Bittner et al. discovered that direct phosphorylation of the inflammasome sensor NLRP3, by the well-established drug target Bruton’s tyrosine kinase (BTK), promotes inflammasome assembly and boosts IL-1β release. NLRP3 phosphorylation by BTK may thus represent a novel target for therapeutic intervention in inflammation.
Abstract
Activity of the NLRP3 inflammasome, a critical mediator of inflammation, is controlled by accessory proteins, posttranslational modifications, cellular localization, and oligomerization. How these factors relate is unclear. We show that a well-established drug target, Bruton’s tyrosine kinase (BTK), affects several levels of NLRP3 regulation. BTK directly interacts with NLRP3 in immune cells and phosphorylates four conserved tyrosine residues upon inflammasome activation, in vitro and in vivo. Furthermore, BTK promotes NLRP3 relocalization, oligomerization, ASC polymerization, and full inflammasome assembly, probably by charge neutralization, upon modification of a polybasic linker known to direct NLRP3 Golgi association and inflammasome nucleation. As NLRP3 tyrosine modification by BTK also positively regulates IL-1β release, we propose BTK as a multifunctional positive regulator of NLRP3 regulation and BTK phosphorylation of NLRP3 as a novel and therapeutically tractable step in the control of inflammation.
Introduction
Inflammation mediated via the NLRP3 inflammasome not only supports the resolution of infections and sterile insults but also contributes to pathology in multiple human diseases, such as cryopyrin-associated periodic fever syndrome, gout, stroke, Alzheimer’s disease, and atherosclerosis (Duewell et al., 2010; Broderick et al., 2015; Magupalli et al., 2020; Franke et al., 2021). Thus, the activity of the NLRP3 inflammasome, a powerful molecular machine maturing IL-1 family cytokines via the activity of caspase-1, is tightly controlled at several levels. At the structural level, recent cryo-electron microscopy (EM) studies demonstrated that the three-dimensional conformation of NLRP3 is critical for NLRP3 oligomerization and may depend on ADP/ATP binding (Sharif et al., 2019). In addition, NLRP3 binding proteins, such as NEK7, have been shown to have an impact on inflammasome activity (He et al., 2016; Schmid-Burgk et al., 2016). Moreover, posttranslational modifications of NLRP3 enhance or reduce its activity by only partially elucidated mechanisms (Song and Li, 2018). Finally, NLRP3 interacts dynamically with subcellular organelles, such as the trans-Golgi network: On the one hand, a polybasic region (PBR) in the linker between the NLRP3 pyrin domain (PYD) and NAIP, CIITA, HET-E and TEP1 (NACHT) domain controls interaction of NLRP3 with negatively charged phosphatidylinositol phosphates (PIPs) at organelles, such as the Golgi, which were shown to disperse upon cell stimulation (Chen and Chen, 2018). On the other hand, dissociation from membranes into the cytosol was proposed as a requirement for the nucleation of larger NLRP3 oligomers (Zhang et al., 2017). The subsequent dynein/HDAC6-dependent trafficking of NLRP3 to the microtubule organizing center (MTOC) prompts the formation of complete inflammasome complexes, which include the adaptor apoptosis-associated speck-like protein containing a caspase activation and recruitment domain (ASC), NEK7, and the IL-1β maturation enzyme caspase-1 (Magupalli et al., 2020). However, cues instigating these shifts in localization remain to be resolved. Generally, how multiple layers of NLRP3 regulation are related or even integrated at the cellular as well as at the molecular level is unclear. If individual regulators were to provide this integration, they could be valuable targets to modulate inflammasome activity.
We and others have recently identified Bruton’s tyrosine kinase (BTK) as a novel and therapeutically relevant NLRP3 regulator (Ito et al., 2015; Liu et al., 2017) that is rapidly activated upon NLRP3 inflammasome stimulation and interacts with NLRP3 and ASC in overexpression systems. Its genetic ablation led to reduced IL-1β secretion in vitro and, importantly, in human ibrutinib-treated patients ex vivo (Liu et al., 2017). BTK is a well-known cancer target for which Food and Drug Administration–approved inhibitors, such as ibrutinib, exist (reviewed in Weber et al., 2017). Furthermore, BTK activation in macrophages was described in COVID-19–related lung inflammation, which was sensitive to BTK kinase inhibitors (Roschewski et al., 2020), and several in vivo models have illustrated BTK’s relevance for promoting NLRP3-mediated inflammation (O’Riordan et al., 2019, 2020; Purvis et al., 2020). Based on the molecular mechanisms and chronic inflammatory processes observed in the pathology of many disorders and cancers, targeting NLRP3 via BTK also appears to be an attractive therapeutic option in other diseases (Banoth and Cassel, 2017; Henrickson, 2017; Weber, 2021)—provided that the molecular basis for BTK’s involvement in the inflammasome process is elucidated.
Here, we report that BTK directly modifies four NLRP3 tyrosine (Y) residues in the PYD-NACHT polybasic linker, altering the charge of the PBR peptide sequence. Tyrosine mutagenesis alters phospholipid interactions, and BTK promotes the shift of NLRP3 from heavy (e.g., intact Golgi) to light (e.g., dispersed Golgi) membranes. Consequently, ablation of BTK kinase activity or tyrosine mutation decreased the formation of cytosolic NLRP3 oligomers, complexes with ASC, and IL-1β release, respectively. Our data suggest that BTK-mediated tyrosine phosphorylation affects the activity of the NLRP3 inflammasome by modifying PBR charge, subcellular localization, inflammasome assembly, and ultimately IL-1β secretion. BTK thus emerges as an important regulation hub for the activation of the NLRP3 inflammasome at multiple levels, which could be simultaneously targeted via BTK kinase inhibitors.
Results
BTK deficiency coincides with reduced NLRP3 tyrosine phosphorylation in vitro and in vivo
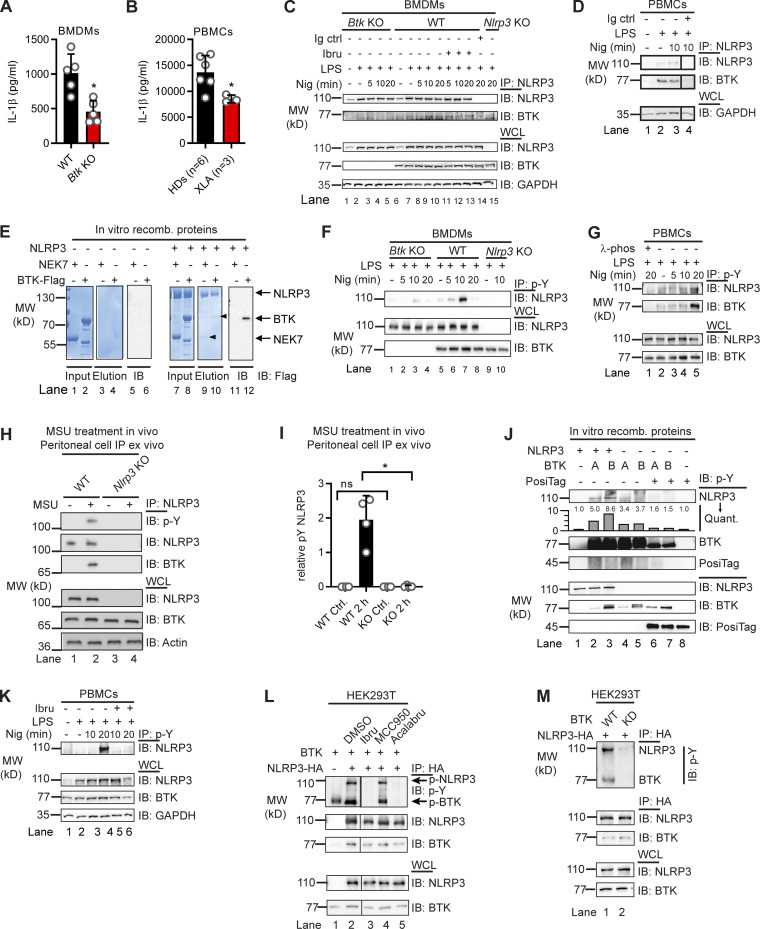
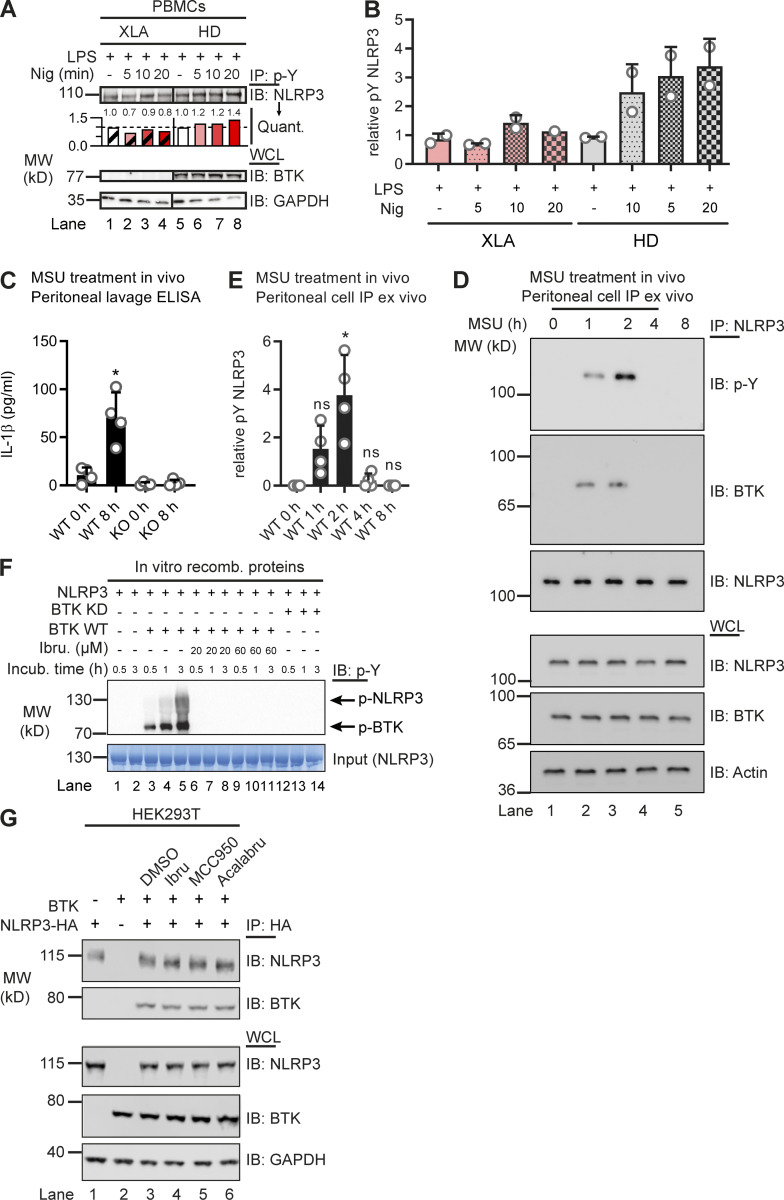
Based on previous work (Liu et al., 2017), we hypothesized that BTK and NLRP3 may engage in a direct kinase–substrate relationship, whose elucidation might unravel novel molecular aspects of NLRP3 inflammasome regulation. As it is the best studied NLRP3 agonist, we primarily used the pore-forming K+-dependent stimulant, nigericin. We sought to test this in Btk-deficient primary murine bone marrow–derived macrophages (BMDMs) and in peripheral blood mononuclear cells (PBMCs) from patients with the genetic BTK deficiency X-linked agammaglobulinemia (XLA). As expected, IL-1β release upon nigericin stimulation was significantly reduced in Btk-deficient BMDMs and patient-derived PBMCs, respectively (Fig. 1, A and B). Interestingly, in BMDMs, endogenous NLRP3 precipitated and interacted with endogenous BTK in WT but not Btk or Nlrp3 KO BMDMs (Fig. 1 C), irrespective of the BTK kinase inhibitor ibrutinib. Similarly, BTK coimmunoprecipitated with NLRP3 in PBMCs from healthy donors (HDs; Fig. 1 D). Thus, in both human and murine primary immune cells, BTK and NLRP3 interact independently of nigericin stimulation. This was also confirmed with a cell-free in vitro pulldown of recombinant purified NLRP3, which precipitated BTK and NEK7 proteins (Fig. 1 E). Being a late NLRP3 interactor (Magupalli et al., 2020), NEK7 was considered of less importance for the early activation events of NLRP3 that were in focus here, and thus only served as a positive binding control for recombinant NLRP3 (Sharif et al., 2019; also see Materials and methods). We next tested whether BTK is able to phosphorylate NLRP3 upon nigericin treatment. In murine BMDMs (Fig. 1 F), immunoprecipitated NLRP3 became rapidly phospho-tyrosine (p-Y) positive in cells expressing Btk but not in Btk or Nlrp3 KO cells. Similarly, NLRP3 phosphorylation was also observed in HD PBMCs (Fig. 1 G), and lower in XLA patients’ PBMCs (Fig. S1, A and B). Treatment with λ-phosphatase abolished p-Y reactivity, further confirming the phospho-antibody specificity. Notably, endogenous NLRP3–BTK interaction and NLRP3 time-dependent tyrosine phosphorylation were also observed upon stimulation with monosodium urate (MSU) crystals, another NLRP3 agonist, in vivo. In brief, when MSU was injected intraperitoneally and lavages analyzed at different time points for IL-1β release (cell-free supernatant, control for stimulation conditions) and NLRP3 immunoprecipitation (lavage cells), an interaction of endogenous NLRP3 with BTK and NLRP3 tyrosine phosphorylation were observed, and these were specific for lavages from WT but not Nlrp3 KO mice, starting at 1 h and peaking at 2 h after injection (Fig. 1, H and I; and Fig. S1, C–E). Thus, BTK and NLRP3 interact endogenously in primary immune cells, in vitro and in vivo, and BTK promotes NLRP3 tyrosine phosphorylation upon NLRP3 stimulation.
Figure 1.
