Abstract
Understanding how differences in animal husbandry practices affect the reproducibility of research results is critical. We sought to understand how different beddings might influence dietary obesity studies. We compared the effects of paper and corncob bedding on weight gain, metabolism, and gut microbiome (GM) of mice fed a high-fat diet (HFD) or a normal diet (ND) and evaluated effects on fecal and cecal microbiomes collected from these cohorts after euthanasia. Male C57BL/6J mice at 5 wk age were allowed to acclimate to the facility and the assigned bedding for one week before being placed on HFD or remaining on the ND for 12 wk. Fecal pellets and cecal samples were collected and frozen for batched 16S sequencing. Mice had similar body weight, visceral gonadal white adipose tissue (GWAT), subcutaneous inguinal white adipose tissue (IWAT), liver and spleen weights and metabolic changes regardless of the bedding type. Baseline microbiota differences were detected one week after bedding assignment. After 12 wk, the GM showed significant differences depending on both bedding and diet. The effects of the bedding were not significantly different between endpoint fecal and cecal GM, despite the inherent differences in microbiota in fecal and cecal samples. A correlation was detected between diet and the relative abundance of Bacteroidetes and Verrucomicrobia:Akkermansia. In conclusion, this study demonstrates the importance of considering bedding type when performing dietary experiments.
Abbreviations: ANOVA, analysis of variance; ATMs, adipose tissue macrophages; DC, dendritic cells; GM, gut microbiome/microbiota; GTT, glucose tolerance test; GWAT, gonadal/visceral white adipose tissue; HFD, high-fat diet (Research Diets D12492 60% calories from fat, New Brunswick, NJ); IWAT, inguinal white adipose tissue; LEfSe, Linear discriminant analysis Effect Size; ND, normal diet (LabDiet 13.5% calories from fat 5L0D, St Louis, MO); OTUs, operational taxonomy units; RO-DI, reverse-osmosis deionized
Recent studies have shown the influence of gut microbiota (GM) in human health and disease conditions.11,29 The microorganisms that are naturally found in the gut microbiome have coevolved with their hosts to provide metabolic benefits, protect against foreign bacterial colonization, and provide immune responses that promote homeostasis.29 The microbiome has also a strong impact on mouse models for human disease.31 This recent understanding has prompted investigations of the GM in disease states affecting metabolism. The GM varies based on where the studies were conducted, and this location effect contributes to lack of reproducibility of findings between facilities.2,14,24 Simple laboratory housing changes can also lead to significant variation in results and metabolic outcomes,1,16,17,22,26,28 making it critical to understand the influence these changes have on metabolism, weight gain, and the microbiome.
Diet has a major role in changing GM profile, and an obesogenic diet is related to dysfunctional microbiota and dysbiosis.26 The GM of mammals is mainly composed of the same 5 phyla (Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria, and Verrucomicrobia). However, Bacteroidetes and Firmicutes, which comprise approximately 90% of the GM, are dominant.5,10 Recent studies have reported that a high-fat diet (HFD) decreases the abundance of Bacteroidetes, but increases the abundance of both Firmicutes and Proteobacteria.17,37 An increase in the Firmicutes to Bacteroidetes (F/B) ratio is associated with obesity and increased food intake.10 Obese mammals have a higher F/B ratio in the GM than do lean mammals, even without an increase in food consumption/dietary intake.10,23 Moreover, such shifts in the GM enhance the ability of the bacteria to harvest energy from ingesta, resulting in a positive feedback loop for body weight gain in obesity.10,23 Thus, any factor capable of altering the ratio of these 2 phyla could potentially initiate a gradual shift toward a phenotype of increased energy storage, such as the expansion of adipose tissue.10
Increased adiposity has also been reported to be negatively associated with the abundance of Akkermansia (phylum Verrucomicrobia),28,30,38 and positively associated with the relative abundances of Lactococcus (phylum Firmicutes) and Allobaculum (phylum Bacteroidetes) in mice.28 However, a few studies have reported that the absence of intestinal microbiota does not protect mice from diet-induced obesity.13,26 These inconsistent findings may partially be due to microbial adaptation to diet over time.26
Studies on diet and GM can provide information on the influence of GM on metabolic pathways, inflammatory-induced obesity, and Type 2 diabetes. Inflammation is associated with metabolic disorders.7,33,36 Excessive calorie intake, increased fat accumulation, and lipotoxicity lead to disproportionate expansion of adipose tissues, tissue inflammation, and proinflammatory cytokines.33 This leads to chronic, low-grade inflammation that further recruits and activates mature immune cells (for example, mast cells, macrophages, and dendritic cells) in adipose and other metabolically active tissues, producing cytokines and leading to tissue dysfunction.33
Studying factors that may alter this inflammatory tone may allow them to be modified in ways that will limit the secondary diseases of obesity. Clinical research into obesity, tissue inflammation, and the microbiome is difficult; therefore, animal models have been used for mechanistic studies to understand the effect of GM on adiposity, obesity, and metabolism.5,7,11,22 However, recent concerns have arisen regarding the reproducibility of these models between institutions and across time.10 Previous rodent studies have shown that several husbandry-associated factors can alter GM.2,3,11 One such factor is bedding. Bedding can vary between animal facilities, and different bedding materials have been linked to changes in the phenotype and relative abundance of the GM.1,3,24 Some changes induced by bedding include body weight, sleeping habits, aggression, lipopolysaccharide levels, and risk of environmental microbial contamination.1,3,11,24 Because all of these changes are associated with dietary patterns in mammals, determining how bedding may affect dietary experiments is important.
A possible cause for some of the inconsistencies among microbiome studies may be the lack of understanding of whether either fecal or cecal material is relevant to the onset and progression of metabolic diseases.27 Previous studies indicate that cecal matter is better than feces for observing differences in the GM.3,11 However, collecting cecal matter requires sacrificing the animal, making it difficult to study changes over time.35 Fecal matter does not require sacrificing the animal, allowing cohort studies of the GM across a lifespan or treatment. However, fecal matter can display either subtle or no differences from the start of experimentation, even when cecal content is changed.35
When our institution considered replacing our standard corncob bedding with paper bedding, we examined how these 2 different beddings might affect the results of dietary experiments. We compared the effects of paper and corncob bedding on weight gain, metabolism, and the GM of mice fed either a HFD or a normal diet (ND).
Materials and Methods
Animal Husbandry.
Thirty-two male C57BL/6J mice were purchased from Jackson Laboratories (Bar Harbor, ME) at 5 wk of age and group-housed (4 mice per cage) in positive-pressure autoclaved IVC (P/NV IVC, Allentown Caging, Allentown, NJ) on either paper (Pure-O’Cel, Andersons Lab Bedding, Maumee, OH) or corncob bedding (1/4’’ Bed-o’Cobs, Andersons Lab Bedding, Maumee, OH). They were allowed to acclimate to the facility for 1 wk on their designated bedding before being assigned to dietary groups. Cages were changed once a week. Eight mice were used per group based on our prior studies demonstrating that this number of mice provides 90% power to detect 10% significant difference of percentage of adipose tissue macrophages (ATMs) in the stromal vascular fraction of visceral fat.34
At 6 wk of age (that is, after 1 wk in the facility on the assigned bedding), half of the mice from each bedding group were switched to a high-fat diet (HFD, Research Diets D12492 60% calories from fat, New Brunswick, NJ), while the rest remained on a normal diet (ND, 5L0D PicoLab Laboratory Rodent Diet, 13.5% calories from fat, St Louis, MO). The ND is a grain-based irradiated diet that consists of high protein and low fat and provides complete life cycle nutrition to mice. It was used as the control diet because of its low-fat content and minimal inherent biologic variations in long-term studies.21 Thus, we used 4 bedding-diet groups (that is, corncob-HFD, corncob-ND, paper-HFD, and paper-ND) with 8 mice per group. Mice were kept in housing rooms with 12:12 h light:dark cycles, 30% to 70% humidity, and 68 ± 4 °F; mice had free access to food and water, with water provided from an automated system with reverse-osmosis deionized (RO-DI, no post-treatment) (Avidity Science, Waterford, WI). Based on our institutional rodent surveillance program, mice were free of mouse hepatitis virus, mouse minute virus, mouse parvovirus, mouse rotavirus, Theiler murine encephalomyelitis virus, pneumonia virus of mice, Sendai virus, lymphocytic choriomeningitis virus, ectromelia virus, Hantaan virus, mouse adenovirus types 1 and 2, mouse cytomegalovirus, pinworms and fur mites. All procedures were performed with the approval of the University of Michigan Institutional Animal Care and Use Committee and are in accordance with the Guide for the Care and Use of Laboratory Animals.19
Metabolic Testing.
