Abstract
Glioblastoma is a highly malignant cancer with no effective treatment. It is vital to elucidate the mechanisms which drive glioblastoma in order to identify therapeutic targets. The differences in protein expression between glioblastoma, grade I–III glioma, and normal brain tissue reflect the functional alterations driving malignancy. However, proteomic analysis of glioblastoma has been hampered by the heterogeneity of glioblastoma and the variety of methodology used in its study. To reduce these inconsistencies, we performed a meta-analysis of the literature published since 2015, including 14 datasets from eight papers comparing the whole proteome of glioblastoma to normal brain or grade I–III glioma. We found that 154 proteins were commonly upregulated and 116 proteins were commonly downregulated in glioblastoma compared to normal brain. Meanwhile, 240 proteins were commonly upregulated and 125 proteins were commonly downregulated in glioblastoma compared to grade I–III glioma. Functional enrichment analysis revealed upregulation of proteins involved in mRNA splicing and the immune system and downregulation of proteins involved in synaptic signaling and glucose and glutamine metabolism. The identification of these altered biological pathways provides a basis for deeper investigation in the pursuit of an effective treatment for glioblastoma.
Introduction
Glioblastoma multiforme (GBM), or glioblastoma, is both the most common and the most aggressive form of malignant brain cancer.1 Current treatments are only minimally effective, resulting in a five-year survival rate of 7%.1 GBM arises from the glial cells of the brain and is classified by the World Health Organization (WHO) as a grade IV astrocytoma.2 Glioblastoma was the first cancer to undergo large-scale genetic analysis by The Cancer Genome Atlas project in 2008.3 This study identified common genetic alterations in GBM, such as amplification and mutation of EGFR, mutation and loss of heterozygosity affecting TP53, loss of NF1, and mutation of P13KCA/PIK3R1.3 These aberrations, along with PTEN deletion, PDGFRA amplification, and IDH1 mutation, were used in the landmark Verhaak et al. study in 2010 to classify GBM tumors into genetically defined subtypes: classical, mesenchymal, proneural, and neural.4 Further transcriptomics analysis has resulted in the removal of the neuronal classification.5 Due to the great advances in the genetic characterization of brain tumors, in 2016 the WHO classification system was updated to include molecular features alongside the traditional histological parameters.6 Glioblastoma tumors are now classified as isocitrate dehydrogenase (IDH)-wild type or IDH-mutant, which tends to correspond to primary (de novo) and secondary (arising from lower grade tumors) GBM, respectively.6
While the genetic alterations in GBM are highly informative, it is vital to gain as deep an understanding of the proteomic changes, as these are the functional consequences of modifications to the genome. For this purpose, mass spectrometry is a valuable tool in the study of GBM, both in basic research and the identification of biomarkers.7,8 Increasingly, mass spectrometry is poised to complement or eventually replace traditional diagnostic methods of GBM presence, stage, and subtype.8 The high sensitivity of mass spectrometry and its ability to investigate multiple protein biomarkers simultaneously has shown it to be a promising diagnostic tool in various biological samples, including biopsy tissue, cerebrospinal fluid (CSF), blood plasma, urine, extracellular vesicles, and even fluid from cavitating ultrasound aspirators used during tumor removal.8−14 Many studies have investigated alterations in protein expression in GBM and several papers have reviewed potential GBM biomarkers in tumor tissue, CSF, plasma, and serum.14−18 However, there is no recent comprehensive review of the literature that determines which proteins and therefore which biological processes are commonly differentially regulated in GBM. This would provide a view of the global functional changes that occur in GBM tumors compared to low-grade glioma (LGG) and normal brain tissue.
A decade ago, Deighton et al. published a review of the GBM proteomics literature.19 This review highlighted the inconsistency between studies in the field, finding only 10 proteins differentially regulated in more than one of the 10 papers included in their analysis and no obvious biological processes implicated.19 They speculated that the discrepancies in methodology between the papers, including the control tissue that the GBM or glioma tissue was compared to, may have contributed to this lack of coherence.19 However, they also cited the limitations of the proteomics technology available at the time. Some of the studies reviewed used only Western blot or 2-D gel electrophoresis (2-DGE) analysis, while others included matrix-assisted laser desorption ionization time of flight mass spectrometry or surface-enhanced laser desorption ionization time of flight mass spectrometry.19 Only two studies used a form of liquid chromatography (LC) separation, which is now standard.19
Mass spectrometry technology has undergone great advances over the last decade, enabling the detection of thousands rather than hundreds of proteins in a single sample.20 These advances arise from increased mass accuracy and resolution, as well as gains in instrument sensitivity as a result of technological advancements such as the growth in the use of hybrid instruments, which allow for methods of ion fractionation and storage and greater scan rates.21 For example, constantly developing technologies such as the Orbitrap developed by Thermo Fisher increase the number of peptides that can be detected simultaneously.7,22 This results in increased coverage of individual proteins, enabling detection of lower abundance proteins and thus increasing coverage of the proteome as a whole.7,22 Furthermore, quantitative analysis has been refined with the development of isobaric tags for relative and absolute quantification (iTRAQ), tandem mass tags (TMT), and dimethyl labeling.23−25 Indeed, proteomic technology and analysis software has now been developed to allow simpler and cheaper label-free quantification.26 These technical improvements have opened up proteomics as a valuable and sensitive tool for a number of clinical applications.27
Deighton et al. were only able to identify 99 differentially regulated proteins across 10 papers published before the end of 2008.19 We hypothesized that improved proteomics techniques over the intervening years have greatly increased the available glioblastoma proteomics data, potentially providing greater coherence. Therefore, we have collated the lists of proteins found to be up- or downregulated in GBM compared to controls published in eight papers between 2015 and the end of 2020. This enabled the identification of proteins commonly up- or downregulated in GBM. These were further analyzed to determine the biological pathways and processes likely to be affected by these changes. We anticipate that this information will provide inspiration for future therapeutic intervention.
Results
Selected Literature
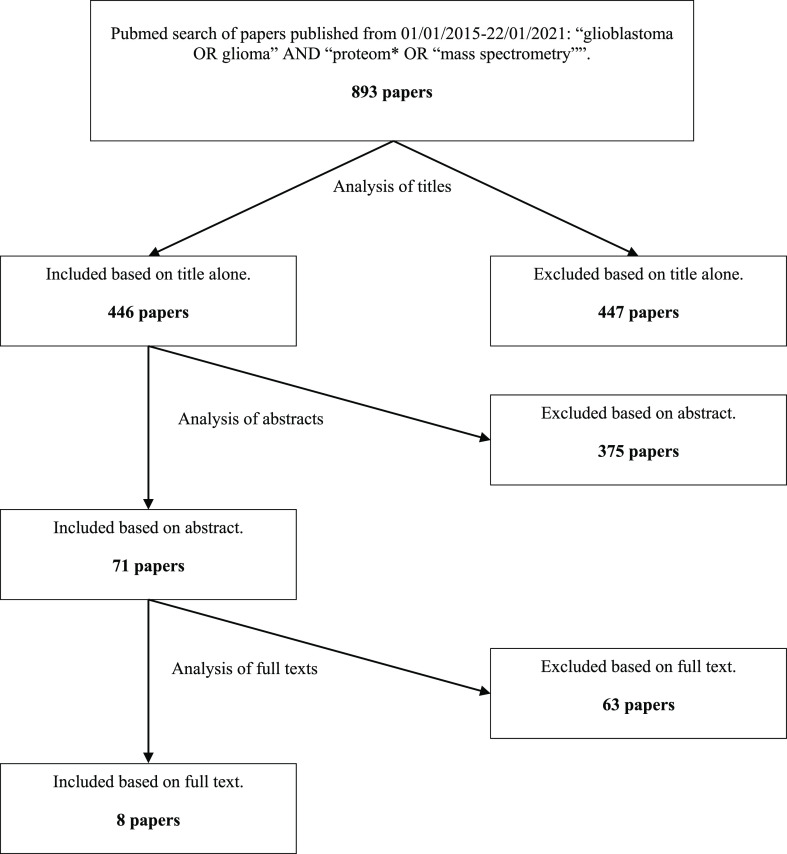
Eight papers published between 2015 and the end of 2020 were selected based on the defined criteria (see Methods and Figure 1).28−35 As in the review by Deighton et al. (2010), these studies varied in their methodology, for example, in the GBM material used, the chosen control, the sample processing, and the analysis.19 These parameters are summarized in Supporting Information File 1, Table S1.
