Abstract
The pheromone response in the yeast Saccharomyces cerevisiae is mediated by a heterotrimeric G protein. The Gβγ subunit (a complex of Ste4p and Ste18p) is associated with both internal and plasma membranes, and a portion is not stably associated with either membrane fraction. Like Ras, Ste18p contains a farnesyl-directing CaaX box motif (C-terminal residues 107 to 110) and a cysteine residue (Cys 106) that is a potential site for palmitoylation. Mutant Ste18p containing serine at position 106 (mutation ste18-C106S) migrated more rapidly than wild-type Ste18p during sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The electrophoretic mobility of wild-type Ste18p (but not the mutant Ste18p) was sensitive to hydroxylamine treatment, consistent with palmitoyl modification at Cys 106. Furthermore, immunoprecipitation of the Gβγ complex from cells cultured in the presence of [3H]palmitic acid resulted in two radioactive species on nonreducing SDS-PAGE gels, with molecular weights corresponding to Gγ and Gβγ. Substitution of serine for either Cys 107 or Cys 106 resulted in the failure of Gβγ to associate with membranes. The Cys 107 substitution also resulted in reduced steady-state accumulation of Ste18p, suggesting that the stability of Ste18p requires modification at Cys 107. All of the mutant forms of Ste18p formed complexes with Ste4p, as assessed by coimmunoprecipitation. We conclude that tight membrane attachment of the wild-type Gβγ depends on palmitoylation at Cys 106 and prenylation at Cys 107 of Ste18p.
Heterotrimeric G proteins (containing Gα, Gβ, and Gγ subunits) are peripheral-membrane proteins that are coupled to cell surface receptors with seven membrane-spanning domains. They mediate response to various extracellular stimuli such as light, odorants, and hormones. Activated receptors stimulate exchange of GTP for GDP in Gα, resulting in dissociation of Gα from the Gβγ dimer. Subsequent events in the signal transduction pathway are elicited either by the action of the GTP-Gα complex or by the free Gβγ. Mounting evidence indicates that lipid modification of Gα and Gγ governs their association with membranes and in some cases promotes specific protein-protein interactions (reviewed in references 38 and 47). The C-terminal cysteine residue of mature Gγ contains an isoprenyl modification (either farnesyl or geranylgeranyl); the isoprenyl group is transferred to the cysteine residue in the CaaX box motif of the Gγ precursor, followed by proteolytic removal of the last three residues and methylation of the C terminus. In the yeast Saccharomyces cerevisiae, the pheromone response is mediated by Gα, Gβ, and Gγ subunits (Gpa1p, Ste4p, and Ste18p, respectively) that are coupled to either of the two pheromone receptors (Ste2p and Ste3p). During the mating of the two haploid cell types, a cells (expressing Ste2p) and α cells (expressing Ste3p) respond to the peptide pheromones (α-factor and a-factor, respectively) produced by cells of the opposite cell type. Free Gβγ leads to stimulation of a nitrogen-activated protein kinase cascade (26, 35). The ultimate physiological responses include arrest of cell division and induction of mating-specific genes. Gpa1p receives both myristoyl (44) and palmitoyl (29, 43) modifications near the N terminus. Like Ras, Ste18p contains two Cys residues near its C terminus. One Cys is contained in the farnesyl-directing CaaX box (CTLM), and the other Cys is a potential site for palmitoylation. Mutants with substitutions for either Cys residue are unresponsive to pheromone (11, 15, 48), raising the possibility that the function of Gβγ depends on lipid modification of these sites.
Our previous work (20) identified three distinct populations of Gβγ: one tightly associated with plasma membranes, a second tightly associated with internal membranes, and a third population that is not associated (or only weakly associated) with membranes. Binding of Ste4p to either membrane fraction requires Ste18p, and plasma membrane localization also requires Gpa1p. Two questions remain: what structural features of Gβγ determine its subcellular localization, and what function does Gβγ serve at these locations? This report addresses the first question by examining how alterations in the residues that specify lipid modification affect the biochemical properties and subcellular localization of the Gβγ complex. We provide evidence for palmitoylation of Ste18p and define the roles for farnesyl and palmitoyl modifications in subcellular localization. Our results are consistent with unpublished results of Manahan and Linder (29), who have found that Ste18p is palmitoylated when expressed in Sf9 insect cells.
MATERIALS AND METHODS
Yeast strains and plasmids.
Strains used in this study are isogenic with strain W303-1A and are described in Table 1. Strains W303-1A, HC106S, HC107S, and BRI1426 were transformed with the SalI-XbaI fragment of plasmid pJR868 (39) containing ram1::HIS3 to generate strains 809-A, 810-A, 811-A, and 814-A, respectively. PCR was used to confirm the genetic structure of the resulting recombinants. Strains 816-1 and 817-1 were generated by selecting for 5-fluoroorotic acid-resistant derivatives of strains BRI1426 and 814-A, respectively. Plasmids M70p2, M70p2C106S, and M70p2C107S (48) are high-copy-number YEp plasmids that contain the STE18+, ste18-C106S, and ste18-C107S alleles, respectively, under transcriptional control of the ADH1 promoter. Plasmid pBH21 contains the STE18 coding sequence under transcriptional control of the ADH1 promoter. It was constructed by digesting high-copy-number plasmid M91p1 (provided by M. Whiteway) with BglII and replacing the URA3-containing BglII fragment with a BglII fragment containing the LEU2 gene from plasmid YEp13 (20). Plasmid pEL37 (provided by E. Leberer) is a single-copy plasmid that contains the HIS3 gene, as well as the STE4 and GPA1 coding sequences under transcriptional control of the GAL1,10 promoter. Plasmid pRS313 is a single-copy vector plasmid containing the HIS3 gene.
TABLE 1.