Fasting glucose measurements were performed at 8 wk after beginning the dietary study. Mice were fasted in the morning for 6 h prior to the measurement of body weight and blood glucose level; whole blood was collected from the tail vein via a tail vein nick procedure. Blood glucose was measured using a FreeStyle Lite blood glucose meter and test strips (Abbott Laboratories, Abbott Park, IL).
Glucose tolerance tests (GTT) were performed at 10 wk after beginning the dietary study. Mice were fasted in the morning for 6 h before receiving an intraperitoneal injection of glucose at a dose of 0.7 g/kg for mice. The glucose injection was diluted using sterile Dulbecco phosphate-buffered saline (ThermoFisher Scientific, Waltham, MA) such that the final concentration was 100 mg/mL glucose. Blood glucose levels were obtained at fasting (0 min) and at 15, 30, 45, 60, 90, and 120 min after injection using blood collected from the tail vein.
Euthanasia.
Mice were anesthetized with 500 µL of isoflurane using a drop-jar method in a fume hood. Mice were euthanized via cervical dislocation while anesthetized.
Flow Cytometry.
Flow cytometry was performed as previously described.33 Gonadal white adipose tissue was digested in RPMI 1640 (Gibco, Indianapolis, IN) with 1 mg/mL collagenase (Clostridiopeptidase A from Clostridium histolyticum type II Sigma–Aldrich, St Louis, MO) on a rocking platform shaker for 25 min at 37 °C. The stromal vascular fraction (SVF) of gonadal white adipose tissue (GWAT) was separated from adipocytes by centrifugation. The following antibodies were used for flow cytometry: antimouse CD45 eFluor 450 (30-F11 monoclonal, Invitrogen, Carlsbad, CA), antimouse CD11c eFluor 780 (N418 monoclonal, Invitrogen, Carlsbad, CA), and antimouse CD64 PE (X54-5/7.1 monoclonal, BD Pharmingen, San Jose, CA). Adipose tissue macrophages were characterized as CD64+ and separated as M2 (CD11c−) or M1 (CD11c+). Dendritic cells are CD64−/CD11c+ cells. Analysis was performed using a BD Biosciences FACSAria and FlowJo v.10 (Treestar) software.
Microbiome Analyses.
Fecal pellets were collected from each mouse at 1 wk after receipt and placement on their designated bedding (baseline) and again at the end of the 12-wk dietary study (endpoint). Briefly, each mouse was restrained, and a fecal stool pellet collected directly from the anus into a tube, or less commonly, after the mouse was placed into a clean, empty, autoclaved cage and the fecal pellet was then removed from the cage floor with an autoclaved wooden toothpick and placed into the tube. All work surfaces were disinfected, and gloves changed between cages. Cecal content was collected aseptically from each mouse after euthanasia.
DNA was extracted from the samples using the Qiagen MagAttract PowerMicrobiome kit (Qiagen, Germantown, MD). The V4 region of the 16S rRNA-encoding gene was then amplified from the extracted DNA using the barcoded dual-index primers20 and sequenced by the Microbiome Core at University of Michigan. The sequence data were used to generate operational taxonomic units (OTUs) on mothur version 1.44.3, as previously described.32 All OTU-based analysis, with the exception for Linear discriminant analysis Effect Size (LEfSe), was conducted in R (R Core Team, Vienna, Austria), as previously described.4 LEfSe was conducted on mothur version 1.44.3, as previously described.32
Statistical Analysis.
Results are presented as mean ± SEM. Analysis was first performed using GraphPad Prism version 8.4.3 (GraphPad Software, San Diego, CA) to detect any significant main effects and interactions of bedding and diet on body weight, organ weights, metabolic parameters (glucose, GTT) and inflammatory parameters (proportional and absolute counts of ATMs, dendritic cells, and monocytes) and concentrations of CD3, CD4, CD19, CD115, and Ly6C).
Comparisons were then made with 2-way analysis of variance (ANOVA) using Tukey method for multiple comparisons. α-diversity was assessed using inverse Simpson index in mothur and compared between treatment groups using Wilcoxon rank-sum test for baseline and 2-way ANOVA for endpoint data in Prism. β-diversity was assessed using principal component analysis (PCA), and significance was determined using permutational 2-way analysis of variance.
Results
Bedding effects on weight gain, adiposity, and metabolism in ND or HFD fed mice.
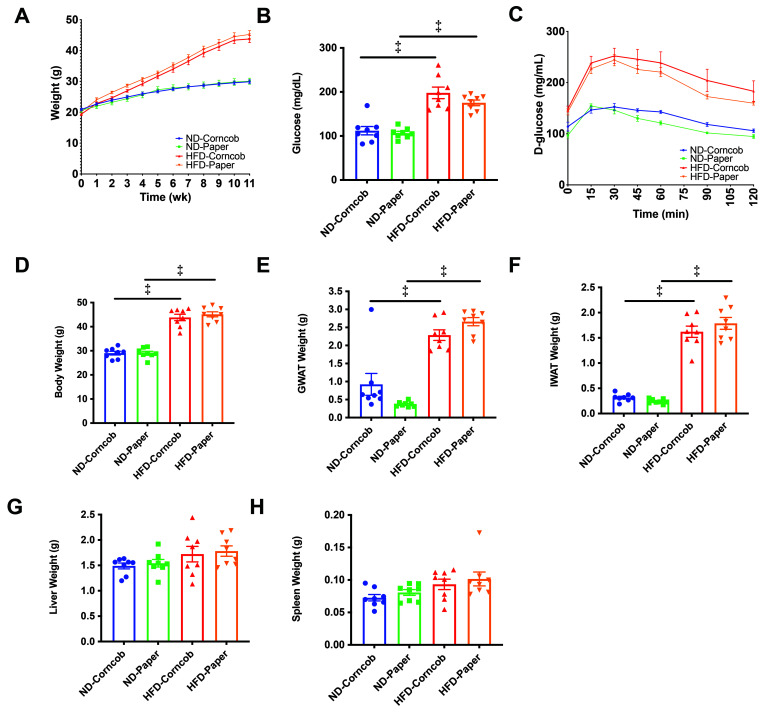
Weekly weights showed that mice had similar weight trajectories regardless of bedding (Figure 1 A). Fasting glucose levels of HFD mice on both corncob bedding (HFD, 198 ± 13 mg/dL; ND, 112 ± 10 mg/dL; P < 0.001) and paper bedding (HFD, 175 ± 6 mg/dL; ND, 106 ± 4 mg/dL; P < 0.001) were significantly higher than those of ND mice (Figure 1 B). GTT was not significantly different for ND and HFD mice housed on paper or corncob (Figure 1 C). After 12 wk of HFD, mice under both bedding conditions had similar weights (Figure 1 D) as well as similar visceral GWAT, subcutaneous IWAT, liver, and spleen weights (Figure 1 E through H).
Figure 1.
Weight gain, metabolism and tissue weights were similar among both beddings. Weekly weights (A) were conducted as well as 8-wk fasting glucose (B) and 10-wk glucose tolerance testing GTT (C). Body weights (D) and (E) GWAT, (F) IWAT, (G) Liver, and (H) Spleen were assessed between corncob and paper bedding. n = 8 per group, data shown as mean ± SEM. Comparisons were made with 2-way ANOVA, using Tukey’s method for multiple comparisons. *P < 0.05, †P < 0.01, +P < 0.005, and ‡P < 0.001.
Effects of bedding and diet on alpha diversity indices.
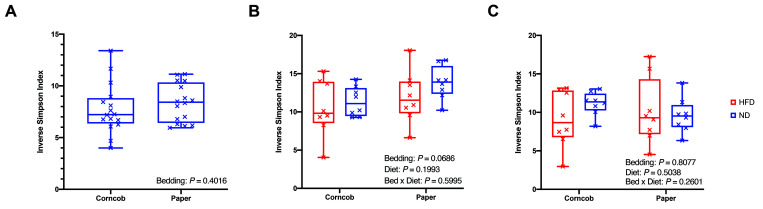
To determine whether bedding material or diet influenced the GM, we first assessed the microbiota from fecal samples. No statistically significant differences of the α diversity-metrics were detected between the 2 bedding conditions at either the basal and endpoint fecal collection times (Figure 2 A and B) nor at endpoint cecal samples (Figure 2 C). Likewise, no statistically significant differences in the α diversity-metrics were detected between mice in ND and HFD groups as determined by endpoint fecal or cecal sample collection (Figure 2 B and C). These findings suggest that bedding and diet did not affect overall GM membership and richness.
Figure 2.
Alpha diversity of gut microbiota with different bedding and HFD challenge. Alpha diversity when grouped by (A) bedding at baseline (n = 16 per group), and when grouped by bedding and diet at endpoint (B) fecal and (C) endpoint cecal (n = 8 per group). Used the Wilcoxon rank-sum test for baseline and 2-way ANOVA for endpoint data.
