Abstract
It has been reported that treatment with β-lactam antibiotics induces leukopenia and candidemia, worsens the clinical response to anticancer immunotherapy and decreases immune response to vaccination. β-lactamases can cleave β-lactam antibiotics by blocking their activity. Two distincts superfamilies of β-lactamases are described, the serine β-lactamases and the zinc ion dependent metallo-β-lactamases. In human, 18 metallo-β-lactamases encoding genes (hMBLs) have been identified. While the physiological role of most of them remains unknown, it is well established that the SNM1A, B and C proteins are involved in DNA repair. The SNM1C/Artemis protein is precisely associated in the V(D)J segments rearrangement, that leads to immunoglobulin (Ig) and T-cell receptor variable regions, which have a crucial role in the immune response. Thus in humans, SNM1C/Artemis mutation is associated with severe combined immunodeficiency characterized by hypogammaglobulinemia deficient cellular immunity and opportunistic infections. While catalytic site of hMBLs and especially that of the SNM1 family is highly conserved, in vitro studies showed that some β-lactam antibiotics, and precisely third generation of cephalosporin and ampicillin, inhibit the metallo-β-lactamase proteins SNM1A & B and the SNM1C/Artemis protein complex. By analogy, the question arises as to whether β-lactam antibiotics can block the SNM1C/Artemis protein in humans inducing transient immunodeficiency. We reviewed here the literature data supporting this hypothesis based on in silico, in vitro and in vivo evidences. Understanding the impact of β-lactam antibiotics on the immune cell will offer new therapeutic clues and new clinical approaches in oncology, immunology, and infectious diseases.
Keywords: ARTEMIS, SNM1, human metallo β-lactamase, metallo β-lactamase, β-lactam antibiotics, cephalosporin, immunity, V(D)J recombination
Introduction
β-lactam antibiotics remain the cornerstone of the antibacterial arsenal, being the most prescribed and the most important class of antibiotics in terms of sales (1–3). In clinical practice, β-lactam antibiotics are increasingly used to treat pulmonary, urinary, skin, osteo-articular and cardio-vascular infections (2, 4). However, parenteral use of β-lactam antibiotics was associated, in human, with an increased risk of complications such as candidemia, especially in the presence of catheters (5). In addition, its utilization prior to anticancer immunotherapy was associated with a worse clinical response (6–8). Nevertheless the direct effect of β-lactam antibiotics on immune cells remains poorly understood (9–15).
β-lactam antibiotics may have multiple activities. First of all, it works by inhibiting the bacterial peptidoglycan cell wall synthesis (16). β-lactamases enzymes hydrolyse the β-lactam antibiotics and may also have different nuclease and hydrolases activities (17–24). Two mechanistically distincts superfamilies of β-lactamases with two distincts ancestors are described, the nucleophilic serine β-lactamases (class A/C/D); and the zinc ion dependent metallo-β-lactamases (class B) (17, 25–27). In human, 18 metallo-β-lactamases (hMBLs) enzymes have been identified (Table 1) (28). While the physiological enzymatic role of most of them remains unknown, mutation of some hMBLs has been identified as linked to clinical disease (Table 1) (28–31). This is the case for the SNM1C/Artemis mutation that induces severe combined immunodeficiency characterized by hypersensitivity to ϒ rays irradiation and severe opportunistic infections (see next chapter) (28).
Table 1.
Human metallo beta lactamases function (28).
| 18 hMBLs | hMBL protein | Gene and chromosome location | Enzymatic activity | Function/Proposed Function | Linked Diseases |
|---|---|---|---|---|---|
| 1 | HAGH (1XM8) | 3029; 16p13.3 | Hydrolase or Glyoxalase II | Glutathione biosynthetic process. | None assigned |
| 2 | HAGHL | 84264; 16p13.3 | Hydrolase or Glyoxalase II | Unknown | None assigned |
| 3 | ETHE1 (4CHL) | 23474; 19q13.31 | Glyoxalase II | Glutathione metabolic process. Hydrogen sulfide metabolic process | Ethylmalonic encephalopathy |
| 4 | LACTB2 (4AD9) | 51110; 8q13.3 | Glyoxalase II | Endoribonuclease activity on ssRNA | None assigned |
| Mitochondrial cell death turnover | |||||
| 5 | MBLAC1 | 255374/153364; 7q22.1/5q14.3 | Glyoxalase II | Cell cycle progression (S phase) | None assigned |
| 6 | MBLAC2 | 255374/153364; 7q22.1/5q14.3 | Glyoxalase II | Penicillin degradation | None assigned |
| B-cell exome biogenesis | |||||
| 7 | SNM1A (4B87) | 9937; 10q25.1 | Exonuclease | Cell cycle, DNA damage/DNA crosslink repair | Mutations: impaired nuclear focus formation, reduced interaction with PIAS and increased sensitivity to cisplatin |
| 8 | PNKD | 25953; 2q35 | Glyoxalase II | Unknown | Dystonia type 8 |
| Tumor cell proliferation (PNKD2) | |||||
| 9 | SNM1B (5AHO) | 64858; 1p13.2 | Exonuclease | DNA damage/DNA crosslink repair | Hoyeraal–Hreidarsson Syndrome |
| 10 | SNM1C | 64421; 10p13 | Exo & endonuclease | Adaptive immunity; DNA damage, recombination, repair/DNA crosslink repair | Severe combined immunodeficiency autosomal recessive T-cell-negative/B cell-negative/NKcell-positive with sensitivity to ionizing radiation (RSSCID), Severe combined immunodeficiency Athabaskan type (SCIDA),Omenn syndrome (OS) |
| 11 | ELAC1 (3ZWF) | 55520; 18q21 | RNase | tRNA processing | None assigned |
| 12 | ELAC2 | 60528; 17p11.2 | RNase | tRNA processing | Prostate cancer, hereditary, 2 (HPC2) |
| 13 | CPSF73 (2I7T) | 51692; 2p25.1 | Endonuclease | mRNA processing | None assigned |
| 14 | CPSF73L | 54973; 1p36.33 | Integrator complex | pre-snRNA processing | None assigned |
| Hydrolase | |||||
| 15 | CPSF100 | 53981; 14q31.1 | Unknown | mRNA processing | None assigned |
| 16 | INTS9 | 55756; 8p21.1 | Integrator complex | snRNA processing | None assigned |
| Hydrolase | |||||
| 17 | NAPE-PLD (4QN9) | 222236; 7q22.1 | Catalyse hydrolysis of NAPE | Lipid/Phospholipid degradation, metabolism | None assigned |
| 18 | CMAHP | 8418; 6p21.32 | Unknown | Unknown | None assigned |
PIAS, Protein inhibitor of Activated STAT; NK, Natural killer; RS-SCID, Radiosensitive severe combined immunodeficiency; SCIDA,severe combined immunodeficiency disease (SCID) occuring among Athabascan-speaking Native Americans (SCIDA) LACT2B, β-lactamase-like protein 2; ELAC1, Zinc phopshodiesterase ELAC protein 1; ELAC2, Zinc phopshodiesterase ELAC protein 2 tRNA, transfert RNA; mRNA, messenger RNA; snRNA, small nuclear RNA; MBLAC1& 2, MBL domain containing protein 1 and 2CPSF73, Cleavage and polyadenylation specificity factor 73; CPSF73L, Cleavage and polyadenylation specificity factor 73 like; CPSF100, Cleavage and polyadenylation specificity factor 100; INTS9, Integrator complex subunit; NAPE-PLD (4QN9), N-acyl-phophatidylethanolamine-hydrolizing phospholipase-D; CMAHP, CMP-N-acetylneuraminic acid hydroxylase pseudogene.
Some β-lactam antibiotics showed direct effect on hMBLs. Cephalosporins were identified as competitive inhibitors of the hMBLs SNM1A, B and C nuclease (32). Ampicillin and cephalosporin, β-lactamin antibiotics, can in vitro inhibit the SNMIC/Artemis protein (33, 34). By analogy, this raises the question whether β-lactam antibiotics can induce reversible humoral deficiency in humans by transiently blocking the SNM1C/Artemis nuclease activity. As the conserved catalytic site is highly similar in SNM1A, B and C proteins, we postulate that the β-lactam antibiotics would be capable of inhibiting the SNM1C/Artemis protein leading to a transient immune deficit (humoral) (33, 35). Here, after introducing the SNM1C/Artemis protein functionality and implications, we will examine the in silico, in vivo and in vitro evidences to support our hypothesis. We searched in Medline and Google scholar for references with no language restrictions and no time limitations nor status. We performed a docking analysis to evaluate in silico the cephalosporin affinity with the SNM1C/Artemis catalytic site.