Figure 1.
Flowchart of paper selection based on defined inclusion and exclusion criteria (see Methods).
Fourteen datasets were extracted from the eight selected papers (see Supporting Information File 1, Tables S2–S4). This was because five of the eight papers selected performed more than one comparative study that was relevant to this meta-analysis.29−32,35 Gollapalli et al. (2017) separately compared the same GBM tumor samples to peritumoral samples and to grade II and III glioma samples.29 Similarly, Ren et al. (2016) separately compared the same GBM tumor samples to grade I, grade II, and grade III glioma tissue.31 In both cases, the three comparisons were treated as three separate datasets. Gimenez et al. (2015) separately compared GBM tumor tissue from patients with a short survival time and from patients with a long survival time to normal brain tissue.30 They also separately compared GBM tumor tissue from patients with a short-survival time and from patients with a long survival time to grade II glioma tissue.30 Up- and downregulated proteins from the two GBM vs normal brain comparisons were merged into a single dataset, as were the GBM vs grade II glioma comparisons. This resulted in two datasets in which the same GBM tumor samples were compared to different controls.
Djuric et al. (2019) did not compare GBM to normal brain or LGG; however, their supporting information contained proteomics datasets which could be put to this use.35 The datasets from whole proteomics analysis of three GBM samples, one grade III anaplastic astrocytoma, and three grade I pilocytic astrocytomas were included for analysis. The three GBM samples were averaged and compared to the grade III astrocytoma and to the three averaged grade I astrocytoma samples separately, resulting in two datasets.
Buser et al. (2019) compared four different GBM tumors to normal brain tissue.32 The expression ratios for each protein in each of the four datasets were collated. Proteins with a value of either ≥2 or effectively infinite value (i.e., present divided by absent, or a large number divided by a very small number) in at least three of the four datasets were considered to be upregulated. Proteins with two effectively infinite values and two values ≤2 or not present across the four datasets were not included due to the conflicting results. Proteins with one effectively infinite value and three values ≤0.5 were considered to be downregulated. The proteins with no effectively infinite values were simply averaged, and proteins with average fold changes ≥2 or ≤0.5 were included as up- and downregulated, respectively, as usual.
Overall, four of the 14 datasets compared GBM to non-cancerous control brain.29,30,32,34 Seven of the 14 datasets compared GBM to grade I or II glioma.28−31,33,35 The remaining three datasets compared GBM to grade III glioma.29,31,35 The datasets comparing GBM to normal brain were analyzed separately to the datasets comparing GBM to LGG (including glioma grades I–III).
Differentially Expressed Proteins
Across the 14 datasets from the eight papers, 8801 proteins were found to have expression increased or decreased by at least 2-fold in GBM samples compared to LGG or normal brain controls in at least one of the datasets. Of these, 3949 (45%) were only identified by Buser et al. (2019) as this paper achieved far more protein identifications than any of the others.32 The increased sensitivity of mass spectrometry technology over the last decade is obvious. Even excluding the Buser et al. paper, 4852 differentially expressed proteins were identified, compared to just 99 in Deighton et al. in 2010.19 Unsurprisingly, the differentially expressed proteins identified by each paper varied. This reflects the known heterogeneity of GBM tumors, as well as the differences in the methodology used in each study.36 Nevertheless, many proteins were commonly up- or downregulated, revealing some consistency between the studies.
Proteins were considered to be commonly upregulated or downregulated in GBM compared to LGG if they were ≥2-fold up- or downregulated, respectively, in at least three of the ten relevant datasets. Due to the small number of studies comparing GBM to normal brain, it was decided that proteins had to be up- or downregulated in at least two of the four relevant datasets to be considered commonly up- or downregulated, respectively.
There were 154 proteins upregulated and 116 proteins downregulated in two or more of the four datasets comparing GBM to normal brain and 240 proteins upregulated and 125 downregulated in three or more of the ten datasets comparing GBM to LGG. Despite the difference between the LGG and normal brain controls, 36 proteins were upregulated in comparison to both controls and 24 proteins were downregulated in comparison to both controls.
The commonly up- and downregulated proteins were interrogated using the Reactome pathway analysis tool and the g:Profiler functional profiling tool.37,38 Twenty-three of the 154 proteins upregulated and 27 of the 116 proteins downregulated in GBM compared to normal brain could not be identified by Reactome. However, all but one of these proteins (P14406) were identified by and included in the g:Profiler analysis. Two of the upregulated proteins could not be identified by g:Profiler but were identified by and included in the Reactome analysis. Similarly, 77 of the 240 proteins upregulated and 42 of the 125 proteins downregulated in GBM compared to LGG could not be identified by Reactome. However, all of the downregulated proteins and all but eight of the upregulated proteins (Q9Y6R7, B4DDM6, B4DE40, B4DU58, B4DXI8, B4E2V5, B7Z8M7, and B3KSY4) were identified by and included in the g:Profiler analysis. One other protein was not identified by g:Profiler but was identified by and included in the Reactome analysis.
For each of the four sets of proteins (up- and downregulated in GBM compared to normal brain and up- and downregulated in GBM compared to LGG), the five most significant parent terms in the analysis results were identified (Figures 2–5, Supporting Information File 1, Tables S5–S20). Terms were not considered unless more than two of the proteins within the term were identified by the meta-analysis.
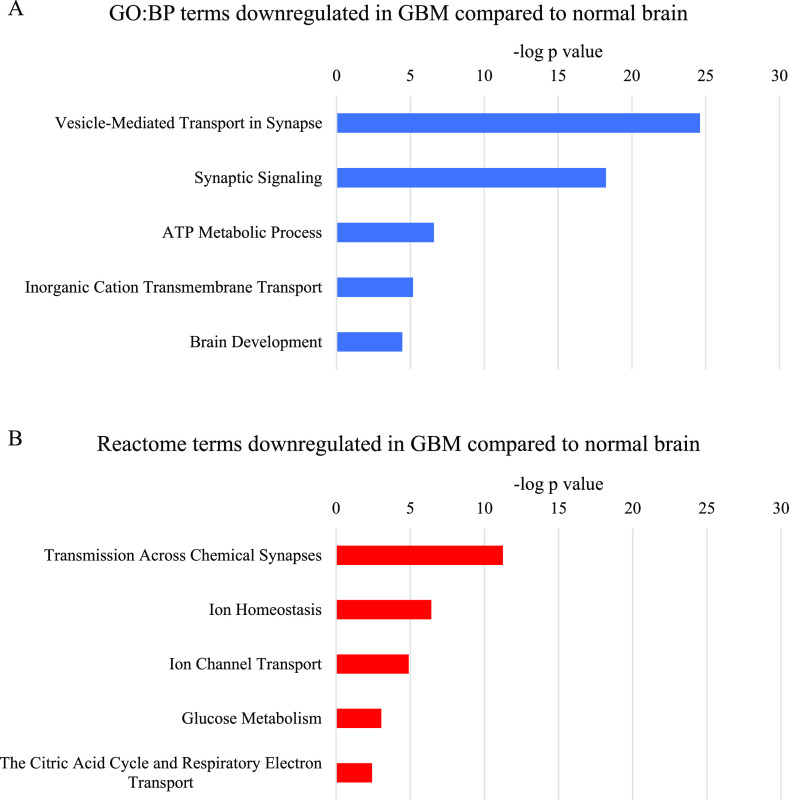
Figure 2.