Microbiota composition after 1 week on assigned bedding.
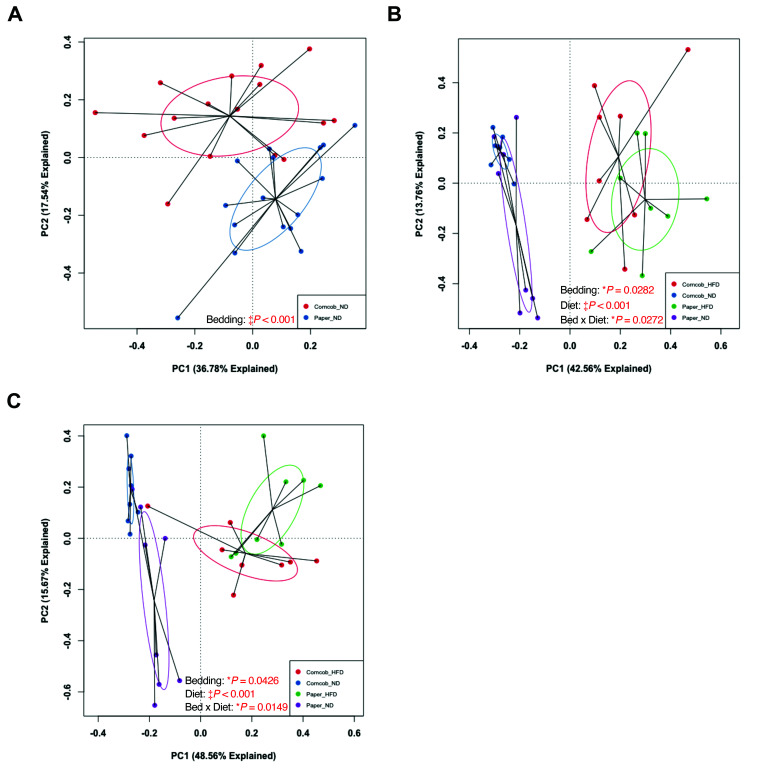
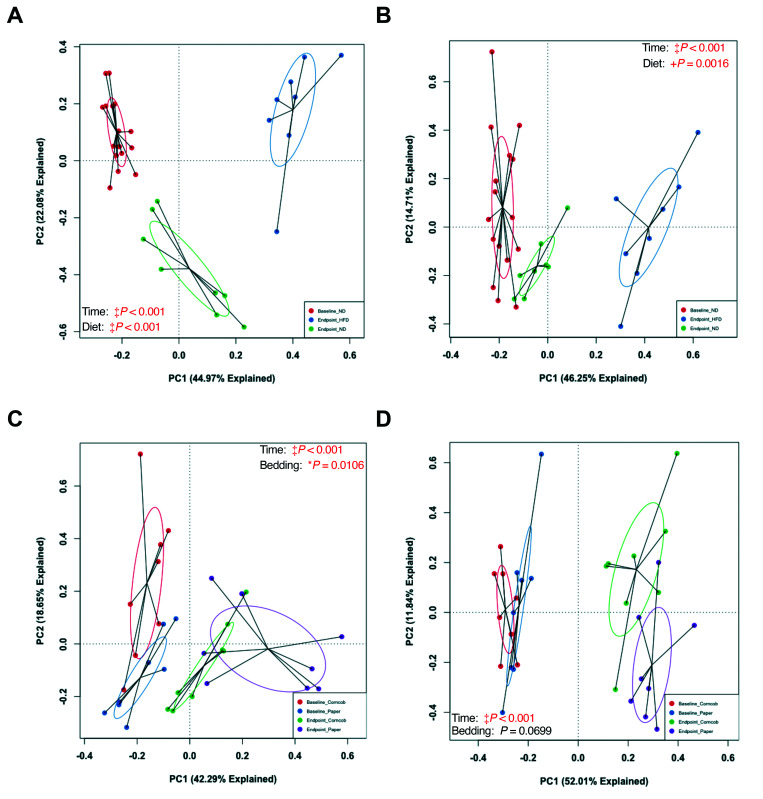
To evaluate broad microbiota changes in baseline and endpoint fecal samples, principal component analysis (PCA) was conducted on fecal samples collected after 1-wk of bedding exposure and before dietary challenge (baseline). PCA revealed a significant difference between the 2 bedding types (Figure 3 A). No difference was found in the baseline fecal samples between the groups starting the different diets.
Figure 3.
Weighted Principal Component Analysis (PCA) plots comparing baseline fecal, and endpoint fecal and cecal samples of the 32 mice. (A) Baseline fecal grouped by bedding (n = 16 per group), (B) endpoint fecal grouped by bedding and diet (n = 8 per group), and (C) endpoint cecal grouped by bedding and diet (n = 8 per group). Baseline fecal was analyzed using PermANOVA; both endpoint data were analyzed using 2-way PermANOVA. *P < 0.05 and ‡P < 0.001.
Principal Component Analysis (PCA) of microbiota changes from baseline and endpoint samples based on diet and bedding.
To further examine how bedding and diet affect the GM, we analyzed endpoint fecal samples and compared them to baseline fecal results. The endpoint fecal GM had significant differences in microbiota composition between groups by both bedding (P = 0.0282) and diet (P < 0.001) (Figure 3 B). Furthermore, the interaction effect of bedding and diet was significant (P = 0.0272), suggesting that these husbandry factors affect each other in terms of effects of bedding or diet on the GM composition. Endpoint cecal samples showed similar significant differences (Figure 3 C).
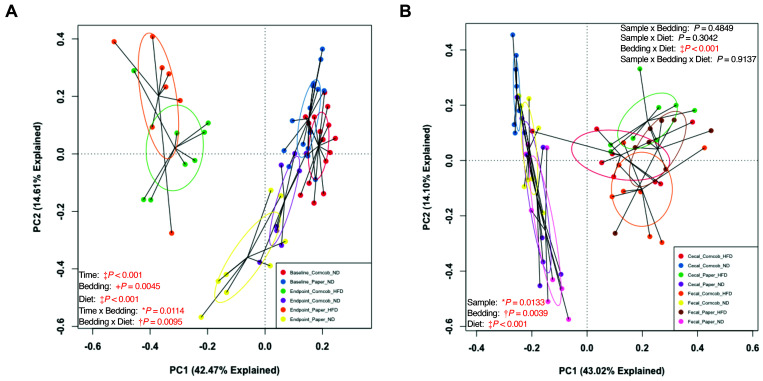
To ensure that bedding and diet truly affected the GM composition, as opposed to an effect of age, we pooled baseline and endpoint fecal data and conducted a permutational 2-way ANOVA comparing time (baseline compared with endpoint), bedding, and diet. We found a significant difference in GM composition between baseline and endpoint fecal samples from both paper and corncob bedding groups (Figure 4 A) (Figure 5 A and B). This finding suggests that the GM did change throughout study as the mice aged. However, the change in GM from baseline to endpoint was not solely due to aging. The interaction effect of bedding with time was significant (P = 0.0114), which suggests that bedding affected how the GM changed due to diet. The mice that continued on ND or were switched to HFD for each bedding group showed a significant difference between baseline and endpoint fecal samples within both ND and HFD cohorts, but only the ND samples showed a significant effect of bedding (P = 0.0106) (Figure 5 C and D).
Figure 4.
Weighted PCAs based on time (left) and fecal compared with cecal samples (right). (A) Comparison of baseline (1 wk of bedding) and endpoint fecal samples from the 32 mice when grouped by time, bedding, and diet. (B) Comparison of endpoint fecal and cecal samples from the 32 mice when grouped by sample collection method, bedding, and diet. Used 2-way PermANOVA. *P < 0.05, †, P < 0.01, +P < 0.005, and ‡P < 0.001.
Figure 5.
Weighted PCA comparisons of baseline and endpoint fecal samples. Grouping by time and diet in (A) paper and (B) corncob bedding. Grouping by time and bedding in (C) ND and (D) HFD mice. Used 2-way PermANOVA. *P < 0.05, +P < 0.005, and ‡P < 0.001.
