Abstract
Background:
A recent kinetic model proposed a new individualized glycaemic marker, calculated HbA1c (cHbA1c), based on kinetic parameters and glucose levels that are specific to each person. The aims of the current work were to validate the accuracy of this glucose metric for clinical use and evaluate data requirements for the estimation of personal kinetic factors.
Methods:
We retrieved HbA1c and glucose data from a group of 51 Japanese T1D patients under sensor-augmented pump (SAP) therapy. Two patient-specific kinetic parameters were identified by data sections, defined as continuous glucose data between two laboratory HbA1c measurements. The cHbA1c was prospectively validated employing subsequent HbA1c data that were not originally used to determine personal kinetic parameters.
Results:
Compared to estimated HbA1c (eHbA1c) and glucose management indicator (GMI), cHbA1c showed clinically relevant accuracy improvement, with 20% or more within ±0.5% (±5.5 mmol/mol) of laboratory HbA1c. The mean absolute deviation of the cHbA1c calculation was 0.11% (1.2 mmol/mol), substantially less than for eHbA1c and GMI at 0.54% (5.9 mmol/mol) and 0.47% (5.1 mmol/mol), respectively.
Conclusion:
Our study shows superior performance of cHbA1c compared with eHbA1c and GMI at reflecting laboratory HbA1c, making it a credible glucose metric for routine clinical use.
Keywords: Glycated hemoglobin, continuous glucose monitoring, kinetic modeling, red blood cell turnover, red blood cell glucose uptake, red blood cell lifespan
Key messages
We validated a novel kinetic model for estimating HbA1c values in an Asian cohort.
The calculated HbA1c has the potential to replace laboratory HbA1c.
The kinetic model provides a method to estimate individual red blood cell lifespan.
Introduction
The role of glycated hemoglobin (HbA1c) to track and estimate the risk of diabetes-related complications has been established by landmark clinical trials,1,2 and has been universally adopted to guide clinical care. However, there are limitations to HbA1c as it is affected by conditions that alter red blood cell (RBC) survival, such as anemia, use of drugs that stimulate erythropoiesis, and kidney disease.3
RBC production and removal are in balance during homeostasis, with the production in the bone marrow4 and removal in the spleen.5,6 These complex mechanisms result in varying RBC survival, and thus their exposure to circulating glucose levels that in turn determine intracellular hemoglobin glycation and hence HbA1c levels. Experimental evidence has shown there is a variation of mean RBC lifespan between hematologically normal individuals,7 but accurate assessment of RBC lifespan is both difficult and time-consuming, beyond the capability of routine diabetes management.8 Further, besides individual variation, there are growing indications for different RBC lifespan across ethnic groups,9,10 making a further understanding of HbA1c glycation processes key for adequate diabetes management.
Beyond RBC survival, the second key variable factor in determining HbA1c is the facilitated cross-membrane transport of glucose into RBCs by GLUT1 transporters.11 The majority of glucose is consumed by the Embden–Meyerhof–Parnas pathway to support the energy requirements of the RBC.4 The fraction of glucose that binds irreversibly to hemoglobin, resulting in “glycated hemoglobin,” is detected via the HbA1c assay.12
Recent work has proposed a novel relationship between the time-course of glucose concentration and HbA1c that takes RBC survival and glucose uptake into account.13 This kinetic model incorporates patient-specific parameters of red blood cell production, elimination (i.e. RBC lifespan), and the apparent hemoglobin glycation rate governed by glucose transport across RBC membrane, with all controlling intracellular glycation of the hemoglobin molecule. The model has been developed and validated using data from European clinical trial cohorts and one specific continuous glucose monitor (CGM) technology (FreeStyle Libre®, Abbott Diabetes Care). Due to the potential to affect clinical decisions, the model needs additional verification across various patient groups and using different CGM technologies. In this study, we validated the model for the first time with data from an Asian cohort and Medtronic MiniMed™ 640G CGM device. Due to the high consistency of the data, we have also been able to evaluate the data requirements for reliable estimation of HbA1c and the kinetic constants.
