfluorononanoic Acid/Sodium Dodecyl Sulfate Mixtures. J Surfactants Deterg. 2013;16(6):903–912. doi: 10.1007/s11743-013-1452-6 [DOI] [Google Scholar]
- 71.Geschwindner S, Ulander J. The current impact of water thermodynamics for small-molecule drug discovery. Expert Opin Drug Discov. 2019;14(12):1221–1225. doi: 10.1080/17460441.2019.1664468 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.