Strain list
| Straina | Genotype |
|---|---|
| W303-1A | MATa ade2 his3 leu2 trp1 ura3 can1 |
| HC106S | W303-1A ste18-C106S |
| HC107S | W303-1A ste18-C107S |
| BRI1426 | W303-1A ste18::URA3 |
| 809-A | W303-1A ram1::HIS3 |
| 810-A | W303-1A ram1::HIS3 ste18-C106S |
| 811-A | W303-1A ram1::HIS3 ste18-C107S |
| 814-A | W303-1A ram1::HIS3 ste18::URA3 |
| 816-1/M70p2 | W303-1A ste18::ura3 containing plasmid M70p2 |
| 816-1/M70p2C106S | W303-1A ste18::ura3 containing plasmid M70p2C106S |
| 816-1/M70p2C107S | W303-1A ste18::ura3 containing plasmid M70p2C107S |
| 817-5/M70p2 | W303-1A ste18::ura3 ram1::HIS3 containing plasmid M70p2 |
| 817-5/M70p2C106S | W303-1A ste18::ura3 ram1::HIS3 containing plasmid M70p2C106S |
| 817-5/M70p2C107S | W303-1A ste18::ura3 ram1::HIS3 containing plasmid M70p2C107S |
| W303 (diploid) | MATa/MATα ade2/ade2 his3/his3 leu2-/leu2 trp1/trp1 ura3/ura3 can1/can1 |
Strains HC106S and HC107S were from M. S. Whiteway (48). Strain BRI146 was also kindly provided by M. S. Whiteway. All other strains were generated during the course of this study.
Reagents and culture media.
YM-1 is a rich liquid medium (17). Minimal glucose medium (18) and minimal galactose medium (20) were supplemented with auxotrophic requirements as described previously. −URA + CAA is supplemented minimal glucose medium containing 0.1% casamino acids (Difco) and lacking uracil (20). Dodecyl-β-d-maltoside, cholesterol hemisuccinate, protein A-Sepharose beads, and hydroxylamine were from Sigma Chemical Co. Primary and secondary antibody preparations were described previously (20).
Preparation of cleared lysates.
Unless indicated otherwise, cleared lysates were prepared from cells growing exponentially in 150 ml of YM-1 medium. Cultures were poured over ice, and the cells were collected by centrifugation. After two washes with ice-cold membrane buffer (10 mM Tris acetate [pH 7.6], 1 mM magnesium acetate, 0.1 mM EDTA, 8% glycerol, 0.1 mM dithiothreitol) containing 100 μg of phenylmethylsulfonyl fluoride (PMSF)/ml and 10 μg of pepstatin A/ml, the cells were resuspended in 0.5 ml of the same buffer and lysed by mechanical disruption with glass beads. Unbroken cells were removed by centrifugation for 5 min at 330 × g. Cells containing plasmids were cultured in −URA + CAA instead of YM-1. Protein concentrations were determined by using the bicinchoninic acid reagent (Pierce).
Immunoblotting methods and quantitation.
Protein samples were diluted 1:3 with sample buffer containing 50% (wt/wt) urea (20), except for the immunoprecipitation procedure. Samples were heated for 10 min at 37°C and resolved on either sodium dodecyl sulfate (SDS)–10% (to detect Ste4p) or SDS–18% (to detect Ste18p) polyacrylamide gels. Gels were processed for immunoblotting, and the proteins were detected by using a chemiluminescent reagent as described previously (20). Results were quantified by using a densitometer (Molecular Dynamics Corp.) and ImageQuant software.
Renografin density gradients.
Membranes were fractionated on Renografin gradients as described previously (40), except that the gradient contained protease inhibitors (100 μg of PMSF and 10 μg of pepstatin A/ml) and additional protease inhibitors were added to the fractions at the same concentrations. The volume of each fraction loaded on the SDS-polyacrylamide gel was proportional to the size of the fraction.
Immunoprecipitation procedure.
Cleared lysates were diluted to 8.5 mg of protein per ml. Samples (60 μl) were adjusted to contain 2 mg of dodecyl-β-d-maltoside/ml, 0.4 mg of cholesterol hemisuccinate/ml, and 0.25 M NaCl. After 160 min on ice, insoluble material was removed by 10 min of centrifugation in an IEC/MicroMax microcentrifuge. One 30-μl aliquot was mixed with 15 μl of packed protein A-Sepharose beads (Sigma) that had been coated with anti-Ste4p antiserum, and a second 30-μl aliquot was mixed with 15 μl of uncoated beads. Beads and lysates were incubated overnight at 4°C with continuous mixing, collected by centrifugation, and then washed three times with a buffer containing 20 mM Tris acetate (pH 7.6), 1 mM magnesium acetate, 250 mM NaCl, 2 mg of dodecyl-β-d-maltoside/ml, and 0.4 mg of cholesterol hemisuccinate/ml. Immune complexes were eluted from the washed beads by incubation in sample buffer (50 mM Tris-Cl [pH 6.8], 2% SDS, 10% glycerol, bromophenol blue) for 5 min at 65°C. The supernatant fraction (containing the proteins that had not bound to the protein A-Sepharose beads) was mixed with one-half volume of 3× SDS sample buffer (25) and incubated for 5 min at 65°C. Ste18p was detected by immunoblotting methods.
[3H]palmitic acid labeling.
Cultures were labeled with [3H]palmitic acid, and Ste18p was analyzed essentially as described by Song and Dohlman (43) for Gpa1p. Strains W303-1A, HC106S, and BRI1426 were cultured overnight in minimal glucose medium at 30°C and concentrated to 108 cells/ml. A 10-ml volume of culture was incubated for 2 h with 5 mCi of [3H]palmitic acid (50 Ci/mmol) in the presence of the fatty acid synthesis inhibitor cerulenin (2 μg/ml). The levels of incorporation of radioactivity into the cells were 54, 51, and 60%, respectively, for the three cultures. As described above for the immunoprecipitation procedure, the cells were disrupted with glass beads, cleared lysates were prepared, membrane proteins were solubilized with detergent, and complexes containing Ste4p were precipitated with anti-Ste4p antiserum. Half of the preparation was resolved on each of two nonreducing SDS–18% polyacrylamide gels. One gel was fixed and treated with 1 M Tris-HCl, pH 7, and the other was treated with 1 M NH2OH, pH 7, as described elsewhere (43). Gels were processed for autoradiography by using Entensify universal autoradiographic enhancer (NEN Life Sciences) and exposed for 3 months at −80°C. Diploid strain W303 containing plasmids pBH21 and pEL37 and the control strain containing plasmids pBH21 and pRS313 were processed identically except that minimal galactose medium was used for both strains and levels of incorporation of radioactivity were 34 and 33%, respectively.
Hydroxylamine treatment of cleared lysates.