Most significant parent terms in (A) Gene Ontology Biological Process domain (GO:BP) and (B) Reactome functional enrichment analysis of proteins downregulated in GBM compared to normal brain, ranked by the −log p value.
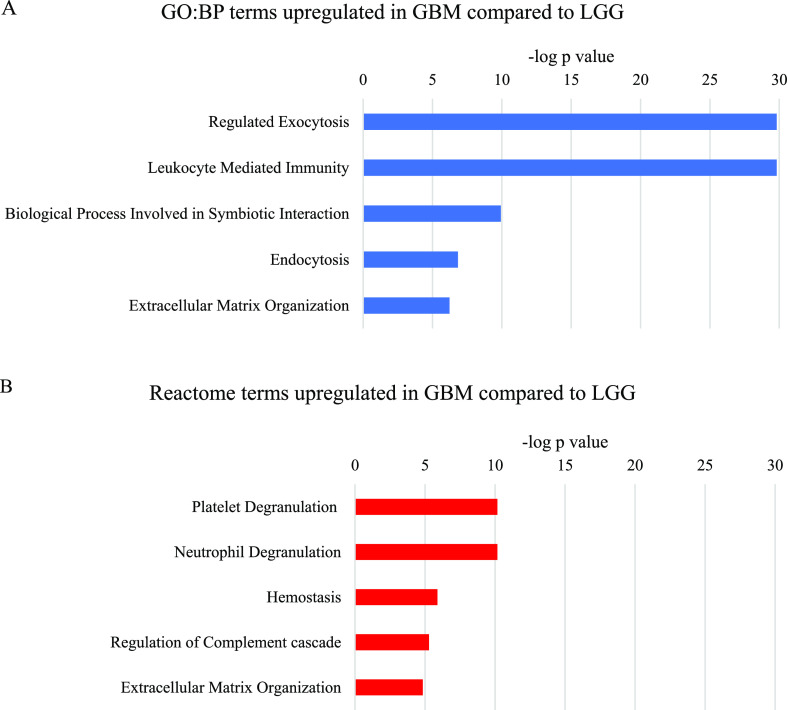
Figure 5.
Most significant parent terms in (A) GO:BP and (B) Reactome functional enrichment analysis of proteins upregulated in GBM compared to LGG, ranked by the −log p value.
Downregulated Proteins
Parent terms related to synaptic signaling were the most significantly over-represented in the analysis of proteins commonly downregulated in GBM compared to normal brain (Figure 2). Significant parent terms relating to glucose and ATP metabolism contained several proteins involved in oxidative phosphorylation, as well as a few involved in the tricarboxylic acid (TCA) cycle and glycolysis. Other proteins in these terms were subunits of ATPases. Ion transport parent terms were also identified; however, these overlapped extensively with the synaptic signaling and ATP metabolism terms. The GO:BP Brain Development term also overlapped with the synaptic signaling terms, however, also contained a few unique neuronal proteins, such as neurofilaments and contactin.
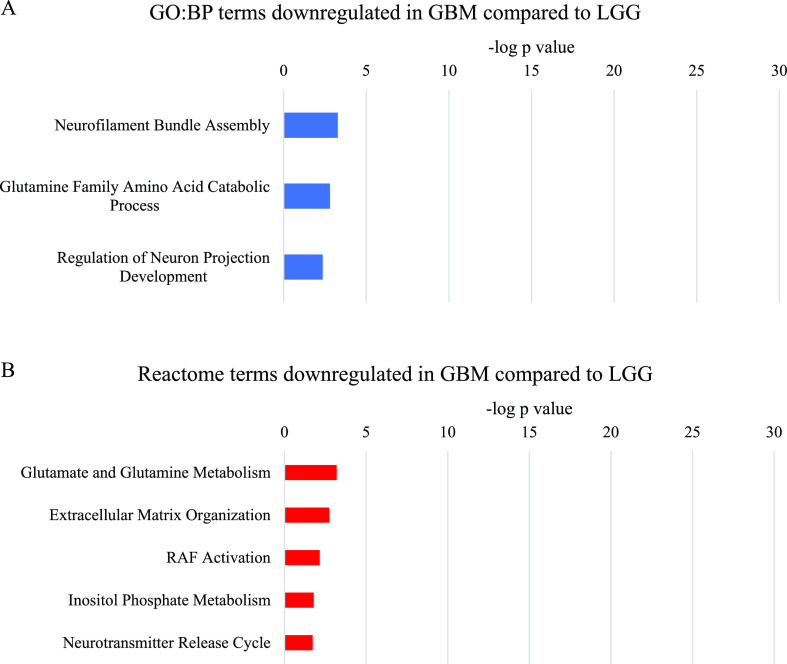
Analysis of the proteins downregulated in GBM compared to LGG also identified over-represented neuronal system terms, however, with much lower significance than in GBM compared to normal brain (Figure 3). Metabolism of the neurotransmitter glutamine was one of the most significant parent terms in the GO:BP and Reactome analyses. The other most significant GO:BP terms were related to cytoskeletal and extracellular matrix components of neurons. Similarly, the Reactome Extracellular Matrix Organization term included both neuron-specific and more general extracellular matrix proteins. RAF activation and inositol phosphate metabolism were also over-represented in the Reactome analysis.
Figure 3.
Most significant parent terms in (A) GO:BP and (B) Reactome functional enrichment analysis of proteins downregulated in GBM compared to LGG, ranked by the −log p value.