Methods
Data acquisition
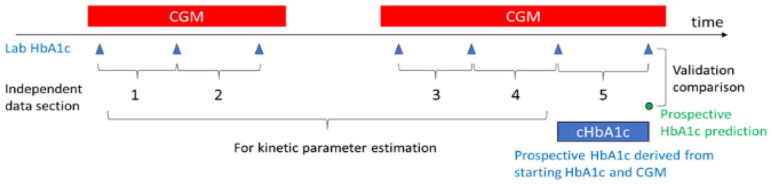
The kinetic model takes one or more data sections to estimate patient-specific kinetic parameters. Each data section consists of a frequent glucose trace (at least every 15 min) between two laboratory HbA1c values at least two weeks apart. To ensure acceptable accuracy of estimates, we required that at least 80% of CGM data points should be present, and any continuous gap should be less than 24-h within a data section. The final data section of each subject was excluded from the parameter estimation. The parameters were then fixed and used to prospectively calculate an HbA1c value (termed “cHbA1c”) for comparison to the final laboratory HbA1c. We required that each individual had a total of three or more data-sections and therefore at least two for parameter estimation. Figure 1 is an example of data sections and prospective evaluation for an individual.
Figure 1.
Illustration of data sections, kinetic parameter estimation, and prospective evaluation.
In this cohort, all subjects had type 1 diabetes treated with the sensor-augmented pump (SAP) from Kobe University Hospital in Japan. All glucose readings were collected by a capillary blood-calibrated CGM sensor (Enlite™, Metronic). HbA1c values were measured by a central laboratory (Kobe University Hospital, HPLC with Arkray HA8181). This analysis received Kobe University’s ethical approval (B190322). Within available data collected by Kobe University, 51 subjects met the pre-specified quality and sufficiency criteria for analysis (Table 1).
Table 1.
Subject and data descriptions.
| Subject count | 51 | |
| Gender M/F | 14/37 | |
| Age (years): median [IQR] [range] | 42 [37–55] [6–73] | |
| CGM usage per subject (days): median [IQR] [range] | 440 [176–489] [112–541] | |
| Data section count per subject: median [IQR] [range] | 13 [6–15] [3–17] | |
| Ending HbA1c (%) | Median [IQR] | 6.9 [6.6–7.5] |
| Mean (STD) | 7.1 (0.96) | |
| Ending 14-day average glucose (mg/dL) | Median [IQR] | 143 [127–160] |
| Mean (STD) | 145 (27) | |
Statistical and computation methods
For each individual, two kinetic parameters were calculated using the kinetic model with all data sections except the last. These parameters are RBC turnover rate kage (or RBC lifespan = 1/ kage) and the apparent hemoglobin glycation rate kgly (dominated by cross-membrane glucose uptake). As shown in Figure 1, the prospective use of the model with the kinetic parameters on the final data section produced cHbA1c throughout the data section and comparison was at the day aligned with laboratory HbA1c. Both kinetic parameter estimation and prospective cHbA1c calculations were performed according to previous publication,13 which is listed in the supplemental materials for convenience.
For comparison to final laboratory HbA1c, the corresponding estimated HbA1c (eHbA1c)14 and glucose management indicator GMI15 values were determined by 14-day average CGM glucose (AG). The performances of these methods were compared by the agreements between estimated and laboratory HbA1c values. Specifically, the absolute deviation and R2 values from Pearson’s correlation of linear regression were compared.
Distributions were characterized by the mean and standard deviation for normally distributed data and by median and interquartile range for non-normally distributed data. Any glucose trace gaps less than 45 min had missing values imputed with the nearest observation or average of nearest observations if both were available (the observations immediately before or after the gap). For a longer gap, each missing value was imputed with the average of the observations at the same time in previous and next days. Python/SciPy16 was used for all analyses.
Based on the model,13 HbA1c is sensitive to kgly and kage during or after a significant day-to-day glucose change. In a period of steady day-to-day glucose, HbA1c is only sensitive to the ratio of kgly and kage. For this reason, it is harder to estimate kinetic parameters than their ratio. As a consequence, a reasonable HbA1c prediction, for steady-state, can be provided when only the ratio of kgly and kage is available. Therefore, fewer data sections are usually required for HbA1c prediction than RBC lifespan (or kage) estimation. Also, since the individual ratio of kgly and kage is usually easy to determine, we can estimate kgly when kage is available and vise versa.
Since the model also assumes no kgly and kage change during the study period, we defined a higher confidence group for subjects with more day-to-day glucose change (between-day glucose CV > 17%), and no major life/therapeutic changes that affect RBC metabolism. These changes include childbirth, iron deficiency treatment, hospitalization, and major drug changes. From the higher confidence group, those with more than 10 data sections were evaluated further to examine the effect of increasing the number of data sections to improve the accuracy of kinetic parameters and HbA1c estimations. By sequentially including additional data sections, we calculated mean absolute deviations to the final RBC lifespan and laboratory HbA1c for each individual. This should set an expectation on the number of data sections required for accurate estimation of HbA1c and individual RBC lifespan.