The hMBLs SNM1C/Artemis Protein Functionality, Clinical Implications and Catalytic Site
The SNM1-family proteins, that belongs to the hMBLs, is characterized by a conserved catalytic site with nuclease activity implicated in the maintenance of the genome stability (36). SNM1 is a member of the SNM1 (or PSO2) gene family that encodes proteins involved in DNA processing, DNA metabolism and cell cycle regulation. Whereas SNM1A and B contribute to the intercross linked repair, SNM1C/Artemis contributes to the double-strand break repair. Indeed, SNM1C/Artemis possesses an exonuclease activity in vitro and when complexed and phosphorylated by DNA PKcs, SNM1C/Artemis’ specificity switched to an endonuclease activity, which is involved in the V(J) and V(D)J segments rearrangement (37).
The hMBLs SNM1C/Artemis Protein and Its Catalytic Site
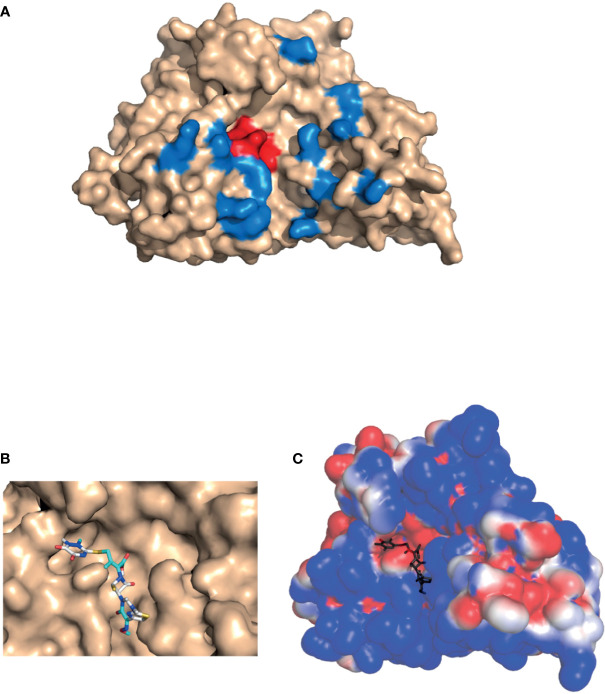
SNM1C/Artemis, a 78 k-Da protein with 692 amino acids, is a member of the metallo-β-lactamase superfamily of nucleases, characterized by a conserved MBL and β-CASP domain. It belongs to a distinct group of the β-CASP family of nucleases that includes 3 proteins : SNM1A/Pso2 related proteins, SNM1B/Apollo, SNM1C/Artemis (38). The 3-dimensional structure of the catalytic domain of recombinant human SNM1C/Artemis has been recently solved (Figure 1A) (35). The SNM1C/Artemis’ active catalytic site contains metallo-β-lactamase, β-CASP domain and a cluster of conserved histidine and aspartate residues capable of binding two metals atoms. These particularities of the active catalytic site are similar to the other members of the DNA cross linked repair gene SNM1 family and mRNA 3’end processing endonuclease (35). As observed in the SNM1A protein, SNM1C/Artemis contains only one zinc ion in its catalytic site, the metallo-β-lactamase domain. The β-CASP domain acts as a plug, covering the active site displayed by the MBL domain, and creates the substrate binding pocket, which should confer substrate selectivity (35, 38). As particularities, the SNM1C/Artemis nuclease presents a second zinc ion in the β-CASP domain reorienting the putative DNA binding surface and extending the substrate binding pocket to a new pocket, the pocket III (Figure 1A) (35). This substrate binding site, pocket III, appears to be deeper and wider in the SNM1C/Artemis nuclease than in SNM1A and SNM1B proteins (35). In addition and contrary to the SNM1A and SNM1B proteins, SNM1C/Artemis exhibits a dominant and extensive positive charge in its substrate-binding surface, probably because of the structurally distinct DNA substrate of this protein (Figure 1A) (35). Mutation on the His-254 residue disrupts the in vivo and in vitro function of SNM1C/Artemis and results in the radiosensitive severe combined immune deficiency in human (see chapter V) (31, 35). His-254 residue is located within the catalytic metallo-β-lactamase domain and has been proposed to be involved at the metal ions coordination. It thus indicates that the unique zinc binding motif in the β-CASP domain of SNM1C/Artemis, which coordonate His-228, His-245, His 256 and Cys 272 residues, is both structurally and functionally important (35). Interestingly, the His-254 residue is conserved within the SNM1 family (31, 38).
Figure 1.
Artemis protein docking. 3-D structure of the catalytic domain of SNM1C/Artemis affinity with β-lactam antibiotics (PDB: 6WO0) (35). (A) Location of the β-lactamase conserved active site shown in red, and of the putative DNA-binding site in the pocket III (described by Karim et al. shown in blue. (B) Predicted binding of ceftriaxone to the SNM1C/Artemis target within its conserved active site. Molecular docking was performed using the AutoDockTools software. (C) Electrostatic potential surface of Artemis with predicted docking of ceftriaxone. An electrostatic potential surface of SNM1C/Artemis protein was generated using the PyMOL1.8.0 software along with the APBS tool plugin. The red color indicates an excess of negative charges near the surface and the blue color arises from a positively charged surface. The ceftriaxone scaffold is shown in dark grey.
Artemis/SNM1C Protein and the V(J) and V(D)J Segments Rearrangement
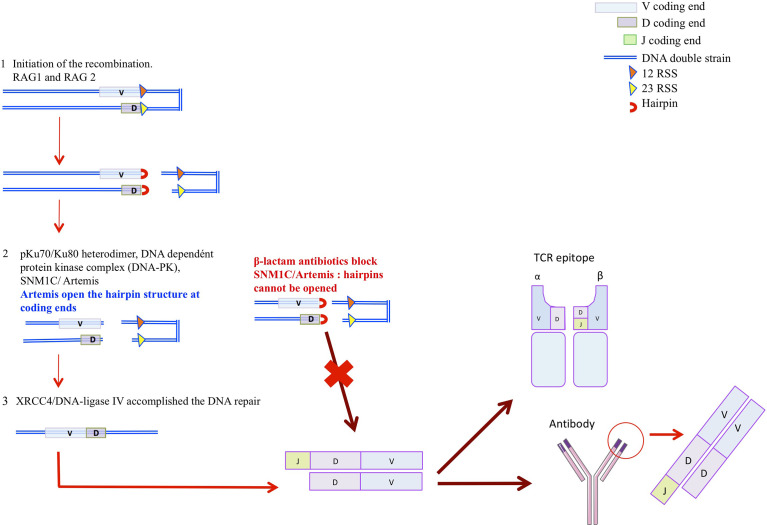
A central part of the immune system is played by antibodies that recognize and distinguish specific molecular patterns of antigens. Structure of antigens are different and the repertoire is currently estimated to be ~1015 members, for the naïve repertoire, and higher than 1018 based on the theorical repertoire combination (39). The variable region of immunoglobulins (VH and VL) and T-cell receptors (Vα, Vβ, Vγ, and Vδ) are encoded by a modest region of gene segment whose diversity is achieved by a series of programmed DNA rearrangment termed the V(J) and V(D)J rearrangements (40–42). Immunoglobulin (Ig) or T-cell receptor variable regions are assembled in developping B or T lymphocytes from germline variable (V), diversity (D), and joining (J) gene units (42–44). While the V(D)J recombination segment is involved in the TCR β and δ and in the Ig heavy chain (IgH) variables exons, the V(J) recombination is involved in the TCRα and γ and Ig light chain (IgL) variables exons (43). These recombinations could be summarized in 3 different phases (Figure 2) (42, 43, 45, 46). The first one is lympoid specific and constitutes the initiation of the recombination. Two protein, RAG1 and RAG2, encode by the recombination activated gene 1 and 2 (RAG1 and RAG 2), recognize the recombination signal sequence (RSS) that flanks all variable (V), diversity (D), and joining (J) gene units and introduce the DNA double strand break (dsb) at the border of the RSS (45). Six proteins are involved in the second and third phases, which are constituted by the ubiquitous DNA-repair machinery (NHEJ), finalizing the DNA gap : Ku70/Ku80, DNA-dependent protein kinase catalytic subunit (DNA-PJcs), Artemis, terminal deoxynucleotidyl transferase (TdT) and XRCCA/LigaseIV (47). In the second phase, the DNA damage is recognized and hairpin are oppened. The pKu70/Ku80 heterodimer binds to the DNA dependent protein kinase complex (DNA-PK) that recognizes the DNA dsb. Then the DNA-PKcs is recruited and phosphoryles the SNM1/Artemis protein that opens the hairpins structures at coding ends (46). Finally, in the terciary phase, terminal deoxynucleotidyl transferase (TdT) ensures the jonctionnal diversity by adding N-nucleotides to the V-(D) and (D)-J jonction. DNA reparation is then achieved since the XRCC4/DNA-ligase IV accomplished the DNA repair. Ig and TCR diversity, is thus due to the inaccuracy in the junction areas V-(J) and V-(D)-J and to the insertion or deletion of a nucleotide under the action of the terminal transferase enzyme (TdT). If mutation in the RAG1/RAG2 compromises the initial phase of the V(D) and V(D)J recombination, defects in SNM1C/Artemis or in the DNA-PKcs induce a non-resolution of the hairpins at coding ends (Figure 2) (48). SNM1C/Artemis has thus a critical role on DNA double strand break repair in eukaryotic cells influencing the V(J) and V(D)J recombination and therefore the humoral and cellular immune diversity and functionality.