Upregulated Proteins
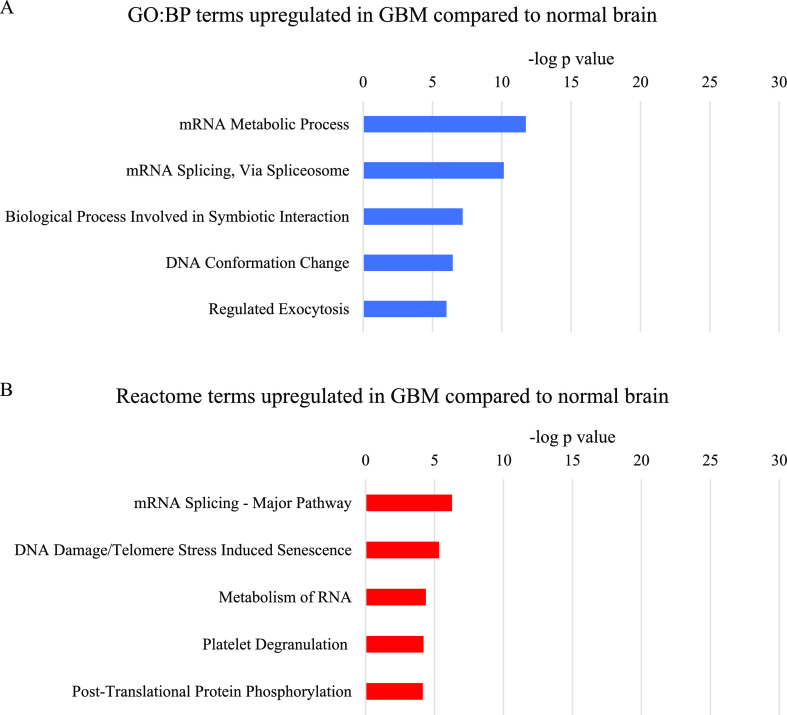
Parent terms related to mRNA metabolism and particularly mRNA splicing were the most significantly over-represented in the analysis of proteins commonly upregulated in GBM compared to normal brain (Figure 4). The more significant mRNA metabolism term consisted mainly of the proteins included in the mRNA splicing term, plus some ribosomal, proteasomal, and cytoskeletal proteins. The GO:BP parent term DNA Conformation Change contained several histone variants, which comprised most of the Reactome term DNA Damage/Telomere Stress Induced Senescence. The GO:BP term Biological Process Involved in Symbiotic Interaction contained proteins with a variety of functions, several of which overlapped with the mRNA metabolism terms. The proteins that were unique to this term were mostly annotated to immune system and stress response terms, although many of the proteins had multiple functions. The GO:BP term Regulated Exocytosis consisted of a variety of proteins, including some involved in the cytoskeleton and in anabolic and catabolic processes.
Figure 4.
Most significant parent terms in (A) GO:BP and (B) Reactome functional enrichment analysis of proteins upregulated in GBM compared to normal brain, ranked by the −log p value.
Analysis of proteins upregulated in GBM compared to LGG revealed processes that were distinct from those seen when GBM was compared to normal brain. The most significantly over-represented parent terms were related to exo- and endocytosis, immune system, platelets, and the extracellular matrix. While the GO:BP term Regulated Exocytosis was significantly over-represented in the analysis of proteins upregulated in GBM compared to normal brain and compared to LGG, the proteins within the term differed in the two analyses. The GBM compared to the LGG Regulated Exocytosis term consisted largely of proteins involved in the immune system, as well as proteins related to the cytoskeleton and blood clotting. This explains the extensive overlap between this term and the GO:BP Leukocyte-Mediated Immunity term, the Reactome Neutrophil Degranulation term and the Reactome Platelet Degranulation and Hemostasis terms. The proteins unique to the GO:BP Leukocyte-Mediated Immunity term are components of immunoglobulin chains, human leukocyte antigen, and the complement system.
The GO:BP term Biological Process Involved in Symbiotic Interaction was significantly over-represented in the analysis of proteins upregulated in GBM compared to normal brain and compared to LGG. However, the proteins annotated to this term in the two analyses barely overlapped. In the GBM compared to LGG analysis, this term overlapped extensively with the immune system terms, but also contained many ribosomal proteins and other proteins involved in protein production.
Implications for Prognosis
Proteins in the parent GO:BP functional enrichment terms identified by the meta-analysis were investigated for prognostic relevance using GEPIA (Supporting Information File 1, Tables S21 and 22).39 Notably, high expression of many proteins in the terms upregulated in GBM compared to LGG was significantly correlated with poorer overall survival or disease-free survival (Supporting Information File 1, Table S22). Most significant were the leukocyte elastase inhibitor (SERPINB1), fermitin family homolog 3 (FERMT3), and matrix metalloproteinase-9 (MMP9), which were significantly correlated with poorer disease-free survival (p = 0.00026, 0.004, and 0.0044, respectively) and NPC intracellular cholesterol transporter 2 (NPC2) and collagen alpha-1(VI) chain (COL6A1), which were significantly correlated with poorer overall survival (p = 0.0044 and 0.0059 respectively) and also less significantly correlated with poorer disease-free survival (p = 0.032 and 0.027, respectively) (Supporting Information File 2, Figures S1–S5). These proteins are mostly involved in cell adhesion and proteolysis. SERPINB1, MMP9, and NPC2 were grouped into both the GO:BP Regulated Exocytosis and Leukocyte Mediated Immunity terms. FERMT3 was grouped into only the GO:BP Regulated Exocytosis term and COL6A1 was in the GO:BP Extracellular Matrix Organization Term. In contrast, overexpression of 60S ribosomal protein L18 (RPL18), found to be commonly upregulated in GBM compared to LGG, was significantly correlated with better prognosis (Supporting Information File 2, Figure S6). This protein was grouped into the GO:BP term Biological Process Involved in Symbiotic Interaction.
Discussion
Analysis of GBM proteomics is hampered by the known intra- and intertumoral heterogeneity of GBM tumors and compounded by the variation in the methodology used by different studies.36 While individual papers identify differentially expressed proteins in GBM, the proteins identified and, therefore, the conclusions drawn vary. The strength of this meta-analysis is that it combined 14 datasets comparing the GBM proteome to the proteomes of normal brain tissue or LGG. This filtered out the noise created by the heterogeneity of the methodology and of GBM tumors themselves to identify proteins that were commonly differentially expressed across several datasets. As only these common proteins were carried through to functional enrichment analysis, we can determine with more certainty which biological pathways are most commonly differentially regulated in GBM.
Just a decade ago, Deighton et al. were unable to identify any biological pathways that were differentially regulated in GBM despite analyzing whole proteome data from 10 papers.19 Their study was limited by the available technology, which identified only 99 differentially expressed proteins. Only 10 were common to more than one paper—all involved in either apoptosis or cellular response to stress. In this meta-analysis, the superior sensitivity of the technology employed in eight more recent papers allowed the identification of 8801 differentially expressed proteins. In at least two of the four datasets comparing GBM to normal brain, 154 proteins were upregulated and 116 proteins were downregulated, while in at least three of the 10 datasets comparing GBM to LGG, 240 proteins were upregulated and 125 proteins were downregulated. The detection of so many differentially expressed proteins made it possible to identify several pathways which are significantly altered in GBM.
One of the most striking alterations in the proteome of GBM was the downregulation of neuronal system proteins, particularly proteins involved in synaptic vesicle release, compared to normal brain. This is reflective of the postsynaptic role of glioma cells in neurogliomal synapses, compared to the pre- and postsynaptic functions of neurons and astrocytes in the normal brain.40−43 Notably, the synaptic signaling term contained glutamate transporter excitatory amino acid transporter 2 (EAAT2). This protein is abundant in normal brain and expression is lost in GBM, disrupting extracellular glutamate homeostasis and enhancing GBM growth, survival, and invasion.44
Proteins involved in glucose and ATP metabolism were downregulated in GBM compared to normal brain. The downregulation of glycolysis proteins hexokinase 1 (HK1) and brain-specific fructose-biphosphate aldolase C (ALDOC) is in line with previous literature; however, the downregulation of ATP-dependent 6-phosphofructokinase (PFKP) is not.45−47 PFKP catalyzes a rate-limiting step in glycolysis, which would be expected to be vital to the Warburg effect often observed in GBM.45−49 Two understudied TCA cycle enzymes, 2-oxoglutarate dehydrogenase (OGDH) and malate dehydrogenase (MDH2), were downregulated, as was IDH3α, which has previously been found to be upregulated in GBM.50 Downregulation of oxidative phosphorylation complex IV (cytochrome c oxidase) subunits in GBM compared to normal brain is concordant with previous literature.49,51 However, expression of complexes III and V (cytochrome b-c1 and ATP synthase), found by this meta-analysis to be downregulated, has previously been shown to be unchanged in GBM compared to normal brain, although a decrease in complex III activity has been observed.49,51
Notably, several of the proteins assigned to the ATP metabolism terms were ATPases, which also made up a large proportion of the ion transport terms. These included subunits of vacuolar ATPases, which have been shown to be differentially expressed between GBM and LGG.52 Subunits of sodium/potassium ion transporter ATPases were also downregulated, including the β2 subunit, which is known to be downregulated in GBM and to inhibit invasion if overexpressed.53
Proteins related to glutamine and glutamate metabolism were downregulated in GBM compared to LGG. This included the downregulation of aspartate aminotransferase (GOT1), glutaminase kidney isoform (GLS), and glutamate dehydrogenase 1 (GLUD1), which are involved in the conversion of glutamine to alpha-ketoglutarate via glutamate.54−56 Although this meta-analysis did not filter by IDH mutation status, it is likely that a higher proportion of the LGG tumors than the GBM tumors included contained an IDH1 mutation.57,58 Rather than converting isocitrate to alpha-ketoglutarate as part of the TCA cycle, mutant IDH1 converts alpha-ketoglutarate to the oncometabolite 2-hydroxyglutarate (2-HG).59 Hence, glutaminolysis is upregulated to feed alpha-ketoglutarate into both the TCA cycle and into the production of 2-HG by mutant IDH1.59 Therefore, it is probable that the downregulation of glutamine and glutamate metabolism in GBM compared to LGG is reflective of the lower proportion of IDH1 mutant tumors in GBM.
