Abstract
The endophytic bacterium Burkholderia contaminans NZ was isolated from jute, which is an important fiber-producing plant. This bacterium exhibits significant growth promotion activity in in vivo pot experiments, and like other plant growth-promoting (PGP) bacteria fixes nitrogen, produces indole acetic acid (IAA), siderophore, and 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity. B. contaminans NZ is considered to exert a promising growth inhibitory effect on Macrophomina phaseolina, a phytopathogen responsible for infecting hundreds of crops worldwide. This study aimed to identify the possibility of B. contaminans NZ as a safe biocontrol agent and assess its effectiveness in suppressing phytopathogenic fungi, especially M. phaseolina. Co-culture of M. phaseolina with B. contaminans NZ on both solid and liquid media revealed appreciable growth suppression of M. phaseolina and its chromogenic aberration in liquid culture. Genome mining of B. contaminans NZ using NaPDoS and antiSMASH revealed gene clusters that displayed 100% similarity for cytotoxic and antifungal substances, such as pyrrolnitrin. GC-MS analysis of B. contaminans NZ culture extracts revealed various bioactive compounds, including catechol; 9,10-dihydro-12’-hydroxy-2’-methyl-5’-(phenylmethyl)- ergotaman 3’,6’,18-trione; 2,3-dihydro-3,5- dihydroxy-6-methyl-4H-pyran-4-one; 1-(1,6-Dioxooctadecyl)- pyrrolidine; 9-Octadecenamide; and 2- methoxy- phenol. These compounds reportedly exhibit tyrosinase inhibitory, antifungal, and antibiotic activities. Using a more targeted approach, an RP-HPLC purified fraction was analyzed by LC-MS, confirming the existence of pyrrolnitrin in the B. contaminans NZ extract. Secondary metabolites, such as catechol and ergotaman, have been predicted to inhibit melanin synthesis in M. phaseolina. Thus, B. contaminans NZ appears to inhibit phytopathogens by apparently impairing melanin synthesis and other potential biochemical pathways, exhibiting considerable fungistatic activity.
Introduction
The continued use of expensive chemical fertilizers and fungicides have adversely affected human health and negatively impacted the environment; hence, the safe use of microorganisms that improve soil fertility, enhance plant growth and limit the growth of phytopathogenic fungi has been receiving immense attention from researchers [1]. Endophytes, mainly bacteria and fungi that spend at least a part of their lifespan within plant tissues without negatively affecting their hosts [2], have been intensely studied for years in terms of their diversity, metagenomics, combinatorial biosynthesis, plant growth promotion, biocontrol, bioremediation, etc. [3]. Plant-associated bacteria that aggressively colonize as symbiotic partners in the plant rhizosphere and roots with beneficial effects on plant growth are considered plant growth-promoting rhizobacteria (PGPR) [4]. In contrast, endophytic bacteria colonize the apoplasm or symplasm of the internal tissues of plants [5]. In general, with respect to growth promotion and protection against microbial infection, the beneficial effects of endophytes are considerably greater than those of several rhizobacteria [6].
The endophytes are grouped into three major clusters based on their different mechanisms of action. These contain: (i) biofertilization, which includes siderophore production, atmospheric nitrogen fixation, phosphate solubilization, and exopolysaccharides production; (ii) phyto-stimulation, which includes production of indole acetic acid, gibberellin, cytokinin, and ethylene; and (iii) phyto-biocontrol, which includes competing for iron, nutrients, and space; production of antibiotics, lytic enzymes, volatile compounds; and induction of systemic resistance [7]. Many studies have emphasized the ability of these microorganisms to promote and protect plant growth through additive/synergistic effects [1]; among them, Streptomyces spp. [8], B. subtilis [9], Pseudomonas parafulva, and Pantoea agglomerans [10] are a few examples.
Jute (Corchorus olitorius var. O-4) is an important natural fiber-producing cash crop in Southeast Asia [11]. Throughout its life cycle, jute is confronted by the destructive necrotrophic fungal pathogen Macrophomina phaseolina (Tassi) Goid., which causes charcoal rot disease in more than 500 plant species from approximately 100 families [12].
While screening for jute endophytes with potential inhibitory effects against M. phaseolina, we isolated an endophytic bacterium, Burkholderia contaminans NZ from the seed, which demonstrated promising antifungal activity [13]. This bacterium contained genes that are characteristic of PGPR, such as siderophore and ACC deaminase activities, phytohormone auxin (IAA) production, and nitrogen-fixing abilities, thereby promoting plant growth.
The genus Burkholderia, which belongs to the subphylum of β-proteobacteria, currently consists of 90 validly named species and several uncultivated candidate species [14]. They are ubiquitous organisms with high genetic versatility and adaptability, and are widespread in water, soil, plants, and animals, including humans [15].
However, the Burkholderia genus also comprises certain pathogenic species that threaten plant, animal, and human health [16]. Burkholderia contaminans is a member of the Burkholderia cepacia complex (Bcc) [17] which includes several closely related Burkholderia species. These opportunistic pathogens frequently colonize the lungs of cystic fibrosis (CF) and immune-compromised patients [18]. Furthermore, Bcc is the most frequently isolated clinical pathogen among Burkholderia spp., followed by B. mallei and B. pseudomallei [19]. Therefore, the environmental release of Burkholderia species as a biocontrol agent in agriculture is severely debated due to difficulties in distinguishing plant growth-promoting and pathogenic bacteria [20]. Recent developments in Burkholderia taxonomy and molecular analysis may help in answering questions concerning differences between the pathogenic and ecological properties of Burkholderia species in an attempt to reconsider the possibility of using selected strains for biocontrol [21].
The present study aimed to determine the safety of B. contaminans NZ as a biocontrol agent and its effectiveness against phytopathogens, especially M. phaseolina. The entire genome sequence was analyzed in this study to unravel information regarding the safety of B. contaminans NZ as a PGPR and biocontrol agent based on their siderophores and secondary metabolites producing capacities, and the absence of virulence genes. In vitro data also revealed the attributes of growth promotion. Moreover, Gas chromatography–mass spectrometry (GC-MS) and liquid chromatography–mass spectrometry (LC-MS) analyses identified several potent antimicrobial compounds and antagonists of melanin synthesis, and the electron transport system produced by the bacterium in the presence of M. phaseolina. These compounds possibly antagonize fungal growth through different modes of action.
Materials and methods
Bacterial, fungal strains and plant materials
The phytopathogenic fungi M. phaseolina was obtained from the Bangladesh Jute Research Institute (BJRI), Dhaka. Nigrospora sphaerica, Xylaria spp., Aspergillus fumigatus, Aspergillus niger, and Penicillium oxalicum were collected from the Molecular Biology Lab, Department of Biochemistry and Molecular Biology, University of Dhaka. Rhizoctonia solani was obtained from the Bangladesh Agricultural University, Mymensingh, Bangladesh. All the fungi were grown and maintained on potato dextrose agar (PDA) and for GC-MS analysis, M. phaseolina was grown in potato dextrose broth (PDB) (HiMedia, India) at 28°C. The antagonistic bacterial strain was isolated from jute seed as an endophytic bacterium as described by Coombs and Franco [22, 23] and identified as B. contaminans by 16S rRNA. Bacterial subculture was maintained on Tryptone Soy Agar (TSA) (HiMedia, India).
Jute seedlings were used to assess plant growth promotion activity of B. contaminans NZ. Fresh seeds of a jute variety (Corchorus olitorius var. O-4) were collected from BJRI. All tests were performed in triplicate if not mentioned otherwise.
Whole genome sequencing of B. contaminans NZ
B. contaminans NZ was cultured overnight in Tryptic Soy Broth (TSB) medium and incubated in an incubator shaker at 37°C. The genomic DNA was extracted using the GenElute™ Bacterial Genomic DNA Kit (Sigma, Germany). A genomic library was constructed and employed for 300-bp paired-end whole-genome sequencing at the Genome Research Institute of North South University, Dhaka, Bangladesh using an IlluminaMiSeq platform (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. The generated raw reads (10xcoverage) were assembled using SPAdes version 3.11 [24]. The generated scaffolds were mapped and ordered using ABACAS–a reference-based assembler [25], considering Burkholderia contaminans CH1 as the reference genome [GenBank accession no. AP018357 to AP018360 (four entries)] [26].
Genome sequence analysis and annotation
For structural annotation of the genome Rapid Annotations using Subsystems Technology (RAST) server [27] was used (http://rast.nmpdr.org) along with SEED database for the functional annotation of predicted gene models. SEED also provides subsystem (collection of functionally related protein families) and derived FIGfams (protein families) which represent the core of RAST annotation engine [28]. Subsequently, genome annotation through PIFAR annotation tool was used for the identification of bacterial genetic factors involved in plant host interaction [29]. Moreover, antiSMASH bacterial version 5.0 (https://antismash.secondarymetabolites.org/#!/start) [30] analysis was carried out to identify the biosynthetic gene clusters for secondary metabolites of B. contaminans NZ.
Examining the presence of proteins fostering virulence and pathogenicity
A total of 147 Burkholderia specific proteins reported for pathogenicity and virulence were collected from Virulence Factor Database (VFDB) (http://www.mgc.ac.cn/VFs/main.htm) [31] and investigated for the presence of virulence and pathogenicity related proteins in B. contaminans NZ with the help of BLASTp considering the e-value of at least 1e-10 and sequence identity, not less than 70%.
Data deposition
This Whole Genome Shotgun project has been deposited in DDBJ/ENA/GenBank under the accession QRBC00000000. The version described in this paper is version QRBC01000000.