Figure 2.
The hypothesis. 1-Lymphoid specific phase is the initiation of the recombination. RAG1 and RAG 2, recognize the recombination signal sequence (RSS) that flank all vairable (V), diversity (D), and joining (J) gene units and introduce the DNA double strand break (dsb) at the border of the RSS 2. This phase results on hairpin formation. 2-DNA damage is recognized and hairpin oppened, pKu70/Ku80 heterodimer bonds to the DNA dependant protein kinase complex (DNA-PK) that recognize the DNA dsb. Then the DNA-PKcs is recruited. DNA-PK recruits and phosphoryles SNM1C/Artemis protein that opens the hairpins structures at coding ends 3-DNA reparation is achieved since the XRCC4/DNA-ligase IV accomplished the DNA repai.
SNM1C/Artemis Mutation in Humans
The Artemis autosomic recessive mutation induces default in the V(J) and V(D)J rearrangement causing humans RS-SCID phenotype characterized by an increase sensitivity to ϒ rays irradiation (Figure 2). SNM1C/Artemis gene mutation is rare in Europe, < 1/500 000 births, but its incidence is much higher in the Athabascan-speaking, Navajo and Apache Native Americans involving 1/2000 births (49). First cases of severe combined immunodeficiency were reported in Athabascan-speaking American Indian Children, who presented B-cells and T-cells deficiency associated with oral and/or genital ulceration (50). Clinical presentations were also characterized by hypogammaglobulinemia and poor antibody response (Table 2) (47, 49, 51, 52, 55–58). These children were dying in the primary year of life because of opportunistics infections caused by cytomegalovirus (CMV), Epstein barr virus (EBV), zooster virus (VZV), and pneumocystis. Reconstitution of the B and T lymphocyte pool was observed after bone marrow transplant therapy (BMT). Studying the prenatal diagnosis of severe combined immunodeficiency disease in the Athabascan population, Li et al. identified that 10% of unaffected carriers presented a polymorphism on the SCIDA mutation, located NspI exon 8 (59). In addition, hypomorphic Artemis mutant were reported as responsible for atypical severe combined immunodeficiencies complicated by autoimmunity, granulomatous inflammation, lymphoproliferative diseases and lymphoma malignancies with later clinical manifestations, in childhood or adulthood (37, 53, 58, 60). While the hMBLs SNM1C/Artemis mutations are associated with immunodeficiency, by analogy, reversible blockage of this protein may induce transient acquired immunodeficiency affecting the humoral response. The impact of cephalosporins on the immune response may have some similarities to the immune deficiencies seen in patients with an innate SNM1C/Artemis mutation. However, it is assumed that the impact of cephalosporins on the immune response is reversible and less severe than that seen in innate SNM1C/Artemis mutations. In the next chapter we will discuss the evidences arguing for a SNM1C/Artemis blockage by cephalosporins. In the next chapter we will discuss the evidences arguing for a SNM1C/Artemis blokage by cephalosporins.
Table 2.
Clinical presentation of genetic SNM1C/Artemis mutation in human.
| Mutation SNM1C/Artemis | Immune deficit | Clinical manifestations | Population | Ref |
|---|---|---|---|---|
| Deletion (exons 1–4, C279T; Genomic Deletion (exons 5–6) | B and T cell deficit | Early onset of severe infection | Athabascan-speaking, Navajo and Apache Native Americans | (47, 50) |
| Exon 11 splice donor (G→C); C279T | Severe oral and genital ulcer | Very high incidence: 1:2000 | ||
| Exon 10 splice donor (G→A) | ||||
| Exon 5 splice donor (G→T) | ||||
| Deletion (exons 1–4) | ||||
| Del G818 | ||||
| Deletion (exons 5–8) | ||||
| Heterozygous and homozygous mutation (genomic exon 3 deletion; 2-nt insertion between T1205 and G1206) | Hypogammaglobulinemia | Otitis/diarrhea/stomatitis | 2 & 3 months | (51) |
| Deletion exons 10, 11, and 12 | B-cell differenciation arrest | Pneumocystis, CMV, VZV | 1 day to 5 months | (52) |
| G>T mutation at position 47 in exon 5 | ||||
| G>T mutation at position 47 in exon 5 homozygous G>A mutation at position 42 in exon 6 | ||||
| Hypomorphic mutation (unprecised) | B and T cell lymphopenia and hypogammaglobulinemia | EBV associated disseminated B-cell lymphoma | NA | (53) |
| Hypomorphic DCLRE1C mutation | Antibody deficiency very low B-cell numbers and serum IgA levels | Recurrent respiratory tract infections, gastroenteritis, mycobacterial skin infection, otitis media, severe varicella, verruca vulgaris, aphtous stomatitis, granulomatous skin lesions, Hashimito thyroiditis, juvenile idiopathic arthritis | 2-10 years | (54) |
| Hypomorphic DCLRE1C mutation | Chronic lymphopenia | Chronic ulcerating intestinal inflammation | 6 years | (43) |
| Deletion in exon 14; c.1464delG; p.Gln488Fs | Hyper IgM syndrome, low IgG and IgA | Diarrhea, CMV, sclerosing cholangitis | 6 & 5 year old | (55) |
| Large granular lymphocytic leukemia, death | ||||
| Homologous deletion of the gene 3 of SNM1C/Artemis, DCLRE1C | NA | Pneumocystis | 5 months | (56) |
| Mutation (c. 194C > T and c. 194C > T) | Poor antibody response | Chronic post vaccinal rubella virus infection (granulomatous dermatitis) | 13 years | (57) |
CMV, cytomegalovirus, VZV, varicella zooster virus, EBV, Epstein Barr virus, Ig, immunoglobulin; DCLRE1C, DNA, cross-link repair 1C.
β-Lactam Antibiotics Can Inhibit the hMBLs SNM1 Fold
Some studies have demonstrated that β-lactam antibiotics inhibit the SNM1 (A, B & C) and MBLAC1.
First, in vitro experiment showed that ampicillin inhibit SNM1C/Artemis nuclease activity (33). Li et al. have evaluated the exo- and endo-nuclease activities of the SNM1C/Artemis protein in presence of ampicillin (33). Incubating the single-stranded DNA substrate with SNM1C/Artemis and ampicillin, they observed low levels of SNM1C/Artemis exonuclease and endonuclease products, in an ampicillin dose-dependent manner (33). Furthermore, as the active architecture site of bacterial MBLs and SNM1 proteins are similars, and as β-lactam antibiotics are the target of bacterial MBLs, Lee et al. screened the β-lactam antibiotics candidates inhibitors of SNM1A/B (32). Incubating 20 nucleotides single–stranded DNA substrates modified with fluorophore in the presence of the SNM1A/B proteins and cephalosporins, they observed, analysing the resultant fluorescent subtrate, that ceftriaxone acts as a competitive reversible inhibitors of the SNM1A/B protein (32). While the in vitro inhibitory concentration of ceftriaxone necessary to block SNM1A and SNM1B is 1mg/L, the plasmatic concentration reached in patients treated with ceftriaxone is much higher (32, 33). Interestingly and even more recently, a glial-expressed Caenorhabditis elegans gene, swip-10, an ortholog of the mammalian MBLAC1 gene, that encodes for a metallo-β-lactamase domain-containing protein limiting glutamate-dependent changes in dopamine neuron excitability, showed high affinity with ceftriaxone (61). At last, using a real time fluorescence-based nuclease assay, ceftriaxone was identified as inhibiting the nuclease activity of the SNM1/C Artemis with a modest IC50 of 65 μM (34).