In the analysis of the commonly upregulated proteins, the most notable alteration in GBM compared to normal brain was the upregulation of mRNA processing, particularly mRNA splicing. Alternative splicing is important in tumors and more than 1000 splicing events have prognostic significance in GBM.60,61 This meta-analysis indicated that splicing machinery is highly upregulated in both GBM and LGG; however, specific mRNA splicing events have previously been shown to differ between GBM and LGG.61 The other proteins commonly upregulated in GBM compared to normal brain showed less obvious trends, with several related to the immune system, stress responses, metabolism, or the cytoskeleton, and many with multiple functions.
The immune system stood out as being commonly upregulated in GBM compared to LGG, with p values as low as 1.5 × 10–30. The significant upregulation of immune system proteins in GBM is likely to reflect differences in tumor infiltration by immune cells rather than changes to the proteome of the tumor cells themselves. As there was no corresponding downregulation of different immune system proteins, this upregulation could indicate that immune infiltration increases in GBM, rather than reflecting a change in the nature of the infiltrate. However, this is a complex area. Myeloid cells can be significantly lower in GBM compared to LGG and tumor-infiltrating CD4 T cells have been shown to decrease in GBM compared to LGG, while CD8 T cells may or may not increase between grade II glioma and GBM.62,63
In summary, the heterogeneity of GBM tumors presents a major challenge for researchers aiming to identify changes that occur during the tumorigenesis of GBM. This is evident in the disparate results obtained by the 14 datasets used in this meta-analysis. Selecting the proteins commonly up- or downregulated across multiple studies allowed identification of the pathways and processes most commonly altered in GBM. This meta-analysis identified significantly altered expression of proteins involved in mRNA splicing, synaptic signaling, glucose and glutamine metabolism, and the immune system, which adds valuable support to the role of these pathways in GBM. Further analysis showed that expression of many of the proteins upregulated in GBM compared to LGG was correlated with poorer survival. Future studies can be guided by these common themes to identify targets for future therapies, which could improve the dire prognosis of GBM.
Methods
Inclusion and exclusion criteria for literature selection.
Inclusion criteria:
-
1
Included in PubMed.
-
2
Published between 1st January 2015 and 22nd January 2021.
-
3
Contained the search terms “glioblastoma OR glioma” AND “proteom* OR “mass spectrometry””.
Exclusion Criteria
-
1
Not a scientific paper.
-
2
Duplicate (i.e., each paper was only considered once even if it appeared more than once in the search results).
-
3
Not a primary research article (e.g., a review article, etc.).
-
4
Not human material studied.
-
5
Not glioblastoma studied.
-
6
Not glioblastoma tissue studied (e.g., cell lines, blood, exosomes, etc.).
-
7
No proteomics performed.
-
8
No mass spectrometry performed.
-
9
No whole proteomics mass spectrometry performed.
-
10
Not glioblastoma compared to normal brain or glioma grades I–III.
-
11
Not the whole tumor studied (e.g., a chosen section).
-
12
Not original publication of proteomics data.
-
13
Proteomics data not available.
-
14
No quantification performed.
Collation of Data
From each paper, proteins found to be at least 2-fold up- or downregulated in GBM compared to the control were collated (Supporting Information File 1, Tables S2–S4). A 2-fold change was selected as this was the most commonly used cut-off in the selected literature and was considered to indicate a definite change in expression levels. Protein names or codes listed in the supporting information were matched to UniProt accession numbers and vice versa as necessary. Some protein names or accession numbers were found to be obsolete and so were excluded from the spreadsheet. Others had been merged into another UniProt entry. In this case, the protein was included in the spreadsheet with its updated name and accession number. Any non-human proteins identified were excluded.
Functional Enrichment Analysis
Functional enrichment analysis was performed using g:Profiler version e102_eg49_p15_7a9b4d6 to search against the Gene Ontology Biological Process (GO:BP) database for Homo Sapiens.38 Statistical domain scope was set to “Only annotated genes”, significance threshold was set to “g:SCS threshold”, and user threshold was set to 0.05. The term size range was set at 2–1000 to avoid broad, uninformative terms. These results were then validated using the “Analyse Data” tool of Reactome version 75.37 The default setting “Project to human”, which converts non-human accession numbers to human equivalents was left selected, although all non-human proteins had already been removed from the data. Interactors were not included. Again, only terms with sizes ≤1000 proteins were included.
Survival Analysis
Survival analysis was performed using the GEPIA survival plot function.39 The Uniprot accession numbers of proteins in the parent terms identified in the functional enrichment analysis were converted to Ensembl gene IDs and entered into the software. The GBM dataset was selected for the analysis. Methods were set to first overall survival and then disease-free survival (RFS) to generate plots for both. The group cutoff was set to median and the axis units to months. The software calculated the hazard ratio based on the Cox PH Model and 95% confidence intervals were included on the plots. Genes were considered to be significantly correlated with survival if the Logrank p value was ≤0.05.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c02991.