In vivo growth promotion study of B. contaminans NZ on jute
Inoculation of jute seeds with the endophyte
At first B. contaminans NZ was cultured on TSA plates and then inoculated in TSB media and incubated at 37°C at a shaking speed of 180 rpm for 24–48 h. At the same time, scarification was done for jute seeds with sandpaper and the seeds surface was sterilized using 5% NaOCl. Sterilized seeds were dipped into B. contaminans NZ suspension. From McFarland standard, bacterial cells of 108 CFU/ml were determined by a serial dilution method [32].
In vivo pot experiment in hydroponic culture
Jute seeds inoculated with a bacterial suspension were grown under controlled environmental conditions in a plant growth chamber (Weiss Technik India Private Limited) at 28°C, 70% relative humidity, and 16-h light/8-h dark cycle. The seeds were allowed to germinate for three days and later grown in a hydroponic culture system. 1.25 ml per litre of a modified Yoshida medium (NH4NO3 91.4g, K2SO4 71.4g, NaH2PO4.2H2O 40.3g, CaCl2.2H2O 88.6g, and MgSO4.7H2O 324.0g per litre) and micronutrients (MnCl2.4H2O 1.5g, (NH4)6Mo7O24.4H2O 0.074g, H3BO3 0.934g, ZnSO4.7H2O 0.035 g, CuSO4 .5H2O 0.31g, FeCl3 .6H2O 7.70 g, citric acid 11.9 g and H2SO450.0g per litre) [33] was added to the seedlings. pH of the solution was adjusted to 5.5 with NaOH. Thirty seeds were sown 1 cm deep in a cork sheet in separate pots. Jute plants were grown for 10 days in hydroponic culture and 5 plants were collected for the measurement of fresh and dry weights, shoot, and root lengths on 4, 7, and 10 days after transfer of the seedlings into the hydroponic solution. The experiment was repeated three times.
Antifungal activity assay
Dual culture assay
In vitro bacteria-fungal dual-culture assays were established by both agar well diffusion and cross streak methods in 9 cm diameter Petri dish systems containing PDA medium [34]. A 5 mm plug taken from the plate of an actively growing fungal colony was inoculated on one side of the Petri dish. Fresh cells of B. contaminans NZ were either streaked in 3 cm length parallel lines on the other side of the fungal plug or 20 μl of the overnight bacterial liquid culture was introduced into a 6 mm diameter well punched aseptically with a pipette tip in an agar plate. Control treatments containing only the fungus were also set up. The plates were incubated at 28°C for 4 to 5 days. The experiment was set up for four replicates. After incubation, the diameters of the fungal colonies were scored and measured.
Antifungal activity of B. contaminans NZ was determined against all the studied phytopathogenic fungi and the toxicity was expressed as a percentage of growth inhibition (PGI) and calculated according to the Zygadlo et al.[35] formula,
where GC represents the average diameter of the fungus grown on PDA (control); GT represents the average diameter of the fungus co-cultivated on the PDA dish with the antagonistic bacterium. A paired t-test was used to check whether the percentage of growth inhibition was significant.
Co-culture of M. phaseolina and B. contaminans NZ
For GC-MS analysis, M. phaseolina was cultured in 20 ml and 100 ml PDB media in two separate conical flasks. B. contaminans NZ was cultured in 80 ml and 100 ml of TSB media in two separate conical flasks.
On the second day of incubation, 20 ml M. phaseolina culture was mixed with 80 ml B. contaminans NZ to form the co-culture and put to incubation again. All the cultures were incubated at 28°C for 5 days for volatile compound extraction and observed for any phenotypic changes in M. phaseolina culture in the presence of B. contaminans NZ.
For HPLC and LC-MS analysis, a separate 1000 ml culture of B. contaminans NZ was cultured under similar conditions described above.
Extraction of organic compounds from culture media
On day 6 of single cultures of fungus, bacterium, and co-cultured media were filtered using Whatman filter paper and centrifuged at 4000 rpm for 7 minutes to remove cell debris. Supernatants were separated into individual conical flasks. 1:1 ethyl acetate (v/v) was added to each flask, shaken, and kept for 4 hours. The organic phases were separated and dried using a rotary evaporator. Finally, the extracts were dissolved in 2 ml of ethyl acetate. A separate extraction (for HPLC and LC-MS analysis) of B. contaminans NZ culture media was also conducted using n-hexane instead of ethyl acetate and shaken for 24 hrs. The extracted fraction was finally dissolved in methanol.
GC-MS analysis of ethyl acetate extract
Ethyl acetate extract of extracellular components was subjected to GC-MS analysis using Shimadzu GCMS-QP2010 Ultra; (Japan) mass detector connected with a capillary column of Rxi-5ms, 30 m long, 0.25 mm i.d.0.25 μm film thickness. 1μL of the sample was injected with splitless mode. Helium was used as the carrier gas with the flow rate was set at 1.0 ml/min. The column oven temperature was programmed from 40°C (1 min hold) at a rate of 10°C/min to 300°C (10 min hold). Injector temperature was maintained at 250°C. Temperatures of the mass interface and ion source were 250°C and 200°C respectively, and detector voltage was 0.4 KV. The analysis was carried out in the EI (electron impact) mode with 70e V of ionization energy. The analysis was performed in full scan mode, ranging from m/z 50 to 400. The compounds were detected after analyzing the mass spectrum of each component using the NIST11 library.
HPLC and LC-MS analysis of methanolic extract
Analytical HPLC Dionex Ultimate 3000 with a C18 column (Nucleodur, 250x4.6 mm, particle size 5μm, pore size 110Å, 100-5C18ec) was used to analyze the bacterial extract dissolved in methanol. After dissolution, the extract was filtered and 200 μL was injected into the column. A multi-step gradient system (solvent system: acetonitrile and water with 0.05% trifluoroacetic acid; flow rate: 1ml/min; temperature: 23°C) was performed with an optimized protocol (0–10 min, 10% acetonitrile; 10–30 min, 10–80% acetonitrile; 30–40 min, 80% acetonitrile; 40–50 min, 80–0% acetonitrile; volume fraction). The wavelength at 225 nm was used to detect compounds eluted from the column. Various fractions were collected containing high peak intensity in the chromatogram and the fractions were dried at low temperature in a lyophilizer (LYOVAC GT 2) and dissolved in methanol to test for antifungal activity and followed by a re-run in HPLC to confirm the reproducibility of the peak. The purified active fraction was then subjected to LC-MS.
For Liquid Chromatography (LC), Agilent 1290 Infinity II instrument was used. C18 column (2.1 mm x 100 mm x 1.8 um) (Zorbax RRHD Eclipse) was used as the stationary phase and 50:50 ratio of water (with 0.1% formic acid) and acetonitrile was used as the mobile phase on isocratic mode. The column temperature and the mobile phase flow rate were kept at 30°C and 0.2 mL/min throughout the program. The sample injection volume was 5 uL. For Mass Spectrometry (MS) analysis, an Agilent 6420 Triple Quad Mass Spectrometer System was used, equipped with an electrospray ionization source operating at positive mode and scanning from m/z 100 to 1000. Nitrogen was used as the nebulizer gas, with pressure and flow set at 45 psi and 11 L/min, respectively. The capillary voltage was maintained at 4000 V, and dry gas temperature at 350°C. Mass Hunter software was used to control and analyze the data. The compound was identified based on its MS fragmentation and analyzed according to literature [36].
Identification of homologous melanin pathway(s) in M. phaseolina
Fungi contain different pathways for melanin biosynthesis; among them two pathways namely 1,8-dihydroxynaphthalene (DHN)-melanin and L-3,4-dihyroxyphenylalanine (DOPA)-melanin pathways, are primarily found in the ascomycota group of fungi [37]. M. phaseolina proteins homologous to this pathway were searched using BLAST. Pathway proteins discovered through literature review and BLAST searches were tested against the M. phaseolina proteome. At least a 30% identity cut-off was taken to consider the homology between the target and query protein [38].
Statistical analysis
The average shoot length, average root length and average height from the three repeated experiment of in vivo pot experiment was taken for statistical analysis. Data obtained were subjected to analysis of variance (ANOVA) test against the control. The results presented as average means, standard deviation (SD), and standard error (SE) were determined by following the standard procedures.
Results
Pot tests for in vivo assay of plant growth promotion
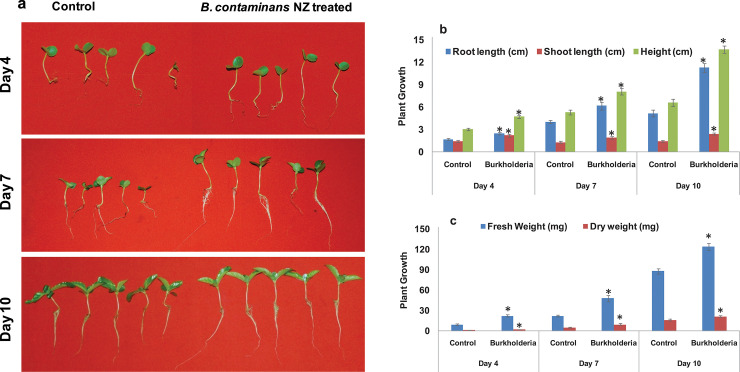
B. contaminans NZ significantly increased the root and shoot lengths, and the average height of jute seedlings (P<0.05) (Fig 1, S1 and S2 Tables). The maximum root length of 11.28 cm, was 116.9% greater than that of the control.
Fig 1. In vivo effect of plant growth promoting endophytic bacteria B. contaminans NZ.
(a) Bacterial effect significantly influences the increase in root and shoot lengths of jute seedlings when compared with the control jute plants at 4, 7, and 10 days. Comparisons of (b) shoot and root lengths, plant heights, and (c) fresh and dry weights of B. contaminans NZ-treated and control jute seeds. Error bars represent the standard error of the mean of the replicates. Shoot and root lengths are represented in cm, and fresh and dry weights in mg. Asterisk (*) denotes the difference between control and endophyte-treated samples at a significance level of P ≤ 0.05, as determined by ANOVA test. Values are mean(s) ± SD.