The ability for β-lactam antibiotics to interact with MBLs, whether in humans or other animals, was thus demonstrated in vitro. We will next review the in silico and in vivo evidences arguing for the possible SNM1C/Artemis inhibition by β-lactam antibiotics (Table 3).
Table 3.
Evidences of the impact of β-lactam antibiotics on SNM1C/Artemis: in vitro and in vivo evidences of a decrease in the immune response especially the humoral response.
| Molecule | Protein/Cell/animal/population | Administration | Evidence | Ref |
|---|---|---|---|---|
| In vitro | ||||
| Ceftriaxone | Metallo-β-lactamase domain-containing protein | High affinity of ceftriaxone to MBLAC1 catalytic site | Glial-expressed Caenorhabditis elegans gene, swip-10, encodes a metallo-β-lactamase domain-containing protein. | (61) |
| MBLAC1 | MBLAC1, a mammalian orthologue of swip-10 | |||
| Ceftriaxone showed high affinity in binding to MBLAC1 | ||||
| Cefotaxime | Resulting substrate fluorescence of SNM1A & B activity | SNM1A : IC50s of 4–7 μM | Competitive inhibitors of SNM1A and SNM1B exonuclease activity | (32) |
| Ceftriaxone | SNM1B : IC50s of 32–129 μM. | |||
| Ampicillin | Evaluation of the SNM1C/Artemis products | 5µM | Inhibition of the SNM1C/Artemis nucleases (endo & exo) activities | (33) |
| Ceftriaxone | SNM1C/Artemis | IC50 : 65 μM | Inhibition of the SNM1C/Artemis nuclease activity | (34) |
| In human | ||||
| Cephalosporin (within the last 3 months) | Adults | Intravenous | Increase risk of candidemia OR = 4, 95% CI 1.3-11 (p=0.01) | (62) |
| Cephalosporin (during ≥ 3 days) | Adults tertiary heath care hospital | Intravenous | Increase risk of candidemia (OR :2.2 IC [1.04-4.77] p=0.04) versus non peripheral line candidemia | (5) |
| Cephalosporin | Extremely low birth weight infants | Intravenous | Increase risk of candidemia (OR :1.77 95 IC[1,31-2,38] p<0.002) | (63) |
| Third-generation cephalosporin use | Neonate | Intravenous | Increase risk of candidemia OR: 4.6 (p <0.01) | (64) |
| In animals | ||||
| Ceftiofur hydrochloride | Pigs | Intravenous | Increase delay in the detection of pseudorabies virus vaccine antibodies | (65) |
| Decrease antibodies levels in response to the swine influenza virus | ||||
| Intravenous cephalosporins (ceftizoxime, ceftezole and DQ2556) | Mice | Intravenous | Decrease response to pneumococcal type III (S3) polysaccharides | (66) |
| β-lactam cefsulodin, cefmenoxime, cefotiam and ceftazidime | Mice | Subcutaneously | Decrease IgG spleen level in mice treated with cephalosporins and immunized with red blood sheet cells than in those treated with penems | (67) |
| Ceftriaxone | Mice | Intragastric | Decrease B cells secreting antibodies in tissues (mesenteric lymph nodes, Peyer’s patches and GALTs) | (68) |
| Decrease secreted IgA and sIgA | ||||
| Ceftriaxone | Mice | Daily gavage (150 days) | Decrease mucus IgG and IgA | (69) |
| Spleen index reduction | ||||
GALT, gut-associated lymphoid tissue; sIgA, secreted IgA; OR, Odd Ratio.
In Silico Proof of Concept: Cephalosporin Can Interact With SNM1C/Artemis
In order to test our hypothesis we evaluate in silico the affinity of the SNM1C/Artemis protein with cephalosporin. The analysis showed a strong affinity of SNM1C/Artemis with cephalosporins predicting their possible interaction.
Docking: Artemis Affinity With Cephalosporin
We performed a docking based on the three-dimensional structure of SNM1C/Artemis catalytic domain that was solved and published by Karim et al. (PDB: 6WO0) (35). They also described the pocket III structural feature of the SNM1C/Artemis as the putative DNA-binding site, that extends the conserved substrate-binding site and is made of mostly positively charged residues on the surface (Figure 1A). The interaction between the SNM1C/Artemis catalytic domain and the ceftriaxone β-lactam antibiotic could be modeled through molecular docking (Figures 1B, C) based on the previously mentioned structure (35). In our model, the cephalosporin appeared to preferentially bind within the conserved active site, mostly constituted of negatively charged residues on the surface, as shown with the electrostatic potential surface (Figure 1C). A binding energy of -3.84 kcal/mol could be predicted using the AutoDockTools software, suggesting promising affinity. This docking analysis is supported by a recent structural proposition, in which ceftriaxone binds to the SNM1C/Artemis protein surface (34).
The In Vivo Impact of β-Lactam Antibiotics on Immunity, a Review of Evidences
Based on a review of the literature, we evaluate the evidences that β-lactam antibiotics have an impact on humoral immunity in animals and humans.
β-Lactam Antibiotics Decrease the Humoral Response in Animals
Reports from veterinarians identified a decrease humoral response in animals treated with β-lactam antibiotics. In pigs, ceftiofur hydrochloride administrated concomitantly to vaccine induced a significant delay in the detection of antibodies in response to pseudorabies virus vaccine and a significant decrease of antibodies levels in response to the swine influenza virus, when compared to controls (65).
Furthermore, the immunotoxicity of intravenous cephalosporins (ceftizoxime, ceftezole and DQ2556) was demonstrated in mice, in which an increased spleen weight and enlargement of splenic germinal centres were observed 5 to 7 days after treatment (66). Cephalosporins are described as affecting cellular and humoral response (66). The response to pneumococcal type III (S3) polysaccharides, a T-cell independent immunogen, and the response to sheep red blood cells, a T-cell dependent immunogen, were decreased in mice treated 5 days with cephalosporins (66). In addition, the IgG level in the spleen of mice treated with cephalosporins and immunized with red blood cells was much lower than that of mice treated with penems (67). Finally, BALB/c mice treated with oral ceftriaxone showed a reduction in activated B and T cells in gut-associated lymphoid tissue (GALT), Peyer’s patches and mesenteric lymph nodes, as well as a significant reduction in mucus IgG and IgA (68, 69). Therefore, animals treated with cephalosporins were not able to mount a sustained protective immune response since a deficient humoral response was observed.
This may result, at least in part, on the blockage of the SNM1C/Artemis protein leading to reduced and delayed production of antibodies.
Clinical and Experimental Reports in Human
In humans, several observations suggest the use of β-lactam antibiotics as a trigger for immunodeficiency, reducing the total number of leukocytes, promoting opportunistic infections and decreasing the response to anticancer therapy with immune checkpoint inhibitors.
In a retrospective analysis of 291 patients treated with antibiotics, leukopenia in addition to neutropenia, thrombopenia and eosinophilia were identified as adverses events of parenteral antibiotics use (70). Interestingly, leukopenia was mainly observed in patients receiving β-lactam (39%) as the only antibiotic (70).
Antibiotics may also facilitate opportunistic infections commonly encountered in immunocompromised patients since the use of β-lactam antibiotics has been reported to be significantly associated with an increased risk of candidemia in neonates, infants and adults (Table 3) (5, 60, 62, 63). If the importance of the humoral response in the control of candidiasis has been discussed, and is supported by several in vitro and in vivo observations (71–74). Anti MP-65 antibodies were related with survival in patients with severe disseminated infection and the administration of human polyvalent IgG mouthwash showed significant reduction of infection in patients with chronic oral candidiasis (62, 71).
In cancer patients, the use of antibiotics before treatment with immune checkpoint inhibitors is significantly associated with relapse and worse clinical outcome (8, 75, 76). Several types of cancer are concerned: renal cell carcinoma, non small cell lung cancer, urothelial carcinoma and melanoma (8, 75, 76). Interestingly, all authors of these studies agreed that β-lactam antibiotics were the most widely used antibiotics (35 to 45%) followed by quinolones and macrolides (8, 75, 77). It would have been interesting to explore the lymphocyte pool of cancer’s patients receiving ceftriaxone to test our hypothesis.
It should be noted that the direct impact of cephalosporin on B-cells is probably not the only risk factor that could explain the occurrence of candida infection since catheters, microbiota composition and TH17 cell response are clearly involved in these infections (5, 63, 78, 79). Nonetheless, these in vivo observations support the possible immunomodulatory and even immunosuppressive role of β-lactam antibiotics on immune cells favouring the occurrence of opportunistic infections.