NLRP3 directly interacts with and is tyrosine phosphorylated by BTK. (A and B) IL-1β release (triplicate ELISA) from WT versus Btk KO BMDMs (A; n = 5 each) or XLA versus HD PBMCs (B; n = 3 or 6 per group). (C and D) Coimmunoprecipitation (Co-IP) of NLRP3 from WT, Btk KO, or Nlrp3 KO BMDM (C; n = 3) or ibrutinib-treated PBMC lysates (D; n = 2). Black lines indicate that intervening lanes have been spliced out. (E) In vitro pulldown of Flag-tagged BTK or His-SUMO–tagged NEK7 by MBP-tagged NLRP3 (n = 3). Arrowheads denote copurified NEK7 and BTK in the eluates. The BTK band was further confirmed by Flag-IP. (F) Co-IP from WT, Btk KO, or Nlrp3 KO BMDMs using anti–p-Y antibodies (n = 3). (G) Co-IP from PBMCs using anti–p-Y (n = 5). λ-Phosphatase was added as a dephosphorylation control where indicated (n = 2). (H and I) Co-IP from peritoneal lavage cells harvested 2 h after MSU i.p. injection (n = 4; quantified in I relative to background in lane 1). (J) In vitro kinase assay using two different commercial suppliers, A and B, of recombinant BTK. Posi-Tag = specificity control. Quantification relative to background in lane 1 (n = 2). (K) Co-IP from PBMCs using anti–p-Y antibodies with prior ibrutinib pretreatment (n = 3). (L and M) IPs from HEK293T cells transfected with NLRP3 and BTK WT or KD constructs or treated with inhibitors (n = 2 each). A, B, and I represent combined data (mean + SD) from n biological replicates (each dot represents one mouse or patient/HD). C–H and J–M are representative of n biological (HD or mouse) or technical replicates. *, P < 0.05 using Student’s t test (A), one-way ANOVA with Dunnett’s correction (B), or Mann–Whitney U test (I). Acalabru, acalabrutinib; ctrl, control; IB, immunoblot; Ibru, ibrutinib; Nig, nigericin; Quant., quantification.
Figure S1.
BTK-dependence of NLRP3 tyrosine phosphorylation. (A and B) Coimmunoprecipitation (Co-IP) of NLRP3 from HD or XLA patient PBMC lysates (n = 2 each), quantified in A relative to lane 1 or 5, respectively, and in B, relative to GAPDH in the respective LPS-only control across experiments. Black lines indicate that intervening lanes have been spliced out. (C–E) In vivo MSU peritonitis model (n = 4 in each group). (C) IL-1β release by triplicate ELISA from peritoneal lavage supernatant 0 or 8 h after MSU-treatment in vivo. (D) Analysis of representative peritoneal lavage cell lysates at different times after MSU-treatment in vivo. (E) Quantification of p-Y–NLRP3 immunoblot (IB) relative to background in NLRP3 IPs from peritoneal lavage cells (n = 3). (F) pNLRP3 occurrence in the in vitro kinase assay with BTK or KD BTK upon incubation with ATP for the indicated time periods, with and without ibrutinib (n = 3). (G) HEK293T cells were transfected with the indicated NLRP3 and BTK WT or mutant constructs and treated with inhibitors, and lysates were subjected to HA-IP and IB (n = 3 each). In A, D, F, and G, one representative example of n biological replicates is shown. B, C, and E represent combined data (mean + SD) from n biological replicates (each dot represents one donor or mouse). *, P < 0.05 according to one-way ANOVA with Šidák (C) or Dunnett’s (E) correction. Acalabru, acalabrutinib; Ibru, ibrutinib; Incub., incubation; Nig, nigericin; recomb., recombinant.
BTK kinase activity is required for NLRP3 tyrosine phosphorylation
We next tested whether BTK kinase activity was required for NLRP3 tyrosine phosphorylation. Two independent cell-free in vitro setups showed that the presence of BTK was necessary and sufficient for NLRP3 p-Y modification (Fig. 1 J and Fig. S1 F). Furthermore, in both PBMCs and the in vitro setup, NLRP3 tyrosine phosphorylation was blocked by ibrutinib treatment (Fig. 1 K and Fig. S1 F) and, thus, was dependent on BTK kinase activity. Next, human embryonic kidney (HEK) 293T cells were transfected with NLRP3 and BTK and treated with or without BTK inhibitors. In this cellular system, NLRP3 tyrosine phosphorylation was abrogated in the presence of both BTK kinase inhibitors (Fig. 1 L), consistent with results in primary BMDMs (Fig. 1 C). In contrast, the NLRP3-specific inhibitor MCC950 (Coll et al., 2015; Tapia-Abellán et al., 2019) failed to prevent BTK-specific interaction and NLRP3 p-Y modification (Fig. 1 L). Interestingly, the presence of NLRP3 seemed to promote BTK phosphorylation, an indicator of activity (Duarte et al., 2020). Notably, the expression of kinase-dead (KD) BTK (K430E mutation; see Weber et al., 2017) was not able to induce NLRP3 tyrosine phosphorylation, despite an intact interaction (Fig. 1 M), ruling out other kinases in this system. Similar results were obtained for the in vitro cell-free setup (Fig. S1 F). Thus, BTK kinase activity appears essential and sufficient for NLRP3 p-Y modification using primary immune cells, the HEK293T system, or purified recombinant proteins, indicative of a direct kinase–substrate relationship.
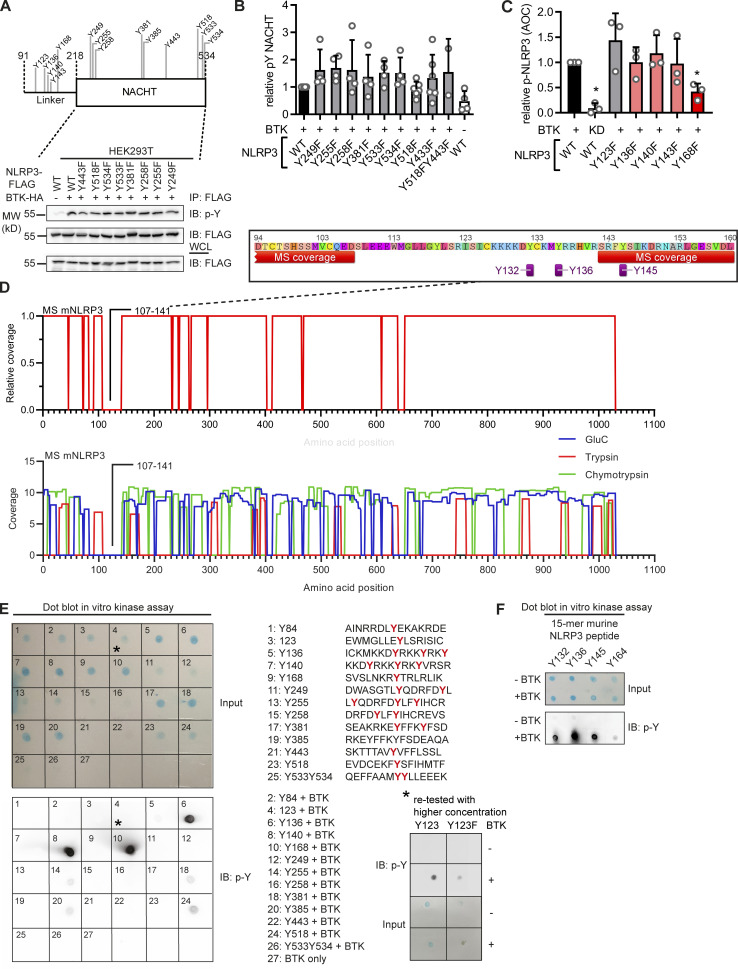
BTK phosphorylates four conserved tyrosine residues in the NLRP3 PYD-NACHT linker domain
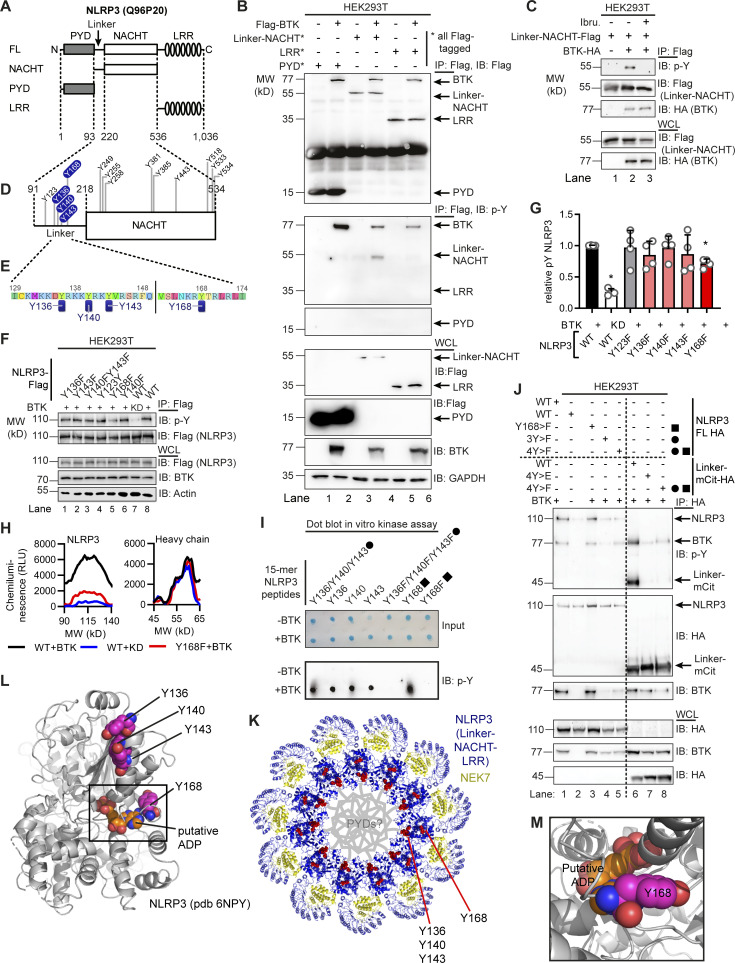
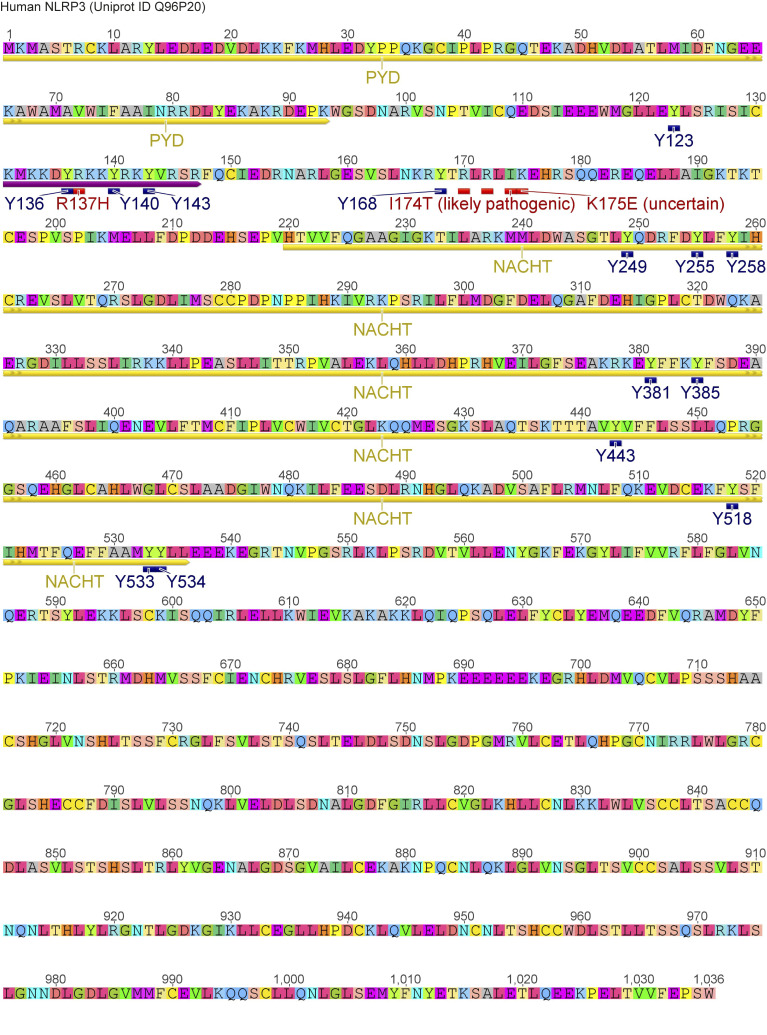
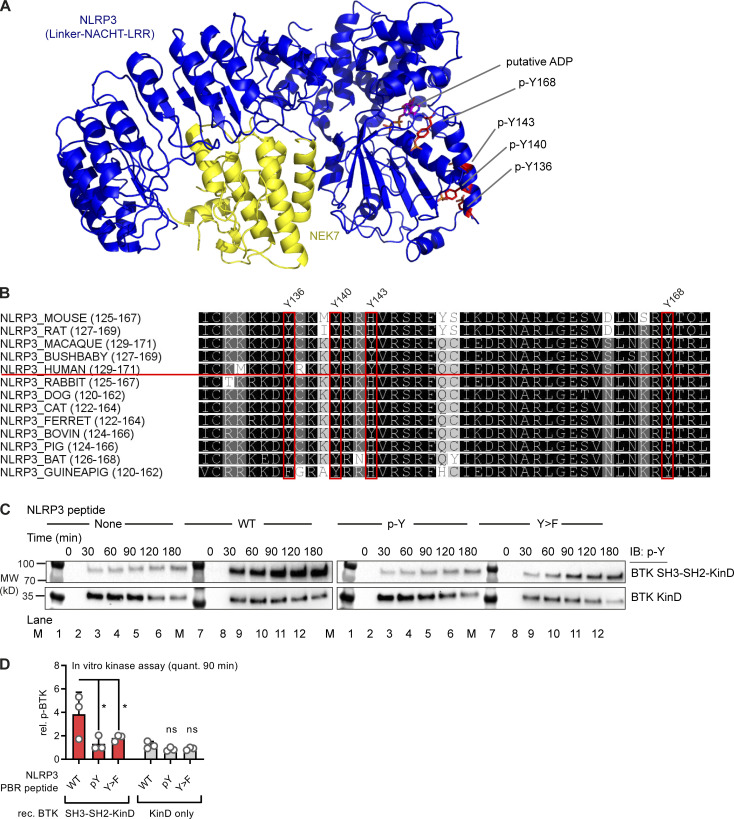
To map the modified tyrosine residues in NLPR3, we compared Flag-tagged full-length with truncated NLRP3 constructs (Mayor et al., 2007) of only the PYD (1–93); the extended NACHT domain (94–534), which includes an N-terminal linker domain (94–219; Sharif et al., 2019); and the leucine-rich repeat (LRR) domain (535–1,036; see Fig. 2 A and Fig. S2). BTK exclusively phosphorylated the extended NACHT construct (Fig. 2, B and C), ruling out Y861 (first identified by Spalinger et al., 2016) as the phosphosite. Individual mutation of the nine tyrosines in the core NACHT domain (220–534; see Fig. 2 D) to phenylalanine (F) did not impact the level of phospho-NLRP3 detected upon BTK coexpression (Fig. S3, A and B). However, when the linker (94–219) tyrosines were targeted (Fig. 2 E), mutated Y168 showed partial, but significant reduction of the p-Y signal, as shown by conventional immunoblotting (Fig. 2, F and G) and automated capillary electrophoresis analysis (Fig. 2 H and Fig. S3 C), respectively. Thus, Y168 emerged as a novel putative p-Y site in NLRP3 specifically modified by BTK. Unfortunately, the linker region is not accessible to mass spectrometric analysis (data acquired from Stutz et al., 2013 and replotted in Fig. S3 D). Therefore, to assess the phosphorylation of Y168 by alternative means, 15-mer peptides covering all linker tyrosines (Fig. 2 E and Table S1) were incubated with His-tagged BTK to assess peptide phosphorylation in a cell-free system. Following BTK removal by anti-His beads, tyrosine phosphorylation of the peptides was visualized by dot blot analysis. The majority of Y-containing peptides and all F-containing corresponding peptides showed little or no phosphorylation (e.g., a weakly phosphorylated Y123; Fig. S3 E). However, the Y168-containing peptide (black square in Fig. 2 I) showed strong tyrosine phosphorylation (Fig. 2 I and Fig. S3 E). Of note, peptides containing Y136, Y140, or Y143—either in combination (black circle) or as single tyrosines—were also phosphorylated (Fig. 2 I), similar to peptides containing the corresponding sequences in mouse NLRP3 (Y132, Y136, Y145, and Y164; see Fig. S3 F). That the linker region contained altogether at least four BTK-modified tyrosines explained the only partial effect of single Y168 mutation observed above (Fig. 2, F–H). In line