Cleared lysates were prepared as described above except that the cells were disrupted in a buffer containing 10 mM Tris acetate (pH 7.6), 2 mM EDTA, 0.1 mM dithiothreitol, 100 μg of PMSF/ml, and 10 μg of pepstatin A/ml. Samples were diluted to 5 mg of protein per ml, treated with an equal volume of 0.5 M hydroxylamine, incubated for 45 min at 30°C, and then processed for SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting.
RESULTS
Electrophoretic mobilities and relative abundance of the ste18 mutant proteins.
Proteins containing the C-terminal CaaX box motif are subject to a series of posttranslational processing steps consisting of prenylation of the cysteine followed by proteolysis of the aaX residues and carboxylmethylation of the prenylated cysteine. When the CaaX motif contains methionine, serine, or glutamine in the X position, it specifies farnesylation of the cysteine residue, whereas sequences containing leucine in the X position specify geranylgeranylation (reviewed in reference 49). As for mammalian Ras, the yeast Ras1p and Ras2p proteins each contain a farnesyl-directing CaaX box. Most Ras proteins are palmitoylated near the C terminus (16); both of the yeast Ras proteins are palmitoylated at a cysteine immediately adjacent to the prenylated cysteine (4). Palmitoylation depends on prior prenylation (8, 16). Ste18p also contains a farnesyl-directing CaaX box and an adjacent cysteine (Fig. 1A), raising the possibility that both cysteine residues receive lipid modifications. Mutations resulting in replacement of either cysteine by serine lead to extreme defects in mating (11, 15, 48). We sought to determine how these mutations in the CaaX motif change the chemical structure and the biochemical properties of Ste18p. To assess changes in chemical structure, we examined the mobilities of the wild-type and mutant proteins on SDS-polyacrylamide gels. We reasoned that mutations affecting farnesylation at Cys 107 or a second modification at Cys 106 might cause detectable changes in the electrophoretic mobility of Ste18p.
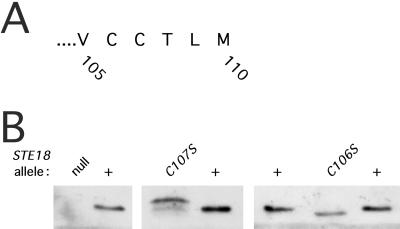
FIG. 1.
SDS-polyacrylamide gel analysis of the ste18 mutant proteins. (A) C-terminal sequence of Ste18p precursor predicted from nucleic acid sequence. (B) Immunoblotting analysis performed on cleared lysates of exponentially growing cells. The STE18 allele designations are listed across the top. Amounts of protein loaded (from left) were as follows: ste18::URA3 (null), 33 μg; STE18+, 5 μg; ste18-C107S, 33 μg; STE18+, 5 μg; STE18+, 5 μg; ste18-C106S, 10 μg; and STE18+, 5 μg. Strains used were BRI1426, W303-1A, HC107S, and HC106S.
Previous studies investigating the electrophoretic mobility of Ste18p were limited to fusion proteins that had been overproduced (11, 13). Under these conditions, the mobility of Ste18p was slower in the ste18-C107Y mutant (13) and in a ram1 mutant defective in farnesyltransferase activity (11). In the present study, we analyzed the mobilities and levels of accumulation of mutant forms of Ste18p that had been expressed under the control of the native promoter at the normal chromosomal location. Because the antiserum was directed against the N terminus of Ste18p (20), it was unnecessary to use an epitope-tagged form of the protein. Cleared lysates were prepared from the various mutant cells, and proteins were resolved by SDS-PAGE and detected by immunoblot analysis (Fig. 1B). The mutant protein encoded by mutant allele ste18-C107S migrated more slowly than wild-type Ste18p. The mobility shift potentially reflected changes in lipid modification, failure to cleave the C-terminal residues, or the amino acid substitution itself. The reduced mobility of the ste18-C107S mutant protein is consistent with the loss of prenylation at Cys 107, as proposed previously (11, 13). In contrast, the ste18-C106S mutant protein migrated more rapidly than wild-type Ste18p, suggesting that this mutation causes a defect in a processing event other than farnesylation.
The relative abundances of the Ste18p proteins in the different mutant and wild-type strains were estimated by comparing the intensities of the Ste18p bands on the immunoblot with respect to the amount of total protein analyzed (Fig. 1B). Only the ste18-C107S mutant showed a consistent reduction in the abundance of Ste18p. In four independent analyses, the ste18-C107S mutant protein was detected at a level that was 10 to 20% of the level of Ste18p from the wild-type control strain. Thus, prenylation of wild-type Ste18p may be essential for maintaining the stability of Gγ; the faster-migrating minor species detected in the ste18-C107S mutant may represent a degradation intermediate. The ste18-C106S mutant protein was relatively abundant (between 25 and 60% of the level of the wild type).
The ste18-C106 mutant protein, but not the ste18-C107 mutant protein, is a substrate for farnesylation.
The electrophoretic mobilities of the Ste18p proteins in the various mutant strains (Fig. 1B) reflect structural differences among the mutant proteins but do not identify the underlying chemical changes. To assess the chemical structure, we examined whether the electrophoretic mobility was sensitive to conditions which either block or remove specific lipid modifications (i.e., farnesylation or palmitoylation). As a test for farnesylation of the ste18-C106S and ste18-C107S mutant proteins, we asked whether the electrophoretic mobility of the mutant protein was sensitive to the presence of the farnesyltransferase encoded by the RAM1 gene. When overexpressed in a ram1 mutant, epitope-tagged Ste18p is known to migrate more slowly on SDS-PAGE gels (11). We examined the effect of the ram1 mutation on the mobilities of overexpressed ste18-C106S and ste18-C107S mutant proteins and wild-type Ste18p (Fig. 2A). High-copy-number derivatives of plasmid M70p2 containing the STE18+, ste18-C106S, and ste18-C107S alleles were introduced into RAM+ and ram1Δ strains that carried a disruption of the chromosomal STE18 gene. Consistent with the previous observation (11), the mobility of wild-type Ste18p was slower in the ram1Δ strain (Fig. 2A). Furthermore, our results indicate that the modification requires Cys 107 since the mobility of the ste18-C107S mutant protein was not affected by the ram1Δ mutation. In contrast, Cys 106 was not required for farnesylation since the mobility of the ste18-C106S mutant protein was strongly affected by the ram1Δ mutation. Thus, when overexpressed, the ste18-C106S but not the ste18-C107S mutant protein is a substrate for farnesylation. Moreover, the modification that occurs at Cys 106 apparently occurs after farnesylation of Cys 107, since the ste18-C106S mutant protein migrates faster than wild-type Ste18p when produced in the RAM1+ strain (Fig. 2A; compare lanes 5 and 7) but not when produced in the ram1Δ strain (compare lanes 4 and 6).