Discussion
β-Lactam Antibiotics May Block the SNM1C/Artemis, hMBLs, Activity
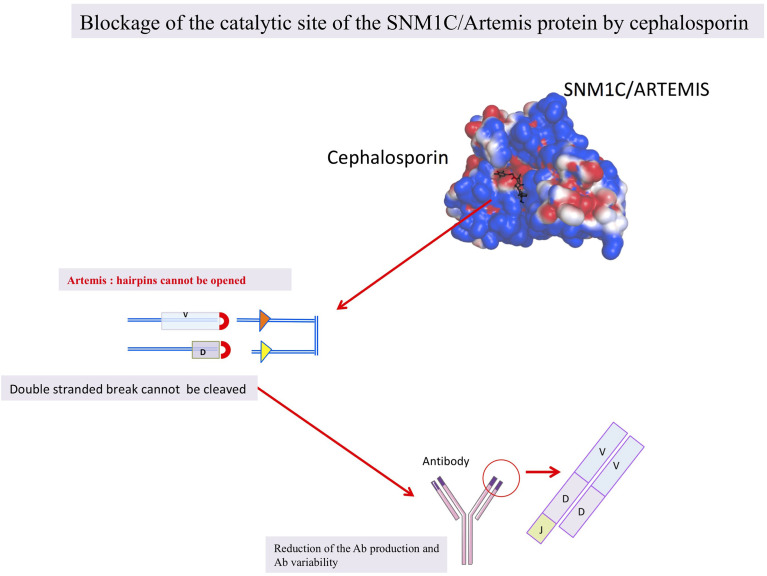
We postulate that β-lactam antibiotics can induce reversible humoral deficiency in humans by transiently blocking the SNM1C/Artemis nuclease activity and our hypothesis is sustained by in silico, in vitro and in vivo observations (Figure 3 and Table 3) (33, 35). β-lactam antibiotics may act reversibly on the hMBLs, SNM1C/Artemis protein, blocking V(J) and V(D)J recombination resulting in a deficient humoral responses. Therefore, the clinical immunocompromized stage observed under β-lactam antibiotics could be linked to the SNM1C/Artemis blockage. Shedding light on this issue would make it possible to improve care. Indeed, if β-lactam antibiotics have an impact on immunity, diminishing the host’s ability to mount a strong vaccinal response, a delay between antibiotic administration and vaccination should be proposed based on further investigations. Moreover, if β-lactam antibiotics confer immunocompromised states and favour opportunistic infections, candidal infections should be actively sought and routinely reported in the event of fever under these antibiotics, the use of catheters should, in addition, be limited.
Figure 3.
The hypothesis shematized: β-lactam antibiotics blocking the SNM1C/Artemis protein.
Further Evidences Are Needed to Confirm This Hypothesis
In vitro experimentations are needed to demonstrate that β-lactam antibiotics, such as cephalosporin, may block the SNM1C/Artemis protein inhibiting the substrates production of this exo and endonuclease. In vivo, testing the ability of mice to produce an effective vaccine response when treated with ceftriaxone, in addition to the evaluation of their B and T cell maturated pool will provide answers to these questions. In humans on cephalosporin, humoral immunity can be studied simply by assessing the vaccine response. Further research evaluating the humoral response, in particular against C. albicans, in patients receiving β-lactam antibiotics, would shed light on our hypothesis. We have focused here on hMBLs but MBLs are present in the world of life, in bacteria, eukaryotes and viruses, so the spectrum of β-lactam may be broader than expected and may also involve some MBLs.
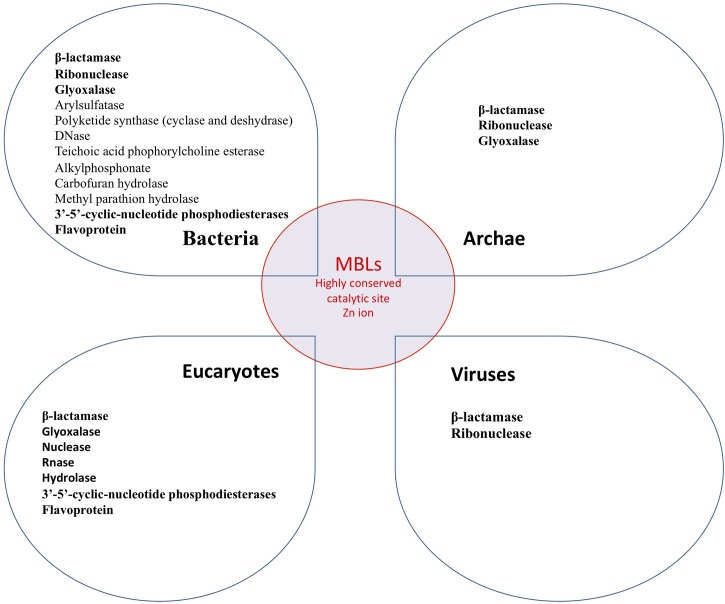
Metallo β-Lactamase in the World of Life
It is important to point out that MBLs are widely distributed in nature. They are found in bacteria, archae, giant viruses, and eukaryotes cells including humans. Thus a landscape of cross enzymatic activities among MBLs enzyme superfamily was reported (31, 77, 80–83). Therefore, MBLs’ cross enzymatic activities (hydrolase and nuclease) among different domains of the living were identified, whereas different enzymatic activities observed may be related to divergent host substrates (17). The interspecies common enzymatic activities can be explained by the highly conserved MBLs catalytic site (Figure 4) (81, 84, 85). Thus, while the catalytic site of MBL is highly conserved, all known enzymatic activities of MBL could possibly be inhibited by β-lactam antibiotics (Figure 3). In humans, blocking the enzymatic activities not only of the SNM1C/Artemis protein but of each of these 18 hMBLs may have a physiological and clinical relevance as yet unknown.
Figure 4.
Known MBLs enzymatic activity in the world of life. All these enzymes may be inhibited by β-lactam antibiotics.
Microbiota and the Indirect Role of β-Lactam on the Immune Response, a Confounding Marker
While our hypothesis supports the direct role of β-lactam on immune cells, the impact of these antibiotics via the microbiota on immunity - i.e. their indirect impact on the immune response - cannot be neglected. In animals, a reduced antibody vaccinal response was observed in germ-free, antibiotic-induced dysbiosis and antibiotic-treated mice. Interestingly, these antibody vaccinal responses were re-increased after oral reconstitution of the gut microbiota (9–11). In humans, the role of the gut microbiota was also highly suspected to modulate the vaccinal response. Bifidobacterium predominance enhanced humoral response to oral and parenteral vaccine (oral polio vaccine, Bacille calmette guerin vaccine, tetanus toxoid) (12, 14, 15). High abundance of Proteobacteria, Clostridium cluster XI, and Streptococcus bovis and low abundance of Bacteroidetes phylum were associated with an increase serological response to oral rotavirus vaccine (12, 14, 15). More recently, a reduced H1N1 vaccinal (IgG and IgA) response was reported in patients treated with neomycin, vancomycin and metronidazole (13). Thus, although the impact of β-lactam on vaccine response has not been specifically studied, antibiotics appear to modulate the humoral response to vaccination, and probably, in part, via the gut microbiota.