with this, the p-Y NLRP3 signal in HEK293T cells was reduced slightly in expression constructs containing an individual Y168 mutation (black square) but drastically so upon simultaneous mutation of Y136, Y140, and Y143 to phenylalanine (“3Y>F,” black circle) or the combination of Y136, Y140, and Y143 with Y168 mutation (“4Y>F,” black square + circle) both in an FL-NLRP3 construct and when only the linker sequence was fused to an mCitrine YFP–hemagglutinin (HA) sequence (here termed “hLinker-Cit-HA”; Fig. 2 J). BTK thus appears to be able to specifically modify not only one but at least four tyrosines—Y136, Y140, Y143, and Y168—in the PYD-NACHT linker of human (and murine) NLRP3 in vitro. To gain structural insights, we mapped the sites in a recent cryo-EM structure of an NLRP3–NEK7 complex (Fig. 2 K and Fig. S4 A; Sharif et al., 2019). Interestingly, Y136, Y140, and Y143 were found in a helical region proposed to contact the PYD for ASC recruitment (Sharif et al., 2019). Furthermore, Y168, which maps to the vicinity of several likely pathogenic cryopyrin-associated periodic fever syndrome mutations (Fig. S2), was adjacent to the putative ADP binding site and might thus influence nucleotide binding (Fig. 2, L and M; and Fig. S4 A). Interestingly, all the BTK-modified Y residues were strongly conserved in other NLRP3 sequences, further highlighting their potential functional relevance (Fig. S4 B). BTK and many other SH3–SH2-containing kinases are thought to select substrate proteins by binding via their kinase domain (KinD) and subsequent phosphorylation. The phosphorylated substrate may, at the same time, allosterically activate kinase activity by binding the SH3–SH2 module (Duarte et al., 2020). We therefore sought to test whether the conserved NLRP3 linker region may bind and affect BTK kinase activity and thereby explore the reason for unequal capacities of the NLRP3 mutants to immunoprecipitate BTK (Fig. 2 J) and enhance BTK Y-phosphorylation in the presence of NLRP3 (Fig. 1 L). BTK activation (as evidenced by BTK Y-phosphorylation) was assessed upon coincubation of NLRP3-derived PBR Y-containing substrate peptides first with a purified truncated BTK protein consisting only of the SH3–SH2 and the KinD (SH3–SH2–BTK construct). In line with an activity-promoting effect of NLRP3 on BTK, WT NLRP3 peptides increased BTK phosphorylation over time (Fig. S4 C, top panels; quantified in D). An F-containing peptide induced much lower BTK phosphorylation, as allosteric activation by binding the SH2 domain is not possible. p-Y–containing peptides failed to induce BTK activity probably because they compete with BTK autophosphorylation (Duarte et al., 2020), which is in line with the evidence that none of the peptides had a direct effect on BTK activity in the context of a purified KinD-only truncated BTK protein (Fig. S4, C and D). This indicates that processive binding and phosphorylation of the NLRP3 linker substrate promote BTK activity via BTK’s SH3–SH2 module. Collectively, our results suggest that BTK directly modifies multiple highly conserved tyrosines within a functionally important domain in NLRP3, which could impact downstream steps in the assembly of the oligomeric inflammasome complexes.
Figure 2.
BTK phosphorylates the PYD-NACHT linker. (A) NLRP3 domains (UniProt ID Q96P20). (B) Flag immunoprecipitation (Flag-IP) from HEK293T cells transfected with Flag-tagged NLRP3 truncation and/or Flag–BTK constructs (n = 3). (C) As in B but including ibrutinib (n = 3). (D) Positions of targeted tyrosine residues. (E) Linker region including polybasic motif. (F) As in B but using NLRP3 Y>F point mutants or WT NLRP3, with WT or KD BTK plasmids (n = 4). (G) Quantification of F (n = 4). (H) WES capillary electrophoresis of NLRP3 Flag-IP from HEK293T cells transfected with WT or mutant Flag-NLRP3 and WT or KD BTK (n = 3). (I) Dot blot of BTK assay with 15-mer NLRP3-derived WT or mutant synthetic peptides (n = 3). After BTK removal using anti-His beads, peptide mixtures were directly spotted and stained with a total peptide stain (input) or anti–p-Y. A circle denotes peptides containing the three PBR tyrosines, and a square denotes Y168-containing peptides. (J) As in F but also NLRP3 linker (WT or Y mutated) fused to mCitrine (mCit)-HA (n = 3). (K) Tyrosines (red) highlighted in the model of NLRP3 (blue)–NEK7 (yellow) complex (PDB: 6NPY). (L and M) Close-up view on dimer interface (L) and putative nucleotide binding site (M). G represents combined data (mean + SD) from n biological replicates (each dot represents one replicate). B, C, F, and H–J are representative of n technical replicates. *, P < 0.05 according to one-sample t test (G). IB, immunoblot; Ibru., ibrutinib; RLU, relative light unit.
Figure S2.
Position of tyrosine residues and disease-associated mutations in the linker and/or NACHT domain of human NLRP3. Annotation of NLRP3 sequence (UniProt accession no. Q96P20).
Figure S3.
Positional mapping of BTK-modified tyrosine residues in NLRP3. (A) Position of all mutated tyrosine residues in linker–NACHT construct and phosphorylation analysis of core-NACHT tyrosine NLRP3 mutants. HEK293T cells were transfected with the indicated NLRP3 mutant constructs and a BTK WT construct as indicated, and lysates were subjected to HA-immunoprecipitation (IP) and immunoblot (IB) as indicated (n = 4 each). (B) Quantification of A combined from n = 4 experiments as the ratio of p-Y–NLRP3 to total NLRP3 in the IP fraction normalized to transfection with WT NLRP3. (C) Quantification of WES capillary electrophoresis of NLRP3 p-Y IPs from HEK293T cells (Fig. 2 H) from n = 3 experiments. (D) Data from MS analysis of purified murine NLRP3 (mNLRP3), digested with different proteases, showing combined (top) and separate coverage (bottom) information extracted from Stutz et al. (2013) and replotted here. (E) Dot blot of in vitro kinase assay of His-BTK and 15-mer synthetic peptides derived from human NLRP3, containing the indicated tyrosines stained with a total protein stain (top grid; input) or anti–p-Y antibodies (bottom grid; n = 3). (F) As in E but with peptides derived from murine NLRP3 ((n = 3). B and C represent combined data (mean + SD) from n biological replicates. In E and F, one representative example of n biological replicates is shown. *, P < 0.05 according to one-sample t test (B and C). AOC, antioxidant capacity.
Figure S4.
Structural positioning, sequence conservation of BTK-modified tyrosine residues in NLRP3, and effect of NLRP3 on BTK kinase activity. (A and B) Structural aspects and conservation of BTK-modified tyrosine residues in NLRP3. (A) NLRP3 model 6NPY showing NLRP3 linker-NACHT-LRR (blue) and NEK7 C-terminal lobe (yellow). A putative bound ADP molecule and selected tyrosines are highlighted. (B) ClustalW multiple sequence alignments of NLRP3 sequences from other species. Coloring according to similarity (black, conserved). BTK-modified tyrosines are highlighted (residue numbering according to human NLRP3). (C and D) Effect of NLRP3 peptides (WT Y sequence, p-Y sequence, or Y>F) on phosphorylation (assessed by p-Y blot) of the SH3-SH2-KinD or KinD-truncated BTK proteins purified from insect cells. BTK phosphorylation as investigated by immunoblot (IB) is indicative of BTK activation (n = 3). C shows one representative example of n technical replicates, combined in D. *, P < 0.05 according to two-way ANOVA (D). M, MW ladder; quant., quantification; rec., recombinant; rel. relative.