The effect on shoot length was also found to be significant; on average, it was 50.7% longer than that of the control jute plants. Fresh weight of 4-, 7-, and 10-day old jute seedlings increased by 141%, 122%, and 41%, respectively, compared to the controls (P<0.05).
In the presence of B. contaminans NZ, the dry weight of jute plants on an average, increased by 49.8% in all three replicates for each of the 4-, 7-, and 10-day measurements.
Whole genome data analysis
A summary of the whole genome annotation of B. contaminans NZ sequenced using Illumina MiSeq technology is presented in S3 Table. Functional analysis of B. contaminans NZ using the RAST server (Rapid Annotation using Subsystem Technology, http://rast.nmpdr.org), antiSMASH, and PIFAR (plant–bacteria information factor resource) revealed numerous genes and gene clusters (Table 1, S4 Table), which are reported to be exclusively associated with plant growth promotion.
Table 1. List of genes involved in plant growth promotion activity detected from RAST, antiSMASH, PIFAR analysis of the B. contaminans NZ whole genome.
| Properties | Name | Biosynthetic genes |
|---|---|---|
| Phytohormone production and stress alleviation | Management of ethylene stress | ACC (1-Aminocyclopropane-1-carboxylic acid) deaminase |
| Production of IAA (Indole Acetic Acid) | Indole-3-glycerate phosphate synthase | |
| Indole pyruvate oxidoreductase | ||
| Indole acetamide hydroxylate | ||
| Tryptophan synthase | ||
| Nitrile hydratase | ||
| Phosphate solubilization | pyrroloquinoline quinone gene | pqq |
| Enolase | eno | |
| Nitrogen fixation | nif gene cluster | nifHDK, nifQ, |
| Others | nodT, nir, nor, nolO | |
| Antibiotic biosynthesis | ||
| Pyrrolnitrin | prnA-prnD | |
| Siderophore Biosynthesis | Polychelin | pchR |
| Ferric siderophore transport | pchD-pchA | |
| ABC-type siderophore export system | feoB | |
| Siderophore pyoverdine | pvd |
As observed in other PGPR Burkholderia strains [39], the nifHDK operon required for nitrogen fixation was detected in B. contaminans NZ, along with the 1-aminocyclopropane-1-carboxylate (ACC) deaminase coding sequence, gene coding for iron(III) ABC transporter substrate-binding protein, phytoene synthase gene, the coding sequence for phosphotransferase system (PTS), and major facilitator superfamily (MFS) transporter genes. In silico analysis using RAST also revealed the presence of genes for indoleacetamide hydrolase involved in IAA production, which is a plant hormone associated with plant growth [40], and the eno gene, which assists in phosphate solubilization. Genome analysis also indicated the presence of pyrroloquinoline quinone synthase and glucose dehydrogenase, which are implicated in gluconic acid and 2-ketogluconic acid production, and required for mineral phosphate solubilization [41]. The annotated genome of B. contaminans NZ also contains 30 siderophore-related genes associated with ferric siderophore transport, ABC-type siderophore export system, arthrobactin, siderophore pyoverdine, and an intact siderophore pyochelin biosynthesis pch gene cluster. From the PIFAR data, in B. contaminans NZ, the biosynthesis of the major siderophore pyochelin is apparently associated with five genes (PMID: 22261733) distributed over five different regions in the bacterial genome.
While examining the core biosynthesis genes, genetic loci related to antibiotic production were also found in B. contaminans NZ, as presented in Table 1. Moreover, genes for pyrrolnitrin, an antifungal secondary metabolite produced by certain Bcc and several other gram-negative bacteria, were also identified. This metabolite inhibits the growth of a wide range of fungi and pathogenic bacteria [42, 43].
Most virulence genes absent in B. contaminans NZ
Various virulence factors reported in pathogenic bacteria and other plant endophytic strains of Burkholderia (B. contaminans CH1, B. contaminans MS14) were screened for in the B. contaminans NZ genome, which failed in identifying major virulence-related genes, such as the biosynthesis genes for cable pili, toxoflavin, cepacian, and O-antigen (O-Ag) biosynthetic cluster (Table 2). This differs from pathogenic strains that contain many virulence genes. However, few virulent genes in B. contaminans NZ are present as incomplete gene clusters.
Table 2. List of major virulence related genes present or absent in B. contaminans NZ.
| Feature name | Relevant gene/gene cluster | Status |
|---|---|---|
| Actin based intracellular motility | BimA | Absent |
| Adherence | BoaA | Absent |
| BoaB | Absent | |
| Type VI pilin system | Incomplete cluster (3 out of 11 genes) | |
| Anti-phagocytosis (O antigens) | Capsule I gene cluster | Absent |
| Secretion system | Bsa T3SS cluster | Absent |
| T6SS-1 cluster | Absent | |
| Signaling | Cdp | Present |
| Quorum sensing | Incomplete cluster (2 out of 8 genes) | |
| Cepacian | bceA-bceK, bceN- bceT | Absent |
| Cable pilin gene | cblA | Absent |
| Toxoflavin | toxR, toxA-toxE | Absent |
| 2-heptyl-3-hydroxy-4(1H)-quinolone | pqsA-pqsE | Absent |
| Hydrogencyanide | hcnA-hcnC | Absent |
| Cu2+ and Zn2+ containing periplasmic SOD | apaH-reG | Present |
| Zinc metalloprotease | Zmp | Present |
Antifungal activity assay
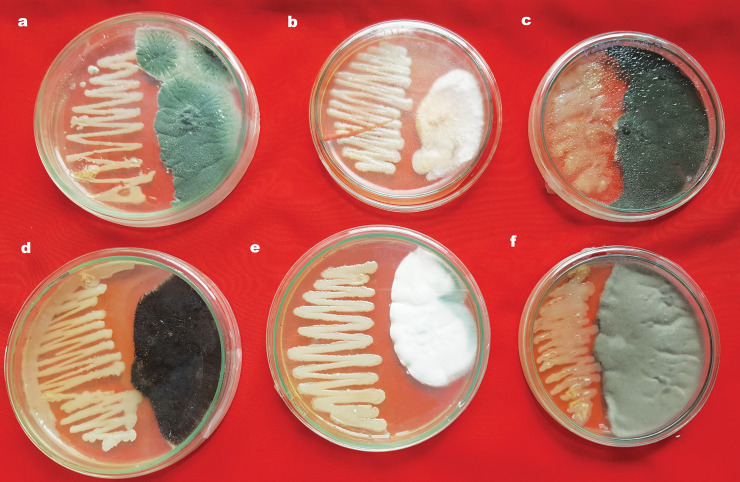
In dual culture, B. contaminans NZ substantially inhibited the growth of six plant fungal pathogens, namely Nigrospora sphaerica, Xylaria spp., Aspergillus fumigatus, Aspergillus niger, Penicillium oxalicum, and Rhizoctonia solani (Fig 2).
Fig 2. Antagonistic properties of B. contaminans NZ against six other phytopathogenic fungi.
(a) Nigrospora sphaerica (b) Xylaria spp.(c) Aspergillus fumigatus (d) Aspergillus niger (e) Penicillium oxalicum, and(f) Rhizoctonia solani.
In dual-culture assay, B. contaminans NZ demonstrated significant growth inhibitory activity, which ranged from 44% to 58% than that of the control (P<0.05), when co-cultured with all the tested fungi. The growth inhibition rates observed for each fungal species are shown in Table 3.
Table 3. Growth inhibition induced by B. contaminans NZ on different plant pathogenic fungi.
| Fungal species | Growth inhibition (%) (Mean ± SD) |
|---|---|
| Xylaria spp. | 57.71 ± 1.88 |
| Aspergillus fumigatus | 45.94 ± 1.45 |
| Aspergillus niger | 54.38 ± 4.10 |
| Penicillium oxalicum | 55.64 ± 2.96 |
| M. phaseolina | 49.04 ± 3.41 |
| Nigrospora sphaerica | 52.99± 3.67 |
| Rhizoctonia solani | 44.31 ± 4.36 |
Growth suppression and chromogenic aberration in B. contaminans NZ challenged M. phaseolina
M. phaseolina inoculated in liquid culture was initially pale in color, which assumes a characteristic black color within two to three days that perpetuates until day 5. However, a M. phaseolina culture challenged by B. contaminans NZ deviates from this usual behavior and retains the initial pale color up to day 5. Furthermore, M. phaseolina growth when subjected to co-culture with the bacterium remained practically static from day 2. Fig 3 shows the phenotypic changes in fungal growth from days 2 to 5.
Fig 3. Chromogenic aberration in B. contaminans NZ challenged M. phaseolina.
(A) M. phaseolina, B. contaminans NZ, and their co-cultures on day 5. (B) M. phaseolina without B. contaminans NZ retains the black color (left) compared to its B. contaminans NZ challenged counterpart (right). Inhibition of M. phaseolina growth and its pigmentation in the presence of B. contaminans NZ is evident compared to the culture containing only M. phaseolina.