Conclusion
β-lactam antibiotics direct impacts on the immune cells should be considered. Since antibiotics can inhibit SNM1A, B and C/Artemis activities in vitro, and since the catalytic site of hMBL, in particular that of the SNM1 family, is very well conserved, we hypothesized that β-lactam antibiotics could reversibly inhibit the SNM1C/Artemis protein and block the V(D)J recombination. Our hypothesis is sustained by in vitro, in silico and in vivo evidences and needs to be further investigated. The putative interference of beta-lactams with SNM1C/Artemis nuclease activity would become clinically relevant in cases of long-term treatment, such as persistent osteoarticular, cardiovascular or cerebro-meningeal infections. Highlighting the physiological role and clinical impact of the hMBLs as the effect of β-lactam antibiotics on these hMBLs could pave the way for new clinical and therapeutic approaches in immunology, infectious diseases and oncology.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
CM and PP wrote the article. LP performed the in silico analysis. CD, J-LM, and DR supervised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
URMITE, IHU Méditerranée Infection. This work was supported by the French Government under the « Investissements d’avenir » (Investments for the Future) programme managed by the Agence Nationale de la Recherche (ANR, fr: National Agency for Research), (reference: Méditerranée Infection 10-IAHU-03). This work was also supported by Région Provence-Alpes-Côte d’Azur and European funding FEDER PRIMMI (Fonds Européen de Développement Régional - Plateformes de Recherche et d’Innovation Mutualisées Méditerranée Infection.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1.Fleming A. On the Antibacterial Action of Cultures of a Penicillium, With Special Reference to Their Use in the Isolation of B. Influenzæ. Br J Exp Pathol (1929) 10(3):226–36. [Google Scholar]
- 2.Klein EY, Van Boeckel TP, Martinez EM, Pant S, Gandra S, Levin SA, et al. Global Increase and Geographic Convergence in Antibiotic Consumption Between 2000 and 2015. Proc Natl Acad Sci USA (2018) 115:E3463–70. doi: 10.1073/pnas.1717295115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tooke CL, Hinchliffe P, Bragginton EC, Colenso CK, Hirvonen VHA, Takebayashi Y, et al. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J Mol Biol (2019) 431:3472–500. doi: 10.1016/j.jmb.2019.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Boeckel TP, Gandra S, Ashok A, Caudron Q, Grenfell BT, Levin SA, et al. Global Antibiotic Consumption 2000 to 2010: An Analysis of National Pharmaceutical Sales Data. Lancet Infect Dis (2014) 14:742–50. doi: 10.1016/S1473-3099(14)70780-7 [DOI] [PubMed] [Google Scholar]
- 5.Ishikane M, Hayakawa K, Kutsuna S, Takeshita N, Ohmagari N. Epidemiology of Blood Stream Infection Due to Candida Species in a Tertiary Care Hospital in Japan Over 12 Years: Importance of Peripheral Line-Associated Candidemia. PloS One (2016) 11:e0165346. doi: 10.1371/journal.pone.0165346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal Bacteria Control Cancer Response to Therapy by Modulating the Tumor Microenvironment. Science (2013) 342:967–70. doi: 10.1126/science.1240527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinato DJ, Howlett S, Ottaviani D, Urus H, Patel A, Mineo T, et al. Association of Prior Antibiotic Treatment With Survival and Response to Immune Checkpoint Inhibitor Therapy in Patients With Cancer. JAMA Oncol (2019) 5:1774. doi: 10.1001/jamaoncol.2019.2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut Microbiome Influences Efficacy of PD-1-Based Immunotherapy Against Epithelial Tumors. Science (2018) 359:91–7. doi: 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- 9.Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, et al. TLR5-Mediated Sensing of Gut Microbiota Is Necessary for Antibody Responses to Seasonal Influenza Vaccination. Immunity (2014) 41:478–92. doi: 10.1016/j.immuni.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jong SE, Olin A, Pulendran B. The Impact of the Microbiome on Immunity to Vaccination in Humans. Cell Host Microbe (2020) 28:169–79. doi: 10.1016/j.chom.2020.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lynn MA, Tumes DJ, Choo JM, Sribnaia A, Blake SJ, Leong LEX, et al. Early-Life Antibiotic-Driven Dysbiosis Leads to Dysregulated Vaccine Immune Responses in Mice. Cell Host Microbe (2018) 23:653–660.e5. doi: 10.1016/j.chom.2018.04.009 [DOI] [PubMed] [Google Scholar]
- 12.Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, et al. Stool Microbiota and Vaccine Responses of Infants. Pediatrics (2014) 134:e362–372. doi: 10.1542/peds.2013-3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagan T, Cortese M, Rouphael N, Boudreau C, Linde C, Maddur MS, et al. Antibiotics-Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans. Cell (2019) 178:1313–1328.e13. doi: 10.1016/j.cell.2019.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris VC, Armah G, Fuentes S, Korpela KE, Parashar U, Victor JC, et al. Significant Correlation Between the Infant Gut Microbiome and Rotavirus Vaccine Response in Rural Ghana. J Infect Dis (2017) 215:34–41. doi: 10.1093/infdis/jiw518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris V, Ali A, Fuentes S, Korpela K, Kazi M, Tate J, et al. Rotavirus Vaccine Response Correlates With the Infant Gut Microbiota Composition in Pakistan. Gut Microbes (2018) 9:93–101. doi: 10.1080/19490976.2017.1376162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandes R, Amador P, Prudêncio C. β-Lactams: Chemical Structure, Mode of Action and Mechanisms of Resistance. Rev Med Microbiol (2013) 24:7–17. doi: 10.1097/MRM.0b013e3283587727 [DOI] [Google Scholar]
- 17.Keshri V, Chabrière E, Pinault L, Colson P, Diene SM, Rolain J-M, et al. Promiscuous Enzyme Activity as a Driver of Allo and Iso Convergent Evolution, Lessons From the β-Lactamases. IJMS (2020) 21:6260. doi: 10.3390/ijms21176260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mikati MO, Miller JJ, Osbourn DM, Barekatain Y, Ghebremichael N, Shah IT, et al. Antimicrobial Prodrug Activation by the Staphylococcal Glyoxalase GloB. ACS Infect Dis (2020) 6:3064–75. doi: 10.1021/acsinfecdis.0c00582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato Y, Takahashi M, Seki M, Nashimoto M, Shimizu-Ibuka A. RNA-Hydrolyzing Activity of Metallo-β-Lactamase IMP-1. PloS One (2020) 15:e0241557. doi: 10.1371/journal.pone.0241557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daiyasu H, Osaka K, Ishino Y, Toh H. Expansion of the Zinc Metallo-Hydrolase Family of the β-Lactamase Fold. FEBS Lett (2001) 503:1–6. doi: 10.1016/S0014-5793(01)02686-2 [DOI] [PubMed] [Google Scholar]
- 21.Solomon JM, Grossman AD. Who’s Competent and When: Regulation of Natural Genetic Competence in Bacteria. Trends Genet (1996) 12:150–5. doi: 10.1016/0168-9525(96)10014-7 [DOI] [PubMed] [Google Scholar]
- 22.Gosink KK, Mann ER, Guglielmo C, Tuomanen EI, Masure HR. Role of Novel Choline Binding Proteins in Virulence of Streptococcus Pneumoniae. Infect Immun (2000) 68:5690–5. doi: 10.1128/iai.68.10.5690-5695.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Topp E, Hanson RS, Ringelberg DB, White DC, Wheatcroft R. Isolation and Characterization of an N-Methylcarbamate Insecticide-Degrading Methylotrophic Bacterium. Appl Environ Microbiol (1993) 59:3339–49. doi: 10.1128/AEM.59.10.3339-3349.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlenzka W, Shaw L, Kelm S, Schmidt CL, Bill E, Trautwein AX, et al. CMP-N-Acetylneuraminic Acid Hydroxylase: The First Cytosolic Rieske Iron-Sulphur Protein to be Described in Eukarya. FEBS Lett (1996) 385:197–200. doi: 10.1016/0014-5793(96)00384-5 [DOI] [PubMed] [Google Scholar]