BTK-mediated phosphorylation sites affect linker charge and PIP binding
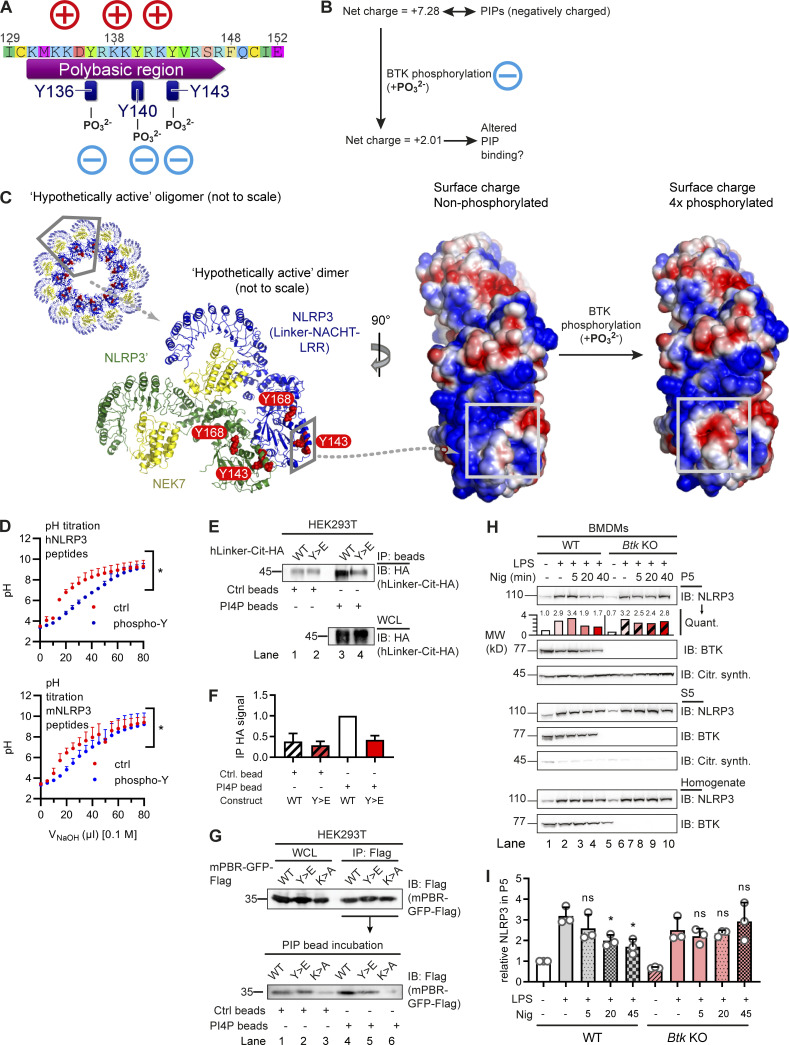
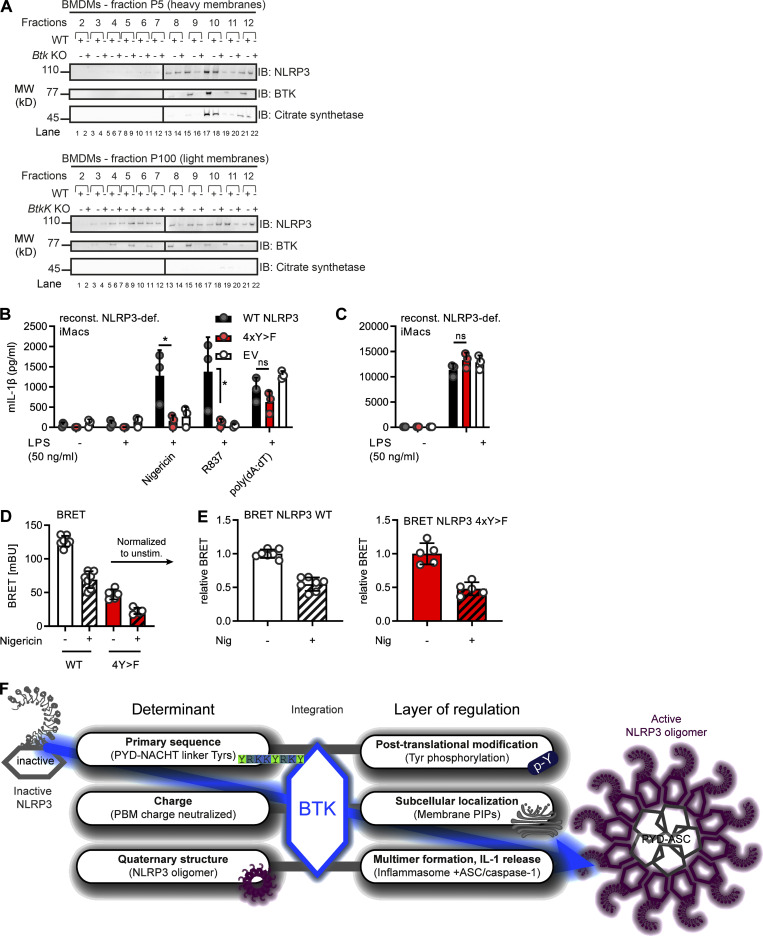
Chen and Chen (2018) identified a PBR (including K127–130 in mouse NLRP3 and K127 and K129–130 in human NLRP3) as critical for NLRP3 charge-dependent binding to organelle PIPs and inflammasome nucleation. Unexpectedly, Y136, Y140, and Y143 precisely mapped to this PBR in the NLRP3 PYD-NACHT linker and interdigitated with the basic residues mediating the proposed NLRP3 interaction with negatively charged membrane phospholipids (Fig. 3 A). The mutation of three positively charged residues (K127, K129, and K130) in the mNLRP3 PBR to alanine (K>A) was shown to abrogate binding (Chen and Chen, 2018) to PI4P, a phospholipid found at the trans-Golgi network and secretory granules (Vicinanza et al., 2008). We therefore hypothesized that BTK phosphorylation of Y136, Y140, and Y143 might generally alter the proposed charge attraction of NLRP3 with membrane phospholipids. Charge computations suggested that at cytoplasmic pH 7.4, the net charge of the isolated PBR sequence of NLRP3 shifts from +7.28 (unphosphorylated) to +2.01 when Y136, Y140, and Y143 are phosphorylated in human NLRP3 (Fig. 3 B) and from +8.33 (unphosphorylated) to +3.06 (3× phosphorylated) in mouse NLRP3. Likewise, in the NLRP3–NEK7 cryo-EM structure, the 3× phospho-modification causes a significant change in the computed surface charge of a “hypothetically active” NLRP3 (Sharif et al., 2019) toward less-positive values (Fig. 3 C). To confirm this charge difference experimentally, synthetic phospho and non-phospho versions of the Y136/Y140/Y143-containing peptides from both human and mouse NLRP3 were studied in pH titrations, demonstrating the p-Y peptide to be significantly less positively charged (Fig. 3 D). We therefore hypothesized that tyrosine phosphorylation by BTK might alter interactions of NLRP3 with PIPs, possibly shifting the binding from monophosphorylated (PI3P, PI4P) toward the more negatively charged diphosphorylated PIPs (PI(3,4)P2, PI(4,5)P2). Taking the same approach as Chen and Chen (2018), the binding of 4xY>E, i.e., phosphomimetic, mutant human NLPR3 linker-Cit-HA fusion protein, to beads coated with a singly charged phospholipid, PI4P, was reduced compared with the corresponding WT fusion protein construct (Fig. 3, E and F). This was further confirmed by the fusion of the murine NLRP3 PBR to GFP-Flag (mPBR-GFP-Flag, as in Chen and Chen, 2018). Despite equal expression, the Y>E construct bound PI4P beads less strongly than WT (Fig. 3 G). Reduced binding was also observed for the corresponding K>A mutant, which was known to be defective in PI4P binding based on charge neutralization (Chen and Chen, 2018). In line with these data, our results indicate that the BTK-modified Y positions could affect NLRP3–PIP interactions. It was beyond the scope of the present study to explore the binding to other PIPs shown to bind NLRP3, namely PI3P, PI3, 5P2, PI4, 5P2, and phosphatidic acid (Chen and Chen, 2018), which differ in charge density, abundance, and distribution across different organelles (Vicinanza et al., 2008). Nevertheless, it seemed important to know whether BTK activity might represent a cue to shift the subcellular localization of NLRP3. To test this by another technique and in primary immune cells, we stimulated WT and Btk KO primary BMDMs with lipopolysaccharide (LPS) and nigericin and fractionated their lysates into P5 (heavy membrane, intact Golgi, and mitochondria) and P100 (light membrane, dispersed Golgi, ER, and polysomes) fractions and probed them for NLRP3 (Fig. S5 A). BTK, whose pleckstrin homology domain also binds PIPs (references in Li et al., 1997), was also analyzed and found to localize similarly to NLRP3 (Fig. S5 A), in keeping with earlier interaction analyses (Fig. 1, C and F). In both WT and Btk KO BMDMs, NLRP3 was detectable in the P5 fraction upon LPS treatment and before nigericin stimulation, indicating that BTK is not essential for initial NLRP3 localization toward P5 membranes (Fig. 3, H and I). However, nigericin stimulation in WT BMDMs coincided with progressive and significant depletion of NLRP3 from the P5 fraction 20–45 min after treatment, consistent with the concepts of Golgi fragmentation and NLRP3 relocalization (Chen and Chen, 2018; Magupalli et al., 2020; Zhang et al., 2017), and the timing of phosphorylation in these cells (Fig. 1 F). Conversely, Btk KO BMDMs did not show significant P5 depletion within this time frame (Fig. 3, H and I), suggesting that the NLRP3 interaction with Golgi membranes may remain more stable in the absence of BTK. Collectively, these experiments show that both the BTK-modified Y positions in the NLRP3 PBR and the presence of BTK may have an impact on NLRP3 PBR charge, PIP binding, and subcellular fractionation of NLRP3.
Figure 3.
BTK phosphorylation of the NLRP3 polybasic motif enables Golgi/PI4P dissociation. (A and B) Charge distribution (A) and ProtPi net charge computation (B) of unmodified and 3× phosphopeptide human NLRP3 PBR. (C) CHARMM surface charge predictions of linker–NACHT–LRR structure in the putative nonphosphorylated (left) and 4× phosphorylated (right) form. Blue, positive charge; red, negative charge. Gray boxes indicate that the area of charge alterations in the monomers maps to a contact area in the hypothetical dimer (center, rotated by 90°; see relative position in oligomer below). (D) pH titration of peptides encompassing the polybasic motifs of human or murine NLRP3 as phospho (blue) or non-phosphorylated control (ctrl; red) peptide (n = 3). (E and F) Human NLRP3 linker-Cit-HA constructs precipitated with PI4P beads (n = 2; quantified in F relative to immunoprecipitation [IP] HA signal in WT transfection [lane 1]). (G) As in E but murine NLRP3 PBR fused to GFP-Flag (mPBR-GFP-Flag; n = 2). (H and I) Subcellular fractionation of nigericin-treated WT or Btk KO BMDM lysates into P5 (heavy membranes) and S5 (light membranes and cytosol; n = 3; quantified relative to untreated [lane 1] in the experiment shown in H or across experiments in I). D, F, and I represent combined data (mean + SD) from n biological replicates. In E, G, and H, one representative example of n technical replicates is shown. *, P < 0.05 according to one-way ANOVA with Šidák correction (I; relative to respective LPS only) or two-way ANOVA (D). Citr. synth., citrate synthetase; Ctrl, control; IB, immunoblot; Nig, nigericin; Quant., quantification.
Figure S5.
Subcellular fractionation of NLRP3 and BTK, effect of tyrosine mutation on NLRP3 activity and conformation and graphical abstract. (A) Sucrose gradient fractionation of WT and Btk KO BMDM lysates upon LPS + nigericin treatment for 5 min. Fractions were analyzed by SDS-PAGE and immunoblot (IB) as indicated (n = 1; pilot fractionation experiment). (B and C) Low concentration (50 ng/ml) LPS priming, nigericin stimulation, and subsequent IL-1β (B) or TNF (C) release from WT or 4xY>F human NLRP3-expressing reconstituted Nlrp3 KO iMacs, measured by ELISA (n = 3). (D and E) Response of NLRP3 BRET sensors to nigericin in HEK293T cells treated 24 h after transfection with 10 μM nigericin for 30 min or left mock treated before subsequent BRET measurement. BRET signals (D) were also normalized to the mean obtained for each construct mock treated (E; both n = 6–8). (F) Graphical summary illustrating multiple roles of BTK as NLRP3 regulator. B–E represent combined data (mean + SD) from n biological replicates. In A, one representative example of n biological replicates is shown. *, P < 0.05 according to one-way ANOVA with (B) or without (C) Šidák correction. def., deficient; EV, empty vector; Nig, nigericin; PBM, polybasic motif; reconst., reconstituted; unstim., unstimulated.
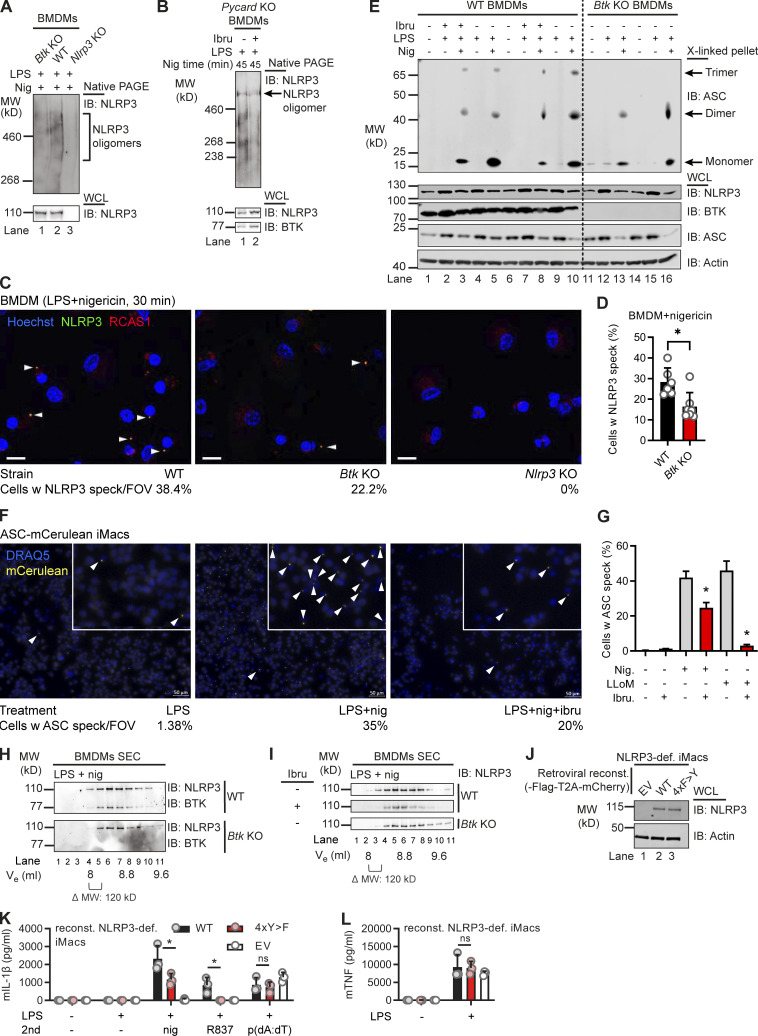
BTK kinase activity affects NLRP3 oligomerization, ASC interaction, and IL-1β release
A recent study proposed that NLRP3 release from the mitochondria-associated ER membranes was required for ASC engagement and full inflammasome assembly (Zhang et al., 2017). As depletion of NLRP3 from the Golgi was lower in the absence of BTK (Fig. 3, H and I), we explored whether BTK kinase activity also affects subsequent full inflammasome assembly, e.g., at the level of NLRP3 oligomerization, and ASC recruitment into the complex. Indeed, native PAGE of nigericin-stimulated WT and Btk KO BMDM lysates (Fig. 4 A) or BTK inhibitor- versus vehicle-treated Pycard (ASC) KO (Fig. 4 B) showed reduced NLRP3 oligomers (typically detectable as nondiscrete “smears”; Green et al., 2018) in Btk-deficient or Btk-inhibited samples. Confocal microscopy of LPS- and nigericin-stimulated primary BMDM showed a lower fraction of cells forming NLRP3 specks costaining positively for the Golgi marker RCAS1 in Btk KO BMDM versus WT BMDM (Fig. 4, C and D). NLRP3 was not detectable in its dispersed state before nigericin addition in all analyzed BMDMs independent of genotype, but nigericin-stimulated Nlrp3 KO showed no staining/specks, confirming the specificity of the staining. The recruitment of ASC into the complex was also assessed in primary BMDM by analyzing ASC cross-linking in whole-cell lysate (WCL) pellets. Evidently, compared with WT BMDM cross-linked pellets, in Btk KO pellets, higher-order oligomers were absent, and in WT BMDM, ibrutinib pretreatment reduced the intensity of recovered ASC staining (Fig. 4 E). Consistent with earlier results, BTK inhibition also reduced speck formation in nigericin or the additional NLRP3 agonist LeuLeu-OMe ASC-mCerulean–expressing immortalized murine macrophages (iMacs; Fig. 4, F and G). Moreover, size-exclusion chromatography of untreated WT cell lysates showed that BTK and NLRP3 coeluted in the high-molecular-weight (MW) fraction (>1,100 kD; Fig. 4 H). Consistent with native PAGE in Btk KO or WT lysates from ibrutinib-treated cells, elution shifted to lower-MW complexes (Fig. 4, H and I). Ablation of BTK activity thus appeared to reduce the subsequent ability of NLRP3 to oligomerize into high MW inflammasomes and to assemble with ASC (Magupalli et al., 2020). To show that BTK-modified Y residues—and the effects of BTK described so far—also had an impact on IL-1β release, Nlrp3-deficient iMacs were retrovirally transduced with WT or tyrosine-mutated (4xY>F) human NLRP3–T2A–mCherry constructs, allowing for cell sorting for equal protein expression (Fig. 4 J). Compared with WT-transduced cells, cells with the 4xY>F construct failed to restore nigericin- and R837 (imiquimod)-dependent IL-1β release (Fig. 4 K). Conversely, TNF release, which is NLRP3 independent (Liu et al., 2017), was comparable between cell lines (Fig. 4 L). To test specificity for the NLRP3 inflammasome and confirm equal capacities of these different cell lines for IL-1β release via another NLRP3- and BTK-independent pathway (Liu et al., 2017), the AIM2 inflammasome, we also stimulated the cells with poly(dA:dT). The resulting IL-1β release was assessed and found to be comparable between WT and mutant cells (Fig. 4, K and L). Of note, the effect of NLRP3 Y mutation was independent of LPS concentration (Fig. S5, B and C; Mao et al., 2020) as 4xY>F cells showed consistently lower IL-1β release when primed with either 50 or 200 ng/ml LPS. To check the folding integrity of the 4xY>F mutant, bioluminescence resonance energy transfer (BRET) was performed (Tapia-Abellán et al., 2019) in HEK293T cells. Compared with WT NLRP3, the 4xY>F mutant NLRP3 showed lower absolute BRET signals (Fig. S5 D) in line with a functional impairment, but similar relative conformational changes to nigericin (Fig. S5 E), indicating that folding was intact. The data show that the BTK-modified Y positions identified here play an important role for full NLRP3 inflammasome activity and subsequent IL-1β release.