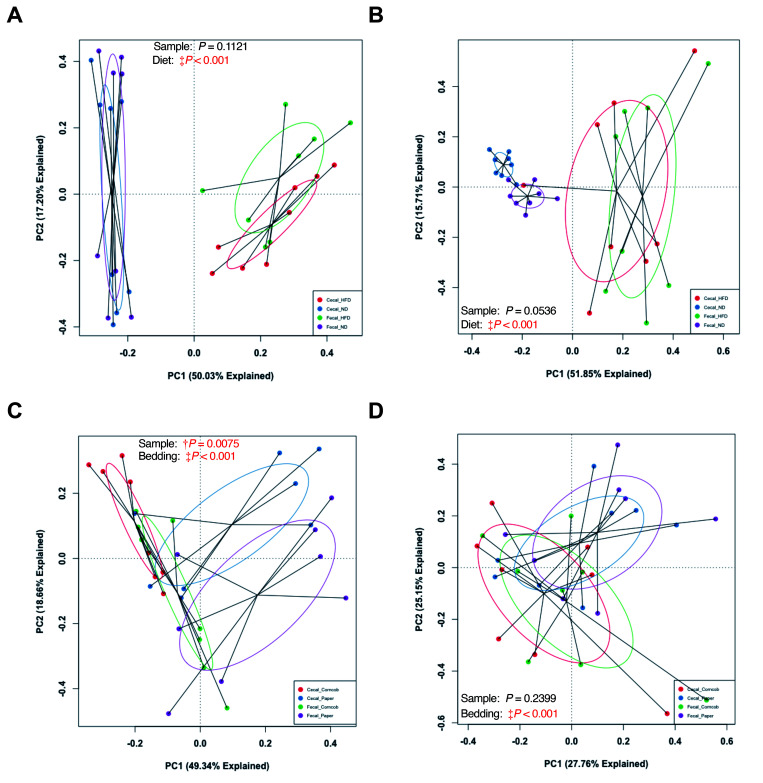
Fecal and cecal microbiota composition.
Although fecal and cecal samples were similar (Figure 3 B and C), to determine whether the effects of bedding and diet were similar, we pooled endpoint cecal and fecal data and conducted a permutational 2-way ANOVA comparing sample (cecal compared with fecal), bedding, and diet. We found a significant difference between fecal and cecal samples (Figure 4 B), as well as a significant difference due to both bedding and diet. However, no significant interactions were found between sample and bedding, sample and diet, or sample, bedding, and diet. However, we did find a significant interaction between bedding and diet. Furthermore, we found no significant difference between fecal and cecal GM in the 32 mice when grouped by bedding (Figure 6 A and B), but we did find a significant difference (P < 0.001) between fecal and cecal samples in ND mice (Figure 4 B, Figure 6 C). HFD mice, however, did not show significant differences between fecal and cecal samples (Figure 4 B, Figure 6 D). Overall, fecal and cecal samples showed an inherent difference in GM composition between fecal and cecal samples, but this difference did not alter the effects of bedding and HFD on the GM.
Figure 6.
Weighted PCA comparisons of fecal and cecal samples. No differences were seen between sample collection methods when grouped by (A) paper and (B) corncob bedding. There were differences in (C) ND mice, but not in (D) HFD mice. Used 2-way PermANOVA. †P < 0.01 and ‡P < 0.001.
Bedding and diet influences on Phylum and Genus GM community membership.
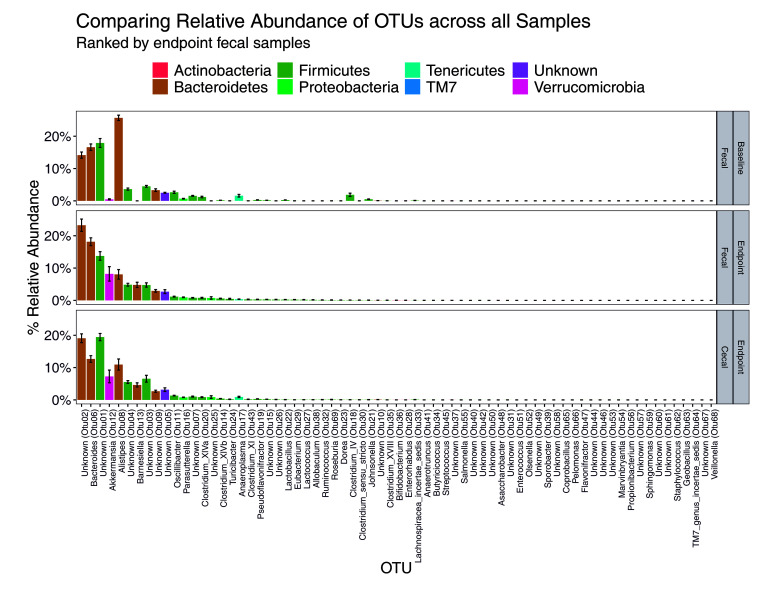
We next sought to evaluate changes in genus and phylum over time and between sample types. We found no significant difference in F/B ratio over time (Figure 7 and 8) but did find differences in endpoint fecal samples, which had a lower F/B ratio than cecal (P < 0.005) (Figure 7 and 8). When comparing by diet and bedding, no overall significant differences in F/B ratio were found between baseline and endpoint fecal, or endpoint fecal and cecal communities (Figure 7 and 8). Bedding was not associated with differences, although in mice on paper bedding, the endpoint fecal F/B ratio was significantly higher in HFD mice than in ND mice (P < 0.05).
Figure 7.
Relative abundance of OTUs for all samples. (A) Baseline fecal, (B) endpoint fecal, and (C) endpoint cecal samples. The OTUs are colored by their identified phyla.
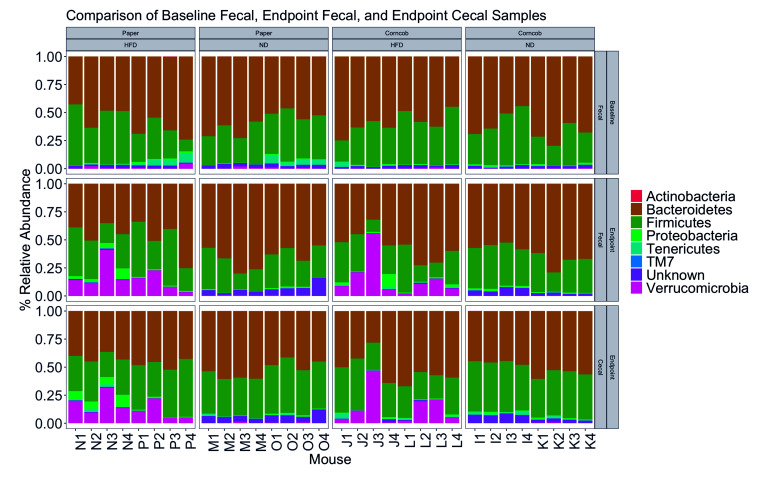
Figure 8.
Comparing relative abundance of GM phyla. Baseline fecal, endpoint fecal, and endpoint cecal samples based on bedding and diet.
Furthermore, we detected an inversion in the relative abundance for OTUs 1, 2, 6, and 8 between baseline and endpoint (Figure 7). OTUs 1, 2, 6, and 8 were identified as Lachnospiraceae family (unknown genus), Porphyromonadaceae family (unknown genus), Bacteroidaceae:Bacteroidetes, and Rikenellaceae:Alistipes, respectively. We also found a spike in the relative abundance of OTU 12 in the endpoint samples as compared with baseline. OTU 12 was identified as Verrucomicrobia:Akkermansia. When comparing samples across all bedding and diet groups, both HFD groups showed a significant shift (P < 0.001) in Verrucomicrobia (Figure 8).
Finally, a significant decrease of Bacteroidetes was detected in HFD as compared with ND mice (P < 0.005), despite the lack of a significant difference in F/B ratio between diets (P = 0.1256). The comparison of diets showed significantly more Verrucomicrobia:Akkermansia in HFD as compared with ND mice (P < 0.001). Thus, a correlation seems to exist between diet, a shift in abundance of Bacteroidetes, and thus changes to F/B ratio, especially with regard to OTUs 2, 6, and 8, and the presence of Verrucomicrobia:Akkermansia. The significant taxa identified as different by bedding and diet in baseline fecal, and endpoint fecal and cecal samples are summarized in Table 1, Table 2, and Table 3, respectively.
Table 1.
List of all differential taxa identified by LEfSe as significant (α-level = 0.05) for baseline fecal. Paper ND compared with Corncob ND.
| OUT | Bedding | LDA | P value | Phylum | Genus | |
| 1 | Otu11 | Corncob | 4.189 | 0.0001 | Firmicutes | Clostridium_IV |
| 2 | Otu25 | Corncob | 2.794 | 0.0090 | Firmicutes | Turicibacter |
| 3 | Otu27 | Purocel | 3.720 | 0.0040 | Verrucomicrobia | Akkermansia |
The list is categorized by the OTU identified, the type of bedding that OTU was found significantly abundant in, the linear discriminant analysis (LDA) effect size (log 10) and corresponding P value, and the phylum and genus that OTU was identified as.
Table 2.