- 25.Mojica MF, Bonomo RA, Fast W. B1-Metallo-β-Lactamases: Where Do We Stand? Curr Drug Targets (2016) 17:1029–50. doi: 10.2174/1389450116666151001105622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bush K, Fisher JF. Epidemiological Expansion, Structural Studies, and Clinical Challenges of New β-Lactamases From Gram-Negative Bacteria. Annu Rev Microbiol (2011) 65:455–78. doi: 10.1146/annurev-micro-090110-102911 [DOI] [PubMed] [Google Scholar]
- 27.Cornaglia G, Giamarellou H, Rossolini GM. Metallo-β-Lactamases: A Last Frontier for β-Lactams? Lancet Infect Dis (2011) 11:381–93. doi: 10.1016/S1473-3099(11)70056-1 [DOI] [PubMed] [Google Scholar]
- 28.Pettinati I, Brem J, Lee SY, McHugh PJ, Schofield CJ. The Chemical Biology of Human Metallo-β-Lactamase Fold Proteins. Trends Biochem Sci (2016) 41:338–55. doi: 10.1016/j.tibs.2015.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Callebaut I, Moshous D, Mornon J-P, de Villartay J-P. Metallo-Beta-Lactamase Fold Within Nucleic Acids Processing Enzymes: The Beta-CASP Family. Nucleic Acids Res (2002) 30:3592–601. doi: 10.1093/nar/gkf470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horsfall LE, Izougarhane Y, Lassaux P, Selevsek N, Liénard BMR, Poirel L, et al. Broad Antibiotic Resistance Profile of the Subclass B3 Metallo-β-Lactamase GOB-1, a Di-Zinc Enzyme. FEBS J (2011) 278:1252–63. doi: 10.1111/j.1742-4658.2011.08046.x [DOI] [PubMed] [Google Scholar]
- 31.Diene SM, Pinault L, Keshri V, Armstrong N, Khelaifia S, Chabrière E, et al. Human Metallo-β-Lactamase Enzymes Degrade Penicillin. Sci Rep (2019) 9:12173. doi: 10.1038/s41598-019-48723-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee SY, Brem J, Pettinati I, Claridge TDW, Gileadi O, Schofield CJ, et al. Cephalosporins Inhibit Human Metallo β-Lactamase Fold DNA Repair Nucleases SNM1A and SNM1B/Apollo. Chem Commun (Camb) (2016) 52:6727–30. doi: 10.1039/c6cc00529b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S, Chang HH, Niewolik D, Hedrick MP, Pinkerton AB, Hassig CA, et al. Evidence That the DNA Endonuclease ARTEMIS Also has Intrinsic 5’-Exonuclease Activity. J Biol Chem (2014) 289:7825–34. doi: 10.1074/jbc.M113.544874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yosaatmadja Y, Baddock HT, Newman JA, Bielinski M, Gavard AE, Mukhopadhyay SMM, et al. Structural and Mechanistic Insights Into the Artemis Endonuclease and Strategies for its Inhibition. Nucleic Acids Res (2021). doi: 10.1093/nar/gkab693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karim MF, Liu S, Laciak AR, Volk L, Koszelak-Rosenblum M, Lieber MR, et al. Structural Analysis of the Catalytic Domain of Artemis Endonuclease/SNM1C Reveals Distinct Structural Features. J Biol Chem (2020) 295:12368–77. doi: 10.1074/jbc.RA120.014136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cattell E, Sengerová B, McHugh PJ. The SNM1/Pso2 Family of ICL Repair Nucleases: From Yeast to Man. Environ Mol Mutagen (2010) 51:635–45. doi: 10.1002/em.20556 [DOI] [PubMed] [Google Scholar]
- 37.Poinsignon C, Moshous D, Callebaut I, de Chasseval R, Villey I, de Villartay J-P. The Metallo-Beta-Lactamase/Beta-CASP Domain of Artemis Constitutes the Catalytic Core for V(D)J Recombination. J Exp Med (2004) 199:315–21. doi: 10.1084/jem.20031142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Villartay J-P, Shimazaki N, Charbonnier J-B, Fischer A, Mornon J-P, Lieber MR, et al. A Histidine in the Beta-CASP Domain of Artemis is Critical for its Full In Vitro and In Vivo Functions. DNA Repair (Amst) (2009) 8:202–8. doi: 10.1016/j.dnarep.2008.10.010 [DOI] [PubMed] [Google Scholar]
- 39.Rees AR. Understanding the Human Antibody Repertoire. MAbs (2020) 12:1729683. doi: 10.1080/19420862.2020.1729683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakano H, Maki R, Kurosawa Y, Roeder W, Tonegawa S. Two Types of Somatic Recombination are Necessary for the Generation of Complete Immunoglobulin Heavy-Chain Genes. Nature (1980) 286:676–83. doi: 10.1038/286676a0 [DOI] [PubMed] [Google Scholar]
- 41.Brack C, Hirama M, Lenhard-Schuller R, Tonegawa S. A Complete Immunoglobulin Gene is Created by Somatic Recombination. Cell (1978) 15:1–14. doi: 10.1016/0092-8674(78)90078-8 [DOI] [PubMed] [Google Scholar]
- 42.Tonegawa S. Somatic Generation of Antibody Diversity. Nature (1983) 302:575–81. doi: 10.1038/302575a0 [DOI] [PubMed] [Google Scholar]
- 43.Bassing CH, Swat W, Alt FW. The Mechanism and Regulation of Chromosomal V(D)J Recombination. Cell (2002) 109:S45–55. doi: 10.1016/S0092-8674(02)00675-X [DOI] [PubMed] [Google Scholar]
- 44.Hesslein DG, Schatz DG. Factors and Forces Controlling V(D)J Recombination. Adv Immunol (2001) 78:169–232. doi: 10.1016/s0065-2776(01)78004-2 [DOI] [PubMed] [Google Scholar]
- 45.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG Proteins and V(D)J Recombination: Complexes, Ends, and Transposition. Annu Rev Immunol (2000) 18:495–527. doi: 10.1146/annurev.immunol.18.1.495 [DOI] [PubMed] [Google Scholar]
- 46.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin Opening and Overhang Processing by an Artemis/DNA-Dependent Protein Kinase Complex in Nonhomologous End Joining and V(D)J Recombination. Cell (2002) 108:781–94. doi: 10.1016/s0092-8674(02)00671-2 [DOI] [PubMed] [Google Scholar]
- 47.Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, et al. Artemis, a Novel DNA Double-Strand Break Repair/V(D)J Recombination Protein, Is Mutated in Human Severe Combined Immune Deficiency. Cell (2001) 105:177–86. doi: 10.1016/S0092-8674(01)00309-9 [DOI] [PubMed] [Google Scholar]
- 48.Le Deist F, Poinsignon C, Moshous D, Fischer A, de Villartay J-P. Artemis Sheds New Light on V(D)J Recombination. Immunol Rev (2004) 200:142–55. doi: 10.1111/j.0105-2896.2004.00169.x [DOI] [PubMed] [Google Scholar]
- 49.Li L, Moshous D, Zhou Y, Wang J, Xie G, Salido E, et al. A Founder Mutation in Artemis, an SNM1-Like Protein, Causes SCID in Athabascan-Speaking Native Americans. J Immunol (2002) 168:6323–9. doi: 10.4049/jimmunol.168.12.6323 [DOI] [PubMed] [Google Scholar]
- 50.Kwong PC, O’Marcaigh AS, Howard R, Cowan MJ, Frieden IJ. Oral and Genital Ulceration: A Unique Presentation of Immunodeficiency in Athabascan-Speaking American Indian Children With Severe Combined Immunodeficiency. Arch Dermatol (1999) 135:927–31. doi: 10.1001/archderm.135.8.927 [DOI] [PubMed] [Google Scholar]
- 51.Kobayashi N. Novel Artemis Gene Mutations of Radiosensitive Severe Combined Immunodeficiency in Japanese Families. Hum Genet (2003) 348–52. doi: 10.1007/s00439-002-0897-x [DOI] [PubMed] [Google Scholar]
- 52.Noordzij JG. Radiosensitive SCID Patients With Artemis Gene Mutations Show a Complete B-Cell Differentiation Arrest at the Pre-B-Cell Receptor Checkpoint in Bone Marrow. Blood (2003) 101:1446–52. doi: 10.1182/blood-2002-01-0187 [DOI] [PubMed] [Google Scholar]
- 53.Moshous D, Callebaut I, de Chasseval R, Poinsignon C, Villey I, Fischer A, et al. The V(D)J Recombination/DNA Repair Factor Artemis Belongs to the Metallo-Beta-Lactamase Family and Constitutes a Critical Developmental Checkpoint of the Lymphoid System. Ann N Y Acad Sci (2003) 987:150–7. doi: 10.1111/j.1749-6632.2003.tb06043.x [DOI] [PubMed] [Google Scholar]
- 54.Volk T, Pannicke U, Reisli I, Bulashevska A, Ritter J, Björkman A, et al. DCLRE1C (ARTEMIS) Mutations Causing Phenotypes Ranging From Atypical Severe Combined Immunodeficiency to Mere Antibody Deficiency. Hum Mol Genet (2015) 24:7361–72. doi: 10.1093/hmg/ddv437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bajin İY, Ayvaz DÇ, Ünal Ş, Özgür TT, Çetin M, Gümrük F, et al. Atypical Combined Immunodeficiency Due to Artemis Defect: A Case Presenting as Hyperimmunoglobulin M Syndrome and With LGLL. Mol Immunol (2013) 56:354–7. doi: 10.1016/j.molimm.2013.05.004 [DOI] [PubMed] [Google Scholar]