Chemical analysis of bio-active compounds using GC-MS
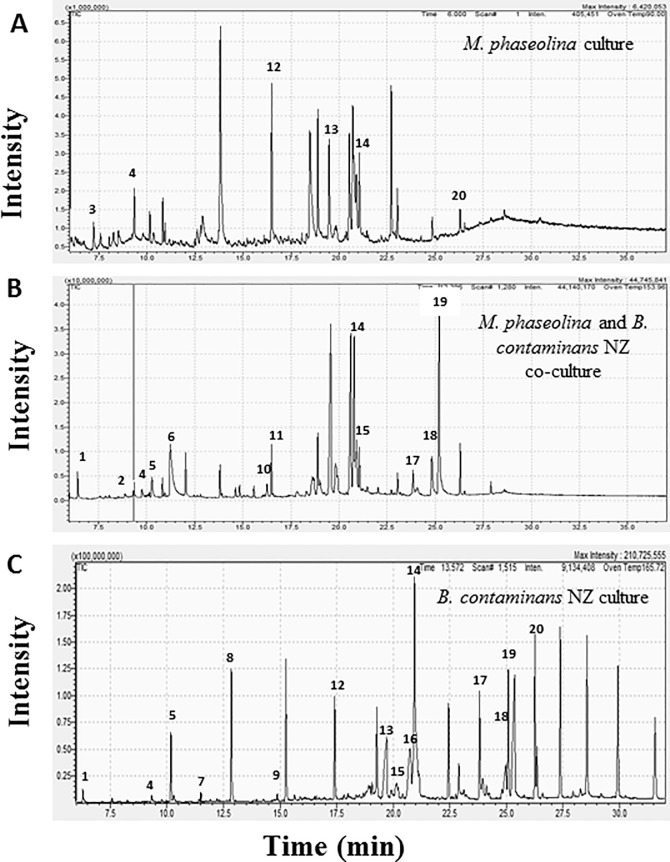
GC coupled with MS was used to identify the organic compounds produced by the fungus, bacteria, and their co-culture, and to identify the chemical nature of the bioactive compounds responsible for inhibiting fungal growth. Three sets of secondary metabolites obtained from (i) B. contaminans NZ, (ii) M. phaseolina, and (iii) B. contaminans NZ -M. phaseolina dual culture aided in identifying more than 72 compounds. Among them, 9,10-dihydro-12’-hydroxy-2’-methyl-5’-(phenylmethyl) ergotaman 3’,6’,18-trione; 2,3-dihydro-3,5-dihydroxy-6-methyl-4H-pyran-4-one; 1-(1,6-dioxooctadecyl)- pyrrolidine; 9-octadecenamide; 3-trifluoroacetoxypentadecane;2-hexyldecanol; and catechol, reportedly exhibit various biological activities, including tyrosinase inhibition, antifungal, and antimicrobial effects (Table 4). For example, catechol acts as a suicide inhibitor of enzyme tyrosinase [44], which forms melanin in fungi. Furthermore, 9, 10-dihydro-12’-hydroxy-2’-methyl-5’-(phenylmethyl) ergotaman 3’, 6’, 18-trione acts as an antimicrobial and anti-inflammatory agent [45]. A partial list of compounds and their corresponding functions are provided in Table 4, and peaks from GC-MS analyses of M. phaseolina (M), B. contaminans NZ (B), and M. phaseolina and B. contaminans NZ co-culture (C) are shown in Fig 4.
Table 4. List of potential compounds with their biological activities and retention times found in GC-MS analysis of B. contaminans NZ (B), M. phaseolina (M), and their co-culture (C).
| Sl. No. | Compound Name | Culture Name | Retention Time (min) | Biological Activity | Reference(s) |
|---|---|---|---|---|---|
| 1 | 2,5-Dimethyl pyrazine | B, C | 6.28 | Fungistatic activity | [46] |
| 2 | 2-Methoxy- phenol | C | 9.30 | Antioxidant, Cytotoxic | [47] |
| 3 | 5-Methyl-furfural | M, C | 7.1 | Antimicrobial, Antioxidant | [48] |
| 4 | 2-Undecanethiol, 2-methyl | M, C, B | 9.01/12.6 | Antimicrobial activity | [49] |
| 5 | 2,3-Dihydro-3,5-dihydroxy-6-methyl-4H-pyran-4-one | B, C | 10.4 | Antifungal, Oxidative stress inducer | [50, 51] |
| 6 | Catechol | C | 11.4 | Antifungal activity, Suicide substrate inhibitors of tyrosinase | [44, 52, 53] |
| 7 | 1,4-Benzenediol / Hydroquinone | B | 11.42 | Cytotoxic activity, Protein kinase inhibition | [54] |
| 8 | 2-(1,1-Dimethylethyl)-4,6-dinitro-phenol | B | 12.8 | Herbicide | [55] |
| 9 | Decahydro-1,4-dimethoxy naphthalene | B | 14.6 | Antimicrobial activity | [56] |
| 10 | 2-Hexyldecanol | C | 16.06 | Suppresses melanin synthesis | [57] |
| 11 | 3-trifluoro acetoxypentadecane | C | 16.6 | Antimicrobial activity | [58] |
| 12 | Methoxyacetic acid, 3-tridecyl ester | M, B | 16.6/17.8 | Cytotoxicity | [59] |
| 13 | Pyrrolo[1,2-a]pyrazine-1,4-dione | M, B | 19.6/19.9 | Antioxidant agent, Antifungal activity, Antibiotic activity | [60–62] |
| 14 | 3,6-Diisopropylpiperazin-2,5-dione | B | 20.1 | Antimalarial activity | [63] |
| 15 | 1-(1,6-Dioxooctadecyl)- pyrrolidine | B, C | 20.23/21.2 | Antioxidant activity | [64] |
| 16 | n-Hexadecanoic acid | B, M, C | 21.04 | Antibacterial, Antifungal, Cytotoxicity, | [65] |
| 17 | Hexadecamethylheptasiloxane | B, C | 23.8 | Antifungal | [66] |
| 18 | 9-Octadecenamide | B, C | 24.8 | Antimicrobial, Anti-inflammatory | [58] |
| 19 | 9,10-Dihydro-12’-hydroxy-2’-methyl-5’-(phenylmethyl)-, (5’ alpha,10 alpha) - ergotaman-`3’,6’,18-trione | B, C | 24.9/25.1 | Antimicrobial, Anti-inflammatory, Alpha-amaylase inhibitory activity | [45] |
| 20 | Di-n-octyl phthalate | B, M | 25.9/ 26.35 | Antifungal and antioxidant | [67, 68] |
Fig 4. Peaks from GC-MS analyses with potent volatile compounds.
(A) M. phaseolina, (B) M. phaseolina and B. contaminans NZ co-culture, and (C) B. contaminans NZ. The number on different peaks corresponds to the serial number of the compounds in Table 4.
Identification of pyrrolnitrin in methanolic extract of B. contaminans NZ culture
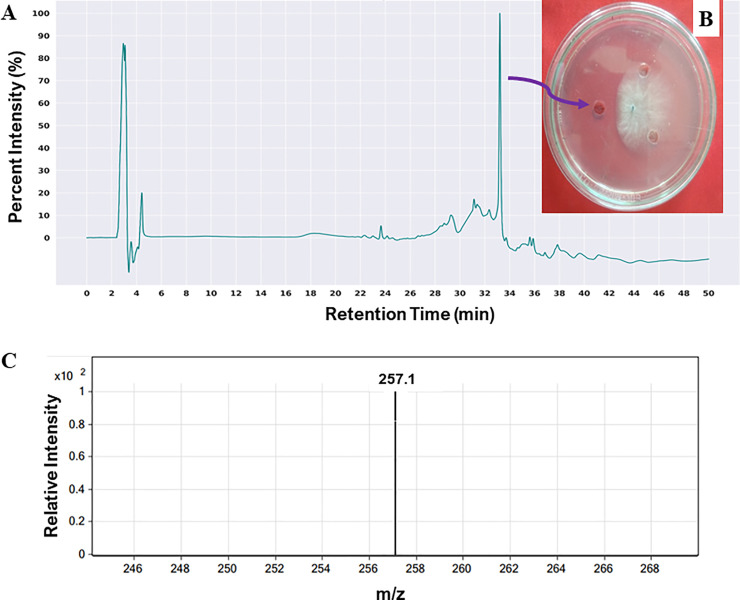
The chromatogram of RP-HPLC analysis of the methanolic crude extract showed various peaks (Fig 5A). RP-HPLC fractions were collected and their activity tested against M. phaseolina, wherein the fraction obtained at a retention time of 33.2 min exhibited antifungal activity (Fig 5B). The compound with a molecular ion peak of 257.1 (m/z) was identified as pyrrolnitrin via LC-MS analysis, after comparing with standard pyrrolnitrin (Fig 5C).
Fig 5. Identification of bioactive compounds in methanolic extract of B. contaminans NZ culture.
(A) HPLC chromatograms of the active fraction (peak at 33.2 min retention time) upon reinjection. (B) Peak which inhibits the mycelial progression of M. phaseolina towards the active fraction when applied on the well, marked by a violet arrow. (C) LC-MS chromatogram of active peak identified to be pyrrolnitrin with a mass of 257.1 Da.
Identification of putative melanin pathways in M. phaseolina through homology-based search
After an extensive literature review of different melanin pathways active in fungi, polyketide synthase, tetrahydroxy naphthalene reductase, trihydroxy naphthalene reductase, scytalone dehydratase, and tyrosinase proteins were selected, and their corresponding homologs in M. phaseolina were identified through a BLAST search. The details are presented in Table 5. In addition, putative pathways proposed in S1 Fig presents the order in which the reactions occur during melanin synthesis in M. phaseolina.
Table 5. Homologous proteins of melanin pathways found in M. phaseolina.
| Query Accession | Query Name | Subject Accession | Subject Name | Percent Identity |
|---|---|---|---|---|
| K2S0W9 | Betaketoacyl synthase | D7RJP3 | Polyketide synthase | 31.25 |
| K2RKI0 | Short-chain dehydrogenase/ reductase SDR | Q12634 | Tetrahydroxy naphthalene reductase | 75 |
| K2RKI0 | Short-chain dehydrogenase/ reductase SDR | W3XC32 | Trihydroxy | 75.5 |
| Naphthalene | ||||
| reductase | ||||
| K2R7Y4 | Scytalone dehydratase | W3XEE6 | Scytalone | 82.7 |
| dehydratase | ||||
| K2R777 | Tyrosinase | U7PL38 | Tyrosinase | 31.132 |
Discussion
Since chemical control and most cultural practices are not effective representative tools that limit the pathogenic fungal growth and their distribution [69], the use of biocontrol agents has become a promising alternative in agriculture. Biocontrol using plant growth-promoting rhizobacteria (PGPR) is a potentially attractive and efficient disease management approach, as it promotes plant growth, enhances tolerance to abiotic stress [70], suppresses pathogens locally, and induces systemic resistance (ISR) against a broad range of crop diseases [71, 72].