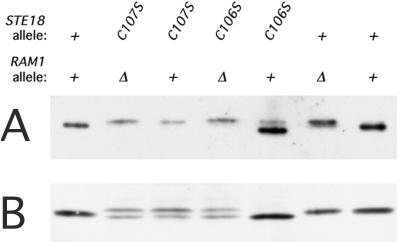
FIG. 2.
Immunoblot analysis of the ste18-C106S and ste18-C107S mutant proteins synthesized in the absence of farnesyltransferase. (A) Mutant and wild-type Ste18p overexpressed in RAM1+ and ram1Δ host strains. The plasmid-bearing strains were grown to exponential phase in 20 ml of selective medium; 0.25-ml volumes of cleared lysates were used for immunoblotting analysis. Lanes were loaded with 3 to 6 μg of total protein. Strains used were 816-1/M70p2, 817-5/M70p2, 816-1/M70p2C106S, 817-5/M70p2C106S, 816-1/M70p2C107S, and 817-5/M70p2C107S. (B) Mutant and wild-type Ste18p expressed at the normal level. Strains containing a single copy of STE18+, ste18-C106S, or ste18-C107S at the normal chromosomal locus were processed as for panel A. Strains used and quantities of total protein analyzed were as follows: W303-1A, 7 μg; 809-A, 12 μg; HC106S, 19 μg; 810-A, 26 μg; HC107S, 41 μg; 811-A, 34 μg; and W303-1A, 7 μg.
We also tested the effect of the ram1Δ mutation on the mutant Ste18p proteins when the allele contained the native promoter at the normal chromosomal location (Fig. 2B). As for the overproduced proteins, the ste18-C106S mutant protein, but not the ste18-C107S mutant protein, showed an electrophoretic pattern that was altered in the ram1 mutant. However, unlike the overproduced protein, relatively minor changes in mobility were observed for wild-type Ste18p from the ram1 mutant. Moreover, the Ste18p from the STE18+ ram1 strain was more abundant than that from the ste18-C107S strain, it did not exhibit the same degradation product, and it did not migrate to the same position. Thus, in the absence of farnesyltransferase activity, wild-type Ste18p received a modification that was not detected when the protein was overproduced; this modification required the presence of both Cys 106 and Cys 107 (Fig. 2B; compare lanes 2, 4, and 6). Possible modifying activities include either geranylgeranyltransferase type I (GGTase I) or GGTase II. Cross-specificity of the farnesyltransferase and GGTase I enzymes has been reported (32, 45, 49).
Palmitoyl modification at Cys 106.
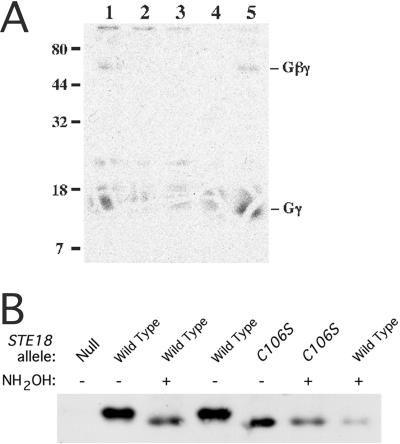
We considered the possibility that Cys 106 is a site for palmitoylation and that the increased mobility of the ste18-C106S mutant protein is due to the loss of this modification. Palmitoylation of cysteine residues occurs through a thioester linkage, which is cleaved by treatment with neutral hydroxylamine (27). As a preliminary test for palmitoylation of Ste18p, we examined whether Ste18p becomes labeled when cells are cultured in the presence of radioactive palmitic acid. The Gβγ complexes that had been solubilized with detergent and precipitated with anti-Ste4p antiserum were resolved by nonreducing SDS-PAGE (Fig. 3A). Nonreducing conditions were maintained to avoid potential cleavage of the thioester linkage by thiol reducing agents. Wild-type cells (lane 1) gave two radioactive species that were not observed for the ste18-C106S and ste18::URA3 control cells (lanes 2 and 3, respectively). The molecular masses of these species were consistent with Gγ (13 kDa) and Gβγ (60 kDa) dimers. Similar labeled species were obtained with cells overproducing Ste4p and Ste18p (lane 5), but not with control cells lacking Ste4p (lane 4). Nearly all of the label was released from these species when the gel was incubated in hydroxylamine (data not shown). These results are consistent with at least some of the Ste18p molecules containing a palmitoyl modification. To determine whether the bulk of the Ste18p molecules contained a hydroxylamine-sensitive modification at Cys 106, we tested whether the electrophoretic mobilities of Ste18p and the ste18-C106S mutant were altered by hydroxylamine treatment (Fig. 3B). If the aberrant mobility of the mutant protein was due to its failure to receive a palmitate moiety, then the mobility of the wild-type protein, but not that of the mutant protein, should increase upon hydroxylamine treatment. Cleared lysates were incubated with neutral hydroxylamine and then processed for SDS-PAGE. The mobility of Ste18p, but not that of the ste18-C106S mutant protein, increased following hydroxylamine treatment; the treated wild-type protein comigrated with the mutant protein. This result indicates that essentially all Ste18p molecules contain a thioester linkage at Cys 106.
FIG. 3.
Evidence for palmitoylation of Ste18p. (A) [3H]palmitate labeling of Ste18p. A nondenaturing detergent was used to extract proteins from cells that had been cultured in the presence of [3H]palmitic acid, Gβγ complexes were precipitated with anti-Ste4p antiserum, and the proteins were resolved by nonreducing SDS-PAGE and then detected by autoradiography. Strains used were as follows: lane 1, wild-type haploid cells (W303-1A); lane 2, ste18-C106S mutant cells (HC106); lane 3, ste18::URA3 cells (BRI1426); lane 4, MATa/MATα diploid control cells (W303) expressing Ste18p (plasmid pBH21) but not Ste4p; and lane 5, MATa/MATα diploid cells (W303) expressing both Ste18p and Ste4p (plasmids pBH21 and pEL37, respectively). (B) Thioester-linked modification of Cys 106. Cleared lysates of strains containing STE18+ (Wild Type), ste18-C106S (C106S), or ste18Δ::URA3 (Null) were either untreated (−) or treated with hydroxylamine (+) as indicated. Ste18p was analyzed by SDS-PAGE and immunoblotting. Strains used were W303-1A, HC106S, and BRI1426.
ste18 mutant proteins form Gβγ complexes with Ste4p.