Results
Prospectively calculated HbA1c and validation of the method
Prospective use of the model with patient-specific kinetic constants produced significantly more accurate predictions of the laboratory HbA1c compared to eHbA1c and GMI. Table 2 lists the comparison metrics of HbA1c estimation using the kinetic model, eHbA1c, and GMI. The kinetic model had the smallest median and mean absolute deviation of 0.10% and 0.11% (1.1 and 1.2 mmol/mol), respectively. The mean absolute deviations from eHbA1c and GMI were significantly larger (p < 0.001), approximately four to five times as large. As an HbA1c difference of 0.5% (5.5 mmol/mol) is usually considered clinically relevant, the rates of correspondence within this range were evaluated. The cHbA1c has minimal clinically relevant deviation with 92.3% of individuals within 0.5% (5.5 mmol/mol), compared to eHbA1c and GMI at 65.5% and 73.1% of individuals, respectively.
Table 2.
Accuracy comparison among HbA1c estimation methods.
| Method | cHbA1c | eHbA1c(AG) | GMI(AG) | ||
|---|---|---|---|---|---|
| Comparing estimated HbA1c against lab HbA1c | Absolute deviation (%) | Mean (STD) | 0.11 (0.06) | 0.54 (0.47) | 0.47 (0.46) |
| Median [IQR] | 0.10 [0.07, 0.13] | 0.42 [0.21, 0.81] | 0.36 [0.18, 0.62] | ||
| Absolute deviation (mmol/mol) | Mean (STD) | 1.2 (0.7) | 5.9 (5.1) | 5.1 (5.0) | |
| Median [IQR] | 1.1 [0.8, 1.4] | 4.6 [2.3, 8.9] | 3.9 [2.0, 6.8] | ||
| MARD (%) | 3.1 | 7.5 | 6.3 | ||
| Fraction of AD < 0.5% (AD < 5.5 mmol/mol) (%) | 92.3 | 65.4 | 73.1 | ||
| Average bias (%) | 0 | −0.4 | −0.3 | ||
| Average bias (mmol/mol) | 0 | −4.4 | −3.3 | ||
| Fraction within ARD (%) | 5 | 79 | 57 | 49 | |
| 10 | 94 | 68 | 82 | ||
| 15 | 100 | 96 | 96 | ||
| Linear regression | R 2 | 0.91 | 0.65 | 0.65 | |
| Slope | 0.94 | 0.84 | 1.22 | ||
| Intercept | 0.49 | 1.5 | −1.17 | ||
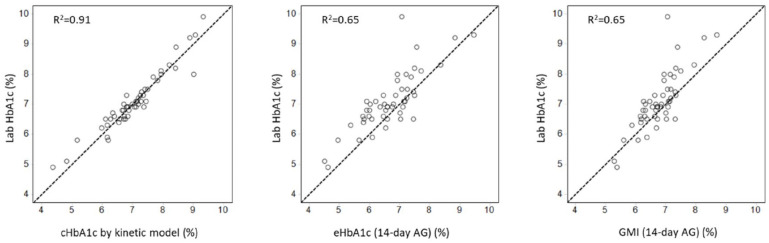
Figure 2 shows the improved agreement between cHbA1c and laboratory HbA1c, compared to eHbA1c and GMI. The cHbA1c had no overall bias, whereas the eHbA1c and GMI had clinically significant biases of −0.4% and −0.3%, respectively. The superior accuracy of cHbA1c was also indicated by a tighter association with laboratory HbA1c, having a coefficient of determination (R2) of 0.91 compared to 0.65 for both eHbA1c and GMI.
Figure 2.
Comparison of correlation between three CGM-derived HbA1c estimates and lab HbA1c.
Laboratory HbA1c ranged between 4.9% and 9.9% (30–85 mmol/mol), with a mean value of 7.1% (54 mmol/mol). At this mean value, cHbA1c had a 95% prediction confidence interval range from 6.9% to 7.3% (52–56 mmol/mol), which is a 78% reduction compared to either eHbA1c (6.5%–8.3% or 48–67 mmol/mol) or GMI (6.5%–8.3% or 48–67 mmol/mol).
Estimation of RBC lifespan
Within this cohort, we were able to calculate RBC lifespan in the higher confidence group of 26 subjects. This subgroup has a similar age distribution to the overall study cohort with a median (IQR) of 44 (37–55) years and a range of 10–70 years. The gender distribution was also similar, with 7 males and 19 females. In this subgroup, the median (IQR) RBC lifespan was 74 (66–88) days with a range of 56–120 days. Two subjects had compromised kidney function measured as eGFR less than 44 (mL/min/1.73m2), and one 14 years old patient. All three individuals showed short RBC lifespans of 55 to 68 days.