Several studies conducted on endophytes have emphasized their ability in promoting plant growth and their additive/synergistic effects on plant growth and protection. In general, such growth-promoting rhizospheric bacteria belong to the following species: Alcaligenes, Arthrobacteria, Azospirillum, Azotobacter, Bacillus, Burkholderia, Enterobacter, Klebsiella, and Pseudomonas [8, 73]. The endophytic strain B. contaminans NZ isolated from jute has also been characterized in vivo for its potential plant growth-promoting (PGP) traits, which significantly increased both yield and biomass when jute seedlings were inoculated with the bacterium, and notably reduced the development of disease symptoms caused by M. phaseolina.
As B. contaminans NZ exhibited growth promotion and apparent protection against the phytopathogen M. phaseolina, its whole genome was sequenced to determine the characteristic genetic features. Genome annotation of B. contaminans NZ assisted in identifying genes related to phytohormone production, stress alleviation, phosphate solubilization, nitrogen fixation, antibiotic biosynthesis, and siderophore biosynthesis. In vitro experiments of the endophyte also displayed significant production of the above-listed compounds (S5 Table, S2 Fig).
Certain Burkholderia species demonstrate clinical associations and plant-pathogenic traits [74, 75]; therefore, genes related to virulence and pathogenicity were also investigated in the B. contaminans NZ genome. Several bacterial species from the B. cepacia complex (Bcc) are opportunistic pathogens [76]. B. cenocepacia strains expressing both cable (Cbl) pili and 22-KDa adhesin proteins have been reported to bind strongly to cytokeratin 13 (CK13), which efficiently infects the host cell by invading the squamous epithelium [77]. However, the cblA (giant cable pili) gene was absent in the symbiotic and legume-nodulating species [78]. B. contaminans NZ also lacks the cable pili biosynthesis gene cluster, indicating their inability to attach to the host cell, which is required to initiate infection. Furthermore, VgrG-5, which is a Burkholderia type VI secretion system 5-associated protein, required for complete mammalian virulence [79], is also absent in B. contaminans NZ genome. Virulence-associated protein sequences (for chemotaxis, attachment, type 3 and type 6 secretion systems; T3SS and T6SS) and typically described hallmark features representing the true and opportunistic pathogenic Burkholderia strains are also absent in B. contaminans NZ. Cystic fibrosis (CF)-related O-antigen of lipopolysaccharides associated with transmitting infections in CF patients [80] and zinc metalloproteases that may be involved in the overall virulence of several Bcc strains [81] are also absent. Among the virulence-associated proteins of Burkholderia, only the sodC gene encoding a Cu2+ and Zn2+ containing periplasmic superoxide dismutase that contributes in intracellular survival, which indicates self-protection ability in CF patients, is present in the genome [82]. Burkholderia species commonly produce plant-toxic secondary metabolites, polysaccharides, and other toxins such as rice grain rot and wilt causal agent, toxoflavin; exo-polysaccharide toxin, cepacian; hydrogen cyanide (HCN); and 2-heptyl-3-hydroxy-4(1H)-quinolone [80]. These pathogenic genes were absent in this jute endophytic bacterium.
The B. contaminans NZ genome also contained antibiotic-biosynthetic genes. Thus, to gather further evidence, M. phaseolina and several other phytopathogenic fungi, namely, Xylaria spp., Aspergillus fumigatus, Aspergillus niger, Penicillium oxalicum, Nigrospora sphaerica, and Rhizoctonia solani were co-cultured with B. contaminans NZ, where all fungi tested were suppressed with varying degrees of inhibition (Fig 2, Table 3), confirming its antifungal activity.
Growth inhibition of M. phaseolina by B. contaminans NZ caused marked morphological changes and chromogenic aberration in the phytofungus [13]. Such morphological changes in the cell membrane have also been reported for B. cepacia-mediated inhibition of F. solani and C. dematium [83]. Burkholderia CF66I noticeably alters R. solani hyphae with multiple branches and swelling [84]. In previous reports, co-culturing M. phaseolina and B. contaminans NZ in solid media caused the fungus to lose its characteristic black color, restricted its growth, and attenuated its infectivity [13]. The present study also observed a similar deviation in pigmentation and growth repression in liquid co-culture (Fig 3).
Various reports have demonstrated that secondary metabolites produced by certain bacteria can influence fungal growth [85, 86]. Therefore, secondary metabolites were analyzed to explain chromogenic aberration and growth suppression of M. phaseolina when co-cultured with B. contaminans NZ. GC-MS analysis of ethyl acetate extracts of culture supernatant revealed over 72 compounds, among which 20 compounds are biologically important; 2,5-dimethyl pyrazine, 2,3-dihydro-3,5-dihydroxy-6-methyl-4H-pyran-4-one, hexadecamethylheptasiloxane, 9-octadecenamide, and 9,10-dihydro-12’- hydroxy-2’-methyl-5’- (phenylmethyl)-, (5’ alpha,10 alpha)- ergotaman-3’,6’,18- trione compounds were discovered in both B. contaminans NZ and its co-cultures, but were absent in the M. phaseolina extract. 2,5-dimethylpyrazine, 2,3-dihydro-3,5-dihydroxy-6-methyl-4H-pyran-4-one, and hexadecamethylheptasiloxane exhibits antifungal activities [46, 50, 51, 87]. In addition, 9-octadecenamide and 9,10-dihydro-12’- hydroxy-2’-methyl-5’- (phenylmethyl)-, (5’ alpha,10 alpha)- ergotaman-3’,6’,18- trione reportedly exhibits antimicrobial properties [58, 88]. The absence of these compounds in M. phaseolina extract implies that they are most likely produced only by B. contaminans NZ, thereby inhibiting the pathogen in the liquid media. 2-hexyldecanol, which suppresses melanin biosynthesis [57], and catechol, which demonstrates antifungal properties [52], are exclusively produced in the co-culture. Catechol, is also known as a suicide substrate for enzyme tyrosinase [53], which is involved in the DOPA-melanin biosynthesis pathway [37]. These compounds apparently produced only by B. contaminans NZ during co-culture condition are expected to play a role in the chromogenic aberration and growth suppression of M. phaseolina. n-Hexadecanoic acid possesses antifungal, antibacterial, and cytotoxic activities [65] and 2-undecanethiol, 2-methyl possesses antimicrobial activity [49]. Both compounds are ubiquitously present in B. contaminans NZ, M. phaseolina, and their co-cultures. Both organisms appear to be armed with antagonistic compounds, ready to inhibit each other in an attempt to avail the available resources.
Burkholderia spp. reportedly produces potent antifungal compound pyrrolnitrin [89]. This compound exhibits inhibitory effects by obstructing the synthesis of vital biomolecules (DNA, RNA, and protein), uncoupling oxidative phosphorylation, impeding mitotic division, and inhibiting several biological mechanisms. The genome of B. contaminans NZ also contained a gene cluster for pyrrolnitrin synthesis (Table 2). Although the GC-MS data revealed the presence of several compounds that implied possible modes for M. phaseolina growth suppression, pyrrolnitrin in the secondary metabolite profile was absent. Therefore, the extraction process was altered by substituting the extraction solvent with n-hexane instead of ethyl acetate to identify pyrrolnitrin, which is in agreement with earlier reports that employed a similar method [43]. Thereafter rest of the method remained similar except that the extract was dissolved in methanol instead of ethyl acetate. RP-HPLC was performed to purify the active compound(s) to detect the appreciable suppression of fungal growth by the crude extract. Only one fraction exhibited considerable inhibitory activity against M. phaseolina, and its mass was identified using LC-MS analysis (257.1 Da), which was identical to the standard pyrrolnitrin (molecular weight: 257.07 g/mol).
A previous iTRAQ proteomic analysis of M. phaseolina challenged with Burkholderia showed the downregulation of beta-ketoacyl synthase, scytalone dehydratase, tyrosinase, enzymes of the DHN-melanin, and DOPA-melanin pathways [13]. Catechol present in the co-culture of B. contaminans NZ and M. phaseolina can explain the downregulation of tyrosinase. Kojic acid [5-hydroxy-2-(hydroxymethyl)-4H-pyran-4-one] is a prominent tyrosinase inhibitor [37], and 2,3-dihydro-3,5-dihydroxy-6-methyl-4H-pyran-4-one, identified by GC-MS analysis, is an oxidized form of kojic acid and is expected to act identical to its reduced counterpart. This derivatization could have occurred during sample processing for GC-MS analysis.
Further investigation of the M. phaseolina enzymes involved in melanin synthesis, similar to those reported earlier in other fungi, led to the identification of SDR–short chain dehydrogenase/reductase, which is similar to both trihydroxy naphthalene reductase [90] and tetrahydroxy naphthalene reductase [91], which are enzymes of the DHN-melanin pathway [92]. Tyrosinase [92, 93] performs multiple catalysis in the DOPA-melanin pathway; it converts phenylalanine to tyrosine, tyrosine to DOPA, and DOPA to DOPA quinone, which is eventually converted to melanin further downstream. However, both DOPA-melanin and DHN-melanin pathways are yet to be fully elucidated. This reduces the scope of finding other homologous pathway proteins in M. phaseolina. The percent identity of beta-ketoacyl synthase and tyrosinase was lower (~31%) than that of tetrahydroxy naphthalene reductase, trihydroxy naphthalene reductase, and scytalone dehydratase (~75% or more). This was possibly due to only few reports that describe the two genes with precise genomic features. Based on this evidence, a tentative melanin pathway was proposed for M. phaseolina in this study (S1 Fig).