Figure 4.
BTK modification affects NLRP3 oligomerization and IL-1β release. (A and B) WT, Btk KO, Nlrp3 KO, or Pycard (ASC) KO BMDMs stimulated (45 min nigericin) and respective lysates analyzed directly by native PAGE (A, n = 2; B, n = 4). (C) Representative fluorescence microscopy images of NLRP3 specks in WT, Btk KO, and Nlrp3 KO primed with LPS, stimulated (30 min nigericin), and stained as indicated (n = 2). Blue, nuclei (Hoechst); green, NLRP3; red, Golgi (RCAS1). Scale bar = 10 μm; arrowheads mark NLRP3 specks. (D) Quantification from multiple 3 × 3 tiles per strain per experiment. (E) As in A or B but ASC in WCLs was cross-linked upon stimulation, with or without ibrutinib pretreatment (n = 4). (F) Representative fluorescence microscopy images of ASC specks in Nlrp3 KO iMacs reconstituted with NLRP3-Flag ASC-mCerulean and stimulated as indicated. Blue, nuclei (DRAQ5); yellow, ASC (mCerulean). Scale bar = 50 μm; arrowheads mark only one ASC speck per overview image for the sake of clarity, but in the insets, all specks are marked. (G) Quantification from multiple images per treatment per experiment (n = 2). (H) As in A but lysates were applied to size-exclusion chromatography (SEC) before fractions were analyzed (n = 3). (I) As in H comparing inhibitor-treated WT BMDM or Btk KO BMDM lysates (n = 3). (J–L) NLRP3 expression levels determined by immunoblot (IB), IL-1β, or TNF release quantified by triplicate ELISA in/from WT or 4xY>F NLRP3-reconstituted NLRP3-deficient iMacs (n = 3). D, G, K, and L represent combined data (mean + SD) from n technical replicates. A–C, E, F, H, and I are representative of n biological (mice) or technical replicates. *, P < 0.05 according to Mann–Whitney U test (D), Student’s t test (G), and one-way ANOVA with (K) or without (L) Šidák correction. def., deficient; EV, empty vector; FOV, field of view; Ibru, ibrutinib; Nig, nigericin; reconst., reconstituted; Ve, elution volume; w, with.
Discussion
The mechanism of activation of the NLRP3 inflammasome has been intensely studied for some time, and recent work has unraveled a critical role of the dynamic localization of NLRP3 to and from organelles in the regulation of its activity (Chen and Chen, 2018; Seoane et al., 2020; Zhang et al., 2017). By using both biochemical and cellular assays in vitro and in human and murine cell lines and primary cells, we found that BTK directly participated in these processes at the level of NLRP3, providing the following novel molecular insights: Direct phosphorylation of four conserved and functionally important tyrosine residues in the NLRP3 polybasic linker motif affected the charge of the PBR sequence, and phosphomimetic mutation altered NLRP3–PIP bead interactions, presumably by neutralization of the linker net surface charge. In line with this, in the presence of BTK—whose subcellular localization overlaps with NLRP3—NLRP3 retention at the Golgi appeared lower, whereas NLRP3 inflammasome oligomerization, ASC association, and IL-1β secretion were higher. Finally, mutation of modified NLRP3 tyrosines to the non-phosphorylatable phenylalanine abrogated full IL-1β release, highlighting the importance of these tyrosines. Our data suggest that BTK-mediated phosphorylation of multiple NLRP3 tyrosines may thus serve as a kind of molecular switch, tuning NLRP3 charge and, subsequently, localization and inflammasome assembly. Modification, localization, and oligomerization of NLRP3 have been recognized to be important. However, they were supposed to be hierarchically separate layers of NLRP3 inflammasome regulation and, hence, of inflammation. Our data indicate that some of these layers may be integrated and interconnected by a multifunctional BTK: By decoding the most basal determinants, such as protein sequence, BTK appears to integrate posttranslational modifications, surface charge, interaction with membranes, and ultimately assembly of a highly oligomeric molecular machinery (Fig. S5 F).
Such a dense and interconnected network of regulation would be in line with the critical need to tightly control excessive IL-1β release to prevent pathologies. As multiple organelles (Seoane et al., 2020; Weber et al., 2020b) and other regulatory proteins (e.g., PKD; Zhang et al., 2017) may also participate in this process, we cannot formally rule out effects of BTK at the level of ASC. Furthermore, the cues that activate BTK activity remain to be deciphered. As NLRP3 and BTK interact before nigericin addition (Fig. 1 D) and show the same localization (Fig. S5 A), we speculate that NLRP3 itself, e.g., upon undergoing functional changes in the wake of K+ efflux (Muñoz-Planillo et al., 2013), may render BTK active through protein–protein interactions. The observation that coexpression of NLRP3 with BTK (Fig. 1 J) and the presence of NLRP3 PBR peptides (Fig. S4, C and D) increase BTK phosphorylation as an indicator of kinase activity (Duarte et al., 2020) is thus in line with the fact that BTK kinase activity is highly dependent on three-dimensional conformation and protein–protein interactions (Duarte et al., 2020). If conformationally “active” NLRP3 (Sharif et al., 2019) itself instigated a positive feed-forward loop by switching BTK on at the Golgi, this would restrict BTK activity to already membrane-associated and oligomerized NLRP3, thereby promoting release of NLRP3 oligomers while not preventing unmodified NLRP3 to associate with PIPs. Furthermore, positioning of BTK at the same level as NLRP3 would be in line with the observation that IL-1β release is boosted by the presence of BTK but is not entirely dependent on it (Fig. 1, A and B; and Fig. 4 I; Ito et al., 2015; Liu et al., 2017). Studies by Chen and Chen (2018), Zhang et al. (2017), and Magupalli et al. (2020) illustrated the staggering complexity of NLRP3 trafficking and its regulation, but due to multiple assays, constructs, and cell types that have been used, a uniform “trafficking-activation mechanism” cannot be established yet. Resolving the remaining ambiguities was outside the scope of this study, but our observations suggest that concerted modification of charge would have an impact on NLRP3–PIP interactions and could be a mechanism for BTK to promote NLRP3 relocalization. Generally, our results highlight BTK as a positive NLRP3 regulator and are congruent with earlier (Ito et al., 2015; Liu et al., 2017) and current studies (O’Riordan et al., 2019, 2020; Purvis et al., 2020). A recent report by Mao et al. (2020) also proposed BTK to act as a positive regulator in murine and human immune cells at LPS priming concentrations of <100 ng/ml, which are physiological (Copeland et al., 2005; Zweigner et al., 2001). However, under nonphysiologically high TLR4 or TLR2 priming, which may blur the lines between priming and actual NLRP3 activation, the lack or inhibition of BTK increased NLRP3 inflammasome IL-1β output. Other results of Mao et al. have already been discussed elsewhere (Weber et al., 2017, 2020a, 2020b), and in our hands and at least in iMacs, LPS concentration in the range of 50–200 ng/ml did not influence the loss-of-function phenotype of NLRP3 phospho mutants (Fig. 4, I and J; and Fig. S5, B and C). Ours and their studies warrant further exploration of a possible “rheostat role” of BTK that may tune NLRP3 activation, depending on TLR signaling intensity. However, our data clearly indicate that BTK is primarily involved in licensing maximal IL-1β release via the regulatory events characterized here.
Due to the complexity of the system, we focused on nigericin as the most widely used NLRP3 agonist. We therefore cannot extrapolate the role of BTK in response to all the many known activators of NLRP3. However, individual datasets shown here or in our previous work (Liu et al., 2017) for the other NLRP3 agonists (ATP, LLoMe, leukocidin A/B, and MSU) indicate that BTK is likely to have similar roles in these pathways, especially since relocalization seems to be a unifying concept of these and even K+-independent NLRP3 stimuli (Chen and Chen, 2018). An additional question for further study is the relationship among NLRP3, BTK, and the additional regulator NEK7, although the precise role of NEK7 in NLRP3 activation remains enigmatic (Schmacke et al., 2019 Preprint).
Nevertheless, our work suggests that NLRP3 phosphorylation may represent an interesting novel biomarker, and possibly a therapeutic target, for early NLRP3 activation. NLRP3 inhibition via BTK blockade may be an unwanted side effect in the treatment of B cell malignancies for which BTK kinase inhibitors have been Food and Drug Administration approved and successfully used (Byrd et al., 2013; reviewed in Weber, 2021). On the other hand, recent investigations showed that macrophage BTK activation in SARS-CoV2–infected patient PBMCs and a beneficial effect of ibrutinib on COVID-19–related lung inflammation are probably attributable to blocking the NLRP3 inflammasome (Roschewski et al., 2020). Additionally, in an experimental in vivo model of sepsis-related heart dysfunction and metabolic inflammation, BTK inhibition directly ameliorated NLRP3-mediated inflammation and pathophysiology (O’Riordan et al., 2019, 2020; Purvis et al., 2020). Based on these studies, our work points to a molecular rationale for targeting strategies that may be applied to block excess IL-1β production in these and other acute inflammasome-related disease states (Weber, 2021).
Materials and methods
Reagents
Nigericin and LPS (from Escherichia coli, namely LPS-EK, catalog no. tlrl-peklps) were purchased from InvivoGen, ATP from Sigma, ibrutinib and acalabrutinib from Selleckchem, recombinant GM-CSF from PeproTech, and Ficoll from Merck Millipore. Coelenterazine h and Lipofectamine 2000 were from Life Technologies. The composition of the physiological (E-Total [ET]) buffer used in BRET experiments was 147 mM NaCl, 10 mM Hepes, 13 mM D-glucose, 2 mM KCl, 2 mM CaCl2, and 1 mM MgCl2 (pH 7.4). Peptides (synthesized in-house) and antibodies are listed in Table S1 and Table S2, respectively.
Peptides
Synthetic peptides were produced by standard 9-fluorenylmethyloxycarbonyl/tert-butyl strategy using peptide synthesizers P11 (Activotec) or Liberty Blue (CEM Corporation). Purity was assessed by reversed phase HPLC (e2695; Waters) and identity affirmed by nano-UHPLC (UltiMate 3000 RSLCnano) coupled online to a hybrid mass spectrometer (LTQ Orbitrap XL; both Thermo Fisher Scientific). Lyophilized peptides were purified by standard HPLC. For certain peptides, a pH titration with 0.1 M NaOH was performed using standard procedures. For in vitro assays, peptides were dissolved at 10 mg/ml in DMSO and diluted 1:10 in bidistilled H2O. Frozen aliquots were further diluted in cell culture medium and sterile filtered if necessary.
Plasmid constructs
ASC, NLRP3, and BTK coding sequences in pENTR clones were generated as described in Wang et al. (2015). BRET sensor constructs consisting of NLRP3 fused N terminally with YFP and C terminally with Renilla luciferase or NLRP3 C terminally fused only to Renilla luciferase as a control in all BRET assays were as described in Tapia-Abellán et al. (2019). Truncated Flag-tagged NLRP3 constructs were a gift of F. Martinon (University of Lausanne, Faculty of Biology and Medicine, Department of Biochemistry, Lausanne, Switzerland; Mayor et al., 2007). Constructs for the human PYD-NACHT linker (residues 94–219) fused to mCitrine-HA or the murine polybasic motif (residues 127–146) in the context of Flag-GFP (as in Chen and Chen, 2018) were synthesized by GENEWIZ. Point mutations in BTK and NLRP3 were subsequently introduced using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies) according to the manufacturer’s instructions. Presence of the desired mutation and absence of unwanted regions in the entire coding DNA sequence was confirmed by automated DNA sequencing. Maltose-binding protein (MBP)–fused NLRP3 dPYD (ΔPYD; deletion of the PYD domain) and NEK-7 constructs used for protein purification were described in Sharif et al. (2019). Full-length SH3–SH2–KinD and KinD constructs used for protein purification were described in Duarte et al. (2020).
Study subjects and blood sample acquisition
XLA patients were recruited at the Centre of Chronic Immunodeficiency, University Hospital Freiburg, and healthy blood donors at the Interfaculty Institute of Cell Biology, Department of Immunology, University of Tübingen. All patients and healthy blood donors included in this study provided their written informed consent before participation. Approval for use of their biomaterials was obtained by the respective local ethics committees in accordance with the principles laid down in the Declaration of Helsinki as well as applicable laws and regulations. XLA patients were clinically identified and genetically characterized as described in Liu et al. (2017).
Mice
Btk KO (originally generated by Khan et al., 1995), NLRP3 KO (stock no. 021302; The Jackson Laboratory), and WT mice, all on a C57BL/6J (The Jackson Laboratory) background, were maintained locally in specific pathogen–free conditions under regular hygiene monitoring. All animal experiments were approved by local authorities and performed in accordance with local institutional guidelines and animal protection laws, including specific locally approved protocols for sacrificing.