List of all differential taxa identified by LEfSe as significant (α-level = 0.05) for endpoint fecal.
| Paper ND compared with Paper HFD | ||||||
| OTU | Diet | LDA | P value | Phylum | Genus | |
| 1 | Otu02 | ND | 5.035 | 0.0087 | Bacteroidetes | Unknown |
| 2 | Otu03 | ND | 3.675 | 0.0357 | Firmicutes | Unknown |
| 3 | Otu04 | HFD | 4.248 | 0.0008 | Firmicutes | Unknown |
| 4 | Otu05 | ND | 4.554 | 0.0008 | Bacteroidetes | Bacteroides |
| 5 | Otu07 | ND | 3.631 | 0.0046 | Firmicutes | Unknown |
| 6 | Otu08 | ND | 3.970 | 0.0033 | Bacteroidetes | Alistipes |
| 7 | Otu09 | HFD | 4.751 | 0.0008 | Verrucomicrobia | Akkermansia |
| 8 | Otu10 | HFD | 4.551 | 0.0008 | Firmicutes | Oscillibacter |
| 9 | Otu11 | HFD | 3.659 | 0.0008 | Bacteroidetes | Unknown |
| 10 | Otu16 | HFD | 3.361 | 0.0003 | Firmicutes | Clostridium_IV |
| 11 | Otu17 | ND | 3.419 | 0.0459 | Firmicutes | Lactobacillus |
| 12 | Otu20 | HFD | 2.475 | 0.0070 | Actinobacteria | Enterorhabdus |
| 13 | Otu21 | HFD | 2.452 | 0.0115 | Firmicutes | Turicibacter |
| 14 | Otu24 | HFD | 3.062 | 0.0016 | Tenericutes | Anaeroplasma |
| 15 | Otu25 | HFD | 3.360 | 0.0008 | Firmicutes | Lactococcus |
| 16 | Otu26 | HFD | 4.237 | 0.0273 | Firmicutes | Unknown |
| 17 | Otu28 | ND | 3.344 | 0.0012 | Firmicutes | Allobaculum |
| 18 | Otu29 | HFD | 2.684 | 0.0020 | Firmicutes | Ruminococcus |
| 19 | Otu31 | HFD | 3.225 | 0.0038 | Firmicutes | Eubacterium |
| 20 | Otu33 | HFD | 2.468 | 0.0107 | Firmicutes | Streptococcus |
| 21 | Otu35 | HFD | 2.702 | 0.0038 | Proteobacteria | Unknown |
| 22 | Otu37 | HFD | 2.961 | 0.0010 | Actinobacteria | Bifidobacterium |
| 23 | Otu38 | HFD | 3.632 | 0.0003 | Firmicutes | Unknown |
| 24 | Otu39 | ND | 2.662 | 0.0012 | Firmicutes | Clostridium_XVIII |
| 25 | Otu41 | ND | 3.592 | 0.0107 | Firmicutes | Unknown |
| 26 | Otu45 | HFD | 2.051 | 0.0107 | Firmicutes | Enterococcus |
| Corncob ND compared with Corncob HFD | ||||||
| OTU | Diet | LDA | P value | Phylum | Genus | |
| 1 | Otu03 | ND | 4.133 | 0.0063 | Firmicutes | Unknown |
| 2 | Otu05 | ND | 4.337 | 0.0008 | Bacteroidetes | Bacteroides |
| 3 | Otu07 | ND | 3.431 | 0.0016 | Firmicutes | Unknown |
| 4 | Otu09 | ND | 4.879 | 0.0008 | Verrucomicrobia | Akkermansia |
| 5 | Otu11 | HFD | 4.711 | 0.0008 | Bacteroidetes | Unknown |
| 6 | Otu13 | ND | 3.694 | 0.0117 | Bacteroidetes | Barnesiella |
| 7 | Otu14 | HFD | 3.642 | 0.0008 | Firmicutes | Clostridium_XlVb |
| 8 | Otu17 | ND | 3.908 | 0.0003 | Firmicutes | Lactobacillus |
| 9 | Otu18 | HFD | 2.551 | 0.0454 | Firmicutes | Clostridium_XlVa |
| 10 | Otu22 | HFD | 3.333 | 0.0011 | Firmicutes | Dorea |
| 11 | Otu26 | ND | 2.601 | 0.0199 | Firmicutes | Unknown |
| 12 | Otu27 | HFD | 3.109 | 0.0003 | Firmicutes | Pseudoflavonifractor |
| 13 | Otu29 | ND | 2.042 | 0.0107 | Firmicutes | Ruminococcus |
| 14 | Otu30 | HFD | 2.810 | 0.0020 | Firmicutes | Clostridium_sensu_stricto |
| 15 | Otu31 | ND | 2.941 | 0.0273 | Firmicutes | Eubacterium |
| 16 | Otu32 | ND | 2.340 | 0.0107 | Firmicutes | Lachnospiracea_incertae_sedis |
| 17 | Otu34 | HFD | 2.167 | 0.0107 | Actinobacteria | Unknown |
| 18 | Otu35 | HFD | 2.964 | 0.0070 | Proteobacteria | Unknown |
| 19 | Otu36 | HFD | 2.202 | 0.0273 | Firmicutes | Butyricicoccus |
| 20 | Otu37 | HFD | 2.975 | 0.0030 | Actinobacteria | Bifidobacterium |
| 21 | Otu49 | ND | 3.282 | 0.0273 | Proteobacteria | Unknown |
| Paper ND compared with Corncob ND | ||||||
| OTU | Bedding | LDA | P value | Phylum | Genus | |
| 1 | Otu04 | Corncob | 3.989 | 0.0181 | Firmicutes | Unknown |
| 2 | Otu06 | Paper | 3.367 | 0.0209 | Unknown | Unknown |
| 3 | Otu09 | Corncob | 3.730 | 0.0274 | Verrucomicrobia | Akkermansia |
| 4 | Otu10 | Paper | 3.864 | 0.0008 | Firmicutes | Oscillibacter |
| 5 | Otu19 | Paper | 2.757 | 0.0357 | Proteobacteria | Parasutterella |
| 6 | Otu20 | Corncob | 3.214 | 0.0356 | Actinobacteria | Enterorhabdus |
| 7 | Otu28 | Corncob | 2.821 | 0.0107 | Firmicutes | Allobaculum |
| 8 | Otu31 | Paper | 2.784 | 0.0273 | Firmicutes | Eubacterium |
| Paper HFD compared with Corncob HFD | ||||||
| OTU | Bedding | LDA | P value | Phylum | Genus | |
| 1 | Otu02 | Corncob | 4.582 | 0.0209 | Bacteroidetes | Unknown |
| 2 | Otu04 | Paper | 3.741 | 0.0460 | Firmicutes | Unknown |
| 3 | Otu08 | Corncob | 4.235 | 0.0209 | Bacteroidetes | Alistipes |
| 4 | Otu15 | Paper | 3.060 | 0.0274 | Firmicutes | Unknown |
| 5 | Otu20 | Paper | 2.385 | 0.0031 | Actinobacteria | Enterorhabdus |
| 6 | Otu29 | Paper | 3.335 | 0.0087 | Firmicutes | Ruminococcus |
| 7 | Otu30 | Paper | 2.819 | 0.0352 | Firmicutes | Clostridium_sensu_stricto |
| 8 | Otu32 | Paper | 2.219 | 0.0107 | Firmicutes | Lachnospiracea_incertae_sedis |
| 9 | Otu34 | Paper | 2.307 | 0.0256 | Actinobacteria | Unknown |
| 10 | Otu37 | Paper | 3.433 | 0.0117 | Actinobacteria | Bifidobacterium |
The list is categorized by bedding and diet groups, providing the OTU identified, the type of bedding or diet that OTU was found significantly abundant in, the linear discriminant analysis (LDA) effect size (log 10) and corresponding P value, and the phylum and genus that OTU was identified as.
Table 3.