Conclusion
Whole genome analysis and secondary metabolite characterization have acknowledged B. contaminans NZ as a good biocontrol agent, in addition to its role in plant growth promotion. It suppresses the growth of multiple fungi, especially the phytopathogen M. phaseolina, by using catechol, pyrrolnitrin, and other antimicrobial agents. Compounds identified in the extracts of B. contaminans NZ or the co-culture of B. contaminans NZ and M. phaseolina contain compounds that inhibit melanin biosynthesis, which possibly contributes to the observed chromogenic aberration and growth suppression of the fungi. B. contaminans NZ can be established as a bio-control agent by conducting toxicity tests to ensure its safety, followed by field experiments to determine its efficacy under different environmental conditions. Hence, further studies are needed to optimize the formulation and application methods of B. contaminans NZ to fully maximize its potential as an effective agent in controlling M. phaseolina.
Supporting information
(TIF)
Qualitative assay for (a) siderophore, (b) ACC deaminase, and (c) nitrogen of B. contaminans NZ.
(TIF)
The data of three replicates per experiment are presented as means and standard deviations.
(DOCX)
The data of three replicates per experiment are presented as mean(s) and standard deviation(s).
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The authors cordially thank NSU Genome Research Institute, North South University, Bangladesh for the whole genome sequencing and Dr. Abdul Baten, AgResearch Ltd. Christchurch, NZ for helping with the whole genome analysis and Enayet Hossain for collecting the GC-MS data. We also acknowledge the Bangladesh Council of Scientific and Industrial Research (BCSIR) for providing support to carry out the GC-MS.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This project is funded by Higher Education Quality Enhancement Project (HEQEP) (Grant number: CP-3250), a World Bank financed development project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Adesemoye AO, Torbert HA, Kloepper JW. Plant growth-promoting rhizobacteria allow reduced application rates of chemical fertilizers. Microb Ecol. 2009;58: 921–929. doi: 10.1007/s00248-009-9531-y [DOI] [PubMed] [Google Scholar]
- 2.Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, Campisano A, et al. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol Mol Biol Rev. 2015;79: 293–320. doi: 10.1128/MMBR.00050-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaul S, Sharma T, Dhar MK. “Omics” tools for better understanding the plant–endophyte interactions. Front Plant Sci. 2016;7: 955. doi: 10.3389/fpls.2016.00955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kloepper JW, Leong J, Teintze M, Schroth MN. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature. 1980;286: 885–886. doi: 10.1038/286885a0 [DOI] [Google Scholar]
- 5.Ma Y, Prasad MNV, Rajkumar M, Freitas H. Plant growth promoting rhizobacteria and endophytes accelerate phytoremediation of metalliferous soils. Biotechnology Advances. Biotechnol Adv; 2011. pp. 248–258. doi: 10.1016/j.biotechadv.2010.12.001 [DOI] [PubMed] [Google Scholar]
- 6.Pillay VK, Nowak J. Inoculum density, temperature, and genotype effects on in vitro growth promotion and epiphytic and endophytic colonization of tomato (Lycopersicon esculentum L.) seedlings inoculated with a pseudomonad bacterium. Can J Microbiol. 1997;43: 354–361. doi: 10.1139/m97-049 [DOI] [Google Scholar]
- 7.Mitter B, Petric A, Shin MW, Chain PSG, Hauberg-Lotte L, Reinhold-Hurek B, et al. Comparative genome analysis of Burkholderia phytofirmans PsJN reveals a wide spectrum of endophytic lifestyles based on interaction strategies with host plants. Front Plant Sci. 2013;4. doi: 10.3389/fpls.2013.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vurukonda SSKP, Giovanardi D, Stefani E. Plant growth promoting and biocontrol activity of streptomyces spp. As endophytes. International Journal of Molecular Sciences. MDPI AG; 2018. doi: 10.3390/ijms19040952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajamanickam S, Karthikeyan G, Kavino M, Manoranjitham SK. Biohardening of micropropagated banana using endophytic bacteria to induce plant growth promotion and restrain rhizome rot disease caused by Pectobacterium carotovorum subsp. carotovorum. Sci Hortic (Amsterdam). 2018;231: 179–187. doi: 10.1016/j.scienta.2017.12.037 [DOI] [Google Scholar]
- 10.Verma SK, Kingsley K, Bergen M, English C, Elmore M, Kharwar RN, et al. Bacterial endophytes from rice cut grass (Leersia oryzoides L.) increase growth, promote root gravitropic response, stimulate root hair formation, and protect rice seedlings from disease. Plant Soil. 2018;422: 223–238. doi: 10.1007/s11104-017-3339-1 [DOI] [Google Scholar]
- 11.Babu BK, Saxena AK, Srivastava AK, Arora DK. Identification and detection of Macrophomina phaseolina by using species-specific oligonucleotide primers and probe. Mycologia. 2007;99: 797–803. doi: 10.3852/mycologia.99.6.797 [DOI] [PubMed] [Google Scholar]
- 12.Islam MS, Haque MS, Islam MM, Emdad EM, Halim A, Hossen QMM, et al. Tools to kill: Genome of one of the most destructive plant pathogenic fungi Macrophomina phaseolina. BMC Genomics. 2012;13: 493. doi: 10.1186/1471-2164-13-493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zaman NR, Kumar B, Nasrin Z, Islam MR, Maiti TK, Khan H. Proteome Analyses Reveal Macrophomina phaseolina’s Survival Tools When Challenged by Burkholderia contaminans NZ. ACS Omega. 2020;5: 1352–1362. doi: 10.1021/acsomega.9b01870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Depoorter E, Bull MJ, Peeters C, Coenye T, Vandamme P, Mahenthiralingam E. Burkholderia: an update on taxonomy and biotechnological potential as antibiotic producers. Applied Microbiology and Biotechnology. Springer Verlag; 2016. pp. 5215–5229. doi: 10.1007/s00253-016-7520-x [DOI] [PubMed] [Google Scholar]
- 15.Mannaa M, Park I, Seo YS. Genomic features and insights into the taxonomy, virulence, and benevolence of plant-associated burkholderia species. International Journal of Molecular Sciences. MDPI AG; 2019. doi: 10.3390/ijms20010121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, Roberts DP, Dery PD, Meyer SLF, Lohrke S, Lumsden RD, et al. Broad spectrum anti-biotic activity and disease suppression by the potential biocontrol agent Burkholderia ambifaria BC-F. Crop Prot. 2002;21: 129–135. doi: 10.1016/S0261-2194(01)00074-6 [DOI] [Google Scholar]
- 17.Degrossi JJ, Merino C, Isasmendi AM, Ibarra LM, Collins C, Bo NE, et al. Whole Genome Sequence Analysis of Burkholderia contaminans FFH2055 Strain Reveals the Presence of Putative β-Lactamases. Curr Microbiol. 2019;76: 485–494. doi: 10.1007/s00284-019-01653-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghequire MGK, De Mot R. Distinct colicin M-like bacteriocin-immunity pairs in Burkholderia. Sci Rep. 2015;5: 17368. doi: 10.1038/srep17368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lupo A, Haenni M, Madec J-Y. Antimicrobial Resistance in Acinetobacter spp. and Pseudomonas spp. Antimicrobial Resistance in Bacteria from Livestock and Companion Animals. American Society of Microbiology; 2018. pp. 377–393. doi: 10.1128/microbiolspec.arba-0007-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellis CN, Cooper VS. Experimental adaptation of burkholderia cenocepacia to onion medium reduces host range. Appl Environ Microbiol. 2010;76: 2387–2396. doi: 10.1128/AEM.01930-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parke JL, Gurian-Sherman D. Diversity of the burkholderia cepacia complex and implications for risk assessment of biological control strains. Annual Review of Phytopathology. Annu Rev Phytopathol; 2001. pp. 225–258. doi: 10.1146/annurev.phyto.39.1.225 [DOI] [PubMed] [Google Scholar]
- 22.Coombs JT, Franco CMM. Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl Environ Microbiol. 2003;69: 5603–5608. doi: 10.1128/AEM.69.9.5603-5608.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Najnin RA, Shafrin F, Polash AH, Zaman A, Hossain A, Taha T, et al. A diverse community of jute (Corchorus spp.) endophytes reveals mutualistic host–microbe interactions. Ann Microbiol. 2015;65: 1615–1626. doi: 10.1007/s13213-014-1001-1 [DOI] [Google Scholar]
- 24.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19: 455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Assefa S, Keane TM, Otto TD, Newbold C, Berriman M. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics. 2009;25: 1968–1969. doi: 10.1093/bioinformatics/btp347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36. doi: 10.1093/nar/gkn179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aziz RK, Bartels D, Best A, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9: 75. doi: 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014;42: D206. doi: 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martínez-García PM, López-Solanilla E, Ramos C, Rodríguez-Palenzuela P. Prediction of bacterial associations with plants using a supervised machine-learning approach. Environ Microbiol. 2016;18: 4847–4861. doi: 10.1111/1462-2920.13389 [DOI] [PubMed] [Google Scholar]