In vivo peritonitis model
Experiments were done as described in Spalinger et al. (2016). In brief, mice were injected with 1.8 mg MSU for the indicated time. Peritoneal cells were collected by intraperitoneal injection of 3 ml PBS and subsequent retrieval of the solution into a sterile conical tube and centrifugation at 300 ×g for 5 min at 4°C. Supernatants were used for IL-1β measurements using a commercially available ELISA kit (#DY401; R&D Systems). The cells were lysed in M-PER lysis buffer (Thermo Fisher Scientific) supplemented with proteinase and phosphatase inhibitors (Roche) and processed for immunoprecipitation as described below.
Cell culture
All cells were cultured at 37°C and 5% CO2 in DMEM or Roswell Park Memorial Institute medium supplemented with 10% fetal calf serum, L-glutamine (2 mM), penicillin (100 U/ml), and streptomycin (100 μg/ml; all from Thermo Fisher Scientific). They were free of mycoplasma contamination and monitored regularly using a PCR-based assay.
Isolation and stimulation of primary human immune cells
PBMCs from HDs and patients were isolated from whole blood using Ficoll density gradient purification; primed with 10 ng/ml LPS for 3 h; and in some cases, treated with 60 μM ibrutinib for 15 min before stimulation with 15 μM nigericin for the indicated time periods.
Generation of primary BMDMs and NLRP3 stimulation
BM cells were isolated from femurs and tibiae of 8–12-wk-old mice, grown, and differentiated using GM-CSF (M1 polarization) as described in Liu et al. (2017). BMDMs were primed with 100 ng/ml LPS for 3 h and either treated with 60 μM ibrutinib for 15 min or stimulated directly with 5 μM nigericin for the indicated time periods.
ASC speck formation assay and confocal microscopy
As described in Stutz et al. (2013) and Liu et al. (2017), Nlrp3 KO iMacs overexpressing NLRP3-FLAG and ASC-mCerulean were pretreated with ibrutinib or solvent control for 10 min before stimulation with either 5 μM nigericin (Life Technologies) for 60 min or 1 mM Leu-Leu-OMe⋅HCl (Chem-Impex) for 90 min. After stimulation, cells were fixed with 4% formaldehyde, and nucleic acids were stained with DRAQ5 (eBioscience). Cells were imaged with a Zeiss Observer.Z1 epifluorescence microscope using a 20× objective as described in Stutz et al. (2013). The number of cells and the number of specks were counted for 10 images per condition using CellProfiler (Carpenter et al., 2006).
NLRP3 speck formation confocal microscopy
BMDM from WT, Btk KO, and Nlrp3 KO mice were generated as described and reseeded in 250,000 cells/well in a 24-well plate containing a 12-mm round coverslip (1.5H thickness, #CG15NH; Thorlabs). The cells were primed with 100 ng/ml LPS (LPS-EK, Ultrapure; InvivoGen) followed by 10 μM nigericin (Life Technologies) for 30 min. The cells were briefly washed with PBS and fixed with freshly prepared 4% paraformaldehyde in prewarmed PEM buffer (80 mM PIPES, 5 mM EGTA, and 1 mM MgCl2 and titrated to pH 6.9 using 5 M KOH) for 15 min at room temperature. To ensure permeabilization, the cells were incubated in PEM + 0.05% saponin for 5 min. This was followed by blocking using 5% BSA in PBS + 0.05% saponin for 1 h at room temperature. The cells were washed three times, with a 5-min interval between the washes, with PBS + 0.05% saponin and followed by primary antibody incubation for 1 h at room temperature (goat polyclonal anti-NLRP3, 1:100, ab4207; Abcam) and rabbit anti-RCAS1 (1:100, #12290; Cell Signaling Technology) and then by secondary antibody (anti-goat Alexa Fluor 488, #A21467, and anti-rabbit Alexa Fluor 647, #A21443; Invitrogen) incubation for 45 min at room temperature. This was followed by staining of the nuclei with Hoechst (1:10,000, #62249; Thermo Fisher Scientific) in PBS. The cells were washed three times for 5 min in PBS + 0.05% saponin after the primary and the secondary antibody incubations. The coverslips were mounted using ProLong Diamond Antifade (#P36965; Thermo Fisher Scientific) and stored for 24 h at room temperature in the dark before imaging. Cells were imaged with a Zeiss LSM 800 Airyscan fluorescence microscope using a 40× objective. Several 3 × 3 tiles were imaged from which the number of cells and the number of specks were counted for three tiles per condition using blinded manual counting.
Expression and purification of recombinant BTK, NEK7, and NLRP3
The plasmids encoding NLRP3 with the deleted PYD (amino acids 134–1034) for MBP fusion protein expression in baculovirus (Bac)-to-Bac system (Thermo Fisher Scientific) and human NEK7 for His-SUMO fusion protein expression in E. coli BL21 (DE3) were described in Sharif et al. (2019). For NLRP3 expression, the baculovirus of NLRP3 was prepared using the Bac-to-Bac system (Thermo Fisher Scientific). Protein expression was induced by infection of Sf9 cells with 1% vol/vol of baculovirus. 48 h after infection, cells were lysed by sonication in buffer containing 30 mM Hepes, 200 mM NaCl, 2 mM 2-mercaptoethanol, and 10% glycerol at pH 7.5 with freshly added protease inhibitor cocktail (Sigma-Aldrich). The supernatant was incubated with 3 ml amylose resin at 4°C for 1 h and subjected to gravity flow. NLRP3 protein was eluted with 50 mM maltose and further purified with size-exclusion chromatography on a Superose 6 10/300 GL column (GE Healthcare) equilibrated with buffer containing 30 mM Hepes, 150 mM NaCl, and 2 mM β-mercaptoethanol at pH 7.5. NEK7 was overexpressed in E. coli BL21 (DE3) overnight at 18°C after induction with 0.1 mM isopropyl-β-d-thio-galacto-pyranoside after optical density at 600 nm reached 0.8. Cells were lysed by sonication in buffer containing 50 mM Hepes, 500 mM NaCl, 5 mM MgCl2, 10 mM imidazole, 10% glycerol, and 2 mM β-mercaptoethanol at pH 7.5 with freshly added protease inhibitor cocktail (Sigma-Aldrich). The His-SUMO-fusion NEK7 was purified by affinity chromatography using Ni-NTA beads (QIAGEN) followed by size-exclusion chromatography on a Superdex 200 10/300 GL column (GE Healthcare), equilibrated with buffer containing 30 mM Hepes, 150 mM NaCl, and 2 mM β-mercaptoethanol at pH 7.5. SH3–SH2–KinD, SH2–KinD, and KinD BTK constructs were expressed in Sf9 cells for 48 h upon infection with 1% vol/vol baculovirus. Cells were lysed by sonication in buffer containing 50 mM Hepes, 150 mM NaCl, 5 mM MgCl2, 10 mM imidazole, 10% glycerol, and 2 mM β-mercaptoethanol at pH 7.5 with freshly added protease inhibitor cocktail. Proteins were purified using Ni-NTA beads, followed by size-exclusion chromatography on a Superdex 200 10/300 GL column equilibrated with buffer containing 30 mM Hepes, 150 mM NaCl, and 2 mM β-mercaptoethanol at pH 7.5. Full-length WT and KD mutant BTK were overexpressed in Expi293 cells (Thermo Fisher Scientific) using transient transfection with polyethylenimine 25K (Polysciences). Cells were harvested 96 h after transfection and lysed in buffer containing 50 mM Hepes, 150 mM NaCl, 2 mM 2-mercaptoethanol, and 10% glycerol at pH 7.5 with freshly added protease inhibitor cocktail. The FLAG-fusion proteins were subjected to affinity chromatography using anti-FLAG M2 affinity gel (Millipore Sigma), eluted with 3×FLAG peptide (Millipore Sigma), and further purified by size-exclusion chromatography on a Superdex 200 10/300 GL column equilibrated with buffer containing 30 mM Hepes, 150 mM NaCl, and 2 mM β-mercaptoethanol at pH 7.5. Proteins were concentrated to 2–7 mg/ml, flash-frozen in liquid nitrogen, and stored at −80°C.
In vitro pulldowns
MBP-tagged NLRP3 (2 μM) was mixed with 4 μM His-SUMO-NEK7 or WT or mutant FLAG-BTK in buffer containing 30 mM Hepes, 150 mM NaCl, and 2 mM β-mercaptoethanol at pH 7.5 and incubated for 30 min at 30°C. The mixture was further incubated for 1 h with 40 μl amylose resin and washed twice with 500 μl of the same buffer, followed by 1-h elution with 50 mM maltose. 30% and 70% of the sample was loaded as input and elution fractions, respectively, and analyzed by SDS-PAGE and immunoblot using monoclonal ANTI-FLAG M2-Peroxidase (HRP) or anti–p-Y antibody (Sigma-Aldrich).
ELISA
Human and murine IL-1β, IL-6, or TNF in supernatants were determined by ELISA using half-area plates and kits by R&D Systems and BioLegend, determining triplicate measurements on a standard plate reader.
Coimmunoprecipitation and immunoblot from primary cells
PBMCs or BMDMs were primed with LPS and stimulated with nigericin, washed with cold PBS, and immediately lysed in radioimmunoprecipitation assay (RIPA) lysis buffer containing protease/phosphatase inhibitors (Roche). A sample of the cleared lysate was taken before addition of the primary antibody (see Table S2). To selected samples, 2,000 U λ-phosphatase (P0753; New England Biolabs) and MnCl2 to 1 mM were added and incubated for 30 min at 30°C. After 18 h of rotation at 4°C, magnetic bead–coupled secondary antibody (Protein G Dynabeads; Thermo Fisher Scientific) was added for another 90 min. The beads were then washed three times with lysis buffer, resuspended in SDS loading buffer, and boiled. Standard SDS-PAGE was performed on Thermo Fisher Scientific precast gels followed by immunoblot according to the antibody manufacturer’s instructions. Membranes were exposed using Fusion FL camera and FusionCapt Advance software (PEQLAB). Quantification was conducted using the same software.
Coimmunoprecipitation and immunoblot from HEK293T cells
HEK293T were transfected using CaPO4 and 24 h later, treated with 1 μM MCC950 or 60 μM acalabrutinib for 6 h or with 60 μM ibrutinib for 2 h followed by 4-h incubation with medium without ibrutinib, where indicated. Cells were lysed 48 h after transfection in RIPA buffer supplemented with protease/phosphatase inhibitors (Roche). Lysates were subjected to immunoprecipitation of the NLRP3-HA or NACHT-FLAG fusion protein with Dynabeads (Sigma-Aldrich) or with agarose beads covered with PI4P (P-B004a; Echelon Biosciences). Washed beads were boiled in loading buffer and applied to standard SDS-PAGE on Thermo Fisher Scientific precast gels, followed by immunoblot according to the antibody manufacturer’s instructions. Membranes were exposed using Fusion FL camera and FusionCapt Advance software (PEQLAB). Quantification was conducted using the same software.
Coimmunoprecipitation and immunoblot from peritoneal cells
Samples from peritoneal cells were first precleared with Sepharose G beads (GE Healthcare) for 1 h, incubated at 4°C with 5 μg mouse anti-NLRP3 (Enzo Life Sciences) under constant agitation overnight, and precipitated with Sepharose G beads. The pellets were washed three times in PBS, 10 μl 1× loading buffer (Thermo Fisher Scientific) was added, the solution was boiled for 10 min at 95°C, beads were pelleted by centrifugation at 300 ×g for 3 min, and the supernatants were loaded on PAGE for electrophoresis. WCLs were mixed with 4× loading buffer. The gels were blotted on nitrocellulose membranes and blocked with 3% milk in washing buffer (Tris-buffered saline with 0.05% Tween), and the membranes were incubated with mouse anti-NLRP3 (Enzo Life Sciences) mouse anti–p-Y (Cell Signaling Technology), rabbit anti-Btk (Cell Signaling Technology), and mouse anti–β-actin antibody (Millipore) overnight followed by incubation with HRP-labeled secondary antibodies for 2 h at room temperature and detection of immunoreactive proteins using an enhanced chemiluminescence kit (Enzo Life Sciences) and exposure on x-ray films (GE Healthcare).
BRET
HEK293T cells were transfected with YFP–NLRP3–Renilla luciferase or NLRP3–Renilla luciferase fusion constructs for WT and 4xY>F mutant using Lipofectamine 2000 according to manufacturer’s instructions for 24 h. 105 cells per well were then reseeded in a poly-L-lysine–coated white opaque 96-well plates; after adhesion, cells were incubated in ET buffer in the presence or absence of nigericin and upon addition of 5 μM coelenterazine h, and BRET signals were detected sequentially with a two-filter setting (Renilla luciferase filter [475 ± 30 nm] and YFP filter [530 ± 30 nm] at 37°C using a FLUOstar OPTIMA microplate reader from BMG LABTECH). In these experiments, a stable BRET signal (15 cycles) was recorded and averaged, and each reading was integrated for 0.5 s. Every condition was measured in duplicate. The BRET ratio in milliBRET units (mBU) was defined as shown below (for details, see Tapia-Abellán et al., 2019). Data were then normalized to the mean mBU for each construct in the unstimulated state. As a control of relative levels of expression for each BRET construct, the same cells were also plated in black, clear-bottom 96-wells to measure YFP fluorescence using an excitation filter at 510 nm and an emission filter at 540 nm.
Whole-exome sequencing (WES) capillary electrophoresis
3 μl of the prepared immunoblot lysates were run on a ProteinSimple WES instrument according to the manufacturer’s instructions. Data were analyzed with Compass for Simple Western software to compare the p-NLRP3 signal with the heavy-chain signal from the same run as an internal control.
Native PAGE
BMDMs were stimulated as described and lysed in RIPA lysis buffer without SDS. Lysates were centrifuged at 2,300 ×g for 10 min to pellet DNA. Supernatant was centrifuged at 16,100 ×g for 25 min, and the pellet was resuspended in native PAGE sample buffer (Thermo Fisher Scientific). The samples were loaded onto NuPage 3%–8% Tris-acetate gels (Thermo Fisher Scientific) without boiling, and native PAGE was conducted using Tris-glycine running buffer (Thermo Fisher Scientific). The gel was soaked in 10% SDS solution for 10 min before performing semidry transfer and continuing with conventional immunoblot.