- 56.Hayase T, Ikeda T, Yoshimoto T, Imai K, Morimoto A. Fatal Idiopathic Pneumonia Syndrome in Artemis Deficiency. Pediatr Int (2019) 61:929–31. doi: 10.1111/ped.13951 [DOI] [PubMed] [Google Scholar]
- 57.Buchbinder D, Hauck F, Albert MH, Rack A, Bakhtiar S, Shcherbina A, et al. Rubella Virus-Associated Cutaneous Granulomatous Disease: A Unique Complication in Immune-Deficient Patients, Not Limited to DNA Repair Disorders. J Clin Immunol (2019) 39:81–9. doi: 10.1007/s10875-018-0581-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rohr J, Pannicke U, Döring M, Schmitt-Graeff A, Wiech E, Busch A, et al. Chronic Inflammatory Bowel Disease as Key Manifestation of Atypical ARTEMIS Deficiency. J Clin Immunol (2010) 30:314–20. doi: 10.1007/s10875-009-9349-x [DOI] [PubMed] [Google Scholar]
- 59.Li L, Zhou Y, Wang J, Hu D, Cowan MJ. Prenatal Diagnosis and Carrier Detection for Athabascan Severe Combined Immunodeficiency Disease. Prenat Diagn (2002) 22:763–8. doi: 10.1002/pd.400 [DOI] [PubMed] [Google Scholar]
- 60.Karaselek MA, Kapaklı H, Keleş S, Güner ŞN, Çelik ŞÇ, Kurar E, et al. Intrauterine Detection of DCLRE1C (Artemis) Mutation by Restriction Fragment Length Polymorphism. Pediatr Allergy Immunol (2019) 30:668–71. doi: 10.1111/pai.13056 [DOI] [PubMed] [Google Scholar]
- 61.Retzlaff CL, Kussrow A, Schorkopf T, Saetear P, Bornhop DJ, Hardaway JA, et al. Metallo-β-Lactamase Domain-Containing Protein 1 (MBLAC1) Is a Specific, High-Affinity Target for the Glutamate Transporter Inducer Ceftriaxone. ACS Chem Neurosci (2017) 8:2132–8. doi: 10.1021/acschemneuro.7b00232 [DOI] [PubMed] [Google Scholar]
- 62.Atamna A, Eliakim-Raz N, Mohana J, Ben-Zvi H, Sorek N, Shochat T, et al. Predicting Candidemia in the Internal Medicine Wards: A Comparison With Gram-Negative Bacteremia—a Retrospectives Study. Diagn Microbiol Infect Dis (2019) 95:80–3. doi: 10.1016/j.diagmicrobio.2019.04.007 [DOI] [PubMed] [Google Scholar]
- 63.Benjamin DK. Neonatal Candidiasis Among Extremely Low Birth Weight Infants: Risk Factors, Mortality Rates, and Neurodevelopmental Outcomes at 18 to 22 Months. PEDIATRICS (2006) 117:84–92. doi: 10.1542/peds.2004-2292 [DOI] [PubMed] [Google Scholar]
- 64.Yu Y, Du L, Yuan T, Zheng J, Chen A, Chen L, et al. Risk Factors and Clinical Analysis for Invasive Fungal Infection in Neonatal Intensive Care Unit Patients. Am J Perinatol (2013) 30:589–94. doi: 10.1055/s-0032-1329688 [DOI] [PubMed] [Google Scholar]
- 65.Pomorska-Mól M, Czyżewska-Dors E, Kwit K, Wierzchosławski K, Pejsak Z. Ceftiofur Hydrochloride Affects the Humoral and Cellular Immune Response in Pigs After Vaccination Against Swine Influenza and Pseudorabies. BMC Vet Res (2015) 11:268. doi: 10.1186/s12917-015-0586-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Furuhama K, Benson W, Knowles J, Roberts DW. Immunotoxicity of Cephalosporins in Mice. Chemotherapy (1993) 39:278–85. doi: 10.1159/000239137 [DOI] [PubMed] [Google Scholar]
- 67.Ogawa M, Goto S, Ishikawa F, Kimura I. Effects of Various Antibiotics on Antibody-Producing Cells of Mouse Spleen. Chemotherapy (1986) 32:464–7. doi: 10.1159/000238452 [DOI] [PubMed] [Google Scholar]
- 68.Luo X, Zheng Y, Wen R, Deng X, Zhou L, Liao H. Effects of Ceftriaxone Induced Intestinal Dysbacteriosis on Lymphocytes in Different Tissues in Mice. Immunobiology (2016) 221:994–1000. doi: 10.1016/j.imbio.2016.04.003 [DOI] [PubMed] [Google Scholar]
- 69.Guo Y, Yang X, Qi Y, Wen S, Liu Y, Tang S, et al. Long-Term Use of Ceftriaxone Sodium Induced Changes in Gut Microbiota and Immune System. Sci Rep (2017) 7:43035. doi: 10.1038/srep43035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoffman-Terry ML, Fraimow HS, Fox TR, Swift BG, Wolf JE. Adverse Effects of Outpatient Parenteral Antibiotic Therapy. Am J Med (1999) 106:44–9. doi: 10.1016/s0002-9343(98)00362-3 [DOI] [PubMed] [Google Scholar]
- 71.Torosantucci A, Tumbarello M, Bromuro C, Chiani P, Posteraro B, Sanguinetti M, et al. Antibodies Against a β-Glucan-Protein Complex of Candida Albicans and its Potential as Indicator of Protective Immunity in Candidemic Patients. Sci Rep (2017) 7:2722. doi: 10.1038/s41598-017-02977-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pedraza-Sánchez S, Méndez-León JI, Gonzalez Y, Ventura-Ayala ML, Herrera MT, Lezana-Fernández JL, et al. Oral Administration of Human Polyvalent IgG by Mouthwash as an Adjunctive Treatment of Chronic Oral Candidiasis. Front Immunol (2018) 9:2956. doi: 10.3389/fimmu.2018.02956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bergeron AC, Barker SE, Brothers KM, Prasad BC, Wheeler RT. Polyclonal Anti-Candida Antibody Improves Phagocytosis and Overall Outcome in Zebrafish Model of Disseminated Candidiasis. Dev Comp Immunol (2017) 68:69–78. doi: 10.1016/j.dci.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gazendam RP, van Hamme JL, Tool ATJ, van Houdt M, Verkuijlen PJJH, Herbst M, et al. Two Independent Killing Mechanisms of Candida Albicans by Human Neutrophils: Evidence From Innate Immunity Defects. Blood (2014) 124:590–7. doi: 10.1182/blood-2014-01-551473 [DOI] [PubMed] [Google Scholar]
- 75.Derosa L, Hellmann MD, Spaziano M, Halpenny D, Fidelle M, Rizvi H, et al. Negative Association of Antibiotics on Clinical Activity of Immune Checkpoint Inhibitors in Patients With Advanced Renal Cell and non-Small-Cell Lung Cancer. Ann Oncol (2018) 29:1437–44. doi: 10.1093/annonc/mdy103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer Immunotherapy by CTLA-4 Blockade Relies on the Gut Microbiota. Science (2015) 350:1079–84. doi: 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim H, Lee JE, Hong SH, Lee MA, Kang JH, Kim I-H. The Effect of Antibiotics on the Clinical Outcomes of Patients With Solid Cancers Undergoing Immune Checkpoint Inhibitor Treatment: A Retrospective Study. BMC Cancer (2019) 19:1100. doi: 10.1186/s12885-019-6267-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bacher P, Hohnstein T, Beerbaum E, Röcker M, Blango MG, Kaufmann S, et al. Human Anti-Fungal Th17 Immunity and Pathology Rely on Cross-Reactivity Against Candida Albicans. Cell (2019) 176:1340–55.e15. doi: 10.1016/j.cell.2019.01.041 [DOI] [PubMed] [Google Scholar]
- 79.Tan CT, Xu X, Qiao Y, Wang Y. A Peptidoglycan Storm Caused by β-Lactam Antibiotic’s Action on Host Microbiota Drives Candida Albicans Infection. Nat Commun (2021) 12:2560. doi: 10.1038/s41467-021-22845-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baier F, Tokuriki N. Connectivity Between Catalytic Landscapes of the Metallo-β-Lactamase Superfamily. J Mol Biol (2014) 426:2442–56. doi: 10.1016/j.jmb.2014.04.013 [DOI] [PubMed] [Google Scholar]
- 81.Zeng X, Lin J. Beta-Lactamase Induction and Cell Wall Metabolism in Gram-Negative Bacteria. Front Microbiol (2013) 4:128. doi: 10.3389/fmicb.2013.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Diene SM, Pinault L, Armstrong N, Azza S, Keshri V, Khelaifia S, et al. Dual RNase and β-Lactamase Activity of a Single Enzyme Encoded in Archaea. Life (Basel) (2020) 10. doi: 10.3390/life10110280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Colson P, Pinault L, Azza S, Armstrong N, Chabriere E, La Scola B, et al. A Protein of the Metallo-Hydrolase/Oxidoreductase Superfamily With Both Beta-Lactamase and Ribonuclease Activity is Linked With Translation in Giant Viruses. Sci Rep (2020) 10:21685. doi: 10.1038/s41598-020-78658-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gao M, Glenn AE, Blacutt AA, Gold SE. Fungal Lactamases: Their Occurrence and Function. Front Microbiol (2017) 8:1775. doi: 10.3389/fmicb.2017.01775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bebrone C. Metallo-Beta-Lactamases (Classification, Activity, Genetic Organization, Structure, Zinc Coordination) and Their Superfamily. Biochem Pharmacol (2007) 74:1686–701. doi: 10.1016/j.bcp.2007.05.021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.