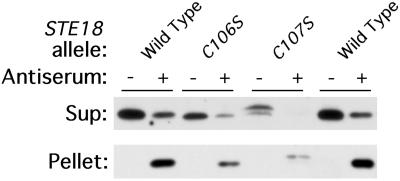
Lipid modifications of proteins potentially influence their interaction with membranes or with other proteins. We used coimmunoprecipitation to test whether formation of the Gβγ complex requires lipid modification of Ste18p. Cleared lysates from the ste18 mutant and wild-type control cells were extracted with the detergent dodecyl-β-d-maltoside and then incubated with anti-Ste4p antiserum. The supernatant fractions and pellet fractions were assayed for Ste18p by immunoblotting methods (Fig. 4). Each mutant Ste18p was found in the pellet fraction after Ste4p had been immunoprecipitated with anti-Ste4p antibody, thus indicating the presence of Gβγ complexes. Quantification of the amount of Ste18p remaining in the supernatant after antibody treatment indicated that immunoprecipitation of each mutant protein was as efficient as that of the wild-type protein (about 80%). This experiment demonstrates that changes in the lipid modification of Ste18p do not prevent Gβγ complex formation. Similar results have been obtained for mammalian Gβγ subunits (22, 31, 42).
FIG. 4.
Mutant ste18 proteins form Gβγ complexes. Detergent-solubilized lysates were incubated with protein A-Sepharose beads that were either uncoupled (−) or had been precoupled with anti-Ste4p antiserum (+). The presence of Ste18p in the supernatant (Sup) and pellet fractions was tested by SDS-PAGE and immunoblotting. Strains used were W303-1A, HC106S, and HC107S.
Both Cys 106 and Cys 107 are required for stable association of Gβγ with membranes.
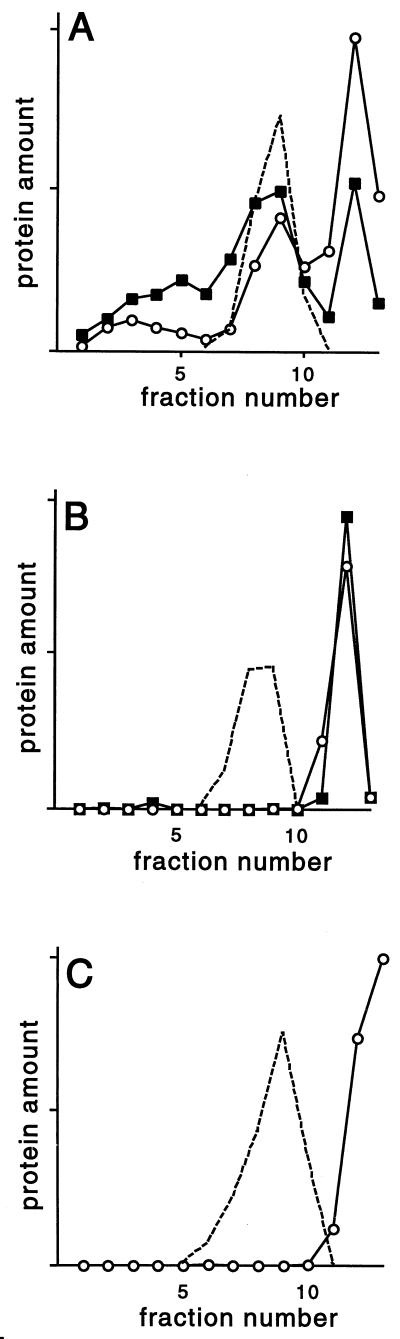
To test whether the ste18 mutations result in changes in the membrane localization of Gβγ, we fractionated membranes from the various ste18 mutants on Renografin density gradients and assayed the fractions for Ste4p and Ste18p. Renografin density gradients resolve plasma membranes, internal membranes, and nonmembrane proteins (23, 40). Previously, we have shown that Ste4p and Ste18p from wild-type cells are associated with all three fractions (20). However, in the ste18-C106S mutant, Ste4p and Ste18p were not associated with either membrane fraction (Fig. 5B). Since the ste18-C106S mutant protein is apparently farnesylated (Fig. 2), our results indicate that farnesylation of Ste18p is not sufficient to promote stable association of Gβγ with membranes; we cannot rule out the possibility that the mutant protein is associated weakly with membranes in vivo and is released during analysis. In the ste18-C107S mutant, the Gβγ complexes were no longer associated with plasma membranes or with internal membranes (Fig. 5C). Because the ste18-C107S mutant protein was not as abundant as the wild-type Ste18p, we were unable to evaluate its distribution in the gradient. When a strain (816-1/M70p2C107S) that overproduces the ste18-C107S mutant protein was examined, Ste4p and the mutant Ste18p were detected only in the dense fractions that contained no membranes (data not shown). The ste18-C107S and ste18-C106S mutant proteins, which formed Gβγ complexes but failed to associate with membranes (Fig. 5), also failed to promote the pheromone response and mating (48).
FIG. 5.
Association of mutant and wild-type Ste18p proteins with internal and plasma membranes. Membranes from mutant and wild-type strains were fractionated on a Renografin gradient. Fractions were assayed for Ste4p (open circles), Ste18p (filled squares), and plasma membrane ATPase (dashed line) by immunoblotting methods. The ste18-C107S mutant protein was below the detectable level in the gradient fractions. (A) Strain W303-1A (STE18+). (B) Strain HC106S (ste18-C106S). (C) Strain HC107S (ste18-C107S).
DISCUSSION
This study explored the role that covalent lipid modifications play in the localization of the yeast Gβγ complex. Previous workers established that residues of the CaaX motif and the neighboring cysteine residue (Cys 106) are important for the activity of Ste18p (15, 48) and that Ste18p receives a farnesyl modification (11). Our results provide evidence for a thioester lipid modification at Cys 106. This lipid modification is likely to be palmitoyl since yeast Ras is palmitoylated at a similar position and since Ste18p can be labeled with [3H]palmitic acid in a manner that is hydroxylamine sensitive and dependent on Cys 106. As for mammalian G proteins (30, 42), prenylation of yeast Gγ is essential for membrane attachment but not for binding Gβ. Unlike other known Gγ subunits, Ste18p apparently requires an additional palmitoyl modification for localization and function.