List of all differential taxa identified by LEfSe as significant (α-level = 0.05) for endpoint cecal.
| Paper ND compared with Paper HFD | ||||||
| OTU | Diet | LDA | P value | Phylum | Genus | |
| 1 | Otu02 | ND | 4.902 | 0.0053 | Bacteroidetes | Unknown |
| 2 | Otu03 | ND | 4.622 | 0.0016 | Firmicutes | Unknown |
| 3 | Otu04 | HFD | 4.289 | 0.0008 | Firmicutes | Unknown |
| 4 | Otu05 | ND | 4.476 | 0.0008 | Unknown | Unknown |
| 5 | Otu06 | HFD | 4.568 | 0.0459 | Firmicutes | Unknown |
| 6 | Otu07 | ND | 4.178 | 0.0063 | Bacteroidetes | Bacteroides |
| 7 | Otu08 | ND | 3.839 | 0.0008 | Bacteroidetes | Alistipes |
| 8 | Otu09 | HFD | 4.406 | 0.0008 | Bacteroidetes | Unknown |
| 9 | Otu10 | HFD | 4.677 | 0.0008 | Firmicutes | Oscillibacter |
| 10 | Otu13 | ND | 3.342 | 0.0033 | Bacteroidetes | Barnesiella |
| 11 | Otu14 | HFD | 3.285 | 0.0033 | Firmicutes | Clostridium_XlVb |
| 12 | Otu18 | HFD | 3.450 | 0.0209 | Firmicutes | Pseudoflavonifractor |
| 13 | Otu25 | HFD | 2.492 | 0.0206 | Firmicutes | Clostridium_sensu_stricto |
| 14 | Otu26 | ND | 3.318 | 0.0003 | Firmicutes | Eubacterium |
| 15 | Otu27 | HFD | 2.075 | 0.0107 | Firmicutes | Unknown |
| 16 | Otu30 | HFD | 3.041 | 0.0004 | Actinobacteria | Enterorhabdus |
| 17 | Otu31 | HFD | 2.944 | 0.0012 | Firmicutes | Turicibacter |
| 18 | Otu32 | ND | 2.148 | 0.0273 | Firmicutes | Lactococcus |
| 19 | Otu33 | ND | 3.291 | 0.0330 | Actinobacteria | Bifidobacterium |
| 20 | Otu35 | ND | 3.137 | 0.0006 | Firmicutes | Lachnospiracea_incertae_sedis |
| 21 | Otu36 | HFD | 2.550 | 0.0012 | Actinobacteria | Unknown |
| 22 | Otu39 | HFD | 3.419 | 0.0003 | Firmicutes | Allobaculum |
| 23 | Otu40 | ND | 3.296 | 0.0038 | Proteobacteria | Unknown |
| Corncob ND compared with Corncob HFD | ||||||
| OTU | Diet | LDA | P value | Phylum | Genus | |
| 1 | Otu03 | ND | 4.038 | 0.0087 | Firmicutes | Unknown |
| 2 | Otu04 | HFD | 4.142 | 0.0016 | Firmicutes | Unknown |
| 3 | Otu05 | ND | 4.325 | 0.0016 | Unknown | Unknown |
| 4 | Otu06 | ND | 4.761 | 0.0063 | Firmicutes | Unknown |
| 5 | Otu07 | HFD | 4.493 | 0.0357 | Bacteroidetes | Bacteroides |
| 6 | Otu08 | ND | 3.671 | 0.0023 | Bacteroidetes | Alistipes |
| 7 | Otu09 | HFD | 3.852 | 0.0357 | Bacteroidetes | Unknown |
| 8 | Otu11 | HFD | 4.401 | 0.0011 | Bacteroidetes | Unknown |
| 9 | Otu13 | ND | 3.682 | 0.0460 | Bacteroidetes | Barnesiella |
| 10 | Otu18 | HFD | 3.190 | 0.0045 | Firmicutes | Pseudoflavonifractor |
| 11 | Otu24 | ND | 3.136 | 0.0061 | Proteobacteria | Unknown |
| 12 | Otu26 | HFD | 2.133 | 0.0273 | Firmicutes | Eubacterium |
| 13 | Otu27 | ND | 2.969 | 0.0206 | Firmicutes | Unknown |
| 14 | Otu28 | ND | 2.095 | 0.0107 | Firmicutes | Butyricicoccus |
| 15 | Otu32 | HFD | 2.830 | 0.0038 | Firmicutes | Lactococcus |
| 16 | Otu35 | HFD | 2.484 | 0.0012 | Firmicutes | Lachnospiracea_incertae_sedis |
| 17 | Otu37 | HFD | 2.559 | 0.0038 | Firmicutes | Anaerotruncus |
| 18 | Otu45 | ND | 3.102 | 0.0273 | Proteobacteria | Unknown |
| 19 | Otu53 | HFD | 2.580 | 0.0038 | Firmicutes | Veillonella |
| Paper ND compared with Corncob ND | ||||||
| OTU | Bedding | LDA | P value | Phylum | Genus | |
| 1 | Otu02 | Paper | 4.693 | 0.0016 | Bacteroidetes | Unknown |
| 2 | Otu07 | Paper | 4.179 | 0.0209 | Bacteroidetes | Bacteroides |
| 3 | Otu08 | Corncob | 4.881 | 0.0063 | Bacteroidetes | Alistipes |
| 4 | Otu09 | Paper | 4.321 | 0.0209 | Bacteroidetes | Unknown |
| 5 | Otu19 | Paper | 2.804 | 0.0199 | Firmicutes | Clostridium_XlVa |
| 6 | Otu23 | Corncob | 2.703 | 0.0435 | Firmicutes | Lactobacillus |
| 7 | Otu30 | Corncob | 2.447 | 0.0435 | Actinobacteria | Enterorhabdus |
| 8 | Otu31 | Paper | 2.681 | 0.0273 | Firmicutes | Turicibacter |
| Paper HFD compared with Corncob HFD | ||||||
| OTU | Bedding | LDA | P value | Phylum | Genus | |
| 1 | Otu08 | Corncob | 4.237 | 0.0063 | Bacteroidetes | Alistipes |
| 2 | Otu25 | Paper | 2.708 | 0.0185 | Firmicutes | Clostridium_sensu_stricto |
The list is categorized by bedding and diet groups, providing the OTU identified, the type of bedding or diet that OTU was found significantly abundant in, the linear discriminant analysis (LDA) effect size (log 10) and corresponding P value, and the phylum and genus that OTU was identified as.
Effects of bedding type on adipose inflammation.
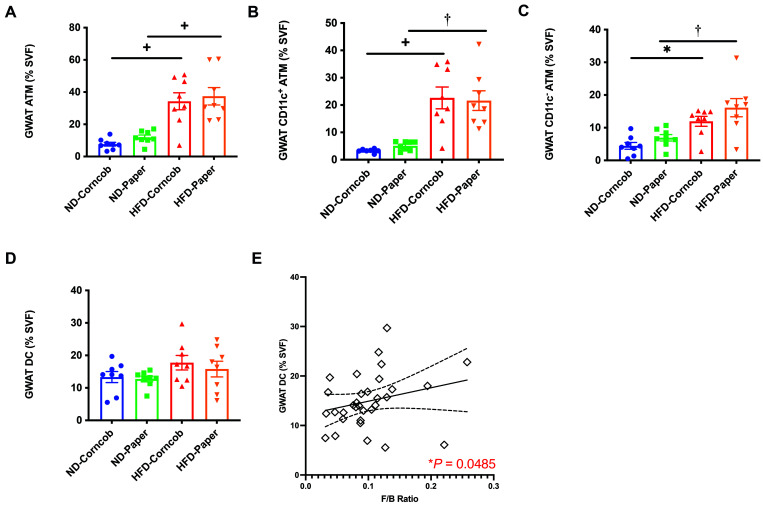
GM diversity has been related to changes in the inflammatory status in metabolic tissues.15 Given that proinflammatory macrophages in visceral GWAT indicate obesity-induced meta-inflammation, we next examined whether bedding and diet affect inflammation. We analyzed GWAT macrophages (ATMs) by flow cytometry. Mice fed ND or HFD on either bedding showed no significant differences in the total number of ATMs. However, HFD on either bedding was associated with an increase in numbers of ATMs (Figure 9 A). Further analysis of ATMs showed a similar pattern in GWAT CD11c+ ATMs and CD11c- ATMs (Figure 9 B and C), with a higher number of ATMs on a HFD with either bedding. However, GWAT dendritic cells (DCs) were not significantly different among groups (Figure 9 D).
Figure 9.
Stromal vascular fraction (SVF) flow cytometry analysis. (A) GWAT ATM, (B) GWAT CD11c+ ATM, (C) GWAT CD11c- ATM, and (D) GWAT DC as a percent of SVF. n = 8 per group (E) Positive correlation between endpoint fecal F/B ratio and GWAT DC across all 32 mice. Data shown as mean ± SEM * P < 0.05, †P < 0.01, +P < 0.005, and ‡P < 0.001.
F/B ratio from baseline and endpoint fecal, as well as endpoint cecal, was compared with body compositions and inflammation parameters. A positive correlation was found between endpoint fecal F/B ratio and GWAT DC when comparing the entire cohort (Figure 9 E). All other comparisons were not significant.