Methodology parameters for selected papers; all proteins up- or downregulated by ≥2-fold in the eight papers (14 datasets) used in the meta-analysis; all proteins up- or downregulated by ≥2-fold in the four papers (four datasets) comparing GBM to normal brain used in the meta-analysis; all proteins up- or downregulated by ≥2-fold in the six papers (10 datasets) comparing GBM to LGG used in the meta-analysis; GO:BP results for proteins downregulated in GBM compared to normal brain; identification of parent terms from GO:BP results for proteins downregulated in GBM compared to normal brain; Reactome results for proteins downregulated in GBM compared to normal brain; identification of parent terms from the Reactome results for proteins downregulated in GBM compared to normal brain; GO:BP results for proteins downregulated in GBM compared to LGG; identification of parent terms from GO:BP results for proteins downregulated in GBM compared to LGG; Reactome results for proteins downregulated in GBM compared to LGG; identification of parent terms from Reactome results for proteins downregulated in GBM compared to LGG; GO:BP results for proteins upregulated in GBM compared to normal brain; identification of parent terms from GO:BP results for proteins upregulated in GBM compared to normal brain; Reactome results for proteins upregulated in GBM compared to normal brain; identification of parent terms from Reactome results for proteins upregulated in GBM compared to normal brain; GO:BP results for proteins upregulated in GBM compared to LGG; identification of parent terms from GO:BP results for proteins upregulated in GBM compared to LGG; Reactome results for proteins upregulated in GBM compared to LGG; identification of parent terms from Reactome results for proteins upregulated in GBM compared to LGG; downregulated parent term proteins with a significant correlation with prognosis; and upregulated parent term proteins with a significant correlation with prognosis (XLSX)
Overall and disease-free survival curves for parent term proteins with the most significant correlation with prognosis (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Ostrom Q. T.; Cioffi G.; Gittleman H.; Patil N.; Waite K.; Kruchko C.; Barnholtz-Sloan J. S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012-2016. Neuro Oncol. 2019, 21, v1–v100. 10.1093/neuonc/noz150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis D. N.; Ohgaki H.; Wiestler O. D.; Cavenee W. K.; Burger P. C.; Jouvet A.; Scheithauer B. W.; Kleihues P. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathol. 2007, 114, 97–109. 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comprehensive Genomic Characterization Defines Human Glioblastoma Genes and Core Pathways. Nature 2008, 455, 1061–1068. 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaak R. G. W.; Hoadley K. A.; Purdom E.; Wang V.; Qi Y.; Wilkerson M. D.; Miller C. R.; Ding L.; Golub T.; Mesirov J. P.; Alexe G.; Lawrence M.; O’Kelly M.; Tamayo P.; Weir B. A.; Gabriel S.; Winckler W.; Gupta S.; Jakkula L.; Feiler H. S.; Hodgson J. G.; James C. D.; Sarkaria J. N.; Brennan C.; Kahn A.; Spellman P. T.; Wilson R. K.; Speed T. P.; Gray J. W.; Meyerson M.; Getz G.; Perou C. M.; Hayes D. N. Integrated Genomic Analysis Identifies Clinically Relevant Subtypes of Glioblastoma Characterized by Abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Hu B.; Hu X.; Kim H.; Squatrito M.; Scarpace L.; deCarvalho A. C.; Lyu S.; Li P.; Li Y.; Barthel F.; Cho H. J.; Lin Y.-H.; Satani N.; Martinez-Ledesma E.; Zheng S.; Chang E.; Sauvé C.-E. G.; Olar A.; Lan Z. D.; Finocchiaro G.; Phillips J. J.; Berger M. S.; Gabrusiewicz K. R.; Wang G.; Eskilsson E.; Hu J.; Mikkelsen T.; DePinho R. A.; Muller F.; Heimberger A. B.; Sulman E. P.; Nam D.-H.; Verhaak R. G. W. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment. Cancer Cell 2017, 32, 42–56. 10.1016/j.ccell.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis D. N.; Perry A.; Reifenberger G.; von Deimling A.; Figarella-Branger D.; Cavenee W. K.; Ohgaki H.; Wiestler O. D.; Kleihues P.; Ellison D. W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- Chen L.; Qin D.; Guo X.; Wang Q.; Li J. Putting Proteomics Into Immunotherapy for Glioblastoma. Front. Immunol. 2021, 12, 593255. 10.3389/fimmu.2021.593255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silantyev A.; Falzone L.; Libra M.; Gurina O.; Kardashova K.; Nikolouzakis T.; Nosyrev A.; Sutton C.; Mitsias P.; Tsatsakis A. Current and Future Trends on Diagnosis and Prognosis of Glioblastoma: From Molecular Biology to Proteomics. Cells 2019, 8, 863. 10.3390/cells8080863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rocca G.; Simboli G. A.; Vincenzoni F.; Rossetti D. V.; Urbani A.; Ius T.; Della Pepa G. M.; Olivi A.; Sabatino G.; Desiderio C. Glioblastoma Cusa Fluid Protein Profiling: A Comparative Investigation of the Core and Peripheral Tumor Zones. Cancers 2021, 13, 30. 10.3390/cancers13010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Yu J.; Shen H.; Zhang J.; Liu W.; Chen Z.; He S.; Zheng S. Mass Spectrometric Analysis of Cerebrospinal Fluid Protein for Glioma and Its Clinical Application. Contemp. Oncol. 2014, 2, 100–105. 10.5114/wo.2014.40455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi E.; Furuta T.; Ohtsuki S.; Tachikawa M.; Uchida Y.; Sabit H.; Obuchi W.; Baba T.; Watanabe M.; Terasaki T.; Nakada M. Identification of Blood Biomarkers in Glioblastoma by SWATH Mass Spectrometry and Quantitative Targeted Absolute Proteomics. PLoS One 2018, 13, e0193799 10.1371/journal.pone.0193799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.; Zhang J.; Wei J.; Zhao Y.; Gao Y. Urinary Biomarker Discovery in Gliomas Using Mass Spectrometry-Based Clinical Proteomics. Chinese Neurosurg. J. 2020, 6, 11. 10.1186/s41016-020-00190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osti D.; Del Bene M.; Rappa G.; Santos M.; Matafora V.; Richichi C.; Faletti S.; Beznoussenko G. V.; Mironov A.; Bachi A.; Fornasari L.; Bongetta D.; Gaetani P.; DiMeco F.; Lorico A.; Pelicci G. Clinical Significance of Extracellular Vesicles in Plasma from Glioblastoma Patients. Clin. Cancer Res. 2019, 25, 266–276. 10.1158/1078-0432.ccr-18-1941. [DOI] [PubMed] [Google Scholar]
- Ghantasala S.; Gollapalli K.; Epari S.; Moiyadi A.; Srivastava S. Glioma Tumor Proteomics: Clinically Useful Protein Biomarkers and Future Perspectives. Expert Rev. Proteomics 2020, 17, 221–232. 10.1080/14789450.2020.1731310. [DOI] [PubMed] [Google Scholar]
- Jayaram S.; Gupta M. K.; Polisetty R. V.; Cho W. C.; Sirdeshmukh R. Towards Developing Biomarkers for Glioblastoma Multiforme: A Proteomics View. Expert Rev. Proteomics 2014, 11, 621–639. 10.1586/14789450.2014.939634. [DOI] [PubMed] [Google Scholar]