Ste18p is similar to the yeast Ras proteins (encoded by RAS1 and RAS2) in several respects. Both proteins contain a cysteine immediately adjacent to a farnesyl-directing CaaX motif, and the palmitoyl moiety is added only after farnesylation (8). Like Ras (1, 4), palmitoylation of Ste18p is essential for localization to the plasma membrane. Both unpalmitoylated Ras (1, 4, 6) and ste18-C107S mutant protein (48) exhibit biological activity that is significantly reduced but not eliminated. Thus, the membrane localization that is afforded by the palmitoyl moiety appears to facilitate interaction with other components of the signal transduction pathway but is not absolutely required. In yeast, the same farnesyltransferase (α and β subunits, encoded by RAM2 and RAM1, respectively) operates on Ras, Ste18p, and the a-factor pheromone (11, 12, 19, 34).
Ste18p is unusual among Gγ proteins in that the C-terminal isoprenyl group is farnesyl. Although transducin γ (13) and, apparently, γ11 (36) are substrates for farnesyltransferase, all other Gγ subunits are modified by GGTase I (49). We are unaware of other Gγ subunits that contain Cys adjacent to the prenylation site. Previous genetic tests (48) suggest that farnesylation is not absolutely required for Ste18p function, since the ram1 mutant, which lacks farnesyltransferase, shows only a minor reduction in pheromone responsiveness. Moreover, mutant Ste18p proteins from these strains showed only slightly slower electrophoretic mobilities than Ste18p from wild-type cells, yet the mobility was faster than the ste18-C107S mutant protein or the Ste18p that had been overproduced in the ram1 mutant (Fig. 1 and 2). Together these results raise the possibility that an enzyme other than farnesyltransferase modifies Cys 107 and that the activity modifies a significant portion of Ste18p when overproduced. Possible modifying activities include either GGTase I or GGTase II. GGTase I potentially modifies Ste18p in the ram1 mutant, since farnesyltransferase and GGTase I exhibit some cross-specificity (32, 45, 49). However, the alternative modification that operates on wild-type Ste18p in the ram1 mutant apparently requires both Cys 106 and Cys 107 (Fig. 2B). This result suggests a role for GGTase II, since this enzyme has been shown to modify both paired cysteines within several C-terminal motifs (−XXCC, −XCXC, or −CCXX) that are found among Rab proteins (9); however, potential substrates containing the motif −CCXXX have not been examined. The partial activity of the ste18 truncation mutant lacking the three C-terminal residues (i.e., containing −XXCC) (48) is consistent with modification by GGTase II. Association of yeast casein kinase I (Yck1p) with the plasma membrane also depends on the motif −XXCC (46). However, a potential problem for the proposal that GGTase II modifies Ste18p and Yck1p is that Rab proteins are substrates for GGTase II only when they have bound the guanine nucleotide dissociation inhibitor-like protein REP1, and short peptides containing the prenylation motif are not recognized by GGTase II (see reference 49). Conceivably, either the paired cysteine motif in Ste18p and Yck1p occurs within a context that permits GGTase II recognition or another, unidentified enzyme or auxiliary factor operates on these substrates.
How does dual lipid modification mediate membrane association of yeast Gβγ? The failure of the ste18-C106S mutant protein to accumulate on membranes indicates that palmitoylation either provides a signal for targeting Ste18p to membranes or contributes to the affinity of Ste18p for membranes. In vitro, synthetic peptides containing both palmitoyl and farnesyl show very slow rates of intermembrane transfer (T1/2 > 50 h) compared with the singly modified peptides (41). According to the bilayer trapping mechanism (2, 38, 41), farnesylation of Ste18p may promote weak membrane interactions, and upon association with a membrane compartment containing palmitoyltransferase, Ste18p may become palmitoylated and, thus, anchored at that site. The location of the palmitoyltransferase for Ste18p is unknown. In mammalian cells, palmitoyltransferase in the plasma membrane modifies Gα (7) whereas an activity in Golgi membranes palmitoylates a farnesylated form of Ras (16). Roles for palmitoylation in both membrane targeting and membrane affinity have been described. Palmitoylation of SNAP-25 is necessary for localization of newly synthesized protein at the plasma membrane but is not required for maintaining fully assembled protein at the membrane, since brefeldin A blocks both palmitoylation and membrane targeting of newly synthesized SNAP-25 and since hydroxylamine hydrolyzes the thioacyl linkage without affecting membrane attachment (14). In contrast, brefeldin A does not block assembly of mammalian Gβγ on the plasma membrane (37), and Ras protein is removed from the plasma membrane upon hydroxylamine treatment (28).
After ligand stimulation and release from Gα, yeast Gβγ (26, 35) as well as many of its mammalian Gβγ isoforms (3, 5, 21, 24) are thought to stimulate the activity of effector molecules located on the plasma membrane. The role that Gβγ plays in signal transduction may be simply to promote assembly of effector molecules at the plasma membrane (35). Thus, dissociation of Gβγ from the plasma membrane provides a possible mechanism for regulating the duration of Gβγ signaling activity. Dissociation could be a consequence of depalmitoylation; evidence for reversible palmitoylation of mammalian Ras exists, and palmitoylation of Ras apparently regulates plasma membrane attachment (28). Alternatively, a carrier protein that binds the Gβγ complex may shield the lipid groups from the aqueous environment and thereby permit dissociation from the membrane. Guanine nucleotide dissociation inhibitor functions as such a carrier during recycling of geranylgeranylated Rab proteins (10, 33). An internal membrane compartment may provide a site where Gβγ can reassociate with Gα before it is reinstated on the plasma membrane. If relevant, repalmitoylation could occur either in this internal compartment or at the plasma membrane. Clearly, evaluation of these models will require determination of the palmitoylation state of Gβγ and the presence of specific binding proteins in the various subcellular compartments.
ACKNOWLEDGMENTS
We thank Malcolm Whiteway for providing strains and plasmids, Ayce Yesilaltay and Aidan Hennigan for comments on the manuscript, and C. L. Manahan and M. E. Linder for communicating results prior to publication.