Discussion
Institutional changes to animal husbandry practices may introduce environmental variations to animal models. We investigated how bedding affects weight gain, metabolism, and GM of mice used in dietary studies. Previous studies have shown that a difference in bedding or diet can alter body weight and GM.1,3,16,24 We found similar effects of diet on weight gain and metabolism, but did not see similar effects regarding bedding. When comparing the effects of paper and corncob bedding in HFD and ND mice, our results showed all measures of weight, including tissue weights (that is GWAT, IWAT, liver, and spleen), were higher in HFD mice as compared with ND mice, but similar weights were seen on both beddings (Figure 1). Likewise, fasting glucose and glucose tolerance levels were higher in HFD mice than in ND mice, with no significant difference between bedding types in fasting glucose or in glucose tolerance levels during the last 30 mins of the GTT (Figure 1). Thus, bedding, unlike diet, did not have a main effect on weight gain and metabolism, although an interaction effect between diet and bedding is possible, as suggested by previous studies.1,24
However, the question remains of whether bedding affects the GM in a dietary study. Studying the influence of animal husbandry on GM is critical, as it may significantly alter study results and interpretation. Here we showed that a simple difference of bedding may not affect the species richness present in the GM (Figure 2) but does influence the composition of the GM (Figure 3 through 8,). Despite the changes in GM composition due to bedding, adipose inflammation was similar, regardless of bedding (Figure 9). In our study, the metabolic differences in the mice were due to the diet, rather than due to the type of bedding used.
Despite this finding, evidence remained for a role of bedding in influencing GM composition. Significant differences in GM at baseline for fecal samples after 1 wk on different bedding (Figure 3). A previous study had also observed baseline differences in fecal community between mice as a function of caging, bedding, and diet at 1 wk after arrival to their facility, although those authors speculated these effects to be due to the relatively young age of the mice at the time of arrival.11 However, our study showed that different bedding was associated with shifts in GM composition even at the end of the 12-wk study (Figure 3 and Figure 4), and that the changes observed in different bedding groups were not solely due to aging over the course of the study (Figure 4). Furthermore, significant differences in GM were detected when bedding types were compared within dietary groups using fecal and cecal samples (Figure 4 through Figure 6), although the effects of bedding were different in HFD and ND mice. The loss of significance for bedding in the HFD group suggest that diet has a larger role in shifting GM composition as compared with bedding over an extended time period. Thus, bedding may have only a transient effect on GM composition in the presence of a HFD, although the timepoint at which the effects of diet overshadow the effect was not determined in our study.
Finally, studies have shown a negative relationship between Akkermansia (Verrucomicrobia phylum) and body fat weight.28,30,38 In comparison, our study found more Verrucomicrobia:Akkermansia in HFD mice than in ND mice (Figure 7 and 8). While we did not test for a negative relationship between Akkermansia and body fat weight in each diet group, our findings do not seem to support the negative correlation seen in the previous study, given that HFD mice typically have more body fat weight than ND mice.
The significance of the findings from our study is that bedding did not affect body and metabolic parameters, despite the shift in GM composition, contrary to a previous study.16 Others have shown that husbandry-associated factors, such as caging, vendor, bedding, water contamination, and even sometimes diet, have little effect on fecal and small intestinal microbiota, with the exception of cecal contents.3,11,24 However, these findings may have been due to the length of these previous studies. Based on our 12 wk study, bedding does not affect body weights or metabolic tissue, while diet does. However, bedding does impact the GM composition, which in turn can influence on adiposity and metabolism. Thus, if conducting shorter-term studies, one may have to consider the effect of bedding on experimental results.
Furthermore, previous studies suggest that cecal sampling is better for detecting husbandry-associated GM changes than is fecal sampling,3,11 which complicates designing cohort studies to determine causal relationships between husbandry-associated factors, GM, and disease status. While longitudinal studies can incorporate cecal communities for analysis by using separate cohorts of mice that are euthanized at predetermined intervals,11 this strategy would likely be cost-prohibitive and time-consuming, and drawing causal rather than correlative relationships would be difficult. However, a strength of our study was the use of fecal and cecal sampling, which showed minimal distinctions between these 2 samples in our evaluation of bedding and diet, thus supporting the use of fecal communities as a proxy to investigate environmental effects on GM.
A limitation of this study was the use of only baseline and endpoint sampling. Future studies should assess intermediate samples to determine the crucial intervals during which significant changes occur and potentially identifying the parameters responsible for the changes. One suggested way of doing this would be to compare fecal communities from samples collected every week during a study to see whether any shifts that occurred in the communities are related to effects of bedding or diet on the GM or any metabolic and inflammatory parameters.
A second limitation of this study is that it used only male mice. A previous dietary study in mice found sex differences in body weight, metabolic, and inflammatory parameters.6 Another study found that the effects of sex on mouse GM persist over time.25 As such, these differences may occur with regard to other animal husbandry-associated factors such as bedding. Thus, the effects of bedding and diet on GM composition should be studied in both female and male mice.
A third limitation is that our study only used C57BL/6J mice. Previous studies have shown that mouse GM varies by strain,9,14,18 vendor,8,9 cage location,8,11 and shipment.8 Thus, further studies of a wider variety of strains and vendor locations are required to capture the total effect of bedding and diet on GM composition.
Finally, the group sizes in our study were small and mice were not analyzed on a per cage basis. In a few cases, combining data from bedding (Figure 5 D) or fecal-cecal (Figure 6 A and B) were borderline significant, and a larger sample size might have either provided the necessary power to determine whether the difference by bedding or fecal-cecal were significant or lessened that possibility. Furthermore, bedding was not analyzed based on cage. This may be problematic because the cage environment may be the primary source of interindividual variation in the mice.12 Although using each mouse as an experimental unit is more appropriate for studies of the effects of diet and for comparing metabolic or inflammatory parameters, larger group sizes would have allowed us to compare bedding using each cage as the experimental unit, which is more appropriate for bedding because bedding is shared by all the mice in each cage, and thus may affect all mice in the cage in an equivalent manner.22 Thus, future studies could evaluate how bedding affects the role of GM in changing phenotypes of mice that are singly housed as compared with group housed.
In conclusion, this study demonstrates the importance of considering bedding in dietary experiments. Although in our study bedding did not affect body and organ weight gains or blood glucose level after fasting and during GTT, it did have varying effects on the GM depending on the diet; these effects may extend even beyond the designated acclimation period. Thus, an important aspect of designing dietary experiments that use mice is consideration of the type of bedding that will be used. Furthermore, fecal matter may be an adequate proxy for determining changes in the GM. Fecal samplings can be collected frequently and then used to identify causal relationships between test parameters and observed changes. Finally, a correlation between diet and the relative abundance of Bacteroidetes and Verrucomicrobia:Akkermansia may explain the crosstalk between the GM, adipose inflammation, and HFD metabolism in the progression toward obesity and disease status for Type 2 diabetes.
Acknowledgments
We would like to thank Dr Patrick Schloss and his team for providing instructions and assistance on how to use mothur. We would also like to thank Dr. Robert Dickson and Christopher Brown for providing instructions and assistance on how to perform OTU-based analysis on R. Finally, we would like to extend our thanks to Mohammed Saqib for helping with the analysis and Gabrielle Huizinga for providing constructive feedback on the research. Funding for this study was provided by the University of Michigan Animal Care and Use Quality Improvement Funds. This work was also carried out with the support of NIH/NIDDK R01DK115583 and Edith Briskin/SKS Foundation Taubman Emerging Scholar support to K.S.
References
- 1.Ambery AG, Tackett L, Penque BA, Hickman DL, Elmendorf JS. 2014.Effect of corncob bedding on feed conversion efficiency in a high-fat diet-induced prediabetic model in C57Bl/6J Mice.J Am Assoc Lab Anim Sci 53:449–451. [PMC free article] [PubMed] [Google Scholar]
- 2.Barnett JA, Gibson DL. 2018.H2 Oh No! The importance of reporting your water source in your in vivo microbiome studies.Gut Microbes 10:261–269. 10.1080/19490976.2018.1539599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bidot WA, Ericsson AC, Franklin CL. 2018.Effects of water decontamination methods and bedding material on the gut microbiota.PLoS One 13:1–16. 10.1371/journal.pone.0198305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown C, Dickson R, Erb-Downward J, Baker J. 2020.Dickson Lab SOP for Analysis of 16S rRNA Gene Amplicon Data.[Cited 7 May 2020]. Available at:https://github.com/cb-42/Dickson_16S_SOP.