- Whittle I. R.; Short D. M.; Deighton R. F.; Kerr L. E.; Smith C.; McCulloch J. Proteomic Analysis of Gliomas. Br. J. Neurosurg. 2007, 21, 576–582. 10.1080/02688690701721691. [DOI] [PubMed] [Google Scholar]
- Niclou S. P.; Fack F.; Rajcevic U. Glioma Proteomics: Status and Perspectives. J. Proteomics 2010, 73, 1823–1838. 10.1016/j.jprot.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Kalinina J.; Peng J.; Ritchie J. C.; Van Meir E. G. Proteomics of Gliomas: Initial Biomarker Discovery and Evolution of Technology. Neuro Oncol. 2011, 13, 926–942. 10.1093/neuonc/nor078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deighton R. F.; McGregor R.; Kemp J.; McCulloch J.; Whittle I. R. Glioma Pathophysiology: Insights Emerging from Proteomics. Brain Pathol. 2010, 20, 691–703. 10.1111/j.1750-3639.2010.00376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Wang W.; Chen J. Recent Progress in Mass Spectrometry Proteomics for Biomedical Research. Sci. China: Life Sci. 2017, 60, 1093–1113. 10.1007/s11427-017-9175-2. [DOI] [PubMed] [Google Scholar]
- Yates J. R. III Recent Technical Advances in Proteomics. F1000Research 2019, 8, F1000. 10.12688/f1000research.16987.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliuk S.; Makarov A. Evolution of Orbitrap Mass Spectrometry Instrumentation. Annu. Rev. Anal. Chem. 2015, 8, 61–80. 10.1146/annurev-anchem-071114-040325. [DOI] [PubMed] [Google Scholar]
- Evans C.; Noirel J.; Ow S. Y.; Salim M.; Pereira-Medrano A. G.; Couto N.; Pandhal J.; Smith D.; Pham T. K.; Karunakaran E.; Zou X.; Biggs C. A.; Wright P. C. An Insight into ITRAQ: Where Do We Stand Now?. Anal. Bioanal. Chem. 2012, 404, 1011–1027. 10.1007/s00216-012-5918-6. [DOI] [PubMed] [Google Scholar]
- Boersema P. J.; Raijmakers R.; Lemeer S.; Mohammed S.; Heck A. J. R. Multiplex Peptide Stable Isotope Dimethyl Labeling for Quantitative Proteomics. Nat. Protoc. 2009, 4, 484–494. 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- Thompson A.; Schäfer J.; Kuhn K.; Kienle S.; Schwarz J.; Schmidt G.; Neumann T.; Hamon C. Tandem Mass Tags: A Novel Quantification Strategy for Comparative Analysis of Complex Protein Mixtures by MS/MS. Anal. Chem. 2003, 75, 1895–1904. 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- Zhao L.; Cong X.; Zhai L.; Hu H.; Xu J.-Y.; Zhao W.; Zhu M.; Tan M.; Ye B.-C. Comparative Evaluation of Label-Free Quantification Strategies. J. Proteomics 2020, 215, 103669. 10.1016/j.jprot.2020.103669. [DOI] [PubMed] [Google Scholar]
- Macklin A.; Khan S.; Kislinger T. Recent Advances in Mass Spectrometry Based Clinical Proteomics: Applications to Cancer Research. Clin. Proteomics 2020, 17, 17. 10.1186/s12014-020-09283-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi B.; Li F.; Guo J.; Li C.; Jing R.; Lv X.; Chen X.; Wang F.; Azadzoi K. M.; Wang L.; Liu Y.; Yang J.-H. Label-Free Quantitative Proteomics Unravels the Importance of RNA Processing in Glioma Malignancy. Neuroscience 2017, 351, 84–95. 10.1016/j.neuroscience.2017.03.023. [DOI] [PubMed] [Google Scholar]
- Gollapalli K.; Ghantasala S.; Atak A.; Rapole S.; Moiyadi A.; Epari S.; Srivastava S. Tissue Proteome Analysis of Different Grades of Human Gliomas Provides Major Cues for Glioma Pathogenesis. OMICS: J. Integr. Biol. 2017, 21, 275–284. 10.1089/omi.2017.0028. [DOI] [PubMed] [Google Scholar]
- Gimenez M.; Marie S. K. N.; Oba-Shinjo S.; Uno M.; Izumi C.; Oliveira J. B.; Rosa J. C. Quantitative Proteomic Analysis Shows Differentially Expressed HSPB1 in Glioblastoma as a Discriminating Short from Long Survival Factor and NOVA1 as a Differentiation Factor between Low-Grade Astrocytoma and Oligodendroglioma. BMC Cancer 2015, 15, 481. 10.1186/s12885-015-1473-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T.; Lin S.; Wang Z.; Shang A. Differential Proteomics Analysis of Low- and High-Grade of Astrocytoma Using ITRAQ Quantification. OncoTargets Ther. 2016, 9, 5883–5895. 10.2147/ott.s111103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buser D. P.; Ritz M.-F.; Moes S.; Tostado C.; Frank S.; Spiess M.; Mariani L.; Jenö P.; Boulay J.-L.; Hutter G. Quantitative Proteomics Reveals Reduction of Endocytic Machinery Components in Gliomas. EBioMedicine 2019, 46, 32–41. 10.1016/j.ebiom.2019.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.; Yeom J.; Cho H. J.; Kim J.-H.; Yoon S.-J.; Kim H.; Sa J. K.; Ju S.; Lee H.; Oh M. J.; Lee W.; Kwon Y.; Li H.; Choi S.; Han J. H.; Chang J. H.; Choi E.; Kim J.; Her N.-G.; Kim S. H.; Kang S.-G.; Paek E.; Nam D.-H.; Lee C.; Kim H. S. Integrated Pharmaco-Proteogenomics Defines Two Subgroups in Isocitrate Dehydrogenase Wild-Type Glioblastoma with Prognostic and Therapeutic Opportunities. Nat. Commun. 2020, 11, 3288. 10.1038/s41467-020-17139-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J.; Sang W.; Su L.-P.; Gao H.-X.; Cui W.-L.; Abulajiang G.; Wang Q.; Zhang J.; Zhang W. Proteomics Reveals Protein Phosphatase 1γ as a Biomarker Associated with Hippo Signal Pathway in Glioma. Pathol., Res. Pract. 2020, 216, 153187. 10.1016/j.prp.2020.153187. [DOI] [PubMed] [Google Scholar]
- Djuric U.; Lam K. H. B.; Kao J.; Batruch I.; Jevtic S.; Papaioannou M.-D.; Diamandis P. Defining Protein Pattern Differences among Molecular Subtypes of Diffuse Gliomas Using Mass Spectrometry. Mol. Cell. Proteomics 2019, 18, 2029–2043. 10.1074/mcp.ra119.001521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inda M.-d. -M.; Bonavia R.; Seoane J. Glioblastoma Multiforme:A Look inside Its Heterogeneous Nature. Cancers 2014, 6, 226–239. 10.3390/cancers6010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassal B.; Matthews L.; Viteri G.; Gong C.; Lorente P.; Fabregat A.; Sidiropoulos K.; Cook J.; Gillespie M.; Haw R.; Loney F.; May B.; Milacic M.; Rothfels K.; Sevilla C.; Shamovsky V.; Shorser S.; Varusai T.; Weiser J.; Wu G.; Stein L.; Hermjakob H.; D’Eustachio P. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2020, 48, D498–D503. 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere U.; Kolberg L.; Kuzmin I.; Arak T.; Adler P.; Peterson H.; Vilo J. Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z.; Li C.; Kang B.; Gao G.; Li C.; Zhang Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res. 2017, 45, W98–W102. 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataramani V.; Tanev D. I.; Strahle C.; Studier-Fischer A.; Fankhauser L.; Kessler T.; Körber C.; Kardorff M.; Ratliff M.; Xie R.; Horstmann H.; Messer M.; Paik S. P.; Knabbe J.; Sahm F.; Kurz F. T.; Acikgöz A. A.; Herrmannsdörfer F.; Agarwal A.; Bergles D. E.; Chalmers A.; Miletic H.; Turcan S.; Mawrin C.; Hänggi D.; Liu H.-K.; Wick W.; Winkler F.; Kuner T. Glutamatergic Synaptic Input to Glioma Cells Drives Brain Tumour Progression. Nature 2019, 573, 532–538. 10.1038/s41586-019-1564-x. [DOI] [PubMed] [Google Scholar]