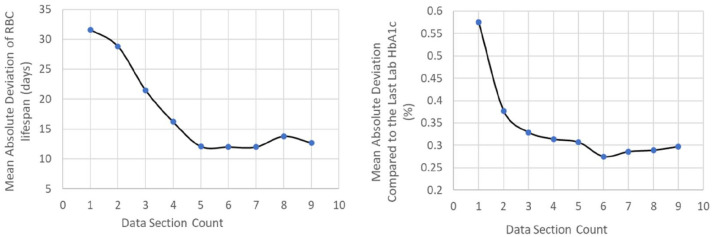
Within the 26 higher confidence subjects with relatively larger day-to-day glucose variability and without major life/therapeutic changes during data collection, 12 patients had at least 10 data sections. Figure 3 shows the prospective absolute deviations of cHbA1c compared with last laboratory HbA1c as well as the absolute deviations of RBC lifespan compared with the final RBC lifespan estimated using all data sections. The average absolute deviations of the cHbA1c predictions decreased sharply and then stabilized after the third data section. The absolute deviations of RBC lifespan also decreased longitudinally, reaching stability after the fifth data section.
Figure 3.
Longitudinal accuracy changes with data section count.
Discussion
The kinetic model evaluated here explains the relationship between glucose levels and HbA1c with two kinetic rate parameters for RBC turnover and intracellular glucose transfer. The performance of the model was previously evaluated in 120 European adults13 and using flash glucose monitoring for glucose levels. In this work, we examined the kinetic model in 51 Japanese patients with diabetes managed with SAP therapy and continuous glucose monitoring using a different device. The superior accuracy of the personalized model has been confirmed in this cohort when compared to established non-personalized methods of eHbA1c and GMI.
The model provides estimates for the kinetic parameters associated with RBC lifespan and glucose uptake. The longitudinal analysis showed that the kinetic parameter estimation usually converges after five data sections. The median RBC lifespan in this cohort was relatively short, around 74 days. In the previous study with the European cohort,13 we observed a similar median RBC lifespan of 78 days (or RBC turnover rate kage = 1.29%/day). These RBC lifespans are within or slightly lower than the reported range of mean RBC age by Cohen et al.7 In their experiment, utilizing data from six individuals with diabetes, the mean RBC age range was 38–56 days, giving RBC lifespans of 76–112 days. The observed shorter RBC lifespans might be related to the disease stage of both Japanese and European cohorts. Notably, in this study, the three subjects expected to have shortened RBC lifespans (either adolescent or with kidney disease) had the lowest RBC lifespans of 55–68 days. Having a routine manner of monitoring RBC lifespan and glucose uptake will aid in accurately predicting the future risks of diabetes complications.
While some conditions are known to affect the reliability of laboratory HbA1c as a marker of average glycaemic control (such as anemia and advanced renal disease),3 our work has the capability of identifying additional individuals in whom laboratory HbA1c can be unreliable. Those with reduced RBC lifespan may be at risk of hyperglycemic damage in tissues affected by diabetes complications, as laboratory HbA1c would underreport hyperglycemic exposure of the organ. Conversely, those with extended RBC lifespan may be at risk of hypoglycemia if treatment is escalated in order to “normalize” HbA1c when tissue exposure to hyperglycaemia is not excessive. Furthermore, RBC lifespan variation may have an impact on the accuracy of HbA1c for the diagnosis of prediabetes and diabetes, which may have major clinical implications.
This study has several strengths. First, it has consistent and high-quality laboratory HbA1c data and this is critical to the accuracy of the model. Second, each individual had high-quality, long term CGM and several concurrent laboratory HbA1c measurements. These longitudinal data were able to confirm the role of additional measurements to improve the accuracy of the personal glycation factors. Third, this is the first analysis in such an ethnic group and also the first analysis using a different CGM technology, further demonstrating the robustness of the method. However, there are limitations to be acknowledged. First, the cohort size is relatively small, which makes further subgroup analyses difficult. Second, only those with type 1 diabetes under SAP therapy were evaluated and therefore generalizability of the results can be questioned.
In conclusion, this study validated the superior performance of an individualized model for glucose-derived HbA1c in Japanese individuals with type 1 diabetes and SAP therapy. The kinetic model offers mechanistic insight into the relationship between glucose levels and glycated hemoglobin with two individualized kinetic parameters for RBC lifespan and glucose uptake. This study extends the model validation to a different CGM technology and further inspires confidence in applying the kinetic model in real-world clinical applications. The work also suggests that two data sections are usually sufficient for accurate HbA1c prediction, while the good estimations of RBC lifespan and glucose uptake likely requires five data sections.