- 5.Castaner O, Goday A, Park Y-M, Lee S-H, Magkos F, Ee Shiow Toh S-A, Schröder H. 2018. The gut microbiome profile in obesity: a systematic review.Int J Endocrinol 2018:1–9. 10.1155/2018/4095789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang E, Varghese M, Singer K. 2018.Gender and sex differences in adipose tissue.Curr Diab Rep 18:1–17. 10.1007/s11892-018-1031-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dabke K, Hendrick G, Devkota S. 2019.The gut microbiome and metabolic syndrome.J Clin Invest 129:4050–4057. 10.1172/JCI129194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickson RP, Erb-Downward JR, Falkowski NR, Hunter EM, Ashley SL, Huffnagle GB. 2018.The Lung microbiota of healthy mice are highly variable, cluster by environment, and reflect variation in baseline lung innate immunity.Am J Respir Crit Care Med 198:497–508. 10.1164/rccm.201711-2180OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ericsson AC, Davis JW, Spollen W, Bivens N, Givan S, Hagan CE, McIntosh M, Franklin CL. 2015.Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice.PLoS One 10:1–19. 10.1371/journal.pone.0116704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ericsson AC, Franklin CL. 2015.Manipulating the gut microbiota: methods and challenges.ILAR J 56:205–217. 10.1093/ilar/ilv021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ericsson AC, Gagliardi J, Bouhan D, Spollen WG, Givan SA, Franklin CL. 2018.The influence of caging, bedding, and diet on the composition of the microbiota in different regions of the mouse gut.Sci Rep 8:1–13. 10.1038/s41598-018-21986-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Festing MFW. 2020.The “completely randomised” and the “randomised block” are the only experimental designs suitable for widespread use in pre-clinical research.Sci Rep 10:1–5. 10.1038/s41598-020-74538-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleissner CK, Huebel N, El-Bary MMA, Loh G, Klaus S, Blaut M. 2010.Absence of intestinal microbiota does not protect mice from diet-induced obesity.Br J Nutr 104:919–929. 10.1017/S0007114510001303. [DOI] [PubMed] [Google Scholar]
- 14.Friswell MK, Gika H, Stratford IJ, Theodoridis G, Telfer B, Wilson ID, McBain AJ. 2010.Site and strain-specific variation in gut microbiota profiles and metabolism in experimental mice.PLoS One 5:1–9. 10.1371/journal.pone.0008584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graessler J, Qin Y, Zhong H, Zhang J, Licinio J, Wong M-L, Xu A, Chavakis T, Bornstein AB, Ehrhart-Bornstein M, Lamounier-Zepter V, Lohmann T, Wolf T, Bornstein SR. 2013.Metagenomic sequencing of the human gut microbiome before and after bariatric surgery in obese patients with type 2 diabetes: correlation with inflammatory and metabolic parameters.Pharmacogenomics J 13:514–522. 10.1038/tpj.2012.43. [DOI] [PubMed] [Google Scholar]
- 16.Gregor A, Fragner L, Trajanoski S, Li W, Sun X, Weckwerth W, König J, Duszka K. 2020.Cage bedding modifies metabolic and gut microbiota profiles in mouse studies applying dietary restriction.Sci Rep 10:1–13. 10.1038/s41598-020-77831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hildebrandt MA, Hoffman C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen Y-Y, Knight R, Ahima RS, Bushman F, Wu GD. 2009.High fat diet determines the composition of the murine gut microbiome independently of obesity.Gastroenterology 137:1–18. 10.1053/j.gastro.2009.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hufeldt MR, Nielsen DS, Vogensen FK, Midtvedt T, Hansen AK. 2010.Variation in the gut microbiota of laboratory mice is related to both genetic and environmental factors.Comp Med 60:336–347. [PMC free article] [PubMed] [Google Scholar]
- 19.Institute for Laboratory Animal Research.2011.Guide for the care and use of laboratory animals. 8th ed.Washington (DC):National Academies Press. [Google Scholar]
- 20.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013.Development of a Dual-Index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform.Appl Environ Microbiol 79:5112–5120. 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LabDiet. [Internet].2020.PicoLab laboratory rodent diet 5L0D*. [Cited 12 January 2021]. Available at:https://www.labdiet.com/cs/groups/lolweb/documents/web_content/ndjf/ndy1/∼edisp/36142_465407.pdf].
- 22.Leone V, Gibbons SM, Martinez K, Hutchison lL, Huang EY, Cham CM, Pierre JF, Heneghan AF, Nadimpalli A, Hubert N, Zale E, Wang Y, Huang Y, Theriault B, Dinner AR, Musch MW, Kudsk KA, Prendergast BJ, Gilbert JA, Chang EB. 2015.Effects of diurnal variation of gut microbes and high fat feeding on host circadian clock function and metabolism.Cell Host Microbe 17:681–689. 10.1016/j.chom.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. 2005.Obesity alters gut microbial ecology.Proc Natl Acad Sci USA102:11070–11075. 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma BW, Bokulich NA, Castillo PA, Kananurak A, Underwood MA, Mills DA, Bevins CL. 2012.Routine habitat change: a source of unrecognized transient alteration of intestinal microbiota in laboratory mice.PLoS One 7:1–11. 10.1371/journal.pone.0047416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyoshi J, Leone V, Nobutani K, Musch MW, Martinez-Guryn K, Wang Y, Miyoshi S, Bobe AM, Eren AM, Chang EB. 2018.Minimizing confounders and increasing data quality in murine models for studies of the gut microbiome.PeerJ 6:1–19. 10.7717/peerj.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy EA, Velazquez KT, Herbert KM. 2015.Influence of high-fat-diet on gut microbiota: a driving force for chronic disease risk.Curr Opin Clin Nutr Metab Care 18:515–520. 10.1097/MCO.0000000000000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panasevich MR, Wankhade UD, Chintapalli SV, Shankar K, Rector RS. 2018.Cecal versus fecal microbiota in Ossabaw swine and implications for obesity.Physiol Genomics 50:355–368. 10.1152/physiolgenomics.00110.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, Pan C, Civelek M, Rau CD, Bennett BJ, Mehrabian M, Ursell LK, He A, Castellani LW, Zinker B, Kirby M, Drake TA, Drevon CA, Knight R, Gargalovic P, Kirchgessner T, Eskin E, Lusis AJ. 2013.Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice.Cell Metab 17:141–152. 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pickard JM, Zeng MY, Caruso R, Núñez G. 2017.Gut microbiota: role in pathogen colonization, immune responses and inflammatory disease.Immunol Rev 279:70–89. 10.1111/imr.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, Myridakis A, Delzenne NM, Klievink J, Bhattacharjee A, van der Ark K C H, Aalvink S, Martinez L, Dumas ME, Maiter D, Loumaye A, Hermans MP, Thissen JP, Belzer C, de Vos WM, Cani PD. 2016.A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice.Nat Med 23:107–113. 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 31.Rosshart SP, Herz J, Vassallo BG, Hunter A, Wall MK, Badger JH, McCulloch JA,, Anastasakis DG, Sarshad AA, Leonardi I, Collins N, Blatter JA, Han S-J, Tamoutounour S,, Potapova S, Foster St. Claire MB, Yuan W, Sen SK, Dreier MS, Hild B, Hafner M, Wang D, Iliev ID, Belkaid Y, Trinchieri G, Rehermann B. 2019.Laboratory mice born to wild mice have natural microbiota and model human immune responses.Science 365:1–29. 10.1126/science.aaw4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ,, Weber CF. 2009.Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities.Appl Environ Microbiol 75:7537–7541. 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singer K, DelProposto J, Lee Morris D, Zamarron B, Mergian T, Maley N, Won Cho K, Geletka L, Subbaiah P, Muir L, Martinez-Santibanez G, Nien-Kai Lumeng C. 2014.Diet-induced obesity promotes myelopoiesis in hematopoietic stem cells.Mol Metab 3:664–675. 10.1016/j.molmet.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singer K, Maley N, Mergian T, DelProposto J, Cho Won K, Zamarron BF, Martin-Santibanez G, Geletka L, Muir L, Wachowiak P, Demirjian C, Lumeng CN. 2015.Differences in hematopoietic stem cells contribute to sexually dimorphic inflammatory responses to high fat diet-induced obesity.J Biol Chem 21:13250–13262. 10.1074/jbc.m114.634568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanley D, Geier MS, Chen H, Hughes RJ, Moore RJ. 2015.Comparison of fecal and cecal microbiotas reveals qualitative similarities but quantitative differences.BMC Microbiol 15:1–11. 10.1186/s12866-015-0388-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weinstock A, Brown EJ, Garabedian ML, Pena S, Sharma M,, Lafaille J, Moore KJ, Fisher EA. 2019.Single-cell RNA sequencing of visceral adipose tissue leukocytes reveals that caloric restriction following obesity promotes the accumulation of a distinct macrophage population with features of phagocytic cells.Immunometabolism 1:1–26. 10.20900/immunometab20190008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang C, Zhang M, Pang X, Zhao Y, Wang L, Zhao L. 2012.Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations.ISME J 6:1848–1857. 10.1038/ismej.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Q, Zhang Y, Wang X, Yang R, Zhu X, Zhang Y, Chen C, Yuan H, Yang Z, Sun L. 2020.Gut bacteria Akkermansia is associated with reduced risk of obesity: evidence from the American Gut Project.Nutr Metab (Lond) 17:1–9. 10.1186/s12986-020-00516-1. [DOI] [PMC free article] [PubMed] [Google Scholar]