This investigation was supported by Public Health Service research grant GM34719 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Bhattacharya S, Chen L, Broach J R, Powers S. Ras membrane targeting is essential for glucose signaling but not for viability in yeast. Proc Natl Acad Sci USA. 1995;92:2984–2988. doi: 10.1073/pnas.92.7.2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cadwallader K A, Paterson H, Macdonald S G, Hancock J F. N-terminally myristoylated Ras proteins require palmitoylation or a polybasic domain for plasma membrane localization. Mol Cell Biol. 1994;14:4722–4730. doi: 10.1128/mcb.14.7.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J Q, Devivo M, Dingus J, Harry A, Li J R, Sui J L, Carty D J, Blank J L, Exton J H, Stoffel R H, Inglese J, Lefkowitz R J, Logothetis D E, Hildebrandt J D, Iyengar R. A region of adenylyl cyclase 2 critical for regulation by G protein βγ subunits. Science. 1995;268:1166–1169. doi: 10.1126/science.7761832. [DOI] [PubMed] [Google Scholar]
- 4.Deschenes R J, Broach J R. Fatty acylation is important but not essential for Saccharomyces cerevisiae RAS function. Mol Cell Biol. 1987;7:2344–2351. doi: 10.1128/mcb.7.7.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeWaard M, Liu H Y, Walker D, Scott V E S, Gurnett C A, Campbell K P. Direct binding of G-protein βγ complex to voltage-dependent calcium channels. Nature. 1997;385:446–450. doi: 10.1038/385446a0. [DOI] [PubMed] [Google Scholar]
- 6.Dudler T, Gelb M H. Palmitoylation of Ha-Ras facilitates membrane binding, activation of downstream effectors, and meiotic maturation in Xenopus oocytes. J Biol Chem. 1996;271:11541–11547. doi: 10.1074/jbc.271.19.11541. [DOI] [PubMed] [Google Scholar]
- 7.Dunphy J T, Greentree W K, Manahan C L, Linder M E. G-protein palmitoyltransferase activity is enriched in plasma membranes. J Biol Chem. 1996;271:7154–7159. doi: 10.1074/jbc.271.12.7154. [DOI] [PubMed] [Google Scholar]
- 8.Farh L, Mitchell D A, Deschenes R J. Farnesylation and proteolysis are sequential, but distinct steps in the CaaX box modification pathway. Arch Biochem Biophys. 1995;318:113–121. doi: 10.1006/abbi.1995.1211. [DOI] [PubMed] [Google Scholar]
- 9.Farnsworth C C, Seabra M C, Ericsson L H, Gelb M H, Glomset J A. Rab geranylgeranyl transferase catalyzes the geranylgeranylation of adjacent cysteines in the small GTPases Rab1A, Rab3A, and Rab5A. Proc Natl Acad Sci USA. 1994;91:11963–11967. doi: 10.1073/pnas.91.25.11963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferro-Novick S, Novick P. The role of GTP-binding proteins in transport along the exocytic pathway. Annu Rev Cell Biol. 1993;9:575–599. doi: 10.1146/annurev.cb.09.110193.003043. [DOI] [PubMed] [Google Scholar]
- 11.Finegold A A, Schafer W R, Rine J, Whiteway M, Tamanoi F. Common modifications of trimeric G proteins and ras protein: involvement of polyisoprenylation. Science. 1990;249:165–169. doi: 10.1126/science.1695391. [DOI] [PubMed] [Google Scholar]
- 12.Fujiyama A, Matsumoto K, Tamanoi F. A novel yeast mutant defective in the processing of ras proteins: assessment of the effect of the mutation on processing steps. EMBO J. 1987;6:223–228. doi: 10.1002/j.1460-2075.1987.tb04742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuda Y, Takao T, Ohguro H, Yoshizawa T, Akino T, Shimonishi Y. Farnesylated γ subunit of photoreceptor G protein indispensable for GTP binding. Nature. 1990;346:658–660. doi: 10.1038/346658a0. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalo S, Linder M E. SNAP-25 palmitoylation and plasma membrane targeting require a functional secretory pathway. Mol Biol Cell. 1998;9:585–597. doi: 10.1091/mbc.9.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grishin A V, Weiner J L, Blumer K J. Biochemical and genetic analysis of dominant-negative mutations affecting a yeast G-protein γ subunit. Mol Cell Biol. 1994;14:4571–4578. doi: 10.1128/mcb.14.7.4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hancock J F, Magee A I, Childs J E, Marshall C J. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 17.Hartwell L H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967;93:1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasson M S, Blinder D, Thorner J, Jenness D D. Mutational activation of the STE5 gene product bypasses the requirement for G protein β and γ subunits in the yeast pheromone response pathway. Mol Cell Biol. 1994;14:1054–1065. doi: 10.1128/mcb.14.2.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He B, Chen P, Chen S Y, Vancura K L, Michaelis S, Powers S. RAM2, an essential gene of yeast, and RAM1 encode the two polypeptide components of the farnesyltransferase that prenylates a-factor and Ras proteins. Proc Natl Acad Sci USA. 1991;88:11373–11377. doi: 10.1073/pnas.88.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirschman J E, DeZutter G S, Simonds W F, Jenness D D. The Gβγ complex of the yeast pheromone response pathway: subcellular fractionation and protein-protein interactions. J Biol Chem. 1997;272:240–248. doi: 10.1074/jbc.272.1.240. [DOI] [PubMed] [Google Scholar]
- 21.Inanobe A, Morishige K I, Takahashi N, Ito H, Yamada M, Takumi T, Nishina H, Takahashi K, Kanaho Y, Katada T, Kurachi Y. Gβγ directly binds to the carboxyl terminus of the G protein-gated muscarinic K+ channel, GIRK1. Biochem Biophys Res Commun. 1995;212:1022–1028. doi: 10.1006/bbrc.1995.2072. [DOI] [PubMed] [Google Scholar]
- 22.Iniguez-Lluhi J A, Simon M I, Robishaw J D, Gilman A G. G protein βγ subunits synthesized in Sf9 cells. Functional characterization and the significance of prenylation of γ. J Biol Chem. 1992;267:23409–23417. [PubMed] [Google Scholar]
- 23.Jenness D D, Li Y, Tipper C, Spatrick S. Elimination of defective α-factor pheromone receptors. Mol Cell Biol. 1997;17:6236–6245. doi: 10.1128/mcb.17.11.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krapivinsky G, Krapivinsky L, Wickman K, Clapham D E. Gβγ binds directly to the G protein-gated K+ channel, IKACh. J Biol Chem. 1995;270:29059–29062. doi: 10.1074/jbc.270.49.29059. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Leeuw T, Wu C, Schrag J D, Whiteway M, Thomas D Y, Leberer E. Interaction of a G-protein β-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature. 1998;391:191–195. doi: 10.1038/34448. [DOI] [PubMed] [Google Scholar]
- 27.Linder M E, Kleuss C, Mumby S M. Palmitoylation of G-protein α subunits. Methods Enzymol. 1995;250:314–330. doi: 10.1016/0076-6879(95)50081-2. [DOI] [PubMed] [Google Scholar]
- 28.Magee A I, Gutierrez L, McKay I A, Marshall C J, Hall A. Dynamic fatty acylation of p21N-ras. EMBO J. 1987;6:3353–3357. doi: 10.1002/j.1460-2075.1987.tb02656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manahan, C. L., and M. E. Linder. Personal communication.