- 30.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, et al. AntiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47: W81–W87. doi: 10.1093/nar/gkz310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, et al. VFDB: A reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33: D325–D328. doi: 10.1093/nar/gki008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Islam S, Akanda AM, Prova A, Islam MT, Hossain MM. Isolation and Identification of Plant Growth Promoting Rhizobacteria from Cucumber Rhizosphere and Their Effect on Plant Growth Promotion and Disease Suppression. Front Microbiol. 2016;6: 1360. doi: 10.3389/fmicb.2015.01360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yuan J, Raza W, Shen Q, Huang Q. Antifungal activity of Bacillus amyloliquefaciens NJN-6 volatile compounds against Fusarium oxysporum f. sp. cubense. Appl Environ Microbiol. 2012;78: 5942–5944. doi: 10.1128/AEM.01357-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balouiri M, Sadiki M, Ibnsouda SK. Methods for in vitro evaluating antimicrobial activity: A review. Journal of Pharmaceutical Analysis. Xi’an Jiaotong University; 2016. pp. 71–79. doi: 10.1016/j.jpha.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elshafie H, Camele I, Racioppi R, Scrano L, Iacobellis N, Bufo S. In Vitro Antifungal Activity of Burkholderia gladioli pv. agaricicola against Some Phytopathogenic Fungi. Int J Mol Sci. 2012;13: 16291–16302. doi: 10.3390/ijms131216291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Navarro-González I, González-Barrio R, García-Valverde V, Bautista-Ortín A, Periago M. Nutritional Composition and Antioxidant Capacity in Edible Flowers: Characterisation of Phenolic Compounds by HPLC-DAD-ESI/MSn. Int J Mol Sci. 2014;16: 805–822. doi: 10.3390/ijms16010805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belozerskaya TA, Gessler NN, Aver‘yanov AA. Melanin Pigments of Fungi. Fungal Metabolites. Springer International Publishing; 2017. pp. 263–291. doi: 10.1007/978-3-319-25001-4_29 [DOI] [Google Scholar]
- 38.Pearson WR. An introduction to sequence similarity (“homology”) searching. Curr Protoc Bioinforma. 2013;Chapter 3: Unit3.1. doi: 10.1002/0471250953.bi0301s42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parra-Cota FI, Peña-Cabriales JJ, de los Santos-Villalobos S, Martínez-Gallardo NA, Délano-Frier JP. Burkholderia ambifaria and B. caribensis Promote Growth and Increase Yield in Grain Amaranth (Amaranthus cruentus and A. hypochondriacus) by Improving Plant Nitrogen Uptake. Freitag NE, editor. PLoS One. 2014;9: e88094. doi: 10.1371/journal.pone.0088094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esmaeel Q, Jacquard C, Clément C, Sanchez L, Ait Barka E. Genome sequencing and traits analysis of Burkholderia strains reveal a promising biocontrol effect against grey mould disease in grapevine (Vitis vinifera L.). World J Microbiol Biotechnol. 2019;35: 1–15. doi: 10.1007/s11274-019-2613-1 [DOI] [PubMed] [Google Scholar]
- 41.Liu C, Mou L, Yi J, Wang J, Liu A, Yu J. The Eno Gene of Burkholderia cenocepacia Strain 71–2 is Involved in Phosphate Solubilization. Curr Microbiol. 2019;76: 495–502. doi: 10.1007/s00284-019-01642-7 [DOI] [PubMed] [Google Scholar]
- 42.Lu SE, Novak J, Austin FW, Gu G, Ellis D, Kirk M, et al. Occidiofungin, a unique antifungal glycopeptide produced by a strain of Burkholderia contaminans. Biochemistry. 2009;48: 8312–8321. doi: 10.1021/bi900814c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jung BK, Hong SJ, Park GS, Kim MC, Shin JH. Isolation of Burkholderia cepacia JBK9 with plant growth-promoting activity while producing pyrrolnitrin antagonistic to plant fungal diseases. Appl Biol Chem. 2018;61: 173–180. doi: 10.1007/s13765-018-0345-9 [DOI] [Google Scholar]
- 44.Land EJ, Ramsden CA, Riley PA. The mechanism of suicide-inactivation of tyrosinase: A substrate structure investigation. Tohoku J Exp Med. 2007;212: 341–348. doi: 10.1620/tjem.212.341 [DOI] [PubMed] [Google Scholar]
- 45.Kumari N, Menghani E, Mithal R. GCMS analysis of compounds extracted from actinomycetes AIA6 isolates and study of its antimicrobial efficacy. Indian J Chem Technol. 2019;26: 362–370. Available: http://nopr.niscair.res.in/handle/123456789/49682 [Google Scholar]
- 46.Chuankun X, Minghe M, Leming Z, Keqin Z. Soil volatile fungistasis and volatile fungistatic compounds. Soil Biol Biochem. 2004;36: 1997–2004. doi: 10.1016/j.soilbio.2004.07.020 [DOI] [Google Scholar]
- 47.Fujisawa S, Ishihara M, Murakami Y, Atsumi T, Kadoma Y, Yokoe I. Predicting the biological activities of 2-methoxyphenol antioxidants: effects of dimers. In Vivo. 2007; 21(2):181–8. . [PubMed] [Google Scholar]
- 48.Mar A, Pripdeevech P. Chemical composition and antibacterial activity of essential oil and extracts of Citharexylum spinosum flowers from Thailand. Nat Prod Commun. 2014;9: 707–710. doi: 10.1177/1934578x1400900532 [DOI] [PubMed] [Google Scholar]
- 49.Meenakshi VK, Gomathy S, Senthamarai S. Paripooranaselvi M. Chamundeswari KP. GC-MS Determination of the Bioactive Components of Microcosmus Exasperatus heller, 1878. J Curr Chem Pharm Sci. 2012;2: 271–276. [Google Scholar]
- 50.Teoh YP, Don MM, Ujang S. Media selection for mycelia growth, antifungal activity against wood-degrading fungi, and GC-MS study by Pycnoporus sanguineus. BioResources. 2011;6: 2719–2731. doi: [DOI] [Google Scholar]
- 51.Hiramoto K, Nasuhara A, Michikoshi K, Kato T, Kikugawa K. DNA strand-breaking activity and mutagenicity of 2,3-dihydro-3,5-dihydroxy-6-methyl-4H-pyran-4-one (DDMP), a Maillard reaction product of glucose and glycine. Mutat Res—Genet Toxicol Environ Mutagen. 1997;395: 47–56. doi: 10.1016/S1383-5718(97)00141-1 [DOI] [PubMed] [Google Scholar]
- 52.Al-Huqail AA, Behiry SI, Salem MZM, Ali HM, Siddiqui MH, Salem AZM. Antifungal, antibacterial, and antioxidant activities of Acacia saligna (Labill.) H. L. Wendl. Flower extract: HPLC analysis of phenolic and flavonoid compounds. Molecules. 2019;24. doi: 10.3390/molecules24040700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang T-S. An Updated Review of Tyrosinase Inhibitors. Int J Mol Sci. 2009;10: 2440–2475. doi: 10.3390/ijms10062440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sladic D, Gasic M. Reactivity and Biological Activity of the Marine Sesquiterpene Hydroquinone Avarol and Related Compounds from Sponges of the Order Dictyoceratida. Molecules. 2006;11: 1–33. doi: 10.3390/11010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beulke S, Malkomes HP. Effects of the herbicides metazachlor and dinoterb on the soil microflora and the degradation and sorption of metazachlor under different environmental conditions. Biol Fertil Soils. 2001;33: 467–471. doi: 10.1007/s003740100354 [DOI] [Google Scholar]
- 56.Upadhayaya RS, Vandavasi JK, Kardile RA, Lahore S V., Dixit SS, Deokar HS, et al. Novel quinoline and naphthalene derivatives as potent antimycobacterial agents. Eur J Med Chem. 2010;45: 1854–1867. doi: 10.1016/j.ejmech.2010.01.024 [DOI] [PubMed] [Google Scholar]
- 57.Hakozaki T, Laughlin T, Zhao S, Wang J, Deng D, Jewell-Motz E, et al. A regulator of ubiquitin-proteasome activity, 2-hexyldecanol, suppresses melanin synthesis and the appearance of facial hyperpigmented spots. Br J Dermatol. 2013;169: 39–44. doi: 10.1111/bjd.12364 [DOI] [PubMed] [Google Scholar]
- 58.Hadi I, Mashkoor Hussein H. Antimicrobial activity and spectral chemical analysis of methanolic leaves extract of adiantum capillus-veneris using GC-MS and FT-IR spectroscopy. Int J Pharmacogn Phytochem Res. 2016;8(3): 369–385. Available: www.ijppr.com [Google Scholar]
- 59.Vivekraj AV P., Anandgideon. and V. Analysis of Phytochemical constituents of the chloroform extracts of Abutilon hirtum (Lam.) Sweet using GC-MS Method. Int J Pharmacol Res. 2019;09: 167–171. doi: 10.7439/ijpr [DOI] [Google Scholar]
- 60.Ser HL, Palanisamy UD, Yin WF, Abd Malek SN, Chan KG, Goh BH, et al. Presence of antioxidative agent, Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro- in newly isolated Streptomyces mangrovisoli sp. nov. Front Microbiol. 2015;6. doi: 10.3389/fmicb.2015.00854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kannabiran DK. Bioactivity of Pyrrolo[1,2-a]pyrazine-1,4-dione,hexahydro-3-(phenylmethyl)- Extracted from Streptomyces sp. VITPK9 Isolated from the Salt Spring Habitat of Manipur, India. Asian J Pharm Free full text Artic from Asian J Pharm. 2016;10: 265. doi: 10.22377/AJP.V10I04.865 [DOI] [Google Scholar]