Cross-linking of ASC oligomers
BMDMs were primed with LPS and treated with ibrutinib and nigericin as described. Cells were lysed in RIPA lysis buffer, and pellets were cross-linked using disuccinimidyl suberate and analyzed as described in Khare et al. (2016).
Size-exclusion chromatography
BMDMs were stimulated and lysed in 50 mM Tris-HCl, pH 7.4, 1% NP-40, and 150 mM NaCl. 100 μl cleared lysate was loaded on a Superdex 200 10/300 GL column (GE Healthcare), and proteins were eluted using an ÄKTA Purifier (GE Healthcare) and buffer (50 mM Tris-HCL, pH 7.4, 150 mM NaCl) with 0.25 ml/min flow. 200 μl fractions were collected and analyzed via Western blot.
In vitro kinase assay
For results in Fig. 1 H, recombinant NLRP3 from Novus Biologicals (H00114548-P01) and BTK from Sino Biological (10578-H08B) or Abcam (ab205800) were incubated at 30°C for 3 h using Cell Signaling Technology kinase buffer (#9802) in the presence of 2 mM ATP. As a negative control, recombinant Posi-Tag Epitope Tag Protein (BioLegend) was used. Before and after kinase assay, samples were boiled and analyzed via SDS-PAGE and Western blot. For results in Fig. S1 F, NLRP3 and BTK (WT or KD) were purified as described above. For reactions, 2 μM MBP-tagged NLRP3 was mixed with or without 0.2 μM purified FLAG-tagged BTK in buffer containing 30 mM Hepes, 150 mM NaCl, 12.5 mM MgCl2, 2.5 mM ATP, and 2 mM β-mercaptoethanol at pH 7.5 in the presence or absence of ibrutinib (catalog no. S2680; Selleckchem). The mixture was incubated at 30°C, and equal aliquots were taken at indicated time points. Samples were analyzed by SDS-PAGE and immunoblot using anti–p-Y antibody (#8954S; Cell Signaling Technology). For results in Fig. S4, C and D, 0.5 μM purified BTK constructs were mixed with 1 μM NLRP3 peptides with WT Y sequence, p-Y, or with Y>F in buffer containing 30 mM Hepes, 150 mM NaCl, 5 mM MgCl2, 2.5 mM ATP, 2 mM β-mercaptoethanol and protease inhibitor cocktail (Sigma) at pH 7.5. The reaction was performed at 30°C, and equal aliquots were taken at indicated time points. For each BTK construct, samples were analyzed by immunoblot using anti–p-Y antibody (Cell signaling, cat. 8954S) at equal exposure times. The quantification was performed with ImageJ software.
Dot blot analysis
Peptides synthetized in-house were incubated with recombinant BTK (Sino Biologicals) for 3 h in Cell Signaling Technology kinase buffer (#9802) supplemented with 2 mM ATP. Next, the samples were boiled, and anti-His magnetic beads (Dynabeads His-Tag Isolation and Pulldown; Thermo Fisher Scientific) were added to deplete the samples of phosphorylated BTK. The samples were cleared from the magnetic beads, and the supernatants were manually spotted on a nitrocellulose membrane. The dried spots were stained using Pierce reversible protein stain to visualize total peptide amounts. The membrane was then blocked with 5% BSA in Tris-buffered saline with 0.1% Tween, and conventional anti–p-Y primary and secondary antibody incubation steps followed.
Subcellular fractionation
Cells were homogenized using a 10-ml syringe and 27G × 19-mm needles in homogenization buffer (0.25 M sucrose, 10 mM Tris HCl, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, protease inhibitor [Roche] and PhosStop [Roche]). Homogenized cells (“homogenate” in Fig. 3 H) were centrifuged at 1,000 ×g for 5 min to remove the nucleus. The supernatant was centrifuged at 5,000 ×g for 10 min to obtain a heavy membrane fraction (pellet, P5, analyzed directly as in Fig. 3 H or fractionated further as described below). The supernatant (S5) was either analyzed directly as in Fig. 3 H or centrifuged 100,000 ×g for 20 min to separate a light membrane fraction (P100) from the cytosol (S100). For Fig. S5 A, P5 and P100 pellets were washed once with homogenization buffer and then used for further sucrose gradient ultracentrifugation separately. For sucrose gradient ultracentrifugation, a continuous 15%–45% (wt/wt) sucrose gradient was prepared in 10 mM Tris-HCl (pH 7.5), 20 mM KCl, and 3 mM MgCl2 using Gradient Station (Biocomp Instruments). P5 or P100 was loaded on top of the gradient and centrifuged at 170,000 ×g for 3 h. The gradient was fractionated into 12 fractions of 1.1 ml using the fraction collector module of Gradient Station.
PI4P bead binding assays
HEK293T cells were transfected with HA-tagged human WT or Y>E PYD/NACHT linker (residues 94–219)–mCitrine–HA constructs. Cells were lysed in RIPA buffer, and PI4P beads (P-B004A; Echelon Biosciences) or the same amount of control beads (P-B000; Echelon Biosciences) were added to cleared lysates and incubated for 1.5 h at 4°C while rotating. Beads were then washed three times with RIPA buffer and boiled, and bound proteins were analyzed via immunoblot. Alternatively, cells were transfected with WT, Y>E, or K>A Flag-tagged murine PBR (residues 127–146)–GFP–Flag constructs adopted from Chen and Chen (2018). PI4P beads or control beads were blocked beforehand in 2% BSA, 0.5% NP-40, and 200 μg/ml Flag peptide (F3290; Sigma-Aldrich) for 2 h at 4°C. Transfected cells were then lysed in RIPA buffer, and the expressed proteins were purified using Anti-FLAG M2 Magnetic Beads (M8823; Merck). Beads were washed three times with RIPA buffer and boiled to elute the purified PBR. Blocked PI4P beads or the same amount of control beads were added to the eluted protein and incubated for 1.5 h at 4°C while rotating. Beads were then washed three times for 8 min with RIPA buffer, resuspended in lithium dodecyl sulfate lithium dodecyl sulfate sample buffer, and boiled, and bound protein was analyzed using immunoblot.
Reconstitution and analysis of Nlrp3-deficient iMacs
Nlrp3-deficient iMacs (Hornung et al., 2008) were retrovirally transduced with human NLRP3 (WT or 4xY>F)–Flag–T2A–mCherry constructs as described in Hornung et al. (2008) and subsequently sorted for similar expression of mCherry. Similar NLRP3 expression was confirmed by anti-Flag immunoblot of cell lysates. Cells were seeded at 105 cells/well in a 96-well plate in 100 μl and primed with LPS (50 or 200 ng/ml) for 3 h, and inflammasome stimuli in OptiMEM were added as follows: nigericin at 8 μM for 1.5 h, R837 (imiquimod) at 20 μg/ml for 2 h, or poly(dA:dT) at 200 ng per well with 0.5 μl lipofectamine 2000 for 4 h. IL-1β and TNF were subsequently determined by IL-1β and TNF homogeneous time-resolved fluorescence assay, respectively (62MIL1BPEH and 62MTNFAPEG; Cisbio).
NLRP3 sequence analysis, structure inspection, and charge prediction
Nod-like receptor sequences were retrieved from UniProt and ClustalW aligned within Geneious R6 software. A hypothetical active conformation of NLRP3 was modeled based on NLRP3–NEK7 structure in an inactive state (Protein Data Bank accession no. 6NPY; Sharif et al., 2019). NACHT domain reorganization and hypothetical NLRP3 oligomerization was generated based on the NLRC4 oligomer (Protein Data Bank accession no. 3JBL) as a homology model template by introduction of a 90° rotation of NBD-HD1 module (Sharif et al., 2019). Phosphorylation of tyrosine residues of interest was performed in PyMOL (Schrödinger) using the PyTMs plugin (Warnecke et al., 2014). Electrostatic potential of the solvent-accessible surface of phosphorylated and nonphosphorylated NLRP3 models was calculated with the PBEQ-Solver online visualization tool (https://www.charmm-gui.org; Jo et al., 2008a; Jo et al., 2008b; MacKerell et al., 1998) and visualized with PyMOL. Protein net charges of the Y136-, Y140-, and Y143-containing linker were conducted with ProtPi (https://www.protpi.ch).
Statistics
Experimental data were analyzed using Excel 2010 (Microsoft) and/or GraphPad Prism 6, 7, or 8; microscopy data with ImageJ/Fiji; and flow cytometry data with FlowJo 10. Normal distribution in each group was always tested using the Shapiro–Wilk test first for the subsequent choice of a parametric (ANOVA, Student’s t test) or nonparametric (e.g., Friedman, Mann–Whitney U, Wilcoxon) test. P values (α = 0.05) were then calculated, and multiple testing was corrected for in Prism, as indicated in the figure legends. P < 0.05 was generally considered as statistically significant and denoted by an asterisk. Comparisons were made to unstimulated control, unless indicated otherwise, and denoted by brackets.
Online supplemental material
Fig. S1 displays additional data related to Fig. 1, showing that in cells from patients with BTK deficiency, NLRP3 phosphorylation is decreased; that in an experimental in vivo mouse model of MSU peritonitis, IL-1β release and NLRP3 phosphorylation are time and BTK dependent; and that in vitro BTK kinase activity is required for NLRP3 phosphorylation. Figs. S2, S3, and S4 show additional data related to Fig. 2. Fig. S2 displays the position of tyrosines in the NLRP3 FISNA and NACHT domains relative to positions of known pathological significance. Fig. S3 shows that the NACHT domain tyrosine mutation does not have an impact on NLRP3 phosphorylation and that for recombinant NLRP3, the linker region is not amenable to mass spectrometric analysis; furthermore, using peptide scanning, individual tyrosines in the linker of human and murine NLRP3, but not tyrosines in the NACHT domain, were found to be BTK modified. Fig. S4 illustrates the position of BTK-modified tyrosines in a recent NEK7-NLRP3 cryo-EM structure and in multiple sequence alignments of different NLRP3 sequences; furthermore, the effect of linker NLRP3 peptides on BTK activity is shown and quantified. Fig. S5 shows data related to Figs. 3 and 4, most notably the fractionation approach employed to probe subcellular localization of NLRP3 in dependence on BTK and the effect of p-Y mutation in reconstituted iMacs primed with different LPS concentrations. Finally, the responsiveness of NLRP3 phospho mutant forms as assessed by BRET and a graphical summary of the entire paper are shown. Table S1 summarizes the peptides used in this study. Table S2 summarizes the antibodies used in this study.
Supplementary Material
shows peptides used in this study.
shows antibodies used in this study.
Acknowledgments
We gratefully acknowledge Pablo Pelegrin (Instituto Murciano de Investigacion Biosanitaria, Murcia, Spain) for provision of the WT NLRP3 BRET construct; Ulrich Wulle for help with peptide synthesis; Teerithveen Pasricha and Jacqueline Berner for help with in vitro kinase assays and BRET measurement, respectively; and Libero Lo Presti for helpful comments. We thank Xiaowu Zhang and Felix Meissner for helpful advice on kinase target residue identification and mass spectrometry, respectively. We thank all study subjects and their families as well as voluntary healthy blood donors for participating in the study.
The study was supported by the Else-Kröner-Fresenius Stiftung (to A.N.R. Weber); Deutsche Forschungsgemeinschaft grants CRC TR156 “The Skin as an Immune Sensor and Effector Organ–Orchestrating Local and Systemic Immunity” (to Z.S. Bittner, F. Herster, F. Bork, C.L. Greve, and A.N.R. Weber) and We-4195/15-1 (to A.N.R. Weber); University Hospital Tübingen (Fortüne Grants 2310-0-0 to X. Liu and A.N.R. Weber, and 2615-0-0 to M. Mateo Tortola and A. Tapia-Abellán); IFM Therapeutics (to S. Shankar and A.N.R. Weber); the E-Rare program of the European Union, managed by the German Research Foundation, grant code GR1617/14-1/iPAD (to B. Grimbacher); the Netzwerke Seltener Erkrankungen of the Federal Ministry of Education and Research (GAIN_01GM1910A, to B. Grimbacher); and the Damon Runyon Cancer Research Foundation (to L. Andreeva). Infrastructural funding was provided by the University of Tübingen; the University Hospital Tübingen; and the Deutsche Forschungsgemeinschaft Clusters of Excellence “iFIT–Image-Guided and Functionally Instructed Tumor Therapies” (EXC 2180 to A.N.R. Weber and M.W. Löffler), “CMFI–Controlling Microbes to Fight Infection (EXC 2124 to X. Liu, A.N.R. Weber, and A. Tapia-Abellán), “CIBSS–Centre for Integrative Signalling Studies” (EXC 2189 to B. Grimbacher), “RESIST–Resolving Infection Susceptibility” (EXC 2155 to B. Grimbacher), “ImmunoSensation2” (EXC 2151, to E. Latz). This work was also supported by (Gefördert durch die) Deutsche Forschungsgemeinschaft im Rahmen der Exzellenzstrategie des Bundes und der Länder–EXC 2180 (390900677), EXC 2124, EXC 2189 (390939984), EXC 2155 (39087428), and EXC 2151 (390873048).
Author contributions: Z.A. Bittner, X. Liu, M. Mateo Tortola, M. Spalinger, Y. Cardona Gloria, T.-H. Chang, F. Bork, C.L. Greve, S. Dickhöfer, H. Kalbacher, K. Bosch, L. Andreeva, A. Marcu, M. Mangan, P. Düwell, M. Lovotti, F. Herster, S. Shankar, A. Tapia-Abellán, O.-O. Wolz, N.A. Schilling, S. Wagner, and A.N.R. Weber performed experiments; Z.A. Bittner, X. Liu, S. Dickhöfer, H. Kalbacher, L. Andreeva, S. Stevanoić, M. Lovotti, M. Mangan, M. Mateo Tortola, M. Spalinger, M. Scharl, O. Hantschel, P. Düwell, F. Herster, N.A. Schilling, S. Wagner, and A.N.R. Weber analyzed data; M.W. Löffler, J.B. Kümmerle-Deschner, M. Scharl, A. Delor, and B. Grimbacher were involved in patient recruitment and sample acquisition; Z.A. Bittner and A.N.R. Weber wrote the manuscript; and L. Andreeva, P. Düwell, M.W. Löffler, S. Shankar, M. Mateo Tortola, A. Tapia-Abellán, O. Hantschel, S. Wagner, H. Wu, and E. Latz provided valuable comments. All authors approved the final manuscript. A.N.R. Weber and Z.A. Bittner conceived and coordinated the study.