- Venkatesh H. S.; Morishita W.; Geraghty A. C.; Silverbush D.; Gillespie S. M.; Arzt M.; Tam L. T.; Espenel C.; Ponnuswami A.; Ni L.; Woo P. J.; Taylor K. R.; Agarwal A.; Regev A.; Brang D.; Vogel H.; Hervey-Jumper S.; Bergles D. E.; Suvà M. L.; Malenka R. C.; Monje M. Electrical and Synaptic Integration of Glioma into Neural Circuits. Nature 2019, 573, 539–545. 10.1038/s41586-019-1563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A. T.; Choi U. B.; Lai Y.; Leitz J.; Zhou Q. Molecular Mechanisms of Fast Neurotransmitter Release. Annu. Rev. Biophys. 2018, 47, 469–497. 10.1146/annurev-biophys-070816-034117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada K.; Kamiya T.; Tsuboi T. Gliotransmitter Release from Astrocytes: Functional, Developmental, and Pathological Implications in the Brain. Front. Neurosci. 2016, 9, 499. 10.3389/fnins.2015.00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert S. M.; Sontheimer H. Glutamate Transporters in the Biology of Malignant Gliomas. Cell. Mol. Life Sci. 2014, 71, 1839–1854. 10.1007/s00018-013-1521-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-H.; Liu R.; Li J.; Zhang C.; Wang Y.; Cai Q.; Qian X.; Xia Y.; Zheng Y.; Piao Y.; Chen Q.; De Groot J. F.; Jiang T.; Lu Z. Stabilization of Phosphofructokinase 1 Platelet Isoform by AKT Promotes Tumorigenesis. Nat. Commun. 2017, 8, 949. 10.1038/s41467-017-00906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf A.; Agnihotri S.; Micallef J.; Mukherjee J.; Sabha N.; Cairns R.; Hawkins C.; Guha A. Hexokinase 2 Is a Key Mediator of Aerobic Glycolysis and Promotes Tumor Growth in Human Glioblastoma Multiforme. J. Exp. Med. 2011, 208, 313–326. 10.1084/jem.20101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. C.; Tsai H. F.; Huang S. P.; Chen C. L.; Hsiao M.; Tsai W. C. Enrichment of Aldolase C Correlates with Low Non- Mutated IDH1 Expression and Predicts a Favorable Prognosis in Glioblastomas. Cancers 2019, 11, 1238. 10.3390/cancers11091238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. The Metabolism of Carcinoma Cells. J. Cancer Res. 1925, 9, 148–163. 10.1158/jcr.1925.148. [DOI] [Google Scholar]
- Strickland M.; Stoll E. A. Metabolic Reprogramming in Glioma. Front. Cell Dev. Biol. 2017, 5, 43. 10.3389/fcell.2017.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May J. L.; Kouri F. M.; Hurley L. A.; Liu J.; Tommasini-Ghelfi S.; Ji Y.; Gao P.; Calvert A. E.; Lee A.; Chandel N. S.; Davuluri R. V.; Horbinski C. M.; Locasale J. W.; Stegh A. H. IDH3α Regulates One-Carbon Metabolism in Glioblastoma. Sci. Adv. 2019, 5, eaat0456 10.1126/sciadv.aat0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feichtinger R. G.; Weis S.; Mayr J. A.; Zimmermann F.; Geilberger R.; Sperl W.; Kofler B. Alterations of Oxidative Phosphorylation Complexes in Astrocytomas. Glia 2014, 62, 514–525. 10.1002/glia.22621. [DOI] [PubMed] [Google Scholar]
- Terrasi A.; Bertolini I.; Martelli C.; Gaudioso G.; Di Cristofori A.; Storaci A. M.; Formica M.; Bosari S.; Caroli M.; Ottobrini L.; Vaccari T.; Vaira V. Specific V-ATPase Expression Sub-Classifies IDHwt Lower-Grade Gliomas and Impacts Glioma Growth in Vivo. EBioMedicine 2019, 41, 214–224. 10.1016/j.ebiom.2019.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M. Z.; Kim J. M.; Oh M. C.; Safaee M.; Kaur G.; Clark A. J.; Bloch O.; Ivan M. E.; Kaur R.; Oh T.; Fouse S. D.; Phillips J. J.; Berger M. S.; Parsa A. T. Na+/K+-ATPase B2-Subunit (AMOG) Expression Abrogates Invasion of Glioblastoma-Derived Brain Tumor-Initiating Cells. Neuro Oncol. 2013, 15, 1518–1531. 10.1093/neuonc/not099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeliga M.; Bogacińska-Karaś M.; Różycka A.; Hilgier W.; Marquez J.; Albrecht J. Silencing of GLS and Overexpression of GLS2 Genes Cooperate in Decreasing the Proliferation and Viability of Glioblastoma Cells. Tumor Biol. 2014, 35, 1855–1862. 10.1007/s13277-013-1247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toney M. D. Aspartate Aminotransferase: An Old Dog Teaches New Tricks. Arch. Biochem. Biophys. 2014, 544, 119–127. 10.1016/j.abb.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.; Sudderth J.; Dang T.; Bachoo R. G.; McDonald J. G.; Deberardinis R. J. Glioblastoma Cells Require Glutamate Dehydrogenase to Survive Impairments of Glucose Metabolism or Akt Signaling. Cancer Res. 2009, 69, 7986–7993. 10.1158/0008-5472.can-09-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H.; Parsons D. W.; Jin G.; McLendon R.; Rasheed B. A.; Yuan W.; Kos I.; Batinic-Haberle I.; Jones S.; Riggins G. J.; Friedman H.; Friedman A.; Reardon D.; Herndon J.; Kinzler K. W.; Velculescu V. E.; Vogelstein B.; Bigner D. D. IDH1 and IDH2 Mutations in Gliomas. N. Engl. J. Med. 2009, 360, 765–773. 10.1056/nejmoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobusawa S.; Watanabe T.; Kleihues P.; Ohgaki H. IDH1 Mutations as Molecular Signature and Predictive Factor of Secondary Glioblastomas. Clin. Cancer Res. 2009, 15, 6002–6007. 10.1158/1078-0432.ccr-09-0715. [DOI] [PubMed] [Google Scholar]
- Dang L.; White D. W.; Gross S.; Bennett B. D.; Bittinger M. A.; Driggers E. M.; Fantin V. R.; Jang H. G.; Jin S.; Keenan M. C.; Marks K. M.; Prins R. M.; Ward P. S.; Yen K. E.; Liau L. M.; Rabinowitz J. D.; Cantley L. C.; Thompson C. B.; Vander Heiden M. G.; Su S. M.; Su M. Cancer-Associated IDH1 Mutations Produce 2-Hydroxyglutarate. Nature 2009, 462, 739–744. 10.1038/nature08617.Cancer-associated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Climente-González H.; Porta-Pardo E.; Godzik A.; Eyras E. The Functional Impact of Alternative Splicing in Cancer. Cell Rep. 2017, 20, 2215–2226. 10.1016/j.celrep.2017.08.012. [DOI] [PubMed] [Google Scholar]
- Li Y.; Ren Z.; Peng Y.; Li K.; Wang X.; Huang G.; Qi S.; Liu Y. Classification of Glioma Based on Prognostic Alternative Splicing. BMC Med. Genomics 2019, 12, 165. 10.1186/s12920-019-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman R. L.Interrogation of the Immune Contexture in Brain Malignancy; Memorial Sloan Kettering Cancer Center, 2016. [Google Scholar]
- Weenink B.; Draaisma K.; Ooi H. Z.; Kros J. M.; Sillevis Smitt P. A. E.; Debets R.; French P. J. Low-Grade Glioma Harbors Few CD8 T Cells, Which Is Accompanied by Decreased Expression of Chemo-Attractants, Not Immunogenic Antigens. Sci. Rep. 2019, 9, 14643. 10.1038/s41598-019-51063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.