- 62.Kiran GS, Priyadharsini S, Sajayan A, Ravindran A, Selvin J. An antibiotic agent pyrrolo[1,2-: A] pyrazine-1,4-dione,hexahydro isolated from a marine bacteria Bacillus tequilensis MSI45 effectively controls multi-drug resistant Staphylococcus aureus. RSC Adv. 2018;8: 17837–17846. doi: 10.1039/c8ra00820e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pérez-Picaso L, Olivo HF, Argotte-Ramos R, Rodríguez-Gutiérrez M, Rios MY. Linear and cyclic dipeptides with antimalarial activity. Bioorganic Med Chem Lett. 2012;22: 7048–7051. doi: 10.1016/j.bmcl.2012.09.094 [DOI] [PubMed] [Google Scholar]
- 64.Nagella P.,. Chemical constituents, larvicidal effects and antioxidant activity of petroleum ether extract from seeds of Coriandrum sativum L. J Med Plants Res. 2012;6. doi: 10.5897/jmpr11.992 [DOI] [Google Scholar]
- 65.Chandrasekaran M, Senthilkumar A, Venkatesalu V. Antibacterial and antifungal efficacy of fatty acid methyl esters from the leaves of Sesuvium portulacastrum L. Eur Rev Med Pharmacol Sci. 2011;15(7): 775–780. . [PubMed] [Google Scholar]
- 66.Mahamuni SV. Antifungal Trait of Burkholderia gladioli Strain VIMP02 (JQ811557). Int J Sci Res. 2015;4: 2059–2064. Available: https://www.ijsr.net/search_index_results_paperid.php?id=SUB157864 [Google Scholar]
- 67.Samling B, John Umaru I. Phytochemical screening, antioxidant, antifungal potentials of Acacia auriculiformis florescent composition. J Anal Pharm Res. 2018;7. doi: 10.15406/japlr.2018.07.00296 [DOI] [Google Scholar]
- 68.Aftab A, Yousaf Z, Javaid A, Riaz N, Younas A, Rashid M, et al. Antifungal activity of vegetative methanolic extracts of Nigella sativa against Fusarium oxysporum and Macrophomina phaseolina and its phytochemical profiling by GC-MS analysis. Int J Agric Biol. 2019;21: 569–576. doi: 10.17957/IJAB/15.0930 [DOI] [Google Scholar]
- 69.Torres MJ, Brandan CP, Petroselli G, Erra-Balsells R, Audisio MC. Antagonistic effects of Bacillus subtilis subsp. subtilis and B. amyloliquefaciens against Macrophomina phaseolina: SEM study of fungal changes and UV-MALDI-TOF MS analysis of their bioactive compounds. Microbiol Res. 2016;182: 31–39. doi: 10.1016/j.micres.2015.09.005 [DOI] [PubMed] [Google Scholar]
- 70.Berendsen RL, Pieterse CMJ, Bakker PAHM. The rhizosphere microbiome and plant health. Trends in Plant Science. Trends Plant Sci; 2012. pp. 478–486. doi: 10.1016/j.tplants.2012.04.001 [DOI] [PubMed] [Google Scholar]
- 71.Shanmugam V, Kanoujia N, Singh M, Singh S, Prasad R. Biocontrol of vascular wilt and corm rot of gladiolus caused by Fusarium oxysporum f. sp. gladioli using plant growth promoting rhizobacterial mixture. Crop Prot. 2011;30: 807–813. doi: 10.1016/j.cropro.2011.02.033 [DOI] [Google Scholar]
- 72.Pineda A, Dicke M, Pieterse CMJ, Pozo MJ. Beneficial microbes in a changing environment: are they always helping plants to deal with insects? Biere A, editor. Funct Ecol. 2013;27: 574–586. doi: 10.1111/1365-2435.12050 [DOI] [Google Scholar]
- 73.Hayat R, Ali S, Amara U, Khalid R, Ahmed I. Soil beneficial bacteria and their role in plant growth promotion: A review. Annals of Microbiology. BioMed Central; 2010. pp. 579–598. doi: 10.1007/s13213-010-0117-1 [DOI] [Google Scholar]
- 74.Holden MTG, Seth-Smith HMB, Crossman LC, Sebaihia M, Bentley SD, Cerdeño-Tárraga AM, et al. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol. 2009;91: 261–277. doi: 10.1128/JB.01230-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lim JY, Lee TH, Baek HN, Yang DC, Kim M, Hwang I. Complete genome sequence of Burkholderia glumae BGR1. J Bacteriol. 2009;191: 3758–3759. doi: 10.1128/JB.00349-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nunvar J, Kalferstova L, Bloodworth RAM, Kolar M, Degrossi J, Lubovich S, et al. Understanding the Pathogenicity of Burkholderia contaminans, an Emerging Pathogen in Cystic Fibrosis. Bevivino A, editor. PLoS One. 2016;11: e0160975. doi: 10.1371/journal.pone.0160975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Urban TA, Goldberg JB, Forstner JF, Sajjan US. Cable pili and the 22-kilodalton adhesin are required for Burkholderia cenocepacia binding to and transmigration across the squamous epithelium. Infect Immun. 2005;73: 5426–5437. doi: 10.1128/IAI.73.9.5426-5437.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Angus AA, Agapakis CM, Fong S, Yerrapragada S, Estrada-de los Santos P, Yang P, et al. Plant-Associated Symbiotic Burkholderia Species Lack Hallmark Strategies Required in Mammalian Pathogenesis. van Schaik W, editor. PLoS One. 2014;9: e83779. doi: 10.1371/journal.pone.0083779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schwarz S, Singh P, Robertson JD, LeRoux M, Skerrett SJ, Goodlett DR, et al. VgrG-5 is a Burkholderia type VI secretion system-exported protein required for multinucleated giant cell formation and virulence. Infect Immun. 2014;82: 1445–1452. doi: 10.1128/IAI.01368-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deng P, Wang X, Baird SM, Showmaker KC, Smith L, Peterson DG, et al. Comparative genome-wide analysis reveals that Burkholderia contaminans MS14 possesses multiple antimicrobial biosynthesis genes but not major genetic loci required for pathogenesis. Microbiologyopen. 2016;5: 353–369. doi: 10.1002/mbo3.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Corbett CR, Burtnick MN, Kooi C, Woods DE, Sokol PA. An extracellular zinc metalloprotease gene of Burkholderia cepacia. Microbiology. Society for General Microbiology; 2003. pp. 2263–2271. doi: 10.1099/mic.0.26243-0 [DOI] [PubMed] [Google Scholar]
- 82.Xu X-H, Su Z-Z, Wang C, Kubicek CP, Feng X-X, Mao L-J, et al. The rice endophyte Harpophora oryzae genome reveals evolution from a pathogen to a mutualistic endophyte. Sci Rep. 2014;4: 5783. doi: 10.1038/srep05783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Narayanasamy P, Narayanasamy P. Mechanisms of Action of Fungal Biological Control Agents. Biological Management of Diseases of Crops. Springer Netherlands; 2013. pp. 99–200. doi: 10.1007/978-94-007-6380-7_3 [DOI] [Google Scholar]
- 84.Quan CS, Zheng W, Liu Q, Ohta Y, Fan SD. Isolation and characterization of a novel Burkholderia cepacia with strong antifungal activity against Rhizoctonia solani. Appl Microbiol Biotechnol. 2006;72: 1276–1284. doi: 10.1007/s00253-006-0425-3 [DOI] [PubMed] [Google Scholar]
- 85.Wheatley RE. The consequences of volatile organic compound mediated bacterial and fungal interactions. Antonie van Leeuwenhoek, Int J Gen Mol Microbiol. 2002;81: 357–364. doi: 10.1023/a:1020592802234 [DOI] [PubMed] [Google Scholar]
- 86.Schalchli H, Hormazabal E, Becerra J, Birkett M, Alvear M, Vidal J, et al. Antifungal activity of volatile metabolites emitted by mycelial cultures of saprophytic fungi. Chem Ecol. 2011;27: 503–513. doi: 10.1080/02757540.2011.596832 [DOI] [Google Scholar]
- 87.Gao Z, Zhang B, Liu H, Han J, Zhang Y. Identification of endophytic Bacillus velezensis ZSY-1 strain and antifungal activity of its volatile compounds against Alternaria solani and Botrytis cinerea. Biol Control. 2017;105: 27–39. doi: 10.1016/j.biocontrol.2016.11.007 [DOI] [Google Scholar]
- 88.Kumari N, Menghani E, Mithal R. Bioactive Compounds characterization and Antibacterial Potentials of Actinomycetes isolated from Rhizospheric soil. J Sci Ind Res. 2019;78: 793–798. Available: http://nopr.niscair.res.in/handle/123456789/51187 [Google Scholar]
- 89.El-Banna N, Winkelmann G. Pyrrolnitrin from Burkholderia cepacia: Antibiotic activity against fungi and novel activities against streptomycetes. J Appl Microbiol. 1998;85: 69–78. doi: 10.1046/j.1365-2672.1998.00473.x [DOI] [PubMed] [Google Scholar]
- 90.Zhang P, Zhou S, Wang G, An Z, Liu X, Li K, et al. Two transcription factors cooperatively regulate DHN melanin biosynthesis and development in Pestalotiopsis fici. Mol Microbiol. 2019;112: 649–666. doi: 10.1111/mmi.14281 [DOI] [PubMed] [Google Scholar]
- 91.VIDAL‐CROS A, VIVIANI F, LABESSE G, BOCCARA M, GAUDRY M. Polyhydroxynaphthalene reductase involved in melanin biosynthesis in Magnaporthe grisea Purification, cDNA cloning and sequencing. Eur J Biochem. 1994;219: 985–992. doi: 10.1111/j.1432-1033.1994.tb18581.x [DOI] [PubMed] [Google Scholar]
- 92.Eisenman HC, Casadevall A. Synthesis and assembly of fungal melanin. Applied Microbiology and Biotechnology. NIH Public Access; 2012. pp. 931–940. doi: 10.1007/s00253-011-3777-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Almeida-Paes R, Frases S, Fialho Monteiro PC, Gutierrez-Galhardo MC, Zancopé-Oliveira RM, Nosanchuk JD. Growth conditions influence melanization of Brazilian clinical Sporothrix schenckii isolates. Microbes Infect. 2009;11: 554–562. doi: 10.1016/j.micinf.2009.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]