References
- Banoth, B., and Cassel S.L.. 2017. Bruton tyrosine kinase inhibition: Clinical relevance beyond B cells. J. Allergy Clin. Immunol. 140:985–987. 10.1016/j.jaci.2017.03.041 [DOI] [PubMed] [Google Scholar]
- Broderick, L., De Nardo D., Franklin B.S., Hoffman H.M., and Latz E.. 2015. The inflammasomes and autoinflammatory syndromes. Annu. Rev. Pathol. 10:395–424. 10.1146/annurev-pathol-012414-040431 [DOI] [PubMed] [Google Scholar]
- Byrd, J.C., Furman R.R., Coutre S.E., Flinn I.W., Burger J.A., Blum K.A., Grant B., Sharman J.P., Coleman M., Wierda W.G., et al. 2013. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N. Engl. J. Med. 369:32–42. 10.1056/NEJMoa1215637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100. 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., and Chen Z.J.. 2018. PtdIns4P on dispersed trans-Golgi network mediates NLRP3 inflammasome activation. Nature. 564:71–76. 10.1038/s41586-018-0761-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll, R.C., Robertson A.A., Chae J.J., Higgins S.C., Muñoz-Planillo R., Inserra M.C., Vetter I., Dungan L.S., Monks B.G., Stutz A., et al. 2015. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 21:248–255. 10.1038/nm.3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland, S., Warren H.S., Lowry S.F., Calvano S.E., and Remick D.. Inflammation and the Host Response to Injury Investigators . 2005. Acute inflammatory response to endotoxin in mice and humans. Clin. Diagn. Lab. Immunol. 12:60–67. 10.1128/CDLI.12.1.60-67.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte, D.P., Lamontanara A.J., La Sala G., Jeong S., Sohn Y.K., Panjkovich A., Georgeon S., Kükenshöner T., Marcaida M.J., Pojer F., et al. 2020. Btk SH2-kinase interface is critical for allosteric kinase activation and its targeting inhibits B-cell neoplasms. Nat. Commun. 11:2319. 10.1038/s41467-020-16128-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duewell, P., Kono H., Rayner K.J., Sirois C.M., Vladimer G., Bauernfeind F.G., Abela G.S., Franchi L., Nuñez G., Schnurr M., et al. 2010. NLRP3 inflammasomes are required for atherogenesis and activated by cholesterol crystals. Nature. 464:1357–1361. 10.1038/nature08938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke, M., Bieber M., Kraft P., Weber A.N.R., Stoll G., and Schuhmann M.K.. 2021. The NLRP3 inflammasome drives inflammation in ischemia/reperfusion injury after transient middle cerebral artery occlusion in mice. Brain Behav. Immun. 92:223–233. 10.1016/j.bbi.2020.12.009 [DOI] [PubMed] [Google Scholar]
- Green, J.P., Yu S., Martín-Sánchez F., Pelegrin P., Lopez-Castejon G., Lawrence C.B., and Brough D.. 2018. Chloride regulates dynamic NLRP3-dependent ASC oligomerization and inflammasome priming. Proc. Natl. Acad. Sci. USA. 115:E9371–E9380. 10.1073/pnas.1812744115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., Zeng M.Y., Yang D., Motro B., and Núñez G.. 2016. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature. 530:354–357. 10.1038/nature16959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrickson, S.E. 2017. Teaching an old pathway new tricks: Targeting BTK to block NLRP3. Sci. Immunol. 2:eaar2548. 10.1126/sciimmunol.aar2548 [DOI] [PubMed] [Google Scholar]
- Hornung, V., Bauernfeind F., Halle A., Samstad E.O., Kono H., Rock K.L., Fitzgerald K.A., and Latz E.. 2008. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9:847–856. 10.1038/ni.1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, M., Shichita T., Okada M., Komine R., Noguchi Y., Yoshimura A., and Morita R.. 2015. Bruton’s tyrosine kinase is essential for NLRP3 inflammasome activation and contributes to ischaemic brain injury. Nat. Commun. 6:7360. 10.1038/ncomms8360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo, S., Kim T., Iyer V.G., and Im W.. 2008a. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29:1859–1865. 10.1002/jcc.20945 [DOI] [PubMed] [Google Scholar]
- Jo, S., Vargyas M., Vasko-Szedlar J., Roux B., and Im W.. 2008b. PBEQ-Solver for online visualization of electrostatic potential of biomolecules. Nucleic Acids Res. 36:W270-5. 10.1093/nar/gkn314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan, W.N., Alt F.W., Gerstein R.M., Malynn B.A., Larsson I., Rathbun G., Davidson L., Müller S., Kantor A.B., Herzenberg L.A., et al. 1995. Defective B cell development and function in Btk-deficient mice. Immunity. 3:283–299. 10.1016/1074-7613(95)90114-0 [DOI] [PubMed] [Google Scholar]
- Khare, S., Radian A.D., Dorfleutner A., and Stehlik C.. 2016. Measuring NLR oligomerization I: Size exclusion chromatography, co-immunoprecipitation, and cross-linking. Methods Mol. Biol. 1417:131–143. 10.1007/978-1-4939-3566-6_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Wahl M.I., Eguinoa A., Stephens L.R., Hawkins P.T., and Witte O.N.. 1997. Phosphatidylinositol 3-kinase-gamma activates Bruton’s tyrosine kinase in concert with Src family kinases. Proc. Natl. Acad. Sci. USA. 94:13820–13825. 10.1073/pnas.94.25.13820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Pichulik T., Wolz O.O., Dang T.M., Stutz A., Dillen C., Delmiro Garcia M., Kraus H., Dickhöfer S., Daiber E., et al. 2017. Human NACHT, LRR, and PYD domain-containing protein 3 (NLRP3) inflammasome activity is regulated by and potentially targetable through Bruton tyrosine kinase. J. Allergy Clin. Immunol. 140:1054–1067.e10. 10.1016/j.jaci.2017.01.017 [DOI] [PubMed] [Google Scholar]
- MacKerell, A.D. Jr., Bashford D., Bellott M., Dunbrack R.L. Jr., Evanseck J.D., Field M.J., Fischer S., Gao J., Guo H., Ha S., et al. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102:3586–3616. 10.1021/jp973084f [DOI] [PubMed] [Google Scholar]
- Magupalli, V.G., Negro R., Tian Y., Hauenstein A.V., Di Caprio G., Skillern W., Deng Q., Orning P., Alam H.B., Maliga Z., et al. 2020. HDAC6 mediates an aggresome-like mechanism for NLRP3 and pyrin inflammasome activation. Science. 369:eaas8995. 10.1126/science.aas8995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, L., Kitani A., Hiejima E., Montgomery-Recht K., Zhou W., Fuss I., Wiestner A., and Strober W.. 2020. Bruton tyrosine kinase deficiency augments NLRP3 inflammasome activation and causes IL-1β-mediated colitis. J. Clin. Invest. 130:1793–1807. 10.1172/JCI128322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor, A., Martinon F., De Smedt T., Pétrilli V., and Tschopp J.. 2007. A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat. Immunol. 8:497–503. 10.1038/ni1459 [DOI] [PubMed] [Google Scholar]
- Muñoz-Planillo, R., Kuffa P., Martínez-Colón G., Smith B.L., Rajendiran T.M., and Núñez G.. 2013. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 38:1142–1153. 10.1016/j.immuni.2013.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Riordan, C.E., Purvis G.S.D., Collotta D., Chiazza F., Wissuwa B., Al Zoubi S., Stiehler L., Martin L., Coldewey S.M., Collino M., and Thiemermann C.. 2019. Bruton’s tyrosine kinase inhibition attenuates the cardiac dysfunction caused by cecal ligation and puncture in mice. Front. Immunol. 10:2129. 10.3389/fimmu.2019.02129 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- O’Riordan, C.E., Purvis G.S.D., Collotta D., Krieg N., Wissuwa B., Sheikh M.H., Ferreira Alves G., Mohammad S., Callender L.A., Coldewey S.M., et al. 2020. X-linked immunodeficient mice with no functional Bruton’s tyrosine kinase are protected from sepsis-induced multiple organ failure. Front. Immunol. 11:581758. 10.3389/fimmu.2020.581758 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Purvis, G.S.D., Collino M., Aranda-Tavio H., Chiazza F., O’Riordan C.E., Zeboudj L., Mohammad S., Collotta D., Verta R., Guisot N.E.S., et al. 2020. Inhibition of Bruton’s TK regulates macrophage NF-κB and NLRP3 inflammasome activation in metabolic inflammation. Br. J. Pharmacol. 177:4416–4432. 10.1111/bph.15182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschewski, M., Lionakis M.S., Sharman J.P., Roswarski J., Goy A., Monticelli M.A., Roshon M., Wrzesinski S.H., Desai J.V., Zarakas M.A., et al. 2020. Inhibition of Bruton tyrosine kinase in patients with severe COVID-19. Sci. Immunol. 5:eabd0110. 10.1126/sciimmunol.abd0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmacke, N.A., Gaidt M.M., Szymanska I., O’Duill F., Stafford C.A., Chauhan D., Fröhlich A.L., Nagl D., Pinci F., Schmid-Burgk J.L., and Hornung V.. 2019. Priming enables a NEK7-independent route of NLRP3 activation. bioRxiv. (Preprint posted October 9, 2019) 10.1101/799320 [DOI]
- Schmid-Burgk, J.L., Chauhan D., Schmidt T., Ebert T.S., Reinhardt J., Endl E., and Hornung V.. 2016. A genome-wide CRISPR (clustered regularly interspaced short palindromic repeats) screen identifies NEK7 as an essential component of NLRP3 Inflammasome activation. J. Biol. Chem. 291:103–109. 10.1074/jbc.C115.700492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane, P.I., Lee B., Hoyle C., Yu S., Lopez-Castejon G., Lowe M., and Brough D.. 2020. The NLRP3-inflammasome as a sensor of organelle dysfunction. J. Cell Biol. 219:e202006194. 10.1083/jcb.202006194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif, H., Wang L., Wang W.L., Magupalli V.G., Andreeva L., Qiao Q., Hauenstein A.V., Wu Z., Núñez G., Mao Y., and Wu H.. 2019. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature. 570:338–343. 10.1038/s41586-019-1295-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, N., and Li T.. 2018. Regulation of NLRP3 inflammasome by phosphorylation. Front. Immunol. 9:2305. 10.3389/fimmu.2018.02305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalinger, M.R., Kasper S., Gottier C., Lang S., Atrott K., Vavricka S.R., Scharl S., Raselli T., Frey-Wagner I., Gutte P.M., et al. 2016. NLRP3 tyrosine phosphorylation is controlled by protein tyrosine phosphatase PTPN22. J. Clin. Invest. 126:1783–1800. 10.1172/JCI83669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz, A., Horvath G.L., Monks B.G., and Latz E.. 2013. ASC speck formation as a readout for inflammasome activation. Methods Mol. Biol. 1040:91–101. 10.1007/978-1-62703-523-1_8 [DOI] [PubMed] [Google Scholar]
- Tapia-Abellán, A., Angosto-Bazarra D., Martínez-Banaclocha H., de Torre-Minguela C., Cerón-Carrasco J.P., Pérez-Sánchez H., Arostegui J.I., and Pelegrin P.. 2019. MCC950 closes the active conformation of NLRP3 to an inactive state. Nat. Chem. Biol. 15:560–564. 10.1038/s41589-019-0278-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicinanza, M., D’Angelo G., Di Campli A., and De Matteis M.A.. 2008. Function and dysfunction of the PI system in membrane trafficking. EMBO J. 27:2457–2470. 10.1038/emboj.2008.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., El Maadidi S., Fischer J., Grabski E., Dickhöfer S., Klimosch S., Flannery S.M., Filomena A., Wolz O.O., Schneiderhan-Marra N., et al. East-German and Swiss Hepatitis C Virus Study Groups . 2015. A frequent hypofunctional IRAK2 variant is associated with reduced spontaneous hepatitis C virus clearance. Hepatology. 62:1375–1387. 10.1002/hep.28105 [DOI] [PubMed] [Google Scholar]
- Warnecke, A., Sandalova T., Achour A., and Harris R.A.. 2014. PyTMs: a useful PyMOL plugin for modeling common post-translational modifications. BMC Bioinformatics. 15:370. 10.1186/s12859-014-0370-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, A.N.R. 2021. Targeting the NLRP3 Inflammasome via BTK. Front. Cell Dev. Biol. 9:630479. 10.3389/fcell.2021.630479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, A.N.R., Bittner Z., Liu X., Dang T.M., Radsak M.P., and Brunner C.. 2017. Bruton’s tyrosine kinase: An emerging key player in innate immunity. Front. Immunol. 8:1454. 10.3389/fimmu.2017.01454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, A., Liu X., Tapia-Abellan A., Shankar S., Bittner Z., and Morita R.. 2020a. BTK promotes NLRP3 inflammasome activation - and acts as rheostat? J. Clin. Invest. https://www.jci.org/eletters/view/128322 (accessed March 23, 2020)
- Weber, A.N.R., Bittner Z.A., Shankar S., Liu X., Chang T.H., Jin T., and Tapia-Abellán A.. 2020b. Recent insights into the regulatory networks of NLRP3 inflammasome activation. J. Cell Sci. 133:jcs248344. 10.1242/jcs.248344 [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Meszaros G., He W.T., Xu Y., de Fatima Magliarelli H., Mailly L., Mihlan M., Liu Y., Puig Gámez M., Goginashvili A., et al. 2017. Protein kinase D at the Golgi controls NLRP3 inflammasome activation. J. Exp. Med. 214:2671–2693. 10.1084/jem.20162040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweigner, J., Gramm H.J., Singer O.C., Wegscheider K., and Schumann R.R.. 2001. High concentrations of lipopolysaccharide-binding protein in serum of patients with severe sepsis or septic shock inhibit the lipopolysaccharide response in human monocytes. Blood. 98:3800–3808. 10.1182/blood.V98.13.3800 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows peptides used in this study.
shows antibodies used in this study.