Supplemental Material
Supplemental material, sj-pdf-1-dvr-10.1177_14791641211013734 for Accurate prediction of HbA1c by continuous glucose monitoring using a kinetic model with patient-specific parameters for red blood cell lifespan and glucose uptake by Yongjin Xu, Yushi Hirota, Ramzi A Ajjan, Akane Yamamoto, Atsuko Matsuoka, Wataru Ogawa and Timothy C Dunn in Diabetes & Vascular Disease Research
Footnotes
Declaration of conflicting interests: The author(s) declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: TD and YX are employees of Abbott Diabetes Care. RAA received other research supports and Honoraria from Abbott Diabetes Care. WO received other research support and honoraria from Abbott Diabetes Care. YH received other lecture fees from Abbott Japan and Medtronic. All other authors declare no conflict of interest.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by Abbott Diabetes Care.
ORCID iD: Yongjin Xu  https://orcid.org/0000-0001-9446-8402
https://orcid.org/0000-0001-9446-8402
Supplemental material: Supplemental material for this article is available online.
References
- 1.DCCT. The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. N Engl J Med 1993; 329: 977–986. [DOI] [PubMed] [Google Scholar]
- 2.UK Prospective Diabetes Study (UKPDS) Group. Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). UK Prospective Diabetes Study (UKPDS) Group. Lancet 1998; 352: 837–853. [PubMed] [Google Scholar]
- 3.American Diabetes Association. Glycemic targets: standards of medical care in diabetes – 2020. Diabetes Care 2020; 43(Suppl. 1): S66–S76. [DOI] [PubMed] [Google Scholar]
- 4.Howard M, Hamilton P.Haemotology: an illustrated colour text. 4th ed.Edinburgh: Elsevier, 2013. [Google Scholar]
- 5.Mebius R, Kraal G.Structure and function of the spleen. Nat Rev Immunol 2005; 5: 606–616. [DOI] [PubMed] [Google Scholar]
- 6.Theurl I, Hilgendorf I, Nairz M, et al. On-demand erythrocyte disposal and iron recycling requires transient macrophages in the liver. Nat Med 2016; 22(8): 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen RM, Franco RS, Khera PK, et al. Red cell life span heterogeneity in hematologically normal people is sufficient to alter HbA1c. Blood 2008; 112(10): 4284–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamdan MA, Hempe JM, Velasco-Gonzalez C, et al. Differences in red blood cell indices do not explain racial disparity in hemoglobin A1c in children with type 1 diabetes. J Pediatr 2016; 176: 197–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergenstal RM, Gal RL, Connor CG, et al. Racial differences in the relationship of glucose concentrations and hemoglobin A1c levels. Ann Intern Med 2017; 167(2): 95–102. [DOI] [PubMed] [Google Scholar]
- 10.Cohen RM, Smith EP, Arbabi S, et al. Do red blood cell indices explain racial differences in the relationship between hemoglobin A1c and blood glucose? J Pediatr 2016; 176: 7–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng D, Xu C, Sun P, et al. Crystal structure of the human glucose transporter GLUT1. Nature 2014; 510: 121–125. [DOI] [PubMed] [Google Scholar]
- 12.Sacks D.Measurement of hemoglobin A(1c). Diabetes Care 2012; 35: 2674–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y, Dunn TC, Ajjan RA.A kinetic model for glucose levels and hemoglobin A1C provides a novel tool for individualized diabetes management. J Diabetes Sci Technol 2021; 15(2): 294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nathan DM, Kuenen J, Borg R, et al. Translating the A1C assay into estimated average glucose values. Diabetes Care 2008; 31: 1473–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergenstal RM, Beck RW, Close KL, et al. Glucose management indicator (GMI): a new term for estimating A1C from continuous glucose monitoring. Diabetes Care 2018; 41(11): 2275–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Virtanen P, Gommers R, Oliphant TE, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 2020; 17(3): 261–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-dvr-10.1177_14791641211013734 for Accurate prediction of HbA1c by continuous glucose monitoring using a kinetic model with patient-specific parameters for red blood cell lifespan and glucose uptake by Yongjin Xu, Yushi Hirota, Ramzi A Ajjan, Akane Yamamoto, Atsuko Matsuoka, Wataru Ogawa and Timothy C Dunn in Diabetes & Vascular Disease Research