- 30.Muntz K H, Sternweis P C, Gilman A G, Mumby S M. Influence of γ subunit prenylation on association of guanine nucleotide-binding regulatory proteins with membranes. Mol Biol Cell. 1992;3:49–61. doi: 10.1091/mbc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nern A, Arkowitz R A. A GTP-exchange factor required for cell orientation. Nature. 1998;391:195–198. doi: 10.1038/34458. [DOI] [PubMed] [Google Scholar]
- 32.Ohya Y, Qadota H, Anraku Y, Pringle J R, Botstein D. Suppression of yeast geranylgeranyl transferase I defect by alternative prenylation of two target GTPases, Rho1p and Cdc42p. Mol Biol Cell. 1993;4:1017–1025. doi: 10.1091/mbc.4.10.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfeffer S R, Dirac-Svejstrup A B, Soldati T. Rab GDP dissociation inhibitor: putting Rab GTPases in the right place. J Biol Chem. 1995;270:17057–17059. doi: 10.1074/jbc.270.29.17057. [DOI] [PubMed] [Google Scholar]
- 34.Powers S, Michaelis S, Broek D, Santa Anna S, Field J, Herskowitz I, Wigler M. RAM, a gene of yeast required for a functional modification of RAS proteins and for production of mating pheromone a-factor. Cell. 1986;47:413–422. doi: 10.1016/0092-8674(86)90598-2. [DOI] [PubMed] [Google Scholar]
- 35.Pryciak P M, Huntress F A. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ray K, Kunsch C, Bonner L M, Robishaw J D. Isolation of cDNA clones encoding eight different human G protein γ subunits, including three novel forms designated the γ4, γ10, and γ11 subunits. J Biol Chem. 1995;270:21765–21771. doi: 10.1074/jbc.270.37.21765. [DOI] [PubMed] [Google Scholar]
- 37.Rehm A, Ploegh H L. Assembly and intracellular targeting of the βγ subunits of heterotrimeric G proteins. J Cell Biol. 1997;137:305–317. doi: 10.1083/jcb.137.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Resh M D. Regulation of cellular signalling by fatty acid acylation and prenylation of signal transduction proteins. Cell Signal. 1996;8:403–412. doi: 10.1016/s0898-6568(96)00088-5. [DOI] [PubMed] [Google Scholar]
- 39.Schafer W R, Trueblood C E, Yang C C, Mayer M P, Rosenberg S, Poulter C D, Kim S H, Rine J. Enzymatic coupling of cholesterol intermediates to a mating pheromone precursor and to the ras protein. Science. 1990;249:1133–1139. doi: 10.1126/science.2204115. [DOI] [PubMed] [Google Scholar]
- 40.Schandel K A, Jenness D D. Direct evidence for ligand-induced internalization of the yeast α-factor pheromone receptor. Mol Cell Biol. 1994;14:7245–7255. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shahinian S, Silvius J R. Doubly-lipid-modified protein sequence motifs exhibit long-lived anchorage to lipid bilayer membranes. Biochemistry. 1995;34:3813–3822. doi: 10.1021/bi00011a039. [DOI] [PubMed] [Google Scholar]
- 42.Simonds W F, Butrynski B J E, Gautam N, Unson C G, Spiegel A M. G-protein βγ dimers. Membrane targeting requires subunit coexpression and intact gamma C-A-A-X domain. J Biol Chem. 1991;266:5363–5366. [PubMed] [Google Scholar]
- 43.Song J P, Dohlman H G. Partial constitutive activation of pheromone responses by a palmitoylation-site mutant of a G protein α subunit in yeast. Biochemistry. 1996;35:14806–14817. doi: 10.1021/bi961846b. [DOI] [PubMed] [Google Scholar]
- 44.Stone D E, Cole G M, Lopes M D, Goebl M, Reed S I. N-Myristoylation is required for function of the pheromone-responsive Gα protein of yeast: conditional activation of the pheromone response by a temperature-sensitive N-myristoyl transferase. Genes Dev. 1991;5:1969–1981. doi: 10.1101/gad.5.11.1969. [DOI] [PubMed] [Google Scholar]
- 45.Trueblood C E, Ohya Y, Rine J. Genetic evidence for in vivo cross-specificity of the CaaX-box protein prenyltransferases farnesyltransferase and geranylgeranyltransferase-I in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:4260–4275. doi: 10.1128/mcb.13.7.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vancura A, Sessler A, Leichus B, Kuret J. Prenylation motif is required for plasma membrane localization and biochemical function of casein kinase I in budding yeast. J Biol Chem. 1994;269:19271–19278. [PubMed] [Google Scholar]
- 47.Wedegaertner P B, Wilson P T, Bourne H R. Lipid modifications of trimeric G proteins. J Biol Chem. 1995;270:503–506. doi: 10.1074/jbc.270.2.503. [DOI] [PubMed] [Google Scholar]
- 48.Whiteway M S, Thomas D Y. Site-directed mutations altering the CAAX box of Ste18, the yeast pheromone-response pathway G gamma subunit. Genetics. 1994;137:967–976. doi: 10.1093/genetics/137.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang F L, Casey P J. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]